- 1Research Institute for Limnology, University of Innsbruck, Mondsee, Austria
- 2Miti Biosystems GmbH, Max F Perutz Laboratories, Wien, Austria
- 3Institute of Virology, Helmholtz Zentrum München, München, Germany
- 4Bioinformatics and Systems Biology, Justus-Liebig-University, Giessen, Germany
Cyanobacteria are frequently involved in the formation of harmful algal blooms wherein, apart from the toxic microcystins, other groups of bioactive peptides are abundant as well, such as anabaenopeptins (APs). The APs are synthesized nonribosomally as cyclic hexapeptides with various amino acids at the exocyclic position. We investigated the presence and recombination of the AP synthesis gene cluster (apnA-E) through comparing 125 strains of the bloom-forming cyanobacterium Planktothrix spp., which were isolated from numerous shallow and deep water habitats in the temperate and tropical climatic zone. Ten ecologically divergent strains were purified and genome sequenced to compare their entire apnA-E gene cluster. In order to quantify apn gene distribution patterns, all the strains were investigated by PCR amplification of 2 kbp portions of the entire apn gene cluster without interruption. Within the 11 strains assigned to P. pseudagardhii, P. mougeotii, or P. tepida (Lineage 3), neither apnA-E genes nor remnants were observed. Within the P. agardhii/P. rubescens strains from shallow waters (Lineage 1, 52 strains), strains both carrying and lacking apn genes occurred, while among the strains lacking the apnA-E genes, the presence of the 5′end flanking region indicated a gene cluster deletion. Among the strains of the more derived deep water ecotype (Lineage 2, 62 strains), apnA-E genes were always present. A high similarity of apn genes of the genus Planktothrix when compared with strains of the genus Microcystis suggested its horizontal gene transfer during the speciation of P. agardhii/P. rubescens. Genetic analysis of the first (A1-) domain of the apnA gene, encoding synthesis of the exocyclic position of the AP molecule, revealed four genotype groups that corresponded with substrate activation. Groups of genotypes were either related to Arginine only, the coproduction of Arginine and Tyrosine or Arginine and Lysine, or even the coproduction of Arginine, Tyrosine, and Lysine in the exocyclic position of the AP-molecule. The increased structural diversity resulted from the evolution of apnA A1 genotypes through a small number of positively selected point mutations that occurred repeatedly and independently from phylogenetic association.
Introduction
The bloom-forming cyanobacteria of Planktothrix agardhii and P. rubescens are frequently involved in cyanotoxin production in lakes and reservoirs. Besides the toxic heptapeptide microcystin, a number of additional bioactive oligopeptides have been elucidated from Planktothrix spp., (e.g., Kurmayer et al., 2016). In particular, the anabaenopeptins (APs) show an impressive diversity in bioactivity. For example, while some AP structural variants inhibit protein phosphatase 1 and 2A, others have serine proteases inhibition activity such as chymotrypsin and trypsin, or they are potent inhibitors of carboxypeptidase A (e.g., in Spoof et al., 2016) and other metallocarboxypeptidases (Halland et al., 2015). APs are cyclic hexapeptides consisting of five amino acid residues forming a ring (pos. 2–6) and an exocyclic residue (pos. 1), which is connected to the ring through an ureido bond (Figure 1). While the D-Lys in pos. 2 and the ureido bond of the AP structure are conserved motifs, different amino acids are found in all other positions of the AP molecule resulting in numerous structural variants (e.g., in Spoof et al., 2016). The first AP structural variants A and B were described from Anabaena flos-aquae (Harada et al., 1995). Other cyanobacteria genera known as prominent AP producers include the planktonic genera Microcystis (e.g., Williams et al., 1996; Fastner et al., 2001), or Nodularia (e.g., Fujii et al., 1997) but also benthic genera such as Lyngbya (e.g., Zi et al., 2012) and Schizothrix (e.g., Reshef and Carmeli, 2002). In general, the AP peptides are the most abundant besides the microcystins in waterbodies of the temperate climate region (Halstvedt et al., 2008; Gkelis et al., 2015). Typically, cellular contents up to 0.5% dry weight are reported in isolated strains (0.9–10 μg AP mg−1 dry weight), (Kosol et al., 2009), and in field samples high concentrations >1 mg L−1 have been observed (e.g., Gkelis et al., 2015).
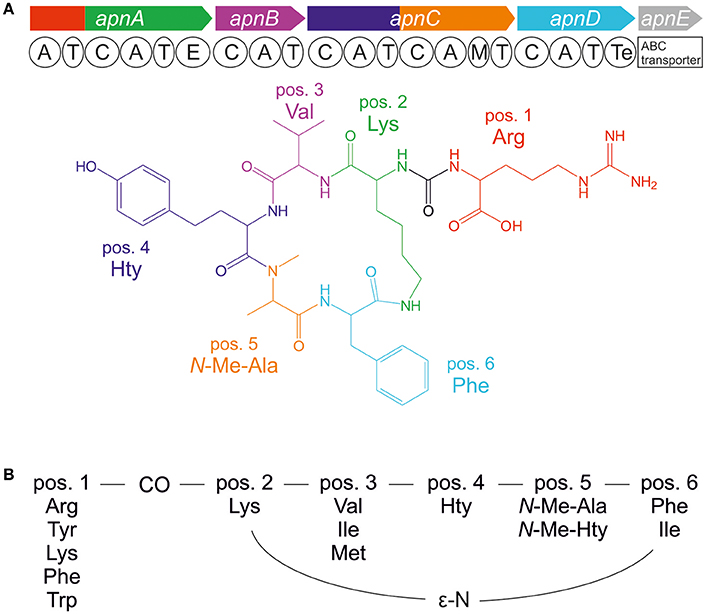
Figure 1. (A) Anabaenopeptin synthesis gene cluster and resulting molecular structure of anabaenopeptin B ([M+H]+ 837) and (B) amino acid variation of anabaenopeptins as observed in the genus Planktothrix.
APs are produced by nonribosomal peptide synthetases (NRPS) that follow a stepwise synthesis pathway using (non) proteinogenic amino acids as substrate (Christiansen et al., 2011). Following the thio-template mechanism, the amino acid substrates are activated as aminoacyl adenylate and condensed to the growing peptide chain. Minimum modules consist of adenylation (A), thiolation (T), and condensation (C) domains. Additional enzyme domains catalyze the epimerization of the conserved D-Lys in pos. 2 and N-methylation in pos. 5 (Figure 1; Christiansen et al., 2011). The first A-domain of ApnA (ApnA A1) activates different amino acids occurring at the variable exocyclic pos. 1. Molecular biological and evolutionary analysis revealed that single point mutations within the first A-domain can result in a remarkable substrate promiscuity since the two chemically divergent amino acids arginine and tyrosine were activated with high selectivity and comparable efficiency (Christiansen et al., 2011). Based on these results, those particular enzyme domains were crystallized and the structural basis of substrate activation describing a bispecific A-domain was elucidated (Kaljunen et al., 2015).
Both of these previous studies, however, were based on the investigation of only a small number of selected strains. In the present study, we aimed to qualitatively and quantitatively analyze the recombination phenomena leading to apnA-E gene cluster evolution in the genus Planktothrix. Planktothrix occurs in shallow and deep water ecosystems of the temperate and tropical climatic zones. Recent phylogenetic and ecological analysis has defined a number of lineages representing ecological diversification (Gaget et al., 2015; Kurmayer et al., 2015). In a first attempt, we compared the apn gene cluster sequence and its flanking regions from 10 ecologically divergent strains for which the genomes were sequenced. In addition, we examined all other Planktothrix strains for the apn gene cluster presence/absence and recombination. In a second step, we analyzed the nucleotide variation of the apnA A1-domain and the resulting AP peptide structural variation to identify the functional consequences of genetic structural recombination in 89 AP-producing strains. If a relationship between apnA A1-genotypes and the occurrence of AP variants exists, the ecological dynamics of specific apnA A1 genotypes can be followed to investigate the evolution of AP synthesis in our water bodies.
Materials and Methods
Organisms
In total, 125 clonal Planktothrix spp. strains, isolated from deep and shallow freshwater habitats, were analyzed in this study (Supplementary Table S1). One hundred twelve strains were previously characterized and assigned to phylogenetic lineages by multilocus sequence analysis (MLSA) and 13 additional strains were added into this earlier phylogeny (Kurmayer et al., 2015). The strains were grown under sterile conditions in BG11 medium with low light intensity (5–10 μmol m−2 s−1 Osram Type L30W/77 Fluora, 16/8 h light-dark cycle, 15° or 23°C).
DNA Isolation
Cells from cultures were harvested by centrifugation and washed in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0 with HCl). DNA was extracted according to Reinard (2008). In brief, the cell pellet was suspended in 0.5 ml of CTAB solution (0.1 M cetyltrimethylammonium bromide, 1.4 M NaCl, 0.1 M Tris, 0.02 M EDTA), incubated at 65°C (30 min) followed by enzymatic treatment (proteinase K, 1 mg ml−1, 60°C, 60 min and RNase A, 0.2 mg ml−1, 20°C) and DNA extraction with chloroform/isoamyl alcohol (24:1, v/v). The DNA was precipitated by mixing the aqueous phase with two volume parts of CTAB precipitation solution (15 mM CTAB, 40 mM NaCl, pH 7.0) at 4°C (1 h) to purify the DNA from polysaccharides. The DNA pellet was dissolved in 1.2 M NaCl solution and extracted again with chloroform/isoamyl alcohol (24:1, v/v). Finally, the DNA was precipitated with 0.6 volume parts isopropanol. The pellet was washed with 70% (v/v) ethanol and the DNA was dissolved in 50 μl EB buffer (Qiagen, Hilden, Germany).
Genome Sequencing
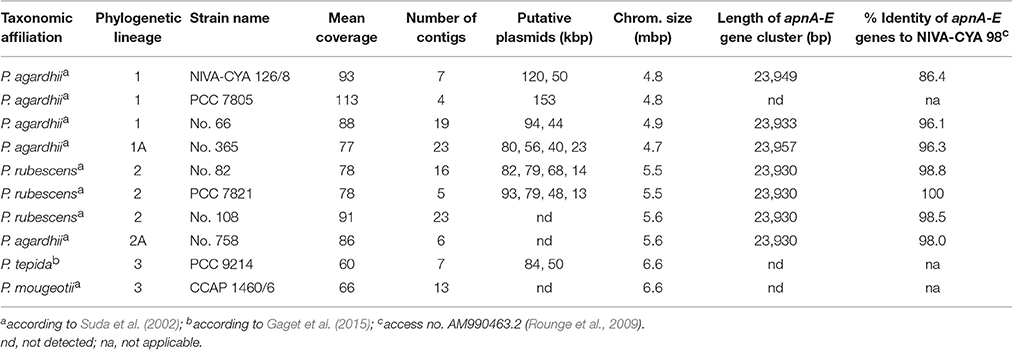
Ten Planktothrix strains from different phylogenetic lineages were axenized (Rippka, 1988). Purity was tested and confirmed using DAPI (4,6-diamidino-2-phenylindole) staining for contaminant bacteria on membrane filters (0.2 μm pore size) and epifluorescence microscopy. High molecular weight DNA was extracted from cells by grinding in liquid nitrogen, purified using anion-exchange columns (Qiagen) according to the manufacturer's protocol and used for single molecule real-time (SMRT) sequencing (GATC Biotech, Constance, Germany). The sequencing efforts aimed to obtain a hundredfold coverage resulting in a relatively low number of contigs (Table 1). The genome sequences were annotated automatically using GenDB (Meyer et al., 2003) with the reference genome of P. agardhii NIVA-CYA 126/8 (Christiansen et al., 2014), access. no. CM002803.1. For all strains with sequenced genome, the automatically annotated chromosomal region carrying the apnA-E gene cluster and/or its flanking regions (approx. 300 kbp) have been manually curated and submitted to DDBJ/EMBL/GenBank under the accession no. KU665235-KU665242.
PCR Analysis and Sequencing
For the MLSA of newly sequenced strains (n = 13), seven gene loci and intergenic spacer (IGS) regions, including the 16S rDNA (301 bp), 16S rDNA-internal transcribed spacer (ITS) region (312 bp), phycocyanin (PC)-IGS (204 bp), psaA, and psaB photosynthesis protein (PSA)-IGS (548 bp), RNaseP gene (299 bp), large subunit of the ribulose bisphosphate carboxylase/oxygenase and rbcX (rbcLX)-IGS (336 bp), and rpoC (492 bp) were amplified by PCR and sequenced (Kurmayer et al., 2015), GenBank accession no. KU574075–KU574122.
In order to estimate the presence/absence of the apnA-E gene cluster in all 125 strains, 17 primer pairs were designed from the reference gene clusters of NIVA-CYA 126/8 (Christiansen et al., 2011; access. no. EF67686) and NIVA-CYA 98 (Rounge et al., 2009; access. no. AM990463.2) and used to amplify overlapping fragments without interruption (approx. 2 kbp for each amplicon), (Supplementary Table S2). Each PCR reaction mixture had a total volume of 10 μl, containing 2 μl (5 ×) Phusion HF Buffer (Thermo Scientific, Vienna, Austria), 500 nM of each primer, 200 μM of each deoxynucleotide triphosphate (Thermo Scientific), 0.1 U of Phusion High-Fidelity DNA Polymerase (Thermo Scientific), and 10 ng of DNA. PCR amplification was performed under the following conditions: Initial denaturation at 98°C for 1 min; followed by 35 cycles of denaturation at 98°C for 10 s, annealing at a variable temperature for 15 s and elongation at 72°C. The PCR amplicon size was determined using gel electrophoresis (0.8% agarose gels in 0.5 × Tris-borate-EDTA buffer and visualized using Midori Green). For the sequencing of adenylation domains, PCR products were extracted from the agarose gel using a gel extraction kit (QIAquick, Gel Extraction Kit, Qiagen) and sequenced (Eurofins Genomics, Ebersberg, Germany). Nucleotide sequences were submitted to GenBank under accession no. KU639970–KU640047.
Nucleotide Sequence Analysis
For the MLSA analysis of all 125 strains, the sequences of the seven gene loci were concatenated. A maximum likelihood tree was calculated using default adjustments in Mega 6.0 (phylogeny test: bootstrap method, 1,000 replicates; substitution model: Tamura 3-parameter model; gaps included). We used the BLASTn algorithm to identify apn genes or homologs in other cyanobacteria genera (access. no. AQPY01000292.1; AZYY01000200.1; CAIH01000020.1; CAIQ01000336.1; CP001037.1; CP003284.1; CP007203.2; JHEG02000042.1; KV757545.1; LJOQ01000025.1; LJOP01000059.1; LWAJ01000278.1) and compared the individual genes by calculating the evolutionary divergence as well by phylogenetic analysis using maximum likelihood (Mega 6.0). The apnA A1 sequences from all 89 AP-producing strains (core motive A4-A6, 510 bp) were aligned and maximum likelihood (ML) was used to construct a phylogenetic tree (Mega 6.0). Default parameters, such as a constant rate variation among the sites as well as a fixed transition/transversion ratio, have been used. In general, sites were not weighted. Statistical significance of the branches was estimated by bootstrap analysis generating 1,000 replicates of the original data set. ApnA A1 domain selectivity was predicted using established bioinformatics tools based on the specificity-conferring code defined by Stachelhaus et al. (1999) and the residues within 8 Å around the substrate as defined by Rausch et al. (2005).
The ratio of non-synonymous (dN) and synonymous (dS) apnA A1 substitution rates per codon site was determined using maximum likelihood estimates as implemented in the PAML package (Version 3.15; Yang, 1997). NS-sites models were applied to identify the sites under potential positive selection. A likelihood ratio test (LRT, df = 2) was constructed to compare the likelihood of the phylogenetic tree, calculated under two different type of models: (1) the null models M1 (nearly neutral), M7 (beta), which do not allow for positively selected sites (dN/dS ≤ 1), and (2) the alternative models M2 (positive selection), M8 (beta & dN/dS > 1), which adds an additional site class that accounts for positive selection (dN/dS > 1). When the LRT was found to be significant, the Bayes empirical Bayes (BEB) approach was used to calculate posterior probabilities that a site comes from the site class with dN/dS > 1 (Yang et al., 2005).
Anabaenopeptin Peptide Structural Identification
Cells were harvested on glass fiber filters (BMC, Ederol, Vienna, Austria) and dried biomass was extracted in 50% (v/v) aqueous methanol on ice according to Kosol et al. (2009). AP structural variants were separated by HPLC (HP 1100, Agilent) using a linear water/acetonitrile (0.05% trifluoroacetic acid) gradient from 80:20 to 50:50 in 45 min at a flow rate of 1 ml min−1 and 30°C oven temperature, LiChrospher 100 octyldecyl silane (ODS) (5 μm particle size) and LiChroCART 250-4 cartridge system (Merck, Darmstadt, Germany) as described previously (Kosol et al., 2009). The HPLC system was coupled to an Electrospray Ionisation (ESI) Mass Spectrometer ion trap (amaZonSL, Bruker) operating in positive ion mode. Nitrogen was used as sheath gas (43 psi, 8 L/min, 300°C) and helium was used as auxiliary gas. Capillary voltage was set to 5 kV. Under these conditions, the following AP structural variants were detected: AP B (836.5 Da), AP A (843.4 Da), AP F (850.5 Da), Oscillamide (Osc) Y (857.4 Da), AP C (808.4 Da), AP D (827.4 Da), AP 908 (908.5 Da), AP 915 (915.5 Da), Osc B (868.4 Da), and ferintoic acid A (866.4 Da), (Harada et al., 1995; Sano and Kaya, 1995; Williams et al., 1996; Shin et al., 1997; Sano et al., 2001; Fujii et al., 2002; Okumura et al., 2009; Supplementary Figures S1, S2). Unknown AP structural variants were assigned according to specific fragmentation patterns (Supplementary Tables S3, S4). In general, for each strain, 2–7 mg of dry weight were extracted. The limit of detection for AP B was 10 ng (corresponding to 0.005–0.0014% of dry weight). All AP structural variants that contributed ≥5% of peak area compared with the most abundant AP structural variant (extracted ion chromatogram) were recorded.
Results
Distribution of the Anabaenopeptin Synthesis Genes among the Genus Planktothrix
Phylogenetic analysis of all 125 Planktothrix strains revealed a diversification comprising three major lineages: Lineage 1 and Lineage 2 represented only the strains assigned to P. agardhii/P. rubescens and Lineage 3 represented P. pseudagardhii, P. mougeotii, and P. tepida (Figure 2). While strains of Lineage 1 were mainly isolated from shallow lakes, most strains of Lineage 2 were isolated from deep lakes in the temperate climatic zone. Strains of Lineage 3 originated from lakes in the tropical climatic zone. From these ecologically divergent lineages, 10 strains were purified and their genomes sequenced (Table 1). Following assembly and annotation, the apnA-E gene cluster structure and flanking regions were elucidated. As reported previously, the apnA-E gene cluster spans 24 kbp and encodes six NRPS modules (two modules in apnA, one in apnB, two in apnC, and one in apnD) as well as an ATP binding cassette (ABC) transporter (apnE), (Christiansen et al., 2011). Except for strain PCC 7805, all sequenced genomes of P. agardhii/P. rubescens strains were found to contain the apnA-E gene cluster in the chromosome (Figure 3). Notably, in all sequenced genomes of strains of Lineage 1+2 (including the PCC 7805 genome), the 5′end flanking region of the apnA-E gene cluster was found to be similar (alignment 6,682 bp, evolutionary divergence 0.010–0.035), and showed a linkage to the cyanopeptolin synthesis gene cluster as described by Rounge et al. (2009). Among the Lineage 1+2 strains with genome sequences, the 3′end flanking region was found variable and either contained another gene cluster encoding microviridin synthesis (Philmus et al., 2008)—thereby forming a meta peptide synthesis gene cluster—or the sequence reported for strain PCC 7805 (strains no. 66 and no. 365). In contrast, strains of Lineage 3 (P. pseudagardhii, P. mougeotii, P. tepida) did not contain any sequence related to the apnA-E gene or flanking region. For all other 115 strains, the presence of the entire apnA-E gene cluster was tested using the PCR amplification of ~2 kbp fragments without interruption (Supplementary Table S2, Figure S3). In total, 90 out of 125 strains (72%) carried the AP synthesis operon. Among Lineages 2 and 2A, all strains were found to contain the apn gene cluster (Figure 2), with only strain CCAP 1459/31 showing a 381 bp-deletion within the A-domain of apnB resulting in the inactivation of AP synthesis. The apnA-E genes also occurred among the strains of Lineage 1 (43% of strains) and 1A (all strains). Corresponding with the genomic results, no evidence for apnA-E gene presence was found among the strains of Lineage 3. The PCR assay amplifying the 5′end flanking region (ociD-apnA) gave the expected specific PCR product (4,719 bp in reference strain NIVA-CYA 126/8) for all of the 90 apnA-E positive strains of Lineage 1 and 2. Three strains (no. 66, 63, 41) showed a longer PCR product due to the insertion of a sequence similar to IS1634, 2,063 bp (access. no. JX 134573). Interestingly, strains lacking the apnA-E gene cluster from Lineage 1 (n = 24) gave the expected PCR product of the 5′end flanking region (5,073 bp, ociD-aspB). In contrast, for the strains of Lineage 3, no PCR products were obtained. The DNA fragment of the 3′end flanking region was amplified less consistently among apnA-E positive strains (77 out of 90). All the strains from Lineage 2 and 2A showed a specific PCR product with the expected size (4,411 bp in reference strain NIVA-CYA 126/8) suggesting the phylogenetic fixation of the meta peptide synthesis gene cluster. However, 18 strains from Lineage 1 (1A) failed to show this PCR product, implying that according to genomic information of strains no. 66 and 365, the mvdA-F gene cluster was not associated with the ociA-D and apnA-E gene cluster (Supplementary Table S1). In summary, the apnA-E gene cluster occurred among all the strains of Lineage 2 (2A) and showed almost identical flanking regions. Among Lineage 1 (1A) strains both carrying and lacking the apn gene cluster were found, but all of them contained at least the 5′end flanking region, suggesting a potential deletion.
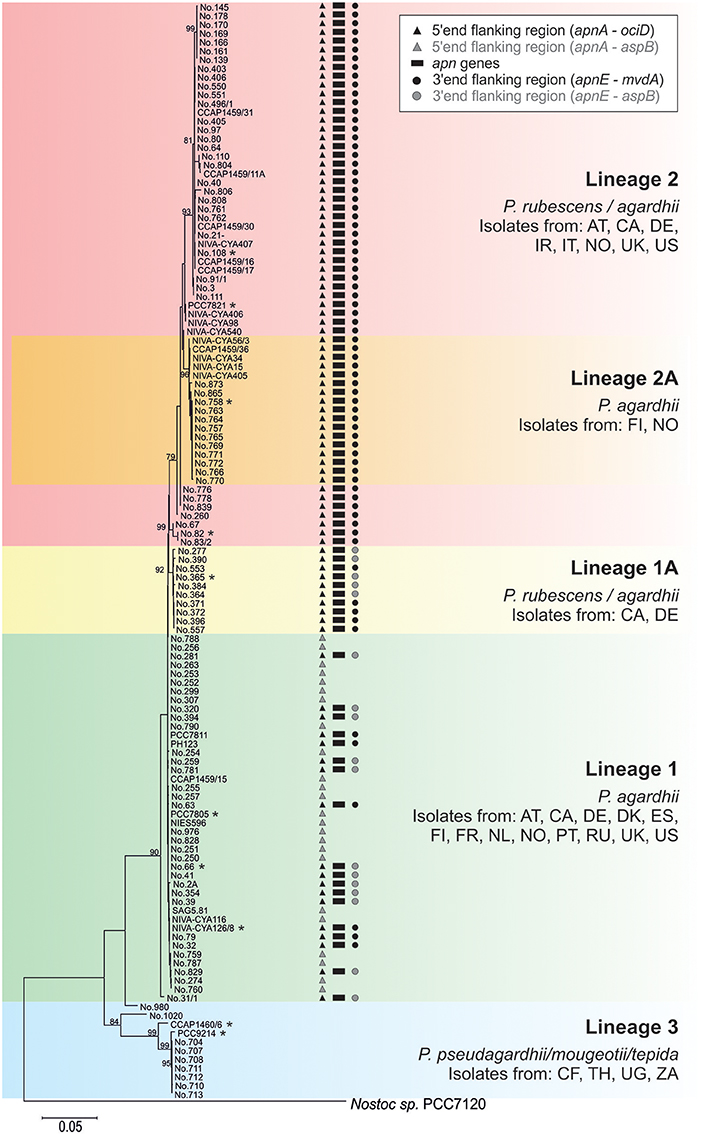
Figure 2. Phylogenetic affiliation of the 125 Planktothrix strains analyzed for the apnA-E gene cluster (genome sequenced strains are marked with a star). The Maximum likelihood tree was constructed from seven gene loci or intergenic spacer regions. Numbers at nodes indicate the percent bootstrap frequency (1,000 replicates). Only bootstrap values >70% are shown. The tree was rooted using Nostoc sp. strain PCC 7120. Countries of origin are indicated by the two-letter code.
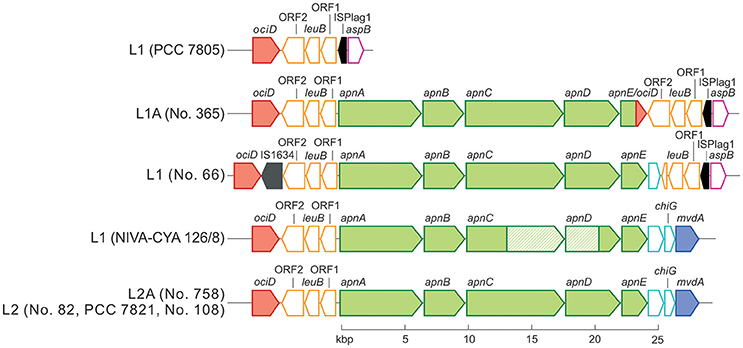
Figure 3. Schematic view of the apnA-E gene cluster (green color) and flanking regions. The microviridin synthesis gene cluster (mvdA-F) and the cyanopeptolin synthetase genes (ociA-D) are shown in blue and red, each. Identical sequences are indicated by the same color. Transposable elements are marked in black.
Comparison of Anabaenopeptin Synthesis Gene Cluster Genes and Flanking Regions among Cyanobacteria
Using the BLASTn algorithm against the Genbank nucleotide database, apn genes or homologs were found for eight genera: Planktothrix, Microcystis, and the heterocystous genera (Anabaena, Nostoc, Nodularia, Aphanizomenon, Tolypothrix, Scytonema), (Figure 4A). With the exception of the large recombination within apnC/D observed for the strain NIVA-CYA 126/8, the apnA-E genes showed low evolutionary divergence (≤0.1) within the genus Planktothrix. Notably, the nucleotide divergence remained low compared to the apn genes of Microcystis, but it increased to >0.3 when comparing with apn genes of other genera. Phylogenetic trees grouped apnA-E genes from both genera Planktothrix and Microcystis into one branch while the apnA-E genes of other heterocystous genera formed a separate lineage (Supplementary Figures S4–S8). Notably, for apnA and apnB genes, Planktothrix strain no. 66 was placed within Microcystis. The high similarity of apnA-E genes between Planktothrix and Microcystis is in contrast to other NRPS genes, such as the microcystin synthesis gene cluster, which is phylogenetically differentiated between Planktothrix and Microcystis (Figure 4B). We conclude that the high similarity of apn genes between Planktothrix and Microcystis stems from horizontal gene transfer.
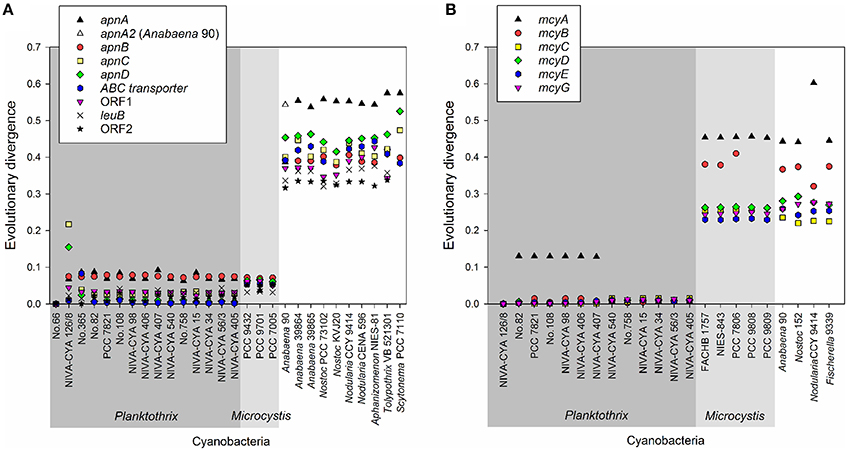
Figure 4. (A) Evolutionary divergence (nucleotide variability) between the individual apnA-E genes of Planktothrix strain no. 66 when compared with other strains Planktothrix and various cyanobacteria; (B) Evolutionary divergence (nucleotide variability) of individual mcyA-G genes as calculated from various cyanobacteria. Planktothrix strain NIVA-CYA 126/8 was used as a reference.
Recombination of Anabaenopeptin Synthesis Genes among Planktothrix
Within the strains with sequenced genome, the apnA-E gene clusters were 96–100% similar to apnA-E genes from the strain NIVA-CYA 98 (Rounge et al., 2009). Only the strain NIVA-CYA 126/8 showed a large recombination of parts of the second module of apnC (3,235 bp) comprising the condensation domain apnC C2 and the A-domains apnC A2 and apnD A (795 bp), which has been previously described (Christiansen et al., 2011). Using PCR amplification, only two other strains of Lineage 1 (nos. 259, 281) showed this large recombination, resulting in the same amino acid variation in the AP molecule as reported for the strain NIVA-CYA 126/8 (Supplementary Table S1; Christiansen et al., 2011).
Notably, the genome of strain no. 365 showed a fusion of the two ABC transporter genes apnE (1,581 bp) with ociD (483 bp), (Figure 3) which is part of the cyanopeptolin synthesis gene cluster (Rounge et al., 2007), at the conserved Walker B motif (amino acid position 526–533 in the apnE of the strain NIVA-CYA 126/8; Pearson et al., 2004). This recombination was found in five additional strains from Lineage 1 and 1A (no. 354, 364, 384, 390, 394; Supplementary Table S1). No functional consequence on AP production was observed due to this recombination. In summary, the recombinations of larger fragments affecting A-domains apnC A2/apnD A occurred only rarely.
Genetic Diversity of the A-Domain apnA A1 Correlates to Structural Variation in the Anabaenopeptin Molecule
For all 89 strains carrying the full apnA-E gene cluster, we observed the synthesis of at least one AP structural variant (Supplementary Table S1). The AP structural variant with Arg (AP B, [M+H]+ 837) in exocyclic pos. 1 of the AP molecule occurred most frequently (93%). Other structural variants with Arg (AP F, [M+H]+ 851) or Tyr (AP A, [M+H]+ 844), (Osc Y, [M+H]+ 858), or Lys (AP C, [M+H]+ 809) in exocyclic pos. 1 of the AP molecule occurred with a median proportion of 45, 55, 33, and 22%, respectively (Figure 5A). In addition, other AP structural variants occurred in a few strains only, e.g., AP D with Phe in pos. 1 ([M+H]+ 828) or ferintoic acid A with Trp in pos. 1 ([M+H]+ 867). Unknown APs were identified by comparing fragmentation patterns with those of known AP structural variants. With regard to the co-production of AP structural variants differing in amino acid (aa) composition in pos. 1 the following chemotypes were observed: Arg (n = 16), Arg + Tyr (n = 48), Arg + Lys (n = 8), Arg + Trp (n = 2), Arg + Tyr + unknown aa (n = 3), Arg + Lys + unknown aa (n = 9), Arg + Tyr + Lys + unknown aa (n = 2), Arg + Tyr + Lys + unknown aa + Phe (n = 1), (Figure 5B).
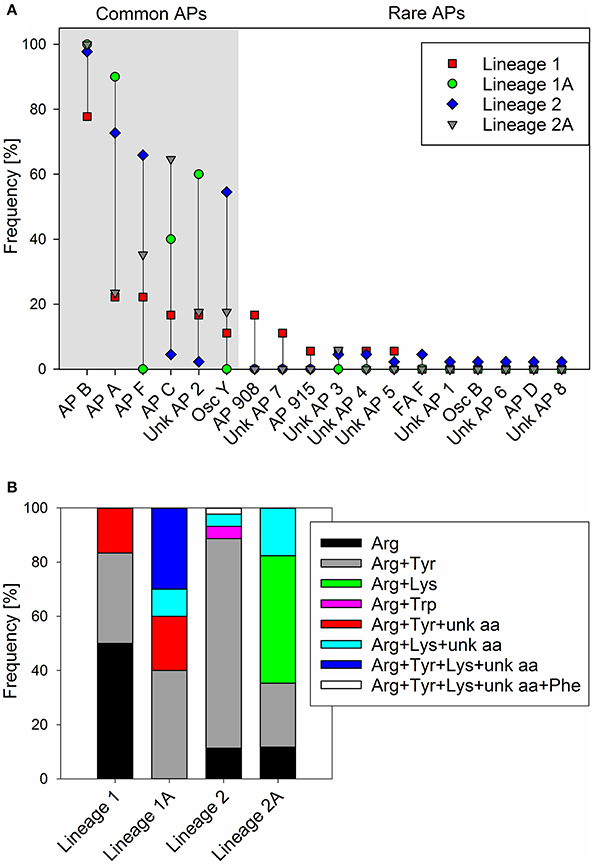
Figure 5. (A) Frequency (proportion) of anabaenopeptin structural variants among Planktothrix phylogenetic lineages 1 (1A), 2 (2A); (B) Frequency (proportion) of co-production of various amino acids in exocyclic pos. 1 of AP structural variants.
To determine the functional consequences of the genetic variability in the apnA A1 domain, sequences of all the AP-producing strains were compared with the structural variation occurring at the exocyclic position 1 of the AP molecule. In total, the maximum nucleotide variability among apnA A1-genotypes was 10.7% (n = 89). Phylogenetic analysis of the catalytic region, including the core motifs A4–A6 (510 bp), revealed four clades which corresponded with substrate activation in pos. 1 of the AP molecule (Figure 6). Clade A contained strains typically co-producing AP structural variants with Arg and Tyr in exocyclic pos. 1. As an exception, two strains (no. 806, 808) produced AP variants with Arg and Trp in this position, but they showed a slightly different apnA A1 sequence compared to the others. Strains among clade B carried only Arg at the exocyclic position, while strains among clade C produced Arg, Lys, and eventually an unknown aa. Clade D contained strains with a variety of AP structural variants with different amino acids in exocyclic pos. 1, either only Arg and Tyr, or combinations of Arg and/or Tyr and/or Lys and/or an unknown aa. Within clade D strain no. 804 co-produced even five AP variants differing in pos. 1 (Arg, Tyr, Lys, Phe, and an unknown aa).
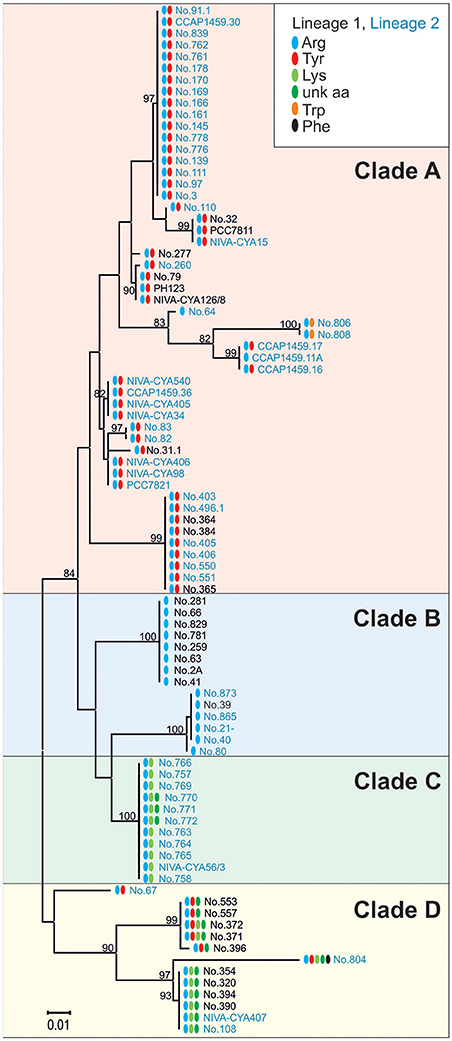
Figure 6. Maximum likelihood tree of the apnA A1 sequences constituting the catalytic domain for substrate activation (core A4-A6). Numbers at the nodes indicate the percent bootstrap frequency (1,000 replicates). Strains of Lineage 1 are colored black, while strains of Lineage 2 are colored blue. Amino acid variability in pos. 1 of the AP molecule is indicated (Arg, blue; Tyr, red; Trp, orange; Lys, lime; unknown aa, green; Phe, black).
Non-synonymous polymorphism within the binding site (core motif A4 and A6) resulted in 21 genotypes (a-u, max. 13.8% aa variability). Notably, different combinations of point substitutions were correlated to the same amino acid substrate activation across all phylogenetic Lineages 1 (1A) and 2 (2A), (Table 2). The bioinformatic predictions of A-domain selectivity (Rausch et al., 2005) resulted in 12 different specificity-conferring codes (Table 2). In addition to the ApnA A1-genotypes (a-c) related to Arg only in exocyclic pos. 1 of the AP molecule, ApnA A1-genotypes (i-t) were related to the co-occurrence of AP structural variants carrying Arg and Tyr in pos. 1 as described previously (Christiansen et al., 2011). Another group of ApnA A1-genotypes (f-h) was related to the co-occurrence of three amino acids Arg, Tyr, Lys in exocyclic pos. 1 of the AP molecule. As part of the specificity-conferring code, pos. 322 (Stachelhaus et al., 1999), located in the core motif A5 (NxYGPTE), is known to play a role in substrate selection (Stachelhaus et al., 1999; Challis et al., 2000). In Planktothrix strains, we found a substitution of Ala or Val vs. Gly at this position (pos. 298 of ApnA in reference genome NIVA-CYA 126/8).
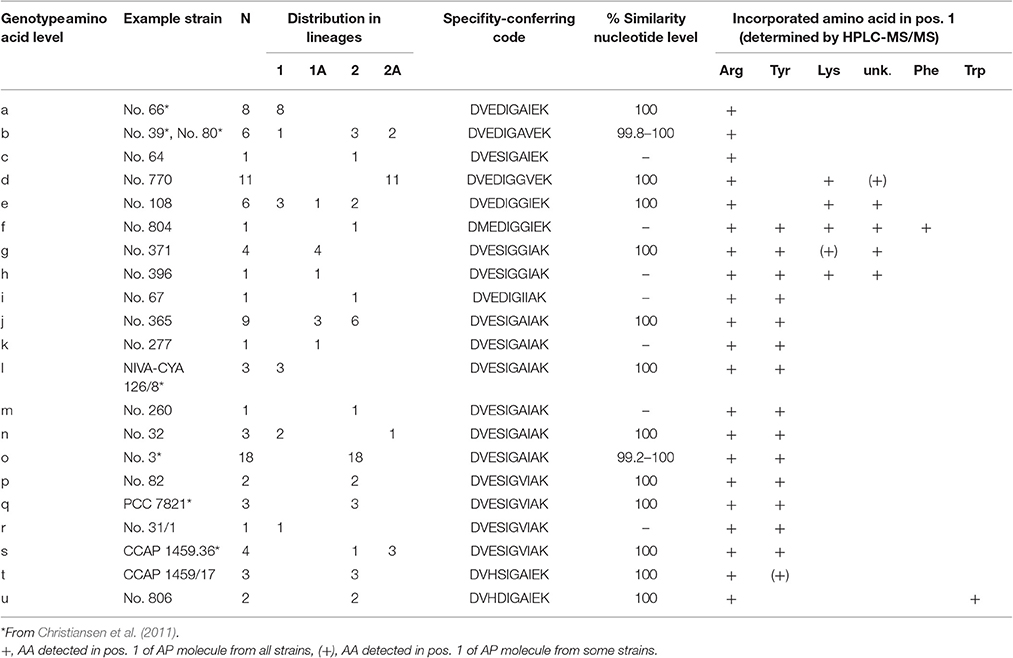
Table 2. ApnA A1 genotypes (core motif A4–A6) and specificity-conferring codes according to Stachelhaus et al. (1999).
The ratio of non-synonymous (dN) and synonymous (dS) apnA A1 substitution rates per codon site for the entire phylogenetic tree (Figure 6) was dN/dS = 0.22. Site models revealed a small number of positively selected sites (W277I/L, S278D, A322/V/I/G, and E331A, numbering following Stachelhaus et al., 1999), (One ratio site model, M7 vs. M8, 2Δl = 6.32, df = 2, p < 0.05; Supplemental Figure S9). These sites were partly found critical for the bispecificity of the A-domain by causing a conformational change (Kaljunen et al., 2015; Ser243, Ala307).
In summary, the close match between bioinformatic prediction, dN/dS site model prediction, and comparison with crystallographic analysis implies that the evolution of apnA A1 genotypes occurred toward increased structural diversity through a small number of positively selected point mutations and independently from phylogenetic association.
Discussion
Evolution of the Anabaenopeptin Peptide Synthesis Gene Cluster
The apnA-E genes from Planktothrix were notably found highly similar to the corresponding apnA-E genes from Microcystis strains when compared to apnA-E gene orthologues of Anabaena, Nodularia, Nostoc (Figure 4A). As opposed to microcystin synthesis (mcy) genes, the high similarity within Microcystis and Planktothrix among apnA-E genes and gene operon structure supports the hypothesis of a HGT event. Furthermore, even the flanking region at the 5′end of the apnA-E gene cluster (4,370 bp) was found to be identical between Planktothrix and Microcystis PCC 9432. As both genera share a planktonic lifestyle and frequently co-occur in high numbers in shallow polymictic habitats of the temperate climatic zone, such HGT events appear reasonable. In addition, no evidence for apnA-E gene cluster remnants in the more distantly related strains P. mougeotii or P. tepida assigned to Lineage 3 were found (Figure 2). In contrast, all of the strains of Lineage 1 lacking the apnA-E gene cluster (i.e., PCC 7805) carried remnants of the apnA-E gene cluster 5′end flanking region. It is thus concluded that strains of Lineage 1 showing this flanking region actually lost the apnA-E gene cluster. Similar to the loss process of microcystin synthesis in Planktothrix strains (Christiansen et al., 2008), this apn gene cluster loss event did not lead to phylogenetic separation but rather strains carrying or lacking the apn genes co-occurred in the same habitat (i.e., strains no. 263, 274 vs. no. 259, 281 isolated from Lake Wannsee, Berlin, Germany).
Recently, Calteau et al. (2014) compared 126 genomes of different cyanobacteria strains and discovered a patchy distribution of various NRPS/polyketide synthase (PKS) gene clusters throughout the whole cyanobacteria phylum. The authors concluded that the history of different NRPS gene clusters can differ and while some of these gene clusters seemed to be transmitted through HGT (as inferred from mobile flanking regions or nucleotide composition), others revealed complex vertical gene cluster evolution (e.g., gene duplication, recombination, inversion, and gene loss). It might be hypothesized that within a specific genus, such as Planktothrix, the history and relative age of various peptide synthesis gene clusters also differ. A hypothetical early ancestor of the genus Planktothrix is unlikely to contain all the different NRPS gene clusters, which would lead to the co-occurrence of at least 9 different NRPS or PKS or NRPS/PKS hybrid or RiPP gene clusters (microcystin, AP, cyanopeptolin, aeruginosin, oscillaginin, microviridin, prenylagaramide, planktocyclin, and luminaolide, e.g., see Ueoka et al., 2015; Kurmayer et al., 2016). Thus, the probability that only vertically inherited gene loss processes resulted in the today's secondary metabolite structural diversity is low. In contrast to vertical inheritance, we hypothesize that the apnA-E gene cluster was introduced into the chromosome of the ancestor of Lineage 1 (P. agardhii) by HGT and became phylogenetically fixed among the strains of Lineage 2 (Figure 2). As the MLSA suggests a more recent evolution of Lineage 2 out of Lineage 1 (Kurmayer et al., 2015), a genotype carrying the apn gene cluster was the hypothetical ancestor of Lineage 2. The same conclusion has been obtained for microcystin synthesis previously (Christiansen et al., 2008). It has been hypothesized by Suda et al. (2002) that red-pigmented Lineage 2 is evolutionary relatively young and, therefore, shows a higher similarity in nucleotide sequences as revealed by DNA-DNA hybridization. In contrast, the phylogenetic tree revealed by MLSA (Figure 2) did not reveal lower genetic divergence among the strains of Lineage 2 when compared with the diversity among the strains of Lineage 1. Thus, the absolute presence of microcystin and anabaenopeptin among the strains of Lineage 2 is striking and the question arises here as to whether the apn gene cluster presence is linked to the ecological diversification of Lineage 2. The first results from the comparison of the average nucleotide identity (ANI) for all of the strains with sequenced genome of Lineage 1 and 2 (Table 1) revealed high similarity within phylogenetic lineages (98.9–100% in Lineage 1 vs. 98.1–100% in Lineage 2) but lower similarity in between them (96.9–98%). In contrast, the nucleotide variability of apn genes is up to 14% among strains of Lineage 1 (no. 66, no. 365, NIVA-CYA 126/8) and only 2% among the strains of Lineage 2 (no. 758, no. 82, PCC 7821, no. 108). Thus, the 100% occurrence of the apnA-E gene cluster combined with the low variability of apn genes among Lineage 2 might indeed imply purifying selection.
Functional Consequences of A-Domain Nucleotide Variability and Structural Diversity
For the different peptide families produced by Planktothrix, structural variants with significant toxicity/bioactivity have been reported (e.g., Kurmayer et al., 2016). Thus, a common function of the different resulting products has been suggested, such as chemical defense through the inhibition of eukaryotic protein phosphatases 1 and 2A (microcystins, APs) and different proteases including metallocarboxypeptidases (APs) or serine proteases (cyanopeptolins, microviridins). In general, the high intracellular concentrations preclude extracellular functions but rather deter parasites such as chytrid fungi (Rohrlack et al., 2013), amoeba (Dirren et al., 2014), and not at least herbivorous crustaceans (Blom et al., 2006). It has been argued that microcystin and other bioactive peptides cannot inhibit the feeding activity of crustaceans, for example when using microcystin-deficient experimental mutants when compared to the wild type (Rohrlack et al., 1999). Under natural conditions, cyanobacteria grow in macroscopic colonies/filaments and predators, such as herbivorous copepods, typically feed by biting off portions of filaments that are not totally consumed (the so-called filament clipping (Schaffner et al., 1994). In addition, protozoans were fed by engulfing the tip of a filament (Dirren et al., 2014) or by breaking of filaments (e.g., the ciliate Obertrumia aurea; Posch et al., 2012). This feeding behavior on large macroscopic Planktothrix is comparable to the grazing behavior by insects on higher plants resulting in the intracellular production of protease inhibitors either found constitutively in various parts of the plant or which may be induced in response to mechanical attack (e.g., Jongsma and Bolter, 1997). Thus, individuals with a more effective chemical defense will survive and the combination of different protease inhibitors is generally considered advantageous to the producer to combat co-evolutionary responses. Intercellular cocktails of toxins/bioactive peptides may provide a better protection due to synergistic interactions but also due to the impediment of a co-evolutionary response in aquatic predators (Schwarzenberger and Von Elert, 2013). In this study, except for the strain CCAP 1459/31, all the strains carrying the apnA-E gene cluster produced at least one AP structural variant. Since all AP producers contained AP carrying Arg in pos. 1, we think that this structural AP variant is the original one. Notably, high inhibitory activity of AP variants with Arg in exocyclic pos. 1 has been reported for carboxypeptidase B, i.e., the activity drops in the order Arg = Lys > Tyr > Phe ≈ Ile (Schreuder et al., 2016). For carboxypeptidase A rather a reverse order of potency has been reported, with aromatic/aliphatic aa in exocyclic pos. 1 showing high activity and Arg with lower activity (references in Schreuder et al., 2016). Thus, it might be that natural enzymatic targets of the most abundant AP B and AP F structural variants resemble more carboxypeptidase B than the frequently tested carboxypeptidase A. Nevertheless, single nucleotide polymorphism led to the co-synthesis of AP structural variants carrying either Arg/Tyr (48 strains), Arg/Lys (8 strains), Arg/Trp (2 strains), or other aa combinations in pos. 1 of the AP molecule. From crystallographic analysis, it is known that ApnA A1-domain genotypes carrying substitutions pos. 278 and pos. 331 (numbering following Stachelhaus et al., 1999) led to conformational change allowing for Arg and Tyr activation (Kaljunen et al., 2015). It remains to be shown whether Arg/Lys or Arg/Trp co-activation is enabled by a similar biochemical mechanism. Notably, even strains co-producing APs with three different amino acids (Arg/Tyr/Lys) in pos. 1 were identified. The occurrence of these potentially functional point mutations within apnA A1 domain does not relate to a specific phylogenetic lineage but is rather distributed across the entire phylogeny. The incongruence with phylogeny can be best explained by a regular exchange of shorter DNA fragments from the NRPS A-domains of different genotypes across phylogenetic lineages, as it was described earlier for similar enzymes of microcystin biosynthesis (Kurmayer and Gumpenberger, 2006). However, the high frequency of promiscuous ApnA A1-domain genotypes points to its ecological significance. In contrast, the recombination affecting the apnC-D genes, leading to the substitution of Ala vs. Hty and Phe vs. Ile in pos. 5, 6 of the AP molecule (Christiansen et al., 2011), was found much less frequent—only in three strains. Thus, this larger fragment recombination is considered selectively neutral. Accordingly, replacing all aa in pos. 1–6 in AP-type peptides (called brunsvicamides; isolated from Tychonema sp.) confirmed the essential role of D-Lys in pos. 2 and Ile in the exocyclic pos. 1 (Walther et al., 2009), but also showed that replacing aa in pos. 3–6 had no effect on carboxypeptidase A inhibitory activity. In summary, an evolutionary process leading to promiscuous ApnA A1-genotypes is plausible.
Author Contributions
EE acquired the data, performed data analysis and interpretation, and wrote parts of the present manuscript. MF and GC assisted in data acquisition, data analysis and data interpretation. LD assisted in manuscript writing. JB assisted in data analysis. RK designed the work, assisted in data analysis and interpretation, and the writing and submission of the present manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Maria Reischauer, Katharina Moosbrugger, and Anneliese Wiedlroither for the excellent assistance that they provided in the laboratory. Rosemarie Rippka assisted in purification of the genome-sequenced strains. Many colleagues are acknowledged for providing strain materials and/or water samples (Reyhan Akcalaan, Olga Babanazarova, Sandra Barnard, David Bird, Alessandra Giani, Frederico Marrone, Luigi Naselli-Flores, Sergio Paulino, and Linda Tonk). We thank two reviewers and the editor for comments on the submitted manuscript. The research has been financed by the Austrian Science Fund (FWF), P24070 to RK, and EE is a recipient of a DOC Fellowship of the Austrian Academy of Sciences. This paper is a contribution to the European Cooperation in Science and Technology, COST Action ES 1105 “CYANOCOST - Cyanobacterial blooms and toxins in water resources: Occurrence, impacts, and management.”
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00219/full#supplementary-material
References
Blom, J. F., Baumann, H. I., Codd, G. A., and Jüttner, F. (2006). Sensitivity and adaptation of aquatic organisms to oscillapeptin J and [D-Asp3,(E)-Dhb7]microcystin-RR. Arch. Hydrobiol. 167, 547–559. doi: 10.1127/0003-9136/2006/0167-0547
Calteau, A., Fewer, D. P., Latifi, A., Coursin, T., Laurent, T., Jokela, J., et al. (2014). Phylum-wide comparative genomics unravel the diversity of secondary metabolism in cyanobacteria. BMC Genomics 15:977. doi: 10.1186/1471-2164-15-977
Challis, G. L., Ravel, J., and Townsend, C. A. (2000). Predictive, structure-based model of amino acid recognition by nonribosomal peptide synthetase adenylation domains. Chem. Biol. 7, 211–224. doi: 10.1016/S1074-5521(00)00091-0
Christiansen, G., Goesmann, A., and Kurmayer, R. (2014). Elucidation of insertion elements carried on plasmids and in vitro construction of shuttle vectors from the toxic cyanobacterium Planktothrix. Appl. Environ. Microbiol. 80, 4887–4897. doi: 10.1128/aem.01188-14
Christiansen, G., Molitor, C., Philmus, B., and Kurmayer, R. (2008). Nontoxic strains of cyanobacteria are the result of major gene deletion events induced by a transposable element. Mol. Biol. Evol. 25, 1695–1704. doi: 10.1093/molbev/msn120
Christiansen, G., Philmus, B., Hemscheidt, T., and Kurmayer, R. (2011). Genetic variation of adenylation domains of the anabaenopeptin synthesis operon and the evolution of substrate promiscuity. J. Bacteriol. 193, 3822–3831. doi: 10.1128/JB.00360-11
Dirren, S., Salcher, M. M., Blom, J. F., Schweikert, M., and Posch, T. (2014). Menage-a-trois: the amoeba Nuclearia sp. from Lake Zurich with its ecto- and endosymbiotic bacteria. Protist 165, 745–758. doi: 10.1016/j.protis.2014.08.004
Fastner, J., Erhard, M., and von Dohren, H. (2001). Determination of oligopeptide diversity within a natural population of Microcystis spp. (cyanobacteria) by typing single colonies by matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol. 67, 5069–5076. doi: 10.1128/aem.67.11.5069-5076.2001
Fujii, K., Sivonen, K., Adachi, K., Noguchi, K., Sano, H., Hirayama, K., et al. (1997). Comparative study of toxic and non-toxic cyanobacterial products: novel peptides from toxic Nodularia spumigena AV1. Tetrahedron Lett. 38, 5525–5528. doi: 10.1016/S0040-4039(97)01192-1
Fujii, K., Sivonen, K., Nakano, T., and Harada, K.-I. (2002). Structural elucidation of cyanobacterial peptides encoded by peptide synthetase gene in Anabaena species. Tetrahedron 58, 6863–6871. doi: 10.1016/S0040-4020(02)00747-0
Gaget, V., Welker, M., Rippka, R., and de Marsac, N. T. (2015). A polyphasic approach leading to the revision of the genus Planktothrix (Cyanobacteria) and its type species, P. agardhii, and proposal for integrating the emended valid botanical taxa, as well as three new species, Planktothrix paucivesiculata sp. nov.ICNP, Planktothrix tepida sp. nov.ICNP, and Planktothrix serta sp. nov.ICNP, as genus and species names with nomenclatural standing under the ICNP. Syst. Appl. Microbiol. 38, 141–158. doi: 10.1016/j.syapm.2015.02.004
Gkelis, S., Lanaras, T., and Sivonen, K. (2015). Cyanobacterial toxic and bioactive peptides in freshwater bodies of Greece: concentrations, occurrence patterns, and implications for human health. Mar. Drugs 13, 6319–6335. doi: 10.3390/md13106319
Halland, N., Bronstrup, M., Czech, J., Czechtizky, W., Evers, A., Follmann, M., et al. (2015). Novel small molecule inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa) from natural product anabaenopeptin. J. Med. Chem. 58, 4839–4844. doi: 10.1021/jm501840b
Halstvedt, C. B., Rohrlack, T., Ptacnik, R., and Edvardsen, B. (2008). On the effect of abiotic environmental factors on production of bioactive oligopeptides in field populations of Planktothrix spp. (Cyanobacteria). J. Plankton Res. 30, 607–617. doi: 10.1093/plankt/fbn025
Harada, K.-I., Fujii, K., Shimada, T., Suzuki, M., Sano, H., Adachi, K., et al. (1995). Two cyclic peptides, anabaenopeptins, a third group of bioactive compounds from the cyanobacterium Anabaena flos-aquae NRC 525-17. Tetrahedron Lett. 36, 1511–1514. doi: 10.1016/0040-4039(95)00073-L
Jongsma, M. A., and Bolter, C. (1997). The adaptation of insects to plant protease inhibitors. J. Insect Physiol. 43, 885–895. doi: 10.1016/S0022-1910(97)00040-1
Kaljunen, H., Schiefelbein, S. H., Stummer, D., Kozak, S., Meijers, R., Christiansen, G., et al. (2015). Structural elucidation of the bispecificity of A-domains as a basis for activating non-natural amino acids. Angew. Chem. Int. Ed. Engl. 54, 8833–8836. doi: 10.1002/anie.201503275
Kosol, S., Schmidt, J., and Kurmayer, R. (2009). Variation in peptide net production and growth among strains of the toxic cyanobacterium Planktothrix spp. Eur. J. Phycol. 44, 49–62. doi: 10.1080/09670260802158659
Kurmayer, R., Blom, J. F., Deng, L., and Pernthaler, J. (2015). Integrating phylogeny, geographic niche partitioning and secondary metabolite synthesis in bloom-forming Planktothrix. ISME J. 9, 909–921. doi: 10.1038/ismej.2014.189
Kurmayer, R., Deng, L., and Entfellner, E. (2016). Role of toxic and bioactive secondary metabolites in colonization and bloom formation by filamentous cyanobacteria Planktothrix. Harmful Algae 54, 69–86. doi: 10.1016/j.hal.2016.01.004
Kurmayer, R., and Gumpenberger, M. (2006). Diversity of microcystin genotypes among populations of the filamentous cyanobacteria Planktothrix rubescens and Planktothrix agardhii. Mol. Ecol. 15, 3849–3861. doi: 10.1111/j.1365-294X.2006.03044.x
Meyer, F., Goesmann, A., McHardy, A. C., Bartels, D., Bekel, T., Clausen, J., et al. (2003). GenDB – an open source genome annotation system for prokaryote genomes. Nucl. Acids Res. 31, 2187–2195. doi: 10.1093/nar/gkg312
Okumura, H. S., Philmus, B., Portmann, C., and Hemscheidt, T. K. (2009). Homotyrosine-containing cyanopeptolins 880 and 960 and anabaenopeptins 908 and 915 from Planktothrix agardhii CYA 126/8. J. Nat. Prod. 72, 172–176. doi: 10.1021/np800557m
Pearson, L. A., Hisbergues, M., Börner, T., Dittmann, E., and Neilan, B. A. (2004). Inactivation of an ABC transporter gene, mcyH, results in loss of microcystin production in the cyanobacterium Microcystis aeruginosa PCC7806. Appl. Environ. Microbiol. 70, 6370–6378. doi: 10.1128/AEM.70.11.6370-6378.2004
Philmus, B., Christiansen, G., Yoshida, W., and Hemscheidt, T. (2008). Posttranslational modification in microviridin biosynthesis. Chembiochem 9, 3066–3073. doi: 10.1002/cbic.200800560
Posch, T., Koster, O., Salcher, M. M., and Pernthaler, J. (2012). Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nat. Clim. Change 2, 809–813. doi: 10.1038/nclimate1581
Rausch, C., Weber, T., Kohlbacher, O., Wohlleben, W., and Huson, D. (2005). Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucl. Acids Res. 33, 5799–5808. doi: 10.1093/nar/gki885
Reshef, V., and Carmeli, S. (2002). Schizopeptin 791, a new anabeanopeptin-like cyclic peptide from the cyanobacterium Schizothrix sp. J. Nat. Prod. 65, 1187–1189. doi: 10.1021/np020039c
Rippka, R. (1988). Isolation and purification of cyanobacteria. Meth. Enzymol. 167, 3–27. doi: 10.1016/0076-6879(88)67004-2
Rohrlack, T., Christiansen, G., and Kurmayer, R. (2013). Putative antiparasite defensive system involving ribosomal and nonribosomal oligopeptides in cyanobacteria of the genus Planktothrix. Appl. Environ. Microbiol. 79, 2642–2647. doi: 10.1128/aem.03499-12
Rohrlack, T., Dittmann, E., Henning, M., Börner, T., and Kohl, J.-G. (1999). Role of microcystins in poisoning and food ingestion inhibition of Daphnia galeata caused by the cyanobacterium Microcystis aeruginosa. Appl. Environ. Microbiol. 65, 737–739.
Rounge, T. B., Rohrlack, T., Nederbragt, A. J., Kristensen, T., and Jakobsen, K. S. (2009). A genome-wide analysis of nonribosomal peptide synthetase gene clusters and their peptides in a Planktothrix rubescens strain. BMC Genomics 10:396. doi: 10.1186/1471-2164-10-396
Rounge, T. B., Rohrlack, T., Tooming-Klunderud, A., Kristensen, T., and Jakobsen, K. S. (2007). Comparison of cyanopeptolin genes in Planktothrix, Microcystis, and Anabaena strains: evidence for independent evolution within each genus. Appl. Environ. Microbiol. 73, 7322–7330. doi: 10.1128/AEM.01475-07
Sano, T., and Kaya, K. (1995). Oscillamide Y, a chymotrypsin inhibitor from toxic Oscillatoria agardhii. Tetrahedron Lett. 36, 5933–5936. doi: 10.1016/0040-4039(95)01198-Q
Sano, T., Usui, T., Ueda, K., Osada, H., and Kaya, K. (2001). Isolation of new protein phosphatase inhibitors from two cyanobacteria species, Planktothrix spp. J. Nat. Prod. 64, 1052–1055. doi: 10.1021/np0005356
Schaffner, W. R., Hairston, N. G., and Howarth, R. W. (1994). Feeding rates and filament clipping by crustacean zooplankton consuming cyanobacteria. Verh. Int. Verein. Limnol. 25, 2375–2381.
Schreuder, H., Liesum, A., Lönze, P., Stump, H., Hoffmann, H., Schiell, M., et al. (2016). Isolation, co-crystallization and structure-based characterization of anabaenopeptins as highly potent inhibitors of activated thrombin activatable fibrinolysis inhibitor (TAFIa). Sci. Rep. 6:32958. doi: 10.1038/srep32958
Schwarzenberger, A., and Von Elert, E. (2013). Cyanobacterial protease inhibitors lead to maternal transfer of increased protease gene expression in Daphnia. Oecologia 172, 11–20. doi: 10.1007/s00442-012-2479-5
Shin, H. J., Matsuda, H., Murakami, M., and Yamaguchi, K. (1997). Anabaenopeptins E and F, two new cyclic peptides from the cyanobacterium Oscillatoria agardhii (NIES 204). J. Nat. Prod. 60, 139–141. doi: 10.1021/np0005356
Spoof, L., Blaszczyk, A., Meriluoto, J., Ceglowska, M., and Mazur-Marzec, H. (2016). Structures and activity of new anabaenopeptins produced by Baltic Sea cyanobacteria. Mar. Drugs 14:8. doi: 10.3390/md14010008
Stachelhaus, T., Mootz, H. D., and Marahiel, M. A. (1999). The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem. Biol. 6, 493–505. doi: 10.1016/S1074-5521(99)80082-9
Suda, S., Watanabe, M. M., Otsuka, S., Mahakahant, A., Yongmanitchai, W., Nopartnaraporn, N., et al. (2002). Taxonomic revision of water-bloom-forming species of oscillatorioid cyanobacteria. Int. J. Syst. Evol. Microbiol. 52, 1577–1595. doi: 10.1099/00207713-52-5-1577
Ueoka, R., Uria, A. R., Reiter, S., Mori, T., Karbaum, P., Peters, E. E., et al. (2015). Metabolic and evolutionary origin of actin-binding polyketides from diverse organisms. Nat. Chem. Biol. 11, 705–712. doi: 10.1038/nchembio.1870
Walther, T., Renner, S., Waldmann, H., and Arndt, H. D. (2009). Synthesis and structure-activity correlation of a brunsvicamide-inspired cyclopeptide collection. Chembiochem 10, 1153–1162. doi: 10.1002/cbic.200900035
Williams, D. E., Craig, M., Holmes, C. F. B., and Andersen, R. J. (1996). Ferintoic acids A and B, new cyclic hexapeptides from the freshwater cyanobacterium Microcystis aeruginosa. J. Nat. Prod. 59, 570–575. doi: 10.1021/np960108l
Yang, Z. (1997). PAML: a program for package for phylogenetic analysis by maximum likelihood. Comp. Appl. Biosci. 13, 555–556.
Yang, Z., Wong, W. S., and Nielsen, R. (2005). Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 22, 1107–1118. doi: 10.1093/molbev/msi097
Keywords: cyanotoxins, cyanoHABs, natural products, chemotype, ecotype, speciation, horizontal gene transfer, gene deletion
Citation: Entfellner E, Frei M, Christiansen G, Deng L, Blom J and Kurmayer R (2017) Evolution of Anabaenopeptin Peptide Structural Variability in the Cyanobacterium Planktothrix. Front. Microbiol. 8:219. doi: 10.3389/fmicb.2017.00219
Received: 29 October 2016; Accepted: 31 January 2017;
Published: 16 February 2017.
Edited by:
Marcelino T. Suzuki, Sorbonne Universities (UPMC) and Centre National de la Recherche Scientifique, FranceReviewed by:
Assaf Sukenik, Israel Oceanographic & Limnological Research, IsraelGabor Vasas, University of Debrecen, Hungary
Copyright © 2017 Entfellner, Frei, Christiansen, Deng, Blom and Kurmayer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rainer Kurmayer, cmFpbmVyLmt1cm1heWVyQHVpYmsuYWMuYXQ=
 Elisabeth Entfellner1
Elisabeth Entfellner1 Jochen Blom
Jochen Blom Rainer Kurmayer
Rainer Kurmayer