- 1Key Open Laboratory of Cold Water Fish Germplasm Resources and Breeding of Heilongjiang Province, Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin, China
- 2College of Life Science, Northeast Agricultural University, Harbin, China
- 3College of Marine Biology and Fisheries, Hainan University, Haikou, China
- 4Heilongjiang Aquatic Animal Resource Conservation Center, Harbin, Heilongjiang, China
Introduction: Cr(VI) is a heavy metal contaminant, can diffuse to ecosystems and harm aquatic animals. Gills, as a vital organ in direct contact with the aquatic environment, have become a key target tissue for assessing the toxicological effects of heavy metal pollution of water bodies due to their sensitivity to heavy metal exposure. However, 3the effects of Cr(VI) on the gill tissues in fish have been less studied. In this study, we revealed the multiple effects of chromium toxicity by assessing the oxidative damage, transcriptomic and metabolomic changes of Cr(VI) on gill tissues of Thymallus grubii.
Methods: A total of 270 fishes were stratified into three experimental groups: control group, low-concentration exposure group (0.2 mg/L), and high-concentration exposure group (1 mg/L). In this study, we revealed the multiple effects of chromium toxicity by assessing the oxidative damage, transcriptomic and metabolomic changes of Cr(VI) on gill tissues of Thymallus grubii.
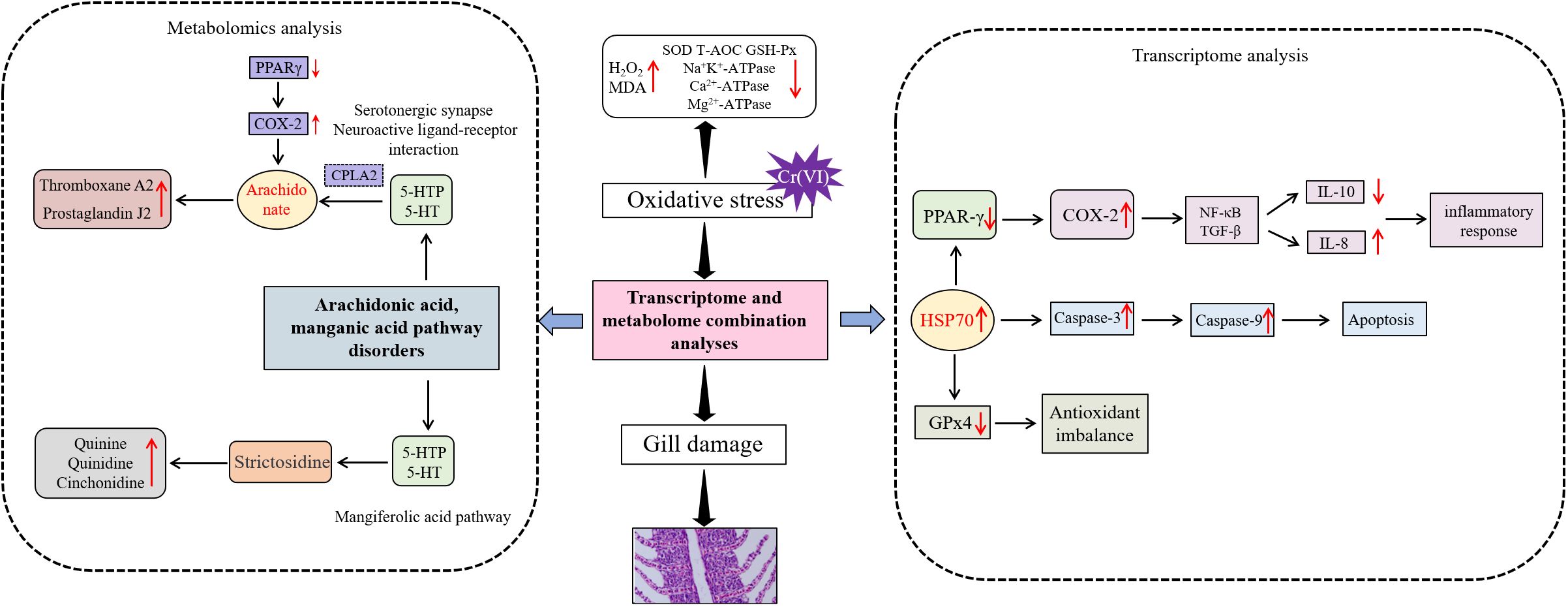
Results: Cr(VI) stress can lead to gill damage with significant reduction in gill filament thickness, significant thinning of gill lamellae, and congestion of epithelial blood vessels. Cr(VI) stress significant increases in H2O2 and MDA levels and significant decreases in antioxidant enzyme activity levels (SOD, GSH-Px, and T-AOC) and energy metabolism-related ATPase activity levels (Na+K+-ATPase, Ca2+-ATPase, and Mg2+-ATPase). Cr(VI) stress induced disturbances in gill arachidonic acid metabolism leading to the release of pro-inflammatory metabolites (e.g., thromboxane A2 and prostaglandin J2) accompanied by the accumulation of oxidised glutathione. However, the synthesis of metabolites with anti-inflammatory/antioxidant functions (e.g. GABA, quinidine and l-artitic acid) was reduced. Transcriptomics and metabolomic coanalyses revealed that Cr(VI) induced PPAR-γ inactivation to deregulate COX-2, which disrupted arachidonic acid metabolic pathways, leading to oxidative stress, apoptosis, and release of inflammatory factors. Disorders of arachidonic acid metabolism led to the release of proinflammatory metabolites (such as thromboxane A2 and prostaglandin J2), and decreased levels of reduced glutathione.
Discussion: The effects of Cr(VI) exposure on gill gene expression and metabolism were analysed using RT-PCR, transcriptomic, and metabolomic approaches. In summary, we better understand the toxic effects of Cr(VI) on gill tissues of aquatic animals. Targeted activation of PPAR-γ and supplementation with anti-inflammatory metabolites such as GABA, quinidine and l- artitic acid may be potential intervention strategies to reverse Cr(VI) toxicity.
1 Introduction
Global water resources are increasingly polluted by heavy metals due to rapid industrial development and agricultural activities, seriously threatening the aquatic animal health and disrupting the balance of ecosystems (1). As a redox-active transition metal ubiquitous in industrial systems, chromium persists in natural ecosystems through multiple valence states, of which Cr3+ and Cr(VI)(Cr6+) are common ionic forms (2). Cr6+ has strong oxidative activity and chemical toxicity and can cause allergic dermatitis, neurotoxicity, genotoxicity, and cancers (3). Cr6+ enters cells in the chromate ionic state, crossing cell membranes through nonspecific phosphate or sulfate anion carriers, leading to mitochondrial damage and cellular DNA damage (4). Owing to human activities, the Cr6+ released from industrial production migrates to groundwater by entering surface water and soil (5). Hexavalent chromium salts are more soluble than Cr3+; thus, Cr6+ is relatively more mobile. In aquatic environments, Cr6+ is present for longer periods of time and is more harmful to biological systems; therefore, Cr6+ is more toxic than Cr3+, which is considered one of the most harmful heavy metals (6). Fish is an important source of protein for humans, and enrichment of Cr6+ in contaminated fish tissue can be transferred to the human body and poses health risks. Therefore, the study of the effects of Cr6+ exposure on fish can provide a basis for the aquaculture industry to formulate aquatic product safety standards, and it is also an inevitable choice to safeguard human food safety and prevent and control environmental pollution.
Studies have shown that chronic exposure of animals to Cr6+ leads to oxidative stress and significantly increases the expression of apoptotic genes; moreover, Cr6+ can affect the normal function of mitochondria, transmembrane potential, and antioxidant enzyme activity, resulting in mitochondrial damage, which triggers a cascade of caspase proteases leading to apoptotic cell death (7–9). In vitro cytotoxicity experiments have shown that Cr6+ ions enter the cell through nonspecific ion channels in the cell membrane and generate large amounts of ROS during their reduction to the low-valent form of chromium and that the continuous accumulation of ROS in cells causes DNA and cellular damage (10, 11). Cr6+ exposure was found to cause neurotoxicity and induce oxidative damage in zebrafish (12). Gills are important organs for gas exchange, osmoregulation and detoxification in fish (13). Gills, characterized by substantial surface area and acute responsiveness to aquatic environmental fluctuations, serve as primary target sites for toxic metal accumulation in aquatic organisms (14). Several studies have confirmed that heavy metal exposure leads to gill damage in fish, with altered morphology and pathology, energy and metabolic imbalances, and impaired antioxidant systems (15–17).
Amur grayling (Thymallus grubii) is an important albino fish in China with high nutritional and economic values. T. grubii has become highly valuable as an economic species for cold Water aquaculture, possessing tasty meat, rich in unsaturated fatty acids, low cholesterol and high protein levels. However, due to environmental pollution and other anthropogenic disturbances, the population of T. grubii has declined sharply (18). In addition, understanding the response mechanism of T. grubii to heavy metal stress can help to provide scientific evidence for monitoring the ecological health of the watershed, and provide toxicological basis for the development of aquatic animal conservation strategies. In heavy metal exposure studies, transcriptomics analyses can comprehensively resolve the dynamics of gene expression in organisms under heavy metal stress. Metabolomics in heavy metal exposure studies can elucidate the details of metabolic processes that occur during environmental adaptation and provide a better understanding of these processes. However, the effects of Cr6+ stress on tissue, gill metabolism, and gene expression in T. grubii have not been reported. In the present study, we assessed the effects of chronic Cr6+ exposure on gill tissue structure, metabolism and gene expression in T. grubii. This study was to elucidate the effects of gill exposure to heavy metals on gene expression, transcriptomics and metabolomics to elucidate the contribution of Cr6+ to T. grubii-induced gill toxicity and the possible underlying mechanisms.
2 Materials and methods
2.1 Animals and diet
This study was approved by the Ethics Committee for Animal Experiments of the Heilongjiang Fisheries Research Institute, Chinese Academy of Fisheries Sciences (20241125–007). The Bohai Experimental Station of the Heilongjiang Provincial Fisheries Research Institute provided the fish for this experiment. Fish were temporarily reared in the aquarium for 30 days to acclimatize to the environment. A total of 270 fish of uniform size were selected and allocated to nine aquaria (three groups), with three biological replicates established in each group. Control fish tanks were filled with tap water. In the aquarium for the low-concentration exposure group (Lcr group: Cr6+ of 0.2 mg/L). In the aquarium for the high-concentration treatment group (Hcr group: Cr6+ of 1 mg/L). Cr6+ exposure levels refer to the results of previous studies and water resources ecosystem surveys (19–21). During the feeding experiment, the fish were handfed twice daily (at 8:30 a.m. and at 16:00 p.m.) to apparent satiation for 4 weeks. Experimental environment of fish: temperature, 10 ± 2°C; pH, 6.8 ± 0.44 dissolved oxygen, 5.57 ± 0.63 mg/L; ammonia concentration < 0.3 mg/L; nitrite concentration < 0.02 mg/L. Potassium dichromate solution was purchased from Merck, China (CAS: 7778-50-9, MDL: MFCD00011367, EC: 231-906-6). Each tank was replaced with 25% water daily.
2.2 Fish sampling
After 24 h of starvation, the fish were anaesthetized with 20.0 mg/L tricaine methanesulfonate (MS-222) at weeks 2 and 4 of the experiment. Eighteen fish per group were randomly selected and prepared for sampling. Fish were euthanized and gills, muscles, livers, and intestines were collected. The collected tissues were frozen in liquid nitrogen and transferred to a −80°C refrigerator.
2.3 Chromium analysis
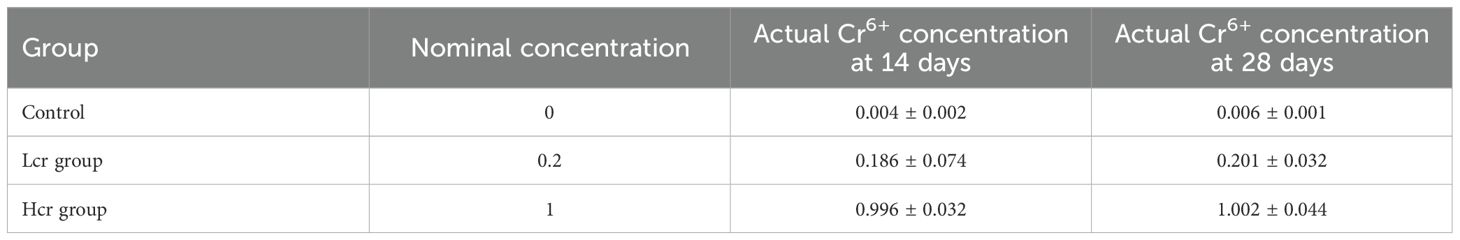
Cr6+ levels in gills, liver, muscle, and intestines were only examined by the methods published in previous studies (22). First 0.1 g of sample, 5.0 mL of nitric acid was mixed in an ablative tube and 3.0 mL of ultrapure water was added. Mineralization of the samples was carried out in a MarXpress microwave ablation system (North Carolina, USA). Cooling 0.5 mL of internal standard was added and the sample was diluted to 50.0 mL with ddH2O and the experiment was repeated 3 times. Cr6+ levels in gills, liver, muscle, and intestines were quantified using an Agilent 7500cx ICP-MS (Agilent Technologies, Santa Clara, CA, USA). Cr6+ levels in water tanks were determined by the same method (Table 1).
2.4 Histopathological examination
Gill samples were dehydrated in increasing concentrations of ethanol (50%, 70%, 80%, 90% and 100%), placed in xylene for 1 minute, dried, and embedded in paraffin. Gill tissue samples were cut into thin slices of 2-6 μm thickness using a rotary slicer (Leica RM2235). Use haematoxylin and eosin and stain according to the manufacturer’s instructions. Observations were made using an Olympus BX53 research-grade biomicroscope (Olympus BX53, Japan), and gill sample changes were observed with cellSens software (cellSens 4.1) and photographed for preservation.
2.5 Histological enzyme activity
Assays of hydrogen peroxide (H2O2), superoxide dismutase (SOD), glutathione peroxidase (GSH-PX), total antioxidant capacity (T-AOC), catalase (CAT), and malondialdehyde (MDA) levels in the gills were performed according to the manufacturer’s instructions. The activities of Na+K+-ATPase, Ca2+-ATPase, and Mg2+-ATPase were determined with an ATPase Assay Kit (Kit No. A016-2-2). All kits were purchased from Nanjing Jiancheng Bioengineering Institute of China (Nanjing, China).
2.6 Gill metabolome analysis
The untargeted metabolomics assay was performed by LC-Bio (Hangzhou, China) with the following procedures: (a) homogenization of 18 fish gills in liquid nitrogen; (b) Metabolite enrichment was performed by methanol-buffer precipitation (50%, v/v). The extracted samples were subjected to random machine sequential testing, and QC samples were inserted before, during and after the samples as a repetitive assessment of the experimental technique. Detection of eluted metabolites in columns by high performance liquid chromatography (HPLC). The data were preprocessed using XCMS (XCMS-v4.7) software. Initial feature annotation via MetaX-driven database comparison (PlantCyc/KEGG/HMDB). Identification of metabolites in product secondary mass spectra using in-house libraries. MetaX software was used to perform univariate and multivariate analyses of the metabolomics data to identify metabolites that were enriched by differences between groups (dm). Differentially abundant features were identified when meeting all criteria: (a) Presence in ≥2 biological replicates or relative abundance ≤50%; (b) Significant inter-group divergence (BH-corrected q < 0.05) via Wilcoxon rank-sum test; (c) OPLS-DA variable importance projection (VIP) score ≥1.0. KEGG pathway enrichment profiling was conducted using a hypergeometric distribution model. Functionally annotated terms with corrected P < 0.05 were designated as significantly enriched clusters for differentially expressed proteins. Gene set enrichment analysis was performed using GSEA (v4.1.0) and MSigDB software programs to determine whether a group of genes were differentially enriched in a specific KEGG pathway.
2.7 Transcriptome analysis
Gill tissue samples from three biological replicates per group were subjected to transcriptome sequencing through Hangzhou Lianchuan Biotechnology Company, Ltd (Hangzhou, China). Total RNA was extracted using TRIzol reagent (TaKaRa, Dalian, China), with RNA integrity verified by Bioanalyzer 2100 Bioanalyzer (RIN > 8.0). First-strand cDNA synthesis was performed using 6-base random hexamer primers with mRNA as template. Second-strand cDNA was generated by adding reaction buffer, RNase H, dNTPs, and DNA polymerase. The products were purified with QIAquick PCR Purification Kit (Qiagen) and eluted in EB buffer. AMPure XP beads (Beckman Coulter) and USER enzyme (NEB) were employed for size selection and degradation of uracil-containing second-strand cDNA, ensuring strand-specificity of the final library. PCR amplification was conducted using Phusion High-Fidelity DNA Polymerase (Thermo Scientific) with indexed primers. Sequencing was performed by LC-Bio Co., Ltd (Hangzhou, China) on Illumina NovaSeq 6000 platform. Then sequence quality was verified using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ , 0.11.9). Genes differential expression analysis was performed by DESeq2 software between two different groups (and by dgeR between two samples). Differentially expressed genes (DEGs) were screened based on DESeq2, with significance thresholds set at |log2(fold change)| ≥ 1 and corrected p value < 0.05. Volcano plots were generated to demonstrate the differential expression distributions by Gplot2, and a hierarchical clustering heatmap was visualized using the pheatmap to visualize hierarchically clustered heatmaps of DEGs. The genes with the parameter of false discovery rate (FDR) below 0.05 and absolute old change ≥ 2 were considered differentially expressed genes. The differentially expressed genes were then analyzed for GO function and KEGG pathway enrichment.
2.8 Gene ontology and enrichment exploration
GO enrichment analysis of DEGs was performed using Wallenius non-central hypergeometric distribution. Pathway enrichment analysis was performed using the KEGG database to further assess significantly enriched metabolic or signaling pathways, with p value < 0.05 being significantly enriched for DGE.
2.9 qRT-PCR validation
Total RNA was isolated from frozen gills using the TRIzol Reagent Kit according to the instructions of the manufacturer and assessed for quality. Samples with A260/A280 RNA ≥ 1.8 were selected for cDNA synthesis according to the manufacturer’s instructions (Beijing Tiangen). The primers used are listed in Supplementary Table S1. Referring to the previous study by Lu et al. (23), specific primers were designed using the online tool Primer 3 plus (https://www.primer3plus.com/) based on the Amur grayling transcriptome sequence (PRJNA907151). qRT-PCR analysis was performed using the SYBR Premix Ex Taq II kit (Tli RNaseH Plus, Takara Bio, Japan) following the manufacturer’s thermal cycling parameters. The β actin served as an internal control to normalize the data. 2-ΔΔCT method was used to calculate the relative expression of target genes (24).
2.10 Statistical analysis
All data are expressed as mean ± standard deviation (SD). One-way statistical analysis of variance (ANOVA) was performed using SPSS 20.0 (SPSS, Chicago, IL, USA). All data were normally distributed and passed the equal variance test. Tukey’s multiple post hoc test, and for the same sampling intervals, different letters indicate that the differences are significant (p < 0.05). Graphical representation of the experimental data was performed using GraphPad Prism 9.0 (GraphPad Software, USA).
3 Results
3.1 Accumulation of Cr in tissues
As shown in Figure 1, Cr6+ accumulated in the gills, liver and intestines of the fish at Days 14 and 28. The level of Cr6+ accumulation in fish increased in a concentration-dependent manner (P < 0.05). The accumulation level of Cr6+ in each tissue was in the order of intestine>liver>gill>muscle. Compared with those in the control group, the Cr6+ accumulation levels in the gill, liver, intestine and muscle in the Hcr group were significantly increased (P < 0.05), whereas Cr6+ accumulation in the muscle did not significantly differ between the Lcr and control groups (P > 0.05).
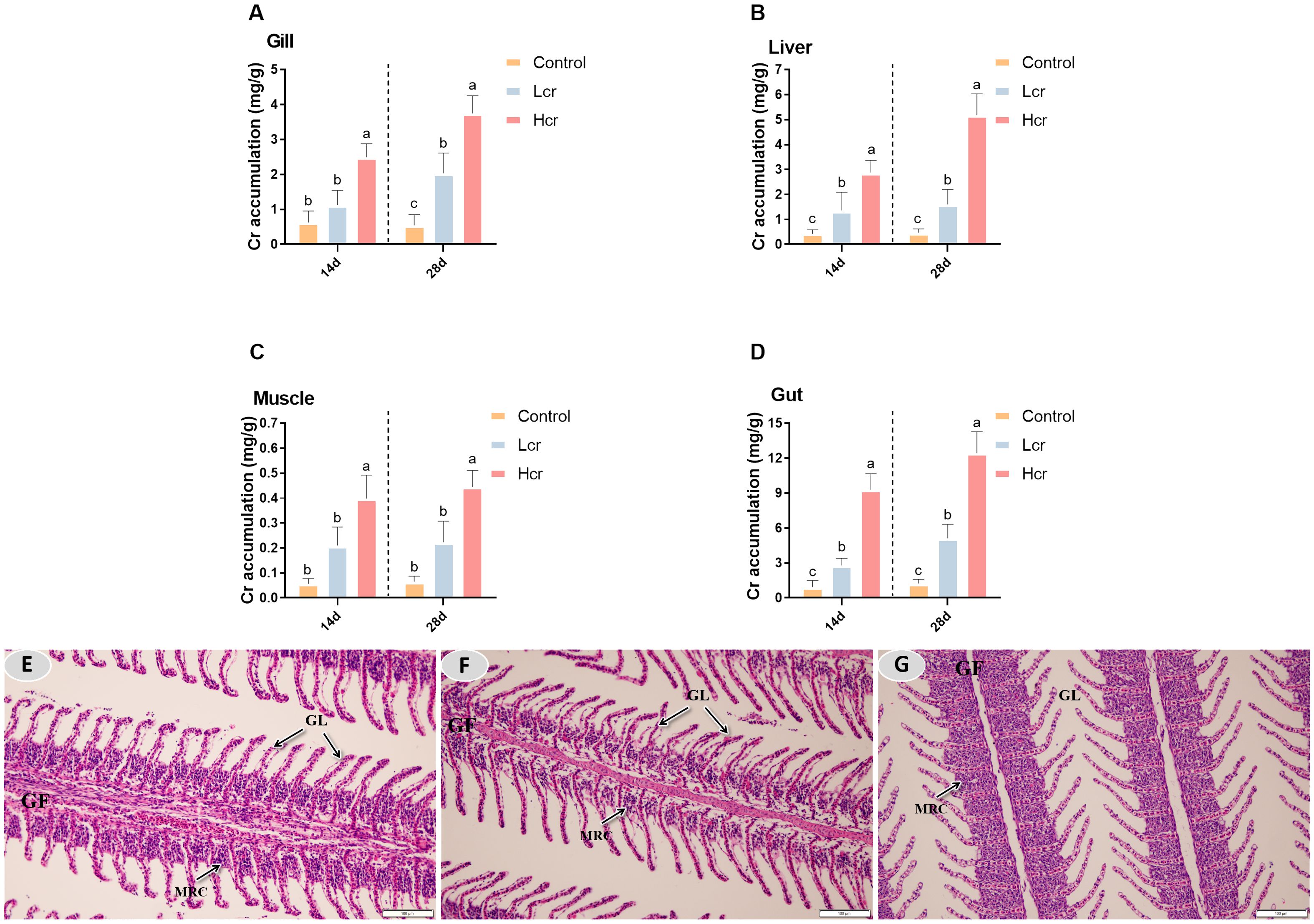
Figure 1. After 14 and 28 days, different levels of Cr6+ accumulated in gills, liver, muscle and intestine of the T. grubii (A-D). The data are expressed as the means ± S.D (n = 6). Bars with different letters are significantly different (P < 0.05) according to Tukey’s test for the same sampling interval. Gill tissue changes in T. grubii after 28 days of Cr6+ exposure (E-G). (E) H&E staining in the control group; scale bars = 100 µm. (F) H&E staining in the Lcr group; scale bars = 100 µm. (G) H&E staining in the Hcr group; scale bars = 50 µm 100 µm. GF (gill filaments), GL (gill lamellae), MRC (mitochondria-rich cell).
3.2 Histological analysis of gill tissues using H&E-stained sections
After 28 days, the gill filaments of the control group remained intact, the gill lamellae were neatly arranged, and complete physiological structures were observed (Figure 1E). Gill filament mitochondria-rich cells (MRCs) were partially vacuolated and gill filament thickness was significantly reduced in the Lcr group compared to the control group (Figure 1F). Compared with those of the control group, the Hcr group gill filaments of MRCs were heavily vacuolated, the gill filament thickness was significantly reduced, gill lamellae were significantly thinner, and epithelium vascular congestion was observed (Figure 1G).
3.3 Exposure to Cr induces changes in enzyme activity
To investigate whether exposure to Cr6+ causes gill damage, we examined the levels of MDA and H2O2 in gill tissue (Figures 2A, B). Compared with the control group, H2O2 levels were significantly greater in both the Lcr and Hcr groups (P < 0.05). Compared with those in the control group, the MDA levels were significantly greater in the Hcr group (P < 0.05). However, there was no significant difference in the MDA levels between the control and Lcr groups (P > 0.05).
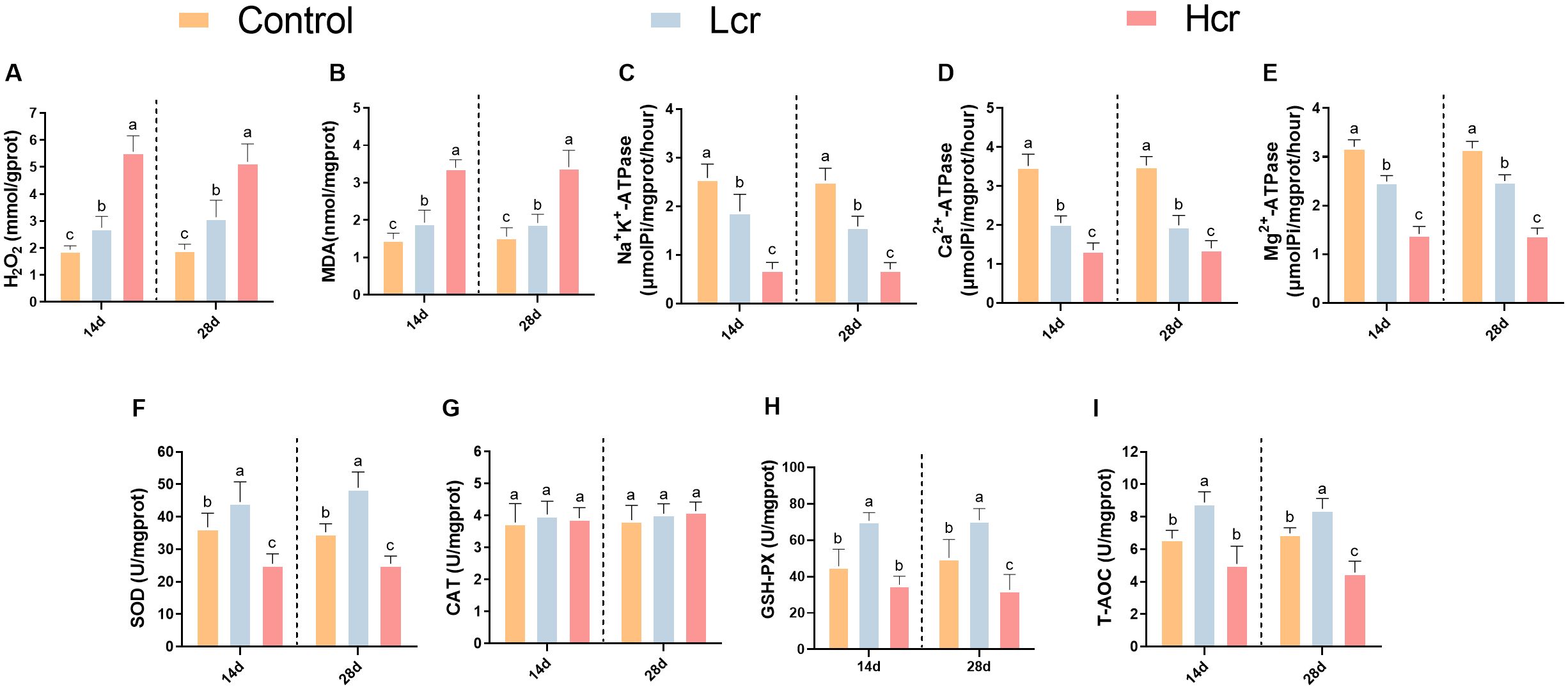
Figure 2. Changes in the gill enzyme activity indices of T. grubii after 14 and 28 days of treatment with different concentrations (0.2 and 1 mg/L) of Cr6+ stress. (A) H2O2; (B) MDA; (C) Na+K+-ATPase; (D) Ca2+-ATPase; (E) Mg2+-ATPase; (F) SOD; (G) CAT; (H) GSH-Px; (I): T-AOC. The data are expressed as the means ± SDs (n=6). Bars with different letters are significantly different (P < 0.05) according to Tukey’s test for the same sampling interval.
We further measured changes in gill enzyme activity indicators using assay kits (Figures 2C-E). Compared with those in the control group, the Na+K+-ATPase, Ca2+-ATPase, and Mg2+-ATPase activities decreased with increasing Cr6+ levels (P < 0.05). Compared with those in the control group (Figures 2F-I), the SOD, GSH-PX, and T-AOC activities increased but then decreased with increasing Cr6+ concentration (P < 0.05). However, the CAT activity did not significantly differ between the groups (P > 0.05).
3.4 Metabolomics analysis of the effects of Cr6+ exposure on gill metabolism
Metabolomics was used to further clarify the negative effects of chronic exposure to Cr6+ on T. grubii gill tissue. The OPLS-DA results revealed significant separation between each Cr6+ treatment group and the control group, suggesting that Cr6+ significantly interfered with the metabolic profile of gill tissues (Figures 3A-C). A permutation test chart was constructed to indicate the degree of model fit. The Q2 values were all <R2 values, and the intercepts of the Q2 values on the y-axis were all <0 (Figures 3D-F), indicating that the OPLS-DA model was not overfitted. Bar charts were constructed to display the number of differentially abundant metabolites (Figure 3G). Supplementary Figure S1. shows heat maps of the top 30 metabolites with the highest differential abundance in the Lcr and C groups. Supplementary Figure S2. shows heatmaps of the top 30 differentially abundant metabolites in the Hcr and C groups. Supplementary Figure S3. shows heat maps of the top 30 metabolites with the highest differential abundance in the Hcr, Lcr, and C groups.
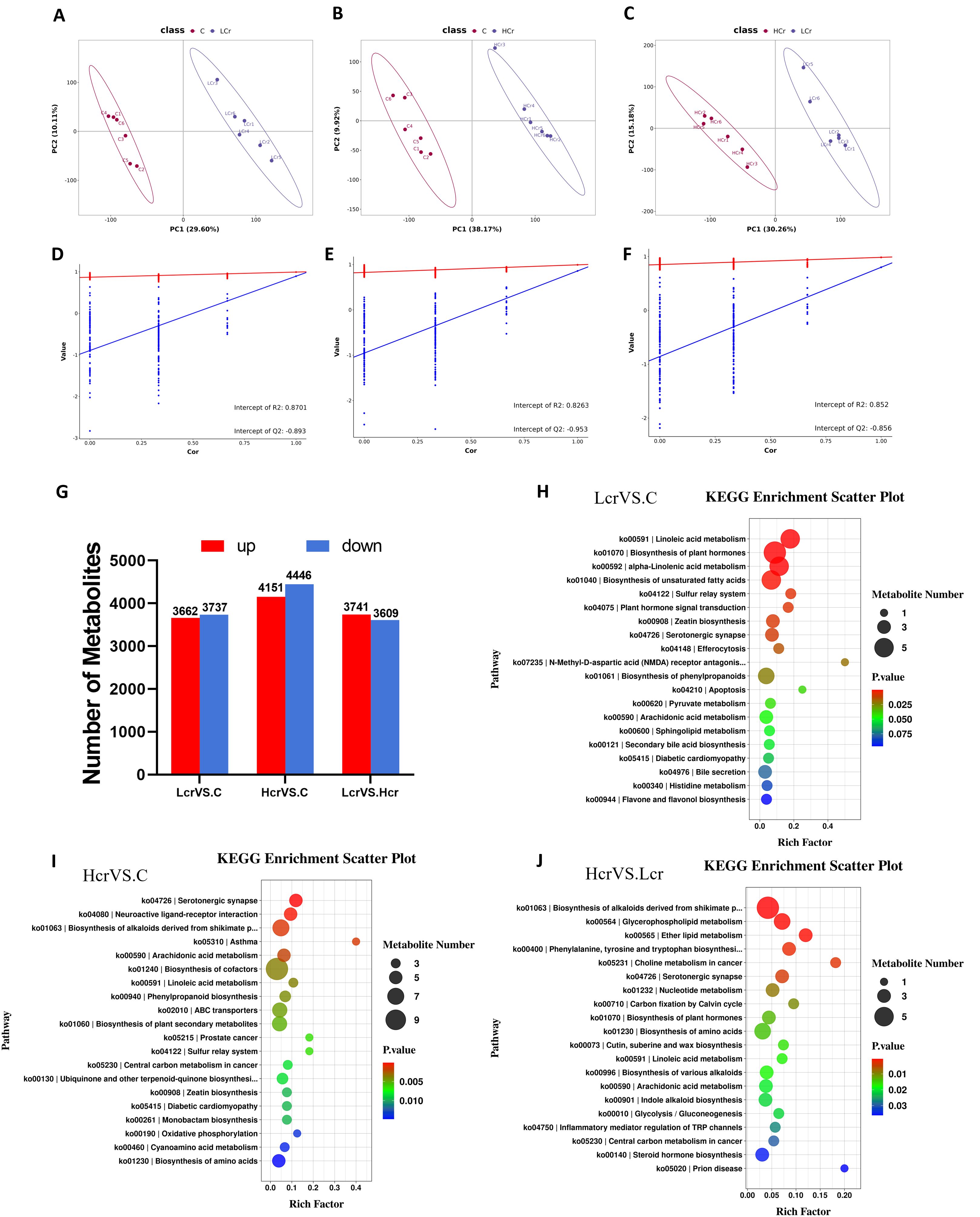
Figure 3. Quality analysis of metabolomics data and KEGG enrichment analysis (n=6). OPLS-DA score plots for metabolites in the C, Lcr and Hcr groups (A–C). OPLS-DA permutation test for the positive and negative ion modes (D) C vs. Lcr; (E) C vs. Hcr; (F) Lcr vs. Hcr). Changes in the total amount of differential metabolites between the two groups (G) KEGG enrichment analysis between the two groups (H) C vs. Lcr; (I) C vs. Hcr; (J) Lcr vs. Hcr).
Enrichment analyses of the KEGG pathway were performed on the differential metabolites (Figures 3H-J). Linoleic acid metabolism, biosynthesis of plant hormones, alpha-linolenic acid metabolism, biosynthesis of plant secondary metabolites, and the sulfur relay system were the five pathways most significantly enriched in the C Vs. Lcr group (Figure 3H). The serotonergic synapse, neuroactive ligand–receptor interaction, biosynthesis of alkaloids derived from the shikimate pathway, asthma, and arachidonic acid metabolism pathways were the five pathways most significantly enriched in the C Vs. Hcr group (Figure 3I). The biosynthesis of alkaloids derived from the shikimate pathway, glycerophospholipid metabolism, ether lipid metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, and choline metabolism in cancer were the five pathways most significantly enriched in the Hcr Vs. Lcr group (Figure 3J).
3.5 Transcriptomic analysis of the effect of Cr6+ exposure on gene expression in fish gills
The levels of DEGs between the different treatment groups are shown in Figure 4, with 107 genes up-regulated and 95 genes down-regulated between the Lcr and C groups after Cr6+ exposure, and 379 genes were up-regulated and 638 genes were down-regulated between the Hcr and C groups. There were 85 genes upregulated and 302 genes downregulated between the Lcr and Hcr groups. Figures 4B-D show volcano plot and heatmap.
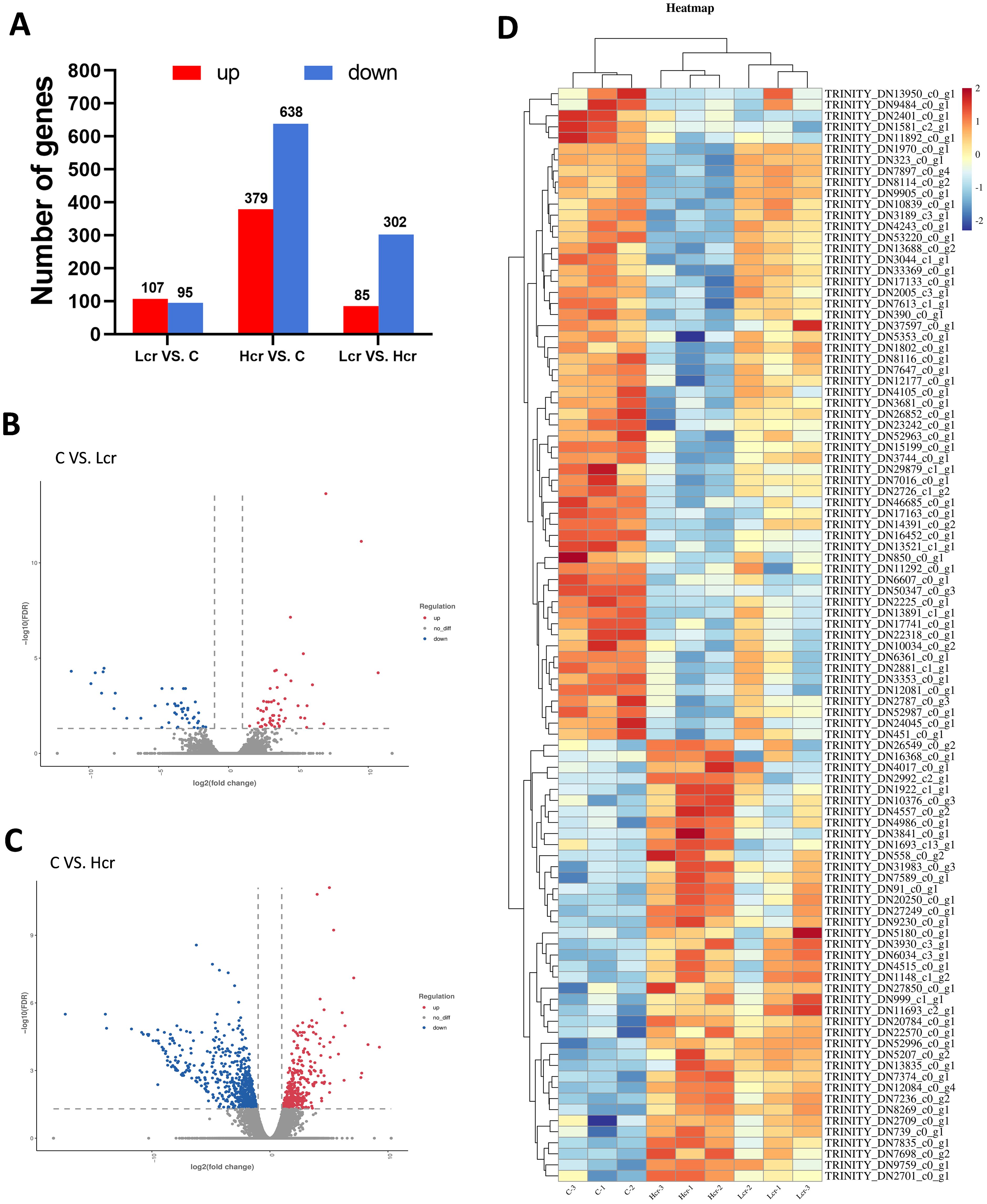
Figure 4. The results of mRNA sequencing. Differentially expressed genes (A), volcano plots (B, C) and heatmaps (D) of the DEGs (P < 0.05). n = 3.
3.6 GO analysis and KEGG pathway enrichment based on DEGs
The results obtained from GO analysis showed that positive regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway, Parkin-FBXW7-Cul1 ubiquitin ligase complex, positive regulation of epidermal growth factor-activated receptor activity, regulation of autophagy of mitochondrion, and positive regulation of ERK1 and ERK2 cascade were the five most significantly enriched terms in the C and Lcr groups (Figure 5A). The sterol biosynthetic process, parkin-cholesterol biosynthetic process, steroid metabolic process, tRNA aminoacylation for protein translation, and endoplasmic reticulum were the five most significantly enriched terms in the C and Hcr groups (Figure 5B). The regulation of dendrite development, dendrite cytoplasm, positive regulation of axon extension, cyclin-dependent protein serine/threonine kinase activity, and solute: proton antiporter activity were the five most significantly enriched terms in the Lcr and Hcr groups (Figure 5C). Supplementary Figure S4. shows the clusters to which each enriched terms in the comparison group belongs.
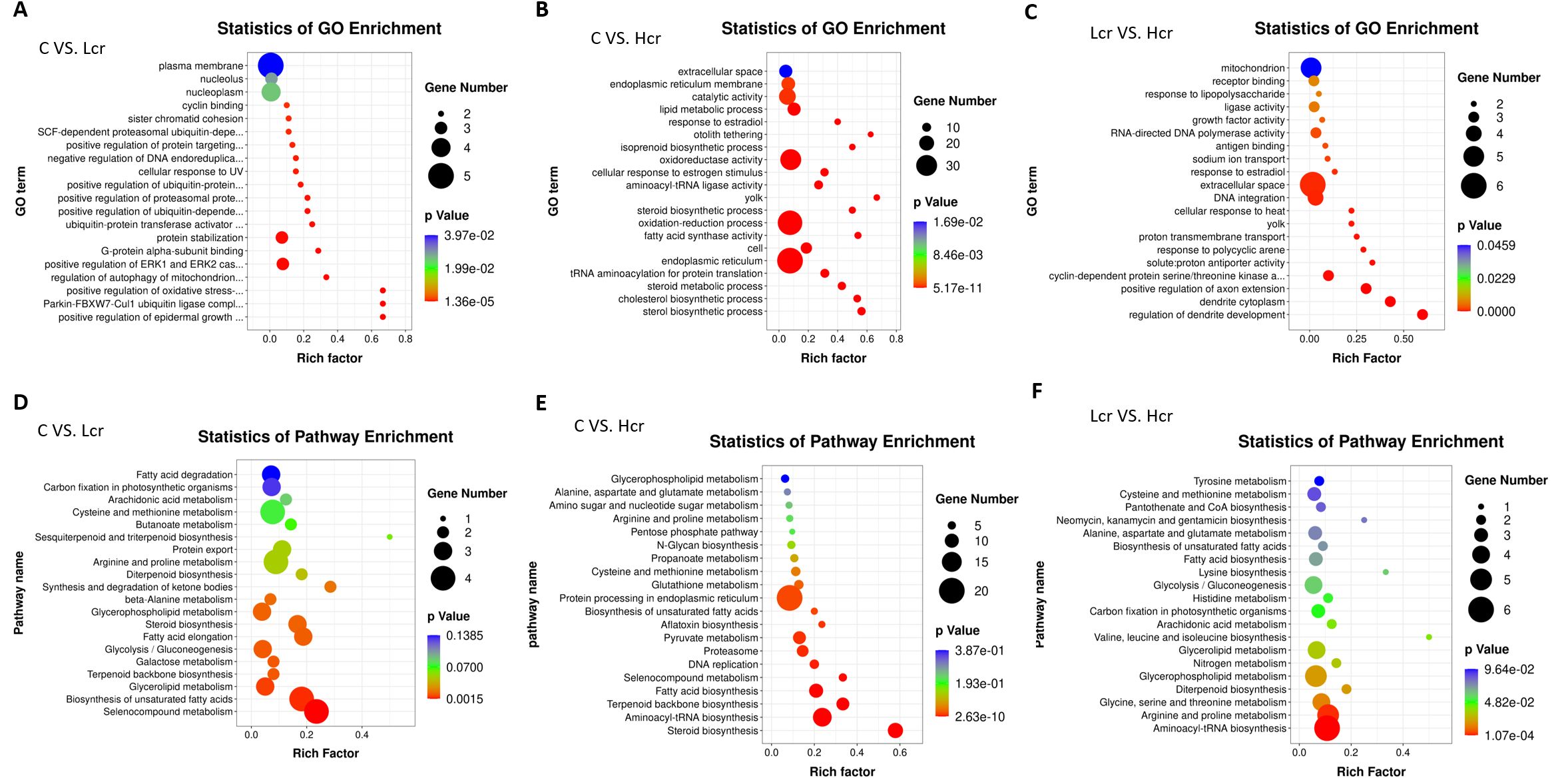
Figure 5. GO enrichment analysis of transcriptomic genes in gills. (A) C vs. Lcr; (B) C vs. Hcr; (C) Lcr vs. Hcr). KEGG enrichment analysis of transcriptomic genes in gills. (D) C vs. Lcr; (E) C vs. Hcr; (F) Lcr vs. Hcr).
This study used the KEGG database to annotate the enrichment pathway of DEGs (Figures 5D-F). The galactose metabolism, steroid biosynthesis, terpenoid backbone biosynthesis, selenocompound metabolism, and biosynthesis of unsaturated fatty acids were the five most significantly enriched signal pathway in the C and Lcr groups. The steroid biosynthesis, aminoacyl-tRNA biosynthesis, fatty acid biosynthesis, proteasome, and pyruvate metabolism were the five most significantly enriched signal pathway in the C and Hcr groups. In addition, signaling pathways such as selenocompound metabolism, aflatoxin biosynthesis, glutathione metabolism, and protein processing in endoplasmic reticulum were similarly significantly enriched. The aminoacyl-tRNA biosynthesis, arginine and proline metabolism, glycine, serine and threonine metabolism, diterpenoid biosynthesis, and glycerophospholipid metabolism were the five most significantly enriched signal pathway in the Lcr and Hcr groups.
3.7 Cr6+ induces changes in the expression of key genes in gills
To further explore the effects of Cr6+-induced oxidative stress on the expression regulation of genes in gill tissues, we validated the key genes by qRT–PCR (Figure 6). Compared with those in the control group, the expression levels of IL-10 and GPx4 were significantly lower, and those of NF-κB, IL-8, and Caspase-9 were significantly greater in the Lcr group (P < 0.05). Compared with those in the control group, the expression levels of PPAR-γ, IL-10, TGF-β and GPx4 were significantly lower, and those of COX-2, NF-κB, IL-8, HSP70, Caspase-3, and Caspase-9 were significantly greater in the Hcr group (P < 0.05).
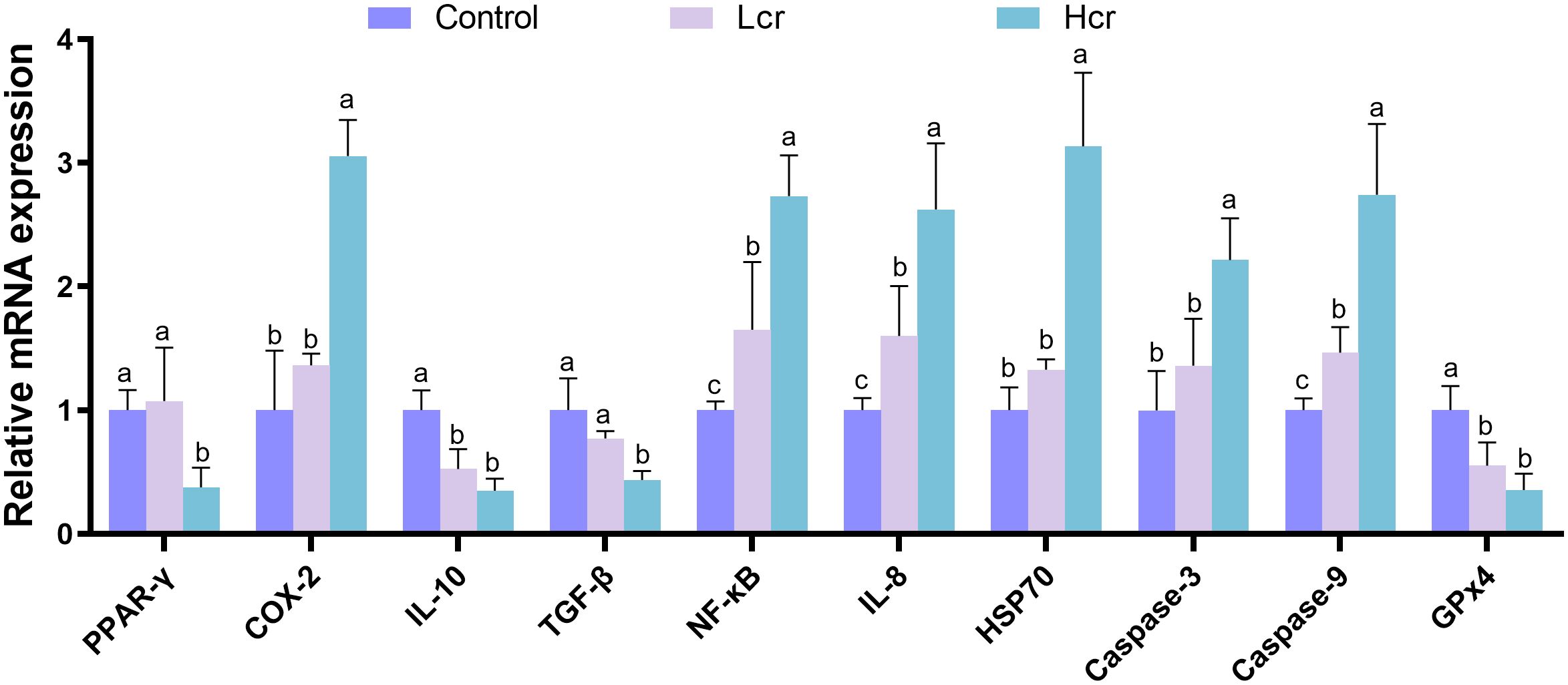
Figure 6. Cr6+ induces changes in the expression of key genes in gills. The data are expressed as the means ± S.Ds. (n = 3). The levels of peroxisome proliferator activated receptor (PPAR)-γ, Cyclooxygenase-2 (COX-2), interleukin 10 (IL-10), transforming growth factor-β (TGF-β), Nuclear factor κB (NF-κB), interleukin 8 (IL-8), 70-kDa heat shock protein (Hsp70), cysteinyl aspartate specific proteinase 3 (Caspase-3), cysteinyl aspartate specific proteinase 9 (Caspase-9), and Glutathione peroxidase 4 (GPx4) were detected. Bars with different letters are significantly different (P < 0.05) according to Tukey’s test for the same sampling interval.
As shown in Figure 7, we selected 10 genes for gill qRT-PCR to verified the reliability and reproducibility of RNA-Seq. The results showed that the upregulation and downregulation trends of the genes in the qRT-PCR results were consistent with the RNA-Seq results, suggesting that the transcriptome sequencing data were reliable.
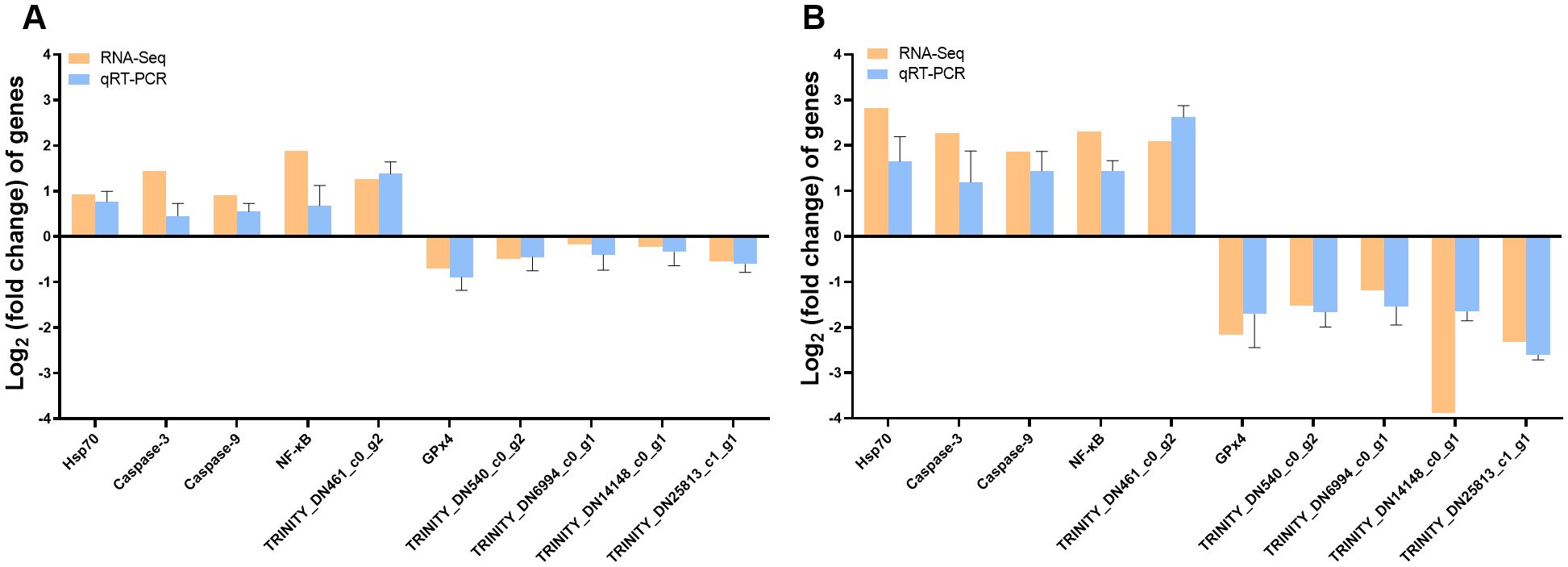
Figure 7. RT–PCR was used to validate transcriptomic gene expression. Comparison of genes for RNA-Seq and qRT–PCR (A) C vs. Lcr; (B) C vs. Hcr).
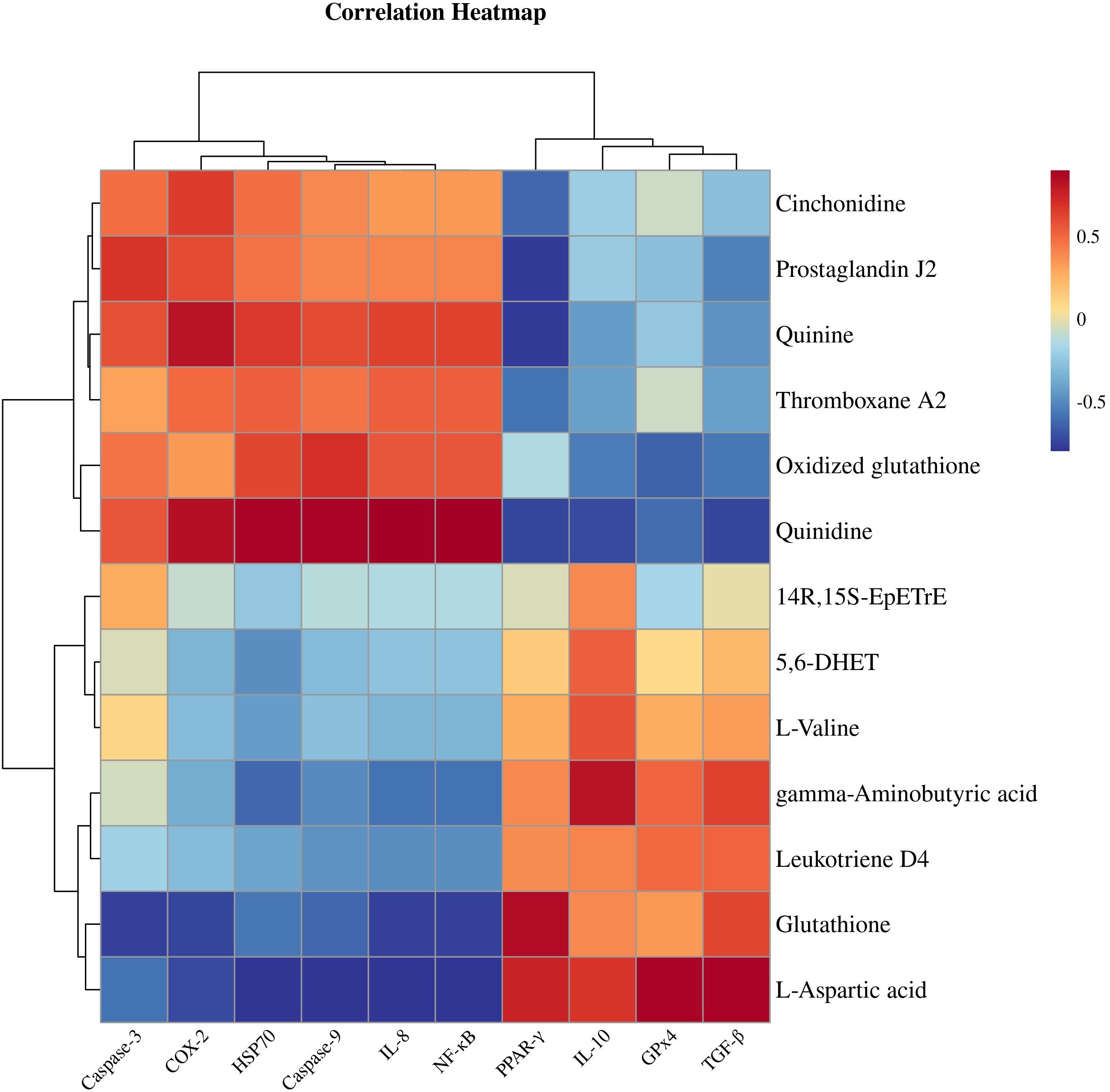
Figure 8 shows a heatmap for gene and metabolite correlation analysis. Correlation analysis in a clustered heatmap revealed that PPAR-γ, IL-10, TGF-β and GPx4 were negatively correlated with the key metabolites cinchonidine, quinidine, quinine, 5-hydroxy-L-tryptophan, thromboxane A2 and prostaglandin J2 and positively correlated with 5,6-DHET, 14R,15S-EpETrE, γ-aminobutyric acid, leukotriene D4, L-aspartic acid, GABA, L- valine, L-tyrosine, and vanillin, whereas COX-2, NF-κB, IL-8, HSP70, Caspase-3, and Caspase-9 were inversely correlated with the above metabolites.
4 Discussion
Aquatic ecosystems are at risk of multiple heavy metal exposures due to increased industrial wastewater discharges and agricultural pollution, of which Cr6+ is a representative persistent pollutant with significant toxic effects on aquatic animals through water column enrichment (1). As a key organ for respiration and ion exchange in fish, the gill is a primary target organ for heavy metal bioaccumulation (25). Therefore, a systematic analysis of the metabolic network remodeling of gill tissues under heavy metal exposure is highly valuable for revealing species-specific detoxification mechanisms and constructing ecological risk threshold models. Hexavalent chromium (CrVI), as a redox-active transition metal, induces ROS-mediated oxidative stress cascades through Fenton-like reactions, specifically depleting reduced glutathione (GSH) while elevating oxidized glutathione (GSSG) ratios (26). In this study, Cr6+ accumulated significantly in all tissues and organs with increasing exposure time. In the present study, Cr6+ accumulated significantly in all tissues and organs with increasing exposure time. Gills were in direct contact with water and were the primary target organ for Cr6+ enrichment. Intestine and liver had high metabolic activities so the Cr6+ accumulation was higher (19, 27). The main reasons for the significantly higher accumulation of Cr6+ in the intestine than in other organs may be twofold: firstly, the relatively limited absorption efficiency of Cr6+ in the intestine leads to the prolonged retention of Cr6+ in the intestinal lumen; secondly, cadmium is able to be transported and redistributed through the trans-organ interactions network (liver-intestinal axis, muscle-intestinal axis, brain-intestinal axis, etc.), which has the intestinal as its core, and ultimately, it forms a high-concentration accumulation in the intestinal tract (17, 28, 29). Two levels of Cr6+ caused damage to the gills of the fish, with the Hcr group being the most severely damaged. We found that Cr6+ can cause gill tissue damage mainly through vacuolization of MRC cells, congestion of epithelial vessels and a significant reduction in gill filament thickness. Previous studies have shown that Cr6+ exposure can lead to oxidative stress in fish through the liver-intestinal axis, resulting in liver damage and gut flora disruption (17). MDA and H2O2 are key indicators used to measure whether tissues are suffering from oxidative damage (30). The present experimental data revealed that Cr6+ exposure induced gill damage, and the concentrations of MDA and H2O2 in gill tissues were significantly and positively correlated with the dose of Cr6+ exposure. Although Cr6+ exposure led to a significant increase in H2O2 levels, CAT activity did not change significantly, probably because the organism had entered an irreversible stage of damage due to chronic Cr6+ stress, which is similar to the results of previous studies (20, 31). SOD, GSH-Px, and T-AOC levels tended to increase but then decreased with increasing Cr6+ content, possibly because T. grubii can adapt to low concentrations of Cr6+ by regulating its metabolism. However, when the Cr6+ concentration exceeded the tolerance threshold, the fish could not adapt to Cr6+ stress through their own metabolism, and the antioxidant defense system was imbalanced, which was similar to the findings of our previous study (17).
Owing to industrial pollution and emissions from agricultural activities, Cr6+ enters water bodies through soil infiltration, seriously threatening the survival of aquatic organisms. Cr6+ can accumulate in aquatic animals to produce toxic effects. In this study, we used a nontargeted metabolomics system to analyze the effects of Cr6+ on the metabolic regulatory network of gill tissues in T. grubii, aiming to elucidate the molecular toxicological mechanisms involved in the induction of oxidative stress, imbalance of lipid metabolism, and cellular inflammatory responses and to provide a theoretical basis for the risk assessment of Cr6+ contamination in aquatic ecosystems. In our study, Cr6+ exposure significantly elevated the metabolites thromboxane A2 and prostaglandin J2. The arachidonic acid metabolic pathway, serotonergic synapses, and neuroactive ligand–receptor interactions are central hubs for the regulation of inflammation, neurotransmitter systems, and cellular homeostasis. Studies have reported that heavy metal exposure can produce an inflammatory response through the metabolism of arachidonic acid to produce excess thromboxane A2 (32, 33). Maria is similar to our results in that activated microglia produce large amounts of PGs after neuronal injury. whereas Prostaglandin J2, the most toxic component of the PGs family, may contribute to many neurodegenerative disorders, PPARγ activation is associated with anti-inflammatory and neuroprotective signaling (34). In addition, cadmium exposure can also lead to significant downregulation of 5,6-DHET and 14R,15S-EpETrE metabolites in rats (35), which is similar to our findings that Cr6+ exposure leads to significant downregulation of these two metabolites. Heavy metals interfere with serotonergic synapses and GAD, reduce γ-aminobutyric acid (GABA) synthesis, lead to neuroexcitotoxicity, and can interact with arachidonic acid metabolism (36, 37). Leukotriene D4 alleviates inflammatory responses by activating the phospholipase C/Ca2+/protein kinase C pathway (38). L-Aspartic acid is another important amino acid that serves as an intermediate in the tricarboxylic acid (TCA) and urea cycles, attenuating external oxidative stress-induced tissue damage and inflammatory responses (39). Gamma-aminobutyric acid (GABA) is also an important amino acid with antioxidant properties that regulates lipid metabolism (40). In our study, chronic Cr6+ exposure resulted in significant increases in the levels of 5-hydroxy-L-tryptophan, thromboxane A2, and prostaglandin J2 and significant decreases in the levels of 5,6-DHET, 14R,15S-EpETrE, GABA, leukotriene D4, L-aspartic acid, and GABA. This suggests that Cr6+ exposure leads to arachidonic acid metabolism disruption and toxic action, whereas elevated levels of 5,6-DHET may be adaptive responses produced by the organism. In our study, the biosynthesis of alkaloids derived from the shikimate pathway was significantly altered, and 5-hydroxy-L-tryptophan, cinchonidine, quinidine, and quinine were significantly elevated, whereas L-alanine, L-tyrosine, and vanillin levels were significantly reduced. Cinchonidine protects against cisplatin-induced oxidative stress by activating the PI3K/AKT pathway (41). In contrast, some drugs, such as quinine and quinidine, have indirect antioxidant effects, thereby mitigating oxidative damage (42, 43). L-Valine promotes fish growth, improves antioxidant capacity and alleviates endoplasmic reticulum stress (44). Thus, the present study demonstrated that chronic Cr6+ exposure leads to disturbances in arachidonic acid metabolism and amino acid metabolism, induces oxidative damage to gill tissues, and regulates redox levels through metabolic reprogramming.
Fish gill tissue is an important osmoregulatory organ (45). When gill tissue is damaged, its osmoregulatory capacity is disrupted, directly affecting ATPase activity and leading to electrolyte imbalance (46). In the present study, we found that Cr6+ exposure resulted that steroid biosynthesis, fatty acid biosynthesis, biosynthesis of unsaturated fatty acids, aflatoxin biosynthesis, and lutathione metabolism signaling pathways were significantly altered. In addition, elevated expression of HSP70, which functions as oxidative stress biomarker, indicated that the cells were in a state of oxidative damage. Studies have shown that heavy metals and physiological stress can induce significant expression of stress proteins such as HSP70 and promote apoptosis by catalyzing Caspase-3 and caspase-9 (47, 48). The experimental data revealed that chronic Cr6+ exposure significantly upregulated the expression of HSP70, activated the apoptotic factors Caspase-3 and Caspase-9 in gill tissues, and inhibited the expression of the key antioxidant regulator GPx4, suggesting that Cr6+ exposure mediates structural damage in gill tissues. The peroxisome proliferator-activated receptor (PPAR) family, classified within the ligand-activated nuclear receptor superfamily, comprises three functionally distinct isoforms designated as α, β/δ, and γ subtypes (49).These nuclear receptors play important roles in metabolism, cell differentiation and inflammatory processes. PPARγ has been shown to be a key factor in the regulation of lipid metabolism and adipocyte differentiation, and increasing evidence suggests that PPARγ exerts anti-inflammatory and neuroprotective functions by regulating the transcription of inflammatory genes (50, 51). In addition, previous studies have found that Cr6+ can cause severe oxidative damage to tissues and organs throughout the body via the blood circulation (52). Alwaili et al. found that Cr6+ exposure inhibits PPARγ expression levels and mediates inflammatory responses in the mouse heart (53). Jin et al. found that Cr6+ exposure inhibited PPARγ expression levels and mediated apoptosis in chicken pancreas (54). Notably, Cr6+ exposure can lead to blood-brain barrier damage and brain-hepatic axis neurotoxicity in zebrafish and snakehead fish (28). The results of the present study showed that chronic Cr6+ exposure resulted in significant inhibition of PPARγ, accompanied by decreases in the expression levels of its downstream anti-inflammatory mediators TGF-β and IL-10. Moreover, Cr6+ promotes the expression of COX-2 and induces the release of the proinflammatory cytokine IL-8 via the NF-κB pathway, which ultimately triggers a cascading inflammatory response. Low PPAR-γ expression deregulates COX-2 inhibition and activates the NF-κB signaling pathway to promote inflammatory progression (55). COX-2 synthesizes and promotes the formation of thromboxane A2 and prostaglandin E2, leading to disorders of arachidonic acid metabolism and causing inflammation and nerve damage (56, 57). Cinchonine inhibits PPARγ expression, downregulates fatty acid synthesis, and modulates inflammatory responses (58). IL-10 and TGF-β1 achieve anti-inflammatory effects through transcriptional silencing of pro-inflammatory cytokines and constitutive inhibition of NF-κB nuclear translocation, thereby attenuating inflammation-driven tissue damage (59, 60). IL-8 can be activated by the NF-κB pathway to promote inflammation (61). These findings are similar to those in our study, where correlation analyses revealed that PPAR-γ was negatively correlated with cinchonidine, quinidine, quinidine, 5-hydroxy-1-tryptophan, thromboxane A2, and prostaglandin J2, whereas the anti-inflammatory factors IL-10, TGF-β, and GPx4 were downregulated, and proinflammatory factors were upregulated. However, COX-2 showed the opposite trends of correlation with the above metabolites. GABA exerts anxiolytic effects as a neurotransmitter while inhibiting the release of proinflammatory factors and regulating lipid metabolism by enhancing PPAR-γ (6, 62). In the present study, we analyzed the adverse effects of Cr6+ exposure on gill tissues by integrating transcriptomics and metabolomics. We found that Cr6+ exposure significantly led to increased glutathione depletion, resulting in decreased GPX4 expression levels. In addition, Cr6+ exposure significantly inhibited the expression level of PPAR-γ, a key factor in lipid metabolism, and induced lipid metabolism disorders, which further activated the NF-κB pro-inflammatory signaling pathway to aggravate the release of inflammatory factors and triggered the Caspase-3 cascade reaction to trigger apoptosis. In this study, we confirmed the mechanism of action of Cr6+ exposure in inducing gill toxicity in Thymallus grubii. Cr6+ accumulated in tissues such as intestine, liver and gill, and these results highlight the multi organ distribution pattern of Cr6+. Based on these results, we will systematically investigate the cross-organ interaction network of Cr6+ to elucidate the mechanism of Cr6+ induced oxidative stress cascade in aquatic animals.
5 Conclusion
In conclusion, this study elucidates for the first time the mechanism of gill tissue damage caused by Cr6+ in aquatic organisms. The data suggest that Cr6+ exposure disrupts the arachidonic acid metabolic pathway by inactivating PPAR-γ to deregulate COX-2, leading to oxidative stress and inflammatory responses. Disturbances in arachidonic acid metabolism led to the release of proinflammatory mediators such as thromboxane A2 and prostaglandin J2, which are accompanied by the accumulation of oxidized glutathione. PPAR-γ activity inhibition leads to blocked synthesis of metabolites with anti-inflammatory/antioxidant functions, such as GABA, quinidine, and L-aspartic acid. Therefore, targeted activation of PPAR-γ combined with COX-2 inhibitors could be a potential intervention strategy to reverse Cr6+ toxicity (Figure 9).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics statement
The animal study was approved by This study was approved by the Ethics Committee for Animal Experiments of the Heilongjiang Fisheries Research Institute, Chinese Academy of Fisheries Sciences (20241125–007). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XS: Conceptualization, Data curation, Funding acquisition, Investigation, Resources, Software, Visualization, Writing – original draft, Writing – review & editing. XC: Conceptualization, Data curation, Investigation, Methodology, Software, Supervision, Writing – original draft, Writing – review & editing. KM: Conceptualization, Investigation, Software, Writing – review & editing. BM: Data curation, Methodology, Validation, Writing – original draft. HS: Conceptualization, Formal analysis, Project administration, Writing – review & editing. WW: Data curation, Formal analysis, Project administration, Writing – review & editing. HH: Methodology, Project administration, Supervision, Writing – review & editing. MX: Formal analysis, Project administration, Visualization, Writing – original draft. WX: Data curation, Project administration, Validation, Writing – original draft. YZ: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Resources, Software, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The work was supported by the Central Public-interest Scientific Institution Basal Research Fund, HRFRI, (No. HSY202513Q); Agricultural Finance Special Project of the Ministry of Agriculture and Rural Affairs, “Normalization Monitoring of Fisheries Resources and Environment in Key Waters of Northeast China”; The Central Public-interest Scientific Institution Basal Research Fund, HRFRI (NO. HSY202414Q); The New Era Longjiang Outstanding Master’s and Doctoral Thesis Project Grant (LJYXL2022-085); Identification and evaluation of characteristic Aquatic Animal Germplasm Resources and creation of improved varieties in Heilongjiang Province; The Central Public-interest Scientific Institution Basal Research Fund, CAFS (2023TD59); Technology Collaborative Innovation and Extension System for the Freshwater Fish Industry of Heilongjiang Province.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Correction note
A correction has been made to this article. Details can be found at: 10.3389/fimmu.2025.1692659.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fimmu.2025.1633174/full#supplementary-material
References
1. RoyChowdhury A, Datta R, and Sarkar D. Heavy metal pollution and remediation. Green Chem. (2018), 359–73. doi: 10.1016/B978-0-12-809270-5.00015-7
2. Barrera-Díaz CE, Lugo-Lugo V, and Bilyeu B. A review of chemical, electrochemical and biological methods for aqueous Cr (VI) reduction. . J Hazard Mater. (2012) 223:1–12. doi: 10.1016/j.jhazmat.2012.04.054
3. Mortada WI, El-Naggar A, Mosa A, Palansooriya KN, Yousaf B, Tang R, et al. Biogeochemical behaviour and toxicology of chromium in the soil-water-human nexus: A review. Chemosphere. (2023) 331:138804. doi: 10.1016/j.chemosphere.2023.138804
4. Salnikow K and Zhitkovich A. Genetic and epigenetic mechanisms in metal carcinogenesis and cocarcinogenesis: nickel, arsenic, and chromium. Chem Res Toxicol. (2008) 21:28–44. doi: 10.1021/tx700198a
5. Loyaux-Lawniczak S, Lecomte P, and Ehrhardt JJ. Behavior of hexavalent chromium in a polluted groundwater: redox processes and immobilization in soils. Environ Sci Technol. (2001) 35:1350–7. doi: 10.1021/es001073l
6. Kim K and Yoon H. Gamma-aminobutyric acid signaling in damage response, metabolism, and disease. Int J Mol Sci. (2023) 24:4584. doi: 10.3390/ijms24054584
7. Emmanouil C, Sheehan TMT, and Chipman JK. Macromolecule oxidation and DNA repair in mussel (Mytilus edulis L.) gill following exposure to Cd and Cr (VI). Aquat Toxicol. (2007) 82:27–35. doi: 10.1016/j.aquatox.2007.01.009
8. Wang XF, Lou XM, Shen Y, Xing ML, and Xu LH. Apoptotic-related protein changes induced by hexavalent chromium in mice liver. Environ Toxicol. (2010) 25:77–82. doi: 10.1002/tox.20478
9. Kopec AK, Thompson CM, Kim S, Forgacs AL, and Zacharewski TR. Comparative toxicogenomic analysis of oral Cr (VI) exposure effects in rat and mouse small intestinal epithelia. Toxicol Appl Pharm. (2012) 262:124–38. doi: 10.1016/j.taap.2012.04.026
10. Zhitkovich A. Chromium in drinking water: sources, metabolism, and cancer risks. Chem Res Toxicol. (2011) 24:1617–29. doi: 10.1021/tx200251t
11. Lou J, Jin L, Wu N, Tan Y, Song Y, Gao M, et al. DNA damage and oxidative stress in human B lymphoblastoid cells after combined exposure to hexavalent chromium and nickel compounds. Food Chem Toxicol. (2013) 55:533–40. doi: 10.1016/j.fct.2013.01.053
12. Li Z, Shah SWA, Zhou Q, Yin X, and Teng X. The contributions of miR-25-3p, oxidative stress, and heat shock protein in a complex mechanism of autophagy caused by pollutant cadmium in common carp (Cyprinus carpio L.) hepatopancreas. Environ pollut. (2021) 287:117554. doi: 10.1016/j.envpol.2021.117554
13. Cappello T, Brandão F, Guilherme S, Santos MA, Maisano M, Mauceri A, et al. Insights into the mechanisms underlying mercury-induced oxidative stress in gills of wild fish (Liza aurata) combining 1H NMR metabolomics and conventional biochemical assays. Sci Total Environ. (2016) 548:13–24. doi: 10.1016/j.scitotenv.2016.01.008
14. Garcia-Reyero N, Thornton C, Hawkins AD, Escalon L, Kennedy AJ, Steevens JA, et al. Assessing the exposure to nanosilver and silver nitrate on fathead minnow gill gene expression and mucus production. Environ nanotechnology Monit management. (2015) 4:58–66. doi: 10.1016/j.enmm.2015.06.001
15. Shobana C, Rangasamy B, Poopal RK, Renuka S, and Ramesh M. Green synthesis of silver nanoparticles using Piper nigrum: tissue-specific bioaccumulation, histopathology, and oxidative stress responses in Indian major carp Labeo rohita. Environ Sci pollut R. (2018) 25:11812–32. doi: 10.1007/s11356-018-1454-z
16. Wang G, Zhang C, and Huang B. Transcriptome analysis and histopathological observations of Geloina erosa gills upon Cr (VI) exposure. Comp Biochem Phys C. (2020) 231:108706. doi: 10.1016/j.cbpc.2020.108706
17. Shang X, Che X, Ma K, Guo W, Wang S, Sun ZP, et al. Chronic Cr (VI) exposure-induced biotoxicity involved in liver microbiota-gut axis disruption in Phoxinus lagowskii Dybowski based on multi-omics technologies. Environ pollut. (2025) 368:125759. doi: 10.1016/j.envpol.2025.125759
18. Zhai C, Li Y, Zhu G, Peng W,EQ, Zhang Y, and Ma B. The effects of water flow speed on swimming capacity and energy metabolism in adult amur grayling (Thymallus grubii). Fishes. (2024) 9:272. doi: 10.3390/fishes9070272
19. Yu Z, Xu SF, Zhao JL, Zhao L, Zhang AZ, and Li MY. Toxic effects of hexavalent chromium (Cr6+) on bioaccumulation, apoptosis, oxidative damage and inflammatory response in Channa asiatica. Environ Toxicol Phar. (2021) 87:103725. doi: 10.1016/j.etap.2021.103725
20. Zhao L, Zhang Y, Yu J, Liu YH, Cai ZH, Li MY, et al. Astaxanthin ameliorates Cr (VI) accumulation, antioxidant, digestive, apoptosis and inflammatory response. Reg Stud Mar Sci. (2025) 87:104240. doi: 10.1016/j.rsma.2025.104240
21. Kamila S, Shaw P, Islam S, and Chattopadhyay A. Ecotoxicology of hexavalent chromium in fish: An updated review. Sci Total Environ. (2023) 890:164395. doi: 10.1016/j.scitotenv.2023.164395
22. Shang X, Geng L, Zhao Z, Luo L, Shi X, Zhang Q, et al. Transcriptomics reveals the mechanism of selenium-enriched Lactobacillus plantarum alleviating brain oxidative stress under cadmium stress in Luciobarbus capito. Ecotox Environ Safe. (2022) 242:113890. doi: 10.1016/j.ecoenv.2022.113890
23. Lu S, Wang CA, Liu Y, Liu B, Zhang Y, Shi H, et al. Effects of dietary fish oil levels on growth performance, lipid metabolism, hepatic health, nonspecific immune response, and intestinal microbial community of juvenile amur grayling (Thymallus grubii). Aquacult Nutr. (2024) 2024:8587410. doi: 10.1155/anu/8587410
24. Pfaffl MW. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. (2001) 29:e45–5. doi: 10.1093/nar/29.9.e45
25. Sharma AK, Sharma M, Sharma S, Malik DS, Sharma M, and Sharma AK. A systematic review on assessment of heavy metals toxicity in freshwater fish species: current scenario and remedial approaches. J Geochem Explor. (2024) 107472. doi: 10.1016/j.gexplo.2024.107472
26. Samrana S, Ali A, Muhammad U, Azizullah A, Ali H, Khan M, et al. Physiological, ultrastructural, biochemical, and molecular responses of glandless cotton to hexavalent chromium (Cr6+) exposure. Environ pollut. (2020) 266:115394. doi: 10.1016/j.envpol.2020.115394
27. Zhao L, Duan H, Liu Y, Wang Y, Li M, and Li M. Long-term exposure of zebrafish (Danio rerio) to Cr (VI): reproductive toxicity and neurotoxicity. Reg Stud Mar Sci. (2024) 74:103559. doi: 10.1016/j.rsma.2024.103559
28. Li MY, Shi YC, Xu WX, Zhao L, and Zhang AZ. Exploring Cr (VI)-induced blood-brain barrier injury and neurotoxicity in zebrafish and snakehead fish, and inhibiting toxic effects of astaxanthin. Environ pollut. (2024) 355:124280. doi: 10.1016/j.envpol.2024.124280
29. Breit S, Kupferberg A, Rogler G, and Hasler G. Vagus nerve as modulator of the brain–gut axis in psychiatric and inflammatory disorders. Front Psychiatry. (2018) 9:44. doi: 10.3389/fpsyt.2018.00044
30. Cui J, Liu Y, Hao Z, Liu Y, Qiu M, Kang L, et al. Cadmium induced time-dependent kidney injury in common carp via mitochondrial pathway: Impaired mitochondrial energy metabolism and mitochondrion-dependent apoptosis. Aquat Toxicol. (2023) 261:106570. doi: 10.1016/j.aquatox.2023.106570
31. Atli G, Alptekin Ö, Tükel S, and Canli M. Response of catalase activity to Ag+, Cd2+, Cr6+, Cu2+ and Zn2+ in five tissues of freshwater fish Oreochromis niloticus. Comp Biochem Phys C. (2006) 143:218–24. doi: 10.1016/j.cbpc.2006.02.003
32. Nanji AA, Khwaja S, Rahemtulla A, Miao L, Zhao S, and Tahan SR. Thromboxane inhibitors attenuate pathological changes in alcoholic liver disease in the rat. Gastroenterology. (1997) 112:200–7. doi: 10.1016/S0016-5085(97)70236-1
33. Stachowicz K. Regulation of COX-2 expression by selected trace elements and heavy metals: Health implications, and changes in neuronal plasticity. A review. J Trace Elem Med Bio. (2023) 79:127226. doi: 10.1016/j.jtemb.2023.127226
34. Figueiredo-Pereira ME, Corwin C, and Babich J. Prostaglandin J2: A potential target for halting inflammation-induced neurodegeneration. Ann Ny Acad Sci. (2016) 1363:125–37. doi: 10.1111/nyas.12987
35. Zeng T, Guo W, Jiang L, Luo Q, Shi Z, Lei B, et al. Integration of omics analysis and atmospheric pressure MALDI mass spectrometry imaging reveals the cadmium toxicity on female ICR mouse. Sci Total Environ. (2021) 801:149803. doi: 10.1016/j.scitotenv.2021.149803
36. Tan TH, Li SW, Chang CW, Chen YC, Liu YH, Ma JT, et al. Rat hair metabolomics analysis reveals perturbations of unsaturated fatty acid biosynthesis, phenylalanine, and arachidonic acid metabolism pathways are associated with amyloid-β-induced cognitive deficits. Mol Neurobiol. (2023) 60:4373–95. doi: 10.1007/s12035-023-03343-6
37. Luo H, Liu Z, Xie F, Bilal M, Liu L, Yang R, et al. Microbial production of gamma-aminobutyric acid: applications, state-of-the-art achievements, and future perspectives. Crit Rev Biotechnol. (2021) 41:491–512. doi: 10.1080/07388551.2020.1869688
38. Rodríguez-Lagunas MJ, Storniolo CE, Ferrer R, and Moreno JJ. 5-Hydroxyeicosatetraenoic acid and leukotriene D4 increase intestinal epithelial paracellular permeability. Int J Biochem Cell Biol. (2013) 45:1318–26. doi: 10.1016/j.biocel.2013.04.005
39. Zeng Y, Deng B, Kang Z, Araujo P, Mjøs SA, Liu R, et al. Tissue accumulation of polystyrene microplastics causes oxidative stress, hepatopancreatic injury and metabolome alterations in Litopenaeus vannamei. Ecotox Environ Safe. (2023) 256:114871. doi: 10.1016/j.ecoenv.2023.114871
40. Xie Z, Xia S, and Le GW. Gamma-aminobutyric acid improves oxidative stress and function of the thyroid in high-fat diet fed mice. J Funct Foods. (2014) 8:76–86. doi: 10.1016/j.jff.2014.03.003
41. Dong W, Jiang Y, Yao Q, Xu M, Jin Y, Dong L, et al. Inhibition of CISD1 attenuates cisplatin-induced hearing loss in mice via the PI3K and MAPK pathways. Biochem Pharmacol. (2024) 223:116132. doi: 10.1016/j.bcp.2024.116132
42. Chattopadhyay A, Biswas S, Bandyopadhyay D, Sarkar C, and Datta AG. Effect of isoproterenol on lipid peroxidation and antioxidant enzymes of myocardial tissue of mice and protection by quinidine. Mol Cell Biochem. (2003) 245:43–9. doi: 10.1023/A:1022808224917
43. Achan J, Talisuna AO, Erhart A, Yeka A, Tibenderana JK, Baliraine FN, et al. Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malaria J. (2011) 10:1–12. doi: 10.1186/1475-2875-10-144
44. Cao Q, Zhao J, He L, Zhang T, Feng L, Jiang W, et al. Evaluation of the dietary L-valine on fish growth and intestinal health after infection with Aeromonas veronii. Aquaculture. (2024) 580:740294. doi: 10.1016/j.aquaculture.2023.740294
45. Karnaky KJ. Structure and function of the chloride cell of Fundulus heteroclitus and other teleosts. Am Zool. (1986) 26:209–24. doi: 10.1093/icb/26.1.209
46. Apiamu A, Osawaru SU, Asagba SO, Evuen UF, and Achuba FI. Exposure of African catfish (Clarias gariepinus) to lead and zinc modulates membrane-bound transport protein: A plausible effect on Na+/K+-ATPase activity. Biol Trace Elem Res. (2022) 200:4160–70. doi: 10.1007/s12011-021-03005-5
47. Rajeshkumar S, Mini J, and Munuswamy N. Effects of heavy metals on antioxidants and expression of HSP70 in different tissues of Milk fish (Chanos chanos) of Kaattuppalli Island, Chennai, India. Ecotox Environ Safe. (2013) 98:8–18. doi: 10.1016/j.ecoenv.2013.07.029
48. Wang B, Feng L, Jiang WD, Wu P, Kuang SY, Jiang J, et al. Copper-induced tight junction mRNA expression changes, apoptosis and antioxidant responses via NF-κB, TOR and Nrf2 signaling molecules in the gills of fish: preventive role of arginine. Aquat Toxicol. (2015) 158:125–37. doi: 10.1016/j.aquatox.2014.10.025
49. Culman J, Zhao Y, Gohlke P, and Herdegen T. PPAR-γ: therapeutic target for ischemic stroke. Trends Pharmacol Sci. (2007) 28:244–9. doi: 10.1016/j.tips.2007.03.004
50. Zhang H, Li Y, Fan Y, Wu J, Zhao B, Guan Y, et al. Klotho is a target gene of PPAR-γ. Kidney Int. (2008) 74:732–9. doi: 10.1038/ki.2008.244
51. Ryan KK, Li B, Grayson BE, Matter EK, Woods SC, and Seeley RJ. A role for central nervous system PPAR-γ in the regulation of energy balance. Nat Med. (2011) 17:623–6. doi: 10.1038/nm.2349
52. Mishra AK and Mohanty B. Acute toxicity impacts of hexavalent chromium on behavior and histopathology of gill, kidney and liver of the freshwater fish, Channa punctatus (Bloch). Environ Toxicol Phar. (2008) 26:136–41. doi: 10.1016/j.etap.2008.02.010
53. Alwaili MA, Elhoby AH, El-Sayed NM, Mahmoud IZ, Alharthi A, El-Nablaway M, et al. Cardioprotective effects of α-asarone against hexavalent chromium-induced oxidative damage in mice. Drug Des Dev Ther. (2024), 383–3397. doi: 10.2147/DDDT.S46433
54. Jin X, Jia T, Liu R, and Xu S. The antagonistic effect of selenium on cadmium-induced apoptosis via PPAR-γ/PI3K/Akt pathway in chicken pancreas. J Hazard Mater. (2018) 357:355–62. doi: 10.1016/j.jhazmat.2018.06.003
55. Du H, Chen X, Zhang J, and Chen C. Inhibition of COX-2 expression by endocannabinoid 2-arachidonoylglycerol is mediated via PPAR-γ. Brit J Pharmacol. (2011) 163:1533–49. doi: 10.1111/j.1476-5381.2011.01444.x
56. Ammu VRK, Garikapati KK, Krishnamurthy PT, Chintamaneni PK, and Pindiprolu SKS. Possible role of PPAR-γ and COX-2 receptor modulators in the treatment of Non-Small Cell lung carcinoma. Med Hypotheses. (2019) 124:98–100. doi: 10.1016/j.mehy.2019.02.024
57. Cheng H, Huang H, Guo Z, Chang Y, and Li Z. Role of prostaglandin E2 in tissue repair and regeneration. Theranostics. (2021) 11:8836. doi: 10.7150/thno.63396
58. Jung SA, Choi M, Kim S, Yu R, and Park T. Cinchonine prevents high-fat-diet-induced obesity through downregulation of adipogenesis and adipose inflammation. PPAR Res. (2012) 2012:541204. doi: 10.1155/2012/541204
59. Bao Y, Shen Y, Li X, Wu Z, Jiao L, Li J, et al. A new insight into the underlying adaptive strategies of euryhaline marine fish to low salinity environment: through cholesterol nutrition to regulate physiological responses. Front Nutr. (2022) 9:855369. doi: 10.3389/fnut.2022.855369
60. Reyes-Cerpa S, Reyes-López F, Toro-Ascuy D, Montero R, Maisey K, Acuña-Castillo C, et al. Induction of anti-inflammatory cytokine expression by IPNV in persistent infection. Fish Shellfish Immun. (2014) 41:172–82. doi: 10.1016/j.fsi.2014.08.029
61. Phuagkhaopong S, Ospondpant D, Kasemsuk T, Sibmooh N, Soodvilai S, Power C, et al. Cadmium-induced IL-6 and IL-8 expression and release from astrocytes are mediated by MAPK and NF-κB pathways. Neurotoxicology. (2017) 60:82–91. doi: 10.1016/j.neuro.2017.03.001
Keywords: gill, metabolome, transcriptome, Cr(VI) stress, inflammatory responses
Citation: Shang X, Che X, Ma K, Ma B, Sun H, Wu W, He H, Xing M, Xu W and Zhang Y (2025) Chronic hexavalent chromium exposure induces oxidative stress-mediated molecular cascades in Thymallus grubii gills: evidence from integrated transcriptomics and metabolomics. Front. Immunol. 16:1633174. doi: 10.3389/fimmu.2025.1633174
Received: 22 May 2025; Accepted: 25 June 2025;
Published: 09 July 2025; Corrected: 04 September 2025.
Edited by:
Doaa M Mokhtar, Assiut University, EgyptReviewed by:
Muyang Li, Heilongjiang Bayi Agricultural University, ChinaKesavan M, SRM UNIVERSITY, India
Copyright © 2025 Shang, Che, Ma, Ma, Sun, Wu, He, Xing, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongquan Zhang, emhhbmd5b25ncXVhbkBocmZyaS5hYy5jbg==
†These authors have contributed equally to this work
 Xinchi Shang
Xinchi Shang Xinghua Che1,3†
Xinghua Che1,3†