- Department of Biochemistry and Biomedical Sciences, McMaster University, Hamilton, CA, Canada
The genus Mycobacterium contains 188 species including several major human pathogens as well as numerous other environmental species. We report here comprehensive phylogenomics and comparative genomic analyses on 150 genomes of Mycobacterium species to understand their interrelationships. Phylogenetic trees were constructed for the 150 species based on 1941 core proteins for the genus Mycobacterium, 136 core proteins for the phylum Actinobacteria and 8 other conserved proteins. Additionally, the overall genome similarity amongst the Mycobacterium species was determined based on average amino acid identity of the conserved protein families. The results from these analyses consistently support the existence of five distinct monophyletic groups within the genus Mycobacterium at the highest level, which are designated as the “Tuberculosis-Simiae,” “Terrae,” “Triviale,” “Fortuitum-Vaccae,” and “Abscessus-Chelonae” clades. Some of these clades have also been observed in earlier phylogenetic studies. Of these clades, the “Abscessus-Chelonae” clade forms the deepest branching lineage and does not form a monophyletic grouping with the “Fortuitum-Vaccae” clade of fast-growing species. In parallel, our comparative analyses of proteins from mycobacterial genomes have identified 172 molecular signatures in the form of conserved signature indels and conserved signature proteins, which are uniquely shared by either all Mycobacterium species or by members of the five identified clades. The identified molecular signatures (or synapomorphies) provide strong independent evidence for the monophyly of the genus Mycobacterium and the five described clades and they provide reliable means for the demarcation of these clades and for their diagnostics. Based on the results of our comprehensive phylogenomic analyses and numerous identified molecular signatures, which consistently and strongly support the division of known mycobacterial species into the five described clades, we propose here division of the genus Mycobacterium into an emended genus Mycobacterium encompassing the “Tuberculosis-Simiae” clade, which includes all of the major human pathogens, and four novel genera viz. Mycolicibacterium gen. nov., Mycolicibacter gen. nov., Mycolicibacillus gen. nov. and Mycobacteroides gen. nov. corresponding to the “Fortuitum-Vaccae,” “Terrae,” “Triviale,” and “Abscessus-Chelonae” clades, respectively. With the division of mycobacterial species into these five distinct groups, attention can now be focused on unique genetic and molecular characteristics that differentiate members of these groups.
Introduction
The genus Mycobacterium encompasses a large group of Gram-positive, rod-shaped, acid-fast organisms in the phylum Actinobacteria (Hartmans et al., 2006; Gao and Gupta, 2012; Magee and Ward, 2012). Many members are well-known human pathogens, most notably Mycobacterium tuberculosis and Mycobacterium leprae are causative agents of tuberculosis and leprosy, respectively (Medjahed et al., 2010; Magee and Ward, 2012; Lory, 2014). In addition, Mycobacterium species are found to inhabit a diverse range of environments including water bodies, soil, and metalworking fluids (Hartmans et al., 2006; Brzostek et al., 2009; Falkinham, 2009; Tortoli, 2012). At the time of writing, the genus Mycobacterium consists of 188 species with validly published names (www.namesforlife.com) (Parte, 2014). In view of the large numbers of both clinically important as well as environmental species present in a single genus, an understanding of the relationships between these organisms is of much importance (Gao and Gupta, 2012; Magee and Ward, 2012; Tortoli, 2012; Lory, 2014; Fedrizzi et al., 2017). Current understanding of the relationships within the genus Mycobacterium is primarily based on analysis of the 16S rRNA gene sequences and other physical and chemotaxonomic characteristics of the species (Runyon, 1965; Rogall et al., 1990; Stahl and Urbance, 1990; Goodfellow and Magee, 1998; Hartmans et al., 2006; Magee and Ward, 2012). Besides the 16S rRNA, the relationships among the mycobacterial species has also been examined using the 16S-23S spacer sequences (Roth et al., 1998) and several housekeeping genes including hsp65 (Kim et al., 2005; Tortoli et al., 2015), gyrB (Kasai et al., 2000), rpoB (Tortoli, 2012), and gyrA (Guillemin et al., 1995). A number of studies have also been performed on a limited number of mycobacterial species using multilocus sequence analysis based on concatenated sequences of nucleotides or amino acid fragments from several gene sequences viz. 16S rRNA, rpoB, and hsp65 (Kim and Shin, 2017); 16S rRNA, hsp65, sodA, recA, rpoB (Adékambi and Drancourt, 2004) and hsp65, tuf, rpoB, smpB, 16S rRNA, sodA, tmRNA (Mignard and Flandrois, 2008). The results of these studies have provided useful insights into the relationships between members of the genus Mycobacterium.
An important difference observed among the mycobacterial species very early was the differences in their growth rates (Tsukamura, 1967a; Wayne and Kubica, 1986; Magee and Ward, 2012). Based on their rates of growth, Mycobacterium species, in general, can be roughly divided into two groups; one group consists of slow-growing bacteria (i.e., requiring more than 7 days to form colonies), while the second group is comprised of rapid-growing bacteria which require <7 days to form colonies (Tsukamura, 1967a; Wayne and Kubica, 1986; Magee and Ward, 2012; Lory, 2014). The clades encompassing most of the slow-growing mycobacteria also branches distinctly from the fast-growing species in the 16S rRNA trees (Rogall et al., 1990; Stahl and Urbance, 1990; Goodfellow and Magee, 1998), and also in phylogenetic trees based on some other genes/proteins sequences (Adékambi and Drancourt, 2004; Kim et al., 2005; Adékambi et al., 2006a; Mignard and Flandrois, 2008; Tortoli, 2012; Tortoli et al., 2015). Although a broad separation of the slow-growing mycobacteria from the rapid-growing species is generally supported, the reliability of the methods used to discern these two groups, particularly the cohesiveness of the rapid-growing mycobacteria, remains of concern (Magee and Ward, 2012; Tortoli, 2012). Recent studies have also identified some distinct groupings within the slow- or rapid-growing mycobacteria. For example, a clade consisting of Mycobacterium terrae and its closely related members, which exhibits slow to intermediate rate of growth, can be differentiated from other slow-growing members by a characteristic 14 nt insert in the helix 18 of 16S rRNA gene and by means of phylogenetic analysis (Mignard and Flandrois, 2008; Kim et al., 2012; Tortoli, 2012; Tortoli et al., 2013; Ngeow et al., 2015; Vasireddy et al., 2016). Another clade of mycobacterial species closely related to Mycobacterium abscessus, can also be differentiated from other rapid-growing members based on phylogenetic branching and unique pathogenicity profile of its members (Adékambi and Drancourt, 2004; Medjahed et al., 2010; Tortoli, 2012; Wee et al., 2017). In light of the increased awareness of the diversity that exists within the mycobacterial species as well the clinical importance of many of the members from this genus, the need for more robust methods of delineation of different groups that exists within this important group of bacteria is warranted (Fedrizzi et al., 2017).
Due to rapid advances in genome sequencing technology, genome sequences for 150 members from the genus Mycobacterium are now publicly available in the NCBI genome database (https://www.ncbi.nlm.nih.gov/genome/). The analysis of whole genome sequences allows for construction of more robust phylogenetic trees providing greater resolution in identifying the relationships at various taxonomic levels (Wu et al., 2009; Segata et al., 2013; Gupta et al., 2015; Adeolu et al., 2016). A number of recent studies have reported phylogenomic analyses based on large datasets of core genes/proteins from the genomes of 28–47 Mycobacterium species in order to elucidate their relationships (Prasanna and Mehra, 2013; Wang et al., 2015; Fedrizzi et al., 2017; Wee et al., 2017). Based on genome sequences, the genomic relatedness among the organisms can also be determined and this approach is now widely applied in taxonomic studies (Konstantinidis and Tiedje, 2005; Thompson et al., 2013; Qin et al., 2014). In addition, the genome sequences provide a unique resource for comparative genomic studies in identifying molecular markers or signatures that are specifically shared by an evolutionarily related group of organisms and are useful in the demarcation of different taxa and for understanding interrelationships (Gao and Gupta, 2012; Gupta, 2014, 2016a; Adeolu et al., 2016). Of the two types of molecular markers that have proven particularly useful for evolutionary/taxonomic studies, conserved signature indels (CSIs) are amino acid insertions or deletions of fixed lengths that are present at a specific position within a conserved region in an evolutionarily related group of species (Gupta, 2014, 2016b; Naushad et al., 2014). Likewise, conserved signature proteins (CSPs) are proteins, whose homologs are exclusively found in a related-group of organisms (Gao et al., 2006; Gao and Gupta, 2012; Gupta et al., 2015; Gupta, 2016b). The presence of these clade-specific marker gene sequences (or synapomorphies) is most parsimoniously accounted by their initial introduction in a common ancestor of the group followed by vertical inheritance (Gupta, 1998, 2016b; Gao and Gupta, 2012; Naushad et al., 2014).
To reliably understand the relationships within the genus Mycobacterium, we have carried out comprehensive phylogenomic and comparative genomic studies on 150 mycobacterial species, whose genome sequences are now available. Based on genome sequences, robust phylogenetic trees have been constructed based on different large datasets of concatenated protein sequences including two trees based on 1941 and 136 core proteins for the genus Mycobacterium and the phylum Actinobacteria, respectively. Based on genome sequences, the pairwise average amino acid identity (AAI) was also determined for the mycobacterial species. Lastly, our detailed comparative genomic studies on mycobacterial genomes have identified 172 highly specific molecular markers in the forms of CSIs and CSPs, which are either uniquely shared by all members of the genus Mycobacterium or for a number of distinct clades within this genus at multiple phylogenetic levels. Based on the results of these comprehensive analyses, it is now possible to reliably divide the species from the genus Mycobacterium into five main monophyletic clades, which are referred to here as the “Tuberculosis-Simiae” clade, the “Terrae” clade, the “Triviale” clade, the “Fortuitum-Vaccae” clade, and the “Abscessus-Chelonae” clade. Based on the large body of evidence presented here which consistently and strongly supports the existence of these five clades, a proposal is made here to divide the genus Mycobacterium into an emended genus Mycobacterium encompassing the members of the “Tuberculosis-Simiae” clade and four new genera Mycolicibacter gen. nov. (“Terrae” clade), Mycolicibacillus gen. nov. (“Triviale” clade), Mycolicibacterium gen. nov. (“Fortuitum-Vaccae” clade), and Mycobacteroides gen. nov. (“Abscessus-Chelonae” clade).
Methods
Phylogenetic and Genomic Analyses of the Genus Mycobacterium
Phylogenetic trees were constructed for 150 members of the genus Mycobacterium whose genomes are now sequenced (some characteristics of these genomes are listed in Supplementary Table 1) and six members from the order Corynebacteriales (viz. Corynebacterium diphtheriae NCTC 11397, Gordonia bronchialis DSM 43247, Nocardia farcinica NCTC 11134, Rhodococcus erythropolis PR4, Segniliparus rotundus DSM 44985 and Tsukamurella paurometabola DSM 20162), which served as outgroups. The first of these trees was based on 1941 core proteins from the genomes of Mycobacterium species and its construction was carried out by using a software pipeline (Adeolu et al., 2016). Briefly, the CD-HIT program was used (Li and Godzik, 2006; Fu et al., 2012) to identify protein families sharing a minimum of 50% in sequence identity and sequence length and which were found in at least 80% of the input genomes. The Clustal Omega (Sievers et al., 2011) algorithm was used to generate multiple sequence alignment (MSA) of these protein families. The aligned protein families were trimmed with TrimAl (Capella-Gutierrez et al., 2009) to remove poorly aligned regions (Talavera and Castresana, 2007) before concatenation to the other core proteins. The concatenated sequence alignment of 1941 core proteins consisted of 624,360 aligned amino acids. Another comprehensive phylogenetic tree was constructed based on concatenated sequences for 136 proteins, which comprise the phyloeco markers set for the phylum Actinobacteria (Wang and Wu, 2013). Information regarding these proteins is provided in Supplementary Table 2. The profile Hidden Markov Models of these protein families were used for the identification of members of these protein families in the input genomes using HMMer 3.1 (Eddy, 2011). The sequence alignments were trimmed using TrimAl (Capella-Gutierrez et al., 2009) before their concatenation into a single file. The combined sequence from the phyloeco set of proteins consisted of a total of 44,976 aligned amino acids. Maximum likelihood (ML) trees based on both these sequence alignments were constructed using the Whelan and Goldman model of protein sequence evolution (Whelan and Goldman, 2001) in FastTree 2 (Price et al., 2010) and the Le and Gascuel model of protein sequence evolution (Le and Gascuel, 2008) in RAxML 8 (Stamatakis, 2014). Optimization of the robustness of the tree was completed by conducting SH tests (Guindon et al., 2010) in RAxML 8 (Stamatakis, 2014). The identification of the conserved protein families and the construction of phylogenetic trees were completed using an internal software pipeline (Adeolu et al., 2016).
In addition to these two comprehensive trees, another phylogenetic tree was constructed based on concatenated sequences for 8 conserved housekeeping proteins (viz. RpoA, RpoB, RpoC, GyrA, GyrB, Hsp65, EF-Tu and RecA). After removal of non-conserved regions, the concatenated sequence alignment in this case consisted of 6052 aligned amino acids. A maximum likelihood phylogenetic tree based on this sequence was constructed as described above.
The sequence alignments of the 1941 core proteins identified by the above methods were also used to measure genome relatedness. Using the amino acid sequences from these conserved protein families, the amino acid sequence identity between each pair of Mycobacterium genomes was calculated (Thompson et al., 2013).
Information regarding branching of all type species from the genus Mycobacterium in a tree based on 16S rRNA sequences was obtained from the SILVA All Species Tree of Life Project 128 (Quast et al., 2013).
Identification of Conserved Signature Indels (CSIs)
The identification of CSIs was carried out as described in earlier work (Gao and Gupta, 2005; Bhandari et al., 2012; Gupta, 2014; Naushad et al., 2014; Sawana et al., 2014). All annotated proteins from the genomes of M. tuberculosis H37Rv and M. sinense JDM601 were used in these analyses. BLASTp (Altschul et al., 1997) searches were conducted on all protein sequences >100 amino acids in length against the NCBI non-redundant (nr) database. Multiple sequence alignments were generated by obtaining 15–25 homologs from diverse Mycobacterium species and 8–10 homologs from other groups of bacteria. The alignments were visually inspected for sequence gaps of fixed lengths which were flanked on both sides by at least 5 conserved amino acids in the neighboring 30–40 amino acids, and appeared to be shared by either some or all mycobacterial homologs. Query sequences encompassing the potential indel and flanking regions (60–100 amino acids long) were collected and subjected to a more detailed BLASTp search (500 or more hits) to determine the group specificities of the observed indels. Signature files for all CSIs of interest were created using SIG_CREATE and SIG_STYLE programs in the GLEANS software package (available on Gleans.net). Unless otherwise noted, the described CSIs are specific for the indicated groups of species.
Identification of Conserved Signature Proteins (CSPs)
The identification of conserved signature proteins was carried out using the protocol described in earlier work (Gao et al., 2006; Adeolu and Gupta, 2014; Naushad et al., 2014). BLASTp (Altschul et al., 1997) searches were conducted on all sequenced proteins from the genomes of M. tuberculosis H37Rv, M. aurum (LSHTM), M. sinense JDM601 (Zhang et al., 2011), M. triviale DSM 44153 (Fedrizzi et al., 2017), and M. abscessus ATCC 19977 (Ripoll et al., 2009) against the NCBI nr database. Proteins of interest were those where either all significant hits were limited to the genus Mycobacterium or the indicated groups/clades of mycobacteria, or where a large increase in E value was observed from the last hit belonging to these groups and the first hit from any other bacteria, and the E-values for the latter hits were >1e−3 (Gao et al., 2006; Gao and Gupta, 2012; Naushad et al., 2014). However, in some cases, a few proteins where an isolated significant hit from an unrelated group of bacteria was observed were also retained as CSPs specific for the group of interest.
Results
Phylogenomic Analysis of the Genus Mycobacterium
In the present work, two comprehensive phylogenomic trees were constructed based on the genome sequences of 150 Mycobacterium species. The first of these trees was a core genome tree of 1941 proteins, whose homologs are present in at least 80% of the input mycobacterial genomes as well as the outgroup species. The second genome sequence tree was based on 136 proteins, which are part of the phyloeco set for the phylum Actinobacteria. The trimmed concatenated sequence alignments for the two sets of core proteins, which were employed for phylogenetic analyses, consisted of 624,360 and 44,976 aligned amino acids, respectively. Although phylogenetic trees based on core genes/proteins for mycobacterial species have also been constructed in earlier studies (Prasanna and Mehra, 2013; Fedrizzi et al., 2017; Wee et al., 2017), they were based only on a small number (between 28 and 47) of Mycobacterium species. In contrast, the trees produced in this work include information for ~80% (150 of the 188) of all known mycobacterial species and thus constitute the most comprehensive phylogenetic trees constructed for the genus Mycobacterium. In addition to the two core genome protein trees, a maximum-likelihood tree was also constructed based on concatenated sequences of 8 conserved housekeeping proteins.
The ML trees based on the core proteins from mycobacterial genomes and for the phylum Actinobacteria are shown in Figures 1A,B, respectively. The tree based on the 8 conserved proteins is provided as Supplementary Figure 1. In all of these phylogenetic trees, which were rooted using the sequences from the Corynebacteriales species, nearly all of the observed nodes were supported with high (100%) bootstrap scores or SH-values. Further, the majority of the interrelationships among the Mycobacterium species were highly similar and consistent in all constructed trees. In all of these trees, members of the genus Mycobacterium consistently grouped into four main clades and a clade consisting of the M. triviale—M. koreense, as indicated in Figure 1. Three of these clades are comprised of the slow-growing species, whereas the other two clades are mostly made up of the fast-growing species. Of the two clades of fast-growing species, the first clade referred to as the “Abscessus-Chelonae” clade, forms the earliest branching lineage within the genus Mycobacterium. The second clade of the fast-growing species designated as the “Fortuitum-Vaccae” clade, encompasses most of the other fast-growing species including those related to M. fortuitum, M. vaccae, M. parafortuitum, and M. mucogenicum (Hartmans et al., 2006; Magee and Ward, 2012; Lory, 2014). Of the three clades of slow-growing mycobacteria, the clade designated as “Tuberculosis-Simiae,” encompasses most of the clinically important Mycobacterium species including those related to M. tuberculosis, M. avium, M. gordonae, M, kansasii and M. simiae (Magee and Ward, 2012). The two other clades of the slow-growing species, often referred to as part of the “M. terrae complex,” group together and they form a sister clade to the “Tuberculosis-Simiae” clade. Of the two clades which form the “M. terrae complex,” most of the species closely related to M. terrae are part of a clade that is designated here as the “Terrae” clade (Magee and Ward, 2012; Tortoli, 2012; Ngeow et al., 2015). Adjacent to the “Terrae” clade, the species M. koreense and M. triviale form a distinct clade (designated here as the “Triviale” clade), which is separated from members of the “Terrae” clade by a long branch. It is important to note that in the phylogenetic trees shown in Figure 1, the two clades of fast-growing species do not form a monophyletic grouping, whereas the clades corresponding to the slow-growing mycobacteria group together and form a monophyletic lineage.
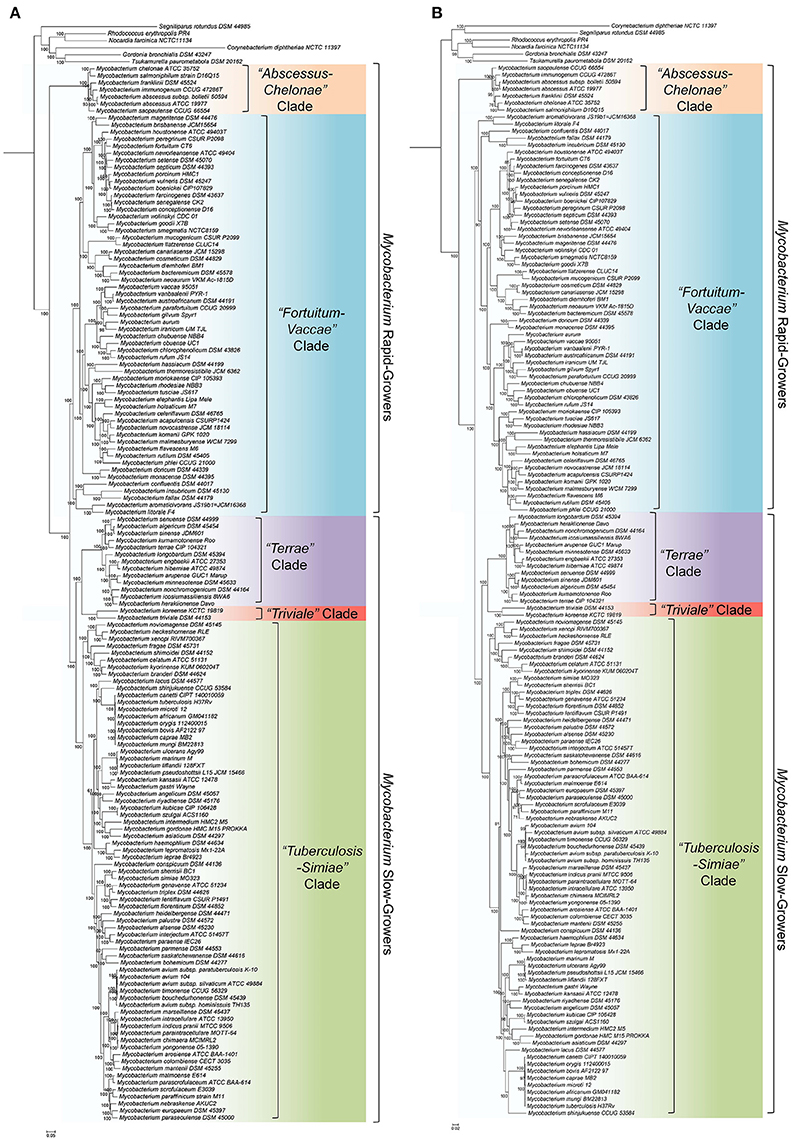
Figure 1. (A) Maximum-likelihood phylogenetic tree for 150 Mycobacterium species based on the concatenated sequence of 1941 core proteins from the genus Mycobacterium. (B) A maximum-likelihood phylogenetic tree based on the 136 proteins consistuting the phyloeco set for the phylum Actinobacteria. Both of these trees were rooted using the sequences from the Corynebacteriales species. Trees were constructed as described in the Methods section. SH-like statistical support values and the bootstrap value are marked on the nodes. The major clades as well as the clusters of slow-growing and fast-growing Mycobacterium species are labeled. Some slow-growing species, which branched within the rapid-growing species are marked with*.
We have also compared the relationships observed in the aforementioned phylogenetic trees with the relationships observed in a tree based on 16S rRNA gene sequences, which was extracted from the SILVA Tree of Life Project 128 (Yarza et al., 2008; Quast et al., 2013). This tree is shown in Supplementary Figure 2 with the analogous groups labeled. Overall, in concordance with the core protein-based phylogenetic trees and the tree based on 8 conserved proteins, the slow-growing mycobacterial species corresponding to the “Tuberculosis-Simiae” clade formed a distinct clade in the 16S rRNA tree. The species corresponding to the “Terrae” clade also branched in the immediate proximity of the “Tuberculosis-Simiae” clade, with members of the “Triviale” clade (viz. M. triviale, M. koreense, and M. parakoreense) forming a deeper-branching lineage. However, in contrast to the different trees based on protein sequences, the rapid-growing Mycobacterium species exhibited extensive polyphyly and their interrelationships were poorly resolved. In particular, the members of the “Abscessus-Chelonae” clade formed a monophyletic lineage within the other rapid-growing Mycobacterium species, whereas the relationships among the other rapid growing species were difficult to discern.
Genome Relatedness of the Members of the Genus Mycobacterium
Based on genome sequences, the average amino acid identity between different species can be calculated to determine the overall genome relatedness of the species (Konstantinidis and Tiedje, 2007; Richter and Rossello-Mora, 2009; Thompson et al., 2013; Qin et al., 2014; Yarza et al., 2014). Pairwise amino acid identity was calculated based on the conserved protein families between each genome used in the analysis and the results of these analyses are presented in the form of a matrix in Figure 2. An expanded version of this matrix is provided in Supplementary Figure 3. As seen from the AAI matrix (Figure 2), the members of the four main clades observed in the phylogenetic trees (Figure 1 and Supplementary Figure 1) showed higher amino acid identity to members within each clade than to the other Mycobacterium species. Further, members of the “Triviale” clade could be clearly distinguished from the “Terrae” clade, based on their much lower amino acid identity to the members of this latter clade. In addition, members of the “Abscessus-Chelonae” clade exhibited a high degree of amino acid identity (Avg. 92%) to other members of this clade, but significantly lower similarity to members of the “Fortuitum-Vaccae” or the “Tuberculosis-Simiae” clades (Avg. 62%). The results of the genome relatedness analysis support the existence of the four main clades observed in the phylogenetic trees and also the distinctness of the “Triviale” clade from members of the “Terrae” clade.
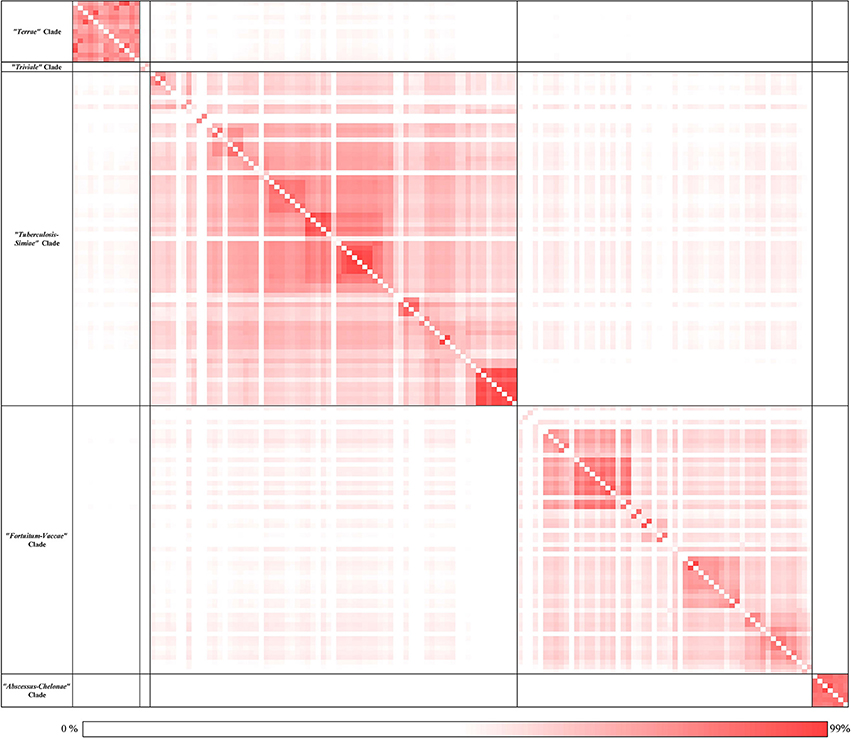
Figure 2. A matrix of the average Amino Acid Identity for the 150 Mycobacterium genomes used in this study. A darker shade represents higher similarity between the pair of genomes. The average amino acid identity between each pair of genomes was calculated as described in the Methods section. The numerical values underlying this matrix are provided in Supplementary Figure 3.
Molecular Signatures Specific for the Genus Mycobacterium and Its Main Clades
The results of phylogenomic studies and genomic similarity analysis indicated that the known mycobacterial species can be divided into five main groups including the “Triviale” clade. However, as the branching of species in phylogenetic trees can be affected by a large number of variables (Stackebrandt, 1992; Ludwig and Klenk, 2005; Klenk and Goker, 2010; Gupta, 2016b), it is important to confirm the genetic cohesiveness of the observed clades by independent means not involving phylogenetic analysis. Rare genetic changes, such as insertions and deletions in genes/proteins as well as novel genes/proteins (viz. CSIs and CSPs) which are uniquely shared by an evolutionary related group of organisms constitute synapomorphic characteristics, whose shared presence in a given group of organisms generally results from the occurrence of the genetic changes in a common ancestor of the group (Gupta, 1998, 2014, 2016b; Rokas and Holland, 2000; Dutilh et al., 2008). In our earlier work on Actinobacteria, we described large numbers of CSIs and CSPs which were distinctive characteristics of either the entire phylum or a number of different clades within this phylum at multiple phylogenetic/taxonomic levels (Gao and Gupta, 2005, 2012; Gao et al., 2006; Gupta et al., 2013b). Although the focus of this earlier study was not on mycobacteria, a limited number of CSIs and CSPs which were then specific for the genus Mycobacterium were also identified (Gao et al., 2006; Gao and Gupta, 2012). Since these earlier studies, genome sequences for a large number of other mycobacterial species have become available (Supplementary Table 1). In the present work, we have carried out comprehensive comparative genomic studies on members of the genus Mycobacterium, to identify molecular markers (CSIs and CSPs) that are specific characteristics of either all mycobacterial species or of the identified main clades within this genus. The results of these analyses have identified 172 molecular markers (CSIs and CSPs) that are uniquely found in either all mycobacteria or by the members of different main clades identified by phylogenomic studies. Brief descriptions of the characteristics of the identified molecular markers and their group specificities are provided below.
Molecular Signatures (CSIs and CSPs) Specific for the Genus Mycobacterium
Our analysis has identified 10 CSIs in proteins involved in diverse functions that are uniquely found in all available mycobacterial homologs. An example of a CSI that is specific for the genus Mycobacterium is shown in Figure 3. In the partial sequence alignment of the protein EgtB (ergothioneine biosynthesis protein), a two amino acid insertion in a conserved region is exclusively found in all members of the genus Mycobacterium, but it is not present in the top 500 homologs of this protein sequence in other bacteria. Ergothionine is a naturally occurring amino acid (thiourea derivative of histidine), whose synthesis is uniquely carried out by only certain groups of actinobacteria as well as some cyanobacteria and fungi (Fahey, 2001). More detailed sequenced information for this CSI as well as sequence information for 9 other CSIs in important proteins, which are also specific for the genus Mycobacterium is provided in Supplementary Figures 4–13 and their main characteristics are summarized in Table 1. Of the described CSIs, the CSI in the protein orotidine 5'-phosphate decarboxylase (Supplementary Figure 7) was identified in our earlier work (Gao and Gupta, 2012). Although the number of sequenced mycobacterial genomes has increased many folds, this CSI is still found only in members of the genus Mycobacterium.
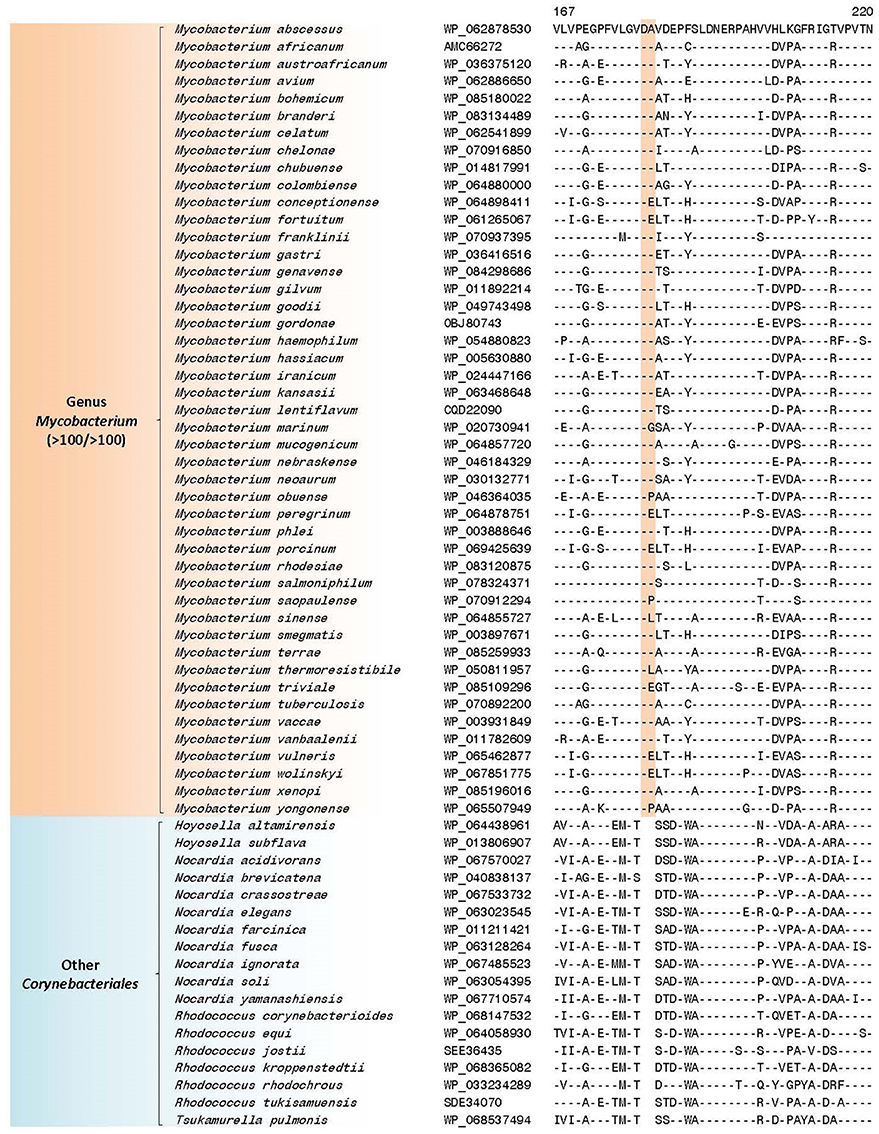
Figure 3. Partial sequence alignment of a conserved region of the ergothioneine biosynthesis protein EgtB showing a two amino acid insertion (boxed) exclusively found in members of the genus Mycobacterium and not present in other Corynebacteriales. Dashes (-) in all alignments denote identity with the amino acid shown in the top sequence. Sequence information for only limited numbers of species is presented in this figure; a detailed alignment for this CSI is shown in Supplementary Figure 4. Information for additional CSIs specific for the genus Mycobacterium are provided in Supplementary Figures 4–13 and summarized in Table 1.
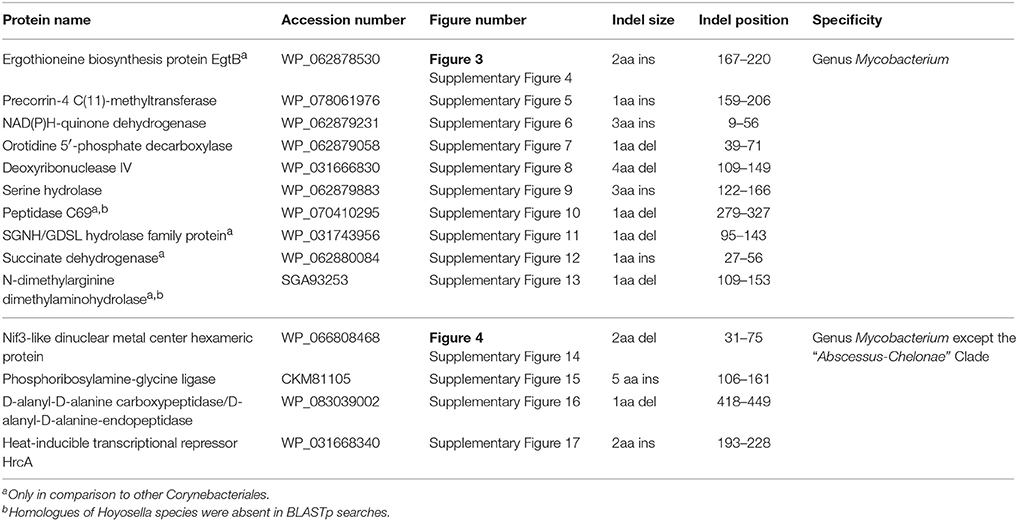
Table 1. Conserved signature indels (CSIs) that are specific for different members of the genus Mycobacterium and those which are lacking only in members of the “Abscessus-Chelonae” clade.
We have previously described a number of CSPs, whose homologs were uniquely found in the then sequenced mycobacterial species (Gao et al., 2006; Gao and Gupta, 2012). In light of the large increase in the number of sequenced mycobacterial genomes, the group specificities of the previously described CSPs were re-examined. Results of these analyses reveal that despite >20-fold increase in the number of sequenced mycobacterial genomes since these CSPs were first identified (Gao et al., 2006), 9 of the CSPs reported in our earlier work are still specific for members of the genus Mycobacterium and no homologs showing significant similarities to these proteins are present in other bacteria (Table 2). In view of the unique shared presence of these 10 CSIs and 9 CSPs by either all or most members of the genus Mycobacterium (except for an isolated exception), the genetic changes leading to these genetic markers most likely initially occurred in a common ancestor of the genus Mycobacterium and then retained by all descendant species.
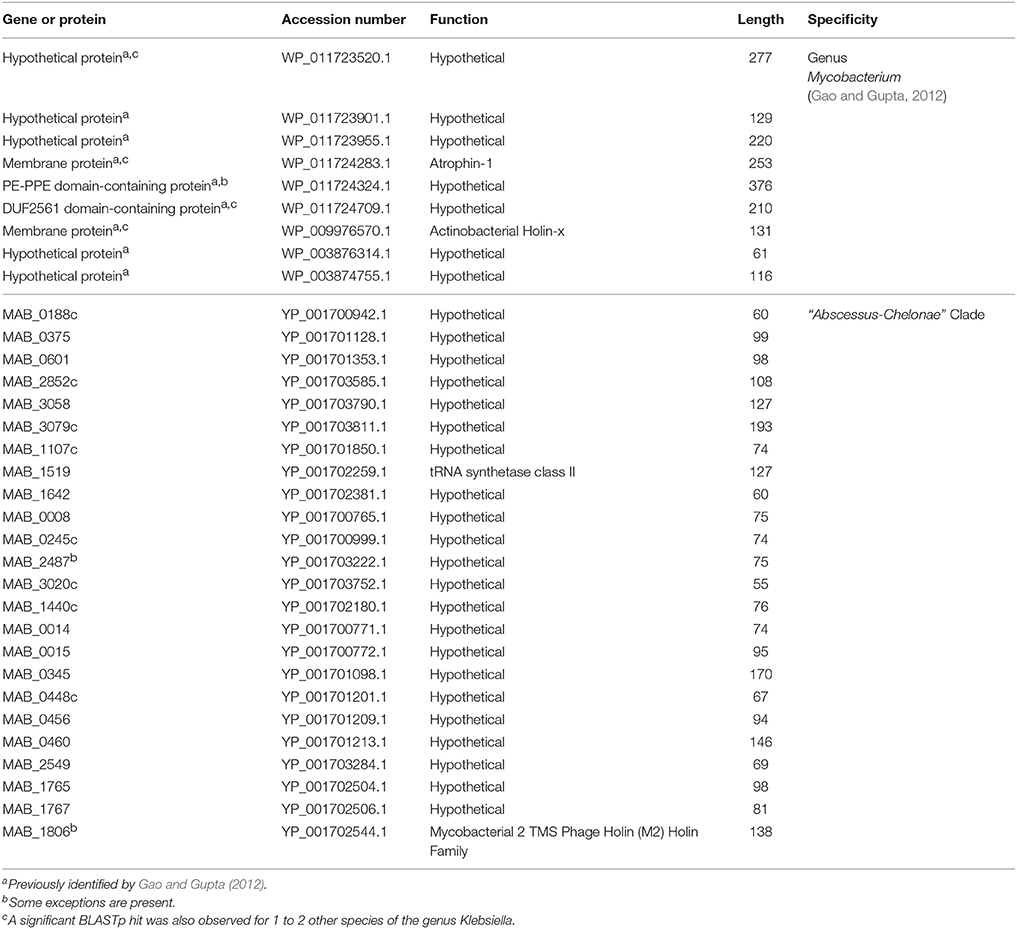
Table 2. Conserved signature proteins (CSPs) specific for the genus Mycobacterium and members of the “Abscessus-Chelonae” clade.
Molecular Signatures Specific for the “Abscessus-Chelonae” Clade and Supporting the Deep Branching of this Group within the Genus Mycobacterium
The “Abscessus-Chelonae” clade, also referred to as M. chelonae or M. abscessus complex (Adékambi and Drancourt, 2004; Medjahed et al., 2010; Tortoli, 2012; Wee et al., 2017), consists of six members and it has recently gained clinical attention in light of its emerging pathogenicity to humans (Medjahed et al., 2010; Tortoli, 2014). In the phylogenetic trees constructed in our work, members of this clade form a monophyletic grouping which comprises the deepest branching lineage among the Mycobacterium species (Figures 1A,B and Supplementary Figure 1). The deep branching of the “Abscessus-Chelonae” clade in comparison to the other Mycobacterium species is also independently supported by 4 CSIs in four different proteins which are commonly shared by the homologs of all other mycobacterial species except those from the “Abscessus-Chelonae” clade. One example of a CSI depicting this pattern is presented in Figure 4, where in the partial sequence alignment of Nif3-like dinuclear metal center hexameric protein, a two amino acid deletion in a conserved region is present in all members of the genus Mycobacterium except members of the “Abscessus-Chelonae” clade. Additional information for this CSI and the sequence information for the three other CSIs exhibiting similar species distributions is provided in Supplementary Figures 14–17 and their main characteristics are summarized in Table 1. Based upon the species distributions of these CSIs, the genetic changes leading to them have likely occurred in a common ancestor of the other Mycobacterium species after the divergence of the “Abscessus-Chelonae” clade.
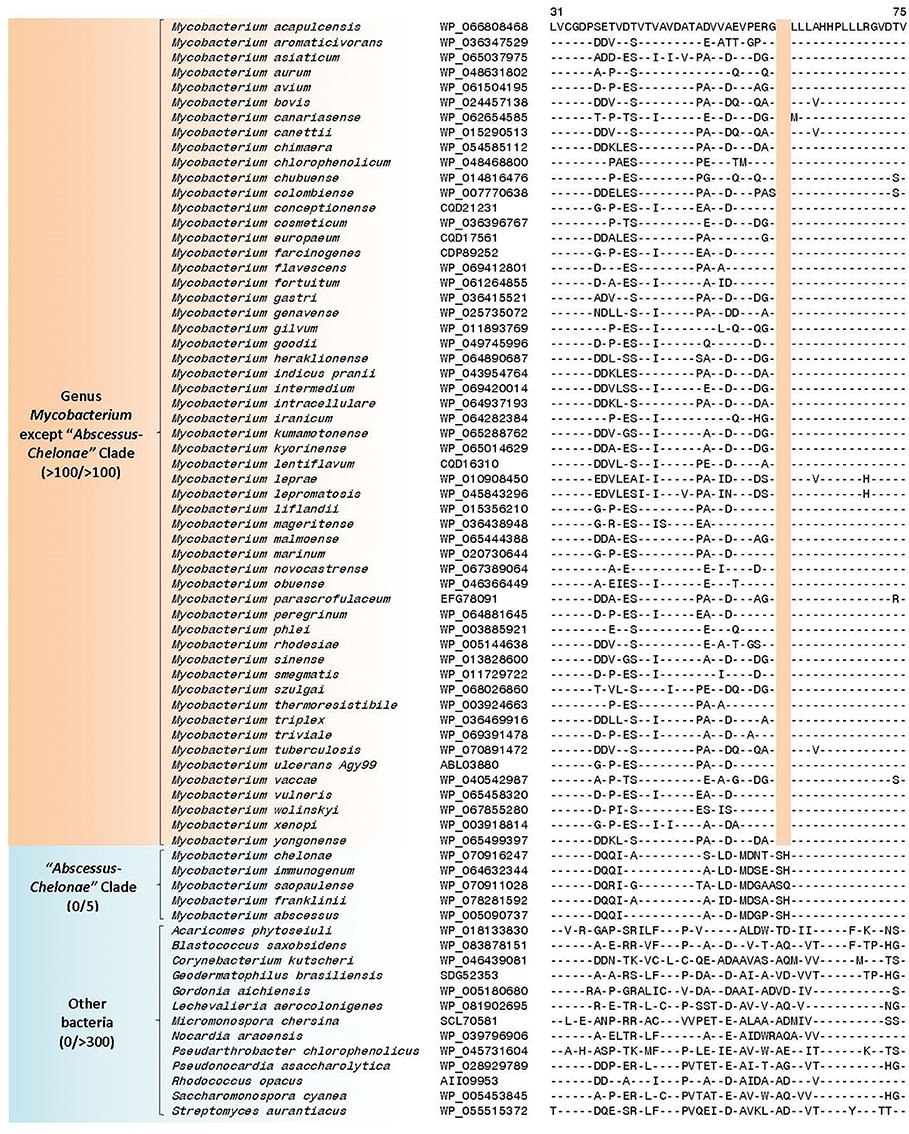
Figure 4. A partial sequence alignment of a conserved region of Nif3-like protein exhibiting a two amino acid deletion that is specific for members of the genus Mycobacterium except members of the “Abscessus-Chelonae” clade; a detailed alignment for this CSI is shown in Supplementary Figure 14. Information for additional CSIs specific for the genus Mycobacterium are provided in Supplementary Figures 14–17 and summarized in Table 1. Dashes (-) in all alignments denote identity with the amino acid shown in the top sequence.
Our analyses have also identified 27 CSIs in proteins involved in diverse functions that are uniquely shared by members of the “Abscessus-Chelonae” clade providing strong evidence of the genetic cohesiveness and distinctness of this group of mycobacteria. Two examples of the CSIs specific for the “Abscessus-Chelonae” clade are shown in Figure 5. Figure 5A shows a partial sequence alignment of the protein uracil phosphoribosyltransferase, where a six amino acid insertion in a conserved region is present in all members of the “Abscessus-Chelonae” clade but absent in the homologs from all other Mycobacterium species as well as other groups of bacteria. Likewise, Figure 5B shows a four amino acid deletion in the sequence alignment of protein L-histidine N(alpha)-methyltransferase, which is also specific for the “Abscessus-Chelonae” clade. More detailed information for these CSIs and the 25 other identified CSIs, which are also specific for the “Abscessus-Chelonae” clade, is provided in Supplementary Figures 15, 18–43 and their main characteristics are summarized in Table 3. In addition to these CSIs, our work has also identified 24 CSPs listed in Table 2, for which homologs exhibiting significant similarity are only found in members of the “Abscessus-Chelonae” clade. Thus, the distinctness of the “Abscessus-Chelonae” clade from all other mycobacteria is strongly supported by 51 highly-specific molecular signatures identified in this work.
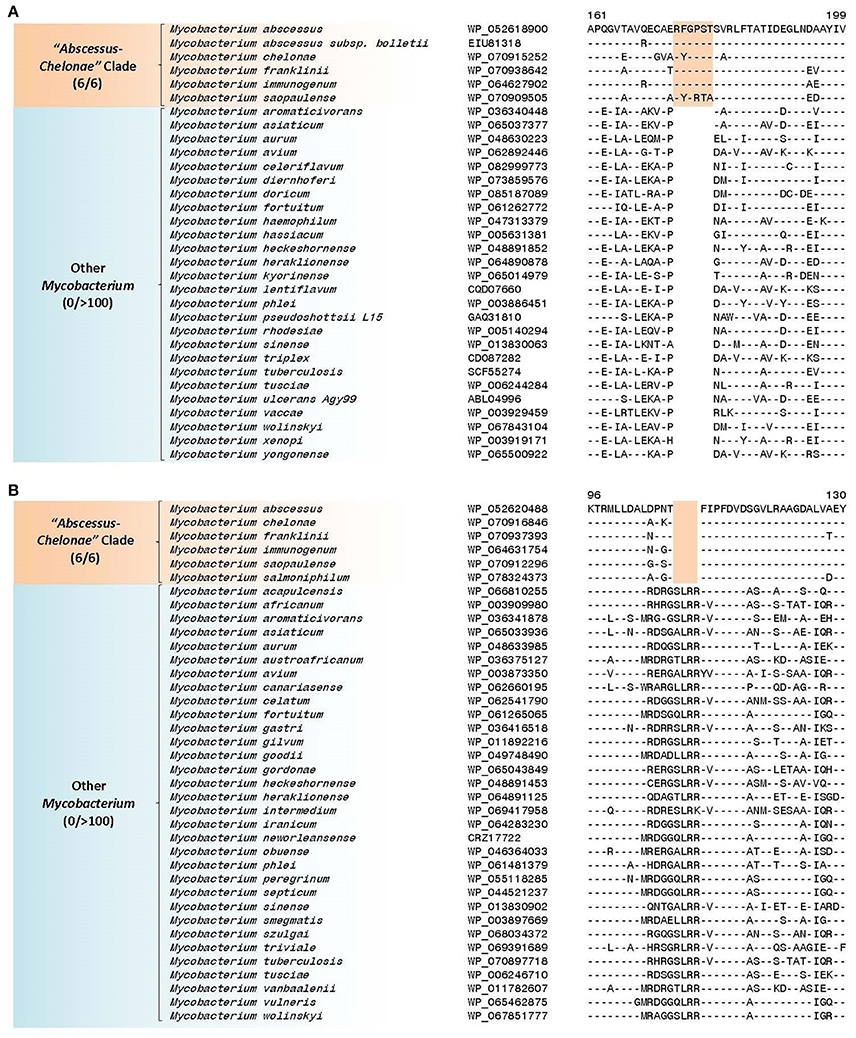
Figure 5. (A) Partial sequence alignment of the protein uracil phosphoribosyltransferase showing a six amino acid insertion that is specific for the “Abscessus-Chelonae” clade; (B) Sequence alignment of L-histidine N(alpha)-methyltransferase showing a four amino acid deletion that is also specific for the “Abscessus-Chelonae” clade. More detailed alignments for these CSIs are shown in Supplementary Figures 18, 19 respectively. Additional CSIs that are specific for this clade are summarized in Table 3 and sequences of these are provided in Supplementary Figures 15, 18–43.
Molecular Signatures Specific for the “Fortuitum-Vaccae” Clade
The “Fortuitum-Vaccae” clade as designated here (see Figure 1) encompasses all rapid-growing mycobacterial species, except those from the “Abscessus-Chelonae” clade. In the present work, 4 CSIs and 10 CSPs have been identified that are specific for either all or most members of the “Fortuitum-Vaccae” clade and support the monophyletic clustering of these species as observed in the phylogenomic trees (Figure 1). One of the identified CSIs, which are specific for the “Fortuitum-Vaccae” clade, is found in the LacI family transcriptional regulator. In the partial sequence alignment of this protein shown in Figure 6, a five amino acid insert in a conserved region is exclusively found in different members of the “Fortuitum-Vaccae” clade but it is not found in any other mycobacteria. Three other CSIs showing similar species specificities are present in three other proteins. Detailed sequence information for all of these CSIs is provided in the Supplementary Figures 44–47 and the main characteristics of all CSIs specific for the “Fortuitum-Vaccae” clade are summarized in Table 4.
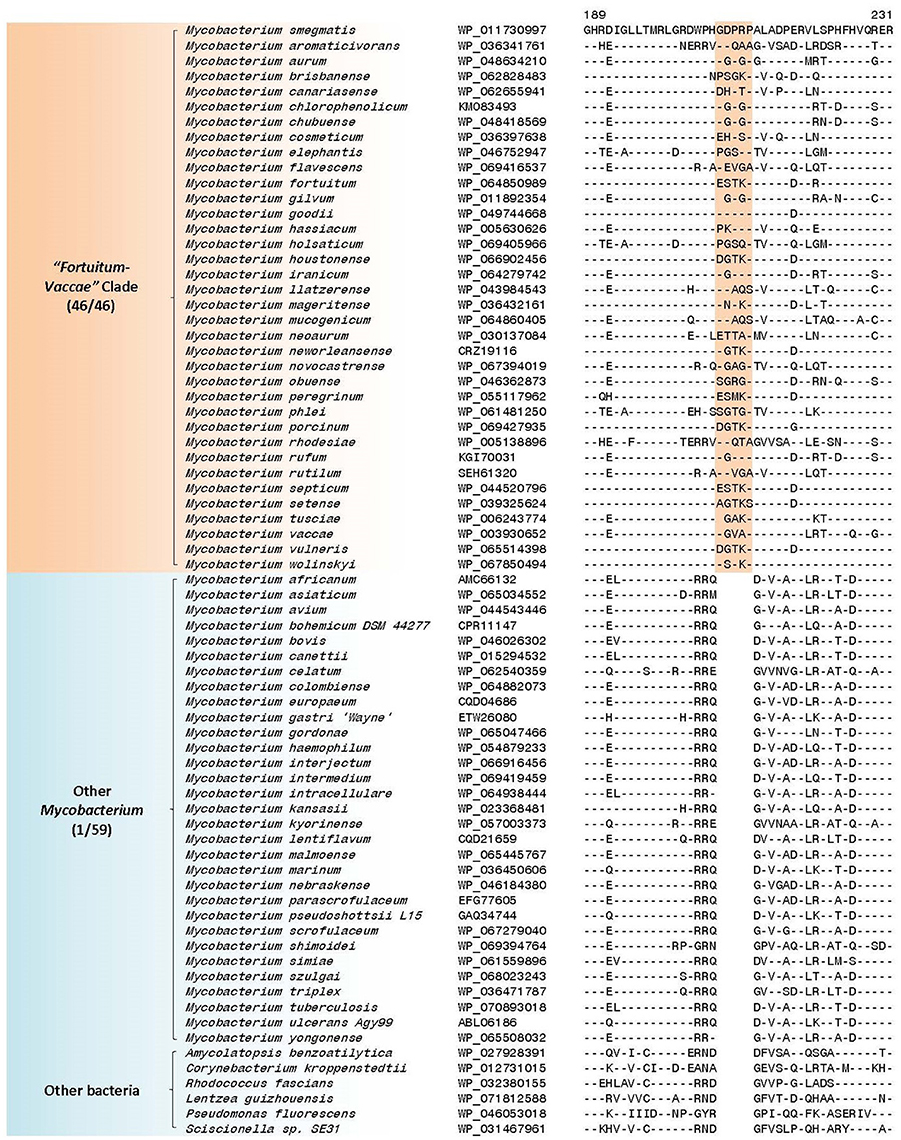
Figure 6. A partial sequence alignment of a conserved region of LacI family transcriptional regulator showing a five amino acid insertion that is specific for the “Fortuitum-Vaccae” clade; a more detailed alignment of this CSI is shown in Supplementary Figure 44. Sequence information for additional CSIs that are specific for this clade is shown in Supplementary Figures 44–47 and summarized in Table 4.
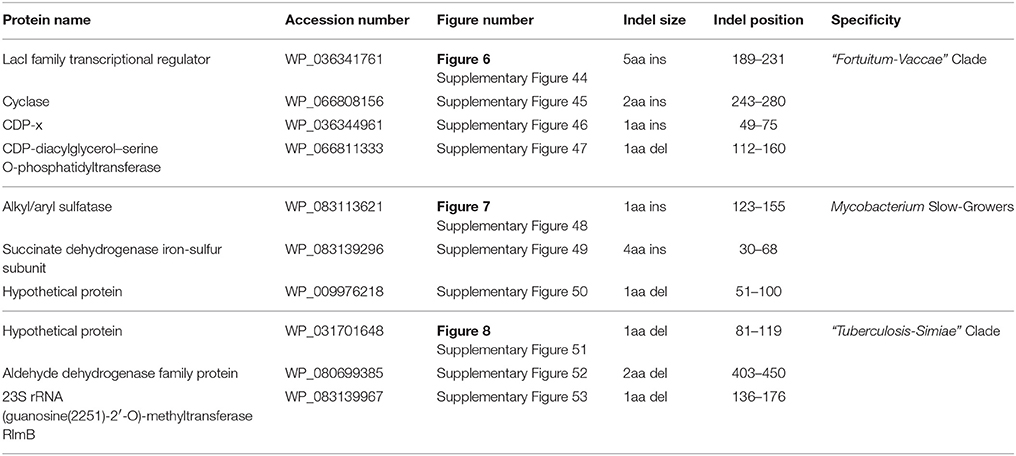
Table 4. Conserved Signature Indels (CSIs) specific for members of the “Fortuitum-Vaccae” clade, Slow-Growing Mycobacterium (“Tuberculosis-Simiae” + “Terrae” clades), and “Tuberculosis-Simiae” clade.
BLASTp searches on the protein sequences from the genome of Mycobacterium aurum (LSHTM) have also identified 10 CSPs, whose homologs, except for rare exceptions, are only found in the “Fortuitum-Vaccae” clade of Mycobacterium species. Most of these CSPs are hypothetical proteins and their characteristics are summarized in Table 5. For the first four CSPs listed in Table 5, the homologs are present in different members of the “Fortuitum-Vaccae” clade, while for the remaining six CSPs, although they are specific for the “Fortuitum-Vaccae” clade, homologs were not detected in some members of this clade. In all, our identification of 14 molecular markers (4 CSIs and 10 CSPs), which are uniquely shared by members of the “Fortuitum-Vaccae” clade support its monophyletic origin and genetic cohesiveness.
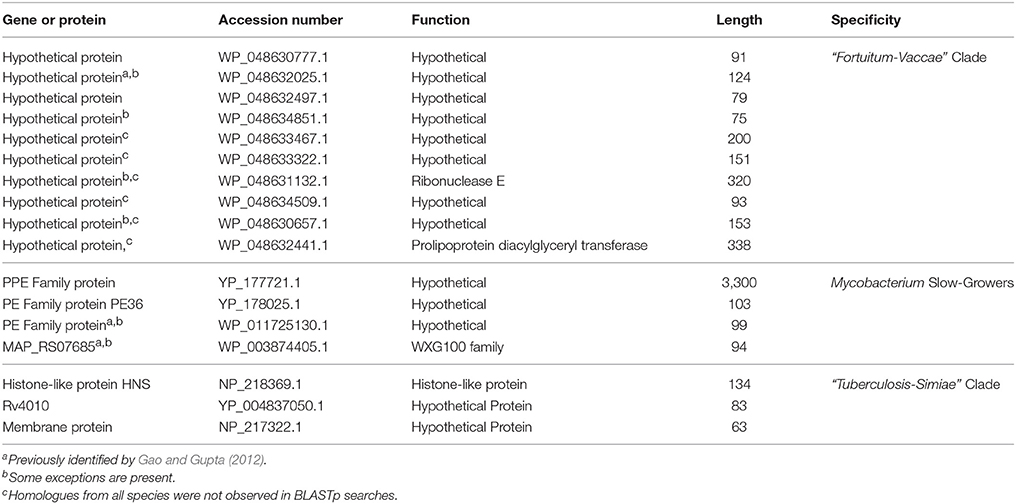
Table 5. Conserved signature proteins (CSPs) specific for members of the “Fortuitum-Vaccae” clade, Slow-Growing Mycobacterium (“Tuberculosis-Simiae” + “Terrae” + “Triviale” clades), and “Tuberculosis-Simiae” clade.
Molecular Signatures that Are Specific for the Slow-Growing Mycobacterium
The slow-growing Mycobacterium species generally form a monophyletic clade in most phylogenetic trees based on protein sequences (see Figure 1 and Supplementary Figure 1) as well as those based on the 16S rRNA gene sequences (see Supplementary Figure 2) (Devulder et al., 2005; Kim et al., 2005; Hartmans et al., 2006; Mignard and Flandrois, 2008; Magee and Ward, 2012; Tortoli, 2012; Quast et al., 2013; Lory, 2014; Wang et al., 2015; Wee et al., 2017). The monophyly of the slow-growing Mycobacterium clade is also supported by 3 CSIs and 4 CSPs that have been identified in this study. One example of a CSI that is largely specific for the slow-growing Mycobacterium clade is shown in Figure 7. In the sequence alignment of alkyl-aryl sulfatase protein, a one amino acid insert in a conserved region is present in all of the homologs from slow-growing Mycobacterium species, but it is not found in the homologs of other Mycobacterium species. Detailed sequence information for this CSI and the two other CSIs showing similar specificities is provided in Supplementary Figures 48–50 and their main characteristics are summarized in Table 4. As noted above, the homologs for four of the identified CSPs (Accession numbers: YP_177721.1, YP_178025.1, WP_011725130.1, WP_003874405.1) are also specifically found in slow-growing Mycobacterium species (Table 5). The last two of these CSPs were identified by our earlier work based on limited number of genomes (Gao and Gupta, 2012) and they continue to be specific for this large clade of mycobacteria. Further, of the identified CSPs, which are specific for the slow-growing mycobacterial clade, three of the CSPs correspond to the PE or PPE family of proteins, which are often involved in mycobacterial virulence (Mukhopadhyay and Balaji, 2011).
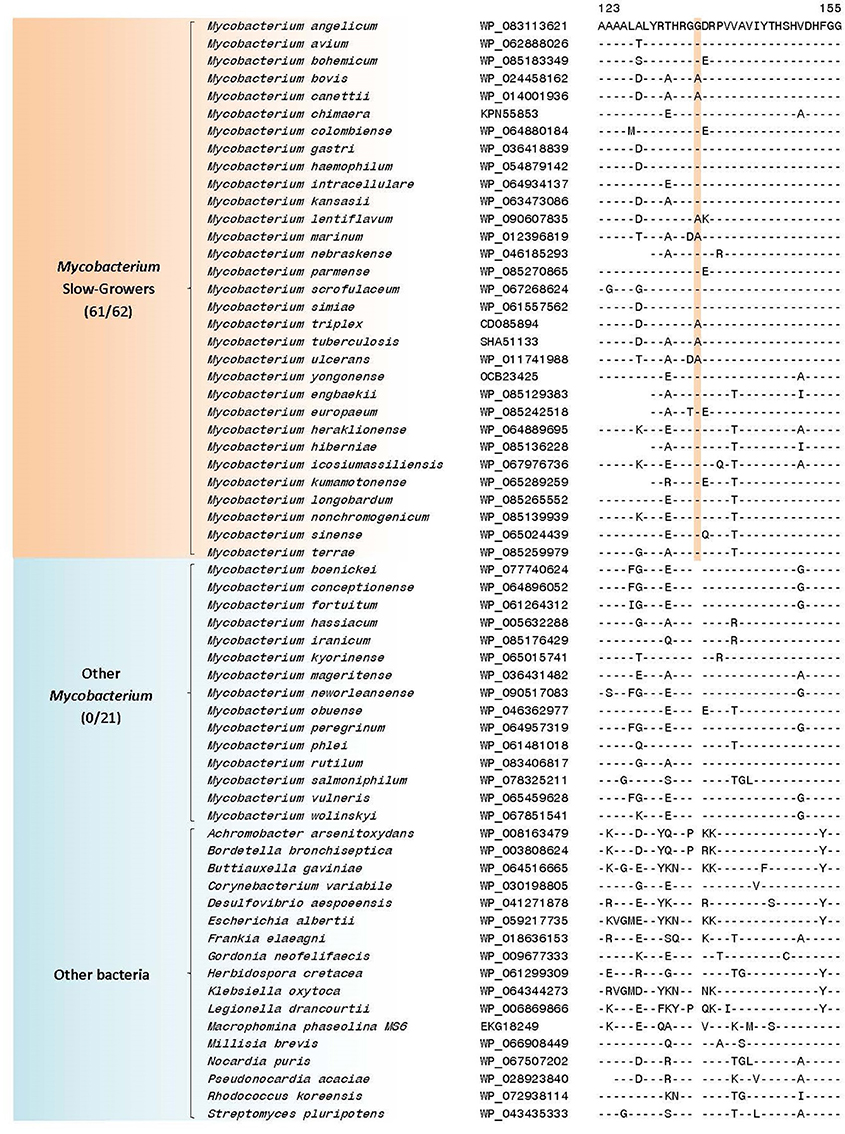
Figure 7. A partial sequence alignment of a conserved region of the protein alkyl/aryl sulfatase showing a one amino acid insertion that is specific for the Mycobacterium slow-growers (i.e., “Tuberculosis-Simiae” + “Terrae”) clade; a detailed alignment of this CSI is shown in Supplementary Figure 48. Additional CSIs that are specific for this clade are summarized in Table 4 and their sequence alignments are shown in Supplementary Figures 48–50.
In our phylogenetic trees, the slow-growing mycobacterial species form three main clades including a clade consisting of M. triviale and M. koreense (“Triviale” clade). The genetic cohesiveness of these clades of slow-growing mycobacteria is also supported by a large number of molecular signatures that are described below.
Molecular Signatures for the “Tuberculosis-Simiae” Clade
The “Tuberculosis-Simiae” clade in our work is comprised of all other slow-growing mycobacteria except those from the “Terrae” and “Triviale” clades. This clade encompasses various pathogenic Mycobacterium species including those from the M. tuberculosis complex, M. avium complex, M. gordonae clade, M. kansasii clade, M. simiae clade, as well as several other slow-growing species (Magee and Ward, 2012; Lory, 2014). We have identified a total of 3 CSIs that are specific for the “Tuberculosis-Simiae” clade (Table 4, Supplementary Figures 51–53). One example of a CSI specific for this clade, which is found in a protein of unknown function is shown in Figure 8, where a single amino acid deletion is found in all members of the “Tuberculosis-Simiae” clade, but it is not present in any other mycobacterial homolog. In addition to these CSIs, BLASTp searches on the proteins found in the genome of Mycobacterium tuberculosis H37Rv have identified 3 CSPs, whose homologs are only found in either all or most members of the “Tuberculosis-Simiae” clade. A summary of the CSPs which are specific for the “Tuberculosis-Simiae” clade is provided in Table 5 and of these CSPs, one protein (Genbank Accession Number NP_218369.1) is annotated as a histone-like protein.
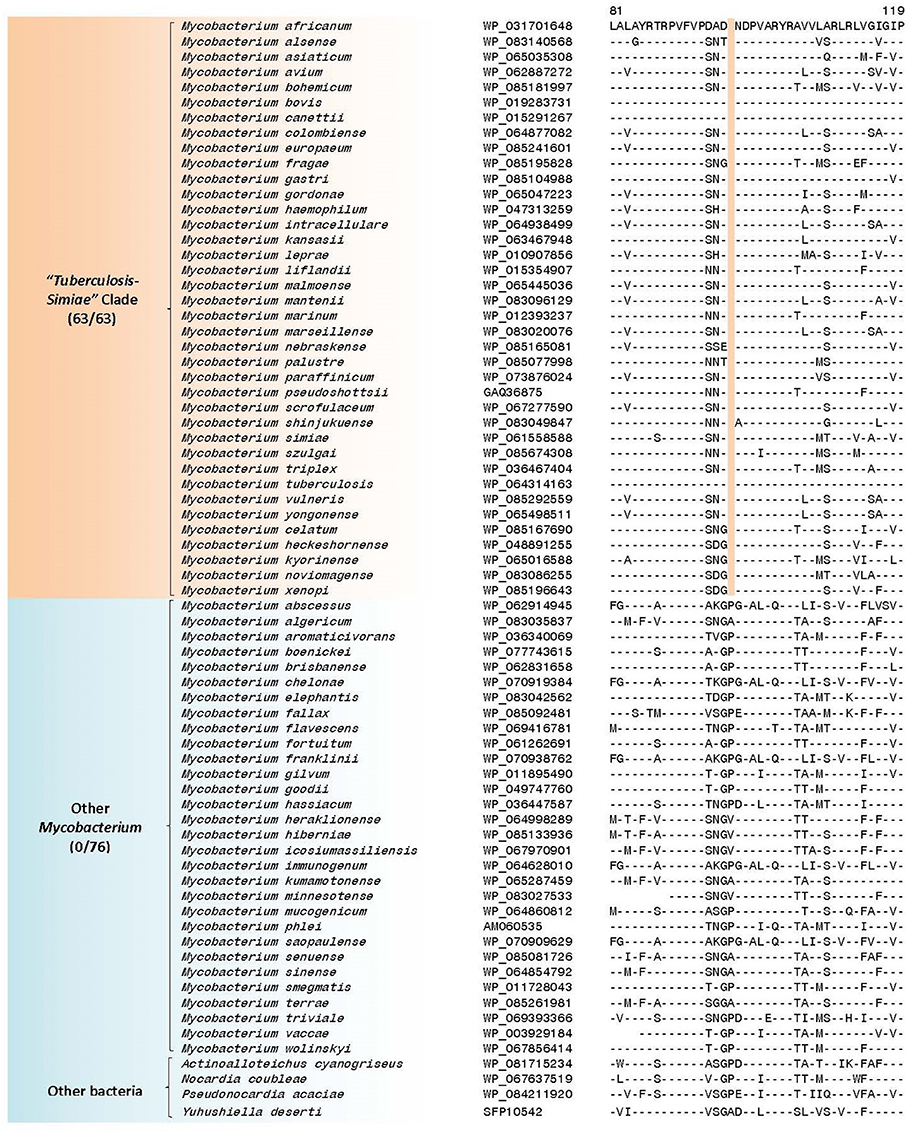
Figure 8. Partial sequence alignment of a conserved region of a hypothetical protein showing a one amino acid deletion exclusively found in members of the “Tuberculosis-Simiae” clade; a detailed alignment of this CSI is shown in Supplementary Figure 51. Additional CSIs that are specific for this clade are shown in Supplementary Figures 51–53 and information for them is summarized in Table 4.
Molecular Signatures Demarcating the “Terrae” and “Triviale” Clades of Mycobacteria
The members of the “M. terrae complex” (Tortoli, 2012; Ngeow et al., 2015) has drawn attention recently as some members of this clade are opportunistic pathogens (Mignard and Flandrois, 2008; Kim et al., 2012, 2013; Tortoli, 2012, 2014; Tortoli et al., 2013; Ngeow et al., 2015; Vasireddy et al., 2016). In the core-genome protein trees and the tree based on 8 conserved proteins, members of the “M. terrae complex” form a monophyletic lineage consisting of two distinct subclades: a larger “Terrae” clade encompassing most of the species from the “M. terrae complex” and a deeper branching “Triviale” clade consisting of M. triviale and M. koreense (M. parakoreense also branches with these species in the 16S rRNA tree, Supplementary Figure 2). The phylogenetic distinctness of this larger “Terrae” + “Triviale” clade is also supported by a number of identified molecular signatures. In this work, we have identified 6 CSIs, which are specific for the larger “Terrae complex” consisting of the “Terrae” + “Triviale” clades (Table 6). Sequence information for one of the CSIs specific for the larger “Terrae complex” is presented in Figure 9A. In this case a four amino acid insertion in the protein ATP-dependent helicase is specifically present in all members of the “Terrae complex,” but it is not present in any other bacteria. Detailed sequence information for this CSI as well as other CSIs specific for this clade is presented in Supplementary Figures 54–59 and summarized in Table 6. In addition to these CSIs, which are commonly shared by the “Terrae” + “Triviale” clades, our analyses have also identified 26 other CSIs listed in Table 6, which are specifically shared by only the members of the “Terrae” clade and not present in M. triviale and M. koreense. An example of such a CSI consisting of a four amino acid insertion found in the protein UDP-N-acetylmuramate–L-alanine ligase is shown in Figure 9B. Sequence information for all the “Terrae” clade CSIs is presented in Supplemntary Figures 35, 60–84 and summarized in Table 6. These CSIs serve to indicate the distinctness of the species from the “Terrae” clade from the deeper branching M. triviale and M. koreense species, which are part of the “Triviale” clade.
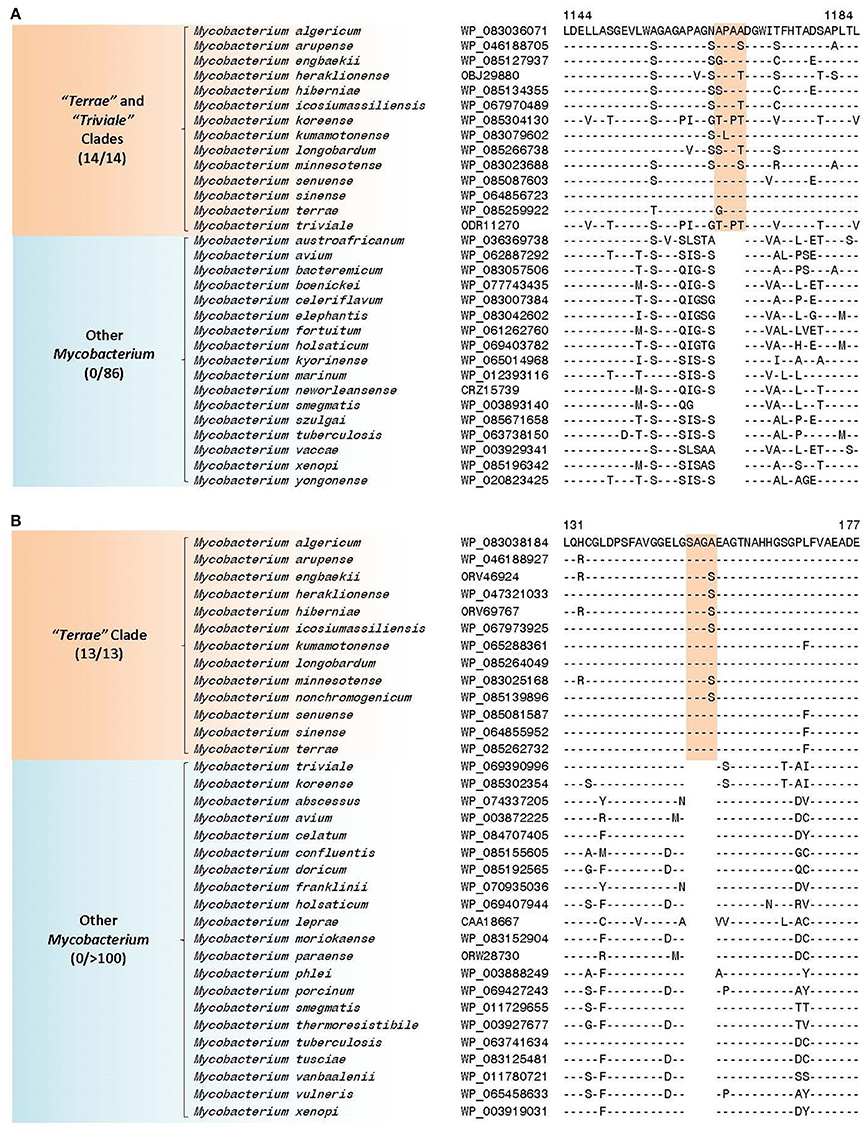
Figure 9. Partial sequence alignment of a conserved region of (A) ATP-dependent helicase showing a four amino acid insertion that is specific for the “Terrae” + “Triviale” clades and (B) UDP-N-acetylmuramate—L-alanine ligase showing a four amino acid insertion that is specific for only the members of the “Terrae” clade but lacking in members of the “Triviale” clade as well as other mycobacteria. More detailed alignments of these CSIs are shown in Supplementary Figures 54 and 74, respectively. Additional CSIs that are specific for this clade are shown in Supplementary Figures 35, 54–84 and summarized in Table 6.
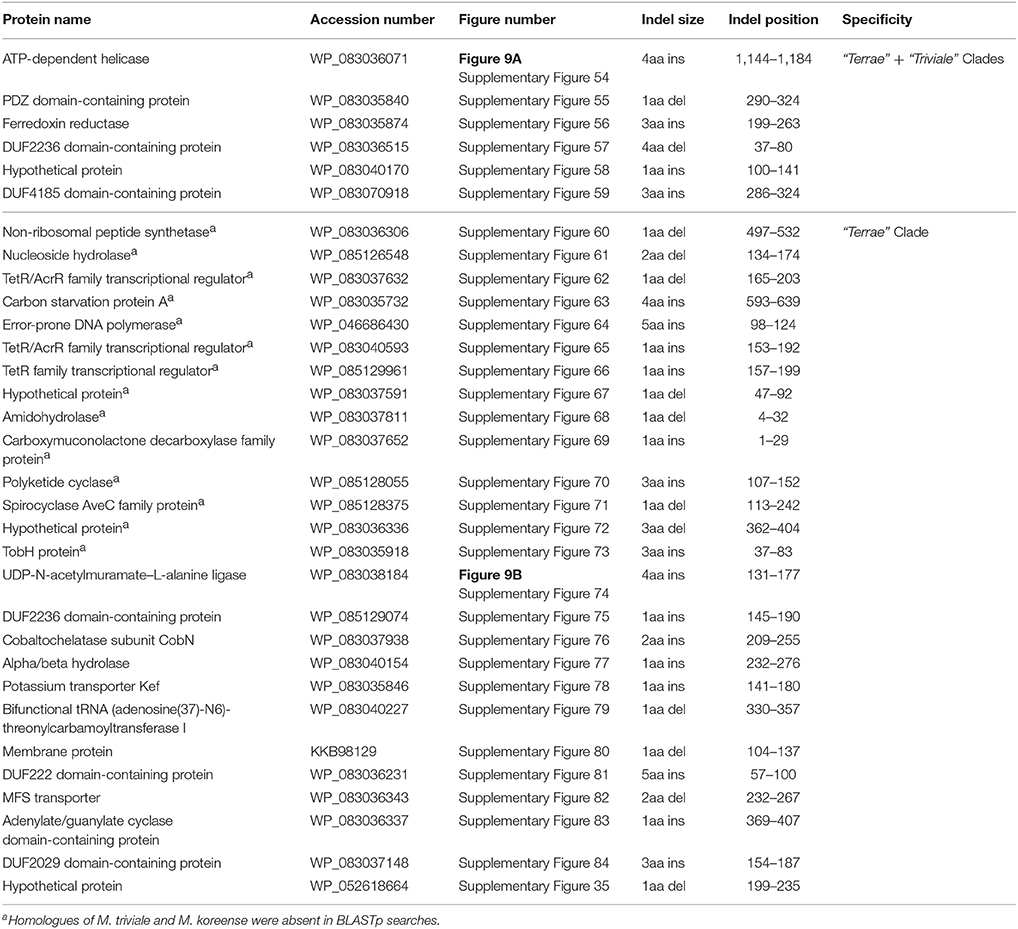
Table 6. Conserved Signature Indels (CSIs) specific for members of the “Terrae” + “Triviale” clades or only the “Terrae” clade.
Our BLASTp searches on the protein sequences from the genome of M. sinense JDM601 (Zhang et al., 2011) and M. triviale DSM 44153 (Fedrizzi et al., 2017) have also identified many CSPs whose homologs are found specifically in either members of the larger “Terrae complex” or uniquely by species which are part of either the “Terrae” clade or the “Triviale” clade. A summary of these CSPs is provided in Table 7. Of the identified CSPs, two CSPs (viz. accession numbers WP_013830140.1 and WP_013827845.1) are uniquely found in most members of the “Terrae” + “Triviale” clades. However, a large number of the other identified CSPs are specific for only either members of the “Terrae” clade (15 CSPs) or members of the “Triviale” clade (22 CSPs) and their homologs are not detected in other mycobacteria. Four of the CSPs specific for the “Triviale” clade included in Table 7 were also previously identified by Ngeow et al. (2015). The identification of a large number of CSPs, which are uniquely found in either all/most members of the “Terrae” clade or those from the “Triviale” clade again serve to clearly differentiate these two groups of mycobacteria and demarcate them in molecular terms.
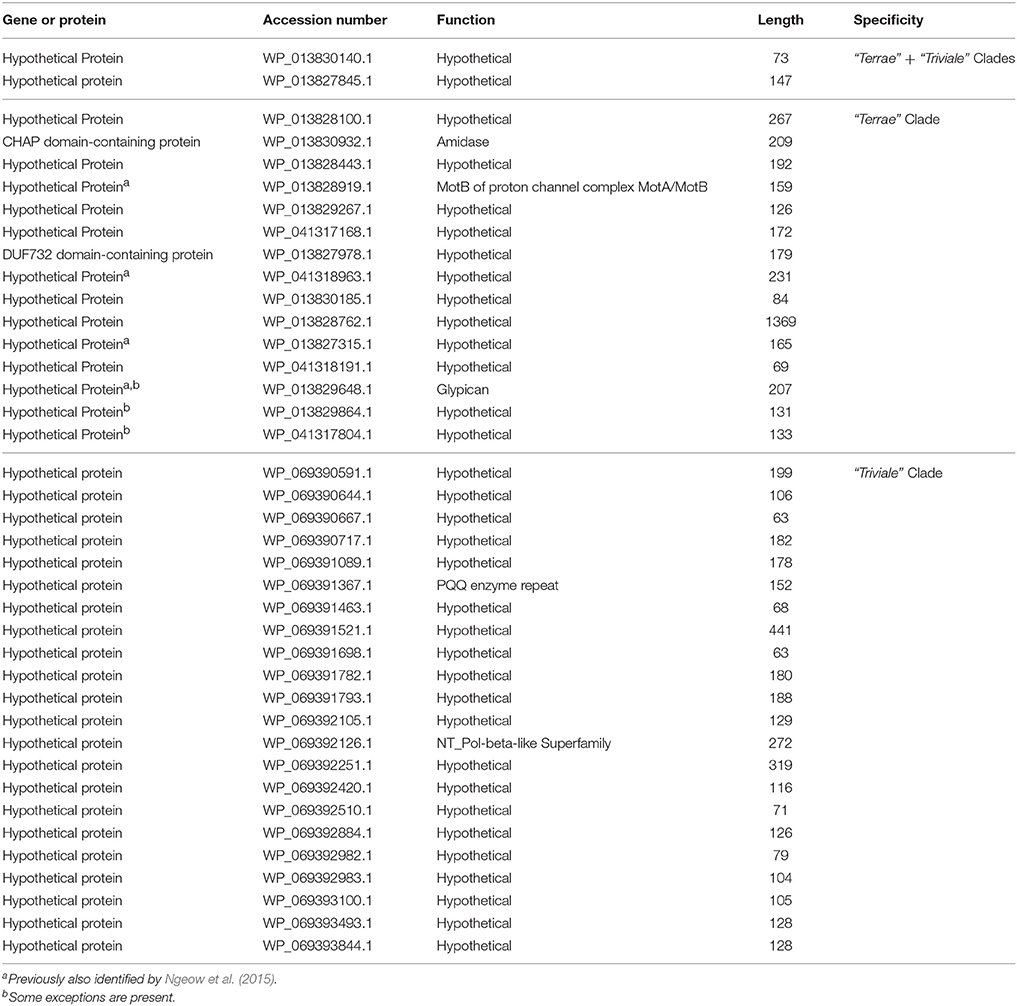
Table 7. Summary of Conserved Signature Proteins (CSPs) that are specific for members of both “Terrae” + “Triviale” clades or only the “Terrae” clade or the “Triviale” clade.
Discussion
The genus Mycobacterium comprises a large group of species (currently 188 species have validly published names), which includes some of the most impactful human pathogens (viz. M. tuberculosis and M. leprae) as well as large numbers of species found in diverse environments (Magee and Ward, 2012; Lory, 2014). In view of the immense clinical importance of certain Mycobacterium species, it is of much interest to have a reliable understanding as to how different species within this large group are related (Tsukamura, 1967a; Rogall et al., 1990; Stahl and Urbance, 1990; Goodfellow and Magee, 1998; Magee and Ward, 2012; Tortoli, 2012; Lory, 2014). However, despite much work (reviewed in Introduction), all known mycobacterial species are currently part of a single genus and their interrelationships are generally poorly understood (Magee and Ward, 2012; Tortoli, 2012; Lory, 2014; Fedrizzi et al., 2017). Genome sequences are now available for 150 of the 188 known mycobacterial species providing a unique opportunity for reliably understanding the relationships among the Mycobacterium species through genomic approaches. Using genome sequences, comprehensive phylogenetic and comparative genome analyses were carried out on Mycobacterium species using multiple independent approaches. In the first approach, phylogenomic trees were constructed for Mycobacterium species based on several large datasets of protein sequences including 1941 core proteins for the genus Mycobacterium, 136 core proteins for the phylum Actinobacteria, and another set of 8 highly conserved essential proteins found in all mycobacteria. Based on the core proteins in mycobacterial genomes, pairwise amino acid identity was also determined amongst different Mycobacterium species, providing a measure of the overall genetic relatedness of the species. In the third approach, exhaustive comparative genomic analyses were carried out on protein sequences of mycobacterial genomes to identify highly specific markers in the forms of CSIs and CSPs that are distinctive characteristics of the genus Mycobacterium as a whole or of different major clades within this genus. The results from all of these comprehensive genomic approaches reveal a consistent picture of the overall evolutionary relationships among the mycobacterial species, a summary of which is presented in Figure 10.
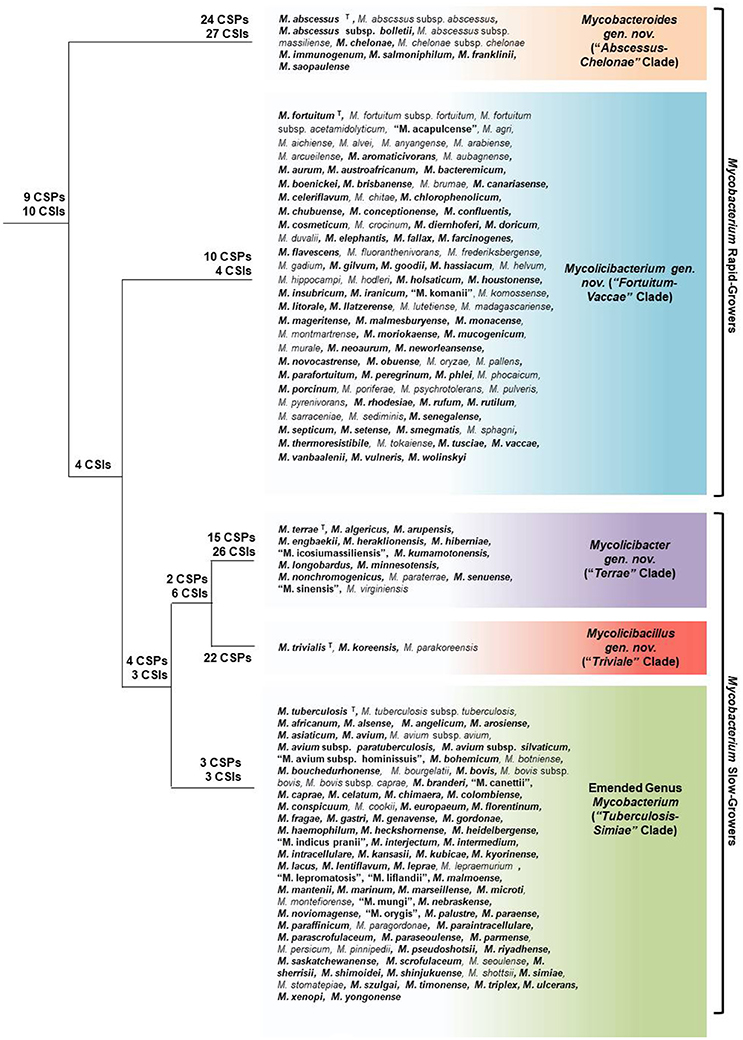
Figure 10. A summary diagram depicting the overall relationships among the major groups of mycobacterial species. The numbers of identified CSIs and CSPs, which are specific for different clades are marked on the nodes. The names of the five main clades of mycobacterial species identified in this work, viz. “Tuberculosis-Simiae,” “Terrae,” “Triviale,” “Fortuitum-Vaccae,” and “Abscessus-Chelonae”, along with their proposed or emended names and the species which are part of these clades are marked. Species which have had their genomes analyzed in this study are bolded. The superscript letter T beside a species indicates that it is the type species of the genus. The placements of other mycobacterial species, whose genomes have not been sequenced into these clades are based on their branching in the 16S rRNA tree (Supplmentary Figure 2). The species whose names are not italicized and are placed within quotation marks have not yet been validly published.
In phylogenetic trees constructed based on different large datasets of protein sequences, the Mycobacterium consistently grouped into four main strongly supported clades at the highest level. Within the larger “Terrae complex,” the species M. triviale and M. koreense also consistently formed a deeper branching “Triviale” clade. The existence of these five clades is also supported by the high degree of genome relatedness amongst the members of each clade, as indicated by the results of average amino acid identity analysis. More importantly, our analyses of protein sequences from Mycobacterium species have resulted in the identification of a total of 172 novel molecular markers (CSIs and CSPs) that are distinctive characteristics of either the entire genus Mycobacterium or of the five clades identified within this genus at various phylogenetic levels. A graphical schematic of the identified molecular markers and the mycobacterial clades for which they are specific for is shown in Figure 10. Thus, the existence as well as the distinctness of the five main clades within the genus Mycobacterium is supported not only by comprehensive phylogenomic studies and by genome relatedness analysis, but also by the identification of large numbers of highly specific molecular markers, which serve to clearly demarcate these clades. Although it is difficult to specify how many characters are sufficient to divide a given taxon into more than one group, as this will depend upon the genetic diversity as well as phylogenetic depth of a taxon, in cases where the monophyly and distinctness of the described clades are strongly supported by multiple genome-scale phylogenetic trees as well as other independent approaches (e.g., AAI or ANI analysis), even 1–2 reliable molecular characters such as the CSIs and CSPs are sufficient for separation of a given group into distinct taxa (Gao and Gupta, 2012; Bhandari et al., 2013; Gupta et al., 2013a,b, 2015; Adeolu and Gupta, 2014; Bhandari and Gupta, 2014; Sawana et al., 2014; Adeolu et al., 2016; Alnajar and Gupta, 2017; Barbour et al., 2017).
It should be noted that molecular markers such as CSIs and CSPs represent synapomorphic characteristics and they provide important means for reliable identification/demarcation of different monophyletic clades of organisms (Baldauf and Palmer, 1993; Gupta, 1998, 2016b; Rokas and Holland, 2000; Dutilh et al., 2008; Chandra and Chater, 2014). Extensive earlier work on these markers show that they are highly reliable characteristics of different groups of organisms and species as relationships based on them are generally not affected by factors such as differences in evolutionary rates or lateral gene transfers (Bhandari et al., 2012; Gupta, 2014, 2016a,b). Further, each of these CSIs or CSPs, which are present in different genes/proteins, provide independent evidence supporting the monophyletic nature of the different identified clades, as well as providing novel and reliable means for the demarcation as well as diagnostics of species from these clades of bacteria (Ahmod et al., 2011; Wong et al., 2014). Extensive earlier work on CSIs/CSPs provides evidence that both large as well as small CSIs (even a one amino acid insert/deletion in protein sequence results from an in frame three nucleotides insertion/deletion within a conserved region) and CSPs provide reliable molecular markers for taxonomic and diagnostic studies, and they also exhibit a high degree of predictive ability to be present in other members of the indicated groups for which sequence information is lacking at present (Gao and Gupta, 2012; Adeolu and Gupta, 2014; Naushad et al., 2014; Sawana et al., 2014; Adeolu et al., 2016; Gupta, 2016b; Alnajar and Gupta, 2017). As noted earlier, some of the CSIs and CSPs specific for the genus Mycobacterium were identified when the sequence information was available for a limited number of mycobacterial genomes (Gao and Gupta, 2005, 2012; Gao et al., 2006). However, despite the large increase in the number of mycobacterial genomes, many of these CSIs and CSPs are still found to be specific for this genus. In view of their demonstrated specificity and reliability for the indicated group of organisms, the CSIs and CSPs in recent years have been used extensively for important taxonomic changes to a number of prokaryotic groups at various phylogenetic levels ranging from description of new classes, orders, families and genera including division of the original Burkholderia, Borrelia and Thermotoga genera into two or more genera (Gao and Gupta, 2012; Bhandari et al., 2013; Gupta et al., 2013a,b, 2015; Adeolu and Gupta, 2014; Bhandari and Gupta, 2014; Sawana et al., 2014; Adeolu et al., 2016; Alnajar and Gupta, 2017; Barbour et al., 2017).
It should be noted that a 12–14 nucleotide insert in the 16S rRNA sequences (in helix 18 between positions 451 and 482 in the E. coli sequence) is often used as a marker to differentiate between rapid-growing and slow-growing mycobacteria (Pitulle et al., 1992; Hartmans et al., 2006; Tortoli, 2012, 2014; Fedrizzi et al., 2017). The presence and absence of this insert in different sequenced mycobacterial species has been examined by us and this information is presented in Supplementary Figure 85. This insert, due to its presence in a conserved region, also represents a CSI. However, in contrast to the large numbers of CSIs described in this work, which are of fixed lengths and highly-specific characteristics of the described clades, this insert is of variable length (9-14 aa insertion) and it is lacking in many members of the slow-growing mycobacteria or the “Tuberculosis-Simiae” clade (Hartmans et al., 2006; Tortoli, 2012, 2014). Thus, unlike the different CSIs identified in the present work, this insert in the 16S RNA is not a distinguishing characteristic of either all slow-growing Mycobacterium species (i.e., “Tuberculosis-Simiae” + “Terrae” + “Triviale” clades) or of the “Tuberculosis-Simiae” clade. However, all of the species belonging to the “Terrae” clade contain a 14 nucleotide insert in this position, which provides a signature CSI for this clade, similar to the large numbers of other CSIs and CSPs reported here (see Figure 9, Tables 6, 7). In contrast to the molecular markers described here, which are discrete and highly specific characteristics of the different indicated clades of mycobacteria, other physical and chemotaxonomic characteristics described in literature for various groups of mycobacteria are not specific for the indicated groups (see Supplementary Table 3; Magee and Ward, 2012). The presence or absence of the described physical and chemotaxonomic characteristics is often based on subjective criteria and information for such characteristics is not available for large numbers of mycobacterial species (Magee and Ward, 2012). This makes it difficult to reliably ascertain the potential usefulness of such characteristics as reliable markers for any particular group of mycobacteria.
The results presented here also strongly indicate that the “Abscessus-Chelonae” clade comprises the earliest branching lineage within the genus Mycobacterium. Its early divergence within the genus Mycobacterium is strongly supported by phylogenetic studies and multiple identified CSIs which are commonly shared by all or most Mycobacterium species, but absent in this clade of species. The deeper branching of the “Abscessus-Chelonae” clade as well as the “Fortuitum-Vaccae” clade of fast-growing mycobacteria, in comparison to the clades of slow-growing mycobacteria, supports the inference from earlier work that the rapid-growing mycobacterial species are ancestral and the slow-growers have evolved from them (Pitulle et al., 1992; Hartmans et al., 2006; Magee and Ward, 2012; Tortoli, 2012, 2014; Fedrizzi et al., 2017). Another important inference from the present work is that while the two clades of slow-growing mycobacteria (i.e., “Tuberculosis-Simiae” and the larger “Terrae + Triviale” clade) group together in phylogenetic trees, the grouping together of the two clades of rapid-growing mycobacteria is not observed in any phylogenetic trees. Further, while in our work 3 CSIs and 4 CSPs were identified that are commonly shared by members of the “Tuberculosis-Simiae” clade plus the “Terrae” + “Triviale” clade, no molecular marker was identified that is uniquely shared by the “Abscessus-Chelonae” and “Fortuitum-Vaccae” clades. It should be noted that while the distribution of most Mycobacterium species into the clades of slow-growing and fast-growing bacteria is generally in concordance with their rate of growth (Hartmans et al., 2006; Magee and Ward, 2012; Fedrizzi et al., 2017), a few exceptions are observed in this regard. In particular, the species M. doricum, M. vulneris and M. tusciae, which are slow-growing mycobacterial species (Magee and Ward, 2012; Fedrizzi et al., 2017), consistently branch within the “Fortuitum-Vaccae” clade of fast-growing mycobacteria. These species are also found to share the molecular signatures specific for the “Fortuitum-Vaccae” clade, but they lack the signatures for the slow-growing clades of mycobacteria. The anomalous branching of M. doricum and M. tusciae with the rapid-growing mycobacteria has also been reported in earlier work (Magee and Ward, 2012; Fedrizzi et al., 2017). This observation in conjunction with our results showing that both the slow-growing and fast-growing Mycobacterium species form at least two distinct clades, and that the rapidly-growing species do not form a monophyletic lineage, indicates that the differentiation of the Mycobacterium species based solely on their growth rate is of limited use for developing a coherent taxonomic framework that is consistent with genomic and phylogenetic characteristics.
Of the main clades of mycobacteria described here, the “Terrae” + “Triviale” and the “Abscessus-Chelonae” clades are recognized from earlier phylogenetic studies (Adékambi and Drancourt, 2004; Mignard and Flandrois, 2008; Tortoli, 2012, 2014; Fedrizzi et al., 2017; Wee et al., 2017). In the present work, distinctness of the “Abscessus-Chelonae” clade is established by 51 molecular markers (CSIs and CSPs) which are specific for this clade. Although our work has identified some molecular markers that are specific for the larger “Terrae” + “Triviale” clade, our results strongly indicate that the species from the “Triviale” clade are phylogenetically and molecularly distinct from those of the “Terrae” clade. The distinctness of these two clades is also strongly supported by larger numbers of molecular markers identified in our work that are uniquely shared by the members of either the “Terrae” clade or the “Triviale” clades. The “Terrae” clade is also distinguished from others by the presence of a 14 nucleotide insertion in the helix 18 of the 16S rRNA gene (Tortoli, 2012, 2014; Ngeow et al., 2015). The other two main clades of mycobacteria described here namely the “Tuberculosis-Simiae” clade and the “Fortuitum-Vaccae” clade, harbor >85% of the known Mycobacterium species and no molecular markers or other characteristics specific for these clades are known from earlier work. However, both these large clades of mycobacteria can now be reliably demarcated on the basis of multiple highly-specific molecular signatures. In addition to the five clades described here, a number of other smaller clades are observed in the phylogenetic trees (Figure 1 and Supplementary Figure 1). However, the work on characterization of these smaller subclades could be undertaken in future studies.
The work presented here based on multiple lines of evidence provide compelling support that the species from the genus Mycobacterium are comprised of five phylogenetically coherent clades, which can now be robustly distinguished from each other based on their branching in phylogenomic trees and multiple highly specific molecular signatures (Figure 10). These results provide a strong phylogenetic and genomic framework for division of the existing genus Mycobacterium into five distinct genera, corresponding to the five main clades described here. On the basis of the presented results, we are proposing that the genus Mycobacterium should be emended to include only members of the “Tuberculosis-Simiae” clade, which includes Mycobacterium tuberculosis, the type species of the genus (Zopf, 1883; Lehmann and Neumann, 1896), (Approved Lists, 1980; Skerman et al., 1980). The species from the other four main clades “Fortuitum-Vaccae”, “Terrae”, “Triviale” and “Abscessus-Chelonae” are transferred to four new genera with the following proposed names, Mycolicibacterium gen. nov., Mycolicibacter gen. nov., Mycolicibacillus gen. nov. and Mycobacteroides gen. nov., respectively. In the proposed classification, all of the major human pathogens are retained within the emended genus Mycobacterium, whereas the genus Mycolicibacterium is primarily comprised of environmental species. Most members of the proposed genera Mycolicibacter and Mycolicibacillus are also non-pathogenic, except occasional association of some species with animal hosts or human patients (Tasler and Hartley, 1981; Smith et al., 2000; Tortoli, 2014). Some members from the proposed genus Mycobacteroides are known to be associated with lung, skin and soft tissue infections (Simmon et al., 2011; Magee and Ward, 2012; Tortoli, 2014), however, none of them are considered as major life-threatening pathogens (Magee and Ward, 2012; Tortoli, 2014). Nonetheless, all five of these genera will remain part of the family Mycobacteriaceae and their proposed names bear close similarity to the original genus name Mycobacterium. Thus, all of them can still be referred to as mycobacterial species or as M. (species name), causing minimum confusion with any other species.
The proposed division of the existing genus Mycobacterium into the five proposed genera will have many benefits in terms of understanding and clarifying the relationships among the known mycobacterial species. The proposed division clearly separates the major human and animal pathogenic species, which are now part of the emended genus Mycobacterium, from all other (i.e., a majority of) mycobacterial species, which are either non-pathogenic or are of lesser clinical significance. With the explicit division of the mycobacterial species into these groups, attention can now be focused on unique genetic and molecular characteristics that differentiate the members of these groups of microbes. For each of these proposed genera, multiple CSIs and CSPs that are specific for these groups have been identified. Based on these molecular markers, it should be possible to develop novel and more reliable diagnostic methods for the identification of members of these groups by either in silico analysis of genomic sequences (based on BLASTp searches examining the presence or absence of these molecular sequences) or by experimental means utilizing PCR-based assays (Ahmod et al., 2011; Wong et al., 2014). Further, although the cellular functions of most of the identified CSIs or CSPs are not known, earlier work on other CSIs/CSPs has shown that these molecular characteristics are essential or play important functional roles in the organisms where they are found (Singh and Gupta, 2009; Schoeffler et al., 2010; Chandra and Chater, 2014; Gupta, 2016c). For example, some of the CSPs which are specific for the slow-growing mycobacterial species belong to the PE or PPE family of proteins, which play a role in virulence determination (Mukhopadhyay and Balaji, 2011). Hence, further functional investigations on the identified CSIs/CSPs are expected to lead to discovery of novel biochemical and/or other properties that are specific for either the entire Mycobacteriaceae family or for members of different genera that are part of this family.
The descriptions of the emended family Mycobacteriaceae, the emended genus Mycobacterium and of the four newly proposed genera viz, Mycolicibacter gen. nov., Mycobacteroides gen. nov., Mycolicibacillus gen. nov. and Mycolicibacterium gen. nov. are given below. Brief descriptions of the new species names combinations as well as some new species names resulting from the proposed taxonomic changes are also given below.
Emended Description of the Family Mycobacteriaceae Chester 1897 (Approved Lists 1980) (Skerman et al., 1980)
Mycobacteriaceae (My.co.bac.te.ri.a.ce´ae. N.L. neut. n. Mycobacterium type genus of the family; suff. -aceae ending to denote a family; N.L. fem. pl. n. Mycobacteriaceae the Mycobacterium family).
The family Mycobacteriaceae contains the type genus Mycobacterium as well as the genera Mycolicibacter gen. nov., Mycolicibacterium gen. nov., Mycolicibacillus gen nov., and Mycobacteroides gen. nov. Additionally, the genus Amycolicoccus is also indicated to be a part of this family (Wang et al., 2010; Parte, 2014). However, the sole type species of this genus, Amycolicoccus subflavus, is now reclassified as Hoyosella subflava (Hamada et al., 2016). The general characteristics of the family Mycobacteriaceae are as described by Magee and Ward (2012) for the genus Mycobacterium. The members of this family are aerobic to microaerophilic, slightly curved or straight rods (0.2–0.6 × 1.0–10 μm), which are acid–alcohol-fast at some stage of growth. Difficult to stain by Gram's-method, but are usually considered Gram-stain-positive. Some species may exhibit filamentous or mycelium-like growth. Cells are nonmotile and asporogenous. Colonies may be white- to cream-colored; some strains produce yellow- or orange-pigmented colonies with or without light stimulation. Whole-organism hydrolysates are rich in meso-diaminopimelic acid, arabinose, and galactose. The peptidoglycan is of the A1g type. Muramic acid moieties are N-glycolated. Cells and cell walls are rich in lipids. These include waxes which have characteristic, chloroform-soluble, mycolic acids with long (60–90 carbon atoms) branched chains. The fatty acid esters released on pyrolysis MS of mycolic acid esters have 22–26 carbon atoms. Cells contain diphosphatidylglycerol, phosphatidyl-ethanolamine, phosphatidylinositol, and phospatidylinositol mannosides as predominant polar lipids, straight-chain saturated, unsaturated, and 10-methyloctadecanoic (tuberculostearic) fatty acids as major fatty acid components, and dihydrogenated menaquinones with nine isoprene units as the predominant isoprenolog. The family includes obligate parasites, saprophytes, and opportunistic forms. The G+C content of genome-sequenced species varies from 57 to 71 (mol %) and genome size ranges from 3.1 to 10.5 Mbp. The members of the family Mycobacteriaceae form a distinct clade in the 16S rRNA tree and they are distinguished from all other members of the order Corynebacteriales by their unique shared presence of conserved signature indels described in this work (Table 1) in the following 10 proteins (viz. serine hydrolase, precorrin-4 C(11)-methyltransferase, NAD(P)H-quinone dehydrogenase, orotidine 5′-phosphate decarboxylase, deoxyribonuclease IV, peptidase C69, SGNH/GDSL hydrolase family protein, succinate dehydrogenase, N-dimethylarginine dimethylaminohydrolase, ergothioneine biosynthesis protein EgtB). Additionally, the homologs of the following nine proteins (accession numbers are in parenthesis) are also uniquely found in members of the family Mycobacteriaceae viz. hypothetical protein (WP_011723520.1), hypothetical protein (WP_011723901.1), MAV_11221(WP_011723955.1), membrane protein (WP_011724283.1), PE-PPE domain-containing protein (WP_011724324.1), DUF2561 domain-containing protein (WP_011724709.1), Membrane protein (WP_009976570.1), hypothetical protein (WP_003876314.1) and hypothetical protein (WP_003874755.1) (see Table 2 in this work).
Emended Description of the Genus Mycobacterium Lehmann and Neuman 1896 (Approved Lists 1980) (Skerman et al., 1980)
Mycobacterium (My.co.bac.te´ri.um. Gr. n. mykes a fungus; N.L. neut. n. bacterium, a small rod; N.L. neut. n. Mycobacterium, a fungus rodlet).
The type species is Mycobacterium tuberculosis (Zopf 1883) Lehmann and Neumann 1896 (Approved Lists 1980) (Skerman et al., 1980).
Members of this genus whose are slow-growing bacteria requiring at least 7 days of incubation at optimal temperatures to form colonies. Several species are obligate parasites of human and animals and the genus harbors a number of important human (e.g., Mycobacterium tuberculosis, M. leprae, M. ulcerans) and animal (e.g., Mycobacterium bovis) pathogens. Other phenotypic and chemotaxonomic characteristics of this genus are similar to that for the family Mycobacteriaceae.
Some species from this clade contain a 9–12 nucleotide long insert in helix 18 of the 16S rRNA gene sequence (Supplementary Figure 85; Hartmans et al., 2006; Tortoli, 2014). Species are indicated to generally lack the LivFGMH operon and the shaACDEFG cluster of genes, which encodes respectively for proteins allowing the transportation of leucine, isoleucine and valine into the bacteria and a Na+/H+ antiporter that is important for the homeostasis of Na+ and H+ (Wee et al., 2017). Presence of the components of Type VII secretion system has been reported in members of this genus (Wee et al., 2017). The members of this genus form a monophyletic clade in phylogenetic trees constructed based on 16S rRNA gene sequences as well as multiple large datasets of protein sequences described in this work including a tree based on 1941 core mycobacterial proteins, a tree based on 136 core proteins for the phylum Actinobacteria, and a tree based on concatenated sequences for eight conserved housekeeping proteins (viz. RpoA, RpoB, RpoC, GyrA, GyrB, Hsp65, EF-Tu, and RecA). Members of the genus Mycobacterium can be clearly distinguished from other genera within the Mycobacteriaceae family based on conserved signature indels described in this study (Table 4) in the following three proteins, a hypothetical protein, aldehyde dehydrogenase family protein and 23S rRNA (guanosine(2251)-2′-O)-methyltransferase, that are uniquely shared by the members of this genus. In addition, the homologs of the following three proteins (accession numbers are in parenthesis): a histone-like protein HNS (NP_218369.1), a hypothetical protein Rv4010 (YP_004837050.1) and a membrane protein (NP_217322.1), are also unique characteristics of the members of this genus.
The G-C content and genome sizes of the member species ranges from 57.8–69.3 (mol %) to 3.2–7.3 Mbp, respectively.
Description of Mycolicibacter gen. nov.
Mycolicibacter (My.co.li.ci.bac´ter. N.L. n. acidum mycolicum, mycolic acid; N.L. masc. n. bacter, rod; N.L. masc. n. Mycolicibacter, a genus of mycolic acid containing rod-shaped bacteria).
The type species is Mycolicibacter terrae.
The members of the genus Mycolicibacter are commonly referred to as the M. terrae complex. This genus contains species that are slow-growing (more than 7 days) and nonchromogenic with some species that show intermediate growth duration (5–15 days) (Tortoli, 2014; Ngeow et al., 2015). In phylogenetic trees, the Mycolicibacter clade forms a sister clade to a clade comprising of the genus Mycobacterium, harboring other slow-growing mycobacteria. Most members of this genus are non-pathogenic, but some species have been isolated from animal hosts (Tasler and Hartley, 1981) and human patients (Smith et al., 2000). Multiple antibiotic resistance has been reported for many of the isolates (Milne et al., 2009; Zhang et al., 2013b).
The members of this genus form a monophyletic clade in phylogenetic trees based on 16S rRNA gene sequences as well as multiple datasets of gene/protein sequences including a tree based on 1941 core mycobacteria proteins and a tree based on 136 core proteins for the phylum Actinobacteria. The members of the genus Mycolicibacter exhibit a closer relationship to members of the genus Mycolicibacillus in phylogenetic trees, which is also supported by a number of CSIs listed (Table 6) in the proteins ATP-dependent helicase, PDZ domain-containing protein, Ferredoxin reductase, DUF2236 domain-containing protein and two hypothetical protein with the accession number WP_083040170 and DUF4185 domain-containing protein, as well as 2 CSPs (viz. accession numbers WP_013830140.1 and WP_013827845.1) that are commonly shared by the members from these two genera. All of the species from this genus contain a 14 nucleotide insertion in the helix 18 of the 16S rRNA gene (Supplementary Figure 85; Tortoli, 2014). Additionally, the members of this genus are distinguished from members of all other genera within the family Mycobacteriaceae due to their possession of 26 conserved signature indels described in this study (Table 6) present in the following proteins, non-ribosomal peptide synthetase, nucleoside hydrolase, three different indels in TetR family transcriptional regulator, carbon starvation protein A, error-prone DNA polymerase, amidohydrolase, carboxymunconolacton decarboxylase family protein, polyketide cyclase, spirocyclase AveC family protein, TobH protein, UDP-N-acetylmuramate–L-alanine ligase, DUF2236 domain-containing protein, cobaltochelatase subunit CobN, alpha/beta hydrolase, potassium transporter Kef, bifunctional tRNA (adenosine(37)-N6)-threonylcarbamoyltransferase complex dimerization subunit Type 1 TsaB/ribosomal protein alanine acetyltransferase RimI, a membrane protein, DUF222 domain-containing protein, MFS transporter, adenylate/guanylate cyclase domain-containing protein, DUF2029 domain-containing protein and the following hypothetical proteins with the accession numbers (WP_083037591, WP_083040170, WP_083036336 and WP_052618664), that are uniquely found in the members of this genus. In addition, the homologs of the 17 conserved signature proteins, whose accession numbers are as follows (viz. WP_013830140.1, WP_013827845.1, WP_013828100.1, WP_013830932.1, WP_013828443.1, WP_013828919.1, WP_013829267.1, WP_041317168.1, WP_013827978.1, WP_041318963.1, WP_013830185.1, WP_013828762.1, WP_013827315.1, WP_041318191.1, WP_013829648.1, WP_013829864.1, and WP_041317804.1) are also distinctive characteristics of either all or most members of this genus (Table 7).
The members of the genus Mycolicibacter are characterized by high G-C content (66.3–70.3 mol %) and they have relatively short genomes (range 3.87–5.11 Mbp).
The description of Mycolicibacter terrae comb. nov. as well as the descriptions of new name combinations for other species which are part of the genus Mycolicibacter are provided in Table 8.
In addition to the new name combinations for species which are part of the genus Mycolicibacter, we also provide below description of two new species that should also be placed in the genus Mycolicibacter.
Description of Mycolicibacter icosiumassiliensis sp. nov. (i.co.si.u.mas.si.li.en´sis; L. masc. n. icosiumassiliensis, from the combination of Icosium, the Latin name of Algiers where the strain was first isolated and Massilia, the Latin name of Marseille, where the strain was described).
The description of this taxon is as given by Djouadi et al. (2016) for “Mycobacterium icosiumassilensis”. The type strain is 8WA6 (= CSUR P1561 = DSM 100711).
Description of Mycolicibacter sinensis sp. nov. (sin.en´sis. N.L. masc. adj. sinensis means “belonging to China,” indicating the source of the type strain).
The description of this taxon is as given by Zhang et al. (2013b) for “Mycobacterium sinense”. The type strain is JDM601.
Description of Mycolicibacillus gen. nov.
Mycolicibacillus (My.co.li.ci.ba.cil´lus. N.L. n. acidum mycolicum, mycolic acid; L. masc. n. bacillus, a small staff or rod; N.L. masc. n. Mycolicibacillus, a genus of mycolic acid containing rod-shaped bacteria).
The type species is Mycolicibacillus trivialis.
The genus Mycolicibacillus is comprised of slow-growing nonchromogenic bacteria requiring more than 7 days of incubation at optimal temperatures to form colonies. In phylogenetic trees, members of this genus form a deep-branching distinct clade that is most closely related to members of the genus Mycolicibacter. A close relationship of the species from the genera Mycolicibacillus and Mycolicibacter is also supported by a number of CSIs listed in Table 6 in the proteins ATP-dependent helicase, PDZ domain-containing protein, ferredoxin reductase, DUF2236 domain-containing protein, non-ribosomal peptide synthetase, hypothetical protein with accession number WP_083040170 and DUF4185 domain-containing protein and CSPs listed in Table 7 (viz. accession numbers WP_013830140.1 and WP_013827845.1) that are commonly shared by these two groups of bacteria. Unlike members of the genus Mycolicibacter, which contain a 14 nucleotide insertion in the helix 18 of the 16S rRNA gene, members of the genus Mycolicibacillus lack an insertion in this position (Tortoli, 2014) (Supplementary Figure 85). In addition, the homologs showing significant sequence similarity for the 22 proteins listed in Table 6 with the accession numbers WP_069390591.1, WP_069390644.1, WP_069390667.1, WP_069390717.1, WP_069391089.1, WP_069391367.1, WP_069391463.1, WP_069391521.1, WP_069391698.1, WP_069391782.1, WP_069391793.1, WP_069392105.1, WP_069392126.1, WP_069392251.1, WP_069392420.1, WP_069392510.1, WP_069392884.1, WP_069392982.1, WP_069392983.1, WP_069393100.1, WP_069393493.1, and WP_069393844.1, are uniquely present in members of this genus. This genus presently contains only three species (M. trivialis, M. koreensis and M. parakoreensis) and their genome sizes (3.89–4.08 Mbp) are among the smallest within the family Mycobacteriaceae. The G+C content of the two sequenced species is 69.4 mol %. Although some members of this genus have been isolated from human patients with pulmonary dysfunction, it is unclear whether they exhibit pathogenicity.
The description of Mycolicibacillus trivialis comb. nov. as well as the descriptions of new name combinations for other species which are part of the genus Mycolicibacillus are provided in Table 9.
Description of Mycobacteroides gen. nov.
Mycobacteroides (My.co.bac.te.ro´i.des. N.L. neut. n. Mycobacterium, a bacterial genus; L. neut. suff. -oides, resembling; N.L. neut. n. Mycobacteroides, a genus resembling Mycobacterium).
The type species is Mycobacteroides abscessus. The genus Mycobacteriodes is comprised of bacteria that are commonly referred to as members of the Abscessus-Chelonae clade. This is another genus within the family Mycobacteriaceae of rapidly-growing bacterial species (besides Mycolicibacterium) which take <7 days to form colonies. Phenotypic characteristics of this genus include a positive 3-day arylsulfatase test, better growth at 30°C than at a 35°C, negative nitrate reductase, negative iron uptake and resistance to polymyxin B (Brown-Elliott and Wallace, 2002). The genome size for the species within this clade ranges from 4.5 to 5.6 Mbp and their G+C content ranges from 63.9 to 64.8 mol %. Phylogenetic studies show that members of the genus Mycobacteriodes form a deep branching monophyletic clade within the family Mycobacteriaceae that is distinct from all other genera within this family. Some members from this genus are known to be involved in causing lung, skin and soft tissue infections (Magee and Ward, 2012; Tortoli, 2014) and some exhibit resistance to multiple antimicrobial drugs (Nessar et al., 2012).
The members of the genus Mycobacteriodes can be reliably distinguished from all other Mycobacteriaceae species as well as other bacteria based upon unique shared presence of 27 CSIs in different proteins listed in Table 3 (viz. uracil phosphoribosyltransferase, L-histidine N(alpha)-methyltransferase, DUF58 domain-containing protein, NADH-quinone oxidoreducatase subunit G, ATP-dependent helicase, tRNA (cytidine(34)-2′-O)-methyltransferase, glutamine-fructose-6-phosphate transaminase (isomerizing), error-prone DNA polymerase, 2-amino-4-hydroxy-6-hydroxymethyldihydropteridine diphosphokinase, DEAD/DEAH box helicase, anion transporter, a membrane protein, nicotinate-nucleotide adenylyltransferase, CoA ester lyase, bifunctional ADP-dependent (S)-NAD(P)H-hydrate dehydratase/NAD(P)H-hydrate epimerase, pyridoxal phosphate-dependent aminotransferase, carotenoid oxygenase, SAM-dependent methyltransferase, phosphoribosylamine-glycine ligase, and hypothetical proteins) and the presence of 24 conserved signature proteins listed in Table 2, (viz. MAB_0188c, MAB_0375, MAB_0601, MAB_2852c, MAB_3058, MAB_3079c, MAB_1107c, MAB_1519, MAB_1642, MAB_0008, MAB_0245c, MAB_2487, MAB_3020c, MAB_1440c, MAB_0014, MAB_0015, MAB_0345, MAB_0448c, MAB_0456, MAB_0460, MAB_2549, MAB_1765, MAB_1767, and MAB_1806) that are also specifically found in these bacteria.
The description of Mycobacteroides abscessus comb. nov. as well as the descriptions of new name combinations for other species which are part of the genus Mycobacteroides are provided in Table 10.
Description of Mycolicibacterium gen. nov.
Mycolicibacterium (My.co.li.ci.bac.te´ri.um. N.L. n. acidum mycolicum, mycolic acid; N.L. neut. n. bacterium, a small rod; N.L. neut. n. Mycolicibacterium, a genus of mycolic acid containing rod-shaped bacteria).
The type species Mycolicibacterium fortuitum.
The genus is comprised of rapidly-growing bacterial species, which take <7 days to form colonies upon primary isolation (Parte, 2014). Some other phenotypic characteristics generally common to the members of this genus include absence of pigmentation, positive 3-day arylsulfatase activity (Brown-Elliott and Wallace, 2002), positive for nitrate reductase and iron uptake (Magee and Ward, 2012). Most species are saprophytic and considered non-pathogenic to humans, however some cases of infections and diseases by members of this group have been reported (Stahl and Urbance, 1990; Brown-Elliott and Wallace, 2002; Ripoll et al., 2009). The members of this genus form a monophyletic clade in phylogenetic trees based on concatenated sequences of multiple large datasets of conserved proteins including a tree based on 1941 core proteins from mycobacterial genomes, a tree based on 136 core proteins for the phylum Actinobacteria, and another tree based on concatenated sequences for 8 conserved proteins described in the present study.
The members of the genus Mycolicibacterium can be distinguished from other genera within the family Mycobacteriaceae as well as other bacteria based upon conserved signature indels in the following four proteins viz. LacI family transcriptional regulator, Cyclase, CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase and CDP-diacylglycerol–serine O-phosphatidyltransferase (Table 4) that are uniquely shared by the members of this genus. Additionally, the homologs of the 10 conserved signature proteins, whose accession numbers are as follows (WP_048630777.1, WP_048632025.1, WP_048632497.1, WP_048634851.1, WP_048633467.1, WP_048633322.1 WP_048631132.1, WP_048634509.1, WP_048630657.1, and WP_048632441.1) are also uniquely found in the members of this genus (Table 5). The genome size for the members of this genus ranges from 3.95 to 8.0 Mbp and their G+C content ranges from 65.4 to 70.3 mol %.
The description of Mycolicibacterium fortuitum comb. nov. as well as the descriptions of new name combinations for other species which are part of the genus Mycolicibacterium are provided in Table 11.
In addition to the new name combinations for species which are part of this genus, we also provide below description of two new species that should also be placed in the genus Mycolicibacterium.
Description of Mycolicibacterium acapulense sp. nov. (a.ce.pul.cen´se. N.L. neut. adj. acapulcense from Acapulco, a town on the Pacific coast of México).
The description of this taxon is as given by Bojalil et al. (1962) for “Mycobacterium acapulensis”. The type strain is AC-103 (= ATCC 14473 = JCM 6402).
Description of Mycolicibacterium komanii sp. nov. (ko.ma´ni.i. N.L. gen. n. komanii named after a town in South Africa where one of the isolates originated from, Komani is the Xhosa name for Queenstown (South Africa)).
The description of this taxon is as given by Gcebe (2015) and Gcebe et al. (2016) for “Mycobacterium komanii”. The type strain is GPK 1020.
Author Contributions
RG was responsible for conceiving the idea of this study, carried out phylogenomic and other analyses reported here, supervised and directed the entire project and obtained funds for carrying out these studies. Involved in the writing and finalizing of the manuscript and all presented data. BL and JS were responsible for analysis and organization of the comparative genomic data on identification of described molecular signatures, under the direction of RG. They also helped in the preparation of a draft version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the research grant No. 249924 from the Natural Science and Engineering Research Council of Canada awarded to RG. We thank T. Vijaykumar for carrying out preliminary work in this regard. Lastly, we express our sincere thanks and the deepest appreciation to Professor Aharon Oren for his valuable input/suggestions regarding the correct etymology and protologues for the names of newly proposed taxa and the new name combinations.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.00067/full#supplementary-material
References
Adékambi, T., and Drancourt, M. (2004). Dissection of phylogenetic relationships among 19 rapidly growing Mycobacterium species by 16S rRNA, hsp65, sodA, recA and rpoB gene sequencing. Int. J. Syst. Evol. Microbiol. 54, 2095–2105. doi: 10.1099/ijs.0.63094-0
Adékambi, T., Berger, P., Raoult, D., and Drancourt, M. (2006a). rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56, 133–143. doi: 10.1099/ijs.0.63969-0
Adékambi, T., Stein, A., Carvajal, J., Raoult, D., and Drancourt, M. (2006b). Description of Mycobacterium conceptionense sp. nov., a Mycobacterium fortuitum group organism isolated from a posttraumatic osteitis inflammation. J. Clin. Microbiol. 44, 1268–1273. doi: 10.1128/JCM.44.4.1268-1273.2006
Adékambi, T., Stein, A., Carvajal, J., Raoult, D., and Drancourt, M. (2006c). Mycobacterium conceptionense sp. nov. in: list of new names and new combinations previously effectively, but not Validly, Published. List no. 111. Int. J. Syst. Evol. Microbiol. 56, 2025–2027. doi: 10.1099/ijs.0.64643-0
Adeolu, M., Alnajar, S., Naushad, S., and Gupta, S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. doi: 10.1099/ijsem.0.001485
Adeolu, M., and Gupta, R. S. (2014). A phylogenomic and molecular marker based proposal for the division of the genus Borrelia into two genera: the emended genus Borrelia containing only the members of the relapsing fever Borrelia, and the genus Borreliella gen. nov. containing the members of the Lyme disease Borrelia (Borrelia burgdorferi sensu lato complex). Antonie van Leeuwenhoek 105, 1049–1072. doi: 10.1007/s10482-014-0164-x
Ahmod, N. Z., Gupta, R. S., and Shah, H. N. (2011). Identification of a Bacillus anthracis specific indel in the yeaC gene and development of a rapid pyrosequencing assay for distinguishing B. anthracis from the B. cereus group. J. Microbiol. Methods 87, 278–285. doi: 10.1016/j.mimet.2011.08.015
Alnajar, S., and Gupta, R. S. (2017). Phylogenomics and comparative genomic studies delineate six main clades within the family Enterobacteriaceae and support the reclassification of several polyphyletic members of the family. Infect. Genet. Evol. 54, 108–127. doi: 10.1016/j.meegid.2017.06.024
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Apajalahti, J. H. A., Karpanoja, P., and Salkinoja-Salonen, M. S. (1986). Rhodococcus chlorophenolicus sp. nov., a chlorophenol-mineralizing actinomycete. Int. J. Syst. Evol. Microbiol. 36, 246–251. doi: 10.1099/00207713-36-2-246
Ausina, V., Luquin, M., García-Barceló, M., Laneelle, M. A., Lévy-Frébault, V. V., Belda, F., et al. (1992). Mycobacterium alvei sp. nov. Int. J. Syst. Bacteriol. 42, 529–535. doi: 10.1099/00207713-42-4-529
Balcázar, J. L., Planas, M., and Pintado, J. (2014a). Mycobacterium hippocampi sp. nov. in Oren A., Garrity G.M. (editors). List of new names and new combinations previously effectively, but not validly, published, list no. 160. Int. J. Syst. Evol. Microbiol. 64, 3603–3606. doi: 10.1099/ijsem.0.001173
Balcázar, J. L., Planas, M., and Pintado, J. (2014b). Mycobacterium hippocampi sp. nov., a rapidly growing scotochromogenic species isolated from a seahorse with tail rot. Curr. Microbiol. 69, 329–333. doi: 10.1007/s00284-014-0588-6
Baldauf, S. L., and Palmer, J. D. (1993). Animals and fungi are each other's closest relatives: congruent evidence from multiple proteins. Proc. Natl. Acad. Sci. U.S.A. 90, 11558–11562. doi: 10.1073/pnas.90.24.11558
Barbour, A. G., Adeolu, M., and Gupta, R. S. (2017). Division of the genus Borrelia into two genera (corresponding to Lyme disease and relapsing fever groups) reflects their genetic and phenotypic distinctiveness and will lead to a better understanding of these two groups of microbes. Int. J. Syst. Evol. Microbiol. 67, 2058–2067. doi: 10.1099/ijsem.0.001815
Bergey, D. H., Harrison, F. C., Breed, R. S., Hammer, B. W., and Huntoon, F. M. (1923). Bergey's Manual of Determinative Bacteriology. Baltimore, MD: The Williams and Wilkins Co.
Bhandari, V., Ahmod, N. Z., Shah, H. N., and Gupta, R. S. (2013). Molecular signatures for Bacillus species: demarcation of the Bacillus subtilis and Bacillus cereus clades in molecular terms and proposal to limit the placement of new species into the genus Bacillus. Int. J. Syst. Evol. Microbiol. 63, 2712–2726. doi: 10.1099/ijs.0.048488-0
Bhandari, V., and Gupta, R. S. (2014). Molecular signatures for the phylum (class) Thermotogae and a proposal for its division into three orders (Thermotogales, Kosmotogales ord. nov. and Petrotogales ord. nov.) containing four families (Thermotogaceae, Fervidobacteriaceae fam. nov., Kosmotogaceae fam. nov. and Petrotogaceae fam. nov.) and a new genus Pseudothermotoga gen. nov. with five new combinations. Antonie van Leeuwenhoek 105, 143–168. doi: 10.1007/s10482-013-0062-7
Bhandari, V., Naushad, H. S., and Gupta, R. S. (2012). Protein based molecular markers provide reliable means to understand prokaryotic phylogeny and support Darwinian mode of evolution. Front. Cell Infect. Microbiol. 2:98. doi: 10.3389/fcimb.2012.00098
Bojalil, L. F., Cerbon, J., and Trujillo, A. (1962). Adansonian classification of mycobacteria. J. Gen. Microbiol. 28, 333–346. doi: 10.1099/00221287-28-2-333
Bonicke, R., and Juhasz, E. (1964). Description of the new species Mycobacterium vaccae n. sp. Zentralblatt für Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. 1. Abt. Medizinisch-hygienische Bakteriologie, Virusforschung und Parasitologie. Originale 192, 133–135.
Brown, B. A., Springer, B., Steingrube, V. A., Wilson, R. W., Pfyffer, G. E., Garcia, M. J., et al. (1999). Mycobacterium wolinskyi sp. nov. and Mycobacterium goodii sp. nov., two new rapidly growing species related to Mycobacterium smegmatis and associated with human wound infections: a cooperative study from the International Working Group on Mycobacterial Taxonomy. Int. J. Syst. Bacteriol. 49, 1493–1511. doi: 10.1099/00207713-49-4-1493
Brown-Elliott, B. A., and Wallace, R. J. (2002). Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria. Clin. Microbiol. Rev. 15, 716–746. doi: 10.1128/CMR.15.4.716-746.2002
Brown-Elliott, B. A., Wallace, R. J. Jr., Petti, C. A., Mann, L. B., McGlasson, M., Chihara, S., et al. (2010). Mycobacterium neoaurum and Mycobacterium bacteremicum sp. nov. as causes of mycobacteremia. J. Clin. Microbiol. 48, 4377–4385. doi: 10.1128/JCM.00853-10
Brown-Elliott, B. A., Wallace, R. J. Jr., Petti, C. A., Mann, L. B., McGlasson, M., Chihara, S., et al. (2012). Mycobacterium bacteremicum sp. nov. in: list of new names and new combinations previously effectively, but not validly, published, list no. 148. Int. J. Syst. Evol. Microbiol. 62, 2549–2554. doi: 10.1099/ijs.0.048033-0
Brzostek, A., Pawelczyk, J., Rumijowska-Galewicz, A., Dziadek, B., and Dziadek, J. (2009). Mycobacterium tuberculosis is able to accumulate and utilize cholesterol. J. Bacteriol. 191, 6584–6591. doi: 10.1128/JB.00488-09
Capella-Gutierrez, S., Silla-Martinez, J. M., and Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Casal, M., and Calero, J. R. (1974). Mycobacterium gadium sp. nov. a new species of rapid-growing scotochromogenic mycobacteria. Tubercle 55, 299–308. doi: 10.1016/0041-3879(74)90039-7
Chamoiseau, G. (1973). Mycobacterium farcinogenes, agent causal du farcin du boeuf en Afrique. Revue d'Elevage et de Médecine Vétérinaire des Pays Tropicaux 27, 61–65. doi: 10.19182/remvt.7975
Chamoiseau, G. (1979). Etiology of farcy in African bovines. Int. J. Syst. Evol. Microbiol. 29, 407–410.
Chandra, G., and Chater, K. F. (2014). Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol. Rev. 38, 345–379. doi: 10.1111/1574-6976.12047
Cloud, J. L., Meyer, J. J., Pounder, J. I., Jost, K. C. Jr., Sweeney, A., Carroll, K. C., et al. (2006). Mycobacterium arupense sp. nov., a non-chromogenic bacterium isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 56, 1413–1418. doi: 10.1099/ijs.0.64194-0
Cooksey, R. C., de Waard, J. H., Yakrus, M. A., Rivera, I., Chopite, M., Toney, S. R., et al. (2004). Mycobacterium cosmeticum sp. nov., a novel rapidly growing species isolated from a cosmetic infection and from a nail salon. Int. J. Syst. Evol. Microbiol. 54, 2385–2391. doi: 10.1099/ijs.0.63238-0
da Costa Cruz, J. C. (1938). Mycobacterium fortuitum, un novo bacilo acido-resistente pathogenico para o human. Acta Med. 1, 297–301.
Derz, K., Klinner, U., Schuphan, I., Stackebrandt, E., and Kroppenstedt, R. M. (2004). Mycobacterium pyrenivorans sp. nov., a novel polycyclic-aromatic-hydrocarbon-degrading species. Int. J. Syst. Evol. Microbiol. 54, 2313–2317. doi: 10.1099/ijs.0.03003-0
Devulder, G., Perouse de Montclos, M., and Flandrois, J. P. (2005). A multigene approach to phylogenetic analysis using the genus Mycobacterium as a model. Int. J. Syst. Evol. Microbiol. 55, 293–302. doi: 10.1099/ijs.0.63222-0
Djouadi, L. N., Levasseur, A., Khalil, J. B., Blanc-Taileur, C., Asmar, S., Ghiloubi, W., et al. (2016). Mycobacterium icosiumassiliensis sp. nov., a new member in the Mycobacterium terrae complex isolated from surface water in Algeria. Curr. Microbiol. 73, 255–264. doi: 10.1007/s00284-016-1062-4
Domenech, P., Jimenez, M. S., Menendez, M. C., Bull, T. J., Samper, S., Manrique, A., et al. (1997). Mycobacterium mageritense sp. nov. Int. J. Syst. Bacteriol. 47, 535–540. doi: 10.1099/00207713-47-2-535
Dutilh, B. E., Snel, B., Ettema, T. J., and Huynen, M. A. (2008). Signature genes as a phylogenomic tool. Mol. Biol. Evol. 25, 1659–1667. doi: 10.1093/molbev/msn115
Eddy, S. R. (2011). Accelerated Profile HMM Searches. PLoS Comput. Biol. 7:e1002195. doi: 10.1371/journal.pcbi.1002195
Fahey, R. C. (2001). Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55, 333–356. doi: 10.1146/annurev.micro.55.1.333
Falkinham, J. O. (2009). Surrounded by mycobacteria: nontuberculous mycobacteria in the human environment. J. Appl. Microbiol. 107, 356–367. doi: 10.1111/j.1365-2672.2009.04161.x
Fedrizzi, T., Meehan, C. J., Grottola, A., Giacobazzi, E., Fregni, S. G., Tagliazucchi, S., et al. (2017). Genomic characterization of Nontuberculous Mycobacteria. Sci. Rep. 7:45258. doi: 10.1038/srep45258
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gao, B., and Gupta, R. S. (2005). Conserved indels in protein sequences that are characteristic of the phylum Actinobacteria. Int. J. Syst. Evol. Microbiol. 55, 2401–2412. doi: 10.1099/ijs.0.63785-0
Gao, B., and Gupta, R. S. (2012). Phylogenetic framework and molecular signatures for the main clades of the phylum Actinobacteria. Microbiol. Mol. Biol. Rev. 76, 66–112. doi: 10.1128/MMBR.05011-11
Gao, B., Paramanathan, R., and Gupta, R. S. (2006). Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie van Leeuwenhoek 90, 69–91. doi: 10.1007/s10482-006-9061-2
Gcebe, N. (2015). The Occurrence and Molecular Characterization of Non-tuberculous Mycobacteria in Cattle, African Buffalo (Syncerus caffer) and Their Environments in South Africa and Genomic Characterization and Proteomic Comparison with Mycobacterium bovis. Ph.D. thesis, University of Pretoria, Pretoria, South Africa.
Gcebe, N., Michel, A., Gey van Pittius, N. C., and Rutten, V. (2016). Comparative genomics and proteomic analysis of four non-tuberculous Mycobacterium species and Mycobacterium tuberculosis complex: occurrence of shared immunogenic proteins. Front. Microbiol. 7:795. doi: 10.3389/fmicb.2016.00795
Gcebe, N., Rutten, V., Pittius, N. G. V., Naicker, B., and Michel, A. (2017). Mycobacterium malmesburyense sp. nov., a non-tuberculous species of the genus Mycobacterium revealed by multiple gene sequence characterization. Int. J. Syst. Evol. Microbiol. 67, 832–838. doi: 10.1099/ijsem.0.001678
Gomila, M., Ramirez, A., Gasco, J., and Lalucat, J. (2008). Mycobacterium llatzerense sp. nov., a facultatively autotrophic, hydrogen-oxidizing bacterium isolated from haemodialysis water. Int. J. Syst. Evol. Microbiol. 58, 2769–2773. doi: 10.1099/ijs.0.65857-0
Goodfellow, M., and Magee, G. M. (1998). “Taxonomy of Mycobacteria,” in Mycobacteria, eds P. A. Gangadharam and P. A. Jenkins (New York, NY: Chapman and Hall), 1–71.
Guillemin, I., Cambau, E., and Jarlier, V. (1995). Sequences of conserved region in the A subunit of DNA gyrase from nine species of the genus Mycobacterium: phylogenetic analysis and implication for intrinsic susceptibility to quinolones. Antimicro. Agents Chemother. 39, 2145–2149. doi: 10.1128/AAC.39.9.2145
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Gupta, R. S. (1998). Protein phylogenies and signature sequences: a reappraisal of evolutionary relationships among Archaebacteria, Eubacteria, and Eukaryotes. Microbiol. Mol. Biol. Rev. 62, 1435–1491.
Gupta, R. S. (2014). “Identification of conserved indels that are useful for classification and evolutionary studies,” in Methods in Microbiology: New Approaches to Prokaryotic Systematics, eds M. Goodfellow, I. Sutcliffe, and J. Chun (Oxford: Academic Press), 153–182.
Gupta, R. S. (2016a). Editorial: Applications of genome sequences for discovering characteristics that are unique to different groups of organisms and provide insights into evolutionary relationships. Front. Genet. 7:27. doi: 10.3389/fgene.2016.00027
Gupta, R. S. (2016b). Impact of genomics on the understanding of microbial evolution and classification: the importance of Darwin's views on classification. FEMS Microbiol. Rev. 40, 520–553. doi: 10.1093/femsre/fuw011
Gupta, R. S. (2016c). Molecular signatures that are distinctive characteristics of the vertebrates and chordates and supporting a grouping of vertebrates with the tunicates. Mol. Phylogenet. Evol. 94, 383–391. doi: 10.1016/j.ympev.2015.09.019
Gupta, R. S., Chander, P., and George, S. (2013a). Phylogenetic framework and molecular signatures for the class Chloroflexi and its different clades; proposal for division of the class Chloroflexia class. nov. [corrected] into the suborder Chloroflexineae subord. nov., consisting of the emended family Oscillochloridaceae and the family Chloroflexaceae fam. nov., and the suborder Roseiflexineae subord. nov., containing the family Roseiflexaceae fam. nov. Antonie van Leeuwenhoek 103, 99–119. doi: 10.1007/s10482-012-9790-3
Gupta, R. S., Chen, W. J., Adeolu, M., and Chai, Y. (2013b). Molecular signatures for the class Coriobacteriia and its different clades; Proposal for division of the class Coriobacteriia into the emended order Coriobacteriales, containing the emended family Coriobacteriaceae and Atopobiaceae fam. nov., and Eggerthellales ord. nov., containing the family Eggerthellaceae fam. nov. Int. J. Syst. Evol. Microbiol. 63, 3379–3397. doi: 10.1099/ijs.0.048371-0
Gupta, R. S., Naushad, S., and Baker, S. (2015). Phylogenomic analyses and molecular signatures for the class Halobacteria and its two major clades: a proposal for division of the class Halobacteria into an emended order Halobacteriales and two new orders, Haloferacales ord. nov. and Natrialbales ord. nov., containing the novel families Haloferacaceae fam. nov. and Natrialbaceae fam. nov. Int. J. Syst. Evol. Microbiol. 65, 1050–1069. doi: 10.1099/ijs.0.070136-0
Hamada, M., Shibata, C., Sakurai, K., Hosoyama, A., Oji, S., Teramoto, K., et al. (2016). Reclassification of Amycolicicoccus subflavus as Hoyosella subflava comb. nov. and emended descriptions of the genus Hoyosella and Hoyosella altamirensis. Int. J. Syst. Evol. Microbiol. 66, 4711–4715. doi: 10.1099/ijsem.0.001415
Hannigan, G. D., Krivogorsky, B., Fordice, D., Welch, J. B., and Dahl, J. L. (2013). Mycobacterium minnesotense sp. nov., a photochromogenic bacterium isolated from sphagnum peat bogs. Int. J. Syst. Evol. Microbiol. 63, 124–128. doi: 10.1099/ijs.0.037291-0
Hartmans, S., de Bont, J. A. M., and Stackebrandt, E. (2006). “Chapter 1.1.18 - The genus Mycobacterium—nonmedical,” in The Prokaryotes: Volume 3: Archaea. Bacteria: Firmicutes, Actinomycetes, eds M. Dworkin, S. Falkow, E. Rosenberg, K. H. Schleifer, and E. Stackebrandt (New York, NY: Springer New York), 889–918.
Hennessee, C. T., Seo, J. S., Alvarez, A. M., and Li, Q. X. (2009). Polycyclic aromatic hydrocarbon-degrading species isolated from Hawaiian soils: Mycobacterium crocinum sp. nov., Mycobacterium pallens sp. nov., Mycobacterium rutilum sp. nov., Mycobacterium rufum sp. nov. and Mycobacterium aromaticivorans sp. nov. Int. J. Syst. Evol. Microbiol. 59, 378–387. doi: 10.1099/ijs.0.65827-0
Hormisch, D., Brost, I., Kohring, G. W., Giffhorn, F., Kroppenstedt, R. M., Stackebrandt, E., et al. (2004). Mycobacterium fluoranthenivorans sp. nov., a fluoranthene and aflatoxin B1 degrading bacterium from contaminated soil of a former coal gas plant. Syst. Appl. Microbiol. 27, 653–660. doi: 10.1078/0723202042369866
Hormisch, D., Brost, I., Kohring, G. W., Giffhorn, F., Kroppenstedt, R. M., Stackebrandt, E., et al. (2006). Mycobacterium fluoranthenivorans sp. nov,” in: list of new names and new combinations previously effectively, but not validly, published, list no. 110. Int. J. Syst. Evol. Microbiol. 56, 1459–1460. doi: 10.1099/ijs.0.64507-0
Jíménez, M. S., Campos-Herrero, M. I., Garcia, D., Luquin, M., Herrera, L., and Garcia, M. J. (2004). Mycobacterium canariasense sp. nov. Int. J. Syst. Evol. Microbiol. 54, 1729–1734. doi: 10.1099/ijs.0.02999-0
Kasai, H., Ezaki, T., and Harayama, S. (2000). Differentiation of phylogenetically related slowly growing mycobacteria by their gyrB sequences. J. Clin. Microbiol. 38, 301–308.
Kazda, J. (1980). Mycobacterium sphagni sp. nov. Int. J. Syst. Evol. Microbiol. 30, 77–81. doi: 10.1099/00207713-30-1-77
Kazda, J., and Müller, K. (1979). Mycobacterium komossense sp. nov. Int. J. Syst. Evol. Microbiol. 29, 361–365. doi: 10.1099/00207713-29-4-361
Kazda, J., Cooney, R., Monaghan, M., Quinn, P. J., Stackebrandt, E., Dorsch, M., et al. (1993). Mycobacterium hiberniae sp. nov. Int. J. Syst. Bacteriol. 43, 352–357. doi: 10.1099/00207713-43-2-352
Kazda, J., Müller, H. J., Stackebrandt, E., Daffé, M., Müller, K., and Pitulle, C. (1992). Mycobacterium madagascariense sp. nov. Int. J. Syst. Evol. Microbiol. 42, 524–528. doi: 10.1099/00207713-42-4-524
Khan, A. A., Kim, S. J., Paine, D. D., and Cerniglia, C. E. (2002). Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1997–2002. doi: 10.1099/00207713-52-6-1997
Kim, B. J., Hong, S. H., Yu, H. K., Park, Y. G., Jeong, J., Lee, S. H., et al. (2013). Mycobacterium parakoreense sp. nov., a slowly growing non-chromogenic species related to Mycobacterium koreense, isolated from a human clinical specimen. Int. J. Syst. Evol. Microbiol. 63, 2301–2308. doi: 10.1099/ijs.0.045070-0
Kim, B. J., Jeong, J., Lee, S. H., Kim, S. R., Yu, H. K., Park, Y. G., et al. (2012). Mycobacterium koreense sp. nov., a slowly growing non-chromogenic species closely related to Mycobacterium triviale. Int. J. Syst. Evol. Microbiol. 62, 1289–1295. doi: 10.1099/ijs.0.033274-0
Kim, B. J., Kim, J. M., Kim, B. R., Lee, S. Y., Kim, G., Jang, Y. H., et al. (2015). Mycobacterium anyangense sp. nov., a rapidly growing species isolated from blood of Korean native cattle, Hanwoo (Bos taurus coreanae). Int. J. Syst. Evol. Microbiol. 65, 2277–2285. doi: 10.1099/ijs.0.000255
Kim, H., Kim, S. H., Shim, T. S., Kim, M. N., Bai, G. H., Park, Y. G., et al. (2005). Differentiation of Mycobacterium species by analysis of the heat-shock protein 65 gene (hsp65). Int. J. Syst. Evol. Microbiol. 55, 1649–1656. doi: 10.1099/ijs.0.63553-0
Kim, S. H., and Shin, J. H. (2017). Identification of nontuberculous mycobacteria using multilocous sequence analysis of 16S rRNA, hsp65, and rpoB. J. Clin. Lab. Anal. 32:e22184. doi: 10.1002/jcla.22184
Kirschner, P., Teske, A., Schroder, K. H., Kroppenstedt, R. M., Wolters, J., and Bottger, E. C. (1992). Mycobacterium confluentis sp. nov. Int. J. Syst. Bacteriol. 42, 257–262. doi: 10.1099/00207713-42-2-257
Kleespies, M., Kroppenstedt, R. M., Rainey, F. A., Webb, L. E., and Stackebrandt, E. (1996). Mycobacterium hodleri sp. nov., a new member of the fast-growing mycobacteria capable of degrading polycyclic aromatic hydrocarbons. Int. J. Syst. Bacteriol. 46, 683–687. doi: 10.1099/00207713-46-3-683
Klenk, H. P., and Goker, M. (2010). En route to a genome-based classification of Archaea and Bacteria? Syst. Appl. Microbiol. 33, 175–182. doi: 10.1016/j.syapm.2010.03.003
Konjek, J., Souded, S., Guerardel, Y., Trivelli, X., Bernut, A., Kremer, L., et al. (2016). Mycobacterium lutetiense sp. nov., Mycobacterium montmartrense sp. nov. and Mycobacterium arcueilense sp. nov., members of a novel group of non-pigmented rapidly growing mycobacteria recovered from a water distribution system. Int. J. Syst. Evol. Microbiol. 66, 3694–3702. doi: 10.1099/ijsem.0.001253
Konstantinidis, K. T., and Tiedje, J. M. (2005). Genomic insights that advance the species definition for prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 2567–2572. doi: 10.1073/pnas.0409727102
Konstantinidis, K. T., and Tiedje, J. M. (2007). Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr. Opin. Microbiol. 10, 504–509. doi: 10.1016/j.mib.2007.08.006
Kubica, G. P., Silcox, V. A., Kilburn, J. O., Smithwick, R. W., Beam, R. E., Jones, W. D., et al. (1970). Differential identification of mycobacteria VI. Mycobacterium triviale Kubica sp. nov. Int. J. Syst. Evol. Microbiol. 20, 161–174. doi: 10.1099/00207713-20-2-161
Kusunoki, S., and Ezaki, T. (1992). Proposal of Mycobacterium peregrinum sp. nov., nom. rev., and elevation of Mycobacterium chelonae subsp. abscessus (Kubica et al.) to species status: Mycobacterium abscessus comb. nov. Int. J. Syst. Bacteriol. 42, 240–245. doi: 10.1099/00207713-42-2-240
Lamy, B., Marchandin, H., Hamitouche, K., and Laurent, F. (2008). Mycobacterium setense sp. nov., a Mycobacterium fortuitum-group organism isolated from a patient with soft tissue infection and osteitis. Int. J. Syst. Evol. Microbiol. 58, 486–490. doi: 10.1099/ijs.0.65222-0
Leao, S. C., Tortoli, E., Euzeby, J. P., and Garcia, M. J. (2011). Proposal that Mycobacterium massiliense and Mycobacterium bolletii be united and reclassified as Mycobacterium abscessus subsp. bolletii comb. nov., designation of Mycobacterium abscessus subsp. abscessus subsp. nov. and emended description of Mycobacterium abscessus. Int. J. Syst. Evol. Microbiol. 61, 2311–2313. doi: 10.1099/ijs.0.023770-0
Le, S. Q., and Gascuel, O. (2008). An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320. doi: 10.1093/molbev/msn067
Lee, H., Lee, S. A., Lee, I. K., Yu, H. K., Park, Y. G., Jeong, J., et al. (2010). Mycobacterium paraterrae sp. nov. recovered from a clinical specimen: novel chromogenic slow growing mycobacteria related to Mycobacterium terrae complex. Microbiol. Immunol. 54, 46–53. doi: 10.1111/j.1348-0421.2009.00184.x
Lee, H., Lee, S. A., Lee, I. K., Yu, H. K., Park, Y. G., Jeong, J., et al. (2016). Mycobacterium parraterrae sp. nov. in: Oren A., Garrity G.M. (editors). List of new names and new combinations previously effectively, but not validly, published, list no. 172. Int. J. Syst. Evol. Microbiol. 66, 4299-4305. doi: 10.1099/ijsem.0.001585
Lehmann, K. B., and Neumann, R. O. (1896). Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik. München: J. F. Lehmann. Available online at: http://www.worldcat.org/title/atlas-und-grundriss-der-bakteriologie-und-lehrbuch-der-speziellen-bakteriologischen-diagnostik/oclc/715501858
Lehmann, K. B., and Neumann, R. O. (1899). Atlas und Grundriss der Bakteriologie und Lehrbuch der speziellen bakteriologischen Diagnostik. München: J. F. Lehmann. Available online at: http://www.worldcat.org/title/atlas-und-grundriss-der-bakteriologie-und-lehrbuch-der-speziellen-bakteriologischen-diagnostik/oclc/715501858
Lévy-Frébault, V. V., Rafidinarivo, A., Prome, J. C., Grandry, J., Boisvert, H., and David, H. L. (1983). Mycobacterium fallax sp. nov. Int. J. Syst. Evol. Microbiol. 33, 336–343. doi: 10.1099/00207713-33-2-336
Li, W., and Godzik, A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659. doi: 10.1093/bioinformatics/btl158
Lory, S. (2014). “The Family Mycobacteriaceae,” in The Prokaryotes- Actinobacteria, eds E. Rosenberg, E. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (New York, NY: Springer), 571–575.
Ludwig, W., and Klenk, H.-P. (2005). “Overview: A phylogenetic backbone and taxonomic framework for prokaryotic systamatics,” in Bergey's Manual of Systematic Bacteriology, eds D. J.Brenner, N. R. Krieg, J. T. Staley, and G. M. Garrity (Berlin: Springer-Verlag), 49–65.
Luquin, M., Ausina, V., Vincent-Lévy-Frébault, V., Lanéelle, M. A., Belda, F., García-Barceló, M., et al. (1993). Mycobacterium brumae sp. nov., a rapidly growing, nonphotochromogenic Mycobacterium. Int. J. Syst. Evol. Microbiol. 43, 405–413. doi: 10.1099/00207713-43-3-405
Magee, G. M., and Ward, A. C. (2012). “Genus I. Mycobacterium Lehmann and Neumann 1896, 363AL,” in Bergey's Manual of Systematic Bacteriology, Vol. 5, Actinobacteria, eds M. Goodfellow, P. Kampfer, H. J. Busse, M. E. Trujillo, K. Suzuki, W. Ludwig, and W. Whitman (New York, NY: Springer), 312–375.
Masaki, T., Ohkusu, K., Hata, H., Fujiwara, N., Iihara, H., Yamada-Noda, M., et al. (2006). Mycobacterium kumamotonense sp. nov. recovered from clinical specimen and the first isolation report of Mycobacterium arupense in Japan: novel slowly growing, nonchromogenic clinical isolates related to Mycobacterium terrae complex. Microbiol. Immunol. 50, 889–897. doi: 10.1111/j.1348-0421.2006.tb03865.x
Masaki, T., Ohkusu, K., Hata, H., Fujiwara, N., Iihara, H., Yamada-Noda, M., et al. (2007). Mycobacterium kumamotonense sp. nov. in: list of new names and new combinations previously effectively, but not validly, published, list no. 114. Int. J. Syst. Evol. Microbiol. 57, 433–434. doi: 10.1099/ijs.0.65052-0
Medjahed, H., Gaillard, J. L., and Reyrat, J. M. (2010). Mycobacterium abscessus: a new player in the mycobacterial field. Trends Microbiol. 18, 117–123. doi: 10.1016/j.tim.2009.12.007
Mignard, S., and Flandrois, J. P. (2008). A seven-gene, multilocus, genus-wide approach to the phylogeny of mycobacteria using supertrees. Int. J. Syst. Evol. Microbiol. 58, 1432–1441. doi: 10.1099/ijs.0.65658-0
Milne, B. W., Arnold, M. H., Hudson, B., and Coolican, M. R. (2009). Infectious arthritis of the knee caused by Mycobacterium terrae: a case report. J. Orthop. Surg. (Hong. Kong.) 17, 103–108. doi: 10.1177/230949900901700123
Moore, M., and Frerichs, J. B. (1953). An unusual acid-fast infection of the knee with subcutaneous, abscess-like lesions of the gluteal region; report of a case with a study of the organism, Mycobacterium abscessus, n. sp. J. Invest. Dermatol. 20, 133–169. doi: 10.1038/jid.1953.18
Mukhopadhyay, S., and Balaji, K. N. (2011). The PE and PPE proteins of Mycobacterium tuberculosis. Tuberculosis. (Edinb.) 91, 441–447. doi: 10.1016/j.tube.2011.04.004
Mun, H. S., Park, J. H., Kim, H., Yu, H. K., Park, Y. G., Cha, C. Y., et al. (2008). Mycobacterium senuense sp. nov., a slowly growing, non-chromogenic species closely related to the Mycobacterium terrae complex. Int. J. Syst. Evol. Microbiol. 58, 641–646. doi: 10.1099/ijs.0.65374-0
Naushad, H. S., Lee, B., and Gupta, R. S. (2014). Conserved signature indels and signature proteins as novel tools for understanding microbial phylogeny and systematics: identification of molecular signatures that are specific for the phytopathogenic genera Dickeya, Pectobacterium and Brenneria. Int. J. Syst. Evol. Microbiol. 64, 366–383. doi: 10.1099/ijs.0.054213-0
Nessar, R., Cambau, E., Reyrat, J. M., Murray, A., and Gicquel, B. (2012). Mycobacterium abscessus: a new antibiotic nightmare. J. Antimicrob. Chemother. 67, 810–818. doi: 10.1093/jac/dkr578
Ngeow, Y. F., Wong, Y. L., Tan, J. L., Hong, K. W., Ng, H. F., Ong, B. L., et al. (2015). Identification of new genomospecies in the Mycobacterium terrae complex. PLoS ONE 10:e0120789. doi: 10.1371/journal.pone.0120789
Nogueira, C. L., Simmon, K. E., Chimara, E., Cnockaert, M., Palomino, J. C., et al. (2015a). Mycobacterium franklinii sp. nov., a species closely related to the members of the Mycobacterium chelonae-Mycobacterium abscessus group. Int. J. Syst. Evol. Microbiol. 65, 2148–2153. doi: 10.1099/ijs.0.000234
Nogueira, C. L., Whipps, C. M., Matsumoto, C. K., Chimara, E., Droz, S., Tortoli, E., et al. (2015b). Mycobacterium saopaulense sp. nov., a rapidly growing Mycobacterium closely related to members of the Mycobacterium chelonae–Mycobacterium abscessus group. Int. J. Syst. Evol. Microbiol. 65, 4403–4409. doi: 10.1099/ijsem.0.000590
Padgitt, P. J., and Moshier, S. E. (1987). Mycobacterium poriferae sp. nov., a scotochromogenic, rapidly growing species isolated from a marine sponge. Int. J. Syst. Evol. Microbiol. 37, 186–191. doi: 10.1099/00207713-37-3-186
Parte, A. C. (2014). LPSN-list of prokaryotic names with standing in nomenclature. Nucleic Acids Res. 42, D613–D616. doi: 10.1093/nar/gkt1111
Pitulle, C., Dorsch, M., Kazda, J., Wolters, J., and Stackebrandt, E. (1992). Phylogeny of rapidly growing members of the genus Mycobacterium. Int. J. Syst. Bacteriol. 42, 337–343. doi: 10.1099/00207713-42-3-337
Prasanna, A. N., and Mehra, S. (2013). Comparative phylogenomics of pathogenic and non-pathogenic Mycobacterium. PLoS ONE 8:e71248. doi: 10.1371/journal.pone.0071248
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Qin, Q. L., Xie, B. B., Zhang, X. Y., Chen, X. L., Zhou, B. C., Zhou, J., et al. (2014). A proposed genus boundary for the prokaryotes based on genomic insights. J. Bacteriol. 196, 2210–2215. doi: 10.1128/JB.01688-14
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramaprasad, E. V. V., Rizvi, A., Banerjee, S., Sasikala, C., and Ramana, C. (2016). Mycobacterium oryzae sp. nov., a scotochromogenic, rapidly growing species is able to infect human macrophage cell line. Int. J. Syst. Evol. Microbiol. 66, 4530–4536. doi: 10.1099/ijsem.0.001386
Reischl, U., Melzl, H., Kroppenstedt, R. M., Miethke, T., Naumann, L., Mariottini, A., et al. (2006). Mycobacterium monacense sp. nov. Int. J. Syst. Evol. Microbiol. 56, 2575–2578. doi: 10.1099/ijs.0.64527-0
Richter, E., Niemann, S., Gloeckner, F. O., Pfyffer, G. E., and Rusch-Gerdes, S. (2002). Mycobacterium holsaticum sp. nov. Int. J. Syst. Evol. Microbiol. 52, 1991–1996. doi: 10.1099/00207713-52-6-1991
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Ripoll, F., Pasek, S., Schenowitz, C., Dossat, C., Barbe, V., Rottman, M., et al. (2009). Non mycobacterial virulence genes in the genome of the emerging pathogen Mycobacterium abscessus. PLoS ONE 4:e5660. doi: 10.1371/journal.pone.0005660
Rogall, T., Wolters, J., Flohr, T., and Bottger, E. C. (1990). Towards a phylogeny and definition of species at the molecular level within the genus Mycobacterium. Int. J. Syst. Bacteriol. 40, 323–330. doi: 10.1099/00207713-40-4-323
Rokas, A., and Holland, P. W. (2000). Rare genomic changes as a tool for phylogenetics. Trends Ecol. Evol. 15, 454–459. doi: 10.1016/S0169-5347(00)01967-4
Roth, A., Fischer, M., Hamid, M. E., Michalke, S., Ludwig, W., and Mauch, H. (1998). Differentiation of phylogenetically related slowly growing mycobacteria based on 16S-23S rRNA gene internal transcribed spacer sequences. J. Clin. Microbiol. 36, 139–147.
Runyon, E. H. (1965). Typical mycobacteria: their classification. Am. Rev. Respir. Dis. 91, 288–289.
Sahraoui, N., Ballif, M., Zelleg, S., Yousfi, N., Ritter, C., Friedel, U., et al. (2011). Mycobacterium algericum sp. nov., a novel rapidly growing species related to the Mycobacterium terrae complex and associated with goat lung lesions. Int. J. Syst. Evol. Microbiol. 61, 1870–1874. doi: 10.1099/ijs.0.024851-0
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front. Genet. 5:429. doi: 10.3389/fgene.2014.00429
Schinsky, M. F., McNeil, M. M., Whitney, A. M., Steigerwalt, A. G., Lasker, B. A., Floyd, M. M., et al. (2000). Mycobacterium septicum sp. nov., a new rapidly growing species associated with catheter-related bacteraemia. Int. J. Syst. Evol. Microbiol. 50, 575–581. doi: 10.1099/00207713-50-2-575
Schinsky, M. F., Morey, R. E., Steigerwalt, A. G., Douglas, M. P., Wilson, R. W., Floyd, M. M., et al. (2004). Taxonomic variation in the Mycobacterium fortuitum third biovariant complex: description of Mycobacterium boenickei sp. nov., Mycobacterium houstonense sp. nov., Mycobacterium neworleansense sp. nov. and Mycobacterium brisbanense sp. nov. and recognition of Mycobacterium porcinum from human clinical isolates. Int. J. Syst. Evol. Microbiol. 54, 1653–1667. doi: 10.1099/ijs.0.02743-0
Schoeffler, A. J., May, A. P., and Berger, J. M. (2010). A domain insertion in Escherichia coli GyrB adopts a novel fold that plays a critical role in gyrase function. Nucleic Acids Res. 38, 7830–7844. doi: 10.1093/nar/gkq665
Schröder, K. H., Naumann, L., Kroppenstedt, R. M., and Reischl, U. (1997). Mycobacterium hassiacum sp. nov., a new rapidly growing thermophilic Mycobacterium. Int. J. Syst. Bacteriol. 47, 86–91. doi: 10.1099/00207713-47-1-86
Segata, N., Bornigen, D., Morgan, X. C., and Huttenhower, C. (2013). PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 4:2304. doi: 10.1038/ncomms3304
Shahraki, A. H., Cavusoglu, C., Borroni, E., Heidarieh, P., Koksalan, O. K., Cabibbe, A. M., et al. (2015). Mycobacterium celeriflavum sp. nov., a rapidly growing scotochromogenic bacterium isolated from clinical specimens. Int. J. Syst. Evol. Microbiol. 65, 510–515. doi: 10.1099/ijs.0.064832-0
Shojaei, H., Daley, C., Gitti, Z., Hashemi, A., Heidarieh, P., Moore, E. R., et al. (2013). Mycobacterium iranicum sp. nov., a rapidly growing scotochromogenic species isolated from clinical specimens on three different continents. Int. J. Syst. Evol. Microbiol. 63, 1383–1389. doi: 10.1099/ijs.0.043562-0
Shojaei, H., Goodfellow, M., Magee, J. G., Freeman, R., Gould, F. K., and Brignall, C. G. (1997). Mycobacterium novocastrense sp. nov., a rapidly growing photochromogenic Mycobacterium. Int. J. Syst. Bacteriol. 47, 1205–1207. doi: 10.1099/00207713-47-4-1205
Shojaei, H., Magee, J. G., Freeman, R., Yates, M., Horadagoda, N. U., and Goodfellow, M. (2000). Mycobacterium elephantis sp. nov., a rapidly growing non-chromogenic Mycobacterium isolated from an elephant. Int. J. Syst. Evol. Microbiol. 50, 1817–1820. doi: 10.1099/00207713-50-5-1817
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Simmon, K. E., Brown-Elliott, B. A., Ridge, P. G., Durtschi, J. D., Mann, L. B., Slechta, E. S., et al. (2011). Mycobacterium chelonae-abscessus complex associated with sinopulmonary disease, Northeastern USA. Emerg. Infect. Dis. 17, 1692–1700. doi: 10.3201/eid1709.101667
Singh, B., and Gupta, R. S. (2009). Conserved inserts in the Hsp60 (GroEL) and Hsp70 (DnaK) proteins are essential for cellular growth. Mol. Genet. Genomics 281, 361–373. doi: 10.1007/s00438-008-0417-3
Skerman, V. B. D., McGowan, V., and Sneath, P. H. A. (1980). Approved lists of bacteria names. Int. J. Syst. Evol. Microbiol. 30, 225–420. doi: 10.1099/00207713-30-1-225
Smith, D. S., Lindholm-Levy, P., Huitt, G. A., Heifets, L. B., and Cook, J. L. (2000). Mycobacterium terrae: case reports, literature review, and in vitro antibiotic susceptibility testing. Clin. Infect. Dis. 30, 444–453. doi: 10.1086/313693
Springer, B., Bottger, E. C., Kirschner, P., and Wallace, R. J. Jr. (1995). Phylogeny of the Mycobacterium chelonae-like organism based on partial sequencing of the 16S rRNA gene and proposal of Mycobacterium mucogenicum sp. nov. Int. J. Syst. Bacteriol. 45, 262–267. doi: 10.1099/00207713-45-2-262
Stackebrandt, E. (1992). “Unifying phylogeny and phenotypic diversity,” in The Prokaryotes, eds A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H.Schleifer (New York, NY: Springer-Verlag), 19–47.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Stahl, D. A., and Urbance, J. W. (1990). The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J. Bacteriol. 172, 116–124. doi: 10.1128/jb.172.1.116-124.1990
Stanford, J. L., and Gunthorpe, W. J. (1971). A study of some fast-growing scotochromogenic mycobacteria including species descriptions of Mycobacterium gilvum (new species) and Mycobacterium duvalii (new species). Br. J. Exp. Pathol. 52, 627–637.
Talavera, G., and Castresana, J. (2007). Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 56, 564–577. doi: 10.1080/10635150701472164
Tasler, G. R. W., and Hartley, W. J. (1981). Foal abortion associated with Mycobacterium terrae infection. Vet. Pathol. 18, 122–125. doi: 10.1177/030098588101800115
Thompson, C. C., Chimetto, L., Edwards, R. A., Swings, J., Stackebrandt, E., and Thompson, F. L. (2013). Microbial genomic taxonomy. BMC genomics 14:913. doi: 10.1186/1471-2164-14-913
Tortoli, E. (2012). Phylogeny of the genus Mycobacterium: many doubts, few certainties. Infect. Genet. Evol. 12, 827–831. doi: 10.1016/j.meegid.2011.05.025
Tortoli, E. (2014). Microbiological features and clinical relevance of new species of the genus Mycobacterium. Clin. Microbiol. Rev. 27, 727–752. doi: 10.1128/CMR.00035-14
Tortoli, E., Baruzzo, S., Heijdra, Y., Klenk, H. P., Lauria, S., Mariottini, A., et al. (2009). Mycobacterium insubricum sp. nov. Int. J. Syst. Evol. Microbiol. 59, 1518–1523. doi: 10.1099/ijs.0.003459-0
Tortoli, E., Fedrizzi, T., Pecorari, M., Giacobazzi, E., De Sanctis, V., Bertorelli, R., et al. (2015). The new phylogenesis of the genus Mycobacterium. Int. J. Mycobacteriol. 4, 77. doi: 10.1016/j.ijmyco.2014.10.017
Tortoli, E., Gitti, Z., Klenk, H. P., Lauria, S., Mannino, R., Mantegani, P., et al. (2013). Survey of 150 strains belonging to the Mycobacterium terrae complex and description of Mycobacterium engbaekii sp. nov., Mycobacterium heraklionense sp. nov. and Mycobacterium longobardum sp. nov. Int. J. Syst. Evol. Microbiol. 63, 401–411. doi: 10.1099/ijs.0.038737-0
Tortoli, E., Kohl, T. A., Brown-Elliott, B. A., Trovato, A., Leao, S. C., Garcia, M. J., et al. (2016). Emended description of Mycobacterium abscessus, Mycobacterium abscessus subsp. abscessus and Mycobacterium abscessus subsp. bolletii and designation of Mycobacterium abscessus subsp. massiliense comb. nov. Int. J. Syst. Evol. Microbiol. 66, 4471–4479. doi: 10.1099/ijsem.0.001376
Tortoli, E., Kroppenstedt, R. M., Bartoloni, A., Caroli, G., Jan, I., Pawlowski, J., et al. (1999). Mycobacterium tusciae sp. nov. Int. J. Syst. Bacteriol. 49, 1839–1844. doi: 10.1099/00207713-49-4-1839
Tortoli, E., Piersimoni, C., Kroppenstedt, R. M., Montoya-Burgos, J. I., Reischl, U., Giacometti, A., et al. (2001). Mycobacterium doricum sp. nov. Int. J. Syst. Evol. Microbiol. 51, 2007–2012. doi: 10.1099/00207713-51-6-2007
Tran, P. M., and Dahl, J. L. (2016). Mycobacterium sarraceniae sp. nov. and Mycobacterium helvum sp. nov., isolated from the pitcher plant Sarracenia purpurea. Int. J. Syst. Evol. Microbiol. 66, 4480–4485. doi: 10.1099/ijsem.0.001377
Trujillo, M. E., Velazquez, E., Kroppenstedt, R. M., Schumann, P., Rivas, R., Mateos, P. F., et al. (2004). Mycobacterium psychrotolerans sp. nov., isolated from pond water near a uranium mine. Int. J. Syst. Evol. Microbiol. 54, 1459–1463. doi: 10.1099/ijs.0.02938-0
Tsukamura, M. (1965a). A group of mycobacteria from soil resources resembling nonphotochromogens (Group 3). Med. Biol. 71, 110–113.
Tsukamura, M. (1965b). Mycobacterium parafortuitum: a new species. J. Gen. Microbiol. 42, 7–12. doi: 10.1099/00221287-42-1-7
Tsukamura, M. (1966). Adansonian classification of mycobacteria. J. Gen. Microbiol. 45, 253–273. doi: 10.1099/00221287-45-2-253
Tsukamura, M. (1967a). Identification of mycobacteria. Tubercle 48, 311–338. doi: 10.1016/S0041-3879(67)80040-0
Tsukamura, M. (1967b). Mycobacterium chitae: a new species. Microbiol. Immunol. 11, 43–47. doi: 10.1111/j.1348-0421.1967.tb00319.x
Tsukamura, M. (1981). Numerical analysis of rapidly growing, nonphotochromogenic mycobacteria, including Mycobacterium agri (Tsukamura 1972) Tsukamura sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 31, 247–258. doi: 10.1099/00207713-31-3-247
Tsukamura, M., Mizuno, S., and Toyama, H. (1983a). Mycobacterium pulveris sp. nov., a nonphotochromogenic Mycobacterium with an intermediate growth rate. Int. J. Syst. Evol. Microbiol. 33, 811–815. doi: 10.1099/00207713-33-4-811
Tsukamura, M., Mizuno, S., and Tsukamura, S. (1981). Numerical analysis of rapidly growing, scotochromogenic mycobacteria, including Mycobacterium obuense sp. nov., nom. rev., Mycobacterium rhodesiae sp. nov., nom. rev., Mycobacterium aichiense sp. nov., nom. rev., Mycobacterium chubuense sp. nov., nom. rev., and Mycobacterium tokaiense sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 31, 263–275. doi: 10.1099/00207713-31-3-263
Tsukamura, M., Nemoto, H., and Yugi, H. (1983b). Mycobacterium porcinum sp. nov., a porcine pathogen. Int. J. Syst. Evol. Microbiol. 33, 162–165. doi: 10.1099/00207713-33-2-162
Tsukamura, M., Van Der Meulen, H. J., and Grabow, W. O. K. (1983c). Numerical taxonomy of rapidly growing, scotochromogenic mycobacteria of the Mycobacterium parafortuitum complex: Mycobacterium austroafricanum sp. nov. and Mycobacterium diernhoferi sp. nov., nom. rev. Int. J. Syst. Bacteriol. 33, 460–469. doi: 10.1099/00207713-33-3-460
Tsukamura, M., Yano, I., and Imaeda, T. (1986a). “Mycobacterium fortuitum subsp. acetamidolyticum sp. Nov,” in Validation of the Publication of New Names and New Combinations Previously Effectively Published Outside the IJSB, List no. 21. Int. J. Syst. Evol. Microbiol. 36, 489.
Tsukamura, M., Yano, I., and Imaeda, T. (1986b). Mycobacterium fortuitum subspecies acetamidolyticum, a new subspecies of Mycobacterium fortuitum. Microbiol. Immunol. 30, 97–110. doi: 10.1111/j.1348-0421.1986.tb00925.x
Tsukamura, M., Yano, I., and Imaeda, T. (1986c). Mycobacterium moriokaense sp. nov., a rapidly growing, nonphotochromogenic Mycobacterium. Int. J. Syst. Evol. Microbiol. 36, 333–338. doi: 10.1099/00207713-36-2-333
van Ingen, J., Boeree, M. J., Kosters, K., Wieland, A., Tortoli, E., Dekhuijzen, P. N., et al. (2009). Proposal to elevate Mycobacterium avium complex ITS sequevar MAC-Q to Mycobacterium vulneris sp. nov. Int. J. Syst. Evol. Microbiol. 59, 2277–2282. doi: 10.1099/ijs.0.008854-0
Vasireddy, R., Vasireddy, S., Brown-Elliott, B. A., Wengenack, N. L., Eke, U. A., Benwill, J. L., et al. (2016). Mycobacterium arupense, Mycobacterium heraklionense, and a newly proposed species, “Mycobacterium virginiense” sp. nov., but not Mycobacterium nonchromogenicum, as species of the Mycobacterium terrae complex causing tenosynovitis and osteomyelitis. J. Clin. Microbiol. 54, 1340–1351. doi: 10.1128/JCM.00198-16
Vasireddy, R., Vasireddy, S., Brown-Elliott, B. A., Wengenack, N. L., Eke, U. A., Benwill, J. L., et al. (2017). Mycobacterium virginiense sp. nov. In: Oren A., Garrity G.M. (editors). list of new names and new combinations previously effectively, but not validly, published, list no. 175. Int. J. Syst. Evol. Microbiol. 67, 1–3. doi: 10.1099/ijsem.0.001733
Vuorio, R., Andersson, M. A., Rainey, F. A., Kroppenstedt, R. M., Kampfer, P., Busse, H. J., et al. (1999). A new rapidly growing mycobacterial species, Mycobacterium murale sp. nov., isolated from the indoor walls of a children's day care centre. Int. J. Syst. Bacteriol. 49, 25–35. doi: 10.1099/00207713-49-1-25
Wang, J., McIntosh, F., Radomski, N., Dewar, K., Simeone, R., Enninga, J., et al. (2015). Insights on the emergence of Mycobacterium tuberculosis from the analysis of Mycobacterium kansasii. Genome Biol. Evol. 7, 856–870. doi: 10.1093/gbe/evv035
Wang, Y. N., Chi, C. Q., Cai, M., Lou, Z. Y., Tang, Y. Q., Zhi, X. Y., et al. (2010). Amycolicicoccus subflavus gen. nov., sp. nov., an actinomycete isolated from a saline soil contaminated by crude oil. Int. J. Syst. Evol. Microbiol. 60, 638–643. doi: 10.1099/ijs.0.010546-0
Wang, Z., and Wu, M. (2013). A phylum-level bacterial phylogenetic marker database. Mol. Biol. Evol. 30, 1258–1262. doi: 10.1093/molbev/mst059
Wayne, L. G. (1966). Classification and identification of mycobacteria. 3. Species within group 3. Am. Rev. Respir. Dis. 93, 919–928.
Wayne, L. G., and Kubica, G. P. (1986). “Genus Mycobacterium,” in Bergey's Manual of Systematic Bacteriology, eds P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (Baltimore, MD: Williams and Wilkins), 1436–1457.
Wee, W. Y., Dutta, A., and Choo, S. W. (2017). Comparative genome analyses of mycobacteria give better insights into their evolution. PLoS ONE 12:e0172831. doi: 10.1371/journal.pone.0172831
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Whipps, C. M., Butler, W. R., Pourahmad, F., Watral, V. G., and Kent, M. L. (2007). Molecular systematics support the revival of Mycobacterium salmoniphilum (ex Ross 1960) sp. nov., nom. rev., a species closely related to Mycobacterium chelonae. Int. J. Syst. Evol. Microbiol. 57, 2525–2531. doi: 10.1099/ijs.0.64841-0
Willumsen, P., Karlson, U., Stackebrandt, E., and Kroppenstedt, R. M. (2001). Mycobacterium frederiksbergense sp. nov., a novel polycyclic aromatic hydrocarbon-degrading Mycobacterium species. Int. J. Syst. Evol. Microbiol. 51, 1715–1722. doi: 10.1099/00207713-51-5-1715
Wilson, R. W., Steingrube, V. A., Bottger, E. C., Springer, B., Brown-Elliott, B. A., Vincent, V., et al. (2001). Mycobacterium immunogenum sp. nov., a novel species related to Mycobacterium abscessus and associated with clinical disease, pseudo-outbreaks and contaminated metalworking fluids: an international cooperative study on mycobacterial taxonomy. Int. J. Syst. Evol. Microbiol. 51, 1751–1764. doi: 10.1099/00207713-51-5-1751
Wong, S. Y., Paschos, A., Gupta, R. S., and Schellhorn, H. E. (2014). Insertion/deletion-based approach for the detection of Escherichia coli O157:H7 in freshwater environments. Environ. Sci. Technol. 48, 11462–11470. doi: 10.1021/es502794h
Wu, D., Hugenholtz, P., Mavromatis, K., Pukall, R., Dalin, E., Ivanova, N. N., et al. (2009). A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature 462, 1056–1060. doi: 10.1038/nature08656
Yarza, P., Richter, M., Peplies, J., Euzeby, J., Amann, R., Schleifer, K. H., et al. (2008). The All-Species Living Tree project: a 16S rRNA-based phylogenetic tree of all sequenced type strains. Syst. Appl. Microbiol. 31, 241–250. doi: 10.1016/j.syapm.2008.07.001
Yarza, P., Yilmaz, P., Pruesse, E., Gloeckner, F. O., Ludwig, W., Schleifer, K. H., et al. (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences 1. Nat. Rev. Microbiol. 12, 635–645. doi: 10.1038/nrmicro3330
Zhang, D. F., Chen, X., Zhang, X. M., Zhi, X. Y., Yao, J. C., Jiang, Y., et al. (2013a). Mycobacterium sediminis sp. nov. and Mycobacterium arabiense sp. nov., two rapidly growing members of the genus Mycobacterium. Int. J. Syst. Evol. Microbiol. 63, 4081–4086. doi: 10.1099/ijs.0.050567-0
Zhang, Y., Zhang, J., Fang, C., Pang, H., and Fan, J. (2012). Mycobacterium litorale sp. nov., a rapidly growing Mycobacterium from soil. Int. J. Syst. Evol. Microbiol. 62, 1204–1207. doi: 10.1099/ijs.0.033449-0
Zhang, Z. Y., Sun, Z. Q., Wang, Z. L., Hu, H. R., Wen, Z. L., Song, Y. Z., et al. (2013b). Identification and pathogenicity analysis of a novel non-tuberculous Mycobacterium clinical isolate with nine-antibiotic resistance. Clin. Microbiol. Infect. 19, 91–96. doi: 10.1111/j.1469-0691.2012.03818.x
Zhang, Z. Y., Sun, Z. Q., Wang, Z. L., Wen, Z. L., Sun, Q. W., Zhu, Z. Q., et al. (2011). Complete genome sequence of a novel clinical isolate, the nontuberculous Mycobacterium strain JDM601. J. Bacteriol. 193, 4300–4301. doi: 10.1128/JB.05291-11
Zopf, W. (1883). Die Spaltpilze: Nachdem neuesten Standpunkte. Breslau: Eduard Trewendt. Available online at: http://www.worldcat.org/title/spaltpilze-nachdem-neuesten-standpunkte/oclc/7445289&referer=brief_results
Keywords: Mycobacterium classification, slow-growing and fast-growing mycobacteria, conserved signature indels and signature proteins, phylogenomic analysis, fortuitum-vaccae clade, abscessus-chelonae clade, terrae clade, triviale clade
Citation: Gupta RS, Lo B and Son J (2018) Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera. Front. Microbiol. 9:67. doi: 10.3389/fmicb.2018.00067
Received: 16 September 2017; Accepted: 11 January 2018;
Published: 13 February 2018.
Edited by:
Antonio Ventosa, Universidad de Sevilla, SpainReviewed by:
Stephanus Nicolaas Venter, University of Pretoria, South AfricaAlice Rebecca Wattam, Virginia Tech, United States
Copyright © 2018 Gupta, Lo and Son. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radhey S. Gupta, Z3VwdGFAbWNtYXN0ZXIuY2E=
 Radhey S. Gupta
Radhey S. Gupta Brian Lo
Brian Lo Jeen Son
Jeen Son