- 1Department of Chemistry, Bose Institute, Kolkata, India
- 2Division of Bioinformatics, Bose Institute, Kolkata, India
Two-component systems (TCSs) are central to the ability of Mycobacterium tuberculosis to respond to stress. One such paired TCS is SenX3-RegX3, which responds to phosphate starvation. Here we show that RegX3 is required for M. tuberculosis to withstand low pH, one of the challenges encountered by the bacterium in the host environment, and that RegX3 activates the cytosolic redox sensor WhiB3 to launch an appropriate response to acid stress. We show that the whiB3 promoter of M. tuberculosis harbors a RegX3 binding motif. Electrophoretic mobility shift assays (EMSAs) show that phosphorylated RegX3 (RegX3-P) (but not its unphosphorylated counterpart) binds to this motif, whereas a DNA binding mutant, RegX3 (K204A) fails to do so. Mutation of the putative RegX3 binding motif on the whiB3 promoter, abrogates the binding of RegX3-P. The significance of this binding is established by demonstrating that the expression of whiB3 is significantly attenuated under phosphate starvation or under acid stress in the regX3-inactivated mutant, ΔregX3. Green fluorescent protein (GFP)-based reporter assays further confirm the requirement of RegX3 for the activation of the whiB3 promoter. The compromised survival of ΔregX3 under acid stress and its increased trafficking to the lysosomal compartment are reversed upon complementation with either regX3 or whiB3, suggesting that RegX3 exerts its effects in a WhiB3-dependent manner. Finally, using an in vitro granuloma model, we show that granuloma formation is compromised in the absence of regX3, but restored upon complementation with either regX3 or whiB3. Our findings provide insight into an important role of RegX3 in the network that regulates the survival of M. tuberculosis under acid stress similar to that encountered in its intracellular niche. Our results argue strongly in favor of a role of the RegX3-WhiB3 axis in establishment of M. tuberculosis infection.
Introduction
Tuberculosis remains a global health problem (World Health Organization, 2019, Global Tuberculosis Report). The causative agent Mycobacterium tuberculosis is endowed with the capacity to remain dormant for years within its host and to relapse under favorable conditions, including drug withdrawal. Efforts to contain the disease have been restricted by the absence of an efficacious vaccine, as well as the development of multi-drug resistance. The latter makes it important to search for new drug targets and approaches for chemotherapy, and specifically to understand how M. tuberculosis subverts the stress imposed by the host environment.
The success of M. tuberculosis rests with its ability to sense extreme conditions within its host such as nutrient deficiency, acid stress, hypoxia, and redox stress. Two-component systems (TCSs) are central to the ability of the bacterium to sense and respond to its environment (Tiwari et al., 2017). A membrane-bound sensor kinase (SK) and a cytosolic response regulator (RR) form two halves of canonical paired TCSs. Once the SK senses a stimulus, it is phosphorylated on a histidine residue. The phosphate is relayed on to an aspartate residue of the cognate RR, which regulates its binding to a distinct set of targets which make up its regulon. M. tuberculosis has twelve paired TCSs, six orphan RRs and two orphan SKs (Parish, 2014; Kundu, 2018). TCSs regulate gene expression to influence mycobacterial biofilm formation (Banerjee et al., 2019) virulence, pathogenesis, intracellular survival and the response to stress within its host (Bretl et al., 2011).
The TCS, SenX3-RegX3 is best characterized as a phosphate starvation responsive TCS which is required for intracellular survival and virulence of M. tuberculosis (Parish et al., 2003). Unlike many other TCSs, which are coordinately transcribed as operons, senX3-regX3 contains a large intergenic region and the two genes can be differentially regulated. Thus, under nutrient starvation, there is bicistronic transcription of the operon, as opposed to phosphate depletion, which induces primarily monocistronic upregulation of regX3 (Rifat and Karakousis, 2014). Additionally, phenotypic studies with mutants containing transposon disruptions in each gene have shown that RegX3 may function independently of its cognate SK, SenX3 (Rifat et al., 2014). RegX3 is inhibited when inorganic phosphate is abundant, in a manner requiring the phosphate-specific Pst transport system (Tischler et al., 2013). The function of the SenX3-RegX3 TCS of mycobacteria has been best studied in relation to phosphate starvation (Rifat et al., 2009; James et al., 2012). For example, it regulates protein secretion dependent on the ESX-5 secretion system (Elliott and Tischler, 2016) and membrane vesicle formation in an ESX-5 –independent manner (White et al., 2018). RegX3 is a regulator of the stringent response (Sanyal et al., 2013) and persister formation (Namugenyi et al., 2017). However, the other stress signals to which RegX3 is responsive, remain to be uncovered.
The seven WhiB proteins of M. tuberculosis are believed to be redox-sensing transcription factors (Larsson et al., 2012). WhiB3 is a virulence regulator of M. tuberculosis which senses host-generated nitric oxide (NO) and low levels of O2 (Steyn et al., 2002; Singh et al., 2007; Saini et al., 2012; Pacl et al., 2018). WhiB3 is required for survival of M. tuberculosis during reactive oxygen species (ROS) and reactive nitrogen species (RNS) stresses (Mehta and Singh, 2019) and during nutrient starvation. It is maximally induced at 2 weeks postinfection in the lungs of wild-type and immunodeficient (gamma interferon receptor-/-, Rag1-/-, and tumor necrosis factor alpha-/-) mice just prior to reaching a peak bacterial burden in the lungs, possibly indicating a role for this gene in quorum sensing in vivo (Banaiee et al., 2006). A whiB3 mutant shows altered colony morphology and growth properties (Steyn et al., 2002; Singh et al., 2007). It modulates virulence lipids including phthioceroldimycocerosate (PDIM), polyacyltrehalose (PAT), sulfolipid (SL-1); and triacylglycerol (Singh et al., 2009). These polyketide lipids in turn, arrest host cell cycle at the G1/S transition (Cumming et al., 2017). The transcription of whiB3 is significantly enhanced under acidic pH (Geiman et al., 2006) and WhiB3 is required to resist acid stress during infection (Mehta et al., 2016). Till date, its transcriptional regulation is incompletely understood. In pathogenic mycobacteria, PhoPR directly activates whiB3 expression in response to low pH (Feng et al., 2018). In addition, a recent study has shown that the nitrogen regulator GlnR directly activates the expression of whiB3 in Mycobacterium smegmatis (You et al., 2019).
Here we uncover a novel role of RegX3 in regulating the M. tuberculosis stress response by demonstrating that the survival of a regX3 mutant is compromised under acid stress. We show that RegX3 is activated under acid stress. We demonstrate conclusively that RegX3 activates WhiB3 by direct binding to its promoter and that whiB3 expression under acid stress is compromised in the absence of RegX3. Further we present evidence that (a) M. tuberculosis subverts lysosomal trafficking in a RegX3-WhiB3 dependent manner, and (b) granuloma formation in an in vitro model of infection of human PBMCs by M. tuberculosis, is regulated by RegX3 in a WhiB3-dependent manner. These observations argue in favor of a role of the RegX3/WhiB3 axis in facilitating M. tuberculosis infection.
Materials and Methods
Bacterial Strains and Growth Conditions
Escherichia coli Top10 and DH5α were used for cloning. E. coli BL21(DE3), and C41(DE3) were used for recombinant protein expression. E. coli was grown in Luria-Bertani (LB) Miller broth or on LB agar (Becton Dickinson, Difco) at 37°C. M. tuberculosis H37Rv or other genetically manipulated M. tuberculosis strains were grown in Middle Brook (MB) 7H9 (Difco) broth supplemented with 10% v/v ADC (Difco or Hi-Media Laboratories, India), 0.05% Tween 80 (Hi-Media Laboratories, India) and appropriate antibiotics where required, at 37°C with shaking. MB7H11 agar (Difco) supplemented with 10% v/v OADC (and antibiotics where required), was used as solid medium for plating M. tuberculosis strains. For E. coli, kanamycin sulfate (Roche Applied Science), ampicillin (Sigma), and hygromycin (Invitrogen) were used at concentrations of 50, 100, and 200 μg/ml, respectively. For M. tuberculosis, kanamycin and hygromycin were used at concentrations of 20 and 50 μg/ml, respectively. All experiments were performed following appropriate biosafety protocols approved by the Institutional Biosafety committee.
Molecular Biology Procedures
Standard procedures were used for cloning and analysis of DNA, PCR, electroporation and transformation. The enzymes used to manipulate DNA were from Roche Applied Science, Fermentas and New England Biolabs.
Generation of Genetically Manipulated Strains of M. tuberculosis H37Rv
Mycobacterium tuberculosis H37Rv lacking regX3 (ΔregX3) was generated previously in our laboratory using temperature sensitive mycobacteriophages (Bardarov et al., 2002) as described by Banerjee et al. (2016). Briefly, approximately 800 bp flanking regions upstream and downstream of regX3 were PCR amplified and cloned into pYUB854 at sites flanking the hygromycin cassette. Each construct was then packaged into temperature sensitive mycobacteriophages and delivered via infection into M. tuberculosis. Double crossovers (DCOs) were screened after 6 weeks by PCR and knockouts confirmed by Western blot using antibodies against RegX3. For complementation of regX3, a 2.5 kb region encompassing the senX3-regX3 promoter, and the senX3-regX3 ORFs were PCR amplified from genomic DNA and cloned into pMV306(Kan), a plasmid that allows integration at the attB site on the genome. A ∼900 bp senX3 region was deleted using Stu1 and the regx3-pMV306 construct was electroporated into the ΔregX3 strain to obtain the regX3 complemented strain (ΔregX3 Comp.regX3), which was selected for growth on plates containing kanamycin as well as hygromycin.
For complementation with whiB3, the whiB3 ORF was PCR amplified using sense and antisense primers 5′-ATGGTACCCATATGCCACAGCCGGAGCAGCTACCG 3′ and 5′ATTAAGCTTCTCGAGAGCTGTGCGGCGGATGCCGCG-3′, respectively, and cloned at the NdeI and XhoI sites of pet20b+ to generate a whiB3 construct (pET20b whiB3-His) in frame with a C-terminal His tag. whiB3 with a C-terminal His tag was then PCR amplified using the sense and antisense primers 5′ TTAGGATCCACCACAGCCGGAGCAGCTACCG3′and 5′ ATTAAGCTTGCTAGCTCAGTGGTGGTGGTGGTGGTGC-3′, respectively, and pET20b whiB3-His as template; and cloned at the BamHI and HindIII sites of pMV261 (an E. coli-mycobacteria shuttle vector in which cloned genes are under the control of the hsp60 promoter). This construct (pMV261 whiB3-His) was electroporated into the ΔregX3 strain to obtain ΔregX3 complemented with whiB3 (ΔregX3 Comp.whiB3), which was selected for growth on plates containing kanamycin as well as hygromycin.
Growth of M. tuberculosis Under Phosphate Starvation
For phosphate starvation M. tuberculosis and its variants were grown in MB7H9 containing MOPS pH 6.6, 17.6 mM Na2HPO4, 7.35 mM KH2PO4, and 0.05% Tween 80, till the O.D.600 reached 0.6. The cells were then centrifuged and washed twice with phosphate-free MB7H9, resuspended in either phosphate containing or phosphate free medium and allowed to grow at 37°C at a shaking speed of 120 rpm for 72 h. Aliquots were removed and cells were stored in RNA Later (Qiagen) at −80°C for isolation of RNA.
Cloning, Expression and Purification of Recombinant Proteins
Escherichia coli BL21(DE3) harboring regX3 cloned in pET28a was grown at 37°C with shaking and induced with isopropyl β-D-thiogalactopyranoside (IPTG) (120 μM) as described by Sanyal et al. (2013). Cells were lysed by sonication and His-RegX3 was purified from the cell-free supernatant by chromatography on Ni2+-NTA agarose. RegX3 (K204A) has been described by Banerjee et al. (2016). A recombinant MalE–EnvZ construct (gift from Dr. M. Igo, University of California, Davis, Davis, CA, United States) was transformed in E. coli BL21(DE3) and used for expression and purification using amylose affinity chromatography as described by Sharma et al. (2015). Briefly, cells were grown and induced with IPTG (100 μM) at 16°C for 20 h. Cells were disrupted by freeze-thaw cycles in the presence of lysozyme and MalE–EnvZ was purified from the cell-free supernatant by amylose affinity chromatography (NEB). EnvZ was autophosphorylated in kinase buffer (50 mM Tris–HCl, pH 8, 50 mM KCl, and 50 mM MgCl2) containing 20 mM ATP for 15 min at 37°C.
Bioinformatic Analyses
The upstream promoter sequence of whiB3 (−500 to +100) was searched for an inverted repeat motif having the following features: a 6–8 bp sequence, followed by a 5 bp spacer, followed by a second 6–8 bp region which is complementary to, and the inverse of the first 6–8 bp sequence. In-house PERL script was used for this search.
In vitro Transphosphorylation Reactions
RegX3 or its DNA binding mutant (K204A) was phosphorylated using phospho-EnvZ as the phosphate donor (Roberts et al., 2011). The reaction was carried out in kinase buffer with 2.5 μM phosphorylated EnvZ and His-RegX3. Phosphorylated RegX3 (RegX3-P) was used for electrophoretic mobility shift assays (EMSAs).
Electrophoretic Mobility Shift Assays
Binding of phospho-RegX3 (or its mutant) to the whiB3 promoter was analyzed by EMSA. A whiB3 promoter region (−307 to −157), containing the RegX3 binding sites was PCR amplified using the sense and antisense primer pair 5′-CAGCTTTCTTTGCGCTAATTTAGG-3′ (A) and 5′-CAATATCGGACCGTTGCGTGAG-3′ (C), respectively (Supplementary Figure S1). The antisense primer was Cy5-labeled. Mutant DNA was generated by overlap extension PCR (Supplementary Figure S1). The Cy5 labeled DNA (30 ng) was incubated with varying concentrations of phospho-RegX3 (or its mutant) in 4 mM Tris–HCl pH 8.0, containing 4 mM MgCl2, 5% (v/v) glycerol, 40 mM NaCl, 0.5 μg salmon sperm DNA, for 30 min at room temperature (RT). The reaction mix was run on a 6% TBE polyacrylamide gel and the DNA-protein complex was visualized using a Typhoon Trio Plus Imager (GE Healthcare). In other assays, EMSAs were carried out with mutated fragments of DNA, and the DNA-protein complexes were visualized using SYBR GOLD staining.
Measurement of Promoter Activity
A DNA fragment encompassing the region –832 to +102 of whiB3 was PCR-amplified using the primer pair 5′-TTAGGATCCCCACCGCCGACGCACCGC-3′ (sense) and 5′-CGGGGTACCCGTCGGGATGGAAGAACATCG-3′ (antisense) and cloned between the BamHI and KpnI restriction sites of pFPV27. The resulting construct or pFPV27 was electroporated separately into wild type M. tuberculosis and ΔregX3 strains. Green fluorescent protein (GFP) fluorescence was measured in a microplate reader (Victor 1420 Multilabel counter) with excitation at 488 nm and emission at 535 nm.
Chromatin Immunoprecipitation (ChIP)
Chromatin Immunoprecipitation (ChIP) was performed as described by Sharma et al. (2015). Briefly, growing M. tuberculosis cells were crosslinked using 1% formaldehyde followed by quenching with 250 μM glycine. The cells were lysed and DNA was sheared on a BIORUPTOR PLUS (Diagenode) with 25 cycles of 30 s/90 s on/off, respectively, so as to generate fragment sizes of approximately 250 bp. Immunoprecipitation (IP) was carried out using RegX3 antibody (raised by Thermo Fisher Scientific) and DNA was purified after decrosslinking with Proteinase K. Amplification of the whiB3 promoter region was quantitated by ChIP-qPCR using the primer pair 5′-CAGCTTTCTTTGCGCTAATTTAGG-3′ (sense) and 5′-CAATATCGGACCGTTGCGTGAG-3′ (antisense).
RNA Isolation From Intracellular Bacteria and qRT-PCR
RNA was prepared from intracellular M. tuberculosis following the method of Rohde et al. (2007) as described by Banerjee et al. (2019). Briefly, RAW264.7 cells were infected at a multiplicity of infection (MOI) of 10 for 4 h followed by treatment with gentamicin for another 2 h to remove extracellular bacteria. The cells were then washed and lysed in guanidine thiocyanate (GTC) buffer containing N-lauryl sarcosine, sodium citrate, and β-mercaptoethanol. Bacteria were pelleted, lysed using lysozyme and Trizol, followed by bead beating. RNA was prepared from the lysate after centrifugation using the Qiagen RNeasy Kit following the manufacturer’s protocol. RNA was treated with Turbo DNAse (Ambion). cDNA was synthesized using the cDNA synthesis kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. For PCR, whiB3 was amplified using sense and antisense primers 5′-AACGCAGACATCTGGAACTG-3′ and 5′-GGTGCCCTTGAGGAGTAGGT-3′, respectively. regX3 was amplified using the sense and antisense primers 5′-CAG CGTTCCGGTGATCATG-3′ and 5′-CAGGCCGACCACCTT GTC-3′, respectively. 16s rRNA was amplified using the sense and antisense primers 5′-TCCCGGGCCTTGTACACA-3′ and 5′-CCACTGGCTTCGGGTGTTA-3′, respectively.
The quantitative PCR assay was performed with KAPA SYBR® FAST Universal Q-PCR Mix (Kapa Biosystems Pty Ltd., Cape Town, South Africa). Melting curve analyses were run after each assay to check PCR specificity. Three serial dilutions were used for each cDNA. PCR was performed in triplicate for each dilution. The relative expression of the target gene was normalized to 16s rRNA The comparative CT (also known as 2–ΔΔCT) method was used for analyzing gene expression, with the assumptions that the difference of one cycle is two-fold and the PCR efficiency of the target gene is similar to that of the internal control (16s rRNA) (Livak and Schmittgen, 2001).
Immunofluorescence Microscopy
RAW264.7 cells were grown on coverslips and infected with fluorescein isothiocyanate (FITC)-labeled M. tuberculosis strains as described by Banerjee et al. (2019). Cells were fixed with 4% (v/v) paraformaldehyde for 10 min permeabilized with 0.01% Triton X-100 in PBS, then treated with 2% BSA in PBS, followed by treatment with anti lysosomal-associated membrane protein 1 (LAMP1) antibody (Abcam) (1:500) overnight at 4°C. Cells were treated with Alexa 546-conjugated goat anti-rabbit antibody (Abcam) and coverslips were mounted with SlowFade (Thermo Scientific). Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Slides were imaged by confocal microscopy.
Acid Stress
Fifty milliliter of starter cultures of M. tuberculosis strains were grown in MB 7H9 broth to early log phase (O.D.600 of 0.3), split into two aliquots and resuspended at pH 5.5 or pH 6.6. For pH 5.5, medium containing 0.02% Tyloxapol and 0.085% NaCl was buffered using 100 mM 2-(N-morpholino)ethanesulfonic acid (MES) (Healy et al., 2016). CFUs were followed over a period of 6 days and the percent reduction in CFUs of cultures grown at pH 5.5 relative to those grown at pH 6.6, was plotted. RNA was isolated from cultures exposed to different pH for different periods of time. Gene expression was evaluated by qRT-PCR using SYBR Green (Kappa Biosystems). The results were normalized relative to 16S rRNA levels.
Granuloma Formation in vitro
Human peripheral blood mononuclear cells (PBMCs) were used for in vitro granuloma formation as described by Mehta and Singh (2019). Briefly, 5–10 ml blood was collected from a healthy human volunteer, diluted 1:1 and layered on Ficoll Paque (GE Healthcare) followed by centrifugation at 480 × g for 40 min at 18°C. The buffy coat containing monocytes was collected and washed twice in sterile cold KRG buffer (120 mM NaCl, 5 mM KCl, 1.5 mM MgCl2, 8.5 mM Na2HPO4, 1.7 mM NaH2PO4, and 10 mM dextrose). Cells were resuspended in complete RPMI medium with glutamine. The monocyte containing suspension was plated at a density of 1.5 × 106 cells/ml in complete RPMI medium in 24 well plates and kept at 37°C in a CO2 incubator for 2 h. Single cell suspensions of each M. tuberculosis strain were made as described by Karim et al. (2011) and added at the required MOI to each well of the 24-well plate containing the PBMCs, centrifuged at 700 rpm for 5 min and incubated at 37°C in a CO2 incubator. After 4 h, 10 μg/ml gentamicin was added to remove the extracellular bacteria. After another 2 h, cells were washed thrice and complete RPMI medium was added to each well. At day 9 post-infection, cells were processed for Giemsa staining.
Ethics Statement
All experiments with human samples were performed with the approval of the Institutional Human Ethics Committee (BIHEC/2017-18/1) and with consent from all donors (healthy volunteers in the laboratory).
Giemsa Staining
Plates were centrifuged and each well was fixed with methanol for 5 min at RT. The fixative was removed, Giemsa stain was added and kept for 30 min at RT. Plates were washed with water to remove excess Giemsa stain. Images were taken in a Zeiss light Microscope with a 10 x objective.
Statistical Analysis
Normality test was performed using GraphPad Prism v5 (applying D’Agostino-Pearson omnibus test and/or Shapiro–Wilk test). All the data distributed normally as inferred by the normality test. Student’s t test was performed for pairwise comparison. In the case of multigroup comparisons, analysis of variance (ANOVA) test was performed. For Anova, Tukey Post hoc test was used to confirm the significant differences between two groups. All analyses were performed using GraphPad Prism v5. A p value ≤ 0.05 was considered to be significant.
Results
RegX3 Is Required for the Survival of M. tuberculosis Under Acid Stress
M. tuberculosis encounters an acidic environment in the phagosomes, and must wire its gene expression program to facilitate growth and persistence in this acidic environment. It is therefore important to understand the repertoire of regulators that enable M. tuberculosis to sense acid stress and to mount an appropriate response. Considering the importance of TCSs in sensing of the extracellular milieu, we searched the literature for regulators that are activated under acid stress or in the intraphagosomal milieu. PhoPR has been documented to be upregulated during acid stress (Bansal et al., 2017). The report that RegX3 is upregulated early during infection of macrophages by M. tuberculosis (Rohde et al., 2007), provided the cue for testing a probable role of RegX3 in mounting a response to acid stress. We tested and validated the upregulation of regX3 following exposure of M. tuberculosis to acid stress (Table 1). Next we tested the survival of the wild type and the mutant ΔregX3 previously reported by us (Banerjee et al., 2016) following exposure to either pH 6.6 or pH 5.5. Survival was monitored at different time intervals post-exposure by enumerating CFUs. There was a decrease in CFUs in wild type M. tuberculosis grown at pH 5.5 compared to cells grown at pH 6.6. However, the reduction in CFUs at pH 5.5 (compared to pH 6.6) was significantly higher in the absence of regX3 (Figure 1A), suggesting that regX3 contributes to the ability of the bacterium to withstand acid pH. This was confirmed by the observation that survival under acid stress was partially restored in the regX3 complemented strain (ΔregX3 Comp.regX3) (Figure 1A). Importantly, there were no significant differences in CFUs among the four strains on day 0 (i.e., at the start of the experiment).
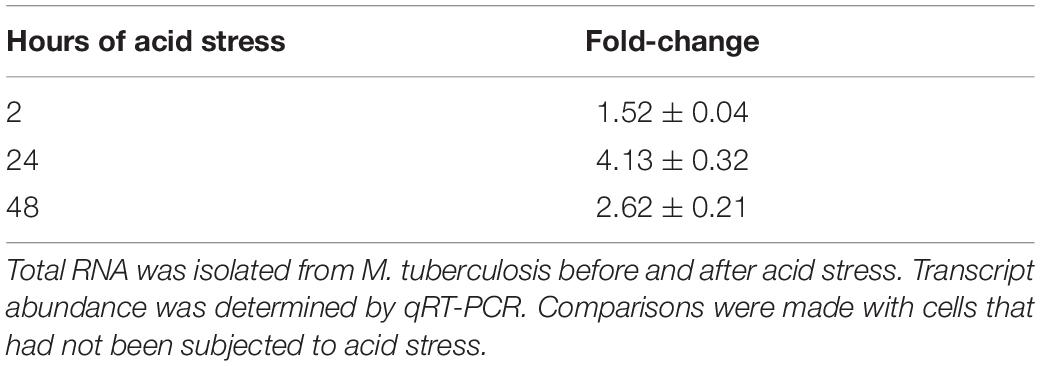
Table 1. Relative fold changes of regX3 in M. tuberculosis subjected to acid stress for different periods of time.
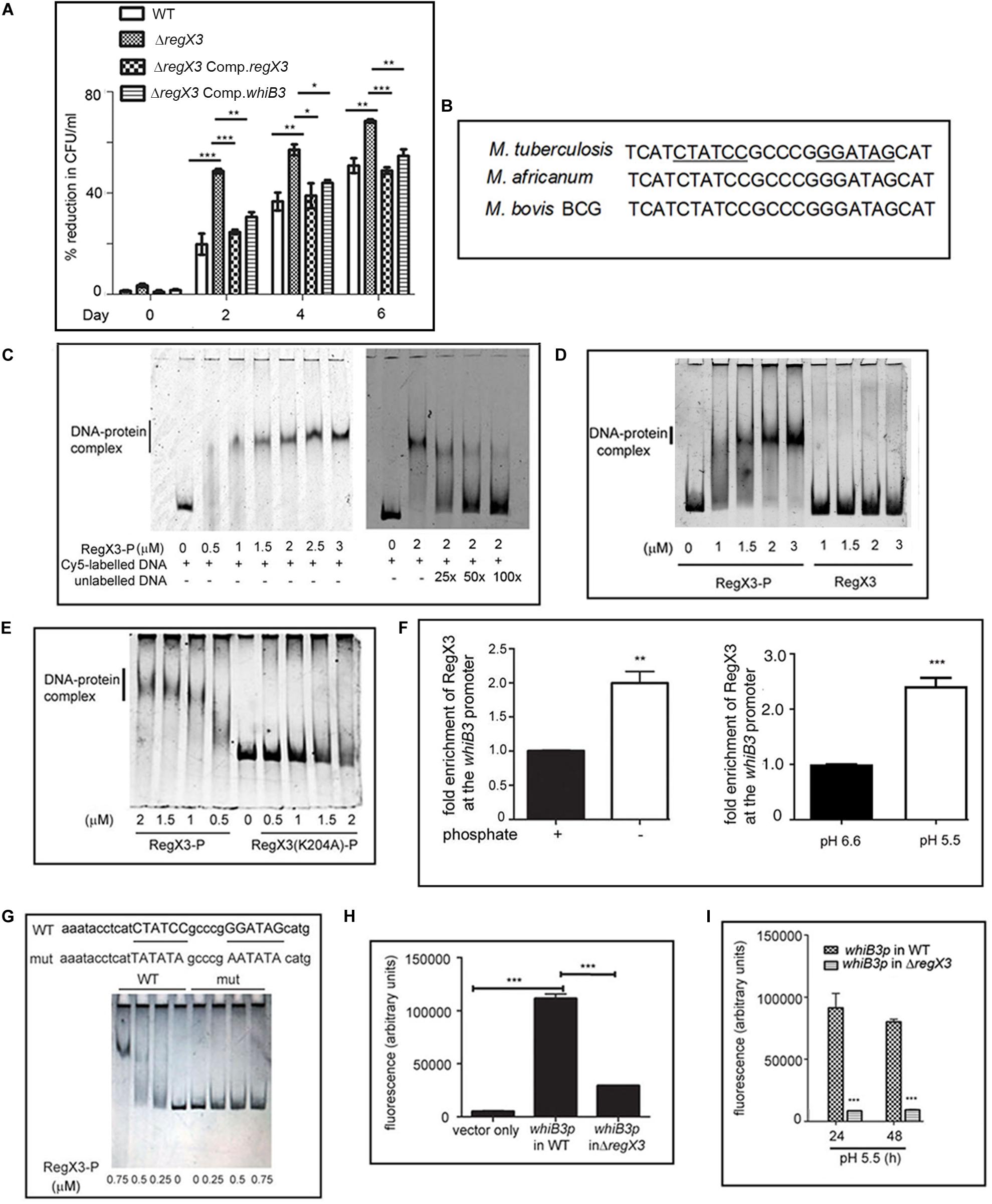
Figure 1. RegX3 is required for the survival of M. tuberculosis under acid stress and it binds to the promoter of whiB3. (A) Wild type M. tuberculosis or its genetically manipulated variants was grown at pH 5.5 or at pH 6.6 for different periods of time. Percent reduction in growth [as determined by enumerating colony forming units (CFUs)] of M. tuberculosis growth at pH 5.5 relative to growth at pH 6.6 is shown. (B) Sequence conservation of RegX3 binding motif of the whiB3 promoter (whiB3p) in M. tuberculosis, M. africanum, and M. bovis BCG. (C,D) EMSA was performed by incubating a Cy5-labeled PCR fragment derived from whiB3p with different concentrations of phosphorylated RegX3 (RegX3-P) (left panel of C). Competitive EMSA was performed by incubating RegX3-P with Cy5-labeled DNA in the presence of increasing concentrations of unlabeled DNA (right panel of C) (D) EMSA was performed with phosphorylated or non-phosphorylated RegX3. (E) EMSA was performed with the DNA binding defective mutant (RegX3 K204A). DNA-protein complexes were visualized under a Typhoon biomolecular imager or by staining with SYBR GOLD. (F) ChIP analysis of RegX3 binding to whiB3p. M. tuberculosis was grown in phosphate enriched (+) or depleted (–) medium (left panel) or grown at different pH (right panel) and subjected to ChIP with RegX3 antibody. The association of RegX3 at the promoter of whiB3 was quantitated by q-PCR of immunoprecipitated DNA or the input (as control) using primers specific for whiB3p or 16s rRNA. The fold enrichment with respect to control [i.e., bacteria grown in phosphate enriched medium (left panel), or at pH 6.6 (right panel)] was set as 1. Data represent means ± S.D., n = 3. (G) EMSA was carried out by incubating wild type DNA or DNA mutated in the RegX3 box, with different concentrations of RegX3-P. Bands were visualized by SYBR GOLD staining. (H,I) M. tuberculosis (WT or ΔregX3) harboring either vector alone or whiB3p were grown either under phosphate stress (H) or acid stress (I) GFP fluorescence was measured in a fluorescence microplate reader. The data are represented as means ± SD, n = 3. ***p < 0.0001; **p < 0.001; *p < 0.05.
We then searched for possible binding of RegX3 to the promoters of genes involved in regulating the response of M. tuberculosis to acid stress. WhiB3 is one such regulator. Its deletion lowers the survival of M. tuberculosis under acidic conditions (Mehta et al., 2016). We fell back on the report of Rustad et al. (2014) documenting that overexpression of RegX3 transcriptionally upregulates whiB3, therefore suggesting its role in regulating whiB3 expression. For the purpose of this communication, we have focused on validating the regulation of WhiB3 by RegX3 under acid stress, and understanding its implications. As expected, the transcription of whiB3 was enhanced when wild type M. tuberculosis was subjected to acid stress (Supplementary Table S1). The acid-induced upregulation of whiB3 was abrogated in ΔregX3 and restored upon complementation of the mutant with regX3 (ΔregX3 Comp.regX3) (Supplementary Table S1), suggesting that RegX3 is required for the transcription of whiB3 under acidic conditions. The importance of a regX3-whiB3 axis in regulating the survival of M. tuberculosis under acidic conditions, was tested by complementing ΔregX3 with whiB3 and assessing CFUs under acidic conditions. The observation that complementation with whiB3 could partly restore the ability of ΔregX3 to withstand acid stress (Figure 1A), argued in favor of a role of the regX3-whiB3 axis in the survival of M. tuberculosis under acid stress.
An inverted repeat (GTGAAC) separated by five to seven unconserved nucleotides constitutes the RegX3 binding motif (Glover et al., 2007). For example, this motif has been identified on the M. tuberculosis polyphosphate kinase 1 (ppk1) promoter in our laboratory (Sanyal et al., 2013). An in silico analysis showed that a putative RegX3-binding motif is present in the M. tuberculosis whiB3 promoter region (Figure 1B). This RegX3 binding palindrome was conserved in Mycobacterium africanum and Mycobacterium bovis BCG, two members of the M. tuberculosis complex. It was absent in Mycobacterium leprae or the fast-growing M. smegmatis.
RegX3 Binds to the Promoter of whiB3 and Regulates whiB3 Promoter Activity
In order to test the binding of RegX3 to the whiB3 promoter region, we amplified the region –307 to −157 of the whiB3 promoter to generate a Cy5-labeled DNA and used this for EMSAs. RegX3-P bound to the aforesaid DNA in a concentration-dependent manner (Figure 1C, left panel). This binding could be competed by unlabeled DNA, suggesting the specificity of the binding (Figure 1C, right panel). Phosphorylation of RegX3 is usually required for optimal DNA binding activity (Sanyal et al., 2013). In line with this, unphosphorylated RegX3 was not able to bind the whiB3 promoter-derived DNA (Figure 1D), suggesting that phosphorylation is required for binding of RegX3 to the whiB3 promoter. In previous studies, we have shown that amino acid lysine 204 (K204) is required for DNA-binding activity of RegX3 (Banerjee et al., 2016). RegX3 (K204A) was unable to bind the whiB3 promoter fragment (Figure 1E) confirming the specificity of the binding.
In order to analyze the physiological relevance of the aforesaid binding, we performed ChIP assays under conditions associated with upregulation of RegX3. RegX3 is best characterized as a phosphate starvation-responsive RR (Rifat et al., 2009). We therefore subjected cells to phosphate starvation and tested the association of RegX3 with the whiB3 promoter region, using an antibody to RegX3. The binding of RegX3 to the whiB3 promoter region was confirmed by ChIP-qPCR (Figure 1F, left panel), arguing in favor of a physiological role of RegX3 in regulating whiB3 expression under stress. In order to test the role of RegX3 in regulating whiB3 under acid stress, we further performed ChIP-qPCR after subjecting cells to acid stress. The association of RegX3 with the whiB3 promoter region was enhanced in cells grown at pH 5.5 compared to cells grown at pH 6.6 (Figure 1F, right panel).
Finally, we confirmed the role of the putative RegX3 binding palindrome identified on the whiB3 promoter by mutational analysis. The sequences CTATCC and GGATAG in the palindrome were substituted with the sequences, TATATA and AATATA, respectively (Figure 1G) to generate a mutant DNA. RegX3 failed to bind to this mutated DNA (Figure 1G), confirming that this region is required for the binding of RegX3 to the whiB3 promoter. To further elucidate the requirement of RegX3 in the expression of whiB3, the whiB3 promoter was cloned in a promoterless GFP vector pFPV27, electroporated in M. tuberculosis (or ΔregX3) and GFP activity was monitored under phosphate starvation (a known trigger for RegX3 expression) or acid stress. Phosphate starvation activated the whiB3 promoter (Figure 1H). However, this was significantly diminished in ΔregX3 (Figure 1H). Promoter activity was also observed in cells exposed to acid stress (Figure 1I), and was significantly diminished in ΔregX3 (Figure 1I).
Intracellular whiB3 Expression Is Regulated by RegX3
We next addressed the question whether the regulation of whiB3 by RegX3 is relevant within the intracellular milieu of the host. It has been reported that whiB3 is upregulated early during infection of macrophages by M. tuberculosis (Rohde et al., 2007). We therefore analyzed the expression of whiB3 during infection of macrophages by either wild type or regX3-inactivated M. tuberculosis. Expression of whiB3 was compromised in the absence of regX3 compared to the wild type (Table 2) and restored upon complementation with regX3. These observations suggested that RegX3 regulates the expression of whiB3 of M. tuberculosis within the intracellular milieu of its host.
RegX3 Regulates Trafficking of M. tuberculosis to Lysosomes in a WhiB3-Dependent Manner
Intracellular trafficking to lysosomes reduces mycobacterial burden in macrophages, and virulent mycobacteria have the ability to evade trafficking to macrophages. ΔregX3 showed reduced survival in macrophages compared to the wild type (Banerjee et al., 2016). We therefore tested whether the ability of M. tuberculosis to evade trafficking to lysosomes is compromised in the absence of regX3. Following infection, the colocalization of FITC-labeled M. tuberculosis with the lysosomal marker LAMP1 was enumerated by fluorescence microscopy. The association of ΔregX3 with lysosomes, was higher than that of wild type M. tuberculosis (Figures 2A,B). This was partly reversed in ΔregX3 Comp.regX3 (Figures 2A,B). Interestingly, complementation with whiB3 (ΔregX3 Comp.whiB3) was equally effective in reversing the trafficking of M. tuberculosis to lysosomes. These results suggested that RegX3 subverts the trafficking of M. tuberculosis to lysosomes in a WhiB3-dependent manner.
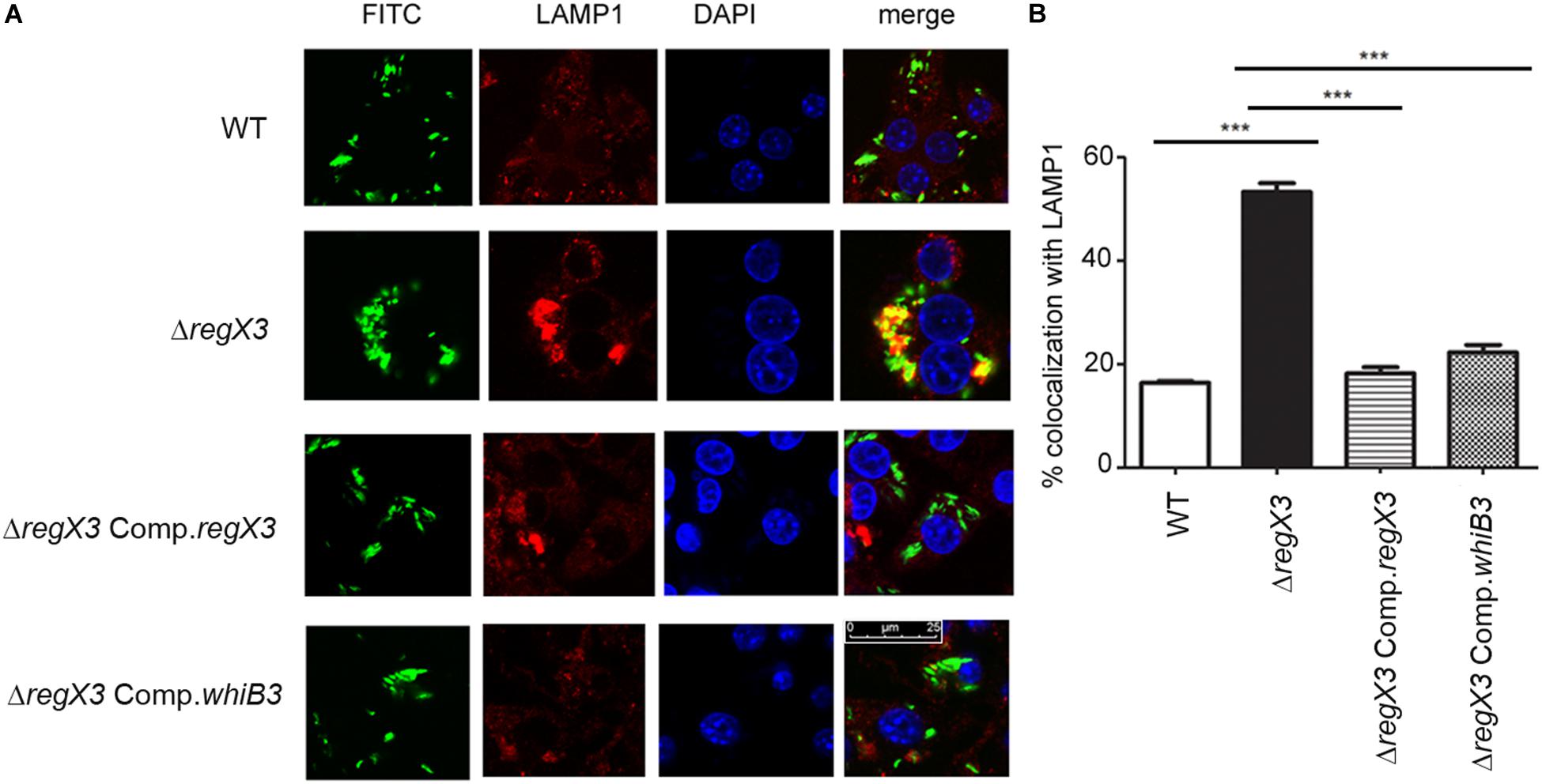
Figure 2. RegX3 and WhiB3 are required for trafficking of M. tuberculosis to lysosomes (A) RAW264.7 cells were infected with FITC-labeled (green) M. tuberculosis (WT, ΔregX3, ΔregX3 Comp.regX3 or ΔregX3 Comp.whiB3), fixed and immunolabeled with anti-LAMP1 followed by Alexa 546-conjugated secondary antibody (red). Cells were stained with DAPI to visualize the nuclei. Colocalization of green and red fluorescence indicates bacterial trafficking to the lysosomal compartment. (B) Percent colocalization of each of the four strains with LAMP1 was calculated by counting at least 100 bacteria from five different fields. Data represents means ± S.D., three independent experiments. ***p < 0.001.
RegX3 Has a Role in Granuloma Formation in M. tuberculosis
Human PBMCs have been previously used to elicit granuloma formation in vitro (Puissegur et al., 2004; Guirado et al., 2015). Previous reports have shown that WhiB3 is required for granuloma formation in vitro (Mehta and Singh, 2019). Considering that whiB3 expression is regulated by RegX3, we tested the role of RegX3 in granuloma formation in vitro 1 million PBMCs were infected with M. tuberculosis at an MOI of 0.01. Aggregates of PBMCs (>100 μm) were observed in the case of infection with the wild type bacterium. However, there was a striking absence of these aggregates when PBMCs were infected with ΔregX3 (Figure 3). Complementation of ΔregX3 with regX3 (ΔregX3 Comp.regX3) restored the ability of the bacterium to elicit aggregation and granuloma formation (Figure 3). Interestingly, ΔregX3 Comp.whiB3 was also able to elicit granuloma formation (Figure 3). These results strongly suggested that the role of RegX3 in granuloma formation is dependent on WhiB3.
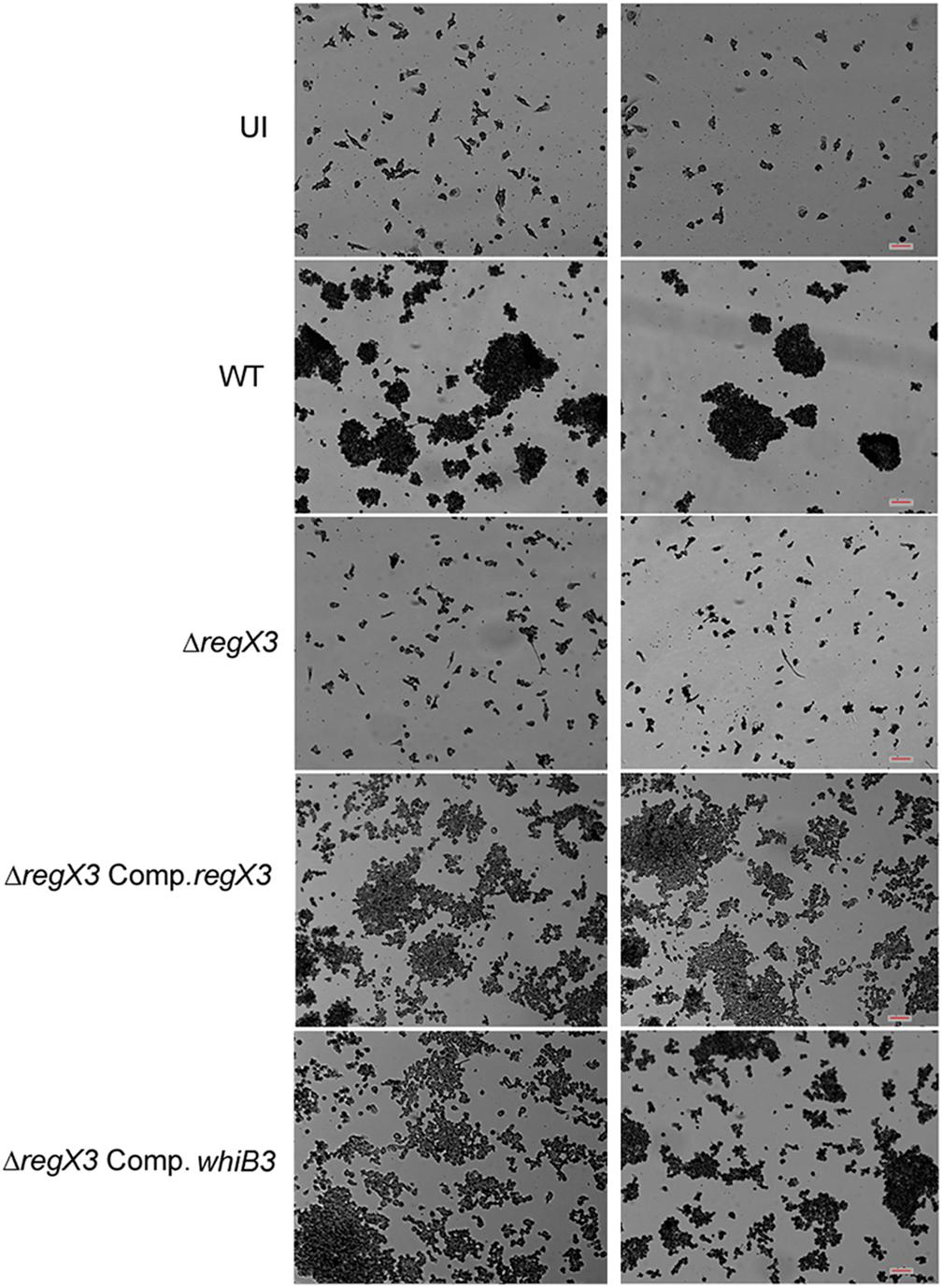
Figure 3. In vitro granuloma formation after infection of PBMCs with different strains of M. tuberculosis. One million human PBMCs were infected with different strains of M. tuberculosis at an MOI of 0.01; or were left uninfected (UI) and stained with May-Grünwald Giemsa stain at day 9 post-infection. Images were taken at 10× magnification. Each left and right panel shows two different fields out of at least five fields captured. Scale bar denotes 100 μm.
Discussion
The TCSs play an important role in regulating the ability of M. tuberculosis to respond to the stressful conditions it encounters during the course of establishing a successful infection. Among the paired TCSs, SenX3-RegX3 is best characterized as a phosphate starvation responsive TCS (Rifat et al., 2009). The other stress signals that it responds to, remain unclear. Its regulon is also incompletely characterized. The intracellular niche for M. tuberculosis is the macrophage, where survival under low pH poses a challenge. We searched for regulators that are activated when M. tuberculosis resides in the intracellular milieu of the macrophage, where the pH is acidic. RegX3 is one such regulator (Rohde et al., 2007). ΔregX3 is compromised in its ability to survive in macrophages compared to the wild type (Banerjee et al., 2016). In the present study we show that RegX3 is upregulated under acid stress (Table 1) and that it is required for the survival of M. tuberculosis under acid stress (Figure 1A).
The 4Fe-4S protein WhiB3 is activated under acid stress (Geiman et al., 2006) and in the response to vitamin C, a trigger of dormancy (Nandi et al., 2019). WhiB3 is required for the induction of immunomodulatory lipids and for blocking phagosomal maturation (Pacl et al., 2018). We show for the first time that RegX3 regulates whiB3 under both phosphate starvation and acid stress.
Previous studies have shown that the whiB3 promoter regions from M. tuberculosis and Mycobacterium marinum contain a PhoP box and that PhoP regulates whiB3 under acid stress (Feng et al., 2018), specifically in slow-growing mycobacteria such as M. tuberculosis. In this study we have identified a RegX3-binding palindrome upstream of whiB3 in M. tuberculosis (Figure 1B). The PhoP binding site on the whiB3 promoter is located between −310 and −271 of the whiB3 upstream region (Feng et al., 2018). The RegX3 binding site is located between −244 and −227. The RegX3 binding sequence is conserved in two members of the M. tuberculosis complex, namely M. africanum and M. bovis BCG. This sequence is not conserved in other mycobacteria such as M. leprae or the fast-growing M. smegmatis. Unlike the PhoP box, the RegX3 box is also absent in Mycobacterium kansasii and M. marinum, suggesting that dual regulation of whiB3 by PhoP and RegX3 under acid stress, may be unique to the M. tuberculosis complex. This suggests that a complex regulation of whiB3 by both PhoP and RegX3 is likely tailored to aid the survival of M. tuberculosis within its intracellular niche.
We observed that RegX3 binds to the whiB3 promoter in a phosphorylation-dependent manner (Figure 1D), and that a DNA-binding mutant of RegX3 fails to do so (Figure 1E), confirming the specificity of the binding. Mutation of a set of nucleotides within the putative RegX3-binding palindrome, abrogated RegX3 binding (Figure 1G), confirming the requirement of the palindromic sequence in the whiB3 promoter for RegX3 binding. We also confirmed binding of RegX3 to the whiB3 promoter region by ChIP using RegX3-specific antibody (Figure 1F). The regX3-deleted strain, ΔregX3, failed to activate the whiB3 promoter and was compromised in its ability to survive under acid stress. It showed compromised levels of whiB3 expression under either phosphate starvation (a known signal for RegX3 activation) or acid stress, suggesting the likely physiological relevance of the RegX3-WhiB3 axis. Taken together, these findings confirmed that RegX3 is a regulator of whiB3 in M. tuberculosis.
During infection, M. tuberculosis colonizes macrophages. We have previously shown that ΔregX3 is compromised in terms of its ability to survive in macrophages (Banerjee et al., 2016). In line with this, it showed increased trafficking to the lysosomal compartment (Figure 2). Concanamycin A, an inhibitor of vacuolar ATPases, blocks acidification of the phagosome in macrophages infected with M. tuberculosis (Rohde et al., 2007). In these conditions, intracellular whiB3 induction is inhibited, suggesting that acidification of the macrophage, triggers whiB3 induction, and that whiB3 probably has a role in subverting the trafficking of M. tuberculosis to macrophages. Interestingly, trafficking of ΔregX3 to the lysosomal compartment was reversed upon complementation with either regX3 or whiB3. These observations suggested that RegX3 functions at acid pH and in macrophages, in a WhiB3-dependent manner.
whiB3 is reportedly required for M. tuberculosis survival under acidic stress (Mehta et al., 2016). Since the RegX3 binding box as well as the PhoP box have been identified only in the whiB3 promoters of M. tuberculosis complex members, we suggest that the PhoPR-whiB3 and the RegX3-whiB3 regulatory pathways may act in concert in a manner unique to the M. tuberculosis complex, facilitating successful infection. In future studies, it would be of interest to evaluate the status of the PhoP and the RegX3 boxes in the whiB3 promoters of clinical strains of M. tuberculosis, and any possible link they may have with mycobacterial virulence. It would also be of importance to dissect the hierarchy of functioning of PhoP and RegX3 in the regulation of whiB3.
M. tuberculosis resides in granulomas within its host. Previous studies have shown that there is reduced lung inflammation in mice infected with ΔwhiB3 compared to wild type M. tuberculosis. There is scant knowledge of the M. tuberculosis factors that facilitate granuloma formation. Considering the physiological differences between animals and humans with respect to M. tuberculosis infection, we chose to study the role of the RegX3-WhiB3 axis in a human in vitro granuloma model. Here we show that granuloma formation in an in vitro model (i.e., the formation of aggregates >100 μm), is compromised in the absence of RegX3 (Figure 3). Interestingly granuloma formation is restored upon complementation of ΔregX3 with either regX3 or with whiB3, underscoring the importance of the RegX3-WhiB3 axis in granuloma formation, possibly through the WhiB3-dependent induction of immunomodulatory lipids. In summary, our present studies show that acid stress induces RegX3, which binds to the promoter of whiB3 to trigger its expression. WhiB3 subsequently mediates subversion of lysosomal trafficking of the bacterium in macrophages, and granuloma formation in vitro (Figure 4). The enhancement of understanding of pathogen biology through the present study, should aid in designing intervention strategies in future. It would be of interest to compare lung pathology elicited in vivo by M. tuberculosis ΔregX3 complemented with either regX3 or whiB3, and to generate a more complete understanding of the genes regulated by the RegX3/WhiB3 axis within the host milieu.
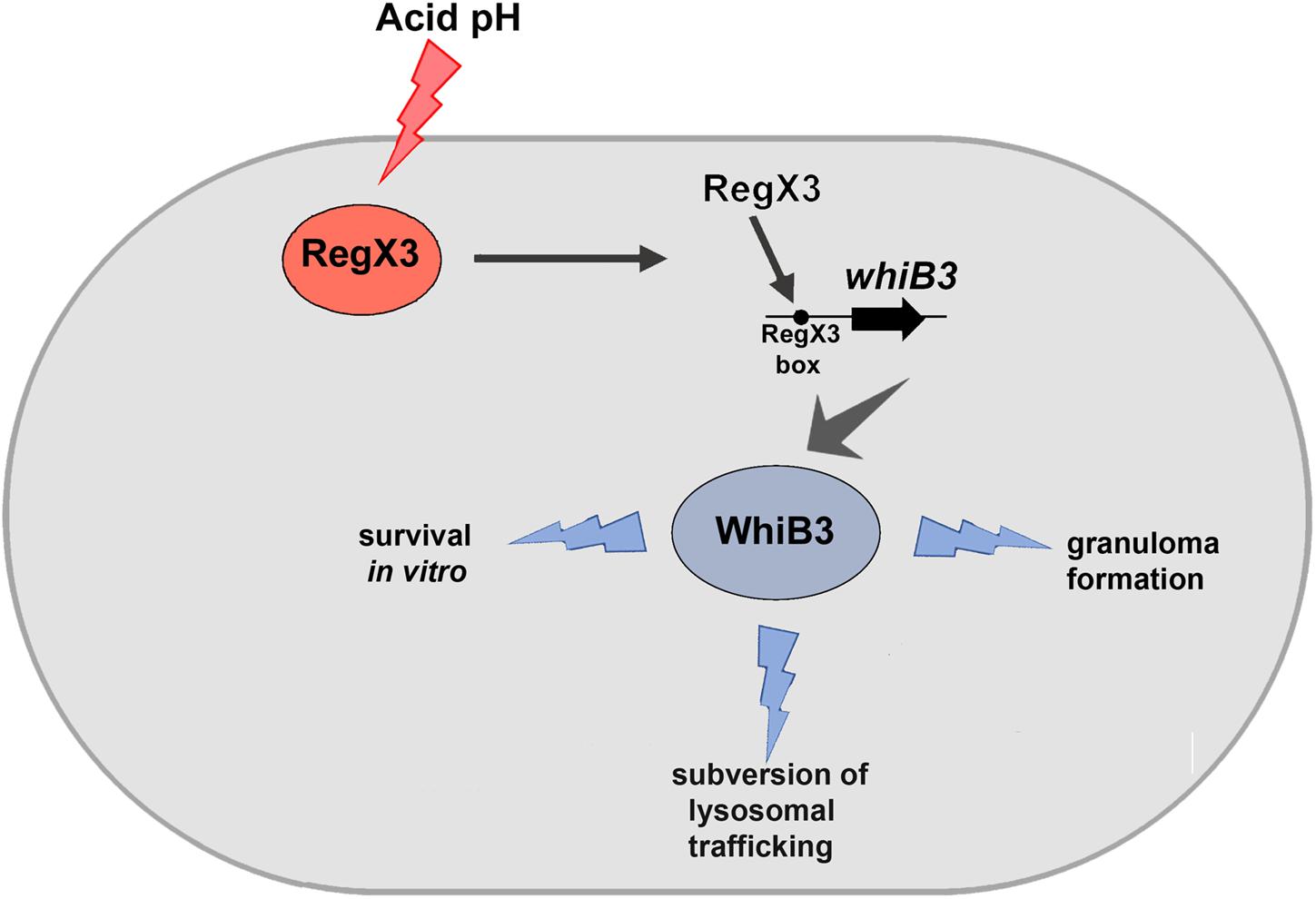
Figure 4. Schematic representation of the effect of RegX3 induction under acid stress in M. tuberculosis. RegX3 is induced upon exposure of M. tuberculosis to acid pH. It binds to the RegX3 box on the whiB3 promoter to trigger whiB3 induction. WhiB3 subsequently mediates subversion of lysosomal trafficking of M. tuberculosis to macrophages and granuloma formation in vitro.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Ethics Statement
The studies involving human participants were reviewed and approved by the Institutional Human Ethics Committee (BIHEC/2017-18/1). The patients/participants provided their written informed consent to participate in this study.
Author Contributions
JB and MK designed the research and analyzed the data. AM performed the experiments and analyzed the data. SM performed fluorescence microscopy. DM was involved in in vitro granuloma formation experiments. AG and SS performed the bioinformatic analyses. All authors contributed to the article and approved the submitted version.
Funding
This work was supported in part by grants from the Science and Engineering Research Board (SB/SO/HS/011/2014) and the Department of Biotechnology (BT/PR20242/MED/29/1062/2016) to MK. JB was supported by funding from the J.C. Bose Fellowship (SB/S2/JCB-049/2016). MK was supported by the Council of Scientific and Industrial Research, Emeritus Scientist Scheme (21(1088)/19/EMR-II).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.572433/full#supplementary-material
References
Banaiee, N., Jacobs, W. R. Jr., and Ernst, J. D. (2006). Regulation of Mycobacterium tuberculosis whiB3 in the mouse lung and macrophages. Infect. Immun. 74, 6449–6457. doi: 10.1128/iai.00190-06
Banerjee, S. K., Kumar, M., Alokam, R., Sharma, A. K., Chatterjee, A., Kumar, R., et al. (2016). Targeting multiple response regulators of Mycobacterium tuberculosis augments the host immune response to infection. Sci. Rep. 16:25851.
Banerjee, S. K., Lata, S., Sharma, A. K., Bagchi, S., Kumar, M., Sahu, S. K., et al. (2019). The sensor kinase MtrB of Mycobacterium tuberculosis regulates hypoxic survival and establishment of infection. J. Biol. Chem. 294, 19862–19876. doi: 10.1074/jbc.ra119.009449
Bansal, R., Anil Kumar, V., Sevalkar, R. R., Singh, P. R., and Sarkar, D. (2017). Mycobacterium tuberculosis virulence-regulator PhoP interacts with alternative sigma factor SigE during acid-stress response. Mol. Microbiol. 104, 400–411. doi: 10.1111/mmi.13635
Bardarov, S., Bardarov, S., Pavelka, M. S., Sambandamurthy, V., Larsen, M., Tufariello, J., et al. (2002). Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017. doi: 10.1099/00221287-148-10-3007
Bretl, D. J., Demetriadou, C., and Zahrt, T. C. (2011). Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582. doi: 10.1128/mmbr.05004-11
Cumming, B. M., Rahman, M. A., Lamprecht, D. A., Rohde, K. H., Saini, V., Adamson, J. H., et al. (2017). Mycobacterium tuberculosis arrests host cycle at the G1/S transition to establish long term infection. PLoS Pathog. 13:e1006389. doi: 10.1371/journal.ppat.1006389
Elliott, S. R., and Tischler, A. D. (2016). Phosphate starvation: a novel signal that triggers ESX-5 secretion in Mycobacterium tuberculosis. Mol. Microbiol. 100, 510–526. doi: 10.1111/mmi.13332
Feng, L., Chen, S., and Hua, Y. (2018). PhoPR positively regulates whiB3 expression in response to low pH in pathogenic mycobacteria. J. Bacteriol. 200:e00766-17.
Geiman, D. E., Raghunand, T. R., Agarwal, N., and Bishai, W. R. (2006). Differential gene expression in response to exposure to antimycobacterial agents and other stress conditions among seven Mycobacterium tuberculosis whiB like genes. Antimicrob. Agents Chemother. 50, 2836–2841. doi: 10.1128/aac.00295-06
Glover, R. T., Kriakov, J., Garforth, S. J., Baughn, A. D., and Jacobs, W. R. Jr. (2007). The two-component regulatory system senX3-regX3 regulates phosphate-dependent gene expression in Mycobacterium smegmatis. J. Bacteriol. 189, 5495–5503. doi: 10.1128/jb.00190-07
Guirado, E., Mbawuike, U., Keiser, T. L., Arcos, J., Azad, A. K., Wang, S.-H., et al. (2015). Characterisation of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 6:e02537-14.
Healy, C., Golby, P., MacHugh, D. E., and Gordon, S. V. (2016). The MarR family transcription factor Rv1404 coordinates adaptation of Mycobacterium tuberculosis to acid stress via controlled expression of Rv1405c, a virulence-associated methyltransferase. Tuberculosis 97, 154–162. doi: 10.1016/j.tube.2015.10.003
James, J. N., Hasan, Z., Iorger, T. R., Brown, A. C., Personne, Y., Carroll, P., et al. (2012). Deletion of SenX3–RegX3, a key two-component regulatory system of Mycobacterium smegmatis, results in growth defects under phosphate-limiting conditions. Microbiology 158, 2724–2731. doi: 10.1099/mic.0.060319-0
Karim, A. F., Chandra, P., Chopra, A., Siddiqui, Z., Bhaskar, A., Singh, A., et al. (2011). Express path analysis identifies a tyrosine kinase Src-centric network regulating divergent host responses to Mycobacterium tuberculosis infection. J. Biol. Chem. 286, 40307–40319. doi: 10.1074/jbc.m111.266239
Kundu, M. (2018). The role of two-component systems in the physiology of Mycobacterium tuberculosis. IUBMB Life 70, 710–717. doi: 10.1002/iub.1872
Larsson, C., Luna, B., Ammerman, N. C., Maiga, M., Agarwal, N., and Bishai, W. R. (2012). Gene expression of Mycobacterium tuberculosis putative transcription factors whiB1-7 in redox environments. PLoS One 7:e37516. doi: 10.1371/journal.pone.0037516
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mehta, M., Rajmani, R. S., and Singh, A. (2016). Mycobacterium tuberculosis WhiB3 responds to vacuolar pH-induced changes in mycothiol redox potential to modulate phagosomal maturation and virulence. J. Biol. Chem. 291, 2888–2903. doi: 10.1074/jbc.m115.684597
Mehta, M., and Singh, A. (2019). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis and survival in response to reactive oxygen and nitrogen species. Free Radic. Biol. Med. 131, 50–58. doi: 10.1016/j.freeradbiomed.2018.11.032
Namugenyi, S. B., Aagesen, A. M., Elliott, S. R., and Tischler, A. D. (2017). Mycobacterium tuberculosis PhoY proteins promote persister formation by mediating Pst/SenX3-RegX3 phosphate sensing. mBio 8:e0494-17.
Nandi, M., Sikri, K., Chaudhary, N., Mande, S. C., Sharma, R. D., and Tyagi, J. S. (2019). Multiple transcription factors co-regulate the Mycobacterium tuberculosis adaptation response to vitamin C. BMC Genomics 20:887. doi: 10.1186/s12864-019-6190-3
Pacl, H. T., Reddy, V. P., Saini, V., Chinta, K. C., and Steyn, A. J. (2018). Host-pathogen redox dynamics modulates Mycobacterium tuberculosis pathogenesis. Pathog. Dis. 76:fty036.
Parish, T. (2014). Two-component regulatory systems of mycobacteria. Microbiol. Spectr. 2:MGM2-0010-2013.
Parish, T., Smith, D. A., Roberts, G., Betts, J., and Stoker, N. G. (2003). The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 149, 1423–1435. doi: 10.1099/mic.0.26245-0
Puissegur, M. P., Botanch, C., Duteyrat, J. L., Delsol, G., Caratero, C., and Altare, F. (2004). An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell. Microbiol. 6, 423–433. doi: 10.1111/j.1462-5822.2004.00371.x
Rifat, D., Belchis, D. A., and Karakousis, P. C. (2014). SenX3-independent contribution of regX3 to Mycobacterium tuberculosis virulence. BMC Microbiol. 14:265. doi: 10.1186/s12866-014-0265-8
Rifat, D., Bishai, W. R., and Karakousis, P. C. (2009). Phosphate depletion: a novel trigger for Mycobacterium tuberculosis persistence. J. Infect. Dis. 200, 1126–1135.
Rifat, D., and Karakousis, P. C. (2014). Differential regulation of the two-component regulatory system senX3-regX3 in Mycobacterium tuberculosis. Microbiology 160, 1125–1133. doi: 10.1099/mic.0.077180-0
Roberts, G., Vadrevu, I. S., Madiraju, M. V., and Parish, T. (2011). Control of CydB and GltA1 expression by the SenX3 RegX3 two component regulatory system of Mycobacterium tuberculosis. PLoS One 6:e21090. doi: 10.1371/journal.pone.0021090
Rohde, K. H., Abramovitch, R. B., and Russell, D. G. (2007). Mycobacterium tuberculosis invasion of macrophages: linking bacterial gene expression to environmental cues. Cell Host Microbe 2, 352–364. doi: 10.1016/j.chom.2007.09.006
Rustad, T. R., Minch, K. J., Ma, S., Winkler, J. K., Hobbs, S., Hickey, M., et al. (2014). Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15:502. doi: 10.1186/s13059-014-0502-3
Saini, V., Farhana, A., and Steyn, A. J. (2012). Mycobacterium tuberculosis WhiB3: a novel iron-sulfur cluster protein that regulates redox homeostasis and virulence. Antioxid. Redox Signal. 16, 687–697. doi: 10.1089/ars.2011.4341
Sanyal, S., Banerjee, S. K., Banerjee, R., Mukhopadhyay, J., and Kundu, M. (2013). Polyphosphate kinase 1, a central node in the stress response network of Mycobacterium tuberculosis, connects the two-component systems MprAB and SenX3–RegX3 and the extracytoplasmic function sigma factor, sigma E. Microbiology 159, 2074–2086. doi: 10.1099/mic.0.068452-0
Sharma, A. K., Chatterjee, A. S., Banerjee, R., Mandal, S., and Mukhopadhyay, J. (2015). MtrA, an essential response regulator of the MtrAB two-component system, regulates the transcription of resuscitation-promoting factor B of Mycobacterium tuberculosis. Microbiology 161, 1271–1281. doi: 10.1099/mic.0.000087
Singh, A., Crossman, D. K., Mai, D., Guidry, L., Voskuil, M. I, Renfrow, M. B., et al. (2009). Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathog. 5:e1000545. doi: 10.1371/journal.ppat.1000545
Singh, A., Guidry, L., Narasimhulu, K. V., Mai, D., Trombley, J., Redding, K. E., et al. (2007). Mycobacterium tuberculosis WhiB3 responds to O2 and nitric oxide via its [4Fe-4S] cluster and is essential for nutrient starvation survival. Proc. Natl. Acad. Sci. U.S.A. 104, 11562–11567. doi: 10.1073/pnas.0700490104
Steyn, A. J. C., Collins, D. M., Hondalus, M. K., Jacobs, W. R., Kawakami, P., and Bloom, B. R. (2002). Mycobacterium tuberculosis WhiB3 interacts with RpoV to affect host survival but is dispensable for in vivo growth. Proc. Natl. Acad. Sci. U.S.A. 99, 3147–3152. doi: 10.1073/pnas.052705399
Tischler, A. D., Leistikow, R. L., Kirksey, M. A., Voskuil, M. I., and McKinney, J. D. (2013). Mycobacterium tuberculosis requires phosphate-responsive gene regulation to resist host immunity. Infect. Immun. 81, 317–328. doi: 10.1128/iai.01136-12
Tiwari, S., Jamal, S. B., Hassan, S. S., Carvalho, P. V. S., Almeida, S., Barh, D., et al. (2017). Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front. Microbiol. 8:1878. doi: 10.3389/fmicb.2017.01878
White, D. W., Elliott, S. R., Odean, E., Bemis, L. T., and Tischler, A. D. (2018). Mycobacterium tuberculosis Pst/SenX3-RegX3 regulates membrane vesicle production independently of ESX-5 activity. mBio 9: e00778-18.
Keywords: Mycobacterium tuberculosis, two-component systems, gene expression, acid stress, lysosomal trafficking, granuloma formation
Citation: Mahatha AC, Mal S, Majumder D, Saha S, Ghosh A, Basu J and Kundu M (2020) RegX3 Activates whiB3 Under Acid Stress and Subverts Lysosomal Trafficking of Mycobacterium tuberculosis in a WhiB3-Dependent Manner. Front. Microbiol. 11:572433. doi: 10.3389/fmicb.2020.572433
Received: 14 June 2020; Accepted: 10 August 2020;
Published: 16 September 2020.
Edited by:
Daniel Pletzer, University of Otago, New ZealandReviewed by:
Mario Alberto Flores-Valdez, CONACYT Centro de Investigación y Asistencia en Tecnología y Diseño del Estado de Jalisco (CIATEJ), MexicoPetros Karakousis, Johns Hopkins University, United States
Yusuf Akhter, Babasaheb Bhimrao Ambedkar University, India
Copyright © 2020 Mahatha, Mal, Majumder, Saha, Ghosh, Basu and Kundu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Manikuntala Kundu, bWFuaWt1bnRhbGEua3VuZHVAZ21haWwuY29t
 Amar Chandra Mahatha
Amar Chandra Mahatha Soumya Mal
Soumya Mal Debayan Majumder
Debayan Majumder Sudipto Saha
Sudipto Saha Abhirupa Ghosh2
Abhirupa Ghosh2 Joyoti Basu
Joyoti Basu Manikuntala Kundu
Manikuntala Kundu