- Department of Pathology, Xinxiang Medical University, Xinxiang, Henan, China
FHOD 1 (Formin homology 2 domain containing protein 1) is a member of Diaphanous-related formins (DRFs) which contains a GTP-binding domain (GBD), formin homology (FH) 1 and FH 2 domains, a coiled-coil, and a diaphanous-like autoregulatory domain. Studies have shown that FHOD1 can not only regulate intracellular signals in tumor cells but also regulate various components of the tumor microenvironment (TME), such as T cells, B cells, cancer-associated fibroblasts (CAFs), some cytokines. Aberrant expression and dysfunction of the FHOD1 protein play a key role in tumor immunosuppression. Specifically, FHOD1 can impair function of chemokine receptors that are supposed to direct immune cells to localize to the tumor site accurately. As a result of this impairment, immune cells cannot migrate efficiently into TME, thereby impairing their ability to attack tumor cells. In addition, FHOD1 activated signaling pathways within the immune cells abnormally, resulting in their inability to recognize and destroy tumor cells effectively. Therefore, FHOD1 ultimately leads to a state of immunosuppression in TME, providing favorable conditions for the growth and spread of tumor cells. Altogether this review provides an in-depth understanding of the role of FHOD1 in tumor immunosuppression.
1 Introduction
Tumors develop within complex tissue microenvironments characterized by dynamic cellar interactions. The initiation of tumorigenesis involves the acquisition of abnormal proliferative capacity through genetic alterations, primarily including activation of proto-oncogenes and inactivation of tumor-suppressor genes. However, a tumor consists not only a group of cancer cells, but also of significant alterations in the surrounding extracellular matrix or tumor microenvironment (TME) (1, 2). These alterations are now recognized as a critical element for tumor development and progression, as well as potential therapeutic targets. Various components of TME, including immune cells and cancer-associated fibroblasts (CAFs), along with diverse cytokines (3), which together impede effective antitumor immunity and promote tumor progression and metastasis.
Formins are multi-domain proteins characterized by the highly conserved formin homology (FH)2 structural domain. This structural domain regulates the nucleation or elongation of actin filaments (4). In addition to the FH2 structural domain, Formins also contains several other structural domains, enabling them to regulate their activation/inactivation state and subcellular localization more finely. For example, some Formins possess specific binding domains at the N-terminal or C-terminal end, which can interact with other molecules (such as signaling molecules, regulatory proteins) to achieve precise regulation of Formin activity (5). In addition, some Formins contain nuclear localization signals or membrane-binding sequences, which enable Formins to function in specific cellular sites, further expanding the diversity of their biological functions. The biochemical properties of individual formins exhibit remarkable diversity and can sometimes be functionally antagonistic: upon activation, they may participate in various actin-related processes, including nucleation, elongation, bundling and capping (6). At the cellular level, individual Formins are involved in the formation of various cell protrusions, and adhesions (7). FHOD1 (Formin homology 2 domain containing protein 1), a 1165-amino acid FH protein, is a potent bundling protein for actin filaments. Unlike most formins, FHOD1 does not elongate actin filaments in vivo (8). FHOD1 contains a GTPase binding domain (GBD), FH1 and FH2 domains, a coiled-coil, and a diaphanous-like autoregulatory domain (DAD) (9). In addition, a large number of studies have shown that FHOD 1 plays an important role in tumor cell migration, invasion, and stress fiber formation (10–13). For example, FHOD1 is frequently overexpressed in triple-negative breast cancer, specifically, overexpression of FHOD1 may promote malignant proliferation and metastasis of cancer cells by regulating cytoskeletal stability, cell migration ability, and modulation of various signaling pathways (10). In addition to triple-negative breast cancer, FHOD1 expression has been observed to be significantly uppregulated in glioma cells, a change that correlated strongly with the degree of malignancy and aggressiveness of gliomas, as well as with the prognosis of patients (14). Moreover, FHOD1 upregulates PDL1 expression in tumors through epithelial-mesenchymal transition (EMT) (15), leading to changes in the immune microenvironment. Specifically, FHOD1 regulates the function and activity of T-lymphocytes, cancer-associated fibroblasts (CAF), and B-cells through PDL1, which in turn affects the process of immune escape and immunosuppression in tumors. In this review, considering the pivotal role of FHOD1 in both tumor cells and TME, we aimed to summarize the functions and underlying mechanisms of FHOD1 acts in the communication between tumor cells and TME.
2 Effect of FHOD1 in tumor cells
2.1 Effect on FHOD1 expression during EMT
Tumor cells acquire invasive and metastasize capabilities through the EMT process (16). This mechanism of cellular plasticity enables malignant cells to remodel their actin cytoskeleton and downregulate cell-cell adhesion proteins. FHOD1 is involved in cytoskeletal remodeling and cell migration in fibroblasts, melanoma cells, and breast cancer cells (15, 17). As actin nucleation- forming are recognized as key regulators of EMT, the expression pattern of FHOD1 in human tissues shows a clear mesenchymal preference, with predominant expression in mesenchymal cells and little expression in epithelial cells. Notably, this expression pattern was significant altered in oral squamous cell carcinoma, where FHOD1 was observed to be upregulated in a PI3K signaling-dependent manner after EMT (Figure 1) (13). In EMT-transformed cells, FHOD 1 promotes the development of spindle-shaped morphology and facilitates the formation of mesenchymal F-actin structures. Functional analyses revealed that FHOD1 significantly enhances cell migration and invasion capabilities. Deletion of FHOD 1 impaired the ability of EMT cancer cells to form invasive nuclear peduncles and degrade the extracellular matrix. Based on this finding, we can further explore the potential role of FHOD1 in the tumor microenvironment. EMT is a complex biological process involving changes in cell morphology, adhesion, migration, and invasive capacity, which plays a critical role in tumorigenesis and metastasis (18–20). Therefore, FHOD1 may indirectly affect tumor progression and immune evasion mechanisms by regulating EMT.
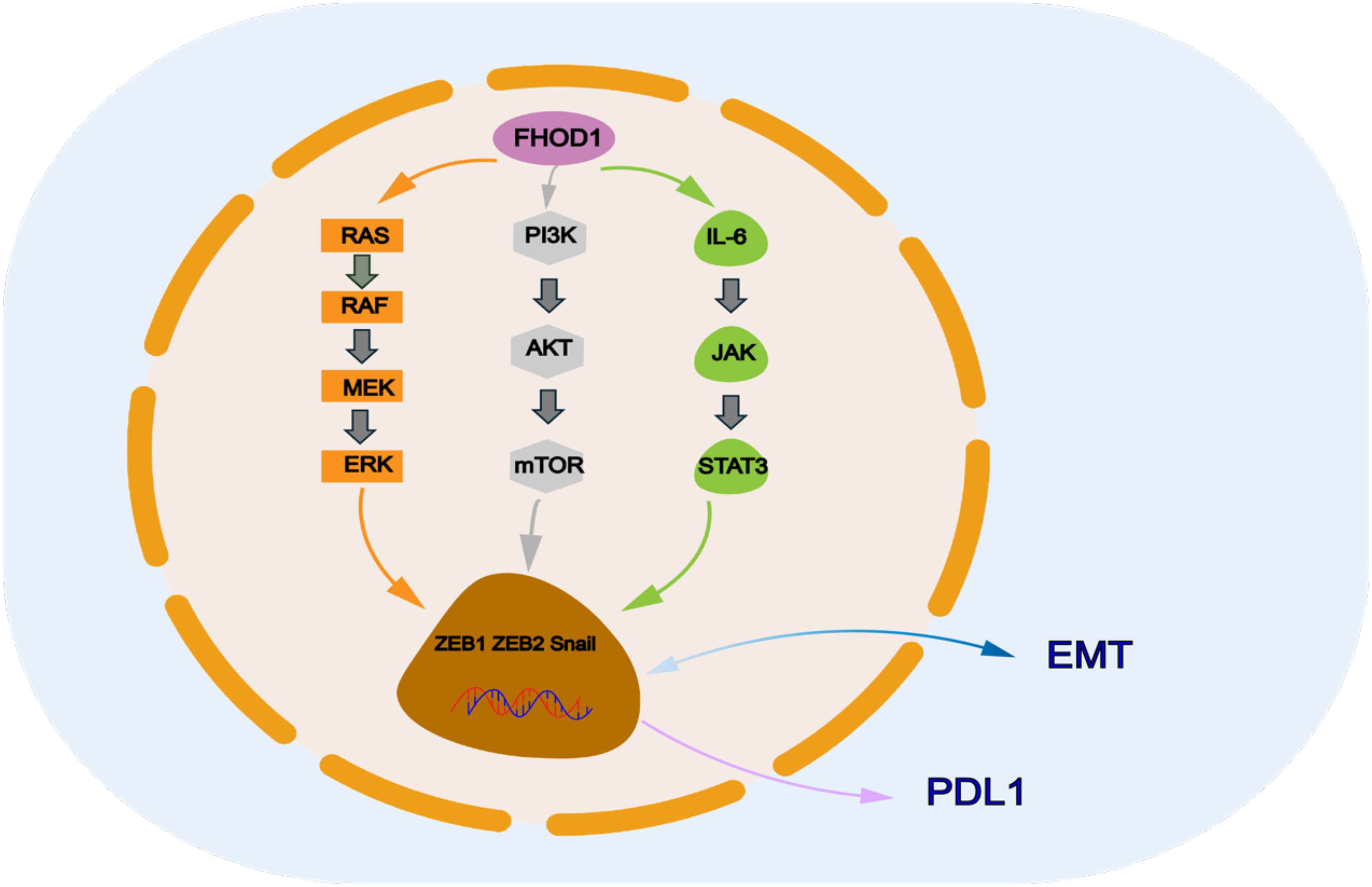
Figure 1. FHOD1 modulates key signaling pathways involved in PDL 1 expression, and downstream signals of these pathways include ZEB and Snail, which are transcription factors of EMT that regulate PDL 1 expression.
2.2 FHOD1 promotes PDL1 expression in tumor EMT
Transcription factors play a key role in inducing FHOD1 expression during EMT, which subsequently promotes tumor cell proliferation, migration, and invasion, thereby accelerating tumourigenesis and progression. For example, it has been shown that the EMT phenotype of oral squamous cell is closely associated with elevated expression levels of ZEB1 and ZEB2 (21). Similarly, in cutaneous squamous cell carcinoma, the EMT process is mainly induced by the transcription factor Snail. FHOD1 is significantly upregulated at both transcriptional and protein levels in various EMT cell lines (13). In addition, transcription factors that promote tissue EMT transformation have also been implicated in the regulation of PDL1 expression (22–25). These transcription factors include but are not limited to Snail, Twist, ZEB1, and ZEB2. They directly regulate the transcriptional level of PDL1 by binding to the promoter or enhancer regions of the PDL1 gene. A representative example is the Snail-induced formation of the CCL2/Lcn2 complex, which establishes an immunosuppressive microenvironment that ultimately leads to upregulation of PDL1 expression (26). Thus, the role of FHOD1 in malignant tumors is not limited to the promotion of EMT, it further exacerbates the immune escape phenomenon in tumors by upregulating PDL1 expression (27, 28). This dual regulatory mechanism confers a unique biological function to FHOD1 in cancer (Figure 1).
3 Role of FHOD1 in tumor immune microenvironment
3.1 Relationship between FHOD1 and tumor-infiltrating T lymphocytes
The TME comprises three major components: tumor cells, tumor stroma (inflammatory cells, fibroblasts, and vascular networks), and the surrounding extracellular matrix (29). The non-malignant cells of the TME play an active role in various steps of tumourigenesis (1), significantly influencing tumor behavior and treatment response (3, 30). Within this complex ecosystem,various cells infiltrate the tumor mass and engage in dynamic interactions with tumor cells through both direct cell-to-cell contact and secreted signaling molecules (31). Tumor-infiltrating T lymphocytes play a pivotal role in tumor immunity (32–34), and they exert complex immunomodulatory effects through different subpopulations such as CD4+ T cells and CD8+ T cells (35, 36), as well as anti-tumor pro-inflammatory T cells, immunosuppressive Th2 cells, Th17 cells, and regulatory T cells. These T cells are capable of secreting various immunosuppressive cytokines, such as IL-10 and transforming growth factor β (TGF-β), which further regulate T cell function (37). However, excessive proliferation of tumor cells alters the supply of nutrients and oxygen in TME, forcing T cells to change their metabolic pathways from a dependence on glycolysis to a reliance on fatty acid oxidation(FAO) and oxidative phosphorylation(OXPHOS) to maintain their effector functions. This metabolic adaptation is critical for T cell survival and function in the tumor microenvironment (38). Studies have shown that FHOD1 expression is associated with lymphocyte infiltration in tumor (39). It is noteworthy that human leukocyte antigens are predominantly localized in lymphoid tissues such as the spleen and thymus, as well as in hematopoietic tissues (39). Subsequent investigations have revealed that the substance is overexpressed in human hematological malignancies, especially in non-Hodgkin’s lymphoma and leukemia cell lines. These findings provide important clues for further investigation of its biological functions and potential herapeutic applications (40, 41).
In tumor cells, FHOD1 exploits the PD-1/PDL1 pathway to evade immune surveillance, through the suppression of T-cell responses within the TME (42, 43). PD-L1, a key ligand of PD-1, is expressed on the surface of immune cells (44, 45) and tumor cells (31). Upon binding to PD-1, which accumulates near the TCR site, recruits the phosphatase SHP2 to its cytoplasmic domain. Subsequently, SHP2 dephosphorylates proximal TCR signaling molecules, leading to reduced T cell proliferation, reduced cytokine secretion (interferon-γ, IL-2, and tumor necrosis factor-α), altered effector function and reduced survival of activated T cells (Figure 2) (31). In summary, FHOD1 enhances the expression level of PDL1 by upregulating its expression during EMT. Therefore, the aberrant expression of PDL1 can be attributed to the upregulation of the oncogenic pathways. These findings elucidate the critical roles of FHOD1 and PDL1 in tumorigenesis, demonstrating their contribution to tumor progression through the regulation of EMT processes (46).
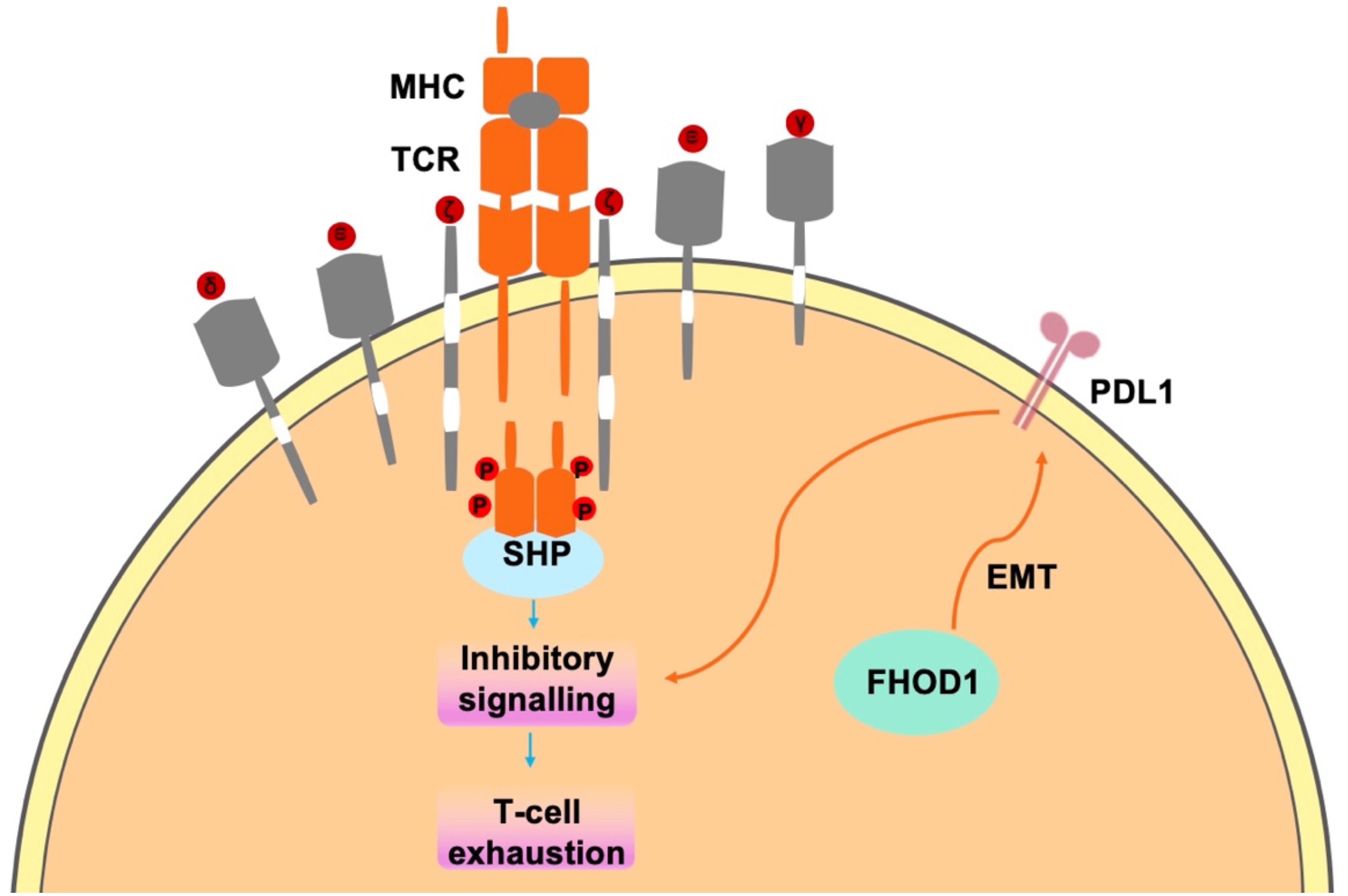
Figure 2. In tumor cells, FHOD1 uses the PD-1/PDL1 pathway to evade immune surveillance mainly by inhibiting t-cell responses in the TME.
3.2 Interaction of FHOD1 and autophagy in CAF
CAFs play a key role in promoting connective tissue proliferation and immunosuppression in TME (47–50), making them potential therapeutic targets for cancer (51–53). Autophagy is a highly regulated, multi-step cellular process that facilitates the transport of cytoplasmic components to lysosomes for degradation and play crucial roles in nutrient recycling and metabolic adaptation (54, 55). FHOD1 is enhanced by the up-regulation of PDL1 expression during EMT, and a large body of literature has shown that upregulation of PDL1 expression causes autophagy in tumor fibroblasts. For example, Xiaozhen Zhang et al. found that in an immunocompetent mouse model, autophagy in CAFs led to up-regulation of PDL1 expression, which resulted tumor immune escape (56). Endo S et al. reported that autophagy of CAFs activates pancreatic stellate cells, promotes the development of pancreatic cancer, and correlates poor prognosis in pancreatic patients cancer (57). In summary, FHOD1 can influence CAF by promoting the up-regulation of PDL1 protein expression (58). This effect was primarily characterized by the enhancement of autophagic activity in CAF cells, a crucial cellular process responsible for intracellular degradation and component recycling. Through autophagy, CAF cells have the potential to help tumor cells evade the surveillance of the immune system, thereby promoting immune escape (53, 59). Immune escape is a key mechanism by which tumor cells evade detection and destruction by the immune system, which is essential for tumor growth and spread. In addition, FHOD1 inhibits TME, including immune cells, extracellular matrix, vascular and other molecular signals, through PDL1-mediated CAF autophagy (60). Inhibition of the TME leads to impaired immune response in the peritumoral milieu, thereby creating a more conducive environment for tumor cell growth and proliferation. Therefore, elucidating the precise role of FHOD1 in this process is important for the development of new tumor therapeutic strategies.
3.3 FHOD1 overexpression in tumors stimulates STAT3 activation
The STAT protein family, comprising STAT1 to STAT6, is an important group of transcription factors that play a key role in cell signaling (61–63). Proliferation, differentiation, apoptosis, and inflammation are important biological mechanisms by which STAT proteins regulate cells (64–66). Among the STAT protein family, STAT1, STAT3, and STAT5 are considered to have important roles in cancer cells (67, 68). Among them, STAT1 is involved in anti-tumor immune response, while STAT3 and STAT5 exhibit pro-tumorigenic properties, and their overexpression is closely associated with tumor progression and malignancy (66, 69–71). STAT3 is a key oncogenic transcription factor that is constitutively activated in tumor cells and immune TMEs, serving as a key signaling hub integrating multiple oncogenic signaling pathways (72–74). Abnormally activated STAT3 inhibits apoptosis, induces cell proliferation (75, 76), upregulates matrix metalloproteinase expression, increase matrix stiffness (77), and promotes EMT (75), with the proinflammatory cytokine interleukin-6 (IL-6) being one of the main culprits. It can drive many cancer “hallmarks” by activating the JAK/STAT3 signaling pathway (78). The IL-6/JAK/STAT3 signaling pathway constitutes a self-sustaining regulatory circuit that plays a key role in both cancer development and progression. This molecular cascade can be triggered by chronic inflammation, which is widely recognized as an important risk factor for tumorigenesis (79–83). IL-6 triggers the activation of CAF (84), and activated CAF up-regulates the expression of markers such as α-smooth muscle actin (α-SMA), fibroblast activation protein, platelet-derived growth factor-β, and N-cadherin (85). In addition, activation of FHOD1 protein has been found to be closely associated with the initiation of the STAT3 signaling pathway (86, 87), a process that has attracted much attention in oncology research. Upon activation, FHOD1 induces STAT3 phosphorylation, initiating a downstream signaling cascade that ultimately leads to a significant upregulation of IL-6 expression. As a key pro-inflammatory cytokine in the tumor microenvironment, IL-6 plays a dual role in promoting CAF activation and creating a favorable environment for tumor cell proliferation and metastasis (85).The activation of CAF is a critical step in the process of tumourigenesis and progression. These activated fibroblasts are capable of secreting a variety of growth factors and extracellular matrix proteins, thus providing the necessary support for tumor cells. Mechanistically, FHOD1-mediated STAT3 activation induces IL-6 production, which subsequently triggers CAF activation (88, 89). This cascade of molecular events establishes a microenvironment that promotes tumorigenesis and progression (90–92).
In addition, the STAT3/EMT axis mediates the invasion of a variety of tumors, including colorectal, lung, breast, and brain tumors (93–95). ZEB1/2 protein, TGF-β, Snail, and other EMT regulators are affected by STAT3 signaling (96–98). It has been reported that Snail-induced up-regulation of CSF1R triggers STAT3 signaling while suppressing the expression of miRNA-34a, which promotes the induction of EMT in colorectal carcinogenesis (99). UBE2S up-regulates the expression level of HIF-1α, which stimulates STAT3 signaling, leading to Snail and Twist1 overexpression and inducing EMT (100). we speculate that FHOD1 could trigger a series of downstream events by activating the STAT3 signaling pathway, which in turn promotes cancer cell invasion and metastasis. Specifically,when the STAT3 signaling pathway is activated, a series of gene expression changes are triggered, two transcription factors known to inhibit the expression of epithelial cell markers while promoting the expression of mesenchymal cell markers, which in turn promotes the EMT process (101–103) (Figure 1). In addition, the expression of TGF-β and Snail proteins, crucial regulators of the EMT process, was significantly upregulated, thereby greatly enhancing cancer cell migration (79). These findings highlight the role of FHOD1 in driving colorectal cancer progression and metastasis, establishing its key molecular role in tumor progression.
3.4 Interaction between FHOD1 and CD21 in human B cells
CD21 is a multifunctional cell surface glycoprotein that is highly expressed in B lymphocytes and follicular dendritic cells (FDCs) (104). However, it is also present in a variety of other cell types.CD21 is a receptor for complement C3dg fragments (105), for CD23 (106), and itself. CD21 consists of an extracellular region consisting of 15–16 short consensus repeat units, a hydrophobic transmembrane region, and an intracellular region of 34 amino acids (107, 108). These three known ligands bind to two N-terminal repeats (106, 107), and crystal structure analysis suggests that they form a highly flexible domain (109). The role of human CD21 in the immune response has been extensively studied and is now well characterized (110, 111). As a key component of the B-cell co-receptor complex, CD21 plays a crucial role in regulating antibody production through multiple mechanisms. These include modulation of B-cell receptor (BCR) signaling through the immune complex (C3d-Ab-Ag), promotion of Fc receptor signaling, and enhancement of B cell memory through the retention of pathogens (including HIV) on FDCs (107, 112–115). The interaction of CD21 and CD23 on B cells is thought to protect B cells from apoptosis (116). However, the function of CD21 on most other cell types remains unclear (117).
The interaction of FHOD1 with CD21 in mammalian cells is realized through the C-terminus, a region containing a dad-like structural domain that binds to its own N-terminus through an intramolecular inhibitory interaction. The binding of phosphorylated Rho GTPase to the n-terminal GTPase-binding domain uncouples this interaction. In colorectal cancer cells, stimulation of CD21 triggers the generation of mechanical forces by Rho GTPase and Rac1, which recruit FHOD1 to the vicinity of CD21. This recruitment facilitates colorectal carcinogenesis and tumor progression. Furthermore, FHOD1 plays a pivotal role in CD21-associated cytoskeletal reorganization and the regulation of intracellular signaling pathways (Figure 3) (117).
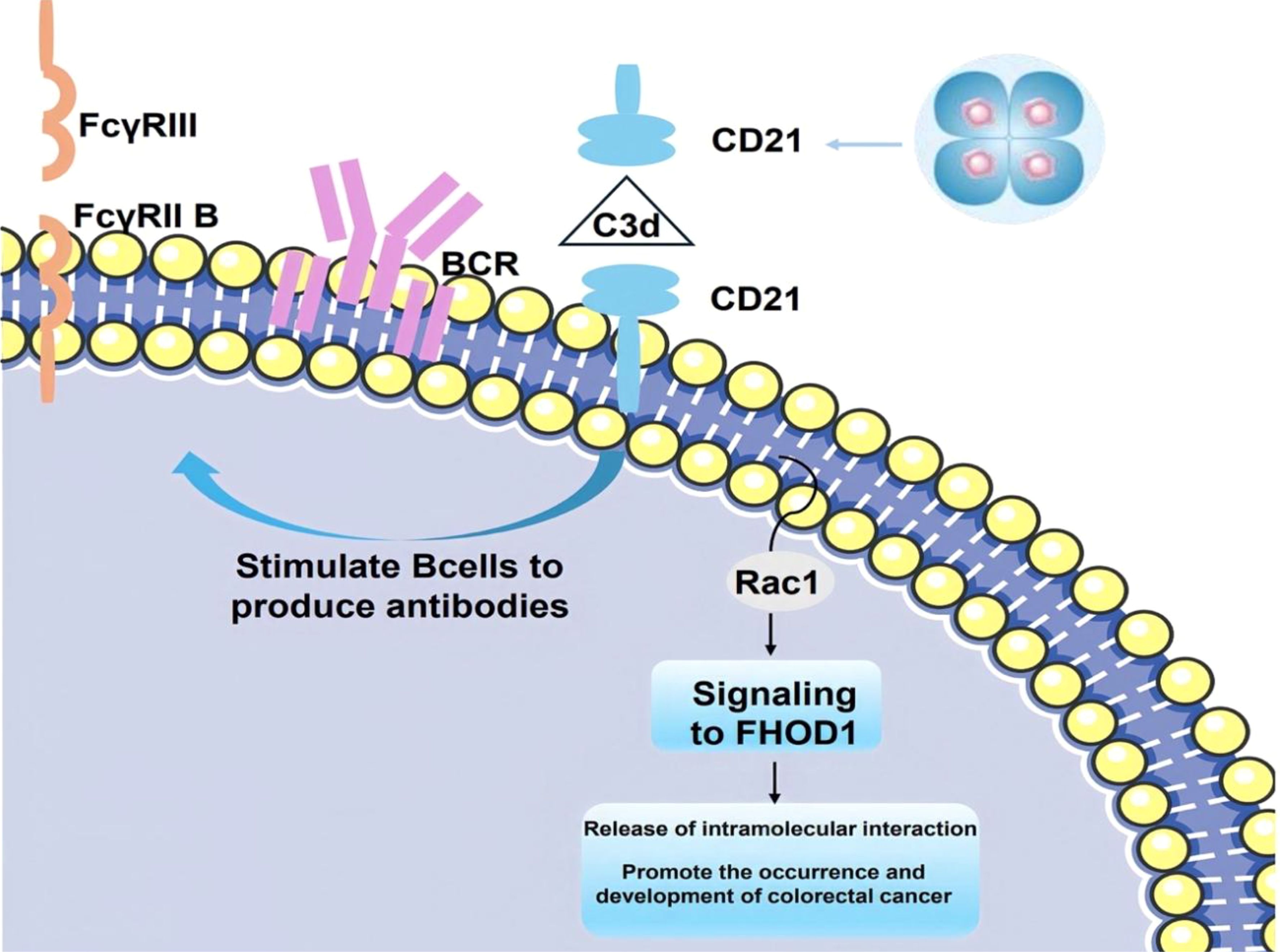
Figure 3. The covalent attachment of activated C3 d to antigen targets the complex to follicular dendritic cells. CD21 stimulates B cells to participate in the regulation of antibody production through BCR signaling mediated by immune complexes (C3d-Ab-Ag) (118). When CD21 is stimulated, the mechanical force generated by Rac1 attracts FHOD1 to the vicinity of CD21, thereby facilitating promoting the development and progression of colorectal cancer.
4 Conclusion
Forins represent a highly conserved family of actin regulatory proteins that play crucial roles in various tumor cell functions (119), such as cell morphogenesis, cell division, and cell polarity (120). In addition, FHOD1 is one of the most highly expressed human forints detected in a variety of tumor cell lines and tissues, underscoring its fundamental importance in cytoskeletal organization and associated cellular processes (12), FHOD1 was also shown to be able to influence immunochemokines within tumor cells through signaling pathways. Immunochemokines are signaling molecules that attract immune cells to specific locations, and they play a key role in immune surveillance and tumor immune escape. The regulatory function of FHOD1 in modulating the accumulation of these immunochemokines may significantly impair the immune system’s capacity to recognize and eliminate tumor cells, consequently fostering an immunosuppressive tumor microenvironment.This phenomenon of immunosuppression is a strategy for tumor cells to escape the surveillance of the host immune system, allowing them to survive and spread in vivo.
In this exhaustive review, we elucidate the pivotal role of FHOD1 protein in tumor biology, with a specific focus on its regulatory mechanism in upregulating PDL1 expression through the EMT pathway (121–123). Our analysis not only reveals the potential mechanism of FHOD1 in tumor cell proliferation and invasion but also demonstrates its importance in tumor microenvironment. Researchers have found that the upregulation of FHOD1 expression is closely related to the invasiveness of tumor cells, which may provide a new target for tumor therapy.
Furthermore, the upregulation of FHOD1 expression in tumor cells in tumor cells is not limited to the promotion of tumor cell proliferation and invasion, but also involves a complex network of signaling pathways that regulate the expression and function of immune molecules. For example, FHOD1 may play an important role in tumor immune escape by regulating certain key signaling molecules, particularly its effect on the expression of the immune checkpoint molecule PD-L1. Increasing evidence suggests that the aberrant expression of FHOD1 is associated with changes in the immune microenvironment of multiple tumor types, which provides new perspectives for understanding the mechanism of tumor immune escape.
In conclusion, the role of FHOD1 in tumorigenesis and development cannot be ignored, as it regulates tumor cell behavior and remodels the immune microenvironment through multiple mechanisms. These findings not only deepen our understanding of tumor biology, but also provide new perspectives and promising therapeutic targets for cancer treatment. With further research, we expect to gain a more comprehensive understanding of the function of FHOD1 in tumor biology, which will help develop innovative and effective therapeutic strategies (124–126).
Author contributions
YY: Conceptualization, Supervision, Writing – original draft. ZK: Writing – original draft. JC: Writing – original draft. SJ: Visualization, Writing – review & editing. SF: Supervision, Writing – review & editing. HZ: Funding acquisition, Visualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was supported by the Science and Technology Development Project of Henan Province (grant number 212102310621).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Bilotta MT, Antignani A, Fitzgerald DJ. Managing the TME to improve the efficacy of cancer therapy. Front Immunol. (2022) 13:954992. doi: 10.3389/fimmu.2022.954992
2. Jin MZ, Jin WL. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct Target Ther. (2020) 5:166. doi: 10.1038/s41392-020-00280-x
3. Sadeghi M, Dehnavi S, Sharifat M, Amiri AM, Khodadadi A. Innate immune cells: Key players of orchestra in modulating tumor microenvironment (TME). Heliyon. (2024) 10:e27480. doi: 10.1016/j.heliyon.2024.e27480
4. Peippo M, Gardberg M, Lamminen T, Kaipio K, Carpén O, Heuser VD. FHOD1 formin is upregulated in melanomas and modifies proliferation and tumor growth. Exp Cell Res. (2017) 350:267–78. doi: 10.1016/j.yexcr.2016.12.004
5. Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. (2005) 30:342–53. doi: 10.1016/j.tibs.2005.04.014
6. Faix J, Grosse R. Staying in shape with formins. Dev Cell. (2006) 10:693–706. doi: 10.1016/j.devcel.2006.05.001
7. Grikscheit K, Grosse R. Formins at the junction. Trends Biochem Sci. (2016) 41:148–59. doi: 10.1016/j.tibs.2015.12.002
8. Schönichen A, Mannherz HG, Behrmann E, Mazur AJ, Kühn S, Silván U, et al. FHOD1 is a combined actin filament capping and bundling factor that selectively associates with actin arcs and stress fibers. J Cell Sci. (2013) 126:1891–901. doi: 10.1242/jcs.126706
9. Westendorf JJ, Koka S. Identification of FHOD1-binding proteins and mechanisms of FHOD1-regulated actin dynamics. J Cell Biochem. (2004) 92:29–41. doi: 10.1002/jcb.20031
10. Heuser VD, Mansuri N, Mogg J, Kurki S, Repo H, Kronqvist P, et al. Formin proteins FHOD1 and INF2 in triple-negative breast cancer: association with basal markers and functional activities. Breast Cancer (Auckl). (2018) 12:1178223418792247. doi: 10.1177/1178223418792247
11. Heuser VD, Kiviniemi A, Lehtinen L, Munthe S, Kristensen BW, Posti JP, et al. Multiple formin proteins participate in glioblastoma migration. BMC Cancer. (2020) 20:710. doi: 10.1186/s12885-020-07211-7
12. Schulze N, Graessl M, Blancke Soares A, Geyer M, Dehmelt L, Nalbant P. FHOD1 regulates stress fiber organization by controlling the dynamics of transverse arcs and dorsal fibers. J Cell Sci. (2014) 127:1379–93. doi: 10.1242/jcs.134627
13. Gardberg M, Kaipio K, Lehtinen L, Mikkonen P, Heuser VD, Talvinen K, et al. FHOD1, a formin upregulated in epithelial-mesenchymal transition, participates in cancer cell migration and invasion. PloS One. (2013) 8:e74923. doi: 10.1371/journal.pone.0074923
14. Zhang F, Wu L, Feng S, Zhao Z, Zhang K, Thakur A, et al. FHOD1 is upregulated in glioma cells and attenuates ferroptosis of glioma cells by targeting HSPB1 signaling. CNS Neurosci Ther. (2023) 29:3351–63. doi: 10.1111/cns.14264
15. Jurmeister S, Baumann M, Balwierz A, Keklikoglou I, Ward A, Uhlmann S, et al. MicroRNA-200c represses migration and invasion of breast cancer cells by targeting actin-regulatory proteins FHOD1 and PPM1F. Mol Cell Biol. (2012) 32:633–51. doi: 10.1128/mcb.06212-11
16. LeBleu VS, Thiery JP. The continuing search for causality between epithelial-to-mesenchymal transition and the metastatic fitness of carcinoma cells. Cancer Res. (2022) 82:1467–9. doi: 10.1158/0008-5472.Can-22-0026
17. Koka S, Neudauer CL, Li X, Lewis RE, McCarthy JB, Westendorf JJ. The formin-homology-domain-containing protein FHOD1 enhances cell migration. J Cell Sci. (2003) 116:1745–55. doi: 10.1242/jcs.00386
18. Fedele M, Sgarra R, Battista S, Cerchia L, Manfioletti G. The epithelial-mesenchymal transition at the crossroads between metabolism and tumor progression. Int J Mol Sci. (2022) 23:800. doi: 10.3390/ijms23020800
19. Zhang N, Ng AS, Cai S, Li Q, Yang L, Kerr D. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol. (2021) 22:e358–68. doi: 10.1016/s1470-2045(21)00343-0
20. Menju T, Date H. Lung cancer and epithelial-mesenchymal transition. Gen Thorac Cardiovasc Surg. (2021) 69:781–9. doi: 10.1007/s11748-021-01595-4
21. Takkunen M, Grenman R, Hukkanen M, Korhonen M, Garcia de Herreros A, Virtanen I. Snail-dependent and -independent epithelial-mesenchymal transition in oral squamous carcinoma cells. J Histochem Cytochem. (2006) 54:1263–75. doi: 10.1369/jhc.6A6958.2006
22. Antonangeli F, Natalini A, Garassino MC, Sica A, Santoni A, Di Rosa F. Regulation of PD-L1 expression by NF-κB in cancer. Front Immunol. (2020) 11:584626. doi: 10.3389/fimmu.2020.584626
23. Zhang H, Qin G, Zhang C, Yang H, Liu J, Hu H, et al. TRAIL promotes epithelial-to-mesenchymal transition by inducing PD-L1 expression in esophageal squamous cell carcinomas. J Exp Clin Cancer Res. (2021) 40:209. doi: 10.1186/s13046-021-01972-0
24. Li H, Wang Z, Liang S, Liu Q, Wang P, Cai L, et al. Radiation induces epithelial to mesenchymal transition via upregulation of PD-L1 in nasopharyngeal carcinoma cell. Transl Cancer Res. (2021) 10:372–81. doi: 10.21037/tcr-20-1899
25. Wudtiwai B, Kodchakorn K, Shwe TH, Pothacharoen P, Phitak T, Suninthaboonrana R, et al. Brazilein inhibits epithelial-mesenchymal transition (EMT) and programmed death ligand 1 (PD-L1) expression in breast cancer cells. Int Immunopharmacol. (2023) 118:109988. doi: 10.1016/j.intimp.2023.109988
26. Kudo-Saito C, Shirako H, Ohike M, Tsukamoto N, Kawakami Y. CCL2 is critical for immunosuppression to promote cancer metastasis. Clin Exp Metastasis. (2013) 30:393–405. doi: 10.1007/s10585-012-9545-6
27. Zhang YC, Zhang YT, Wang Y, Zhao Y, He LJ. What role does PDL1 play in EMT changes in tumors and fibrosis? Front Immunol. (2023) 14:1226038. doi: 10.3389/fimmu.2023.1226038
28. Wang YL, Gong Y, Lv Z, Li L, Yuan Y. Expression of PD1/PDL1 in gastric cancer at different microsatellite status and its correlation with infiltrating immune cells in the tumor microenvironment. J Cancer. (2021) 12:1698–707. doi: 10.7150/jca.40500
29. Mu Q, Najafi M. Modulation of the tumor microenvironment (TME) by melatonin. Eur J Pharmacol. (2021) 907:174365. doi: 10.1016/j.ejphar.2021.174365
30. Wang M, Zhao J, Zhang L, Wei F, Lian Y, Wu Y, et al. Role of tumor microenvironment in tumorigenesis. J Cancer. (2017) 8:761–73. doi: 10.7150/jca.17648
31. Al Zein M, Boukhdoud M, Shammaa H, Mouslem H, El Ayoubi LM, Iratni R, et al. Immunotherapy and immunoevasion of colorectal cancer. Drug Discov Today. (2023) 28:103669. doi: 10.1016/j.drudis.2023.103669
32. Qayoom H, Sofi S, Mir MA. Targeting tumor microenvironment using tumor-infiltrating lymphocytes as therapeutics against tumorigenesis. Immunol Res. (2023) 71:588–99. doi: 10.1007/s12026-023-09376-2
33. Hua S, Gu X, Jin H, Zhang X, Liu Q, Yang J. Tumor-infiltrating T lymphocytes: A promising immunotherapeutic target for preventing immune escape in cholangiocarcinoma. BioMed Pharmacother. (2024) 177:117080. doi: 10.1016/j.biopha.2024.117080
34. Imazu Y, Matsuo Y, Hokuto D, Yasuda S, Yoshikawa T, Kamitani N, et al. Distinct role of tumor-infiltrating lymphocytes between synchronous and metachronous colorectal cancer. Langenbecks Arch Surg. (2023) 408:72. doi: 10.1007/s00423-023-02815-6
35. Farhood B, Najafi M, Mortezaee K. CD8(+) cytotoxic T lymphocytes in cancer immunotherapy: A review. J Cell Physiol. (2019) 234:8509–21. doi: 10.1002/jcp.27782
36. Huang Y, Jia A, Wang Y, Liu G. CD8(+) T cell exhaustion in anti-tumour immunity: The new insights for cancer immunotherapy. Immunology. (2023) 168:30–48. doi: 10.1111/imm.13588
37. Kumagai S, Koyama S, Itahashi K, Tanegashima T, Lin YT, Togashi Y, et al. Lactic acid promotes PD-1 expression in regulatory T cells in highly glycolytic tumor microenvironments. Cancer Cell. (2022) 40:201–218 e209. doi: 10.1016/j.ccell.2022.01.001
38. Faas MM, de Vos P. Mitochondrial function in immune cells in health and disease. Biochim Biophys Acta Mol Basis Dis. (2020) 1866:165845. doi: 10.1016/j.bbadis.2020.165845
39. Mansuri N, Heuser VD, Birkman EM, Lintunen M, Algars A, Sundstrom J, et al. FHOD1 and FMNL1 formin proteins in intestinal gastric cancer: correlation with tumor-infiltrating T lymphocytes and molecular subtypes. Gastric Cancer. (2021) 24:1254–63. doi: 10.1007/s10120-021-01203-7
40. Favaro P, Traina F, MaChado-Neto JA, Lazarini M, Lopes MR, Pereira JK, et al. FMNL1 promotes proliferation and migration of leukemia cells. J Leukoc Biol. (2013) 94:503–12. doi: 10.1189/jlb.0113057
41. Favaro PM, de Souza Medina S, Traina F, Basseres DS, Costa FF, Saad ST. Human leukocyte formin: a novel protein expressed in lymphoid Malignancies and associated with Akt. Biochem Biophys Res Commun. (2003) 311:365–71. doi: 10.1016/j.bbrc.2003.10.012
42. Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat BioMed Eng. (2019) 3:306–17. doi: 10.1038/s41551-019-0375-6
43. Yao H, Lan J, Li C, Shi H, Brosseau JP, Wang H, et al. Author Correction: Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat BioMed Eng. (2019) 3:414. doi: 10.1038/s41551-019-0402-7
44. Wu J, Wang N. Current progress of anti−PD−1/PDL1 immunotherapy for glioblastoma (Review). Mol Med Rep. (2024) 30:221. doi: 10.3892/mmr.2024.13344
45. Aydın EM, Demir TD, Seymen N, Said SS, Oktem-Okullu S, Tiftikci A, et al. The crosstalk between H. pylori virulence factors and the PD1:PD-L1 immune checkpoint inhibitors in progression to gastric cancer. Immunol Lett. (2021) 239:1–11. doi: 10.1016/j.imlet.2021.06.009
46. Chen QY, Chen YX, Han QY, Zhang JG, Zhou WJ, Zhang X, et al. Prognostic significance of immune checkpoints HLA-G/ILT-2/4 and PD-L1 in colorectal cancer. Front Immunol. (2021) 12:679090. doi: 10.3389/fimmu.2021.679090
47. Kennel KB, Bozlar M, De Valk AF, Greten FR. Cancer-associated fibroblasts in inflammation and antitumor immunity. Clin Cancer Res. (2023) 29:1009–16. doi: 10.1158/1078-0432.Ccr-22-1031
48. Zhang H, Yue X, Chen Z, Liu C, Wu W, Zhang N, et al. Define cancer-associated fibroblasts (CAFs) in the tumor microenvironment: new opportunities in cancer immunotherapy and advances in clinical trials. Mol Cancer. (2023) 22:159. doi: 10.1186/s12943-023-01860-5
49. Liao Z, Tan ZW, Zhu P, Tan NS. Cancer-associated fibroblasts in tumor microenvironment - Accomplices in tumor Malignancy. Cell Immunol. (2019) 343:103729. doi: 10.1016/j.cellimm.2017.12.003
50. Raudenska M, Balvan J, Hanelova K, Bugajova M, Masarik M. Cancer-associated fibroblasts: Mediators of head and neck tumor microenvironment remodeling. Biochim Biophys Acta Rev Cancer. (2023) 1878:188940. doi: 10.1016/j.bbcan.2023.188940
51. Whittle MC, Hingorani SR. Fibroblasts in pancreatic ductal adenocarcinoma: biological mechanisms and therapeutic targets. Gastroenterology. (2019) 156:2085–96. doi: 10.1053/j.gastro.2018.12.044
52. Sahai E, Astsaturov I, Cukierman E, DeNardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. (2020) 20:174–86. doi: 10.1038/s41568-019-0238-1
53. Ge W, Yue M, Lin R, Zhou T, Xu H, Wang Y, et al. PLA2G2A(+) cancer-associated fibroblasts mediate pancreatic cancer immune escape via impeding antitumor immune response of CD8(+) cytotoxic T cells. Cancer Lett. (2023) 558:216095. doi: 10.1016/j.canlet.2023.216095
54. Kimmelman AC, White E. Autophagy and tumor metabolism. Cell Metab. (2017) 25:1037–43. doi: 10.1016/j.cmet.2017.04.004
55. Amaravadi RK, Kimmelman AC, Debnath J. Targeting autophagy in cancer: recent advances and future directions. Cancer Discov. (2019) 9:1167–81. doi: 10.1158/2159-8290.Cd-19-0292
56. Zhang X, Lao M, Yang H, Sun K, Dong Y, He L, et al. Targeting cancer-associated fibroblast autophagy renders pancreatic cancer eradicable with immunochemotherapy by inhibiting adaptive immune resistance. Autophagy. (2024) 20:1314–34. doi: 10.1080/15548627.2023.2300913
57. Endo S, Nakata K, Ohuchida K, Takesue S, Nakayama H, Abe T, et al. Autophagy is required for activation of pancreatic stellate cells, associated with pancreatic cancer progression and promotes growth of pancreatic tumors in mice. Gastroenterology. (2017) 152:1492–1506.e1424. doi: 10.1053/j.gastro.2017.01.010
58. Zhang Z, Yu Y, Zhang Z, Li D, Liang Z, Wang L, et al. Cancer-associated fibroblasts-derived CXCL12 enhances immune escape of bladder cancer through inhibiting P62-mediated autophagic degradation of PDL1. J Exp Clin Cancer Res. (2023) 42:316. doi: 10.1186/s13046-023-02900-0
59. Gao D, Fang L, Liu C, Yang M, Yu X, Wang L, et al. Microenvironmental regulation in tumor progression: Interactions between cancer-associated fibroblasts and immune cells. BioMed Pharmacother. (2023) 167:115622. doi: 10.1016/j.biopha.2023.115622
60. Chen D, Zhang X, Li Z, Zhu B. Metabolic regulatory crosstalk between tumor microenvironment and tumor-associated macrophages. Theranostics. (2021) 11:1016–30. doi: 10.7150/thno.51777
61. Pham TH, Park HM, Kim J, Hong JT, Yoon DY. STAT3 and p53: dual target for cancer therapy. Biomedicines. (2020) 8:637. doi: 10.3390/biomedicines8120637
62. Arora L, Kumar AP, Arfuso F, Chng WJ, Sethi G. The role of signal transducer and activator of transcription 3 (STAT3) and its targeted inhibition in hematological Malignancies. Cancers (Basel). (2018) 10:327. doi: 10.3390/cancers10090327
63. Garg M, Shanmugam MK, Bhardwaj V, Goel A, Gupta R, Sharma A, et al. The pleiotropic role of transcription factor STAT3 in oncogenesis and its targeting through natural products for cancer prevention and therapy. Med Res Rev. (2020) 2020:1–46. doi: 10.1002/med.21761
64. Chun KS, Jang JH, Kim DH. Perspectives regarding the intersections between STAT3 and oxidative metabolism in cancer. Cells. (2020) 9:2202. doi: 10.3390/cells9102202
65. Lee JH, Kim C, Sethi G, Ahn KS. Brassinin inhibits STAT3 signaling pathway through modulation of PIAS-3 and SOCS-3 expression and sensitizes human lung cancer xenograft in nude mice to paclitaxel. Oncotarget. (2015) 6:6386–405. doi: 10.18632/oncotarget.3443
66. Kim C, Lee SG, Yang WM, Arfuso F, Um JY, Kumar AP, et al. Formononetin-induced oxidative stress abrogates the activation of STAT3/5 signaling axis and suppresses the tumor growth in multiple myeloma preclinical model. Cancer Lett. (2018) 431:123–41. doi: 10.1016/j.canlet.2018.05.038
67. Lee JH, Chiang SY, Nam D, Chung WS, Lee J, Na YS, et al. Capillarisin inhibits constitutive and inducible STAT3 activation through induction of SHP-1 and SHP-2 tyrosine phosphatases. Cancer Lett. (2014) 345:140–8. doi: 10.1016/j.canlet.2013.12.008
68. Wong GL, Manore SG, Doheny DL, Lo HW. STAT family of transcription factors in breast cancer: Pathogenesis and therapeutic opportunities and challenges. Semin Cancer Biol. (2022) 86:84–106. doi: 10.1016/j.semcancer.2022.08.003
69. Garces de Los Fayos Alonso I, Zujo L, Wiest I, Kodajova P, Timelthaler G, Edtmayer S, et al. PDGFRβ promotes oncogenic progression via STAT3/STAT5 hyperactivation in anaplastic large cell lymphoma. Mol Cancer. (2022) 21:172. doi: 10.1186/s12943-022-01640-7
70. Fasouli ES, Katsantoni E. Age-associated myeloid Malignancies - the role of STAT3 and STAT5 in myelodysplastic syndrome and acute myeloid leukemia. FEBS Lett. (2024) 598:2809–28. doi: 10.1002/1873-3468.14985
71. Guru SA, Sumi MP, Mir R, Waza AA, Bhat MA, Zuberi M, et al. Ectopic PD-L1 expression in JAK2 (V617F) myeloproliferative neoplasm patients is mediated via increased activation of STAT3 and STAT5. Hum Cell. (2020) 33:1099–111. doi: 10.1007/s13577-020-00370-6
72. Avalle L, Camporeale A, Camperi A, Poli V. STAT3 in cancer: A double edged sword. Cytokine. (2017) 98:42–50. doi: 10.1016/j.cyto.2017.03.018
73. Dong J, Cheng XD, Zhang WD, Qin JJ. Recent update on development of small-molecule STAT3 inhibitors for cancer therapy: from phosphorylation inhibition to protein degradation. J Med Chem. (2021) 64:8884–915. doi: 10.1021/acs.jmedchem.1c00629
74. Mohan CD, Rangappa S, Preetham HD, Chandra Nayaka S, Gupta VK, Basappa S, et al. Targeting STAT3 signaling pathway in cancer by agents derived from Mother Nature. Semin Cancer Biol. (2022) 80:157–82. doi: 10.1016/j.semcancer.2020.03.016
75. Yu H, Pardoll D, Jove R. STATs in cancer inflammation and immunity: a leading role for STAT3. Nat Rev Cancer. (2009) 9:798–809. doi: 10.1038/nrc2734
76. Demaria M, Misale S, Giorgi C, Miano V, Camporeale A, Campisi J, et al. STAT3 can serve as a hit in the process of Malignant transformation of primary cells. Cell Death Differ. (2012) 19:1390–7. doi: 10.1038/cdd.2012.20
77. Laklai H, Miroshnikova YA, Pickup MW, Collisson EA, Kim GE, Barrett AS, et al. Author Correction: Genotype tunes pancreatic ductal adenocarcinoma tissue tension to induce matricellular fibrosis and tumor progression. Nat Med. (2024) 30:908. doi: 10.1038/s41591-023-02694-w
78. Wang SW, Sun YM. The IL-6/JAK/STAT3 pathway: potential therapeutic strategies in treating colorectal cancer (Review). Int J Oncol. (2014) 44:1032–40. doi: 10.3892/ijo.2014.2259
79. Sadrkhanloo M, Entezari M, Orouei S, Ghollasi M, Fathi N, Rezaei S, et al. STAT3-EMT axis in tumors: Modulation of cancer metastasis, stemness and therapy response. Pharmacol Res. (2022) 182:106311. doi: 10.1016/j.phrs.2022.106311
80. Hu Z, Sui Q, Jin X, Shan G, Huang Y, Yi Y, et al. IL6-STAT3-C/EBPβ-IL6 positive feedback loop in tumor-associated macrophages promotes the EMT and metastasis of lung adenocarcinoma. J Exp Clin Cancer Res. (2024) 43:63. doi: 10.1186/s13046-024-02989-x
81. Chuangchot N, Jamjuntra P, Yangngam S, Luangwattananun P, Thongchot S, Junking M, et al. Enhancement of PD-L1-attenuated CAR-T cell function through breast cancer-associated fibroblasts-derived IL-6 signaling via STAT3/AKT pathways. Breast Cancer Res. (2023) 25:86. doi: 10.1186/s13058-023-01684-7
82. Eskiler GG, Bezdegumeli E, Ozman Z, Ozkan AD, Bilir C, Kucukakca BN, et al. IL-6 mediated JAK/STAT3 signaling pathway in cancer patients with cachexia. Bratisl Lek Listy. (2019) 66:819–26. doi: 10.4149/bll_2019_136
83. Rapp J, Jung M, Klar RFU, Wolf J, Arnold J, Gorka O, et al. STAT3 signaling induced by the IL-6 family of cytokines modulates angiogenesis. J Cell Sci. (2023) 136:jcs260182. doi: 10.1242/jcs.260182
84. Heichler C, Scheibe K, Schmied A, Geppert CI, Schmid B, Wirtz S, et al. STAT3 activation through IL-6/IL-11 in cancer-associated fibroblasts promotes colorectal tumour development and correlates with poor prognosis. Gut. (2020) 69:1269–82. doi: 10.1136/gutjnl-2019-319200
85. Avalle L, Raggi L, Monteleone E, Savino A, Viavattene D, Statello L, et al. STAT3 induces breast cancer growth via ANGPTL4, MMP13 and STC1 secretion by cancer associated fibroblasts. Oncogene. (2022) 41:1456–67. doi: 10.1038/s41388-021-02172-y
86. Yang J, Wang L, Guan X, Qin JJ. Inhibiting STAT3 signaling pathway by natural products for cancer prevention and therapy: In vitro and in vivo activity and mechanisms of action. Pharmacol Res. (2022) 182:106357. doi: 10.1016/j.phrs.2022.106357
87. Li Q, Yang F, Shi X, Bian S, Shen F, Wu Y, et al. MTHFD2 promotes ovarian cancer growth and metastasis via activation of the STAT3 signaling pathway. FEBS Open Bio. (2021) 11:2845–57. doi: 10.1002/2211-5463.13249
88. Guo Z, Zhang H, Fu Y, Kuang J, Zhao B, Zhang L, et al. Cancer-associated fibroblasts induce growth and radioresistance of breast cancer cells through paracrine IL-6. Cell Death Discov. (2023) 9:6. doi: 10.1038/s41420-023-01306-3
89. Jia C, Wang G, Wang T, Fu B, Zhang Y, Huang L, et al. Cancer-associated Fibroblasts induce epithelial-mesenchymal transition via the Transglutaminase 2-dependent IL-6/IL6R/STAT3 axis in Hepatocellular Carcinoma. Int J Biol Sci. (2020) 16:2542–58. doi: 10.7150/ijbs.45446
90. Albrengues J, Bertero T, Grasset E, Bonan S, Maiel M, Bourget I, et al. Epigenetic switch drives the conversion of fibroblasts into proinvasive cancer-associated fibroblasts. Nat Commun. (2015) 6:10204. doi: 10.1038/ncomms10204
91. Hendrayani SF, Al-Khalaf HH, Aboussekhra A. The cytokine IL-6 reactivates breast stromal fibroblasts through transcription factor STAT3-dependent up-regulation of the RNA-binding protein AUF1. J Biol Chem. (2014) 289:30962–76. doi: 10.1074/jbc.M114.594044
92. Zheng X, Xu M, Yao B, Wang C, Jia Y, Liu Q. IL-6/STAT3 axis initiated CAFs via up-regulating TIMP-1 which was attenuated by acetylation of STAT3 induced by PCAF in HCC microenvironment. Cell Signal. (2016) 28:1314–24. doi: 10.1016/j.cellsig.2016.06.009
93. Li Q, Zhang H, He Y, Zhang H, Han C. Inhibition of Colorectal Cancer Metastasis by Total Flavones of Abelmoschus manihot via LncRNA AL137782-mediated STAT3/EMT Pathway Regulation. Curr Pharm Des. (2025) 31:219–32. doi: 10.2174/0113816128298998240828060306
94. Park SJ, Jung HJ. Bufotalin suppresses proliferation and metastasis of triple-negative breast cancer cells by promoting apoptosis and inhibiting the STAT3/EMT axis. Molecules. (2023) 28:6783. doi: 10.3390/molecules28196783
95. Wu S, Lu J, Zhu H, Wu F, Mo Y, Xie L, et al. A novel axis of circKIF4A-miR-637-STAT3 promotes brain metastasis in triple-negative breast cancer. Cancer Lett. (2024) 581:216508. doi: 10.1016/j.canlet.2023.216508
96. Yuan W, Wei F, Ouyang H, Ren X, Hang J, Mo X, et al. CMTM3 suppresses chordoma progress through EGFR/STAT3 regulated EMT and TP53 signaling pathway. Cancer Cell Int. (2021) 21:510. doi: 10.1186/s12935-021-02159-5
97. Zhao C, Hu X, Tong S, Mo M, He W, Wang L, et al. MEST promotes bladder cancer cell proliferation, migration and invasion via STAT3/Twist-1-mediated EMT. Transl Cancer Res. (2020) 9:6178–88. doi: 10.21037/tcr-20-1006
98. Peng N, Lu M, Kang M, Liu X, Li B, Dong C. Recombinant human IL-11 promotes lung adenocarcinoma A549 cell growth and EMT through activating STAT3/HIF-1α/EMT signaling pathway. Anticancer Agents Med Chem. (2021) 21:1996–2003. doi: 10.2174/1871520621666201207091248
99. Shi X, Kaller M, Rokavec M, Kirchner T, Horst D, Hermeking H. Characterization of a p53/miR-34a/CSF1R/STAT3 feedback loop in colorectal cancer. Cell Mol Gastroenterol Hepatol. (2020) 10:391–418. doi: 10.1016/j.jcmgh.2020.04.002
100. Wang L, Liang Y, Li P, Liang Q, Sun H, Xu D, et al. Oncogenic activities of UBE2S mediated by VHL/HIF-1α/STAT3 signal via the ubiquitin-proteasome system in PDAC. Onco Targets Ther. (2019) 12:9767–81. doi: 10.2147/ott.S228522
101. Sun Z, Shao B, Liu Z, Dang Q, Guo Y, Chen C, et al. LINC01296/miR-141-3p/ZEB1-ZEB2 axis promotes tumor metastasis via enhancing epithelial-mesenchymal transition process. J Cancer. (2021) 12:2723–34. doi: 10.7150/jca.55626
102. Wang J, Farkas C, Benyoucef A, Carmichael C, Haigh K, Wong N, et al. Interplay between the EMT transcription factors ZEB1 and ZEB2 regulates hematopoietic stem and progenitor cell differentiation and hematopoietic lineage fidelity. PloS Biol. (2021) 19:e3001394. doi: 10.1371/journal.pbio.3001394
103. Li T, Tang C, Huang Z, Yang L, Dai H, Tang B, et al. miR-144-3p inhibited the growth, metastasis and epithelial-mesenchymal transition of colorectal adenocarcinoma by targeting ZEB1/2. Aging (Albany NY). (2021) 13:17349–69. doi: 10.18632/aging.203225
104. Li D, Xu Q, Hu Y, Wang W, Xie S, Zhao C, et al. Tumor-intrinsic CD21 expression impacts the response of B-cell Malignancy cells to CD19-CAR-T cells. J Leukoc Biol. (2022) 112:913–8. doi: 10.1002/jlb.5ma0122-474r
105. Iida K, Nadler L, Nussenzweig V. Identification of the membrane receptor for the complement fragment C3d by means of a monoclonal antibody. J Exp Med. (1983) 158:1021–33. doi: 10.1084/jem.158.4.1021
106. Aubry JP, Pochon S, Gauchat JF, Nueda-Marin A, Holers VM, Graber P, et al. CD23 interacts with a new functional extracytoplasmic domain involving N-linked oligosaccharides on CD21. J Immunol. (1994) 152:5806–13. doi: 10.4049/jimmunol.152.12.5806
107. Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. (2000) 18:393–422. doi: 10.1146/annurev.immunol.18.1.393
108. Moore MD, Cooper NR, Tack BF, Nemerow GR. Molecular cloning of the cDNA encoding the Epstein-Barr virus/C3d receptor (complement receptor type 2) of human B lymphocytes. Proc Natl Acad Sci U.S.A. (1987) 84:9194–8. doi: 10.1073/pnas.84.24.9194
109. Prota AE, Sage DR, Stehle T, Fingeroth JD. The crystal structure of human CD21: Implications for Epstein-Barr virus and C3d binding. Proc Natl Acad Sci U.S.A. (2002) 99:10641–6. doi: 10.1073/pnas.162360499
110. Bergman P, Wullimann D, Gao Y, Wahren Borgström E, Norlin AC, Lind Enoksson S, et al. Elevated CD21(low) B cell frequency is a marker of poor immunity to pfizer-bioNTech BNT162b2 mRNA vaccine against SARS-coV-2 in patients with common variable immunodeficiency. J Clin Immunol. (2022) 42:716–27. doi: 10.1007/s10875-022-01244-2
111. Freudenhammer M, Voll RE, Binder SC, Keller B, Warnatz K. Naive- and memory-like CD21(low) B cell subsets share core phenotypic and signaling characteristics in systemic autoimmune disorders. J Immunol. (2020) 205:2016–25. doi: 10.4049/jimmunol.2000343
112. Cherukuri A, Cheng PC, Sohn HW, Pierce SK. The CD19/CD21 complex functions to prolong B cell antigen receptor signaling from lipid rafts. Immunity. (2001) 14:169–79. doi: 10.1016/s1074-7613(01)00098-x
113. Moir S, Malaspina A, Li Y, Chun TW, Lowe T, Adelsberger J, et al. B cells of HIV-1-infected patients bind virions through CD21-complement interactions and transmit infectious virus to activated T cells. J Exp Med. (2000) 192:637–46. doi: 10.1084/jem.192.5.637
114. Poe JC, Hasegawa M, Tedder TF. CD19, CD21, and CD22: multifaceted response regulators of B lymphocyte signal transduction. Int Rev Immunol. (2001) 20:739–62. doi: 10.3109/08830180109045588
115. Prodinger WM. Complement receptor type two (CR2,CR21): a target for influencing the humoral immune response and antigen-trapping. Immunol Res. (1999) 20:187–94. doi: 10.1007/bf02790402
116. Bonnefoy JY, Henchoz S, Hardie D, Holder MJ, Gordon JA. subset of anti-CD21 antibodies promote the rescue of germinal center B cells from apoptosis. Eur J Immunol. (1993) 23:969–72. doi: 10.1002/eji.1830230432
117. Gill MB, Roecklein-Canfield J, Sage DR, Zambela-Soediono M, Longtine N, Uknis M, et al. EBV attachment stimulates FHOS/FHOD1 redistribution and co-aggregation with CD21: formin interactions with the cytoplasmic domain of human CD21. J Cell Sci. (2004) 117:2709–20. doi: 10.1242/jcs.01113
118. Carroll MC. CD21/CD35 in B cell activation. Semin Immunol. (1998) 10:279–86. doi: 10.1006/smim.1998.0120
119. Antoku S, Schwartz TU, Gundersen GG. FHODs: Nuclear tethered formins for nuclear mechanotransduction. Front Cell Dev Biol. (2023) 11:1160219. doi: 10.3389/fcell.2023.1160219
120. Shi X, Zhao S, Cai J, Wong G, Jiu Y. Active FHOD1 promotes the formation of functional actin stress fibers. Biochem J. (2019) 476:2953–63. doi: 10.1042/bcj20190535
121. Messeha SS, Zarmouh NO, Soliman KFA. Polyphenols modulating effects of PD-L1/PD-1 checkpoint and EMT-mediated PD-L1 overexpression in breast cancer. Nutrients. (2021) 13:1718. doi: 10.3390/nu13051718
122. Lems CM, Burger GA, Beltman JB. Tumor-mediated immunosuppression and cytokine spreading affects the relation between EMT and PD-L1 status. Front Immunol. (2023) 14:1219669. doi: 10.3389/fimmu.2023.1219669
123. Morelli MB, Amantini C, Rossi de Vermandois JA, Gubbiotti M, Giannantoni A, Mearini E, et al. Correlation between high PD-L1 and EMT/invasive genes expression and reduced recurrence-free survival in blood-circulating tumor cells from patients with non-muscle-invasive bladder cancer. Cancers (Basel). (2021) 13:5989. doi: 10.3390/cancers13235989
124. Jiang C, Yuan B, Hang B, Mao JH, Zou X, Wang P. FHOD1 is upregulated in gastric cancer and promotes the proliferation and invasion of gastric cancer cells. Oncol Lett. (2021) 22:712. doi: 10.3892/ol.2021.12973
125. Du M, Sun L, Guo J, Lv H. Macrophages and tumor-associated macrophages in the senescent microenvironment: From immunosuppressive TME to targeted tumor therapy. Pharmacol Res. (2024) 204:107198. doi: 10.1016/j.phrs.2024.107198
Keywords: FHOD1, tumor immune microenvironment, epithelial-mesenchymal transition, PDL1, PD-1
Citation: Yang Y, Kang Z, Cai J, Jia S, Fan S and Zhu H (2025) Role of FHOD1 in tumor cells and tumor immune microenvironment. Front. Immunol. 16:1514488. doi: 10.3389/fimmu.2025.1514488
Received: 21 October 2024; Accepted: 11 April 2025;
Published: 29 April 2025.
Edited by:
Youqiong Ye, Shanghai Jiao Tong University, ChinaReviewed by:
Dongyao Wang, University of Science and Technology of China, ChinaDmitry Aleksandrovich Zinovkin, Gomel State Medical University, Belarus
Copyright © 2025 Yang, Kang, Cai, Jia, Fan and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Huifang Zhu, emhmODM4MkAxNjMuY29t
 Yang Yang
Yang Yang Zhanting Kang
Zhanting Kang Huifang Zhu
Huifang Zhu