- Department of Biology, Ecology and Earth Sciences, University of Calabria, Rende, Italy
Background: Antimicrobial resistance is emerging as a significant threat to public health, prompting the search for novel natural molecules, such as Essential Oils (EOs), that can affect, alone or in combination with conventional antibiotics, growth and various biological activities in microorganisms.
Methods: First, the effects of ten essential oils extracted from aromatic plants grown in Calabria (Southern Italy) and seven conventional antibiotics against Staphylococcus aureus cells were studied individually, determining the Minimum Inhibitory Concentrations (MICs) through broth microdilutions. Subsequently, limited to Origanum vulgare EO (OEO) only, the compounds were evaluated in combination through checkerboard and time kill assays. ZIP synergy scores and Fractional Inhibitory Concentrations Indexes (FICI) were calculated to determine the interactive effects of the combinations. At 0.5 x MIC concentration values of OEO-antibiotic combinations, the biofilm and the expression of genes involved in the Quorum Sensing (QS) process were determined by the crystal violet method and quantitative real-time PCR reactions, respectively. At the same concentrations, adenine and cytosine methylation levels were quantified through ELISA.
Results: The results showed that S. aureus was highly sensitive only to OEO, in which a small MIC value was noticed (0.312 mg/mL). Synergistic effects were observed when combining OEO and ampicillin, gentamicin, tetracycline, and tobramycin, resulting in reductions of antibiotic MICs. An inhibition of biofilm formation and a general down-regulation of the expression of agrA, hld, RNAIII, and rot genes were observed. Similarly, up- and down-methylation of cytosines and adenines, respectively, compared to antibiotics alone was noticed.
Conclusions: Taken together, our observations provide evidence on the role of the OEO-antibiotic combinations in enhancing the action of antibiotics on the growth and suggest that these combinations could influence biological processes such as biofilm formation, QS, and epigenetic changes.
Introduction
Antimicrobial resistance (AMR) is rapidly becoming a major global health concern, accounting for approximately 9% of global deaths due to critical challenges in the management and eradication of an ever-increasing range of infections caused by bacteria, fungi, parasites, and viruses (Salam et al., 2023; Tang et al., 2023). In this context, Staphylococcus aureus stands out as one of the prevalent bacterial pathogens due to its great adaptability and increasing ability to develop or acquire resistance traits to antibiotic drugs over time, to the point of being included in the Bacterial Priority Pathogens List (BPPL) by the World Health Organization (Sati et al., 2025; Touaitia et al., 2025). S. aureus is a Gram-positive human commensal microorganism that, under certain conditions, can become an opportunistic pathogen capable of causing infections ranging from superficial skin to life-threatening systemic diseases, including bacteremia, infective endocarditis, pulmonary infections, meningitis, and toxic shock syndrome, especially when the host’s immune system is compromised or the bacteria breach protective barriers (Cheung et al., 2021). This is mainly due to the complex regulatory network of the Quorum Sensing (QS) system which manages virulence factors leading to toxin production, adhesion, and biofilm formation, allowing the pathogen to thrive in different environmental conditions (Yamazaki et al., 2024; Touaitia et al., 2025). What is more, mainly through genetic changes, S. aureus may develop resistance against a variety of clinically relevant antibiotics to which it was previously sensitive (Brdová et al., 2024; Shao et al., 2025). This poses an ever-increasing challenge not only in the development/research of new antibiotics but also in the exploration and optimization of alternative/combinatorial therapeutic approaches to overcome the limitations of conventional antibiotics and ensure more effective treatments for life-threatening S. aureus infections.
In this frame, interesting results come from natural bioactive compounds which exhibit a multitude of biological activities against bacteria (Arrigoni et al., 2024; Hamad et al., 2024). More particularly, essential oils (EOs), concentrated mixtures of volatile, hydrophobic and liposoluble compounds obtained from various parts of aromatic plants, have drawn considerable attention due to their chemically diverse composition rich in terpenes, terpenoids, and phenolic compounds which are responsible for antimicrobial properties against a wide range of bacteria, including antibiotic-resistant strains. Particularly, depending on the microbial strain and their composition, EOs may primarily act at the membrane level by modifying the cell wall biogenesis and membrane hydrophobicity and polarity, resulting in the destabilization of the cell membrane potential and the breakdown of membrane integrity (Swamy et al., 2016; Tariq et al., 2019; Bacińska et al., 2025; Ma et al., 2025a). In addition, the interaction between EO components and cell wall receptors leads to a dysregulation of vital bacterial metabolic pathways as well as the disruption of the signalling network of cell-to-cell communication, thus inhibiting Quorum Sensing and biofilm formation which ultimately leads to cell growth arrest and/or bacterial death (Gonçalves et al., 2023; D’Aquila et al., 2024; Maggio et al., 2025). Being a mixture of bioactive molecules, each acting on several targets, on the one hand this makes EOs effective against a variety of microorganisms, and on the other, the risk of inducing bacterial resistance is significantly reduced.
The most widely studied OE for its antimicrobial properties is certainly that obtained from Origanum vulgare. Mainly due to its high content of carvacrol and thymol, oregano essential oil (OEO) has shown particularly strong antibacterial activity against Gram-positive and Gram-negative and even multidrug-resistant bacterial and fungal microorganisms, including S. aureus, Bacillus subtilis, Enterococcus faecalis, Escherichia coli, Pseudomonas aeruginosa, and Candida albicans, exhibiting inhibitory effects on microbial growth, biofilm formation, swarming motility, and QS signaling (Fikry et al., 2019; Soltani et al., 2021; Merghni et al., 2022; D’Aquila et al., 2022, D’Aquila et al., 2023; Nurzyńska-Wierdak and Walasek-Janusz, 2025). What is more, a body of evidence reports that EOs can enhance the effectiveness of traditional antibiotics for improved therapeutic efficacy, overcoming/increasing the efficacy of clinical antimicrobials that have become ineffective due to the antimicrobial resistance issue as well as avoiding the undesirable toxic effects by lowering the dose of the antimicrobial treatment (Langeveld et al., 2014; Liu et al., 2023; Zhao et al., 2024; Ma et al., 2025a; Jilani et al., 2025). For instance, the antibiotic-potentiating effectiveness of a variety of EO, such as Origanum compactum, Origanum elongatum, O. vulgare, Rosmarinus officinalis L, Thymus broussonnetii, Thymus pallidus to combat planktonic cells, biofilms, and efflux pump activities were recently demonstrated in several bacterial and fungal strains, also including highly prioritized multidrug-resistant strain of E. coli, E. faecalis, Klebsiella pneumoniae, P. aeruginosa, S. aureus, Salmonella enterica and Acinetobacter baumannii (Amassmoud et al., 2023; Khashei et al., 2025; Drioiche et al., 2024; Sena et al., 2024).
Accordingly, the purpose of this study is to investigate the potential impact of Essential Oils extracted from ten aromatic plants in combination with seven commercial antibiotics against S. aureus growth, biofilm formation, Quorum Sensing regulation, as well as epigenetic modifications.
Materials and methods
Bacterial strains and growth conditions
This study was carried out on S. aureus strain (Merck NCTC 6571 Lenticule® discs) kept frozen in stock cultures at −80°C in cryovials. Bacterial cells were cultured in Luria-Bertani medium containing 10 g of Bacto-Tryptone, 5 g of yeast extract, and 0.5 g of sodium chloride at 37°C under gentle agitation.
EOs and antibiotics
Ten EOs were obtained from a commercial producer located in Calabria (Italy). They were extracted from the following plants: Clinopodium nepeta L. (Kuntze, Carl Ernst Otto), Citrus bergamia (Risso, Joseph Antoine & Poiteau, Pierre Antoine), Citrus limon L. (Osbeck, Pehr), Citrus reticulata (Blanco, Francisco Manuel), Foeniculum vulgare subsp. piperitum, Laurus nobilis L., Myrtus communis L., Origanum vulgare L. subsp. viridulum (Martrin-Donos, Julien Victor) Nyman, Carl Frederik, Salvia officinalis L., and Salvia rosmarinus (Spenn, Fridolin Carl). In particular, the fruit peel of C. bergamia and C. limon, the flower, leaf, and terminal branches of C. nepeta, F. vulgare, M. communis, O. vulgare, S. officinalis, and S. Rosmarinus, and only the leaf and terminal branches of C. reticulata and L. nobilis were used. The extraction procedure and composition analysis were carried out as described in D’Aquila et al. (2023). Since EOs are highly lipophilic organic mixtures, we dissolved them in 2% DMSO in growth medium in a working solution of 10 mg EO/mL.
Ampicillin (AMP), ciprofloxacin (CIP), erythromycin (ERY), gentamicin (GEN), levofloxacin (LVX), tetracycline (TET), and tobramycin (TOB) (Sigma-Aldrich, St Louis, MO, USA) were analyzed in the study. Working solutions of all antibiotics were prepared at a final concentration of 500 μg/mL.
Determination of minimum inhibitory concentration of the EOs and antibiotics
The Minimum Inhibitory Concentration (MIC) of the EOs and antibiotics was determined by the broth microdilution method carried out in multi-well microplate. 10 μL of bacterial suspension from overnight cultures of S. aureus were inoculated into 96-well plates (~5 x 104 CFU/well) containing 1:2 serial dilutions of the working solution of EOs or antibiotics at final volume of 150 μL (final DMSO concentrations from 1% to 0.00098%). Microplates were incubated for 22 h at 37°C. In all experiments, positive (medium with cells) and negative (medium without cells) control free of EOs and antibiotics were also analyzed to verify the appropriateness of microbial growth and sterility, respectively. In addition, two further controls were represented by the cell-free medium in the presence of each EO to discern the turbidity background, as well as by the medium-containing cells in the presence of 2% DMSO free of EOs, to confirm that this concentration has no effect on bacterial growth. Turbidity measurement was performed at 600 nm in a spectrophotometer. MIC values were determined as the lowest concentrations of EO and antibiotics corresponding to values of optical density (OD) comparable to those of the cell-free medium. Each experiment was carried out in triplicate, with three independent repetitions.
Checkerboard assay
The interaction between the EO extracted from oregano (OEO) and the different antibiotics was analyzed using the checkerboard assay, as previously described.
10 μL of bacterial suspension from overnight cultures of S. aureus were inoculated into 96-well plates (~5 x 104 CFU/well) containing twofold dilutions of OEO and antibiotics at concentrations ranging from five values below and one above the MIC values at final volume of 150 μL (final DMSO concentrations from 0.125% to 0.0019%). After 22 h of incubation at 37°C, the OD600 values were measured using a microplate reader to determine the new MIC values for the mixtures and to calculate vitality percentages as follows: (ODuntreated - ODtreated/ODuntreated) x 100. Each experiment was carried out in triplicate.
The interactions between OEO and antibiotics combinations over the entire range of concentrations were analyzed through the free web-based SynergyFinder software (https://synergyfinder.org/) that exploits the Zero Interaction Potency (ZIP) calculation model which examines the drug interaction relationships by comparing the change in the potency of the dose-response curves between individual drugs and their combinations (Zheng et al., 2022).
The ZIP scores of the OEO and antibiotic combinations were interpreted as follows: synergism (>10), additive (<10 and >–10), and antagonistic (<–10) (Ianevski et al., 2017; 2020).
The lowest concentrations of OEO and antibiotic that showed an inhibition greater than 99% were used to calculate the Fractional Inhibitor Concentration Index (FICI) through the following formula: FICI = FICOEO + FICantibiotic, with FICOEO = (MIC of OEO in combination with antibiotic)/(MIC of OEO) and FIC antibiotic = (MIC of antibiotic in combination with OEO)/(MIC of antibiotic alone). The results were interpreted according to Fratini et al. (2017) as follow: total synergism when FICI < 1; FICI =1 indicates additivity; 1 < FICI ≤ 2 no interaction; and FICI > 2 antagonism. The MIC gain of the antibiotics was calculated according to the following formula: MIC gain = MICantibiotic/MIC antibiotic+OEO inferred by considering the lowest sub-MIC concentration values at which an inhibition in bacterial growth of >99% was observed.
Time-kill tests
Time-kill experiments were performed in 96-well plates containing combinations of OEO and antibiotics that elicit a synergistic effect on checkboard dilution analysis (0.16 mg/mL of the OEO and 0.063 μg/mL of AMP, 0.08 mg/mL of the OEO and 0.25 μg/mL of GEN, 0.16 mg/mL of the OEO and 0.03125 μg/mL of TET, and 0.08 mg/mL of the OEO and 1 μg/mL of TOB). The effect of OEO and antibiotics alone was also monitored. Briefly, bacterial cells were placed on microtiter plates (∼5 × 104 CFU/well), and incubated at 37°C. At time intervals of 0, 2, 4, 6, and 24 h, cultures were serially diluted at 1:10 in sterile saline solution and 10 μL were plated onto LB agar plates and incubated for 24 h at 37°C. Then, the viable colonies were counted and represented as log10 (CFU/mL) The combination is considered synergic when it causes a ≥2log10 reduction in CFU/mL with respect antibiotic alone at 24 hours (NCCLS, 1999).
Biofilm assay
Combinations of OEO-antibiotics were assessed for their potential on bacterial biofilm. 10 μL of suspension from overnight cultures of S. aureus were inoculated into 96-well plates (~5 x 104 CFU/well) containing 0.5x MIC of the synergic OEO-antibiotic combinations at final volume of 150 μL. After 22 h of incubation at 37°C, the planktonic cells were removed, and adherent cells were fixed with 150 μL of methanol and then stained with 150 μL of crystal violet solution 0.2%. After 5 min, the excess of the stain was removed by three repeated washes with 150 μL of isotonic saline solution (0.9% sodium chloride). 150 μL of acetic acid at a concentration of 33% was added to each well and the optical density at 570 nm was determined spectrophotometrically. As controls of experimental procedure, wells containing only the inoculated bacteria and the cell-free growth medium supplemented with OEO, respectively, were analyzed. Each experiment was carried out in triplicate. Effects of treatment on biofilm are expressed as Fold Change in the optical density values of samples treated with OEO-antibiotic combinations compared to that of the antibiotic alone.
DNA and RNA extraction
50 μL of bacterial suspension from overnight cultures of S. aureus were inoculated into tubes (~5 x 107 CFU/tube) containing 0.5x MIC of the synergic OEO-antibiotic combinations at final volume of 3 mL. After 22 h of incubation at 37°C, DNA and total RNA were be extracted by using the “ZymoBIOMICS DNA/RNA Miniprep Kit” (Zymo Research). Briefly, cell culture was collected and centrifuged for 10 minutes. Isolated pellets were resuspended in 750 µl DNA/RNA Shield, transferred to a ZR BashingBead Lysis Tube and vortexed for 5 minutes. After centrifugation at 12,000 x g for 30 seconds, 400 µl of DNA/RNA Lysis Buffer was added to the supernatant, and the solution was transferred to a Spin-Away™ Filter and centrifuged at 12,000 x g for 30 seconds. After adding an equal volume of 70% ethanol, the eluate, containing the RNA fraction, was transferred to a ZymoSpin™ IIICG Column and centrifuged at 12,000 x g for 30 seconds, while 400 µl of DNA/RNA Prep Buffer were transferred into the Spin-Away™ Filter column, containing the DNA. After centrifugation at 12,000 x g for 30 seconds, two consecutive washes were performed in the presence of DNA/RNA Wash Buffer. 100 µl of ZymoBIOMICS™ DNase/RNase-Free water was transferred to the matrices of the two columns which, after incubation for 5 minutes at room temperature, were centrifuged. The DNA and RNA present in the two eluates were finally transferred onto previously reconstituted Zymo-Spin™ III-HRC Filter columns and isolated following a centrifugation at 16,000 x g for 3 minutes. The concentration and purity of the nucleic acids were determined spectrophotometrically using the absorbance ratio of 260/280 nm.
Expression profiles of genes encoding enzymes involved in quorum sensing processes
The reverse transcriptase-PCR (RT-PCR) reaction was performed using the “High-Capacity cDNA Reverse Transcription Kit” (Thermofisher). Briefly, in a final volume of 20 μl, 250 ng of total RNA were mixed with a reaction mixture consisting of RT Buffer 1X, dNTP mix 4 mM, RT Random Primers 1X, MultiScribe™ Reverse Transcriptase 50 U, and RNase Inhibitor 20 U. Then, the samples were incubated at 25°C for 10 minutes, then at 37°C for 120 minutes and, successively, at 85°C for 5 minutes to inactivate the reverse transcriptase.
The obtained cDNAs were then used as template in Real-Time PCR reactions performed in a QuantStudio3 Real-Time PCR system (Thermo Fisher Scientific). Forward and reverse primers specific for the genes involved in the Quorum Sensing process in S. aureus are reported in Table 1 (Mootz et al., 2015; Sharifi et al., 2021; Martínez et al., 2023).

Table 1. Nucleotide sequence (5’→3’) of primers used in the quantification of mRNA levels of agrA, hld, RNAIII, and rot genes encoding components of the QS system in S. aureus, as well as of the 16S ribosomal gene (16S RNA), used as an internal control.
Primer efficiencies were determined using five 10-fold serial dilutions of cDNA, and all primer pairs (agrA, hld, RNAIII, rot, and 16S rRNA) showed amplification efficiencies within the acceptable range (98.3% to 99.66%) and high linearity (0.985≥R2 ≤ 1).
The final PCR mixture (15 µL) contained 1 µL of cDNA, SensiFAST SYBR Hi-ROX Mix 1X (Bioline, London, UK) and 0.2 µM of each primer. The thermal profile used for the reaction included a 2-minutes heat activation of the enzyme at 95°C, followed by 35 cycles of denaturation at 95°C for 15 seconds and annealing/extension at 60°C for 60 seconds, followed by melt analysis ramping at 60–95°C with a melt peak level of 10%. The presence of single melting peaks was verified. All measurements were taken in the log phase of amplification. Negative controls (in which water instead of cDNA was added) were also run in each plate. Real-time PCR data were analyzed by Diomni™ Design and Analysis (RUO)3.0.2 (Applied Biosystem) through the comparative Ct method, according to which the 2^dCt values of each target gene were normalized to that of 16S ribosomal gene (16S RNA). Then, the relative quantification of gene expression was determined by using the values determined in the cells subjected to the antibiotic alone as Reference Values.
Quantification of global N6-methyladenosine and 5-methylcytosine levels
Global DNA methylation levels of N6-methyladenosines (m6A) and 5-methylcytosines (5mC) were determined by using the MethylFlash m6A DNA Methylation ELISA Kit (Epigentek, Farmingdale, Nassau County, NY, USA) and the MethylFlash Global DNA Methylation (5mC) ELISA Easy Kit, respectively, following the manufacturer’s instructions. Shortly, the methylated fraction of bacterial genomic DNA, through ELISA-like reactions, was recognized by the m6A or 5mC antibodies and quantified in a microplate spectrophotometer by reading the absorbance at 450 nm. In each experiment, the percentage of m6A and 5mC was calculated using the second-order regression equation of a standard curve that was constructed by mixing equivalent molar concentrations at different ratios of full unmethylated and methylated control DNA. Each sample was analyzed in triplicate. The methylation values of DNA samples extracted from cells treated with antibiotic alone were used as reference values (Relative Quantification, RQ) for the corresponding samples treated with combinations of OEO and antibiotics.
Statistical analysis
Statistical analyses were performed using SPSS 28.0 statistical software (SPSS Inc., Chicago, IL, USA). Kruskal–Wallis one-way analysis of variance and Mann-Whitney tests were adopted. Significance level was defined as p ≤ 0.05.
Results
Antimicrobial activity of the EOs
In the present study, ten EOs belonging to Apiaceae (F. vulgare subsp. Piperitum), Lamiaceae (C. nepeta, O. vulgare L. subsp. Viridulum, S. officinalis L., S. rosmarinus), Lauraceae (L. nobilis L.), Myrtaceae (M. communis L.) and Rutaceae (C. bergamia, C. limon, C. reticulata) family plants were analyzed. Plant family characteristics and a complete description of all EOs are shown in Supplementary Table S1 and Supplementary Table S2 (Sena et al., 2024).
The results from the broth microdilution assay in terms of MIC are reported in Table 2.
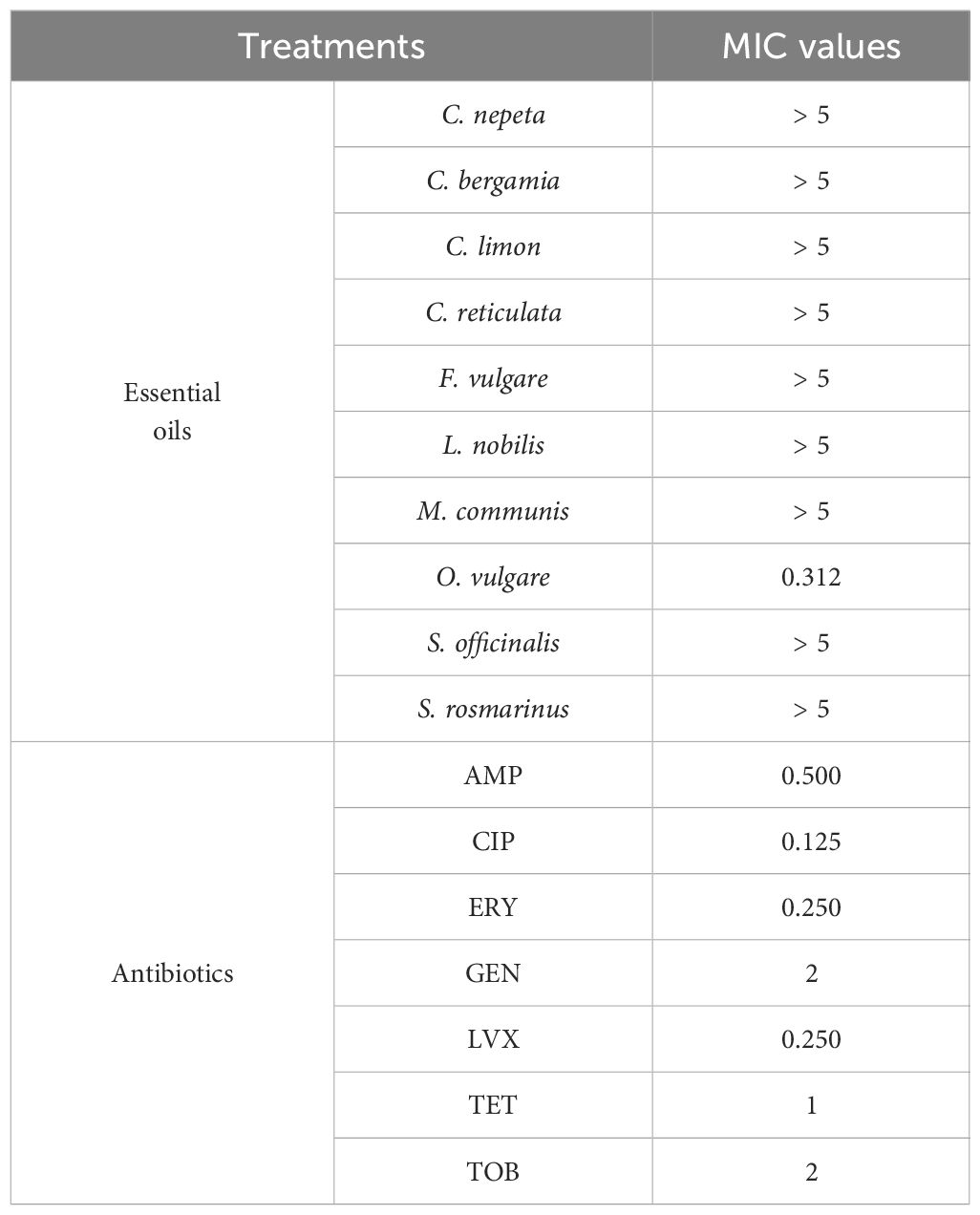
Table 2. MIC values of the essential oils (mg/mL) and the antibiotics (µg/mL) examined in the study.
The analysis revealed that the only EO that showed significant antibacterial activity was the one extracted from O. vulgare, as deduced from the MIC values equal to 0.312 mg/mL. Under our laboratory conditions, the remaining EOs did not show antibacterial activity, so they were not further investigated.
Regarding antibiotics, the MIC values observed are in line with those reported in the MIC European Committee on Antimicrobial Susceptibility Testing (EUCAST) database (https://www.eucast.org”) in which MIC distributions for individual microorganisms are reported.
The MIC values of OEO and antibiotics were then used to evaluate the effects against bacteria when they were analyzed in combination with each other.
Effects of the combination of the OEO and antibiotics against S. aureus
Combinations of AMP, CIP, ERY, GEN, LVX, TET, and TOB with OEO against S. aureus were tested through checkerboard assays, and ZIP model was used to analyze the OEO-antibiotic interactions over the entire range of concentrations.
The resulting matrices of interaction show that only the combinations of OEO with AMP, GEN, TET, and TOB increase antibacterial activity against S. aureus when compared to OEO or antibiotic alone (p<0.001) (Figure 1; Supplementary Table S3).
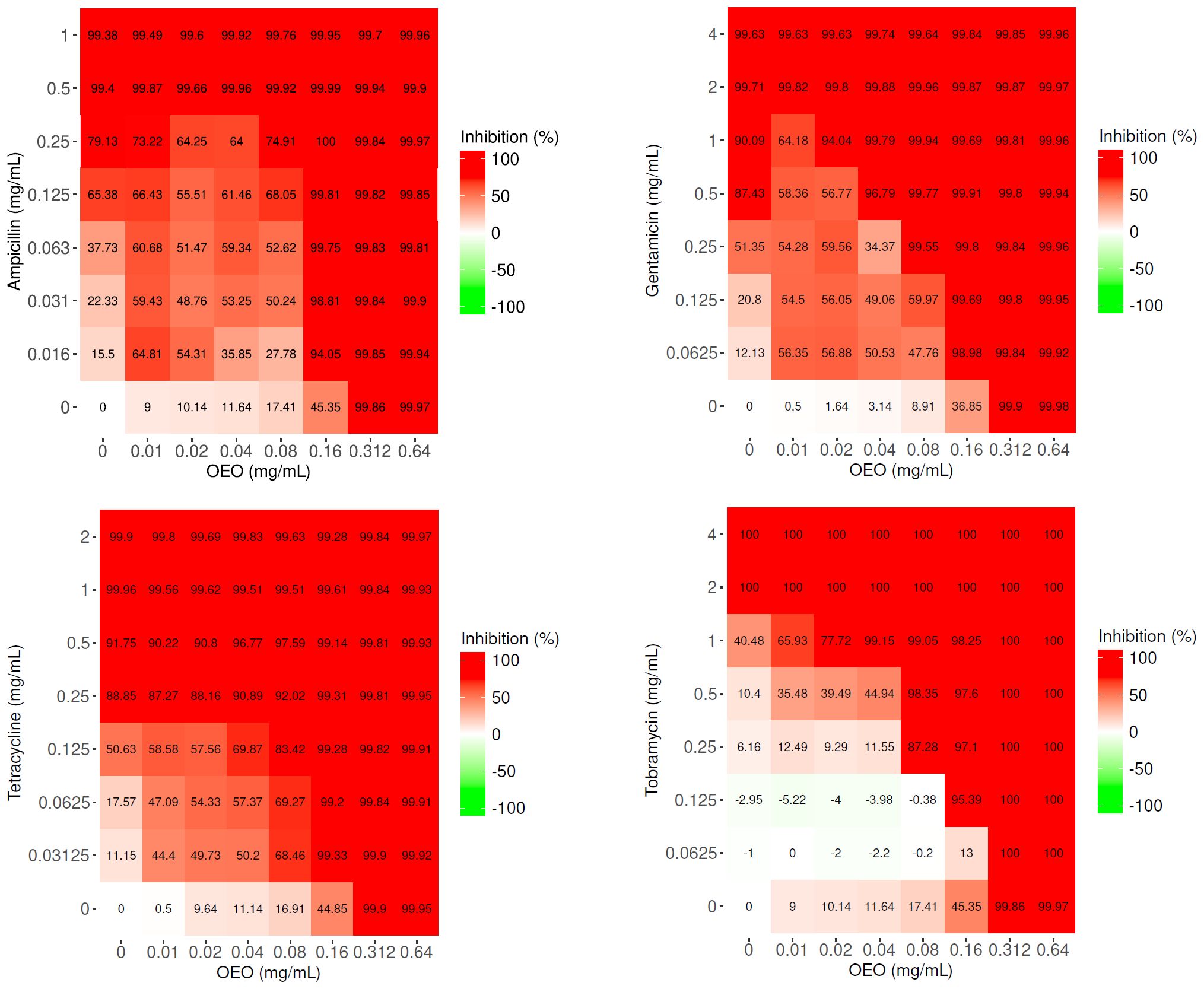
Figure 1. Matrices of interaction relative to the combinations of OEO (mg/mL) with ampicillin, gentamicin, tetracycline, and tobramycin (μg/mL). Values represent the average of three independent experiments.
If we consider the lowest sub-MIC concentration values of OEO and antibiotics at which an inhibition in bacterial growth of >99% occurs, the association of the OEO with AMP determined a gain in the antibiotic MIC of about 8-fold (0.16 mg/mL of the OEO and 0.063 μg/mL of the AMP), with GEN of 8-fold (0.08 mg/mL of the OEO and 0.25 μg/mL of the GEN), and with TOB of 2-fold (0.08 mg/mL of the OEO and 1 μg/mL of the TOB). Notable, a reduction of MIC value was appreciated, with a gain of up to 32-fold, in the case of TET (0.16 mg/mL of the OEO and 0.03125 μg/mL of the TET) (Figure 1).
The ZIP scores, graphically represented as surface plots in Figure 2 and reported as data in Supplementary Table S4, highlight a synergism occurring between OEO and the four antibiotics. In particular, different combinations of OEO and antibiotics showed ZIP scores >10. Accordingly, these conditions of synergy were also confirmed by determining FICI values (Supplementary Table S4).
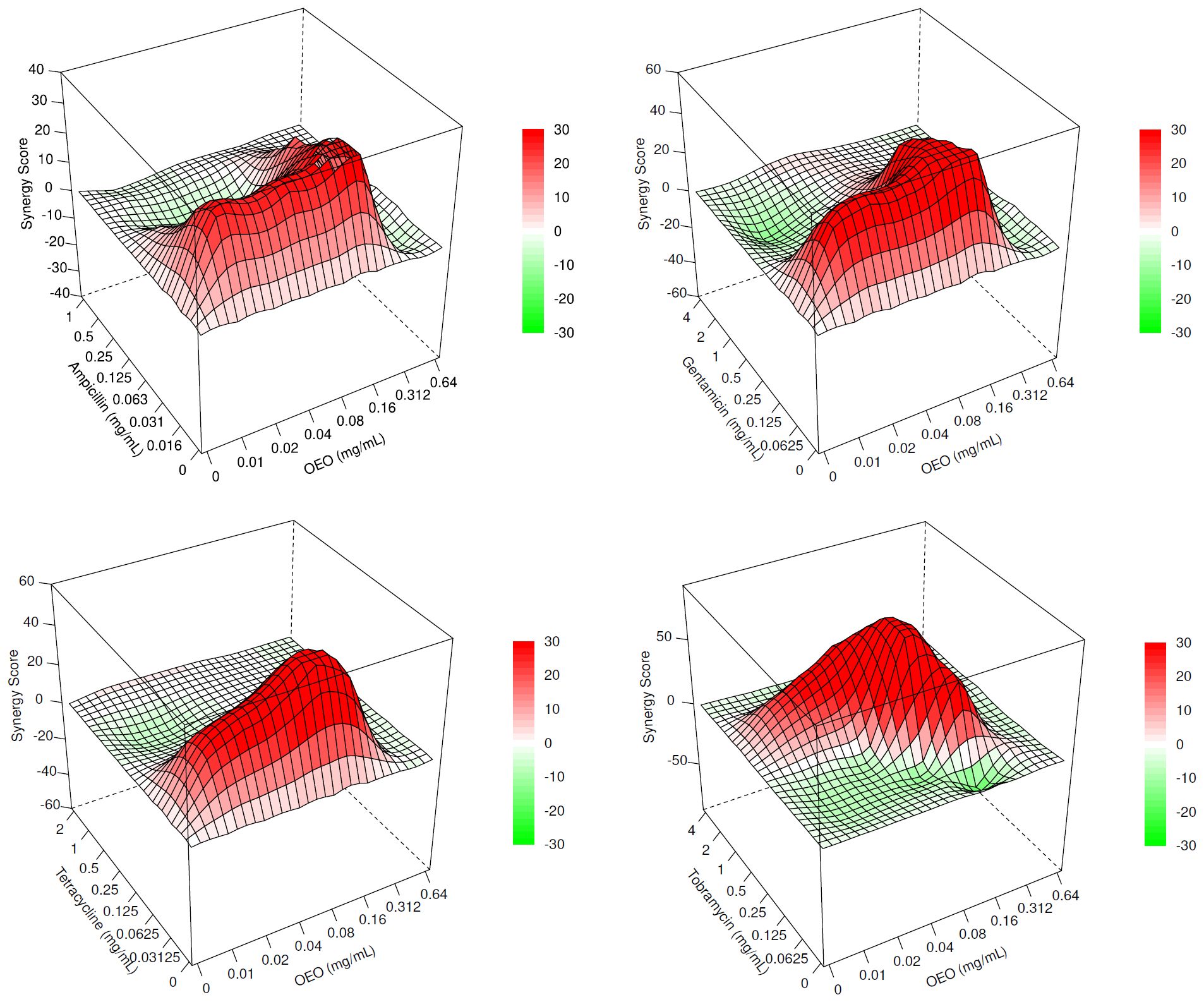
Figure 2. Surface plots representing the ZIP scores highlighting the interaction occurring between OEO and ampicillin, gentamicin, tetracycline, and tobramycin. Synergistic (>10), additive (<10 and 638 >–10), and antagonistic (<–10) effects.
CIP, ERY, and LVX did not exhibit significant change of the antibacterial activity when tested with OEO at various concentrations and were, therefore, not investigated further.
Time-kill curves
The best synergistic combinations from checkerboard assay (0.16 mg/mL of the OEO and 0.063 μg/mL of AMP, 0.08 mg/mL of the OEO and 0.25 μg/mL of GEN, 0.16 mg/mL of the OEO and 0.03125 μg/mL of TET, and 0.08 mg/mL of the OEO and 1 μg/mL of TOB) were evaluated through time-kill assay. We compared the bacterial killing kinetics of antibiotics and OEO antibiotic combinations. According to the checkerboard assay, all the OEO antibiotic combinations kill S. aureus as emerged by a decrease greater of 2log10 in CFU/mL cells within 24 hours. OEO combining with AMP showed the antibacterial activity of 2.8log10 in CFU/mL from 6 hours towards, combining with GEN of 2.5log10 in CFU/mL from 4 hours towards, with TOB of 4log10 in CFU/mL from 2 hours towards(Figure 3). These findings demonstrated that OEO combining antibiotics exhibit different efficacy over time depending on the specific antibiotic.
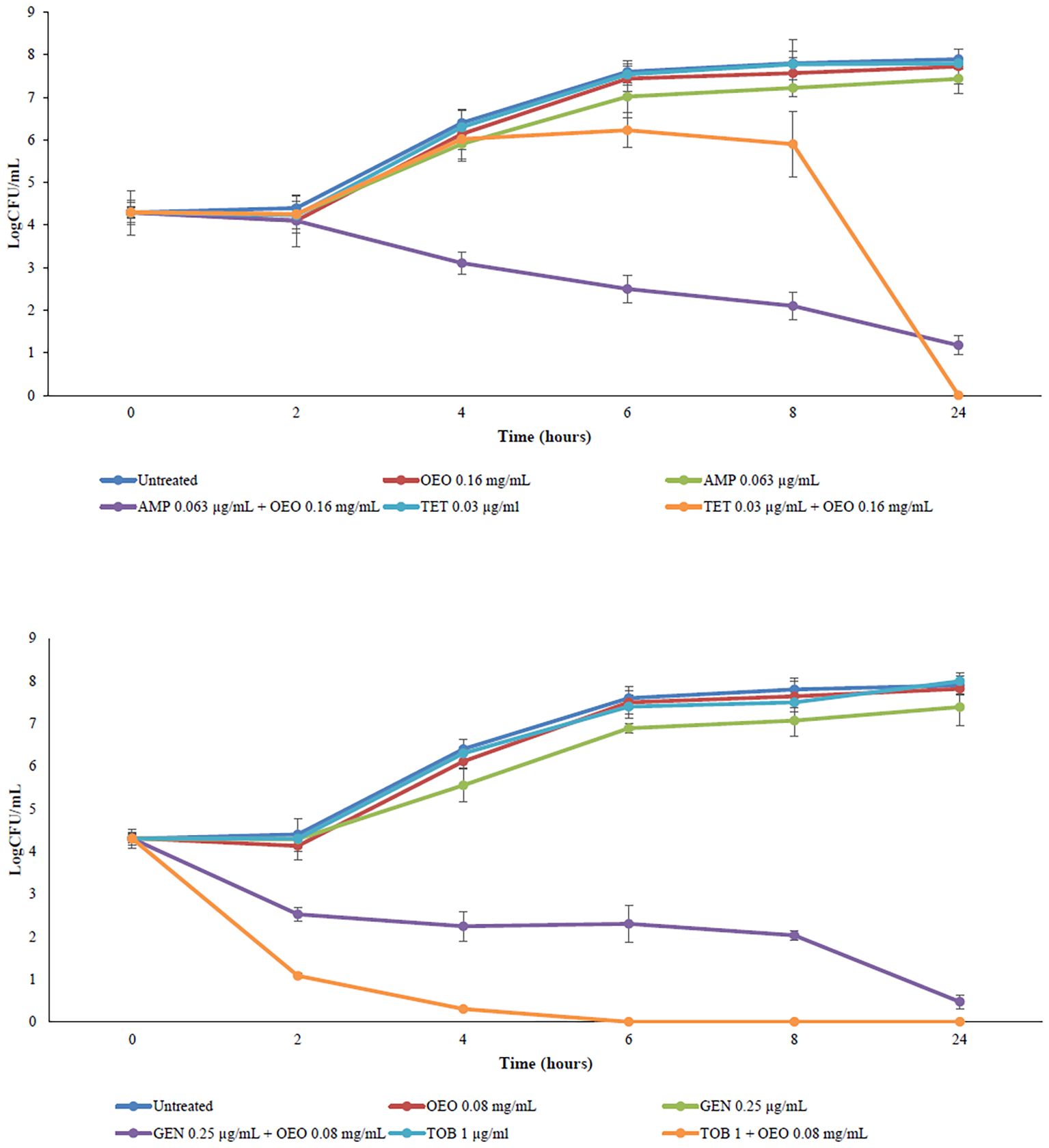
Figure 3. Time-kill curves of OEO combining with ampicillin, gentamicin, tetracycline, and tobramycin against S. aureus.
Effects on biofilm induced by combinations of OEO with antibiotics
Figure 4 reports the effects on biofilm inhibition in S. aureus exerted by combinations of OEO with antibiotics. All the combinations of the OEO with antibiotics were able to inhibit biofilm with inhibition percentages of 53% in the combination of OEO with ampicillin (p<0.01), 73% with gentamicin (p<0.001), 71% with tetracycline (p<0.001), and 91% with tobramycin (p<0.001)(Supplementary Table S5).
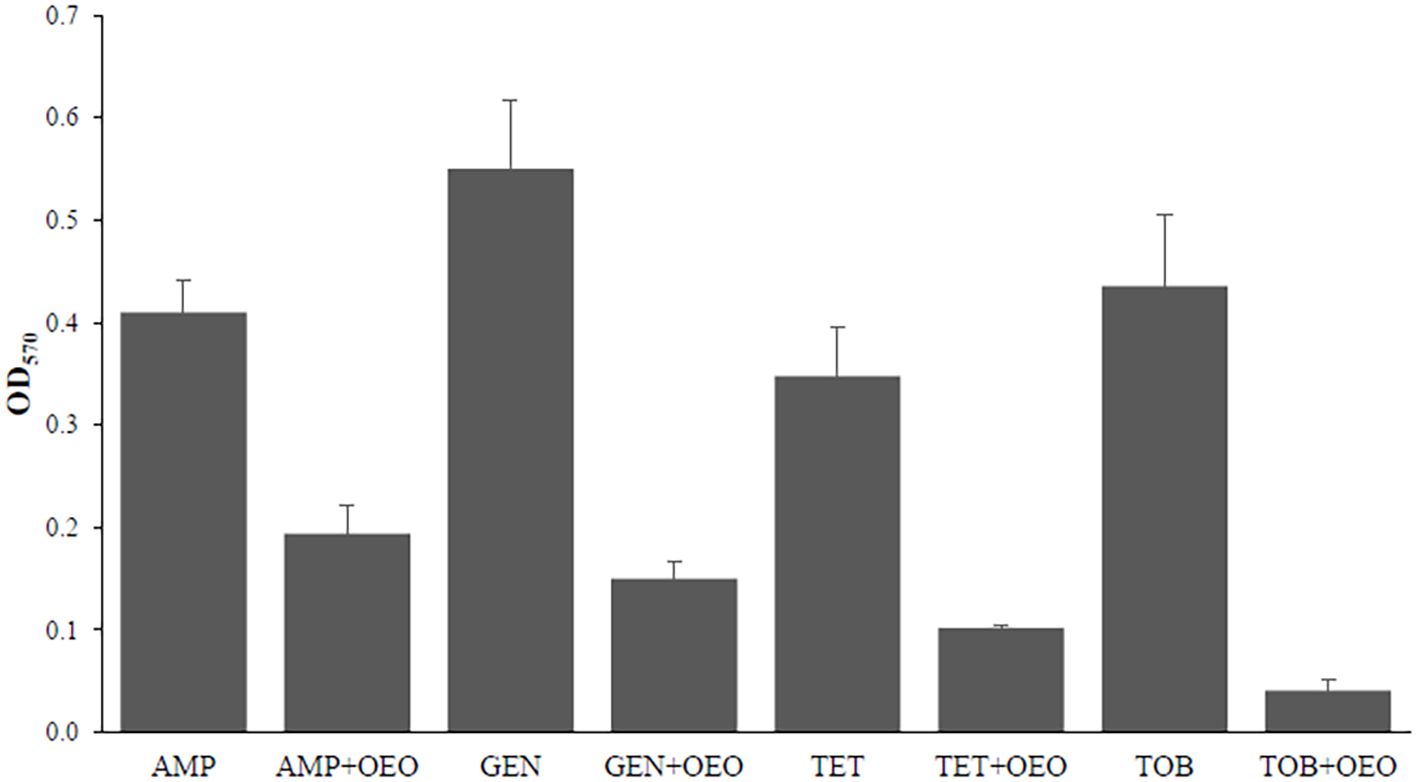
Figure 4. Effects induced by the combination of OEO and antibiotics on biofilm in S. aureus. Values are reported in terms of Fold Change determined using cells treated with the antibiotic alone as reference. Values represent the average of three independent experiments with a standard error of the mean. AMP, ampicillin; GEN, gentamicin; TET, tetracycline; TOB, tobramycin.
Effects on quorum sensing induced by combinations of OEO with antibiotics
The expression levels of agrA, hld, RNAIII, and rot genes encoding components of the QS system in S. aureus were evaluated following combined treatment of cells with OEO and antibiotics. We can observe a more marked decrease in expression for the genes agrA at the combination of OEO with tetracycline (p<0.01), hld with ampicillin (p<0.001), gentamicin (p<0.001), tetracycline (p<0.001), and tobramicin (p<0.01), RNAIII with gentamicin (p<0.001), and tetracycline (p<0.001) and rot with all four antibiotics (p<0.001) (Figure 5; Supplementary Table S6). No significant changes in gene expression were observed in the other combinations.
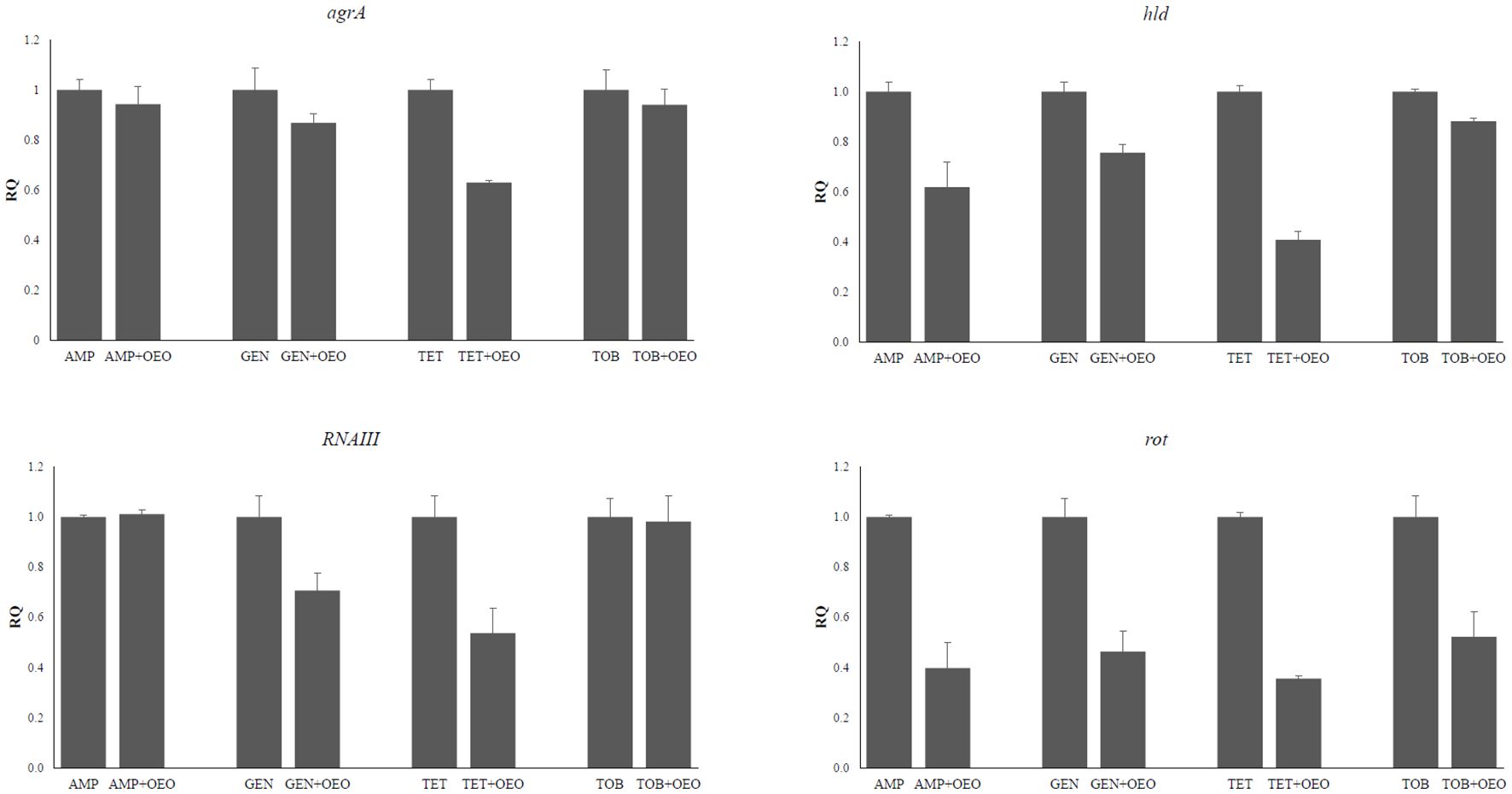
Figure 5. Effects induced by the combinations of OEO and antibiotics on the expression of agra, hld, RNAIII, and rot genes. Values are reported as Relative Quantification of gene expression determined by using the values determined in the cells subjected to the antibiotic alone as reference. Values represent the average of three independent experiments with a standard error of the mean. AMP, ampicillin; GEN, gentamicin; TET, tetracycline; TOB, tobramycin.
Effects of the synergism of OEO with antibiotics on global methylation profiles
The global methylation levels of adenine and cytosine residues were evaluated in DNA samples extracted from S. aureus cells and treated with OEO combined with antibiotics. Standard curves were generated by plotting the absorbance values for each combination versus percentage of m6A (detection range 0.02-0.2 ng) and 5-mC (detection range 0.1-5%) observing a logarithmic relationship with a correlation of 0.97 and 0.99, respectively, thus confirming the effectiveness of the experimental conditions (Supplementary Figure S1). By comparing global DNA methylation levels of adenines (m6A) quantified at the different combinations of OEO with antibiotics with those in presence of the antibiotic alone, we can observe significant hypo-methylation in all OEO and antibiotic combinations (p<0.01). As concerns 5mC levels, a hypermethylation is evident in the combinations of OEO with ampicillin, gentamicin, and tetracycline meanwhile hypomethylation occurs when the cells are treated with OEO combined with tobramycin (P<0.01) (Figure 6 and Supplementary Table S7).
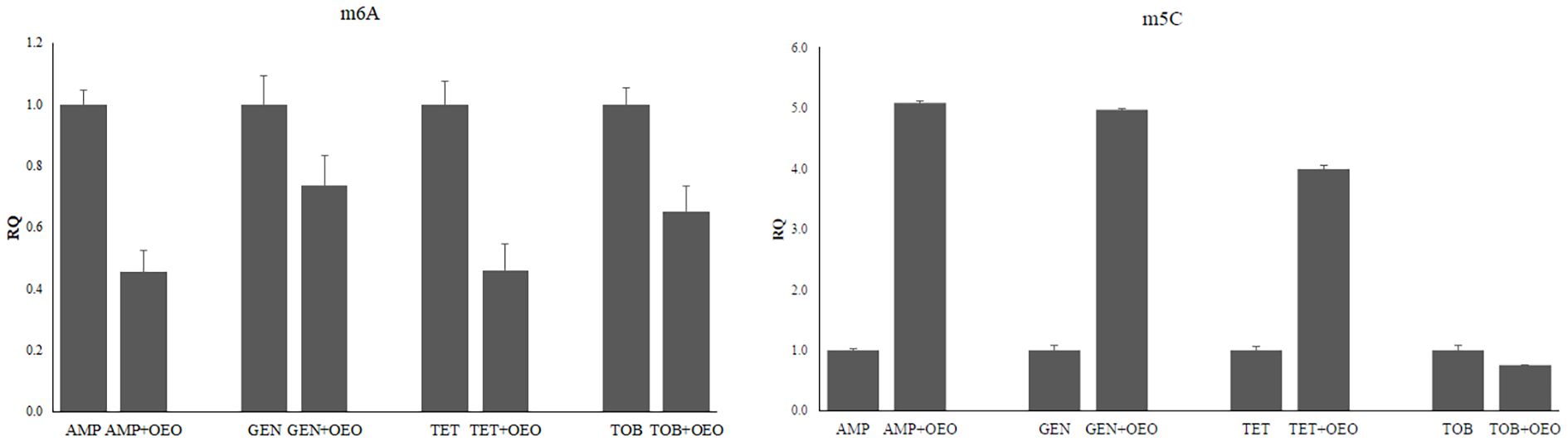
Figure 6. Effects induced by the combinations of OEO and antibiotics on methylation of adenines (m6A) and cytosines (m5C). Values are reported as Relative Quantification (RQ) determined by using cells treated with the antibiotic alone as reference. Values represent the average of three independent experiments with a standard error of the mean. AMP, ampicillin; GEN, gentamicin; TET, tetracycline; TOB, tobramycin.
Discussion
The global increase in antimicrobial resistance is driving the urgent identification of alternative antimicrobial approaches to improve treatment of infectious diseases and health care outcomes. In this context, the recent discovery of plant-derived components with effective antibacterial properties suggests as they can serve, alone or in combination with other products as new and effective antibacterial therapies.
The present study explored the antibacterial properties of ten EOs extracted from aromatic plants (C. nepeta, C. bergamia, C. limon, C. reticulata, F. vulgare, L. nobilis, M. communis, O. vulgare, S. officinalis, and S. rosmarinus) against S. aureus, considering both its very high pathogenic potential and its strong ability to develop antibiotic resistance, making it an extremely problematic pathogen in clinical settings. Among the EOs analyzed, the one extracted from O. vulgare (OEO) showed the highest antimicrobial activity (MIC = 0.312 mg/mL), further confirming its role as effective antibacterial against a whole series of both antibiotic-resistant and non-resistant bacterial strains, such as A. baumannii, E. faecalis, E. coli, K. pneumoniae, Listeria monocytogenes, P. aeruginosa, Salmonella enteritidis, Salmonella Typhimurium, Streptococcus mutans, and S. aureus (Sakkas and Papadopoulou, 2017; Yuan et al., 2023; Tejada-Muñoz et al., 2024; Ezzariga et al., 2025; Tao et al., 2025). As reported in Saoudi et al. (2024), the mechanism of action of EOs is strictly dependent on the quality and quantity of their chemical compounds. In fact, their antibacterial activity does not result from a single mechanism, but rather from a cascade of reactions involving the entire bacterial cell. The remarkable efficacy of the OEO we analysed appears to be attributed to the presence of high percentages of thymol, terpinene, and p-cymene, which are the main components of the oil analyzed in this study. In particular, thymol disrupts bacterial cell membranes, causing leakage of cellular contents and ultimately leading to cell death. It can also inhibit efflux pumps which are involved in antibiotic resistance (Langeveld et al., 2014; Sakkas and Papadopoulou, 2017; Walsh et al., 2003).
Diversity in the antibacterial activity of the EO from O. vulgare is mainly ascribed to the abundance percentage of its principal constituents, which vary considerably depending on altitude, temperature, harvest season, and geographical location, and extraction methods (Piasecki et al., 2023; Tejada-Muñoz et al., 2024).
Few studies have reported that OEO can synergize with certain antibiotics, enhancing their effectiveness against bacteria of clinical importance and potentially reducing the required dosage of the antibiotics. For further expanding the data of synergistic effects, the interactions between the OEO and conventional antibiotics were assayed through checkerboard method. It emerged that the combination of OEO with ampicillin, gentamicin, tetracycline, and tobramycin significantly enhance the antibacterial activity of the antibiotics against S. aureus when compared to antibiotic or OEO alone, as demonstrated by the application of effective models describing the synergism between different compounds and by the low FICI values. Depending to the antibiotic, these combinations decrease antibiotic dosage able to inhibit S. aureus growth from a minimum of 2 to a maximum of 32 folds.
Results obtained from time-kill kinetics assays provide more complementary evidence for the observed synergy effect of OEO antibiotic combination generated by our checkerboard assay since the combinations were more effective against S. aureus than the antibiotic alone, displaying an overtime bactericidal action antibiotic-dependent.
As reported by Drioiche et al. (2024), these synergistic effects could result from their ability to target the same bacterial proteins or facilitate access to target sites, as suggested by molecular docking simulations. Results we observed are in line with some studies, such as that of Xiao et al. (2020) who conducted a large-scale study analyzing the activity of 139 essential oils, demonstrating that, when combined with some currently recommended antibiotics for S. aureus infections, OEO showed a positive enhancement effect in increasing the activity of some antibiotics (Rosato et al., 2010; Amassmoud et al., 2023; Sena et al., 2024; Drioiche et al., 2024; Khashei et al., 2025). What is more, our results appear particularly interesting because they prove that the synergistic effect of OEO is antibiotic specific. Indeed, OEO is most effective when combined with β-lactam and tetracyclines antibiotics, less effective with aminoglycosides, and ineffective in combination with fluoroquinolones and macrolides. Furthermore, the reduction of the minimum dose necessary for each drug to exert its antibacterial effect is particularly relevant, if we consider the need to reduce not only the use of antibiotics, but also their possible toxic effects, such nephrotoxicity and ototoxicity, often associated with their high dosage and long-term exposure (Tyers and Wright, 2019; Coates et al., 2020). It is difficult to propose general antibacterial mechanisms of essential oil-antibiotic combinations owing to those specific biological properties of the OEO, which we mentioned earlier, and to distinct sensitivity of the microorganisms. However, as reported in literature, it seems that one of the most recognized mechanisms that could lead to this synergistic effect between OEO and intracellular-targeted antibiotics, such as gentamycin and tobramycin, is the destabilization of the bacterial membrane by oil components, which facilitates the penetration of the antibiotics towards the cytoplasm and better reaches their targets. With β-lactam and tetracycline antibiotics, the synergy could be due to the ability of certain oil components to inhibit bacterial resistance mechanisms such as efflux pumps that can pump antibiotics out of the bacterial cell before they can have effect. Lastly, combination of the OEO with β-lactams, which interfere with peptidoglycan formation, may have synergistic effects enhancing the disruption of bacterial cell walls (Aelenei et al., 2016; Saoudi et al., 2024).
Interestingly, the synergic OEO-antibiotic combinations exhibited antibiofilm activity since they are more effective at interfere with biofilm formation than either the OEO or antibiotic alone, as demonstrated by inhibition percentages of up to 91%. This activity could be attributed to the ability of OEO components to synergistically act with antibiotics in interfering with the various processes involved in biofilm formation, such as QS. This would seem to be confirmed by the downregulation we observed of agrA, hld, RNAIII, and tot genes encoding key components of the QS system in S. aureus. The agr locus encodes a two-component QS system that modulates the synthesis of the transcriptional regulator RNA III and the autoregulation of the agr system. The fact that OEO-antibiotic combinations influence the expression of the RNA III gene, significantly reducing its transcription, demonstrates its role in S. aureus biofilm formation. It is responsible for the post-transcriptional regulation of several virulence factors, which in turn influence the expression of cell surface-related proteins and extracellular toxins such as alpha-hemolysin (hla) and delta-hemolysin (hld) (Martínez et al., 2023). Negative regulation of the hld gene by the OEO-antibiotic combinations suggests that they affect not only hemolysin synthesis but also QS signal recognition proteins, causing inhibition on the expression of transcriptional regulators, toxin production, and biofilm formation.
As we have already reported in a previous study, combinations of EOs and antibiotics induce changes of global methylation levels of cytosines and adenines compared to EO or antibiotics alone. Results we obtained in the present study confirmed that overall, the OEO-antibiotic combinations induce a hypermethylation of cytosines and a downregulation of adenines even if these changes are specific for each antibiotic analyzed, thus shedding light on the molecular mechanisms through which the combinations exert their effects at the intracellular level. As we stated previously whether these changes are correlated to the inhibition or activation of gene expression even today remains unclear. One thing is certain: in different experimental conditions based on different antibiotics in different bacterial strains, epigenetic changes seem to recur in mediating the effects of the EO-antibiotic combinations in bacteria. Similarly, although a number of literature data report the enzymes involved in establishing and maintaining bacterial DNA methylation, it remains to be experimentally determined which of these enzymes are affected by oils and their combinations with antibiotics (Oliveira and Fang, 2021; Gao et al., 2023; Ma et al., 2025b).
Results we obtained in this study support the use of essential oils in combination with antibiotics as new treatment modalities to the bacterial infections opening the door for more research into the related field. If these data will be further confirmed, the adoption of novel therapeutic approaches might be used not exclusively against pathogenic bacteria, but primarily against multidrug-resistant bacteria, which are progressively increasing and against which conventional antibiotics appear to be ineffective. We are aware that, despite the promising results obtained so far in basic research which are confirmed in a plethora of works, challenges remain, such as the stability, selectivity, and bioavailability of these natural products in the human body and any potential adverse reactions. These factors need to be further explored to provide sufficient evidence for their effective application in the clinical field.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
ED: Investigation, Methodology, Writing – review & editing. PD: Data curation, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing. GS: Investigation, Methodology, Writing – review & editing. AP: Methodology, Writing – review & editing. GP: Project administration, Resources, Supervision, Writing – review & editing. DB: Conceptualization, Formal Analysis, Supervision, Validation, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This work was supported by Gruppo Baffa (Sadel Spa, Sadel San Teodoro srl, Sadel CSsrl, Casa di Cura Madonna dello Scoglio, AGI srl, Casa di Cura Villa del Rosario srl, Savelli Hospital srl) and Casa di Cura Villa Ermelinda in the frame of the agreements Tecnologie Avanzate per il Miglioramento dei Servizi alla Persona and Tecnologie Avanzate per l’Indagine delle Relazioni tra Uomo ed Ambienti di Vita. The authors declare that this study received the Essential Oils from Michele Crudo Farm, Vibo valentia (Italy). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2025.1684624/full#supplementary-material
Supplementary Figure 1 | Representative standard curves generated with m6A and m5C standard controls.
References
Aelenei, P., Miron, A., Trifan, A., Bujor, A., Gille, E., and Aprotosoaie, A. C. (2016). Essential oils and their components as modulators of antibiotic activity against gram-negative bacteria. Medicines (Basel). 3, 19. doi: 10.3390/medicines3030019
Amassmoud, O., Abbad, I., Iriti, M., Hassani, L., Mezrioui, N., and Abbad, A. (2023). Antibacterial Activity of Essential Oils Combinations based on Thymus broussonnetii, and Their Synergism with some Antibiotics. CurrMicrobiol. 80, 398. doi: 10.1007/s00284-023-03510-x
Arrigoni, R., Ballini, A., Jirillo, E., and Santacroce, L. (2024). Current view on major natural compounds endowed with antibacterial and antiviral effects. Antibiotics (Basel). 13, 603. doi: 10.3390/antibiotics13070603
Bacińska, Z., Strub, D. J., and Balcerzak, L. (2025). Antibacterial potential of essential oils against oral pathogenic bacteria: a literature and clinical review. J. Appl. Microbiol. 136, lxaf161. doi: 10.1093/jambio/lxaf161
Brdová, D., Ruml, T., and Viktorová, J. (2024). Mechanism of staphylococcal resistance to clinically relevant antibiotics. Drug Resist. Updat. 77, 101147. doi: 10.1016/j.drup.2024.101147
Cheung, G. Y. C., Bae, J. S., and Otto, M. (2021). Pathogenicity and virulence of Staphylococcus aureus. Virulence. 12, 547–569. doi: 10.1080/21505594.2021.1878688
Coates, A. R. M., Hu, Y., Holt, J., and Yeh, P. (2020). Antibiotic combination therapy against resistant bacterial infections: synergy, rejuvenation and resistance reduction. Expert Rev. Anti InfectTher. 18, 5–15. doi: 10.1080/14787210.2020.1705155
D’Aquila, P., De Rose, E., Sena, G., Scorza, A., Cretella, B., Passarino, G., et al. (2024). Quorum quenching approaches against bacterial-biofilm-induced antibiotic resistance. Antibiotics (Basel). 13, 619. doi: 10.3390/antibiotics13070680
D’Aquila, P., Paparazzo, E., Crudo, M., Bonacci, S., Procopio, A., Passarino, G., et al. (2022). Antibacterial activity and epigenetic remodeling of essential oils from calabrian aromatic plants. Nutrients. 14, 391. doi: 10.3390/nu14020391
D’Aquila, P., Sena, G., Crudo, M., Passarino, G., and Bellizzi, D. (2023). Effect of Essential Oils of Apiaceae, Lamiaceae, Lauraceae, Myrtaceae, and Rutaceae Family Plants on Growth, Biofilm Formation, and Quorum Sensing in Chromobacteriumviolaceum, Pseudomonas aeruginosa, and Enterococcus faecalis. Microorganisms. 11, 1150. doi: 10.3390/microorganisms11051150
Drioiche, A., Baammi, S., Zibouh, K., Al Kamaly, O., Alnakhli, A. M., Remok, F., et al. (2024). A Study of the Synergistic Effects of Essential Oils from Origanum compactum and Origanum elongatum with Commercial Antibiotics against Highly Prioritized Multidrug-Resistant Bacteria for the World Health Organization. Metabolites. 14, 210. doi: 10.3390/metabo14040210
Ezzariga, N., Moukal, A., Asdadi, A., Lemkhente, Z., Moustaoui, F., Kaaya, A., et al. (2025). Evaluation of the antimicrobial activity of 20 essential oils and their combinations on bacterial and fungal strains. Cureus. 17, e79499. doi: 10.7759/cureus.79499
Fikry, S., Khalil, N., and Salama, O. (2019). Chemical profiling, biostatic and biocidal dynamics of Origanum vulgare L. essential oil. AMB Express. 9, 41. doi: 10.1186/s13568-019-0764-y
Fratini, F., Mancini, S., Turchi, B., Friscia, E., Pistelli, L., Giusti, G., et al. (2017). A novel interpretation of the Fractional Inhibitory Concentration Index: The case Origanum vulgare L. and Leptospermum scoparium J. R. et G. Forst essential oils against Staphylococcus aureus strains. Microbiol. Res. 195, 11–17. doi: 10.1016/j.micres.2016.11.005
Gao, Q., Lu, S., Wang, Y., He, L., Wang, M., Jia, R., et al. (2023). Bacterial DNA methyltransferase: A key to the epigenetic world with lessons learned from proteobacteria. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1129437
Gonçalves, A. S. C., Leitão, M. M., Simões, M., and Borges, A. (2023). The action of phytochemicals in biofilm control. Nat. Prod. Rep. 40, 595–627. doi: 10.1039/D2NP00053A
Hamad, M., Al-Marzooq, F., Srinivasulu, V., Sulaiman, A., Menon, V., Ramadan, W. S., et al. (2024). Antimicrobial activity of nature-inspired molecules against multidrug-resistant bacteria. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1336856
Ianevski, A., Giri, A. K., and Aittokallio, T. (2020). SynergyFinder 2.0: visual analytics of multi-drugcombination synergies. Nucleic Acids Res. 48, W488–W493. doi: 10.1093/nar/gkaa216
Ianevski, A., He, L., Aittokallio, T., and Tang, J. (2017). SynergyFinder: a web application for analyzingdrug combination dose-response matrix data. Bioinformatics. 33. doi: 10.3389/fmicb.2023.1336856
Jilani, S., Ferjeni, M., Al-Shammery, K., Rashid Mohammed AlTamimi, H., Besbes, M., Ahmed Lotfi, S., et al. (2025). The synergistic effect of Thymus vulgaris essential oil and carvacrol with imipenem against carbapenem-resistant Acinetobacter baumannii: in vitro, molecular docking, and molecular dynamics studies. Front. Pharmacol. 16. doi: 10.3389/fphar.2025.1582102
Khashei, S., Fazeli, H., Rahimi, F., and Karbasizadeh, V. (2025). Antibiotic-potentiating efficacy of Rosmarinus officinalis L. @ to combat planktonic cells, biofilms, and efflux pump activities of extensively drug-resistant Acinetobacter baumannii clinical strains. Front. Pharmacol. 16. doi: 10.3389/fphar.2025.1558611
Langeveld, W. T., Veldhuizen, E. J., and Burt, S. A. (2014). Synergy between essential oil components and antibiotics: a review. Crit. RevMicrobiol. 40, 76–94. doi: 10.3109/1040841X.2013.763219
Liu, H., Hu, D., Wang, D., Wu, H., Pan, Y., Chen, X., et al. (2023). In vitro analysis of synergistic combination of polymyxin B with 12 other antibiotics against MDR Acinetobacter baumannii isolated from a Chinese tertiary hospital. J. Antibiot (Tokyo). 76, 20–26. doi: 10.1038/s41429-022-00573-z
Ma, W. Y., Shen, K. S., Wang, Z., Liu, Q., Diao, X. J., and Liu, G. R. (2025a). Synergistic antimicrobial effect and mechanism of enterocin Gr17 and cinnamaldehyde against Escherichia coli and Candida albicans. Arch. Microbiol. 207, 195. doi: 10.1007/s00203-025-04391-3
Ma, Y.-X., Wang, X.-D., and Li, X.-M. (2025b). The emerging role of DNA methylation in the pathogenicity of bacterial pathogens. J. bacteriology 207, e0010825. doi: 10.1128/jb.00108-25
Maggio, F., Rossi, C., Serio, A., Chaves-Lopez, C., Casaccia, M., and Paparella (2025). A. Anti-biofilm mechanisms of action of essential oils by targeting genes involved in quorum sensing, motility, adhesion, and virulence: A review. Int. J. Food Microbiol. 426, 110874. doi: 10.1016/j.ijfoodmicro.2024.110874
Martínez, A., Stashenko, E. E., Sáez, R. T., Zafra, G., and Ortiz, C. (2023). Effect of Essential Oil from Lippiaoriganoides on the Transcriptional Expression of Genes Related to Quorum Sensing, Biofilm Formation, and Virulence of Escherichia coli and Staphylococcus aureus. Antibiotics (Basel). 12, 845. doi: 10.3390/antibiotics12050845
Merghni, A., Haddaji, N., Bouali, N., Alabbosh, K. F., Adnan, M., Snoussi, M., et al. (2022). Comparative Study of Antibacterial, Antibiofilm, Antiswarming and Antiquorum Sensing Activities of Origanum vulgare Essential Oil and Terpinene-4-ol against Pathogenic Bacteria. Life (Basel). 12, 1616. doi: 10.3390/life12101616
Mootz, J. M., Benson, M. A., Heim, C. E., Crosby, H. A., Kavanaugh, J. S., Dunman, P. M., et al. (2015). Rot is a key regulator of Staphylococcus aureus biofilm formation. Mol. Microbiol. 96, 388–404. doi: 10.1111/mmi.12943
NCCLS (1999). Methods for determining bactericidal activity of antimi- crobial agents; approved guideline (Wayne, PA: Document M26-A. National Commit- tee for Clinical Laboratory Standards).
Nurzyńska-Wierdak, R. and Walasek-Janusz, M. (2025). Chemical composition, biological activity, and potential uses of oregano (Origanum vulgare L.) and oregano essential oil. Pharm. (Basel). 18, 267. doi: 10.3390/ph18020267
Oliveira, P. H. and Fang, G. (2021). Conserved DNA methyltransferases: A window into fundamental mechanisms of epigenetic regulation in bacteria. Trends Microbiol. 29, 28–40. doi: 10.1016/j.tim.2020.04.007
Piasecki, B., Balázs, V. L., Kieltyka-Dadasiewicz, A., Szabó, P., Kocsis, B., Horváth, G., et al. (2023). Microbiological studies on the influence of essential oils from several origanum species on respiratory pathogens. Molecules. 28, 3044. doi: 10.3390/molecules28073044
Rosato, A., Piarulli, M., Corbo, F., Muraglia, M., Carone, A., Vitali, M. E., et al. (2010). In vitro synergistic antibacterial action of certain combinations of gentamicin and essential oils. Curr. Med. Chem. 17, 3289–3295. doi: 10.2174/092986710792231996
Sakkas, H. and Papadopoulou, C. (2017). Antimicrobial activity of basil, oregano, and thyme essential oils. J. Microbiol. Biotechnol. 27, 429–438. doi: 10.4014/jmb.1608.08024
Salam, M. A., Al-Amin, M. Y., Salam, M. T., Pawar, J. S., Akhter, N., Rabaan, A. A., et al. (2023). Antimicrobial resistance: A growing serious threat for global public health. Healthcare (Basel). 11, 1946. doi: 10.3390/healthcare11131946
Saoudi, B., Bariz, K., Saci, S., Belounis, Y., Ait Issad, H., Abbaci, M., et al. (2024). Enhancing Antibiotic Efficacy and Combating Biofilm Formation: Evaluating the Synergistic Potential of Origanum vulgare Essential Oil against Multidrug-Resistant Gram-Negative Bacteria. Microorganisms. 12, 1651. doi: 10.3390/microorganisms12081651
Sati, H., Carrara, E., Savoldi, A., Hansen, P., Garlasco, J., Campagnaro, E., et al. (2025). WHO Bacterial Priority Pathogens List Advisory Group. The WHO Bacterial Priority Pathogens List 2024: a prioritisation study to guide research, development, and public health strategies against antimicrobial resistance. Lancet InfectDis. 25, 1033–1043. doi: 10.1016/S1473-3099(25)00118-5
Sena, G., De Rose, E., Crudo, M., Filippelli, G., Passarino, G., Bellizzi, D., et al. (2024). Essential Oils from Southern Italian Aromatic Plants Synergize with Antibiotics against Escherichia coli, Pseudomonas aeruginosa and Enterococcus faecalis Cell Growth and Biofilm Formation. Antibiotics (Basel). 13, 605. doi: 10.3390/antibiotics13070605
Shao, K., Yang, Y., Gong, X., Chen, K., Liao, Z., and Ojha, S. C. (2025). Staphylococcal drug resistance: mechanisms, therapies, and nanoparticle interventions. Infect. Drug Resist. 18, 1007–1033. doi: 10.2147/IDR.S510024
Sharifi, A., Mohammadzadeh, A., Salehi, T. Z., Mahmoodi, P., and Nourian, A. (2021). Cuminum cyminum L. Essential Oil: A Promising Antibacterial and Antivirulence Agent Against Multidrug-Resistant Staphylococcus aureus. Front. Microbiol. 12. doi: 10.3389/fmicb.2021.667833
Soltani, S., Shakeri, A., Iranshahi, M., and Boozari, M. (2021). A review of the phytochemistry and antimicrobial properties of origanum vulgare L. and subspecies. Iran J. Pharm. Res. Spring. 20, 268–285. doi: 10.22037/ijpr.2020.113874.14539
Swamy, M. K., Akhtar, M. S., and Sinniah, U. R. (2016). Antimicrobial properties of plant essential oils against human pathogens and their mode of action: an updated review. Evid Based Complement Alternat Med. 2016, 3012462. doi: 10.1155/2016/3012462
Tang, K. W. K., Millar, B. C., and Moore, J. E. (2023). Antimicrobial resistance (AMR). Br. J. BioMed. Sci. 80. doi: 10.3389/bjbs.2023.11387
Tao, L., Liang, Y., Xia, Z., Wang, X., Wang, X., Chao, Z., et al. (2025). Antibacterial activities of oregano essential oils and their active components. Front. Pharmacol. 16. doi: 10.3389/fphar.2025.1579283
Tariq, S., Wani, S., Rasool, W., Shafi, K., Bhat, M. A., Prabhakar, A., et al. (2019). A comprehensive review of the antibacterial, antifungal and antiviral potential of essential oils and their chemical constituents against drug-resistant microbial pathogens. MicrobPathog. 134, 103580. doi: 10.1016/j.micpath.2019.103580
Tejada-Muñoz, S., Cortez, D., Rascón, J., Chavez, S. G., Caetano, A. C., Díaz-Manchay, R. J., et al. (2024). Antimicrobial Activity of Origanum vulgare Essential Oil against Staphylococcus aureus and Escherichia coli. Pharm. (Basel). 17, 1430. doi: 10.3390/ph17111430
Touaitia, R., Mairi, A., Ibrahim, N. A., Basher, N. S., Idres, T., and Touati, A. (2025). Staphylococcus aureus: A review of the pathogenesis and virulence mechanisms. Antibiotics (Basel). 14, 470. doi: 10.3390/antibiotics14050470
Tyers, M. and Wright, G. D. (2019). Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 17, 141–155. doi: 10.1038/s41579-018-0141-x
Walsh, S. E., Maillard, J. Y., Russell, A. D., Catrenich, C. E., Charbonneau, D. L., and Bartolo, R. G. (2003). Activity and mechanisms of action of selected biocidal agents on Gram-positive and -negative bacteria. J. ApplMicrobiol. 94, 240–247. doi: 10.1046/j.1365-2672.2003.01825.x
Xiao, S., Cui, P., Shi, W., and Zhang, Y. (2020). Identification of essential oils with activity against stationary phase Staphylococcus aureus. BMC Complementary Med. Therapies. 20, 99. doi: 10.1186/s12906-020-02898-4
Yamazaki, Y., Ito, T., Tamai, M., Nakagawa, S., and Nakamura, Y. (2024). The role of Staphylococcus aureus quorum sensing in cutaneous and systemic infections. Inflammation Regen. 44, 9. doi: 10.1186/s41232-024-00323-8
Yuan, Y., Sun, J., Song, Y., Raka, R. N., Xiang, J., Wu, H., et al. (2023). Antibacterial activity of oregano essential oils against Streptococcus mutans in vitro and analysis of active components. BMC Complement Med. Ther. 23, 61. doi: 10.1186/s12906-023-03890-4
Zhao, K., Jiang, Y., Dev, K., He, X., Sharma, V., and Pang, X. (2024). Terpenoids as principal bioactive compound of Cissampelosoppositifolia essential oils: enhancing synergistic efficacy with conventional antibiotics. Front. Cell Infect. Microbiol. 14. doi: 10.3389/fcimb.2024.1481656
Keywords: essential oil, Origanum vulgare, Staphylococcus aureus, minimum inhibitory concentration (MIC), antibiotics, biofilm, quorum sensing, DNA methylation
Citation: De Rose E, D’Aquila P, Sena G, Perrotta A, Passarino G and Bellizzi D (2025) Antimicrobial effects of essential oil from Origanum vulgare in combination with conventional antibiotics against Staphylococcus aureus. Front. Cell. Infect. Microbiol. 15:1684624. doi: 10.3389/fcimb.2025.1684624
Received: 12 August 2025; Accepted: 26 September 2025;
Published: 23 October 2025.
Edited by:
Emanuela Roscetto, Federico II University Hospital, ItalyReviewed by:
Drioiche Aziz, Moulay Ismail University, MoroccoHikmat Douhri, Abdelmalek Essaadi University, Morocco
Copyright © 2025 De Rose, D’Aquila, Sena, Perrotta, Passarino and Bellizzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dina Bellizzi, ZGluYS5iZWxsaXp6aUB1bmljYWwuaXQ=
†These authors have contributed equally to this work and share first authorship
 Elisabetta De Rose
Elisabetta De Rose Patrizia D’Aquila
Patrizia D’Aquila Giada Sena
Giada Sena Annamaria Perrotta
Annamaria Perrotta Giuseppe Passarino
Giuseppe Passarino Dina Bellizzi
Dina Bellizzi