- 1Department of Food Science, Faculty of Food Science and Technology, Universiti Putra Malaysia, Selangor, Malaysia
- 2Department of Livestock and Avian Science, Faculty of Livestock, Fisheries and Nutrition, Wayamba University of Sri Lanka, Makandura, Sri Lanka
- 3Center for Southeast Asian Studies, Kyoto University, Kyoto, Japan
- 4Laboratory of Food Safety and Food Integrity, Institute of Tropical Agriculture and Food Security, Universiti Putra Malaysia, Selangor, Malaysia
Numerous prevalence studies and outbreaks of Vibrio parahaemolyticus infection have been extensively reported in shellfish and crustaceans. Information on the quantitative detection of V. parahaemolyticus in finfish species is limited. In this study, short mackerels (Rastrelliger brachysoma) obtained from different retail marketplaces were monitored with the presence of total and pathogenic strains of V. parahaemolyticus. Out of 130 short mackerel samples, 116 (89.2%) were detected with the presence of total V. parahaemolyticus and microbial loads of total V. parahaemolyticus ranging from <3 to >105 MPN/g. Prevalence of total V. parahaemolyticus was found highest in wet markets (95.2%) followed by minimarkets (89.1%) and hypermarkets (83.3%). Pathogenic V. parahaemolyticus strains (tdh+ and/or trh+) were detected in 16.2% (21 of 130) of short mackerel samples. The density of tdh+ V. parahaemolyticus strains were examined ranging from 3.6 to >105 MPN/g and microbial loads of V. parahaemolyticus strains positive for both tdh and trh were found ranging from 300 to 740 MPN/g. On the other hand, antibiotic susceptibility profiles of V. parahaemolyticus strains isolated from short mackerels were determined through disc diffusion method in this study. Assessment of antimicrobial susceptibility profile of V. parahaemolyticus revealed majority of the isolates were highly susceptible to ampicillin sulbactam, meropenem, ceftazidime, and imipenem, but resistant to penicillin G and ampicillin. Two isolates (2.99%) exhibited the highest multiple antibiotic resistance (MAR) index value of 0.41 which shown resistance to 7 antibiotics. Results of the present study demonstrated that the occurrence of pathogenic V. parahaemolyticus strains in short mackerels and multidrug resistance of V. parahaemolyticus isolates could be a potential public health concerns to the consumer. Furthermore, prevalence data attained from the current study can be further used to develop a microbial risk assessment model to estimate health risks associated with the consumption of short mackerels contaminated with pathogenic V. parahaemolyticus.
Introduction
Vibrio parahaemolyticus is found naturally in the marine environment and may accumulate in seafood especially shellfish at high concentrations. Consumption of foods contaminated with high concentration of total V. parahaemolyticus and/or pathogenic V. parahaemolyticus can cause gastrointestinal infections. An open wound in skin comes in contact with V. parahaemolyticus is suggested as an infection pathway as well. Major syndromes caused by V. parahaemolyticus include gastroenteritis, wound infection, and septicaemia. Typical symptoms of the gastroenteritis may include abdominal pain, diarrhea, nausea, and fever. Based on the Foodborne Diseases Active Surveillance Network (FoodNet) data and Morbidity and Mortality Weekly Report (MMWR) published by Centers for Disease Control and Prevention (CDC) in the United States during the year 2016, V. parahaemolyticus is the major foodborne pathogen compared to other Vibrio isolates and it is estimated V. parahaemolyticus causes about 34,664 episodes of domestically acquired foodborne illness annually in the United States (Scallan et al., 2011; Huang et al., 2016).
In term of pathogenicity, thermostable direct hemolysin (TDH) and thermostable direct-related hemolysin (TRH) are the well-known virulence factors in V. parahaemolyticus. TDH is a pore-forming and heat stable protein which remains unharmed even heating at 100°C for 10 min (Yanagihara et al., 2010). Two distinct biological characteristics of TDH are the capability of inducing hemolysis and cytotoxicity (Baba et al., 1992; Honda et al., 1992; Fabbri et al., 1998; Raimondi et al., 2000). TRH was first identified and isolated from the gastroenteritis outbreak in Maldives in 1985 (Honda et al., 1988). The isolated V. parahaemolyticus strains from the outbreak shown Kanagawa negative on Wagatsuma agar and did not possess the TDH genes (Nishibuchi et al., 1989). Unlike TDH thermostability, TRH is a heat labile protein at 60°C for 10 min (Honda et al., 1988). TRH shares similar biological, immunological, and physicochemical characteristics with TDH have been described (Honda and Iida, 1993). For instance, TRH induces Cl− secretion in cultured human colonic epithelial cells by a similar mechanism with TDH (Takahashi et al., 2000).
Antimicrobial resistance (AMR) has now been recognized as a significant global threat issue to global public health and food security (FAO, 2016). Many frequently used antibiotics are no longer effective to control infections. Extensive use and misuse of antibiotics in agriculture, aquaculture, and livestock production are considered to be the main factor leads to the emergence and spread of AMR. Multidrug-resistant (MDR) bacterial strain is another emerging challenge when a bacterial cell becomes resistant toward multiple antibiotics. Development of MDR bacterial strains can be achieved through several mechanisms such as chromosomal DNA mutations, enzymatic inactivation, transformation as well as conjugation (Van Hoek et al., 2011). Antimicrobial residues present in the environment may also resultant in selection pressure for AMR bacteria. In aquaculture, antibiotics are used in fish farming to promote the growth of aquatic organisms and control bacterial infections. Administration of antibiotics into feed and water is a usual practice to improve production growth and for the treatment of infection diseases caused by pathogenic bacteria. Several Vibrio species are known to cause infections in aquatic fish and extensive use of antibiotics in the past have now led to significant increases in the occurrence of AMR Vibrio species (Letchumanan et al., 2015b).
Quinolones, cephalosporins, tetracycline, cefotaxime, ceftazidime, and penicillins are some commonly recommended clinical antibiotics used for the treatment of non-cholerae Vibrio spp. infections (Han et al., 2007; Wong et al., 2015). The use of quinolone is generally effective to against all Vibrio species; while the use of cephalosporin and tetracycline antibiotics was reported ineffective and associated with higher mortality in patients with vibriosis (Wong et al., 2015). In aquaculture industry, tetracyclines, erythromycin, sulfonamides, oxytetracyclines, chlortetracycline, and amoxicilin are allowed to be used in some of the ASEAN countries including Malaysia, Myanmar, and Philippines; while other antimicrobial drugs such as nitrofurans, chloramphenicol, and dimetridazole/metronidazole are banned in most countries (ASEAN, 2013; Weese et al., 2015). For controlling Vibrio spp., sulfonamides sold under the trade name Dimeton is an example of veterinary antibiotic commonly used in hatcheries to control infections (Shariff et al., 2000).
Short mackerel (Rastrelliger brachysoma) is an important commercial small pelagic fish species and have a high preference among consumers because of its affordable price and widely available throughout the year in many of the Southeast Asia countries including Malaysia, Thailand, Cambodia, Philippines, and Indonesia. Numerous prevalence studies and outbreaks of V. parahaemolyticus infection have been extensively reported in oysters, clams, cockles, mussels, crabs, and shrimps (Newton et al., 2014; Rodgers et al., 2014; Xu et al., 2014; Malcolm et al., 2015). In finfish species, qualitative detection of V. parahaemolyticus in the anchovies (Engraulis spp.), gray mullet (Mugil cephalus), red mullet (Mullus surmuletus), sardines (Sardina spp.), and Atlantic mackerel (Scomber scombrus) have been examined by Baffone et al. (2000) with the used of selective medium and biochemical tests. Hara-kudo et al. (2003) examined 15 horse mackerels purchased from the marketplaces in Japan, for tdh positive and total V. parahaemolyticus by using PCR and CHROMagar Vibrio (CV) agar. Reports focused on the detection and enumeration of V. parahaemolyticus in finfish species such as mackerel is very limited in extent. Available data on the quantitative detection of V. parahaemolyticus in different parts of the mackerel's body can only be found in one study reported in a Japanese-language literature (Ohno et al., 1993). Hence, the main purpose of this study was to determine the prevalence of total and pathogenic V. parahaemolyticus in short mackerels purchased from different wet markets, hypermarkets, and minimarkets in the state of Selangor of Malaysia by using MPN-PCR methods and to evaluate the antibiotic susceptibility profiles of V. parahaemolyticus isolates obtained from the short mackerels.
Materials and Methods
Sample Collection
A total of 130 short mackerel (R. brachysoma) samples were collected from 67 sampling date within a period of 6 months from Jan 2016 to June 2016 in four wet markets (n = 42), five hypermarkets (n = 42), and five minimarkets (n = 46) in Selangor, Malaysia. A maximum of two short mackerels were collected at each sampling date from the sampling location. All the samples were transported to the laboratory and processed immediately on the day of sampling.
Sample Processing and Most Probable Number (MPN) Method
Flesh (with skin), gills, and intestines excised from each short mackerel were used as the test sample. A total of 390 test samples including of 130 fleshes, 130 gills, and 130 intestines were analyzed in this study. Ten grams of flesh was transferred into 90 ml of alkaline peptone water (APW) (Merck, Darmstadt, Germany) in a sterile stomacher bag. One gram of gills and one gram of intestines, each was transferred into 9 ml of APW in two separate sterile stomacher bags. Samples were homogenized for 1 min using a Stomacher Lab-Blender 400 (Seward Medical, UK). MPN preparation was followed the US FDA Bacteriological Analytical Manual (BAM) three tubes MPN methodology with some modifications (Kaysner and DePaola, 2004). Briefly, a serial dilution was carried out up to 10−5 by transferring 1 ml suspension mixture into 9 ml of APW. One milliliters of each dilution sample was pipetted into three microcentrifuge tubes and tubes were incubated at 37°C for 18–24 h.
Genomic DNA Extraction
A number of tubes with growth at each dilution was subjected to genomic DNA extraction through physical cell disruption methods. Samples with growth or turbidity were pelleted by centrifugation at 13,400 × g for 3 min. The supernatant was discarded and the remained pellet was suspended in 200 μl TE buffer. The suspension mixture was heated at 100°C for 15 min in a dry bath (Labnet, USA) and immediately kept at −20°C for another 15 min. After the heat and cold-shock cell lysis, samples were centrifuged at 13,400 × g for 1 min and the supernatant was used as DNA template for multiplex PCR.
Multiplex PCR
Multiplex PCR was performed by using 3 set of primers: (i) toxR (F: 5′-GTCTTCTGACGCAATCGTTG-3′ and R: 5′-ATACGAGTGGTTGCTGTCATG-3′) for species-specific detection of V. parahaemolyticus (Kim et al., 1999); (ii) tdh (F: 5′-CCACTACCACTCTCATATGC-3′ and R: 5′-GGTACTAAATGGCTGACATC-3′) for the detection of pathogenic tdh gene (Tada et al., 1992); and (iii) trh (F: 5′-TTGGCTTCGATATTTTCAGTATCT-3′ and R: 5′-CATAACAAACATATGCCCATTTCCG-3′) for the detection of pathogenic trh gene (Bej et al., 1999). All primers were synthesized by Sigma-Aldrich (USA) and PCR protocol was carried out according to the method as described by Malcolm et al. (2015). Briefly, PCR reagents (Promega, USA) were prepared by mixing 1.4 × PCR buffer, 2.5 mM of MgCl2, 0.2 mM of dNTPs, 0.2 μM of each primer, 2.5U of Taq polymerase, 2 μL of DNA template and top up to the final volume of 25 μL with sterilized ultrapure water. Amplification was performed in a Kyratec SuperCycler Trinity (Australia) with the following thermocycling conditions: initial denaturation at 95°C for 5 min for 1 cycle, 30 cycles consisting of denaturation at 95°C for 30 s, annealing at 60°C for 45 s, extension at 68°C for 1 min, and a final extension at 72°C for 3 min.
Antibiotic Susceptibility Test
One V. parahaemolyticus colony from each sampling date was collected for antibiotic susceptibility test. A total of 67 V. parahaemolyticus colonies isolated from short mackerel on 67 sampling date were tested for antibiotic susceptibility by using disc diffusion method. Isolates were cultured in 5 mL of Mueller-Hinton broth (Merck, Darmstadt, Germany) supplemented with 3% (w/v) of NaCl (Merck, Darmstadt, Germany) and incubated at 37°C, 120 rpm for 24 h. Inoculums were swabbed with the sterile cotton swab on the entire surface of Mueller-Hinton agar (Merck, Darmstadt, Germany) supplemented with 3% (w/v) of NaCl and left to dry for 3–5 min. Antimicrobial susceptibility test discs (Oxoid, UK) were placed on the inoculated agar plate with a disc dispenser and incubated at 37°C for 24 h. A total of 17 antimicrobial susceptibility test discs impregnated with ampicillin (10 μg), ampicillin sulbactam (20 μg), amikacin (30 μg), amoxicillin/clavulanic acid (30 μg), ceftazidime (30 μg), cefotaxime (30 μg), cephalothin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), doxycycline (30 μg), gentamicin (10 μg), imipenem (10 μg), levofloxacin (5 μg), meropenem (10 μg), penicillin G (10 unit), streptomycin (10 μg), and tetracycline (30 μg) were used in this study. Escherichia coli ATCC 25922 was used as quality control organism for this antibiotic susceptibility test. After incubation, the diameter of inhibition zone was measured in nearest whole millimeter. Antibiotic susceptibility profile of the isolate was interpreted as sensitivity, intermediate, and resistance based on the Clinical and Laboratory Standards Institute (CLSI) M45 guideline for Vibrio spp. (CLSI, 2010). Interpretive criteria for doxycycline, streptomycin and penicillin G not available in the M45 guidelines was referred to CLSI M100 guideline (CLSI, 2016). Multiple antibiotic resistance (MAR) index value was calculated according to Krumperman (1983) by using the formula, a/b, where “a” is the number of antibiotics to which the particular isolate was resistant and “b” is the total number of antibiotics tested.
Statistical Analysis
Statistically significant differences among the sample microbial loads and sampling locations were analyzed with analysis of variance (ANOVA) tests using Minitab statistical package version 16.2 (Minitab Inc., State College, PA). The level of significance was set at P ≤ 0.05. Significant differences between the microbial concentrations in the flesh, gills, and intestines of short mackerel were examined as well.
Results
Total V. parahaemolyticus in Short Mackerels
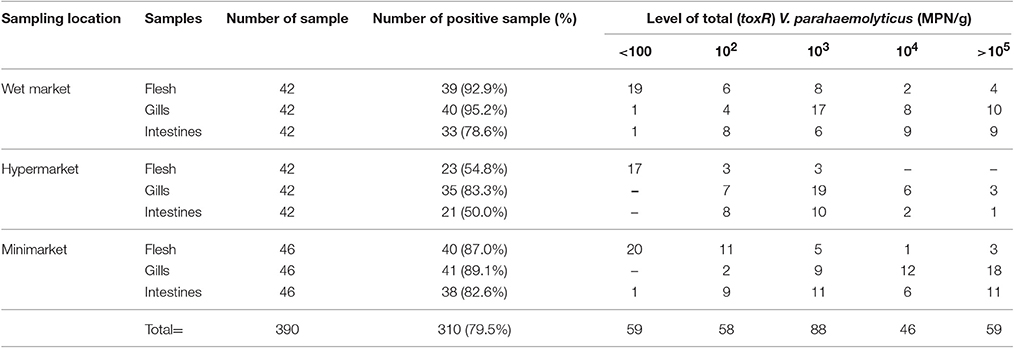
Prevalence and microbial loads of total V. parahaemolyticus in short mackerels are summarized in Table 1. DNA fragments of 368 bp in size were produced from the amplification of V. parahaemolyticus species-specific gene (toxR) indicating the presence of total V. parahaemolyticus. Out of 390 short mackerel tested samples, 310 (79.5%) samples were detected with the presence of total V. parahaemolyticus (toxR) and the microbial loads was between <3 to >105 MPN/g. Highest prevalence of total V. parahaemolyticus was detected in the samples obtained from wet markets (78.6–95.2%), followed by the minimarkets (82.6–89.1%) and hypermarkets (50.0–83.3%). Highest density of total V. parahaemolyticus were found in short mackerel gills with mean concentration of 3.66 log MPN/g, followed by intestines with mean concentration of 2.67 log MPN/g and flesh with mean concentration of 1.74 log MPN/g. Besides, 33.9% (105/310) of the samples were detected with high levels (≥104 MPN/g) of total V. parahaemolyticus.
Overall, the prevalence of total V. parahaemolyticus in short mackerel whole-body was 89.2% (116/130). A total of 95.2% (40/42), 89.1% (41/46), and 83.3% (35/42) of the short mackerel samples obtained from the wet markets, minimarkets, and hypermarkets, respectively, were detected with the presence of total V. parahaemolyticus. For statistical analyses, samples collected from the hypermarkets was detected significant lower (P < 0.05) from the wet markets and minimarkets. The level of total V. parahaemolyticus in short mackerel flesh, gills, and intestines were also showed significantly different (P < 0.05) from each other.
Pathogenic V. parahaemolyticus Strains in Short Mackerels
Prevalence and microbial loads of pathogenic V. parahaemolyticus (tdh+ and/or trh+) strains in short mackerels are shown in Tables 2, 3. DNA fragments of 484 and 251 bp in size were produced from the amplification of V. parahaemolyticus pathogenic trh and tdh genes, respectively. A total of 33 out of 390 (8.5%) tested samples were detected positive for the tdh gene (tdh+ and trh−) and the microbial loads ranged from 3.6 to >105 MPN/g. Strains of V. parahaemolyticus harbored both the tdh and trh genes (tdh+ and trh+) were detected in 4 out of 390 (1.0%) tested samples and the density of tdh+ and trh+ V. parahaemolyticus strains found in the samples ranged from 300 to 740 MPN/g. V. parahaemolyticus strain carrying only the trh gene (trh+ and tdh-) was not detected in this study.
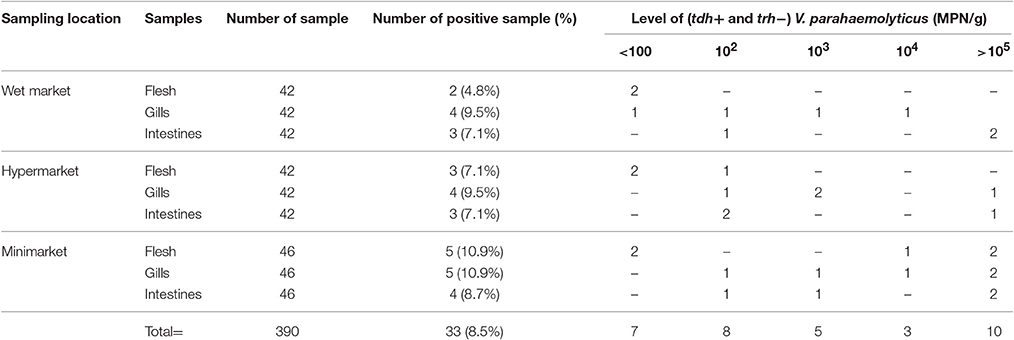
Table 2. Prevalence and microbial loads of pathogenic tdh+ and trh− V. parahaemolyticus strains in short mackerels.
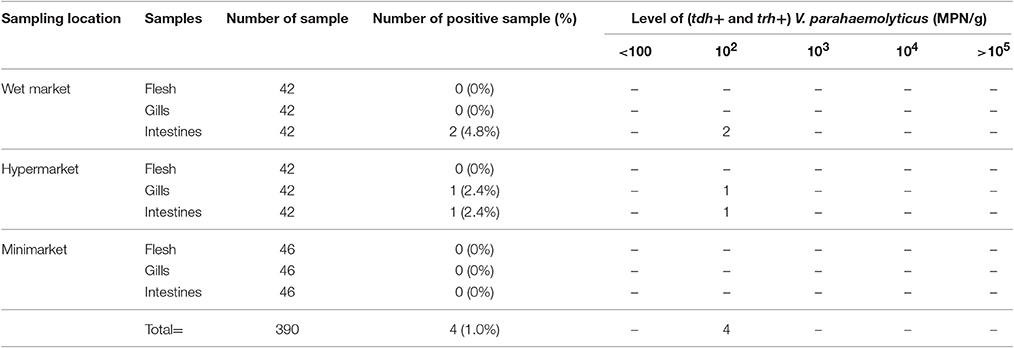
Table 3. Prevalence and microbial loads of pathogenic tdh+ and trh+ V. parahaemolyticus strains in short mackerels.
Overall, the prevalence of pathogenic V. parahaemolyticus strain (tdh+ and/or trh+) in short mackerel whole-body was 16.2% (21/130). The presence of pathogenic V. parahaemolyticus strains can be found in either combination or alone in each part of short mackerel flesh, gills, and intestines. For statistical analyses, the level of pathogenic (tdh+ and/or trh+) V. parahaemolyticus was detected non-significantly different (P > 0.05) in various sampling locations and tested samples.
Antibiotics Susceptibility Profile of V. parahaemolyticus Isolates
Antibiotic susceptibility profiles of V. parahaemolyticus isolated from short mackerel samples are shown in Table 4. The majority of isolates tested were susceptible to all of the antibiotics. Isolates tested were highly susceptible to antibiotics such as ampicillin sulbactam (100%), meropenem (100%), ceftazidime (98.5%), and imipenem (98.5%). High level of resistance was observed to penicillin G (92.5%) and ampicillin (82.1%). Of 67 isolates, 60 (89.6%) showed resistant to two or more antibiotics. MAR index value of V. parahaemolyticus isolates from short mackerel samples are summarized in Table 5. Most of the isolates (40.3%) were detected with MAR index value of 0.12 followed by 16.4 and 13.4% of isolates with MAR index value of 0.29 and 0.24, respectively. Two (3.0%) isolates exhibited the highest MAR index value of 0.41 which shown resistance to 7 types of antibiotics.
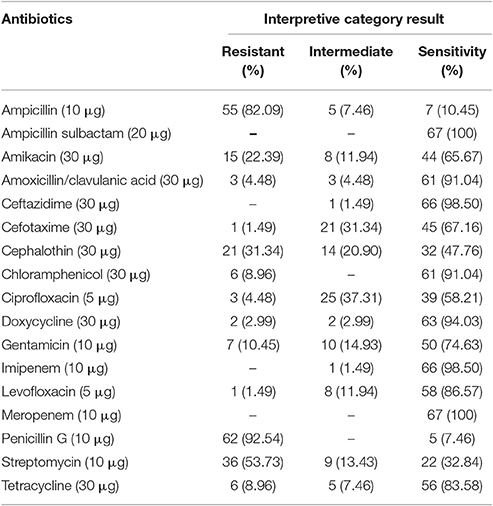
Table 4. Antibiotic susceptibility profiles of V. parahaemolyticus isolated from short mackerels by disc diffusion method.
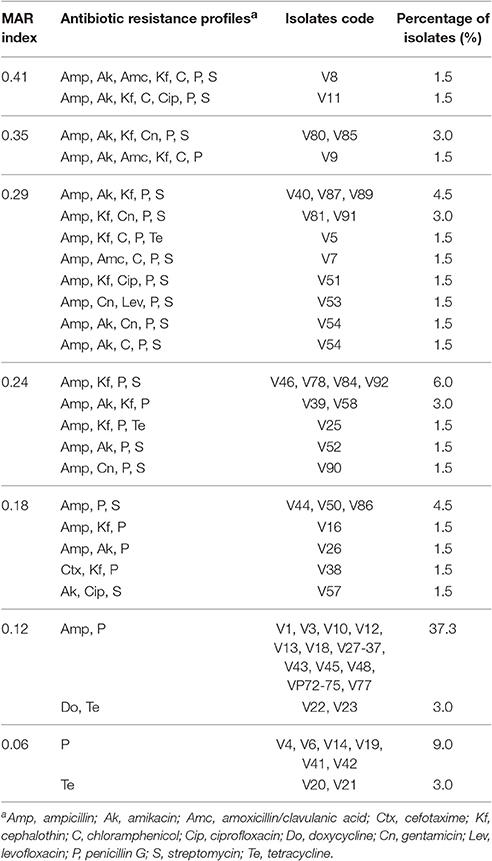
Table 5. Multiple antibiotic resistance (MAR) index value of V. parahaemolyticus isolates from short mackerel samples.
Discussion
High prevalence and microbial loads of total V. parahaemolyticus were detected in 89.2% (116/130) of short mackerel samples. Similar results were found in the Hara-kudo et al. (2003) study where 15 out of 17 (88.2%) horse mackerel samples in Japan were detected with the presence of total V. parahaemolyticus. However, the incidence rate of total V. parahaemolyticus in short mackerels was comparatively higher than many other prevalence studies in other countries. For instance, Baffone et al. (2000) revealed 2.6% (3/114) fish samples including of anchovies, gray mullet, red mullet, sardines, Atlantic mackerel, and other species common to the Adriatic Sea were detected with the presence of V. parahaemolyticus. Wong et al. (1999) reported 29.3% of a total 92 fish samples imported from Indonesia were detected with total V. parahaemolyticus. Nakaguchi (2013) reported an average of 57.4% fish samples obtained from Asia countries including Vietnam, Indonesia, and Malaysia were found with total V. parahaemolyticus. Total V. parahaemolyticus were found more prevalently in the samples obtained from wet markets (78.6–97%) followed by minimarkets (82.6–89.1%) and hypermarkets (50.0–83.3%) in this study. Similar study findings reported by Letchumanan et al. (2015a) also found a high level of V. parahaemolyticus contamination in shrimp samples purchased from wet markets compared to supermarkets. The reason for these findings may due to the poor storage handling practices by the wet market retailers. For instance, seafood especially cheaper fishes such as short mackerels in the wet markets were usually displayed on a tray with little or no ice left amongst the fish. Additionally, finfish are generally sent directly from the farm to wet markets retailers under minimum temperature controlled.
Microbial loads of total V. parahaemolyticus in short mackerel tested samples including flesh, gills, and intestines obtained from the hypermarkets were detected significant lower (P < 0.05) from the wet markets and minimarkets. The study findings are in agreement with another study reported the surf clams obtained from hypermarkets were found to contain lower density of V. parahaemolyticus (Malcolm et al., 2015). In general, seafood is extremely perishable which needs strictly controlled storage requirements to maintain their freshness and limit the growth of harmful microorganisms. In this study, samples obtained from hypermarkets were found to contain significant lower number of total V. parahaemolyticus (P < 0.05). This might due to the well-established cold chains and cold storage facilities available in the hypermarkets. Low temperature controlled in supply chains and storage facilities available in the hypermarkets which therefore help reduce the growth of total V. parahaemolyticus in short mackerels. Su and Liu (2007) also reported that V. parahaemolyticus are cold sensitive thus preserving seafood on low temperature may limit or reduce their growth.
Short mackerel gills were detected with the highest total V. parahaemolyticus microbial loads with a mean concentration of 3.66 log MPN/g compared to intestines with a mean concentration of 2.67 log MPN/g and flesh with a mean concentration of 1.74 log MPN/g. Hairy structures of gills provided large surface areas for the bacteria adsorption is suggested one of the reasons where short mackerel gills were detected with the highest density of total V. parahaemolyticus. In Malaysia, there is no standard safety limit or minimum allowable level of V. parahaemolyticus in seafood. Safety regulations and standards for fish and fishery product established from other countries such as the United States are referred. According to the FDA microbiological safety limits for V. parahaemolyticus in ready-to-eat fish products must be <1 × 104 per gram (FDA, 2011). In this study, 33.9% of short mackerels exceed the microbiological safety levels but fish evisceration, washing, rinsing, and cooking process done by food handlers during food preparation is suggested capable of achieving certain reductions of V. parahaemolyticus. For examples, Watanabe et al. (1994) demonstrated the effectiveness of washing of the horse mackerel eviscerated cavity with clean water resulted in 1.99 log reduction of V. parahaemolyticus. Ye et al. (2012) reported mild heat treatment at 50°C for 20 min was capable of reducing the number of V. parahaemolyticus to below detection limit (<3 MPN/g).
Most of the V. parahaemolyticus clinical isolates exhibit Kanagawa positive or negative possessing the hemolysin tdh and/or trh genes. Only small percentage of V. parahaemolyticus isolates from food and environmental samples carrying tdh and/or trh genes. This statement was in agreement with our findings as V. parahaemolyticus strains with tdh gene and strains with both tdh and trh genes were detected in low level, 8.5 and 1.0%, respectively, in short mackerels. No trh+ V. parahaemolyticus strains was found in all samples can be associated with the warmer climate in Malaysia as Rodriguez-Castro et al. (2010) reported that trh+ strains more prevailing in the coldest water and tdh+ V. parahaemolyticus tends to disseminate in the warmer water. Even in clinical V. parahaemolyticus strains only minority of the isolates carried the trh genes. Suthienkul et al. (1995) reported merely 10 out of 489 (2%) V. parahaemolyticus isolates isolated from patients with acute gastroenteritis possessed the trh genes and 27 out of 489 (6%) isolates harbored both the trh and tdh genes. Bhoopong et al. (2007) revealed only 0.5% (3/629) of the clinical V. parahaemolyticus isolates from the 63 patients in Thailand carried the trh gene alone, compared to 87.4% (550/629) and 7% (44/629) of the isolates possessed the tdh gene and both genes, respectively. Chen et al. (2016) reported 93% and 1% of the 501 clinical V. parahaemolyticus isolates from southeastern China were carried tdh gene and trh gene, respectively. However, distributions of tdh+ and/or trh+ strains may vary depend on the geographical region, sample source, and detection method (Raghunath, 2015).
V. parahaemolyticus isolates obtained from short mackerels displayed a high level of resistance to penicillin and ampicillin. This finding was consistent with the results reported by Letchumanan et al. (2015a) where 82% of the isolates from shrimp samples were resistant to ampicillin. Besides, Elexson et al. (2014) reported all of the V. parahaemolyticus isolates from cultured seafood products were resistant to both penicillin and ampicillin. Based on these findings, resistant to penicillin and ampicillin could be probably due to extensively used of antibiotics in aquaculture and the impact of antimicrobial residues in aquatic systems. It is also likely due to the complexity of the Gram-negative bacteria outer membrane which inhibits antibiotic compounds to pass through the outer membrane (IFT, 2006; Blair et al., 2014). Penicillin and ampicillin are therefore ineffective for the treatment of V. parahaemolyticus infections. Nevertheless, the majority of the isolates were susceptible to most antibiotics tested in this study. Susceptibility profiles of V. parahaemolyticus isolates to antibiotic classes such as tetracycline, phenicols, quinolone, and cephalosporin were comparable with many other studies reported in several countries and sample sources (Lesmana et al., 2001; Han et al., 2007; Yano et al., 2011; Ottaviani et al., 2013). In this study, 37.31% of isolates with MAR value more than 0.2 indicating samples originated from a high risk source of contamination where several antibiotics are used (Krumperman, 1983). Widespread usage of antibiotics in clinical, agriculture, aquaculture, and livestock production can result in antimicrobial residues present in the environment and dispersion of antimicrobial residues that reach the marine environment could lead to selective pressure on the marine bacteria and the emergence of MDR bacterial strains in marine life.
Misuse and overuse of antibiotics are recognized as two of the major factors contribute to the development of resistance genes in bacteria and widespread dissemination of MDR bacterial strains. An alternative to antibiotics is urgently needed in order to overcome the continuous emergence of MDR bacterial strains in the environment (Tan et al., 2016). Research on alternative approaches such as the use of bacteriophage and probiotics has become possible solution to replace or reduce the use of antibiotics. Bacteriophages are bacteria viruses with the ability to attack and destroy bacteria cells. Jun et al. (2014) demonstrated bacteriophage-based therapies displays effectual protection against multiple antibiotics resistant V. parahaemolyticus strains infection in mice. Lomelí-Ortega and Martínez-Díaz (2014) reported two isolated lytic bacteriophages can be used to control vibriosis in whiteleg shrimp larvae (Litopenaeus vannamei). Probiotics are organisms or substances that bring beneficial effect to the host (Hai, 2015). Tan et al. (2016) proposed the feed supplemented with the genus of Streptomyces bacteria as probiotics could protect aquaculture livestock from pathogens and enhance the growth performance of the aquatic cultured organisms. Augustine et al. (2016) demonstrated Streptomyces rubrolavendulae M56 biogranules showed a competitive exclusion effect on V. alginolyticus, V. parahaemolyticus, V. fluvialis, and V. harveyi can be used as a promising alternative to the use of antibiotics in the prawn larval production systems. Although the application of bacteriophages and probiotics offer promising alternative to antibiotics, efforts for continuous monitoring of V. parahaemolyticus antibiotic resistance patterns and risk assessment on the use of antibiotics in therapeutic and non-therapeutic purposes is still needed to overcome the development of MDR bacterial strains.
Conclusion
V. parahaemolyticus not only accumulates in shellfish and crustaceans but also can be found in the short mackerels at high concentrations. V. parahaemolyticus multiply more rapidly and reach the highest concentrations during warmer months. In Malaysia, warm temperatures remain fairly constant all year round thus it is expected high initial concentrations of V. parahaemolyticus can be found in the seafood. Preserving seafood at low temperature conditions can limit and control the growth of V. parahaemolyticus. Unbroken cold chain and cold storage facilities available in the hypermarkets maintained seafood products freshness as well as minimized the growth of foodborne pathogen such as V. parahaemolyticus. The low detection rate of pathogenic V. parahaemolyticus strains in the short mackerels may still at risk for food poisoning if storage conditions and preparation procedures are not receiving proper attention.
Author Contributions
CT is the corresponding author for this work. TM, CK, TT, WC, YL, JP, OR, MN, and NY provided assistance and guidance in throughout the research. YR, YN, MN, and SR are the mentor in the research study and assist in manuscript checking.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was funded by a Research University Grant Scheme Initiative Six (RUGS 6) of Universiti Putra Malaysia (GP-IPS 9438703) and Fundamental Research Grant Scheme (FRGS) of Ministry of Higher Education (MOHE), Malaysia (02-01-14-1475FR) and, in part, by the Kakenhi Grant-in-Aid for Scientific Research (KAKENHI 24249038), Japan Society for the Promotion of Sciences and grant-in-aid of Ministry of Health, Labor and Welfare, Japan.
References
ASEAN (2013). Appendix II: List of Chemicals Used in Aquaculture by ASEAN Member States. Available online at: http://www.asean.org/wp-content/uploads/images/Community/AEC/AMAF/UpdateApr2014/ASEAN%20Guideliness%20for%20Chemicals%20Final%20Draft%20Malaysia%20OK.pdf
Augustine, D., Jacob, J. C., and Philip, R. (2016). Exclusion of Vibrio spp. by an antagonistic marine actinomycete Streptomyces rubrolavendulae M56. Aquacult. Res. 47, 2951–2960. doi: 10.1111/are.12746
Baba, K., Yamasaki, S., Nishibuchi, M., and Takeda, Y. (1992). Examination by site-directed mutagenesis of the amino acid residues of the thermostable direct hemolysin of Vibrio parahaemolyticus required for its hemolytic activity. Microb. Pathog. 12, 279–287. doi: 10.1016/0882-4010(92)90046-Q
Baffone, W., Pianetti, A., Bruscolini, F., Barbieri, E., and Citterio, B. (2000). Occurrence and expression of virulence-related properties of Vibrio species isolated from widely consumed seafood products. Int. J. Food Microbiol. 54, 9–18. doi: 10.1016/S0168-1605(99)00189-0
Bej, A. K., Patterson, D. P., Brasher, C. W., Vickery, M. C. L., Jones, D. D., and Kaysner, C. A. (1999). Detection of total and hemolysin-producing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tlh, tdh and trh. J. Microbiol. Methods 36, 215–225. doi: 10.1016/S0167-7012(99)00037-8
Bhoopong, P., Palittapongarnpim, P., Pomwised, R., Kiatkittipong, A., Kamruzzaman, M., Nakaguchi, Y., et al. (2007). Variability of properties of Vibrio parahaemolyticus strains isolated from individual patients. J. Clin. Microbiol. 45, 1544–1550. doi: 10.1128/JCM.02371-06
Blair, J. M. A., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. V. (2014). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Chen, Y., Chen, X., Yu, F., Wu, M., Wang, R., Zheng, S., et al. (2016). Serology, virulence, antimicrobial susceptibility and molecular characteristics of clinical Vibrio parahaemolyticus strains circulating in southeastern China from 2009 to 2013. Clin. Microbiol. Infect. 22, 258.e9–258.e16. doi: 10.1016/j.cmi.2015.11.003
CLSI (2010). Methods for Antimicrobial Dilution and Disk Susceptibility Testing of Infrequently Isolated or Fastidious Bacteria; Approved Guideline, 2nd Edn. CLSI document M45-A2. Wayne, PA: Clinical and Laboratory Standards Institute.
CLSI (2016). Performance Standards for Antimicrobial Susceptibility Testing, 26th Edn. CLSI supplement M100S. Wayne, PA: Clinical and Laboratory Standards Institute.
Elexson, N., Afsah-Hejri, L., Rukayadi, Y., Soopna, P., Lee, H. Y., Tuan Zainazor, T. C., et al. (2014). Effect of detergents as antibacterial agents on biofilm of antibiotics-resistant Vibrio parahaemolyticus isolates. Food Control. 35, 378–385. doi: 10.1016/j.foodcont.2013.07.020
Fabbri, A., Falzano, L., Frank, C., Donelli, G., Matarrese, P., Raimondi, F., et al. (1998). Vibrio parahaemolyticus thermostable direct hemolysin modulates cytoskeletal organization and calcium homeostasis in intestinal cultured cells. Infect. Immun. 67, 1139–1148.
FAO (2016). Drivers, Dynamics and Epidemiology of Antimicrobial Resistance in Animal Production. Available online at: http://www.fao.org/3/a-i6209e.pdf
FDA (2011). Appendix 5: FDA and EPA Safety Levels in Regulations and Guidance. Available online at: http://www.fda.gov/downloads/Food/GuidanceRegulation/UCM251970.pdf
Hai, N. V. (2015). The use of probiotics in aquaculture. J. Appl. Microbiol. 119, 917–935. doi: 10.1111/jam.12886
Han, F., Walker, R. D., Janes, M. E., Prinyawiwatkul, W., and Ge, B. (2007). Antimicrobial susceptibilities of Vibrio parahaemolyticus and Vibrio vulnificus isolates from Louisiana Gulf and retail raw oysters. Appl. Environ. Microbiol. 73, 7096–7098. doi: 10.1128/AEM.01116-07
Hara-kudo, Y., Sugiyama, K., Nishibuchi, M., Chowdhury, A., Yatsuyanagi, J., Saito, A., et al. (2003). Prevalence of pandemic thermostable direct hemolysin-producing Vibrio parahaemolyticus O3:K6 in seafood and the coastal environment in Japan. Appl. Environ. Microbiol. 69, 3883–3891. doi: 10.1128/AEM.69.7.3883-3891.2003
Honda, T., and Iida, T. (1993). The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev Med Microbiol. 4, 106–113. doi: 10.1097/00013542-199304000-00006
Honda, T., Ni, Y. X., and Miwatani, T. (1988). Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect. Immun. 56, 961–965.
Honda, T., Ni, Y., Miwatani, T., Adachi, T., and Kim, J. (1992). The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can. J. Microbiol. 38, 1175–1180. doi: 10.1139/m92-192
Huang, J. Y., Henao, O. L., Griffin, P. M., Vugia, D. J., Cronquist, A. B., Hurd, S., et al. (2016). Infection with pathogens transmitted commonly through food and the effect of increasing use of culture-independent diagnostic tests on surveillance – foodborne diseases active surveillance network, 10 U.S. sites, 2012-2015. Morb. Mortal. Wkly. Rep. 65, 368–371. doi: 10.15585/mmwr.mm6514a2
IFT (2006). Antimicrobial resistance: implications for the food system. Compr. Rev. Food Sci. Food Saf. 5, 71–137. doi: 10.1111/j.1541-4337.2006.00004.x
Jun, J. W., Shin, T. H., Kim, J. H., Shin, S. P., Han, J. E., Heo, G. J., et al. (2014). Bacteriophage therapy of a Vibrio parahaemolyticus infection caused by a multiple-antibiotic-resistant O3:K6 pandemic clinical strain. J. Infect. Dis. 210, 72–78. doi: 10.1093/infdis/jiu059
Kaysner, C. A., and DePaola, A. J. (2004). BAM: Vibrio. Available online at: http://www.fda.gov/Food/FoodScienceResearch/LaboratoryMethods/ucm070830.htm
Kim, Y. B. U., Okuda, J. U. N., Matsumoto, C., Takahashi, N., Hashimoto, S., and Nishibuchi, M. (1999). Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J. Clin. Microbiol. 37, 1173–1177.
Krumperman, P. H. (1983). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46, 165–170.
Lesmana, M., Subekti, D., Simanjuntak, C. H., Tjaniadi, P., Campbell, J. R., and Oyofo, B. A. (2001). Vibrio parahaemolyticus associated with cholera-like diarrhea among patients in North Jakarta, Indonesia. Diagn. Microbiol. Infect. Dis. 39, 71–75. doi: 10.1016/S0732-8893(00)00232-7
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015a). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015b). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in Selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Lomelí-Ortega, C. O., and Martínez-Díaz, S. F. (2014). Phage therapy against Vibrio parahaemolyticus infection in the whiteleg shrimp (Litopenaeus vannamei) larvae. Aquaculture 434, 208–211. doi: 10.1016/j.aquaculture.2014.08.018
Malcolm, T. T. H., Cheah, Y. K., Radzi, C. W. J. W. M., Kasim, F. A., Kantilal, H. K., John, T. Y. H., et al. (2015). Detection and quantification of pathogenic Vibrio parahaemolyticus in shellfish by using multiplex PCR and loop-mediated isothermal amplification assay. Food Control. 47, 664–671. doi: 10.1016/j.foodcont.2014.08.010
Nakaguchi, Y. (2013). Contamination by Vibrio parahaemolyticus and its virulent strains in seafood marketed in Thailand, Vietnam, Malaysia, and Indonesia. Trop. Med. Health 41, 95–102. doi: 10.2149/tmh.2011-06
Newton, A. E., Garrett, N., Stroika, S. G., Halpin, J. L., Turnsek, M., and Mody, R. K. (2014). Increase in Vibrio parahaemolyticus infections associated with consumption of Atlantic Coast shellfish - 2013. Morb. Mortal. Wkly. Rep. 63, 335–336.
Nishibuchi, M., Taniguchi, T., Misawa, T., Khaeomanee-Iam, V., Honda, T., and Miwatani, T. (1989). Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect. Immun. 57, 2691–2697.
Ohno, S., Tazawa, T., Kon, M., Uno, Y., Terao, M., and Goto, K. (1993). Contamination by Vibrio parahaemolyticus of Fish Landed at Fishery Markets in Niigata Prefecture in Japanese. Niigataken Eisei Kogai Kenkyusyo Nenpo [Annual Report of Niigata Prefectural Research Laboratory for Health and Environment] 9, 77–82.
Ottaviani, D., Leoni, F., Talevi, G., Masini, L., Santarelli, S., Rocchegiani, E., et al. (2013). Extensive investigation of antimicrobial resistance in Vibrio parahaemolyticus from shellfish and clinical sources, Italy. Int. J. Antimicrob. Agents 42, 191–193. doi: 10.1016/j.ijantimicag.2013.05.003
Raghunath, P. (2015). Roles of thermostable direct hemolysin (TDH) and TDH-related hemolysin (TRH) in Vibrio parahaemolyticus. Front. Microbiol. 5:805. doi: 10.3389/fmicb.2014.00805
Raimondi, F., Kao, J. P. Y., Fiorentini, C., Fabbri, A., Donelli, G., Gasparini, N., et al. (2000). Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect. Immun. 68, 3180–3185. doi: 10.1128/IAI.68.6.3180-3185.2000
Rodgers, C., Parveen, S., Chigbu, P., Jacobs, J., Rhodes, M., and Harter-Dennis, J. (2014). Prevalence of Vibrio parahaemolyticus, and Vibrio vulnificus in blue crabs (Callinectes sapidus), seawater and sediments of the Maryland Coastal Bays. J. Appl. Microbiol. 117, 1198–1209. doi: 10.1111/jam.12608
Rodriguez-Castro, A., Ansede-Bermejo, J., Blanco-Abad, V., Varela-Pet, J., Garcia-Martin, O., and Martinez-Urtaza, J. (2010). Prevalence and genetic diversity of pathogenic populations of Vibrio parahaemolyticus in coastal waters of Galicia, Spain. Environ Microbiol Rep. 2, 58–66. doi: 10.1111/j.1758-2229.2009.00064.x
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States - major pathogens. Emerging Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Shariff, M., Nagaraj, G., Chua, F. H. C., and Wang, Y. G. (2000). “The use of chemicals in aquaculture in Malaysia and Singapore,” in Proceedings of the Meeting on the Use of Chemicals in Aquaculture in Asia (Iloilo), 127–140.
Su, Y. C., and Liu, C. (2007). Vibrio parahaemolyticus: a concern of seafood safety. Food Microbiol. 24, 549–558. doi: 10.1016/j.fm.2007.01.005
Suthienkul, O., Ishibashi, M., Iida, T., Nettip, N., Supavej, S., Eampokalap, B., et al. (1995). Urease production correlates with possession of the trh gene in Vibrio parahaemolyticus strains isolated in Thailand. J. Infect. Dis. 172, 1405–1408. doi: 10.1093/infdis/172.5.1405
Tada, J., Ohashi, T., Nishimura, N., Shirasaki, Y., Ozaki, H., Fukushima, S., et al. (1992). Detection of the thermostable direct hemolysin gene (tdh) and the thermostable direct hemolysin-related hemolysin gene (trh) of Vibrio parahaemolyticus by polymerase chain reaction. Mol. Cell. Probes 6, 477–487. doi: 10.1016/0890-8508(92)90044-X
Takahashi, A., Kenjyo, N., Imura, K., Myonsun, Y., and Honda, T. (2000). Cl- secretion in colonic epithelial cells induced by the Vibrio parahaemolyticus hemolytic toxin related to thermostable direct hemolysin. Infect. Immun. 68, 5435–5438. doi: 10.1128/IAI.68.9.5435-5438.2000
Tan, L. T. H., Chan, K. G., Lee, L. H., and Goh, B. H. (2016). Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol. 7:79. doi: 10.3389/fmicb.2016.00079
Van Hoek, A. H. A. M., Mevius, D., Guerra, B., Mullany, P., Roberts, A. P., and Aarts, H. J. M. (2011). Acquired antibiotic resistance genes: an overview. Front. Microbiol. 2:203. doi: 10.3389/fmicb.2011.00203
Watanabe, T., Tazawa, T., Kon, M., Ohno, S., Uno, Y., Terao, M., et al. (1994). “Growth of Vibrio parahaemolyticus in different methods of cooking fish,” in Proceeding of the National Conference of Food Safety Inspection, 113–116.
Weese, J. S., Giguère, S., Guardabassi, L., Morley, P. S., Papich, M., Ricciuto, D. R., et al. (2015). ACVIM consensus statement on therapeutic antimicrobial use in animals and antimicrobial resistance. J. Vet. Intern. Med. 29, 487–498. doi: 10.1111/jvim.12562
Wong, H. C., Chen, M. C., Liu, S. H., and Liu, D. P. (1999). Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int. J. Food Microbiol. 52, 181–188. doi: 10.1016/S0168-1605(99)00143-9
Wong, K. C., Brown, A. M., Luscombe, G. M., Wong, S. J., and Mendis, K. (2015). Antibiotic use for Vibrio infections: important insights from surveillance data. BMC Infect. Dis. 15:226. doi: 10.1186/s12879-015-0959-z
Xu, X., Wu, Q., Zhang, J., Cheng, J., Zhang, S., and Wu, K. (2014). Prevalence, pathogenicity, and serotypes of Vibrio parahaemolyticus in shrimp from Chinese retail markets. Food Control 46, 81–85. doi: 10.1016/j.foodcont.2014.04.042
Yanagihara, I., Nakahira, K., Yamane, T., Kaieda, S., Mayanagi, K., Hamada, D., et al. (2010). Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin. J. Biol. Chem. 285, 16267–16274. doi: 10.1074/jbc.M109.074526
Yano, Y., Hamano, K., Satomi, M., and Tsutsui, I. (2011). Diversity and characterization of oxytetracycline-resistant bacteria associated with non-native species, white-leg shrimp (Litopenaeus vannamei), and native species, black tiger shrimp (Penaeus monodon), intensively cultured in Thailand. J. Appl. Microbiol. 110, 713–722. doi: 10.1111/j.1365-2672.2010.04926.x
Keywords: Vibrio parahaemolyticus, finfish, MPN, antibiotic susceptibility, MAR
Citation: Tan CW, Malcolm TTH, Kuan CH, Thung TY, Chang WS, Loo YY, Premarathne JMKJK, Ramzi OB, Norshafawatie MFS, Yusralimuna N, Rukayadi Y, Nakaguchi Y, Nishibuchi M and Radu S (2017) Prevalence and Antimicrobial Susceptibility of Vibrio parahaemolyticus Isolated from Short Mackerels (Rastrelliger brachysoma) in Malaysia. Front. Microbiol. 8:1087. doi: 10.3389/fmicb.2017.01087
Received: 11 April 2017; Accepted: 30 May 2017;
Published: 13 June 2017.
Edited by:
Giovanna Suzzi, University of Teramo, ItalyReviewed by:
Learn-Han Lee, Monash University Malaysia, MalaysiaDapeng Wang, Shanghai Jiao Tong University, China
Copyright © 2017 Tan, Malcolm, Kuan, Thung, Chang, Loo, Premarathne, Ramzi, Norshafawatie, Yusralimuna, Rukayadi, Nakaguchi, Nishibuchi and Radu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chia Wanq Tan, Y2hpYXdhbnFAZ21haWwuY29t
 Chia W. Tan
Chia W. Tan Tan T. H. Malcolm
Tan T. H. Malcolm Chee H. Kuan
Chee H. Kuan Tze Y. Thung1
Tze Y. Thung1 Jayasekara M. K. J. K. Premarathne
Jayasekara M. K. J. K. Premarathne Yoshitsugu Nakaguchi
Yoshitsugu Nakaguchi Mitsuaki Nishibuchi
Mitsuaki Nishibuchi