- 1Protein Engineering Lab, School of Biosciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India
- 2Department of Pharmaceutical Sciences, Pharmacy Program, Batterjee Medical College, Jeddah, Saudi Arabia
- 3Institute of Pharmaceutical Research, GLA University, Mathura, Uttar Pradesh, India
- 4Department of Chemistry and Biochemistry, School of Sciences, JAIN (Deemed to Be University), Bengaluru, Karnataka, India
- 5Department of General Medicine, IMS & SUM Hospital, Siksha ‘O’ Anusandhan (Deemed to be University), Bhubaneswar, Odisha, India
- 6Department of Pharmacology, Teerthanker Mahaveer College of Pharmacy, Teerthanker Mahaveer University, Moradabad, Uttar Pradesh, India
- 7Department of Pharmacology, Kyrgyz State Medical College, Bishkek, Kyrgyzstan
- 8Uttaranchal Institute of Pharmaceutical Sciences, Uttaranchal University, Dehradun, Uttarakhand, India
- 9Centre for Research Impact and Outcome-Chitkara College of Pharmacy, Chitkara University, Rajpura, Punjab, India
- 10Centre of Medical and Bio-allied Health Sciences Research, Ajman University, Ajman, United Arab Emirates
- 11Department of Integrative Biology, School of Biosciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India
Recent studies on macrophages showed their contribution to tumorigenesis, progression, metastasis, and chemoresistance by influencing the local tumor microenvironment and cancer cells. Exosomes form a subset of extracellular vesicles and have played a major role in the interaction between cancer cells and macrophages. This review intends to discuss the existing literature on employing macrophage-derived exosomes as a vehicle for microRNA (miRNA) delivery in oncological applications. It will evaluate the molecular principles of this therapeutic approach and its capacity to enhance cancer therapy by elucidating problems like drug and radio-resistance. This review uniquely emphasizes the diagnostic and therapeutic potential of macrophage-derived exosomal miRNAs, summarizing current understandings into their molecular processes, tumor specificity, and strategies to overcome therapeutic resistance. This review synthesizes recent studies and evaluates how macrophage-derived exosomes and their miRNAs contribute to cancers. These vesicles are multipurpose tools that regulate tumor behavior, considering they can regulate it through post-transcriptional regulation and protein phosphorylation. Such exosomes that are engineered can potentially introduce a novel dimension because they have the capability of delivering targeted oncogenic or tumor-suppressive miRNAs to overcome limitations of current cancer therapeutics, particularly drug and radioresistance. Engineered macrophage-derived exosomes may thus have the potential as a novel approach for cancer treatment and overcoming therapeutic resistance.
1 Introduction
Exosomes are extracellular vesicles (EVs), typically 30–150 nm in diameter, secreted by various cells, such as cancer and immune cells or stromal cells (1). Proteins, nucleic acids, and lipids carried by these vesicles are biologically active and mediate intercellular communication (2). Exosomes have been in the limelight in the field of oncology and are already regarded as significant contributors to the tumor microenvironment (TME) (3). They participate in numerous processes essential to tumorigenesis, e.g., tumor growth, metastasis, immune evasion, and the development of drug resistance (4). The fact that exosomes can induce plasticity in the recipient cells, mostly through inciting malignant transformation, makes them an indispensable to cancer development and a target for therapeutic intervention (5, 6). Exosomes form as intraluminal vesicles (ILVs) in multivesicular bodies (MVBs) and are released by fusion with the plasma membrane; their proteins, lipids, and nucleic acid cargo can reprogram recipient cells within the TME (7). In cancers, tumor-associated macrophages (TAMs) tend to be biased towards immunoregulatory M2-like phenotypes, with the M1-like macrophages being pro-inflammatory. Meanwhile, macrophage-derived exosomes (MDEs) are exosomes that are released by macrophages; their microRNA (miRNA) cargo (MDE-miRNAs) can be suggestive of macrophage polarization and local stimuli and reorganize tumor growth, metastasis, immune evasion, and therapeutic resistance (8, 9). MDEs may play an important role in cancer biology (10). The TAMs are primarily polarized to a pro-tumorigenic phenotype (11). These TAMs will be involved in circuits maintaining tumor growth, angiogenesis, or invasion/metastasis, which may reprogram cancer behavior (12). By transmitting such a load, MDEs stimulate tumor cell survival and growth and invasion, and the development of an immunosuppressed microenvironment hostile to immune editing by tumor cells (13, 14).
Recent advancements in studies have navigated this avenue of MDE therapeutics, particularly with regard to the delivery of miRNA to cancer cells. Selective treatment based on the affinity of MDEs to areas of tumors creates an appealing treatment strategy (15). Targeted delivery of therapeutic miRNAs resulting in gene modulation via MDEs has the potential to prevent the advancement of cancer and overcome the limitations present in conventional nanocarriers (16). In addition, individual variability in MDEs across cancers is the rationale for personalized medicine approaches (17). MiRNA content personalization in MDEs with the help of the tumor environment signature can lead to amelioration and personalized intervention (18). Developments in our understanding of the complexity of MDEs in cancer hold great possibilities to be further exploited as potential markers and as therapeutic vehicles in this context to allow the creation of specific new treatment paradigms (19). Even though exosome biogenesis is common across cell types, it is the cell of origin that leaves a mark on cargo and cellular functions (20). Relative to tumor-derived exosomes (TDEs) that tend to exacerbate oncogenic messaging in cancer cells, MDEs predominantly inform the TME and remodel cancer cells, stromal cells, and immunity by miRNAs associated with the macrophage state. They also exhibit tropism for hypoxic tumor subregions, and MDEs carry immunomodulatory signals that are less noticeable in alternative stromal exosomes. Although there is growing evidence on the diverse functions, there is inadequate systematic knowledge on how macrophage-derived exosomal miRNAs determine tumor biology and respond to therapy. The review is a summary of contemporary information on their mechanistic contributions, clinical associations with multiple large-scale cancer types, and obstacles to utilizing them in translation research. Given their dual potential role in tumor promotion and suppression, these exosomes represent a promising yet underexplored therapeutic avenue. The primary focus in this review is MDEs; other exosomes released by tumor cells or other stromal cells are only mentioned in the form of contrast where necessary. Understanding their molecular mechanisms and translational potential could unlock innovative strategies to overcome drug and radioresistance, paving the way for more precise and effective cancer treatments. In this review, we have explored the potential of Macrophage-derived exosomal miRNAs in tumor progression and resistance.
2 Biogenesis of macrophage-derived exosomes
2.1 Mechanisms of exosome formation
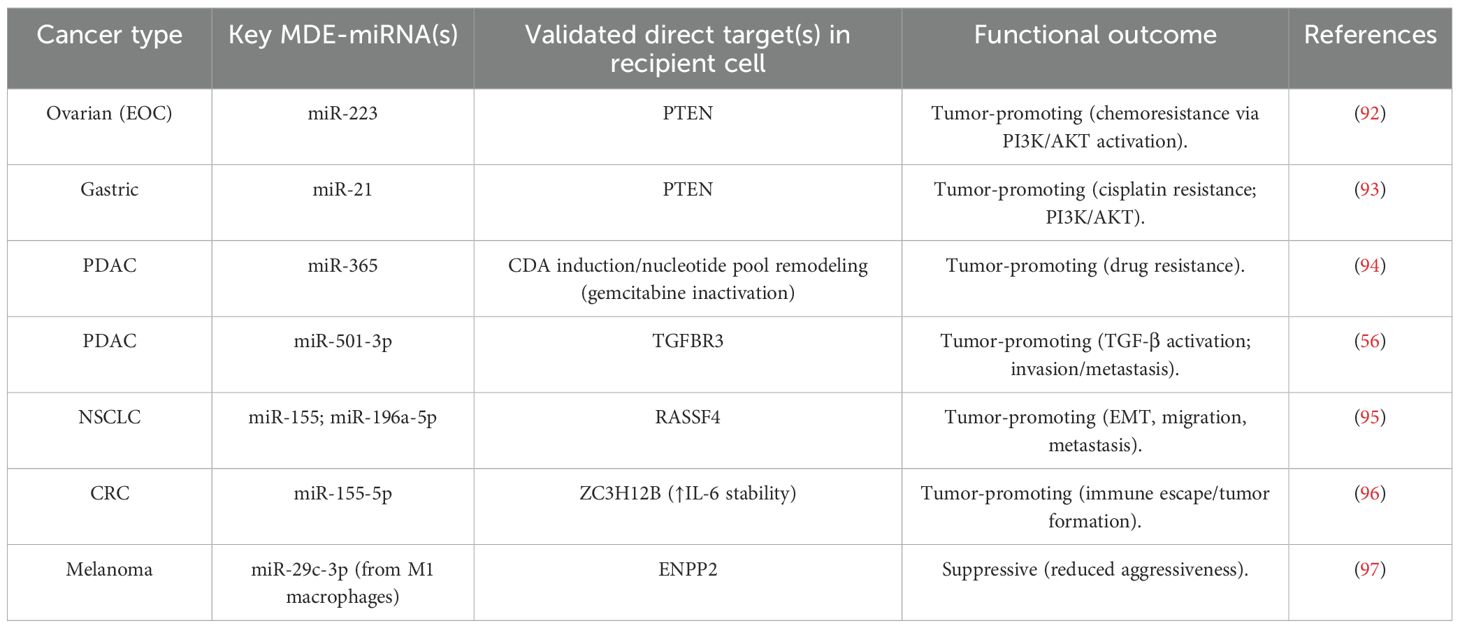
The biogenesis of exosomes initiates with the internal budding of the cellular plasma membrane, leading to small vesicle-like structures called early endosomes; this is followed by a highly complex process known as MVBs formation (21). Subsequent invagination of these early endosomes leads to the formation of ILVs within MVBs (22). One of these steps is the formation of ILVs, which are important precursor vesicles containing bioactive cargo that will ultimately be delivered to recipient cells in exosomes (23). The membrane of these endosomes is then remodeled and organized into discrete domains that are enriched in particular proteins, lipids, or nucleic acids (24). This sorting process is mostly selective, by which only specific molecules are included in exosomes (25). After MVBs reach full maturation, they either fuse with lysosomes for degradation or fuse with the plasma membrane and release ILVs as exosomes into the extracellular space (26). The choice between these fates is controlled by several signaling pathways and molecule interactions that are the subject matter of ongoing research (27). At the heart of this endosomal maturation process are Ras-associated binding (Rab) GTPases that direct MVBs to fuse with the cell surface through SNARE protein-dependent final fusion (28). Exosomes are released into the extracellular space to act as a mode of intercellular communication, particularly within the TME, thus influencing cancer progression (Figure 1) (29). The polarization state (M1 vs. M2/TAM) of macrophages in particular alters miRNA abundance and loading in exosomes through established sorting pathways to remodel the exosomal repertoire.
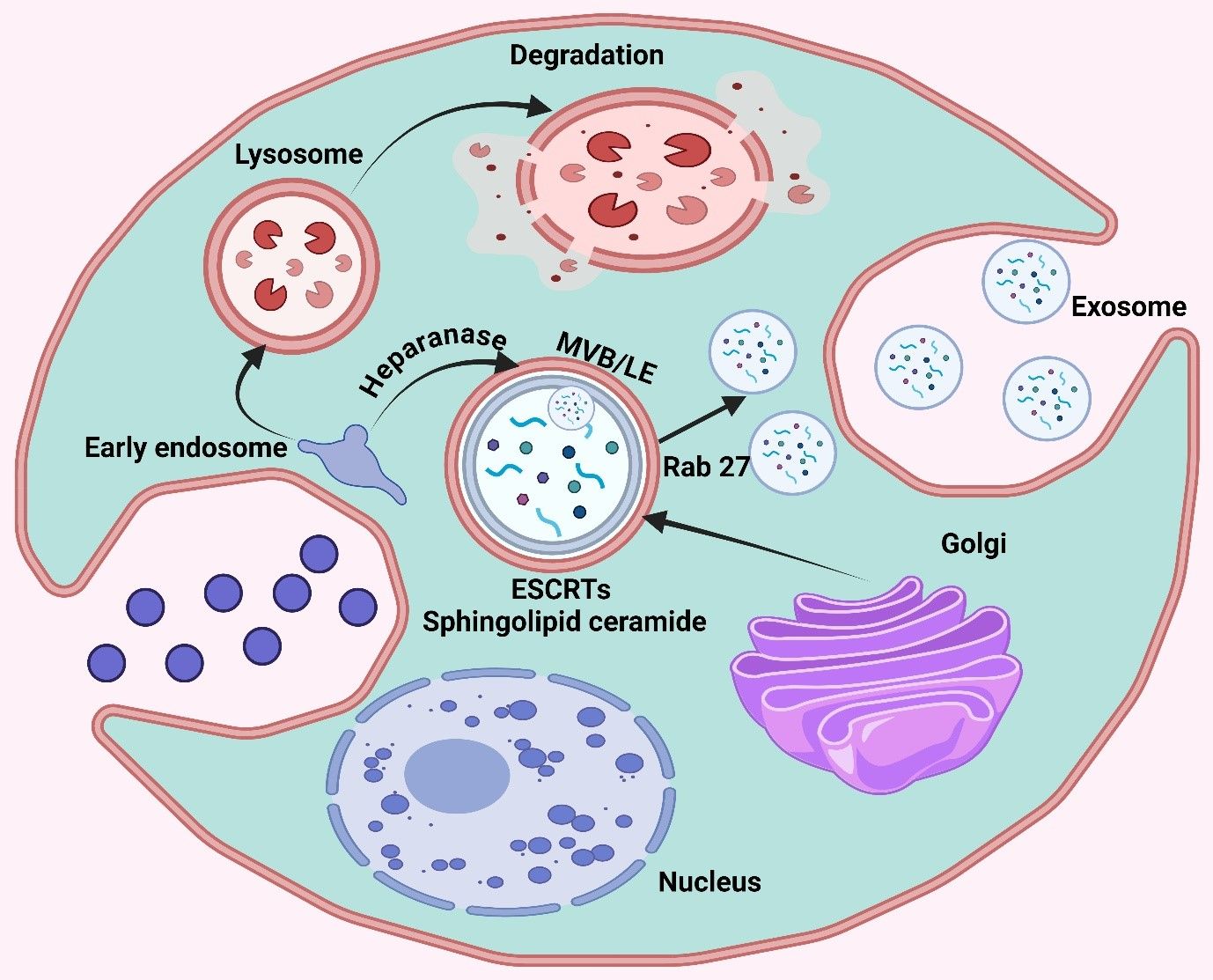
Figure 1. The graphic illustrates the cellular process of exosome production. This diagram depicts the process of early endosome creation leading to the production of multivesicular bodies (MVB/LE) and the eventual release of exosomes, as well as the routes involved in lysosomal degradation. Important compounds include ESCRT, Rab27, and sphingolipid ceramide.
Polarization leaves different miRNA signatures on the MDEs with functional implications in the tumor. M1-polarized macrophages specifically pack miR-155-5p into exosomes (greater in M1 than in M0/M2), and M1-exosomal miRNA can have anti-tumor effects in several models. Conversely, the reports of M2/TAM exosomes being repeatedly highly enriched with oncogenic miRNAs that engage cancer cells and trigger activated pro-survival signatures and invasion, and chemoresistance are recurrently mentioned (30). Mechanistically, such polarization-specific differences in cargo fold into general pathways of exosome sorting, hnRNPA2B1 attachment to EXOmotifs (31), YBX1-selection (e.g., of miR-223) (32), relative enrichment of 3′-uridylated isoforms in exosomes (33), and ceramide/neutral sphingomyelinase 2 (nSMase2)-mediated miRNA export (34) to provide a macrophage-specific basis to how M1 versus M2 states bias the exosomes-mediated miRNA positing.
2.2 Role of ESCRT machinery and other molecular players
The endosomal sorting complexes required for transport (ESCRT) machinery is an ATP-dependent mechanism that functions in the packaging of cargo into intraluminal vesicles (ILVs) within multivesicular bodies (MVBs), a critical step during exosome biogenesis (35). The ESCRT machine consists of four large complexes that include ESCRT-0, I, II, and III. Various complexes perform different roles in the process of exosome production (36). The major role of ESCRT-0 is to identify and enclose the cargo tagged with ubiquitin at the endosome membrane. During the formation of the MVBs, ILVs are generated by the process catalyzed by ESCRT-I and ESCRT-II. Meanwhile, ESCRT-III performs a last separation by breaking membranes to release nascent ILVs inside the lumen of MVB (37).
A wide variety of accessory proteins has also been shown to be involved in exosome biogenesis in addition to the ESCRT complexes (38). Alix and TSG101 are significant regulators that get involved with the ESCRT apparatus in orchestrating proper cargo sorting/packaging. Alix plays a critical role in the process of budding, and TSG101 associates with the ubiquitinated protein to mark them in the immature exosomes (39). Along with this, other molecular elements such as Rab GTPases are also necessary to release exosomes in the plasma membrane through transport and docking of MVBs (40). Two of the Rab family members, namely Rab27a and Rab27b have been demonstrated as being critical in the localization of MVBs near the plasma membrane through exosome release. Newer studies have also indicated the orchestration of the lipid nanodomains (also called as room rafts) in the exosome biogenesis (41). These membrane lipid-rich areas serve as protein complex scaffolds that participate in exosome formation (42). Consequently, the biophysical nature of such microdomains is significantly modulated by cholesterol, sphingomyelin, and ceramide, and hence various abilities to convert them to exosomes (43).
2.3 Influence of external stimuli
The generation and secretion of MDE are induced by external factors such as hypoxia, inflammation, or oxidative stress. EV content changes in the hypoxic, acidic, and low glucose TME, which leads to alterations in their function (44). Hypoxia is a hallmark of solid tumors known to promote the secretion of exosomes from cancer cells and TAMs (45). Exosomal cargo is likewise altered under hypoxic conditions that tend to enrich for pro-angiogenic factors such as miRNA and vascular endothelial growth factor (VEGF), enhancing metastasis. These observations suggest that not only does hypoxia produce exosomes, but it also modifies their cargo to the advantage of tumor progression (46). In line with its inflammatory status, which is well characterized in cancer, exosome biogenesis was also altered. The released exosomes contain additional protein and RNA cargo when macrophages are exposed to inflammatory cytokines such as TNF-α or IL-6 (47, 48). Studies have demonstrated that exosomes secreted from inflamed cells can deliver immunosuppressive molecules to dampen the activation of cytotoxic T cells and facilitate immune evasion (49). On the other hand, cellular stress, such as that induced by oxidative stress and nutrient deprivation or provoked by treatment with chemotherapeutic agents, may result in increased exosome secretion (50, 51). This stress-induced exosomal secretion is believed to represent a cytoprotective process that removes deleterious proteins and RNAs from cells, as well as modulates the TME, which further influences cancer cell proliferation/survival in response to treatment (52).
3 Functions of MDEs in cancer
The MDEs significantly contribute to tumor biology through intercellular communication in the TME. These vesicles convey regulatory miRNAs, proteins, and lipids that significantly affect cancer cells’ surrounding stroma. MDEs can lead to tumor progression, angiogenesis, invasion, and immune suppression and therapy resistance depending on their molecular cargo (53). The subsections below provide an overview of the key MDE-regulated processes in cancer progression.
3.1 MDEs promote tumor proliferation
One of the major functionalities that MDEs carry out in case of cancer is to create a suitable condition for tumor growth and proliferation (54). Oncogenic dysregulation of MDEs often involves oncogenic miRNAs and proteins to instigate signaling pathways in human cancer cells, e.g., PI3K/AKT or Wnt/β-catenin pathway that brings about increased cell proliferation/survival (55). MiR-501-3p in M2-exosomes favors tumor progression by triggering the transforming growth factor-β (TGF-β) cascade and suppressing the tumor suppressor gene TGFBR3 (56). In human epithelial ovarian cancer (EOC), exosomal miR-221−3p derived from M2 macrophages promotes cancer growth by reducing the cyclin-dependent kinase inhibitor 1B (CDKN1B) (57). A separate investigation indicates that exosomal miR-29a-3p and miR-21−5p derived from M2 macrophages increase the proportion of T regulatory cells (Tregs) to T helper cell 17 (Th17), thereby contributing to a tumor immune inhibitory TME and facilitating the development of cancer and metastasis (58). The data indicate that macrophage-derived exosomal miRNAs target not only cancerous cells directly but also the immune systems, thereby influencing cancer cells indirectly.
3.2 MDEs promote metastasis
Moreover, MDEs modulate the TME by modifying stromal cell behavior, favoring angiogenesis, and establishing a permissive niche for many phases of tumorigenesis. MDEs are also involved in promoting the metastatic ability of cancer cells (59, 60). Through the transmission of pro-metastatic molecules, MDEs can reshape extracellular matrices to promote cancer cell invasiveness and assist in developing pre-metastatic niches at target organs (61). M2-Exos carry lncRNA AFAP1-AS1, downregulating miR-26a and upregulating activating transcription factor 2 (ATF2), hence facilitating esophageal cancer (EC) penetration and metastasis. Engaging M2 macrophages and the lncRNA AFAP1AS1/miR-26a/ATF2 pathway is a promising treatment option for EC (62). Lan et al. observed that M2-Exos regulate Brahma-related gene 1 (BRG1) by delivering miR-21 and miR-155-5p, leading to the downregulation of BRG1 and promoting colorectal cancer (CRC) metastasis (63).
3.3 MDEs confer drug resistance
Furthermore, MDEs are also involved in drug resistance to cancer. They can take up and transmit miRNAs, as well as other molecules that regulate the expression of drug-resistance genes in cancer cells, which helps reduce both chemotherapeutic response and response to targeted therapies (64, 65). Such exosome-mediated communication develops a multidrug-resistant and aggressive tumor phenotype. Recent data indicate that MDEs-derived miRNAs exert an inhibitory effect on cancer. MDEs, rich in miR-7, are internalized by EOC cells following treatment with tumor necrosis factor-like weak inducer of apoptosis (TWEAK). This internalization inhibits cell invasion by targeting the EGFR-mediated AKT/ERK system (66). MDEs miR-let-7a-5p can be transferred to lung cancer cells, resulting in the inhibition of cell growth, migration, and invasion through the downregulation of Bcl2-like 1 (BCL2L1) expression (67).
3.4 MDEs regulate immune responses
MDEs partake in the immune regulation within the TME (68). Consequently, these cells can modulate anti-tumor immune responses by affecting the function of different types of innate and adaptive immunity effector cells, leading to immunostimulants and cancer progression (69).
4 miRNA loading and cargo selection in MDEs
MiRNA loading and cargo selection in MDEs is a tightly regulated transcriptional program that dictates response to various stimuli, including cancer (70). Besides, the selective incorporation of specific miRNAs and other bioactive molecules into MDEs is not random, as specific cellular mechanisms mediate this process to provide directed signaling to recipient cells. MiRNA incorporation into MDEs occurs during the biogenesis of these vesicles in parent mesenchymal stromal/stem cells (MSCs) (71, 72). Many important proteins and pathways participate in modulating the selective packaging of miRNAs (73, 74). Key players for recognizing and mediating the loading of selected miRNAs into MDEs are those represented by RNA-binding proteins (RBPs), such as Argonaute 2 (Ago2), heterogeneous nuclear ribonucleoproteins, and endosomal sorting complexes required for transport (75, 76). Exosome-targeting motifs (EXO-motifs; GGAG/UGGA-like motifs) and U/CA-rich elements are preferentially bound by hnRNP family proteins, in macrophages, enhancing their export in budding intraluminal vesicles. The miRNAs that interact with Ago2 are selectively targeted to exosomes by a combination of their stable seed imaginary pairing potential and their preference for 5’-nucleotides to interact with Ago2. Additional sequence-independent sorting bias modulates RBP affinity/retention, versus export decisions are affected by secondary structure and 3-end modifications. A combination of these features can be used to operationalize the selective, non-random packaging of miRNAs into macrophage-derived exosomes. In practice, these RBPs read the above EXO-motifs, U/CA-rich elements, 3′-end marks, and exposed hairpins to triage individual miRNAs for vesicular export, committing them as cargo precursors during ILV budding within maturing MVBs (77). The miRNAs are then loaded into the intraluminal vesicles, which, after the bulbs are fused to the plasma membrane, lead to the release of MDEs out of the cell. The miRNA content of MDEs may reflect the physiological status of MSCs and even stimuli from external surroundings (78, 79). Under hypoxic conditions or in response to inflammatory signals, MSCs may selectively furnish MDEs with different miRNA cargos (i.e., pro-angiogenic miRs for TME redirecting recipient cancer cells and immunomodulatory miRs preventing anti-tumoral immune cell activation) (80). Hypoxia, which is a characteristic of solid tumors, enhances exosome release by TAMs and cargo composition, in part by altering the expression and post-translational status of RBPs, favoring certain motifs and diminishing others (81). Cellular stress and inflammatory cytokines also reprogram exosomes’ biogenesis and cargo selection, enriching pro-angiogenic or immunomodulatory miRNAs and reconfiguring the TME (82). Such context-dependent modifications justify the reason why exosomes of hypoxia-related, macrophage-produced exosomes often carry pro-metastatic/therapy-resistance miRNAs, which are evident throughout Section 5.
In addition to miRNAs, MDEs also selectively incorporate other molecular cargo, such as proteins, lipids, and lncRNA, under specific cellular contexts to mediate a targeted biological effect (83–85). The cargo sorting is indispensable to the function of MDEs in regulating the TME, promoting metastasis, and inducing drug resistance (86). The interaction between miRNAs and other cargo components in MDEs may enhance their potency on recipient cells synergistically (87). The miRNA cargo of MDEs in cancer can downregulate tumor suppressor genes or upregulate oncogenes in recipient cells, leading to the promotion of tumorigenesis and metastasis. Additionally, MDEs could be artificially designed to carry tumor-suppressive miRNAs and serve as a therapeutic strategy (88–90). Elucidating these processes in MDEs will be critical for the future design of targeted therapies that either suppress the pathological functions of MDEs or use them to deliver therapeutic molecules in cancer and potentially other diseases (91).
5 Applications of miRNA-loaded macrophage-derived exosomes as delivery vehicles
For a cross-comparison of key MDE-miRNAs, their validated targets, and net functional outcome across cancers, refer to Table 1.
5.1 Lung cancer
Lung cancer, the most lethal of all cancers worldwide, arises principally in cells that line the airways. This is generally divided into non-small cell lung cancer (NSCLC), which represents around 85% of cases, and small cell lung cancer, which is more aggressive (98). This disease is associated with risk factors like smoking, carcinogen exposure, and genetic predisposition. The early stage of lung cancer is often asymptomatic; thus, most patients present with advanced disease at diagnosis, and they usually have a poor prognosis (99, 100). Lung cancer is the leading cause of cancer deaths, with more than 2 million new cases reported every year worldwide, especially in developing countries where smoking rates are high. It occurs much more often among men, although it is becoming an increasingly common problem with women (101). Currently, despite increasingly effective surgical and chemotherapeutic treatments, the five-year survival rate is only around 20%, mainly due to the late detection of disease with its aggressive nature (102, 103). There is a growing realization that miRNAs are key players in gene expression closely associated with lung tumor development, progression to advanced disease, and therapeutic resistance (104, 105). All these miRNAs can work as oncomiRs or tumor suppressor genes, influencing key regulators implicated in cell proliferation, apoptosis, and metastasis (106, 107). Considering the ability of miRNAs to serve as both blood-based markers for early detection and prognosis, in addition to being therapeutic targets themselves, they represent a novel target group in personalized treatment strategies for lung cancer (108). For instance, let-7a-5p downregulates the oncogene BCL2L1 by· thereby targeting it for gene silencing through association with a specific mRNA encoding protein activity that affects multiple signaling transcripts and inhibiting cell proliferation and migration/invasive phenotype in lung cancer (109). The miRNA activates the PI3Kγ signaling pathway and induces autophagy and apoptosis in human lung cancer cells (110). Duan et al. utilized let-7a-5p as exosome cargo because high levels of this miRNA in macrophages hinder lung cancer, and the cells also treated A549 NSCLC with BCL2L1 are targets. Surprisingly, let-7a-5p upregulation induced autophagy and cell death in lung cancer cells without triggering apoptosis or pyroptosis (Figure 2) (111).
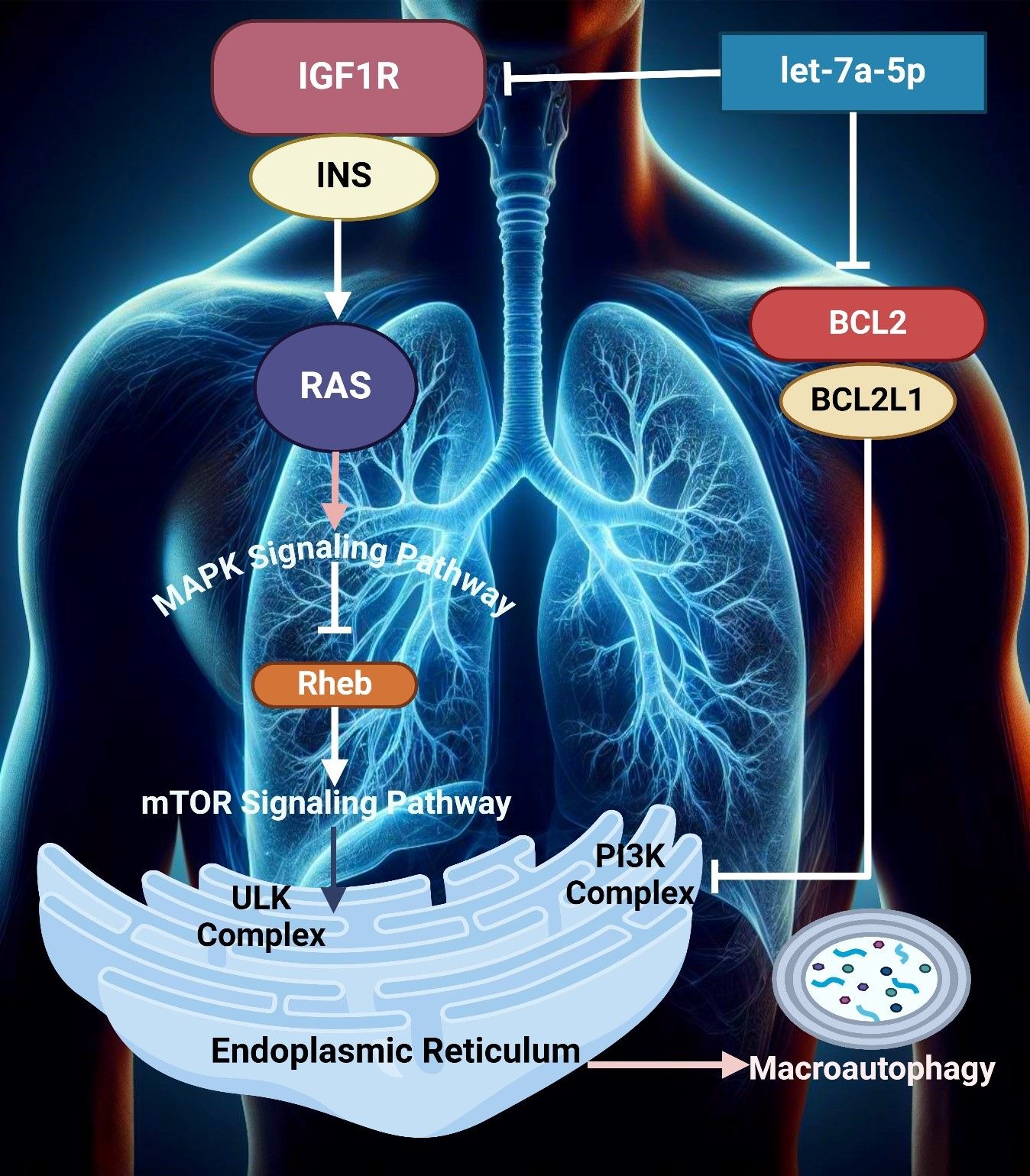
Figure 2. The image illustrates key signaling pathways involved in lung cellular processes, highlighting the MAPK and mTOR pathways. It depicts the interaction between IGF1R, INS, and RAS and the downstream effects on autophagy through the endoplasmic reticulum, mediated by Rheb and PI3K complexes.
The biological role of TNF-α in lung cancer is paradoxical, being capable of both averting tumor proliferation and also facilitating it (112, 113). On the one hand, this cytokine can mediate inflammation and promote tumor growth through activating transduction pathways that enhance cell survival processes like nuclear factor-κB (NF-κB). It acts alone or together with other agents under various conditions, selectively kills cancer cells by apoptosis cascade clearance mediated through ligating its receptor pathway (114, 115). Jiao et al. identified that TNF-α-stimulated exosomes of PMNs were implicated in sepsis-related ALI, promoting M1 macrophage activation and pyroptosis via the NF-κB signal pathway. Inhibition of this pathway is regulated by exosomal miR-30d-5p directly targeting SOCS1/SIRT1 in macrophages. Knocking down miR-30d-5p decreased the pyroptosis, macrophage activation, and lung injury in sepsis-related acute lung injury (ALI) rats by inhibiting PMN-M interaction via a novel mechanism; these results suggest new potential therapeutic targets for treating sepsis (116).
Moreover, the use of certain microRNAs as diagnostic biomarkers led to progress in the early detection of lung cancer (117). Cazzoli et al. discovered microRNAs able to distinguish lung adenocarcinomas from benign lung conditions with sensitivities and specificities exceeding 90 percent. Their screening test was 97.5% sensitive and 72.0% specific, whereas, for lung adenocarcinoma versus granulomas, their diagnostic assay had a sensitivity of 96.0%, with a specificity of only 60%. This implies a promise of using circulating exosomal miRNAs as early diagnostic markers for lung cancer (118). In addition, Munagala et al. showed that miR-21 and miR-155 exhibited higher levels of the two sequences in serum exosomes from recurrent lung tumors compared to primary ones. These miRNAs are proposed to be potential biomarkers for non-invasive diagnosis of recurrent lung cancer, as their expression patterns in exosomes overlap with those of pathological tissues from primary and metastatic lung tumors. Consequently, miRNAs serve as potential markers of disease progression and relapse monitoring (119). Exosomal miRNAs including let-7a-5p, miR-21, and miR-155 that are released by macrophages influence both the proliferation of tumors and development of drug-resistance as well as function as non-invasive diagnostic and predictive biomarkers that can be detected in serum exosomes (120, 121), which has a practical value in screening and monitoring drug-resistance lung cancer in early diagnosis and the chosen therapeutic response and treatment.
5.2 Breast cancer
Breast cancer is the most frequently diagnosed of all cancers among women, with the highest incidence rates in North America and Europe; it originates in ducts (ductal carcinoma) or lobules (lobular carcinoma) (122). With the capacity to metastasize, this cancer can spread all over, and hence its early detection matters (112, 113, 123). The prognosis has substantially improved with the advent of screening methods such as mammography, which allows early detection. There are many treatments for breast cancer, which vary depending on the stage of the disease and its characteristics (124–126). Although the disease is much more common in women over 50, even men can have breast cancer. Age, genetic mutations (e.g., BRCA1/2), family history, reproductive history, and lifestyle factors. Although advancements in detection and treatment have improved survival rates, disparities persist, influenced by geographic, socioeconomic, and racial factors (127).
MiRNAs are involved in the post-transcriptional regulation of gene expression and play a fundamental role in breast cancer by controlling genes related to growth, metastasis, or resistance to therapy. These miRNAs exert oncogenic or tumor suppressor roles modulating key signaling pathways that lead to cancer development (128–130). Due to their potential as biomarkers for diagnosis and prognosis or even therapeutic targets, miRNAs are of great interest in breast cancer research workflow development. In breast cancer, bone marrow stromal macrophages exert a dual effect on both cancer cell dormancy and chemoresistance (131). The M2 macrophages are mainly associated with the induction of dormancy and chemoresistance to cancer cells. The M1 macrophages, however, counteract the dormancy, reactivating the cancer cells and making them more susceptible to chemotherapy (132). This balance is essential in the determination of both dormancy and susceptibility to the treatment of breast cancer in the bone marrow (133). Walker et al. revealed that bone marrow stromal macrophages induced breast cancer cell dormancy, with M2 cells enhancing quiescence and resistance to carboplatin via a gap junctional communication. However, the process was reversed by the M1 cells, which activated NF-κB, increased cellular sensitivity to carboplatin, and improved the survival of the hosts. Driving the modulation of the macrophage phenotype could be the preferred treatment option to control breast cancer dormancy. This will hinder cancer cells from spreading to other parts of the body (134).
Exosomes, as one of the types of microvesicles, are of paramount importance in breast cancer, as they help cells communicate with each other (135). Cells are capable of transferring oncogenic molecules, including several forms of miRNA, proteins, and lipids responsible for the cells’ promotion of tumor growth, invasion, and metastasis (136). Exosomes are of essential importance due to their role in forming the TME (137). According to the study by Yang et al., IL-4-activated macrophages enhance the invasion of breast cancer cells by transferring miR-223. The latter, in turn, raises the invasiveness of cancer cells through the Mef2c-β-catenin pathway. Inhibition of miR-223 in macrophages decreases the suppressive effect of these cells on the invasiveness of breast cancer cells in co-culture, demonstrating the pivotal role of the exosomal transfer of miRNAs in the interaction of macrophages with breast cancer cells that promotes metastasis (138).
One of the leading accelerators of breast cancer development is TAMs. They provide favorable conditions for tumors by enhancing growth, invasion, and metastasis and promoting angiogenesis, immune escape, and tissue remodeling in the TME (139, 140). In most cases, TAMs are oriented toward the M2 type, as it is closely related to the poor prognosis of breast cancer patients (141). The study of Li et al. shows that miR-146a and miR-222 are downregulated, which contributes to tumor progression. Specifically, miR-146a promotes M2 macrophage polarization, which accelerates tumor progression. Secondly, miR-222 inhibits TAM chemotaxis by targeting C-X-C chemokine receptor type 4 (CXCR4) and C-X-C motif chemokine ligand 12 (CXCL12), which ultimately suppresses tumor growth in vivo. Thus, miRNAs are a significant factor in regulating TAM functioning and the rate of breast cancer progression (142). MDEs’ miRNAs such as miR-223 and miR-146a affect metastasis and macrophage polarization and hence connect macrophage polarization with tumor aggressiveness and immune evasion (120, 143). Their constancy is contributed by their exosomal state that allows food to nurture them as floating biomarkers and possible agents to reverse this chemoresistance within breast cancer.
5.3 Ovarian cancer
Ovarian cancer is a malignancy that starts in the ovaries and often goes undetected until it has spread within the pelvis and abdomen. Therefore, ovarian cancer is the most malignant of gynecologic cancers due to late diagnosis (144, 145). The disease reportedly has several subtypes, including EOC, which is the most common. Generally, however, ovarian cancer is usually manifested by non-specific symptoms such as bloating, pelvic pain, and changes in bowel habits, which makes its detection at early stages quite challenging (146). Moreover, the disease is the eighth most common type of cancer among women worldwide, in addition to being the greatest cause of death from all gynecologic malignancies (147, 148). Ovarian cancer is more commonly reported in Europe and North America than in other parts of the world (149). The disease is mainly encountered among postmenopausal women, with a median age at diagnosis standing at 63 years. Despite the rapid increase in the number of ovarian cancer treatment options, the overall 5-year survival rate remains less than 50% due to its tendency to be diagnosed in late stages (150). In particular, risk factors are common and include age, family history of ovarian cancer, family history of breast cancer, a personal history of breast or ovarian cancer, BRCA1 or BRCA2 mutation, never having given birth, and estrogen hormone therapy (151).
Ovarian cancer depends on oncogenic or tumor suppressor miRNAs that control gene expression involved in tumor development, invasion, and resistance to chemotherapy. Some miRNAs have been strongly linked to poor clinical outcomes in ovarian cancer and have been proposed as early diagnostic or therapeutic targets (152, 153). Li et al. found that exosomal miR-221-3p originating in TAMs drives late EOC progression via CDKN1B inhibition and is selectively enriched in M2 exosomes. miR-221-3p is highly enriched in both the cytoplasm and nucleus of M2 exosomes and appears to stimulate the G1/S phase transmission of EOC cells. The results of this study identify exosomal miR-221-3p as a potential diagnostic serum EOC biomarker and a novel M2-derived target for EOC therapy (57).
GATA3 is a transcription factor involved in breast cancer. However, its role in ovarian cancer cells is also emerging as an important finding. These vital factors are often found in cells, which contribute to the progression of the tumor (154). Tumor cells become aggressive by modulating the TME. GATA3 can also activate other associated pathways (155) in a study conducted by El-Arabey et al. GATA3 is demonstrated to be released from TAMs in an exosomal form. This also activates macrophage polarization and its interaction with High-grade serous carcinoma (HGSOC) cells. It facilitates tumor growth and epithelial-mesenchymal transformation. Major tumor-stimulating effects are reduced by the use of siRNA in the GATA3-targeted TAMs. Therefore, GATA3 acts as an important marker for prognosis and a better therapeutic technique for HGSOC (Figure 3) (156). The interaction between regulatory T cells and Th17 cells within the TME serves to define the type and the intensity of immune response-resistant EOC, which, as a rule, defines the clinical course and treatment results (157, 158). Tregs play a rather valuable role in boosting tumors, offering a suppressive effect on anti-tumor immunity. In contrast, Th17 cells accumulate in tumors in response to the immune response to regulation (159). The study by Zhou et al. investigated the functions of the exosomal miRNAs from TAMs, which are termed miR-29a-3p and miR-21-5p, since they suppress the CD4+ T cells in a STAT3 manner. This suppresses the balance of the Treg/Th17 and promotes the progression of EOC in an immunosuppressive atmosphere. This remains clear as targeting the exosomes by the miRNAs may be a novel manner of treating EOC (58).
Figure 3. The image illustrates the role of mutant TP53 in high-grade serous ovarian cancer (HGSOC), highlighting GATA3 expression’s impact on tumor growth, angiogenesis, and migration. It also depicts exosome-mediated communication, contributing to chemoresistance, EMT, and epigenetic regulation in the tumor microenvironment.
In ovarian cancer, TWEAK double communicates as it promotes tumor progression and immune perturbation. Namely, TWEAK serves to enhance the proliferation, migration, and resistance of cancer cells by activating the NF-κB signaling pathway (160). Moreover, according to Hu et al., exosomes collected by TWEAK-stimulated macrophages impede the metastasis of ovarian cancer by transmitting miR-7 to EOC cells to reduce passage through the EGFR/AKT/ERK1/2 pathway. Using antagomiR-7 transfection, the levels of miR-7 in the macrophages, exosomes, and EOC cells were reduced, and metastasis was enhanced. Studies in a mixed xenograft rodent model showed that TAMs’ exosomal miR-7 prohibits EOC metastasis (66). Besides, Zhu et al. showed that exosomal miR-223 secreted by hypoxic macrophages accelerated the resistance of EOC to chemotherapy by inhibiting the PTEN-PI3K/AKT signaling pathway. Hypoxic exosomes highly expressed miR-223, and they were transferred to EOC to increase its drug resistance. In addition, higher levels of miR-223 contained in the exosomes of patients with EOC were identified in the course of the disease recurrence, which confirms the interaction between macrophages and EOC, which increases the resistance of EOC to chemotherapy (92). M2-macrophage-derived exosomal miR-221-3p, miR-29a-3p, and miR-21-5p are actively amplified to facilitate EOC growth and immune suppression (161). They serve as both clinical agents (correlation in tumor progression prevention) and clinical biomarkers (early detection or monitoring of disease).
5.4 Hepatocellular carcinoma
Hepatocellular carcinoma (HCC) is the prime variation of primary liver cancer, usually presenting in the background of chronic liver diseases, such as hepatitis B and C, alcoholic liver disease, and non-alcoholic fatty liver disease (NAFLD). Of note, HCC is characterized by its notorious aggressiveness and extremely poor prognosis (162). Since HCC frequently manifests at an advanced stage, making early detection difficult and curable treatment scarce, this disease is followed by a high rate of death. Regarding the disease frequency, worldwide HCC ranks as the fifth most frequent cancer and the third major cause of cancer death (163). It predominantly affects the male population and has been notably recognized in East Asia and Sub-Saharan African regions, wherein hepatitis B and C infections are common. Nevertheless, in Western countries, this phenomenon is continuously increasing as a result of the burgeoning rates of obesity, diabetes, and NAFLD (164, 165). Notably, the function of miRNAs in HCC is pivotal. It is related to their ability to control gene expression associated with tumor development, progression, and metastasis in either a stimulatory or repressive manner (166).
CD90 is one of the important markers in HCC, and it is Thymus cell antigen 1 (Thy-1), a stem cell antigen that plays an essential role as a marker for cancer stem cells (CSCs). A variety of lines of evidence suggest that CD90+ cells are associated with tumor initiation, progression, and resistance to therapy. CD90+ cells in HCCs exhibit higher self-renewal, invasion, and metastasis properties (167, 168). Wang et al. demonstrated that exosomes from TAMs promote HCC cell proliferation and enhance the relative CSC traits by delivering miR-125a/b. The down-regulation of miR-125a/b in TAM-derived exosomes enhanced CSC traits associated primarily targeting the stem cell marker CD90. These results suggest that miR-125a and miR-125 b play a critical role in the regulation of CSCs in HCC through TAM-derived exosomes (169). Apart from miR-125a/b, miR-142 and miR-223 transfer between the cells substantially contributes to HCC (170). Aucher et al. showed that human macrophages transfer miR-142 and miR-223 to HCC cells via gap junctions. The transfer inhibits the proliferation of HCC since it regulates the expression of the key proteins, including stathmin-1 and insulin-like growth factor 1 (IGF-1) receptor (IGF1R); thus, a novel defense mechanism of immune cells from tumor proliferation. Multiple mechanisms and processes keep cell proliferation under control, and some of them have been discovered only recently, such as the transfer of small RNA molecules between cells (171).
Alcoholic liver disease is a disorder that is caused by excessive consumption of alcohol. This results in damage to the liver, which could range from fatifying of liver to more severe forms of the disorder, such as alcoholic hepatitis or cirrhosis (172). The chronic consumption of alcohol disrupts the normal functions of the liver cells and is associated with inflammation, which may cause an individual to develop scar tissue on the liver. It increases the risk of liver failure and death and is regarded as a major cause of liver-related incidences of morbidity and mortality in most parts of the world (172). Babuta et al. found that alcohol disrupts autophagy in alcoholic liver disease (ALD) by impairing the functions of autophagosomes and lysosomes. The latter is attributed to the downregulation of Lysosome-Associated Membrane Protein 1 & Protein 2 (LAMP1 and LAMP2). This is a result of miR-155, which targets key oncogenes in the process and causes an increase in the production of exosomes. Notably, the mice that lacked the miR-155 molecule were not affected by the effects, and this proves its role in the disruption of autophagy and the release of exosomes by alcohol in ALD (173). TAM-derived exosomal miR-125a/b, miR-142, and miR-223 control the expression of cancer-stem-cell-associated genes like CD90 that boost tumor recurrence and resistance in HCC (174). Their practical usefulness and their identification in plasma justify their possible use as biomarkers that predict relapse and treat therapeutic nodes in precision-based management of HCC.
5.5 Pancreatic cancer
Pancreatic cancer is a highly aggressive malignancy that originates in the pancreatic tissues and is often detected late, as it is asymptomatic and has rapid progress and a lack of response to any treatments (175). This is the unfinished business of most malignant neoplasms in the pancreas and perhaps one of the most lethal types of cancer. Pancreatic ductal adenocarcinoma (PDAC) is the most common form of pancreatic cancer (176). Risk factors associated with this blockage include cigarette smoking, chronic pancreatitis, being overweight, and having a family history of pancreatic cancer. There are only a several drugs to deal with this diagnosis, which is why the five-year percentile has a very low rate of 5-10% pancreatic cancer (177). Compared to other types of cancer, this one is the twelfth different cancer.
On the other hand, it has a high mortality rate and occupies the seventh place among the deadliest forms of neoplasms. The biggest number of incidence falls in prosperous countries where a large number of men were affected by it (178). People who are obese and have diabetes are also in the high-risk group. One of the critical roles of miRNAs in pancreatic cancer is the involvement in signaling pathways that control the growth, metastasis, and drug resistance of tumors. This is achieved by miRNAs either as oncogenes or as tumor-suppressive genes (179). Binenbaum et al. discovered that miR-365, which is found in TAMs-derived exosomes in PDAC, impairs gemcitabine activity, which is a drug used in chemotherapy, thus inducing cancer cell resistance to this medicine. The study showed that restoring the latter’s sensitivity was possible by blocking miR-365, which provides yet more evidence for the fact that MDEs are among the primary regulators of chemotherapy resistance in PDAC (94).
X-linked inhibitor of apoptosis protein (XIAP) is also a critical factor in pancreatic cancer. It inhibits apoptosis and promotes cell survival. Hence, the XIAP’s overexpression in cancer cells guarantees survival despite chemotherapy and other treatments (180). Overexpression of the factor is associated with poor prognosis and the aggressiveness of cancer. Interestingly, XIAP seems to be a practical therapeutic target in pancreatic cancer (181) in the study. Yin et al. show that M2 macrophage-derived exosomes promote pancreatic cancer by transmitting long non-coding RNA SBF2-AS1 that suppresses MiR-122-5p and upregulates XIAP. Suppression of XIAP restrains the tumor from growing, helping treat the disease (182). Similarly, Yin et al., revealed that exosomes in M2 TAMs consist of miR-501-3p to advance PDAC via targeting the suppression gene TGFBR3. This downregulation, in turn, activates the TGF-β signaling pathway that will foster tumor growth and metastasis. Depletion of miR-501-3p in these exosomes acted to suppress tumorigenesis and metastasis, together with their upstream TGF-β signaling pathway, might be viable therapeutic targets for PDAC treatment (56). Macrophage-derived exosomal miR-365 silenced the effect of gemcitabine by interfering with nucleotide metabolism in PDAC, and miR-501-3p and lncRNA SBF2-AS1 changed TGF-β and XIAP to promote chemoresistance (94). These results reveal MDE cargoes as potent drug resistance mediators and promising therapeutic targets tested in in vivo xenograft studies.
5.6 Colorectal cancer
CRC is a type of cancer that develops in the colon or rectum. It is often preceded by the growth of benign polyps, which may later become malignant. It is the third most common cancer and the second leading cause of cancer mortality globally (183). The common symptoms experienced are changes in defecation rhythm, bloody stool, and abdominal pain. CRC is monitored by colonoscopy, and when detected, early survival rates are high (184). It is treated with surgery, chemotherapy, and radiation, often in combination. The incidence rates of CRC are third in the most often diagnosed cancers and second in terms of deaths worldwide, with higher rates in the more developed areas, particularly North America, Europe, and Australia (185). There are numerous risk factors, such as age, family history, diet, smoking and alcohol consumption, obesity, and inflammatory bowel diseases. There has been a downward trend in the mortality rates associated with CRC directly linked to the implementation of screening programs, as the introduction of early detection methods has been decisive (186).
MiRNAs play an essential role in CRC by controlling the expression of genes. These genes are responsible for the growth, metastasis, and response to treatment of the tumor. When miRNAs are not functioning properly, they can become oncogenic or mutated (187). Undeveloped miRNAs have attracted consideration as potential indicators for the first detection of CRC. They are additionally appealing targets for therapeutic intervention and TSR catalysts. There has been an observation that miR-15a and miR-16 are significantly under-expressed in CRC tissues in contrast to the normal mucosa (188). According to the research study of Xiao et al., the low expression of these miRNAs is dependent on the advanced disease degree, poor histological evaluation, and the presence of nodes. The cumulative low level of these two miRNAs can securely guarantee a deficient general and disease-free existence of CRC patients. On this note, the delivery of these miRNAs, which are not well controlled, to the agitated tumors through exosomes may be difficult. The miR-15a and miR-16 are often used as markers for treatment and recovery (189).
The p53 protein is a key player in the mechanism of colon cancer as a tumor suppressor, orchestrating the arrest of the cell cycle, DNA repair, or apoptosis, when necessary, in response to cellular stress or damage to DNA. However, the TP53 gene, which encodes the p53 protein, is mutated in numerous cases of colon cancer, thus losing its function of suppressing tumors (190). In their studies, Cooks et al. found that colon cancer cells with gain-of-function mutp5–3 secrete exosomes containing a high concentration of miR-1246. These exosomes reprogram the TAMs into an anti-inflammatory and cancer-promoting state in CRC through the action of miR-1246. Thus, miR-1246 sustains the immunosuppression, as well as the activity of TGF-β, thus promoting the inflammatory state, as well as the progression of cancer, and poor survival of patients with CRC (191). One of the proteins is ZC3H12B, which is a tumor suppressor in colon cancer, regulating inflammation and cell proliferation (192). The downregulation or loss of ZC3H12B in colon cancer cells results in a more aggressive tumor, increased growth, invasion, and worse prognosis. Such a suppression degrades pro-inflammatory mRNAs, reducing the inflammatory environment that allows cancer to progress (193). Another protein that was identified by Ma et al. is M2 macrophage-derived exosomal miR-155-5p. It fosters immune escape in CRC, interacting with ZC3H12B, degrading its expression, and raising IL-6 levels. As a result, CRC proliferation and anti-apoptotic were supported, and immune escape was achieved. It is possible to regard that such a miR-155-5p in exosomes might be one of the CRC progressors and an anti-cancer target (96). MDE-mediated miR-155-5p and miR-1246 drive immune escape and pro-inflammatory signaling in CRC by regulating ZC3H12B and TGF-β cascade (194). Their consistent detection in patient exosomes underscores their value as liquid biopsy biomarkers and targets to re-sensitize tumors to therapy.
5.7 Other cancers
MDEs have shown potential to be a great inducer in numerous cancers other than lung, pancreatic, colorectal, ovarian, and breast cancers. These exosomes promote tumorigenesis by promoting disease progression. Moreover, MDEs transfer oncogenic miRNAs and proteins that have implications for the upregulation of TME alteration-related genes (195). The upregulated genes promote glioblastoma (GBM) due to the interaction and alteration of the molecular pathways conducive to GBM growth and enhancement of therapy resistance. These exosomes were shown to promote gastric cancer (GC) by activating new metastasis-related genes (196). It seems that MDEs promote cancer progression and metastasis in all organs of a living cell, although they have not been isolated from all organs to date. In EC, miRNAs seem to play a significant role in shifting gene equilibrium to produce changes in the rate of cancer development (197).
Moreover, the altered miRNAs may act as oncogenes or tumor suppressors. Changes in the pathway characteristic of these types of miRNAs, such as cell proliferation, apoptosis, and invasion, have made the miRNAs popular in the hunt for diagnostic biomarkers and treatments (198). Mi et al. showed that M2 macrophage-derived exosomes are involved in the migration and invasion of EC. The exosomes contained the long non-coding RNA AFAP1-AS1 that inhibited miR-26a and promoted ATF2. EC was found to be able to migrate and invade due to the expression of ATF2 alone or together with the miRNA. The results showed that a therapeutic strategy could be initiated by targeting this signaling pathway, supporting more advanced EC (62). MDEs are involved in GC via the transfer of miRNAs that regulate tumor growth, invasion, and metastasis.
As the miRNAs act on the gene expression of the TME, they are one target for cancer therapeutic approaches (199). Zheng et al. discovered that TAMs activate the migration of GC through exosomes by polarizing into M2 macrophages. This exosome transfer not only involves lipid transfer but also the transfer of Apolipoprotein E (ApoE) and specific miRNAs. After the delivery of the M2-derived exosomes into the cancer cells, their migration ability is enhanced by the PI3K-AKT signaling pathway. The cancer cells from Apoe-/- mice lacked the effect of the exosomes on their migration ability, and this indicates the importance of ApoE and miRNAs in driving the exosomal transfer by TAMs in the progression of cancer (200). Li et al. found that miR-16-5p loaded into exosomes derived from M1 macrophages suppresses the development of GC via the activation of T-cell-dependent immunity by targeting programmed death-ligand 1 expression in GC cells. The delivery of miR-16-5p from M1 macrophages to GC cells induced an immune response against the tumor in vitro and in vivo, specifically. Hence, the authors concluded the proposal that M1 macrophages could serve as a cellular treatment agent for GC by facilitating miR-16-5p delivery in exosomes (201). Gao et al. discovered that macrophage-derived exosomal miR-223 inhibited tumor-suppressive ubiquitin ligase substrate specificity of F-box and WD-40 repeat domain-containing 7 to promote doxorubicin resistance in GC. The transfer of miR-223 from macrophages to GC cells was found to occur using exosomes, and knockdown of miR-223 in macrophages appeared to reduce the resistance. As for clinical settings, the presence of high levels of miR-223 in both GC tissues and plasma exosomes has been linked to doxorubicin resistance. Therefore, due to this link, targeting exosome-mediated miR-223 transfer is likely to become a helpful therapy for GC patients (202).
In GBM, miRNAs serve vital functions that facilitate the regulation of targeted genes, such as gene growth, invasion, and resistance to therapy. MiRNAs could be oversensitive or work as inhibitors of these processes, leading to their function as oncogenes or tumor suppressors that can further control major pathways and be involved in GBM (203). The role of miRNAs as biomarkers for early diagnosis, prognosis, and therapeutic targets is promising for those affected by GBM (204). Moreover, it was emphasized by Qian et al. that hypoxic glioma-derived exosomes significantly promote the M2 polarization of macrophages, resulting in enhanced glioma growth, movements, and aggressive invasion. The microRNA sequencing identified miR-1246 as the leading miRNA in H-GDEs, positively correlated with the activity of the STAT3 signaling pathway operated by the targeting recognition of TERF2IP. It was concluded that miR-1246 levels in the cerebrospinal fluid of GB patients are novel targeted chemotherapy antitumor factors (205). In other malignancies, in GC, GBM, and EC, the MDEs miRNAs, e.g., miR-223, miR-16-5p, and miR-1246, coordinate metastasis, chemoresistance, and immune modulation. The fact that they are retained regulators in a wide variety of tumor types points to their potential in becoming pan-cancer exosomal biomarkers and pan-cancer targets of therapeutic interest (206). The dynamic interplay between macrophages and the tumor microenvironment via macrophage-derived exosomes was depicted (Figure 4).
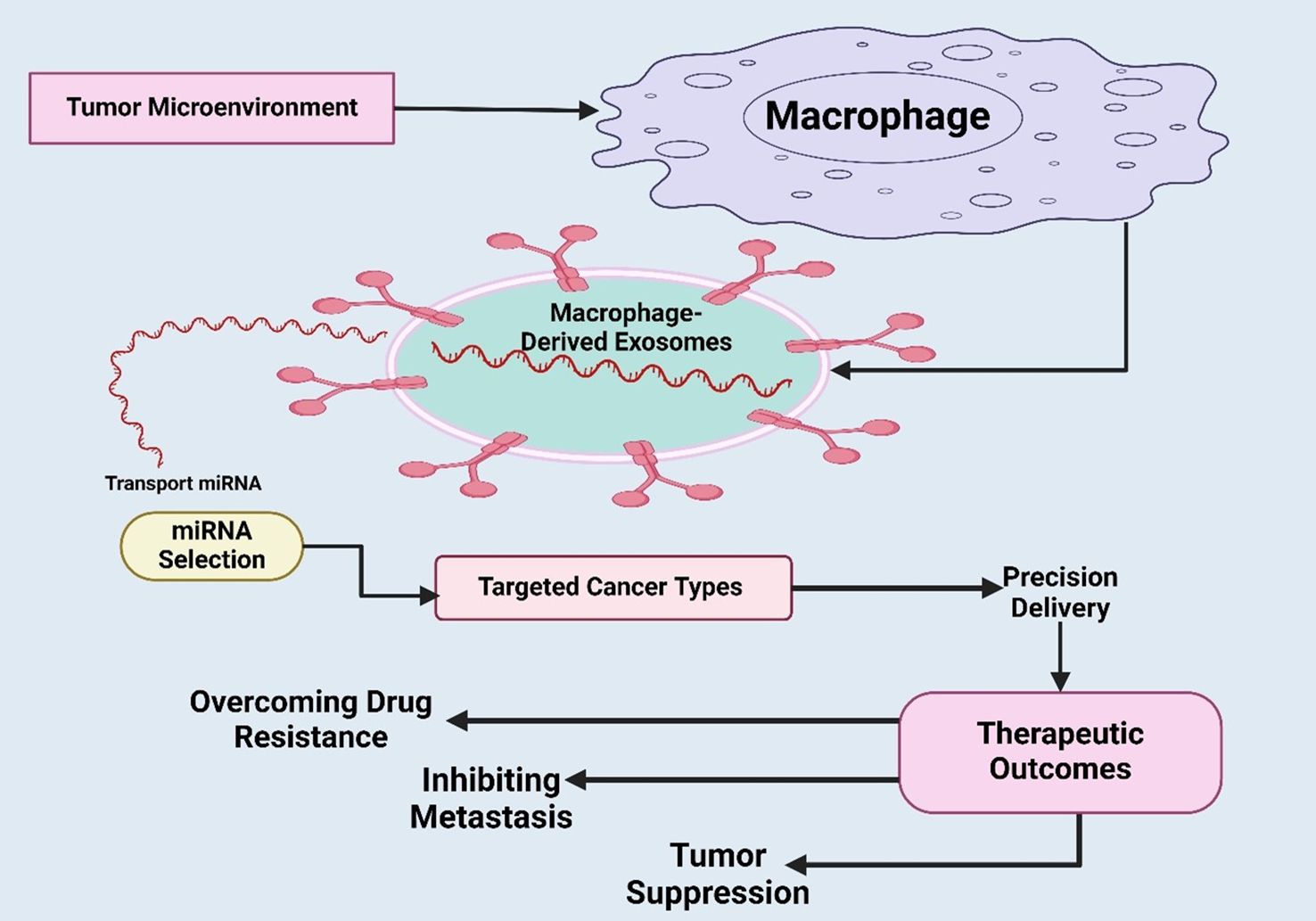
Figure 4. Illustration of dynamic interplay between macrophages and the tumor microenvironment via macrophage-derived exosomes were depicted. It also highlights how selected miRNAs are transported within exosomes to specific cancer targets, enabling precision delivery of therapeutic molecules. Overall, it underscores the translational significance of exosome-based strategies for targeted cancer therapy.
6 Limitations, safety concerns, and translational barriers of engineered MDEs
Although MDEs containing therapeutic miRNAs hold enormous potential, a series of biological and translational limitations need to be overcome before using these in clinical practice.
First, the problem of off-target effects must stay significant: the non-target tissues have the potential to absorb exosomal miRNAs, causing unwanted gene silencing and possibly a change in physiological or immune functions (207). On the same note, MDEs have inherent immunomodulatory functions, potentially leading to undesirable immunosuppression or a balance in cytokines, thus facilitating tumor immune evasion over regression (208).
In terms of bioengineering, heterogeneity of exosome isolation and miRNA loading capacity does not help with the reproducibility and dose standardization in production batches. Moreover, the large-scale production of exosomes, the absence of potent purification procedures, and the inconsistency of switching between the polarization states of the macrophages considerably affect therapy consistency (209). A further significant roadblock in the translation is related to biodistribution and clearance: when injected intravenously, MDEs are quickly taken away by the liver and the spleen, minimizing their accumulation and therapeutic efficacy to tumor properties (210, 211).
Besides, the regulatory environment of exosome-based therapeutics remains to be developed, and there is a lack of agreed quality control standards, long-term safety evaluation, and immunogenicity examination. Therefore, even with the promising preclinical research, the development of engineered MDE-miRNA systems into the clinic would demand stringent pharmacokinetic, toxicological, and immunological studies in a variety of animal models and controlled early-phase human trials. The standardized manufacturing protocols, specific surface engineering, and the selection of the miRNA will play a key role in the implementation of reproducible, safe, and effective MDE-based cancer therapies. Also, we have summarized the overview of macrophage-derived exosome engineering and modification for cancer therapeutics and diagnostics in Table 2.
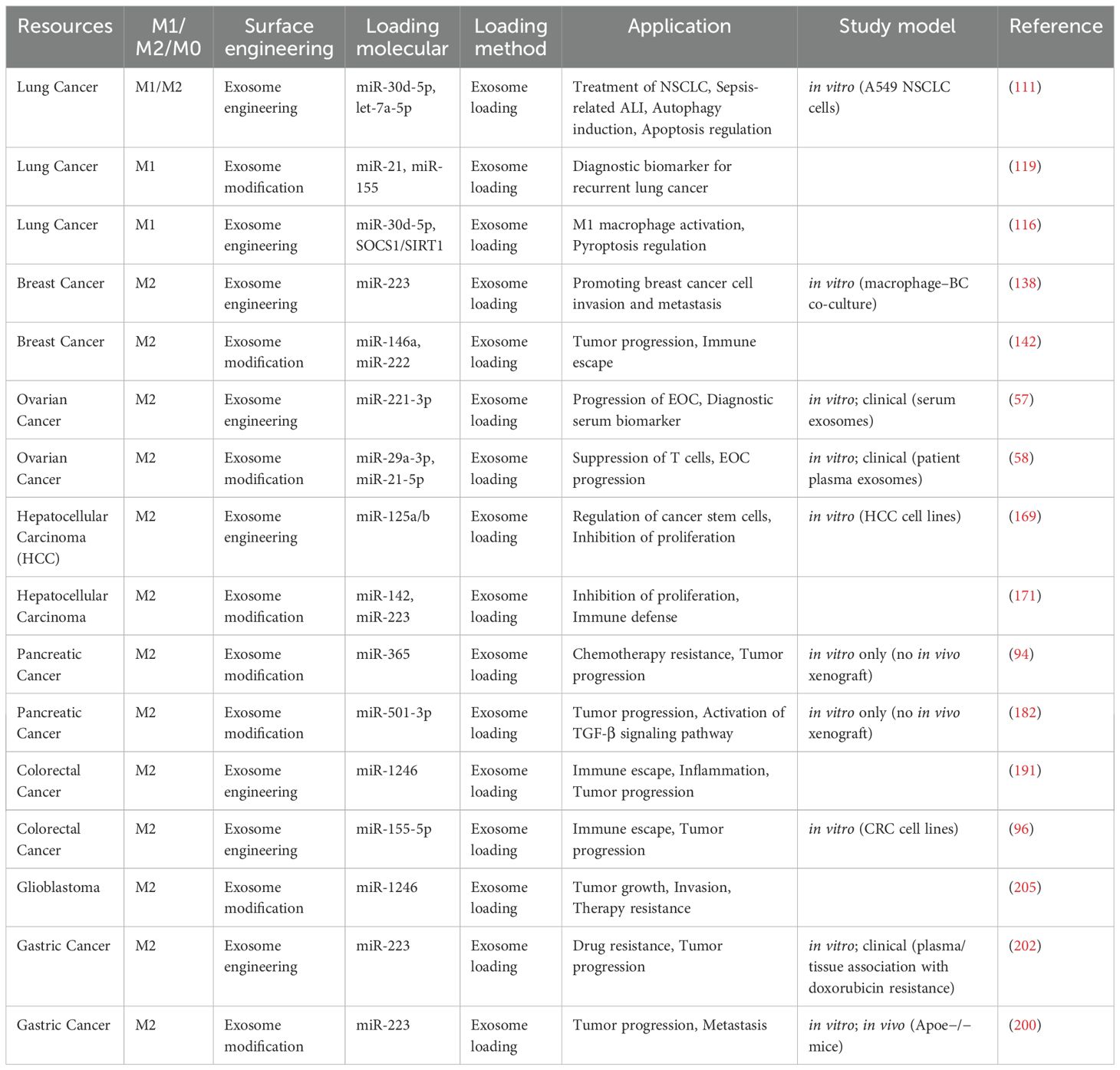
Table 2. Overview of macrophage-derived exosome engineering and modification for cancer applications.
7 Conclusion and future prospects
Collectively, these findings support clear near-term opportunities for translational studies while highlighting specific avenues for optimization and clinical validation. MDEs have a vital function in cancer biology by serving as intermediaries for communication between macrophages and tumor cells. These exosomes play a crucial role in several processes that are vital for the advancement of cancer, such as stimulating the development of tumors, aiding in the spread of cancer to other parts of the body, causing resistance to drugs, and influencing the immune response. MDEs can greatly modify the behavior of cancer cells and the TME by transporting specific cargo, especially miRNAs. The results presented in this context demonstrate that MDEs include miRNAs that may either facilitate or impede cancer development, depending on their unique composition. For instance, the introduction of cancer-causing miRNAs via MDEs may amplify the development, invasion, and resistance to the treatment of tumors. In contrast, miRNAs that decrease tumor growth can hinder these processes. The ability to manipulate MDEs for the specific administration of therapeutic miRNAs presents new opportunities for precision medicine in the field of cancer, providing enhanced and individualized therapy alternatives.
Advances in the MDE-miRNA regimens rely on standardized work on the isolation and characterization of exosomes. Spike-in controls, cross-platform reproducibility, appropriate normalization, and common QC standards for miRNA profiling should be implemented. To be delivered safely, engineered MDEs need constructive evaluation of biodistribution, immunogenicity, complement activation, thrombogenicity, and off-target effects, preferably on two vertebrates before clinical translation. Embracing these standards will make it possible to achieve a realistic, reproducible, and safe route to clinical testing of MDE-miRNA-based therapies.
Nevertheless, several critical constraints still need to be addressed to facilitate clinical translation. These include improving the precision of miRNA loading into MDEs and enhancing their tumor-specific delivery while minimizing off-target effects. Future studies should focus on optimizing ESCRT-dependent and -independent sorting pathways to enable selective and reproducible miRNA encapsulation. Parallel efforts should evaluate engineered MDEs functionalized with tumor-homing ligands or stimuli-responsive coatings in preclinical and early-phase (Phase I) clinical trials to assess biodistribution, immunogenicity, and therapeutic efficacy. Moreover, integrating multi-omics analyses with bioengineering and nanotechnology platforms could allow real-time monitoring of exosomal cargo loading and release kinetics, offering a rational framework for precision MDE-based therapeutics. Addressing these challenges through interdisciplinary strategies will accelerate the safe and effective translation of MDE-miRNA delivery systems into personalized cancer care.
Author contributions
PR: Conceptualization, Writing – review & editing, Writing – original draft, Project administration. MA: Methodology, Writing – original draft, Data curation. MB: Validation, Writing – review & editing. RM: Data curation, Writing – original draft. SS: Data curation, Writing – review & editing. SP: Formal analysis, Writing – original draft, Data curation. HA: Writing – review & editing, Formal analysis. MH: Formal analysis, Data curation, Writing – review & editing, Writing – original draft. GG: Writing – review & editing, Investigation, Conceptualization. JM: Writing – review & editing, Formal analysis, Supervision. SA: Validation, Writing – review & editing, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to thank their respective institutes/organizations for providing the necessary facilities to carry out this work. Prasanna Srinivasan Ramalingam would like to thank the Council for Scientific and Industrial Research (CSIR) for providing him the Senior Research Fellowship (File No: 09/0844(18240)/2024-EMR-I). Janaki Ramaiah Mekala and Sivakumar Arumugam specially thank Vellore Institute of Technology, Vellore for providing the necessary facilities for the work.
Conflict of interest
The authors declare that this review was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Generative AI was used in the creation of this manuscript. During the preparation of this work, the authors used Generative AI and AI-assisted tools solely to improve the language, grammar, and readability of the manuscript. No AI tools were used for generating substantive content, data analysis, interpretation of results, or drawing scientific conclusions. All intellectual contributions, study design, data collection, analysis, and final interpretations were conceived and executed by the authors. The authors reviewed and verified the accuracy and appropriateness of all AI-assisted edits and take full responsibility for the content of this publication.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Abe C, Bhaswant M, Miyazawa T, and Miyazawa T. The potential use of exosomes in anti-cancer effect induced by polarized macrophages. Pharmaceutics. (2023) 15:1024. doi: 10.3390/pharmaceutics15031024
2. Zhang Z, Liu L, Ti H, Chen M, Chen Y, Du D, et al. Synovial fibroblast derived small extracellular vesicles miRNA15–29148 promotes articular chondrocyte apoptosis in rheumatoid arthritis. Bone Res. (2025) 13:61. doi: 10.1038/s41413-025-00430-3
3. Lu JY, Guo Z, Huang WT, Bao M, He B, Li G, et al. Peptide-graphene logic sensing system for dual-mode detection of exosomes, molecular information processing and protection. Talanta. (2024) 267:125261. doi: 10.1016/j.talanta.2023.125261
4. Baig MS, Roy A, Rajpoot S, Liu D, Savai R, Banerjee S, et al. Tumor-derived exosomes in the regulation of macrophage polarization. Inflammation Res. (2020) 69:435–51. doi: 10.1007/s00011-020-01318-0
5. Tai YL, Chen KC, Hsieh JT, and Shen TL. Exosomes in cancer development and clinical applications. Cancer Sci. (2018) 109:2364–74. doi: 10.1111/cas.13697
6. Huang H, Huang F, Liang X, Fu Y, Cheng Z, Huang Y, et al. Afatinib reverses EMT via inhibiting CD44-stat3 axis to promote radiosensitivity in nasopharyngeal carcinoma. Pharm (Basel). (2022) 16:37. doi: 10.3390/ph16010037
7. Khan Y, Hussain MS, Ramalingam PS, Fatima R, Maqbool M, Ashique S, et al. Exploring extracellular RNA as drivers of chemotherapy resistance in cancer. Mol Biol Rep. (2025) 52:142. doi: 10.1007/s11033-025-10263-2
8. Ammendola M, Curcio S, Ammerata G, Luposella M, Battaglia C, Laface C, et al. Macrophages in tumor microenvironment: from molecular aspects to clinical applications. Eurasian J Med Oncol. (2023) 7:201. doi: 10.14744/ejmo.2023.26480
9. Zhang Y, Zhu L, Li X, Ge C, Pei W, Zhang M, et al. M2 macrophage exosome-derived lncRNA AK083884 protects mice from CVB3-induced viral myocarditis through regulating PKM2/HIF-1α axis mediated metabolic reprogramming of macrophages. Redox Biol. (2024) 69:103016. doi: 10.1016/j.redox.2023.103016
10. Beeraka NM, Doreswamy SH, Sadhu SP, Srinivasan A, Pragada RR, Madhunapantula SV, et al. The role of exosomes in stemness and neurodegenerative diseases-chemoresistant-cancer therapeutics and phytochemicals. Int J Mol Sci. (2020) 21:6818. doi: 10.3390/ijms21186818
11. Chen Q, Li Y, Gao W, Chen L, Xu W, and Zhu X. Exosome-mediated crosstalk between tumor and tumor-associated macrophages. Front Mol Biosci. (2021) 8:764222. doi: 10.3389/fmolb.2021.764222
12. Fabris L, Sato K, Alpini G, and Strazzabosco M. The tumor microenvironment in cholangiocarcinoma progression. Hepatology. (2021) 73 Suppl 1:75–85. doi: 10.1002/hep.31410
13. Chatterjee B, Saha P, Bose S, Shukla D, Chatterjee N, Kumar S, et al. MicroRNAs: as critical regulators of tumor- associated macrophages. Int J Mol Sci. (2020) 21:7117. doi: 10.3390/ijms21197117
14. Song Y, Huang Y, Zhou F, Ding J, and Zhou W. Macrophage-targeted nanomedicine for chronic diseases immunotherapy. Chin Chem Lett. (2022) 33:597–612. doi: 10.1016/j.cclet.2021.08.090
15. Abd-Aziz N, Kamaruzman NI, and Poh CL. Development of microRNAs as potential therapeutics against cancer. J Oncol. (2020) 2020:8029721. doi: 10.1155/2020/8029721
16. Fang X, Lan H, Jin K, and Qian J. Pancreatic cancer and exosomes: role in progression, diagnosis, monitoring, and treatment. Front Oncol. (2023) 13:1149551. doi: 10.3389/fonc.2023.1149551
17. Iqbal MJ, Javed Z, Sadia H, Mehmood S, Akbar A, Zahid B, et al. Targeted therapy using nanocomposite delivery systems in cancer treatment: highlighting miR34a regulation for clinical applications. Cancer Cell Int. (2023) 23:84. doi: 10.1186/s12935-023-02929-3
18. Chen Q, Shan X, Shi S, Jiang C, Li T, Wei S, et al. Tumor microenvironment-responsive polydopamine-based core/shell nanoplatform for synergetic theranostics. J Mater Chem B. (2020) 8:4056–66. doi: 10.1039/D0TB00248H
19. Chakrabortty A, Patton DJ, Smith BF, and Agarwal P. miRNAs: potential as biomarkers and therapeutic targets for cancer. Genes (Basel). (2023) 14:1375. doi: 10.3390/genes14071375
20. Yan J, Liu H, Yang W, Liu N, Wang J, Li Z, et al. Small-molecule-induced liquid-liquid phase separation suppresses the carcinogenesis of β-catenin. Nat Commun. (2025) 16:5997. doi: 10.1038/s41467-025-61112-6
21. Arya SB, Collie SP, and Parent CA. The ins-and-outs of exosome biogenesis, secretion, and internalization. Trends Cell Biol. (2024) 34:90–108. doi: 10.1016/j.tcb.2023.06.006
22. Lee YJ, Shin KJ, Jang HJ, Ryu JS, Lee CY, Yoon JH, et al. GPR143 controls ESCRT-dependent exosome biogenesis and promotes cancer metastasis. Dev Cell. (2023) 58:320–334.e328. doi: 10.1016/j.devcel.2023.01.006
23. Mashouri L, Yousefi H, Aref AR, Ahadi AM, Molaei F, and Alahari SK. Exosomes: composition, biogenesis, and mechanisms in cancer metastasis and drug resistance. Mol Cancer. (2019) 18:75. doi: 10.1186/s12943-019-0991-5
24. Wen B, Tao R, Liu Y, and Zhang Z. Investigating the role of exosomal microRNA-5703 in modulating tumor-associated endothelial cells in lung cancer. Cytojournal. (2024) 21:77. doi: 10.25259/Cytojournal_99_2024
25. Raposo G and Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. (2013) 200:373–83. doi: 10.1083/jcb.201211138
26. Wei H, Chen Q, Lin L, Sha C, Li T, Liu Y, et al. Regulation of exosome production and cargo sorting. Int J Biol Sci. (2021) 17:163–77. doi: 10.7150/ijbs.53671
27. Zhang L and Yu D. Exosomes in cancer development, metastasis, and immunity. Biochim Biophys Acta Rev Cancer. (2019) 1871:455–68. doi: 10.1016/j.bbcan.2019.04.004
28. Zhang X, Xu Y, Ma L, Yu K, Niu Y, Xu X, et al. Essential roles of exosome and circRNA_101093 on ferroptosis desensitization in lung adenocarcinoma. Cancer Commun (Lond). (2022) 42:287–313. doi: 10.1002/cac2.12275
29. Zhao S, Mi Y, Guan B, Zheng B, Wei P, Gu Y, et al. Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. J Hematol Oncol. (2020) 13:156. doi: 10.1186/s13045-020-00991-2
30. Luo G, Zhou Z, Cao Z, Huang C, Li C, Li X, et al. M2 macrophage-derived exosomes induce angiogenesis and increase skin flap survival through HIF1AN/HIF-1α/VEGFA control. Arch Biochem Biophys. (2024) 751:109822. doi: 10.1016/j.abb.2023.109822
31. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, Pérez-Hernández D, Vázquez J, Martin-Cofreces N, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun. (2013) 4:2980. doi: 10.1038/ncomms3980
32. Shurtleff MJ, Temoche-Diaz MM, Karfilis KV, Ri S, and Schekman R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife. (2016) 5:e19276. doi: 10.7554/eLife.19276
33. Koppers-Lalic D, Hackenberg M, Bijnsdorp IV, Van Eijndhoven M, Sadek P, Sie D, et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. (2014) 8:1649–58. doi: 10.1016/j.celrep.2014.08.027
34. Kosaka N, Iguchi H, Hagiwara K, Yoshioka Y, Takeshita F, and Ochiya T. Neutral sphingomyelinase 2 (nSMase2)-dependent exosomal transfer of angiogenic microRNAs regulate cancer cell metastasis. J Biol Chem. (2013) 288:10849–59. doi: 10.1074/jbc.M112.446831
35. Han Q, Zhao H, Jiang Y, Yin C, and Zhang J. HCC-derived exosomes: critical player and target for cancer immune escape. Cells. (2019) 8:558. doi: 10.3390/cells8060558
36. Krylova SV and Feng D. The machinery of exosomes: biogenesis, release, and uptake. Int J Mol Sci. (2023) 24:1337. doi: 10.3390/ijms24021337
37. Zhou Z, Sui X, Cao Z, Li X, Qing L, and Tang J. Substance P promote macrophage M2 polarization to attenuate secondary lymphedema by regulating NF-kB/NLRP3 signaling pathway. Peptides. (2023) 168:171045. doi: 10.1016/j.peptides.2023.171045
38. Jeffries J, Zhou W, Hsu AY, and Deng Q. miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. (2019) 451:136–41. doi: 10.1016/j.canlet.2019.02.051
39. Lee YJ, Shin KJ, and Chae YC. Regulation of cargo selection in exosome biogenesis and its biomedical applications in cancer. Exp Mol Med. (2024) 56:877–89. doi: 10.1038/s12276-024-01209-y
40. Jo H, Shim K, and Jeoung D. Exosomes: diagnostic and therapeutic implications in cancer. Pharmaceutics. (2023) 15:1465. doi: 10.3390/pharmaceutics15051465
41. Blanc L and Vidal M. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases. (2018) 9:95–106. doi: 10.1080/21541248.2016.1264352
42. Khalaf K, Hana D, Chou JT, Singh C, Mackiewicz A, and Kaczmarek M. Aspects of the tumor microenvironment involved in immune resistance and drug resistance. Front Immunol. (2021) 12:656364. doi: 10.3389/fimmu.2021.656364
43. Pollet H, Conrard L, Cloos AS, and Tyteca D. Plasma membrane lipid domains as platforms for vesicle biogenesis and shedding? Biomolecules. (2018) 8:94. doi: 10.3390/biom8030094
44. Chen Z, Han F, Du Y, Shi H, and Zhou W. Hypoxic microenvironment in cancer: molecular mechanisms and therapeutic interventions. Signal Transduct Target Ther. (2023) 8:70. doi: 10.1038/s41392-023-01332-8
45. Lai X, Zhong J, Zhang B, Zhu T, and Liao R. Exosomal non-coding RNAs: novel regulators of macrophage-linked intercellular communication in lung cancer and inflammatory lung diseases. Biomolecules. (2023) 13:536. doi: 10.3390/biom13030536
46. To KKW and Cho WCS. Exosome secretion from hypoxic cancer cells reshapes the tumor microenvironment and mediates drug resistance. Cancer Drug Resist. (2022) 5:577–94. doi: 10.20517/cdr.2022.38
47. Li X, Liu Y, Zheng S, Zhang T, Wu J, Sun Y, et al. Role of exosomes in the immune microenvironment of ovarian cancer. Oncol Lett. (2021) 21:377. doi: 10.3892/ol.2021.12638
48. Munir MT, Kay MK, Kang MH, Rahman MM, Al-Harrasi A, Choudhury M, et al. Tumor-associated macrophages as multifaceted regulators of breast tumor growth. Int J Mol Sci. (2021) 22:6526. doi: 10.3390/ijms22126526
49. Othman N, Jamal R, and Abu N. Cancer-derived exosomes as effectors of key inflammation-related players. Front Immunol. (2019) 10:2103. doi: 10.3389/fimmu.2019.02103
50. Padoan A, Plebani M, and Basso D. Inflammation and pancreatic cancer: focus on metabolism, cytokines, and immunity. Int J Mol Sci. (2019) 20:676. doi: 10.3390/ijms20030676
51. Nowak M and Klink M. The role of tumor-associated macrophages in the progression and chemoresistance of ovarian cancer. Cells. (2020) 9:1299. doi: 10.3390/cells9051299
52. Salimi L, Akbari A, Jabbari N, Mojarad B, Vahhabi A, Szafert S, et al. Synergies in exosomes and autophagy pathways for cellular homeostasis and metastasis of tumor cells. Cell Biosci. (2020) 10:64. doi: 10.1186/s13578-020-00426-y
53. Huang R, Kang T, and Chen S. The role of tumor-associated macrophages in tumor immune evasion. J Cancer Res Clin Oncol. (2024) 150:238. doi: 10.1007/s00432-024-05777-4
54. Wang J, Long R, and Han Y. The role of exosomes in the tumour microenvironment on macrophage polarisation. Biochim Biophys Acta Rev Cancer. (2022) 1877:188811. doi: 10.1016/j.bbcan.2022.188811
55. Nogueras Pérez R, Heredia-Nicolás N, De Lara-Peña L, López De Andrés J, Marchal JA, Jiménez G, et al. Unraveling the potential of miRNAs from CSCs as an emerging clinical tool for breast cancer diagnosis and prognosis. Int J Mol Sci. (2023) 24:16010. doi: 10.3390/ijms242116010.
56. Yin Z, Ma T, Huang B, Lin L, Zhou Y, Yan J, et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-β signaling pathway. J Exp Clin Cancer Res. (2019) 38:310. doi: 10.1186/s13046-019-1313-x
57. Li X and Tang M. Exosomes released from M2 macrophages transfer miR-221-3p contributed to EOC progression through targeting CDKN1B. Cancer Med. (2020) 9:5976–88. doi: 10.1002/cam4.3252
58. Zhou J, Li X, Wu X, Zhang T, Zhu Q, Wang X, et al. Exosomes released from tumor-associated macrophages transfer miRNAs that induce a treg/th17 cell imbalance in epithelial ovarian cancer. Cancer Immunol Res. (2018) 6:1578–92. doi: 10.1158/2326-6066.CIR-17-0479
59. Xu S, Xu H, Wang W, Li S, Li H, Li T, et al. The role of collagen in cancer: from bench to bedside. J Transl Med. (2019) 17:309. doi: 10.1186/s12967-019-2058-1
60. Cheng YQ, Wang SB, Liu JH, Jin L, Liu Y, Li CY, et al. Modifying the tumour microenvironment and reverting tumour cells: New strategies for treating Malignant tumours. Cell Prolif. (2020) 53:e12865. doi: 10.1111/cpr.12865
61. Chitty JL, Filipe EC, Lucas MC, Herrmann D, Cox TR, and Timpson P. Recent advances in understanding the complexities of metastasis. F1000Res. (2018) 7:169. doi: 10.12688/f1000research.15064.1
62. Mi X, Xu R, Hong S, Xu T, Zhang W, and Liu M. M2 macrophage-derived exosomal lncRNA AFAP1-AS1 and microRNA-26a affect cell migration and metastasis in esophageal cancer. Mol Ther Nucleic Acids. (2020) 22:779–90. doi: 10.1016/j.omtn.2020.09.035
63. Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res. (2019) 79:146–58. doi: 10.1158/0008-5472.CAN-18-0014
64. Liang Y, Liang Q, Qiao L, and Xiao F. MicroRNAs modulate drug resistance-related mechanisms in hepatocellular carcinoma. Front Oncol. (2020) 10:920. doi: 10.3389/fonc.2020.00920
65. Xu Z, Chen Y, Ma L, Chen Y, Liu J, Guo Y, et al. Role of exosomal non-coding RNAs from tumor cells and tumor-associated macrophages in the tumor microenvironment. Mol Ther. (2022) 30:3133–54. doi: 10.1016/j.ymthe.2022.01.046
66. Hu Y, Li D, Wu A, Qiu X, Di W, Huang L, et al. TWEAK-stimulated macrophages inhibit metastasis of epithelial ovarian cancer via exosomal shuttling of microRNA. Cancer Lett. (2017) 393:60–7. doi: 10.1016/j.canlet.2017.02.009
67. Duan S, Yu S, Yuan T, Yao S, and Zhang L. Corrigendum: Exogenous let-7a-5p induces A549 lung cancer cell death through BCL2L1-mediated PI3Kγ signaling pathway. Front Oncol. (2024) 14:1513956. doi: 10.3389/fonc.2024.1513956
68. Zhang Y, Tang S, Gao Y, Lu Z, Yang Y, Chen J, et al. Application of exosomal miRNA mediated macrophage polarization in colorectal cancer: Current progress and challenges. Oncol Res. (2023) 32:61–71. doi: 10.32604/or.2023.043481
69. Maacha S, Bhat AA, Jimenez L, Raza A, Haris M, Uddin S, et al. Extracellular vesicles-mediated intercellular communication: roles in the tumor microenvironment and anti-cancer drug resistance. Mol Cancer. (2019) 18:55. doi: 10.1186/s12943-019-0965-7
70. Bolzoni M, Toscani D, Storti P, Marchica V, Costa F, and Giuliani N. Possible targets to treat myeloma-related osteoclastogenesis. Expert Rev Hematol. (2018) 11:325–36. doi: 10.1080/17474086.2018.1447921
71. Chistiakov DA, Orekhov AN, and Bobryshev YV. Extracellular vesicles and atherosclerotic disease. Cell Mol Life Sci. (2015) 72:2697–708. doi: 10.1007/s00018-015-1906-2
72. Ali Syeda Z, Langden SSS, Munkhzul C, Lee M, and Song SJ. Regulatory mechanism of microRNA expression in cancer. Int J Mol Sci. (2020) 21:1723. doi: 10.3390/ijms21051723
73. Entezari M, Sadrkhanloo M, Rashidi M, Asnaf SE, Taheriazam A, Hashemi M, et al. Non-coding RNAs and macrophage interaction in tumor progression. Crit Rev Oncol Hematol. (2022) 173:103680. doi: 10.1016/j.critrevonc.2022.103680
74. Wang X, Xie N, Zhang H, Zhou W, and Lei J. Isoorientin ameliorates macrophage pyroptosis and atherogenesis by reducing KDM4A levels and promoting SKP1-cullin1-F-box E3 ligase-mediated NLRP3 ubiquitination. Inflammation. (2025) 48(5):3629–3648. doi: 10.1007/s10753-025-02289-2
75. Herrington CS, Poulsom R, and Coates PJ. Recent advances in pathology: the 2020 annual review issue of the journal of pathology. J Pathol. (2020) 250:475–9. doi: 10.1002/path.5425
76. Hashemi M, Khosroshahi EM, Chegini MK, Abedi M, Matinahmadi A, Hosnarody YSD, et al. miRNAs and exosomal miRNAs in lung cancer: New emerging players in tumor progression and therapy response. Pathol Res Pract. (2023) 251:154906. doi: 10.1016/j.prp.2023.154906
77. Wani S, Man Law IK, and Pothoulakis C. Role and mechanisms of exosomal miRNAs in IBD pathophysiology. Am J Physiol Gastrointest Liver Physiol. (2020) 319:G646–g654. doi: 10.1152/ajpgi.00295.2020
78. Jia Y and Wei Y. Modulators of microRNA function in the immune system. Int J Mol Sci. (2020) 21:2357. doi: 10.3390/ijms21072357
79. Jiang H, Zhao H, Zhang M, He Y, Li X, Xu Y, et al. Hypoxia induced changes of exosome cargo and subsequent biological effects. Front Immunol. (2022) 13:824188. doi: 10.3389/fimmu.2022.824188
80. Boon RA and Vickers KC. Intercellular transport of microRNAs. Arterioscler Thromb Vasc Biol. (2013) 33:186–92. doi: 10.1161/ATVBAHA.112.300139
81. Wu T, Zhao Y, Zhang X, Wang Y, Chen Q, Zhang M, et al. Short-chain acyl post-translational modifications in cancers: Mechanisms, roles, and therapeutic implications. Cancer Commun (Lond). (2025) 45:1247–84. doi: 10.1002/cac2.70048
82. Zhang S, Chen W, Zhou J, Liang Q, Zhang Y, Su M, et al. The benefits and safety of monoclonal antibodies: implications for cancer immunotherapy. J Inflammation Res. (2025) 18:4335–57. doi: 10.2147/JIR.S499403
83. Khan MI, Alsayed R, Choudhry H, and Ahmad A. Exosome-mediated response to cancer therapy: modulation of epigenetic machinery. Int J Mol Sci. (2022) 23:6222. doi: 10.3390/ijms23116222
84. Kovaleva O, Sorokin M, Egorova A, Petrenko A, Shelekhova K, and Gratchev A. Macrophage - tumor cell interaction beyond cytokines. Front Oncol. (2023) 13:1078029. doi: 10.3389/fonc.2023.1078029
85. Wang L, Yu Q, Xiao J, Chen Q, Fang M, and Zhao H. Cigarette Smoke Extract-Treated Mouse Airway Epithelial Cells-Derived Exosomal LncRNA MEG3 Promotes M1 Macrophage Polarization and Pyroptosis in Chronic Obstructive Pulmonary Disease by Upregulating TREM-1 via m(6)A Methylation. Immune Netw. (2024) 24:e3. doi: 10.4110/in.2024.24.e3
86. Lathigara D, Kaushal D, and Wilson RB. Molecular mechanisms of western diet-induced obesity and obesity-related carcinogenesis-A narrative review. Metabolites. (2023) 13:675. doi: 10.3390/metabo13050675
87. Jiang C, Zhang J, Wang W, Shan Z, Sun F, Tan Y, et al. Extracellular vesicles in gastric cancer: role of exosomal lncRNA and microRNA as diagnostic and therapeutic targets. Front Physiol. (2023) 14:1158839. doi: 10.3389/fphys.2023.1158839
88. Momen-Heravi F and Bala S. miRNA regulation of innate immunity. J Leukoc Biol. (2018). doi: 10.1002/JLB.3MIR1117-459R
89. Lone SN, Bhat AA, Wani NA, Karedath T, Hashem S, Nisar S, et al. miRNAs as novel immunoregulators in cancer. Semin Cell Dev Biol. (2022) 124:3–14. doi: 10.1016/j.semcdb.2021.04.013
90. Nail HM, Chiu CC, Leung CH, Ahmed MMM, and Wang HD. Exosomal miRNA-mediated intercellular communications and immunomodulatory effects in tumor microenvironments. J BioMed Sci. (2023) 30:69. doi: 10.1186/s12929-023-00964-w
91. İlhan A, Golestani S, Shafagh SG, Asadi F, Daneshdoust D, Al-Naqeeb BZT, et al. The dual role of microRNA (miR)-20b in cancers: Friend or foe? Cell Communication Signaling. (2023) 21:26. doi: 10.1186/s12964-022-01019-7
92. Zhu X, Shen H, Yin X, Yang M, Wei H, Chen Q, et al. Macrophages derived exosomes deliver miR-223 to epithelial ovarian cancer cells to elicit a chemoresistant phenotype. J Exp Clin Cancer Res. (2019) 38:81. doi: 10.1186/s13046-019-1095-1
93. Zheng P, Chen L, Yuan X, Luo Q, Liu Y, Xie G, et al. Exosomal transfer of tumor-associated macrophage-derived miR-21 confers cisplatin resistance in gastric cancer cells. J Exp Clin Cancer Res. (2017) 36:53. doi: 10.1186/s13046-017-0528-y
94. Binenbaum Y, Fridman E, Yaari Z, Milman N, Schroeder A, Ben David G, et al. Transfer of miRNA in macrophage-derived exosomes induces drug resistance in pancreatic adenocarcinoma. Cancer Res. (2018) 78:5287–99. doi: 10.1158/0008-5472.CAN-18-0124
95. Li X, Chen Z, Ni Y, Bian C, Huang J, Chen L, et al. Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer. Transl Lung Cancer Res. (2021) 10:1338–54. doi: 10.21037/tlcr-20-1255
96. Ma YS, Wu TM, Ling CC, Yu F, Zhang J, Cao PS, et al. M2 macrophage-derived exosomal microRNA-155-5p promotes the immune escape of colon cancer by downregulating ZC3H12B. Mol Ther Oncolytics. (2021) 20:484–98. doi: 10.1016/j.omto.2021.02.005
97. An B, Shin CH, Kwon JW, Tran NL, Kim AH, Jeong H, et al. M1 macrophage-derived exosomal microRNA-29c-3p suppresses aggressiveness of melanoma cells via ENPP2. Cancer Cell Int. (2024) 24:325. doi: 10.1186/s12935-024-03512-0
98. Rudin CM, Brambilla E, Faivre-Finn C, and Sage J. Small-cell lung cancer. Nat Rev Dis Primers. (2021) 7:3. doi: 10.1038/s41572-020-00235-0
99. Dela Cruz CS, Tanoue LT, and Matthay RA. Lung cancer: epidemiology, etiology, and prevention. Clin Chest Med. (2011) 32:605–44. doi: 10.1016/j.ccm.2011.09.001
100. Nazri HM, Greaves E, Quenby S, Dragovic R, Tapmeier TT, and Becker CM. The role of small extracellular vesicle-miRNAs in endometriosis. Hum Reprod. (2023) 38:2296–311. doi: 10.1093/humrep/dead216
101. Thandra KC, Barsouk A, Saginala K, Aluru JS, and Barsouk A. Epidemiology of lung cancer. Contemp Oncol (Pozn). (2021) 25:45–52. doi: 10.5114/wo.2021.103829
102. Takeda Y, Kobayashi S, Kitakaze M, Yamada D, Akita H, Asai A, et al. Immuno-surgical management of pancreatic cancer with analysis of cancer exosomes. Cells. (2020) 9:1645. doi: 10.3390/cells9071645
103. Saleh RO, Hjazi A, Bansal P, Ahmad I, Kaur H, Ali SHJ, et al. Mysterious interactions between macrophage-derived exosomes and tumors; what do we know? Pathol Res Pract. (2024) 256:155261. doi: 10.1016/j.prp.2024.155261
104. Yu W, Liang X, Li X, Zhang Y, Sun Z, Liu Y, et al. MicroRNA-195: a review of its role in cancers. Onco Targets Ther. (2018) 11:7109–23. doi: 10.2147/OTT.S183600
105. Thapa R, Afzal O, Bhat AA, Goyal A, Alfawaz Altamimi AS, Almalki WH, et al. New horizons in lung cancer management through ATR/CHK1 pathway modulation. Future medicinal Chem. (2023) 15:1807–18. doi: 10.4155/fmc-2023-0164
106. Yang C, Kim HS, Song G, and Lim W. The potential role of exosomes derived from ovarian cancer cells for diagnostic and therapeutic approaches. J Cell Physiol. (2019) 234:21493–503. doi: 10.1002/jcp.28905
107. Yoshida K, Yokoi A, Kato T, Ochiya T, and Yamamoto Y. The clinical impact of intra- and extracellular miRNAs in ovarian cancer. Cancer Sci. (2020) 111:3435–44. doi: 10.1111/cas.14599
108. Pekarek L, Torres-Carranza D, Fraile-Martinez O, García-Montero C, Pekarek T, Saez MA, et al. An overview of the role of microRNAs on carcinogenesis: A focus on cell cycle, angiogenesis and metastasis. Int J Mol Sci. (2023) 24:7268. doi: 10.3390/ijms24087268
109. Zeng Y and Fu BM. Resistance mechanisms of anti-angiogenic therapy and exosomes-mediated revascularization in cancer. Front Cell Dev Biol. (2020) 8:610661. doi: 10.3389/fcell.2020.610661
110. Song Y, Kelava L, and Kiss I. MiRNAs in lung adenocarcinoma: role, diagnosis, prognosis, and therapy. Int J Mol Sci. (2023) 24:13302. doi: 10.3390/ijms241713302
111. Duan S, Yu S, Yuan T, Yao S, and Zhang L. Exogenous let-7a-5p induces A549 lung cancer cell death through BCL2L1-mediated PI3Kγ Signaling pathway. Front Oncol. (2019) 9:808. doi: 10.3389/fonc.2019.00808
112. Ahmad A. Epigenetic regulation of immunosuppressive tumor-associated macrophages through dysregulated microRNAs. Semin Cell Dev Biol. (2022) 124:26–33. doi: 10.1016/j.semcdb.2021.09.001
113. Ashrafizadeh M, Kumar AP, Aref AR, Zarrabi A, and Mostafavi E. Exosomes as promising nanostructures in diabetes mellitus: from insulin sensitivity to ameliorating diabetic complications. Int J Nanomedicine. (2022) 17:1229–53. doi: 10.2147/IJN.S350250
114. Chen C, Liu JM, and Luo YP. MicroRNAs in tumor immunity: functional regulation in tumor-associated macrophages. J Zhejiang Univ Sci B. (2020) 21:12–28. doi: 10.1631/jzus.B1900452
115. Mercogliano MF, Bruni S, Mauro F, Elizalde PV, and Schillaci R. Harnessing tumor necrosis factor alpha to achieve effective cancer immunotherapy. Cancers (Basel). (2021) 13:564. doi: 10.3390/cancers13030564
116. Jiao Y, Zhang T, Zhang C, Ji H, Tong X, Xia R, et al. Exosomal miR-30d-5p of neutrophils induces M1 macrophage polarization and primes macrophage pyroptosis in sepsis-related acute lung injury. Crit Care. (2021) 25:356. doi: 10.1186/s13054-021-03775-3
117. Metcalf G. MicroRNAs: circulating biomarkers for the early detection of imperceptible cancers via biosensor and machine-learning advances. Oncogene. (2024) 43:2135–42. doi: 10.1038/s41388-024-03076-3
118. Cazzoli R, Buttitta F, Di Nicola M, Malatesta S, Marchetti A, Rom WN, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol. (2013) 8:1156–62. doi: 10.1097/JTO.0b013e318299ac32
119. Munagala R, Aqil F, and Gupta RC. Exosomal miRNAs as biomarkers of recurrent lung cancer. Tumor Biol. (2016) 37:10703–14. doi: 10.1007/s13277-016-4939-8
120. Wang C, Wang X, Zhang D, Sun X, Wu Y, Wang J, et al. The macrophage polarization by miRNAs and its potential role in the treatment of tumor and inflammation (Review). Oncol Rep. (2023) 50:190. doi: 10.3892/or.2023.8627
121. Bazi A, Rashidi-Juybari FZ, Hosseini SM, Khazaee-Nasirabadi MH, Ghorbani Biregani K, Peymaninezhad F, et al. Extracellular vesicle–derived microRNAs: A deep review of the latest literature investigating their role in drug resistance, prognosis, and microenvironment interactions in hematologic Malignancies. Eur J Cancer Care. (2025) 2025:5512907. doi: 10.1155/ecc/5512907
122. Li X, Xiang J, Wang J, Li J, Wu FX, and Li M. FUNMarker: fusion network-based method to identify prognostic and heterogeneous breast cancer biomarkers. IEEE/ACM Trans Comput Biol Bioinform. (2021) 18:2483–91. doi: 10.1109/TCBB.2020.2973148
123. Feng Y, Spezia M, Huang S, Yuan C, Zeng Z, Zhang L, et al. Breast cancer development and progression: Risk factors, cancer stem cells, signaling pathways, genomics, and molecular pathogenesis. Genes Dis. (2018) 5:77–106. doi: 10.1016/j.gendis.2018.05.001
124. Syed SN, Frank AC, Raue R, and Brüne B. MicroRNA-A tumor trojan horse for tumor-associated macrophages. Cells. (2019) 8:1482. doi: 10.3390/cells8121482
125. Ginsburg O, Yip CH, Brooks A, Cabanes A, Caleffi M, Dunstan Yataco JA, et al. Breast cancer early detection: A phased approach to implementation. Cancer. (2020) 126 Suppl 10:2379–93. doi: 10.1002/cncr.32887
126. Xu WX, Wang DD, Zhao ZQ, Zhang HD, Yang SJ, Zhang Q, et al. Exosomal microRNAs shuttling between tumor cells and macrophages: cellular interactions and novel therapeutic strategies. Cancer Cell Int. (2022) 22:190. doi: 10.1186/s12935-022-02594-y
127. Łukasiewicz S, Czeczelewski M, Forma A, Baj J, Sitarz R, and Stanisławek A. Breast cancer-epidemiology, risk factors, classification, prognostic markers, and current treatment strategies-an updated review. Cancers (Basel). (2021) 13:4287. doi: 10.3390/cancers13174287
128. Santos P and Almeida F. Role of exosomal miRNAs and the tumor microenvironment in drug resistance. Cells. (2020) 9:1450. doi: 10.3390/cells9061450
129. Syed SN and Brüne B. Exosomal and non-exosomal microRNAs: new kids on the block for cancer therapy. Int J Mol Sci. (2022) 23:4493. doi: 10.3390/ijms23094493
130. Muñoz JP, Pérez-Moreno P, Pérez Y, and Calaf GM. The role of microRNAs in breast cancer and the challenges of their clinical application. Diagnostics (Basel). (2023) 13:3072. doi: 10.3390/diagnostics13193072
131. Baylie T, Kasaw M, Getinet M, Getie G, Jemal M, Nigatu A, et al. The role of miRNAs as biomarkers in breast cancer. Front Oncol. (2024) 14:1374821. doi: 10.3389/fonc.2024.1374821
132. Strizova Z, Benesova I, Bartolini R, Novysedlak R, Cecrdlova E, Foley LK, et al. M1/M2 macrophages and their overlaps - myth or reality? Clin Sci (Lond). (2023) 137:1067–93. doi: 10.1042/CS20220531
133. Clements ME and Johnson RW. Breast cancer dormancy in bone. Curr Osteoporos Rep. (2019) 17:353–61. doi: 10.1007/s11914-019-00532-y
134. Walker ND, Elias M, Guiro K, Bhatia R, Greco SJ, Bryan M, et al. Exosomes from differentially activated macrophages influence dormancy or resurgence of breast cancer cells within bone marrow stroma. Cell Death Dis. (2019) 10:59. doi: 10.1038/s41419-019-1304-z
135. Neophytou CM, Panagi M, Stylianopoulos T, and Papageorgis P. The role of tumor microenvironment in cancer metastasis: molecular mechanisms and therapeutic opportunities. Cancers (Basel). (2021) 13:2053. doi: 10.3390/cancers13092053
136. Liu J, Ren L, Li S, Li W, Zheng X, Yang Y, et al. The biology, function, and applications of exosomes in cancer. Acta Pharm Sin B. (2021) 11:2783–97. doi: 10.1016/j.apsb.2021.01.001
137. Jin Y, Xing J, Xu K, Liu D, and Zhuo Y. Exosomes in the tumor microenvironment: Promoting cancer progression. Front Immunol. (2022) 13:1025218. doi: 10.3389/fimmu.2022.1025218
138. Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. (2011) 10:117. doi: 10.1186/1476-4598-10-117
139. Moradi-Chaleshtori M, Hashemi SM, Soudi S, Bandehpour M, and Mohammadi-Yeganeh S. Tumor-derived exosomal microRNAs and proteins as modulators of macrophage function. J Cell Physiol. (2019) 234:7970–82. doi: 10.1002/jcp.27552
140. Huang X, Cao J, and Zu X. Tumor-associated macrophages: An important player in breast cancer progression. Thorac Cancer. (2022) 13:269–76. doi: 10.1111/1759-7714.14268
141. Allison E, Edirimanne S, Matthews J, and Fuller SJ. Breast cancer survival outcomes and tumor-associated macrophage markers: A systematic review and meta-analysis. Oncol Ther. (2023) 11:27–48. doi: 10.1007/s40487-022-00214-3
142. Li Y, Zhao L, Shi B, Ma S, Xu Z, Ge Y, et al. Functions of miR-146a and miR-222 in Tumor-associated Macrophages in Breast Cancer. Sci Rep. (2015) 5:18648. doi: 10.1038/srep18648
143. Zare E, Yaghoubi SM, Khoshnazar M, Jafari Dargahlou S, Machhar JS, Zheng Z, et al. MicroRNAs in cancer immunology: master regulators of the tumor microenvironment and immune evasion, with therapeutic potential. Cancers (Basel). (2025) 17:2172. doi: 10.3390/cancers17132172
144. Jelovac D and Armstrong DK. Recent progress in the diagnosis and treatment of ovarian cancer. CA Cancer J Clin. (2011) 61:183–203. doi: 10.3322/caac.20113
145. Mohan A, Agarwal S, Clauss M, Britt NS, and Dhillon NK. Extracellular vesicles: novel communicators in lung diseases. Respir Res. (2020) 21:175. doi: 10.1186/s12931-020-01423-y
146. Konstantinopoulos PA, Norquist B, Lacchetti C, Armstrong D, Grisham RN, Goodfellow PJ, et al. Germline and somatic tumor testing in epithelial ovarian cancer: ASCO guideline. J Clin Oncol. (2020) 38:1222–45. doi: 10.1200/JCO.19.02960
147. Malla RR and Kiran P. Tumor microenvironment pathways: Cross regulation in breast cancer metastasis. Genes Dis. (2022) 9:310–24. doi: 10.1016/j.gendis.2020.11.015
148. Matsuzaka Y and Yashiro R. Regulation of extracellular vesicle-mediated immune responses against antigen-specific presentation. Vaccines (Basel). (2022) 10:1691. doi: 10.3390/vaccines10101691
149. Momenimovahed Z, Tiznobaik A, Taheri S, and Salehiniya H. Ovarian cancer in the world: epidemiology and risk factors. Int J Womens Health. (2019) 11:287–99. doi: 10.2147/IJWH.S197604
150. Ali AT, Al-Ani O, and Al-Ani F. Epidemiology and risk factors for ovarian cancer. Prz Menopauzalny. (2023) 22:93–104. doi: 10.5114/pm.2023.128661
151. Kotsopoulos J, Lubinski J, Gronwald J, Cybulski C, Demsky R, Neuhausen SL, et al. Factors influencing ovulation and the risk of ovarian cancer in BRCA1 and BRCA2 mutation carriers. Int J Cancer. (2015) 137:1136–46. doi: 10.1002/ijc.29386
152. Li Y, Jin D, Xie W, Wen L, Chen W, Xu J, et al. Mesenchymal stem cells-derived exosomes: A possible therapeutic strategy for osteoporosis. Curr Stem Cell Res Ther. (2018) 13:362–8. doi: 10.2174/1574888X13666180403163456
153. Fasoulakis Z, Psarommati MZ, Papapanagiotou A, Pergialiotis V, Koutras A, Douligeris A, et al. MicroRNAs can influence ovarian cancer progression by dysregulating integrin activity. Cancers (Basel). (2023) 15:4449. doi: 10.3390/cancers15184449
154. Chou J, Provot S, and Werb Z. GATA3 in development and cancer differentiation: cells GATA have it! J Cell Physiol. (2010) 222:42–9. doi: 10.1002/jcp.21943
155. Ben Hamouda S and Essafi-Benkhadir K. Interplay between signaling pathways and tumor microenvironment components: A paradoxical role in colorectal cancer. Int J Mol Sci. (2023) 24:5600. doi: 10.3390/ijms24065600
156. El-Arabey AA, Denizli M, Kanlikilicer P, Bayraktar R, Ivan C, Rashed M, et al. GATA3 as a master regulator for interactions of tumor-associated macrophages with high-grade serous ovarian carcinoma. Cell Signalling. (2020) 68:109539. doi: 10.1016/j.cellsig.2020.109539
157. Khatun M and Ray RB. Mechanisms underlying hepatitis C virus-associated hepatic fibrosis. Cells. (2019) 8:1249. doi: 10.3390/cells8101249
158. Kwon Y, Kim M, Kim Y, Jung HS, and Jeoung D. Exosomal microRNAs as mediators of cellular interactions between cancer cells and macrophages. Front Immunol. (2020) 11:1167. doi: 10.3389/fimmu.2020.01167
159. Whiteside TL. What are regulatory T cells (Treg) regulating in cancer and why? Semin Cancer Biol. (2012) 22:327–34. doi: 10.1016/j.semcancer.2012.03.004
160. Holmberg R, Robinson M, Gilbert SF, Lujano-Olazaba O, Waters JA, Kogan E, et al. TWEAK-fn14-relB signaling cascade promotes stem cell-like features that contribute to post-chemotherapy ovarian cancer relapse. Mol Cancer Res. (2023) 21:170–86. doi: 10.1158/1541-7786.MCR-22-0486
161. Xu C, Chen J, Tan M, and Tan Q. The role of macrophage polarization in ovarian cancer: from molecular mechanism to therapeutic potentials. Front Immunol. (2025) 16:1543096. doi: 10.3389/fimmu.2025.1543096
162. Gallo P, Silletta M, Prinzi FL, Farolfi T, and Coppola A. Hepatocellular carcinoma and non-alcoholic fatty liver disease: A modern context for an ancient disease. J Clin Med. (2023) 12:4605. doi: 10.3390/jcm12144605
163. Chidambaranathan-Reghupaty S, Fisher PB, and Sarkar D. Hepatocellular carcinoma (HCC): Epidemiology, etiology and molecular classification. Adv Cancer Res. (2021) 149:1–61. doi: 10.1016/bs.acr.2020.10.001
164. El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. (2012) 142:1264–1273.e1261. doi: 10.1053/j.gastro.2011.12.061
165. Hadad S, Khalaji A, Sarmadian AJ, Sarmadian PJ, Janagard EM, and Baradaran B. Tumor-associated macrophages derived exosomes; from pathogenesis to therapeutic opportunities. Int Immunopharmacol. (2024) 136:112406. doi: 10.1016/j.intimp.2024.112406
166. Morishita A, Oura K, Tadokoro T, Fujita K, Tani J, and Masaki T. MicroRNAs in the pathogenesis of hepatocellular carcinoma: A review. Cancers (Basel). (2021) 13:514. doi: 10.3390/cancers13030514
167. Fedele M, Cerchia L, and Chiappetta G. The epithelial-to-mesenchymal transition in breast cancer: focus on basal-like carcinomas. Cancers (Basel). (2017) 9:134. doi: 10.3390/cancers9100134
168. Luong AB, Do HQ, Tarchi P, Bonazza D, Bottin C, Cabral LKD, et al. The mRNA distribution of cancer stem cell marker CD90/thy-1 is comparable in hepatocellular carcinoma of eastern and western populations. Cells. (2020) 9:2672. doi: 10.3390/cells9122672
169. Wang Y, Wang B, Xiao S, Li Y, and Chen Q. miR-125a/b inhibits tumor-associated macrophages mediated in cancer stem cells of hepatocellular carcinoma by targeting CD90. J Cell Biochem. (2019) 120:3046–55. doi: 10.1002/jcb.27436
170. Wang HC, Yin WX, Jiang M, Han JY, Kuai XW, Sun R, et al. Function and biomedical implications of exosomal microRNAs delivered by parenchymal and nonparenchymal cells in hepatocellular carcinoma. World J Gastroenterol. (2023) 29:5435–51. doi: 10.3748/wjg.v29.i39.5435
171. Aucher A, Rudnicka D, and Davis DM. MicroRNAs transfer from human macrophages to hepato-carcinoma cells and inhibit proliferation. J Immunol. (2013) 191:6250–60. doi: 10.4049/jimmunol.1301728
172. Singal AK, Bataller R, Ahn J, Kamath PS, and Shah VH. ACG clinical guideline: alcoholic liver disease. Am J Gastroenterol. (2018) 113:175–94. doi: 10.1038/ajg.2017.469
173. Babuta M, Furi I, Bala S, Bukong TN, Lowe P, Catalano D, et al. Dysregulated autophagy and lysosome function are linked to exosome production by micro-RNA 155 in alcoholic liver disease. Hepatology. (2019) 70:2123–41. doi: 10.1002/hep.30766
174. Chung KPS, Leung RWH, and Lee TKW. Hampering stromal cells in the tumor microenvironment as a therapeutic strategy to destem cancer stem cells. Cancers (Basel). (2021) 13:3191. doi: 10.3390/cancers13133191
175. McGuigan A, Kelly P, Turkington RC, Jones C, Coleman HG, and Mccain RS. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J Gastroenterol. (2018) 24:4846–61. doi: 10.3748/wjg.v24.i43.4846
176. Sarantis P, Koustas E, Papadimitropoulou A, Papavassiliou AG, and Karamouzis MV. Pancreatic ductal adenocarcinoma: Treatment hurdles, tumor microenvironment and immunotherapy. World J Gastrointest Oncol. (2020) 12:173–81. doi: 10.4251/wjgo.v12.i2.173
177. Ke TM, Lophatananon A, and Muir KR. Risk factors associated with pancreatic cancer in the UK biobank cohort. Cancers (Basel). (2022) 14:4991. doi: 10.3390/cancers14204991
178. Rawla P and Barsouk A. Epidemiology of gastric cancer: global trends, risk factors and prevention. Prz Gastroenterol. (2019) 14:26–38. doi: 10.5114/pg.2018.80001
179. Kozłowska M and Śliwińska A. The link between diabetes, pancreatic tumors, and miRNAs-new players for diagnosis and therapy? Int J Mol Sci. (2023) 24:10252. doi: 10.3390/ijms241210252
180. Tu H and Costa M. XIAP’s profile in human cancer. Biomolecules. (2020) 10:1493. doi: 10.3390/biom10111493
181. Li S, Sun J, Yang J, Zhang L, Wang L, Wang X, et al. XIAP expression is associated with pancreatic carcinoma outcome. Mol Clin Oncol. (2013) 1:305–8. doi: 10.3892/mco.2013.58
182. Yin Z, Zhou Y, Ma T, Chen S, Shi N, Zou Y, et al. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J Cell Mol Med. (2020) 24:5028–38. doi: 10.1111/jcmm.15125
183. Sawicki T, Ruszkowska M, Danielewicz A, Niedźwiedzka E, Arłukowicz T, and Przybyłowicz KE. A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis. Cancers (Basel). (2021) 13:2025. doi: 10.3390/cancers13092025
184. Nittayaboon K, Leetanaporn K, Sangkhathat S, Roytrakul S, and Navakanitworakul R. Proteomic analysis of butyrate-resistant colorectal cancer-derived exosomes reveals potential resistance to anti-cancer drugs. Discov Med. (2024) 36:1306–15. doi: 10.24976/Discov.Med.202436185.121
185. Rawla P, Sunkara T, and Barsouk A. Epidemiology of colorectal cancer: incidence, mortality, survival, and risk factors. Prz Gastroenterol. (2019) 14:89–103. doi: 10.5114/pg.2018.81072
186. Ionescu VA, Gheorghe G, Bacalbasa N, Chiotoroiu AL, and Diaconu C. Colorectal cancer: from risk factors to oncogenesis. Med (Kaunas). (2023) 59:1646. doi: 10.3390/medicina59091646
187. Sado AI, Batool W, Ahmed A, Zafar S, Patel SK, Mohan A, et al. Role of microRNA in colorectal carcinoma (CRC): a narrative review. Ann Med Surg (Lond). (2024) 86:308–18. doi: 10.1097/MS9.0000000000001494
188. Wai Hon K, Zainal Abidin SA, Othman I, and Naidu R. Insights into the Role of microRNAs in Colorectal Cancer (CRC) Metabolism. Cancers (Basel). (2020) 12:2462. doi: 10.3390/ijms241210252
189. Xiao G, Tang H, Wei W, Li J, Ji L, and Ge J. Aberrant expression of microRNA-15a and microRNA-16 synergistically associates with tumor progression and prognosis in patients with colorectal cancer. Gastroenterol Res Pract. (2014) 2014:364549. doi: 10.1155/2014/364549
190. Liebl MC and Hofmann TG. The role of p53 signaling in colorectal cancer. Cancers (Basel). (2021) 13:2125. doi: 10.3390/cancers13092125
191. Cooks T, Pateras IS, Jenkins LM, Patel KM, Robles AI, Morris J, et al. Mutant p53 cancers reprogram macrophages to tumor supporting macrophages via exosomal miR-1246. Nat Commun. (2018) 9:771. doi: 10.1038/s41467-018-03224-w
192. Wawro M, Wawro K, Kochan J, Solecka A, Sowinska W, Lichawska-Cieslar A, et al. ZC3H12B/MCPIP2, a new active member of the ZC3H12 family. Rna. (2019) 25:840–56. doi: 10.1261/rna.071381.119
193. Klampfer L. Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets. (2011) 11:451–64. doi: 10.2174/156800911795538066
194. Yimin E, Lu C, Zhu K, Li W, Sun J, Ji P, et al. Function and mechanism of exosomes derived from different cells as communication mediators in colorectal cancer metastasis. iScience. (2024) 27:109350. doi: 10.1016/j.isci.2024.109350
195. Sinha D, Roy S, Saha P, Chatterjee N, and Bishayee A. Trends in research on exosomes in cancer progression and anticancer therapy. Cancers (Basel). (2021) 13:326. doi: 10.3390/cancers13020326
196. Onciul R, Brehar FM, Toader C, Covache-Busuioc RA, Glavan LA, Bratu BG, et al. Deciphering glioblastoma: fundamental and novel insights into the biology and therapeutic strategies of gliomas. Curr Issues Mol Biol. (2024) 46:2402–43. doi: 10.3390/cimb46030153
197. Baranwal S and Alahari SK. miRNA control of tumor cell invasion and metastasis. Int J Cancer. (2010) 126:1283–90. doi: 10.1002/ijc.25014
198. Peng Y and Croce CM. The role of MicroRNAs in human cancer. Signal Transduction Targeted Ther. (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
199. Sell MC, Ramlogan-Steel CA, Steel JC, and Dhungel BP. MicroRNAs in cancer metastasis: biological and therapeutic implications. Expert Rev Mol Med. (2023) 25:e14. doi: 10.1017/erm.2023.7
200. Zheng P, Luo Q, Wang W, Li J, Wang T, Wang P, et al. Tumor-associated macrophages-derived exosomes promote the migration of gastric cancer cells by transfer of functional Apolipoprotein E. Cell Death Dis. (2018) 9:434. doi: 10.1038/s41419-018-0465-5
201. Li Z, Suo B, Long G, Gao Y, Song J, Zhang M, et al. Exosomal miRNA-16-5p derived from M1 macrophages enhances T cell-dependent immune response by regulating PD-L1 in gastric cancer. Front Cell Dev Biol. (2020) 8:572689. doi: 10.3389/fcell.2020.572689
202. Gao H, Ma J, Cheng Y, and Zheng P. Exosomal transfer of macrophage-derived miR-223 confers doxorubicin resistance in gastric cancer. Onco Targets Ther. (2020) 13:12169–79. doi: 10.2147/OTT.S283542
203. Buruiană A, Florian ŞI, Florian AI, Timiş TL, Mihu CM, Miclăuş M, et al. The roles of miRNA in glioblastoma tumor cell communication: diplomatic and aggressive negotiations. Int J Mol Sci. (2020) 21:1950. doi: 10.3390/ijms21061950
204. Ordóñez-Rubiano EG, Rincón-Arias N, Espinosa S, Shelton WJ, Salazar AF, Cómbita A, et al. The potential of miRNA-based approaches in glioblastoma: An update in current advances and future perspectives. Curr Res Pharmacol Drug Discov. (2024) 7:100193. doi: 10.1016/j.crphar.2024.100193
205. Qian M, Wang S, Guo X, Wang J, Zhang Z, Qiu W, et al. Hypoxic glioma-derived exosomes deliver microRNA-1246 to induce M2 macrophage polarization by targeting TERF2IP via the STAT3 and NF-κB pathways. Oncogene. (2020) 39:428–42. doi: 10.1038/s41388-019-0996-y
206. Guo L, Fu Z, Li H, Wei R, Guo J, Wang H, et al. Smart hydrogel: A new platform for cancer therapy. Adv Colloid Interface Sci. (2025) 340:103470. doi: 10.1016/j.cis.2025.103470
207. Zhang M, Hu S, Liu L, Dang P, Liu Y, Sun Z, et al. Engineered exosomes from different sources for cancer-targeted therapy. Signal Transduct Target Ther. (2023) 8:124. doi: 10.1038/s41392-023-01382-y
208. Gharavi AT, Hanjani NA, Movahed E, and Doroudian M. The role of macrophage subtypes and exosomes in immunomodulation. Cell Mol Biol Lett. (2022) 27:83. doi: 10.1186/s11658-022-00384-y
209. Yadav A, Xuan Y, Sen CK, and Ghatak S. Standardized reporting of research on exosomes to ensure rigor and reproducibility. Adv Wound Care (New Rochelle). (2024) 13:584–99. doi: 10.1089/wound.2024.0093
210. Nieuwland R, Falcón-Pérez JM, Théry C, and Witwer KW. Rigor and standardization of extracellular vesicle research: Paving the road towards robustness. J Extracell Vesicles. (2020) 10:e12037. doi: 10.1002/jev2.12037
Keywords: cancer biomarkers, exosomal miRNAs, macrophage-derived exosomes, therapeutic resistance, tumor-associated macrophages, tumor microenvironment
Citation: Ramalingam PS, Afzal M, Babu MA, M. M. R, Sahoo S, Pandey SN, Ali H, Hussain MS, Gupta G, Mekala JR and Arumugam S (2025) Targeting cancer via macrophage-derived exosomal miRNAs: implications for tumor progression and resistance. Front. Immunol. 16:1683799. doi: 10.3389/fimmu.2025.1683799
Received: 11 August 2025; Accepted: 03 November 2025;
Published: 20 November 2025.
Edited by:
Abdullah Saeed, City of Hope National Medical Center, United StatesReviewed by:
Sheefa Mirza, University of Witwatersrand, South AfricaYing Liang, Air Force Medical University, China
Copyright © 2025 Ramalingam, Afzal, Babu, M. M., Sahoo, Pandey, Ali, Hussain, Gupta, Mekala and Arumugam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sivakumar Arumugam, c2l2YV9rdW1hci5hQHZpdC5hYy5pbg==; Janaki Ramaiah Mekala, amFuYWtpcmFtYWlhaC5tQHZpdC5hYy5pbg==
 Prasanna Srinivasan Ramalingam
Prasanna Srinivasan Ramalingam Muhammad Afzal
Muhammad Afzal M. Arockia Babu3
M. Arockia Babu3 Md Sadique Hussain
Md Sadique Hussain Sivakumar Arumugam
Sivakumar Arumugam