- 1Department of Microbiology and Fermentation Technology, CSIR-Central Food Technological Research Institute, Mysuru, India
- 2DBT-AIST International Centre for Translational and Environmental Research and Bioinformatics Centre, Department of Microbiology, School of Life Sciences, Sikkim University, Gangtok, India
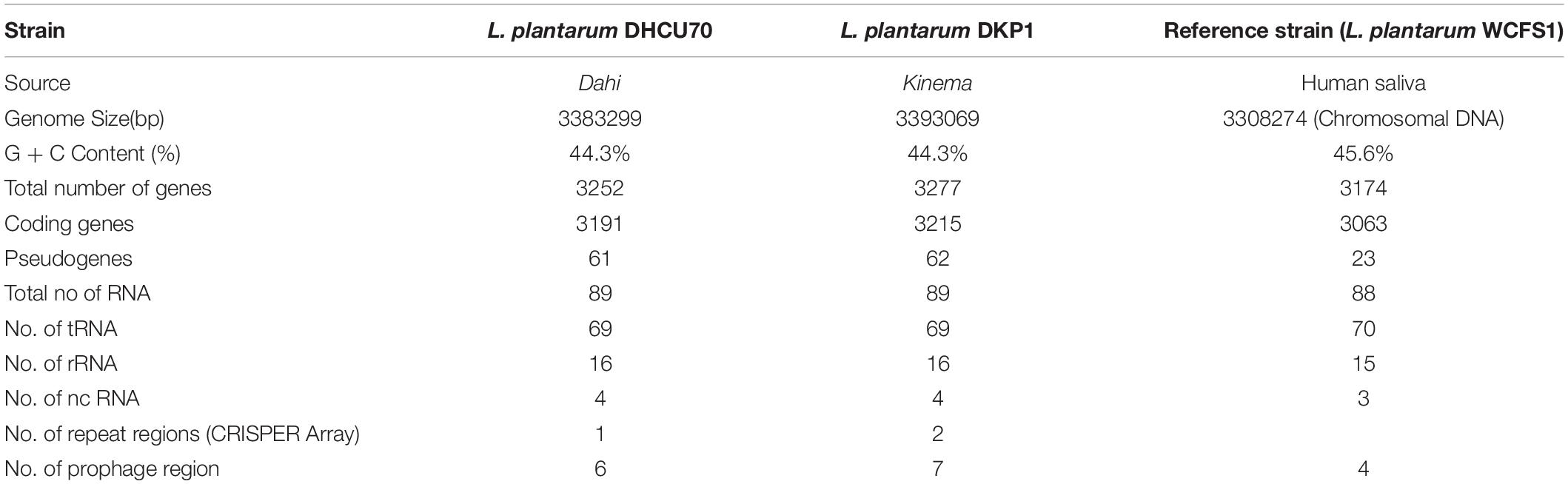
In this study, Lactobacillus plantarum strain DHCU70 isolated from dahi, a fermented milk product and L. plantarum strain DKP1 isolated from kinema, a fermented soybean food of India, respectively were evaluated for their bacteriocin production and probiotic properties. Both strains of L. plantarum (DHCU70 and DKP1) were found to have potent antimicrobial activity against Kocuria rhizophila ATCC 9341. Bacteriocin produced by L. plantarum strains DHCU70 and DKP1 did not exhibit inhibition of cell wall, DNA and fatty acids biosynthesis mechanisms as evaluated by whole cell reporter assays. We characterized the bacteriocin encoding genes in L. plantarum strains DHCU70 and DKP1 by whole genome sequence which consisted of a single and circular chromosome with genome size of 3.38 Mb (GC content of 44.3%) and 3.39 Mb, respectively and a GC content of 44.3%. L. plantarum DHCU70 has 3252 number of protein encoding genes comprising 89 number of RNA genes (69tRNA, 16rRNA, 4nc RNA) whereas L. plantarum DKP1 has total of 3277 number of protein encoding genes with 89 number. of RNA genes (69tRNA, 16S rRNA, 4nc RNA). Analysis revealed the presence of 20.5 kb long and 23 numbers of plantaricin encoding locus (pln locus) for production of antimicrobial compound. BAGEL analysis has shown that the pln locus of both the strains of L. plantarum showed maximum sequence similarity with plantaricin NC8 of L. plantarum NC8, originally isolated from grass silage. Annotated whole genome sequence of both strains DHCU70 and DKP1 was analyzed for the presence of probiotic marker genes. The probiotic properties of these strains of were also evaluated in vitro. Due to the presence of genes responsible for antimicrobial activity and probiotic properties, both strains of L. plantarum may be considered as a suitable probiotic candidate in food industry.
Introduction
Fermented foods and beverages have several functional microorganisms including many species of lactic acid bacteria (LAB) which are well known for antimicrobial and probiotic properties (Tamang et al., 2016b; Rezac et al., 2018). LAB are certified as generally regarded as safe (GRAS) bacteria granted by the American Food and Drug Agency (FDA)1. Most of the genera belong to LAB such as Lactococcus, Lactobacillus, Leuconostoc, Pediococcus, and few Streptococcus have also received the Qualified Presumption of Safety (QPS) status by European Food Safety Authority (EFSA)2. LAB mostly act as protective cultures due to the production of antimicrobial compounds bacteriocin, nisin, enterocin, etc. (Gaggia et al., 2011; Tamang et al., 2016a) which are present in many fermented dairy and vegetable (Tamang et al., 2009; Jiang et al., 2012; Grosu-Tudor and Zamfir, 2013; Mokoena, 2017; Silva et al., 2018). Among the various species of Lactobacillus, L. plantarum is well known for its probiotic application due to its natural habitation in human gastro intestinal tract (Zhang et al., 2012) and has several functional properties such as antioxidant activity, anti-cholesterol effect, bio-protective and immune-modulation (Ren et al., 2014; Devi et al., 2015, 2016; Tamang et al., 2016a). L. plantarum has both narrow and broad spectra of antibacterial activity against Gram-positive pathogenic bacteria viz Listeria monocytogenes, Staphylococcus aureus and also against Gram-negative pathogenic bacteria such as Salmonella Typhimurium, Klebsiella, Rhizophila, E. coli, Aeromonas hydrophila, and Yersinia (Behera et al., 2018; Silva et al., 2018; Spangler et al., 2019).
Lactobacillus plantarum produces antimicrobial compound known as plantaricin (Song et al., 2014), which has a bactericidal mode of action against other closely related microorganisms by dissipating the proton motive force or by creating the pores in the cell membrane that permits the efflux of relatively large molecules (Todorov, 2009). Most of the experiments on the mode of action is being done by conventional microbiological methods involving screening of antimicrobial compound by agar well diffusion assay, which are tedious and unwieldy (Balouiri et al., 2016). Nowadays, the application of high-throughput screening (HTS)with targeted cell-based assays, that carry reporters such as β-galactosidase or luciferase genes (Shobharani and Halami, 2016) for microbial compounds, is getting more popular and reliable (Baker et al., 2016).
Different strains of L. plantarum are genetically diverse with respect to genome size, number of proteins and diversity in plantaricin encoding locus (Devi and Halami, 2019). The genome size of L. plantarum varies from 3.0 to 3.3 Mb (Li et al., 2015). L. plantarum P8 (Accession no. NC_021224) has genome size of 3 Mb with total no of 2892 protein encoding genes whereas L. plantarum WCFS1 has genome size of 3.3 Mb with 3057 number of total protein encoding genes (Accession no. NC 004567). L. plantarum ZJ316 (Accession no. NC_020229) found to have 3159 number of protein encoding genes. Devi and Halami (2019) characterized the plantaricin produced by L. plantarum in different plantari-types based on presence or absence of various pln genes. Pln locus of plantaricin encoding genes is genetically organized in either simple or complex operon (Diep et al., 2009). Plantaricin 423, S and W are found on one simple operon (Nissen-Meyer et al., 2009) whereas, L. plantarum C11, WCFS1, JDM1, J23, J51 and NC8 strains are organized in complex pln locus consisting of 25–28 genes in the mosaic-like structure of 5–6 operons encoding Class IIb bacteriocin (Diep et al., 2009; Tsapieva et al., 2011; Tai et al., 2015). All bacteriocin related information collected from bacteriocin databases, that have been created to compile the increasing number of bacteriocins characterized from both Gram-positive and Gram-negative bacteria, may be used for the automated screening of bacteriocin gene clusters (Blin et al., 2013; Van Heel et al., 2013).
Some species of Lactobacillus isolated from ethnic fermented foods of Sikkim in India were reported to have antimicrobial activities (Tamang et al., 2005, 2008, 2009; Dewan and Tamang, 2007; Tamang and Tamang, 2009). However, there has been no report of whole genome sequencing of Lactobacillus species, isolated from ethnic fermented foods of Sikkim in India, targeting the bacteriocin producing gene for characterization of bacteriocin and identification of probiotic genes. The present work aims to study the whole genome sequence of Lactobacillus strains isolated from some ethnic fermented foods of Sikkim, India to characterize the bacteriocin producing gene and evaluation of its probiotic properties.
Materials and Methods
Bacterial Strains and Culture Condition
Different strains of LAB were isolated from two common ethnic fermented foods of Sikkim in India viz. kinema, fermented soybean food (Tamang, 2015) and dahi, fermented milk product (Rai et al., 2016), respectively. Isolates were sub-cultured in de Man, Rogosa and Sharpe (MRS) media (HiMedia, Mumbai, India) and incubated at 37°C for 18 h. Kocuria rhizophila ATCC 9341, used as an indicator strain to check the antimicrobial activity of the isolates, was cultured in BHI broth (HiMedia, India) and incubated at 37°C for 24 h under aerobic conditions. Study on mode of action was carried out by using reporter strains of Bacillus subtilis BSF2470 (cell wall inhibition), AUT1 (Nisin specific reporter), yorB promoter (DNA inhibition) and fabHB (fatty acid inhibition) (Shobharani et al., 2015), which were stored at −20°C in Luria Bertani (LB) broth containing 16% (v/v) glycerol following the method of Nithya and Halami (2012). Reporter cultures were sub-cultured to antibiotics (erythromycin at a concentration of 5 mg/ml for BSF and chloramphenicol at a concentration of 5 mg/ml for all other reporters) containing fresh LB broth and incubated aerobically at 37°C, 150 rpm for 24 h (Shobharani et al., 2015).
Evaluation of the Antimicrobial Activity of L. plantarum
Previously isolated bacterial isolates from various fermented foods of North East India was evaluated for bacteriocin production by agar well diffusion method (Li et al., 2015). Cells were grown in MRS for 16–18 h and then centrifuged at 8000 rpm, 4°C for 15 min to remove the cells. The supernatant was neutralized to the pH of 7.0 with 0.1 N NaOH followed by filter-sterilization through 0.2 μm membrane. Consequently agar well diffusion assay was performed against indicator strain K. rhizophila. Briefly, 50 μL of cell free supernatants were placed into 6 mm wells on BHI agar plates seeded with the above indicator strains. After incubation at 37°C for 12 h, the diameters of inhibition zones were measured. Proteinaceous nature of antibacterial substance was checked by incubating the cell-free supernatant (CFS) with 1 mg/ml of proteinase K (Hi Media) at 37°C for 2 h (Devi and Halami, 2011). Both protease-treated CFS were assayed for activity as indicated above (Devi and Halami, 2011). CFS was also tested for heat resistance at boiling temperature for 15 min (Sure et al., 2016).
Characterization of Isolates
Isolates of LAB (DHCU70 strain from dahi and DKP1 strain from kinema) showing prominent anti bacterial activity against Kocuria rhizophila were characterized by using conventional methods involving Gram-staining, SEM analysis followed by various Biochemical tests – Catalase Test, Growth at various temperature, pH and salt concentrations(Li et al., 2015). Sugar fermentation profile was checked by incubating the cultures at 37°C for 48 h in Bromo Cresol purple broth with different sugars as described by Schillinger and Lücke (1987). Identity of cultures was confirmed by 16S rRNA gene sequencing using one set of bacterial universal primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTT ACGACTT-3′) (Lane, 1991). Subsequently, taxonomy of the cultures was confirmed by whole genome sequencing analysis.
Chromogenic Plate Assay for the Mode of Action
Study on mode of action was carried out by using whole cell reporter assay (Nithya and Halami, 2012). Briefly, overnight grown reporter bacteria in antibiotic containing LB Broth was sub-cultured again in LB Broth without antibiotic and incubated at 37°C aerobically till the absorbance reached to 0.7 OD600. Subsequently, LB agar plates containing reporter bacterial strain supplemented with 50 μg/ml of X-Gal were prepared. Then isolates were spotted on the LB agar plate and incubated for 24–36 h at room temperature until a blue coloration was observed due to the induction of lacZ resulting in the production of β-galactosidase.
DNA Extraction
Extraction and purification of genomic DNA of isolates was carried out using Qiagen DNeasy blood and tissue kit (Qiagen, Hilden, Germany). The concentration and purity of genomic DNA was quantified by previously calibrated Nano-drop spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States) and Qubit fluorometer (Invitrogen, United States).
Genome Sequencing
Whole genome sequencing of two tentatively identified L. plantarum cultures: DHCU70 and DKP1 was carried out following the method of Tanizawa et al. (2015) using Illumina Miseq 300 × 2 Platform (Illumina, San Diego, CA, United States) at the facility of Genotypic Technology Pvt. Ltd., Bangalore. A total number of 55143 and 567922 reads were obtained for L. plantarum DHCU70 and DKP1, respectively, with N50 value 135,312 and 135,926, respectively.
Bioinformatics Analysis
High-quality reads were assembled in contigs using SPADES 3.9.1 assembler. Later the order of contigs was determined by aligning the contigs with the genome sequence of originally published strain L. plantarum Wcfs1 (GenBank Accession no. AL935263). Gene prediction for assembled genome was carried out using Genmark which were annotated by similarity searched against UniProt bacterial protein database using DIAMOND BLAST with an e-value of1e-5 for gene ontology and annotation (Buchfink et al., 2015). Prophage insert regions were detected with an on-line phage search tool, PHASTER (Arndt et al., 2016). The CRISPR regions were identified with a CRISPR on-line detection tool, CRISPR finder (Grissa et al., 2007). Information about plasmids was obtained by using online tool Plasmid Finder.
Phylogenetic Analysis
The genetic presence and comparison of 16S rRNA and recA genes were performed by analyzing the nucleotide sequence data available at National Center for Biotechnology Information (NCBI) database. The 16S rRNA and recA gene sequences of related organisms were obtained from NCBI database and compared with the sequences of our L. plantarum strains to know the closest neighbor in the evolutionary tree. Neighbor-joining (NJ) phylogenetic tree with p-distance model was constructed by MEGA 6 software (Tamura et al., 2011).
Identification of Bacteriocin Encoding Genes
Different pln genes were identified in WGS of L. plantarum strains DHCU70 and DKP1 by sequence similarity search using BLASTP and compared with already known plantaricin (Diep et al., 2009). Subsequently, organization of plantaricin encoding genes of individual strain of L. plantarum was analyzed using bacteriocin database BAGEL4 (Van Heel et al., 2013).
Probiotic Functionalities
Identification of Probiotic Genes
The sequence information for different probiotic genes (Lebeer et al., 2008) of related strain was obtained from the NCBI database and used to find out the probiotic genes present in our L. plantarum strains by sequence similarity search using BLASTP.
Acid and Bile Salt Tolerance
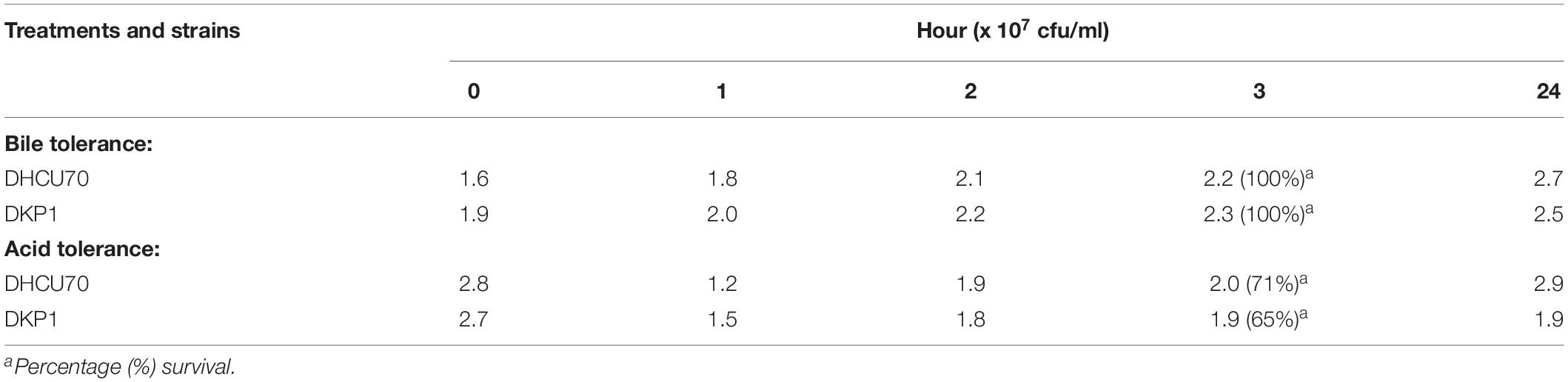
Acid and bile tolerance of L. plantarum cultures was evaluated as described by Archer and Halami (2015). For acid tolerance MRS broth was adjusted to pH 3 with 1N HCl; and for bile salt tolerance test, 0.3% (w/v)bile salts (MP Biomedicals, India Pvt. Ltd.) was added to MRS broth. The broth with adjusted pH values and bile salt concentration were inoculated with 109CFU/mL of O/N grown cultures of L. plantarum DHCU70 and DKP1 strains and incubated at 37°C. Each tube containing 1 ml of culture was taken at 0, 1, 2, 3, and 24 h interval and absorbance was measured at 600 nm. Subsequently cultures were serially diluted in 0.8% saline water and plated on MRS agar followed by incubation at 37°C for 48 h. The viable bacterial cell counts in terms of the colony forming units (cfu/ml) were recorded after 24 h. All the experiments were repeated twice. MRS broth with neutral pH 7.0 and without bile was served as a control, respectively.
DPPH Radical-Scavenging Assay
Antioxidant activity was measured by DPPH assay as described by Son and Lewis (2002) and Archer and Halami (2015). Briefly overnight grown cultures were centrifuged at 8000 rpm for 10 min at 4°C and the CFS was collected. 100 μl of CFS was mixed with 1.9 ml of methanol. Later, 2 ml of 2,2-diphenyl-1-picrylhydrazyl (DPPH) (6 mg/100 ml of methanol)was added to CFS. DPPH without addition of CFS was used as control while only methanol was used as blank. The tubes were mixed properly and incubated at room temperature for 30 min in dark. After incubation absorbance was measured at 517 nm and DPPH activity was measured by following formula:
Cell Surface Hydrophobicity
Bacterial adhesion to hydrocarbons was determined according to the method described by Rosenberg et al. (2006). Bacterial cells were grown in MRS broth at 37°C for 18 h and centrifuged at 8000 rpm for 10 min. The cell pellets were washed twice with phosphate buffer, pH 7.0, resuspended in phosphate buffer and the initial absorbance was adjusted to 0.7 OD at 600 nm (Abi). Cell suspension was mixed with n-hexadecane or xylene (3:1), vortexed and incubated at 37°C for 10 min. The mixture was again vortexed and kept at 37°C for 1 h for phase separations. The aqueous phase was removed gently and its absorbance (AbF) was measured at 600 nm. The surface hydrophobicity (%) was calculated as per the following formula:
Cellular Auto-Aggregation
Auto-aggregation was performed as described by Archer and Halami (2015). The 5 ml of cultures were mixed properly and incubated at 15°C for 2–3 h. One ml of upper suspension was taken from undisturbed incubated tube; OD was measured at 600 nm and percentage auto aggregation was calculated as follow:
Results and Discussion
Evaluation of Antimicrobial Activity
We isolated 683 bacterial isolates from various fermented foods of North East India, out of which 129 lactic bacteria bacterial isolates were found to produce an antimicrobial compound (data not shown). All 129 isolates were phenotypically characterized on the basis of physiological and biochemical tests out of which, only seven isolates were tentatively identified as Lactobacillus plantarum. Out of seven strains of L. plantarum, only two strains- DHCU70 isolated from dahi, a fermented milk product of Sikkim and DKP1 isolated from kinema, a fermented soybean food of Sikkim, India, showed prominent inhibition zones against Kocuria rhizophila ATCC 9341for untreated CFS, CFS at acidic pH, CFS at basic pH and CFS treated at boiling temperature (Supplementary Table S1 and Supplementary Figure S1). Antimicrobial activity of both strains of L. plantarum was found to be stable at boiling temperature, acidic (pH 3) and basic (pH 9) (Supplementary Table S1 and Supplementary Figure S1). Identification of Lactobacillus plantarum strains DHCU70 and DKP1 was confirmed by 16S rRNA gene sequences (Figure 1A). Antimicrobial compound produced by L. plantarum strains DHCU70 and DKP1 was found to be proteinaceous in nature, as when CFS was treated with proteinase K it showed complete loss of antimicrobial activity (Yang et al., 2012).
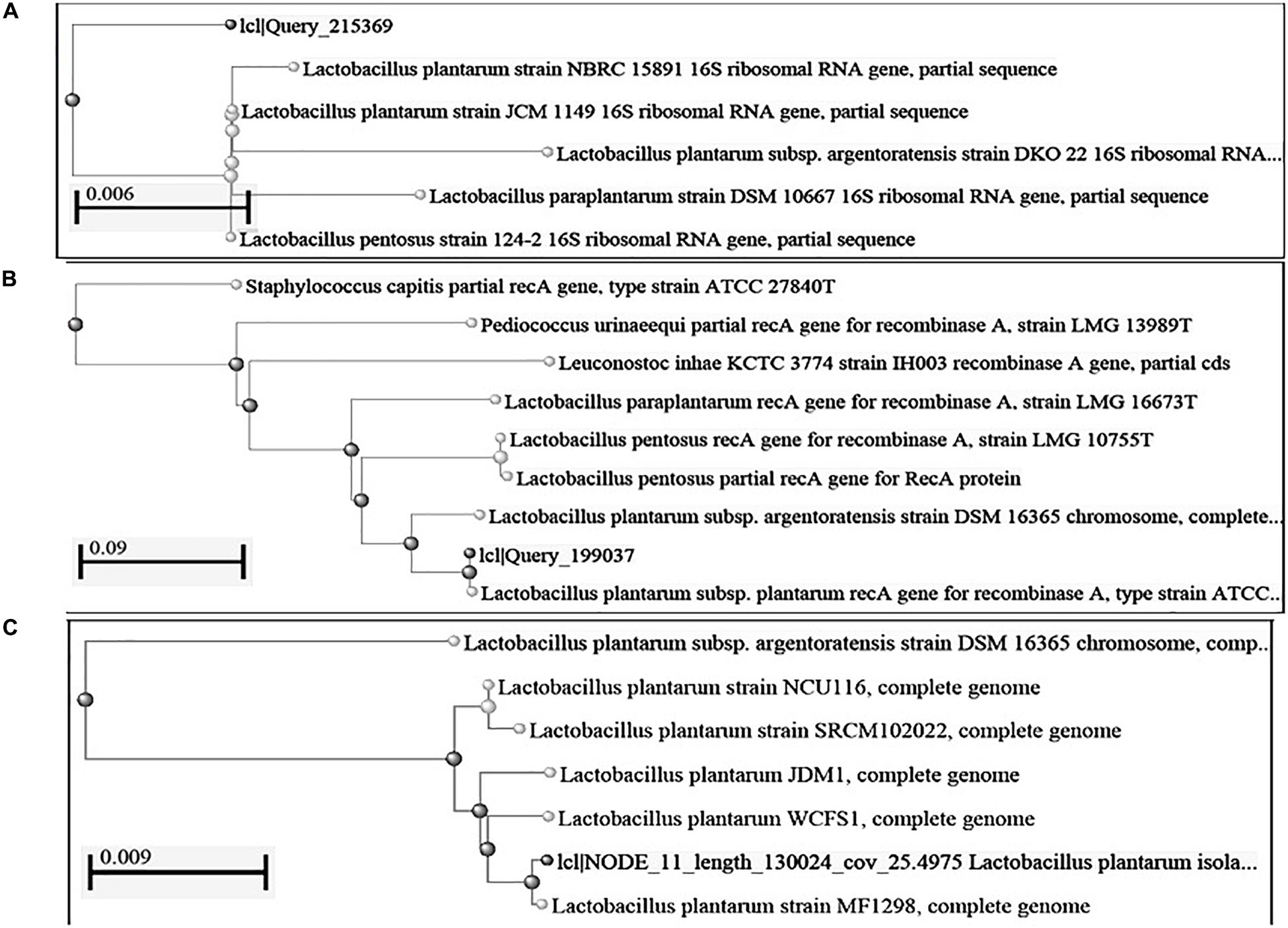
Figure 1. Construction of phylogenetic tree using NCBI Database by Neighbor-Joining method (A) 16S rRNA gene; (B): recA gene sequence; (C): complete genome sequence.
Similar observations were earlier reported on broad spectrum of activity of plantaricin LR14 produced by L. plantarum against Kocuria rhizophila, Listeria monocytogenes, Salmonella, Yersinia, enterocolitica, Bacillus lichniformis, and E. coli (Tiwari and Srivastava, 2008). Plantaricin LPL-1 isolated from fermented fish was active against S. aureus, Listeria. monocytogenes, B. pumilus, B. amyloliquefaciens, E. faecalis, L. plantarum, L. delbrueckii, L. bulgaricus, L. salivarius, and L. lactis (Wang et al., 2018). L. plantarum strains isolated from fermented vegetables products of India showed antibacterial activity against K. rhizophila (Devi and Halami, 2019). Besides, antibacterial activity plantaricins also showed thermo-stability (Behera et al., 2018; Lim et al., 2018).
The mode of actions was carried out by using whole cell reporter assay. L. plantarum strains DHCU70 and DKP1 induced the production of β-galactosidase in reporter strains of Bacillus subtilis BSF2470 specific for cell wall inhibition (Figure 2). Bacteriocin produced by DHCU70 and DKP1 strains of L. plantarum did not show any positive response toward DNA specific reporter (yorB) as well as fatty acid specific reporter (fabHB) indicating that bacteriocin produced by both the strains of L. plantarum did not act on DNA and fatty acids (data not shown). So it may be concluded that bacteriocin produced by DHCU70 and DKP1 acts through inhibition of cell wall. In general, class II bacteriocins show bactericidal mode of action by dissipating the proton motive force by disrupting the trans-membrane potential or pH gradient of indicator strains (Todorov, 2009). Specifically in class IIb bacteriocins, two peptide bacteriocin acts by creating pores which results in dissipating trans membrane potential (Zhang et al., 2015). As most of the L. plantarum known till date found to act on cell membrane (Arena et al., 2019). Interestingly, L. plantarum strains DHCU70 and DKP1 did not show positive result on Bacillus subtilis BSF2470 reporter. So we repeated the assay with concentrated bacteriocin preparation and observed that the AMC produced by L. plantarum strains are acting on BSF2470 reporter (Figure 2). These results found to be consistent in every repetition.
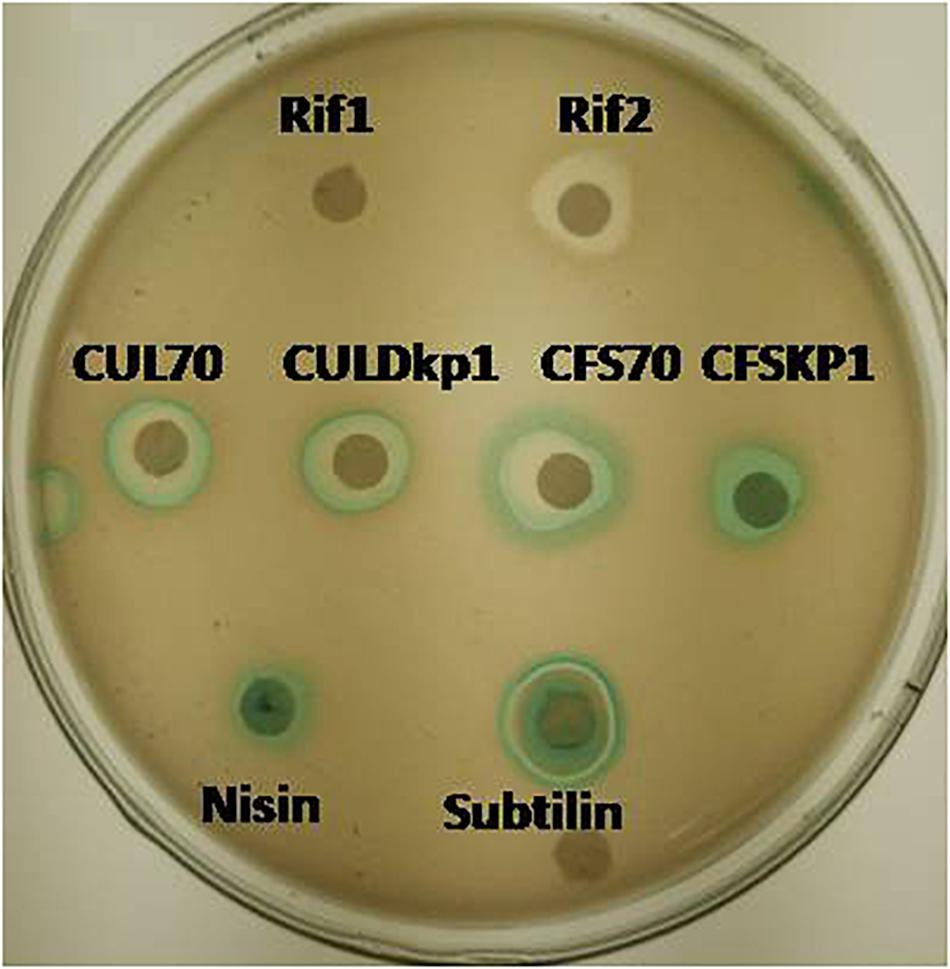
Figure 2. Mode of action studies using whole cell reporter assay for Lactobacillus plantarum strains DHCU70 and DKP1. Indicators: Rif1-Rifampicin (–ve control) 1 μg/ml, Rif2-Rifampicin (–ve control) 0.1 μg/ml, CUL70- culture of dhcu70, CULDKP1- culture of DKP1, CFS70 – Cell filtrate supernatant of DHCU70, CFSDKP1 – cell filtrate supernatant of DKP1; Nisin and Subtilin (+ve control).
Genome Analysis of L. plantarum
We obtained a complete genome sequence of Lactobacillus plantarum strains DHCU70 and DKP1, which showed a single circular chromosome (Figures 3A,B). Both strains shared maximum sequence similarity with L. plantarum strain WCFS1 (Kleerebezem et al., 2003) having a genome size of 3.38 and 3.39 Mb, and G + C content of 44.3%, respectively (Table 1). A total of 3252 and 3277 protein-coding sequences (CDSs) were identified in L. plantarum strains DHCU70 and DKP1, respectively, The 3191 and 3215 number of proteins present in strains DHCU70 and DKP1, respectively, were found to be functionally categorized among the predicted coding sequence for protein. Kleerebezem et al. (2003) have noted 3052 protein encoding sequences in L. plantarum strain Wcfs1. Both the strains of L. plantarum have no plasmid showing its significance in transformation and conjugation experiments in our study. Aukrust and Blom (1992) used L. plantarum and L. sake strains isolated from meat and vegetable for transformation. Similarly, Shrago et al. (1986) transferred erythromycin resistant streptococcal plasmid pAM β1 to L. plantarum via conjugation. The chromosome of L. plantarum strains DHCU70 and DKP1 contained 89 RNA genes including 69 tRNA, 16 rRNA and 4 ncRNA. Many strains of L. plantarum isolated from fermented vegetables and milk products have already been sequenced which include L. plantarum K25 from Tibetan Kefir (Jiang et al., 2018), L. plantarum SK151 from kimchi (Amoranto et al., 2018), and L. plantarum LL441 from dairy cheese (Flórez and Mayo, 2018) having genome size of 3.1 Mb (GC content of 44.6%), 3.2 Mb (GC content of 44.6%), 3.1 Mb (GC content of 44.5%), respectively. Zhang et al. (2009) reported 62 tRNA and 16 rRNA encoding genes in L. plantarum JDM1. However in our study we characterized bacteriocin encoding genes in L. plantarum strains DHCU70 and DKP1 through whole genome sequencing.
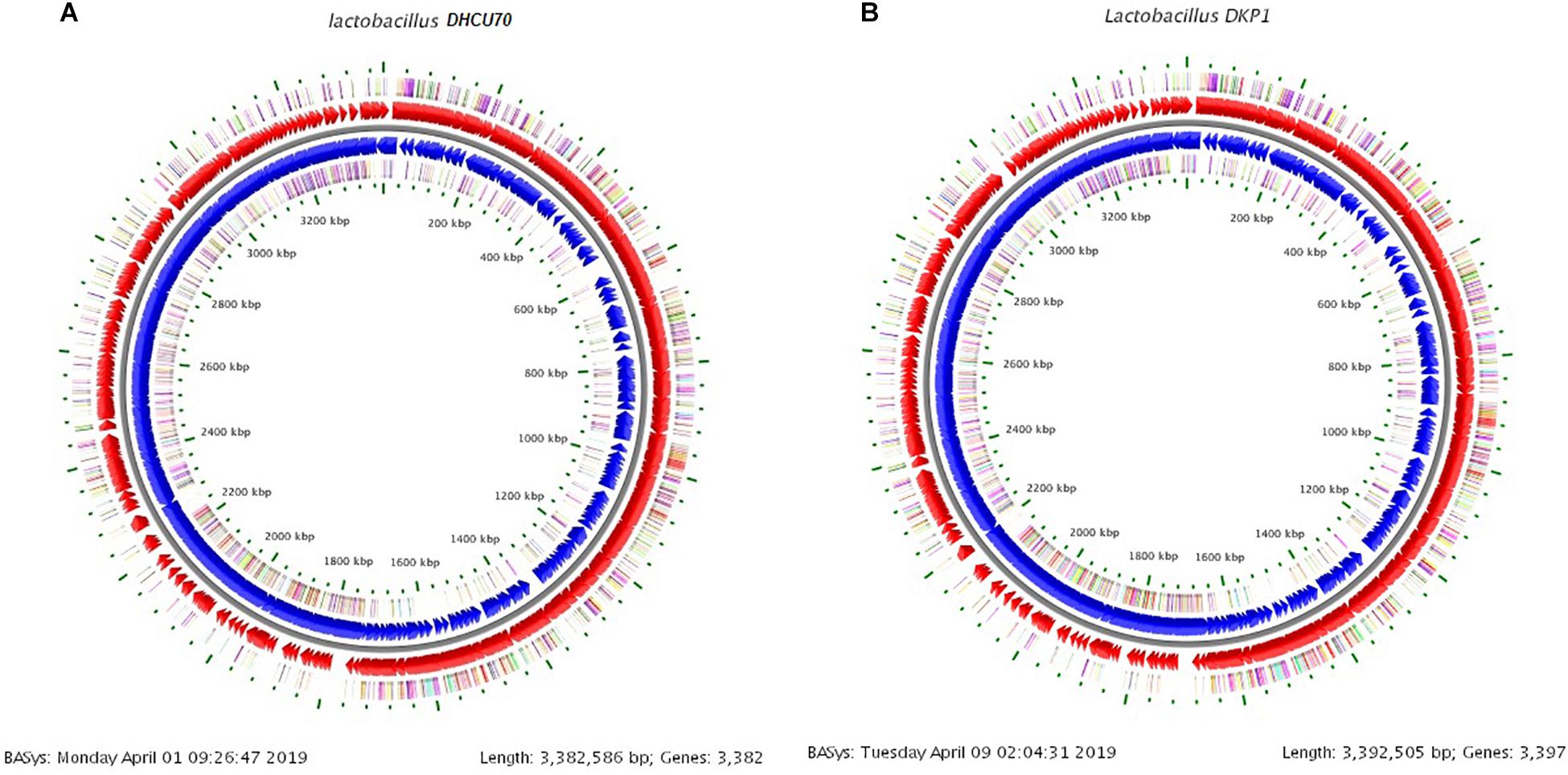
Figure 3. (A) Circular genome map of Lactobacillus plantarum DHCU70; (B) Circular genome map of Lactobacillus plantarum DKP1.
Based on the phylogenetic analysis of 16S rRNA gene and recA gene (Devi et al., 2016) through BLASTn, strains DHCU70 and DKP1 were found to display more than 99% similarity with L. plantarum group strains (Figure 1B). However, genomes of different L. plantarum group strains were difficult to distinguish by 16S rRNA gene sequence similarity since current taxonomy of L. plantarum group has closely related with species of L. paraplantarum, L. pentosus, L. arixonensis, L. plantarum subsp. plantarum, L. plantarum subsp. argentoratensis, L. xiangfangensis, and L. fabifermentans (Torriani et al., 2001; Kostinek et al., 2005; Devi et al., 2016). Phylogenetic tree was constructed based on the whole genome sequence to understand the phylogenetic relationship among L. plantarum strains which showed a close relationship with L. plantarum subsp. plantarum strain WCFS1 and strain MF1298 (Figure 1C). This observation suggests that the strains DHCU70 and DKP1 belonged to L. plantarum subsp. plantarum.
Characterization of Bacteriocin Locus
Screening of the entire genome of L. plantarum strains DHCU70 and DKP1 revealed that bacteriocin encoding locus (pln locus) was located in a 20.5 kb long region consisting of 23 genes (pln) organized in operon-like structure (Figure 4). Strains DHCU70 and DKP1 have genes encoding two peptides plantaricin plnJK (classIIb), plnEF (classI) and inducible classII plantaricin NC8βα. The presence of two pln loci encoding plantaricins from two different classes contributes to a broad inhibitory spectrum of L. plantarum (Tai et al., 2015). Precursor peptides of NC8α and NC8β were made up of 47 and 55 amino acids, respectively, comprising leader sequences of the double-glycine type at N-terminal (Maldonado et al., 2003). However, the mature alpha and beta peptides contained 29 and 34 amino acids, respectively. Both the strains have a regulatory operon which included the inducing peptide encoding genes plnc8IF and histidine protein kinase plnc8K, as reported earlier in L. plantarum NC8 (Maldonado et al., 2003). The presence of plantaricin secretary genes plnG and plnH has also been confirmed in L. plantarum strains DHCU70 and DKP1 (Table 2), which are involved in the ABC transport system (Rizzello et al., 2014). The pln locus of the strains appeared to form several operons. Their production is regulated by secreted peptide pheromone, a membrane-located sensor and transcription regulators (Diep et al., 2009). Comparative analysis of pln locus with other known pln locus showed that both L. plantarum strains DHCU70 and DKP1 matched with class IIb two peptide antibiotics with L. plantarum NC8 as the closest neighbor (Axelsson et al., 2012).
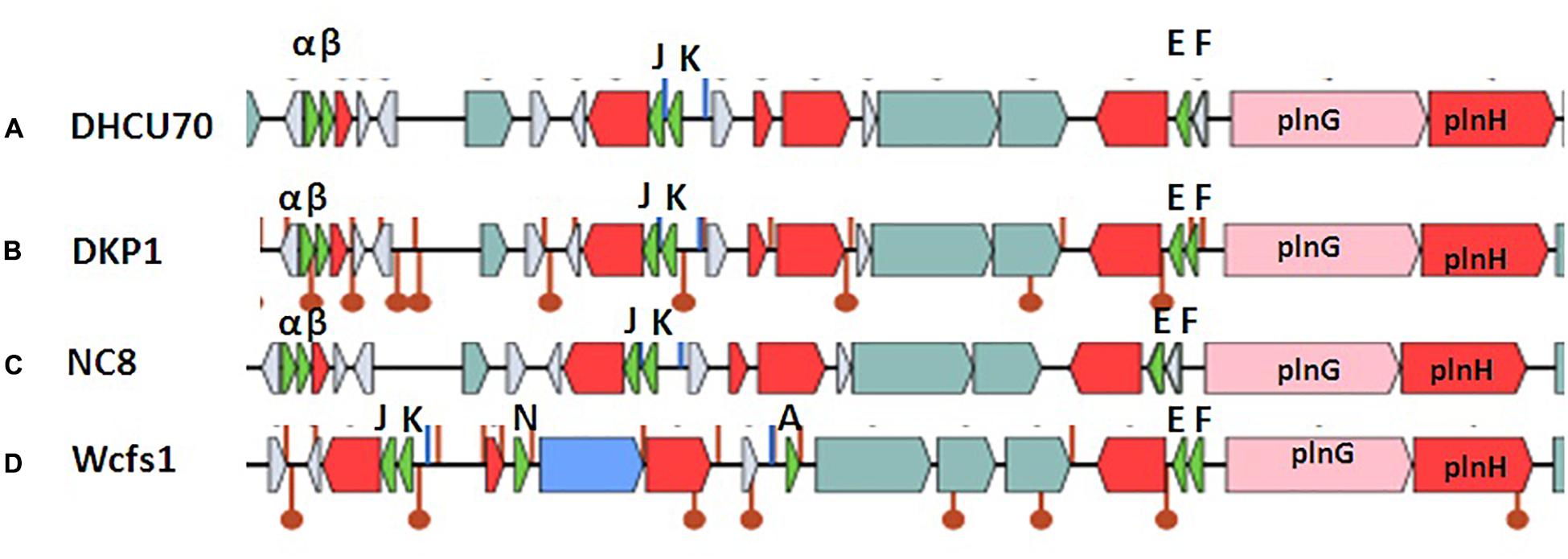
Figure 4. Genetic organization of pln locus of (A): Lactobacillus plantarum DHCU70; (B) L. plantarum DKP1; (C): Lactobacillus plantarum NC8; (D): Lactobacillus plantarum WCFS1; Indicators: α – NC8α, β– NC8β, J – pln J, K- plnK, E-plnE, F-plnF, Red color indicated immunity protein.
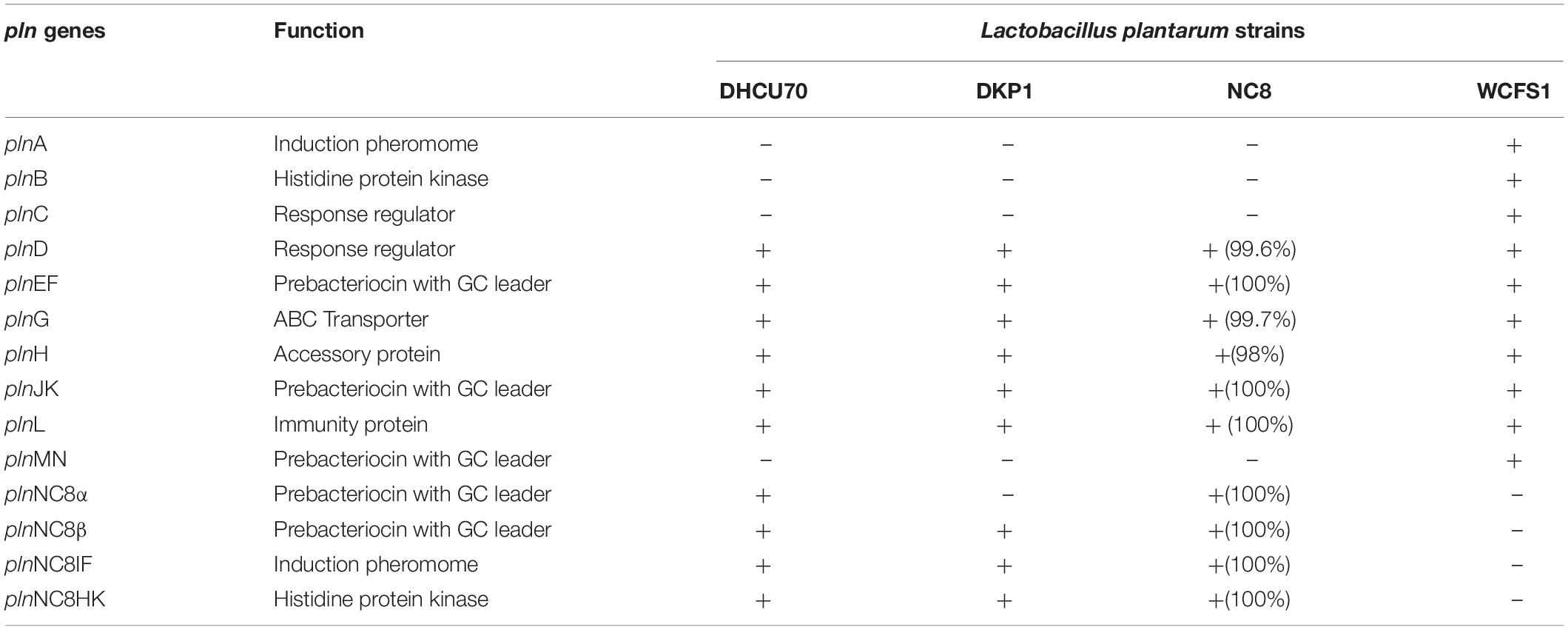
Table 2. Comparison of pln genes of Lactobacillus plantarum strains DHCU70 and DKP1 with reference strains (NC8 and WCFS1).
Lactobacillus plantarum strains were reported to have diversity in pln locus. Lb plantarum strain C11 is found to be organized in five operons-regulatory operon plnABCD, plnEFI, plnJKLR, plnMNOP, plnGHSTUVWYXY (Daeschel et al., 1990). Whereas, organization of pln operon in L. plantarum strain NC8 was found to be similar to that of C11 with different regulatory operons consisting of plnNC8IF, plnNC8HK and plnD (Maldonado et al., 2004). L. plantarum type strains J23 and J51 also shows common feature of both strains C11 and NC8 with new ORFs (Rojo-Bezares et al., 2008; Navarro et al., 2008).
Probiotic Characters
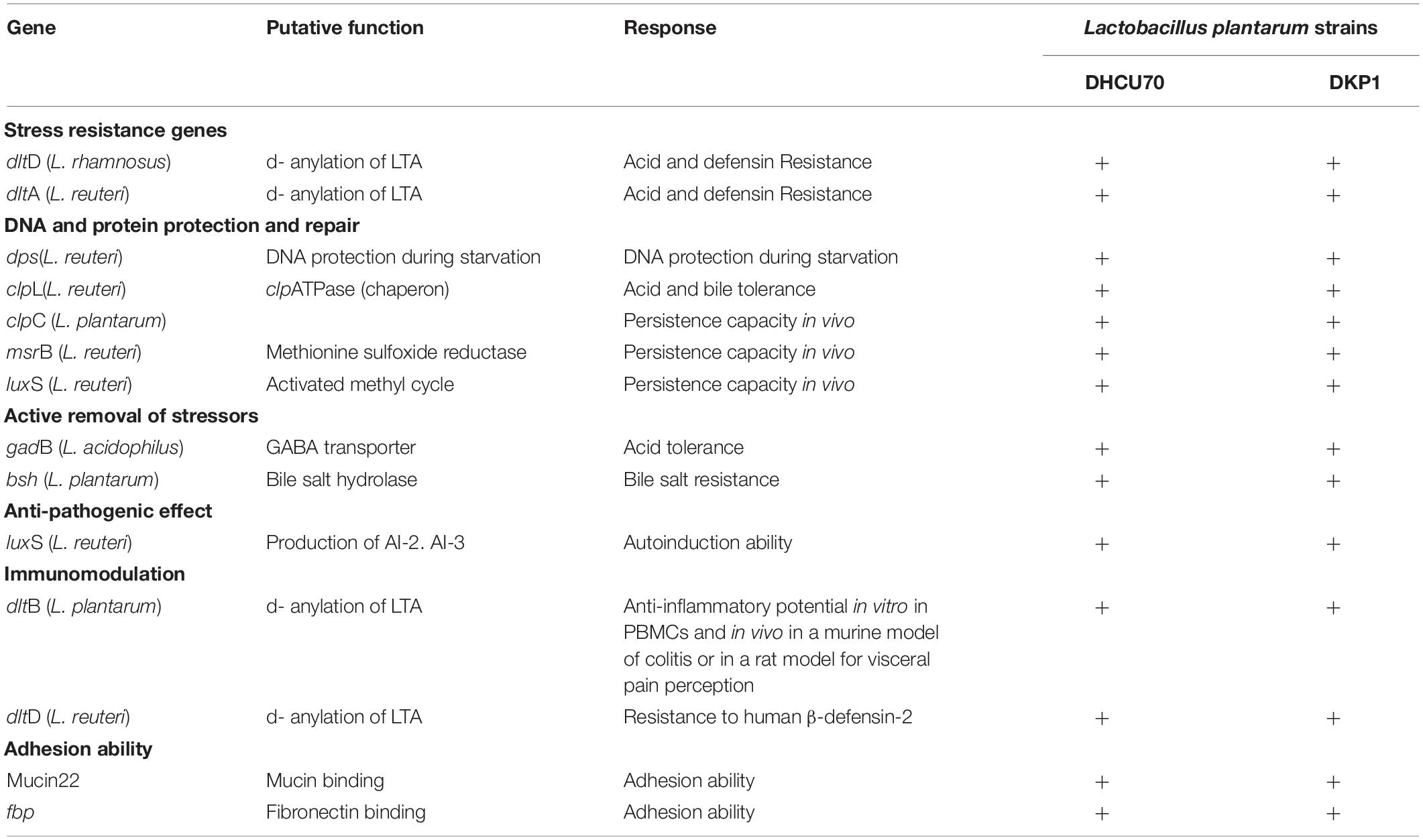
We analyzed various probiotic genes responsible for stress resistance, active removal of stressors, bile salt hydrolase activity, adhesion ability and immunomodulatory activity to find out the probiotic potential of bacteriocin producing L. plantarum strains DHCU70 and DKP1 (Table 3). Both L. plantarum strains DHCU70 and DKP1 were found to have dltA&D and gadB genes, through whole genome sequencing analysis (Figure 3), which are responsible for acid tolerance, bsh gene for bile tolerance, clpL gene for acid and bile tolerance and dltB gene for anti-inflammatory potential (Yunes et al., 2016; Bustos et al., 2018). We have also identified the genes Mucin22 and fbp responsible for adhesion ability to the intestinal epithelial layer (Turpin et al., 2012), probably to exclude the pathogenic species (Garcia-Gonzalez et al., 2018). Probiotics attributes shown by L. plantarum strains DHCU70 and DKP1 may be responsible for the potential capability to survive in vitro environmental stresses and in vivo human GIT conditions (Varankovich et al., 2015). L. plantarum strains isolated from different plant or animal (milk)-origins have antioxidant activity, probiotic effect, and protein fortification (Behera et al., 2018). Probiotic Lactobacillus shows probiotic factors such as stress response and adherence ability (Lebeer et al., 2008), and also to adaptation factors which include microbe-microbe interaction, epithelial barrier protection and immune modulatory effects (Llewellyn and Foey, 2017; Song et al., 2018).
Lactobacillus plantarum strains DHCU70 and DKP1 were subjected to acid tolerance at bile tolerance and pH3. Strains DHCU70 and DKP1 were found to have high tolerance rate to bile at 3 h (Figure 5A and Table 4) and pH 3 (Figure 5B and Table 4). Strain DHCU70 also showed growth in the given conditions to tolerate the stress till 24 h. The pH of human stomach varies from 1.5 (before food) to 5.0 (after food) due to the secretion of gastric juices (Cotter and Hill, 2003). Small intestine also receives various ranges of bile from liver (Staley et al., 2017). So, it is important for a probiotic bacterium to tolerate such conditions in order to survive in human GIT tract. Our findings showed that both the cultures are able to withstand the acidic and bile conditions of human GIT tract.
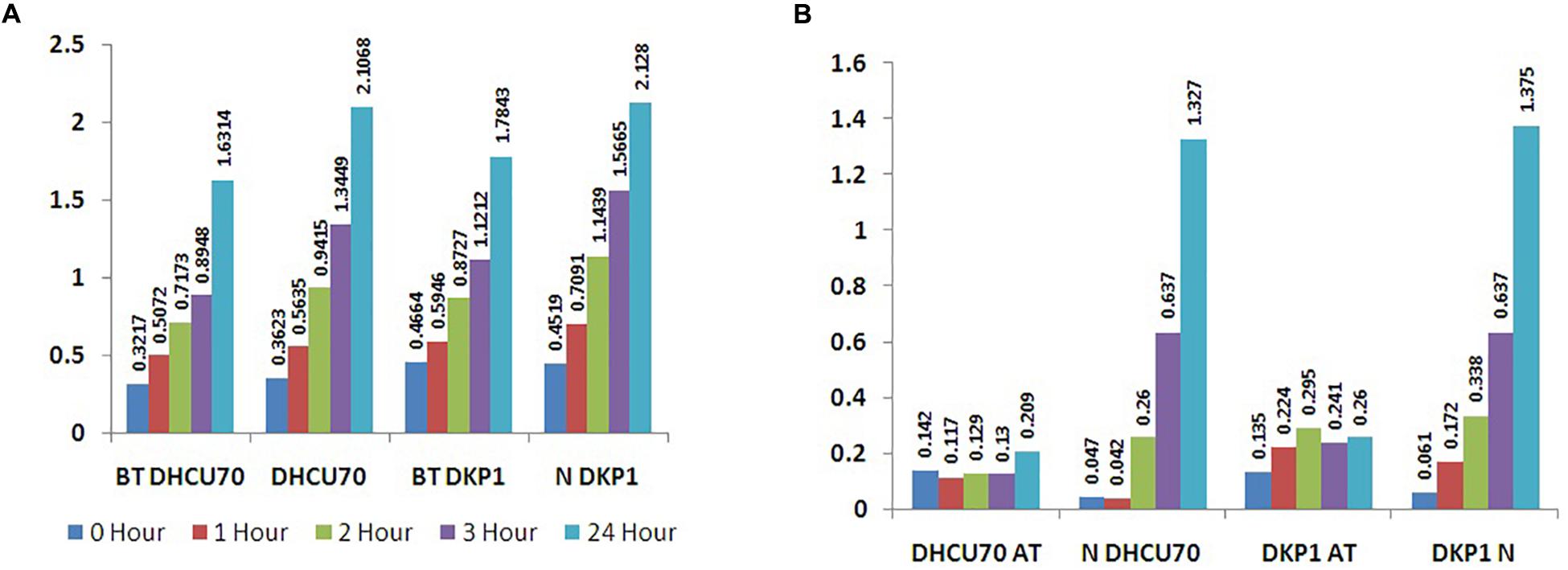
Figure 5. (A) Bile tolerance of L. plantarum strains DHCU70 and DKP1; (B) Acid tolerance of L. plantarum strains DHCU70 and DKP1.
Lactobacillus plantarum strains DHCU70 and DKP1 were analyzed for antioxidant activity through DPPH radical-scavenging assay and showed 77.57 and 68% antioxidant activity (Supplementary Table S2a), respectively. Many Lactobacillus strains isolated from fermented foods were reported to have antioxidant activity ranging from 40 to 90% (Kuda et al., 2014). Such probiotic isolates with high antioxidant ability helps to reduce the oxidative stress (Li et al., 2012). This suggests that our L. plantarum cultures can be used as a suitable probiotic candidate in food industry. Bacterial adhesion was evaluated using hydrocarbons like xylene and n-hexadecane. Auto-aggregation ability of strain DHCU70 was 72.84% whereas for strain DKP1 was 52.91% (Supplementary Table S2b). The percentage hydrophobicity for L. plantarum strains DHCU70 and DKP1 was 55.57% and 5.40% for xylene, respectively, and 42.9 and 40.88% for hexadecane, respectively (Supplementary Table S2c). L. plantarum shows a defense mechanism against the pathogen due to its hydrophobic and auto aggregative ability (Honey Chandran and Keerthi, 2018).
Conclusion
Lactobacillus plantarum strains DHCU70 and DKP1 isolated from fermented foods dahi and kinema, respectively, found to have inducing peptides, immunity peptide and ABC transporter proteins. Originally analyzed L. plantarum strain NC8 having NC8-type bacteriocin was isolated from grass silage. However L. plantarum DHCU70 having the same NC8 type of bacteriocin has been isolated from dairy origin, dahi (fermented milk product) which indicates that L. plantarum DHCU70 of dairy origin may have better adaptability to the GIT conditions. Moreover, the mode of action studies showed that both the strains of L. plantarum DHCU70 and DKP1 have novel mode of actions that may help in resolving the problem of antibiotic resistance. Along with antibacterial properties, strains L. plantarum DHCU70 and DKP1 are found to have probiotic genes which may help them to survive in vitro environmental stresses and in vivo human GIT conditions, indicating as potential probiotic candidates.
Data Availability Statement
Genome sequence of the L. plantarum strains DHCU70 and DKP1 was submitted to NCBI under accession numbers RZJQ00000000 and SDIE00000000, respectively.
Author Contributions
AG, PH, and JT formulated and designed the study. AG and PH performed, analyzed, and interpreted the experiments along with the preparation of the manuscript draft. JT and PH have finalized the manuscript for publication.
Funding
The authors are grateful to the Department of Biotechnology (DBT), Government of India, New Delhi for financial support. AG is grateful to UGC for the award of Junior Research Fellowship. This manuscript has been approved by the Planning, Monitoring and Coordination Department of CSIR-CFTRI, stating void no. PMC/2019-20/73.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00040/full#supplementary-material
FIGURE S1 | Antimicrobial activity under various environmental conditions. (a) AMA at pH 1, (b) AMA at pH 9, (c) AMA at 121°C, and (d) AMA of PK treated CFS.
TABLE S1 | AMA under various environmental conditions.
TABLE S2 | (a) Antioxidant activity (DPPH Assay), (b) Autoaggregation, and (c) Hydrophobicity.
Footnotes
References
Amoranto, M. B. C., Oh, J. K., Bagon, B. B., Hwang, I. C., Kim, S. H., Cho, C. S., et al. (2018). Complete genome sequence of Lactobacillus plantarum SK151 isolated from kimchi. Korean J. Microbiol. 54, 295–298. doi: 10.7845/kjm.2018.8038
Archer, A. C., and Halami, P. M. (2015). Probiotic attributes of Lactobacillus fermentum isolated from human feces and dairy products. Appl. Microbiol. Biotechnol. 99, 8113–8123. doi: 10.1007/s00253-015-6679-x
Arena, M. P., Capozzi, V., Longo, A., Russo, P., Weidmann, S., Rieu, A., et al. (2019). The phenotypic analysis of Lactobacillus plantarum shsp mutants reveals a potential role for hsp1 in cryotolerance. Front. Microbiol. 10:838. doi: 10.3389/fmicb.2019.00838
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, 16–21. doi: 10.1093/nar/gkw387
Aukrust, T., and Blom, H. (1992). Transformation of Lactobacillus strains used in meat and vegetable fermentations. Food Res. Int. 25, 253–261. doi: 10.1016/0963-9969(92)90121
Axelsson, L., Rud, I., Naterstad, K., Blom, H., Renckens, B., Boekhorst, J., et al. (2012). Genome sequence of the naturally plasmid-free Lactobacillus plantarum strain NC8 (CCUG 61730). J. Bacteriol. 194, 2391–2392. doi: 10.1128/JB.00141-12
Baker, K. R., Jana, B., Franzyk, H., and Guardabassi, L. (2016). A high-throughput approach to identify compounds that impair envelope integrity in Escherichia coli. Antimicrob. Agents Chem. 60, 5995–6002. doi: 10.1128/AAC.00537-16
Balouiri, M., Sadiki, M., and Ibnsouda, S. K. (2016). Methods for in vitro evaluating antimicrobial activity: a review. J. Pharm. Anal. 6, 71–79. doi: 10.1016/j.jpha.2015.11.005
Behera, S. S., Ray, R. C., and Zdolec, N. (2018). Lactobacillus plantarum with functional properties: an approach to increase safety and shelf-life of fermented foods. Biomed Res. Int. 2018:9361614. doi: 10.1155/2018/9361614
Blin, K., Medema, M. H., Kazempour, D., Fischbach, M. A., Breitling, R., Takano, E., et al. (2013). antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 41, 204–212.
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Bustos, A. Y., deValdez, G. F., Fadda, S., and Taranto, M. P. (2018). New insights into bacterial bile resistance mechanisms: the role of bile salt hydrolase and its impact on human health. Food Res. Int. 112, 250–262. doi: 10.1016/j.foodres.2018.06.035
Cotter, P. D., and Hill, C. (2003). Surviving the acid test: responses of Gram-positive bacteria to low pH. Microbiol. Mol. Biol. Rev. 67, 429–453. doi: 10.1128/mmbr.67.3.429-453.2003
Daeschel, M. A., McKenney, M. C., and McDonald, L. C. (1990). Bacteriocidal activity of Lactobacillus plantarum C-11. Food Microbiol. 7, 91–98. doi: 10.1016/0740-0020(90)90014-9
Devi, S. M., Aishwarya, S., and Halami, P. M. (2016). Discrimination and divergence among Lactobacillus plantarum group (LPG) isolates with reference to their probiotic functionalities from vegetable origin. Syst. Appl. Microbiol. 39, 562–570. doi: 10.1016/j.syapm.2016.09.005
Devi, S. M., Archer, A. C., and Halami, P. M. (2015). Screening, characterization and in vitro evaluation of probiotic properties among lactic acid bacteria through comparative analysis. Probiotics Antimicrob. Protein 7, 181–192. doi: 10.1007/s12602-015-9195-5
Devi, S. M., and Halami, P. M. (2011). Detection and characterization of pediocin PA-1/AcH like bacteriocin producing lactic acid bacteria. Curr. Microbiol. 63, 181–185. doi: 10.1007/s00284-011-9963-8
Devi, S. M., and Halami, P. M. (2019). Genetic variation of pln loci among probiotic Lactobacillus plantarum group strains with antioxidant and cholesterol-lowering ability. Probiotics Antimicrob. Proteins 11, 11–22. doi: 10.1007/s12602-017-9336-0
Dewan, S., and Tamang, J. P. (2007). Dominant lactic acid bacteria and their technological properties isolated from the Himalayan ethnic fermented milk products. Antonie Van Leeuwenhoek 92, 343–352. doi: 10.1007/s10482-007-9163-5
Diep, D. B., Straume, D., Kjos, M., Torres, C., and Nes, I. F. (2009). An overview of the mosaic bacteriocin pln loci from 433 Lactobacillus plantarum. Peptides 30, 1562–1574. doi: 10.1016/j.peptides.2009.05.014
Flórez, A. B., and Mayo, B. (2018). Genome analysis of Lactobacillus plantarum LL441 and genetic characterization of the locus for the lantibiotic plantaricin C. Front. Microbiol. 9:1916. doi: 10.3389/fmicb.2018.01916
Gaggia, F., Di Gioia, D., Baffoni, L., and Biavati, B. (2011). The role of protective and probiotic cultures in food and feed and their impact on food safety. Trends Food Sci. Technol. 22, 58–66. doi: 10.1016/j.tifs.2011.03.003
Garcia-Gonzalez, N., Prete, R., Battista, N., and Corsetti, A. (2018). Adhesion properties of food-associated Lactobacillus plantarum strains on human intestinal epithelial cells and modulation of IL-8 release. Front. Microbiol. 9:2392. doi: 10.3389/fmicb.2018.02392
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPR finder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, 52–57. doi: 10.1093/nar/gkm360
Grosu-Tudor, S. S., and Zamfir, M. (2013). Functional properties of LAB isolated from Romanian fermented vegetables. Food Biotechnol. 27, 235–248. doi: 10.1080/08905436.2013.811082
Honey Chandran, C., and Keerthi, T. R. (2018). Probiotic potency of Lactobacillus plantarum KX519413 and KX519414 isolated from honey bee gut. FEMS Microbiol. Lett. 365:fnx285. doi: 10.1093/femsle/fnx285
Jiang, J., Shi, B., Zhu, D., Cai, Q., Chen, Y., Li, J., et al. (2012). Characterization of a novel bacteriocin produced by Lactobacillus sakei LSJ618 isolated from traditional Chinese fermented radish. Food Control 23, 338–344. doi: 10.1016/j.foodcont.2011.07.027
Jiang, Y., Zhang, J., Zhao, X., Zhao, W., Yu, Z., Chen, C., et al. (2018). Complete genome sequencing of exopolysaccharide-producing Lactobacillus plantarum K25 provides genetic evidence for the probiotic functionality and cold endurance capacity of the strain. Biosci. Biotechnol. Biochem. 82, 1225–1233. doi: 10.1080/09168451.2018.1453293
Kleerebezem, M., Boekhorst, J., van Kranenburg, R., Molenaar, D., Kuipers, O. P., Leer, R., et al. (2003). Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. U.S.A. 100, 1990–1995. doi: 10.1073/pnas.0337704100
Kostinek, M., Specht, I., Edward, V. A., Schillinger, U., Hertel, C., Holzapfel, W. H., et al. (2005). Diversity and technological properties of predominant lactic acid bacteria from fermented cassava used for the preparation of Gari, a traditional African food. Syst. Appl. Microbiol. 28, 527–540. doi: 10.1016/j.syapm.2005.03.001
Kuda, T., Kawahara, M., Nemoto, M., Takahashi, H., and Kimura, B. (2014). In vitro antioxidant and anti-inflammation properties of lactic acid bacteria from fish intestines and fermented fish from the Sanriku Satoumi region in Japan. Food Res. Int. 64, 248–255. doi: 10.1016/j.foodres.2014.06.028
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt, and M. Goodfellow (New York, NY: John Wiley & Sons), 115–175.
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. J. (2008). Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764. doi: 10.1128/MMBR.00017-08
Li, D., Ni, K., Pang, H., Wang, Y., Cai, Y., and Jin, Q. (2015). Identification and antimicrobial activity detection of lactic acid bacteria isolated from corn Stover silage. Asian Australas. J. Anim. Sci. 28, 620–631. doi: 10.5713/ajas.14.0439
Li, S., Zhao, Y., Zhang, L., Zhang, X., Huang, L., Li, D., et al. (2012). Antioxidant activity of Lactobacillus plantarum strains isolated from traditional Chinese fermented foods. Food Chem. 135, 1914–1919. doi: 10.1016/j.foodchem.2012.06.048
Lim, J. H., Yoon, S. M., Tan, P. L., Yang, S., Kim, S. H., and Park, H. J. (2018). Probiotic properties of Lactobacillus plantarum LRCC5193, a plant-origin lactic acid bacterium isolated from kimchi and its use in chocolates. J. Food Sci. 83, 2802–2811. doi: 10.1111/1750-3841.14364
Llewellyn, A., and Foey, A. (2017). Probiotic modulation of innate cell pathogen sensing and signalling events. Nutrients 9:E1156. doi: 10.3390/nu9101156
Maldonado, A., Jiménez-Díaz, R., and Ruiz-Barba, J. L. (2004). Induction of plantaricin production in Lactobacillus plantarum NC8 after coculture with specific gram-positive bacteria is mediated by an autoinduction mechanism. J. Bacteriol. 186, 1556–1564. doi: 10.1128/JB.186.5.1556-1564.2004
Maldonado, A., Ruiz-Barba, J. L., and Jiménez-Díaz, R. (2003). Purification and genetic characterization of plantaricin NC8, a novel coculture-inducible two-peptide bacteriocin from Lactobacillus plantarum NC8. Appl. Environ. Microbiol. 69, 383–389. doi: 10.1128/AEM.69.1.383-389.2003
Mokoena, M. P. (2017). Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uro-pathogens. Molecules 22:E1255. doi: 10.3390/molecules22081255
Navarro, L., Rojo-Bezares, B., Sáenz, Y., Díez, L., Zarazaga, M., Ruiz-Larrea, F., et al. (2008). Comparative study of the pln locus of the quorum-sensing regulated bacteriocin-producing L. plantarum J51 strain. Int. J. Food Microbiol. 128, 390–394. doi: 10.1016/j.ijfoodmicro.2008.08.004
Nissen-Meyer, J., Oppegård, C., Rogne, P., Haugen, H. S., and Kristiansen, P. E. (2009). Structure and mode-of-action of the two-peptide (Class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2, 52–60. doi: 10.1007/s12602-009-9021-z
Nithya, V., and Halami, P. M. (2012). Novel whole-cell reporter assay for stress-based classification of antibacterial compounds produced by locally isolated Bacillus spp. Indian J. Microbiol. 52, 180–184. doi: 10.1007/s12088-012-0256-0
Rai, R., Shangpliang, H. N. J., and Tamang, J. P. (2016). Naturally fermented milk products of the Eastern Himalayas. J. Ethn. Foods 3, 270–275. doi: 10.1016/j.jef.2016.11.006
Ren, D., Li, C., Qin, Y., Yin, R., Du, S., Ye, F., et al. (2014). In vitro evaluation of the probiotic and functional potential of Lactobacillus strains isolated from fermented food and human intestine. Anaerobe 30, 1–10. doi: 10.1016/j.anaerobe.2014.07.004
Rezac, S., Kok, C. R., Heermann, M., and Hutkins, R. (2018). Fermented foods as a dietary source of live organisms. Front. Microbiol. 9:1785. doi: 10.3389/fmicb.2018.01785
Rizzello, C. G., Filannino, P., Di Cagno, R., Calasso, M., and Gobbetti, M. (2014). Quorum-sensing regulation of constitutive plantaricin by Lactobacillus plantarum strains under a model system for vegetables and fruits. Appl. Environ. Microbiol. 80, 777–787. doi: 10.1128/AEM.03224-13
Rojo-Bezares, B., Saenz, Y., Navarro, L., Jimenez-Diaz, R. W., Zarazaga, M., Ruiz-Larrea, F., et al. (2008). Characterization of a new organization of the plantaricin locus in the inducible bacteriocin-producing Lactobacillus plantarum J23 of grape must origin. Arch. Microbiol. 189, 491–499. doi: 10.1007/s00203-007-0342-6
Rosenberg, M., Gutnick, D., and Rosenberg, E. (2006). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9, 29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x
Schillinger, U., and Lücke, F. K. (1987). Identification of lactobacilli from meat and meat products. Food Microbiol. 4, 199–208. doi: 10.1016/0740-0020(87)90002-5
Shobharani, P., and Halami, P. M. (2016). In vitro evaluation of the cholesterol-reducing ability of a potential probiotic Bacillus spp. Annl. Microbiol. 66, 643–651. doi: 10.1007/s13213-015-1146-6
Shobharani, P., Padmaja, R. J., and Halami, P. M. (2015). Diversity in the antibacterial potential of probiotic cultures Bacillus licheniformis MCC2514 and Bacillus licheniformis MCC2512. Res. Microbiol. 166, 546–554. doi: 10.1016/j.resmic.2015.06.003
Shrago, A. W., Chassy, B. M., and Dobrogosz, W. J. (1986). Conjugal plasmid transfer (pAM beta 1) in Lactobacillus plantarum. Appl. Environ. Microbiol. 52, 574–576. doi: 10.1128/aem.52.3.574-576.1986
Silva, C., Silva, S., and Ribeiro, S. C. (2018). Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 9:594. doi: 10.3389/fmicb.2018.00594
Son, S., and Lewis, B. A. (2002). Free radical scavenging and antioxidative activity of caffeic acid amide and ester analogues: structure-activity relationship. J. Agric. Food Chem. 50, 468–472. doi: 10.1021/jf010830b
Song, D. F., Zhu, M. Y., and Gu, Q. (2014). Purification and characterization of plantaricin ZJ5, a new bacteriocin produced by Lactobacillus plantarum ZJ5. PLoS One 9:e105549. doi: 10.1371/journal.pone.0105549
Song, Y., He, Q., Zhang, J., Qiao, J., Xu, H., Zhong, Z., et al. (2018). Genomic variations in probiotic Lactobacillus plantarum P-8 in the human and rat gut. Front. Microbiol. 9:893. doi: 10.3389/fmicb.2018.00893
Spangler, J. R., Dean, S. N., Leary, D. H., and Walper, S. A. (2019). Response of Lactobacillus plantarum WCFS1 to the Gram-negative pathogen-associated quorum sensing molecule N-3-oxododecanoyl homoserine lactone. Front. Microbiol. 10:715. doi: 10.3389/fmicb.2019.00715
Staley, C., Weingarden, A. R., Khoruts, A., and Sadowsky, M. J. (2017). Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 101, 47–64. doi: 10.1007/s00253-016-8006-6
Sure, K. P., Kotnis, P. V., Bhagwat, P. K., Ranveer, R. C., Dandge, P. B., and Sahoo, A. K. (2016). Production and characterization of bacteriocin produced by Lactobacillus viridescence (NICM 2167). Braz. Arch. Biol. Technol. 59:e16150518.
Tai, H. F., Foo, H. L., Abdul Rahim, R., Loh, T. C., Abdullah, M. P., and Yoshinobu, K. (2015). Molecular characterization of new organisation of plnEF and plw loci of bacteriocin genes harbour concomitantly in Lactobacillus plantarum I-UL4. Microb. Cell Fact. 14:89. doi: 10.1186/s12934-015-0280-y
Tamang, B., and Tamang, J. P. (2009). Lactic acid bacteria isolated from indigenous fermented bamboo products of Arunachal Pradesh in India and their functionality. Food Biotechnol. 23, 133–147. doi: 10.1080/08905430902875945
Tamang, B., Tamang, J. P., Schillinger, U., Franz, C. M. A. P., Gores, M., and Holzapfel, W. H. (2008). Phenotypic and genotypic identification of lactic acid bacteria isolated from ethnic fermented tender bamboo shoots of North East India. Int. J. Food Microbiol. 121, 35–40. doi: 10.1016/j.ijfoodmicro.2007.10.009
Tamang, J. P. (2015). Naturally fermented ethnic soybean foods of India. J. Ethn. Foods 2, 8–17. doi: 10.3389/fmicb.2019.01744
Tamang, J. P., Shin, D. H., Jung, S. J., and Chae, S. W. (2016a). Functional properties of microorganisms in fermented foods. Front. Microbiol. 7:578. doi: 10.3389/fmicb.2016.00578
Tamang, J. P., Tamang, B., Schillinger, U., Franz, C. M. A. P., Gores, M., and Holzapfel, W. H. (2005). Identification of predominant lactic acid bacteria isolated from traditional fermented vegetable products of the Eastern Himalayas. Int. J. Food Microbiol. 105, 347–356. doi: 10.1016/j.ijfoodmicro.2005.04.024
Tamang, J. P., Tamang, B., Schillinger, U., Guigas, C., and Holzapfel, W. H. (2009). Functional properties of lactic acid bacteria isolated from ethnic fermented vegetables of the Himalayas. Int. J. Food Microbiol. 135, 28–33. doi: 10.1016/j.ijfoodmicro.2009.07.016
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016b). Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanizawa, Y., Tohno, M., Kaminuma, E., Nakamura, Y., and Arita, M. (2015). Complete genome sequence and analysis of Lactobacillus hokkaidonensis LOOC260(T), a psychrotrophic lactic acid bacterium isolated from silage. BMC Genomics 16:240. doi: 10.1186/s12864-015-1435-2
Tiwari, S., and Srivastava, S. (2008). Purification and characterization of plantaricin LR14: a novel bacteriocin produced by Lactobacillus plantarum LR/14. Appl. Microbiol. Biotechnol. 79, 759–767. doi: 10.1007/s00253-008-1482-6
Todorov, S. D. (2009). Bacteriocins from Lactobacillus plantarum production, genetic organization and mode of action. Braz. J. Microbiol. 40, 209–221. doi: 10.1590/S1517-83822009000200001
Torriani, S., Felis, G. E., and Dellaglio, F. (2001). Differentiation of Lactobacillus plantarum, L. pentosus, and L. paraplantarum by recA gene sequence analysis and multiplex PCR assay with recA gene-derived primers. Appl. Environ. Microbiol. 67, 3450–3454. doi: 10.1128/AEM.67.8.3450-3454.2001
Tsapieva, A., Duplik, N., and Suvorov, A. (2011). Structure of plantaricin locus of Lactobacillus plantarum 8P-A3. Benef. Microbes 2, 255–261. doi: 10.3920/BM2011.0030
Turpin, W., Humblot, C., Noordine, M. L., Thomas, M., and Guyot, J. P. (2012). Lactobacillaceae and cell adhesion: genomic and functional screening. PLoS One 7:e38034. doi: 10.1371/journal.pone.0038034
Van Heel, A. J., De Jong, A., Montalbán-López, M., Kok, J., and Kuipers, O. P. (2013). BAGEL3: automated identification of genes encoding bacteriocins and (non-) bactericidal post-translationally modified peptides. Nucleic Acids Res. 41, 448–453. doi: 10.1093/nar/gkt391
Varankovich, N. V., Nickerson, M. T., and Korber, D. R. (2015). Probiotic-based strategies for therapeutic and prophylactic use against multiple gastrointestinal diseases. Front. Microbiol. 6:685. doi: 10.3389/fmicb.2015.00685
Wang, Y., Qin, Y., Xie, Q., Zhang, Y., Hu, J., and Li, P. (2018). Purification and characterization of plantaricin LPL-1, a novel class IIa bacteriocin produced by Lactobacillus plantarum LPL-1 isolated from fermented fish. Front. Microbiol. 9:2276. doi: 10.3389/fmicb.2018.02276
Yang, E., Fan, L., Jiang, Y., Doucette, C., and Fillmore, S. (2012). Antimicrobial activity of bacteriocin-producing lactic acid bacteria isolated from cheeses and yogurts. AMB Express 2:48. doi: 10.1186/2191-0855-2-48
Yunes, R. A., Poluektova, E. U., Dyachkova, M. S., Klimina, K. M., Kovtun, A. S., Averina, O. V., et al. (2016). GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 42, 197–204. doi: 10.1016/j.anaerobe.2016.10.011
Zhang, X., Wang, Y., Liu, L., Wei, Y., Shang, N., Zhang, X., et al. (2015). Two-peptide bacteriocin PlnEF causes cell membrane damage to Lactobacillus plantarum. Biochem. Biophys. Acta 1858, 274–280. doi: 10.1016/j.bbamem.2015.11.018
Zhang, Z. Y., Liu, C., Zhu, Y. Z., Zhong, Y., Zhu, Y. Q., Zheng, H. J., et al. (2009). Complete genome sequence of Lactobacillus plantarum JDM1. J. Bacteriol. 191, 5020–5021. doi: 10.1128/JB.00587-09
Keywords: antimicrobial activity, reporter bacteria, plantaricin, bacteriocin, probiotics
Citation: Goel A, Halami PM and Tamang JP (2020) Genome Analysis of Lactobacillus plantarum Isolated From Some Indian Fermented Foods for Bacteriocin Production and Probiotic Marker Genes. Front. Microbiol. 11:40. doi: 10.3389/fmicb.2020.00040
Received: 09 October 2019; Accepted: 09 January 2020;
Published: 29 January 2020.
Edited by:
Gianluigi Mauriello, Università degli Studi di Napoli Federico II, ItalyReviewed by:
Alex Galanis, Democritus University of Thrace, GreeceGunjan Goel, Central University of Haryana, India
Copyright © 2020 Goel, Halami and Tamang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Prakash M. Halami, cHJha2FzaGFsYW1pQGNmdHJpLnJlcy5pbg==; a2FzaDEzQHlhaG9vLmNvbQ==; Jyoti Prakash Tamang, anlvdGlfdGFtYW5nQGhvdG1haWwuY29t
 Aditi Goel
Aditi Goel Prakash M. Halami1*
Prakash M. Halami1* Jyoti Prakash Tamang
Jyoti Prakash Tamang