- 1Department of Immunology, Niigata University Graduate School of Medical and Dental Sciences, Niigata, Japan
- 2Department of Pharmacology, Wakayama Medical University, Wakayama, Japan
- 3Department of Physiological Sciences, School of Pharmaceutical Sciences, Wakayama Medical University, Wakayama, Japan
- 4Department of Immunology, Tokyo Medical University, Tokyo, Japan
- 5Department of Experimental Genome Research, Research Institute for Microbial Diseases, Osaka University, Suita, Japan
- 6Department of Radiological Sciences, Faculty of Medical Science Technology, Morinomiya University of Medical Sciences, Osaka, Japan
- 7Department of Child Development and Molecular Brain Science, United Graduate School of Child Development, Suita, Japan
Type-2 bitter taste receptors (Tas2Rs) are a large family of G protein-coupled receptors that are expressed in the oral cavity and serve to detect substances with bitter tastes in foods and medicines. Recent evidence suggests that Tas2Rs are also expressed extraorally, including in immune cells. However, the role of Tas2Rs in immune cells remains controversial. Here, we demonstrate that Tas2R126, Tas2R135, and Tas2R143 are expressed in mouse neutrophils, but not in other immune cells such as macrophages or T and B lymphocytes. Treatment of bone marrow-derived neutrophils from wild-type mice with the Tas2R126/143 agonists arbutin and D-salicin led to enhanced C-X-C motif chemokine ligand 2 (CXCL2)-stimulated migration in vitro, but this response was not observed in neutrophils from Tas2r126/135/143-deficient mice. Enhancement of CXCL2-stimulated migration by Tas2R agonists was accompanied by increased phosphorylation of myosin light chain 2 (MLC2) and was blocked by pretreatment of neutrophils with inhibitors of Rho-associated coiled-coil-containing protein kinase (ROCK), but not by inhibitors of the small GTPase RhoA. Taken together, these results demonstrate that mouse neutrophils express functional Tas2R126/143 and suggest a role for Tas2R126/143–ROCK–MLC2-dependent signaling in the regulation of neutrophil migration.
Introduction
Type-2 bitter taste receptors (TAS2Rs (human) or Tas2Rs (mouse)) are a large family of G protein-coupled receptors that are expressed in taste receptor cells in the oral cavity. The Tas2R family comprises approximately 30 members in humans and rodents (1, 2). In the oral cavity, activation of Tas2Rs (or TAS2Rs) induced by bitter taste substances transduces downstream molecules such as specific G proteins and TRPM5 (3, 4), and the bitter taste are recognized. Some of these substances are subsequently rejected from the oral cavity due to their bitter taste being potentially harmful to our health. Increasing evidence suggests that Tas2Rs are also expressed extraorally, including in airway smooth muscle cells, intestinal tuft cells, and immune cells, and their roles at these extraoral sites have been studied intensively in recent years (5).
Several groups have recently reported on the roles of Tas2Rs in the immune response. For example, mouse Tas2R138 (ortholog of human TAS2R38) in neutrophils binds with acyl-homoserine lactone (AHL)-12, a quorum sensing molecule (QSM) produced by Pseudomonas aeruginosa (P. aeruginosa). The study has indicated that the Tas2R138/AHL-12 binding promotes the degradation of lipid droplets to facilitate AHL-12 clearance from neutrophils in vitro and also shown suggestive evidence that the binding plays a role in protecting against bacterial infection using P. aeruginosa infection model mice (6). In addition, human TAS2R14 (ortholog of mouse Tas2R140) signaling induced by QSMs was found to enhance phagocytosis by macrophages via nitric oxide production and to promote the secretion of cytokines including IL-8, TNF-α, and IL-6 from gingival epithelial cells in vitro (7, 8). In contrast, other studies have suggested that Tas2R (or TAS2R) signaling suppresses immune responses (9–12), raising the possibility that each Tas2R subtype may play a different role in the immune system. Thus, further work is necessary to determine the pattern of expression of Tas2R subtypes in immune cells and to examine the contribution of these receptors in the immune response. Such an undertaking would be facilitated by the availability of Tas2R-deficient animal models. However, to the best of our knowledge, only two Tas2R-deficient mouse lines have been established: Tas2R105-deficient mice (13) and, more recently, triple-deficient mice that lack Tas2R126, Tas2R135, and Tas2R143 (Tas2r126/135/143−/−) (14). The Tas2r105−/− mouse strain was shown to exhibit strong and selective impairment in the ability to taste one specific bitter taste substance, cycloheximide (15), whereas the Tas2r126/135/143−/− strain exhibited impaired responses to the Tas2R126/143 agonist D-salicin (16, 17) and the Tas2R135 agonist acesulfame potassium (16). However, whether these receptors are functionally expressed extraorally, including in immune cells, is unknown.
In the present study, we examined the expression profile of mouse Tas2R family members in immune cells in silico and then confirmed that Tas2r126/135/143 mRNA is expressed at high levels in neutrophils relative to other immune cell types. In addition, we show that Tas2R126/143-mediated signaling enhances C-X-C motif chemokine ligand 2 (CXCL2)-stimulated migration of mouse neutrophils in a Rho-associated coiled-coil-containing protein kinase (ROCK)-dependent manner. This enhancement of migration was not observed in neutrophils from Tas2r126/135/143−/− mice, confirming that these Tas2R family members play a functional role in mouse neutrophils.
Materials and methods
Reagents
Arbutin, D-salicin, phenyl-β-D-glucopyranoside (PBD-Gluco), denatonium, and 5-propyl-2-thiouracil were purchased from Sigma-Aldrich (MO, USA). Recombinant CXCL2 was obtained from BioLegend (CA, USA), CXCL1 was from R&D system (MN, USA), N-formylmethionyl-leucyl-phenylalanine (fMLP) and Y-27632 were from Nacalai-Tesque (Kyoto, Japan), KD025 was from MedChemExpress (NJ, USA), and Tat-C3 was from Cytoskeleton (CO, USA). FITC-conjugated anti-Ly6G (1A8), APC/eFluor 780-anti-Ly6G (1A8), APC-anti-Ly6G (1A8), and PE-anti-CD11b (M1/70) monoclonal antibodies (mAbs) were purchased from Thermo Fisher Scientific (MA, USA). BV421-anti-CD8 (53-6.7), BV421-anti-F4/80 (BM8), Alexa Fluor 647-anti-CD4 (GK1.5), PE-anti-B220 (RA3-6B2), APC/Cy7-anti-Ly6G (1A) mAbs, and Dylight 649-anti-rabbit IgG polyclonal antibody (pAb) (Poly4064) were purchased from BioLegend. Anti-phospho-myosin light chain 2 (MLC2; Ser19) pAb was purchased from Cell Signaling Technology (MA, USA).
Animals
Wild-type C57BL/6 mice were purchased from Japan SLC (Shizuoka, Japan) or Charles River Laboratories (Yokohama, Japan) and housed under specific pathogen-free conditions. Tas2r126/135/143 heterozygous (+/−) or triple-deficient (−/−) mice were produced by introducing guide (g) RNAs and Cas9 enzyme (Thermos Fisher Scientific) into fertilized eggs using an electroporator (NEOA21, Nepagene, Chiba, Japan) as described previously (18). On-target and potential off-target sequences were screened using Benchling (https://benchling.com) or CRISPRdirect software (https://crispr.dbcls.jp/). The gRNA on-target sequences are listed in Supplementary Table 1. The resulting mice were screened by polymerase chain reaction (PCR) of genomic DNA followed by direct sequencing. Detailed genotypes are shown in Figure 4, and the PCR primer sequences are listed in Supplementary Table 1. The animal experimental protocols were approved by the Animal Research Committees of Wakayama Medical University, Niigata University, and Osaka University. Tas2r126/135/143-/- mouse line is available through the Riken BioResource Center (Riken BRC, Tsukuba, Japan; #11633) and the Center for Animal Resources and Development, Kumamoto University (CARD, Kumamoto, Japan; #3187).
Cell purification
Macrophages were purified from the peritoneal cavity, neutrophils from bone marrow (BM), and T and B lymphocytes from spleens. For this, single cell suspensions were prepared in RPMI-1640 medium by gentle dissociation using 40-or 35-μm cell strainers. The cells were then stained by incubation with specific antibodies as described in section 2-1 for 30 min at 4°C, washed, and sorted using a FACSAria III flow cytometer (BD Biosciences, CA, USA). For some experiments, neutrophils were also purified from BM samples using an Easysep Mouse Neutrophil Enrichment Kit (STEMCELL Technologies, BC, Canada) or MojoSort Mouse Neutrophil Isolation Kit (BioLegend) in accordance with the manufacturers’ instructions. The purity of neutrophils isolated using the kits was approximately 80% (Supplementary Figure 1).
Reverse transcription quantitative polymerase chain reaction (RT-qPCR)
RT-qPCR was performed as described previously (19) with several modifications. Total RNA was isolated from purified cell populations using a PureLink RNA isolation kit (Thermo Fisher Scientific) with PureLink DNase (Thermo Fisher Scientific), and first-strand cDNA was synthesized using M-MLV Reverse Transcriptase (Promega, WI, USA). qPCR was performed using a QuantStudio 7 Real-Time PCR system (Thermo Fisher Scientific) with TB Green® Premix Ex Taq™ II (Takara Bio, Shiga, Japan) or SYBR® Green Master Mix (Thermo Fisher Scientific). The qPCR primers have been described previously (19, 20) and are listed in Supplementary Table 1.
Neutrophil migration assay
This assay was performed as described previously (21, 22) with minor modifications. Briefly, total BM cells were treated with ACK buffer to lyse red blood cells, then serum-starved in RPMI-1640 medium containing 0.1% bovine serum albumin (BSA) for 30 min, and then BM cells were stimulated with the indicated concentrations of the Tas2R agonists arbutin, D-salicin, denatonium, 5-propyl-2-thiouracil, and/or PBD-Gluco for 30 min. The cells were placed in the upper chamber of a 24-well Transwell plate containing 3.0-μm pore inserts (Corning, NY, USA). The chemoattractant CXCL2 (20 ng/ml), CXCL1 (20 ng/ml) or fMLP (3 μM) in RPMI-1640/0.1% BSA were then placed in the lower chambers and the plates were incubated for 1 h at 37°C in a CO2 incubator. The migrated cells were collected from the lower chamber, stained with APC/eFluor 780-anti-Ly6G and PE-anti-CD11b mAbs for 30 min at 4°C, and enumerated using a FACSVerse or FACSCelesta flow cytometer (BD Biosciences). Purified neutrophils and peritoneal neutrophils were also prepared for neutrophil migration assay. For purified neutrophils, the cells were collected from BM samples using the Easysep Mouse Neutrophil Enrichment Kit and used for migration assay as described above. For peritoneal neutrophils, mice were injected intraperitoneally (IP) with 200 μg zymosan (Wako pure Chemica, Osaka, Japan) in 0.5 ml sterile saline. 2 h after injection, the peritoneal cells were collected and used for migration assay as described above. For the ROCK inhibitor experiments, serum-starved BM cells were treated with 10 μM Y-27632 or 20 μM KD025 in the presence or absence of arbutin for 30 min at 37°C before being placed in the upper chamber. For the RhoA inhibitor experiments, serum-starved BM cells were treated with 0.5 μg/ml Tat-C3 for 4 h at 37°C before pretreatment of arbutin.
Zymosan-induced peritonitis model in vivo
Mice were injected IP with zymosan (Wako pure Chemica) in 0.5 ml sterile saline. 6 h after injection, the peritoneal cavities were washed with 2 ml ice cold PBS. The neutrophils in the peritoneal lavage fluid were stained with APC/eFlour 780-anti-Ly6G and PE-anti-CD11b for analysis by flow cytometry. Subsequently, the number of neutrophils was determined using Count Bright Absolute Counting Beads (Thermo Fischer Scientific).
FACS analysis of CXCR2 and phosphorylated MLC2 expression levels
CXCR2 expression was analyzed by staining of total BM cells with FITC-anti-Ly6G, PE-anti-CD11b, and Alexa Fluor 647-anti-CXCR2 mAbs for 30 min at 4°C. Phosphorylated MLC2 levels were determined using neutrophils purified with kits as described above. The purified cells were serum-starved in RPMI-1640/0.1% BSA for 4 h, treated with arbutin for 30 min, and then stimulated with CXCL2 for 2 min. The cells were fixed with Fixation Buffer (BioLegend) for 30 min, permeabilized with 0.1% Triton-X for 10 min, and stained with anti-phospho-MLC2 pAb for 18 h at 4°C. After washing, the cells were incubated with Dylight 649-anti-rabbit IgG pAb for 60 min at room temperature. Data were acquired using a FACSCelesta flow cytometer and analyzed with FlowJo software (Tree Star Inc. OR, USA).
Neutrophil polarization assay
Neutrophil polarization assay was performed using neutrophils purified with kits as described above. The purified neutrophils were serum-starved in RPMI-1640/0.1% BSA for 4 h, treated with or without arbutin for 30 min at 37°C in an 8-well chamber slide (Merck, CA, USA), and then stimulated with CXCL2 for 2 min at 37°C. The cells were fixed with 4% PFA (Wako pure Chemica) for 10 min, permeabilized with 0.1% Triton-X for 10 min, and then stained with Flash Phalloidin Green 488 (BioLegend) for 20 min at room temperature. The nucleus was stained with DAPI (Nacalai-Tesque). The images were randomly acquired using an FV1200 confocal laser microscope with a 40x objective lens (Olympus, Tokyo, Japan). The number of total neutrophils was counted with Duolink Image Tool software (Sigma). Polarized neutrophils were determinized by nonuniform phalloidin staining (F-actin).
Statistical analysis
Data are presented as the mean ± standard deviation (SD) or standard error of the mean (SEM). Statistical analysis was performed as described previously (22) using Prism software (GraphPad, CA, USA). Briefly, differences between groups were evaluated by Student’s t-test for single comparisons or by one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test for multiple comparisons.
Results
Neutrophils express Tas2r126, Tas2r135, and Tas2r143
To explore the expression of Tas2Rs in immune cells, we surveyed 36 mouse Tas2r genes by analysis of the Immunological Genome Project (ImmGen, https://www.immgen.org/) RNA-seq database. As shown in Figure 1A and Supplementary Data Set 1, Tas2r126, Tas2r135, and Tas2r143 were highly expressed in neutrophils compared with other immune cell types, whereas other Tas2rs were low expressed in neutrophils. To confirm Tas2r126, Tas2r135, and Tas2r143 expression in neutrophils, we performed RT-qPCR analysis of FACS-sorted neutrophils isolated from BM, macrophages isolated from the peritoneal cavity, and T and B cells from the spleen of wild-type C57BL/6 mice. We detected Tas2r126, Tas2r135, and Tas2r143 mRNAs in neutrophils, whereas expression was undetectable in the other cells examined (Figure 1B). Thus, Tas2r126/135/143 are expressed in mouse neutrophils, consistent with a previous report (23).
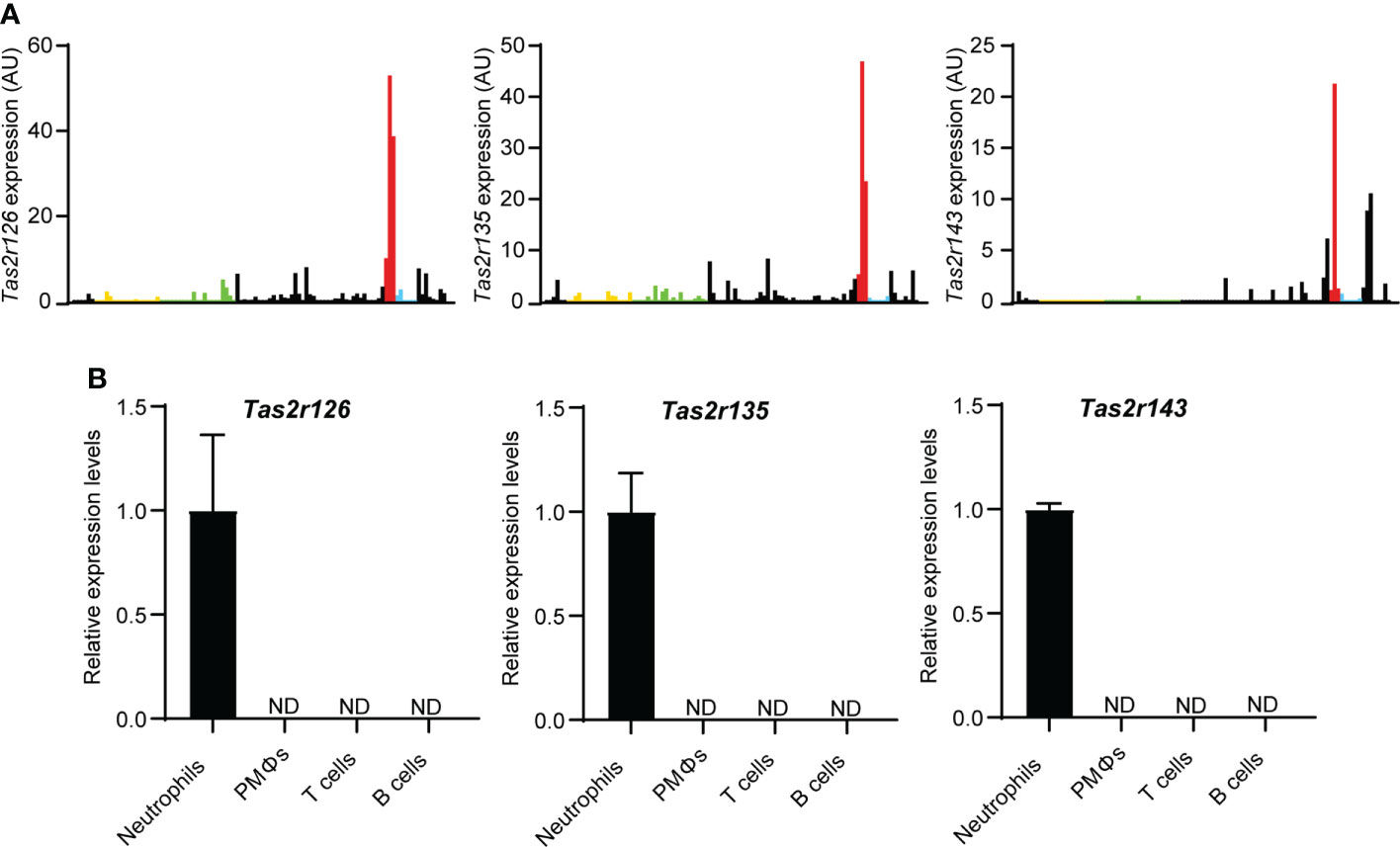
Figure 1 Neutrophils express Tas2r126, Tas2r135, and Tas2r143. (A) Analysis of Tas2r expression in immune cells from the ImmGen RNA-seq dataset. The data shown is Tas2r126, Tas2r135, and Tas2r143 mRNA levels in immune cells. Yellow, green, red, and blue bars show Tas2r126, Tas2r135, or Tas2r143 mRNA expression in B cells, T cells, neutrophils, or macrophages. Black bars show the mRNA expression in immune cells other than B cells, T cells, neutrophils, and macrophages. (B) RT-qPCR analysis of Tas2r126, Tas2r135, and Tas2r143 mRNA levels in purified spleen-derived B220+ B cells or CD4+/CD8+ T cells; F4/80+/CD11b+ peritoneal macrophages (PMΦs); and bone marrow-derived CD11b+/Ly6G+ neutrophils. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. ND, not detected.
Tas2R126/143 agonists augment chemoattractant-induced neutrophil migration
Neutrophils are the most motile cells in higher organisms and efficiently migrate in response to a chemoattractant gradient (24). Therefore, we next performed classical Transwell migration assays to investigate the effects of Tas2R agonists on BM-derived neutrophil migration. We selected the agonists arbutin, D-salicin (both Tas2R126/143 agonists), PBD-Gluco (Tas2R126 agonist), denatonium, and 5-propyl-2-thiouracil (both Tas2R135 agonists) for this study on the basis of a previous report of the 50% effective concentrations (EC50) for these compounds (16). However, our preliminary data suggested that treatment of neutrophils with these Tas2R agonists alone had very little effect on migration (Supplementary Figure 2). Therefore, we examined whether pretreatment with the agonists affected migration induced by the chemoattractant CXCL2. Indeed, the Tas2R126/143 agonists arbutin (16, 25) and D-salicin (16, 17), and the Tas2R126 agonist PBD-Gluco (16) all significantly enhanced CXCL2-induced neutrophil migration (Figure 2A), whereas the Tas2R135 agonists denatonium and 5-propyl-2-thiouracil had no significant effect (Figure 2B). We next examined whether Tas2R126/143 agonists affect CXCL2 receptor CXCR2 expression. The most potent agonist, arbutin, had no effect on expression of CXCR2 on the surface of neutrophils (Figure 2C). In addition, arbutin did not affect CXCL2-induced neutrophil polarization (Supplementary Figure 3). Thus, our results suggest that agonists of Tas2R126/143, but not Tas2R135, enhance CXCL2-stimulated neutrophil migration without modulating CXCR2 expression and cell polarity.
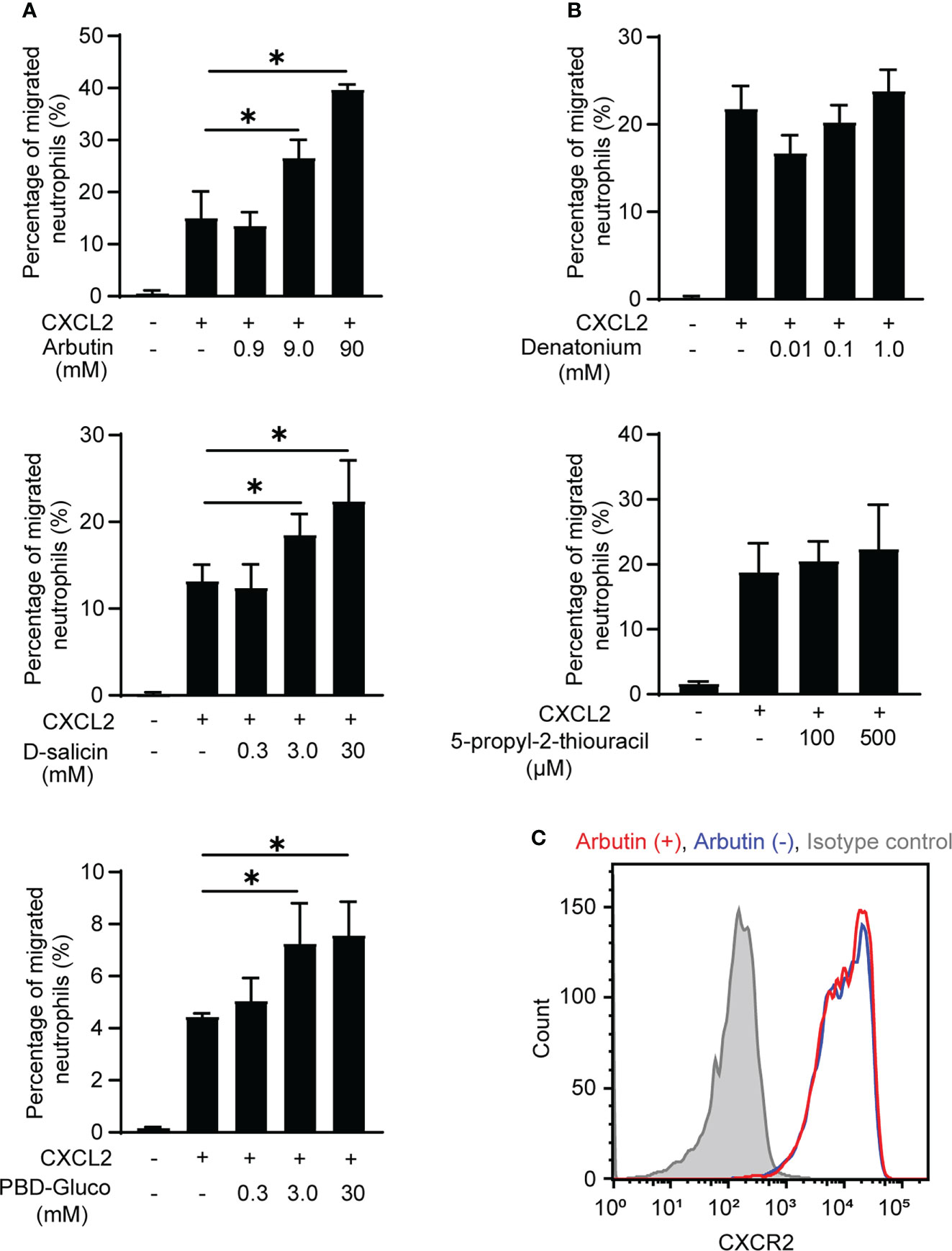
Figure 2 Effects of Tas2R126/135/143 agonists on CXCL2-induced neutrophil migration. (A) Transwell migration assays of bone marrow (BM)-derived neutrophils from wild-type C57BL/6 mice after pretreatment with the indicated concentrations of the Tas2R126/143 agonists arbutin and D-salicin or the Tas2R126 agonist PBD-Gluco for 30 min followed by stimulation with 20 ng/ml CXCL2. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. *p < 0.05 by Student’s t test. (B) Transwell migration assays of BM-derived neutrophils from wild-type C57BL/6 mice after pretreatment with the Tas2R135 agonists denatonium and 5-propyl-2-thiouracil for 30 min followed by stimulation with 20 ng/ml CXCL2. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. (C) FACS analysis of CXCR2 expression on purified neutrophils incubated with (red) or without (blue) arbutin. Gray histogram shows the isotype control-stained cells. Data are representative of three independent experiments.
To rule out the possibility that BM cells other than neutrophils were involved in the enhancement of neutrophil migration by Tas2R126/143 agonists, we used purified neutrophils. As we expected, arbutin enhanced CXCL2-induced purified neutrophil migration (Figure 3A). We also found that arbutin enhanced peritoneal neutrophil migration (Figure 3B). These results support the idea that arbutin directly acts on neutrophils and enhances CXCL2-induced neutrophil migration. To address whether arbutin affected other chemoattractants-induced neutrophil migration, we analyzed the effect of arbutin on fMLP- or CXCL1-induced neutrophil migration. As shown in Figures 3C, D, pretreatment of arbutin significantly enhanced fMLP- or CXCL1-induced neutrophil migration. These observations suggest that arbutin generally enhances chemoattractant-induced neutrophil migration.
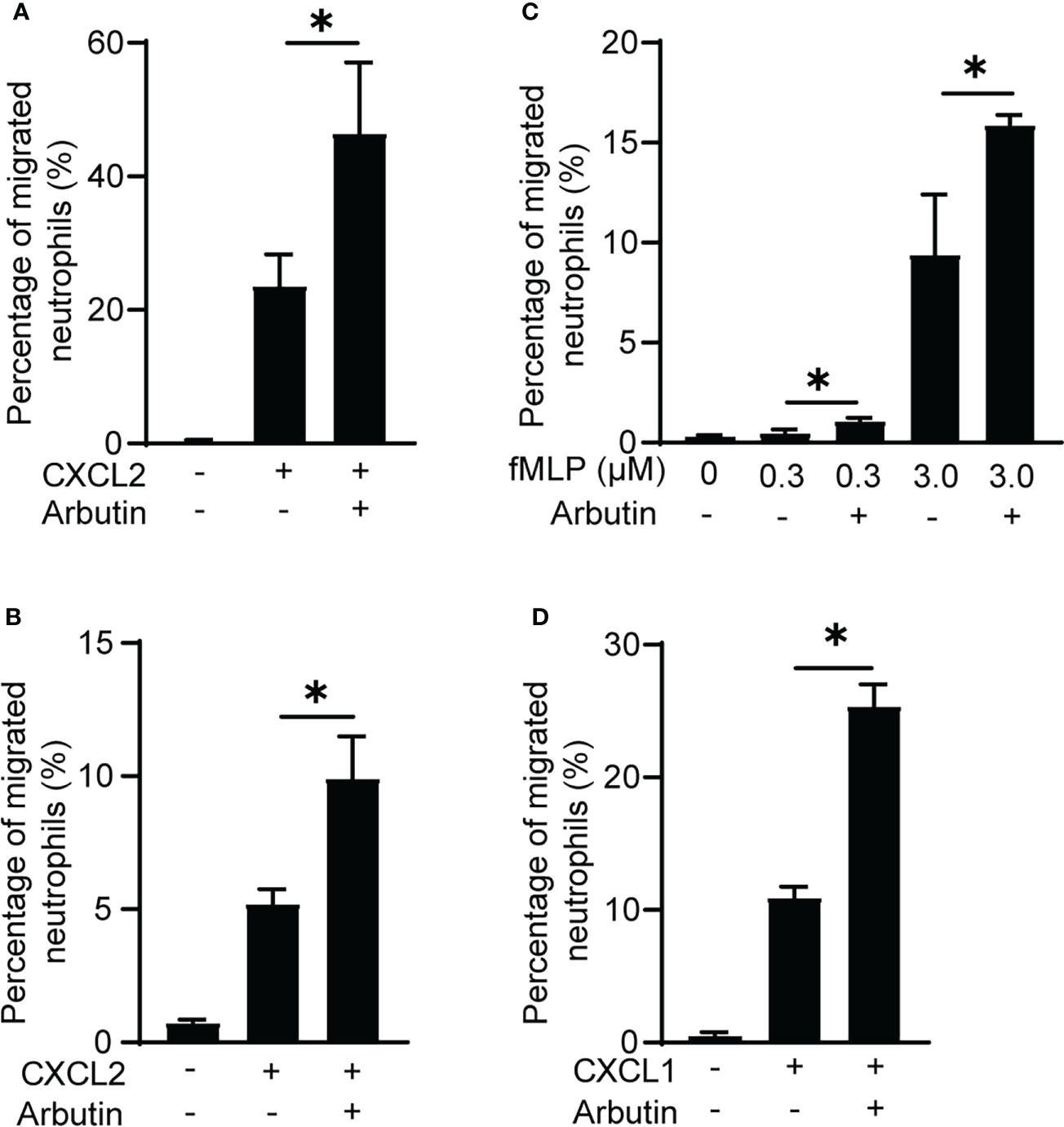
Figure 3 Arbutin enhances neutrophil migration. (A, B) Effects of arbutin on purified neutrophil (A) or peritoneal neutrophil (B) migration to CXCL2 was assessed by Transwell assay. Data are presented as the mean ± SD of triplicates from one experiment and are representative of two independent experiments. *p < 0.05 by Student’s t test. (C, D) Neutrophil migration in response to fMLP (C) or CXCL1 (D) with or without arbutin was evaluated by Transwell assay. Data are presented as the mean ± SD of triplicates from one experiment and are representative of two independent experiments. *p < 0.05 by Student’s t test.
Tas2R agonist-mediated enhancement of neutrophil migration is abolished by deletion of Tas2r126/135/143 expression
To exclude the possibility that arbutin enhanced neutrophil migration in a Tas2R126/143-independent manner, we generated lines of triple-deficient Tas2r126/135/143−/− mice. These three Tas2Rs are all located on mouse chromosome 6qB2.1; therefore, we deleted the region between Tas2r143 and Tas2r126 by introducing two gRNAs and Cas9 into fertilized eggs of C57BL/6 mice (Figure 4A). Effective deletion of the genes was confirmed by PCR of genomic DNA (Figure 4B), RT-qPCR of BM-derived neutrophils (Figure 4C), and direct gene sequencing (Figure 4D). As shown in Supplementary Figure 4, the expression of Tas2rs in heterozygous (+/−) and wild-type control (+/+) were almost comparable levels. Importantly, arbutin-mediated augmentation of CXCL2-stimulated migration was abolished in neutrophils from homozygous Tas2r126/135/143−/− mice but not those from heterozygous (+/−) or wild-type control (+/+) mice (Figure 4E). These results confirm that arbutin promotes chemoattractant-induced neutrophil migration via Tas2R126 and/or Tas2R143 signaling.
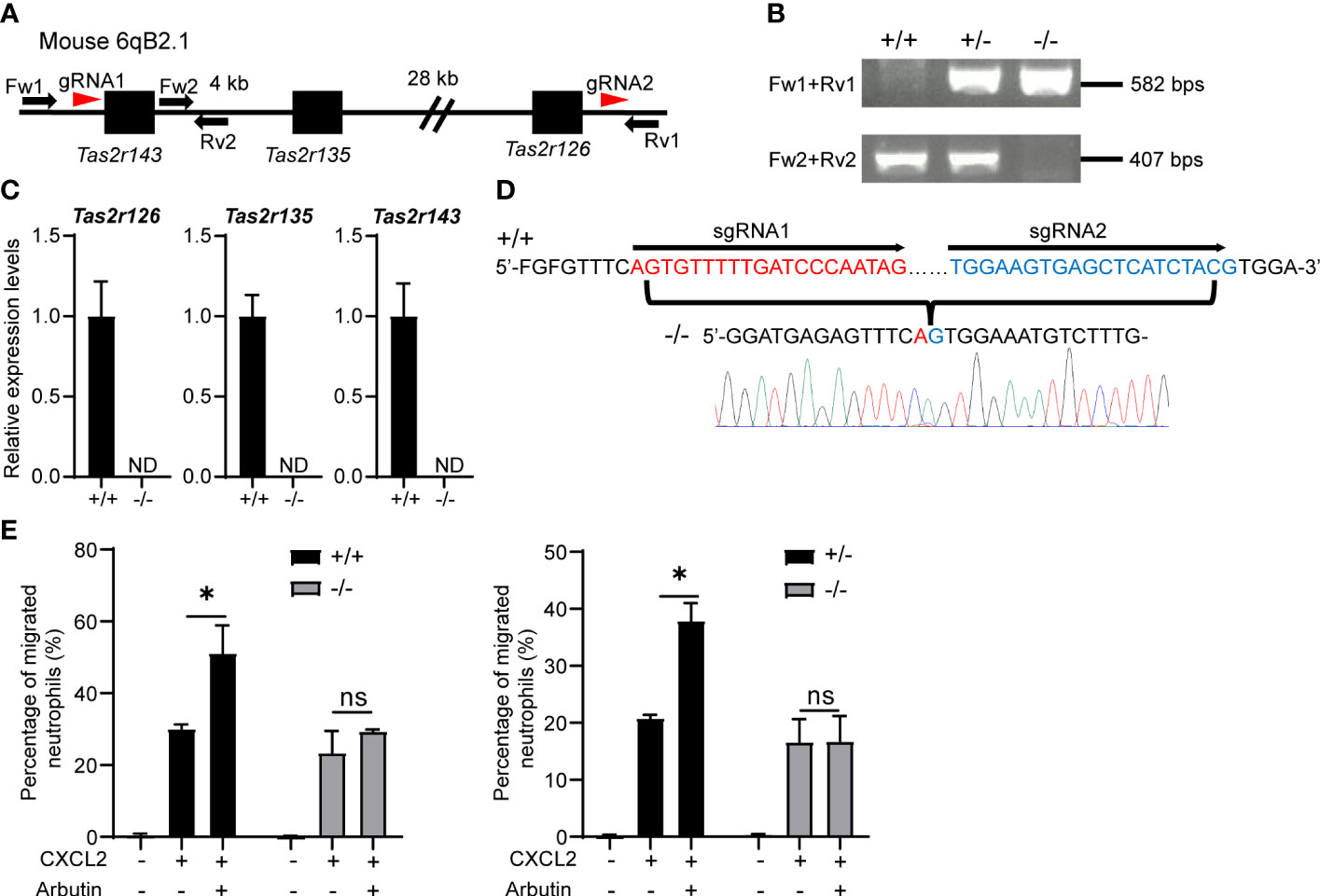
Figure 4 Arbutin-mediated enhancement of CXCL2-stimulated neutrophil migration is attenuated by deletion of Tas2r126, Tas2r135, and Tas2r143. (A) Schematic showing the location of Tas2r126, Tas2r135, and Tas2r143 genes on mouse chromosome 6qB2.1 and the position of gRNAs designed for deletion of the gene cluster between Tas2r143 and Tas2r126 (red arrowheads). Fw1, Fw2, Rv1, and Rv2 indicate the primers used for PCR genotyping of the mice. (B) DNA fragments amplified from Tas2r126/135/143+/+, +/−, and −/− mice. (C) RT-qPCR analysis of Tas2r126, Tas2r135, and Tas2r143 mRNA expression levels in bone marrow (BM)-derived neutrophils from Tas2r126/135/143+/+ or −/− mice. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. ND, not detected. (D) DNA sequences of PCR amplicons. A total of 35 kb of the genomic region between Tas2r143 and Tas2r126 was deleted in the Tas2r126/135/143−/− mice. (E) Transwell migration assays of BM-derived neutrophils from Tas2r126/135/143+/+ and −/− mice (left panel) or +/− and −/− mice (right panel). Cells were pretreated with 90 mM arbutin for 30 min and then stimulated with 20 ng/ml CXCL2. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. *p < 0.05 by Student’s t test. ns, not significant.
ROCK signaling is involved in Tas2R agonist-mediated enhancement of neutrophil migration
Previous studies have shown that bitter taste substances induce smooth muscle contraction and the studies have indicated the involvement of ROCK signaling (26) and MLC activation (27) in the smooth muscle contraction. In addition, we and others have reported that ROCK-dependent MLC2 activation plays a critical role in immune cell migration (28, 29). Therefore, we examined whether the ROCK–MLC2 pathway is involved in Tas2R126/143 signaling for enhanced neutrophil migration. Phosphorylated MLC2 (pMLC2) levels were analyzed and quantified by intracellular staining and flow cytometry of treated neutrophils. We found that pMLC2 levels were not affected by treatment of cells with arbutin alone and were slightly but not significantly increased by CXCL2 alone; however, combination treatment with arbutin and CXCL2 resulted in a striking increase in pMLC2 levels (Figure 5A). Moreover, this increase in pMLC2 was competently inhibited by co-treatment of neutrophils with the ROCK1/2 inhibitor, Y-27632 (Figure 5A). Notably, the arbutin-mediated increase in pMLC2 levels was not observed in neutrophils from Tas2r126/135/143−/− mice (Figure 5B), indicating that arbutin activates ROCK-mediated phosphorylation of MLC2 in neutrophils in a CXCL2- and Tas2R126/143-dependent manner.
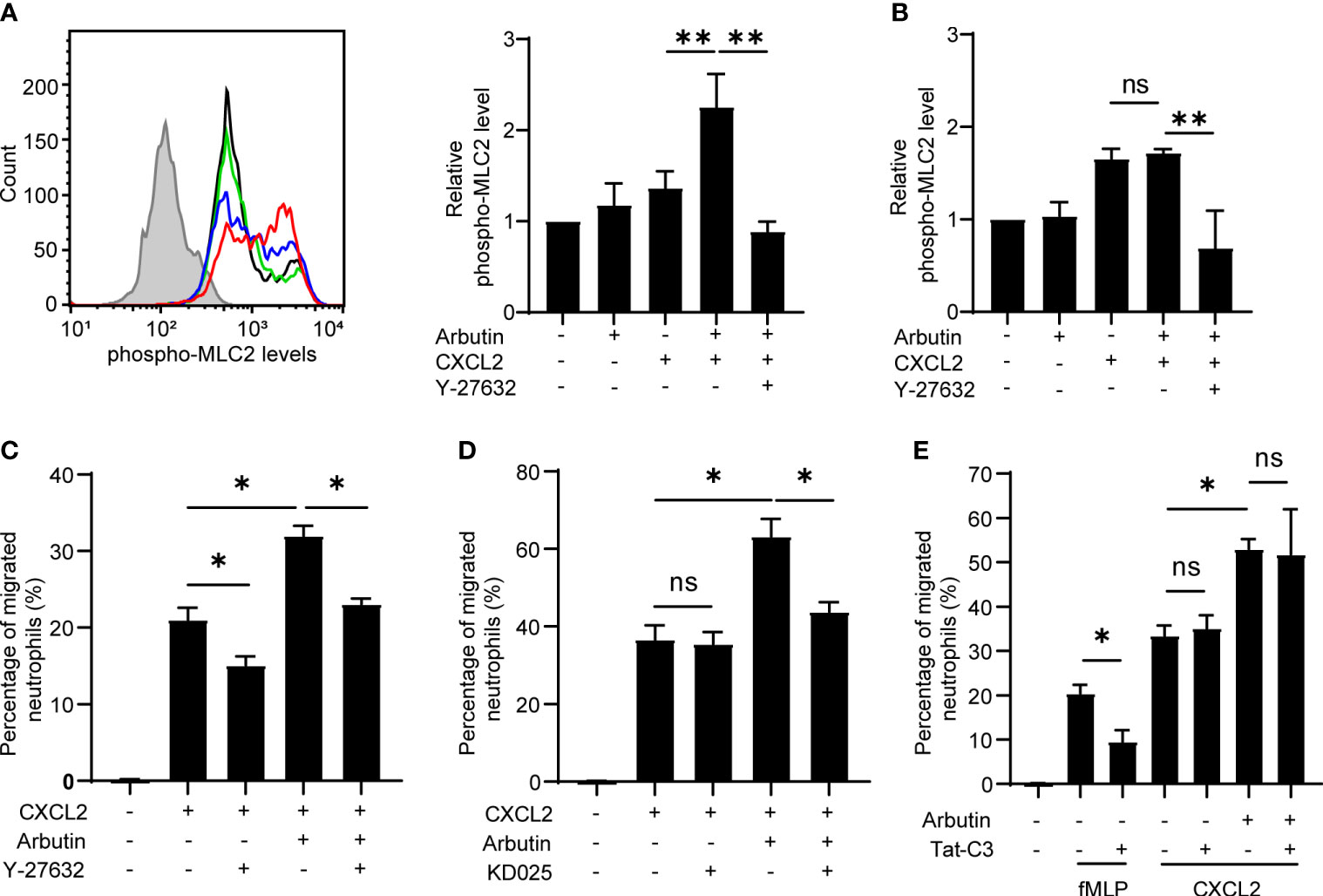
Figure 5 The ROCK–MLC2 pathway is involved in arbutin-mediated enhancement of CXCL2-stimulated neutrophil migration. (A) Left panel: FACS histograms of phosphorylated MLC2 expression levels in bone marrow (BM)-derived neutrophils from wild-type C57BL/6 mice. Neutrophils were untreated (black line) or incubated with CXCL2 alone (blue line), arbutin (green line), or both arbutin and CXCL2 (red line), Gray histogram shows control cells (anti-phospho-MLC2 pAb omitted). Right panel: Quantification of phosphorylated MLC2 expression (median fluorescence intensity) in the treated cells. Cells were also analyzed after stimulation with CXCL2 in the presence of Y-27632. Data are presented as the mean ± SD of three to five independent experiments. **p < 0.01 by Student’s t test. (B) Quantification of phosphorylated MLC2 expression (median fluorescence intensity) in BM-derived neutrophils from Tas2r126/135/143−/− mice after treatment with arbutin and CXCL2. Data are presented as the mean ± SD of three to six independent experiments. **p < 0.01 by Student’s t test. (C, D) Transwell migration assays of BM-derived neutrophils from Tas2r126/135/143+/+ mice after pretreatment with arbutin and either Y-27632 (C) or KD025 (D) (both ROCK inhibitors) for 30 min followed by stimulation with CXCL2. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. *p < 0.05 by Student’s t test. (E) Transwell migration assay of BM-derived neutrophils from Tas2r126/135/143+/+ mice after pretreatment with arbutin and/or the RhoA inhibitor Tat-C3 followed by stimulation with fMLP or CXCL2. Data are presented as the mean ± SD of triplicates from one experiment and are representative of three independent experiments. ns, not significant; *p < 0.05 by Student’s t test.
To assess the involvement of ROCK signaling in arbutin-enhanced neutrophil migration, we pretreated cells with the ROCK1/2 inhibitor Y-27632 or the ROCK2-selective inhibitor KD025 and examined CXCL2-stimulated migration in the presence or absence of arbutin. Notably, Y-27632, but not KD025, significantly attenuated migration induced by CXCL2 alone, whereas both compounds significantly suppressed arbutin-enhanced CXCL2-dependent migration (Figures 5C, D). These results suggest that CXCL2-induced neutrophil migration is likely mediated via ROCK1 alone, whereas arbutin-dependent augmentation most likely involves ROCK2. Because ROCK signaling is regulated by the upstream small G protein RhoA (30), we also examined the effect of RhoA inhibition on arbutin-enhanced neutrophil migration. As shown in Figure 5E, treatment with the RhoA inhibitor Tat-C3 inhibited migration of cells stimulated by fMLP, but not by CXCL2, similar to a previous report (31). Moreover, Tat-C3 also had no effect on arbutin-enhanced neutrophil migration in response to CXCL2 (Figure 5E). Based on these results, we propose a model in which Tas2R126/143 signaling enhances CXCL2-stimulated neutrophil migration via a ROCK2-dependent and RhoA-independent mechanism.
Discussion
Tas2Rs are expressed in the oral cavity where they detect bitter taste substances that may be harmful to health. Increasing evidence indicates that Tas2Rs are also expressed extraorally (5) and, although Tas2Rs expressed in immune cells are assumed to play roles in host defense, this has not been studied in detail. Here, we determined that neutrophils express Tas2r126, Tas2r135, and Tas2r143, and Tas2R126/143 play a role in promoting neutrophil migration. Specifically, we found that Tas2R126/143 agonists enhanced the chemoattractant-induced neutrophil migration via a mechanism involving RhoA-independent ROCK activation and MLC2 phosphorylation. This function of Tas2R126/143 was confirmed by demonstrating that the effects of Tas2R agonists on neutrophil migration and pMLC2 production were markedly diminished in neutrophils isolated from Tas2r126/135/143-deficient mice.
Currently, little is known about Tas2R signaling (32). Our results indicate that arbutin facilitates neutrophil migration via a Tas2R126/143–ROCK2–MLC2 pathway, which is consistent with findings that bitter taste substances induce ROCK-dependent smooth muscle contraction and MLC phosphorylation in smooth muscle cells (26, 27). As shown in Figure 5A, compared to CXCL2 alone, the combination treatment with arbutin and CXCL2 increased pMLC2 levels in a specific neutrophil population rather than total neutrophils. These results likely show that both Tas2R agonist-responsive and nonresponsive neutrophil populations exist. Although our data indicate that Tas2R signaling regulates ROCK2 and pMLC2 in a CXCL2-dependent manner, the precise molecular mechanisms underlying this sequence of events remain to be clarified.
Recent evidence suggests that bitter taste substances produced by pathogens can elicit immune responses. For example, a QSM, heptyl-hydroxyquinoline secreted by Pseudomonas was shown to enhance bacterial phagocytosis by human macrophages via TAS2R14-dependent nitric oxide production (8). In addition, other QSMs, competence-stimulating peptides secreted by Streptococcus mutans can induce secretion of proinflammatory cytokines from human gingival epithelial cells via TAS2R14 (7). Another group showed that substances excreted/secreted by the parasitic helminth Trichinella spiralis can induce IL-25 production by mouse tuft cells via Tas2R-mediated signaling, and, in turn, IL-25 triggers a type 2 immune response and leads to a “weep and sweep” response in the gut (17). Taking these reports into consideration, we speculate that several bitter taste substances produced by pathogens may be potent Tas2R126/143 agonists. The Tas2R126 agonists arbutin, D-salicin, and PBD-Gluco are commonly classified as β-D glucopyranosides (16), and several pathogens, especially fungi, are known to produce β-D glucopyranosides (33, 34). Thus, fungal β-D glucopyranosides could be considered to be members of the pathogen-associated molecular pattern family of molecules that promote responses by innate immune cells; as such, Tas2R126/143 would function as pattern recognition receptors.
To the best of our knowledge, only low-affinity agonists for Tas2R126/143 have been reported to date. Previous studies have reported EC50s of 30 mM, 10 mM, and 10 mM arbutin, D-salicin, and PBD-Gluco, respectively, for activation of Tas2R126, and of 3 mM and 0.3 mM arbutin and D-salicin, respectively, for activation of Tas2R143 (16, 17, 25). However, high-affinity agonists have been reported for human TAS2R14; they include heptyl-hydroxyquinoline and competence-stimulating peptides, which activated human TAS2R14 at concentrations of 20 μM (8) and 50 μM (7), respectively. Thus, we speculate that additional pathogen-derived substances may be identified that are high-affinity agonists of Tas2R126/143 and might play additional roles in promoting neutrophil functions, including migration. However, further work will be necessary to establish this.
In humans, β-D glucopyranosides including arbutin, D-salicin, and PBD-Gluco are recognized by TAS2R16 (ortholog of mouse Tas2R118) (35). While the ortholog of mouse Tas2r143 is a pseudogene in humans, the ortholog of mouse Tas2r126 is considered to be human TAS2R41 (16, 36). As TAS2R41 agonist, an antibacterial drug, chloramphenicol, is the only reported (37), and is not classified as a β-D glucopyranoside. Using the neutrophil RNA-seq database (Gene Expression in Neutrophil-like Cell Lines, https://collinslab.ucdavis.edu/neutrophilgeneexpression/) (38), we analyzed mouse Tas2r126/135, human TAS2R41, human TAS2R16, and human TAS2R60 (ortholog of mouse Tas2r135). We found the expression of TAS2R41 and TAS2R60 in human neutrophils, though the expression levels of TAS2R41/60 were lowered compared with mouse Tas2r counterparts (Supplementary Figure 5). In contrast, TAS2R16 was at undetectable levels in human neutrophils. These data suggest that the enhancement of neutrophil migration by β-D glucopyranosides may be restricted in specific species such as mice. Given that chloramphenicol is produced by several gram-positive soil actinomycetes (39), TAS2R41 expressed on human neutrophils might sense the bacteria products such as chloramphenicol. These points are fascinating, and future studies should be focused on them.
There are several limitations to this study. First, although we evaluated the effect of arbutin on neutrophil infiltration in vivo using the zymosan-induced peritonitis model, we observed no effect of Tas2r126/135/143 deletion in this model (Supplementary Figure 6), suggesting that other mechanisms of arbutin-facilitated neutrophil infiltration exist, at least in vivo. Second, we examined neutrophils from triple-deficient Tas2r126/135/143−/− mice, and it is unclear whether Tas2R126, Tas2R143, or both receptors participate in arbutin-induced enhancement of neutrophil migration. Third, it is unclear whether Tas2R126/143-mediated signaling also affects other crucial neutrophil functions, including phagocytosis, production of reactive oxygen species, and neutrophil extracellular trap formation. Since our current study assessed the effect of Tas2R agonists in a short period (0.5-1.5 h), the longer-term effects of Tas2R agonists on neutrophil function also remains uncertain. Finally, although we showed that neutrophils express Tas2r135 mRNA, the Tas2R135 agonists denatonium and 5-propyl-2-thiouracil had no effect on CXCL2-induced migration; therefore, it is unclear whether neutrophil-expressed Tas2R135 protein is functional. As another possibility, although neutrophils expressed Tas2R135 at functional levels, the downstream molecules of Tas2R135 might differ from Tas2R126/143 in neutrophils, and thus Tas2R135 agonists might have no effect on neutrophil migration.
Collectively, the results of this study provide evidence that Tas2R126 and/or Tas2R143 signaling induced by exogenous bitter taste substances enhance chemoattractant-induced mouse neutrophil migration via ROCK and MLC2-dependent signaling.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was reviewed and approved by Osaka University, Wakayama Medical University, and Niigata University.
Author contributions
DK conceived and designed the study; acquired, analyzed, and interpreted the data; and wrote the first draft of the article. TK analyzed, and interpreted the data; and wrote the first draft of the article. TW, MO, and YK acquired, analyzed and interpreted the data. FS, NK, AT, MI, and SM analyzed and interpreted the data. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by funding from the Japan Dairy Association (J-milk), a Niigata University Interdisciplinary Research (U-go) Grant, the Smoking Research Foundation, Fuji Foundation for Protein Research, and JSPS KAKENHI grants numbers 19K16697, 22K17823, and JP16H06276 (AdAMS).
Acknowledgments
We thank Dr. Masayuki Miyasaka (Osaka University) for his support of this research. We thank Ms. Eri Hosoyamada (Osaka University) for her technical assistance. We thank Anne M. O’Rourke, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2022.973880/full#supplementary-material
Supplementary Figure 1 | The purity of neutrophils after isolation from BM samples. Neutrophils were purified from BM samples by Easysep Mouse Neutrophil Enrichment Kit (A) or MojoSort Mouse Neutrophil Isolation Kit (B). The purity of neutrophils was assessed by FACS analysis.
Supplementary Figure 2 | Tas2R agonists alone do not affect neutrophil migration. Neutrophil migration in the presence or absence of Tas2R agonists in the upper chamber was examined by Transwell assay. Neutrophil migration to CXCL2 added to the lower chamber was used as a positive control. Data are presented as the mean ± SD of triplicates from one experiment and are representative of two independent experiments. *p < 0.05 by Student’s t test.
Supplementary Figure 3 | Effect of arbutin on CXCL2-induced neutrophil polarization. (A) Polarized neutrophils after stimulation with CXCL2 in the presence or absence of arbutin were counted (Green; F-actin, Blue; nucleus). White arrowheads show polarized neutrophils. (B) Quantification of the percentage of polarized neutrophils. Data are presented as the mean ± SD of four fields. The result shown is representative of two independent experiments, and per experiment 203 or 388 neutrophils (control), 299 or 630 neutrophils (arbutin), 1542 or 2452 neutrophils (CXCL2), and 759 or 1528 neutrophils (arbutin and CXCL2) were evaluated. NS, not significant. Scale bar, 20 μm.
Supplementary Figure 4 | Expression levels of Tas2r126/135/143 in Tas2r126/135/143 +/+ and +/− mice derived neutrophils. RT-qPCR analysis of Tas2r126, Tas2r135, and Tas2r143 mRNA expression levels in purified neutrophils from Tas2r126/135/143+/+ or +/− mice. Data are presented as the mean ± SD of triplicates from one experiment and are representative of two independent experiments.
Supplementary Figure 5 | Ortholog relationships between mouse Tas2rs and human TAS2Rs. (A) Human TASR41 and human TAS2R60 are ortholog of mouse Tas2r126 and mouse Tas2r135, respectively. Mouse Tas2r143 is a pseudogene in human (TAS2R62P). Human TAS2R41/60/62P are all located on human chromosome 7q35. (B) Expression levels of TAS2R41 and TAS2R60 in human neutrophils were analyzed by using the RNA-seq database.
Supplementary Figure 6 | Arbutin enhances neutrophil infiltration in a zymosan-induced peritonitis model in a Tas2R126/135/143-independent manner. (A) Wild-type C57BL/6 mice were injected IP with the indicated amount of zymosan in 0.5 ml sterile saline. Infiltrated neutrophils in peritoneal lavage fluid were collected 6 h later and enumerated by FACS analysis. Data represent the mean ± SEM of three mice. (B, C) Wild-type C57BL/6 mice (B) and Tas2r126/135/143−/− mice (C) were injected IP with saline, 2 μg zymosan, or 2 μg zymosan with 25 mg arbutin, and infiltered neutrophils were enumerated 6 h later by FACS analysis. Data represent the mean ± SEM of 5–9 mice. *p < 0.05 by Student’s t test. **p < 0.05 by one-way ANOVA followed by Tukey’s multiple comparison test.
Supplementary Table 1 | List of gRNA on-target sequences and PCR primer sequences.
Supplementary Data Set 1 | Analysis of 36 mouse Tas2r gene expression levels in immune cells by ImmGen RNA-seq database.
References
1. Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. A novel family of mammalian taste receptors. Cell (2000) 100(6):693–702. doi: 10.1016/s0092-8674(00)80705-9
2. Matsunami H, Montmayeur JP, Buck LB. A family of candidate taste receptors in human and mouse. Nature (2000) 404(6778):601–4. doi: 10.1038/35007072
3. Huang YA, Roper SD. Intracellular Ca2+ and TRPM5-mediated membrane depolarization produce ATP secretion from taste receptor cells. J Physiol (2010) 20:70–6. doi: 10.1113/jphysiol.2010.191106
4. Wong GT, Gannon KS, Margolskee RF. Transduction of bitter and sweet taste by gustducin. Nature (1996) 381(6585):796–800. doi: 10.1038/381796a0
5. Harmon CP, Deng D, Breslin PAS. Bitter taste receptors (T2Rs) are sentinels that coordinate metabolic and immunological defense responses. Curr Opin Physiol (2021) 20:70–6. doi: 10.1016/j.cophys.2021.01.006
6. Pu Q, Guo K, Lin P, Wang Z, Qin S, Gao P, et al. Bitter receptor TAS2R138 facilitates lipid droplet degradation in neutrophils during pseudomonas aeruginosa infection. Signal Transduction Targeted Ther (2021) 6(1):210. doi: 10.1038/s41392-021-00602-7
7. Medapati MR, Singh N, Bhagirath AY, Duan K, Triggs-Raine B, Batista EL Jr., et al. Bitter taste receptor T2R14 detects quorum sensing molecules from cariogenic streptococcus mutans and mediates innate immune responses in gingival epithelial cells. FASEB J (2021) 35(3):e21375. doi: 10.1096/fj.202000208R
8. Gopallawa I, Freund JR, Lee RJ. Bitter taste receptors stimulate phagocytosis in human macrophages through calcium, nitric oxide, and cyclic-gmp signaling. Cell Mol Life Sci (2020) 78(1):271–86. doi: 10.1007/s00018-020-03494-y
9. Orsmark-Pietras C, James A, Konradsen JR, Nordlund B, Söderhäll C, Pulkkinen V, et al. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J (2013) 42(1):65–78. doi: 10.1183/09031936.00077712
10. Sharma P, Yi R, Nayak AP, Wang N, Tang F, Knight MJ, et al. Bitter taste receptor agonists mitigate features of allergic asthma in mice. Sci Rep (2017) 7(1):46166. doi: 10.1038/srep46166
11. Zhou Z, Xi R, Liu J, Peng X, Zhao L, Zhou X, et al. TAS2R16 activation suppresses lps-induced cytokine expression in human gingival fibroblasts. Front Immunol (2021) 12:726546(5440). doi: 10.3389/fimmu.2021.726546
12. Grassin-Delyle S, Salvator H, Mantov N, Abrial C, Brollo M, Faisy C, et al. Bitter taste receptors (TAS2Rs) in human lung macrophages: Receptor expression and inhibitory effects of TAS2R agonists. Front Physiol (2019) 10:1267(1267). doi: 10.3389/fphys.2019.01267
13. Mueller KL, Hoon MA, Erlenbach I, Chandrashekar J, Zuker CS, Ryba NJP. The receptors and coding logic for bitter taste. Nature (2005) 434(7030):225–9. doi: 10.1038/nature03352
14. Lu P, ElMallah MK, Liu Z, Wu C, Chen J, Lifshitz LM, et al. Genetic deletion of the Tas2r143/Tas2r135/Tas2r126 cluster reveals that Tas2rs may not mediate bitter tastant-induced bronchodilation. J Cell Physiol (2021) 236(9):6407–23. doi: 10.1002/jcp.30315
15. Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, et al. T2Rs function as bitter taste receptors. Cell (2000) 100(6):703–11. doi: 10.1016/s0092-8674(00)80706-0
16. Lossow K, Hübner S, Roudnitzky N, Slack JP, Pollastro F, Behrens M, et al. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J Biol Chem (2016) 291(29):15358–77. doi: 10.1074/jbc.M116.718544
17. Luo XC, Chen ZH, Xue JB, Zhao DX, Lu C, Li YH, et al. Infection by the parasitic helminth trichinella spiralis activates a Tas2r-mediated signaling pathway in intestinal tuft cells. Proc Natl Acad Sci United States America (2019) 116(12):5564–9. doi: 10.1073/pnas.1812901116
18. Fujihara Y, Noda T, Kobayashi K, Oji A, Kobayashi S, Matsumura T, et al. Identification of multiple Male reproductive tract-specific proteins that regulate sperm migration through the oviduct in mice. Proc Natl Acad Sci (2019) 116(37):18498–506. doi: 10.1073/pnas.1908736116
19. Kobayashi D, Kiguchi N, Saika F, Kishioka S, Matsuzaki S. Insufficient efferocytosis by M2-like macrophages as a possible mechanism of neuropathic pain induced by nerve injury. Biochem Biophys Res Commun (2020) 525(1):216–23. doi: 10.1016/j.bbrc.2020.02.032
20. Liu S, Lu S, Xu R, Atzberger A, Günther S, Wettschureck N, et al. Members of bitter taste receptor cluster Tas2r143/Tas2r135/Tas2r126 are expressed in the epithelium of murine airways and other non-gustatory tissues. Front Physiol (2017) 8:849(849). doi: 10.3389/fphys.2017.00849
21. Kobayashi D, Endo M, Ochi H, Hojo H, Miyasaka M, Hayasaka H. Regulation of CCR7-dependent cell migration through CCR7 homodimer formation. Sci Rep (2017) 7(1):8536. doi: 10.1038/s41598-017-09113-4
22. Kobayashi D, Sugiura Y, Umemoto E, Takeda A, Ueta H, Hayasaka H, et al. Extracellular ATP limits homeostatic T cell migration within lymph nodes. Front Immunol (2021) 12:786595(5518). doi: 10.3389/fimmu.2021.786595
23. Lee N, Jung YS, Lee HY, Kang N, Park YJ, Hwang JS, et al. Mouse neutrophils express functional umami taste receptor T1R1/T1R3. BMB Rep (2014) 47(11):649–54. doi: 10.5483/bmbrep.2014.47.11.185
24. Xu W, Wang P, Petri B, Zhang Y, Tang W, Sun L, et al. Integrin-induced Pip5k1c kinase polarization regulates neutrophil polarization, directionality, and in vivo infiltration. Immunity (2010) 33(3):340–50. doi: 10.1016/j.immuni.2010.08.015
25. Foster SR, Blank K, Hoe LES, Behrens M, Meyerhof W, Peart JN, et al. Bitter taste receptor agonists elicit G-Protein-Dependent negative inotropy in the murine heart. FASEB J (2014) 28(10):4497–508. doi: 10.1096/fj.14-256305
26. Avau B, Rotondo A, Thijs T, Andrews CN, Janssen P, Tack J, et al. Targeting extra-oral bitter taste receptors modulates gastrointestinal motility with effects on satiation. Sci Rep (2015) 5(1):15985. doi: 10.1038/srep15985
27. Upadhyaya JD, Singh N, Sikarwar AS, Chakraborty R, Pydi SP, Bhullar RP, et al. Dextromethorphan mediated bitter taste receptor activation in the pulmonary circuit causes vasoconstriction. PloS One (2014) 9(10):e110373. doi: 10.1371/journal.pone.0110373
28. Takeda A, Kobayashi D, Aoi K, Sasaki N, Sugiura Y, Igarashi H, et al. Fibroblastic reticular cell-derived lysophosphatidic acid regulates confined intranodal T-cell motility. eLife (2016) 5:e10561. doi: 10.7554/eLife.10561
29. Niggli V. Rho-kinase in human neutrophils: A role in signalling for myosin light chain phosphorylation and cell migration. FEBS Lett (1999) 445(1):69–72. doi: 10.1016/S0014-5793(99)00098-8
30. Hartmann S, Ridley AJ, Lutz S. The function of rho-associated kinases Rock1 and Rock2 in the pathogenesis of cardiovascular disease. Front Pharmacol (2015) 6:276(276). doi: 10.3389/fphar.2015.00276
31. Jennings RT, Strengert M, Hayes P, El-Benna J, Brakebusch C, Kubica M, et al. RhoA determines disease progression by controlling neutrophil motility and restricting hyperresponsiveness. Blood (2014) 123(23):3635–45. doi: 10.1182/blood-2014-02-557843
32. Jalševac F, Terra X, Rodríguez-Gallego E, Beltran-Debón R, Blay MT, Pinent M, et al. The hidden one: What we know about bitter taste receptor 39. Front Endocrinol (2022) 13:854718. doi: 10.3389/fendo.2022.854718
33. Lin J, Pillay B, Singh S. Purification and biochemical characteristics of β-D-Glucosidase from a thermophilic fungus, thermomyces lanuginosus-ssbp. Biotechnol Appl Biochem (1999) 30(1):81–7. doi: 10.1111/j.1470-8744.1999.tb01163.x
34. Otsuka K, Yadomae T, Miyazaki T. Fungal glycosidases. v. purification and properties of an extracellular exo-(1→3)-β-D-Glucosidase from trichophyton mentagrophytes. Chem Pharm Bull (1979) 27(9):2042–7. doi: 10.1248/cpb.27.2042
35. Imai H, Suzuki N, Ishimaru Y, Sakurai T, Yin L, Pan W, et al. Functional diversity of bitter taste receptor Tas2r16 in primates. Biol Lett (2012) 8(4):652–6. doi: 10.1098/rsbl.2011.1251
36. Dainat J, Paganini J, Pontarotti P, Gouret P. Gladx: An automated approach to analyze the lineage-specific loss and pseudogenization of genes. PloS One (2012) 7(6):e38792. doi: 10.1371/journal.pone.0038792
37. Thalmann S, Behrens M, Meyerhof W. Major haplotypes of the human bitter taste receptor Tas2r41 encode functional receptors for chloramphenicol. Biochem Biophys Res Commun (2013) 435(2):267–73. doi: 10.1016/j.bbrc.2013.04.066
38. Rincón E, Rocha-Gregg BL, Collins SR. A map of gene expression in neutrophil-like cell lines. BMC Genomics (2018) 19(1):573. doi: 10.1186/s12864-018-4957-6
Keywords: Tas2R, cell migration, bitter taste substances, ROCK, neutrophils
Citation: Kobayashi D, Watarai T, Ozawa M, Kanda Y, Saika F, Kiguchi N, Takeuchi A, Ikawa M, Matsuzaki S and Katakai T (2022) Tas2R signaling enhances mouse neutrophil migration via a ROCK-dependent pathway. Front. Immunol. 13:973880. doi: 10.3389/fimmu.2022.973880
Received: 20 June 2022; Accepted: 29 July 2022;
Published: 18 August 2022.
Edited by:
Valentin P. Yakubenko, East Tennessee State University, United StatesReviewed by:
Alan Y. Hsu, Boston Children’s Hospital and Harvard Medical School, United StatesRobert J. Lee, University of Pennsylvania, United States
Copyright © 2022 Kobayashi, Watarai, Ozawa, Kanda, Saika, Kiguchi, Takeuchi, Ikawa, Matsuzaki and Katakai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daichi Kobayashi, a29iYXlAbWVkLm5paWdhdGEtdS5hYy5qcA==; a29iYXkuaW1tdW5vbG9neUBnbWFpbC5jb20=; Tomoya Katakai, a2F0YWthaUBtZWQubmlpZ2F0YS11LmFjLmpw
 Daichi Kobayashi
Daichi Kobayashi Tomoya Watarai1
Tomoya Watarai1 Madoka Ozawa
Madoka Ozawa Yasuhiro Kanda
Yasuhiro Kanda Fumihiro Saika
Fumihiro Saika Norikazu Kiguchi
Norikazu Kiguchi Arata Takeuchi
Arata Takeuchi Masahito Ikawa
Masahito Ikawa Shinsuke Matsuzaki
Shinsuke Matsuzaki Tomoya Katakai
Tomoya Katakai