Abstract
Introduction:
Mesoderm-specific transcript (Mest), a paternally expressed imprinted gene, is involved in the modulation of adipose tissue expansion. Mest is also highly expressed in the developing and adult brain, suggesting a role in behavioral phenotypes. Previously, we showed that female mice with paternal Mest inactivation (MestpKO) exhibit no discernible behavioral impairments compared to wild-type mice. In this study, we performed metabolic phenotyping of female MestpKO mice in response to a dietary challenge.
Methods:
Eight-week-old female and male wild-type and MestpKO mice were fed a control or Western diet (40 kcal% fat) until 24 weeks of age. Body weight, body composition, and metabolic parameters were measured during the course of the feeding regimens, and gene expression and type-2-deiodinase (DIO2) activity were examined in white adipose tissue and brain at the end of the study.
Results:
Mest pKO female mice fed a Western diet were protected against diet-induced obesity. Strikingly, these mice showed increased ambulatory activity and speed, coupled with reduced resting parameters, suggesting a role for MEST in the regulation of spontaneous physical activity, a form of nonexercise activity thermogenesis. When considering body mass (control diet) and lean mass (Western diet), energy expenditure was increased in the female MestpKO mice. Male MestpKO mice did not exhibit these changes. Analyses of hypothalamic gene expression revealed upregulation of Dio2, and RNA-seq highlighted differential expression of numerous thyroid hormone-responsive genes in MestpKO female mice.
Conclusion:
Mechanistically, our results suggest that MEST directly or indirectly regulates thyroid hormone-responsive genes in the hypothalamus, thereby modulating the neurobiological control of nonexercise activity thermogenesis in Western diet-fed female mice.
1 Introduction
The global increase in the prevalence of obesity substantially raises the risk of its related comorbidities, such as type 2 diabetes, metabolic dysfunction-associated steatotic liver disease (MASLD), hypertension, cardiovascular disease, dementia, joint pain, and various cancers, imposing a significant burden on public health (1, 2). The development of obesity results from a combination of genetic, environmental, lifestyle, and epigenetic factors (3). Although monozygotic twin studies have revealed that the heritability of obesity is more than 40%, the identification of > 1,000 obesity-linked genetic variants through genome-wide association studies still accounts for a small fraction of the variance of body mass index (4, 5). A significant component of obesity remains unaccounted for, further reinforcing the complexity of the condition.
Mesoderm-specific transcript (MEST), a paternally expressed imprinted gene with an unknown molecular function, belongs to the alpha/beta hydrolase protein family, which exhibits a broad range of activities (6, 7). Mest mRNA and protein levels vary greatly—up to ~ 50-fold—in white adipose tissue (WAT) from individual C57BL/6J mice fed an obesogenic diet, and variations in WAT Mest show a positive association with fat mass deposition (8). The role of MEST in adipose tissue has been well characterized (9–11). Transgenic overexpression of Mest in mice results in enlarged adipocytes (10), whereas mice with global and adipocyte-specific inactivation of Mest show reduced diet-induced obesity (12), further highlighting a role for Mest in WAT expansion. Moreover, Mest is localized to the endoplasmic reticulum of adipocytes, in close proximity to the membranes of lipid droplets (13).
Human total daily energy expenditure is composed of basal metabolic rate, the thermic effect of food, and activity thermogenesis, which includes exercise and “nonexercise activity thermogenesis” (NEAT) (14). Purposeful human exercise is volitional, whereas NEAT encompasses all activities outside of chosen exercise (15), including the drive to stand and walk, fidgeting, and gesticulating (16). NEAT-associated activities can account for 100 to 800 kcal of energy expenditure (EE) per day (17). Harnessing NEAT as a mechanism to increase total daily energy expenditure is an attractive strategy to dissipate energy and provide protection against diet-induced obesity during times of excess caloric intake. Weight gain in obesity results from a chronic imbalance between caloric intake and energy expenditure. Excessive caloric intake combined with physical inactivity contributes to the development of obesity. Increasing physical activity through NEAT represents a potential strategy to enhance the energy expenditure component of body weight regulation (18, 19).
The significance of MEST in the central nervous system is highlighted by its expression in the embryonic midbrain, as well as in the early postnatal and adult brain (20–22). Mest is one of the most highly upregulated genes during early mesodiencephalic dopaminergic neuronal development in the midbrain (22) and during the morphological transition of primary neurons during formation of the mammalian neocortex (23). Mice with inactivation of Mest (Mesttm1.2Rkz) generated in our laboratory show no discernible impairments in object recognition memory, social behavior, or maternal behavior (20). Paradoxically, abnormal maternal behavior (21) and reduced climbing behavior (22) were reported by others using a different Mest knockout mouse (Mesttm1Masu). Phenotypic differences between these two models may result from differences in genetic background or gene-targeting approaches (20).
This study focused on metabolic phenotyping of female mice with paternal inactivation of Mest (MestpKO) fed a control or obesogenic diet. Remarkably, female MestpKO mice exhibited increased spontaneous physical activity (SPA) and were protected against diet-induced obesity. RNA-sequencing (RNA-seq) analysis of the hypothalamus from MEST-deficient female mice revealed upregulation of several thyroid hormone (TH)-responsive genes, suggesting a mechanism involving TH and MEST in the neurobiological regulation of increased SPA and protection against obesity in female MestpKO mice.
2 Materials and methods
2.1 Animals, diets, and experimental design
The Institutional Animal Care and Use Committee at MaineHealth Institute for Research approved all animal experiments in accordance with National Institutes of Health guidelines for the care and use of laboratory animals. We used Mesttm1.2Rkz mice (12) with a global inactivation of Mest and littermate wild-type (WT) mice for our studies. All mice were congenic on the C57BL/6J genetic background. Mice were fed the 2018 Teklad Global 18% protein rodent diet (Envigo, Indianapolis, IN, USA) and housed in a barrier facility with standard light and humidity conditions at an ambient temperature of 23 °C–24 °C. For dietary studies, female mice were fed a control (CD; D14042701i, Research Diets, New Brunswick, NJ, USA) or a Western diet (WD; 40 kcal% fat; D12079Bi, Research Diets, New Brunswick, NJ, USA). Singly housed 8-week-old, age-matched female and male WT mice and MestpKO mice were fed a CD or WD until 24 weeks of age, as illustrated in Figure 1A. Body weight (BW), body composition (NMR), glucose and insulin tolerance, and metabolic parameters (indirect calorimetry) were measured as indicated (Figure 1A). Detailed analyses were performed on mutant female mice that exhibited a pronounced increase in SPA. Male data are presented in the Supplementary Material as indicated in the Results section.
Figure 1

Phenotypic analyses of WT and MestpKO (pKO) female mice fed control (CD) and Western diet (WD). (A) Schematic showing the study design used for dietary studies. BWs of WT and MestpKO female mice fed CD (B) and WD (C) from 8 to 24 weeks of age. (D) Fat mass, (E) % fat mass, (F) lean mass, and (G) % lean mass of CD-fed female WT and MestpKO mice at 8, 16, and 23 weeks of age measured by NMR. (H) Fat mass, (I) % fat mass, (J) lean mass, and (K) % lean mass of WD-fed female mice at 8, 16, and 24 weeks of age. (L) Glucose tolerance test (GTT) and (M) area under the curve (AUC) for WT and MestpKO female mice fed WD at 16 weeks of age. (N) Insulin tolerance test of WD-fed WT and MestpKO female mice at 17 weeks of age. (O) Analyses of Mest gene expression show it to be almost completely inactivated in the hypothalamus (Hyp), pituitary (Pit), amygdala (Amy), motor cortex (MC), and inguinal (Ing) and gonadal (Gon) white adipose tissue of female MestpKO mice. Data in figure panels (B–K) were analyzed by two-way ANOVA, and (L–O) by unpaired t-tests. Annotations with a–d indicate p-values lower than 0.05, 0.01, 0.001, and 0.0001 for data in (B, C). p-values are numerically indicated for the remaining figures, with < 0.001 indicating a p-value of less than 0.0005. Schematic (A) was created in https://BioRender.com.
2.2 Body composition and metabolic cage analyses
Body composition of mice was measured using a Minispec LF50 TD-NMR analyzer (Bruker, Massachusetts) as previously described (12). Indirect calorimetry was performed using the Promethion metabolic cage system (Sable Systems International, North Las Vegas, NV, USA). Metabolic variables—oxygen consumption (VO2), carbon dioxide production (VCO2), EE, respiratory exchange ratio (RER), and locomotor activity—were analyzed using web-based CalR software (24). Generalized linear modeling, using BW, fat mass (FM), and lean mass (LM) as covariates, was performed for VO2, VCO2, and EE. Analysis of variance (ANOVA) was used for mass-independent variables, including locomotor activity and RER. Fine movements, such as grooming and scratching, were calculated as the difference between AllMeters and PedMeters (Sable Systems). NEAT was calculated as previously described (25). Total EE (TEE; kcal/h) was divided by two to obtain TEE values every 30 min (TEE-30). The lowest TEE-30 value for each day was considered the basal metabolic rate (BMR). NEAT was then calculated by subtracting BMR from each TEE-30, yielding NEAT values every 30 min (NEAT-30) for each day.
2.3 Glucose and insulin tolerance tests
Glucose tolerance tests (GTT) were performed after an overnight fast, followed by intraperitoneal administration of 2 g/kg glucose. Insulin tolerance tests (ITT) were performed after a 4-h fast, with intraperitoneal administration of 0.5 IU/kg insulin (Humulin R U-100, Eli Lilly and Company, Indianapolis, IN, USA). Blood glucose levels in the tail vein were monitored using a glucometer (Accu-check Aviva Plus, Roche Diagnostics, Indianapolis, IN, USA).
2.4 Serum corticosterone assay
At the end of the study, blood was collected from isoflurane-anesthetized mice by intracardiac puncture into BD SST™ tubes (Becton Dickinson, Franklin Lakes, NJ, USA). The tubes were inverted 10 × and allowed to clot at room temperature for 30 min. Samples were centrifuged at 1,300×g for 15 min, and serum was collected and stored at − 70 °C. Serum corticosterone levels were measured using the Corticosterone AssayMax ELISA Kit (EC3001-1, AssayPro, St. Charles, MO, USA).
2.5 DIO2 and DIO3 enzymatic assays
Type-2-deiodinase (DIO2) and type-3-deiodinase (DIO3) enzymatic activities were determined as previously described (26). In brief, frozen tissues were homogenized on ice in a buffer containing 20 mM Tris-HCl, 0.25 M sucrose, and 5 mM dithiothreitol (Sigma-Aldrich, St. Louis, MO, USA) (pH = 7.0 for DIO2 and 7.4 for DIO3). A suitable volume of tissue homogenate containing 50–200 μg of protein was used in the enzymatic reaction to ensure that total deiodination did not exceed 40% and remained proportional to the protein content. Tissue homogenates were incubated at 37 °C for an hour in the presence of 25 mM dithiothreitol (Sigma-Aldrich) with either 1 nM of 125I-labeled thyroxine (T4; Catalog No. NEX111X, Revvity, Waltham, MA, USA) for the DIO2 assay, or 2 nM 125I-labeled triiodothyronine (T3; Catalog No. NEX110X, Revvity) for the DIO3 assay. Unlabeled T3 (1 μM) was added to the DIO2 enzymatic reaction to prevent interference from DIO3 in the DIO2 assay. The enzymatic reactions were stopped with 1 vol of ethanol, and an aliquot was subjected to paper chromatography (Catalog No. 3001-614, Whatman, GE Healthcare Life Sciences, Chicago, IL, USA) as previously described (27), using ammonia-saturated 2-methyl-2-butanol as the eluent. Chromatograms were autoradiographed, and the bands corresponding to different metabolites were excised and counted using a gamma counter. Deiodination was determined based on the percentage of 125I-T3 produced from T4 (for DIO2 assay), accounting for unlabeled T3, or the percentage of 125I-3,3′-diiodothyronine produced from T3 (for the DIO3 assay).
2.6 Adipose tissue histology
Subcutaneous inguinal (iWAT) and gonadal white adipose tissue (gWAT) were fixed in Bouin’s solution (Harleco, Millipore Sigma, Darmstadt, Germany) for 48 h, paraffin-embedded, and processed for H&E staining.
2.7 RNA isolation and quantitative reverse transcription PCR
Tissue total RNA was extracted as previously described (12) using the RNeasy Mini Kit (Qiagen, Germantown, MD, USA). Briefly, tissues were homogenized in TriReagent (Molecular Research Center, Cincinnati, OH, USA), and RNA was purified using the RNeasy Mini Kit and RNAse-free DNAse (Qiagen). One-step quantitative reverse transcription PCR (RT-qPCR) was performed using the TaqMan RNA-to-CT Reagent (Thermo Fisher Scientific, Waltham, MA, USA) on a CFX384 Real-Time Platform (BioRad, Hercules, CA, USA). Two-step RT-qPCR was performed using SYBR Select Master Mix (Thermo Fisher Scientific) with cDNA synthesized using the High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific). Gene-specific TaqMan probes (LGC, BioSearch Technologies, Middlesex, TW11 0LY, UK) and PrimeTime primers (Integrated DNA Technologies, Coralville, IA, USA), as well as primer/probe combinations, were used for gene quantification. Gene expression was normalized to TATA box binding protein (Tbp) for TaqMan assays. The relative abundance of genes of interest measured using SYBR green was quantified and normalized to Tbp using the 2-ΔΔCT method. Probe and primer sequences for TaqMan assays, primer sequences for SYBR green assays, and stock numbers for primer and probe sets (Integrated DNA Technologies, Coralville, IA, USA) are listed in Supplementary File S1.
2.8 RNA-sequencing
Bulk RNA-seq of the whole mouse hypothalamus was performed at the Pennington Biomedical Research Center Genomics Core Facility (Baton Rouge, LA, USA). Total RNA quality was assessed using the Agilent Bioanalyzer RNA 6000 Assay. RNA-seq library construction was performed using the Lexogen QuantSeq 3’ mRNA-seq V2 Library Prep Kit FWD with UDI (Catalog No. 191.96; Lexogen, Greenland, NH, USA). Libraries were validated using the Agilent BioAnalyzer High Sensitivity DNA Assay (Agilent, Santa Clara, CA, USA). The ~ 275-bp libraries were pooled in equimolar amounts and sequenced on the Illumina NextSeq2000 (Illumina, San Diego, CA, USA) at 75 bp read length. Quality control of raw sequencing data was checked using FastQC (28), and Cutadapt (29) was used to trim adapter sequences and low-quality bases, ensuring cleaner data for downstream analysis. The cleaned reads were aligned to the reference genome (GRCm39 release 110) using the STAR aligner (30). Following alignment, HTSeq (31) was used to count the number of reads mapping to each gene, generating a count matrix. Differential analysis of RNA read count data was then performed using DESeq (32), which models the total counts as a negative binomial distribution and applies an empirical Bayes shrinkage-based method to estimate signal dispersion and fold changes. Gene expression signals were logarithmically transformed (base 2) for all downstream analyses, with the lowest expression value being set to 1. Genes with an absolute log fold change ≥ 1 and a false discovery rate (FDR) of 5% were considered differentially expressed. Analysis of differentially expressed genes was performed using the online enrichment analysis tool Enrichr (33). Gene ontology data were obtained from the GO Biological Process 2023, GO Cellular Component 2023, GO Molecular Function 2023, and MGI Mammalian Phenotype Level 4 libraries. Pathway analysis terms were retrieved from the Reactome 2022 library. Statistical significance was defined as an adjusted p-value ≤ 0.05.
2.9 Data analyses
All results are expressed as mean ± SEM. Data were tested for normality to assess Gaussian distribution, and differences between datasets were analyzed using unpaired parametric t-tests, one- or two-way ANOVA, or simple linear regression in GraphPad Prism 10.0.2.
3 Results
3.1 Female mice with paternal inactivation of Mest show resistance to diet-induced obesity
Using the experimental design in Figure 1A, female WT and MestpKO mice were fed a CD (10 kcal% fat) or WD (40 kcal% fat) from 8 to 24 weeks of age. BW measurements showed that CD-fed MestpKO female mice had slightly reduced BW compared to WT mice throughout the 8–14-week period (Figure 1B), due to small reductions in both FM and LM (Figures 1D–G). When fed WD from 8 to 24 weeks of age, mice showed considerable divergence in BW (Figure 1C), with WT mice gaining weight approximately twofold faster than MestpKO (0.51 g/week vs. 0.24 g/week; p = 0.0013). Indices of FM prior to WD feeding at 8 weeks of age (Figures 1H, I) and average weekly WD intake measured from 14 to 16 weeks of age (WT, 2.40 g/week vs. MestpKO, 2.28 g/week; p = 0.23), showed no difference between genotypes. However, divergence in FM occurred between genotypes by 16 weeks of age (Figures 1H, I). Analyses using two-way ANOVA showed that time and genotype contributed to differences in FM between WT and MestpKO mice fed WD (Supplementary Table S1A). While LM was similar between genotypes at 8 weeks of age, significant differences emerged after mice were fed WD (Figure 1J). Percentage LM relative to BW decreased by ~ 5%–6% in WT mice fed WD from 8 to 24 weeks of age, whereas it remained consistent in MestpKO mice throughout the period and was significantly higher than in WT mice at 16 and 24 weeks of age (Figure 1K). Since ~ 25% of adipose tissue mass consists of fat-free mass—often referred to as the quarter fat-free mass rule (34)—our results suggest that this component contributes to overall LM in WT mice (Figure 1J). Additionally, anal–nasal length measurements of 24-week-old WT and MestpKO female mice fed a 10-kcal% fat diet showed no differences between genotypes (WT, 91.2 mm; MestpKO, 90.2 mm; n = 8 mice per genotype; p = 0.20), suggesting that Mest inactivation does not significantly affect longitudinal growth. Glucose tolerance in CD-fed mice was comparable between genotypes (Supplementary Table S1B); however, when fed WD, MestpKO female mice exhibited significantly improved blood glucose levels after glucose injection compared to WT mice (Figure 1L). This was reflected by marginally significant differences (p = 0.062) in the area under the curve (Figure 1M). While circulating insulin levels were not measured in our study, an insulin tolerance test assessing whole-body insulin action showed improved insulin sensitivity in MestpKO female mice compared to WT (Figure 1N).
Male MestpKO mice fed CD or WD from 8 to 16 weeks of age showed significantly reduced BW gain associated with LM compared to WT mice (Supplementary Figure S1). Thus, reduced BW gain in response to dietary intervention is a phenotype consistent across both sexes.
Inactivation of the paternal allele of Mest has been shown to ablate Mest in WAT (12). In this study, we demonstrate similar monoallelic expression of Mest in various regions of the brain, as evidenced by inactivation of Mest in the hypothalamus (Hyp), pituitary (Pit), amygdala (Amy), and motor cortex (MC) of MestpKO mice (Figure 1O).
3.2 Female MestpKO mice fed a Western diet show increased energy expenditure and physical activity compared to WT mice
Indirect calorimetry of female WT and MestpKO mice fed CD or WD for 16 weeks showed no significant differences in average daily EE between genotypes (Figure 2A). However, analysis using generalized linear modeling (24) indicated that BW (total mass) was the primary determinant of 24-h EE between genotypes in CD-fed mice (Figure 2B; p = 0.040). The leftward shift in the regression plot for EE of MestpKO mice (Figure 2B) indicates a significant genotype effect associated with a higher metabolic rate. Furthermore, VO2, VCO2, and EE all exhibited significant genotype effects during dark hours when BW was included as a covariate (Supplementary Table S2A). In mice fed WD, LM was the primary determinant of 24-h EE (Figure 2C; p = 0.044). LM was also a significant covariate for VO2, VCO2, and EE during the light hours, with no observable genotype effect (Supplementary Table S2B). RER (VCO2/VO2) was comparable between genotypes fed CD, except that MestpKO showed significantly higher RER than WT mice during dark hours (Figure 2D, Supplementary Table S2A), suggesting increased carbohydrate metabolism at night. No differences in RER were observed between WT and MestpKO female mice fed WD (Figure 1D, Supplementary Table S2B).
Figure 2

Metabolic and physical activity measurements of female WT and MestpKO (pKO) mice fed CD or WD. (A–C) Energy expenditure (EE), (D) respiratory exchange ratio (RER), and (E–M) indices of activity and sleep were measured in female WT and MestpKO mice fed CD or WD for 16 weeks. (N, O) Nonexercise activity thermogenesis (NEAT) and (P) fine movements were calculated as described in Supplementary File Methods. Data in (A, D–P) were analyzed via unpaired t-tests for each circadian period comparing WT and MestpKO female mice fed CD or WD. Data in (B, C) were calculated to show significant differences between genotypes when adjusted for (B) total mass or (C) lean mass. p-values are numerically indicated in all figures, with < 0.001 denoting a p-value of less than 0.0005.
Average ambulatory distance (Figure 2E) and speed within the cage (Figure 2F) were consistent between MestpKO and WT female mice fed CD. Generalized linear modeling using one-way ANOVA for mass-independent variables determined significant genotype effects for pedestrian locomotion and total distance in the cage for all time periods in MestpKO (Supplementary Table S2A). When fed WD, MestpKO female mice showed significantly increased pedestrian distance during the day, night, and 24 h compared to WT (Figure 2E, Supplementary Table S2B). Average pedestrian speed was significantly increased for MestpKO female mice compared with WT female mice fed WD only at night (Figure 2F). Measurements of X- and Y-breaks showed no differences between genotypes when fed CD but were significantly increased in MestpKO mice during all time periods compared to WT when fed WD (Figures 2G, H). No differences in Z-breaks were observed between WT and MestpKO female mice fed either CD or WD (Figure 2I). Measurements of % time spent still or asleep, and total sleep hours, showed no differences between genotypes fed CD but significant differences in mice fed WD, with MestpKO mice showing reduced still and sleep percentages and times (Figures 2J–L). Correspondingly, the time spent walking was also significantly increased for all time periods, only for MestpKO female mice compared to WT female mice when fed WD (Figure 2M).
In the context of dietary fat, MestpKO female mice show increased SPA compared to WT mice, including pedestrian locomotion (PedMeters), X- and Y-breaks, and time spent walking (Figures 2E, G, H, M), which correlate with reduced time spent still and sleeping (Figures 2J–L). The energy associated with SPA, referred to as NEAT, shows no differences between genotypes when fed CD but is higher in the MestpKO compared to WT at all time periods when fed WD (Figure 2N). Further analysis revealed that WT mice show reduced NEAT when fed WD compared to MestpKO, whereas both genotypes maintained comparable levels of NEAT when fed CD (Figure 2O). Additionally, MestpKO female mice fed WD were more engaged in fine movements, such as grooming and scratching, particularly during daylight, compared to WT mice when fed WD (Figure 2P). Thus, increased SPA and NEAT, in the context of an obesogenic diet and paternal inactivation of Mest, may protect MestpKO female mice from diet-induced obesity. Resistance to dietary obesity in female MestpKO mice, to some extent, parallels that previously observed in male mice (12); however, no changes in physical activity were observed in MestpKO male mice fed CD or WD (Supplementary Tables S3A, B).
3.3 Hypothalamic gene expression is altered in control diet-fed female MestpKO mice
Analysis of Mest mRNA in multiple brain regions of WT and MestpKO mice, including the hypothalamus, pituitary, amygdala, and motor cortex, confirms that transcription occurs exclusively from the paternal allele, with the highest expression in the pituitary (Figure 1O). To investigate the neurobiological basis for increased physical activity without the influence of dietary fat, several potential candidate genes were measured in the different brain regions of CD-fed WT and MestpKO female mice. Initial studies focused on the hypothalamus and pituitary because of their well-recognized role in metabolism. MestpKO female mice showed significantly reduced hypothalamic expression of corticotropin-releasing hormone (Crh), oxytocin (Oxt), thyrotropin-releasing hormone (Trh), and the primary central melanocortin signaling gene melanocortin 4 receptor (Mc4r), compared to WT mice (Figure 3A). Agouti-related neuropeptide (Agrp), an antagonist of melanocortin receptor signaling, also trended lower in MestpKO female mice (Figure 3A). Hypocretin (Hcrt) and the G-protein-coupled receptor hypocretin receptor 1 (Hcrtr1), involved in orexigenic signaling and regulation of sleep and arousal, showed no differences in expression between genotypes (Figure 3A). Since Trh is associated with thyroid-stimulating hormone release and is subject to negative feedback by TH, additional TH-related genes were measured in the hypothalamus (Figure 3B). Results showed increased expression of Dio2 in MestpKO female mice, which correlated with increased hairless (Hr), a recognized TH-responsive gene, possibly indicating a localized TH effect in the hypothalamus (Figure 3B). While Kruppel-like factor 9 (Klf9), another TH-responsive gene, was marginally elevated in the MestpKO female hypothalamus, the expression of other recognized TH-responsive genes—uncoupling protein 2 (Ucp2), glycerol phosphate dehydrogenase 2 (Gpd2), and thyroid hormone responsive (Thrsp)—showed no difference in expression between genotypes (Figure 3B). Dio3, which converts active T3 to inactive metabolites, also showed comparable hypothalamic expression between genotypes (Figure 3B). The pituitary showed no differences in gene expression between genotypes (Figures 3C, D), whereas the amygdala showed increased expression of Dio3 in MestpKO mice, while Dio2 and Klf9 were unchanged (Figure 3E). The motor cortex showed marginally increased expression of Dio2 and a small but significant increase in Klf9 mRNA in MestpKO female mice, whereas Dio3 expression was similar for both genotypes (Figure 3F). DIO2 and DIO3 enzymatic activity measured in the amygdala and motor cortex correlated with gene expression patterns and showed increased Dio3 mRNA and DIO3 activity in the amygdala of MestpKO mice (Figures 3E–G). Levels of circulating corticosterone in female WT and MestpKO mice were comparable (Supplementary Figure S2), suggesting that perceived stress is unlikely to be linked with differences in physical activity between genotypes.
Figure 3

Key thyroid hormone metabolic and responsive genes show increased expression in the hypothalamus of CD-fed female Mestpko (pKO) mice. (A, B) Data show hypothalamic (Hyp), (C, D) pituitary (Pit), (E) amygdala (Amy), and (F) motor cortex (MC) gene expression in CD-fed WT and MestpKO mice measured using RT-qPCR. (G) Enzymatic measurements of type 2 (DIO2) and type 3 (DIO3) deiodinases were measured in the Amy and MC from female WT and MestpKO mice fed CD. Unpaired t-tests were used to compare gene expression data between genotypes, and p-values are numerically in all figures.
Since we previously demonstrated that the conception rate between WT (84.2%) and MestpKO (82.4%) female mice was comparable (20), we did not assess estrous cyclicity or estrogen levels in this study. In addition, measurements of pituitary expression of luteinizing hormone beta (Lhb) and follicle-stimulating hormone beta (Fshb)—genes associated with ovulation, estrogen production, follicular development, and control of the estrous cycle—were similar between genotypes (Supplementary Figure S3), suggesting normal hormonal patterns.
In contrast to the results in female mice, the expression of the hypothalamic genes Dio2 and Hr in male mice showed no differences between genotypes (Supplementary Figure S4). Overall, patterns of gene expression suggested that the increased physical activity of MestpKO female mice could be at least partially due to increased hypothalamic TH signaling.
A limitation of our study is that circulating thyroid hormones were not measured. However, while not as direct or convincing as measurements of T3 and T4 in serum or plasma, we show that hepatic gene expression of Dio1 and Thrsp—which have been shown to correlate with levels of circulating thyroid hormones (35, 36)—showed no differences between female WT and MestpKO mice after being fed CD or WD (Supplementary Figure S5). These data suggest that localized conversion of T4 to T3 by DIO2 in the hypothalamus contributes to the phenotypic differences in physical activity observed between genotypes.
3.4 The hypothalamic transcriptome is altered in control diet-fed female MestpKO mice
The RNA-seq transcriptome of WT and MestpKO female mice, selected from the CD-fed cohort (Figure 1A), revealed 866 differentially expressed genes (DEGs; padj < 0.05), with 580 upregulated and 286 downregulated in MestpKO compared to WT female mice (Figure 4A, Supplementary Table S4). Functional annotation of upregulated genes showed enrichment in pathways associated with the neuronal system, neurexins and neuroligins, and protein–protein interactions at synapses. Downregulated genes showed enrichment in axon guidance, eukaryotic translation termination, and nonsense-mediated decay independent of the exon junction complex (Figure 4B). Gene ontology was consistent with pathway analysis and emphasized enrichment for biological process terms associated with axonogenesis, positive regulation of excitatory postsynaptic potential, and chemical and glutamatergic synaptic transmission, and was consistent with cellular component (neuron projection, axon, dendrite, etc.) and molecular function (voltage-gated potassium and monoatomic cation channel activity) ontologies (Figure 4C). MGI Mammalian Phenotype (level 4) analyses showed enrichment for hyperactivity, which closely matches the phenotype observed in MestpKO female mice (Figure 4C).
Figure 4

Transcriptomic analyses reveal differential hypothalamic expression patterns between CD-fed WT and MestpKO (pKO) female mice. (A) Venn diagram showing the numbers of differentially expressed genes between WT and MestpKO female mice and the top 10 Reactome pathways (B) associated with genes expressed at higher or lower levels in MestpKO mice compared to WT. (C) Ontology analyses of genes upregulated in the hypothalamus of MestpKO mice compared to WT. (D) Venn diagram showing the overlap between thyroid hormone (TH)-responsive genes from a published compendium (17) and genes differentially expressed between WT and MestpKO mice. (E) Heatmap of TH-responsive genes showing significant differences in expression between WT and MestpKO mice. (F) Differentially expressed TH-responsive genes identified by RNA-seq were validated in hypothalamic RNA from WT (n = 9) and MestpKO (n = 9) mice using RT-qPCR. Data in (F) were analyzed using unpaired t-tests, and p-values are numerically indicated with < 0.001 representing a value less than 0.0005.
We next explored the extent of TH involvement in the hypothalamus of CD-fed female mice and identified 99 DEGs (padj ≤ 0.05) from a total of 866 DEGs (Supplementary Table S4) that overlapped with a published compendium of 734 top TH-responsive genes (37) (Figure 4D), with 69 upregulated and 30 downregulated in MestpKO compared to WT female mice (Figures 4D, E). Fifteen upregulated genes were selected, and their expression was validated by RT-qPCR (Figure 4F). While three of the downregulated genes in MestpKO (Crh, Oxt, and Trh) were validated (Figure 3A), an additional five failed to validate by RT-qPCR, with two (Cnr1, Dcx) showing significantly higher expression in MestpKO compared to WT mice (Figure 4F). Overall, several well-established TH-responsive genes were significantly upregulated in the hypothalamus of female MestpKO mice, suggesting a role for TH signaling in increased activity-associated behavior. An analysis comparing hypothalamic DEGs in our study with TH-target genes identified in astrocytes and hypothalamus in two additional studies (38, 39) showed an overlap with 21 TH-upregulated and six TH-downregulated genes TH (39), and eight TH-upregulated and eight TH-downregulated genes (38), with Klf9 and Hr being upregulated across all three studies. Hypergeometric analyses (http://nemates.org/MA/progs/overlap_stats.html) of overlap between TH-responsive DEGs from our study identified a representation factor of 2.8 (p < 1.3e−06) compared to Zekri et al. (39), and 2.4 (p < 9.4e−04) compared to Wu et al. (38), providing further evidence for a central role of TH in the activity behavior of female MestpKO mice.
Because loss of Mest was thought to mediate mouse behavior via abnormal development of mesodiencephalic dopaminergic neurons, resulting in reduced tyrosine hydrolase (Th) protein and dopamine release (22), we investigated DEGs associated with dopamine biosynthesis in our study. Our analyses revealed no downregulated expression of DEGs involved in dopamine biosynthesis in the hypothalamus of female MestpKO mice; however, three DEGs—Th (padj < 0.001; 1.5F), Slc6a3 (padj = 0.0017; 5.6F), and Gpr37 (padj = 0.0037; 1.3F)—showed significant upregulation of expression compared to female WT mice. Gene ontology analyses of DEGS with elevated expression in MestpKO did not identify significant pathways associated with dopamine synthesis or function. Similarly, pathways associated with catecholamine secretion, transport, uptake, and biosynthesis showed no significant differences between genotypes. Our results using Mesttm1.2RKz are contradictory to past studies using Mest knockout mice (e.g., Mesttm1Masu), which could be due to differences in the design of the targeted allele (20, 21).
Loss of Mest, a paternally expressed imprinted gene, is associated with differential expression of other imprinted genes, including Klf14, a maternally expressed gene (MEG), and several paternally expressed genes (PEGs; Peg3, Ndn, Dlk1, Magel2; Supplementary Figure S6). It is possible that Mest disruption acts in trans on the expression of other MEGs or PEGs as a component of a network of imprinted genes (40).
3.5 Increased adipose expression of Dio2 and Ucp1 in female MestpKO mice fed WD
We previously determined that upregulation of Mest in iWAT and gWAT in WT female and male mice fed a high-fat diet shows coordinated expression patterns with Krüpple-like factor 14 (Klf14), a maternally expressed gene ~ 230 kb downstream of Mest on mouse Chr 6 (41, 42). Herein, similar coregulation between Mest and Klf14 was observed in WAT depots of female WT mice fed CD (iWAT, R2 = 0.75; gWAT, R2 = 0.64) and WD (iWAT, R2 = 0.82; gWAT, R2 = 0.81), and Mest was absent in the WAT of female MestpKO mice (Figures 5A, B). Lep showed comparable expression between genotypes in iWAT and gWAT in mice fed CD, but its expression was significantly lower in WAT depots of MestpKO mice compared to WT when fed WD, consistent with reduced adipose mass between genotypes (Figure 5C). While Ucp1 was comparable between genotypes when fed CD, WD-fed MestpKO female mice showed significantly elevated Ucp1 in both WAT depots compared to WT mice (Figure 5D). In contrast, Tfam, a mitochondrial transcription factor, and Cpt1b, which plays a role in mitochondrial fatty acid oxidation, were significantly downregulated in both WAT depots of mice fed WD compared to CD, with no differences observed between genotypes (Supplementary Figures S5E, F). Dio2 showed similar patterns of expression as Ucp1 and was upregulated in both iWAT and gWAT of MestpKO mice fed WD (Figure 5G). Measurements of DIO2 activity in WAT depots corresponded poorly with gene expression but showed increased activity in both WAT depots in mice fed WD, with significantly higher levels in gWAT of MestpKO mice compared with controls (Figure 5H). Additional screening of thermogenesis-associated genes in WAT (Ppargc1a, Cidea, Ppara, and Gpd2) showed expression patterns consistent with Ucp1 in WAT depots of WD-fed MestpKO mice (Figures 5I–L), which were strongly influenced by genotype (Supplementary Table S5). These data are further supported by increased “beiging” of iWAT and gWAT in histological sections of WD-fed MestpKO female mice compared with WT (Supplementary Figure S7), suggestive of increased UCP1 thermogenesis and EE. In total, our data suggest that increased WAT thermogenesis and SPA likely contribute to the reduced susceptibility of MestpKO female mice to dietary obesity.
Figure 5
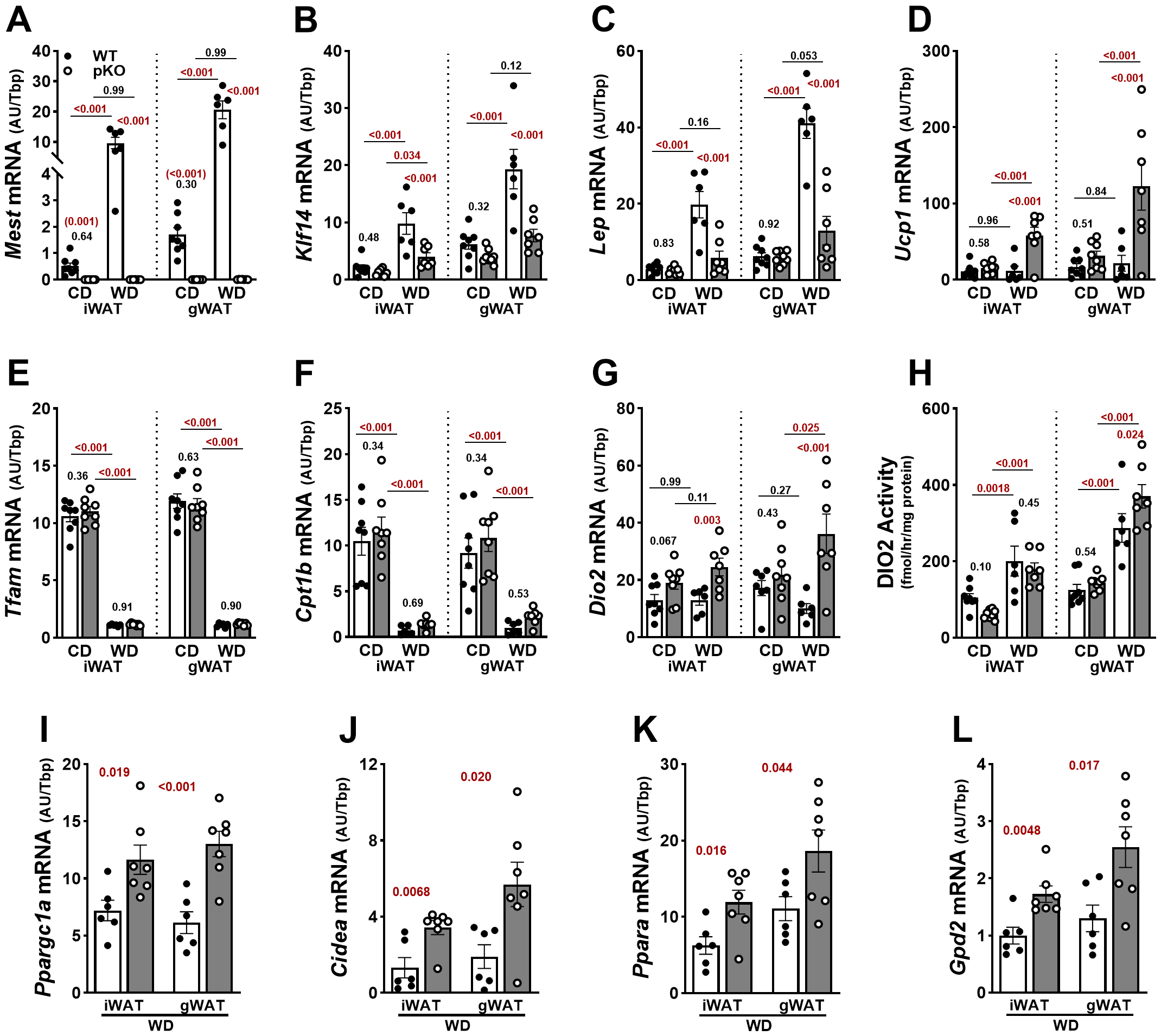
Increased adipose tissue expression of Ucp1 and Dio2 in female MestpKO (pKO) mice fed a Western diet. (A) RT-qPCR measurements of gene expression in inguinal (iWAT) and gonadal (gWAT) white adipose tissue show inactivation of Mest in female MestpKO mice. (B, C) Genes associated with fat mass expansion showed reduced expression in both WAT depots of MestpKO mice fed WD compared with WT. (D) While the thermogenic gene Ucp1 showed increased expression in MestpKO mice when fed WD compared to WT, genes associated with (E) mitochondrial gene transcription and (F) fatty acid transport and oxidation showed no differences between genotypes, but expression was reduced in WD-fed mice. (G)Dio2 expression and (H) enzymatic activity were higher in gWAT of MestpKO mice fed WD compared with WT. (I–L) Additional genes associated with adipose thermogenesis and glycerol-3-phosphate cycling measured in iWAT and gWAT showed increased expression in MestpKO mice when fed WD. Data in (A–H) were analyzed using two-way ANOVA, and in (I–L) by unpaired t-tests. p-values are numerically indicated, and data annotated with < 0.001 indicate a p-value of less than 0.0005.
4 Discussion
Previous studies in our laboratory and others show Mest is predominantly expressed from the paternal allele in WAT (12, 43, 44), with concomitant levels of WAT Mest mRNA and MEST protein correlating with dietary fat-induced FM accretion (8, 9, 41), and reduced diet-induced obesity in mice with global or adipocyte-specific inactivation of the paternal allele of Mest (12). While our earlier studies focused on the role of Mest in male B6 mice, we recently determined that female B6 mice show dietary-fat inducible Mest in WAT (~ 27-fold), which is comparable to male mice (~ 24-fold), albeit with lower baseline expression levels (42). Therefore, dietary fat-induced Mest expression in WAT is a phenomenon common to both sexes.
Herein, we focused on understanding the metabolic phenotypes of female mice with paternal inactivation of Mest under two dietary conditions. MestpKO female mice fed CD were significantly leaner than WT, with reduced FM and LM, which was further accentuated after feeding WD. Protection of MestpKO female mice from dietary obesity was associated with improved glucose tolerance and insulin sensitivity. MestpKO female mice fed CD show noteworthy adaptations in energy metabolism, including increased EE and a trend toward increased locomotor activity compared to WT, enabling maintenance of a lean phenotype over time. Remarkably, when fed WD, MestpKO female mice responded to increased caloric intake by substantially increasing locomotor activity and SPA, translating to increased NEAT compared to WT female mice. In addition, MestpKO female mice allocated more time moving at increased speed, engaged in fine movements, and spent less time staying still and sleeping compared to WT when fed WD.
A notable phenotype of MestpKO female mice, the interaction of diet and SPA, highlights an important strategy to counteract dietary obesity. Increased physical activity of MestpKO female mice corresponded to fine movements such as walking, grooming, and scratching that are not associated with exercise. This was reflected in significantly elevated values for NEAT and fine movements in WD-fed MestpKO female mice compared to WT (Figures 2N, O). In human terms, this corresponds to the energy utilized to walk, perform housework, undertake agricultural tasks, and fidget (45). Fidgeting and other small movements are spontaneous and random, triggering body and limb movements and locomotion (46). In industrialized societies, human locomotion, essential for daily activities, is low compared to that of our ancestors (47). However, harnessing this “fidget” factor in humans has proven beneficial in resisting fat gain caused by increased caloric intake (46, 48). Individuals who gained less weight under caloric excess were observed to have increased EE associated with nonexercise movements, which was equivalent to up to 700 kcal/day above usual EE (17).
The specifics of neural control of SPA and NEAT are still being resolved. In contrast to exercise, which is governed by higher-level cortical function, SPA is likely facilitated by more primitive areas of the brain, such as the hypothalamus (16). Neuromediators implicated in SPA, and consequently NEAT, include hypocretin/orexin, Agrp, ghrelin (Ghrl), neuromedin U (Nmu), Crh, cholecystokinin (Cck), estrogen, leptin, and dopamine (16, 19, 49), some of which exhibit altered expression in the hypothalamus of MestpKO female mice. Furthermore, studies have demonstrated that rats supplemented with T3 show increased SPA and NEAT (50).
Thyroid hormones are well recognized for their profound effects on energy expenditure and body weight regulation. The changes mediated by thyroid hormones in energy expenditure require a signal from the brain, which alters local hypothalamic Dio2 expression, with consequent changes in the regulation of energy balance (51). The enhanced SPA phenotype in WD-fed MestpKO female mice presents a model to investigate neural mechanisms of NEAT in relation to TH signaling and energy metabolism. TH controls an array of developmental and physiological processes in the brain (37). We observe upregulation of Dio2 in the MestpKO female hypothalamus, which suggests a possible TH-regulated mechanism in the control of physical activity. TH that reaches the brain through the circulation undergoes tightly controlled metabolism, leading to hormonal activation or inactivation. Dio2 can locally increase TH signaling by converting the inactive prohormone T4 into the biologically active T3 in a tissue- and temporal-specific fashion, independent of circulating hormone levels (52). Dio2-generated T3 from glial cells can influence neighboring neural cell types, acting in a paracrine fashion to modulate T3-responsive genes (53). In humans, TH dysfunction is linked to sedentary behavior in hypothyroidism, as opposed to increased activity in hyperthyroidism (54). Pharmacologic mouse models of hypo- and hyperthyroidism recapitulate human phenotypes, including alterations in overall physical activity (55). Interestingly, male mice with astrocyte-specific Dio2 inactivation have normal serum T3 but exhibit anxiety-depressive behavior linked to decreased hippocampal expression of classic TH-responsive genes (56). The increased hypothalamic Dio2 expression and the large number of TH-responsive genes upregulated in the hypothalamus of the MestpKO female mice suggest a local increase in T3 levels, which may augment SPA. In addition, peripheral tissue T3 effects elicited by Dio2 upregulation in MestpKO female mice are demonstrated by increased WAT Ucp1, a recognized T3-responsive gene (57). MestpKO female mice fed WD showed markedly increased Dio2 and Ucp1 in WAT, in addition to genes associated with thermogenesis and fatty acid oxidation. Although a mechanism for the functional association between MEST and DIO2 remains to be resolved, it is of marked interest that both are colocalized subcellularly in the endoplasmic reticulum membrane (13, 58), unlike DIO1 and DIO3, which are found in the plasma membrane (58). This further supports the possibility that disruption of Mest leads to DIO2-dependent local activation of TH signaling in the brain and WAT.
Imprinted genes are highly expressed in the brain and influence synaptic function and plasticity, neural development and wiring, social behaviors, energy balance, and cognition, among other processes (59, 60). In the adult mouse brain, MEST has been reported in neuron-rich areas by lacZ staining (21), and more recent advanced technologies have described its expression in brain regions such as the pituitary (61), hypothalamus (62–64), and mouse forebrain (65). Single-cell RNA sequencing data indicate that Mest is expressed in neurons and nonneuronal populations, e.g., astrocytes and tanycytes (64), and it was shown to be expressed in neurons and tanycytes of female mice regardless of age (63). Mest is also highly expressed in the lateral ganglionic eminence of the mouse forebrain, which gives rise to all forebrain GABAergic cells (65), is among the top 1% of enriched imprinted genes in the embryonic and adult mouse pituitary, and scRNA sequencing showed high Mest expression in lactotrophs and thyrotrophs in the postnatal d4 and d49 anterior pituitary (61). The unique increased physical activity phenotype observed in Mest mutant female mice further supports the essential role of imprinted genes in the brain.
Since male mice with adipocyte-specific inactivation of Mest show similar resistance to diet-induced obesity as mice with global inactivation of Mest (12), it is possible that metabolic imbalance caused by the reduced capacity for lipid storage in adipocytes in the absence of Mest in female mice may drive central nervous system regulation of SPA. Alternatively, reduced dietary fat-induced obesity in female mice with global inactivation of Mest could result from an additive contribution from both adipose tissue and the central nervous system (CNS), or from a sex-specific CNS-dominant mechanism. Future investigation of a probable mechanistic link between Mest and TH metabolism in the hypothalamus will determine the effects of adipocyte- or neural cell-specific inactivation of Mest, distinguishing between peripheral and central effects of Mest inactivation on metabolism and SPA in female mice. Moreover, differences in physical activity between male and female MestpKO mice are not well understood but may involve developmental programming of the neonatal brain by reproductive hormones such as estrogen, which is recognized for motivating behavioral changes in female mice (66).
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by Institutional Animal Care and Use Committee at MaineHealth Institute for Research. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RA-K: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Project administration, Validation, Writing – original draft, Writing – review & editing. MM: Data curation, Formal Analysis, Investigation, Methodology, Writing – review & editing. VD: Formal Analysis, Investigation, Methodology, Resources, Writing – review & editing. AH: Conceptualization, Formal Analysis, Investigation, Methodology, Resources, Writing – review & editing. RK: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. The Molecular Phenotyping and Physiology Core Facilities at MaineHealth Institute for Research used for studies herein are supported by U54GM115516 and P20GM121301 from the National Institute of General Medical Sciences. The Genomics Core Facility at the Pennington Biomedical Research Center is supported in part by COBRE (P20 GM135002 & P20 GM103528) and NORC (NIH P30 DK072476) center grants from the National Institutes of Health.
Acknowledgments
We thank Morgan–Carter Moulton for assistance in sample processing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2025.1680158/full#supplementary-material
Supplementary Figure 1Phenotypic analyses of WT and MestpKO (pKO) male mice fed control (CD) and western diet (WD). (A) BWs of WT and MestpKO male mice fed CD and (B) WD from 8 to 16 weeks of age. (C) Fat mass, (D) % fat mass, (E) lean mass and (F) % lean mass of CD fed male WT and MestpKO mice at 8, 12 and 16 weeks of age measured by NMR. (G) Fat mass, (H) % fat mass, (I) lean mass and (J) % lean mass of WD fed male mice at 8, 12 and 16 weeks of age. All data were analyzed by two-way ANOVA. Annotation with a, b, c, or d indicate p-values lower than 0.05, 0.01, 0.001 and 0.0001 for data in figure panels (A, B) and indicated numerically in figure panels (C-J).
Supplementary Figure 2Serum corticosterone levels are similar between genotypes when fed either control (CD) or Western diet (WD). Serum corticosterone was measured in WT and MestpKO (pKO) mice fed CD (n=7 per genotype) or WD (n=15–17 per genotype). Unpaired t-tests were used to measure significance between genotypes for each diet.
Supplementary Figure 3Pituitary expression of Lhb and Fshb is comparable in the pituitary of female WT and MestpKO (pKO) mice fed CD. Gene expression measured in RNA from pituitary of WT (n=7) and MestpKO (n=7) mice. Unpaired t-tests were used to measure significance between genotypes for each diet. P-values are numerically indicated.
Supplementary Figure 4Hypothalamic expression of metabolic and thyroid-hormone responsive genes is similar between male WT and MestpKO (pKO) mice fed WD. Gene expression measured in RNA isolated from hypothalamus of WT (n=8) and MestpKO (n=7) mice showed no significant differences in thyroid metabolic (Dio2, Dio3), regulatory (Crh) or responsive (Hr, Klf9, Mbp) genes between genotypes. Mest mRNA is mostly absent in hypothalamus of MestpKO mice. Unpaired t-tests were used to measure significance between genotypes. P-values are numerically indicated and data annotated with <0.001 indicates a p-value of less than 0.0005.
Supplementary Figure 5Hepatic thyroid hormone responsive genes show similar expression in female WT and MestpKO (pKO) mice fed CD or WD. Dio1 and Thrsp gene expression was measured in RNA isolated from liver of WT (n=7-8) and MestpKO (n=7-8) mice. Unpaired t-tests were used to measure significance between genotypes for each diet. P-values are numerically indicated.
Supplementary Figure 6Hypothalamic imprinted genes are differentially expressed between female WT and MestpKO (pKO) mice fed CD. Gene expression measured in RNA from hypothalamus of WT (n=8) and MestpKO (n=8) mice. The letters in parentheses below each gene indicate the imprinting status as maternal-expressed (m) or paternal-expressed (p). Unpaired t-tests were used to measure significance between genotypes for each diet. P-values are numerically indicated.
Supplementary Figure 7Adipocyte morphology of gonadal (gWAT) and inguinal (iWAT white adipose tissue (WAT) of wildtype (WT; n=4) and MestpKO (pKO; n=4) female mice after being fed Western diet (WD) for 4 weeks. Data shows smaller adipocytes in both WAT depots in WD-fed female MestpKO mice compared to WT.
Abbreviations
Mest, mesoderm-specific transcript; WAT, white adipose tissue; iWAT, inguinal WAT; gWAT, gonadal WAT; BW, body weight; SPA, spontaneous physical activity; NEAT, nonexercise activity thermogenesis; TH, thyroid hormone.
References
1
Bluher M . Obesity: global epidemiology and pathogenesis. Nat Rev Endocrinol. (2019) 15:288–98. doi: 10.1038/s41574-019-0176-8
2
Ruggiero-Ruff RE Coss D . Neuroendocrinology and the genetics of obesity. Endocrinology. (2025) 166. doi: 10.1210/endocr/bqaf121
3
Trang K Grant SFA . Genetics and epigenetics in the obesity phenotyping scenario. Rev Endocr Metab Disord. (2023) 24:775–93. doi: 10.1007/s11154-023-09804-6
4
Loos RJF Yeo GSH . The genetics of obesity: from discovery to biology. Nat Rev Genet. (2022) 23:120–33. doi: 10.1038/s41576-021-00414-z
5
Stunkard AJ Harris JR Pedersen NL McClearn GE . The body-mass index of twins who have been reared apart. New Engl J Med. (1990) 322:1483–7. doi: 10.1056/NEJM199005243222102
6
Holmquist M . Alpha/Beta-hydrolase fold enzymes: structures, functions and mechanisms. Curr Protein Pept Sci. (2000) 1:209–35. doi: 10.2174/1389203003381405
7
Lenfant N Hotelier T Velluet E Bourne Y Marchot P Chatonnet A . ESTHER, the database of the alpha/beta-hydrolase fold superfamily of proteins: tools to explore diversity of functions. Nucleic Acids Res. (2013) 41:D423–9. doi: 10.1093/nar/gks1154
8
Koza RA Nikonova L Hogan J Rim JS Mendoza T Faulk C et al . Changes in gene expression foreshadow diet-induced obesity in genetically identical mice. PloS Genet. (2006) 2:e81. doi: 10.1371/journal.pgen.0020081
9
Nikonova L Koza RA Mendoza T Chao PM Curley JP Kozak LP . Mesoderm-specific transcript is associated with fat mass expansion in response to a positive energy balance. FASEB J: Off Publ Fed Am Soc Exp Biol. (2008) 22:3925–37. doi: 10.1096/fj.08-108266
10
Takahashi M Kamei Y Ezaki O . Mest/Peg1 imprinted gene enlarges adipocytes and is a marker of adipocyte size. Am J Physiol Endocrinol Metab. (2005) 288:E117–24. doi: 10.1152/ajpendo.00244.2004
11
Voigt A Ribot J Sabater AG Palou A Bonet ML Klaus S . Identification of Mest/Peg1 gene expression as a predictive biomarker of adipose tissue expansion sensitive to dietary anti-obesity interventions. Genes Nutr. (2015) 10:477. doi: 10.1007/s12263-015-0477-z
12
Anunciado-Koza RP Manuel J Mynatt RL Zhang J Kozak LP Koza RA . Diet-induced adipose tissue expansion is mitigated in mice with a targeted inactivation of mesoderm specific transcript (Mest). PloS One. (2017) 12:e0179879. doi: 10.1371/journal.pone.0179879
13
Prudovsky I Anunciado-Koza RP Jacobs CG Kacer D Siviski ME Koza RA . Mesoderm-specific transcript localization in the ER and ER-lipid droplet interface supports a role in adipocyte hypertrophy. J Cell Biochem. (2018) 119:2636–45. doi: 10.1002/jcb.26429
14
Levine JA . Nonexercise activity thermogenesis–liberating the life-force. J Intern Med. (2007) 262:273–87. doi: 10.1111/j.1365-2796.2007.01842.x
15
Levine JA Kotz CM . NEAT–non-exercise activity thermogenesis–egocentric & geocentric environmental factors vs. biological regulation. Acta Physiol Scand. (2005) 184:309–18. doi: 10.1111/j.1365-201X.2005.01467.x
16
Kotz CM Teske JA Billington CJ . Neuroregulation of nonexercise activity thermogenesis and obesity resistance. Am J Physiol Regulatory Integr Comp Physiol. (2008) 294:R699–710. doi: 10.1152/ajpregu.00095.2007
17
Ravussin E Lillioja S Anderson TE Christin L Bogardus C . Determinants of 24-hour energy expenditure in man. Methods and results using a respiratory chamber. J Clin Invest. (1986) 78:1568–78. doi: 10.1172/JCI112749
18
Bonilla DA Peralta-Alzate JO Bonilla-Henao JA Cannataro R Cardozo LA Vargas-Molina S et al . Insights into non-exercise physical activity on control of body mass: A review with practical recommendations. J Funct Morphol Kinesiol. (2023) 8. doi: 10.3390/jfmk8020044
19
Novak CM Levine JA . Central neural and endocrine mechanisms of non-exercise activity thermogenesis and their potential impact on obesity. J Neuroendocrinol. (2007) 19:923–40. doi: 10.1111/j.1365-2826.2007.01606.x
20
Anunciado-Koza RP Stohn JP Hernandez A Koza RA . Social and maternal behavior in mesoderm specific transcript (Mest)-deficient mice. PloS One. (2022) 17:e0271913. doi: 10.1371/journal.pone.0271913
21
Lefebvre L Viville S Barton SC Ishino F Keverne EB Surani MA . Abnormal maternal behaviour and growth retardation associated with loss of the imprinted gene Mest. Nat Genet. (1998) 20:163–9. doi: 10.1038/2464
22
Mesman S van Hooft JA Smidt MP . Mest/peg1 is essential for the development and maintenance of a SNc neuronal subset. Front Mol Neurosci. (2016) 9:166. doi: 10.3389/fnmol.2016.00166
23
Ji L Bishayee K Sadra A Choi S Choi W Moon S et al . Defective neuronal migration and inhibition of bipolar to multipolar transition of migrating neural cells by Mesoderm-Specific Transcript, Mest, in the developing mouse neocortex. Neuroscience. (2017) 355:126–40. doi: 10.1016/j.neuroscience.2017.05.003
24
Mina AI LeClair RA LeClair KB Cohen DE Lantier L Banks AS . CalR: A web-based analysis tool for indirect calorimetry experiments. Cell Metab. (2018) 28:656–66 e1. doi: 10.1016/j.cmet.2018.06.019
25
Bunney PE Zink AN Holm AA Billington CJ Kotz CM . Orexin activation counteracts decreases in nonexercise activity thermogenesis (NEAT) caused by high-fat diet. Physiol Behav. (2017) 176:139–48. doi: 10.1016/j.physbeh.2017.03.040
26
Hernandez A Martinez ME Fiering S Galton VA St Germain D . Type 3 deiodinase is critical for the maturation and function of the thyroid axis. J Clin Invest. (2006) 116:476–84. doi: 10.1172/JCI26240
27
Galton VA Hiebert A . Hepatic iodothyronine 5-deiodinase activity in Rana catesbeiana tadpoles at different stages of the life cycle. Endocrinology. (1987) 121:42–7. doi: 10.1210/endo-121-1-42
28
Andrews S . FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics (2010). Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (Accessed October 21, 2025).
29
Martin M . Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. (2011) 17:10–2. doi: 10.14806/ej.17.1.200
30
Dobin A Davis CA Schlesinger F Drenkow J Zaleski C Jha S et al . STAR: ultrafast universal RNA-seq aligner. Bioinformatics. (2013) 29:15–21. doi: 10.1093/bioinformatics/bts635
31
Putri GH Anders S Pyl PT Pimanda JE Zanini F . Analysing high-throughput sequencing data in Python with HTSeq 2.0. Bioinformatics. (2022) 38:2943–5. doi: 10.1093/bioinformatics/btac166
32
Love MI Huber W Anders S . Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
33
Chen EY Tan CM Kou Y Duan Q Wang Z Meirelles GV et al . Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. (2013) 14:128. doi: 10.1186/1471-2105-14-128
34
Heymsfield SB Gonzalez MC Shen W Redman L Thomas D . Weight loss composition is one-fourth fat-free mass: a critical review and critique of this widely cited rule. Obes Rev: An Off J Int Assoc Study Obes. (2014) 15:310–21. doi: 10.1111/obr.12143
35
Ritter MJ Amano I Hollenberg AN . Thyroid hormone signaling and the liver. Hepatology. (2020) 72:742–52. doi: 10.1002/hep.31296
36
Zavacki AM Ying H Christoffolete MA Aerts G So E Harney JW et al . Type 1 iodothyronine deiodinase is a sensitive marker of peripheral thyroid status in the mouse. Endocrinology. (2005) 146:1568–75. doi: 10.1210/en.2004-1392
37
Chatonnet F Flamant F Morte B . A temporary compendium of thyroid hormone target genes in brain. Biochim Biophys Acta. (2015) 1849:122–9. doi: 10.1016/j.bbagrm.2014.05.023
38
Wu S Dellinger J Markossian S Dusabyinema Y Guyot R Hughes S et al . An atlas of thyroid hormone responsive genes in adult mouse hypothalamus. Endocrinology. (2025) 166. doi: 10.1210/endocr/bqaf084
39
Zekri Y Guyot R Flamant F . An atlas of thyroid hormone receptors’ Target genes in mouse tissues. Int J Mol Sci. (2022) 23. doi: 10.3390/ijms231911444
40
Patten MM Cowley M Oakey RJ Feil R . Regulatory links between imprinted genes: evolutionary predictions and consequences. Proc Biol Sci. (2016) 283. doi: 10.1098/rspb.2015.2760
41
Anunciado-Koza RP Manuel J Koza RA . Molecular correlates of fat mass expansion in C57BL/6J mice after short-term exposure to dietary fat. Ann New York Acad Sci. (2016) 1363:50–8. doi: 10.1111/nyas.12958
42
Anunciado-Koza RVP Yin H Bilodeau CL Cooke D Ables GP Ryzhov S et al . Interindividual differences of dietary fat-inducible Mest in white adipose tissue of C57BL/6J mice are not heritable. Obes (Silver Spring). (2024) 32:1144–55. doi: 10.1002/oby.24020
43
Kamei Y Suganami T Kohda T Ishino F Yasuda K Miura S et al . Peg1/Mest in obese adipose tissue is expressed from the paternal allele in an isoform-specific manner. FEBS Lett. (2007) 581:91–6. doi: 10.1016/j.febslet.2006.12.002
44
Koza RA Rogers P Kozak LP . Inter-individual variation of dietary fat-induced mesoderm specific transcript in adipose tissue within inbred mice is not caused by altered promoter methylation. Epigenetics. (2009) 4:512–8. doi: 10.4161/epi.4.7.10031
45
Levine JA . Nonexercise activity thermogenesis (NEAT): environment and biology. Am J Physiol Endocrinol Metab. (2004) 286:E675–85. doi: 10.1152/ajpendo.00562.2003
46
Levine JA . The Fidget Factor and the obesity paradox. How small movements have big impact. Front Sports Act Living. (2023) 5:1122938. doi: 10.3389/fspor.2023.1122938
47
Garland T Jr. Schutz H Chappell MA Keeney BK Meek TH Copes LE et al . The biological control of voluntary exercise, spontaneous physical activity and daily energy expenditure in relation to obesity: human and rodent perspectives. J Exp Biol. (2011) 214:206–29. doi: 10.1242/jeb.048397
48
Ravussin E . Physiology. A NEAT way to control weight? Science. (2005) 307:530–1. doi: 10.1126/science.1108597
49
Teske JA Billington CJ Kotz CM . Neuropeptidergic mediators of spontaneous physical activity and non-exercise activity thermogenesis. Neuroendocrinology. (2008) 87:71–90. doi: 10.1159/000110802
50
Levine JA Nygren J Short KR Nair KS . Effect of hyperthyroidism on spontaneous physical activity and energy expenditure in rats. J Appl Physiol. (2003) 94:165–70. doi: 10.1152/japplphysiol.00499.2002
51
Herwig A Ross AW Nilaweera KN Morgan PJ Barrett P . Hypothalamic thyroid hormone in energy balance regulation. Obes Facts. (2008) 1:71–9. doi: 10.1159/000123428
52
Gereben B Zavacki AM Ribich S Kim BW Huang SA Simonides WS et al . Cellular and molecular basis of deiodinase-regulated thyroid hormone signaling. Endocr Rev. (2008) 29:898–938. doi: 10.1210/er.2008-0019
53
Freitas BC Gereben B Castillo M Kallo I Zeold A Egri P et al . Paracrine signaling by glial cell-derived triiodothyronine activates neuronal gene expression in the rodent brain and human cells. J Clin Invest. (2010) 120:2206–17. doi: 10.1172/JCI41977
54
Mullur R Liu YY Brent GA . Thyroid hormone regulation of metabolism. Physiol Rev. (2014) 94:355–82. doi: 10.1152/physrev.00030.2013
55
Niedowicz DM Wang WX Price DA Nelson PT . Modulating thyroid hormone levels in adult mice: impact on behavior and compensatory brain changes. J Thyroid Res. (2021) 2021:9960188. doi: 10.1155/2021/9960188
56
Bocco BM Werneck-de-Castro JP Oliveira KC Fernandes GW Fonseca TL Nascimento BP et al . Type 2 deiodinase disruption in astrocytes results in anxiety-depressive-like behavior in male mice. Endocrinology. (2016) 157:3682–95. doi: 10.1210/en.2016-1272
57
Cohen P Spiegelman BM . Brown and beige fat: molecular parts of a thermogenic machine. Diabetes. (2015) 64:2346–51. doi: 10.2337/db15-0318
58
Sabatino L Vassalle C Del Seppia C Iervasi G . Deiodinases and the three types of thyroid hormone deiodination reactions. Endocrinol Metab (Seoul). (2021) 36:952–64. doi: 10.3803/EnM.2021.1198
59
Perez JD Rubinstein ND Dulac C . New perspectives on genomic imprinting, an essential and multifaceted mode of epigenetic control in the developing and adult brain. Annu Rev Neurosci. (2016) 39:347–84. doi: 10.1146/annurev-neuro-061010-113708
60
Wilkinson LS Davies W Isles AR . Genomic imprinting effects on brain development and function. Nat Rev Neurosci. (2007) 8:832–43. doi: 10.1038/nrn2235
61
Scagliotti V Costa Fernandes Esse R Willis TL Howard M Carrus I Lodge E et al . Dynamic expression of imprinted genes in the developing and postnatal pituitary gland. Genes (Basel). (2021) 12. doi: 10.3390/genes12040509
62
Chen R Wu X Jiang L Zhang Y . Single-cell RNA-seq reveals hypothalamic cell diversity. Cell Rep. (2017) 18:3227–41. doi: 10.1016/j.celrep.2017.03.004
63
Hajdarovic KH Yu D Hassell LA Evans S Packer S Neretti N et al . Single-cell analysis of the aging female mouse hypothalamus. Nat Aging. (2022) 2:662–78. doi: 10.1038/s43587-022-00246-4
64
Steuernagel L Lam BYH Klemm P Dowsett GKC Bauder CA Tadross JA et al . HypoMap-a unified single-cell gene expression atlas of the murine hypothalamus. Nat Metab. (2022) 4:1402–19. doi: 10.1038/s42255-022-00657-y
65
Lee DR Rhodes C Mitra A Zhang Y Maric D Dale RK et al . Transcriptional heterogeneity of ventricular zone cells in the ganglionic eminences of the mouse forebrain. Elife. (2022) 11. doi: 10.7554/eLife.71864
66
Krause WC Rodriguez R Gegenhuber B Matharu N Rodriguez AN Padilla-Roger AM et al . Oestrogen engages brain MC4R signalling to drive physical activity in female mice. Nature. (2021) 599:131–5. doi: 10.1038/s41586-021-04010-3
Summary
Keywords
obesity, adipose tissue, hypothalamus, epigenetics, mouse models, physical activity, thyroid hormones
Citation
Anunciado-Koza RVP, Martinez ME, DeMambro V, Hernandez A and Koza RA (2025) Increased spontaneous physical activity in female MEST-deficient mice protects against diet-induced obesity. Front. Endocrinol. 16:1680158. doi: 10.3389/fendo.2025.1680158
Received
05 August 2025
Accepted
15 October 2025
Published
29 October 2025
Volume
16 - 2025
Edited by
Alma Martelli, University of Pisa, Italy
Reviewed by
Sruti Chandra, Tulane University, United States
Michael Freemark, Duke University, United States
Yutaro Kumagai, National Institute of Advanced Industrial Science and Technology (AIST), Japan
Updates
Copyright
© 2025 Anunciado-Koza, Martinez, DeMambro, Hernandez and Koza.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robert A. Koza, Robert.Koza@mainehealth.org
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.