- Center for Cellular Engineering, Clinical Center, National Institutes of Health, Bethesda, MD, United States
This review explores recent advances in the characteristics and manufacturing of CAR T-cell products. Traditional potency assays have been designed based on well-established CAR T-cell functionalities. However, the advent of innovative tools and methodologies has revealed a broader spectrum of important CAR T-cell characteristics that correlate with function. Furthermore, as manufacturing strategies continue to evolve, conventional potency assays may no longer fully capture the complexity of these products. Therefore, it is essential to examine these emerging characteristics and manufacturing approaches and consider the development of tailored potency assays to ensure products are fully characterized.
1 Introduction
Chimeric antigen receptor (CAR) T-cells demonstrate promising clinical outcomes (1–7), as the indications for the use of these therapies are growing there is a need to develop appropriate and robust potency assays that can accurately assess their therapeutic potential. Potency assays are generally designed to measure the biological activities of CAR T-cells based on their mechanism of action (MoA) (8, 9). The MoA of CAR T-cells is a multifaceted process that underlies their therapeutic effects on target cells. CAR T-cells are designed to express chimeric antigen receptors that specifically recognize and bind to antigens on the surface of target cells (1–4, 10–14). Upon antigen recognition, CAR T-cells become activated, initiating a cascade of cellular responses that ultimately lead to the destruction of target cells (15). Beyond their immediate cytotoxic functions, their viability and in vivo expansion and persistence are critical for sustained therapeutic effect (15–17).
Understanding the key components of MoA is essential for developing potency assays that accurately reflect the functional capabilities of CAR T-cells and ideally, these will correlate with clinical outcomes. The potency of the FDA-approved CAR T-cell products is primarily assessed by measuring the release of IFN-γ in response to target cells, along with other factors, such as cell viability, and product-specific attributes, such as the expression of the specific target CAR (8, 9).
Over the past decade, advances in CAR T-cell research have led to the identification of new cellular characteristics associated with clinical responses and innovative manufacturing procedures have been developed to enhance these characteristics, driven by emerging tools and methodologies. These advances raise concerns that conventional potency assays may not fully capture the complexity of manufactured products. Given these developments, it is crucial to comprehensively review recent progress in the design and production of CAR T-cells and explore the need for tailored potency assays that fully define manufactured products.
2 Advances in CAR T-cell product profiling
Evaluating CAR T-cell potency requires a comprehensive matrix of assays that fully profile the key activities and characteristics of the cells (Figure 1). Over the past decade, advanced multi-omics approaches, including genomics, epigenomics, transcriptomics, proteomics, and metabolomics at both bulk and single-cell resolution, have significantly enhanced our understanding of CAR T-cell function at the molecular level (Table 1). In this section, we review these advancements and highlight key insights that may guide the development of next-generation potency assays (Figure 2).
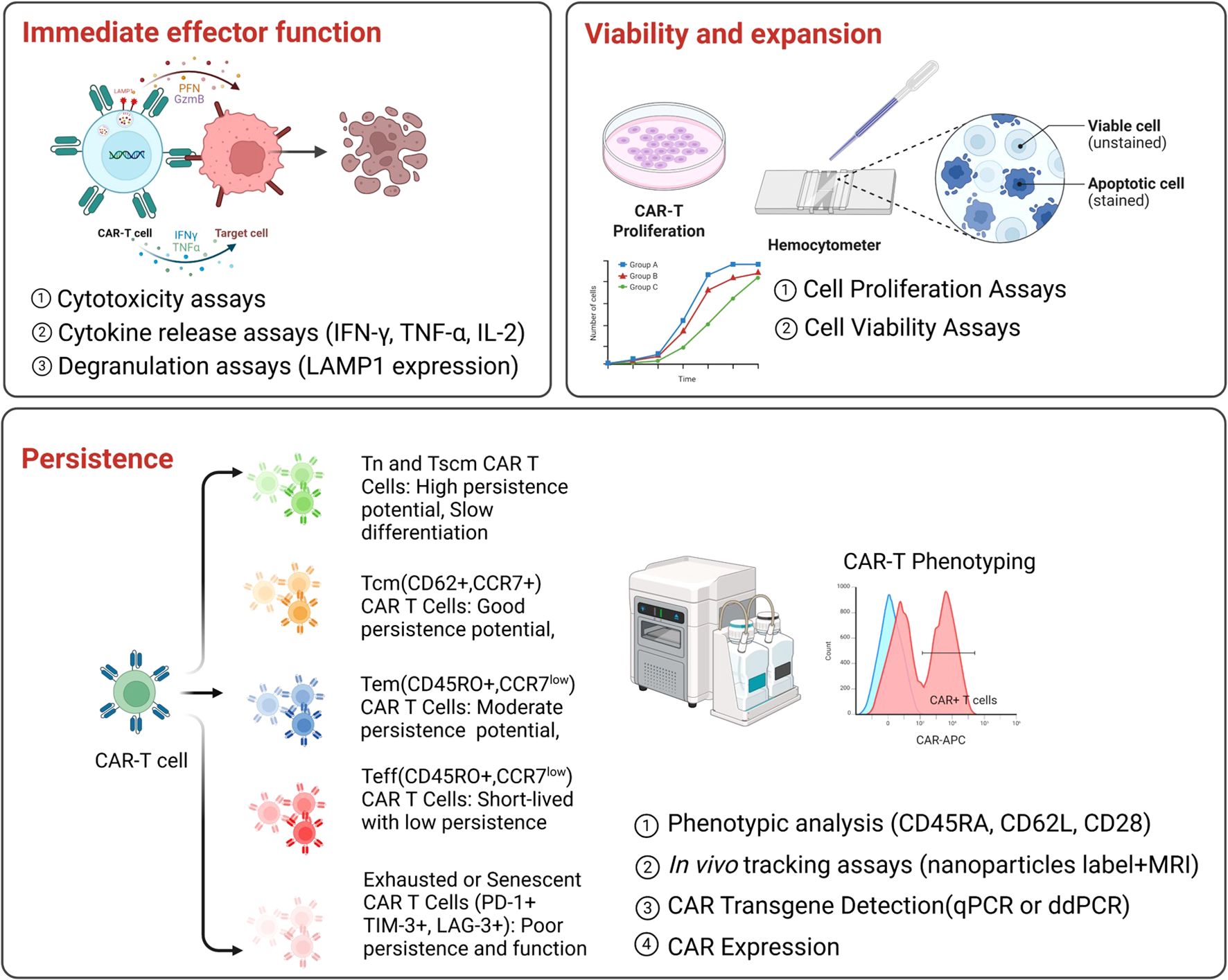
Figure 1. Current potency assays for CAR T-cell products. Key functional assays used to evaluate CAR T-cell potency, categorized into three main aspects.Upper panel (left): Evaluation of immediate effector function, by measuring cytotoxicity, cytokine release (e.g., IFN-g, TNF-a, IL-2), and degranulation (e.g., LAMP1 expression). Upper panel (right): Evaluation of viability and expansion by assessing cell proliferation and viability. Bottom panel: Evaluation of persistence by analyzing CAR T-cell phenotypes, performing in vivo tracking, and assessing CAR transgene expression at pre-infusion and post-infusion.
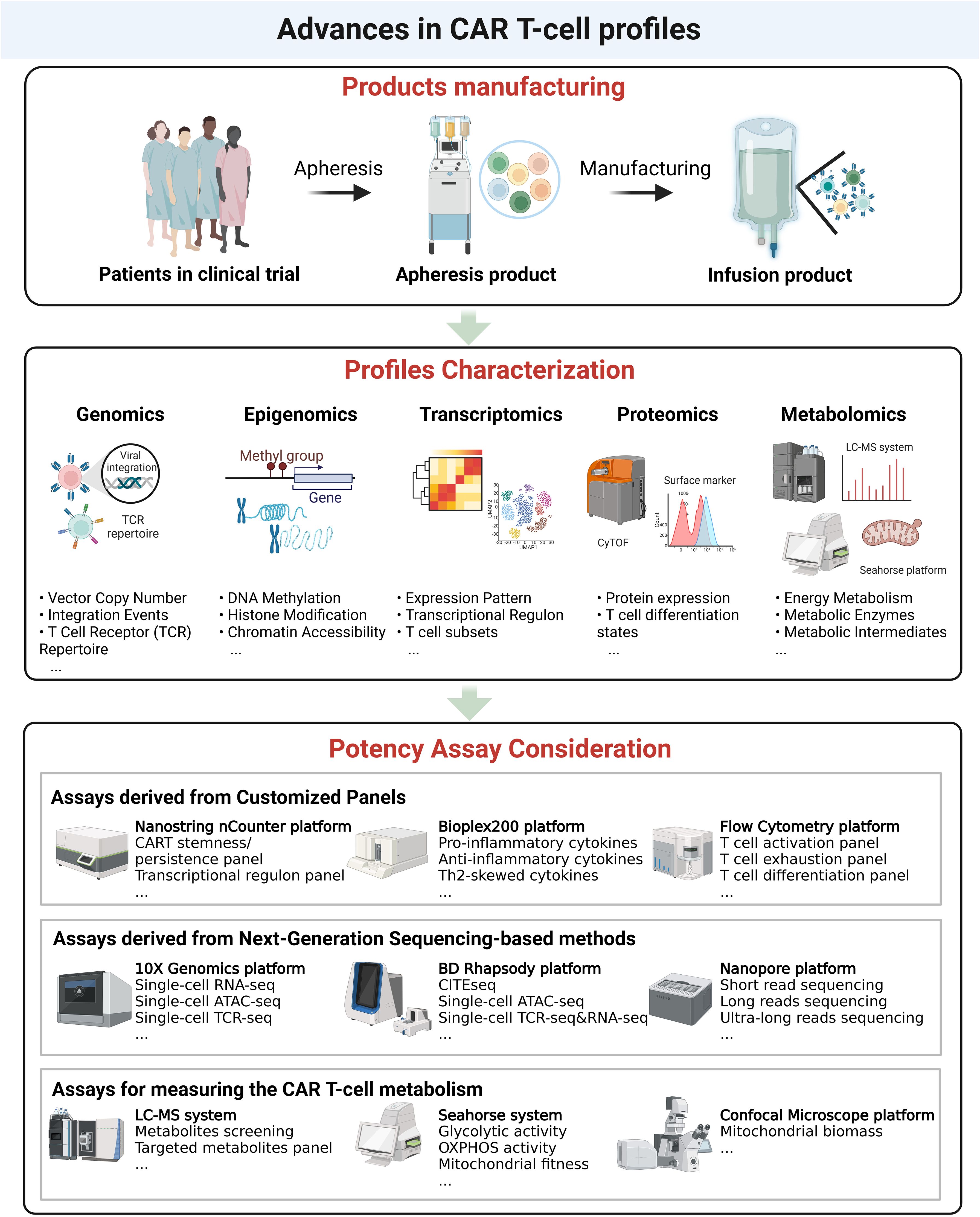
Figure 2. Advances in CAR T-cell analysis. Upper panel. Overview of CAR T-cell product manufacturing, from apheresis collection to infusion of the final CAR T-cell product. Middle panel. Characterization of CAR T-cell profiles at multiple molecular levels, including genomics (vector copy number, integration events, TCR repertoire), epigenomics (DNA methylation, histone modifications, chromatin accessibility), transcriptomics (expression patterns, transcriptional regulation, T-cell subsets), proteomics (protein expression, T-cell differentiation states), and metabolomics (energy metabolism, metabolic enzymes, intermediates). Bottom panel. Potency assay considerations, including customized panel-based assays (e.g., cytokine profiling, T-cell activation), next-generation sequencing-based approaches (e.g., single-cell RNA-seq, ATAC-seq, TCR-seq), and metabolic assessment methods (e.g., glycolytic activity, mitochondrial fitness).
2.1 Genomic profiles in CAR T-cell products
In recent years, the genomic profiling of CAR T-cell products has primarily focused on vector copy number (VCN) (9, 18, 19), and vector integration sites (8, 20, 21). Additionally, immunogenomic analyses, such as bulk and single-cell T cell receptor (TCR) sequencing (TCR-seq), have been applied to assess the TCR repertoire in CAR T-cells (22–25). For FDA-approved CAR T-cell products, VCN quantification is a mandatory component of lot-release testing, with droplet digital PCR (ddPCR) being widely used as a routine safety assay to measure VCN in most quality control (QC) laboratories (8, 9). Here, we focus on recent advancements in vector integration and TCR repertoire profiling.
On November 28, 2023, the U.S. FDA announced an investigation into cases of secondary malignancies in patients who received CAR T-cell therapy (20). The potential risk for secondary malignancies caused by insertional mutagenesis has long been a concern in CAR T-cell therapy, as viral vector transduction is required for CAR expression. However, the exact nature and frequency of genotoxicity risk associated with retroviral or lentiviral insertion remains unclear and deserves thorough investigation and transparency. Recent large-cohort follow-up studies report the incidence of secondary malignancies after CAR infusion ranging from 2-16% (26–31). Insertional mutagenesis occurs when viral vector integrates into a gene associated with cancer development, inadvertently activating an oncogene or inactivating a tumor suppressor gene and potentially leading to oncogenesis. Beyond this risk, vector integration has, in certain circumstances, been associated with a selective growth advantage, resulting in clonal CAR T-cell expansion, dominance, and persistence (32–34).
Research by Carl June’s team revealed that the integration events at the TET2 gene enhanced CAR T-cell potency (32). Similarly, an NCI research group identified clonal expansion of CAR T-cells harboring lentivector integration in the CBL gene following CAR T-cell therapy (33). Christopher et al. demonstrated that both the number and genomic loci of integration events correlate with clinical outcome in CD19 CAR T-cell products (35). Their study found that genes with integration sites enriched in responders were commonly involved in cell-signaling and chromatin modification pathways, suggesting that insertional mutagenesis in these genes promoted therapeutic T-cell proliferation. However, the consequences of viral vector integration into these reported genes have not been consistently reproducible (32). A study on the clonal dynamics of CAR T-cells over time found that not all T-cells with TET2 integration exhibit expansion, either during CAR T-cell production or after infusion (34). These findings underscore the importance of monitoring vector integration sites with potency assays in order to address safety and efficacy concerns. Our team and other research groups have developed robust pipelines for detecting viral insertion events (21, 36–40). The Bushman lab initially applied the Illumina sequencing method to investigate viral integration events in cellular products and developed the INSPIIRED pipeline (36), which enables measurement of integration events at bulk-cell resolution. Furthermore, Wenliang Wang and colleagues developed the EpiVIA pipeline, which enables detection of integration sites at the single-cell level (39). These advancements have significantly improved the feasibility of detecting integration events and facilitate their incorporation into mechanistic and safety evaluations. However, in contrast to vector copy number (VCN), which has a defined regulatory cutoff, integration site analysis currently lacks standardized criteria for determining which insertion events are definitively oncogenic and should be excluded from infusion products. Even in the case of well-characterized oncogenes such as TP53, additional mutations are often required to drive malignant transformation, as demonstrated by Perica et al. (41). As such, integration site analysis is presently better suited for informational purposes rather than serving as a standalone lot-release assay.
Another critical genomic feature is the TCR repertoire, which has been increasingly recognized as a crucial factor influencing the treatment efficacy in immunotherapies (42–45). While CAR T-cell therapy primarily relies on the target CAR expression, endogenous TCR diversity, characterized by oligoclonality and polyclonality, within the infusion products may also contribute to treatment outcomes. Recent studies have characterized the kinetic profiling of different TCR clonotypes throughout the CAR T-cell treatment process (25, 46, 47), demonstrating the cytotoxic and proliferative features of highly expanded CAR T-cell clonotypes in patients. Paired single-cell RNA analysis and TCR repertoire profiling allow for the identification of individual CAR T-cells with distinct transcriptional phenotypes (48, 49), enabling the use of TCR clonotypes as surrogate for the expansion and persistence of functional T-cell states. Qing et al. applied this paired single-cell approach to 24 infusion products and found products associated with poor clinical responses exhibited moderately reduced TCR clonotypic diversity and showed exhaustion signatures (50).
γδ T-cells present another potentially advantageous subset in the infusion products. While the majority of CAR T-cell infusion products consist of αβ T-cells, γδ CAR T-cells have demonstrated resistance to exhaustion, exhibiting lower levels of TIM3 and PD1 expression following activation (51, 52). A longitudinal analysis of CD19 CAR T-cell therapy in a chronic lymphocytic leukemia patient who achieved a durable complete response revealed the expansion of a γδ CAR T-cell population, accounting for up to 33% of all CAR+ cells three months post-infusion (47). Our previous work also suggests that γδ T cell in CAR T-cell products may enhance cytotoxicity and be associated with favorable clinical responses (53). Collectively, these findings highlight the importance of TCR repertoire assessment in determining the potency of CAR T-cell infusion products.
2.2 Epigenomic profiles in CAR T-cell products
Preclinical and clinical trial data highlight the critical role of CAR T-cell differentiation states in determining therapeutic efficacy (54–56). CAR T-cell differentiation states refer to the developmental stages that T cells progress through, such as naïve, stem-cell like memory, central memory, effector memory, and terminally differentiated effector cells (57). These states are characterized by distinct gene expression profiles, functional properties, and persistence potential (58). T-cell differentiation is epigenetically programmed and maintained in progeny cells through chromatin states and DNA methylation (59). Insights from epigenomics studies have expanded our understanding of factors influencing CAR T-cell potential beyond transcriptomic profiling alone (60–70). Carlos et al. analyzed DNA methylation profiles in 114 CD19 CAR T-cell products and identified 18 distinct epigenetic loci associated with complete response (CR), event-free survival (EFS), and overall survival (OS) post-infusion. Using these CR-associated sites, they developed and validated an epigenetic signature, termed the EPICART signature, across different cohorts, demonstrating its potential as a predictor of CAR T-cell efficacy (62). Caitlin and colleagues performed a longitudinal DNA methylation assessment of CD8+ CD19 CAR T-cells from patients with B-cell acute lymphoblastic leukemia (B-ALL), revealing DNA methylation programs linked to a decline in CD19 CAR T-cell memory potential and the establishment of an exhaustion trajectory (63). These findings that have also been reported by others (64–66).
Epigenetic modulation has also been explored as a method to enhance CAR T-cell functionality. Yao et al. found CAR T-cells treated with low-dose decitabine (DAC, a de novo DNA methylation inhibitor) maintained higher memory-associated and lower exhaustion-associated gene expression profiles (64). Brooke and colleagues found that deleting de novo DNA methyltransferase 3 alpha (DNMT3A) in CAR T-cells prevented exhaustion and enhanced antitumor activity (65).
Beyond DNA methylation, histone modifications have been implicated in CAR T-cell function. Research has identified distinct histone markers that distinguish CD8+ T-cell subsets within CAR T-cell products (61). In preclinical investigations, Michel Sadelain’s group demonstrated that disrupting SUV39H1-mediated H3K9 methylation enhances the functional persistence of CD28-based CAR T-cells (67). Similarly, Mackall and colleagues restored functionality in exhausted CAR T-cells through epigenetic remodeling (68). Collectively, these findings underscore the importance of epigenomic profiling in understanding and optimizing CAR T-cell functionality. Beyond assessing product characteristics, epigenomic insights could inform potency assay development, guiding strategies to refine CAR T-cell manufacturing and enhance therapeutic efficacy. In the future, QC laboratories should consider implementing sequencing-based DNA methylation panels as potency assays, provided they can be robustly correlated with functional outputs. Alternatively, a single PCR-based assay targeting key epigenetic loci, such as those from the EPICART signature (62), could offer a more cost-effective and accessible option for routine potency testing.
2.3 Transcriptomics profiles in CAR T-cell products
Transcriptomics is a widely applied tool for analyzing gene expression (71–73), including in CAR T-cell products (5, 74–77). Both bulk and single-cell RNA sequencing (scRNA-seq) have emerged as powerful techniques for deciphering the molecular mechanisms governing CAR T-cell functionality, persistence, dysfunction, and therapeutic efficacy (50, 54, 78–82). To date, findings from transcriptomic profiling can be summarized into several key aspects.
2.3.1 Distinct expression patterns correlate with clinical outcomes
Studies have reported that CD19 CAR T-cells from complete responders are enriched in memory-related gene signatures, including IL-6/STAT3 signatures, whereas CAR T-cells from non-responders exhibit upregulated programs associated with effector differentiation, exhaustion and apoptosis (50, 82–84), which correlates with FACS data concerning CAR-T subsets. Additionally, CD19 CAR T-cells from CR patients demonstrate significantly higher expression of genes involved in glycolysis (82, 85). Preclinical studies further suggest that increased glycolytic activity in CAR T-cells is linked to enhanced potency, which may contribute to favorable efficacy (86, 87).
2.3.2 Key transcriptional factors and regulons are associated with CAR T-cell function
Transcription factors (TFs) serve as master regulators of T-cell differentiation, expansion, fitness, and anti-tumor activity (88–90). Transcriptomic studies have identified several key TFs and associated regulons that play crucial roles in shaping CAR T-cell functionality (54, 91–96). One of the most well-characterized TFs in CAR T-cell biology is TCF7 and its regulatory network, the TCF7 regulon, which serves as a master regulator of T-cell memory. High TCF7 expression has been linked to enhanced persistence and long-term efficacy in CAR T-cell therapy by maintaining a less-differentiated, stem-like phenotype associated with sustained antitumor activity (54, 93, 97). Another critical transcriptional network is FOXO1 and its regulon (94, 95), which has been identified as a key enhancer of CAR T-cell function that boosts stemness, metabolic fitness, and antitumor activity. Additionally, AP-1 family members play a significant role in modulating CAR T-cell exhaustion (98–100). BATF and IRF4 cooperate to counter exhaustion in CAR T-cells (98), while c-Jun overexpression has been shown to induce resistance to exhaustion, thereby improving CAR T-cell functionality (99).
2.3.3 Subsets of CAR T-cell populations are associated with clinical outcomes and long-term event-free survival
The advancement of single-cell RNA sequencing (scRNA-seq) has significantly enhanced our ability to uncovered previously unappreciated T-cell subsets in CAR T-cell infusion products, enabling the identification of minor yet functionally distinct CAR T-cell populations associated with clinical efficacy, highlighting their potential as biomarkers for potency assessment (22, 78, 81, 85). Less differentiated populations of CD8⁺ CAR T-cells, such as those with stem-like memory T-cell (Tscm) and central memory T-cell (Tcm) phenotypes, are associated with superior expansion, sustained tumor clearance, and prolonged EFS (50, 82, 101).
Emerging evidence suggests that a subset of cytotoxic CD4⁺ T cells in both infusion products and post-infusion samples, characterized by high expression of cytotoxic markers (PRF1, GZMK, GZMB, NKG7, and GNLY), correlates with clinical response (47, 50, 82, 102). Deng et al. found this subtype to be enriched in products with partial response (PR) and progressive disease (PD) (50). Maus’s team also reported CD4+NKG7+ cells were more prominent in non-responders (82). Moreover, Melenhorst and colleagues observed that in two patients who experienced decade-long remissions, cytotoxic CD4+ T-cells dominated the persistent population 5 to 10 years post-infusion (47). Regulatory T-cells (Tregs), expressing FOXP3, IKZF2, and CTLA4, have been observed in commercial CAR T-cell products and are suspected to contribute to CAR T-cell therapeutic resistance (82, 85, 102). Studies by Nicholas and colleagues and Good and colleagues found CAR-Treg cells were more frequent in non-responders and may contribute to relapse in vivo (82, 102). Bai and colleagues studied CD19 CAR T-cell infusion products from 82 pediatric patients with B-ALL using scRNA-seq and CITE-seq. They found that Th2 function deficiency was associated with CD19-positive relapse, whereas Th2 functionality correlated with ultra-long-term event-free survival (EFS > 96 months) (79, 101, 103). A unique subset of CD8+ CAR T-cells termed CD8-fit T-cells, characterized by enhanced migration capacity, serial killing ability, and balanced mitochondrial and lysosomal volumes, has been identified (104). Infusion products with a higher proportion of CD8-fit T-cells correlated with favorable outcomes and long-term persistence in patients (104). Developing strategies to enrich CAR T-cell products with CD8-fit T-cells may significantly enhance clinical efficacy. Additionally, a double-negative T-cell phenotype was recently reported as a unique subset in infusion products that is associate with long-lived CAR T-cells (47, 78). Collectively, there is a need to develop manufacturing processes that select for desired CAR T-cell phenotypes and to establish potency assays that characterize gene expression patterns associated with positive clinical outcomes. These assays should be designed to provide actionable results in a timely manner, ensuring they effectively capture the unique characteristics of CAR T-cells and their therapeutic potential.
2.4 Proteomics profiles in CAR T-cell products
Advancements in proteomics profiling of CAR T-cell products have been driven primarily by mass cytometry, enabling the simultaneous characterization of intracellular signaling, activation, proliferation, cytokine production, and phenotype within a single assay (81, 105–111). Several studies have focused on deciphering CAR T-cell mechanisms of action using mass cytometry.
Goldberg and colleagues developed an integrative mass cytometry panel to analyze trafficking and functional protein expression in CD19 CAR T-cells (109), identifying upregulation of activation markers (CD27, GZMB, CD69 and CD25), proliferation marker (Ki-67), and glycolysis markers (Glut1, and LDHA) in infusion products compared to baseline leukapheresis T-cells (81, 109). Salter and colleagues used mass spectrometry (MS)-based proteomics to reveal CAR T-cell activation pathways, including MAPKs (110). Hegde et al. using cytometry by time-of-flight (CyTOF) in HER-2 CAR T-cells, found lower frequencies of CD8+ T cells expressing PD-1+TIM-3+, PD-1+LAG-3+ or PD-1+CD39+ in patients achieving CR, while higher and more variable levels were observed in those with SD and PD (108). Additionally, single-cell CyTOF analysis of day 7 circulating CAR T-cells in axi-cel-treated (a commercial CAR T-cell product) patients with large B-cell lymphoma identified three metaclusters associated with long-term clinical response (102).
2.5 Metabolomics profiles in CAR T-cell products
Unlike previous omics approaches, metabolomics focuses on cellular metabolism, including energy metabolism (glycolysis, oxidative phosphorylation, fatty acid oxidation, etc.), mitochondrial metabolism (biogenesis, fitness, ROS production, etc.), and the analysis of other metabolites. Recently, increasing attention has been given to metabolomics profiling of CAR T-cell products, which has provided crucial insights into how metabolic fitness influences persistence, cytotoxicity, and how this correlates with overall therapeutic efficacy (80, 86, 112–115). Oxidative phosphorylation (OXPHOS) is the predominant metabolic program in memory T-cells, while aerobic glycolysis characterizes effector T-cells (116, 117). Kawalekar and colleagues demonstrated that CAR T-cells with a CD28ζ costimulatory domain primarily rely on aerobic glycolysis, whereas those with BBζ preferentially utilize fatty acid oxidation (FAO), contributing to their enhanced persistence and central memory differentiation (118). Additionally, Cappabianca and colleagues found that metabolic priming by reducing aerobic glycolysis and increasing bound NAD(P)H activity was associated with lower cytokine production, including IFN-γ, IL-2, IL-17, and TGF-β, while promoting central memory CAR T-cell expansion and persistence in GD2 CAR T-cells (119).
As for the metabolites, Paul Renauer and colleagues identified ADA and PDK1 as key metabolic regulators that enhance CAR19 T-cell cytolysis against leukemia cells (120). ADA (adenosine deaminase) catalyzes the conversion of adenosine to inosine within the purine metabolism pathway, and inosine has been reported to induce stemness features in CAR T-cells, enhancing their potency (121). Additionally, Ye and colleagues screened 27 differentially abundant metabolites in CD22 CAR T-cells with varying efficacy, identifying proline metabolism as the most significant contributor to CAR T-cell function (112). These findings suggest that detecting inosine or other metabolites in CAR T-cell supernatants could serve as a novel potency assay.
Mitochondrial properties have also been investigated in CAR T-cell products beyond energy metabolism. CAR T-cells from patients with a complete response exhibited increased mitochondrial biomass and volume compared to non-responders, correlating positively with expansion and persistence (104, 122). This enhanced persistence may be attributed to reserved bioenergetic potential, also known as spare respiratory capacity (SRC), which enables CAR T-cells to meet metabolic demand upon activation (112, 123). Additionally, mitochondrial quantities, ATP content, and the NADH/NAD ratio were found to be higher in Tscm, which are associated with superior efficacy compared to effector memory (Tem) and central memory (Tcm) T-cells (124). Overall, glycolysis, OXPHOS activity, metabolites in supernatant and mitochondrial indices could serve as potential markers for potency assay development.
2.6 Insights on developing potency assays
As CAR T-cell therapies advance, potency assays must evolve to accurately assess the complex characteristics of these cellular products. Insights from multi- omics approaches emphasize the need for more refined potency assays that effectively capture CAR T-cells efficacy and functionality (Figure 2). While further mechanistic studies are required to establish correlations between specific certain characteristics with a functional output, such as epigenomic profiles to IFN-γ secretion or cytotoxicity, it remains valuable to broadly explore CAR T-cell characteristics for their potential for potency assay development. Some of these characteristics require several days to complete which may prevent them from being used for potency assessment at this time. However, the field is progressing rapidly and if a specific profile is found to be useful for assessing CAR T-cell potency, alternative platforms that allow for more rapid testing will likely be developed. Here, we summarize key considerations for optimizing potency assays based on recent advancements.
2.6.1 Adoption of customized panels in potency assay development
The implementation of customized panels for assessing key functional attributes, including gene expression (CAR T-cell stemness panel, CAR T-cell exhaustion panel, CAR T-cell persistence panel, etc.), DNA methylation [EPICART signature (62)], cytokine secretion (cytotoxic cytokines, Th2-skewed cytokines), surface marker expression (activation panel, exhaustion panel, stemness panel, etc.), and metabolite profiling, offers a promising approach for potency evaluation. Platforms like NanoString nCounter provide a rapid and robust solution for gene expression analysis using predefined panels. For cytokine detection, commercial panels from Bio-Rad, ProteinSimple, and MSD (Meso Scale Discovery) offer efficient and time-saving options. Additionally, surface marker detection has become more streamlined with high-throughput technologies such as flow cytometry, CyTOF, and Cytek platforms. Metabolite analysis can be performed using both targeted and untargeted panels available from providers like Metabolon Inc.
2.6.2 Routine genomic CAR T-cell evaluation
NGS-based methods provide a powerful approach to evaluating CAR T-cells, enabling comprehensive analysis from vector integration sites to TCR repertoire, as well as from bulk and single-cell gene expression to chromatin accessibility. Integrating vector integration sites (Targeted DNA-seq) and TCR repertoire profiling (bulk TCR-seq and scTCR-seq) with gene expression data (RNA-seq, scRNA-seq, CITE-seq, etc.) allows for tracking clonal expansion and composition in CAR T-cells. Additionally, assessing histone modifications (ChIP-seq, CUT&Run-seq, etc.) and chromatin accessibility (ATAC-seq, DNase-seq, scATAC-seq, etc.) could be incorporated into QC pipelines to provide additional potency metrics. However, given the time-consuming nature of these methods, they may not be suitable for use as a “Lot-releasing assay”. However, the data could be used to engineer new manufacturing processes to enhance the desired characteristics identified by these assays.
2.6.3 Routine monitoring of CAR T-cell metabolism
Metabolic fitness plays a critical role in CAR T-cell persistence and function, highlighting the need to integrate metabolic assessments into potency assays. Developing assays that evaluate glycolysis and OXPHOS activity in CAR T-cells, particularly before and after antigen stimulation in vitro, should be considered. The Seahorse XF Analyzer from Agilent provides a robust and efficient platform for real-time monitoring of these metabolic activities, offering insights into energy metabolism, mitochondrial function, and overall, CAR T-cell fitness. Routine metabolic monitoring could enhance CAR T-cell product characterization and contribute to guiding the development manufacturing of processes that produce products with the desired phenotypes.
3 Consideration in potency assays for unconventional CAR T-cells
Real-world experiences with CAR T-cell therapies have highlighted the limitations of conventional manufacturing processes, which are often low-throughput, resource-intensive, and time-consuming (125–127). Traditionally, following apheresis, cells undergo activation, ex vivo modification, expansion, and rigorous quality control testing before infusion. Aiming to overcome these manufacturing challenges, in recent years, several unconventional manufacturing procedures have emerged, including non-viral CAR T-cell generation (128–131), in vivo CAR T-cell manufacturing (132–137), and rapid manufacturing protocols (138, 139). While these approaches offer advantages in efficiency, they also introduce new complexities for potency assays. In this section, we summarize recent advancements in CAR T-cell manufacturing and discuss key considerations for evaluating the potency of these unconventional CAR T-cells (Figure 3).
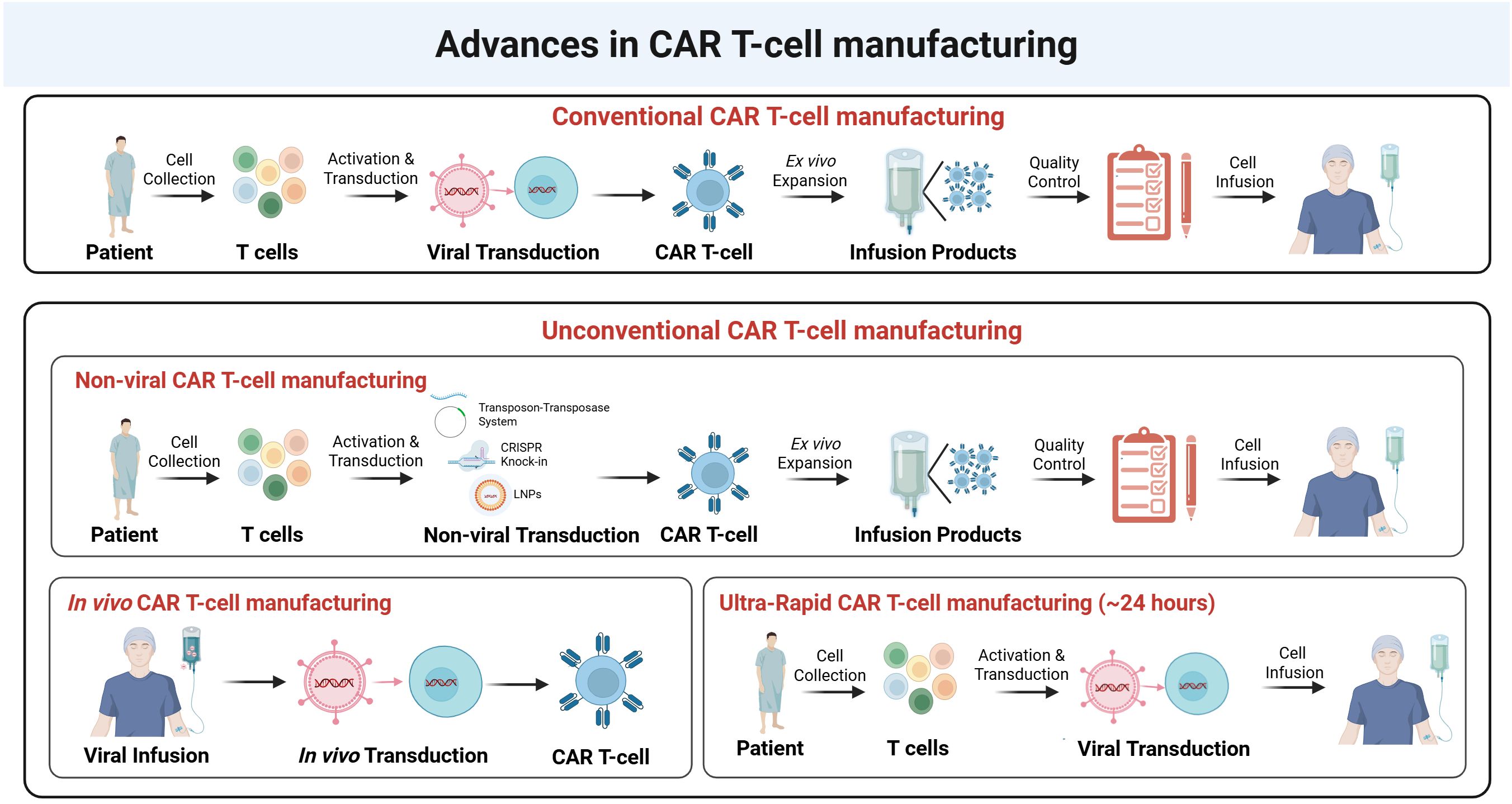
Figure 3. Advances in CAR T-cell manufacturing. Upper panel. Conventional CAR T-cell manufacturing involves the collection of T-cells from patients, activation, and viral transduction to introduce the CAR construct. The transduced CAR T-cells undergo ex vivo expansion, followed by quality control assessment before infusion into patients. Middle panel. Unconventional CAR T-cell manufacturing includes non-viral approaches, such as transposon-transposase systems, CRISPR knock-in, and lipid nanoparticle (LNP)-mediated delivery, as alternative strategies for CAR gene insertion. These non-viral methods follow a similar workflow of activation, transduction, expansion, and quality control before infusion. Bottom panel (left). In vivo CAR T-cell manufacturing eliminates the need for ex vivo manipulation by directly infusing viral vectors into the patient, allowing in vivo transduction and CAR T-cell generation within the body. Bottom panel (right). Ultra-rapid CAR T-cell manufacturing (~24 hours) aims to accelerate the process by minimizing ex vivo expansion steps, allowing rapid viral transduction and direct infusion into patients.
3.1 Non-viral CAR T-cells
The emergence of non-viral gene delivery methods (e.g., transposon systems, CRISPR-mediated knock-in) has provided alternative strategies for generating CAR T-cells without the need for viral vectors (140–142). These methods generally exhibit lower transduction efficiency than viral vector methods due to reduced knock-in rates, leading to a smaller proportion of CAR-expressing cells (140). In transposon-based systems (Sleeping beauty and PiggyBac), VCN varies widely due to uncontrolled transgene integration, resulting in heterogenous CAR expression across cells. Unlike viral vector-based CAR T-cells (VCN: 1–2 copies/cell), transposon-based CAR T-cells can exhibit 0-10+ copies/cell (18, 19, 130, 143, 144), raising concerns regarding product consistency and regulatory compliance. Given the FDA’s recommendation that the VCN should remain <5 copies/genome in infusion products (8), potency assays for transposon-based CAR T-cells must account for VCN thresholds to ensure safety and efficacy. For CRISPR-mediated CAR knock-in, off-target genome edits remain a significant concern, potentially affecting T-cell function and stability. Therefore, potency assays for these CAR T-cells should include whole-genome sequencing (WGS) (131, 145, 146) or GUIDE-seq (147, 148) to accurately identify and characterize these off-target sites.
3.2 In-vivo manufactured CAR T-cells
In vivo CAR T-cell generation eliminates the need for ex vivo expansion, shifting potency assessment from traditional pre-infusion characterization to real-time in vivo monitoring (149–151). In this instance, the viral vector itself is the primary product, making vector characterization an essential component of potency assay assessment. Conventional potency assays, such as those measuring transduction efficiency, cytotoxicity, and cytokine secretion in vitro, are no longer applicable. Future efforts in vitro potency analysis should focus on linking the characteristics of viral vectors and their functional outputs.
3.3 Ultra-rapid manufactured CAR T-cells
Ultra-rapid CAR T-cell manufacturing (3 days or less) (138, 152–154) significantly shortens the ex vivo expansion phase, introducing unique challenges for potency assessment. With some protocols completing the process in as little as 24 hours, the limited cell yield poses a challenge for conducting potency assays. Additionally, it remains uncertain whether these cells achieve sufficient CAR expression and vector copy number to be reliably detected by flow cytometry and ddPCR. Moreover, the shortened manufacturing time may result in a higher proportion of less-differentiated and less cytotoxic T-cells (153, 155), potentially biasing cytotoxicity potency assays for these CAR T-cells.
4 Summary
Ensuring the quality, consistency, and therapeutic efficacy of CAR T-cell products requires robust potency assays. Traditional potency assessments have been well-established for conventional CAR T-cell products. These assays focus on key parameters such as transduction efficiency, cytokine secretion, cytotoxicity. However, as CAR T-cell therapies continue to evolve, the emergence of new characteristics and manufacturing platforms necessitates a reassessment of current potency assays to ensure they remain accurate and relevant.
A critical aspect of potency assessment is the ability to accurately measure CAR T-cell functionality. While current in vitro assays provide valuable insights into cytotoxic activity and cytokine production, they may not fully capture the breadth of relevant cellular profiles. Additional profiles, such as vector integration events, T-cell differentiation state, and metabolic profiles, should also be considered when evaluating CAR T-cell potency. Advanced analytical techniques, including single-cell transcriptomics, high-dimensional flow cytometry, and metabolic analysis, offer more precise assessments of CAR T-cell function. These approaches help elucidate the complex interplay between CAR T-cell phenotype, functionality, and clinical outcomes, forming the foundation for developing robust potency assays.
The emergence of unconventional CAR T-cell manufacturing strategies, including non-viral gene delivery, in vivo CAR T-cell generation, and ultra-rapid manufacturing protocols, introduces new considerations for potency assessments. Non-viral CAR T-cells, while eliminating the need for viral vectors, exhibit greater variability in vector copy number and transgene integration, requiring refined potency assays to ensure product safety and efficacy. In vivo generated CAR T-cells shift potency evaluation from pre-infusion characterization to real-time in vivo monitoring, requiring novel biomarkers and functional assays to track their expansion and persistence post infusion. Ultra-rapid manufacturing, which significantly shortens ex vivo expansion time, poses challenges in achieving sufficient CAR expression and cell yield for traditional potency assays, necessitating innovative assay adaptations. Additionally, the development and usage of “off-the-shelf” allogeneic CAR T-cell products (129, 156, 157) introduces unique challenges related to donor suitability and ethical oversight. The application of genomic assays in this context may uncover clinically significant genomic lesions in donor cells, raising concerns about how to manage such findings in a way that ensures donor well-being while maintaining product quality. These evolving platforms highlight the need for a flexible and ethically informed potency assessment framework that can adapt to the changing landscape of CAR T-cell therapy.
Furthermore, the regulatory landscape for CAR T-cell potency testing continues to evolve. Regulatory agencies emphasize the importance of comprehensive characterization to ensure safety and efficacy while allowing for flexibility in adapting potency assays to emerging technologies. Establishing standardized potency criteria for diverse profiles and various CAR T-cell platforms will be essential for streamlining clinical translation and regulatory approval processes.
In addition to assessing the final CAR T-cell products, some omics methods can also be applied to the starting leukapheresis material (158). This upstream application is particularly valuable given the growing recognition that the functional fitness of the starting T-cell population strongly influences the quality and potency of the final CAR T-cell product. For example, profiling metabolic or protein expression signatures in pre-manufacture T cells may help predict manufacturing outcomes or therapeutic efficacy. Leveraging such assays early in the process could enable better donor or patient stratification, identification of optimal manufacturing candidates, and potentially guide pre-conditioning strategies to enhance T-cell fitness. Incorporating these omics approaches at the leukapheresis stage may therefore offer significant advantages for improving both the consistency and clinical performance of CAR T-cell therapies.
In conclusion, the rapid advancements in CAR T-cell therapy demand continuous refinement of potency assays to align with newly discovered characteristics and evolving manufacturing technologies. A multi-faceted approach that integrates traditional functional assays with cutting-edge analytical techniques will be crucial for accurately assessing CAR T-cell potency. By addressing these challenges, researchers and manufacturers can enhance the development of next-generation CAR T-cell therapies, ultimately improving patient outcomes in hematologic malignancies and solid tumors.
Author contributions
LS: Visualization, Conceptualization, Writing – original draft, Writing – review & editing. YZ: Visualization, Writing – review & editing. RS: Writing – review & editing, Supervision, Conceptualization. DS: Resources, Funding acquisition, Conceptualization, Supervision, Writing – review & editing. PJ: Supervision, Writing – review & editing, Writing – original draft, Conceptualization, Resources.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Intramural Research Program, Clinical Center, NIH
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
CAR T-cells: Chimeric Antigen Receptor T-cells
MoA: Mechanism of Action
FDA: Food and Drug Administration
NCI: National Cancer Institute
VCN: Vector Copy Number
ddPCR: Droplet Digital PCR
QC: Quality Control
CLL: Chronic Lymphoblastic Leukemia
ALL: Acute lymphoblastic Leukemia
LBCL: Large B-Cell Lymphoma
CR: Complete Response
PR: Partial Response
SD: Stable Disease
PD: Progressive Disease
EFS: Event-Free Survival
OS: Overall Survival
RNA-seq: RNA Sequencing
TCR-seq: T-cell Receptor Sequencing
scRNA-seq: Single-cell RNA-Sequencing
CITE-seq: Cellular Indexing of Transcriptomes and Epitopes Sequencing
ATAC-seq: Assays for Transposase-Accessible Chromatin with Sequencing
ChIP-seq: Chromatin Immunoprecipitation Sequencing
DNase-seq: DNase I Hypersensitive Sites Sequencing
CUT&Run-seq: Cleavage Under Targets and Release Using Nuclease Sequencing
Tn: Naïve T cells
Teff: Effector T cells
Tscm: Stem-like Memory T Cells
Tcm: Central Memory T Cells
Tem: Effector Memory T Cells
CyTOF: Cytometry by Time of Flight
MS: Mass Spectrometry
MC: Mass Cytometry
OXPHOS: Oxidative Phosphorylation
FAO: Fatty Acid Oxidation
SRC: Spare Respiratory Capacity
References
1. Wang N, Hu X, Cao W, Li C, Xiao Y, Cao Y, et al. Efficacy and safety of CAR19/22 T-cell cocktail therapy in patients with refractory/relapsed B-cell Malignancies. Blood. (2020) 135:17–27. doi: 10.1182/blood.2019000017
2. Lee DW, Kochenderfer JN, Stetler-Stevenson M, Cui YK, Delbrook C, Feldman SA, et al. T cells expressing CD19 chimeric antigen receptors for acute lymphoblastic leukaemia in children and young adults: a phase 1 dose-escalation trial. Lancet. (2015) 385:517–28. doi: 10.1016/S0140-6736(14)61403-3
3. Raje N, Berdeja J, Lin Y, Siegel D, Jagannath S, Madduri D, et al. Anti-BCMA CAR T-cell therapy bb2121 in relapsed or refractory multiple myeloma. New Engl J Med. (2019) 380:1726–37. doi: 10.1056/NEJMoa1817226
4. Shah NN, Highfill SL, Shalabi H, Yates B, Jin J, Wolters PL, et al. CD4/CD8 T-cell selection affects chimeric antigen receptor (CAR) T-cell potency and toxicity: updated results from a phase I anti-CD22 CAR T-cell trial. J Clin Oncol. (2020) 38:1938–50. doi: 10.1200/JCO.19.03279
5. Lichtenstein DA, Schischlik F, Shao L, Steinberg SM, Yates B, Wang H-W, et al. Characterization of HLH-like manifestations as a CRS variant in patients receiving CD22 CAR T cells. Blood. (2021) 138:2469–84. doi: 10.1182/blood.2021011898
6. Müller F, Taubmann J, Bucci L, Wilhelm A, Bergmann C, Völkl S, et al. CD19 CAR T-cell therapy in autoimmune disease - A case series with follow-up. N Engl J Med. (2024) 390:687–700. doi: 10.1056/NEJMoa2308917
7. English EP, Swingler RN, Patwa S, Tosun M, Howard JF Jr., Miljković MD, et al. Engineering CAR-T therapies for autoimmune disease and beyond. Sci Trans Med. (2024) 16 :eado2084. doi: 10.1126/scitranslmed.ado2084
8. FDA: considerations for the development of chimeric antigen receptor (CAR) T cell products. Rockville, MD: Dockets Management Food and Drug Administration (2024).
10. Larson SM, Walthers CM, Ji B, Ghafouri SN, Naparstek J, Trent J, et al. CD19/CD20 bispecific chimeric antigen receptor (CAR) in naive/memory T cells for the treatment of relapsed or refractory non-hodgkin lymphoma. Cancer Discovery. (2023) 13:580–97. doi: 10.1158/2159-8290.CD-22-0964
11. Yan Z, Sheng L, Wang L, Wu W, Zhang Y, Shen R, et al. A phase 1 study of LY007, a novel anti-CD20 CAR-T cell therapy in patients with relapsed or refractory B-cell non-Hodgkin lymphoma. J Clin Oncol. (2024) 42:7027–7. doi: 10.1200/JCO.2024.42.16_suppl.7027
12. Li C, Xu J, Luo W, Liao D, Xie W, Wei Q, et al. Bispecific CS1-BCMA CAR-T cells are clinically active in relapsed or refractory multiple myeloma. Leukemia. (2024) 38:149–59. doi: 10.1038/s41375-023-02065-x
13. Mathur R, Zhang Z, He J, Galetto R, Gouble A, Chion-Sotinel I, et al. Universal SLAMF7-specific CAR T-cells as treatment for multiple myeloma. Blood. (2017) 130:502. doi: 10.1182/blood.V130.Suppl_1.502.502
14. Steffin D, Ghatwai N, Montalbano A, Rathi P, Courtney AN, Arnett AB, et al. Interleukin-15-armoured GPC3 CAR T cells for patients with solid cancers. Nature. (2025) 637:940–6. doi: 10.1038/s41586-024-08261-8
15. Korell F, Berger TR, Maus MV. Understanding CAR T cell-tumor interactions: Paving the way for successful clinical outcomes. Med. (2022) 3:538–64. doi: 10.1016/j.medj.2022.05.001
16. Maude SL, Frey N, Shaw PA, Aplenc R, Barrett DM, Bunin NJ, et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N Engl J Med. (2014) 371:1507–17. doi: 10.1056/NEJMoa1407222
17. Lionel AC, Neelapu SS. CAR T-cell expansion: harmful or helpful? Blood Adv. (2024) 8:3311–3. doi: 10.1182/bloodadvances.2024013146
18. Murphy LA, Marians RC, Miller K, Brenton MD, Mallo RLV, Kohler ME, et al. Digital polymerase chain reaction strategies for accurate and precise detection of vector copy number in chimeric antigen receptor T-cell products. Cytotherapy. (2023) 25:94–102. doi: 10.1016/j.jcyt.2022.09.004
19. Lu A, Liu H, Shi R, Cai Y, Ma J, Shao L, et al. Application of droplet digital PCR for the detection of vector copy number in clinical CAR/TCR T cell products. J Trans Med. (2020) 18:191. doi: 10.1186/s12967-020-02358-0
20. FDA. FDA investigating serious risk of T-cell Malignancy following BCMA-directed or CD19-directed autologous chimeric antigen receptor (CAR) T cell immunotherapies. (2023).
21. Shao L, Shi R, Zhao Y, Liu H, Lu A, Ma J, et al. Genome-wide profiling of retroviral DNA integration and its effect on clinical pre-infusion CAR T-cell products. J Trans Med. (2022) 20:514. doi: 10.1186/s12967-022-03729-5
22. Rade M, Grieb N, Weiss R, Sia J, Fischer L, Born P, et al. Single-cell multiomic dissection of response and resistance to chimeric antigen receptor T cells against BCMA in relapsed multiple myeloma. Nat Cancer. (2024) 5:1318–33. doi: 10.1038/s43018-024-00763-8
23. Ledergor G, Fan Z, Wu K, McCarthy E, Hyrenius-Wittsten A, Starzinski A, et al. CD4+ CAR T-cell exhaustion associated with early relapse of multiple myeloma after BCMA CAR T-cell therapy. Blood Adv. (2024) 8:3562–75. doi: 10.1182/bloodadvances.2023012416
24. Wilson TL, Kim H, Chou C-H, Langfitt D, Mettelman RC, Minervina AA, et al. Common trajectories of highly effective CD19-specific CAR T cells identified by endogenous T-cell receptor lineages. Cancer Discovery. (2022) 12:2098–119. doi: 10.1158/2159-8290.CD-21-1508
25. Ikegawa S, Sperling AS, Ansuinelli M, Nikiforow S, Quinn D, Bu D, et al. T-charge™ Manufacturing of the durcabtagene autoleucel (PHE885), promotes expansion and persistence of CAR-T cells with high TCR repertoire diversity. Blood. (2023) 142:3469–9. doi: 10.1182/blood-2023-177721
26. Elsallab M, Ellithi M, Lunning MA, D’Angelo C, Ma J, Perales M-A, et al. Second primary Malignancies after commercial CAR T-cell therapy: analysis of the FDA Adverse Events Reporting System. Blood. (2024) 143:2099–105. doi: 10.1182/blood.2024024166
27. Ghilardi G, Fraietta JA, Gerson JN, Van Deerlin VM, Morrissette JJD, Caponetti GC, et al. T cell lymphoma and secondary primary Malignancy risk after commercial CAR T cell therapy. Nat Med. (2024) 30:984–9. doi: 10.1038/s41591-024-02826-w
28. Zhao WH, Wang BY, Chen LJ, Fu WJ, Xu J, Liu J, et al. Four-year follow-up of LCAR-B38M in relapsed or refractory multiple myeloma: a phase 1, single-arm, open-label, multicenter study in China (LEGEND-2). J Hematol Oncol. (2022) 15:86. doi: 10.1186/s13045-022-01301-8
29. Cordeiro A, Bezerra ED, Hirayama AV, Hill JA, Wu QV, Voutsinas J, et al. Late events after treatment with CD19-targeted chimeric antigen receptor modified T cells. Biol Blood Marrow Transplant. (2020) 26:26–33. doi: 10.1016/j.bbmt.2019.08.003
30. Chong EA, Ruella M, Schuster SJ. Five-year outcomes for refractory B-cell lymphomas with CAR T-cell therapy. N Engl J Med. (2021) 384:673–4. doi: 10.1056/NEJMc2030164
31. Jadlowsky JK, Hexner EO, Marshall A, Grupp SA, Frey NV, Riley JL, et al. Long-term safety of lentiviral or gammaretroviral gene-modified T cell therapies. Nat Med. (2025) 31:1134–1144. doi: 10.1038/s41591-024-03478-6
32. Fraietta JA, Nobles CL, Sammons MA, Lundh S, Carty SA, Reich TJ, et al. Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature. (2018) 558:307–12. doi: 10.1038/s41586-018-0178-z
33. Shah NN, Qin H, Yates B, Su L, Shalabi H, Raffeld M, et al. Clonal expansion of CAR T cells harboring lentivector integration in the CBL gene following anti-CD22 CAR T-cell therapy. Blood Adv. (2019) 3:2317–22. doi: 10.1182/bloodadvances.2019000219
34. Biasco L, Izotova N, Rivat C, Ghorashian S, Richardson R, Guvenel A, et al. Clonal expansion of T memory stem cells determines early anti-leukemic responses and long-term CAR T cell persistence in patients. Nat Cancer. (2021) 2:629–42. doi: 10.1038/s43018-021-00207-7
35. Nobles CL, Sherrill-Mix S, Everett JK, Reddy S, Fraietta JA, Porter DL, et al. CD19-targeting CAR T cell immunotherapy outcomes correlate with genomic modification by vector integration. J Clin Invest. (2020) 130:673–85. doi: 10.1172/JCI130144
36. Sherman E, Nobles C, Berry CC, Six E, Wu Y, Dryga A, et al. INSPIIRED: A pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol Ther - Methods Clin Dev. (2017) 4:39–49. doi: 10.1016/j.omtm.2016.11.002
37. Timms RT, Tchasovnikarova IA, Lehner PJ. Differential viral accessibility (DIVA) identifies alterations in chromatin architecture through large-scale mapping of lentiviral integration sites. Nat Protoc. (2019) 14:153–70. doi: 10.1038/s41596-018-0087-5
38. Wells DW, Guo S, Shao W, Bale MJ, Coffin JM, Hughes SH, et al. An analytical pipeline for identifying and mapping the integration sites of HIV and other retroviruses. BMC Genomics. (2020) 21:216. doi: 10.1186/s12864-020-6647-4
39. Wang W, Fasolino M, Cattau B, Goldman N, Kong W, Frederick MA, et al. Joint profiling of chromatin accessibility and CAR-T integration site analysis at population and single-cell levels. Proc Natl Acad Sci U.S.A. (2020) 117:5442–52. doi: 10.1073/pnas.1919259117
40. Mikkilineni L, Natrakul DA, Lam N, Manasanch EE, Mann J, Weissler KA, et al. Rapid anti-myeloma activity by T cells expressing an anti-BCMA CAR with a human heavy-chain-only antigen-binding domain. Mol Ther. (2024) 32:503–26. doi: 10.1016/j.ymthe.2023.12.018
41. Perica K, Jain N, Scordo M, Patel R, Eren OC, Patel U, et al. CD4+ T-cell lymphoma harboring a chimeric antigen receptor integration in TP53. New Engl J Med. (2025) 392:577–83. doi: 10.1056/NEJMoa2411507
42. Frank ML, Lu K, Erdogan C, Han Y, Hu J, Wang T, et al. T-cell receptor repertoire sequencing in the era of cancer immunotherapy. Clin Cancer Res. (2023) 29:994–1008. doi: 10.1158/1078-0432.CCR-22-2469
43. Cascone T, William WN, Weissferdt A, Leung CH, Lin HY, Pataer A, et al. Neoadjuvant nivolumab or nivolumab plus ipilimumab in operable non-small cell lung cancer: the phase 2 randomized NEOSTAR trial. Nat Med. (2021) 27:504–14. doi: 10.1038/s41591-020-01224-2
44. Abed A, Beasley AB, Reid AL, Law N, Calapre L, Millward M, et al. Circulating pre-treatment T-cell receptor repertoire as a predictive biomarker in advanced or metastatic non-small-cell lung cancer patients treated with pembrolizumab alone or in combination with chemotherapy. ESMO Open. (2023) 8:102066. doi: 10.1016/j.esmoop.2023.102066
45. Han J, Duan J, Bai H, Wang Y, Wan R, Wang X, et al. TCR repertoire diversity of peripheral PD-1+CD8+ T cells predicts clinical outcomes after immunotherapy in patients with non–small cell lung cancer. Cancer Immunol Res. (2020) 8:146–54. doi: 10.1158/2326-6066.CIR-19-0398
46. Sheih A, Voillet V, Hanafi L-A, DeBerg HA, Yajima M, Hawkins R, et al. Clonal kinetics and single-cell transcriptional profiling of CAR-T cells in patients undergoing CD19 CAR-T immunotherapy. Nat Commun. (2020) 11:219. doi: 10.1038/s41467-019-13880-1
47. Melenhorst JJ, Chen GM, Wang M, Porter DL, Chen C, Collins MA, et al. Decade-long leukaemia remissions with persistence of CD4+ CAR T cells. Nature. (2022) 602:503–9. doi: 10.1038/s41586-021-04390-6
48. Abbas HA, Hao D, Tomczak K, Barrodia P, Im JS, Reville PK, et al. Single cell T cell landscape and T cell receptor repertoire profiling of AML in context of PD-1 blockade therapy. Nat Commun. (2021) 12:6071. doi: 10.1038/s41467-021-26282-z
49. Gao S, Wu Z, Arnold B, Diamond C, Batchu S, Giudice V, et al. Single-cell RNA sequencing coupled to TCR profiling of large granular lymphocyte leukemia T cells. Nat Commun. (2022) 13:1982. doi: 10.1038/s41467-022-29175-x
50. Deng Q, Han G, Puebla-Osorio N, Ma MCJ, Strati P, Chasen B, et al. Characteristics of anti-CD19 CAR T cell infusion products associated with efficacy and toxicity in patients with large B cell lymphomas. Nat Med. (2020) 26:1878–87. doi: 10.1038/s41591-020-1061-7
51. Deniger DC, Switzer K, Mi T, Maiti S, Hurton L, Singh H, et al. Bispecific T-cells expressing polyclonal repertoire of endogenous γδ T-cell receptors and introduced CD19-specific chimeric antigen receptor. Mol Ther. (2013) 21:638–47. doi: 10.1038/mt.2012.267
52. Capsomidis A, Benthall G, Van Acker HH, Fisher J, Kramer AM, Abeln Z, et al. Chimeric antigen receptor-engineered human gamma delta T cells: enhanced cytotoxicity with retention of cross presentation. Mol Ther. (2018) 26:354–65. doi: 10.1016/j.ymthe.2017.12.001
53. Song HW, Benzaoui M, Dwivedi A, Underwood S, Shao L, Achar S, et al. Manufacture of CD22 CAR T cells following positive versus negative selection results in distinct cytokine secretion profiles and γδ T cell output. Mol Ther Methods Clin Dev. (2024) 32. doi: 10.1016/j.omtm.2023.101171
54. Chen GM, Chen C, Das RK, Gao P, Chen C-H, Bandyopadhyay S, et al. Integrative bulk and single-cell profiling of premanufacture T-cell populations reveals factors mediating long-term persistence of CAR T-cell therapy. Cancer Discovery. (2021) 11:2186–99. doi: 10.1158/2159-8290.CD-20-1677
55. Wang Y, Tong C, Lu Y, Wu Z, Guo Y, Liu Y, et al. Characteristics of premanufacture CD8+ T cells determine CAR-T efficacy in patients with diffuse large B-cell lymphoma. Signal Transduction Targeted Ther. (2023) 8:409. doi: 10.1038/s41392-023-01659-2
56. Zhaoyun LC, Fu R. CAR-tscm cells as an effective immune treatment to multiple myeloma patients. Blood. (2024) 144:6852–2. doi: 10.1182/blood-2024-208878
57. Bulliard Y, Andersson BS, Baysal MA, Damiano J, Tsimberidou AM. Reprogramming T cell differentiation and exhaustion in CAR-T cell therapy. J Hematol Oncol. (2023) 16:108. doi: 10.1186/s13045-023-01504-7
58. Tantalo DGM, Oliver AJ, von Scheidt B, Harrison AJ, Mueller SN, Kershaw MH, et al. Understanding T cell phenotype for the design of effective chimeric antigen receptor T cell therapies. J ImmunoTherapy Cancer. (2021) 9:e002555. doi: 10.1136/jitc-2021-002555
59. Henning AN, Roychoudhuri R, Restifo NP. Epigenetic control of CD8+ T cell differentiation. Nat Rev Immunol. (2018) 18:340–56. doi: 10.1038/nri.2017.146
60. Fiorenza S, Cao Y, Zheng Y, Purushe J, Bock T, Kimble EL, et al. Prediction of therapeutic potential of CD19 CAR-T cells for LBCL by histone mark analyses of core epigenetic programming. Blood. (2024) 144:2028–8. doi: 10.1182/blood-2024-200589
61. Fiorenza S, Zheng Y, Purushe J, Bock TJ, Sarthy J, Janssens DH, et al. Histone marks identify novel transcription factors that parse CAR-T subset-of-origin, clinical potential and expansion. Nat Commun. (2024) 15:8309. doi: 10.1038/s41467-024-52503-2
62. Garcia-Prieto CA, Villanueva L, Bueno-Costa A, Davalos V, González-Navarro EA, Juan M, et al. Epigenetic profiling and response to CD19 chimeric antigen receptor T-cell therapy in B-cell Malignancies. J Natl Cancer Inst. (2022) 114:436–45. doi: 10.1093/jnci/djab194
63. Zebley CC, Brown C, Mi T, Fan Y, Alli S, Boi S, et al. CD19-CAR T cells undergo exhaustion DNA methylation programming in patients with acute lymphoblastic leukemia. Cell Rep. (2021) 37:110079. doi: 10.1016/j.celrep.2021.110079
64. Wang Y, Tong C, Dai H, Wu Z, Han X, Guo Y, et al. Low-dose decitabine priming endows CAR T cells with enhanced and persistent antitumour potential via epigenetic reprogramming. Nat Commun. (2021) 12:409. doi: 10.1038/s41467-020-20696-x
65. Prinzing B, Zebley CC, Petersen CT, Fan Y, Anido AA, Yi Z, et al. Deleting DNMT3A in CAR T cells prevents exhaustion and enhances antitumor activity. Sci Trans Med. (2021) 13:eabh0272. doi: 10.1126/scitranslmed.abh0272
66. Salz L, Seitz A, Schäfer D, Franzen J, Holzer T, Garcia-Prieto CA, et al. Culture expansion of CAR T cells results in aberrant DNA methylation that is associated with adverse clinical outcome. Leukemia. (2023) 37:1868–78. doi: 10.1038/s41375-023-01966-1
67. Jain N, Zhao Z, Koche RP, Antelope C, Gozlan Y, Montalbano A, et al. Disruption of SUV39H1-mediated H3K9 methylation sustains CAR T-cell function. Cancer Discovery. (2024) 14:142–57. doi: 10.1158/2159-8290.CD-22-1319
68. Weber EW, Parker KR, Sotillo E, Lynn RC, Anbunathan H, Lattin J, et al. Transient rest restores functionality in exhausted CAR-T cells through epigenetic remodeling. Science. (2021) 372:eaba1786. doi: 10.1126/science.aba1786
69. Gennert DG, Lynn RC, Granja JM, Weber EW, Mumbach MR, Zhao Y, et al. Dynamic chromatin regulatory landscape of human CAR T cell exhaustion. Proc Natl Acad Sci. (2021) 118:e2104758118. doi: 10.1073/pnas.2104758118
70. Jiang P, Zhang Z, Hu Y, Liang Z, Han Y, Li X, et al. Single-cell ATAC-seq maps the comprehensive and dynamic chromatin accessibility landscape of CAR-T cell dysfunction. Leukemia. (2022) 36:2656–68. doi: 10.1038/s41375-022-01676-0
71. Mamo T, Cox CA, Demorest C, Fontaine MJ, Hubel A, Kelley L, et al. Cryopreservation of mesenchymal stem/stromal cells using a DMSO-free solution is comparable to DMSO-containing cryoprotectants: results of an international multicenter PACT/BEST collaborative study. Cytotherapy. (2024) 26:1522–31. doi: 10.1016/j.jcyt.2024.07.001
72. Shao L, Boothby A, Bailey SL, Chauhan A, Boechenek M, Gernsheimer TB, et al. T-cell transcriptomic signatures in adults with primary untreated immune thrombocytopenia segregate by age, revealing distinct druggable pathways. Blood. (2024) 144:124. doi: 10.1182/blood-2024-203670
73. Peidli S, Nouailles G, Wyler E, Adler JM, Kunder S, Voß A, et al. Single-cell-resolved interspecies comparison shows a shared inflammatory axis and a dominant neutrophil-endothelial program in severe COVID-19. Cell Rep. (2024) 43. doi: 10.1016/j.celrep.2024.114328
74. Underwood S, Jin J, Shao L, Prochazkova M, Shi R, Song HW, et al. T cell activators exhibit distinct downstream effects on chimeric antigen receptor T cell phenotype and function. ImmunoHorizons. (2024) 8:404–14. doi: 10.4049/immunohorizons.2400008
75. Prochazkova M, Dreyzin A, Shao L, Garces P, Cai Y, Shi R, et al. Deciphering the importance of culture pH on CD22 CAR T-cells characteristics. J Trans Med. (2024) 22:384. doi: 10.1186/s12967-024-05197-5
76. Shao L, Shi R, Cai Y, Dreyzin A, Pelayo A, Ma J, et al. Single-cell transcriptome analysis reveals distinct characteristics of anti-cd22 car T-cell infusion products associated with efficacy and toxicity. Cytotherapy. (2024) 26:S12–3. doi: 10.1016/j.jcyt.2024.03.023
77. Song HW, Prochazkova M, Shao L, Traynor R, Underwood S, Black M, et al. CAR-T cell expansion platforms yield distinct T cell differentiation states. Cytotherapy. (2024) 26:757–68. doi: 10.1016/j.jcyt.2024.03.003
78. Anderson ND, Birch J, Accogli T, Criado I, Khabirova E, Parks C, et al. Transcriptional signatures associated with persisting CD19 CAR-T cells in children with leukemia. Nat Med. (2023) 29:1700–9. doi: 10.1038/s41591-023-02415-3
79. Bai Z, Feng B, McClory SE, de Oliveira BC, Diorio C, Gregoire C, et al. Single-cell CAR T atlas reveals type 2 function in 8-year leukaemia remission. Nature. (2024) 634:702–11. doi: 10.1038/s41586-024-07762-w
80. Bai Z, Lundh S, Kim D, Woodhouse S, Barrett DM, Myers RM, et al. Single-cell multiomics dissection of basal and antigen-specific activation states of CD19-targeted CAR T cells. J ImmunoTherapy Cancer. (2021) 9:e002328. doi: 10.1136/jitc-2020-002328
81. Goldberg L, Haas ER, Vyas V, Urak R, Forman SJ, Wang X. Single-cell analysis by mass cytometry reveals CD19 CAR T cell spatiotemporal plasticity in patients. OncoImmunology. (2022) 11:2040772. doi: 10.1080/2162402X.2022.2040772
82. Haradhvala NJ, Leick MB, Maurer K, Gohil SH, Larson RC, Yao N, et al. Distinct cellular dynamics associated with response to CAR-T therapy for refractory B cell lymphoma. Nat Med. (2022) 28:1848–59. doi: 10.1038/s41591-022-01959-0
83. Fraietta JA, Lacey SF, Orlando EJ, Pruteanu-Malinici I, Gohil M, Lundh S, et al. Determinants of response and resistance to CD19 chimeric antigen receptor (CAR) T cell therapy of chronic lymphocytic leukemia. Nat Med. (2018) 24:563–71. doi: 10.1038/s41591-018-0010-1
84. Sarén T, Ramachandran M, Gammelgård G, Lövgren T, Mirabello C, Björklund ÅK, et al. Single-cell RNA analysis reveals cell-intrinsic functions of CAR T cells correlating with response in a phase II study of lymphoma patients. Clin Cancer Res. (2023) 29:4139–52. doi: 10.1158/1078-0432.CCR-23-0178
85. Li X, Henderson J, Gordon MJ, Sheikh I, Nastoupil LJ, Westin J, et al. A single-cell atlas of CD19 chimeric antigen receptor T cells. Cancer Cell. (2023) 41:1835–7. doi: 10.1016/j.ccell.2023.08.015
86. Guerrero JA, Klysz DD, Chen Y, Malipatlolla M, Lone J, Fowler C, et al. GLUT1 overexpression in CAR-T cells induces metabolic reprogramming and enhances potency. Nat Commun. (2024) 15:8658. doi: 10.1038/s41467-024-52666-y
87. Toledano Zur R, Atar O, Barliya T, Hoogi S, Abramovich I, Gottlieb E, et al. Genetically engineering glycolysis in T cells increases their antitumor function. J ImmunoTherapy Cancer. (2024) 12:e008434. doi: 10.1136/jitc-2023-008434
88. Chen Z, Ji Z, Ngiow SF, Manne S, Cai Z, Huang AC, et al. TCF-1-centered transcriptional network drives an effector versus exhausted CD8 T cell-fate decision. Immunity. (2019) 51:840–855.e845. doi: 10.1016/j.immuni.2019.09.013
89. Shifrut E, Carnevale J, Tobin V, Roth TL, Woo JM, Bui CT, et al. Genome-wide CRISPR screens in primary human T cells reveal key regulators of immune function. Cell. (2018) 175:1958–1971.e1915. doi: 10.1016/j.cell.2018.10.024
90. Khan O, Giles JR, McDonald S, Manne S, Ngiow SF, Patel KP, et al. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. (2019) 571:211–8. doi: 10.1038/s41586-019-1325-x
91. Teng X, Shao M, Guo X, Cheng T, Qian P, Huang H. Autophagy inhibition prevents CAR-T exhaustion and terminal differentiation via TCF7 accumulation. Blood. (2023) 142:4823. doi: 10.1182/blood-2023-189441
92. Zheng W, Wei J, Zebley CC, Jones LL, Dhungana Y, Wang Y-D, et al. Regnase-1 suppresses TCF-1+ precursor exhausted T-cell formation to limit CAR–T-cell responses against ALL. Blood. (2021) 138:122–35. doi: 10.1182/blood.2020009309
93. Jung I-Y, Narayan V, McDonald S, Rech AJ, Bartoszek R, Hong G, et al. BLIMP1 and NR4A3 transcription factors reciprocally regulate antitumor CAR T cell stemness and exhaustion. Sci Trans Med. (2022) 14:eabn7336. doi: 10.1126/scitranslmed.abn7336
94. Doan AE, Mueller KP, Chen AY, Rouin GT, Chen Y, Daniel B, et al. FOXO1 is a master regulator of memory programming in CAR T cells. Nature. (2024) 629:211–8. doi: 10.1038/s41586-024-07300-8
95. Chan JD, Scheffler CM, Munoz I, Sek K, Lee JN, Huang Y-K, et al. FOXO1 enhances CAR T cell stemness, metabolic fitness and efficacy. Nature. (2024) 629:201–10. doi: 10.1038/s41586-024-07242-1
96. Seo H, Chen J, González-Avalos E, Samaniego-Castruita D, Das A, Wang YH, et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci U.S.A. (2019) 116:12410–5. doi: 10.1073/pnas.1905675116
97. Jiang Z, Chu Y-Y, Lee H-H, Hung M-C, Yang L, Lin C. Abstract 33: Revitalizing CAR-T cells for TNBC: Targeting phosphorylation of TCF7 to overcome T cell exhaustion. Cancer Res. (2024) 84:33–3. doi: 10.1158/1538-7445.AM2024-33
98. Seo H, González-Avalos E, Zhang W, Ramchandani P, Yang C, Lio C-WJ, et al. BATF and IRF4 cooperate to counter exhaustion in tumor-infiltrating CAR T cells. Nat Immunol. (2021) 22:983–95. doi: 10.1038/s41590-021-00964-8
99. Lynn RC, Weber EW, Sotillo E, Gennert D, Xu P, Good Z, et al. c-Jun overexpression in CAR T cells induces exhaustion resistance. Nature. (2019) 576:293–300. doi: 10.1038/s41586-019-1805-z
100. Zhang X, Zhang C, Qiao M, Cheng C, Tang N, Lu S, et al. Depletion of BATF in CAR-T cells enhances antitumor activity by inducing resistance against exhaustion and formation of central memory cells. Cancer Cell. (2022) 40:1407–1422.e1407. doi: 10.1016/j.ccell.2022.09.013
101. Bai Z, Woodhouse S, Zhao Z, Arya R, Govek K, Kim D, et al. Single-cell antigen-specific landscape of CAR T infusion product identifies determinants of CD19-positive relapse in patients with ALL. Sci Adv. (2022) 8:eabj2820. doi: 10.1126/sciadv.abj2820
102. Good Z, Spiegel JY, Sahaf B, Malipatlolla MB, Ehlinger ZJ, Kurra S, et al. Post-infusion CAR TReg cells identify patients resistant to CD19-CAR therapy. Nat Med. (2022) 28:1860–71. doi: 10.1038/s41591-022-01960-7
103. Bai Z, Feng B, McClory SE, Diorio C, Zhao Z, Tang L, et al. Single-cell multi-omics reveals type-2 functionality in maintaining CAR T cell longevity associated with 8-year leukemia remission. Blood. (2023) 142:352. doi: 10.1182/blood-2023-179480
104. Rezvan A, Romain G, Fathi M, Heeke D, Martinez-Paniagua M, An X, et al. Identification of a clinically efficacious CAR T cell subset in diffuse large B cell lymphoma by dynamic multidimensional single-cell profiling. Nat Cancer. (2024) 5:1010–23. doi: 10.1038/s43018-024-00768-3
105. Michelozzi IM, Sufi J, Adejumo TA, Amrolia PJ, Tape CJ, Giustacchini A. High-dimensional functional phenotyping of preclinical human CAR T cells using mass cytometry. STAR Protoc. (2022) 3:101174. doi: 10.1016/j.xpro.2022.101174
106. Corneau A, Parizot C, Cherai M, Todesco E, Blanc C, Litvinova E, et al. Mass Cytometry: a robust platform for the comprehensive immunomonitoring of CAR-T-cell therapies. Br J Haematology. (2021) 194:779–92. doi: 10.1111/bjh.v194.4
107. Michelozzi IM, Gomez-Castaneda E, Pohle RVC, Cardoso Rodriguez F, Sufi J, Puigdevall Costa P, et al. Activation priming and cytokine polyfunctionality modulate the enhanced functionality of low-affinity CD19 CAR T cells. Blood Adv. (2023) 7:1725–38. doi: 10.1182/bloodadvances.2022008490
108. Hegde M, Navai S, DeRenzo C, Joseph SK, Sanber K, Wu M, et al. Autologous HER2-specific CAR T cells after lymphodepletion for advanced sarcoma: a phase 1 trial. Nat Cancer. (2024) 5:880–94. doi: 10.1038/s43018-024-00749-6
109. Goldberg L, Haas ER, Urak R, Vyas V, Pathak KV, Garcia-Mansfield K, et al. Immunometabolic adaptation of CD19-targeted CAR T cells in the central nervous system microenvironment of patients promotes memory development. Cancer Res. (2024) 84:1048–64. doi: 10.1158/0008-5472.CAN-23-2299
110. MacMullan MA, Dunn ZS, Qu Y, Wang P, Graham NA. Phospho-proteomic analysis of CAR-T cell signaling following activation by antigen-presenting cancer cells. bioRxiv. (2022) 2022.2002.2024.481820. doi: 10.1101/2022.02.24.481820
111. Mahamed D, Awong G, Wang L, Impey G. 287 Functional characterization of CAR T cell activation, exhaustion and cytokine production with a 52-plex CyTOF assay. J ImmunoTherapy Cancer. (2024) 12:A326–6. doi: 10.1136/jitc-2024-SITC2024.0287
112. Ye L, Park JJ, Peng L, Yang Q, Chow RD, Dong MB, et al. A genome-scale gain-of-function CRISPR screen in CD8 T cells identifies proline metabolism as a means to enhance CAR-T therapy. Cell Metab. (2022) 34:595–614.e514. doi: 10.1016/j.cmet.2022.02.009
113. Fultang L, Booth S, Yogev O, Martins da Costa B, Tubb V, Panetti S, et al. Metabolic engineering against the arginine microenvironment enhances CAR-T cell proliferation and therapeutic activity. Blood. (2020) 136:1155–60. doi: 10.1182/blood.2019004500
114. Shen L, Xiao Y, Zhang C, Li S, Teng X, Cui L, et al. Metabolic reprogramming by ex vivo glutamine inhibition endows CAR-T cells with less-differentiated phenotype and persistent antitumor activity. Cancer Lett. (2022) 538:215710. doi: 10.1016/j.canlet.2022.215710
115. Hirabayashi K, Du H, Xu Y, Shou P, Zhou X, Fucá G, et al. Dual-targeting CAR-T cells with optimal co-stimulation and metabolic fitness enhance antitumor activity and prevent escape in solid tumors. Nat Cancer. (2021) 2:904–18. doi: 10.1038/s43018-021-00244-2
116. van der Windt GJ, Everts B, Chang C-H, Curtis JD, Freitas TC, Amiel E, et al. Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity. (2012) 36:68–78. doi: 10.1016/j.immuni.2011.12.007
117. MacIver NJ, Michalek RD, Rathmell JC. Metabolic regulation of T lymphocytes. Annu Rev Immunol. (2013) 31:259–83. doi: 10.1146/annurev-immunol-032712-095956
118. Kawalekar OU, O’Connor RS, Fraietta JA, Guo L, McGettigan SE, Posey AD Jr., et al. Distinct signaling of coreceptors regulates specific metabolism pathways and impacts memory development in CAR T cells. Immunity. (2016) 44:380–90. doi: 10.1016/j.immuni.2016.01.021
119. Cappabianca D, Pham D, Forsberg MH, Bugel M, Tommasi A, Lauer A, et al. Metabolic priming of GD2 TRAC-CAR T cells during manufacturing promotes memory phenotypes while enhancing persistence. Mol Ther Methods Clin Dev. (2024) 32. doi: 10.1016/j.omtm.2024.101249
120. Renauer P, Park JJ, Bai M, Acosta A, Lee W-H, Lin GH, et al. Immunogenetic metabolomics reveals key enzymes that modulate CAR T-cell metabolism and function. Cancer Immunol Res. (2023) 11:1068–84. doi: 10.1158/2326-6066.CIR-22-0565
121. Klysz DD, Fowler C, Malipatlolla M, Stuani L, Freitas KA, Chen Y, et al. Inosine induces stemness features in CAR-T cells and enhances potency. Cancer Cell. (2024) 42:266–282.e268. doi: 10.1016/j.ccell.2024.01.002
122. van Bruggen JAC, Martens AWJ, Fraietta JA, Hofland T, Tonino SH, Eldering E, et al. Chronic lymphocytic leukemia cells impair mitochondrial fitness in CD8(+) T cells and impede CAR T-cell efficacy. Blood. (2019) 134:44–58. doi: 10.1182/blood.2018885863
123. Jena B, Rushworth D, McNamara GT, Cooper LJN. Mitochondrial biomass as a measure of fitness for T cells expressing chimeric antigen receptors. Blood. (2015) 126:3242. doi: 10.1182/blood.V126.23.3242.3242
124. Kondo T, Ando M, Nagai N, Tomisato W, Srirat T, Liu B, et al. The NOTCH–FOXM1 axis plays a key role in mitochondrial biogenesis in the induction of human stem cell memory–like CAR-T cells. Cancer Res. (2020) 80:471–83. doi: 10.1158/0008-5472.CAN-19-1196
125. Ayala Ceja M, Khericha M, Harris CM, Puig-Saus C, Chen YY. CAR-T cell manufacturing: Major process parameters and next-generation strategies. J Exp Med. (2024) 221:e20230903. doi: 10.1084/jem.20230903
126. Mikhael J, Fowler J, Shah N. Chimeric antigen receptor T-cell therapies: barriers and solutions to access. JCO Oncol Pract. (2022) 18:800–7. doi: 10.1200/OP.22.00315
127. Litvinova Y, Merkur S, Allin S, Angulo-Pueyo E, Behmane D, Bernal-Delgado E, et al. Availability and financing of CAR-T cell therapies: A cross-country comparative analysis. Health Policy. (2024) 149:105153. doi: 10.1016/j.healthpol.2024.105153
128. Prommersberger S, Reiser M, Beckmann J, Danhof S, Amberger M, Quade-Lyssy P, et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther. (2021) 28:560–71. doi: 10.1038/s41434-021-00254-w
129. Tipanee J, Samara-Kuko E, Gevaert T, Chuah MK, VandenDriessche T. Universal allogeneic CAR T cells engineered with Sleeping Beauty transposons and CRISPR-CAS9 for cancer immunotherapy. Mol Ther. (2022) 30:3155–75. doi: 10.1016/j.ymthe.2022.06.006
130. Lock D, Monjezi R, Brandes C, Bates S, Lennartz S, Teppert K, et al. Automated, scaled, transposon-based production of CAR T cells. J ImmunoTherapy Cancer. (2022) 10:e005189. doi: 10.1136/jitc-2022-005189
131. Zhang J, Hu Y, Yang J, Li W, Zhang M, Wang Q, et al. Non-viral, specifically targeted CAR-T cells achieve high safety and efficacy in B-NHL. Nature. (2022) 609:369–74. doi: 10.1038/s41586-022-05140-y
132. Short L, Holt RA, Cullis PR, Evgin L. Direct in vivo CAR T cell engineering. Trends Pharmacol Sci. (2024) 45:406–18. doi: 10.1016/j.tips.2024.03.004
133. Pfeiffer A, Thalheimer FB, Hartmann S, Frank AM, Bender RR, Danisch S, et al. In vivo generation of human CD19-CAR T cells results in B-cell depletion and signs of cytokine release syndrome. EMBO Mol Med. (2018) 10:e9158. doi: 10.15252/emmm.201809158
134. Smith TT, Stephan SB, Moffett HF, McKnight LE, Ji W, Reiman D, et al. In situ programming of leukaemia-specific T cells using synthetic DNA nanocarriers. Nat Nanotechnology. (2017) 12:813–20. doi: 10.1038/nnano.2017.57
135. Parayath NN, Stephan SB, Koehne AL, Nelson PS, Stephan MT. In vitro-transcribed antigen receptor mRNA nanocarriers for transient expression in circulating T cells. vivo. Nat Commun. (2020) 11:6080. doi: 10.1038/s41467-020-19486-2
136. Rurik JG, Tombácz I, Yadegari A, Méndez Fernández PO, Shewale SV, Li L, et al. CAR T cells produced in vivo to treat cardiac injury. Science. (2022) 375:91–6. doi: 10.1126/science.abm0594
137. Hamilton JR, Chen E, Perez BS, Sandoval Espinoza CR, Kang MH, Trinidad M, et al. In vivo human T cell engineering with enveloped delivery vehicles. Nat Biotechnol. (2024) 42:1684–92. doi: 10.1038/s41587-023-02085-z
138. Yang J, He J, Zhang X, Li J, Wang Z, Zhang Y, et al. Next-day manufacture of a novel anti-CD19 CAR-T therapy for B-cell acute lymphoblastic leukemia: first-in-human clinical study. Blood Cancer J. (2022) 12:104. doi: 10.1038/s41408-022-00694-6
139. Agarwalla P, Ogunnaike EA, Ahn S, Froehlich KA, Jansson A, Ligler FS, et al. Bioinstructive implantable scaffolds for rapid in vivo manufacture and release of CAR-T cells. Nat Biotechnol. (2022) 40:1250–8. doi: 10.1038/s41587-022-01245-x
140. Balke-Want H, Keerthi V, Cadinanos-Garai A, Fowler C, Gkitsas N, Brown AK, et al. Non-viral chimeric antigen receptor (CAR) T cells going viral. Immuno-Oncology Technol. (2023) 18:100375. doi: 10.1016/j.iotech.2023.100375
141. Mueller KP, Piscopo NJ, Forsberg MH, Saraspe LA, Das A, Russell B, et al. Production and characterization of virus-free, CRISPR-CAR T cells capable of inducing solid tumor regression. J ImmunoTherapy Cancer. (2022) 10:e004446. doi: 10.1136/jitc-2021-004446
142. Kebriaei P, Singh H, Huls MH, Figliola MJ, Bassett R, Olivares S, et al. Phase I trials using Sleeping Beauty to generate CD19-specific CAR T cells. J Clin Invest. (2016) 126:3363–76. doi: 10.1172/JCI86721
143. Kaštánková I, Štach M, Žižková H, Ptáčková P, Šmilauerová K, Mucha M, et al. Enzymatically produced piggyBac transposon vectors for efficient non-viral manufacturing of CD19-specific CAR T cells. Mol Ther - Methods Clin Dev. (2021) 23:119–27. doi: 10.1016/j.omtm.2021.08.006
144. Ma J, Shao L, Fuksenko T, Liu H, Shi R, Dinh A, et al. Reference gene selection for clinical chimeric antigen receptor T-cell product vector copy number assays. Cytotherapy. (2023) 25:598–604. doi: 10.1016/j.jcyt.2023.02.010
145. Kaeuferle T, Stief TA, Canzar S, Kutlu NN, Willier S, Stenger D, et al. Genome-wide off-target analyses of CRISPR/Cas9-mediated T-cell receptor engineering in primary human T cells. Clin Trans Immunol. (2022) 11:e1372. doi: 10.1002/cti2.v11.1
146. Georgiadis C, Preece R, Nickolay L, Etuk A, Petrova A, Ladon D, et al. Long terminal repeat CRISPR-CAR-coupled “Universal” T cells mediate potent anti-leukemic effects. Mol Ther. (2018) 26:1215–27. doi: 10.1016/j.ymthe.2018.02.025
147. Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. (2015) 33:187–97. doi: 10.1038/nbt.3117
148. Malinin NL, Lee G, Lazzarotto CR, Li Y, Zheng Z, Nguyen NT, et al. Defining genome-wide CRISPR–Cas genome-editing nuclease activity with GUIDE-seq. Nat Protoc. (2021) 16:5592–615. doi: 10.1038/s41596-021-00626-x
149. Pinto E, Lione L, Compagnone M, Paccagnella M, Salvatori E, Greco M, et al. From ex vivo to in vivo chimeric antigen T cells manufacturing: new horizons for CAR T-cell based therapy. J Trans Med. (2025) 23:10. doi: 10.1186/s12967-024-06052-3
150. Nicolai CJ, Parker MH, Qin J, Tang W, Ulrich-Lewis JT, Gottschalk RJ, et al. In vivo CAR T-cell generation in nonhuman primates using lentiviral vectors displaying a multidomain fusion ligand. Blood. (2024) 144:977–87. doi: 10.1182/blood.2024024523
151. Michels KR, Sheih A, Hernandez SA, Brandes AH, Parrilla D, Irwin B, et al. Preclinical proof of concept for VivoVec, a lentiviral-based platform for in vivo CAR T-cell engineering. J ImmunoTherapy Cancer. (2023) 11:e006292. doi: 10.1136/jitc-2022-006292
152. Deng C, Caimi PF, Farooq U, Idippily N, Giraudo MF, Reese JS, et al. Phase I study results of UF-kure19, a CAR-T product manufactured in less than 1 day, in patients with relapsed/refractory non-hodgkin’s lymphoma. Blood. (2024) 144:94. doi: 10.1182/blood-2024-206596
153. Ghassemi S, Durgin JS, Nunez-Cruz S, Patel J, Leferovich J, Pinzone M, et al. Rapid manufacturing of non-activated potent CAR T cells. Nat BioMed Eng. (2022) 6:118–28. doi: 10.1038/s41551-021-00842-6
154. Zhang C, He J, Liu L, Wang J, Wang S, Liu L, et al. Novel CD19 chimeric antigen receptor T cells manufactured next-day for acute lymphoblastic leukemia. Blood Cancer J. (2022) 12:96. doi: 10.1038/s41408-022-00688-4
155. Ahmadi M, Putnam N, Dotson M, Hayoun D, Padilla J, Fatima N, et al. Accelerating CAR T cell manufacturing with an automated next-day process. Curr Res Trans Med. (2025) 73:103489. doi: 10.1016/j.retram.2024.103489
156. Shahid S, Prockop SE, Flynn GC, Mauguen A, White CO, Bieler J, et al. Allogeneic off-the-shelf CAR T-cell therapy for relapsed or refractory B-cell Malignancies. Blood Adv. (2025) 9:1644–57. doi: 10.1182/bloodadvances.2024015157
157. Li Y-R, Fang Y, Niu S, Chen Y, Lyu Z, Yang L. Managing allorejection in off-the-shelf CAR-engineered cell therapies. Mol Ther. (2024) 33. doi: 10.1016/j.ymthe.2024.11.035
Keywords: potency assay development, CAR T cell therapy, multi-omics, advances and discoveries, quality control
Citation: Shao L, Zheng Y, Somerville RP, Stroncek DF and Jin P (2025) New insights on potency assays from recent advances and discoveries in CAR T-cell therapy. Front. Immunol. 16:1597888. doi: 10.3389/fimmu.2025.1597888
Received: 21 March 2025; Accepted: 15 April 2025;
Published: 08 May 2025.
Edited by:
Brian H. Johnstone, Ossium Health, Inc., United StatesReviewed by:
Gaurav Sutrave, The University of Sydney, Darlington, AustraliaClaudia Manriquez Roman, Mayo Clinic, Rochester, United States
Copyright © 2025 Shao, Zheng, Somerville, Stroncek and Jin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: David F. Stroncek, ZHN0cm9uY2VrQGNjLm5paC5nb3Y=; Ping Jin, cGppbkBtYWlsLmNjLm5paC5nb3Y=
 Lipei Shao
Lipei Shao Yanyan Zheng
Yanyan Zheng Robert P. Somerville
Robert P. Somerville David F. Stroncek
David F. Stroncek