- 1Plant Systems Engineering Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, South Korea
- 2Department of Biosystems and Bioengineering, University of Science and Technology, Daejeon, South Korea
Viruses are obligate intracellular parasites that have small genomes with limited coding capacity; therefore, they extensively use host intracellular machinery for their replication and infection in host cells. In recent years, it was elucidated that plants have evolved intricate defense mechanisms to prevent or limit damage from such pathogens. Plants employ two major strategies to counteract virus infections: resistance (R) gene-mediated and RNA silencing-based defenses. In this review, plant defenses and viral counter defenses are described, as are recent studies examining the cross-talk between different plant defense mechanisms.
Introduction
Plant viruses comprise an important group of pathogens causing a range of plant diseases that are often responsible for significant losses in crop production. Among the wide range of known plant viruses, most viruses have a very limited host range and only a few viruses cause severe disease symptoms (Dawson and Hilf, 1992). Even though viruses contain relatively simple genomes, the molecular basis of the mechanisms by which plant viruses infect their hosts and the signaling components involved in host resistance are not well defined.
The immune response against bacterial or fungal pathogens often relies on recognition of the conserved molecules associated with a group of pathogens, designated pathogen-associated molecular patterns (PAMPs), by pattern recognition receptors (PRRs) (Boller and He, 2009). Upon PAMP recognition, activated PRRs induce PAMP-triggered immunity (PTI) (Monaghan and Zipfel, 2012). PTI against viral pathogens has been primarily described in mammalian cells, but not in plant cells (Calil and Fontes, 2016). However, several recent studies provided evidence that PTI and related components are also involved in antiviral defense responses in plants (Korner et al., 2013; Nicaise, 2014; Iriti and Varoni, 2015; Calil and Fontes, 2016; Nicaise and Candresse, 2016; Niehl et al., 2016). In general, plants defense responses triggered against viral pathogens are based on RNA- or protein-mediated resistance. The RNA-mediated resistance response is a basal defense response to viral invasion that mainly involves the RNA silencing pathway of the host, which mediates the cleavage of viral RNA. Compared to this basal defense response, the host resistance (R) protein-mediated defense response against viral pathogens is far more robust, in most cases limiting viral replication and spread to inoculated leaves (Zhou and Chai, 2008; Verlaan et al., 2013; Nakahara and Masuta, 2014). In this review, we summarize molecular mechanisms underlying two major defense pathways employed during plant resistance to viral pathogens and highlight a few studies addressing the cross-talk between these defense pathways.
RNA Silencing in Viral Defense
RNA gene silencing, also termed RNA interference (RNAi), which acts as a basal defense mechanism against viruses, is one of the main plant immune responses against viral pathogens (Vaucheret, 2006; Ding and Voinnet, 2007). Most viruses that cause disease in plants have RNA genomes containing imperfect regulatory stem-loops, which are copied into complementary double-stranded RNA (dsRNA) replication intermediates by virus-encoded RNA-dependent RNA polymerases (RDRs) (Ruiz-Ferrer and Voinnet, 2009). The dsRNAs are then recognized by a host ribonuclease III-like protein, namely, Dicer-like (DCL), and then processed into 21–24-nucleotide short interfering RNAs (siRNAs). The siRNAs are recruited to a functional RNA-induced silencing complex (RISC) and then act as guides to direct RISC to their target viral RNA molecules, which have complementary sequences (Ruiz-Ferrer and Voinnet, 2009). Consequently, viral RNAs are degraded by the core components of RISC, which are members of the Argonaut (AGO) protein family (Vaucheret, 2008). The antiviral RNAi response is effective in various species (Katiyar-Agarwal and Jin, 2010), even though it is slow and thus viral infections are often not completely cleared.
The concept of PTI against viral pathogens is currently confined to animals because receptors that sense RNA or DNA viruses as ligands have only been identified in animals (Takeuchi and Akira, 2009). In plants, dsRNAs produced during virus infection are also regarded as viral PAMPs (Ding, 2010; Jensen and Thomsen, 2012). The RNA silencing pathway was assumed to play a role in the immune responses that recognize such dsRNAs in plants, unlike in animals (Ding and Voinnet, 2007). However, a few recent publications indicate that the known PTI components are involved in dsRNA recognition and that the reaction is an immune response distinct from the RNA silencing pathway. Therefore, these studies claim that PTI against viral pathogens is preserved in plants and animals (Korner et al., 2013; Nicaise, 2014; Nicaise and Candresse, 2016; Niehl et al., 2016). However, there is no direct evidence to explain how dsRNAs are recognized in plants; therefore, further studies are needed to determine whether an animal-like mechanism underlies dsRNA-mediated PTI in plants.
To overcome RNAi-mediated host defense, plant viruses frequently encode viral suppressors of RNAi (VSRs) that perturb the plant RNA silencing pathway (Ding and Voinnet, 2007). VSRs have been isolated from nearly all plant virus families. In addition to suppressing RNAi silencing during viral pathogenesis, most VSRs identified to date play important roles in replication, assembly, or movement of viruses. Although the primary sequences and structures of these VSR proteins vary considerably, most VSR-mediated suppression is thought to occur via two general mechanisms (Figure 1). Some VSRs, such as potyviral HcPro, Beet Yellow Virus P21 protein, Peanut Clump Virus P15 protein, and TCV coat protein (CP or P38), sequester small RNA duplexes by binding to short or long viral dsRNAs, which then leads to impaired assembly of AGOs into RISCs (Lakatos et al., 2006; Carbonell and Carrington, 2015). Alternatively, some VSRs impede the activity of AGO proteins that have a central role in the anti-viral RNA silencing pathway (Carbonell and Carrington, 2015). For example, Cucumber Mosaic Virus 2b protein suppresses RISC activity through a physical interaction with the PAZ domain of AGO1 (Duan et al., 2012). Similarly, two other viral VSR proteins, Sweet Potato Mild Mottle Virus (SMMV) P1 and TCV CP, also directly interact with AGO proteins through glycine/tryptophan (GW/WG) repeat motifs, which resemble the AGO1-binding peptides on RISC (Azevedo et al., 2010; Giner et al., 2010). These findings demonstrate that VSR suppression of RNAi silencing might involve independently evolved VSR proteins that show functional overlap (Carbonell and Carrington, 2015). Studies on VSRs will not only improve our understanding of plant–virus interactions, but they will also help elucidate the signaling mechanism underlying host RNA silencing pathways.
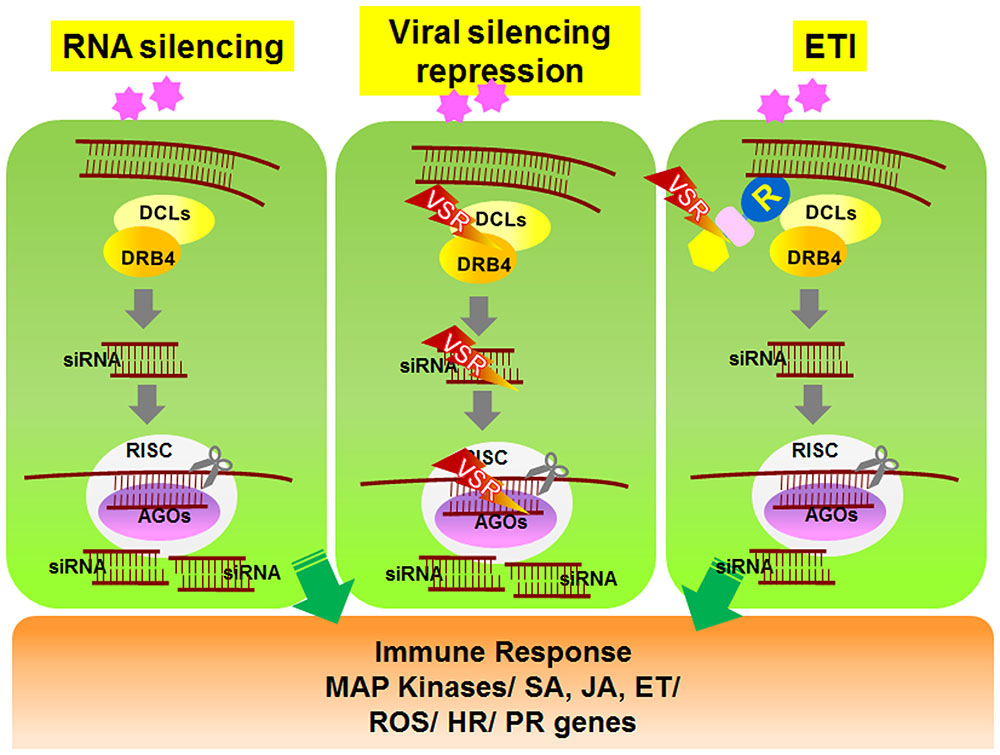
FIGURE 1. Schematic model of RNA silencing- and R-mediated responses in plant cells. Upon amplification of viruses in plant cells, viral double-stranded RNAs (dsRNAs) activate RNA silencing mechanisms. Viral dsRNAs are processed into small RNA fragments (siRNAs) by DCL1 and its cofactor DRB4. The siRNAs are recruited to RISC, which is associated with AGO protein. RISC/AGO/siRNA then targets and degrades complementary viral transcripts (left panel). Viruses express genes encoding VSR proteins that inhibit the regulation and activation of gene silencing mechanisms (center panel). In response, several R proteins recognize the VSRs and induce downstream ETI responses (right panel). DCL, Dicer-like; DRB, dsRNA-binding protein; siRNA, small interfering RNA; AGO, Argonaute; RISC, RNA-induced silencing complex; VSR, viral suppressors of RNA silencing.
Resistance Gene-Mediated Defense Responses Against Viral Pathogens
To circumvent PTI, pathogens produce effectors that suppress immune responses triggered by active PRRs (Deslandes and Rivas, 2012). The bacterial pathogens usually encode ∼20–30 highly regulated effectors that are secreted directly into the host cytoplasm. Although individual effectors from closely related bacterial strains exhibit functional diversity, they possess highly redundant activities and extensive interchangeability (Cunnac et al., 2009; Deslandes and Rivas, 2012). This also applies to viral proteins such as movement proteins (MPs), and replicase proteins, which act as avirulent (Avr) factors (Kachroo et al., 2006).
Resistance (R) genes have evolved in plants as a countermeasure to the effect of pathogen effectors on PTI (Jones and Dangl, 2006). R genes mediate effector-triggered immunity (ETI), which is a highly amplified version of PTI (Jones and Dangl, 2006). Many R genes have been identified, which confer resistance to diverse pathogens including bacteria, fungi, oomycetes, insects, and viruses (Jones and Dangl, 2006; Kachroo et al., 2006). Notable examples of R genes conferring resistance against viral pathogens include tobacco “N” against TMV, “Rx1/2” in potato against Potato Virus X (PVX), and “HRT” and “RCY1” against TCV and CMV in Arabidopsis, respectively (Whitham et al., 1994; Bendahmane et al., 1999, 2000; Cooley et al., 2000; Takahashi et al., 2001). The R genes are largely dominant, whereas some genes exhibit recessive, tolerance, or partial resistance characteristics. Moreover, dominant R genes HRT and RCY1 require recessive factors to confer resistance (Cooley et al., 2000; Takahashi et al., 2001). Since viruses require host factors for their infection (termed susceptibility factors), loss of these can also confer resistance to viral pathogens. Such resistance is often recessive (Truniger and Aranda, 2009). Notably, most such recessive R genes have been analyzed in potyviruses and encode translation initiation factors of the 4E or 4G family (eIF4E/eIF4G) (Kang et al., 2005; Truniger and Aranda, 2009). Interestingly, EF1A is required for Soybean Mosaic virus (SMV)-induced endoplasmic reticulum (ER) stress and, therefore, replication of SMV (Luan et al., 2016). Consequently, silencing of EF1A inhibits SMV replication and confers resistance against SMV.
The majority of dominant R proteins contain nucleotide-binding site (NBS) and leucine rich repeat (LRR) domains (Collier and Moffett, 2009), which is also the case for R genes that confer resistance against viral pathogens (de Ronde et al., 2014). The NBS-LRR R proteins can be further subcategorized as putative coiled-coil- or toll-interleukin-1 receptor-like (TIR)-type proteins based on the presence of these domains at their N-termini (Collier and Moffett, 2009). TIR, NBS, and LRR domains are also found in Drosophila and human receptor proteins involved in innate immunity (Nürnberger et al., 2004), suggesting that the animal and plant proteins evolutionarily diverged from a common ancestor and that and that similar modules were selected to regulate innate immune responses.
While only selected R proteins show direct interactions with Avr factors (Dodds et al., 2006; Ueda et al., 2006; Cesari et al., 2013), most R proteins are thought to act indirectly via other intermediary host proteins. This is further explained by the “guard/decoy” model, which describes how R proteins guard host accessory proteins (guardees), and pathogen effector-mediated alteration of the guardees results in the activation of R protein (Jones and Dangl, 2006; Collier and Moffett, 2009; Dodds and Rathjen, 2010). For example, N protein from tobacco indirectly recognizes a p50 helicase fragment of the TMV replicase protein via a chloroplast-localized N receptor-interacting protein 1 (NRIP1) (Caplan et al., 2008). Upon TMV infection, NRIP1 residing in the chloroplast translocates to the cytoplasm and nucleus. Cytosolic NRIP1 associates with TMV replicase and then recruits N protein through a direct interaction between NRIP1 and the TIR domain of N (Ueda et al., 2006).
Unlike viral pathogens, both fungal and bacterial genomes encode multiple Avr factors, which are thought to play a role in the suppression of PTI. However, viruses appear to compensate for the presence of a single Avr effector by undergoing frequent alterations in the critical amino acid sequences without drastically changing the protein structure. Host R protein-mediated recognition of the modified Avr factor then depends on the relative affinity between R protein and the modified Avr factor. For instance, several hypervirulent strains of TCV isolated from in planta-propagated TCV are able to escape HRT-mediated recognition and cause disease in resistant plants (Wobbe et al., 1998; Zhu et al., 2013).
Cross-Talk Between RNAi- and R Gene-Mediated Anti-Viral Defense Responses
Since both RNAi and R gene-mediated pathways participate in antiviral defense, it is plausible that these pathways undergo cross-talk to maximize the efficiency of defense responses against viral infections (Nakahara and Masuta, 2014). Indeed, viral pathogens often encode a single protein that functions as a suppressor of RNAi as well as an Avr effector (Figure 2) (Palanichelvam et al., 2000; Eggenberger et al., 2008; Katiyar-Agarwal and Jin, 2010; Wen et al., 2012; Zhu et al., 2013). For example, TMV replicase and TCV CP function as VSRs and are recognized by N and HRT, respectively, to induce the HR (Wang et al., 2012; Zhu et al., 2013). However, it is currently unclear how they communicate with each other and whether they assist each other to increase disease resistance or have sequential defense functions and thereby act individually.
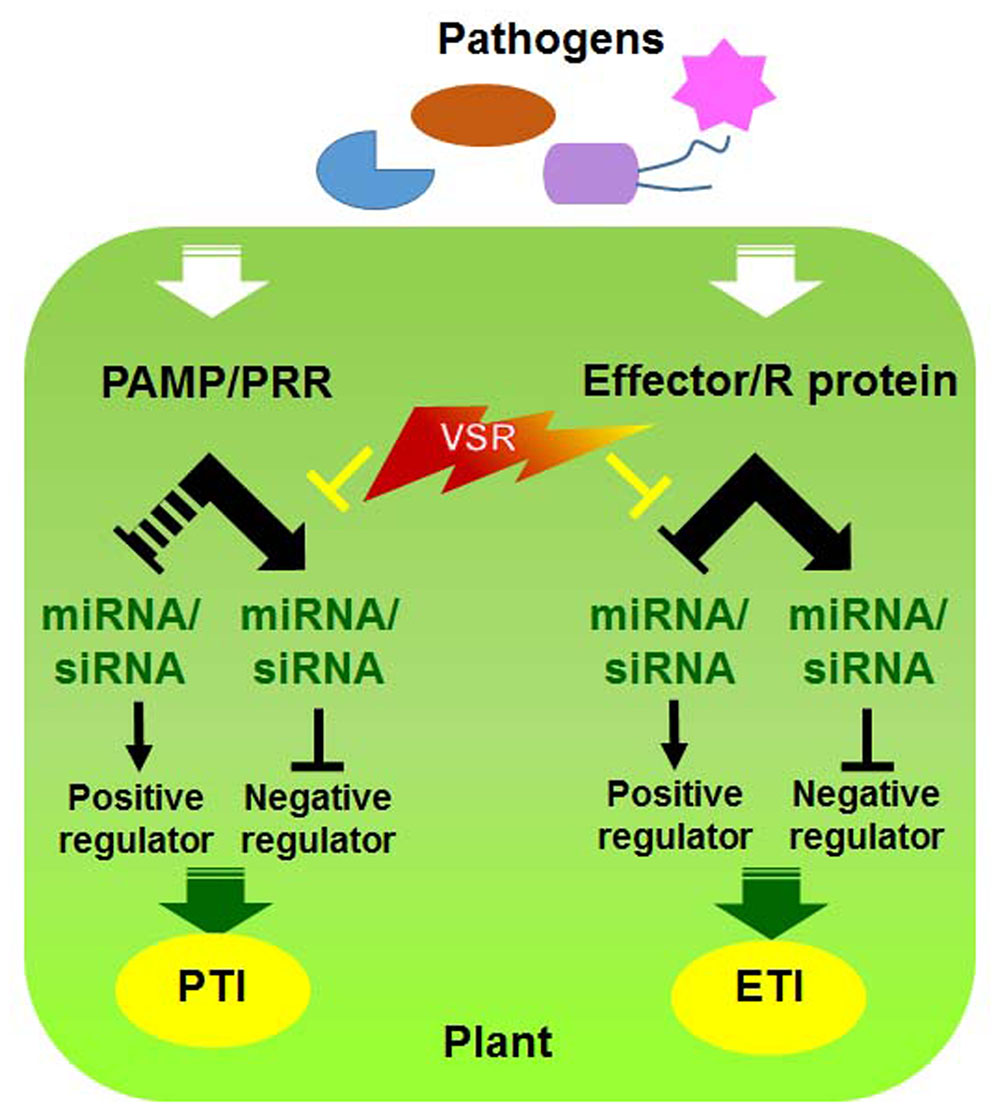
FIGURE 2. Interactions between viral silencing suppression and host factors involved in PTI and ETI. Model of molecular virus–host interactions in RNA silencing and PRR/R-mediated resistance [modified from (Katiyar-Agarwal and Jin, 2010)].
Recently, a few studies provided molecular evidence that these two defense mechanisms are associated with each other (Li et al., 2012; Shivaprasad et al., 2012; Verlaan et al., 2013; Zhu et al., 2013). Several components involved in host RNA silencing mechanisms have recently been shown to be required for R gene-mediated defense. For example, double-stranded RNA binding protein (DRB) four interacts with HRT and is required for HRT stability (Zhu et al., 2013). In addition, R genes against Tomato yellow leaf curl virus were recently shown to encode DFDGD-class RDR (Verlaan et al., 2013). Interestingly, activation of HRT-mediated resistance is not dependent on the RNA silencing suppressor activity of CP and is not associated with the accumulation of TCV-specific small RNA. This finding suggests that the HRT-mediated signaling pathway recruits components of the RNA silencing pathway, but this resistance response is not associated with the cleavage of viral RNA.
It is likely that alteration of small RNAs derived from viral infections plays a role in regulating R gene expression levels, thereby regulating resistance signaling (Li et al., 2012; Shivaprasad et al., 2012), rather than direct regulation by VSR activity. For instance, Li et al. (2012) observed that miR6019 and miR6020 in tobacco cause specific cleavage of transcripts of N and its homologs by binding to the complementary sequence of a conserved region encoding the TIR domain of the N protein. Furthermore, phasiRNA synthesis from the N coding sequence via overexpression of miR6019 was accompanied by reductions in N transcript levels and N-mediated resistance against TMV (Li et al., 2012). In addition, a group of 22 nt miRNAs from the miR482/2118 superfamily targets numerous NLRs within Solanaceae species. These miRNAs target highly conserved sequences in the genes encoding predicted NLR proteins (Zhai et al., 2011; Shivaprasad et al., 2012). Activation of VSR induces quantitative changes of whole small RNA species in host cells. Interestingly, VSRs upregulate the transcript levels of the targeted NLRs by attenuating the production or activity of miR482/2118 family members. The miR482/2118 family members are thought to ordinarily down-regulate their target NLR genes but upregulate these genes only when they are required for plant resistance via the VSRs of viral pathogens (Padmanabhan and Dinesh-Kumar, 2014). Altogether, these studies suggest that the RNA silencing response is integrated with R gene-mediated anti-viral resistance responses; however, it is not yet clear whether degradation of the viral genome via host RNA silencing-mediated defense is necessary for R gene-mediated defense.
Concluding Remarks
Since the zig-zag model was first proposed by Jones and Dangl (2006), many interactions between plant and bacterial pathogens have been reported, in which a pathogen suppresses or alters PTI by effectors, and plants have developed induced ETI, a stronger type of defense against effectors, during evolution (Boller and He, 2009). Long-term plant disease resistance studies of viral pathogens have revealed RNA silencing and R gene-mediated defense responses. In recent years, studies of the relationship between these two resistance responses have enhanced understanding of the interaction between plants and viruses. As genome analysis techniques are developed, understanding of plant–virus interactions increases. Kontra et al. (2016) recently reported that the tombusviral P19 suppressor preferentially affects virus-derived small interfering RNAs rather than endogenous host miRNAs in virus-infected plants. The authors suggested that the relationship between VSRs and host RNA silencing, as well as their contribution to the virulence of viruses, should be reconsidered. In parallel, Li et al. (2013) revealed a role for miRNAs in translational inhibition as well as silencing in plants and demonstrated that this process occurs in the ER. It would be interesting to integrate our knowledge of the roles of the ER in viral pathogenesis and in R gene-mediated defense responses (Jheng et al., 2014; Verchot, 2014; Moon et al., 2016). Uncovering the subcellular localization of small RNAs, VSR, and R protein will be critical for understanding how the two antiviral pathways interact. Although the concept of PTI and ETI is less clear in viral pathogenesis than in bacterial pathogenesis at present, future in-depth studies of the two anti-viral defenses and cross-talk between them will enhance understanding of plant immune responses, as well as to bacteria and fungi.
Author Contributions
JM wrote the paper. JP wrote and edited the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the KRIBB Research Initiative Program, Ministry of Science, ICT, and Future Planning, Republic of Korea.
References
Azevedo, J., Garcia, D., Pontier, D., Ohnesorge, S., Yu, A., Garcia, S., et al. (2010). Argonaute quenching and global changes in Dicer homeostasis caused by a pathogen-encoded GW repeat protein. Genes Dev. 24, 904–915. doi: 10.1101/gad.1908710
Bendahmane, A., Kanyuka, K., and Baulcombe, D. C. (1999). The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11, 781–792. doi: 10.1105/tpc.11.5.781
Bendahmane, A., Querci, M., Kanyuka, K., and Baulcombe, D. C. (2000). Agrobacterium transient expression system as a tool for the isolation of disease resistance genes: application to the Rx2 locus in potato. Plant J. 21, 73–81. doi: 10.1046/j.1365-313x.2000.00654.x
Boller, T., and He, S. Y. (2009). Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science 324, 742–744. doi: 10.1126/science.1171647
Calil, I. P., and Fontes, E. P. (2016). Plant immunity against viruses: antiviral immune receptors in focus. Ann. Bot. doi: 10.1093/aob/mcw200 [Epub ahead of print],
Caplan, J. L., Mamillapalli, P., Burch-Smith, T. M., Czymmek, K., and Dinesh-Kumar, S. P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132, 449–462. doi: 10.1016/j.cell.2007.12.031
Carbonell, A., and Carrington, J. C. (2015). Antiviral roles of plant ARGONAUTES. Curr. Opin. Plant Biol. 27, 111–117. doi: 10.1016/j.pbi.2015.06.013
Cesari, S., Thilliez, G., Ribot, C., Chalvon, V., Michel, C., Jauneau, A., et al. (2013). The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1-CO39 by direct binding. Plant Cell 25, 1463–1481. doi: 10.1105/tpc.112.107201
Collier, S. M., and Moffett, P. (2009). NB-LRRs work a “bait and switch” on pathogens. Trends Plant Sci. 14, 521–529. doi: 10.1016/j.tplants.2009.08.001
Cooley, M. B., Pathirana, S., Wu, H. J., Kachroo, P., and Klessig, D. F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12, 663–676. doi: 10.1105/tpc.12.5.663
Cunnac, S., Lindeberg, M., and Collmer, A. (2009). Pseudomonas syringae type III secretion system effectors: repertoires in search of functions. Curr. Opin. Microbiol. 12, 53–60. doi: 10.1016/j.mib.2008.12.003
Dawson, W. O., and Hilf, M. E. (1992). Host-range determinants of plant viruses. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 527–555. doi: 10.1146/annurev.pp.43.060192.002523
de Ronde, D., Butterbach, P., and Kormelink, R. (2014). Dominant resistance against plant viruses. Front. Plant Sci. 5:307. doi: 10.3389/fpls.2014.00307
Deslandes, L., and Rivas, S. (2012). Catch me if you can: bacterial effectors and plant targets. Trends Plant Sci. 17, 644–655. doi: 10.1016/j.tplants.2012.06.011
Ding, S. W. (2010). RNA-based antiviral immunity. Nat. Rev. Immunol. 10, 632–644. doi: 10.1038/nri2824
Ding, S. W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi: 10.1016/j.cell.2007.07.039
Dodds, P. N., Lawrence, G. J., Catanzariti, A. M., Teh, T., Wang, C. I., Ayliffe, M. A., et al. (2006). Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc. Natl. Acad. Sci. U.S.A. 103, 8888–8893. doi: 10.1073/pnas.0602577103
Dodds, P. N., and Rathjen, J. P. (2010). Plant immunity: towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 11, 539–548. doi: 10.1038/nrg2812
Duan, C.-G., Fang, Y.-Y., Zhou, B.-J., Zhao, J.-H., Hou, W.-N., Zhu, H., et al. (2012). Suppression of Arabidopsis ARGONAUTE1-mediated slicing, transgene-induced RNA silencing, and DNA methylation by distinct domains of the Cucumber mosaic virus 2b protein. Plant Cell 24, 259–274. doi: 10.1105/tpc.111.092718
Eggenberger, A. L., Hajimorad, M. R., and Hill, J. H. (2008). Gain of virulence on Rsv1-genotype soybean by an avirulent soybean mosaic virus requires concurrent mutations in both P3 and HC-Pro. Mol. Plant Microbe Interact. 21, 931–936. doi: 10.1094/MPMI-21-7-0931
Giner, A., Lakatos, L., Garcia-Chapa, M., Lopez-Moya, J. J., and Burgyan, J. (2010). Viral protein inhibits RISC activity by argonaute binding through conserved WG/GW motifs. PLoS Pathog. 6:e1000996. doi: 10.1371/journal.ppat.1000996
Iriti, M., and Varoni, E. M. (2015). Chitosan-induced antiviral activity and innate immunity in plants. Environ. Sci. Pollut. Res. Int. 22, 2935–2944. doi: 10.1007/s11356-014-3571-7
Jensen, S., and Thomsen, A. R. (2012). Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. J. Virol. 86, 2900–2910. doi: 10.1128/JVI.05738-11
Jheng, J.-R., Ho, J.-Y., and Horng, J.-T. (2014). ER stress, autophagy, and RNA viruses. Front. Microbiol. 5:388. doi: 10.3389/fmicb.2014.00388
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kachroo, P., Chandra-Shekara, A. C., and Klessig, D. F. (2006). “Plant signal transduction and defense against viral pathogens,” in Advances in Virus Research, eds K. Maramososch and A. J. Shatkin (San Diego, CA: Academic Press), 161–191.
Kang, B.-C., Yeam, I., and Jahn, M. M. (2005). Genetics of plant virus resistance. Annu. Rev. Phytopathol. 43, 581–621. doi: 10.1146/annurev.phyto.43.011205.141140
Katiyar-Agarwal, S., and Jin, H. (2010). Role of small RNAs in host-microbe interactions. Annu. Rev. Phytopathol. 48, 225–246. doi: 10.1146/annurev-phyto-073009-114457
Kontra, L., Csorba, T., Tavazza, M., Lucioli, A., Tavazza, R., Moxon, S., et al. (2016). Distinct effects of p19 RNA silencing suppressor on small RNA mediated pathways in plants. PLoS Pathog. 12:e1005935. doi: 10.1371/journal.ppat.1005935
Korner, C. J., Klauser, D., Niehl, A., Dominguez-Ferreras, A., Chinchilla, D., Boller, T., et al. (2013). The immunity regulator BAK1 contributes to resistance against diverse RNA viruses. Mol. Plant Microbe Interact. 26, 1271–1280. doi: 10.1094/MPMI-06-13-0179-R
Lakatos, L., Csorba, T., Pantaleo, V., Chapman, E. J., Carrington, J. C., Liu, Y. P., et al. (2006). Small RNA binding is a common strategy to suppress RNA silencing by several viral suppressors. EMBO J. 25, 2768–2780. doi: 10.1038/sj.emboj.7601164
Li, F., Pignatta, D., Bendix, C., Brunkard, J. O., Cohn, M. M., Tung, J., et al. (2012). MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. U.S.A. 109, 1790–1795. doi: 10.1073/pnas.1118282109
Li, S., Liu, L., Zhuang, X., Yu, Y., Liu, X., Cui, X., et al. (2013). MicroRNAs inhibit the translation of target mRNAs on the endoplasmic reticulum in Arabidopsis. Cell 153, 562–574. doi: 10.1016/j.cell.2013.04.005
Luan, H., Shine, M., Cui, X., Chen, X., Ma, N., Kachroo, P., et al. (2016). The potyviral P3 protein targets EF1A to promote the unfolded protein response and viral pathogenesis. Plant Physiol. 172, 221–234. doi: 10.1104/pp.16.00505
Monaghan, J., and Zipfel, C. (2012). Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. doi: 10.1016/j.pbi.2012.05.006
Moon, J. Y., Lee, J. H., Oh, C.-S., Kang, H.-G., and Park, J. M. (2016). Endoplasmic reticulum stress responses function in the HRT-mediated hypersensitive response in Nicotiana benthamiana. Mol. Plant Pathol. 17, 1382–1397. doi: 10.1111/mpp.12369
Nakahara, K. S., and Masuta, C. (2014). Interaction between viral RNA silencing suppressors and host factors in plant immunity. Curr. Opin. Plant Biol. 20, 88–95. doi: 10.1016/j.pbi.2014.05.004
Nicaise, V. (2014). Crop immunity against viruses: outcomes and future challenges. Front. Plant Sci. 5:660. doi: 10.3389/fpls.2014.00660
Nicaise, V., and Candresse, T. (2016). Plum pox virus capsid protein suppresses plant pathogen-associated molecular pattern (PAMP)-triggered immunity. Mol. Plant Pathol. doi: 10.1111/mpp.12447 [Epub ahead of print].
Niehl, A., Wyrsch, I., Boller, T., and Heinlein, M. (2016). Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 211, 1008–1019. doi: 10.1111/nph.13944
Nürnberger, T., Brunner, F., Kemmerling, B., and Piater, L. (2004). Innate immunity in plants and animals: striking similarities and obvious differences. Immunol. Rev. 198, 249–266. doi: 10.1111/j.0105-2896.2004.0119.x
Padmanabhan, M. S., and Dinesh-Kumar, S. P. (2014). The conformational and subcellular compartmental dance of plant NLRs during viral recognition and defense signaling. Curr. Opin. Microbiol. 20, 55–61. doi: 10.1016/j.mib.2014.05.003
Palanichelvam, K., Cole, A. B., Shababi, M., and Schoelz, J. E. (2000). Agroinfiltration of Cauliflower mosaic virus gene VI elicits hypersensitive response in Nicotiana Species. Mol. Plant Microbe Interact. 13, 1275–1279. doi: 10.1094/MPMI.2000.13.11.1275
Ruiz-Ferrer, V., and Voinnet, O. (2009). Roles of plant small RNAs in biotic stress responses. Annu. Rev. Plant Biol. 60, 485–510. doi: 10.1146/annurev.arplant.043008.092111
Shivaprasad, P. V., Chen, H. M., Patel, K., Bond, D. M., Santos, B. A., and Baulcombe, D. C. (2012). A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 24, 859–874. doi: 10.1105/tpc.111.095380
Takahashi, H., Suzuki, M., Natsuaki, K., Shigyo, T., Hino, K., Teraoka, T., et al. (2001). Mapping the virus and host genes involved in the resistance response in cucumber mosaic virus-Infected Arabidopsis thaliana. Plant Cell Physiol. 42, 340–347. doi: 10.1093/pcp/pce039
Takeuchi, O., and Akira, S. (2009). Innate immunity to virus infection. Immunol. Rev. 227, 75–86. doi: 10.1111/j.1600-065X.2008.00737.x
Truniger, V., and Aranda, M. A. (2009). “Chapter 4 – recessive resistance to plant viruses,” in Advances in Virus Research, eds L. Gad and P. C. John (San Diego, CA: Academic Press), 119–231.
Ueda, H., Yamaguchi, Y., and Sano, H. (2006). Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol. Biol. 61, 31–45. doi: 10.1007/s11103-005-5817-8
Vaucheret, H. (2006). Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes Dev. 20, 759–771. doi: 10.1101/gad.1410506
Vaucheret, H. (2008). Plant ARGONAUTES. Trends Plant Sci. 13, 350–358. doi: 10.1016/j.tplants.2008.04.007
Verchot, J. (2014). The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 5:66. doi: 10.3389/fpls.2014.00066
Verlaan, M. G., Hutton, S. F., Ibrahem, R. M., Kormelink, R., Visser, R. G. F., Scott, J. W., et al. (2013). The tomato yellow leaf curl virus resistance genes < italic > Ty-1 < /italic > and < italic > Ty-3 < /italic > are allelic and code for DFDGD-class RNA–dependent RNA polymerases. PLoS Genet. 9:e1003399. doi: 10.1371/journal.pgen.1003399
Wang, L. Y., Lin, S. S., Hung, T. H., Li, T. K., Lin, N. C., and Shen, T. L. (2012). Multiple domains of the tobacco mosaic virus p126 protein can independently suppress local and systemic RNA silencing. Mol. Plant Microbe Interact. 25, 648–657. doi: 10.1094/MPMI-06-11-0155
Wen, R. H., Khatabi, B., Ashfield, T., Maroof, M. A. S., and Hajimorad, M. R. (2012). The HC-Pro and P3 cistrons of an avirulent Soybean mosaic virus are recognized by different resistance genes at the complex Rsv1 locus. Mol. Plant Microbe Interact. 26, 203–215. doi: 10.1094/MPMI-06-12-0156-R
Whitham, S., Dinesh-Kumar, S. P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: similarity to toll and the interleukin-1 receptor. Cell 78, 1101–1115. doi: 10.1016/0092-8674(94)90283-6
Wobbe, K. K., Akgoz, M., Dempsey, D. M. A., and Klessig, D. F. (1998). A single amino acid change in Turnip crinkle virus movement protein p8 affects RNA binding and virulence on Arabidopsis thaliana. J. Virol. 72, 6247–6250.
Zhai, J., Jeong, D. H., De Paoli, E., Park, S., Rosen, B. D., Li, Y., et al. (2011). MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 25, 2540–2553. doi: 10.1101/gad.177527.111
Zhou, J. M., and Chai, J. (2008). Plant pathogenic bacterial type III effectors subdue host responses. Curr. Opin. Microbiol. 11, 179–185. doi: 10.1016/j.mib.2008.02.004
Keywords: defense responses, R gene, RNAi, viral suppressor of RNAi, virus
Citation: Moon JY and Park JM (2016) Cross-Talk in Viral Defense Signaling in Plants. Front. Microbiol. 7:2068. doi: 10.3389/fmicb.2016.02068
Received: 30 September 2016; Accepted: 07 December 2016;
Published: 21 December 2016.
Edited by:
Yule Liu, Tsinghua University, ChinaReviewed by:
Pradeep Kachroo, University of Kentucky, USAElizabeth P. B. Fontes, Universidade Federal de Viçosa, Brazil
Copyright © 2016 Moon and Park. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeong M. Park, am1wYXJrQGtyaWJiLnJlLmty
 Ju Y. Moon
Ju Y. Moon Jeong M. Park
Jeong M. Park