- The First Affiliated Hospital of Zhejiang Chinese Medical University (Zhejiang Provincial Hospital of Chinese Medicine), Hangzhou, Zhejiang, China
Carbapenem-resistant Enterobacteriaceae (CRE) present an escalating threat to global health due to their high transmissibility, limited treatment options, and high mortality rates. The gastrointestinal tract serves as both a major reservoir and a transmission hub for CRE, especially under conditions of antibiotic-induced dysbiosis. This review highlights the growing interest in the gut microbiome as a potential target for preventing and managing CRE infections. Building upon the understanding of CRE pathogenesis, we examine how commensal microbiota contribute to colonization resistance through mechanisms such as nutrient competition, spatial niche exclusion, immune modulation, and the production of antimicrobial metabolites. We further discuss microbiome-based therapeutic strategies, including probiotic administration, fecal microbiota transplantation (FMT), and supplementation with short-chain fatty acids (SCFAs), that have shown encouraging results in reducing intestinal CRE colonization. In addition, we explore emerging microbiome engineering approaches, particularly CRISPR-Cas9-mediated systems, which enable the selective elimination of resistant strains while maintaining microbial homeostasis. Current microbiome-based approaches have shown promise in the treatment and prevention of CRE infections, but further research is still needed to clarify their mechanisms, evaluate long-term safety, and determine their effectiveness in different clinical settings. With continued studies and thoughtful integration into existing infection control and antibiotic stewardship practices, these strategies may gradually contribute to a more practical and sustainable way to manage CRE.
1 Introduction
Carbapenem-resistant Enterobacteriaceae (CRE) have become a serious global public health concern. The World Health Organization (WHO) lists carbapenem-resistant Klebsiella pneumoniae (K. pneumoniae) and Escherichia coli (E. coli) as critical priority pathogens in its 2024 Bacterial Priority Pathogens List (BPPL), emphasizing their significance in antimicrobial resistance (Sati et al., 2025). These bacteria exhibit a high capacity for acquiring and disseminating resistance genes, making containment and treatment increasingly difficult. CRE cause severe infections such as bloodstream infections, pneumonia, and urinary tract infections. These infections often result in high death rates, longer hospital stays, and higher healthcare costs (Chen et al., 2024; Bhat et al., 2025). Although agents such as polymyxins, tigecycline, and ceftazidime-avibactam are used in the treatment of CRE infections, their roles vary. Polymyxins and tigecycline are often reserved for multidrug-resistant K. pneumoniae, whereas ceftazidime-avibactam is mainly applied to KPC-producing strains. Their clinical utility, however, is increasingly compromised by rising resistance rates, rapid dissemination of resistance genes, and drug-related toxicities (Gao et al., 2025; Hou et al., 2025).
In addition to overt infections, CRE can persist silently in the intestinal tract, particularly in hospitalized and immunocompromised patients (Wu et al., 2023; Xiao et al., 2024; Han et al., 2025). Increasing evidence shows that antibiotic exposure, underlying diseases, and immune suppression can disturb the gut microbiota, lowering colonization resistance and facilitating CRE expansion (Kang et al., 2022; Wang Z. et al., 2024). A disrupted microbial environment not only weakens host defense but also promotes overgrowth of resistant strains. As host immunity declines, CRE may translocate across the intestinal barrier, leading to severe bloodstream infections and other life-threatening complications. The gastrointestinal tract is both a key site of susceptibility and a potential target for new strategies in prevention and treatment. In this context, targeting the gut microbiome offers a promising addition to traditional antibiotics. Methods such as probiotics, fecal microbiota transplantation (FMT), and microbiota-derived metabolites have shown potential in reducing CRE colonization and preventing transmission in both animal models and clinical trials (Gutiérrez-Fernández et al., 2024; Merrick et al., 2025). Key questions remain regarding their mechanisms, long-term effects, and practical use in clinical settings.
This review provides an overview of the challenges posed by CRE in terms of resistance, epidemiology, and colonization, while also evaluating microbiome-based strategies such as probiotics, FMT, and microbial metabolites. The aim is to identify interventions that can complement conventional therapies and inform future clinical management.
2 Resistance mechanisms and clinical burden of CRE
CRE are members of the Enterobacteriaceae order that exhibit resistance to at least one carbapenem-class antibiotic, such as imipenem, meropenem, or ertapenem (Smith et al., 2025). K. pneumoniae and E. coli are the most clinically important CRE species. These pathogens are commonly responsible for infections of the bloodstream, respiratory tract, and urinary tract, especially in healthcare-associated settings (Li Y. et al., 2024; Zhong et al., 2025). Carbapenem resistance in Enterobacteriaceae is mainly caused by the production of carbapenemases. Among them, KPC, NDM, and OXA-48-like enzymes are the most common. These enzymes are increasingly detected in clinical isolates (Ma et al., 2023; Alvisi et al., 2025). In many cases, the resistance genes are located on plasmids, which promote horizontal gene transfer and often carry virulence factors as well (Heng et al., 2025; Li et al., 2025). In addition to enzyme production, membrane-associated mechanisms also play a critical role. The loss of outer membrane porins, such as OmpK36, limits antibiotic entry. At the same time, efflux systems like the tripartite antimicrobial metabolism system actively pump drugs out of the cell, reducing their effectiveness (Jung et al., 2021; Meekes et al., 2025). These mechanisms frequently act together, leading to broad resistance that extends beyond β-lactams, Aminoglycosides, fluoroquinolones, and even last-line agents like colistin may also be rendered ineffective (Wu et al., 2024; Song et al., 2025).
The global prevalence of CRE continues to rise. Surveillance reports show ongoing transmission of dominant clones, such as ST258 in the Mediterranean and ST11 in Asia, especially within hospital environments (Wang Q. et al., 2024; García-González et al., 2025; Zhang et al., 2025). In intensive care units, colonization rates exceed 20%. Long-term care facilities often struggle with persistent environmental contamination (Wu et al., 2023; Elton et al., 2024). Underdiagnosis is common due to limited surveillance infrastructure and the shortcomings of current screening strategies, allowing ongoing silent transmission (Kedišaletše et al., 2023; Pople et al., 2023). The organism can survive on surfaces and equipment, making infection control difficult. In overcrowded healthcare settings, this persistence helps resistant strains spread more easily (Salomão et al., 2023; Elton et al., 2024). Clinical outcomes of CRE infections remain poor. Mortality often exceeds 40% in bloodstream infections and ventilator-associated pneumonia, especially when appropriate therapy is delayed (Chen et al., 2024; Ruvinsky et al., 2024; Kim et al., 2025). Treatment options are limited and frequently complicated by toxicity. Polymyxins and tigecycline serve as last-resort agents but carry risks of nephrotoxicity and rising resistance, including plasmid-mediated mechanisms (Wu et al., 2024; Xie et al., 2024; Jiang et al., 2025). Newer drugs such as ceftazidime-avibactam and cefiderocol offer broader coverage. Still, treatment failures are common due to rapid emergence of resistance caused by porin mutations and novel carbapenemase variants (Li JW. et al., 2024; Tang et al., 2024; Faxén et al., 2025). The rise of hypervirulent carbapenem-resistant strains further worsens outcomes and limits therapeutic success (Lei et al., 2024; Wang et al., 2025).
Recent efforts in drug discovery have turned to repurposing established antibiotics and testing combination regimens. Fosfomycin has demonstrated synergistic activity with agents such as meropenem, polymyxin B, and colistin, and both experimental and clinical evidence suggest improved outcomes compared with monotherapy (Ribeiro et al., 2023; Katip et al., 2024). Novel therapeutic strategies are also being explored for hypervirulent carbapenem-resistant K. pneumoniae. Some isolates carrying both multidrug resistance and hypervirulent traits show unexpectedly attenuated pathogenicity, reflecting the complex relationship between resistance and virulence that may guide future drug development (Ni et al., 2022; Kochan et al., 2023).
3 Role of the gut microbiota in CRE colonization
3.1 Antibiotic-induced dysbiosis and CRE colonization
Antibiotic exposure is a major risk factor for CRE colonization in the gastrointestinal tract. Short-term, targeted oral antibiotics such as rifampicin can help rapidly decolonize CRE in acute clinical settings, while promoting the enrichment of antagonistic commensals and supporting immune recovery (Ni et al., 2024). However, prolonged or inappropriate use of antibiotics can cause lasting alterations to the gut microbiota, reducing diversity and depleting beneficial bacteria, judicious use of certain agents may help restore a healthier microbial community. Broad-spectrum antibiotics, especially those targeting anaerobic bacteria, disrupt the gut microbiota, reducing commensals and microbial diversity, which weakens colonization resistance (Macareño-Castro et al., 2022). This allows CRE to occupy vacant ecological niches and proliferate. Clinical studies have shown that patients treated with carbapenem, cephalosporin, or fluoroquinolone antibiotics experience significantly higher CRE colonization rates (Sindi et al., 2022; Yuan et al., 2022). Antibiotics also create an environment favorable for CRE growth by depleting microbial metabolites that inhibit its proliferation, while enriching nutrients that CRE can use (Yip et al., 2023). Moreover, dysbiosis promotes the horizontal transfer of resistance genes, turning the gut into a reservoir of multidrug resistance (Rooney et al., 2019; He et al., 2025). The biofilm environment in the gut promotes resistance gene transfer, accelerating the rapid spread of resistance among members of the Enterobacteriaceae family, including gene transfer from E. coli to Klebsiella and from commensals to pathogens (Kent et al., 2020; Michaelis and Grohmann, 2023).
Gastrointestinal colonization plays a central role in the persistence and dissemination of CRE. Long-term shedding of CRE is common in asymptomatic carriers, facilitating its ongoing transmission within hospital wards and intensive care units (Sindi et al., 2022; Baek et al., 2023). The carrier state can persist for up to one year, with approximately 33% of CRE carriers remaining positive after one year (Ciobotaro et al., 2016). Surfaces, devices, and healthcare worker hands often become secondary reservoirs (Deng et al., 2022; Han et al., 2025). Colonization risk is heightened in patients with antibiotic-induced dysbiosis, immunosuppression, or frequent invasive procedures (Khachab et al., 2024; Lee et al., 2024). Repeated antimicrobial exposure further complicates eradication. Standard decolonization approaches are often ineffective, and recolonization occurs frequently. These factors facilitate silent persistence and recurrent infections, even after apparent clearance. Among patients with CRE colonization, approximately 21% developed secondary infections within 180 days of initial colonization, with most occurring within 30 days (Tubb et al., 2025). Consequently, the gut remains a stable reservoir for both endogenous infection and nosocomial transmission (Liu et al., 2022; Sim et al., 2022).
3.2 Gut microbiota defense mechanisms against CRE colonization
Healthy gut microbiota confer resistance to colonization by CRE by occupying both nutritional and spatial niches, limiting the resources and ecological space required for pathogen expansion. Commensal bacteria, including species such as Bacteroides, Clostridia and Lactobacillus, engage in exploitative competition by rapidly consuming available monosaccharides, amino acids, and micronutrients, restricting the supply of metabolic substrates necessary for CRE proliferation (Djukovic et al., 2022; Isaac et al., 2022). Meanwhile, mucosa-associated microbial communities form structured biofilms within the inner mucus layer and intestinal crypts, where densely packed bacterial cells and extracellular matrix components create a physical barrier that effectively blocks pathogen access to epithelial adhesion sites (Zhao and Maynard, 2022). These spatially organized structures are stabilized through dynamic interactions between commensal microbes and host-derived mucus, contributing to immune tolerance and sustained exclusion of pathogenic bacteria.
The gut microbiota also plays a crucial role in modulating host immune responses to combat CRE infection. Pattern recognition receptors (PRRs), including NOD1, NOD2, and Toll-like receptors (TLRs), recognize microbial-associated molecular patterns (MAMPs) derived from commensal bacteria, activating downstream signaling pathways that promote the production of antimicrobial peptides (Zhao et al., 2018; Martin-Gallausiaux et al., 2022). In addition, the microbiota regulates cytokine responses by promoting the expression of cytokines such as IL-1β and IL-22, thereby enhancing epithelial barrier function and modulating inflammatory responses (Wu et al., 2022; Zhao et al., 2025).
The metabolic activity of the gut microbiota profoundly shapes the chemical environment of the intestinal lumen, creating conditions that are unfavorable for CRE survival and colonization. Short-chain fatty acids (SCFAs), including acetate, propionate, and butyrate, lower luminal pH and enhance epithelial oxygen consumption, thereby eliminating oxygen-rich niches that favor CRE colonization (Sorbara et al., 2019; Yip et al., 2023). Moreover, butyrate and propionate function as histone deacetylase (HDAC) inhibitors, inducing epigenetic modifications that regulate host gene expression (Korsten et al., 2022). These changes upregulate genes involved in antimicrobial defense, mucin production, and barrier integrity (Pace et al., 2021; Korsten et al., 2023), collectively reducing CRE adhesion to and invasion of intestinal epithelial cells. Studies have shown that certain commensal strains produce narrow-spectrum bacteriocins, particularly microcins, which penetrate the outer membrane of Gram-negative Enterobacteriaceae via receptor-mediated uptake, exerting targeted antimicrobial activity and interfering with essential cellular processes such as peptidoglycan synthesis and nucleic acid metabolism (Telhig et al., 2020; Telhig et al., 2022) (Figure 1).
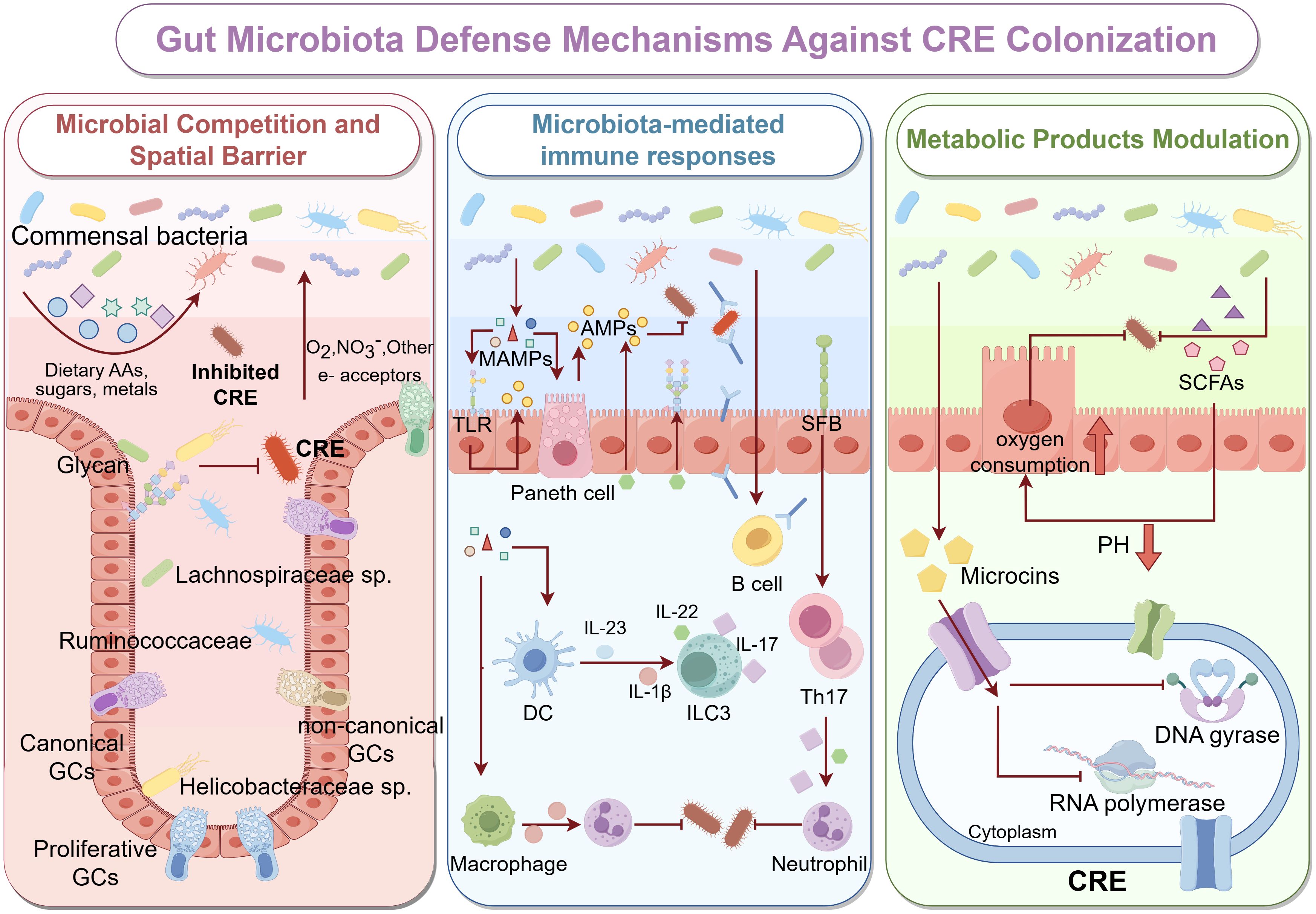
Figure 1. Gut Microbiota Defense Mechanisms Against CRE Colonization The healthy gut microbiota protects against colonization by CRE through three major mechanisms: microbial competition and spatial exclusion (left panel), microbiota-mediated immune responses (middle panel), and metabolic modulation (right panel). left panel: Healthy gut commensals, including Lachnospiraceae, Ruminococcaceae, and non-canonical GC-associated taxa, establish dense, biofilm-like communities along the intestinal epithelium. These bacteria occupy specific ecological niches and utilize dietary amino acids, complex glycans, and essential micronutrients, thereby reducing the resources available for CRE to proliferate. Spatial organization of microbial aggregates, along with mucin layers secreted by goblet cells, creates physical barriers that limit CRE adherence to epithelial surfaces and hinder their colonization. middle panel: MAMPs, such as lipopolysaccharides, peptidoglycans, and flagellin, are sensed by host PRRs, including TLRs on epithelial and immune cells. This recognition stimulates Paneth cells to secrete AMPs, including defensins and regenerating islet-derived proteins, which directly inhibit CRE growth. Commensal bacteria further modulate innate and adaptive immunity by activating DCs, macrophages, B cells, and type ILC3s, promoting the release of cytokines such as IL-22 and IL-17. These cytokines enhance mucosal barrier integrity, stimulate epithelial proliferation, and recruit neutrophils to sites of potential CRE invasion. right panel: Gut microbiota produce metabolites, such as SCFAs, which lower luminal pH, enhance epithelial oxygen consumption, and create metabolically unfavorable conditions for CRE survival. Additionally, microbiota-derived bacteriocins, including microcins, can penetrate CRE membranes and inhibit key cellular processes, such as DNA gyrase, RNA polymerase activity, and cell wall biosynthesis, leading to targeted suppression.
4 Therapeutic strategies targeting the microbiome against CRE
4.1 Probiotic therapy
Probiotic therapy represents a promising microbiome-based strategy against CRE. The antimicrobial effects of probiotics are primarily mediated through the production of acidic metabolites and reprogramming of the gut microbial community. The anti-CRE effect of probiotics was related to the pH-dependent mechanism, and the antibacterial effect was eliminated at pH 7.0 in the upper layer of the cell membrane, but the antibacterial effect remained unchanged after heat treatment (Tang et al., 2023). Lactic acid bacteria produce lactate, organic acids, CO2, exopolysaccharides, bacteriocins, and enzymes, which lower the intestinal pH and exert direct antimicrobial effects (Tang et al., 2023; Hatem et al., 2024). Multiple probiotics have been shown to modulate the gut microbiota by exhibiting strong bile salt hydrolase deconjugation and 7α-dehydroxylation activity, leading to increased levels of deoxycholic acid and lithocholic acid, while simultaneously reducing the production of isobutyric acid, isovaleric acid, hydrogen sulfide, and ammonia (Foley et al., 2021; Liu et al., 2024).
Probiotics also enhance host defenses by strengthening the intestinal barrier and modulating mucosal immunity. They promote the gene and protein expression of tight junction proteins Occludin, Claudin, and ZO-1 and stimulate mucin secretion, thereby reinforcing epithelial barrier integrity and reducing pathogen adhesion and invasion (di Vito et al., 2022; Bu et al., 2023). At the same time, probiotics activate the gut mucosal immune system, markedly increasing the levels of secretory IgA, IgA, and IgG in the intestine, while enhancing the functions of CD11c positive dendritic cells and CD4 positive T cells (Lin et al., 2021; Bu et al., 2023). These effects help maintain gut microbial homeostasis and reduce the risk of pathogen translocation and systemic inflammation.
In one screening study of 57 strains, five candidates (LUC0180, LUC0219, LYC0289, LYC0413, and LYC1031) produced inhibition zones larger than 15 mm and sustained suppression of carbapenem-resistant E. coli (CRE316) and K. pneumoniae (CRE632) (Chen et al., 2019). In addition to these strains, other species including Bifidobacterium longum (B. longum), Lactiplantibacillus plantarum (L. plantarum), and Lacticaseibacillus rhamnosus (L. rhamnosus) have also demonstrated notable antimicrobial activity against CRE, with inhibition zones exceeding 20 mm in representative isolates (Chornchoem et al., 2025). Clinically, a retrospective analysis of ICU patients showed that among 474 individuals receiving probiotics, the incidence of new CRE colonization was significantly reduced, with only 13 patients developing new CRE colonization, compared to a markedly higher rate in the control group (Lee et al., 2023).
4.2 FMT
FMT restores colonization resistance against CRE by reestablishing a diverse and balanced gut microbiome following disruption by antibiotics. Antibiotic exposure depletes commensals such as Bifidobacteriaceae and Bacteroidales, exhausts inhibitory metabolites, and enriches the intestinal environment with fermentable nutrients that CRE can exploit for growth (Yip et al., 2023). FMT introduces a complex microbial consortium from healthy donors to restore ecological competition, metabolic inhibition, and spatial exclusion (Millan et al., 2016). Transkingdom interactions between the virome and bacteriome induced by FMT may play a critical role in CRE clearance. Studies have observed a striking increase in E. coli phages in carriers of CRE E. coli following FMT, as well as concurrent CRE elimination and similar evolutionary patterns of Klebsiella phages in mouse models (Liu et al., 2022).
In a study of 10 carriers with prolonged CP-CRE carriage, FMT achieved decolonization rates of 40.0%, 50.0%, and 90.0% within 1, 3, and 5 months after the initial treatment, respectively, especially in patients whose gut microbiota rapidly shift toward the donor composition and have lower baseline Klebsiella abundance (Lee et al., 2021). Consistently, another cohort study of 35 patients reported that 68.6% were decolonized within one year after FMT (Shin et al., 2022). Microbiota analyses revealed significant increases in α- and β-diversity metrics in patients with successful CRE decolonization, whereas no such changes were observed in non-responders (Bar-Yoseph et al., 2021). FMT also shows a favorable safety profile. Among the 209 patients reviewed, including immunocompromised individuals, no serious adverse events were attributed to FMT (Macareño-Castro et al., 2022).
4.3 Metabolite supplementation therapy
Microbial metabolites, particularly SCFAs, play a key role in the prevention and treatment of CRE. Studies have found that the levels of isobutyric acid and valeric acid are significantly reduced in CRE carriers (Baek et al., 2023). Propionate shows dose-dependent growth inhibition against various multidrug-resistant bacteria, including E. coli, with minimum inhibitory concentrations ranging from 10 to 25 mM (Ormsby et al., 2020). Butyrate enhances macrophage antimicrobial activity through HDAC3 inhibition, increasing antimicrobial peptide expression and resistance to enteropathogens (Schulthess et al., 2019). SCFAs also suppress plasmid-mediated resistance gene transfer, with conjugation fully suppressed at concentrations of 0.1–1 M and significant reductions observed even at 0.01 M (Ott and Mellata, 2024). Combining SCFAs with antibiotics yields synergistic effects. SCFAs restore the susceptibility of resistant Enterobacteriaceae to β-lactam/β-lactamase inhibitor combinations and downregulate virulence genes including fliC, ipaH, fimH, and bssS (Kadry et al., 2023).
4.4 Emerging microbiome engineering technologies
Synthetic biology offers new tools for precisely engineering the gut microbiota to prevent CRE. CRISPR/Cas9 gene editing technology has shown great potential in the prevention and treatment of CRE by targeting carbapenemase genes to reverse resistance. The CRISPR-Cas9-mediated plasmid clearing system has been developed to effectively eliminate carbapenemase genes such as blaKPC, blaNDM, and blaOXA-48, with an efficiency exceeding 94% (Hao et al., 2020). This system has demonstrated excellent results across various clinical isolates of Enterobacteriaceae, including K. pneumoniae, E. coli, and Enterobacter cloacae (E. cloacae) (Hao et al., 2020; Tao et al., 2022). This strategy can be used for in situ microbiome modification to eradicate targeted resistant and/or pathogenic bacteria without affecting other non-targeted bacterial species. In addition, the delivery of CRISPR-Cas9 by engineered probiotics has achieved over 99.9% elimination of targeted antibiotic-resistant E. coli in the mouse gut microbiota with a single dose (Neil et al., 2021). CRISPR-armed phages, which integrate the CRISPR-Cas system, enable precise targeting and killing of E. coli, targeting bacteria in biofilms and reducing the emergence of antibiotic-resistant strains (Gencay et al., 2024) (Table 1).
5 Conclusion
The global rise of CRE presents a critical challenge to infection control, driven by complex resistance mechanisms, asymptomatic gastrointestinal colonization, and limited treatment options. The gut serves as both a reservoir and a transmission hub for CRE, particularly under conditions of antibiotic-induced dysbiosis that impair colonization resistance and facilitate horizontal gene transfer. In this context, the gut microbiota has emerged as a promising therapeutic target.
Microbiome-based interventions such as probiotics, FMT, and SCFA supplementation have shown potential to restore microbial balance and suppress CRE colonization. Most current evidence, however, is derived from in vitro experiments and animal models, with only limited support from small-scale or retrospective clinical studies. These findings suggest potential preventive and therapeutic value, but their clinical efficacy and safety remain to be rigorously validated in large, well-designed randomized controlled trials. While traditional approaches may provide broad-spectrum benefits, precision tools such as CRISPR/Cas9 gene editing and engineered probiotics represent a highly innovative frontier. These technologies hold the promise of selectively removing resistance genes while minimizing collateral disruption to commensal microbes. Yet, their translation into clinical practice is still at the proof-of-concept stage, with substantial barriers including the development of reliable delivery systems, managing potential off-target effects, and navigating complex regulatory pathways for live biotherapeutics.
Moreover, inter-individual variability in baseline microbiota may influence treatment outcomes, underscoring the importance of personalized approaches. For these advanced strategies to succeed, they must ultimately be integrated into established infection control frameworks, including patient screening, contact precautions, environmental hygiene, and antimicrobial stewardship. A coordinated and evidence-based strategy that bridges microbiome-targeted therapies with existing infection control practices will be essential to move from reactive treatment toward proactive and sustainable CRE containment.
Author contributions
LZ: Writing – original draft. TX: Writing – original draft. WC: Writing – original draft. YC: Writing – original draft. YW: Writing – original draft. XD: Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Acknowledgments
Figures in this manuscript were created using the Figdraw platform (https://www.figdraw.com/), and we gratefully acknowledge its comprehensive support and functionality.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRE, Carbapenem-resistant Enterobacteriaceae; WHO, World Health Organization; BPPL, Bacterial Priority Pathogens List; PRRs, Pattern Recognition Receptors; TLRs, Toll-like Receptors; NOD, Nucleotide-binding Oligomerization Domain; MAMPs, Microbial-associated Molecular Patterns; SCFAs, Short-chain Fatty Acids; HDAC, Histone Deacetylase; FMT, Fecal Microbiota Transplantation; B. longum, Bifidobacterium longum; E. coli, Escherichia coli; E. cloacae, Enterobacter cloacae; K. pneumoniae, Klebsiella pneumonia; L. plantarum, Lactiplantibacillus plantarum; L. rhamnosus, Lacticaseibacillus rhamnosus.
References
Alvisi, G., Curtoni, A., Fonnesu, R., Piazza, A., Signoretto, C., Piccinini, G., et al. (2025). Epidemiology and genetic traits of carbapenemase-producing enterobacterales: A global threat to human health. Antibiotics (Basel) 14 (2), 141. doi: 10.3390/antibiotics14020141
Baek, M. S., Kim, S., Kim, W. Y., Kweon, M. N., and Huh, J. W. (2023). Gut microbiota alterations in critically Ill patients with carbapenem-resistant Enterobacteriaceae colonization: A clinical analysis. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1140402
Bar-Yoseph, H., Carasso, S., Shklar, S., Korytny, A., Even Dar, R., Daoud, H., et al. (2021). Oral capsulized fecal microbiota transplantation for eradication of carbapenemase-producing enterobacteriaceae colonization with a metagenomic perspective. Clin. Infect. Dis. 73, e166–ee75. doi: 10.1093/cid/ciaa737
Bhat, D., Rajan, A. K., Aditya, V., Eshwara, V. K., Varma, M., Umakanth, S., et al. (2025). Does carbapenem resistance influence clinical outcomes in ICU patients with Klebsiella pneumoniae bacteraemia? A comparative analysis of Carbapenem-resistant and susceptible isolates. Acta Trop. 269, 107744. doi: 10.1016/j.actatropica.2025.107744
Bu, Y., Liu, Y., Zhang, T., Liu, Y., Zhang, Z., and Yi, H. (2023). Bacteriocin-producing lactiplantibacillus plantarum YRL45 enhances intestinal immunity and regulates gut microbiota in mice. Nutrients 15 (15), 3437. doi: 10.3390/nu15153437
Chen, Y., Huang, J., Dong, L., Xu, B., Li, L., Zhao, Z., et al. (2024). Clinical and genomic characterization of carbapenem-resistant Enterobacterales bloodstream infections in patients with hematologic Malignancies. Front. Cell Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1471477
Chen, C. C., Lai, C. C., Huang, H. L., Huang, W. Y., Toh, H. S., Weng, T. C., et al. (2019). Antimicrobial activity of lactobacillus species against carbapenem-resistant enterobacteriaceae. Front. Microbiol. 10. doi: 10.3389/fmicb.2019.00789
Chornchoem, P., Tandhavanant, S., Saiprom, N., Preechanukul, A., Thongchompoo, N., Sensorn, I., et al. (2025). Metagenomic evaluation, antimicrobial activities, and immune stimulation of probiotics from dietary supplements and dairy products. Sci. Rep. 15, 11537. doi: 10.1038/s41598-025-95664-w
Ciobotaro, P., Flaks-Manov, N., Oved, M., Schattner, A., Hoshen, M., Ben-Yosef, E., et al. (2016). Predictors of persistent carbapenem-resistant enterobacteriaceae carriage upon readmission and score development. Infect. Control Hosp Epidemiol. 37, 188–196. doi: 10.1017/ice.2015.278
Deng, J., Liao, Q., Zhang, W., Wu, S., Liu, Y., Xiao, Y., et al. (2022). Molecular epidemiology characteristics and detecting transmission of carbapenemase-producing enterobacterales in southwestern China. J. Infect. Public Health 15, 1047–1052. doi: 10.1016/j.jiph.2022.08.010
di Vito, R., Conte, C., and Traina, G. (2022). A multi-strain probiotic formulation improves intestinal barrier function by the modulation of tight and adherent junction proteins. Cells 11 (16), 2617. doi: 10.3390/cells11162617
Djukovic, A., Garzón, M. J., Canlet, C., Cabral, V., Lalaoui, R., García-Garcerá, M., et al. (2022). Lactobacillus supports Clostridiales to restrict gut colonization by multidrug-resistant Enterobacteriaceae. Nat. Commun. 13, 5617. doi: 10.1038/s41467-022-33313-w
Elton, L., Williams, A., Ali, S., Heaphy, J., Pang, V., Commins, L., et al. (2024). Tracing the transmission of carbapenem-resistant Enterobacterales at the patient: ward environmental nexus. Ann. Clin. Microbiol. Antimicrob. 23, 108. doi: 10.1186/s12941-024-00762-8
Faxén, L., Müller, V., Chatzopoulou, M., Karlsson Lindsjö, O., Lagerbäck, P., Westmo, K., et al. (2025). Antibiotic susceptibility to new antibiotics and genetic characterisation of carbapenemase-producing Enterobacterales: Low activity of cefiderocol against NDM-producing isolates. Int. J. Antimicrob. Agents. 66, 107553. doi: 10.1016/j.ijantimicag.2025.107553
Foley, M. H., O’Flaherty, S., Allen, G., Rivera, A. J., Stewart, A. K., Barrangou, R., et al. (2021). Lactobacillus bile salt hydrolase substrate specificity governs bacterial fitness and host colonization. Proc. Natl. Acad. Sci. U.S.A. 118 (6), e2017709118. doi: 10.1073/pnas.2017709118
Gao, H., Wang, B., Li, M., Zhou, P., Wu, C., Wan, C., et al. (2025). Emergence and dissemination of multidrug-resistant Klebsiella pneumoniae harboring the novel tmexCD-toprJ RND efflux pump operon. Front. Cell Infect. Microbiol. 15. doi: 10.3389/fcimb.2025.1579880
García-González, N., Beamud, B., Sevilla-Fortuny, J., Sánchez-Hellín, V., Vidal, I., Rodríguez, J. C., et al. (2025). Genomic surveillance reveals different transmission patterns between third-generation cephalosporin and carbapenem resistance in Klebsiella pneumoniae in the Comunidad Valenciana (Spain), 2018-2020. Antimicrob. Resist. Infect. Control. 14, 44. doi: 10.1186/s13756-025-01553-2
Gencay, Y. E., Jasinskytė, D., Robert, C., Semsey, S., Martínez, V., Petersen, A., et al. (2024). Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 42, 265–274. doi: 10.1038/s41587-023-01759-y
Gutiérrez-Fernández, J., Cerezo-Collado, L., Garcés, V., Alarcón-Guijo, P., Delgado-López, J. M., and Dominguez-Vera, J. M. (2024). Probiotic-loaded bacterial cellulose as an alternative to combat carbapenem-resistant bacterial infections. Antibiotics (Basel). 13 (11), 1003. doi: 10.3390/antibiotics13111003
Han, X., Song, M., Yu, Q., Zhou, J., Hu, H., Jiang, Y., et al. (2025). Unveiling the clonal dynamics and transmission mechanism of carbapenem-resistant Klebsiella pneumoniae in the ICU environment. Int. J. Antimicrob. Agents. 66, 107532. doi: 10.1016/j.ijantimicag.2025.107532
Hao, M., He, Y., Zhang, H., Liao, X. P., Liu, Y. H., Sun, J., et al. (2020). CRISPR-cas9-mediated carbapenemase gene and plasmid curing in carbapenem-resistant enterobacteriaceae. Antimicrob. Agents Chemother. 64 (9), e00843–20. doi: 10.1128/aac.00843-20
Hatem, E. A., Hashad, N. A., Bayomi, E. B., and Kamal, A. A. (2024). Antimicrobial effect of isolated lactic acid bacteria and Bacillus spp. Sci. J. Faculty Sci. 28, 49–67. doi: 10.21608/sjfsmu.2024.326749.1009
He, Y., Hong, Q., Chen, S., Zhou, J., and Qiu, S. (2025). Reprogramming tumor-associated macrophages in gastric cancer: a pathway to enhanced immunotherapy. Front. Immunol. 16. doi: 10.3389/fimmu.2025.1558091
Heng, H., Sun, R., Yang, X., Ye, L., Chen, K., Li, J., et al. (2025). Profiling the landscape of carbapenem resistance and hypervirulence in Klebsiella pneumoniae: A global epidemiological analysis of the plasmidome. Drug Resist. Updat. 81, 101254. doi: 10.1016/j.drup.2025.101254
Hou, B., Niu, X., Yu, Q., and Wang, W. (2025). Epidemiological trends and drug resistance patterns of carbapenem-resistant gram-negative bacteria: A retrospective study in a tertiary hospital in China (2019-2024). Infect. Drug Resist. 18, 2867–2880. doi: 10.2147/idr.S518461
Isaac, S., Flor-Duro, A., Carruana, G., PuChades-Carrasco, L., Quirant, A., Lopez-Nogueroles, M., et al. (2022). Microbiome-mediated fructose depletion restricts murine gut colonization by vancomycin-resistant Enterococcus. Nat. Commun. 13, 7718. doi: 10.1038/s41467-022-35380-5
Jiang, J., Long, T., Porter, A. R., Lovey, A., Lee, A., Jacob, J. T., et al. (2025). Carbapenem-resistant, virulence plasmid-harboring klebsiella pneumoniae, United States. Emerg. Infect. Dis. 31, 761–771. doi: 10.3201/eid3104.241396
Jung, H. J., Sorbara, M. T., and Pamer, E. G. (2021). TAM mediates adaptation of carbapenem-resistant Klebsiella pneumoniae to antimicrobial stress during host colonization and infection. PloS Pathog. 17, e1009309. doi: 10.1371/journal.ppat.1009309
Kadry, A. A., El-Antrawy, M. A., and El-Ganiny, A. M. (2023). Impact of short chain fatty acids (SCFAs) on antimicrobial activity of new β-lactam/β-lactamase inhibitor combinations and on virulence of Escherichia coli isolates. J. Antibiot (Tokyo). 76, 225–235. doi: 10.1038/s41429-023-00595-1
Kang, J. T. L., Teo, J. J. Y., Bertrand, D., Ng, A., Ravikrishnan, A., Yong, M., et al. (2022). Long-term ecological and evolutionary dynamics in the gut microbiomes of carbapenemase-producing Enterobacteriaceae colonized subjects. Nat. Microbiol. 7, 1516–1524. doi: 10.1038/s41564-022-01221-w
Katip, W., Rayanakorn, A., Oberdorfer, P., Taruangsri, P., Nampuan, T., and Okonogi, S. (2024). Comparative effectiveness and mortality of colistin monotherapy versus colistin-fosfomycin combination therapy for the treatment of carbapenem-resistant Enterobacteriaceae (CRE) infections: A propensity score analysis. J. Infect. Public Health 17, 727–734. doi: 10.1016/j.jiph.2024.03.010
Kedišaletše, M., Phumuzile, D., Angela, D., Andrew, W., and Mae, N. F. (2023). Epidemiology, risk factors, and clinical outcomes of carbapenem-resistant Enterobacterales in Africa: A systematic review. J. Glob Antimicrob. Resist. 35, 297–306. doi: 10.1016/j.jgar.2023.10.008
Kent, A. G., Vill, A. C., Shi, Q., Satlin, M. J., and Brito, I. L. (2020). Widespread transfer of mobile antibiotic resistance genes within individual gut microbiomes revealed through bacterial Hi-C. Nat. Commun. 11, 4379. doi: 10.1038/s41467-020-18164-7
Khachab, Y., El Shamieh, S., and Sokhn, E. S. (2024). Gram-negative bacterial colonization in the gut: Isolation, characterization, and identification of resistance mechanisms. J. Infect. Public Health 17, 102535. doi: 10.1016/j.jiph.2024.102535
Kim, M., Jeon, K., Kym, D., Jung, J., Jang, Y. J., and Han, S. B. (2025). Carbapenem-resistant Enterobacterales infection and colonization in patients with severe burns: a retrospective cohort study in a single burn center. Antimicrob. Resist. Infect. Control. 14, 3. doi: 10.1186/s13756-025-01514-9
Kochan, T. J., Nozick, S. H., Valdes, A., Mitra, S. D., Cheung, B. H., Lebrun-Corbin, M., et al. (2023). Klebsiella pneumoniae clinical isolates with features of both multidrug-resistance and hypervirulence have unexpectedly low virulence. Nat. Commun. 14, 7962. doi: 10.1038/s41467-023-43802-1
Korsten, S., Peracic, L., van Groeningen, L. M. B., Diks, M. A. P., Vromans, H., Garssen, J., et al. (2022). Butyrate prevents induction of CXCL10 and non-canonical IRF9 expression by activated human intestinal epithelial cells via HDAC inhibition. Int. J. Mol. Sci. 23 (7), 3980. doi: 10.3390/ijms23073980
Korsten, S., Vromans, H., Garssen, J., and Willemsen, L. E. M. (2023). Butyrate protects barrier integrity and suppresses immune activation in a caco-2/PBMC co-culture model while HDAC inhibition mimics butyrate in restoring cytokine-induced barrier disruption. Nutrients 15 (12), 2760. doi: 10.3390/nu15122760
Lee, I., Jo, J. W., Woo, H. J., Suk, K. T., Lee, S. S., and Kim, B. S. (2024). Proton pump inhibitors increase the risk of carbapenem-resistant Enterobacteriaceae colonization by facilitating the transfer of antibiotic resistance genes among bacteria in the gut microbiome. Gut Microbes 16, 2341635. doi: 10.1080/19490976.2024.2341635
Lee, J. H., Shin, J., Park, S. H., Cha, B., Hong, J. T., Lee, D. H., et al. (2023). Role of probiotics in preventing carbapenem-resistant enterobacteriaceae colonization in the intensive care unit: risk factors and microbiome analysis study. Microorganisms 11 (2), 2970. doi: 10.3390/microorganisms11122970
Lee, J. J., Yong, D., Suk, K. T., Kim, D. J., Woo, H. J., Lee, S. S., et al. (2021). Alteration of gut microbiota in carbapenem-resistant enterobacteriaceae carriers during fecal microbiota transplantation according to decolonization periods. Microorganisms 9 (2), 352. doi: 10.3390/microorganisms9020352
Lei, T. Y., Liao, B. B., Yang, L. R., Wang, Y., and Chen, X. B. (2024). Hypervirulent and carbapenem-resistant Klebsiella pneumoniae: A global public health threat. Microbiol. Res. 288, 127839. doi: 10.1016/j.micres.2024.127839
Li, J. W., Mao, Y. M., Chen, S. L., Ye, R., Fei, Y. R., Li, Y., et al. (2024). The interplay between metal ions and immune cells in glioma: pathways to immune escape. Discov. Oncol. 15, 348. doi: 10.1007/s12672-024-01229-0
Li, J., Wu, W., Wu, H., Huang, J., Li, Z., Wang, J., et al. (2025). Rapid emergence, transmission, and evolution of KPC and NDM coproducing carbapenem-resistant Klebsiella pneumoniae. Microbiol. Res. 293, 128049. doi: 10.1016/j.micres.2025.128049
Li, Y., Zhang, Y., Sun, X., Wu, Y., Yan, Z., Ju, X., et al. (2024). National genomic epidemiology investigation revealed the spread of carbapenem-resistant Escherichia coli in healthy populations and the impact on public health. Genome Med. 16, 57. doi: 10.1186/s13073-024-01310-x
Lin, S., Mukherjee, S., Li, J., Hou, W., Pan, C., and Liu, J. (2021). Mucosal immunity-mediated modulation of the gut microbiome by oral delivery of probiotics into Peyer’s patches. Sci. Adv. 7 (20), eabf0677. doi: 10.1126/sciadv.abf0677
Liu, W., Li, Z., Ze, X., Deng, C., Xu, S., and Ye, F. (2024). Multispecies probiotics complex improves bile acids and gut microbiota metabolism status in an in vitro fermentation model. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1314528
Liu, Q., Zuo, T., Lu, W., Yeoh, Y. K., Su, Q., Xu, Z., et al. (2022). Longitudinal evaluation of gut bacteriomes and viromes after fecal microbiota transplantation for eradication of carbapenem-resistant enterobacteriaceae. mSystems 7, e0151021. doi: 10.1128/msystems.01510-21
Ma, J., Song, X., Li, M., Yu, Z., Cheng, W., Yu, Z., et al. (2023). Global spread of carbapenem-resistant Enterobacteriaceae: Epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 266, 127249. doi: 10.1016/j.micres.2022.127249
Macareño-Castro, J., Solano-Salazar, A., Dong, L. T., Mohiuddin, M., and Espinoza, J. L. (2022). Fecal microbiota transplantation for Carbapenem-Resistant Enterobacteriaceae: A systematic review. J. Infect. 84, 749–759. doi: 10.1016/j.jinf.2022.04.028
Martin-Gallausiaux, C., Garcia-Weber, D., Lashermes, A., Larraufie, P., Marinelli, L., Teixeira, V., et al. (2022). Akkermansia muciniphila upregulates genes involved in maintaining the intestinal barrier function via ADP-heptose-dependent activation of the ALPK1/TIFA pathway. Gut Microbes 14, 2110639. doi: 10.1080/19490976.2022.2110639
Meekes, L. M., Heikema, A. P., Tompa, M., Astorga Alsina, A. L., Hiltemann, S. D., Stubbs, A. P., et al. (2025). Proteogenomic analysis demonstrates increased bla(OXA-48) copy numbers and OmpK36 loss as contributors to carbapenem resistance in Klebsiella pneumoniae. Antimicrob. Agents Chemother. 69, e0010725. doi: 10.1128/aac.00107-25
Merrick, B., Prossomariti, D., Allen, E., Bisnauthsing, K., Kertanegara, M., Sergaki, C., et al. (2025). Faecal microbiota transplant to ERadicate gastrointestinal carriage of Antibiotic-Resistant Organisms (FERARO): A feasibility randomised controlled trial. J. Infect. 91, 106504. doi: 10.1016/j.jinf.2025.106504
Michaelis, C. and Grohmann, E. (2023). Horizontal gene transfer of antibiotic resistance genes in biofilms. Antibiotics (Basel). 12 (2), 328. doi: 10.3390/antibiotics12020328
Millan, B., Park, H., Hotte, N., Mathieu, O., Burguiere, P., Tompkins, T. A., et al. (2016). Fecal microbial transplants reduce antibiotic-resistant genes in patients with recurrent clostridium difficile infection. Clin. Infect. Dis. 62, 1479–1486. doi: 10.1093/cid/ciw185
Neil, K., Allard, N., Roy, P., Grenier, F., Menendez, A., Burrus, V., et al. (2021). High-efficiency delivery of CRISPR-Cas9 by engineered probiotics enables precise microbiome editing. Mol. Syst. Biol. 17, e10335. doi: 10.15252/msb.202110335
Ni, H., Chan, K. B., Cheng, Q., Chen, K., Xie, M., Wang, H., et al. (2022). A novel clinical therapy to combat infections caused by Hypervirulent Carbapenem-Resistant Klebsiella pneumoniae. J. Infect. 85, 174–211. doi: 10.1016/j.jinf.2022.05.004
Ni, H., Chan, B. K., Ye, L., Wu, H., Heng, H., Xu, Q., et al. (2024). Lowering mortality risk in CR-HvKP infection in intestinal immunohistological and microbiota restoration. Pharmacol. Res. 206, 107254. doi: 10.1016/j.phrs.2024.107254
Ormsby, M. J., Johnson, S. A., Carpena, N., Meikle, L. M., Goldstone, R. J., McIntosh, A., et al. (2020). Propionic acid promotes the virulent phenotype of crohn’s disease-associated adherent-invasive escherichia coli. Cell Rep. 30, 2297–305.e5. doi: 10.1016/j.celrep.2020.01.078
Ott, L. C. and Mellata, M. (2024). Short-chain fatty acids inhibit bacterial plasmid transfer through conjugation in vitro and in ex vivo chicken tissue explants. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1414401
Pace, F., Rudolph, S. E., Chen, Y., Bao, B., Kaplan, D. L., and Watnick, P. I. (2021). The Short-Chain Fatty Acids Propionate and Butyrate Augment Adherent-Invasive Escherichia coli Virulence but Repress Inflammation in a Human Intestinal Enteroid Model of Infection. Microbiol. Spectr. 9, e0136921. doi: 10.1128/Spectrum.01369-21
Pople, D., Kypraios, T., Donker, T., Stoesser, N., Seale, A. C., George, R., et al. (2023). Model-based evaluation of admission screening strategies for the detection and control of carbapenemase-producing Enterobacterales in the English hospital setting. BMC Med. 21, 492. doi: 10.1186/s12916-023-03007-1
Ribeiro, A., Chikhani, Y., Valiatti, T. B., Valêncio, A., Kurihara, M. N. L., Santos, F. F., et al. (2023). In Vitro and In Vivo Synergism of Fosfomycin in Combination with Meropenem or Polymyxin B against KPC-2-Producing Klebsiella pneumoniae Clinical Isolates. Antibiotics (Basel) 12 (2), 237. doi: 10.3390/antibiotics12020237
Rooney, C. M., Sheppard, A. E., Clark, E., Davies, K., Hubbard, A. T. M., Sebra, R., et al. (2019). Dissemination of multiple carbapenem resistance genes in an in vitro gut model simulating the human colon. J. Antimicrob. Chemother. 74, 1876–1883. doi: 10.1093/jac/dkz106
Ruvinsky, S., Voto, C., Roel, M., Portillo, V., Naranjo Zuñiga, G., Ulloa-Gutierrez, R., et al. (2024). Carbapenem-resistant enterobacteriaceae bacteremia in pediatric patients in latin america and the caribbean: A systematic review and meta-analysis. Antibiotics (Basel) 13 (12), 1117. doi: 10.3390/antibiotics13121117
Salomão, M. C., Freire, M. P., Lázari, C. S., Cury, A. P., Rossi, F., Segurado, A. A. C., et al. (2023). Transmission of carbapenem-resistant enterobacterales in an overcrowded emergency department: controlling the spread to the hospital. Clin. Infect. Dis. 77, S46–s52. doi: 10.1093/cid/ciad263
Sati, H., Carrara, E., Savoldi, A., Hansen, P., Garlasco, J., Campagnaro, E., et al. (2025). The WHO Bacterial Priority Pathogens List 2024: a prioritisation study to guide research, development, and public health strategies against antimicrobial resistance. Lancet Infect. Dis. 25 (9), 1033–1043 doi: 10.1016/s1473-3099(25)00118-5
Schulthess, J., Pandey, S., Capitani, M., Rue-Albrecht, K. C., Arnold, I., Franchini, F., et al. (2019). The short chain fatty acid butyrate imprints an antimicrobial program in macrophages. Immunity 50, 432–45.e7. doi: 10.1016/j.immuni.2018.12.018
Shin, J., Lee, J. H., Park, S. H., Cha, B., Kwon, K. S., Kim, H., et al. (2022). Efficacy and safety of fecal microbiota transplantation for clearance of multidrug-resistant organisms under multiple comorbidities: A prospective comparative trial. Biomedicines 10 (10), 2404. doi: 10.3390/biomedicines10102404
Sim, C. K., Kashaf, S. S., Stacy, A., Proctor, D. M., Almeida, A., Bouladoux, N., et al. (2022). A mouse model of occult intestinal colonization demonstrating antibiotic-induced outgrowth of carbapenem-resistant Enterobacteriaceae. Microbiome 10, 43. doi: 10.1186/s40168-021-01207-6
Sindi, A. A., Alsayed, S. M., Abushoshah, I., Bokhary, D. H., and Tashkandy, N. R. (2022). Profile of the gut microbiome containing carbapenem-resistant enterobacteriaceae in ICU patients. Microorganisms 10 (7), 1309. doi: 10.3390/microorganisms10071309
Smith, H. Z., Hollingshead, C. M., and Kendall, B. (2025). “Carbapenem-resistant enterobacterales,” in StatPearls (StatPearls Publishing Copyright ©, Treasure Island (FL).
Song, X., Xu, C., Zhu, Z., Zhang, C., Qin, C., Liu, J., et al. (2025). Multidrug-resistant Klebsiella pneumoniae coinfection with multiple microbes: a retrospective study on its risk factors and clinical outcomes. mSystems. 10 (8), e0175724. doi: 10.1128/msystems.01757-24
Sorbara, M. T., Dubin, K., Littmann, E. R., Moody, T. U., Fontana, E., Seok, R., et al. (2019). Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J. Exp. Med. 216, 84–98. doi: 10.1084/jem.20181639
Tang, H., Huang, W., and Yao, Y. F. (2023). The metabolites of lactic acid bacteria: classification, biosynthesis and modulation of gut microbiota. Microb. Cell. 10, 49–62. doi: 10.15698/mic2023.03.792
Tang, C., Shen, S., Yang, W., Shi, Q., Ding, L., Han, R., et al. (2024). Dynamic evolution of ceftazidime-avibactam resistance from a single patient through the IncX3_NDM-5 plasmid transfer and bla(KPC) mutation. Int. J. Antimicrob. Agents. 64, 107228. doi: 10.1016/j.ijantimicag.2024.107228
Tao, S., Chen, H., Li, N., and Liang, W. (2022). The application of the CRISPR-cas system in antibiotic resistance. Infect. Drug Resist. 15, 4155–4168. doi: 10.2147/idr.S370869
Telhig, S., Ben Said, L., Torres, C., Rebuffat, S., Zirah, S., and Fliss, I. (2022). Evaluating the potential and synergetic effects of microcins against multidrug-resistant enterobacteriaceae. Microbiol. Spectr. 10, e0275221. doi: 10.1128/spectrum.02752-21
Telhig, S., Ben Said, L., Zirah, S., Fliss, I., and Rebuffat, S. (2020). Bacteriocins to thwart bacterial resistance in gram negative bacteria. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.586433
Tubb, C. M., Tubb, M., Hooijer, J., Chomba, R., and Nel, J. (2025). Carbapenem-resistant Enterobacterales (CRE) colonisation as a predictor for subsequent CRE infection: A retrospective surveillance study. S Afr J. Infect. Dis. 40, 687. doi: 10.4102/sajid.v40i1.687
Wang, M., Jin, L., Wang, R., Wang, Q., Wang, S., Wu, X., et al. (2025). KpnK48 clone driving hypervirulent carbapenem-resistant Escherichia coli epidemics: Insights into its evolutionary trajectory similar to Klebsiella pneumoniae. Drug Resist. Updat. 81, 101243. doi: 10.1016/j.drup.2025.101243
Wang, Z., Shao, C., Shao, J., Hao, Y., and Jin, Y. (2024). Risk factors of Carbapenem-resistant Enterobacterales intestinal colonization for subsequent infections in hematological patients: a retrospective case-control study. Front. Microbiol. 15. doi: 10.3389/fmicb.2024.1355069
Wang, Q., Wang, R., Wang, S., Zhang, A., Duan, Q., Sun, S., et al. (2024). Expansion and transmission dynamics of high risk carbapenem-resistant Klebsiella pneumoniae subclones in China: An epidemiological, spatial, genomic analysis. Drug Resist. Updat. 74, 101083. doi: 10.1016/j.drup.2024.101083
Wu, Y., Chen, J., Zhang, G., Li, J., Wang, T., Kang, W., et al. (2024). In-vitro activities of essential antimicrobial agents including aztreonam/avibactam, eravacycline, colistin and other comparators against carbapenem-resistant bacteria with different carbapenemase genes: A multi-centre study in China, 2021. Int. J. Antimicrob. Agents. 64, 107341. doi: 10.1016/j.ijantimicag.2024.107341
Wu, Y. L., Hu, X. Q., Wu, D. Q., Li, R. J., Wang, X. P., Zhang, J., et al. (2023). Prevalence and risk factors for colonisation and infection with carbapenem-resistant Enterobacterales in intensive care units: A prospective multicentre study. Intensive Crit. Care Nurs. 79, 103491. doi: 10.1016/j.iccn.2023.103491
Wu, W. H., Kim, M., Chang, L. C., Assie, A., Saldana-Morales, F. B., Zegarra-Ruiz, D. F., et al. (2022). Interleukin-1β secretion induced by mucosa-associated gut commensal bacteria promotes intestinal barrier repair. Gut Microbes 14, 2014772. doi: 10.1080/19490976.2021.2014772
Xiao, Y., Duan, J., Tan, C., Zou, J., Chen, S., Liu, T., et al. (2024). Correlation between intestinal CRE colonization and consequent systemic infection in hospitalized patients in China. Sci. Rep. 14, 26017. doi: 10.1038/s41598-024-76261-9
Xie, M., Ye, L., Chen, K., Xu, Q., Yang, C., Chen, X., et al. (2024). Clinical use of tigecycline may contribute to the widespread dissemination of carbapenem-resistant hypervirulent Klebsiella pneumoniae strains. Emerg. Microbes Infect. 13, 2306957. doi: 10.1080/22221751.2024.2306957
Yip, A. Y. G., King, O. G., Omelchenko, O., Kurkimat, S., Horrocks, V., Mostyn, P., et al. (2023). Antibiotics promote intestinal growth of carbapenem-resistant Enterobacteriaceae by enriching nutrients and depleting microbial metabolites. Nat. Commun. 14, 5094. doi: 10.1038/s41467-023-40872-z
Yuan, W., Xu, J., Guo, L., Chen, Y., Gu, J., Zhang, H., et al. (2022). Clinical risk factors and microbiological and intestinal characteristics of carbapenemase-producing enterobacteriaceae colonization and subsequent infection. Microbiol. Spectr. 10, e0190621. doi: 10.1128/spectrum.01906-21
Zhang, F., Liu, X., Li, Z., Li, Z., Lei, Z., Fan, Y., et al. (2025). Tracking international and regional dissemination of the KPC/NDM co-producing Klebsiella pneumoniae. Nat. Commun. 16, 5574. doi: 10.1038/s41467-025-60765-7
Zhao, N., Geng, P., Perez, A. G., Maya, A. C., Yadav, B., Du, Y., et al. (2025). Genomic and functional characterization of a Butyricicoccus porcorum strain isolated from human gut microbiota. mSystems. 10 (8), e0079025. doi: 10.1128/msystems.00790-25
Zhao, Q. and Maynard, C. L. (2022). Mucus, commensals, and the immune system. Gut Microbes 14, 2041342. doi: 10.1080/19490976.2022.2041342
Zhao, K., Yu, L., Wang, X., He, Y., and Lu, B. (2018). Clostridium butyricum regulates visceral hypersensitivity of irritable bowel syndrome by inhibiting colonic mucous low grade inflammation through its action on NLRP6. Acta Biochim. Biophys. Sin. (Shanghai). 50, 216–223. doi: 10.1093/abbs/gmx138
Keywords: carbapenem-resistant Enterobacteriaceae, gut microbiota, colonization resistance, microbiome-targeted therapy, probiotics, fecal microbiota transplantation, short-chain fatty acids
Citation: Zhang L, Xu T, Chen W, Chai Y, Wu Y and Du X (2025) The potential of the microbiome as a target for prevention and treatment of carbapenem-resistant Enterobacteriaceae infections. Front. Cell. Infect. Microbiol. 15:1674534. doi: 10.3389/fcimb.2025.1674534
Received: 28 July 2025; Accepted: 24 September 2025;
Published: 08 October 2025.
Edited by:
Xuebin Tian, Shandong Provincial Hospital, ChinaReviewed by:
Ritam Sinha, National Institute of Cholera and Enteric Diseases (ICMR), IndiaHamed Tahmasebi, Shahroud University of Medical Sciences, Iran
Hongyuhang Ni, Carnegie Mellon University, United States
Copyright © 2025 Zhang, Xu, Chen, Chai, Wu and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xinghai Du, ZHV4aW5naGFpMTZAMTYzLmNvbQ==
†These authors have contributed equally to this work and share first authorship
 Lu Zhang†
Lu Zhang† Tinghui Xu
Tinghui Xu Yinying Chai
Yinying Chai