- 1State Key Laboratory of Chemical Resource Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, China
- 2Key Laboratory of Molecular Pathology, School of Basic Medical Science, Inner Mongolia Medical University, Hohhot, China
- 3Key Laboratory of Receptors-Mediated Gene Regulation and Drug Discovery, Hebi People’s Hospital, School of Medicine, Henan University, Kaifeng, China
Gut microbiota, an integral part of the human body, comprise bacteria, fungi, archaea, and protozoa. There is consensus that the disruption of the gut microbiota (termed “gut dysbiosis”) is influenced by host genetics, diet, antibiotics, and inflammation, and it is closely linked to the pathogenesis of inflammatory diseases, such as obesity and inflammatory bowel disease (IBD). Macrophages are the key players in the maintenance of tissue homeostasis by eliminating invading pathogens and exhibit extreme plasticity of their phenotypes, such as M1 or M2, which have been demonstrated to exert pro- and anti-inflammatory functions. Microbiota-derived metabolites, short-chain fatty acids (SCFAs) and Gram-negative bacterial lipopolysaccharides (LPS), exert anti-inflammatory or pro-inflammatory effects by acting on macrophages. Understanding the role of macrophages in gut microbiota-inflammation interactions might provide us a novel method for preventing and treating inflammatory diseases. In this review, we summarize the recent research on the relationship between gut microbiota and inflammation and discuss the important role of macrophages in this context.
Introduction
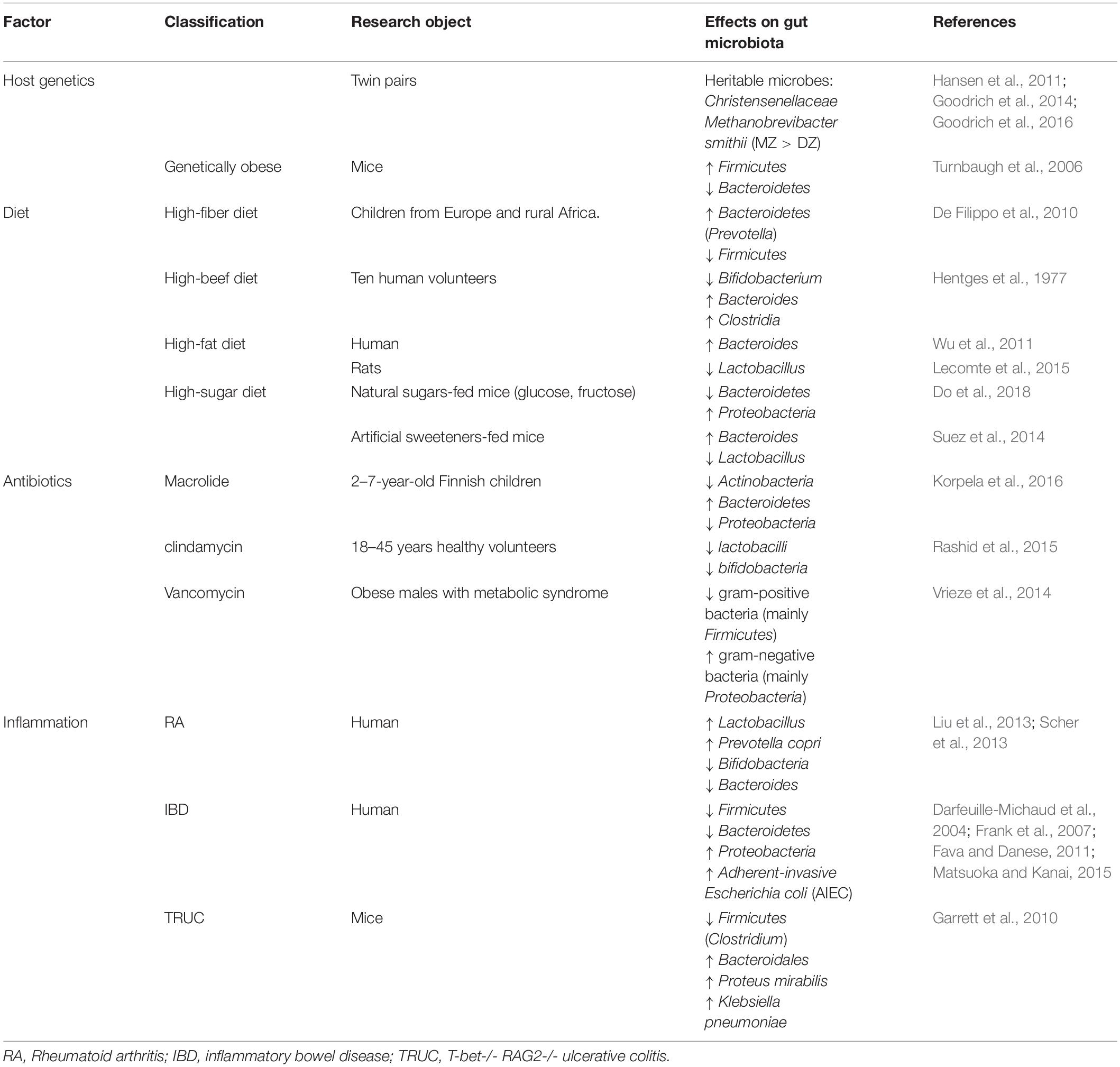
Gut microbes, an essential part of the microbiota ecosystem, outnumbers human cells by 10-fold (Zhang et al., 2015). The gut microbiota contains bacteria, fungi, archaea, and protozoa and can be altered by the host genetics, overuse of antibiotics, and changes in diet, as described in the accompanying review (Belkaid and Hand, 2014; Pickard et al., 2017). The most abundant phyla in human intestine were Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Tap et al., 2009). Firmicutes are gram-positive bacteria including the large class of Clostridia and the lactic acid bacteria. Lactic acid bacteria are marketed as probiotic which are benefit for human health. Another sort of gram-positive bacteria are Actinobacteria, which include Collinsella and Bifidobacterium spp. Bifidobacteria is the other probiotic, which has been made in functional foods. Conversely, Bacteroidetes, and Proteobacteria are gram-negative bacteria, and LPS on their surface can induce activation of macrophages toward pro-inflammatory phenotype. Both can cause infection or diseases under certain conditions. The gut microbiota plays such a critical role in human health and disease that it has been called the “forgotten organ” (O’Hara and Shanahan, 2006). During millions of years of coevolution, the gut microbiota has been living in a symbiotic relationship with the host and affecting the energy balance (Backhed et al., 2004). In addition, symbiotic bacteria promote the intestinal immune system maturity (Mazmanian et al., 2005) and protect against pathogen colonization (Kaiser et al., 2012). Changes in the gut microbial composition result in chronic inflammation and metabolic dysfunction, as has been reviewed elsewhere (Sommer and Backhed, 2013). It is worth noting that the microbiota metabolites, short-chain fatty acids (SCFAs), play a key role in colonic inflammation (Zeng et al., 2019). Many studies have shown that not only epithelial cells or neutrophils but also monocytes and macrophages are modulated by SCFAs (Correa-Oliveira et al., 2016).
Inflammation is a normal physiological response of the body to the foreign pathogen invasion and plays two conflicting roles in human health (Xie et al., 2013). On the one hand, inflammation is the body’s automatic defense response, which also promotes wound healing. On the other hand, excessive inflammatory response results in a series of diseases such as obesity, atherosclerosis, and cancer, which has been reviewed in elsewhere (Wellen and Hotamisligil, 2005; Galkina and Ley, 2009; Crusz and Balkwill, 2015). During acute inflammation, neutrophils are recruited to the inflamed tissue sites, while during chronic inflammation, lymphocytes, macrophages, and plasma cells accumulate and infiltrate the junction tissue (Hakansson and Molin, 2011). There is growing awareness that many prevalent diseases are linked to chronic inflammation. Thus, it is important to regulate inflammation in a timely manner to control the morbidity from disease (Tracey, 2002).
Macrophages are regarded as critical effectors of inflammation. Resident tissue macrophages perform specific functions in response to their local environment (Hume et al., 2002). For example, macrophages are Kupffer cells in the liver and microglia in the central nervous system (CNS). Historically, blood monocytes exit the blood, enter tissues and undergo terminal differentiation to become tissue-resident macrophages (Geissmann et al., 2010). More recently, evidence has shown that tissue-resident macrophages, including lung macrophages and Kupffer cells, are established before birth and complemented by recruited monocytes under inflammatory conditions (Yona et al., 2013). They express pattern recognition molecules, such as Toll-like Receptor (TLR) 4, to recognize foreign pathogens, remove foreign molecules, and protect against infection (Gordon, 2002). In addition, they respond to the inflammatory stimuli and differentiate into classically (“M1”) or alternatively (“M2”) activated macrophages. As reviewed by Hakansson and Molin (2011) macrophages infiltrate tissues during inflammation and perform major functions, including antigen presentation, phagocytosis, and production of various cytokines and growth factors to participate in immune regulation. It is worth mentioning that macrophages are pro-inflammatory under the Lipopolysaccharides (LPS) stimulation (Fujihara et al., 2003).
In this review, we summarize the current understanding of the link between gut microbiota and inflammation focusing on the roles of macrophages. In particular, we discuss two major inflammatory diseases, obesity and inflammatory bowel disease (IBD), and provide a description of the macrophages as the immune and inflammatory effector cells.
Gut Microbiota and Inflammation
The gut microbiota and its metabolites may regulate the host inflammatory conditions (Yang et al., 2017; Feng et al., 2018). Numerous studies have linked the gut microbiota to inflammatory diseases. Forbes et al. (2018) have demonstrated that the immune-mediated inflammatory diseases, such as Crohn’s disease (CD), ulcerative colitis (UC), multiple sclerosis (MS), and rheumatoid arthritis (RA), change the composition of the gut microbiota. In addition, there are abundant reports highlighting the gut microbiota role in the pathogenesis of the inflammatory diseases such as asthma, type 1 and type 2 diabetes mellitus (DM), and obesity (Arrieta et al., 2015; Knip and Siljander, 2016; Meijnikman et al., 2018).
The 16S ribosomal RNA (rRNA) sequencing paved the way for comparing the gut microbiota composition among individuals, which revealed the correlation between specific microbes and disease (Supplementary Table S1). Enterobacteriaceae, a large class of gram-negative facultative bacteria, are commonly linked to many inflammation diseases like IBD and obesity (Zeng et al., 2017). For example, adherent-invasive E. coli (AIEC) has been associated with CD, while diffusely adherent E. coli (DAEC) has been linked to UC (Mirsepasi-Lauridsen et al., 2019). In addition, the depletion of the phyla Firmicutes and the increase of the Proteobacteria populations have been linked to human IBD (Matsuoka and Kanai, 2015). Moreover, an increase in the ratio of Firmicutes to Bacteroidetes populations has been associated with obesity (Turnbaugh et al., 2006). Zhang et al. (2013) reported that the high levels of the phylum Firmicutes and the class Clostridia are found in diabetic patients. Zhu et al. (2013) found that children with non-alcoholic steatohepatitis (NASH) had a significantly higher proportion of Proteobacteria. Additionally, the gut microbiota of the RA patients contained higher levels of Lactobacillus and Prevotella copri and, correspondingly, the numbers of Bifidobacteria and Bacteroides decreased (Liu et al., 2013; Scher et al., 2013; Supplementary Table S1).
Short-chain fatty acids the major metabolic products of the gut microbiota digestion of the non-absorbable dietary fiber and resistant starches, included acetate, propionate, and butyrate (Kles and Chang, 2006). In addition to providing energy for the host, SCFAs exhibit anti-inflammatory effects via binding to G-protein-coupled receptor 43 (GPR43) (Maslowski et al., 2009), which is expressed in immune cells, including macrophages (Le Poul et al., 2003; Sivaprakasam et al., 2016). Butyrate is the most important SCFA due to its role in antagonizing colonic inflammation (Hamer et al., 2008) and the ability to meet 6–10% of the daily human energy requirements (Bergman, 1990). Ruminococcaceae, Eubacterium, Clostridia, and Firmicutes were identified as the main producers of butyrate (Ohira et al., 2017). Moreover, butyrate can exert an anti-inflammatory effect in part by suppressing the activation of NF-κB (Luhrs et al., 2002), a transcription factor that regulates the inflammatory and innate immune responses (Karin et al., 2002). In addition, butyrate strongly inhibits the interferon-gamma (IFN-γ) signaling to ameliorate inflammation (Klampfer et al., 2003). Butyrate also targets peroxisome proliferator-activated receptor-γ (PPARγ) to prevent colon inflammation (Kinoshita et al., 2002).
Macrophages and Inflammation
Macrophages have been considered to be the central effector cells in many chronic inflammatory diseases (Moore and Tabas, 2011). Macrophages have high functional plasticity (Stout and Suttles, 2004) and exhibit pro- or anti-inflammatory properties in response to various cytokines and microbial products (Mantovani et al., 2004). In the early 1990s, scientists found that interleukin (IL)-4 exerts different effects on macrophage phenotypes compared to IFN-γ and/or LPS, and the concept of alternatively activated macrophages has been proposed (Stein et al., 1992). Since then, macrophages have been divided into two groups: classically activated M1 phenotypes, which are stimulated by IFN-γ and LPS and exert pro-inflammatory effects, and alternatively activated M2 phenotypes, which are stimulated by the IL-4 or IL-13 and perform anti-inflammatory functions, as reviewed by Tang et al. (2019).
Many studies have been focused on applying the macrophage phenotype changes to the treatment of inflammatory diseases. For instance, atherosclerosis is a chronic inflammatory disease in which macrophages play a major role at all stages (Tabas and Bornfeldt, 2016). Ouimet et al. (2015) reported that miR-33 antagonizes atherosclerosis partly through promoting the M2 macrophage polarization. In addition, macrophages are associated with obesity-associated inflammatory diseases of the adipose tissue (Weisberg et al., 2003). A study by Lumeng et al. (2007) illustrated that adipose tissue macrophages (ATMs) switch from the M1 polarized macrophage to the M2 polarized macrophage, thereby reducing the adipose tissue-derived inflammatory signals (Lumeng et al., 2007). Additionally, the RA was found to be closely related to the imbalance of the M1 and M2 macrophages and could be attenuated by reestablishing the macrophage equilibrium (Li et al., 2012). Identically, IBD could be ameliorated via the M1 to M2 switch in the colitis mouse model (Zhu et al., 2016). These findings suggest a strong correlation between macrophages and the inflammatory diseases and imply that macrophages participate in the inflammatory process mainly by shifting from the pro-inflammatory (M1) to anti-inflammatory (M2) phenotype (Porcheray et al., 2005).
Macrophages Are Involved in the Interaction Between the Gut Microbiota and Obesity
Obesity is a state of low-grade chronic inflammation, characterized by the expanded adipose tissue (AT) in which macrophages account for 1–30% of its composition (Hauner, 2005). As reviewed by Russo and Lumeng (2018), adipose tissue macrophages (ATMs) include tissue-resident and monocyte-derived macrophages, and the increased number of ATMs in AT is mainly due to the recruitment of blood-macrophages and proliferation of resident macrophages. ATMs are the major effectors of the adipose tissue inflammation; ATMs accumulate in AT to participate in the inflammatory pathways, such as increasing the adipose tissue production of pro-inflammatory cytokines (Weisberg et al., 2003). Since scientists have found that the composition of the gut microbiota is significantly different between the lean mice and the obese mice, the latter showing lower Bacteroidetes and higher Firmicutes bacteria (Ley et al., 2005) levels, people began to realize the close connection between the gut microbiota and obesity. Subsequently, the gut microbiota has been found not only responsible for the weight gain and energy harvest (Samuel et al., 2008), but also involved in the development of the low-grade inflammation associated with obesity (Cani and Delzenne, 2009).
Accumulating evidence indicates that alterations in the gut microbiota are responsible for the progression of obesity. There are two possible pathways of the gut microbiota affecting obesity and the obesity-related diseases, such as diabetes and cardiovascular diseases: the circulation of bacterial components (e.g., LPS) and metabolites SCFAs (Harris et al., 2012) (Figure 1). The obesity-associated decrease in the Bifidobacterium levels leads to the reduced production of GLP-2, a key molecule that promotes the intestinal barrier function, eventually destroying the tight junction integrity of the epithelial barrier and enhancing the intestinal permeability (Cani et al., 2009). Under these circumstances, LPS enters circulation through passive diffusion across the intestinal mucosa (Harris et al., 2012). In addition, Caesar et al. (2012) found that the conventionally raised (CONV-R) mice showed increased plasma LPS concentrations compared with the germ-free (GF) mice, both mildly obese. Moreover, gut-derived LPS promoted macrophage accumulation in the adipose tissue (Caesar et al., 2012) and increased the proliferation of the ATMs via a CD14-dependent pathway (Luche et al., 2013). In AT, macrophages recognized LPS through the TLR4 receptor, which is expressed on the cell surface, thereby converting from the M2 phenotype to the M1 phenotype and subsequently secreting pro-inflammatory cytokines, such as IL-1β and TNF-α, to participate in the inflammatory response (Weisberg et al., 2003; Park et al., 2009; Harford et al., 2011). Similarly, some reports reveal that the lack of TLR4 attenuates the adipose tissue inflammation by shifting the ATM polarization toward the M2 phenotype (Orr et al., 2012). In addition, the level of SCFAs was found to be higher in the obese compared with the normal-weight children due to the differences in the gut microbiota (Rahat-Rozenbloom et al., 2014; Riva et al., 2017). Conversely, the HFD-fed mice displayed decreased SCFAs levels (Lu et al., 2016). Abundant reports indicate that macrophages are involved in the SCFA-mediated anti-inflammatory effects in obesity. For example, propionic acid, a major SCFA, prevents the obesity-related inflammation partially through acting on macrophages (Al-Lahham et al., 2012). In vitro studies have demonstrated that butyrate could inhibit the LPS-mediated macrophage migration via the suppression of Src (Maa et al., 2010). Also, the circulating acetate level was increased by the antibiotic use in HFD-fed mice, reducing macrophage infiltration via activating AMPK (Carvalho et al., 2012).
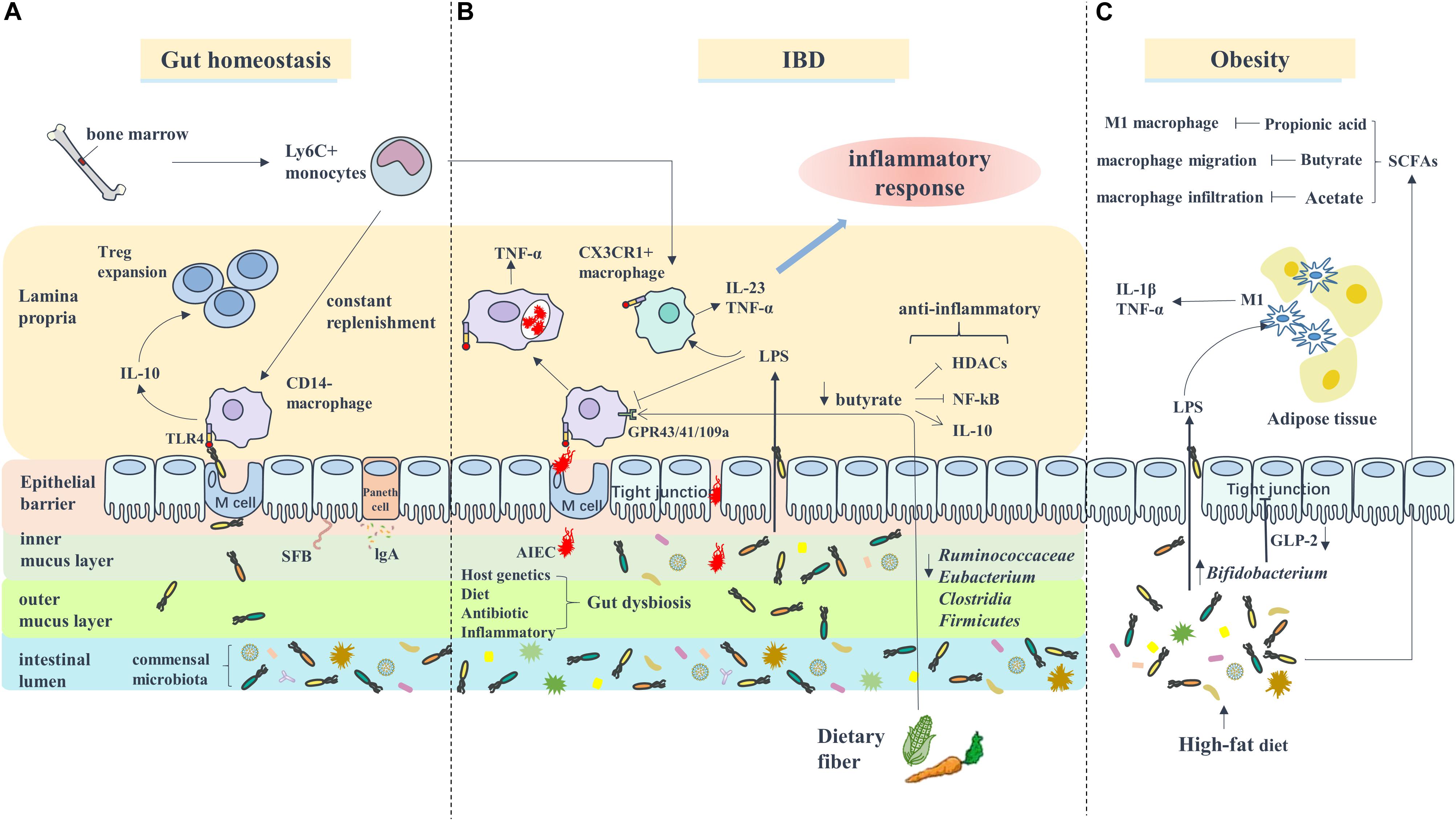
Figure 1. Macrophages are involved in the interaction between gut microbiota and IBD or Obesity. (A) Under gut homeostasis, bone marrow derived Ly6C+ monocytes are constantly recruited to gut to replenish the intestinal macrophages (CD14– macrophages) which recognize the pathogen through TLR4 receptor and secret anti-inflammatory cytokine IL-10 to promote Treg cell expansion. (B) In IBD, the inflammatory environment lead to gut dysbiosis included increased number of AIEC which can survive and replicate in macrophages, and decreased butyrate bacteria like Ruminococcaceae, Eubacterium, Clostridia, and Firmicutes, which impairs the ability of butyrate exert anti-inflammatory role through inhibiting HDACs/NF-κB (GPR43/41) or promoting IL-10 secretion (GPR109a). Additionally, blood Ly6C+ monocytes are recruited to intestinal to become inflammatory macrophages (CX3CR1+ macrophages) and secrete pro-inflammatory cytokines such as IL-23 and TNF-α to participated in inflammatory response. (C) In Obesity, the alteration of gut microbiota composition caused by HFD-diet lead to an increase of intestinal permeability, therefore LPS enter system circulation (i.e., metabolic endotoxemia). Adipose tissue macrophages are response to LPS activation and transform to M1 phenotype.
Macrophages Are Involved in the Interaction Between the Gut Microbiota and IBD
Inflammatory bowel disease, which includes UC and CD, is a persistent inflammatory disorder of the intestine. It is becoming increasingly appreciated that the gut dysbiosis is linked to the pathogenesis of IBD (Ni et al., 2017). Under normal conditions, the mucosal immune system is precisely controlled to protect the intestines from the pathogenic bacteria infections and simultaneously maintain the immunological tolerance to benefit the gut microbiota, as described in the accompanying review (Agace and McCoy, 2017). The disruption of normal mucosal immunity toward the commensal microbiota leads to continuous microbial antigenic stimulation and contributes to chronic intestine inflammation (Strober et al., 2007). In IBD, the numbers of the commensal bacteria (predominantly Firmicutes and Bacteroidetes) were reduced (Frank et al., 2007), and the numbers of the facultative anaerobic bacteria of the family Enterobacteriaceae were increased (Zuo and Ng, 2018). Furthermore, the levels of the microbiota metabolites, the SCFAs, decreased along with the reduction in the population of Clostridium IXa and IV groups, the main butyrate-producing bacteria (Fava and Danese, 2011), limiting intestinal inflammation by regulating the function of colonic T cells (Tregs) (Smith et al., 2013). In a word, the inflammation-related disruption of the host-microbe interactions and decrease in the SCFA levels are involved in the development of IBD (Figure 1).
Intestinal macrophages, which reside in the lamina propria, are among the most abundant immune cells in the gut (Lee et al., 1985) and the first line of defense against the invasion of foreign pathogens (Smythies et al., 2005). Unlike blood monocytes or other tissue-resident macrophages, intestinal macrophages do not respond to LPS due to the absence of CD14, a surface receptor that plays a key role in the LPS-induced cell activation; hence, the LPS-induced pro-inflammatory cytokine production (IL-1, IL-6, IL-8, and TNF-α) is markedly reduced (Ulevitch and Tobias, 1995; Smith et al., 2001; Smythies et al., 2005). Moreover, intestinal macrophages produce anti-inflammatory cytokines like IL-10 and further regulate T-cell differentiation to prevent mucosal auto-inflammation (Denning et al., 2007). Although human intestinal macrophages exhibit profound “inflammatory anergy,” they retain strong phagocytic activity and perform defense functions (Smythies et al., 2005). Since intestinal macrophages perform such essential functions, they are integral to maintaining intestinal homeostasis. Kamada et al. (2008) identified a unique CD-specific macrophage subset that expresses high levels of CD14 and produces pro-inflammatory cytokines, such as IL-23 and TNF-α, leading to the accumulation of pro-inflammatory macrophages. In other words, there are “resident” and “inflammatory” macrophages in the gut that share the same Ly6Chi monocyte precursors under different conditions (Bain et al., 2013).
Although there is a current consensus that gut dysbiosis is closely linked to IBD, it should not be concluded that there is a direct causal relationship between them (Ni et al., 2017). However, it is clear that intestinal inflammation can alter the composition of the gut microbiota, which further exacerbates inflammation (Pickard et al., 2017). Macrophages are well established as the innate immune system cells that recognize invading microbiota using pattern recognition receptors and exhibit efficient phagocytic and bactericidal activity (Janeway and Medzhitov, 2002). Under normal conditions, intestinal macrophages rely on the commensal microbiota for the immune response, while in IBD, inflammatory macrophages are recruited to the inflamed tissue (Grainger et al., 2017). Moreover, the impaired acute inflammatory response in CD leads to the delayed clearance of bacteria (Smith et al., 2009). Adherent-invasive E. coli (AIEC), the aggressive functionally altered resident strains in IBD, have the ability to invade intestinal epithelial cells (IEC) (Sartor and Wu, 2017; Palmela et al., 2018). Subsequently, AIEC is swallowed by macrophages and replicates inside them due to the defect in autophagy (Lapaquette et al., 2010), which is involved in the pathogenesis of IBD. Additionally, the SCFA butyrate can exert anti-inflammatory effects via regulating the intestinal macrophage function as a histone deacetylase (HDAC) inhibitor (Chang et al., 2014) or suppressing the NF-κB activation (Luhrs et al., 2002). Furthermore, butyrate has the ability to exhibit anti-inflammatory activity on intestinal macrophages, which express Gpr109a by inducing the expression of IL-10 (Singh et al., 2014), while these functions are attenuated in IBD due to the decrease of butyrate concentration.
Conclusion
This review provides current understanding of the role of macrophages in gut microbiota and inflammation interactions. On the one hand, the polarization of macrophages, mediated by gut-derived LPS and its metabolites SCFA, plays a key role in the regulation of inflammatory diseases. On the other hand, macrophages are the main players in ensuring intestinal homeostasis through pathogen recognition and elimination and production of anti-inflammatory cytokines. Besides, a recent report by Earley et al. (2018) revealed that a loss of intestinal macrophages influenced the establishment of the gut microbiota in zebrafish, which suggests a critical role of macrophages in shaping the gut microbiota. Taking into account the important role of macrophages in the relationship between gut microbiota and inflammation, developing macrophage-targeting approaches in the prevention and therapy of inflammatory diseases is an appealing strategy. However, the related molecular signaling pathways involved in the roles of macrophages between gut microbiota and inflammation diseases are remained to be defined. Further work is needed to apply macrophages as a perspective target in the treatment of gut microbiota-related inflammatory diseases, like obesity and non-alcoholic fatty liver.
Author Contributions
JW wrote the manuscript. Y-DW and W-DC initiated the idea for writing, revised the manuscript, and secured the funding for this work.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81672433 and 81970551), the Fundamental Research Funds for the Central Universities and Research projects on biomedical transformation of China-Japan Friendship Hospital (Grant No. PYBZ1803) to Y-DW, the National Natural Science Foundation of China (Grant Nos. 81970726 and 81472232), Henan Provincial Natural Science Foundation (Grant No. 182300410323), Program for Science and Technology Innovation Talents in Universities of Henan Province (HASTIT, Grant No. 13HASTIT024), and Plan for Scientific Innovation Talent of Henan Province to W-DC.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01065/full#supplementary-material
References
Agace, W. W., and McCoy, K. D. (2017). Regionalized development and maintenance of the intestinal adaptive immune landscape. Immunity 46, 532–548. doi: 10.1016/j.immuni.2017.04.004
Al-Lahham, S., Roelofsen, H., Rezaee, F., Weening, D., Hoek, A., Vonk, R., et al. (2012). Propionic acid affects immune status and metabolism in adipose tissue from overweight subjects. Eur. J. Clin. Invest. 42, 357–364. doi: 10.1111/j.1365-2362.2011.02590.x
Arrieta, M. C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Backhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. U.S.A. 101, 15718–15723. doi: 10.1073/pnas.0407076101
Bain, C. C., Scott, C. L., Uronen-Hansson, H., Gudjonsson, S., Jansson, O., Grip, O., et al. (2013). Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 6, 498–510. doi: 10.1038/mi.2012.89
Belkaid, Y., and Hand, T. W. (2014). Role of the microbiota in immunity and inflammation. Cell 157, 121–141. doi: 10.1016/j.cell.2014.03.011
Bergman, E. N. (1990). Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70, 567–590. doi: 10.1152/physrev.1990.70.2.567
Caesar, R., Reigstad, C. S., Backhed, H. K., Reinhardt, C., Ketonen, M., Lunden, G. O., et al. (2012). Gut-derived lipopolysaccharide augments adipose macrophage accumulation but is not essential for impaired glucose or insulin tolerance in mice. Gut 61, 1701–1707. doi: 10.1136/gutjnl-2011-301689
Cani, P. D., and Delzenne, N. M. (2009). Interplay between obesity and associated metabolic disorders: new insights into the gut microbiota. Curr. Opin. Pharmacol. 9, 737–743. doi: 10.1016/j.coph.2009.06.016
Cani, P. D., Possemiers, S., Van de Wiele, T., Guiot, Y., Everard, A., Rottier, O., et al. (2009). Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 58, 1091–1103. doi: 10.1136/gut.2008.165886
Carvalho, B. M., Guadagnini, D., Tsukumo, D. M. L., Schenka, A. A., Latuf-Filho, P., Vassallo, J., et al. (2012). Modulation of gut microbiota by antibiotics improves insulin signalling in high-fat fed mice. Diabetologia 55, 2823–2834. doi: 10.1007/s00125-012-2648-4
Chang, P. V., Hao, L., Offermanns, S., and Medzhitov, R. (2014). The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. U.S.A. 111, 2247–2252. doi: 10.1073/pnas.1322269111
Correa-Oliveira, R., Fachi, J. L., Vieira, A., Sato, F. T., and Vinolo, M. A. (2016). Regulation of immune cell function by short-chain fatty acids. Clin. Transl. Immunol. 5:e73. doi: 10.1038/cti.2016.17
Crusz, S. M., and Balkwill, F. R. (2015). Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 12, 584–596. doi: 10.1038/nrclinonc.2015.105
Darfeuille-Michaud, A., Boudeau, J., Bulois, P., Neut, C., Glasser, A. L., Barnich, N., et al. (2004). High prevalence of adherent-invasive Escherichia coli associated with ileal mucosa in Crohn’s disease. Gastroenterology 127, 412–421. doi: 10.1053/j.gastro.2004.04.061
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. U.S.A. 107, 14691–14696. doi: 10.1073/pnas.1005963107
Denning, T. L., Wang, Y. C., Patel, S. R., Williams, I. R., and Pulendran, B. (2007). Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat. Immunol. 8, 1086–1094. doi: 10.1038/ni1511
Do, M. H., Lee, E., Oh, M. J., Kim, Y., and Park, H. Y. (2018). High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight change. Nutrients 10:761. doi: 10.3390/nu10060761
Earley, A. M., Graves, C. L., and Shiau, C. E. (2018). Critical role for a subset of intestinal macrophages in shaping gut microbiota in adult zebrafish. Cell Rep. 25, 424–436. doi: 10.1016/j.celrep.2018.09.025
Fava, F., and Danese, S. (2011). Intestinal microbiota in inflammatory bowel disease: Friend of foe? World J. Gastroenterol. 17, 557–566. doi: 10.3748/wjg.v17.i5.557
Feng, Q., Chen, W. D., and Wang, Y. D. (2018). Gut microbiota: an integral moderator in health and disease. Front. Microbiol. 9:151. doi: 10.3389/fmicb.2018.00151
Forbes, J. D., Chen, C. Y., Knox, N. C., Marrie, R. A., El-Gabalawy, H., de Kievit, T., et al. (2018). A comparative study of the gut microbiota in immune-mediated inflammatory diseases-does a common dysbiosis exist? Microbiome 6:221. doi: 10.1186/s40168-018-0603-4
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U.S.A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Fujihara, M., Muroi, M., Tanamoto, K., Suzuki, T., Azuma, H., and Ikeda, H. (2003). Molecular mechanisms of macrophage activation and deactivation by lipopolysaccharide: roles of the receptor complex. Pharmacol. Ther. 100, 171–194. doi: 10.1016/j.pharmthera.2003.08.003
Galkina, E., and Ley, K. (2009). Immune and inflammatory mechanisms of atherosclerosis (∗). Annu. Rev. Immunol. 27, 165–197. doi: 10.1146/annurev.immunol.021908.132620
Garrett, W. S., Gallini, C. A., Yatsunenko, T., Michaud, M., DuBois, A., Delaney, M. L., et al. (2010). Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe 8, 292–300. doi: 10.1016/j.chom.2010.08.004
Geissmann, F., Manz, M. G., Jung, S., Sieweke, M. H., Merad, M., and Ley, K. (2010). Development of monocytes, macrophages, and dendritic cells. Science 327, 656–661. doi: 10.1126/science.1178331
Goodrich, J. K., Davenport, E. R., Beaumont, M., Jackson, M. A., Knight, R., Ober, C., et al. (2016). Genetic determinants of the gut microbiome in UK twins. Cell Host Microbe 19, 731–743. doi: 10.1016/j.chom.2016.04.017
Goodrich, J. K., Waters, J. L., Poole, A. C., Sutter, J. L., Koren, O., Blekhman, R., et al. (2014). Human genetics shape the gut microbiome. Cell 159, 789–799. doi: 10.1016/j.cell.2014.09.053
Gordon, S. (2002). Pattern recognition receptors: doubling up for the innate immune response. Cell 111, 927–930.
Grainger, J. R., Konkel, J. E., Zangerle-Murray, T., and Shaw, T. N. (2017). Macrophages in gastrointestinal homeostasis and inflammation. Pflugers Arch. 469, 527–539. doi: 10.1007/s00424-017-1958-2
Hakansson, A., and Molin, G. (2011). Gut microbiota and inflammation. Nutrients 3, 637–682. doi: 10.3390/nu3060637
Hamer, H. M., Jonkers, D., Venema, K., Vanhoutvin, S., Troost, F. J., and Brummer, R. J. (2008). Review article: the role of butyrate on colonic function. Aliment. Pharmacol. Ther. 27, 104–119. doi: 10.1111/j.1365-2036.2007.03562.x
Hansen, E. E., Lozupone, C. A., Rey, F. E., Wu, M., Guruge, J. L., Narra, A., et al. (2011). Pan-genome of the dominant human gut-associated archaeon, Methanobrevibacter smithii, studied in twins. Proc. Natl. Acad. Sci. U.S.A. 108(Suppl. 1), 4599–4606. doi: 10.1073/pnas.1000071108
Harford, K. A., Reynolds, C. M., McGillicuddy, F. C., and Roche, H. M. (2011). Fats, inflammation and insulin resistance: insights to the role of macrophage and T-cell accumulation in adipose tissue. Proc. Nutr. Soc. 70, 408–417. doi: 10.1017/s0029665111000565
Harris, K., Kassis, A., Major, G., and Chou, C. J. (2012). Is the gut microbiota a new factor contributing to obesity and its metabolic disorders? J. Obes. 2012, 879151. doi: 10.1155/2012/879151
Hauner, H. (2005). Secretory factors from human adipose tissue and their functional role. Proc. Nutr. Soc. 64, 163–169. doi: 10.1079/pns2005428
Hentges, D. J., Maier, B. R., Burton, G. C., Flynn, M. A., and Tsutakawa, R. K. (1977). Effect of a high-beef diet on the fecal bacterial flora of humans. Cancer Res. 37, 568–571.
Hume, D. A., Ross, I. L., Himes, S. R., Sasmono, R. T., Wells, C. A., and Ravasi, T. (2002). The mononuclear phagocyte system revisited. J. Leukoc. Biol. 72, 621–627.
Janeway, C. A. Jr., and Medzhitov, R. (2002). Innate immune recognition. Annu. Rev. Immunol. 20, 197–216. doi: 10.1146/annurev.immunol.20.083001.084359
Kaiser, P., Diard, M., Stecher, B., and Hardt, W. D. (2012). The streptomycin mouse model for Salmonella diarrhea: functional analysis of the microbiota, the pathogen’s virulence factors, and the host’s mucosal immune response. Immunol. Rev. 245, 56–83. doi: 10.1111/j.1600-065X.2011.01070.x
Kamada, N., Hisamatsu, T., Okamoto, S., Chinen, H., Kobayashi, T., Sato, T., et al. (2008). Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J. Clin. Invest. 118, 2269–2280. doi: 10.1172/jci34610
Karin, M., Cao, Y., Greten, F. R., and Li, Z. W. (2002). NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer 2, 301–310. doi: 10.1038/nrc780
Kinoshita, M., Suzuki, Y., and Saito, Y. (2002). Butyrate reduces colonic paracellular permeability by enhancing PPARgamma activation. Biochem. Biophys. Res. Commun. 293, 827–831. doi: 10.1016/s0006-291x(02)00294-2
Klampfer, L., Huang, J., Sasazuki, T., Shirasawa, S., and Augenlicht, L. (2003). Inhibition of interferon gamma signaling by the short chain fatty acid butyrate. Mol. Cancer Res. 1, 855–862.
Kles, K. A., and Chang, E. B. (2006). Short-chain fatty acids impact on intestinal adaptation, inflammation, carcinoma, and failure. Gastroenterology 130(2 Suppl. 1), S100–S105. doi: 10.1053/j.gastro.2005.11.048
Knip, M., and Siljander, H. (2016). The role of the intestinal microbiota in type 1 diabetes mellitus. Nat. Rev. Endocrinol. 12, 154–167. doi: 10.1038/nrendo.2015.218
Korpela, K., Salonen, A., Virta, L. J., Kekkonen, R. A., Forslund, K., Bork, P., et al. (2016). Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat. Commun. 7:10410. doi: 10.1038/ncomms10410
Lapaquette, P., Glasser, A. L., Huett, A., Xavier, R. J., and Darfeuille-Michaud, A. (2010). Crohn’s disease-associated adherent-invasive E. coli are selectively favoured by impaired autophagy to replicate intracellularly. Cell. Microbiol. 12, 99–113. doi: 10.1111/j.1462-5822.2009.01381.x
Le Poul, E., Loison, C., Struyf, S., Springael, J. Y., Lannoy, V., Decobecq, M. E., et al. (2003). Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 278, 25481–25489. doi: 10.1074/jbc.M301403200
Lecomte, V., Kaakoush, N. O., Maloney, C. A., Raipuria, M., Huinao, K. D., Mitchell, H. M., et al. (2015). Changes in gut microbiota in rats fed a high fat diet correlate with obesity-associated metabolic parameters. PLoS One 10:e0126931. doi: 10.1371/journal.pone.0126931
Lee, S. H., Starkey, P. M., and Gordon, S. (1985). Quantitative analysis of total macrophage content in adult mouse tissues. Immunochemical studies with monoclonal antibody F4/80. J. Exp. Med. 161, 475–489. doi: 10.1084/jem.161.3.475
Ley, R. E., Backhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Li, J., Hsu, H. C., and Mountz, J. D. (2012). Managing macrophages in rheumatoid arthritis by reform or removal. Curr. Rheumatol. Rep. 14, 445–454. doi: 10.1007/s11926-012-0272-4
Liu, X., Zou, Q., Zeng, B., Fang, Y., and Wei, H. (2013). Analysis of fecal Lactobacillus community structure in patients with early rheumatoid arthritis. Curr. Microbiol. 67, 170–176. doi: 10.1007/s00284-013-0338-1
Lu, Y., Fan, C., Li, P., Lu, Y., Chang, X., and Qi, K. (2016). Short chain fatty acids prevent high-fat-diet-induced obesity in mice by regulating G protein-coupled receptors and gut microbiota. Sci. Rep. 6:37589. doi: 10.1038/srep37589
Luche, E., Cousin, B., Garidou, L., Serino, M., Waget, A., Barreau, C., et al. (2013). Metabolic endotoxemia directly increases the proliferation of adipocyte precursors at the onset of metabolic diseases through a CD14-dependent mechanism. Mol. Metab. 2, 281–291. doi: 10.1016/j.molmet.2013.06.005
Luhrs, H., Gerke, T., Muller, J. G., Melcher, R., Schauber, J., Boxberge, F., et al. (2002). Butyrate inhibits NF-kappaB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 37, 458–466. doi: 10.1080/003655202317316105
Lumeng, C. N., Bodzin, J. L., and Saltiel, A. R. (2007). Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 117, 175–184. doi: 10.1172/jci29881
Maa, M. C., Chang, M. Y., Hsieh, M. Y., Chen, Y. J., Yang, C. J., Chen, Z. C., et al. (2010). Butyrate reduced lipopolysaccharide-mediated macrophage migration by suppression of Src enhancement and focal adhesion kinase activity. J. Nutr. Biochem. 21, 1186–1192. doi: 10.1016/j.jnutbio.2009.10.004
Mantovani, A., Sica, A., Sozzani, S., Allavena, P., Vecchi, A., and Locati, M. (2004). The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 25, 677–686. doi: 10.1016/j.it.2004.09.015
Maslowski, K. M., Vieira, A. T., Ng, A., Kranich, J., Sierro, F., Yu, D., et al. (2009). Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286. doi: 10.1038/nature08530
Matsuoka, K., and Kanai, T. (2015). The gut microbiota and inflammatory bowel disease. Semin. Immunopathol. 37, 47–55. doi: 10.1007/s00281-014-0454-4
Mazmanian, S. K., Liu, C. H., Tzianabos, A. O., and Kasper, D. L. (2005). An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 122, 107–118. doi: 10.1016/j.cell.2005.05.007
Meijnikman, A. S., Gerdes, V. E., Nieuwdorp, M., and Herrema, H. (2018). Evaluating causality of gut microbiota in obesity and diabetes in humans. Endocr. Rev. 39, 133–153. doi: 10.1210/er.2017-00192
Mirsepasi-Lauridsen, H. C., Vallance, B. A., Krogfelt, K. A., and Petersen, A. M. (2019). Escherichia coli pathobionts associated with inflammatory bowel disease. Clin. Microbiol. Rev. 32:e00060-18. doi: 10.1128/cmr.00060-18
Moore, K. J., and Tabas, I. (2011). Macrophages in the pathogenesis of atherosclerosis. Cell 145, 341–355. doi: 10.1016/j.cell.2011.04.005
Ni, J., Wu, G. D., Albenberg, L., and Tomov, V. T. (2017). Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 14, 573–584. doi: 10.1038/nrgastro.2017.88
O’Hara, A. M., and Shanahan, F. (2006). The gut flora as a forgotten organ. EMBO Rep. 7, 688–693. doi: 10.1038/sj.embor.7400731
Ohira, H., Tsutsui, W., and Fujioka, Y. (2017). Are short chain fatty acids in gut microbiota defensive players for inflammation and atherosclerosis? J. Atheroscler. Thromb. 24, 660–672. doi: 10.5551/jat.RV17006
Orr, J. S., Puglisi, M. J., Ellacott, K. L., Lumeng, C. N., Wasserman, D. H., and Hasty, A. H. (2012). Toll-like receptor 4 deficiency promotes the alternative activation of adipose tissue macrophages. Diabetes 61, 2718–2727. doi: 10.2337/db11-1595
Ouimet, M., Ediriweera, H. N., Gundra, U. M., Sheedy, F. J., Ramkhelawon, B., Hutchison, S. B., et al. (2015). MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Invest. 125, 4334–4348. doi: 10.1172/jci81676
Palmela, C., Chevarin, C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., et al. (2018). Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587. doi: 10.1136/gutjnl-2017-314903
Park, B. S., Song, D. H., Kim, H. M., Choi, B. S., Lee, H., and Lee, J. O. (2009). The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458, 1191–1195. doi: 10.1038/nature07830
Pickard, J. M., Zeng, M. Y., Caruso, R., and Nunez, G. (2017). Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. 279, 70–89. doi: 10.1111/imr.12567
Porcheray, F., Viaud, S., Rimaniol, A. C., Leone, C., Samah, B., Dereuddre-Bosquet, N., et al. (2005). Macrophage activation switching: an asset for the resolution of inflammation. Clin. Exp. Immunol. 142, 481–489. doi: 10.1111/j.1365-2249.2005.02934.x
Rahat-Rozenbloom, S., Fernandes, J., Gloor, G. B., and Wolever, T. M. (2014). Evidence for greater production of colonic short-chain fatty acids in overweight than lean humans. Int. J. Obes. 38, 1525–1531. doi: 10.1038/ijo.2014.46
Rashid, M. U., Zaura, E., Buijs, M. J., Keijser, B. J., Crielaard, W., Nord, C. E., et al. (2015). Determining the long-term effect of antibiotic administration on the human normal intestinal microbiota using culture and pyrosequencing methods. Clin. Infect. Dis. 60(Suppl. 2), S77–S84. doi: 10.1093/cid/civ137
Riva, A., Borgo, F., Lassandro, C., Verduci, E., Morace, G., Borghi, E., et al. (2017). Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 19, 95–105. doi: 10.1111/1462-2920.13463
Russo, L., and Lumeng, C. N. (2018). Properties and functions of adipose tissue macrophages in obesity. Immunology 155, 407–417. doi: 10.1111/imm.13002
Samuel, B. S., Shaito, A., Motoike, T., Rey, F. E., Backhed, F., Manchester, J. K., et al. (2008). Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. U.S.A. 105, 16767–16772. doi: 10.1073/pnas.0808567105
Sartor, R. B., and Wu, G. D. (2017). Roles for intestinal bacteria, viruses, and fungi in pathogenesis of inflammatory bowel diseases and therapeutic approaches. Gastroenterology 152, 327–339.e4. doi: 10.1053/j.gastro.2016.10.012
Scher, J. U., Sczesnak, A., Longman, R. S., Segata, N., Ubeda, C., Bielski, C., et al. (2013). Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. eLife 2:e01202. doi: 10.7554/eLife.01202
Singh, N., Gurav, A., Sivaprakasam, S., Brady, E., Padia, R., Shi, H., et al. (2014). Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 40, 128–139. doi: 10.1016/j.immuni.2013.12.007
Sivaprakasam, S., Prasad, P. D., and Singh, N. (2016). Benefits of short-chain fatty acids and their receptors in inflammation and carcinogenesis. Pharmacol. Ther. 164, 144–151. doi: 10.1016/j.pharmthera.2016.04.007
Smith, A. M., Rahman, F. Z., Hayee, B., Graham, S. J., Marks, D. J., Sewell, G. W., et al. (2009). Disordered macrophage cytokine secretion underlies impaired acute inflammation and bacterial clearance in Crohn’s disease. J. Exp. Med. 206, 1883–1897. doi: 10.1084/jem.20091233
Smith, P. D., Smythies, L. E., Mosteller-Barnum, M., Sibley, D. A., Russell, M. W., Merger, M., et al. (2001). Intestinal macrophages lack CD14 and CD89 and consequently are down-regulated for LPS- and IgA-mediated activities. J. Immunol. 167, 2651–2656. doi: 10.4049/jimmunol.167.5.2651
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573. doi: 10.1126/science.1241165
Smythies, L. E., Sellers, M., Clements, R. H., Mosteller-Barnum, M., Meng, G., Benjamin, W. H., et al. (2005). Human intestinal macrophages display profound inflammatory anergy despite avid phagocytic and bacteriocidal activity. J. Clin. Invest. 115, 66–75. doi: 10.1172/jci19229
Sommer, F., and Backhed, F. (2013). The gut microbiota–masters of host development and physiology. Nat. Rev. Microbiol. 11, 227–238. doi: 10.1038/nrmicro2974
Stein, M., Keshav, S., Harris, N., and Gordon, S. (1992). Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 176, 287–292. doi: 10.1084/jem.176.1.287
Stout, R. D., and Suttles, J. (2004). Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J. Leukoc. Biol. 76, 509–513. doi: 10.1189/jlb.0504272
Strober, W., Fuss, I., and Mannon, P. (2007). The fundamental basis of inflammatory bowel disease. J. Clin. Invest. 117, 514–521. doi: 10.1172/jci30587
Suez, J., Korem, T., Zeevi, D., Zilberman-Schapira, G., Thaiss, C. A., Maza, O., et al. (2014). Artificial sweeteners induce glucose intolerance by altering the gut microbiota. Nature 514, 181–186. doi: 10.1038/nature13793
Tabas, I., and Bornfeldt, K. E. (2016). Macrophage phenotype and function in different stages of atherosclerosis. Circ. Res. 118, 653–667. doi: 10.1161/circresaha.115.306256
Tang, P. M., Nikolic-Paterson, D. J., and Lan, H. Y. (2019). Macrophages: versatile players in renal inflammation and fibrosis. Nat. Rev. Nephrol. 15, 144–158. doi: 10.1038/s41581-019-0110-2
Tap, J., Mondot, S., Levenez, F., Pelletier, E., Caron, C., Furet, J. P., et al. (2009). Towards the human intestinal microbiota phylogenetic core. Environ. Microbiol. 11, 2574–2584. doi: 10.1111/j.1462-2920.2009.01982.x
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Ulevitch, R. J., and Tobias, P. S. (1995). Receptor-dependent mechanisms of cell stimulation by bacterial endotoxin. Annu. Rev. Immunol. 13, 437–457. doi: 10.1146/annurev.iy.13.040195.002253
Vrieze, A., Out, C., Fuentes, S., Jonker, L., Reuling, I., Kootte, R. S., et al. (2014). Impact of oral vancomycin on gut microbiota, bile acid metabolism, and insulin sensitivity. J. Hepatol. 60, 824–831. doi: 10.1016/j.jhep.2013.11.034
Weisberg, S. P., McCann, D., Desai, M., Rosenbaum, M., Leibel, R. L., Ferrante, A. W. Jr., et al. (2003). Obesity is associated with macrophage accumulation in adipose tissue. J. Clin. Invest. 112, 1796–1808. doi: 10.1172/jci19246
Wellen, K. E., and Hotamisligil, G. S. (2005). Inflammation, stress, and diabetes. J. Clin. Invest. 115, 1111–1119. doi: 10.1172/jci25102
Wu, G. D., Chen, J., Hoffmann, C., Bittinger, K., Chen, Y. Y., Keilbaugh, S. A., et al. (2011). Linking long-term dietary patterns with gut microbial enterotypes. Science 334, 105–108. doi: 10.1126/science.1208344
Xie, W., Li, M., Xu, N., Lv, Q., Huang, N., He, J., et al. (2013). MiR-181a regulates inflammation responses in monocytes and macrophages. PLoS One 8:e58639. doi: 10.1371/journal.pone.0058639
Yang, H. E., Li, Y., Nishimura, A., Jheng, H. F., Yuliana, A., Kitano-Ohue, R., et al. (2017). Synthesized enone fatty acids resembling metabolites from gut microbiota suppress macrophage-mediated inflammation in adipocytes. Mol. Nutr. Food Res. 61:1700064. doi: 10.1002/mnfr.201700064
Yona, S., Kim, K. W., Wolf, Y., Mildner, A., Varol, D., Breker, M., et al. (2013). Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91. doi: 10.1016/j.immuni.2012.12.001
Zeng, H., Umar, S., Rust, B., Lazarova, D., and Bordonaro, M. (2019). Secondary bile acids and short chain fatty acids in the colon: a focus on colonic microbiome, cell proliferation, inflammation, and cancer. Int. J. Mol. Sci. 20:1214. doi: 10.3390/ijms20051214
Zeng, M. Y., Inohara, N., and Nunez, G. (2017). Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26. doi: 10.1038/mi.2016.75
Zhang, X., Shen, D., Fang, Z., Jie, Z., Qiu, X., Zhang, C., et al. (2013). Human gut microbiota changes reveal the progression of glucose intolerance. PLoS One 8:e71108. doi: 10.1371/journal.pone.0071108
Zhang, Y. J., Li, S., Gan, R. Y., Zhou, T., Xu, D. P., and Li, H. B. (2015). Impacts of gut bacteria on human health and diseases. Int. J. Mol. Sci. 16, 7493–7519. doi: 10.3390/ijms16047493
Zhu, L., Baker, S. S., Gill, C., Liu, W., Alkhouri, R., Baker, R. D., et al. (2013). Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609. doi: 10.1002/hep.26093
Zhu, Y., Li, X., Chen, J., Chen, T., Shi, Z., Lei, M., et al. (2016). The pentacyclic triterpene Lupeol switches M1 macrophages to M2 and ameliorates experimental inflammatory bowel disease. Int. Immunopharmacol. 30, 74–84. doi: 10.1016/j.intimp.2015.11.031
Keywords: gut microbiota, inflammatory diseases, macrophage, obesity, inflammatory bowel disease
Citation: Wang J, Chen W-D and Wang Y-D (2020) The Relationship Between Gut Microbiota and Inflammatory Diseases: The Role of Macrophages. Front. Microbiol. 11:1065. doi: 10.3389/fmicb.2020.01065
Received: 14 February 2020; Accepted: 29 April 2020;
Published: 09 June 2020.
Edited by:
Laurel L. Lenz, University of Colorado, United StatesReviewed by:
Rohit Gundamaraju, University of Tasmania, AustraliaSiddappa N. Byrareddy, University of Nebraska Omaha, United States
Copyright © 2020 Wang, Chen and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei-Dong Chen, d2RjaGVuNjY2QDE2My5jb20=; Yan-Dong Wang, eWR3YW5nYnVjdDIwMDlAMTYzLmNvbQ==
 Ji Wang
Ji Wang Wei-Dong Chen
Wei-Dong Chen Yan-Dong Wang
Yan-Dong Wang