- 1Department of Geological Sciences, University of Delaware, Newark, DE, United States
- 2Project Team for Development of New-Generation Research Protocol for Submarine Resources, Japan Agency for Marine-Earth Science and Technology, Kanagawa, Japan
- 3School of Marine Science and Policy, University of Delaware, Newark, DE, United States
- 4Department of Biology, East Carolina University, Greenville, NC, United States
Chemolithotrophic iron-oxidizing bacteria (FeOB) could theoretically inhabit any environment where Fe(II) and O2 (or nitrate) coexist. Until recently, marine Fe-oxidizing Zetaproteobacteria had primarily been observed in benthic and subsurface settings, but not redox-stratified water columns. This may be due to the challenges that a pelagic lifestyle would pose for Zetaproteobacteria, given low Fe(II) concentrations in modern marine waters and the possibility that Fe oxyhydroxide biominerals could cause cells to sink. However, we recently cultivated Zetaproteobacteria from the Chesapeake Bay oxic–anoxic transition zone, suggesting that they can survive and contribute to biogeochemical cycling in a stratified estuary. Here we describe the isolation, characterization, and genomes of two new species, Mariprofundus aestuarium CP-5 and Mariprofundus ferrinatatus CP-8, which are the first Zetaproteobacteria isolates from a pelagic environment. We looked for adaptations enabling strains CP-5 and CP-8 to overcome the challenges of living in a low Fe redoxcline with frequent O2 fluctuations due to tidal mixing. We found that the CP strains produce distinctive dreadlock-like Fe oxyhydroxide structures that are easily shed, which would help cells maintain suspension in the water column. These oxides are by-products of Fe(II) oxidation, likely catalyzed by the putative Fe(II) oxidase encoded by the cyc2 gene, present in both CP-5 and CP-8 genomes; the consistent presence of cyc2 in all microaerophilic FeOB and other FeOB genomes supports its putative role in Fe(II) oxidation. The CP strains also have two gene clusters associated with biofilm formation (Wsp system and the Widespread Colonization Island) that are absent or rare in other Zetaproteobacteria. We propose that biofilm formation enables the CP strains to attach to FeS particles and form flocs, an advantageous strategy for scavenging Fe(II) and developing low [O2] microenvironments within more oxygenated waters. However, the CP strains appear to be adapted to somewhat higher concentrations of O2, as indicated by the presence of genes encoding aa3-type cytochrome c oxidases, but not the cbb3-type found in all other Zetaproteobacteria isolate genomes. Overall, our results reveal adaptations for life in a physically dynamic, low Fe(II) water column, suggesting that niche-specific strategies can enable Zetaproteobacteria to live in any environment with Fe(II).
Introduction
Chemolithotrophic Fe-oxidizing bacteria (FeOB) use Fe(II) oxidation for energy and growth, and are therefore thought to play important roles in Fe cycling. Fe is practically ubiquitous, raising the question of whether FeOB are active in every environment with Fe redox cycling, which would likely require a variety of niche-specific adaptations. Fe cycling is particularly important at coasts, where Fe transformations affect the chemistry of waters in coastal sediments and estuaries, and ultimately the concentrations of nutrients (e.g., Fe, P) and other metals (e.g., As) transported to the ocean (Charette et al., 2005; Jung et al., 2009). Significant redox activity occurs in stratified marine waters, such as the Chesapeake Bay, which experience seasonal anoxia in bottom waters (Officer et al., 1984). In our previous studies of the Chesapeake, water samples from the oxic–anoxic transition zone always yielded enrichments of chemolithotrophic FeOB (MacDonald et al., 2014; Field et al., 2016). From these enrichments, we isolated two FeOB, which represent the first known marine FeOB from the water column (isolate strain CP-8 previously reported in Field et al., 2016). The presence of FeOB was somewhat surprising given the relatively low (micromolar) concentrations of Fe, and the strong tidal mixing, which may frequently expose FeOB to higher O2 concentrations, making it harder for them to compete with abiotic Fe(II) oxidation. Further study of these isolates may reveal their distinct adaptations to life in the estuarine water column, while also showing commonalities shared among all marine FeOB across different environments.
The Chesapeake FeOB isolates are members of the Zetaproteobacteria, all of which are marine neutrophilic chemolithotrophic FeOB. The other Zetaproteobacteria isolated to date primarily originate from deep sea hydrothermal microbial mats and sediments (Emerson et al., 2007; McAllister et al., 2011; Field et al., 2015; Makita et al., 2016), with some from coastal sediment (Laufer et al., 2016, 2017). Zetaproteobacteria sequences have also been found in coastal groundwater and worm burrows (16S rRNA gene analysis; McAllister et al., 2015) and briny terrestrial groundwater (metagenomics; Emerson et al., 2016). Steel coupon incubation experiments provide sequence and culture-based evidence that Zetaproteobacteria inhabit coastal waters (Dang et al., 2011; McBeth et al., 2011; Mumford et al., 2016), but the Chesapeake isolates are the first Zetaproteobacteria isolated directly from a coastal redox-stratified water column. In total, previous studies show that Zetaproteobacteria grow at oxic–anoxic interfaces where Fe(II) and O2 are available, typically preferring lower O2 concentrations (Chan et al., 2016), though Mariprofundus sp. DIS-1 is an exception in that it tolerates saturated O2 conditions (Mumford et al., 2016). The molecular mechanism of neutrophilic Fe(II) oxidation is not well-known; comparative analysis of six existing Zetaproteobacteria isolate genomes with freshwater FeOB genomes has resulted in hypothesized pathways (Singer et al., 2011; Liu et al., 2012; Barco et al., 2015), but differences in single amplified genomes (SAGs) and metagenomes suggest that the pathway has some variants (Field et al., 2015; Fullerton et al., 2017). Fe(II) oxidation by the Zetaproteobacteria results in Fe(III) oxyhydroxides, typically in the form of twisted ribbon-like stalks, which form the framework of Fe microbial mats (Chan et al., 2016). Such large, dense stalk structures would make it difficult for a pelagic FeOB to maintain buoyancy. In sum, our knowledge of benthic Zetaproteobacteria may not necessarily be representative of FeOB in the water column.
Here we detail the isolation, physiological characterization, and genomic analysis of two new Fe-oxidizing Zetaproteobacteria from the Chesapeake Bay, Mariprofundus aestuarium CP-5 and Mariprofundus ferrinatatus CP-8. We compare the CP strains to the other Zetaproteobacteria and propose that physiological and genomic distinctions represent adaptive strategies for the Chesapeake Zetaproteobacteria to scavenge Fe in low Fe(II) waters and to withstand highly variable oxygen conditions associated with physically dynamic redoxclines.
Materials and Methods
Sampling, Enrichments, and Isolation
The redox-stratified waters of the Chesapeake Bay at Station 858 (38°58.600 N, 076°22.080 W) were sampled aboard the R/V Hugh R Sharp in August, 2014. Details of sampling and the geochemical conditions can be found in Field et al. (2016). Water samples collected from the oxic–anoxic transition zone were used to inoculate FeOB enrichment cultures. Agarose-stabilized gradient tube cultures (Emerson and Floyd, 2005) were set up with a FeCO3 plug (1% w/v high-melt agarose) and simulated estuary medium (0.15% w/v low-melt agarose), which is a 50:50 mixture of modified Wolfe’s mineral medium (MWMM) and artificial seawater (ASW). Per liter, estuary medium contains 13.75 g NaCl, 2.69 g MgCl2-6H2O, 3.49 g MgSO4-7H2O, 0.36 g KCl, 0.75 g CaCl2-2H2O, 1 g NH4Cl, 0.05 g KH2PO4, 0.42 g NaHCO3. After autoclaving, estuary medium was amended with 1 μL/mL Wolfe’s trace mineral solution and 1 μL/mL vitamin solution and adjusted to pH 6.2 with CO2. The headspace of all tube cultures contained a low O2 gas mixture (N2/CO2/O2; 95:4:1).
Strains CP-5 and CP-8 were isolated by serial dilution-to-extinction from water samples CTD12-5 and IS8-11.3 respectively (water geochemistry in Supplementary Table ST1; further details in Field et al., 2016). Growth was confirmed by the development of colonies or distinct growth bands in agarose-stabilized tubes (Figure 1) and by microscopy. Purity was checked by microscopic observation, absence of heterotrophic growth on R2A-estuary medium agar plates, and sequencing of the 16S rRNA genes amplified with the bacterial-universal primer sets Bac27F and Uni1492R (Lane, 1991).
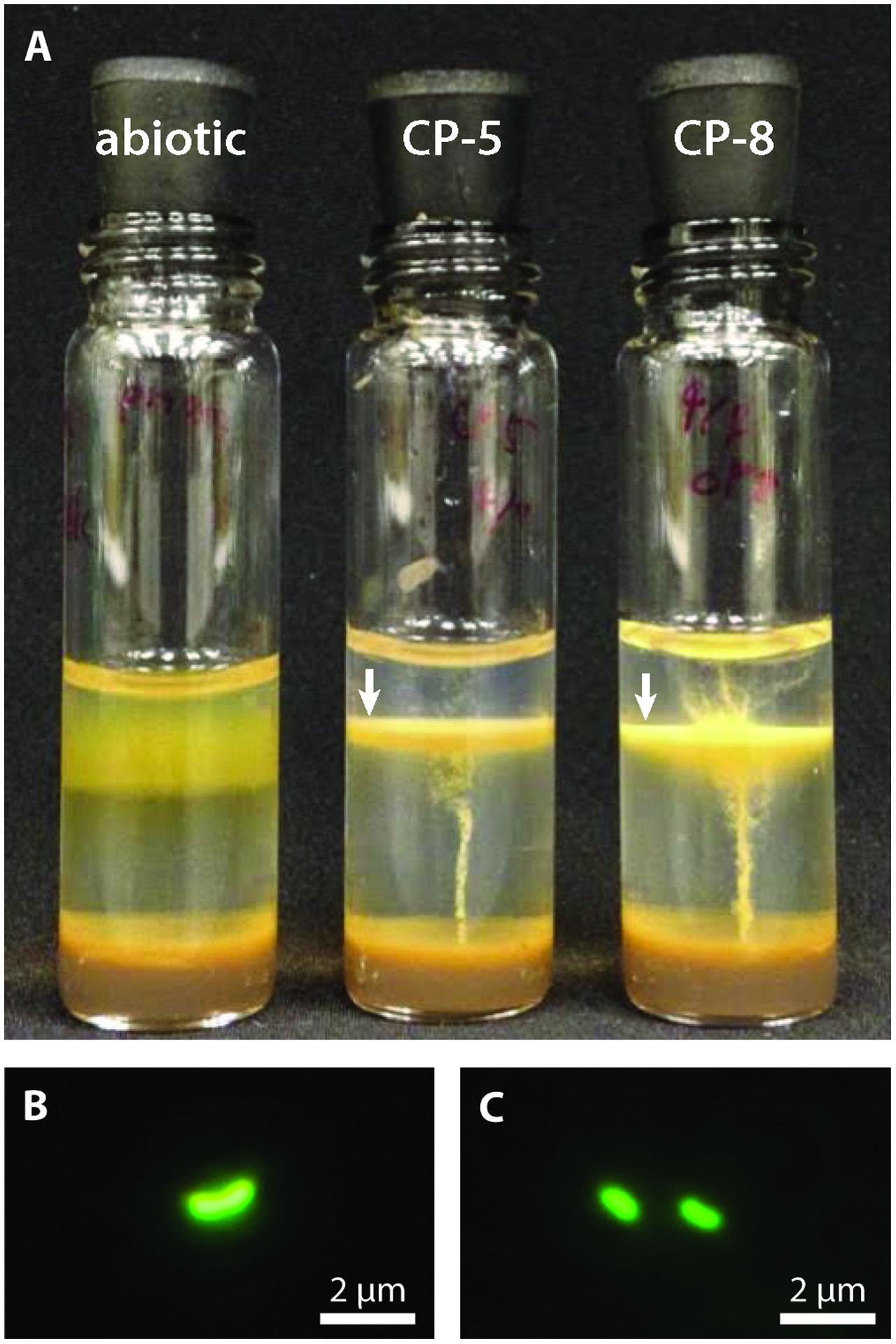
FIGURE 1. (A) CP strain gradient tube cultures with FeCO3 plugs (8 days old). CP strain-inoculated tubes display distinct orange growth bands (indicated by white arrows) in contrast to the diffuse oxide cloud in the abiotic control. The inoculum for the CP strain tubes is visible as thin orange vertical lines. Fluorescence micrographs of CP-5 (B) and CP-8 (C) cells.
Physiological Characterization
To assess alternate substrate usage and optimal growth conditions of strains CP-5 and CP-8, growth tests were carried out in agarose-stabilized gradient tubes as described above, but buffered to pH 7.0 except for pH testing. To test if the strains could use non-Fe(II) substrates, we tested growth on 5 mM sodium thiosulfate, 5 mM sodium sulfide, 10 mM sodium pyruvate, 10 mM glucose, 10 mM sodium acetate, and 0.2% w/v yeast extract. The pH range of growth was determined using several buffers: acetate-acetic acid (pH 5.0 and 5.2, 10 mM), MES (pH 5.5, 6.0, 6.4 and 6.9, 10 mM), and HEPES (pH 7.2, 7.4, 7.7, 8.0, 8.3 and 8.5, 10 mM). pH measurements taken before and after cultivation confirmed minimal (0.1–0.2) decreases during cultivation periods. Preferred growth temperature was determined by incubating cultures at 5, 10, 15, 20, 25, 30, 35, and 40°C, and preferred salinity was determined using different ratios of MWMM (0‰): ASW (35‰): 0:10, 1:9, 2:8, 3:7, 4:6, 5:5, 6:4, 7:3, 8:2, 9:1, and 10:0. All cultures were assessed for growth after 2 weeks based on the development of growth bands and observation by fluorescent microscopy.
To determine the preferred oxygen concentration for growth, the dissolved oxygen (DO) within strain CP-5 and CP-8 growth bands was measured 48 h after inoculation using a Firesting optical oxygen probe with a needle-type sensor (PyroScience, Aachen, Germany) mounted on a micromanipulator (Narishige International, Amityville, NY, United States). Attempts to test growth under anoxic conditions were also set up by preparing deoxygenated media (bubbling with N2 and autoclaving in an N2-flushed vessel), setting up gradient tubes under a stream of N2, and using a 100% N2 headspace. However, the Firesting optical oxygen probe detected trace O2 in the gradient tube medium (∼250 nM) indicating that this procedure did not yield completely anoxic cultures.
To measure the growth rate of each strain, growth bands from triplicate gradient tube cultures (buffered with PIPES) were harvested daily over the course of the experiment (10 and 9 days for strain CP-5 and CP-8 respectively). Samples were stained with Syto 13 for cell counting using a Petroff-Hausser counting chamber. Aliquots of harvested growth bands were also used to measure total Fe concentrations in cultures over time. Fe concentrations in abiotic control gradient tubes were measured as well, using samples at the same height as biotic growth bands.
We used total Fe measurements to follow Fe(II) oxidation because nearly all Fe accumulated in developing growth bands was shown to be Fe(III) in the strain CP-5 growth experiment (data not shown). Samples for total Fe analysis were reduced with 200 mM hydroxylamine for 22–24 h and measured using the ferrozine method (modified from Stookey, 1970).
Microscopy
Phase contrast and fluorescent micrographs of cultures (stained with SYBR green I, in the case of fluorescence) were captured on an Olympus BH-2 microscope with 400x total magnification. For these analyses, we used liquid cultures (without agarose) grown for 24 h. Samples for scanning electron microscopy (SEM) were gently mounted on a 0.2-μm-pore-size polycarbonate filter, air dried, and coated with gold/palladium for observation, or with carbon for energy dispersive spectroscopy (EDS) analysis. Samples for transmission electron microscopy (TEM) were gently mounted on a Formvar-coated copper grid, air dried, and coated with gold/palladium. Electron microscopy was performed at the Delaware Biotechnology Institute Bioimaging Center, using a Hitachi S-4700 field emission SEM with an Oxford INCA EDS system and a Zeiss LIBRA 120 TEM.
Genome Sequencing and Analysis
For DNA extraction, strains CP-5 and CP-8 were grown using 25 mL FeCO3 gradient plates (1 L total volume per strain) under microaerobic conditions (N2/CO2/O2; 95:4:1; Emerson and Floyd, 2005). Genomic DNA was isolated from these cultures using the FastDNA Spin Kit for Soil (MP Biomedicals, Santa Ana, CA, United States). We used the PowerClean Pro DNA kit (MO BIO Laboratories, Carlsbad, CA, United States) to remove remaining inhibitors. The purified DNA (2.5 and 0.5 μg of CP-5 and CP-8, respectively) was size-selected using electrophoresis (BluePippin, Sage Science, Beverly, MA, United States) to a minimum size of 6 kb, resulting in an average size of 12 kb. The genomes were sequenced using PacBio RSII technology at the University of Delaware Sequencing and Genotyping Center. Size-selected DNA was prepared for sequencing using the SMRTbell Template Prep Kit 1.0 (PacBio, Menlo Park, CA, United States) as per the manufacturer’s instructions. One SMRT cell per genome was sequenced with P6-C4 chemistry and a 6-h movie. For strain CP-5, sequencing generated 1.37 Gbp of raw data (mean read length 15,263 bp; N50 26,034 bp); for strain CP-8, sequencing generated 0.84 Gbp of raw data (mean read length 8,877 bp; N50 19,311 bp). Assembly was completed on the PacBio SMRT Portal. Subreads were filtered to a minimum length of 1 kb (CP-5) or 2 kb (CP-8) with a polymerase quality score minimum of 0.8. The hierarchical genome assembly process 3 (HGAP-3) was used to assemble a single high quality contig from each of the sequencing runs. The average coverage over the entire sequenced contigs was 382x for strain CP-5 and 300x for strain CP-8. Gepard (v.1.40; Krumsiek et al., 2007) was used to compare each genome against itself to check for inverted repeats and to close each contig into a complete circular genome.
The complete genomes of strains CP-5 and CP-8 were annotated using the pipeline of the Integrated Microbial Genome Expert Review (IMG/ER) system (Markowitz et al., 2012). Manual verification of predicted genes of interest was completed using MUSCLE alignments in MEGA (v.7.0.14) against reference gene sequences from UniProt or the RSCB Protein Data Bank (Edgar, 2004; Kumar et al., 2016). The Rapid Annotation using Subsystem Technology (RAST) platform (Aziz et al., 2008; Overbeek et al., 2014) was used to identify possible frameshifts (none were detected) and to help find genes unique to the CP strain genomes, with respect to the other Zetaproteobacteria. Average amino acid identities (AAIs) of bidirectional best hit proteins were calculated using a web-based calculator1. Reported AAI values are the average of the separate calculations run in both directions for each pair (standard deviation < 1.18%). Average nucleotide identities (ANIs) were calculated using OrthoANI (Yoon et al., 2017). An AAI heatmap was made using the R package gplots heatmap.2 (v 3.0.1); hierarchical clustering using complete agglomeration was used to calculate the dendrogram.
16S rRNA Gene Analysis
The CP strain 16S rRNA genes were found in their completed genomes and aligned to the arb-SILVA database using the SINA online web tool (v.1.2.11; Pruesse et al., 2012). Aligned sequences were masked to unambiguously aligned base positions and a maximum-likelihood tree was constructed using RAxML with the GTR-gamma nucleotide substitution model (v.8.2.8; Stamatakis, 2014). Bootstrap values were estimated from 500 replicates. To calculate pairwise percent nucleotide identity, we calculated the Similarity score metric on the Ribosomal Database Project (RDP) website (Cole et al., 2014).
Genome Accession Numbers and Culture Availability
GenBank accession numbers for Mariprofundus aestuarium CP-5 and Mariprofundus ferrinatatus CP-8 are CP018799 and CP018800 respectively. IMG taxon IDs for strains CP-5 and CP-8 are 267118011 and 267180111 respectively. Both isolates are available on request from C.S. Chan (University of Delaware, United States) and at the Provasoli-Guillard National Center for Marine Algae and Microbiota (NCMA; Bigelow Laboratory for Ocean Sciences, United States).
Results and Discussion
Isolation and Physiological Characterization
Strains CP-5 and CP-8 were both successfully isolated using Fe(II)/O2 gradient tubes after five transfers of the 10-5 serial dilutions. Growth consistently appeared as a sharp orange band typical of microaerophilic FeOB (Figure 1A) and cells appeared as curved rods under fluorescent microscopy (Figures 1B,C). Strain CP-5 cells are 0.43 ± 0.05 μm × 1.01 ± 0.18 μm, and strain CP-8 cells are 0.45 ± 0.04 μm × 0.91 ± 0.08 μm. Purity was demonstrated by the lack of growth on R2A-estuary medium plates (no contaminant oligotrophs) and by a single unambiguous full length 16S rRNA gene sequence amplified from each culture.
Strains CP-5 and CP-8 have doubling times of 19.5 and 27 h, respectively. These generation times are slower than M. ferrooxydans PV-1 (12 h), but similar to the 24 h doubling time reported for the closely related Mariprofundus micogutta. During growth, strains CP-5 and CP-8 both accelerated Fe(II) oxidation, compared to uninoculated controls (Figure 2). The O2 concentration in the growth bands of inoculated gradient tubes was <2 μM O2 (Supplementary Figure S1), comparable to or lower than M. ferrooxydans PV-1 (Krepski et al., 2013). Strains CP-5 and CP-8 appear to be obligate Fe(II)-oxidizers as neither grew on reduced S or organic carbon substrates (Table 1). Overall, our experiments suggest that strains CP-5 and CP-8 are microaerophilic chemolithoautotrophic Fe(II)-oxidizers, consistent with all other Zetaproteobacteria isolates.
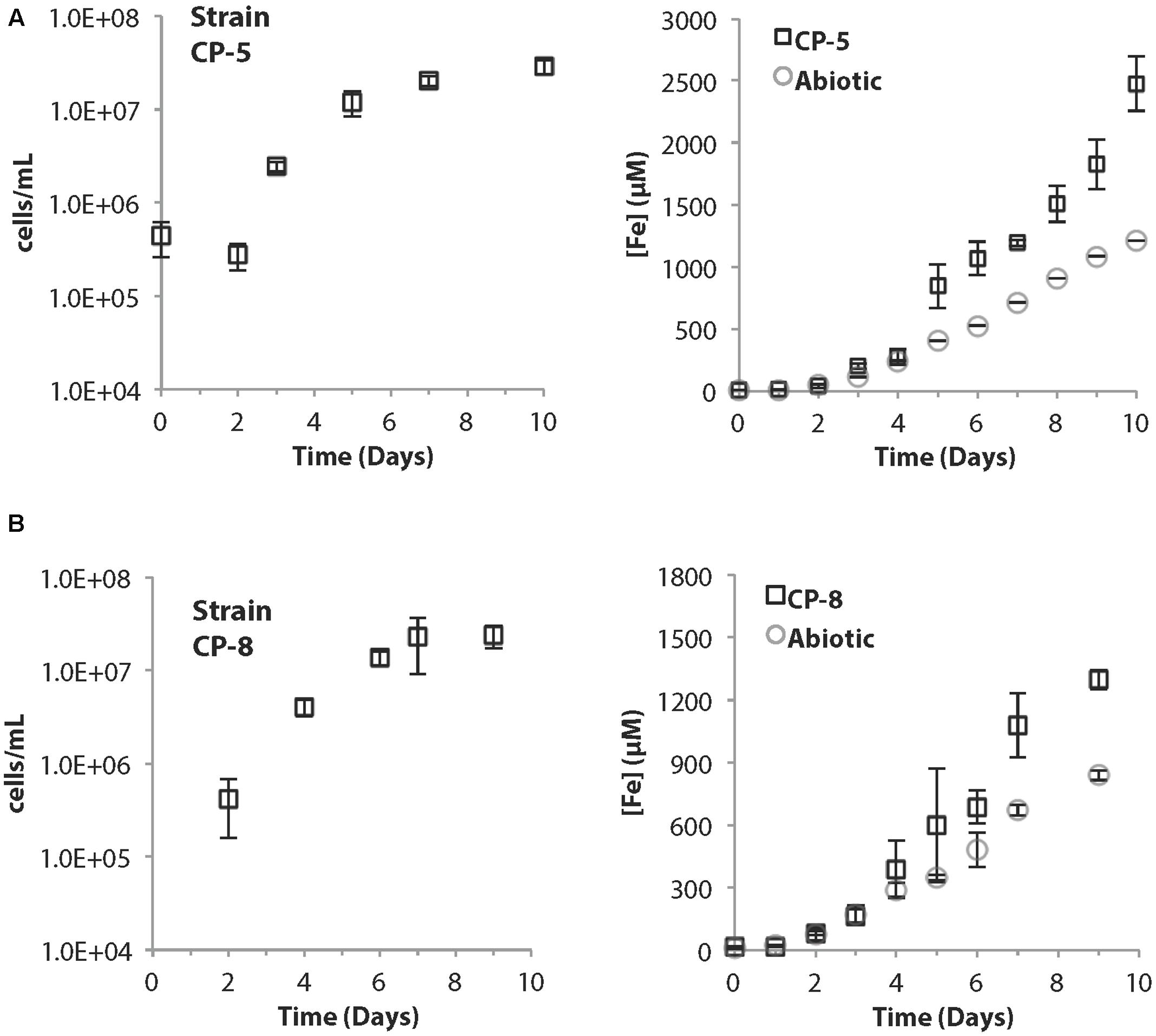
FIGURE 2. (A) Strain CP-5 growth curve (left) and corresponding total Fe curve (right) including abiotic controls. (B) Strain CP-8 growth curve (left) and corresponding total Fe curve (right) including abiotic controls. All cell concentrations are an average of direct cell counts from triplicate samples per time point. Error bars represent one standard deviation from the mean.
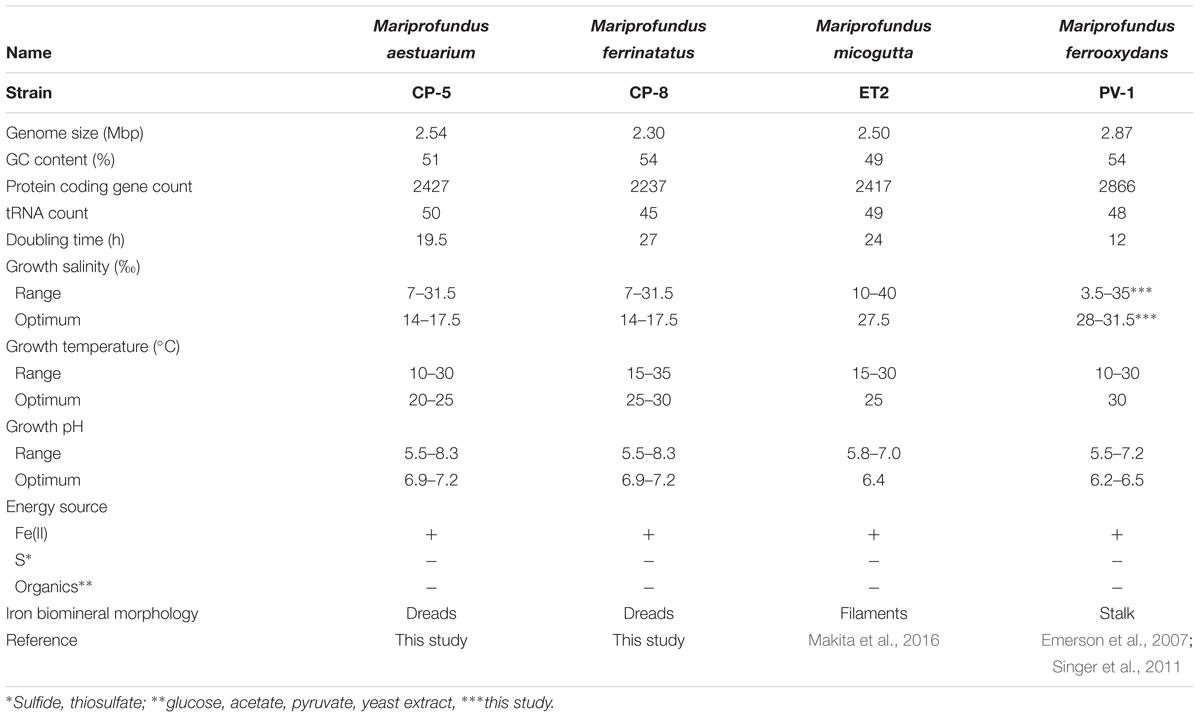
TABLE 1. Summary of strains CP-5 and CP-8 genomic and physiological characteristics in comparison to other selected Zetaproteobacteria.
To optimize culturing of the CP strains, growth was tested over a range of salinity and pH. The preferred salinity was brackish, 14–17.5‰, with no growth at 0‰ (freshwater) or 35‰ (normal seawater). The preferred pH range was 6.9–7.2, and both strains grew at pH up to 8.3, unusually high for neutrophilic FeOB isolates, which typically prefer pH between 6.0 and 6.5 (e.g., M. ferrooxydans PV-1 and M. micogutta, Table 1; freshwater FeOB Gallionella capsiferriformans ES-2, Sideroxydans lithotrophicus ES-1, and Ferriphaselus amnicola OYT-1; Emerson and Moyer, 1997; Kato et al., 2014). One exception is Mariprofundus sp. DIS-1, which can grow at pH 8.0 (Mumford et al., 2016). The CP strain salinity and pH preferences reflect the brackish seawater from which they were sampled.
Phylogenetic Analyses
Strains CP-5 and CP-8 are representative of the Chesapeake Bay environment, as their 16S rRNA gene sequences match the dominant 16S rRNA sequences of the original FeOB enrichment cultures from which each strain was isolated (Supplementary Figure S2). Phylogenetic analysis of 16S rRNA gene sequences shows that strains CP-5 and CP-8 are Zetaproteobacteria within OTUs 18 and 37 respectively (as defined by McAllister et al., 2011 and determined using ZetaHunter2) and cluster with nearly all other isolated Zetaproteobacteria (Figure 3). Among the Zetaproteobacteria isolates and SAGs, strains CP-5 and CP-8 are most similar to each other based on ANI, average AAI, and 16S rRNA gene identity (Table 2). Because both strains share less than 97% 16S rRNA gene identity (Stackebrandt and Goebel, 1994) and have less than 95% ANI (Konstantinidis and Tiedje, 2005) with all other Zetaproteobacteria isolates and SAGs, including each other, strains CP-5 and CP-8 are two new species, with proposed names Mariprofundus aestuarium CP-5 and Mariprofundus ferrinatatus CP-8.
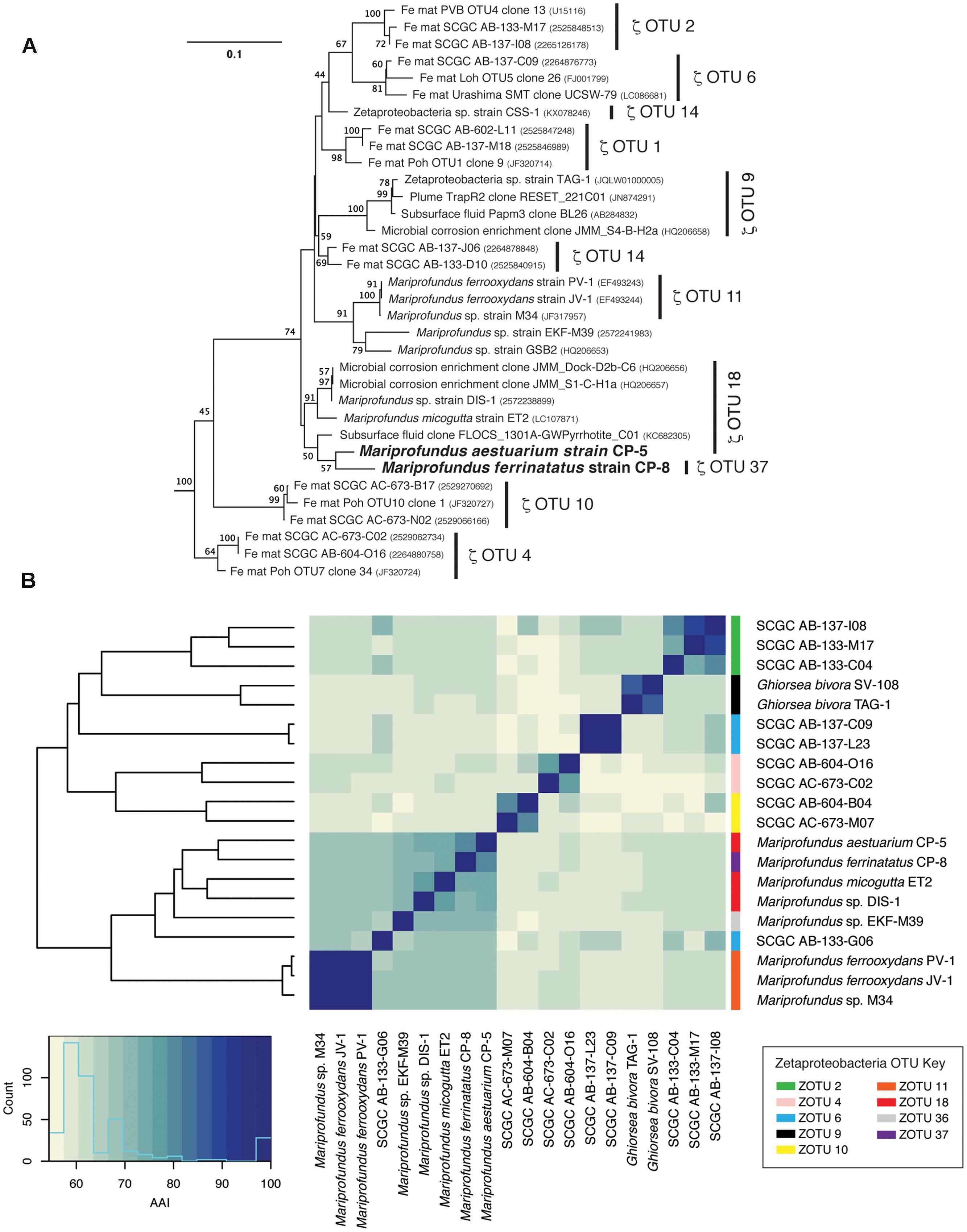
FIGURE 3. (A) 16S rRNA gene phylogenetic tree of Mariprofundus ferrinatatus CP-8 and Mariprofundus aestuarium CP-5 and other Zetaproteobacteria. Thermotoga maritima (AJ401021) and Aquifex pyrophilus (M83548) were used as outgroups (not shown). All sequences were masked to 1275 bp. (B) Heatmap showing pairwise comparisons of AAI between all Zetaproteobacteria isolates and the most complete SAGs.
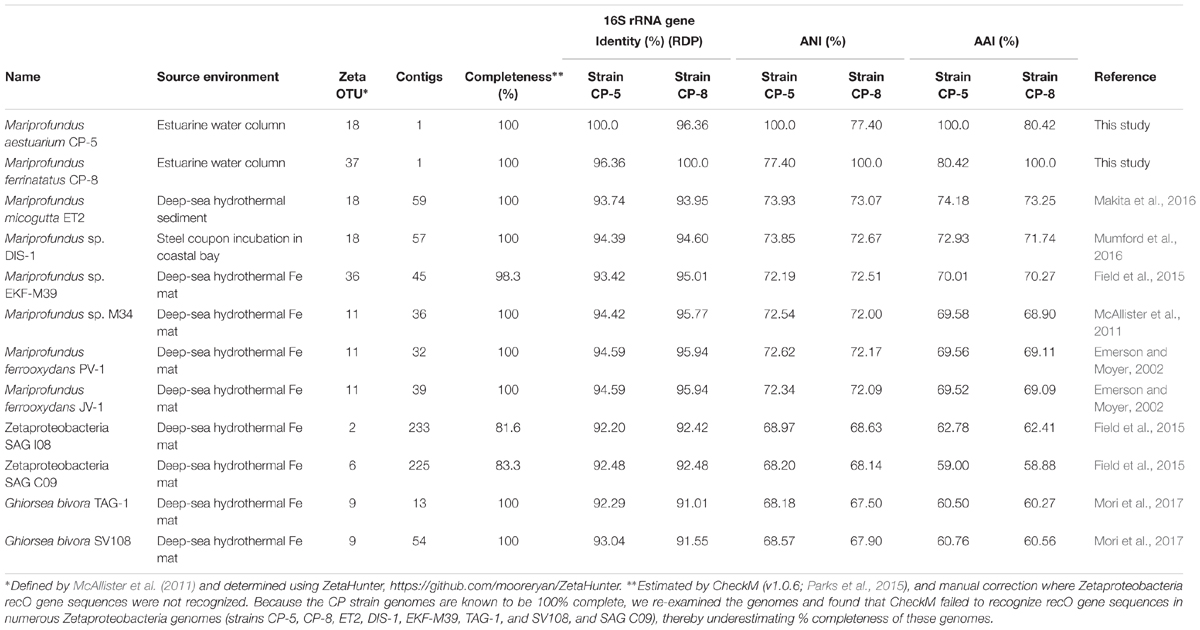
TABLE 2. Source environment, 16S rRNA gene identity, ANI, and AAI comparisons of strains CP-5 and CP-8 to other Zetaproteobacteria.
Comparisons of 16S rRNA sequences and %AAI among Zetaproteobacteria show that most of the isolates fall within a closely related group, i.e., the genus Mariprofundus (Figure 3). By 16S rRNA gene identity and %AAI, the CP strains are most closely related to Mariprofundus sp. DIS-1, isolated from a steel coupon incubated in a coastal bay (Mumford et al., 2016), and M. micogutta, isolated from marine hydrothermal sediment (Table 2 and Figure 3; Makita et al., 2016). These close relationships show that Mariprofundus is a cosmopolitan genus that inhabits a variety of environments, coastal and deep sea, as well as planktonic, benthic, and subsurface.
Iron Oxyhydroxide Biomineral Morphology
To investigate how suspended FeOB manage Fe oxyhydroxide precipitation to avoid sinking, we examined the Fe biominerals produced by strains CP-5 and CP-8. Both strains produce bundles of stubby rod-shaped extracellular structures (Figure 4), confirmed to be Fe-rich by SEM-EDX (Supplementary Figure S3) and morphologically distinct from abiotic mineral precipitates (Supplementary Figures S4, S5). This morphology has previously been identified in freshwater FeOB and referred to as dreadlocks (or dreads) given their resemblance to the dreadlock hairstyle (Figure 5; Kato et al., 2015). Dreads are somewhat similar to the fibrillar twisted Fe stalks produced by other microaerophilic FeOB (Figure 5B; Chan et al., 2011; Kato et al., 2014), in that they are bundles of elongated Fe oxyhydroxides (referred to as oxides from here on). However, dreads are short, never exceeding 10 μm in length, and many dreads can radiate from, and surround a single cell. In contrast, Fe oxide stalks range in length from 10’s of μm to mm, extend from one side of the cell, and are used by mat-forming FeOB to anchor themselves to surfaces (Chan et al., 2011, 2016). Dreads were closely associated with CP cells observed by fluorescent microscopy (Figure 4A) while the radiating arrangement observed under SEM made it apparent that CP strain cells were once attached (Figure 5A). In fact, the lack of cell-attached dreads under SEM suggests they are easily shed. In total, these observations suggest that the CP strains produce short Fe oxide dreads as an adaptation to shed their biominerals to maintain suspension within the water column.
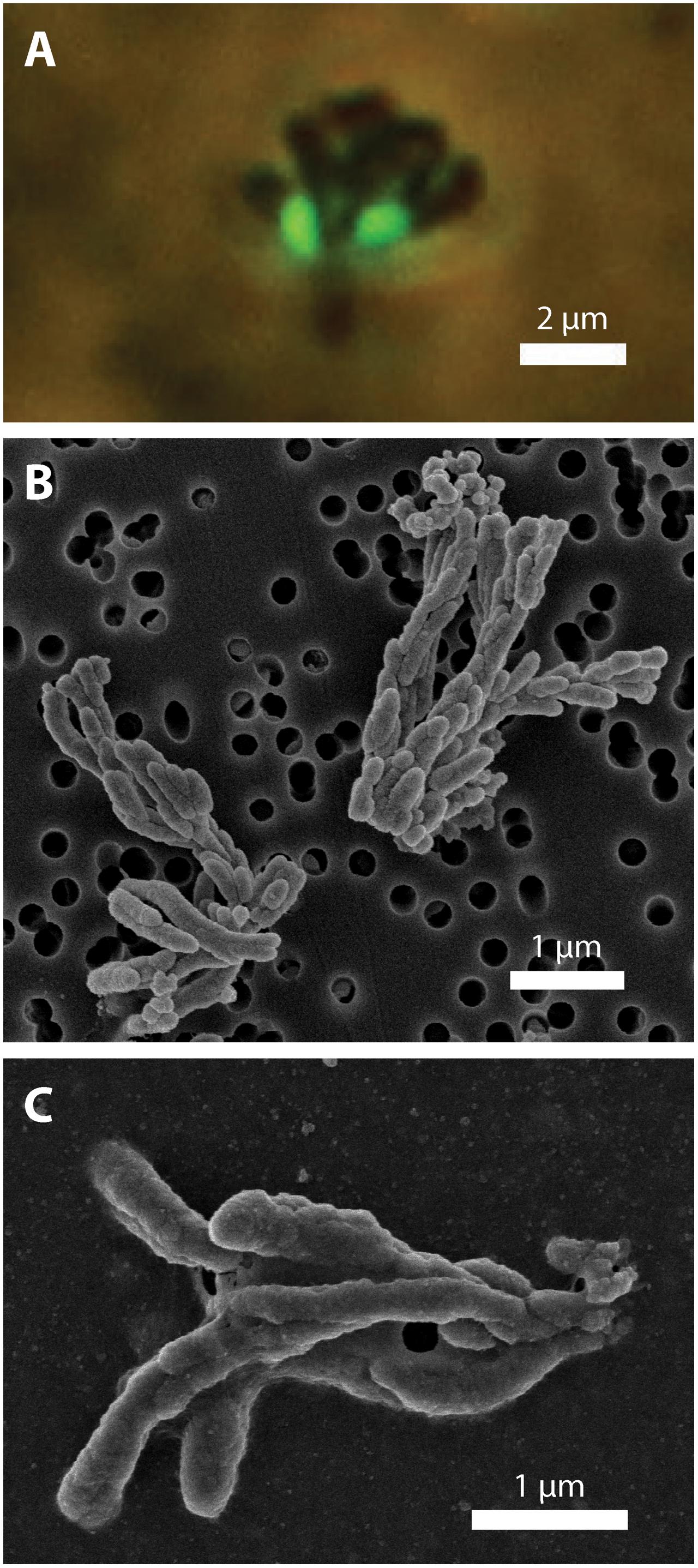
FIGURE 4. CP strain cell and biomineral micrographs. (A) Phase contrast and fluorescence micrograph (overlay) of strain CP-5 showing bean-shaped cells (green), stained with SYBR Green I, and iron oxide dreads. (B) Scanning electron micrograph of bundles of iron oxide dreads produced by strain CP-8. (C) Scanning electron micrograph of bundles of iron oxide dreads produced by strain CP-5.
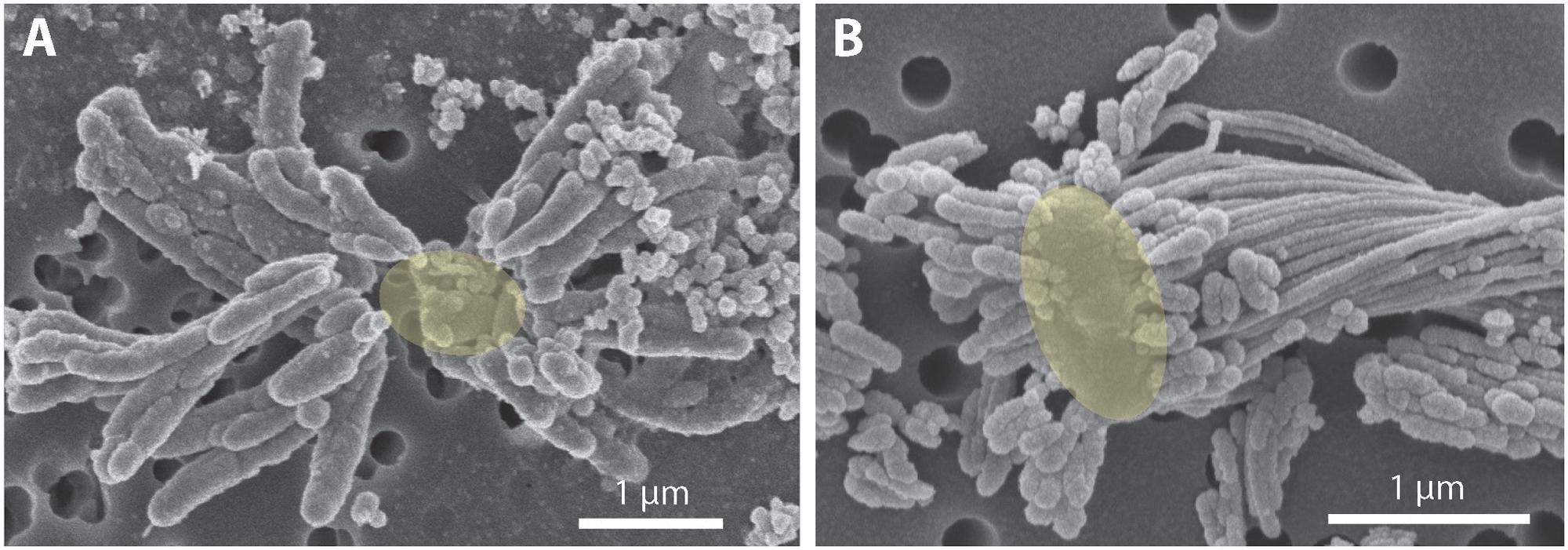
FIGURE 5. Scanning electron micrograph of extracellular iron oxide biomineral structures. (A) Dreads produced by strain CP-8, with likely location of missing cell denoted as a yellow oval. (B) Dreads surrounding a freshwater FeOB Ferriphaselus R-1 cell, highlighted in yellow (modified from Kato et al., 2015). To the right of the cell, a longer iron oxide stalk produced by R-1 is also visible.
General Genome Features of Strains CP-5 and CP-8
The CP-5 and CP-8 strain genomes are both single circular chromosomes, which make them the first and only closed Zetaproteobacteria genomes. High consensus read coverage (382x for strain CP-5; 300x for strain CP-8) led to significant overlap of the ends of each CP strain genome assembly (15 and 9 kb, respectively), overall providing confidence in genome accuracy and completion. The CP-5 and CP-8 strain genomes are 2.54 and 2.30 Mbp, respectively; sizes, GC contents, and COG distributions are comparable to the other sequenced Zetaproteobacteria isolates (Table 1 and Supplementary Tables ST2, ST3). The COG distribution of the two CP strains is highly similar (Supplementary Table ST3) and there are no obvious major metabolic or physiological differences apparent in the genes distinguishing the two strains from one another (Supplementary Tables ST4, ST5). The CP-5 and CP-8 strain genomes contain 258 and 211 genes without homologs in other Zetaproteobacteria isolates. As described below, the CP strains share several genes that are absent or rare in the other sequenced Zetaproteobacteria and may represent adaptations to life in the water column.
Electron Transport Chain Analysis
Based on the electron transport-related genes identified in the genomes (Supplementary Table ST6), strains CP-5 and CP-8 appear to have an electron transport system similar to other Zetaproteobacteria (Figure 6), with some key differences, described below. Like all microaerophilic FeOB, including Zetaproteobacteria, the CP strains have genes encoding the putative Fe oxidase, outer membrane cytochrome Cyc2 (e.g., Barco et al., 2015; Kato et al., 2015; Mumford et al., 2016), which has been proven to oxidize Fe(II) in Acidithiobacillus ferrooxidans (Castelle et al., 2008). The CP strain cyc2 gene sequences are homologous to characterized cyc2 sequences from PV-1 (e-values: 10-72 to 10-73; Supplementary Table ST6; Barco et al., 2015) and each contain a predicted signal sequence, one CXXCH heme-binding motif, and an outer membrane beta barrel domain as with other cyc2 gene sequences (White et al., 2016). The CP strains both lack the putative outer membrane Fe oxidase MtoA (Liu et al., 2012), consistent with our observation that Cyc2 is common amongst microaerophilic and other FeOB, while MtoA is rare (Kato et al., 2015).
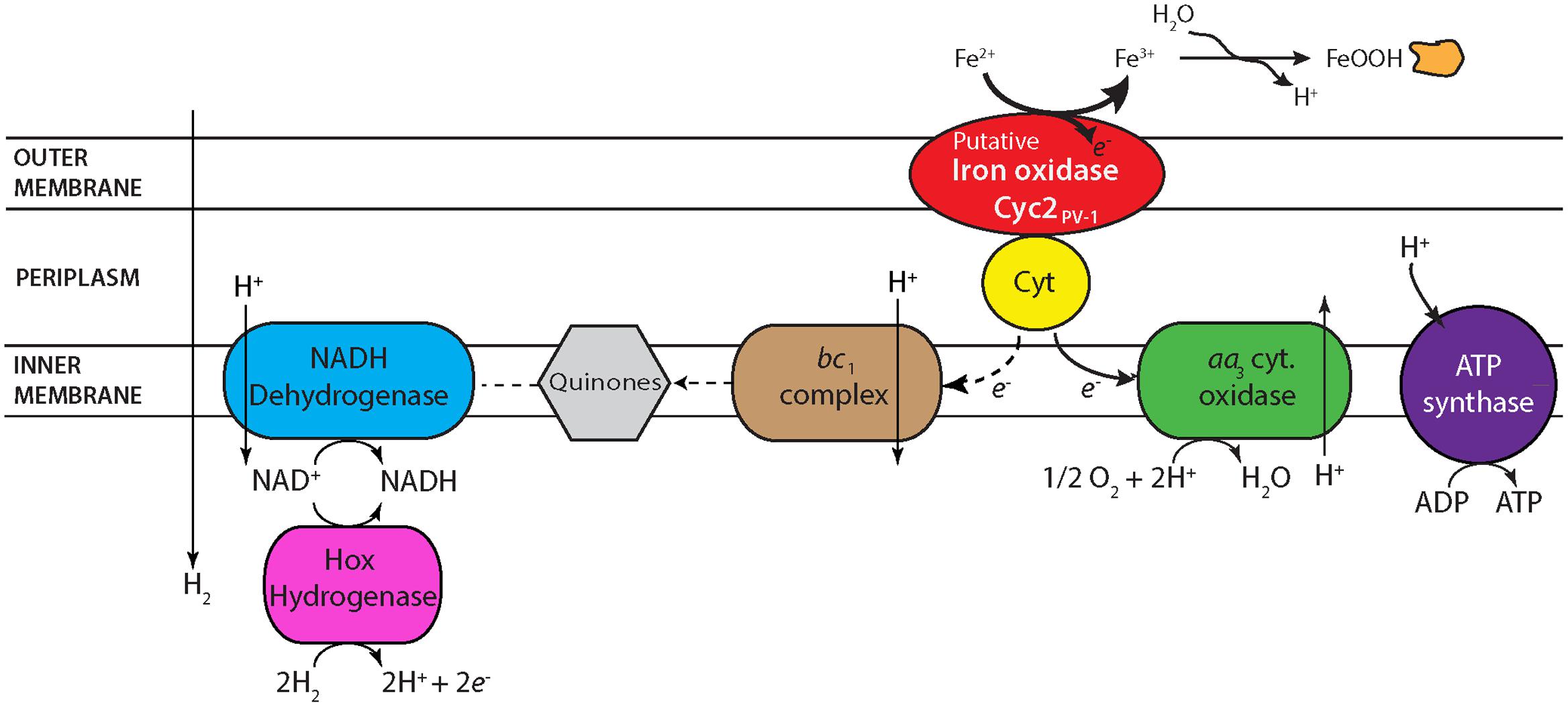
FIGURE 6. Proposed electron transport system in strains CP-5 and CP-8 based on genomic analysis. See text for further description.
One unusual feature in the CP strain genomes is the possession of aa3-type cytochrome c oxidases (Supplementary Table ST6) in place of the cbb3-type cytochrome c oxidases present in all other Zetaproteobacteria genomes to date. Several SAGs and isolate M. micogutta have both aa3 and cbb3-type oxidases (Field et al., 2015), but no other Zetaproteobacteria has only the aa3-type. Between the aa3 and cbb3-type oxidases, the cbb3-type oxidase is considered to be better adapted for low O2 conditions given its higher affinity for oxygen (Arai et al., 2014), consistent with the association of Zetaproteobacteria with low O2 habitats. Conversely, the lower oxygen affinity of the aa3-type oxidase suggests adaptation to somewhat higher O2 conditions. Though the Km values of both oxidases would be considered low O2 (Km, cbb3 on the order of nanomolar and Km, aa3 on the scale of micromolar O2; Arai et al., 2014), the difference suggests that the CP strains may have a higher O2 niche. Curiously, the single Zetaproteobacteria isolate shown capable of growing in O2-saturated waters, DIS-1, possesses only the cbb3-type oxidase, suggesting other genetic adaptations contribute to its O2 tolerance. Still, the uncommon possession of only aa3-type oxidases in the CP strains likely represents an adaptation to frequent exposure to high O2 waters.
Periplasmic electron carriers are required for electron transport between Cyc2 and the terminal oxidase. Because of the high redox potential of Fe(II)/Fe(III)OOH (24 mV for ferrihydrite; Majzlan, 2012), these electron carriers are most likely cytochromes. In A. ferrooxidans, the cytochrome Cyc1 is one of these intermediate electron carriers (Malarte et al., 2005; Castelle et al., 2008). While there are homologs to cyc1 in several Zetaproteobacteria isolate genomes, the CP strain genomes lack homologs. However, Cyc1 is suggested to interact specifically with the cbb3-type oxidase in M. ferrooxydans PV-1 (Barco et al., 2015), making the lack of cyc1 homologs in the CP strains consistent with the absence of the cbb3-type oxidase. There is a different predicted periplasmic cytochrome found in the CP strains, which may transfer electrons between Cyc2 and the aa3-type terminal oxidase. This potential periplasmic cytochrome gene in both strains CP-5 and CP-8 codes for a 127aa protein, with a signal sequence and one CXXCH heme-binding motif (Supplementary Table ST6). In the strain CP-8 genome, this gene is located near the genes encoding the terminal aa3-type oxidase, but it is in a different genomic neighborhood in strain CP-5. Homologs of this periplasmic cytochrome are found in several Zetaproteobacteria isolates (PV-1, JV-1, M34, and EKF-M39; e-values 10-23 to 10-21) and are also near terminal oxidases. The genomic context and presence in several Zetaproteobacteria (including seven SAGs) suggests that this predicted cytochrome plays a role in Fe(II) oxidation and energy conservation.
The high potential of Fe(II)/Fe(III)OOH requires FeOB to regenerate NADH using either reverse electron transport, or an alternate reductant. Like other Zetaproteobacteria, the CP strains have the components for reverse electron transport: a bc1 complex, ubiquinone synthesis genes, and NADH dehydrogenase (Figure 6). However, the CP strains are the only Zetaproteobacteria isolates that definitively lack an alternative complex III, indicating that it is not a necessary component for neutrophilic Fe(II) oxidation, despite its conservation in other FeOB (Singer et al., 2011; Kato et al., 2015). Both CP strains have a cytochrome b/diheme cytochrome c gene cluster (Supplementary Table ST6) that likely also plays an electron transport role. Present in all Zetaproteobacteria isolates and several SAGs, these two genes in each of the CP strains are also homologous to the fused cytbc gene in the Fe-oxidizing KS culture Gallionellaceae, which was proposed to pass electrons from periplasmic cytochromes to quinones and on toward denitrification (He et al., 2016). The CP strains lack a dissimilatory nitrate reductase, but this novel bc complex may still function to reduce quinones for reverse electron transport. Both CP strains have genes coding sulfide quinone oxidoreductases (Supplementary Table ST6), which would allow them to take advantage of the high sulfide concentrations in the Chesapeake Bay to reduce quinones. The CP strains also have hoxWHYUF genes, which could allow them to use H2 to reduce NAD+ to NADH (Tran-Betcke et al., 1990; Thiemermann et al., 1996), relieving at least some of the need for reverse electron transport. In sum, the CP strain genomes show multiple options for regenerating NADH for carbon fixation and biosynthetic pathways.
Carbon Metabolism Analysis
The CP strain genomes are consistent with autotrophy in these organisms. The CP strains each possess complete gene sets for the Calvin–Benson–Bassham (CBB) cycle, including form II ribulose 1,5-bisphosphate carboxylase (RuBisCO) for fixing inorganic carbon (Supplementary Table ST7). Also present are the genes to convert the chief product of the CBB cycle, glycerate 3P, to pyruvate, which can then be utilized in the predicted, complete tricarboxylic acid (TCA) cycle to generate energy and biosynthetic precursors (Supplementary Table ST7).
The CP strain genomes each contain form II RuBisCO and lack form I RuBisCO, as observed in several other Zetaproteobacteria (e.g., EKF-M39, SV108 M. micogutta, Zetaproteobacteria SAGs). CO2 concentrations were ∼70–80 μM in the waters from which the CP strains were isolated (Cai et al., 2017). These concentrations are within the Km,CO2 ranges for both Form I and Form II RuBisCO (Badger and Bek, 2008), so either should be functional in this environment. However, the absence of form I RuBisCO is somewhat unexpected in the CP strains given that form I is considered to be better adapted to higher O2 conditions than form II (Badger and Bek, 2008) and would provide a potential adaptation for more efficient carbon fixation during exposure to higher O2 waters. Indeed, Mumford et al. (2016) suggest that the presence of both forms of RuBisCO in DIS-1 helps adapt this strain to a larger range of oxygenated environments. In any case, the form II RuBisCO genes in the CP strain genomes support Fe(II) oxidation chemolithoautotrophy, consistent with all other Zetaproteobacteria.
Support for strict autotrophy comes from the apparent lack of transporters for organic carbon substrates. Close analysis of a cluster of genes annotated as phosphotransferase (PTS) system genes in each CP strain genome suggests they do not make up a complete system for carbohydrate uptake, but may instead play a role in nitrogen regulation (Supplementary Table ST7). The CP strain genomes also lack complete ABC transport systems for sugars, peptides, and amino acids, making heterotrophy unlikely.
Unusual Genomic Features for Biofilm Formation
We surveyed the CP genomes for genes that could represent adaptations to life in the Chesapeake Bay redoxcline, focusing on ones that were rare or absent in other Zetaproteobacteria. We found two gene clusters related to biofilm formation: the Wsp system, a chemosensory system that produces the biofilm-inducing signal molecule cyclic dimeric guanosine monophosphate (c-di-GMP), and the widespread colonization island (WCI), a pilus assembly system that enables surface attachment.
Each CP strain genome includes a complete wsp gene cluster (wspABCDEFR), which encodes the Wsp chemosensory system (Figure 7 and Supplementary Table ST8). Genetic and protein functional studies have demonstrated the role of these genes in biofilm formation in Pseudomonas, the model organism for the Wsp system (D’Argenio et al., 2002; Hickman et al., 2005). The Wsp system is homologous to the Che chemotaxis system; both contain a methyl-accepting chemotaxis protein (MCP) chemoreceptor and a complex of signal transduction proteins (Bantinaki et al., 2007). However, the Wsp system regulates biofilm formation rather than flagellar motor switching as in the Che system. The major distinguishing feature of the Wsp system is subunit WspR, a diguanylate cyclase response regulator required for Wsp system-induced biofilm production. Phosphorylation stimulates WspR to synthesize the signal molecule cyclic di-GMP (c-di-GMP), which induces biofilm formation pathways, including the production of extracellular polymeric substances (EPSs; D’Argenio et al., 2002; Hickman et al., 2005; Malone et al., 2007). The signal activating the Wsp system MCP, WspA, remains unclear, but has been shown to be related to physical and/or chemical signals associated with growth on surfaces (Güvener and Harwood, 2007; O’Connor et al., 2012). This suggests that given a mechanism for initial surface attachment, the Wsp system could enable the CP strains to form biofilms to colonize particles in the water column.
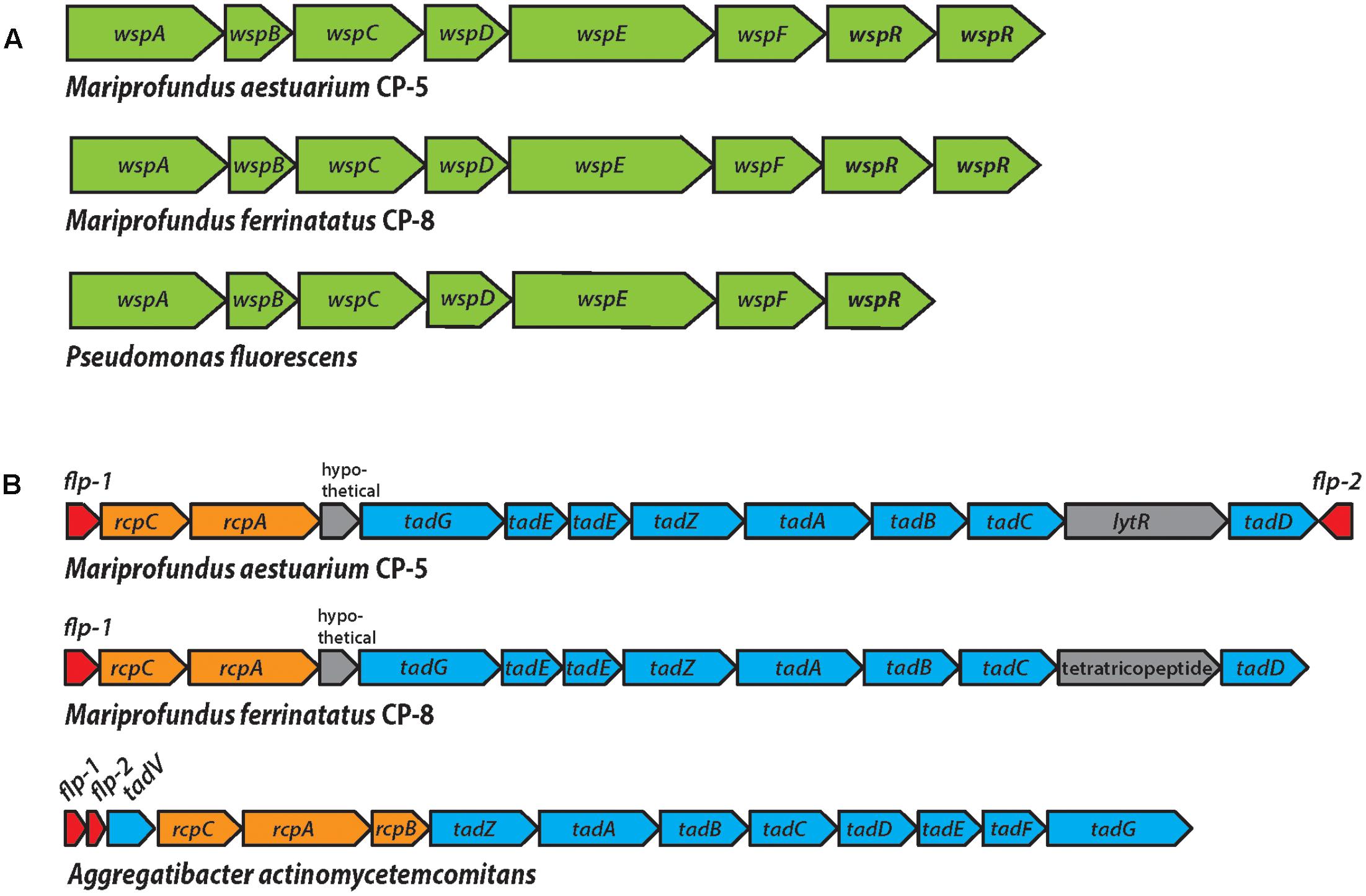
FIGURE 7. (A) The Wsp operon in strains CP-5 and CP-8, showing synteny with the well-studied Pseudomonas fluorescens Wsp operon. (B) The WCI genes in strains CP-5 and CP-8, showing similar gene content, but somewhat different gene order compared to Aggregatibacter actinomycetemcomitans.
Each CP strain genome contains two predicted copies of wspR that are homologs of the gene sequences of the functionally and structurally characterized WspR of Pseudomonas aeruginosa (e-values: 10-111 to 10-93; Supplementary Table ST8; De et al., 2008). Like Pseudomonas wspR, all CP strain wspR sequences contain the conserved C-terminal diguanylate cyclase domain GGEEF in the active site loop and the RxxD motif making up the conserved inhibitory site in GGEEF domain-containing proteins (De et al., 2008, 2009). The remaining Wsp system subunit genes (wspABCDEF) are also present in the CP strain genomes and homologous to the wsp counterparts in Pseudomonas species (e-values 0 to 10-35; Supplementary Table ST8). Other Zetaproteobacteria genomes either lack wsp gene homologs or only contain single subunits (TAG-1, wspR; SV108, wspR; M34, wspE). The exception is Zetaproteobacteria SAG C09, a Loihi Seamount Fe mat single cell genome (2.45 Mb; Field et al., 2015), which contains wspABCDEF, but clearly lacks wspR, with the gene cluster in the middle of a contig. Instead, the immediately adjacent features are a pseudouridine synthase and a tRNA, which are not obviously related to biofilm formation, though further downstream in the cluster (6 ORFs away from wspF), there is an adenylate cyclase gene. Adenylate cyclase forms the signal molecule cAMP, which is associated with many processes (Gancedo, 2013), one of which may be initial cell attachment to surfaces (O’Toole and Wong, 2016). Nevertheless, the lack of wspR in Zetaproteobacteria C09 genome suggests that the wspABCDEF homologs in C09 have a different role and output than in the CP strains. The absence of complete Wsp systems in Zetaproteobacteria genomes other than the CP strains suggests that Wsp-related biofilm formation may be an adaptation specific to pelagic Zetaproteobacteria for particle colonization.
The second genomic feature of the CP strains that is rare among Zetaproteobacteria is the WCI, a gene cluster responsible for tight attachment to surfaces (Figure 7 and Supplementary Table ST9). The WCI includes flp-1, a gene encoding for the major structural component of the type IV Flp (fimbrial low-molecular-weight protein) pili, as well as the tad (tight adherence) genes, and rcp pilus assembly genes (Planet et al., 2003). First characterized in Aggregatibacter actinomycetemcomitans, but studied in numerous other organisms including Caulobacter and Pseudomonas (Skerker and Shapiro, 2000; Bentzmann et al., 2006), the WCI genes assemble adhesive Flp pili that mediate tenacious surface adherence and biofilm formation (Kachlany et al., 2000, 2001; Planet et al., 2003). The CP strain genomes each contain flp-1 gene sequences that were confirmed to contain the conserved processing site motif GXXXXEY (Inoue et al., 1998; Kachlany et al., 2001), as well as tadABCDEGZ and rcpAC (Supplementary Table ST9). Both CP strain genomes have two predicted copies of tadE, one of which is likely tadF given the high sequence similarity of these two subunits (Tomich et al., 2006). Two WCI genes, tadV and rcpB, are not present in the CP genomes, which may be due to the general variability of WCI organization across bacteria or the potential for individual species to possess novel genes in place of individual WCI components (Tomich et al., 2007). For example, P. aeruginosa was demonstrated to encode a novel prepilin peptidase, FppA, instead of TadV (Bentzmann et al., 2006), suggesting that the CP strains could possess different versions of TadV and RcpB that would not be recognized by genomic analysis alone. The set of WCI genes in the CP strain genomes is also found in EKF-M39, but entirely absent from all other Zetaproteobacteria isolates, suggesting that only a small subset of Zetaproteobacteria can produce Flp pili. The mechanisms thought to regulate WCI Flp pilus production vary across species (Tomich et al., 2007) and could be controlled by c-di-GMP signaling in the CP strains. In each CP strain genome, predicted WCI components are adjacent to pilZ domain-containing genes (Supplementary Table ST9), which have been linked to c-di-GMP-regulated fimbriae production (Johnson et al., 2011; Wilksch et al., 2011). The CP strain pilZ sequences contain the c-di-GMP binding motifs RxxxR-(D/ N)x(S/A)xxG (Ryjenkov et al., 2006; Christen et al., 2007), and thus may connect Wsp system c-di-GMP synthesis to WCI Flp pilus production to promote a surface-attached biofilm lifestyle.
Adaptations of Pelagic Zetaproteobacteria in Estuarine Water Columns
In many ways, the Chesapeake strains are like other Zetaproteobacteria isolates: they are autotrophic, obligate Fe(II)-oxidizers, with similar electron transport and carbon fixation mechanisms. However, the CP strains differ in that they produce distinctive dreadlock-shaped oxides that are much smaller than the twisted stalks common to other Zetaproteobacteria. This is likely a key difference between pelagic and benthic FeOB. Our previous work has shown that Fe biomineral stalks are the building blocks of Fe microbial mats (Chan et al., 2016). Millimeters-long stalks create a highly porous framework unlike any other biofilm or mat, in that the bulk is made of mineral, without much interstitial EPS. This architecture allows Fe(II)-bearing fluids to flow through mats, enabling FeOB to biomineralize and position themselves at a benthic Fe(II)/O2 interface (e.g., hydrothermal vent on the seafloor). In contrast, for FeOB that need to maintain position in a water column, large stalk structures would be undesirable because of their weight. Instead, the CP strains make smaller dreads that can be constantly shed, thereby eliminating the heavy oxide by-products to avoid sinking out of the oxic–anoxic transition zone.
Instead of making Fe mats, it appears that the CP strains have the genes to form standard, EPS-bound biofilms, as is typical of organisms possessing the WCI and Wsp system. EPS production enables marine bacteria to colonize suspended particle surfaces (Decho, 1990), and is likely key for CP strains to attach to and access nutrients from Fe(II)-bearing minerals such as FeS. In seasonally stratified Chesapeake Bay waters, particulate FeS is formed by an O2-Fe-H2S catalytic cycle where sulfidic (H2S) bottom waters reduce solid Fe(III) oxyhydroxides to dissolved Fe(II) and react further to precipitate solid FeS particles (Figure 8A; MacDonald et al., 2014; Hansel et al., 2015; Field et al., 2016). Indeed, a majority of the Fe(II) in the Chesapeake Bay oxic–anoxic transition zone was found to be particulate (Field et al., 2016), likely as FeS. Still, FeS particles are likely to be sparse given the low overall Fe(II) concentration, necessitating strategies for the CP strains to recognize and firmly attach to these Fe(II)-bearing particles. The c-di-GMP produced by the Wsp system could stimulate WCI Flp pilus production and other pathways involved in biofilm production to facilitate tight cell attachment to suspended FeS particles (Figure 8B). These small cell-mineral aggregates could further assemble and grow into larger flocs, incorporating other suspended particles and cells (Figure 8C). These flocs could trap more FeS for consumption (Figure 8D), and any trapped dreads could be recycled back to FeS if flocs settle into sulfidic bottom waters (Figure 8E). Floc formation can further benefit the CP strains by effectively increasing their O2 tolerance, as previously described for freshwater floc-dwelling FeOB (Elliott et al., 2014). Diffusion of O2 into flocs is slowed by the EPS matrix (Brezonik, 1993). If floc-dwelling bacteria consume O2 faster than it can diffuse in, low oxic or anoxic microenvironments develop within the floc structure (Flemming and Wingender, 2001; Han et al., 2012). Should a CP strain floc be mixed into oxic layers of the water column, low [O2] microenvironments would provide a niche where the CP strains could still compete with abiotic oxidation for matrix-bound Fe(II). In all, biofilm-related genes would give the CP Zetaproteobacteria multiple advantages for persisting in the water column despite low Fe(II) and fluctuating O2 conditions.
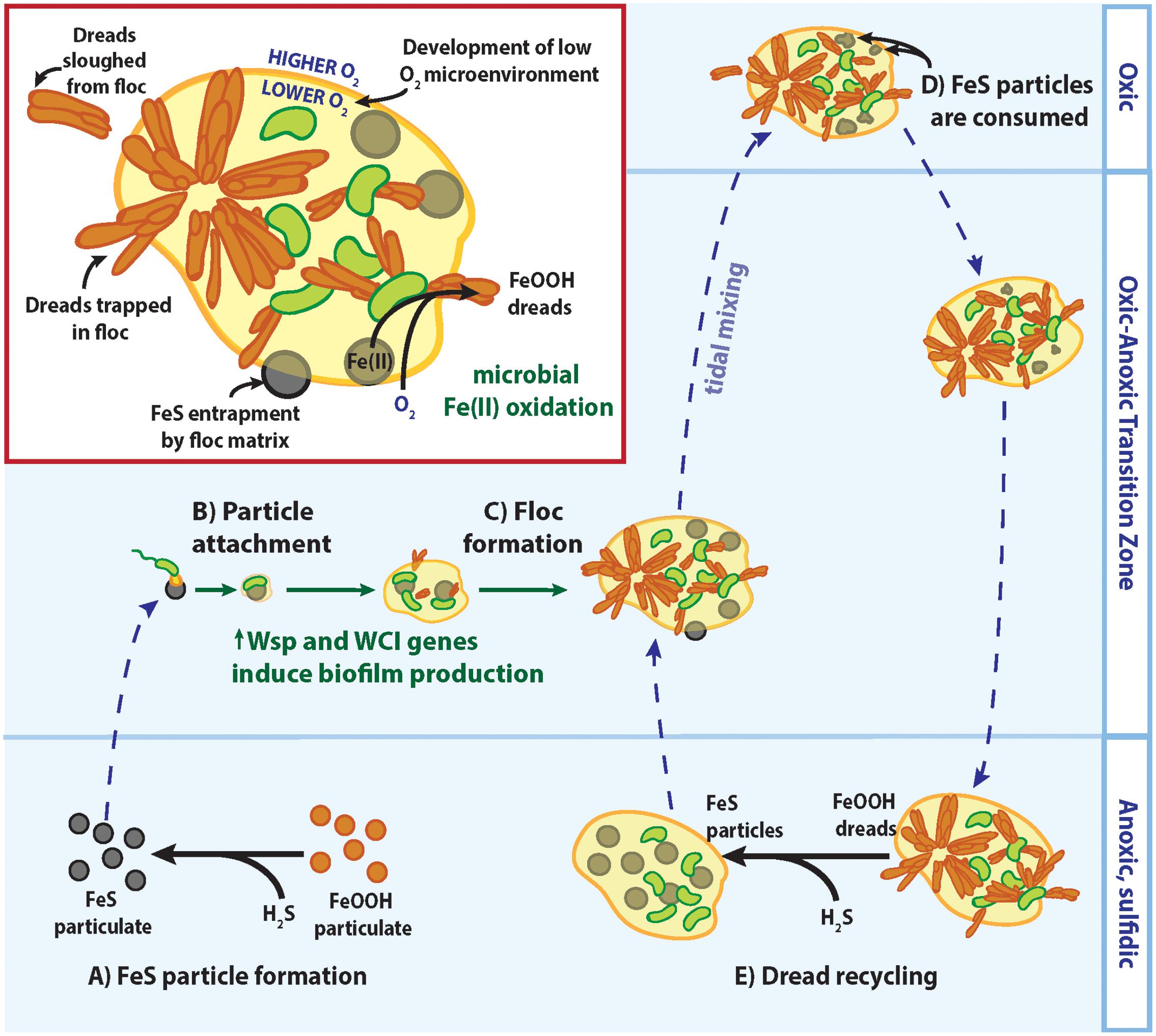
FIGURE 8. Model of CP strain floc formation and interactions with Fe/S biogeochemical cycling in redox-stratified waters. See text for detailed descriptions of (A–E).
Mariprofundus aestuarium CP-5 and Mariprofundus ferrinatatus CP-8 add to the growing number of Zetaproteobacteria isolates that form a closely related phylogenetic cluster despite differing environmental origins and lifestyles. Our analyses suggest that key genes can confer specialized strategies for these organisms to live in diverse environmental niches. If broadly true, the Fe-oxidizing Zetaproteobacteria would be expected to live anywhere that Fe(II) and O2 are available, and thereby be a widespread driver of marine Fe cycling.
Description of Mariprofundus aestuarium sp. nov.
Mariprofundus aestuarium [aes.tu.a’ri.um. L. n. aestuarium an estuary].
Cells are slightly curved, short rods (0.43 ± 0.05 μm × 1.01 ± 0.18 μm). Does not form spores. Mesophilic and neutrophilic. Microaerobic, growing with opposing gradients of Fe(II) and O2. Autotrophic. Grows at 10–30°C (optimally at 20–25°C), pH 5.5–8.3 (optimally at pH 6.9–7.2), and 7–31.5‰ salinity (optimally at 14–17.5‰ salinity). Utilizes ferrous iron as an energy source for lithotrophic growth. Does not utilize thiosulfate, sulfide, pyruvate, glucose, or acetate as an energy source. Produces extracellular dreadlock-like iron oxides around the cell. The doubling time under optimal conditions is 19.5 h. The type strain is CP-5T, isolated from redox-stratified waters in the Chesapeake Bay, United States. The total DNA G+C content of the type strain is 51.5 mol%.
Description of Mariprofundus ferrinatatus sp. nov.
Mariprofundus ferrinatatus [fer.ri.na’ta.tus. L. neut. n. ferrum iron; L. masc. n. natatus floating; N.L. masc. n. ferrinatatus floating iron].
Cells are slightly curved, short rods (0.45 ± 0.04 μm × 0.91 ± 0.08 μm). Does not form spores. Mesophilic and neutrophilic. Microaerobic, growing with opposing gradients of Fe(II) and O2. Autotrophic. Grows at 15–35°C (optimally at 25–30°C), pH 5.5–8.3 (optimally at pH 6.9–7.2), and 7–31.5‰ salinity (optimally at 14–17.5‰ salinity). Utilizes ferrous iron as an energy source for lithotrophic growth. Does not utilize thiosulfate, sulfide, pyruvate, glucose, or acetate as an energy source. Produces extracellular dreadlock-like iron oxides around the cell. The doubling time under optimal conditions is 27 h. The type strain is CP-8T, isolated from redox-stratified waters in the Chesapeake Bay, United States. The total DNA G+C content of the type strain is 53.7 mol%.
Author Contributions
CC conceived of and directed the study, contributed to genome analysis, and wrote the paper. BC performed Fe(II) oxidation growth curve experiments, analyzed genomes, and wrote this paper. SK isolated strains CP-5 and CP-8, performed physiological growth tests and microscopy, prepared the strains for sequencing, and wrote portions of the paper. SM helped plan and prepare DNA for sequencing, performed genome quality control, and performed phylogenetic analyses. EF initiated and advised on genome analysis. All coauthors edited the manuscript.
Funding
This research was supported by NASA Exobiology Program Grant NNX12AG20G, NSF CAREER Geobiology grant EAR-1151682, and NSF Biological Oceanography grant OCE-1155290 (to CC), and a Grant-in-Aid for JSPS Overseas Research Fellow and Young Scientists (A)-16H06180 (to SK).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Deborah Powell and Shannon Modla for their assistance with SEM and TEM sample preparation and analysis. We also thank Brewster Kingham and Olga Shevchenko for advice and assistance in genome sequencing and analysis.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01280/full#supplementary-material
Footnotes
References
Arai, H., Kawakami, T., Osamura, T., Hirai, T., Sakai, Y., and Ishii, M. (2014). Enzymatic characterization and in vivo function of five terminal oxidases in Pseudomonas aeruginosa. J. Bacteriol. 196, 4206–4215. doi: 10.1128/JB.02176-14
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75
Badger, M. R., and Bek, E. J. (2008). Multiple Rubisco forms in proteobacteria: their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 59, 1525–1541. doi: 10.1093/jxb/erm297
Bantinaki, E., Kassen, R., Knight, C. G., Robinson, Z., Spiers, A. J., and Rainey, P. B. (2007). Adaptive divergence in experimental populations of Pseudomonas fluorescens. III. Mutational origins of wrinkly spreader diversity. Genetics 176, 441–453. doi: 10.1534/genetics.106.069906
Barco, R. A., Emerson, D., Sylvan, J. B., Orcutt, B. N., Jacobson Meyers, M. E., Ramírez, G. A., et al. (2015). New insight into microbial iron oxidation as revealed by the proteomic profile of an obligate iron-oxidizing chemolithoautotroph. Appl. Environ. Microbiol. 81, 5927–5937. doi: 10.1128/AEM.01374-15
Bentzmann, S., Aurouze, M., Ball, G., and Filloux, A. (2006). FppA, a novel Pseudomonas aeruginosa prepilin peptidase involved in assembly of type IVb pili. J. Bacteriol. 188, 4851–4860. doi: 10.1128/JB.00345-06
Brezonik, P. L. (1993). Chemical Kinetics and Process Dynamics in Aquatic Systems. Boca Raton, FL: CRC Press.
Cai, W. J., Huang, W. J., Luther, G. W., Pierrot, D., Li, M., Testa, J., et al. (2017). Redox reactions and weak buffer capacity lead to acidification in the Chesapeake Bay. Nat. Commun. (in press).
Castelle, C., Guiral, M., Malarte, G., Ledgham, F., Leroy, G., Brugna, M., et al. (2008). A new iron-oxidizing/O2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 283, 25803–25811. doi: 10.1074/jbc.M802496200
Chan, C. S., Fakra, S. C., Emerson, D., Fleming, E. J., and Edwards, K. J. (2011). Lithotrophic iron-oxidizing bacteria produce organic stalks to control mineral growth: implications for biosignature formation. ISME J. 5, 717–727. doi: 10.1038/ismej.2010.173
Chan, C. S., McAllister, S. M., Leavitt, A. H., Glazer, B. T., Krepski, S. T., and Emerson, D. (2016). The architecture of iron microbial mats reflects the adaptation of chemolithotrophic iron oxidation in freshwater and marine environments. Front. Microbiol. 7:796. doi: 10.3389/fmicb.2016.00796
Charette, M. A., Sholkovitz, E. R., and Hansel, C. M. (2005). Trace element cycling in a subterranean estuary: Part 1. Geochemistry of the permeable sediments. Geochim. Cosmochim. Acta 69, 2095–2109. doi: 10.1016/j.gca.2004.10.024
Christen, M., Christen, B., Allan, M. G., Folcher, M., Jeno, P., Grzesiek, S., et al. (2007). DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U.S.A. 104, 4112–4117. doi: 10.1073/pnas.0607738104
Cole, J. R., Wang, Q., Fish, J. A., Chai, B., McGarrell, D. M., Sun, Y., et al. (2014). Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42, D633–D642. doi: 10.1093/nar/gkt1244
Dang, H., Chen, R., Wang, L., Shao, S., Dai, L., Ye, Y., et al. (2011). Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ. Microbiol. 13, 3059–3074. doi: 10.1111/j.1462-2920.2011.02583.x
D’Argenio, D. A., Worth Calfee, M., Rainey, P. B., and Pesci, E. C. (2002). Autolysis and autoaggregation in Pseudomonas aeruginosa colony morphology mutants. J. Bacteriol. 184, 6481–6489. doi: 10.1128/JB.184.23.6481
De, N., Navarro, M. V. A. S., Raghavan, R. V., and Sondermann, H. (2009). Determinants for the activation and autoinhibition of the diguanylate cyclase response regulator WspR. J. Mol. Biol. 393, 619–633. doi: 10.1016/j.jmb.2009.08.030
De, N., Pirruccello, M., Krasteva, P. V., Bae, N., Raghavan, R. V., and Sondermann, H. (2008). Phosphorylation-independent regulation of the diguanylate cyclase WspR. PLoS Biol. 6:e67. doi: 10.1371/journal.pbio.0060067
Decho, A. W. (1990). “Microbial exopolymer secretions in ocean environments: their role(s) in food webs and marine processes,” in Oceanography and Marine Biology an Annual Review, Vol. 28, eds H. Barnes and M. Barnes (Aberdeen: Aberdeen University Press), 73–153.
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113
Elliott, A. V. C., Plach, J. M., Droppo, I. G., and Warren, L. A. (2014). Collaborative microbial Fe-redox cycling by pelagic floc bacteria across wide ranging oxygenated aquatic systems. Chem. Geol. 366, 90–102. doi: 10.1016/j.chemgeo.2013.11.017
Emerson, D., and Floyd, M. M. (2005). Enrichment and isolation of iron-oxidizing bacteria at neutral pH. Methods Enzymol. 397, 112–123. doi: 10.1016/S0076-6879(05)97006-7
Emerson, D., and Moyer, C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792.
Emerson, D., and Moyer, C. L. (2002). Neutrophilic Fe-oxidizing bacteria are abundant at the Loihi Seamount hydrothermal vents and play a major role in Fe oxide deposition. Appl. Environ. Microbiol. 68, 3085–3093. doi: 10.1128/AEM.68.6.3085-3093.2002
Emerson, D., Rentz, J. A., Lilburn, T. G., Davis, R. E., Aldrich, H., Chan, C., et al. (2007). A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2:e667. doi: 10.1371/journal.pone.0000667
Emerson, J. B., Thomas, B. C., Alvarez, W., and Banfield, J. F. (2016). Metagenomic analysis of a high carbon dioxide subsurface microbial community populated by chemolithoautotrophs and bacteria and archaea from candidate phyla. Environ. Microbiol. 18, 1686–1703. doi: 10.1111/1462-2920.12817
Field, E. K., Kato, S., Findlay, A. J., MacDonald, D. J., Chiu, B. K., Luther, G. W., et al. (2016). Planktonic marine iron oxidizers drive iron mineralization under low-oxygen conditions. Geobiology 14, 499–508. doi: 10.1111/gbi.12189
Field, E. K., Sczyrba, A., Lyman, A. E., Harris, C. C., Woyke, T., Stepanauskas, R., et al. (2015). Genomic insights into the uncultivated marine Zetaproteobacteria at Loihi Seamount. ISME J. 9, 857–870. doi: 10.1038/ismej.2014.183
Flemming, H. C., and Wingender, J. (2001). Relevance of microbial extracellular polymeric substances (EPSs)–Part I: structural and ecological aspects. Water Sci. Technol. 43, 1–8.
Fullerton, H., Hager, K. W., McAllister, S. M., and Moyer, C. L. (2017). Hidden diversity revealed by genome-resolved metagenomics of iron-oxidizing microbial mats from Loihi Seamount, Hawai’i. ISME J. doi: 10.1038/ismej.2017.40 [Epub ahead of print].
Gancedo, J. M. (2013). Biological roles of cAMP: variations on a theme in the different kingdoms of life. Biol. Rev. 88, 645–668. doi: 10.1111/brv.12020
Güvener, Z. T., and Harwood, C. S. (2007). Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol. Microbiol. 66, 1459–1473. doi: 10.1111/j.1365-2958.2007.06008.x
Han, Y., Liu, J., Guo, X., and Li, L. (2012). Micro-environment characteristics and microbial communities in activated sludge flocs of different particle size. Bioresour. Technol. 124, 252–258. doi: 10.1016/j.biortech.2012.08.008
Hansel, C. M., Lentini, C. J., Tang, Y., Johnston, D. T., Wankel, S. D., and Jardine, P. M. (2015). Dominance of sulfur-fueled iron oxide reduction in low-sulfate freshwater sediments. ISME J. 9, 2400–2412. doi: 10.1038/ismej.2015.50
He, S., Tominski, C., Kappler, A., Behrens, S., and Roden, E. E. (2016). Metagenomic analyses of the autotrophic Fe(II)-oxidizing, nitrate-reducing enrichment culture KS. Appl. Environ. Microbiol. 82, 2656–2668. doi: 10.1128/AEM.03493-15
Hickman, J. W., Tifrea, D. F., and Harwood, C. S. (2005). A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. PNAS 102, 14422–14427. doi: 10.1073/pnas.0507170102
Inoue, T., Tanimoto, I., Ohta, H., Kato, K., Murayama, Y., and Fukui, K. (1998). Molecular characterization of low-molecular-weight component protein, Flp, in Actinobacillus actinomycetemcomitans fimbriae. Microbiol. Immunol. 42, 253–258. doi: 10.1111/j.1348-0421.1998.tb02280.x
Johnson, J. G., Murphy, C. N., Sippy, J., Johnson, T. J., and Clegg, S. (2011). Type 3 fimbriae and biofilm formation are regulated by the transcriptional regulators MrkHI in Klebsiella pneumoniae. J. Bacteriol. 193, 3453–3460. doi: 10.1128/JB.00286-11
Jung, H. B., Charette, M. A., and Zheng, Y. (2009). Field, laboratory, and modeling study of reactive transport of groundwater arsenic in a coastal aquifer. Environ. Sci. Technol. 43, 5333–5338. doi: 10.1021/es900080q
Kachlany, S. C., Planet, P. J., Bhattacharjee, M. K., Kollia, E., DeSalle, R., Fine, D. H., et al. (2000). Nonspecific adherence by Actinobacillus actinomycetemcomitans requires genes widespread in Bacteria and Archaea. J. Bacteriol. 182, 6169–6176. doi: 10.1128/JB.182.21.6169-6176.2000
Kachlany, S. C., Planet, P. J., DeSalle, R., Fine, D. H., Figurski, D. H., and Kaplan, J. B. (2001). Flp-1, the first representative of a new pilin gene subfamily, is required for non-specific adherence of Actinobacillus actinomycetemcomitans. Mol. Microbiol. 40, 542–554. doi: 10.1046/j.1365-2958.2001.02422.x
Kato, S., Krepski, S., Chan, C., Itoh, T., and Ohkuma, M. (2014). Ferriphaselus amnicola gen. nov., sp. nov., a neutrophilic, stalk-forming, iron-oxidizing bacterium isolated from an iron-rich groundwater seep. Int. J. Syst. Evol. Microbiol. 64, 921–925. doi: 10.1099/ijs.0.058487-0
Kato, S., Ohkuma, M., Powell, D. H., Krepski, S. T., Oshima, K., Hattori, M., et al. (2015). Comparative genomic insights into ecophysiology of neutrophilic, microaerophilic iron oxidizing bacteria. Front. Microbiol. 6:1265. doi: 10.3389/fmicb.2015.01265
Konstantinidis, K. T., and Tiedje, J. M. (2005). Towards a genome-based taxonomy for prokaryotes. J. Bacteriol. 187, 6258–6264. doi: 10.1128/JB.187.18.6258
Krepski, S. T., Emerson, D., Hredzak-Showalter, P. L., Luther, G. W., and Chan, C. S. (2013). Morphology of biogenic iron oxides records microbial physiology and environmental conditions: toward interpreting iron microfossils. Geobiology 11, 457–471. doi: 10.1111/gbi.12043
Krumsiek, J., Arnold, R., and Rattei, T. (2007). Gepard: a rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23, 1026–1028. doi: 10.1093/bioinformatics/btm039
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (Chichester: Wiley), 115–117.
Laufer, K., Nordhoff, M., Halama, M., Martinez, R. E., Obst, M., Nowak, M., et al. (2017). Microaerophilic Fe(II)-oxidizing Zetaproteobacteria isolated from low-Fe marine coastal sediments – physiology and characterization of their twisted stalks. Appl. Environ. Microbiol. 83. doi: 10.1128/AEM.03118-16
Laufer, K., Nordhoff, M., Schmidt, C., Behrens, S., Jørgensen, B. B., and Kappler, A. (2016). Co-existence of microaerophilic, nitrate-reducing, and phototrophic Fe(II)-oxidizers and Fe(III)-reducers in coastal marine sediment. Appl. Environ. Microbiol. 82, 1433–1447. doi: 10.1128/AEM.03527-15.Editor
Liu, J., Wang, Z., Belchik, S. M., Edwards, M. J., Liu, C., Kennedy, D. W., et al. (2012). Identification and characterization of MtoA: a decaheme c-type cytochrome of the neutrophilic Fe(ll)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 3:37. doi: 10.3389/fmicb.2012.00037
MacDonald, D. J., Findlay, A. J., McAllister, S. M., Barnett, J. M., Hredzak-Showalter, P., Krepski, S. T., et al. (2014). Using in situ voltammetry as a tool to identify and characterize habitats of iron-oxidizing bacteria: from fresh water wetlands to hydrothermal vent sites. Environ. Sci. Process. Impacts 16, 2117–2126. doi: 10.1039/c4em00073k
Majzlan, J. (2012). “Minerals and aqueous species of iron and manganese as reactants and products of microbial metal respiration,” in Microbial Metal Respiration: From Geochemistry to Potential Applications, 1st Edn, eds J. Gescher and A. Kappler (Berlin: Springer-Verlag), 1–28.
Makita, H., Tanaka, E., Mitsunobu, S., Miyazaki, M., Nunoura, T., Uematsu, K., et al. (2016). Mariprofundus micogutta sp. nov., a novel iron-oxidizing zetaproteobacterium isolated from a deep-sea hydrothermal field at the Bayonnaise knoll of the Izu-Ogasawara arc, and a description of Mariprofundales ord. nov. and Zetaproteobacteria classis nov. Arch. Microbiol 199, 335–346. doi: 10.1007/s00203-016-1307-4
Malarte, G., Leroy, G., Lojou, E., Abergel, C., Bruschi, M., and Giudici- Orticoni, M. T. (2005). Insight into molecular stability and physiological properties of the diheme cytochrome CYC41 from the acidophilic bacterium Acidithiobacillus ferrooxidans. Biochemistry 44, 6471–6481. doi: 10.1021/bi048425b
Malone, J. G., Williams, R., Christen, M., Jenal, U., Spiers, A. J., and Rainey, P. B. (2007). The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153, 980–994. doi: 10.1099/mic.0.2006/002824-0
Markowitz, V. M., Chen, I. M. A., Palaniappan, K., Chu, K., Szeto, E., Grechkin, Y., et al. (2012). IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 40, 115–122. doi: 10.1093/nar/gkr1044
McAllister, S. M., Barnett, J. M., Heiss, J. W., Findlay, A. J., Macdonald, D. J., Dow, C. L., et al. (2015). Dynamic hydrologic and biogeochemical processes drive microbially enhanced iron and sulfur cycling within the intertidal mixing zone of a beach aquifer. Limnol. Oceanogr. 60, 329–345. doi: 10.1111/lno.10029
McAllister, S. M., Davis, R. E., McBeth, J. M., Tebo, B. M., Emerson, D., and Moyer, C. L. (2011). Biodiversity and emerging biogeography of the neutrophilic iron-oxidizing Zetaproteobacteria. Appl. Environ. Microbiol. 77, 5445–5457. doi: 10.1128/AEM.00533-11
McBeth, J. M., Little, B. J., Ray, R. I., Farrar, K. M., and Emerson, D. (2011). Neutrophilic iron-oxidizing “Zetaproteobacteria” and mild steel corrosion in nearshore marine environments. Appl. Environ. Microbiol. 77, 1405–1412. doi: 10.1128/AEM.02095-10
Mori, J. F., Scott, J. J., Hager, K. W., Moyer, C. L., Küsel, K., and Emerson, D. (2017). Physiological and ecological implications of an iron- or hydrogen-oxidizing member of the Zetaproteobacteria, Ghiorsea bivora, gen. nov., sp. nov.ISME J. (in press).
Mumford, A. C., Adaktylou, I. J., and Emerson, D. (2016). Peeking under the iron curtain: development of a microcosm for imaging the colonization of steel surfaces by Mariprofundus sp. strain DIS-1, an oxygen-tolerant Fe-oxidizing bacterium. Appl. Environ. Microbiol. 82, 6799–6807. doi: 10.1128/AEM.01990-16
O’Connor, J. R., Kuwada, N. J., Huangyutitham, V., Wiggins, P. A., and Harwood, C. S. (2012). Surface sensing and lateral subcellular localization of WspA, the receptor in a chemosensory-like system leading to c-di-GMP production. Mol. Microbiol. 86, 720–729. doi: 10.1111/mmi.12013s
Officer, C. B., Biggs, R. B., Taft, J. L., Cronin, L. E., Tyler, M. A., and Boynton, W. R. (1984). Chesapeake Bay anoxia: origin, development, and significance. Science 223, 22–27. doi: 10.1126/science.223.4631.22
O’Toole, G. A., and Wong, G. C. (2016). Sensational biofilms: surface sensing in bacteria. Curr. Opin. Microbiol. 30, 139–146. doi: 10.1016/j.mib.2016.02.004
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, 206–214. doi: 10.1093/nar/gkt1226
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Planet, P. J., Kachlany, S. C., Fine, D. H., DeSalle, R., and Figurski, D. H. (2003). The widespread colonization island of Actinobacillus actinomycetemcomitans. Nat. Genet. 34, 193–198. doi: 10.1038/ng1154
Pruesse, E., Peplies, J., and Glöckner, F. O. (2012). SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829. doi: 10.1093/bioinformatics/bts252
Ryjenkov, D. A., Simm, R., Römling, U., and Gomelsky, M. (2006). The PilZ domain is a receptor for the second messenger c-di-GMP: the PilZ domain protein YcgR controls motility in enterobacteria. J. Biol. Chem. 281, 30310–30314. doi: 10.1074/jbc.C600179200
Singer, E., Emerson, D., Webb, E. A., Barco, R. A., Kuenen, J. G., Nelson, W. C., et al. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6:e25386. doi: 10.1371/journal.pone.0025386
Skerker, J. M., and Shapiro, L. (2000). Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19, 3223–3234. doi: 10.1093/emboj/19.13.3223
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stookey, L. L. (1970). Ferrozine–a new spectrophotometric reagent for iron. Anal. Chem. 42, 779–781. doi: 10.1021/ac60289a016
Thiemermann, S., Dernedde, J., Bernhard, M., Schroeder, W., Massanz, C., and Friederich, B. (1996). Carboxyl-terminal processing of the cytoplasmic NAD-reducing hydrogenase of Alcaligenes eutrophus requires the hoxW gene product. J. Bacteriol. 178, 2368–2374. doi: 10.1128/jb.178.8.2368-2374.1996
Tomich, M., Fine, D. H., and Figurski, D. H. (2006). The TadV protein of Actinobacillus actinomycetemcomitans is a novel aspartic acid prepilin peptidase required for maturation of the Flp1 pilin and TadE and TadF pseudopilins. J. Bacteriol. 188, 6899–6914. doi: 10.1128/JB.00690-06
Tomich, M., Planet, P. J., and Figurski, D. H. (2007). The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5, 363–375. doi: 10.1038/nrmicro1636
Tran-Betcke, A., Warnecke, U., Böcker, C., Zaborosch, C., and Friedrich, B. (1990). Cloning and nucleotide sequences of the genes for the subunits of NAD-reducing hydrogenase. J. Bacteriol. 172, 2920–2929. doi: 10.1128/jb.172.6.2920-2929.1990
White, G. F., Edwards, M. J., Gomez-Perez, L., Richardson, D. J., Butt, J. N., and Clarke, T. A. (2016). “Chapter three - Mechanisms of bacterial extracellular electron exchange,” in Advances in Bacterial Electron Transport Systems and Their Regulation: Advances in Microbial Physiology, Vol. 68, ed. R. K. Poole (Cambridge, MA: Academic Press), 87–138. doi: 10.1016/bs.ampbs.2016.02.002
Wilksch, J. J., Yang, J., Clements, A., Gabbe, J. L., Short, K. R., Cao, H., et al. (2011). MrkH, a novel c-di-GMP-dependent transcriptional activator, controls Klebsiella pneumoniae biofilm formation by regulating type 3 fimbriae expression. PLoS Pathog. 7:e1002204. doi: 10.1371/journal.ppat.1002204
Keywords: iron-oxidizing bacteria, Zetaproteobacteria, biominerals, iron oxides, biofilm, pelagic bacteria
Citation: Chiu BK, Kato S, McAllister SM, Field EK and Chan CS (2017) Novel Pelagic Iron-Oxidizing Zetaproteobacteria from the Chesapeake Bay Oxic–Anoxic Transition Zone. Front. Microbiol. 8:1280. doi: 10.3389/fmicb.2017.01280
Received: 27 March 2017; Accepted: 26 June 2017;
Published: 18 July 2017.
Edited by:
Stefan M. Sievert, Woods Hole Oceanographic Institution, United StatesReviewed by:
Esther Singer, Joint Genome Institute, United StatesBeth Orcutt, Bigelow Laboratory for Ocean Sciences, United States
Copyright © 2017 Chiu, Kato, McAllister, Field and Chan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clara S. Chan, Y3NjaGFuQHVkZWwuZWR1
 Beverly K. Chiu
Beverly K. Chiu Shingo Kato
Shingo Kato Sean M. McAllister
Sean M. McAllister Erin K. Field
Erin K. Field Clara S. Chan
Clara S. Chan