- College of Horticulture and Landscape Architecture, Northeast Agricultural University, Harbin, China
The gene encoding a putative phosphatidate phosphatase (PAP) from tolerant saline-alkali (TSA) Chlorella, ChPAP, was identified from a yeast cDNA library constructed from TSA Chlorella after a NaCl treatment. ChPAP expressed in yeast enhanced its tolerance to NaCl and sorbitol. The ChPAP protein from a GFP-tagged construct localized to the plasma membrane and the lumen of vacuoles. The relative transcript levels of ChPAP in Chlorella cells were strongly induced by NaCl and sorbitol as assessed by northern blot analyses. Thus, ChPAP may play important roles in promoting Na-ion movement into the cell and maintaining the cytoplasmic ion balance. In addition, ChPAP may catalyze diacylglycerol pyrophosphate to phosphatidate in vacuoles.
Introduction
Phosphatidate phosphatase (PAP) has the effects of catalyzing the dephosphorylation of phosphatidate (PA), and generating diacylglycerol and inorganic phosphate (Smith et al., 1957). It is also an essential enzyme in lipid metabolism, plays important roles in lipid synthesis, and is involved in the generation or degradation of lipid signaling molecules (Brindley and Pilquil, 1984; Carman, 1997; Nanjundan and Possmayer, 2001). The PAP enzymes are divided into Mg2+-dependent PAP1 or Mg2+-independent PAP2 [also be called lipid phosphate phosphatase (LPP) or diacylglycerol pyrophosphate (DGPP) phosphatase] based on the cofactor requirement for catalytic activity (Jamal et al., 1991; Brindley et al., 2002). The PAP1 enzymes play roles in cell homeostasis and lipid synthesis (Han et al., 2006; Sherr et al., 2017; Hassaninasab et al., 2019), and PAP1 enzyme, PAH1, performs catalytic function to regulate phospholipid synthesis on the nuclear and endoplasmic reticulum (Eastmond et al., 2010; Hassaninasab et al., 2019). The absence of Pahp1 (encoded PAH1) leads to the upregulated of V-ATPase (Sherr et al., 2017). The expression of PAH1 is induced in the absence of Zn (Soto-Cardalda et al., 2012). The PAH1 protein of the fungal pathogen Candida albicans restricts viral replication by affecting phospholipid synthesis and plays significant roles in hyphal growth and environmental stress regulation (Mu et al., 2019). In this study, the PAP2 enzymes are highlighted. PAP2, encoded by DPP1 and LPP1, is a vacuole membrane-associated enzyme that catalyzes DGPP to form PA and then catalyzes PA to form diacylglycerol by removing the phosphate (Wu et al., 1996; Han et al., 2001). PAP2 enzymes contain a three-domain lipid phosphatase catalytic motif containing the conserved sequences KxxxxxxRP (domain 1), PSGH (domain 2), and SRxxxxxHxxxD (domain 3). The conserved arginine residue in domain 1 and the conserved histidine residues in domains 2 and 3 are essential for the catalytic activities of PAP2 enzymes (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997; Toke et al., 1998, 1999a,b; Zhang et al., 2000; Han et al., 2004). The PAP2 enzymes are localized on the hydrophilic surfaces of the membrane (Stukey and Carman, 1997; Toke et al., 1998, 1999a,b; Zhang et al., 2000; Han et al., 2004), the vacuole (Han et al., 2001, 2004), and Golgi (Huh et al., 2003) and have broad substrate specificity levels and may function under stress conditions (Oshiro et al., 2003). In the early studies, PAP2 enzymes are found that are responsible for lipid signaling in yeast and mammals (Carman, 1997; Nanjundan and Possmayer, 2001; Brindley, 2004; Pyne et al., 2005). In plants, Arabidopsis thaliana AtLPP1 appears to be more highly expressed in the leaves and roots compared with other tissues, and the expression level of AtLPP1 increased in Arabidopsis after ionization and UV-B irradiation (Brindley, 2004; Pierrugues et al., 2011). In addition, the AtLPP2 appears to be expressed at similar levels in all the plant’s tissues, and AtLPP2 is involved with abscisic acid signaling and regulation of stomatal movements (Paradis et al., 2011).
The PAP2 gene has been also identified in microalgae (eukaryotic microbes), including Chlorella variabilis (Blanc et al., 2010), Chlorella protothecoides (Gao et al., 2014), Chlamydomonas reinhardtii (Deng et al., 2013), and Coccomyxa subellipsoidea (Blanc et al., 2010). However, there are limited reports on the cloning and functional analyses of PAP2 genes of microalgae. Some studies indicated that the expression levels of citrate synthase and phosphoenolpyruvate carboxylase 1 in Chlamydomonas reinhardtii are decreased, but the PAP2 has higher expression in the RNAi transgenic Chlamydomonas strains (Deng et al., 2013, 2014). In our previous study, we determined that tolerant saline-alkali (TSA) Chlorella can survive in an environment containing 600-mm NaCl, and the TSA Chlorella PAP gene was isolated from a TSA Chlorella full-length cDNA yeast library constructed under 1-M NaCl-stress conditions (Qiao et al., 2015). Here, we determined the growth rates of transgenic yeast on a solid medium under high salinity and drought conditions. The subcellular localization of the ChPAP protein in yeast cells was detected using confocal microscopy, and the effects of high salinity and drought conditions on ChPAP expression were investigated.
Materials and Methods
Chlorella Source, Culture, and Gene
The TSA Chlorella was previously isolated from extreme saline-alkali soil on the Songnen Plain, Heilongjiang Province, China (Wang et al., 2011), which is rich in different salt types, including NaCl and NaHCO3, and grown in liquid Bold’s basal medium (BBM, Bold and Wynne, 1984). The culture conditions were 23°C under a 16-h light/8-h dark photoperiod. The illumination intensity was 40-mmol photons m−1 s−1. The TSA Chlorella cells were maintained in solid BBM, and the sub-culturing and rapid propagation of TSA Chlorella was cultured in liquid BBM. A full-length cDNA yeast library of TSA Chlorella was constructed (Qiao et al., 2018). A sequence screened from the 1-M NaCl-treated TSA Chlorella library had close similarity levels to sequences of other species’ PAP genes. Accordingly, it was named ChPAP.
Sequence Analysis
The full-length ChPAP sequence was analyzed using BlastX and ORFfinder on the NCBI Web site.1 GeneDoc 3.0 software was used to align the sequences of the ChPAP protein and other species. The maximum-likelihood-based phylogenetic tree was constructed using MEGA 5.1 software. The transmembrane domains in the ChPAP sequence were predicted using the TMHMM server v. 2.0.2
Plasmid Construction, Yeast Transformation, and Stress-Tolerance Assays
The ChPAP cDNA fragment harboring the open reading frame was amplified from TSA Chlorella using PCR with the ChPAP-forward (5'-
Subcellular Localization of the ChPAP Protein in Yeast
To determine the subcellular localization of ChPAP, the expression plasmid pYES2-ChPAP-EGFP was constructed. The ChPAP full-length sequence with restriction sites was cloned using PCR with EGFP-specific forward GFP-F (5'-
The pYES2-ChPAP-EGFP and pYES2-EGFP plasmids were transferred independently into yeast cells. The transgenic yeast cells were pre-cultured in liquid medium containing 1% yeast extract, 2% peptone, and 2% glucose at 30°C for 2days. Afterward, they were washed three times to remove the remaining glucose. The EGFP and ChPAP-EGFP plasmids in yeast cells were induced to express in liquid yeast (1% yeast extract, 2% peptone, and 2% galactose) medium at 30°C for 6h, and then, ChPAP-EGFP yeast cells were incubated at 30°C with 20-μm FM4-64 dye for 3h. The remaining dye was removed by washing three times with sterile H2O before samples were observed. The fluorescence was detected using laser-scanning confocal imaging system (Olympus Fluoview, FV500). The EGFP and FM4-64 signals were excited at 488nm and 543nm, respectively.
Expression Analysis of ChPAP
To investigate the ChPAP transcript levels under high salinity and drought stresses, the TSA Chlorella samples were grown on medium supplemented independently with 200-mm NaCl and 300-mm sorbitol. The samples were collected at 0, 3, 6, 12, 24, and 48h and then ground with a mortar and pestle in liquid nitrogen for RNA isolation.
The total RNA of TSA Chlorella was extracted using RNAiso Plus reagent (TaKaRa, Japan). The ChPAP-specific forward (5'-ATGGGCCTCAAGGAAGAC-3') and reverse (5'-TCAAGCGTACTTCGCCTTCAG-3') primers were used to amplify the cDNA probes using a PCR Digoxigenin Probe Synthesis kit (Roche, Switzerland). The northern blot analysis was performed in accordance with a previously published protocol (Qiao et al., 2018).
Results and Discussion
Characterization of ChPAP Gene
The ChPAP nucleic acid sequence contained an open reading frame of 1,002 nucleotides that translated into 333 amino acids. The ChPAP amino acid sequence shared close similarities with the previously reported PAP sequences of other species, such as 70% similarity with C. variabilis and 53% similarity with Micractinium conductrix. A comparison of the ChPAP domains with those in PAPs of other species revealed the presence of conserved domains 1, 2, and 3 (Figure 1), which contained two arginine, one histidine, and two histidine residues, respectively. These results were consistent with previously reported PAP2 protein structures (Hemrika et al., 1997; Neuwald, 1997; Stukey and Carman, 1997). The phylogenetic analysis showed that ChPAP was closely related to the PAP of C. variabilis (XP_005848979.1). The TSA Chlorella firstly clustered with unicellular microalgae, and then clustered with microbes, animals, and plants (Figure 2). Thus, ChPAP2 is the PAP gene of TSA Chlorella.

Figure 1. Sequence alignment of the phosphatidate phosphatase (PAP) domains from tolerant saline-alkali (TSA) Chlorella ChPAP2 with those of other species. The conserved residues, including the KxxxxxxRP (domain 1), PSGH (domain 2), and SRxxxxxHxxxD (domain 3) motif, are indicated by boldface in boxes. The ChPAP2 sequence data have been deposited in the GenBank database and assigned accession no. KT750011. The other PAP protein sequences were downloaded from GenBank.
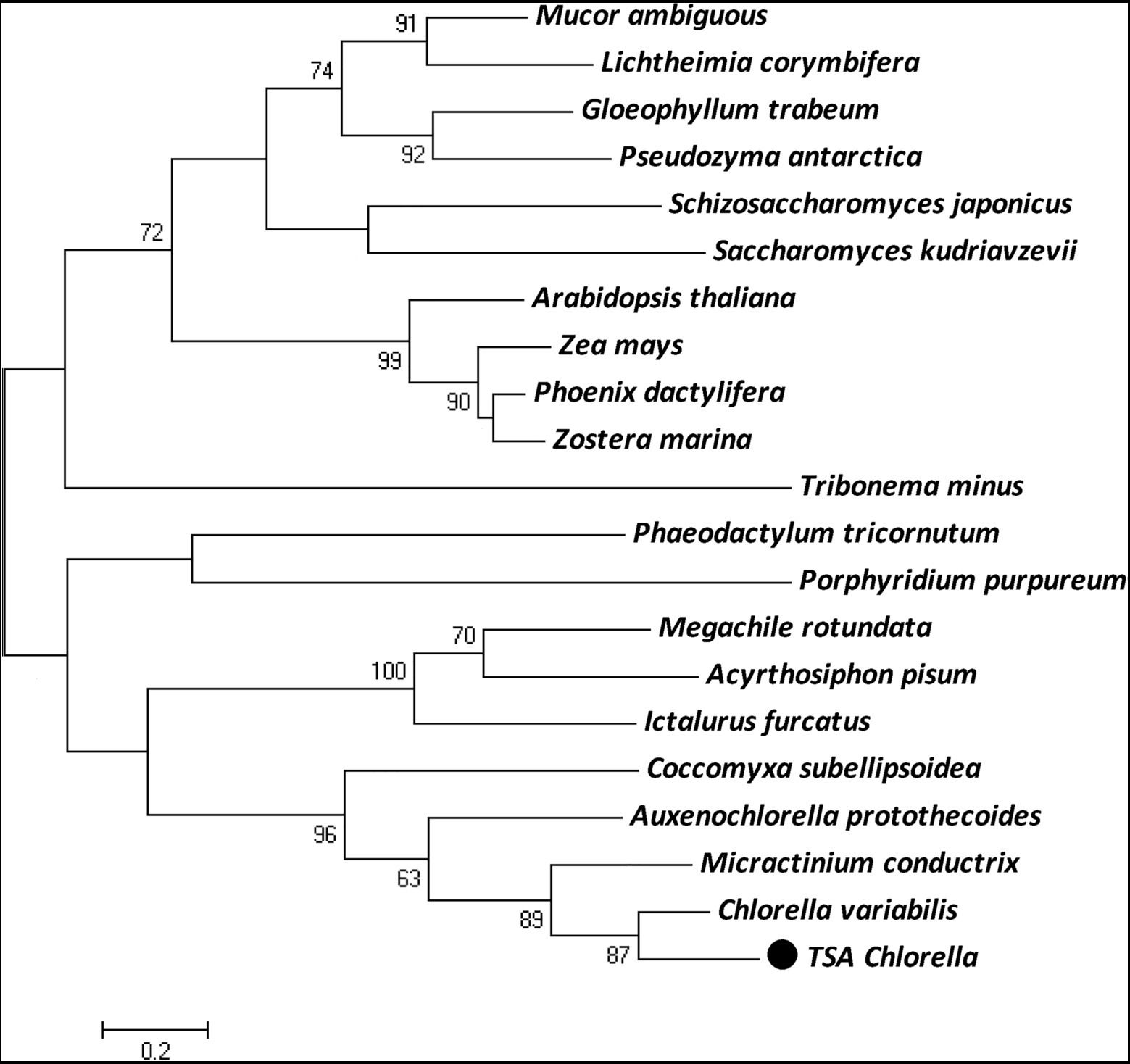
Figure 2. The maximum-likelihood (ML) phylogenetic relationships of PAP proteins between TSA Chlorella sp. and those of other species. Bootstrap values were calculated 1,000 times, and values below 50% were not included. The TSA Chlorella sp. is indicated by boldface in a box.
Overexpression of ChPAP2 in Yeast Enhanced Tolerance to NaCl and Sorbitol
The five serial dilutions of transgenic yeast cells were spotted into a solid yeast medium. The growth of yeast cells harboring the empty pYES2 vector was similar to that of the ChPAP2 transgenic yeast in medium containing 1% yeast extract, 2% peptone, and 2% glucose without any stress. The ChPAP2 transgenic yeast grew better than the controls in the presence of 0.8-M NaCl, 1-M NaCl, or 1.6-M sorbitol (Figure 3). Thus, the expression of ChPAP2 in yeast cells improved the tolerance to NaCl and sorbitol, which indicated that ChPAP2 functions as lipid signaling molecule during abiotic stress (Munnik et al., 1996). PAH1 encoded PAP1 was regulated to express under Zn deficiency and enhanced the activity of PAP enzyme (Soto-Cardalda et al., 2012). In addition, Arabidopsis PAP2 was found that was involved with ABA signaling and regulating the stomatal movements (Paradis et al., 2011). However, there was no relevant report on the investigation of PAP2 under abiotic stress.
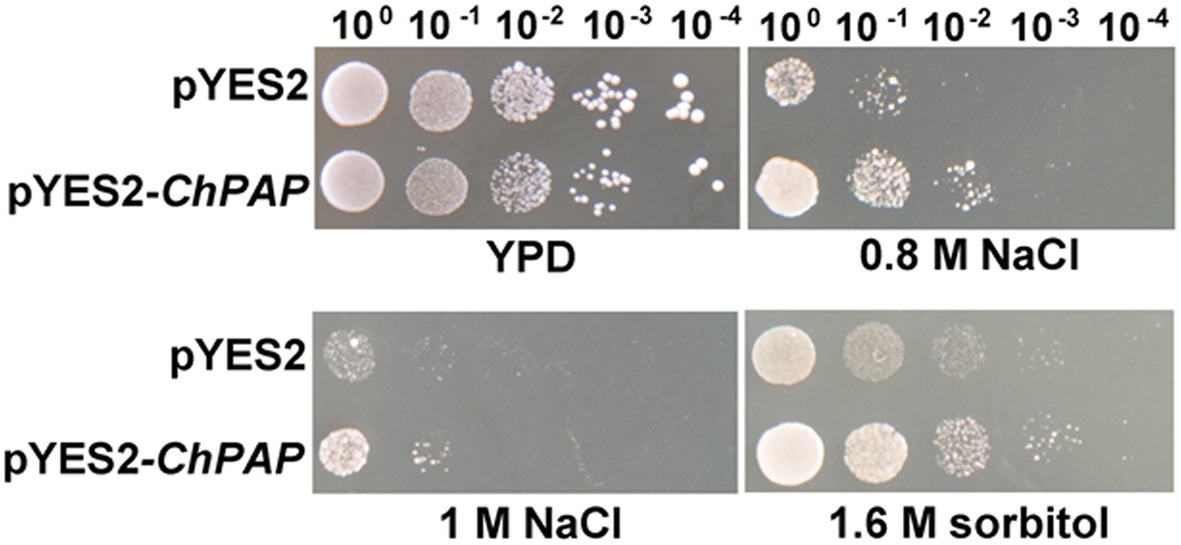
Figure 3. ChPAP2-overexpressing yeast cells exposed to NaCl and sorbitol stresses. Serial dilutions of yeast cells containing the pYES2 empty vector or pYES2-ChPAP2 were independently spotted into solid yeast (1% yeast extract, 2% peptone, and 2% galactose) medium containing plates supplemented independently with 0.8-M NaCl, 1.0-M NaCl, and 1.6-M sorbitol.
The Expression of ChPAP2 Was Inducible Under NaCl and Sorbitol Stresses
A northern blot analysis was used to detect ChPAP2 expression patterns. Total RNA was used to analyze the effects of high salinity and drought stresses on ChPAP2 expression. The TSA Chlorella cells were exposed independently to NaCl and sorbitol for 0, 3, 6, 12, 24, and 48h. The ChPAP2 mRNA expression dramatically increased with the 200-mm NaCl treatment from 6 to 48h compared with the control (0h), indicating that ChPAP2 was upregulated (Figure 4A). Thus, in yeast, the increased ChPAP2 expression level may increase NaCl resistance. The PA and DGPP levels also increase under hyperosmotic stress conditions (Munnik et al., 2000). The presence of NaCl in liquid media not only leads to high salinity stress, but also to hyperosmotic stress in plants. We hypothesized that the ChPAP2 expression level may be upregulated, allowing it to catalyze the excess DGPP into PA.
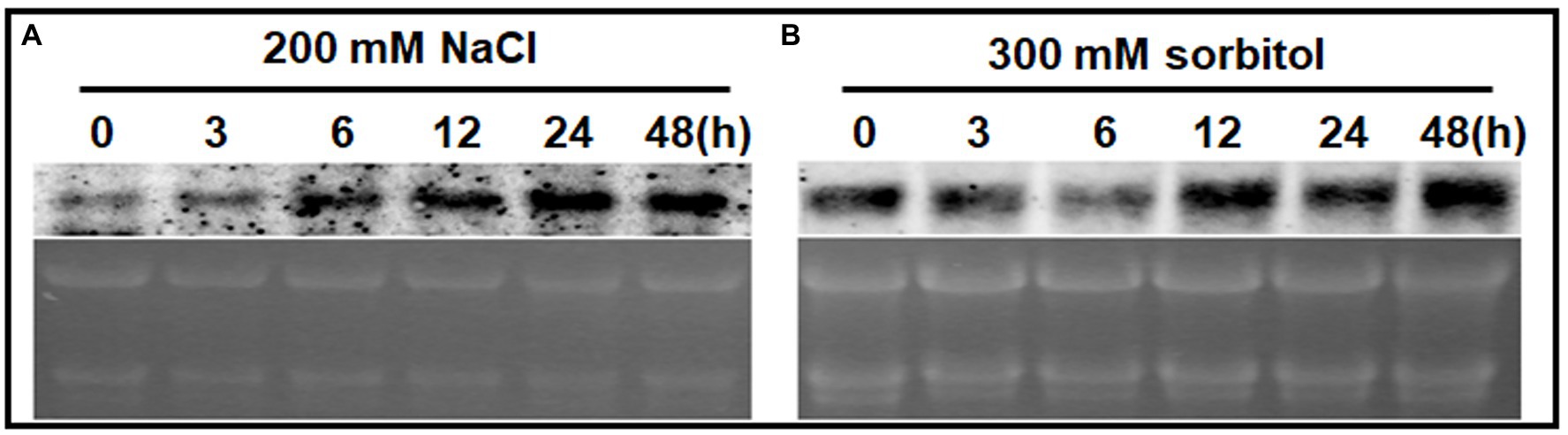
Figure 4. The mRNA expression levels of ChPAP2 at various time points after exposure to NaCl- and sorbitol-stress treatments. Northern blot analyses of the ChPAP2 gene’s expression levels in TSA Chlorella cells using a digoxigenin-labeled ChPAP2 cDNA probe. The total RNA (5μg) was extracted from Chlorella cells treated independently with 200-mm NaCl (A) and 300-mm sorbitol (B).
In this study, a sorbitol solution was used as the drought agent (Bocchini et al., 2018; Lu et al., 2019). The ChPAP2 expression levels did not differ significantly compared with those of the control after 3–6h of exposure to 300-mm sorbitol. The ChPAP2 mRNA level began to rise at 12h, and the level was the most significantly different from that of the control at 48h after treatment (Figure 4B). In Saccharomyces cerevisiae, PAP accumulates during exposure to hyperosmotic and dehydration stresses (Munnik et al., 2000). Thus, the upregulated ChPAP2 expression may enhance drought tolerance.
ChPAP2 Localized at the Plasma Membrane and in the Lumen of Vacuoles
The deduced ChPAP2 amino acid sequence was predicted to contain six transmembrane domains (Figure 5A). To determine its subcellular localization, GFP was fused to the C-terminus of ChPAP2 (ChPAP2-GFP). The green fluorescence of the GFP protein alone was almost evenly distributed throughout the yeast cells (Figures 5B1,2). FM4-64 stained the vacuole membrane, and its localization signal was consistent with the vacuolar membrane signal (red fluorescence area; Figure 5B4). Thus, the ChPAP2 protein appeared to localize on the plasma membrane and in the lumen of vacuoles in yeast cells (Figures 5B3,5). The results suggested that ChPAP2 might play important roles in transporting lipids through the plasma membrane and in catalyzing DGPP into PA in the vacuoles.
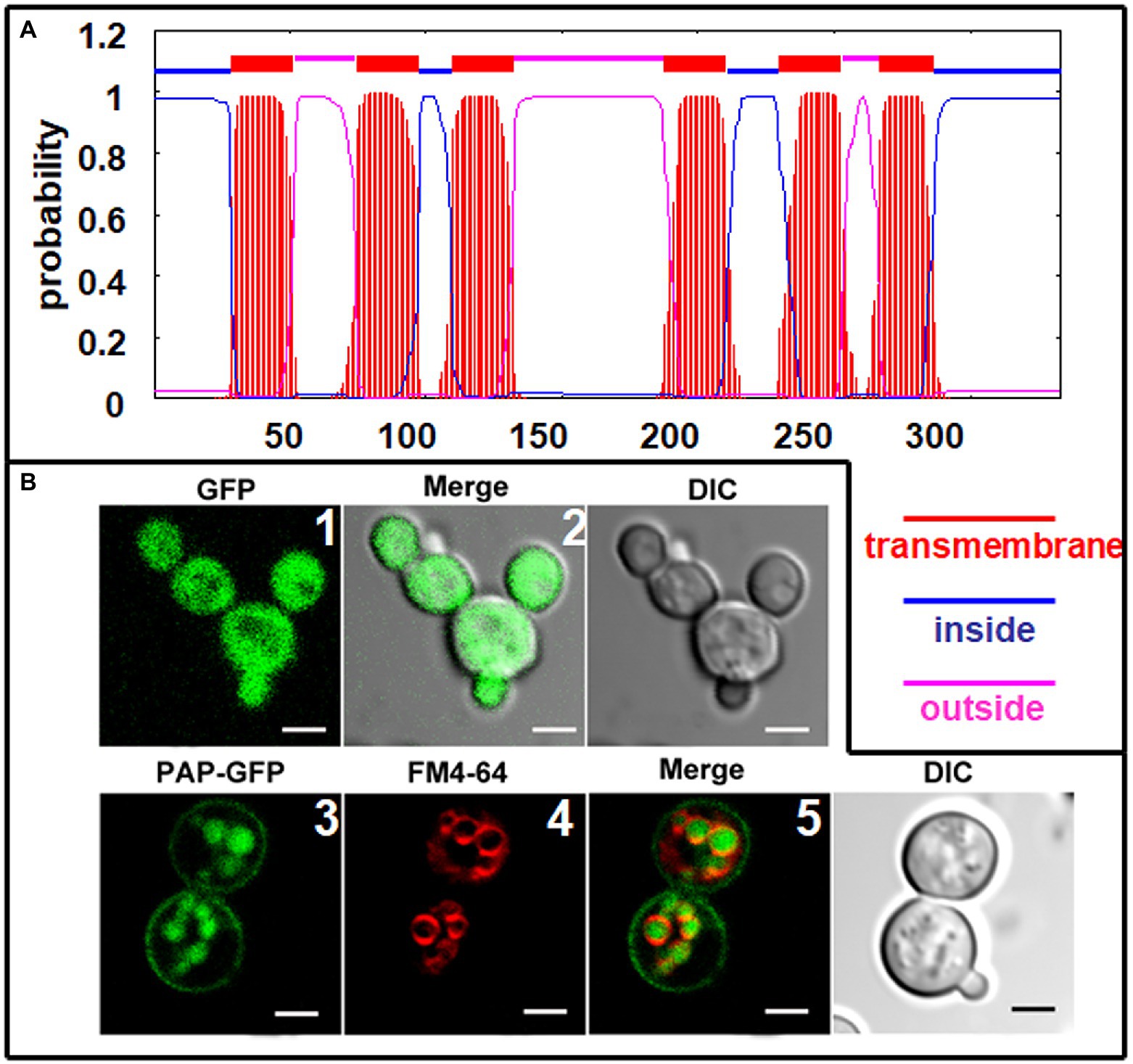
Figure 5. Subcellular localization of ChPAP2 in yeast cells. (A) Transmembrane helices in ChPAP2 (GenBank no. KT750011) were predicted using the TMHMM server v. 2.0; (B) the green fluorescence was observed using a confocal microscope. Upper: GFP gene expressed in the pYES2 vector and in the yeast strain INVSc1. Left, middle, and right panels show fluorescence, merged, and bright-field (DIC) images. Lower: ChPAP2-EGFP fusion gene expressed in the pYES2 vector and in the yeast strain INVSc1. Live cells overexpressing ChPAP2-EGFP were incubated with FM4-64. Left, second, third, and right panels show fluorescence, FM4-64, merged, and bright-field (DIC) images. Bars=5μm.
ChPAP2, as a catalytic enzyme, participates in lipid metabolism. ChPAP2 localized on the plasma membrane may function in maintaining cell membrane stability and the ion balance of the cytoplasm in response to abiotic stresses. The ChPAP2 protein also is localized in the lumen of vacuoles, where it may be involved with lipid translocation and DGPP catalysis to form PA. Therefore, we hypothesized that ChPAP2 participates in catalyzing DGPP in the lumen of vacuoles.
The PAP protein may function as a signaling molecule in planta under stress conditions, and its levels accumulate during hyperosmotic and dehydration stresses (Munnik et al., 1996). Therefore, the PAP enzyme may play a role in regulating specific cellular DGPP and PA pools under stress conditions (Oshiro et al., 2003).
In summary, the ChPAP in TSA Chlorella was upregulated expression in treated with high salinity and drought, and the ChPAP in yeasts could tolerate high salinity and drought stresses. Its protein was localized at the plasma membrane and in the lumen of vacuoles. The ChPAP might translocate excess NaCl and sorbitol from plasma membrane and then segregate them into vacuole to regulate ion balance in the cytoplasm. As a consequence, the PAP as a novel transporter can enhance high salinity tolerance and accumulate excess high salinity. These characteristics make ChPAP for the bioremediation of saline-alkali soil.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author Contributions
JW designed the research. QS, YR, and DS performed the experiments. HZ, JZ, and SG analyzed the data. KQ and AZ wrote the manuscript. All authors revised and approved the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (grant nos. 31800200, 31902052, and 31972450), the Natural Science Foundation of Heilongjiang Province of China (grant no. YQ2020C002), and the Postdoctoral Research Initiation Funding Project of Heilongjiang Province (grant no. LBH-Q19084).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Blanc, G., Duncan, G., Agarkova, I., Borodovsky, M., Gurnon, J., Kuo, A., et al. (2010). The Chlorella variabilis NC64A genome reveals adaptation to photosymbiosis, coevolution with viruses, and cryptic sex. Plant Cell 22, 2943–2955. doi: 10.1105/tpc.110.076406
Bocchini, M., D’Amato, R., Ciancaleoni, S., Fontanella, M. C., Palmerini, C. A., Beone, G. M., et al. (2018). Soil selenium (se) biofortification changes the physiological, biochemical and epigenetic responses to water stress in Zea mays L. by inducing a higher drought tolerance. Front. Plant Sci. 9:389. doi: 10.3389/fpls.2018.00389
Bold, H. C., and Wynne, M. J. (1984). (Bold) introduction to the algae prentice hall. Inc. New Jersey 720, 573–574.
Brindley, D. N. (2004). Lipid phosphate phosphatases and related proteins: signaling functions in development, cell division, and cancer. J. Cell. Biochem. 92, 900–912.
Brindley, D. N., English, D., Pilquil, C., Buri, K., and Ling, Z. C. (2002). Lipid phosphate phosphatases regulate signal transduction through glycerolipids and sphingolipids. Biochim. Biophys. Acta 1582, 33–44. doi: 10.1016/s1388-1981(02)00135-x
Brindley, D. N., and Pilquil, C. (1984). Lipid phosphate phosphatases and signaling. Prog. Lipid Res. 23, 115–133.
Carman, G. M. (1997). Phosphatidate phosphatases and diacylglycerol pyrophosphate phosphatases in Saccharomyces cerevisiae and Escherichia coli. Biochim. Biophys. Acta 1348, 45–55. doi: 10.1016/s0005-2760(97)00095-7
Deng, X. D., Cai, J. J., and Fei, X. W. (2013). Effect of the expression and knockdown of citrate synthase gene on carbon flux during triacylglycerol biosynthesis by green algae Chlamydomonas reinhardtii. BMC Biochem. 14:38. doi: 10.1186/1471-2091-14-38
Deng, X. D., Cai, J. J., Li, Y. J., and Fei, X. W. (2014). Expression and knockdown of the PEPC1 gene affect carbon flux in the biosynthesis of triacylglycerols by the green alga Chlamydomonas reinhardtii. Biotechnol. Lett. 36, 2199–2208. doi: 10.1007/s10529-014-1593-3
Eastmond, P. J., Quettier, A. L., Kroon, J. T. M., Craddock, C., Adams, N., and Slabasb, A. R. (2010). Phosphatidic acid phosphohydrolases 1 and 2 regulate phospholipid synthesis at the endoplasmic reticulum in Arabidopsis. Plant Cell 22, 2796–2811. doi: 10.1105/tpc.109.071423
Gao, C. F., Wang, Y., Shen, Y., Yan, D., He, X., Dai, J. B., et al. (2014). Oil accumulation mechanisms of the oleaginous microalga Chlorella protothecoides revealed through its genome, transcriptomes, and proteomes. BMC Genomics 15:582. doi: 10.1186/1471-2164-15-582
Gietz, D., Stjean, A., Woods, R. A., and Schiestl, R. H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425. doi: 10.1093/nar/20.6.1425
Han, G. S., Johnston, C. N., and Carman, G. M. (2004). Vacuole membrane topography of the DPP1-encoded diacylglycerol pyrophosphate phosphatase catalytic site from Saccharomyces cerevisiae. J. Biol. Chem. 279, 5338–5345. doi: 10.1074/jbc.M311779200
Han, G. S., Johnston, C. N., Chen, X., Athenstaedt, K., Daum, G., and Carman, G. M. (2001). Regulation of the Saccharomyces cerevisiae DPP1-encoded diacylglycerol pyrophosphate phosphatase by zinc. J. Biol. Chem. 276, 10126–10133. doi: 10.1074/jbc.M011421200
Han, G. S., Wu, W. I., and Carman, G. M. (2006). The Saccharomyces cerevisiae lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218. doi: 10.1074/jbc.M600425200
Hassaninasab, A., Hsieh, L. S., Su, W. M., Han, G. S., and Carman, G. M. (2019). Yck1 casein kinase I regulates the activity and phosphorylation of Pah1 phosphatidate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 294, 18256–18268. doi: 10.1074/jbc.RA119.011314
Hemrika, W., Renirie, R., Dekker, H. L., Barnett, P., and Wever, R. (1997). From phosphatases to vanadium peroxidases: a similar architecture of the active site. Proc. Natl. Acad. Sci. U. S. A. 94, 2145–2149. doi: 10.1073/pnas.94.6.2145
Huh, W. K., Falvo, J. V., Gerke, L. C., Carroll, A. S., Howson, R. W., Weissman, J. S., et al. (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. doi: 10.1038/nature02026
Jamal, Z., Martin, A., Gomez-Muiioz, A., and Brindley, D. N. (1991). Plasma membrane fractions from rat liver contain a phosphatidate phosphohydrolase distinct from that in the endoplasmic reticulum and cytosol. J. Biol. Chem. 266, 2988–2996. doi: 10.1016/S0021-9258(18)49945-0
Lu, X. P., Gao, H. J., Zhang, L., Wang, Y. P., Shao, K. Z., Zhao, Q., et al. (2019). Dynamic responses of Haloxylon ammodendron to various degrees of simulated drought stress. Plant Physiol. Biochem. 139, 121–131. doi: 10.1016/j.plaphy.2019.03.019
Mu, C. H., Pan, C. Y., Han, Q., Liu, Q. Z., Wang, Y., and Sang, J. L. (2019). Phosphatidate phosphatase Pah1 has a role in the hyphal growth and virulence of Candida albicans. Fungal Genet. Biol. 124, 47–58. doi: 10.1016/j.fgb.2018.12.010
Munnik, T., deVrije, T., Irvine, R. F., and Musgrave, A. (1996). Identification of diacylglycerol pyrophosphate as a novel metabolic product of phosphatidic acid during G-protein activation in plants. J. Biol. Chem. 271, 15708–15715. doi: 10.1074/jbc.271.26.15708
Munnik, T., Meijer, H. J. G., Ter Riet, B., Hirt, H., Frank, W., Bartels, D., et al. (2000). Hyperosmotic stress stimulates phospholipase D activity and elevates the levels of phosphatidic acid and diacylglycerol pyrophosphate. Plant J. 22, 147–154. doi: 10.1046/j.1365-313x.2000.00725.x
Nanjundan, M., and Possmayer, F. (2001). Pulmonary lipid phosphate phosphohydrolase in plasma membrane signalling platforms. Biochem. J. 358, 637–646. doi: 10.1042/bj3580637
Neuwald, A. F. (1997). An unexpected structural relationship between integral membrane phosphatases and soluble haloperoxidases. Protein Sci. 6, 1764–1767. doi: 10.1002/pro.5560060817
Oshiro, J., Han, G. S., and Carman, G. M. (2003). Diacylglycerol pyrophosphate phosphatase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1635, 1–9. doi: 10.1016/j.bbalip.2003.10.002
Paradis, S., Villasuso, A. L., Aguayo, S. S., Maldiney, R., Habricot, Y., Zalejski, C., et al. (2011). Arabidopsis thaliana lipid phosphate phosphatase 2 is involved in abscisic acid signalling in leaves. Plant Physiol. Biochem. 49, 357–362. doi: 10.1016/j.plaphy.2011.01.010
Pierrugues, O., Brutesco, C., Oshiro, J., Gouy, M., Deveaux, Y., Carman, G. M., et al. (2001). Lipid phosphate phosphatases in Arabidopsis regulation of the AtLPP1 gene in response to stress. J. Biol. Chem. 276, 20300–20308. doi: 10.1074/jbc.M009726200
Pyne, S., Long, J. S., Ktistakis, N. T., and Pyne, N. J. (2005). Lipid phosphate phosphatases and lipid phosphate signaling. Biochem. Soc. Trans. 33, 1370–1374. doi: 10.1042/BST0331370
Qiao, K., Takano, T., and Liu, S. K. (2015). Discovery of two novel highly tolerant NaHCO3 Trebouxiophytes: identification and characterization of microalgae from extreme saline-alkali soil. Algal Res. 9, 245–253. doi: 10.1016/j.algal.2015.03.023
Qiao, K., Wang, M., Takano, T., and Liu, S. K. (2018). Overexpression of acyl-CoA-binding protein 1 (ChACBP1) from saline-alkali-tolerant chlorella sp. enhances stress tolerance in Arabidopsis. Front. Plant Sci. 9:1772. doi: 10.3389/fpls.2018.01772
Sherr, G. L., LaMassa, N., Li, E., Phillips, G., and Shen, C. H. (2017). Pah1p negatively regulates the expression of V-ATPase genes as well as vacuolar acidification. Biochem. Bioph. Res. Co. 491, 693–700. doi: 10.1016/j.bbrc.2017.07.127
Smith, S. W., Weiss, S. B., and Kennedy, E. P. (1957). The enzymatic dephosphorylation of phosphatidic acids. J. Biol. Chem. 228, 915–922. doi: 10.1016/S0021-9258(18)70670-4
Soto-Cardalda, A., Fakas, S., Pascual, F., Choi, H. S., and Carman, G. M. (2012). Phosphatidate phosphatase plays role in zinc-mediated regulation of phospholipid synthesis in yeast. J. Biol. Chem. 287, 968–977. doi: 10.1074/jbc.M111.313130
Stukey, J., and Carman, G. M. (1997). Identification of a novel phosphatase sequence motif. Protein Sci. 6, 469–472. doi: 10.1002/pro.5560060226
Toke, D. A., Bennett, W. L., Dillon, D. A., Wu, W. I., Chen, X. M., Ostrander, D. B., et al. (1998). Isolation and characterization of the Saccharomyces cerevisiae DPP1 gene encoding for diacylglycerol pyrophosphate phosphatase. J. Biol. Chem. 273, 3278–3284. doi: 10.1074/jbc.273.6.3278
Toke, D. A., Bennett, W. L., Oshiro, J., Wu, W. I., Voelker, D. R., and Carman, G. M. (1999a). Isolation and characterization of the Saccharomyces cerevisiae LPP1 gene encoding a Mg2+-independent phosphatidate phosphatase. J. Biol. Chem. 273, 14331–14338. doi: 10.1074/jbc.273.23.14331
Toke, D. A., Mcclintick, M. L., and Carman, G. M. (1999b). Mutagenesis of the phosphatase sequence motif in diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. Biochemistry 38, 14606–14613. doi: 10.1021/bi991472x
Wang, J., Shi, W., Takano, T., and Liu, S. K. (2011). Purification and resistance analysis of algae of soda soil in Northeast China. Mol. Soil Biol. 3, 1–4. doi: 10.5376/msb.2011.03.0001
Wu, W. I., Liu, Y. S., Riedel, B., Wissing, J. B., Fischl, A. S., and Carman, G. M. (1996). Purification and characterization of diacylglycerol pyrophosphate phosphatase from Saccharomyces cerevisiae. J. Biol. Chem. 271, 1868–1876. doi: 10.1074/jbc.271.4.1868
Keywords: tolerant saline-alkali Chlorella, NaCl, sorbitol, subcellular localization, gene expression
Citation: Wang J, Shan Q, Ran Y, Sun D, Zhang H, Zhang J, Gong S, Zhou A and Qiao K (2021) Molecular Characterization of a Tolerant Saline-Alkali Chlorella Phosphatidate Phosphatase That Confers NaCl and Sorbitol Tolerance. Front. Microbiol. 12:738282. doi: 10.3389/fmicb.2021.738282
Edited by:
Kianoush KHosravi-Darani, National Nutrition and Food Technology Research Institute, IranReviewed by:
Shiguo Li, Research Center for Eco-environmental Sciences (CAS), ChinaQuanyu Zhao, Nanjing Tech University, China
Swarnendu Roy, University of North Bengal, India
Santosh Kumar, Agriculture and Agri-Food Canada, Canada
Copyright © 2021 Wang, Shan, Ran, Sun, Zhang, Zhang, Gong, Zhou and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Aimin Zhou, YWltaW56aG91QG5lYXUuZWR1LmNu Kun Qiao, a3VucWlhb0BuZWF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Jingang Wang
Jingang Wang Qinghua Shan†
Qinghua Shan† Jinzhu Zhang
Jinzhu Zhang Aimin Zhou
Aimin Zhou Kun Qiao
Kun Qiao