- 1Department of Thoracic Surgery, Tangdu Hospital, Air Force Medical University, Xi’an, China
- 2College of Life Sciences, Northwest University, Xi’an, China
- 3Department of Pathology, Tangdu Hospital, Air Force Medical University, Xi’an, China
- 4Department of Pharmacy, Tangdu Hospital, Air Force Medical University, Xi’an, China
- 5Department of Thoracic Surgery, The First People’s Hospital of Xianyang, Xianyang, China
- 6Basic Medical College, Air Force Medical University, Xi’an, China
- 7Department of Thoracic Surgery, Air Force Medical Center, Xi’an, China
Background: Esophageal squamous cell carcinoma (ESCC) treatment often involves neoadjuvant therapy combining chemotherapy and immune checkpoint inhibitors. However, the effectiveness of these treatments is limited by immune infiltration in the tumor microenvironment.
Methods: We analyzed single-cell transcriptomic data from 22 patients with resectable ESCC, collected before and after neoadjuvant therapy. Differences in gene expression between patients achieving a complete pathological response (pCR) and those who did not were assessed. We further validated our findings using RNAseq data from The Cancer Genome Atlas (TCGA), and conducted quantitative qRT-PCR and Western blot analyses on tumor tissues from a clinical cohort.
Results: Significant differences in gene expression related to T cell activation, natural killer cell activity, and cytokine signaling were observed between pCR and non-pCR patients. Notable genes included CXCL10, CXCL11, ME1, MT1X, FAT1, OAS2, and MT2A. TCGA data confirmed a correlation between high gene expression and increased tumor mutational burden as well as improved survival rates, particularly for CXCL10. qRT-PCR revealed significant upregulation of CXCL10, CXCL11, ME1, MT1X, FAT1, OAS2, and MT2A in tumor tissues compared to normal tissues. Western blot analysis showed increased protein levels of CXCL10, CXCL11, OAS2, MT1E, and MT1X, while FAT1 was downregulated.
Conclusion: Our study highlights the critical role of immune infiltration and associated molecular pathways in the efficacy of neoadjuvant immunotherapy for ESCC. Specific genes, such as CXCL10, are promising as predictive markers for treatment response and survival.
1 Introduction
Esophageal squamous cell carcinoma (ESCC) represents the predominant histologic type globally, particularly prevalent in developing countries. Despite incremental advances in diagnostics and therapeutics, ESCC presents a grim 5-year survival rate ranging from 12-20% (1). The standard treatment for locally advanced ESCC involves neoadjuvant chemoradiotherapy combined with surgery, following the success of the CROSS trial (2). However, this approach can heighten toxicity levels, leading to severe side effects such as pneumonia and myocardial injury (3).
In recent years, the integration of immune checkpoint inhibitors (ICIs), targeting programmed cell death 1 (PD-1) and its ligand PD-L1, has emerged as a promising neoadjuvant treatment strategy for early-stage solid tumors, including breast cancer, lung cancer, and ESCC (4, 5). Clinical trials such as KEYNOTE 590 and CheckMate 649 have shown promising antitumor activity and safety of immunotherapies with or without chemotherapy in advanced ESCC patients (6, 7). These results provide a strong rationale for utilizing ICIs in the preoperative treatment setting for ESCC.
Our study aims to investigate the impact of immune infiltration-related genes on the efficacy of neoadjuvant immunotherapy for esophageal cancer. Through the SCALE-1 exploratory study (8), we conducted an in-depth analysis of biomarkers associated with PD-1 treatment response. We found distinct gene expression profiles between patients achieving pathological complete response (pCR) and those without pCR, indicating the potential role of immune-related genes in treatment outcomes. Notably, genes involved in T cell activation, natural killer cell activity, cytokine and chemokine signaling, and IFN-γ response pathways showed significant differential expression, suggesting their importance in modulating the tumor microenvironment.
Furthermore, integrating our findings with data from The Cancer Genome Atlas (TCGA) provided additional insights into the molecular landscape of esophageal cancer. Consistent upregulation of identified genes in esophageal cancer samples, along with their association with higher tumor mutational burden (9) and survival rates, supports their consideration as potential predictive markers for immunotherapy response.
In summary, our study highlights the critical role of immune infiltration intensity and its molecular determinant — CXCL10 in shaping the efficacy of neoadjuvant immunotherapy for esophageal cancer. These findings deepen our understanding of tumor-immune interactions and offer implications for personalized treatment strategies in this challenging malignancy.
2 Materials and methods
2.1 The origin of single-cell RNA sequencing data
The data for our article were derived from the single-cell data of the previous study (10) and subjected to conventional cell annotation analysis based on the methods outlined in the previous article.
2.2 scRNA-seq data processing
Based on scRNA-seq data and H&E pathological examination results from esophageal squamous cell carcinoma (ESCC) patients undergoing neoadjuvant chemoimmunotherapy, we categorized pre-treatment (T_B) and post-treatment (T_A) ESCC tumors into three distinct groups: pathological complete response (pCR_T_B, pCR_T_A), major pathological response (MPR_T_B, MPR_T_A), and incomplete pathological response (IPR_T_B, IPR_T_A) groups. Combined with the previous comprehensive analysis of the tumor immune microenvironment (TIME) (using the data in Appendix Table A1 of the SCALE-1 trial, which lists 289 immune-related genes), we used the R package “Seurat” (11) to perform differential expression analysis and obtained 9 genes (CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD27, CD79A, KLRB1, and TNFRSF17) (12–18).
2.3 Analyzing the expression of key 9 genes based on ESCA sample data from TCGA
We obtained 182 esophageal carcinoma (ESCA) samples and 13 normal samples from the TCGA database (https://portal.gdc.cancer.gov/). Comparing normal samples to ESCA samples, we analyzed the mRNA expression levels of CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD79A, KLRB1, and TNFRSF17 in ESCA cases using the R package “DESeq2” (19).
2.4 Analysis of tumor mutation burden about those key 9 genes
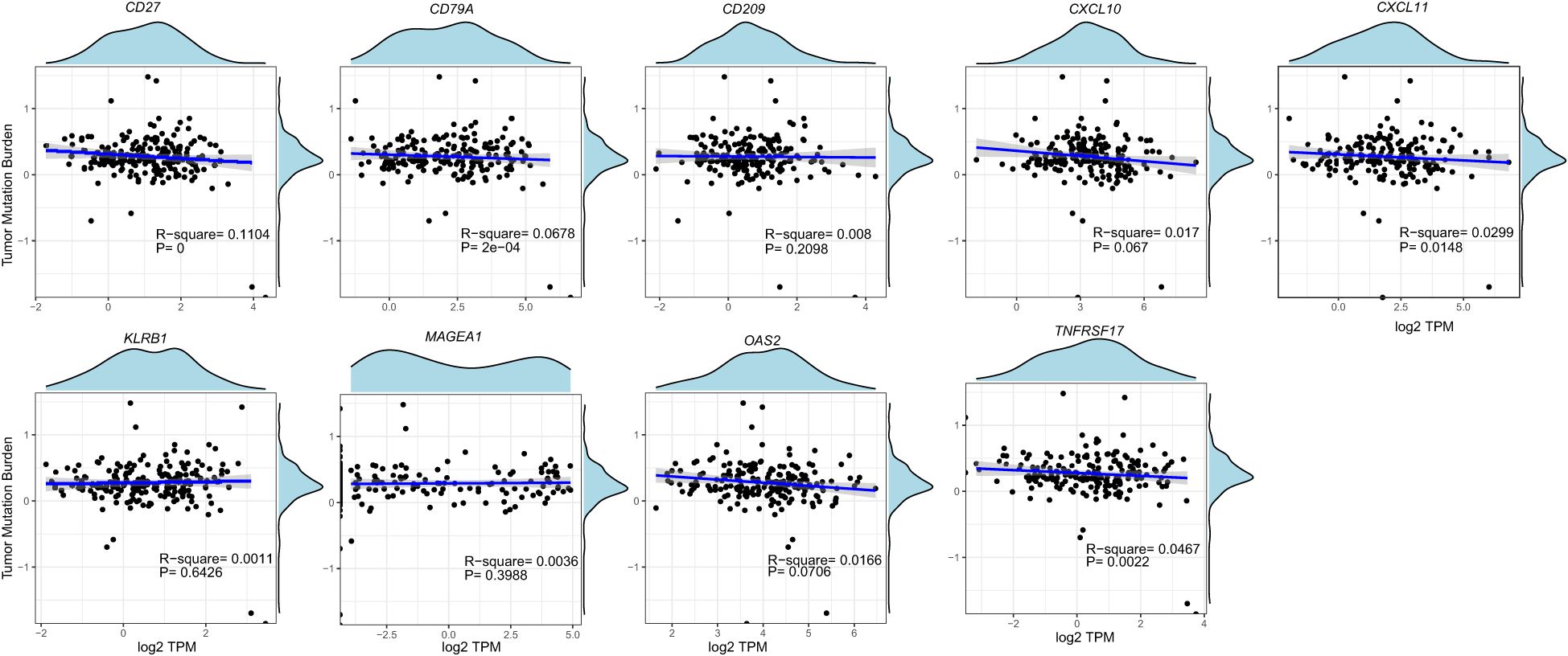
Mutation data of ESCA cases were retrieved from the TCGA database, and tumor mutational burden (TMB) was calculated using the R package “maftools” (20). Pearson correlation analysis (21) was employed to examine the relationship among CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD27, CD79A, KLRB1, and TNFRSF17.
2.5 Survival curve analysis
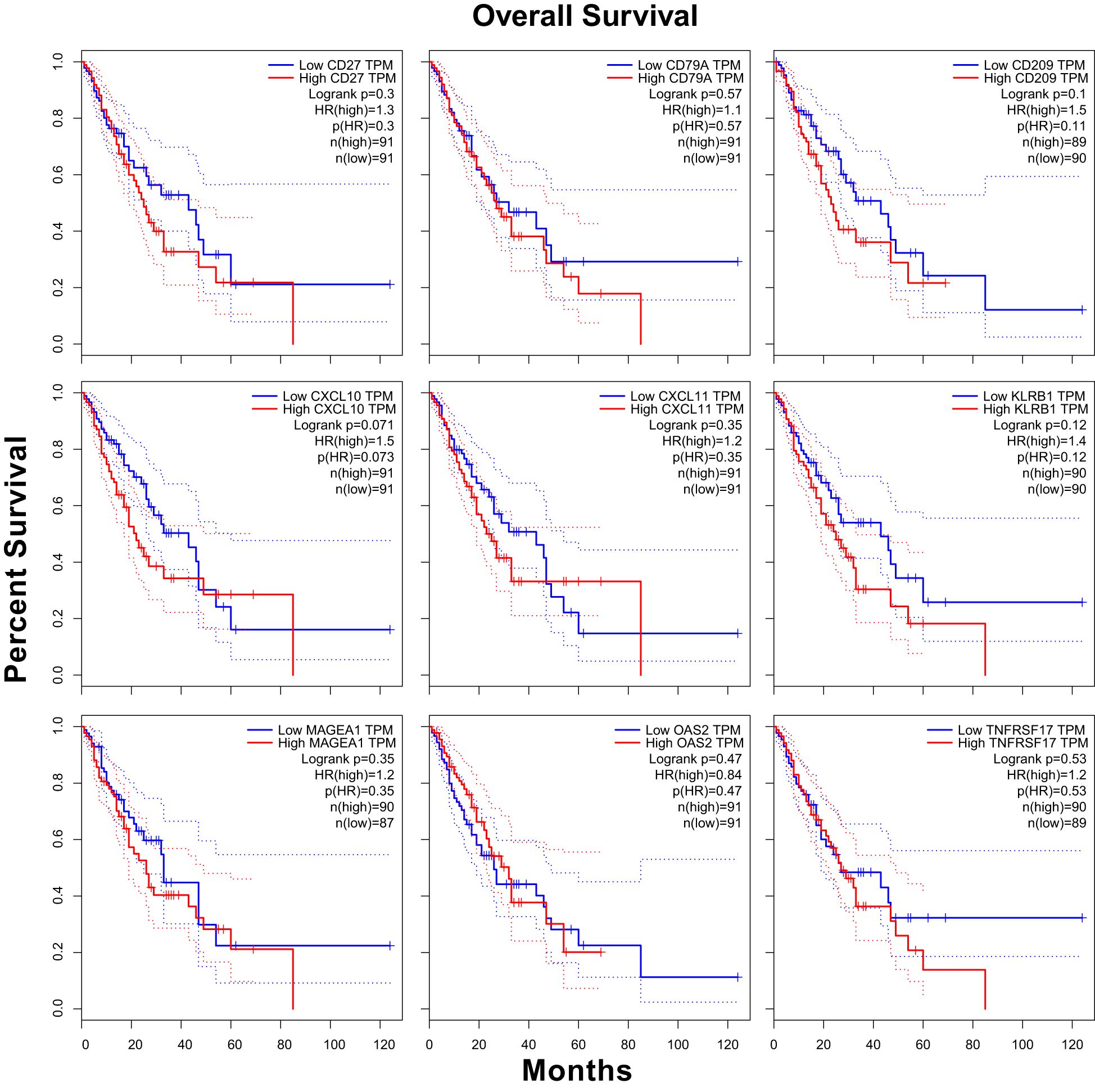
In this study, 182 ESCA cases were stratified into high-expression and low-expression groups based on the median values of CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD27, CD79A, KLRB1, and TNFRSF17 mRNA expression. Kaplan-Meier survival curves (22) were generated using the R package “survminer”.
2.6 Analysis of half maximal inhibitory concentration (IC50)
The Cyclopamine sensitivity (23) of CXCL10 in the high-expression and low-expression groups was analyzed using the GEPIA2 (24) web tool (http://gepia2.cancer-pku.cn/#index), and relevant sensitivity bar graphs were generated.
2.7 Real-time quantitative PCR and western blot of nine genes
Total RNA was extracted from culture cells using TRI pure based on the manufacturer’s instructions. The OD value of total RNA was detected and used for subsequent RT-PCR quantification. Then, a Prime Script TM reagent Kit with gDNA Eraser was used to reverse-transcribe 1 µg of total RNA in a 20 µL volume into cDNA. Quantitative real-time PCR was performed using the Quantitative Real-time PCR Kit. All primers were designed and synthesized by Shanghai Integrated Biotech Solutions Co., Ltd. (Shanghai, China) Supplementary Table S1. The results were normalized using GAPDH as an internal control. Western blot analysis was conducted as previously described. 6 Samples were probed with anti-genes (9) (Abcam, Cambridge, UK), or anti-actin (Sigma-Aldrich). Densitometric analysis was carried out using Image J software (Version 1.44o; NIH).
2.8 Statistical analysis
Statistical analyses were performed using R software (version 4.1.2, https://www.r-project.org/). To compare continuous variables between two groups, an independent Student’s t-test was used for normally distributed data, while the Mann–Whitney U-test was applied for non-normally distributed data. All p-values were calculated using a two-tailed approach, with significance defined as P < 0.05.
3 Results
3.1 The expression of nine key genes in ESCA before/after neoadjuvant chemoimmunotherapy
Using previously collected single-cell RNA sequencing (scRNA-seq) data from ESCA diseased tissues, we investigated the expression levels of CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD79A, KLRB1, and TNFRSF17 in various groups before and after treatment. Our study revealed that expression of OAS2 (Supplementary Figure 1c) was significantly increased in the IPR_T_B group.
3.2 Expression of nine genes in various subtypes of different samples
Based on early cell subtype annotation, we conducted differential analysis on the same single-cell subtypes across different samples, primarily observing changes in the expression of the nine genes mentioned above. We identified significant differential expression in seven genes: CD27, CD79A, KLRB1, CD209, CXCL10, OAS2, and TNFRSF1. Notably, CD79A showed consistent changes across all differential analyses in the B_cells subtype (Figure 1). Similarly, CD209 displayed changes in both the MP and Mural_cells subtypes. In the Plasma_cells subtype, CD27, CD79A, and TNFRSF1 demonstrated consistent changes. Lastly, in the T_cells subtype, CD27 and KLRB1 showed consistent alterations (Figure 2).
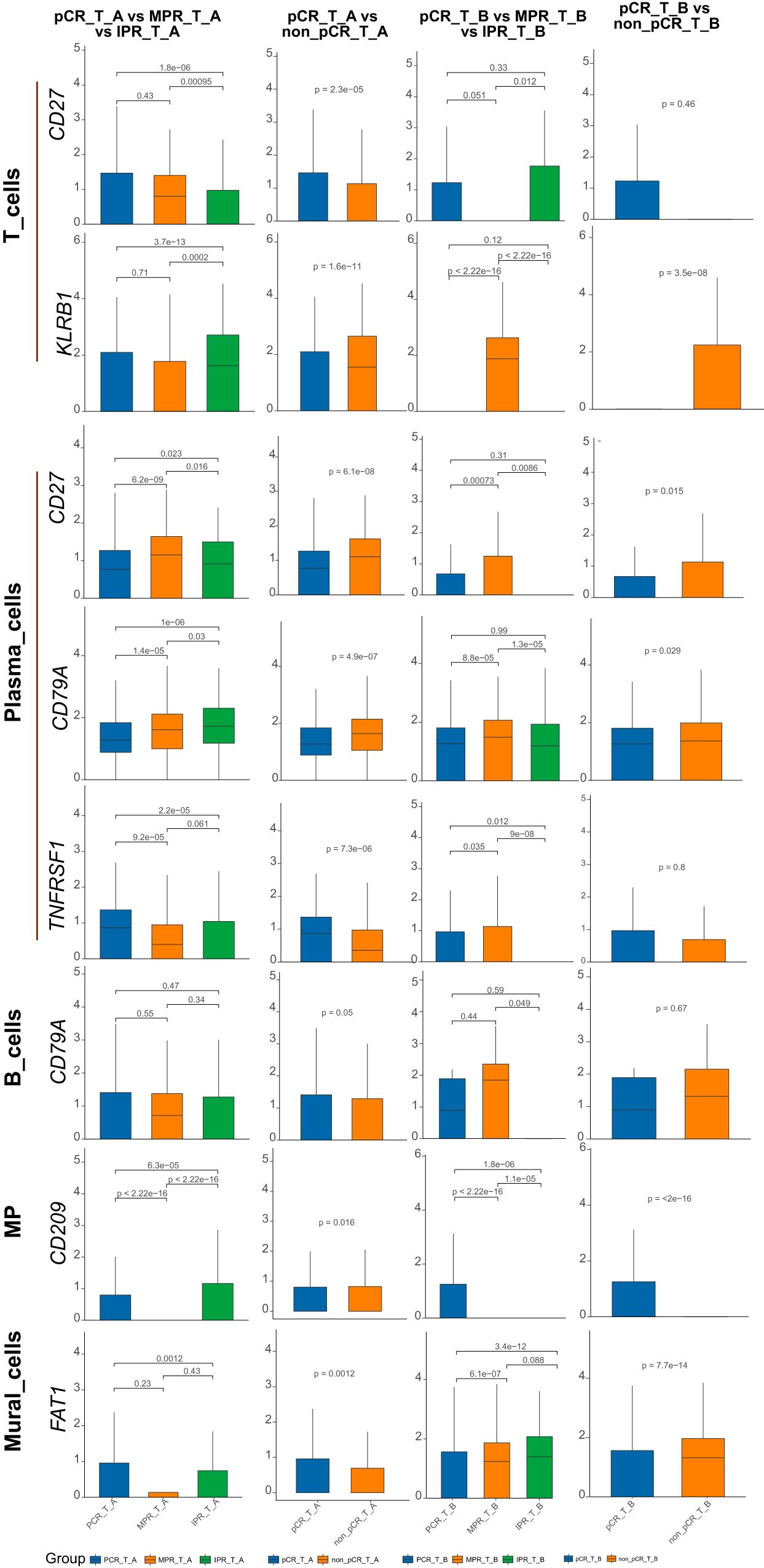
Figure 1. Nine genes expression in ESCA subtype cells before/after neoadjuvant chemoimmunotherapy. CD27, CD79A, FAT1, KLRB1, CD209, CXCL10, OAS2, TNFRSF1 showed significant differential expression across the subtypes in the above barplot. Specifically, in all differential analyses, CD79A exhibited changes in the B_cells subtype. CD209 displayed alterations in both the MP and Mural_cells subtypes. CD27, CD79A, and TNFRSF1 showed changes in the Plasma_cells subtype, while CD27 and KLRB1 exhibited alterations in the T_cells subtype.
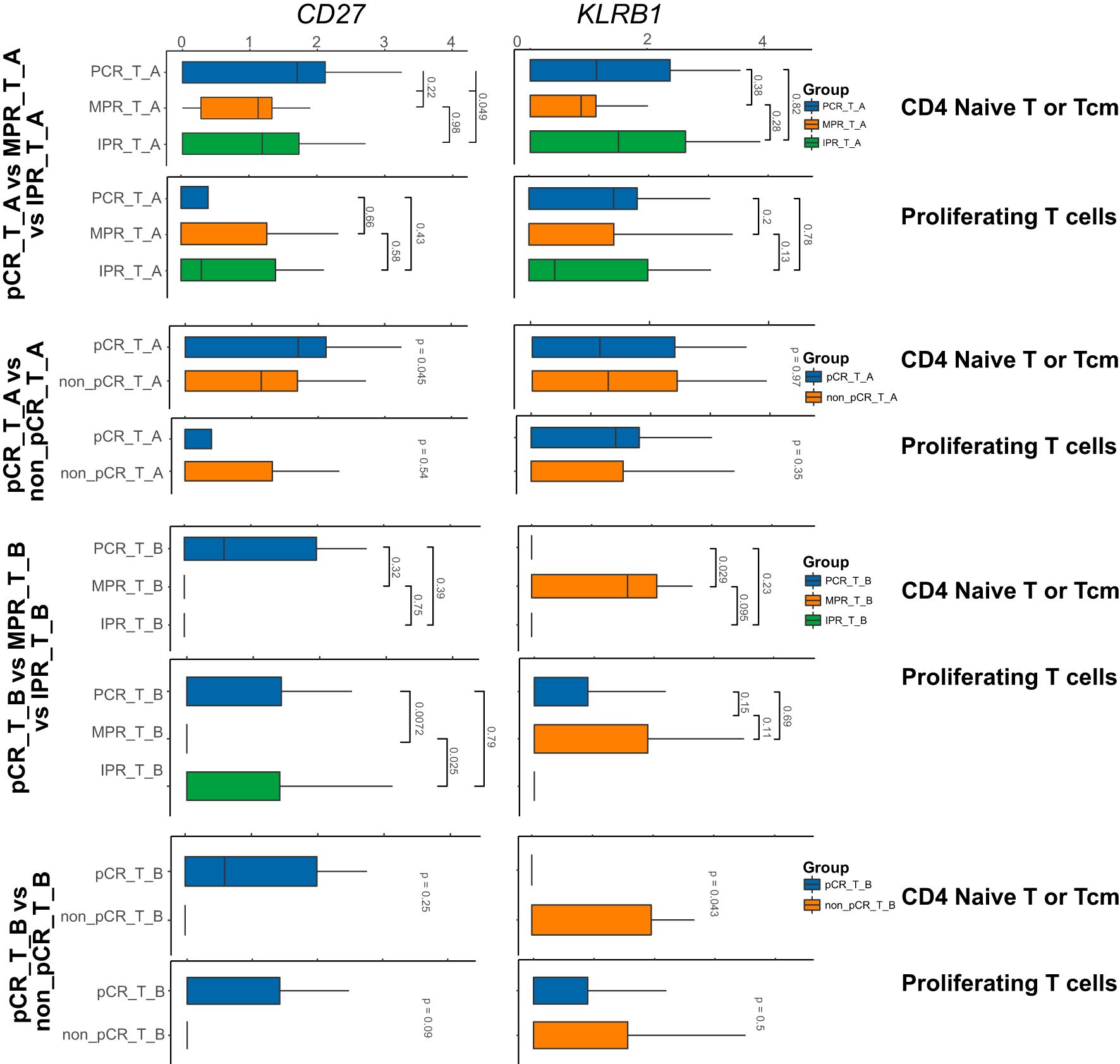
Figure 2. Nine genes expression in the subtype cells of T subtype-cell before/after neoadjuvant chemoimmunotherapy. In the above barplot, CD4_Naive_T and ProlifT subtypes exhibited significant differences in CD27 and KLRB1 expression. In the CD8_exhausted_T subtype, significant differences were observed in CD27, KLRB1, and OAS2 expression. Additionally, except for Treg, KLRB1 was identified as a significantly differentially expressed gene in the remaining subtypes.
The expression of CXCL10 showed significant differences in the MP subtype of PCR_T_B, MPR_T_B, and IPR_T_B samples, with a notable increase in IPR_T_B and extremely low expression in MPR_T_B. This pattern was similarly observed in the MP subtype of PCR_T_B and non_PCR_T_B samples. However, CXCL10 exhibited a significant increase in expression in the pCR_T_B subtype and extremely low expression in the non_pCR_T_B subtype (Supplementary Figures 2a, b).
We further investigated the expression differences of these genes in the T cell subtypes of different samples through group differential analysis. The results revealed that CD27, KLRB1, OAS2, and FAT1 exhibited relatively significant changes across various T cell subtypes, with distinct genes showing significant changes in different subtypes. Specifically, CD27 and KLRB1 showed significant differences in CD4_Naive_T and ProlifT subtypes, while CD27, KLRB1, and OAS2 exhibited significant differences in the CD8_exhausted_T subtype. Additionally, apart from Treg, KLRB1 was identified as a significant differentially expressed gene in the remaining subtypes (Figure 3).
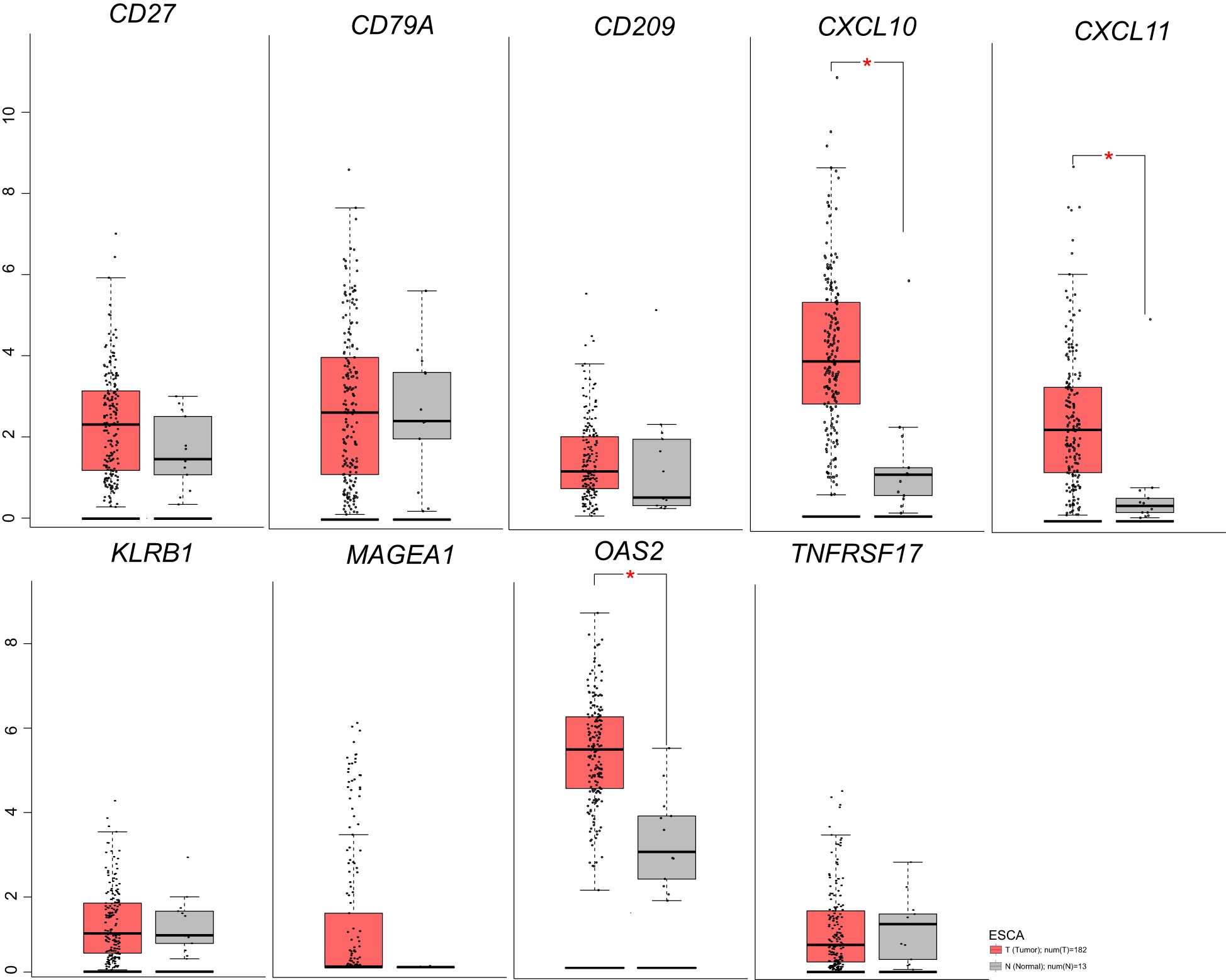
Figure 3. Nine genes expression in ESCA before/after neoadjuvant chemoimmunotherapy based on TGCA database.
3.3 Analysis of nine genes expression in ESCA via the TCGA Database
We analyzed ESCA data from the TCGA database to investigate the differential expression of nine genes. Our findings revealed an overall increase in the expression of CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD27, CD79A, KLRB1, and TNFRSF17 in ESCA patients compared to the normal group (Figure 3). Specifically, CXCL10, CXCL11, and OAS2 exhibited significant elevation, while the expression of other genes showed a slight increase. Additionally, our analysis unveiled that, except for the CXCL10 gene, there was no significant correlation between the expression of these genes and the tumor mutational burden (TMB) (Figure 4).
3.4 The association between the expression of nine genes and prognosis in ESCA
We conducted survival curve analysis for CXCL10, CXCL11, MAGEA1, OAS2, CD209, CD27, CD79A, KLRB1, and TNFRSF17 in a sample of 182 ESCA cases from the TCGA database. Our findings indicated that the group with high CXCL10 expression exhibited a significantly lower survival probability and relatively poorer prognosis compared to the group with low CXCL10 expression (HR = 1.5, p(HR) = 0.073) (25, 26) (Figure 5).
3.5 Analysis of half maximal inhibitory concentration (IC50) on CXCL10 gene
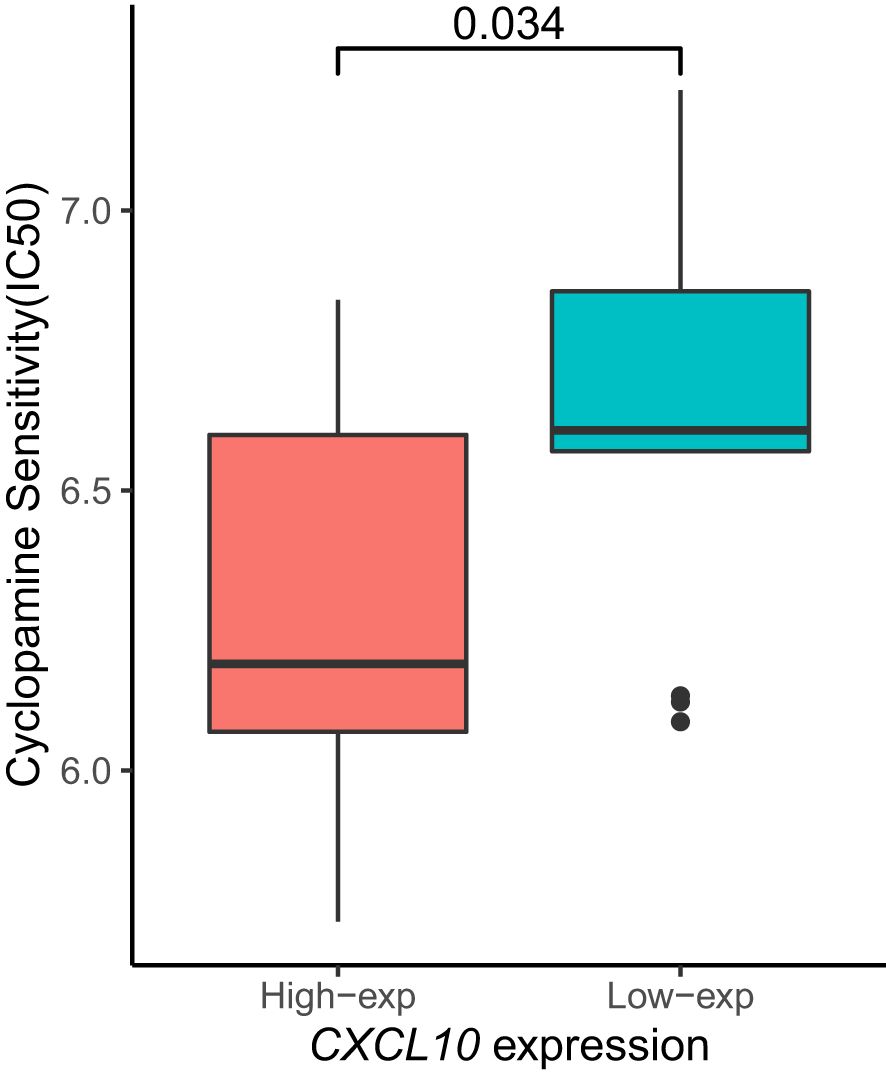
Subsequently, we aimed to investigate the sensitivity of patients stratified by high or low expression of the CXCL10 gene to specific therapeutic drugs, thus performing an analysis of the half maximal inhibitory concentration (IC50). We found that the disease group with low CXCL10 expression exhibited higher sensitivity to Cyclopamine (Figure 6).
3.6 GeneQuantitative PCR and WB to detect the expression of 9 genes
In order to further validate the expression of these nine genes in esophageal cancer, we conducted a study involving clinical cases where we assessed the transcriptome and protein levels of these genes. Our qRT-PCR analysis revealed significant differences in 8 genes between tumor tissues and normal tissues (Figure 7), including CXCL10, CXCL11, ME1E, MT1X, FAT1, OAS2, MT2A and CD209. These findings were consistent across different samples, showing a notable increase in expression levels in tumor tissues compared to normal tissues, except for CD209 which displayed higher expression in normal tissues. This trend was similarly observed at the protein level, as Western blot results indicated significant upregulation of CXCL10, CXCL11, OAS2, MTIE, and MTIX in tumor tissues, although FAT1 exhibited an opposite trend compared to the qPCR results (Figure 8). Notably, CD209 expression mirrored the qPCR results, showing higher levels in normal tissues.
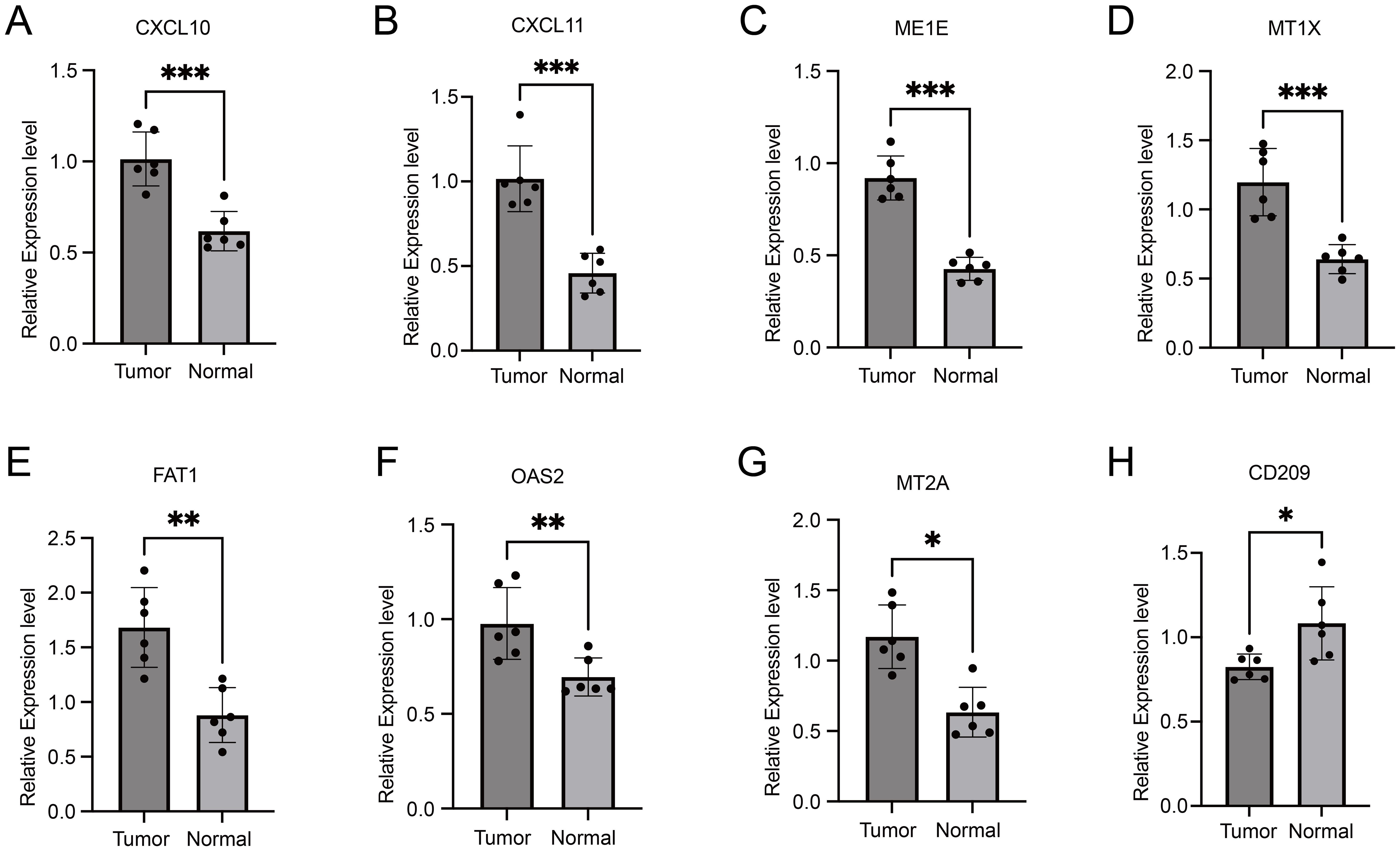
Figure 7. Comparative qRT-PCR analysis of gene expression levels in tumor tissues versus normal tissues. Asterisks indicating levels of significance: ***p < 0.001, **p < 0.01, *p < 0.05.

Figure 8. The expression of nine proteins by western blot in normal and tumor tissues. Western blot analysis revealed a significant upregulation of CXCL10, CXCL11, OAS2, MTIE, and MTIX proteins in tumor tissues, which is consistent with the qPCR data. In contrast, FAT1 exhibited a downregulation in tumor tissues, presenting an opposite trend to the qPCR results. The expression of CD209, mirrored the qPCR results, with higher levels observed in normal tissues.
4 Discussion
Based on our findings, we propose that CXCL10 could serve as a potential prognostic marker to guide therapeutic interventions for esophageal cancer. Neoadjuvant therapy, comprising chemotherapy and immune checkpoint inhibitors, represents a promising approach for treating esophageal cancer, particularly squamous cell carcinoma. However, the efficacy of this treatment modality is often compromised by immune infiltration in the tumor microenvironment.
Our study utilized single-cell transcriptomic analysis of resectable esophageal cancer patients before and after neoadjuvant therapy, revealing significant differences in gene expression between those achieving a complete pathological response (pCR) (27) and those who did not. Notably, genes associated with T cell activation, natural killer cell activity, and cytokine signaling exhibited (28–31) substantial alterations, suggesting their potential as predictive markers for treatment response.
Further analysis of RNAseq data from The Cancer Genome Atlas (TCGA) corroborated our findings, indicating a correlation between high gene expression levels, particularly CXCL10, with greater tumor mutational burden and improved survival rates. This underscores the importance of considering immune-related gene expression profiles in determining patient prognosis and treatment outcomes.
Moreover, our investigation into the sensitivity of patients with varying CXCL10 expression levels to specific therapeutic drugs revealed that those with low CXCL10 expression exhibited increased sensitivity to Cyclopamine. This highlights the potential utility of CXCL10 expression as a biomarker for predicting treatment response and guiding personalized therapeutic strategies in esophageal cancer.
In conclusion, our study contributes to a deeper understanding of tumor-immune interactions and offers valuable insights for optimizing treatment strategies in esophageal cancer. By identifying CXCL10 as a potential prognostic marker and elucidating its role in therapeutic sensitivity, we pave the way for the development of more tailored and effective treatment approaches for this challenging malignancy.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
YZ: Data curation, Resources, Writing – original draft. XX: Data curation, Resources, Writing – original draft. XM: Data curation, Resources, Writing – original draft. JW: Data curation, Resources, Writing – original draft. JZ: Formal Analysis, Software, Visualization, Writing – original draft. GX: Formal Analysis, Software, Visualization, Writing – original draft. JL: Formal Analysis, Software, Visualization, Writing – original draft. CZ: Formal Analysis, Software, Visualization, Writing – original draft. HW: Formal Analysis, Software, Visualization, Writing – original draft. QL: Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by National Natural Fund Youth Project of China (82002422) and Key Research and Development Program Project of Shaanxi Province (2024SF-YBXM-112).
Acknowledgments
This study partially utilized data from the research by Li C, Song W, Zhang J et al. (DOI: 10.3389/fimmu.2023.1322147). We sincerely thank the authors for their valuable contribution and for providing the data. Additionally, data from The Cancer Genome Atlas (TCGA) were also used in this study. We are grateful to the TCGA project team and all data contributors for their generous data sharing, which was crucial for the successful completion of this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1543283/full#supplementary-material
Supplementary Figure 1 | (a) Nine genes expression in ESCA samples containing PCR_T_A, MPR_T_A, and IPR_T_A. (b) Nine genes expression in ESCA samples containing pCR_T_A, and non_pCR_T_A. (c) Nine genes expression in ESCA samples containing PCR_T_B, MPR_T_B, and IPR_T_B. (d) Nine genes expression in ESCA samples containing pCR_T_B, and non_pCR_T_B.
Supplementary Figure 2 | (a) Three genes expression in ESCA subtype cells before/after neoadjuvant chemoimmunotherapy. (b) One gene expression in ESCA subtype cells before/after neoadjuvant chemoimmunotherapy.
References
1. Napier KJ, Scheerer M, and Misra S. Esophageal cancer: A Review of epidemiology, pathogenesis, staging workup and treatment modalities. World J Gastrointestinal Oncol. (2014) 6:112–20. doi: 10.4251/wjgo.v6.i5.112
2. van Hagen P, Hulshof MCCM, van Lanschot JJB, Steyerberg EW, Henegouwen MIV, Wijnhoven BPL, et al. Preoperative chemoradiotherapy for esophageal or junctional cancer. New Engl J Med. (2012) 366:2074–84. doi: 10.1056/NEJMoa1112088
3. Zhang Z and Zhang H. Impact of neoadjuvant chemotherapy and chemoradiotherapy on postoperative cardiopulmonary complications in patients with esophageal cancer. Dis Esophagus. (2017) 30:1–7. doi: 10.1093/dote/dox002
4. Shu CA, Gainor JF, Awad MM, Chiuzan C, Grigg CM, Pabani A, et al. Neoadjuvant atezolizumab and chemotherapy in patients with resectable non-small-cell lung cancer: an open-label, multicentre, single-arm, phase 2 trial. Lancet Oncol. (2020) 21:786–95. doi: 10.1016/S1470-2045(20)30140-6
5. Schmid P, Salgado R, Park YH, Muñoz-Couselo E, Kim SB, Sohn J, et al. Pembrolizumab plus chemotherapy as neoadjuvant treatment of high-risk, early-stage triple-negative breast cancer: results from the phase 1b open-label, multicohort KEYNOTE-173 study. Ann Oncol. (2020) 31:569–81. doi: 10.1016/j.annonc.2020.01.072
6. Sun JM, Shen L, Shah MA, Enzinger P, Adenis A, Doi T, et al. Pembrolizumab plus chemotherapy versus chemotherapy alone for first-line treatment of advanced oesophageal cancer (KEYNOTE-590): a randomised, placebo-controlled, phase 3 study. Lancet. (2021) 398:759–71. doi: 10.1016/S0140-6736(21)01234-4
7. Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet. (2021) 398:27–40. doi: 10.1016/S0140-6736(21)00797-2
8. Zhang Y, Li HT, Yu B, Sun S, Hu ZH, Wu XH, et al. Neoadjuvant chemoimmunotherapy for locally advanced esophageal squamous cell carcinoma: Data from literature review and a real-world analysis. Thorac Cancer. (2024) 15:1072–81. doi: 10.1111/1759-7714.15291
9. Gocher AM, Workman CJ, and Vignali DAA. Interferon-gamma: teammate or opponent in the tumour microenvironment? Nat Rev Immunol. (2022) 22:158–72. doi: 10.1038/s41577-021-00566-3
10. Li C, Song W, Zhang J, and Luo Y. Single-cell transcriptomics reveals heterogeneity in esophageal squamous epithelial cells and constructs models for predicting patient prognosis and immunotherapy. Front Immunol. (2023) 14:1322147. doi: 10.3389/fimmu.2023.1322147
11. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. (2023) 42:1–12. doi: 10.1038/s41587-023-01767-y
12. Zhou X, Guo S, and Shi Y. Comprehensive analysis of the expression and significance of CXCLs in human diffuse large B-cell lymphoma. Sci Rep. (2022) 12:2817. doi: 10.1038/s41598-022-06877-2
13. Zhang X, Peng L, Luo Y, Zhang S, Pu Y, Chen Y, et al. Dissecting esophageal squamous-cell carcinoma ecosystem by single-cell transcriptomic analysis. Nat Commun. (2021) 12:5291. doi: 10.1038/s41467-021-25539-x
14. Li CY, Zhang WW, Xiang JL, Wang XH, Wang JL, and Li J. Integrated analysis highlights multiple long non-coding RNAs and their potential roles in the progression of human esophageal squamous cell carcinoma. Oncol Rep. (2019) 42:2583–99. doi: 10.3892/or.2019.7377
15. Li JY, Chen SZ, Li Y, Zhu Z, Huang HY, Wang WD, et al. Comprehensive profiling analysis of CD209 in Malignancies reveals the therapeutic implication for tumor patients infected with SARS-CoV-2. Front Genet. (2022) 13:883234. doi: 10.3389/fgene.2022.883234
16. Shan TT, Zhao X, Zhang Z, Wang JP, Zhang Y, Yang Y, et al. Clinical significance of down-Regulated CD70 and CD27 expression in poor prognosis of esophageal squamous cell carcinoma (Retracted article). Cancer Manage Res. (2020) 12:6909–20. doi: 10.2147/Cmar.S241377
17. Chen ZC, Huang YW, Hu ZY, Zhao MN, Bian YY, Chen ZW, et al. Dissecting the single-cell transcriptome network in patients with esophageal squamous cell carcinoma receiving operative paclitaxel plus platinum chemotherapy. Oncogenesis. (2021) 10:71. doi: 10.1038/s41389-021-00359-2
18. Jiang N, Zhang J, Guo Z, Wu Y, Zhao L, Kong C, et al. Short-course neoadjuvant radiotherapy combined with chemotherapy and toripalimab for locally advanced esophageal squamous cell carcinoma (SCALE-1): a single-arm phase Ib clinical trial. J Immunother Cancer. (2024) 12:e008229. doi: 10.1136/jitc-2023-008229
19. Love MI, Huber W, and Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. (2014) 15:550. doi: 10.1186/s13059-014-0550-8
20. Mayakonda A, Lin DC, Assenov Y, Plass C, and Koeffler HP. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. (2018) 28:1747–56. doi: 10.1101/gr.239244.118
21. Garren ST. Maximum likelihood estimation of the correlation coefficient in a bivariate normal model with missing data. Stat Probability Lett. (1998) 38:281–8. doi: 10.1016/S0167-7152(98)00035-2
22. Yang SR, Pi JY, Ma WF, Gu WY, Zhang HX, Xu AY, et al. Prognostic value of the fibrinogen-to-albumin ratio (FAR) in patients with chronic heart failure across the different ejection fraction spectrum. Libyan J Med. (2024) 19:2309757. doi: 10.1080/19932820.2024.2309757
23. Zhang XM, Harrington N, Moraes RC, Wu MF, Hilsenbeck SG, and Lewis MT. Cyclopamine inhibition of human breast cancer cell growth independent of. Breast Cancer Res Treat. (2009) 115:505–21. doi: 10.1007/s10549-008-0093-3
24. Tang ZF, Kang BX, Li CW, Chen TX, and Zhang ZM. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. (2019) 47:W556–60. doi: 10.1093/nar/gkz430
25. Zhang WA, Li LF, Zhou Z, Liu Q, Wang G, and Liu D. Cost-effectiveness of Paxlovid in reducing severe COVID-19 and mortality in China. Front Public Health. (2023) 11:1174879. doi: 10.3389/fpubh.2023.1174879
26. Barraclough H, Simms L, and Govindan R. Biostatistics primer what a clinician ought to know: hazard ratios. J Thorac Oncol. (2011) 6:978–82. doi: 10.1097/JTO.0b013e31821b10ab
27. Sasanpour P, Sandoughdaran S, Mosavi-Jarrahi A, and Malekzadeh M. Predictors of pathological complete response to neoadjuvant chemotherapy in Iranian breast cancer patients. Asian Pac J Cancer Prev. (2018) 19:2423–7. doi: 10.22034/APJCP.2018.19.9.2423
28. Zheng YX, Chen ZY, Han YC, Han L, Zou X, Zhou BQ, et al. Immune suppressive landscape in the human esophageal squamous cell carcinoma microenvironment. Nat Commun. (2020) 11:6268. doi: 10.1038/s41467-020-20019-0
29. Wu J, Gao FX, Wang C, Qin M, Han F, Xu T, et al. IL-6 and IL-8 secreted by tumour cells impair the function of NK cells via the STAT3 pathway in oesophageal squamous cell carcinoma. J Exp Clin Cancer Res. (2019) 38:321. doi: 10.1186/s13046-019-1310-0
30. Hu K, Zhang GM, and Zhang WY. A new evaluation and prediction model of sound quality of high-speed permanent magnet motor based on genetic algorithm-radial basis function artificial neural network. Sci Prog. (2021) 104:368504211031114. doi: 10.1177/00368504211031114
Keywords: immune infiltration, neoadjuvant immunotherapy, esophageal cancer, PD-1, CXCL10
Citation: Zhang Y, Xu X, Mu X, Wang J, Zhang J, Xiang G, Li J, Zheng C, Wang H and Lu Q (2025) Effect of immune infiltration intensity on the efficacy of neoadjuvant immunotherapy for esophageal cancer. Front. Immunol. 16:1543283. doi: 10.3389/fimmu.2025.1543283
Received: 11 December 2024; Accepted: 19 May 2025;
Published: 12 June 2025.
Edited by:
Monde Ntwasa, University of South Africa, South AfricaReviewed by:
Hermann Frieboes, University of Louisville, United StatesHesong Wang, Fourth Hospital of Hebei Medical University, China
Copyright © 2025 Zhang, Xu, Mu, Wang, Zhang, Xiang, Li, Zheng, Wang and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiang Lu, bHVxaWFuZ0BmbW11LmVkdS5jbg==; Huaiyu Wang, YWxleHBlcHNpQDEyNi5jb20=
 Yong Zhang
Yong Zhang Xinyao Xu1,2
Xinyao Xu1,2 Qiang Lu
Qiang Lu