- Department of Cell Biology and Biochemistry, Texas Tech University Health Science Center, Lubbock, TX, United States
1 Introduction
Leishmaniasis is a neglected disease, widespread throughout the world. It represents a major global health challenge due to its economic and social implications. It is caused by protozoan parasites of the genus Leishmania (Mann et al., 2021; Mathison and Bradley Benjamin, 2022). Unlike most eukaryotes, Leishmania has an atypical genome, characterized by its high plasticity (Thomas et al., 2009; Rogers et al., 2011; Iantorno et al., 2017; Glans et al., 2021), the absence of introns, polycistronic constitutive transcription of its genes and lack of gene regulation at the transcriptional level (Martinez-Calvillo et al., 2003; Ivens et al., 2005; Peacock et al., 2007; Rogers et al., 2011). Because of this, the regulation of gene expression in Leishmania and trypanosomatids in general occurs at the post-transcriptional level. Recent studies have found that non-coding RNAs (ncRNAs) play an important role in these regulatory mechanisms in trypanosomatids, however, the precise function and mechanisms associated with them are poorly understood (Rajan et al., 2020; Fort et al., 2022; Guegan et al., 2022; Espada et al., 2025; Quilles et al., 2025).
In general, ncRNAs are a class of RNA transcripts that are not translated into proteins but serve essential regulatory functions in a variety of biological processes. ncRNA are typically categorized based on length or functions. Based on length, they are classified into small ncRNAs (<200 nucleotides) and long ncRNAs (>200 nucleotides) (Quinn and Chang, 2016; Zhang et al., 2019; Chen and Kim, 2024; Jouravleva and Zamore, 2025). Based on function, they are divided into two major categories: 1) housekeeping ncRNAs (ribosomal RNAs (rRNAs), transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), small nucleolar RNAs (snoRNAs) and telomerase RNA (TERC); these ncRNAs are ubiquitously expressed and participate in fundamental cellular activities and 2) regulatory ncRNAs (microRNAs (miRNAs), small interfering RNAs (siRNAs), PIWI-interacting RNAs (piRNAs), tRNA-derived fragments (tRFs), and tRNA halves (tiRNAs), enhancer RNAs (eRNAs), long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs)); these are involved in fine-tuning gene expression at multiple levels — epigenetic, transcriptional, and post-transcriptional (Pasquinelli and Ruvkun, 2002; Ambros, 2003; Zhang et al., 2019; Chen and Kim, 2024).
Similar to other eukaryotes, Leishmania parasites carry same types of housekeeping ncRNAs. A diverse repertoire of regulatory ncRNAs has been identified across various Leishmania species including siRNAs (Atayde et al., 2013; Lambertz et al., 2015; Brettmann et al., 2016), tRNA- and rRNA-derived small RNAs (Lambertz et al., 2015; Kusakisako et al., 2023), snoRNAs (Liang et al., 2007; Saxena et al., 2007; Eliaz et al., 2015; Piel et al., 2022; Rajan et al., 2024), and lncRNAs (Dumas et al., 2006; Emond-Rheault et al., 2025; Espada et al., 2025). Although many Leishmania species lost the canonical RNA interference (RNAi) pathway, however, several species in the Viannia subgenus (Leishmania braziliensis and Leishmania guyanensis) (Lye et al., 2022) retain a functional RNAi machinery capable of producing siRNAs (Atayde et al., 2013; Brettmann et al., 2016). Importantly, while canonical miRNAs have not been identified in Leishmania, some in silico studies have proposed the existence of miRNA-like molecules with potential regulatory function (Sahoo et al., 2014; Nimsarkar et al., 2020; Martinez-Hernandez et al., 2025). However, these findings remain speculative and require in vitro validation before any definitive conclusions can be drawn. Complementing these predictions, various RNA-seq datasets have revealed a wide range of ncRNAs encoded in the Leishmania genome. In many cases these transcripts remain functionally uncharacterized; they have been identified in species such as L. braziliensis (Torres et al., 2017; Ruy et al., 2019; Martinez-Hernandez et al., 2025; Quilles et al., 2025), L. amazonensis (Aoki et al., 2017; Goes et al., 2023), L. major (Liang et al., 2007; Eliaz et al., 2015; Freitas Castro et al., 2017; Rajan et al., 2024; Martinez-Hernandez et al., 2025), L. donovani (Saxena et al., 2007; Freitas Castro et al., 2017; Piel et al., 2022; Martinez-Hernandez et al., 2025), L. infantum (Dumas et al., 2006; Emond-Rheault et al., 2025), L. mexicana (Kalesh et al., 2022).
Across a wide range of organisms, ncRNAs have been implicated in mRNA processing, mRNA stability and emerge as key players in a variety of regulatory processes, such as DNA replication, chromosome maintenance, transcriptional regulation, translation, protein stability, the translocation of regulatory proteins and host-parasite interactions (Freitas Castro et al., 2017). Understanding the role and mechanisms of ncRNAs actions in Leishmania and host may lead to new avenues in the search for strategies to control leishmaniasis.
2 Post-transcriptional regulation and translational control
Leishmania parasites are characterized by the absence of classic genetic control at the transcriptional level, therefore, one of the main roles of ncRNAs could be at the post-transcriptional level (Figure 1A), through regulation of mRNA stability, processing, transport, and degradation; these processes at the post-transcriptional level have been seen in other protozoa (Li et al., 2020; Simantov et al., 2022). Recent evidence suggests a significant presence of regulatory ncRNAs derived from untranslated regions (UTRs) of mRNAs (Freitas Castro et al., 2017). RNA-Binding Proteins (RBPs) are central to post-transcriptional regulation (Glisovic et al., 2008). Over 2,400 RBPs, including non-poly(A)-binding proteins, form complexes with ncRNAs, suggesting roles in RNA transport and stability in L. mexicana (Kalesh et al., 2022).
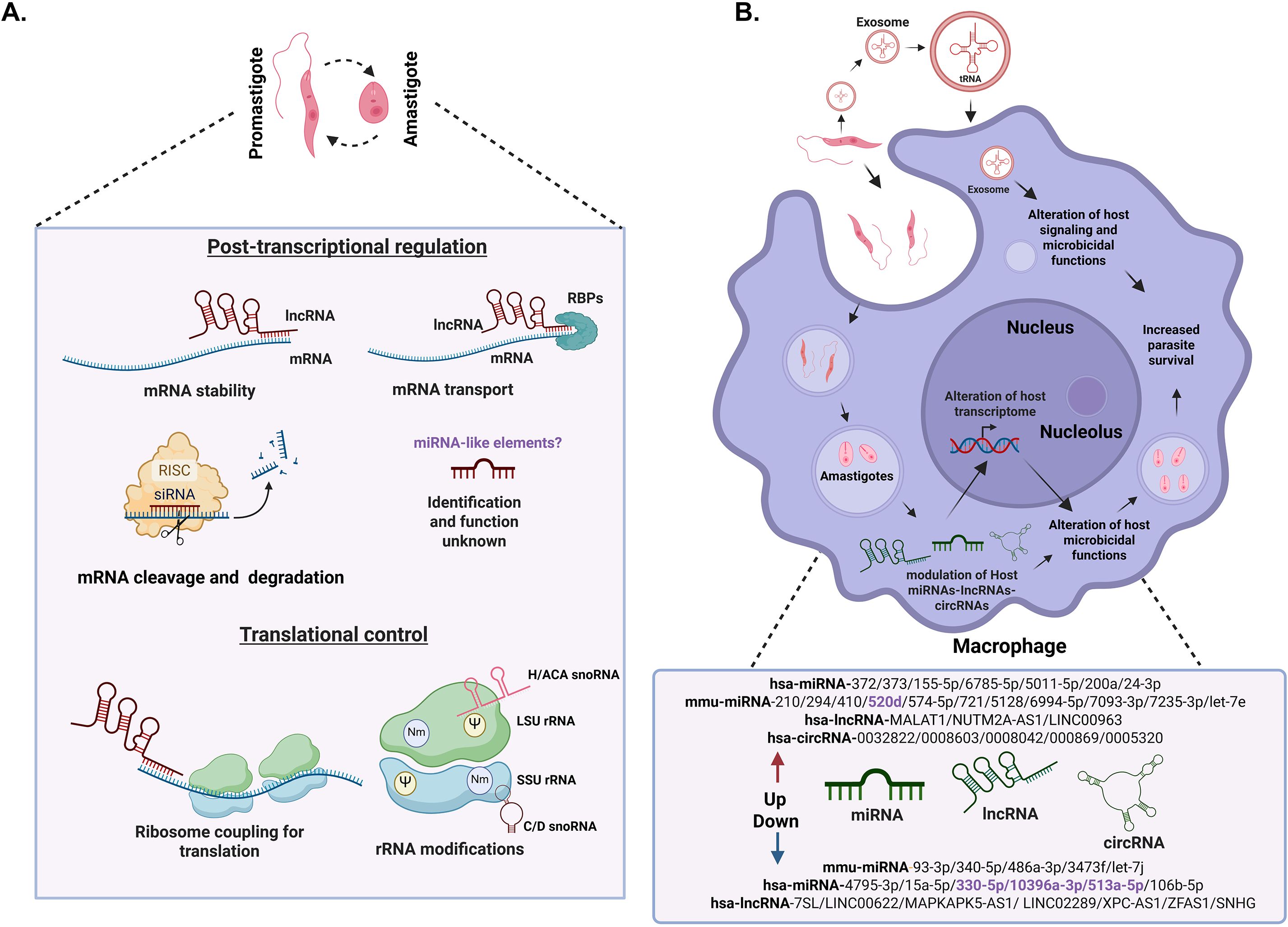
Figure 1. The role of ncRNAs in Leishmania parasites and host cells. (A). Modulation of ncRNAs in Leishmania parasites during development: we show the role of ncRNAs in different steps of gene expression in Leishmania promastigotes and amastigotes. At the post-transcriptional level, expression level is regulated through mRNA stability, translation, transport and degradation, as well as the possible presence of microRNA-like elements. Ribosome RNA (rRNA) modifications such as 2’-O-methylation (Nm) (Purple circle) and pseudouridylation (Ψ) (Yellow circle) by snoRNAs modulate mRNA translation. These modifications are essential for the parasite, as they allow it to adapt and survive in different environments through modifications of ribosomal RNA (rRNA), influencing ribosome biogenesis and gene expression regulation at multiple levels. (B) This figure shows schematic changes in a macrophage during the phagocytosis of Leishmania and is not to the scale. Modulation of host ncRNAs during Leishmania infection. Leishmania can mainly modulate micro RNAs (miRNAs), long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) in the host. These changes lead to the alterations in the host transcriptome, immune response evasion and increased parasite survival. Host transcripts are shown in green and Leishmania parasite transcripts in red. The ncRNAs in black correspond to those validated with RT-qPCR, while in purple correspond to those that have not yet been validated. In the description of ncRNAs, “hsa” refers to ncRNAs from humans (Homo sapiens) while “mmu” refers to ncRNAs from mice (Mus musculus).Biorender software was used to create this figure under an academic license.
In the absence of transcriptional regulation translational control plays a crucial role in Leishmania gene expression supporting survival and adaptation to dramatically different environments during change of host (Jaramillo et al., 2011; Cloutier et al., 2012; Karamysheva et al., 2020; Gutierrez Guarnizo et al., 2023; Rodriguez-Almonacid et al., 2023). In eukaryotes ncRNAs play direct roles in modulating protein synthesis, either by interacting with ribosomes, regulating the availability of mRNAs for translation or governing modifications of ribosomal proteins (Godet et al., 2024). rRNAs facilitate the peptidyl transfer reaction during protein synthesis (Moore and Steitz, 2011). Recent studies support importance of rRNA modifications by snoRNAs in L. major (Eliaz et al., 2015). snoRNAs are organized in gene clusters containing both C/D and H/ACA types, guiding rRNA modifications like 2’-O-methylation (Nm) and pseudouridylation (Eliaz et al., 2015; Rajan et al., 2024). These modifications occur in conserved rRNA domains and are critical for rRNA maturation, stability and mRNA translation.
In L. infantum, a class of lncRNAs (300–600 nucleotides) was mainly identified in amastigotes, showing a preferential association with the small ribosomal subunit (40S) (Dumas et al., 2006). These findings indicate a possible role in regulation at the translation level, although a direct effect on translational initiation has not been demonstrated. In L. braziliensis, the lncRNA45 was functionally characterized, demonstrating possible roles in RNA processing and modulation of translation rates, either enhancing or impairing protein synthesis (Espada et al., 2025). Additionally, a small ncRNA called ncRNA97, was found to be preferentially expressed in the amastigote form of L. braziliensis (Quilles et al., 2025). This ncRNA modulates gene expression through the control of the stability of the mRNAs that are involved in metacyclogenesis and responses to nutritional stress, indicating a role in developmental adaptation.
These examples underscore the intricate and multifaceted roles of ncRNAs in regulating gene expression in Leishmania parasites. However, the number of examples is limited, so the specific functions and mechanisms are still areas to be explored.
3 Modulation of host ncRNAs by Leishmania parasites
Leishmania, being an intracellular pathogen, has machinery that allows it to adapt and survive the hostile environment within the hosts. One of the main mechanisms of Leishmania to alter the host’s responses favoring parasite survival involves host transcriptome remodeling that includes modulating the expression of both coding RNAs and ncRNAs such as miRNAs (Lemaire et al., 2013; Pandey et al., 2016; Singh et al., 2016; Muxel et al., 2017; Kumar et al., 2018; Muxel et al., 2018; Kumar et al., 2020; Acuña et al., 2022; Lago et al., 2023; Fernandes et al., 2024; Hadifar et al., 2024; Tabrez et al., 2024; Akand et al., 2025; Atri et al., 2025; Masoudsinaki et al., 2025; Roy et al., 2025), lncRNAs (Misra et al., 2005; Maruyama et al., 2022; Fernandes et al., 2023; Li et al., 2023; Hadifar et al., 2024; Sirekbasan and Gurkok-Tan, 2025) and circRNAs levels (Li et al., 2022; Alizadeh et al., 2025), (Figure 1B). Interestingly, 30% of differentially expressed transcripts in infected macrophages correspond to lncRNAs supporting their importance to control macrophage function during infection (Fernandes et al., 2023).
Leishmania modulates host immune responses via alteration of host ncRNA expression profiles, affecting processes such as apoptosis, phagocytosis, and immune signaling (Fernandes et al., 2023; Scaramele et al., 2024; Tabrez et al., 2024; Akand et al., 2025). L. donovani infection in CD4+ T cells upregulates certain miRNAs (miR-6994-5p, miR-5128, miR-7093-3p, miR-574-5p and miR-7235) which interferes with the expression of the pro-inflammatory cytokine IFN-γ (Kumar et al., 2020). Moreover, the downregulation of miR-340-5p, miR-93-3p, let 7j, 486a-3p and miR-3473f promotes the differentiation of macrophages towards a Th2 phenotype, favoring the survival of the parasite. In L. braziliensis the upregulation of miR-2940-3p and miR-5100 caused suppression of TNF and NF-κB pathways, reducing inflammatory responses (Lago et al., 2023). L. amazonensis parasites are able to change the TLR signaling pathways by modulating the expression level of miRNA-let-7e (Muxel et al., 2018). The upregulation of this miRNA decreases the inflammatory response of host cells. Also, L. amazonensis induces an upregulation of miR-294 and miR-721 in macrophages (Muxel et al., 2017). This upregulation causes a repression of inducible nitric oxide synthase (NOS2) leading to reduced production of nitric oxide and subsequent decrease in the capability of the macrophage to kill the parasites. L donovani and L. major causes an upregulation of miR-210 in macrophages under hypoxic conditions, leading to downregulation of pro-inflammatory cytokines and enhancing parasite survival (Lemaire et al., 2013; Kumar et al., 2018).
L. infantum infection has been shown to alter the expression of numerous lncRNAs in human neutrophils, leading to the impairment of key antimicrobial responses such as phagocytosis and nitric oxide production (Scaramele et al., 2024). This contributes to immune evasion and the survival of the parasite. In THP-1 macrophages, the infection with L. amazonensis, L. braziliensis, and L. infantum led to a differential expression of different host lncRNAs upon infection suggesting a mechanism by which Leishmania can control macrophages and evade the immune response (Fernandes et al., 2023). A test performed on peripheral blood from patients infected with visceral leishmaniasis cured patients; asymptomatic infected individuals and healthy controls showed that L. infantum alters the expression of host lncRNAs (Maruyama et al., 2022). These lncRNAs are co-expressed with immune-related protein-coding genes and may regulate immune pathways, potentially influencing the host’s ability to respond to infection. Leishmania infection of macrophages leads to downregulation of host lncRNA 7SL RNA, an essential component of the signal recognition particle (SRP), which is responsible for targeting newly synthesized proteins to the endoplasmic reticulum (ER). This generates a downregulation of protein targeting and secretion altering trafficking of immune effectors and antigen presentation, favoring parasite persistence (Misra et al., 2005).
Leishmania influences host cellular metabolism to create a more favorable environment for its survival. miR-210 has been linked to altered L-arginine metabolism, leading to a reduction in nitric acid production. Certain miRNAs (miR-372/373/520d family) are upregulated in human macrophages during infection with L. amazonensis, leading to changes in arginine metabolism and increased polyamine production, which supports parasite survival (Fernandes et al., 2024). Inhibiting these miRNAs reduces parasite survival. During infection of bone marrow-derived macrophages with L. amazonensis the miRNAs miR-294 and miR-410 were upregulated; these miRNAs can interfere with the production of L-arginine and the immune response in the macrophages (Acuña et al., 2022). L. donovani also modulates host miRNAs that regulate cholesterol and sphingolipid biosynthesis which are crucial for the parasite’s survival (Tabrez et al., 2024; Akand et al., 2025). While the functional impact of lnRNAs and miRNAs is only beginning to be uncovered, host circRNAs have emerged as another relevant class of ncRNAs in leishmaniasis. A recent study found a large number of circRNAs differentially expressed in the serum of patients with leishmaniasis compared with healthy controls (Li et al., 2022) while in THP-1 cells infected with L. tropica and L. infantum distinct circRNAs profiles were found depending on the parasite strain (Alizadeh et al., 2025).
In addition to directly modulating host ncRNAs, Leishmania also influences the host environment through the secretion of extracellular vesicles, particularly exosomes (Figure 1B). During infection, those exosomes can modulate the host immune response (Silverman and Reiner, 2011; Peng et al., 2022; Sharma and Singh, 2025). tRFs and other small RNAs have been detected in exosomes secreted by Leishmania, suggesting a role in intercellular communication and possibly in the modulation of host translation (Lambertz et al., 2015; Kusakisako et al., 2023). These vesicles can modulate immune responses in different ways. It has been found that the parasite is able to release exosomes in sand flies and host cells, which can stimulate an inflammatory response leading to exacerbated cutaneous leishmaniasis (da Silva Lira Filho et al., 2022). In vivo studies have demonstrated that treatment of mice with L. donovani exosomes prior to challenge with the parasite exacerbates infection, promotes IL-10 and inhibits TNF-α production (Silverman et al., 2010). These findings indicate that Leishmania exosomes, with their ncRNA cargo, are predominantly immunosuppressive and play a significant role in shaping the host immune response to favor parasite survival.
Together, the modulation of host miRNAs, lncRNAs and circRNAs as well as the production of exosomes in the parasite uncovers the strategy by which Leishmania manipulates host immune responses and cellular functions for its own benefit. While progress has been made in studying how the parasite alters host ncRNAs, the possible functions of Leishmania’s own ncRNAs (beyond those secreted into exosomes) remain largely unexplored. Understanding these mechanisms of host-parasite interactions would allow to identify new therapeutic targets.
4 Discussion
Although recent studies have expanded our understanding of the roles of ncRNAs in Leishmania, their precise functions and mechanisms remain unclear, largely because most findings are based on transcriptomic descriptions rather than functional validation. Current solid evidence indicates that host miRNA modulation plays a major role in the survival of the parasite, mainly through the modulation of effective host immune response. However, growing evidence shows that other host ncRNAs, including lncRNAs and most recently circRNAs are also altered during infection and may contribute to parasite survival (Figure 1B).
It is clear that parasite is able to modulate host ncRNAs, however, the role of the parasite’s own ncRNAs in this modulation is poorly understood. This represents a significant gap and an opportunity for future research. To move beyond simple characterization, we must fully understand the mechanism of their action and what they target in the host and parasite. Advanced tools such as single-cell transcriptomics, polysome profiling and CRISPR-based gene editing could help to validate their role and functions in both the host and parasites.
This research can open new avenues for combating leishmaniasis. ncRNAs could serve as therapeutic targets, be used as biomarkers for diagnosis and prognosis, or even be developed as a tool to restore or enhance the host’s immune response. The growing body of evidence highlights the essential roles of ncRNAs in parasite biology and host-pathogen interactions. As the field advances, ncRNAs may deepen our understanding of Leishmania pathogenesis.
Author contributions
EQ: Visualization, Investigation, Writing – original draft, Writing – review & editing. ZK: Writing – original draft, Supervision, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Start-up funds from Texas Tech University Health Sciences Center to ZK.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Acuña, S. M., Zanatta, J. M., de Almeida Bento, C., Floeter-Winter, L. M., and Muxel, S. M. (2022). miR-294 and miR-410 Negatively Regulate Tnfa, Arginine Transporter Cat1/2, and Nos2 mRNAs in Murine Macrophages Infected with Leishmania amazonensis. Non-Coding. RNA. 8, 17. doi: 10.3390/ncrna8010017
Akand, S. K., Rahman, A., Masood, M., Tabrez, S., Jawed, J. J., Ahmed, M. Z., et al. (2025). Leishmania donovani alters the host sphingolipid biosynthetic pathway regulatory microRNA hsa-miR-15a-5p for its survival. Microb. Pathog. 208, 108019. doi: 10.1016/j.micpath.2025.108019
Alizadeh, H., Muftuoglu, C., Omondi, Z. N., Mert, U., Asadi, M., Ozbilgin, A., et al. (2025). Circular RNAs as a new perspective in the diagnosis and mechanism of Leishmania infections. Acta Trop. 261, 107509. doi: 10.1016/j.actatropica.2024.107509
Ambros, V. (2003). MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell 113, 673–676. doi: 10.1016/S0092-8674(03)00428-8
Aoki, J. I., Muxel, S. M., Zampieri, R. A., Laranjeira-Silva, M. F., Müller, K. E., Nerland, A. H., et al. (2017). RNA-seq transcriptional profiling of Leishmania amazonensis reveals an arginase-dependent gene expression regulation. PloS Negl. Trop. Dis. 11, e0006026. doi: 10.1371/journal.pntd.0006026
Atayde, V. D., Shi, H., Franklin, J. B., Carriero, N., Notton, T., Lye, L. F., et al. (2013). The structure and repertoire of small interfering RNAs in Leishmania (Viannia) Braziliensis reveal diversification in the trypanosomatid RNAi pathway. Mol. Microbiol. 87, 580–593. doi: 10.1111/mmi.12117
Atri, C., Mkannez, G., Attia, H., Sghaier, R. M., Bali, A., Ben-Cheikh, A., et al. (2025). Host-parasite interactions after in vitro infection of human macrophages by Leishmania major: Dual analysis of microRNA and mRNA profiles reveals regulation of key processes through time kinetics. Microbes Infect. 27, 105502. doi: 10.1016/j.micinf.2025.105502
Brettmann, E. A., Shaik, J. S., Zangger, H., Lye, L. F., Kuhlmann, F. M., Akopyants, N. S., et al. (2016). Tilting the balance between RNA interference and replication eradicates Leishmania RNA virus 1 and mitigates the inflammatory response. Proc. Natl. Acad. Sci. U. S. A. 113, 11998–12005. doi: 10.1073/pnas.1615085113
Chen, L. L. and Kim, V. N. (2024). Small and long non-coding RNAs: Past, present, and future. Cell 187, 6451–6485. doi: 10.1016/j.cell.2024.10.024
Cloutier, S., Laverdière, M., Chou, M.-N., Boilard, N., Chow, C., and Papadopoulou, B. (2012). Translational Control through eIF2alpha Phosphorylation during the Leishmania Differentiation Process. PloS One 7, e35085. doi: 10.1371/journal.pone.0035085
da Silva Lira Filho, A., Fajardo, E. F., Chang, K. P., Clément, P., and Olivier, M. (2022). Leishmania exosomes/extracellular vesicles containing GP63 are essential for enhance cutaneous leishmaniasis development upon co-inoculation of leishmania amazonensis and its exosomes. Front. Cell. Infect. Microbiol. 11 - 2021. doi: 10.3389/fcimb.2021.709258
Dumas, C., Chow, C., Müller, M., and Papadopoulou, B. (2006). A novel class of developmentally regulated noncoding RNAs in leishmania. Eukaryot. Cell. 5, 2033–2046. doi: 10.1128/EC.00147-06
Eliaz, D., Doniger, T., Tkacz, I. D., Biswas, V. K., Gupta, S. K., Kolev, N. G., et al. (2015). Genome-wide analysis of small nucleolar RNAs of Leishmania major reveals a rich repertoire of RNAs involved in modification and processing of rRNA. RNA Biol. 12, 1222–1255. doi: 10.1080/15476286.2015.1038019
Emond-Rheault, J. G., Ferreira, G. R., Lavoie-Ouellet, C., Smith, M. A., and Papadopoulou, B. (2025). Novel insights into the Leishmania infantum transcriptome diversity of protein-coding and non-coding sequences in both stages of parasite development using nanopore direct RNA sequencing. BMC Genomics 26, 573. doi: 10.1186/s12864-025-11767-8
Espada, C. R., Anthon, C., Magalhães, R. D. M., Quilles Junior, J. C., Teles, N. M. M., Pais, F. S., et al. (2025). Computational discovery of conserved RNA structures and functional characterization of a structured lncRNA in Leishmania Braziliensis. Non-coding. RNA Res. 14, 51–64. doi: 10.1016/j.ncrna.2025.05.010
Fernandes, J. C. R., Gonçalves, A. N. A., Floeter-Winter, L. M., Nakaya, H. I., and Muxel, S. M. (2023). Comparative transcriptomic analysis of long noncoding RNAs in Leishmania-infected human macrophages. Front. Genet. 13 - 2022. doi: 10.3389/fgene.2022.1051568
Fernandes, J. C. R., Muxel, S. M., López-Gonzálvez, M. A., Barbas, C., and Floeter-Winter, L. M. (2024). Early Leishmania infectivity depends on miR-372/373/520d family-mediated reprogramming of polyamines metabolism in THP-1-derived macrophages. Sci. Rep. 14, 996. doi: 10.1038/s41598-024-51511-y
Fort, R. S., Chavez, S., Trinidad Barnech, J. M., Oliveira-Rizzo, C., Smircich, P., Sotelo-Silveira, J. R., et al. (2022). Current status of regulatory non-coding RNAs research in the tritryp. Non-Coding. RNA. 8, 54. doi: 10.3390/ncrna8040054
Freitas Castro, F., Ruy, P. C., Nogueira Zeviani, K., Freitas Santos, R., Simões Toledo, J., and Kaysel Cruz, A. (2017). Evidence of putative non-coding RNAs from Leishmania untranslated regions. Mol. Biochem. Parasitol. 214, 69–74. doi: 10.1016/j.molbiopara.2017.04.002
Glans, H., Lind Karlberg, M., Advani, R., Bradley, M., Alm, E., Andersson, B., et al. (2021). High genome plasticity and frequent genetic exchange in Leishmania tropica isolates from Afghanistan, Iran and Syria. PloS Negl. Trop. Dis. 15, e0010110. doi: 10.1371/journal.pntd.0010110
Glisovic, T., Bachorik, J. L., Yong, J., and Dreyfuss, G. (2008). RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 582, 1977–1986. doi: 10.1016/j.febslet.2008.03.004
Godet, A.-C., Roussel, E., Laugero, N., Morfoisse, F., Lacazette, E., Garmy-Susini, B., et al. (2024). Translational control by long non-coding RNAs. Biochimie 217, 42–53. doi: 10.1016/j.biochi.2023.08.015
Goes, W. M., Brasil, C. R. F., Reis-Cunha, J. L., Coqueiro-Dos-Santos, A., Grazielle-Silva, V., de Souza Reis, J., et al. (2023). Complete assembly, annotation of virulence genes and CRISPR editing of the genome of Leishmania amazonensis PH8 strain. Genomics 115, 110661. doi: 10.1016/j.ygeno.2023.110661
Guegan, F., Rajan, K. S., Bento, F., Pinto-Neves, D., Sequeira, M., Guminska, N., et al. (2022). A long noncoding RNA promotes parasite differentiation in African trypanosomes. Sci. Adv. 8, eabn2706. doi: 10.1126/sciadv.abn2706
Gutierrez Guarnizo, S. A., Tikhonova, E. B., Karamyshev, A. L., Muskus, C. E., and Karamysheva, Z. N. (2023). Translational reprogramming as a driver of antimony-drug resistance in Leishmania. Nat. Commun. 14, 2605. doi: 10.1038/s41467-023-38221-1
Hadifar, S., Masoudzadeh, N., Andersson, B., Heydari, H., Mashayekhi Goyonlo, V., Kerachian, M., et al. (2024). Integrated analysis of lncRNA and mRNA expression profiles in cutaneous leishmaniasis lesions caused by Leishmania tropica. Front. Cell Infect. Microbiol. 14, 1416925. doi: 10.3389/fcimb.2024.1416925
Iantorno, S. A., Durrant, C., Khan, A., Sanders, M. J., Beverley, S. M., Warren, W. C., et al. (2017). Gene expression in leishmania is regulated predominantly by gene dosage. mBio 8. doi: 10.1128/mBio.01393-17
Ivens, A. C., Peacock, C. S., Worthey, E. A., Murphy, L., Aggarwal, G., Berriman, M., et al. (2005). The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442. doi: 10.1126/science.1112680
Jaramillo, M., Gomez, M. A., Larsson, O., Shio, M. T., Topisirovic, I., Contreras, I., et al. (2011). Leishmania repression of host translation through mTOR cleavage is required for parasite survival and infection. Cell Host Microbe 9, 331–341. doi: 10.1016/j.chom.2011.03.008
Jouravleva, K. and Zamore, P. D. (2025). A guide to the biogenesis and functions of endogenous small non-coding RNAs in animals. Nat. Rev. Mol. Cell Biol. 26, 347–370. doi: 10.1038/s41580-024-00818-9
Kalesh, K., Wei, W., Mantilla Brian, S., Roumeliotis Theodoros, I., Choudhary, J., and Denny Paul, W. (2022). Transcriptome-wide identification of coding and noncoding RNA-binding proteins defines the comprehensive RNA interactome of leishmania mexicana. Microbiol. Spectrum. 10, e02422–e02421. doi: 10.1128/spectrum.02422-21
Karamysheva, Z. N., Gutierrez Guarnizo, S. A., and Karamyshev, A. L. (2020). Regulation of translation in the protozoan parasite leishmania. Int. J. Mol. Sci. 21, 2981. doi: 10.3390/ijms21082981
Kumar, V., Das, S., Kumar, A., Tiwari, N., Kumar, A., Abhishek, K., et al. (2020). Leishmania donovani infection induce differential miRNA expression in CD4+ T cells. Sci. Rep. 10, 3523. doi: 10.1038/s41598-020-60435-2
Kumar, V., Kumar, A., Das, S., Kumar, A., Abhishek, K., Verma, S., et al. (2018). Leishmania donovani Activates Hypoxia Inducible Factor-1α and miR-210 for Survival in Macrophages by Downregulation of NF-κB Mediated Pro-inflammatory Immune Response. Front. Microbiol. 9 - 2018. doi: 10.3389/fmicb.2018.00385
Kusakisako, K., Nakao, R., and Katakura, K. (2023). Detection of parasite-derived tRNA and rRNA fragments in the peripheral blood of mice experimentally infected with Leishmania donovani and Leishmania amazonensis using next-generation sequencing analysis. Parasitol. Int. 93, 102716. doi: 10.1016/j.parint.2022.102716
Lago, T., Medina, L., Lago, J., Santana, N., Cardoso, T., Rocha, A., et al. (2023). MicroRNAs regulating macrophages infected with Leishmania L. (V.) Braziliensis isolated from different clinical forms of American tegumentary leishmaniasis. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1280949
Lambertz, U., Oviedo Ovando, M. E., Vasconcelos, E. J. R., Unrau, P. J., Myler, P. J., and Reiner, N. E. (2015). Small RNAs derived from tRNAs and rRNAs are highly enriched in exosomes from both old and new world Leishmania providing evidence for conserved exosomal RNA Packaging. BMC Genomics 16, 151. doi: 10.1186/s12864-015-1260-7
Lemaire, J., Mkannez, G., Guerfali, F. Z., Gustin, C., Attia, H., Sghaier, R. M., et al. (2013). MicroRNA expression profile in human macrophages in response to leishmania major infection. PloS Negl. Trop. Dis. 7, e2478. doi: 10.1371/journal.pntd.0002478
Li, Y., Baptista, R. P., and Kissinger, J. C. (2020). Noncoding RNAs in apicomplexan parasites: an update. Trends Parasitol. 36, 835–849. doi: 10.1016/j.pt.2020.07.006
Li, Z., Fang, Y., Zhang, Y., and Zhou, X. (2023). RNA-seq analysis of differentially expressed LncRNAs from leishmaniasis patients compared to uninfected humans. Acta Trop. 238, 106738. doi: 10.1016/j.actatropica.2022.106738
Li, Z., Zeng, W., Yang, Y., Zhang, P., Zhou, Z., Li, Y., et al. (2022). Expression profile analysis of circular RNAs in leishmaniasis. Trop. Med. Infect. Dis. 7. doi: 10.3390/tropicalmed7080176
Liang, X. H., Hury, A., Hoze, E., Uliel, S., Myslyuk, I., Apatoff, A., et al. (2007). Genome-wide analysis of C/D and H/ACA-like small nucleolar RNAs in Leishmania major indicates conservation among trypanosomatids in the repertoire and in their rRNA targets. Eukaryot. Cell. 6, 361–377. doi: 10.1128/EC.00296-06
Lye, L. F., Owens, K. L., Jang, S., Marcus, J. E., Brettmann, E. A., and Beverley, S. M. (2022). An RNA interference (RNAi) toolkit and its utility for functional genetic analysis of leishmania (Viannia). Genes (Basel). 14. doi: 10.3390/genes14010093
Mann, S., Frasca, K., Scherrer, S., Henao-Martínez, A. F., Newman, S., Ramanan, P., et al. (2021). A review of leishmaniasis: current knowledge and future directions. Curr. Trop. Med. Rep. 8, 121–132. doi: 10.1007/s40475-021-00232-7
Martinez-Calvillo, S., Yan, S., Nguyen, D., Fox, M., Stuart, K., and Myler, P. J. (2003). Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol. Cell. 11, 1291–1299. doi: 10.1016/S1097-2765(03)00143-6
Martinez-Hernandez, J. E., Aliaga-Tobar, V., González-Rosales, C., Monte-Neto, R., Martin, A. J. M., and Maracaja-Coutinho, V. (2025). Comparative and systems analyses of Leishmania spp. non-coding RNAs through developmental stages. PloS Negl. Trop. Dis. 19, e0013108. doi: 10.1371/journal.pntd.0013108
Maruyama, S. R., Fuzo, C. A., Oliveira, A. E. R., Rogerio, L. A., Takamiya, N. T., Pessenda, G., et al. (2022). Insight Into the Long Noncoding RNA and mRNA Coexpression Profile in the Human Blood Transcriptome Upon Leishmania infantum Infection. Front. Immunol. 13 - 2022. doi: 10.3389/fimmu.2022.784463
Masoudsinaki, T., Hadifar, S., Sarvnaz, H., Farahmand, M., Masoudzadeh, N., Mashayekhi Goyonlo, V., et al. (2025). Altered miRNA expression in the lesions of cutaneous leishmaniasis caused by L. major and L. tropica with insights into apoptosis regulation. Sci. Rep. 15, 20680. doi: 10.1038/s41598-025-03802-1
Mathison, B. A. and Bradley Benjamin, T. (2022). Review of the clinical presentation, pathology, diagnosis, and treatment of leishmaniasis. Lab. Med. 54, 363–371. doi: 10.1093/labmed/lmac134
Misra, S., Tripathi, M. K., and Chaudhuri, G. (2005). Down-regulation of 7SL RNA expression and impairment of vesicular protein transport pathways by leishmania infection of macrophages*. J. Biol. Chem. 280, 29364–29373. doi: 10.1074/jbc.M504162200
Moore, P. B. and Steitz, T. A. (2011). The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 3, a003780. doi: 10.1101/cshperspect.a003780
Muxel, S. M., Acuña, S. M., Aoki, J. I., Zampieri, R. A., and Floeter-Winter, L. M. (2018). Toll-Like Receptor and miRNA-let-7e Expression Alter the Inflammatory Response in Leishmania amazonensis-Infected Macrophages. Front. Immunol. 9 - 2018. doi: 10.3389/fimmu.2018.02792
Muxel, S. M., Laranjeira-Silva, M. F., Zampieri, R. A., and Floeter-Winter, L. M. (2017). Leishmania (Leishmania) amazonensis induces macrophage miR-294 and miR-721 expression and modulates infection by targeting NOS2 and L-arginine metabolism. Sci. Rep. 7, 44141. doi: 10.1038/srep44141
Nimsarkar, P., Ingale, P., and Singh, S. (2020). Systems Studies Uncover miR-146a as a Target in Leishmania major Infection Model. ACS Omega. 5, 12516–12526. doi: 10.1021/acsomega.0c01502
Pandey, R. K., Sundar, S., and Prajapati, V. K. (2016). Differential expression of miRNA regulates T cell differentiation and plasticity during visceral leishmaniasis infection. Front. Microbiol. 7, 206. doi: 10.3389/fmicb.2016.00206
Pasquinelli, A. E. and Ruvkun, G. (2002). Control of developmental timing by micrornas and their targets. Annu. Rev. Cell Dev. Biol. 18, 495–513. doi: 10.1146/annurev.cellbio.18.012502.105832
Peacock, C. S., Seeger, K., Harris, D., Murphy, L., Ruiz, J. C., Quail, M. A., et al. (2007). Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 39, 839–847. doi: 10.1038/ng2053
Peng, R., Santos, H. J., and Nozaki, T. (2022). Transfer RNA-derived small RNAs in the pathogenesis of parasitic protozoa. Genes 13, 286. doi: 10.3390/genes13020286
Piel, L., Rajan, K. S., Bussotti, G., Varet, H., Legendre, R., Proux, C., et al. (2022). Experimental evolution links post-transcriptional regulation to Leishmania fitness gain. PloS Pathog. 18, e1010375. doi: 10.1371/journal.ppat.1010375
Quilles, J. C., Espada, C. R., Orsine, L. A., Defina, T. A., Almeida, L., Holetz, F., et al. (2025). A short ncRNA modulates gene expression and affects stress response and parasite differentiation in Leishmania Braziliensis. Front. Cell. Infect. Microbiol. 15 - 2025. doi: 10.3389/fcimb.2025.1513908
Quinn, J. J. and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Rajan, K. S., Aryal, S., Hiregange, D. G., Bashan, A., Madmoni, H., Olami, M., et al. (2024). Structural and mechanistic insights into the function of Leishmania ribosome lacking a single pseudouridine modification. Cell Rep. 43, 114203. doi: 10.1016/j.celrep.2024.114203
Rajan, K. S., Doniger, T., Cohen-Chalamish, S., Rengaraj, P., Galili, B., Aryal, S., et al. (2020). Developmentally Regulated Novel Non-coding Anti-sense Regulators of mRNA Translation in Trypanosoma b rucei. iScience 23, 101780. doi: 10.1016/j.isci.2020.101780
Rodriguez-Almonacid, C. C., Kellogg, M. K., Karamyshev, A. L., and Karamysheva, Z. N. (2023). Ribosome specialization in protozoa parasites. Int. J. Mol. Sci. 24. doi: 10.3390/ijms24087484
Rogers, M. B., Hilley, J. D., Dickens, N. J., Wilkes, J., Bates, P. A., Depledge, D. P., et al. (2011). Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res. 21, 2129–2142. doi: 10.1101/gr.122945.111
Roy, R., Hudachek, C. L., Bhushan Chauhan, S., Kumar, S., Kumar, A., Zhanbolat, B., et al. (2025). The circulating plasma microRNA signature in human visceral leishmaniasis. mSphere 10, e0064624. doi: 10.1128/msphere.00646-24
Ruy, P. C., Monteiro-Teles, N. M., Miserani Magalhães, R. D., Freitas-Castro, F., Dias, L., Aquino Defina, T. P., et al. (2019). Comparative transcriptomics in Leishmania Braziliensis: disclosing differential gene expression of coding and putative noncoding RNAs across developmental stages. RNA Biol. 16, 639–660. doi: 10.1080/15476286.2019.1574161
Sahoo, G. C., Ansari, M. Y., Dikhit, M. R., Gupta, N., Rana, S., and Das, P. (2014). Computational Identification of microRNA-like Elements in Leishmania major. Microrna 2, 225–230. doi: 10.2174/2211536602666131203232422
Saxena, A., Lahav, T., Holland, N., Aggarwal, G., Anupama, A., Huang, Y., et al. (2007). Analysis of the Leishmania donovani transcriptome reveals an ordered progression of transient and permanent changes in gene expression during differentiation. Mol. Biochem. Parasitol. 152, 53–65. doi: 10.1016/j.molbiopara.2006.11.011
Scaramele, N. F., Troiano, J. A., Felix, J., Costa, S. F., Almeida, M. C., Florencio de Athayde, F. R., et al. (2024). Leishmania infantum infection modulates messenger RNA, microRNA and long non-coding RNA expression in human neutrophils in vitro. PloS Negl. Trop. Dis. 18, e0012318. doi: 10.1371/journal.pntd.0012318
Sharma, M. and Singh, U. (2025). Role of tRNA-derived fragments in protozoan parasite biology. Cells 14, 115. doi: 10.3390/cells14020115
Silverman, J. M., Clos, J., Horakova, E., Wang, A. Y., Wiesgigl, M., Kelly, I., et al. (2010). Leishmania exosomes modulate innate and adaptive immune responses through effects on monocytes and dendritic cells. J. Immunol. 185, 5011–5022. doi: 10.4049/jimmunol.1000541
Silverman, J. M. and Reiner, N. E. (2011). Leishmania exosomes deliver preemptive strikes to create an environment permissive for early infection. Front. Cell Infect. Microbiol. 1, 26. doi: 10.3389/fcimb.2011.00026
Simantov, K., Goyal, M., and Dzikowski, R. (2022). Emerging biology of noncoding RNAs in malaria parasites. PloS Pathogens. 18, e1010600. doi: 10.1371/journal.ppat.1010600
Singh, A. K., Pandey, R. K., Shaha, C., and Madhubala, R. (2016). MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy 12, 1817–1831. doi: 10.1080/15548627.2016.1203500
Sirekbasan, S. and Gurkok-Tan, T. (2025). Temporal expression dynamics of lncRNAs and cis-target gene interactions in Leishmania major-infected human macrophages. Med. (Baltimore). 104, e44129. doi: 10.1097/MD.0000000000044129
Tabrez, S., Akand, S. K., Ali, R., Naqvi, I. H., Soleja, N., Mohsin, M., et al. (2024). Leishmania donovani modulates host miRNAs regulating cholesterol biosynthesis for its survival. Microbes Infect. 26, 105379. doi: 10.1016/j.micinf.2024.105379
Thomas, S., Green, A., Sturm, N. R., Campbell, D. A., and Myler, P. J. (2009). Histone acetylations mark origins of polycistronic transcription in Leishmania major. BMC Genomics 10, 152. doi: 10.1186/1471-2164-10-152
Torres, F., Arias-Carrasco, R., Caris-Maldonado, J. C., Barral, A., Maracaja-Coutinho, V., and De Queiroz, A. T. L. (2017). LeishDB: a database of coding gene annotation and non-coding RNAs in Leishmania Braziliensis. Database (Oxford). 2017. doi: 10.1093/database/bax047
Keywords: non-coding RNA, Leishmania, post-transcriptional regulation, translational control, immune response, host-parasite interactions
Citation: Quiceno E and Karamysheva ZN (2025) Non-coding RNAs as emerging players in Leishmania development and host-parasite interactions. Front. Cell. Infect. Microbiol. 15:1682470. doi: 10.3389/fcimb.2025.1682470
Received: 08 August 2025; Accepted: 22 October 2025;
Published: 04 November 2025.
Edited by:
Shahid Karim, University of Southern Mississippi, United StatesReviewed by:
Alexandre F. Marques, University of Southern Mississippi, United StatesZhongqiu Li, Chinese Center For Disease Control and Prevention, China
Copyright © 2025 Quiceno and Karamysheva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zemfira N. Karamysheva, emVtZmlyYS5rYXJhbXlzaGV2YUB0dHVoc2MuZWR1
 Eyson Quiceno
Eyson Quiceno Zemfira N. Karamysheva
Zemfira N. Karamysheva