- 1State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, Heilongjiang, China
- 2College of Veterinary Medicine, Shanxi Agricultural University, Taigu, Shanxi, China
The rapid advancement of vaccines and immunotherapies has significantly improved public health. However, a significant translational gap remains between basic research and clinical application, largely attributed to the disconnect between in vitro studies and in vivo models. To bridge this gap, in vitro models of immune organs, including bone marrow, thymus, spleen, lymph nodes, and tonsils, have emerged as a promising solution. By integrating cutting-edge technologies such as ex vivo culture, microfluidic chips, engineered tissues, and organoid models, researchers have successfully established a new-generation in vitro immune simulation platform. This review systematically summarizes recent progress in immune organ-based in vitro models, outlines the current technological landscape and highlights the unique advantages of immune organoids within this field. Notably, we classify immune organoids into strictly and broadly defined categories based on their origin and construction methodology, while emphasizing the importance of multi-model integration. This platform provides a novel framework for advancing translational immunology research, particularly in the fields of adaptive immunity and vaccine development.
1 Introduction
The successful development of mRNA vaccines during the COVID-19 pandemic has further advanced immunology research (1). The pandemic has shown that infectious diseases remain a global threat (2, 3), with persistent infectious pathogens and new epidemic risks, which highlights the need to explore disease mechanisms for better prevention and treatment strategies (4). This phenomenon is influenced by various environmental and host factors, such as pollution and aging, among which immune cell aging is particularly significant. The impacts of these factors on human immune functions deserve in-depth investigation (5, 6).
In human cell research, two-dimensional cultured cell lines remain the primary focus. These classical cell lines are cost-effective, easy to manipulate, and compatible with various experimental techniques. However, establishing these cell lines is inefficient, and extensive genetic and phenotypic alterations occur under culture conditions (7, 8). Currently, research predominantly focuses on in vivo immune responses, including cell engineering, vaccine development, and immunotherapies (9, 10). Since most human data are confined to cells and molecules found in the blood, our understanding of immunity at the tissue level mainly comes from in vivo, ex vivo, and in vitro immune models.
Laboratory model systems are essential for elucidating disease mechanisms, with in vivo, in vitro and ex vivo models serving as the foundation of our understanding of infectious diseases (11). Ex vivo culture methods, including explants and slices, bridge the gap between in vivo animal models and in vitro cell cultures (12, 13). These tissue samples preserve the original structural and functional characteristics of organs while enhancing experimental operability. However, removing an organ from its natural environment disrupts physiological fluid flow dynamics. Microfluidic chip systems dynamically regulate the introduction of antigens, cells, and other elements (such as cytokines or growth factors) (14, 15). By introducing fluid flow and controlled microenvironments under ex vivo and in vitro conditions, microfluidic chips serve as versatile platforms for studying immune responses. Nevertheless, many chips are customized for specific functions, requiring tailored designs for different biological problems. Engineered tissues provide highly flexible platform for investigating the functions of various immune cells (14, 16). By constructing model tissues from scratch, researchers can explore specific cell-cell and cell-matrix interactions under tightly controlled conditions. However, most models rely on immortalized cell lines, lacking the diversity of primary cells. Although technologies such as ex vivo cultures, microfluidic chips, and engineered tissues exhibit certain advantages over animal models, they also have inherent limitations. Current systems fail to fully recapitulate the multicellular architecture of immune organs, thereby limiting the study of regulatory mechanisms in the adaptive immune system.
Organoids are three-dimensional (3D) multicellular self-organizing tissues that mimic the structure, function, and complexity of their corresponding internal organs (17, 18). Over the past five years, immune organoids have been developed and utilized to advance immunological research (19–22). They are primarily derived from lymphoid tissues, including the bone marrow, thymus, spleen, lymph nodes (LNs), and tonsils (21, 23, 24),. Immune organoids hold significant potential to overcome the limitations of animal models, thereby advancing human immunology research (25). For example, bone marrow organoids derived from pluripotent stem cells encompass diverse cell types and exhibit vascularized architectures that faithfully replicates the bone marrow’s physiological microenvironment (26). Similarly, tonsil organoids crafted from discarded tonsil tissue following tonsillectomy demonstrate remarkable capabilities in mimicking germinal center attributes, including somatic hypermutation (SHM), antigen-specific antibody production, affinity maturation, and class switching (22). This innovative technology represents a significant breakthrough, enabling the development of physiologically relevant adaptive immune system models and addressing certain constraints of alternative clinical methodologies.
Despite considerable advancements in immunological research, many intricate mechanisms of the immune system remain poorly understood. Additionally, the development of relevant in vitro models remains insufficient or nonexistent. This review systematically examines the critical roles of key immune organs models, including the bone marrow, thymus, spleen, LNs, and tonsils, in human adaptive immunity, integrating recent technological advances with immunological insights to demonstrate their translational applications (Table 1). Although other immune-associated tissues such as Peyer’s patches and the appendix also contribute to mucosal immunity, their corresponding models remain underdeveloped and are therefore outside the scope of this review, primarily due to limited research progress and technical challenges in replicating their specialized microenvironments.
2 Immune organs and tissues: indispensable systems for immunity homeostasis
The physiological functions of immune organs are fundamentally determined by their specialized tissue architectures and cellular ecosystems. These structural and compositional features create unique microenvironments that directly influence immune cell development, activation, and functional responses. Below we systematically examine both the physiological roles of major immune organs and the tissue-level organization enabling these functions.
2.1 Physiological functions of immune organs and tissues
As the body’s primary defense mechanism against foreign pathogens, the immune system plays an indispensable role in maintaining health. Within this intricate defense network, the adaptive immune response, primarily mediated by T and B cells, constitutes a pivotal mechanism for eliminating infections. Naive T cells become activated upon recognition and binding to their specific antigens, subsequently undergoing a series of complex yet orderly differentiation and proliferation processes that result in effector immune responses (35). In the context of antibody-mediated humoral immunity, follicular helper T cells, a specialized subset of antigen-specific T cells, facilitate the maturation of B cells through intimate interactions (36, 37), leading to the differentiation of B cells into memory B cells or plasma cells capable of secreting antibodies. Once this activation cascade is initiated, T cells and B cells traverse the circulatory system to various peripheral tissues, primed to respond swiftly to potential threats (12, 35, 38). Following the resolution of the initial threat, these activated memory cells do not dissipate; instead, they persist in infected tissues, LNs, or bone marrow, forming an immune reservoir that can be rapidly reactivated upon encountering the same or similar pathogens (39–42), thereby ensuring long-term and effective immunity against previously encountered pathogens.
2.2 Immune cell development of distinct organs
Immune cell development begins in the bone marrow, where T cell precursors migrate to the thymus for further differentiation, while B cells, natural killer cells, and myeloid cells, including macrophages and dendritic cells (DCs), complete their development in the bone marrow and then migrate to secondary lymphoid organs (SLOs), such as LNs and the spleen (12). In SLOs, immune cells are activated in response to specific pathogens or molecules and then migrate back into the bloodstream and sites of infection, cancer, or disease. These migration patterns suggest that immune cells programmed in different organs, such as spleen or LNs, can migrate throughout the host to initiate systemic and specific responses (16, 43).
In the bone marrow, hematopoietic stem cells (HSCs) develop into common precursor cells that differentiate into immune cells. Myeloid precursors give rise to APCs, including macrophages and DCs, which are responsible for detecting foreign molecules in blood and tissues (16, 44). Simultaneously, the bone marrow, as the center of proliferation and differentiation of HSCs, not only supports the entire maturation process of B cells but also produces T cell precursor cells (Figure 1A). These T cell precursors then migrate to the thymus and undergo rigorous positive selection (ensuring functional T cell receptor expression) and negative selection (clearing autoreactive T cells), ultimately forming a mature T cell repertoire with appropriate antigen specificity and affinity (12, 45, 46) (Figure 1B). These processes ensure that T cells express relevant molecules and integrate the corresponding signaling mechanisms required for immune signals from APCs. Meanwhile, through rigorous screening and regulation, they avoid inappropriate binding to autoantigens, thereby preventing abnormal responses that may lead to autoimmune diseases (47, 48). The spleen, LNs, and MALT, including the tonsils, constitute the principal sLOs where lymphocytes execute their immunological functions. These strategically distributed organs function as an integrated filtration network, systematically screening and processing the molecular and cellular constituents of extracellular fluids, including lymph, blood and interstitial fluid, thereby orchestrating targeted immune responses against potential pathogens (49, 50). The tonsils, as crucial components of MALT, play pivotal roles in orchestrating mucosal immune responses (51). Their distinctive crypt architecture, characterized by deep invaginations of the epithelial surface, creates an optimized microenvironment for efficient antigen capture, processing, and presentation, thereby facilitating the initiation of robust immune responses against respiratory and gastrointestinal pathogens (51–53). Within this specialized lymphoid structure, B lymphocytes undergo antigen-driven differentiation into IgA-secreting plasma cells, which subsequently migrate to mucosal surfaces to provide localized, antigen-specific immune protection through the production of secretory IgA (Figure 1C). Functioning as a critical filtration hub within the lymphatic circulatory system, LNs execute immune surveillance through their highly organized compartmental architecture. Afferent lymphatic vessels deliver tissue-derived antigens and professional APCs (particularly DCs) to the LNs, where T cell-mediated immune responses are initiated within the paracortical T cell zones, while follicular B cell activation and germinal center formation occur in the cortical regions. Specialized high endothelial venules within LNs facilitate lymphocyte recirculation, maintaining systemic immune cell homeostasis and ensuring rapid deployment of effector cells throughout the organism (54, 55). Complementing this network, the spleen, as the largest secondary lymphoid organ, serves as a central regulator of systemic immunity. Its distinctive microanatomical organization, comprising white pulp (supporting adaptive immunity) and red pulp (mediating innate immune functions), enables comprehensive immune surveillance. The marginal zone (MZ) of the spleen serves as the gateway for antigens and lymphocytes in the blood to enter the white pulp, where immune responses against blood-borne pathogens are initiated (55, 56) (Figure 1D). Through their distinct structural and functional properties, various organs meticulously regulate the development and functionality of immune cells, thereby enabling the host to mount effective responses against diverse pathogens and disease conditions.
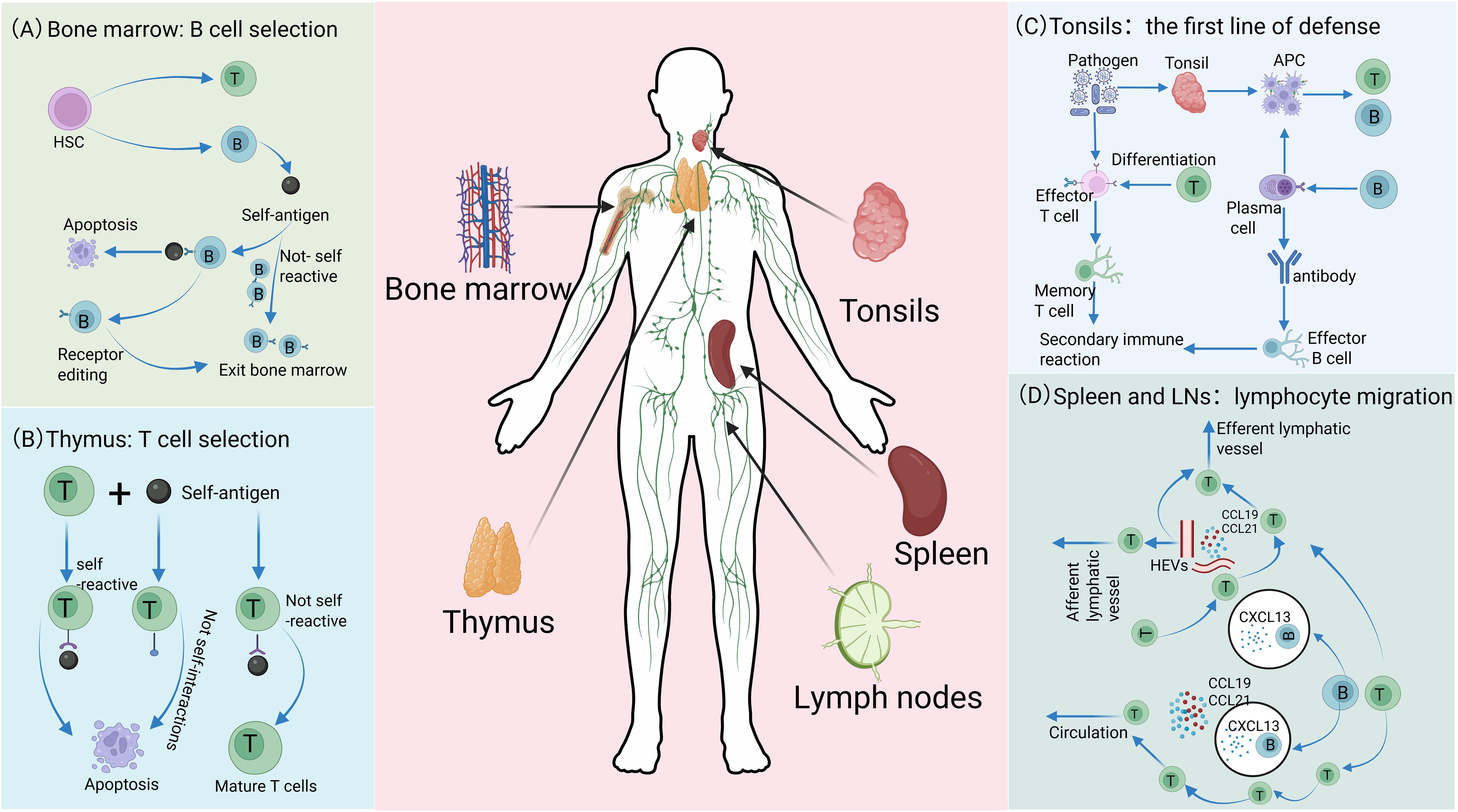
Figure 1. Distinct roles of immune organs in cellular development. (A) In the bone marrow, lymphocytes originate from HSCs. Among these, the majority of B cells matures and migrates to SLOs, while a small subset exhibiting high self-antigen affinity is eliminated through clonal deletion or receptor editing, thereby ensuring the maintenance of autoimmune tolerance. (B) After T cell precursors are generated in the bone marrow, they migrate to the thymus for maturation and selection. T cells with strong self-antigen binding or ineffective interaction with self-molecules are eliminated, whereas non-self-reactive precursors successfully mature and differentiate into functional T cells. (C) The tonsil serves as the first line of defense, capturing pathogens. Its APCs process antigens and activate T and B cells, promoting T cell differentiation into effector and memory T cells, and B cell differentiation into plasma and memory B cells. Plasma cells secrete antibodies, effector T cells eliminate pathogens, and memory cells establish immune memory to enable robust secondary responses. (D) Lymphocytes enter SLOs through distinct migration mechanisms and achieve region-specific localization. In the spleen, lymphocytes access the white pulp via MZ, with migration precisely regulated by chemokine receptor signaling: B cells migrate to follicles guided by CXCL13, while T cells are recruited to T cell zones in response to CCL19 and CCL21. Although the mechanisms of lymphocyte egress from the white pulp remain incompletely understood, this process is critical for maintaining systemic immune homeostasis. In LNs, lymphocytes primarily enter through HEVs rather than afferent lymphatic vessels. Guided by CXCL13, CCL19, and CCL21, they home to B cell follicles or T cell zones, respectively, and eventually exit via efferent lymphatics to re-enter systemic circulation.
Many critical aspects of the lymphoid microenvironment, including dynamic cell migration and the regulatory mechanisms of stromal cells in T/B cell interactions, remain challenging to accurately model using conventional approaches. While humanized mouse models (Figure 2A) and 3D tumor organoids incorporating immune components provide partial research platforms, they fail to fully recapitulate the complex spatial architecture and functional characteristics of LOs. Recently developed ex vivo culture systems (Figure 2B), microfluidic chip technologies (Figure 2C), and engineered tissues (Figure 2D) have demonstrated advantages in simulating immune responses. However, these systems still cannot completely preserve the native multicellular composition of immune organs, precisely reconstruct tissue spatial organization, or faithfully reproduce the sophisticated regulatory networks of human adaptive immunity. In this context, immune organoid technology has emerged as a breakthrough solution. Through 3D self-organizing culture systems, it achieves functional reconstitution of stem cells or primary tissues. This advancement not only overcomes the technical limitations of existing models but also provides a revolutionary platform for studying immune cell interactions. More importantly, it opens new avenues for investigating adaptive immune mechanisms and developing translational applications.
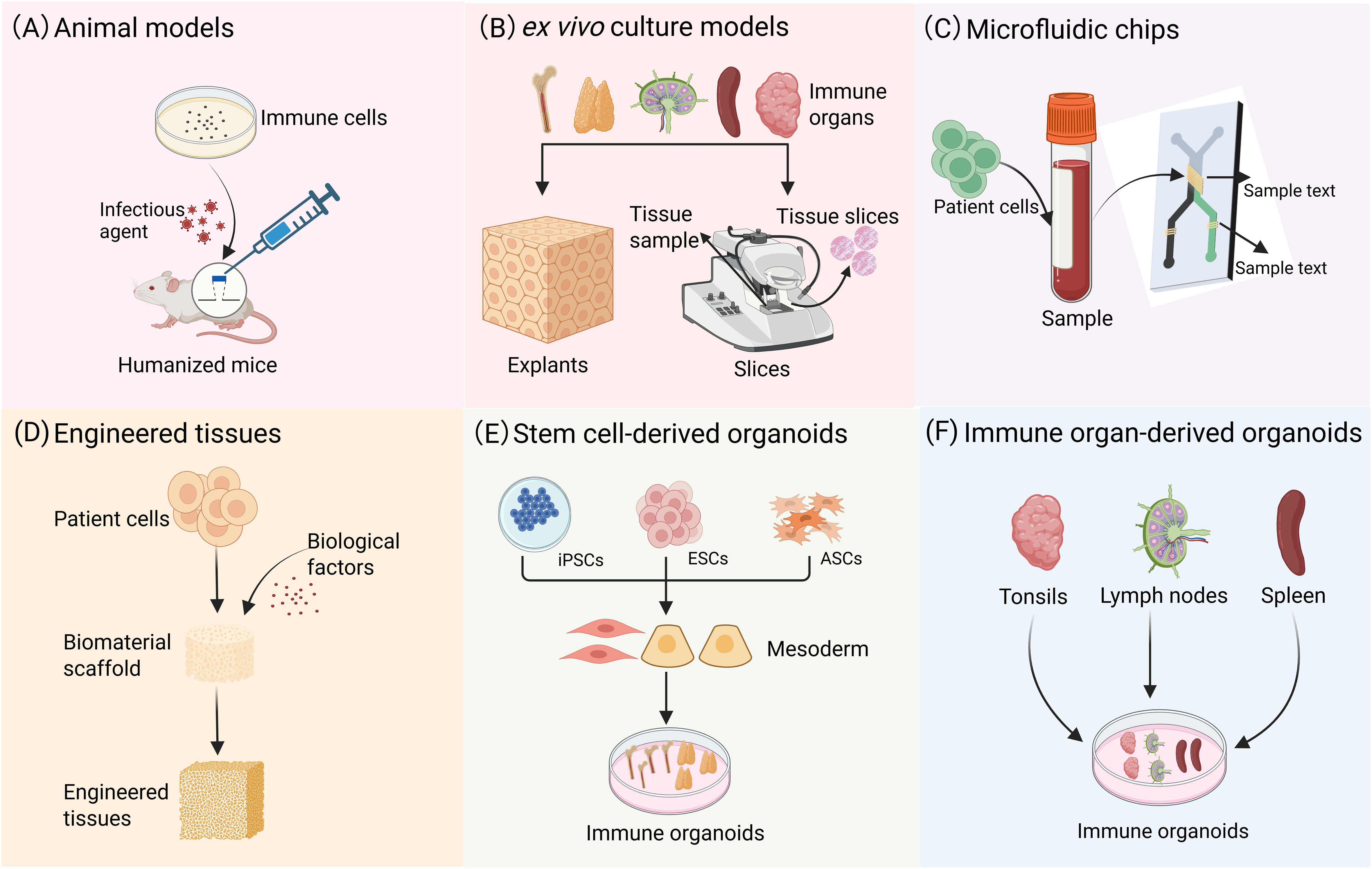
Figure 2. Modeling immune system: from animal models to organoids. (A) Animal models are capable of replicating comprehensive biological systems, including organ-organ interactions, the immune system, and the neuroendocrine system. (B) In vitro culture preserves the original cellular composition and spatial architecture of tissues, and this well-established technique is suitable for short-term studies. (C) Microfluidic chips can replicate complex microenvironments, such as blood flow and shear stress, and facilitate the simulation of interactions between different organs. (D) Engineered tissues provide a highly adaptable experimental platform for investigating the functions of diverse immune cells. (E) Tissue analogues with definite spatial structures formed through the 3D directional differentiation of ASCs or PSCs. (F) Organoid models can be derived from the direct culture of primary tissues, such as tonsils, spleen, and LNs.
3 Immune organoids: bridging the gap between traditional models and immune organs
Immune organoids can be categorized into strictly and broadly defined organoids based on their source and construction methodology (Table 2). Strictly defined organoids are spatially organized tissue analogs generated through the 3D differentiation of adult stem cells (ASCs) or pluripotent stem cells (PSCs), such as bone marrow and thymus organoids (Figure 2E). Their defining feature is stem cell-driven self-assembly and biomimetic tissue reconstruction. Broadly defined organoids generally refer to organoid models derived directly from primary tissues, such as spleen, LNs, and tonsils (Figure 2F). Notably, the formation of tonsil organoids does not rely on the differentiation of ASCs but is achieved through the spontaneous reorganization of tissue fragments. This characteristic makes them a unique model for studying tissue regeneration and the immune microenvironment. Below, we briefly outline the fundamental principles to consider when creating models for specific aspects of the immune system. We then summarize existing techniques and recent advances, with a focus on the advantages demonstrated by immune organoids.
3.1 Bone marrow organoids
As the central organ of the human hematopoietic system, the bone marrow plays a critical role in generating blood cells. Within this specialized tissue, HSCs exhibit remarkable characteristics, such as unlimited self-renewal capacity and the ability to continuously produce identical daughter cells, ensuring the ongoing hematopoietic process. Typically, HSCs reside in the G0 phase of the cell cycle, indicating a quiescent state where they are temporarily detached from active cell cycling. This quiescence endows HSCs with high differentiation potential, enabling them to rapidly activate and differentiate into various mature blood cells with specific functions—such as erythrocytes, leukocytes, and platelets—thereby maintaining the stability and normal function of the human blood system (59).
Most experimental work on bone marrow typically commences with cell suspensions acquired through bone marrow aspiration or biopsy. However, for immunohistochemical analysis, entire bone marrow samples are generally fixed (60). Although in vitro cultures of bone marrow have proven beneficial for investigating hematopoietic function, live slices and explants have not been extensively utilized for studying bone marrow immune function (61).
Current research on bone marrow-on-a-chip models primarily focuses on the development of biomimetic scaffolds and HSCs culture techniques, but there are still significant gaps in replicating the core biological functions of bone marrow—the hematopoietic differentiation process (62–65). Torizawa’s team pioneered the groundbreaking development of a biomimetic chip platform with comprehensive bone marrow microenvironment features, which innovatively integrates the cellular heterogeneity of bone marrow tissue and functional hematopoietic niches (65). However, it should be noted that their validation experiments were solely based on a murine model system, and the translatability of their findings to human bone marrow systems still requires rigorous experimental verification. To address this critical scientific challenge, the team achieved significant progress in 2020 by successfully constructing a vascularized human bone marrow biomimetic chip model (66). This model not only accurately recapitulates the dynamic features of hematopoietic processes but also effectively simulates pathological phenotypes of bone marrow dysfunction. In 2021, Nelson’s team achieved a technological breakthrough with the development of a high-throughput 96-well format bone marrow-on-a-chip system (67). The key design highlight lies in its simultaneous integration of microenvironmental features from critical hematopoietic niches, including the endosteal, central marrow, and perivascular regions. This sophisticated model exhibits not only remarkable architectural fidelity to native tissue organization, but more critically, establishes unprecedented physiological relevance at the functional level—most notably through its capacity to reliably sustain long-term in vitro expansion and maintain the self-renewal potential of primitive CD34+ HSCs.
Bone marrow engineered tissues modeling technologies have evolved from traditional static culture systems to dynamic biomimetic systems (68–70). The field primarily relies on static 3D culture systems using HSCs or mesenchymal stem cells, which have successfully validated immune cell generation. Notably, Mortera-Blanco’s team developed an innovative bone marrow biomimetic model using poly lactic-co-glycolic acid and polyurethane composite scaffold materials, achieving long-term expansion of cord blood mononuclear cells under cytokine-free conditions (69). Recent advancements have introduced bioreactor perfusion technology, enabling models to simulate physiological conditions in dynamic environments. For example, Nichol’s team successfully cultured HSCs in a rotating wall vessel bioreactor system using 3D polyacrylamide scaffolds with inverse opal crystal structures, where B lymphocyte differentiation was observed (70). These technological breakthroughs not only refine the bone marrow engineered tissues system but also provide new research platforms for investigating hematopoietic differentiation mechanisms and constructing blood disease models.
Current human ex vivo bone marrow models face significant technical limitations. A core issue is the lack of authentic sinusoidal endothelial structures, forcing many studies to rely on non-physiological human umbilical vein endothelial cells as substitutes (71). Although organ-on-a-chip technologies have made some progress in simulating certain bone marrow components, these systems still suffer from critical deficiencies. For example, there is an absence of functional stromal networks, and vascular formation is defective, leading to insufficient active hematopoiesis (66, 71–74). To overcome these technical barriers, Khan’s team pioneered the successful construction in 2022 of 3D bone marrow organoids containing mesenchymal stroma, myeloid cells, and sinusoidal vascular networks using human induced pluripotent stem cells (iPSCs) (75) (Figure 3A). This model accurately recapitulates key bone marrow components, including HSCs, myeloid cells, megakaryocytes, endothelial cells, and MSCs. Single-cell transcriptomic analysis confirmed the model’s high molecular-level similarity to native bone marrow tissue. Subsequently, Frenz-Wiessner’s team further developed more advanced in vitro models of the bone marrow microenvironment. By optimizing culture systems, they constructed complex organoids containing multilineage hematopoietic cells, functional mesenchymal stroma, and vascular networks, achieving high-fidelity simulation of key features of the bone marrow niche (26) (Figure 3A). However, current organoid models still face technical challenges, such as the absence of functional immune cells and insufficient vascular network maturation. These factors limit their ability to fully replicate the in vivo bone marrow microenvironment and represent key directions for future research breakthroughs.
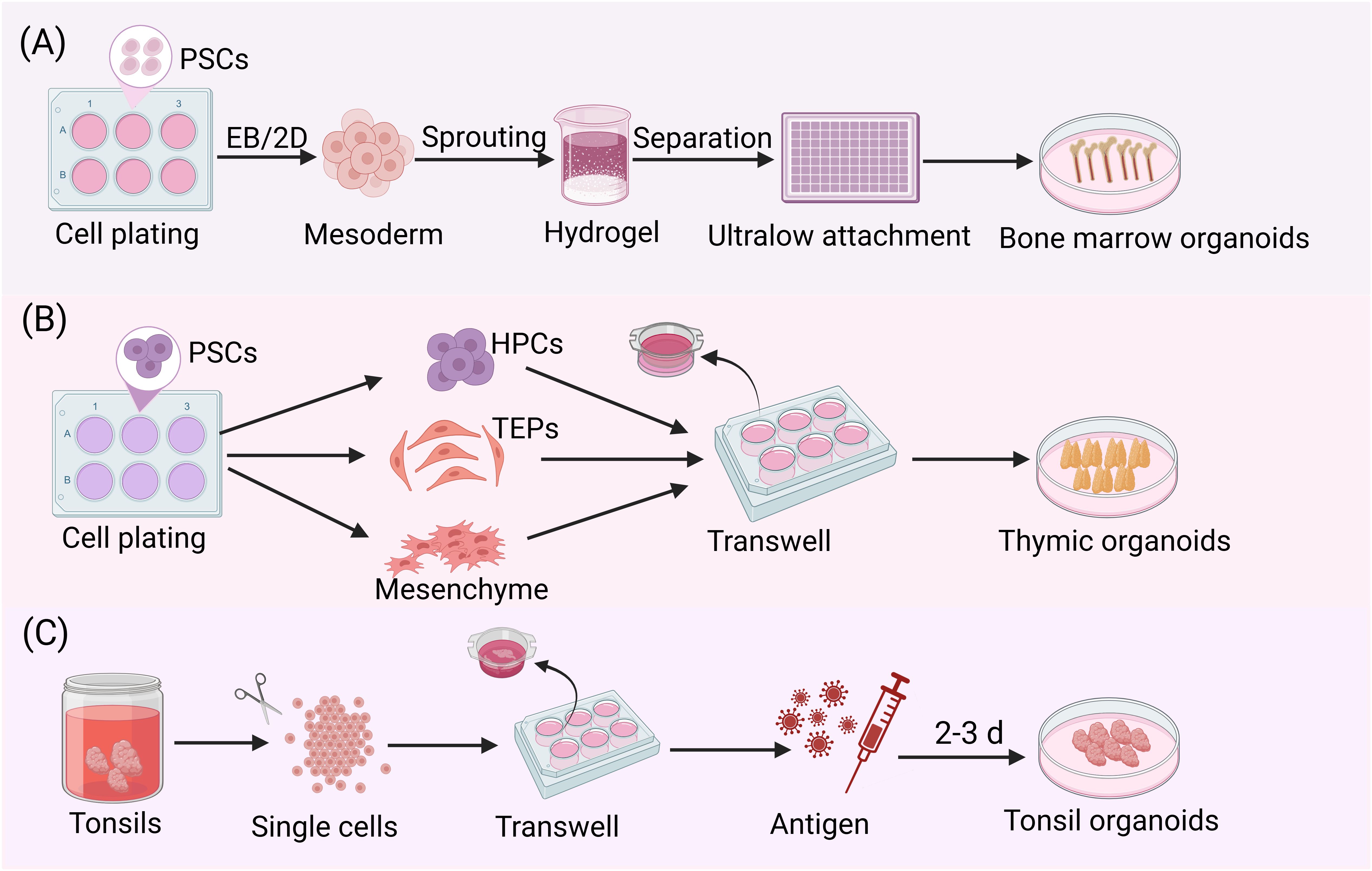
Figure 3. Stepwise protocols for culturing immune organoids. (A) After seeding iPSCs, mesodermal aggregates were formed either by direct addition of mesoderm induction medium or by first generating embryoid bodies followed by mesodermal differentiation. These aggregates were then embedded in Matrigel. Following the formation of vascular sprouts, individual sprouts were carefully isolated and cultured in 96-well ultralow attachment plate to ultimately generate bone marrow organoids. (B) hPSCs were plated and simultaneously differentiated into three critical progenitor populations: thymic epithelial progenitors (TEPs), HPCs and mesenchymal stromal cells. These cells were populations at an optimized ratio to generate stem cell-derived thymic organoids (sTOs) through 3D co-culture. Under air-liquid interface conditions, sTOs further differentiated into functionally competent TECs. (C) After mechanical dissociation of tonsil tissue into single-cell suspensions, the cells were co-seeded with the antigen of interest at high density on transwell inserts. Following several days in culture, tonsil organoids could be observed.
3.2 Thymic organoids
As a vital lymphoid organ in the human body, the thymus plays an indispensable role in the adaptive immune response and serves as a specialized site for the generation and maturation of T lymphocytes (76). It modulates positive selection, negative selection, and tolerance establishment of T cells through interactions between its cortical and medullary regions, as well as with thymic epithelial cells (TECs) (77–79). Since no microfluidic models of the thymus have been identified to date, the following sections focus on models of in vitro cultures, engineered tissues, and thymic organoids.
The in vitro thymic culture system provides an important research platform for in-depth investigation of the key regulatory mechanisms of T cell development and the formation of central tolerance. Among these systems, the fetal thymus organ culture (FTOC) serves as a classical model that has played a foundational role in elucidating the microenvironmental requirements and molecular regulatory mechanisms underlying normal T cell development (32, 80–83). In recent years, Campinoti’s team has optimized the FTOC system and combined with whole-thymus perfusion technology to successfully obtain naturally decellularized extracellular matrix (ECM), achieving precise simulation and functional reconstruction of the thymic microenvironment (32). Meanwhile, the establishment of thymic tissue slice culture technology has enabled researchers to dynamically analyze the formation process of T cell tolerance under near-physiological conditions (84–88). Ross’s team utilized this technology to demonstrate that thymic precursors recognize self-peptides bound to the major histocompatibility complex, undergo positive and negative selection, and ultimately develop into a functional T cell repertoire with both immune responsiveness and self-tolerance, while systematically elucidating the molecular characteristics of each selection stage in thymic slices (87). Notably, with the advancement of research, thymic slice technology has been extended to human thymic tissue, providing new experimental approaches for exploring the unique features of human T cell development (89).
Successful thymic bioengineering models must precisely reconstruct key features of the thymic microenvironmental to support thymocyte maturation, positive selection, and terminal differentiation into functional single-positive CD4+ or CD8+ T cells, ultimately generating a diverse and self-tolerant T cell repertoire (90). The field of thymic tissue bioengineering remains in its nascent stages, with current research primarily focused on developing in vitro T-cell generation models. In a pioneering study, Poznansky’s team implanted murine thymic tissue fragments into porous cellular scaffolds. Upon achieving 80% confluence of stromal cells, they introduced human bone marrow-derived progenitor cells, demonstrating their efficient differentiation into functional T cells within 14 days (91). However, conventional in vitro T-cell differentiation systems (e.g., the OP9-DL1 co-culture system) still exhibit significant limitations in promoting terminal T-cell maturation and positive selection. Recent breakthroughs in human thymic modeling have led to the development of novel artificial thymic organoid systems that demonstrate remarkable robustness, effectively overcoming inherent limitations of animal models. Notably, researchers have successfully achieved complete differentiation from human HSCs to mature T cells under serum-free culture conditions (92). This advancement provides a transformative technological platform for both thymic development studies and T-cell-based immunotherapy. In 2024, thymic organoid research achieved significant breakthroughs. Scientists successfully established long-term expandable 3D TECs organoids from adult mouse thymic tissue and generated thymic organoids derived the stromal compartments of LOs (93, 94). Although the T-cell output levels in these bioengineered thymic models remain relatively low, potentially limiting in-depth analysis of the composition and functional diversity of the TCR repertoire, studies have shown that, where feasible, these models still exhibit a relatively broad coverage of TCR gene segments and comparable representative distributions to those observed in natural thymus tissue or blood samples (90, 92, 95).
While humanized mouse models hold significant value for immune system research and disease modeling, their inherent limitations cannot be overlooked: the absence of a physiological human thymic microenvironment prevents human T cells developed in mice from establishing functional interaction networks with other human immune cell subsets, thereby failing to reconstruct a complete human immune response. To overcome this bottleneck, researchers have focused on reconstructing the 3D thymic microenvironment in vitro to mimic its critical physiological functions (96, 97). In recent years, through pluripotent stem cell differentiation technology, scientists have successfully established transformation systems from human embryonic stem cells to functional TECs (98, 99). These culture systems can self-organize into thymus-like structures, supporting not only in vitro T cell generation but also maintaining functional activity after transplantation into immunodeficient mice. Notably, the team led by Stephan A. Ramos optimized the differentiation protocols for iPSCs into thymic epithelial progenitor cells and, by combining hematopoietic progenitor cells (HPCs) with mesenchymal cells in co-culture, successfully constructed the first functional homologous stem cell-derived thymic organoids (sTOs) in vitro (100) (Figure 3B). This advancement not only provides a powerful tool for investigating thymic development and mechanisms of T-cell differentiation but also establishes a critical foundation for development of novel therapies targeting immune-related diseases and strategies for thymus regeneration.
3.3 Spleen organoids
The spleen, as the largest secondary lymphoid organ in the body, performs a broad spectrum of immune functions while also plays a crucial role in hematopoiesis and red blood cell clearance (56, 101). Anatomically and functionally, the spleen is divided into the red pulp and the white pulp. In rodents, the interface between these two regions is designated as the MZ, whereas in humans, this transitional area is referred to as the perifollicular zone (102, 103). The red pulp houses macrophages, which primarily function to filter blood and recycle iron from senescent red blood cells. Structurally analogous to LNs, the white pulp comprises T-cell and B-cell regions (the latter also referred to as follicles) and is capable of mounting antigen-specific immune responses to safeguard the body against diseases caused by blood-borne bacterial, viral, and fungal pathogens (103). Moreover, the spleen plays crucial roles in modulating potentially harmful immune responses to the host. Despite advances in engineered tissues and stem cell research, engineered tissue models and stem cell-derived spleen organoids remain underexplored in this field. Therefore, the following sections mainly focus on ex vivo culture and microfluidic chips.
In the field of spleen research, traditional approaches have primarily relied on autopsy and animal models, but these methods fail to accurately replicate organ function under physiological conditions. As early as the 1970s, researchers attempted to develop mouse spleen slicing techniques, but these early methods were significantly limited in maintaining tissue viability and functionality due to technological constraints (104). With technological advancements, this field has achieved critical breakthroughs. James’s team pioneered the application of precision-cutting technology to human spleen modeling (105). This technique preserves the complete ECM structure and faithfully recreates the complex cellular interaction networks within the tissue microenvironment, providing a more accurate model for studying spleen function under both physiological and pathological conditions. Building upon this foundation, recent studies have further refined an innovative precision-cutting protocol for mouse spleen. By optimizing vibratome parameters and agarose embedding techniques, researchers can now efficiently prepare structurally intact and highly viable spleen slices suitable for organotypic culture for up to 48 hours (106). This technological breakthrough not only overcomes the limitations of traditional methods in maintaining tissue viability but also establishes a physiologically relevant experimental platform for spleen research.
Efforts to model the spleen on microfluidic devices have predominantly concentrated on its principal function of filtering red blood cells rather than its immunological role. A microengineering model termed the human spleen chip, developed by Rigat-Brugarolas’s team, effectively emulates the filtering function of the human spleen (107). This multi-layer microfluidic device was meticulously designed to replicate the microcirculation and physical characteristics of the spleen, including rapid closure and gradual reopening of microcirculation, a reticular structure with elevated hematocrit levels, and intercellular spaces between endothelial cells. The future incorporation of immune function components into these models may significantly enhance our capability to investigate the outcomes and mechanisms associated with blood-borne infections. In another study of microfluidic systems, researchers developed an oxygen-regulated spleen chip platform to simulate the splenic interendothelial gap (S-filter) and macrophage (M-filter) through two functional modules, S-Chip and M-Chip, respectively, to study the mechanical retention and phagocytosis processes of red blood cells under hypoxic conditions (108).
Engineered tissues has opened new avenues for spleen functional reconstruction and immunological research. Purwada’s team innovatively co-cultured primary mouse splenic B cells with transgenic 40LB stromal cells in a 3D system, successfully constructing a functional spleen organoid model (19, 20). This landmark achievement laid the foundation for subsequent human studies. In 2020, significant progress was made when scientists established a continuously proliferative human spleen organoid culture system by optimizing the processing protocols for human spleen samples (109). Most remarkably, these engineered human spleen organoids demonstrated remarkable therapeutic potential in transplantation experiments—when implanted into splenectomized mouse models, they self-assembled into spleen-like tissue structures and effectively restored the host’s erythrocyte clearance function within 4 weeks post-transplantation. These findings validate the feasibility of engineered tissues spleen and advance its clinical translation.
3.4 Lymph nodes organoids
The LN is structurally divided into three primary regions: the cortex, paracortex, and medulla. The cortex, as the outermost layer of the LN, primarily houses B cell follicles, which are niches enriched with B cells and follicular DCs, along with the interfollicular zone that delineates these follicles. The paracortex, an internal region also known as the T-cell zone, is predominantly occupied by fibroblastic reticular cells forming a ductal network throughout this area. Proximal to the efferent lymphatic vessels, the medulla contains the medullary sinuses (110). LNs are critical components of the lymphatic and immune systems, playing a pivotal role in detecting, responding to, and eliminating harmful substances efficiently (111). Developing a model of LNs could significantly enhance our understanding of the mechanisms underlying immune response generation.
As crucial immune organs, LNs exhibit intricate cellular interaction networks, dynamic lymphocyte migration, and antigen recognition mechanisms, which are primarily studied using live imaging techniques (112–114). To investigate the deep tissue architecture of LNs, vibratome-sectioned live slices with thicknesses ranging from 250 to 400 μm can be prepared for real-time imaging monitoring (115, 116). This tissue slice platform enables temporal visualization of individual cells while simultaneously monitoring global changes in surface marker expression and quantifying bulk cytokine secretion from intact slices or explants to assess immune responses (29, 117, 118).
Microfluidic chip technology has been effectively utilized to emulate the intricate environment and functionality of LNs. This technology not only integrates seamlessly with precision slice culture techniques but also establishes an experimental platform that more closely mimics in vivo physiological conditions for cell research. Moreover, it offers robust technical support for elucidating the interaction mechanisms between cells and the precise regulation of cellular behavior. Ross’s team employed microfluidic integrated optical imaging to investigate cytokine diffusion within LNs, thereby quantifying the diffusion of bioactive molecules in living tissues (119). In a study conducted by Moura Rosa’s team, a microfluidic device was utilized to investigate the interaction between infused T cells and adherent DC cells, as well as the effects of varying shear stresses within the device (120). To date, LN on-chip models have only partially recapitulated select features of human LNs (121).
Unlike other engineering systems that focus on tissue architecture, engineered tissues for immunity research predominantly aim to replicate specific immune functions (16). Giese’s team developed a novel in vitro model of human LNs, which can be maintained continuously for several weeks (122). This extended operational period enables prolonged and repeated drug exposures, facilitating the induction and monitoring of both cellular and humoral immune responses. Tomei’s team encapsulated FRCs within macroporous polyurethane scaffolds composed of type I collagen and Matrigel (123). They applied a gap flow ranging from 1 to 23 μL/min in this in vitro 3D system. This experimental setup successfully recapitulates the in vivo morphology of FRCs and demonstrates the critical role of lymphatic flow in LN function. The results demonstrate that lymphatic flow significantly enhances the organization of FRCs, whereas FRCs in the absence of flow do not produce CCL21.
3.5 Tonsil organoids
The palatine tonsils, as MALT located in the upper respiratory tract, together with the adenoids and lingual tonsils, form part of the Waldeyer’s ring, which constitutes the first line of immune defense against inhaled or ingested pathogens (51, 52). As prominent components of the Waldeyer’s ring, tonsils and adenoids are functionally closely associated with nasopharynx-associated lymphoid tissues in rodents and other species. The cellular architecture of these tissues is intricate, featuring germinal centers within B-cell follicles and T-cell-rich regions, closely resembling that of LNs. Notably, the tonsil lacks afferent lymphatic vessels, enabling direct contact with antigens from the external environment. Importantly, the tonsil plays a crucial role in inducing B cell immune responses, particularly following direct antigen stimulation (124, 125). Although tonsil tissue is relatively accessible, research on engineered tissues applications remains at an exploratory stage. To date, no studies have reported the successful generation of stem cell-derived tonsillar organoids. Therefore, the following sections will primarily focus on research progress regarding broadly defined organoids.
Currently, research on tonsils primarily utilizes live slices or explant culture techniques. Studies have shown that human tonsil tissue blocks can be effectively maintained in vitro for up to four days, providing an important model for investigating the dynamic behavior of immune cells within their microenvironment (126). Additionally, Grevel’s team employed a tonsil explant culture model to deeply analyze the molecular mechanisms of host cell interactions and their interplay with pathogens (118). These studies confirm that this in vitro culture system effectively supports comprehensive research on microbiome characteristics, histopathological properties, and viral susceptibility of patient-derived tonsil tissues (127, 128).
The in vitro culture of tonsil sections suffers from inherent limitations, such as short preservation time and high contamination risk. These limitations severely restrict its applications. Wagar’s team innovatively developed a tonsil organoid model based on high-density culture on low-attachment surfaces. By maintaining lymphoid tissue cells derived from tonsils, spleen, and LNs, this model successfully preserves the structural and functional characteristics of the original tissue (22) (Figure 3C). This system can effectively induce B cells to undergo SHM, class switch recombination, and affinity maturation while producing specific antibodies, providing a controlled, high-throughput, and cost-effective platform for studying adaptive immune responses. However, the model still exhibits significant limitations: first, its reliance on spontaneous cell aggregation leads to poor reproducibility in organoid size and morphology, severely impacting its application in vaccine and drug screening. Second, whether the model contains critical lymphoid stromal cells remains unclear, potentially compromising the simulation of a complete immune microenvironment.
4 Application and perspectives of immune organoids in immunological research
4.1 Current applications of immune organoids
In vaccine development (Figure 4A), previous studies have attempted to simulate vaccine-induced adaptive immune responses using organ-on-a-chip systems (13), synthetic biomaterial-based immune organoids (129, 130), or LN tissue slices (117), however, all these systems exhibit significant limitations. A research team utilized the high-throughput advantage of tonsil organoids to systematically compare the differences in immune responses induced by inactivated vaccines, live attenuated vaccines, and wild-type viruses, thereby providing critical technical support for investigations into vaccine mechanisms (22, 131). In disease modeling (Figure 4B), lymphoma organoids and bone marrow organoids (75, 132), as novel in vitro models, have demonstrated significant value in hematological disease research. Lymphoma organoids can stably maintain tumor genetic characteristics and interactions within the immune microenvironment, offering a reliable platform for studying the pathogenesis of follicular lymphoma and evaluating drug efficacy. Bone marrow organoids, in contrast, accurately mimic the physiological and pathological states of the bone marrow microenvironment, making them suitable for studying diseases such as leukemia and myelofibrosis. By incorporating patient-derived cells or specific factors (e.g., TGF-β), these models can reconstruct disease microenvironment features, thus serving as ideal tools for mechanistic exploration and therapeutic optimization. In drug screening and toxicity assessment (Figure 4C), patient-derived lymphoid organoid models retain patient-specific tumor microenvironments, enabling precise evaluation of immunotherapies effects (e.g., bispecific antibodies) on tumor cell killing and the activation properties of T-cell. Bone marrow organoids can systematically assess the toxic effects of drugs on hematopoietic function, offering reliable preclinical safety data for evaluating the myelosuppressive risks of chemotherapy and targeted therapies (75, 132). Notably, immune organoids are compatible with many traditional and cutting-edge technologies (Figure 4D). For instance, in the case of tonsil organoids (22), single-cell RNA sequencing (scRNA-seq) successfully identified germinal center-like B cells (CD38+/CD27+), while transcriptomic profiling confirmed stable gene expression patterns across different culture durations. Flow cytometry analysis precisely quantified shifts in immune cell populations following stimulation with live-attenuated influenza vaccine. Notably, ELISA assays detected influenza-specific IgG secretion as early as 7 days post-vaccination, demonstrating the functional maturation of naive B cells into hemagglutinin-specific antibody producers.
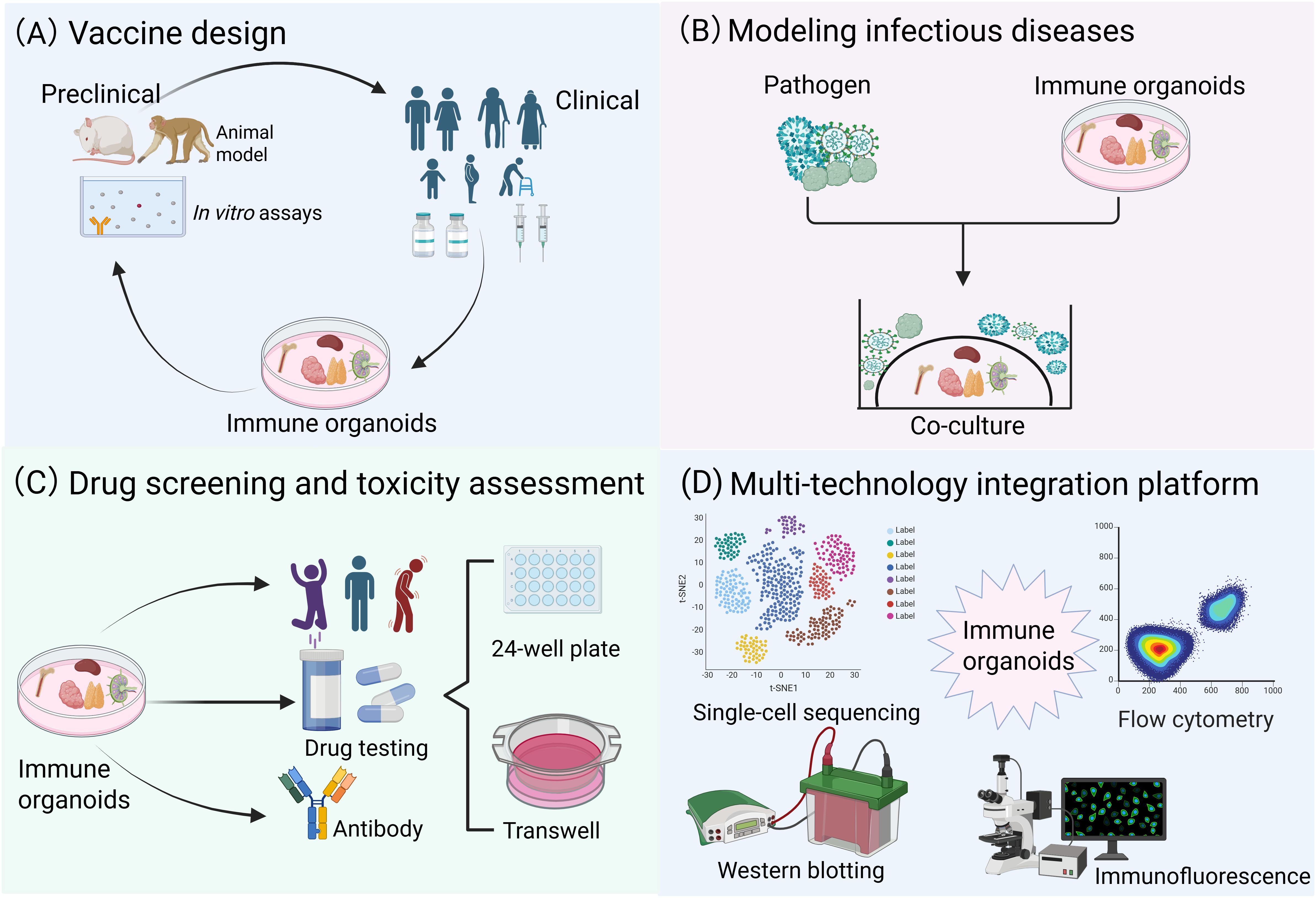
Figure 4. Multidisciplinary convergence platforms: translational applications in biomedical research. (A) Immune organoids can be rationally designed for vaccine development, enabling a more precise simulation of the human immune response mechanism and supporting the evaluation of vaccine immunogenicity and safety. (B) Immune organoids co-culture systems for modeling infectious diseases and cancer. (C) Immune organoids exhibit significant potential in drug screening and toxicity assessment. (D) Immune organoids are compatible with many traditional and cutting-edge technologies.
4.2 Future challenges and prospects of immune organoids
While organoid technology has made remarkable progress, the lack of standardized protocols continues to limit experimental reproducibility and consistency. Notably, China recently issued its first national standard for organ-on-a-chip technology — “General Technical Requirements for Skin-on-a-Chip” (GB/T 44831-2024), which sets a crucial benchmark for quality control in this field (133). Furthermore, disruptive technologies like 3D bioprinting and automation are bringing revolutionary opportunities to organoid research. 3D bioprinting platforms can efficiently construct complex structures such as liver organoids and lung cancer organoids (134, 135), with researchers having successfully simulated liver lobule-like structures and alveolar tissues in vitro. The automated microfluidic platform developed by Tay’s team has enabled high-throughput dynamic drug screening using pancreatic tumor organoids (136). The application of gene-editing tools like CRISPR/Cas9 has further facilitated genotype-phenotype relationship studies in organoids (137). Regrettably, these cutting-edge technologies remain largely unexplored in the field of immune organoids. Looking forward, immune organoid research urgently requires breakthroughs in several key areas: first, standardized culture systems and evaluation metrics must be established to enhance experimental reproducibility. Second, integration of microfluidics and 3D printing technologies is essential to construct more physiologically relevant immune microenvironments. Most importantly, comprehensive utilization of single-cell sequencing, spatial transcriptomics and other omics technologies will be essential to deeply analyze the dynamic regulatory mechanisms underlying immune responses. Only through the interdisciplinary convergence of multiple technologies can immune organoids truly evolve into an ideal platform for vaccine development, tumor immunotherapy, and autoimmune disease research.
5 Concluding remarks
In recent years, significant advancements have been achieved in the development and application of in vitro and ex vivo immune models. This review highlights several experimental systems utilized to investigate and emulate immune interactions, including ex vivo culture systems, organ-on-a-chip platforms, engineered tissue technologies, and stem cell-derived organoids. These technological platforms demonstrate unique value in adaptive immunity research and clinical translation applications (such as vaccine development) by integrating engineering advancements with immunological theories. Notably, these systems exhibit remarkable complementarity in functionality and application, with the tissue slice culture system combining engineered tissues with microfluidic technology being particularly outstanding. This multidisciplinary integrated platform can more accurately simulate the complex immune interaction networks observed in vivo, providing novel technological approaches for immunological research. Looking ahead, the integration of immune organoids with other organoid systems will open new research directions. For instance, coupling tonsil organoids with airway mucosal organoids could simulate respiratory mucosal immune responses, while combining LN organoids with intestinal organoids may help elucidate the regulatory mechanisms of gut-associated lymphoid tissue. Although current organoid technology still faces challenges related to functional completeness and technical standardization, its future development will undoubtedly break through the limitations of single-model paradigms, driving immunological research to new heights through multi-system integration.
Author contributions
TL: Writing – original draft. DP: Writing – review & editing. MY: Writing – review & editing. ML: Writing – review & editing. YW: Writing – review & editing. SL: Writing – review & editing. DZ: Writing – review & editing. BY: Funding acquisition, Writing – review & editing. H-JQ: Conceptualization, Supervision, Writing – review & editing. L-FL: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (grant 32372983), the Central Public-interest Scientific Institution Basal Research Fund (grant Y2025YC117), and the special fund for Science and Technology Innovation Teams of Shanxi Province (202304051001041).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Hotaling NA, Tang L, Irvine DJ, and Babensee JE. Biomaterial strategies for immunomodulation. Annu Rev BioMed Eng. (2015) 17:317–49. doi: 10.1146/annurev-bioeng-071813-104814
2. COVID-19 Excess Mortality Collaborators. Estimating excess mortality due to the COVID-19 pandemic: A systematic analysis of COVID-19-related mortality, 2020-21. Lancet. (2022) 399:1513–36. doi: 10.1016/s0140-6736(21)02796-3
3. Peng D, Li M, Yu Z, Yan T, Yao M, Li S, et al. Synergy between pluripotent stem cell-derived macrophages and self-renewing macrophages: Envisioning a promising avenue for the modelling and cell therapy of infectious diseases. Cell Prolif. (2025) 58:e13770. doi: 10.1111/cpr.13770
4. Blutt SE and Estes MK. Organoid models for infectious disease. Annu Rev Med. (2022) 73:167–82. doi: 10.1146/annurev-med-042320-023055
5. Aiello A, Farzaneh F, Candore G, Caruso C, Davinelli S, Gambino CM, et al. Immunosenescence and its hallmarks: How to oppose aging strategically? A review of potential options for therapeutic intervention. Front Immunol. (2019) 10:2247. doi: 10.3389/fimmu.2019.02247
6. Yousefzadeh MJ, Flores RR, Zhu Y, Schmiechen ZC, Brooks RW, Trussoni CE, et al. An aged immune system drives senescence and ageing of solid organs. Nature. (2021) 594:100–5. doi: 10.1038/s41586-021-03547-7
7. Schutgens F and Clevers H. Human organoids: Tools for understanding biology and treating diseases. Annu Rev Pathol. (2020) 15:211–34. doi: 10.1146/annurev-pathmechdis-012419-032611
8. Lee SY, Koo IS, Hwang HJ, and Lee DW. In vitro three-dimensional (3D) cell culture tools for spheroid and organoid models. SLAS Discov. (2023) 28:119–37. doi: 10.1016/j.slasd.2023.03.006
9. Barbier AJ, Jiang AY, Zhang P, Wooster R, and Anderson DG. The clinical progress of mRNA vaccines and immunotherapies. Nat Biotechnol. (2022) 40:840–54. doi: 10.1038/s41587-022-01294-2
10. Vishwakarma A, Bhise NS, Evangelista MB, Rouwkema J, Dokmeci MR, Ghaemmaghami AM, et al. Engineering immunomodulatory biomaterials to tune the inflammatory response. Trends Biotechnol. (2016) 34:470–82. doi: 10.1016/j.tibtech.2016.03.009
11. Aguilar C, Alves da Silva M, Saraiva M, Neyazi M, Olsson IAS, and Bartfeld S. Organoids as host models for infection biology - a review of methods. Exp Mol Med. (2021) 53:1471–82. doi: 10.1038/s12276-021-00629-4
12. Hammel JH, Cook SR, Belanger MC, Munson JM, and Pompano RR. Modeling immunity in vitro: Slices, chips, and engineered tissues. Annu Rev BioMed Eng. (2021) 23:461–91. doi: 10.1146/annurev-bioeng-082420-124920
13. Goyal G, Prabhala P, Mahajan G, Bausk B, Gilboa T, Xie L, et al. Ectopic lymphoid follicle formation and human seasonal influenza vaccination responses recapitulated in an organ-on-a-chip. Adv Sci (Weinh). (2022) 9:e2103241. doi: 10.1002/advs.202103241
14. Giese C and Marx U. Human immunity in vitro - solving immunogenicity and more. Adv Drug Delivery Rev. (2014) 69-70:103–22. doi: 10.1016/j.addr.2013.12.011
15. Ramadan Q and Ting FC. In vitro micro-physiological immune-competent model of the human skin. Lab Chip. (2016) 16:1899–908. doi: 10.1039/c6lc00229c
16. Gosselin EA, Eppler HB, Bromberg JS, and Jewell CM. Designing natural and synthetic immune tissues. Nat Mater. (2018) 17:484–98. doi: 10.1038/s41563-018-0077-6
17. Corrò C, Novellasdemunt L, and Li VSW. A brief history of organoids. Am J Physiol Cell Physiol. (2020) 319:C151–c65. doi: 10.1152/ajpcell.00120.2020
18. Zhao Z, Chen X, Dowbaj AM, Sljukic A, Bratlie K, Lin L, et al. Organoids. Nat Rev Methods Primers. (2022) 2(1):49. doi: 10.1038/s43586-022-00174-y
19. Purwada A, Jaiswal MK, Ahn H, Nojima T, Kitamura D, Gaharwar AK, et al. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials. (2015) 63:24–34. doi: 10.1016/j.biomaterials.2015.06.002
20. Purwada A and Singh A. Immuno-engineered organoids for regulating the kinetics of B-cell development and antibody production. Nat Protoc. (2017) 12:168–82. doi: 10.1038/nprot.2016.157
21. Lenti E, Bianchessi S, Proulx ST, Palano MT, Genovese L, Raccosta L, et al. Therapeutic regeneration of lymphatic and immune cell functions upon lympho-organoid transplantation. Stem Cell Rep. (2019) 12:1260–8. doi: 10.1016/j.stemcr.2019.04.021
22. Wagar LE, Salahudeen A, Constantz CM, Wendel BS, Lyons MM, Mallajosyula V, et al. Modeling human adaptive immune responses with tonsil organoids. Nat Med. (2021) 27:125–35. doi: 10.1038/s41591-020-01145-0
23. Tajima A, Pradhan I, Geng X, Trucco M, and Fan Y. Construction of thymus organoids from decellularized thymus scaffolds. Methods Mol Biol. (2019) 1576:33–42. doi: 10.1007/7651_2016_9
24. Kobayashi Y and Watanabe T. Artificial construction of immune tissues/organoids and their application for immunological intervention. Curr Top Microbiol Immunol. (2020) 426:143–60. doi: 10.1007/82_2020_215
25. Suhito IR, Sunil C, and Tay A. Engineering human immune organoids for translational immunology. Bioact Mater. (2025) 44:164–83. doi: 10.1016/j.bioactmat.2024.10.010
26. Frenz-Wiessner S, Fairley SD, Buser M, Goek I, Salewskij K, Jonsson G, et al. Generation of complex bone marrow organoids from human induced pluripotent stem cells. Nat Methods. (2024) 21:868–81. doi: 10.1038/s41592-024-02172-2
27. Robinson NB, Krieger K, Khan FM, Huffman W, Chang M, Naik A, et al. The current state of animal models in research: A review. Int J Surg. (2019) 72:9–13. doi: 10.1016/j.ijsu.2019.10.015
28. Saeidnia S, Manayi A, and Abdollahi M. From in vitro experiments to in vivo and clinical studies; pros and cons. Curr Drug Discov Technol. (2015) 12:218–24. doi: 10.2174/1570163813666160114093140
29. Groff BD, Kinman AWL, Woodroof JF, and Pompano RR. Immunofluorescence staining of live lymph node tissue slices. J Immunol Methods. (2019) 464:119–25. doi: 10.1016/j.jim.2018.10.010
30. Saorin G, Caligiuri I, and Rizzolio F. Microfluidic organoids-on-a-chip: The future of human models. Semin Cell Dev Biol. (2023) 144:41–54. doi: 10.1016/j.semcdb.2022.10.001
31. Yu F and Choudhury D. Microfluidic bioprinting for organ-on-a-chip models. Drug Discov Today. (2019) 24:1248–57. doi: 10.1016/j.drudis.2019.03.025
32. Campinoti S, Gjinovci A, Ragazzini R, Zanieri L, Ariza-McNaughton L, Catucci M, et al. Reconstitution of a functional human thymus by postnatal stromal progenitor cells and natural whole-organ scaffolds. Nat Commun. (2020) 11:6372. doi: 10.1038/s41467-020-20082-7
33. Dao V, Yuki K, Lo YH, Nakano M, and Kuo CJ. Immune organoids: From tumor modeling to precision oncology. Trends Cancer. (2022) 8:870–80. doi: 10.1016/j.trecan.2022.06.001
34. Ye W, Luo C, Li C, Huang J, and Liu F. Organoids to study immune functions, immunological diseases and immunotherapy. Cancer Lett. (2020) 477:31–40. doi: 10.1016/j.canlet.2020.02.027
35. Kumar BV, Connors TJ, and Farber DL. Human T cell development, localization, and function throughout life. Immunity. (2018) 48:202–13. doi: 10.1016/j.immuni.2018.01.007
36. Vinuesa CG, Linterman MA, Yu D, and MacLennan IC. Follicular helper T cells. Annu Rev Immunol. (2016) 34:335–68. doi: 10.1146/annurev-immunol-041015-055605
37. Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. (2014) 41:529–42. doi: 10.1016/j.immuni.2014.10.004
38. Pieper K, Grimbacher B, and Eibel H. B-cell biology and development. J Allergy Clin Immunol. (2013) 131:959–71. doi: 10.1016/j.jaci.2013.01.046
39. Sallusto F, Geginat J, and Lanzavecchia A. Central memory and effector memory T cell subsets: Function, generation, and maintenance. Annu Rev Immunol. (2004) 22:745–63. doi: 10.1146/annurev.immunol.22.012703.104702
40. Xing Z, Afkhami S, Bavananthasivam J, Fritz DK, D’Agostino MR, Vaseghi-Shanjani M, et al. Innate immune memory of tissue-resident macrophages and trained innate immunity: Re-vamping vaccine concept and strategies. J Leukoc Biol. (2020) 108:825–34. doi: 10.1002/jlb.4mr0220-446r
41. Inoue T and Kurosaki T. Memory B cells. Nat Rev Immunol. (2024) 24:5–17. doi: 10.1038/s41577-023-00897-3
42. Steinbach K, Vincenti I, and Merkler D. Resident-memory T cells in tissue-restricted immune responses: For better or worse? Front Immunol. (2018) 9:2827. doi: 10.3389/fimmu.2018.02827
43. Yao M, Li M, Peng D, Wang Y, Li S, Zhang D, et al. Unraveling macrophage polarization: Functions, mechanisms, and “double-edged sword” roles in host antiviral immune responses. Int J Mol Sci. (2024) 25(22):12078. doi: 10.3390/ijms252212078
44. Kapsenberg ML. Dendritic-cell control of pathogen-driven T-cell polarization. Nat Rev Immunol. (2003) 3:984–93. doi: 10.1038/nri1246
45. Shichkin VP and Antica M. Key factors for thymic function and development. Front Immunol. (2022) 13:926516. doi: 10.3389/fimmu.2022.926516
46. Thompson EC. Focus issue: Structure and function of lymphoid tissues. Trends Immunol. (2012) 33:255. doi: 10.1016/j.it.2012.05.001
47. Hogquist KA, Baldwin TA, and Jameson SC. Central tolerance: Learning self-control in the thymus. Nat Rev Immunol. (2005) 5:772–82. doi: 10.1038/nri1707
48. Takahama Y. Journey through the thymus: Stromal guides for T-cell development and selection. Nat Rev Immunol. (2006) 6:127–35. doi: 10.1038/nri1781
49. Buettner M and Lochner M. Development and function of secondary and tertiary lymphoid organs in the small intestine and the colon. Front Immunol. (2016) 7:342. doi: 10.3389/fimmu.2016.00342
50. Jones GW, Hill DG, and Jones SA. Understanding immune cells in tertiary lymphoid organ development: It is all starting to come together. Front Immunol. (2016) 7:401. doi: 10.3389/fimmu.2016.00401
51. Brandtzaeg P. Function of mucosa-associated lymphoid tissue in antibody formation. Immunol Invest. (2010) 39:303–55. doi: 10.3109/08820131003680369
52. Carrasco A, Sjölander I, Van Acker A, Dernstedt A, Fehrm J, Forsell M, et al. The tonsil lymphocyte landscape in pediatric tonsil hyperplasia and obstructive sleep apnea. Front Immunol. (2021) 12:674080. doi: 10.3389/fimmu.2021.674080
53. Aydin S and Uner C. Normal palatine tonsil size in healthy children: A sonographic study. Radiol Med. (2020) 125:864–9. doi: 10.1007/s11547-020-01168-0
54. Liao S and von der Weid PY. Lymphatic system: An active pathway for immune protection. Semin Cell Dev Biol. (2015) 38:83–9. doi: 10.1016/j.semcdb.2014.11.012
55. Mebius RE and Kraal G. Structure and function of the spleen. Nat Rev Immunol. (2005) 5:606–16. doi: 10.1038/nri1669
56. Lewis SM, Williams A, and Eisenbarth SC. Structure and function of the immune system in the spleen. Sci Immunol. (2019) 4(33):eaau6085. doi: 10.1126/sciimmunol.aau6085
57. Azar J, Bahmad HF, Daher D, Moubarak MM, Hadadeh O, Monzer A, et al. The use of stem cell-derived organoids in disease modeling: An update. Int J Mol Sci. (2021) 22(14):7667. doi: 10.3390/ijms22147667
58. Bartfeld S and Clevers H. Stem cell-derived organoids and their application for medical research and patient treatment. J Mol Med (Berl). (2017) 95:729–38. doi: 10.1007/s00109-017-1531-7
59. Ribeiro-Filho AC, Levy D, Ruiz JLM, Mantovani MDC, and Bydlowski SP. Traditional and advanced cell cultures in hematopoietic stem cell studies. Cells. (2019) 8(12):1628. doi: 10.3390/cells8121628
60. Mokhtari Z, Mech F, Zehentmeier S, Hauser AE, and Figge MT. Quantitative image analysis of cell colocalization in murine bone marrow. Cytomet A. (2015) 87:503–12. doi: 10.1002/cyto.a.22641
61. Bellido T and Delgado-Calle J. Ex vivo organ cultures as models to study bone biology. JBMR Plus. (2020) 4(3):10. doi: 10.1002/jbm4.10345
62. Luo Y, Li X, Zhao Y, Zhong W, Xing M, and Lyu G. Development of organs-on-chips and their impact on precision medicine and advanced system simulation. Pharmaceutics. (2023) 15(8):2094. doi: 10.3390/pharmaceutics15082094
63. Sharipol A, Lesch ML, Soto CA, and Frisch BJ. Bone marrow microenvironment-on-chip for culture of functional hematopoietic stem cells. Front Bioeng Biotechnol. (2022) 10:855777. doi: 10.3389/fbioe.2022.855777
64. Sieber S, Wirth L, Cavak N, Koenigsmark M, Marx U, Lauster R, et al. Bone marrow-on-a-chip: Long-term culture of human hematopoietic stem cells in a three-dimensional microfluidic environment. J Tissue Eng Regener Med. (2018) 12:479–89. doi: 10.1002/term.2507
65. Torisawa YS, Spina CS, Mammoto T, Mammoto A, Weaver JC, Tat T, et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods. (2014) 11:663–9. doi: 10.1038/nmeth.2938
66. Chou DB, Frismantas V, Milton Y, David R, Pop-Damkov P, Ferguson D, et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat BioMed Eng. (2020) 4:394–406. doi: 10.1038/s41551-019-0495-z
67. Nelson MR, Ghoshal D, Mejías JC, Rubio DF, Keith E, and Roy K. A multi-niche microvascularized human bone marrow (hBM) on-a-chip elucidates key roles of the endosteal niche in hBM physiology. Biomaterials. (2021) 270:120683. doi: 10.1016/j.biomaterials.2021.120683
68. Bourgine PE, Klein T, Paczulla AM, Shimizu T, Kunz L, Kokkaliaris KD, et al. In vitro biomimetic engineering of a human hematopoietic niche with functional properties. Proc Natl Acad Sci U.S.A. (2018) 115:E5688–e95. doi: 10.1073/pnas.1805440115
69. Mortera-Blanco T, Mantalaris A, Bismarck A, Aqel N, and Panoskaltsis N. Long-term cytokine-free expansion of cord blood mononuclear cells in three-dimensional scaffolds. Biomaterials. (2011) 32:9263–70. doi: 10.1016/j.biomaterials.2011.08.051
70. Nichols JE, Cortiella J, Lee J, Niles JA, Cuddihy M, Wang S, et al. In vitro analog of human bone marrow from 3D scaffolds with biomimetic inverted colloidal crystal geometry. Biomaterials. (2009) 30:1071–9. doi: 10.1016/j.biomaterials.2008.10.041
71. Janagama D and Hui SK. 3-D cell culture systems in bone marrow tissue and organoid engineering, and BM phantoms as in vitro models of hematological cancer therapeutics-a review. Mater (Basel). (2020) 13(24):5609. doi: 10.3390/ma13245609
72. Di Buduo CA, Wray LS, Tozzi L, Malara A, Chen Y, Ghezzi CE, et al. Programmable 3D silk bone marrow niche for platelet generation ex vivo and modeling of megakaryopoiesis pathologies. Blood. (2015) 125:2254–64. doi: 10.1182/blood-2014-08-595561
73. Raic A, Naolou T, Mohra A, Chatterjee C, and Lee-Thedieck C. 3D models of the bone marrow in health and disease: Yesterday, today and tomorrow. MRS Commun. (2019) 9:37–52. doi: 10.1557/mrc.2018.203
74. Tozzi L, Laurent PA, Di Buduo CA, Mu X, Massaro A, Bretherton R, et al. Multi-channel silk sponge mimicking bone marrow vascular niche for platelet production. Biomaterials. (2018) 178:122–33. doi: 10.1016/j.biomaterials.2018.06.018
75. Khan AO, Rodriguez-Romera A, Reyat JS, Olijnik AA, Colombo M, Wang G, et al. Human bone marrow organoids for disease modeling, discovery, and validation of therapeutic targets in hematologic Malignancies. Cancer Discov. (2023) 13:364–85. doi: 10.1158/2159-8290.Cd-22-0199
76. Thapa P and Farber DL. The role of the thymus in the immune response. Thorac Surg Clin. (2019) 29:123–31. doi: 10.1016/j.thorsurg.2018.12.001
77. Bleul CC, Corbeaux T, Reuter A, Fisch P, Mönting JS, and Boehm T. Formation of a functional thymus initiated by a postnatal epithelial progenitor cell. Nature. (2006) 441:992–6. doi: 10.1038/nature04850
78. Anderson G and Takahama Y. Thymic epithelial cells: Working class heroes for T cell development and repertoire selection. Trends Immunol. (2012) 33:256–63. doi: 10.1016/j.it.2012.03.005
79. Bar-Ephraim YE, Kretzschmar K, and Clevers H. Organoids in immunological research. Nat Rev Immunol. (2020) 20:279–93. doi: 10.1038/s41577-019-0248-y
80. Mandel T and Russell PJ. Differentation of fetal mouse thymus. Ultrastructure of organ cultures and of subcapsular grafts. Immunology. (1971) 21:659–74.
81. Oh SH and Kim K. Expression of interleukin-1 receptors in the later period of fetal thymic organ culture and during suspension culture of thymocytes from aged mice. Immunol Cell Biol. (1999) 77:491–8. doi: 10.1046/j.1440-1711.1999.00852.x
82. Sahni H, Ross S, Barbarulo A, Solanki A, Lau CI, Furmanski A, et al. A genome wide transcriptional model of the complex response to pre-TCR signaling during thymocyte differentiation. Oncotarget. (2015) 6:28646–60. doi: 10.18632/oncotarget.5796
83. Takeuchi Y, Horiuchi T, Hamamura K, Sugimoto T, Yagita H, and Okumura K. Role of CD4 molecule in intrathymic t-cell development. Immunology. (1991) 74:183–90.
84. Kurd N and Robey EA. T-cell selection in the thymus: A spatial and temporal perspective. Immunol Rev. (2016) 271:114–26. doi: 10.1111/imr.12398
85. Lancaster JN and Ehrlich LI. Analysis of thymocyte migration, cellular interactions, and activation by multiphoton fluorescence microscopy of live thymic slices. Methods Mol Biol. (2017) 1591:9–25. doi: 10.1007/978-1-4939-6931-9_2
86. Lancaster JN, Thyagarajan HM, Srinivasan J, Li Y, Hu Z, and Ehrlich LIR. Live-cell imaging reveals the relative contributions of antigen-presenting cell subsets to thymic central tolerance. Nat Commun. (2019) 10:2220. doi: 10.1038/s41467-019-09727-4
87. Ross JO, Melichar HJ, Au-Yeung BB, Herzmark P, Weiss A, and Robey EA. Distinct phases in the positive selection of CD8+ t cells distinguished by intrathymic migration and t-cell receptor signaling patterns. Proc Natl Acad Sci U.S.A. (2014) 111:E2550–8. doi: 10.1073/pnas.1408482111
88. Ross JO, Melichar HJ, Halkias J, and Robey EA. Studying T cell development in thymic slices. Methods Mol Biol. (2016) 1323:131–40. doi: 10.1007/978-1-4939-2809-5_11
89. Hale LP, Neff J, Cheatham L, Cardona D, Markert ML, and Kurtzberg J. Histopathologic assessment of cultured human thymus. PloS One. (2020) 15:e0230668. doi: 10.1371/journal.pone.0230668
90. Michaels YS, Buchanan CF, Gjorevski N, and Moisan A. Bioengineering translational models of lymphoid tissues. Nat Rev Bioeng. (2023) 1:731–48. doi: 10.1038/s44222-023-00101-0
91. Poznansky MC, Evans RH, Foxall RB, Olszak IT, Piascik AH, Hartman KE, et al. Efficient generation of human T cells from a tissue-engineered thymic organoid. Nat Biotechnol. (2000) 18:729–34. doi: 10.1038/77288
92. Seet CS, He C, Bethune MT, Li S, Chick B, Gschweng EH, et al. Generation of mature T cells from human hematopoietic stem and progenitor cells in artificial thymic organoids. Nat Methods. (2017) 14:521–30. doi: 10.1038/nmeth.4237
93. Hübscher T, Lorenzo-Martín LF, Barthlott T, Tillard L, Langer JJ, Rouse P, et al. Thymic epithelial organoids mediate T-cell development. Development. (2024) 151(17):dev202853. doi: 10.1242/dev.202853
94. Lim S, J F van Son G, Wisma Eka Yanti NL, Andersson-Rolf A, Willemsen S, Korving J, et al. Derivation of functional thymic epithelial organoid lines from adult murine thymus. Cell Rep. (2024) 43:114019. doi: 10.1016/j.celrep.2024.114019
95. Zeleniak A, Wiegand C, Liu W, McCormick C, K R, Alavi A, et al. De novo construction of T cell compartment in humanized mice engrafted with iPSC-derived thymus organoids. Nat Methods. (2022) 19:1306–19. doi: 10.1038/s41592-022-01583-3
96. Hashi H, Yoshida H, Honda K, Fraser S, Kubo H, Awane M, et al. Compartmentalization of Peyer’s patch anlagen before lymphocyte entry. J Immunol. (2001) 166:3702–9. doi: 10.4049/jimmunol.166.6.3702
97. Mebius RE, Streeter PR, Michie S, Butcher EC, and Weissman IL. A developmental switch in lymphocyte homing receptor and endothelial vascular addressin expression regulates lymphocyte homing and permits CD4+ CD3- cells to colonize lymph nodes. Proc Natl Acad Sci U.S.A. (1996) 93:11019–24. doi: 10.1073/pnas.93.20.11019
98. Parent AV, Russ HA, Khan IS, LaFlam TN, Metzger TC, Anderson MS, et al. Generation of functional thymic epithelium from human embryonic stem cells that supports host T cell development. Cell Stem Cell. (2013) 13:219–29. doi: 10.1016/j.stem.2013.04.004
99. Sun X, Xu J, Lu H, Liu W, Miao Z, Sui X, et al. Directed differentiation of human embryonic stem cells into thymic epithelial progenitor-like cells reconstitutes the thymic microenvironment in vivo. Cell Stem Cell. (2013) 13:230–6. doi: 10.1016/j.stem.2013.06.014
100. Ramos SA, Armitage LH, Morton JJ, Alzofon N, Handler D, Kelly G, et al. Generation of functional thymic organoids from human pluripotent stem cells. Stem Cell Rep. (2023) 18:829–40. doi: 10.1016/j.stemcr.2023.02.013
101. Ehimwenma O and Tagbo MT. Determination of normal dimension of the spleen by ultrasound in an endemic tropical environment. Niger Med J. (2011) 52:198–203. doi: 10.4103/0300-1652.86141
102. Bronte V and Pittet MJ. The spleen in local and systemic regulation of immunity. Immunity. (2013) 39:806–18. doi: 10.1016/j.immuni.2013.10.010
103. Franken L, Schiwon M, and Kurts C. Macrophages: Sentinels and regulators of the immune system. Cell Microbiol. (2016) 18:475–87. doi: 10.1111/cmi.12580
104. Willerson JT, Wakeland JR, Stone MJ, and Wildenthal K. Maintenance of mouse spleen in organ culture and assessment of certain functional capabilities. Cytol (Tokyo). (1975) 40:433–40. doi: 10.1508/cytologia.40.433
105. James K, Skibinski G, and Hoffman P. A comparison of the performance in vitro of precision cut tissue slices and suspensions of human spleen with special reference to immunoglobulin and cytokine production. Hum Antibodies Hybridomas. (1996) 7:138–50. doi: 10.3233/HAB-1996-7401
106. Finetti F, Capitani N, Manganaro N, Tatangelo V, Libonati F, Panattoni G, et al. Optimization of organotypic cultures of mouse spleen for staining and functional assays. Front Immunol. (2020) 11:471. doi: 10.3389/fimmu.2020.00471
107. Rigat-Brugarolas LG, Elizalde-Torrent A, Bernabeu M, De Niz M, Martin-Jaular L, Fernandez-Becerra C, et al. A functional microengineered model of the human splenon-on-a-chip. Lab Chip. (2014) 14:1715–24. doi: 10.1039/c3lc51449h
108. Qiang Y, Sissoko A, Liu ZL, Dong T, Zheng F, Kong F, et al. Microfluidic study of retention and elimination of abnormal red blood cells by human spleen with implications for sickle cell disease. Proc Natl Acad Sci U.S.A. (2023) 120:e2217607120. doi: 10.1073/pnas.2217607120
109. Gee K, Isani MA, Fode A, Maselli KM, Zuber SM, Fowler KL, et al. Spleen organoid units generate functional human and mouse tissue-engineered spleen in a murine model. Tissue Eng Part A. (2020) 26:411–8. doi: 10.1089/ten.TEA.2019.0178
110. Grant SM, Lou M, Yao L, Germain RN, and Radtke AJ. The lymph node at a glance - how spatial organization optimizes the immune response. J Cell Sci. (2020) 133(5):jcs241828. doi: 10.1242/jcs.241828
111. Shou Y, Johnson SC, Quek YJ, Li X, and Tay A. Integrative lymph node-mimicking models created with biomaterials and computational tools to study the immune system. Mater Today Bio. (2022) 14:100269. doi: 10.1016/j.mtbio.2022.100269
112. Miller MJ, Wei SH, Parker I, and Cahalan MD. Two-photon imaging of lymphocyte motility and antigen response in intact lymph node. Science. (2002) 296:1869–73. doi: 10.1126/science.1070051
113. Stein JV and F Gonzalez S. Dynamic intravital imaging of cell-cell interactions in the lymph node. J Allergy Clin Immunol. (2017) 139:12–20. doi: 10.1016/j.jaci.2016.11.008
114. Huang JH, Cárdenas-Navia LI, Caldwell CC, Plumb TJ, Radu CG, Rocha PN, et al. Requirements for T lymphocyte migration in explanted lymph nodes. J Immunol. (2007) 178:7747–55. doi: 10.4049/jimmunol.178.12.7747
115. Katakai T. Live imaging of interstitial T cell migration using lymph node slices. Methods Mol Biol. (2018) 1763:29–42. doi: 10.1007/978-1-4939-7762-8_4
116. Salmon H, Rivas-Caicedo A, Asperti-Boursin F, Lebugle C, Bourdoncle P, and Donnadieu E. Ex vivo imaging of T cells in murine lymph node slices with widefield and confocal microscopes. J Vis Exp. (2011) 53):e3054. doi: 10.3791/3054
117. Belanger MC, Ball AG, Catterton MA, Kinman AWL, Anbaei P, Groff BD, et al. Acute lymph node slices are a functional model system to study immunity ex vivo. ACS Pharmacol Transl Sci. (2021) 4:128–42. doi: 10.1021/acsptsci.0c00143
118. Grivel JC and Margolis L. Use of human tissue explants to study human infectious agents. Nat Protoc. (2009) 4:256–69. doi: 10.1038/nprot.2008.245
119. Ross AE and Pompano RR. Diffusion of cytokines in live lymph node tissue using microfluidic integrated optical imaging. Anal Chim Acta. (2018) 1000:205–13. doi: 10.1016/j.aca.2017.11.048
120. Moura Rosa P, Gopalakrishnan N, Ibrahim H, Haug M, and Halaas Ø. The intercell dynamics of t cells and dendritic cells in a lymph node-on-a-chip flow device. Lab Chip. (2016) 16:3728–40. doi: 10.1039/c6lc00702c
121. Shanti A, Hallfors N, Petroianu GA, Planelles L, and Stefanini C. Lymph nodes-on-chip: Promising immune platforms for pharmacological and toxicological applications. Front Pharmacol. (2021) 12:711307. doi: 10.3389/fphar.2021.711307
122. Giese C, Lubitz A, Demmler CD, Reuschel J, Bergner K, and Marx U. Immunological substance testing on human lymphatic micro-organoids in vitro. J Biotechnol. (2010) 148:38–45. doi: 10.1016/j.jbiotec.2010.03.001
123. Tomei AA, Siegert S, Britschgi MR, Luther SA, and Swartz MA. Fluid flow regulates stromal cell organization and ccl21 expression in a tissue-engineered lymph node microenvironment. J Immunol. (2009) 183:4273–83. doi: 10.4049/jimmunol.0900835
124. Boyaka PN, Wright PF, Marinaro M, Kiyono H, Johnson JE, Gonzales RA, et al. Human nasopharyngeal-associated lymphoreticular tissues. Functional analysis of subepithelial and intraepithelial B and T cells from adenoids and tonsils. Am J Pathol. (2000) 157:2023–35. doi: 10.1016/s0002-9440(10)64841-9
125. Morris MC, Kozara K, Salamone F, Benoit M, and Pichichero ME. Adenoidal follicular T helper cells provide stronger B-cell help than those from tonsils. Laryngoscope. (2016) 126:E80–5. doi: 10.1002/lary.25536
126. Giger B, Bonanomi A, Odermatt B, Ladell K, Speck RF, Kojic D, et al. Human tonsillar tissue block cultures differ from autologous tonsillar cell suspension cultures in lymphocyte subset activation and cytokine gene expression. J Immunol Methods. (2004) 289:179–90. doi: 10.1016/j.jim.2004.04.015
127. Kostić M, Ivanov M, Babić SS, Tepavčević Z, Radanović O, Soković M, et al. Analysis of tonsil tissues from patients diagnosed with chronic tonsillitis-microbiological profile, biofilm-forming capacity and histology. Antibiot (Basel). (2022) 11(12):1747. doi: 10.3390/antibiotics11121747
128. Langlois M, Bounou S, Tremblay MJ, and Barbeau B. Infection of the ex vivo tonsil model by htlv-1 envelope-pseudotyped viruses. Pathogens. (2023) 12(2):182. doi: 10.3390/pathogens12020182
129. Moeller TD, Shah SB, Lai K, Lopez-Barbosa N, Desai P, Wang W, et al. Profiling germinal center-like B cell responses to conjugate vaccines using synthetic immune organoids. ACS Cent Sci. (2023) 9:787–804. doi: 10.1021/acscentsci.2c01473
130. Béguelin W, Rivas MA, Calvo Fernández MT, Teater M, Purwada A, Redmond D, et al. Ezh2 enables germinal center formation through epigenetic silencing of cdkn1a and an RB-E2F1 feedback loop. Nat Commun. (2017) 8:877. doi: 10.1038/s41467-017-01029-x
131. Kastenschmidt JM, Sureshchandra S, Jain A, Hernandez-Davies JE, de Assis R, Wagoner ZW, et al. Influenza vaccine format mediates distinct cellular and antibody responses in human immune organoids. Immunity. (2023) 56:1910–26.e7. doi: 10.1016/j.immuni.2023.06.019
132. Kastenschmidt JM, Schroers-Martin JG, Sworder BJ, Sureshchandra S, Khodadoust MS, Liu CL, et al. A human lymphoma organoid model for evaluating and targeting the follicular lymphoma tumor immune microenvironment. Cell Stem Cell. (2024) 31:410–20.e4. doi: 10.1016/j.stem.2024.01.012
133. Zhu S, Chen D, Yang X, Yang L, and Han Y. Organoid models to study human infectious diseases. Cell Prolif. (2025) 0:e70004. doi: 10.1111/cpr.70004
134. Jian H, Li X, Dong Q, Tian S, and Bai S. In vitro construction of liver organoids with biomimetic lobule structure by a multicellular 3D bioprinting strategy. Cell Prolif. (2023) 56:e13465. doi: 10.1111/cpr.13465
135. Wang H, Wen T, Zhu W, Li K, Gong X, and Li Z. Microfluidic strategies for biomimetic lung chip establishment and SARS-CoV-2 study. Mater Today Bio. (2024) 24:100905. doi: 10.1016/j.mtbio.2023.100905
136. Schuster B, Junkin M, Kashaf SS, Romero-Calvo I, Kirby K, Matthews J, et al. Automated microfluidic platform for dynamic and combinatorial drug screening of tumor organoids. Nat Commun. (2020) 11:5271. doi: 10.1038/s41467-020-19058-4
Keywords: immune organoids, ex vivo culture, microfluidic chips, engineered tissues, immune organs
Citation: Li T, Peng D, Yao M, Li M, Wang Y, Li S, Zhang D, Yang B, Qiu H-J and Li L-F (2025) Immune organoids: emerging platforms for modeling and analyzing human adaptive immunity. Front. Immunol. 16:1632117. doi: 10.3389/fimmu.2025.1632117
Received: 20 May 2025; Accepted: 18 July 2025;
Published: 06 August 2025.
Edited by:
Laurent Rénia, Nanyang Technological University, SingaporeReviewed by:
Garima Kaushik, Champions Oncology, Inc., United StatesLauriane Cabon, Roche (Switzerland), Switzerland
Forrest Lee Baker, University of Arizona, United States
Copyright © 2025 Li, Peng, Yao, Li, Wang, Li, Zhang, Yang, Qiu and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hua-Ji Qiu, cWl1aHVhamlAY2Fhcy5jbg==; Lian-Feng Li, bGlsaWFuZmVuZ0BjYWFzLmNu
 Tianlong Li1,2
Tianlong Li1,2 Su Li
Su Li Ding Zhang
Ding Zhang Hua-Ji Qiu
Hua-Ji Qiu Lian-Feng Li
Lian-Feng Li