- 1Affiliated Hospital of Gansu University of Traditional Chinese Medicine, Lanzhou, China
- 2Gansu University of Traditional Chinese Medicine, Lanzhou, China
Rheumatoid arthritis is a chronic autoimmune disorder characterized by destructive, symmetric joint inflammation and synovitis, resulting in substantial disability that profoundly compromises patients’ quality of life. Its pathogenesis encompasses complex interactions between genetic and environmental factors. Recent advances in bacterial DNA sequencing technologies have uncovered a significant correlation between the human gut microbiota composition and rheumatoid arthritis progression. Growing clinical and experimental evidence establishes the gut-joint axis as a crucial mediator in rheumatoid arthritis pathogenesis. Comprehensive investigation of gut microbial communities and their metabolites’ influence on rheumatoid arthritis mechanisms, coupled with the elucidation of microbiome’s bidirectional regulatory effects in disease development, not only deepens our understanding of pathological processes but also establishes a theoretical framework for developing novel diagnostic biomarkers and personalized therapeutic interventions to enhance patient outcomes.
1 Introduction
Rheumatoid arthritis (RA) is a chronic inflammatory autoimmune disease characterized primarily by polyarticular synovitis, leading to bone and cartilage erosion and ultimately resulting in joint deformity and functional impairment (1). This condition affects approximately 1% of the global adult population (2). While the precise etiology of RA remains elusive, evidence suggests that its pathogenesis involves complex interactions between genetic and environmental factors, triggering aberrant T-cell immune responses and subsequent progressive inflammatory reactions in synovial joints (3–5).
The human microbiome constitutes a sophisticated ecological system essential for health maintenance and disease progression (6–10). The gastrointestinal tract is the most densely populated microbial habitat in humans, with the gut microbiota comprising approximately 70% of the total microbial load (11). The oral cavity contains the second largest microbial community, representing approximately 10% of the total. This oral microbiota plays a crucial role in maintaining oral health. The remaining microbial populations are distributed across other niches, including the skin, respiratory tract, and urogenital tract, where their combined abundance is lower, contributing to the overall diversity of the human microbiome. The human gut microbiome is predominantly composed of phyla including Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria (12), orchestrating essential physiological processes including nutrient metabolism, toxin degradation, immune regulation, and intestinal barrier maintenance (13). Evidence indicates that gut microbiota critically modulate host immune responses, with their dysbiosis strongly correlating with various autoimmune disorders’ pathogenesis (14). In dysbiotic conditions, metabolites—including short-chain fatty acids, polyphenols, vitamins, and tryptophan—produced by proliferating pathogenic strains potentially contribute to RA pathogenesis (15). Moreover, alterations in gut microbiota composition and their metabolic products influence both RA treatment efficacy and drug-induced adverse effects. Thus, elucidating the gut microbiota-RA relationship has profound implications for understanding disease mechanisms, enhancing diagnostic approaches, and developing optimal therapeutic strategies.
2 Gut microbiome ecology and its interplay with rheumatoid arthritis pathogenesis
Clinical and experimental studies have established a significant role for the gut-joint axis in the pathogenesis of rheumatoid arthritis (RA) (16, 17). Specifically, Collinsella spp. are significantly elevated in patients with stage 1 RA, while Faecalibacterium is associated with stage 3 RA, and Bifidobacterium longum and Eggerthella are enriched in stage 4 RA patients (18). Characterization of the gut microbiome in Chinese RA cohorts has revealed a signature marked by the enrichment of Lactobacillus salivarius and a reduction in Haemophilus spp (19). Furthermore, meta-analysis has demonstrated a significant alteration in the gut microbial composition of RA patients compared to healthy controls (16). Microbial community diversity is commonly assessed using α-diversity (species richness within a single sample) and β-diversity (differences in microbial composition between samples). Analyses of microbial diversity in RA patients generally indicate reduced or unchanged α-diversity, while β-diversity analyses reveal significant shifts in microbial community structure (20, 21). At the phylum level, the gut microbiota of RA patients is characterized by a significant increase in Proteobacteria and Verrucomicrobia, and a decrease in Firmicutes. Although the Firmicutes/Bacteroidetes (F/B) ratio has not consistently reached statistical significance, its imbalance has been implicated in activating pro-inflammatory pathways by disrupting intestinal barrier integrity (22). At the genus level, RA patients exhibit a significant increase in Klebsiella, Escherichia, Eisenbergiella, and Flavonifractor, alongside a significant decrease in Clostridium, Megamonas, and Enterococcus (21, 23–26). Specifically, Prevotella spp. have been identified as key pathobionts in RA, with significant enrichment observed in patient fecal samples (21, 27). A 27 kDa protein-derived peptide produced by Prevotella can mediate Th1 cell immune responses in early RA patients via HLA-DR (28, 29). Gut microbes and their metabolites exert a dual regulatory role in RA. Metabolomic analyses have revealed elevated levels of glycerophospholipids, benzene and its derivatives, and cholesterol in RA patients, coupled with decreased levels of sphingolipids and tryptophan-derived downstream metabolites (21). Furthermore, fecal butyrate levels are significantly reduced in RA patients. Butyrate, known for its anti-inflammatory properties, can inhibit arthritis progression, a finding supported by animal studies showing that exogenous butyrate supplementation suppresses arthritis (21). The reduced levels of tryptophan-derived downstream metabolites in RA patients, along with the correlation between serum tryptophan and its metabolites with disease activity and rheumatoid factor, suggest that gut microbes and their metabolites influence RA development. Systematically elucidating the molecular mechanisms underlying the gut microbiome’s association with RA pathogenesis will provide critical insights for understanding disease mechanisms, developing diagnostic markers, and optimizing therapeutic strategies.
3 Mechanistic insights into gut microbiome and metabolite-driven pathogenesis of rheumatoid arthritis
Gut microbiota dysbiosis is increasingly recognized as a pivotal factor in the pathogenesis of RA, primarily by modulating intestinal barrier integrity and immune system function (30, 31). Immune dysregulation resulting from microbial imbalance affects both innate and adaptive immunity (16). Within innate immunity, a disrupted microbiota can lead to aberrant activation of pattern recognition receptors, subsequently inducing the upregulation of pro-inflammatory cytokines and the downregulation of anti-inflammatory mediators, thereby disrupting local immune homeostasis. In adaptive immunity, a dysbiotic microbiota mediates the initiation and perpetuation of autoimmune responses by modulating antigen-presenting cell function, influencing T cell subset differentiation, and regulating B cell activation states. Microbiota-driven inflammation can compromise the tight junctions between intestinal epithelial cells, leading to increased intestinal permeability (32). However, while increased epithelial barrier permeability is a prerequisite for substance translocation, it is not the sole determinant. Although heightened permeability allows for greater translocation of microbes, their metabolites, and antigenic components into the systemic circulation, the specific substances critically involved in RA pathogenesis depend on the precise composition of the barrier-crossing metabolites and bacterial constituents. Specific bacterial components, such as lipopolysaccharide, can elicit significant systemic inflammation due to their potent immunostimulatory properties, even in small quantities. Similarly, imbalances in the levels of specific microbial metabolites, such as short-chain fatty acids (SCFAs), can contribute to the RA pathological process by influencing immune cell function (33). Therefore, while increased intestinal barrier permeability facilitates the entry of microbes and their metabolites into the host, the etiology of RA is contingent upon the specific types and concentrations of metabolites and bacterial components that traverse the barrier (34). Further elucidation of RA pathogenesis requires a deeper understanding of the precise mechanisms underlying increased intestinal barrier permeability, the identification of key metabolites and bacterial components that effectively translocate across the epithelial barrier, and the assessment of their relative importance in RA development. It can be hypothesized that increased intestinal barrier permeability, the composition of specific metabolites, and the presence of particular bacterial components act synergistically to mediate the translocation of substances across the epithelial barrier (35), ultimately influencing systemic immune responses and promoting the onset and progression of RA. Figure 1 illustrates the proposed role of gut microbes and their metabolites in promoting rheumatoid arthritis development.
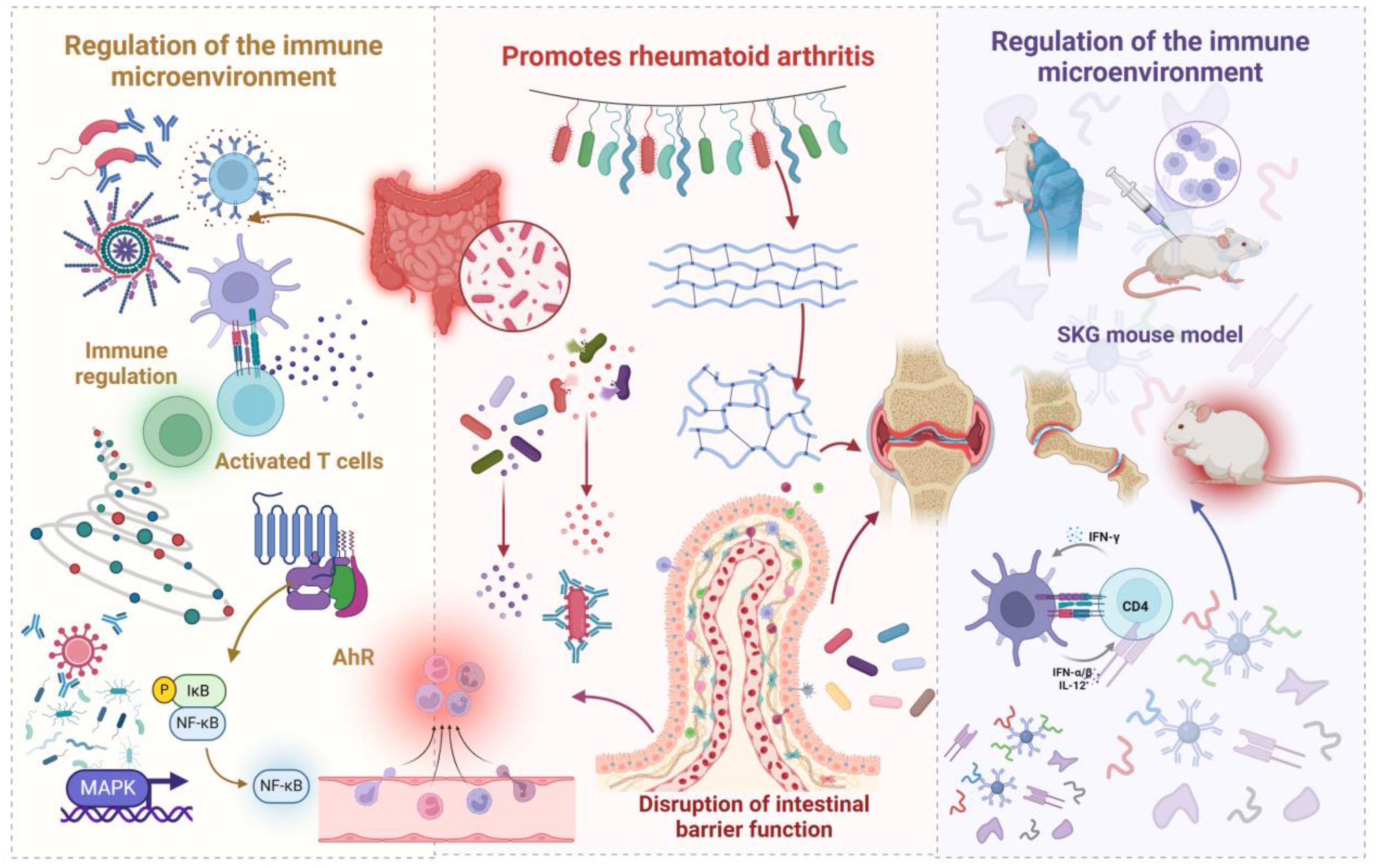
Figure 1. Schematic illustration depicting how gut microbiota and their metabolites promote the development of RA. Dysbiosis of the gut microbiome leads to an increase in harmful bacteria, which, by modulating the immune microenvironment, triggers inflammation. This microbial imbalance also enhances intestinal permeability, facilitating bacterial translocation that activates immune responses and initiates a cascade of events leading to the destruction of bone and cartilage. The immune cross-reactivity mechanism, supported by molecular simulations, elucidates how microbial antigens, due to their structural similarity to self-proteins, induce autoimmunity, subsequently causing inflammatory damage to joint tissues.
3.1 Immunological microenvironment modulation
The gut microbiota orchestrates RA pathogenesis through complex immunological networks. In innate immunity, dynamic signaling between gut-associated lymphoid tissue immune cells and intestinal microbiota establishes critical immune barriers (36). Microbial dysbiosis initiates aberrant innate immune cell activation, elevating pro-inflammatory cytokines (Interleukin-6 (IL-6), tumor necrosis factor - alpha (TNF-α), Interleukin-beta (IL-1β), Interleukin-12, Interleukin-23 (IL-23)) while suppressing anti-inflammatory mediators (transforming growth factor beta (TGF-β), Interleukin-10 (IL-10)) (31, 37). The gastrointestinal tract, as the body’s largest immune organ, engages in bidirectional communication with the host immune system to precisely balance immune homeostasis and inflammatory responses (38).
In adaptive immunity, microbiota primarily modulates the T helper 17/regulatory T cells (Th17/Treg) axis. After microbial antigen presentation by dendritic cells and macrophages to CD4+ T cells, these lymphocytes differentiate into distinct functional subsets (39, 40). RA patients display elevated peripheral Th17 cells, secreting pro-inflammatory cytokines (Interleukin-17 (IL-17), Interleukin-21, Interleukin-22 (IL-22), granulocyte-macrophage colony-stimulating factor, TNF-α) that induce synovial fibroblast receptor activator of nuclear factor kappa-B ligand (RANKL) expression. RANKL/osteoprote gerin imbalance drives osteoclastogenesis and bone erosion while promoting pannus formation and synovial angiogenesis (41, 42). Conversely, Treg cells maintain immunological homeostasis by secreting IL-10 and TGF-β1, thereby suppressing Th17 activity (43–45). The Th17/Treg balance regulation exhibits strain-specific microbial control. Clinical investigations reveal inverse correlations between Firmicutes abundance and Th17 cells in RA patients (46), while Bacteroides fragilis colonization enhances immunomodulation through Treg activation (47, 48). Faecalibacterium prausnitzii exerts protective effects by maintaining intestinal barrier integrity, modulating the Th17/Treg balance, and suppressing inflammatory responses (49). Studies in germ-free K/BxN mice demonstrate that Segmented filamentous bacteria colonization promotes functional Th17 cell differentiation via two mechanisms: elevated ileal serum amyloid A mediating dendritic cell-driven Th17 differentiation, and ATP-activated CD70highCD11clow cells inducing IL-6, IL-23p19, and TGF-β expression through integrin-αV/-β8 signaling (50–52).
Microbiota further modulates T follicular helper and regulatory cell equilibrium. Decreased T follicular regulatory cells correlate with enhanced RA activity and autoantibody production, inversely relating to Ruminococcus levels (53). Microbial antigens cooperate with T follicular helper cells to trigger B cell hyperactivation, enhancing pathogenic autoantibody production (18). Bacterial peptidoglycan detection in RA synovium (29, 54) confirms altered microbiota’s role in immune dysregulation (19, 55, 56). The aryl hydrocarbon receptor (AhR) pathway critically influences RA immunoregulation, particularly in RANKL-mediated osteoclastogenesis through nuclear factor kappa-light-chain-enhancer of activated B cells/mitogen-activated protein kinase (NF-κB/MAPK) signaling (57, 58). AhR additionally regulates immune surveillance via Th17/Treg modulation (59), with pathway disruption promoting autoimmunity and systemic inflammation.
3.2 Enhanced intestinal mucosal barrier permeability
The intestinal barrier system consists of multiple structural components: mucus layer, epithelial cell layer, basement membrane, vascular endothelium, and immune molecules. This system maintains precise regulation of innate and adaptive immune responses under physiological conditions, enabling selective barrier function while facilitating nutrient absorption (44). Zonulin, a protein secreted by intestinal epithelial cells, primarily functions to modulate tight junction protein complexes between these cells, thereby influencing intestinal barrier permeability. Increased expression or activity of zonulin can alter the structure and function of tight junction proteins, leading to enlarged intercellular spaces and a consequent increase in intestinal barrier permeability, which allows larger molecules to translocate into the bloodstream via the paracellular pathway. Clinical studies have demonstrated a concurrent increase in serum and fecal zonulin levels, providing evidence for the association between intestinal barrier dysfunction and RA (60). Gut microbiota dysbiosis is a critical factor in inducing intestinal barrier dysfunction. Studies have revealed an abnormal increase in the abundance of Collinsella spp. in RA patients (27, 61), and the molecular mechanisms by which these bacteria disrupt the intestinal barrier include the downregulation of tight junction protein expression in intestinal epithelial cells and the activation of the NF-κB1-mediated inflammatory pathway. Furthermore, metabolites produced by Collinsella, such as α-aminoadipic acid and asparagine, synergize with IL-17A to further exacerbate intestinal barrier damage (62). The regulatory relationship between microbial composition and intestinal permeability has been validated in HLA-DQ8 transgenic mouse models. Experimental evidence indicates that overexpression of Collinsella aerofaciens increases the incidence of collagen-induced arthritis by suppressing the expression of tight junction proteins in intestinal epithelial cells (63). Increased intestinal permeability facilitates the translocation of microbes and their metabolites across the compromised barrier into the systemic circulation, subsequently activating local tissue immune responses (37). This can initiate a cascade of destructive events in bone and cartilage tissues (64–66).
3.3 Molecular simulation insights into self-antigen recognition and rheumatoid arthritis pathogenesis
Molecular mimicry constitutes a mechanism wherein structural or sequence homology between pathogenic microorganisms and host molecules generates cross-reactive immune responses that trigger autoimmunity. In RA pathogenesis, this process operates through cross-recognition between microbial antigens and self-protein epitopes. Clinical investigations demonstrate that RA patient T and B cells recognize N-acetylglucosamine-6-sulfatase and filamin A as autoantigens (52% and 56%, respectively) (67) These autoantigens share extensive sequence homology with Prevotella-derived bacterial proteins within human leukocyte antigen-DR presentation regions. Through cross-recognition, anti-P. copri immune responses activate autoantigen-specific lymphocyte responses, inducing autoantibody production. These autoantibodies display marked RA specificity, occurring significantly less frequently in healthy individuals and patients with other autoimmune conditions (67). The spleen tyrosine kinase deficient mouse model has illuminated molecular mimicry’s role in RA pathogenesis. This model harbors zeta-chain-associated protein kinase 70 gene mutations that impair T cell receptor signaling, potentiating T cell autoreactivity (68). In specific pathogen-free environments, P. copri colonization enhances Th17 responses and arthritis manifestations in spleen tyrosine kinase deficient mice. Notably, germ-free mice maintain arthritis resistance despite fungal exposure, underscoring the gut microbiota’s essential role in autoreactive T cell activation (68). Mechanistic studies reveal that P. copri monocolonization amplifies immune responses to the arthritis-associated autoantigen 60S ribosomal protein L23a in lymphoid tissues (69). The molecular mimicry-based mechanism of immune cross-reactivity elucidates how microbial antigens, through their structural resemblance to self-proteins, can trigger autoimmune responses, subsequently leading to inflammatory damage in joint tissues. Beyond the immune-inflammatory responses mediated by molecular mimicry, gut microbial metabolites may also play a pathogenic role in RA. Paralleling the mechanism of microbial metabolite-induced bone loss in periodontitis, we hypothesize that gut microbiota dysbiosis in RA patients leads to the aberrant production of specific metabolites, such as SCFAs and lipopolysaccharide, which may translocate to the joints via a compromised intestinal barrier. Locally within the joints, these metabolites may directly activate osteoclasts or indirectly induce synovial cells to release pro-inflammatory cytokines and bone-resorbing factors, thereby promoting the destruction of joint bone. Thus, microbial metabolite-induced bone resorption may constitute another significant pathway through which the gut microbiota influences RA pathological progression, acting in concert with molecular mimicry mechanisms to promote disease onset and development. This finding offers a novel perspective on understanding the pathogenesis of RA.
4 Mechanistic insights into gut microbiome and metabolites’ inhibitory effects on rheumatoid arthritis pathogenesis
Gut microbiota dysbiosis exerts bidirectional effects on RA pathogenesis. Beneficial microbes maintain intestinal homeostasis through immune modulation and barrier preservation, particularly Lactobacillus, Bifidobacterium, and Bacteroides fragilis species, which attenuate RA manifestation and progression (70, 71). Clinical studies have characterized distinct alterations in RA patients’ gut microbiota, revealing depletion of multiple genera including Bacteroides, Streptococcus, Rhodococcus, Prevotella, Haemophilus, and Parabacteroides (72–74). These compositional shifts influence host immunity through altered metabolic profiles. The tryptophan metabolic pathway emerges as a critical mediator of RA suppression, where microbe-derived tryptophan metabolites function as aryl hydrocarbon receptor ligands to regulate both innate and adaptive immunity (75). Metabolomic profiling has identified reduced concentrations of tryptophan-derived compounds in RA patients’ fecal samples, specifically N-methyl-5-hydroxytryptamine, 5-hydroxyindoleacetic acid, kynurenic acid, xanthurenic acid, and 3-hydroxyanthranilic acid, underscoring tryptophan metabolism’s pivotal role in RA pathogenesis (21). Figure 2 illustrates how gut microbiota and their metabolites inhibit the development and progression of RA.
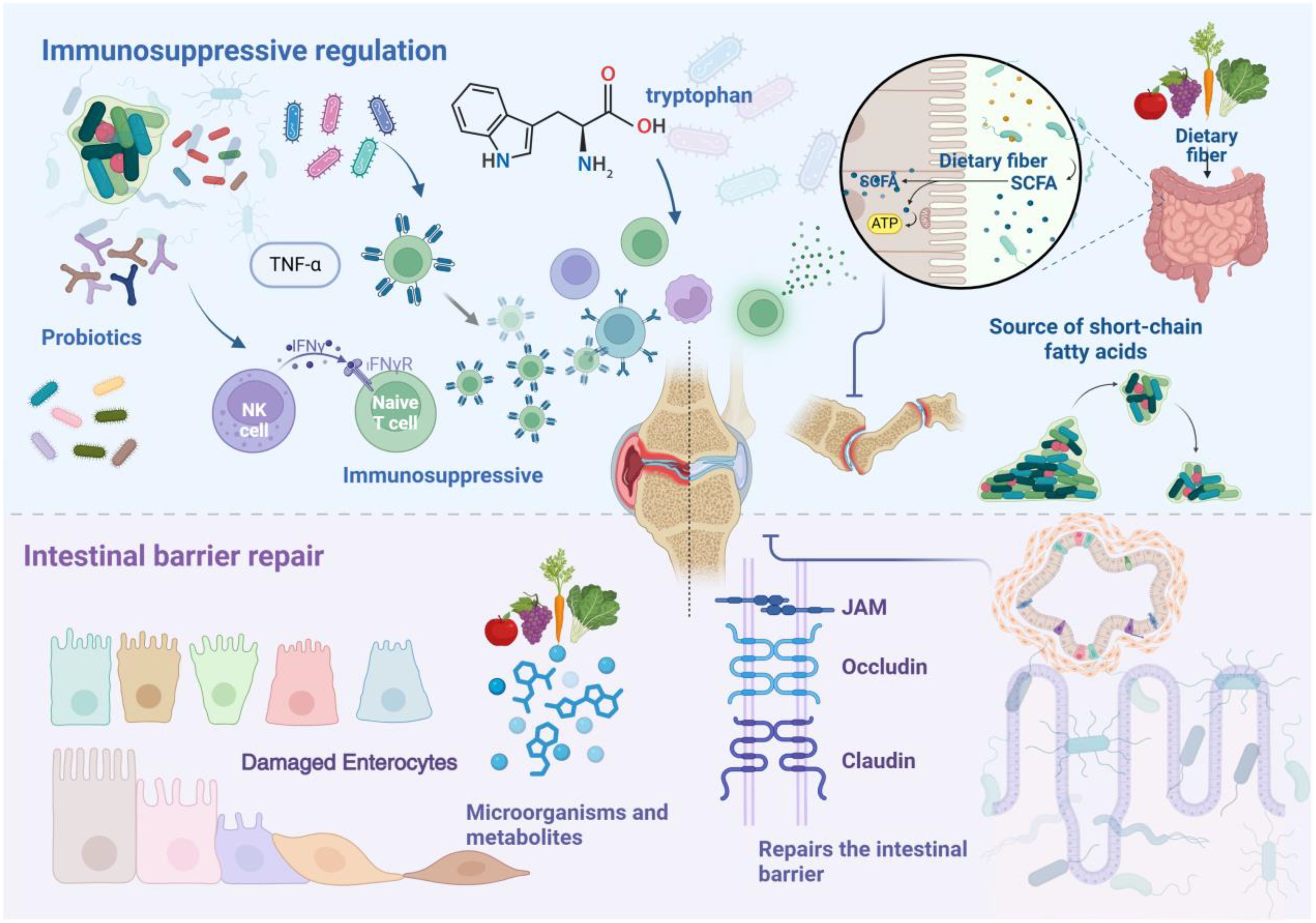
Figure 2. Schematic illustration showing how gut microbiota and their metabolites inhibit the development and progression of RA. Probiotics and their metabolites exert inhibitory effects on RA by modulating immune cell functions and regulating the expression of inflammatory cytokines. Short-chain fatty acids play a key role in this process. Additionally, various nutritional factors contribute to the repair and maintenance of the intestinal barrier by modulating microbiota composition and metabolic pathways. Dietary fiber, an essential nutrient for maintaining barrier integrity, when adequately consumed, reduces serum levels of Zonulin and calprotectin, thereby inhibiting the onset and progression of RA.
4.1 Immunosuppressive regulatory mechanisms
Gut microbiota and their metabolites attenuate RA progression through modulation of immune responses and inflammatory pathways. Probiotic strains exhibit distinct immunosuppressive properties, with Lactobacillus casei, L. rhamnosus, and Lactococcus lactis demonstrating therapeutic potential (76, 77). Mechanistic studies suggest that Lactobacillus casei may induce anti-inflammatory responses by influencing the maturation and function of dendritic cells, upregulating the expression of the anti-inflammatory cytokine IL-10, and downregulating the levels of pro-inflammatory cytokines IL-6 and TNF-α, thereby leading to reduced C-reactive protein levels and improved arthritis symptoms. Lactobacillus rhamnosus, on the other hand, primarily mediates immunosuppressive effects by modulating the expression of the pro-inflammatory cytokine IL-1 (27). Despite elevated detection in severe RA patients (78, 79), L. salivarius exhibits anti-inflammatory activity in collagen-induced arthritis models, mitigating bone erosion and neutrophil infiltration. Falcatibacterium prausnitzii reduces pro-inflammatory cytokines IL-17, IL-1β, TNF-α while enhancing commensal microbiota proliferation, particularly Akkermansia and Bilophila (27).
SCFAs and bile acid metabolites serve as key immunomodulators. In arthritis models, butyrate drives follicular regulatory T cell development, attenuating autoimmune progression (80). Propionate may influence the balance between Th17 cells and regulatory T cells. Metabolites produced by Parabacteroides distasonis, including lithocholic acid, deoxycholic acid, isoallolithocholic acid, and 3-oxolithocholic acid, exert anti-arthritic effects through synergistic action. Specifically, 3-oxolithocholic acid and isoallolithocholic acid act as TGR5 receptor agonists, inducing macrophage polarization towards the M2 phenotype and inhibiting Th17 cell differentiation (81). The tryptophan metabolic pathway emerges as a critical immunoregulator in RA, with clinical studies revealing diminished tryptophan metabolites in RA synovial fluid versus osteoarthritis samples (82). The serotonin metabolite 5-hydroxyindoleacetic acid promotes regulatory T cell development via AhR activation (83). Microbial SCFAs enhance regulatory B cell function through elevated 5-hydroxyindoleacetic acid levels, reducing arthritis manifestations (84). Additionally, kynurenic acid limits synovial proliferation (21), while 3-hydroxyanthranilic acid attenuates lipopolysaccharide-induced macrophage inflammation through NF-κB inhibition (85). Dietary composition significantly influences the composition of the gut microbiota and the production of its metabolites, thereby indirectly modulating these immunosuppressive mechanisms. Signaling molecules generated by the gut microbiota and their metabolites can traverse the bloodstream to impact the systemic immune system, including immune responses in distal sites such as the joints.
4.2 Intestinal mucosal barrier restoration
The intestinal barrier functions as a crucial regulator of organismal homeostasis, with its impairment contributing to autoimmune disease pathogenesis. Diverse nutritional factors orchestrate barrier maintenance and repair by modulating microbial ecosystems and metabolic networks. Dietary fiber constitutes an essential nutritional element in preserving barrier integrity, reducing serum concentrations of both the permeability regulator Zonulin and the inflammatory biomarker calprotectin (86). Furthermore, dietary fiber maintains mucus layer stability, thereby influencing microbial community structure (87). SCFAs fortify intestinal barrier function while serving as microbial energy substrates and community regulators. These metabolites enhance barrier integrity through three primary mechanisms: tight junction protein induction, epithelial cell regeneration, and antimicrobial peptide production. Butyrate demonstrates particular efficacy in promoting tight junction protein expression (88). Vitamins contribute to barrier homeostasis through epithelial cell regulation: vitamin D coordinates tight junction assembly and cell survival (88), while vitamin E strengthens barrier function by supporting butyrate-producing bacteria and maintaining microbial homeostasis (89). Select amino acids emerge as critical mediators of barrier function. Glutamine and tryptophan insufficiency directly compromises barrier integrity, elevating intestinal permeability (90, 91). Plant polyphenols quercetin, myricetin, kaempferol, and curcumin reinforce barrier function by augmenting transepithelial electrical resistance and enhancing tight junction protein expression zonula occludens-1 and claudin-1 (92), consequently attenuating RA progression.
5 Translational applications of gut microbiome in rheumatoid arthritis diagnosis and treatment
Dysregulation of gut microbial composition and diversity drives RA pathogenesis and progression through immune system modulation (65). Therapeutic strategies targeting intestinal microbiota represent an emerging frontier in RA treatment. Methotrexate, while established as the primary RA therapeutic, exhibits marked inter-patient variability in clinical outcomes. Such heterogeneity encompasses both pharmacodynamic responses sensitivity and tolerability and pharmacokinetic parameters distribution and clearance across renal, hepatic, and serosal compartments. Gut microbiota and their enzymatic products modulate drug efficacy through multiple mechanisms, influencing the bioavailability, therapeutic response, and adverse effects of various medications, particularly methotrexate. Reciprocally, these therapeutics and their metabolites reshape microbial communities, thereby altering immune function. Interventions targeting intestinal homeostasis have demonstrated promising clinical outcomes in RA management, with preclinical studies validating the therapeutic potential of microbiome modulation and barrier function enhancement (93). Furthermore, leveraging immune reactivity against selected gut bacterial species holds potential clinical utility. Exploring the detection of specific antibodies or T cell responses against particular gut bacteria in RA patients could lead to the development of novel diagnostic biomarkers, enabling earlier or more precise RA diagnosis. Moreover, immune reactivity against these specific gut bacteria may also offer new insights into therapeutic strategies (94). Developing specific immunotherapies, such as vaccines or antibodies, targeting pathogenic bacteria to inhibit their growth or mitigate their induced inflammatory responses warrants consideration. Inducing immune tolerance to specific gut bacteria could also be a strategy to alleviate inflammation driven by these bacteria. These advances suggest that targeted manipulation of microbial ecosystems may yield novel diagnostic markers and personalized interventions. Figure 3 shows the translational applications of the gut microbiome in the diagnosis and treatment of RA. This microbiome-centric approach establishes new paradigms for precision medicine in RA treatment.
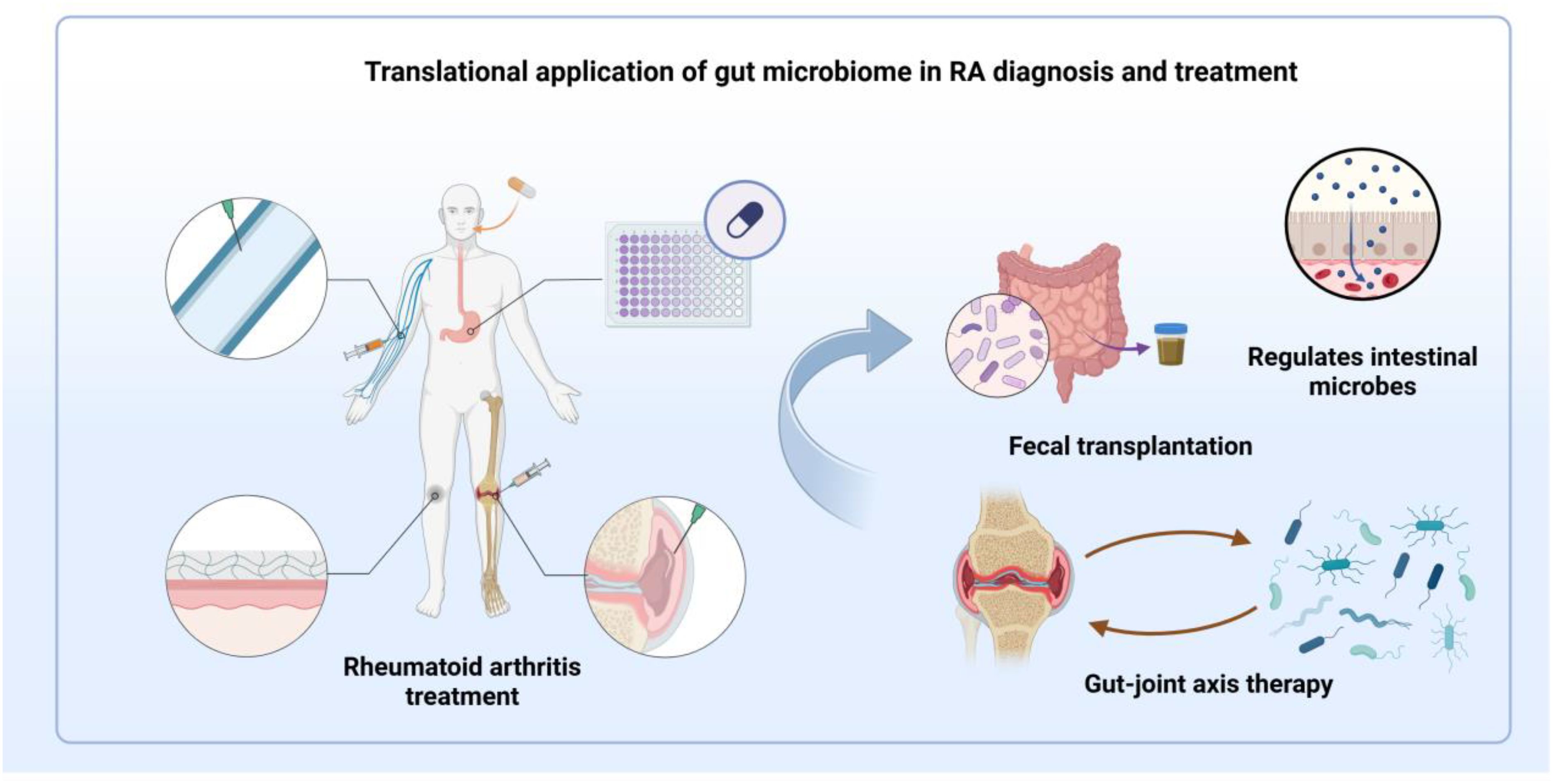
Figure 3. Schematic illustration of the translational applications of the gut microbiome in the diagnosis and treatment of RA. While RA can be managed through various methods, including oral medications, clinical outcomes exhibit significant individual variability. Interventions such as probiotic supplementation and stabilization of the intestinal barrier show potential therapeutic benefits in RA. By precisely modulating microbiota composition, interventions like probiotics, fecal microbiota transplantation, and therapies targeting the gut-joint axis may pave the way for the development of novel diagnostic biomarkers and personalized treatment strategies.
5.1 Biomarker discovery and diagnostic tool development
The composition of the gut microbiota and serum cytokine profiles are emerging as valuable molecular biomarkers for the early diagnosis of rheumatoid arthritis (RA), monitoring disease activity, and evaluating treatment responses. Microbiome sequencing analyses have revealed characteristic alterations in the gut microbiota of RA patients, including an increased ratio of Bacteroidetes to Firmicutes (24, 95). Community structure analysis further indicates that early RA patients exhibit an increased abundance of Prevotella and a decreased abundance of Bacteroides, a pattern with potential diagnostic utility. The pathogenic role of the microbiota in RA development has been validated in germ-free SKG mouse transplantation models. Patients with active disease display specific microbial signatures, such as reduced levels of Haemophilus and increased Streptococcus salivarius. Compared to patients in remission, those with active disease show a significant increase in the relative abundance of Collinsella and Akkermansia, suggesting that these microbial shifts may serve as effective indicators for monitoring disease activity (3, 96).
RA-associated autoantibodies, such as rheumatoid factor, anti-citrullinated peptide antibodies, and anti-carbamylated antibodies, can be detected in serum years before the clinical onset of RA (97–99). Although established RA typically involves the joints, the disease’s pathogenesis may originate beyond the articular structures, potentially linking to citrullination in the lungs or periodontal tissues, the gut microbiota, or adipose tissue. The normal appearance of synovial tissue in anti-citrullinated peptide antibodies-positive individuals with arthralgia supports this notion (100). The oral microbiome in patients with rheumatoid arthritis exhibits significant dysbiosis, which can be restored with RA treatment, and the extent of this restoration correlates with the patient’s response to therapy (101). Epidemiological studies have shown a high prevalence of periodontitis in RA patients. Periodontitis, a chronic inflammatory disease of the oral cavity, suggests a close relationship between the oral microbiome and RA. The periodontal anaerobe Porphyromonas gingivalis can synthesize bacterial peptidylarginine deiminase involved in RA pathogenesis (102). P. gingivalis is currently the only known bacterium that expresses peptidylarginine deiminase and is significantly associated with RA. Continuous peptidylarginine deiminase secretion by P. gingivalis leads to the citrullination of α- and β-fibrin chains in the joint synovium, generating autoantigens that drive anti-citrullinated peptide antibodies production. anti-citrullinated peptide antibodies then forms immune complexes with citrullinated proteins, binding to immune-inflammatory cells via Fc and C5a receptors, triggering a complex cascade of reactions and the release of inflammatory mediators, ultimately leading to local synovial inflammation and the development of RA (103).
Analysis of serum cytokine profiles provides crucial insights for evaluating disease activity and treatment responses. Elevated levels of pro-inflammatory cytokines TNF-α and IL-6 reflect increased disease activity, while decreased levels of the anti-inflammatory cytokine IL-10 suggest impaired immunosuppressive function. Integrating the characteristic changes in microbiota composition and cytokine profiles can facilitate the construction of a multi-dimensional diagnostic system based on the gut-joint axis (3, 96). Next-generation high-throughput sequencing technologies and metabolomics analysis platforms have enhanced the precision of identifying gut microbiota and their metabolites. The application of multi-omics data combined with machine learning algorithms offers robust technical support for developing more accurate and sensitive diagnostic tools for RA. Machine learning algorithms can integrate gut microbiota data, cytokine profiles, and other clinical data to improve the accuracy and sensitivity of RA diagnosis. Furthermore, multi-omics data can be used to identify different subtypes of RA patients, enabling personalized treatment strategies. Next-generation high-throughput sequencing technologies allow for the precise analysis of gut microbiota composition and diversity, while metabolomics analysis platforms provide a more comprehensive analysis of gut metabolites, aiding in the discovery of novel biomarkers.
5.2 Microbiome modulatory mechanisms in existing therapeutic paradigms
Disease-modifying antirheumatic drugs modulate RA progression through restructuring intestinal microbial communities. Clinical studies reveal elevated relative expression units of Bacteroides and Prevotella species in disease-modifying antirheumatic drugs-treated patients’ fecal samples, concurrent with decreased Clostridium abundance (25). Antibiotic intervention studies establish the microbiome’s causative role in RA pathogenesis: four-week clindamycin treatment selectively depletes anaerobic bacteria, intensifying both incidence and severity of collagen-induced arthritis in murine models (104).
Natural compounds demonstrate therapeutic efficacy through regulation of microbial ecosystems and metabolic networks. Berberine administration specifically reduces Prevotella while enriching butyrate-producing bacteria, attenuating collagen-induced arthritis symptoms via enhanced butyrate synthesis, intestinal hypoxia stabilization, and nitrate metabolism modulation (105). The parasitic immunomodulator excretory/secretory products of Acanthocheilonema viteae, derived from Acanthocheilonema, preserves microbiota homeostasis and barrier function when administered subcutaneously, preventing arthritic manifestations in collagen-induced arthritis models (106). Tripterygium wilfordii extract restores microbial equilibrium by suppressing pro-inflammatory taxa (Desulfovibrio, Mucor, Helicobacter, Spirochaetaceae), thereby attenuating tissue damage and inflammatory cytokine production such as TNF-α, IL-1β, IL-6, IL-7, IL-23 (88).
Paeoniae radix glycosides normalize gut microbial taxonomy in collagen-induced arthritis models, enhancing commensal populations while suppressing inflammatory mediators, including secretory Immunoglobulin A and Interferon gamma (107). Microbial metabolites emerge as therapeutic targets, with SCFAs mitigating RA progression through concurrent inhibition of T helper 1 differentiation and promotion of Treg proliferation (108). Clematis saponins regulate SCFAs homeostasis, with linear discriminant analysis effect size validating their role in maintaining Gram-negative/positive bacterial balance (109), consequently ameliorating RA symptoms.
5.3 Microbiome-mediated mechanisms of therapeutic efficacy modulation
The gut microbiota emerges as a potential therapeutic target in RA, with targeted modulation strategies encompassing probiotic interventions, prebiotic supplementation, and fecal microbiota transplantation. These microbiome-based therapeutic approaches ameliorate disease progression through restoration of intestinal microbial homeostasis. Such microbiota-mediated interventions, operating through multiple regulatory layers, establish novel paradigms for precision RA treatment.
5.3.1 Therapeutic strategies modulated by microbiome regulation
Probiotics demonstrate therapeutic potential through diverse mechanisms. They suppress inflammatory signaling pathways by downregulating Toll-like receptor expression. These pattern recognition receptors mediate immune cell activation and inflammatory mediator production through pathogen-associated molecular pattern recognition. Probiotics additionally induce immunoregulat forkhead box P3 ory factor secretion by antigen-presenting cells. Experimental studies confirm their promotion of forkhead box P3-positive regulatory T cell differentiation, thereby inhibiting inflammatory cascades. Furthermore, Lactobacillus and Bifidobacterium species maintain intestinal barrier integrity through short-chain fatty acid production (71). Clinical investigations validate probiotic therapeutic efficacy. RA patients in remission (DMARD-naïve) demonstrate significantly improved health scores following 12-month L. rhamnosus intervention (110). Combined supplementation with L. acidophilus, L. casei, and B. bifidum reduces disease activity scores and high-sensitivity C-reactive protein levels (111, 112). Multi-strain formulations, including LC-11, LA-14, LL-23, BL-04, and BB-06, exhibit significant efficacy in active RA patients (113).
Microbial metabolites play pivotal roles in maintaining intestinal barrier function. Butyrate and short-chain fatty acids ameliorate inflammation associated with leaky gut syndrome by enhancing tight junction protein expression (114, 115). Lithocholic acid demonstrates anti-inflammatory activity in collagen-induced arthritis models (116, 117), while Bacteroides fragilis and propionate serve as adjuvant therapeutic factors, enhancing conventional treatment efficacy (118). Indole-3-methanol inhibits adjuvant-induced arthritis progression (119). Sinomenine attenuates RA-associated inflammation by elevating microbiota-derived indolic tryptophan metabolites, activating AhR, and modulating NF-κB/MAPK signaling pathways (16).
5.3.2 Microecosystem reconstruction
Fecal Microbiota Transplantation (FMT) constitutes a therapeutic approach that rehabilitates recipient microbial ecosystems via healthy donor microbiota transfer. Mechanistic studies reveal that normal microbiota transplantation enhances bone mass regeneration through orchestration of microbial metabolism and immune networks (120). The interplay between dysbiosis and mucosal immunity emerges as a critical initiating factor in RA pathogenesis (121). Investigations using germ-free BALB/c ZAP-70W163C mutant models demonstrate that RA patient-derived FMT intensifies arthritic manifestations via Th17-mediated signaling cascades (122). FMT, which involves the transfer of gut microbiota from a healthy donor, aims to reconstruct the recipient’s intestinal microecosystem. Its mechanisms of action extend beyond the direct supplementation of beneficial bacteria to encompass complex modulation of the host immune system. FMT reduces systemic inflammation by regulating the production of gut metabolites, influencing host immune cell function, and repairing the compromised intestinal barrier, thereby limiting the translocation of bacteria and their metabolites into the systemic circulation. Furthermore, FMT can modulate the balance of immune cells within the intestine of RA patients, increasing the number of Tregs and decreasing the proportion of pro-inflammatory Th17 cells. With the continuous advancement of FMT techniques, its application in clinical translational research for RA is gaining momentum (123). Preliminary clinical studies have observed some symptom relief in a subset of RA patients; however, larger-scale clinical trials are needed to validate its efficacy due to the limited sample sizes of current studies. The efficacy of FMT is influenced by multiple factors, including donor selection, microbial composition of the transplant, and the route of administration (124). Therefore, precise analysis of the gut microbiota composition of healthy donors and optimization of delivery methods (e.g., oral capsules, enemas) are crucial for enhancing the clinical efficacy of FMT. Nevertheless, FMT carries potential risks of infection, particularly for immunocompromised RA patients. The long-term effects of microbiota transplantation on the host remain largely unknown and necessitate long-term follow-up studies. Further refinement is required regarding the optimization of FMT administration routes, long-term efficacy, and safety assessments. Systemic FMT treatment holds promise for providing a novel therapeutic target for the precision treatment of RA by reshaping the diversity and functional network of the gut microbiota in affected individuals. This strategy underscores a new research direction for RA treatment by modulating RA-related immune responses from the perspective of restoring microecological balance.
5.4 Translational clinical strategies for microbiome-based therapeutics
Triptolide orchestrates anti-arthritic effects by modulating microbial communities and metabolic networks. Mechanistic investigations demonstrate its therapeutic action through three primary pathways: enhancement of Lactobacillus populations, upregulation of microbiota-derived tryptophan metabolites, and AhR signaling activation for inflammatory suppression (81). Moreover, triptolide-enriched Paracasei and L. casei communities mitigate RA manifestations through potentiated tryptophan metabolism and AhR pathway stimulation. Photobiomodulation therapy attenuates TNF-mediated RA progression via gut-joint axis modulation (125). This intervention reconstructs microbial ecosystems, facilitating beneficial bacterial expansion - particularly Clostridium, Lactobacillus, and Escherichia species - thus reinforcing symbiotic network integrity. Microbial amino acid metabolites emerge as key mediators in photobiomodulation’s regulation of the gut-joint axis, with efficacy determined by precise intervention protocols, metabolomic signatures, and tissue-specific enzymatic activities (125). Although non-steroidal anti-inflammatory drugs suppress RA symptoms, they commonly disrupt intestinal mucosal integrity and microbial homeostasis (74). Evidence indicates that probiotic supplementation counteracts non-steroidal anti-inflammatory drugs-induced enteropathy, with Bifidobacterium, Lactobacillus, and Prevotella species demonstrating robust mucosal protection in preclinical models (126), underscoring the microbiome’s therapeutic potential.
6 Conclusion
Gut microbiota and their metabolites orchestrate critical regulatory networks in RA pathogenesis. RA patients demonstrate enrichment of Klebsiella, Escherichia, Eisenbergiella, and Flavobacterium, concurrent with depletion of beneficial microorganisms, particularly Bifidobacterium, Streptococcus, and SCFA-producers. Pathogenic bacteria accelerate RA progression through inflammatory cascade activation and barrier compromise, while beneficial microbes attenuate disease development via anti-inflammatory mechanisms and barrier reinforcement. The microbiome emerges as a promising therapeutic and diagnostic frontier, with specific microbial populations representing potential intervention targets.
Therapeutic strategies aimed at modulating the gut microbiome and restoring intestinal microecological balance have demonstrated the potential to regulate the immune system, offering new avenues for the treatment of RA. However, several limitations remain. Current research is largely correlational, observing alterations in gut microbial composition and metabolite levels in RA patients. Whether these changes are a direct cause or a consequence of RA requires further investigation. Furthermore, the gut microbiota and metabolite profiles of RA patients exhibit significant inter-individual variability, potentially influenced by genetic, environmental, and lifestyle factors. The application of gut microbiota modulation strategies in RA treatment is still in its early stages, necessitating more extensive clinical trials to validate their efficacy and safety. The development of diagnostic and therapeutic tools for RA based on the gut microbiota and its metabolites also warrants further research. Future endeavors should prioritize more clinical studies to verify the effects and mechanisms of gut homeostasis regulation in RA prevention and treatment. Exploring the mechanisms of specific probiotics or metabolites, developing novel therapies targeting the gut microbiome, and integrating these approaches with traditional pharmacological treatments and lifestyle interventions will offer more comprehensive strategies for RA management. With a deeper understanding of the intricate relationship between the gut microbiota and its metabolites and the immune system, future research should emphasize the integrated analysis of multi-omics data, the combined use of animal models and clinical trials, and the development of personalized treatment strategies. Notably, exploring the interplay between the gut microbiota and drug metabolism in RA patients, and investigating the therapeutic potential of modulating gut microbes and their metabolites holds promise for novel insights and therapeutic targets for the clinical prevention and treatment of RA, ultimately leading to improved patient outcomes and enhanced quality of life.
Author contributions
XX: Funding acquisition, Methodology, Writing – original draft, Writing – review & editing. XC: Writing – original draft, Writing – review & editing. XW: Writing – review & editing. SW: Writing – review & editing. PQ: Methodology, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Nature Fund Regional Project (82160911, 81860864); Central University Basic Scientific Research Business Expenses Project (31920210041); The special project of science and technology development under the guidance of the central government (YDZX20206200002356).
Acknowledgments
The authors acknowledge the use of Biorender to create schematic representations in Figures 1–3.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
RA, rheumatoid arthritis; NF-κB, nuclear factor kappa-light-chain-enhancer of activated B cells; MAPK, mitogen-activated protein kinase; FMT, fecal microbiota transplantation; IL-6, Interleukin-6; TNF-α, tumor necrosis factor - alpha; IL-1β, Interleukin-beta; IL-23, Interleukin-23; TGF-β, transforming growth factor beta; IL-10, Interleukin-10; Th17, T helper 17; IL-17, Interleukin-17; IL-22, Interleukin-22; AhR, aryl hydrocarbon receptor; SCFAs, short-chain fatty acids; RANKL, receptor activator of nuclear factor kappa-B ligand; Treg, regulatory T cells.
References
1. McInnes IB, Schett G. The pathogenesis of rheumatoid arthritis. N Engl J Med. (2011) 365:2205–19. doi: 10.1056/NEJMra1004965
2. Firestein GS. Evolving concepts of rheumatoid arthritis. Nature. (2003) 423:356–61. doi: 10.1038/nature01661
3. Zhao T, Wei Y, Zhu Y, Xie Z, Hai Q, Li Z, et al. Gut microbiota and rheumatoid arthritis: From pathogenesis to novel therapeutic opportunities. Front Immunol. (2022) 13:1007165. doi: 10.3389/fimmu.2022.1007165
4. Brandl C, Bucci L, Schett G, Zaiss MM. Crossing the barriers: Revisiting the gut feeling in rheumatoid arthritis. Eur J Immunol. (2021) 51:798–810. doi: 10.1002/eji.202048876
5. Edilova MI, Akram A, Abdul-Sater AA. Innate immunity drives pathogenesis of rheumatoid arthritis. BioMed J. (2021) 44:172–82. doi: 10.1016/j.bj.2020.06.010
6. Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, et al. A new genomic blueprint of the human gut microbiota. Nature. (2019) 568:499–504. doi: 10.1038/s41586-019-0965-1
7. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
8. Zheng D, Liwinski T, Elinav E. Interaction between microbiota and immunity in health and disease. Cell Res. (2020) 30(6):492–506. doi: 10.1038/s41422-020-0332-7
9. Gomaa EZ. Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek. (2020) 113:2019–40. doi: 10.1007/s10482-020-01474-7
10. Durack J, Lynch SV. The gut microbiome: Relationships with disease and opportunities for therapy. J Exp Med. (2019) 216:20–40. doi: 10.1084/jem.20180448
11. Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. (2010) 90:859–904. doi: 10.1152/physrev.00045.2009
12. Song M, Chan AT, Sun J. Influence of the gut microbiome, diet, and environment on risk of colorectal cancer. Gastroenterology. (2020) 158:322–40. doi: 10.1053/j.gastro.2019.06.048
13. Sun J, Chang EB. Exploring gut microbes in human health and disease: Pushing the envelope. Genes Dis. (2014) 1:132–9. doi: 10.1016/j.gendis.2014.08.001
14. Attur M, Scher JU, Abramson SB, Attur M. Role of intestinal dysbiosis and nutrition in rheumatoid arthritis. Cells. (2022) 11:2436. doi: 10.3390/cells11152436
15. Johnson CH, Spilker ME, Goetz L, Peterson SN, Siuzdak G. Metabolite and microbiome interplay in cancer immunotherapy. Cancer Res. (2016) 76:6146–52. doi: 10.1158/0008-5472.CAN-16-0309
16. Jiang ZM, Zeng SL, Huang TQ, Lin Y, Wang FF, Gao XJ, et al. Sinomenine ameliorates rheumatoid arthritis by modulating tryptophan metabolism and activating aryl hydrocarbon receptor via gut microbiota regulation. Sci Bull (Beijing). (2023) 68:1540–55. doi: 10.1016/j.scib.2023.06.027
17. Paul AK, Paul A, Jahan R, Jannat K, Bondhon TA, Hasan A, et al. Probiotics and Amelioration of Rheumatoid Arthritis: Significant Roles of Lactobacillus casei and Lactobacillus acidophilus. Microorganisms. (2021) 9:1070. doi: 10.3390/microorganisms9051070
18. Bakinowska E, Stańska W, Kiełbowski K, Szwedkowicz A, Boboryko D, Pawlik A. Gut dysbiosis and dietary interventions in rheumatoid arthritis-A narrative review. Nutrients. (2024) 16:3215. doi: 10.3390/nu16183215
19. Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. (2015) 21:895–905. doi: 10.1038/nm.3914
20. Chu XJ, Cao NW, Zhou HY, Meng X, Guo B, Zhang HY, et al. The oral and gut microbiome in rheumatoid arthritis patients: a systematic review. Rheumatol (Oxford). (2021) 60:1054–66. doi: 10.1093/rheumatology/keaa835
21. Yu D, Du J, Pu X, Zheng L, Chen S, Wang N, et al. The gut microbiome and metabolites are altered and interrelated in patients with rheumatoid arthritis. Front Cell Infect Microbiol. (2022) 11:763507. doi: 10.3389/fcimb.2021.763507
22. Khan R, Sharma A, Ravikumar R, Parekh A, Srinivasan R, George RJ, et al. Association between gut microbial abundance and sight-threatening diabetic retinopathy. Invest Ophthalmol Vis Sci. (2021) 62:19. doi: 10.1167/iovs.62.7.19
23. Vaahtovuo J, Munukka E, Korkeamäki M, Luukkainen R, Toivanen P. Fecal microbiota in early rheumatoid arthritis. J Rheumatol. (2008) 35:1500–5.
24. Lee JY, Mannaa M, Kim Y, Kim J, Kim GT, Seo YS. Comparative analysis of fecal microbiota composition between rheumatoid arthritis and osteoarthritis patients. Genes (Basel). (2019) 10:748. doi: 10.3390/genes10100748
25. Rodrigues GSP, Cayres LCF, Gonçalves FP, Takaoka NNC, Lengert AH, Tansini A, et al. Detection of increased relative expression units of bacteroides and prevotella, and decreased clostridium leptum in stool samples from Brazilian rheumatoid arthritis patients: A pilot study. Microorganisms. (2019) 7:413. doi: 10.3390/microorganisms7100413
26. Chen Y, Ma C, Liu L, He J, Zhu C, Zheng F, et al. Analysis of gut microbiota and metabolites in patients with rheumatoid arthritis and identification of potential biomarkers. Aging (Albany NY). (2021) 13:23689–701. doi: 10.18632/aging.203641
27. Dagar S, Singh J, Saini A, Kumar Y, Chhabra S, Minz RW, et al. Gut bacteriome, mycobiome and virome alterations in rheumatoid arthritis. Front Endocrinol (Lausanne). (2022) 13:1044673. doi: 10.3389/fendo.2022.1044673
28. Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. (2013) 2:e01202. doi: 10.7554/eLife.01202
29. Pianta A, Arvikar S, Strle K, Drouin EE, Wang Q, Costello CE, et al. Evidence of the immune relevance of prevotella copri, a gut microbe, in patients with rheumatoid arthritis. Arthritis Rheumatol. (2017) 69:964–75. doi: 10.1002/art.40003
30. Fan Z, Ross RP, Stanton C, Hou B, Zhao J, Zhang H, et al. Lactobacillus casei CCFM1074 Alleviates Collagen-Induced Arthritis in Rats via Balancing Treg/Th17 and Modulating the Metabolites and Gut Microbiota. Front Immunol. (2021) 12:680073. doi: 10.3389/fimmu.2021.680073
31. Horta-Baas G, Romero-Figueroa MDS, Montiel-Jarquín AJ, Pizano-Zárate ML, García-Mena J, Ramírez-Durán N. Intestinal dysbiosis and rheumatoid arthritis: A link between gut microbiota and the pathogenesis of rheumatoid arthritis. J Immunol Res. (2017) 2017:4835189. doi: 10.1155/2017/4835189
32. Liebing E, Krug SM, Neurath MF, Siegmund B, Becker C. Wall of resilience: how the intestinal epithelium prevents inflammatory onslaught in the gut. Cell Mol Gastroenterol Hepatol. (2025) 19:101423. doi: 10.1016/j.jcmgh.2024.101423
33. Kim CH. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol Immunol. (2023) 20:341–50. doi: 10.1038/s41423-023-00987-1
34. Nagpal R, Yadav H. Bacterial translocation from the gut to the distant organs: an overview. Ann Nutr Metab. (2017) 71 Suppl 1:11–6. doi: 10.1159/000479918
35. Wang Z, Shen J. The role of goblet cells in Crohn’ s disease. Cell Biosci. (2024) 14:43. doi: 10.1186/s13578-024-01220-w
36. Dong Y, Yao J, Deng Q, Li X, He Y, Ren X, et al. Relationship between gut microbiota and rheumatoid arthritis: A bibliometric analysis. Front Immunol. (2023) 14:1131933. doi: 10.3389/fimmu.2023.1131933
37. Zhang X, Chen B, LD Z, Li H. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol Med. (2020) 26:862–73. doi: 10.1016/j.molmed.2020.04.001
38. Shin C, Kim YK. Autoimmunity in microbiome-mediated diseases and novel therapeutic approaches. Curr Opin Pharmacol. (2019) 49:34–42. doi: 10.1016/j.coph.2019.04.018
39. Flannigan KL, Denning TL. Segmented filamentous bacteria-induced immune responses: a balancing act between host protection and autoimmunity. Immunology. (2018) 154:537–46. doi: 10.1111/imm.2018.154.issue-4
40. Wang Y, Yin Y, Chen X, Zhao Y, Wu Y, Li Y, et al. Induction of intestinal th17 cells by flagellins from segmented filamentous bacteria. Front Immunol. (2019) 10:2750. doi: 10.3389/fimmu.2019.02750
41. Lubberts E, van den Bersselaar L, Oppers-Walgreen B, Schwarzenberger P, Coenen-de Roo CJJ, Kolls JK, et al. IL-17 promotes bone erosion in murine collagen-induced arthritis through loss of the receptor activator of NF-kappa B ligand/osteoprotegerin balance. J Immunol. (2003) 170:2655–62. doi: 10.4049/jimmunol.170.5.2655
42. Kim KW, Kim HR, Park JY, Park JS, Oh HJ, Woo YJ, et al. Interleukin-22 promotes osteoclastogenesis in rheumatoid arthritis through induction of RANKL in human synovial fibroblasts. Arthritis Rheumatol. (2012) 64:1015–23. doi: 10.1002/art.33446
43. Lina C, Conghua W, Nan L, Ping Z. Combined treatment of etanercept and MTX reverses Th1/Th2, Th17/Treg imbalance in patients with rheumatoid arthritis. J Clin Immunol. (2011) 31:596–605. doi: 10.1007/s10875-011-9542-6
44. Chen X, Oppenheim JJ. Th17 cells and Tregs: unlikely allies. J Leukoc Biol. (2014) 95:723–31. doi: 10.1189/jlb.1213633
45. Xu H, Zhao H, Lu C, Qiu Q, Wang G, Huang J, et al. Triptolide inhibits osteoclast differentiation and bone resorption in vitro via enhancing the production of IL-10 and TGF-β1 by regulatory T cells. Mediators Inflamm. (2016) 2016:8048170. doi: 10.1155/2016/8048170
46. Wang Q, Zhang SX, Chang MJ, Qiao J, Wang CH, Li XF, et al. Characteristics of the gut microbiome and its relationship with peripheral CD4+ T cell subpopulations and cytokines in rheumatoid arthritis. Front Microbiol. (2022) 13:799602. doi: 10.3389/fmicb.2022.799602
47. Round JL, Lee SM, Li J, Tran G, Jabri B, Chatila TA, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. (2011) 332:974–7. doi: 10.1126/science.1206095
48. Telesford KM, Yan W, Ochoa-Reparaz J, Pant A, Kircher C, Christy MA, et al. A commensal symbiotic factor derived from Bacteroides fragilis promotes human CD39(+)Foxp3(+) T cells and Treg function. Gut Microbes. (2015) 6:234–42. doi: 10.1080/19490976.2015.1056973
49. Zhou L, Zhang M, Wang Y, Dorfman RG, Liu H, Yu T, et al. Faecalibacterium prausnitzii produces butyrate to maintain th17/treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflammation Bowel Dis. (2018) 24:1926–40. doi: 10.1093/ibd/izy182
50. Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. (2009) 139:485–98. doi: 10.1016/j.cell.2009.09.033
51. Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. (2010) 32:815–27. doi: 10.1016/j.immuni.2010.06.001
52. Atarashi K, Nishimura J, Shima T, Umesaki Y, Yamamoto M, Onoue M, et al. ATP drives lamina propria T(H)17 cell differentiation. Nature. (2008) 455:808–12. doi: 10.1038/nature07240
53. Wu R, Wang D, Cheng L, Su R, Li B, Fan C, et al. Impaired immune tolerance mediated by reduced Tfr cells in rheumatoid arthritis linked to gut microbiota dysbiosis and altered metabolites. Arthritis Res Ther. (2024) 26:21. doi: 10.1186/s13075-023-03260-y
54. Schrijver IA, Melief MJ, Tak PP, Hazenberg MP, Laman JD. Antigen-presenting cells containing bacterial peptidoglycan in synovial tissues of rheumatoid arthritis patients coexpress costimulatory molecules and cytokines. Arthritis Rheumatol. (2000) 43:2160–8. doi: 10.1002/1529-0131(200010)43:10<2160::AID-ANR3>3.0.CO;2-T
55. Kinashi Y, Hase K. Partners in leaky gut syndrome: intestinal dysbiosis and autoimmunity. Front Immunol. (2021) 12:673708. doi: 10.3389/fimmu.2021.673708
56. Chen B, Sun L, Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J Autoimmun. (2017) 83:31–42. doi: 10.1016/j.jaut.2017.03.009
57. Izawa T, Arakaki R, Mori H, Tsunematsu T, Kudo Y, Tanaka E, et al. The nuclear receptor ahR controls bone homeostasis by regulating osteoclast differentiation via the RANK/c-fos signaling axis. J Immunol. (2016) 197:4639–50. doi: 10.4049/jimmunol.1600822
58. Dong F, Perdew GH. The aryl hydrocarbon receptor as a mediator of host-microbiota interplay. Gut Microbes. (2020) 12:1859812. doi: 10.1080/19490976.2020.1859812
59. Nguyen NT, Nakahama T, Nguyen CH, Tran TT, Le VS, Chu HH, et al. Aryl hydrocarbon receptor antagonism and its role in rheumatoid arthritis. J Exp Pharmacol. (2015) 7:29–35. doi: 10.2147/JEP.S63549
60. Heidt C, Kämmerer U, Fobker M, Rüffer A, Marquardt T, Reuss-Borst M. Assessment of intestinal permeability and inflammation bio-markers in patients with rheumatoid arthritis. Nutrients. (2023) 15:2386. doi: 10.3390/nu15102386
61. Tomofuji Y, Kishikawa T, Maeda Y, Ogawa K, Nii T, Okuno T, et al. Whole gut virome analysis of 476 Japanese revealed a link between phage and autoimmune disease. Ann Rheum Dis. (2022) 81:278–88. doi: 10.1136/annrheumdis-2021-221267
62. Yutin N, Benler S, Shmakov SA, Wolf YI, Tolstoy I, Rayko M, et al. Analysis of metagenome-assembled viral genomes from the human gut reveals diverse putative CrAss-like phages with unique genomic features. Nat Commun. (2021) 12:1044. doi: 10.1038/s41467-021-21350-w
63. Chen J, Wright K, Davis JM, Jeraldo P, Marietta EV, Murray J, et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. (2016) 8:43. doi: 10.1186/s13073-016-0299-7
64. Pacifici R. T cells, osteoblasts, and osteocytes: interacting lineages key for the bone anabolic and catabolic activities of parathyroid hormone. Ann N Y Acad Sci. (2016) 1364:11–24. doi: 10.1111/nyas.2016.1364.issue-1
65. Tajik N, Frech M, Schulz O, Schälter F, Lucas S, Azizov V, et al. Targeting zonulin and intestinal epithelial barrier function to prevent onset of arthritis. Nat Commun. (2020) 11:1995. doi: 10.1038/s41467-020-15831-7
66. Matei DE, Menon M, Alber DG, Smith AM, Nedjat-Shokouhi B, Fasano A, et al. Intestinal barrier dysfunction plays an integral role in arthritis pathology and can be targeted to ameliorate disease. Med. (2021) 2:864–883.e9. doi: 10.1016/j.medj.2021.04.013
67. Jahreis S, Kuhn S, Madaj AM, Bauer M, Polte T. Mold metabolites drive rheumatoid arthritis in mice via promotion of IFN-gamma- and IL-17-producing T cells. Food Chem Toxicol. (2017) 109:405–13. doi: 10.1016/j.fct.2017.09.027
68. van Hamburg JP, Tas SW. Molecular mechanisms underpinning T helper 17 cell heterogeneity and functions in rheumatoid arthritis. J Autoimmun. (2018) 87:69–81. doi: 10.1016/j.jaut.2017.12.006
69. Carding SR, Davis N, Hoyles L. Review article: the human intestinal virome in health and disease. Aliment Pharmacol Ther. (2017) 46:800–15. doi: 10.1111/apt.2017.46.issue-9
70. Fan Z, Yang B, Ross RP, Stanton C, Zhao J, Zhang H, et al. The prophylactic effects of different Lactobacilli on collagen-induced arthritis in rats. Food Funct. (2020) 11:3681–94. doi: 10.1039/C9FO02556A
71. Bungau SG, Behl T, Singh A, Sehgal A, Singh S, Chigurupati S, et al. Targeting probiotics in rheumatoid arthritis. Nutrients. (2021) 13:3376. doi: 10.3390/nu13103376
72. Ferreira-Halder CV, Faria AV de S, Andrade SS. Action and function of Faecalibacterium prausnitzii in health and disease. Best Pract Res Clin Gastroenterol. (2017) 31:643–8. doi: 10.1016/j.bpg.2017.09.011
73. Popescu M, Van Belleghem JD, Khosravi A, Bollyky PL. Bacteriophages and the immune system. Annu Rev Virol. (2021) 8:415–35. doi: 10.1146/annurev-virology-091919-074551
74. Wu X, He B, Liu J, Feng H, Ma Y, Li D, et al. Molecular insight into gut microbiota and rheumatoid arthritis. Int J Mol Sci. (2016) 17:431. doi: 10.3390/ijms17030431
75. Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. (2013) 39:372–85. doi: 10.1016/j.immuni.2013.08.003
76. Guo R, Li S, Zhang Y, Zhang Y, Wang G, Ullah H, et al. Dysbiotic oral and gut viromes in untreated and treated rheumatoid arthritis patients. Microbiol Spectr. (2022) 10:e0034822. doi: 10.1128/spectrum.00348-22
77. Krych Ł, Nielsen DS, Hansen AK, Hansen CHF. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-γ level in NOD mice. Gut Microbes. (2015) 6:101–9. doi: 10.1080/19490976.2015.1011876
78. Lv Z, Xiong D, Shi J, Long M, Chen Z. The interaction between viruses and intestinal microbiota: A review. Curr Microbiol. (2021) 78:3597–608. doi: 10.1007/s00284-021-02623-5
79. Sovran B, Planchais J, Jegou S, Straube M, Lamas B, Natividad JM, et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome. (2018) 6:152. doi: 10.1186/s40168-018-0538-9
80. Takahashi D, Hoshina N, Kabumoto Y, Maeda Y, Suzuki A, Tanabe H, et al. Microbiota-derived butyrate limits the autoimmune response by promoting the differentiation of follicular regulatory T cells. EBioMedicine. (2020) 58:102913. doi: 10.1016/j.ebiom.2020.102913
81. Sun H, Guo Y, Wang H, Yin A, Hu J, Yuan T, et al. Gut commensal Parabacteroides distasonis alleviates inflammatory arthritis. Gut. (2023) 72:1664–77. doi: 10.1136/gutjnl-2022-327756
82. Kang KY, Lee SH, Jung SM, Park SH, Jung BH, Ju JH. Downregulation of tryptophan-related metabolomic profile in rheumatoid arthritis synovial fluid. J Rheumatol. (2015) 42:2003–11. doi: 10.3899/jrheum.141505
83. Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe. (2018) 23:716–24. doi: 10.1016/j.chom.2018.05.003
84. Rosser EC, Piper CJM, Matei DE, Blair PA, Rendeiro AF, Orford M, et al. Microbiota-derived metabolites suppress arthritis by amplifying aryl-hydrocarbon receptor activation in regulatory B cells. Cell Metab. (2020) 31:837–851.e10. doi: 10.1016/j.cmet.2020.03.003
85. Lee K, Kwak JH, Pyo S. Inhibition of LPS-induced inflammatory mediators by 3-hydroxyanthranilic acid in macrophages through suppression of PI3K/NF-κB signaling pathways. Food Funct. (2016) 7:3073–82. doi: 10.1039/C6FO00187D
86. Sturgeon C, Fasano A. Zonulin, a regulator of epithelial and endothelial barrier functions, and its involvement in chronic inflammatory diseases. Tissue Barriers. (2016) 4:e1251384. doi: 10.1080/21688370.2016.1251384
87. Harre U, Georgess D, Bang H, Bozec A, Axmann R, Ossipova E, et al. Induction of osteoclastogenesis and bone loss by human autoantibodies against citrullinated vimentin. J Clin Invest. (2012) 122:1791–802. doi: 10.1172/JCI60975
88. Xiao M, Fu X, Ni Y, Chen J, Jian S, Wang L, et al. Protective effects of Paederia scandens extract on rheumatoid arthritis mouse model by modulating gut microbiota. J Ethnopharmacol. (2018) 226:97–104. doi: 10.1016/j.jep.2018.08.012
89. Guerreiro CS, Calado Â, Sousa J, Fonseca JE. Diet, microbiota, and gut permeability-the unknown triad in rheumatoid arthritis. Front Med (Lausanne). (2018) 5:349. doi: 10.3389/fmed.2018.00349
90. Lima AAM, Brito LFB, Ribeiro HB, Martins MCV, Lustosa AP, Rocha EM, et al. Intestinal barrier function and weight gain in malnourished children taking glutamine supplemented enteral formula. J Pediatr Gastroenterol Nutr. (2005) 40:28–35. doi: 10.1002/j.1536-4801.2005.tb00922.x
91. De Santis S, Cavalcanti E, Mastronardi M, Jirillo E, Chieppa M. Nutritional keys for intestinal barrier modulation. Front Immunol. (2015) 6:612. doi: 10.3389/fimmu.2015.00612
92. Finamore A, Massimi M, Conti Devirgiliis L, Mengheri E. Zinc deficiency induces membrane barrier damage and increases neutrophil transmigration in Caco-2 cells. J Nutr. (2008) 138:1664–70. doi: 10.1093/jn/138.9.1664
93. Pan H, Guo R, Ju Y, Wang Q, Zhu J, Xie Y, et al. A single bacterium restores the microbiome dysbiosis to protect bones from destruction in a rat model of rheumatoid arthritis. Microbiome. (2019) 7:107. doi: 10.1186/s40168-019-0719-1
94. Cheng W, Zhu N, Wang J, Yang R. A role of gut microbiota metabolites in HLA-E and NKG2 blockage immunotherapy against tumors: new insights for clinical application. Front Immunol. (2024) 15:1331518. doi: 10.3389/fimmu.2024.1331518
95. Hanchi H, Mottawea W, Sebei K, Hammami R. The genus enterococcus: between probiotic potential and safety concerns-an update. Front Microbiol. (2018) 9:1791. doi: 10.3389/fmicb.2018.01791
96. Feng J, Zhao F, Sun J, Lin B, Zhao L, Liu Y, et al. Alterations in the gut microbiota and metabolite profiles of thyroid carcinoma patients. Int J Cancer. (2019) 144:2728–45. doi: 10.1002/ijc.v144.11
97. Shi J, van de Stadt LA, Levarht EWN, Huizinga TWJ, Hamann D, van Schaardenburg D, et al. Anti-carbamylated protein (anti-CarP) antibodies precede the onset of rheumatoid arthritis. Ann Rheum Dis. (2014) 73:780–3. doi: 10.1136/annrheumdis-2013-204154
98. Raposo B, Klareskog L, Robinson WH, Malmström V, Grönwall C. The peculiar features, diversity and impact of citrulline-reactive autoantibodies. Nat Rev Rheumatol. (2024) 20:399–416. doi: 10.1038/s41584-024-01124-6
99. Scherer HU, van der Woude D, Toes REM. From risk to chronicity: evolution of autoreactive B cell and antibody responses in rheumatoid arthritis. Nat Rev Rheumatol. (2022) 18:371–83. doi: 10.1038/s41584-022-00786-4
100. de Hair MJH, van de Sande MGH, Ramwadhdoebe TH, Hansson M, Landewé R, van der Leij C, et al. Features of the synovium of individuals at risk of developing rheumatoid arthritis: implications for understanding preclinical rheumatoid arthritis. Arthritis Rheumatol. (2014) 66:513–22. doi: 10.1002/art.38273
101. Bodkhe R, Balakrishnan B, Taneja V. The role of microbiome in rheumatoid arthritis treatment. Ther Adv Musculoskelet Dis. (2019) 11:1759720X19844632. doi: 10.1177/1759720X19844632
102. Potempa J, Mydel P, Koziel J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat Rev Rheumatol. (2017) 13:606–20. doi: 10.1038/nrrheum.2017.132
103. Liao F, Li Z, Wang Y, Shi B, Gong Z, Cheng X. Porphyromonas gingivalis may play an important role in the pathogenesis of periodontitis-associated rheumatoid arthritis. Med Hypotheses. (2009) 72(6):732–5. doi: 10.1016/j.mehy.2008.12.040
104. Yang S, Chen H, Wei B, Xiang M, Hu Z, Peng Z, et al. Clindamycin administration increases the incidence of collagen-induced arthritis in mice through the prolonged impact of gut immunity. Inflammation. (2018) 41:1900–11. doi: 10.1007/s10753-018-0833-4
105. Yue M, Tao Y, Fang Y, Lian X, Zhang Q, Xia Y, et al. The gut microbiota modulator berberine ameliorates collagen-induced arthritis in rats by facilitating the generation of butyrate and adjusting the intestinal hypoxia and nitrate supply. FASEB J. (2019) 33:12311–23. doi: 10.1096/fj.201900425RR
106. Doonan J, Tarafdar A, Pineda MA, Lumb FE, Crowe J, Khan AM, et al. The parasitic worm product ES-62 normalises the gut microbiota bone marrow axis in inflammatory arthritis. Nat Commun. (2019) 10:1554. doi: 10.1038/s41467-019-09361-0
107. Peng J, Lu X, Xie K, Xu Y, He R, Guo L, et al. Dynamic alterations in the gut microbiota of collagen-induced arthritis rats following the prolonged administration of total glucosides of paeony. Front Cell Infect Microbiol. (2019) 9:204. doi: 10.3389/fcimb.2019.00204
108. Mizuno M, Noto D, Kaga N, Chiba A, Miyake S. The dual role of short fatty acid chains in the pathogenesis of autoimmune disease models. PloS One. (2017) 12:e0173032. doi: 10.1371/journal.pone.0173032
109. Guo LX, Wang HY, Liu XD, Zheng JY, Tang Q, Wang XN, et al. Saponins from Clematis mandshurica Rupr. regulates gut microbiota and its metabolites during alleviation of collagen-induced arthritis in rats. Pharmacol Res. (2019) 149:104459. doi: 10.1016/j.phrs.2019.104459
110. Hatakka K, Martio J, Korpela M, Herranen M, Poussa T, Laasanen T, et al. Effects of probiotic therapy on the activity and activation of mild rheumatoid arthritis–a pilot study. Scand J Rheumatol. (2003) 32:211–5. doi: 10.1080/03009740310003695
111. Vaghef-Mehrabany E, Alipour B, Homayouni-Rad A, Sharif SK, Asghari-Jafarabadi M, Zavvari S. Probiotic supplementation improves inflammatory status in patients with rheumatoid arthritis. Nutrition. (2014) 30:430–5. doi: 10.1016/j.nut.2013.09.007
112. Alipour B, Homayouni-Rad A, Vaghef-Mehrabany E, Sharif SK, Vaghef-Mehrabany L, Asghari-Jafarabadi M, et al. Effects of Lactobacillus casei supplementation on disease activity and inflammatory cytokines in rheumatoid arthritis patients: a randomized double-blind clinical trial. Int J Rheum Dis. (2014) 17:519–27. doi: 10.1111/apl.2014.17.issue-5
113. Cannarella LAT, Mari NL, Alcântara CC, Iryioda TMV, Costa NT, Oliveira SR, et al. Mixture of probiotics reduces inflammatory biomarkers and improves the oxidative/nitrosative profile in people with rheumatoid arthritis. Nutrition. (2021) 89:111282. doi: 10.1016/j.nut.2021.111282
114. de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. (2017) 152:1–12. doi: 10.1111/imm.2017.152.issue-1
115. Marietta E, Mangalam AK, Taneja V, Murray JA. Intestinal Dysbiosis in, and Enteral Bacterial Therapies for, Systemic Autoimmune Diseases. Front Immunol. (2020) 11:573079. doi: 10.3389/fimmu.2020.573079
116. Bruusgaard A, Andersen RB. Abnormal bile acid metabolism in rheumatoid arthritis. Preliminary communication. Dan Med Bull. (1976) 23:95–8.
117. Li ZY, Zhou JJ, Luo CL, Zhang LM. Activation of TGR5 alleviates inflammation in rheumatoid arthritis peripheral blood mononuclear cells and in mice with collagen II−induced arthritis. Mol Med Rep. (2019) 20:4540–50. doi: 10.3892/mmr.2019.10711
118. Chen H, Fu X, Wu X, Zhao J, Qiu F, Wang Z, et al. Gut microbial metabolite targets HDAC3-FOXK1-interferon axis in fibroblast-like synoviocytes to ameliorate rheumatoid arthritis. Bone Res. (2024) 12:31. doi: 10.1038/s41413-024-00336-6
119. Hasan H, Ismail H, El-Orfali Y, Khawaja G. Therapeutic benefits of Indole-3-Carbinol in adjuvant-induced arthritis and its protective effect against methotrexate induced-hepatic toxicity. BMC Complement Altern Med. (2018) 18:337. doi: 10.1186/s12906-018-2408-1
120. Sjögren K, Engdahl C, Henning P, Lerner UH, Tremaroli V, Lagerquist MK, et al. The gut microbiota regulates bone mass in mice. J Bone Miner Res. (2012) 27:1357–67. doi: 10.1002/jbmr.1588
121. Hvatum M, Kanerud L, Hällgren R, Brandtzaeg P. The gut-joint axis: cross reactive food antibodies in rheumatoid arthritis. Gut. (2006) 55:1240–7. doi: 10.1136/gut.2005.076901
122. Maeda Y, Kurakawa T, Umemoto E, Motooka D, Ito Y, Gotoh K, et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. (2016) 68:2646–61. doi: 10.1002/art.v68.11
123. Li Y, Ouyang Y, He C. Research trends on clinical fecal microbiota transplantation: A biliometric analysis from 2001 to 2021. Front Immunol. (2022) 13:991788. doi: 10.3389/fimmu.2022.991788
124. Danne C, Rolhion N, Sokol H. Recipient factors in faecal microbiota transplantation: one stool does not fit all. Nat Rev Gastroenterol Hepatol. (2021) 18:503–13. doi: 10.1038/s41575-021-00441-5
125. Meng Q, Lin M, Song W, Wu J, Cao G, Huang P, et al. The gut-joint axis mediates the TNF-induced RA process and PBMT therapeutic effects through the metabolites of gut microbiota. Gut Microbes. (2023) 15:2281382. doi: 10.1080/19490976.2023.2281382
Keywords: gut microbiota, intestinal metabolites, gut-joint axis, rheumatoid arthritis, bidirectional regulation
Citation: Xie X, Chen X, Wang X, Wang S and Qi P (2025) Dual regulatory effects of gut microbiota and their metabolites in rheumatoid arthritis: balancing pathogenic and protective mechanisms. Front. Immunol. 16:1584023. doi: 10.3389/fimmu.2025.1584023
Received: 26 February 2025; Accepted: 07 April 2025;
Published: 30 April 2025.
Edited by:
Francisco Jose Roig, Universidad San Jorge, SpainCopyright © 2025 Xie, Chen, Wang, Wang and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peng Qi, ODIxODEwMTQ2QHFxLmNvbQ==
†These authors have contributed equally to this work
 Xingwen Xie
Xingwen Xie Xin Chen1†
Xin Chen1† Peng Qi
Peng Qi