- Food College, Shihezi University, Shihezi, China
Petit Manseng is widely used for fermenting sweet wine and is popular among younger consumers because of its sweet taste and attractive flavor. To understand the mechanisms underlying spontaneous fermentation of Petit Manseng sweet wine in Xinjiang, the dynamic changes in the microbial population and volatile compounds were investigated through high-throughput sequencing (HTS) and headspace solid-phase microextraction (HS-SPME) coupled to gas chromatography-mass spectrometry (GC-MS) technology, respectively. Moreover, the relationship between the microbial population and volatile compounds was deduced via multivariate data analysis. Candida and Mortierella were dominant genera in Petit Manseng wine during spontaneous fermentation. Many fermentative aroma compounds, including ethyl octanoate, isoamyl acetate, ethyl butyrate, ethyl decanoate, isoamyl alcohol, ethyl laurate, isopropyl acetate, hexanoic acid, and octanoic acid, were noted and found to be responsible for the strong fruity and fatty aroma of Petit Manseng sweet wine. Multivariate data analysis indicated that the predominant microorganisms contributed to the formation of these fermentative aroma compounds. Hannaella and Neomicrosphaeropsis displayed a significantly positive correlation with the 6-methylhept-5-en-2-one produced. The current results provide a reference for producing Petit Manseng sweet wine with desirable characteristics.
Introduction
Wine is an alcoholic beverage derived from grapes, and its production involves chemistry and biology (Belda et al., 2017). Based on its sugar content, wine can be classified into dry, semi-dry, sweet, and semi-sweet wine (Brainina et al., 2004). Because of their taste and rich flavor, sweet wines are popular (Swami et al., 2014). The grape varieties used to make sweet wine mainly include Riesling, Sauvignon Blanc, and Petit Manseng (Englezos et al., 2018; Lan et al., 2021). Of these, Petit Manseng is a thick-skinned, white grape variety grown in southwest France, and it is widely used to produce high-quality sweet wines rich in a fruity, slightly spicy aroma (Gardner et al., 2017). As a late-harvest cultivar, Petit Manseng has a high sugar and acid content, and consequently, it is strongly resistant to several fungal diseases (Robinson et al., 2013).
A wine’s quality is determined based on its taste (Loureiro et al., 2016), color (Zhang et al., 2018), and aroma (Sherman et al., 2020). An important component of wine, the aroma is considered to be indicative of the regional and cultivar characteristics of a wine. This is because aroma is affected by natural elements (e.g., grape cultivars and climatic condition) (Mirás-Avalos et al., 2017), wine-making techniques (e.g., oak maturation and fermentation method) (Crump et al., 2015; Shi et al., 2019), and indigenous microorganism (e.g., bacteria, yeast, and fungi) (Liu et al., 2017). Wine aroma can be divided into varietal, fermentative, and aging (Belda et al., 2017). Varietal aromas, derived from wine grapes, are responsible for most of the aromas in a wine (Waterhouse et al., 2016). For example, when homologous, Cabernet Franc, Cabernet Sauvignon, Sauvignon Blanc, Merlot, and Carmenère have a typical bell pepper, vegetal, and earthy aroma (Ruiz et al., 2019).
High-throughput sequencing (HTS) is useful for evaluating dynamic changes in microbial populations during fermentation (Guo et al., 2020). Therefore, HTS has enabled the identification of a variety of dominant microorganisms, especially culture-independent microorganisms, in wine (Bučková et al., 2018). Wine is fermented by various indigenous microorganisms, and these vary based on the wine grape cultivar (Francesca et al., 2016). For instance, Hanseniaspora, Metschnikowia, Saccharomyces, Lactococcus, and Leuconostoc are predominant in Grüner Veltliner ice wine (Alessandria et al., 2013), whereas Leuconostoc, Lactococcus, and Saccharomyces are dominant in “Marselan” red wine (Lu et al., 2020). Among the aforementioned indigenous microorganisms, Saccharomyces cerevisiae not only promotes alcoholic fermentation but also increases phenol and antioxidant content through alcoholic fermentation (Caridi et al., 2017). Non-Saccharomyces yeasts are gaining increasing attention because they can improve the aroma complexity or mouth-feel of wines (Padilla et al., 2016).
Based on whether or not a starter culture is used, wine fermentation can be divided into inoculated fermentation and spontaneous fermentation. Compared with inoculated fermentation, spontaneous fermentation, which is caused by complex indigenous microorganisms, can provide a more complex and richer flavor in wine (Lu et al., 2021). Spontaneous fermentation makes full use of diverse microorganisms, especially the specific autochthonous yeasts (de Celis et al., 2019). The spontaneous fermentation by non-Saccharomyces yeasts has been found to enhance the aroma character of Cabernet Sauvignon, Chardonnay, and Vidal wines (Lu et al., 2020).
High-throughput sequencing (HTS) combined with multivariate data analysis has been widely used to investigate microbial diversity and its impact on wine quality (Böhmer et al., 2020). An increasing number of microorganisms, contributing to potential wine flavors, including Hanseniaspora uvarum, Wickerhamomyces anomalus, and Lachancea thermotolerans, have been identified (Belda et al., 2016).
Petit Manseng sweet wines are popular among young consumers (Chaplin, 2011). Although the sensory characteristics of Petit Manseng wines have been investigated via headspace solid-phase microextraction (HS-SPME) coupled to gas chromatography-mass spectrometry (GC-MS) and by using a polymer electronic nose (Gardner et al., 2017), the relationship of volatile compounds with the microbial community in these wines during spontaneous fermentation remains unclear. To understand the mechanisms underlying the spontaneous fermentation of Petit Manseng sweet wine in Xinjiang, dynamic changes in the microbial community and volatile compounds during spontaneous fermentation were investigated through HTS and HS-SPME/GC-MS, respectively. The relationship between the microbial populations and the volatile compounds was elucidated using multivariate data analysis. The current results provide a theoretical reference for producing Petit Manseng sweet wines with typical characteristics.
Materials and Methods
Wine Making and Sampling
Petit Manseng was harvested from Changji County, Xinjiang Province, China in October 2019. After strict screening, healthy grapes were immediately crushed. The biochemistry of these grapes was then analyzed: residual sugar content, 347.35 g/L; yeast assimilable nitrogen (YAN): 347 mg N/L; pH, 3.49; soluble solids, 32.6 °Brix; total acidity, 5.17 g/L. This was followed by the addition of 20 mg/L pectinase. The grapes were macerated at 8−10°C for 48 h; then, the separated grape supernatants were subjected to alcoholic fermentation. All fermentation was performed in 50-L fermenters at 15–17°C (factory ambient temperature) under static conditions for 14°days. The alcohol fermentation was considered to be completed when residual sugar content was <110 g/L. The fermentation liquid (250 mL) was collected on fermentation days 0, 1, 4, 7, 11, and 14. All samples were centrifuged at 1,000 × g at 4°C for 10 min. The precipitates were then collected and used for HTS analysis, whereas the supernatants were used to assess volatile compound analysis. All samples were stored at −80°C until analysis. Each sample was analyzed in triplicate.
Determination of Physicochemical Properties
During alcoholic fermentation, some fermentation parameters were measured including pH, total acidity, and residual sugar and ethanol content. The pH of wine was measured using a calibrated pH meter (PHS-3C; Shanghai Jingke, Shanghai, China). Residual sugar content was assessed using dinitrosalicylic acid (DNS) method (Miller, 1959). Total acidity and ethanol content were determined according to the national standard of China: GB/T 15038-2006, “Analytical methods of wine and fruit wine.” Organic acids in the wine samples were analyzed through high-performance liquid chromatography (HPLC) using previously described methods with some modifications (Murtaza et al., 2017). Each sample was centrifuged and filtered through a 0.22-μm filter, and the organic acids were identified through HPLC equipped with a Spursil C18 column (250 mm × 4.6 mm × 5 μm; Dima Technology, Guangzhou, China). A mixture of 0.1% phosphoric acid-methanol was regarded as the mobile phase, with a flow rate of 1 mL/min. The UV detection wavelength was maintained at 210 nm, and the column temperature was 40°C. Each sample was measured in three replicates.
Fungal Isolation and Identification
The wine sample was serially diluted from 1 × 10 to 1 × 105 times by using sterile water and plated on yeast extract-peptone-dextrose (YPD) agar, and all plates were cultured at 28°C for 3 days under sterile conditions. After incubation, the fungal colonies on each plate were selected and isolated as representative fungal populations present during spontaneous fermentation. All representative strains were incubated on the WL nutrient agar (Haibo, Qingdao, China) at 28°C for 5 days. Then, fungal colonies with different morphological types were selected for the next step of identification. Single colonies of fungi were purified and stored in YPD liquid medium with 40% glycerol at −20°C. Fungal strains were analyzed and identified by comparing nucleotide sequences in the GenBank database (NCBI: https://blast.ncbi.nlm.nih.gov/blast.cgi).
HTS
The total DNA of fungal colonies isolated and selected from our wine samples was extracted using the CTAB/SDS method (Lin et al., 1994; Brandfass and Karlovsky, 2008). Next, the DNA of microorganisms were sent to Novegene Company (China) for PCR amplification and HTS. The primers used were ITS5-1737F (5′-GGAAGTAAAAGTCGTAACAAGG-3′) and ITS2-2043R (5′-GCTGCGTTCTTCATCGATGC-3′). For PCR, the total reaction volume was 25 μL, comprising 15 μL of the PCR Master Mix, 2 μL of the forward and reverse primers, 10 ng of the template DNA, and water (PCR-grade) to adjust the volume. We used the following thermal cycling program: initial denaturation at 98°C for 1 min, followed by 35 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and elongation at 72°C for 30 s, and finally, extension at 72°C for 5 min. All PCR products were detected through 2% agarose gel electrophoresis and purified using the Qiagen Gel Extraction Kit (Qiagen, Germany). Next, sequencing libraries were generated using TruSeq DNA PCR-Free Sample Preparation Kit (Illumina, United States). The library was assessed on a Qubit 2.0 Fluorometer (Thermo Fisher Scientific) and Agilent Bioanalyzer 2100. The Illumina NovaSeq platform was used to sequence 250-bp paired-end reads.
Processing of Sequence Analysis
Paired-end reads were assigned to samples based on their unique barcodes and truncated by cutting off the barcodes and primer sequences. Paired-end reads were merged using FLASH (version 1.2.7). The raw reads were filtered to obtain high-quality clean reads using QIIME (version 1.9.1). Moreover, the chimeric sequences were filtered out by comparing them with those in the SILVA database (a reference database). Moreover, the same operational taxonomic units (OTUs) were clustered from sequences with 97% sequence similarity by using Uparse (version 7.0.1001). The most abundant sequence in each OTU was selected as the representative sequence. Taxonomic units were assigned to the representative sequence (i.e., to each OTU) using the SILVA database. MUSCLE (version 3.8.31) was used for multiple sequence alignment and studied the phylogenetic relationship of different OTUs. Finally, alpha diversity indexes such as Shannon, Simpson, Chao1, and ACE were calculated on QIIME (version 1.9.1).
Determination of Volatile Compounds
Headspace solid-phase microextraction (HS-SPME)/gas chromatography-mass spectrometry (GC-MS) was used to extract and analyze volatile compounds in our wine, by using Kong’s et al. (2019) method with some modifications. In brief, 1 g of NaCl was added to 5 mL of sample supernatants. This mixture was then transferred to a 20-mL HS vial, followed by 1 μL of 3-octanol (165 mg/mL) as the internal standard. The vial was sealed immediately with a polytetrafluoroethene (PTFE) silicone diaphragm and balanced at 40°C for 10 min. An SPME fiber (DVB/CAR/PDMS 50/30 μm; Supelco, Bellefonte, PA, United States) was then inserted into the sealed glass vial and exposed to the HS for resolution for 40 min at 40°C. After extraction, the fiber was inserted into the GC injection port at an interface temperature of 250°C for 5 min. Each volatile compound was analyzed using an HP INNOWAX column (30 m × 0.25 mm; Agilent). Helium was the carrier gas, circulated at 1 mL/min, and the electron ionization energy was maintained at 70 eV. The scanning range of the total ion chromatographs was 35–350 m/z. The temperature program was as follows: 3 min at 50°C, followed by 2°C/min to 100°C, 4°C/min to 180°C, and 10°C/min to 230°C.
The volatile aroma compounds were identified by matching their retention indexes with those in the mass spectra library (NIST). Moreover, the retention indexes were evaluated using the Kovats system (Zenkevich, 2002). All compounds were semiquantified as 3-octanol equivalents. The semi-quantitative data of the aroma compounds were calculated as follows:
where RCAC is the relative concentration of the volatile compounds, PAVC is the peak area of volatile compounds, PAIS is the peak area of internal standard, and CIS is the final concentration of internal standard.
Sensory Analysis of Wine
In this study, the Petit Manseng grapes were used as raw materials for vinification, we also compared them with the Petit Manseng wine. Sensory analysis was performed using Belda’s et al. (2015) method with some modifications. In brief, the wine was assessed by 10 trained panelists (6 female and 4 male) from Shihezi University. During the formal sensory analysis process, the team members were asked to form consistent terminology by consensus and described the sensory notes. Sensory descriptions included fruity, floral, green, acidity, sweetness, fatty, and bitterness, all scored from 0 (weak) to 10 (intense).
Statistical Analysis
For each sample group, three parallel samples were analyzed. The differences between our samples were analyzed using analysis of variance with Duncan multiple tests, with the significance level set at 5% (P < 0.05) on SPSS (version 20; IBM, Chicago, United States). R (version 4.0.4) was used to produce a heat-map for the microbial populations and volatile compounds. The correlation among microorganisms and volatile compounds was determined multivariate data analysis, including bidirectional partial least squares (O2PLS), on the software program SIMCA (version 14.1). The linear correlation coefficient between the selected microbial genera and volatile compounds was visualized via O2PLS on Cyto-scape (version 3.6.1).
Results
Physicochemical Properties During Spontaneous Fermentation
Dynamic changes in the ethanol, total sugar, and total acid content and the pH of zymotic fluid from six periods during Petit Manseng spontaneous fermentation were detected (Figure 1). The total sugar content of Petit Manseng juice reached up to 345.35 g/L. During Petit Manseng spontaneous fermentation, sugar in the zymotic fluid was converted to ethanol. Thus, total sugar content sharply decreased after 4 days of fermentation and remained at 110.84 ± 1.69 g/L on the 14th day. Then, ethanol concentrations increased after 4 days of fermentation and reached up to 12.01 ± 0.09% (v/v) on the 14th day. Total acid content fluctuated from 4.97 ± 0.02 to 6.00 ± 0.26 g/L. The pH remained relatively stable at 3.2 throughout the fermentation process.
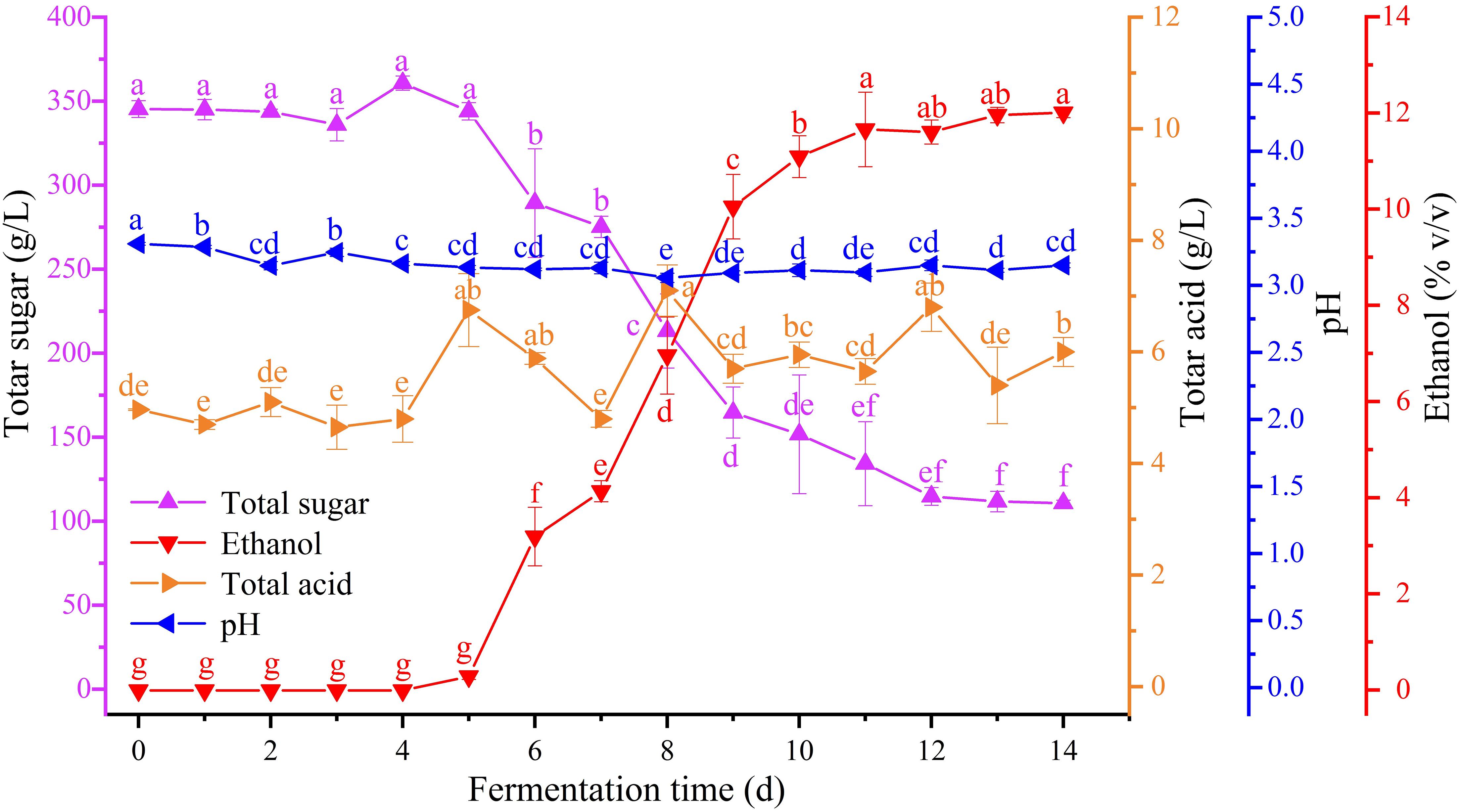
Figure 1. Change of physicochemical indexes in different fermentation time: total sugar (purple), ethanol (red), total acid (orange), and pH (blue) (for interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
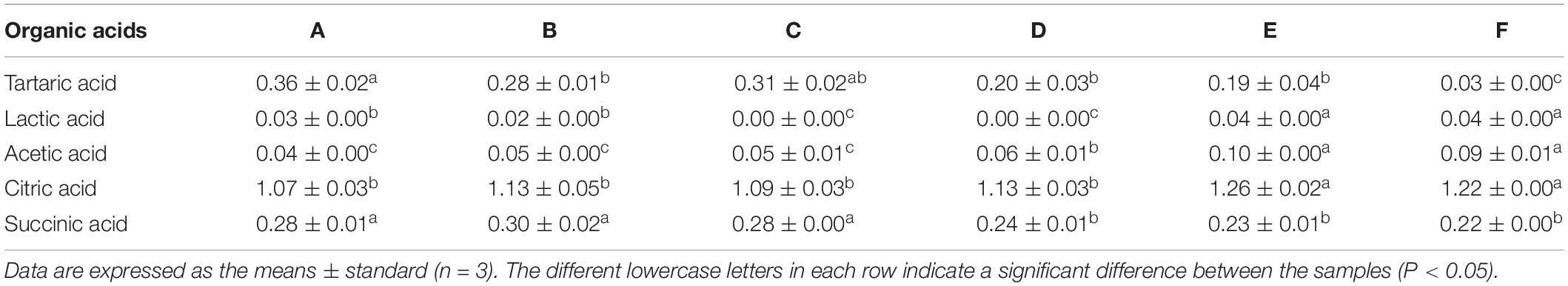
The organic acid content directly affects the flavor and taste of white wine (Nie et al., 2017). In this study, the organic acids detected from Petit Manseng were tartaric acid, lactic acid, acetic acid, citric acid, and succinic acid (Table 1). Tartaric acid content sharply decreased throughout the fermentation process, with it almost depleted at the end of fermentation. In contrast, lactic acid content showed a slight increase, with a final value of 0.04 ± 0.00 mg/mL. Acetic acid content showed an upward trend overall, increasing sharply on the 11th day and then slowly decreasing to finally reach a concentration of 0.09 ± 0.01 mg/mL. Citric acid showed an upward trend overall. In particular, citric acid was continuously consumed and synthesized during spontaneous fermentation. Finally, succinic acid content remained relatively stable throughout the fermentation process.
Volatile Compounds During Spontaneous Fermentation
Volatile compounds from six fermentation periods of Petit Manseng wine during spontaneous fermentation included 17 esters, 19 alcohols, 9 acids, 4 aldehydes, 4 ketones, and 3 other volatile compounds (Table 2).
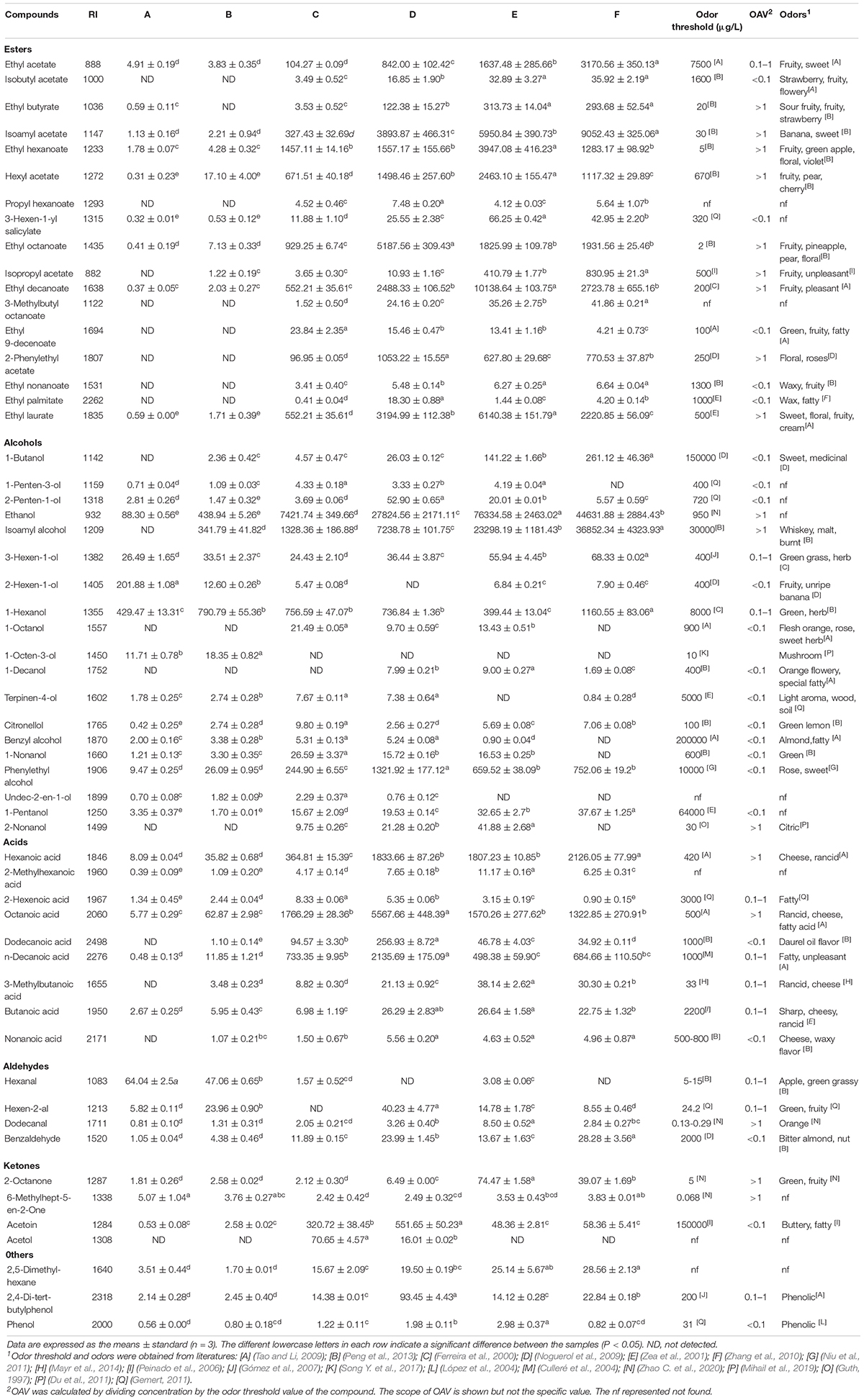
Table 2. Dynamic changes of flavor compound content during Petit Manseng fermentation process (μg/L).
The esters mainly included isoamyl acetate, ethyl lurate, ethyl decanoate, ethyl acetate, hexyl acetate, ethyl hexanoate, and ethyl octanoate. A large amount of acetate esters and ethyl esters were produced because a high concentration of alcohol and the related acetyl-CoA and acyltransferases interacted during spontaneous fermentation. Moreover, the produced wine contained high levels of ethanol, isoamyl alcohol, 1-hexanol, and phenylethyl alcohol. Our wine was also rich in hexanoic acid, octanoic acid, and decanoic acid. These esters, alcohols, and acids mainly conferred the overall flavor of our Petit Manseng wine. Aldehydes, ketones, and other volatiles may have also contributed to its flavors.
To better understand the dynamic changes in these volatile compounds, we analyzed the aroma components throughout the spontaneous fermentation process (Figure 2). The volatile compounds were divided into two classes based on the trends during the fermentation process: Class I contained 26 aromatic compounds (mainly including higher alcohols, C6 compounds, and fatty acids) and Class II contained 30 aromatic compounds (mainly including benzene derivatives, ethyl esters, acetic esters, and other volatiles). The high content of acetic and ethyl esters is responsible for the fruity flavor in the wine, whereas excessive concentrations of higher alcohols and fatty acids lead to unpleasant green, pungent, and rancid flavor in the wine. Based on our results, the spontaneous fermentation of Petit Manseng might produce wines with a more coordinated wine aroma (Shi et al., 2019).
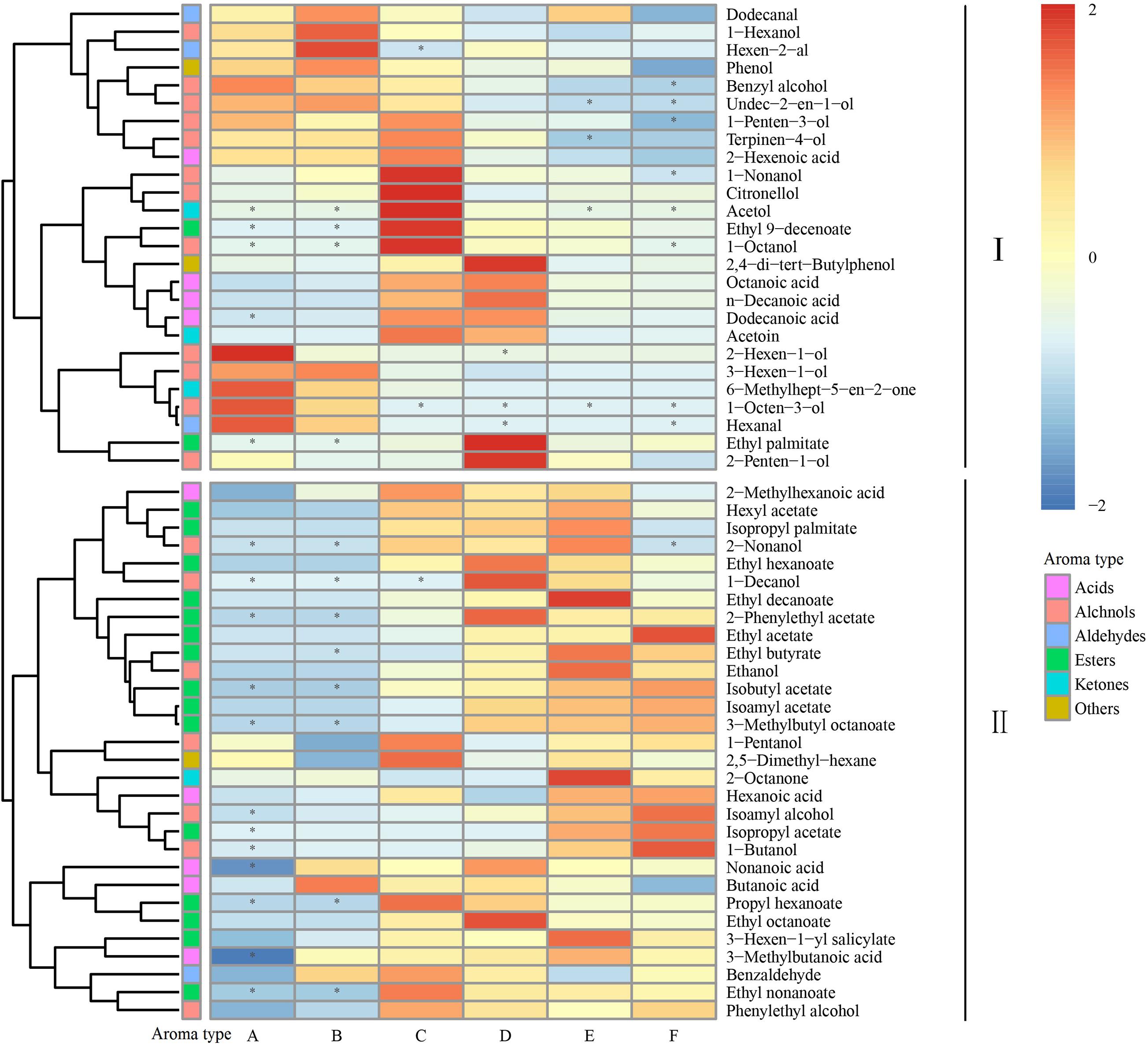
Figure 2. Heatmap cluster analysis of the volatile components during the fermentation. The asterisk indicates the substance with P < 0.05. Samples collected on fermentation days 0, 1, 4, 7, 11, and 14, respectively (for interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article).
We next used principal component analysis (PCA) to understand the correlation and segregation of aroma compounds in different wine samples (Figure 3). Here, 76.2% of the variance was explained by 56 different components, with PC1 and PC2 accounted for 51.3 and 24.9% of the variance, respectively. Most volatile compounds clustered at stages D and F, which corroborated the results indicating higher concentrations of volatile compounds in the wine. The rest of the wine samples clustered with only a few volatile compounds, also consistent with their lower concentration of volatile compounds in the respective stages. The variations in the location indicated that the aroma of Petit Manseng wine differed by the fermentation stage. Notably, half of the volatile compounds at stage D (i.e., alcohols and esters) were all located in the upper right quadrant.
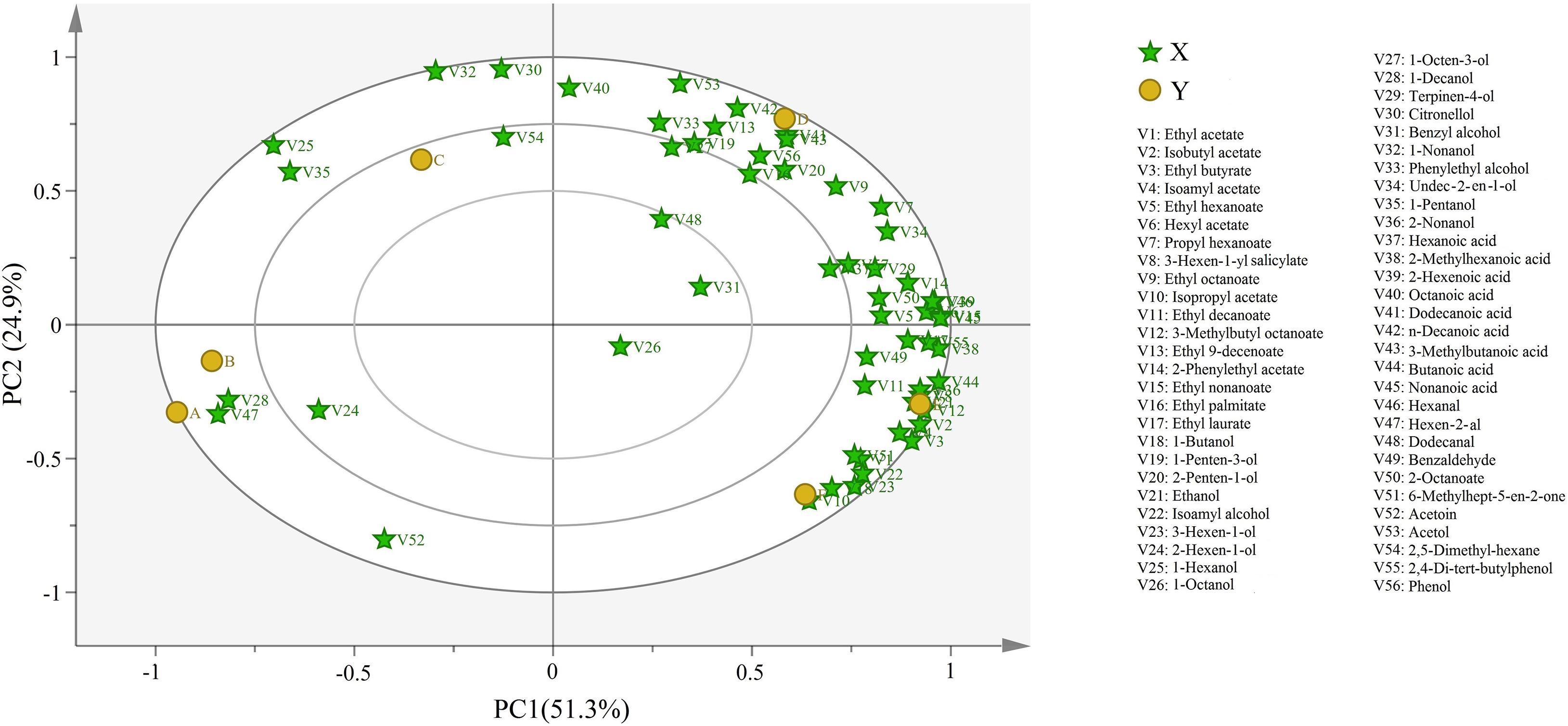
Figure 3. Bioplot of PCA for volatile compound of wine samples. A, B, C, D, E, and F represented samples collected on fermentation days 0, 1, 4, 7, 11, and 14, respectively.
Sensory Analysis of Wine
The average scores of the sensory descriptions are presented in Figure 4D. Obvious differences were recorded in Petit Manseng grapes and wine fermented by Petit Manseng. The bitterness, green, acidity, and sweetness of the grapes scored higher than wine. The scores were almost similar of Petit Manseng grape and wine. Interestingly, the wine was noted to produce slightly stronger floral sensations than our grapes after spontaneous fermentation. These results indicated that our Petit Manseng wine has great market potential.
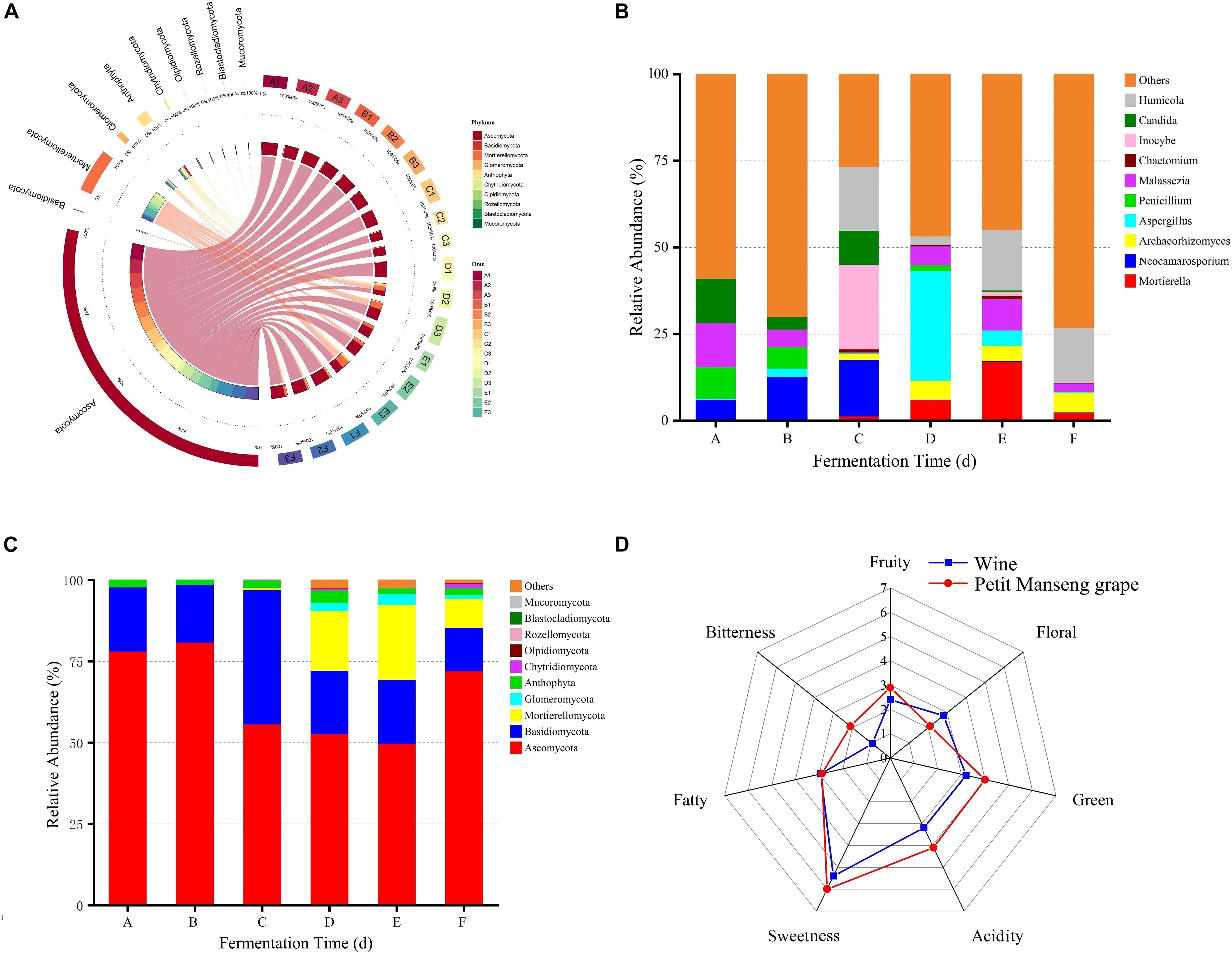
Figure 4. The relative abundance between fungal groups at the phylum (A) level, the relative abundance of the top 10 fungi at the genus (B) level, the relative abundance of top 10 fungi at the phylum (C) level, and sensory analysis of wine samples (D). A, B, C, D, E, and F represented samples collected on fermentation days 0, 1, 4, 7,11, and 14, respectively.
Richness and Diversity Analysis
A total of 844,661 sequences for the fungal communities were clustered into 643 OTUs at a 97% identity threshold. The rarefaction curves of all samples tended to be flat, indicating that the amount of our sequencing data was reasonable (Supplementary Figure 1). For fungal communities, the richness in stage F was the highest according to the Chao1 and ACE indexes; stage F also showed the highest diversity based on the Shannon and Simpson indexes (Supplementary Table 1). Moreover, the values of the Chao1, ACE, Shannon, and Simpson indexes at stages E and F were higher than those at other stages, whereas Robbins index was the highest at stage C among all the stages (Supplementary Figure 2). Therefore, the fungal richness and diversity of the wine samples from different fermentation stages showed differences according to the alpha indexes. In addition, this study described the common and unique OTUs of the microorganisms from different fermentation stages. There were 37, 9, 11, 65, 54, and 67 fungal OTUs noted in stages A, B, C, D, E, and F, respectively, and 3 OTUs were shared among all six fermentation stages (Figure 5A). Therefore, the structure of the fungal communities in the sweet wine of Petit Manseng demonstrated differences based on the fermentation stage. Moreover, we investigated the dynamic changes in fungal communities during different fermentation periods. Although the original OTU data showed that the total number of OTUs increased as fermentation continued, the number of cultivable fungi gradually decreased (Supplementary Table 2). The changes corresponding to the fermentation stages, along with changes in environmental factors, led to changes in fungal adaptability. Four colony micrographs (Supplementary Figure 4), five genera, and seven species of yeast were isolated in the spontaneous fermentation process based on the phylogenetic tree (Supplementary Figure 7).
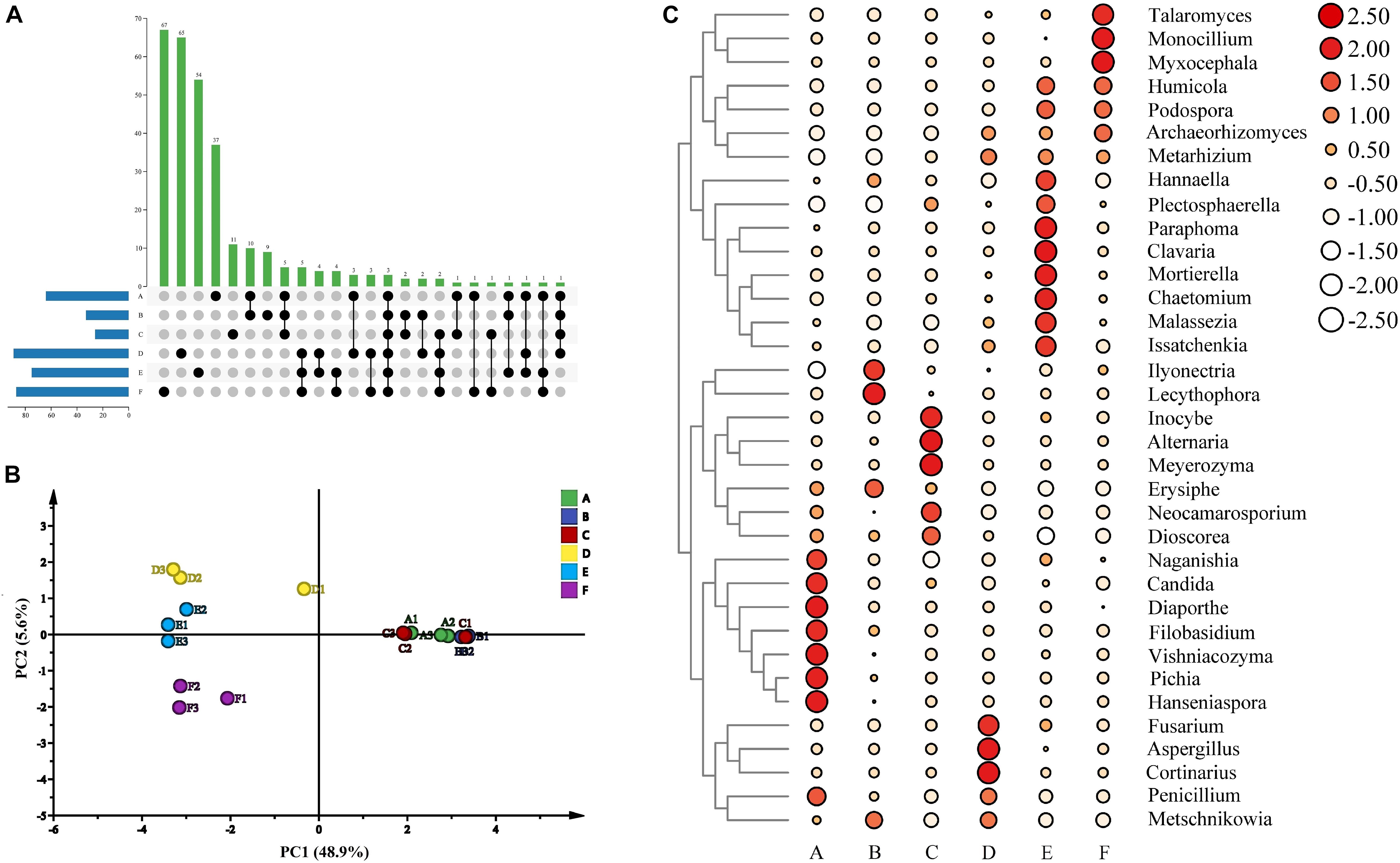
Figure 5. The upset diagram of the fungi genus among samples (A), PCA of wine samples (B), abundance clustering heatmap of major fungi genera in zymotic fluids collected from different fermentation times (C). The scale bar shows the relative abundance (log scale) of each genus within a sample. A, B, C, D, E, and F represented samples collected on fermentation days 0, 1, 4, 7,11, and 14, respectively.
Our beta diversity analysis of the fungal structure based on PCA during wine fermentation indicated that the contribution rate of PC1 was 48.9%, whereas that of PC2 was 5.6% (Figure 5B). We found that the structure of fungal composition was clustered together at stages A, B, and C and was very large at stages D, E, and F. These results contradicted the previous results, possibly because the previous study performed incomplete fermentation of Petit Manseng wine (Chen et al., 2020). We also noted some unusual dissimilarities. For example, our D1 deviated from the main clusters. In summary, our data indicated that fungal community abundance was dependent on the fermentation stage.
Microbial Abundance During Spontaneous Fermentation
By comparing the fungal communities in wine samples from different fermentation stages, we found that the main phyla appeared in all samples, but their content varied (Figure 4A). In particular, the top 10 relatively abundant fungal phyla included Ascomycota, Basidiomycota, Mortierellomycota, Glomeromycota, Anthophyta, Chytridiomycota, Olpidiomycota, Rozellomycota, Blastocladiomycota, and Mucoromycota (Figure 4C). The distinction of fungal abundance was obvious in different fermentation stages. For example, the relative abundance of Ascomycota was 78, 81, 56, 53, 50, and 72% in stages A, B, C, D, E, and F, respectively. Moreover, fungal abundance was confirmed to be affected by the fermentation stage.
The extracted high abundance of fungal OTUs as well as the distribution of six different phylum-to-species species is illustrated in Supplementary Figure 3. The result showed that Ascomycota had a denser species distribution than other phyla, whereas Glomeromycota showed the opposite trend. At the fungal genus level, Candida and Malassezia had the highest percent relative abundance at stage A. These results differed from those reported previously, possibly because of differences in the grape varieties and origins used (Sirén et al., 2019). Moreover, Candida and Malassezia, the abundant genera, dominated in the early and middle stages of fermentation (Figure 4B), whereas Mortierella and Humicola dominated the later fermentation stages. The relative abundance of Aspergillus demonstrated notable fluctuations, which may be the reason for the increased alcohol concentration. The relative abundance of Penicillium peaked at stage A and then decreased rapidly. Moreover, Vishniacozyma, Hanseniaspora, and Pichia, as dominant fungi present on the grapes in China’s vine-growing regions, were found in our wine during spontaneous fermentation (Li et al., 2010). Regarding the fungal communities, Hanseniaspora, Pichia, Vishniacozyma, Filobasidium, Diaporthe, and Candida were the dominant taxa at stage A (Figure 5C). In addition, Lecythophora, Inocybe, Alternaria, Meyerozyma, and Erysiphe dominated stages B and C, whereas at stages E and F, the dominant taxa were Humicola, Podospora, Archaeorhizomyces, and Metarhizium. Furthermore, Metschnikowia and Issatchenkia were detected in the fermentation process, with a strong positive correlation at stages D and E. Metschnikowia and Issatchenkia may also be responsible for the increased alcohol contents (Liu et al., 2016).
Co-occurrence and Exclusion Analysis Reveals the Relationships Among Different Microorganisms
The interaction of microorganisms is considered an important factor supporting the structure of microbial communities (Huang et al., 2018). We calculated Pearson’s rank correction coefficients to represent beneficial or antagonistic relationships between the dominant microorganisms. The correlations of different fungi are displayed in Figure 6. Issatchenkia showed weak exclusion from other fungal genera except for Hannaella, Naganishia, Fusarium, Aspergillus, and Cortinarius (P < 0.05). In addition, Dioscorea and Erysiphe showed obvious exclusion toward Talaromyces, Humicola, Podospora, Archaeorhizomyces, and Metarhizium. In contrast, Paraphoma, Clavaria, Mortierella, and Chaetomium demonstrated strong co-occurrence with Malassezia and Issatchenkia, and Pichia and Hanseniaspora presented strong co-occurrence with Diaporthe, Filobasidium, and Vishniacozyma (P < 0.05). Thus, most genera present during spontaneous fermentation presented weak co-occurrence patterns.
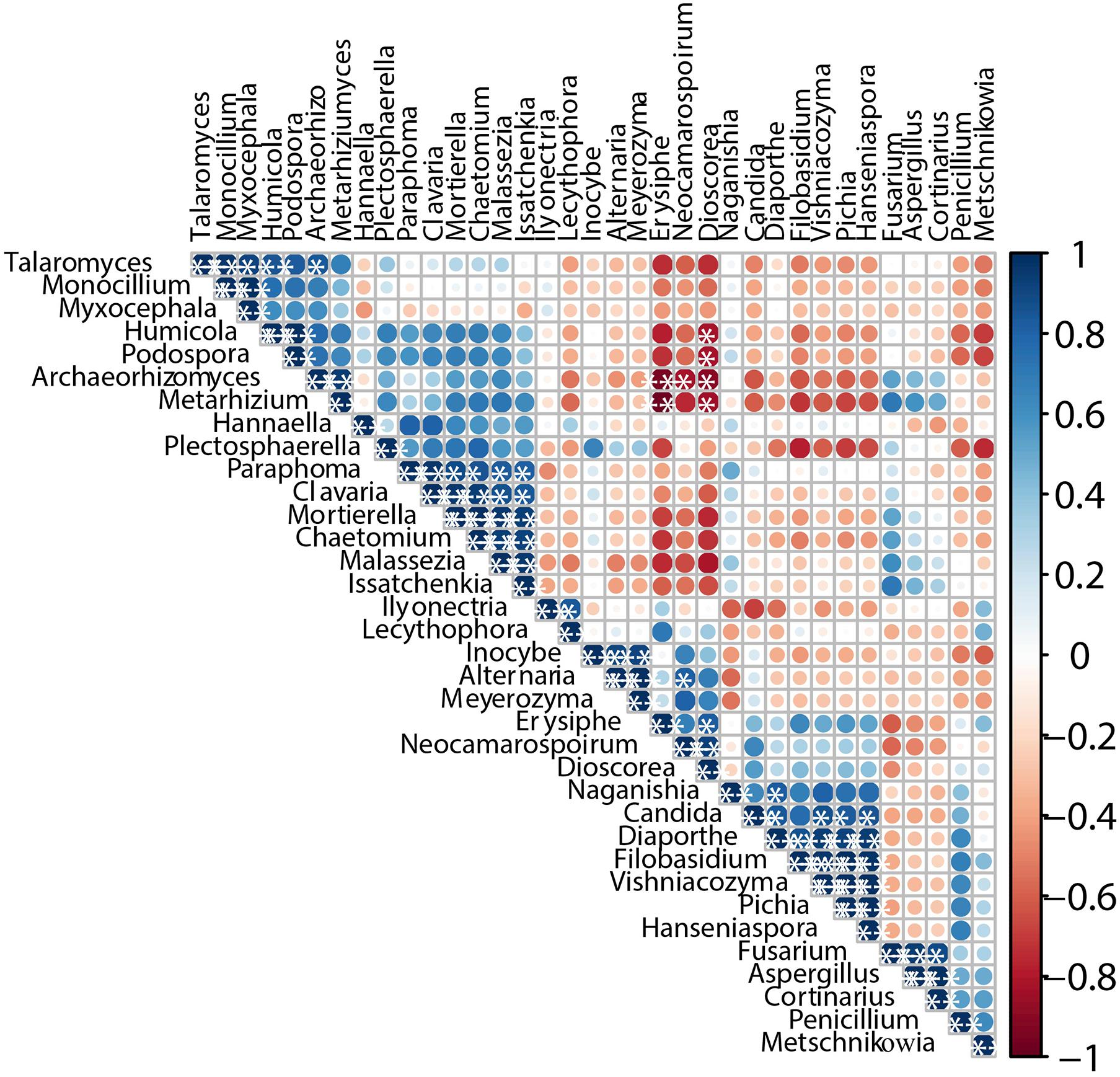
Figure 6. Co-occurrence and co-exclusion relationship analysis of fungi. The figure presents a Pearson’s rank correlation matrix of the top 35 fungi genera abundance. Strong correlations are indicated by large circles, whereas weak correlations are indicated by small circles. The color of the scale bar denotes the nature of the correlation, with 1 indicating a perfect positive correlation (dark blue) and -1 indicating a perfect negative correlation (dark red). Only significant correlations (| r| > 0.7, FDR < 0.05) are shown with *.
Correlation Analysis Between Core Microbiota and Volatile Compounds
The correlation of core microbiota and volatile compounds were analyzed during the Petit Manseng wine fermentation process, as shown in Figure 7. Three criteria were used to discover the relationship between the microbiota and the aroma compounds: (1) a relatively stable abundance was maintained throughout the fermentation process, (2) the variable importance for the predictive component (VIP) values of microorganisms and volatiles were >1.0, and (3) the absolute values of linear correlation coefficient (R) between the concentration of volatile compounds and the relative abundance of microbial were >0.7 (He et al., 2020). The changes in VIP values for the fungal group at the genus level and the volatile compounds are shown in Supplementary Figures 5, 6, respectively.
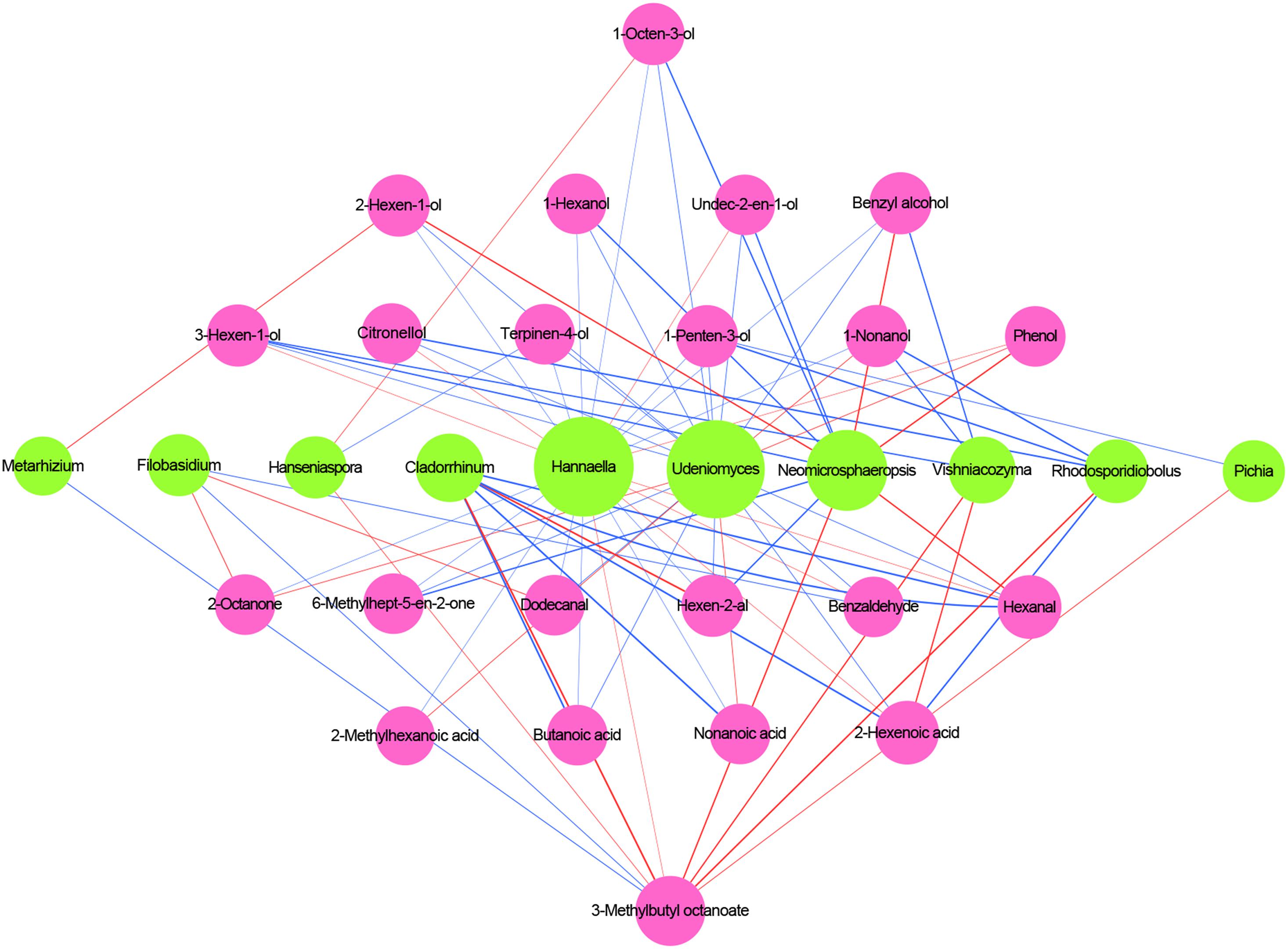
Figure 7. Correlation analysis of microbiota and volatile compounds during spontaneous fermentation. Core fungal genera are shown in green and core volatile compounds are shown in purple. Blue lines indicate positive correlations, and red lines indicate negative correlations.
According to the aforementioned criteria, 10 microbial genera were regarded as key microorganisms in spontaneous fermentation. Udeniomyces, Hannaella, and Neomicrosphaeropsis presented high correlations with alcohol and acids, such as 1-octen-3-ol and nonanoic acid. Vishniacozyma correlated with five core volatile compounds, of which it was positively correlated with 3-hexen-1-ol, 1-nonanol, and benzyl alcohol. Cladosporium negatively correlated with hexen-2-al and 3-methylbutyl octanoate. Non-Saccharomyces cerevisiae yeasts commonly found in wine, such as Hanseniaspora and Pichia, were positively correlated with some alcohols. Metarhizium presented characteristics similar to Filobasidium concerning 3-methylbutyl octanoate. In general, most fungal genera, except Metarhizium and Filobasidium, were negatively correlated with 3-methylbutyl octanoate. Whether the aforementioned microorganisms can produce and absorb related volatile compounds warrants further study.
Overall, the changes in the microbial community differed throughout the spontaneous fermentation process of Petit Manseng. As reported previously, wine quality is related to the metabolic activity and fermentation behaviors of the yeasts (Antonelli et al., 1999). Compared with S. cerevisiae, non-Saccharomyces yeasts have gained more attention because they produce esterase, β-glucosidase, lipase, and protease and thus increase the amount of active aroma substance (Escribano et al., 2017).
In this study, Hanseniaspora, Vishniacozyma, and Pichia were significantly correlated with at least one volatile compound during spontaneous fermentation. Based on our multivariate analysis results, the correlation between microorganisms and volatile compounds was evident. This requires further research using multiomics methods to verify the association between core microorganisms and specific flavors (Huang et al., 2018).
Discussion
Our Petit Manseng wine was produced via spontaneous fermentation and had a final alcohol concentration of 12.01% (v/v), accompanied by a quick decrease in the total sugar content from 345.35 to 110.84 g/L. The gradually accumulated ethanol inhibited yeast metabolism and the ability to produce ethanol.
With regard to organic acids, Pichia can metabolize lactic acid to pyruvate and then to acetyl-CoA and acetaldehyde during liquor production (Song Z. et al., 2017). The microorganisms that accumulated citric acid mainly include some fungi from the genera Aspergillus, Candida, and Penicillium (Papagianni, 2007). Tartaric acid is resistant to degradation and metabolism by wine microorganisms (Torija et al., 2003). In this study, the wine tartaric acid concentration decreased slowly because it was precipitated as potassium and calcium salts during fermentation.
In the present study, we identified 56 volatile compounds linked to spontaneous wine fermentation. Esters, which can increase the complexity of volatiles with the extension of the fermentation stage, are indispensable components of wine (Guo et al., 2020). The concentrations of fatty acid ethyl esters, ethyl butyrate, ethyl hexanoate, ethyl octanoate, ethyl decanoate, and ethyl laurate initially increased and then showed a downward trend. In contrast, the contents of the acetic esters like isoamyl acetate, isopropyl acetate, and 2-phenylethyl acetate increased during the fermentation process. The odor activity values (OAVs) of these compounds were >1 at the end of spontaneous fermentation; this observation may be related to high acyltransferase and alcohol acetyltransferase expression during spontaneous fermentation (Saerens et al., 2006; Procopio et al., 2011).
Alcohols represented one of the largest groups of volatile compounds noted during spontaneous fermentation, and they included 1-hexanol, phenylethyl alcohol, isoamyl alcohol, and ethanol. The contents of all alcohols except ethanol gradually increased during the fermentation process. However, these alcohols (1-hexanol and phenylethyl alcohol) do not positively enhance the wine flavor wines because of their relatively high odor threshold. In contrast, ethanol with a low threshold showed an OAV of >1. Moreover, citronellol’s OAVs were also >1 and influenced the fruity character of the wine (Pérez et al., 2018). Acids, considered important for wine fermentation, were produced via yeast metabolism during spontaneous fermentation. These acids primarily comprised hexanoic acid and octanoic acid and flocked in the late stage of spontaneous fermentation, with a total content of 3448.90 μg/L.
Aldehydes, the significant source of herbaceous in wine, mainly came from fatty acid oxidation and amino acid degradation due to microbial fermentation (Zhao G. et al., 2020). Benzaldehyde was a widely used aromatic aldehyde, with an OAV of <0.1 at the end of fermentation. Fatty acid release and flavor substance catabolism via β-oxidation led to ketone formation (Collins et al., 2003). 2-Octanone actively contributed to the fruity of wine, with an OAV of >1.
The OAVs of the trace compounds formed during spontaneous fermentation were <1. In addition, most of these volatile compounds showed a concentration increase throughout wine fermentation. Although these compounds were present at a relatively low concentration, some of them had an indirect effect on the flavor and countered the organoleptic properties of the wine (Asproudi et al., 2018).
Changes were also noted in the indigenous microorganisms during the spontaneous fermentation process. The microbial communities participating in initial spontaneous alcoholic fermentation are inherently present in Petit Manseng grapes. Nevertheless, many other microorganisms also participate in this process. Therefore, the relative abundance of fungal communities during spontaneous fermentation demonstrated obvious fluctuations, possibly also reflecting the effects of the ethanol fermentation environment. Wang and Liu (2013) reported fungal communities were inhibited by harsh conditions until they became tolerant or adapted to the harsh fermentation environments. In the current study, the abundance of Candida reduced during spontaneous fermentation due to its sensitivity to ethanol, similar to that described in previous studies (Heard and Fleet, 1985). Notably, Humicola began to appear at stage C and gradually increased in abundance at the end fermentation. This result was expected because Humicola was resistant to ethanol (Bassey et al., 2017). Some Humicola species positively contributed flavor by producing various enzymes, thus indicating many of their potential applications in various industries (Bokhari et al., 2010). Aspergillus was present throughout the fermentation process with different relative abundance values. Their abundance peaked in the middle stage of the fermentation. Aspergillus has powerful environmental adaptability, and it is resistant to acids and ethanol (Freire et al., 2017; Sheng et al., 2018). Some fungi such as Pichia, the important producers of various secondary metabolites, occurred in the early spontaneous fermentation period of our wine. This result indicated a potential application of these microorganisms in producing many aroma substances to improve the quality of Petit Manseng wine.
The relationship between volatile compounds and microbial succession dynamics during the spontaneous fermentation of Petit Manseng wine remains unclear. We thus investigated the key functional microorganism responsible for the generation of aroma compounds. We found that Hannaella, Udeniomyces, and Neomicrosphaeropsis were the highest contributors to the generation of volatile compounds, especially during alcohol production. In addition, Filobasidium and Hannaella abundance was negatively correlated with 2-octanone production. Moreover, Vishniacozyma produced antibiotic compounds or enzymes to maintain their niches (Gramisci et al., 2018). This was related to the formation of aldehydes, such as hexanal, and acids, such as 2-hexanoic acid, during fermentation.
In this study, the indigenous microbial of wine grapes reflect the health of grapes and play an indispensable role in wine flavor and quality. To the best of our knowledge, this is the first study of microbial succession during the spontaneous fermentation of sweet wine fermented by Petit Manseng using HTS technology. Additional studies on the indigenous non-Saccharomyces species detected in this study may aid in understanding their role during spontaneous fermentation, their contribution to the sensory quality of sweet wine, and microbial safety.
Conclusion
Spontaneous fermentation of Petit Manseng could strengthen its aromatic profile. Fungi, as a significant part of wine, were responsible for the formation of the characteristic aroma and volatile compounds (including many esters and alcohols) in the wine. The results of this study may be used as a reference for producing Petit Manseng sweet wine with typical characteristics.
Data Availability Statement
The data presented in the study are deposited in the National Center for Biotechnology Information (NCBI) repository, accession number PRJNA748614 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA748614). The accession numbers for dataset of our 18 samples that have been generated or analyzed in the study are: SRX11512770, SRX11513000, SRX11513001, SRX11513002, SRX11513003, SRX11513004, SRX11513005, SRX11513006, SRX11513007, SRX11513008, SRX11513- 009, SRX11513010, SRX11513011, SRX11513012, SRX11513013, SRX11513014, SRX11513015, and SRX11513016.
Author Contributions
YM wrote the main text of the manuscript and performed the experiment. BW analyzed the fungi data. TL, XX, YJ, and XJ analyzed the volatile component data. XS supervised the research activities. All authors reviewed the manuscript.
Funding
We are grateful for the Science and Technology Research Project of the Xinjiang Production and Construction Corps (Nos. 2019AB025 and 2020AB014), the Science and Technology Research Project of the Eighth Division (No. 2020GY07), the Science and Technology Research Project of the Fifth Division (No. 20GY01), and the National Natural Science Foundation of China (No. 31960465).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.717387/full#supplementary-material
References
Alessandria, V., Giacosa, S., Campolongo, S., Rolle, L., Rantsiou, K., and Cocolin, L. (2013). Yeast population diversity on grapes during on-vine withering and their dynamics in natural and inoculated fermentations in the production of icewines. Food Res. Int. 54, 139–147. doi: 10.1016/j.foodres.2013.06.018
Antonelli, A., Castellari, L., Zambonelli, C., and Carnacini, A. (1999). Yeast influence on volatile composition of wines. J. Agr. Food Chem. 47, 1139–1144. doi: 10.1021/jf9807317
Asproudi, A., Ferrandino, A., Bonello, F., Vaudano, E., Pollon, M., and Petrozziello, M. (2018). Key norisoprenoid compounds in wines from early-harvested grapes in view of climate change. Food Chem. 268, 143–152. doi: 10.1016/j.foodchem.2018.06.069
Bassey, N. S., Whong, C., and Ado, S. A. (2017). Determination of fermentative properties of yeast strains isolated from spoilt fruits and beverages. Sci. J. Microbiol. 6, 166–173. doi: 10.14196/sjm.v6i7.2552
Belda’s, I., Navascués, E., Marquina, D., Santos, A., Calderon, F., and Benito, S. (2015). Dynamic analysis of physiological properties of Torulaspora delbrueckii in wine fermentations and its incidence on wine quality. Appl. Microbiol. Biot. 99, 1911–1922. doi: 10.1007/s00253-014-6197-2
Belda, I., Ruiz, J., Alastruey, A., Navascués, E., Marquina, D., and Santos, A. (2016). Unraveling the enzymatic basis of wine “flavorome”: a phylo-functional study of wine related yeast species. Front. Microbiol. 7:12. doi: 10.3389/fmicb.2016.00012
Belda, I., Ruiz, J., Esteban-Fernández, A., Navascués, E., Marquina, D., Santos, A., et al. (2017). Microbial contribution to wine aroma and its intended use for wine quality improvement. Molecules. 22:189. doi: 10.3390/molecules22020189
Böhmer, M., Smol’ak, D., Ženišová, K., Čaplová, Z., Pangallo, D., Puškárová, A., et al. (2020). Comparison of microbial diversity during two different wine fermentation processes. FEMS Microbiol. Lett. 367:fnaa150. doi: 10.1093/femsle/fnaa150
Bokhari, S. A. I., Latif, F., Akhtar, M. W., and Rajoka, M. I. (2010). Characterization of a β-xylosidase produced by a mutant derivative of Humicola lanuginosa in solid state fermentation. Ann. Microbiol. 60, 21–29. doi: 10.1007/s13213-010-0026-3
Brainina, K. Z., Stozhko, N. Y., Belysheva, G. M., Inzhevatova, O. V., Kolyadina, L. I., Cremisini, C., et al. (2004). Determination of heavy metals in wines by anodic stripping voltammetry with thick-film modified electrode. Anal. Chim. Acta. 514, 227–234. doi: 10.1016/j.aca.2004.03.047
Brandfass, C., and Karlovsky, P. (2008). Upscaled CTAB-based DNA extraction and real-time PCR assays for Fusarium culmorum and F. graminearum DNA in plant material with reduced sampling error. Int. J. Mol Sci. 9, 2306–2321. doi: 10.3390/ijms9112306
Bučková, M., Puškárová, A., Ženišová, K., Kraková, L., Piknová, L., Kuchta, T., et al. (2018). Novel insights into microbial community dynamics during the fermentation of central European ice wine. Int. J. Food Microbiol. 266, 42–51. doi: 10.1016/j.ijfoodmicro.2017.11.010
Caridi, A., De Bruno, A., De Salvo, E., Piscopo, A., Poiana, M., and Sidari, R. (2017). Selected yeasts to enhance phenolic content and quality in red wine from low pigmented grapes. Eur. Food Res Technol. 243, 367–378. doi: 10.1007/s00217-016-2750-9
Chaplin, B. (2011). South-west france: the wines and winemakers. Gastronomica 11, 119–120. doi: 10.1525/gfc.2011.11.2.119
Chen, Y., Zhang, W., Yi, H., Wang, B., Xiao, J., Zhou, X., et al. (2020). Microbial community composition and its role in volatile compound formation during the spontaneous fermentation of ice wine made from Vidal grapes. Process Biochem. 92, 365–377. doi: 10.1016/j.procbio.2020.01.027
Collins, Y. F., McSweeney, P. L. H., and Wilkinson, M. G. (2003). Lipolysis and free fatty acid catabolism in cheese: a review of current knowledge. Int. Dairy J. 13, 841–866. doi: 10.1016/S0958-6946(03)00109-2
Crump, A. M., Johnson, T. E., Wilkinson, K. L., and Bastian, S. E. P. (2015). Influence of oak maturation regimen on composition, sensory properties, quality, and consumer acceptability of Cabernet Sauvignon wines. J. Agr. Food Chem. 63, 1593–1600. doi: 10.1021/jf5044025
Culleré, L., Escudero, A., Cacho, J., and Ferreira, V. (2004). Gas chromatography- olfactometry and chemical quantitative study of the aroma of six premium quality Spanish aged red wines. J. Agr. Food Chem 52, 1653–1660. doi: 10.1021/jf0350820
de Celis, M., Ruiz, J., Martín-Santamaría, M., Alonso, A., Marquina, D., Navascués, E., et al. (2019). Diversity of Saccharomyces cerevisiae yeasts associated to spontaneous and inoculated fermenting grapes from Spanish vineyards. Lett. Appl. Microbiol. 68, 580–588. doi: 10.1111/lam.13155
Du, X., Plotto, A., Baldwin, E., and Rouseff, R. (2011). Evaluation of volatiles from two subtropical strawberry cultivars using GC–olfactometry, GC-MS odor activity values, and sensory analysis. J. Agr. Food Chem. 59, 12569–12577. doi: 10.1021/jf2030924
Englezos, V., Rantsiou, K., Cravero, F., Torchio, F., Pollon, M., Fracassetti, D., et al. (2018). Volatile profile of white wines fermented with sequential inoculation of Starmerella bacillaris and Saccharomyces cerevisiae. Food Chem. 257, 350–360. doi: 10.1016/j.foodchem.2018.03.018
Escribano, R., González, L., Garijo, P., Berlanas, C., López, I., López, R., et al. (2017). Screening of enzymatic activities within different enological non-Saccharomyces yeasts. J. Food Sci. Tech. 54, 1555–1564. doi: 10.1007/s13197-017-2587-7
Ferreira, V., López, R., and Cacho, J. F. (2000). Quantitative determination of the odorants of young red wines from different grape varieties. J. Sci. Food Agr. 80, 1659–1667. doi: 10.1002/1097-0010(20000901)80:11<1659::AID-JSFA693<3.0.CO;2-6
Francesca, N., Gaglio, R., Alfonzo, A., Settanni, L., Corona, O., Mazzei, P., et al. (2016). The wine: typicality or mere diversity? The effect of spontaneous fermentations and biotic factors on the characteristics of wine. Agric. Sci. Proc. 8, 769–773. doi: 10.1016/j.aaspro.2016.02.064
Freire, L., Passamani, F. R. F., Thomas, A. B., Nassur, R. D. C. M. R., Silva, L. M., Paschoal, F. N., et al. (2017). Influence of physical and chemical characteristics of wine grapes on the incidence of Penicillium and Aspergillus fungi in grapes and ochratoxin A in wines. Int. J. Food Microbiol. 241, 181–190. doi: 10.1016/j.ijfoodmicro.2016.10.027
Gardner, D. M., Duncan, S. E., and Zoecklein, B. W. (2017). Aroma characterization of Petit Manseng wines using sensory consensus training, SPME GC-MS, and electronic nose analysis. Am. J. Enol. Viticul. 68, 112–119. doi: 10.5344/ajev.2016.15099
Gemert, L. J. V. (2011). Compilations of Odour Threshold Values in Air, Water and Other Media (Second Enlarged and Revised Edition). Utrecht: Oliemans Punter & Partners BV. 1–648.
Gómez, M. J., Cacho, J. F., Ferreira, V., Vicario, I. M., and Heredia, F. J. (2007). Volatile components of Zalema white wines. Food Chem. 100, 1464–1473. doi: 10.1016/j.foodchem.2005.11.045
Gramisci, B. R., Lutz, M. C., Lopes, C. A., and Sangorrín, M. P. (2018). Enhancing the efficacy of yeast biocontrol agents against postharvest pathogens through nutrient profiling and the use of other additives. Bio. Control. 121, 151–158. doi: 10.1016/j.biocontrol.2018.03.001
Guo, L., Luo, Y., Zhou, Y., Bianba, C., Guo, H., Zhao, Y., et al. (2020). Exploring microbial dynamics associated with flavours production during highland barley wine fermentation. Food Res. Int. 130:108971. doi: 10.1016/j.foodres.2019.108971
Guth, H. (1997). Quantitation and sensory studies of character impact odorants of different white wine varieties. J. Agar. Food Chem. 45, 3027–3032. doi: 10.1021/jf970280a
He, G., Huang, J., Wu, C., Jin, Y., and Zhou, R. (2020). Bioturbation effect of fortified Daqu on microbial community and flavor metabolite in Chinese strong-flavor liquor brewing microecosystem. Food Res. Int. 129:108851. doi: 10.1016/j.foodres.2019.108851
Heard, G. M., and Fleet, G. H. (1985). Growth of natural yeast flora during the fermentation of inoculated wines. Appl. Environ Microb. 50, 727–728. doi: 10.1128/AEM.50.3.727-728.1985
Huang, Z., Hong, J., Xu, J., Li, L., Guo, W., Pan, Y., et al. (2018). Exploring core functional microbiota responsible for the production of volatile flavour during the traditional brewing of Wuyi Hong Qu glutinous rice wine. Food Microbiol. 76, 487–496. doi: 10.1016/j.fm.2018.07.014
Kong’s, C., Li, A., Jin, G., Zhu, X., and Tao, Y. (2019). Evolution of volatile compounds treated with selected non-Saccharomyces extracellular extract during Pinot noir winemaking in monsoon climate. Food Res. Int. 119, 177–186. doi: 10.1016/j.foodres.2019.01.036
Lan, Y., Guo, J., Qian, X., Zhu, B., Shi, Y., Wu, G., et al. (2021). Characterization of key odor-active compounds in sweet petit manseng (Vitis vinifera L.) wine by gas chromatography-olfactometry, aroma reconstitution, and omission tests. J. Food Sci. 86, 1258–1272. doi: 10.1111/1750-3841.15670
Li, S., Cheng, C., Li, Z., Chen, J., Yan, B., Han, B., et al. (2010). Yeast species associated with wine grapes in China. Int. J. Food Microbiol. 138, 85–90. doi: 10.1016/j.ijfoodmicro.2010.01.009
Lin, Z., Cai, J. J., Scriven, L. E., and Davis, H. T. (1994). Spherical-to-wormlike micelle transition in CTAB solutions. J. Phys. Chem. 98, 5984–5993. doi: 10.1021/j100074a027
Liu, P., Lu, L., Duan, C., and Yan, G. (2016). The contribution of indigenous non-Saccharomyces wine yeast to improved aromatic quality of Cabernet Sauvignon wines by spontaneous fermentation. Lwt Food Sci. Technol. 71, 356–363. doi: 10.1016/j.lwt.2016.04.031
Liu, Y., Rousseaux, S., Tourdot-Maréchal, R., Sadoudi, M., Gougeon, R., Schmitt-Kopplin, P., et al. (2017). Wine microbiome: a dynamic world of microbial interactions. Crit. Rev. Food Sci. 57, 856–873. doi: 10.1080/10408398.2014.983591
López, R., Ezpeleta, E., Sánchez, I., Cacho, J., and Ferreira, V. (2004). Analysis of the aroma intensities of volatile compounds released from mild acid hydrolysates of odourless precursors extracted from Tempranillo and Grenache grapes using gas chromatography-olfactometry. Food Chem. 88, 95–103. doi: 10.1016/j.foodchem.2004.01.025
Loureiro, V., Brasil, R., and Malfeito-Ferreira, M. (2016). A new wine tasting approach based on emotional responses to rapidly recognize classic European wine styles. Beverages. 2:6. doi: 10.3390/beverages2010006
Lu, Y., Guan, X., Li, R., Wang, J., Liu, Y., Ma, Y., et al. (2021). Comparative study of microbial communities and volatile profiles during the inoculated and spontaneous fermentation of persimmon wine. Process Biochem. 100, 49–58. doi: 10.1016/j.procbio.2020.09.023
Lu, Y., Sun, F., Wang, W., Liu, Y., Wang, J., Sun, J., et al. (2020). Effects of spontaneous fermentation on the microorganisms diversity and volatile compounds during ‘Marselan’from grape to wine. LWT 134:110193. doi: 10.1016/j.lwt.2020.110193
Mayr, C. M., Geue, J. P., Holt, H. E., Pearson, W. P., Jeffery, D. W., and Francis, I. L. (2014). Characterization of the key aroma compounds in Shiraz wine by quantitation, aroma reconstitution, and omission studies. J. Agr. Food Chem. 62, 4528–4536. doi: 10.1021/jf405731v
Mihail, M., Cǎlugǎr, A., Pop, N., Gal, E., and Pop, T. I. (2019). Odor activity value in red wines aroma from three wine regions in Romania. Agricultura 109, 29–38. doi: 10.15835/agrisp.v109i1-2.13245
Miller, G. L. (1959). Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anall Chem. 31, 426–428. doi: 10.1021/ac60147a030
Mirás-Avalos, J. M., Fandiño, M., Trigo-Córdoba, E., Rey, B. J., Orriols, I., and Cancela, J. J. (2017). Influence of irrigation on consumer acceptability of Albariño and Godello wines. Lwt Food Sci. Technol. 85, 345–352. doi: 10.1016/j.lwt.2017.01.076
Murtaza, M. A., Huma, N., Shabbir, M. A., Murtaza, M. S., and Anees-ur-Rehman, M. (2017). Survival of micro-organisms and organic acid profile of probiotic Cheddar cheese from buffalo milk during accelerated ripening. Int. J. Dairy Technol. 70, 562–571. doi: 10.1111/1471-0307.12406
Nie, Z., Zheng, Y., Xie, S., Zhang, X., Song, J., Xia, M., et al. (2017). Unraveling the correlation between microbiota succession and metabolite changes in traditional Shanxi aged vinegar. Sci. Rep. 7, 1–12. doi: 10.1038/s41598-017-09850-6
Niu, Y., Zhang, X., Xiao, Z., Song, S., Eric, K., Jia, C., et al. (2011). Characterization of odor-active compounds of various cherry wines by gas chromatography–mass spectrometry, gas chromatography–olfactometry and their correlation with sensory attributes. J. Chromatogr. B. 879, 2287–2293. doi: 10.1016/j.jchromb.2011.06.015
Noguerol, R., González, C., Cancho, B., and Simal, J. (2009). Quantitative determination and characterisation of the main odourants of Mencía monovarietal red wines. Food Chem. 117, 473–484. doi: 10.1016/j.foodchem.2009.04.014
Padilla, B., Gil, J. V., and Manzanares, P. (2016). Past and future of non-Saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity. Front. Microbiol. 7:411. doi: 10.3389/fmicb.2016.00411
Papagianni, M. (2007). Advances in citric acid fermentation by Aspergillus niger: Biochemical aspects, membrane transport and modeling. Biotechnol. Adv. 25, 244–263. doi: 10.1016/j.biotechadv.2007.01.002
Peinado, R. A., Mauricio, J. C., and Moreno, J. (2006). Aromatic series in sherry wines with gluconic acid subjected to different biological aging conditions by Saccharomyces cerevisiae var. capensis. Food Chem. 94, 232–239. doi: 10.1016/j.foodchem.2004.11.010
Peng, C. T., Wen, Y., Tao, Y. S., and Lan, Y. Y. (2013). Modulating the formation of Meili wine aroma by prefermentative freezing process. J. Agr. Food Chem. 61, 1542–1553. doi: 10.1021/jf3043874
Pérez, D., Assof, M., Bolcato, E., Sari, S., and Fanzone, M. (2018). Combined effect of temperature and ammonium addition on fermentation profile and volatile aroma composition of Torrontés Riojano wines. LWT 87, 488–497. doi: 10.1016/j.lwt.2017.09.020
Procopio, S., Qian, F., and Becker, T. (2011). Function and regulation of yeast genes involved in higher alcohol and ester metabolism during beverage fermentation. Eur. Food Res. Technol. 233, 721–729. doi: 10.1007/s00217-011-1567-9
Robinson, J., Harding, J., and Vouillamoz, J. (2013). Wine Grapes: a Complete Guide to 1,368 Vine Varieties, Including Their Origins and Flavours. New York, NY: Penguin.
Ruiz, J., Kiene, F., Belda, I., Fracassetti, D., Marquina, D., Navascués, E., et al. (2019). Effects on varietal aromas during wine making: a review of the impact of varietal aromas on the flavor of wine. Appl. Microbiol. Biot. 103, 7425–7450. doi: 10.1007/s00253-019-10008-9
Saerens, S. M. G., Verstrepen, K. J., Van Laere, S. D. M., Voet, A. R. D., Van Dijck, P., Delvaux, F. R., et al. (2006). The Saccharomyces cerevisiae EHT1 and EEB1 genes encode novel enzymes with medium-chain fatty acid ethyl ester synthesis and hydrolysis capacity. J. Biol. Chem. 281, 4446–4456. doi: 10.1074/jbc.M512028200
Sheng, X. D., Zeng, X., Lin, D., and Yao, S. (2018). Ethanol tolerant endoglucanase from Aspergillus niger isolated from wine fermentation cellar. Biocatal. Agric. Biotechnol. 15, 19–24. doi: 10.1016/j.bcab.2018.04.016
Sherman, E., Coe, M., Grose, C., Martin, D., and Greenwood, D. R. (2020). Metabolomics approach to assess the relative contributions of the volatile and non-volatile composition to expert quality ratings of Pinot Noir wine quality. J. Agr. Food Chem. 68, 13380–13396. doi: 10.1021/acs.jafc.0c04095
Shi, W. K., Wang, J., Chen, F. S., and Zhang, X. Y. (2019). Effect of Issatchenkia terricola and Pichia kudriavzevii on wine flavor and quality through simultaneous and sequential co-fermentation with Saccharomyces cerevisiae. LWT 116:108477. doi: 10.1016/j.lwt.2019.108477
Sirén, K., Mak, S. S. T., Melkonian, C., Carøe, C., Swiegers, J. H., Molenaar, D., et al. (2019). Taxonomic and functional characterization of the microbial community during spontaneous in vitro fermentation of Riesling must. Front. Microbiol. 10:697. doi: 10.3389/fmicb.2019.00697
Song, Y., Zhang, Y. J., Liu, N., Ye, D., Gong, X., Qin, Y., et al. (2017). Volatile compounds in wild strawberry and their odorants of wild strawberry wines: effects of different stages of fermentation. Int. J. Food Prop. 20, S399–S415. doi: 10.1080/10942912.2017.1297951
Song, Z., Du, H., Zhang, Y., and Xu, Y. (2017). Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing. Front. Microbiol. 8:1294. doi: 10.3389/fmicb.2017.01294
Swami, S. B., Thakor, N. J., and Divate, A. D. (2014). Fruit wine production: a review. J. Food Res. Technol. 2, 93–100. doi: 10.17508/CJFST.2018.10.2.19
Tao, Y. S., and Li, H. (2009). Active volatiles of cabernet sauvignon wine from Changli County. Health. 1, 176–182. doi: 10.4236/health.2009.13028
Torija, M. J., Beltran, G., Novo, M., Poblet, M., Rozès, N., Mas, A., et al. (2003). Effect of organic acids and nitrogen source on alcoholic fermentation: study of their buffering capacity. J. Agr. Food. Chem. 51, 916–922. doi: 10.1021/jf020094r
Wang, C., and Liu, Y. (2013). Dynamic study of yeast species and Saccharomyces cerevisiae strains during the spontaneous fermentations of Muscat blanc in Jingyang. China. Food Microbiol. 33, 172–177. doi: 10.1016/j.fm.2012.09.014
Waterhouse, A. L., Sacks, G. L., and Jeffery, D. W. (2016). Understanding Wine Chemistry. Hoboken, NJ: John Wiley & Sons.
Zea, L., Moyano, L., Moreno, J., Cortes, B., and Medina, M. (2001). Discrimination of the aroma fraction of sherry wines obtained by oxidative and biological ageing. Food Chem. 75, 79–84. doi: 10.1016/S0308-8146(01)00190-X
Zenkevich, I. G. (2002). “Kovats retention index system,” in Encyclopedia of Chromatography, ed. J. Cazes (New York: Marcel Dekker Inc).
Zhang, X., He, F., Zhang, B., Reeves, M. J., Liu, Y., Zhao, X., et al. (2018). The effect of prefermentative addition of gallic acid and ellagic acid on the red wine color, copigmentation and phenolic profiles during wine aging. Food Res. Int. 106, 568–579. doi: 10.1016/j.foodres.2017.12.054
Zhang, Y., Li, X., Lo, C. K., and Guo, S. T. (2010). Characterization of the volatile substances and aroma components from traditional soypaste. Molecules 15, 3421–3427. doi: 10.3390/molecules15053421
Zhao, C., Su, W., Mu, Y., Jiang, L., and Mu, Y. (2020). Correlations between microbiota with physicochemical properties and volatile flavor components in black glutinous rice wine fermentation. Food Res. Int. 138:109800. doi: 10.1016/j.foodres.2020.109800
Keywords: Petit Manseng, spontaneous fermentation, fungal community, volatile composition, correlation analysis
Citation: Ma Y, Li T, Xu X, Ji Y, Jiang X, Shi X and Wang B (2021) Investigation of Volatile Compounds, Microbial Succession, and Their Relation During Spontaneous Fermentation of Petit Manseng. Front. Microbiol. 12:717387. doi: 10.3389/fmicb.2021.717387
Received: 30 May 2021; Accepted: 21 July 2021;
Published: 12 August 2021.
Edited by:
Van-Tuan Tran, Vietnam National University, Hanoi, VietnamReviewed by:
Eduardo Dellacassa, Universidad de la República, UruguayTao Feng, Shanghai Institute of Technology, China
Copyright © 2021 Ma, Li, Xu, Ji, Jiang, Shi and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuewei Shi, c2hpeHVld2VpQHNoenUuZWR1LmNu; Bin Wang, Ymlud2FuZzAyMjhAc2h6dS5lZHUuY24=
 Yanqin Ma
Yanqin Ma Xuewei Shi
Xuewei Shi Bin Wang
Bin Wang