- 1Neonatal Intensive Care Unit, Department of Surgical Sciences, University of Cagliari, Monserrato, Italy
- 2Laboratory Medicine, Department of Surgical Sciences, School of Medicine, University of Cagliari, Monserrato, Italy
The integrity of the gastrointestinal tract structure and function is seriously compromised by two pathological conditions sharing, at least in part, several pathogenetic mechanisms: inflammatory bowel diseases (IBD) and coronavirus disease 2019 (COVID-19), caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. IBD and COVID-19 are marked by gut inflammation, intestinal barrier breakdown, resulting in mucosal hyperpermeability, gut bacterial overgrowth, and dysbiosis together with perturbations in microbial and human metabolic pathways originating changes in the blood and fecal metabolome. This review compared the most relevant metabolic and microbial alterations reported from the literature in patients with IBD with those in patients with COVID-19. In both diseases, gut dysbiosis is marked by the prevalence of pro-inflammatory bacterial species and the shortfall of anti-inflammatory species; most studies reported the decrease in Firmicutes, with a specific decrease in obligately anaerobic producers short-chain fatty acids (SCFAs), such as Faecalibacterium prausnitzii. In addition, Escherichia coli overgrowth has been observed in IBD and COVID-19, while Akkermansia muciniphila is depleted in IBD and overexpressed in COVID-19. In patients with COVID-19, gut dysbiosis continues after the clearance of the viral RNA from the upper respiratory tract and the resolution of clinical symptoms. Finally, we presented and discussed the impact of gut dysbiosis, inflammation, oxidative stress, and increased energy demand on metabolic pathways involving key metabolites, such as tryptophan, phenylalanine, histidine, glutamine, succinate, citrate, and lipids.
Introduction
Since the onset of the pandemic outbreak caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), it emerged that frailty, elderly, and pre-existing chronic diseases, such as chronic kidney disease, hypertension, cardiovascular disease, and diabetes, are risk factors for the development of severe and/or fatal coronavirus disease 2019 (COVID-19; Grasselli et al., 2020; Falandry et al., 2021). Theoretically, patients with immune-mediated inflammatory diseases, such as inflammatory bowel disease (IBD), might be at increased risk of developing severe COVID-19. However, current knowledge on the pathophysiology of IBD and COVID-19 points out that patients with IBD are not at increased risk or have adverse outcomes for COVID-19 (Neurath, 2020). Strong evidence supporting this conclusion emerge from clinical studies published elsewhere (Allocca et al., 2020; D'Amico et al., 2020), including the discovery that biological therapies (e.g., monoclonal antibodies) may play a protective role against the cytokine storm observed in the course of the SARS-CoV-2 infection (Allocca and Craviotto, 2021). IBD and COVID-19 may share many alterations in molecular mechanisms, microbial communities, and biochemical pathways; “omics” technologies may considerably contribute to decipher mechanisms inducing these alterations, improving patient care and outcome. Microbiome and metabolome were primarily investigated in IBD, and similar but relatively few studies were conducted in patients with COVID-19; in this review, we examined analogies and differences in gut microbiota and body fluids metabolome between IBD and COVID-19 with the aim to identify microbial and metabolic hallmarks linking IBD, COVID-19 and SARS-CoV-2 infection.
SARS-CoV-2 Infection in Patients With IBD
IBD is an umbrella term encompassing a group of disorders, namely, Crohn’s disease (CD), ulcerative colitis (UC), and inflammatory bowel disease, type unclassified (IBDU; Satsangi et al., 2006). IBD is marked by chronic relapsing–remitting or continuously active idiopathic inflammation and bowel injuries; both adults and children exhibit an immunological dysregulation. The etiology of IBD is multifactorial, including the contribution of genetic, environmental, host factors and their reciprocal interactions (Flynn and Eisenstein, 2019); recent data indicate a worldwide 0.3% incidence and prevalence of IBD (Ng et al., 2017). The therapeutic treatment of IBD with immunomodulators (Park et al., 2020) and biologics (Neurath, 2019) may activate a transient or persistent immunocompromised state inducing opportunistic infections, especially when multiple drugs are prescribed simultaneously (Bonovas et al., 2016; Shah et al., 2017; Irving et al., 2021). Several research groups investigated whether patients with IBD may be or not more susceptible to developing SARS-CoV-2 infection (Monteleone and Ardizzone, 2020), how they should be managed in the context of the COVID-19 pandemic, and the risks and benefits of the therapeutic treatment with immunomodulators, especially in the pediatric age (Dipasquale et al., 2020; Sultan et al., 2020). An early analysis of data collected from the international registry Surveillance Epidemiology of Coronavirus Under Research Exclusion for Inflammatory Bowel Disease (SECURE-IBD) showed that among 525 pediatric and adult patients with IBD and confirmed COVID-19, 31% were hospitalized, 7% developed severe COVID-19, and 3% died (Brenner et al., 2021). Data from the registry evidenced that among patients with IBD, corticosteroids treatment may be a key risk factor for severe COVID-19 (Brenner et al., 2020), confirming results previously reported elsewhere (Mazza et al., 2020). A recent multicenter study enrolling 1816 patients with IBD treated with biologic therapy over the first 2 months of the pandemic reported an overall COVID-19 incidence of 3.9 per 1,000 patients with a 57% hospitalization rate and 29% case fatality rate (CFR; Ardizzone et al., 2021). In a cohort of 1912 patients with an IBD median duration of 17 years, the crude incidence rate of COVID-19 was at 6.2 cases per 1,000 patients, lower than that found in the general population (6.6 cases per 1,000 individuals); the mortality rate was 0.9 per 1,000 and 1 per 1,000 in patients with IBD and the general population, respectively (Taxonera et al., 2020). Although the CFR for IBD cases with COVID-19 was higher than in the general population (16.7 vs. 13.2%, respectively), the statistical difference was not significant. Finally, a meta-analysis including 9,177 patients with IBD from eight studies reported an incidence of 0.3% for COVID-19; 8.6% required admission to the intensive care unit, and the mortality rate was 6.3% (Aziz et al., 2020). In a cohort of patients with IBD, the rate of positive results for anti-SARS-CoV-2 antibodies (approximately 4.6% for IgG and IgM, and 6% for IgA) was found higher than that in healthcare professionals without inflammatory diseases (approximately 1.6% for IgG and IgM, and 1% for IgA); interestingly, no SARS-CoV-2-infected patients with IBD developed symptomatic COVID-19 (Łodyga et al., 2021). Further studies reported similar results, confirming that SARS-CoV-2 seroprevalence among individuals with IBD is closely comparable to that in subjects without IBD (Norsa et al., 2020; Bertè et al., 2021).
Gut Microbiota in IBD
Gut microbiota plays a key role in health and disease; it actively impacts multiple host systems and organs. A balanced gut microbial ecosystem with high biodiversity is associated with the beneficial effects of a myriad of symbiotic interactions between intra- and inter-microbial species, genera, families, phyla, and between microbes and host systems and organs, such as the immune system (Zheng et al., 2020), the brain, and the lung (Morais et al., 2021; Sencio et al., 2021). Conversely, perturbations in gut microbial communities, namely, dysbiosis, induce detrimental effects on these networks and are associated with diseases (Durack and Lynch, 2019). Gut dysbiosis can be defined as the loss of the overall microbial biodiversity with the imbalance between beneficial commensal and opportunistic pathogens, resulting in excessive production of pro-inflammatory mediators (Wei et al., 2021). A large body of literature investigated and evaluated extensively gut dysbiosis in individuals with IBD; the most frequently observed alterations are the overgrowth of pro-inflammatory bacterial species (e.g., Escherichia coli) associated with the shortfall of anti-inflammatory species (e.g., Faecalibacterium prausnitzii). The latter are involved in the generation of short-chain fatty acids (SCFAs), namely, butyrate, propionate, and acetate (Zuo and Ng, 2018; Khan et al., 2019; Aldars-García et al., 2021; Alshehri et al., 2021). Regrettably, data on gut microbiota composition are partially heterogeneous between studies, and results could be categorized as (a) fully concordant between studies; (b) roughly concordant with some exceptions; (c) discordant between studies. Table 1 recapitulates the most relevant data on gut dysbiosis in IBD, obtained from a great proportion of available studies from the literature (Favier et al., 1997; Seksik et al., 2003; Macfarlane et al., 2004; Martin et al., 2004; Ott et al., 2004; Gophna et al., 2006; Manichanh et al., 2006; Scanlan et al., 2006; Frank et al., 2007; Andoh et al., 2009; Kang et al., 2010; Rehman et al., 2010; Schwiertz et al., 2010; Willing et al., 2010; Joossens et al., 2011; Mondot et al., 2011; Rausch et al., 2011; Walker et al., 2011; Michail et al., 2012; Morgan et al., 2012; Nemoto et al., 2012; Vigsnæs et al., 2012; Fujimoto et al., 2013; Kabeerdoss et al., 2013, 2015; Kumari et al., 2013; Prideaux et al., 2013; Sha et al., 2013; Tong et al., 2013; Varela et al., 2013; Gevers et al., 2014; Hedin et al., 2014; Machiels et al., 2014; Walters et al., 2014; Wang et al., 2014; Hoarau et al., 2016; Jacobs et al., 2016; Mar et al., 2016; Takahashi et al., 2016; Chen et al., 2017; Halfvarson et al., 2017; Pascal et al., 2017; Santoru et al., 2017; Sokol et al., 2017; Vrakas et al., 2017; Zhang et al., 2017, 2021; de Meij et al., 2018; Laserna-Mendieta et al., 2018; Nishino et al., 2018; Franzosa et al., 2019; Heidarian et al., 2019; Lloyd-Price et al., 2019; Yilmaz et al., 2019; Alam et al., 2020; Ryan et al., 2020; Shahir et al., 2020; Clooney et al., 2021). No specific pattern of dysbiosis in patients with IBD has been definitively established; nevertheless, there is a broad agreement between studies on the imbalance of gut bacterial abundance in IBD. In particular, most studies report the depletion of Clostridium genus, C. leptum (cluster IV), C. coccoides (cluster XIVa) groups, F. prausnitzii, E. rectale, R. bromii species, Ruminococcaceae, Lachnospiraceae families, and the overgrowth of Enterococcus and Fusobacterium genera, E. coli and F. nucleatum species, Enterobacteriaceae, Veillonellaceae families. Controversial results may derive from many variables affecting gut microbiota composition, including the pre-existence of chronic diseases, the intensive therapeutic treatment in critically ill patients, especially with antibiotics to prevent secondary bacterial infections, and sudden and radical changes to eating habits (Table 2). Gut microbiome composition in IBD is strongly influenced by complex interactions between microbial communities and genetically altered host functional pathways (Huang et al., 2014; Knights et al., 2014). In CD, gut dysbiosis is more pronounced than in UC and is marked by a lower microbial diversity, a more altered microbiome composition, and a more unstable microbial community (Pascal et al., 2017). Microbial diversity and abundance significantly differ between feces and gut mucosa, as reported in early studies (Lepage et al., 2005; Gillevet et al., 2010; Morgan et al., 2012) and confirmed in more recently published papers (Lo Presti et al., 2019; Ryan et al., 2020). In IBD, inflammation alters the mucosal barrier inducing bacterial translocation; in patients with CD, bacterial translocation is revealed by the increase in several bacterial families within the submucosa compared to the corresponding superjacent mucosa at the advancing disease margin (Chiodini et al., 2016). Mucosal and fecal microbiome differences may explain, at least in part, some discrepancies between studies; for example, the increase in fecal F. prausnitzii (Table 1) corresponds to the decreased proportion of this bacterium at the mucosal surface (Walters et al., 2014). Differences in the abundance of various bacterial species and families (e.g., Lactobacilli, C. leptum group, E. coli, and F. prausnitzii) were observed between ulcerated (inflamed) and non-ulcerated (non-inflamed) mucosa (Zhang et al., 2007; Li et al., 2012) as well as between patients with high clinical activity indexes and/or sigmoidoscopy scores and patients with low clinical activity indexes and/or sigmoidoscopy scores (Fite et al., 2013). Other studies reported no difference in microbiota composition and enrichment between inflamed and non-inflamed mucosa (Kabeerdoss et al., 2015; Nishino et al., 2018). The abundance of some bacteria, such as the genus of Faecalibacterium and the family of Enterobacteriaceae, significantly differs between ileal CD and colonic CD (Dicksved et al., 2008; Naftali et al., 2016); for example, F. prausnitzii is markedly reduced in CD localized in the ileum compared with colonic localization (Willing et al., 2010). Interestingly, in current smokers with CD, the abundance of Bacteroides–Prevotella genera is higher than in non-smokers with CD (Benjamin et al., 2012a). Significant differences in several microbial taxa can be observed between young adults with IBD and IBD adults aged 60 years or older; in particular, Bifidobacterium genus decrease with age, and Bacteroides genus increase with age, probably reflecting body mass index and diet changes over time (Morgan et al., 2012). Researchers have a unanimous consensus on the E. coli overgrowth in IBD. E. coli overgrowth has been found in children with severe IBD (Schwiertz et al., 2010; Michail et al., 2012; Gevers et al., 2014; de Meij et al., 2018) and in adults with CD (Mondot et al., 2011). In patients with CD, the high prevalence of E. coli strictly adhering to the ileal mucosa has led to the identification of a new group of E. coli strains (Nadalian et al., 2021). This pathogenic group, called adherent-invasive E. coli (AIEC), has the ability to adhere and colonize enterocytes as well as to internalize into macrophages and replicate within their cytoplasm, inducing the release of tumor necrosis factor-α (TNF-α) and the cytotoxic response of Th17 and CD8+ (Lee et al., 2019). Thus, AIEC is involved in the pathogenesis of IBD, specifically CD (Palmela et al., 2018; Chervy et al., 2020), by promoting inflammatory diseases that originated from the adaptative evolution of the genome (Nash et al., 2010; Ellermann et al., 2015). The recognition of AIEC is unusual in patients with UC; rather, UC is associated with the intestinal enrichment of a heterogeneous, diarrheagenic group of E. coli strains, termed diffusely adherent E. coli (DAEC); this group was found expressed not only in children and young adults with UC but even in those with CD (Walczuk et al., 2019).

Table 1. Gut dysbiosis in patients with IBD and COVID-19 compared with healthy subjects (s, stool sample; m, mucosal biopsy; e, endoscopic lavage).
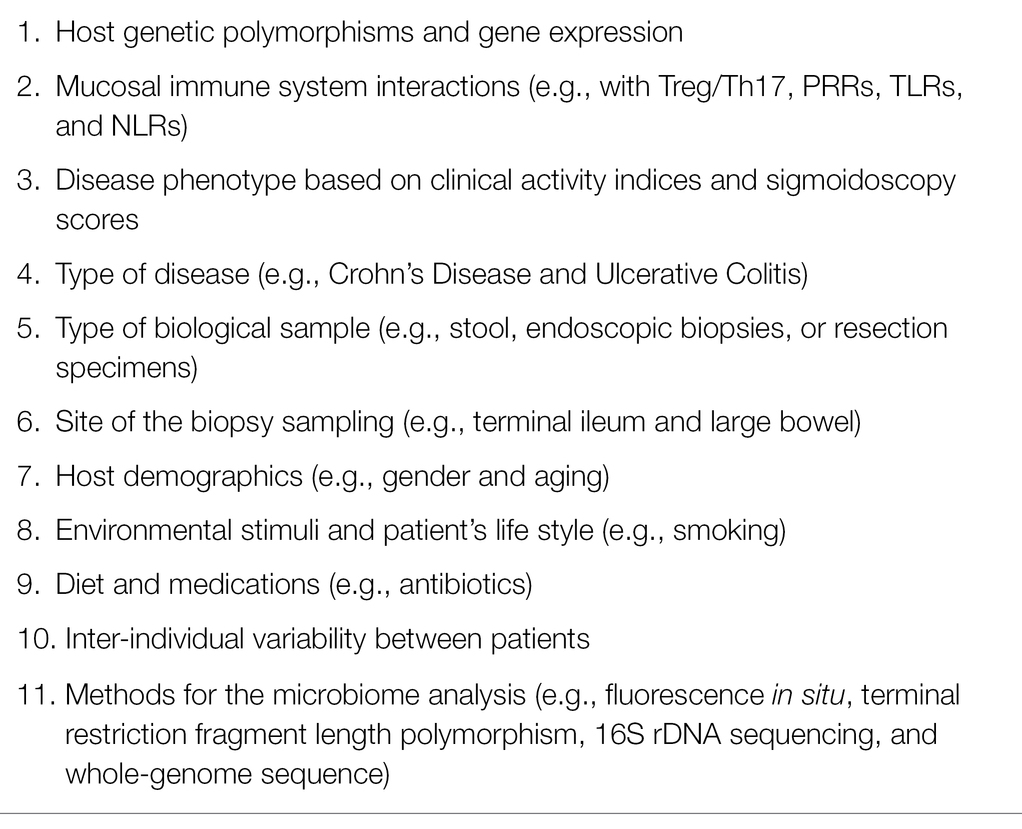
Table 2. Main factors affecting the variability of the results between published studies on gut microbiota composition in IBD.
Gut Microbiota in SARS-CoV-2 Infection
Although the lung is considered the main entry route for SARS-CoV-2, the gastrointestinal tract is equally a key target of the virus (Xiao et al., 2020; Qian et al., 2021). Indeed, the brush border of human enterocytes exhibits the highest expression of the SARS-CoV-2 receptor angiotensin-converting enzyme 2 (ACE2; Qi et al., 2020); even the transmembrane serine protease 2 (TMPRSS2), mediating the entry of SARS-CoV-2, is expressed on the luminal surface of enterocytes from ileum and colon as well as on the epithelial and gland cells of the esophagus (Knyazev et al., 2021). Thus, it is not surprising that SARS-CoV-2 infects human gut enterocytes (Lamers et al., 2020), promoting gut mucosal inflammatory infiltration with activated immune cells and cytokines (Lehmann et al., 2021). These findings support (a) the frequently observed gastrointestinal symptoms in patients with COVID-19, including severe abdominal pain, diarrhea, nausea, and vomiting (Devaux et al., 2021); (b) the persistence of viral RNA in patient’s stool even after the virus clearance from oropharyngeal swab (Morone et al., 2020); (c) the likelihood of SARS-CoV-2 transmission by the fecal–oral route (Cheung et al., 2020; Guo et al., 2021); (d) gut dysbiosis in asymptomatic infected individuals and patients with COVID-19 (Yamamoto et al., 2021). SARS-CoV-2 gut colonization significantly alters the gut microbial ecosystem, leading to dysbiosis; on the other hand, gut dysbiosis, due to aging, unhealthy lifestyle habits, and pre-existing chronic diseases (e.g., hypertension, type-2 diabetes, autoimmune diseases, and metabolic syndrome), is a key risk factor for developing COVID-19 (Magalhães et al., 2021). In hospitalized patients with COVID-19, the reduction in gut microbiota diversity, the depletion of beneficial bacterial symbionts, and the enrichment of opportunistic pathogens closely correlate with the host immune response and, in turn, with the disease severity and the clinical outcome (Villapol, 2020). The variety of the therapeutic treatment for COVID-19 may impact changes in gut microbiome composition, as highlighted in a recent review (Aktas and Aslim, 2021). Gut dysbiosis continues after the clearance of the viral RNA from the upper respiratory tract and the resolution of clinical symptoms. These associations result from complex interactions between the gut and the lung microbiota, the so-called gut–lung axis (de Oliveira et al., 2021). In particular, a balanced gut microbial ecosystem enhances the pulmonary defense against viral infections, for example, by stimulating the lung’s synthesis of type I interferons (Cyprian et al., 2021). Conversely, gut dysbiosis negatively influences the progression of the viral infection, COVID-19 development, and patient outcome (Hussain et al., 2021).
Since SCFAs inhibit the overgrowth of opportunistic pathogens, activate the adaptive immune response by enhancing antiviral immunity, and contribute to maintaining the integrity of the intestinal mucosal barrier, the depletion of SCFAs producer bacteria is closely associated with COVID-19 severity and adverse outcome; therefore, the number of SCFAs producer bacteria could predict the severity of the disease (Tang et al., 2020). Based on the observation that the Enterococcus/Enterobacteriaceae ratio is altered in approximately 74% of patients with severe/critical COVID-19, being significantly increased in non-survivors compared with survivors, it was proposed that this index may be useful to predict death in critically ill patients (Tang et al., 2020). However, a strong limitation may be the heterogeneity of this ratio. Enterococcus is a genus belonging to the Enterococcaceae family (Firmicutes phylum), while Enterobacteriaceae family belongs to Proteobacteria phylum. Despite the limited number of studies on the gut microbiome in patients with COVID-19, the pattern of gut dysbiosis associated with the disease has been partially defined (Table 1). When compared with non-infected individuals, gut dysbiosis in patients with COVID-19 is marked by the depletion of C. leptum (cluster IV) group, F. prausnitzii, and E. rectale species, Ruminococcaceae, and Lachnospiraceae families, in conjunction with the overgrowth of Enterococcus genus, Veillonellaceae, and Enterobacteriaceae families (Tang et al., 2020; Tao et al., 2020; Zuo et al., 2020, 2021; Gaibani et al., 2021; He et al., 2021; Wu et al., 2021; Yeoh et al., 2021). The abundance of Coprobacillus, C. ramosum, C. hathewayi (Zuo et al., 2020), and Enterococcus (Gaibani et al., 2021) was found positively correlated with COVID-19 severity; conversely, an inverse correlation was observed between the disease severity and the abundance of C. leptum (cluster IV) group, Lactobacillus, Bifidobacterium, C. butyricum (Tang et al., 2020), Bilophila, Citrobacter (Tao et al., 2020), Bacteroides (Gaibani et al., 2021), F. prausnitzii, E. rectale (Tang et al., 2020; Yeoh et al., 2021), B. bifidum, and B. adolescentis (Yeoh et al., 2021). In addition, the abundance of some microbial taxa, including Erysipelotrichaceae bacterium 2_2_44A (Zuo et al., 2020), P. copri, E. dolichum (Wu et al., 2021), C. aerofaciens, C. tanakaei, S. infantis (Zuo et al., 2021) positively correlates with the fecal SARS-CoV-2 load. Conversely, the abundance of B. dorei, B. thetaiotaomicron, B. massiliens, B. ovatus (Zuo et al., 2020), S. anginosus, Dialister, Alistipes, Ruminococcus, C. citroniae, Bifidobacterium, Haemophilus, H. parainfluenzae (Wu et al., 2021), P. merdae, B. stercoris, A. onderdonkii, and Lachnospiraceae bacterium 1_1_57FAA (Zuo et al., 2021) was inversely correlated with the fecal viral load. The increased gut colonization of bacteria usually resident in the oral and respiratory tracts, such as Actinomyces (Gu et al., 2020; Zuo et al., 2020; Gaibani et al., 2021; Wu et al., 2021) and Granulicatella (Wu et al., 2021), is associated with COVID-19 and its severity, confirming the active interchange between the gut, oral, and respiratory tract microbiota. The close relationship between gut microbiota and the immune-mediate response to SARS-CoV-2 infection and COVID-19 progression and outcome is supported by the correlation between the abundance of some gut microbial taxa and biomarkers of inflammation. For example, in critical patients with COVID-19, a negative correlation was observed between serum C-reactive protein (CRP) levels and the gut abundance of C. butyricum and F. prausnitzii (Tang et al., 2020). Fecal inflammatory cytokine IL-18 concentration was positively correlated with the abundance of Peptostreptococcus, Fusobacterium, and Citrobacter in patients with COVID-19 (Tao et al., 2020). Similarly, B. dorei and A. muciniphila abundance was found positively correlated with the serum level of IL-1β, IL-6, and C-X-C motif ligand 8 (CXCL8), whereas F. prausnitzii, E. rectale, and B. adolescentis were negatively correlated with serum level of TNF-α, IL-10, C-C motif ligand 2 (CCL2), and CXCL10 (Yeoh et al., 2021).
Metabolomics in IBD and COVID-19
Metabolomics is an evolving “omic” discipline allowing the identification and the quantification of endogenous and exogenous products of the cellular metabolism, namely, metabolites, within a biological system in a high-throughput manner (Liu and Locasale, 2017). The set of metabolites recognizable in a biological matrix is called metabolome or metabolic profile; it is a highly personalized readout of the current metabolism and metabolic activity that occurred in the past (Zamboni et al., 2015). Qualitative and quantitative data on metabolites reveal basic information on changes and perturbations of metabolic pathways deriving from interactions between genome, environment, microbiome, nutrients, and the intake of drugs and toxicants in health and disease states (Ashrafian et al., 2020). An updated PubMed literature search, querying the keyword metabolomics, results in approximately 26,200 studies, including in vitro experimental studies, studies on animal models, and clinical studies on patients with various diseases (Kang et al., 2021). Approximately 200 studies used the metabolomic approach in patients with IBD (Gallagher et al., 2021) and 25 in patients with COVID-19 (Mussap and Fanos, 2021). The most relevant findings are reported below.
Tryptophan Metabolism
Tryptophan (TRP) is an essential amino acid mainly derived from the diet and involved in serotonin, melatonin, and niacin biosynthesis, as detailed in Figure 1. More than 95% of TRP is metabolized along the kynurenine pathway; in the liver, the enzyme tryptophan dioxygenase (TDO) converts tryptophan into kynurenine. In the brain and the immunocompetent cells, the conversion is catalyzed by indoleamine 2,3-dioxygenase (IDO-1 and 2; Gao et al., 2018; Agus et al., 2018). Kynurenine can be converted either into neurotoxic metabolites, namely, 3-hydroxykynurenine, 3-hydroxyanthranilic acid, quinolinic acid, or neuroprotective metabolites, such as kynurenic acid, anthranilic acid, xanthurenic acid, and picolinic acid (Savitz, 2020). Depending on gut microbiota composition, approximately 4–6% of tryptophan is converted into various intermediates. For example, the prevalence of Clostridium sporogenes and Ruminococcus gnavus spp. originates tryptamine (Williams et al., 2014), whereas the prevalence of Lactobacillus, Bacteroides, and Clostridium genera originates indole derivatives (Roager and Licht, 2018). Among bacterial metabolites, indoles play a crucial role in the regulation of gastrointestinal barrier function and integrity by their binding with the pregnane X receptor (PXR), also known as steroid and xenobiotic receptor (SXR; Venkatesh et al., 2014; Oladimeji and Chen, 2018). Finally, 1–2% tryptophan is converted into serotonin (5-hydroxytryptamine), a neurotransmitter and key regulator of intestinal secretion and motility. More than 90% of serotonin is synthesized by the rate-limiting enzyme tryptophan hydroxylase (Tph/TPH) 1 within enterochromaffin cells of the gut (Stasi et al., 2019); notably, gut microbiota may considerably affect serotonergic regulation via microbiota-derived SCFA (Reigstad et al., 2015).
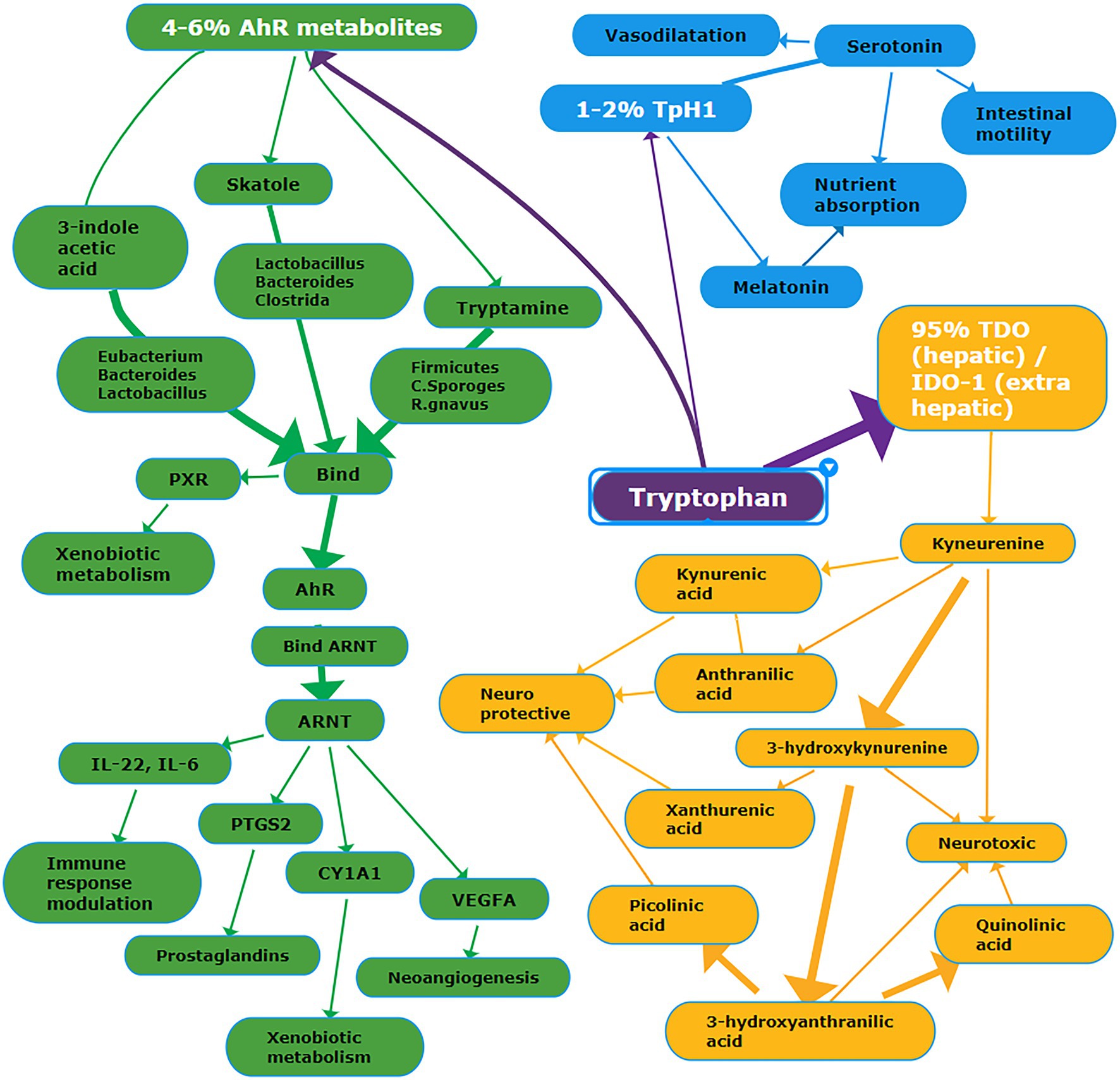
Figure 1. Schema summarizing the main metabolic pathways deriving from tryptophan. Details are reported in the text. Abbreviations: PXR, Pregnane X Receptor; AhR, Aryl Hydrocarbon Receptor; ARNT, AhR Nuclear Translocator protein; PTGS2, Prostaglandin G/H Synthase 2; CYP1A1, Cytochrome P450 1A1; VEGFA, vascular endothelial growth factor A; TpH1, tryptophan hydroxylase-1; TDO, tryptophan dioxygenase; IDO-1, indoleamine 2,3-dioxygenase-1. White font = name of the metabolic pathway; purple background = tryptophan pathway; yellow background = TDO/IDO-1 pathway; green background = AhR metabolic pathway; blue background = TpH1 pathway.
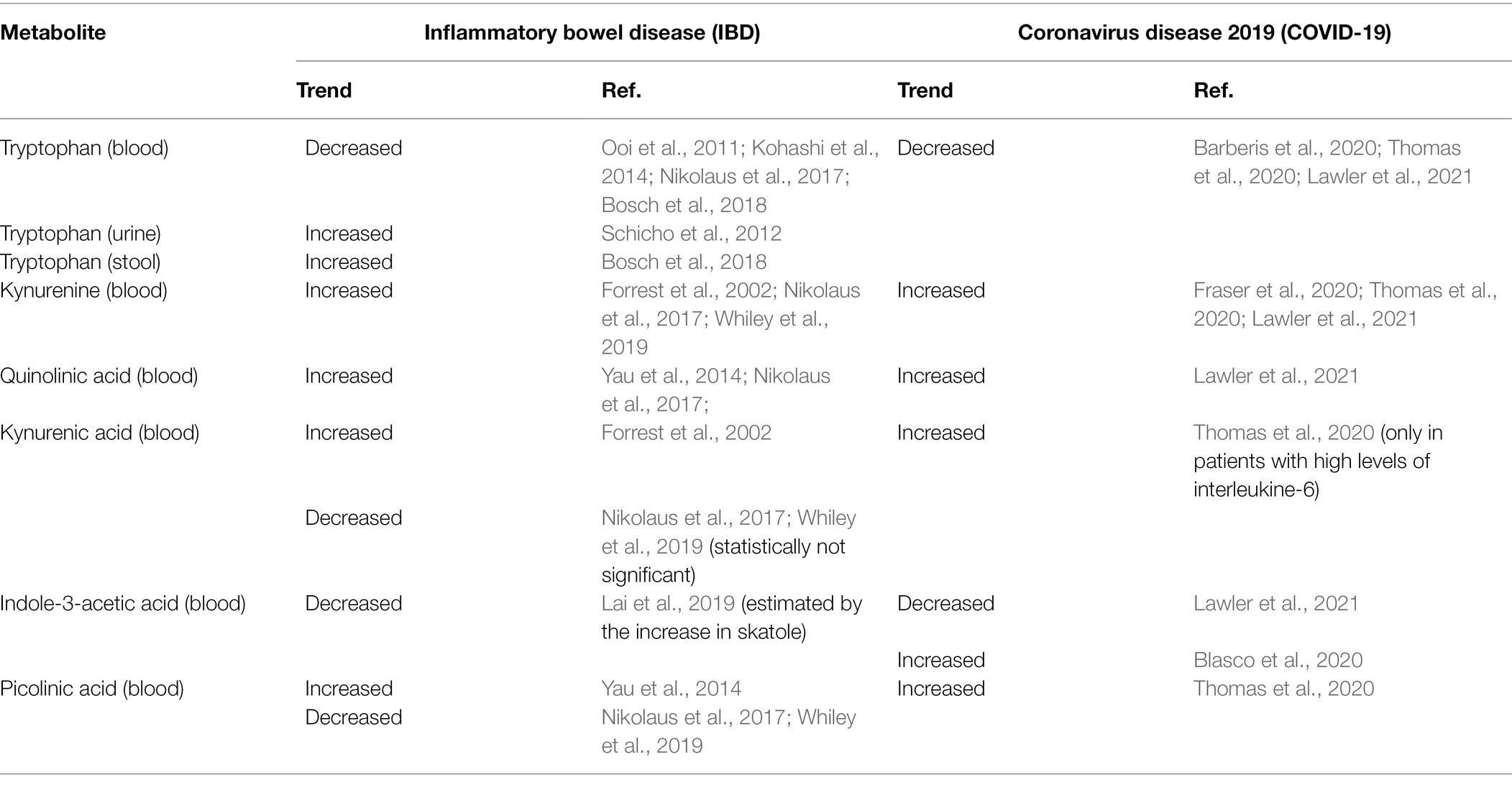
Tryptophan and indole-3-acetic are decreased in the blood of patients with IBD (Table 3; Ooi et al., 2011; Kohashi et al., 2014; Nikolaus et al., 2017; Bosch et al., 2018; Lai et al., 2019). Conversely, kynurenine and quinolinic acid are increased (Forrest et al., 2002; Yau et al., 2014; Nikolaus et al., 2017; Whiley et al., 2019). In the urine and stool of patients with IBD, tryptophan is increased (Schicho et al., 2012; Bosch et al., 2018). Kynurenic acid blood levels were increased in IBD (Forrest et al., 2002); similarly, picolinic acid was increased (Yau et al., 2014). More recently, kynurenic acid and picolinic acid were found decreased in patients with IBD (Nikolaus et al., 2017; Whiley et al., 2019). These alterations seem to be closely related to gut dysbiosis (de Meij et al., 2018), promoting the massive activation of pro-inflammatory cytokines (e.g., INF-γ and TNF-α), and the upregulation of the IDO expression (Wu et al., 2018). Gut dysbiosis significantly affects the conversion of tryptophan into indole derivatives, such as indole-3 acetic acid and indole-3-acetaldehyde. Low blood levels of indole-3-acetic acid have been associated with the overgrowth of Clostridium and Lactobacillus genera; both bacterial genera decarboxylate indole-3-acetic acid in 3-methylindole, also known as skatole. As a result, indole-3-acetic acid is metabolized with the concomitant accumulation of skatole. In patients with IBD, skatole blood levels were significantly increased (Lai et al., 2019). Most indole derivatives are ligands of the Aryl Hydrocarbon Receptor (AHR), a cytosolic ligand-dependent transcription factor widely expressed by cells of the immune system and involved in antimicrobial activity and gut immune homeostasis (Stockinger et al., 2014; Lamas et al., 2018). The indole derivatives-induced AHR activation promotes the local synthesis of the anti-inflammatory cytokine IL-22 by the innate lymphoid cells (Monteleone et al., 2011; Qiu et al., 2012; Yang et al., 2020); IL-22 protects the mucosa integrity against fungal infection by Candida albicans, commonly observed in patients with IBD (Sokol et al., 2017; Gronke et al., 2019). In healthy subjects with gut eubiosis, the activation of AHR modulates local IL-22 production; conversely, in IBD patients, the decrease of indole-3 acetic acid due to the imbalance of gut flora reduces the AHR activity and hence IL-22 synthesis, as observed in an animal model (Lamas et al., 2016). In patients with COVID-19, most metabolomics-based studies reported a significant decrease in blood tryptophan level in conjunction with the significant increase of kynurenine, kynurenic acid, and downstream metabolites of the kynurenine pathway (Barberis et al., 2020; Fraser et al., 2020; Thomas et al., 2020; Lawler et al., 2021). The decrease of tryptophan was inversely correlated with biomarkers of inflammation, such as IL-6 and CRP (Thomas et al., 2020). The increase of kynurenine is significantly associated with the increase of several cytokines, including interferon γ-induced protein 10 (IP-10), the mitogen-inducible cytokine macrophage inflammatory protein-1 β (MIP-1-β), also known as CCL4, TNF-α, interleukin-1 receptor antagonist (IL-1RA), IL-7, IL-18, and IL-8 (Lawler et al., 2021). Controversial results on indole-3-acetic acid in patients with COVID-19 have been published. On the one hand, it was found decreased (Lawler et al., 2021), and this finding remains unclear. A possible explanation may be the decrease of indole-3 acetic acid produced by the host because of the upregulation of the kynurenine pathway. This assumption is plausible, taking into account that a fraction of indole-3 acetic acid is produced by mammalian cells (Zhang et al., 2020). On the other hand, indole-3 acetic acid has been found increased (Blasco et al., 2020); this finding is coherent with gut dysbiosis marked by the prevalence of bacteria converting tryptophan into indoles.
Glutamine
Glutamine is an L-α gluconeogenic and proteogenic amino acid containing five carbons and two amino groups. L-glutamine is considered a conditionally essential amino acid; it is obtained mainly through the diet, as well as it is synthesized de novo from glutamate and ammonia in almost all the human cells by the activity of glutamine synthase (E.C.: 6.3.1.2.). In rapidly diving cells, such as enterocytes of the small intestine (Windmueller and Spaeth, 1974), lymphocytes, neutrophils, macrophages, and tumor cells, as well as under catabolic stressed conditions, including severe infections and sepsis, the endogenous synthesis does not meet the energy cell demand and the amount deriving from digested food absorbed through the small intestine becomes vitally important (Cruzat et al., 2018). Glutamine is involved in many cytoplasmatic and mitochondrial pathways, such as (a) the preservation of the reactive oxygen species (ROS) homeostasis, by contributing to the synthesis of the glutathione (Matés et al., 2002); (b) the biosynthesis of hexosamine, nucleotides, asparagine; and (c) the activation of glutaminolysis (Yoo et al., 2020). In the gut, the role of L-glutamine is crucial (Kim and Kim, 2017). Firstly, glutamine exerts an anti-inflammatory activity, preventing the expression of pro-inflammatory cytokines through the inhibition of both nuclear factor k light chain-enhancer of activated B cells (NF-kB) and signal transducer and activator of transcription (STAT) proteins (Kretzmann et al., 2008). Secondly, L-glutamine enhances tight junction integrity, as demonstrated in animal models (Wang et al., 2016), triggering the mitogen-activated protein kinase (MAPK) function (Basuroy et al., 2006; Perna et al., 2019); in addition, L-glutamine is pivotal for gut cells proliferation (Rhoads et al., 1997). Finally, L-glutamine modulates NO synthetase expression (Hecker et al., 1990; Swierkosz et al., 1990). Thus, it is not surprising that L-glutamine is the most abundant free amino acid in humans. L-glutamine provides energy as a substitute fuel to the tricarboxylic acid (TCA) cycle to produce adenosine triphosphate (ATP; Curi et al., 2005); recently, it was postulated that L-glutamine is the fuel for the immune system, generating the concept of immunometabolism (Wang et al., 2019).
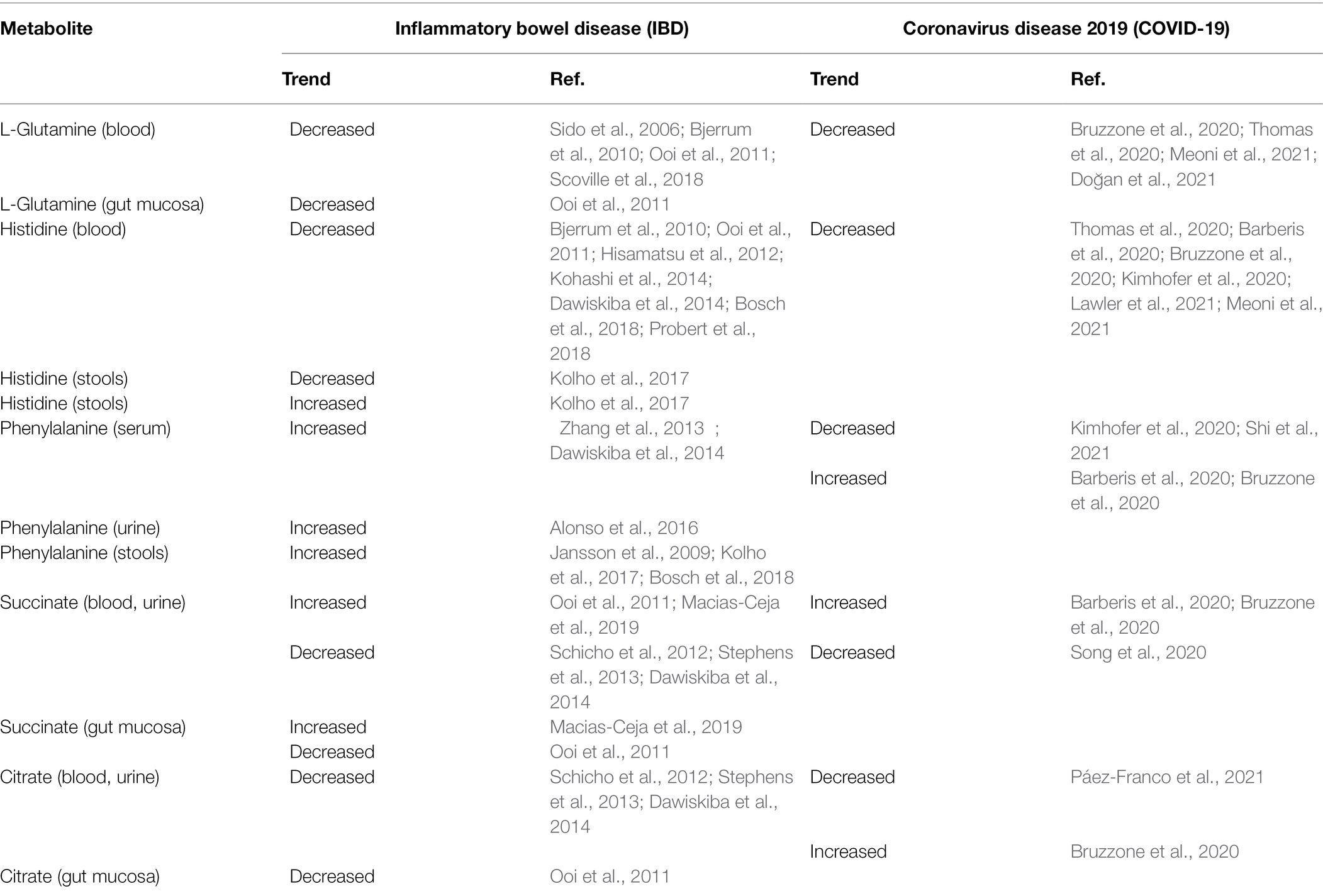
In IBD, L-glutamine plasma levels are decreased (Table 4), especially during the acute exacerbation of CD (Sido et al., 2006; Bjerrum et al., 2010; Ooi et al., 2011; Scoville et al., 2018). Supplementation improves inflammation (Sugihara et al., 2019) and the mucosal barrier integrity in patients with remissive CD (Benjamin et al., 2012b). Glutamine is reduced in the blood of patients with non-severe COVID19 and much more in severe forms (Thomas et al., 2020; Doğan et al., 2021; Meoni et al., 2021; Table 4). In patients with COVID-19, it was observed a 19% reduction in glutamine blood level compared to that before the onset of the disease (Bruzzone et al., 2020). This finding may be related to higher consumption of gluconeogenic amino acids, especially in patients with severe forms, because of the significant scarcity of amino acids (Fanos et al., 2021). A further possible explanation may be the increased conversion of L-glutamine into glutamate, supported by the blood Krebs cycle’s intermediates elevation and by the increase in oxidative stress (Páez-Franco et al., 2021).
Histidine
Histidine is an essential amino acid playing a crucial role as a ROS scavenger and anti-inflammatory mediator (Son et al., 2005; Holeček, 2020). The decarboxylation of histidine, catalyzed by histidine decarboxylase, originates histamine, a primary mediator in allergic diseases and a neurotransmitter involved in the control of food intake and sleep biorhythm. Histidine decarboxylase is expressed in bacteria of the large intestine and human muscle, liver, lung, and gastric mucosa (Moro et al., 2020). Blood histidine has been found significantly decreased in patients with IBD (Bjerrum et al., 2010; Ooi et al., 2011; Dawiskiba et al., 2014; Kohashi et al., 2014; Bosch et al., 2018; Probert et al., 2018). An early study found that histidine was significantly decreased in a large cohort (n = 387) of IBD patients (Hisamatsu et al., 2012); interestingly, plasma histidine was significantly lower in patients with active disease than in those in remission. Furthermore, a significant inverse correlation was observed between plasma histidine and serum CRP in patients with UC and CD. Authors postulated that the decrease in plasma histidine may reflect chronic inflammation in patients with IBD, suggesting supplementation with histidine as a novel therapy. In a subsequent 1-year follow-up of patients with UC in clinical remission, the same research group found that the decrease in histidine plasma level over time was associated with the increased risk of clinical relapse (Hisamatsu et al., 2015). Previous in vitro studies and in animal models suggested that low levels of histidine do not allow the effective suppression of NF-kB (Andou et al., 2009; Hasegawa et al., 2011); thus, LPS-induced TNF-α expression cannot be inhibited, resulting in the exacerbation of inflammation. This was confirmed by the amelioration of intestinal inflammation after oral administration of histidine. A recent metabolomics-based study on serum and stool samples in pediatric patients with IBD found controversial results. Compared with healthy controls, fecal histidine was significantly decreased in children with CD and significantly increased in those with UC (Kolho et al., 2017). Moreover, in the group of children with UC, serum histidine inversely correlated with erythrocyte sedimentation rate (ESR). Histidine fecal levels have been positively associated with more extended disease in UC but not in CD pediatric patients (Jagt et al., 2021). A possible explanation may be either the colonic leakage of histidine and other amino acids or malabsorption. However, the overgrowth of Bacteroides vulgatus in patients with UC leads to an increased fecal proteolytic and elastase activity (Galipeau et al., 2021). Therefore, it is reasonable to assume that the increased proteolytic and elastase activity might be the main factor promoting the high concentrations of fecal histidine rather than the protein-losing enteropathy. Several studies found L-histidine significantly decreased in the serum/plasma of patients with COVID-19 (Table 4; Barberis et al., 2020; Bruzzone et al., 2020; Thomas et al., 2020); the magnitude of L-histidine decline correlated with the disease severity (Lawler et al., 2021; Meoni et al., 2021). COVID-19 is marked by the activation of gluconeogenesis that is positively correlated with the severity of the disease. In patients with COVID-19, it was observed a 16% reduction in L-histidine blood levels compared to those before the onset of the disease, similarly to glutamine (Bruzzone et al., 2020). In COVID-19, low L-histidine serum levels may be related to the skeletal muscle breakdown (Kimhofer et al., 2020).
Phenylalanine
Phenylalanine is an aromatic, hydrophobic essential amino acid involved in the biosynthesis of catecholamines (Matthews, 2007). Phenylalanine is hydroxylated to tyrosine by phenylalanine 4-hydroxylase; this reaction primarily occurs in the liver and the kidney. In turn, tyrosine is hydroxylated to L-DOPA (3,4-Dihydroxyphenylalanine) by tyrosine-5 hydrolase, and the enzyme DOPA-decarboxylase converts L-DOPA into Dopamine. A restricted number of gut bacteria, mainly belonging to the phylum Firmicutes, such as Clostridium sporogenes and C. botulinum spp., metabolize aromatic amino acids tryptophan, phenylalanine, and tyrosine to their corresponding propionic acid derivatives, namely, phenylpropionic acid and 4-hydroxyphenylpropionic acid (Elsden et al., 1976). In C. sporogens, this metabolic pathway can produce nine metabolites; they accumulate in host serum and exhibit specificity by engaging receptors and altering host biology, especially systemic immunity and gut permeability (Dodd et al., 2017). Although an early metabolomics-based study focused on amino acids did not report any data on phenylalanine level in stool samples from adults with IBD (Marchesi et al., 2007), further studies found a significant increase of phenylalanine in serum (Zhang et al., 2013; Dawiskiba et al., 2014), urine (Alonso et al., 2016), and stools (Jansson et al., 2009; Kolho et al., 2017; Bosch et al., 2018). Interestingly, the magnitude of fecal phenylalanine increase differed between CD and UC, and no correlation was found between the disease activity and fecal phenylalanine concentration (Bosch et al., 2018). Given that the activity of phenylalanine-4-hydroxylase is impaired during immune activation and inflammation (Scholl-Bürgi et al., 2013), it is reasonable to assume that the increase of phenylalanine in patients with IBD may originate from the accumulation of this amino acid. A further contribution to the fecal phenylalanine increase in IBD may be related to the microbial biosynthesis of aromatic amino acids via the shikimate pathway (Sprenger, 2006). The condensation of phosphoenolpyruvate with erythrose 4-phosphate, deriving from the glycolysis pathway and the non-oxidative branch of the pentose phosphate pathway, respectively, yields 3-deoxy-d-arabino-heptulosonate-7-phosphate (DAHP), which is converted into chorismic acid and then into aromatic amino acids L-phenylalanine, L-tyrosine, and L-tryptophan. Approximately one-third of gut bacteria expresses all the transcripts coded from the genes of the shikimate pathway, including A. muciniphila (Mesnage and Antoniou, 2020); the remaining bacteria species do not exhibit a complete shikimate pathway, having lost either one enzyme (nearly 22%) or five or more enzymes (nearly 74%; Zucko et al., 2010).
In COVID-19, the considerable generation of ROS, due to cytokine activation, induces the irreversible non-enzymatic oxidation of 5, 6, 7, 8-tetrahydrobiopterin (BH4), a cofactor of phenylalanine 4-hydroxylase (PAH), the enzyme catalyzing the conversion of phenylalanine to tyrosine. BH4 shortage induces the loss of PAH activity, with the reduced biosynthesis of tyrosine and the accumulation of phenylalanine. Data on phenylalanine emerging from metabolomic studies in patients with COVID-19 are heterogeneous and discordant: both phenylalanine and tyrosine were decreased, especially in severe and fatal outcomes (Barberis et al., 2020) or increased (Kimhofer et al., 2020; Shi et al., 2021). Interestingly, in non-survivors, phenylalanine serum levels are significantly higher than in survivors (Shi et al., 2021). Moreover, the increase in phenylalanine was associated with the decrease in tyrosine (Bruzzone et al., 2020). The role of phenylalanine in SARS-CoV-2 infection seems to be crucial. In a multicenter study on elderly patients with COVID-19, phenylalanine and tyrosine metabolisms were highly upregulated in 132 deceased patients (median age 74 years) compared with 91 survivors (median age 70 years), suggesting that these amino acids contribute as building blocks for the production of internal SARS-CoV-2 protein and its subsequent assembly into viral particles (Mei et al., 2021).
Succinate and Citrate
Succinate and citrate are intermediates of the TCA cycle. Succinate is generated within mitochondria via the TCA cycle from succinyl-CoA; then, succinate is oxidized to fumarate by succinate dehydrogenase. Succinate is also a product of the glyoxylate cycle, a pathway active in many bacteria, plants, and fungi. Overall, succinate accumulation is a metabolic signature of anoxia, asphyxia, and ischemia; the dysregulation of succinate metabolism can lead to pathological conditions, such as malignant transformation, inflammation, and tissue injury. Metabolomics-based studies carried out in serum, plasma, and urine of patients with IBD found that succinate and citrate significantly decreased in CD and UC compared to non-IBD individuals (Schicho et al., 2012; Stephens et al., 2013; Dawiskiba et al., 2014). Succinate and citrate are strongly involved in energy metabolism, and their depletion confirms the increased demand and the rapid utilization of cellular energy in IBD. In this context, citrate depletion may be associated with the increase in fatty acids biosynthesis. Indeed, citrate is essential for carrying acetyl-CoA from mitochondria to the cytosol. The significant increase in circulating triglycerides, observed in several studies on IBD, can be considered further evidence (Levy et al., 2000; Sappati Biyyani et al., 2010; Tefas et al., 2020). Beyond the role of succinate as an energy supplier, this amino acid is an inflammation mediator; it selectively binds to and activates the succinate receptor-1 (SUCNR1), which promotes pro-inflammatory signaling pathways (Mills and O'Neill, 2014). In patients with CD, hypoxia, inflammation, and necrosis promote the accumulation of succinate in gut inflamed areas, with a further activation and infiltration of macrophages and fibroblasts, the overexpression of pro-inflammatory cytokines, and ultimately the acceleration of fibrosis (Macias-Ceja et al., 2019).
Data on succinate emerging from metabolomics-based studies in COVID-19 are controversial (Table 4); the concentration of this amino acid has been found increased (Barberis et al., 2020; Bruzzone et al., 2020) decreased in moderate and severe disease (Song et al., 2020), or unchanged (Thomas et al., 2020). Differences between patients normo-oxygenate and patients undergoing intense respiratory treatment may affect results obtained in different studies. In the paper of Bruzzone, the increase in succinate and citrate (156 and 12%, respectively) has been associated with central carbon metabolism dysfunction (Bruzzone et al., 2020). In severe COVID-19, the association of citrate decrease with succinate increase could be related to mitochondrial dysfunction due to hypoxia (Páez-Franco et al., 2021). Hypoxia inhibits oxidative phosphorylation, and thus energy is supplied by the anaerobic glycolysis, which is activated by the accumulation of the hypoxia-inducible factor 1α (HIF 1α; Majmundar et al., 2010). As a result, the TCA is blocked, with the consequent accumulation of succinate, depletion of citrate, and increased demand for glucose. In turn, the latter induces the decreased availability of gluconeogenic amino acids and oxaloacetate, being utilized as substrates for gluconeogenesis. Oxaloacetate is converted into glucose, whereas mitochondrial acetyl-CoA oxidation is drastically reduced, and acetyl-CoA is redirected to the synthesis of ketone bodies.
Ketone Bodies
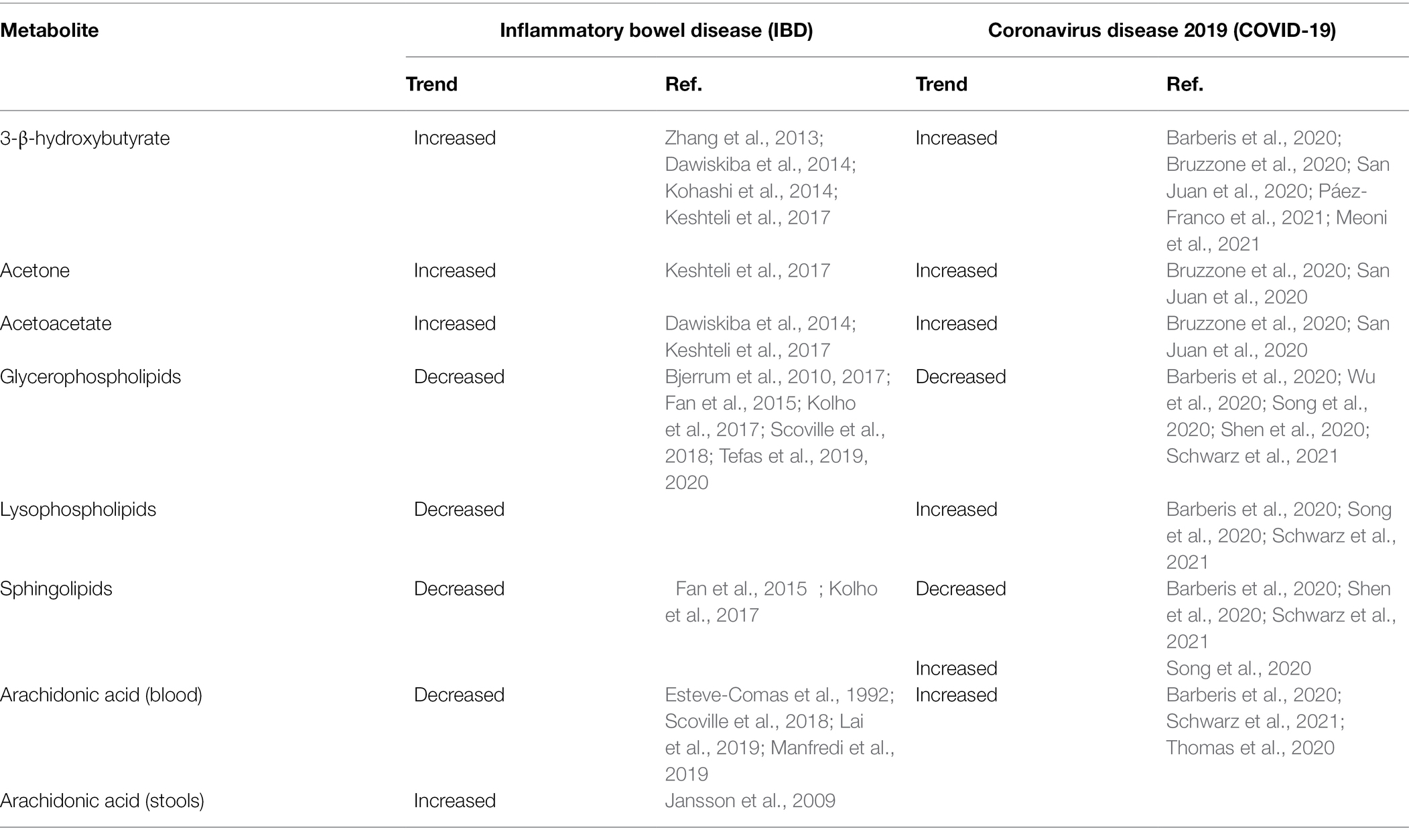
Ketone bodies are small lipid-derived molecules, namely, 3-β-hydroxybutyrate, acetone, and acetoacetate (Laffel, 1999). During fasting or prolonged exercise, the liver converts fatty acids mobilized from adipocytes into ketone bodies; then, they enter circulation, serving as an energy source. Ketone bodies are crucial regulators of metabolic health and longevity; in fact, they are neuroprotective and cytoprotective (Yang et al., 2019), having the ability to inhibit histone deacetylase activity and thereby epigenetic gene regulation (Newman and Verdin, 2014). On the one hand, ketone bodies were increased in the serum of patients with IBD (Table 5), and this finding was correlated to the increased energy demand (Zhang et al., 2013; Dawiskiba et al., 2014; Kohashi et al., 2014; Keshteli et al., 2017). On the other hand, ketone bodies may play a strategic role in IBD when supplemented with diet, thanks to their capacity to protect from toxic effects of chronic inflammation. An experimental study demonstrated that 3-β-hydroxybutyrate suppresses the activation of the NLRP3 inflammasome in response to urate crystals, ATP, and lipotoxic fatty acids (Youm et al., 2015). Then, in a mouse model of NLRP3-mediated diseases, authors observed that 3-β-hydroxybutyrate attenuates caspase-1 activation and the release of pro-inflammatory cytokines IL-1β and IL-18 from macrophages, reducing in definitive the severity of NLRP3-mediated chronic inflammatory diseases. The same effect was obtained by applying a ketogenic diet or supplementing 3-β-hydroxybutyrate (Youm et al., 2015). The accumulation of ketone bodies following a ketogenic diet strongly impacts gut microbiota composition; in vivo and in vitro experiments demonstrated that ketone bodies selectively inhibit the growth of several Bifidobacterial spp., with downstream consequences for gut immune cells, especially Th17, and induce the decrease in the relative abundance of Actinobacteria (Ang et al., 2020). In an animal model of inflammatory colitis, it was observed that the ketogenic diet alters gut microbiota and serum metabolome, alleviating colitis (Kong et al., 2021). In particular, Akkermansia, Roseburia, and Ruminococcaceae genera were enriched. After colitis induction, the ketogenic diet protected intestinal barrier function and reduced the presence of RORgt+CD3− group 3 innate lymphoid cells and related inflammatory cytokines (IL-17α, IL-18, IL-22, and Ccl4). As a result, the ketogenic diet in patients with IBD may substantially contribute to control inflammation and shape gut microbiota.
Ketone bodies accumulate in the serum of patients with COVID-19, mimicking diabetic ketoacidosis (Li et al., 2020). Further studies confirmed the elevation of circulating ketone bodies in COVID-19 (Table 5), suggesting their role as an alternative energy source during SARS-CoV-2 replication (Barberis et al., 2020; Bruzzone et al., 2020; San Juan et al., 2020; Meoni et al., 2021; Páez-Franco et al., 2021). The increase in 3-β-hydroxybutyrate could interfere with viral replication by upregulating the expression of antioxidant genes, the cytoplasmic NADPH, and directly scavenging free radicals (Stubbs et al., 2020). In addition, 3-β-hydroxybutyrate may induce the closing of mitochondrial permeability transition pore, apoptosis, and the inhibition of glycolysis. It was postulated that ketone bodies inactivate the extracellular virions (Shaheen, 2021). Ketone bodies have carbonyl groups reacting with the ε-amino group of lysine to form a Schiff base. Given that SARS-CoV-2 spike protein contains approximately 4.5% lysine residues, almost equally distributed between the two subunits, the reaction between the ketone body, mainly acetoacetate, and the lysine residues of the spike protein forms Schiff bases, altering the conformation of the spike protein. This change promotes the separation of the spike protein from the virion, either by the separation between S1 and S2 subunits or promoting its bending. As a result, acetoacetate induces protein conformational changes by altering the secondary structure, namely, reducing the α-helix content (Bohlooli et al., 2016). The sum of these researches has raised the question of whether or not it may be effective to induce ketosis both in asymptomatic individuals infected with SARS-CoV-2 and in patients with COVID-19 (Bradshaw et al., 2020).
Phospholipids
Glycerophospholipids, commonly termed phospholipids, their by-product lysophospholipids, and sphingolipids are key components of the cellular membrane; remarkably, phospholipids are involved in the metabolism of cell signaling. Phospholipids are essential for the biosynthesis of lipoproteins. As reported in Table 5, in patients with IBD, circulating phospholipids were found decreased in various studies (Bjerrum et al., 2010, 2017; Fan et al., 2015; Kolho et al., 2017; Scoville et al., 2018; Tefas et al., 2019, 2020). Factors, such as the compromised integrity of the intestinal mucosa, TNF-α, NF-kB, mitogen-activated protein kinase (MAPK) pathway, and peroxisome proliferator-activated receptor (PPAR) signaling, seem to be closely implicated in phospholipids depletion. Glycerophosphocholine was significantly decreased in two subsequent studies (Bjerrum et al., 2010, 2017); more recently, in patients with extensive UC and colonic CD, tetracosanoic acid, phosphatidylcholine, (PC) lysophosphatidylcholine (LPC), sphingomyelin (SM), and diacylglycerol were found decreased compared with healthy controls; interestingly, saturated LPC (18:2) was found decreased whereas polyunsaturated LPC (20:4) and LPC (22:6) increased (Tefas et al., 2019, 2020). It is likely that the anti-inflammatory polyunsaturated LPC effectively antagonized the pro-inflammatory activity of saturated LPC. Two studies found sphingolipids significantly reduced in patients with IBD (Fan et al., 2015; Kolho et al., 2017). The significant depletion in sphingolipids may be the result of the increased activity of sphingomyelinases, which are activated in IBD by the combined action of TNF-α, NF-kB, and IFN-γ (Schütze et al., 1992).
Phospholipids metabolism is strongly influenced by COVID-19 and by the severity of the disease (Mussap and Fanos, 2021); data emerging from the literature suggest that SARS-CoV-2 infection promotes the downregulation of most phospholipids and sphingolipids (Table 5), while various lysophospholipids are either overexpressed or unchanged (Barberis et al., 2020; Shen et al., 2020; Song et al., 2020; Wu et al., 2020; Schwarz et al., 2021). The downregulation of phospholipids may originate from the liver impairment in patients with severe COVID-19, whereas lysophospholipids upregulation is the result of the increased activity of phospholipase A2. Low-risk infected patients are well discriminated from non-infected individuals by high levels of phosphatidylcholine (PC38:8), phosphatidylethanolamine (PE38:4), and phosphatidylglycerol (PG20:5). In patients with COVID-19, the predominance of inflammation over the macrophage-driven anti-inflammatory response leads to the underexpression of sphingosine 1-phosphate (Shen et al., 2020; Song et al., 2020). The considerable number of lipids belonging to any lipid class gives a limited value to the definition of upregulation and downregulation of phospholipids, lysophospholipids, and sphingolipids in COVID-19; recently, a targeted lipidomic analysis measured a considerable number of phospholipids (n = 90), lysophospholipids (n = 14), and sphingolipids (n = 15), discovering that certain lipids decrease and other increase within the same lipid class (Caterino et al., 2021).
Arachidonic Acid and Phospholipases
Arachidonic acid is a 20-carbon chain belonging to the ω-6 (n-6) polyunsaturated fatty acids (PUFAs); it is obtained from food and then incorporated in phospholipids (Tallima and El Ridi, 2017). Arachidonic acid is the primary precursor for the biosynthesis of eicosanoids, a complex family of lipid signaling mediators including but not limited to leukotrienes, prostaglandins, thromboxane, and prostacyclin A2 (Calder, 2020). By the cleavage of arachidonic acid from membrane phospholipids, phospholipases A2-IID, -IIF, -III, and -X initiate the arachidonic acid cascade and eicosanoid production (Murakami et al., 2020). Eicosanoids are generated by three main pathways, namely, cyclooxygenases, lipoxygenases, and cytochrome P-450 epoxygenase pathways (Dennis and Norris, 2015). Beyond their crucial role in a broad range of physiological processes, eicosanoids are involved in the pathogenesis of IBD (Alhouayek et al., 2021). During the acute phase of the disease, their concentration significantly increases within the inflamed intestinal mucosa (Wallace, 2019). In patients with IBD, arachidonic acid was increased in feces (Jansson et al., 2009) and decreased in the blood (Table 5); these variations were often associated with the exacerbation of the disease activity (Esteve-Comas et al., 1992; Scoville et al., 2018; Lai et al., 2019; Manfredi et al., 2019; Gallagher et al., 2021). Based on current knowledge, the decrease of circulating arachidonic acid in IBD might be related to the increased synthesis of eicosanoids in the gut (Shores et al., 2011). SARS-CoV-2 infection induces the overexpression of phospholipase A2, especially in macrophages, T, and B cells; as a consequence, the biosynthesis of arachidonic acid is upregulated in patients with COVID-19 (Table 5). The arachidonic acid upregulation positively correlates with the IL-6 concentration and the disease’s severity (Barberis et al., 2020; Thomas et al., 2020; Schwarz et al., 2021). In patients with COVID-19 and severe liver injury, however, arachidonic acid may be downregulated (Shen et al., 2020). Arachidonic acid is a potent antiviral PUFA that can inactivate the enveloped viruses, including SARS-CoV-2 (Hoxha, 2020); human cells infected by HCoV-229E and MERS-CoV are inhibited by arachidonic acid (Yan et al., 2019). The increased activity of the cytosolic phospholipase A2α (cPLA2α) in cells infected by SARS-CoV-2 generates the overproduction of lysophospholipids that are essential for the viral replication (Casari et al., 2021); the pharmacological inhibition of cPLA2α in human Huh-7 cells infected with coronavirus 229E drastically reduces the viral RNA synthesis, blocking an early step in the replication cycle (Müller et al., 2018). This finding opens new perspectives on the effective treatment of SARS-CoV-2 infection. A recent study performed a targeted lipidomic analysis of bronchoalveolar lavages fluids (BALs) from patients with severe COVID-19 (Archambault et al., 2021). The most relevant finding was the significant increase in several bioactive lipids, such as thromboxane, leukotrienes, and 15-lipoxygenase metabolites derived from linoleic acid, linolenic acid, and dihomo-γ-linolenic acid (Table 5). During the early stage of inflammation, the enzymatic oxygenation of essential fatty acids, including arachidonic acid, eicosapentaenoic acid, docosapentaenoic acid, and docosahexaenoic acid, generates a class of bioactive lipids, the so-called specialized pro-resolving mediators (SPMs; Serhan et al., 2000). This class includes lipoxins, resolvins, maresins, and protectins (Basil and Levy, 2016). Interestingly, BALs from patients with severe COVID-19 were also marked by the increase in SPMs. Increased levels of pro-inflammatory bioactive lipids and anti-inflammatory SPMs have also been reported in serum samples collected from hospital inpatients with a confirmed diagnosis of COVID-19 (Turnbull et al., 2022).
Conclusive and Prospective Remarks
IBD and COVID-19 are characterized by gut dysbiosis associated with impaired gut barrier function and immune-mediated chronic inflammation. However, COVID-19 can rapidly evolve to multisystemic organ damage due to a dysregulated, fulminant inflammatory response, the so-called “cytokine storm” (Jain, 2020; Hu et al., 2021). The evident imbalance between the number of studies on gut microbiota in IBD and that in SARS-CoV-2 does not exclude a reliable data analysis; we noticed more similarities than differences in gut microbial alterations between IBD and COVID-19 (Table 1). The depletion in F. prausnitzii, E. rectale, R, bromii, Lachnospiraceae, C. leptum (cluster IV), and the overgrowth of Enterococcus, E. coli, Shigella, P. mirabilis, Fusobacterium, Veillonellaceae are common patterns of dysbiosis, playing a key role in the severity and clinical outcome of IBD and COVID-19. An intriguing dissimilarity between IBD and COVID-19 is the abundance of A. muciniphila (Table 1), a Gram-negative bacterium belonging to the Verrucomicrobia phylum. A. muciniphila colonizes the mucus layer close to gut epithelial cells and is able to degrade mucin sugars and the protein backbone by specific enzymes, such as sialidases and fucosidases, providing carbon and nitrogen for bacteria unable to produce these enzymes (van Passel et al., 2011). Mucus degradation by A. muciniphila generates SCFAs, which are strongly involved in promoting host metabolic health. Therefore, A. muciniphila has several beneficial effects on host health by reducing inflammation, stimulating mucin biosynthesis and mucus thickness, preserving the integrity of the mucosal barrier, increasing the expression of tight junction proteins (e.g., occluding), and modulating the intestinal adaptative immune response (Ottman et al., 2017; Ansaldo et al., 2019; Ashrafian et al., 2019; Liu et al., 2021). The depletion of A. muciniphila in IBD (Table 1), reported by several studies, confirms the well-known inverse relationship between this bacterium and IBD (Rajilić-Stojanović et al., 2013); on the other hand, the enrichment in A. muciniphila in patients with COVID-19 has been associated with that of opportunistic pathogens, such as Enterococcus, Staphylococcus, Serratia, Collinsella, Actinomyces, and many others (Gaibani et al., 2021). Concerns emerge about results on the probiotic strains Lactobacilli and Bifidobacteria, especially in patients with COVID-19 (Table 1). These strains are Gram-positive, non-spore-forming, lactic acid producer bacteria with the antiviral activity performed by various mechanisms, including the synthesis of antiviral inhibitory metabolites, the upregulation of the protective immune responses, and by competing for nutrients and colonization sites with the virus and, more extensively, with pathogens (Kesika et al., 2021). Five studies reported Lactobacillus enrichment in patients with COVID-19 (Table 1), confuting the general notion that gut dysbiosis due to severe infections promotes the depletion of these strains (Harper et al., 2021). Very few data support the depletion of Lactobacillus and Bifidobacterium in patients with COVID-19. Two articles cited in the PubMed library, written in Chinese with the same English abstract and the same digital object identifier (doi), described gut dysbiosis in “some patients” with COVID-19 marked by the decrease in Lactobacillus and Bifidobacterium abundance (Xu et al., 2020a,b). Clearly, these data are unreliable. Therefore, further studies are required to elucidate the significance of Lactobacillus and Bifidobacterium enrichment in COVID-19.
Metabolomics reveals considerable similarities in the tryptophan metabolism between IBD and COVID-19 (Table 3). In IBD, the increase in quinolinic acid is associated with the decrease in picolinic acid, a non-selective metal ion chelating agent with antimicrobial, antiviral, and antifungal activity formed by a non-enzymic cyclization of aminomuconic acid semialdehyde. In COVID-19, the increase in quinolinic acid is associated with the increase of kynurenic acid; the biochemical mechanism underlying this unusual association should be clarified. In fact, quinolinic acid and kynurenic acid are closely related to each other by an inverse relationship that becomes imbalanced in various diseases. In COVID-19, the increase in picolinic acid reflects the activation of the enzymatic conversion of 2-amino-3-carboxymuconate-6-semialdehyde (ACMS) to 2-aminomuconic-6-semialdehyde. In turn, the latter undergoes either non-enzymatic cyclization to form picolinic acid or enzymatic transformation to 2-aminomuconic acid, yielding acetyl-CoA (Badawy, 2017). It is unclear why picolinic acid is increased in COVID-19, taking into account the alteration of brain functions during the disease (Chou et al., 2021; Marshall, 2021). It is reasonable to assume that the limited number of metabolic differences between IBD and COVID-19 (Tables 4, 5), for example, blood arachidonic acid, originates from the acute systemic damage and impairment of organs, tissues, and biological systems (e.g., coagulation) in COVID-19, while IBD remains a chronic disease localized in the gastrointestinal tract with a broad spectrum of extraintestinal symptoms and comorbidities (Argollo et al., 2019). Emerging evidence indicates the role of the microbiome in modulating the immune response to vaccination and the role of metabolic profile in predicting vaccination outcome (Hagan et al., 2019; Alexander et al., 2021). Therefore, microbiomics and metabolomics may be considered powerful tools for the early identification and monitoring of individuals at risk of adverse events, for example, fragile individuals or patients with severe chronic diseases. Since the COVID-19 pandemic is far from over, SARS-CoV-2 vaccination is the mainstay for preventing the COVID-19 spread. As a result, there is the need to extend SARS-CoV-2 vaccination to any adult or young subject, even if affected by pre-existing chronic diseases, such as IBD (Alexander et al., 2021). Therefore, deciphering the individual metabolic and microbial fingerprint in IBD is crucial to define an effective strategy for the safe administration of the vaccine (e.g., to discover any metabolic/microbial alteration induced by recent steroid or rituximab therapy), to manage vaccinated individuals, and to avoid any possible adverse effect in individuals at risk (Ferretti et al., 2021). On the other hand, deciphering the individual metabolic and microbial fingerprint in patients with IBD with COVID-19 may considerably improve the therapeutic approach, preventing the risk of adverse outcomes.
Author Contributions
GC and FB contributed to the conception and design of the study. VF conceptualized and performed the modeling. GC wrote the first draft of the manuscript. MM supervised the manuscript and wrote the final version of the paper. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agus, A., Planchais, J., and Sokol, H. (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724. doi: 10.1016/j.chom.2018.05.003
Aktas, B., and Aslim, B. (2021). Neuropathy in COVID-19 associated with dysbiosis-related inflammation. Turk. J. Biol. 45, 390–403. doi: 10.3906/biy-2105-53
Alam, M. T., Amos, G. C. A., Murphy, A. R. J., Murch, S., Wellington, E. M. H., and Arasaradnam, R. P. (2020). Microbial imbalance in inflammatory bowel disease patients at different taxonomic levels. Gut Pathog. 12:1. doi: 10.1186/s13099-019-0341-6
Aldars-García, L., Chaparro, M., and Gisbert, J. P. (2021). Systematic review: the gut microbiome and its potential clinical application in inflammatory bowel disease. Microorganisms 9:977. doi: 10.3390/microorganisms9050977
Alexander, J. L., Moran, G. W., Gaya, D. R., Raine, T., Hart, A., Kennedy, N. A., et al. (2021). SARS-CoV-2 vaccination for patients with inflammatory bowel disease: a British Society of Gastroenterology Inflammatory Bowel Disease section and IBD clinical research group position statement. Lancet Gastroenterol. Hepatol. 6, 218–224. doi: 10.1016/S2468-1253(21)00024-8
Alhouayek, M., Ameraoui, H., and Muccioli, G. G. (2021). Bioactive lipids in inflammatory bowel diseases - From pathophysiological alterations to therapeutic opportunities. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1866:158854. doi: 10.1016/j.bbalip.2020.158854
Allocca, M., Chaparro, M., Gonzalez, H. A., Bosca-Watts, M. M., Palmela, C., D'Amico, F., et al. (2020). Patients with inflammatory bowel disease are not at increased risk of COVID-19: a large multinational cohort study. J. Clin. Med. 9:3533. doi: 10.3390/jcm9113533
Allocca, M., and Craviotto, V. (2021). Low risk of severe COVID-19 in patients with inflammatory bowel disease: keep calm and take stock. Pol. Arch. Intern. Med. 131, 222–223. doi: 10.20452/pamw.15903
Alonso, A., Julià, A., Vinaixa, M., Domènech, E., Fernández-Nebro, A., Cañete, J. D., et al. (2016). Urine metabolome profiling of immune-mediated inflammatory diseases. BMC Med. 14:133. doi: 10.1186/s12916-016-0681-8
Alshehri, D., Saadah, O., Mosli, M., Edris, S., Alhindi, R., and Bahieldin, A. (2021). Dysbiosis of gut microbiota in inflammatory bowel disease: current therapies and potential for microbiota-modulating therapeutic approaches. Bosn. J. Basic Med. Sci. 21, 270–283. doi: 10.17305/bjbms.2020.5016
Andoh, A., Tsujikawa, T., Sasaki, M., Mitsuyama, K., Suzuki, Y., Matsui, T., et al. (2009). Faecal microbiota profile of Crohn's disease determined by terminal restriction fragment length polymorphism analysis. Aliment. Pharmacol. Ther. 29, 75–82. doi: 10.1111/j.1365-2036.2008.03860.x
Andou, A., Hisamatsu, T., Okamoto, S., Chinen, H., Kamada, N., Kobayashi, T., et al. (2009). Dietary histidine ameliorates murine colitis by inhibition of proinflammatory cytokine production from macrophages. Gastroenterology 136, 564–574.e2. doi: 10.1053/j.gastro.2008.09.062
Ang, Q. Y., Alexander, M., Newman, J. C., Tian, Y., Cai, J., Upadhyay, V., et al. (2020). Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell 181, 1263–275.e16. doi: 10.1016/j.cell.2020.04.027
Ansaldo, E., Slayden, L. C., Ching, K. L., Koch, M. A., Wolf, N. K., Plichta, D. R., et al. (2019). Akkermansia muciniphila induces intestinal adaptive immune responses during homeostasis. Science 364, 1179–1184. doi: 10.1126/science.aaw7479
Archambault, A. S., Zaid, Y., Rakotoarivelo, V., Turcotte, C., Doré, É., Dubuc, I., et al. (2021). High levels of eicosanoids and docosanoids in the lungs of intubated COVID-19 patients. FASEB J. 35:e21666. doi: 10.1096/fj.202100540R
Ardizzone, S., Ferretti, F., Monico, M. C., Carvalhas Gabrielli, A. M., Carmagnola, S., Bezzio, C., et al. (2021). Lower incidence of COVID-19 in patients with inflammatory bowel disease treated with non-gut selective biologic therapy. J. Gastroenterol. Hepatol. doi: 10.1111/jgh.15591 [Epub ahead of print].
Argollo, M., Gilardi, D., Peyrin-Biroulet, C., Chabot, J. F., Peyrin-Biroulet, L., and Danese, S. (2019). Comorbidities in inflammatory bowel disease: a call for action. Lancet Gastroenterol. Hepatol. 4, 643–654. doi: 10.1016/S2468-1253(19)30173-6
Ashrafian, F., Shahriary, A., Behrouzi, A., Moradi, H. R., Keshavarz Azizi Raftar, S., Lari, A., et al. (2019). Akkermansia muciniphila-derived extracellular vesicles as a mucosal delivery vector for amelioration of obesity in mice. Front. Microbiol. 10:2155. doi: 10.3389/fmicb.2019.02155
Ashrafian, H., Sounderajah, V., Glen, R., Ebbels, T., Blaise, B. J., Kalra, D., et al. (2020). Metabolomics: The stethoscope for the twenty-first century. Med. Princ. Pract. 30, 301–310. doi: 10.1159/000513545
Aziz, M., Fatima, R., Haghbin, H., Lee-Smith, W., and Nawras, A. (2020). The incidence and outcomes of COVID-19 in IBD patients: A rapid review and meta-analysis. Inflamm. Bowel Dis. 26, e132–e133. doi: 10.1093/ibd/izaa170
Badawy, A. A. (2017). Tryptophan availability for kynurenine pathway metabolism across the life span: control mechanisms and focus on aging, exercise, diet and nutritional supplements. Neuropharmacology 112, 248–263. doi: 10.1016/j.neuropharm.2015.11.015
Barberis, E., Timo, S., Amede, E., Vanella, V. V., Puricelli, C., Cappellano, G., et al. (2020). Large-scale plasma analysis revealed new mechanisms and molecules associated with the host response to SARS-CoV-2. Int. J. Mol. Sci. 21:8623. doi: 10.3390/ijms21228623
Basil, M. C., and Levy, B. D. (2016). Specialized pro-resolving mediators: endogenous regulators of infection and inflammation. Nat. Rev. Immunol. 16, 51–67. doi: 10.1038/nri.2015.4
Basuroy, S., Seth, A., Elias, B., Naren, A. P., and Rao, R. (2006). MAPK interacts with occludin and mediates EGF-induced prevention of tight junction disruption by hydrogen peroxide. Biochem. J. 393, 69–77. doi: 10.1042/BJ20050959
Benjamin, J. L., Hedin, C. R., Koutsoumpas, A., Ng, S. C., McCarthy, N. E., Prescott, N. J., et al. (2012a). Smokers with active Crohn's disease have a clinically relevant dysbiosis of the gastrointestinal microbiota. Inflamm. Bowel Dis. 18, 1092–1100. doi: 10.1002/ibd.21864
Benjamin, J., Makharia, G., Ahuja, V., Anand Rajan, K. D., Kalaivani, M., and Gupta, S. D. (2012b). Glutamine and whey protein improve intestinal permeability and morphology in patients with Crohn's disease: a randomized controlled trial. Dig. Dis. Sci. 57, 1000–1012. doi: 10.1007/s10620-011-1947-9
Bertè, R., Mazza, S., Stefanucci, M. R., Noviello, D., Costa, S., Ciafardini, C., et al. (2021). Seroprevalence of SARS-CoV2 in IBD patients treated with biologic therapy. J. Crohns Colitis 15, 864–868. doi: 10.1093/ecco-jcc/jjaa237
Bjerrum, J. T., Nielsen, O. H., Hao, F., Tang, H., Nicholson, J. K., Wang, Y., et al. (2010). Metabonomics in ulcerative colitis: diagnostics, biomarker identification, and insight into the pathophysiology. J. Proteome Res. 9, 954–962. doi: 10.1021/pr9008223
Bjerrum, J. T., Steenholdt, C., Ainsworth, M., Nielsen, O. H., Reed, M. A., Atkins, K., et al. (2017). Metabonomics uncovers a reversible proatherogenic lipid profile during infliximab therapy of inflammatory bowel disease. BMC Med. 15:184. doi: 10.1186/s12916-017-0949-7
Blasco, H., Bessy, C., Plantier, L., Lefevre, A., Piver, E., Bernard, L., et al. (2020). The specific metabolome profiling of patients infected by SARS-COV-2 supports the key role of tryptophan-nicotinamide pathway and cytosine metabolism. Sci. Rep. 10:16824. doi: 10.1038/s41598-020-73966-5
Bohlooli, M., Ghaffari-Moghaddam, M., Khajeh, M., Shahraki-Fallah, G., Haghighi-Kekhaiye, B., and Sheibani, N. (2016). The role of acetoacetate in Amadori product formation of human serum albumin. J. Photochem. Photobiol. B 163, 345–351. doi: 10.1016/j.jphotobiol.2016.09.004
Bonovas, S., Fiorino, G., Allocca, M., Lytras, T., Nikolopoulos, G. K., Peyrin-Biroulet, L., et al. (2016). Biologic therapies and risk of infection and malignancy in patients with inflammatory bowel disease: a systematic review and network meta-analysis. Clin. Gastroenterol. Hepatol. 14, 1385–97.e10. doi: 10.1016/j.cgh.2016.04.039
Bosch, S., Struys, E. A., van Gaal, N., Bakkali, A., Jansen, E. W., Diederen, K., et al. (2018). Fecal amino acid analysis can discriminate De novo treatment-Naïve pediatric inflammatory bowel disease from controls. J. Pediatr. Gastroenterol. Nutr. 66, 773–778. doi: 10.1097/MPG.0000000000001812
Bradshaw, P. C., Seeds, W. A., Miller, A. C., Mahajan, V. R., and Curtis, W. M. (2020). COVID-19: proposing a ketone-based metabolic therapy as a treatment to blunt the cytokine storm. Oxidative Med. Cell. Longev. 2020:6401341. doi: 10.1155/2020/6401341
Brenner, E. J., Ungaro, R. C., and Colombel, J. F. (2021). SECURE-IBD database public data update. Available at: https://covidibd.org/ (Accessed September 9, 2021).
Brenner, E. J., Ungaro, R. C., Gearry, R. B., Kaplan, G. G., Kissous-Hunt, M., Lewis, J. D., et al. (2020). Corticosteroids, but not TNF antagonists, are associated with adverse COVID-19 outcomes in patients with inflammatory bowel diseases: results from an international registry. Gastroenterology 159, 481–91.e3. doi: 10.1053/j.gastro.2020.05.032
Bruzzone, C., Bizkarguenaga, M., Gil-Redondo, R., Diercks, T., Arana, E., García de Vicuña, A., et al. (2020). SARS-CoV-2 infection dysregulates the metabolomic and lipidomic profiles of serum. iScience 23:101645. doi: 10.1016/j.isci.2020.101645
Casari, I., Manfredi, M., Metharom, P., and Falasca, M. (2021). Dissecting lipid metabolism alterations in SARS-CoV-2. Prog. Lipid Res. 82:101092. doi: 10.1016/j.plipres.2021.101092
Caterino, M., Gelzo, M., Sol, S., Fedele, R., Annunziata, A., Calabrese, C., et al. (2021). Dysregulation of lipid metabolism and pathological inflammation in patients with COVID-19. Sci. Rep. 11:2941. doi: 10.1038/s41598-021-82426-7
Chen, G. L., Zhang, Y., Wang, W. Y., Ji, X. L., Meng, F., Xu, P. S., et al. (2017). Partners of patients with ulcerative colitis exhibit a biologically relevant dysbiosis in fecal microbial metacommunities. World J. Gastroenterol. 23, 4624–4631. doi: 10.3748/wjg.v23.i25.4624
Chervy, M., Barnich, N., and Denizot, J. (2020). Adherent-invasive E. coli: update on the lifestyle of a troublemaker in Crohn's disease. Int. J. Mol. Sci. 21, 3734. doi: 10.3390/ijms21103734
Cheung, K. S., Hung, I. F. N., Chan, P. P. Y., Lung, K. C., Tso, E., Liu, R., et al. (2020). Gastrointestinal manifestations of SARS-CoV-2 infection and virus load in fecal samples from a Hong Kong cohort: systematic review and meta-analysis. Gastroenterology 159, 81–95. doi: 10.1053/j.gastro.2020.03.065
Chiodini, R. J., Dowd, S. E., Galandiuk, S., Davis, B., and Glassing, A. (2016). The predominant site of bacterial translocation across the intestinal mucosal barrier occurs at the advancing disease margin in Crohn's disease. Microbiology 162, 1608–1619. doi: 10.1099/mic.0.000336
Chou, S. H., Beghi, E., Helbok, R., Moro, E., Sampson, J., Altamirano, V., et al. (2021). Global incidence of neurological manifestations Among patients hospitalized With COVID-19 - a report for the GCS-NeuroCOVID consortium and the ENERGY consortium. JAMA Netw. Open 4:e2112131. doi: 10.1001/jamanetworkopen.2021.12131
Clooney, A. G., Eckenberger, J., Laserna-Mendieta, E., Sexton, K. A., Bernstein, M. T., Vagianos, K., et al. (2021). Ranking microbiome variance in inflammatory bowel disease: a large longitudinal intercontinental study. Gut 70, 499–510. doi: 10.1136/gutjnl-2020-321106
Cruzat, V., Macedo Rogero, M., Noel Keane, K., Curi, R., and Newsholme, P. (2018). Glutamine: metabolism and immune function, supplementation and clinical translation. Nutrients 10:1564. doi: 10.3390/nu10111564
Curi, R., Lagranha, C. J., Doi, S. Q., Sellitti, D. F., Procopio, J., Pithon-Curi, T. C., et al. (2005). Molecular mechanisms of glutamine action. J. Cell. Physiol. 204, 392–401. doi: 10.1002/jcp.20339
Cyprian, F., Sohail, M. U., Abdelhafez, I., Salman, S., Attique, Z., Kamareddine, L., et al. (2021). SARS-CoV-2 and immune-microbiome interactions: lessons from respiratory viral infections. Int. J. Infect. Dis. 105, 540–550. doi: 10.1016/j.ijid.2021.02.071
D'Amico, F., Danese, S., and Peyrin-Biroulet, L. (2020). Systematic review on inflammatory bowel disease patients with coronavirus disease 2019: it is time to take stock. Clin. Gastroenterol. Hepatol. 18, 2689–2700. doi: 10.1016/j.cgh.2020.08.003
Dawiskiba, T., Deja, S., Mulak, A., Ząbek, A., Jawień, E., Pawełka, D., et al. (2014). Serum and urine metabolomic fingerprinting in diagnostics of inflammatory bowel diseases. World J. Gastroenterol. 20, 163–174. doi: 10.3748/wjg.v20.i1.163
de Meij, T. G. J., de Groot, E. F. J., Peeters, C. F. W., de Boer, N. K. H., Kneepkens, C. M. F., Eck, A., et al. (2018). Variability of core microbiota in newly diagnosed treatment-naïve paediatric inflammatory bowel disease patients. PLoS One 13:e0197649. doi: 10.1371/journal.pone.0197649
de Oliveira, G. L. V., Oliveira, C. N. S., Pinzan, C. F., de Salis, L. V. V., and Cardoso, C. R. B. (2021). Microbiota modulation of the gut-lung axis in COVID-19. Front. Immunol. 12:635471. doi: 10.3389/fimmu.2021.635471
Dennis, E. A., and Norris, P. C. (2015). Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 15, 511–523. doi: 10.1038/nri3859
Devaux, C. A., Lagier, J. C., and Raoult, D. (2021). New insights into the physiopathology of COVID-19: SARS-CoV-2-associated gastrointestinal illness. Front. Med. 8:640073. doi: 10.3389/fmed.2021.640073
Dicksved, J., Halfvarson, J., Rosenquist, M., Järnerot, G., Tysk, C., Apajalahti, J., et al. (2008). Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J. 2, 716–727. doi: 10.1038/ismej.2008.37
Dipasquale, V., Cucchiara, S., Martinelli, M., Miele, E., Aloi, M., and Romano, C. (2020). Challenges in paediatric inflammatory bowel diseases in the COVID-19 time. Dig. Liver Dis. 52, 593–594. doi: 10.1016/j.dld.2020.03.015
Dodd, D., Spitzer, M. H., Van Treuren, W., Merrill, B. D., Hryckowian, A. J., Higginbottom, S. K., et al. (2017). A gut bacterial pathway metabolizes aromatic amino acids into nine circulating metabolites. Nature 551, 648–652. doi: 10.1038/nature24661
Doğan, H. O., Şenol, O., Bolat, S., Yıldız, Ş. N., Büyüktuna, S. A., Sarıismailoğlu, R., et al. (2021). Understanding the pathophysiological changes via untargeted metabolomics in COVID-19 patients. J. Med. Virol. 93, 2340–2349. doi: 10.1002/jmv.26716
Durack, J., and Lynch, S. V. (2019). The gut microbiome: relationships with disease and opportunities for therapy. J. Exp. Med. 216, 20–40. doi: 10.1084/jem.20180448
Ellermann, M., Huh, E. Y., Liu, B., Carroll, I. M., Tamayo, R., and Sartor, R. B. (2015). Adherent-invasive Escherichia coli production of cellulose influences iron-induced bacterial aggregation, phagocytosis, and induction of colitis. Infect. Immun. 83, 4068–4080. doi: 10.1128/IAI.00904-15
Elsden, S. R., Hilton, M. G., and Waller, J. M. (1976). The end products of the metabolism of aromatic amino acids by clostridia. Arch. Microbiol. 107, 283–288. doi: 10.1007/BF00425340
Esteve-Comas, M., Ramírez, M., Fernández-Bañares, F., Abad-Lacruz, A., Gil, A., Cabré, E., et al. (1992). Plasma polyunsaturated fatty acid pattern in active inflammatory bowel disease. Gut 33, 1365–1369. doi: 10.1136/gut.33.10.1365
Falandry, C., Malapert, A., Roche, M., Subtil, F., Berthiller, J., Boin, C., et al. (2021). Risk factors associated with day-30 mortality in patients over 60 years old admitted in ICU for severe COVID-19: the senior-COVID-Rea multicentre survey protocol. BMJ Open 11:e044449. doi: 10.1136/bmjopen-2020-044449
Fan, F., Mundra, P. A., Fang, L., Galvin, A., Moore, X. L., Weir, J. M., et al. (2015). Lipidomic profiling in inflammatory bowel disease: comparison Between ulcerative colitis and Crohn's disease. Inflamm. Bowel Dis. 21, 1511–1518. doi: 10.1097/MIB.0000000000000394
Fanos, V., Pintus, R., Pintus, M. C., Mussap, M., and Marcialis, M. A. (2021). Seven secrets of COVID-19: fever, ACE2 receptors, gut-lung axis, metabolomics, microbiomics, probiotics, diet. J. Pediatr. Neon Ind. Med. 10:e100145. doi: 10.7363/100145
Favier, C., Neut, C., Mizon, C., Cortot, A., Colombel, J. F., and Mizon, J. (1997). Fecal beta-D-galactosidase production and bifidobacteria are decreased in Crohn's disease. Dig. Dis. Sci. 42, 817–822. doi: 10.1023/A:1018876400528
Ferretti, F., Cannatelli, R., Benucci, M., Carmagnola, S., Clementi, E., Danelli, P., et al. (2021). How to manage COVID-19 vaccination in immune-mediated inflammatory diseases: an expert opinion by IMIDs study group. Front. Immunol. 12:656362. doi: 10.3389/fimmu.2021.656362
Fite, A., Macfarlane, S., Furrie, E., Bahrami, B., Cummings, J. H., Steinke, D. T., et al. (2013). Longitudinal analyses of gut mucosal microbiotas in ulcerative colitis in relation to patient age and disease severity and duration. J. Clin. Microbiol. 51, 849–856. doi: 10.1128/JCM.02574-12
Flynn, S., and Eisenstein, S. (2019). Inflammatory bowel disease presentation and diagnosis. Surg. Clin. North Am. 99, 1051–1062. doi: 10.1016/j.suc.2019.08.001
Forrest, C. M., Youd, P., Kennedy, A., Gould, S. R., Darlington, L. G., and Stone, T. W. (2002). Purine, kynurenine, neopterin and lipid peroxidation levels in inflammatory bowel disease. J. Biomed. Sci. 9, 436–442. doi: 10.1007/BF02256538
Frank, D. N., St Amand, A. L., Feldman, R. A., Boedeker, E. C., Harpaz, N., and Pace, N. R. (2007). Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. U. S. A. 104, 13780–13785. doi: 10.1073/pnas.0706625104
Franzosa, E. A., Sirota-Madi, A., Avila-Pacheco, J., Fornelos, N., Haiser, H. J., Reinker, S., et al. (2019). Gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat. Microbiol. 4, 293–305. doi: 10.1038/s41564-018-0306-4
Fraser, D. D., Slessarev, M., Martin, C. M., Daley, M., Patel, M. A., Miller, M. R., et al. (2020). Metabolomics profiling of critically ill coronavirus disease 2019 patients: identification of diagnostic and prognostic biomarkers. Crit. Care Explor. 2:e0272. doi: 10.1097/CCE.0000000000000272
Fujimoto, T., Imaeda, H., Takahashi, K., Kasumi, E., Bamba, S., Fujiyama, Y., et al. (2013). Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn's disease. J. Gastroenterol. Hepatol. 28, 613–619. doi: 10.1111/jgh.12073
Gaibani, P., D'Amico, F., Bartoletti, M., Lombardo, D., Rampelli, S., Fornaro, G., et al. (2021). The gut microbiota of critically ill patients With COVID-19. Front. Cell. Infect. Microbiol. 11:670424. doi: 10.3389/fcimb.2021.670424
Galipeau, H. J., Caminero, A., Turpin, W., Bermudez-Brito, M., Santiago, A., Libertucci, J., et al. (2021). Novel fecal biomarkers That precede clinical diagnosis of ulcerative colitis. Gastroenterology 160, 1532–1545. doi: 10.1053/j.gastro.2020.12.004
Gallagher, K., Catesson, A., Griffin, J. L., Holmes, E., and Williams, H. R. T. (2021). Metabolomic analysis in inflammatory bowel disease: a systematic review. J. Crohns Colitis 15, 813–826. doi: 10.1093/ecco-jcc/jjaa227
Gao, J., Xu, K., Liu, H., Liu, G., Bai, M., Peng, C., et al. (2018). Impact of the gut microbiota on intestinal immunity mediated by tryptophan metabolism. Front. Cell. Infect. Microbiol. 8:13. doi: 10.3389/fcimb.2018.00013
Gevers, D., Kugathasan, S., Denson, L. A., Vázquez-Baeza, Y., Van Treuren, W., Ren, B., et al. (2014). The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe 15, 382–392. doi: 10.1016/j.chom.2014.02.005
Gillevet, P., Sikaroodi, M., Keshavarzian, A., and Mutlu, E. A. (2010). Quantitative assessment of the human gut microbiome using multitag pyrosequencing. Chem. Biodivers. 7, 1065–1075. doi: 10.1002/cbdv.200900322
Gophna, U., Sommerfeld, K., Gophna, S., Doolittle, W. F., and Veldhuyzen van Zanten, S. J. (2006). Differences between tissue-associated intestinal microfloras of patients with Crohn's disease and ulcerative colitis. J. Clin. Microbiol. 44, 4136–4141. doi: 10.1128/JCM.01004-06
Grasselli, G., Zangrillo, A., Zanella, A., Antonelli, M., Cabrini, L., Castelli, A., et al. (2020). Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA 323, 1574–1581. doi: 10.1001/jama.2020.5394
Gronke, K., Hernández, P. P., Zimmermann, J., Klose, C. S. N., Kofoed-Branzk, M., Guendel, F., et al. (2019). Interleukin-22 protects intestinal stem cells against genotoxic stress. Nature 566, 249–253. doi: 10.1038/s41586-019-0899-7
Gu, S., Chen, Y., Wu, Z., Chen, Y., Gao, H., Lv, L., et al. (2020). Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 71, 2669–2678. doi: 10.1093/cid/ciaa709
Guo, M., Tao, W., Flavell, R. A., and Zhu, S. (2021). Potential intestinal infection and faecal-oral transmission of SARS-CoV-2. Nat. Rev. Gastroenterol. Hepatol. 18, 269–283. doi: 10.1038/s41575-021-00416-6
Hagan, T., Cortese, M., Rouphael, N., Boudreau, C., Linde, C., Maddur, M. S., et al. (2019). Antibiotics-driven gut microbiome perturbation alters immunity to vaccines in humans. Cell 178, 1313–328.e13. doi: 10.1016/j.cell.2019.08.010
Halfvarson, J., Brislawn, C. J., Lamendella, R., Vázquez-Baeza, Y., Walters, W. A., Bramer, L. M., et al. (2017). Dynamics of the human gut microbiome in inflammatory bowel disease. Nat. Microbiol. 2:17004. doi: 10.1038/nmicrobiol.2017.4
Harper, A., Vijayakumar, V., Ouwehand, A. C., Ter Haar, J., Obis, D., Espadaler, J., et al. (2021). Viral infections, the microbiome, and probiotics. Front. Cell. Infect. Microbiol. 10:596166. doi: 10.3389/fcimb.2020.596166
Hasegawa, S., Ichiyama, T., Sonaka, I., Ohsaki, A., Hirano, R., Haneda, Y., et al. (2011). Amino acids exhibit antiinflammatory effects in human monocytic leukemia cell line, THP-1 cells. Inflamm. Res. 60, 1013–1019. doi: 10.1007/s00011-011-0362-1
He, F., Zhang, T., Xue, K., Fang, Z., Jiang, G., Huang, S., et al. (2021). Fecal multi-omics analysis reveals diverse molecular alterations of gut ecosystem in COVID-19 patients. Anal. Chim. Acta 1180:338881. doi: 10.1016/j.aca.2021.338881
Hecker, M., Sessa, W. C., Harris, H. J., Anggård, E. E., and Vane, J. R. (1990). The metabolism of L-arginine and its significance for the biosynthesis of endothelium-derived relaxing factor: cultured endothelial cells recycle L-citrulline to L-arginine. Proc. Natl. Acad. Sci. U. S. A. 87, 8612–8616. doi: 10.1073/pnas.87.21.8612
Hedin, C. R., McCarthy, N. E., Louis, P., Farquharson, F. M., McCartney, S., Taylor, K., et al. (2014). Altered intestinal microbiota and blood T cell phenotype are shared by patients with Crohn's disease and their unaffected siblings. Gut 63, 1578–1586. doi: 10.1136/gutjnl-2013-306226
Heidarian, F., Alebouyeh, M., Shahrokh, S., Balaii, H., and Zali, M. R. (2019). Altered fecal bacterial composition correlates with disease activity in inflammatory bowel disease and the extent of IL8 induction. Curr. Res. Transl. Med. 67, 41–50. doi: 10.1016/j.retram.2019.01.002
Hisamatsu, T., Okamoto, S., Hashimoto, M., Muramatsu, T., Andou, A., Uo, M., et al. (2012). Novel, objective, multivariate biomarkers composed of plasma amino acid profiles for the diagnosis and assessment of inflammatory bowel disease. PLoS One 7:e31131. doi: 10.1371/journal.pone.0031131
Hisamatsu, T., Ono, N., Imaizumi, A., Mori, M., Suzuki, H., Uo, M., et al. (2015). Decreased plasma histidine level predicts risk of relapse in patients with ulcerative colitis in remission. PLoS One 10:e0140716. doi: 10.1371/journal.pone.0140716
Hoarau, G., Mukherjee, P. K., Gower-Rousseau, C., Hager, C., Chandra, J., Retuerto, M. A., et al. (2016). Bacteriome and Mycobiome interactions underscore microbial dysbiosis in familial Crohn's disease. MBio 7, e01250–e01216. doi: 10.1128/mBio.01250-16
Holeček, M. (2020). Histidine in health and disease: metabolism, physiological importance, and use as a supplement. Nutrients 12:848. doi: 10.3390/nu12030848
Hoxha, M. (2020). What about COVID-19 and arachidonic acid pathway? Eur. J. Clin. Pharmacol. 76, 1501–1504. doi: 10.1007/s00228-020-02941-w
Hu, B., Huang, S., and Yin, L. (2021). The cytokine storm and COVID-19. J. Med. Virol. 93, 250–256. doi: 10.1002/jmv.26232
Huang, H., Vangay, P., McKinlay, C. E., and Knights, D. (2014). Multi-omics analysis of inflammatory bowel disease. Immunol. Lett. 162, 62–68. doi: 10.1016/j.imlet.2014.07.014
Hussain, I., Cher, G. L. Y., Abid, M. A., and Abid, M. B. (2021). Role of gut microbiome in COVID-19: An insight into pathogenesis and therapeutic potential. Front. Immunol. 12:765965. doi: 10.3389/fimmu.2021.765965
Irving, P. M., de Lusignan, S., Tang, D., Nijher, M., and Barrett, K. (2021). Risk of common infections in people with inflammatory bowel disease in primary care: a population-based cohort study. BMJ Open Gastroenterol. 8:e000573. doi: 10.1136/bmjgast-2020-000573
Jacobs, J. P., Goudarzi, M., Singh, N., Tong, M., McHardy, I. H., Ruegger, P., et al. (2016). A disease-associated microbial and metabolomics state in relatives of pediatric inflammatory bowel disease patients. Cell. Mol. Gastroenterol. Hepatol. 2, 750–766. doi: 10.1016/j.jcmgh.2016.06.004
Jagt, J. Z., Struys, E. A., Ayada, I., Bakkali, A., Jansen, E. E. W., Claesen, J., et al. (2021). Fecal amino acid analysis in newly diagnosed pediatric inflammatory bowel disease: a multicenter case-control study. Inflamm. Bowel Dis. izab256. doi: 10.1093/ibd/izab256 [Epub ahead of print].
Jansson, J., Willing, B., Lucio, M., Fekete, A., Dicksved, J., Halfvarson, J., et al. (2009). Metabolomics reveals metabolic biomarkers of Crohn's disease. PLoS One 4:e6386. doi: 10.1371/journal.pone.0006386
Joossens, M., Huys, G., Cnockaert, M., De Preter, V., Verbeke, K., Rutgeerts, P., et al. (2011). Dysbiosis of the faecal microbiota in patients with Crohn's disease and their unaffected relatives. Gut 60, 631–637. doi: 10.1136/gut.2010.223263
Kabeerdoss, J., Jayakanthan, P., Pugazhendhi, S., and Ramakrishna, B. S. (2015). Alterations of mucosal microbiota in the colon of patients with inflammatory bowel disease revealed by real time polymerase chain reaction amplification of 16S ribosomal ribonucleic acid. Indian J. Med. Res. 142, 23–32. doi: 10.4103/0971-5916.162091
Kabeerdoss, J., Sankaran, V., Pugazhendhi, S., and Ramakrishna, B. S. (2013). Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol. 13:20. doi: 10.1186/1471-230X-13-20
Kang, S., Denman, S. E., Morrison, M., Yu, Z., Dore, J., Leclerc, M., et al. (2010). Dysbiosis of fecal microbiota in Crohn's disease patients as revealed by a custom phylogenetic microarray. Inflamm. Bowel Dis. 16, 2034–2042. doi: 10.1002/ibd.21319
Kang, P., Kalloniatis, M., and Doig, G. S. (2021). Using updated PubMed: new features and functions to enhance literature searches. JAMA 326, 479–480. doi: 10.1001/jama.2021.12021
Keshteli, A. H., van den Brand, F. F., Madsen, K. L., Mandal, R., Valcheva, R., Kroeker, K. I., et al. (2017). Dietary and metabolomic determinants of relapse in ulcerative colitis patients: a pilot prospective cohort study. World J. Gastroenterol. 23, 3890–3899. doi: 10.3748/wjg.v23.i21.3890
Kesika, P., Sivamaruthi, B. S., Thangaleela, S., and Chaiyasut, C. (2021). The antiviral potential of probiotics—a review on scientific outcomes. Appl. Sci. 11:8687. doi: 10.3390/app11188687
Khan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., et al. (2019). Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8:126. doi: 10.3390/pathogens8030126
Kim, M. H., and Kim, H. (2017). The roles of glutamine in the intestine and its implication in intestinal diseases. Int. J. Mol. Sci. 18:1051. doi: 10.3390/ijms18051051
Kimhofer, T., Lodge, S., Whiley, L., Gray, N., Loo, R. L., Lawler, N. G., et al. (2020). Integrative modeling of quantitative plasma lipoprotein, metabolic, and amino acid data reveals a multiorgan pathological signature of SARS-CoV-2 infection. J. Proteome Res. 19, 4442–4454. doi: 10.1021/acs.jproteome.0c00519
Knights, D., Silverberg, M. S., Weersma, R. K., Gevers, D., Dijkstra, G., Huang, H., et al. (2014). Complex host genetics influence the microbiome in inflammatory bowel disease. Genome Med. 6:107. doi: 10.1186/s13073-014-0107-1
Knyazev, E., Nersisyan, S., and Tonevitsky, A. (2021). Endocytosis and Transcytosis of SARS-CoV-2 across the intestinal epithelium and other tissue barriers. Front. Immunol. 12:636966. doi: 10.3389/fimmu.2021.636966
Kohashi, M., Nishiumi, S., Ooi, M., Yoshie, T., Matsubara, A., Suzuki, M., et al. (2014). A novel gas chromatography mass spectrometry-based serum diagnostic and assessment approach to ulcerative colitis. J. Crohns Colitis 8, 1010–1021. doi: 10.1016/j.crohns.2014.01.024
Kolho, K. L., Pessia, A., Jaakkola, T., de Vos, W. M., and Velagapudi, V. (2017). Faecal and serum metabolomics in paediatric inflammatory bowel disease. J. Crohns Colitis 11, 321–334. doi: 10.1093/ecco-jcc/jjw158
Kong, C., Yan, X., Liu, Y., Huang, L., Zhu, Y., He, J., et al. (2021). Ketogenic diet alleviates colitis by reduction of colonic group 3 innate lymphoid cells through altering gut microbiome. Signal Transduct. Target. Ther. 6:154. doi: 10.1038/s41392-021-00549-9
Kretzmann, N. A., Fillmann, H., Mauriz, J. L., Marroni, C. A., Marroni, N., González-Gallego, J., et al. (2008). Effects of glutamine on proinflammatory gene expression and activation of nuclear factor kappa B and signal transducers and activators of transcription in TNBS-induced colitis. Inflamm. Bowel Dis. 14, 1504–1513. doi: 10.1002/ibd.20543
Kumari, R., Ahuja, V., and Paul, J. (2013). Fluctuations in butyrate-producing bacteria in ulcerative colitis patients of North India. World J. Gastroenterol. 19, 3404–3414. doi: 10.3748/wjg.v19.i22.3404
Laffel, L. (1999). Ketone bodies: a review of physiology, pathophysiology and application of monitoring to diabetes. Diabetes Metab. Res. Rev. 15, 412–426. doi: 10.1002/(SICI)1520-7560(199911/12)15:6<412::AID-DMRR72>3.0.CO;2-8
Lai, Y., Xue, J., Liu, C. W., Gao, B., Chi, L., Tu, P., et al. (2019). Serum metabolomics identifies altered bioenergetics, signaling cascades in parallel with exposome markers in Crohn's disease. Molecules 24:449. doi: 10.3390/molecules24030449
Lamas, B., Natividad, J. M., and Sokol, H. (2018). Aryl hydrocarbon receptor and intestinal immunity. Mucosal Immunol. 11, 1024–1038. doi: 10.1038/s41385-018-0019-2
Lamas, B., Richard, M. L., Leducq, V., Pham, H. P., Michel, M. L., Da Costa, G., et al. (2016). CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 22, 598–605. doi: 10.1038/nm.4102
Lamers, M. M., Beumer, J., van der Vaart, J., Knoops, K., Puschhof, J., Breugem, T. I., et al. (2020). SARS-CoV-2 productively infects human gut enterocytes. Science 369, 50–54. doi: 10.1126/science.abc1669
Laserna-Mendieta, E. J., Clooney, A. G., Carretero-Gomez, J. F., Moran, C., Sheehan, D., Nolan, J. A., et al. (2018). Determinants of reduced genetic capacity for butyrate synthesis by the gut microbiome in Crohn's disease and ulcerative colitis. J. Crohns Colitis 12, 204–216. doi: 10.1093/ecco-jcc/jjx137
Lawler, N. G., Gray, N., Kimhofer, T., Boughton, B., Gay, M., Yang, R., et al. (2021). Systemic perturbations in amine and kynurenine metabolism associated with acute SARS-CoV-2 infection and inflammatory cytokine responses. J. Proteome Res. 20, 2796–2811. doi: 10.1021/acs.jproteome.1c00052
Lee, J. G., Han, D. S., Jo, S. V., Lee, A. R., Park, C. H., Eun, C. S., et al. (2019). Characteristics and pathogenic role of adherent-invasive Escherichia coli in inflammatory bowel disease: potential impact on clinical outcomes. PLoS One 14:e0216165. doi: 10.1371/journal.pone.0216165
Lehmann, M., Allers, K., Heldt, C., Meinhardt, J., Schmidt, F., Rodriguez-Sillke, Y., et al. (2021). Human small intestinal infection by SARS-CoV-2 is characterized by a mucosal infiltration with activated CD8+ T cells. Mucosal Immunol. 14, 1381–1392. doi: 10.1038/s41385-021-00437-z
Lepage, P., Seksik, P., Sutren, M., de la Cochetière, M. F., Jian, R., Marteau, P., et al. (2005). Biodiversity of the mucosa-associated microbiota is stable along the distal digestive tract in healthy individuals and patients with IBD. Inflamm. Bowel Dis. 11, 473–480. doi: 10.1097/01.MIB.0000159662.62651.06
Levy, E., Rizwan, Y., Thibault, L., Lepage, G., Brunet, S., Bouthillier, L., et al. (2000). Altered lipid profile, lipoprotein composition, and oxidant and antioxidant status in pediatric Crohn disease. Am. J. Clin. Nutr. 71, 807–815. doi: 10.1093/ajcn/71.3.807
Li, J., Wang, X., Chen, J., Zuo, X., Zhang, H., and Deng, A. (2020). COVID-19 infection may cause ketosis and ketoacidosis. Diabetes Obes. Metab. 22, 1935–1941. doi: 10.1111/dom.14057
Li, Q., Wang, C., Tang, C., Li, N., and Li, J. (2012). Molecular-phylogenetic characterization of the microbiota in ulcerated and non-ulcerated regions in the patients with Crohn's disease. PLoS One 7:e34939. doi: 10.1371/journal.pone.0034939
Liu, X., and Locasale, J. W. (2017). Metabolomics: A primer. Trends Biochem. Sci. 42, 274–284. doi: 10.1016/j.tibs.2017.01.004
Liu, X., Zhao, F., Liu, H., Xie, Y., Zhao, D., and Li, C. (2021). Transcriptomics and metabolomics reveal the adaption of Akkermansia muciniphila to high mucin by regulating energy homeostasis. Sci. Rep. 11:9073. doi: 10.1038/s41598-021-88397-z
Lloyd-Price, J., Arze, C., Ananthakrishnan, A. N., Schirmer, M., Avila-Pacheco, J., Poon, T. W., et al. (2019). Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662. doi: 10.1038/s41586-019-1237-9
Lo Presti, A., Zorzi, F., Del Chierico, F., Altomare, A., Cocca, S., Avola, A., et al. (2019). Fecal and mucosal microbiota profiling in irritable bowel syndrome and inflammatory bowel disease. Front. Microbiol. 10:1655. doi: 10.3389/fmicb.2019.01655
Łodyga, M., Maciejewska, K., Eder, P., Waszak, K., Stawczyk-Eder, K., Michalak, M., et al. (2021). Inflammatory bowel disease is associated with higher seroprevalence rates of antibodies against SARS-CoV-2. Pol. Arch. Intern. Med. 131, 226–232. doi: 10.20452/pamw.15796
Macfarlane, S., Furrie, E., Cummings, J. H., and Macfarlane, G. T. (2004). Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin. Infect. Dis. 38, 1690–1699. doi: 10.1086/420823
Machiels, K., Joossens, M., Sabino, J., De Preter, V., Arijs, I., Eeckhaut, V., et al. (2014). A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283. doi: 10.1136/gutjnl-2013-304833
Macias-Ceja, D. C., Ortiz-Masiá, D., Salvador, P., Gisbert-Ferrándiz, L., Hernández, C., Hausmann, M., et al. (2019). Succinate receptor mediates intestinal inflammation and fibrosis. Mucosal Immunol. 12, 178–187. doi: 10.1038/s41385-018-0087-3
Magalhães, N. S., Savino, W., Silva, P. M. R., Martins, M. A., and Carvalho, V. F. (2021). Gut microbiota dysbiosis is a crucial player for the poor outcomes for COVID-19 in elderly, diabetic and hypertensive patients. Front. Med. 8:644751. doi: 10.3389/fmed.2021.644751
Majmundar, A. J., Wong, W. J., and Simon, M. C. (2010). Hypoxia-inducible factors and the response to hypoxic stress. Mol. Cell 40, 294–309. doi: 10.1016/j.molcel.2010.09.022
Manfredi, M., Conte, E., Barberis, E., Buzzi, A., Robotti, E., Caneparo, V., et al. (2019). Integrated serum proteins and fatty acids analysis for putative biomarker discovery in inflammatory bowel disease. J. Proteome 195, 138–149. doi: 10.1016/j.jprot.2018.10.017
Manichanh, C., Rigottier-Gois, L., Bonnaud, E., Gloux, K., Pelletier, E., Frangeul, L., et al. (2006). Reduced diversity of faecal microbiota in Crohn's disease revealed by a metagenomic approach. Gut 55, 205–211. doi: 10.1136/gut.2005.073817
Mar, J. S., LaMere, B. J., Lin, D. L., Levan, S., Nazareth, M., Mahadevan, U., et al. (2016). Disease severity and immune activity relate to distinct Interkingdom gut microbiome states in ethnically distinct ulcerative colitis patients. MBio 7, e01072–e01016. doi: 10.1128/mBio.01072-16
Marchesi, J. R., Holmes, E., Khan, F., Kochhar, S., Scanlan, P., Shanahan, F., et al. (2007). Rapid and noninvasive metabonomic characterization of inflammatory bowel disease. J. Proteome Res. 6, 546–551. doi: 10.1021/pr060470d
Marshall, M. (2021). COVID and the brain: researchers zero in on how damage occurs. Nature 595, 484–485. doi: 10.1038/d41586-021-01693-6
Martin, H. M., Campbell, B. J., Hart, C. A., Mpofu, C., Nayar, M., Singh, R., et al. (2004). Enhanced Escherichia coli adherence and invasion in Crohn's disease and colon cancer. Gastroenterology 127, 80–93. doi: 10.1053/j.gastro.2004.03.054
Matés, J. M., Pérez-Gómez, C., Núñez de Castro, I., Asenjo, M., and Márquez, J. (2002). Glutamine and its relationship with intracellular redox status, oxidative stress and cell proliferation/death. Int. J. Biochem. Cell Biol. 34, 439–458. doi: 10.1016/S1357-2725(01)00143-1
Matthews, D. E. (2007). An overview of phenylalanine and tyrosine kinetics in humans. J. Nutr. 137, 1549S–1555S; discussion 1573S-1575S. doi: 10.1093/jn/137.6.1549S
Mazza, S., Sorce, A., Peyvandi, F., Vecchi, M., and Caprioli, F. (2020). A fatal case of COVID-19 pneumonia occurring in a patient with severe acute ulcerative colitis. Gut 69, 1148–1149. doi: 10.1136/gutjnl-2020-321183
Mei, Q., Wang, A. Y., Bryant, A., Yang, Y., Li, M., Wang, F., et al. (2021). Survival factors and metabolic pathogenesis in elderly patients (≥65) with COVID-19: A multi-center study. Front. Med. 7:595503. doi: 10.3389/fmed.2020.595503
Meoni, G., Ghini, V., Maggi, L., Vignoli, A., Mazzoni, A., Salvati, L., et al. (2021). Metabolomic/lipidomic profiling of COVID-19 and individual response to tocilizumab. PLoS Pathog. 17:e1009243. doi: 10.1371/journal.ppat.1009243
Mesnage, R., and Antoniou, M. N. (2020). Computational modelling provides insight into the effects of glyphosate on the shikimate pathway in the human gut microbiome. Curr. Res. Toxicol. 1, 25–33. doi: 10.1016/j.crtox.2020.04.001
Michail, S., Durbin, M., Turner, D., Griffiths, A. M., Mack, D. R., Hyams, J., et al. (2012). Alterations in the gut microbiome of children with severe ulcerative colitis. Inflamm. Bowel Dis. 18, 1799–1808. doi: 10.1002/ibd.22860
Mills, E., and O'Neill, L. A. (2014). Succinate: a metabolic signal in inflammation. Trends Cell Biol. 24, 313–320. doi: 10.1016/j.tcb.2013.11.008
Mondot, S., Kang, S., Furet, J. P., Aguirre de Carcer, D., McSweeney, C., Morrison, M., et al. (2011, 2011). Highlighting new phylogenetic specificities of Crohn's disease microbiota. Inflamm. Bowel Dis. 17, 185–192. doi: 10.1002/ibd.21436
Monteleone, G., and Ardizzone, S. (2020). Are patients with inflammatory bowel disease at increased risk for Covid-19 infection? J. Crohns Colitis 14, 1334–1336. doi: 10.1093/ecco-jcc/jjaa061
Monteleone, I., Rizzo, A., Sarra, M., Sica, G., Sileri, P., Biancone, L., et al. (2011). Aryl hydrocarbon receptor-induced signals up-regulate IL-22 production and inhibit inflammation in the gastrointestinal tract. Gastroenterology 141, 237–248.e1. doi: 10.1053/j.gastro.2011.04.007
Morais, L. H., Schreiber, H. L., and Mazmanian, S. K. (2021). The gut microbiota-brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 19, 241–255. doi: 10.1038/s41579-020-00460-0
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol. 13:R79. doi: 10.1186/gb-2012-13-9-r79
Moro, J., Tomé, D., Schmidely, P., Demersay, T. C., and Azzout-Marniche, D. (2020). Histidine: a systematic review on metabolism and physiological effects in human and different animal species. Nutrients 12:1414. doi: 10.3390/nu12051414
Morone, G., Palomba, A., Iosa, M., Caporaso, T., De Angelis, D., Venturiero, V., et al. (2020). Incidence and persistence of viral shedding in COVID-19 post-acute patients With Negativized pharyngeal swab: a systematic review. Front. Med. 7:562. doi: 10.3389/fmed.2020.00562
Müller, C., Hardt, M., Schwudke, D., Neuman, B. W., Pleschka, S., and Ziebuhr, J. (2018). Inhibition of cytosolic phospholipase A2α impairs an early step of coronavirus replication in cell culture. J. Virol. 92, e01463–e01417. doi: 10.1128/JVI.01463-17
Murakami, M., Sato, H., and Taketomi, Y. (2020). Updating phospholipase A2 biology. Biomol. Ther. 10:1457. doi: 10.3390/biom10101457
Mussap, M., and Fanos, V. (2021). Could metabolomics drive the fate of COVID-19 pandemic? A narrative review on lights and shadows. Clin. Chem. Lab. Med. 59, 1891–1905. doi: 10.1515/cclm-2021-0414
Nadalian, B., Yadegar, A., Houri, H., Olfatifar, M., Shahrokh, S., Asadzadeh Aghdaei, H., et al. (2021). Prevalence of the pathobiont adherent-invasive Escherichia coli and inflammatory bowel disease: a systematic review and meta-analysis. J. Gastroenterol. Hepatol. 36, 852–863. doi: 10.1111/jgh.15260
Naftali, T., Reshef, L., Kovacs, A., Porat, R., Amir, I., Konikoff, F. M., et al. (2016). Distinct microbiotas are associated with ileum-restricted and colon-involving Crohn's disease. Inflamm. Bowel Dis. 22, 293–302. doi: 10.1097/MIB.0000000000000662
Nash, J. H., Villegas, A., Kropinski, A. M., Aguilar-Valenzuela, R., Konczy, P., Mascarenhas, M., et al. (2010). Genome sequence of adherent-invasive Escherichia coli and comparative genomic analysis with other E. coli pathotypes. BMC Genomics 11:667. doi: 10.1186/1471-2164-11-667
Nemoto, H., Kataoka, K., Ishikawa, H., Ikata, K., Arimochi, H., Iwasaki, T., et al. (2012). Reduced diversity and imbalance of fecal microbiota in patients with ulcerative colitis. Dig. Dis. Sci. 57, 2955–2964. doi: 10.1007/s10620-012-2236-y
Neurath, M. F. (2019). Targeting immune cell circuits and trafficking in inflammatory bowel disease. Nat. Immunol. 20, 970–979. doi: 10.1038/s41590-019-0415-0
Neurath, M. F. (2020). COVID-19 and immunomodulation in IBD. Gut 69, 1335–1342. doi: 10.1136/gutjnl-2020-321269
Newman, J. C., and Verdin, E. (2014). Ketone bodies as signaling metabolites. Trends Endocrinol. Metab. 25, 42–52. doi: 10.1016/j.tem.2013.09.002
Ng, S. C., Shi, H. Y., Hamidi, N., Underwood, F. E., Tang, W., Benchimol, E. I., et al. (2017). Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet 390, 2769–2778. doi: 10.1016/S0140-6736(17)32448-0
Nikolaus, S., Schulte, B., Al-Massad, N., Thieme, F., Schulte, D. M., Bethge, J., et al. (2017). Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 153, 1504–516.e2. doi: 10.1053/j.gastro.2017.08.028
Nishino, K., Nishida, A., Inoue, R., Kawada, Y., Ohno, M., Sakai, S., et al. (2018). Analysis of endoscopic brush samples identified mucosa-associated dysbiosis in inflammatory bowel disease. J. Gastroenterol. 53, 95–106. doi: 10.1007/s00535-017-1384-4
Norsa, L., Cosimo, P., Indriolo, A., Sansotta, N., D'Antiga, L., and Callegaro, A. (2020). Asymptomatic severe acute respiratory syndrome coronavirus 2 infection in patients with inflammatory bowel disease under biologic treatment. Gastroenterology 159, 2229–31.e2. doi: 10.1053/j.gastro.2020.08.046
Oladimeji, P. O., and Chen, T. (2018). PXR: more than just a master xenobiotic receptor. Mol. Pharmacol. 93, 119–127. doi: 10.1124/mol.117.110155
Ooi, M., Nishiumi, S., Yoshie, T., Shiomi, Y., Kohashi, M., Fukunaga, K., et al. (2011). GC/MS-based profiling of amino acids and TCA cycle-related molecules in ulcerative colitis. Inflamm. Res. 60, 831–840. doi: 10.1007/s00011-011-0340-7
Ott, S. J., Musfeldt, M., Wenderoth, D. F., Hampe, J., Brant, O., Fölsch, U. R., et al. (2004). Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53, 685–693. doi: 10.1136/gut.2003.025403
Ottman, N., Reunanen, J., Meijerink, M., Pietilä, T. E., Kainulainen, V., Klievink, J., et al. (2017). Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One 12:e0173004. doi: 10.1371/journal.pone.0173004
Páez-Franco, J. C., Torres-Ruiz, J., Sosa-Hernández, V. A., Cervantes-Díaz, R., Romero-Ramírez, S., Pérez-Fragoso, A., et al. (2021). Metabolomics analysis reveals a modified amino acid metabolism that correlates with altered oxygen homeostasis in COVID-19 patients. Sci. Rep. 11:6350. doi: 10.1038/s41598-021-85788-0
Palmela, C., Chevarin, C., Xu, Z., Torres, J., Sevrin, G., Hirten, R., et al. (2018). Adherent-invasive Escherichia coli in inflammatory bowel disease. Gut 67, 574–587. doi: 10.1136/gutjnl-2017-314903
Park, A., Kim, S., Jung, I. H., and Byun, J. H. (2020). An immune therapy model for effective treatment on inflammatory bowel disease. PLoS One 15:e0238918. doi: 10.1371/journal.pone.0238918
Pascal, V., Pozuelo, M., Borruel, N., Casellas, F., Campos, D., Santiago, A., et al. (2017). A microbial signature for Crohn's disease. Gut 66, 813–822. doi: 10.1136/gutjnl-2016-313235
Perna, S., Alalwan, T. A., Alaali, Z., Alnashaba, T., Gasparri, C., Infantino, V., et al. (2019). The role of glutamine in the complex interaction between gut microbiota and health: a narrative review. Int. J. Mol. Sci. 20:5232. doi: 10.3390/ijms20205232
Prideaux, L., Kang, S., Wagner, J., Buckley, M., Mahar, J. E., De Cruz, P., et al. (2013). Impact of ethnicity, geography, and disease on the microbiota in health and inflammatory bowel disease. Inflamm. Bowel Dis. 19, 2906–2918. doi: 10.1097/01.MIB.0000435759.05577.12
Probert, F., Walsh, A., Jagielowicz, M., Yeo, T., Claridge, T. D. W., Simmons, A., et al. (2018). Plasma nuclear magnetic resonance metabolomics discriminates between high and low endoscopic activity and predicts progression in a prospective cohort of patients with ulcerative colitis. J. Crohns Colitis 12, 1326–1337. doi: 10.1093/ecco-jcc/jjy101
Qi, F., Qian, S., Zhang, S., and Zhang, Z. (2020). Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 526, 135–140. doi: 10.1016/j.bbrc.2020.03.044
Qian, Q., Fan, L., Liu, W., Li, J., Yue, J., Wang, M., et al. (2021). Direct evidence of active SARS-CoV-2 replication in the intestine. Clin. Infect. Dis. 73, 361–366. doi: 10.1093/cid/ciaa925
Qiu, J., Heller, J. J., Guo, X., Chen, Z. M., Fish, K., Fu, Y. X., et al. (2012). The aryl hydrocarbon receptor regulates gut immunity through modulation of innate lymphoid cells. Immunity 36, 92–104. doi: 10.1016/j.immuni.2011.11.011
Rajilić-Stojanović, M., Shanahan, F., Guarner, F., and de Vos, W. M. (2013). Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflamm. Bowel Dis. 19, 481–488. doi: 10.1097/MIB.0b013e31827fec6d
Rausch, P., Rehman, A., Künzel, S., Häsler, R., Ott, S. J., Schreiber, S., et al. (2011). Colonic mucosa-associated microbiota is influenced by an interaction of Crohn disease and FUT2 (secretor) genotype. Proc. Natl. Acad. Sci. U. S. A. 108, 19030–19035. doi: 10.1073/pnas.1106408108
Rehman, A., Lepage, P., Nolte, A., Hellmig, S., Schreiber, S., and Ott, S. J. (2010). Transcriptional activity of the dominant gut mucosal microbiota in chronic inflammatory bowel disease patients. J. Med. Microbiol. 59, 1114–1122. doi: 10.1099/jmm.0.021170-0
Reigstad, C. S., Salmonson, C. E., Rainey, J. F. 3rd, Szurszewski, J. H., Linden, D. R., Sonnenburg, J. L., et al. (2015). Gut microbes promote colonic serotonin production through an effect of short-chain fatty acids on enterochromaffin cells. FASEB J. 29, 1395–1403. doi: 10.1096/fj.14-259598
Rhoads, J. M., Argenzio, R. A., Chen, W., Rippe, R. A., Westwick, J. K., Cox, A. D., et al. (1997). L-glutamine stimulates intestinal cell proliferation and activates mitogen-activated protein kinases. Am. J. Phys. 272, G943–G953. doi: 10.1152/ajpgi.1997.272.5.G943
Roager, H. M., and Licht, T. R. (2018). Microbial tryptophan catabolites in health and disease. Nat. Commun. 9:3294. doi: 10.1038/s41467-018-05470-4
Ryan, F. J., Ahern, A. M., Fitzgerald, R. S., Laserna-Mendieta, E. J., Power, E. M., Clooney, A. G., et al. (2020). Colonic microbiota is associated with inflammation and host epigenomic alterations in inflammatory bowel disease. Nat. Commun. 11:1512. doi: 10.1038/s41467-020-15342-5
San Juan, I., Bruzzone, C., Bizkarguenaga, M., Bernardo-Seisdedos, G., Laín, A., Gil-Redondo, R., et al. (2020). Abnormal concentration of porphyrins in serum from COVID-19 patients. Br. J. Haematol. 190, e265–e267. doi: 10.1111/bjh.17060
Santoru, M. L., Piras, C., Murgia, A., Palmas, V., Camboni, T., Liggi, S., et al. (2017). A. Cross sectional evaluation of the gut-microbiome metabolome axis in an Italian cohort of IBD patients. Sci. Rep. 7:9523. doi: 10.1038/s41598-017-10034-5
Sappati Biyyani, R. S., Putka, B. S., and Mullen, K. D. (2010). Dyslipidemia and lipoprotein profiles in patients with inflammatory bowel disease. J. Clin. Lipidol. 4, 478–482. doi: 10.1016/j.jacl.2010.08.021
Satsangi, J., Silverberg, M. S., Vermeire, S., and Colombel, J. F. (2006). The Montreal classification of inflammatory bowel disease: controversies, consensus, and implications. Gut 55, 749–753. doi: 10.1136/gut.2005.082909
Savitz, J. (2020). The kynurenine pathway: a finger in every pie. Mol. Psychiatry 25, 131–147. doi: 10.1038/s41380-019-0414-4
Scanlan, P. D., Shanahan, F., O'Mahony, C., and Marchesi, J. R. (2006). Culture-independent analyses of temporal variation of the dominant fecal microbiota and targeted bacterial subgroups in Crohn's disease. J. Clin. Microbiol. 44, 3980–3988. doi: 10.1128/JCM.00312-06
Schicho, R., Shaykhutdinov, R., Ngo, J., Nazyrova, A., Schneider, C., Panaccione, R., et al. (2012). Quantitative metabolomic profiling of serum, plasma, and urine by (1)H NMR spectroscopy discriminates between patients with inflammatory bowel disease and healthy individuals. J. Proteome Res. 11, 3344–3357. doi: 10.1021/pr300139q
Scholl-Bürgi, S., Schroecksnadel, S., Jenny, M., Karall, D., and Fuchs, D. (2013). Chronic immune stimulation may cause moderate impairment of phenylalanine 4-hydroxylase. Pteridines 22, 120–125. doi: 10.1515/pteridines.2011.22.1.120
Schütze, S., Potthoff, K., Machleidt, T., Berkovic, D., Wiegmann, K., and Krönke, M. (1992). TNF activates NF-kappa B by phosphatidylcholine-specific phospholipase C-induced "acidic" sphingomyelin breakdown. Cell 71, 765–776. doi: 10.1016/0092-8674(92)90553-O
Schwarz, B., Sharma, L., Roberts, L., Peng, X., Bermejo, S., Leighton, I., et al. (2021). Cutting edge: severe SARS-CoV-2 infection in humans is defined by a shift in the serum lipidome, resulting in dysregulation of eicosanoid immune mediators. J. Immunol. 206, 329–334. doi: 10.4049/jimmunol.2001025
Schwiertz, A., Jacobi, M., Frick, J. S., Richter, M., Rusch, K., and Köhler, H. (2010). Microbiota in pediatric inflammatory bowel disease. J. Pediatr. 157, 240–244.e1. doi: 10.1016/j.jpeds.2010.02.046
Scoville, E. A., Allaman, M. M., Brown, C. T., Motley, A. K., Horst, S. N., Williams, C. S., et al. (2018). Alterations in lipid, amino acid, and energy metabolism distinguish Crohn's disease from ulcerative colitis and control subjects by serum metabolomic profiling. Metabolomics 14:17. doi: 10.1007/s11306-017-1311-y
Seksik, P., Rigottier-Gois, L., Gramet, G., Sutren, M., Pochart, P., Marteau, P., et al. (2003). Alterations of the dominant faecal bacterial groups in patients with Crohn's disease of the colon. Gut 52, 237–242. doi: 10.1136/gut.52.2.237
Sencio, V., Machado, M. G., and Trottein, F. (2021). The lung-gut axis during viral respiratory infections: the impact of gut dysbiosis on secondary disease outcomes. Mucosal Immunol. 14, 296–304. doi: 10.1038/s41385-020-00361-8
Serhan, C. N., Clish, C. B., Brannon, J., Colgan, S. P., Chiang, N., and Gronert, K. (2000). Novel functional sets of lipid-derived mediators with antiinflammatory actions generated from omega-3 fatty acids via cyclooxygenase 2-nonsteroidal antiinflammatory drugs and transcellular processing. J. Exp. Med. 192, 1197–1204. doi: 10.1084/jem.192.8.1197
Sha, S., Xu, B., Wang, X., Zhang, Y., Wang, H., Kong, X., et al. (2013). The biodiversity and composition of the dominant fecal microbiota in patients with inflammatory bowel disease. Diagn. Microbiol. Infect. Dis. 75, 245–251. doi: 10.1016/j.diagmicrobio.2012.11.022
Shah, E. D., Farida, J. P., Siegel, C. A., Chong, K., and Melmed, G. Y. (2017). Risk for overall infection with anti-TNF and antiintegrin agents used in IBD: A systematic review and meta-analysis. Inflamm. Bowel Dis. 23, 570–577. doi: 10.1097/MIB.0000000000001049
Shaheen, A. (2021). Can ketone bodies inactivate coronavirus spike protein? The potential of biocidal agents against SARS-CoV-2. BioEssays 43:e2000312. doi: 10.1002/bies.202000312
Shahir, N. M., Wang, J. R., Wolber, E. A., Schaner, M. S., Frank, D. N., Ir, D., et al. (2020). Crohn's disease differentially affects region-specific composition and aerotolerance profiles of mucosally adherent bacteria. Inflamm. Bowel Dis. 26, 1843–1855. doi: 10.1093/ibd/izaa103
Shen, B., Yi, X., Sun, Y., Bi, X., Du, J., Zhang, C., et al. (2020). Proteomic and metabolomic characterization of COVID-19 patient sera. Cell 182, 59–72.e15. doi: 10.1016/j.cell.2020.05.032
Shi, D., Yan, R., Lv, L., Jiang, H., Lu, Y., Sheng, J., et al. (2021). The serum metabolome of COVID-19 patients is distinctive and predictive. Metabolism 118:154739. doi: 10.1016/j.metabol.2021.154739
Shores, D. R., Binion, D. G., Freeman, B. A., and Baker, P. R. (2011). New insights into the role of fatty acids in the pathogenesis and resolution of inflammatory bowel disease. Inflamm. Bowel Dis. 17, 2192–2204. doi: 10.1002/ibd.21560
Sido, B., Seel, C., Hochlehnert, A., Breitkreutz, R., and Dröge, W. (2006). Low intestinal glutamine level and low glutaminase activity in Crohn's disease: a rational for glutamine supplementation? Dig. Dis. Sci. 51, 2170–2179. doi: 10.1007/s10620-006-9473-x
Sokol, H., Leducq, V., Aschard, H., Pham, H. P., Jegou, S., Landman, C., et al. (2017). Fungal microbiota dysbiosis in IBD. Gut 66, 1039–1048. doi: 10.1136/gutjnl-2015-310746
Son, D. O., Satsu, H., and Shimizu, M. (2005). Histidine inhibits oxidative stress- and TNF-alpha-induced interleukin-8 secretion in intestinal epithelial cells. FEBS Lett. 579, 4671–4677. doi: 10.1016/j.febslet.2005.07.038
Song, J. W., Lam, S. M., Fan, X., Cao, W. J., Wang, S. Y., Tian, H., et al. (2020). Omics-driven systems interrogation of metabolic dysregulation in COVID-19 pathogenesis. Cell Metab. 32, 188–202.e5. doi: 10.1016/j.cmet.2020.06.016
Sprenger, G. A. (2006). “Aromatic amino acids,” in Amino Acid Biosynthesis - Pathways, Regulation and Metabolic Engineering. Microbiology Monographs. Vol. 5. ed. V. F. Wendisch (Berlin, Heidelberg: Springer), 93–127.
Stasi, C., Sadalla, S., and Milani, S. (2019). The relationship Between the serotonin metabolism, gut-microbiota and the gut-brain axis. Curr. Drug Metab. 20, 646–655. doi: 10.2174/1389200220666190725115503
Stephens, N. S., Siffledeen, J., Su, X., Murdoch, T. B., Fedorak, R. N., and Slupsky, C. M. (2013). Urinary NMR metabolomic profiles discriminate inflammatory bowel disease from healthy. J. Crohns Colitis 7, e42–e48. doi: 10.1016/j.crohns.2012.04.019
Stockinger, B., Di Meglio, P., Gialitakis, M., and Duarte, J. H. (2014). The aryl hydrocarbon receptor: multitasking in the immune system. Annu. Rev. Immunol. 32, 403–432. doi: 10.1146/annurev-immunol-032713-120245
Stubbs, B. J., Koutnik, A. P., Goldberg, E. L., Upadhyay, V., Turnbaugh, P. J., Verdin, E., et al. (2020). Investigating ketone bodies as immunometabolic countermeasures against respiratory viral infections. Med 1, 43–65. doi: 10.1016/j.medj.2020.06.008
Sugihara, K., Morhardt, T. L., and Kamada, N. (2019). The role of dietary nutrients in inflammatory bowel disease. Front. Immunol. 9:3183. doi: 10.3389/fimmu.2018.03183
Sultan, K., Mone, A., Durbin, L., Khuwaja, S., and Swaminath, A. (2020). Review of inflammatory bowel disease and COVID-19. World J. Gastroenterol. 26, 5534–5542. doi: 10.3748/wjg.v26.i37.5534
Swierkosz, T. A., Mitchell, J. A., Sessa, W. C., Hecker, M., and Vane, J. R. (1990). L-glutamine inhibits the release of endothelium-derived relaxing factor from the rabbit aorta. Biochem. Biophys. Res. Commun. 172, 143–148. doi: 10.1016/S0006-291X(05)80184-6
Takahashi, K., Nishida, A., Fujimoto, T., Fujii, M., Shioya, M., Imaeda, H., et al. (2016). Reduced abundance of butyrate-producing bacteria species in the fecal microbial Community in Crohn's disease. Digestion 93, 59–65. doi: 10.1159/000441768
Tallima, H., and El Ridi, R. (2017). Arachidonic acid: physiological roles and potential health benefits - a review. J. Adv. Res. 11, 33–41. doi: 10.1016/j.jare.2017.11.004
Tang, L., Gu, S., Gong, Y., Li, B., Lu, H., Li, Q., et al. (2020). Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering 6, 1178–1184. doi: 10.1016/j.eng.2020.05.013
Tao, W., Zhang, G., Wang, X., Guo, M., Zeng, W., Xu, Z., et al. (2020). Analysis of the intestinal microbiota in COVID-19 patients and its correlation with the inflammatory factor IL-18. Med. Microecol. 5:100023. doi: 10.1016/j.medmic.2020.100023
Taxonera, C., Sagastagoitia, I., Alba, C., Mañas, N., Olivares, D., and Rey, E. (2020). 2019 novel coronavirus disease (COVID-19) in patients with inflammatory bowel diseases. Aliment. Pharmacol. Ther. 52, 276–283. doi: 10.1111/apt.15804
Tefas, C., Ciobanu, L., Tanțău, M., Moraru, C., and Socaciu, C. (2020). The potential of metabolic and lipid profiling in inflammatory bowel diseases: A pilot study. Bosn. J. Basic Med. Sci. 20, 262–270. doi: 10.17305/bjbms.2019.4235
Tefas, C., Socaciu, C., Moraru, C., and Tanțău, M. (2019). Lipidomic signatures of colonic inflammatory bowel diseases: a pilot study. J. Gastrointestin. Liver Dis. 28, 246–247. doi: 10.15403/jgld-188
Thomas, T., Stefanoni, D., Reisz, J. A., Nemkov, T., Bertolone, L., Francis, R. O., et al. (2020). COVID-19 infection alters kynurenine and fatty acid metabolism, correlating with IL-6 levels and renal status. JCI Insight 5:e140327. doi: 10.1172/jci.insight.140327
Tong, M., Li, X., Wegener Parfrey, L., Roth, B., Ippoliti, A., Wei, B., et al. (2013). A modular organization of the human intestinal mucosal microbiota and its association with inflammatory bowel disease. PLoS One 8:e80702. doi: 10.1371/journal.pone.0080702
Turnbull, J., Jha, R., Ortori, C. A., Lunt, E., Tighe, P. J., Irving, W. L., et al. (2022). Serum levels of pro-inflammatory lipid mediators and specialised pro-resolving molecules are increased in SARS-CoV-2 patients and correlate with markers of the adaptive immune response. J. Infect. Dis. jiab632 doi: 10.1093/infdis/jiab632, [Epub ahead of print]
van Passel, M. W., Kant, R., Zoetendal, E. G., Plugge, C. M., Derrien, M., Malfatti, S. A., et al. (2011). The genome of Akkermansia muciniphila, a dedicated intestinal mucin degrader, and its use in exploring intestinal metagenomes. PLoS One 6:e16876. doi: 10.1371/journal.pone.0016876
Varela, E., Manichanh, C., Gallart, M., Torrejón, A., Borruel, N., Casellas, F., et al. (2013). Colonisation by Faecalibacterium prausnitzii and maintenance of clinical remission in patients with ulcerative colitis. Aliment. Pharmacol. Ther. 38, 151–161. doi: 10.1111/apt.12365
Venkatesh, M., Mukherjee, S., Wang, H., Li, H., Sun, K., Benechet, A. P., et al. (2014). Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and toll-like receptor 4. Immunity 41, 296–310. doi: 10.1016/j.immuni.2014.06.014
Vigsnæs, L. K., Brynskov, J., Steenholdt, C., Wilcks, A., and Licht, T. R. (2012). Gram-negative bacteria account for main differences between faecal microbiota from patients with ulcerative colitis and healthy controls. Benefic. Microbes 3, 287–297. doi: 10.3920/BM2012.0018
Villapol, S. (2020). Gastrointestinal symptoms associated with COVID-19: impact on the gut microbiome. Transl. Res. 226, 57–69. doi: 10.1016/j.trsl.2020.08.004
Vrakas, S., Mountzouris, K. C., Michalopoulos, G., Karamanolis, G., Papatheodoridis, G., Tzathas, C., et al. (2017). Intestinal bacteria composition and translocation of bacteria in inflammatory bowel disease. PLoS One 12:e0170034. doi: 10.1371/journal.pone.0170034
Walczuk, U., Sobieszczańska, B., Turniak, M., Rzeszutko, M., Duda-Madej, A., and Iwańczak, B. (2019). The prevalence of mucosa-associated diffusely adherent Escherichia coli in children with inflammatory bowel disease. Adv. Clin. Exp. Med. 28, 899–905. doi: 10.17219/acem/94149
Walker, A. W., Sanderson, J. D., Churcher, C., Parkes, G. C., Hudspith, B. N., Rayment, N., et al. (2011). High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 11:7. doi: 10.1186/1471-2180-11-7
Wallace, J. L. (2019). Eicosanoids in the gastrointestinal tract. Br. J. Pharmacol. 176, 1000–1008. doi: 10.1111/bph.14178
Walters, S. S., Quiros, A., Rolston, M., Grishina, I., Li, J., Fenton, A., et al. (2014). Analysis of gut microbiome and diet modification in patients with Crohn's disease. SOJ Microbiol. Infect. Dis. 2, 1–13. doi: 10.15226/sojmid/2/3/00122
Wang, W., Chen, L., Zhou, R., Wang, X., Song, L., Huang, S., et al. (2014). Increased proportions of bifidobacterium and the lactobacillus group and loss of butyrate-producing bacteria in inflammatory bowel disease. J. Clin. Microbiol. 52, 398–406. doi: 10.1128/JCM.01500-13
Wang, A., Luan, H. H., and Medzhitov, R. (2019). An evolutionary perspective on immunometabolism. Science 363:eaar3932. doi: 10.1126/science.aar3932
Wang, B., Wu, Z., Ji, Y., Sun, K., Dai, Z., and Wu, G. (2016). L-glutamine enhances tight junction integrity by activating CaMK kinase 2-AMP-activated protein kinase signaling in intestinal porcine epithelial cells. J. Nutr. 146, 501–508. doi: 10.3945/jn.115.224857
Wei, S., Bahl, M. I., Baunwall, S. M. D., Hvas, C. L., and Licht, T. R. (2021). Determining gut microbial dysbiosis: a review of applied indexes for assessment of intestinal microbiota imbalances. Appl. Environ. Microbiol. 87, e00395–e00321. doi: 10.1128/AEM.00395-21
Whiley, L., Nye, L. C., Grant, I., Andreas, N., Chappell, K. E., Sarafian, M. H., et al. (2019). Ultrahigh-performance liquid chromatography tandem mass spectrometry with electrospray ionization quantification of tryptophan metabolites and markers of gut health in serum and plasma-application to clinical and epidemiology cohorts. Anal. Chem. 91, 5207–5216. doi: 10.1021/acs.analchem.8b05884
Williams, B. B., van Benschoten, A. H., Cimermancic, P., Donia, M. S., Zimmermann, M., Taketani, M., et al. (2014). Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 16, 495–503. doi: 10.1016/j.chom.2014.09.001
Willing, B. P., Dicksved, J., Halfvarson, J., Andersson, A. F., Lucio, M., Zheng, Z., et al. (2010). A pyrosequencing study in twins shows that gastrointestinal microbial profiles vary with inflammatory bowel disease phenotypes. Gastroenterology 139, 1844–1854.e1. doi: 10.1053/j.gastro.2010.08.049
Windmueller, H. G., and Spaeth, A. E. (1974). Uptake and metabolism of plasma glutamine by the small intestine. J. Biol. Chem. 249, 5070–5079. doi: 10.1016/S0021-9258(19)42329-6
Wu, Y., Cheng, X., Jiang, G., Tang, H., Ming, S., Tang, L., et al. (2021). Altered oral and gut microbiota and its association with SARS-CoV-2 viral load in COVID-19 patients during hospitalization. NPJ Biofilms Microbiomes 7:61. doi: 10.1038/s41522-021-00232-5
Wu, H., Gong, J., and Liu, Y. (2018). Indoleamine 2, 3-dioxygenase regulation of immune response (review). Mol. Med. Rep. 17, 4867–4873. doi: 10.3892/mmr.2018.8537
Wu, D., Shu, T., Yang, X., Song, J.-X., Zhang, M., Yao, C., et al. (2020). Plasma metabolomic and lipidomic alterations associated with COVID-19. Natl. Sci. Rev. 7, 1157–1168. doi: 10.1093/nsr/nwaa086
Xiao, F., Tang, M., Zheng, X., Liu, Y., Li, X., and Shan, H. (2020). Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 158, 1831–833.e3. doi: 10.1053/j.gastro.2020.02.055
Xu, K., Cai, H., Shen, Y., Ni, Q., Chen, Y., Hu, S., et al. (2020a). Management of corona virus disease-19 (COVID-19): the Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban 49, 147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02
Xu, K., Cai, H., Shen, Y., Ni, Q., Chen, Y., Hu, S., et al. (2020b). Management of COVID-19: the Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban 49, 147–157. doi: 10.3785/j.issn.1008-9292.2020.02.02
Yamamoto, S., Saito, M., Tamura, A., Prawisuda, D., Mizutani, T., and Yotsuyanagi, H. (2021). The human microbiome and COVID-19: A systematic review. PLoS One 16:e0253293. doi: 10.1371/journal.pone.0253293
Yan, B., Chu, H., Yang, D., Sze, K. H., Lai, P. M., Yuan, S., et al. (2019). Characterization of the Lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses 11:73. doi: 10.3390/v11010073
Yang, H., Shan, W., Zhu, F., Wu, J., and Wang, Q. (2019). Ketone bodies in neurological diseases: focus on neuroprotection and underlying mechanisms. Front. Neurol. 10:585. doi: 10.3389/fneur.2019.00585
Yang, W., Yu, T., Huang, X., Bilotta, A. J., Xu, L., Lu, Y., et al. (2020). Intestinal microbiota-derived short-chain fatty acids regulation of immune cell IL-22 production and gut immunity. Nat. Commun. 11:4457. doi: 10.1038/s41467-020-18262-6
Yau, Y. Y., Leong, R. W., Shin, S., Bustamante, S., Pickford, R., Hejazi, L., et al. (2014). Bimodal plasma metabolomics strategy identifies novel inflammatory metabolites in inflammatory bowel diseases. Discov. Med. 18, 113–124.
Yeoh, Y. K., Zuo, T., Lui, G. C., Zhang, F., Liu, Q., Li, A. Y., et al. (2021). Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 70, 698–706. doi: 10.1136/gutjnl-2020-323020
Yilmaz, B., Juillerat, P., Øyås, O., Ramon, C., Bravo, F. D., Franc, Y., et al. (2019). Microbial network disturbances in relapsing refractory Crohn's disease. Nat. Med. 25, 323–336. doi: 10.1038/s41591-018-0308-z
Yoo, H. C., Yu, Y. C., Sung, Y., and Han, J. M. (2020). Glutamine reliance in cell metabolism. Exp. Mol. Med. 52, 1496–1516. doi: 10.1038/s12276-020-00504-8
Youm, Y. H., Nguyen, K. Y., Grant, R. W., Goldberg, E. L., Bodogai, M., Kim, D., et al. (2015). The ketone metabolite b-hydroxybutyrate blocks NLRP3 inflammasome-mediated inflammatory disease. Nat. Med. 21, 263–269. doi: 10.1038/nm.3804
Zamboni, N., Saghatelian, A., and Patti, G. J. (2015). Defining the metabolome: size, flux, and regulation. Mol. Cell 58, 699–706. doi: 10.1016/j.molcel.2015.04.021
Zhang, J., Chen, S. L., and Li, L. B. (2017). Correlation between intestinal flora and serum inflammatory factors in patients with Crohn's disease. Eur. Rev. Med. Pharmacol. Sci. 21, 4913–4917.
Zhang, X., Gan, M., Li, J., Li, H., Su, M., Tan, D., et al. (2020). Endogenous Indole pyruvate pathway for tryptophan metabolism mediated by IL4I1. J. Agric. Food Chem. 68, 10678–10684. doi: 10.1021/acs.jafc.0c03735
Zhang, J., Hoedt, E. C., Liu, Q., Berendsen, E., The, J. J., Hamilton, A., et al. (2021). Elucidation of Proteus mirabilis as a key bacterium in Crohn's disease inflammation. Gastroenterology 160, 317–330.e11. doi: 10.1053/j.gastro.2020.09.036
Zhang, Y., Lin, L., Xu, Y., Lin, Y., Jin, Y., and Zheng, C. (2013). 1H NMR-based spectroscopy detects metabolic alterations in serum of patients with early-stage ulcerative colitis. Biochem. Biophys. Res. Commun. 433, 547–551. doi: 10.1016/j.bbrc.2013.03.012
Zhang, M., Liu, B., Zhang, Y., Wei, H., Lei, Y., and Zhao, L. (2007). Structural shifts of mucosa-associated lactobacilli and Clostridium leptum subgroup in patients with ulcerative colitis. J. Clin. Microbiol. 45, 496–500. doi: 10.1128/JCM.01720-06
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Zucko, J., Dunlap, W. C., Shick, J. M., Cullum, J., Cercelet, F., Amin, B., et al. (2010). Global genome analysis of the shikimic acid pathway reveals greater gene loss in host-associated than in free-living bacteria. BMC Genomics 11:628. doi: 10.1186/1471-2164-11-628
Zuo, T., Liu, Q., Zhang, F., Lui, G. C., Tso, E. Y., Yeoh, Y. K., et al. (2021). Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 70:gutjnl-2020-322294. doi: 10.1136/gutjnl-2020-322294
Zuo, T., and Ng, S. C. (2018). The gut microbiota in the pathogenesis and therapeutics of inflammatory bowel disease. Front. Microbiol. 9:2247. doi: 10.3389/fmicb.2018.02247
Keywords: inflammatory bowel disease, Crohn’s disease, ulcerative colitis, SARS-CoV-2, COVID-19, metabolomics, microbiomics
Citation: Cortes GM, Marcialis MA, Bardanzellu F, Corrias A, Fanos V and Mussap M (2022) Inflammatory Bowel Disease and COVID-19: How Microbiomics and Metabolomics Depict Two Sides of the Same Coin. Front. Microbiol. 13:856165. doi: 10.3389/fmicb.2022.856165
Edited by:
Roshan Kumar, Magadh University, IndiaReviewed by:
Younes Zaid, Université de Montréal, CanadaAndrew S. Day, University of Otago, New Zealand
Busra Aktas, Mehmet Akif Ersoy University, Turkey
Copyright © 2022 Cortes, Marcialis, Bardanzellu, Corrias, Fanos and Mussap. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michele Mussap, bXVtaWtlMTUzQGdtYWlsLmNvbQ==
 Gian Mario Cortes
Gian Mario Cortes Maria Antonietta Marcialis
Maria Antonietta Marcialis Flaminia Bardanzellu
Flaminia Bardanzellu Angelica Corrias1
Angelica Corrias1 Vassilios Fanos
Vassilios Fanos Michele Mussap
Michele Mussap