- 1Department of Emergency, the Affiliated Hospital of Qingdao University, Qingdao, Shandong, China
- 2Department of Neurosurgery, the Affiliated Hospital of Qingdao University, Qingdao, Shandong, China
Regulatory immune cells are pivotal in maintaining immune homeostasis and modulating immune responses to prevent pathologies. While T regulatory cells (Tregs) are extensively recognized for their immunosuppressive roles, emerging subsets of regulatory cells, including regulatory CD8+ cells (CD8+Tregs) regulatory B cells (Bregs), myeloid-derived suppressor cells (MDSCs), regulatory dendritic cells (DCregs), regulatory innate lymphoid cells (ILCregs), and regulatory natural killer cells (NKregs), are garnering increased attention. This review delves into the phenotypic characteristics, mechanisms of action, and immune-regulatory functions of these lesser-known but crucial immune cell subsets. The review provides a comprehensive examination of each cell type, detailing their origins, unique functionalities, and contributions to immune homeostasis. It emphasizes the complex interplay among these cells and how their coordinated regulatory activities influence immune responses in diverse pathological and therapeutic contexts, including autoimmunity, cancer immunotherapy, chronic inflammation, and transplant tolerance. By unraveling these mechanisms, the review outlines novel therapeutic avenues, such as targeting these regulatory cells to modulate immune activity and enhance precision medicine approaches. The future of immunotherapy and immune modulation lies in leveraging the expanded knowledge of these regulatory immune cells, presenting challenges and opportunities in clinical applications.
Introduction
Recent advancements in immunology have underscored the pivotal role of regulatory immune cells in orchestrating immune responses and maintaining immune homeostasis (1–5). These specialized subsets of immune cells possess unique immunoregulatory functions, modulating the activity of various immune effectors to prevent autoimmunity, limit inflammation, and facilitate tissue repair (2, 6–9). Understanding the intricate interplay between regulatory immune cells and the broader immune system is paramount for deciphering the pathogenesis of immune-related disorders and devising novel therapeutic strategies.
T regulatory cells (Tregs) represent a cornerstone in the realm of regulatory immune cells. Initially identified for their role in immune tolerance and prevention of autoimmunity, Tregs have garnered substantial attention due to their diverse functional repertoire and plasticity (10–12). Historic discoveries elucidating the crucial function of Tregs in maintaining immune balance have been complemented by recent insights into their heterogeneity, tissue-specific localization, and crosstalk with other immune cell subsets. The evolving landscape of Tregs biology continues to unravel novel mechanisms underlying immune regulation and their implications in health and disease (13).
Beyond Tregs, a myriad of other regulatory immune cell populations has emerged as key players in immune modulation. CD8+ Tregs are a specialized subset of T lymphocytes expressing the CD8 co-receptor and they suppress immune responses through mechanisms such as targeted cytotoxicity against activated immune cells, secretion of anti-inflammatory cytokines (e.g., IL-10, TGF-β), and direct inhibition of effector T cells, uniquely regulating CD8+ T cell-driven immunity while maintaining peripheral tolerance (14). Regulatory B cells, characterized by their ability to produce anti-inflammatory cytokines and induce T cell tolerance, represent a burgeoning field of study with implications in autoimmune diseases and cancer immunotherapy (15). The Treg-of-B cells are a unique subset of regulatory T cells generated by B cells, notable for their lack of FOXP3 (Forkhead box P3) expression, setting them apart from conventional regulatory T cells (16). These cells are characterized by markers such as LAG3 (Lymphocyte-activation gene 3), ICOS (Inducible T-cell co-stimulator), PD1 (Programmed cell death protein 1), GITR(Glucocorticoid-induced TNFR-related protein), and CTLA4 (Cytotoxic T-lymphocyte-associated protein 4) and primarily exert their regulatory function through cell-cell contact mechanisms, rather than cytokines like IL-10 (Interleukin-10). While they do produce IL-10, it is not essential for their suppressive activity, which operates through both IL-10-dependent and independent pathways. This unique profile suggests that Treg-of-B cells contribute to immune tolerance through mechanisms distinct from conventional Tregs (17, 18). MDSCs, DCregs, ILCregs, and NKregs collectively contribute to the intricate network of immune regulation, each exerting unique suppressive functions and immune-modulating properties in diverse pathological contexts (19–23).
As research into regulatory immune cells advances, several challenges and opportunities lie ahead. Unraveling the complexities of regulatory cell subsets, deciphering their precise mechanisms of action, and elucidating their crosstalk within the immune microenvironment pose formidable tasks. Moreover, translating fundamental insights into clinical applications necessitates overcoming hurdles related to cell-based therapies, biomarker identification, and patient stratification. Nonetheless, the burgeoning field of regulatory immune cells holds promise for revolutionizing immunotherapy and ushering in a new era of precision medicine in immune-mediated disorders.
Advancements in understanding regulatory immune cells
In recent years, significant strides have been made in elucidating the intricate roles of regulatory immune cells in modulating immune responses and maintaining immune homeostasis. These advancements have revolutionized our understanding of the immune system and its regulatory mechanisms, shedding light on diverse cell populations beyond Tregs. Research efforts have uncovered a myriad of regulatory immune cell subsets, each endowed with distinct functions and regulatory capacities. From the discovery of regulatory B cells to the emerging insights into the regulatory potential of innate lymphoid cells, our comprehension of these cells continues to evolve rapidly (24). We will provide an overview of the recent advancements in understanding regulatory immune cells, highlighting their diverse functions, regulatory mechanisms, and implications for immune-related diseases and therapeutic interventions. In exploring the landscape of regulatory immune cells, one cannot ignore the significant contributions and insights derived from Treg research, which continue to shape our understanding of immune regulation.
Regulatory T cells
Although Tregs are not the central theme here, their pivotal role in the broader context of regulatory cells cannot be overlooked in any comprehensive research of immune regulation.
Conventional Tregs have long been recognized for their pivotal role in maintaining immune homeostasis by suppressing inappropriate immune responses (25). This capacity to modulate immune responses is critical not only in preventing autoimmune diseases but also in controlling inflammation and promoting tolerance across various biological systems (13) (Figure 1). Recent studies have expanded our understanding of Tregs functions beyond their traditional roles (31). These cells are now known to engage in several non-immune functions that are crucial for maintaining tissue homeostasis. For instance, Tregs have been implicated in metabolic regulation, particularly in adipose tissues where they influence insulin sensitivity and lipid metabolism (32). Tregs are alos influenced by lipid metabolism, particularly through the PPAR-γ (Peroxisome proliferator-activated receptor-γ) receptor, which is sensitive to lipid interactions that can impair Tregs functionality (33). Moreover, Tregs contribute to the maintenance of stem cell niches, such as those found in the bone marrow, skin, and intestines (34, 35). By modulating the local microenvironment, Tregs can protect stem cells from oxidative stress and promote their quiescence, which is crucial for long-term tissue regeneration (36). This interaction also highlights the broader role of Tregs in tissue repair and regeneration, where they can directly affect tissue cells to promote healing and restoration after injury. Furthermore, Tregs have been shown to facilitate tissue repair by producing growth factors and cytokines that directly interact with tissue cells. These molecules help in the proliferation and function of cells involved in tissue repair, such as epithelial cells in the skin and lung, and satellite cells in muscle tissue (2, 37).
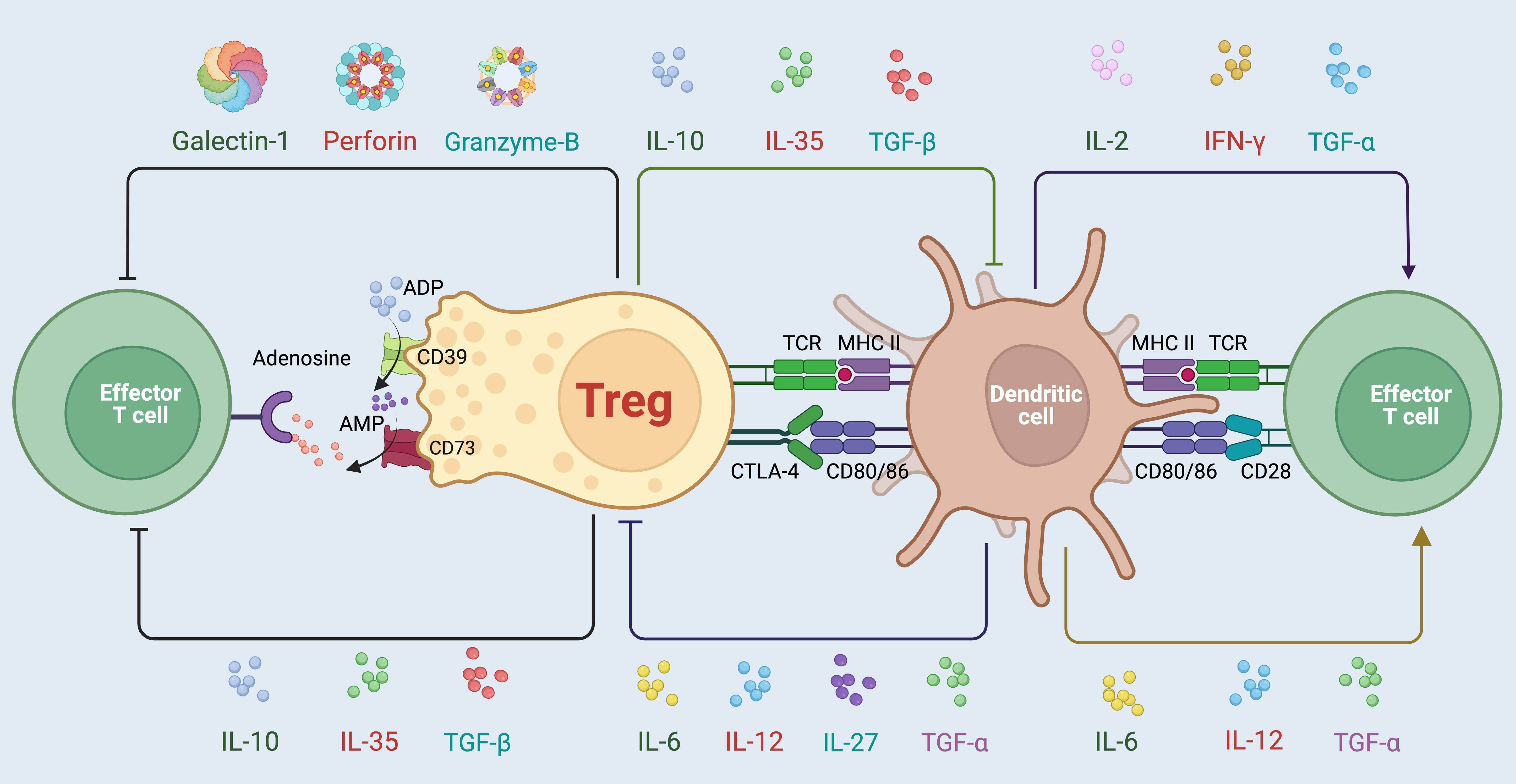
Figure 1. Variable mechanisms involved in the immunosuppressive activities mediated by Tregs. Tregs employ a diverse array of strategies to maintain immune homeostasis and suppress overactive immune responses. One primary mechanism involves the secretion of immunosuppressive cytokines such as IL-10, TGF-β, and IL-35, which directly inhibit the activation and proliferation of effector T cells (13). Additionally, Tregs modulate immune responses through cell surface interactions, notably via the expression of CTLA-4, which competes with CD28 for CD80/86 on dendritic cells, dampening their ability to activate effector T cells (26). Tregs also exert influence through metabolic disruption, utilizing CD39 and CD73 to convert extracellular ATP to adenosine, further suppressing effector cell function via adenosine receptor signaling (27, 28). Furthermore, Tregs can release cytotoxic molecules like Galectin-1, Perforin, and granzyme B, contributing to the direct elimination of pathologically active cells (29, 30). Through these complex and coordinated actions, Tregs play a critical role in preventing autoimmune diseases, controlling chronic inflammation, and promoting tolerance to self-antigens and transplanted tissues.
In the context of clinical applications, the expanded understanding of Treg functions opens new avenues for therapeutic interventions aimed at modulating Treg activity. By targeting the non-canonical functions of Tregs, it is possible to develop treatments that enhance their regulatory capabilities, thereby improving outcomes in diseases characterized by inflammation, autoimmunity, or impaired tissue repair. These insights into the multifaceted roles of Tregs underscore their importance not only in immune regulation but also in broader physiological processes, making them a key target for future research and therapeutic development (1, 38).
When discussing regulatory T cells, it is increasingly imperative not to overlook the presence and significance of CD8+ Tregs. These cells represent a unique subset of T cells with critical implications in maintaining immunological tolerance and modulating the immune environment in various diseases, including cancer (39–41). They exhibit a capacity to suppress immune responses, which is pivotal in preventing autoimmunity but can also facilitate tumor immune evasion by weakening antitumor immune attacks (42). Research has elucidated distinct properties and mechanisms of CD8+ Tregs that differentiate them from their CD4+ counterparts (43). Predominantly, CD8+ Tregs exert their regulatory functions through the expression of molecules like Foxp3, CTLA-4, and others which are shared with CD4+ Tregs, yet they also display unique markers such as CD122 and CD28⁻, reflecting their distinct regulatory pathways (44). These cells are found elevated in various cancers, where they contribute to an immunosuppressive microenvironment that promotes tumor growth and survival by inhibiting effective immune surveillance and clearance. Interestingly, these cells not only modulate immune responses through direct cell-cell interactions but also through the secretion of immunosuppressive cytokines and metabolic disruption of effector cells (45–48). The induction of these Tregs can be driven by the tumor-derived factors and by the tumor microenvironment itself, which alters T cell metabolism and differentiation paths, skewing them towards a regulatory phenotype. The profound impact of CD8+ Tregs in cancer suggests that they could serve as a potential target for therapeutic intervention (49). Modulating the function or abundance of these cells could enhance the efficacy of cancer immunotherapies, providing new avenues to improve patient outcomes in oncological treatments. Understanding and manipulating the balance of Treg activity in the tumor context is therefore crucial for developing more effective cancer immunotherapies (50).
Regulatory B cells
Bregs (Regulatory B cells) are a specialized subset of B lymphocytes that play a crucial role in immune regulation and maintaining immune homeostasis. They are known for their ability to suppress immune responses and promote immune tolerance. Researchers have made significant progress in identifying and characterizing Bregs. They have identified specific cell surface markers and functional properties that distinguish Bregs from other B cell subsets, which helps in understanding their unique regulatory functions (51).
Establishing the existence of one or multiple subsets of Bregs has been challenging due to the absence of specific markers that are analogous to the Treg marker FoxP3 (51, 52). The presence of multiple reported phenotypes for Bregs suggests the potential existence of distinct subsets. However, there are common surface markers shared among the proposed Breg subsets, which suggests the possibility that all Bregs originate from a common precursor (53, 54). The observed differences in phenotype could be attributed to the activation of this common Breg precursor in different microenvironments.
Specific markers for Bregs include (1). CD19: It is important to note that CD19 expression alone is not specific to Bregs, as it is also present on other B cell subsets. It is typically used in combination with other markers to define and isolate regulatory B cell subsets more precisely (51). (2). CD1d: CD1d, a molecule involved in lipid antigen presentation, plays a significant role in the context of Bregs. CD1d allows Bregs to directly interact with NKT cells through the presentation of lipid antigens via CD1d. Then Bregs can influence the function and activity of NKT cells (55). (3). CD5: In some studies, CD5 expression on B cells is commonly associated with B1 cells, a subset of B cells involved in innate-like immune responses and regulatory functions (56). CD5+ Bregs can produce regulatory cytokines such as IL-10 and could suppress immune responses and promote immune tolerance. While CD5+ Bregs have been well-characterized in mice, their presence and significance in human Bregs are still under investigation (57). (4). CD24: High expression of CD24 is often used as a defining feature and a marker for identifying Bregs in various contexts. CD24+ Bregs have been shown to possess potent immunosuppressive properties, including the production of regulatory cytokines such as IL-10 and the ability to suppress excessive immune responses. Modulating the function or increasing the numbers of CD24+ Bregs could be explored as a strategy for treating immune-related disorders, including autoimmune diseases and transplantation rejection (58). (5). CD38: Bregs with regulatory properties have been found to exhibit higher levels of CD38 compared to other B cell subsets. This suggests that CD38 can be used as a phenotypic marker to identify Bregs with immunosuppressive potential. CD38-expressing Bregs have been implicated in various disease contexts (59–61). For example, in autoimmune diseases like RA and SLE (systemic lupus erythematosus), decreased numbers or impaired function of CD38+ Bregs have been observed (62, 63). This suggests that CD38-expressing Bregs may play a role in maintaining immune tolerance and preventing excessive inflammatory responses (58). (6). CD27: The relationship between CD27 expression and Bregs is complex and can vary depending on the context and species studied. These CD27+ Bregs are believed to possess regulatory functions and contribute to immune tolerance. CD27 expression on Bregs has been associated with enhanced suppressive activity and the production of anti-inflammatory cytokines such as IL-10 and TGF-β. Some memory B cells with regulatory functions have been identified within the CD27+ B cell subset. These memory Bregs have been implicated in the regulation of immune responses and the maintenance of immune homeostasis (52).
Bregs employ various mechanisms to suppress immune responses and promote immune tolerance (Figure 2). Initially, Bregs are known to produce immunosuppressive cytokines, such as IL-10 and TGF-β. These cytokines play a crucial role in dampening immune responses by inhibiting the activation and function of effector immune cells. IL-10, in particular, is a potent anti-inflammatory cytokine that can inhibit pro-inflammatory cytokine production and immune cell activation (65, 66). Furthermore, Bregs can act as APCs and present antigens to T cells. However, instead of inducing T cell activation, Bregs often promote tolerance by inducing T cell anergy or Treg differentiation (67, 68). Moreover, Bregs have been shown to interact with and promote the generation and function of Tregs which are key mediators of immune tolerance and can suppress immune responses. Bregs can induce the differentiation and expansion of Tregs through cell-cell contact and the production of regulatory cytokines like IL-10 and TGF-β (65, 69, 70). Additionally, Bregs can exert suppressive effects through direct contact with immune cells. For example, Bregs can engage with and inhibit the activation of APCs such as DCs, thereby reducing their ability to stimulate immune responses. Bregs also directly interact with and suppress the function of effector T cells (71). Finally, Bregs can regulate immune responses by modulating the expression of co-stimulatory molecules. They can downregulate the expression of co-stimulatory molecules on APCs, which is essential for efficient T cell activation. By dampening the co-stimulatory signals, Bregs contribute to the suppression of immune responses (72, 73). Apart from IL-10 and TGF-β, Bregs can secrete other anti-inflammatory factors such as IL-35 and granulocyte-macrophage colony-stimulating factor (GM-CSF). These factors can suppress the activity of immune cells and promote immunosuppression (74, 75).
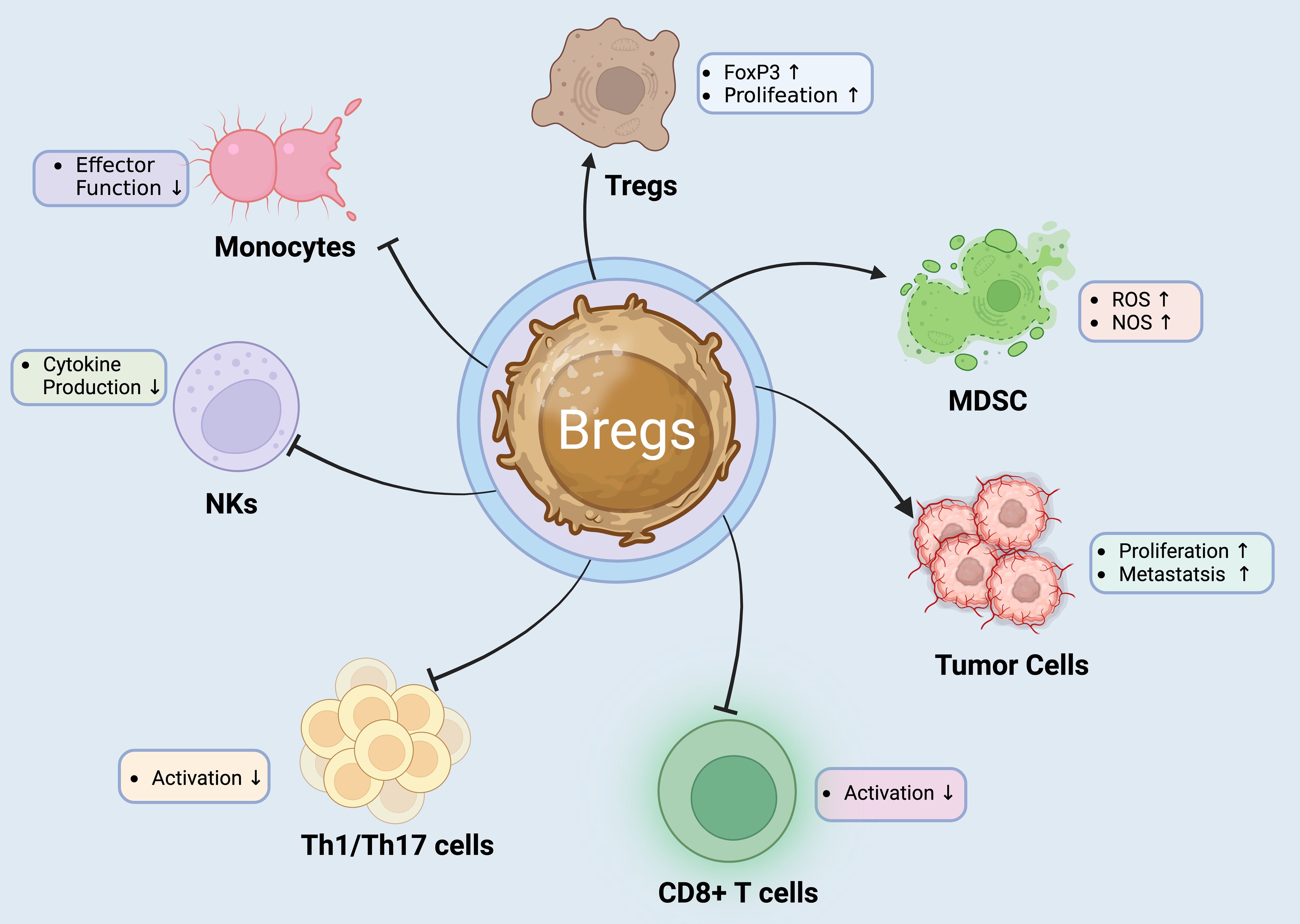
Figure 2. Regulatory B Cells: Orchestrators of Immune Suppression and Modulation. Bregs play a critical role in immune regulation by exerting suppressive effects on various immune cell types. They promote the proliferation and FoxP3 expression of Tregs, enhancing their immunosuppressive functions. Bregs also impact the tumor microenvironment by increasing ROS and NOS levels in MDSCs, which can aid tumor progression (55, 64). Additionally, Bregs inhibit the effector functions of NK cells and monocytes, reducing cytokine production and thus dampening inflammatory responses. Furthermore, Bregs decrease the activation of CD8+ T cells and Th1/Th17 cells, which are crucial for mediating autoimmune and inflammatory reactions. This multifunctional regulatory capacity makes Bregs a target of interest in developing therapies for autoimmune diseases and cancer immunotherapy (55).
Myeloid-derived suppressor cells
MDSCs are a heterogeneous population of immature myeloid cells with potent immunosuppressive capabilities. They play a crucial role in regulating immune responses in various pathological conditions, including cancer, infections, autoimmune diseases, and chronic inflammation (76–81). MDSCs are characterized by their myeloid origin, early differentiation stage, and ability to suppress immune responses through multiple mechanisms.
MDSCs arise from myeloid progenitor cells in the bone marrow. Under certain pathological conditions, such as cancer or inflammation, the differentiation of these cells is disrupted, leading to the accumulation of immature myeloid cells with suppressive functions in peripheral tissues (82, 83). MDSCs lack mature markers of myeloid cells and often express a combination of markers associated with early myeloid progenitors, including CD11b and Gr-1 in mice (84, 85). In humans, MDSCs are commonly identified by the expression of CD11b and CD33, as well as the lack of markers associated with mature immune cells, such as HLA-DR (86, 87). However, it is essential to note that MDSC phenotypes can vary depending on the context and the specific tissue involved.
MDSCs suppress immune responses through various mechanisms, contributing to immune evasion in cancer and other diseases. To begin with, MDSCs can secrete factors like IL-10, IL-6, IL-1β, TGF-β, and arginase-1, which dramatically increase the rate of accumulation and T cell suppressive activity of MDSC (88–92). Furthermore, MDSCs can trigger apoptosis in activated T cells through the production of reactive oxygen species (ROS) and other pro-apoptotic factors (93–95). Additionally, MDSCs consume and deplete essential nutrients such as arginine, tryptophan and cysteine, restricting their availability for T cell function (96–99). Moreover, MDSCs can impair the function of dendritic cells and macrophages, leading to decreased antigen presentation and diminished T cell activation (100–102) (Figure 3).
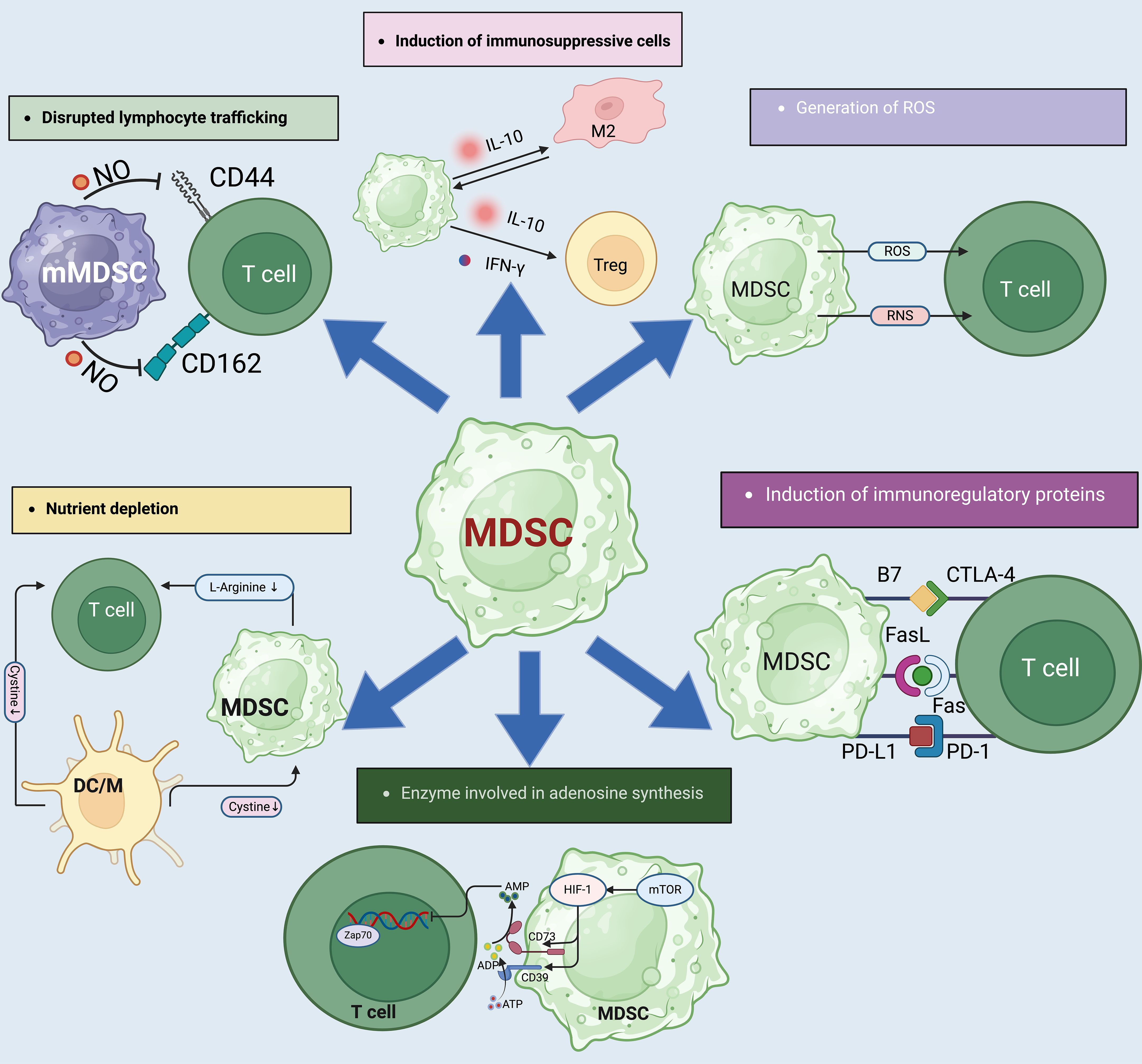
Figure 3. The complex mechanisms by which MDSCs influence immune suppression. (1). MDSCs also contribute to immune suppression by the impairment of T-cell homing to lymphoid tissues via interactions with selectins and cellular adhesion molecules like CD44 and CD62L. This impairment is facilitated by NO which alters T-cell migration, impacting immune surveillance and response. (2). MDSCs express ARG1 which depletes L-arginine, a critical molecule for T-cell receptor (TCR) expression and T-cell function. MDSCs can lead to reduced protein synthesis and glutathione production in T-cells, weakening the immune response. (4). Adenosine production is regulated by CD39 and CD73 ectoenzymes on MDSCs, which convert ATP to adenosine under hypoxic conditions. This adenosine then inhibits T-cell activation via suppression of kinase pathways, further contributing to the immunosuppressive microenvironment. (5). MDSCs contribute to immune suppression by producing IL-10 and IFN-γ. In addition, MDSCs downregulate pro-inflammatory cytokines like IL-6 and TNF-α in M2 macrophages, reinforcing a suppressive environment. (6). Free radicals, particularly reactive oxygen species (ROS) and reactive nitrogen species (RNS), are produced by MDSCs through enzymes like arginase-1 (ARG1), NOX2, and NOS2. These radicals inhibit T-cell function by inducing T-cell energy loss and promoting apoptosis, further contributing to immunosuppression. (7). MDSCs and Tregs express and activate inhibitory molecules like PD-L1, CTLA-4, and B7, which interact with PD-1 and CD28 on T cells to inhibit their activation and induce apoptosis.
MDSCs can be further classified into two main subsets based on their phenotype in mice: Granulocytic MDSCs (G-MDSCs) express high levels of Ly6G and Ly6C and are morphologically similar to neutrophils (103, 104). Monocytic MDSCs (M-MDSCs) cells are characterized by high expression of Ly6C and low expression of Ly6G in mice, resembling monocytes (105, 106). In humans, the classification of MDSC subsets is more complex and remains an area of ongoing research.
MDSCs play a critical role in promoting tumor progression and immune evasion in cancer. Their accumulation is associated with poor prognosis in cancer patients (107–110). Moreover, MDSCs have been implicated in the pathogenesis of various inflammatory and autoimmune diseases, contributing to the dysregulation of immune responses (111–119).
Regulatory DC cells
DCs (dendritic cells) are specialized antigen-presenting cells derived from the bone marrow that play a crucial role in initiating and regulating innate and adaptive immune responses. Regulatory or “tolerogenic” DCs are particularly important especially in the context of maintaining self-tolerance in a healthy state. These Dcregs employ various mechanisms to suppress or redirect the responses of naïve or memory T cells. In animal models of autoimmune diseases and transplant rejection, DCregs have demonstrated the ability to induce or restore T cell tolerance (120–124). Moreover, there is compelling evidence indicating that the transfer of DCregs can effectively modulate T cell responses not only in non-human primates but also in human subjects (120, 125). Building upon insights gained from in vitro experiments and animal models, efforts have been made to develop clinical-grade DCregs for the treatment of autoimmune diseases (120, 126–128). Clinical trials in Phase I evaluating the use of regulatory dendritic cell therapy in type-1 diabetes, rheumatoid arthritis and Crohn’s disease have shown promising results, demonstrating the feasibility and safety of this approach (129–132).
DCregs possess unique suppressive mechanisms that allow them to actively regulate immune responses and promote immune tolerance (see Figure 4).
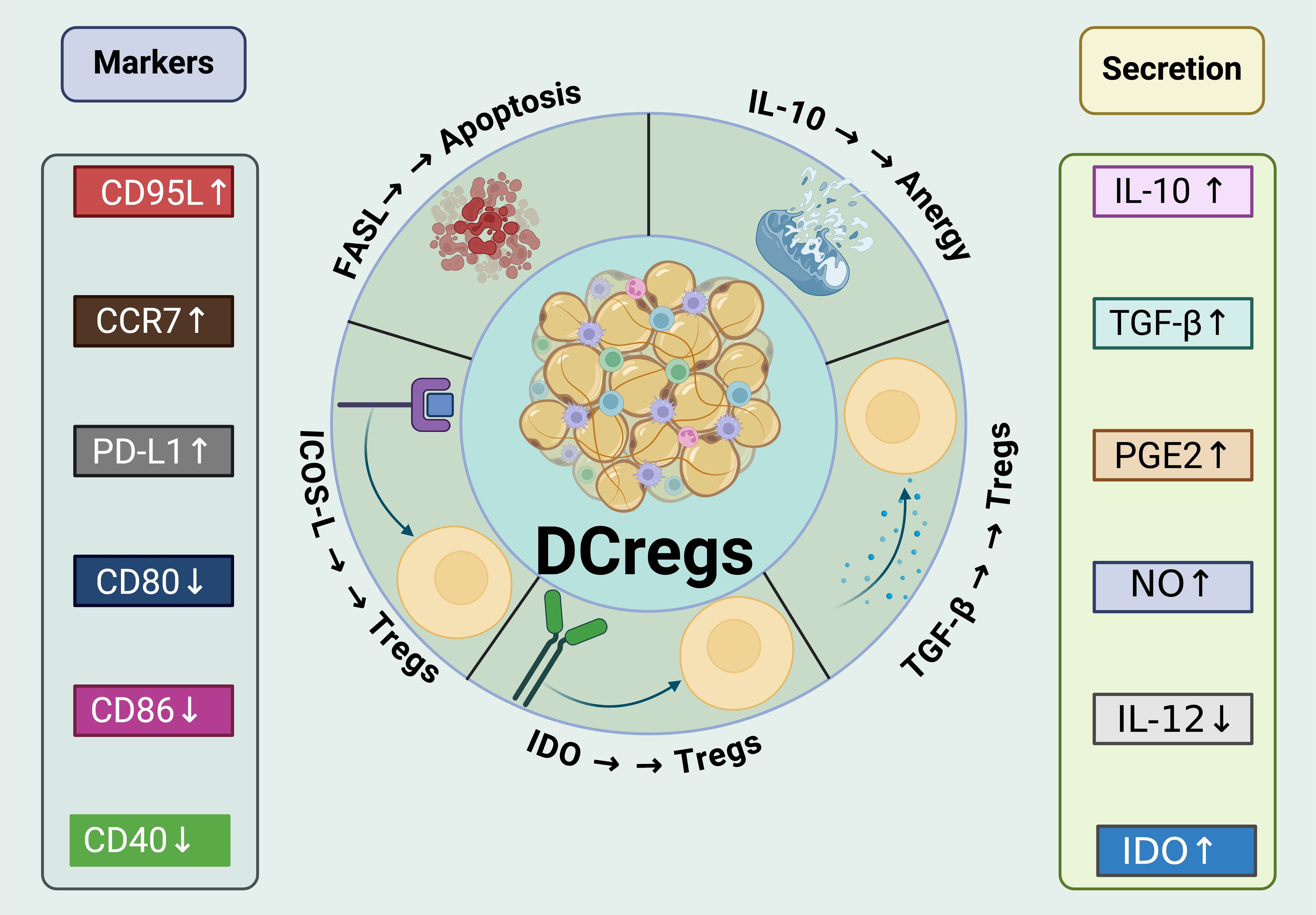
Figure 4. Mechanisms of immunosuppressive and tolerogenic activities of DCregs. DCregs express a variety of surface markers that are critical for their interaction with T cells and other immune cells. Markers like CD95L, CCR7, PD-L1, CD80, and CD86 are differentially expressed to modulate the immune response. The increased expression of CD95L and PD-L1, for example, enhances the ability of DCregs to induce apoptosis and anergy in T cells. These cells also show an elevated secretion of immunoregulatory cytokines such as IL-10 and TGF-β, which are known to promote Treg expansion and contribute to the suppression of effector T cell functions. In addition,DCregs produce various molecules like prostaglandin E2 (PGE2), nitric oxide (NO), and IDO (Indoleamine 2,3-dioxygenase), all of which have profound effects on the immune environment. PGE2 and NO contribute to the overall suppressive milieu, while IDO activity leads to metabolic depletion that inhibits effector T cell functions and supports Treg cell maintenance. Moreover, DCregs interact with Tregs to enhance their suppressive function and stability through mechanisms such as the expression of IDO. They also directly inhibit the activation and proliferation of CD8+ T cells and Th1/Th17 cells, pivotal in controlling inflammation and autoimmunity.
Induction of Tregs
DCregs could induce the differentiation and expansion of Tregs, particularly Foxp3+ Tregs. By presenting antigens in a tolerogenic manner and providing co-stimulatory signals, DCregs promote the development of Tregs that suppress immune responses and maintain self-tolerance (133–138).
Immune checkpoint molecules
Similar to conventional DCs, DCregs express immune checkpoint molecules such as PD-L1and CTLA-4. The engagement of these molecules with their respective receptors on T cells inhibits their activation and proliferation, thereby suppressing immune responses.
Production of immunomodulatory cytokines
DCregs secrete immunomodulatory cytokines such as IL-10 and TGF-β (139, 140). These cytokines have potent immunosuppressive effects, including the inhibition of pro-inflammatory cytokine production, suppression of effector T cell responses, and promotion of regulatory T cell function.
Indoleamine 2,3-dioxygenase expression
DCregs express the enzyme IDO, which catabolizes tryptophan, an essential amino acid required for T cell proliferation. By depleting tryptophan and generating tryptophan metabolites, DCregs induce a state of tryptophan starvation that inhibits T cell activation and promotes the differentiation of regulatory T cells (139, 141, 142).
Modulation of co-stimulatory molecules
DCregs exhibit reduced expression of co-stimulatory molecules such as CD80 and CD86, resulting in impaired T cell activation. This modulation of co-stimulatory signals contributes to the induction of T cell anergy or tolerance (143).
Antigen presentation in lymphoid organs
DCregs preferentially migrate to lymphoid organs and present antigens to T cells in a tolerogenic manner. This leads to the induction of antigen-specific tolerance and suppression of immune responses (144).
The extensive data significantly enhances our present comprehension of different subsets of DCregs in the regulation of different conditions. Nevertheless, the primary challenge at present is how to translate our knowledge of DCregs in mouse models to manipulation of human immune system and reveal therapeutic potential of DCregs in human diseases. Promisingly, several research have initiated investigations into the characteristics of DCregs in patients with autoimmune and inflammatory diseases. These studies aim to explore the therapeutic potential of DCregs in the treatment of AID, offering an exciting avenue for further research and potential clinical applications.
The initial investigation on tolerogenic dendritic cells in humans was conducted by Ralph Steinman’s laboratory. Their study involved the subcutaneous administration of antigen-loaded immature dendritic cells to study subjects, with a dosage of 2×106 cells per subject. The treatment was well tolerated by the participants, and the findings showed that the therapy could effectively suppress antigen-specific CD8+ T cell responses for a duration of up to 6 months (145, 146). In a more recent clinical trial, 10 individuals with type 1 diabetes participated, and each subject received four intradermal administrations of 1 × 107 autologous dendritic cells at 2-week intervals. The dendritic cells used in the treatment were modified through the transduction of anti-sense oligonucleotides, which aimed to silence the expression of co-stimulatory molecules such as CD40, CD80, and CD86. However, no specific data regarding the efficacy of the silencing process was reported in the study (129).
The researchers had previously established their silencing protocols in a mouse model of type 1 diabetes (147–149). They demonstrated that the dendritic cell treatments, which involved the silencing of co-stimulatory molecules, had resulted in statistically significant, albeit modest, effects in sparing the progression of the disease in the mouse model (150, 151). Similar to the earlier study conducted by Steinman, no adverse events associated with the dendritic cell treatments were reported in this subsequent study (152–154). However, there were limited or no detectable immunological signs of tolerance that could be attributed to the dendritic cell treatments. In conclusion, the study highlights that dendritic cells can acquire a tolerogenic phenotype through various mediators, and these play a significant role in shaping interactions between dendritic cells and naive or effector T cells. Tolerogenic dendritic cells utilize both secreted molecules like IL-10 and retinoic acid, as well as inhibitory receptors, to promote the induction of regulatory T cells (155, 156). Additionally, they provide supplementary signals such as integrins to guide the localization of these developing regulatory T cells to the appropriate anatomical sites. A major challenge in the future application of tolerogenic dendritic cells in immunotherapy will be to carefully select or optimize the specific type(s) of tolerogenic dendritic cells to be used, considering the clinical targets and desired outcomes.
Regulatory innate lymphoid cells
ILCregs are a subset of innate lymphoid cells that possess immunosuppressive functions and play a crucial role in maintaining immune homeostasis. While the concept of ILCregs is still relatively new and evolving, their importance in immune regulation is becoming increasingly recognized (see Figure 5).
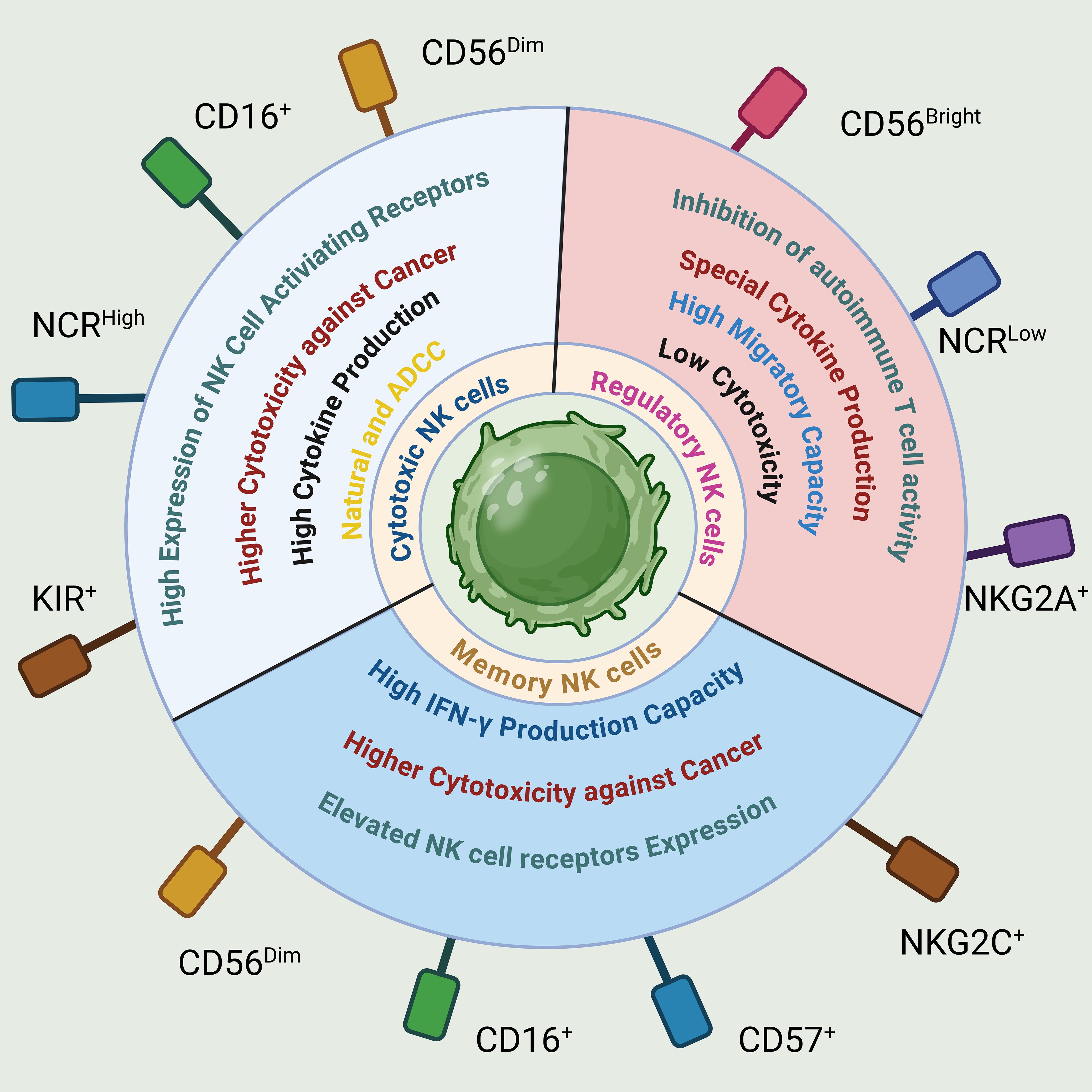
Figure 5. Phenotypical and functional properties of NK cells. NK cells are categorized based on their phenotypic and functional traits, linked to specific receptors like adhesion molecules (CD56, CD57), activating receptors (CD16, NCR, KIR, NKG2C), and inhibitory receptors (NKG2A). These classifications lead to distinct NK cell subsets with regulatory, cytotoxic, or memory functions, each showing unique operational characteristics.
ILCregs have been identified in various tissues and organs, including kidneys and intestines (21, 157). They are characterized by their ability to produce immuno-suppressive cytokines such as IL-10 and TGF-β, which help dampen excessive immune responses and promote tolerance (21). Similar to other ILC subsets, ILCregs can be classified based on the expression of specific transcription factors and surface markers. Though they lacked expression of the Treg transcription factor Foxp3, ILCregs exert their immune-suppressive effects through multiple mechanisms. In addition to secreting immunosuppressive cytokines, they can interact with other immune cells, such as dendritic cells and T cells to modulate their functions (158). ILCregs can inhibit dendritic cell maturation and antigen presentation, leading to decreased activation of effector T cells. They can also directly interact with T cells, promoting the development of regulatory T cells and suppressing the activity of pro-inflammatory T cell subsets (157) (Figure 6).
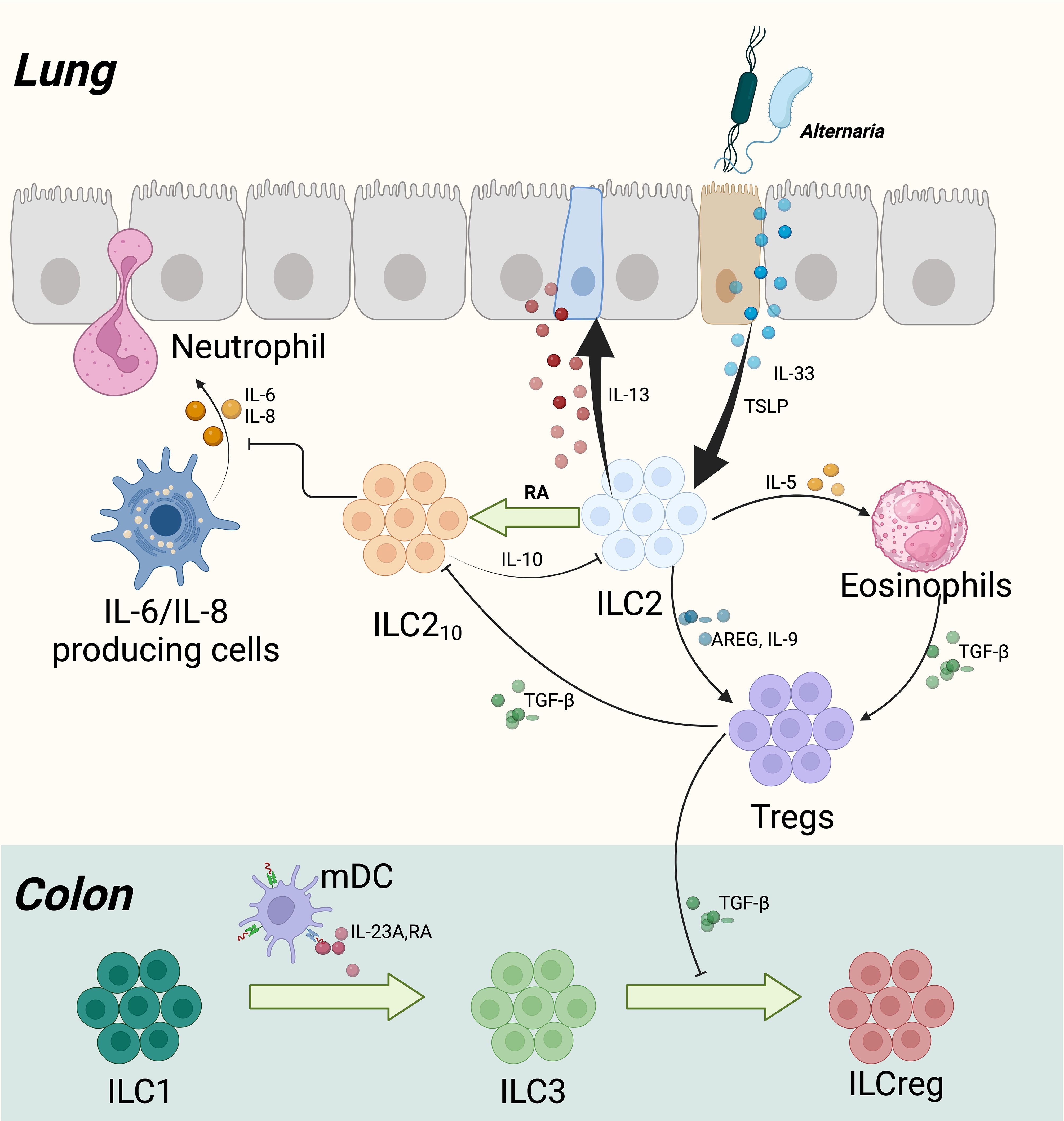
Figure 6. Development of IL-10+ ILCs in the Lung and Colon of Humans. In human lung and colon development, exposure to the fungus Alternaria alternata initiates a sequence of immunological responses starting with activation of the airway epithelium. This activation leads to the release of cytokines such as TSLP (thymic stromal lymphopoietin) and IL-33, which in turn activate type 2 innate lymphoid cells (ILC2s). These cells respond by producing IL-5 and IL-13, contributing to eosinophil recruitment and goblet cell hyperplasia, respectively. Additionally, TSLP is implicated in reducing corticosteroid responsiveness. IL-13 also stimulates the release of retinoic acid (RA) from the epithelium, which facilitates the transformation of ILC2s into a subtype producing IL-10 (ILC210s). These ILC210s play a critical role in dampening type 2 inflammatory responses and enhancing epithelial barrier integrity by reducing IL-6 and IL-8 levels, thereby inhibiting neutrophil migration. Concurrently, Tregs develop and secrete transforming growth factor-beta (TGF-β) to further regulate inflammation and influence the activity of ILC210s. In the colon, CD103+ myeloid dendritic cells (mDCs) release RA and IL-23A, which promote the transformation of CD127+ ILC1s into ILC3s. Tregs support this transition by releasing TGF-β, further promoting the formation of ILCregs.
Unlike the previously described ILCregs, there is also evidence that ILC2s have the capacity to produce IL-10 and may have immunoinhibitory potential (159). Additionally, several pieces of evidence suggest that ILC3s are plastic and can become ILCregs (160). Indeed, the recent findings reveal a regulatory plasticity within all ILC subtypes, and potential crosstalk between DCs and ILCs which should be further investigated in future research.
The study of ILCregs is still in its early stages, and further research is needed to fully understand their ontogeny, functional diversity, and specific roles in different immune contexts. However, their potential as therapeutic targets for immune-mediated diseases and their ability to regulate immune responses make them an exciting area of investigation in immunology.
Regulatory nature kill cells
While NK cells were initially classified as a uniform population of innate lymphocytes, emerging evidence suggests that NK cells comprise diverse subsets with varying functions, distributions, and developmental origins. Human NK cells in peripheral blood can be categorized into at least two functional subsets based on their expression of CD56 and CD16 (161) (Figure 7). CD56dimCD16+ NK cells make up approximately 90% of the total NK cells present in the blood. These cells are highly efficient in killing target cells and secrete lower levels of cytokines. On the other hand, regulatory NK cells, namely CD56bright CD16- NK cells, account for less than 10% of total blood NK cells but are enriched in secondary lymphoid tissues (162).
NK cells have been recognized for their ability to carry out effector functions through both direct cytotoxicity and the release of IFN-γ. However, a study conducted by Perona-Wright and colleagues has shed light on an additional role of NK cells in attenuating inflammatory processes (163). They demonstrated that NK cells can dampen inflammation by producing IL-10. Therefore, NK cells possess the capacity to not only exert their cytotoxic and pro-inflammatory activities but also contribute to the regulation and resolution of immune responses. Some of the key functions of regulatory NK cells include: (1). Immunomodulation: Regulatory NK cells can interact with other immune cells, such as T cells and dendritic cells, and modulate their activation and function. They can suppress the proliferation and activation of T cells, thereby regulating the adaptive immune response (164–166). (2). Cytokine Production: Regulatory NK cells produce various immunomodulatory cytokines, including IL-10 and TGF-β (167–171). These cytokines have anti-inflammatory properties and can suppress immune responses. (3). Interaction with Dendritic Cells: Regulatory NK cells can interact with dendritic cells and influence their maturation and antigen presentation. This interaction can impact the activation and differentiation of other immune cells, such as T cells (163, 169). (4). Induction of Tolerance: Regulatory NK cells contribute to the induction of immune tolerance, particularly in the context of transplantation and autoimmunity. They can suppress excessive immune responses and promote immune tolerance to self-antigens (172, 173).
Although regulatory NK cells are a relatively less studied subset compared to conventional NK cells, their role in immune regulation is increasingly recognized. Further research is needed to fully understand their precise mechanisms and contribution to immune tolerance and regulation. It’s important to note that regulatory NK cells are still an area of active research, and their clinical applications are being explored in various ongoing studies and clinical trials. As our understanding of their functions and therapeutic potential advances, regulatory NK cells may become an integral part of personalized and targeted immunotherapies in the future.
Conclusion
In the realm of regulatory immune cells, Tregs, Bregs, MDSCs, DCregs, NKregs, and ILCregs demonstrate their importance in maintaining immune homeostasis and preventing immune-related diseases (13, 49). The interactions and functional overlaps among these cells are manifested in several aspects: First, all these regulatory cells can modulate immune responses by secreting anti-inflammatory and immunosuppressive cytokines like IL-10 and TGF-β. For example, Tregs suppress effector T cells and inflammatory responses by secreting these cytokines, while Bregs similarly use them to suppress excessive humoral responses and promote immune tolerance (13, 28, 49). Moreover, regulatory cells can interact through molecules on their surfaces to jointly suppress immune responses. For instance, Bregs can interact with Tregs or other cells through expressing CD40, CD80, and CD86, thus regulating T cell activation and differentiation (174). DCregs can modulate T cells by engaging PD-1 through expressed PDL-1 (175). Furthermore, regulatory cells can directly or indirectly affect the function of effector cells, such as effector T cells and B cells. Tregs can directly interact with effector T cells to inhibit their activation and proliferation. Similarly, Bregs and DCregs can regulate humoral responses by inhibiting the differentiation and IgG production of effector B cells (128, 176). Additionally, in various disease models, these regulatory cells exhibit similar roles, such as in cancer, autoimmune diseases, and transplantation tolerance. For instance, MDSCs promote tumor immune escape by inhibiting NK and CD8+ T cell activity in the tumor microenvironment, a function similarly exhibited by Tregs (177, 178). Finally, these regulatory cells share signaling pathways in cytokine networks, such as STAT3 and NF-κB, which play key roles in regulating immune responses (179–181). Through these pathways, regulatory cells collaboratively maintain immune homeostasis and prevent overactivation of the immune system. Not only do regulatory immune cells overlap in function, but they also interact through various mechanisms to collectively maintain immune homeostasis and tolerance. A deeper understanding of these cells and their interactions is crucial for developing new immunoregulatory therapeutic strategies.
The expanding research into regulatory immune cells offers a promising frontier for developing novel therapeutic strategies. These diverse roles of immune cells in immune modulation present both opportunities and challenges in translating their functions into effective treatments (182). The clinical applications of regulatory immune cells are vast, ranging from enhancing cancer immunotherapy to preventing autoimmune diseases and improving transplant outcomes (183, 184). As we delve deeper into understanding these cells’ mechanisms and interactions, the potential for innovative treatments grows. These types of cells are pivotal in modulating immune homeostasis, where they prevent autoimmune diseases, mitigate chronic inflammation, and enhance graft tolerance. Therapeutic potential of these cells extends across various domains, including cancer treatment, where manipulation of their numbers and functions can improve outcomes. It underscores the ongoing research into their regulatory mechanisms and their application in treating a wide array of immune-related conditions, necessitating sophisticated strategies to harness their full potential while maintaining the delicate balance of immune responses. However, the clinical application of regulatory immune cells faces significant challenges, including ensuring specificity and selectivity in immune modulation to avoid global immunosuppression, managing long-term safety concerns such as potential chronic infections or oncogenic transformation, and maintaining rigorous standardization and quality control in cell production (49, 185, 186). Ethical and regulatory considerations also play a crucial role, requiring careful management of patient-derived cell manipulations. To overcome these challenges, strategies such as developing targeted therapies that home to specific tissues, leveraging synthetic biology to enhance cell functionality, and optimizing cell preparation and infusion protocols are essential (186, 187). Collaborative efforts across disciplines are required to refine these therapies, ensuring they are safe, effective, and ethically developed. These concerted actions will be pivotal in harnessing the full therapeutic potential of regulatory immune cells, offering innovative treatments for a range of immune-mediated diseases. Future research needs to focus on refining the specificity and safety of therapies that modulate regulatory immune cells, ensuring that they can be integrated effectively into patient care (188–192). This ongoing exploration holds the key to unlocking the full potential of immune regulation in treating a wide array of diseases, potentially revolutionizing our approach to immunotherapy and transplantation medicine.
Author contributions
PS: Software, Writing – original draft. YY: Resources, Writing – revised draft. HX: Supervision, Writing – review & editing. XY: Writing – original draft, Conceptualization, Investigation. XC: Writing – review & editing, Supervision, Validation. YZ: Writing – review & editing. HZ: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the Shandong Province Natural Science Foundation grants ZR2022QH372.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Iglesias-Escudero M, Arias-González N, and Martínez-Cáceres E. Regulatory cells and the effect of cancer immunotherapy. Mol Cancer (2023) 22:26. doi: 10.1186/s12943-023-01714-0
2. Dikiy S and Rudensky AY. Principles of regulatory T cell function. Immunity (2023) 56:240–55. doi: 10.1016/j.immuni.2023.01.004
3. Tay C, Tanaka A, and Sakaguchi S. Tumor-infiltrating regulatory T cells as targets of cancer immunotherapy. Cancer Cell (2023) 41:450–65. doi: 10.1016/j.ccell.2023.02.014
4. Gocher-Demske AM, Cui J, Szymczak-Workman AL, Vignali KM, Latini JN, Pieklo GP, et al. IFNγ-induction of TH1-like regulatory T cells controls antiviral responses. Nat Immunol (2023) 24:841–54. doi: 10.1038/s41590-023-01453-w
5. Chen WJ. TGF-β regulation of T cells. Annu Rev Immunol (2023) 41:483–512. doi: 10.1146/annurev-immunol-101921-045939
6. Sanz AB, Sanchez-Niño MD, Ramos AM, and Ortiz A. Regulated cell death pathways in kidney disease. Nat Rev Nephrol (2023) 19:281–99. doi: 10.1038/s41581-023-00694-0
7. Wegrzyn AS, Kedzierska AE, and Obojski A. Identification and classification of distinct surface markers of T regulatory cells. Front Immunol (2023) 13:1055805. doi: 10.3389/fimmu.2022.1055805
8. Pisetsky DS. Pathogenesis of autoimmune disease. Nat Rev Nephrol (2023) 19:509–24. doi: 10.1038/s41581-023-00720-1
9. Bittner S, Hehlgans T, and Feuerer M. Engineered Treg cells as putative therapeutics against inflammatory diseases and beyond. Trends Immunol (2023) 44:468–83. doi: 10.1016/j.it.2023.04.005
10. Damle NK and Gupta S. Heterogeneity of concanavalin a-induced suppressor T cells in man defined with monoclonal antibodies. Clin Exp Immunol (1982) 48:581.
11. Fehérvari Z and Sakaguchi S. CD4+ Tregs and immune control. J Clin Invest (2004) 114:1209–17. doi: 10.1172/JCI200423395
12. Hori S, Nomura T, and Sakaguchi S. Control of regulatory T cell development by the transcription factor Foxp3. J Immunol (2017). doi: 10.1126/science.1079490
13. Zhao H, Liao X, and Kang Y. Tregs: Where we are and what comes next? Front Immunol (2017) 8:1578. doi: 10.3389/fimmu.2017.01578
14. Zhang S, Wu M, and Wang F. Immune regulation by CD8+ Treg cells: novel possibilities for anticancer immunotherapy. Cell Mol Immunol (2018) 15:805–7. doi: 10.1038/cmi.2018.170
15. Rosser EC and Mauri C. Regulatory B cells: Origin, phenotype, and function. Immunity (2015) 42:607–12. doi: 10.1016/j.immuni.2015.04.005
16. Chu K-H and Chiang B-L. A novel subset of regulatory T cells induced by B cells alleviate the severity of immunological diseases. Clin Rev Allergy Immunol (2024), 1–10. doi: 10.1007/s12016-024-09009-y
17. Chien C-H and Chiang B-L. Regulatory T cells induced by B cells: a novel subpopulation of regulatory T cells. J BioMed Sci (2017) 24:86. doi: 10.1186/s12929-017-0391-3
18. Huang JH, Lin YL, Wang LC, and Chiang BL. M2-like macrophages polarized by Foxp3– Treg-of-B cells ameliorate imiquimod-induced psoriasis. J Cell Mol Med (2023) 27:1477–92. doi: 10.1111/jcmm.17748
19. Ostrand-Rosenberg S, Lamb TJ, and Pawelec G. Here, there, and everywhere: Myeloid-derived suppressor cells in immunology. J Immunol (2023) 210:1183–97. doi: 10.4049/jimmunol.2200914
20. Pittet MJ, Di Pilato M, Garris C, and Mempel TR. Dendritic cells as shepherds of T cell immunity in cancer. Immunity (2023) 56:2218–30. doi: 10.1016/j.immuni.2023.08.014
21. Wang S, Xia P, Chen Y, Qu Y, Xiong Z, Ye B, et al. Regulatory innate lymphoid cells control innate intestinal inflammation. Cell (2017) 171:201–216.e18. doi: 10.1016/j.cell.2017.07.027
22. Jegatheeswaran S, Mathews JA, and Crome SQ. Searching for the elusive regulatory innate lymphoid cell. J Immunol (2021) 207:1949–57. doi: 10.4049/jimmunol.2100661
23. Fu B, Tian Z, and Wei H. Subsets of human natural killer cells and their regulatory effects. Immunology (2014) 141:483–9. doi: 10.1111/imm.12224
24. Zhao H, Feng R, Peng A, Li G, and Zhou L. The expanding family of noncanonical regulatory cell subsets. J Leukoc Biol (2019) 106:369–83. doi: 10.1002/JLB.6RU0918-353RRRR
25. Vignali DAA, Collison LW, and Workman CJ. How regulatory T cells work. Nat Rev Immunol (2008) 8:523–32. doi: 10.1038/nri2343
26. Wing K, Onishi Y, Prieto-Martin P, Yamaguchi T, Miyara M, Fehervari Z, et al. CTLA-4 control over Foxp3+ regulatory T cell function. Sci (80- ) (2008) 322:271–5. doi: 10.1126/science.1160062
27. Deaglio S, Dwyer KM, Gao W, Friedman D, Usheva A, Erat A, et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J Exp Med (2007) 204:1257–65. doi: 10.1084/jem.20062512
28. Zhao H, Bo C, Kang Y, and Li H. What else can CD39 tell us? Front Immunol (2017) 8:727. doi: 10.3389/fimmu.2017.00727
29. Garín MI, Chu NC, Golshayan D, Cernuda-Morollón E, Wait R, and Lechler RI. Galectin-1: A key effector of regulation mediated by CD4 +CD25+ T cells. Blood (2007) 109:2058–65. doi: 10.1182/blood-2006-04-016451
30. Cao X, Cai SF, Fehniger TA, Song J, Collins LI, Piwnica-Worms DR, et al. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity (2007) 27:635–46. doi: 10.1016/j.immuni.2007.08.014
31. Astarita JL, Dominguez CX, Tan C, Guillen J, Pauli ML, Labastida R, et al. Treg specialization and functions beyond immune suppression. Clin Exp Immunol (2023). doi: 10.1093/cei/uxac123
32. Grusdat M and Brenner D. Adipose Treg cells in charge of metabolism. Nat Immunol (2024) 25:392–3. doi: 10.1038/s41590-024-01762-8
33. Meng F, Hao P, and Du H. Regulatory T cells differentiation in visceral adipose tissues contributes to insulin resistance by regulating JAZF-1/PPAR-γ pathway. J Cell Mol Med (2023) 27:553–62. doi: 10.1111/jcmm.17680
34. Hicks MR and Pyle AD. The emergence of the stem cell niche. Trends Cell Biol (2023) 33:112–23. doi: 10.1016/j.tcb.2022.07.003
35. Cohen JN, Gouirand V, Macon CE, Lowe MM, Boothby IC, Moreau JM, et al. Regulatory T cells in skin mediate immune privilege of the hair follicle stem cell niche. Sci Immunol (2024) 9:eadh0152. doi: 10.1126/sciimmunol.adh0152
36. Wang Y, Huang T, Gu J, and Lu L. Targeting the metabolism of tumor-infiltrating regulatory T cells. Trends Immunol (2023) 44:598–612. doi: 10.1016/j.it.2023.06.001
37. Hanna BS, Wang G, Galván-Peña S, Mann AO, Ramirez RN, Muñoz-Rojas AR, et al. The gut microbiota promotes distal tissue regeneration via RORγ+ regulatory T cell emissaries. Immunity (2023). doi: 10.1016/j.immuni.2023.01.033
39. Smith TRF and Kumar V. Revival of CD8+ Treg-mediated suppression. Trends Immunol (2008) 29:337–42. doi: 10.1016/j.it.2008.04.002
40. Wang RF. CD8+ regulatory T cells, their suppressive mechanisms, and regulation in cancer. Hum Immunol (2008) 69:811–4. doi: 10.1016/j.humimm.2008.08.276
41. Zhang S, Wu M, and Wang F. Immune regulation by CD8+ Treg cells: novel possibilities for anticancer immunotherapy. Cell Mol Immunol (2018), 1–3. doi: 10.1038/cmi.2018.170
42. Shi Z, Okuno Y, Rifa’i M, Endharti AT, Akane K, Isobe K-I, et al. Human CD8+CXCR3+ T cells have the same function as murine CD8+CD122+ Treg. Eur J Immunol (2009) 39:2106–19. doi: 10.1002/eji.200939314
43. Vuddamalay Y, Attia M, Vicente R, Pomié C, Enault G, Leobon B, et al. Mouse and human CD8 + CD28 low regulatory T lymphocytes differentiate in the thymus. Immunology (2016). doi: 10.1111/imm.12600
44. Churlaud G, Pitoiset F, Jebbawi F, Lorenzon R, Bellier B, Rosenzwajg M, et al. Human and mouse CD8+CD25+FOXP3+ regulatory T cells at steady state and during interleukin-2 therapy. Front Immunol (2015) 6:171. doi: 10.3389/fimmu.2015.00171
45. Gupta S and Agrawal S. In vitro effects of CD8+ regulatory T cells on human B cell subpopulations. Int Arch Allergy Immunol (2020) 181:476–80. doi: 10.1159/000506806
46. Gupta S, Su H, and Agrawal S. CD8 Treg cells inhibit B-cell proliferation and immunoglobulin production. Int Arch Allergy Immunol (2020) 181:947–55. doi: 10.1159/000509607
47. Niederlova V, Tsyklauri O, Chadimova T, and Stepanek O. CD8+ Tregs revisited: A heterogeneous population with different phenotypes and properties. Eur J Immunol (2021) 51:512–30. doi: 10.1002/eji.202048614
48. Kasahara TM and Gupta S. CD8 Treg-mediated suppression of naive CD4+ T cell differentiation into follicular helper T cells. Int Arch Allergy Immunol (2022) 183:682–92. doi: 10.1159/000521427
49. Zhao H, Feng R, Peng A, Li G, and Zhou L. The expanding family of noncanonical regulatory cell subsets. J Leukoc Biol (2019) 106:369–83. doi: 10.1002/JLB.6RU0918-353RRRR
50. Gupta S, Demirdag Y, and Gupta AA. Members of the regulatory lymphocyte club in common variable immunodeficiency. Front Immunol (2022) 13:864307. doi: 10.3389/fimmu.2022.864307
51. Mauri C and Menon M. The expanding family of regulatory B cells. Int Immunol (2015) 27:479–86. doi: 10.1093/intimm/dxv038
52. Matsumura Y, Watanabe R, and Fujimoto M. Suppressive mechanisms of regulatory B cells in mice and humans. Int Immunol (2023) 35:55–65. doi: 10.1093/intimm/dxac048
53. Krummey SM and Li XC. Multiple human B cell subsets give rise to regulatory B cells. Am J Transplant (2023) 23:3–4. doi: 10.1016/j.ajt.2022.11.010
54. Satitsuksanoa P, Iwasaki S, Boersma J, Imam MB, Schneider SR, Chang I, et al. B cells: The many facets of B cells in allergic diseases. J Allergy Clin Immunol (2023). doi: 10.1016/j.jaci.2023.05.011
55. Catalán D, Mansilla MA, Ferrier A, Soto L, Oleinika K, Aguillón JC, et al. Immunosuppressive mechanisms of regulatory B cells. Front Immunol (2021) 12:611795. doi: 10.3389/fimmu.2021.611795
56. Berland R and Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol (2002). doi: 10.1146/annurev.immunol.20.100301.064833
57. Dasgupta S, Dasgupta S, and Bandyopadhyay M. Regulatory B cells in infection, inflammation, and autoimmunity. Cell Immunol (2020). doi: 10.1016/j.cellimm.2020.104076
58. Mickael ME, Bieńkowska I, and Sacharczuk M. An update on the evolutionary history of bregs. Genes (Basel) (2022) 13. doi: 10.3390/genes13050890
59. Dwivedi S, Rendón-Huerta EP, Ortiz-Navarrete V, and Montaño LF. CD38 and regulation of the immune response cells in cancer. J Oncol (2021) 2021:1–11. doi: 10.1155/2021/6630295
60. Tompa A and Faresjö M. Shift in the B cell subsets between children with type 1 diabetes and/or celiac disease. Clin Exp Immunol (2024). doi: 10.1093/cei/uxad136
61. Liu Z, Zhao X, Shen H, Liu X, Xu X, and Fu R. Cellular immunity in the era of modern multiple myeloma therapy. Int J Cancer (2023) 153:1436–47. doi: 10.1002/ijc.34609
62. HSIEH T-Y, LUI S-W, LU J-W, CHEN Y-C, LIN T-C, JHENG W-L, et al. Using Treg, Tr1, and breg expression levels to predict clinical responses to csDMARD treatment in drug-naive patients with rheumatoid arthritis. In Vivo (Brooklyn) (2023) 37:2018–27. doi: 10.21873/invivo.13299
63. Pérez-Lara JC, Espinosa E, Santos-Argumedo L, Romero-Ramírez H, López-Herrera G, García-García F, et al. CD38 correlates with an immunosuppressive Treg phenotype in lupus-prone mice. Int J Mol Sci (2021) 22. doi: 10.3390/ijms222111977
64. Morris G, Puri BK, Olive L, Carvalho AF, Berk M, and Maes M. Emerging role of innate B1 cells in the pathophysiology of autoimmune and neuroimmune diseases: Association with inflammation, oxidative and nitrosative stress and autoimmune responses. Pharmacol Res (2019) 148:104408. doi: 10.1016/j.phrs.2019.104408
65. Murray SE, Toren KG, and Parker DC. Peripheral CD4+ T-cell tolerance is induced in vivo by rare antigen-bearing B cells in follicular, marginal zone, and B-1 subsets. Eur J Immunol (2013) 43:1818–27. doi: 10.1002/eji.201242784
66. Huai G, Markmann JF, Deng S, and Rickert CG. TGF-β-secreting regulatory B cells: unsung players in immune regulation. Clin Transl Immunol (2021) 10. doi: 10.1002/cti2.1270
67. Rastogi I, Jeon D, Moseman JE, Muralidhar A, Potluri HK, and McNeel DG. Role of B cells as antigen presenting cells. Front Immunol (2022) 13:954936. doi: 10.3389/fimmu.2022.954936
68. Lee-Chang C and Lesniak MS. Next-generation antigen-presenting cell immune therapeutics for gliomas. J Clin Invest (2023) 133. doi: 10.1172/JCI163449
69. Morlacchi S, Soldani C, Viola A, and Sarukhan A. Self-antigen presentation by mouse B cells results in regulatory T-cell induction rather than anergy or clonal deletion. Blood (2011) 118:984–91. doi: 10.1182/blood-2011-02-336115
70. Mann MK, Maresz K, Shriver LP, Tan Y, and Dittel BN. B cell regulation of CD4+CD25+ T regulatory cells and IL-10 Via B7 is essential for recovery from experimental autoimmune encephalomyelitis. J Immunol (2007) 178:3447–56. doi: 10.4049/jimmunol.178.6.3447
71. Hermankova B, Zajicova A, Javorkova E, Chudickova M, Trosan P, Hajkova M, et al. Suppression of IL-10 production by activated B cells via a cell contact-dependent cyclooxygenase-2 pathway upregulated in IFN-γ-treated mesenchymal stem cells. Immunobiology (2016) 221:129–36. doi: 10.1016/j.imbio.2015.09.017
72. Blair PA, Noreña LY, Flores-Borja F, Rawlings DJ, Isenberg DA, and Michael R Ehrenstein CM. CD19+ CD24 hi CD38 hi B cells exhibit regulatory capacity in healthy individuals but are functionally impaired in systemic lupus erythematosus patients. Immunity (2010) 32:129–40. doi: 10.1016/j.immuni.2009.11.009
73. Hu HT, Ai X, Lu M, Song Z, and Li H. Characterization of intratumoral and circulating IL-10-producing B cells in gastric cancer. Exp Cell Res (2019) 384. doi: 10.1016/j.yexcr.2019.111652
74. Wang RX, Yu CR, Dambuza IM, Mahdi RM, Dolinska MB, Sergeev YV, et al. Interleukin-35 induces regulatory B cells that suppress autoimmune disease. Nat Med (2014) 20:633–41. doi: 10.1038/nm.3554
75. Yu CR, Choi JK, Uche AN, and Egwuagu CE. Production of IL-35 by bregs is mediated through binding of BATF-IRF-4-IRF-8 complex to il12a and ebi3 promoter elements. J Leukoc Biol (2018) 104:1147–57. doi: 10.1002/JLB.3A0218-071RRR
76. Ma T, Renz BW, Ilmer M, Koch D, Yang Y, Werner J, et al. Myeloid-derived suppressor cells in solid tumors. Cells (2022) 11:310.
77. Jiménez-Cortegana C, Galassi C, Klapp V, Gabrilovich DI, and Galluzzi L. Myeloid-derived suppressor cells and radiotherapy. Cancer Immunol Res (2022) 10:545–57.
78. Wu Y, Yi M, Niu M, Mei Q, and Wu K. Myeloid-derived suppressor cells: an emerging target for anticancer immunotherapy. Mol Cancer (2022) 21:184.
79. Veglia F, Perego M, and Gabrilovich D. Myeloid-derived suppressor cells coming of age. Nat Immunol (2018) 19:108–19.
80. Veglia F, Sanseviero E, and Gabrilovich DI. Myeloid-derived suppressor cells in the era of increasing myeloid cell diversity. Nat Rev Immunol (2021) 21:485–98.
82. Millrud CR, Bergenfelz C, and Leandersson K. On the origin of myeloid-derived suppressor cells. Oncotarget (2017) 8:3649–65. doi: 10.18632/oncotarget.12278
83. Swatler J, Turos-Korgul L, Kozlowska E, and Piwocka K. Immunosuppressive cell subsets and factors in myeloid leukemias. Cancers (Basel) (2021) 13:1203. doi: 10.3390/cancers13061203
84. Xia S, Sha H, Yang L, Ji Y, Ostrand-Rosenberg S, and Qi L. Gr-1+ CD11b+ myeloid-derived suppressor cells suppress inflammation and promote insulin sensitivity in obesity. J Biol Chem (2011) 286:23591–9. doi: 10.1074/jbc.M111.237123
85. Ribechini E, Greifenberg V, Sandwick S, and Lutz MB. Subsets, expansion and activation of myeloid-derived suppressor cells. Med Microbiol Immunol (2010) 199:273–81. doi: 10.1007/s00430-010-0151-4
86. Verschoor CP, Johnstone J, Millar J, Dorrington MG, Habibagahi M, Lelic A, et al. Blood CD33(+)HLA-DR(–) myeloid-derived suppressor cells are increased with age and a history of cancer. J Leukoc Biol (2013) 93:633–7. doi: 10.1189/jlb.0912461
87. Greten TF, Manns MP, and Korangy F. Myeloid derived suppressor cells in human diseases. Int Immunopharmacol (2011) 11:802–7. doi: 10.1016/j.intimp.2011.01.003
88. Bunt SK, Sinha P, Clements VK, Leips J, and Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol (2006) 176:284–90. doi: 10.4049/jimmunol.176.1.284
89. Bunt SK, Clements VK, Hanson EM, Sinha P, and Ostrand-Rosenberg S. Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through toll-like receptor 4. J Leukoc Biol (2009) 85:996–1004. doi: 10.1189/jlb.0708446
90. Ibrahim ML, Klement JD, Lu C, Redd PS, Xiao W, Yang D, et al. Myeloid-derived suppressor cells produce IL-10 to elicit DNMT3b-dependent IRF8 silencing to promote colitis-associated colon tumorigenesis. Cell Rep (2018) 25:3036–3046.e6. doi: 10.1016/j.celrep.2018.11.050
91. Lee CR, Lee W, Cho SK, and Park SG. Characterization of multiple cytokine combinations and TGF-β on differentiation and functions of myeloid-derived suppressor cells. Int J Mol Sci (2018) 19. doi: 10.3390/ijms19030869
92. Highfill SL, Rodriguez PC, Zhou Q, Goetz CA, Koehn BH, Veenstra R, et al. Bone marrow myeloid-derived suppressor cells (MDSCs) inhibit graft-versus-host disease (GVHD) via an arginase-1-dependent mechanism that is up-regulated by interleukin-13. Blood (2010) 116:5738–47. doi: 10.1182/blood-2010-06-287839
93. Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, et al. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol (2009) 182:5693–701. doi: 10.4049/jimmunol.0900092
94. Andrés C, Pérez de la Lastra J, Juan C, Plou F, and Pérez-Lebeña E. Myeloid-derived suppressor cells in cancer and COVID-19 as associated with oxidative stress. Vaccines (2023) 11:218. doi: 10.3390/vaccines11020218
95. Zhao T, Liu S, Ding X, Johnson EM, Hanna NH, Singh K, et al. Lysosomal acid lipase, CSF1R, and PD-L1 determine functions of CD11c+ myeloid-derived suppressor cells. JCI Insight (2022) 7. doi: 10.1172/jci.insight.156623
96. Darcy CJ, Minigo G, Piera KA, Davis JS, McNeil YR, Chen Y, et al. Neutrophils with myeloid derived suppressor function deplete arginine and constrain T cell function in septic shock patients. Crit Care (2014) 18:R163. doi: 10.1186/cc14003
97. Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, et al. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res (2004) 64:5839–49. doi: 10.1158/0008-5472.CAN-04-0465
98. Raber PL, Thevenot P, Sierra R, Wyczechowska D, Halle D, Ramirez ME, et al. Subpopulations of myeloid-derived suppressor cells impair T cell responses through independent nitric oxide-related pathways. Int J Cancer (2014) 134:2853–64. doi: 10.1002/ijc.28622
99. Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, et al. Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J Clin Invest (2011) 121:4015–29. doi: 10.1172/JCI45862
100. Ugolini A, Tyurin VA, Tyurina YY, Tcyganov EN, Donthireddy L, Kagan VE, et al. Polymorphonuclear myeloid-derived suppressor cells limit antigen crosspresentation by dendritic cells in cancer. JCI Insight (2020) 5. doi: 10.1172/jci.insight.138581
101. Pramanik A and Bhattacharyya S. Myeloid derived suppressor cells and innate immune system interaction in tumor microenvironment. Life Sci (2022) 305:120755. doi: 10.1016/j.lfs.2022.120755
102. De Sanctis F, Adamo A, Canè S, and Ugel S. Targeting tumour-reprogrammed myeloid cells: the new battleground in cancer immunotherapy. Semin Immunopathol (2023) 45:163–86. doi: 10.1007/s00281-022-00965-1
103. Raskov H, Orhan A, Gaggar S, and Gögenur I. Neutrophils and polymorphonuclear myeloid-derived suppressor cells: An emerging battleground in cancer therapy. Oncogenesis (2022) 11:22.
104. Youn J-I, Collazo M, Shalova IN, Biswas SK, and Gabrilovich DI. Characterization of the nature of granulocytic myeloid-derived suppressor cells in tumor-bearing mice. J Leukoc Biol (2011) 91:167–81. doi: 10.1189/jlb.0311177
105. Dorhoi A and Du Plessis N. Monocytic myeloid-derived suppressor cells in chronic infections. Front Immunol (2018) 8:1895.
106. Ren X, Tao Q, Wang H, Zhang Q, Zhou M, Liu L, et al. Monocytic myeloid-derived suppressor cells but not monocytes predict poor prognosis of acute myeloid leukemia. Turkish J Hematol (2022) 39:230–6. doi: 10.4274/tjh.galenos.2022.2022.0137
107. Tomiyama T, Itoh S, Iseda N, Toshida K, Morinaga A, Yugawa K, et al. Myeloid-derived suppressor cell infiltration is associated with a poor prognosis in patients with hepatocellular carcinoma. Oncol Lett (2022) 23. doi: 10.3892/ol.2022.13213
108. Kohada Y, Kuromoto A, Takeda K, Iwamura H, Atobe Y, Ito J, et al. Circulating PMN-MDSC level positively correlates with a poor prognosis in patients with metastatic hormone-sensitive prostate cancer. Front Urol (2022) 2:967480. doi: 10.3389/fruro.2022.967480
109. Fan R, De Beule N, Maes A, De Bruyne E, Menu E, Vanderkerken K, et al. The prognostic value and therapeutic targeting of myeloid-derived suppressor cells in hematological cancers. Front Immunol (2022) 13:1016059. doi: 10.3389/fimmu.2022.1016059
110. Wang Y, Wang J, Zhu F, Wang H, Yi L, Huang K, et al. Elevated circulating myeloid-derived suppressor cells associated with poor prognosis in B-cell non-hodgkin’s lymphoma patients. Immunity Inflammation Dis (2022) 10. doi: 10.1002/iid3.616
111. Guo C, Hu F, Yi H, Feng Z, Li C, Shi L, et al. Myeloid-derived suppressor cells have a proinflammatory role in the pathogenesis of autoimmune arthritis. Ann Rheum Dis (2016) 75:278–85. doi: 10.1136/annrheumdis-2014-205508
112. Zhang H, Wang S, Huang Y, Wang H, Zhao J, Gaskin F, et al. Myeloid-derived suppressor cells are proinflammatory and regulate collagen-induced arthritis through manipulating Th17 cell differentiation. Clin Immunol (2015) 157:175–86. doi: 10.1016/j.clim.2015.02.001
113. Rahman S, Sagar D, Hanna RN, Lightfoot YL, Mistry P, Smith CK, et al. Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis (2019) 78:957–66. doi: 10.1136/annrheumdis-2018-214620
114. Huang A, Zhang B, Wang B, Zhang F, Fan K-X, and Guo Y-J. Increased CD14+ HLA-DR-/low myeloid-derived suppressor cells correlate with extrathoracic metastasis and poor response to chemotherapy in non-small cell lung cancer patients. Cancer Immunol Immunother (2013) 62:1439–51.
115. Florez-Pollack S, Tseng Lc, Kobayashi M, Hosler GA, Ariizumi K, and Chong BF. Expansion of myeloid-derived suppressor cells in the peripheral blood and lesional skin of cutaneous lupus patients. J Invest Dermatol (2019) 139:478–81. doi: 10.1016/j.jid.2018.08.023
116. Glenn JD, Liu C, and Whartenby KA. Frontline science: Induction of experimental autoimmune encephalomyelitis mobilizes Th17-promoting myeloid derived suppressor cells to the lung. J Leukoc Biol (2019) 105:829–41. doi: 10.1002/JLB.4HI0818-335R
117. Xue F, Yu M, Li L, Zhang W, Ma Y, Dong L, et al. Elevated granulocytic myeloid-derived suppressor cells are closely related with elevation of Th17 cells in mice with experimental asthma. Int J Biol Sci (2020) 16:2072–83. doi: 10.7150/ijbs.43596
118. Cripps JG and Gorham JD. MDSC in autoimmunity. Int Immunopharmacol (2011) 11:789–93. doi: 10.1016/j.intimp.2011.01.026
119. Park Y and Kwok S-K. Recent advances in cell therapeutics for systemic autoimmune diseases. Immune Netw (2022) 22. doi: 10.4110/in.2022.22.e10
120. Gordon JR, Ma Y, Churchman L, Gordon SA, and Dawicki W. Regulatory dendritic cells for immunotherapy in immunologic diseases. Front Immunol (2014) 5:7. doi: 10.3389/fimmu.2014.00007
121. Schülke S. Induction of interleukin-10 producing dendritic cells as a tool to suppress allergen-specific T helper 2 responses. Front Immunol (2018) 9:455. doi: 10.3389/fimmu.2018.00455
122. Du X, Chang S, Guo W, Zhang S, and Chen ZK. Progress in liver transplant tolerance and tolerance-inducing cellular therapies. Front Immunol (2020) 11:1326. doi: 10.3389/fimmu.2020.01326
123. Thomson AW, Zahorchak AF, Ezzelarab MB, Butterfield LH, Lakkis FG, and Metes DM. Prospective clinical testing of regulatory dendritic cells in organ transplantation. Front Immunol (2016) 7:15. doi: 10.3389/fimmu.2016.00015
124. Ritprajak, Kaewraemruaen, and Hirankarn. Current paradigms of tolerogenic dendritic cells and clinical implications for systemic lupus erythematosus. Cells (2019) 8:1291. doi: 10.3390/cells8101291
125. Li X, Yang A, Huang H, Zhang X, Town J, Davis B, et al. Induction of type 2 T helper cell allergen tolerance by IL-10-differentiated regulatory dendritic cells. Am J Respir Cell Mol Biol (2010) 42:190–9. doi: 10.1165/rcmb.2009-0023OC
126. Nayyar A, Dawicki W, Huang H, Lu M, Zhang X, and Gordon JR. Induction of prolonged asthma tolerance by IL-10–differentiated dendritic cells: Differential impact on airway hyperresponsiveness and the Th2 immunoinflammatory response. J Immunol (2012) 189:72–9. doi: 10.4049/jimmunol.1103286
127. Passeri L, Marta F, Bassi V, and Gregori S. Tolerogenic dendritic cell-based approaches in autoimmunity. Int J Mol Sci (2021) 22:8415. doi: 10.3390/ijms22168415
128. Ghobadinezhad F, Ebrahimi N, Mozaffari F, Moradi N, Beiranvand S, Pournazari M, et al. The emerging role of regulatory cell-based therapy in autoimmune disease. Front Immunol (2022) 13:1075813. doi: 10.3389/fimmu.2022.1075813
129. Giannoukakis N, Phillips B, Finegold D, Harnaha J, and Trucco M. Phase I (Safety) study of autologous tolerogenic dendritic cells in type 1 diabetic patients. Diabetes Care (2011) 34:2026–32. doi: 10.2337/dc11-0472
130. Benham H, Nel HJ, Law SC, Mehdi AM, Street S, Ramnoruth N, et al. Citrullinated peptide dendritic cell immunotherapy in HLA risk genotype–positive rheumatoid arthritis patients. Sci Transl Med (2015) 7. doi: 10.1126/scitranslmed.aaa9301
131. Bell GM, Anderson AE, Diboll J, Reece R, Eltherington O, Harry RA, et al. Autologous tolerogenic dendritic cells for rheumatoid and inflammatory arthritis. Ann Rheum Dis (2017) 76:227–34. doi: 10.1136/annrheumdis-2015-208456
132. Jauregui-Amezaga A, Cabezón R, Ramírez-Morros A, España C, Rimola J, Bru C, et al. Intraperitoneal administration of autologous tolerogenic dendritic cells for refractory crohn’s disease: A phase I study. J Crohn’s Colitis (2015) 9:1071–8. doi: 10.1093/ecco-jcc/jjv144
133. Akbari O, DeKruyff RH, and Umetsu DT. Pulmonary dendritic cells producing IL-10 mediate tolerance induced by respiratory exposure to antigen. Nat Immunol (2001) 2:725–31. doi: 10.1038/90667
134. Jaensson E, Uronen-Hansson H, Pabst O, Eksteen B, Tian J, Coombes JL, et al. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J Exp Med (2008) 205:2139–49. doi: 10.1084/jem.20080414
135. Yokota A, Takeuchi H, Maeda N, Ohoka Y, Kato C, Song S-Y, et al. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int Immunol (2009) 21:361–77. doi: 10.1093/intimm/dxp003
136. Enk AH. Dendritic cells in tolerance induction. Immunol Lett (2005) 99:8–11. doi: 10.1016/j.imlet.2005.01.011
137. Li R, Li H, Yang X, Hu H, Liu P, and Liu H. Crosstalk between dendritic cells and regulatory T cells: Protective effect and therapeutic potential in multiple sclerosis. Front Immunol (2022) 13:970508. doi: 10.3389/fimmu.2022.970508
138. Johnson BA, Baban B, and L’Mellor A. Targeting the immunoregulatory indoleamine 2,3 dioxygenase pathway in immunotherapy. Immunotherapy (2009) 1:645–61. doi: 10.2217/IMT.09.21
139. Gordon JR, Li F, Nayyar A, Xiang J, and Zhang X. CD8α+, but not CD8α–, dendritic cells tolerize Th2 responses via contact-dependent and -independent mechanisms, and reverse airway hyperresponsiveness, Th2, and eosinophil responses in a mouse model of asthma. J Immunol (2005) 175:1516–22. doi: 10.4049/jimmunol.175.3.1516
140. Lu X, Oh-Hora M, Takeda K, and Yamasaki S. Selective suppression of IL-10 transcription by calcineurin in dendritic cells through inactivation of CREB. Int Immunol (2022) 34:197–206. doi: 10.1093/intimm/dxab112
141. Wang F, Liu M, Ma D, Cai Z, Liu L, Wang J, et al. Dendritic cell-expressed IDO alleviates atherosclerosis by expanding CD4+CD25+Foxp3+Tregs through IDO-Kyn-AHR axis. Int Immunopharmacol (2023) 116:109758. doi: 10.1016/j.intimp.2023.109758
142. Safdarian AR, Farhangnia P, and Rezaei N. Indoleamine 2, 3-dioxygenase (IDO) and cancerous cells. In: Handbook of cancer and immunology. Springer (2023). p. 1–23.
143. Huang H, Dawicki W, Lu M, Nayyar A, Zhang X, and Gordon JR. Regulatory dendritic cell expression of MHCII and IL-10 are jointly requisite for induction of tolerance in a murine model of OVA-asthma. Allergy Eur J Allergy Clin Immunol (2013) 68:1126–35. doi: 10.1111/all.12203
144. Liu J, Zhang X, and Cao X. Dendritic cells in systemic lupus erythematosus: From pathogenesis to therapeutic applications. J Autoimmun (2022) 132:102856. doi: 10.1016/j.jaut.2022.102856
145. Dhodapkar MV and Steinman RM. Antigen-bearing immature dendritic cells induce peptide-specific CD8+ regulatory T cells in vivo in humans. Blood (2002) 100:174–7. doi: 10.1182/blood.V100.1.174
146. Dhodapkar MV, Steinman RM, Krasovsky J, Munz C, and Bhardwaj N. Antigen-specific inhibition of effector T cell function in humans after injection of immature dendritic cells. J Exp Med (2001) 193:233–8. doi: 10.1084/jem.193.2.233
147. Yuan H, Lanting L, Xu ZG, Li SA, Swiderski P, Putta S, et al. Effects of cholesterol-tagged small interfering RNAs targeting 12/15-lipoxygenase on parameters of diabetic nephropathy in a mouse model of type 1 diabetes. Am J Physiol - Ren Physiol (2008). doi: 10.1152/ajprenal.90268.2008
148. Singh R, Gholipourmalekabadi M, and Shafikhani SH. Animal models for type 1 and type 2 diabetes: advantages and limitations. Front Endocrinol (Lausanne) (2024) 15:1359685. doi: 10.3389/fendo.2024.1359685
149. Van Belle TL, Taylor P, and Von Herrath MG. Mouse models for type 1 diabetes. Drug Discovery Today Dis Model (2009) 6:41–5.
150. Khan FU, Khongorzul P, Raki AA, Rajasekaran A, Gris D, and Amrani A. Dendritic cells and their immunotherapeutic potential for treating type 1 diabetes. Int J Mol Sci (2022). doi: 10.3390/ijms23094885
151. Mukherjee G and Dilorenzo TP. The immunotherapeutic potential of dendritic cells in type 1 diabetes. Clin Exp Immunol (2010). doi: 10.1111/j.1365-2249.2010.04157.x
152. Steinman RM, Lustig DS, and Cohn ZA. IDENTIFICATION OF a NOVEL CELL TYPE IN PERIPHERAL LYMPHOID ORGANS OF MICE. J Exp Med (1974). doi: 10.1084/jem.139.6.1431
153. Giannoukakis N. Tolerogenic dendritic cells in type 1 diabetes: no longer a concept. Front Immunol (2023) 14:1212641. doi: 10.3389/fimmu.2023.1212641
154. Najafi S and Mortezaee K. Advances in dendritic cell vaccination therapy of cancer. BioMed Pharmacother (2023) 164:114954. doi: 10.1016/j.biopha.2023.114954
155. Huang C, Lyu C, Mok H-L, Xu Y, Cheng K-W, Zhang C, et al. Tolerogenic dendritic cell-mediated regulatory T cell differentiation by chinese herbal formulation attenuates colitis progression. J Adv Res (2024).
156. Bourque J and Hawiger D. Life and death of tolerogenic dendritic cells. Trends Immunol (2023) 44:110–8. doi: 10.1016/j.it.2022.12.006
157. Cao Q, Wang R, Wang Y, Niu Z, Chen T, Wang C, et al. Regulatory innate lymphoid cells suppress innate immunity and reduce renal ischemia/reperfusion injury. Kidney Int (2020) 97:130–42. doi: 10.1016/j.kint.2019.07.019
158. Tynecka M, Radzikowska U, and Eljaszewicz A. IL-10-producing innate lymphoid cells: Did we find a missing piece of the puzzle? Allergy Eur J Allergy Clin Immunol (2021) 76:3849–51. doi: 10.1111/all.14980
159. Thomas CM and Peebles RS. Development and function of regulatory innate lymphoid cells. Front Immunol (2022) 13:1014774. doi: 10.3389/fimmu.2022.1014774
160. Wang S, Qu Y, Xia P, Chen Y, Zhu X, Zhang J, et al. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res (2020) 30:610–22. doi: 10.1038/s41422-020-0312-y
161. Ferlazzo G, Thomas D, Lin S-L, Goodman K, Morandi B, Muller WA, et al. The abundant NK cells in human secondary lymphoid tissues require activation to express killer cell ig-like receptors and become cytolytic. J Immunol (2004) 172:1455–62. doi: 10.4049/jimmunol.172.3.1455
162. Strowig T, Brilot F, and Münz C. Noncytotoxic functions of NK cells: Direct pathogen restriction and assistance to adaptive immunity. J Immunol (2008) 180:7785–91. doi: 10.4049/jimmunol.180.12.7785
163. Perona-Wright G, Mohrs K, Szaba FM, Kummer LW, Madan R, Karp CL, et al. Systemic but not local infections elicit immunosuppressive IL-10 production by natural killer cells. Cell Host Microbe (2009) 6:503–12. doi: 10.1016/j.chom.2009.11.003
164. Martín-Fontecha A, Thomsen LL, Brett S, Gerard C, Lipp M, Lanzavecchia A, et al. Induced recruitment of NK cells to lymph nodes provides IFN-γ for TH1 priming. Nat Immunol (2004) 5:1260–5. doi: 10.1038/ni1138
165. Mailliard RB, Alber SM, Shen H, Watkins SC, Kirkwood JM, Herberman RB, et al. IL-18-induced CD83+CCR7+ NK helper cells. J Exp Med (2005) 202:941–53. doi: 10.1084/jem.20050128
166. Wong JL, Berk E, Edwards RP, and Kalinski P. IL-18-primed helper NK cells collaborate with dendritic cells to promote recruitment of effector CD8+ T cells to the tumor microenvironment. Cancer Res (2013) 73:4653–62. doi: 10.1158/0008-5472.CAN-12-4366
167. Viganò P, Gaffuri B, Somigliana E, Infantino M, Vignali M, and Di Blasio AM. Interleukin-10 is produced by human uterine natural killer cells but does not affect their production of interferon-γ. Mol Hum Reprod (2001) 7:971–7. doi: 10.1093/molehr/7.10.971
168. Deniz G, Erten G, Kücüksezer UC, Kocacik D, Karagiannidis C, Aktas E, et al. Regulatory NK cells suppress antigen-specific T cell responses. J Immunol (2008) 180:850–7. doi: 10.4049/jimmunol.180.2.850
169. Jinushi M, Takehara T, Tatsumi T, Kanto T, Miyagi T, Suzuki T, et al. Negative regulation of NK cell activities by inhibitory receptor CD94/NKG2A leads to altered NK cell-induced modulation of dendritic cell functions in chronic hepatitis C virus infection. J Immunol (2004) 173:6072–81. doi: 10.4049/jimmunol.173.10.6072
170. De Maria A, Fogli M, Mazza S, Basso M, Picciotto A, Costa P, et al. Increased natural cytotoxicity receptor expression and relevant IL-10 production in NK cells from chronically infected viremic HCV patients. Eur J Immunol (2007) 37:445–55. doi: 10.1002/eji.200635989
171. Ostapchuk YO, Cetin EA, Perfilyeva YV, Yilmaz A, Skiba YA, Chirkin AP, et al. Peripheral blood NK cells expressing HLA-g, IL-10 and TGF-β in healthy donors and breast cancer patients. Cell Immunol (2015) 298:37–46. doi: 10.1016/j.cellimm.2015.09.002
172. Tian Z, Gershwin ME, and Zhang C. Regulatory NK cells in autoimmune disease. J Autoimmun (2012) 39:206–15. doi: 10.1016/j.jaut.2012.05.006
173. Mikami N and Sakaguchi S. Regulatory T cells in autoimmune kidney diseases and transplantation. Nat Rev Nephrol (2023). doi: 10.1038/s41581-023-00733-w
174. Ibrahim EH, Aly M, Morath C, Sayed DM, Ekpoom N, Opelz G, et al. Relationship of transitional regulatory B and regulatory T cells and immunosuppressive drug doses in stable renal transplant recipients. Immunity Inflammation Dis (2021) 9:1252–71. doi: 10.1002/iid3.473
175. Ness S, Lin S, and Gordon JR. Regulatory dendritic cells, T cell tolerance, and dendritic cell therapy for immunologic disease. Front Immunol (2021) 12:633436. doi: 10.3389/fimmu.2021.633436
176. Gavrilova MV, Snegireva NA, and Sidorova EV. Influence of breg and IL-10 upon humoral immune response. Med Immunol (2016). doi: 10.15789/1563-0625-2016-4-331-338
177. Tumino N, Di Pace AL, Besi F, Quatrini L, Vacca P, and Moretta L. Interaction between MDSC and NK cells in solid and hematological malignancies: Impact on HSCT. Front Immunol (2021). doi: 10.3389/fimmu.2021.638841
178. Hanson EM, Clements VK, Sinha P, Ilkovitch D, and Ostrand-Rosenberg S. Myeloid-derived suppressor cells down-regulate l-selectin expression on CD4+ and CD8+ T cells. J Immunol (2009) 183:937–44. doi: 10.4049/jimmunol.0804253
179. Yu H, Kortylewski M, and Pardoll D. Crosstalk between cancer and immune cells: role of STAT3 in the tumour microenvironment. Nat Rev Immunol (2007) 7:41–51.
180. Hövelmeyer N, Schmidt-Supprian M, and Ohnmacht C. NF-κB in control of regulatory T cell development, identity, and function. J Mol Med (2022) 100:985–95.
181. McNee A, Kannan A, Jull P, and Shankar S. Expanding human breg for cellular therapy in transplantation: Time for translation. Transplantation (2024), 10–1097.
182. Lee KM, Stott RT, Zhao G, SooHoo J, Xiong W, Lian MM, et al. TGF-β-producing regulatory B cells induce regulatory T cells and promote transplantation tolerance. Eur J Immunol (2014) 44:1728–36.
183. Najar M, Raicevic G, Fayyad-Kazan H, Bron D, Toungouz M, and Lagneaux L. Mesenchymal stromal cells and immunomodulation: A gathering of regulatory immune cells. Cytotherapy (2016) 18:160–71. doi: 10.1016/j.jcyt.2015.10.011
184. Wood KJ, Bushell A, and Hester J. Regulatory immune cells in transplantation. Nat Rev Immunol (2012) 12:417–30. doi: 10.1038/nri3227
185. Neurath MF, Sands BE, and Rieder F. Cellular immunotherapies and immune cell depleting therapies in inflammatory bowel diseases: the next magic bullet? Gut (2025) 74:9–14.
186. Fenis A, Demaria O, Gauthier L, Vivier E, and Narni-Mancinelli E. New immune cell engagers for cancer immunotherapy. Nat Rev Immunol (2024), 1–16.
187. Ho P, Cahir-McFarland E, Fontenot JD, Lodie T, Nada A, Tang Q, et al. Harnessing regulatory T cells to establish immune tolerance. Sci Transl Med (2024) 16:eadm8859.
188. Yang Z, Liu Y, and Zhao H. CAR T treatment beyond cancer: Hope for immunomodulatory therapy of non-cancerous diseases. Life Sci (2024) 344:122556. doi: 10.1016/j.lfs.2024.122556
189. Feng Y, Yang Z, Wang J, and Zhao H. Cuproptosis: unveiling a new frontier in cancer biology and therapeutics. Cell Commun Signal (2024) 22. doi: 10.1186/s12964-024-01625-7
190. Che S, Yan Z, Feng Y, and Zhao H. Unveiling the intratumoral microbiota within cancer landscapes. Iscience (2024).
191. Wang H, Liu Y, Che S, Li X, Tang D, Lv S, et al. Deciphering the link: ferroptosis and its role in glioma. Front Immunol. (2024) 15:1346585.
Keywords: Tregs, CD8+ Tregs, Bregs, MDSC, DCregs, ILCregs, NKregs
Citation: Shi P, Yu Y, Xie H, Yin X, Chen X, Zhao Y and Zhao H (2025) Recent advances in regulatory immune cells: exploring the world beyond Tregs. Front. Immunol. 16:1530301. doi: 10.3389/fimmu.2025.1530301
Received: 18 November 2024; Accepted: 31 March 2025;
Published: 16 May 2025.
Edited by:
Sudhir Gupta, University of California, Irvine, United StatesReviewed by:
Hridesh Banerjee, University of Pittsburgh, United StatesGul Ahmad, Peru State College, United States
Copyright © 2025 Shi, Yu, Xie, Yin, Chen, Zhao and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Zhao, eWlkYW9AcWR1LmVkdS5jbg==
 Peng Shi1
Peng Shi1 Hai Zhao
Hai Zhao