- 1Department of Science, Roma Tre University, Rome, Italy
- 2Department of Veterinary and Animal Sciences, University of Copenhagen, Frederiksberg C, Denmark
- 3Department of Biosciences, University of Milan, Milan, Lombardy, Italy
Editorial on the Research Topic
Deciphering antimicrobial resistance: genetic insights and perspectives
Antimicrobial agents have propelled medicine into the era of modern healthcare. However, the efficacy of antimicrobial therapies is progressively weakening as antimicrobial resistance (AMR) escalates, representing a profound threat to global health that transcends human, veterinary, and environmental boundaries. The development of AMR arises from multiple drivers, including i) antimicrobial overuse and misuse, ii) insufficient diagnostic practices, and iii) the intrinsic adaptability of microbial genomes. As AMR continues to spread, our ability to combat common infections and undertake routine medical interventions safely is increasingly compromised (Antimicrobial Resistance Collaborators, 2022). Addressing this challenge requires comprehensive research into the genetic basis of resistance mechanisms, surveillance of microbial communities across multiple reservoirs, and the development of novel strategies to counteract AMR (Jauneikaite et al., 2023).
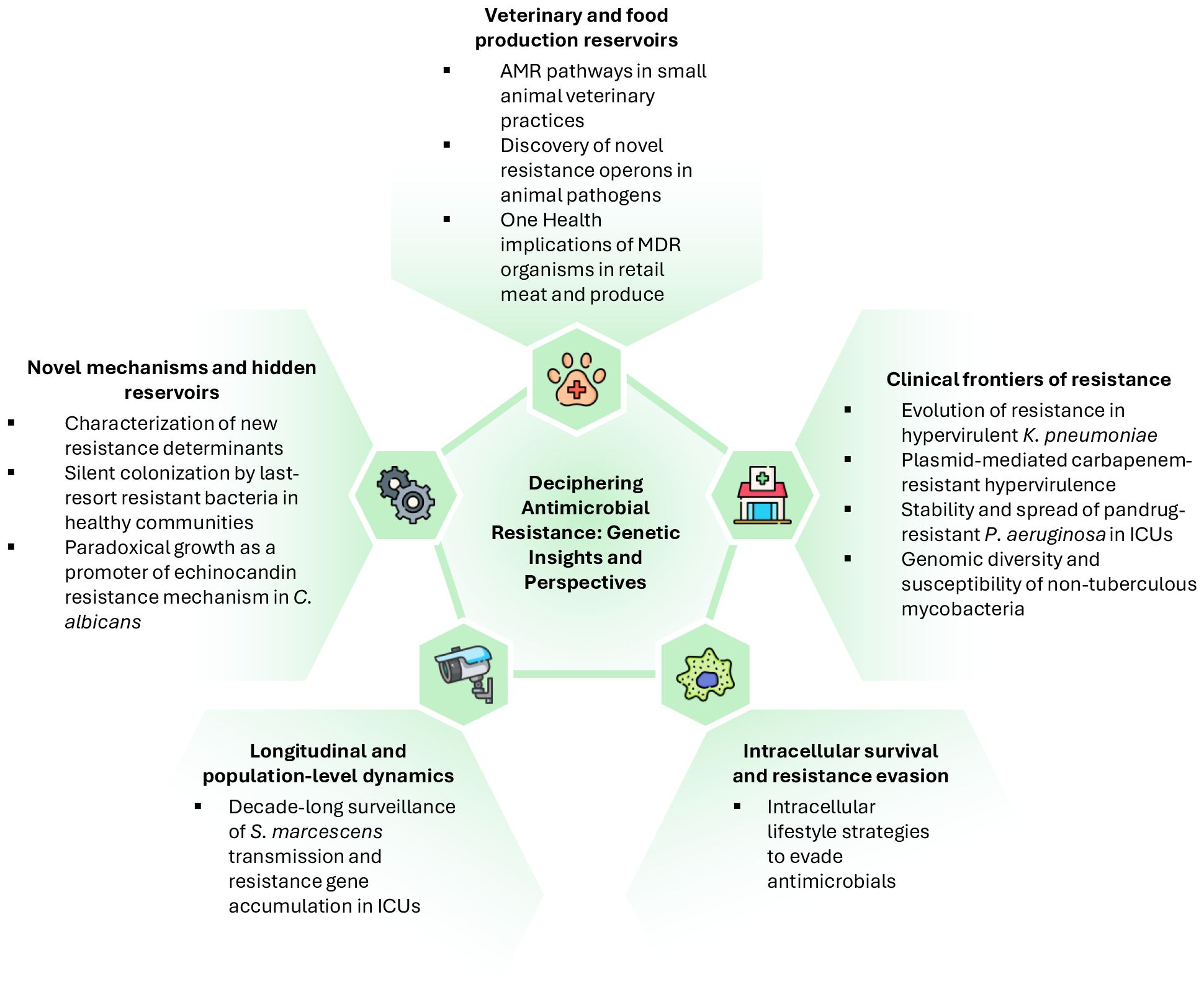
The Research Topic presented in Frontiers in Cellular and Infection Microbiology provides a comprehensive view of the genetic determinants, epidemiology, and functional consequences of AMR across clinical, veterinary, and environmental settings. These studies highlight the need for One Health strategies, integrating surveillance and interventions across humans, animals, and the environment to curb the global spread of AMR.
In veterinary settings, the study of AMR reveals both environmental and human-mediated pathways for the dissemination of resistant organisms. Moreira da Silva et al. demonstrated that small animal veterinary practices in Portugal are contaminated by multidrug-resistant (MDR) microorganisms, including carbapenem- and methicillin-resistant strains, emphasizing the urgent need for robust infection prevention and control measures. Concerning resistant pathogens, Glambek et al. identified a novel trexAB operon in Streptococcus dysgalactiae, elucidating the genetic basis of low-grade tetracycline resistance and revealing previously unrecognized mechanisms of AMR in both clinical and veterinary settings. The relevance of veterinary reservoirs extends into food production, as illustrated by Habib et al., who reported widespread MDR and toxigenic methicillin-resistant Staphylococcus aureus (MRSA) strains in retail meat and produce in the United Arab Emirates, emphasizing the One Health implications of AMR and the potential for community exposure.
Mechanistic insights into novel resistance determinants further broaden our understanding of AMR evolution and its dissemination across microbial species. Lin et al. characterized aac(6’)-Iaq, a new aminoglycoside acetyltransferase from Brucella intermedia, which confers resistance to multiple aminoglycosides. At the community level, Peng et al. identified silent intestinal colonization by tet(X4)-positive, tigecycline-resistant MDR Escherichia coli in healthy people, highlighting hidden reservoirs of last-resort antibiotic resistance beyond clinical settings.
Concerning the hospital settings, clinical isolates continue to offer critical insights into AMR dynamics. Liu et al. described the evolution of ceftazidime-avibactam resistance in hypervirulent ST11-K64 Klebsiella pneumoniae, driven by the blaKPC-190 variant that simultaneously confers resistance and partially restores carbapenem susceptibility. Zhang et al. traced the emergence of NDM-1-positive ST592 K. pneumoniae, illustrating plasmid-mediated evolution toward carbapenem-resistant hypervirulence. Similarly, Zheng et al. examined the coexistence of blaKPC-2 and blaVIM-2 plasmids in intensive care unit (ICU)-derived pandrug-resistant Pseudomonas aeruginosa strains, pointing out their stability and minimal fitness cost, which facilitate dissemination within healthcare environments. Expanding the scope to non-tuberculous mycobacteria, Zhao et al. demonstrated that Mycobacterium seoulense isolates exhibit variable susceptibility and genomic diversity, reinforcing the importance of targeted susceptibility testing. In the fungal realm, Wei et al. reported that Candida albicans can exhibit paradoxical growth under caspofungin exposure, a phenomenon in which fungal cells resume growth at drug concentrations above the minimum inhibitory concentration. This response is driven by activation of stress-response pathways coupled with segmental aneuploidy, which together confer reversible echinocandin resistance, illustrating how dynamically fungi can adapt their genomes under antimicrobial pressure. Intracellular lifestyle is one strategy some microbes use to evade antimicrobials. In this context, Bao et al. offered a lysosome-centered view of Mycobacterium tuberculosis pathogenesis, revealing intracellular survival strategies that indirectly influence therapeutic efficacy and potentially contribute to resistance development.
Finally, longitudinal studies, such as that from Zhu et al., can provide an essential perspective to enhance our understanding of AMR evolution and population dynamics. Indeed, this study analysed the Serratia marcescens complex in ICU patients over 11 years, identifying inter-ICU transmission and the accumulation of β-lactamase and carbapenemase genes, highlighting the importance of continuous genomic surveillance
Figure 1 highlights the valuable scientific contributions of this Research Topic, exemplifying the breadth and depth of current AMR research. The studies demonstrate that resistance emerges through adaptation to intracellular lifestyle and diverse genetic mechanisms, including plasmid-mediated gene acquisition, point mutations, efflux operons, and genome-scale rearrangements. Surveillance across clinical, community, veterinary, and environmental reservoirs is indispensable to identify emerging threats, while mechanistic studies guide the development of targeted interventions. Additionally, the works highlight the importance of combining genomic, phenotypic, and epidemiological approaches to fully understand the evolution and fitness costs associated with resistance.
Altogether, this Research Topic underscores the urgent need for One Health approaches to combat AMR. It advocates investment in diagnostics, genomic surveillance, and strategies to limit its spread, as deepening our understanding of AMR sharpens surveillance and therapeutic precision while informing policies that bridge human, animal, and environmental health. By fostering interdisciplinary and mechanistic research, the field moves closer to preserving the effectiveness of antimicrobials that underpin modern medicine.
Author contributions
ML: Writing – review & editing, Writing – original draft. MP: Writing – review & editing. VB: Writing – review & editing. DV: Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Keywords: antibiotic resistance genes (ARGs), infectious disease epidemiology, mobile genetic elements (MBEs), plasmids, phages, regulatory perspectives, resistance mechanisms, surveillance
Citation: Lucidi M, Pirolo M, Baldelli V and Visaggio D (2025) Editorial: Deciphering antimicrobial resistance: genetic insights and perspectives. Front. Cell. Infect. Microbiol. 15:1720241. doi: 10.3389/fcimb.2025.1720241
Received: 07 October 2025; Accepted: 16 October 2025;
Published: 22 October 2025.
Edited and reviewed by:
Vincent Cattoir, University of Rennes, FranceCopyright © 2025 Lucidi, Pirolo, Baldelli and Visaggio. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Massimiliano Lucidi, bWFzc2ltaWxpYW5vLmx1Y2lkaUB1bmlyb21hMy5pdA==
 Massimiliano Lucidi
Massimiliano Lucidi Mattia Pirolo
Mattia Pirolo Valerio Baldelli
Valerio Baldelli Daniela Visaggio
Daniela Visaggio