- 1Department of Botany, DDU Gorakhpur University, Gorakhpur, Uttar Pradesh, India
- 2Centre of Advanced Study in Botany, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 3School of Life Sciences, Jawaharlal Nehru University, New Delhi, India
- 4Department of Chemistry, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 5National Fungal Culture Collection of India (NFCCI), Biodiversity and Palaeobiology Group, MACS Agharkar Research Institute, Pune, Maharashtra, India
- 6Forest Pathology Department, KSCSTE-Kerala Forest Research Institute, Thrissur, Kerala, India
- 7Martin-Luther-Universität, Institut für Biologie, Bereich Geobotanik and Botanischer Garten, Herbarium, Halle (Saale), Germany
During a survey of foliicolous fungi in India, two interesting anamorphic hyphomycetous fungal specimens were collected from infected leaves of Calotropis spp. and Mallotus philippensis. Calotropis spp. produce fascicles of conidiophores from stromata, accompanied by secondary superficial hyphae bearing solitary conidiophores. The specimen on Mallotus philippensis resembled Mycovellosiella, characterized by secondary superficial hyphae bearing micronematous to semi-macronematous, mononematous, unbranched, and aseptate conidiophores. A polyphasic approach—including morphological, cultural, and multilocus phylogenetic analyses (LSU-Rpb2-ITS), coupled with genealogical concordance phylogenetic species recognition—identified its relationship with cercosporoid fungi within the family Mycosphaerellaceae. The analysis confirmed that these fungal specimens represent distinct lineages without known morphological or DNA sequence counterparts. Consequently, two new genera are proposed: Marcstadlera and Neoclypeosphaerella, with M. malloti comb. nov. and N. calotropidis comb. nov. as their respective type species. Additionally, Clypeosphaerella calotropidis, Clypeosphaerella quasiparkii, and Pseudocercospora malloti are recognized as new synonyms. Several genera in the Mycosphaerellaceae, including Marcstadlera and Neoclypeosphaerella, are monophyletic. The ultrastructure of the conidiogenous loci and hila differs between these two genera. In Marcstadlera, the loci are cylindrical or peg-like, truncate at the apex, while the conidial base is narrowly obconically truncate. In Neoclypeosphaerella, the loci are slightly protuberant and surrounded by a circular rim-like structure, forming a truncated apex with a centrally positioned small apical depression. The conidial base is obconically truncated and also surrounded by a circular rim-like structure.
Introduction
Mycosphaerellaceae Lindau is a diverse family of fungi in the order Mycosphaerellales (Ascomycota), comprising over 3,000 species (Crous and Braun, 2003; Braun et al., 2013, Braun et al., 2015, Braun et al., 2016). They thrive in diverse habitats and exhibit diverse life modes, including pathogenic, endophytic, saprophytic, and epiphytic modes of existence in various hosts worldwide (Videira et al., 2017). They have garnered significant research attention due to their association with a wide range of economically and ornamentally important host plants (Videira et al., 2017; Abdollahzadeh et al., 2020; Bakhshi et al., 2021; Bakhshi and Braun, 2022).
Members of Mycosphaerellaceae exhibit both sexual (teleomorphic) and asexual (anamorphic) stages (Crous et al., 2009). However, many, or perhaps most, species in this complex are asexual holomorphs—that is, they have lost the ability to produce sexual morphs. Within this complex, the sexual morphs of Mycosphaerella are morphologically quite uniform and offer few distinguishing features to justify further division into smaller genera. In contrast, the asexual morphs exhibit significant morphological diversity, which has led to the establishment of numerous asexual genera based on specific morphological traits (Videira et al., 2017).
Currently, more than 120 genera have been accepted in Mycosphaerellaceae (Wijayawardene et al., 2014; Videira et al., 2017; Crous et al., 2020b; Bakhshi et al., 2020, Bakhshi and Braun, 2022; Yadav et al., 2022, Yadav et al., 2023; Bakhshi and Crous, 2024; Melo et al., 2025). Advances in molecular phylogenetics have significantly reshaped the taxonomy of this family, uncovering cryptic species and refining classification (Crous et al., 2007, Crous et al., 2013a, Crous et al., 2013b; Verkley et al., 2013; Quaedvlieg et al., 2014; Videira et al., 2017; Bakhshi et al., 2021; Bakhshi and Braun, 2022). Beyond their pathogenic roles, Mycosphaerellaceae species play a crucial part in ecological dynamics, influencing plant health and ecosystem stability.
Clypeosphaerella Guatim. et al. and Mycovellosiella Rangel are the two notable genera within the Mycosphaerellaceae, and the members of these genera typically cause leaf spot diseases. The genus Clypeosphaerella exhibits both sexual and asexual morphs. The sexual morph is distinguished by its thicker upper ascomatal wall, resembling a pseudoclypeus. In contrast, the asexual morph develops fasciculate conidiophores from stromata, as well as solitary conidiophores arising from secondary superficial hyphae, with conidia forming singly or in chains (Chupp, 1954; Kamal et al., 1990; Braun, 2000a; Wilkinson et al., 2005; Haldar and Ray, 2001; Guatimosim et al., 2016; Videira et al., 2017). Similarly, the genus Mycovellosiella is characterized by the absence or poor development of stromata. It produces secondary superficial hyphae that give rise to solitary or fasciculate conidiophores as lateral branches, with conidia forming either singly or in chains (Videira et al., 2017).
During a 2023–2024 survey of foliicolous fungi in Uttar Pradesh, India, two anamorphic hyphomycetous fungal specimens were collected from diseased leaves. The first specimen was found on Calotropis spp., where it developed fascicles of conidiophores from stromata, accompanied by secondary superficial hyphae bearing solitary conidiophores. Molecular phylogenetic analyses revealed that the isolate forms an independent lineage within Mycosphaerellaceae, clustering with Clypeosphaerella calotropidis but remaining distinct from C. sticheri, the type species of Clypeosphaerella. To accommodate this unique lineage, the novel genus Neoclypeosphaerella is proposed, emphasizing its uniqueness within Mycosphaerellaceae.
Similarly, another specimen, Mycovellosiella malloti—the basionym of Pseudocercospora malloti—was isolated from Mallotus philippensis, where it developed secondary superficial hyphae with micronematous to semi-macronematous solitary conidiophores. Phylogenetic analyses revealed that this isolate segregates from Mycovellosiella, forming an independent lineage within Mycosphaerellaceae. As a result, the novel genus Marcstadlera is proposed to accommodate this fungus, underscoring its distinct evolutionary trajectory within the family. Mycovellosiella was previously distinguished from closely related genera, Passalora Fr. and Phaeoramularia Munt.-Cvetk. based on the formation of superficial mycelium with solitary conidiophores formed in vivo. However, these traits are phylogenetically and taxonomically insignificant and appear unreliable (Videira et al., 2017). Therefore, species exhibiting mycovellosiella-like morphology should be tentatively maintained in or assigned to Passalora s. lat., unless their phylogenetic affinity is thoroughly investigated (Videira et al., 2017).
These taxonomic revisions, driven by molecular phylogenetics and morphological analyses, refine the classification and relationships of these taxa and are discussed in detail in this manuscript.
Materials and methods
Sample collection and fungal isolation
During a field survey of phytopathogenic fungi conducted between 2023 and 2024, infected leaves of Calotropis spp. were collected from Kushmi Forest, while infected leaves of Mallotus philippensis were collected from Varanasi and Mirzapur, Uttar Pradesh, India. These locations are part of tropical dry deciduous forest ecosystems, primarily consisting of dense forests dominated by Sal and Teak trees. This region is characterized by hot, dry summers, mild winters, undulating hilly terrain, and rain-fed streams. Herbs and shrubs are commonly found during the rainy season. The collected infected leaves were placed in sterilized polybags along with relevant collection details and transported to the laboratory for further processing. Standard techniques as described by Hawksworth (1974); Savile (1962), and Verma et al. (2021c) were followed. In the lab, the samples were dried for approximately 1 week between fresh sheets of blotting paper. The dried and pressed leaf specimens were then sealed in airtight polyethylene bags and stored in paper envelopes, accompanied by the corresponding collection information.
Slides were mounted in a 1:1 mixture of glycerine and lactophenol cotton-blue from the infected part of the leaves. Observations were made with a stereo zoom microscope (Magnus: MSZ-TR) with an attached camera (CatCam300EF) and an Olympus compound microscope (BX53) equipped with differential interference contrast (DIC) illumination, and images were captured using an Olympus DP28 camera with associated software. Scanning electron microscopy (SEM) was conducted using a field emission scanning electron microscope (FEI Nova Nano SEM-450). For SEM micrographs, specimens were coated with gold-palladium using a POLARON Sputter coater and examined with a LEO-430 scanning electron microscope. Detailed observations of morphological characters were carried out at different magnifications through light microscopy (×450 and ×1,000) and scanning electron microscopy (up to ~×55K). Size ranges of morphological features were determined from at least 25 measurements, and 95% confidence intervals were calculated for the measurements, with the extreme values given in parentheses. The examined reference specimens were deposited in the fungarium of Ajrekar Mycological Herbarium (AMH), MACS, Agharkar Research Institute (ARI), Pune, India, and duplicates were retained in the Mycological Herbarium of the Department of Botany of Banaras Hindu University, Varanasi, U.P., India (MH-BHU). For in vitro isolation, conidia were transferred to Petri dishes containing malt extract agar (MEA), potato dextrose agar (PDA), and agar media supplemented with undefined vegetable peelings. The petri dishes were incubated at 25°C ± 5°C and diffused daylight. The ex-type living cultures were deposited at the National Fungal Culture Collection of India (NFCCI), MACS, Agharkar Research Institute, Pune, India.
DNA extraction, PCR, and sequencing
The genomic DNA was extracted from mycelia and conidia freshly scraped from PDA plates using a sterile scalpel blade. Approximately 200 mg of wet weight was transferred to 2-mL microcentrifuge tubes kept in liquid nitrogen for 2 min and then grinded to a fine powder using a pestle and mortar. DNA was extracted using a modified CTAB method using the protocol of Van Burik et al. (1998). The internal transcribed spacer (ITS) region was amplified using ITS1/ITS4 (White et al., 1990), large subunit nuclear ribosomal DNA (LSU) gene with LROR/LR7 (Vilgalys and Hester, 1990; Rehner and Samuels, 1994), and partial DNA-directed RNA polymerase II subunit (Rpb2) with RPB2-5F2/RPB2-7cR (Liu et al., 1999; Sung et al., 2007) primer pairs. Amplification reaction mixtures and conditions described by Yadav et al. (2022, 2023) were followed for standard amplification and subsequent sequencing of the ITS, LSU, and Rpb2 by Eurofins Genomics (Bengaluru, India).
Sequence alignment and phylogenetic analysis
The obtained ITS, LSU, and RBP2 sequences from the isolates NFCCI 5818, NFCCI 5819, NFCCI 5983, and NFCCI 5984 were assembled and edited using Chromas v.2.6.6. The manually edited sequences were submitted to NCBI GenBank (Table 1) and were subjected to a megablast search of the NCBI GenBank nucleotide database to retrieve the most closely matched sequences of related strains. Reference sequences were also selected from relevant published literature (Table 1). Sequence alignments were generated using MAFFT v.7 (Katoh et al., 2019). The alignments of individual loci were concatenated using Mesquite v. 3.61 (Maddison and Maddison, 2018) and deposited as electronic supplementary materials in TreeBASE (http://www.treebase.org/), under the accession number 32049 and URL http://purl.org/phylo/treebase/phylows/study/TB2:S32049?x-access-code=b06647353af0bee2d49542a8bb895832&format=html.
Phylogenetic trees were constructed using Bayesian inference (BI) performed with MrBayes v. 3.2.7 (Ronquist et al., 2012) and maximum likelihood (ML) analysis performed with RAxML v.8.2.10 (Stamatakis, 2014) as explained in Yadav et al. (2022, 2023). The phylogenetic analyses were individually applied to two datasets as different combinations were used as barcodes and could provide valuable information for understanding evolutionary relationships at the genus and species level in Mycosphaerellaceae (Videira et al., 2017; Chen et al., 2022). Dataset 1 consisted of LSU-Rpb2 sequences, and dataset 2 consisted of LSU-Rpb2-ITS sequences from 32 genera currently known to the Mycosphaerellaceae. All trees were rooted with Ramichloridium apiculatum (CBS 156.59) and Uwebraunia australiensis (CBS 120729).
The phylogenetic trees of ML were visualized using FigTree v.1.4.4 and edited using Adobe® Illustrator v. CC 2017 (Figures 1, 2).
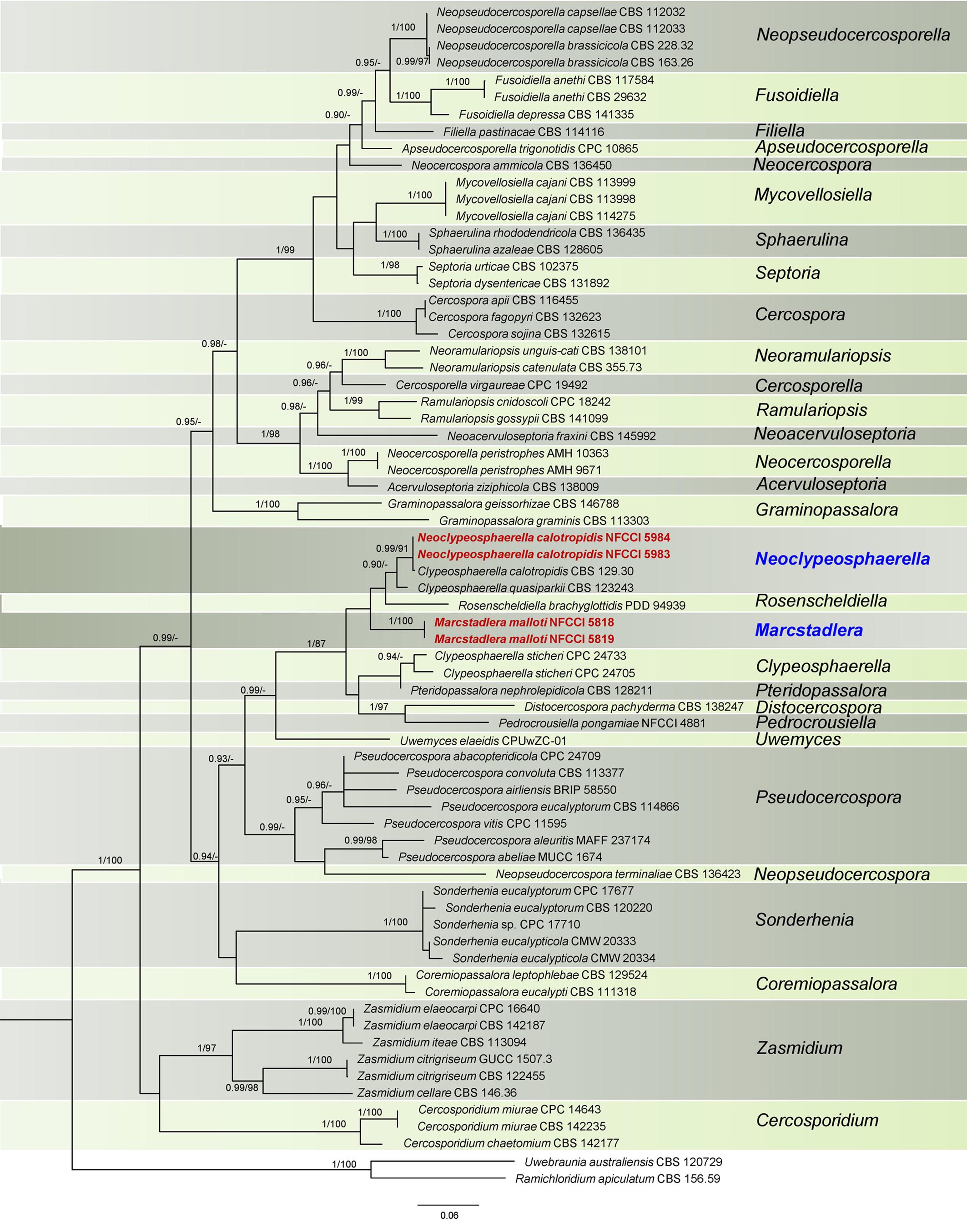
Figure 1. Phylogenetic tree resulting from a RAxML analysis of the combined LSU-Rpb2 sequence alignment (dataset 1). The Bayesian posterior probabilities (≥0.90; BI-PP) and maximum likelihood bootstrap support values (≥85%; ML-BS) are given at the nodes (BI-PP/ML-BS). The newly introduced lineage is represented in red bold and novel genera denoted in blue. The tree is rooted to Ramichloridium apiculatum CBS 156.59 and Uwebraunia australiensis CBS 120729.
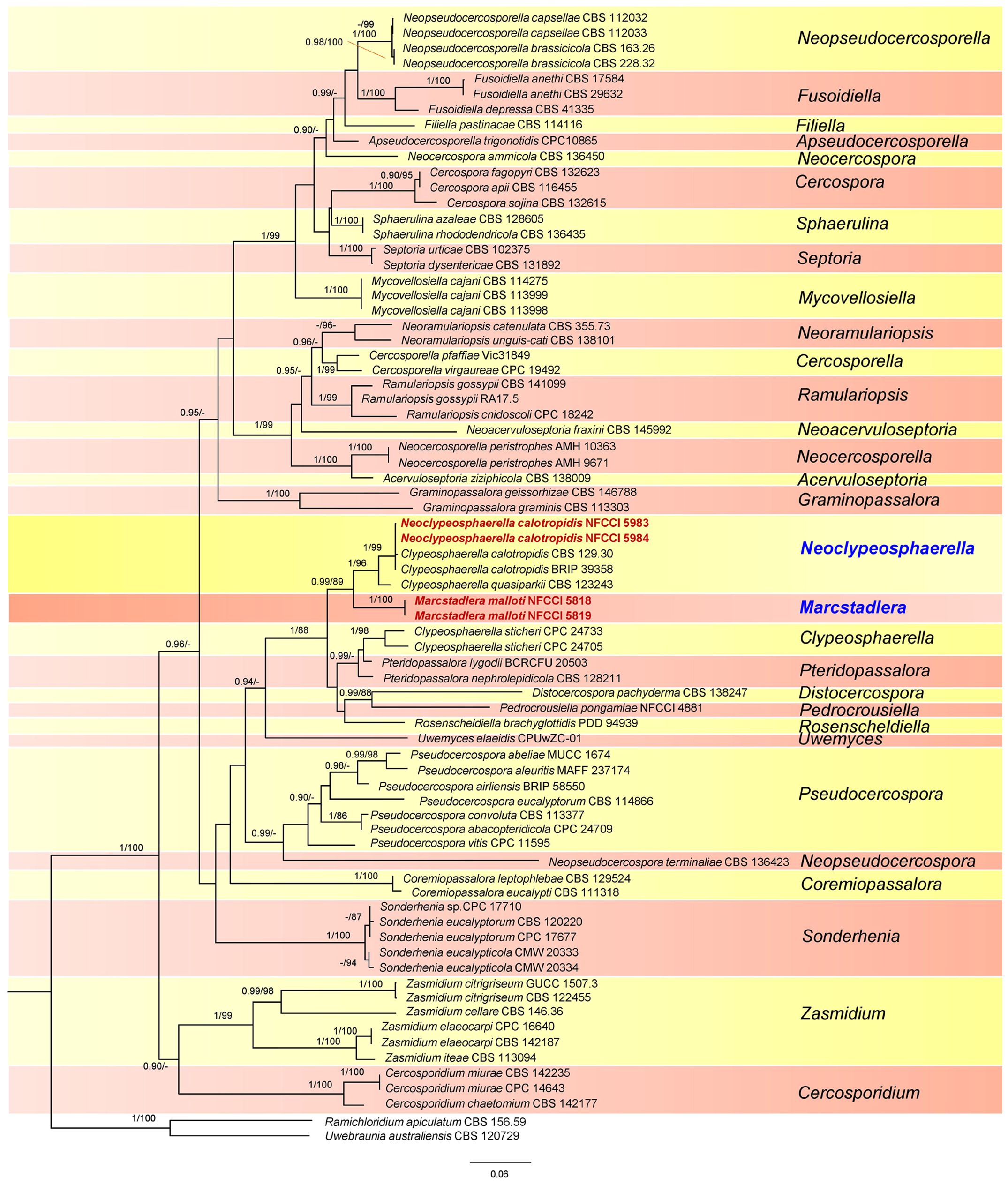
Figure 2. Phylogenetic tree resulting from a RAxML analysis of the combined LSU-Rpb2-ITS sequence alignment (dataset 2). The Bayesian posterior probabilities (≥0.90; BI-PP) and maximum likelihood bootstrap support values (≥85%; ML-BS) are given at the nodes (BI-PP/ML-BS). The newly introduced lineage is represented in red bold and novel genera denoted in blue. The tree is rooted to Ramichloridium apiculatum CBS 156.59 and Uwebraunia australiensis CBS 120729.
Genealogical concordance phylogenetic species recognition analysis
The genealogical concordance phylogenetic species recognition (GCPSR) model (as described by Taylor et al., 2000) was used to clarify species boundaries among closely related and potentially ambiguous taxa by using a pairwise homoplasy index (Φw) test, a statistical test to evaluate genetic data. GCPSR is valued for its ability to synthesize information from multiple genes, evaluate gene flow, operate within an evolutionary timescale, and provide practical insights into species delimitation. It underscores the complexity of species boundaries and offers a robust framework for understanding evolutionary relationships among organisms (Koufopanou et al., 1997; Geiser et al., 1998; Taylor et al., 2000; Starkey et al., 2007). A pairwise homoplasy index (PHI) test (Philippe and Bryant, 2006) was performed in SplitsTree4 (Huson, 1998; Huson and Bryant, 2006) to determine the recombination level within phylogenetically closely related species using a three-locus concatenated dataset of closely related species. If the PHI value exceeds the threshold of 0.05 (Φw ≥ 0.05), it signifies the absence of significant recombination in the dataset. The relationships between these 15, closely related, species were visualized by constructing split graphs (Figure 3) from the three-locus concatenated datasets, using both the Log-Det transformation and splits decomposition options.
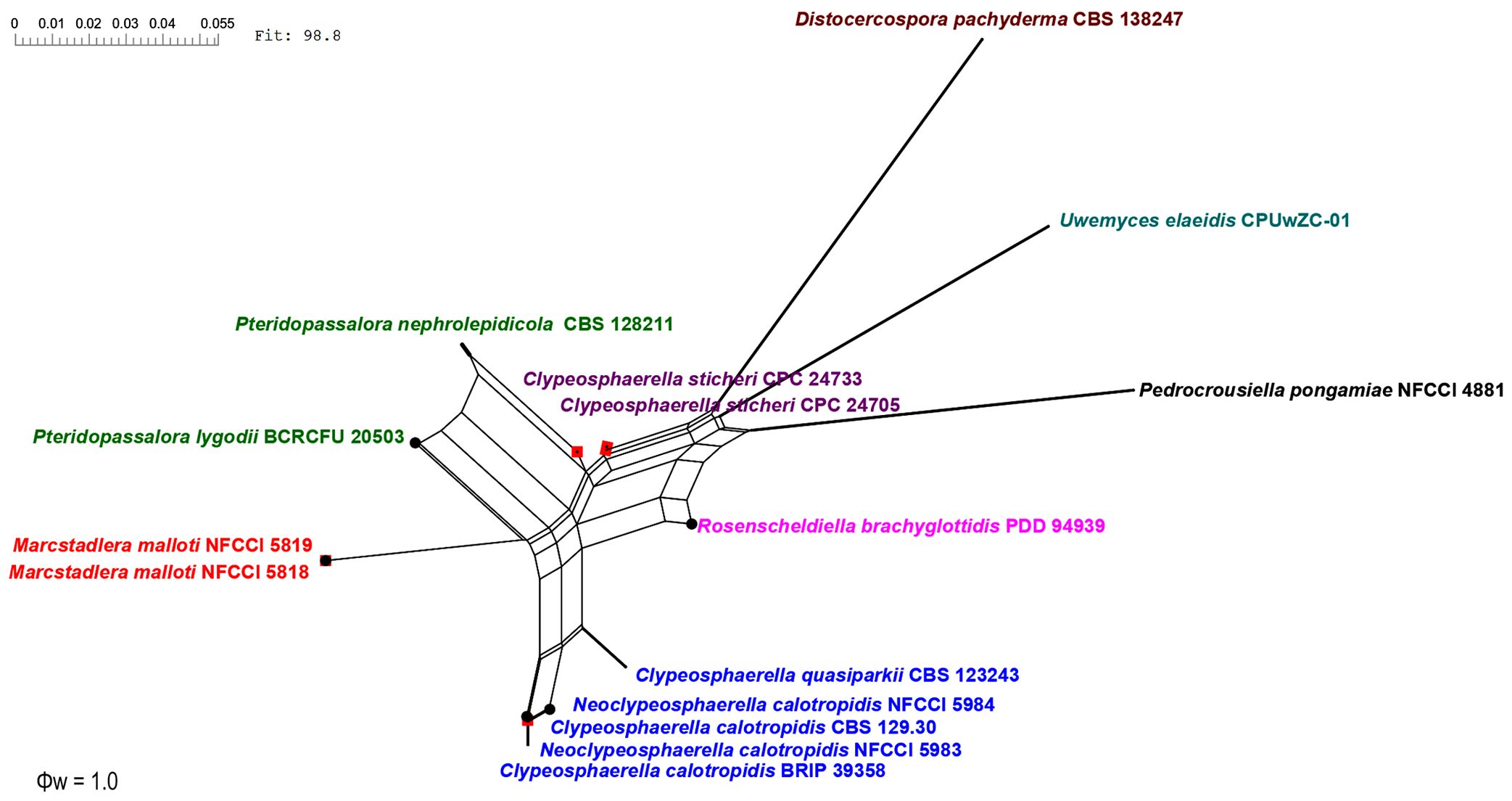
Figure 3. Split graphs showing the results of the pairwise homoplasy index (PHI) test of closely related species using both LogDet transformation and splits decomposition. PHI test results (Φw) ≤0.05 from the PHI test denotes the presence of significant recombination within the dataset. The newly identified taxa are shown in red and blue.
Results
The sequences from specimens NFCCI 5818 and NFCCI 5819 were 100% identical across all regions. Likewise, the sequences from specimens NFCCI 5983 and NFCCI 5984 were also 100% identical in each region. The data for the trees conducted in the different analyses are shown in Table 1. Phylogenetic trees obtained from the combined gene analyses are supplied below (Figures 1, 2).
Dataset 1 (LSU-Rpb2 phylogeny)
This dataset consisted of a concatenated alignment of two loci: LSU and Rpb2. The final alignment has a total of 1,235 characters, with LSU contributing 692 characters and Rpb2 contributing 543 characters, inclusive of alignment gaps. The phylogenetic trees generated from Bayesian analyses (BI) and maximum parsimony (MP) have shown similar overall topology, indicating consistent results across these methods (Videira et al., 2017). The best scoring RAxML tree is presented in Figure 1, with the likelihood value of −14,653.366735. Estimated base frequencies were as follows: A = 0.235837, C = 0.305805, G = 0.252457, T = 0.205901; substitution rates AC = 1.073482, AG = 3.362976, AT = 0.756117, CG = 0.718572, CT = 5.587844, GT = 1.000000; gamma distribution shape parameter α = 0.547561. In this analysis, Clypeosphaerella calotropidis (CBS 129.30) and C. quasiparkii (CBS 123243) are now separated from the Clypeosphaerella (type species: C. sticheri) clade and are placed in a separate sister branch of Rosenscheldiella brachyglottidis (PDD 94939) along with NFCCI 5983 and NFCCI 5984 (Figure 1). Clypeosphaerella calotropidis, C. quasiparkii, and C. sticheri form a paraphyletic group. Marcstadlera is identified as a sister group to both Neoclypeosphaerella and Rosenscheldiella. However, the statistical support for this relationship is very low (BI-PP/ML-BS: 0.51/37).
Dataset 2 (LSU-Rpb2-ITS phylogeny)
This dataset consisted of a concatenated alignment of three loci: LSU, Rpb2, and ITS. The final alignment of this dataset contained a total of 1,675 characters divided into three partitions containing 692 (LSU), 543 (Rpb2), and 440 (ITS) characters, including alignment gaps. The phylogenetic trees generated from BI and MP have shown similar overall topology, indicating consistent results across these methods. The best scoring RAxML tree is presented in Figure 2, with the likelihood value of −19,399.308522. Estimated base frequencies were as follows: A = 0.201081, C = 0.284874, G = 0.278655, T = 0.235390; substitution rates AC = 1.857707, AG = 4.458699, AT = 1.107011, CG = 0.007891, CT = 6.730028, GT = 1.000000; gamma distribution shape parameter α = 0.484710. The results of the analysis of dataset 2 (Figure 2) strongly supported the dataset 1 analysis, except for the placement of Rosenscheldiella brachyglottidis (PDD 94939) (Figure 1). Marcstadlera is identified as a sister group to Neoclypeosphaerella with high statistical support (BI-PP/ML-BS: 0.99/89), suggesting a close evolutionary relationship between these two genera.
In both datasets, C. calotropidis (BRIP 39358 and CBS 129.30) and C. quasiparkii (CBS 123243) are separately clustered from the type species of Clypeosphaerella, C. sticheri (CPC 24705 and CPC 24733), and form a paraphyletic group. Both C. calotropidis and C. quasiparkii are grouped with NFCCI 5983 and NFCCI 5984 (Neoclypeosphaerella calotropidis) in a distinct sister branch of the newly introduced genus Marcstadlera (Figure 2), forming a statistically supported monophyletic group (BI-PP/ML-BS: 0.99/89).
Clypeosphaerella, Distocercospora, Marcstadlera, Neoclypeosphaerella, Pedrocrousiella, Pteridopassalora, and Rosenscheldiella form a statistically supported monophyletic group in both datasets (BI-PP/ML-BS: 1/87 or 1/88).
Genealogical concordance phylogenetic species recognition analysis
The PHI tests were carried out to calculate the recombination level within two novel genera and their phylogenetically closely related taxa. The PHI tests showed that there was no significant recombination (Фw = 1.0) between closely related taxa, viz., Clypeosphaerella, Distocercospora, Marcstadlera, Neoclypeosphaerella, Pedrocrousiella, Pteridopassalora, Rosenscheldiella, and Uwemyces (Figure 3).
Taxonomy
Marcstadlera Gargee Singh & Raghv. Singh, gen. nov. (Figures 4–6).

Figure 4. Marcstadlera malloti (AMH 10726) on Mallotus philippensis (Euphorbiaceae). (A) Mallotus philippensis in natural habitat. (B) Symptoms on upper leaf surface. (C) Symptom on lower leaf surface. (D) Close-up of leaf surface showing fungal fructifications. (E) Germinating conidium. (F) Top view of ex-epitype culture on PDA. (G) Reverse view of ex-epitype culture on PDA. (H–J) Mycelia from ex-epitype culture showing formation of chlamydospores and conidia (showing arrows for conidia). Scale bars: (B, C, F, G) 20 mm, (H–J) 10 µm.

Figure 5. Microphotographs of Marcstadlera malloti (AMH 10726). (A–C) Superficial hyphae with developing conidiogenous cells (green arrows for conidiogenous loci). (D–F) Conidia in chain (red arrows for catenation). (G) Ramoconidia with branched catenation (pink arrow for branched catenation). (H, I) Superficial hyphae with conidiogenous loci bearing conidia (yellow arrows for conidiogenous loci). (J–S) Conidia (blue arrows for the development of conidiogenous loci). (T) Monopolar germination in conidium. Bars: 10 µm.
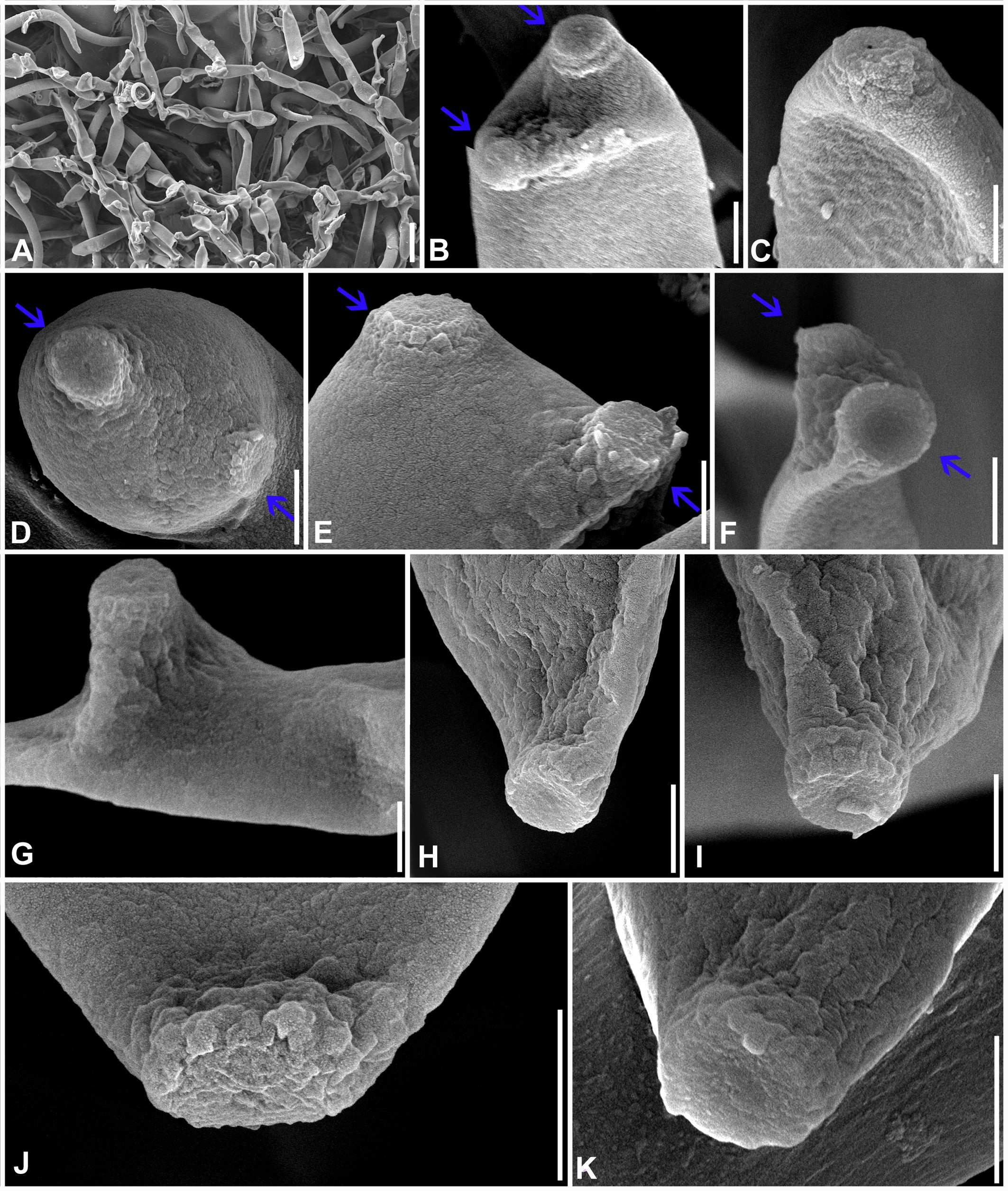
Figure 6. Scanning electron microphotographs of Marcstadlera malloti (AMH 10726). (A) Superficial hyphae with conidia and developing conidiogenous cells. (B–G) Top and lateral views of conidiogenous loci (blue arrows for polyblastic nature of conidiogenous cells). (H–K) Top and lateral view of hila of conidia. Scale bars: (A) 10 µm, (B–K) 1 µm.
MycoBank number: MB854795.
Etymology: This is derived from the name of Professor Dr. Marc Stadler (Helmholtz Centre for Infection Research, Braunschweig, Germany), a globally renowned expert in industrial microbiology and mycology, as well as fungal biodiversity research and natural product chemistry.
Diagnosis: This differs from the genus Neoclypeosphaerella by developing conidiophores reduced to conidiogenous cells, arising singly from external hyphae as intercalary or terminal cells of superficial hyphae, micronematous to semi-macronematous, mononematous, unbranched, aseptate, and mostly catenate conidia.
Description: Phytopathogenic, causing leaf spots. Stromata absent. Mycelium mostly external and superficial, septate, branched, smooth to slightly roughened, light brown or olivaceous brown. Conidiophores micronematous, reduced to conidiogenous cells or semi-macronematous, mononematous, developing individually from intercalary or terminal cells of external hyphae, unbranched, aseptate, light brown or olivaceous brown. Conidiogenous cells mono- to polyblastic, cylindrical or peg-like, integrated, conidiogenous loci (scars) flattened (ultrastructure), unthickened, to slightly thickened and darkened. Ramoconidia obclavate-cylindrical, smooth to slightly roughened, light brown to pale olivaceous brown. Conidia blastocatenate, obclavate-cylindrical, transversely septate, straight to curved, thick-walled, tip subacute to rounded, base narrowly obconical, truncate (ultrastructure), smooth to slightly roughened, pale brown to pale olivaceous-brown, hilum unthickened to slightly thickened and darkened.
Type species: Marcstadlera malloti (Kharwar, P.N. Singh and R.K. Chaudhary) Gargee Singh, Raghv. Singh, and Sahana.
Notes: Based on a megablast search of NCBI’s GenBank nucleotide database, the closest hits using the ITS sequence had the highest similarity to Clypeosphaerella quasiparkii [strain CBS 123243, GenBank MH863287; identities = 416/434 (96%), 3 gaps (0%)], Clypeosphaerella calotropidis [strain BRIP 39358, GenBank AY303969; identities = 402/418 (96%), 2 gaps (0%)], and Ramulariopsis gossypii [strain RA17.5, GenBank KR265337; identities = 417/441 (95%), 14 gaps (3%)]. Closest hits using the LSU sequence are Clypeosphaerella sticheri [strain CPC 24733, GenBank KT037577; identities = 516/527 (98%), 0 gap (0%)], Pteridopassalora nephrolepidicola [strain CBS 128211, GenBank HQ599591; identities = 516/527 (98%), 0 gap (0%)], and Clypeosphaerella quasiparkii [strain CBS 123243, GenBank MH874811; identities = 516/529 (98%), 2 gap (0%)]. Closest hits using the Rpb2 sequence had the highest similarity to Clypeosphaerella calotropidis [strain CBS 129.30, GenBank MF951477; identities = 517/586 (88%), 0 gaps (0%)], Clypeosphaerella quasiparkii [strain CBS 123243, GenBank MF951478; identities = 515/586 (88%), 0 gaps (0%)], and Pteridopassalora nephrolepidicola [strain CBS 128211, GenBank KX462646; identities = 492/576 (85%), 0 gaps (0%)].
Marcstadlera malloti (Kharwar, P.N. Singh & R.K. Chaudhary) Gargee Singh, Raghv. Singh, & Sahana comb. nov. (Figures 4-6).
MycoBank number: MB854796.
≡ Mycovellosiella malloti Kharwar, P.N. Singh and R.K. Chaudhary, Mycol. Res. 100(6), 689 (1996).
= Pseudocercospora malloti (Kharwar, P.N. Singh and R.K. Chaudhary) U. Braun, Schlechtendalia 19, 69 (2009).
Description: Leaf spots amphiphyllous, angular, grayish brown to dark brown, vein-limited, 1–2.5 mm wide, sometimes coalescing. Colonies effuse, hypogenous, grayish brown, velvety. Stromata absent. Mycelium mostly external and superficial, septate, branched, smooth to slightly roughened, light brown or olivaceous brown, 2–4.5 μm wide. Conidiophores micronematous, reduced to conidiogenous cells or semi-macronematous, mononematous, developing individually from intercalary or terminal cells of external hyphae, unbranched, aseptate, light brown or olivaceous brown, (10–)12–15(–20) × (2–)2.5–3(–3.5) μm. Conidiogenous cells mono to polyblastic, determinate or sympodial elongated, cylindrical or peg-like, integrated, 1.5–3 × 1–1.5 μm. Conidiogenous loci (scars) complanate, unthickened to slightly thickened and darkened, flattened at the apex. Ramoconidia intercalary, obclavate-cylindrical, smooth to slightly roughened, light brown to pale olivaceous brown. Conidia blastocatenate, obclavate-cylindrical, apex subacute to rounded, base narrowly obconical truncate, straight to curved, 1–8-euseptate, smooth to slightly roughened, pale brown to pale olivaceous brown, dry, (10–)45–78(–117) × (2.5–)3–4(–5.5) μm, thick-walled, hilum unthickened to slightly thickened and darkened, 0.8–1.5 μm wide.
Culture characteristics: Colonies on PDA slow-growing and attained a diameter of approximately 30 mm after 21 days at 25 °C ± 5 °C, raised, irregular, aerial mycelium velvety, upper surface dark gray to black centrally and white fluffy at the periphery, reverse brown to black. Cultures fertile. Hyphae 1.5–2.5 μm wide, branched, septate, smooth to slightly roughened, subhyaline to very light olivaceous brown. Conidiophores micronematous, reduced to conidiogenous cells or semi-macronematous, mononematous, unbranched, aseptate, hyaline to very light olivaceous brown, (12–)20–22(–25) × (2–)2.5–3(–3.5) μm. Conidiogenous cells monoblastic, cylindrical or peg-like, intercalary and terminal, determinate. Conidiogenous loci (scars) unthickened to slightly thickened and darkened, loci cylindrical or peg-like, 1.5–3 × 1–1.5 μm. Conidia solitary, obclavate-cylindrical, base narrowly obconical truncate, apex subacute to rounded, straight to curved, 1–8-euseptate, smooth to slightly roughened, light brown to pale olivaceous brown, (25–)70–100(–110) × (3–)4–4.5(–5) μm, hilum unthickened to slightly thickened and darkened, 1–1.5 μm wide. Chlamydospores spherical to oval, light brown to mid brown, smooth, 2–7 × 2–5 μm.
Specimens examined: NEPAL, Chitwan, Narayangarh, on living leaves of Mallotus philippensis (Lam.) Müll. Arg. (Euphorbiaceae), January 1995, Kamal (GPU 3008, HCIO 41505 isotype, IMI 366204 holotype); INDIA, Uttar Pradesh, Gorakhpur, Kushmi Forest, 26.749748°N 83.468645°E, on living leaves of Mallotus philippensis, 8 February 2023, Gargee Singh, MH-BHU 114 (AMH 10726, epitype designated here, MycoBank MBT10024805), ex-type culture NFCCI 5818, gene sequence GenBank: PQ012587 (ITS), PQ012588 (LSU), PQ034553 (Rpb2); INDIA, Uttar Pradesh, Gorakhpur, Kushmi Forest, on living leaves of M. philippensis, 25 March 2024, Raghvendra Singh, MH-BHU 115(AMH 10727), culture NFCCI 5819, gene sequence GenBank: PQ013688 (ITS), PQ013689 (LSU), PQ034554 (Rpb2).
Note: Currently, five species of Pseudocercospora have been described on Mallotus, namely, P. bakeriana Deighton [≡ Cercospora bakeriana Saccardo 1914] (Deighton, 1976), P. malloti (Kharwar, P.N. Singh and R.K. Chaudhary) U. Braun [≡ Mycovellosiella malloti Kharwar, P.N. Singh and R.K. Chaudhary] (Kharwar et al., 1996; Braun, 2009), P. malloti-repandi (Bhalla, S.K. Singh and A.K. Srivast.) U. Braun [≡ Mycovellosiella malloti-repandi Bhalla, S.K. Singh and A.K. Srivast.] (Bhalla et al., 1997; Braun, 2000b), P. melanolepidis Goh and W.H. Hsieh (Goh and Hsieh, 1987), and P. pampangensis (Petr.) U. Braun [≡ Cercospora pampangensis Petr.] (Petrak, 1956; Braun, 1996).
In comparison to Marcstadlera malloti, P. bakeriana exhibits several distinctive features in its development of conidiophores. Notably, P. bakeriana forms fascicles of conidiophores that arise from both external and internal hyphae. These conidiophores can range from simple to highly branched ones. They are septate and can reach lengths of up to 130 μm with widths varying between 3 and 6 μm. This morphological variability in conidiophore structure is a significant distinguishing characteristic between the two species.
Marcstadlera malloti appears to be most closely related to P. malloti and P. malloti-repandi. Both species develop superficial hyphae that give rise to micronematous to semi-macronematous, mononematous conidiophores, either terminally or as lateral branches. The solitary to catenate nature of conidia in P. malloti closely resembles those of M. malloti, making them morphologically indistinguishable. As a result, P. malloti is used as the type species for Marcstadlera. In contrast, P. malloti-repandi can be differentiated by its branched, septate, and longer, wider conidiophores (2.5–65 × 2.5–6 μm).
In addition to the formation of stromata and fascicles of primary conidiophores (20–65 × 3–4 μm), P. melanolepidis develops secondary external hyphae that bear secondary conidiophores (up to 10 μm long) both terminally and laterally. Notably, both types of conidiophores are septate.
Pseudocercospora pampangensis develops stromata that bear large clusters (fascicles) of subsynnematous conidiophores. These conidiophores are occasionally branched, pluriseptate, and relatively larger in size (15–250 × 3–6 μm).
Neoclypeosphaerella S. Rajwar & Raghv. Singh, gen. nov. (Figures 7–12).
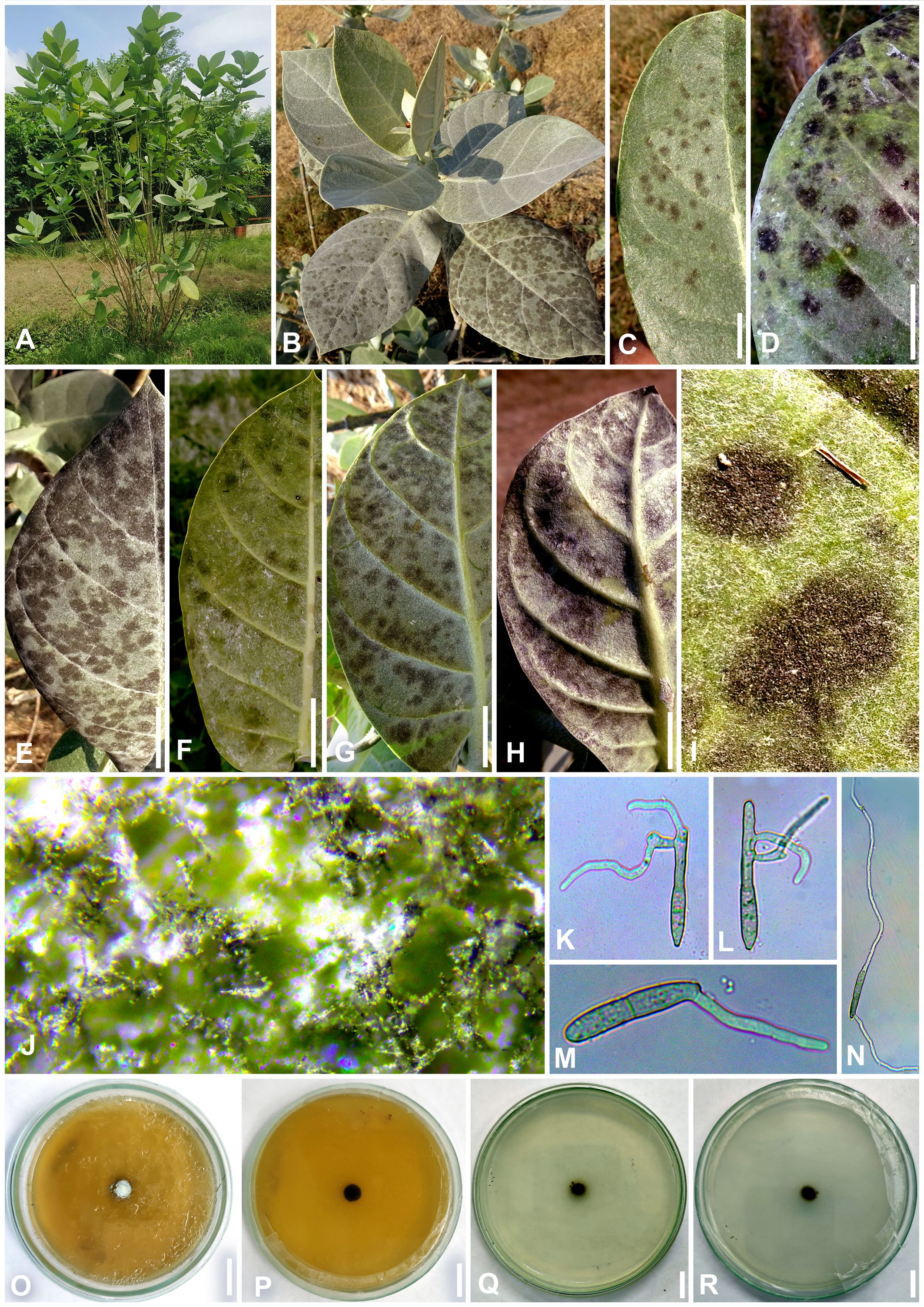
Figure 7. Neoclypeosphaerella calotropidis (AMH 10781) on Calotropis gigantea (Apocynaceae). (A, B) Calotropis gigantea in natural habitat. (C–E) Symptoms on the upper surfaces of leaves. (F–H) Symptom on the lower surfaces of leaves. (I) Stereoscopic view of infection spots. (J) Close-up of leaf symptoms showing fungal fructifications. (K–N) Germinating conidia. (O) Top view of ex-epitype culture on MEA. (P) Reverse view of ex-epitype culture on MEA. (Q) Top view of ex-epitype culture on PDA. (R) Reverse view of ex-epitype culture on PDA. Scale bars: (C–H) 20 mm, (O–R) 10 mm.
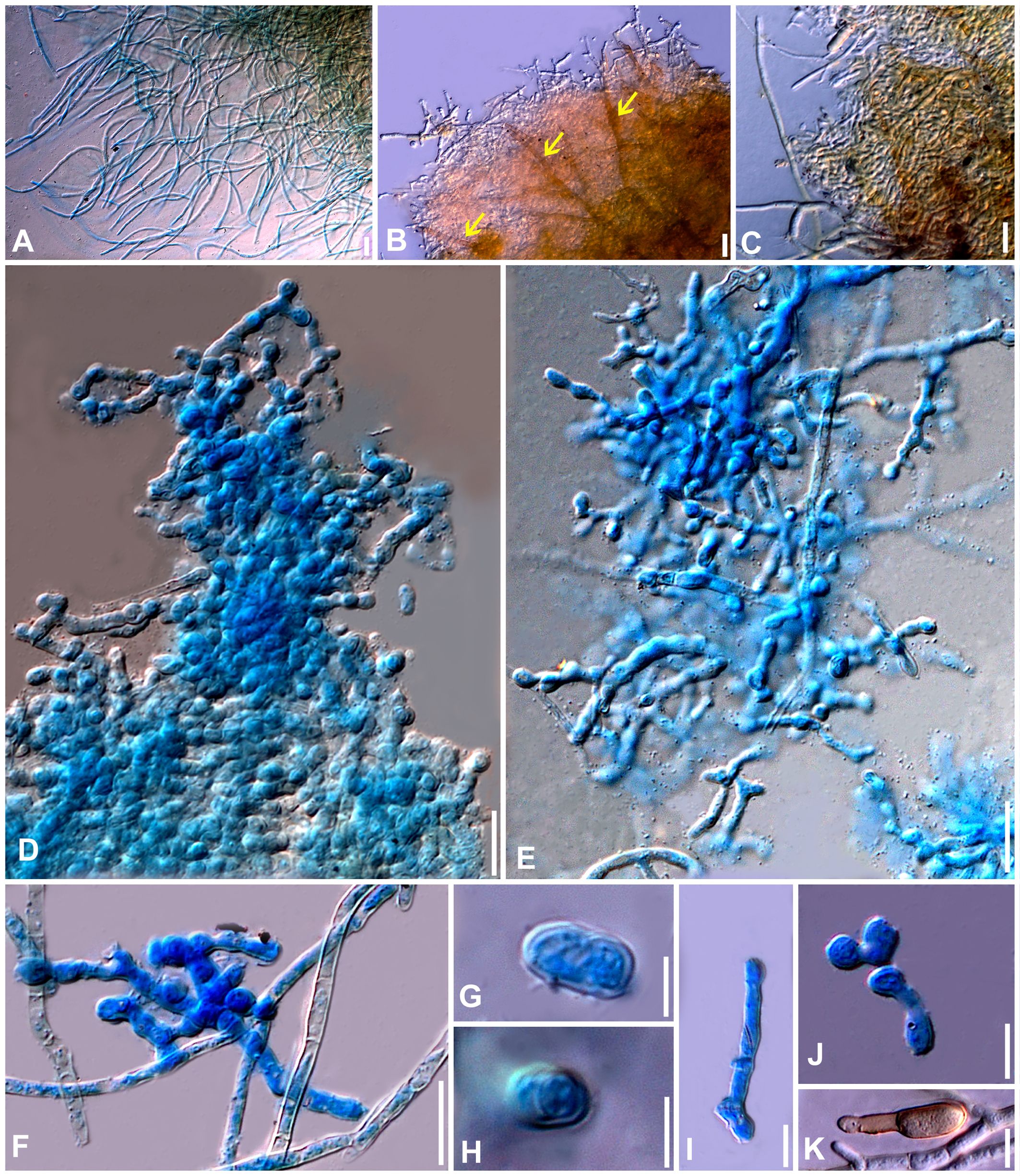
Figure 8. Microphotographs of the ex-type culture of Neoclypeosphaerella calotropidis (NFCCI 5983) on MEA. (A) Hyphae. (B, C) Fructification showing formation of chlamydospores with setae-like structures (yellow arrows for setae-like structures). (D–F) Developing chains of intercalary and terminal chlamydospores. (G, H) Chlamydospores. (I–K) Germinating chlamydospores. Scale bars: (A–F) 20 µm, (G, H) 5 µm, (I–K) 10 µm.

Figure 9. Microphotographs of the ex-type culture of Neoclypeosphaerella calotropidis (NFCCI 5983) on agar media supplemented with undefined vegetable peelings. (A) Top view of ex-epitype culture. (B, C) Stromata with fascicles of conidiophores (red arrows for developing conidia). (D–F) Highly branched conidiophores. (G–I) Conidiogenous cells with loci (yellow arrows). (J, K) Ampulliform conidiogenous cells (pink arrows). (L–U) Conidia. (V–Z) Chlamydospores in chain. Scale bars: (B–T) 20 µm, (U–Z) 10 µm.

Figure 10. Microphotographs of Neoclypeosphaerella calotropidis (AMH 10781). (A) Stromata with fascicles of conidiophores. (B) Stromata with fascicles of conidiophores and superficial hyphae. (C–E) Fascicles of conidiophores emerges through stomata. (F, G) Erumpent stromata bearing conidiophores with swollen basal cell. (H, I) Highly branched conidiophores. (J–L) Superficial hyphae with conidiophores. (M–P) Conidiophores with developing conidia. (Q–T) Polyblastic nature of conidiogenous cells (blue arrows). Scale bars: (A–L) 20 µm, (M–T) 10 µm.

Figure 11. Microphotographs of Neoclypeosphaerella calotropidis (AMH 10781). (A1–A23) Conidia. (B1–B3) Ramoconidia with apical hila and developing conidium (yellow arrows). (C1–C3) Intercalary conidia. (D1–D3) Conidia in catenation (orange arrows for point of catenation). (E1–E4) Conidiogenous nature of conidial cells (green arrows). (F1–F6) Conidia with developing conidium (blue arrows). (G1, G2) Development of conidiophores from conidial cells (pink arrows). Bars: 10 µm.
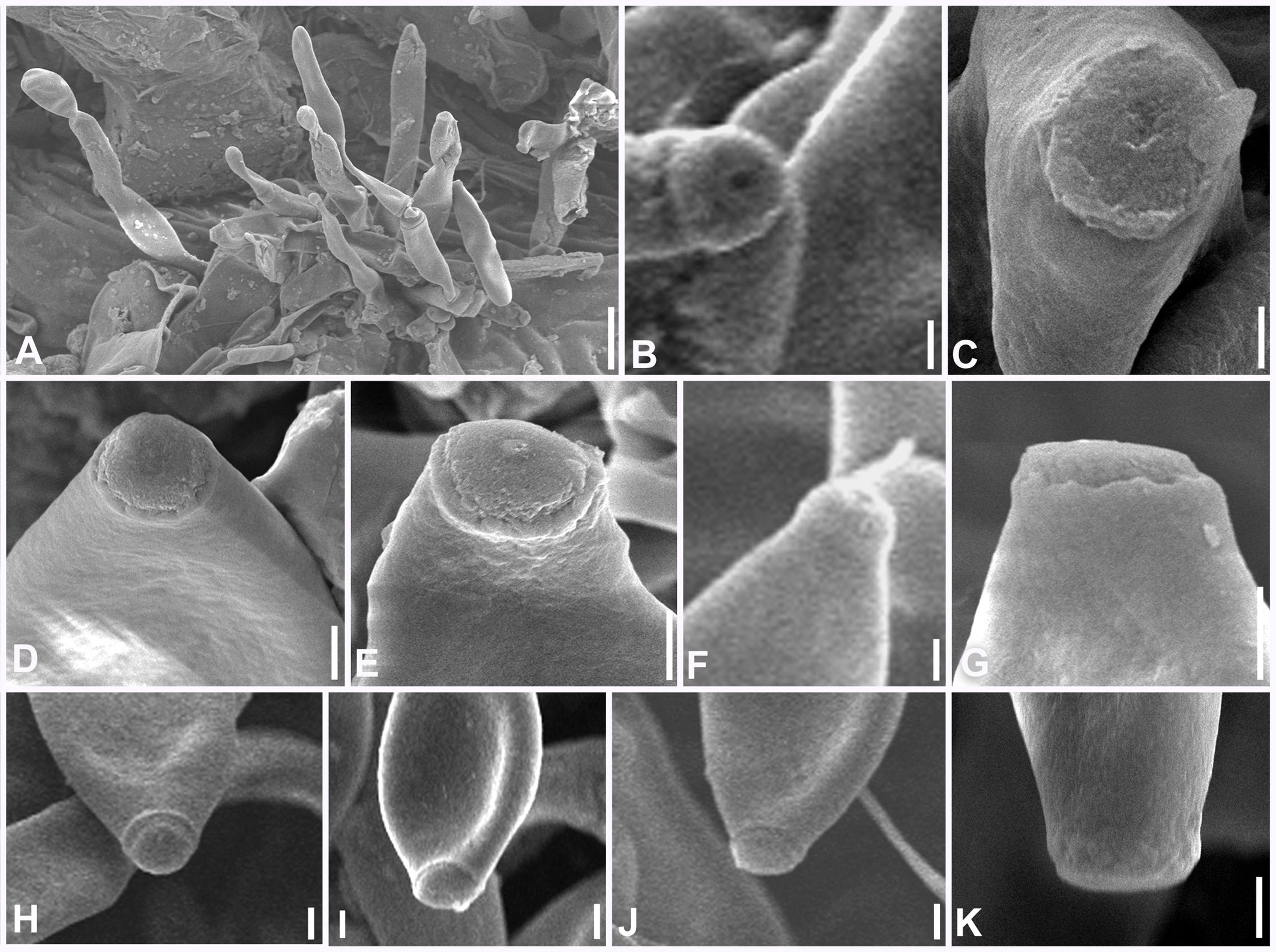
Figure 12. Scanning electron microphotographs of Neoclypeosphaerella calotropidis (AMH 10781). (A) Fascicle of conidiophores with conidia. (B–G) Top and lateral view of loci of conidiogenous cells. (H–K) Top and lateral view of hila of conidia. Scale bars: (A) 10 µm, (B–K) 1 µm.
MycoBank number: MB854797.
Etymology: This is composed of Neo- (new) and the genus name Clypeosphaerella.
Diagnosis: This differs from the genus Marcstadlera by developing fascicles of conidiophores emerging from stromata and conidia that are rarely catenate.
Description: Plant pathogenic. Ascomata epiphyllous, black, subepidermal to erumpent, subglobose, wall of 3–4 layers of medium to dark brown textura angularis, apical ostiole central. Asci aparaphysate, fasciculate, bitunicate, subsessile, broad ellipsoid to obclavate, straight to slightly curved, 8-spored. Ascospores bi- to multiseriate, overlapping, hyaline, guttulate, thin-walled, straight to slightly curved, ellipsoidal to obovoid with obtuse ends, widest in the middle of the apical cell, 1-septate, not constricted at the septum, tapering toward both ends, with a thin mucilaginous sheath. Conidiophores macronematous, mostly arising in fascicles from stromata, occasionally as lateral branches of superficial secondary hyphae or conidial cells, erect to slightly curved, divergent, subcylindrical to geniculate-sinuous at the tip, basal cell slightly swollen, mostly unbranched, rarely branched, smooth, sometimes slightly roughened, light brown to brown, septate, thick-walled. Conidiogenous cells polyblastic, cylindrical, integrated, terminal and intercalary, sympodial elongated, conidiogenous loci slightly protuberant, surrounded by a circular rim-like structure, forming a truncated apex with a centrally positioned small apical depression, loci thickened and darkened. Ramoconidia cylindrical to subcylindrical, rarely sickle-shaped. Intercalary conidia sometimes present, cylindrical to subcylindrical, sometimes curved, occurring in chains. Conidia sometimes blastocatenate, polymorph, cylindrical to subcylindrical, obclavate-cylindrical, acicular, ovoid to obovoid, doliiform, elliptical, L-shaped to sickle-shaped, rarely V-shaped, obtuse apex, base obconical, truncate at the base, surrounded by a circular rim-like structure, euseptate, smooth to slightly roughened, light olivaceous brown to brown, hilum thickened and darkened.
Type species: Neoclypeosphaerella calotropidis (Ellis and Everh.) Raghv. Singh, S. Rajwar, Sanjay, P.N. Singh and U. Braun.
Neoclypeosphaerella calotropidis (Ellis & Everh.) Raghv. Singh, S. Rajwar, Sanjay, P.N. Singh & U. Braun, comb. nov. (Figures 7-12).
MycoBank number: MB854798.
≡ Cercospora calotropidis Ellis and Everh., Rep. (Annual) Missouri Bot. Gard. 120 (1898).
= Phaeoramularia calotropidis (Ellis and Everh.) Kamal, A.S. Moses and R. Chaudhary, Mycol. Res. 94, 716 (1990).
= Passalora calotropidis (Ellis and Everh.) U. Braun, Schlechtendalia 5, 60 (2000).
= Pseudocercospora calotropidis (Ellis and Everh.) Haldar and J.B. Ray, J. Mycopathol. Res. 39(1), 43 (2001).
= Clypeosphaerella calotropidis (Ellis and Everh.) Videira and Crous, Stud. Mycol. 87, 314 (2017).
For additional synonyms see Crous and Braun (2003) and MycoBank (https://www.mycobank.org/).
Description: Leaf spots amphiphyllous, initially circular to subcircular, 6–7 mm diam., later irregular and spread over the entire leaf surface, brown to dark blackish brown. Colonies amphigenous, effuse, brown to dark brown, velvety. Mycelium mostly internal, sometimes superficial secondary hyphae developing from stromata, branched, septate, smooth, thin-walled, hyaline to very light olivaceous, (2−)2.5–3.5(−4) μm. Stromata present, globose to subglobose, mostly substomatal, later erumpent, pseudoparenchymatous, light olivaceous brown to mid brown, 20−25 × 15−25 μm. Conidiophores macronematous, mostly arising in fascicles from stromata, occasionally as lateral branches of superficial secondary hyphae or conidial cells, erect to slightly curved, divergent, subcylindrical to geniculate-sinuous at the tip, basal cell slightly swollen, mostly unbranched, rarely branched, smooth, sometimes slightly roughened, light brown to brown, 0−8-septate, thick-walled, (17−)25−85(−100) × (3−)3.5−5.5(−6.5) μm. Conidiogenous cells integrated, terminal as well as intercalary, polyblastic, cylindrical, conidiogenous loci slightly protuberant, surrounded by a circular rim-like structure, forming a truncated apex with a centrally positioned small apical depression (ultrastructure), loci thickened and darkened, 1.5−2 μm wide. Ramoconidia cylindrical to subcylindrical, rarely sickle-shaped, (40−)45–75(−115) × (3−)5–6(−6.5) μm, with 2 apical hila. Intercalary conidia sometimes present, cylindrical to subcylindrical, sometimes curved, occurring in chains of up to 4 conidia, (32−)40−108(−136) × (3−)4−5(−5.5) μm. Conidia mostly solitary, sometimes blastocatenate, polymorph [acicular, ovoid to obovoid, cylindrical or obclavate-cylindrical, doliiform, elliptical, Lshaped to sickle-shaped, rarely V-shaped], (14−)25–215(−250) × (2.5−)3–6(−7.5)] µm, obtuse apex, base obconical, truncate at the base, surrounded by circular rim-like structure, 0−12-euseptate, smooth to slightly roughened, light olivaceous brown to brown, hilum thickened and darkened, 1.5−2 μm diam.
Culture characteristics: Colonies slow-growing, reaching a diameter of approximately 6 mm on MEA and 7 mm on PDA after 14 days at 25°C ± 5°C. The colonies were circular in outline with a velvety aerial mycelium. On MEA, the upper surface white and fluffy, while the reverse was black. On PDA, the upper surface ranged from dark gray to black, with a brown to black reverse. On MEA: Hyphae (1.5–)2.5–4(–5) μm wide, branched, septate, smooth to slightly roughened and subhyaline to very light olivaceous brown. Fructification occurred with the formation of chlamydospores accompanied by seta-like structures. Setae branched, septate, smooth to slightly roughened, light brown to dark brown and (2–)2.5–3.5(–4) μm diam. Chlamydospores developed in chains, occurring intercalarily and terminally. They were spherical to oval, subhyaline to mid brown, thick-walled, smooth to slightly roughened, (5–)6–17(–23) × (4–)5–7(–8) μm. Germinating chlamydospores were also observed.
Sporulation takes place on agar media supplemented with undefined vegetable peelings. The colonies were whitish gray to smoky black. Stromata well developed, hard, irregular, and light olivaceous brown to blackish brown. Hyphae branched, septate, smooth-walled, subhyaline to light olivaceous, 2–3 μm wide. Conidiophores macronematous, mostly arising in fascicles from the stromata, occasionally solitary, sometimes reduced to a single-celled ampulliform conidiogenous cell, erect to slightly curved, divergent, subcylindrical, basal cell slightly swollen, mostly unbranched, rarely branched, smooth, sometimes slightly roughened, subhyaline to light olivaceous brown, 0−9-septate, thick-walled, (16–)25–70(–90) × (3–)4–5.5(–8.5) μm. Conidiogenous cells integrated, terminal as well as intercalary, mono- to polyblastic, cylindrical, conidiogenous loci slightly protuberant, loci thickened and darkened, (1.5−)2–2.5(–3) μm wide. Conidia solitary, simple, dry, subhyaline to light olivaceous brown, mostly cylindrical or obclavate-cylindrical, ovoid to obovoid, sometimes curved, smooth-walled, sometimes slightly roughened, thin to thick-walled, tapering toward an obtuse apex, sometimes apical cell swollen, 0−6-septate, constricted at the septa, (13–)18–50(–75) × (3–)4–5.5(–8) μm, base obconically truncated, hilum slightly thickened and darkened, 1.5−2.5 μm diam. Chlamydospores developed in chains, occurring intercalarily and terminally, spherical to oval, mostly horizontally but sometimes vertically and obliquely septate, subhyaline to mid brown, thick-walled, smooth to slightly roughened, (6–)8–10(–13) × (7–)9–13(–15) μm. Germinating chlamydospores were also observed.
Specimens examined: INDIA, Uttar Pradesh, Gorakhpur, on Calotropis procera, A. S. Moses (Herb. GPU No. KRNC 64, IMI 337033); INDIA, Uttar Pradesh, Gorakhpur, on Calotropis procera, Kamal (Herb. GPU No. KK 300, IMI 314694); INDIA, Uttar Pradesh, Gorakhpur, on Calotropis procera, C. Gupta (Herb. GPU No. KC-126, IMI 314110); INDIA, Uttar Pradesh, Gorakhpur, Caltropis procera, R. K. Verma (Herb. GPU No. KK 213, IMI 300481); INDIA, Uttar Pradesh, Varanasi, 25.2685°N 82.9905°E, on living leaves of Calotropis gigantea, 10 September 2024, Sanjay Yadav, MH-BHU 128 (AMH 10781), culture NFCCI 5983, gene sequence GenBank: PV112567 (ITS), PQ816342 (LSU), PV125517 (Rpb2); INDIA, Uttar Pradesh, Mirzapur, 25.1337°N 82.5644°E, on living leaves of Calotropis procera, 01 December 2024, Soumyadeep Rajwar, MH-BHU 129 (AMH 10782), culture NFCCI 5984, gene sequence GenBank: PV112568 (ITS), PQ816341 (LSU), PV125518 (Rpb2).
Notes: The genus Clypeosphaerella was established by Guatimosim et al. (2016) with the type species of Clypeosphaerella sticheri. This genus is morphologically similar to species of Mycosphaerella s. lat. but differs mainly in having a thicker upper wall of the ascomata, which resembles a pseudoclypeus. Moreover, Clypeosphaerella is phylogenetically distinct from other mycosphaerella-like fungi (Figures 1, 2) and forms a well-supported clade as determined by Guatimosim et al. (2016).
Three Clypeosphaerella species have been reported across the world, namely, C. calotropidis (Ellis and Everh.) Videira and Crous (Videira et al., 2017), C. quasiparkii (Cheew. et al.) Guatim. et al (Guatimosim et al., 2016), and C. sticheri Guatim. et al (Guatimosim et al., 2016). Clypeosphaerella calotropidis is the only species in this genus represented by an asexual morph, while the other two species are only known from their sexual morphs.
Basionym of Clypeosphaerella calotropidis is Cercospora calotropidis Ellis and Everh. Braun (2000a) transferred Cercospora calotropidis to the genus Passalora based on the morphological observations. He noted that this species was highly variable and exhibited characteristics that were intermediate between several genera, viz., Passalora (known for having fasciculate conidiophores and conidia formed singly), Phaeoramularia (characterized by conidia formed in chains), and Mycovellosiella (identified by secondary superficial hyphae with solitary conidiophores). This intermediate nature justified the transfer to Passalora, reflecting its closest alignment with the morphological traits of this genus. Furthermore, Braun (2000a) cited C. calotropidis as an example to demonstrate that the genera Passalora, Phaeoramularia, and Mycovellosiella should be merged, a view supported by Crous et al. (2001). A similar diagnostic approach was followed by Wilkinson et al. (2005) for Passalora calotropidis (Braun, 2000a) as the phylogenetic analysis based on ITS placed this species in a single-strain lineage closely related to Pseudocercospora (Wilkinson et al., 2005).
Based on a multigene analysis (LSU-Rpb2-ITS), Passalora calotropidis (CBS 129.30) clustered with Clypeosphaerella quasiparkii (CBS 123243) with high statistical support, which was found to be closely related to Pseudocercospora and separated as a sister lineage of Distocercospora pachyderma (CBS 138247) with high statistical support (Videira et al., 2017). Therefore, Passalora calotropidis was recombined as Clypeosphaerella calotropidis (CBS 129.30). However, this analysis did not incorporate the type species of Clypeosphaerella, C. sticheri.
Rajeshkumar et al. (2021) introduced the new genus Pedrocrousiella based on LSU-Rpb2 sequence data, which formed a sister lineage to Distocercospora pachyderma (CBS 138247) with high statistical support (BI-PP/ML-BS: 1/98). In this analysis, the inclusion of all three species of Clypeosphaerella, along with their type species, forming a monophyletic group, suggested a significant finding in their monophyletic evolutionary relationships. However, in the parsimony analysis, the relationship between Clypeosphaerella sticheri and other Clypeosphaerella species was unresolved. This unresolved relationship might be due to the missing Rpb2 data for Clypeosphaerella sticheri (Rajeshkumar et al., 2021).
Pteridopassalora was introduced by C. Nakash. and Crous (Chen et al., 2022). It was established based on LSU-Rpb2-ITS sequence data, which clustered closely with the genus Clypeosphaerella and formed a sister lineage of Distocercospora pachyderma (CBS 138247). The analysis included two species of Clypeosphaerella, namely, C. calotropidis and C. quasiparkii, but not the type species, C. sticheri.
Based on both datasets (Figures 1, 2), the type species of Clypeosphaerella, C. sticheri, is segregated from the other two Clypeosphaerella species, C. calotropidis and C. quasiparkii, which cluster together with the newly generated sequences obtained from the cultures NFCCI 5983 and NFCCI 5984, isolated from Calotropis spp., with strong statistical support (BI-PP/ML-BS: 1/96). Consequently, a new genus, Neoclypeosphaerella, is introduced to accommodate C. calotropidis and C. quasiparkii. The significant nucleotide differences between Clypeosphaerella sticheri and Neoclypeosphaerella calotropidis (ITS: 28 differences with 11 gaps, LSU: 12 differences with 3 gaps) suggest that they do not belong to the same genus and should be maintained as separate, independent genera.
Several cercosporoid fungi have been described from Calotropis spp., namely, Cercospora baroipurensis Purkay. and Mallik (Purkay. and Mallik, 1978), Clypeosphaerella calotropidis (Ellis and Everh.) Videira and Crous (Chupp, 1954; Kamal et al., 1990; U. Braun, 2000a; Wilkinson et al., 2005; Haldar and Ray, 2001; Videira et al., 2017), Mycosphaerella calotropidis T.S. Viswan. (Viswan. and Tilak, 1960), Paracercosporidium microsorum (Sacc.) U. Braun et al (Videira et al., 2017), and Pseudocercospora peronosporoidea (Pat. and Har.) Deighton (Deighton, 1981).
Cercospora baroipurensis and Pseudocercospora peronosporoidea can be easily distinguished from N. calotropidis based on conidial and conidiophore characteristics. In C. baroipurensis, the conidia are hyaline, while the conidiophores are colored with thickened and darkened loci and hila. In contrast, P. peronosporoidea has both conidia and conidiophores that are colored, without any thickened or darkened loci and hila.
Clypeosphaerella calotropidis closely resembles our two collected samples (NFCCI 5983, NFCCI 5984) on Calotropis spp., exhibiting several similarities. Both samples exhibit indefinite leaf spots and large circular to irregular blotches merging into black patches. The immersed, subhyaline mycelium produces amphigenous fruiting with stromata filling stomatal openings. Conidiophores, in fascicles, are pigmented, sparingly septate, occasionally branched, and mildly geniculate near the tip, with a blunt or conic apex bearing a conspicuous conidiogenous locus (scar). Conidia are almost straight to slightly curved, cylindrical to obclavate, pigmented, sparingly catenate, septate, with an obconic base and rounded apex. In our collected samples (NFCCI 5983, NFCCI 5984), some additional features were developed only at a very late stage of infection, including the formation of slightly longer mature acicular conidia (up to 250 μm), the occasional development of superficial secondary hyphae, and the catenation of conidia. These features were not observed during the development of the early stages. Our phylogenetic analysis, providing strong statistical support (BI-PP/ML-BS: 1/99), corroborated these morphological findings and confirmed C. calotropidis as the type species of a new genus, Neoclypeosphaerella.
Paracercosporidium microsorum can be clearly distinguished from N. calotropidis by its internal hyphae and solitary and cylindrical to obclavate conidia.
The asexual morph of Mycosphaerella calotropidis is unknown, making it impossible to compare this name with N. calotropidis. Additionally, the absence of molecular sequence data makes it impossible to determine whether it represents the perfect state of N. calotropidis. However, this detail is irrelevant in terms of nomenclatural implications for the current case because Cercospora calotropidis, the name of the basionym, is much older than M. calotropidis. Hence, it only remains open whether the later name is a synonym of N. Calotropidis or not.
Based on both datasets, Marcstadlera could not be placed within any of the currently described genera of the Mycosphaerellaceae (Figures 1, 2) and is positioned as a sister lineage to Neoclypeosphaerella. Marcstadlera is represented by its asexual morph and belongs to the cercosporoid group of fungi in Mycosphaerellaceae based on both datasets. Many asexual morphs linked to mycosphaerella-like sexual morphs exhibit cercosporoid morphology (Videira et al., 2017). Since sexual morphs are morphologically conserved, genera within Mycosphaerellaceae are primarily distinguished based on their asexual morphs (Crous et al., 2009). The type species of Neoclypeosphaerella, N. calotropidis, is represented by its asexual morph and is morphologically distinct from Marcstadlera. In vivo, Neoclypeosphaerella primarily develops internal mycelium and forms well-developed stromata bearing fascicles of conidiophores that are geniculate-sinuous at the tip, mostly simple, occasionally branched, and septate. The conidiogenous cells are both terminal and intercalary, with slightly protuberant, thickened, and darkened loci. In contrast, Marcstadlera exhibits significant morphological differences. In vivo, it develops predominantly external mycelium, lacks stromata entirely, and produces conidiophores that are micronematous to semi-macronematous, mononematous, unbranched, and aseptate. These conidiophores arise individually from intercalary or terminal cells of external hyphae and are reduced to conidiogenous cells. The conidiogenous loci (scars), formed on cylindrical or peg-like conidiogenous cells, are unthickened to slightly thickened and darkened. These differences justify the introduction of a new genus, Marcstadlera, for this monotypic lineage.
The significant nucleotide differences between Marcstadlera and Neoclypeosphaerella (ITS: 16, LSU: 14, Rpb2: 69) indicate that they cannot belong to the same genus and should be maintained as separate, independent genera.
Although Marcstadlera morphologically resembles Mycovellosiella species, as both develop secondary superficial hyphae with solitary conidiophores, the two genera are phylogenetically distant (Figures 1, 2). The Mycovellosiella-like morphological traits are considered phylogenetically and taxonomically insignificant and appear unreliable (Videira et al., 2017).
In a megablast search of LSU sequences for Marcstadlera in NCBI’s GenBank nucleotide database, Rosenscheldiella brachyglottidis (PDD 94939) appeared with 96% sequence similarity (508/527) with no gaps. The phylogenetic analysis based on LSU-Rpb2 (Figure 1) placed R. brachyglottidis as a sister lineage to Neoclypeosphaerella, though with very low statistical support. Rosenscheldiella brachyglottidis is represented by its sexual morph, which can be easily differentiated from the closely related sexual morph N. quasiparkii (CBS 123243) by forming pseudothecia with fissitunicate asci, which develop externally to the host leaf on small pads of stromatic tissue growing superficially from hyphae that penetrate through the stomata (Sultan et al., 2011). Therefore, the significant morphological differences between R. brachyglottidis and N. quasiparkii suggest that R. brachyglottidis should be tentatively retained in the genus Rosenscheldiella rather than being reclassified under N. quasiparkii, reflecting uncertainties in the taxonomy of these organisms.
Based on both datasets, it has been confirmed that Clypeosphaerella, Marcstadlera, Neoclypeosphaerella, and Rosenscheldiella are distinct genera, forming separate clades (Figures 1, 2).
Neoclypeosphaerella quasiparkii (Cheew. et al.) Raghv. Singh & Sham. Kumar, comb. nov.
MycoBank number: MB854799.
≡ Mycosphaerella quasiparkii Cheew. et al., Persoonia 21, 85 (2008).
= Clypeosphaerella quasiparkii (Cheew. et al.) Guatim. et al. Persoonia 37, 121 (2016).
Description and illustration: Cheewangkoon et al. (2008).
Notes: Based on dataset 1, N. calotropidis and N. quasiparkii are clustered together with very low statistical support (BI-PP/ML-BS: 0.90/-) (Figure 1). When LSU, ITS, and Rpb2 are used as barcodes, they provide valuable insights into evolutionary relationships at the species level in Mycosphaerellaceae (Chen et al., 2022). In dataset 2, both species are clustered together with high statistical support (BI-PP/ML-BS: 1/96) (Figure 2), indicating a close relationship. Therefore, N. quasiparkii is accommodated in Neoclypeosphaerella along with N. calotropidis, despite being represented by different morphs.
Discussion
Taxonomic challenges within the family Mycosphaerellaceae are numerous and arise from both historical classification practices and current methodological limitations. One of the primary issues is the morphological similarity among species—their fruiting bodies and spores are often small, conserved, and difficult to distinguish. This is compounded by morphological convergence across unrelated taxa occupying similar ecological niches, frequently leading to misidentifications. Historically, the genus Mycosphaerella served as a “dumping ground” for numerous unrelated species that had minimal or overlapping morphological characteristics. This practice created significant confusion, especially in light of uncertain or incorrect anamorph–teleomorph associations (Crous et al., 2007, Crous et al., 2009; Groenewald et al., 2013). The problem is further compounded by the presence of cryptic species complexes; notably, Mycosphaerella graminicola (now Zymoseptoria tritici), the causal agent of Septoria tritici blotch in wheat, was revealed to consist of several genetically distinct but morphologically similar species (Quaedvlieg et al., 2011).
Modern molecular phylogenetic studies have shown that many traditional genera within Mycosphaerellaceae are paraphyletic or polyphyletic, indicating that current classifications often fail to reflect true evolutionary relationships (Videira et al., 2017). However, progress is limited by the lack of molecular data for many species, especially ex-type sequences and material from undersampled regions. Furthermore, different gene regions used in phylogenetic analyses can yield conflicting results (Videira et al., 2017).
Culturing remains a significant challenge, particularly for obligate or poorly sporulating taxa, which restricts the ability to extract DNA, conduct reproductive studies, or perform pathogenicity and fungicide sensitivity tests (Simon et al., 2009; Hunter et al., 2011). In plant disease diagnostics, multiple species can coexist in a single lesion, and high but variable host specificity further complicates accurate identification and understanding of host–pathogen dynamics (Braun et al., 2013, Braun et al., 2015, Braun et al., 2016). Additional complications stem from poorly understood mating systems and the frequent renaming or redefinition of genera, leading to nomenclatural instability (Aylward et al., 2022). This instability hinders effective communication in applied disciplines such as plant pathology and quarantine regulation.
To overcome the challenges posed by the Mycosphaerellaceae family, such as taxonomic ambiguity, diagnostic limitations, and fungicide resistance, several modern approaches have been proposed. Molecular phylogenetics and genomics, particularly multilocus sequence typing (MLST) and whole-genome sequencing (WGS), have significantly enhanced the resolution of phylogenetic relationships and revealed cryptic species diversity (Stengel et al., 2022; Menicucci et al., 2025). DNA barcoding using markers like ITS, LSU, Rpb2, EF-1α, and β-tubulin further aids in accurate species identification (Videira et al., 2017; Abdollahzadeh et al., 2020; Bakhshi et al., 2021; Bakhshi and Braun, 2022; Chen et al., 2022). Species boundaries within species complexes were further resolved using sequence data combined with several clades found to be host-specific—supporting the concept of host-driven speciation in the Mycosphaerellaceae (Crous et al., 2008; Arzanlou et al., 2009; Li et al., 2020; Ko et al., 2025). These findings highlight the value of a polyphasic approach in fungal taxonomy, integrating molecular, morphological, and ecological data to refine species concepts and improve classification systems (Crous et al., 2013a, Crous et al., 2013b).
Advanced diagnostics like quantitative PCR (qPCR) and loop-mediated isothermal amplification (LAMP) allow rapid pathogen detection, while integrated disease management and genomic-assisted breeding enhance control (Patel et al., 2022). Global collaboration via platforms like MycoBank and GenBank supports data sharing, aiding in managing Mycosphaerellaceae challenges (Robert et al., 2013; Hariharan and Prasannath, 2021; Zhang et al., 2023).
This study clarified the phylogenetic placement of the newly introduced genera Marcstadlera and Neoclypeosphaerella within the Mycosphaerellaceae family using an integrative taxonomic approach that combined multigene phylogenetic analysis (based on ITS, LSU, and Rpb2 barcode genes), morphological characterization, and host/ecological data. The results also confirmed the paraphyly of Clypeosphaerella and the polyphyly of Mycovellosiella, with isolates clustering into distinct genera consistent with current taxonomic frameworks. These findings support the reliability of DNA-based classification, particularly when morphology alone proves insufficient for distinguishing Mycosphaerellaceaean fungi at the generic level (Videira et al., 2017; Chen et al., 2022).
In the Indian context, fungal diversity is remarkably high, with new species being reported annually. Historically, studies on phytopathogenic fungi related to Mycosphaerellaceae in India have primarily relied on morphological data (Singh et al., 2007, Singh et al., 2008, Singh and Kumar, 2011, Singh et al., 2012, Singh et al., 2013, Singh et al., 2014a, Singh et al., 2014b, Singh et al., 2020a, Singh et al., 2020b, Singh et al., 2022; Kamal, 2010; Kumar et al., 2014; Kumar and Singh, 2015, Kumar and Singh, 2016; Singh and Kumar, 2017; Kushwaha et al., 2020; Verma et al., 2023). However, recent investigations (Singh et al., 2020b; Verma et al., 2021a, Verma et al., 2021b; Yadav et al., 2021, Yadav et al., 2022, Yadav et al., 2023) demonstrate a significant shift toward the use of cultures, SEM imaging, and DNA sequencing to support taxonomic conclusions.
Overall, this research contributes to the ongoing revision of Mycosphaerellaceae classification using molecular tools, while also uncovering hidden diversity and enhancing our understanding of species and generic boundaries, as well as evolutionary relationships.
Data availability statement
The specimen studied in this work was deposited in the Ajrekar Mycological Herbarium (AMH), Agharkar Research Institute (ARI), Pune and National Fungal Culture Collection of India (NFCCI), Pune, Maharashtra, India. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, ITS: PQ012587, PQ013688, PV112567, and PV112568; LSU: PQ012588, PQ013689, PQ816342, and PQ816341; Rpb2: PQ034553, PQ034554, PV125517, and PV125518.
Author contributions
GS: Data curation, Methodology, Formal Analysis, Writing – review & editing, Investigation. SR: Data curation, Methodology, Investigation, Formal Analysis, Writing – review & editing. SKk: Formal Analysis, Writing – review & editing. SY: Methodology, Formal Analysis, Writing – review & editing. PK: Writing – review & editing, Formal Analysis, Methodology. RS: Investigation, Software, Methodology, Conceptualization, Formal Analysis, Writing – original draft. KK: Investigation, Writing – review & editing, Formal Analysis, Methodology. SM: Writing – review & editing, Formal Analysis. PS: Writing – review & editing, Formal Analysis, Validation, Supervision. SKu: Writing – review & editing, Validation, Formal Analysis. UB: Validation, Writing – review & editing, Supervision.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. Raghvendra Singh thanks the Science & Engineering Research Board (SERB), Department of Science & Technology (DST), Govt. of India (Scheme No. CRG/2020/006053); Institution of Eminence (R/Dev./D/IoE/Incentive/2021-22/32387), BHU, Varanasi; Bridge Grant (No. SRICC/Bridge Grant/2024-25/3151), BHU, Varanasi; and Sanjay Yadav thanks Raja Jwala Prasad Post-Doctoral Fellowship (No. SRICC/RJP-PDF/2023-24/6158) under Institution of Eminence, BHU, Varanasi for providing financial support.
Acknowledgments
The authors are indebted to reviewers for helpful comments and the curators of AMH and NFCCI for accepting material and providing accession numbers. They are also thankful to the Head, CAS in Botany, Banaras Hindu University, Varanasi, for the instrumental facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdollahzadeh, J., Groenewald, J. Z., Coetzee, M. P. A., Wingfield, M. J., and Crous, P. W. (2020). Evolution of lifestyles in Capnodiales. Stud. Mycol. 95, 381–414. doi: 10.1016/j.simyco.2020.02.004
An, Y. Y., Zeng, X. Y., Geng, K., Hyde, K. D., and Wang, Y. (2021). One new species and one new record of Zasmidium in China. Biodivers. Data. J. 9, e59001. doi: 10.3897/BDJ.9.e59001
Arzanlou, M., Crous, P. W., and Zwiers, L.-H. (2009). Evolutionary dynamics of mating-type loci of Mycosphaerella spp. occurring on banana. Eukaryot. Cell 9, 164–172. doi: 10.1128/EC.00194-09
Aylward, J., Havenga, M., Wingfield, B. D., Wingfield, M. J., Dreyer, L. L., Roets, F., et al. (2022). Novel mating-type-associated genes and gene fragments in the genomes of Mycosphaerellaceae and Teratosphaeriaceae fungi. Mol. Phylogenet. Evol. 177, 107456. doi: 10.1016/j.ympev.2022.107456
Bakhshi, M. and Braun, U. (2022). Acericercospora hyrcanica gen. et sp. nov. (Mycosphaerellaceae) and Paramycocentrospora acericola gen. et sp. nov. (Dothidotthiaceae) on maple trees in Hyrcanian forests. Mycol. Prog. 21, 71. doi: 10.1007/s11557-022-01824-x
Bakhshi, M. and Crous, P. W. (2024). The genera of fungi - G7: Hirudinaria. Fungal Syst. Evol. 14, 1–8. doi: 10.3114/fuse.2024.14.01
Bakhshi, M., Zare, R., Arzanlou, M., and Narmani, A. (2020). Occurrence of Cercospora leaf spot disease on Cucumis melo in Guilan province and characterization of the causal agent(s) using molecular and morphological approaches. Iran J. Plant Pathol. 56, 17–31. doi: 10.22034/ijpp.2020.43566
Bakhshi, M., Zare, R., Braun, U., and Taheri, H. (2021). Polyphasic taxonomy of four passalora-like taxa occurring on fruit and forest trees. Mycol. Prog. 20, 1157–1173. doi: 10.1007/s11557-021-01725-5
Bhalla, K., Singh, S. K., and Srivastava, A. K. (1997). Further Mycovellosiella species from the Indian sub-continent. Mycol. Res. 101, 1496–1498. doi: 10.1017/S0953756297004310
Braun, U. (1996). Taxonomic notes on some species of the Cercospora complex (IV). Sydowia 48, 205–217.
Braun, U. (2000a). Annotated list of Cercospora spp. described by C. Spegazzini. Schlechtendalia, 5, 57–79.
Braun, U. (2009). New species, combinations, and records of hyphomycetes. Schlechtendalia 19, 63–71.
Braun, U., Crous, P. W., and Nakashima, C. (2013). Cercosporoid fungi (Mycosphaerellaceae) 1. Species on other fungi, Pteridophyta and Gymnospermae. IMA Fungus 4, 265–345. doi: 10.5598/imafungus.2013.04.02.12
Braun, U., Crous, P. W., and Nakashima, C. (2015). Cercosporoid fungi (Mycosphaerellaceae) 4. Species on dicots (Acanthaceae to Amaranthaceae). IMA Fungus 6, 373–469. doi: 10.5598/imafungus.2015.06.02.09
Braun, U., Crous, P. W., and Nakashima, C. (2016). Cercosporoid fungi (Mycosphaerellaceae) 5. Species on dicots (Anacardiaceae to Annonaceae). IMA Fungus 7, 161–216. doi: 10.5598/imafungus.2016.07.01.10
Cheewangkoon, R., Crous, P. W., Hyde, K. D., Groenewald, J. Z., and To-anan, C. (2008). Species of Mycosphaerella and related anamorphs on Eucalyptus leaves from Thailand. Persoonia 21, 77–91. doi: 10.3767/003158508X370857
Chen, Q., Bakhshi, M., Balci, Y., Broders, K. D., Cheewangkoon, R., Chen, S. F., et al. (2022). Genera of phytopathogenic fungi: GOPHY 4. Stud. Mycol. 101, 417–564. doi: 10.3114/sim.2022.101.06
Crous, P. W. and Braun, U. (2003). “Mycosphaerella and its anamorphs: 1. Names published in Cercospora and Passalora,” in CBS biodiversity series no. 1 (Westerdijk Fungal Biodiversity Institute, Utrecht, the Netherlands).
Crous, P. W., Braun, U., and Groenewald, J. Z. (2007). Mycosphaerella is polyphyletic. Stud. Mycol. 58, 1–32. doi: 10.3114/sim.2007.58.01
Crous, P. W., Braun, U., Hunter, G. C., Wingfield, M. J., Verkley, G. J. M., Shin, H. D., et al. (2013a). Phylogenetic lineages in pseudocercospora. Stud. Mycol. 75, 37–114. doi: 10.3114/sim0005
Crous, P. W., Cowan, D. A., Maggs-Kölling, G., Yilmaz, N., Larsson, E., Angelini, C., et al. (2020a). Fungal Planet description sheets: 1112–1181. Persoonia 46, 313–528. doi: 10.3767/persoonia.2020.45.10
Crous, P. W., Groenewald, J. Z., and Shivas, R. G. (2010). Fungal planet 59. Pseudocercospora nephrolepidicola sp. nov. Persoonia 25, 138–139.
Crous, P. W., Kang, J. C., and Braun, U. (2001). A phylogenetic redefinition of anamorph genera in Mycosphaerella based on ITS rDNA sequence and morphology. Mycologia 93, 1081–1101. doi: 10.1080/00275514.2001.12063243
Crous, P. W., Shivas, R. G., Quaedvlieg, W., van der Bank, M., Zhang, Y., Summerell, B. A., et al. (2014). Fungal Planet description sheets: 214–280. Persoonia 32, 184–306. doi: 10.3767/003158514X682395
Crous, P. W., Summerell, B. A., Carnegie, A. J., Wingfield, M. J., Hunter, G. C., Burgess, T. I., et al. (2009). Unravelling Mycosphaerella: Do you believe in genera? Persoonia 23, 99–118. doi: 10.3767/003158509X479487
Crous, P. W., Summerell, B. A., Mostert, L., and Groenewald, J. Z. (2008). Host specificity and speciation of Mycosphaerella and Teratosphaeria species associated with leaf spots of Proteaceae. Persoonia 20, 59–86. doi: 10.3767/003158508X323949
Crous, P. W., Wingfield, M. J., Cheewangkoon, R., Carnegie, A. J., Burgess, T. I., Summerell, B. A., et al. (2019). Foliar pathogens of eucalypts. Stud. Mycol. 94, 125–298. doi: 10.1016/j.simyco.2019.08.001
Crous, P. W., Wingfield, M. J., Guarro, J., Cheewangkoon, R., van der Bank, M., Swart, W. J., et al. (2013b). Fungal Planet description sheets: 154–213. Persoonia 31, 188–296. doi: 10.3767/003158513X675925
Crous, P. W., Wingfield, M. J., Schumacher, R. K., Akulov, A., Bulgakov, T. S., Carnegie, A. J., et al. (2020b). New and interesting fungi. 3. Fung. Syst. Evol. 6, 157–231. doi: 10.3114/fuse.2020.06.09
Deighton, F. C. (1976). Studies on Cercospora and allied genera. VI. Pseudocercospora Speg., Pantospora Cif., and Cercoseptoria Petr. Mycol. Pap. 140, 1–168.
Deighton, F. C. (1981). Pseudocercospora peronosporoidea (Pat. and Har.) comb. nov. a little-known species from Chad. Trans. Br. Mycol. Soc 77, 200–202. doi: 10.1016/S0007-1536(81)80200-8
Geiser, D. M., Pitt, J. I., and Taylor, J. W. (1998). Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc. Natl. Acad. Sci. U.S.A. 95, 388–393. doi: 10.1073/pnas.95.1.388
Goh, T.-K. and Hsieh, W. H. (1987). Studies on Cercospora and allied genera of Taiwan (V). T. Mycol. Soc R. China. 2, 125–148. doi: 10.1079/9780851983653.0000
Groenewald, J. Z., Nakashima, C., Nishikawa, J., Shin, H. D., Park, J. H., Jama, A. N., et al. (2013). Species concepts in Cercospora: spotting the weeds among the roses. Stud. Mycol. 75, 115–170. doi: 10.3114/sim0012
Guatimosim, E., Schwartsburd, P. B., Barreto, R. W., and Crous, P. W. (2016). Novel fungi from an ancient niche: cercosporoid and related sexual morphs on ferns. Persoonia 37, 106–141. doi: 10.3767/003158516X690934
Haldar, D. and Ray, J. B. (2001). Leaf inhabiting fungi from West Bengal, India. J. Mycopathol. Res. 39, 43–47.
Hariharan, G. and Prasannath, K. (2021). Recent advances in molecular diagnostics of fungal plant pathogens: a mini review. Front. Cell. Infect. Microbiol. 10. doi: 10.3389/fcimb.2020.600234
Hawksworth, D. L. (1974). Mycologist’s handbook (Kew: Commonwealth Mycological Institute), 231. doi: 10.1016/S0007-1528(74)80047-7
Hunter, G. C., Crous, P. W., Carnegie, A. J., and Wingfield, M. J. (2011). Mycosphaerella and Teratosphaeria diseases of Eucalyptus: easily confused and with serious consequences. Fungal Divers. 50, 145–166. doi: 10.1007/s13225-011-0131-z
Hunter, G. C., Wingfield, B. D., Crous, P. W., and Wingfield, M. J. (2006). A multi-gene phylogeny for species of Mycosphaerella occurring on Eucalyptus leaves. Stud. Mycol. 55, 147–161. doi: 10.3114/sim.55.1.147
Huson, D. H. (1998). SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics 14, 68–73. doi: 10.1093/bioinformatics/14.1.68
Huson, D. H. and Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Mol. Biol. Evol. 23, 254–267. doi: 10.1093/molbev/msj030
Kamal, A. (2010). Cercosporoid fungi of India (Dehra Dun, India: Bishen Singh Mahendra Pal Singh), 1–351.
Kamal, A., Rai, A. N., Moses, A. S., and Chaudhary, R. (1990). Two new species and a new combination in Phaeoramularia from Uttar Pradesh, India. Mycol. Res. 94, 714–717. doi: 10.1016/S0953-7562(09)80676-X
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kharwar, R. N., Singh, P. N., and Chaudhary, R. K. (1996). New species of Mycovellosiella associated with foliar spots in Nepal. Mycol. Res. 100, 689–692. doi: 10.1016/S0953-7562(96)80200-0
Kirschner, R. and Wang, H. (2015). New species and records of mycosphaerellaceous fungi from living fern leaves in East Asia. Mycol. Prog. 14, 1–10. doi: 10.1007/s11557-015-1085-4
Ko, Y.-Z., Shih, H.-C., Shiao, M.-S., and Chiang, Y.-C. (2025). Cryptic host-associated differentiation and diversity: unravelling the evolutionary dynamics of the plant pathogen Lasiodiplodia. IMA Fungus 16, e147543. doi: 10.3897/imafungus.16.147543
Koufopanou, V., Burt, A., and Taylor, J. W. (1997). Concordance of gene genealogies reveals reproductive isolation in the pathogenic fungus Coccidioides immitis. Proc. Natl. Acad. Sci. U.S.A. 94, 5478–5482. doi: 10.1073/pnas.94.10.5478
Kumar, S. and Singh, R. (2015). Passalora musicola, sp. nov. – a new Indian hyphomycete. Sydowia 67, 21–23. doi: 10.12905/0380.sydowia67-2015-0021
Kumar, S. and Singh, R. (2016). Passalora caesalpiniicola sp. nov. from India on Caesalpinia bonduc. Mycotaxon 131, 25–30. doi: 10.5248/131.25
Kumar, S., Singh, R., Upadhyaya, P. P., and Saini, D. C. (2014). First report of foliar disease caused by Cercospora apii s. lat. on Nymphaea nouchali from Uttar Pradesh, India. J. New. Biol. Rep. 3, 252–254.
Kushwaha, P., Singh, R., and Chaurasia, B. (2020). Ramularia titarpaniensis—a new species of ramularioid complex from central India. Phytotaxa 429, 274–280. doi: 10.11646/phytotaxa.429.4.3
Li, J., Cornelissen, B., and Rep, M. (2020). Host-specificity factors in plant pathogenic fungi. Fungal Genet. Biol. 144, 103447. doi: 10.1016/j.fgb.2020.103447
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
MaChado, A. R., Pinho, D. B., Silva, M., and Pereira, O. L. (2012). First report of leaf spot disease caused by Cercosporella pfaffiae on Brazilian ginseng (Pfaffia glomerata) in Brazil. Plant Dis. 96, 1702–1702. doi: 10.1094/PDIS-06-12-0614-PDN
Maddison, W. P. and Maddison, D. R. (2018). Mesquite: A modular system for evolutionary analysis. Version 3.61. Available online at: http://www.mesquiteproject.org (Accessed March 5, 2025).
Mehta, Y. R., Galbieri, R., Marangoni, M. S., Borsato, L. C., Rodrigues, H. P., Pereira, J., et al. (2016). Mycosphaerella areola–the teleomorph of Ramularia areola of cotton in Brazil, and its epidemiological significance. Am. J. Plant Sci. 7, 1415–1422. doi: 10.4236/ajps.2016.710135
Melo, J. A. S., Silva, E. S. O., Brito, A. C. Q., Mello, J. F., Souza, A. E. A., Souza-Motta, C. M., et al. (2025). A new genus of Mycosphaerellaceae associated with leaf spots in Philodendron sp. from the Brazilian Atlantic forest. Mycol. Prog. 24, 14. doi: 10.1007/s11557-025-02034-x
Menicucci, A., Iacono, S., Ramos, M., Fiorenzani, C., Peres, N. A., Timmer, L. W., et al. (2025). Can whole genome sequencing resolve taxonomic ambiguities in fungi? The case study of Colletotrichum associated with ferns. Front. Fungal Biol. 6. doi: 10.3389/ffunb.2025.1540469
Nakashima, C., Motohashi, K., Chen, C. Y., Groenewald, J. Z., and Crous, P. W. (2016). Species diversity of Pseudocercospora from far East Asia. Mycol. Prog. 15, 1093–1117. doi: 10.1007/s11557-016-1231-7
Patel, R., Mitra, B., Vinchurkar, M., Adami, A., Patkar, R., Giacomozzi, F., et al. (2022). A review of recent advances in plant–pathogen detection systems. Heliyon 8, e11855. doi: 10.1016/j.heliyon.2022.e11855
Philippe, H. and Bryant, D. (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681. doi: 10.1534/genetics.105.048975
Quaedvlieg, W. G. J. M., Binder, M., Groenewald, J. Z., Summerell, B. A., Carnegie, A. J., Burgess, T. I., et al. (2014). Introducing the consolidated species concept to resolve species in the Teratosphaeriaceae. Persoonia-Molecular Phylogeny Evol. Fungi. 33, 1–40. doi: 10.3767/003158514X681981
Quaedvlieg, W., Kema, G. H. J., Groenewald, J. Z., Verkley, G. J. M., Seifbarghi, S., Razavi, M., et al. (2011). Zymoseptoria gen. nov.: a new genus to accommodate Septoria-like species occurring on graminicolous hosts. Persoonia 26, 57–69. doi: 10.3767/003158511X571841
Rajeshkumar, K. C., Braun, U., Groenewald, J. Z., Lad, S. S., Ashtekar, N., Fatima, S., et al. (2021). Phylogenetic placement and reassessment of Asperisporium pongamiae as Pedrocrousiella pongamiae gen. et comb. nov. (Mycosphaerellaceae). Fungal Syst. Evol. 7, 165–176. doi: 10.3114/fuse.2021.07.08
Rehner, S. A. and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/S0953-7562(09)80409-7
Robert, V., Vu, D., Amor, A. B. H., van de Wiele, N., Brouwer, C., Jabas, B., et al. (2013). MycoBank gearing up for new horizons. IMA Fungus 4, 371–379. doi: 10.5598/imafungus.2013.04.02.16
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D., Darling, A., Ohna, S. H., et al. (2012). MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Savile, D. B. O. (1962). Collection and care of botanical specimens Vol. 1113 (Canada, Ottawa: Canadian Department of Agriculture, Research Branch, Publication), 1–124. doi: 10.5962/bhl.title.53755
Shivas, R. G., Marney, T. S., Tan, Y. P., and McTaggart, A. R. (2015). Novel species of Cercospora and Pseudocercospora (Capnodiales, mycosphaerellaceae) from Australia. Fungal Biol. 119, 362–369. doi: 10.1016/j.funbio.2014.09.004
Simon, U. K., Groenewald, J. Z., and Crous, P. W. (2009). Cymadothea trifolii, an obligate biotrophic leaf parasite of Trifolium, belongs to Mycosphaerellaceae as shown by nuclear ribosomal DNA analyses. Persoonia 22, 49–55. doi: 10.3767/003158509X425350
Singh, R., Chaurasia, B., Shukla, K., and Upadhyaya, P. P. (2012). Passalora aseptata, a new cercosporoid fungus from northeastern Uttar Pradesh, India. Mycotaxon 120, 461–463. doi: 10.5248/120.461
Singh, A., Kharwar, R. N., Singh, R., and Kumar, S. (2014a). A new species of Zasmidium (Mycosphaerellaceae) from India. Sydowia 66, 309–312. doi: 10.12905/0380.sydowia66(2)2014-0309
Singh, R. and Kumar, S. (2011). Two new species of Passalora and Pseudocercospora from northeastern Uttar Pradesh, India. Mycotaxon 117, 137–143. doi: 10.5248/117.137
Singh, R. and Kumar, S. (2017). Passalora rhamnaecearum comb. nov. (Capnodiales, Mycosphaerellaceae) from India. Kavaka 48, 50–51.
Singh, R., Kumar, S., Pal, V. K., Upadhyaya, P. P., and Agrawal, D. K. (2007). New taxa of foliicolous hyphomycetes—Cercospora, Corynespora, and Phaeotrichochonis from North-Eastern U.P. Indian Phytopath. 60, 506–512.
Singh, R., Kumar, S., Saini, D. C., Upadhyaya, P. P., and Kamal Braun, U. (2013). Diversity of Passalora on Ficus. Mycol. Prog. 12, 637–643. doi: 10.1007/s11557-012-0870-6
Singh, A., Kumar, S., Singh, R., and Agrawal, D. K. (2008). Two new species of Ramularia from Indian sub-continents. Indian Phytopath. 61, 348–352.
Singh, R., Singh, A., Kumar, S., Upadhyaya, P. P., and Castañeda-Ruíz, R. F. (2014b). Two new species of Zasmidium from northeastern Uttar Pradesh India. Nova Hedwigia 98, 257–263. doi: 10.1127/0029-5035/2013/0137
Singh, R., Verma, S. K., Yadav, S., Bhojak, P., and Kumar, S. (2020b). Morphology and phylogeny of Pseudocercospora hamiltoniani—a new species comparable to Sirosporium from Uttarakhand, India. Phytotaxa 58, 281–293. doi: 10.11646/phytotaxa.458.4.4
Singh, R., Verma, S. K., Yadav, S., and Kumar, S. (2020a). Cercosporella bundelkhandae comb. nov. from India. Mycotaxon 135, 315–320. doi: 10.5248/135.315
Singh, G., Yadav, S., Singh, R., and Kumar, S. (2022). Passalora golaghati comb. nov. from India. Mycotaxon 137, 89–94. doi: 10.5248/137.89
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Starkey, D. E., Ward, T. J., Aoki, T., Gale, L. R., Kistler, H. C., Geiser, D. M., et al. (2007). Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 44, 1191–1204. doi: 10.1016/j.fgb.2007.03.001
Stengel, A., Stanke, K. M., Quattrone, A. C., and Herr, J. R. (2022). Improving taxonomic delimitation of fungal species in the age of genomics and phenomics. Front. Microbiol. 13. doi: 10.3389/fmicb.2022.847067
Sultan, A., Johnston, P. R., Park, D., and Robertson, A. W. (2011). Two new pathogenic ascomycetes in Guignardia and Rosenscheldiella on New Zealand’s pygmy mistletoes (Korthalsella: Viscaceae). Stud. Mycol. 68, 237–247. doi: 10.3114/sim.2011.68.11
Summerbell, B. A., Groenewald, J. Z., Carnegie, A. J., Summerbell, R. C., and Crous, P. W. (2006). Eucalyptus microfungi known from culture. 2. Alysidiella, Fusculina and Phlogicylindrium genera nova, with notes on some other poorly known taxa. Fungal Divers. 23, 323–350.
Sung, G.-H., Sung, J.-M., Hywel-Jones, N. L., and Spatafora, J. W. (2007). A multigene phylogeny of Clavicipitaceae (Ascomycota, Fungi): identification of localized incongruence using a combinational bootstrap approach. Mol. Phylogenet. Evol. 44, 1204–1223. doi: 10.1016/j.ympev.2007.03.011
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., et al. (2000). Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31, 21–32. doi: 10.1006/fgbi.2000.1228
Van Burik, J.-A., Schreckhise, R., White, T. C., Bowden, R., and Myerson, D. (1998). Comparison of six extraction techniques for isolation of DNA from filamentous fungi. Med. Mycol. 36, 299–303. doi: 10.1046/j.1365-280X.1998.00161.x
Verkley, G. J., Quaedvlieg, W., Shin, H. D., and Crous, P. W. (2013). A new approach to species delimitation in Septoria. Stud. Mycol. 75, 213–305. doi: 10.3114/sim0018
Verma, S. K., Kushwaha, P., Yadav, S., and Singh, R. (2021a). Morphology and phylogeny of Teratoramularia rumicis—a new foliar pathogen of Rumex crispus from India and diversity of Ramularioid complex on. Rumex Phytotaxa 523, 208–228. doi: 10.11646/phytotaxa.523.3.2
Verma, S. K., Yadav, S., and Singh, R. (2021c). Molecular phylogeny of Aplosporella abexaminans: a novel species revealing the second report of sexual-asexual connection in Aplosporellaceae (Botryosphaeriales) from India. Phytotaxa 525, 205–222. doi: 10.11646/phytotaxa.525.3.3
Verma, S. K., Yadav, S., and Singh, R. (2023). Distocercospora curvulata sp. nov. from northern India. Mycotaxon 137, 679–686. doi: 10.5248/137.679
Verma, S. K., Yadav, S., Singh, G., and Singh, R. (2021b). Pseudocercospora cappadocici, a new Stigmina-like Pseudocercospora species on Acer cappadocicum from India. Sydowia 74, 79–91. Available online at: http://www.sydowia.at/syd74/T6-Verma.htm.
Videira, S. I., Groenewald, J. Z., Braun, U., Shin, H. D., and Crous, P. W. (2016). All that glitters is not Ramularia. Stud. Mycol. 83, 49–163. doi: 10.1016/j.simyco.2016.06.001
Videira, S. I., Groenewald, J. Z., Nakashima, C., Braun, U., Barreto, R. W., de Wit, P. J., et al. (2017). Mycosphaerellaceae–chaos or clarity? Stud. Mycol. 87, 257–421. doi: 10.1016/j.simyco.2017.09.003
Vilgalys, R. and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Vu, D., Groenewald, M., De Vries, M., Gehrmann, T., Stielow, B., Eberhardt, U., et al. (2019). Large–scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001
White, T. J., Bruns, T., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in A guide to molecular methods and applications. Eds. Innis, M. A., Gelfand, D. H., and Sninsky, J. J. (Academic Press, New York), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Crous, P. W., Kirk, P. M., Hawksworth, D. L., Boonmee, S., Braun, U., et al. (2014). Naming and outline of Dothideomycetes–2014 including proposals for the protection or suppression of generic names. Fungal Divers. 69, 1–55. doi: 10.1007/s13225-014-0309-2
Wilkinson, P. M., Thomas-Hall, S., Marney, T. S., and Shivasc, R. G. (2005). First record of Passalora calotropidis in Australia and its generic position. Australas. Plant Pathol. 34, 95–98. doi: 10.1071/AP04074
Yadav, S., Singh, R., Verma, S. K., Singh, G., and Kushwaha, P. (2023). Addition of three new lineages in Mycosphaerellaceae: Neoacervuloseptoria gen. nov., Neocercosporella gen. nov. and Neoramulariopsis gen. nov. Mycol. Prog. 22, 1–19. doi: 10.1007/s11557-023-01871-y
Yadav, S., Verma, S. K., Singh, R., Singh, V. K., Chaurasia, B., Singh, P. N., et al. (2022). Neokamalomyces indicus gen. nov., sp. nov. (Mycosphaerellaceae)—a Septoria-like genus from India. Phytotaxa 571, 141–168. doi: 10.11646/phytotaxa.571.2.3
Yadav, S., Verma, S. K., Singh, V. K., Singh, R., Singh, A., and Kumar, S. (2021). Morphology and phylogeny of a new species, Pseudocercospora haldinae (Mycosphaerellaceae) on Haldina cordifolia from India. Phytotaxa 501, 281–292. doi: 10.11646/phytotaxa.501.2.3
Keywords: anamorph, Dothideomycetes, multigene-phylogeny, Mycosphaerellales, new taxa, nomenclature
Citation: Singh G, Rajwar S, Khatoon S, Yadav S, Kumari P, Singh R, Kumar K, Mall S, Singh PN, Kumar S and Braun U (2025) Addition of two new genera—Marcstadlera gen. nov. and Neoclypeosphaerella gen. nov. (Mycosphaerellaceae)—based on polyphasic evidence. Front. Cell. Infect. Microbiol. 15:1668928. doi: 10.3389/fcimb.2025.1668928
Received: 18 July 2025; Accepted: 29 September 2025;
Published: 23 October 2025.
Edited by:
Joseph Nickels, Jr, Genesis Biotechnology Group, United StatesReviewed by:
Marasinghe Wadige Diana Sandamali, Guizhou University, ChinaRafael F. Castañeda-Ruiz, Instituto de Investigaciones de Sanidad Vegetal - INISAV, Cuba
Copyright © 2025 Singh, Rajwar, Khatoon, Yadav, Kumari, Singh, Kumar, Mall, Singh, Kumar and Braun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raghvendra Singh, ZHJzaW5naHRheG9uQGdtYWlsLmNvbQ==; c2luZ2hyLmJvdEBiaHUuYWMuaW4=
†ORCID: Gargee Singh, orcid.org/0000-0001-9827-5907
Soumyadeep Rajwar, orcid.org/0000-0001-8298-7969
Sahana Khatoon, orcid.org/0009-0006-2754-232X
Sanjay Yadav, orcid.org/0000-0003-1850-7566
Pooja Kumari, orcid.org/0009-0001-4760-7518
Raghvendra Singh, orcid.org/0000-0002-8672-6149
Kamalesh Kumar, orcid.org/0000-0001-7374-5941
Smriti Mall, orcid.org/0000-0001-7894-3576
Paras N. Singh, orcid.org/0000-0002-3555-819X
Shambhu Kumar, orcid.org/0000-0001-9534-9309
Uwe Braun, orcid.org/0000-0002-2205-9180
 Gargee Singh1†
Gargee Singh1† Raghvendra Singh
Raghvendra Singh Smriti Mall
Smriti Mall Paras N. Singh
Paras N. Singh Shambhu Kumar
Shambhu Kumar Uwe Braun
Uwe Braun