- 1Department of Biology, Faculty of Science, University of Copenhagen, Copenhagen, Denmark
- 2Department of Microbiology and Biotechnology, Max Rubner-Institut (Federal Research Institute of Nutrition and Food), Kiel, Germany
- 3Department of Food Science, Faculty of Science, University of Copenhagen, Frederiksberg, Denmark
Leuconostoc (Ln.), Weissella, and Oenococcus form a group of related genera of lactic acid bacteria, which once all shared the name Leuconostoc. They are associated with plants, fermented vegetable products, raw milk, dairy products, meat, and fish. Most of industrially relevant Leuconostoc strains can be classified as either Ln. mesenteroides or Ln. pseudomesenteroides. They are important flavor producers in dairy fermentations and they initiate nearly all vegetable fermentations. Therefore, bacteriophages attacking Leuconostoc strains may negatively influence the production process. Bacteriophages attacking Leuconostoc strains were first reported in 1946. Since then, the majority of described Leuconostoc phages was isolated from either dairy products or fermented vegetable products. Both lytic and temperate phages of Leuconostoc were reported. Most of Leuconostoc phages examined using electron microscopy belong to the Siphoviridae family and differ in morphological details. Hybridization and comparative genomic studies of Leuconostoc phages suggest that they can be divided into several groups, however overall diversity of Leuconostoc phages is much lower as compared to, e.g., lactococcal phages. Several fully sequenced genomes of Leuconostoc phages have been deposited in public databases. Lytic phages of Leuconostoc can be divided into two host species-specific groups with similarly organized genomes that shared very low nucleotide similarity. Phages of dairy Leuconostoc have rather limited host-ranges. The receptor binding proteins of two lytic Ln. pseudomesenteroides phages have been identified. Molecular tools for detection of dairy Leuconostoc phages have been developed. The rather limited data on phages of Oenococcus and Weissella show that (i) lysogeny seems to be abundant in Oenococcus strains, and (ii) several phages infecting Weissella cibaria are also able to productively infect strains of other Weissella species and even strains of the genus Lactobacillus.
Introduction
Taxonomy of Leuconostoc, Oenococcus, and Weissella
Leuconostoc (Ln.), Weissella (W.), Oenococcus (O.), and Fructobacillus (F.) form a group of related genera of lactic acid bacteria. Based on 16S rRNA sequencing, Collins et al. (1993) proposed that Ln. paramesenteroides and related species (Lactobacillus (Lb.) confusus, Lb. halotolerans, Lb. kandleri, Lb. minor, and Lb. viridescens) should be reclassified in the new genus Weissella. Dicks et al. (1995) assigned Ln. oenos to the new genus Oenococcus. Endo and Okada (2008) proposed to allocate several Leuconostoc species to the new genus Fructobacillus. Schleifer (2009), on the basis of 16S rRNA sequences, transferred the three genera Leuconostoc (including those species synonymous with Fructobacillus), Weissella, and Oenococcus into the newly formed family Leuconostocaceae. Members of the family show highest similarity to the genus Lactobacillus: they all are Gram-positive, catalase-negative, facultative anaerobes, and are characterized by heterofermentative lactic acid fermentation. While all members of the genera Leuconostoc and Oenococcus exhibit ovoid-shaped morphology, members of the genus Fructobacillus are rod-shaped. Species within the genus Weissella show two different (i.e., rod-shaped and ovoid-shaped) morphotypes.
Species in the Family Leuconostocaceae
According to information presented on the web-site of the “List of prokaryotic names with standing in nomenclature” (http://www.bacterio.net) the genus Leuconostoc is represented by 23 species and 4 subspecies for Ln. mesenteroides. However, several of the species names are synonyms within the genus Leuconostoc (like, e.g., Ln. argentinum and Ln. lactis, or Ln. mesenteroides subsp. cremoris and Ln. cremoris) or within different genera (like, e.g., Ln. pseudoficulneus and F. pseudoficulneus, Ln. paramesenteroides and W. paramesenteroides, Ln. oenos and O. oeni). Besides being recognized as meat spoilage organisms, Leuconostoc species have been described to be involved in several fermentation processes (Björkroth and Holzapfel, 2006). Ln. mesenteroides subsp. mesenteroides, Ln. mesenteroides subsp. cremoris, Ln. lactis, and Ln. pseudomesenteroides are regular constituents of aroma-producing starter-cultures applied in dairy fermentations (Farrow et al., 1989). In addition, Ln. mesenteroides subsp. mesenteroides is an important component of vegetable fermentations: it is involved in fermentation of coffee beans and (together with Ln. fallax) in sauerkraut fermentation. Ln. mesenteroides, Ln. citreum, Ln. gelidum and Ln. kimchii are dominant species in early kimchi fermentation, and Ln. mesenteroides subsp. dextranicum plays a key role in sourdough fermentations (Schleifer, 2009).
The genus Weissella comprises 18 species. As already mentioned for the genus Leuconostoc, some of the species names are synonyms within the genus (like, e.g., W. cibaria and W. kimchii) or within different genera (like the above mentioned Lactobacillus species proposed to be reclassified as Weissella). The species W. cibaria, W. confuse, and W. koreensis have been described to be associated with vegetable fermentations (Schleifer, 2009). Recently, W. fabalis and W. paramesenteroides have been detected in cocoa bean fermentation (Snauwaert et al., 2013) and in traditional Caciocavallo cheese (Settanni et al., 2012), respectively.
The genus Oenococcus comprises just two species: O. oeni originally described as Ln. oenos, and O. kitaharae isolated from composting residues of schochu distillation (Endo and Okada, 2006). O. oeni plays an important role in wine fermentation, where it decarboxylates malic acid to lactic acid (Schleifer, 2009).
The genus Fructobacillus is represented by five species, all of which except F. tropaeoli are synonyms of Leuconostoc species (Endo et al., 2011). Species of this genus have been described to be involved in spontaneous cocoa bean fermentations (Snauwaert et al., 2013). So far, no bacteriophages have been described for this genus. Therefore, the fructobacilli will not be further addressed in this review.
History of Bacteriophages in the Family Leuconostocaceae
The first description of bacteriophages affecting Leuconostoc was published in 1946 (Mosimann and Ritter, 1946). In this publication already, the negative impact on butter aroma of bacteriophages infecting Leuconostoc strains was shown. Just 1 year later, Leiva-Quiros and McCleskey (1947) isolated phages infecting Ln. mesenteroides for phage-typing purposes. From late 1970's to beginning of 2000's only dairy Leuconostoc phages have been reported on with the exception of one report also included phages from coffee fermentations (Table 1) (Boizet et al., 1992). From 2002 to 2012 a number of reports on Leuconostoc phages from sauerkraut fermentations have been published, and since 2010–2012 genomes of Ln. mesenteroides and Ln. pseudomesenteroides phages have been published (Table 1). A thorough classification of dairy Leuconostoc phages has been presented recently (Ali et al., 2013).
Lu et al. (2003) reported on bacteriophages infecting Weissella sp. Later, several studies described Podoviridae-phages infecting W. cibaria (Pringsulaka et al., 2011; Kleppen et al., 2012b).
Sozzi et al. (1976) were the first to describe phage infecting lactic acid bacteria in wine, which were later identified as O. oeni (Sozzi et al., 1982; Dicks et al., 1995). Lysogeny appears to be rather frequent in O. oeni, with 45–60% of O. oeni strains reported to be lysogenic (Arendt et al., 1991; Poblet-Icart et al., 1998). Pan-genome comparisons have confirmed these results and have demonstrated that apparently six different bacterial tRNA genes are involved as targets for prophage DNA integration of temperate bacteriophages in different strains of O. oeni (Borneman et al., 2012).
Fermentations Affected by Phages Infecting Species of Leuconostocaceae
Dairy fermentations are the most frequently described fermentations affected by bacteriophages (Samson and Moineau, 2013). This may be due to two major reasons: (i) milk is a liquid substrate in which phage are easily distributed, and (ii) most dairy fermentations involve application of starter culture mixtures, thus variations in acidification performance become readily evident. Presence of phages infecting dairy Leuconostoc strains has only been described occasionally (Sozzi et al., 1978; Boizet et al., 1992; Davey et al., 1995). However, the publications never acquired attention similar to those describing phages causing disturbances of acidification. This is probably due to the fact that during fermentation acidification failures are much easier and much earlier detectable than aroma defects. As a consequence, while the negative impact of phage on starter strains and acidification is well documented, the impact of phage on starter strains and aroma development is much less well known and is only beginning to be investigated systematically (Samtlebe et al., 2012). In recent years Swedish and Danish dairies have reported problems related to lack of diacetyl and CO2 in fermented milks (similar to report by Mosimann and Ritter, 1946) that could be correlated to phages attacking Leuconostoc strains (Anon. meeting reports, Vogensen, not published). Similarly, in several cases phage attacks on Leuconostoc strains in blue-mold cheeses have been correlated with lack of mold growth probably due to less openness in the cheese structure (Kot et al., 2014; Pujato et al., 2014).
Highest Leuconostoc phage titers in dairy products or in whey samples can vary significantly within a range from approximately 102 to 107 plaque-forming units (PFUs) per gram or per ml (Atamer et al., 2011) (Figure 1). The maximum numbers of PFUs for Leuconostoc phages in dairy samples are approximately 2 log units lower than maximum lactococcal phage numbers (approximately 109 PFU/ml). The lower Leuconostoc phage numbers are therefore probably reflecting the use of Leuconostoc as a minor starter component (1–10%) in undefined complex cultures consisting mainly of lactococcal strains. While the homo-lactic lactococci mainly contribute to acidification, the heterolactic and only weakly acidifying leuconostocs contribute to aroma by production of acetate, acetoin, and diacetyl (Farkye and Vedamuthu, 2002).
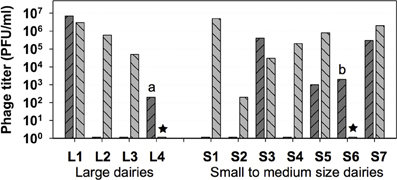
Figure 1. Highest Leuconostoc phage titers in whey ( ) and in brine (
) and in brine ( ) samples obtained from 4 large and from 7 small to medium size dairies. ★, no brine samples (dairies L4 and S6); a, butter milk sample; b, butter cream sample. (Modified from Atamer et al., 2011).
) samples obtained from 4 large and from 7 small to medium size dairies. ★, no brine samples (dairies L4 and S6); a, butter milk sample; b, butter cream sample. (Modified from Atamer et al., 2011).
The Leuconostoc lytic phages involved in dairy fermentations have generally been shown to be members of the Siphoviridae group of phages (Davey et al., 1995; Kleppen et al., 2012a; Ali et al., 2013; Kot et al., 2014; Pujato et al., 2014). However, for Leuconostoc phages isolated from sauerkraut fermentations also Myoviridae phages are seen (Barrangou et al., 2002; Yoon et al., 2002; Lu et al., 2003). Other than in dairy fermentations, in sauerkraut fermentations phages may even play an important role by affecting the development of different lactic acid bacteria species over fermentation time (Lu et al., 2003). However, when defined starter strains are supposed to be applied, phage infection may negatively affect quality parameters of the final product (Mudgal et al., 2006). Applying metagenomic analysis to kimchi, a traditional Korean fermented cabbage, evidence for presence of phage infecting Leuconostoc was obtained (Jung et al., 2011). So far, the only phage/host pair characterized for kimchi is a Podoviridae phage infecting Weissella cibaria (Kleppen et al., 2012b). A similar pair, Podoviridae phage and W. cibaria host, has been described for Nham, a Thai fermented pork sausage (Pringsulaka et al., 2011). Recently, several phages infecting W. cibaria and W. paramesenteroides were isolated from commercial cucumber fermentations and one phage for each host was shown to belong to the Siphoviridae family of phages (Lu et al., 2012). The only fermentation known to be affected by phages infecting O. oeni is wine fermentation, due to the exclusive involvement of these host bacteria in this type of fermentation (Schleifer, 2009). The phages have been shown to belong to the Siphoviridae family of phages (Poblet-Icart et al., 1998).
Leuconostoc Phages
Phages attacking Leuconostoc are best documented among all phages of the Leuconostocaceae family. The majority of reports on Leuconostoc phages are connected to problems in dairy fermentations, however few of the reports are dealing with Leuconostoc phages in vegetable or in coffee fermentations (Table 1). However, Leuconostoc gelidum is a known meat spoilage organism (Sakala et al., 2002). Interestingly, it was proposed to use Ln. gelidum phages to prevent bacterial spoilage of the meat products, pointing toward a different angle of phage-host interactions, i.e., phage bioprotection in fermented foods (Greer et al., 2007).
Morphology of Leuconostoc Phages
Recently, the morphotypes of dairy Ln. pseudomesenteroides and of Ln. mesenteroides phages from the dairy environment have been studied extensively with a set of 83 phage isolates (Ali et al. (2013). Although the phages were isolated from various sources (11 dairies, 3 phage collections), a low degree of variation was documented for their morphotypes. All phages were small isometric-headed Siphoviridae phages with non-contractile 140-nm long tails, however, according to their baseplate structure, these phages were differentiated into 6 different subgroups with six globular baseplate appendices or with peculiar Y-shaped baseplate structures (Ln. mesenteroides phages of morphotypes Ia or Ib), with plain baseplates but with or without characteristic collar structures or with uncommon tail striations (Ln. pseudomesenteroides phages of morphotypes IIa, b and d), or with undefined “fluffy” baseplate appendices (morphotype IIc Ln. pseudomesenteroides phages) (Figure 2). Dairy Leuconostoc phages of morphotypes Ia and IIb had been reported occasionally, i.e., Ia type phages: (Neve et al., 1988); IIb type phages: (Saxelin et al., 1986; Davey et al., 1995; Kleppen et al., 2012a). Siphoviridae phages of Leuconostoc with longer phage tails have also been described previously, indicating a broader biodiversity (Saxelin et al., 1986) within Leuconostoc phage populations. This correlates well with the establishment of 6 DNA homology groups for Ln. mesenteroides phages (Boizet et al., 1992). Leuconostoc phages isolated from sauerkraut fermentations did also reveal different morphotypes Siphoviridae phages with different tail lengths and Myoviridae phages (Barrangou et al., 2002; Lu et al., 2003, 2012). Temperate Siphoviridae phages from lysogenic Ln. mesenteroides and Ln. pseudomesenteroides strains with different tail lengths have been shown by Shin and Sato (1979), Lu et al. (2012), and Jang et al. (2010).
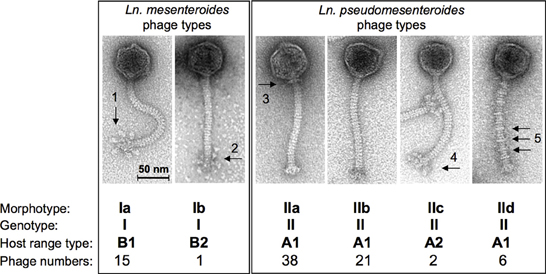
Figure 2. Overview on the current taxonomy of dairy Leuconostoc phages based on transmission electron microscopy, genotyping, and host range profiles. Arrows indicate structural details as follows: globular baseplate appendices (1), non-globular (Y-shaped) baseplate appendages (2), collar or neck passage structure (3), “fluffy” baseplate appendices (4), tail striations (5). (modified from Ali et al., 2013).
Genetics of Leuconostoc Phages
Currently, there are 12 full genomes of phages infecting Leuconostoc sp. present in publically available databases. All phages have dsDNA genomes with sizes from 25.7 to 38.7 kb (Table 2). Genomic G + C content varies from 36.1% for phage Φ-A4 to 38.7% for phage ΦMH1. All described lytic phages of Leuconostoc exhibit high similarity in regard to genome organization. Five modules can be distinguished in the genomes specifying replication, packaging, morphogenesis, host cell lysis, and regulation and modification. Moreover, high similarity of putative proteins encoded in the genomes of lytic Leuconostoc phages suggests that they originated from a common ancestor (Kot et al., 2014). Ali et al. (2013) reported two groups of hybridization patterns among lytic Leuconostoc phages of dairy origin; one for Ln. mesenteroides phages and one for Ln. pseudomesenteroides phages, although all phage members of the two groups share a short, cross-hybridizing genome region. The cross-hybridizing region codes for tail proteins, e.g., major tail protein (mtp) and tape measure protein (tmp), however higher similarities were found within mtp gene. This conserved region was used as target for developing a universal PCR-based detection system for lytic phages of Ln. mesenteroides and Ln. pseudomesenteroides (Ali et al., 2013). The PCR assay resulted in 322-bp long fragments and was validated with all reported 83 lytic phages of Leuconostoc (Ali et al., 2013). The recent sequencing data confirms that the selected region is indeed the only region that can be used for PCR-based detection for both phage species (Kot et al., 2014).
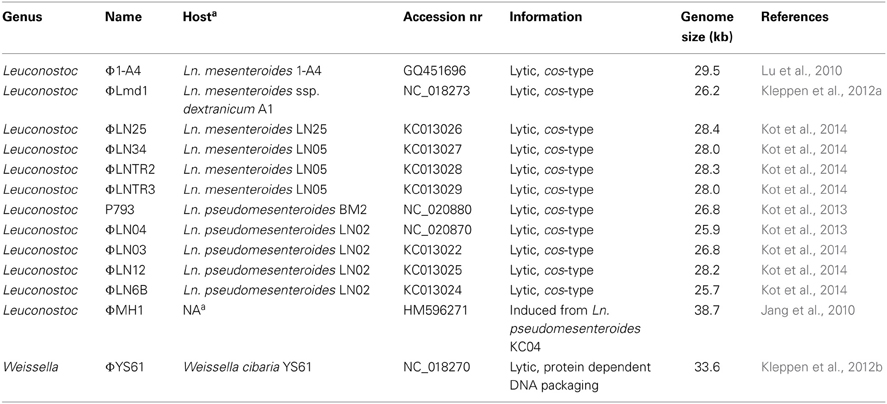
Table 2. Bacteriophages of Leuconostoc and Weissella with complete genome sequences deposited in public databases.
Genomes of lytic phages of Leuconostoc contain from 38 to 50 predicted genes. Some of us were involved in biological characterization of one of the genes present in Ln. pseudomesenteroides phages, namely the receptor binding protein (RBP) (Kot et al., 2013). Construction of chimeric phages resulted in the transition in host range allowing the identification of the receptor binding protein genes to be ORF21P793 and ORF23Φ LN04, respectively. Until now, the host-encoded receptor for Leuconostoc phages remains unknown.
Currently, there is only one complete genome sequence of a temperate phage attacking Leuconostoc deposited in public databases. The phage is designated ΦMH1 and it was obtained from a UV-induced lysate of Ln. pseudomesenteroides strain KC04 (Jang et al., 2010). No host for ΦMH1 phage was reported. ΦMH1 has a dsDNA genome with a length of 38.7 kb with 65 putative ORFs identified. ΦMH1 did not show significant similarities with other described phages of Leuconostoc (Jang et al., 2010). Besides of ΦMH1 phages, several predicted prophages can be identified in the sequenced genomes of Leuconostoc (Table 3). Analysis of complete translatome of fully sequenced phages and prophages shows that diversity of prophage elements is higher than within sequenced two groups of lytic phages (Figure 3).
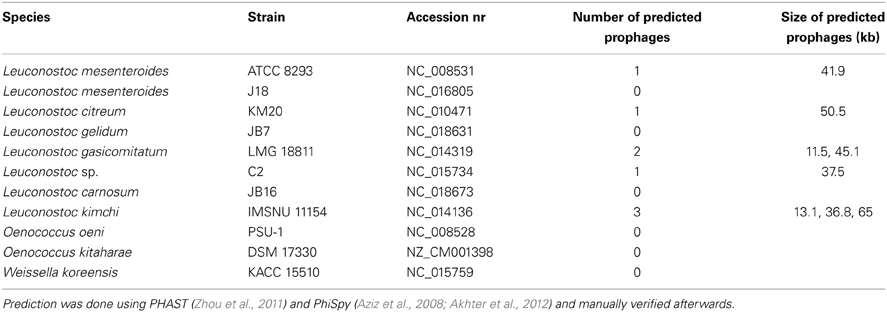
Table 3. Predicted prophage sequences found in fully assembled chromosomes of Leuconostoc, Oenococcus, and Weissella available in GenBank.
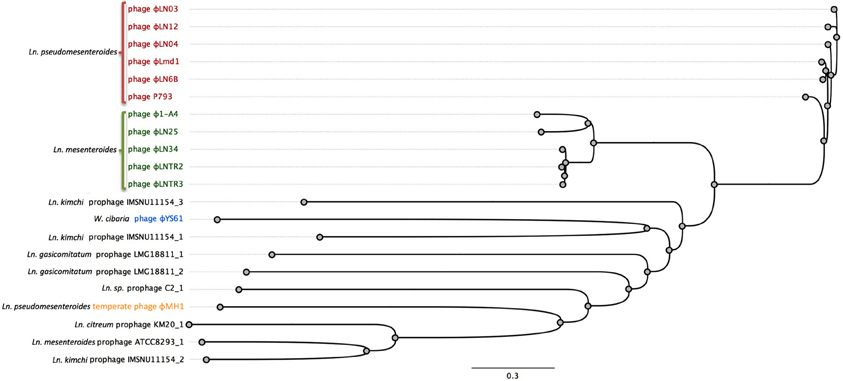
Figure 3. Comparison of the complete translatome of fully sequenced phages and predicted prophages from complete genomes of Leuconostoc, Weissella, and Oenococcus. Genomes were prepared as described before (Mahony et al., 2013). The concatemeric sequences were aligned using Geneious 7.1.2 software using BLOSUM62 cost matrix. The phylogenetic tree was constructed and visualized with bootstrapped neighbor-joining method with 500 replicates in Geneious 7.1.2 software.
Oenococcus Phages
Phages attacking O. oeni were reported already in the late 1960's and beginning of 1970's (Sozzi et al., 1976), and 3 phage morphotypes were described for phages isolated from wine. They all had isometric heads and 3 distinct non-contractile tail lengths, i.e., belonging to Siphoviridae. Later, lytic phages from four Australian wine areas attacking approximately 40% of O. oeni isolates from the same regions were isolated (Davis et al., 1985). These also were belonging to Siphoviridae with isometric heads and long non-contractile tails (approximately 300 nm). Subsequently, phage P58I was isolated from a phage carrying culture of O. oeni 58N (Arendt et al., 1990). This phage was able to plaque on strain 58N as well as on 58PF, which was a phage-free derivative of 58N strain. A similar phage P58II isolated after mitomycin C induction of the 58N strain was not able to plaque on any of the two strains. Surprisingly no DNA homology was detected between the two phages genomes and the chromosome of O. oeni 58N.
Huang et al. isolated a temperate phage Φ1002 that was able to grow lytically on approximately 46% of all O. oeni isolates from Australian wine (Huang et al., 1996). The phage belonged to the Siphoviridae family with a 52 nm isometric head and a 210 nm non-contractile tail. A set of 17 prophages were induced from O. oeni isolated from Portuguese wines (Santos et al., 1998). They all had a similar morphology with isometric heads of approximately 40–50 nm and non-contractile tails of approximately 220–240 nm. The cos-type phages were divided into 6 groups based on restriction enzyme digestion profiles. These could further be divided into 2 main groups α and β based on restriction maps (Santos et al., 1998). Cross-hybridization between the α and β group was located in the central part of the genomes and included the phage attachment site (attP). This part was later sequenced (Parreira et al., 1999) and revealed the presence of lysin and holin genes.
The lysin (Lys44) from O. oeni phage fOg44 was described in greater detail. Interestingly, secretion of the lysin seems to occur with the aid of a signal peptide and independent of the holin, also encoded in the phage genome. A potential role of the holin as a triggering factor for lytic activity is discussed (São-José et al., 2000).
Screening of 167 isolates of O. oeni for lysogeny by mitomycin C resulted in the identification of approximately 45% of lysogenic strains and for some of these propagating hosts were identified as indicator strains (Poblet-Icart et al., 1998).
Until now, there is no complete genome sequence of a phage attacking O. oeni, however a number of partial sequences derived from phages of O. oeni have been deposited in public databases. Borneman et al. (2012) reported several prophage sequences in the O. oeni pan genome. Prophage-like sequences were integrated into six different tRNA genes, with some of these sequences representing presumably functional phages (Borneman et al., 2012). Recently, Doria et al. (2013) communicated a PCR-based method for detection and identification of lysogenic strains of O. oeni. The assay allowed detection of a target sequence within the prophage lysin gene in 25 out of 37 isolates tested. Furthermore, the majority of the lysogenic isolates could be prophage induced (Doria et al., 2013). Shortly after, Jaomanjaka et al. (2013) analyzed oenococcal prophages based on integrase gene polymorphism and classified them into four groups (A–D). Remarkably, in the two fully assembled chromosomes of Oenococcus sp. no prophage sequences were detected using PHAST and PhiSpy program (Aziz et al., 2008; Zhou et al., 2011; Akhter et al., 2012) (Table 3). Absence of prophage-like sequences in the O. oeni PSU-1 strain had been reported before by Mills et al. (2005).
Weissella Phages
Pringsulaka et al. (2011) isolated phage Φ22 attacking Weissella cibaria N22 from a Thai fermented pork sausage Nham. This phage belonged to the Podoviridae family with morphotype C2 with a prolate head of approximately 92 × 50 nm and a non-contractile tail of 37 nm. Phage Φ22 had a narrow host-range attacking only one of 40 W. cibaria strains.
Lu et al. (2012) also isolated phages attacking W. cibaria from the initial phase of cucumber fermentation. Interestingly, the host range of some of these phages crossed the species barrier and in some cases also the genus barrier. Phage Φ3.8.18 belonging to the Myoviridae family attacked two isolates of W. cibaria, one isolate of Lb. plantarum and one isolate of Lb. brevis. Phage Φ3.8.18 had an isometric head of approximately 80 nm and an approximately 200 nm tail with indication of a baseplate structure. Another Myoviridae phage Φ7.2.50 attacked same two isolates of W. cibaria and 24 isolates of Lb. brevis. Also two Siphoviridae phages crossed the species/genus barrier. Phage Φ3.8.43 attacked, beside four W. cibaria isolates, one isolate of Lb. plantarum, and one isolate of Lb. brevis (both of which were also attacked by Φ3.8.18). Phage Φ3.8.43 had an isometric head of approximately 50–60 nm and an approximately 250 nm long non-contractile tail. Besides two isolates of W. cibaria, phage Φ3.8.48 also attacked one isolate of W. paramesenteroides. Kleppen et al. (2012b) determined the genome sequence of ΦYS61 attacking W. cibaria (Table 2). This phage isolated from 1-week old kimchi fermentation belonged to the Podoviridae family of morphotype C2 (Ackermann, 1998) with a prolate head of 85 × 36 nm and a short non-contractile tail.
Phage ΦYS61 is infecting W. cibaria (Kleppen et al., 2012b) and has a 33.6 kb dsDNA genome, which is similar to the estimated genome size of another podovirus of W. cibaria isolated recently from a Thai sausage (Pringsulaka et al., 2011). The genome of the ΦYS61 phage codes for 48 putative ORFs. It is very likely that ΦYS61 utilizes a protein-dependent DNA replication mechanism similarly to Φ29 phage from Bacillus subtilis (Kleppen et al., 2012b). Very few putative genes of ΦYS61 show significant similarities to the sequences present in public databases. No prophages were detected in W. koreensis KACC 15510 strain (Table 3).
Conclusion
Phages of Leuconostoc, Oenococcus, and Weissella are present in many types of food-related fermentations, where they are responsible for various defects in production. The majority of described phages were isolated from dairy samples, where they attack Leuconostoc starter strains and subsequently contribute to aroma- and CO2-production defects. Another large reservoir of Leuconostoc and Weissella phages are various vegetable fermentations, most importantly kimchi and sauerkraut fermentations. All phages of Oenococcus described so far are solely reported in connection to wine production, where they can disturb the malolactic fermentation.
All phages of Leuconostoc, Oenococcus, and Weissella belong to Caudovirales order with members of the Siphoviridae, Podoviridae, and Myoviridae families. Thirteen complete genomes of phages infecting Leuconostoc and Weissella have been reported. Among them, lytic phages of Leuconostoc belonging to Siphoviridae exhibit high similarities in overall composition, regardless on the environment they were isolated from. PCR-based assays for detecting lytic Leuconostoc and Oenococcus phages have been established so far, however further detailed knowledge of the genetic diversity of Leuconostoc, Oenococcus, and Weissella phages, e.g., Myoviridae phages from sauerkraut fermentations as well as temperate phages is needed in order to provide better taxonomy, control, and detection strategies for these groups of phages.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Witold Kot was the recipient of a Ph.D. grant from University of Copenhagen (UoC). The work on dairy Leuconostoc phages done in our own labs at UoC and at Max Rubner-Institut (MRI) was partially financed by MetaPhageLAB (UoC: FTP project no. 11-106991) and was supported by a grant from the Forschungskreis der Ernährungsindustrie e.V. (Bonn, Germany), the AiF, and the Ministry of Economics and Technology (MRI: project no. AiF-FV 15886N). MRI authors sincerely acknowledge the cooperation with University of Hohenheim (J. Hinrichs and Z. Atamer) within project AiF-FV 15886N.
References
Ackermann, H. W. (1998). Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51, 135–201. doi: 10.1016/S0065-3527(08)60785-X
Akhter, S., Aziz, R. K., and Edwards, R. A. (2012). PhiSpy: a novel algorithm for finding prophages in bacterial genomes that combines similarity- and composition-based strategies. Nucleic Acids Res. 40, 126. doi: 10.1093/nar/gks406
Ali, Y., Kot, W., Atamer, Z., Hinrichs, J., Vogensen, F. K., Heller, K. J., et al. (2013). Classification of lytic bacteriophages attacking dairy Leuconostoc starter strains. Appl. Environ. Microbiol. 79, 3628–3636. doi: 10.1128/AEM.00076-13
Arendt, E. K., Lonvaud, A., and Hammes, W. P. (1991). Lysogeny in Leuconostoc oenos. J. Gen. Microbiol. 137, 2135–2139. doi: 10.1099/00221287-137-9-2135
Arendt, E. K., Neve, H., and Hammes, W. P. (1990). Characterization of phage isolates from a phage-carrying culture of Leuconostoc oenos 58n. Appl. Microbiol. Biotechnol. 34, 220–224. doi: 10.1007/BF00166784
Atamer, Z., Ali, Y., Neve, H., Heller, K. J., and Hinrich, J. (2011). Thermal resistance of bacteriophages attacking flavour-producing dairy Leuconostoc starter cultures. Int. Dairy J. 21, 327–334. doi: 10.1016/j.idairyj.2010.11.005
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Barrangou, R., Yoon, S.-S., Breidt, F. Jr., Flemming, H. P., and Klaenhammer, T. R. (2002). Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 68, 5452–5458. doi: 10.1128/AEM.68.11.5452-5458.2002
Björkroth, J., and Holzapfel, W. (2006). “Genera Leuconostoc, Oenococcus and Weissella,” in The Prokaryotes, 3rd Edn. eds M. Dworkin, S. Falkow, E. Rosenberg, K. Schleifer, and E. Stackebrandt (New York, NY: Springer Science + Business Media), 267–319.
Boizet, B., Mata, M., Mignot, O., Ritzenthaler, P., and Sozzi, T. (1992). Taxonomic characterization of Leuconostoc mesenteroides and Leuconostoc oenos bacteriophage. FEMS Microbiol. Lett. 90, 211–216. doi: 10.1111/j.1574-6968.1992.tb05154.x
Borneman, A. R., McCarthy, J. M., Chambers, P. J., and Bartowsky, E. J. (2012). Comparative analysis of the Oenococcus oeni pan genome reveals genetic diversity in industrially-relevant pathways. BMC Genomics 13:373. doi: 10.1186/1471-2164-13-373
Collins, M. D., Samelis, J., Metaxopoulos, J., and Wallbanks, S. (1993). Taxonomic studies on some Leuconostoc−like organisms from fermented sausages: description of a new genus Weissella for the Leuconostoc paramesenteroides group of species. J. Appl. Microbiol. 75, 595–603.
Davey, G. P., Ward, L. J. H., and Brown, J. C. S. (1995). Characterisation of four Leuconostoc bacteriophages isolated from dairy fermentations. FEMS Microbiol. Lett. 128, 21–25. doi: 10.1111/j.1574-6968.1995.tb07494.x
Davis, C., Davis, C., Silveira, N. F., Silveira, N. F., Fleet, G. H., and Fleet, G. H. (1985). Occurrence and properties of bacteriophages of Leuconostoc oenos in Australian wines. Appl. Environ. Microbiol. 50, 872–876.
Dicks, L., Dellaglio, F., and Collins, M. D. (1995). Proposal to reclassify Leuconostoc oenos as Oenococcus oeni [corrig.] gen. nov., comb. nov. Int. J. Syst. Bacteriol. 45, 395–397. doi: 10.1099/00207713-45-2-395
Doria, F., Napoli, C., Costantini, A., Berta, G., Saiz, J. C., and Garcia-Moruno, E. (2013). Development of a new method for detection and identification of Oenococcus oeni bacteriophages based on endolysin gene sequence and randomly amplified polymorphic DNA. Appl. Environ. Microbiol. 79, 4799–4805. doi: 10.1128/AEM.01307-13
Endo, A., Irisawa, T., Futagawa-Endo, Y., Sonomoto, K., Itoh, K., Takano, K., et al. (2011). Fructobacillus tropaeoli sp. nov., a fructophilic lactic acid bacterium isolated from a flower. Int. J. Syst. Bacteriol. 61, 898–902. doi: 10.1099/ijs.0.023838-0
Endo, A., and Okada, S. (2006). Oenococcus kitaharae sp. nov., a non-acidophilic and non-malolactic-fermenting oenococcus isolated from a composting distilled shochu residue. Int. J. Syst. Bacteriol. 56, 2345–2348. doi: 10.1099/ijs.0.64288-0
Endo, A., and Okada, S. (2008). Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov., Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int. J. Syst. Bacteriol. 58, 2195–2205. doi: 10.1099/ijs.0.65609-0
Farkye, N. Y. and Vedamuthu, E. R. (2002). “Microbiology of soft cheeses,” in Dairy Microbiology Handbook, 3rd Edn, ed R. K. Robinson (New York, NY: Wiley Interscience) 479–513.
Farrow, J. A. E., Facklam, R. R., and Collins, M. D. (1989). Nucleic acid homologies of some vancomycin-resistant Leuconostocs and description of Leuconostoc citreum sp. nov and Leuconostoc pseudomesenteroides sp. nov. Int. J. Syst. Bacteriol. 39, 279–283. doi: 10.1099/00207713-39-3-279
Greer, G. G., Dilts, B. D., and Ackermann, H. W. (2007). Characterization of a Leuconostoc gelidum bacteriophage from pork. Int. J. Food Microbiol. 114, 370–375. doi: 10.1016/j.ijfoodmicro.2006.09.021
Huang, C. M., Asmundson, R. V., and Kelly, W. J. (1996). Characterization of a temperate phage isolated from Leuconostoc oenos strain 1002. Appl. Microbiol. Biotech. 45, 472–476. doi: 10.1007/s002530050715
Jang, S. H., Hwang, M. H., and Chang, H.-I. (2010). Complete genome sequence of ΦMH1, a Leuconostoc temperate phage. Arch. Virol. 155, 1883–1885. doi: 10.1007/s00705-010-0799-5
Jaomanjaka, F., Ballestra, P., Dols-Lafargue, M., and Le Marrec, C. (2013). Expanding the diversity of oenococcal bacteriophages: insights into a novel group based on the integrase sequence. Int. J. Food Microbiol. 166, 331–340. doi: 10.1016/j.ijfoodmicro.2013.06.032
Johansen, E., and Kibenich, A. (1992). Characterization of Leuconostoc isolates from commercial mixed strain mesophilic starter cultures. J. Dairy Sci. 75, 1186–1191. doi: 10.3168/jds.S0022-0302(92)77865-5
Jung, J. Y., Lee, S. H., Kim, J. M., Park, M. S., Bae, J. W., Hahn, Y., et al. (2011). Metagenomic analysis of kimchi, a traditional Korean fermented food. Appl. Environ. Microbiol. 77, 2264–2274. doi: 10.1128/AEM.02157-10
Kleppen, H. P., Holo, H., Jeon, S. R., Nes, I. F., and Yoon, S. S. (2012a). Characterization of a Leuconostoc bacteriophage infecting flavor producers of cheese starter cultures. Appl. Environ. Microbiol. 78, 6769–6772. doi: 10.1128/AEM.00562-12
Kleppen, H. P., Holo, H., Jeon, S.-R., Nes, I. F., and Yoon, S.-S. (2012b). Novel Podoviridae family bacteriophage infecting Weissella cibaria isolated from kimchi. Appl. Environ. Microbiol. 78, 7299–7308. doi: 10.1128/AEM.00031-12
Kot, W., Hammer, K., Neve, H., and Vogensen, F. K. (2013). Identification of the receptor-binding protein in lytic Leuconostoc pseudomesenteroides bacteriophages. Appl. Environ. Microbiol. 79, 3311–3314. doi: 10.1128/AEM.00012-13
Kot, W., Hansen, L. H., Neve, H., Hammer, K., Jacobsen, S., Pedersen, P. D., et al. (2014). Sequence and comparative analysis of Leuconostoc dairy bacteriophages. Int. J. Food Microbiol. 176, 29–37. doi: 10.1016/j.ijfoodmicro.2014.01.019
Leiva-Quiros, A., and McCleskey, C. S. (1947). The application of bacteriophage and serology in the differentiation of strains of Leuconostoc mesenteroides. J. Bacteriol. 54, 709–713.
Lu, Z., Altermann, E., Breidt, F., and Kozyavkin, S. (2010). Sequence analysis of Leuconostoc mesenteroides bacteriophage 1-A4 Isolated from an industrial vegetable fermentation. Appl. Environ. Microbiol. 76, 1955–1966. doi: 10.1128/AEM.02126-09
Lu, Z., Breidt, F., Plengvidhya, V., and Fleming, H. P. (2003). Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 69, 3192–3202. doi: 10.1128/AEM.69.6.3192-3202.2003
Lu, Z., Perez-Diaz, I. M., Hayes, J. S., and Breidt, F. (2012). Bacteriophage ecology in a commercial cucumber fermentation. Appl. Environ. Microbiol. 78, 8571–8578. doi: 10.1128/AEM.01914-12
Mahony, J., Martel, B., Tremblay, D. M., Neve, H., Heller, K. J., Moineau, S., et al. (2013). Identification of a new P335 subgroup through molecular analysis of lactococcal phages Q33 and BM13. Appl. Environ. Microbiol. 79, 4401–4409. doi: 10.1128/AEM.00832-13
Mills, D. A., Rawsthorne, H., Parker, C., Tamir, D., and Makarova, K. (2005). Genomic analysis of Oenococcus oeni PSU-1 and its relevance to winemaking. FEMS Microbiol. Rev. 29, 465–475. doi: 10.1016/j.femsre.2005.04.011
Mosimann, W., and Ritter, W. (1946). Bacteriophages as cause of loss of aroma in butter cultures (Bakteriophagen als Ursache von Aromaschwund in Rahmsäuerungskulturen). Schweizerische Milchzeitung 72, 211–212.
Mudgal, P., Breidt, F., Lubkin, S. R., and Sandeep, K. P. (2006). Quantifying the significance of phage attack on starter cultures: a mechanistic model for population dynamics of phage and their hosts isolated from fermenting sauerkraut. Appl. Environ. Microbiol. 72, 3908–3915. doi: 10.1128/AEM.02429-05
Neve, H., Lilischkis, R., and Teuber, M. (1988). Characterisation of a virulent bacteriophage of Leuconostoc mesenteroides subsp. cremoris. Kiel. Milchwirtschaftl. Forschungsber. 40, 205–215.
Parreira, R., Saxelin, M.-L., São-José, C., Nurmiaho-Lassila, E.-L., Isidro, A., Meriläinen, V. T., et al. (1999). Gene organization in a central DNA fragment of Oenococcus oeni bacteriophage fOg44 encoding lytic, integrative and non-essential functions. Gene 226, 83–93. doi: 10.1016/S0378-1119(98)00554-X
Poblet-Icart, M., Bordons, A., and Lonvaud-Funel, A. (1998). Lysogeny of Oenococcus oeni (syn. Leuconostoc oenos) and study of their induced bacteriophages. Curr. Microbiol. 36, 365–369. doi: 10.1007/s002849900324
Pringsulaka, O., Patarasinpaiboon, N., Suwannasai, N., Atthakor, W., and Rangsiruji, A. (2011). Isolation and characterisation of a novel Podoviridae-phage infecting Weissella cibaria N 22 from Nham, a Thai fermented pork sausage. Food Microbiol. 28, 518–525. doi: 10.1016/j.fm.2010.10.011
Pujato, S. A., Guglielmotti, D. M., Ackermann, H. W., Patrignani, F., Lanciotti, R., Reinheimer, J. A., et al. (2014). Leuconostoc bacteriophages from blue cheese manufacture: long-term survival, resistance to thermal treatments, high pressure homogenization and chemical biocides of industrial application. Int. J. Food Microbiol. 177, 1–8. doi: 10.1016/j.ijfoodmicro.2014.02.012
Sakala, R. M., Hayashidani, H., Kato, Y., Hirata, T., Makino, Y., Fukushima, A., et al. (2002). Change in the composition of microflora on vacuum-packaged beef durin chiller storage. Int. J. Food Microbiol. 74, 87–99. doi: 10.1016/S0168-1605(01)00732-2
Samson, J. E., and Moineau, S. (2013). Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 4, 347–368. doi: 10.1146/annurev-food-030212-182541
Samtlebe, M., Neve, H., Heller, K. J., Hinrichs, J., and Atamer, Z. (2012). Leuconostoc bacteriophages and their effects on the organoleptic properties of fermented milk products. Eur. Dairy Mag. 5, 20–23.
Santos, R., São-José, C., Vieira, G., Paveia, H., and Santos, M. A. (1998). Genome diversity in temperate bacteriophages of Oenococcus oeni. Arch. Virol. 143, 523–536. doi: 10.1007/s007050050308
São-José, C., Parreira, R., Vieira, G., and Santos, M. A. (2000). The N-Terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on Oenococcal cells. J. Bacteriol. 182, 5823–5831. doi: 10.1128/JB.182.20.5823-5831.2000
Saxelin, M.-L., Nurmiaho-Lassila, E.-L., Meriläinen, V., and Forsén, R. (1986). Ultrastructure and host specificity of bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and Leuconostoc cremoris from Finnish fermented milk viili. Appl. Environ. Microbiol. 52, 771–777.
Schleifer, K. (2009). “Family V. Leuconostocaceae fam. nov,” in Bergey's Manual of Systematic Bacteriology, Vol. 3, 2nd Edn, (The Firmicutes), eds P. De Vos, G. Garrity, J. Jones, N. R. Krieg, W. Ludwig, F. A. Rainey, K. Schleifer, and W. B. Whitman (New York; Dordrecht; Heidelberg; London: Springer), 624.
Settanni, L., Di Grigoli, A., Tornambé, G., Bellina, V., Francesca, N., Moschetti, G., et al. (2012). Persistence of wild Streptococcus thermophilus strains on wooden vat and during the manufacture of a traditional Caciocavallo type cheese. Int. J. Food Microbiol. 155, 73–81. doi: 10.1016/j.ijfoodmicro.2012.01.022
Shin, C. (1983). Some characteristics of Leuconostoc cremoris bacteriophage isolated from blue cheese. Jpn. J. Zootech. Sci. 54, 481–486.
Shin, C., and Sato, Y. (1979). Isolation of Leuconostoc bacteriophages from dairy products. Jpn. J. Zootech. Sci. 50, 419–422. doi: 10.2508/chikusan.50.419
Snauwaert, I., Papalexandratou, Z., De Vuyst, L., and Vandamme, P. (2013). Characterization of strains of Weissella fabalis sp nov and Fructobacillus tropaeoli from spontaneous cocoa bean fermentations. Int. J. Syst. Evol. Microbiol. 63, 1709–1716. doi: 10.1099/ijs.0.040311-0
Sozzi, T., D'Amico, N., Hose, H., and Gnaegi, F. (1982). Difficultes de fermentation malolactique du vin dues a des bacteriophage de Leuconostoc oenos. Revue Suisse de Viticulture 32, 568–569.
Sozzi, T., Maret, R., and Poulin, J. M. (1976). Observation of bacteriophages in wine. Experientia 32, 568–569. doi: 10.1007/BF01990165
Sozzi, T., Poulin, J. M., Maret, R., and Pousaz, R. (1978). Isolation of a bacteriophage of Leuconostoc mesenteroides from dairy products. J. Appl. Microbiol. 44, 159–161. doi: 10.1111/j.1365-2672.1978.tb00787.x
Yoon, S. S., Barrangou-Poueys, R., Breidt, F., Klaenhammer, T. R., and Fleming, H. P. (2002). Isolation and characterization of bacteriophages from fermenting sauerkraut. Appl. Environ. Microbiol. 68, 973–976. doi: 10.1128/AEM.68.2.973-976.2002
Keywords: bacteriophages, Leuconostoc, Oenococcus, Weissella, morphogenesis, DNA sequence analysis
Citation: Kot W, Neve H, Heller KJ and Vogensen FK (2014) Bacteriophages of Leuconostoc, Oenococcus, and Weissella. Front. Microbiol. 5:186. doi: 10.3389/fmicb.2014.00186
Received: 27 December 2013; Accepted: 04 April 2014;
Published online: 28 April 2014.
Edited by:
Jennifer Mahony, University College Cork, IrelandReviewed by:
Eric Altermann, AgResearch Ltd, New ZealandBeatriz Martínez, Consejo Superior de Investigaciones Científicas, Spain
Copyright © 2014 Kot, Neve, Heller and Vogensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Finn K. Vogensen, Department of Food Science, Faculty of Science, University of Copenhagen, Rolighedsvej 30, DK-1958 Frederiksberg, Denmark e-mail:Zmt2QGZvb2Qua3UuZGs=
 Witold Kot
Witold Kot Horst Neve
Horst Neve Knut J. Heller
Knut J. Heller Finn K. Vogensen
Finn K. Vogensen