- 1IBD Center, Department of Gastroenterology, IRCCS Humanitas Research Hospital, Milan, Italy
- 2Department of Biomedical Sciences, Humanitas University, Milan, Italy
- 3Gastroenterology, Department of Public Health, University of Naples Federico II, Naples, Italy
- 4Gastroenterology Department, Hospital Clinico Universitario de Santiago de Compostela, Santiago de Compostela, Spain
- 5Fundacion Instituto de Investigacion Sanitaria de Santiago (IDIS), Santiago de Compostela, Spain
- 6Radiology Department, IRCCS Humanitas Research Hospital, Milan, Italy
Background and aims: Artificial intelligence (AI) is rapidly gaining traction in gastroenterology, particularly in the management of inflammatory bowel disease (IBD). Given the complexity of IBD care, AI offers the potential to enhance diagnosis, monitoring, and treatment. This review aims to summarize recent developments in AI applications for IBD and identify key challenges and opportunities for future research and clinical implementation.
Methods: A narrative literature review was conducted, incorporating recent studies utilizing AI —including machine learning (ML) and deep learning (DL) — across various aspects of IBD care.
Results: AI has demonstrated utility in multiple domains of IBD management, including endoscopic disease activity assessment, histological evaluation, imaging interpretation, prediction of disease course, treatment response, and real-world data integration. Despite promising accuracy and utility, most models remain in early development stages and lack widespread clinical validation. Major barriers include data heterogeneity, limited generalizability, and regulatory uncertainties.
Conclusion: AI has significant potential to revolutionize IBD care. Continued multidisciplinary collaboration, validation in diverse clinical settings, and integration into clinical workflows are critical for realizing its full impact.
1 Introduction
Inflammatory Bowel Disease (IBD), encompassing Crohn’s disease (CD) and ulcerative colitis (UC), is a chronic, immune-mediated and relapsing condition that significantly impacts on patients’ quality of life (1, 2). IBD is a multifactorial disease influenced by genetic, immune, and environmental factors. While UC typically presents with bloody diarrhea, CD is more often associated with watery diarrhea and nonspecific symptoms. Diagnosis relies primarily on colonoscopy with terminal ileum intubation. Due to their low sensitivity and specificity, laboratory tests are mainly supportive in ambiguous cases. Among imaging techniques, magnetic resonance enterography (MRE) is preferred for its enhanced sensitivity in perianal disease and its avoidance of radiation exposure (3).
Despite significant advances in therapeutics, accurate disease monitoring and personalized treatment strategies remain major challenges in clinical practice. Patients frequently encounter unpredictable disease flares, variable responses to therapy, and a substantial burden related to frequent clinical visits and invasive monitoring techniques (4).
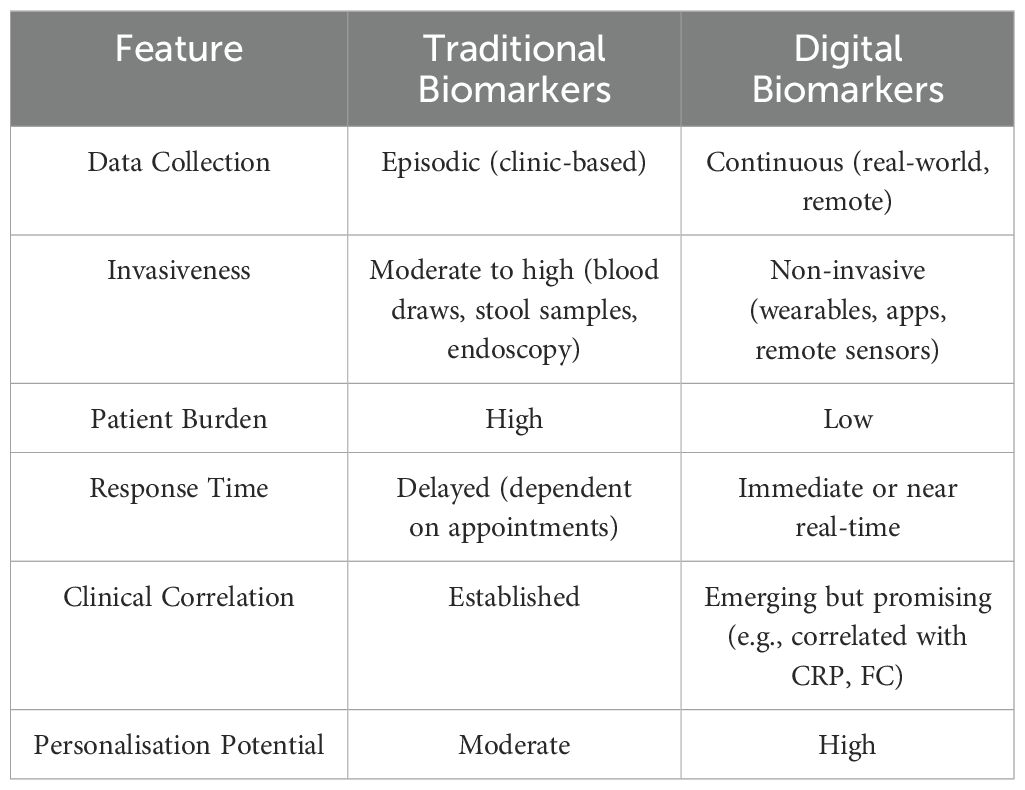
Traditional biomarkers, such as C-reactive protein (CRP) and fecal calprotectin (FC), endoscopy, imaging techniques (e.g. ultrasonography, magnetic resonance), and histology play a role in disease diagnosis and monitoring but have limitations in providing real-time, patient-specific insights (5). However, their utility is limited by poor accuracy, delayed response to inflammatory changes, and an inability to provide continuous real-time insights into disease activity predicting flares or treatment success. Endoscopic evaluation remains the gold standard for assessing mucosal healing (6), yet it is invasive, costly, and impractical for frequent monitoring (7, 8).
Although mucosal healing is a major therapeutic target in IBD and a reliable predictor of clinical outcomes, it does not necessarily reflect histological remission. The clinical relevance of including histological healing as an additional target is still under investigation, and its incremental benefits remain to be clearly defined (9). These limitations highlight the urgent need for more advanced, patient-centered tools capable of offering dynamic disease tracking (10).
The rapid evolution of digital health technologies, including wearable sensors, smartphone applications, and remote monitoring systems, has paved the way for digital biomarkers-quantifiable, patient-generated data that provide objective insights into disease status (11). When coupled with artificial intelligence (AI) and machine learning (ML), these biomarkers can revolutionize IBD management by enabling early detection of disease activity, predicting treatment response, facilitating personalized interventions and decreasing unnecessary healthcare utilization and costs (12). AI-driven models can integrate multi-source data (e.g., symptom tracking, physiological parameters) to refine risk stratification and therapeutic decision-making, reducing the reliance on invasive procedures and episodic clinical assessments (13).
This review aims to critically explore the potential role of AI-powered digital biomarkers in transforming IBD management. We discuss current advancements, clinical applications, potential benefits, and existing challenges, highlighting how these innovations can contribute to a more precise, patient-centered approach to IBD care. Furthermore, we address the limitations, ethical considerations, and future research directions necessary to fully harness the power of digital health in gastroenterology.
2 Digital biomarkers
While traditional clinical indices, biomarkers, and endoscopic evaluations remain integral to patient care, they often fail to fully capture the complex, fluctuating, and multidimensional nature of IBD at the individual level. The advent of digital biomarkers, particularly when integrated with AI, is revolutionizing personalized care by facilitating continuous, real-time monitoring and enabling earlier, more precise interventions (14).
Digital biomarkers are defined as objective, quantifiable physiological and behavioral data collected and measured through digital devices such as smartphones, wearable sensors, remote monitoring tools, and advanced imaging technologies (15). In contrast to conventional biomarkers, which provide episodic, point-in-time data, digital biomarkers offer a dynamic, continuous view of disease activity, detecting subtle physiological changes that may precede clinical symptoms.
FC is a specific marker for intestinal mucosal inflammation and is routinely measured in stool samples, while CRP and interleukin-6 (IL-6) are nonspecific markers of inflammation associated with inflammatory bowel disease–related inflammation (16).
IBD management has historically relied on traditional biomarkers such as FC, CRP, and endoscopic findings. While these biomarkers remain fundamental, they provide episodic snapshots of disease activity rather than a continuous picture (17). Digital biomarkers can offer a powerful complement by filling critical gaps in real-time disease monitoring and patient engagement.
Traditional biomarker assessment is usually performed during scheduled clinical visits, which can miss fluctuations in disease activity between appointments. In contrast, digital biomarkers — collected via wearables, smartphone apps, or remote sensors — enable continuous, real-time monitoring of physiological and behavioral changes. This dynamic tracking captures subtle shifts in disease activity, facilitating earlier detection of disease flares, enabling proactive interventions, and allowing for more precise, individualized therapy adjustments (Table 1). However, continuous real-time recording of physiological and behavioral changes may cause some patients to become overly fixated on their data, potentially increasing anxiety and undermining the intended benefits of digital tracking by decreasing adherence (e.g. checking CF at home).
In a study conducted by Hirten et al. 309 participants from 36 different states wore devices (Apple Watch, Fitbit, Oura Ring) capable of non-invasively and passively acquiring longitudinal heart rate (HR), resting heart rate (RHR), heart rate variability (HRV), steps and oxygenation. HR and RHR are higher during inflammatory and symptomatic phases while daily steps are lower during inflammatory phases. Wearable metrics identify subclinical inflammation and the presence of inflammation up to 7 weeks before flare during symptomatic phases (18). In a one-year prospective study on the use of biosensors by Yvellez et al., 91 outpatients and inpatients with IBD were analyzed. Daily steps, HR and sleep data were collected with a Fitbit device and patients entered daily information on a smart phone app using the Wong-Baker FACES™ pain rating scale (WB) and visual analogue scale questions related to sleep quality and general well-being. No association was found between median HR variability, steps or number of awakenings and next-day WB score (OR 9.7, p = 0.685; OR 0.89, p = 0.51; OR 1.05, p-value = 0.84 respectively). However, resting HR was significantly associated with reported pain the next day (OR 1.05, p = < 0.001); each 1 bpm increase in daily resting HR increased the odds of experiencing pain the next day by 5% (19).
The reliance on periodic blood tests, stool sampling, and invasive endoscopic procedures imposes both logistical and psychological burdens on patients, potentially affecting adherence and quality of life. Digital biomarkers provide a non-invasive, passive alternative, collected without disrupting daily activities. This patient-centered approach may promote adherence to monitoring protocols, enhancing patient engagement and supporting the principles of individualized care.
2.1 Correlation with standard biomarkers
Emerging evidence indicates strong correlations between specific digital biomarkers and traditional indicators of IBD activity. For example, continuous monitoring of physiological parameters such as heart rate variability, sleep patterns, and localized skin temperature has demonstrated potential to reflect systemic inflammatory states associated with IBD (20, 21). Additionally, wearable devices capable of measuring CRP and IL-6 in sweat demonstrated feasibility as real-time inflammatory monitors, correlating well with serum-based assays (22). These data streams enable continuous and remote disease monitoring, shifting from sporadic clinic-based evaluations to dynamic, personalized care.
Shahub et al. in 2024 enrolled 33 IBD patients who were monitored for 40–130 minutes with a proprietary wearable sensor device used to measure CRP, IL-6 and calprotectin. The analysis of the linear relationship between sweating and serum calprotectin (R2 = 0.7195), C-reactive protein (R2 = 0.615) and IL-6 (R2 = 0.5411) demonstrated a strong to moderate relationship between the various means supporting the clinical utility of sweating as a non-invasive means for continuous measurement correlated with standard inflammatory markers in serum and feces (22). Sossenheimer et al., in another one-year prospective study on the use of biosensors in IBD, provided 194 outpatients and inpatients with IBD with a Fitbit and a proprietary smartphone app for data collection and compilation of patient-reported outcomes. Patients recorded a lower number of daily steps (mean 6062 vs. 8541, p < 0.001) in the week prior to CRP or HR elevation, predictive of elevated biomarker collection within 7 days (area under the curve [AUC] for steps = 0.70, 95% CI = 0.65-0.75). In contrast, there was no difference in daily resting heart rate (mean 66.9 vs. 66.3, p = 0.42) (23).
By integrating digital biomarkers with conventional laboratory and imaging data, clinicians can enhance diagnostic precision, refine disease phenotyping, and strengthen predictive models for flare-ups and treatment response. This convergence supports a shift from static, visit-based assessments toward dynamic, remote disease management - enabling earlier detection of relapses, timely therapeutic adjustments, and more proactive, individualized care strategies (24).
3 The role of AI in IBD: unlocking hidden patterns
Artificial intelligence refers to computational systems capable of performing tasks traditionally requiring human intelligence, such as learning, problem-solving, and prediction. AI encompasses ML and its subfields, including supervised, unsupervised, reinforcement, and deep learning (DL) (Figure 1). AI algorithms are trained on diverse datasets; the larger and more heterogeneous the dataset, the more accurate and generalizable the models are in clinical settings. In the context of IBD, AI has the potential to advance precision medicine by enhancing diagnostics and informing therapeutic decisions (25). As illustrated in the Figure 1, ML is a key subfield of AI, focused on algorithms that learn from structured data. Unlike traditional rule-based AI systems, ML algorithms improve their performance over time through data exposure, enabling them to identify meaningful patterns and build predictive models. The figure also provides a brief overview of ML applications in the context of IBD, highlighting its potential to support digital biomarker discovery and personalized disease monitoring.
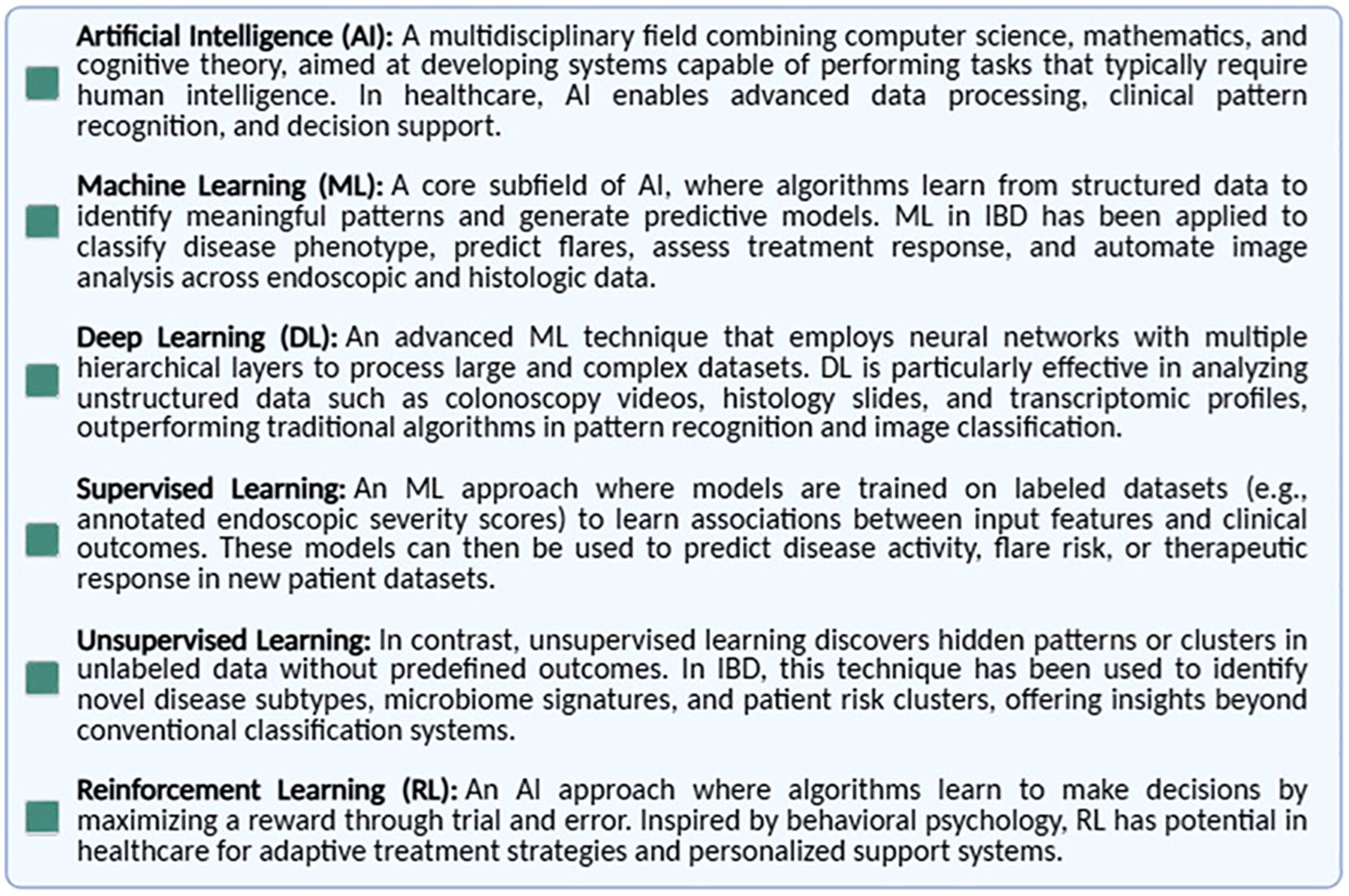
Figure 1. Key definitions in artificial intelligence and machine learning. Overview of the most commonly used terms in the field of AI as applied to healthcare, including artificial intelligence (AI), machine learning (ML), supervised and unsupervised learning, deep learning (DL), and neural networks. These definitions provide a conceptual framework for understanding how AI technologies can be developed and applied in clinical research and practice.
The convergence of Digital Health (DH) and AI in the management of IBD marks a paradigm shift, opening unprecedented opportunities to enhance patient care and outcomes. DH includes innovations such as mobile health platforms, wearable devices, telehealth, and telemedicine, which make healthcare more accessible, efficient, and patient-centered (26).
Elkjaer et al. in the largest web-based intervention RCT, randomized 333 patients (233 Danes and 100 Irish) with mild/moderate UC and being treated with 5-aminosalicylic acid to a web group that received disease-specific education and “Constant-care” via http://www.constant-care.dk or to a control group that continued standard of care (SoC) for 12 months. Overall, there was no notable difference in hospitalizations, surgeries, or adverse events. In a secondary analysis, the authors observed a numerically higher frequency but significantly shorter duration of relapses in the intervention group compared to the SoC group [Denmark: median 18 days (95% CI, 10-21) vs 77 days (95% CI, 46-108), p<0.001; Ireland: median 30 days (95% CI, 2-37) vs 70 days (95% CI, 7-217), p<0.03]. The number of acute and routine visits to the outpatient clinic was lower in the web group than in the control group, saving 189 euros/patient/year (27). Using the same “Constant-care” platform, Carlsen et al. evaluated the effectiveness of web-based management versus SoC in two different cohorts. The first included 53 non-biological treatment patients (27 eHealth/26 control) focused on monitoring disease activity in children/adolescents with IBD (young.constant-care.com, YCC). They found no differences between the groups in treatment escalation and disease activity (e.g. symptoms, biomarkers). The number of total outpatient visits (mean: eHealth 3.26, SEM 0.51; control 7.31, SEM 0.69; P < 0.0001) and IBD-related school absences (mean days: eHealth 1.6, SEM 0.5; control 16.5, SEM 4.4; P < 0.002) were significantly lower in the eHealth group. No differences were found in medical adherence and QoL, and none of the patients or parents felt insecure in using the eHealth system (28). In the second cohort, patients with IBD (19 CD and 10 UC in the eHealth group/16 CD, 4 UC and 1 IBD-Unclassified in the control one) aged 10 to 17 years treated with infliximab (IFX) were prospectively included. Starting 4 weeks after the last infusion, patients reported weekly symptom scores (via the abbreviated pediatric CD activity index, abbrPCDAI, and the pediatric UC activity index, PUCAI) and provided a stool sample for FC analysis, defining a new total inflammatory load scoring algorithm (TIBS). Based on the scores obtained, the eHealth program calculated a total inflammatory burden value that determined the timing of the next IFX infusion with 94 infusions in the eHealth group (mean interval 9.5 weeks; SD 2.3) compared with 105 infusions in the control group (mean interval 6.9 weeks; SD 1.4); treatment intervals were longer in the eHealth group (P < 0.001) (29).
For individuals living with IBD, DH not only expands access to care but also empowers them to take an active role in their health journey, promoting preventive strategies, facilitating earlier diagnosis, optimizing chronic disease management, and easing the long-term financial burden. At the same time, AI is reshaping the landscape of medical research and clinical practice (30).
3.1 Machine learning and predictive analytics
ML, a core component of AI, decodes complex patterns within large datasets, transforming raw data into actionable insights. ML models trained on real-world diagnostic and outcome data can accurately predict disease trajectories, offering significant potential for early intervention, personalized treatment, and continuous monitoring in IBD (31).
Given the intricate and evolving nature of IBD, such predictive capabilities could redefine early intervention, personalized treatment planning, and continuous disease monitoring. Together, DH and AI represent more than technological advances: they embody a new era in which precision medicine and patient empowerment converge to transform the future of IBD care.
ML methodologies include supervised and unsupervised learning. In supervised learning, models are trained on labeled datasets, learning input-output relationships to predict novel outcomes. Common approaches include Random Forest (RF) and Support Vector Machines (SVM), widely used in biomedical fields (31). Unsupervised learning, on the other hand, identifies patterns autonomously in unlabeled data, revealing new disease phenotypes and predictors.
DL, an advanced subset of ML, eliminates manual feature engineering, using layered neural networks to extract and amplify critical features. DL models excel in handling complex, high-dimensional biomedical data, enhancing predictive power and scalability (32).
For example, Gardiner et al. used an explainable ML approach to integrate demographic, clinical, and multi-omic data (genomic and transcriptomic) to predict differences in drug response among patients. Their model highlighted how factors specific to each patient — such as gender, age, and disease phenotype — affect drug efficacy. Additionally, it identified genetic polymorphisms linked to therapeutic responses, offering valuable insights for developing personalized treatment strategies (33). Sahoo et al. instead demonstrated how ML models offer the opportunity to identify barrier-protective therapies and predict candidate agents for clinical trials. They showed that AI can predict genes related to epithelial barriers, such as PRKAB1, the β1 subunit of the metabolic master regulator, AMPK, which might represent a novel target for gut barrier-protective therapies (34).
In 2024, two notable studies examined the use of ML in histological analysis for IBD. Peyrin-Biroulet et al. used an automated image analysis approach combined with ML to evaluate histological activity based on the Nancy Histological Index (NHI) in 200 images from UC patients. Their AI system’s performance was compared with that of four histopathologists, showing strong correlations despite limitations in the training dataset (35). In another study, Liu et al. explored AI-assisted histology for predicting therapeutic responses in pediatric patients with UC. Their ML model, through 18 histologic features, accurately predicted steroid-free remission in patients receiving mesalamine therapy, highlighting AI’s potential to customize treatment strategies for IBD (36).
A growing body of research has explored the application of AI in the management of IBD, with promising results across diagnostic tasks, disease activity assessment, and prediction of therapeutic response.
3.2 The expanding role of AI in IBD detection and classification
AI is rapidly transforming the diagnostic landscape of IBD, with ML algorithms and convolutional neural networks (CNNs) increasingly applied to genomic, imaging, endoscopic, and proteomic data to enhance diagnostic accuracy and efficiency (37). A key challenge in IBD diagnosis is distinguishing CD from UC, traditionally based on anatomical and clinical features. AI models trained on molecular and omics data are showing promise in improving differential diagnosis (38).
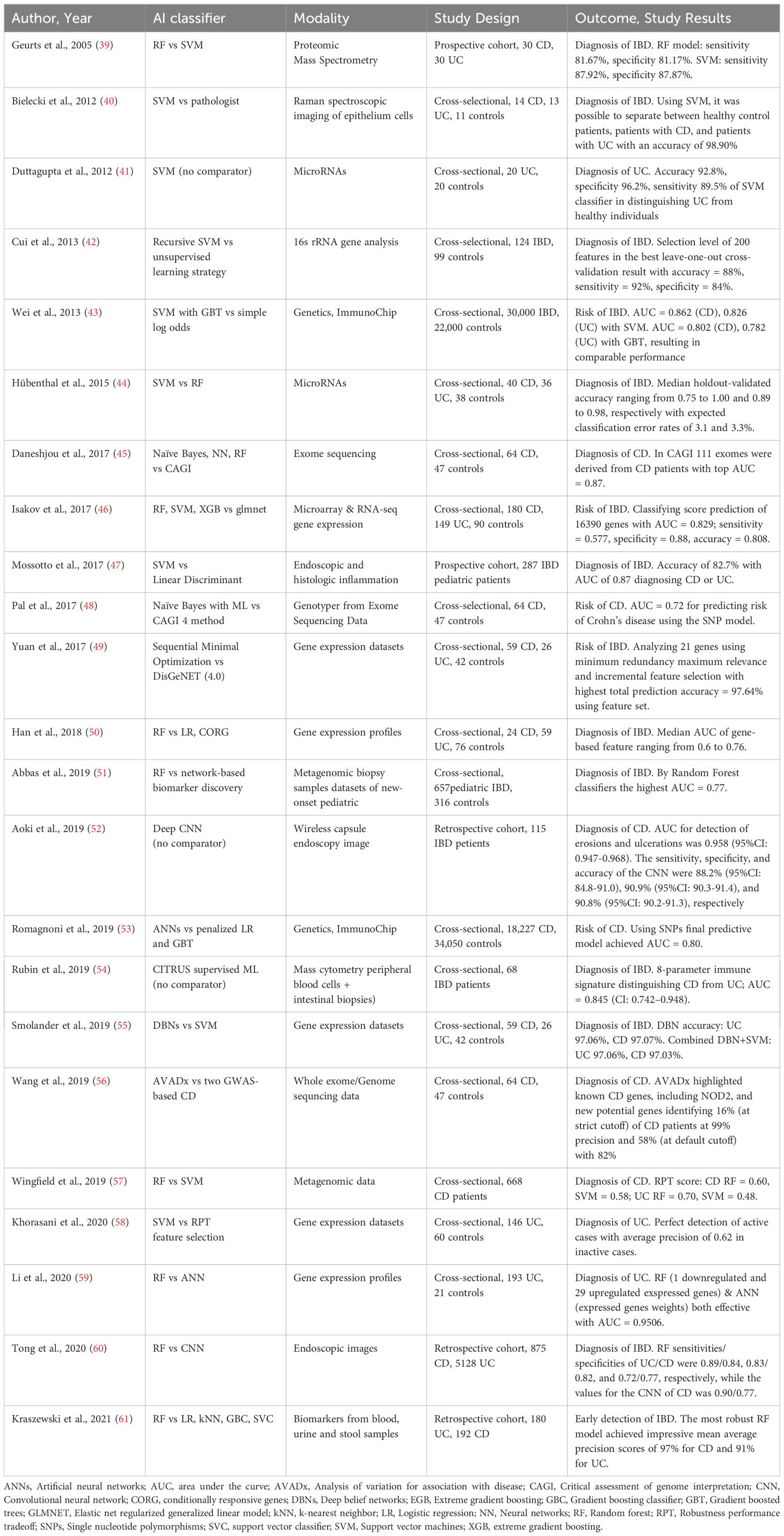
AI and ML are also being used to explore genetic variants’ role in disease pathophysiology, supporting the shift toward molecularly informed, precision diagnostics in IBD. Studies highlight the potential of AI in improving diagnosis and risk prediction by analyzing molecular and imaging data. Table 2 summarizes 23 studies, ordered chronologically, that employed various AI techniques (e.g., support vector machines, random forest, artificial neural networks, and deep learning) to assess IBD diagnosis and risk prediction. Of 18 studies focused on IBD diagnosis, 11 focus on both UC and CD (39, 40, 42, 44, 50, 54, 55, 60, 61), 4 only on CD (45, 52, 56, 57), 3 only on UC (41, 58, 59) and 2 on pediatric IBD (47, 51). Regarding risk prediction, 5 studies address both UC and CD (43, 46, 49), with 2 focusing solely on CD (48, 53).
Studies such as that by Mossotto et al. achieved diagnostic accuracies above 80% in differentiating CD from UC (47), while image-based approaches (e.g., Tong et al.) reached precision values as high as 99% for UC when using CNNs (60). In addition, multi-omics integration and immunophenotyping strategies have demonstrated high discriminatory power in classifying IBD as in the study by Rubin et al. in which using CITRUS, a supervised ML algorithm, they analyzed single-cell immunophenotyping data from peripheral blood mononuclear cells and distinguished CD from UC with an AUC of 0.845 (95% CI: 0.742-0.948) in a cohort of 68 patients with IBD (54). Romagnoni et al. analyzed gene expression profiles from a large cross-sectional cohort (18,227 CD patients and 34,050 healthy controls) using gradient-boosted trees and artificial neural networks. Their predictive model, based on single nucleotide polymorphism data, yielded an AUC of 0.80 for CD diagnosis (53). In a smaller study, Duttagupta et al. used a SVM classifier to analyze microRNA expression profiles from 20 patients with UC and 20 healthy individuals. They achieved an impressive predictive accuracy of 92.8%, with a specificity of 96.2% and sensitivity of 89.5% in distinguishing UC patients from healthy controls (41).
These models, incorporating diverse data such as transcriptomics, microRNA profiles, immunogenetics, and endoscopic imaging, demonstrate the feasibility of AI in distinguishing between CD, UC, and healthy controls, achieving high accuracy and AUC across various populations and study designs. However, performance varies depending on AI methodology and data modality (molecular vs. imaging), underscoring the need to tailor AI tools to specific clinical contexts.
By the other hand, Five-Nations multinational survey study evaluated the consistency of gastroenterologists in applying the Montreal classification for Crohn’s disease. Involving 59 IBD experts from five countries, the study revealed substantial inter-rater variability: agreement on disease location was only 59.4%, and on disease behavior just 46.8%. When the same case scenarios were analyzed using an AI-based algorithm, agreement levels improved modestly (location 68.1%, behavior 59.4%). These findings highlight both the promise and current limitations of IA in IBD classification. While AI tools can enhance standardization and reduce human variability, experienced gastroenterologists still achieved higher accuracy, especially when considering clinical subtleties. Thus, AI should be viewed as a valuable complement (but not a replacement) for expert clinical judgment in complex diagnostic scenarios (62).
3.3 AI-driven assessment in IBD
Assessment of disease activity in IBD requires integration of clinical, biochemical, endoscopic, and histologic parameters. Traditional tools include clinical indices (e.g., Harvey-Bradshaw Index for CD, Mayo Score for UC), biomarkers (CRP, FC), endoscopic scores (e.g., Mayo Endoscopic Score for UC, Simple Endoscopic Score for CD), and histologic indices (e.g., Nancy Histological Index, Robarts Histopathology Index) (9, 63). While foundational, these tools are limited by subjectivity, recall bias, and interobserver variability. In this context, AI offers enhanced precision, reproducibility, and integration of multidimensional data.
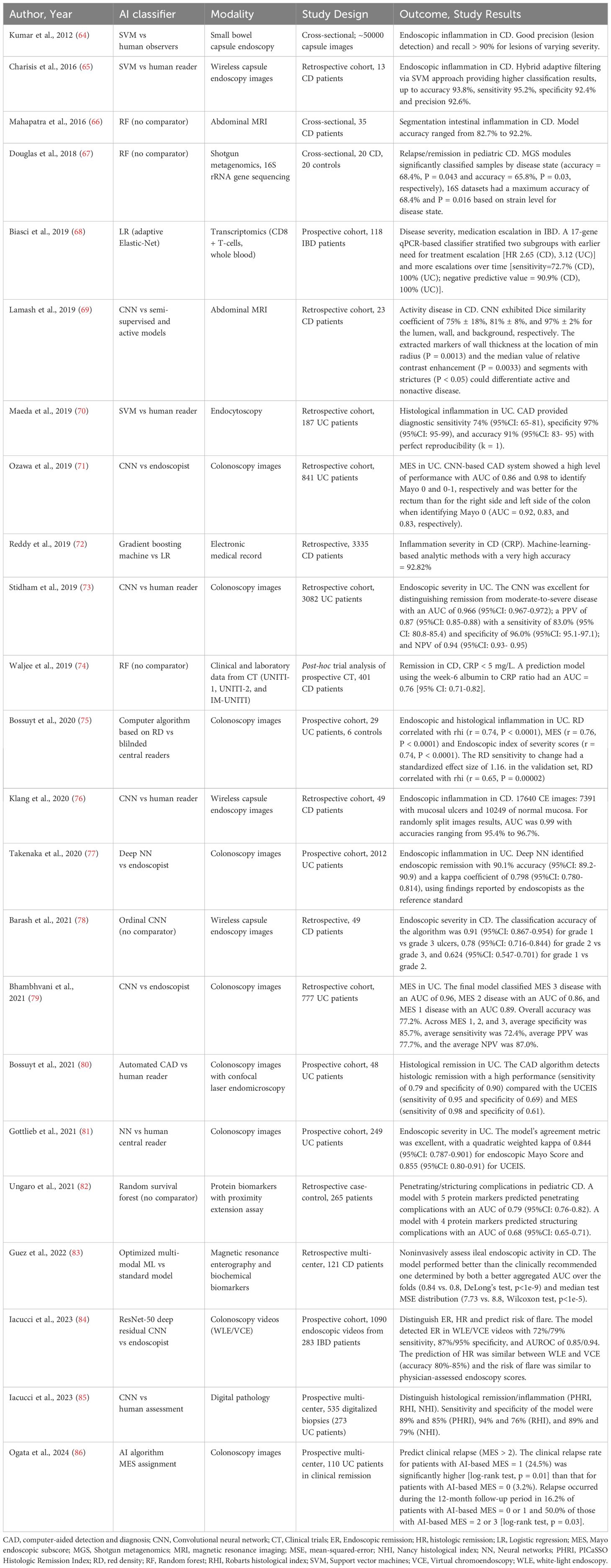
Table 3 summarizes 23 studies applying AI to IBD activity and severity assessment. There were 12 studies that focused on endoscopic inflammation in IBD (84) of which 4 focused only on CD (64, 65, 76, 78) and 7 on UC (71, 73, 75, 77, 79, 81, 86). Other focuses were clinical disease activity (67), 4 studies that assessed disease activity by biomarkers (68, 72, 74, 82), 3 studies on radiological activity of disease (66, 69, 83), and 3 studies that focused on histological inflammation (70, 80, 85). Data sources included electronic health records, molecular datasets, endoscopy, imaging, and histology from confocal endomicroscopy.
Iacucci et al. have developed a CNN from 1090 endoscopic videos of 283 patients with IBD. The endoscopic activity of the UC has been classified by experts using the Ulcerative Colitis Endoscopic Index of Severity (UCEIS) and the Paddington International virtual ChromoendoScopy ScOre (PICaSSO). The AI system detected endoscopic remission (ER) (UCEIS ≤ 1) in white light endoscopy (WLE) videos with a sensitivity of 72%, specificity of 87% and area of receiver characteristic operating curve (AUROC) of 0.85; for the detection of ER in virtual chromo-endoscopy (VCE) videos (PICaSSO ≤ 3), sensitivity was 79%, specificity 95% and AUROC 0.94. Histological remission prediction was similar between WLE and VCE videos (accuracy between 80% and 85%) and the stratification of flare risk performed by the model was similar to that of endoscopic scores evaluated by the physician (84). Likewise, in an ordinal CNN analysis of wireless capsule endoscopy images in a retrospective cohort of 49 CD patients by Barash et al., the classification accuracy of the algorithm was 0.91 for grade 1 vs grade 3 ulcers, 0.78 for grade 2 vs grade 3, and 0.624 for grade 1 vs grade 2 (78). Guez et al. developed and evaluated a multimodal ML model to assess the endoscopic activity of ileal CD by integrating information from MRE and the biochemical biomarkers from the 121 subjects of the multi-center database of the ImageKids study. Determined by both a better median test mean-squared-error distribution (7.73 vs. 8.8, Wilcoxon test, p < 1e-5) and a better aggregated AUC over the folds (0.84 vs. 0.8, DeLong’s test, p < 1e-9), the optimized fusion model performed better than the clinically recommended model (83).
CNNs and DL models have automated endoscopic scoring, mucosal healing assessment, and histologic remission classification, demonstrating high diagnostic accuracy and interobserver consistency. AI-enhanced endoscopy, in particular, shows potential for real-time decision support, standardization, and reduced interpretive variability (38, 87).
In a single-center retrospective study Stidhman et al. developed a NLP system to automatically identify and determine the activity status of extraintestinal manifestations (EIMs) in IBD using outpatient clinical notes. The NLP tool demonstrated high accuracy (94.1%) and strong agreement with human chart review (κ = 0.76), significantly outperforming administrative coding. By enabling automated, patient-level extraction of granular EIM data, this approach may enhance individualized care, support biomarker discovery, and improve prognostic precision in IBD (88).
These studies highlight AI’s clinical utility in evaluating IBD activity across modalities. However, performance may vary with inflammation location and subtle disease features, underscoring the need for context-specific model optimization.
3.4 AI-Based prediction of treatment response
Despite expanding therapeutic options, the heterogeneity of IBD limits the effectiveness of one-size-fits-all treatment strategies. Combination therapies targeting multiple inflammatory pathways may improve long-term disease control; however, this requires a deeper understanding of subtype-specific pathogenesis and molecular signatures. In this context, precision medicine — powered by AI and ML —offers the potential to predict therapeutic response and tailor treatment based on individual biological profiles.
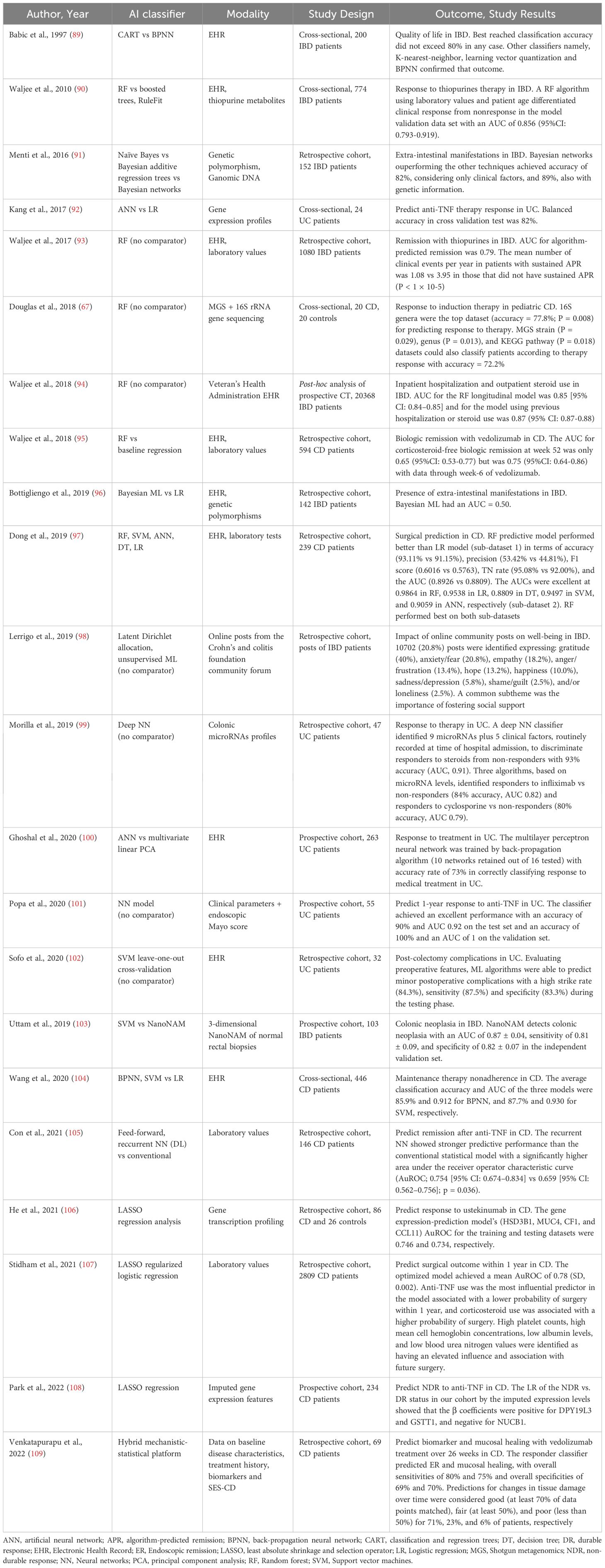
Table 4 summarizes 22 studies evaluating AI applications in predicting treatment response and disease prognosis in IBD. Thirteen studies assessed therapeutic response (biologics and non-biologics), including 7 in CD (67, 95, 104–106, 108, 109) and 4 in UC only (92, 99–101). Additional studies focused on extraintestinal manifestations (91, 96), quality of life of affected patients (89, 98), surgical risk in CD (97, 107), hospitalization risk (94), colorectal cancer risk (103) and post-colectomy complications in UC in UC (102).
Venkatapurapu et al. developed a mechanistic-statistical hybrid platform to predict biomarkers and tissue health time in patients (n = 69) with CD. The respondent’s classifier predicted endoscopic remission and mucosal healing for vedolizumab treatment over 26 weeks, with overall sensitivities of 80% and 75% and overall specificity of 69% and 70%, respectively (109). ML models incorporating routinely collected laboratory studies to predict surgical outcomes in U.S. Veterans with CD were evaluated from Stidham et al. Their optimized model from 2809 patients, among whom 256 had surgery, achieved a mean AUROC of 0.78 (SD, 0.002). Anti-tumor necrosis factor use was linked to a lower likelihood of surgery within one year, making it the strongest predictor. Conversely, corticosteroid use increased the probability of surgery. Key laboratory variables associated with future surgery included high platelet counts, elevated mean cell hemoglobin concentrations, low albumin levels, and low blood urea nitrogen values (107). In a prospective study Uttam et al. recruited 103 IBD patients undergoing surveillance colonoscopy and measured submicroscopic alterations in aberrant intrinsic nuclear architecture of epithelial cells from normal-appearing rectal biopsies with nanoscale nuclear architecture mapping (nanoNAM). Using nanoNAM-based structural characterization as input features into a soft margin-based ν-SVM risk classifier, it has been shown to detect colon neoplasia with AUC of 0.87 ± 0.04, sensitivity of 0.81 ± 0.09, and specificity of 0.82 ± 0.07 in the independent validation set. In addition, projecting nanoNAM features onto a 2-sphere reveals patients with low-risk and high-risk IBD colitis existing on separate hemispheres (103).
The endo-omics study evaluated the predictive value of a computer aided confocal laser endomicroscopy (pCLE) image analysis and fluorescent-labeled biologic binding in predicting therapeutic response in IBD patients starting anti-TNF or anti-integrin therapy. In vivo pCLE features—such as vessel tortuosity and fluorescein leakage — were highly predictive of response in both UC and CD. Ex vivo, increased mucosal binding of labeled biologics predicted response in UC but not CD. Thus, the use of pCLE and mucosal drug-binding profiles as tools for individualized treatment strategies in IBD (110).
A recent review by Sedano et al. emphasizes the impact of AI in improving IBD clinical trials. AI enhances patient recruitment (boosting efficiency by up to 30%) and supports more accurate analysis of complex clinical data. It also enables prediction of individual treatment responses and allows real-time adjustments in adaptive trial designs. These advances demonstrate AI’s potential to optimize trial methodology and improve outcomes in IBD research (111).
These models integrate clinical, laboratory, and omics data to predict outcomes such as corticosteroid or anti-TNF response, need for surgery, and risk of complications. ML algorithms — including LASSO regression, random forest, and gradient boosting — have shown robust predictive performance, supporting their role in individualized treatment planning and risk stratification.
4 Remote monitoring and predictive analytics in IBD
Managing IBD remains challenging due to its fluctuating course, frequent flares, and the frequent mismatch between symptoms and underlying inflammation (112).
Conventional monitoring — via patient-reported outcomes, biomarkers (serologic and fecal), imaging, and endoscopy — is episodic, invasive, and often dependent on patient compliance, offering only static assessments of disease activity. These limitations underscore the need for continuous, non-invasive, and real-time monitoring tools. Digital health technologies — including wearable devices, mobile health apps, and telemedicine — are emerging as key components of proactive disease management (Figure 2). By enabling continuous physiological and behavioral monitoring, they facilitate earlier clinical interventions, improve adherence, and support better long-term outcomes (18).
Figure 2. Wearable technologies and mobile health applications for remote monitoring in IBD. Illustration of digital health tools used for remote disease monitoring in inflammatory bowel disease (IBD), including wearable devices (e.g., smartwatches, biosensors) and mobile health (mHealth) applications. These technologies enable real-time tracking of physiological parameters, symptom reporting, medication adherence, and patient-reported outcomes, supporting personalized and proactive disease management.
4.1 Wearable technologies: continuous monitoring for proactive IBD care
Wearable devices enable non-invasive, passive, and continuous acquisition of physiological data, supporting early detection of inflammatory activity and potential preclinical diagnosis of IBD. Data collection may be active (user-driven) or fully passive after device application.
Jagannath et al. (2020) introduced a forearm-mounted sensor (SWEATSENSER) capable of detecting interleukin-1β (IL-1β) and CRP in sweat, demonstrating feasibility for real-time inflammatory monitoring in IBD. The sensor device can detect IL-1β and CRP in sweat over a dynamic range of 3 log orders with Pearson correlation of r = 0.99 and r = 0.95 achieved for IL-1β and CRP, respectively, with ELISA (113). Shahub et al. (2024) validated a similar device (IBD AWARE) measuring CRP, IL-6, and FC, with expression of FC that was significantly elevated in the active cohort compared with the remission cohort in perspiration (P < 0.05; median = 906.69 ng/mL; active 95% confidence interval [CI], 466.0–1833 ng/mL; remission 95% CI, 328.4-950.8 ng/mL), serum (median = 1860.82 ng/mL; active 95% CI, 1705–2985 ng/mL; remission 95% CI, 870.2–1786 ng/mL), and stool (P <.05; median = 126.74 µg/g; active 95% CI, 77.08-347.1 µg/g; remission 95% CI, 5.038-190.4 µg/g) (22).
In the IBD Forecast study, Hirten et al. showed that wearables (Apple Watch, Fitbit, Oura Ring) could predict flares up to 7 weeks in advance via changes in HR, HRV, RHR, oxygenation, and circadian HRV patterns (18). These findings suggest wearable-derived digital biomarkers can enable early, proactive interventions and individualized disease management.
Cleveland et al. described the first use of handheld ultrasound by a patient with UC for at-home monitoring during change of therapy. By offering real-time insights into treatment response, the handheld ultrasound by patients may support more timely and informed therapeutic decisions by both patients and clinicians. However, further studies and validation are awaited to determine its broader applicability (114).
4.2 Mobile health applications
Mobile health (mHealth) applications play a pivotal role in IBD management by facilitating self-monitoring, enhancing medication adherence, improving disease literacy, and enabling early clinical intervention (Figure 3).

Figure 3. Key functions of mobile health (mHealth) applications in IBD care. mHealth applications can support patients with inflammatory bowel disease (IBD) by facilitating self-monitoring, enhancing medication adherence, improving disease literacy, and enabling early clinical intervention. These digital tools empower patients and promote a more proactive, personalized approach to disease management.
Symptom tracking apps allow real-time logging of stool frequency, abdominal pain, and fatigue—core indicators of disease activity. Some incorporate AI to correlate dietary patterns with symptoms, aiding identification of individual triggers and differentiating from functional overlap. Adherence-focused platforms use AI-driven reminders and behavioral prompts to improve compliance — crucial in chronic disease care. Apps like HealthPROMISE and TELE-IBD have shown reductions in hospitalizations and emergency visits compared to standard care (115). IBD-Home users had increased care engagement, highlighting the value of remote monitoring (24). IBDoc and IBDsmart reduced outpatient visits without compromising outcomes (116).
However, a 2022 systematic review of 14 RCTs by Nguyen et al. found that while digital interventions improved healthcare utilization and cost metrics, impacts on disease activity, adherence, and quality of life were variable (117). MHealth apps thus serve as integral tools in digital IBD care, enhancing patient autonomy, clinician oversight, and system efficiency.
4.3 Tele-medicine in IBD
Telemedicine has become an integral component of IBD care, accelerated by the COVID-19 pandemic as a viable alternative to in-person visits. Virtual consultations enhance accessibility, reduce geographic and logistical barriers, and improve chronic disease management efficiency (118). Recent platforms increasingly integrate data from wearables and mHealth apps, enabling real-time analysis of physiological and patient-reported outcomes to support data-driven clinical decision-making.
The MyIBDcoach platform, evaluated by de Jong et al., demonstrated reduced outpatient visits and hospitalizations over 12 months compared to standard care, without compromising disease monitoring (119). Similarly, Del Hoyo et al. showed that the TECCU telemonitoring system significantly decreased clinic visits and improved disease activity, achieving clinical remission in 81% of complex IBD patients, versus 71.4% and 66.7% with standard and telephone care, respectively (120).
The CRONICA-UC study validated remote self-assessment of disease activity in UC using the SCCAI, demonstrating strong correlation with physician scores (ρ=0.79; κ=0.66; 85% agreement) (109). Li et al. further showed that virtual IBD clinics reduce costs (average $62 per visit saved) and time burden, while maintaining care quality (121).
Recently, a prospective study evaluated the accuracy and completeness of ChatGPT-3.5 responses to 38 real-world questions from IBD patients, using ECCO guidelines as a reference. Fourteen IBD experts assessed responses across topics including disease management, pregnancy, vaccination, and complementary therapies. While most replies were rated as largely accurate (mean score 3.87/5), completeness was more limited (mean score 2.24/3), with variability across questions. Highest accuracy and completeness were seen in responses about smoking, while the lowest were for malignancy screening and vaccination in immunosuppressed patients (122).
With widespread mobile technology and improved connectivity, telemedicine offers scalable, patient-centered care for IBD, supporting continuous monitoring and timely interventions. ChatGPT may be a useful adjunct for patient education, though caution is warranted in complex clinical areas.
5 Challenges and future directions
ML models have been utilized to stratify patients based on longitudinal digital health data, effectively identifying individuals at high risk for disease flares who may benefit from early therapeutic escalation, while distinguishing those in sustained remission. These predictive analytics enable timely, individualized treatment decisions, reducing overtreatment and enhancing clinical outcomes. As these tools mature, they hold the potential to transition IBD management from episodic, clinic-based encounters to continuous, precision-guided care (Figure 4). However, despite the promise of AI and digital biomarkers, key barriers — such as data standardization, validation across diverse populations, regulatory approval, and integration into clinical workflows — must be addressed before widespread implementation is achieved.
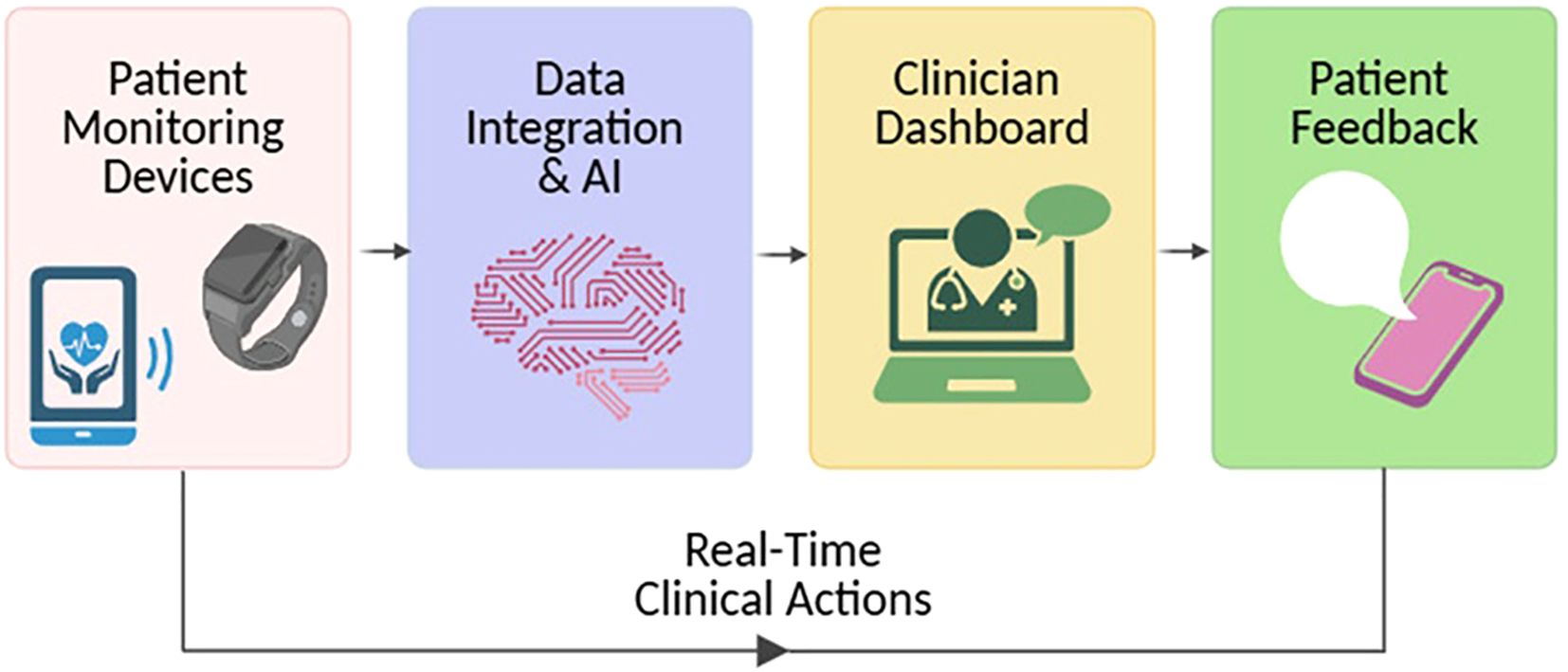
Figure 4. The evolving model of IBD care through AI and remote monitoring. Artificial intelligence (AI) tools and remote monitoring technologies are reshaping inflammatory bowel disease (IBD) management by shifting from episodic, clinic-centered visits to a model of continuous, precision-guided care. This transition supports earlier intervention, personalized treatment adjustments, and improved long-term outcomes.
5.1 Challenges in implementing digital biomarkers in IBD
DH tools require consistent and sustained patient engagement to achieve their full potential, yet long-term adherence often declines. Contributing factors include digital fatigue, perceived depersonalization, and limited perceived clinical benefits. Designing interfaces that are intuitive, gamified, and personalized may mitigate attrition and enhance user engagement.
As DH solutions become increasingly integrated into IBD care pathways, concerns surrounding data privacy, security, and ethical oversight have emerged as key barriers to widespread adoption. These concerns are particularly pronounced in IBD, where patients routinely share sensitive clinical, behavioral, and biometric data across interconnected platforms (123). Wearable devices, mobile apps and artificial intelligence algorithms continuously collect data on symptoms, biometrics, medication adherence and even geolocation (30). The scale and granularity of this data amplify the risks of unauthorized access, data breaches, and secondary misuse. Notably, healthcare cybersecurity incidents are rising, underscoring the urgent need for end-to-end encryption, secure data storage, and robust authentication protocols (124).
Although frameworks such as the General Data Protection Regulation (GDPR, EU) and Health Insurance Portability and Accountability Act (HIPAA, US) provide foundational protections, they were not designed for real-time, adaptive DH and AI applications. A regulatory gap persists, particularly concerning ML algorithms that evolve via unsupervised learning and lack fixed rule sets. Adaptive and context-specific compliance strategies are under development, but global regulatory harmonization remains elusive (125).
Moreover, the question of data ownership remains unresolved — whether it resides with the patient, healthcare provider, or digital platform. In some instances, aggregated data are leveraged for secondary purposes such as pharmaceutical marketing or insurance risk modeling, often without explicit patient consent. Ethical implementation of AI in IBD must prioritize data sovereignty, enforce transparent secondary use policies, and guarantee informed patient control over personal data. In summary, he ethical integration of DH and AI in IBD care necessitates more than technological innovation — it demands comprehensive regulatory reform, secure data governance, and a patient-centered approach. Sustaining patient trust will be critical, as ethical design becomes as important as clinical efficacy in shaping the future of IBD management.
6 Discussion
The future of IBD management lies in the integration of AI, remote monitoring technologies, and digital phenotyping into routine clinical practice. This evolution should not be perceived as a threat by patients or clinicians but embraced as an opportunity to enhance individualized care. ML-driven clinical decision support systems are poised to assist gastroenterologists in therapeutic decision-making, early risk stratification, and outcome prediction, enabling more timely and accurate interventions (26).
As mucosal healing is a central therapeutic target in IBD, AI-based tools may offer less invasive and more continuous assessment of disease activity. However, since mucosal healing does not always reflect histological remission, future models may also help explore digital correlates of histological healing, whose clinical value remains under investigation (5).
To address challenges related to centralized data storage and privacy, federated learning enables decentralized model training across multiple institutions without the need to exchange raw patient data. This approach preserves data confidentiality while enhancing the generalizability and performance of predictive algorithms. Moreover, the advent of edge computing — processing data directly on or near the patient’s device—will enhance the speed, scalability, and responsiveness of remote IBD monitoring systems. Conversational AI tools, such as chatbots and virtual health assistants, are also being deployed to support patient self-management by addressing medication adherence, stress reduction, dietary modifications, and other behavioral health components (31).
These tools offer continuous, scalable, and personalized support aligned with integrative care principles. Looking ahead, a fully AI-integrated ecosystem is anticipated — one that synthesizes genomic data, wearable-derived digital biomarkers, and real-time analytics to enable predictive, preventive, and precision-guided management of IBD.
7 Conclusion
The integration of digital biomarkers and AI in IBD marks a shift toward personalized, proactive, real-time medicine. Wearable devices detecting subclinical inflammation and AI-driven algorithms for patient stratification and treatment optimization show considerable promise in terms of accuracy, scalability, and patient engagement in early studies.
However, challenges persist, including data source heterogeneity, lack of standardization, and limited large-scale validation of predictive models. The reproducibility and generalizability of AI solutions across diverse clinical settings and populations remain uncertain.
To ensure broad access to these innovations, further prospective, multicenter trials are needed to assess the real-world efficacy of digital biomarkers and AI tools. Additionally, standardized data collection protocols and robust regulatory and ethical frameworks are critical to balancing patient privacy with technological advancement.
Moving forward, it is essential to validate, standardize, and responsibly implement these tools in clinical practice.
Author contributions
DD: Writing – original draft, Methodology, Investigation, Data curation. AD: Writing – original draft, Writing – review & editing, Conceptualization. RG: Supervision, Writing – review & editing. ON: Supervision, Writing – original draft, Writing – review & editing. RF-I: Writing – original draft, Writing – review & editing, Supervision. GP: Writing – review & editing, Writing – original draft. CBo: Supervision, Writing – review & editing. MB-d: Supervision, Writing – review & editing. CBe: Writing – review & editing, Supervision. AA: Writing – review & editing, Supervision, Validation, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Conflict of interest
AA has received consulting fees from AbbVie, Allergan, Amgen, Arena, Biogen, AA has received consulting fees from AbbVie, Allergan, Amgen, Arena, Biogen, Boehringer Ingelheim, Bristol-Myers Squibb, Celgene, Celltrion, Eli-Lilly, Ferring, Galapagos, Gilead, Janssen, MSD, Mylan, Pfizer, Protagonist Therapeutics, Roche, Samsung Bioepis, Sandoz and Takeda; speaker’s fees from AbbVie, Amgen, Arena, Biogen, Bristol-Myers Squibb, Eli-Lilly, Ferring, Galapagos, Gilead, Janssen,MSD, Novartis, Pfizer, Roche, Samsung Bioepis, Sandoz, Takeda, and Tigenix; and research support from Biogen, MSD, Takeda, and Pfizer. CBe received lecture fees and served as a consultant for Takeda, MSD, Ferring, Abbvie, Galapagos, Celltrion, Janssen, and Eli-Lilly. GP has received speaker’s and consulting fees from Janssen and Alphasigma. RG has received speaker’s fees from Pfizer, MSD, Eli-Lilly, Janssen, Takeda, and Celltrion and consulting fees from Pfizer. AD has received speaker’s fees from AbbVie, Galapagos, Eli-Lilly, Janssen, and Celltrion, and consulting fees from Ferring. DD declares no conflict of interest. ON has received speaker fee from Janssen, Abbvie, Ferring, Pfizer, Alfa Sigma, Eli Lilly; consulting fee from Nestlé, Janssen, Eli Lilly. RF-I has served as a speaker, consultant and advisory member for or has received research funding from MSD, AbbVie, Janssen, Kern Pharma, Takeda, Pfizer, Lilly, Ferring, Faes Farma, Dr. Falk Pharma, Chiesi, Adacyte, Diasorin and TillotsPharma. MBarreiro-d has received financial support for traveling and educational activities from Pfizer, MSD, Takeda, Abbvie, Kern, Janssen, Fresenius Kabi, Galapagos, Lilly, BMS, Faes Pharma, Chiesi and Adacyte.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Dolinger M, Torres J, and Vermeire S. Crohn’s disease. Lancet (London England). (2024) 403:1177–91. doi: 10.1016/S0140-6736(23)02586-2
2. Le Berre C, Honap S, and Peyrin-Biroulet L. Ulcerative colitis. Lancet (London England). (2023) 402:571–84. doi: 10.1016/S0140-6736(23)00966-2
3. Flynn S and Eisenstein S. Inflammatory bowel disease presentation and diagnosis. Surg Clin North Am. (2019) 99:1051–62. doi: 10.1016/j.suc.2019.08.001
4. Majidova K, Handfield J, Kafi K, Martin RD, and Kubinski R. Role of digital health and artificial intelligence in inflammatory bowel disease: A scoping review. Genes (Basel). (2021) 12. doi: 10.3390/genes12101465
5. Colombel JF, D’Haens G, Lee WJ, Petersson J, and Panaccione R. Outcomes and strategies to support a treat-to-target approach in inflammatory bowel disease: A systematic review. J Crohn’s Colitis. (2020) 14:254–66. doi: 10.1093/ecco-jcc/jjz131
6. Maaser C, Sturm A, Vavricka SR, Kucharzik T, Fiorino G, Annese V, et al. ECCO-ESGAR Guideline for Diagnostic Assessment in IBD Part 1: Initial diagnosis, monitoring of known IBD, detection of complications. J Crohns Colitis. (2019) 13:144–64. doi: 10.1093/ecco-jcc/jjy113
7. Goodsall TM, Noy R, Nguyen TM, Costello SP, Jairath V, and Bryant RV. Systematic review: patient perceptions of monitoring tools in inflammatory bowel disease. J Can Assoc Gastroenterol. (2021) 4:e31–41. doi: 10.1093/jcag/gwaa001
8. Barsky M, Meserve J, Le H, Collins A, Singh S, Boland B, et al. Understanding determinants of patient preferences between stool tests and colonoscopy for the assessment of disease activity in inflammatory bowel disease. Dig. Dis Sci. (2021) 66:2564–9. doi: 10.1007/s10620-020-06568-w
9. Magro F, Sabino J, Rosini F, Tripathi M, Borralho P, Baldin P, et al. ECCO position on harmonisation of crohn’s disease mucosal histopathology. J Crohns Colitis. (2022) 16:876–83. doi: 10.1093/ecco-jcc/jjac006
10. Adamina M, Feakins R, Iacucci M, Spinelli A, Cannatelli R, D’Hoore A, et al. ECCO topical review optimising reporting in surgery, endoscopy, and histopathology. J Crohn’s Colitis. (2021) 15:1089–105. doi: 10.1093/ecco-jcc/jjab011
11. Harindranath S and Desai D. Wearable technology in inflammatory bowel disease: current state and future direction. Expert Rev Med Devices. (2025) 22:121–6. doi: 10.1080/17434440.2025.2453561
12. Burisch J, Zhao M, Odes S, De Cruz P, Vermeire S, Bernstein CN, et al. The cost of inflammatory bowel disease in high-income settings: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol. (2023) 8:458–92. doi: 10.1016/S2468-1253(23)00003-1
13. Kröner PT, Engels MML, Glicksberg BS, Johnson KW, Mzaik O, van Hooft JE, et al. Artificial intelligence in gastroenterology: A state-of-the-art review. World J Gastroenterol. (2021) 6794–6824. doi: 10.3748/wjg.v27.i40.6794
14. Atreya R and Neurath MF. Biomarkers for personalizing IBD therapy: the quest continues. Clin Gastroenterol Hepatol. (2024) 22:1353–64. doi: 10.1016/j.cgh.2024.01.026
15. Spiegel B. 2015 American Journal of Gastroenterology Lecture: How digital health will transform gastroenterology. Am J Gastroenterol. (2016) 111:624–30. doi: 10.1038/ajg.2016.68
16. Mosli MH, Zou G, Garg SK, Feagan SG, MacDonald JK, Chande N, et al. C-reactive protein, fecal calprotectin, and stool lactoferrin for detection of endoscopic activity in symptomatic inflammatory bowel disease patients: A systematic review and meta-analysis. Am J Gastroenterol. (2015) 110:802–19. doi: 10.1038/ajg.2015.120
17. Liu D, Saikam V, Skrada KA, Merlin D, and Iyer SS. Inflammatory bowel disease biomarkers. Med Res Rev. (2022) 42:1856–87. doi: 10.1002/med.21893
18. Hirten RP, Danieletto M, Sanchez-Mayor M, Whang JK, Lee KW, Landell K, et al. Physiological data collected from wearable devices identify and predict inflammatory bowel disease flares. Gastroenterology. (2025) 168:939–951.e5. doi: 10.1053/j.gastro.2024.12.024
19. Yvellez OV, Sossenheimer PH, Andersen M, El Jurdi K, Mayampurath A, and Rubin DT. P411 Using wearable devices to assess pain in inflammatory bowel disease. J Crohn’s Colitis. (2019) 13:S315–5. doi: 10.1093/ecco-jcc/jjy222.535
20. Gunterberg V, Simrén M, Öhman L, Friberg P, Jones MP, Van Oudenhove L, et al. Autonomic nervous system function predicts the inflammatory response over three years in newly diagnosed ulcerative colitis patients. Neurogastroenterol. Motil. (2016) 28:1655–62.
21. Engel T, Ben-Horin S, and Beer-Gabel M. Autonomic dysfunction correlates with clinical and inflammatory activity in patients with crohn’s disease. Inflammation Bowel Dis. (2015) 21:2320–6. doi: 10.1097/MIB.0000000000000508
22. Shahub S, Kumar RM, Lin K-C, Banga I, Choi NK, Garcia NM, et al. Continuous monitoring of CRP, IL-6, and calprotectin in inflammatory bowel disease using a perspiration-based wearable device. Inflammation Bowel Dis. (2024), 647–54. doi: 10.1093/ibd/izae054
23. Sossenheimer PH, Yvellez OV, Andersen M, Pearl T, El Jurdi K, Rubin DB, et al. P579 Wearable Devices Can Predict Disease Activity in inflammatory bowel disease Patients. J Crohn’s Colitis. (2019) 13:S404–4. doi: 10.1093/ecco-jcc/jjy222.703
24. Östlund I, Werner M, and Karling P. Self-monitoring with home based fecal calprotectin is associated with increased medical treatment. A randomized controlled trial on patients with inflammatory bowel disease. Scand J Gastroenterol. (2021) 56:38–45. doi: 10.1080/00365521.2020.1854342
25. Pinton P. Computational models in inflammatory bowel disease. Clin Transl Sci. (2022) 15:824–30. doi: 10.1111/cts.13228
26. Atreja A, Otobo E, Ramireddy K, and Deorocki A. Remote patient monitoring in IBD: current state and future directions. Curr Gastroenterol Rep. (2018) 20:1–11. doi: 10.1007/s11894-018-0611-3
27. Elkjaer M, Shuhaibar M, Burisch J, Bailey Y, Scherfig H, Laugesen B, et al. E-health empowers patients with ulcerative colitis: a randomised controlled trial of the web-guided “Constant-care” approach. Gut. (2010) 59:1652–61. doi: 10.1136/gut.2010.220160
28. Carlsen K, Jakobsen C, Houen G, Kallemose T, Paerregaard A, Riis LB, et al. Self-managed eHealth disease monitoring in children and adolescents with inflammatory bowel disease: A randomized controlled trial. Inflammation Bowel Dis. (2017) 23:357–65. doi: 10.1097/MIB.0000000000001026
29. Carlsen K, Houen G, Jakobsen C, Kallemose T, Paerregaard A, Riis LB, et al. Individualized infliximab treatment guided by patient-managed eHealth in children and adolescents with inflammatory bowel disease. Inflammation Bowel Dis. (2017) 23:1473–82. doi: 10.1097/MIB.0000000000001170
30. Kelso M and Feagins LA. Can smartphones help deliver smarter care for patients with inflammatory bowel disease? Inflammation Bowel Dis. (2018) 24:1453–8. doi: 10.1093/ibd/izy162
31. Stidham RW and Takenaka K. Artificial intelligence for disease assessment in inflammatory bowel disease: how will it change our practice? Gastroenterology. (2022) 162:1493–506. doi: 10.1053/j.gastro.2021.12.238
32. Panch T, Szolovits P, and Atun R. Artificial intelligence, machine learning and health systems. J Glob. Health. (2018) 8:20303. doi: 10.7189/jogh.08.020303
33. Gardiner L-J, Carrieri AP, Bingham K, Macluskie G, Bunton D, McNeil M, et al. Combining explainable machine learning, demographic and multi-omic data to inform precision medicine strategies for inflammatory bowel disease. PloS One. (2022) 17:e0263248. doi: 10.1371/journal.pone.0263248
34. Sahoo D, Swanson L, Sayed IM, Katkar GD, Ibeawuchi S-R, Mittal Y, et al. Artificial intelligence guided discovery of a barrier-protective therapy in inflammatory bowel disease. Nat Commun. (2021) 12:4246. doi: 10.1038/s41467-021-24470-5
35. Peyrin-Biroulet L, Adsul S, Stancati A, Dehmeshki J, and Kubassova O. An artificial intelligence-driven scoring system to measure histological disease activity in ulcerative colitis. United Eur Gastroenterol J. (2024) 12:1028–33. doi: 10.1002/ueg2.12562
36. Liu X, Prasath S, Siddiqui I, Walters TD, Denson LA, and Dhaliwal J. Machine learning-based prediction of pediatric ulcerative colitis treatment response using diagnostic histopathology. Gastroenterology. (2024) 166:921–924.e4. doi: 10.1053/j.gastro.2024.01.033
37. Gubatan J, Levitte S, Patel A, Balabanis T, Wei MT, and Sinha SR. Artificial intelligence applications in inflammatory bowel disease: Emerging technologies and future directions. World J Gastroenterol. (2021) 27:1920–35. doi: 10.3748/wjg.v27.i17.1920
38. Iacucci M, Santacroce G, Maeda Y, and Ghosh S. AI-driven personalized medicine: transforming clinical practice in inflammatory bowel disease. Gastroenterology. (2025), 1–16. doi: 10.1053/j.gastro.2025.03.005
39. Geurts P, Fillet M, de Seny D, Meuwis M-A, Malaise M, Merville M-P, et al. Proteomic mass spectra classification using decision tree based ensemble methods. Bioinformatics. (2005) 21:3138–45. doi: 10.1093/bioinformatics/bti494
40. Bielecki C, Bocklitz TW, Schmitt M, Krafft C, Marquardt C, Gharbi A, et al. Classification of inflammatory bowel diseases by means of Raman spectroscopic imaging of epithelium cells. J BioMed Opt. (2012) 17:76030. doi: 10.1117/1.JBO.17.7.076030
41. Duttagupta R, DiRienzo S, Jiang R, Bowers J, Gollub J, Kao J, et al. Genome-wide maps of circulating miRNA biomarkers for Ulcerative Colitis. PloS One. (2012) 7. doi: 10.1371/journal.pone.0031241
42. Cui H and Zhang X. Alignment-free supervised classification of metagenomes by recursive SVM. BMC Genomics. (2013) 14:641. doi: 10.1186/1471-2164-14-641
43. Wei Z, Wang W, Bradfield J, Li J, Cardinale C, Frackelton E, et al. Large sample size, wide variant spectrum, and advanced machine-learning technique boost risk prediction for inflammatory bowel disease. Am J Hum Genet. (2013) 92:1008–12. doi: 10.1016/j.ajhg.2013.05.002
44. Hübenthal M, Hemmrich-Stanisak G, Degenhardt F, Szymczak S, Du Z, Elsharawy A, et al. Sparse modeling reveals miRNA signatures for diagnostics of inflammatory bowel disease. PloS One. (2015) 10:e0140155. doi: 10.1371/journal.pone.0140155
45. Daneshjou R, Wang Y, Bromberg Y, Bovo S, Martelli PL, Babbi G, et al. Working toward precision medicine: Predicting phenotypes from exomes in the Critical Assessment of Genome Interpretation (CAGI) challenges. Hum Mutat. (2017) 38:1182–92. doi: 10.1002/humu.23280
46. Isakov O, Dotan I, and Ben-Shachar S. Machine learning-based gene prioritization identifies novel candidate risk genes for inflammatory bowel disease. Inflammation Bowel Dis. (2017) 23:1516–23. doi: 10.1097/MIB.0000000000001222
47. Mossotto E, Ashton JJ, Coelho T, Beattie RM, MacArthur BD, and Ennis S. Classification of paediatric inflammatory bowel disease using machine learning. Sci Rep. (2017) 7:2427. doi: 10.1038/s41598-017-02606-2
48. Pal LR, Kundu K, Yin Y, and Moult J. CAGI4 Crohn’s exome challenge: Marker SNP versus exome variant models for assigning risk of Crohn disease. Hum Mutat. (2017) 38:1225–34. doi: 10.1002/humu.23256
49. Yuan F, Zhang Y-H, Kong X-Y, and Cai Y-D. Identification of candidate genes related to inflammatory bowel disease using minimum redundancy maximum relevance, incremental feature selection, and the shortest-path approach. BioMed Res Int. (2017) 2017:5741948. doi: 10.1155/2017/5741948
50. Han L, Maciejewski M, Brockel C, Gordon W, Snapper SB, Korzenik JR, et al. A probabilistic pathway score (PROPS) for classification with applications to inflammatory bowel disease. Bioinformatics. (2018) 34:985–93. doi: 10.1093/bioinformatics/btx651
51. Abbas M, Matta J, Le T, Bensmail H, Obafemi-Ajayi T, Honavar V, et al. Biomarker discovery in inflammatory bowel diseases using network-based feature selection. PloS One. (2019) 14:e0225382. doi: 10.1371/journal.pone.0225382
52. Aoki T, Yamada A, Aoyama K, Saito H, Tsuboi A, Nakada A, et al. Automatic detection of erosions and ulcerations in wireless capsule endoscopy images based on a deep convolutional neural network. Gastrointest Endosc. (2019) 89:357–363.e2. doi: 10.1016/j.gie.2018.10.027
53. Romagnoni A, Jégou S, Van Steen K, Wainrib G, Hugot JP, Peyrin-Biroulet L, et al. Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Rep. (2019) 9:1–18. doi: 10.1038/s41598-019-46649-z
54. Rubin SJS, Bai L, Haileselassie Y, Garay G, Yun C, Becker L, et al. Mass cytometry reveals systemic and local immune signatures that distinguish inflammatory bowel diseases. Nat Commun. (2019) 10. doi: 10.1038/s41467-019-10387-7
55. Smolander J, Dehmer M, and Emmert-Streib F. Comparing deep belief networks with support vector machines for classifying gene expression data from complex disorders. FEBS Open Bio. (2019) 9:1232–48. doi: 10.1002/2211-5463.12652
56. Wang Y, Miller M, Astrakhan Y, Petersen B-S, Schreiber S, Franke A, et al. Identifying Crohn’s disease signal from variome analysis. Genome Med. (2019) 11:59. doi: 10.1186/s13073-019-0670-6
57. Wingfield B, Coleman S, McGinnity TM, and Bjourson AJ. Robust microbial markers for non-invasive inflammatory bowel disease identification. IEEE/ACM Trans Comput Biol Bioinform. (2019) 16:2078–88. doi: 10.1109/TCBB.2018.2831212
58. Khorasani HM, Usefi H, and Peña-Castillo L. Detecting ulcerative colitis from colon samples using efficient feature selection and machine learning. Sci Rep. (2020) 10:13744. doi: 10.1038/s41598-020-70583-0
59. Li H, Lai L, and Shen J. Development of a susceptibility gene based novel predictive model for the diagnosis of ulcerative colitis using random forest and artificial neural network. Aging (Albany NY). (2020) 12:20471–82. doi: 10.18632/aging.v12i20
60. Tong Y, Lu K, Yang Y, Li J, Lin Y, Wu D, et al. Can natural language processing help differentiate inflammatory intestinal diseases in China? Models applying random forest and convolutional neural network approaches. BMC Med Inform Decis. Mak. (2020) 20:1–9. doi: 10.1186/s12911-020-01277-w
61. Kraszewski S, Szczurek W, Szymczak J, Reguła M, and Neubauer K. Machine learning prediction model for inflammatory bowel disease based on laboratory markers. Working model in a discovery cohort study. J Clin Med. (2021) 10. doi: 10.3390/jcm10204745
62. Ukashi O, Amiot A, Laharie D, Menchén L, Gutiérrez A, Fernandes S, et al. Inter-rater disagreements in applying the montreal classification for crohn’s disease: the five-nations survey study. United Eur Gastroenterol J. (2025). doi: 10.1093/ecco-jcc/jjae190.0425
63. SpIceland CM and Lodhia N. Endoscopy in inflammatory bowel disease: Role in diagnosis, management, and treatment. World J Gastroenterol. (2018) 24:4014–20. doi: 10.3748/wjg.v24.i35.4014
64. Kumar R, Zhao Q, Seshamani S, Mullin G, Hager G, and Dassopoulos T. Assessment of Crohn’s disease lesions in wireless capsule endoscopy images. IEEE Trans BioMed Eng. (2012) 59:355–62. doi: 10.1109/TBME.2011.2172438
65. Charisis VS and Hadjileontiadis LJ. Potential of hybrid adaptive filtering in inflammatory lesion detection from capsule endoscopy images. World J Gastroenterol. (2016) 22:8641–57. doi: 10.3748/wjg.v22.i39.8641
66. Mahapatra D, Vos FM, and Buhmann JM. Active learning based segmentation of Crohns disease from abdominal MRI. Comput Methods Programs Biomed. (2016) 128:75–85. doi: 10.1016/j.cmpb.2016.01.014
67. Douglas GM, Hansen R, Jones CMA, Dunn KA, Comeau AM, Bielawski JP, et al. Multi-omics differentially classify disease state and treatment outcome in pediatric Crohn’s disease. Microbiome. (2018) 6:13. doi: 10.1186/s40168-018-0398-3
68. Biasci D, Lee JC, Noor NM, Pombal DR, Hou M, Lewis N, et al. A blood-based prognostic biomarker in IBD. Gut. (2019) 68:1386–95. doi: 10.1136/gutjnl-2019-318343
69. Lamash Y, Kurugol S, Freiman M, Perez-Rossello JM, Callahan MJ, Bousvaros A, et al. Curved planar reformatting and convolutional neural network-based segmentation of the small bowel for visualization and quantitative assessment of pediatric Crohn’s disease from MRI. J Magn Reson Imaging. (2019) 49:1565–76. doi: 10.1002/jmri.26330
70. Maeda Y, Kudo S-E, Mori Y, Misawa M, Ogata N, Sasanuma S, et al. Fully automated diagnostic system with artificial intelligence using endocytoscopy to identify the presence of histologic inflammation associated with ulcerative colitis (with video). Gastrointest Endosc. (2019) 89:408–15. doi: 10.1016/j.gie.2018.09.024
71. Ozawa T, Ishihara S, Fujishiro M, Saito H, Kumagai Y, Shichijo S, et al. Novel computer-assisted diagnosis system for endoscopic disease activity in patients with ulcerative colitis. Gastrointest Endosc. (2019) 89:416–421.e1. doi: 10.1016/j.gie.2018.10.020
72. Reddy BK, Delen D, and Agrawal RK. Predicting and explaining inflammation in Crohn’s disease patients using predictive analytics methods and electronic medical record data. Health Inf J. (2019) 25:1201–18. doi: 10.1177/1460458217751015
73. Stidham RW, Liu W, Bishu S, Rice MD, Higgins PDR, Zhu J, et al. Performance of a deep learning model vs human reviewers in grading endoscopic disease severity of patients with ulcerative colitis. JAMA Netw Open. (2019) 2:e193963. doi: 10.1001/jamanetworkopen.2019.3963
74. Waljee AK, Wallace BI, Cohen-Mekelburg S, Liu Y, Liu B, Sauder K, et al. Development and validation of machine learning models in prediction of remission in patients with moderate to severe crohn disease. JAMA Netw Open. (2019) 2:e193721. doi: 10.1001/jamanetworkopen.2019.3721
75. Bossuyt P, Nakase H, Vermeire S, de Hertogh G, Eelbode T, Ferrante M, et al. Automatic, computer-aided determination of endoscopic and histological inflammation in patients with mild to moderate ulcerative colitis based on red density. Gut. (2020) 69:1778–86. doi: 10.1136/gutjnl-2019-320056
76. Klang E, Barash Y, Margalit RY, Soffer S, Shimon O, Albshesh A, et al. Deep learning algorithms for automated detection of Crohn’s disease ulcers by video capsule endoscopy. Gastrointest Endosc. (2020) 91:606–613.e2. doi: 10.1016/j.gie.2019.11.012
77. Takenaka K, Ohtsuka K, Fujii T, Negi M, Suzuki K, Shimizu H, et al. Development and validation of a deep neural network for accurate evaluation of endoscopic images from patients with ulcerative colitis. Gastroenterology. (2020) 158:2150–7. doi: 10.1053/j.gastro.2020.02.012
78. Barash Y, Azaria L, Soffer S, Margalit Yehuda R, Shlomi O, Ben-Horin S, et al. Ulcer severity grading in video capsule images of patients with Crohn’s disease: an ordinal neural network solution. Gastrointest Endosc. (2021) 93:187–92. doi: 10.1016/j.gie.2020.05.066
79. Bhambhvani HP and Zamora A. Deep learning enabled classification of Mayo endoscopic subscore in patients with ulcerative colitis. Eur J Gastroenterol Hepatol. (2021) 33:645–9. doi: 10.1097/MEG.0000000000001952
80. Bossuyt P, De Hertogh G, Eelbode T, Vermeire S, and Bisschops R. Computer-aided diagnosis with monochromatic light endoscopy for scoring histologic remission in ulcerative colitis. Gastroenterology. (2021) 160:23–5. doi: 10.1053/j.gastro.2020.09.053
81. Gottlieb K, Requa J, Karnes W, Gudivada Chandra R, Shen J, Rael E, et al. Central reading of ulcerative colitis clinical trial videos using neural networks. Gastroenterology. (2021) 160:710–719.e2. doi: 10.1053/j.gastro.2020.10.024
82. Ungaro RC, Hu L, Ji J, Nayar S, Kugathasan S, Denson LA, et al. Machine learning identifies novel blood protein predictors of penetrating and stricturing complications in newly diagnosed paediatric Crohn’s disease. Aliment Pharmacol Ther. (2021) 53:281–90. doi: 10.1111/apt.16136
83. Guez I, Focht G, Greer MLC, Cytter-Kuint R, Pratt LT, Castro DA, et al. Development of a multimodal machine-learning fusion model to non-invasively assess ileal Crohn’s disease endoscopic activity. Comput Methods Programs BioMed. (2022) 227:107207. doi: 10.1016/j.cmpb.2022.107207
84. Iacucci M, Cannatelli R, Parigi TL, Nardone OM, Tontini GE, Labarile N, et al. A virtual chromoendoscopy artificial intelligence system to detect endoscopic and histologic activity/remission and predict clinical outcomes in ulcerative colitis. Endoscopy. (2022) 55:332–41. doi: 10.1055/a-1960-3645
85. Iacucci M, Parigi TL, Del Amor R, Meseguer P, Mandelli G, Bozzola A, et al. Artificial intelligence enabled histological prediction of remission or activity and clinical outcomes in ulcerative colitis. Gastroenterology. (2023) 164:1180–1188.e2. doi: 10.1053/j.gastro.2023.02.031
86. Ogata N, Maeda Y, Misawa M, Takenaka K, Takabayashi K, Iacucci M, et al. Artificial intelligence-assisted video colonoscopy for disease monitoring of ulcerative colitis: A prospective study. J Crohn’s Colitis. (2024). doi: 10.1093/ecco-jcc/jjae080
87. Iacucci M, Santacroce G, Zammarchi I, Maeda Y, Del Amor R, Meseguer P, et al. Artificial intelligence and endo-histo-omics: new dimensions of precision endoscopy and histology in inflammatory bowel disease. Lancet Gastroenterol Hepatol. (2024) 9:758–72. doi: 10.1016/S2468-1253(24)00053-0
88. Stidham RW, Yu D, Zhao X, Bishu S, Rice M, Bourque C, et al. Identifying the presence, activity, and status of extraintestinal manifestations of inflammatory bowel disease using natural language processing of clinical notes. Inflammation Bowel Dis. (2023) 29:503–10. doi: 10.1093/ibd/izac109
89. Babic A, Ster B, Pavesic N, and Wigertz O. Machine learning for the quality of life in inflammatory bowel disease. Stud Health Technol Inform. (1997) 43 Pt B:661–5.
90. Waljee AK, Joyce JC, Wang S, Saxena A, Hart M, Zhu J, et al. Algorithms outperform metabolite tests in predicting response of patients with inflammatory bowel disease to thiopurines. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. (2010) 8:143–50. doi: 10.1016/j.cgh.2009.09.031
91. Menti E, Lanera C, Lorenzoni G, Giachino DF, Marchi De M, Gregori D, et al. Bayesian Machine Learning Techniques for revealing complex interactions among genetic and clinical factors in association with extra-intestinal Manifestations in IBD patients. AMIA. Annu Symp Proc AMIA Symp. (2016) 2016:884–93.
92. Kang T, Ding W, Zhang L, Ziemek D, and Zarringhalam K. A biological network-based regularized artificial neural network model for robust phenotype prediction from gene expression data. BMC Bioinf. (2017) 18:565. doi: 10.1186/s12859-017-1984-2
93. Waljee AK, Sauder K, Patel A, Segar S, Liu B, Zhang Y, et al. Machine learning algorithms for objective remission and clinical outcomes with thiopurines. J Crohns Colitis. (2017) 11:801–10. doi: 10.1093/ecco-jcc/jjx014
94. Waljee AK, Lipson R, Wiitala WL, Zhang Y, Liu B, Zhu J, et al. Predicting hospitalization and outpatient corticosteroid use in inflammatory bowel disease patients using machine learning. Inflammation Bowel Dis. (2017) 24:45–53. doi: 10.1093/ibd/izx007
95. Waljee AK, Liu B, Sauder K, Zhu J, Govani SM, Stidham RW, et al. Predicting corticosteroid-free biologic remission with vedolizumab in crohn’s disease. Inflammation Bowel Dis. (2018) 24:1185–92. doi: 10.1093/ibd/izy031
96. Bottigliengo D, Berchialla P, Lanera C, Azzolina D, Lorenzoni G, Martinato M, et al. The role of genetic factors in characterizing extra-intestinal manifestations in crohn’s disease patients: are bayesian machine learning methods improving outcome predictions? J Clin Med. (2019) 8. doi: 10.3390/jcm8060865
97. Dong Y, Xu L, Fan Y, Xiang P, Gao X, Chen Y, et al. A novel surgical predictive model for Chinese Crohn’s disease patients. Med (Baltimore). (2019) 98:e17510. doi: 10.1097/MD.0000000000017510
98. Lerrigo R, Coffey JTR, Kravitz JL, Jadhav P, Nikfarjam A, Shah NH, et al. The emotional toll of inflammatory bowel disease: Using machine learning to analyze online community forum discourse. Crohn’s Colitis 360. (2019) 1:1–8. doi: 10.1093/crocol/otz011
99. Morilla I, Uzzan M, Laharie D, Cazals-Hatem D, Denost Q, Daniel F, et al. Colonic microRNA profiles, identified by a deep learning algorithm, that predict responses to therapy of patients with acute severe ulcerative colitis. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. (2019) 17:905–13. doi: 10.1016/j.cgh.2018.08.068
100. Ghoshal UC, Rai S, Kulkarni A, and Gupta A. Prediction of outcome of treatment of acute severe ulcerative colitis using principal component analysis and artificial intelligence. J Gastroenterol Hepatol. (2020) 4:889–97. doi: 10.1002/jgh3.12342
101. Popa IV, Burlacu A, Mihai C, and Prelipcean CC. A machine learning model accurately predicts ulcerative colitis activity at one year in patients treated with anti-tumour necrosis factor α Agents. Med. (Kaunas). (2020) 56. doi: 10.3390/medicina56110628
102. Sofo L, Caprino P, Schena CA, Sacchetti F, Potenza AE, and Ciociola A. New perspectives in the prediction of postoperative complications for high-risk ulcerative colitis patients: machine learning preliminary approach. Eur Rev Med Pharmacol Sci. (2020) 24:12781–7. doi: 10.26355/eurrev_202012_24178
103. Uttam S, Hashash JG, LaFace J, Binion D, Regueiro M, Hartman DJ, et al. Three-dimensional nanoscale nuclear architecture mapping of rectal biopsies detects colorectal neoplasia in patients with inflammatory bowel disease. Cancer Prev Res (Phila). (2019) 12:527–38. doi: 10.1158/1940-6207.CAPR-19-0024
104. Wang L, Fan R, Zhang C, Hong L, Zhang T, Chen Y, et al. Applying machine learning models to predict medication nonadherence in crohn’s disease maintenance therapy. Patient Prefer. Adherence. (2020) 14:917–26. doi: 10.2147/PPA.S253732
105. Con D, van Langenberg DR, and Vasudevan A. Deep learning vs conventional learning algorithms for clinical prediction in Crohn’s disease: A proof-of-concept study. World J Gastroenterol. (2021) 27:6476–88. doi: 10.3748/wjg.v27.i38.6476
106. He M, Li C, Tang W, Kang Y, Zuo Y, Wang Y, et al. Machine learning gene expression predicting model for ustekinumab response in patients with Crohn’s disease. Immun. Inflammation Dis. (2021) 9:1529–40. doi: 10.1002/iid3.506
107. Stidham RW, Liu Y, Enchakalody B, Van T, Krishnamurthy V, Su GL, et al. The use of readily available longitudinal data to predict the likelihood of surgery in crohn disease. Inflammation Bowel Dis. (2021) 27:1328–34. doi: 10.1093/ibd/izab035
108. Park SK, Kim YB, Kim S, Lee CW, Choi CH, Kang SB, et al. Development of a machine learning model to predict non-durable response to anti-TNF therapy in crohn’s disease using transcriptome imputed from genotypes. J Pers Med. (2022) 12:1–12. doi: 10.3390/jpm12060947
109. Venkatapurapu SP, Iwakiri R, Udagawa E, Patidar N, Qi Z, Takayama R, et al. A computational platform integrating a mechanistic model of crohn’s disease for predicting temporal progression of mucosal damage and healing. Adv Ther. (2022) 39:3225–47. doi: 10.1007/s12325-022-02144-y
110. Iacucci M, Jeffery L, Acharjee A, Grisan E, Buda A, Nardone OM, et al. Computer-aided imaging analysis of probe-based confocal laser endomicroscopy with molecular labeling and gene expression identifies markers of response to biological therapy in IBD patients: the endo-omics study. Inflammation Bowel Dis. (2023) 29:1409–20. doi: 10.1093/ibd/izac233
111. Soiza RL, Donaldson AIC, and Myint PK. Artificial intelligence to revolutionize IBD clinical trials: a comprehensive review. Ther Adv Vaccines. (2018) 9:259–61.
112. Colombel J-F, Keir ME, Scherl A, Zhao R, de Hertogh G, Faubion WA, et al. Discrepancies between patient-reported outcomes, and endoscopic and histological appearance in UC. Gut. (2017) 66:2063–8. doi: 10.1136/gutjnl-2016-312307
113. Jagannath B, Lin KC, Pali M, Sankhala D, Muthukumar S, and Prasad S. A Sweat-based wearable enabling technology for realtime monitoring of il-1β and crp as potential markers for nflammatory bowel disease. Inflammation Bowel Dis. (2020) 26:1533–42. doi: 10.1093/ibd/izaa191
114. Krugliak Cleveland N, Miyatani Y, Picker EA, and Rubin DT. At-home disease monitoring by patient-performed intestinal ultrasound in severe ulcerative colitis. Inflammation Bowel Dis. (2023) 29:1997–8. doi: 10.1093/ibd/izad237
115. Zhen J, Marshall JK, Nguyen GC, Atreja A, and Narula N. Impact of digital health monitoring in the management of inflammatory bowel disease. J Med Syst. (2021) 45. doi: 10.1007/s10916-021-01706-x
116. McCombie A, Walmsley R, Barclay M, Ho C, Langlotz T, Regenbrecht H, et al. A noninferiority randomized clinical trial of the use of the smartphone-based health applications IBDsmart and IBDoc in the care of inflammatory bowel disease patients. Inflammation Bowel Dis. (2020) 26:1098–109. doi: 10.1093/ibd/izz252
117. Nguyen NH, Martinez I, Atreja A, Sitapati AM, Sandborn WJ, Ohno-Machado L, et al. Digital health technologies for remote monitoring and management of inflammatory bowel disease: A systematic review. Am J Gastroenterol. (2022) 117:78–97. doi: 10.14309/ajg.0000000000001545
118. Riaz MS and Atreja A. Personalized technologies in chronic gastrointestinal disorders: self-monitoring and remote sensor technologies. Clin Gastroenterol Hepatol Off Clin Pract J Am Gastroenterol Assoc. (2016) 14:1697–705. doi: 10.1016/j.cgh.2016.05.009
119. de Jong MJ, van der Meulen-de Jong AE, Romberg-Camps MJ, Becx MC, Maljaars JP, Cilissen M, et al. Telemedicine for management of inflammatory bowel disease (myIBDcoach): a pragmatic, multicentre, randomised controlled trial. Lancet (London England). (2017) 390:959–68. doi: 10.1016/S0140-6736(17)31327-2
120. Del Hoyo J, Nos P, Faubel R, Muñoz D, Domínguez D, Bastida G, et al. A web-based telemanagement system for improving disease activity and quality of life in patients with complex inflammatory bowel disease: pilot randomized controlled trial. J Med Internet Res. (2018) 20:e11602. doi: 10.2196/11602
121. Marín-Jiménez I, Nos P, Domènech E, Riestra S, Gisbert JP, Calvet X, et al. Diagnostic performance of the simple clinical colitis activity index self-administered online at home by patients with ulcerative colitis: CRONICA-UC study. Am J Gastroenterol. (2016) 111:261–8. doi: 10.1038/ajg.2015.403
122. Sciberras M, Farrugia Y, Gordon H, Furfaro F, Allocca M, Torres J, et al. Accuracy of information given by chatGPT for patients with inflammatory bowel disease in relation to ECCO guidelines. J Crohns Colitis. (2024) 18:1215–21. doi: 10.1093/ecco-jcc/jjae040
123. Bajwa J, Munir U, Nori A, and Williams B. Artificial intelligence in healthcare: transforming the practice of medicine. Futur Healthc. J. (2021) 8:e188–94. doi: 10.7861/fhj.2021-0095
124. Giebel GD, Speckemeier C, Abels C, Plescher F, Börchers K, Wasem J, et al. Problems and barriers related to the use of digital health applications: scoping review. J Med Internet Res. (2023) 25:e43808. doi: 10.2196/43808
Keywords: artificial intelligence, inflammatory bowel disease, machine learning, digital biomarkers, personalized medicine artificial neural network, APR: algorithm-predicted remission, BPNN: back-propagation neural network, CART: classification and regression trees
Citation: De Deo D, Dal Buono A, Gabbiadini R, Nardone OM, Ferreiro-Iglesias R, Privitera G, Bonifacio C, Barreiro-de Acosta M, Bezzio C and Armuzzi A (2025) Digital biomarkers and artificial intelligence: a new frontier in personalized management of inflammatory bowel disease. Front. Immunol. 16:1637159. doi: 10.3389/fimmu.2025.1637159
Received: 28 May 2025; Accepted: 11 July 2025;
Published: 04 August 2025.
Edited by:
Anamelia Lorenzetti Bocca, University of Brasilia, BrazilReviewed by:
Luiz Gustavo Gardinassi, Department of Health, BrazilPu Wang, University of Electronic Science and Technology, China
Copyright © 2025 De Deo, Dal Buono, Gabbiadini, Nardone, Ferreiro-Iglesias, Privitera, Bonifacio, Barreiro-de Acosta, Bezzio and Armuzzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arianna Dal Buono, YXJpYW5uYS5kYWxidW9ub0BodW1hbml0YXMuaXQ=
 Diletta De Deo
Diletta De Deo Arianna Dal Buono
Arianna Dal Buono Roberto Gabbiadini
Roberto Gabbiadini Olga Maria Nardone
Olga Maria Nardone Rocio Ferreiro-Iglesias
Rocio Ferreiro-Iglesias Giuseppe Privitera1,2
Giuseppe Privitera1,2 Manuel Barreiro-de Acosta
Manuel Barreiro-de Acosta Alessandro Armuzzi
Alessandro Armuzzi