- 1School of Medicine, Huzhou University; Key Laboratory of Vector Biology and Pathogen Control of Zhejiang Province, Huzhou, Zhejiang, China
- 2Department of Clinical Laboratory, the First Affiliated Hospital of Wenzhou Medical University, Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province, Wenzhou, Zhejiang, China
- 3Department of Clinical Laboratory, the First Affiliated Hospital of Ningbo University, Ningbo, Zhejiang, China
Objectives: The emergence and spread of carbapenem-resistant Morganella morganii (M. morganii) pose a serious global challenge. This study aimed to investigate the clinical characteristics, resistance patterns, and molecular mechanisms of carbapenem-resistant M. morganii.
Methods: A total of 170 M. morganii clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between January 2016 and December 2017. Carbapenem MICs were determined by antimicrobial susceptibility testing. Carbapenem resistance determinants, including carbapenemase genes (blaKPC-2, blaVIM, blaIMP, blaNDM, and blaOXA-48) and extended-spectrum β-lactamase (ESBL) genes (blaTEM, blaCTX-M-1, and blaSHV), were analyzed by polymerase chain reaction (PCR). PCR and sequencing assays were performed to detect penicillin-binding protein (PBP) mutations. Efflux pump activity was also assessed in carbapenem-resistant isolates. Quantitative real-time PCR (qRT-PCR) was used to determine the relative mRNA expression levels of outer membrane porin-encoding gene ompC and PBP activator-encoding genes lpoA and lpoB.
Results: Twenty-six imipenem-resistant and 108 imipenem-intermediate M. morganii isolates were identified, accounting for 15.29% and 63.53% of cases, respectively. No isolates were resistant to meropenem or ertapenem. Among the 26 carbapenem-resistant isolates, the prevalence of ESBL genes blaTEM and blaCTX-M-1 was 30.77% and 11.54%, respectively, while carbapenemase genes were not detected. Resistant isolates carried more specific PBP mutations than carbapenem-susceptible and carbapenem-intermediate isolates. Efflux pump phenotypes were associated with reduced imipenem susceptibility in 13 carbapenem-resistant isolates. qRT-PCR revealed no significant differences in ompC expression among the resistant, intermediate, and susceptible groups; however, significant differences were observed in lpoA and lpoB expression. Isolates in the imipenem-resistant group carried more PBP mutations.
Conclusion: M. morganii isolates were commonly non-susceptible to imipenem but remained susceptible to meropenem and ertapenem. Low expression of PBP activator genes (lpoA and lpoB), along with the presence of specific PBP mutations, appeared to be the primary mechanisms of resistance. In addition, efflux pump overexpression may contribute to imipenem resistance in M. morganii.
1 Introduction
Morganella morganii (M. morganii), a facultative anaerobic Gram-negative bacterium, is the only species in the genus Morganella of the Enterobacteriaceae family. It is divided into two subspecies: Morganella subspecies and siboni subspecies (Liu et al., 2016; Chen et al., 2024; Liu et al., 2025). This bacterium is widely found in the natural environment and in the intestines of humans, mammals, and reptiles. M. morganii is an important opportunistic pathogen in clinical settings, often causing urinary tract infections after catheterization and postoperative wound infections. It has also been reported to cause sepsis, meningitis, pneumonia, arthritis, and other nosocomial infections (Erlanger et al., 2019; Bandy, 2020; Zaric et al., 2021; Laupland et al., 2022). Due to increasing bacterial resistance to third-generation cephalosporins, aminoglycosides, and fluoroquinolones, carbapenems have gradually become the last option for treating multidrug-resistant M. morganii infections, which pose life-threatening health risks (Guo et al., 2019; Bandy, 2020; Park et al., 2020). However, with the wide application of carbapenems in clinical practice, the global spread of carbapenem-resistant pathogens has brought great challenges to public health worldwide (Bonnin et al., 2024; Yao et al., 2025).
Several studies have reported that carbapenem resistance in M. morganii is associated not only with harboring carbapenemases, including KPC, NDM, IMP, VIM, and OXA, and extended-spectrum β-lactamases (ESBLs), but also with the overexpression of efflux pumps (Guo et al., 2019; Shrestha et al., 2020; Bonnin et al., 2024). In addition, the outer membrane structure of this pathogen can effectively prevent harmful substances from entering bacterial cells. However, the loss or decreased expression of outer membrane porins in M. morganii contributes to the multidrug-resistant (MDR) phenotype (Nordmann et al., 2012; Logan and Weinstein, 2017).
In Gram-positive bacteria, β-lactamases and permeability barriers play a limited role in drug resistance, so penicillin-binding protein (PBP)-related mechanisms have been studied extensively in this group. By contrast, the role of PBPs in drug resistance among Gram-negative bacteria has often been ignored. In recent years, increasing attention has been paid to PBP-related resistance in Gram-negative bacteria, with studies focusing on PBPs as antibiotic targets from different perspectives, which has important implications for the treatment of infectious diseases (Aissa et al., 2016; Aitken et al., 2024; Tellapragada et al., 2024; Fabrizio and Tascini, 2025). Carbapenems inhibit cell wall synthesis by binding to bacterial PBPs, including the high-molecular-weight enzymes PBP1a, PBP1b, PBP2, and PBP3 (Shalaby et al., 2020; Bertonha et al., 2023; Hu and Wang, 2024). In Enterobacteriaceae, different carbapenems have varying affinities for PBPs. Imipenem has higher affinity for PBP1a and PBP1b, binding two to four times more strongly than other carbapenems (Aissa et al., 2016; Fabrizio and Tascini, 2025). Substitution of amino acids in PBPs or the acquisition of new PBPs can lead to bacterial resistance to carbapenems. PBP modification often contributes to resistance to β-lactam antibiotics in both Gram-positive and Gram-negative bacteria; however, PBP modification alone rarely results in high levels of resistance to carbapenems (Cherkaoui et al., 2017; Malik et al., 2020). With the increasing prevalence of carbapenem-resistant M. morganii worldwide, it is of great clinical importance to explore their specific resistance patterns and analyze molecular epidemiology to prevent and control the occurrence and transmission of resistance, as well as to guide antimicrobial therapy (Guo et al., 2019; Xiang et al., 2021).
In this study, 170 M. morganii strains clinically isolated at our hospital in southeastern China between January 2016 and December 2017 were retrospectively analyzed. We characterized the specific resistance patterns and molecular mechanisms of 26 imipenem-resistant isolates that remained susceptible to meropenem and ertapenem.
2 Materials and methods
2.1 Bacterial strains
A total of 170 M. morganii clinical isolates were collected from the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, China) between January 2016 and December 2017. All isolates were identified as M. morganii using the VITEK®2 mass spectrometry (MS) system (bioMérieux, France). After collection, isolates were stored at −80 °C in Luria–Bertani (LB) broth with 30% sterilized glycerol. Relevant clinical data, including date of isolation, patient age, sex, sample type, and ward, were retrieved from medical records.
2.2 Minimum inhibitory concentration determination
According to the latest guidelines version CLSI M100–2025 recommendedby the Clinical and Laboratory Standards Institute (CLSI, 2025), we measured the MICs by agar dilution method, including imipenem, meropenem, and ertapenem in this study. Briefly, after overnight culture of single colonies, the suspension was adjusted to 0.5 McFarland (approximately 1.5 × 108 CFU/mL) in sterilized NaCl. Following a 10-fold dilution, suspensions were evenly spread on medicated Mueller–Hinton agar plates and incubated at 37 °C for 16–18 h. Results were recorded after incubation. Meropenem and ertapenem were dissolved in sterile water, while imipenem was dissolved in sterile phosphate-buffered saline (PBS, 0.01 mol/L, pH 7.2), and tested over a concentration range of 0.0125–16 µg/mL. Escherichia coli (E. coli) strain ATCC25922 was used as the quality control strain. MIC values were determined in at least three independent experiments. Twenty-six imipenem-resistant M. morganii strains that remained susceptible to meropenem and ertapenem were selected for further analysis of resistance mechanisms.
2.3 Determinations of carbapenemases and extended-spectrum β-lactamase
Polymerase chain reaction (PCR) and sequencing assays were performed to detect carbapenem resistance determinants, including carbapenemase genes (blaKPC-2, blaVIM, blaIMP, blaNDM, and blaOXA-48), and ESBL genes (blaTEM, blaCTX-M-1, and blaSHV), in the 26 imipenem-resistant M. morganii isolates. Genomic DNA was extracted using a commercial Genomic DNA Extraction Kit (Qiagen). Supplementary Table S1 showed the primers used to amplify DNA templates in this study. Electrophoresis was performed using 1% agarose gels. Subsequently, these resulted positive amplifications were commissioned to Shanghai BGI Technology Co. (China) for sequencing. The nucleotide sequences were compared by searching the GenBank using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.4 Efflux pump activity on imipenem-resistant M. morganii
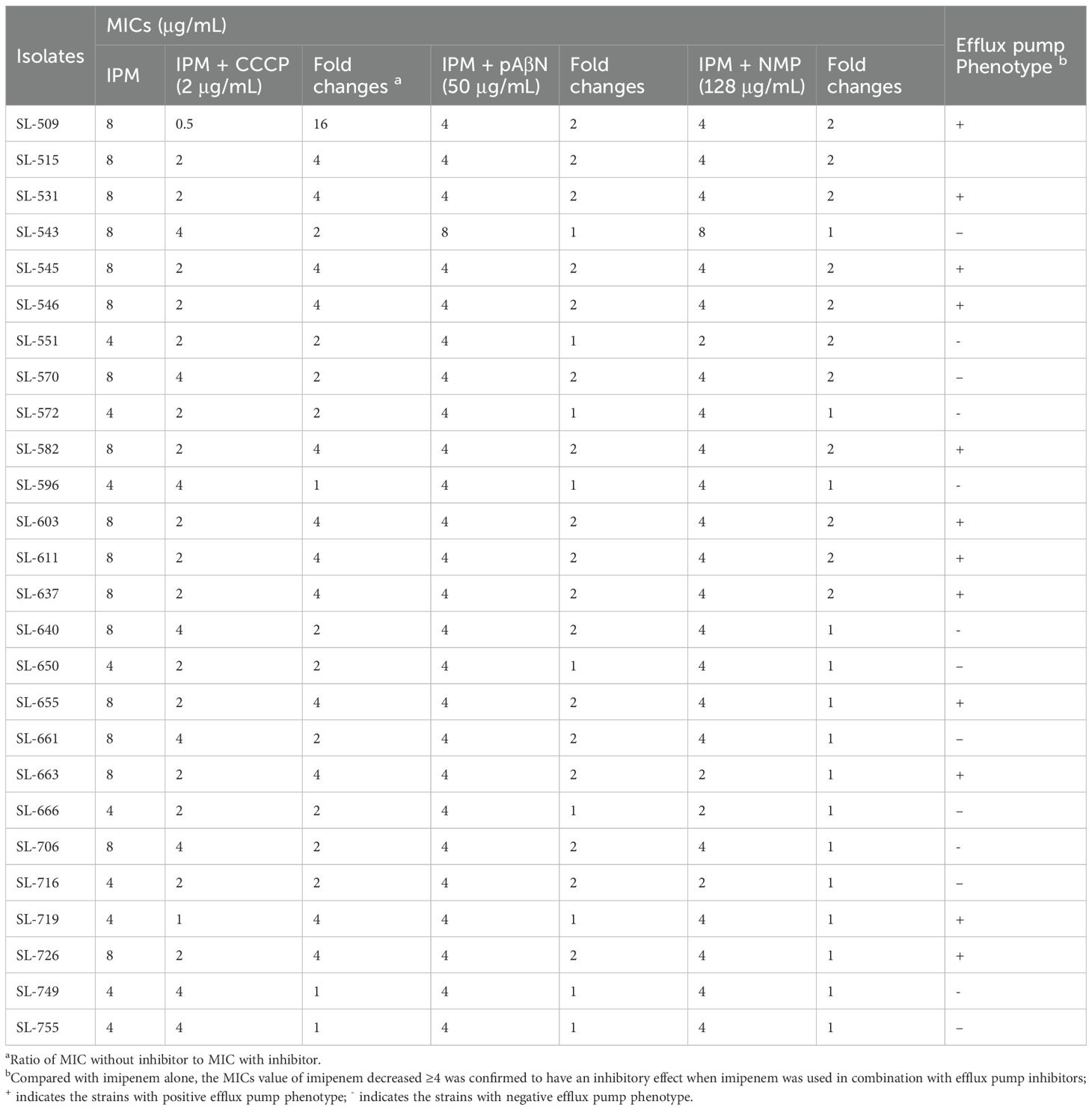
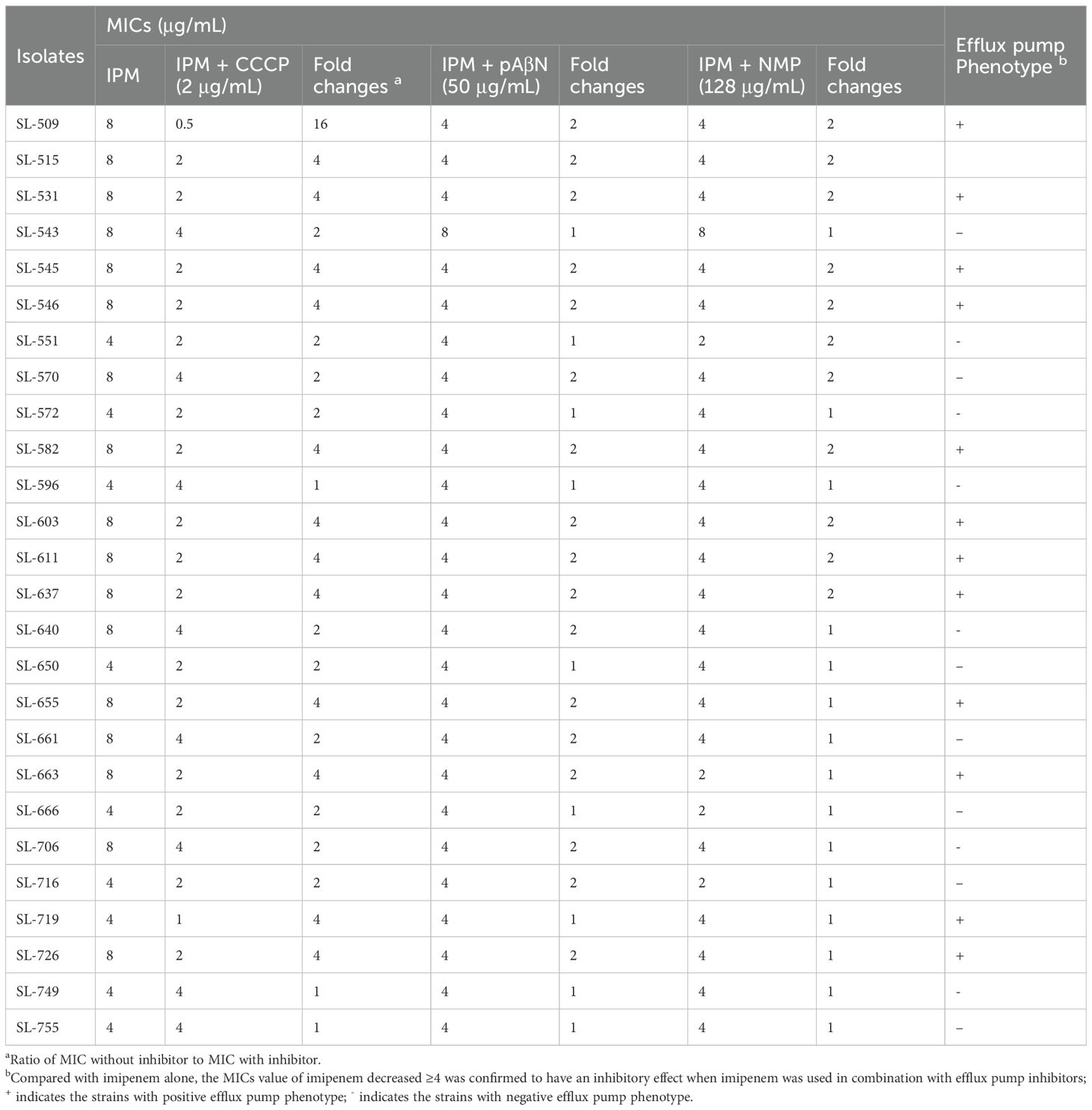
The methodology for the efflux pump inhibition test has been described in detail in a previous publication (Zheng et al., 2022). Efflux pump inhibitors carbonyl cyanide m-chlorophenylhydrazone (CCCP), N-methylpyrrolidone (NMP), and Phe-Arg-β-naphthylamide (pAβN) were used. First, the agar dilution method was applied to determine the appropriate concentrations of CCCP, pAβN, and NMP that inhibited efflux activity without affecting bacterial growth. The final concentrations were 2 μg/mL, 50 μg/mL, and 128 μg/mL, respectively. MICs of imipenem against 26 imipenem-resistant M. morganii strains were then determined on Mueller–Hinton agar plates with or without the inhibitors. A ≥4-fold reduction in imipenem MIC in the presence of an inhibitor was considered evidence of efflux pump activity (Zheng et al., 2022).
2.5 PBP mutation analysis
Mutations in PBP1a, PBP1c, and PBP2 of all 26 imipenem-resistant M. morganii and M. morganii ATCC25830 strains were analyzed by PCR, while 15 imipenem-intermediate and 15 imipenem-susceptible M. morganii strains were analyzed in parallel. Genomic DNA was extracted using a commercial Genomic DNA Extraction Kit (Qiagen). The primers used for amplification are listed in Supplementary Table S1. Electrophoresis was performed on 1% agarose gels. Positive amplicons were sequenced by Shanghai BGI Technology Co. (China). Nucleotide sequences were compared with reference sequences in GenBank using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
2.6 Quantitative real-time polymerase chain reaction
All 26 imipenem-resistant M. morganii and M. morganii ATCC25830 strains were cultured to an OD600 of 0.5~0.6 (the logarithmic growth phase) in fresh Luria–Bertani (LB) broth at 37°C/180 rpm. The total RNA of 26 imipenem-resistant M. morganii and M. morganii ATCC25830 strains were extracted using the Bacterial RNA Miniprep Kit (Biomiga, Shanghai, China). In addition, 15 imipenem-intermediate and 15 imipenem-susceptible M. morganii strains were analyzed in parallel. A total of 100 ng RNA was reverse-transcribed into first-strand cDNA using the PrimeScript™ RT reagent Kit (Perfect Real Time) (Takara, Japan). The primers used in this study are listed in Supplementary Table S1. The 16S rRNA gene was used as an endogenous control. Expression levels of the outer membrane porin-encoding gene ompC and the PBP activator-encoding genes lpoA and lpoB were determined by qRT-PCR using TB Green Premix Ex Taq II (Tli RNase H Plus) (2×) (Takara, Japan). Gene transcript levels were compared among resistant, intermediate, and susceptible groups using Student’s t-test. Expression levels of each gene in M. morganii ATCC25830 strains were normalized to a value of 1.0. The 2-ΔΔCt method was applied for data analysis.
2.7 Statistical analysis
All experimental procedures were performed in triplicate with independent biological replicates. Quantitative data are presented as mean ± standard deviation (SD). Intergroup comparisons were analyzed using one-way ANOVA. Statistical analyses were performed using GraphPad Prism, version 9.02 (GraphPad Software Inc., San Diego, CA, USA). Two-tailed tests were used throughout, and P < 0.05 was considered statistically significant.
3 Results
3.1 Antimicrobial susceptibility profiles and clinical characteristics
The results of antimicrobial susceptibility testing revealed that 26 imipenem-resistant and 108 imipenem-intermediate M. morganii isolates were identified among the 170 isolates tested, accounting for 15.29% and 63.53%, respectively. However, all 170 M. morganii isolates were susceptible to meropenem and ertapenem (Table 1). In addition, 26 carbapenem-resistant M. morganii isolates were mainly from wound samples (38.46%, 10/26), followed by urine (19.23%, 5/26) and pus (15.38%, 4/26). There were more isolates from males than females (76.92% (20/26) vs 23.08% (6/26), respectively). Isolates were collected from patients aged 21 to 91 years (mean age, 68.27 years). The majority of the isolates were from patients in the wound center, urology, and endocrinology departments.
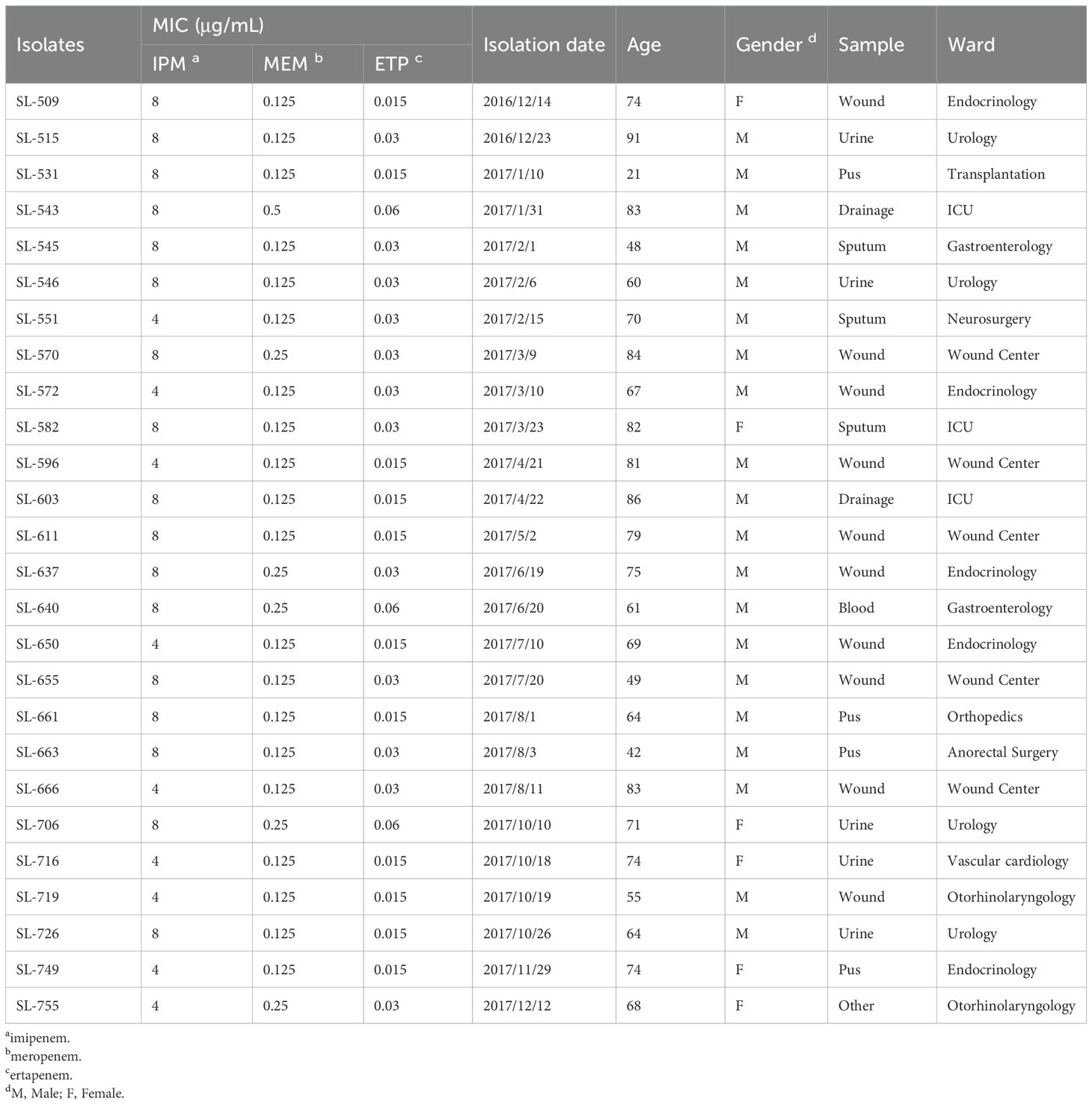
Table 1. Antimicrobial susceptibility profiles and clinical characteristics of 26 carbapenem-resistant M. morganella isolates.
3.2 Carbapenemases and β-lactamase genes prevalence
The mechanisms of carbapenem resistance in the 26 imipenem-resistant M. morganii isolates were investigated by PCR. No carbapenemase-encoding genes (blaKPC-2, blaNDM, etc.) were detected. By contrast, ESBL genes were identified: blaTEM was present in 30.77% of isolates and blaCTX-M-1 in 11.54%, while blaSHV was not detected (Figure 1).
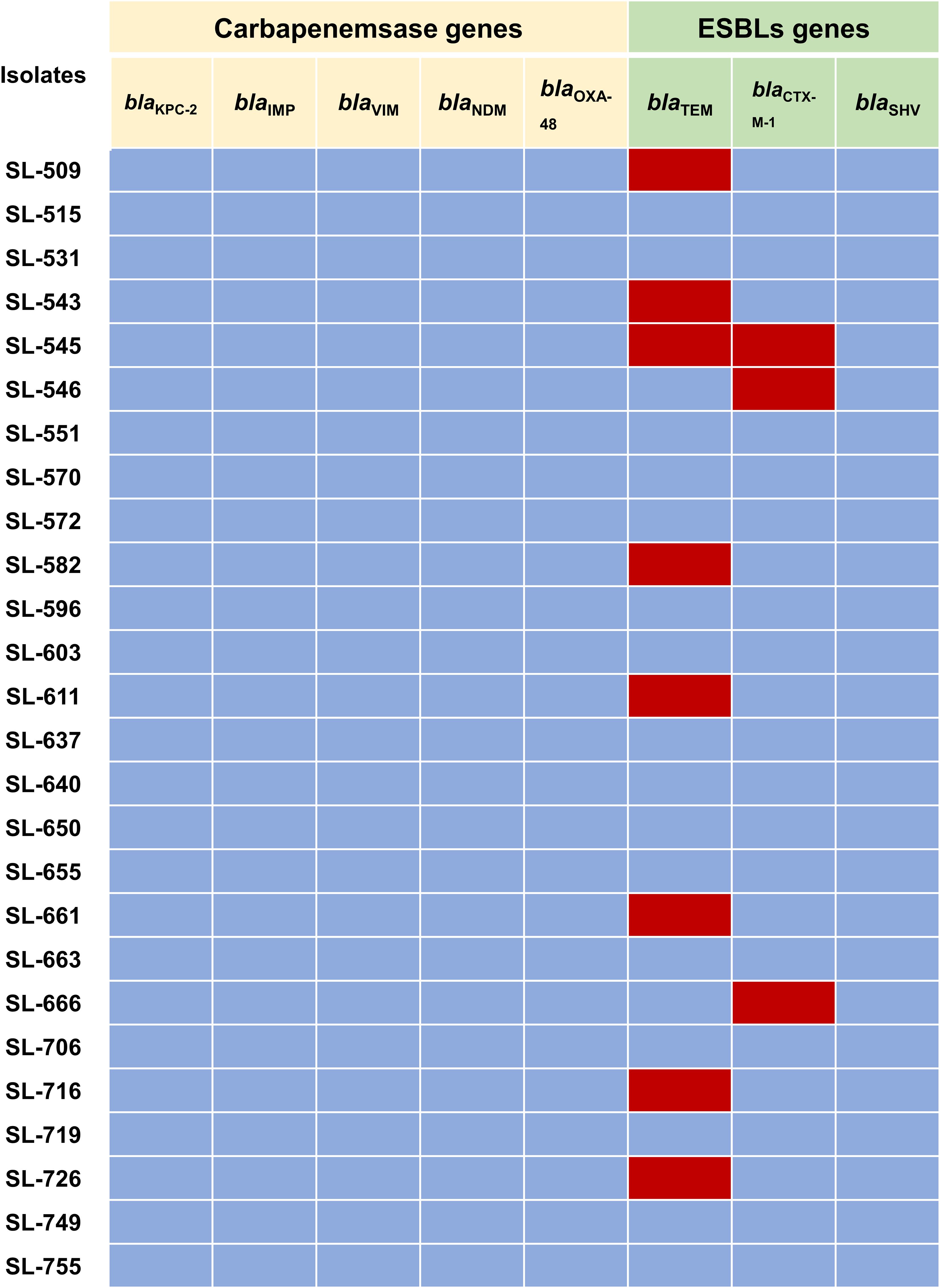
Figure 1. Prevalence of carbapenemases-encoding genes and ESBLs-encoding genes in 26 imipenem-resistant M. morganii isolates. Red colours indicate presence while blue colours represent absence.
3.3 Phenotypic detection of the efflux pump overexpression
A series of efflux pump inhibitors was tested for their effect on imipenem susceptibility, including CCCP, pAβN, and NMP. Imipenem MICs of the 26 imipenem-resistant M. morganii isolates decreased by less than twofold when exposed to pAβN and NMP. However, imipenem MICs decreased by more than fourfold in the presence of 2 μg/mL CCCP 13 imipenem-resistant isolates, confirming an association between efflux pump phenotypes and reduced imipenem susceptibility in these strains (Table 2).
3.4 Determination of PBP mutations
The genes mrcA, pbpC, and mdrA encode the three primary PBPs in M. morganii: PBP1a, PBP1c, and PBP2, respectively. PCR results demonstrated that imipenem-resistant M. morganii isolates carried more specific PBP mutations than the imipenem-susceptible and imipenem-intermediate strains, such as PBP1c-Glu76Gln, PBP1c-Asn235Asp, PBP1c-Thr475Ile, PBP1c-Val783Met, and PBP2-Gly487Ser (Table 3). Supplementary Table S2 details all PBP missense mutations in the imipenem-resistant, intermediate, and susceptible isolate groups.
3.5 Expression of gene encoding outer membrane porin
A study was conducted to examine the relationship between imipenem resistance and ompC (encoding the outer membrane porin) expression. Expression levels were assessed in 26 imipenem-resistant M. morganii, 15 imipenem-intermediate strains, 15 imipenem-susceptible strains, and the reference strain M. morganii ATCC 25830. qRT-PCR results showed that expression levels of ompC did not differ significantly among the resistant, intermediate, and susceptible groups compared with the reference strain (Figure 2).
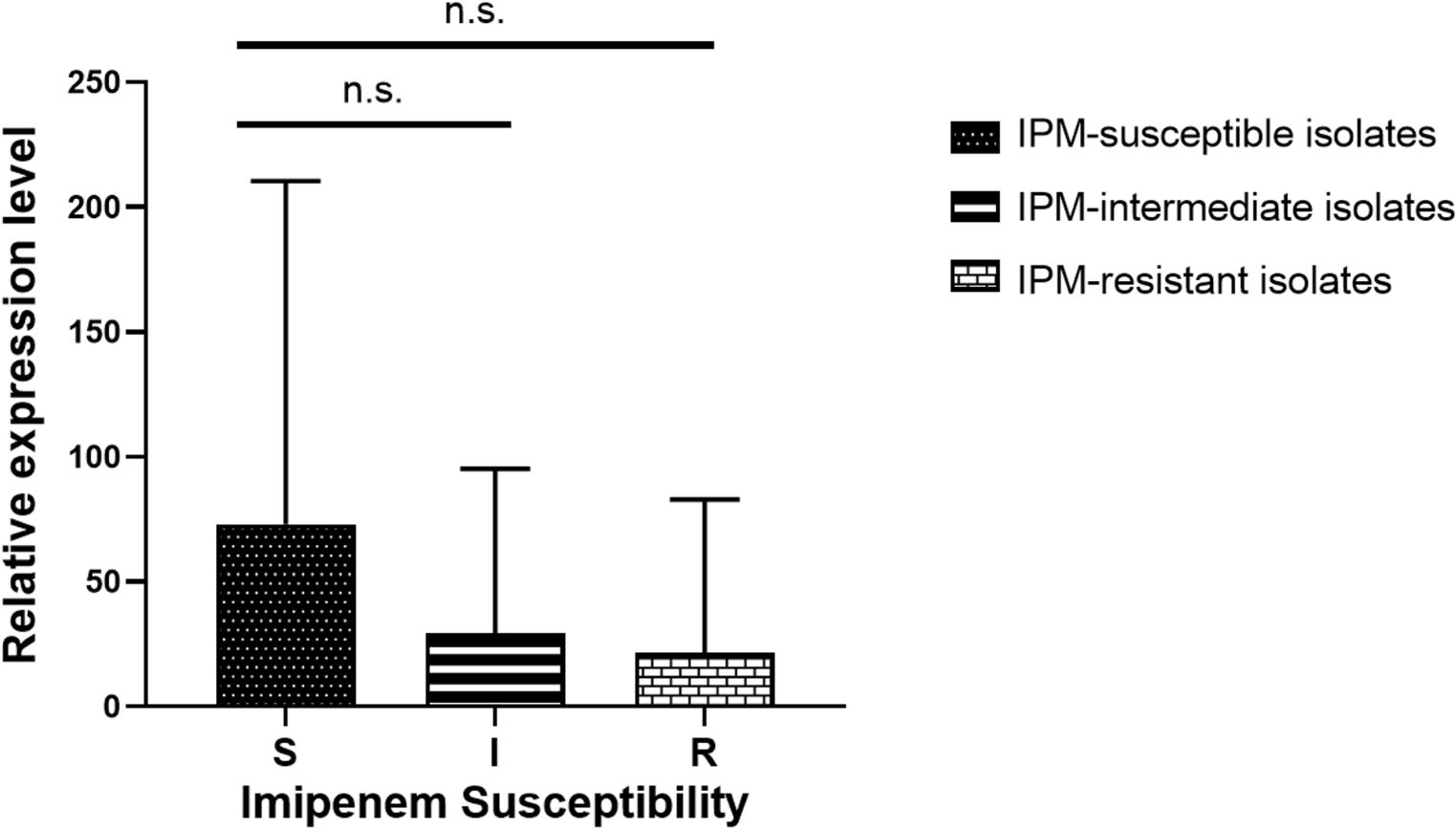
Figure 2. Transcript levels of outer membrane porin-encoding gene ompC among the 26 imipenem-resistant M. morganii, 15 imipenem-intermediate M. morganii strains, 15 imipenem-susceptible M. morganii groups. n.s., P >0.05.
3.6 PBP activator-encoding gene expressions
The expression of the PBP activator-encoding genes lpoA and lpoB was examined in 26 imipenem-resistant isolates, 15 imipenem-intermediate isolates, 15 imipenem-susceptible isolates, and the reference strain M. morganii ATCC25830. qRT-PCR analysis showed that, compared with the reference strain, lpoA and lpoB expression levels were reduced in the imipenem-resistant isolates. Significant differences in lpoA and lpoB expression were also observed among the resistant, intermediate, and susceptible groups (Figure 3).
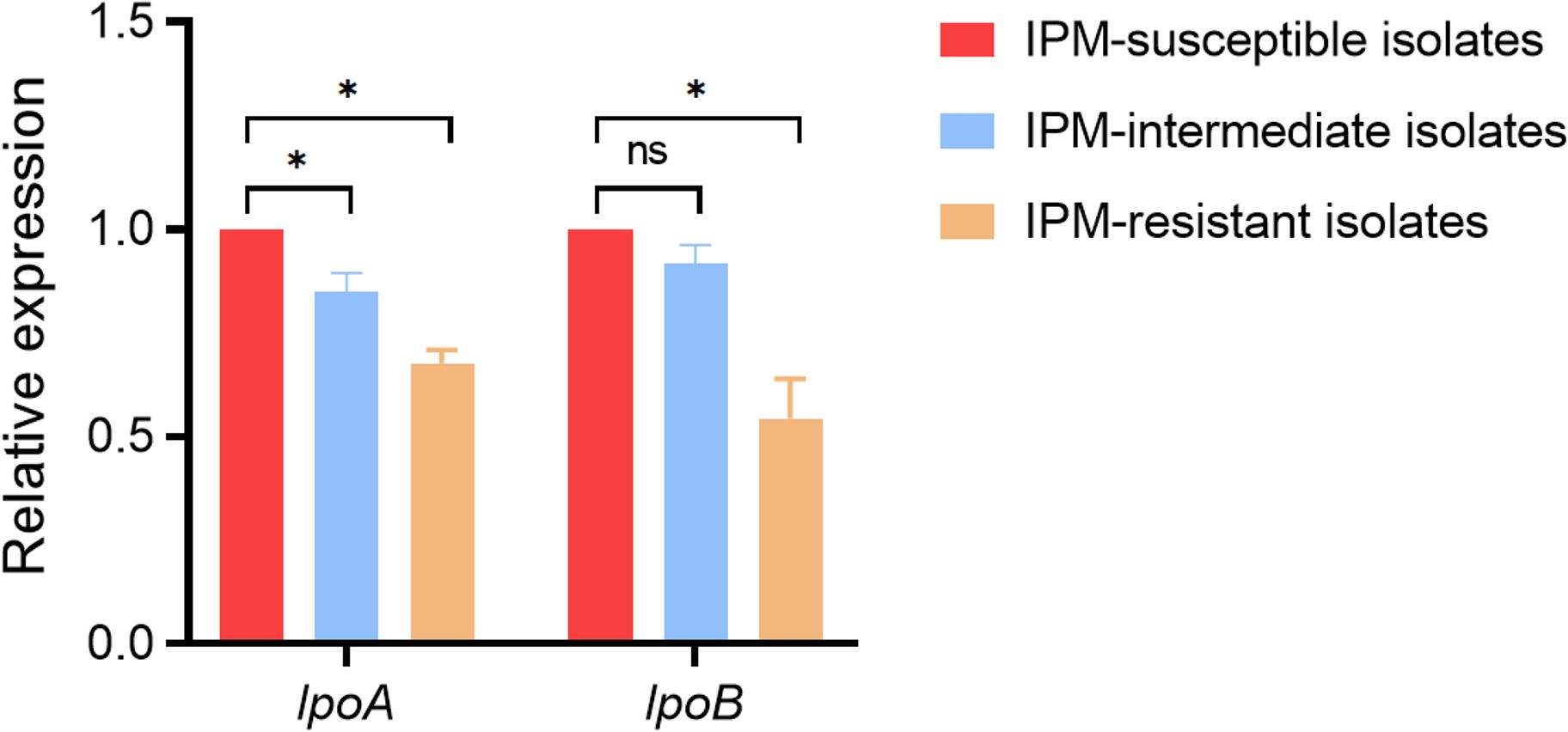
Figure 3. Relative expression level of PBP activator-encoding genes lpoA and lpoB among the 26 imipenem-resistant M. morganii, 15 imipenem-intermediate M. morganii strains, 15 imipenem-susceptible M. morganii groups. *P <0.05; n.s., P >0.05.
4 Discussion
Infection by M. morganii, a rod-shaped Gram-negative bacterial pathogen, is characterized by its prevalence in nosocomial infections. First isolated in 1906 from the feces of a child, M. morganii belongs to the family Proteae, a subfamily of enteric bacteria (Liu et al., 2016; Laupland et al., 2022). The genome length of M. morganii is approximately 4 Mbp, encoding about 4,000 potential protein sequences. Historically, M. morganii was not considered clinically significant in relation to summer diarrhea, but since it has been detected in a variety of conditions, including urinary tract infections, it has been reclassified as an opportunistic pathogen capable of causing rare infections (Sakaguchi et al., 2014).
According to the latest analysis from the SENTRY antimicrobial resistance surveillance program, M. morganii ranks 12th among Gram-negative organisms responsible for bloodstream infections (Zaric et al., 2021). Typically, M. morganii acquires resistance through mobile genetic elements; however, resistance may also result from gene alterations. There are many types of mobile genetic elements in bacteria, including plasmids, transposons, integrons, and insertion sequences. This mode of transfer increases the likelihood of acquiring antimicrobial resistance and leads to wider transmission of resistance in clinical settings (Bandy, 2020). In addition, resistance of M. morganii to fluoroquinolones, most aminoglycosides, and azithromycin increases the difficulty of anti-infective therapeutic management. Since carbapenems possess a broad spectrum of antimicrobial activity, they are extensively used in clinical settings to treat MDR Gram-negative pathogenic microorganisms (Timofte et al., 2016; Yao et al., 2025). Yet, several surveillance programs have reported that carbapenem resistance is increasing rapidly, making clinical treatment more challenging. Resistance of M. morganii to carbapenems is mainly attributed to the production of carbapenemases, including KPC-2 and New Delhi metallo-β-lactamase 1 (NDM-1) (Guo et al., 2019; Bonnin et al., 2024; Munir et al., 2025). It has been reported that blaKPC-2, located within the same mobile genetic element (an integrative structure consisting of a Tn3-based transposon and a partial fragment of Tn4401), may be transmitted between different plasmids in three M. morganii isolates by M Huang et al (Shen et al., 2009; Shi et al., 2012).
As a result, it is important to stress the need for monitoring carbapenem resistance in M. morganii. Moreover, clinical experience with treatment regimens for aggressive MDR or extensively drug-resistant (XDR) M. morganii infections should continue to be developed.
The type and content of PBPs vary among bacterial species, including high-molecular-weight enzymes PBP1a, PBP1b, PBP2, and PBP3, but PBPs of different bacteria share similar structures and functions. PBPs are the main targets of β-lactam antibiotics (Shalaby et al., 2020; Bertonha et al., 2023; Hu and Wang, 2024). These antibiotics specifically bind to PBPs on the inner membrane of bacterial cells, interfere with the normal enzymatic activity of PBPs, and consequently disrupt peptidoglycan synthesis. As a result, cell wall synthesis is blocked, ultimately leading to bacterial death (Lessel, 1996). Different β-lactam antibiotics bind to different PBPs, and changes in PBP structure or abundance are important mechanisms leading to bacterial resistance (Aissa et al., 2016; Cherkaoui et al., 2017). Even the same antibiotic can act on different PBPs depending on the bacterial species. For example, a study comparing the binding affinities of Escherichia coli PBPs with three antibiotics—methicillin, cephalexin, and cefradine—showed that methicillin bound specifically to PBP2, whereas cephalexin and cefradine had high affinity for PBP1a (Katayama et al., 2004). Doripenem also exhibits high affinity for PBPs in various species, such as PBP3 in Pseudomonas, PBP1, PBP2, and PBP4 in Staphylococcus aureus, and PBP2 in E. coli. Compared with imipenem, the stronger antipseudomonal activity of doripenem and meropenem against Pseudomonas aeruginosa may be attributed to their stronger binding affinity for PBP2 and PBP3 (Goffin and Ghuysen, 2002; Malik et al., 2020). It has also been reported that doripenem binds more efficiently to PBP1a and PBP1b, followed by PBP2, while imipenem is more likely to penetrate the bacterial cell wall and reach its target PBPs at a faster rate than other antibiotics in Streptococcus pneumonia (Cherkaoui et al., 2017). There is evidence that carbapenem resistance is caused by a reduction in expression or absence of the two main porins, combined with β-lactamase activity and PBP alterations (Breilh et al., 2013). However, PBP modification alone rarely leads to high levels of carbapenem resistance. In this study, we detected 26 imipenem-resistant M. morganii isolates; however, these strains remained susceptible to both meropenem and ertapenem and had low MIC values. In addition, the imipenem-resistant strains carried more specific PBP mutations than the imipenem-susceptible and imipenem-intermediate isolates, a finding consistent with previous studies.
It was reported many years ago that PBP enzymes EcPBP1a and EcPBP1b from E. coli may be specifically activated by the outer-membrane lipoproteins EcLpoA (activating EcPBP1a) and EcLpoB (activating EcPBP1b), respectively (Paradis-Bleau et al., 2010; Typas et al., 2010). Both activators interact directly with their cognate homologs to form a transenvelope complex (Egan et al., 2014; Jean et al., 2014; King et al., 2014). In Gammaproteobacteria, the expression level of LpoA was found to be relatively conserved (Typas et al., 2010). By contrast, LpoB and the UB2H regulatory domain of EcPBP1b to which it binds appeared to be largely restricted to the Enterobacteriaceae, despite the relatively broad distribution of PBP1b sequences (Typas et al., 2010). Imipenem exhibits uniquely high affinity for PBP1a and PBP1b in Enterobacteriaceae, relying critically on their activation by LpoA/LpoB for bactericidal activity. Downregulation of lpoA/lpoB impairs the transenvelope complex formation required for PBP1a/1b function, directly compromising imipenem binding. In contrast, meropenem and ertapenem primarily target PBP2/3 with minimal dependence on PBP1a/1b activation. This mechanistic distinction explains why lpoA/lpoB downregulation selectively confers imipenem resistance while preserving susceptibility to other carbapenems. In agreement with other studies (Dorr et al., 2014; Yin et al., 2015; Greene et al., 2018), we found that the expression levels of lpoA and lpoB in 26 imipenem-resistant M. morganii isolates were decreased compared with those in the reference strain M. morganii ATCC25830. In addition, significant differences in lpoA and lpoB expression were observed among the resistant, intermediate, and susceptible groups. These findings confirm that reduced expression of the PBP activator genes lpoA and lpoB may contribute to imipenem resistance in clinical M. morganii isolates.
5 Conclusion
In conclusion, this study is the first to investigate the clinical characteristics, specific resistance patterns, and molecular mechanisms of carbapenem-resistant M. morganii clinical isolates. These findings indicate that M. morganii clinical isolates showed distinct patterns and mechanisms of resistance to carbapenem antibiotics, which were different from those of other Enterobacteriaceae. M. morganii clinical isolates were commonly non-susceptible to imipenem, while imipenem-non-susceptible strains remained susceptible to meropenem and ertapenem. The low expression of the PBP activator-encoding genes lpoA and lpoB was the main underlying mechanism among the imipenem-resistant M. morganii isolates that remained susceptible to meropenem and ertapenem, along with the presence of specific PBP mutations. Meanwhile, the overexpression of efflux pump might also contribute to imipenem resistance in M. morganii. With carbapenems being increasingly used as therapeutic options, it is urgent to establish monitoring programs to prevent the spread of carbapenem resistance.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by The Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The ethics committee/institutional review board waived the requirement of written informed consent for participation from the participants or the participants’ legal guardians/next of kin because Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Author contributions
XZ: Data curation, Formal Analysis, Funding acquisition, Methodology, Software, Writing – original draft. WZ: Data curation, Formal Analysis, Methodology, Software, Writing – review & editing. YL: Data curation, Investigation, Validation, Writing – review & editing. LF: Investigation, Validation, Writing – review & editing. JZ: Methodology, Visualization, Writing – review & editing. TZ: Funding acquisition, Resources, Visualization, Writing – review & editing. CQ: Conceptualization, Project administration, Supervision, Writing – review & editing. CZ: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This research was funded by Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022) and the General Scientific Research Project of Education Department of Zhejiang Province (Y202147847).
Acknowledgments
The authors acknowledge the financial support of the Key Laboratory of Clinical Laboratory Diagnosis and Translational Research of Zhejiang Province (2022E10022) and the General Scientific Research Project of Education Department of Zhejiang Province (Y202147847).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor JZ declared a past co-authorship with the author TZ.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1672736/full#supplementary-material
References
Aissa, N., Mayer, N., Bert, F., Labia, R., Lozniewski, A., and Nicolas-Chanoine, M. H. (2016). A new mechanism to render clinical isolates of Escherichia coli non-susceptible to imipenem: substitutions in the PBP2 penicillin-binding domain. J. Antimicrob. Chemother. 71, 76–79. doi: 10.1093/jac/dkv318
Aitken, S. L., Pierce, V. M., Pogue, J. M., Kline, E. G., Tverdek, F. P., and Shields, R. K. (2024). The growing threat of NDM-producing escherichia coli with penicillin-binding protein 3 mutations in the United States-is there a potential role for durlobactam? Clin. Infect. Dis. 79, 834–837. doi: 10.1093/cid/ciae229
Bandy, A. (2020). Ringing bells: Morganella morganii fights for recognition. Public Health 182, 45–50. doi: 10.1016/j.puhe.2020.01.016
Bertonha, A. F., Silva, C. C. L., Shirakawa, K. T., Trindade, D. M., and Dessen, A. (2023). Penicillin-binding protein (PBP) inhibitor development: A 10-year chemical perspective. Exp. Biol. Med. (Maywood) 248, 1657–1670. doi: 10.1177/15353702231208407
Bonnin, R. A., Creton, E., Perrin, A., Girlich, D., Emeraud, C., Jousset, A. B., et al. (2024). Spread of carbapenemase-producing Morganella spp from 2013 to 2021: a comparative genomic study. Lancet Microbe 5, e547–e558. doi: 10.1016/S2666-5247(23)00407-X
Breilh, D., Texier-Maugein, J., Allaouchiche, B., Saux, M. C., and Boselli, E. (2013). Carbapenems. J. Chemother. 25, 1–17. doi: 10.1179/1973947812Y.0000000032
Chen, J., Wu, Y., Zhang, G., Kang, W., Wang, T., Li, J., et al. (2024). Tracing the possible evolutionary trends of Morganella morganii: insights from molecular epidemiology and phylogenetic analysis. mSystems 9, e0030624. doi: 10.1128/msystems.00306-24
Cherkaoui, A., Diene, S. M., Fischer, A., Leo, S., Francois, P., and Schrenzel, J. (2017). Transcriptional Modulation of Penicillin-Binding Protein 1b, Outer Membrane Protein P2 and Efflux Pump (AcrAB-TolC) during Heat Stress Is Correlated to Enhanced Bactericidal Action of Imipenem on Non-typeable Haemophilus influenzae. Front. Microbiol. 8. doi: 10.3389/fmicb.2017.02676
CLSI (2025). “Performance standard for antimicrobial susceptibility testing,” in CLSI supplement M100, 35th ed (Clinical and Laboratory Standards Institute, Wayne, PA).
Dorr, T., Moll, A., Chao, M. C., Cava, F., Lam, H., Davis, B. M., et al. (2014). Differential requirement for PBP1a and PBP1b in in vivo and in vitro fitness of Vibrio cholerae. Infect. Immun. 82, 2115–2124. doi: 10.1128/IAI.00012-14
Egan, A. J., Jean, N. L., Koumoutsi, A., Bougault, C. M., Biboy, J., Sassine, J., et al. (2014). Outer-membrane lipoprotein LpoB spans the periplasm to stimulate the peptidoglycan synthase PBP1B. Proc. Natl. Acad. Sci. U.S.A. 111, 8197–8202. doi: 10.1073/pnas.1400376111
Erlanger, D., Assous, M. V., Wiener-Well, Y., Yinnon, A. M., and Ben-Chetrit, E. (2019). Clinical manifestations, risk factors and prognosis of patients with Morganella morganii sepsis. J. Microbiol. Immunol. Infect. 52, 443–448. doi: 10.1016/j.jmii.2017.08.010
Fabrizio, C. and Tascini, C. (2025). Chances/challenges of the role of new beta-lactamase inhibitors against the growing threat of NDM-producing escherichia coli with penicillin-binding protein 3 mutations. Clin. Infect. Dis. 80, 925. doi: 10.1093/cid/ciae343
Goffin, C. and Ghuysen, J. M. (2002). Biochemistry and comparative genomics of SxxK superfamily acyltransferases offer a clue to the mycobacterial paradox: presence of penicillin-susceptible target proteins versus lack of efficiency of penicillin as therapeutic agent. Microbiol. Mol. Biol. Rev. 66, 702–738. doi: 10.1128/MMBR.66.4.702-738.2002
Greene, N. G., Fumeaux, C., and Bernhardt, T. G. (2018). Conserved mechanism of cell-wall synthase regulation revealed by the identification of a new PBP activator in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 115, 3150–3155. doi: 10.1073/pnas.1717925115
Guo, X., Rao, Y., Guo, L., Xu, H., Lv, T., Yu, X., et al. (2019). Detection and genomic characterization of a morganella morganii isolate from China that produces NDM-5. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.01156
Hu, J. J. and Wang, J. P. (2024). Production of natural penicillin-binding protein 2 and development of a signal-amplified fluorescence polarization assay for the determination of 28 beta-lactam antibiotics in milk. J. Agric. Food Chem. 72, 27528–27537. doi: 10.1021/acs.jafc.4c07028
Jean, N. L., Bougault, C. M., Lodge, A., Derouaux, A., Callens, G., Egan, A. J., et al. (2014). Elongated structure of the outer-membrane activator of peptidoglycan synthesis LpoA: implications for PBP1A stimulation. Structure 22, 1047–1054. doi: 10.1016/j.str.2014.04.017
Katayama, Y., Zhang, H. Z., and Chambers, H. F. (2004). PBP 2a mutations producing very-high-level resistance to beta-lactams. Antimicrob. Agents Chemother. 48, 453–459. doi: 10.1128/AAC.48.2.453-459.2004
King, D. T., Lameignere, E., and Strynadka, N. C. (2014). Structural insights into the lipoprotein outer membrane regulator of penicillin-binding protein 1B. J. Biol. Chem. 289, 19245–19253. doi: 10.1074/jbc.M114.565879
Laupland, K. B., Paterson, D. L., Edwards, F., Stewart, A. G., and Harris, P. N. A. (2022). Morganella morganii, an emerging cause of bloodstream infections. Microbiol. Spectr. 10, e0056922. doi: 10.1128/spectrum.00569-22
Lessel, J. (1996). Penicillin–binding protein: the target for beta-lactam antibiotics, beta-lactamases and their inhibitors. Pharm. Unserer Zeit 25, 17–27. doi: 10.1002/pauz.19960250107
Liu, Q., Shen, H., Wei, M., Chen, X., Gu, L., and Zhu, W. (2025). Global phylogeography and antibiotic resistance characteristics of Morganella: An epidemiological, spatial, comparative genomic study. Drug Resist. Update 78, 101180. doi: 10.1016/j.drup.2024.101180
Liu, H., Zhu, J., Hu, Q., and Rao, X. (2016). Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 50, 10–17. doi: 10.1016/j.ijid.2016.07.006
Logan, L. K. and Weinstein, R. A. (2017). The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J. Infect. Dis. 215, S28–S36. doi: 10.1093/infdis/jiw282
Malik, S., Kaminski, M., Landman, D., and Quale, J. (2020). Cefiderocol Resistance in Acinetobacter baumannii: Roles of beta-Lactamases, Siderophore Receptors, and Penicillin Binding Protein 3. Antimicrob. Agents Chemother. 64(11), e01221-20. doi: 10.1128/AAC.01221-20
Munir, A., Lv, Y., Song, R., Lu, X., Li, Z., Wang, Z., et al. (2025). Emergence of carbapenem resistant Escherichia coli, Morganella morganii, Providencia rettgeri harboring bla(NDM) genes in minks from China. Vet. Microbiol. 307, 110585. doi: 10.1016/j.vetmic.2025.110585
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Paradis-Bleau, C., Markovski, M., Uehara, T., Lupoli, T. J., Walker, S., Kahne, D. E., et al. (2010). Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell 143, 1110–1120. doi: 10.1016/j.cell.2010.11.037
Park, S. Y., Lee, K., Cho, Y., Lim, S. R., Kwon, H., Han, J. E., et al. (2020). Emergence of third-generation cephalosporin-resistant morganella morganii in a captive breeding dolphin in South Korea. Anim. (Basel) 10 (11), 2052-2058. doi: 10.3390/ani10112052
Sakaguchi, S., Nishi, K., Yamashita, Y., Hiratsuka, T., Hara, S., and Okayama, A. (2014). White urine due to urinary tract infection. Kidney Int. 86, 655. doi: 10.1038/ki.2014.23
Shalaby, M. W., Dokla, E. M. E., Serya, R. A. T., and Abouzid, K. A. M. (2020). Penicillin binding protein 2a: An overview and a medicinal chemistry perspective. Eur. J. Med. Chem. 199, 112312. doi: 10.1016/j.ejmech.2020.112312
Shen, P., Wei, Z., Jiang, Y., Du, X., Ji, S., Yu, Y., et al. (2009). Novel genetic environment of the carbapenem-hydrolyzing beta-lactamase KPC-2 among Enterobacteriaceae in China. Antimicrob. Agents Chemother. 53, 4333–4338. doi: 10.1128/AAC.00260-09
Shi, D. S., Wang, W. P., Kuai, S. G., Shao, H. F., and Huang, M. (2012). Identification of bla KPC-2 on different plasmids of three Morganella morganii isolates. Eur. J. Clin. Microbiol. Infect. Dis. 31, 797–803. doi: 10.1007/s10096-011-1377-9
Shrestha, S., Tada, T., Sherchan, J. B., Uchida, H., Hishinuma, T., Oshiro, S., et al. (2020). Highly multidrug-resistant Morganella morganii clinical isolates from Nepal co-producing NDM-type metallo-beta-lactamases and the 16S rRNA methylase ArmA. J. Med. Microbiol. 69, 572–575. doi: 10.1099/jmm.0.001160
Tellapragada, C., Razavi, M., Peris, P. S., Jonsson, P., Vondracek, M., and Giske, C. G. (2024). Resistance to aztreonam-avibactam among clinical isolates of Escherichia coli is primarily mediated by altered penicillin-binding protein 3 and impermeability. Int. J. Antimicrob. Agents 64, 107256. doi: 10.1016/j.ijantimicag.2024.107256
Timofte, D., Panzaru, C. V., Maciuca, I. E., Dan, M., Mare, A. D., Man, A., et al. (2016). Active surveillance scheme in three Romanian hospitals reveals a high prevalence and variety of carbapenamase-producing Gram-negative bacteria: a pilot study, December 2014 to May 2015. Euro Surveill 21 (25), pii=30262. doi: 10.2807/1560-7917.ES.2016.21.25.30262
Typas, A., Banzhaf, M., van den Berg van Saparoea, B., Verheul, J., Biboy, J., Nichols, R. J., et al. (2010). Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell 143, 1097–1109. doi: 10.1016/j.cell.2010.11.038
Xiang, G., Lan, K., Cai, Y., Liao, K., Zhao, M., Tao, J., et al. (2021). Clinical molecular and genomic epidemiology of morganella morganii in China. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.744291
Yao, J., Hu, Y., Wang, X., Sheng, J., Zhang, Y., Zhao, X., et al. (2025). Carbapenem-resistant Morganella morganii carrying bla(KPC-2) or bla(NDM-1) in the clinic: one-decade genomic epidemiology analysis. Microbiol. Spectr. 13, e0247624. doi: 10.1128/spectrum.02476-24
Yin, J., Sun, Y., Mao, Y., Jin, M., and Gao, H. (2015). PBP1a/LpoA but not PBP1b/LpoB are involved in regulation of the major beta-lactamase gene blaA in Shewanella oneidensis. Antimicrob. Agents Chemother. 59, 3357–3364. doi: 10.1128/AAC.04669-14
Zaric, R. Z., Jankovic, S., Zaric, M., Milosavljevic, M., Stojadinovic, M., and Pejcic, A. (2021). Antimicrobial treatment of Morganella morganii invasive infections: Systematic review. Indian J. Med. Microbiol. 39, 404–412. doi: 10.1016/j.ijmmb.2021.06.005
Zheng, X., Zhang, X., Zhou, B., Liu, S., Chen, W., Chen, L., et al. (2022). Clinical characteristics, tolerance mechanisms, and molecular epidemiology of reduced susceptibility to chlorhexidine among Pseudomonas aeruginosa isolated from a teaching hospital in China. Int. J. Antimicrob. Agents 60, 106605. doi: 10.1016/j.ijantimicag.2022.106605
Keywords: Morganella morganii, carbapenem-resistant, imipenem, penicillin-binding protein, penicillin-binding protein activator
Citation: Zheng X, Zeng W, Liu Y, Feng L, Zheng J, Zhou T, Qian C and Zhou C (2025) Clinical characteristics, specific resistance patterns, and molecular mechanisms of carbapenem-resistant Morganella morganii isolates. Front. Cell. Infect. Microbiol. 15:1672736. doi: 10.3389/fcimb.2025.1672736
Received: 24 July 2025; Accepted: 18 August 2025;
Published: 05 September 2025.
Edited by:
Jinxin Zhao, Monash University, AustraliaReviewed by:
Jia Wei Chen, Zhejiang University, ChinaRongrong Li, First Affiliated Hospital of Anhui Medical University, China
Copyright © 2025 Zheng, Zeng, Liu, Feng, Zheng, Zhou, Qian and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cui Zhou, emhvdWN1aTA4MjZAMTYzLmNvbQ==; Changrui Qian, cWlhbmNoYW5ncnVpQDE2My5jb20=
 Xiangkuo Zheng
Xiangkuo Zheng Weiliang Zeng
Weiliang Zeng Yan Liu
Yan Liu Luozhu Feng
Luozhu Feng Jiayin Zheng
Jiayin Zheng Tieli Zhou2
Tieli Zhou2 Changrui Qian
Changrui Qian Cui Zhou
Cui Zhou