- 1Department of Hematology, Puyang Oilfield General Hospital Affiliated with Xinxiang Medical College, Puyang, Henan, China
- 2Department of Hematology and Central Hematology Laboratory, Inselspital, Bern University Hospital, University of Bern, Bern, Switzerland
- 3Institute of Hematology and Transfusion, Department of Experimental Hematology, Warsaw, Poland
- 4Department of Medicine, Sanford Stem Cell Institute, Moores Cancer Center, University of California San Diego, San Diego, CA, United States
Primary immune thrombocytopenia (ITP) is an autoimmune disorder characterized by platelet destruction and impaired production, leading to bleeding risk. While immunosuppressive therapies are standard, many patients experience relapses or refractory disease, highlighting the need for novel approaches. Emerging evidence suggests the gut microbiota plays a role in immune regulation, yet its impact on ITP remains unclear. Dysbiosis has been linked to immune dysfunction in other autoimmune diseases, but whether it drives or results from immune dysregulation in ITP is debated. This review explores the gut-immune axis in ITP, focusing on microbiota-driven immune modulation, cytokine signaling, and platelet homeostasis. We assess microbiota-targeted interventions, including fecal microbiota transplantation (FMT), probiotics, and dietary modifications, while addressing key controversies and knowledge gaps. Advances in microbiome sequencing and artificial intelligence may facilitate personalized interventions. Standardizing microbiota-based diagnostics and validating their efficacy in clinical trials are crucial for their integration into ITP management. Bridging these gaps may lead to microbiota-driven strategies that enhance immune regulation and improve patient outcomes.
Highlights
● The gut-immune axis influences ITP pathogenesis and platelet homeostasis.
● Dysbiosis disrupts immune regulation and drives disease progression.
● FMT, probiotics, and dietary interventions offer potential ITP therapies.
● Microbiome sequencing and AI may advance personalized treatments.
● Standardization and clinical validation are crucial for microbiota-based strategies.
1 Introduction
Primary immune thrombocytopenia (ITP) is an acquired autoimmune disorder characterized by a persistent low platelet count due to immune-mediated platelet destruction and impaired platelet production (1, 2). While immunosuppressive therapies, such as corticosteroids and intravenous immunoglobulin (IVIG), are commonly used, their effectiveness varies, and long-term management remains challenging (3, 4). In particular, the frequent need for alternative therapeutic strategies in refractory cases underscores the need for a deeper understanding of ITP pathogenesis and the development of novel treatment approaches.
Traditionally, ITP is associated with autoantibody-mediated platelet destruction, where antibodies target platelet surface glycoproteins such as GPIIb/IIIa and GPIb/IX (5–7). However, emerging evidence reveals a more complex pathophysiology involving dysregulated T-cell responses, pro-inflammatory cytokines, and defective megakaryopoiesis. An imbalance between regulatory T cells (Tregs) and effector T cells (Th1 and Th17) drives persistent inflammation and immune-mediated platelet destruction, underscoring the multifaceted nature of ITP pathogenesis (8, 9). Notably, a reduction or dysfunction of Tregs is associated with the induction of ITP, whereas their expansion or restoration is considered immunoprotective, restoring immune tolerance and suppressing autoreactive responses against platelets (10).
Beyond these immune mechanisms, recent research suggests that gut microbiota plays a crucial role in shaping immune responses in ITP (11). The gut-immune axis, which governs interactions between intestinal microbiota and systemic immunity, has been implicated in various autoimmune diseases, including systemic lupus erythematosus and rheumatoid arthritis (RA) (12–14). Dysbiosis, an imbalance in gut microbial composition, has been shown to drive immune dysregulation, promote inflammation, and influence hematologic conditions (15–17). However, the precise role of the gut microbiota in ITP pathogenesis remains unclear, presenting a significant knowledge gap.
A key controversy center on whether alterations in the gut microbiota are a consequence of immune dysfunction in ITP or an independent driver of disease progression (18). Additionally, while microbiota-targeted interventions such as fecal microbiota transplantation (FMT) and probiotics have shown promise in other autoimmune conditions, their therapeutic potential in ITP remains largely unexplored (19, 20). These gaps highlight the need for further investigation into the mechanisms linking gut dysbiosis to ITP pathogenesis and treatment response.
This review therefore comprehensively analyzes the gut-immune axis in ITP, critically evaluating current knowledge and identifying key unanswered questions. We explore the potential of microbiota-targeted therapies to restore immune homeostasis, highlight areas of consensus and controversy, and propose directions for future research. By synthesizing available evidence and pinpointing knowledge gaps, we aim to advance the understanding of gut microbiota’s role in ITP and its implications for novel therapeutic strategies.
2 The gut-immune axis: a new perspective in ITP pathogenesis
Recent advancements in immunology and microbiome research have revealed the intricate interplay between the gut microbiota and systemic immune regulation (21, 22). This relationship is particularly relevant in autoimmune disorders such as primary ITP, where immune dysregulation leads to platelet destruction. The gut-immune axis serves as a dynamic interface between microbial communities and immune homeostasis, influencing inflammation and hematologic balance (22). Although disruptions in this axis have been implicated in several autoimmune diseases, their precise role in hematologic diseases like ITP remains poorly understood, underscoring significant gaps in current knowledge (19, 23).
2.1 Overview of the gut microbiota and immune system interactions
The gut microbiota, comprising a diverse and dynamic ecosystem of bacteria, viruses, fungi, and archaea, interacts closely with the immune system to maintain immune homeostasis. Gut-associated lymphoid tissue (GALT) plays a pivotal role in sensing microbial antigens and orchestrating immune responses (24). Dendritic cells (DCs) sample microbial metabolites and antigens, directing the differentiation of naïve T cells into Tregs or effector T cells (Th1 and Th17) (25, 26). This interaction promotes immune tolerance while maintaining a controlled inflammatory response.
However, the precise role of the gut microbiota in modulating hematologic immune responses remains controversial (22, 27). While some studies suggest that gut microbial communities influence immune regulation through cytokine production, others propose that the immune system primarily shapes microbiota composition (28, 29). This bidirectional relationship underscores the complexity of gut-immune interactions and necessitates further investigation in the context of ITP.
The intricate interactions between the gut microbiota and the immune system, including the influence of both commensal and pathogenic microbes in shaping immune tolerance, modulating inflammatory responses, and regulating systemic immune regulation, is critical to understanding hematologic disorders such as ITP. These mechanisms are visually summarized in Figure 1.
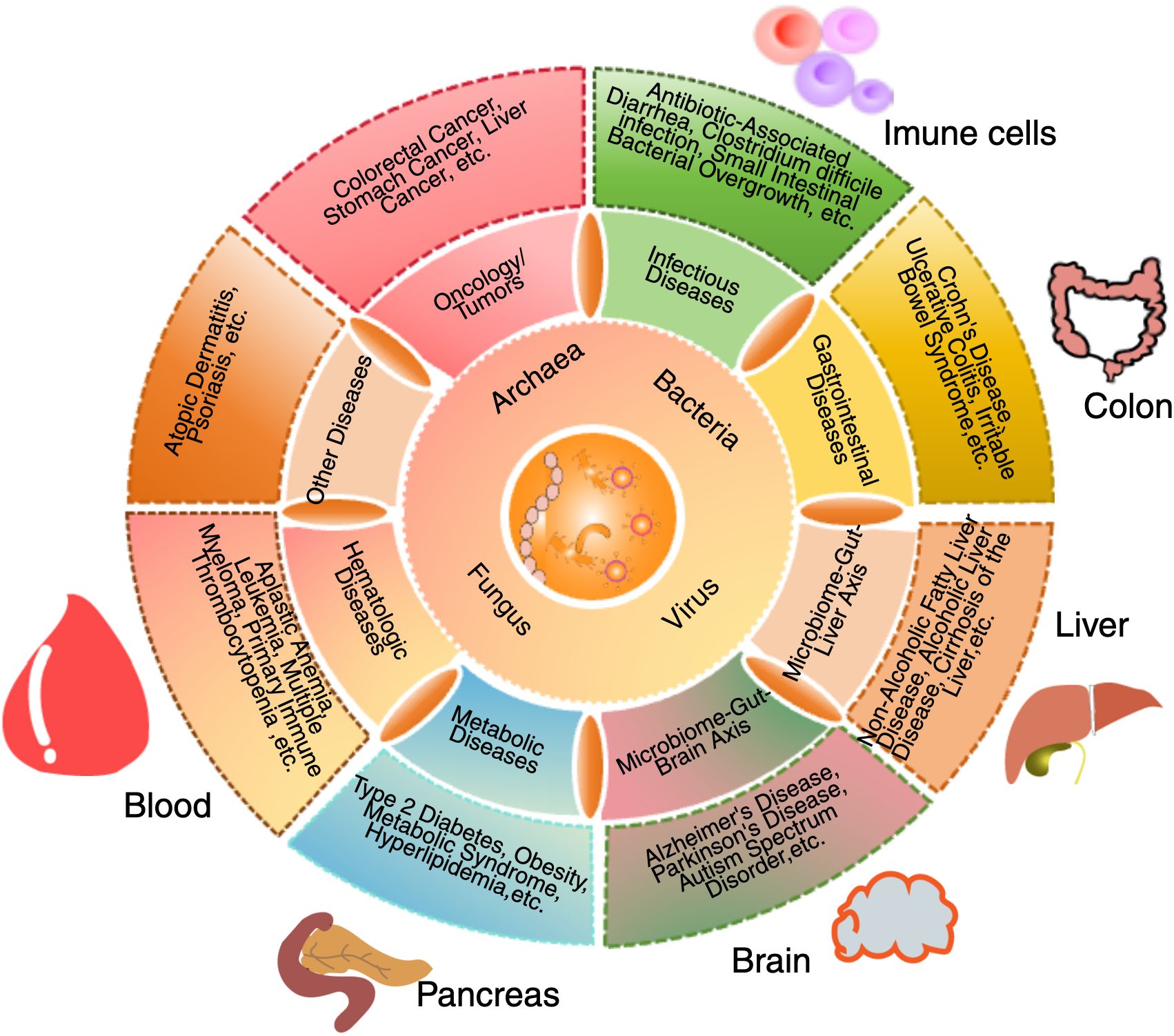
Figure 1. Interactions between gut microbiota, systemic organs and the immune system. The gut microbiota plays a central role in regulating immune function, metabolism, and overall systemic health through interactions with multiple organ systems. This schematic illustration depicts how bacteria, viruses, fungi, and archaea influence host physiology by modulating immune responses, metabolic processes, and disease development. Dysbiosis has been implicated in various conditions, including autoimmune diseases, neurodegenerative disorders, metabolic syndromes, and inflammatory diseases. Key organ systems affected by alterations in microbiota include the immune system, colon, liver, and pancreas. In immune system, the gut microbiota regulates Tregs, Th1/Th17 responses, and inflammation. In the colon, microbial communities influence gut barrier integrity and are associated with inflammatory bowel diseases. In the liver, the gut-derived metabolites affect metabolic conditions such as non-alcoholic fatty liver disease (NAFLD). In the pancreas, microbiota alterations have been associated with type 2 diabetes and other metabolic disorders. In the bloodstream, microbial dysbiosis is linked to hematologic conditions, including immune thrombocytopenia (ITP). In the brain, microbiota-derived metabolites have been implicated in neurological conditions such as Alzheimer’s disease, depression, and neuroinflammation. By modulating microbial composition and function, the gut microbiota exerts local and systemic effects that contribute to immune homeostasis, disease pathogenesis, and potential therapeutic interventions.
2.2 Mechanisms of gut microbiota in immune homeostasis and the role of B cells in ITP
The gut microbiota plays a pivotal role in immune regulation through multiple interconnected mechanisms, influencing both systemic and hematologic immune responses (12). Key pathways include microbial metabolite production, pattern recognition receptor (PRR) signaling, and maintenance of intestinal barrier integrity, each impacting immune homeostasis and ITP disease progression (30).
One essential pathway involves the production of microbial metabolites, particularly short-chain fatty acids (SCFAs) such as butyrate, propionate, and acetate, generated by commensal bacteria fermenting dietary fiber (31). SCFAs enhance regulatory T cell (Treg) differentiation, suppress inflammatory cytokines (IL-6, TNF-α, IFN-γ), and promote anti-inflammatory responses both systemically and within the gut (32). A reduction in SCFA-producing bacteria has been linked to impaired immune tolerance, heightened inflammation, and increased risk of autoimmunity, suggesting a potential link between dysbiosis and ITP pathogenesis (33, 34). Importantly, diminished Treg frequency or function is associated with ITP induction, whereas restoration or expansion of Tregs has been shown to confer protective effects by re-establishing immune homeostasis and attenuating platelet destruction (35).
Another critical mechanism is PRR signaling, where Toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) recognize microbe-associated molecular patterns (MAMPs) to distinguish commensal bacteria from pathogens (36, 37). This interaction normally regulates immune tolerance by limiting unnecessary immune activation (38). However, in gut dysbiosis, aberrant PRR signaling may trigger chronic immune activation, disrupting the balance between Tregs/effector T cells (Th1/Th17) and promoting a pro-inflammatory milieu that exacerbates platelet destruction in ITP (39, 40).
Furthermore, the gut microbiota maintains intestinal barrier integrity, preventing the translocation of microbial components into systemic circulation (41). Tight junction proteins (occludin, claudin, zonulin) regulate intestinal permeability, preventing leakage of bacterial endotoxins e.g., lipopolysaccharides (LPS) into the bloodstream (41, 42). Dysbiosis-induced compromise of the gut barrier can elevate circulating LPS levels, trigger systemic inflammation, and aberrantly activate monocytes and DCs factors that may contribute to immune dysregulation and platelet destruction in ITP (11, 43).
Although these mechanisms highlight the immunomodulatory potential of the gut microbiota, key controversies remain regarding its specific role in ITP (12, 44). For instance, SCFA’s effects on immune tolerance appear to be context-dependent, varying with disease state, microbial composition, and host genetic factors (45, 46). Similarly, while PRR signaling is crucial for immune surveillance, its dysbiosis-related overactivation has been associated with both immune suppression and chronic inflammation, resulting in conflicting findings in hematologic disorders (22). This lack of consensus underscores the need for further research to clarify how gut microbiota alterations influence platelet homeostasis and immune regulation in ITP (19, 47).
In addition to T cell-mediated pathways, B cells, particularly regulatory B cells (Bregs) have emerged as important players in maintaining immune tolerance in autoimmune disorders, including ITP (48). Bregs exert their immunosuppressive effects primarily through IL-10 production, which inhibits pro-inflammatory T cell responses and promotes Treg development (49). Dysregulation of Bregs has been observed in ITP patients, suggesting that impaired Breg function may contribute to loss of peripheral tolerance and heightened platelet destruction (50). Gut microbiota has been shown to modulate Breg development and function, likely via microbial metabolites and PRR signaling. Thus, altered microbial composition in ITP may impair Breg-mediated suppression of autoimmunity, further implicating the gut-immune axis in disease pathogenesis (51).
2.3 Dysbiosis and its impact on autoimmunity and hematologic diseases
Dysbiosis, defined as a disruption in gut microbial composition, has been implicated in numerous autoimmune and hematologic disorders (16, 52). Studies have demonstrated that conditions such as systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), and multiple sclerosis (MS) exhibit characteristic microbial imbalances, often marked by an overrepresentation of pro-inflammatory species (e.g., Prevotella, Ruminococcus) and a concurrent depletion of beneficial bacteria (e.g., Bifidobacterium, Lactobacillus) (53, 54). These alterations are associated with immune dysregulation, excessive cytokine production, and chronic inflammation, all of which contribute to disease progression (55).
In hematologic disorders, gut dysbiosis has been increasingly associated with the breakdown of immune tolerance, platelet dysregulation, and systemic inflammation (19, 52). Certain bacterial taxa, such as Enterobacteriaceae and Prevotella, have been associated with an elevated pro-inflammatory Th17 response, promoting systemic inflammation and disrupting hematologic homeostasis (41, 56). Conversely, commensal bacteria like Bifidobacterium and Lactobacillus have been shown to enhance regulatory T cell (Treg) activity, mitigate excessive immune activation, and support immune balance (57, 58). The depletion of these beneficial microbes in patients with ITP suggests a potential role of the gut microbiota in modulating platelet homeostasis; however, direct causal relationships have yet to be confirmed (19, 47).
Beyond immune cell modulation, dysbiosis is associated with metabolic shifts that further influence immune responses (59, 60). A decrease in SCFA-producing bacteria, such as Bacteroides and Firmicutes, correlates with reduced Treg activity, impaired immune regulation, and increased inflammation, all of which are observed in autoimmune conditions (32, 61). Additionally, altered bile acid metabolism and tryptophan catabolism can modulate T cell differentiation, cytokine production, and systemic immune responses, potentially exacerbating platelet destruction in ITP (32, 62). However, whether these microbial and metabolic changes are a cause or consequence of immune dysregulation in ITP remains unclear, necessitating longitudinal studies and mechanistic research to establish causality.
A growing body of evidence suggests that distinct microbiota alterations are shared among various autoimmune diseases, highlighting common patterns of dysbiosis that may drive immune dysregulation across multiple conditions (12, 44). Table 1 summarizes key microbial alterations in autoimmune diseases, including ITP, and their associated immune effects.
2.4 Evidence linking gut microbiota alterations to ITP development and severity
Emerging research suggests that alterations in gut microbiota may contribute to the pathogenesis of ITP; however, findings remain inconsistent (18, 69). Comparative studies of the gut microbiomes of ITP patients and healthy controls have identified notable microbial differences (52, 70). Specifically, a reduction in SCFA-producing bacteria, such as Bacteroides and Firmicutes, may impair immune tolerance mechanisms, potentially exacerbating autoimmune responses (12, 46). Concurrently, an increase in pro-inflammatory taxa, including Escherichia coli and Enterobacteriaceae, has been associated with heightened Th17 responses and systemic inflammation (71). Additionally, experimental models suggest that antibiotic-induced dysbiosis can alter platelet counts and immune responses, reinforcing a potential link between gut microbiota and ITP progression (19, 72).
Despite these compelling findings, several critical gaps remain in the literature (11, 47, 72–74). Many studies are limited by small sample sizes, cross-sectional designs, and a lack of longitudinal analyses, making it difficult to establish causality between gut dysbiosis and ITP development (74). To clarify the causal relationship between gut dysbiosis and ITP, recent proposals emphasize the need for prospective longitudinal studies that track microbiota composition before and after disease onset or treatment (72). Additionally, mechanistic investigations using gnotobiotic mouse models may help elucidate how specific microbial taxa and metabolites influence immune regulation and platelet homeostasis (75, 76). Furthermore, while microbiota-targeted therapies such as FMT and probiotics have shown promise in other autoimmune diseases, their efficacy and applicability in ITP remain largely unexplored (77). Emerging studies indicate that microbiota-driven systemic metabolic changes can influence immune regulation in diseases beyond the gut, as seen in brain metastasis, where alterations in the gut microbiome affect tumor progression via the gut-to-brain axis (78).
To address these knowledge gaps, future research should focus on three key areas. First, longitudinal studies are needed to investigate how changes in gut microbiota correlate with the onset, severity, and treatment response of ITP over time (19). Second, mechanistic studies should explore the causal relationships between specific microbial taxa, microbial metabolites, immune dysregulation, and platelet homeostasis (47). Lastly, controlled clinical trials are essential to assess the therapeutic potential of microbiota-targeted interventions, including probiotics, prebiotics, dietary modifications, and FMT, in the management of ITP (11, 79).
By addressing these critical gaps, researchers can determine whether modulating the gut microbiota represents a viable therapeutic avenue for improving immune regulation and clinical outcomes in patients with ITP (80).
3 Gut microbiota modulation in ITP: a novel therapeutic strategy
Given the emerging evidence linking gut microbiota dysbiosis to immune dysregulation in ITP, researchers have begun exploring microbiota-targeted interventions as potential therapeutic strategies (19, 81). By restoring gut microbial balance, these approaches aim to modulate immune responses, promote immune tolerance, and mitigate the pathogenic mechanisms underlying ITP. Several microbiota-based interventions, including fecal FMT, probiotics, dietary modifications, and microbiome-targeted pharmacologic strategies, have shown promise in modulating the gut-immune axis (82, 83). However, the clinical translation of these therapies remains challenging due to a limited mechanistic understanding and the need for well-designed clinical trials.
Various microbiota-targeted therapies have been proposed as potential interventions for ITP, aiming to restore microbial balance and modulate immune function (11, 84). These strategies include FMT, probiotics, prebiotics, SCFA supplementation, and bile acid modulation (85, 86). While some have shown promise in autoimmune diseases, their application in ITP remains underexplored. The following Table 2 summarizes the mechanisms, current evidence, and challenges associated with these microbiota-based therapies.
3.1 Fecal microbiota transplantation: mechanisms, clinical applications, and emerging evidence in ITP
FMT has gained attention as a promising approach to modulating gut microbiota composition (87). This procedure involves transferring fecal material from a healthy donor to a recipient, aiming to restore microbial diversity and improve immune homeostasis. Studies suggest that FMT can replenish beneficial taxa such as Bacteroides and Firmicutes, which are associated with immune tolerance (88). Originally developed for treating recurrent Clostridioides difficile infections, FMT has shown potential in various autoimmune and inflammatory diseases, including ITP (81).
Restoring gut microbial diversity and modulating immune responses through fecal microbiota transplantation (FMT) has emerged as a promising therapeutic strategy to improve disease outcomes, as illustrated in Figure 2. This schematic highlights the key mechanisms through which FMT may contribute to immune homeostasis, including shifts in microbial composition, an increase in the activity of Tregs, and a reduction in systemic inflammation.
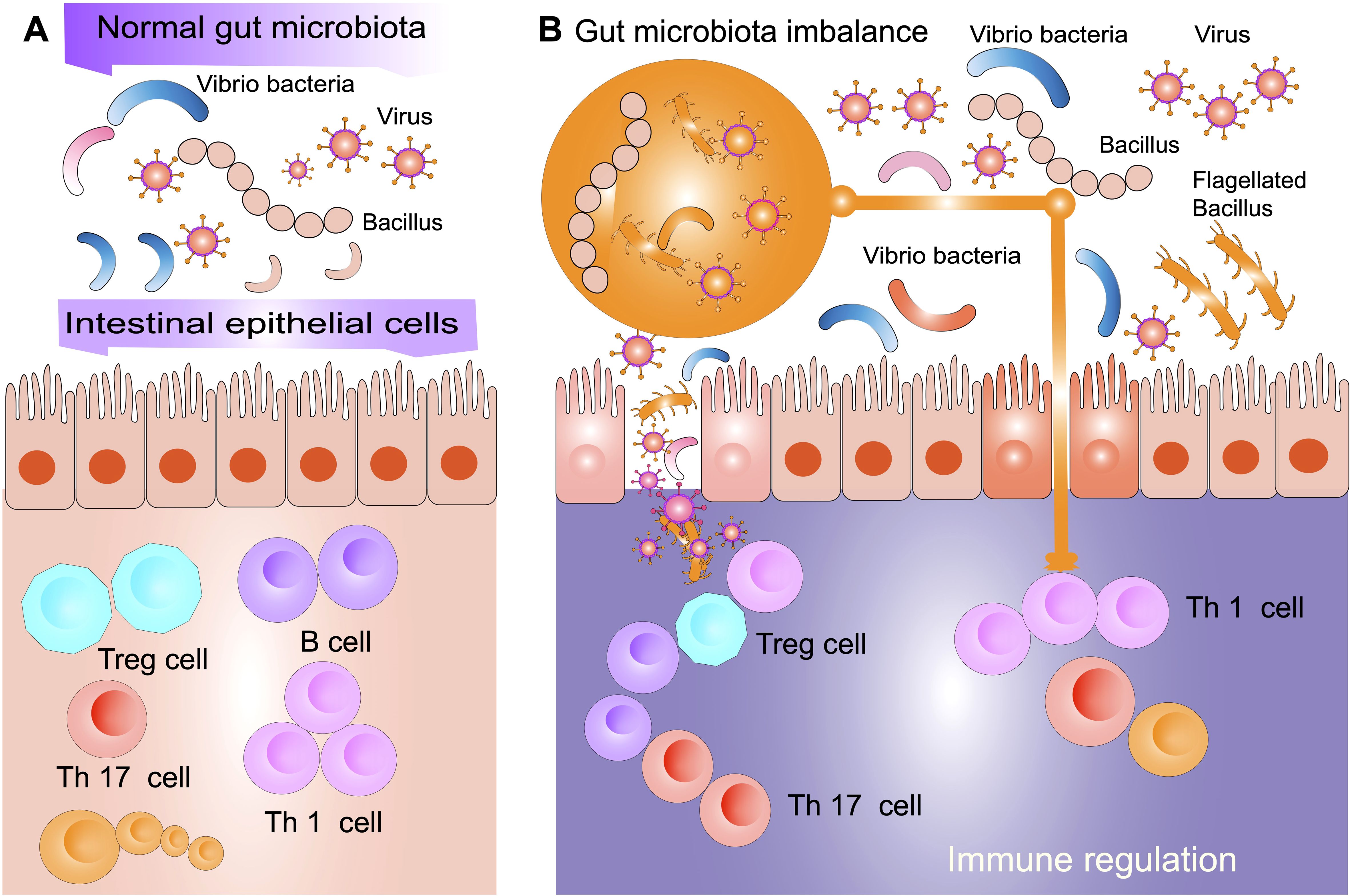
Figure 2. Gut microbiota balance and immune modulation. (A) In a normal gut microbiota environment, commensal bacteria such as Vibrio and Bacillus interact with intestinal epithelial cells to maintain immune homeostasis. This balance promotes the differentiation of regulatory T cells (Treg) and immune tolerance while preventing excessive inflammation driven by Th1 and Th17 cells. A balanced gut microbiota contributes to a well-regulated immune system by enhancing gut barrier integrity and modulating host immune responses. (B) In a state of gut microbiota imbalance (known as dysbiosis), there is an overrepresentation of pathogenic bacteria and viruses, along with a reduction in beneficial microbial populations. This microbial shift disrupts gut barrier function, leading to increased bacterial translocation and heightened immune activation. Dysbiosis skews immune regulation by reducing Treg activity and increasing Th1 and Th17 responses, promoting systemic inflammation and immune dysregulation. Such alterations in the gut microbiota may contribute to autoimmune conditions, including ITP, by exacerbating inflammatory pathways and impairing immune tolerance.
FMT modulates immune responses through several mechanisms, including the enhancement of Tregs, increased Bifidobacterium abundance, and suppression of pro-inflammatory Th1 and Th17 responses, as observed in autoimmune conditions such as multiple sclerosis and inflammatory bowel disease (IBD) (12, 46). Additionally, microbial metabolites such as SCFAs contribute to immune modulation by strengthening the intestinal barrier and suppressing excessive immune activation (89, 90).
The gut microbiota also influences systemic immune function, as demonstrated by recent findings on the gut-brain axis, where microbiota-derived metabolites, such as kynurenic acid, impact immune interactions at distant sites. This highlights the broader systemic effects of gut microbiota and underscores the need for further investigation into microbiota-targeted therapies in autoimmune conditions like ITP (78).
Emerging evidence suggests that gut microbiota may directly influence platelet regulation. Certain microbial metabolites, including SCFAs and secondary bile acids, have been shown to modulate platelet function by affecting megakaryocyte differentiation and platelet activation (11, 91). Moreover, gut dysbiosis has been associated with increased platelet aggregation and altered hemostatic balance in other diseases, suggesting that restoring microbial equilibrium through FMT could contribute to improved platelet homeostasis (92). While direct evidence of FMT’s effect on platelet regulation in ITP is limited, case reports and small clinical studies have observed platelet count improvement following microbiota restoration therapies (19, 72).
Despite these promising findings, several challenges remain in translating FMT into a standardized treatment for ITP. Donor microbiota variability and long-term engraftment pose significant challenges, as microbial compositions differ significantly between donors and recipients, potentially affecting therapeutic outcomes. Additionally, safety concerns include the risk of pathogen transmission, unintended immune activation, and unpredictable long-term effects (93). Therefore, well-controlled, randomized clinical trials are necessary to confirm the efficacy and safety of FMT in ITP management and to determine optimal protocols for donor selection, microbiota preparation, and recipient response monitoring. Addressing these issues is critical for integrating FMT into mainstream ITP treatment (88).
3.2 Probiotics and prebiotics: potential to restore gut balance and regulate immune responses
Probiotics and prebiotics represent another avenue for modulating gut microbiota in ITP (21, 47). Probiotics, including specific Lactobacillus and Bifidobacterium strains, enhance beneficial bacterial populations, while prebiotics, such as inulin and fructooligosaccharides, selectively stimulate microbial growth, promoting gut-immune homeostasis (94). However, the immunomodulatory effects of probiotics are highly strain-dependent, with some strains exerting potent anti-inflammatory properties, while others may have limited or even opposing effects in different individuals (95).
Clinical studies suggest that Lactobacillus rhamnosus and Bifidobacterium breve increase Treg activity while reducing IL-6 and TNF-α, leading to decreased systemic inflammation in autoimmune diseases (96). Additionally, Lactobacillus plantarum has been shown to enhance gut barrier integrity by upregulating tight junction proteins, thereby preventing bacterial translocation and reducing systemic immune activation (97). Similarly, prebiotics, including inulin and fructooligosaccharides, serve as metabolic substrates for beneficial bacteria, fostering a gut environment that supports immune homeostasis (98). The combination of specific probiotic strains with targeted prebiotic supplementation (synbiotics) may offer enhanced therapeutic potential by optimizing microbial colonization and metabolic activity (99).
Despite promising preclinical and clinical evidence supporting probiotic use in other autoimmune diseases, their application in ITP remains underexplored (100). One of the major challenges is the variability in probiotic efficacy, which stems from strain-specific effects, inter-individual differences in microbiota composition, and inconsistencies in host immune responses (101). For instance, while Bifidobacterium adolescentis has been shown to reduce Th17-mediated inflammation in rheumatoid arthritis, similar effects have not been confirmed in ITP (102). Additionally, the interaction between probiotics and endogenous microbial communities can lead to variable colonization success, limiting the predictability of therapeutic outcomes (103).
Future research should focus on defining optimal probiotic formulations, identifying microbial signatures predictive of response, and evaluating their impact on platelet homeostasis and immune regulation (69). Advancements in microbiome sequencing and metabolomic profiling may enable the development of personalized microbiota-based interventions, tailoring probiotic and prebiotic therapies to individual ITP patients for improved efficacy (104).
3.3 Dietary and metabolomic interventions: role of SCFAs, bile acids, and tryptophan metabolites in immune modulation
Dietary interventions play a pivotal role in shaping gut microbiota composition and function (87). Specific dietary components influence the production of key microbial metabolites, which, in turn, modulate immune responses (105). Among these, SCFAs, bile acids, and tryptophan-derived metabolites are particularly significant in maintaining immune homeostasis and resolving inflammation (105). However, while their immunoregulatory effects have been well characterized in autoimmune diseases, their specific impact on platelet regulation in ITP remains an emerging area of research.
SCFAs, including butyrate and propionate, are microbial fermentation products that exert profound immunomodulatory effects (53). These metabolites enhance regulatory T cell (Treg) differentiation and inhibit pro-inflammatory cytokines such as IL-6, TNF-α, and IFN-γ, thereby reducing autoimmune activity (106). Butyrate, in particular, acts as a histone deacetylase inhibitor, promoting Treg expansion and suppressing Th17-mediated inflammation, mechanism relevant to platelet destruction in ITP (69, 107). Additionally, SCFAs strengthen gut barrier integrity by upregulating tight junction proteins, thereby reducing microbial translocation and systemic immune activation factors that may contribute to excessive immune responses in ITP (62, 69).
Recent studies suggest that lower SCFA levels in autoimmune conditions are linked to immune dysregulation and altered thrombopoiesis (108, 109). Butyrate has been shown to reduce megakaryocyte apoptosis, potentially impacting platelet production and turnover (110). Additionally, SCFA supplementation has demonstrated protective effects on platelet homeostasis in inflammatory conditions, indicating a possible therapeutic avenue for ITP (32, 108).
Bile acids, traditionally recognized for their role in lipid metabolism, have recently emerged as key modulators of immune responses, influencing T-cell differentiation via interactions with gut microbiota (111). Secondary bile acids, such as deoxycholic acid and lithocholic acid, regulate immune pathways through host receptors, including the Farnesoid X receptor (FXR) and Takeda G-protein-coupled receptor 5 (TGR5) (112). FXR activation suppresses Th17 cell differentiation, thereby reducing inflammatory cytokine production, a process that could mitigate platelet autoantibody formation in ITP (113). TGR5 signaling has been shown to enhance Treg function, contributing to immune tolerance and reducing autoimmunity in conditions such as SLE and RA (8, 114).
Alterations in bile acid metabolism have been identified in ITP patients, suggesting a potential role in immune dysregulation (47). Recent studies indicate that bile acid supplementation can modulate thrombopoiesis, potentially linking gut microbiota-derived bile acids to platelet production and function (115). These findings support the investigation of bile acid-targeted therapies in ITP as a novel immunomodulatory approach (11).
Tryptophan metabolism also plays a crucial role in immune homeostasis, with its metabolites modulating immune responses via the aryl hydrocarbon receptor (AhR) and indoleamine 2,3-dioxygenase (IDO) pathways (116). Kynurenine, an AhR ligand, has been shown to promote Treg differentiation, enhancing immune tolerance and reducing autoimmune activity (117). Indole derivatives regulate Th17 differentiation, thereby controlling inflammatory responses that may drive platelet destruction in ITP (2, 118). Additionally, serotonin, a tryptophan metabolite, has been implicated in platelet aggregation, further underscoring the potential gut-immune-thrombosis axis in ITP (11, 43). Dysregulation of tryptophan metabolism in autoimmune conditions suggests a possible link to ITP pathophysiology, with recent findings highlighting microbiota-mediated tryptophan metabolism as a key factor in hematologic disorders (119).
Despite growing interest in dietary interventions, their role in ITP remains speculative (120). Limited clinical data exist regarding the effects of specific dietary modifications on platelet counts and immune function in ITP patients (121). Additionally, dietary interventions may have variable effects depending on individual microbiota composition and metabolic responses (122). Future research should prioritize characterizing microbial metabolite profiles in ITP patients to identify potential therapeutic targets, investigating the roles of SCFAs, bile acids, and tryptophan metabolites in modulating platelet regulation and immune responses. It should also assess the clinical efficacy of dietary interventions, including probiotic and prebiotic supplementation, as well as bile acid modulation, through well-designed randomized controlled trials (47, 123). By integrating microbiome sequencing and metabolomic profiling, researchers can better define the role of microbial metabolites in platelet function and immune modulation, leading to potential novel therapeutic approaches in ITP (52, 79).
3.4 Pharmacologic strategies targeting the gut: microbiome-based drug development
Pharmacologic approaches to modulating the gut microbiota are emerging as a potential strategy for treating autoimmune diseases, including ITP (12). These approaches include microbiome-based small molecules, postbiotics, and engineered probiotics designed to selectively modulate microbial communities and immune function (21).
Postbiotics, which are bioactive compounds produced by beneficial bacteria, have shown promise in immune modulation without the need for live microorganisms (124). Microbiome-based small molecules are being developed to target specific microbial metabolic pathways that influence immune responses (125). Additionally, engineered probiotics are being designed to deliver immunomodulatory molecules directly within the gut, offering a targeted approach to restoring immune balance (126).
While microbiome-based drug development is still in its early stages, these strategies hold great potential for providing precise and effective treatments for ITP (127). However, key challenges include ensuring microbial stability, understanding long-term safety, and optimizing drug delivery systems (128). Future studies should investigate the pharmacokinetics and pharmacodynamics of these microbiome-based therapies in ITP, as well as their potential for integration with standard treatments (129).
4 Challenges and limitations in gut microbiota therapy for ITP
Despite the growing interest in microbiota-targeted therapies for ITP, several challenges and limitations must be addressed before these strategies can be effectively translated into clinical practice. While preclinical and early clinical studies suggest that microbiota modulation may help restore immune tolerance and regulate platelet homeostasis (130), inconsistencies in research findings, methodological limitations, and unresolved safety concerns remain key barriers. This section critically examines the current knowledge, debates ongoing controversies, and identifies gaps that future research should address.
4.1 Variability in gut microbiota composition among individuals
One of the fundamental challenges in microbiota-based therapies is the high degree of inter-individual variability in gut microbial composition (131). Factors such as genetics, diet, medication history (including prior antibiotic use), and environmental influences contribute to significant differences in microbial diversity and function (132). This variability complicates the standardization of microbiota-based interventions, as a treatment effective for one patient may not yield similar benefits for another (133). Additionally, baseline microbiota differences may influence therapeutic responses, highlighting the need for patient-specific approaches. Future research should focus on stratifying patient populations based on microbiome profiling to optimize treatment efficacy (134).
4.2 Standardization and safety concerns in FMT and microbiota-based interventions
FMT has shown promise as a microbiota-based intervention in autoimmune diseases, but its application in ITP remains largely experimental (135). One major concern is the lack of standardization in FMT protocols, including donor selection, preparation methods, and delivery routes (87). Donor microbiota composition can vary significantly, leading to inconsistent therapeutic outcomes (136). Additionally, potential risks associated with FMT include the transmission of infectious agents, unintended immune activation, and long-term alterations in gut microbiota that may have unpredictable consequences (137).
Beyond FMT, the safety profile of probiotics and prebiotics in ITP patients has not been rigorously evaluated (69). While some probiotic strains exhibit immunomodulatory properties, others may provoke excessive immune activation or lead to bacterial overgrowth, particularly in immunocompromised individuals (138). To address these safety concerns, further clinical trials with well-defined protocols are needed to assess the risks and benefits of microbiota-targeted therapies in ITP.
4.3 Need for robust clinical trials and biomarker discovery in ITP-microbiota research
Although emerging studies suggest a link between gut dysbiosis and ITP, most existing research relies on small-scale, cross-sectional studies with limited statistical power (139). Longitudinal studies are needed to investigate how changes in gut microbiota correlate with the onset, severity, and treatment response of ITP over time. Second, mechanistic studies should explore the causal relationships determine whether gut microbiota alterations precede ITP onset or arise as a consequence of the disease and its treatments (18). Furthermore, the identification of reliable microbial biomarkers for disease progression and treatment response remains a significant gap in current research (140). Developing standardized methods for microbiome analysis, including metagenomic sequencing and metabolomic profiling, could help establish microbiota-based diagnostic and prognostic tools for ITP (141).
Clinical trials evaluating microbiota-targeted interventions in ITP are also lacking (52). While probiotics, prebiotics, dietary modifications, and FMT have been explored in other autoimmune conditions, few studies have directly assessed their impact on platelet counts and immune regulation in ITP patients. Large-scale, randomized controlled trials are essential to determine the efficacy, safety, and durability of these interventions in a hematologic context (142).
4.4 Ethical and regulatory considerations in applying microbiota therapies to hematologic disorders
The integration of microbiota-based therapies into hematologic disease management raises several ethical and regulatory challenges (143). Unlike conventional pharmacologic agents, microbiota-based interventions involve live organisms, making it difficult to define consistent dosing, manufacturing processes, and quality control standards (69). Regulatory agencies, such as the FDA and EMA, currently classify FMT as an investigational therapy, necessitating rigorous oversight before its widespread adoption in ITP treatment (77).
Additionally, ethical concerns related to FMT donor selection, consent processes, and long-term safety monitoring must be addressed (87). Patients undergoing microbiota-based treatments should be informed of potential risks, including unforeseen immune complications or persistent microbiome alterations (144). Establishing regulatory frameworks that balance innovation with patient safety will be crucial in advancing microbiota-targeted therapies in ITP (129).
To overcome these limitations, future research should focus on developing standardized microbiota-based protocols by establishing guidelines for donor screening, sample preparation, and treatment administration to improve the reproducibility and safety of microbiota-based therapies (93). Additionally, personalizing microbiota interventions through microbiome sequencing and precision medicine approaches can help tailor treatments based on individual microbiota profiles, thereby increasing therapeutic efficacy (145). Conducting large-scale, controlled clinical trials will be essential for evaluating the long-term impact of microbiota-targeted therapies in ITP and ensuring their clinical adoption (84). Furthermore, clarifying the mechanisms of action through further research is necessary to elucidate how specific microbial taxa and metabolites influence platelet regulation and immune responses in ITP (11). Addressing these aspects will provide a stronger foundation for the integration of microbiota-based therapies into ITP management (96). By tackling these challenges and knowledge gaps, microbiota-targeted interventions may become a viable and evidence-based approach for managing ITP, complementing existing immunomodulatory treatments and improving patient outcomes.
5 Future directions and clinical translation
While significant strides have been made in understanding the gut-immune axis in ITP, translating these findings into effective clinical applications remains a challenge (18). Several critical gaps must be addressed, including the lack of standardized microbiota profiling in clinical practice, variability in treatment responses, and the need for robust clinical trials to validate microbiota-based interventions (146). This section highlights the key areas of future research that could bridge these gaps and facilitate the integration of microbiota-targeted therapies into mainstream ITP management.
5.1 Integrating microbiota profiling in ITP diagnosis and prognosis
One promising avenue for advancing ITP management is the incorporation of microbiota profiling into diagnostic and prognostic assessments. Given the increasing evidence that gut microbiota composition influences immune responses and disease severity, recent studies have identified microbial signatures linked to hematologic diseases, where altered Bacteroides and Enterobacteriaceae profiles predict disease progression, suggesting potential biomarker applications for ITP (19, 147).
Recent advancements in microbiome sequencing have facilitated the identification of microbial alterations linked to systemic disease progression (140). For instance, studies in brain metastases have demonstrated how gut microbiota composition can influence disease dynamics, underscoring the potential of microbiota profiling as a valuable tool for diagnosing and monitoring immune-mediated disorders such as ITP (78). Metagenomic sequencing enables high-resolution characterization of microbial taxa, revealing microbiota-related risk factors predictive of autoimmune disease severity (148).
An overview of microbiota-based biomarkers and diagnostic approaches in ITP, including key microbial alterations, sequencing methodologies, and their potential clinical applications, is presented in Figure 3. This schematic highlights how microbiota profiling could be integrated into routine diagnostic workflows to enhance precision medicine approaches in ITP.
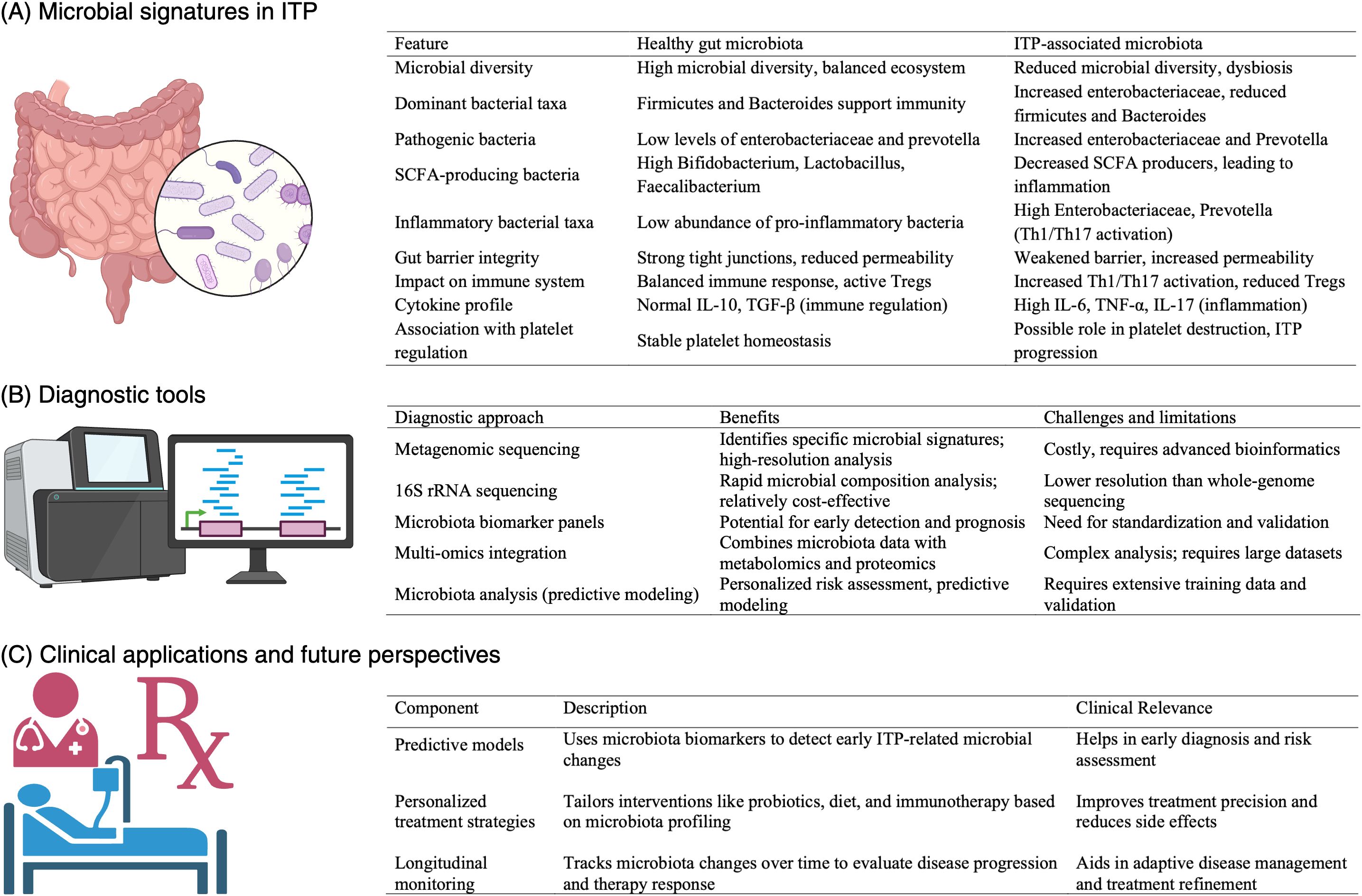
Figure 3. Overview of microbiota-based biomarkers and diagnostic approaches in ITP. This figure illustrates the role of microbiota profiling in the diagnosis and management of immune thrombocytopenia (ITP). (A) Microbial signatures in ITP highlight key alterations in bacterial taxa, including decreased Bacteroides and Firmicutes, and increased Enterobacteriaceae, which are associated with immune dysregulation and platelet destruction. (B) A comparison of diagnostic tools—metagenomic sequencing, 16S rRNA sequencing, microbiota biomarker panels, and multi-omics integration—highlighting their benefits and limitations for clinical application in ITP. (C) Clinical applications and future perspectives include predictive models for early detection, personalized treatment strategies integrating probiotics and dietary interventions, and longitudinal monitoring to assess disease progression and therapeutic responses. This schematic underscores the potential of microbiota-based diagnostics in improving precision medicine approaches for ITP management.
Despite these advances, specific microbial signatures distinguishing ITP from other autoimmune and hematologic diseases remain unclear. While some studies suggest that gut dysbiosis may contribute to platelet regulation and immune modulation in ITP, no definitive microbial biomarkers have been validated for ITP diagnosis or disease stratification (11, 19). Furthermore, the high inter-individual variability in gut microbiota composition complicates the reproducibility and clinical application of microbiome-based diagnostics (12, 131).
The application of microbiota profiling in ITP diagnosis presents both opportunities and challenges (18, 19). Table 3 summarizes the advantages and limitations of various microbiome-based diagnostic approaches, highlighting their potential role in disease monitoring and personalized treatment strategies.
While microbiota profiling holds promise, its clinical relevance in ITP remains uncertain. Studies suggest an association between gut dysbiosis and platelet regulation in ITP, but causal mechanisms remain unclear, necessitating longitudinal studies to establish microbiota-driven disease modulation (19, 47). Additionally, the standardization of sampling methods, data interpretation, and validation across different populations is needed before microbiota profiling can be reliably incorporated into clinical practice (149).
To advance microbiota profiling as a diagnostic tool in ITP, future research should focus on identifying robust microbial biomarkers specific to ITP that distinguish it from other hematologic and autoimmune disorders (150); conducting large-scale, multi-cohort microbiome studies to evaluate consistent microbial signatures across diverse populations (151); integrating microbiome data with clinical parameters and multi-omics approaches (e.g., metabolomics, proteomics) to refine diagnostic accuracy (152); and developing standardized guidelines for microbiome-based clinical diagnostics to ensure reproducibility and regulatory approval for ITP diagnosis (153).
5.2 Personalized microbiota-based therapies for ITP management
The concept of personalized microbiota-based therapies is gaining traction as an alternative or adjunctive strategy for ITP management (19). Given the variability in microbiota composition among individuals, tailoring interventions based on a patient’s specific microbial profile may enhance treatment efficacy (82). Personalized approaches may include selective probiotic formulations, prebiotic-enriched diets, or customized FMT protocols designed to restore microbial balance and immune homeostasis (154).
However, significant challenges remain. The efficacy of probiotics and prebiotics in ITP has not been rigorously tested, and the optimal strains or formulations for modulating immune responses remain undefined (138). Moreover, inter-individual differences in microbiota composition may influence therapeutic outcomes, necessitating a precision medicine approach (145). Future studies should focus on characterizing microbiota profiles associated with positive treatment responses and developing predictive models to guide personalized microbiome-based interventions (155).
5.3 Advances in microbiome sequencing and artificial intelligence for targeted interventions
Technological advancements in microbiome sequencing and artificial intelligence (AI) are revolutionizing the development of microbiota-targeted interventions. High-throughput sequencing techniques, such as 16S rRNA sequencing and shotgun metagenomics, allow for comprehensive profiling of gut microbial communities, facilitating the identification of microbial alterations linked to ITP (156). These tools could be leveraged to refine microbiota-based diagnostics and therapeutic strategies.
AI-driven approaches further enhance our ability to analyze complex microbiota datasets, predict treatment responses, and develop targeted therapeutic interventions (157). Machine learning models can identify microbial patterns associated with disease states and recommend tailored microbiota-based therapies based on an individual’s gut microbiota composition. Integrating AI with microbiome research holds great potential for optimizing precision medicine approaches in ITP and improving clinical outcomes (158).
5.4 Potential for gut microbiota manipulation in combination with standard ITP therapies
Combining microbiota-targeted therapies with conventional ITP treatments represents an exciting avenue for improving patient outcomes (159). Current standard therapies, such as corticosteroids, thrombopoietin receptor agonists, and immunosuppressive agents, exhibit variable efficacy and often associated with significant side effects (160). Modulating the gut microbiota may serve as a complementary strategy to enhance treatment efficacy, reduce immune-related side effects, and improve long-term disease management (69).
Certain microbiota-targeted interventions, such as probiotics and SCFA-based dietary strategies, may help regulate immune responses, reduce inflammation, and promote platelet production, thereby decreasing reliance on long-term immunosuppressive therapies (108). Additionally, microbiota-based strategies may facilitate immune tolerance in refractory ITP cases, improving the likelihood of sustained remission. However, the challenge remains in identifying the most effective microbiota-based combinations and understanding their interactions with existing therapies. Future clinical trials should investigate the synergistic effects of microbiota-targeted therapies with current ITP treatments to optimize patient care (129).
Future directions should prioritize robust Phase II/III clinical trials, standardized microbiome diagnostics, longitudinal microbiota-immune tracking, and integration of AI-driven predictive models to personalize treatment. These efforts will advance precision microbiome therapeutics for ITP and improve long-term outcomes.
While microbiota-targeted therapies hold great promise for ITP, several obstacles must be addressed before they can be effectively implemented in clinical practice (161). Future research should prioritize large-scale, randomized controlled trials to validate the efficacy of microbiota-based interventions, establish standardized diagnostic protocols, and further explore the mechanistic pathways linking gut dysbiosis to ITP pathogenesis (162). Integrating microbiota profiling into precision medicine approaches and leveraging AI-driven strategies may pave the way for innovative and personalized treatment options (145).
Robust Phase II/III clinical trials are urgently needed to validate the clinical efficacy of microbiota-targeted interventions, including fecal microbiota transplantation, SCFA supplementation, and strain-specific probiotics (163). These studies should incorporate standardized microbiome profiling and immune monitoring to establish reproducible outcomes (164). In parallel, recent proposals emphasize the importance of prospective longitudinal studies and gnotobiotic mouse models to establish causality and elucidate the underlying microbial mechanisms that regulate immune responses and platelet homeostasis in ITP (165).
By addressing these challenges, the field can move toward more targeted and effective microbiota-based therapies for ITP, ultimately improving patient outcomes and transforming disease management (127).
6 Conclusion
The growing recognition of the gut-immune axis in primary ITP represents a paradigm shift in understanding and managing this autoimmune disorder (19). Traditional treatment strategies have primarily focused on immunosuppressive approaches; however, emerging evidence highlights the pivotal role of gut microbiota in immune regulation and disease progression (12). The interplay between gut microbiota, immune tolerance, and platelet homeostasis offers new avenues for therapeutic intervention, shifting the focus toward microbiota-targeted strategies.
Microbiota-based therapies, including FMT, probiotics, prebiotics, and dietary interventions, hold significant promise in modulating immune responses and restoring microbial balance in ITP (166). These approaches can enhance immune tolerance, reduce inflammation, and complement existing treatment modalities (167). However, while early studies provide compelling insights, further clinical trials are essential to validate the safety, efficacy, and long-term effects of these interventions (168).
Despite these promising developments, several challenges must be addressed before microbiota-targeted therapies can be fully integrated into clinical practice. Standardizing microbiome profiling methods, identifying reliable microbial biomarkers, and optimizing therapeutic strategies tailored to individual microbiota compositions remain critical research priorities (154, 169). Importantly, clarifying the temporal relationship between dysbiosis and immune dysfunction in ITP requires longitudinal microbiome studies and mechanistic experiments, including the use of gnotobiotic animal models (165). Additionally, long-term studies are needed to evaluate the durability of microbiota modulation and its sustained effects on disease outcomes.
Continued research is crucial for translating gut-immune insights into effective clinical strategies. Advancements in microbiome sequencing, artificial intelligence-driven microbiota profiling, and biomarker discovery may facilitate the development of personalized, precision-based therapies for ITP (104). Integrating microbiota modulation with standard immunosuppressive therapies and thrombopoietin receptor agonist therapies may lead to synergistic treatment effects and improved patient outcomes (82).
In conclusion, leveraging the gut-immune axis for ITP management represents an exciting frontier in autoimmune disease research. As our understanding of gut microbiota expands, microbiota-targeted interventions may pave the way for more effective, sustainable, and personalized treatment strategies for ITP, ultimately improving patients’ quality of life and long-term disease management.
Author contributions
XG: Writing – original draft, Data curation, Resources, Writing – review & editing, Funding acquisition, Investigation, Validation, Project administration, Visualization, Conceptualization, Supervision. WK: Writing – original draft, Methodology, Formal Analysis, Software, Investigation, Data curation, Validation. QL: Formal Analysis, Data curation, Methodology, Writing – original draft, Investigation. NB: Funding acquisition, Formal Analysis, Writing – review & editing, Validation, Investigation. WM: Validation, Data curation, Resources, Visualization, Formal Analysis, Project administration, Conceptualization, Software, Methodology, Writing – review & editing, Supervision, Writing – original draft, Investigation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Henan Provincial Science and Technology Research Project in Biomedicine (Grant No. 182102311233, X.G.)National Science Centre, Poland (Grant No. 2021/43/B/NZ5/03345, N.B.).
Conflict of interest
WM is a scientific and medical advisor at Calidi Biotherapeutics Inc. San Diego, CA, USA.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
WM holds the position of associate editor of Frontiers in Immunology at the time of submission. This had no impact on the peer review process or the final decision.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
AhR, aryl hydrocarbon receptor; AI, artificial intelligence; Bregs, regulatory B cells; FMT, fecal microbiota transplantation; GALT, gut-associated lymphoid tissue; GPIb/IX, glycoprotein Ib/IX; GPIIb/IIIa, glycoprotein IIb/IIIa; IBD, inflammatory bowel disease; IFN-γ, interferon-gamma; IL-6, interleukin-6; ITP, immune thrombocytopenia; IVIG, intravenous immunoglobulin; MAMPs, microbial-associated molecular patterns; NK, natural killer; PRRs, pattern recognition receptors; RA, rheumatoid arthritis; SCFA, short-chain fatty acid; SLE, systemic lupus erythematosus; Th1, T helper 1 cells; Th17, T helper 17 cells; TLRs, toll-like receptors; TME, tumor microenvironment; TNF-α, tumor necrosis factor-alpha; TPO-RA, thrombopoietin receptor agonists; Tregs, regulatory T cells.
References
1. David P, Santos GM, Patt YS, Orsi FA, and Shoenfeld Y. Immune thrombocytopenia (Itp) - could it be part of autoimmune/inflammatory syndrome induced by adjuvants (Asia)? Autoimmun Rev. (2024) 23:103605. doi: 10.1016/j.autrev.2024.103605
2. Liu XG, Hou Y, and Hou M. How we treat primary immune thrombocytopenia in adults. J Hematol Oncol. (2023) 16:4. doi: 10.1186/s13045-023-01401-z
3. Danieli MG, Antonelli E, Gammeri L, Longhi E, Cozzi MF, Palmeri D, et al. Intravenous immunoglobulin as a therapy for autoimmune conditions. Autoimmun Rev. (2025) 24:103710. doi: 10.1016/j.autrev.2024.103710
4. Wang Y, Sheng L, Han F, Guo Q, Zhang Z, Hou Y, et al. Efficacy and safety of treatments in newly diagnosed adult primary immune thrombocytopenia: A systematic review and network meta-analysis. EClinicalMedicine. (2023) 56:101777. doi: 10.1016/j.eclinm.2022.101777
5. Roeser A, Lazarus AH, and Mahévas M. B cells and antibodies in refractory immune thrombocytopenia. Br J Haematol. (2023) 203:43–53. doi: 10.1111/bjh.18773
6. Schoenaker JM, Nelson VS, Henderickx JGE, Terveer EM, Jansen AJG, Porcelijn L, et al. The intestinal flora: the key to unraveling heterogeneity in immune thrombocytopenia? Blood Rev. (2025) 69:101252. doi: 10.1016/j.blre.2024.101252
7. Skoric D, Krcunovic J, Svorcan J, Krstovski N, Rodic P, Lazic J, et al. Pediatric immune thrombocytopenia: the impact of antithyroid antibodies on the treatment outcomes. Clin Pediatr (Phila). (2025) 64:264–70. doi: 10.1177/00099228241262173
8. Hardtke-Wolenski M and Landwehr-Kenzel S. Tipping the balance in autoimmunity: are regulatory T cells the cause, the cure, or both? Mol Cell Pediatr. (2024) 11:3. doi: 10.1186/s40348-024-00176-8
9. Mackie J, Suan D, McNaughton P, Haerynck F, O’Sullivan M, Guerin A, et al. Functional validation of a novel Stat3 ‘Variant of unknown significance’ Identifies a new case of Stat3 Gof syndrome and reveals broad immune cell defects. Clin Exp Immunol. (2025) 219(1):uxaf005. doi: 10.1093/cei/uxaf005
10. Elsayh KI, Saad K, Osman NS, Mahmoud KH, Ahmad FA, Khalaf SM, et al. Regulatory T-lymphocyte subsets in children with chronic immune thrombocytopenia after high-dose of dexamethasone. Pediatr Res. (2022) 92:1432–6. doi: 10.1038/s41390-022-01978-0
11. Zhu G, Yan L, Fang L, Fan C, Sun H, Zhou X, et al. Possible immune mechanisms of gut microbiota and its metabolites in the occurrence and development of immune thrombocytopenia. Front Microbiol. (2024) 15:1426911. doi: 10.3389/fmicb.2024.1426911
12. Wang X, Yuan W, Yang C, Wang Z, Zhang J, Xu D, et al. Emerging role of gut microbiota in autoimmune diseases. Front Immunol. (2024) 15:1365554. doi: 10.3389/fimmu.2024.1365554
13. Qi P, Chen X, Tian J, Zhong K, Qi Z, Li M, et al. The gut homeostasis-immune system axis: novel insights into rheumatoid arthritis pathogenesis and treatment. Front Immunol. (2024) 15:1482214. doi: 10.3389/fimmu.2024.1482214
14. Li J, Xie Z, Yang L, Guo K, and Zhou Z. The impact of gut microbiome on immune and metabolic homeostasis in type 1 diabetes: clinical insights for prevention and treatment strategies. J Autoimmun. (2025) 151:103371. doi: 10.1016/j.jaut.2025.103371
15. Lu Q, Zhu R, Zhou L, Zhang R, Li Z, Xu P, et al. Gut dysbiosis contributes to the development of Budd-Chiari syndrome through immune imbalance. mSystems. (2024) 9:e0079424. doi: 10.1128/msystems.00794-24
16. Fakharian F, Thirugnanam S, Welsh DA, Kim WK, Rappaport J, Bittinger K, et al. The role of gut dysbiosis in the loss of intestinal immune cell functions and viral pathogenesis. Microorganisms. (2023) 11:1849. doi: 10.3390/microorganisms11071849
17. Yang X, Huang J, Peng J, Wang P, Wong FS, Wang R, et al. Gut microbiota from B-cell-specific Tlr9-deficient nod mice promote Il-10(+) Breg cells and protect against T1d. Front Immunol. (2024) 15:1413177. doi: 10.3389/fimmu.2024.1413177
18. Wang JG, Dou HH, and Liang QY. Impact of gut microbiota and inflammatory cytokines on immune thrombocytopenia. Eur J Haematol. (2025) 114:120–8. doi: 10.1111/ejh.14310
19. Sun H, Yan L, Fang L, and Shi Z. A novel approach to immune thrombocytopenia intervention: modulating intestinal homeostasis. BMC Immunol. (2024) 25:71. doi: 10.1186/s12865-024-00660-w
20. Napiorkowska-Baran K, Bilinski J, Pujanek M, Halakuc P, Pietryga A, Szymczak B, et al. Fecal microbiota transplantation in a patient with chronic diarrhea and primary and secondary immunodeficiency (Common variable immunodeficiency and splenectomy). Front Cell Infect Microbiol. (2024) 14:1456672. doi: 10.3389/fcimb.2024.1456672
21. Zhou P, Chen C, Patil S, and Dong S. Unveiling the therapeutic symphony of probiotics, prebiotics, and postbiotics in gut-immune harmony. Front Nutr. (2024) 11:1355542. doi: 10.3389/fnut.2024.1355542
22. Li Z, Xiong W, Liang Z, Wang J, Zeng Z, Kołat D, et al. Critical role of the gut microbiota in immune responses and cancer immunotherapy. J Hematol Oncol. (2024) 17:33. doi: 10.1186/s13045-024-01541-w
23. Schifferli A, Cavalli F, Godeau B, Liebman HA, Recher M, Imbach P, et al. Understanding immune thrombocytopenia: looking out of the box. Front Med (Lausanne). (2021) 8:613192. doi: 10.3389/fmed.2021.613192
24. Chakraborty P, Banerjee D, Majumder P, and Sarkar J. Gut microbiota nexus: exploring the interactions with the brain, heart, lungs, and skin axes and their effects on health. Med Microecol. (2024) 20:100104. doi: 10.1016/j.medmic.2024.100104
25. Ma S, Ming Y, Wu J, and Cui G. Cellular metabolism regulates the differentiation and function of T-cell subsets. Cell Mol Immunol. (2024) 21:419–35. doi: 10.1038/s41423-024-01148-8
26. Kedmi R and Littman DR. Antigen-presenting cells as specialized drivers of intestinal T cell functions. Immunity. (2024) 57:2269–79. doi: 10.1016/j.immuni.2024.09.011
27. Xie Y and Liu F. The role of the gut microbiota in tumor, immunity, and immunotherapy. Front Immunol. (2024) 15:1410928. doi: 10.3389/fimmu.2024.1410928
28. Colella M, Charitos IA, Ballini A, Cafiero C, Topi S, Palmirotta R, et al. Microbiota revolution: how gut microbes regulate our lives. World J Gastroenterol. (2023) 29:4368–83. doi: 10.3748/wjg.v29.i28.4368
29. Han K, Xu J, Xie F, Crowther J, and Moon JJ. Engineering strategies to modulate the gut microbiome and immune system. J Immunol. (2024) 212:208–15. doi: 10.4049/jimmunol.2300480
30. Wilhelm G, Mertowska P, Mertowski S, Przysucha A, Struzyna J, Grywalska E, et al. The crossroads of the coagulation system and the immune system: interactions and connections. Int J Mol Sci. (2023) 24:12563. doi: 10.3390/ijms241612563
31. Zhang D, Jian Y-P, Zhang Y-N, Li Y, Gu L-T, Sun H-H, et al. Short-chain fatty acids in diseases. Cell Communication Signaling. (2023) 21:212. doi: 10.1186/s12964-023-01219-9
32. Du Y, He C, An Y, Huang Y, Zhang H, Fu W, et al. The role of short chain fatty acids in inflammation and body health. Int J Mol Sci. (2024) 25:7379. doi: 10.3390/ijms25137379
33. Yan Q, Jia S, Li D, and Yang J. The role and mechanism of action of microbiota-derived short-chain fatty acids in neutrophils: from the activation to becoming potential biomarkers. Biomedicine Pharmacother. (2023) 169:115821. doi: 10.1016/j.biopha.2023.115821
34. He M, Wei W, Zhang Y, Xiang Z, Peng D, Kasimumali A, et al. Gut microbial metabolites Scfas and chronic kidney disease. J Trans Med. (2024) 22:172. doi: 10.1186/s12967-024-04974-6
35. Bu S, Liu M, Yang L, Lee P, Miller H, Park CS, et al. The function of T cells in immune thrombocytopenia. Front Immunol. (2025) 16:1499014. doi: 10.3389/fimmu.2025.1499014
36. Wicherska-Pawlowska K, Wrobel T, and Rybka J. Toll-like receptors (Tlrs), nod-like receptors (Nlrs), and rig-I-like receptors (Rlrs) in innate immunity. Tlrs, Nlrs, and Rlrs ligands as immunotherapeutic agents for hematopoietic diseases. Int J Mol Sci. (2021) 22:13397. doi: 10.3390/ijms222413397
37. Sundaram B, Tweedell RE, Prasanth Kumar S, and Kanneganti T-D. The Nlr family of innate immune and cell death sensors. Immunity. (2024) 57:674–99. doi: 10.1016/j.immuni.2024.03.012
38. Kawai T, Ikegawa M, Ori D, and Akira S. Decoding toll-like receptors: recent insights and perspectives in innate immunity. Immunity. (2024) 57:649–73. doi: 10.1016/j.immuni.2024.03.004
39. Ullah H, Arbab S, Tian Y, Chen Y, Liu CQ, Li Q, et al. Crosstalk between gut microbiota and host immune system and its response to traumatic injury. Front Immunol. (2024) 15:1413485. doi: 10.3389/fimmu.2024.1413485
40. Kato R, Yamamoto T, Ogata H, Miyata K, Hayashi S, Gershon MD, et al. Indigenous gut microbiota constitutively drive release of ciliary neurotrophic factor from mucosal enteric glia to maintain the homeostasis of enteric neural circuits. Front Immunol. (2024) 15:1372670. doi: 10.3389/fimmu.2024.1372670
41. Di Vincenzo F, Del Gaudio A, Petito V, Lopetuso LR, and Scaldaferri F. Gut microbiota, intestinal permeability, and systemic inflammation: A narrative review. Intern Emerg Med. (2024) 19:275–93. doi: 10.1007/s11739-023-03374-w
42. Dmytriv TR, Storey KB, and Lushchak VI. Intestinal barrier permeability: the influence of gut microbiota, nutrition, and exercise. Front Physiol. (2024) 15:1380713. doi: 10.3389/fphys.2024.1380713
43. Yan M, Wang Z, Qiu Z, Cui Y, and Xiang Q. Platelet signaling in immune landscape: comprehensive mechanism and clinical therapy. biomark Res. (2024) 12:164. doi: 10.1186/s40364-024-00700-y
44. Zhou H, Balint D, Shi Q, Vartanian T, Kriegel MA, and Brito I. Lupus and inflammatory bowel disease share a common set of microbiome features distinct from other autoimmune disorders. Ann Rheumatic Dis. (2024) 84(1):93–105. doi: 10.1136/ard-2024-225829
45. Van Hul M, Cani PD, Petitfils C, De Vos WM, Tilg H, and El-Omar EM. What defines a healthy gut microbiome? Gut. (2024) 73:1893–908. doi: 10.1136/gutjnl-2024-333378
46. Sadeghpour Heravi F. Gut microbiota and autoimmune diseases: mechanisms, treatment, challenges, and future recommendations. Curr Clin Microbiol Rep. (2024) 11:18–33. doi: 10.1007/s40588-023-00213-6
47. Hong Y, Zhang C, Shen K, Dong X, and Chen B. Genetically predicted plasma metabolites mediate the causal relationship between gut microbiota and primary immune thrombocytopenia (Itp). Front Microbiol. (2024) 15:1447729. doi: 10.3389/fmicb.2024.1447729
48. Zhu Q, Rui K, Wang S, and Tian J. Advances of regulatory B cells in autoimmune diseases. Front Immunol. (2021) 12:592914. doi: 10.3389/fimmu.2021.592914
49. Zhang Q, Liao J, Liu Z, Song S, Tian L, and Wang Y. The immune tolerance role of Bregs in inhibiting human inflammatory diseases, with a focus on diabetes mellitus. Front Immunol. (2025) 16:1565158. doi: 10.3389/fimmu.2025.1565158
50. Provan D and Semple JW. Recent advances in the mechanisms and treatment of immune thrombocytopenia. EBioMedicine. (2022) 76:103820. doi: 10.1016/j.ebiom.2022.103820
51. He L, Li X, Jiang S, Ou Y, Wang S, Shi N, et al. The influence of the gut microbiota on B cells in autoimmune diseases. Mol Med. (2025) 31:149. doi: 10.1186/s10020-025-01195-5
52. Saki N, Hadi H, Keikhaei B, Mirzaei A, and Purrahman D. Gut microbiome composition and dysbiosis in immune thrombocytopenia: A review of literature. Blood Rev. (2024) 67:101219. doi: 10.1016/j.blre.2024.101219
53. Kim CH. Complex regulatory effects of gut microbial short-chain fatty acids on immune tolerance and autoimmunity. Cell Mol Immunol. (2023) 20:341–50. doi: 10.1038/s41423-023-00987-1
54. Gui L, Zuo X, Feng J, Wang M, Chen Z, Sun Y, et al. Outgrowth of Escherichia is susceptible to aggravation of systemic lupus erythematosus. Arthritis Res Ther. (2024) 26:191. doi: 10.1186/s13075-024-03413-7
55. Lathakumari RH, Vajravelu LK, Satheesan A, Ravi S, and Thulukanam J. Antibiotics and the gut microbiome: understanding the impact on human health. Med Microecol. (2024) 20:100106. doi: 10.1016/j.medmic.2024.100106
56. Singh S, Giron LB, Shaikh MW, Shankaran S, Engen Phillip A, Bogin ZR, et al. Distinct intestinal microbial signatures linked to accelerated systemic and intestinal biological aging. Microbiome. (2024) 12:31. doi: 10.1186/s40168-024-01758-4
57. Bhutta NK, Xu X, Jian C, Wang Y, Liu Y, Sun J, et al. Gut microbiota mediated T cells regulation and autoimmune diseases. Front Microbiol. (2024) 15:1477187. doi: 10.3389/fmicb.2024.1477187
58. Li C, Peng K, Xiao S, Long Y, and Yu Q. The role of lactobacillus in inflammatory bowel disease: from actualities to prospects. Cell Death Discov. (2023) 9:361. doi: 10.1038/s41420-023-01666-w
59. Thapa R, Magar AT, Shrestha J, Panth N, Idrees S, Sadaf T, et al. Influence of gut and lung dysbiosis on lung cancer progression and their modulation as promising therapeutic targets: A comprehensive review. MedComm (2020). (2024) 5:e70018. doi: 10.1002/mco2.70018
60. Alagiakrishnan K, Morgadinho J, and Halverson T. Approach to the diagnosis and management of dysbiosis. Front Nutr. (2024) 11:1330903. doi: 10.3389/fnut.2024.1330903
61. Qu S, Gao Y, Ma J, and Yan Q. Microbiota-derived short-chain fatty acids functions in the biology of B lymphocytes: from differentiation to antibody formation. Biomedicine Pharmacother. (2023) 168:115773. doi: 10.1016/j.biopha.2023.115773
62. Druszczynska M, Sadowska B, Kulesza J, Gasienica-Gliwa N, Kulesza E, and Fol M. The intriguing connection between the gut and lung microbiomes. Pathogens. (2024) 13. doi: 10.3390/pathogens13111005
63. Yao K, Xie Y, Wang J, Lin Y, Chen X, and Zhou T. Gut microbiota: A newly identified environmental factor in systemic lupus erythematosus. Front Immunol. (2023) 14:1202850. doi: 10.3389/fimmu.2023.1202850
64. Chasov V, Gilyazova E, Ganeeva I, Zmievskaya E, Davletshin D, Valiullina A, et al. Gut microbiota modulation: A novel strategy for rheumatoid arthritis therapy. Biomolecules. (2024) 14:1653. doi: 10.3390/biom14121653
65. Taneja V. Gut microbes as the major drivers of rheumatoid arthritis: our microbes are our fortune! Microorganisms. (2025) 13:255. doi: 10.3390/microorganisms13020255
66. Campagnoli LIM, Marchesi N, Varesi A, Morozzi M, Mascione L, Ricevuti G, et al. New therapeutic avenues in multiple sclerosis: is there a place for gut microbiota-based treatments? Pharmacol Res. (2024) 209:107456. doi: 10.1016/j.phrs.2024.107456
67. Turner T-A, Lehman P, Ghimire S, Shahi SK, and Mangalam A. Game of microbes: the battle within – gut microbiota and multiple sclerosis. Gut Microbes. (2024) 16:2387794. doi: 10.1080/19490976.2024.2387794
68. Rui X, Fu Y, Cai J, Zhang Y, Fu Q, and He C. Gut microbiota were altered with platelet count and red blood cell count in immune thrombocytopenia patients with different treatments. Front Cell Infect Microbiol. (2023) 13:1168756. doi: 10.3389/fcimb.2023.1168756
69. Zhao L-Y, Mei J-X, Yu G, Lei L, Zhang W-H, Liu K, et al. Role of the gut microbiota in anticancer therapy: from molecular mechanisms to clinical applications. Signal Transduction Targeted Ther. (2023) 8:201. doi: 10.1038/s41392-023-01406-7
70. Saini S, Koh AY, and Zia A. Identifying gut microbiota and immune host factors associated with bleeding risk in children with immune thrombocytopenic purpura. Blood. (2024) 144:2551–. doi: 10.1182/blood-2024-199928
71. Gilliland A, Chan JJ, De Wolfe TJ, Yang H, and Vallance BA. Pathobionts in inflammatory bowel disease: origins, underlying mechanisms, and implications for clinical care. Gastroenterology. (2024) 166:44–58. doi: 10.1053/j.gastro.2023.09.019
72. Guo D, Chen Q, Wang G, Li C, and FinnGen c. Causal relationship between gut microbiota and immune thrombocytopenia: A Mendelian randomization study of two samples. Front Microbiol. (2023) 14:1190866. doi: 10.3389/fmicb.2023.1190866
73. Wang H, Bi H, Yang M, Wang X, Song C, Yang W, et al. Intestinal flora altered and correlated with interleukin-2/4 in patients with primary immune thrombocytopenia. Hematology. (2023) 28:2277501. doi: 10.1080/16078454.2023.2277501
74. Mei Z, Wang F, Bhosle A, Dong D, Mehta R, Ghazi A, et al. Strain-specific gut microbial signatures in type 2 diabetes identified in a cross-cohort analysis of 8,117 metagenomes. Nat Med. (2024) 30:2265–76. doi: 10.1038/s41591-024-03067-7
75. Park JC and Im SH. Of men in mice: the development and application of a humanized gnotobiotic mouse model for microbiome therapeutics. Exp Mol Med. (2020) 52:1383–96. doi: 10.1038/s12276-020-0473-2
76. Mars RAT, Yang Y, Ward T, Houtti M, Priya S, Lekatz HR, et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell. (2020) 182:1460–73.e17. doi: 10.1016/j.cell.2020.08.007
77. Al-Samkari H. 2025 Update on clinical trials in immune thrombocytopenia. Am J Hematol. (2024) 99:2178–90. doi: 10.1002/ajh.27448
78. Massara M, Ballabio M, Dolfi B, Morad G, Wischnewski V, Lamprou E, et al. The bacterial microbiome modulates the initiation of brain metastasis by impacting the gut-to-brain axis. iScience. (2025) 28:111874. doi: 10.1016/j.isci.2025.111874
79. Jiang C, Deng S, Ma X, Song J, Li J, and Yuan E. Mendelian randomization reveals association of gut microbiota with Henoch–Schönlein Purpura and immune thrombocytopenia. Int J Hematol. (2024) 120:50–9. doi: 10.1007/s12185-024-03777-1
80. Simpson RC, Shanahan ER, Scolyer RA, and Long GV. Towards modulating the gut microbiota to enhance the efficacy of immune-checkpoint inhibitors. Nat Rev Clin Oncol. (2023) 20:697–715. doi: 10.1038/s41571-023-00803-9
81. Ebrahimi R, Farsi Y, and Nejadghaderi SA. Fecal microbiota transplantation for glaucoma; a potential emerging treatment strategy. Curr Res Microb Sci. (2024) 7:100314. doi: 10.1016/j.crmicr.2024.100314
82. Kang X, Lau HC, and Yu J. Modulating gut microbiome in cancer immunotherapy: harnessing microbes to enhance treatment efficacy. Cell Rep Med. (2024) 5:101478. doi: 10.1016/j.xcrm.2024.101478
83. Loh JS, Mak WQ, Tan LKS, Ng CX, Chan HH, Yeow SH, et al. Microbiota-gut-brain axis and its therapeutic applications in neurodegenerative diseases. Signal Transduct Target Ther. (2024) 9:37. doi: 10.1038/s41392-024-01743-1
84. Li J, Li J, Liu Y, Zeng J, Liu Y, and Wu Y. Large-scale bidirectional Mendelian randomization study identifies new gut microbiome significantly associated with immune thrombocytopenic purpura. Front Microbiol. (2024) 15:1423951. doi: 10.3389/fmicb.2024.1423951
85. Qin L, Fan B, Zhou Y, Zheng J, Diao R, Wang F, et al. Targeted gut microbiome therapy: applications and prospects of probiotics, fecal microbiota transplantation and natural products in the management of type 2 diabetes. Pharmacol Res. (2025), 107625. doi: 10.1016/j.phrs.2025.107625
86. Gao Y, Li W, Huang X, Lyu Y, and Yue C. Advances in gut microbiota-targeted therapeutics for metabolic syndrome. Microorganisms. (2024) 12:851. doi: 10.3390/microorganisms12050851
87. Yadegar A, Bar-Yoseph H, Monaghan TM, Pakpour S, Severino A, Kuijper EJ, et al. Fecal microbiota transplantation: current challenges and future landscapes. Clin Microbiol Rev. (2024) 37:e0006022. doi: 10.1128/cmr.00060-22
88. Karimi M, Shirsalimi N, Hashempour Z, Salehi Omran H, Sedighi E, Beigi F, et al. Safety and efficacy of fecal microbiota transplantation (Fmt) as a modern adjuvant therapy in various diseases and disorders: A comprehensive literature review. Front Immunol. (2024) 15:1439176. doi: 10.3389/fimmu.2024.1439176
89. Rasouli-Saravani A, Jahankhani K, Moradi S, Gorgani M, Shafaghat Z, Mirsanei Z, et al. Role of microbiota short-chain fatty acids in the pathogenesis of autoimmune diseases. Biomedicine Pharmacother. (2023) 162:114620. doi: 10.1016/j.biopha.2023.114620
90. Facchin S, Bertin L, Bonazzi E, Lorenzon G, De Barba C, Barberio B, et al. Short-chain fatty acids and human health: from metabolic pathways to current therapeutic implications. Life. (2024) 14:559. doi: 10.3390/life14050559
91. Zheng K, Wei Z, and Li W. Ecological insights into hematopoiesis regulation: unraveling the influence of gut microbiota. Gut Microbes. (2024) 16:2350784. doi: 10.1080/19490976.2024.2350784
92. Singh P, Meenatchi R, Ahmed ZHT, Thacharodi A, R M, Kumar RRS, et al. Implications of the gut microbiome in cardiovascular diseases: association of gut microbiome with cardiovascular diseases, therapeutic interventions and multi-omics approach for precision medicine. Med Microecol. (2024) 19:100096. doi: 10.1016/j.medmic.2023.100096
93. Rasmussen TS, Mao X, Forster S, Larsen SB, Von Münchow A, Tranæs KD, et al. Overcoming donor variability and risks associated with fecal microbiota transplants through bacteriophage-mediated treatments. Microbiome. (2024) 12:119. doi: 10.1186/s40168-024-01820-1
94. Yoo S, Jung SC, Kwak K, and Kim JS. The role of prebiotics in modulating gut microbiota: implications for human health. Int J Mol Sci. (2024) 25:4834. doi: 10.3390/ijms25094834
95. Mazziotta C, Tognon M, Martini F, Torreggiani E, and Rotondo JC. Probiotics mechanism of action on immune cells and beneficial effects on human health. Cells. (2023) 12:184. doi: 10.3390/cells12010184
96. Zeng L, Yang K, He Q, Zhu X, Long Z, Wu Y, et al. Efficacy and safety of gut microbiota-based therapies in autoimmune and rheumatic diseases: A systematic review and meta-analysis of 80 randomized controlled trials. BMC Med. (2024) 22:110. doi: 10.1186/s12916-024-03303-4
97. Chandrasekaran P, Weiskirchen S, and Weiskirchen R. Effects of probiotics on gut microbiota: an overview. Int J Mol Sci. (2024) 25:6022. doi: 10.3390/ijms25116022
98. Kumari A, GR K, Sudhakaran VA, Warrier AS, and Singh NK. Unveiling the health benefits of prebiotics: A comprehensive review. Indian J Microbiol. (2024) 64:376–88. doi: 10.1007/s12088-024-01235-4
99. Al-Habsi N, Al-Khalili M, Haque SA, Elias M, Olqi NA, and Al Uraimi T. Health benefits of prebiotics, probiotics, synbiotics, and postbiotics. Nutrients. (2024) 16:3955. doi: 10.3390/nu16223955
100. Fneish FH, Abd El Galil KH, and Domiati SA. Evaluation of single and multi-strain probiotics with gentamicin against E. Coli O157:H7: insights from in vitro and in vivo studies. Microorganisms. (2025) 13:460. doi: 10.3390/microorganisms13020460
101. Tufail MA and Schmitz RA. Exploring the probiotic potential of Bacteroides spp. Within one health paradigm. Probiotics Antimicrob Proteins. (2024) 17(2):681–704. doi: 10.1007/s12602-024-10370-9
102. Jia X, Wang J, Ren D, Zhang K, Zhang H, Jin T, et al. Impact of the gut microbiota-Th17 cell axis on inflammatory depression. Front Psychiatry. (2024) 15:1509191. doi: 10.3389/fpsyt.2024.1509191
103. Wang X, Peng J, Cai P, Xia Y, Yi C, Shang A, et al. The emerging role of the gut microbiota and its application in inflammatory bowel disease. Biomedicine Pharmacother. (2024) 179:117302. doi: 10.1016/j.biopha.2024.117302
104. Manrique P, Montero I, Fernandez-Gosende M, Martinez N, Cantabrana CH, and Rios-Covian D. Past, present, and future of microbiome-based therapies. Microbiome Res Rep. (2024) 3:23. doi: 10.20517/mrr.2023.80
105. Arifuzzaman M, Collins N, Guo CJ, and Artis D. Nutritional regulation of microbiota-derived metabolites: implications for immunity and inflammation. Immunity. (2024) 57:14–27. doi: 10.1016/j.immuni.2023.12.009
106. Cao S, Budina E, Raczy MM, Solanki A, Nguyen M, Beckman TN, et al. A serine-conjugated butyrate prodrug with high oral bioavailability suppresses autoimmune arthritis and neuroinflammation in mice. Nat Biomed Eng. (2024) 8:611–27. doi: 10.1038/s41551-024-01190-x
107. Zhang C, Liu X, Gu C, Su Y, Lv J, Liu Y, et al. Histone deacetylases facilitate Th17-cell differentiation and pathogenicity in autoimmune uveitis via Cdk6/Id2 axis. J Advanced Res. (2024). doi: 10.1016/j.jare.2024.07.029
108. Liu XF, Shao JH, Liao YT, Wang LN, Jia Y, Dong PJ, et al. Regulation of short-chain fatty acids in the immune system. Front Immunol. (2023) 14:1186892. doi: 10.3389/fimmu.2023.1186892
109. Zhang W, Mackay CR, and Gershwin ME. Immunomodulatory effects of microbiota-derived short-chain fatty acids in autoimmune liver diseases. J Immunol. (2023) 210:1629–39. doi: 10.4049/jimmunol.2300016
110. Hua T, Yao F, Wang H, Liu W, Zhu X, and Yao Y. Megakaryocyte in sepsis: the trinity of coagulation, inflammation and immunity. Crit Care. (2024) 28:442. doi: 10.1186/s13054-024-05221-6
111. Larabi AB, Masson HLP, and Baumler AJ. Bile acids as modulators of gut microbiota composition and function. Gut Microbes. (2023) 15:2172671. doi: 10.1080/19490976.2023.2172671
112. Fleishman JS and Kumar S. Bile acid metabolism and signaling in health and disease: molecular mechanisms and therapeutic targets. Signal Transduction Targeted Ther. (2024) 9:97. doi: 10.1038/s41392-024-01811-6
113. Spivak I, Fluhr L, and Elinav E. Local and systemic effects of microbiome-derived metabolites. EMBO Rep. (2022) 23:e55664. doi: 10.15252/embr.202255664
114. Yoon JH, Do JS, Velankanni P, Lee CG, and Kwon HK. Gut microbial metabolites on host immune responses in health and disease. Immune Netw. (2023) 23:e6. doi: 10.4110/in.2023.23.e6
115. Zhang Z, Lv T, Wang X, Wu M, Zhang R, Yang X, et al. Role of the microbiota–gut–heart axis between bile acids and cardiovascular disease. Biomedicine Pharmacother. (2024) 174:116567. doi: 10.1016/j.biopha.2024.116567
116. Yan J, Chen D, Ye Z, Zhu X, Li X, Jiao H, et al. Molecular mechanisms and therapeutic significance of tryptophan metabolism and signaling in cancer. Mol Cancer. (2024) 23:241. doi: 10.1186/s12943-024-02164-y
117. Liu X, Yang M, Xu P, Du M, Li S, Shi J, et al. Kynurenine-Ahr reduces T-cell infiltration and induces a delayed T-cell immune response by suppressing the Stat1-Cxcl9/Cxcl10 axis in tuberculosis. Cell Mol Immunol. (2024) 21:1426–40. doi: 10.1038/s41423-024-01230-1
118. Moulinet T, Moussu A, Pierson L, and Pagliuca S. The many facets of immune-mediated thrombocytopenia: principles of immunobiology and immunotherapy. Blood Rev. (2024) 63:101141. doi: 10.1016/j.blre.2023.101141
119. Xu X, Zhang J, Xing H, Han L, Li X, Wu P, et al. Identification of metabolism-related key genes as potential biomarkers for pathogenesis of immune thrombocytopenia. Sci Rep. (2024) 14:9040. doi: 10.1038/s41598-024-59493-7
120. Wiggins RW, Woo J, Cauba JN, and Mito S. Evaluating the potential of herbal extracts as treatment in immune thrombocytopenia: A review of evidence and limitations. Appl Biosci. (2025) 4:1. doi: 10.3390/applbiosci4010001
121. Kuter DJ. The treatment of immune thrombocytopenia (Itp)—Focus on thrombopoietin receptor agonists. Ann Blood. (2021) 6:7. doi: 10.21037/aob-21-23
122. Ben-Yacov O, Godneva A, Rein M, Shilo S, Lotan-Pompan M, Weinberger A, et al. Gut microbiome modulates the effects of a personalised postprandial-targeting (Ppt) diet on cardiometabolic markers: A diet intervention in pre-diabetes. Gut. (2023) 72:1486–96. doi: 10.1136/gutjnl-2022-329201
123. Datta S, Pasham S, Inavolu S, Boini KM, and Koka S. Role of gut microbial metabolites in cardiovascular diseases-current insights and the road ahead. Int J Mol Sci. (2024) 25:10208. doi: 10.3390/ijms251810208
124. Zhao X, Liu S, Li S, Jiang W, Wang J, Xiao J, et al. Unlocking the power of postbiotics: A revolutionary approach to nutrition for humans and animals. Cell Metab. (2024) 36:725–44. doi: 10.1016/j.cmet.2024.03.004
125. Yang X and Hang HC. Chemical genetic approaches to dissect microbiota mechanisms in health and disease. Science. (2024) 386:eado8548. doi: 10.1126/science.ado8548
126. Heavey MK, Hazelton A, Wang Y, Garner M, Anselmo AC, Arthur JC, et al. Targeted delivery of the probiotic saccharomyces Boulardii to the extracellular matrix enhances gut residence time and recovery in murine colitis. Nat Commun. (2024) 15:3784. doi: 10.1038/s41467-024-48128-0
127. Madkhali MA. Recent advances in the management of immune thrombocytopenic purpura (Itp): A comprehensive review. Med (Baltimore). (2024) 103:e36936. doi: 10.1097/MD.0000000000036936
128. Ezike TC, Okpala US, Onoja UL, Nwike CP, Ezeako EC, Okpara OJ, et al. Advances in drug delivery systems, challenges and future directions. Heliyon. (2023) 9:e17488. doi: 10.1016/j.heliyon.2023.e17488
129. Lambert MP. On the horizon: upcoming new agents for the management of itp. Hematol Am Soc Hematol Educ Program. (2024) 2024:692–9. doi: 10.1182/hematology.2024000596
130. Brown JA, Bashir H, and Zeng MY. Lifelong partners: gut microbiota-immune cell interactions from infancy to old age. Mucosal Immunol. (2025) 25. doi: 10.1016/j.mucimm.2025.01.006
131. Zhao Q, Chen Y, Huang W, Zhou H, and Zhang W. Drug-microbiota interactions: an emerging priority for precision medicine. Signal Transduction Targeted Ther. (2023) 8:386. doi: 10.1038/s41392-023-01619-w
132. Huang H, Jiang J, Wang X, Jiang K, and Cao H. Exposure to prescribed medication in early life and impacts on gut microbiota and disease development. eClinicalMedicine. (2024) 68:102428. doi: 10.1016/j.eclinm.2024.102428
133. Crudele L, Gadaleta RM, Cariello M, and Moschetta A. Gut microbiota in the pathogenesis and therapeutic approaches of diabetes. eBioMedicine. (2023) 97:104821. doi: 10.1016/j.ebiom.2023.104821
134. Arnone AA, Ansley K, Heeke AL, Howard-McNatt M, and Cook KL. Gut microbiota interact with breast cancer therapeutics to modulate efficacy. EMBO Mol Med. (2025) 17:219–34. doi: 10.1038/s44321-024-00185-0
135. Beyi AF, Wannemuehler M, and Plummer PJ. Impacts of gut microbiota on the immune system and fecal microbiota transplantation as a re-emerging therapy for autoimmune diseases. Antibiotics (Basel). (2022) 11:1093. doi: 10.3390/antibiotics11081093
136. Chen Q, Wu C, Xu J, Ye C, Chen X, Tian H, et al. Donor-recipient intermicrobial interactions impact transfer of subspecies and fecal microbiota transplantation outcome. Cell Host Microbe. (2024) 32:349–65.e4. doi: 10.1016/j.chom.2024.01.013
137. Porcari S, Benech N, Valles-Colomer M, Segata N, Gasbarrini A, Cammarota G, et al. Key determinants of success in fecal microbiota transplantation: from microbiome to clinic. Cell Host Microbe. (2023) 31:712–33. doi: 10.1016/j.chom.2023.03.020
138. Sarita B, Samadhan D, Hassan MZ, and Kovaleva EG. A comprehensive review of probiotics and human health-current prospective and applications. Front Microbiol. (2024) 15:1487641. doi: 10.3389/fmicb.2024.1487641
139. Kaltsas A, Giannakodimos I, Markou E, Adamos K, Stavropoulos M, Kratiras Z, et al. The role of gut microbiota dysbiosis in erectile dysfunction: from pathophysiology to treatment strategies. Microorganisms. (2025) 13:250. doi: 10.3390/microorganisms13020250
140. Ma Z, Zuo T, Frey N, and Rangrez AY. A systematic framework for understanding the microbiome in human health and disease: from basic principles to clinical translation. Signal Transduct Target Ther. (2024) 9:237. doi: 10.1038/s41392-024-01946-6
141. Kumar B, Lorusso E, Fosso B, and Pesole G. A comprehensive overview of microbiome data in the light of machine learning applications: categorization, accessibility, and future directions. Front Microbiol. (2024) 15:1343572. doi: 10.3389/fmicb.2024.1343572
142. Han R, Acosta JN, Shakeri Z, Ioannidis JPA, Topol EJ, and Rajpurkar P. Randomised controlled trials evaluating artificial intelligence in clinical practice: A scoping review. Lancet Digital Health. (2024) 6:e367–e73. doi: 10.1016/S2589-7500(24)00047-5
143. Guevara-Ramirez P, Cadena-Ullauri S, Paz-Cruz E, Ruiz-Pozo VA, Tamayo-Trujillo R, Cabrera-Andrade A, et al. Gut microbiota disruption in hematologic cancer therapy: molecular insights and implications for treatment efficacy. Int J Mol Sci. (2024) 25:10255. doi: 10.3390/ijms251910255
144. Zhang Y, Zhao X, Zhang J, Zhang Y, and Wei Y. Advancements in the impact of human microbiota and probiotics on leukemia. Front Microbiol. (2024) 15:1423838. doi: 10.3389/fmicb.2024.1423838
145. Shukla V, Singh S, Verma S, Verma S, Rizvi AA, and Abbas M. Targeting the microbiome to improve human health with the approach of personalized medicine: latest aspects and current updates. Clin Nutr ESPEN. (2024) 63:813–20. doi: 10.1016/j.clnesp.2024.08.005
146. Davoutis E, Gkiafi Z, and Lykoudis PM. Bringing gut microbiota into the spotlight of clinical research and medical practice. World J Clin cases. (2024) 12:2293–300. doi: 10.12998/wjcc.v12.i14.2293
147. Hasan R, Kumar A, Bose S, Roy R, Arora A, Sharma P, et al. Novel microbial signatures in the faecal microbiome associated with severe alcoholic hepatitis: Bacteroides finegoldii and Veillonella dispar. Open Microbiol J. (2023) 17. doi: 10.2174/18742858-v17-e230803-2023-7
148. Zheng J, Sun Q, Zhang M, Liu C, Su Q, Zhang L, et al. Noninvasive, microbiome-based diagnosis of inflammatory bowel disease. Nat Med. (2024) 30:3555–67. doi: 10.1038/s41591-024-03280-4
149. Lee D, Maaskant A, Ngo H, Montijn RC, Bakker J, Langermans JAM, et al. A rapid, affordable, and reliable method for profiling microbiome biomarkers from fecal images. iScience. (2024) 27:111310. doi: 10.1016/j.isci.2024.111310
150. Kapur R. Broadening the horizon of immune thrombocytopenia through omics approaches. Br J Haematol. (2024) 204:2159–61. doi: 10.1111/bjh.19514
151. Chen Y-C, Su Y-Y, Chu T-Y, Wu M-F, Huang C-C, and Lin C-C. Prelect: prevalence leveraged consistent feature selection decodes microbial signatures across cohorts. NPJ Biofilms Microbiomes. (2025) 11:3. doi: 10.1038/s41522-024-00598-2
152. Chetty A and Blekhman R. Multi-omic approaches for host-microbiome data integration. Gut Microbes. (2024) 16:2297860. doi: 10.1080/19490976.2023.2297860
153. Stumptner C, Stadlbauer V, O’Neil D, Gessner A, Hiergeist A, Zatloukal K, et al. The pre-analytical Cen/Ts standard for microbiome diagnostics-how can research and development benefit? Nutrients. (2022) 14:1976. doi: 10.3390/nu14091976
154. Kamel M, Aleya S, Alsubih M, and Aleya L. Microbiome dynamics: A paradigm shift in combatting infectious diseases. J Personalized Med. (2024) 14:217. doi: 10.3390/jpm14020217
155. Barone M, Martucci M, Sciara G, Conte M, Medina LSJ, Iattoni L, et al. Towards a personalized prediction, prevention and therapy of insomnia: gut microbiota profile can discriminate between paradoxical and objective insomnia in post-menopausal women. EPMA J. (2024) 15:471–89. doi: 10.1007/s13167-024-00369-1
156. Bars-Cortina D, Ramon E, Rius-Sansalvador B, Guinó E, Garcia-Serrano A, Mach N, et al. Comparison between 16s Rrna and shotgun sequencing in colorectal cancer, advanced colorectal lesions, and healthy human gut microbiota. BMC Genomics. (2024) 25:730. doi: 10.1186/s12864-024-10621-7
157. D’Urso F and Broccolo F. Applications of artificial intelligence in microbiome analysis and probiotic interventions—an overview and perspective based on the current state of the art. Appl Sci. (2024) 14:8627. doi: 10.3390/app14198627
158. Danieli MG, Brunetto S, Gammeri L, Palmeri D, Claudi I, Shoenfeld Y, et al. Machine learning application in autoimmune diseases: state of art and future prospectives. Autoimmun Rev. (2024) 23:103496. doi: 10.1016/j.autrev.2023.103496
159. He J, Chen Y, Zhao H, and Li Y. The interplay between gut bacteria and targeted therapies: implications for future cancer treatments. Mol Med. (2025) 31:58. doi: 10.1186/s10020-025-01108-6
160. Soulard M, Galicier L, Mahlaoui N, Fieschi C, Deshayes S, Gobert D, et al. Efficacy and safety of Tpo receptor agonists in treatment of Itp associated with predominantly antibody deficiencies. Blood Adv. (2024) 8:6171–82. doi: 10.1182/bloodadvances.2024014370
161. Gebrayel P, Nicco C, Al Khodor S, Bilinski J, Caselli E, Comelli EM, et al. Microbiota medicine: towards clinical revolution. J Trans Med. (2022) 20:111. doi: 10.1186/s12967-022-03296-9
162. LeVine DN, Goggs R, Kohn B, Mackin AJ, Kidd L, Garden OA, et al. Acvim consensus statement on the treatment of immune thrombocytopenia in dogs and cats. J Vet Intern Med. (2024) 38:1982–2007. doi: 10.1111/jvim.17079
163. Allegretti JR, Khanna S, Mullish BH, and Feuerstadt P. The progression of microbiome therapeutics for the management of gastrointestinal diseases and beyond. Gastroenterology. (2024) 167:885–902. doi: 10.1053/j.gastro.2024.05.004
164. Metwaly A, Kriaa A, Hassani Z, Carraturo F, Druart C, Consortium I, et al. A consensus statement on establishing causality, therapeutic applications and the use of preclinical models in microbiome research. Nat Rev Gastroenterol Hepatol. (2025) 22:343–56. doi: 10.1038/s41575-025-01041-3
165. Darnaud M, De Vadder F, Bogeat P, Boucinha L, Bulteau AL, Bunescu A, et al. A standardized gnotobiotic mouse model harboring a minimal 15-member mouse gut microbiota recapitulates Sopf/Spf phenotypes. Nat Commun. (2021) 12:6686. doi: 10.1038/s41467-021-26963-9
166. Noguera-Fernandez N, Candela-Gonzalez J, and Orenes-Pinero E. Probiotics, prebiotics, fecal microbiota transplantation, and dietary patterns in inflammatory bowel disease. Mol Nutr Food Res. (2024) 68:e2400429. doi: 10.1002/mnfr.202400429
167. Song Y, Li J, and Wu Y. Evolving understanding of autoimmune mechanisms and new therapeutic strategies of autoimmune disorders. Signal Transduction Targeted Ther. (2024) 9:263. doi: 10.1038/s41392-024-01952-8
168. Kishon R, Modlin NL, Cycowicz YM, Mourtada H, Wilson T, Williamson V, et al. A rapid narrative review of the clinical evolution of psychedelic treatment in clinical trials. NPJ Ment Health Res. (2024) 3:33. doi: 10.1038/s44184-024-00068-9
Keywords: primary immune thrombocytopenia (ITP), gut-immune axis, immune dysregulation, microbiota-based therapy, platelet homeostasis
Citation: Guo X, Wang K, Liu Q, Baran N and Ma W (2025) The gut-immune axis in primary immune thrombocytopenia (ITP): a paradigm shifts in treatment approaches. Front. Immunol. 16:1595977. doi: 10.3389/fimmu.2025.1595977
Received: 19 March 2025; Accepted: 26 May 2025;
Published: 12 June 2025.
Edited by:
Nar Singh Chauhan, Maharshi Dayanand University, IndiaReviewed by:
Yean Kong Yong, Malaysia, MalaysiaAvinash Lomash, Medanta The Medicity Hospital, India
Rohit Patel, Harvard Medical School, United States
Copyright © 2025 Guo, Wang, Liu, Baran and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuejun Guo, cHlneGpAMTYzLmNvbQ==; Wenxue Ma, d21hQGhlYWx0aC51Y3NkLmVkdQ==
 Xuejun Guo
Xuejun Guo Ke Wang
Ke Wang Qianhui Liu1
Qianhui Liu1 Natalia Baran
Natalia Baran Wenxue Ma
Wenxue Ma