- 1State Key Laboratory of Veterinary Etiological Biology, National Professional Laboratory for Animal Echinococcosis, Key Laboratory of Veterinary Parasitology of Gansu Province, Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Lanzhou, China
- 2Department of Life Sciences, Natural History Museum, London, United Kingdom
- 3London Centre for Neglected Tropical Disease Research, London, United Kingdom
- 4Guizhou Provincial Center for Animal Disease Control and Prevention, Guiyang, China
- 5State Key Laboratory of Plateau Ecology and Agriculture, Qinghai Academy of Animal Science and Veterinary Medicine, Qinghai University, Xining, China
The Cyclophyllidea comprises the most species-rich order of tapeworms (Platyhelminthes, Cestoda) and includes species with some of the most severe health impact on wildlife, livestock, and humans. We collected seven Cyclophyllidea specimens from rodents in Qinghai-Tibet Plateau (QTP) and its surrounding mountain systems, of which four specimens in QTP were unsequenced, representing “putative new species.” Their complete mitochondrial (mt) genomes were sequenced and annotated. Phylogenetic reconstruction of partial 28S rDNA, cox1 and nad1 datasets provided high bootstrap frequency support for the categorization of three “putative new species,” assigning each, respectively, to the genera Mesocestoides, Paranoplocephala, and Mosgovoyia, and revealing that some species and families in these three datasets, which contain 291 species from nine families, may require taxonomic revision. The partial 18S rDNA phylogeny of 29 species from Taeniidae provided high bootstrap frequency support for the categorization of the “putative new species” in the genus Hydatigera. Combined with the current investigation, the other three known Taeniidae species found in this study were Taenia caixuepengi, T. crassiceps, and Versteria mustelae and may be widely distributed in western China. Estimates of divergence time based on cox1 + nad1 fragment and mt protein-coding genes (PCGs) showed that the differentiation rate of Cyclophyllidea species was strongly associated with the rate of change in the biogeographic scenarios, likely caused by the uplift of the QTP; i.e., species differentiation of Cyclophyllidea might be driven by host-parasite co-evolution caused by the uplift of QTP. We propose an “out of QTP” hypothesis for the radiation of these cyclophyllidean tapeworms.
Introduction
Cestoda is a class of parasitic worms in the flatworm phylum Platyhelminthes that parasitize the intestines of all major groups of vertebrates, including fish (Kuchta et al., 2020), reptiles (Yudhana et al., 2019), birds (Okulewicz, 2014), and mammals (Carlson et al., 2020), and cause severe, mild or no symptoms of infection. Their larvae usually achieve development in one or two intermediate hosts (Kuchta et al., 2020) and can cause severe symptoms and even death in animals and humans; species of Taenia and Echinococcus cause the highest health and economic impact.
To date, there are approximately 4,800 described species in the class Cestoda, belonging to 833 genera and 19 orders, of which the Cyclophyllidea is the most species-rich order in the class, with more than 3,100 valid species, distributed among 16 families (Sharma et al., 2016; Caira and Jensen, 2017; Kuchta et al., 2020). Each free-living metazoan species is considered to harbor at least one protozoan or metazoan parasite species (Poulin and Morand, 2004) with which they interact and usually co-evolve (Ebert and Fields, 2020). Parasites are probably one of the largest groups of undescribed organisms, as most are cryptic whether in their parasitic or free-living forms (Dobson et al., 2008; Larsen et al., 2017; Okamura et al., 2018; Carlson et al., 2020). It is estimated that there are a total of about 100,000–350,000 helminth species in vertebrates around the world, of which 85–95% are undiscovered or recorded to science (Carlson et al., 2020).
Many of the world’s biodiversity hotspots are located in large mountain systems, and their role in the evolutionary diversification of organisms is manifold (Hoorn et al., 2018; Rahbek et al., 2019a,b). The Qinghai-Tibet Plateau (QTP) and its surrounding mountain systems of the Eurasian continent, have yielded arguably the biggest and probably the most biologically diverse area of montane species (Päckert et al., 2020). In the past decade, several new Taeniidae species have been described from wild rodents on the QTP (Xiao et al., 2005; Wu et al., 2021). In terms of species diversity and sheer population sizes, rodents are perhaps the most important intermediate and definitive hosts of tapeworms, and are the most widely distributed and diverse group of mammals; about 43% of all species (Singla et al., 2008; Wu et al., 2018). Changes in climate and vegetation caused by the uplift of the QTP may have promoted local adaptations such as the evolution of cold- and hypoxic-tolerant rodent species, which in turn may have led to the co-evolution and radiation of their parasites (Wang et al., 2018; Wu et al., 2021). We hypothesize that there may be many undiscovered tapeworm species parasitizing the rodents in the QTP. Although the number of tapeworm species has been underestimated in general, some practices may lead to the erroneous proposal of new species (Carlson et al., 2020; Kuchta et al., 2020).
Morphological distinctions have been used for the description of many tapeworms, however, the homoplasy in morphology poses quite a challenge to infer their evolutionary lineage (Scholz et al., 2021), where morphological features may be significantly affected by different intermediate host sources (Lymbery, 1998). Many undescribed species may be genetically different but morphologically indistinguishable. The inability to distinguish such cryptic species affects accurate assessment of host range and estimates of total diversity (Carlson et al., 2020). The 28S and 18S nuclear ribosomal RNA genes (rDNA) are relatively conserved within species and are often used to differentiate different species (Wickström et al., 2005; Nakao et al., 2013; Scholz et al., 2021). The mitochondrial (mt) genome is largely haploid and uniparentally inherited, so their effective population size is four times smaller than that of the nuclear genome (Toews and Brelsford, 2012). Since the process of lineage sorting is inversely proportional to the effective population size, this means mitochondrial (mt) genomes will complete this process faster than nuclear genomes (Toews and Brelsford, 2012). Thus, the genetic nature of a mt genome makes it likely more sensitive than any single nuclear marker to distinguish closely related species and study their phylogenetic relationships (Lee et al., 2007).
Therefore, in this study, we sampled parasites in rodents from the QTP and its surrounding areas. In total, seven Cyclophyllidea specimens were collected and characterized using molecular tools. By comparing new (mitochondrial, nuclear 28SrDNA, 18S rDNA) with published homologs (GenBank) we found four isolates to be markedly different in DNA sequence thus likely representing ‘‘putative new species.’’ The complete mt genomes of these unknown taxa were sequenced and annotated, and their taxonomic status was analyzed and verified through phylogenetic reconstruction of five datasets containing a total of 320 species from 10 families. By combining evolutionary divergence time analyses of mt genes of classified cyclophyllideans in NCBI Taxonomy Database1 and the paleogeography of QTP, we thus speculate an “out of QTP” theory for cyclophyllideans.
Materials and Methods
Sample Collection
Rodents were live-trapped in meadows in Tibet, Qinghai, Sichuan, Gansu and Xinjiang province or autonomous region of China in 2013 and from 2018 to 2020. Rodents were euthanized and dissected according to the Ethics Statement mentioned below, cysticerci and host livers were collected from the enterocoelia and thorax, and adults were extracted from the intestines. Detailed sample collection data and host identities are described in Supplementary Table 1. After detaching the lesions, parasite specimens and host livers were kept in 75% (v/v) ethanol for molecular identification.
DNA Isolation, Amplification, and Sequencing
DNA samples of hosts and parasites were extracted using Blood and Tissue Kit (Cat. No. 69504, Qiagen, Germany) as instructed by the manufacturer, and were amplified and sequenced for identification by conserved primers of cytb gene of small mammals (Fan et al., 2011) and cox1 gene of tapeworm (Bowles et al., 1992), respectively. By means of highly similar BLAST search in the nucleotide collection (nr/nt) database,2 four of the Cyclophyllidea specimens with less than 95% identity of the most similar cox1 sequence were identified as putatively unknown species (Xiao et al., 2005). The mt genomes of four of the putatively unknown specimens were sequenced and assembled according to the following procedure: firstly, the DNA of the four species was amplified and sequenced using primers published in Wu et al. (2021); missing fragments were amplified with newly designed primers using the program Oligo 6.0 with the method described in Wu et al. (2021), until entire circular mtDNAs were amplified and sequenced. A list of primers for each species can be found in Supplementary Table 2. The 18S and 28S rDNA fragments of these four species were also amplified and sequenced with conserved primers (Littlewood et al., 2000; Yan et al., 2013) for further species identification and phylogenetic analyses. Primers were synthesized by Tsingke Biotechnology (Xi’an, China). Standard 25 μl PCR protocol was used to amplify the DNA fragments. The PCR products were purified and sequenced according to protocols in Wu et al. (2021). SeqMan software was used to assemble the cytb gene sequences, 18S and 28S rDNA partial sequences and mt genomes (see Supplementary Table 3 for GenBank accession nos.).
Mitochondrial Genome Annotation
The four new mtDNAs were annotated preliminarily by Geseq3 with the reference of the most closely related species (Mesocestoides corti, Anoplocephala magna, Hydatigera krepkogorski, and Moniezia expansa) identified by the phylogenetic analyses in Figures 1, 2, whose mt genome annotations are available in GenBank. Putative tRNA genes were verified using ARWEN4 using default parameters (Laslett and Canbäck, 2008). The positions of their open reading frames (ORF) and rRNA genes were further checked and modified using SnapGene (v3.2.1) based on alignments with the reference of the most closely related species mentioned above. SnapGene (v3.2.1) was used to translate the protein-coding genes (PCGs) into their amino acid sequence with echinoderm/flatworm mitochondrial code (NCBI translation table 9) and to illustrate the annotated mt genomes.
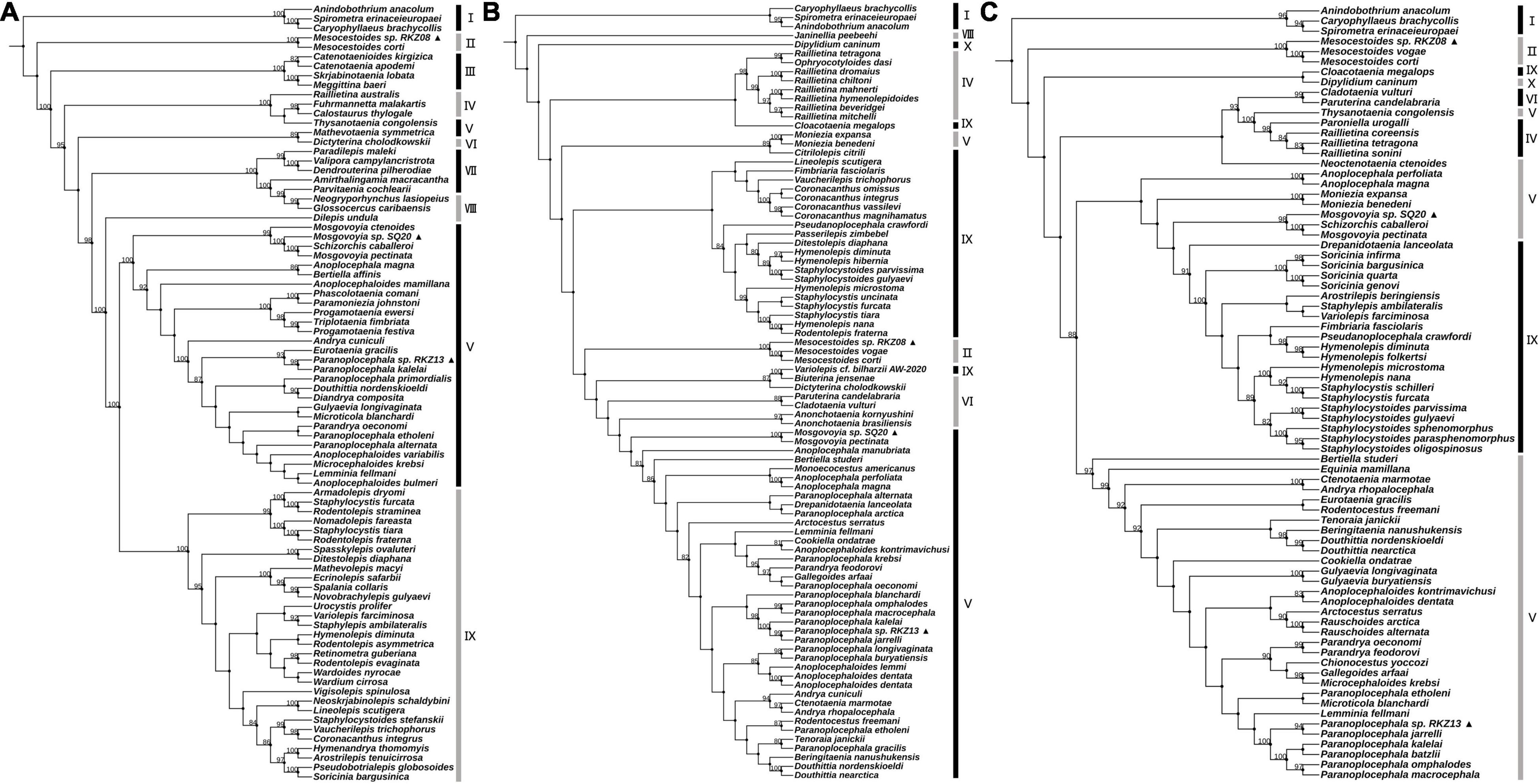
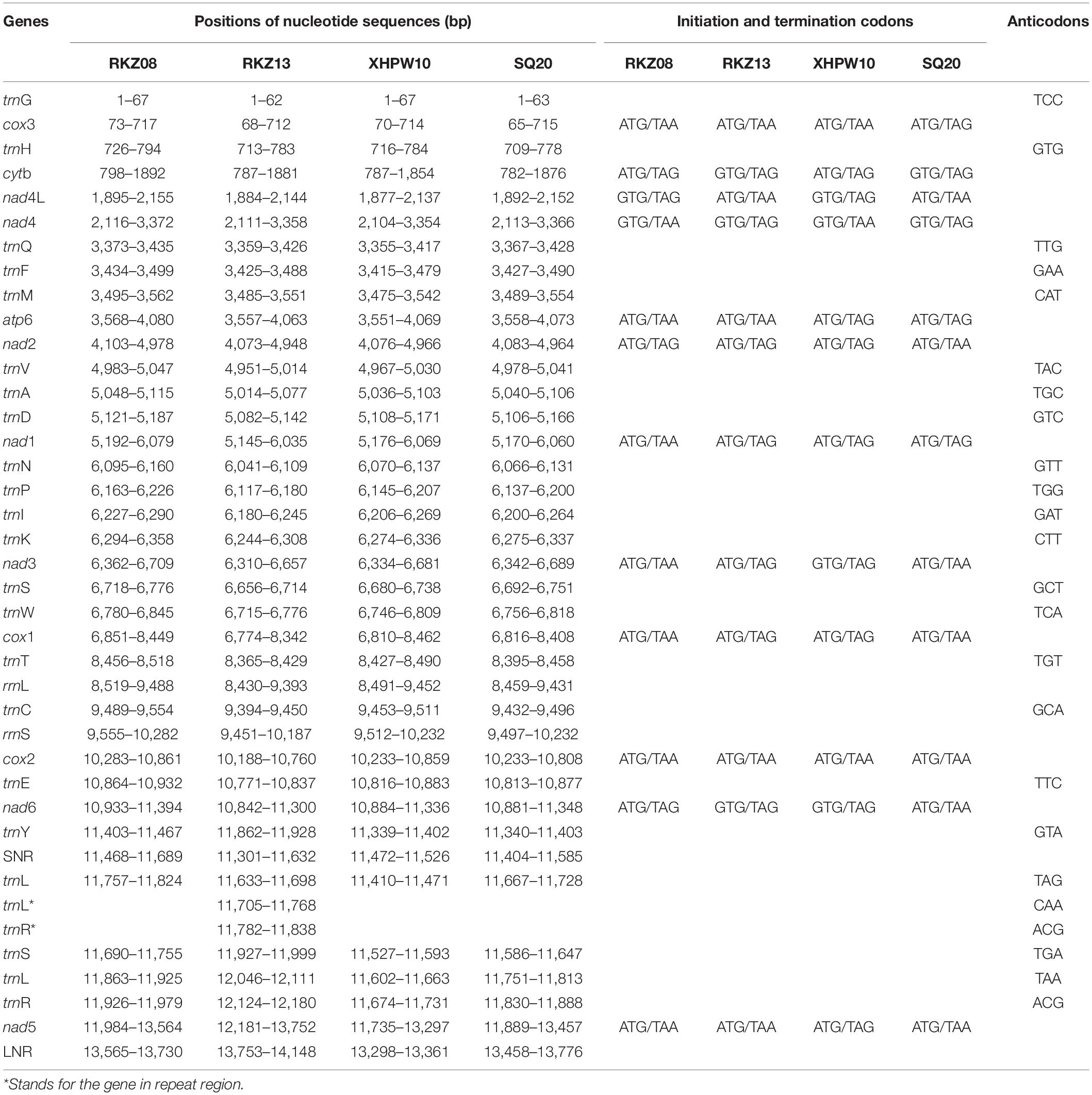
Figure 1. Maximum likelihood analyses of the four “putative new species” with other Cyclophyllidea species, except for Taeniidae, based on simplified 28S rDNA (A), cox1 (B) and nad1 (C) fragments. Alternating black and gray bands are used to classify different genera in the Taxonomy of NCBI, The Roman numerals to the right of the bands represent outgroup and different genus names (i.e., I: outgroup, II: Mesocestoididae, III: Catenotaeniidae, IV: Dipylidiidae, V: Anoplocephalidae, VI: Paruterinidae, VII: Gryporhynchidae, VIII: Dilepididae, IX: Hymenolepididae, X: Dipylidiidae). The ▲ after the species name represents “putative new species”. Bootstrap frequency support values are stated only for nodes where > 80%.
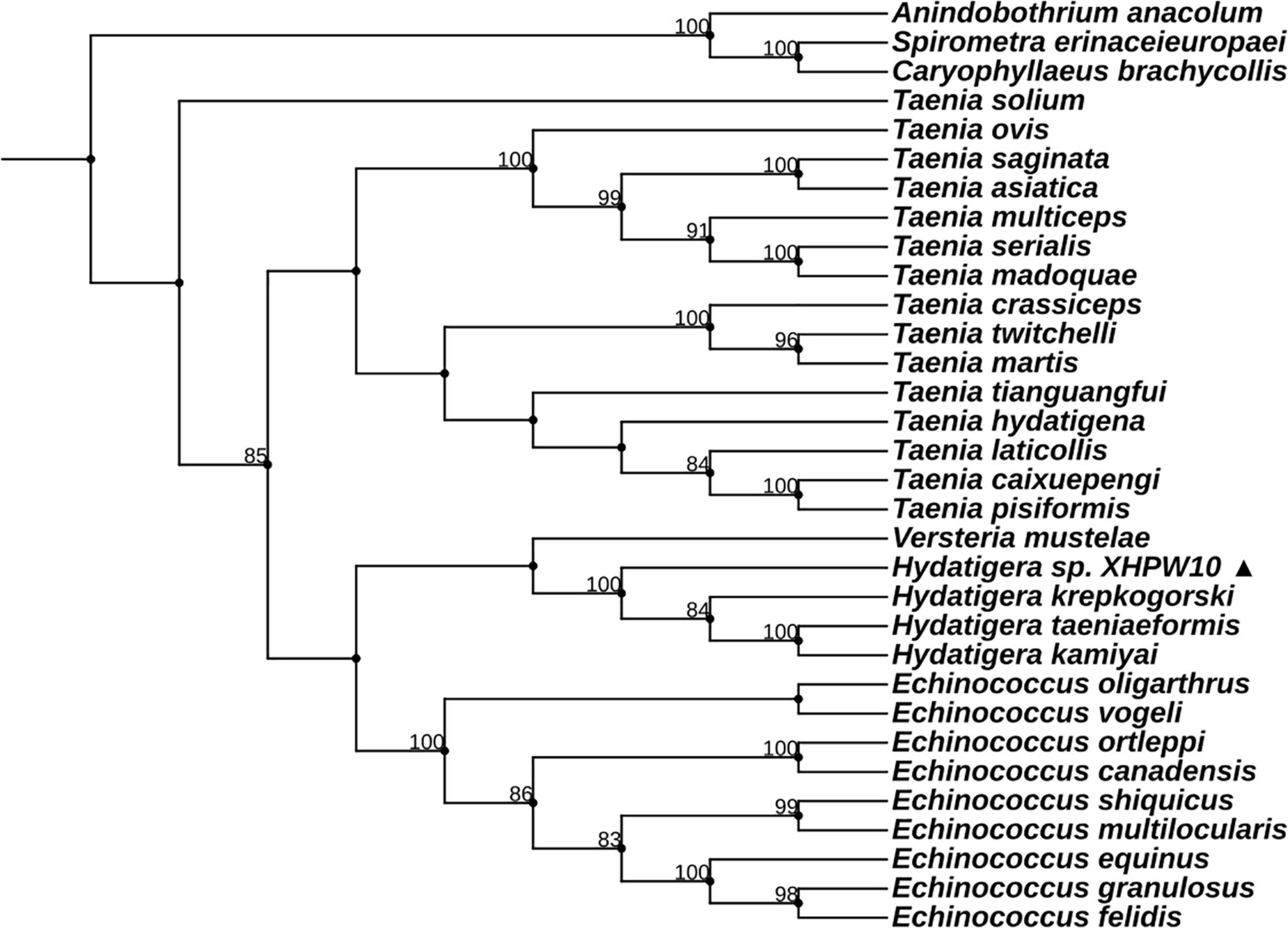
Figure 2. Maximum likelihood analysis of 18S rDNA of family Taeniidae. The ▲ after the species name represents “putative new species.” The outgroup is the same as Figure 1. Bootstrap frequency support values are stated only for nodes where > 80%.
Phylogenetic Analyses and Sequence Identity
As ingroups for phylogenetic reconstruction, we combined mt genes and 18S or 28S rDNA sequenced in this study with those of other classified cyclophyllideans available in GenBank. Three species (Caryophyllaeus brachycollis, Anindobothrium anacolum, Spirometra erinaceieuropaei) of Eucestoda belonging to different orders were chosen as outgroups.
The 28S rDNA fragments of classified cyclophyllideans in NCBI Taxonomy Database are available for most families in GenBank except Taeniidae, while that of the 18S rDNA were only available for Taeniidae, so here the evolutionary trees of 28S and 18S rDNA fragments were constructed separately. Based on the results of the 28S rDNA phylogeny, a simplified 28S evolutionary tree was reconstructed without affecting the topological structure of the tree by removing the species in the same genus and clade. To complement and validate the inferred evolutionary relationship between Cyclophyllidea species, except the species of family Taeniidae, the phylogenies with cox1 and nad1 fragments were reconstructed.
In summary, 5 datasets containing 320 species in 10 families were assembled for phylogenetic analyses (see Supplementary Tables 4–8 for GenBank accession nos. and species, genera and families used in each dataset). Since the limited availability of data on different genes of the same species, we performed separate phylogenetic reconstruction for each dataset to cover more species. Datasets were aligned using MAFFT v7.487 with auto option (Katoh and Standley, 2013). The alignments were trimmed by using Trimal v1.2 under the automated1 option (Capella-Gutiérrez et al., 2009) to preserve the same sequence regions and exclude the ambiguously aligned sites. All phylogenetic trees were constructed with maximum likelihood (ML) inference using IQ-TREE v2.1.4 (Nguyen et al., 2015) with ultrafast bootstrap 1,000 replicates in Ubuntu 20.04.2 LTS operating system, where the best-fit models were automatically selected by ModelFinder (Supplementary Table 9; Kalyaanamoorthy et al., 2017) and the best number of threads were also selected under AUTO option (using the command line: iqtree -s alignment_file -m -MFP -bb 1000 -nt AUTO). All other parameters were set to their default values. The ML trees were then visualized on the IToL web server (Letunic and Bork, 2016).
The related species of the putatively unknown species were identified based on phylogenetic reconstruction, and the percentage identity of sequences in the 5 datasets between the putatively unknown species and their related species was calculated by BLAST (Supplementary Table 10).
Analysis of Divergence Times
Species divergence times were calculated using BEAST v2.6.2 (Bouckaert et al., 2014) based on two datasets without shared species: all available mt PCGs dataset of 54 species (Supplementary Table 11) and cox1 + nad1 fragments dataset of other classified 54 species (Supplementary Table 12). These two datasets were aligned and trimmed with MAFFT v7.487 and Trimal v1.2 as described above, and were partitioned according to different genes. Model selection of partitions was identified by Partition Finder v2.1.1 (Lanfear et al., 2012) with the set of “linked” branch lengths, “beast” models, “aicc” model selection, and “greedy” search. Partition schemes and substitution models can be found in Supplementary Table 9. The Strict Clock model was chosen to ignore the rate differences between the branches in the mode and the gamma category count was set to 4. Other settings, such as substitution rate and shape, in the site model were evaluated in the analysis. The Calibrated Yule model (Heled and Drummond, 2015) was used as the tree prior, as it is a simple model of speciation that is generally appropriate when considering sequences from different species. Time calibration was calibrated with the divergence date between T. saginata and T. asiatica (∼1.14 Mya) and divergence date between Schistosoma japonicum and S. mansoni (∼56.10 Mya), which agreed with reported fossil evidence from shark coprolites and previously estimated dates based on mt genes (Wang et al., 2016). Posterior probability estimates were sampled every 1,000 iterations over a total of 10,000,000 iterations per MCMC run. Other options were run on their default values. Tracer (v1.7.1) was used to summarize posterior probabilities. Trees were annotated via TreeAnnotator (v2.1.2) using a maximum clade credibility tree and median heights settings with 10% burn-in. The number of divergence nodes in every 2 Mya was summarized based on the evolutionary divergence time trees of two datasets, respectively.
Results
Species Identification and Phylogenetic Relationships
The hosts of parasites were confirmed by BLAST searches (Supplementary Table 1; see Supplementary Table 3 for GenBank accession nos. of host cytb gene). Two of the parasite species were cysticerci and adult worms, each retrieved from the abdominal cavity and intestinal tract of two vole hosts (Neodon leucurus) collected from the same pasture location near the Shigatse City of Tibet Autonomous Region (Supplementary Table 1). Phylogenetic analyses shown in Supplementary Figure 1 and Figures 1A–C confirmed that these two species likely belong to the genera Mesocestoides and Paranoplocephala, and were labeled as Mesocestoides sp. RKZ08 and Paranoplocephala sp. RKZ13, respectively. The cysticerci collected from the liver of plateau pika (Ochotona curzoniae) from Xietongmen county of Tibet and zokors (Eospalax fontanierii) from Xiahe county of Gansu province were confirmed to be the same species by alignment of cox1 and 18S rDNA segments, which was in the monophyletic group of the genus Hydatigera in the phylogenetic trees (Figure 2), and was marked as Hydatigera sp. XHPW10. The final species was an adult tapeworm collected from the intestine of plateau pikas from Shiqu county, Sichuan province, and occurred within the genus Mosgovoyia (Supplementary Figure 1 and Figures 1A–C), and was thus named Mosgovoyia sp. SQ20. These four species were identified as newly sequenced and “putative new species” as their cox1 and nad1 sequences having less than 95% identity with available related taxa (Supplementary Table 10; Xiao et al., 2005). The degree of differentiation of their mt genes was higher than that of the nuclear genes 18S and 28S rDNA (Supplementary Table 10), reflecting their differentiation likely results from the so-called deep mitochondrial divergence (DMD, Zhang et al., 2019). In addition, three known cysticerci of Taeniidae, T. caixuepengi, T. crassiceps, and Versteria mustelae, were also identified in the present study (see Supplementary Table 13 for their cox1 fragments).
General Features of the Mitochondrial Genome of “Putative New Species”
The complete mt genomes of the four “putative new species” were 13,361 bp (GenBank ID: MW808979), 13,730 bp (GenBank ID: MW808980), 14,148 bp (GenBank ID: MW808981), and 13,776 bp (GenBank ID: MW808982) in length. Each of them contains two rRNA genes (rrnS and rrnL) and 12 protein-encoding genes (atp6, cytb, nad4L, cox1-3, and nad1-6), which are arrayed in the typical order of mt genomes of cestodes. They each contain the 22 typical tRNA genes set of cestodes, and share a common set of anticodons. The order of tRNA genes is roughly the same, except between nad6 and nad5 genes. Species Paranoplocephala sp. RKZ13 had a repeat sequence of trnL and trnR in the highly variable region between nad6 and nad5, suggesting that it had an insertion sequence in this region that made its mt genome longer than the other three species (Figure 3 and Table 1).
Figure 3. The diagram of complete mitochondrial genome of Mesocestoides sp. RKZ08 (A), Paranoplocephala sp. RKZ13 (B), Hydatigera sp. XHPW10 (C), and Mosgovoyia sp. SQ20 (D). The protein-encoding genes are depicted in plum, the tRNAs are depicted in green, the rRNAs are depicted in light green and the non-coding mitochondrial regions (NCRs including LNR and SNR) are depicted in gray. The inferred gene boundaries of them are shown in Table 1.
Flatworms use a unique set of mt code for protein translation (Nakao et al., 2000; Telford et al., 2000). In addition to ATG, GTG was also used as an initiation codon in a small fraction of coding genes in their mt genomes. For the termination codon, all species used only TAA and TAG; TGA was not identified as a termination codon (Table 1).
Divergence Times Analysis
The divergence time based on the two datasets used for time calibration is consistent with previous genome-based analysis results (Figure 4; Wang et al., 2016). Three of the “putative new species,” Mesocestoides sp. RKZ08, Mosgovoyia sp. SQ20, and Hydatigera sp. XHPW10, might have originated from a similar phase in the late Miocene, while Paranoplocephala sp. RKZ13 diverged during the Pleistocene (Figure 4). By summarizing the number of divergent time nodes in the divergence time trees over time, it was found that the trees of mt PCGs and cox1 + nad1 produced similar differentiation rate trends: there was a slight acceleration of the evolutionary rate in the period 14–24 Mya, and a marked acceleration during the period 4–10 Mya (Figure 5). However, compared with cox1 + nad1, the differentiation rate of mt PCGs was faster in the 14–24 Mya but slower in 4–10 Mya, which may have been caused by the species bias used in both datasets.
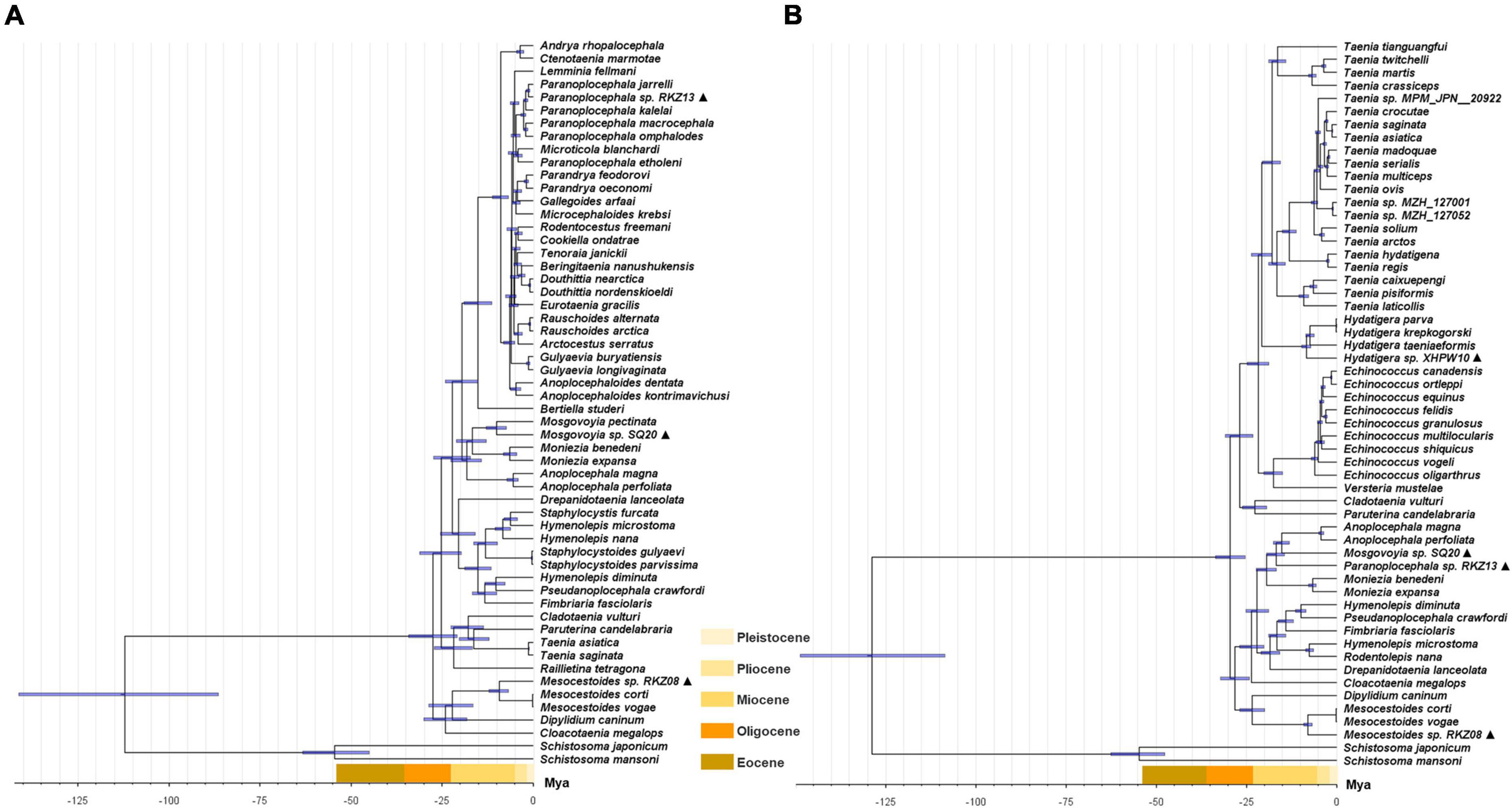
Figure 4. Divergence time construction of concatenated cox1 + nad1 gene (A) and 12 mt PCGs (B) of Cyclophyllidea species. The ▲ after the species name represents “putative new species”. The blue bar represents interval of 95% highest probability density. The time scale bars in different colors shows the extent of the Eocene, Oligocene, Miocene, Pliocene, and Pleistocene period.
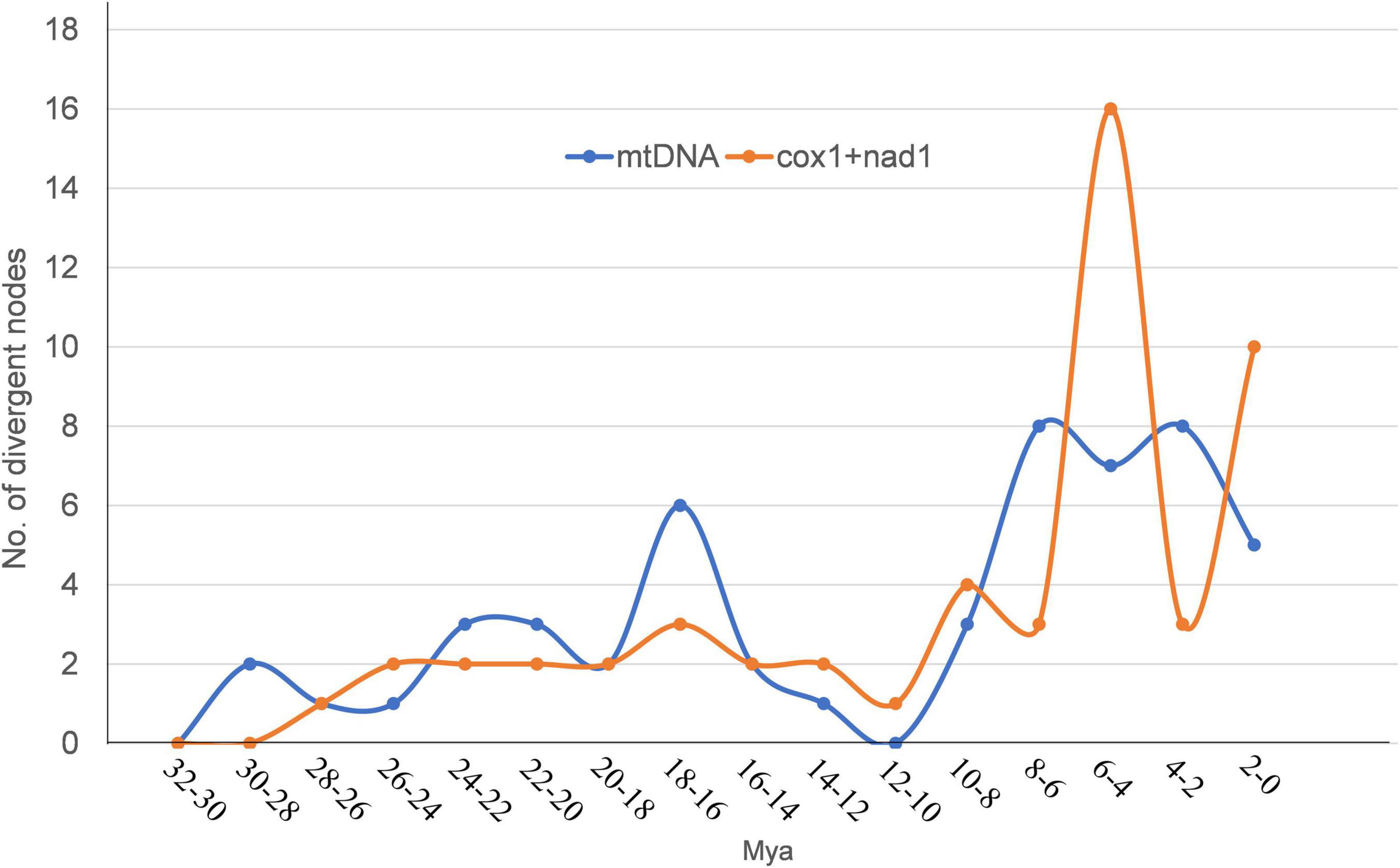
Figure 5. The number of divergent nodes in every 2 Mya over time based on the divergence time trees of Figure 4. The orange curve represents the change of divergence nodes number based on Figure 4A. The blue curve represents the change of divergence nodes number based on Figure 4B.
Discussion
Given the rich diversity and the large rodent population in western China and the few published reports of tapeworms in rodents, except for Taeniidae (Xiao et al., 2005; Zhao et al., 2014; Wang et al., 2018; Zhang et al., 2018; Wu et al., 2021), the present knowledge of tapeworm biodiversity in rodents in western China suggests far greater biodiversity yet to be uncovered.
Here, the mt genes and 18S or 28S rDNA fragments of four (two larvae and two adults) unidentified Cyclophyllidea species differed from their related species (Supplementary Table 10). However, due to the specimen distortion and insufficient specimen encountered in this study, it is not clear whether they have been described morphologically. All four species showed apparent discordance percentage identity with the related species in their mtDNA and 18S or 28S rDNA, which is the common DMD pattern across the animal kingdom, and demonstrated in Echinococcus granulosus sensu stricto (Kinkar et al., 2017), possibly due to the parthenogenetic inheritance of mitochondria, gene flow and recombination in the nuclear genome (Toews and Brelsford, 2012).
The larvae Hydatigera sp. XHPW10 was found to live in the livers of both zokors from northeast of QTP and plateau pika from southwest of QTP (Supplementary Table 1). This adds to the recently described cysticerci species in plateau pika from Qinghai province (Wu et al., 2021). Here the T. caixuepengi larva was also found to be parasitic in plateau pikas from Xietongmen, Saga and Sa’gya county of Tibet and Qilian county of Qinghai (Supplementary Table 1), and so far, not found in other sympatric rodent species. Due to the absence of comparative studies, it is currently not clear if this species has preference only for plateau pikas as its intermediate host. The larvae of T. crassiceps and V. mustelae were, respectively, found in Jeminay and Xinyuan county of Xinjiang autonomous region (Supplementary Table 1), which were also distributed on the northeast QTP (Li et al., 2013; Zhao et al., 2014). Wide geographic distributions of identical species suggest that their endemic geographic range should be far beyond the available survey data. Except for Mesocestoides sp. RKZ08 identified in this study, the Me. litteratus is another species of the genus Mesocestoides reported in Qinghai and Heilongjiang provinces of China (Wang et al., 2006; Li et al., 2013). However, there is no previous record of Paranoplocephala spp. and Mosgovoyia spp. available in China except for Paranoplocephala sp. RKZ13 and Mosgovoyia sp. SQ20 found in this study.
Complete mt genomes of the four “putative new species” were sequenced and annotated, and the sequences were clearly different from all available mt genomes sequences; however, they were similar in length, gene order and composition as other cyclophyllideans with respect to rRNA, tRNA, and protein-encoding genes (Le et al., 2000; Jeon et al., 2005, 2007; Wu et al., 2021). Different arrangement of genes occurred in tRNA genes between nad5 and nad6 genes, but was consistent with their respective most relative species, matching the expectation that the arrangement of mt genes partly determines the genetic relationship of parasites (Gazi et al., 2016). Moreover, repeat copies of tRNA gene between the nad5 and nad6 genes in the mt genome of E. granulosus sequenced by third-generation sequencing have been reported (Kinkar et al., 2019). We also observed a repeat sequence of trnL and trnR genes between nad5 and nad6 genes of the Paranoplocephala sp. RKZ13 mt genome (Figure 3 and Table 1), which indicates that there may be hidden tRNA gene repeats in the mt genome of tapeworms that are hard to identify due to errors in PCR amplification and Sanger sequencing, techniques which often fail to recover repeat regions (Kinkar et al., 2019).
Genetic drift and adaptive differentiation between allopatric populations is responsible for most speciation amongst plants and animals (Turelli et al., 2001). For parasites, however, host association is a key driver in their evolution. Host switching among sympatric populations may lead to ecological isolation, so sympatric speciation of parasites is common (de Meeûs et al., 1998; Paul, 2002; Huyse et al., 2005; Wu et al., 2021). These two models of evolution are not in conflict: the adaptation of parasite to specific host is like the adaptation of animal to specific living environment; so, the evolution of the host, especially its immune system, may be viewed as equivalent to the change of the living environment for the parasite. The environmental and climatic changes caused by the uplift of the QTP are a major driving force for the evolution of associated biotas (Favre et al., 2015). Paleobotanical data suggest that the southeastern margin of the QTP was dominated by a warm and humid subtropical or tropical climate during the Miocene due to the influence of South Asian and East Asian monsoons (Sun and Wang, 2005; Jacques et al., 2011). Since the mid-Miocene, the significant rise of the Himalayas and the Tianshan Mountains, together with worldwide cooling, incurred dramatic changes in air circulation, leading to gradual aridification of the QTP and Central Asia (Miao et al., 2012; Favre et al., 2015). Finally, in the Late Miocene and Early Pliocene, QTP uplift resulted in the accumulation of global ice and the eventual disappearance of the Tethys Sea, which also contributed to the drying of Central Asia (Lu and Guo, 2013).
These timelines of climatic and environmental changes caused by the uplift of the QTP are highly consistent with the timelines of the differentiation rate of Cyclophyllidea species analyzed in this study (Figures 4, 5) and that of plateau pika analyzed in Wang et al. (2020). We speculated in this study that the differentiation of cyclophyllideans may have been driven by host evolution caused by the uplift of the QTP. During the tropical period of QTP, the optimum living environment created the biological diversity (Cai et al., 2020; Päckert et al., 2020), and the species of Cyclophyllidea gradually differentiated. With the rapid uplift of the QTP, the environment changed into a dry and cold climate (Lu and Guo, 2013), and species differentiation of Cyclophyllidea was accelerated by the rapid adaptive evolution of their hosts and geographical isolation caused by the radiation of hosts to the Palaearctic (Favre et al., 2015; Xing and Ree, 2017; Päckert et al., 2020). Finally, in the last 2 million years, Cyclophyllidea differentiation demonstrated an accelerated diversification based on cox1 + nad1 divergence tree (Figure 5), which may be related to the evolution and broad distribution of mammals in Eurasia and the associated population expansion and migration of hominids from Africa to Asia (Dennell, 2004; Rohland et al., 2005; Brugal and Croitor, 2007; Turner et al., 2008; Klein, 2009; Terefe et al., 2014; Wang et al., 2016). The Host-parasite Database of Natural History Museum (HPDNHM) found that most tapeworm were prevalent mainly in the Palaearctic,5 which is also consistent with the viewpoint that the order Cyclophyllidea originated from the QTP. The Nearctic is another major endemic area where Cyclophyllidea species may have spread over land Bridges across the Bering Strait. However, some species parasitizing birds and other economic and companion animals tend to show a global epidemic, which may be due to the long-distance migration of birds and the spread of human trade and activities.
Phylogenetic reconstruction reveals that some classifications of Cyclophyllidea species may need to be redefined. Closely related species of tapeworm parasites often have similar host specificities and life history (Scholz et al., 2021), a pattern common in the HPDNHM and in our evolutionary analyses, and may provide a basis for revising the classification of Cyclophyllidea species; for example, the family Hymenolepididae and Anoplocephalidae can be divided into multiple families (Figures 1A–C), and Thysanotaenia congolensis should be reclassified into family Davaineidae (Figures 1A,C). However, considering that there is still a large number of undiscovered species, which may provide better support for classification, the current classification status is likely to remain for some time.
Conclusion
In conclusion, this study expands the biodiversity of Cyclophyllidea in rodents in QTP and its surrounding mountain systems, and suggests an “out of QTP” hypothesis for the Cyclophyllidea, wherein species differentiation was driven by the uplift of the QTP. Although beyond the scope of this study to consider the evolutionary relationships and history of the whole cyclophyllideans, the species analyzed represent 10 of all 16 families, making this the most extensive study of the evolution of Cyclophyllidea order to date. Verifying the taxonomic revision and the “out of QTP” hypothesis requires more sampling and investigation, including data on a wider geographic and host range, and molecular studies uncovering patterns of host-parasite co-evolution.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MZ476188–MZ476193; https://www.ncbi. nlm.nih.gov/genbank/, MW808979–MW808982; https://www. ncbi.nlm.nih.gov/genbank/, OM140661–OM140665.
Ethics Statement
The animal study was reviewed and approved by the Animal Ethics Procedures and Guidelines of the People’s Republic of China. Animal Ethics Committee of Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences (No. LVRIAEC2012-007).
Author Contributions
Y-DW, LL, H-BY, and W-ZJ conceived and designed the experiments. Y-DW, G-DD, L-SZ, A-MG, Y-TW, and YF conducted the sample collection. Y-DW, G-DD, X-WN, and NS performed the experiments. Y-DW and G-DD performed the data analyses. Y-DW prepared the figures and wrote the manuscript. DTJL, JO, W-HL, N-ZZ, and B-QF provided very constructive suggestions for revisions. All authors read and approved the final manuscript.
Funding
This work was funded by the National Key Research and Development Plan (2021YFE0191600), Cultivation of Achievements (SKLVEB2020CGPY01), and Open Project (SKLVEB2020KFKT008) of State Key Laboratory of Veterinary Etiological Biology, and the Applied Basic Research of Qinghai Province (2021-ZJ-724).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.747484/full#supplementary-material
Supplementary Figure 1 | Maximum likelihood analysis based on the 28S rDNA fragments of classified cyclophyllideans in NCBI Taxonomy Database. The ▲ after the species name represents “putative new species”. The outgroup is the same as Figure 1. Bootstrap frequency support values are stated only for nodes where > 80%.
Footnotes
- ^ https://www.ncbi.nlm.nih.gov/taxonomy
- ^ https://blast.ncbi.nlm.nih.gov/Blast.cgi
- ^ https://chlorobox.mpimp-golm.mpg.de/geseq.html
- ^ http://130.235.46.10/ARWEN/
- ^ https://www.nhm.ac.uk/research-curation/scientific-resources/taxonomy-systematics/host-parasites/database/index.jsp
References
Bouckaert, R., Heled, J., Kuhnert, D., Vaughan, T., Wu, C. H., Xie, D., et al. (2014). BEAST 2: a software platform for Bayesia evolutionary analysis. PLoS Comput. Biol. 10:e1003537. doi: 10.1371/journal.pcbi.1003537
Bowles, J., Blair, D., and McManus, D. P. (1992). Genetic variants within the genus Echinococcus identified by mitochondrial DNA sequencing. Mol. Biochem. Parasitol. 54, 165–173. doi: 10.1016/0166-6851(92)90109-w
Brugal, J. P., and Croitor, R. (2007). Evolution, ecology and biochronology of herbivore associations in Europe during the last 3 million years. Quaternaire 18, 129–152.
Cai, T., Shao, S., Kennedy, J. D., Alström, P., Moyle, R. G., Qu, Y., et al. (2020). The role of evolutionary time, diversification rates and dispersal in determining the global diversity of a large radiation of passerine birds. J. Biogeogr. 47, 1612–1625.
Caira, J. N., and Jensen, K. (eds) (2017). Planetary Biodiversity Inventory (2008–2017): Tapeworms from Vertebrate Bowels of the Earth. USA: University of Kansas. 463.
Capella-Gutiérrez, S., Silla-Martínez, J. M., and Gabaldón, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Carlson, C. J., Dallas, T. A., Alexander, L. W., Phelan, A. L., and Phillips, A. J. (2020). What would it take to describe the global diversity of parasites? Proc. Biol. Sci. 287:20201841. doi: 10.1098/rspb.2020.1841
de Meeûs, T., Michalakis, Y., and Renaud, F. (1998). Santa Rosalia revisited: or why are there so many kinds of parasites in the garden of earthly delights’? Parasitol. Today 14, 10–13. doi: 10.1016/s0169-4758(97)01163-0
Dennell, R. W. (2004). Hominid dispersals and Asian biogeography during lower and early middle Pleistocene, c. 2.0–0.5 Mya. Asian Perspect. 43, 205–226.
Dobson, A., Lafferty, K. D., Kuris, A. M., Hechinger, R. F., and Jetz, W. (2008). Colloquium paper: homage to Linnaeus: how many parasites? How many hosts? Proc. Natl. Acad. Sci. U. S. A. 105, 11482–11489. doi: 10.1073/pnas.0803232105
Ebert, D., and Fields, P. D. (2020). Host-parasite co-evolution and its genomic signature. Nat. Rev. Genet. 21, 754–768. doi: 10.1038/s41576-020-0269-1
Fan, Z., Liu, S., Liu, Y., Zhang, X., and Yue, B. (2011). How Quaternary geologic and climatic events in the southeastern margin of the Tibetan Plateau influence the genetic structure of small mammals: inferences from phylogeography of two rodents, Neodon irene and Apodemus latronum. Genetica 139, 339–351. doi: 10.1007/s10709-011-9553-5
Favre, A., Päckert, M., Pauls, S. U., Jähnig, S. C., Uhl, D., Michalak, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan Plateau for the evolution of Tibetan biotas. Biol. Rev. Camb. Philos. Soc. 90, 236–253. doi: 10.1111/brv.12107
Gazi, M., Kim, J., García-Varela, M., Park, C., Littlewood, D. T. J., and Park, J. K. (2016). Mitogenomic phylogeny of Acanthocephala reveals novel Class relationships. Zoologica Scripta 45, 437–454.
Heled, J., and Drummond, A. J. (2015). Calibrated birth-death phylogenetic time-tree priors for Bayesian inference. Syst. Biol. 64, 369–383. doi: 10.1093/sysbio/syu089
Hoorn, C., Perrigo, A., and Antonelli, A. (eds) (2018). Mountains, Climate, and Biodiversity (1st ed.). Hoboken: Wiley.
Huyse, T., Poulin, R., and Théron, A. (2005). Speciation in parasites: a population genetics approach. Trends. Parasitol. 21, 469–475. doi: 10.1016/j.pt.2005.08.009
Jacques, F. M. B., Guo, S. X., Su, T., Xing, Y. W., Huang, Y. J., Liu, Y. S., et al. (2011). Quantitative reconstruction of the Late Miocene monsoon climates of southwest China: a case study of the Lincang flora from Yunnan Province. Palaeogeogr. Palaeoclimatol. Palaeoecol. 304, 318–327. doi: 10.1016/j.palaeo.2010.04.014
Jeon, H. K., Kim, K. H., and Eom, K. S. (2007). Complete sequence of the mitochondrial genome of Taenia saginata: comparison with T. solium and T. asiatica. Parasitol. Int. 56, 243–246. doi: 10.1016/j.parint.2007.04.001
Jeon, H. K., Lee, K. H., Kim, K. H., Hwang, U. W., and Eom, K. S. (2005). Complete sequence and structure of the mitochondrial genome of the human tapeworm, Taenia asiatica (Platyhelminthes; Cestoda). Parasitology 130, 717–726. doi: 10.1017/s0031182004007164
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kinkar, L., Korhonen, P. K., Cai, H., Gauci, C. G., Lightowlers, M. W., Saarma, U., et al. (2019). Long-read sequencing reveals a 4.4 kb tandem repeat region in the mitogenome of Echinococcus granulosus (sensu stricto) genotype G1. Parasit. Vectors 12:238. doi: 10.1186/s13071-019-3492-x
Kinkar, L., Laurimäe, T., Sharbatkhori, M., Mirhendi, H., Kia, E. B., Ponce-Gordo, F., et al. (2017). New mitogenome and nuclear evidence on the phylogeny and taxonomy of the highly zoonotic tapeworm Echinococcus granulosus sensu stricto. Infect. Genet. Evol. 52, 52–58. doi: 10.1016/j.meegid.2017.04.023
Klein, R. G. (2009). Darwin and the recent African origin of modern humans. Proc Natl. Acad. Sci. U. S. A. 106, 16007–16009. doi: 10.1073/pnas.0908719106
Kuchta, R., Řehulková, E., Francová, K., Scholz, T., Morand, S., and Šimková, A. (2020). Diversity of monogeneans and tapeworms in cypriniform fishes across two continents. Int. J. Parasitol. 50, 771–786. doi: 10.1016/j.ijpara.2020.06.005
Lanfear, R., Calcott, B., Ho, S. Y., and Guindon, S. (2012). PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol. Biol. Evol. 29, 1695–1701. doi: 10.1093/molbev/mss020
Larsen, B. B., Miller, E. C., Rhodes, M. K., and Wiens, J. J. (2017). Inordinate fondness multiplied and redistributed: the number of species on earth and the new pie of life. Q. Rev. Biol. 92, 229–265. doi: 10.1086/693564
Laslett, D., and Canbäck, B. (2008). ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics 24, 172–175. doi: 10.1093/bioinformatics/btm573
Le, T. H., Blair, D., Agatsuma, T., Humair, P. F., Campbell, N. J., Iwagami, M., et al. (2000). Phylogenies inferred from mitochondrial gene orders-a cautionary tale from the parasitic flatworms. Mol. Biol. Evol. 17, 1123–1125. doi: 10.1093/oxfordjournals.molbev.a026393
Lee, S. U., Chun, H. C., and Huh, S. (2007). Molecular phylogeny of parasitic Platyhelminthes based on sequences of partial 28S rDNA D1 and mitochondrial cytochrome c oxidase subunit I. Korean J. Parasitol. 45, 181–189. doi: 10.3347/kjp.2007.45.3.181
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Li, W., Guo, Z., Duo, H., Fu, Y., Peng, M., Shen, X., et al. (2013). Survey on helminths in the small intestine of wild foxes in Qinghai, China. J. Vet. Med. Sci. 75, 1329–1333. doi: 10.1292/jvms.13-0187
Littlewood, D. T., Curini-Galletti, M., and Herniou, E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Mol. Phylogenet. Evol. 16, 449–466. doi: 10.1006/mpev.2000.0802
Lu, H. Y., and Guo, Z. T. (2013). Evolution of the monsoon and dry climate in East Asia during late Cenozoic: a review. Sci. China Earth Sci. 57, 70–79. doi: 10.1007/s11430-013-4790-3
Lymbery, A. J. (1998). Combining data from morphological traits and genetic markers to determine transmission cycles in the tape worm, Echinococcus granulosus. Parasitology 117, 185–192. doi: 10.1017/s0031182098002911
Miao, Y. F., Herrmann, M., Wu, F. L., Yan, X. L., and Yang, S. L. (2012). What controlled Mid-Late Miocene long-term aridification in Central Asia? – Global cooling or Tibetan Plateau uplift: a review. Earth Sci. Rev. 112, 155–172. doi: 10.1016/j.earscirev.2012.02.003
Nakao, M., Lavikainen, A., Iwaki, T., Haukisalmi, V., Konyaev, S., Oku, Y., et al. (2013). Molecular phylogeny of the genus Taenia (Cestoda: Taeniidae): proposals for the resurrection of Hydatigera Lamarck, 1816 and the creation of a new genus Versteria. Int. J. Parasitol. 43, 427–437. doi: 10.1016/j.ijpara.2012.11.014
Nakao, M., Sako, Y., Yokoyama, N., Fukunaga, M., and Ito, A. (2000). Mitochondrial genetic code in cestodes. Mol. Biochem. Parasitol. 111, 415–424. doi: 10.1016/s0166-6851(00)00334-0
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Okamura, B., Hartigan, A., and Naldoni, J. (2018). Extensive uncharted biodiversity: the parasite dimension. Integr. Comp. Biol. 58, 1132–1145. doi: 10.1093/icb/icy039
Okulewicz, A. (2014). Helminths in migrating and wintering birds recorded in Poland. Ann. Parasitol. 60, 19–24.
Päckert, M., Favre, A., Schnitzler, J., Martens, J., Sun, Y. H., Tietze, D. T., et al. (2020). “Into and Out of” the Qinghai-Tibet Plateau and the Himalayas: centers of origin and diversification across five clades of Eurasian montane and alpine passerine birds. Ecol. Evol. 10, 9283–9300. doi: 10.1002/ece3.6615
Paul, R. (2002). Species concepts versus species criteria. Trends Parasitol. 18, 439–440. doi: 10.1016/s1471-4922(02)02319-x
Poulin, R., and Morand, S. (2004). Parasite Biodiversity. Washington: Smithsonian Institution Press.
Rahbek, C., Borregaard, M. K., Antonelli, A., Colwell, R. K., Holt, B. G., Nogues-Bravo, D., et al. (2019a). Building mountain biodiversity: geological and evolutionary processes. Science 365, 1114–1119. doi: 10.1126/science.aax0151
Rahbek, C., Borregaard, M. K., Colwell, R. K., Dalsgaard, B., Holt, B. G., Morueta-Holme, N., et al. (2019b). Humboldt’s enigma: what causes global patterns of mountain biodiversity? Science 365, 1108–1113. doi: 10.1126/science.aax0149
Rohland, N., Pollack, J. L., Nagel, D., Beauval, C., Airvaux, J., Pääbo, S., et al. (2005). The population history of extant and extinct hyenas. Mol. Biol. Evol. 22, 2435–2443. doi: 10.1093/molbev/msi244
Scholz, T., Waeschenbach, A., Oros, M., Brabec, J., and Littlewood, D.T.J. (2021). Phylogenetic reconstruction of early diverging tapeworms (Cestoda: Caryophyllidea) reveals ancient radiations in vertebrate hosts and biogeographic regions. Int. J. Parasitol. 51, 263–277. doi: 10.1016/j.ijpara.2020.09.009
Sharma, S., Lyngdoh, D., Roy, B., and Tandon, V. (2016). Molecular phylogeny of Cyclophyllidea (Cestoda: Eucestoda): an in-silico analysis based on mtCOI gene. Parasitol. Res. 115, 3329–3335. doi: 10.1007/s00436-016-5092-4
Singla, L. D., Singla, N., Parshad, V. R., Juyal, P. D., and Sood, N. K. (2008). Rodents as reservoirs of parasites in India. Integr. Zool. 3, 21–26. doi: 10.1111/j.1749-4877.2008.00071.x
Sun, X. J., and Wang, P. X. (2005). How old is the Asian monsoon system? Palaeobotanical records from China. Palaeogeogr. Palaeoclimatol. Palaeoecol. 222, 181–222. doi: 10.1016/j.palaeo.2005.03.005
Telford, M. J., Herniou, E. A., Russell, R. B., and Littlewood, D. T. (2000). Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proc. Natl. Acad. Sci. U. S. A. 97, 11359–11364. doi: 10.1073/pnas.97.21.11359
Terefe, Y., Hailemariam, Z., Menkir, S., Nakao, M., Lavikainen, A., Haukisalmi, V., et al. (2014). Phylogenetic characterisation of Taenia tapeworms in spotted hyenas and reconsideration of the “Out of Africa” hypothesis of Taenia in humans. Int. J. Parasitol. 44, 533–541. doi: 10.1016/j.ijpara.2014.03.013
Toews, D. P., and Brelsford, A. (2012). The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 21, 3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x
Turelli, M., Barton, N. H., and Coyne, J. A. (2001). Theory and speciation. Trends Ecol. Evol. 16, 330–343. doi: 10.1016/s0169-5347(01)02177-2
Turner, A., Antón, M., and Werdelin, L. (2008). Taxonomy and evolutionary patterns in the fossil Hyaenidae of Europe. Geobios 41, 677–687.
Wang, C. R., Qiu, J. H., Zhao, J. P., Xu, L. M., Yu, W. C., and Zhu, X. Q. (2006). Prevalence of helminthes in adult dogs in Heilongjiang Province, the People’s Republic of China. Parasitol. Res. 99, 627–630. doi: 10.1007/s00436-006-0219-7
Wang, S., Wang, S., Luo, Y., Xiao, L., Luo, X., Gao, S., et al. (2016). Comparative genomics reveals adaptive evolution of Asian tapeworm in switching to a new intermediate host. Nat. Commun. 7:12845. doi: 10.1038/ncomms12845
Wang, X., Liang, D., Jin, W., Tang, M., Liu, S., and Zhang, P. (2020). Out of Tibet: genomic perspectives on the evolutionary history of extant pikas. Mol. Biol. Evol. 37, 1577–1592. doi: 10.1093/molbev/msaa026
Wang, X., Liu, J., Zuo, Q., Mu, Z., Weng, X., Sun, X., et al. (2018). Echinococcus multilocularis and Echinococcus shiquicus in a small mammal community on the eastern Tibetan Plateau: host species composition, molecular prevalence, and epidemiological implications. Parasit. Vectors 11:302. doi: 10.1186/s13071-018-2873-x
Wickström, L. M., Haukisalmi, V., Varis, S., Hantula, J., and Henttonen, H. (2005). Molecular phylogeny and systematics of anoplocephaline cestodes in rodents and lagomorphs. Syst. Parasitol. 62, 83–99. doi: 10.1007/s11230-005-5488-5
Wu, Y. D., Li, L., Fan, Y. L., Ni, X. W., Ohiolei, J. A., Li, W. H., et al. (2021). Genetic evolution and implications of the mitochondrial genomes of two newly identified Taenia spp. in rodents from Qinghai-Tibet Plateau. Front. Microbiol. 12:647119. doi: 10.3389/fmicb.2021.647119
Wu, Z., Lu, L., Du, J., Yang, L., Ren, X., Liu, B., et al. (2018). Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome 6:178. doi: 10.1186/s40168-018-0554-9
Xiao, N., Qiu, J., Nakao, M., Li, T., Yang, W., Chen, X., et al. (2005). Echinococcus shiquicus n. sp., a taeniid cestode from Tibetan fox and plateau pika in China. Int. J. Parasitol. 35, 693–701. doi: 10.1016/j.ijpara.2005.01.003
Xing, Y., and Ree, R. H. (2017). Uplift-driven diversification in the Hengduan Mountains, a temperate biodiversity hotspot. Proc. Natl. Acad. Sci. U. S. A. 114, E3444–E3451. doi: 10.1073/pnas.1616063114
Yan, H., Lou, Z., Li, L., Ni, X., Guo, A., Li, H., et al. (2013). The nuclear 18S ribosomal RNA gene as a source of phylogenetic information in the genus Taenia. Parasitol. Res. 112, 1343–1347. doi: 10.1007/s00436-012-3199-9
Yudhana, A., Praja, R. N., and Supriyanto, A. (2019). The medical relevance of Spirometra tapeworm infection in Indonesian Bronzeback snakes (Dendrelaphis pictus): a neglected zoonotic disease. Vet. World 12, 844–848. doi: 10.14202/vetworld.2019.844-848
Zhang, D., Tang, L., Cheng, Y., Hao, Y., Xiong, Y., Song, G., et al. (2019). ‘Ghost introgression’ as a cause of deep mitochondrial divergence in a bird species complex. Mol. Biol. Evol. 36, 2375–2386. doi: 10.1093/molbev/msz170
Zhang, X. Y., Jian, Y. N., Ma, L. Q., Li, X. P., and Karanis, P. (2018). A case of coenurosis in a wild rabbit (Lepus sinensis) caused by Taenia serialis metacestode in Qinghai Tibetan Plateau area, China. Korean J. Parasitol. 56, 195–198. doi: 10.3347/kjp.2018.56.2.195
Keywords: Cyclophyllidea, phylogeny, species differentiation, biogeography, Qinghai-Tibet Plateau, rodents
Citation: Wu Y-D, Dai G-D, Li L, Littlewood DTJ, Ohiolei JA, Zhang L-S, Guo A-M, Wu Y-T, Ni X-W, Shumuye NA, Li W-H, Zhang N-Z, Fu B-Q, Fu Y, Yan H-B and Jia W-Z (2022) Expansion of Cyclophyllidea Biodiversity in Rodents of Qinghai-Tibet Plateau and the “Out of Qinghai-Tibet Plateau” Hypothesis of Cyclophyllideans. Front. Microbiol. 13:747484. doi: 10.3389/fmicb.2022.747484
Received: 26 July 2021; Accepted: 10 January 2022;
Published: 08 February 2022.
Edited by:
Rodrigo Pulgar Tejo, University of Chile, ChileReviewed by:
Denis Jacob Machado, University of North Carolina at Charlotte, United StatesDaxi Wang, Beijing Genomics Institute (BGI), China
Guo-Hua Liu, Hunan Agricultural University, China
Copyright © 2022 Wu, Dai, Li, Littlewood, Ohiolei, Zhang, Guo, Wu, Ni, Shumuye, Li, Zhang, Fu, Fu, Yan and Jia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan-Zhong Jia, amlhd2FuemhvbmdAY2Fhcy5jbg==; Hong-Bin Yan, eWFuaG9uZ2JpbkBjYWFzLmNu
†These authors have contributed equally to this work
 Yao-Dong Wu
Yao-Dong Wu Guo-Dong Dai1†
Guo-Dong Dai1† D. Timothy J. Littlewood
D. Timothy J. Littlewood John Asekhaen Ohiolei
John Asekhaen Ohiolei Ai-Min Guo
Ai-Min Guo Nigus Abebe Shumuye
Nigus Abebe Shumuye Nian-Zhang Zhang
Nian-Zhang Zhang Hong-Bin Yan
Hong-Bin Yan Wan-Zhong Jia
Wan-Zhong Jia