- 1ithree institute, University of Technology Sydney, Sydney, NSW, Australia
- 2Australian Centre for Genomic Epidemiological Microbiology, University of Technology Sydney, Sydney, NSW, Australia
- 3NSW Department of Primary Industries, Elizabeth MacArthur Agricultural Institute, Menangle, NSW, Australia
aEPEC are associated with persistent diarrhea, and diarrheal outbreaks in both humans and animals worldwide. They are differentiated from typical EPEC by the lack of bundle-forming pili, and from EHEC by the lack of phage-mediated stx toxins. However, phylogenetic analyses often associate aEPEC with EHEC, promoting the hypothesis that aEPEC are the progenitors of EHEC, which is supported by aEPEC conversion to EHEC by stx-carrying phages. While aEPEC can cause disease outright, the potential to acquire stx, one of the most potent bacterial toxins known, merits close monitoring. Escherichia coli ST302 (O108:H9, O182:H9, O45:H9) are aEPEC that have been isolated from diarrheic human, pig and rabbit hosts, as well as in healthy pigs, however, no study to date has focused on E. coli ST302 strains. Through WGS and hybrid assembly we present the first closed chromosome, and two circularized plasmids of an ST302 strain - F2_18C, isolated from a healthy pig in Australia. A phylogenetic analysis placed E. coli ST302 strains in proximity to EHEC ST32 (O145:H28) strains. Public databases were interrogated for WGSs of E. coli ST302 strains and short-read gene screens were used to compare their virulence-associated gene (VAG) and antimicrobial resistance gene (ARG) cargo. E. coli ST302 strains carry diverse VAGs, including those that typically associated with extraintestinal pathogenic E. coli (ExPEC). Plasmid comparisons showed that pF2_18C_FIB shared homology with EHEC virulence plasmids such as pO103 while pF2_18C_HI2 is a large multidrug resistance IncHI2:ST3 plasmid. A comparison of 33 HI2:ST3 plasmids demonstrated that those of Australian origin have not acquired resistances to extended-spectrum beta-lactams, colistin, fosfomycin or rifampicin, unlike those originating from Asia. F2_18C was shown to carry two additional pathogenicity islands – ETT2, and the STEC-associated PAICL3, plasmid-associated heavy metal resistance genes, as well as several unoccupied stx-phage attachment sites. This study sheds light on the virulence and AMR potential of E. coli ST302 strains and informs AMR genomic surveillance.
Introduction
Escherichia coli is a versatile Gram-negative bacterium whose genome is shaped by lateral gene transfer. The plasticity of E. coli genomes allows for some strains to exist as gastrointestinal tract commensals, while the acquisition of different combinations of virulence-associated genes (VAGs) has generated clades that cause a diverse range of intestinal and extraintestinal illnesses (Tenaillon et al., 2010). Pathogenic E. coli that induce intestinal disease are known as diarrheagenic E. coli (DEC) and are classified into pathotypes according to specific virulence factors and ensuing clinical manifestations (Gomes et al., 2016).
Enterohemorrhagic E. coli (EHEC) and enteropathogenic E. coli (EPEC) are DEC pathotypes that share a common mechanism of pathogenesis defined in part by the carriage of a chromosomally located pathogenicity island (PAI) termed the locus of enterocyte effacement (LEE). The LEE enables EPEC and EHEC to adhere intimately to intestinal epithelial cells and cause attaching and effacing (A/E) lesions leading to diarrheal illness (Kaper et al., 2004). The genes located within the LEE are sufficient for A/E lesion formation, as transferring LEE to commensal E. coli confers A/E lesion activity (McDaniel and Kaper, 1997). In addition to the LEE, EHEC and EPEC possess various non-LEE (nle)-encoded effector genes. nle genes are frequently carried on prophage elements and contribute to virulence by interfering with host signaling pathways, apoptosis and phagocytosis (Wong et al., 2011) as well as disrupting host cell cytoskeleton and tight junctions (Gomes et al., 2016). However, EHEC differ from EPEC in that they possess phage-associated Shiga toxins (stx), which contribute to the hemorrhagic colitis and hemolytic uremic syndrome (HUS) associated with EHEC infections (Mayer et al., 2012). EHEC also fall under the category of Shiga toxin-producing E. coli (STEC); however, STEC encompass both LEE-positive (typical EHEC) and LEE-negative (atypical EHEC) E. coli.
EPEC are further classified into typical EPEC (tEPEC) and atypical EPEC (aEPEC) by the presence, in the former, of the EPEC adherence factor plasmid (pEAF) encoding bundle-forming pili (Gomes et al., 2016). tEPEC and aEPEC also differ in terms of serogroups, reservoirs and virulence factors. tEPEC generally belong to the twelve classical EPEC serogroups recognized by the World Health Organization (O26, O55, O111, O114, O119, O126, O127, O128, O142, O158) (World Health Organization [WHO], 1987). Humans are their principal reservoir (Trabulsi et al., 2002), and their virulence factors are mainly encoded by LEE and pEAF (Abreu et al., 2013). Conversely, aEPEC strains belong to more than 4200 different serotypes (Gomes et al., 2016), and include tEPEC and EHEC that have lost pEAF and stx genes, respectively (Afset et al., 2008; Bielaszewska et al., 2008). aEPEC infect both human and animal hosts (Hu and Torres, 2015) and are also more heterogeneous than tEPEC in their virulence factors; frequently carrying genes associated with other DEC pathotypes, including enterohemolysin (ehxA) and heat-stable enterotoxin (astA) (Bando et al., 2009).
aEPEC strains have been associated with persistent diarrhea in both humans (Hu and Torres, 2015) and animals (Watson et al., 2017; Arais et al., 2018) and in diarrheal outbreaks in Brazil, China, Finland, Japan, and the United States (Hu and Torres, 2015; Gomes et al., 2016). While recent epidemiological studies have indicated that aEPEC is more prevalent than tEPEC in both industrialized and developing countries (Hu and Torres, 2015), aEPEC pathogenicity remains controversial due to similar isolation rates from both diarrheic and non-diarrheic hosts (Gomes et al., 2016). Establishing a baseline for which virulence factors may or may not contribute to disease can help elucidate the mechanisms behind aEPEC pathogenicity.
Escherichia coli is not only an agent of disease, but also a driving force behind antimicrobial resistance (AMR). In fact, E. coli is one of the most significant global concerns in human and animal health sectors, the food industry and in the environment (Paitan, 2018). AMR surveillance programs have indicated that resistance to all the major classes of antibiotics now circulate among E. coli strains (Pitout, 2012), including extended-spectrum β-lactams (ESBL), carbapenems, and more recently, plasmid-mediated colistin resistance (mcr-1), particularly in food animals from China and Vietnam (Liu et al., 2016; Huang et al., 2017; Caruso, 2018). The ubiquitous nature of E. coli means it constitutes a shared reservoir for AMR across a One Health framework, and concerns have been raised about the possible transmission of AMR E. coli between animals and humans through direct contact or via the food chain (Poirel et al., 2018). As inter- and intraspecies horizontal gene transfer (HGT) and mobile genetic elements (MGE) are considered the prevailing mechanisms that drive AMR (Tzouvelekis et al., 2012; Partridge et al., 2018), close genomic surveillance of AMR cargo within E. coli populations is warranted.
aEPEC from sequence type (ST) 302 (serotype O108:H9) have been isolated from both healthy and diseased hosts, and the environment. They have been isolated from healthy pigs (Fröhlicher et al., 2008; Malik et al., 2017; Reid et al., 2017), water tanks in a poultry slaughterhouse (Alonso et al., 2014) and pork products made for human consumption (Lugsomya et al., 2018), and have been associated with diarrheic rabbits (Zhao X. et al., 2018), pigs (Kleta et al., 2014), and humans (Foster et al., 2015). Yet to date, no genomic comparisons of E. coli ST302 strains, nor a complete ST302 genome, has been published. Here we sequenced and annotated the complete genome of E. coli aEPEC strain F2_18C (ST302, O108:H9), isolated from a healthy pig not previously treated with antibiotics on an Australian farm. We performed comparative analyses with other E. coli ST302 strains, screening for virulence and AMR cargo, and compared F2_18C’s three plasmids – an HI2:ST3 plasmid with homology to colistin-resistant plasmids, an FIB plasmid with homology to the EHEC virulence plasmid, and a colRNAI plasmid carrying colicin-10 – to other plasmids with high sequence identity.
Materials and Methods
Ethics Statement
NSW Department of Primary Industries did not require the study to be reviewed or approved by an ethics committee as E. coli strain F2_18C was isolated from a veterinary sample provided by the farm veterinarian as part of routine diagnostics.
Isolation and Storage of E. coli Strain F2_18C
Strain F2_18C was isolated from a commercial-raised, healthy piglet (weaner), not previously treated with antibiotics, on an Australian farm as previously described (Reid et al., 2017). The strain was cultured in LB medium, frozen as a glycerol stock (500 μl M9 salts solution, 500 μl 50% v/v glycerol) and stored at −80°C. The strain was again cultured in LB medium before total cellular DNA isolation for sequencing.
DNA Extraction, Whole-Genome Sequencing and Assembly
For short-read sequencing, DNA was extracted using the ISOLATE II genomic DNA kit (Bioline) and stored at −20°C. Short-read sequencing was performed using an Illumina HiSeq 2500 v4 sequencer in rapid PE150 mode as previously described (Reid et al., 2017). For long-read sequencing, DNA was extracted from mid-log phase sub-cultures of strains grown overnight in LB broth using a gentle phenol-chloroform extraction protocol. Long-read sequencing was performed by the Ramaciotti Centre for Genomics using a Pacific Bioscience (PacBio) RSII sequencer with P6-C4 chemistry. Sequence read quality was assessed using FastQC version 0.111. A hybrid assembly, using both Illumina and PacBio reads, was produced using Unicycler (Wick et al., 2017).
Genome Annotation
Automated annotations were generated by RASTtk (Brettin et al., 2015) and managed using SnapGene v4.1.9. Manual annotations were added using publicly available databases and tools: insertion sequences (IS) were annotated manually using ISfinder (Siguier et al., 2006)2; genomic islands (GIs) were identified using IslandViewer 4 (Bertelli et al., 2017)3; functional assignments of ORFs were designated using the Comprehensive Genome Analysis service available through Pathosystems Resource Integration Center (PATRIC v3.5.41) (Wattam et al., 2017)4 and; phage elements were identified using Phage Search Tool Enhanced Release (PHASTER) (Arndt et al., 2016)5. Sequences for the F2_18C chromosome, pF2_18C_HI2, pF2_18C_FIB, pF2_18C_Col have been deposited to NCBI under the accession numbers CP043542, CP043545, CP043544, and CP043543, respectively, and annotated using the NCBI automated pipeline. Authors’ annotations (GenBank files) can be viewed in Supplementary Data S1.
Phylogenetic Analysis
Maximum-likelihood phylogenetic distances between genomes were analyzed using Phylosift (Darling et al., 2014), generated using FastTree2 (Price et al., 2010), and visualized using both FigTree v1.4.26 and the Interactive Tree of Life (iTOL v4) (Letunic and Bork, 2019)7. SNP-based phylogenetic analysis was performed using Snplord, an automated pipeline that utilizes snippy8, Gubbins9, SNP-sites10, and FastTree 2. Snplord is available at https://github.com/CJREID/snplord. The ST302 pangenome was calculated by Roary v3.11.2 (Page et al., 2015).
Genotyping, Serotyping, and Phylogenetic Classification
F2_18Cs ST, serogroup and phylogroup were determined in silico using MLST 2.0 (Larsen et al., 2012)11, SerotypeFinder 2.0 (Joensen et al., 2015)12 and ClermontTyping (Clermont et al., 2013)13, respectively. To compare VAG, AMR gene and plasmid replicon carriage between F2_18C and other E. coli ST302 strains, short-reads of publicly available E. coli ST302 strains were first downloaded from Enterobase (Alikhan et al., 2018; downloaded 01/09/18)14 and then screened using ARIBA read-mapping tool (Hunt et al., 2017), using the following reference databases: ResFinder (Zankari et al., 2012)15, CARD (Jia et al., 2017)16, VirulenceFinder (Joensen et al., 2014)17, VFDB (Liu et al., 2019)18, and PlasmidFinder (Carattoli et al., 2014)19. ARIBA data was processed using a bespoke script accessible at https://github.com/maxlcummins/ARIBAlord.
Plasmid and Pathogenicity Island Comparisons
Plasmids with high sequence identity (>98%) were found through the PLSDB v2019_06_03 (Galata et al., 2019)20 and comparative analyses were achieved using BRIG (Alikhan et al., 2011)21 and Mauve (Darling, 2004)22. Mauve and Easyfig (Sullivan et al., 2011) were used to compare PAICL3 and ETT2 pathogenicity islands.
Phenotypic Resistance Testing
F2_18C phenotypic resistance was tested using the calibrated dichotomous susceptibility (CDS) test (Bell et al., 2016) against the following antibiotics: ampicillin (25 μg), azithromycin (15 μg), cefoxitin (30 μg), ciprofloxacin (2.5 μg), chloramphenicol (30 μg), gentamicin (10 μg), imipenem (10 μg), kanamycin (50 μg), nalidixic acid (30 μg), neomycin (30 μg), streptomycin (25 μg), sulphafurazole (300 μg), tetracycline (10 μg), and trimethoprim (5 μg).
Results
General Genomic Features of E. coli Strain F2_18C
Escherichia coli strain F2_18C has a circular genome of 5,439,436 bp consisting of 5436 CDS, 158 repeat regions, 91 tRNA and 22 rRNA. Virulence Factor Database (VFDB) identified 114 VAGs, and the Comprehensive Antibiotic Resistance Database (CARD) identified 85 antimicrobial resistance (AMR) genes. The National Database of Antibiotic Resistance Organisms (NDARO) database identified 15 AMR genes (Figure 1A). In terms of functional properties, strain F2_18C has 4753 genes encoding proteins with functional assignments and 683 hypothetical proteins. The three categories in which the most genes are attributed to are metabolism (n = 972), energy (n = 333), and protein processing (n = 265) (Figure 1B).
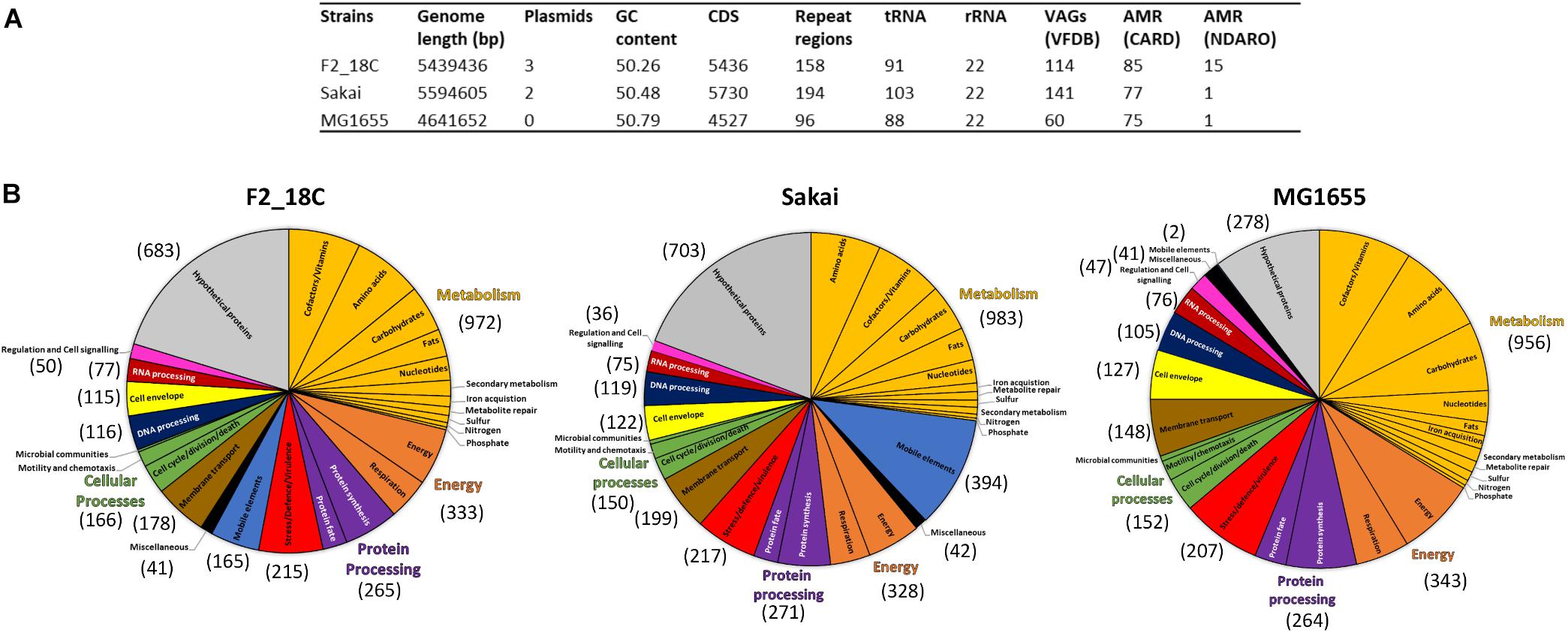
Figure 1. General features of strain F2_18C compared to E. coli strains Sakai and MG1655. (A) Table listing general genomic features. (B) Subsystem class gene distributions. Number in brackets represent number of genes assigned to each functional category.
When compared to a highly pathogenic E. coli strain (O157:H7 strain Sakai), and a non-pathogenic E. coli strain (laboratory strain MG1655), strain F2_18C had fewer VAGs than Sakai (n = 141) but more than MG1655 (n = 60). However, F2_18C had more AMR genes than both Sakai (77 CARD/1 NDARO) and MG1655 (75 CARD/1 NDARO). An interesting observation regarding differences in functional properties between the three strains was the number of genes associated with mobile elements (prophages, transposable elements and plasmids) – 165 for F2_18C, 394 for Sakai, and 2 for MG1655. A complete list of F2_18C gene functional assignments (GO assignments, subsystems, and PL/PGfam) and specialty genes (VAGs, transporters, drug targets, and AMR genes) can be viewed in Supplementary Table S1.
Phylogenetic Analysis
Porcine E. coli strain F2_18C was determined to be ST ST302, serotype O108:H9 and phylogroup E or clade I by Achtman 7 MLST, in silico serotype prediction and in silico Clermont typing, respectively. To determine the genetic relatedness of F2_18C to other E. coli ST302 strains and to other E. coli strains in general, all publicly available ST302 genomes (n = 14), and 67 E. coli genomes of varying STs, serotypes, and phylogroups were used to construct a phylogenetic tree (Figure 2; details of analyzed genomes found in Supplementary Table S2). Where known, the ST302 genomes analyzed in this study were from a range of hosts, including pigs, poultry, companion animals and humans, and from broad geographical locations, including Australia, Canada, Kenya, Slovakia, the United Kingdom, and the United States. Tree topology demonstrated consonance between STs, serotypes and phylogroups. The analysis showed that E. coli ST302 strains are most closely related to a branch of ST32s (serotype O145:H28, phylogroup D) and that they all belong to the E or cryptic clade 1 phylogroup and carry the H9 antigen. However, three ST302 serogroups were identified – O182, O45, and O108 – the latter being the most common (n = 11; 73%). Additionally, F2_18C and two O182:H9 strains (29396 and 29563) formed an offshoot branch from the other ST302 strains. An ST302 SNP analysis demonstrated that F2_18C was separated from its closest relative (29396) by 684 SNPs, and its most distant relative (FSIS11706110) by 2139 SNPs. SNP analysis details, including SNP sites and distribution, can be viewed in Supplementary Data S2. The ST302 pangenome comprised a total of 10262 genes, with 3558 genes forming the core genome (genes present in 100% of ST302 strains), and 6704 genes forming the accessory genome (shell genome [15–95% ST302 strains] = 2587 genes; cloud genome [<15% ST302 strains] = 4117 genes). Pangenome details, including gene absence/presence table and Roary-generated graphs, can be viewed in Supplementary Table S3.
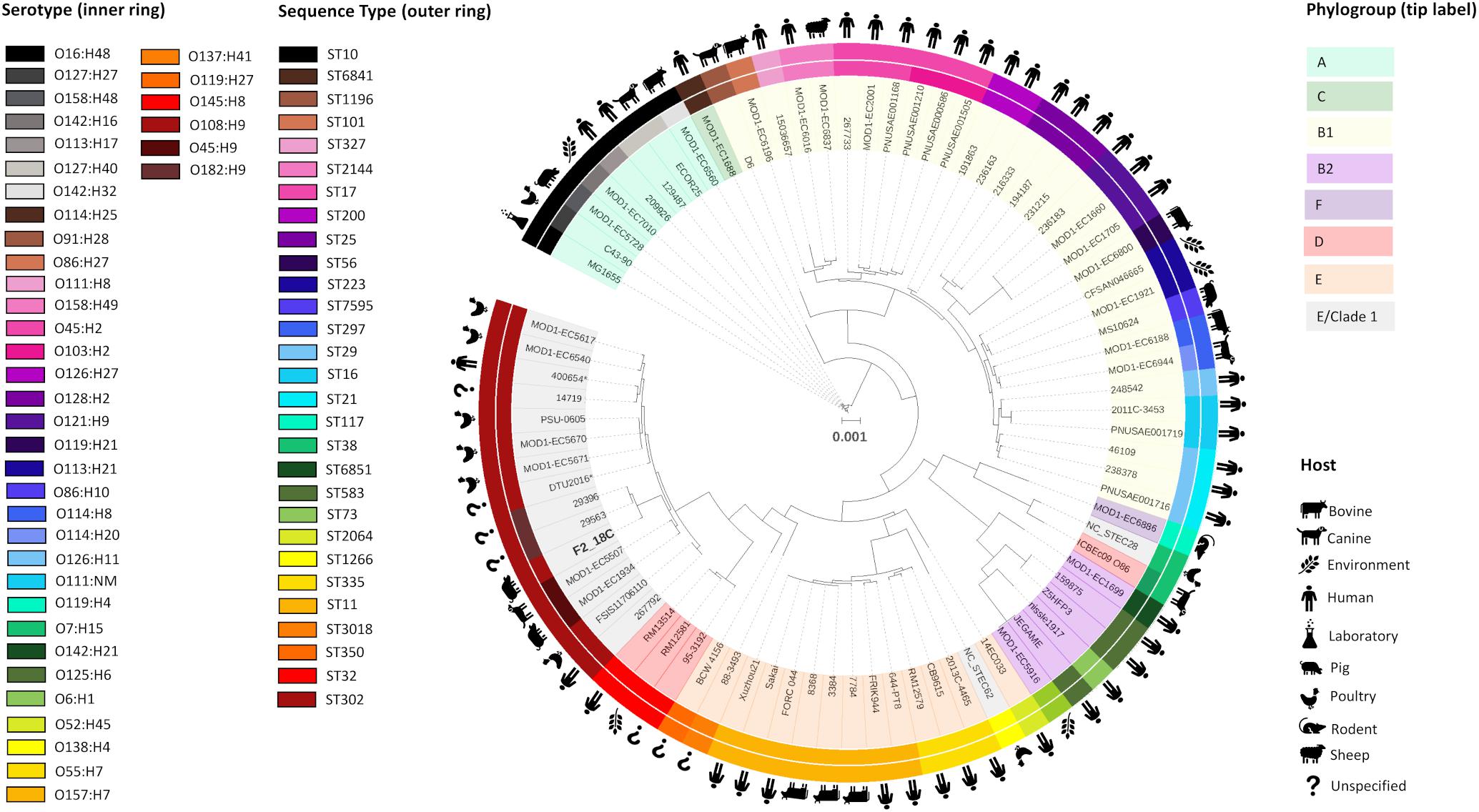
Figure 2. Maximum-likelihood phylogenetic tree showing genetic relatedness of E. coli strain F2_18C. Phylosift, FastTree2, FigTree and iTOL were used to generate and visualize a maximum-likelihood phylogenetic tree, rooted on E. coli laboratory strain MG1655. A total of 91 E. coli strains were used, including all publicly available E. coli ST302 strains. The colored bands that form the outer ring relate to each strains ST and the second ring refers to serotype. Icons around the circumference refer to the host or environment from which the strains were isolated. Shading over tip labels indicates phylogroups. F2_18C is shown in bold. ∗Indicates an abbreviated strain name: 400654∗ = Escherichia_coli_400654-sc-2012-01-13T20:11:31Z-1332520; DTU2016∗ = DTU2016_ 526_ PRJ1050_ Eschericha_coli_Isol133.
ST302 Gene Screening
All E. coli ST302 strains, including strain F2_18C, were screened for acquired antimicrobial resistance genes (ARGs), heavy metal resistance genes, VAGs and plasmid replicons using Ariba. The text below refers to genes presented in Figure 3, which is a selection of significant VAG and ARG, the full list of screened genes can be viewed in Supplementary Table S4.
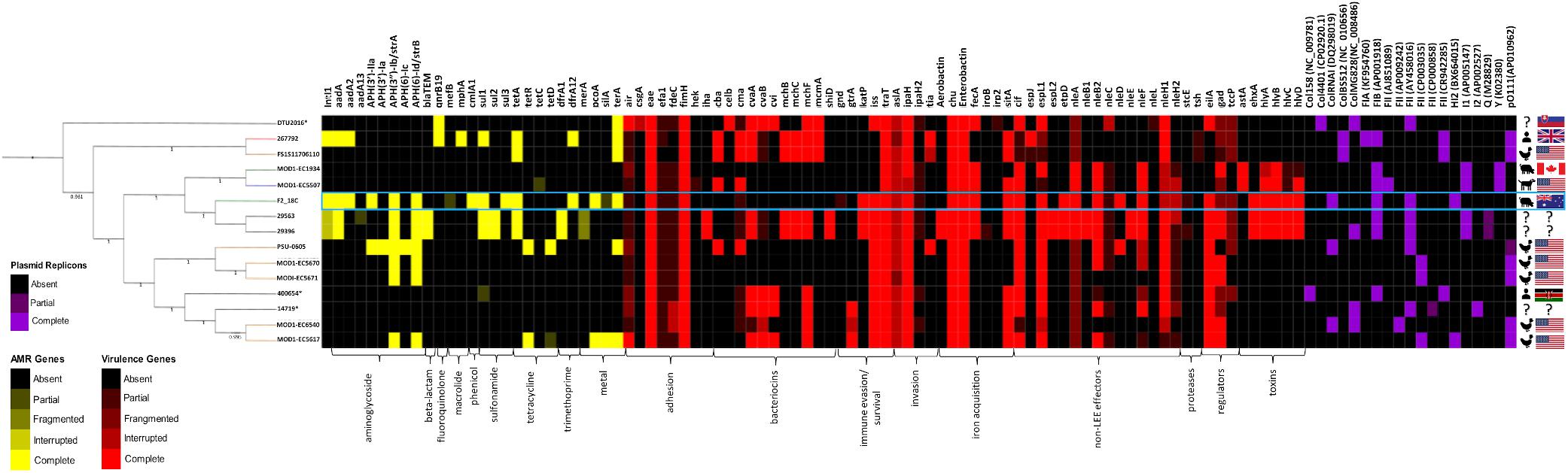
Figure 3. Heatmap and SNP tree of ST302 virulence factors, antimicrobial resistance and plasmid replicons. Intact genes are represented in solid colors, while partial, fragmented or interrupted genes are less opaque. Partial, fragmented and interrupted genes encompass all non-intact genes. ARGs and heavy metal resistance genes are shown in yellow, VAGs in red, and plasmid replicons in purple. Host and country of origin depicted on the right. F2_18C boxed in blue. Tree branch colors are representative of host – red = human; yellow = poultry; green = canine; blue = porcine; black = unspecified. ∗Indicates an abbreviated strain name: 400654∗ = Escherichia_coli_400654-sc-2012-01-13T20:11:31Z-1332520; DTU2016∗ = DTU2016_526_ PRJ1050_ Eschericha_coli_Isol133. The phylogenetic tree was produced by SNP-based phylogenetic analysis.
Acquired ARGs and Heavy Metal Resistance Genes
In E. coli ST302 strains, genes conferring resistance to aminoglycosides, beta-lactams, fluoroquinolones, macrolides, phenicols, sulfonamides, tetracycline and trimethoprim were detected (Figure 3). No genes conferring resistance to extended-spectrum beta-lactams and carbapenems were detected in any E. coli ST302 strains. Strain F2_18C had the greatest number of ARGs (11 versus an average of 3), was 1 of 4 strains carrying resistance genes to 3 or more classes of antibiotics, and was 1 of 2 strains with an intact intI1, the other belonging to strain 267792, isolated from a human in the United Kingdom. The most common acquired ARGs across E. coli ST302 strains were streptomycin resistance genes strA and strB (n = 7; 47%), tetracycline resistance gene tetA (n = 5; 33%), aminoglycoside resistance gene aadA1 (n = 4; 27%), and sulfonamide resistance gene sul1 (n = 4; 27%). In terms of heavy metal resistance genes, E. coli ST302 strains carry intact copper resistance gene pcoA (n = 2; 13%), silver resistance gene silA (n = 1; 7%), and tellurite resistance gene terA (n = 6; 40%).
Strain F2_18C phenotypic resistances and ARGs were congruent. F2_18C was resistant to sulphafurazole (sul1, sul3), tetracycline (tetA), streptomycin (strA, strB), chloramphenicol (cmlA), kanamycin and neomycin [aph(3’)-IIa], and trimethoprim (dfrA12). F2_18C also possesses two additional aminoglycoside resistance genes aadA1 and aadA2.
Virulence-Associated Genes
All E. coli ST302 strains possess the LEE pathogenicity island adhesin gene eae (β variant in F2_18C) but lack Shiga toxin genes (stx), and bundle-forming pili (BFP) genes, broadly classifying them as aEPEC. Strain F2_18C had the most VAGs of all the O108:H2 serotypes (n = 23, versus an average of 14) (Figure 3). However, the two O182:H9 strains, 29563 and 29396, which together with F2_18C, form an offshoot from other E. coli ST302 strains (Figure 1), had the highest number of VAGs overall (n = 33 each). The additional VAGs seen in these two strains, but not in F2_18C include adhesin iha, bacteriocins cvaA, mchB, mchC, mchF, mcmA, shiD, aerobactin iron acquisition genes (iucABCD, iutA), and non-LEE effectors espL2, nleB1 and nleE. Strains F2_18C, 29563 and 29396 were the only E. coli ST302 strains to possess EHEC virulence plasmid associated genes ehxA, etpD and katP.
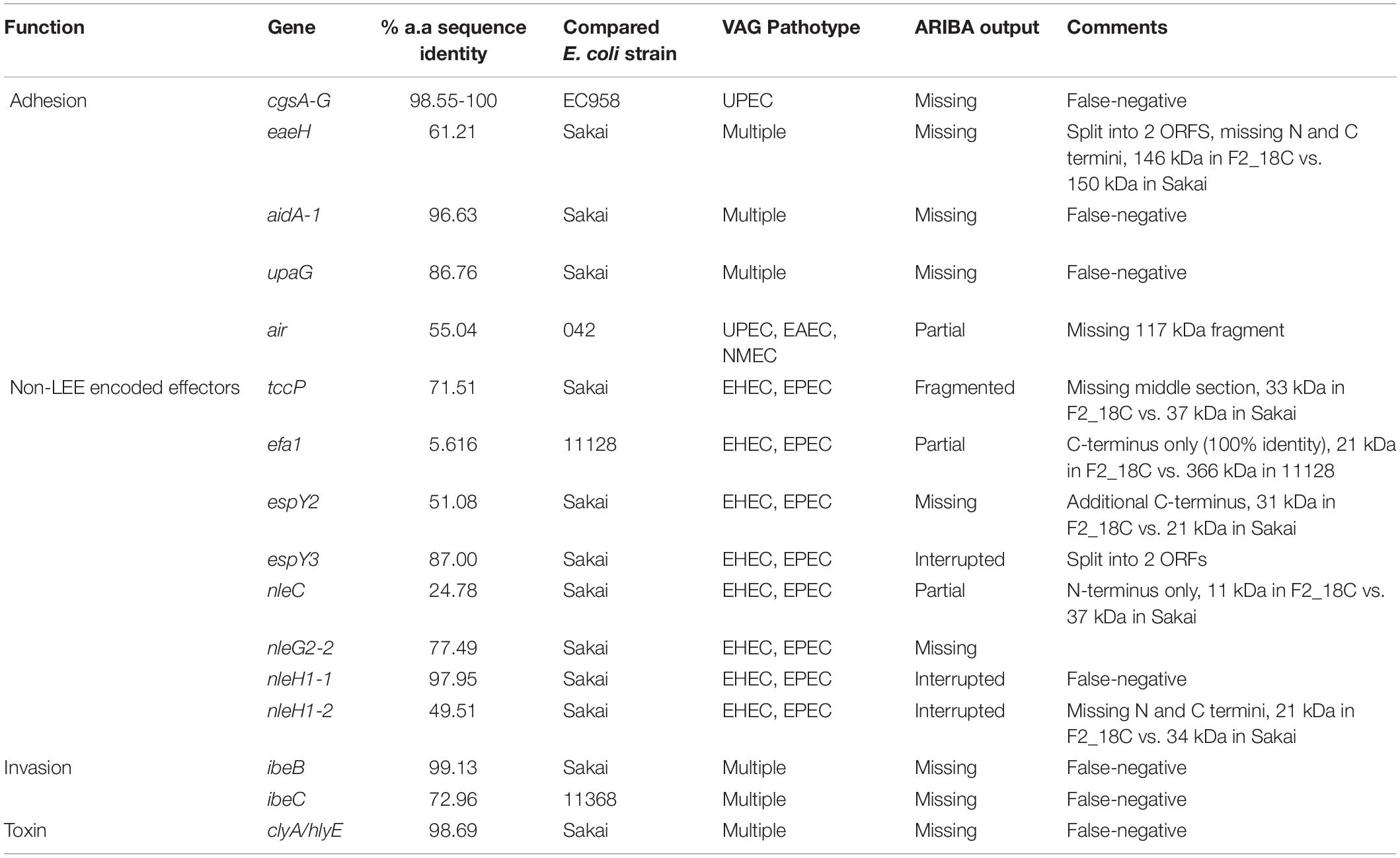
Regarding adherence, in addition to aforementioned eae, iha and fimH, two strains, DTU2016 and MODI-EC5617, possess an intact EAEC adhesin encoding gene air, and all other strains have at least partial hits to this gene. Most strains also have at least partial efa1 (EHEC factor for adherence; n = 14; 93%) and fdeC (factor adherence E. coli; n = 13; 87%) genes. However, they all lack paa (porcine attaching-effacing associated protein) and plasmid-associated toxB genes. Regarding iron uptake, all E. coli ST302 strains possess E. coli heme uptake (chu) and enterobactin genes (ent, fep). Only the aforementioned O182:H9 strains had aerobactin genes, however several strains, including F2_18C, also possess fecA (n = 6; 40%) and sitA (n = 10; 67%). E. coli ST302 strains carry an array of non-LEE effector genes, the most common being cif (cycle inhibiting factor; n = 12; 80%), espL1 (n = 12; 80%), and nleH1 (n = 10; 67%), and the least common being nleD (n = 2; 13%), which F2_18C possesses, and nleE (n = 2; 13%). In terms of immune evasion and survival, E. coli ST302 strains possess iss (increases serum survival; n = 11; 73%), traT (n = 15, 100%), and gtrA (n = 2; 13%). For invasion, ST302 have ipaH (n = 14; 93%), tia (n = 2; 13%), and all have at least partial aslA. Several E. coli ST302 strains have hemolysin toxin genes, and two strains (MOD1-EC1934 and MOD1-EC5507) also possess astA (heat-stable enterotoxin 1/EAST-1). As ARIBA gene screening utilizes Illumina short reads only, the F2_18C chromosome was interrogated manually, using BLASTp and VFDB for reference sequeneces, for gene products flagged as not intact (only VAGs absent in laboratory E. coli strain MG1655 were searched). The amino acid (a.a) sequence identity of these ORFs compared to pathogenic E. coli strains are shown in Table 1, and EPEC and/or EHEC VAGs absent in F2_18C are shown in Table 2.
Plasmid Replicons
All E. coli ST302 strains were identified as possessing at least two plasmid replicons (Figure 3). Various Col plasmids were identified in 9 strains (60%), FIA in 1 strain, FIB in 7 strains (47%), various FII in 14 strains (93%), HI2 in 2 strains (13%), I1 in 4 (27%), I2 in a single strain, Y in 2 strains (13%) and pO111 in 6 (40%). FII, FIB, HI2 and ColRNAI replicons were identified in F2_18C.
F2_18C Mobile Genetic Elements
F2_18C Chromosomal MGEs
The F2_18C chromosome has 52 intact ISs with IS1203 (and IS1203-like) occurring the most frequently (n = 23), followed by a novel IS with some homology to ISKpn28 (n = 7, 85% sequence identity to ISKpn28), and IS4 (n = 6). Several IS1203s disrupt ORFs, including a DNA protecting protein DprA (458183.459396), N-acetylgalactosamine-6-phosphate deacetylase (600929.602142), D-glucarate transporter (1032798.1034011), lipoprotein YnfC (2369004.2370217) and invasion plasmid antigen (2788262.2789475).
When comparing F2_18C with other ST302 strains, MGEs contribute significantly to chromosomal variations (Figure 4). PHASTER identified four intact prophages, three questionable, and six incomplete prophages within the F2_18C chromosome and these carry several VAGs such as non-LEE effector genes (nleB2-1, nleC-like, nleF, nleD, partial nleG, nleH, nleH-1), Tir-cytoskeleton coupling protein tccP2, increased serum survival iss, cycle inhibiting factor cif, and Z2121 (pathogenicity island OI-57 protein). nleA nleF and nleH1-1 are marker genes for pathogenicity island OI-71, however in F2_18C they are carried by various phage elements (Figure 4). The phage elements also carry several heavy metal-associated genes such as sitABCD (iron transport), fecABCDR (iron transport), tonB and fiu (iron transport), zinT (zinc binding), zntB (cadmium, zinc, and copper resistance), copR (copper resistance), and trkG (potassium uptake). PHASTER output is provided in Supplementary Data S3.
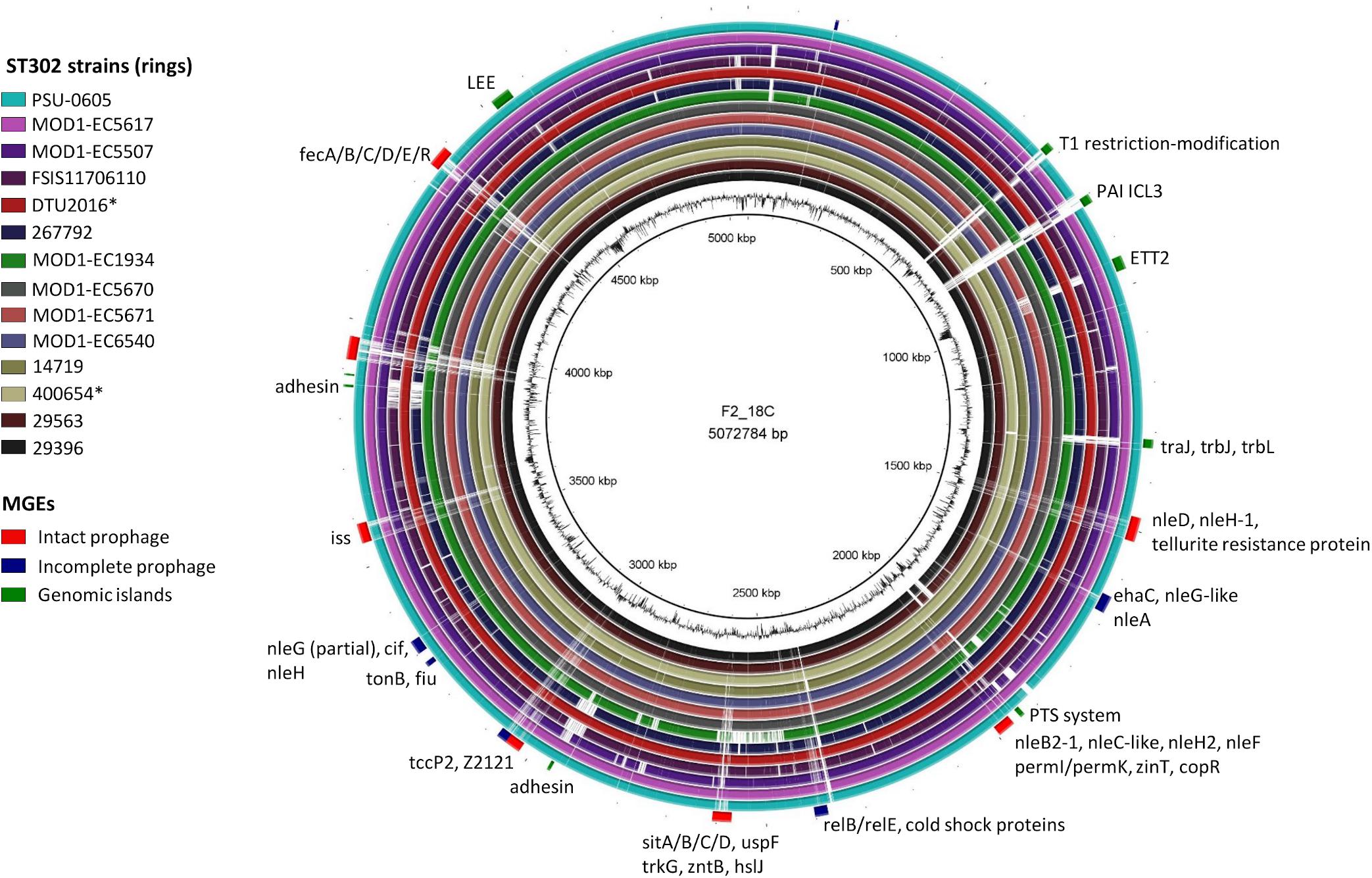
Figure 4. BRIG diagram of MGE distribution in the F2_18C chromosome compared to other ST302 strains. Along the periphery, intact prophages are shown in red, incomplete prophages in navy and GIs in green. Some notable genes present in these regions are labeled accordingly. ∗Indicates an abbreviated strain name: 400654∗ = Escherichia_coli_400654-sc-2012-01-13T20:11:31Z-1332520; DTU2016∗ = DTU2016_526_ PRJ1050_ Escherichia _coli_Isol133.
IslandViewer4 was used to identify genomic islands (GIs). The GIs predicted by at least two algorithms are depicted in Figure 4, and a full list can be viewed in Supplementary Table S5. Notably, F2_18C possesses an intact LEE pathogenicity island, with β1-eae and β-tir, and LEE-encoded T3SS effectors. The PAI is attached at tRNA-Phe. F2_18C also possesses a near-intact E. coli type 3 secretion system island 2 (ETT2) (Figure 5A). This island in F2_18C has fewer pseudogenes than in E. coli strain Sakai (3 vs. 5) however epaS is split into two ORFs, EivC is truncated at the C-terminus by 13.7 kDa (120 amino acids), and an IS1F insertion which interrupts eivG, removing 18 kDa (163 amino acids) off the product’s N-terminus. Furthermore, F2_18C possesses a near-complete PAICL3 pathogenicity island containing adhesin, hemolysin and pagC-like genes. When compared to the original PAICL3 found in E. coli O113:H21 (STEC) strain CL3, F2_18C PAICL3 is smaller by 3629 bp (23668 bp vs. 27297 bp). This is largely due to missing the right-side boundary genes S15 and Z1644, and only possessing a fragment of Z1643 (Figure 5B). Other notable differences are as follows: the Yersinia pestis ShlA/HecA/FhaA family adhesin (S3/S4 in CL3) appears to be split into 3 ORFs in F2_18C and is larger by 277 amino acids than in strain CL3; the transposase-like factor is truncated at the N-terminus and is smaller by 6.8 kDa (58 amino acids) than the equivalent S6 in CL3.; integrase (S7 in CL3) is extended at the N-terminus making it 18.7 kDa (160 amino acids) larger than S7 and; type VI secretion related protein ImpA does not appear truncated at the N- or C-terminus as it does in CL3, making it 25.7 kDa (227 amino acids) larger than S14. Immediately downstream to the F2_18C PAICL3 right-side boundary is a phage T7 exclusion protein, NTPase KAP (Figure 5B), which may contribute to the aforementioned missing S15 and Z1644 ORFS. The F2_18C PAICL3 isoform is not present in any other ST302 (Figure 4).
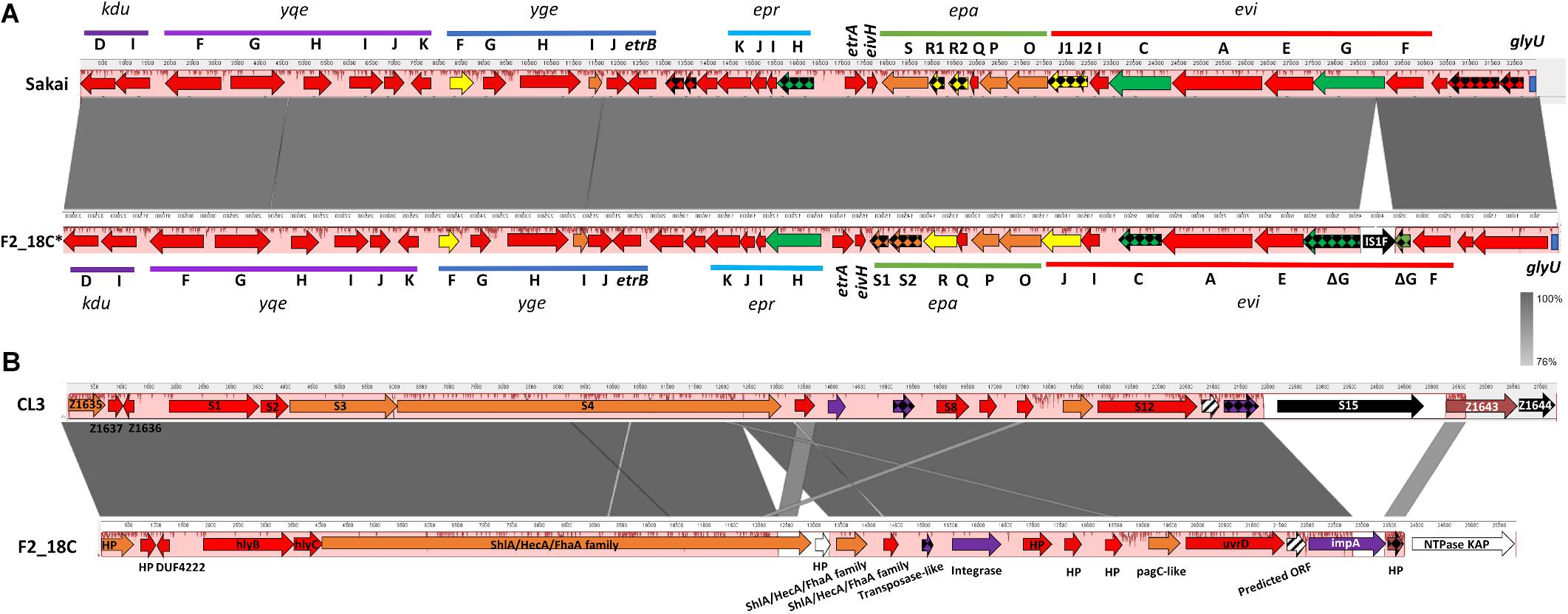
Figure 5. F2_18C pathogenicity island comparisons. Mauve and Easyfig alignments and SnapGene annotations depicting (A) the ETT2 regions in E. coli strains Sakai and F2_18C (F2_18C sequence is depicted in a reversed orientation) and (B) the PAICL3 regions in strains CL3 and F2_18C. The arrow colors represent% amino acid similarity: red = > 95%, orange = > 90%, yellow = > 85%, green = > 60%, purple = > 45%, brown = < 34%, white = ORF not present in CL3, black = ORF not present in F2_18C, checkered = pseudogenes, striped = ORF predicted in F2_18C.
As aEPEC can be converted to EHEC by the presence of stx-carrying bacteriophages, the F2_18C chromosome was also checked for potential stx1 and stx2 integration sites (Bonanno et al., 2015), and was found to have the following unoccupied: wrbA (2984895.2985491), yehV (1780350.1781081), yecE (2076464.2077282), sbcB (1921208.1922635), Z2577 (2354785.2354802) and the torS/torT intergenic region (2996339.2996467). Additionally, F2_18C possesses the lambda phage receptor gene LamB (4667691.4669028) required for lambda bacteriophage adsorption (Bertozzi Silva et al., 2016). F2_18C also possesses the argW integration site (1521099.152173); however this site is occupied by a non-stx-carrying lambdoid prophage.
F2_18C AMR Plasmid
Unicycler hybrid assemblies resolved plasmid pF2_18C_HI2 as a 274, 059 bp circular sequence with 343 CDSs (163 with functional assignments and 180 hypothetical proteins). It carries 17 IS elements, with IS26 variant IS15DI being the most common (n = 8), followed by IS903B (n = 3). The plasmid was typed using pMLST as incompatibility group HI2, sequence type 3 (HI2:ST3). A full plasmid map is presented in Figure 6A, along with content comparisons to high sequence identity (>98%) matched HI2:ST3 plasmids publicly available (n = 33). pF2_18C_HI2 has three regions of AMR carriage. One region comprises a class 1 integron (intI1) located within a Tn172/Tn21 hybrid tetracycline resistance (tetA, tetR) transposon that has acquired dfrA12 (trimethoprim resistance) and aadA2 (aminoglycoside resistance) cassettes with qacEΔ1 (small multidrug resistance efflux transporter), and sul1 (sulfonamide resistance) genes (Figure 6Ci). A second region contains a truncated sul3-associated class 1 integron structure, located near a Tn7-like transposon mobilizing copper and silver (sil/pco) resistance genes at one end and a tellurium resistance genes (ter) at the other. The atypical sul3-associated integron encodes estX (aminoglycoside resistance), psp (phosphoserine phosphatase), aadA2 (aminoglycoside resistance), cmlA (phenicol resistance) and aadA1 (aminoglycoside resistance) cassettes. Further upstream lies a novel truncated version of macrolide resistance gene ΔmefB150, sul3 (sulfonamide resistance) and strA and strB (aminoglycoside resistance) genes (Figure 6Cii). The third region (Figure 6Ciii) is flanked by direct IS15DI and contains aph(3′)-IIa (neomycin, kanamycin, paromomycin, butirosin, gentamicin B and ribostamycin resistance), bleomycin resistance gene blemS, and a NimC/NimA family gene (nitroimidazole reductase) as well as relE/relB toxin/antitoxin genes and three conjugation proteins, traO, traF, and traG. This third region appears unique to three other Australian isolates (Figure 6A) and may represent a new IS15DI-mediated transposon that has evolved in Australia. However, these regions have undergone some inversions and rearrangements, and acquired probable IS15DI-mediated fluoroquinolone resistance genes oqxABR in pSDC-F2_12BHI2 (Figure 6D).
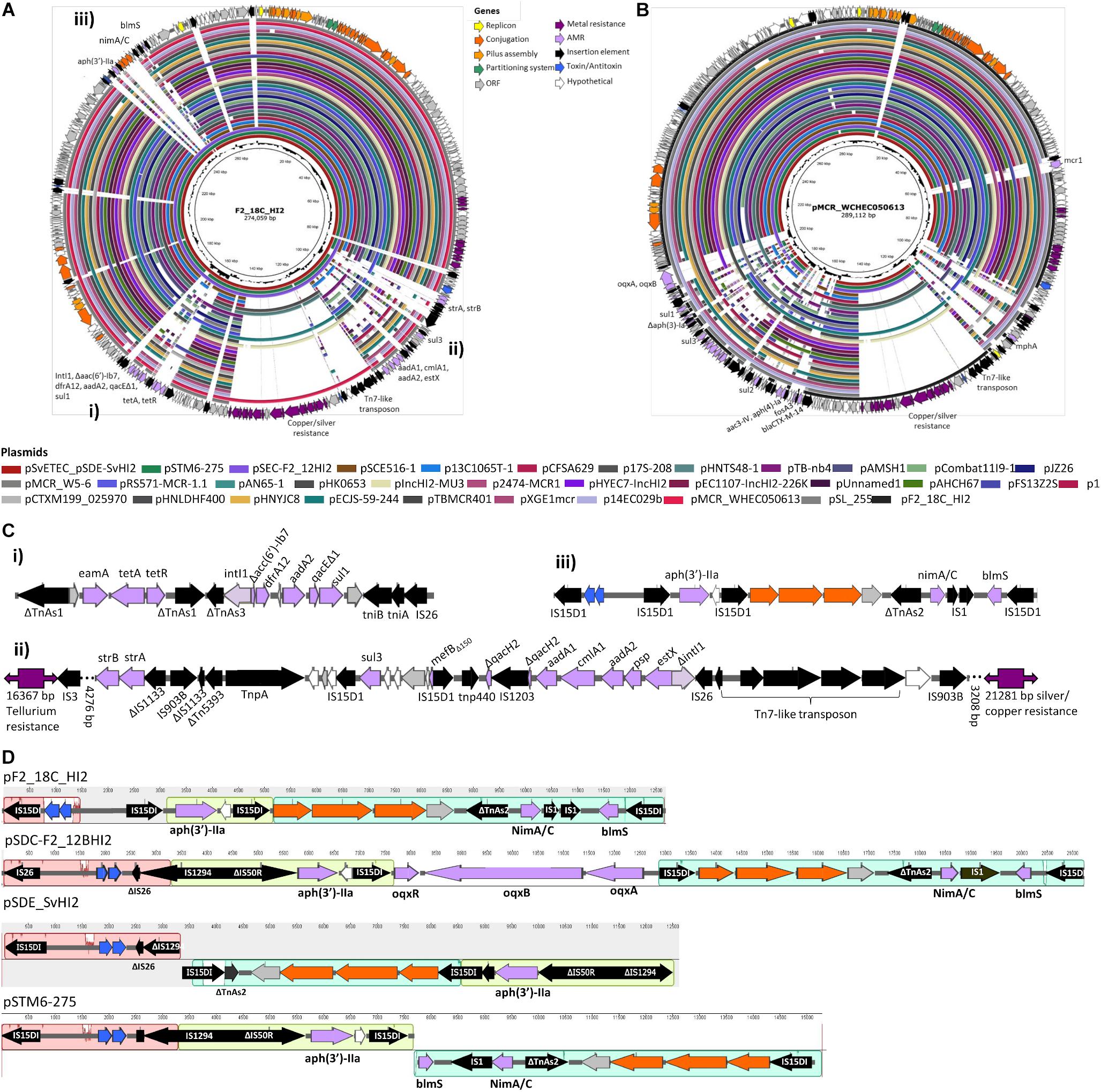
Figure 6. Plasmid maps and BRIG comparison of HI2:ST3 plasmids. (A) Plasmid map of pF2_18C_HI2 and alignments of other HI2:ST3 plasmids. (B) Plasmid map and alignments of pMCR_WHEC050613 from E. coli strain WCHECO50613 isolated from sewerage in China. Plasmid maps show gene annotations and are color coded by function (legend top center). (C) Gene structures of F2_18C AMR regions, flagged in (A). (D) Mauve alignment of region iii found in other HI2:ST3 isolates originating from Australia.
The three AMR regions account for most of the structural difference between pF2_18C_HI2 and other HI2:ST3 plasmids (Figure 6A). However, HI2:ST3s carry a smorgasbord of diverse AMR genes (Figures 6B, 7). Currently, no HI2:ST3 plasmid originating from Australia carries any extended-spectrum beta lactams (ESBLs), colistin, fosfomycin or rifampicin resistance genes, unlike HI2:ST3 plasmids originating from Asia (Figure 7).
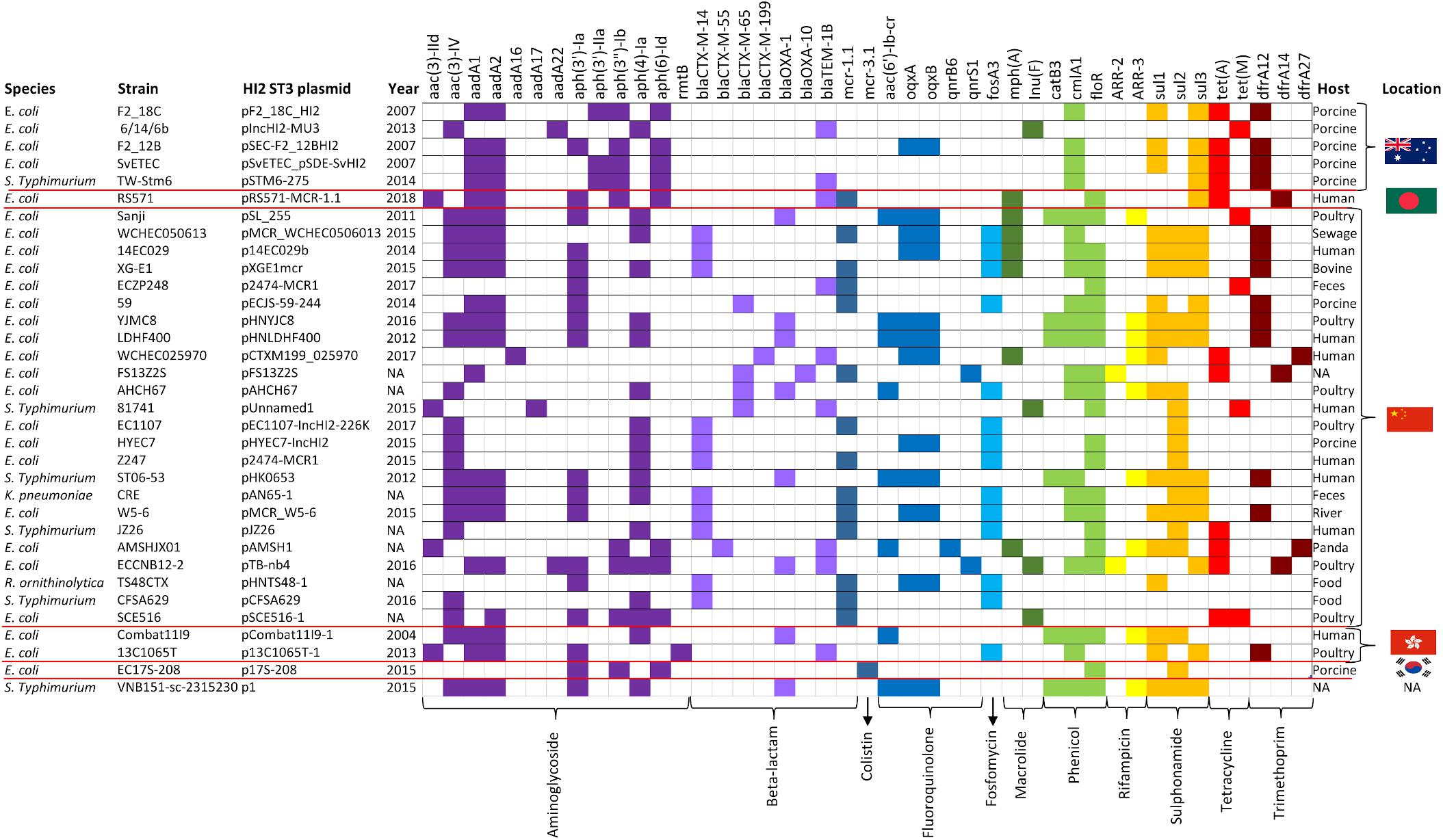
Figure 7. HI2:ST3 AMR gene carriage profiles. AMR genes present in the 33 HI2:ST3 plasmids analyzed in Figure 6. Flags on the right represent isolation location and from top to bottom are Australia, Bangladesh, China, Hong Kong, and South Korea.
F2_18C Virulence Plasmid
Unicycler hybrid assemblies resolved pF2_18C_FIB as a circular sequence of 82, 910 bp containing 100 CDSs (76 with functional assignments, and 24 encoding hypothetical proteins). pF2_18C_FIB contains 6 IS elements with IS1203 (IS629 variant) being the most common. This plasmid could not be typed by pMLST, however, it has >99% sequence identity with four FIB13 plasmids, all of which associate with E. coli ST17 but of varying serotypes (O45:H2, O153:H2 and two O103:H2). The difficulties in typing may be attributed to an additional RepA replicon in pF2_18C_FIB, which is also seen in pO157 found in O157:H7 strains (Figure 8). In terms of virulence, like the FIB13 plasmids, pF2_18C_FIB carries hemolysin encoding genes (ehxA, hlyB, hlyC, hlyD), lipid A modification/synthesis genes (including msbB), stcE (secreted protease of C1 esterase inhibitor from EHEC), and EHEC associated gene etpD. pF2_18C_FIB also carries the catalase gene katP, only seen in one other FIB13 plasmid; however, the F2_18C katP gene is more homologous to katP from pO157 (Figure 8). These plasmids have undergone considerable inversions and rearrangements, which may be the result of the IS elements that flank most regions (Figure 8). pF2_18C_FIB and the FIB13 plasmids share homology to EHEC plasmid pO157 (66% coverage at 90% identity); however, pO157 (from strain Sakai) is 12,688 bp larger than pF2_18C_FIB and contains additional virulence factors including the toxB gene and serine protease etpP (Figure 8).
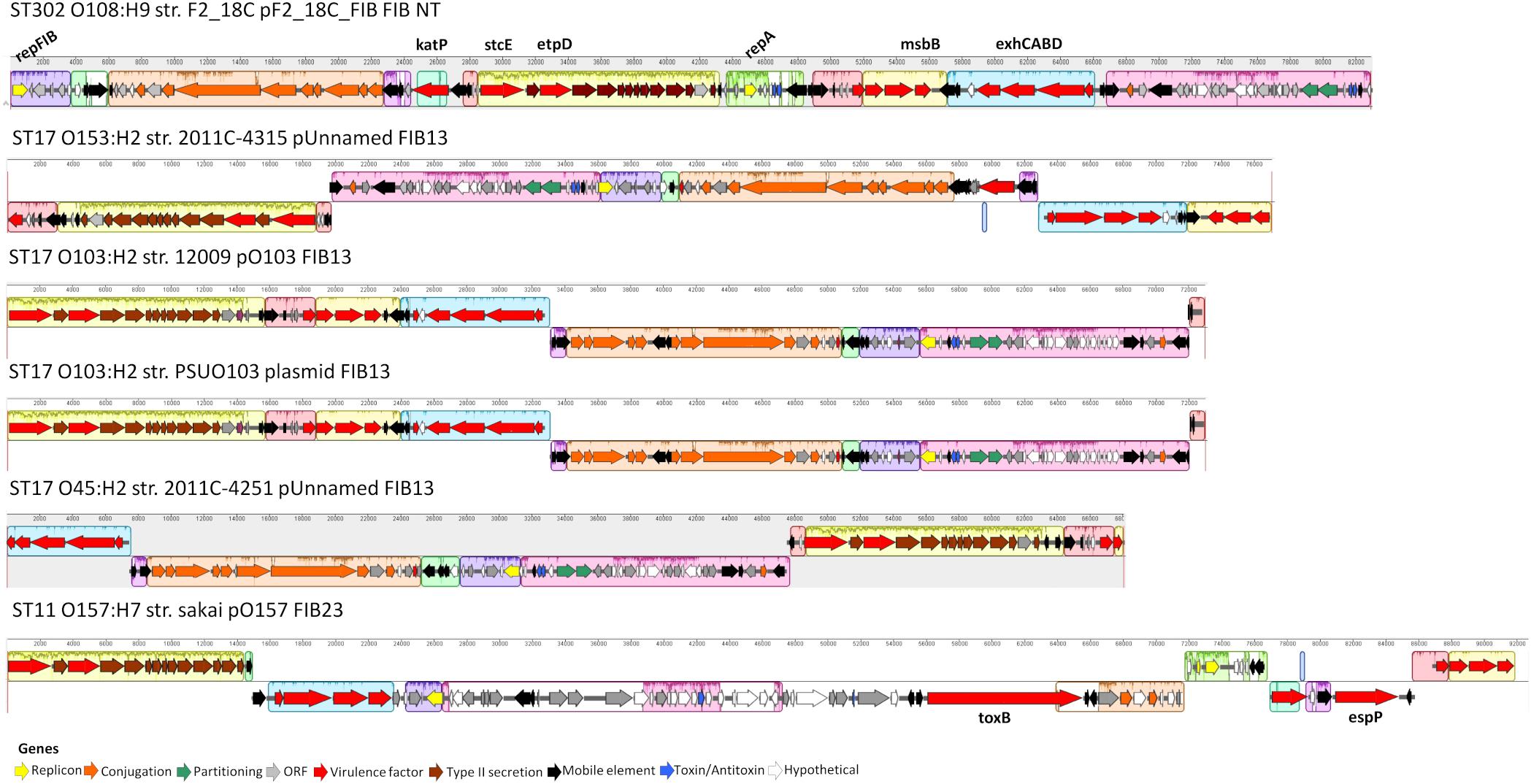
Figure 8. pF2_18C_F plasmid map and FIB13 plasmid alignments. Plasmid maps showing gene annotations for pF2_18C_FIB and four FIB13 plasmids and one FIB23 plasmid over a mauve alignment to demonstrate homologous regions.
F2_18C Col Plasmid
Three F2_18C contigs did not circularize; however, they have homology to Shigella sonnei ColRNAI plasmids (Figure 9). Like the S. sonnei plasmids, F2_18C contig 4 carries colicin-10, the colicin transporter, colicin lysis protein, a partial replication initiation protein and two plasmid exclusion proteins, but additional carries a mobC family plasmid mobilization relaxome protein (100% amino acid sequence identity to E. coli mobC WP_00613379.1) and a nuclease (100% sequence identity to E. coli nuclease WP_00635769.1). Contig 5 carries the nickel ABC transporter protein NikA, common to the S. sonnei plasmids but has additional hypothetical proteins, and contig 6 consists of non-coding DNA common to the S. sonnei plasmids.
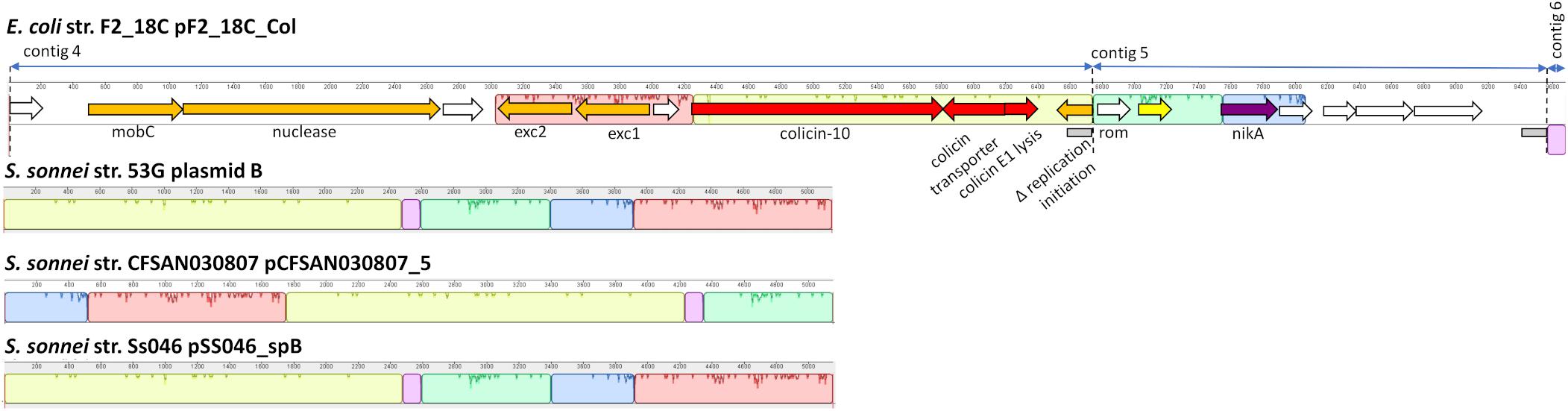
Figure 9. F2_18C contig annotation with mauve alignments to ColRNAI plasmids. Three F2_18C contigs were aligned to ColRNAI_1 plasmids from S. sonnei strains (∼98% identity through PLSDB). Homologous regions are shown in color. Two gray boxes within the annotation denote repeat regions.
Discussion
aEPEC are emerging enteropathogens that have been detected in both humans and animals globally. However, aEPEC are a heterogenous pathotype regarding their serotypes, STs, virulence factors and ability to cause disease. E. coli ST302 strains are aEPEC that have been isolated from both healthy and diseased animal and human hosts, yet no study to date has focused on this ST. Here we present the first closed chromosome and two circularized plasmids (pMLST types HI2:ST3 and FIB) from an ST302 strain; F2_18C, isolated from a healthy pig from Australia. Additionally, we compare F2_18C virulence and AMR gene cargo with other E. coli ST302 strains from a range of hosts including a human isolate, using Illumina short-read gene screening and compare F2_18C plasmids to those with high sequence identity.
Enteric colibacillosis is a significant cause of economic losses to pig industries worldwide (Fairbrother et al., 2005). The major causative pathotype is enterotoxigenic E. coli (ETEC), which is routinely screened for by veterinary diagnostic laboratories, and characterized by their fimbriae and enterotoxins (Fairbrother et al., 2005). However, though not routinely screened for, aEPEC (eae+, stx−, bfp−) are also known to cause enteric colibacillosis in swine (Fairbrother et al., 2005; Luppi, 2017) and García-Meniño et al. (2018) found aEPEC outnumbered ETEC as the causative agents in suckling piglets. Nevertheless, eae+ E. coli has been isolated from both healthy and diarrheic pigs (Krause et al., 2005; Fröhlicher et al., 2008; Malik et al., 2017), implying that a combination of additional VAGs, as well as host-related factors, and poor herd management and husbandry practices, are required to establish disease.
Despite being isolated from a healthy pig, aEPEC strain F2_18C possesses almost twice as many VAGs than non-pathogenic E. coli strain MG1655 (Figure 1). Many of these VAGs are carried on MGEs, including plasmids, prophage-like elements, and genomic islands. F2_18C carries an 82,910 bp plasmid, pF2_18C_FIB, with high sequence identity to FIB13 plasmids from EHEC ST17 strains (Figure 8). These plasmids carry VAGs implicated in EHEC virulence, including the pore-forming toxin ehxA, T2SS gene etpD, C1 esterase inhibitor cleaving protease stcE, and lipid A synthesis gene msbB (Somerville et al., 1999; Noll et al., 2018). Additionally, pF2_18C_FIB and one other ST17 plasmid carry the katP gene. katP encodes for a catalase-peroxidase thought to be expressed by pathogenic E. coli as a defensive mechanism against host-mounted oxidative stress (Uhlich, 2009). katP was also shown to be significantly associated with ST17 human isolates, rather than animal isolates (Söderlund et al., 2016), though in ST11s (O157:H7) katP has been found equally distributed among human and bovine isolates (González et al., 2017). Nevertheless, EHEC strains carrying both katP and stcE have been linked to high virulence (Kobayashi et al., 2013). pF2_18C_FIB and the ST17 FIB13 plasmids share homology with pO157 found in the most notorious of food-borne pathogens O157:H7 (Figure 8). However, they lack the adhesin toxB and the serine protease espP, which cleaves pepsin A and human coagulation factor V, hence thought to contribute to hemorrhagic colitis symptoms (Brunder et al., 1997).
In addition to pF2_18C_FIB, F2_18C has three contigs that did not circularize, but nonetheless have high sequence identity to S. sonnei ColRNAI plasmids. These plasmids carry colicin-10, and the colicin-10 immunity gene, which are also present in F2_18C contig 4 (Figure 9). Colicins are a type of bacteriocin produced by and active against E. coli (Fredericq, 1957). In the advent of plasmid-mediated colistin resistance, particularly in swine, colicins are currently being given consideration as viable alternatives to antibiotics (Ben Lagha et al., 2017). In particular, colicin E1 has been shown to inhibit the growth of swine ETEC strains in vitro (Gillor et al., 2004), and the addition of colicin E1 to pig feeds has been shown to decrease the severity and incidence of ETEC infections (Cutler et al., 2007). However, the colicin-10 immunity protein grants full protection against colicin E1 activity (Pilsl and Braun, 1995). Thus, a potential colistin-10 plasmid circulating amongst a pig population may dampen efforts in implementing colicin-E1 as an effective agent against ETEC and growing AMR in swine.
Like all aEPEC, F2_18C harbors a chromosomal LEE pathogenicity island that includes intimin (eae), an adhesin with a critical role in intestinal colonization. There are at least 30 distinct subtypes of intimin (Ramachandran et al., 2003; Xu et al., 2016), and it is thought that specific subtypes may be involved in tissue tropism and host specificity (Hartland et al., 1999; Torres et al., 2005). The ability of intimin to also bind β–integrins may be significant in this regard (Phillips and Frankel, 2000). The eae gene in F2_18C is subtype β1, which has been significantly correlated to disease in human patients (Xu et al., 2016), and is common in cattle and in healthy pigs (Ramachandran et al., 2003; Krause et al., 2005; Malik et al., 2006; Fröhlicher et al., 2008). In addition to LEE, F2_18C carries two other pathogenicity islands, ETT2, and PAICL3 (Figure 5). ETT2 shares homology to the Salmonella type 3 secretion system located on the Salmonella pathogenicity island 1 (SPI-1). Though it is thought unlikely that ETT2 functions as a secretion system, it is known to influence virulence by regulating VAGs expression (Zhou et al., 2014; Luzader et al., 2016), promoting motility, adhesion, biofilm formation and serum survival (Zhou et al., 2014; Shulman et al., 2018), and enhancing invasion and intracellular survival (Yao et al., 2009). While most E. coli strains possess ETT2 gene clusters, the vast majority have undergone extensive deletions and mutations (Ren et al., 2004; Zhou et al., 2014). However, intact ETT2s are prevalent in pathogenic porcine EHEC strains, particularly those associated with stx2e (Cheng et al., 2012). F2_18C possesses the full complement of ETT2 genes (Figure 5A), however an IS1F insertion interrupts the eivG gene, and eivC gene is truncated. The latter observation may be particularly important as eivC is involved in motility, serum resistance and intra-macrophage survival, at least in avian pathogenic E. coli (APEC) (Wang et al., 2016).
Like ETT2, PAICL3 is rarely found in its entirety, and in most cases, all that remains are boundary genes (Girardeau et al., 2009). The complete PAICL3 locus is a hybrid genomic region of 35 ORFs, composed in part, of a Yersinia pestis-like hemolysin/adhesin genes cluster, OI-122 genes, including the VAG pagC, and OI-48 genes (Girardeau et al., 2009). PAICL3 was originally believed to be restricted to LEE-negative STEC only and a significant predictor of a virulent status (Girardeau et al., 2009). However, Bardiau et al. (2012) identified PAICL3 in 8 (11.3%) O26 EHEC and EPEC strains of human and bovine origins. These strains had the three left-side boundary OI-48 genes (Z1635-7), the hemolysin/adhesin cluster (S1–S4), and the three right boundary OI-48 genes (S15, Z1643-4), however most lacked S5–S14 genes (10 genes), including the five OI-122 genes. Conversely, the F2_18C PAICL3 isoform remains near intact, lacking only two right-side boundary genes (Figure 5B). To our knowledge, this is the first time PAICL3 has been described in swine and has potentially negative implications for its use as a marker for highly virulent strains.
The F2_18C chromosome is infected with 13 prophage elements (Figure 4), which carry a number of VAGs including non-LEE encoded effectors, iron acquisition genes and EHEC-associated gene Z2121 (Delannoy et al., 2013). The aidA gene was also found on a prophage element. This gene encodes for a membrane-bound autotransporter protein that functions as an adhesin that self-associates and also interacts with Ag43, another surface adhesin, to promote biofilm formation (Sherlock et al., 2004). An aidA-deletion mutant ETEC strain was shown to lose the ability to colonize and induce disease in swine emphasizing the important role it plays in pathogenesis (Fairbrother et al., 2005). Importantly, aidA was one a several genes that resulted in a false-negative identification by short-read gene screening (Table 1). Bacteriophages are the most abundant life-form on earth (Brüssow and Hendrix, 2002) and are widespread in sewage effluent. Given the propensity of aEPEC to be converted to EHEC by the introduction of Shiga toxin-carrying lambdoid bacteriophages (Eichhorn et al., 2018), F2_18C was checked for known stx1 and stx2-phage integration sites and was found to have several vacant. It is theorized that aEPEC are the progenitors of EHEC (Kyle et al., 2012; Eichhorn et al., 2018), and so it is therefore prudent to monitor their VAG cargo, particularly EHEC-associated VAGs, and factor in aEPEC detection into EHEC control measures. That being said, though the virulence potential of F2_18C appears high, no STEC ST302 strains have been observed, and F2_18C was isolated from a healthy pig, therefore it was also prudent to take note of EPEC/EHEC VAGs missing in this strain (Table 2). Nonetheless, serotype O108:H9 E. coli such as F2_18C are frequently isolated from pigs with gastrointestinal disease (Malik et al., 2006; Kleta et al., 2014) and from healthy swine (Krause et al., 2005; Fröhlicher et al., 2008) suggesting that they may be opportunistic pathogens that cause diarrheal disease in swine during stressful episodes such as weaning.
When compared to other E. coli ST302 strains, F2_18C had more VAGs than most, with the notable exception of two O182:H9 strains 29563 and 29396. These two strains were highly homologous in their VAG profile (Figure 3) and were the only two in addition to F2_18C to carry the EHEC plasmid marker genes ehxA, etpD, and katP. However, unlike F2_18C, 29563 and 29396 appear to have acquired several extraintestinal E. coli (ExPEC) VAGs, including cvaA, iha, shiD and genes encoding aerobactin iron acquisition. Additionally, 29563 and 29396 uniquely carry nleB1, nleE and espL2/ent, which together with eae, represent a strong signature for human pathogenic EHEC (Bugarel et al., 2011). Incidentally, both maximum-likelihood and SNP-based phylogenetic analyses (Figures 2, 3), place F2_18C, 29563 and 29396 in a distinct off-shoot from other E. coli ST302 strains and may represent an evolutionary pathway shaped by VAG acquisition. When compared to 76 E. coli genomes of varying STs, serotypes, phylogroups and hosts, E. coli ST302 strains were most closely related to EHEC O145:H28 strains, two of which, RM12581 and RM12761, were associated with major food-borne outbreaks (Cooper et al., 2014) (Figure 2).
As well as being an ST302 with one of the highest virulence potentials, F2_18C also carries the most AMR genes acquired through HGT (Figure 3). This is, in all likelihood, due to F2_18C being only one of two ST302 strains to carry an intact class 1 integron marker gene intI1. Integrons are MGEs capable of capturing and mobilizing functional gene cassettes, and class 1 integrons have played a pivotal role in disseminating antibiotic resistance worldwide (Hall and Collis, 1998). F2_18C carries ARGs conferring resistance to aminoglycoside (aadA1, aadA2), chloramphenicol (cmlA), kanamycin and neomycin [aph(3′)-IIa], streptomycin (strA, strB), sulphonoamide (sul1, sul3), tetracycline (tetA, tetR) and trimethoprim (drfA12). Strain 267792, the only other ST302 to carry an intact intI1 gene, was isolated from a human in the United Kingdom, and possesses a different AMR profile to F2_18C, carrying additional genes conferring resistance to macrolides (mphA) and quinolone (qnrB19), but lacking resistance genes to chloramphenicol, streptomycin, kanamycin and neomycin. Limitations of our ST302 VAG and ARG comparisons include the reliance of short-read data, which we previously mentioned can provide false-negatives, the limited number of ST302 genomes currently available and the limited metadata provided for strains, including the host, isolation date and location for some strains, and host disease status for all strains, except F2_18C. We were able to map all F2_18C horizontally transferred ARGs to a 274, 059 bp circular plasmid, pF2_18C_HI2 (Figure 6A). This plasmid was typed by pMLST as HI2:ST3, a lineage known as a major contributor in transferring complex class 1 integrons conferring AMR (Zhao H. et al., 2018) pF2_18C_HI2 is very similar to three HI2:ST3 plasmids circulating in Australian pig farms (Dyall-Smith et al., 2017; Billman-Jacobe et al., 2018; Wyrsch et al., 2019) (Figure 6A). pF2_18C_H12 carries the same AMR profile as pSDE-SvHI2, isolated from a severe ETEC strain (Wyrsch et al., 2019), and differs to a pSDC-F2_12BHI2, isolated from a commensal E. coli strain (Wyrsch et al., 2019), and pSTM6-275, isolated from Salmonella enterica subsp. Enterica Typhimurium (Dyall-Smith et al., 2017), by lacking aminoglycoside resistance gene aph(3′)-Ia and fluoroquinolone resistance genes oqxAB in the former, and beta-lactam resistance gene blaTEM-1B in the latter.
We compared the AMR cargo of 34 HI2:ST3 plasmids (Figure 7), five from Australia, one from an unspecified origin, and 28 from Asia, the vast majority of which originated in China. Perhaps due to restrictions on live animal imports and stringent antibiotic usage practices, HI2:ST3 originating from Australia do not, yet, carry any ARG conferring resistance to extended spectrum beta-lactams (ESBLs), colistin, fosfomycin or rifampicin. This is in stark contrast to HI2:ST3 plasmids originating from Asia, in which ESBL genes, such as blaCTX–M–14,–55,–65,–199, blaOXA–1, –10, and fosfomycin gene fosA3, are prevalent. Moreover, 54% of HI2:ST3s from China carry colistin-resistance gene mcr-1. The increased global usage of colistin in agriculture coincided with the rising prevalence of multidrug resistant Enterobacteriaceae (Falagas and Michalopoulos, 2006). China, being the world’s largest producer of pork, is incidentally also the world’s largest user and producer of colistin, making up to 73% of global colistin production and driving global demands for agricultural colistin usage to an anticipated 11,942 tons per annum in 2015 (Liu et al., 2016). Resistance to colistin was originally believed to only occur from relatively rare mutations in a lipid A modification gene (Falagas et al., 2010). Although, the first plasmid mediated-colistin resistance was identified in swine from China in 2015 (Liu et al., 2016), it has now been identified in over 31 countries across five continents from a range of animal and human hosts, sparking worldwide concern, as colistin is used clinically as a last resort for multidrug resistant Gram-negative infections (Holmes et al., 2016). The colistin resistance gene mcr-1 is mobilized by the transposon Tn6330 (ISApl1-mcr-1-orf-ISApl1) which can generate a circular intermediate (IsApl1-mcr-1-orf) (Li et al., 2017; Wang et al., 2018). This circular intermediate is capable of inserting itself into the highly conserved HI2:ST3 backbone (Figure 6B). China banned the use of colistin in pig feeds in 2016 (Walsh and Wu, 2016), however, due to the sheer number of ARG carried by these plasmids, as well as large metal-resistance regions (Figure 6), researchers fear that selective-pressures, through the use of other antibiotics and metal feed additives, will still favor these plasmids, and colistin resistance will continue to spread.
Conclusion
Despite the limited number of publicly available genome sequences it is evident that ST302 E. coli should be broadly considered aEPEC strains that may reside in healthy hosts or cause opportunistic disease. Of concern is the apparent divergence of strains encoding additional virulence traits including those typical of EHEC and ExPEC, which may yield more severe disease presentation. It would be of great benefit to phenotypically characterize these strains in order to fully understand their potential for pathogenicity in humans and animals. Furthermore, the acquisition of MDR plasmids in lineages with pathogenic potential is a major issue for global public health as antimicrobial use may co-select for virulence traits. Our study highlights E. coli ST302 as a potentially emerging pathogen that should be monitored in the context of public health. However, further genomic epidemiology studies of E. coli ST302 within a One Health framework are required to fully understand this lineage, its virulence potential, and inform genomic surveillance in the future.
Data Availability Statement
The datasets generated for this study can be found in the GenBank repository under the assession numbers CP043542, CP043545, CP043544, and CP043543.
Author Contributions
VJ performed the genomic analyses, created the figures, and drafted the manuscript. CR performed the genome assemblies and SNP phylogeny, and assisted in the analyses. TC provided the strain F2_18C and performed the antibiotic sensitivity testing. SD initiated, funded, and supervised the study. All authors contributed to the manuscript revisions and gave final approval for publication.
Funding
This work was partly funded by the Australian Research Council, linkage grant LP150100912, and was supported by the Australian Centre for Genomic Epidemiological Microbiology (Ausgem), a collaborative partnership between the NSW Department of Primary Industries and the University of Technology Sydney.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Max Cummins and Dr. Ethan Wyrsch from the University of Technology Sydney for their guidance in performing the genomic analyses.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.03098/full#supplementary-material
DATA S1 | E. coli strain F2_18C genome annotations.
DATA S2 | ST302 SNP analysis.
DATA S3 | E. coli straim F2_18C PHASTER analysis.
TABLE S1 | E. coli strain F2_18C gene functional assignments and specialty genes provided by PATRIC.
TABLE S2 | E. coli strains used in ST302 phylogenetic analysis.
TABLE S3 | ST302 pangenome analysis.
TABLE S4 | ST302 short-read gene screening output by Ariba.
TABLE S5 | IslandViewer output for E. coli strain F2_18C.
Footnotes
- ^ https://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- ^ http://www-is.biotoul.fr
- ^ http://www.pathogenomics.sfu.ca/islandviewer/
- ^ https://www.patricbrc.org/
- ^ http://phaster.ca/
- ^ http://tree.bio.ed.ac.uk/software/figtree/
- ^ https://itol.embl.de/
- ^ https://github.com/tseemann/snippy
- ^ https://github.com/sanger-pathogens/gubbins
- ^ https://github.com/sanger-pathogens/snp-sites
- ^ https://cge.cbs.dtu.dk/services/MLST/
- ^ https://cge.cbs.dtu.dk/services/SerotypeFinder/
- ^ http://enterobase.warwick.ac.uk/
- ^ http://enterobase.warwick.ac.uk/
- ^ https://cge.cbs.dtu.dk/services/ResFinder/
- ^ https://card.mcmaster.ca/home
- ^ https://cge.cbs.dtu.dk/services/VirulenceFinder/
- ^ http://www.mgc.ac.cn/VFs/
- ^ https://cge.cbs.dtu.dk/services/PlasmidFinder/
- ^ https://ccb-microbe.cs.uni-saarland.de/plsdb/
- ^ http://brig.sourceforge.net/
- ^ http://darlinglab.org/mauve/mauve.html
References
Abreu, A. G., Bueris, V., Porangaba, T. M., Sircili, M. P., Navarro-Garcia, F., and Elias, W. P. (2013). Autotransporter protein-encoding genes of diarrheagenic Escherichia coli are found in both typical and atypical enteropathogenic E. coli Strains. Appl. Environ. Microbiol. 79, 411–414. doi: 10.1128/AEM.02635-12
Afset, J. E., Anderssen, E., Bruant, G., Harel, J., Wieler, L., and Bergh, K. (2008). Phylogenetic backgrounds and virulence profiles of atypical enteropathogenic Escherichia coli strains from a case-control study using multilocus sequence typing and DNA microarray analysis. J. Clin. Microbiol. 46, 2280–2290. doi: 10.1128/JCM.01752-07
Alikhan, N.-F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Alikhan, N.-F., Zhou, Z., Sergeant, M. J., and Achtman, M. (2018). A genomic overview of the population structure of Salmonella. PLoS Genet. 14:e1007261. doi: 10.1371/journal.pgen.1007261
Alonso, M. Z., Sanz, M. E., Padola, N. L., and Lucchesi, P. M. (2014). [Characterization of enteropathogenic Escherichia coli (EPEC) strains isolated during the chicken slaughtering process]. Rev. Argent. Microbiol. 46, 122–125. doi: 10.1016/S0325-7541(14)70060-4
Arais, L. R., Barbosa, A. V., Andrade, J. R. C., Gomes, T. A. T., Asensi, M. D., Aires, C. A. M., et al. (2018). Zoonotic potential of atypical enteropathogenic Escherichia coli (aEPEC) isolated from puppies with diarrhoea in Brazil. Vet. Microbiol. 227, 45–51. doi: 10.1016/j.vetmic.2018.10.023
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Bando, S. Y., Andrade, F. B., Guth, B. E. C., Elias, W. P., Moreira-Filho, C. A., and Pestana de Castro, A. F. (2009). Atypical enteropathogenic Escherichia coli genomic background allows the acquisition of non-EPEC virulence factors. FEMS Microbiol. Lett. 299, 22–30. doi: 10.1111/j.1574-6968.2009.01735.x
Bardiau, M., Taminiau, B., Duprez, J.-N., Labrozzo, S., and Mainil, J. G. (2012). Comparison between a bovine and a human enterohaemorrhagic Escherichia coli strain of serogroup O26 by suppressive subtractive hybridization reveals the presence of atypical factors in EHEC and EPEC strains. FEMS Microbiol. Lett. 330, 132–139. doi: 10.1111/j.1574-6968.2012.02542.x
Bell, S. M., Pham, J. N., Rafferty, D. L., and Allerton, J. K. (2016). Antibiotic Sensitivity Testing by the CDS Method: A Manual for Medical and Veterinary Laboratories, 8th Edn. Kogarah, NSW: South Eastern Area Laboratory Services.
Ben Lagha, A., Haas, B., Gottschalk, M., and Grenier, D. (2017). Antimicrobial potential of bacteriocins in poultry and swine production. Vet. Res. 48:22. doi: 10.1186/s13567-017-0425-6
Bertelli, C., Laird, M. R., Williams, K. P., Lau, B. Y., Hoad, G., Winsor, G. L., et al. (2017). IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res. 45, W30–W35. doi: 10.1093/nar/gkx343
Bertozzi Silva, J., Storms, Z., and Sauvageau, D. (2016). Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 363:fnw002. doi: 10.1093/femsle/fnw002
Bielaszewska, M., Middendorf, B., Köck, R., Friedrich, A. W., Fruth, A., Karch, H., et al. (2008). Shiga toxin-negative attaching and effacing Escherichia coli: distinct clinical associations with bacterial phylogeny and virulence traits and inferred in-host pathogen evolution. Clin. Infect. Dis. 47, 208–217. doi: 10.1086/589245
Billman-Jacobe, H., Liu, Y., Haites, R., Weaver, T., Robinson, L., Marenda, M., et al. (2018). pSTM6-275, a conjugative IncHI2 plasmid of Salmonella enterica that confers antibiotic and heavy-metal resistance under changing physiological conditions. Antimicrob. Agents Chemother. 62:e02357-17. doi: 10.1128/AAC.02357-17
Bonanno, L., Loukiadis, E., Mariani-Kurkdjian, P., Oswald, E., Garnier, L., Michel, V., et al. (2015). Diversity of shiga toxin-producing Escherichia coli (STEC) O26:H11 strains examined via stx subtypes and insertion sites of Stx and EspK bacteriophages. Appl. Environ. Microbiol. 81, 3712–3721. doi: 10.1128/AEM.00077-15
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Brunder, W., Schmidt, H., and Karch, H. (1997). EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol. Microbiol. 24, 767–778. doi: 10.1046/j.1365-2958.1997.3871751.x
Brüssow, H., and Hendrix, R. W. (2002). Phage genomics: small is beautiful. Cell 108, 13–16. doi: 10.1016/s0092-8674(01)00637-7
Bugarel, M., Martin, A., Fach, P., and Beutin, L. (2011). Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiol. 11:142. doi: 10.1186/1471-2180-11-142
Carattoli, A., Zankari, E., García-Fernández, A., Larsen, M. V., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using plasmidfinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Caruso, G. (2018). Antibiotic resistance in Escherichia coli from farm livestock and related analytical methods: a review. J. AOAC Int. 101, 916–922. doi: 10.5740/jaoacint.17-0445
Cheng, D., Zhu, S., Su, Z., Zuo, W., and Lu, H. (2012). Prevalence and isoforms of the pathogenicity island ETT2 among Escherichia coli isolates from colibacillosis in pigs and mastitis in cows. Curr. Microbiol. 64, 43–49. doi: 10.1007/s00284-011-0032-0
Clermont, O., Christenson, J. K., Denamur, E., and Gordon, D. M. (2013). The clermont Escherichia coli phylo-typing method revisited: improvement of specificity and detection of new phylo-groups. Environ. Microbiol. Rep. 5, 58–65. doi: 10.1111/1758-2229.12019
Cooper, K. K., Mandrell, R. E., Louie, J. W., Korlach, J., Clark, T. A., Parker, C. T., et al. (2014). Complete genome sequences of two Escherichia coli O145:H28 outbreak strains of food origin. Genome Announc. 2:e00482-14. doi: 10.1128/genomeA.00482-14
Cutler, S. A., Lonergan, S. M., Cornick, N., Johnson, A. K., and Stahl, C. H. (2007). Dietary inclusion of colicin E1 is effective in preventing postweaning diarrhea caused by F18-Positive Escherichia coli in pigs. Antimicrob. Agents Chemother. 51, 3830–3835. doi: 10.1128/AAC.00360-07
Darling, A. C. E. (2004). Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 14, 1394–1403. doi: 10.1101/gr.2289704
Darling, A. E., Jospin, G., Lowe, E., Iv, F. A. M., Bik, H. M., and Eisen, J. A. (2014). PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2:e243. doi: 10.7717/peerj.243
Delannoy, S., Beutin, L., and Fach, P. (2013). Towards a molecular definition of enterohemorrhagic Escherichia coli (EHEC): detection of genes located on O island 57 as markers to distinguish EHEC from closely related enteropathogenic E. coli strains. J. Clin. Microbiol. 51, 1083–1088. doi: 10.1128/JCM.02864-12
Dyall-Smith, M. L., Liu, Y., and Billman-Jacobe, H. (2017). Genome sequence of an Australian monophasic Salmonella enterica subsp. enterica Typhimurium isolate (TW-Stm6) carrying a large plasmid with multiple antimicrobial resistance genes. Genome Announc. 5:e00793-17. doi: 10.1128/genomeA.00793-17
Eichhorn, I., Heidemanns, K., Ulrich, R. G., Schmidt, H., Semmler, T., Fruth, A., et al. (2018). Lysogenic conversion of atypical enteropathogenic Escherichia coli (aEPEC) from human, murine, and bovine origin with bacteriophage Φ3538 Δstx2::cat proves their enterohemorrhagic E. coli (EHEC) progeny. Int. J. Med. Microbiol. 308, 890–898. doi: 10.1016/j.ijmm.2018.06.005
Fairbrother, J. M., Nadeau, É, and Gyles, C. L. (2005). Escherichia coli in postweaning diarrhea in pigs: an update on bacterial types, pathogenesis, and prevention strategies. Anim. Health Res. Rev. 6, 17–39. doi: 10.1079/AHR2005105
Falagas, M. E., and Michalopoulos, A. (2006). Polymyxins: old antibiotics are back. Lancet 367, 633–634. doi: 10.1016/S0140-6736(06)68241-X
Falagas, M. E., Rafailidis, P. I., and Matthaiou, D. K. (2010). Resistance to polymyxins: mechanisms, frequency and treatment options. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 13, 132–138. doi: 10.1016/j.drup.2010.05.002
Foster, M. A., Iqbal, J., Zhang, C., McHenry, R., Cleveland, B. E., Romero-Herazo, Y., et al. (2015). Enteropathogenic and enteroaggregative E. coli in stools of children with acute gastroenteritis in Davidson County, Tennessee. Diagn. Microbiol. Infect. Dis. 83, 319–324. doi: 10.1016/j.diagmicrobio.2015.07.016
Fredericq, P. (1957). Colicins. Annu. Rev. Microbiol. 11, 7–22. doi: 10.1146/annurev.mi.11.100157.000255
Fröhlicher, E., Krause, G., Zweifel, C., Beutin, L., and Stephan, R. (2008). Characterization of attaching and effacing Escherichia coli (AEEC) isolated from pigs and sheep. BMC Microbiol. 8:144. doi: 10.1186/1471-2180-8-144
Galata, V., Fehlmann, T., Backes, C., and Keller, A. (2019). PLSDB: a resource of complete bacterial plasmids. Nucleic Acids Res. 47, D195–D202. doi: 10.1093/nar/gky1050
García-Meniño, I., García, V., Mora, A., Díaz-Jiménez, D., Flament-Simon, S. C., Alonso, M. P., et al. (2018). Swine enteric colibacillosis in spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates. Front. Microbiol. 9:2659. doi: 10.3389/fmicb.2018.02659
Gillor, O., Kirkup, B. C., and Riley, M. A. (2004). Colicins and microcins: the next generation antimicrobials. Adv. Appl. Microbiol. 54, 129–146. doi: 10.1016/S0065-2164(04)54005-4
Girardeau, J. P., Bertin, Y., and Martin, C. (2009). Genomic analysis of the PAI ICL3 locus in pathogenic LEE-negative Shiga toxin-producing Escherichia coli and Citrobacter rodentium. Microbiology 155, 1016–1027. doi: 10.1099/mic.0.026807-0
Gomes, T. A. T., Elias, W. P., Scaletsky, I. C. A., Guth, B. E. C., Rodrigues, J. F., Piazza, R. M. F., et al. (2016). Diarrheagenic Escherichia coli. Braz. J. Microbiol. 47, 3–30. doi: 10.1016/j.bjm.2016.10.015
González, J., Sanso, A. M., Cadona, J. S., and Bustamante, A. V. (2017). Virulence traits and different nle profiles in cattle and human verotoxin-producing Escherichia coli O157:H7 strains from Argentina. Microb. Pathog. 102, 102–108. doi: 10.1016/j.micpath.2016.11.022
Hall, R. M., and Collis, C. M. (1998). Antibiotic resistance in gram-negative bacteria: the role of gene cassettes and integrons. Drug Resist. Updat. Rev. Comment. Antimicrob. Anticancer Chemother. 1, 109–119. doi: 10.1016/s1368-7646(98)80026-5
Hartland, E. L., Batchelor, M., Delahay, R. M., Hale, C., Matthews, S., Dougan, G., et al. (1999). Binding of intimin from enteropathogenic Escherichia coli to Tir and to host cells. Mol. Microbiol. 32, 151–158. doi: 10.1046/j.1365-2958.1999.01338.x
Holmes, A. H., Moore, L. S. P., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Hu, J., and Torres, A. G. (2015). Enteropathogenic Escherichia coli: foe or innocent bystander? Clin. Microbiol. Infect. Off. Publ. Eur. Soc. Clin. Microbiol. Infect. Dis. 21, 729–734. doi: 10.1016/j.cmi.2015.01.015
Huang, X., Yu, L., Chen, X., Zhi, C., Yao, X., Liu, Y., et al. (2017). High prevalence of colistin resistance and mcr-1 Gene in Escherichia coli isolated from food animals in China. Front. Microbiol. 8:562. doi: 10.3389/fmicb.2017.00562
Hunt, M., Mather, A. E., Sánchez-Busó, L., Page, A. J., Parkhill, J., Keane, J. A., et al. (2017). ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb. Genomics 3:e000131. doi: 10.1099/mgen.0.000131
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Joensen, K. G., Scheutz, F., Lund, O., Hasman, H., Kaas, R. S., Nielsen, E. M., et al. (2014). Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 52, 1501–1510. doi: 10.1128/JCM.03617-13
Joensen, K. G., Tetzschner, A. M. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Kaper, J. B., Nataro, J. P., and Mobley, H. L. (2004). Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140. doi: 10.1038/nrmicro818
Kleta, S., Nordhoff, M., Tedin, K., Wieler, L. H., Kolenda, R., Oswald, S., et al. (2014). Role of F1C Fimbriae, flagella, and secreted bacterial components in the inhibitory effect of probiotic Escherichia coli Nissle 1917 on atypical enteropathogenic E. coli infection. Infect. Immun. 82, 1801–1812. doi: 10.1128/IAI.01431-13
Kobayashi, N., Lee, K., Yamazaki, A., Saito, S., Furukawa, I., Kono, T., et al. (2013). Virulence gene profiles and population genetic analysis for exploration of pathogenic serogroups of shiga toxin-producing Escherichia coli. J. Clin. Microbiol. 51, 4022–4028. doi: 10.1128/JCM.01598-13
Krause, G., Zimmermann, S., and Beutin, L. (2005). Investigation of domestic animals and pets as a reservoir for intimin- (eae) gene positive Escherichia coli types. Vet. Microbiol. 106, 87–95. doi: 10.1016/j.vetmic.2004.11.012
Kyle, J. L., Cummings, C. A., Parker, C. T., Quiñones, B., Vatta, P., Newton, E., et al. (2012). Escherichia coli serotype O55:H7 diversity supports parallel acquisition of bacteriophage at Shiga toxin phage insertion sites during evolution of the O157:H7 lineage. J. Bacteriol. 194, 1885–1896. doi: 10.1128/JB.00120-12
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, R., Xie, M., Lv, J., Wai-Chi Chan, E., and Chen, S. (2017). Complete genetic analysis of plasmids carrying mcr-1 and other resistance genes in an Escherichia coli isolate of animal origin. J. Antimicrob. Chemother. 72, 696–699. doi: 10.1093/jac/dkw509
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Liu, Y.-Y., Wang, Y., Walsh, T. R., Yi, L.-X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lugsomya, K., Yindee, J., Niyomtham, W., Tribuddharat, C., Tummaruk, P., Hampson, D. J., et al. (2018). Antimicrobial resistance in commensal Escherichia Coli isolated from pigs and pork derived from farms either routinely using or not using in-feed antimicrobials. Microb. Drug Resist. 24, 1054–1066. doi: 10.1089/mdr.2018.0154
Luppi, A. (2017). Swine enteric colibacillosis: diagnosis, therapy and antimicrobial resistance. Porc. Health Manag. 3:16. doi: 10.1186/s40813-017-0063-4
Luzader, D. H., Willsey, G. G., Wargo, M. J., and Kendall, M. M. (2016). The type three secretion system 2-encoded regulator EtrB modulates enterohemorrhagic Escherichia coli virulence gene expression. Infect. Immun. 84, 2555–2565. doi: 10.1128/IAI.00407-16
Malik, A., Nagy, B., Kugler, R., and Szmolka, A. (2017). Pathogenic potential and virulence genotypes of intestinal and faecal isolates of porcine post-weaning enteropathogenic Escherichia coli. Res. Vet. Sci. Oxf. 115, 102–108. doi: 10.1016/j.rvsc.2017.02.002
Malik, A., Tóth, I., Beutin, L., Schmidt, H., Taminiau, B., Dow, M. A., et al. (2006). Serotypes and intimin types of intestinal and faecal strains of eae+Escherichia coli from weaned pigs. Vet. Microbiol. 114, 82–93. doi: 10.1016/j.vetmic.2005.11.044
Mayer, C. L., Leibowitz, C. S., Kurosawa, S., and Stearns-Kurosawa, D. J. (2012). Shiga toxins and the pathophysiology of hemolytic uremic syndrome in humans and animals. Toxins 4, 1261–1287. doi: 10.3390/toxins4111261
McDaniel, T. K., and Kaper, J. B. (1997). A cloned pathogenicity island from enteropathogenic Escherichia coli confers the attaching and effacing phenotype on E. coli K-12. Mol. Microbiol. 23, 399–407. doi: 10.1046/j.1365-2958.1997.2311591.x
Noll, L. W., Worley, J. N., Yang, X., Shridhar, P. B., Ludwig, J. B., Shi, X., et al. (2018). Comparative genomics reveals differences in mobile virulence genes of Escherichia coli O103 pathotypes of bovine fecal origin. PLoS One 13:e0191362. doi: 10.1371/journal.pone.0191362
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T. G., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paitan, Y. (2018). “Current trends in antimicrobial resistance of Escherichia coli,” in Escherichia Coli, a Versatile Pathogen, Current Topics in Microbiology and Immunology, eds G. Frankel and E. Z. Ron, (Cham: Springer International Publishing), 181–211. doi: 10.1007/82_2018_110
Partridge, S. R., Kwong, S. M., Firth, N., and Jensen, S. O. (2018). Mobile genetic elements associated with antimicrobial resistance. Clin. Microbiol. Rev. 31:e00088-17. doi: 10.1128/CMR.00088-17
Phillips, A. D., and Frankel, G. (2000). Intimin-mediated tissue specificity in enteropathogenic Escherichia coli interaction with human intestinal organ cultures. J. Infect. Dis. 181, 1496–1500. doi: 10.1086/315404
Pilsl, H., and Braun, V. (1995). Novel colicin 10: assignment of four domains to TonB- and TolC-dependent uptake via the Tsx receptor and to pore formation. Mol. Microbiol. 16, 57–67. doi: 10.1111/j.1365-2958.1995.tb02391.x
Pitout, J. D. D. (2012). Extraintestinal pathogenic Escherichia coli: an update on antimicrobial resistance, laboratory diagnosis and treatment. Expert Rev. Anti Infect. Ther. 10, 1165–1176. doi: 10.1586/eri.12.110
Poirel, L., Madec, J.-Y., Lupo, A., Schink, A.-K., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 6, doi: 10.1128/microbiolspec.ARBA-0026-2017
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Ramachandran, V., Brett, K., Hornitzky, M. A., Dowton, M., Bettelheim, K. A., Walker, M. J., et al. (2003). Distribution of intimin subtypes among Escherichia coli isolates from ruminant and human sources. J. Clin. Microbiol. 41, 5022–5032. doi: 10.1128/JCM.41.11.5022-5032.2003
Reid, C. J., Wyrsch, E. R., Roy Chowdhury, P., Zingali, T., Liu, M., Darling, A. E., et al. (2017). Porcine commensal Escherichia coli: a reservoir for class 1 integrons associated with IS26. Microb. Genomics 3:e000143. doi: 10.1099/mgen.0.000143
Ren, C.-P., Chaudhuri, R. R., Fivian, A., Bailey, C. M., Antonio, M., Barnes, W. M., et al. (2004). The ETT2 gene cluster, encoding a second Type III secretion system from Escherichia coli, is present in the majority of strains but has undergone widespread mutational attrition. J. Bacteriol. 186, 3547–3560. doi: 10.1128/JB.186.11.3547-3560.2004
Sherlock, O., Schembri, M. A., Reisner, A., and Klemm, P. (2004). Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186, 8058–8065. doi: 10.1128/JB.186.23.8058-8065.2004
Shulman, A., Yair, Y., Biran, D., Sura, T., Otto, A., Gophna, U., et al. (2018). The Escherichia coli Type III secretion system 2 has a global effect on cell surface. mBio 9:e01070-18. doi: 10.1128/mBio.01070-18
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Söderlund, R., Hurel, J., Jinnerot, T., Sekse, C., Aspán, A., Eriksson, E., et al. (2016). Genomic comparison of Escherichia coli serotype O103:H2 isolates with and without verotoxin genes: implications for risk assessment of strains commonly found in ruminant reservoirs. Infect. Ecol. Epidemiol. 6:10.3402/iee.v6.30246. doi: 10.3402/iee.v6.30246
Somerville, J. E., Cassiano, L., and Darveau, R. P. (1999). Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67, 6583–6590.
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Tenaillon, O., Skurnik, D., Picard, B., and Denamur, E. (2010). The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 8, 207–217. doi: 10.1038/nrmicro2298
Torres, A. G., Zhou, X., and Kaper, J. B. (2005). Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect. Immun. 73, 18–29. doi: 10.1128/IAI.73.1.18-29.2005
Trabulsi, L. R., Keller, R., and Gomes, T. A. T. (2002). Typical and Atypical enteropathogenic Escherichia coli. Emerg. Infect. Dis. 8, 508–513. doi: 10.3201/eid0805.010385
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Uhlich, G. A. (2009). KatP contributes to OxyR-regulated hydrogen peroxide resistance in Escherichia coli serotype O157:H7. Microbiology 155, 3589–3598. doi: 10.1099/mic.0.031435-0
Walsh, T. R., and Wu, Y. (2016). China bans colistin as a feed additive for animals. Lancet Infect. Dis. 16, 1102–1103. doi: 10.1016/S1473-3099(16)30329-2
Wang, R., van Dorp, L., Shaw, L. P., Bradley, P., Wang, Q., Wang, X., et al. (2018). The global distribution and spread of the mobilized colistin resistance gene mcr-1. Nat. Commun. 9:1179. doi: 10.1038/s41467-018-03205-z
Wang, S., Liu, X., Xu, X., Yang, D., Wang, D., Han, X., et al. (2016). Escherichia coli Type III secretion system 2 ATPase EivC is involved in the motility and virulence of avian pathogenic Escherichia coli. Front. Microbiol. 7:1387. doi: 10.3389/fmicb.2016.01387
Watson, V. E., Jacob, M. E., Flowers, J. R., Strong, S. J., DebRoy, C., and Gookin, J. L. (2017). Association of atypical enteropathogenic Escherichia coli with diarrhea and related mortality in kittens. J. Clin. Microbiol. 55, 2719–2735. doi: 10.1128/JCM.00403-17
Wattam, A. R., Davis, J. J., Assaf, R., Boisvert, S., Brettin, T., Bun, C., et al. (2017). Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res. 45, D535–D542. doi: 10.1093/nar/gkw1017
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, A. R. C., Pearson, J. S., Bright, M. D., Munera, D., Robinson, K. S., Lee, S. F., et al. (2011). Enteropathogenic and enterohaemorrhagic Escherichia coli: even more subversive elements. Mol. Microbiol. 80, 1420–1438. doi: 10.1111/j.1365-2958.2011.07661.x
World Health Organization [WHO] (1987). Program for Control of Diarrheal Diseases (CDD/83.3 Rev1), in Manual for Laboratory Investigations of Acute Enteric Infections. Geneva: World Health Organization, 27.
Wyrsch, E. R., Reid, C. J., DeMaere, M. Z., Liu, M. Y., Chapman, T. A., Roy Chowdhury, P., et al. (2019). Complete sequences of multiple-drug resistant IncHI2 ST3 plasmids in Escherichia coli of porcine origin in Australia. Front. Sustain. Food Syst. 3:18. doi: 10.3389/fsufs.2019.00018
Xu, Y., Bai, X., Zhao, A., Zhang, W., Ba, P., Liu, K., et al. (2016). Genetic diversity of intimin gene of atypical enteropathogenic Escherichia coli isolated from human, animals and raw meats in China. PLoS One 11:e0152571. doi: 10.1371/journal.pone.0152571
Yao, Y., Xie, Y., Perace, D., Zhong, Y., Lu, J., Tao, J., et al. (2009). The type III secretion system is involved in the invasion and intracellular survival of Escherichia coli K1 in human brain microvascular endothelial cells. FEMS Microbiol. Lett. 300, 18–24. doi: 10.1111/j.1574-6968.2009.01763.x
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhao, H., Chen, W., Xu, X., Zhou, X., and Shi, C. (2018). Transmissible ST3-IncHI2 plasmids are predominant carriers of diverse complex IS26-Class 1 integron arrangements in multidrug-resistant Salmonella. Front. Microbiol. 9:2492. doi: 10.3389/fmicb.2018.02492
Zhao, X., Yang, J., Ju, Z., Chang, W., and Sun, S. (2018). Molecular characterization of antimicrobial resistance in Escherichia coli from rabbit farms in Tai’an, China. BioMed Res. Int. 2018, 8607647. doi: 10.1155/2018/8607647
Keywords: aEPEC, ST302, mobile genetic elements, IncHI2 plasmid, IncFIB, ETT2, Escherichia coli
Citation: Jarocki VM, Reid CJ, Chapman TA and Djordjevic SP (2020) Escherichia coli ST302: Genomic Analysis of Virulence Potential and Antimicrobial Resistance Mediated by Mobile Genetic Elements. Front. Microbiol. 10:3098. doi: 10.3389/fmicb.2019.03098
Received: 08 September 2019; Accepted: 20 December 2019;
Published: 21 January 2020.
Edited by:
Costas C. Papagiannitsis, University of Thessaly, GreeceReviewed by:
Frederic Auvray, Ecole Nationale Veterinaire de Toulouse, FranceAriadnna Cruz-Córdova, Children’s Hospital of Mexico Federico Gómez, Mexico
Copyright © 2020 Jarocki, Reid, Chapman and Djordjevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven P. Djordjevic, c3RldmVuLmRqb3JkamV2aWNAdXRzLmVkdS5hdQ==
 Veronica M. Jarocki
Veronica M. Jarocki Cameron J. Reid
Cameron J. Reid Toni A. Chapman
Toni A. Chapman Steven P. Djordjevic
Steven P. Djordjevic