- 1School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 2State Key Laboratory of Infectious Disease Prevention and Control, National Institute for Communicable Disease Control and Prevention, Chinese Center for Disease Control and Prevention, Beijing, China
- 3Laboratory of Clinical Microbiology and Infectious Diseases, Department of Pulmonary and Critical Care Medicine, Beijing Key Laboratory of Surveillance, Early Warning and Pathogen Research on Emerging Infectious Diseases, National Center for Respiratory Medicine, China-Japan Friendship Hospital, Beijing, China
Background: Antimicrobial resistance is prevalent across Nocardia species, with varying resistance profiles among different species, which poses significant challenges to effective treatment strategies. Our study aimed to assess the antimicrobial susceptibility profiles of various Nocardia species and investigate the potential correlation between resistance phenotypes and their underlying genotypes.
Methods: This study analyzed 148 clinical Nocardia isolates from 13 provinces in China. Minimum inhibitory concentrations (MICs) for 15 antimicrobial agents were determined using the microbroth dilution method. Whole genome sequencing (WGS) was performed for all isolates, followed by bioinformatics analyses integrated with 70 human-sourced Nocardia genomes in the National Center for Biotechnology Information (NCBI), encompassing species verification, phylogenetic analysis, and the identification of antimicrobial resistance genes (ARGs).
Results: Average Nucleotide Identity (ANI) analysis reclassified several misidentified isolates and revealed 14 potentially novel Nocardia species, underscoring the taxonomic complexity within this genus. Nocardia species exhibited distinct resistance profiles: Nocardia farcinica demonstrated elevated resistance to cephalosporins and tobramycin; Nocardia otitidiscaviarum showed broad resistance to β-lactams and quinolones; and Nocardia cyriacigeorgica exhibited resistance to quinolones, cefepime, and cefoxitin. Notably, clarithromycin resistance was consistently high across all species. Moreover, 38.51% of Nocardia isolates are resistant to two or more commonly used antibiotics, which revealed widespread multidrug resistance (MDR). Strong genotype–phenotype correlations were observed, including sul1 in sulfamethoxazole/trimethoprim-resistant N. farcinica, blaAST-1 in β-lactam-resistant N. otitidiscaviarum, and tetA/B(58) across tetracycline-intermediate isolates. Additionally, resistance mechanisms beyond ARGs were observed, including species-specific presence of warA and aph(2’’), and gyrA mutations largely correlating with ciprofloxacin resistance. Nonetheless, resistance in some strains lacking known resistance determinants indicates the presence of uncharacterized mechanisms.
Conclusions: These findings provide critical insights into the drug resistance patterns of Nocardia strains and antimicrobial resistance genes, highlighting the importance of ongoing genomic surveillance and personalized treatments.
1 Introduction
Nocardia, a member of the Actinobacteria order, primarily infects immunocompromised individuals, such as those with Human Immunodeficiency Virus (HIV), organ transplants, or chronic lung disease, but can also cause infections in immunocompetent individuals (Shen et al., 2025). If Nocardia infection at the primary site is not promptly and effectively treated, it may spread to other parts of the body, causing severe infections and potentially leading to death (Fatahi-Bafghi, 2018). In recent years, with the increasing number of elderly individuals, immunocompromised patients, organ transplant recipients, and HIV-infected patients, the incidence of Nocardia infections has been gradually rising (Ji et al., 2022; Shen et al., 2025). Studies suggest that the mortality rate for Nocardia bacteremia patients is approximately 50% (Kontoyiannis et al., 1998), while the mortality rate for patients with disseminated infection ranges from 44% (Beaman and Beaman, 1994) to 85% (Presant et al., 1973). As a life-threatening infection, Nocardia infection poses significant clinical challenges. However, given the extensive diversity within the Nocardia genus, identification of rare species by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) remains challenging for clinical laboratories, despite some species being readily identifiable (Conville et al., 2017). Furthermore, varying drug susceptibility patterns among Nocardia species may result in treatment failures (Zhao et al., 2017; Wang et al., 2022; Hershko et al., 2023b). Accurate species identification is essential for understanding the epidemiology and pathogenicity of Nocardia. The global rise in antimicrobial resistance (AMR) represents a significant public health threat (GBD 2021 Antimicrobial Resistance Collaborators, 2024). While most Nocardia strains remain sensitive to amikacin, linezolid, and trimethoprim-sulfamethoxazole, their susceptibility profiles vary when it comes to β-lactam antibiotics, quinolones, aminoglycosides, and other antibiotics (Zhao et al., 2017; Wang et al., 2022; Hershko et al., 2023b).
Reports on the antibiotic sensitivity of Nocardia species in various regions of China have been published (Wang et al., 2022; Yi et al., 2019; Wei et al., 2017; Han et al., 2024), but these reports have been limited to the phenotypic aspects of drug resistance and have not explored the genetic basis of drug resistance.
Whole-genome sequencing (WGS), a high-throughput sequencing technology, offers a comprehensive approach to detecting AMR genes in clinical bacterial strains (Balloux et al., 2018). Additionally, WGS facilitates the prediction of bacterial genetic profiles and functional phenotypes (Balloux et al., 2018). Consequently, the application of this technology significantly enhances the efficiency of clinical microbiological testing and provides critical information for species identification of Nocardia and guiding therapeutic interventions.
Understanding genetic diversity of Nocardia isolates and their resistance mechanisms is essential for informing empirical antibiotic therapy. This study aimed to perform antimicrobial susceptibility testing and WGS on 148 clinical Nocardia isolates collected from 13 provinces in China, to assess their drug resistant phenotype, AMR genes, and genetic characteristics. The findings will deepen our understanding of the genetic factors driving resistance in Nocardia, as well as the relationship between genetic background and drug resistance. This valuable information will contribute to more effective clinical management of nocardiosis.
2 Materials and methods
2.1 Strains collection and initial identification
Nocardia spp. strains in this study were isolated from clinical specimens collected across 13 provincial-level administrative divisions in China, including provinces, autonomous regions, and municipalities (Beijing, Gansu Province, Guangdong Province, Guangxi Zhuang Autonomous Region, Hebei Province, Henan Province, Heilongjiang Province, Hunan Province, Inner Mongolia Autonomous Region, Ningxia Hui Autonomous Region, Shandong Province, Zhejiang Province, Jiangsu Province). After strain purification, bacterial colonies were cultured on Brain Heart Infusion (BHI) blood agar for at least 48 hours. Freshly picked colonies were then subjected to MALDI-TOF MS (Zybio EXS3000, Zybio Inc., Chongqing, China) analysis using the formic acid/ethanol extraction method.
2.2 Antibiotic susceptibility testing
Minimum inhibitory concentration (MIC) was determined using the Sensititre RAPMYCO microdilution panel (Thermo Fisher Scientific, Sunnyvale, CA, USA) according to the manufacturer’s protocol, with results visualized using the R package ggplot2. Additionally, MIC50 and MIC90 were calculated using R Studio software, version 4.3.3. The AST plate included 15 antibiotics: amikacin, amoxicillin/clavulanic acid, cefepime, cefoxitin, ceftriaxone, ciprofloxacin, clarithromycin, doxycycline, imipenem, linezolid, minocycline, moxifloxacin, tigecycline, tobramycin, trimethoprim/sulfamethoxazole. Quality control was performed using Staphylococcus aureus ATCC 29213 and Escherichia coli ATCC 25922 as reference strains. Results were interpreted according to the Nocardia spp. interpretive criteria specified in Clinical and Laboratory Standards Institute (CLSI) standard M24-A2 (Woods et al., 2011). As the MIC for sulfamethoxazole/trimethoprim is defined as the lowest concentration inhibiting 80% of growth, additional verification was conducted using the MIC test strip (MTS) (Liofilchem Ltd., Roseto Degli Abruzzi, TE, Italy) method to ensure the accuracy of AST results.
2.3 DNA extraction and WGS
The strains were subcultured for two consecutive generations in BHI liquid medium. Bacterial cells were then collected by centrifugation to form a pellet. Genomic DNA was extracted from the pellet using the Wizard genomic DNA purification kit (Promega, Madison, WI, USA). Following extraction, DNA purity and concentration were measured using a NanoDrop instrument (Thermo Fisher Scientific, Sunnyvale, CA, USA). Then, the DNA samples were sequenced using the Illumina Novaseq platform in the PE150 mode (Shanghai Majorbio Bio-pharm Technology Co., Ltd., Shanghai, China), ensuring all sequencing depths exceeded 100-fold. The quality of reads was checked and trimmed by Fastp v0.23.4 (Chen, 2023). The cleaned reads were de novo assembled by SPAdes v3.15.5 (Bankevich et al., 2012), and the draft genomes were annotated by Prokka v1.12 (Seemann, 2014). The genome size, number of the coding sequences (CDSs), and GC content were estimated by Quast v5.0.2 (Gurevich et al., 2013) and CheckM v1.1.3 (Parks et al., 2015).
2.4 Phylogenetic tree construction and average nucleotide identity analysis
To gain a more comprehensive understanding of the classification and genetic evolution of Nocardia strains, we downloaded the whole genome sequences of 70 Nocardia strains isolated from human origin from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/datasets/genome/). Pangenome and core genome analysis was conducted using Roary v3.12.0 (Page et al., 2015). Single-copy core genes were identified by BLASTn. Then, the phylogenetic tree based on the core genome was constructed using FastTree and visualized by iTOL (https://itol.embl.de/). Pairwise ANI values across all genomes were calculated by Pyani v0.2.12 (Pritchard et al., 2016) and visualized by the R package pheatmap. The in silico DNA-DNA hybridization (isDDH) values were calculated through the Genome-to-Genome Distance Calculator 2.1 (GGDC) (http://ggdc.dsmz.de/ggdc.php) using “Formula 2” (Auch et al., 2010).
2.5 Antibiotic resistance gene analysis
The ARGs were identified using Rgi v6.0.2 with a cutoff of 60% identity and 70% coverage against the Comprehensive Antibiotic Resistance Database (CARD) (https://card.mcmaster.ca/analyze/rgi). AGRs was visualized with the R package pheatmap. Gene presence/absence was analyzed using BLASTn, gene mutations were analyzed via sequence alignment with Clustal Omega (http://lilab2.sysu.edu.cn/Tools/msa/clustalo/). The associations between phenotypic resistance and resistance genes were assessed using Cramér’s V statistic, a measure of association between two categorical variables, ranging from 0 (no association) to 1 (perfect association) (*P < 0.05, **P < 0.01, ***P < 0.001). The results were visualized as a heatmap, where each cell contains the Cramér’s V value along with its statistical significance. The color intensity reflects the strength of association: yellow shades indicate weak or no association (values close to 0), while deep purple shades indicate strong association (values close to 1).
3 Results
3.1 Distribution of Nocardia species
Among the 148 clinical isolates preliminarily identified by MALDI-TOF MS (Supplementary Table S1), 10 Nocardia species were characterized, most of which were isolated from the respiratory tract. Specifically, the predominant species were N. farcinica (60.8%, 90/148), followed by N. cyriacigeorgica (16.2%, 24/148), N. otitidiscaviarum (10.8%, 16/148), and N. wallacei (5.4%, 8/148). In contrast, less frequently isolated species included N. brasiliensis (2.0%, 3/148), N. nova (1.4%, 2/148), N. blacklockiae (1.4%, 2/148), N. transvalensis (0.7%, 1/148), N. beijingensis (0.7%, 1/148), and N. aobensis (0.7%, 1/148). Notably, N. farcinica accounted for over half of all isolates.
3.2 Verification of species by ANI
A 95-96% ANI threshold was applied to define species boundaries between pairwise genomes (Richter and Rosselló-Móra, 2009). A total of 218 Nocardia strains, 148 clinical isolates from this study and 70 clinically isolated strains (comprising 42 species) retrieved from NCBI, were subjected to pairwise ANI comparison. Based on ANI analysis (Figure 1 and Supplementary Table S1), four species without a type strain in NCBI were reclassified (Supplementary Table S2), and four Nocardia strains from this study were reclassified: N. brasiliensis was reclassified as N. cyriacigeorgica in two cases; N. farcinica as N. sputorum in one case; and N. beijingensis as N. sputi in one case. Additionally, 14 isolates potentially represented taxonomically unclassified species (i.e. novel species) within the genus Nocardia (Supplementary Table S3). These strains, initially identified by MALDI-TOF MS to N. cyriacigeorgica (n=4), N. wallacei (n=6), N. farcinica (n=1) and N. blacklockiae (n=1), exhibited genomic divergence exceeding established species thresholds. Additionally, N. cyriacigeorgica 105602, CDC381, CDC481, CDC519, CDC528, CDC546, and CDC579 exhibited the closest ANI with N. cyriacigeorgica. However, their ANI values with other N. cyriacigeorgica strains were below the 95% species delineation threshold. Consequently, these strains were identified as representing novel/unidentified Nocardia species. Phylogenetic analysis further revealed that strains 105602, CDC381, and CDC546 constitute one distinct species, while strains CDC481, CDC519, CDC528, and CDC579 constitute another. Furthermore, six unidentified Nocardia strains demonstrated the closest ANI with N. wallacei. However, as their ANI values fell below the 95% species delineation threshold, they likely represent novel species phylogenetically related to N. wallacei. MALDI-TOF MS correctly identified the majority of Nocardia isolates. Except for the unidentified Nocardia species, the technique achieved a species-level identification accuracy of 97.1% (132/136) for the isolated strains. This high accuracy provides reliable guidance for the initial clinical screening of Nocardia infections.
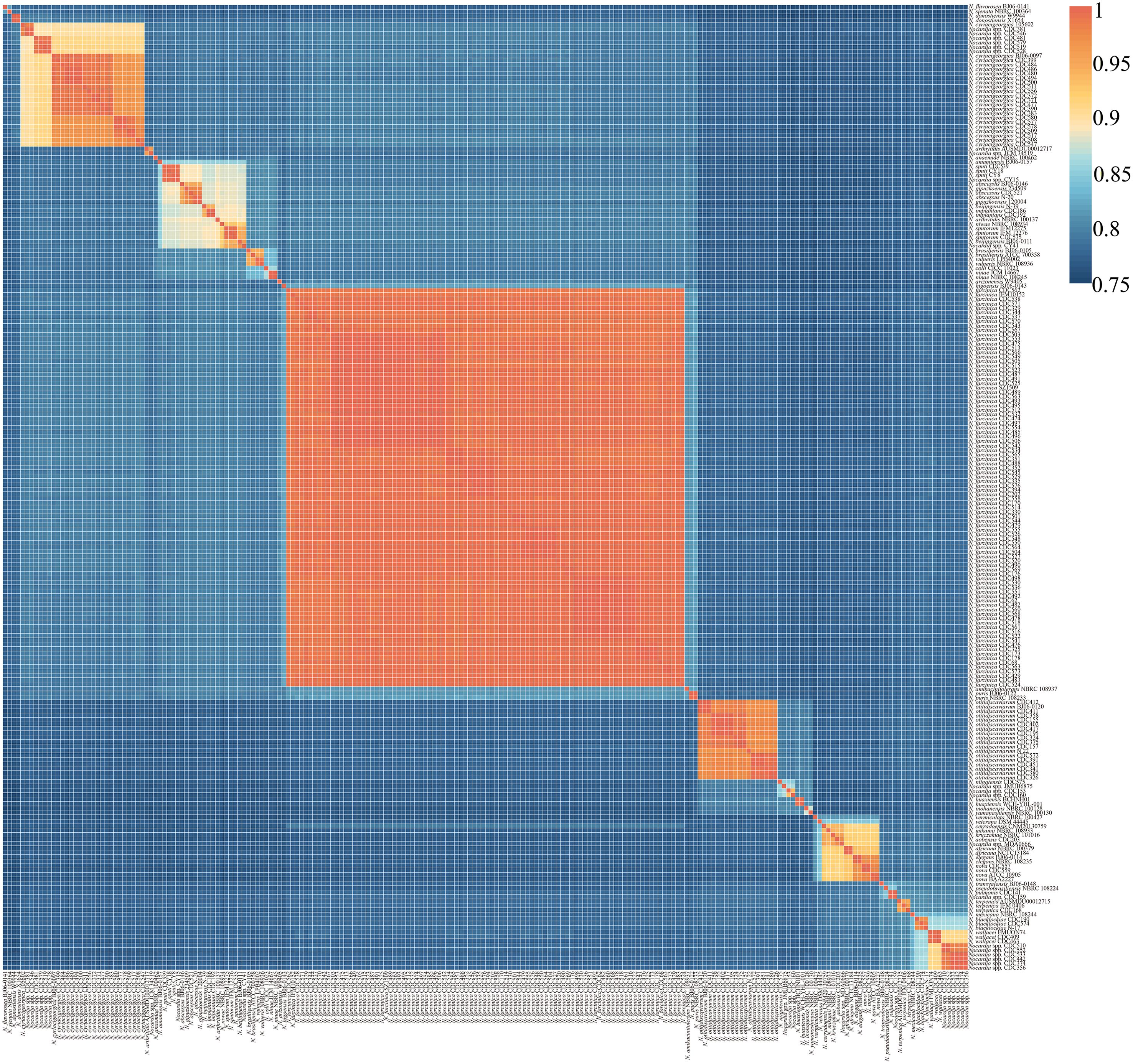
Figure 1. Heatmap of pairwise average nucleotide identity (ANI) values for 218 genome assemblies of Nocardia (148 of this study, 70 of NCBI).
3.3 Genomic characteristics and whole-genome phylogenetic analysis
The genomic characteristics of 148 clinical Nocardia isolates and 70 reference strains from NCBI are detailed in Supplementary Table S4. The predominant species, N. farcinica, N. otitidiscaviarum, and N. cyriacigeorgica, exhibited mean genome sizes of 6.36 Mb, 7.52 Mb, and 6.44 Mb, respectively, with mean coding sequences (CDSs) of 5,956, 6,892 and 5,820, and mean GC contents of 70.7%, 69.0%, and 68.3%, respectively (Supplementary Figure S1).
To further assess the taxonomic relationship among Nocardia strains, a circular whole-genome phylogenetic tree was constructed using the whole genome sequence of 668 single-copy genes from 218 Nocardia strains (Figure 2). The phylogenomic tree, encompassing 46 species, revealed that most strains of the same Nocardia species clustered within the same clade. The 148 strains in this study and the 70 strains sourced from NCBI were all isolated from clinical patients. Among the 12 unidentified Nocardia strains in this study, 6 exhibited close phylogenetic relationships with N. cyriacigeorgica, while the remaining 6 clustered within the N. wallacei clade and likely represent novel species within the N. transvalensis complex (Conville et al., 2008).
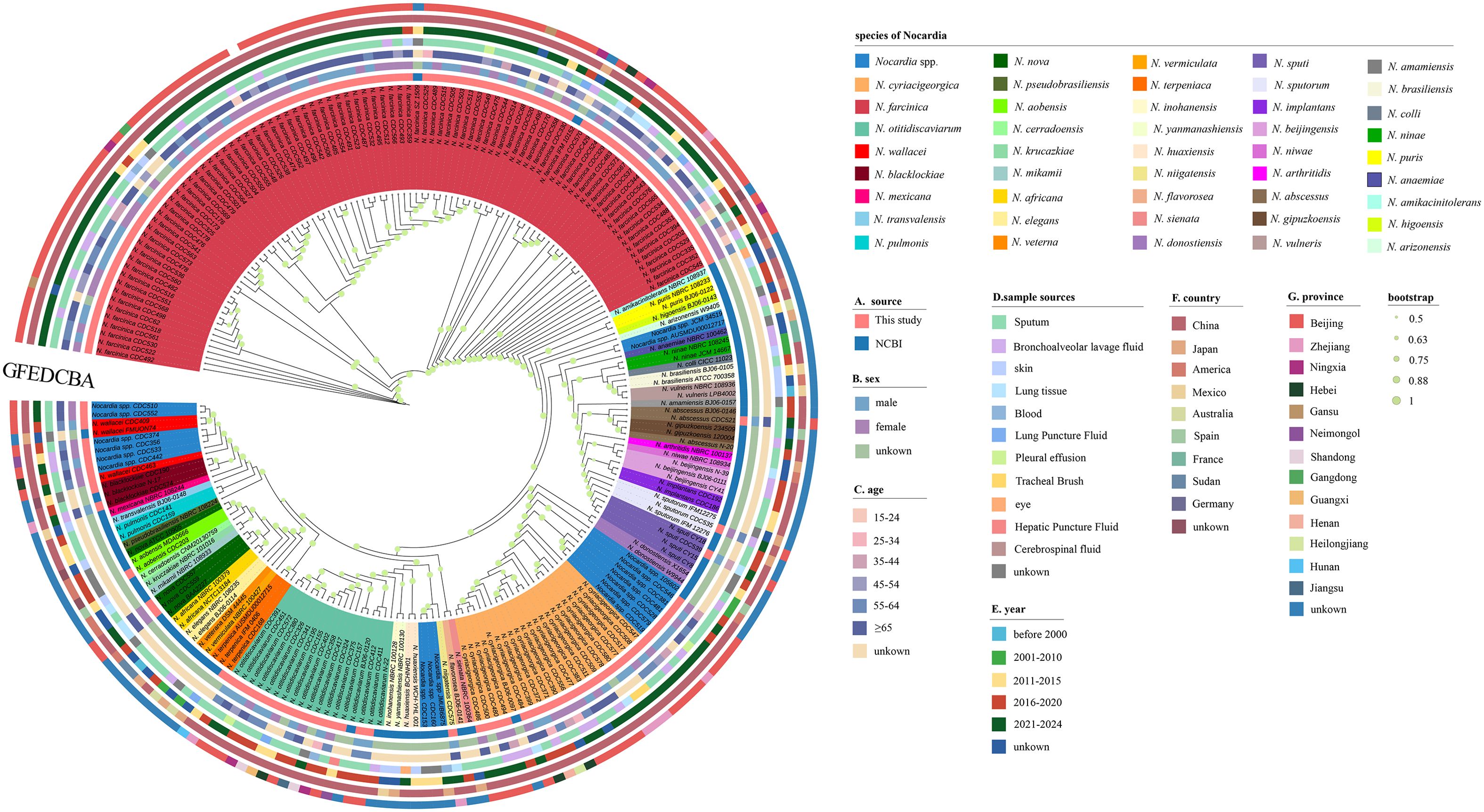
Figure 2. Nocardia genome phylogenetic relationships based on the concatenation of thenucleotide sequences of 668 single-copy core genes. The innermost colored bands represent 46 different species of Nocardia and Nocardia spp. (unable to identify species). (A) A colored ring showing different sources of genomic information of Nocardia. (B) A colored ring showing the gender of patients with Nocardia infections. (C) A colored ring showing the age of patients with Nocardia infections. (D) A colored ring showing sample sources of Nocardia isolates. (E) A colored ring showing five different periods of collection years and the unknown collection years. (F) A colored ring showing different country origins of Nocardia. (G) A colored ring showing different province origins of Nocardia.
3.4 Phenotypic analysis of drug resistance
Figure 3 presents the MIC distributions for 15 AST against the three most prevalent Nocardia species in this study, including the MIC50, MIC90, and resistance rates, AST results for other Nocardia species can be found in Supplementary Table S5. AST results for sulfamethoxazole/trimethoprim was performed using the MTS method to ensure the accuracy (Supplementary Table S6). All clinically isolated Nocardia strains demonstrated 100% susceptibility to linezolid, followed by trimethoprim/sulfamethoxazole (140/148, 94.6%) and amikacin (138/148, 93.2%). In contrast, susceptibility to clarithromycin was markedly lower at 4.7% (7/148). For trimethoprim/sulfamethoxazole, MIC values exceeding 8/152 mg/L were exclusively observed in N. farcinica. Furthermore, amikacin resistance was detected only in the N. transvalensis complex, including N. wallacei, N. blacklockiae, and some novel/unidentified closely related species of N. wallacei.
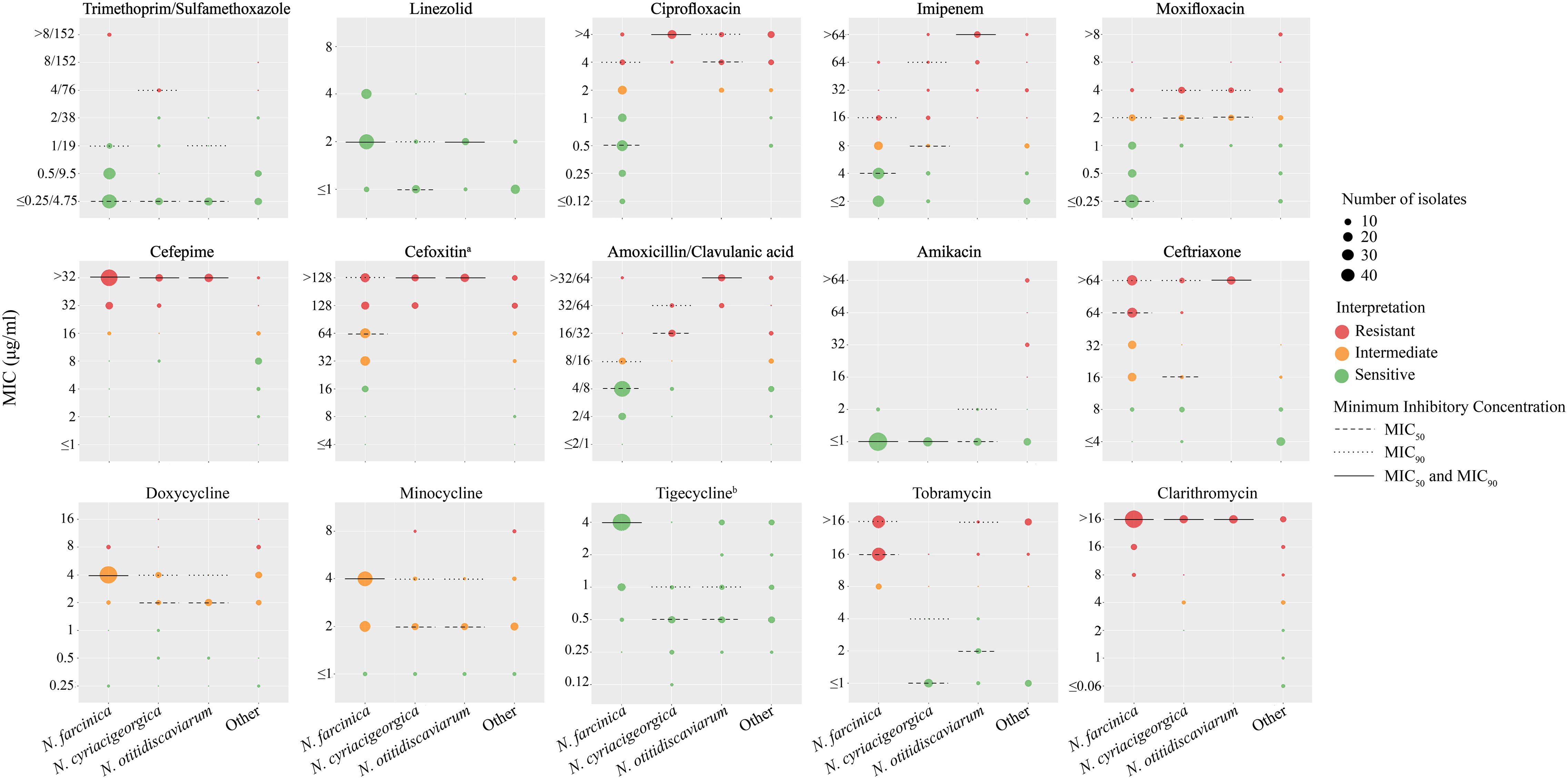
Figure 3. The MIC Minimum Inhibitory Concentrations (MICs) of 15 antimicrobials for Nocardia species. The red, orange and green dots indicate the resistant, intermediate and susceptible breakpoints. The size of the points represents the corresponding number of strains. The dashed line represents MIC50, the dotted line represents MIC90, and the solid black line indicates where MIC50 and MIC90 coincide. a:The breakpoint for cefotaxime is referenced from the breakpoint for rapidly growing mycobacteria in CLSI-M24-A2. b: The clinical breakpoint for tigecycline is not provided in CLSI M24-A2.
For β-lactam antibiotics, including imipenem, cefepime, cefoxitin, amoxicillin/clavulanic acid, and ceftriaxone, Nocardia species exhibited low susceptibility to cephalosporins: cefepime (14.2%), cefoxitin (8.8%), and ceftriaxone (23.0%). Significant interspecies variations in antimicrobial susceptibility were noted. Imipenem susceptibility were 89.8% for N. farcinica, compared to 35.0% for N. cyriacigeorgica and 0% for N. otitidiscaviarum. Similarly, amoxicillin/clavulanic susceptibility varied markedly, with N. farcinica at 85.2%, while N. cyriacigeorgica and N. otitidiscaviarum showed 20.0% and 0% susceptibility, respectively. Moreover, N. otitidiscaviarum strains were completely resistance to all β-lactam antibiotics, with correspondingly high MIC values.
For tetracyclines, doxycycline and minocycline exhibited high intermediate susceptibility rates of 83.1% and 87.8%. Nocardia species displayed distinct susceptibility profiles to quinolone antibiotics, ciprofloxacin and moxifloxacin. N. farcinica demonstrated susceptibility rates of 69.3% to ciprofloxacin and 84.1% to moxifloxacin, whereas N. cyriacigeorgica and N. otitidiscaviarum were completely resistance to ciprofloxacin (100% non-susceptibility) but showed 15.0% and 12.5% susceptibility to moxifloxacin, respectively. Except for N. farcinica, other Nocardia species have lower MIC values for tigecycline. Tobramycin susceptibility varied significantly: N. farcinica exhibited complete resistance (100% non-susceptibility), whereas N. cyriacigeorgica and N. otitidiscaviarum demonstrated susceptibility rates of 90.0% and 68.8%, respectively.
In this study, we followed the definition proposed by Schlaberg et al (Schlaberg et al., 2014), defining multidrug resistance (MDR) Nocardia as isolates that are resistant or intermediate resistant to two or more of the most commonly used empirical antibiotics (amikacin, ceftriaxone, imipenem, and trimethoprim/sulfamethoxazole). The overall MDR rate for clinical Nocardia isolates in this study was 38.51% (57/148), with 29.55% (26/88) for N. farcinica, 45% (9/20) for N. cyriacigeorgica, 100% (16/16) for N. otitidiscaviarum, and 100% (2/2) for N. wallacei.
3.5 Antibiotic resistance genes
Antibiotic resistance profiling based on WGS of 148 clinical Nocardia isolates was visualized through a dot plot (Figure 4A), 27 ARGs were identified and these ARGs are involved at least 9 antimicrobial classes of drugs. The rifampicin resistance gene rbpA and the tetracycline resistance genes tetA(58) and tetB(58) were universally present across Nocardia species. Additionally, the β-lactamase gene blaFAR-1, conferring partial β-lactams antibiotics resistance, predominated in N. farcinica, while vanRO and vanSO, conferring vancomycin resistance, were absent in this species (Figure 4A). blaAST-1 demonstrated species-specific distribution, prevalent in N. cyriacigeorgica and N. otitidiscaviarum, with its homolog in N. otitidiscaviarum linked to pan-β-lactam resistance (Figure 4B). The lincosamide antibiotic resistance gene lmrC was exclusive present in N. cyriacigeorgica, while the aminocoumarin antibiotic resistance gene novA was unique to N. otitidiscaviarum. Strikingly, three N. farcinica strains carrying the sulfonamide-resistant gene sul1 exhibited elevated sulfamethoxazole/trimethoprim MIC values (>8/152 μg/mL), while sul1 was absent in other sulfamethoxazole/trimethoprim-resistant Nocardia species (Figure 4B).
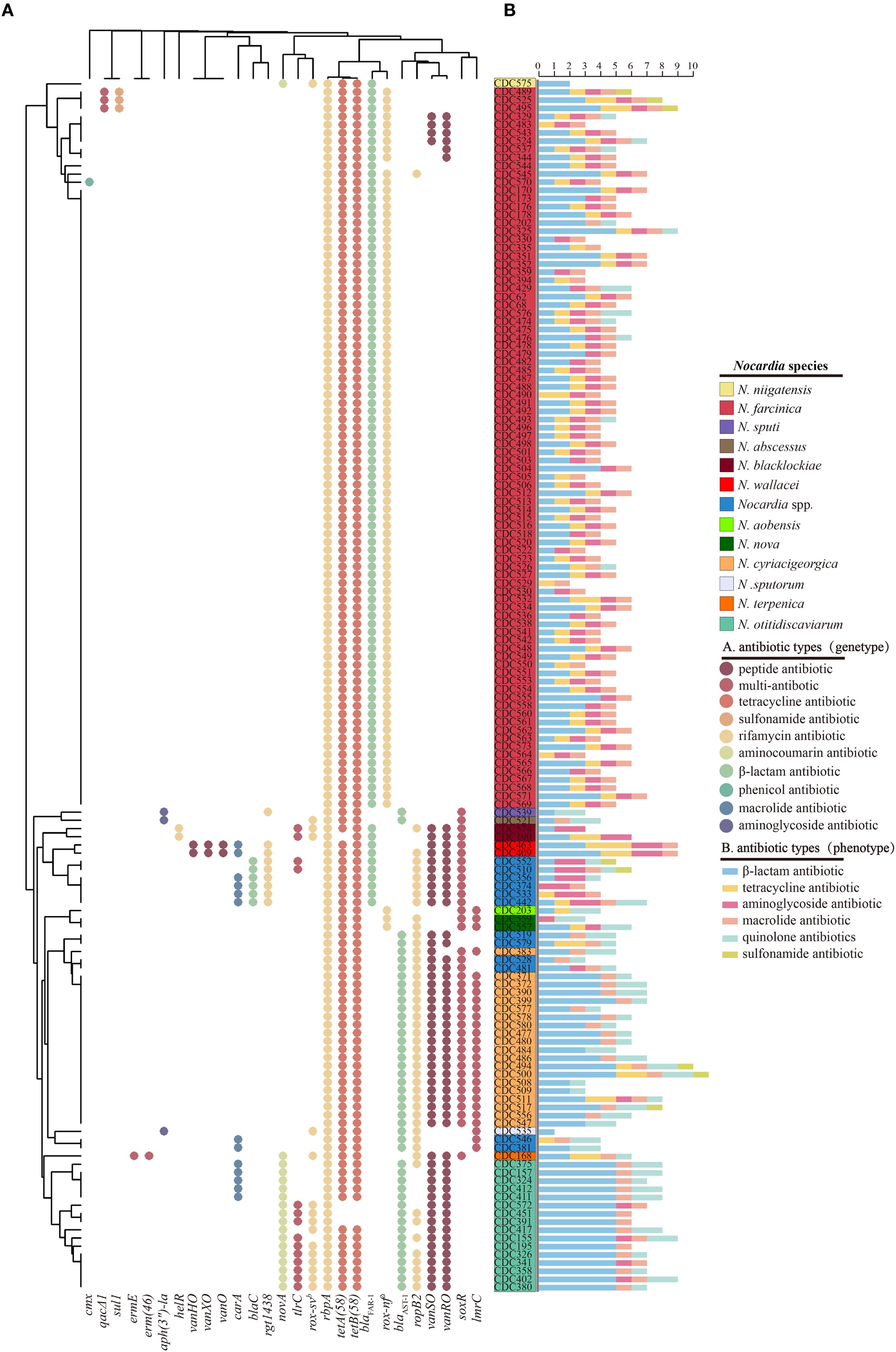
Figure 4. Distribution of Antibiotics Resistance Genes (ARGs) in the genomes of 148 multiple Nocardia strains. Each dot indicates the presence of a gene. Different colors indicate different types of ARGs. rox-nf a: Nocardia farcinica rox; rox-sv b: Streptomyces venezuelae rox (A). Number of antibiotic-resistant phenotypes exhibited by Nocardia spp. across six antimicrobial categories (B).
For genes conferring resistance beyond ARGs, we analyzed warA, aph(2’’), and gyrA using multiple sequence alignment (Supplementary Figure S2). warA, which confers resistance to aminoglycoside antibiotics, was detected in N. wallacei and its related species but was absent in amikacin-resistant N. blacklockiae. The aph(2’’) gene, encoding aminoglycoside 2’’-O-phosphotransferase and mediating tobramycin resistance, was widely detected in N. farcinica and N. blacklockiae, consistent with their tobramycin-resistant phenotype; however, it was absent in other tobramycin-resistant Nocardia species. Ciprofloxacin resistance is potentially caused by mutations in the gyrA gene. In this study, we identified five amino acid substitutions in gyrA: Ser83Ala in N. cyriacigeorgica, Ser83Thr in N. otitidiscaviarum and N. terpenica, Ser83Val in N. sputi, and both Ser83Leu and Ser83Trp in N. wallacei-related new species. These mutations were generally consistent with the ciprofloxacin resistance phenotype. Moreover, no gyrA mutation was detected in ciprofloxacin-resistant N. farcinica (Supplementary Figure S2).
The Cramér’s V analysis identified significant associations between phenotypic resistance and resistance genes. sul1 and qacEΔ1 were strongly associated with the sulfamethoxazole resistance phenotype (both V = 0.60, P < 0.001). Similarly, blaAST-1 and blaFAR-1 showed strong associations with amoxicillin resistance (V = 0.64 and 0.63, respectively; both P < 0.001) (Supplementary Figure S3). gyrA mutations exhibited a strong positive correlation with ciprofloxacin resistance (Cramer’s V = 0.77, P <0.001), while the presence of the aph(2’’) gene significantly correlated with tobramycin resistance (V = 0.67, P <0.001), and warA gene carriage demonstrated the strongest association with amikacin resistance (V = 0.89, P <0.001) (Supplementary Figure S4).
4 Discussion
This study combined species identification, AST, and WGS of clinical Nocardia isolates to systematically explore their phylogenetic architecture, characterize antimicrobial resistance patterns, and elucidate inter-species differences in resistance determinants genomic features with implications for both taxonomy and clinical management.
In this study, clinical Nocardia isolates were predominantly composed of three species: N. farcinica (59.5%, 88/148), N. cyriacigeorgica (13.5%, 20/148) and N. otitidiscaviarum (10.8%, 16/148), together accounting for 83.8% of all isolates. In large-scale Nocardia studies conducted in New Zealand (McKinney et al., 2023) and the United States (Hamdi et al., 2020), N. nova, N. cyriacigeorgica, N. farcinica were dominant, while in largest-scale Nocardia study in China (Wang et al., 2022), N. farcinica, N. cyriacigeorgica, N. abscess, and N. otitidiscaviarum were dominant, similar to this study, indicating that the prevalence trends of different species vary across regions. N. farcinica and N. cyriacigeorgica demonstrate higher clinical prevalence compared to other species in this genus.
Mass spectrometry demonstrated high accuracy in species-level identification, successfully identifying most clinically prevalent Nocardia species. These findings suggest its considerable potential for preliminary Nocardia screening in clinical settings (Carrasco et al., 2016). Consequently, primary mass spectrometric analysis of clinical isolates is an essential step in the diagnostic workflow for suspected nocardiosis. Nevertheless, it misidentified several genetically divergent isolates, particularly novel taxa, underscoring the limitations of Mass spectrometry-based approaches for complex genera. Mass spectrometry identification is based on databases, which need to be continuously improved and updated to cope with the large number of Nocardia species (Conville et al., 2017). ANI analysis seems to have resolved these ambiguities, leading to the reclassification of four isolates and the discovery of 14 potentially novel species, primarily within the N. cyriacigeorgica and N. transvalensis lineages. Although 130 species of Nocardia have been successfully named to date (https://lpsn.dsmz.de/genus/Nocardia), this study shows that there are still many unnamed species of Nocardia in clinical practice. These findings reflect ongoing diversification within clinically relevant Nocardia species and underscore the need for continual taxonomic refinement based on WGS. This study suggests that the MALDI-TOF database should be updated based on the reclassification of ANI to better address the species identification of Nocardia.
AST revealed substantial interspecies variability in Nocardia species with direct implications for therapeutic strategies. Linezolid and amikacin were highly effective against the majority of clinical Nocardia isolates. However, their dose-dependent toxicities and adverse effects preclude their use as first-line treatments (Green et al., 2001; Sack et al., 1976). Amikacin resistance appears to be restricted to the N. transvalensis complex (including N. wallacei, N. blacklockiae, and N. transvalensis), may mediated by the warA mechanism, with a concordance rate of 92.2% (Hershko et al., 2024). In this study, the warA gene was detected exclusively in N. wallacei and its related species but was absent in N. blacklockiae. This absence suggests the potential existence of alternative resistance-conferring genes in N. blacklockiae, which requires further exploration.
Sulfamethoxazole/trimethoprim have been the antimicrobials of choice to treat nocardiosis (Wilson, 2012), remains the cornerstone of clinical management. The overall concordance rate between sul1 gene and sulfamethoxazole/trimethoprim resistance was 37.5%, while the concordance rate with high MIC (>8/152 µg/mL) sulfamethoxazole/trimethoprim resistance was 100%. In our study, three N. farcinica strains with elevated MICs (>8/152 µg/mL) uniquely harbored the sul1 gene, confirming sul1-mediated horizontal gene transfer drives high-level resistance. Previous studies on the diversity of antimicrobial resistance determinants in 76 sulfamethoxazole/trimethoprim-resistant Nocardia strains (MIC values ≥ 32/608 μg/mL) showed that these isolates carried one or more resistance determinants (sul1 accounted for 93.42%, sul2 accounted for 78.94%, and dfrA (S1) accounted for 14.70%) (Valdezate et al., 2015). Transposon-mediated sulfamethoxazole/trimethoprim resistance in N. farcinica has been elucidated in previous studies conducted in our laboratory (Che et al., 2022). In this study of N. farcinica, the sul1-mediated resistance rate was 3.4% (3/88). As the drug of choice for treating Nocardia, we should pay attention to the prevalence and relevance of this resistance determinant to provide effective clinical management of the resulting disease. No sul1, sul2, or dfrA genes were detected in other sulfamethoxazole/trimethoprim-resistant isolates (MIC values of 4/76 and 8/152 μg/mL) likely developed resistance through mutations in dihydropteroate synthase (DHPS/folP) and dihydropteroate reductase (DHPR/folA) genes (Mehta et al., 2018). Although a prior study (Hershko et al., 2023b) detected a potentially significant Ala146Val mutation in folP2 among trimethoprim-sulfamethoxazole -resistant Nocardia, our analysis of folP and its homologs folP2, folA, folC (encoding dihydrofolate synthase) across 148 Nocardia isolates via multiple sequence alignment revealed no aberrant mutations within these genes in trimethoprim-sulfamethoxazole-resistant strains. Current understanding suggests that the precise underlying mechanisms have yet to be fully elucidated (Patamatamkul, 2025). This partial genotype-phenotype correlation highlights the complex resistance landscape for this first-line agent and warrants further molecular investigation.
β-lactam antibiotics are sometimes used as alternatives to sulfamethoxazole/trimethoprim for nocardiosis treatment (Wilson, 2012). Species-specific β-lactam resistance was evident both phenotypically and genotypically in Nocardia species. The overall consistency of blaAST-1 from N.otitidiscaviarum with all β-lactamase antibiotics was 100%, whereas in N. cyriacigeorgica, it was 50% for imipenem, 85% for cefepime, 100% for cefoxitin, 75% for amoxicillin/clavulanic acid, and 40% for ceftriaxone. N.otitidiscaviarum displayed extensive resistance to all β-lactam antibiotics, with universally high MIC values, consistent with previous studies (Hamdi et al., 2020). This phenotype correlated with the presence of the blaAST-1 β-lactamase gene homolog (74.02-74.47% nucleotide identity to class A β-lactamases) exclusive to this species, potentially encoding a novel broad-spectrum metallo-β-lactamase responsible for complete β-lactam resistance—a previously unreported mechanism in the Nocardia genus, while nucleotide identity in N. cyriacigeorgica was 85.52-87.54%, the blaAST-1 genes in these two Nocardia species may represent distinct β-lactamase genes. In contrast, blaFAR-1, another β-lactamase gene, was predominantly detected in N. farcinica and likely contributed to partial resistance to cephalosporins, although susceptibility to imipenem and amoxicillin/clavulanic acid remained relatively high (Laurent et al., 1999). These results emphasize the importance of distinguishing between different β-lactamase gene families in predicting resistance phenotypes. Our data suggest that sulfamethoxazole/trimethoprim, amikacin, and linezolid may be a reasonable empirical choice for N. otitidiscaviarum, though clinical validation is needed. At the same time, we must also be vigilant against the emergence of sulfamethoxazole/trimethoprim-resistant N. otitidiscaviarum, this is likely to lead to the limited use of antibiotics and result in death (Fu et al., 2023; Barry et al., 2022).
Tobramycin, an aminoglycoside, exhibited complete resistance in N. farcinica (100% non-susceptibility), this differs slightly from the 83.6% (107/128) reported in previously study (Valdezate et al., 2017) and is similar to the 99–99.5% reported by Schlaberg et al. and Hamdi et al (Schlaberg et al., 2014; Hamdi et al., 2020), whereas N. cyriacigeorgica and N. otitidiscaviarum demonstrated susceptibility rates of 90.0% and 68.8%, respectively, highlighting significant interspecies variability that precludes the empirical tobramycin use for N. farcinica infections. The aph(2’’) gene, potentially mediating tobramycin resistance (Hershko et al., 2023a), was detected exclusively in N. farcinica and N. blacklockiae in this study, showing a strong association with resistance in these species; however, it was not detected in other tobramycin-resistant Nocardia isolates, with a concordance rate of 92.2%. In Tobramycin-resistant N. wallacei and N. nova, there may be a potential gene similar to aph(2’’). Fluoroquinolone susceptibility was similarly variable. N. farcinica showed high susceptibility to ciprofloxacin and moxifloxacin, whereas N. cyriacigeorgica and N. otitidiscaviarum were uniformly resistant to ciprofloxacin and minimally susceptible to moxifloxacin (Brown-Elliott et al., 2006). While a prior study (Hershko et al., 2023b) detected the Ser83Ala mutation in ciprofloxacin-resistant Nocardia, our investigation identified four additional mutations across distinct Nocardia species, all resulting from nucleotide substitutions at position 83 of the gyrA gene. Fluoroquinolones should be used with caution when treating N. cyriacigeorgica and N. otitidiscaviarum. These patterns highlight the need for precise species identification prior to antibiotic selection.
The universal presence of rbpA, tetA(58), and tetB(58) across all isolates aligns with the widespread reduced susceptibility to rifampicin and intermediate susceptibility (>80%) to tetracyclines (e.g., doxycycline and minocycline). This resistance is likely mediated by the efflux pump systems encoded by tetA/tetB(58) (Ishikawa et al., 2004). Additionally, the lmrC gene, associated with lincosamide resistance, was exclusively detected in N. cyriacigeorgica, whereas novA, encoding aminocoumarin resistance, was unique to N. otitidiscaviarum, despite the absence of phenotypic validation. This correlation analysis revealed several consistent genotype-phenotype links, while also exposing gaps in current resistance databases, particularly in cases where resistance was observed without known genetic determinants. These findings reinforce the need to expand Nocardia-specific resistance gene repositories.
sul1 and qacEΔ1 were strongly associated with the sulfamethoxazole resistance phenotype (both V = 0.60, P < 0.001). Similarly, blaAST-1 and blaFAR-1 showed strong associations with amoxicillin resistance (V = 0.64 and 0.63, respectively; both P < 0.001). Interestingly, some resistance genes exhibited strong statistical associations (high Cramér’s V values) with resistance phenotypes that are not mechanistically related. Such discrepancies may arise from co-occurrence patterns driven by clonal spread, geographical clustering, sampling bias or other epidemiological factors rather than direct functional interactions. Cramér’s V captures the degree of association in the observed data but does not imply causality; therefore, these associations should be interpreted cautiously.
There are several strengths in this study. A major strength is its combined analysis of drug-resistant phenotypes and genotypes, elucidating the molecular basis of resistance through detailed examination of drug-resistance genes, thereby linking genotypic mechanisms to observed resistance patterns. This study further validates the utility of MALDI-TOF MS, benchmarked against ANI, as a highly accurate method for identifying clinically relevant Nocardia species, affirming its value for preliminary clinical screening. Additionally, it provides species-specific insights into antimicrobial resistance profiles, identifying effective antibiotics like linezolid and amikacin. Moreover, we present the first documented evidence of complete β-lactam resistance in N. otitidiscaviarum may mediated by a blaAST-1 β-lactamase gene homolog. This will deepen our understanding of these bacteria and enhance our ability to combat Nocardia infections. Our study still has several limitations. First, the diversity of strains in this study was limited, mainly focusing on N. farcinica, and the isolates were mainly collected from a few provinces in China. Future studies need to include a more balanced sample containing multiple clinically relevant Nocardia species to validate the broad applicability of these findings and explore intrageneric differences. Second, we performed antimicrobial susceptibility testing and obtained phenotypic resistance data; however, we did not carry out additional mechanistic experiments to directly link resistance phenotypes with specific resistance genes. Such experiments (e.g., gene knockout, overexpression, or transcriptomic validation) require dedicated molecular biology approaches and resources that were beyond the scope of the present study. In future work, we aim to perform these functional validations to elucidate the genetic basis of resistance and provide a more comprehensive understanding of genotype–phenotype correlations. Third, this study is the absence of follow-up clinical outcome data. Due to the prolonged treatment course required for Nocardia infections and the fact that all isolates were collected as baseline bacterial samples from the clinical microbiology laboratory, subsequent patient information—such as infection severity, treatment response, and prognosis—was not available. This precluded direct assessment of associations between resistance genotypes or MIC values and clinical outcomes. Future studies integrating genomic and phenotypic analyses with detailed longitudinal clinical data will be important to clarify the real-world implications of these findings. Furthermore, beyond WGS, some innovative technologies can be applied to Nocardia studies (Fan et al., 2024; Liu et al., 2023; Fan et al., 2025).
5 Conclusion
In conclusion, our study demonstrates the value of integrating phenotypic and genotypic approaches for the comprehensive characterization of Nocardia species. WGS proved essential not only for species delineation and novel taxon discovery but also for revealing key antimicrobial resistance determinants. The integration of genome-based diagnostics into clinical microbiology laboratories will enhance the accuracy of Nocardia identification and resistance prediction, ultimately guiding more effective, species-specific therapy.
Data availability statement
The raw sequencing data generated for this study have been deposited in the Genome Sequence Archive (GSA) in National Genomics Data Center under accession number CRA028987 (https://ngdc.cncb.ac.cn/gsa/browse/CRA028987), CRA011633 (https://ngdc.cncb.ac.cn/gsa/browse/CRA011633), CRA005399 (https://ngdc.cncb.ac.cn/gsa/browse/CRA005399), CRA003805 (https://ngdc.cncb.ac.cn/gsa/browse/CRA003805) (Wang et al., 2017).
Author contributions
ZS: Writing – original draft, Writing – review & editing, Conceptualization, Formal Analysis, Visualization. BD: Conceptualization, Methodology, Writing – review & editing. MY: Supervision, Writing – review & editing. JS: Supervision, Writing – review & editing. SX: Supervision, Writing – review & editing. WG: Project administration, Writing – review & editing. BL: Resources, Writing – review & editing. ZL: Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2024YFC2309300).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1672889/full#supplementary-material
References
Auch, A. F., von Jan, M., Klenk, H. P., and Göker, M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2, 117–134. doi: 10.4056/sigs.531120
Balloux, F., Brønstad Brynildsrud, O., van Dorp, L., Shaw, L. P., Chen, H., Harris, K. A., et al. (2018). From theory to practice: translating whole-genome sequencing (WGS) into the clinic. Trends Microbiol. 26, 1035–1048. doi: 10.1016/j.tim.2018.08.004
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Barry, M., AlShehri, S., Alguhani, A., Barry, M., Alhijji, A., Binkhamis, K., et al. (2022). A fatal case of disseminated nocardiosis due to Nocardia otitidiscaviarum resistant to trimethoprim-sulfamethoxazole: case report and literature review. Ann. Clin. Microbiol. Antimicrob. 21, 17. doi: 10.1186/s12941-022-00511-9
Beaman, B. L. and Beaman, L. (1994). Nocardia species: Host-parasite relationships. Clin. Microbiol. Rev. 7, 213–264. doi: 10.1128/CMR.7.2.213
Brown-Elliott, B. A., Brown, J. M., Conville, P. S., and Wallace, R. J., Jr. (2006). Clinical and laboratory features of the Nocardia spp. based Curr. Mol. taxonomy. Clin. Microbiol. Rev. 19, 259–282. doi: 10.1128/CMR.19.2.259-282.2006
Carrasco, G., de Dios Caballero, J., Garrido, N., Valdezate, S., Cantón, R., and Sáez-Nieto, J. A. (2016). Shortcomings of the commercial MALDI-TOF MS database and use of MLSA as an arbiter in the identification of nocardia species. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00542
Che, Y., Xu, S., Kang, Y., Liu, X., Yue, Y., Han, L., et al. (2022). Complete genome sequencing of transposon-mediated sulfamethoxazole resistance encoded by the Sul1 gene in multidrug-resistant Nocardia farcinica SZ 1509. J. Glob Antimicrob. Resist. 30, 60–65. doi: 10.1016/j.jgar.2022.03.004
Chen, S. (2023). Ultrafast one-pass FASTQ data preprocessing, quality control, and deduplication using fastp. Imeta. 2, e107. doi: 10.1002/imt2.107
Conville, P. S., Brown, J. M., Steigerwalt, A. G., Brown-Elliott, B. A., and Witebsky, F. G. (2008). Nocardia wallacei sp. nov. and Nocardia blacklockiae sp. nov., human pathogens and members of the “Nocardia transvalensis Complex. J. Clin. Microbiol. 46, 1178–1184. doi: 10.1128/JCM.02011-07
Conville, P. S., Brown-Elliott, B. A., Smith, T., and Zelazny, A. M. (2017). The complexities of nocardia taxonomy and identification. J. Clin. Microbiol. 56, e01419–e01417. doi: 10.1128/JCM.01419-17
Fan, R., Baysoy, A., Tian, X., Zhang, F., Renauer, P., Bai, Z., et al. (2025). Spatially Resolved Panoramic in vivo CRISPR Screen via Perturb-DBiT. Res. Sq. doi: 10.21203/rs.3.rs-6481967/v1
Fan, R., Zhang, D., Rodrıguez-Kirby, L., Lin, Y., Song, M., Wang, L., et al. (2024). Spatial dynamics of mammalian brain development and neuroinflammation by multimodal tri-omics mapping. Res. Sq. doi: 10.21203/rs.3.rs-4814866/v1
Fatahi-Bafghi, M. (2018). Nocardiosis from 1888 to 2017. Microb. Pathog. 114, 369–384. doi: 10.1016/j.micpath.2017.11.012
Fu, K., White, K., Ramaniuk, A., Kollu, V., and Urbine, D. (2023). Manifestations and management of trimethoprim/sulfamethoxazole-resistant nocardia otitidiscaviarum infection. Emerg. Infect. Dis. 29, 1266–1267. doi: 10.3201/eid2906.221854
GBD 2021 Antimicrobial Resistance Collaborators (2024). Global burden of bacterial antimicrobial resistance 1990-2021: A systematic analysis with forecasts to 2050. Lancet. 404, 1199–1226. doi: 10.1016/S0140-6736(24)01867-1
Green, S. L., Maddox, J. C., and Huttenbach, E. D. (2001). Linezolid and reversible myelosuppression. JAMA. 285, 1291. doi: 10.1001/jama.285.10.1291
Gurevich, A., Saveliev, V., Vyahhi, N., and Tesler, G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics. 29, 1072–1075. doi: 10.1093/bioinformatics/btt086
Hamdi, A. M., Fida, M., Deml, S. M., Abu Saleh, O. M., and Wengenack, N. L. (2020). Retrospective analysis of antimicrobial susceptibility profiles of nocardia species from a tertiary hospital and reference laboratory 2011 to 2017. Antimicrob. Agents Chemother. 64, e01868–e01819. doi: 10.1128/AAC.01868-19
Han, Y., Cheng, M., Li, Z., Chen, H., Xia, S., Zhao, Y., et al. (2024). Clinical characteristics and drug resistance of Nocardia in Henan, China 2017-2023. Ann. Clin. Microbiol. Antimicrob. 23, 23. doi: 10.1186/s12941-024-00677-4
Hershko, Y., Adler, A., and Barkan, D. (2023a). The nocardial aph(2’’) gene confers tobramycin and gentamicin resistance and is an effective positive selection marker in mycobacteria and nocardia. Microorganisms. 11, 1697. doi: 10.3390/microorganisms11071697
Hershko, Y., Levytskyi, K., Rannon, E., Assous, M. V., Ken-Dror, S., Amit, S., et al. (2023b). Phenotypic and genotypic analysis of antimicrobial resistance in Nocardia species. J. Antimicrob. Chemother. 78, 2306–2314. doi: 10.1093/jac/dkad236
Hershko, Y., Rannon, E., Adler, A., Burstein, D., and Barkan, D. (2024). WarA, a remote homolog of NpmA and KamB from Nocardia wallacei, confers broad spectrum aminoglycoside resistance in Nocardia and Mycobacteria. Int. J. Antimicrob. Agents 63, 107089. doi: 10.1016/j.ijantimicag.2024.107089
Ishikawa, J., Yamashita, A., Mikami, Y., Hoshino, Y., Kurita, H., Hotta, K., et al. (2004). The complete genomic sequence of Nocardia farcinica IFM 10152. Proc. Natl. Acad. Sci. U S A. 101, 14925–14930. doi: 10.1073/pnas.0406410101
Ji, X., Han, L., Zhang, W., Sun, L., Xu, S., Qiu, X., et al. (2022). Molecular, cellular and neurological consequences of infection by the neglected human pathogen Nocardia. BMC Biol. 20, 251. doi: 10.1186/s12915-022-01452-7
Kontoyiannis, D. P., Ruoff, K., and Hooper, D. C. (1998). Nocardia bacteremia. Rep. 4 cases Rev. literature. Med. 77, 255–267. doi: 10.1097/00005792-199807000-00004
Laurent, F., Poirel, L., Naas, T., Chaibi, E., Labia, R., Boiron, P., et al. (1999). Biochemical-genetic analysis and distribution of FAR-1, a class A beta-lactamase from Nocardia farcinica. Antimicrob. Agents Chemother. 43, 1644–1650. doi: 10.1128/AAC.43.7.1644
Liu, Y., DiStasio, M., Su, G., Asashima, H., Enninful, A., Qin, X., et al. (2023). High-plex protein and whole transcriptome co-mapping at cellular resolution with spatial CITE-seq. Nat. Biotechnol. 41, 1405–1409. doi: 10.1038/s41587-023-01676-0
McKinney, W. P., Smith, M. R., Roberts, S. A., and Morris, A. J. (2023). Species distribution and susceptibility of Nocardia isolates in New Zealand 2002-2021. Pathology. 55, 680–687. doi: 10.1016/j.pathol.2023.03.008
Mehta, H., Weng, J., Prater, A., Elworth, R. A. L., Han, X., and Shamoo, Y. (2018). Pathogenic Nocardia cyriacigeorgica and Nocardia nova Evolve To Resist Trimethoprim-Sulfamethoxazole by both Expected and Unexpected Pathways. Antimicrob. Agents Chemother. 62, e00364–e00318. doi: 10.1128/AAC.00364-18
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T.G., et al. (2015). Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Patamatamkul, S. (2025). Troubleshooting the pitfalls of TMP-SMX susceptibility interpretation in nocardia species. Clin. Infect. Dis. doi: 10.1093/cid/ciaf373
Presant, C. A., Wiernik, P. H., and Serpick, A. A. (1973). Factors affecting survival in nocardiosis. Am. Rev. Respir. Dis. 108, 1444–1448. doi: 10.1164/arrd.1973.108.6.1444
Pritchard, L., Glover, R. H., Humphris, S., Elphinstone, J. G., and Toth, L. K. (2016). Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal. Methods 8, 12–24. doi: 10.1039/c5ay02550h
Richter, M. and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Sack, K., Freiesleben, H., Züllich, B., Beck, H., and Schulz, E. (1976). Nebenwirkungen von Aminoglykosiden: Nephrotoxizität [Side effects of aminoglycosides: nephrotoxicity (author’s transl). Infection. 4, 231–238. doi: 10.1007/BF01638932
Schlaberg, R., Fisher, M. A., and Hanson, K. E. (2014). Susceptibility profiles of Nocardia isolates based on current taxonomy. Antimicrob. Agents Chemother. 58, 795–800. doi: 10.1128/AAC.01531-13
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shen, J., Du, B., Liu, Z., Song, Z., Yuan, M., Qiu, X., et al. (2025). Multicenter systematic review of clinical characteristics, diagnostic optimization, and personalized treatment for brain Nocardia infections. Microb. Pathog. 198, 107147. doi: 10.1016/j.micpath.2024.107147
Valdezate, S., Garrido, N., Carrasco, G., Medina-Pascual, M., Villalón, P., Navarro, A., et al. (2017). Epidemiology and susceptibility to antimicrobial agents of the main Nocardia species in Spain. J. Antimicrob. Chemother. 72, 754–761. doi: 10.1093/jac/dkw489
Valdezate, S., Garrido, N., Carrasco, G., Villalón, P., Medina-Pascual, M. J., and Saéz-Nieto, J. A. (2015). Resistance gene pool to co-trimoxazole in non-susceptible Nocardia strains. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.00376
Wang, Y., Song, F., Zhu, J., Zhang, S., Yang, Y., Chen, T., et al. (2017). GSA: genome sequence archive. Genomics Proteomics Bioinf. 15, 14–18. doi: 10.1016/j.gpb.2017.01.001
Wang, H., Zhu, Y., Cui, Q., Wu, W., Li, G., Chen, D., et al. (2022). Epidemiology and antimicrobial resistance profiles of the nocardia species in China 2009 to 2021. Microbiol. Spectr. 10, e0156021. doi: 10.1128/spectrum.01560-21
Wei, M., Wang, P., Qu, J., Li, R., Liu, Y., Gu, L., et al. (2017). Identification and antimicrobial susceptibility of clinical Nocardia species in a tertiary hospital in China. J. Glob Antimicrob. Resist. 11, 183–187. doi: 10.1016/j.jgar.2017.08.011
Wilson, J. W. (2012). Nocardiosis: updates and clinical overview. Mayo Clin. Proc. 87, 403–407. doi: 10.1016/j.mayocp.2011.11.016
Woods, G. L., Brown-Elliott, B. A., Conville, P. S., et al(2011). Susceptibility Testing of Mycobacteria, Nocardiae, and Other Aerobic Actinomycetes. 2nd ed. Wayne (PA), Clinical and Laboratory Standards Institute.
Yi, M., Wang, L., Xu, W., Sheng, L., Jiang, L., Yang, F., et al. (2019). Species distribution and antibiotic susceptibility of nocardia isolates from yantai, China. Infect. Drug Resist. 12, 3653–3661. doi: 10.2147/IDR.S232098
Keywords: Nocardia, whole genome sequencing, antimicrobial resistance, average nucleotide identity, minimum inhibitory concentration
Citation: Song Z, Du B, Yuan M, Shen J, Xu S, Guan W, Lu B and Li Z (2025) Genomic and phenotypic characterization of antimicrobial resistance in clinical Nocardia species isolates. Front. Cell. Infect. Microbiol. 15:1672889. doi: 10.3389/fcimb.2025.1672889
Received: 25 July 2025; Accepted: 29 August 2025;
Published: 18 September 2025.
Edited by:
Xin Du, University of California, San Diego, United StatesReviewed by:
Fu Gao, Yale University, United StatesSixuan Pan, University of California, San Francisco, United States
Ibidunni Bode-Sojobi, Beth Israel Deaconess Medical Center Cancer Center, United States
Copyright © 2025 Song, Du, Yuan, Shen, Xu, Guan, Lu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenjun Li, bGl6aGVuanVuQGljZGMuY24=; Binghuai Lu, enMyNTA0MUAxMjYuY29t; Wanchun Guan, Z3djQHdtdS5lZHUuY24=
†These authors have contributed equally to this work
 Ziyu Song
Ziyu Song Bingqian Du
Bingqian Du Min Yuan
Min Yuan Jirao Shen
Jirao Shen Shuai Xu
Shuai Xu Wanchun Guan
Wanchun Guan Binghuai Lu
Binghuai Lu Zhenjun Li
Zhenjun Li