- 1Department of Laboratory Medicine, The First Affiliated Hospital of Chongqing Medical University, Chongqing, China
- 2Department of Clinical Laboratory, Institute of Translational Medicine, Renmin Hospital of Wuhan University, Wuhan, China
Objectives: The rapid dissemination of carbapenem-resistant Enterobacteriaceae (CRE) poses a severe global health threat due to limited treatment options. Polymyxin is currently considered a last-resort therapy for human infections caused by CRE. The increasing clinical use of polymyxin has resulted in alarming resistance rates. This study investigated the clinical characteristics, molecular epidemiological characteristics, and resistance mechanisms of polymyxin-resistant CRE (PR-CRE) in Chongqing, China.
Methods: Antimicrobial susceptibility and resistance genes were analyzed by PCR-based amplification and sequence analysis. Molecular epidemiological characteristics were analyzed using Enterobacterial Repetitive Intergenic Consensus (ERIC)-PCR and multilocus sequence typing (MLST). The gene expression of pmrC and pmrK was analyzed using qRT-PCR. Conjugation experiments were performed to determine plasmid transferability. Whole-genome sequencing (WGS) was performed to analyze their genetic environment.
Results: Thirty PR-CRE isolates, including 21 Klebsiella pneumoniae and 9 Escherichia coli, exhibited multidrug resistance, with three pan-resistant K. pneumoniae strains. The predominant lineage was ST11-K64 K. pneumoniae (n=14/21), with four genetically identical isolates from ICUs, confirming clonal transmission. In contrast, all E. coli displayed high genetic diversity. Hypervirulence determinants were detected in 38.1% (n=8/21) of K. pneumoniae. Two rare K. pneumoniae strains were identified: one hypervirulent ST3984-KL64 strain co-harboring blaKPC-2 and blaNDM-1, and one ST2383-KL81 strain harboring blaOXA-48. Species-specific resistance mechanisms emerged: K. pneumoniae relied on chromosomal mutations in mgrB, phoPQ, and pmrAB, especially mgrB inactivation (57.1%, n=12/21) via ISKpn26/IS903B/ISAeme19/ISKpn14 and pmrK upregulation (95.2%, n=20/21), while E. coli exclusively used plasmid-borne mcr-1 with 55.6% (n=5/9) conjugation efficiency, conferring low-level resistance. Genomic sequencing revealed that four identical ISAeme19 copies were first identified in ST11-KL64 hypervirulent CRKP-5: two on the chromosome (mgrB and kdsD), and two on plasmids (IncFII/IncR pkp2007-KPC and recombinant pkp2007-D). Transposition of ISAeme19 from pkp2007-KPC to mgrB was evidenced by the inverted orientation and matching flanking repeats. Crucially, a pan-resistant ST11-KL64 K. pneumoniae harbored a fusion plasmid with dual blaKPC-2 and catA2 copies bracketed by IS26, a previously unreported configuration. Additionally, four novel deleterious mutations were detected: mgrB-Asn25Thr, phoP-Lys199Met, phoQ-Tyr89His, and RamR-Ala17Thr.
Conclusion: These findings reveal species-divergent resistance mechanisms to polymyxin, necessitating enhanced surveillance of these high-risk clones, mobile elements, and emergent resistance mechanisms.
1 Introduction
In recent years, carbapenem-resistant Enterobacteriaceae (CRE) isolates, particularly carbapenem-resistant Klebsiella pneumoniae (CRKP) and Escherichia coli (CRECO), have emerged as formidable nosocomial pathogens (van Duin et al., 2020). Infections caused by CRE are associated with high mortality rates due to limited therapeutic options (Stewardson et al., 2019). Polymyxin is regarded as a last-resort agent for CRE infections; however, extensive clinical use has driven an alarming increase in resistance rates (Puljko et al., 2024; Xie et al., 2025). According to data from the China Antimicrobial Surveillance Network (CHINET, https://www.chinets.com/), the rate of polymyxin resistance among CRKP isolates has increased from 3.6% in 2020 to 11.8% in 2023. Furthermore, a systematic review revealed that the global prevalence of polymyxin-resistant K. pneumoniae was about 11.64% from 1987 to 2020, and the resistance rates reported in America, Europe, and Asia were 18.67%, 16.16%, and 10.17% (Sameni et al., 2022).
The primary mechanism of polymyxin resistance involves the modification of lipopolysaccharide (LPS) through the addition of cationic groups, specifically 4-amino-4-deoxy-L-arabinose (L-Ara4N) and/or phosphoethanolamine (PEtN). These modifications reduce the binding affinity of polymyxin and elevate the minimal inhibitory concentrations (MICs) (Binsker et al., 2021). This process is regulated by chromosomally encoded two-component regulatory systems (phoPQ, pmrAB, and crrAB) and the negative regulator mgrB (El-Sayed Ahmed et al., 2020). Additionally, plasmid-mediated mobile polymyxin resistance genes (mcr-1 to mcr-10) induce outer membrane modifications by adding PEtN to lipid A (Rodríguez-Santiago et al., 2021; Liu et al., 2024). These genes demonstrate efficient horizontal transfer among humans, animals, and environments in more than 60 countries (Huang et al., 2020; Mmatli et al., 2022).
Furthermore, the convergence of polymyxin resistance and hypervirulent genes in CRKP isolates has fueled fatal hospital outbreaks globally, making molecular epidemiology essential for containment (Liu et al., 2022b). Given the complexity and diversity of these resistance mechanisms, our study aimed to investigate the molecular epidemiological characteristics and resistance mechanisms of polymyxin-resistant CRE (PR-CRE) isolates from Chongqing, China.
2 Materials and methods
2.1 Bacterial isolates and antimicrobial susceptibility testing
Clinical CRE isolates were collected from hospitalized patients at a tertiary teaching hospital in Chongqing, China. Species identification was initially performed using MALDI-TOF MS (BioMérieux, France). All isolates were stored at −80°C for subsequent analyses. MICs of polymyxin, tigecycline, and ceftazidime/avibactam were determined by broth microdilution according to the Clinical and Laboratory Standards Institute M100 (2024) guidelines. Additional antibiotics were tested using agar dilution. Tigecycline breakpoints followed the Food and Drug Administration criteria.
2.2 Detection of resistance genes, virulence genes and capsular serotyping
Carbapenemase genes (blaKPC, blaNDM, blaOXA-48, blaVIM, and blaIMP), β-lactamase genes (blaTEM, blaSHV, and blaCTX-M), virulence-associated genes (iucA, iroB, p-rmpA, p-rmpA2, and peg-344) and capsular serotyping in K. pneumoniae were detected by PCR and Sanger sequencing. The primers used can be found in Supplementary Table 1.
2.3 Multilocus sequence typing and enterobacterial repetitive intergenic consensus-PCR fingerprinting
Molecular epidemiological characteristics were assessed using MLST and ERIC-PCR. Cluster analysis was performed at 90% similarity using BioNumerics v7.6.3. MLST employed standard schemes: seven housekeeping genes (ropB, gapA, mdh, pgi, phoE, infB, and tonB) for K. pneumoniae and seven (gyrB, icd, adk, mdh, fumC, purA, and recA) for E. coli. Sequence types were assigned using the Pasteur Institute (http://bigsdb.pasteur.fr/Klebsiella/Klebsiella.html) and PubMLST (Carattoli et al., 2014) databases. The primers used are provided in Supplementary Table 2.
2.4 Polymyxin resistance mechanism characterization
Multiplex PCR was performed to detect mcr-1 to mcr-10. Meanwhile, compared with the reference genomes of K. pneumoniae MGH78578 (NC_009648.1) and E. coli K12 MG1655 (NC_000913.3), chromosomal regulators (pmrAB, phoPQ, crrAB, mgrB) were amplified and sequenced. The impact of mutations was predicted using PROVEAN v1.1.5, with scores ≤ −2.5 considered deleterious. Insertion sequences disrupting the mgrB gene were identified using ISfinder (https://www-is.biotoul.fr/). Quantitative real-time PCR (qRT-PCR) was used to measure the expression of pmrC and pmrK, normalized to rpsL using the 2−ΔΔCT method. A polymyxin-susceptible K. pneumoniae strain ATCC BAA-1705 was used as reference for the gene expression analysis. The primers used are provided in Supplementary Table 3. Conjugation assays transferred mcr-1 to rifampicin-resistant E. coli EC600 on Mueller-Hinton agar containing polymyxin (2 mg/L) and rifampicin (400 mg/L). Subsequent antimicrobial susceptibility testing and PCR amplification confirmed plasmid transfer to the recipient.
2.5 Whole-genome sequencing and bioinformatics analysis
Genomic DNA was extracted and sequenced using PacBio RS II SMRT technology (Pacific Biosciences, USA) and Illumina HiSeq X Ten (Illumina, USA). Hybrid assembly was performed using Unicycler v0.4.8 (Wick et al., 2017), followed by error correction based on Pilon v1.24 (Walker et al., 2014). Gene annotation was conducted using Glimmer v3.0 (Delcher et al., 2007) and Prokka v1.11 (Seemann, 2014). Resistance genes, plasmid replicon types, and MLST were identified using the CGE platform (https://cge.cbs.dtu.dk/services/). Virulence factors, mobile elements, and plasmid transfer regions were detected using VFDB (Liu et al., 2022a), ISfinder (Siguier et al., 2006), and OriTfinder (Li et al., 2018) databases. Comparative genomics analysis was performed by BLASTn, BRIG (v1.3.0, https://sourceforge.net/projects/brig/) , and Easyfig (v2.2.5, https://mjsull.github.io/Easyfig/).
2.6 Statistical analysis
Statistical analysis was performed using GraphPad Prism 8 (GraphPad Software, San Diego, CA). With K. pneumoniae ATCC BAA-1705 as the reference, the expression levels of pmrC and pmrK in polymyxin-resistant K. pneumoniae strains were analyzed using unpaired two-tailed Student’s t-test. Statistical significance thresholds were defined as follows: *P < 0.05; **P < 0.01; ***P < 0.001.
3 Results
3.1 Antimicrobial susceptibility and clinical characteristics of PR-CRE isolates
All 30 polymyxin-resistant CRE isolates, including 21 polymyxin-resistant CRKP (PR-CRKP) and 9 polymyxin-resistant CRECO (PR-CRECO), exhibited multidrug resistance (Figure 1A). Polymyxin MICs for PR-CRKP isolates ranged from 4 to 128 µg/mL (MIC50 = 16 µg/mL, MIC90 = 64 µg/mL), with 71.4% (n=15/21) of the isolates exhibiting high-level resistance (MIC≥16 µg/mL). Three isolates (CRKP-1/8/12) exhibited co-resistance to tigecycline and polymyxin (Supplementary Table 4). Notably, most PR-CRKP isolates remained susceptible to ceftazidime/avibactam (90.5%, n=19/21) and tigecycline (85.7%, n=18/21). All PR-CRECO isolates displayed low-level polymyxin resistance (MIC range: 4–8 µg/mL) and were fully susceptible to tigecycline (Supplementary Table 4).
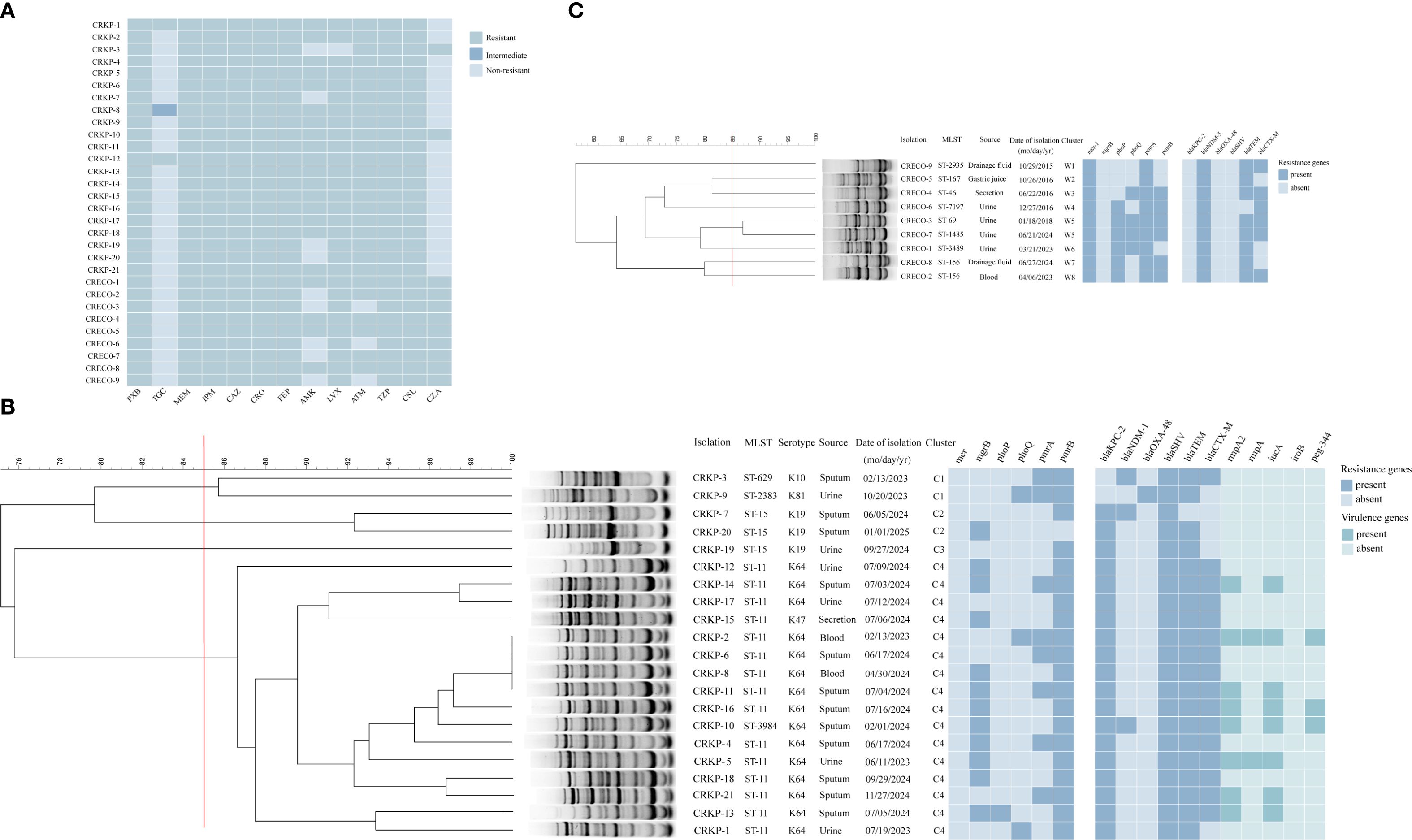
Figure 1. Characteristics of 30 polymyxin-resistant CRE isolates. (A) Heatmap showing polymyxin resistance profiles of 30 polymyxin-resistant CRE isolates (21 Klebsiella pneumoniae, 9 Escherichia coli); (B) ERIC-PCR dendrogram (≥85% similarity) of polymyxin-resistant CRKP strains. Strain isolations, multilocus sequence typing (MLST), setrotype, source of initial isolation, and data of isolation are included along each lane. The heatmap depict antimicrobial resistance genes and virulence genes of polymyxin-resistant CRKP isolates. (C) ERIC-PCR dendrogram (≥85% similarity) of polymyxin-resistant CRECO strains. Strain isolations, multilocus sequence typing (MLST) and data of isolation are included along each lane. The heatmap depict antimicrobial resistance genes of polymyxin-resistant CRECO isolates. The names of antimicrobial resistance genes are labeled at the top of this figure. Abbreviations: PXB, polymyxin B; TGC, tigecycline; MEM, meropenem; IPM, imipenem; CAZ,ceftazidime; CRO, ceftriaxone; FEP, cefepime; AMK, amikacin; LVX, levofloxacin; ATM, aztreonam; TZP, piperacillin-tazobactam; CSL, cefoperazone-sulbactam; CZA, ceftazidime-avibactam. Numbers in boldface type indicate susceptibility according to CLSI/EUCAST breakpoints.
Patients infected with PR-CRKP were predominantly elderly males in ICUs (mean age: 66.7 years; 81.0% male). Sputum (57.1%, n=12/21) and urine (28.6%, n=6/21) were the primary specimen sources. Prior polymyxin exposure occurred in 66.7% (n=14/21) of cases, and 28.6% (n=6/21) experienced treatment failure (Table 1). Patients infected with PR-CRECO had a mean age of 54.1 years, with 44.4% (n=4/9) originating from urology departments and urine specimens accounting for 44.4% (n=4/9) of isolates (Table 1).
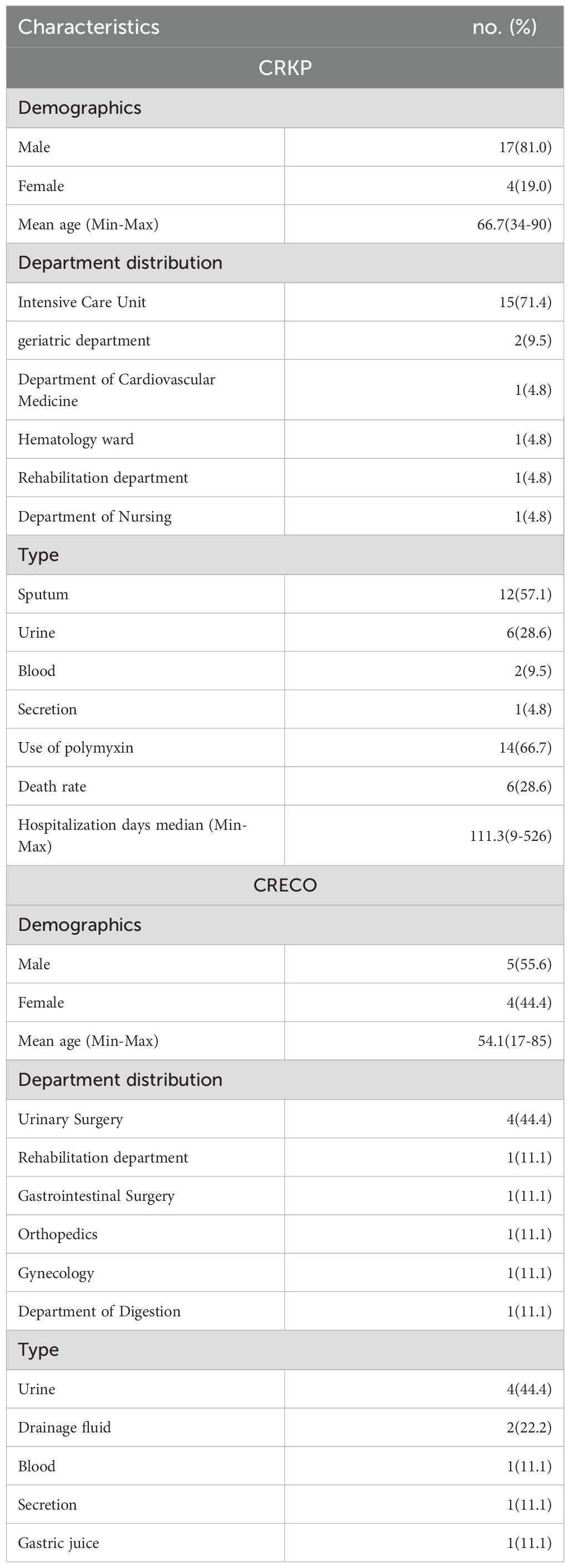
Table 1. Clinical characteristics of 30 patients infected with polymyxin-resistant CRKP isolates (n=21) and CRECO isolates (n=9).
3.2 Molecular characteristics of polymyxin-resistant CRE isolates
3.2.1 Resistance determinants, virulence genes, and molecular epidemiological characteristics of K. pneumoniae
All 21 PR-CRKP isolates produced carbapenemases: blaKPC-2 (n=18), blaNDM-1 (n=1), blaOXA-48 (n=1), or co-harbored blaKPC-2 and blaNDM-1 (n=1). Furthermore, extended-spectrum β-lactamase (ESBL) genes blaSHV, blaTEM, and blaCTX-M were found in 100% (n=21), 95.2% (n=20), and 76.2% (n=16) of the isolates. Hypervirulent biomarkers were identified in seven PR-CRKP isolates, with distributions as follows: rmpA2 (n=8), iucA (n=8), peg344 (n=3) and rmpA (n=2) (Figure 1B).
MLST identified five distinct sequence types (STs) among the 21 isolates: ST11 (n=15), ST15 (n=3), ST629 (n=1), ST2383(n=1), and ST3984 (n=1) (Figure 1B). ST2383 and ST3984 represent rare lineages. Genetic analysis indicated that ST3984 was a single-locus variant of ST11 in rpoB, suggesting that ST3984 may be derived from ST11. Capsular genotyping revealed five KL types: KL64 (n=15), KL19 (n=3), KL47 (n=1), KL10 (n=1), and KL81 (n=1) (Figure 1B). Fourteen of fifteen ST11 isolates harbored KL64, while one ST11 isolate harbored KL47. Interestingly, KL64 was also present in the ST3984 isolate (CRKP-10). All ST15 isolates possessed KL19, whereas ST2383 and ST629 corresponded to KL81 and KL10, respectively.
ERIC-PCR clustering analysis (Figure 1B) delineated four major clusters (C1-C4) at >85.0% similarity. Cluster C4 (15 ST11 and 1 ST3984) was predominantly ICU-associated, which has emerged as the dominant and widely disseminated lineage regionally, with most isolates sharing >90.0% similarity. Within C4, four ICU isolates (CRKP-2/6/8/11) exhibited 100.0% genetic identity, confirming clonal transmission.
3.2.2 Antimicrobial resistance determinants and molecular epidemiological characteristics of E. coli
PCR analysis confirmed that all PR-CRECO isolates produced blaNDM-5 carbapenemase (Figure 1C). Among β-lactamase genes, blaTEM (n=8) and blaCTX-M (n=6) were prevalent, while blaSHV was undetected. The nine PR-CRECO isolates belonged to eight distinct STs and formed eight ERIC-PCR groups, with a similarity threshold of >85.0%, indicating high genetic diversity (Figure 1C).
3.3 Species-divergent resistance mechanisms to polymyxin
3.3.1 Chromosome-mediated polymyxin resistance in K. pneumoniae
Given the absence of mcr genes among all PR-CRKP isolates, we focused on investigating chromosomal resistance mechanisms. Sequencing of key regulatory genes revealed missense mutations at seven sites in pmrB, four in pmrA, three in phoQ, and one in phoP (Table 2). Among these mutations, PROVEAN analysis predicted three pmrB mutations (Asp313Asn, Asp150Tyr, and Thr157Pro), one pmrA mutation (G53S), one phoP mutation (Lys199Met), and one phoQ mutation (Tyr89His) as deleterious. Notably, the phoP Lys199Met mutation in ST11 CRKP-13 strain and the phoQ Tyr89His mutation in ST11 CRKP-2 strain are previously unreported.
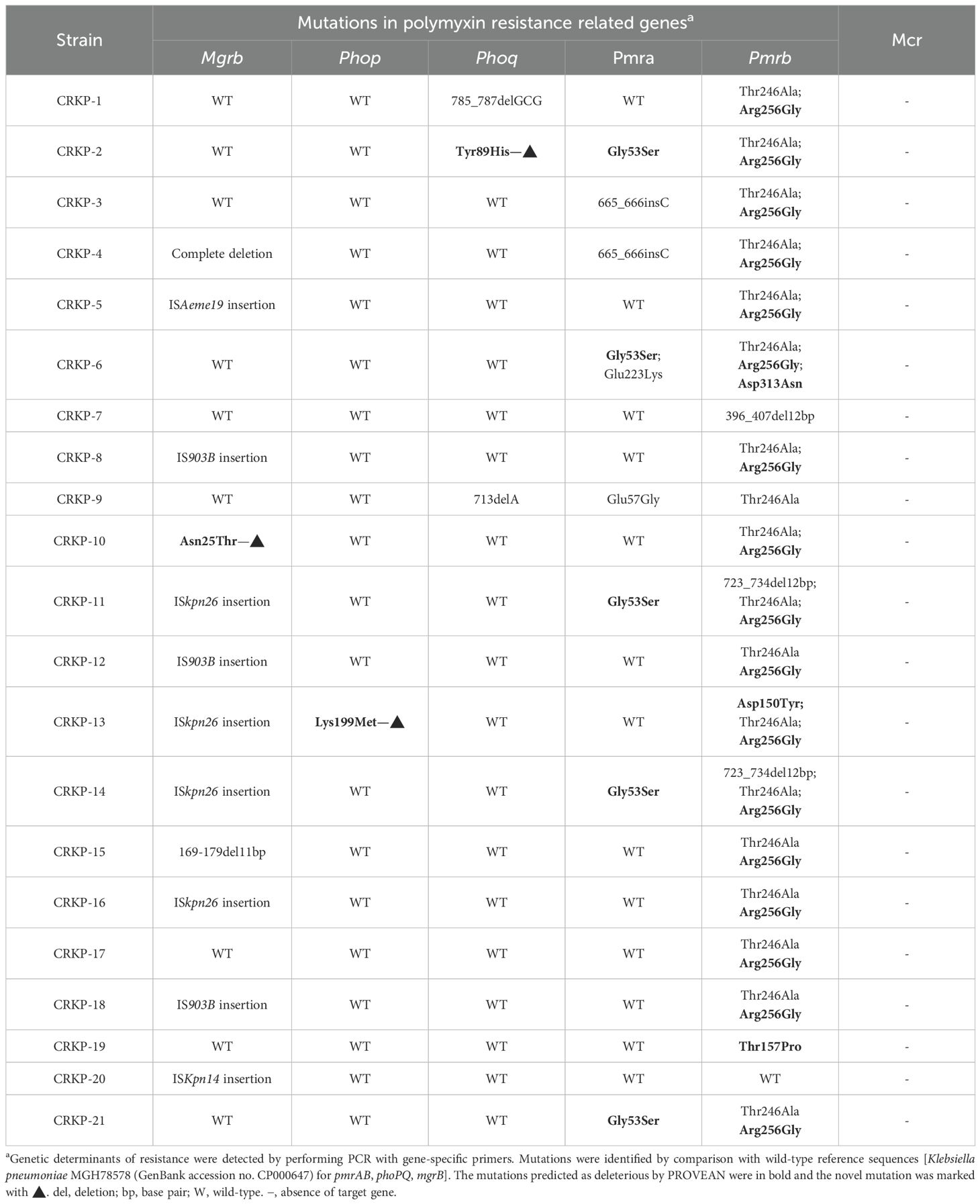
Table 2. Chromosomal mutations and plasmid-mediated genes related to polymyxin resistance in CRKP isolates.
Additionally, 57.1% (n=12/21) of isolates carried modified mgrB, including Asn25Thr mutation (n=1), complete or partial deletions (n=2), and insertional inactivation mediated by ISKpn26 (n=4), IS903B (n=3), ISAeme19 (n=1), and ISKpn14 (n=1) (Table 2; Figure 2A). Among the three isolates with IS903B (IS5 family), one occurred at position + 36 and two at +69 (Figure 2B; Figure 2D). ISKpn26 (IS5 family) disrupted mgrB at nucleotide +74 in four isolates (Figure 2C). ISAeme19 (ISL3 family) disrupted mgrB at nucleotide +12 in one isolate, while ISKpn14 (IS1 family) caused upstream disruption (-28 nt) in one isolate (Figure 2E).
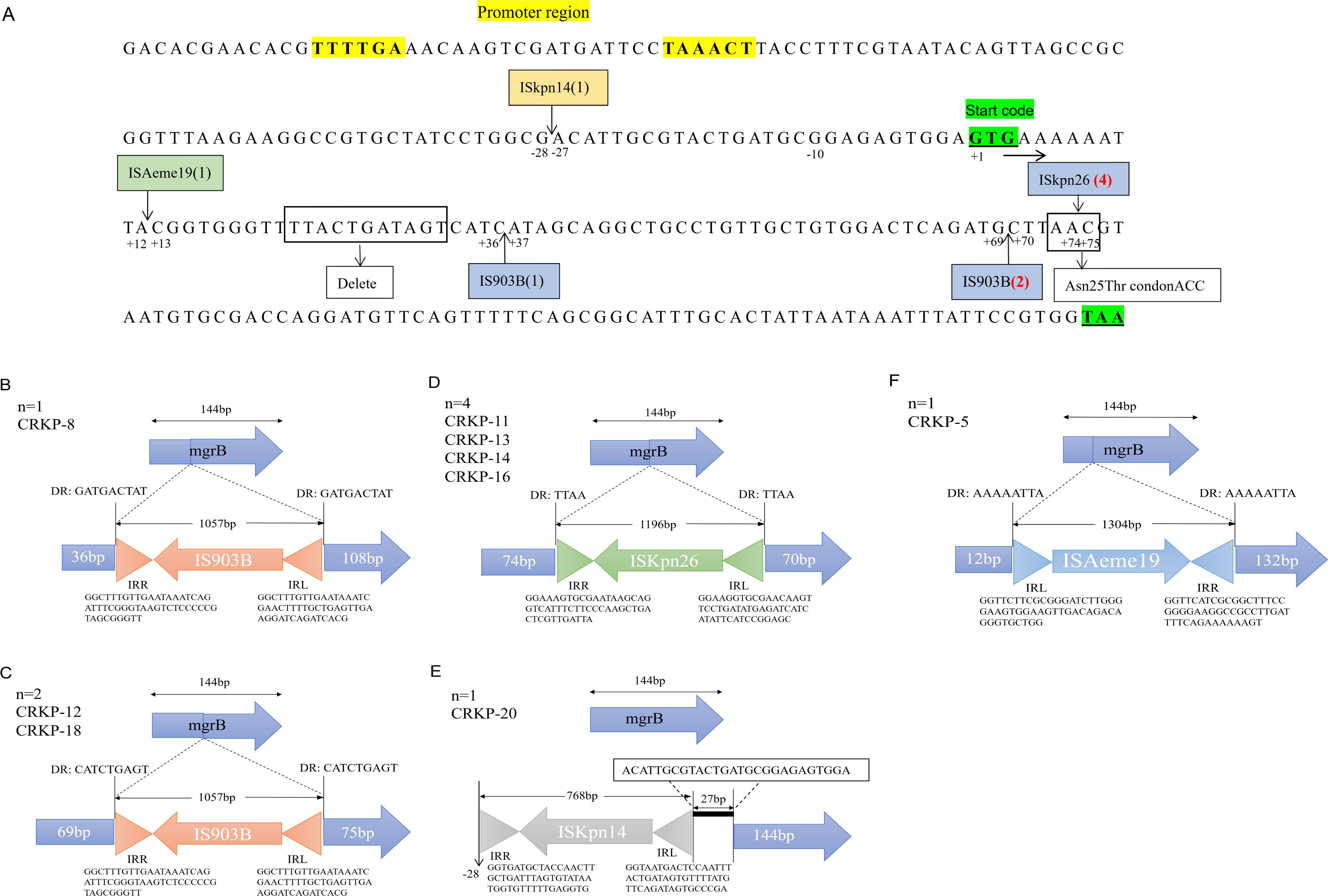
Figure 2. Alterations in the mgrB gene of polymyxin-resistant CRKP isolates. (A) Arrows indicate the positions of insertion mediate by IS elements. The number of insertions for each IS is given in parentheses. IS series are highlighted in different colors: blue, IS5; green, ISL3; orange, IS1. Boldsequences highlight the promoter region (without underline) and the start and stopcodons (both underlined). (B) One isolate with the IS903B insertion at position + 36. (C) Four isolates harboring the ISKpn26 insertion had insertions at position + 74. (D) Two isolates with the IS903B insertion at position + 69. (E) One had an insertion at -28 bp, located in the promoter region upstream of the start codon. (F) One isolate with the ISAeme19 insertion at position + 12. DR: direct repeat sequences; IRL/IRR: left/right inverted repeats (triangles).
To assess the functional impact of these genetic alterations, we measured the expression of pmrC and pmrK (components of the pmrHFIJKLM operon), which are key genes involved in LPS modification by adding PEtN and L-Ara4N. Compared with K. pneumoniae ATCC BAA-1705, 38.1% (n=8/21) of the isolates showed increased pmrC expression (1.29- to 29.86-fold) (Figure 3A), while 95.2% (n=20/21) exhibited upregulation of pmrK (5.89- to 104.85-fold) (Figure 3B).
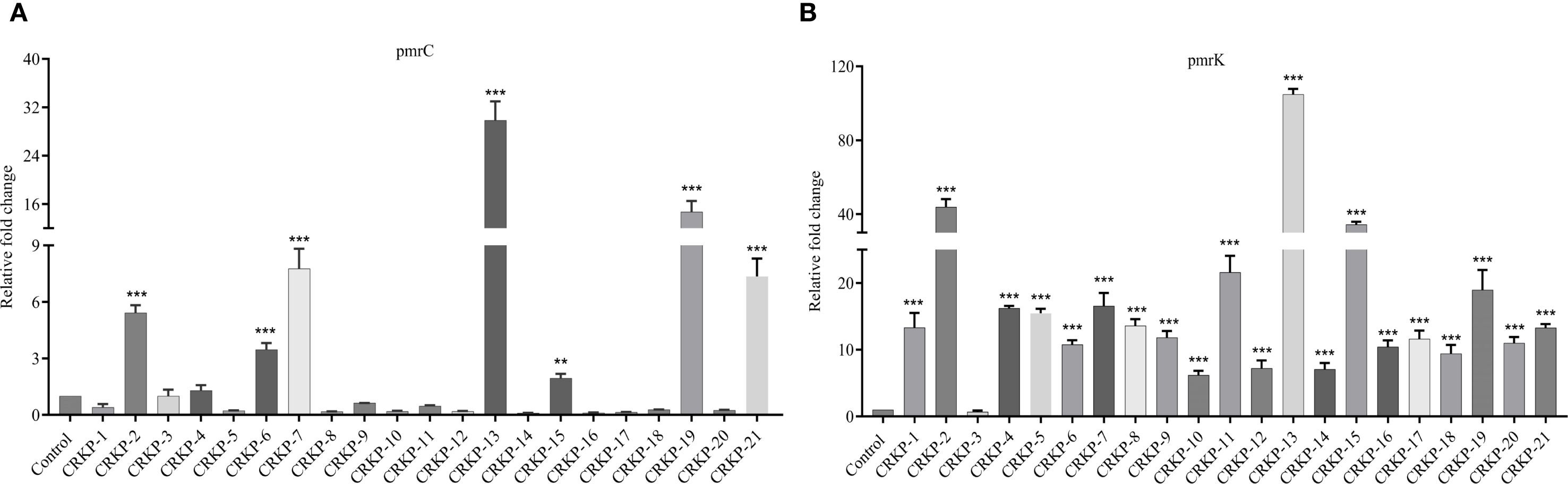
Figure 3. Expression analysis of LPS modification genes. Relative expressions of the pmrC (A) and pmrK (B) genes in polymyxin-resistant CRKP. Expression was normalized to rpsL using the susceptible K. pneumoniae ATCC BAA-1705 as control (expression = 1). (*P <0.05; **P < 0.01; ***P < 0.001).
3.3.2 Plasmid-mediated resistance and conjugation assay of mcr in E. coli
All PR-CRECO isolates exhibiting low-level resistance harbored the mcr-1 gene (Figure 1C). Notably, these isolates possessed the wild-type mgrB gene. PROVEAN analysis indicated that all amino acid substitutions in pmrAB and phoPQ were neutral (Supplementary Table 5). Conjugation assays confirmed the successful transfer of mcr-1-bearing IncI2 and IncHI2 plasmids from five isolates (CRECO-3, CRECO-4, CRECO-5, CRECO-6, and CRECO-7) to E. coli EC600, resulting in 4-fold to 8-fold increases in polymyxin MICs among transconjugants (Table 3). These findings suggest that IncHI2 and IncI2 plasmids are key vectors for the dissemination of mcr-1 in PR-CRECO.
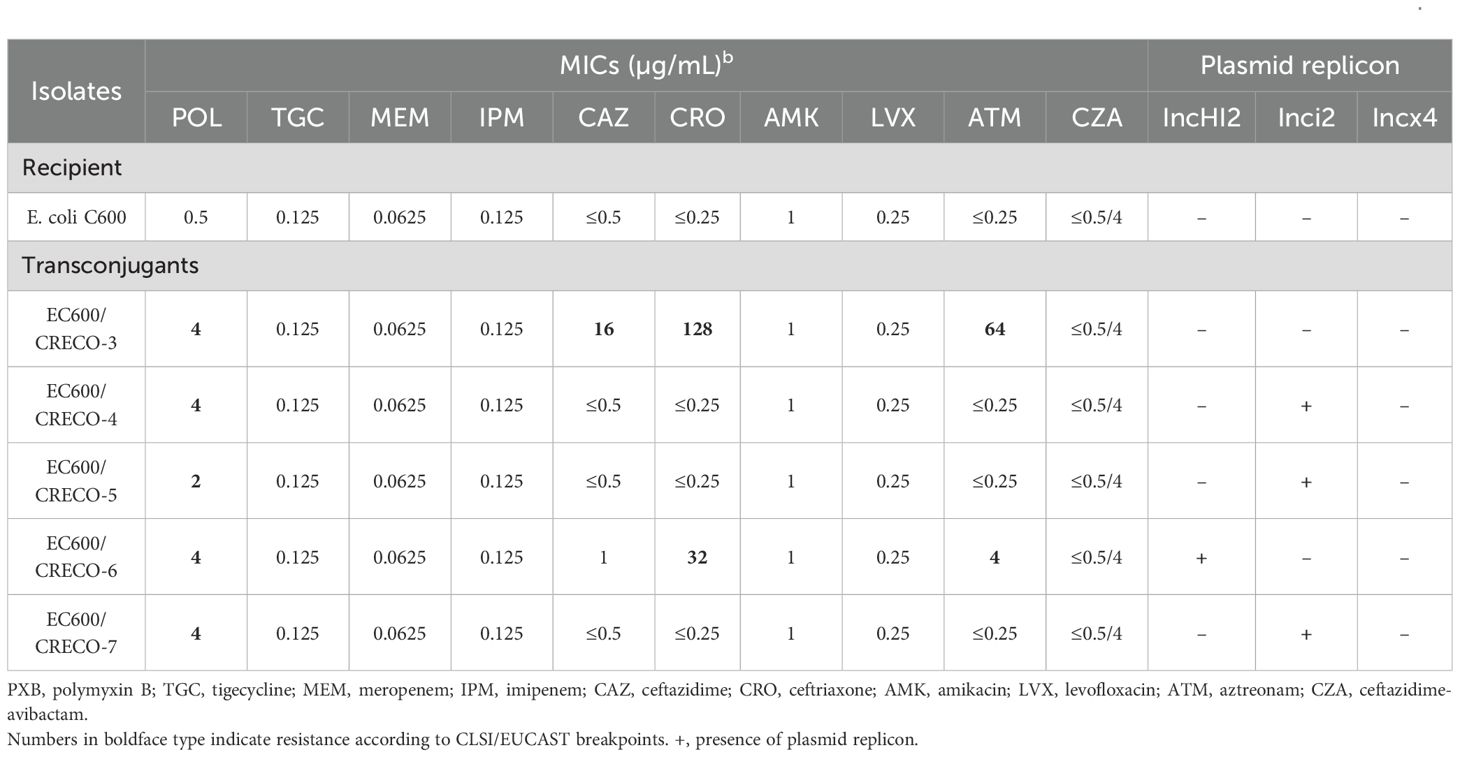
Table 3. Antimicrobial susceptibility profiles and plasmid replicon of recipient and transconjugants that received the polymyxin resistance gene mcr-1.
3.4 Genomic features of ISAeme19-mediated transposition and plasmid-chromosome exchange in CRKP-5
The CRKP-5 (ST11, KL64) isolate harbored a 5.49-Mb chromosome and six plasmids (5–195 kb), mainly including one virulence plasmid (pkp2007-VIR) and two multiple resistance plasmids (pkp2007-KPC and pkp2007-C) (Supplementary Table 6). The 195-kb IncHI1B(pNDM-MAR)/repB virulence plasmid pkp2007-VIR showed >99.9% identity to the prototypical pLVPK reference, carrying the aerobactin biosynthesis cluster (iucABCD-iutA) and mucoid phenotype regulators (rmpA, rmpA2) while lacking conjugation-related genes (Supplementary Figure 1A). The IncFII(pHN7A8)/IncR plasmid pkp2007-KPC closely resembled many published IncFII(pHN7A8)/IncR-type plasmids. Genetic environment analysis identified three multidrug resistance regions within this plasmid, which were composed of four IS26 elements: (i) Tn3-IS26-blaCTX-M-65-IS903B; (ii) ISAeme19-rmtB-blaTEM-1B-Tn3-IS26; (iii) IS26-blaSHV-12-TnAs1-ISKpn6-blaKPC-2-ISKpn27-Tn3-IS26 (Figure 4B). The IncFII (pCRY) plasmid pkp2007-C backbone exhibited high conservation. It harbored two fused resistance regions (dfrA14-sul2-tetA-catA2-qnrS1-blaLAP-2) with complete modules characteristic of self-transmissible plasmids (Supplementary Figure 1B).
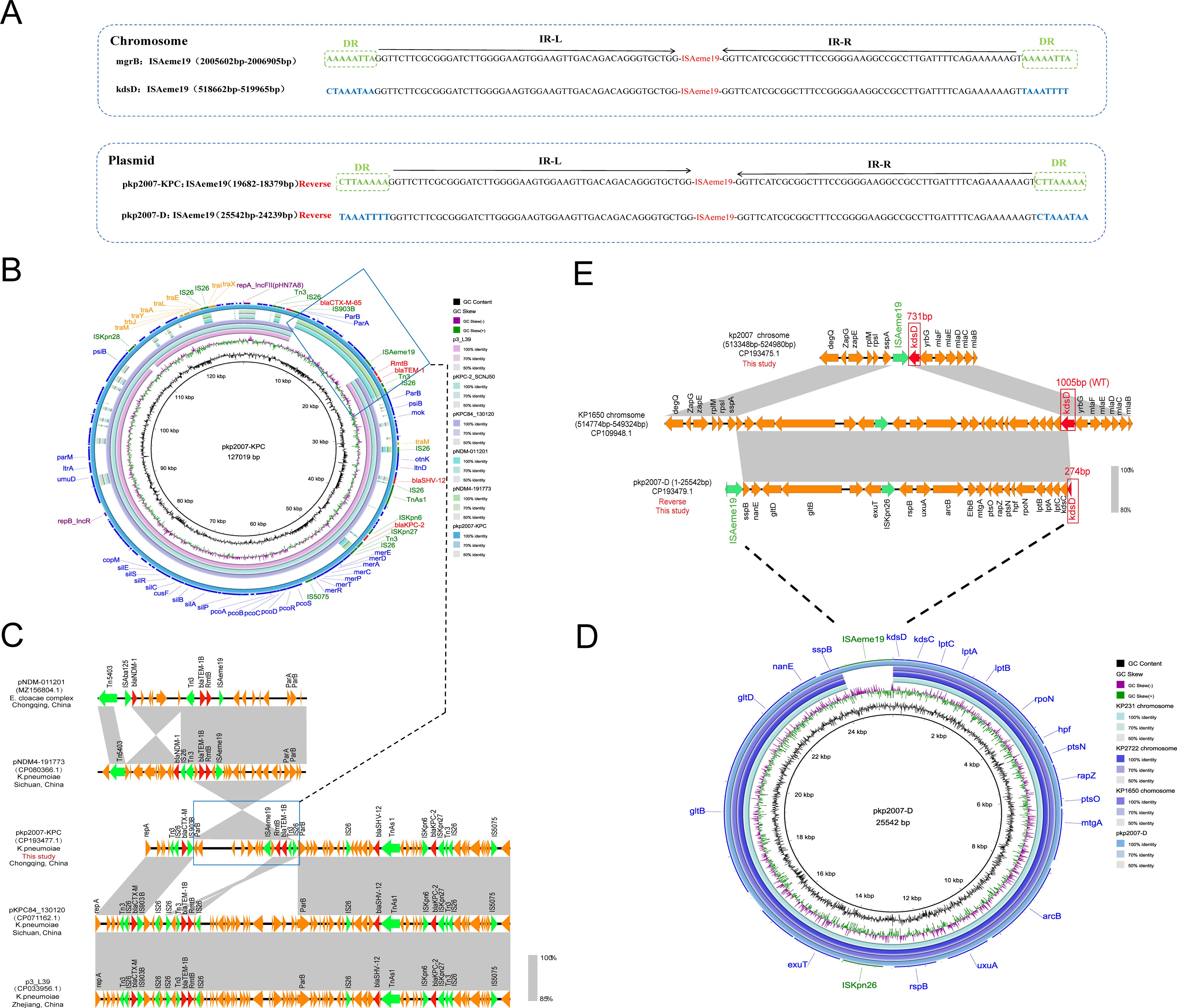
Figure 4. Genomic architecture and dynamics of ISAeme19 in CRKP-5. (A) Structural organization of ISAeme19 elements. Each element contains 19-bp imperfect terminal inverted repeats (IR-L/R). Chromosomal copies (inserted in mgrB and kdsD) and plasmid copies (pkp2007-KPC, pkp2007-D) exhibit inverted orientations. Flanking sequences: 8-bp direct repeats (DRs) at mgrB and pkp2007-KPC sites vs. AT-rich regions at kdsD and pkp2007-D sites. Resistance genes (red), insertion sequences (green). (B) Circular map of plasmid pkp2007-KPC. BLASTn alignment reveals >99% identity with epidemic IncFII(pHN7A8)/IncR plasmids: pKPC84_130120 (Sichuan, China, CP071162.1), p3_L39 (Zhejiang, China, CP033956.1), and pKPC_SCNJ50 (Hunan, China, PP746481.1). Resistance genes (red arrows), insertion sequences (green arrows). (C) Homology analysis of the variable resistance region (ISAeme19-RmtB-blaTEM-1B-Tn3-IS26) showing 99% coverage and 99.95% identity with blaNDM-harboring plasmids: pNDM-011201 (Chongqing, China, MZ156804.1) from the Enterobacter cloacae complex and pNDM4-19773 (Sichuan, China, CP080366.1) from ST16 Klebsiella pneumonia. Resistance genes are portrayed by red arrows. Green arrows indicate insertion sequences. (D) Formation mechanism of plasmid pkp2007-D via ISAeme19-mediated excision of a 24-kb chromosomal segment. The excised region shares >99% identity with homologous segments in K. pneumoniae chromosomes KP1650/KP231/KP2722. (E) Sequence alignment analysis revealed that compared with KP1650 chromosome harboring the wild type kdsD gene (1005bp) between 514774bp and 549324bp, chromosome sequence of the kp2007 strain between 518657 bp and 518658 bp had a large fragment deletion and carried the truncated kdsD gene (731bp), which overlaps the gene fragments in pkp2007-D containing the deletion fragment (274bp) of the kdsD gene on the chromosome sequence of the kp2007. Mobile genetic elements (green), truncated/WT kdsD (red).
3.4.1 Distribution and plasmid-chromosome transposition of ISAeme19
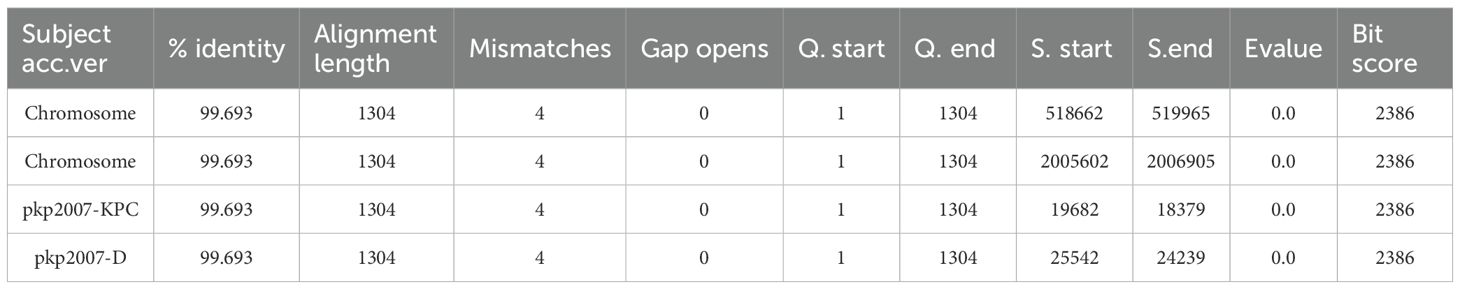
Sequence comparison identified that CRKP-5 harbored four identical ISAeme19 copies: two on the chromosome (mgrB and kdsD), and two on plasmids (IncFII/IncR plasmid pkp2007-KPC and plasmid pkp2007-D), suggesting the active transposition capability of the ISAeme19 element (Figure 4A; Table 4). Crucially, the ISAeme19 element disrupting mgrB exhibited 100% sequence identity among the 1304 nucleotides, inverted orientation and identical 8-bp flanking direct repeats (DRs) to its counterpart on the multidrug resistance plasmid pkp2007-KPC, providing strong evidence for plasmid-to-chromosome transposition (Figure 4A). Evolutionary analysis of pkp2007-KPC further revealed inversion of a composite transposon (ISAeme19-rmtB-blaTEM-1B-Tn3-IS26) relative to homologous blaNDM-harboring plasmids with inverted orientation and same 8-bp DRs on both sides of ISAeme19, including pNDM-011201 (MZ156804.1, Enterobacter cloacae complex, Chongqing) and pNDM4-19773 (CP080366.1, Klebsiella pneumoniae, Sichuan) (Figure 4B; Figure 4C). These findings indicated that ISAeme19 may be inserted on pkp2007-KPC plasmid prior to its integration into the chromosome.
3.4.2 Chromosomal excision and plasmid formation mediated by ISAeme19
The pkp2007-D was a 25,542 bp plasmid, containing 24 open reading frames, which did not contain any resistance genes and plasmid replicons. Genomic alignment confirmed pkp2007-D originated from the precise excision and recircularization of a 24-kb chromosomal segment (coordinates 518,657–542,201 bp) flanked by ISAeme19 elements, encompassing the truncated kdsD locus and adjacent genes (Figure 4D; Figure 4E), providing mechanistic evidence for ISAeme19-driven excision and recircularization of chromosomal DNA into a stable episomal form lacking autonomous replication functions.
3.5 Genomic features of a fusion plasmid with dual blaKPC-2 copies and novel mutations in pan-resistant CRKP-1
To characterize polymyxin-tigecycline co-resistance, the pan-resistant CRKP-1 strain (ST11, KL64) was selected for WGS. A comprehensive summary of antimicrobial resistance genes and virulence-associated genes is presented in Supplementary Table 7.
We identified that pan-resistant CRKP-1 strain harbored a large fusion plasmid pkp2020 (287,004 bp). BLASTn analysis revealed near-identical regions in pkp2020: the IncFIIpHN7A8/IncR region shared 99.9–100.0% identity and 49.0–58.0% query coverage with plasmids pKPC-2_SCNJ50 (PP746481.1), p3_L39 (CP033956.1), and pZHKPC-1 (OM928502.1), while the IncFIB(K) segment exhibited 99.9–100.0% identity and 62.0% query coverage with plasmids pSH2-KPC (MH643791.1) and pWYKP586-1 (OQ801413.1) (Figure 5A). Sequence alignment indicated pkp2020 was a fusion product through recombination (Figure 5B). Crucially, pkp2020 contained an additional 15,048 bp multidrug resistance unit carrying blaKPC-2. Two direct-repeat blaKPC-2 copies resided 14,349 bp apart within the IncFIIpHN7A8/IncR scaffold (Figure 5C).
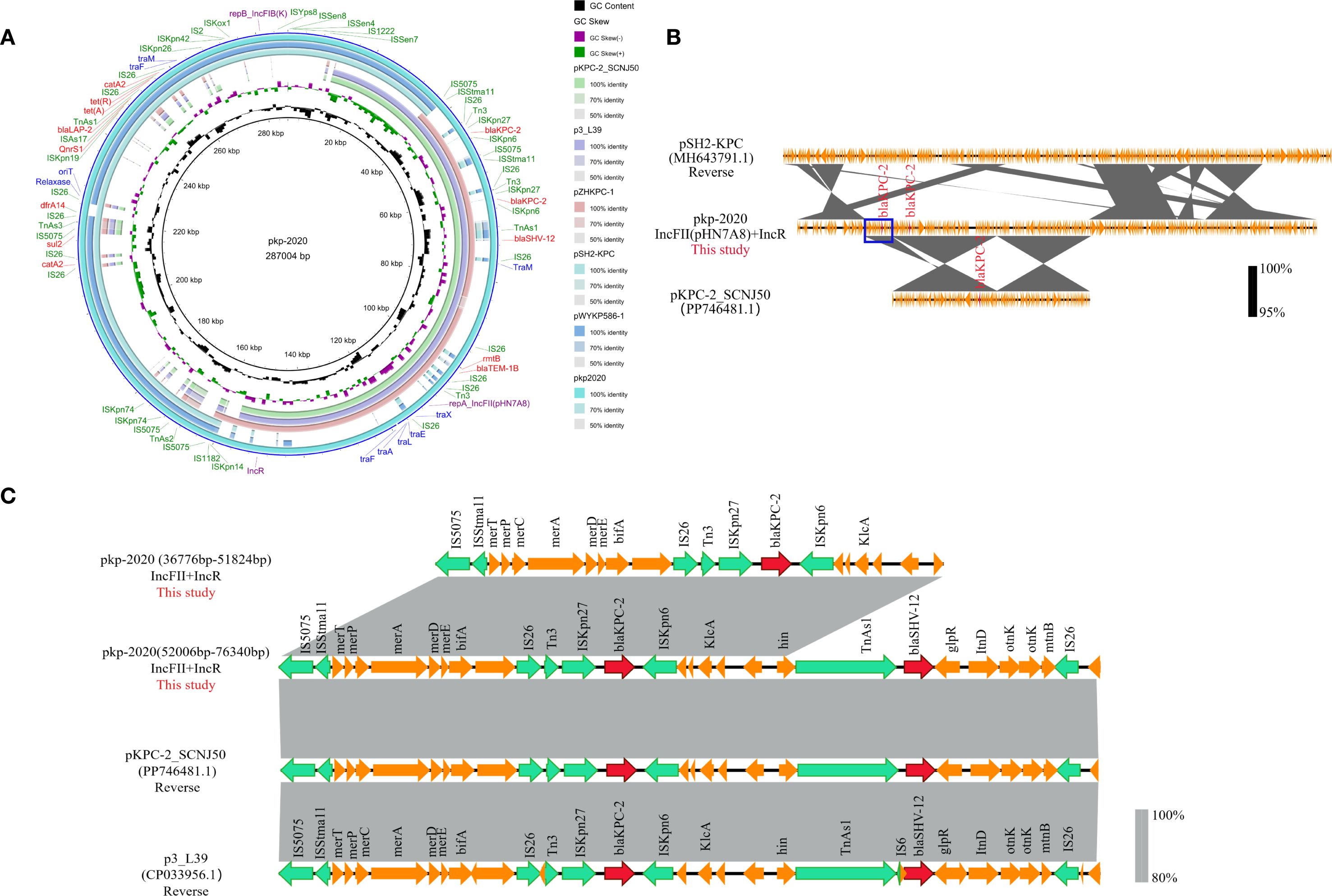
Figure 5. Genomic features of the fusion plasmid pKP2020 from CRKP-1. (A) Circular maps showing resistance genes (red), insertion sequences (green), replicons (purple), and conjugative modules (blue). (B) BLAST alignment of fusion plasmid pKP2020 harboring dual blaKPC-2 copies with related plasmids: pSH2-KPC and pKPC-2_SCNJ50. (C) Comparison of the regions harboring dual blaKPC-2 copies in pKP2020, pKPC-2_SCNJ50, and p3_L39. ORFs are colored: resistance (red), mobile elements (green), other accessory genes (yellow).
In addition, this fusion plasmid carried 47 IS elements and 12 resistance genes, especially IS26 (n=12/47), which mediates the insertion or recombination of resistance genes. Genetic environment analysis identified four multidrug resistance regions within this plasmid, composed of eleven IS26 elements: (i) IS26-Tn3-ISKpn27-blaKPC-2-ISKpn6-ORF-IS26-Tn3-ISKpn27-blaKPC-2-ISKpn6-ORF-TnAs1-blaSHV-12-ORF-IS26; (ii) IS26-rmtB-blaTEM-1B-IS26; (iii) IS26-catA2-IS26-sul2-IS5075-TnAs3-IS26-dfrA14-IS26; (iv) ISKpn19-qnrS1-ISAs17-blaLAP-2-TnAs1-tetA-tetR-IS26-catA2-IS26 (Figure 5A). Notably, pkp2020 lacked part of the tra gene region due to IS26-mediated truncation of plasmid conjugation genes during evolution.
Compared with the reference sequences from E. coli plasmid RP1 (X00006) for tet(A) and K. pneumoniae MGH78578 (CP000647) for other genes, CRKP-1 harbored tet(A) variants with Type 1 substitutions (I5R, V55M, I75V, T84A, S201A, F202S, V203F), ISKpn26-mediated acrR inactivation, and unreported RamR mutations (Ala17Thr and *194Lys). PROVEAN predicted Ala17Thr as deleterious (Supplementary Table 8).
4 Discussion
Our findings reveal a striking divergence in polymyxin resistance trajectories: while K. pneumoniae evolved high-level resistance predominantly through chromosomal mutations (particularly ISKpn26/IS903B/ISAeme19/ISKpn14-mediated mgrB inactivation) and the LPS modification gene (particularly pmrK) upregulation, E. coli evolved low-level resistance that relied exclusively on horizontally acquired mcr-1. Furthermore, the near-universal upregulation of pmrK (95.2% of isolates) versus pmrC (38.1% of isolates) in PR-CRKP isolates suggests that L-Ara4N addition is the dominant mechanism, with pEtN modification providing synergistic enhancement in a subset of CRKP strains. Previous studies have demonstrated that the Leu26Pro mutation in PhoQ (Cheng et al., 2015), Asp191Tyr mutation in PhoP (Jayol et al., 2015), and Thr157Pro mutation in pmrB (Jayol et al., 2014) were associated with polymyxin resistance. In our study, previously unreported deleterious substitutions (mgrB-Asn25Thr, phoP-Lys199Met, phoQ-Tyr89His, and RamR-Ala17Thr) expand the known mutational repertoire. These findings align with established evidence that chromosome-mediated mechanism is a primary cause of polymyxin resistance (Cannatelli et al., 2014; Zhu et al., 2023), and that polymyxin resistance likely occurs through increased expression of pmrCAB and pmrHFIJKLM operons, leading to lipid A modification (Nang et al., 2021).
This dichotomy underscores two distinct molecular epidemiological characteristics. First, the predominance of ST11-KL64 PR-CRKP aligns with its role as a major reservoir of carbapenem resistance in China (Liu et al., 2023), which was accelerated by IS element-mediated mgrB disruption (particularly ISKpn26) under polymyxin selection pressure. Clonal transmission was confirmed by four genetically identical isolates from ICU patients, indicating persistent intra-hospital transmission despite infection controls. Second, plasmid-driven dissemination of mcr-1 via IncHI2/IncI2 vectors occurred in genetically diverse E. coli isolates, with 55.6% conjugation efficiency underscoring the ongoing risk of horizontal transfer in community settings (Wang et al., 2017). This epidemiological bifurcation necessitates tailored infection control strategies: genomic surveillance should prioritize ST11-KL64 containment in ICUs while monitoring mcr-plasmid mobility.
Prior studies have associated virulence-resistance hybrids with elevated mortality in China (Hu et al., 2024b). In our study, two of six patients who experienced fatal outcomes were infected by ST11-KL64 strains that exhibited both high-level polymyxin resistance (MIC ≥ 16 μg/mL) and carried hypervirulence determinants. This suggests that the synergy between virulence and resistance contributes to unfavorable outcomes. Furthermore, two of three pan-resistant strains (CRKP-8, CRKP-12) were associated with treatment failure. These findings highlight an evolutionary trajectory toward untreatable infections, necessitating enhanced screening for virulence-resistance hybrids and pan-resistant strains in high-risk settings.
A recent study showed that the dissemination of ISKpn25 (ISL3 family), which inserts into the mgrB gene at a specific site on pKpQIL plasmids, could explain the polymyxin resistance observed in clonally unrelated isolates harboring identical mgrB mutations (Giordano et al., 2018). ISAeme19 (ISL3 family) is frequently located on plasmids carrying blaNDM (Zhang et al., 2022; Sy et al., 2024). Although ISAeme19-mediated mgrB inactivation has previously been reported only in ST16 Klebsiella pneumoniae from Southeast Asia (Sy et al., 2024; Nguyen et al., 2021), here we report the first case of ISAeme19-mediated polymyxin resistance in an epidemic ST11-KL64 hypervirulent CRKP strain, involving a dual-targeting mechanism (disrupting both mgrB and kdsD) and a plasmid-chromosome recombination event. Critically, kdsD perturbation may compromise lipopolysaccharide core biosynthesis (Gunn and Ernst, 2007), potentially synergizing with mgrB-dysregulated lipid A modifications to establish a dual barrier against polymyxin. Given ISAeme19’s transposition activity evidenced by identical 8-bp DRs flanking and inverted orientation of ISAeme19 between plasmid pkp2007-KPC and chromosomal mgrB, we propose that carbapenemase plasmids serve as Trojan horses for polymyxin resistance determinants in high-risk clones. This mechanism likely originated from blaNDM-bearing plasmids circulating in Southwest China (Figure 4C). Notably, pkp2007-KPC is an epidemic IncFII(pHN7A8)/IncR plasmid harboring blaKPC-2, indicating that ISAeme19 may co-disseminate with carbapenemase genes. Polymyxin therapy likely accelerates this process, as suggested by the emergence of resistance following treatment in our clinical case.
The fusion plasmid pKP2020 in pan-resistant ST11-KL64 CRKP-1 exemplifies alarming resistance consolidation. This fusion plasmid harbored duplicated blaKPC-2 and catA2 copies bracketed by IS26. This unreported structure may amplify resistance through gene dosage effects during β-lactam exposure, potentially compromising ceftazidime/avibactam efficacy (Hu et al., 2024a). Concurrently, IS26 recombinogenic hotspots facilitate modular acquisition of resistance genes (e.g., catA2, rmtB, and qnrS1), representing a critical evolutionary pathway for last-line antibiotic failure (He et al., 2015; Feng et al., 2019). Moreover, chromosomal mutations in CRKP-1, including the novel deleterious RamR-Ala17Thr substitution that likely dysregulates efflux pumps, synergized with plasmid-borne resistance, culminating in pan-drug resistance.
This study has several limitations. First, the small sample size of bacterial isolates necessitates expanded regional surveillance to validate the epidemiological patterns. Second, although the observed chromosomal mutations were associated with polymyxin resistance, their mechanistic roles require further functional validation.
5 Conclusions
This study reveals species-divergent resistance mechanisms to polymyxin. The persistence of mcr-1 plasmids in diverse E. coli backgrounds highlights the ongoing risk of horizontal resistance spread. Meanwhile, the dominance of ST11-KL64 K. pneumoniae reliant on chromosomal adaptations and the emergence of novel genetic mechanisms (ISAeme19 transposition, fusion plasmids) underscore the continuous evolution of high-risk clones. While ceftazidime/avibactam retained activity against most PR-CRKP isolates, its vulnerability to porin mutations and β-lactamase amplification (e.g., dual blaKPC-2 copies in pkp2020) necessitates ongoing vigilance. Future studies should screen for ISAeme19-like elements in high-risk plasmids and strengthen surveillance for these high-risk clones, mobile elements, and emergent resistance mechanisms to curb the spread of polymyxin-resistant CRE.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material. The complete genome sequences of K. pneumoniae strains CRKP-1 and CRKP-5 have been deposited in the NCBI database under BioProject accession numbers PRJNA1286129 and PRJNA1268731.
Author contributions
TS: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. HL: Data curation, Methodology, Writing – review & editing. PX: Conceptualization, Investigation, Software, Writing – review & editing. NH: Data curation, Methodology, Writing – review & editing. ST: Formal Analysis, Investigation, Methodology, Writing – review & editing. YQL: Conceptualization, Investigation, Validation, Writing – review & editing. QH: Data curation, Methodology, Writing – review & editing. YS: Conceptualization, Investigation, Writing – review & editing. YYL: Data curation, Methodology, Writing – review & editing. YX: Funding acquisition, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1678719/full#supplementary-material
Supplementary Figure 1 | Comparative analysis of pkp2007-VIR and pkp2007-C with other similar plasmids in CRKP-5. Circular maps and genomic analysis of all insertion sequences distribution and antibiotic resistance genes. (A) BLASTn analysis showed that pkp2007-VIR backbone was similar to the classic virulence plasmid pLVPK,with over 99.94% nucleotide identity and 99% coverage ((e.g. CP103316.1, CP033955.1, CP054769.1 and CP146191.1). Virulence genes are indicated in red. Green arrows indicate insertion sequences. (B) BLASTn analysis showed that pkp2007-C was almost identical (100% query coverage, over 99.97% identity) to the plasmids pKPTCM2-3 (Zhejiang, China, CP118694.1), pKP12_4 (Zhejiang, China, CP082768.1), pHSKP43-3 (Shanghai, China, CP100101.1) and pHSKP8-3 (Shanghai, China, CP100090.1). ORFs encoding resistance genes are portrayed by red arrows. Green arrows indicate insertion sequences.
References
Binsker, U., Käsbohrer, A., and Hammerl, J. A. (2021). Global colistin use: a review of the emergence of resistant Enterobacterales and the impact on their genetic basis. FEMS Microbiol. Rev. 46, fuab049. doi: 10.1093/femsre/fuab049
Cannatelli, A., Giani, T., D’Andrea, M. M., di Pilato, V., Arena, F., Conte, V., et al. (2014). MgrB inactivation is a common mechanism of colistin resistance in KPC-producing klebsiella pneumoniae of clinical origin. Antimicrob. Agents Chemother. 58, 5696–5703. doi: 10.1128/aac.03110-14
Carattoli, A., Zankari, E., García-Fernández, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cheng, Y.-H., Lin, T.-L., Pan, Y.-J., Wang, Y.-P., Lin, Y.-T., and Wang, J.-T. (2015). Colistin resistance mechanisms in klebsiella pneumoniae strains from Taiwan. Antimicrob. Agents Chemother. 59, 2909–2913. doi: 10.1128/AAC.04763-14
Delcher, A. L., Bratke, K. A., Powers, E. C., and Salzberg, S. L. (2007). Identifying bacterial genes and endosymbiont DNA with Glimmer. Bioinforma. Oxf. Engl. 23, 673–679. doi: 10.1093/bioinformatics/btm009
El-Sayed Ahmed, M. A. E.-G., Zhong, L.-L., Shen, C., Yang, Y., Doi, Y., and Tian, G.-B. (2020). Colistin and its role in the Era of antibiotic resistance: an extended review, (2000–2019). Emerg. Microbes Infect. 9, 868–885. doi: 10.1080/22221751.2020.1754133
Feng, Y., Liu, L., McNally, A., and Zong, Z. (2019). Coexistence of three blaKPC-2 genes on an IncF/IncR plasmid in ST11 Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 17, 90–93. doi: 10.1016/j.jgar.2018.11.017
Giordano, C., Barnini, S., Tsioutis, C., Chlebowicz, M. A., Scoulica, E. V., Gikas, A., et al. (2018). Expansion of KPC-producing Klebsiella pneumoniae with various mgrB mutations giving rise to colistin resistance: the role of ISL3 on plasmids. Int. J. Antimicrob. Agents 51, 260–265. doi: 10.1016/j.ijantimicag.2017.10.011
Gunn, J. S. and Ernst, R. K. (2007). The structure and function of francisella lipopolysaccharide. Ann. N. Y. Acad. Sci. 1105, 202–218. doi: 10.1196/annals.1409.006
He, S., Hickman, A. B., Varani, A. M., Siguier, P., Chandler, M., Dekker, J. P., et al. (2015). Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6, e00762–e00715. doi: 10.1128/mBio.00762-15
Hu, F., Pan, Y., Li, H., Han, R., Liu, X., Ma, R., et al. (2024b). Carbapenem-resistant Klebsiella pneumoniae capsular types, antibiotic resistance and virulence factors in China: a longitudinal, multi-centre study. Nat. Microbiol. 9, 814–829. doi: 10.1038/s41564-024-01612-1
Hu, D., Wang, S., Xu, M., Zhang, J., Luo, X., Zhou, W., et al. (2024a). Double blaKPC-2 copies quadrupled minimum inhibitory concentration of ceftazidime-avibactam in hospital-derived Klebsiella pneumoniae. Microbiol. Spectr. 12, e0033124. doi: 10.1128/spectrum.00331-24
Huang, H., Dong, N., Shu, L., Lu, J., Sun, Q., Chan, E. W.-C., et al. (2020). Colistin-resistance gene mcr in clinical carbapenem-resistant Enterobacteriaceae strains in China–2019. Emerg. Microbes Infect. 9, 237–245. doi: 10.1080/22221751.2020.1717380
Jayol, A., Nordmann, P., Brink, A., and Poirel, L. (2015). Heteroresistance to colistin in Klebsiella pneumoniae associated with alterations in the PhoPQ regulatory system. Antimicrob. Agents Chemother. 59, 2780–2784. doi: 10.1128/AAC.05055-14
Jayol, A., Poirel, L., Brink, A., Villegas, M.-V., Yilmaz, M., and Nordmann, P. (2014). Resistance to Colistin Associated with a Single Amino Acid Change in Protein PmrB among Klebsiella pneumoniae Isolates of Worldwide Origin. Antimicrob. Agents Chemother. 58, 4762–4766. doi: 10.1128/aac.00084-14
Li, X., Xie, Y., Liu, M., Tai, C., Sun, J., Deng, Z., et al. (2018). oriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46, W229–W234. doi: 10.1093/nar/gky352
Liu, J.-H., Liu, Y.-Y., Shen, Y.-B., Yang, J., Walsh, T. R., Wang, Y., et al. (2024). Plasmid-mediated colistin-resistance genes: mcr. Trends Microbiol. 32, 365–378. doi: 10.1016/j.tim.2023.10.006
Liu, L., Lou, N., Liang, Q., Xiao, W., Teng, G., Ma, J., et al. (2023). Chasing the landscape for intrahospital transmission and evolution of hypervirulent carbapenem-resistant Klebsiella pneumoniae. Sci. Bull. 68, 3027–3047. doi: 10.1016/j.scib.2023.10.038
Liu, X., Wu, Y., Zhu, Y., Jia, P., Li, X., Jia, X., et al. (2022b). Emergence of colistin-resistant hypervirulent Klebsiella pneumoniae (CoR-HvKp) in China. Emerg. Microbes Infect. 11, 648–661. doi: 10.1080/22221751.2022.2036078
Liu, B., Zheng, D., Zhou, S., Chen, L., and Yang, J. (2022a). VFDB 2022: a general classification scheme for bacterial virulence factors. Nucleic Acids Res. 50, D912–D917. doi: 10.1093/nar/gkab1107
Mmatli, M., Mbelle, N. M., and Osei Sekyere, J. (2022). Global epidemiology, genetic environment, risk factors and therapeutic prospects of mcr genes: A current and emerging update. Front. Cell. Infect. Microbiol. 12. doi: 10.3389/fcimb.2022.941358
Nang, S. C., Azad, M. A. K., Velkov, T., Zhou, Q., and Li, J. (2021). Rescuing the last-line polymyxins: achievements and challenges. Pharmacol. Rev. 73, 679–728. doi: 10.1124/pharmrev.120.000020
Nguyen, T. N. T., Nguyen, P. L. N., Le, N. T. Q., Nguyen, L. P. H., Duong, T. B., Ho, N. D. T., et al. (2021). Emerging carbapenem-resistant Klebsiella pneumoniae sequence type 16 causing multiple outbreaks in a tertiary hospital in southern Vietnam. Microb. Genomics 7, mgen000519. doi: 10.1099/mgen.0.000519
Puljko, A., Barišić, I., Dekić Rozman, S., Križanović, S., Babić, I., Jelić, M., et al. (2024). Molecular epidemiology and mechanisms of carbapenem and colistin resistance in Klebsiella and other Enterobacterales from treated wastewater in Croatia. Environ. Int. 185, 108554. doi: 10.1016/j.envint.2024.108554
Rodríguez-Santiago, J., Cornejo-Juárez, P., Silva-Sánchez, J., and Garza-Ramos, U. (2021). Polymyxin resistance in Enterobacterales: overview and epidemiology in the Americas. Int. J. Antimicrob. Agents 58, 106426. doi: 10.1016/j.ijantimicag.2021.106426
Sameni, F., Ghazi, M., Dadashi, M., Bostanshirin, N., Al-Dahmoshi, H. O. M., Khosravi-Dehaghi, N., et al. (2022). Global distribution, genotypes and prevalent sequence types of colistin-resistant Klebsiella pneumoniae isolated from clinical samples; A systematic review. Gene Rep. 28, 101635. doi: 10.1016/j.genrep.2022.101635
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stewardson, A. J., Marimuthu, K., Sengupta, S., Allignol, A., El-Bouseary, M., Carvalho, M. J., et al. (2019). Effect of carbapenem resistance on outcomes of bloodstream infection caused by Enterobacteriaceae in low-income and middle-income countries (PANORAMA): a multinational prospective cohort study. Lancet Infect. Dis. 19, 601–610. doi: 10.1016/S1473-3099(18)30792-8
Sy, B. T., Boutin, S., Kieu Linh, L. T., Weikert-Asbeck, S., Eger, E., Hauswaldt, S., et al. (2024). Heterogeneity of colistin resistance mechanism in clonal populations of carbapenem-resistant Klebsiella pneumoniae in Vietnam. Lancet Reg. Health West. Pac. 51, 101204. doi: 10.1016/j.lanwpc.2024.101204
van Duin, D., Arias, C. A., Komarow, L., Chen, L., Hanson, B. M., Weston, G., et al. (2020). Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the (CRACKLE-2): a prospective cohort study. Lancet Infect. Dis. 20, 731–741. doi: 10.1016/S1473-3099(19)30755-8
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PloS One 9, e112963. doi: 10.1371/journal.pone.0112963
Wang, X., Liu, Y., Qi, X., Wang, R., Jin, L., Zhao, M., et al. (2017). Molecular epidemiology of colistin-resistant Enterobacteriaceae in inpatient and avian isolates from China: high prevalence of mcr-negative Klebsiella pneumoniae. Int. J. Antimicrob. Agents 50, 536–541. doi: 10.1016/j.ijantimicag.2017.05.009
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PloS Comput. Biol. 13, e1005595. doi: 10.1371/journal.pcbi.1005595
Xie, M., Zhang, Y., Chen, K., Dong, N., Zhou, H., Huang, Y., et al. (2025). Increasing polymyxin resistance in clinical carbapenem-resistant Klebsiella pneumoniae strains in China between 2000 and 2023. Commun. Med. 5, 73. doi: 10.1038/s43856-025-00748-3
Zhang, B., Hu, R., Liang, Q., Liang, S., Li, Q., Bai, J., et al. (2022). Comparison of two distinct subpopulations of klebsiella pneumoniae ST16 co-occurring in a single patient. Microbiol. Spectr. 10, e0262421. doi: 10.1128/spectrum.02624-21
Keywords: carbapenem-resistant Enterobacteriaceae, polymyxin resistance, mgrB inactivation, MCR-1, fusion plasmid, clonal transmission
Citation: Si T, Liu H, Xia P, Huang N, Tang S, Li Y, Han Q, Song Y, Liu Y and Xia Y (2025) Clonal transmission and species-specific mechanisms of polymyxin resistance in carbapenem-resistant Enterobacteriaceae from Southwest China. Front. Cell. Infect. Microbiol. 15:1678719. doi: 10.3389/fcimb.2025.1678719
Received: 03 August 2025; Accepted: 29 September 2025;
Published: 13 October 2025.
Edited by:
Jaroslav Hrabak, Charles University, CzechiaReviewed by:
Trojan Rugira, Yale University, United StatesQi Wang, Peking University People’s Hospital, China
Yujiao Wang, The First Affiliated Hospital of Shandong First Medical University, China
Copyright © 2025 Si, Liu, Xia, Huang, Tang, Li, Han, Song, Liu and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yun Xia, eGlheXVuMTJjbkBhbGl5dW4uY29t
 Tingting Si1
Tingting Si1 Hang Liu
Hang Liu Na Huang
Na Huang Yuqiong Li
Yuqiong Li Yun Xia
Yun Xia