- 1State Key Laboratory of Marine Environmental Science, College of Ocean and Earth Sciences, Xiamen University, Xiamen, China
- 2Department of Ocean Science, The Hong Kong University of Science and Technology, Kowloon, Hong Kong SAR, China
- 3Southern Marine Science and Engineering Guangdong Laboratory, Zhuhai, China
Vibriosis is one of the most common bacterial diseases that cause high rates of mortality and considerable economic losses in aquaculture. Phage therapy has been considered as a promising alternative method to antibiotics in the biocontrol of infectious diseases. Genome sequencing and characterization of the phage candidates are prerequisites before field applications to ensure environmental safety. In this study, a lytic phage, named vB_VhaS-R18L (R18L), was isolated from the coastal seawater of Dongshan Island, China. The phage was characterized in terms of morphology, genetic content, infection kinetics, lytic profile, and virion stability. Transmission electronic microscopy indicated that R18L is siphovirus-like, comprising an icosahedral head (diameter 88.6 ± 2.2 nm) and a long noncontractile tail (225 × 11 nm). Genome analysis indicated R18L to be a double-stranded DNA virus with a genome size of 80,965 bp and a G + C content of 44.96%. No genes that encode known toxins or genes implicated in lysogeny control were found in R18L. A one-step growth experiment showed that R18L had a latent period of approximately 40 min and a burst size of 54 phage particles per infected cell. R18L showed lytic activity against a wide range of at least five Vibrio species (V. alginolyticus, V. cholerae, V. harveyi, V. parahemolyticus, and V. proteolyticus). R18L was relatively stable at pH 6–11 and at temperatures ranging from 4°C to 50°C. The broad lytic activity across Vibrio species and the stability in the environment make R18L a potential candidate for phage therapy in controlling vibriosis in aquaculture systems.
Introduction
Vibriosis is a major bacterial disease of aquaculture that is associated with high mortality rates among marine animals and considerable economic losses to the seafood industry (Lafferty et al., 2015). Vibriosis can be caused by a number of Vibrio species, among which V. harveyi is a notifiable and highly prevalent pathogen in marine environments (Austin and Zhang, 2006; Zhang et al., 2020). Marine vertebrates (mainly fish) and invertebrates (mainly penaeid shrimp) infected by V. harveyi show vasculitis, gastroenteritis, eye lesions, and luminous vibriosis. These diseases have severely affected seafood production in Asia and South America, including China, Japan, India, Thailand, Java Island, Philippines, Kuwait, northern Chile, etc. (Austin and Zhang, 2006; Defoirdt et al., 2007). Antibiotics and sanitizers have traditionally been used in the prevention and control of vibriosis in aquaculture (Sano, 1998). However, the overuse of drugs has resulted in antibiotic resistance, chemical residues in aquatic products, a microecological imbalance, and environmental pollution (Defoirdt et al., 2007; Larsson and Flach, 2022). A variety of antibiotic-resistant pathogens have been increasingly reported (Kang et al., 2014; Stalin and Srinivasan, 2016). The emergence of antimicrobial resistance in pathogens highlights the urgent need for alternative therapeutic methods to reduce mortality and minimize the impact on human health and the environment (Defoirdt et al., 2007).
Bacteriophage (or phage) therapy, which has the advantages of specific targeting, self-replication, and low inherent toxicity, has been historically employed as a biological control strategy and has been proposed as an eco-friendly method to control bacterial disease (Defoirdt et al., 2007; Nobrega et al., 2015; Wang et al., 2017; Yen et al., 2017; Gordillo Altamirano and Barr, 2019). To date, a number of studies have reported the use of phage therapy against V. harveyi pathogens (Vinod et al., 2006; Shivu et al., 2007; Crothers-Stomps et al., 2010; Wu et al., 2021). Phages infecting V. harveyi have been isolated from various environments and tested in terms of their potential application (Vinod et al., 2006; Shivu et al., 2007; Crothers-Stomps et al., 2010; Wu et al., 2021). For example, phage treatment of V. harveyi-infected Penaeus monodon larvae resulted in higher survival rates (80%) compared with the control larvae (25%) (Vinod et al., 2006). Lytic phages P4A and P4F, isolated from the seawater of an abalone farm, significantly reduced the population of pathogenic V. harveyi (Luo et al., 2016). It is now accepted that phage therapy, after careful selection and extensive studies of phage candidates, will eventually become an effective alternative to antibiotics (Defoirdt et al., 2007; Nobrega et al., 2015; Gordillo Altamirano and Barr, 2019; Nachimuthu et al., 2021). However, comprehensive studies must be undertaken in selecting phage candidates because some phages may encode toxins and/or lead to altered bacterial virulence, and others may be inefficient when applied in the field. One such example is bacteriophage VHML (V. harveyi myovirus-like), which was shown to confer virulence in various V. harveyi strains (Munro et al., 2003). Similarly, two isolated myoviruses were reported to integrate as prophages into the host genome and induce bacteriocin production when infecting V. harveyi strains, which excludes their usage in phage therapy (Crothers-Stomps et al., 2010).
In general, the suitability of a particular phage to control bacterial pathogens is determined by the presence of toxic genes, the host range, the length of viral infection, the number of progeny produced, and, importantly, the stability of the phage in the environment (Hyman, 2019). Considering the high degree of phenotypic and genotypic diversity among Vibrio pathogens, a phage with a wide host range is potentially valuable. When a disease is caused by a mixed bacterial infection, the use of a broad-host-range phage with the ability to kill multiple strains would be preferable to a mixture of different therapeutic phages (Hyman, 2019). Taking this into consideration, the present study aimed to isolate bacteriophages with a broad host range and evaluate their efficiency as potential biocontrol agents against vibriosis. Different V. harveyi-specific phages (a total of 12 phages) were first isolated and screened to determine their host range. On the basis of its broad host range, one of these phages, vB_VhaS-R18L (hereafter, R18L), was selected for further analysis of its genomic and morphological properties, as well as its burst size and latent period. Furthermore, the virion stability of R18L was determined under different temperature and pH conditions to determine its suitability for potential therapeutic applications in the future.
Materials and methods
Phage isolation and purification
The host strain Vibrio harveyi BYK0632 used in this study was purchased from the National Pathogen Collection Center for Aquatic Animals, Shanghai Ocean University (Shanghai, China). V. harveyi BYK0632 was grown in rich organic (RO) medium (1 M peptone, 1 M yeast extract, 1 M sodium acetate, artificial seawater, pH 7.5) at 28°C with shaking at 180 rpm/min. Surface seawater samples for phage isolation were collected in April 2016 at the coast of Dongshan Island (Fujian, China) and filtered through a 0.2 μm membrane. Before being mixed with the host strain, the virus-containing filtrate was concentrated using a 30 kDa cartridge (Millipore, MA, USA) by tangential flow filtration to improve the probability of successful phage infection (Cai et al., 2015, 2019). The concentrated seawater samples were mixed with exponentially-growing V. harveyi BYK0632 (OD600: 0.1–0.2) using a double-layer agar method according to previous studies (Yang et al., 2017; Cai et al., 2019). After 18–24 h incubation at 28°C, individual lytic plaques were picked from the agarose plate and dissolved in SM buffer (50 mM Tris–HCl, 0.1 M NaCl, 8 mM MgSO4, 0.1 g/L gelatin, pH 7.5). This double-layer agar plating was repeated five times to ensure the purity of the phage.
Preparation of high-titer phage suspensions
To obtain high-titer phage suspensions for morphological observation and genome sequencing, phages were propagated in one liter of V. harveyi BYK0632. After cell lysis, the culture was centrifuged at 10,000 × g for 10 min and filtered through 0.2 μm filters to obtain the phage-containing suspension. The phage suspension was precipitated with polyethylene glycol 8,000 (100 g L˗1 final concentration) overnight at 4°C and collected by centrifugation at 10,000 × g for 30 min at 4°C. The phage pellet was re-suspended in SM buffer and then concentrated by CsCl (1.5 g/mL in SM buffer) gradient ultracentrifugation (200,000 × g, 4°C, 24 h). The clear phage band was extracted and dialyzed against SM buffer at 4°C.
Host range
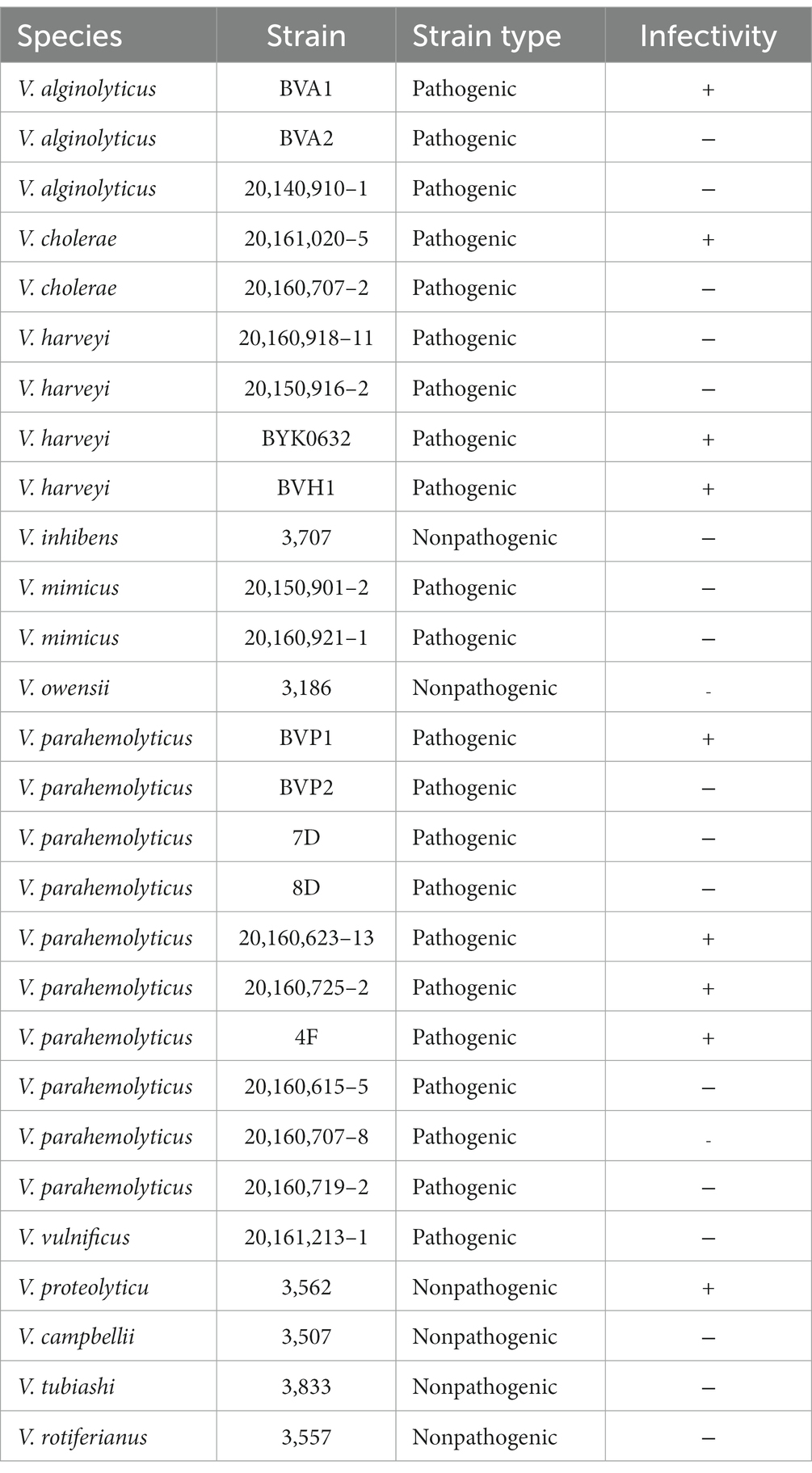
The lytic profiles of purified vibriophages were determined using spot assay (Yang et al., 2017; Cai et al., 2019). Briefly, different phages were challenged against 28 Vibrio strains from 12 Vibrio species (V. alginolyticus, V. campbellii, V. cholera, V. harveyi, V. inhibens, V. mimicus, V. owensii, V. parahemolyticus, V. proteolyticus, V. tubiashii, V. vulnificus, and V. rotiferianus), which were originally isolated from the aquatic environment and diseased shrimp and fish (Table 1) and purchased from the National Pathogen Collection Center for Aquatic Animals (China). Each of these exponentially growing bacterial cultures was mixed with molten RO agar medium (0.5% w/v agar) and poured onto solid RO agar medium (1.5% w/v agar). After the agarose plates solidified, 5 μL of diluted phage lysate was added onto the surface of each bacterial plate and incubated at 28°C for more than 12 h. The formation of clear plaques where lysates were added indicated successful phage infection of the test strains. Tests were repeated at least three times. One of these isolated phages, R18L, showing a broad host range (see results below), was selected for further characterization.
Transmission electron microscopy (TEM)
The morphology of R18L was determined by TEM. In brief, 3 μL of high-titer phage was adsorbed onto a carbon-coated copper microscopy grid for 10 min, followed by negative staining with 2% (w/v) phosphotungstic acid for 1 min. After the grid was air-dried for 30 min, the sample was observed by TEM using a JEM-2100 microscope (JEOL, Tokyo, Japan) at 80 kV. Images were acquired by a CCD image transmission system (Gatan Inc., Pleasanton, CA, United States).
Lipid test
To determine whether the capsid of R18L contained lipids, a chloroform sensitivity test was conducted (Yang et al., 2017; Cai et al., 2019). Briefly, 1 mL of phage lysate was incubated with 20 μL and 200 μL of chloroform for 30 min at room temperature in the dark. Control aliquots were included without the addition of chloroform. After incubation, chloroform was removed by centrifugation at 5,000 × g for 5 min. The titers of the phage were then determined by a spot assay. Each treatment was tested in triplicate.
One-step growth curve
The life cycle of R18L was examined by a one-step growth experiment (Middelboe et al., 2010; Yang et al., 2017). Briefly, the freshly prepared phage lysate was added to 1 mL of exponentially growing V. harveyi BYK0632 culture with a multiplicity of infection of 0.001 in triplicate, then incubated for 5 min at room temperature (24°C) in the dark for phage adsorption. To remove unabsorbed phage particles, the culture was centrifuged for 5 min (10,000 × g, 4°C) and resuspended in RO medium. This procedure was repeated twice. The suspension was incubated at 28°C in the dark. Samples were taken every 10 min, and the viral abundance was determined by a plaque assay.
Thermal stability and pH sensitivity
The effects of environmental factors on the phage were determined by testing its thermal stability and pH sensitivity. For the thermal stability test, 1.5 mL of aliquots of freshly prepared phage lysate (~107 plaque-forming units/mL) was incubated at different temperatures (4°C, 24°C, 37°C, 50°C, 55°C, and 60°C) in triplicate. Subsamples were collected at 3, 24, and 48 h, and the phage titer was determined by a plaque assay. For the pH stability test, SM buffer with a pH ranging from 2 to 12 was prepared using HCl or NaOH as required. Then, 100 μL of freshly prepared phage lysate was inoculated and incubated under different pH conditions at room temperature (24°C) in triplicate. Subsamples were collected at 3 h and 24 h, and the phage titer was determined by a plaque assay.
DNA extraction
Prior to DNA extraction, the high-titer phage concentrate was treated with DNase I and RNase A to remove possible contamination of free host DNA and RNA. The DNase was inactivated at 65°C for 15 min. Phages were lysed with 1.5 μL of proteinase K (100 mg/mL), 10 μL of EDTA (0.5 M, pH 8.0), and 100 μL of sodium dodecyl sulfate (SDS; 10% w/v) at 55°C for 3 h. The phage DNA was extracted with a phenol/chloroform/isoamyl alcohol mixture, which promotes the partitioning of cellular debris into the organic phase, leaving isolated DNA in the aqueous phase. The purified DNA was further precipitated with isoamyl alcohol. The quality of the DNA was checked via agarose gel electrophoresis and analysis using a NanoDrop 2000 Spectrophotometer (Thermo Fisher Scientific, MA, United States).
Genome sequencing and analysis
The extracted DNA was sheared into 300-bp fragments in a Covaris ultrasonicator (KBiosciences, United Kingdom) before preparing the Illumina paired-end sequencing library using the NEBNext Ultra II DNA library prep kit. The quality and size of the libraries were analyzed using the Agilent 2,100 Bioanalyzer. The concentration of the libraries was determined using the Qubit 2.0 dsDNA HS Assay kit (Life Technologies, Germany). Sequencing was performed on the MiSeq platform (Illumina, San Diego, CA, United States). Raw reads were trimmed and filtered using Trimmomatic v0.36 to remove adaptor sequences and low-quality reads (Bolger et al., 2014). The sequences were assembled using A5-miseq (version 20,150,522). Intergenomic similarities between phages were calculated using VIRIDIC (Moraru et al., 2020). The open reading frames (ORFs) were predicted using GeneMark (Besemer and Borodovsky, 2005) and ORF Finder (Rombel et al., 2002). Gene annotation was performed using BLASTP against the NCBI nonredundant (nr) database1 with an e-value <10˗3. tRNA sequences in the R18L genome were analyzed by tRNAscan-SE (Chan and Lowe, 2019). The spacer of the R18L genomic sequence was searched against the viral spacer database of IMG/VR (Roux et al., 2021). Phylogenetic analyses of specific genes were performed using the maximum-likelihood method with 1,000 bootstrap replicates and MEGA software (Tamura et al., 2021). The complete genome sequence of vibriophage vB_VhaS-R18L has been deposited in the GenBank database under accession number MT451873.
Results and discussion
Biological features of R18L
In this study, a total of 12 phages against V. harveyi were isolated from the coastal surface seawater of Dongshan Island, China. A novel V. harveyi phage, designated vB_VhaS-R18L (R18L), showing a broad host range (see below), was selected for further detailed analysis. The lysis of R18L formed semitransparent plaques of 2.0 mm in diameter on host lawn plates (Figure 1A). As shown in the TEM micrograph, R18L has an icosahedral capsid (diameter 88.6 ± 2.2 nm) and a long noncontractile tail (225 ± 2.2 nm in length and 11 ± 1.0 nm in width), being a siphovirus-like phage (Figure 1B). Different vibriophages have been isolated and characterized using V. harveyi as the host, with most belonging to siphoviruses (Vinod et al., 2006; Wang et al., 2017; Wu et al., 2021; Droubogiannis and Katharios, 2022; Kang et al., 2022), followed by myoviuses (Surekhamol et al., 2014; Lal et al., 2017) and podoviruses (Thiyagarajan et al., 2011). The reason why siphoviruses are the predominant viral group infecting Vibrio is currently unknown. However, as more vibriophage genomes and features become available, we will be better able to assess the abundance and role of different vibriophage groups. The capsid size and tail length of R18L were relatively large compared with those previously reported for siphoviruses with diameters of 40–92 nm and tail lengths of 60–277 nm (Vinod et al., 2006; Crothers-Stomps et al., 2010; Thiyagarajan et al., 2011; Raghu Patil et al., 2014; Stalin and Srinivasan, 2017). The chloroform sensitivity test demonstrated that the infectivity of R18L was not affected by different concentrations of chloroform, suggesting the absence of lipids outside of the R18L capsid. To date, lipids have been considered to be a rare feature for bacteriophages, representing less than 5% of the published isolates (Atanasova et al., 2015; Mantynen et al., 2019).
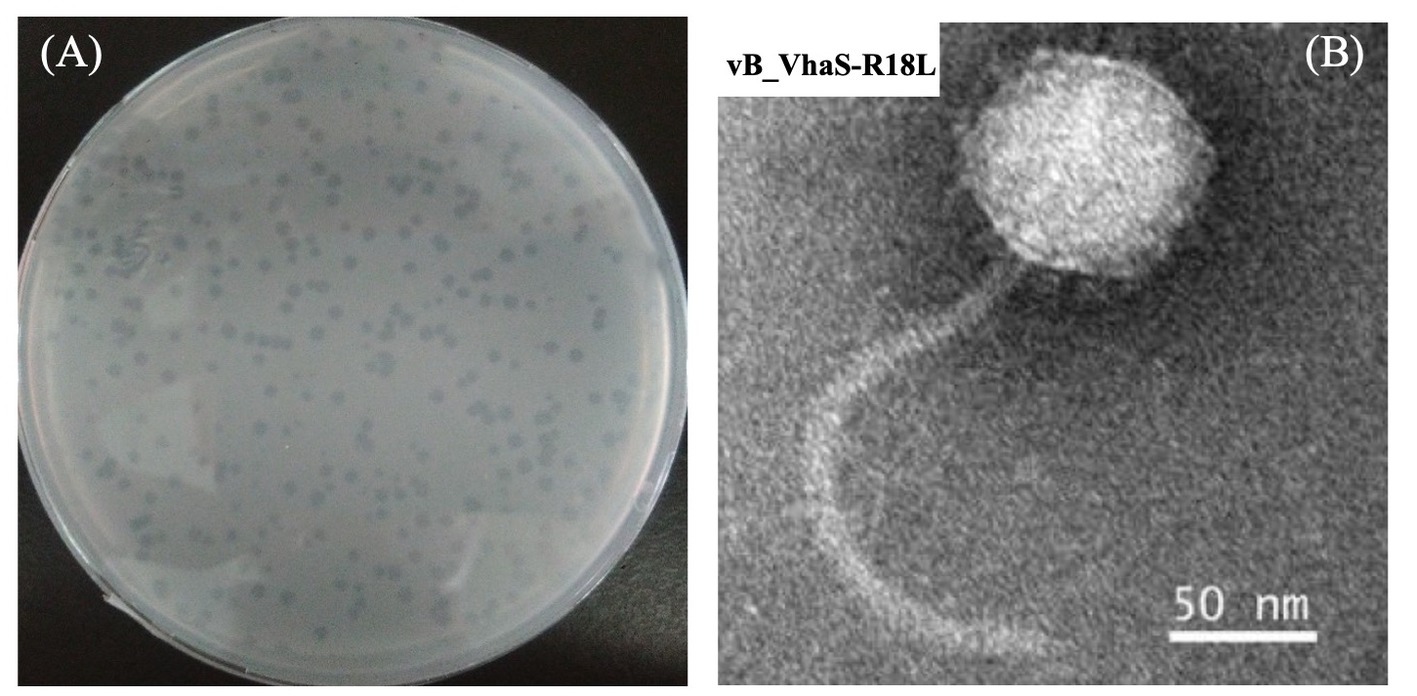
Figure 1. Plaques (A) and transmission electron microscopic image (B) of vibriophage vB_VhaS-R18L. Scale bar, 50 nm.
The latent period and burst size of R18L, as important characteristics of the phage infection process, were determined by the one-step growth curve (Figure 2). R18L exhibited a latent period of 40 min and a rise period of 30 min. The burst size of R18L was calculated at approximately 54 phage particles per infected cell. Recent reports have shown that the latent period and burst size of other Vibrio phages were 10–70 min and 2–180 phage particles/cell (Baudoux et al., 2012; Lal et al., 2017; Stalin and Srinivasan, 2017). The latent period and burst size of R18L were within the documented range for vibriophages.
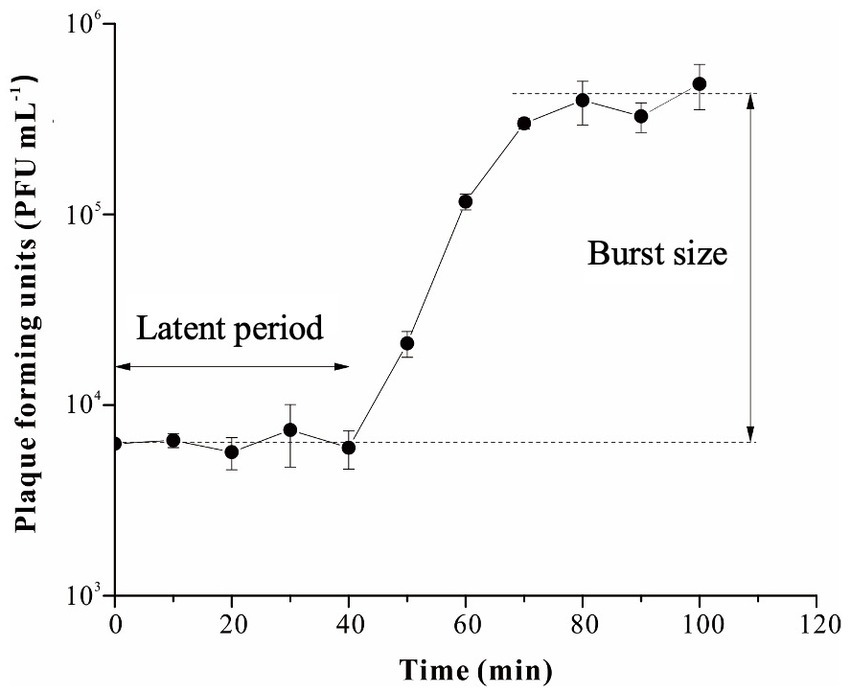
Figure 2. One-step growth curve of vibriophage vB_VhaS-R18L. Data points represent the mean values and standard deviations of three independent experiments.
The host range of R18L was tested on 28 Vibrio strains, including 22 pathogenic strains isolated from diseased animals and six nonpathogenic strains from seawater (Table 1). Nine (32%) of the 28 Vibrio strains tested were lysed by R18L, including strains from V. alginolyticus, V. cholerae, V. harveyi, V. parahemolyticus, and V. proteolyticus. Of these nine strains, eight were pathogenic. R18L could infect bacteria across at least five Vibrio species, even including V. proteolyticus and V. cholerae, which are not members of the Harveyi clade (Sawabe et al., 2007). Interestingly, two species that are closely related to V. harveyi (V. campbellii and V. rotiferianus), belonging to the Harveyi clade (Sawabe et al., 2007), were not susceptible to phage R18L. Therefore, the genetic similarity between Vibrio species does not necessarily correlate with the lytic spectrum of R18L. Previously reported vibriophages have shown different lytic abilities against Vibrio species. Phage PW2 infected different strains of V. harveyi but not 13 other Vibrio species (V. alginolyticus, V. cholera, V. campbelli, V. logei, etc.) (Phumkhachorn and Rattanachaikunsopon, 2010). Furthermore, phage SIO-2 could only infect two strains from relatively closely-related Vibrio species (V. harveyi and V. campbellii) when tested against 17 Vibrio species (Baudoux et al., 2012). Whereas phages with a broad lytic spectrum have also been reported (Shivu et al., 2007; Thiyagarajan et al., 2011). For example, phages φVh1, φVh2, and φVh3 showed a relatively broad lytic spectrum involving four Vibrio species (V. harveyi, V. parahemolyticus, V. alginolyticus, and V. logei) (Thiyagarajan et al., 2011). In phage therapy, when bacterial diseases are caused by polymicrobial infections, a therapeutic phage mixture or phages with a broader host range would be needed for treatment (de Jonge et al., 2019; Hyman, 2019). Given that phage cocktails (mixtures) require the individual phage targeting different pathogens to be isolated and studied, broader-host-range phages might be preferable for complex vibriosis. Hence, R18L, possessing a broad spectrum of infectivity against different pathogenic Vibrio spp., provides a promising potential biocontrol agent for bacterial diseases in aquaculture. However, the estimation of the efficiency of R18L in treating different pathogens is needed before field applications in the future.
Genome features of R18L
The genome of R18L was a double-stranded DNA comprising 80,965 bp and a G + C content of 44.96%. The R18L genome consisted of 118 putative ORFs, of which 31 ORFs (26.2%) have known functions, while the other 87 ORFs (73.7%) were assigned as genes with unknown functions (Figure 3). All of the predicted ORFs are oriented on the positive strand (in a rightward direction). The total gene length of all coding sequences was 76,620 bp, comprising ~94.6% of the genome. Gene annotation using BLASTP (e-value <10˗3) identified different functional clusters, including structural genes and genes involved in DNA metabolism, DNA replication, DNA packaging, cell lysis, and additional functions (see below). Among the 31 ORFs of known function, eight were related to the structure of R18L, while 15 were associated with DNA replication, metabolism, and packaging, cell lysis (ORF 93). In addition, seven uncategorized ORFs had a wide range of functions, including a serine protease XkdF (ORF 8), which is a protease frequently found in phage genomes. The function of virally encoded serine proteases is currently unknown, but they have been found to be strongly expressed during the late stage of viral infection, suggesting their potential role in virion assembly or maturation (Baum et al., 2021). Tyrosine phosphatase (ORF 53) was involved in signaling by controlling the phosphorylation state of proteins (Walchli et al., 2004). Pyruvate phosphate dikinase (ORF 51) and pyruvate decarboxylase (ORF 55) were two potential auxiliary enzymes that were involved in the host glycolytic metabolism (DelVecchio et al., 2002). No tRNA gene was detected in the R18L genome sequence, suggesting that R18L depended on the translation machinery of its host. Furthermore, R18L did not possess lysogeny-related genes (transposase or integrase, excisionase, and repressor), and no antibiotic resistance genes or virulence factor-related genes were detected in the genome of R18L using the ARDB (Liu and Pop, 2009) and VFDB (Chen et al., 2005) databases, which is beneficial for its potential application in phage therapy.
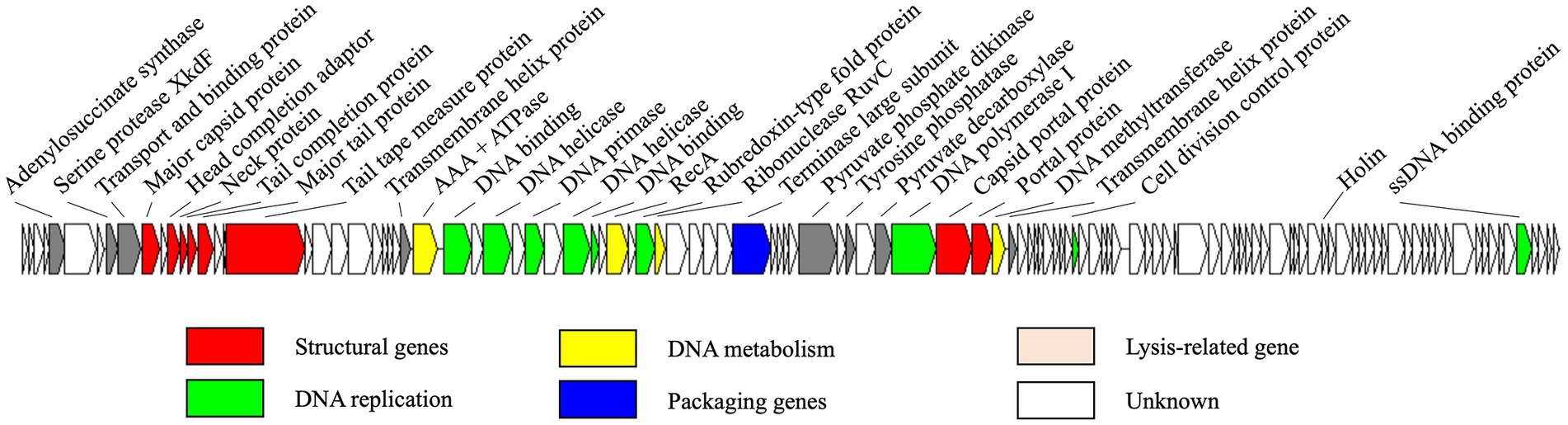
Figure 3. Genome map of vB_VhsS-R18L. Colored arrows represent different putative functions predicted from BLASTP similarity. The direction of each arrow represents the direction of transcription.
A nucleotide-based genome comparison revealed that the genome of R18L shared the most similarity to that of Vibrio phage SIO-2 (Baudoux et al., 2012), with an identity of 97.33% and a coverage of 99%, consistent with the intergenomic similarity of 96.95% between them based on VIRIDIC calculation (Moraru et al., 2020). However, R18L shows a broader host range than SIO-2. R18L could infect strains of V. harveyi, V. alginolyticus, V. cholerae, V. parahemolyticus, and V. proteolyticus among 12 Vibrio species, while SIO-2 only lysed strains of V. harveyi and V. campbellii among 17 Vibrio species tested. The host range of phages might correlate with variations in their tail-related genes (de Jonge et al., 2019). Three ORFs (ORFs 14, 15, 18) of R18L were identified as tail-related genes, while ORF 15 (major tail protein) of R18L exhibited relatively low identity (93%) with the corresponding gene of SIO-2, which might explain the different host ranges of the two phages. Additionally, seven predicted proteins with unknown function (ORFs 48, 50, 78, 79, 95, 117, 118) of R18L were distinct from those of SIO-2, with 53–88% identity at the amino acid level, which may contribute to the difference in the lytic spectrum of these two phages.
Phylogenetic trees of DNA polymerase I and terminase large subunit (TerL) were constructed to analyze the evolutionary relationships of R18L. As demonstrated in Figure 4, R18L clustered with three vibriophages, SIO-2 (NC_016567.1), vB_VhaS-a (KX198615.1), and ValSw4_1 (MH925091.1), which were all isolated using V. harveyi as the host, in both the DNA polymerase I and TerL trees. These four phages are clearly related to another branch, where four vibriophages were isolated from different Vibrio species. All eight vibriophages in the phylogenetic analysis were siphoviruses, with similar genome sizes (79.6–82.2 kb) and G + C contents (45.0–47.6%).
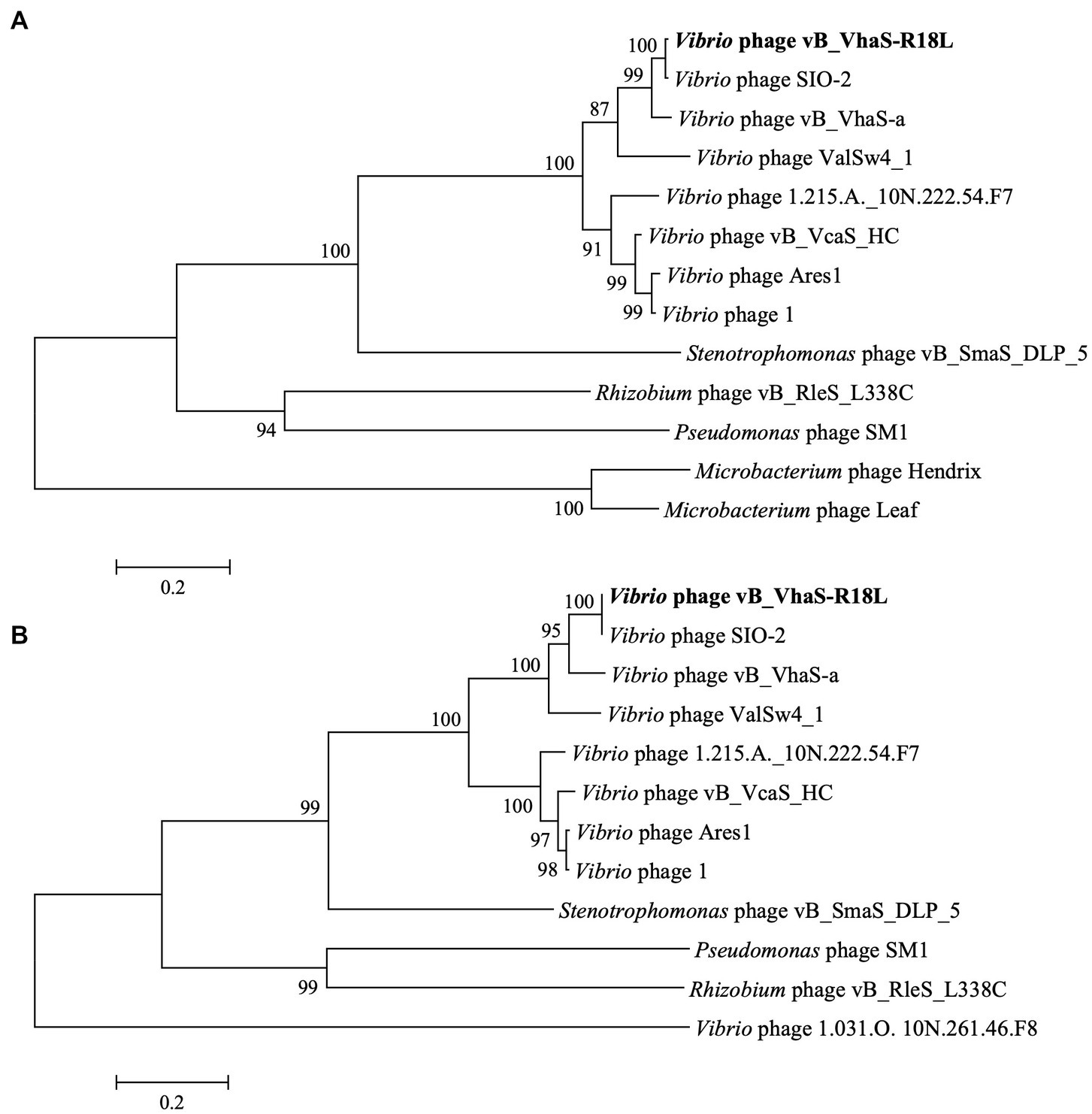
Figure 4. Phylogenetic tree of Vibrio phage R18L-related phages based on the amino acid sequences of DNA polymerase I (A) and the terminase large subunit (B) using a maximum likelihood method with 1,000 bootstraps (percentage value given on nodes). The scale bar represents 0.2 fixed mutations per amino acid position. R18L isolated in this study is shown in bold.
Virion stability of R18L
The thermal stability test showed that R18L was highly stable, with its activity remaining fairly constant at 4–40°C for 3 h (Figure 5A). Furthermore, greater than 50% of R18L phage remained active after incubation at 4–40°C for 24–48 h. The stability of R18L below 40°C simplifies the storage and transport requirements for this phage. Most phages become inactive when the temperature reaches 55°C, with survival percentages lower than 15% within 3 h. R18L did not show any activity when the temperature increased to 60°C, showing better thermal tolerance than previously reported vibriophages IME271 (40°C) (Li et al., 2019) and V-YDF132 (Kang et al., 2022). R18L exhibited stability over a wide range of pH (pH 6–11) for 3 h (Figure 5B). The survival percentage of phages decreased dramatically when the pH decreased to 5. When the incubation time reached 24 h, more than 60% of phages were still infective within the pH range 6–9, which indicated that R18L was relatively stable under such pH conditions.
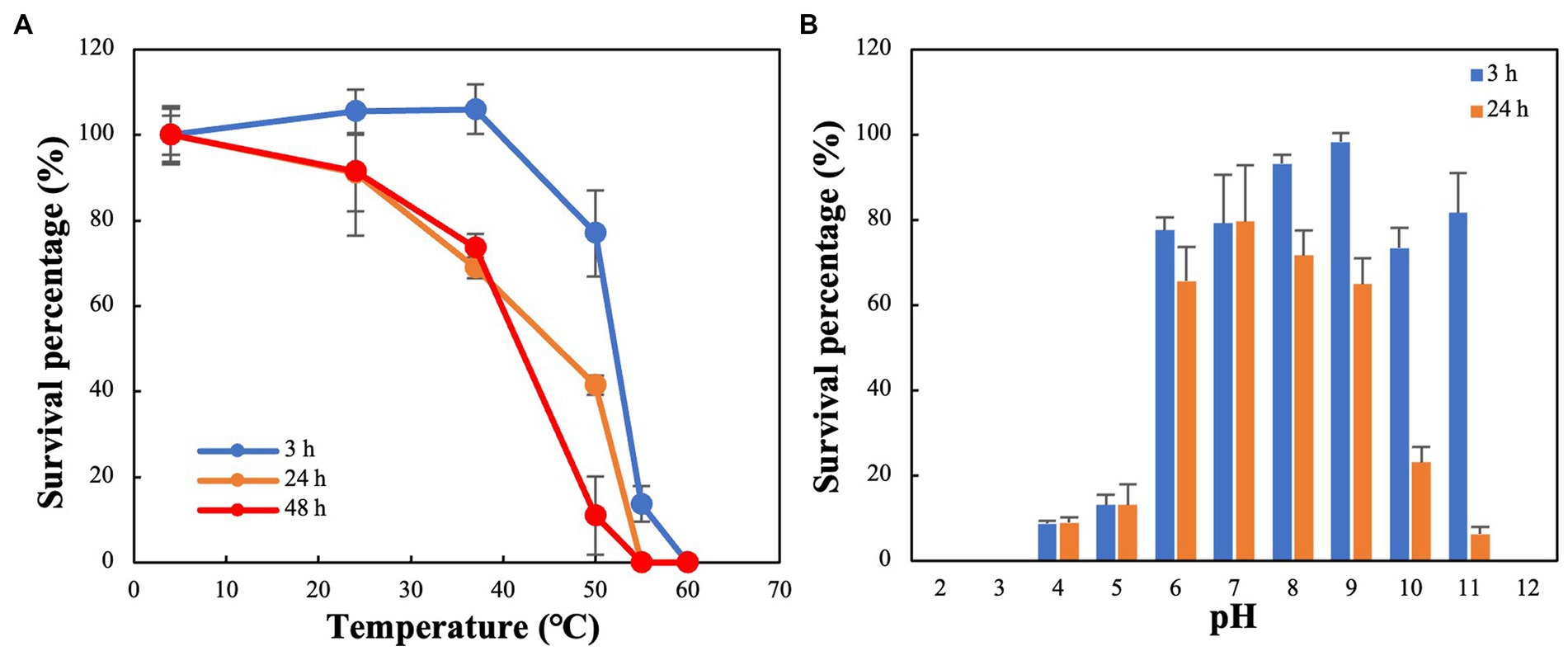
Figure 5. Stability of vibriophage vB_VhaS-R18L under different temperature (A) and pH (B) conditions. Data points represent the mean values and standard deviations of three independent experiments.
Conclusion
Vibrio is a major pathogen of various aquatic animals and causes vibriosis outbreaks in the aquaculture industry. Phage therapy is gaining increasing attention as a potentially effective strategy for controlling pathogenic bacteria. In this study, lytic phage R18L was isolated and characterized in terms of genomic organization, and phylogenetic and microbiological characteristics. R18L was able to infect bacteria across at least five pathogenic Vibrio species, thereby indicating its potential application as a biocontrol agent to control vibriosis. No antibiotic resistance, lysogeny-related, or virulence genes were detected in the R18L genome, suggesting the safety of this phage in biocontrol applications. Furthermore, R18L may be a good candidate for phage therapy because of its stability across a wide range of pH (6.0–11.0) and thermal (up to 55°C) conditions. In the field, one key feature of various Vibrio strains is their ability to form biofilm. Biofilm destruction by phages has been revealed to be more effective than antibiotics (Khalifa et al., 2016). Numerous experiments have been performed using single phages or phage cocktails against biofilms (Pires et al., 2017). However, the demonstration of the biofilm removal ability of R18L is necessary to determine its application in the biocontrol of vibriosis. Furthermore, other crucial questions that remain to be answered before field applications include testing the effectiveness of R18L in saline environments in future studies.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
RZ, LC, and CA designed the project. ZL performed seawater sampling and phage isolation. LC, YT, ZL, and YY performed experiments and data analyses. LC, YT, and RZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the National Key Research and Development Program of China (2021YFE0193000 and 2020YFA0608300) and National Natural Science Foundation of China (42188102 and 91951209).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Atanasova, N. S., Sencilo, A., Pietila, M. K., Roine, E., Oksanen, H. M., and Bamford, D. H. (2015). Comparison of lipid-containing bacterial and archaeal viruses. Adv. Virus Res. 92, 1–61. doi: 10.1016/bs.aivir.2014.11.005
Austin, B., and Zhang, X. H. (2006). Vibrio harveyi: a significant pathogen of marine vertebrates and invertebrates. Lett. Appl. Microbiol. 43, 119–124. doi: 10.1111/j.1472-765X.2006.01989.x
Baudoux, A. C., Hendrix, R., Lander, G., Bailly, X., Podell, S., Paillard, C., et al. (2012). Genomic and functional analysis of Vibrio phage SIO-2 reveals novel insights into ecology and evolution of marine siphoviruses. Environ. Microbiol. 14, 2071–2086. doi: 10.1111/j.1462-2920.2011.02685.x
Baum, L., Nguyen, M., Jia, Y., Biazik, J., and Thomas, T. (2021). Characterization of a novel roseophage and the morphological and transcriptional response of the sponge symbiont Ruegeria AU67 to infection. Environ. Microbiol. 23, 2532–2549. doi: 10.1111/1462-2920.15474
Besemer, J., and Borodovsky, M. (2005). GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res. 33, W451–W454. doi: 10.1093/nar/gki487
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cai, L., Ma, R., Chen, H., Yang, Y., Jiao, N., and Zhang, R. (2019). A newly isolated roseophage represents a distinct member of Siphoviridae family. Virol. J. 16:128. doi: 10.1186/s12985-019-1241-6
Cai, L., Yang, Y., Jiao, N., and Zhang, R. (2015). Evaluation of tangential flow filtration for the concentration and separation of bacteria and viruses in contrasting marine environments. PLoS One 10:e0136741. doi: 10.1371/journal.pone.0136741
Chan, P. P., and Lowe, T. M. (2019). tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol. Biol. 1962, 1–14. doi: 10.1007/978-1-4939-9173-0_1
Chen, L., Yang, J., Yu, J., Yao, Z., Sun, L., Shen, Y., et al. (2005). VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328. doi: 10.1093/nar/gki008
Crothers-Stomps, C., Hoj, L., Bourne, D. G., Hall, M. R., and Owens, L. (2010). Isolation of lytic bacteriophage against Vibrio harveyi. J. Appl. Microbiol. 108, 1744–1750. doi: 10.1111/j.1365-2672.2009.04578.x
de Jonge, P. A., Nobrega, F. L., Brouns, S. J. J., and Dutilh, B. E. (2019). Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. 27, 51–63. doi: 10.1016/j.tim.2018.08.006
Defoirdt, T., Boon, N., Sorgeloos, P., Verstraete, W., and Bossier, P. (2007). Alternatives to antibiotics to control bacterial infections: luminescent vibriosis in aquaculture as an example. Trends Biotechnol. 25, 472–479. doi: 10.1016/j.tibtech.2007.08.001
DelVecchio, V. G., Kapatral, V., Redkar, R. J., Patra, G., Mujer, C., Los, T., et al. (2002). The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. U. S. A. 99, 443–448. doi: 10.1073/pnas.221575398
Droubogiannis, S., and Katharios, P. (2022). Genomic and biological profile of a novel bacteriophage, Vibrio phage Virtus, which improves survival of Sparus aurata larvae challenged with Vibrio harveyi. Pathogens 11:630. doi: 10.3390/pathogens11060630
Gordillo Altamirano, F. L., and Barr, J. J. (2019). Phage therapy in the postantibiotic era. Clin. Microbiol. Rev. 32, e00066–e00018. doi: 10.1128/CMR.00066-18
Hyman, P. (2019). Phages for phage therapy: isolation, characterization, and host range breadth. Pharmaceuticals 12:35. doi: 10.3390/ph12010035
Kang, C. H., Kim, Y., Oh, S. J., Mok, J. S., Cho, M. H., and So, J. S. (2014). Antibiotic resistance of Vibrio harveyi isolated from seawater in Korea. Mar. Pollut. Bull. 86, 261–265. doi: 10.1016/j.marpolbul.2014.07.008
Kang, S., Zhang, L., Liao, J., Zhang, D., Wu, S., Zhang, X., et al. (2022). Isolation and characterization of a newly discovered phage, V-YDF132, for lysing Vibrio harveyi. Viruses 14:1802. doi: 10.3390/v14081802
Khalifa, L., Shlezinger, M., Beyth, S., Houri-Haddad, Y., Coppenhagen-Glazer, S., Beyth, N., et al. (2016). Phage therapy against Enterococcus faecalis in dental root canals. J. Oral Microbiol. 8:32157. doi: 10.3402/jom.v8.32157
Lafferty, K. D., Harvell, C. D., Conrad, J. M., Friedman, C. S., Kent, M. L., Kuris, A. M., et al. (2015). Infectious diseases affect marine fisheries and aquaculture economics. Annu. Rev. Mar. Sci. 7, 471–496. doi: 10.1146/annurev-marine-010814-015646
Lal, T. M., Sano, M., and Ransangan, J. (2017). Isolation and characterization of large marine bacteriophage (Myoviridae), VhKM4 infecting Vibrio harveyi. J. Aquat. Anim. Health 29, 26–30. doi: 10.1080/08997659.2016.1249578
Larsson, D. G. J., and Flach, C. F. (2022). Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269. doi: 10.1038/s41579-021-00649-x
Li, F., Xing, S., Fu, K., Zhao, S., Liu, J., Tong, Y., et al. (2019). Genomic and biological characterization of the Vibrio alginolyticus-infecting "Podoviridae" bacteriophage, vB_ValP_IME271. Virus Genes 55, 218–226. doi: 10.1007/s11262-018-1622-8
Liu, B., and Pop, M. (2009). ARDB--antibiotic resistance genes database. Nucleic Acids Res. 37, D443–D447. doi: 10.1093/nar/gkn656
Luo, Z.-H., Yu, Y.-P., Jost, G., Liu, W.-H., Huang, X.-L., and Gu, L. (2016). Characterization of two bacteriophages for specific treatment of biofilm formed by a Vibrio sp. isolated from an abalone farm. Aquac. Res. 47, 3964–3972. doi: 10.1111/are.12846
Mantynen, S., Sundberg, L. R., Oksanen, H. M., and Poranen, M. M. (2019). Half a century of research on membrane-containing bacteriophages: bringing new concepts to modern virology. Viruses 11:76. doi: 10.3390/v11010076
Middelboe, M., Chan, A. M., and Bertelsen, S. K. (2010). Isolation and life cycle characterization of lytic viruses infecting heterotrophic bacteria and cyanobacteria. MAVE 13, 118–133. doi: 10.4319/mave.2010.978-0-9845591-0-7.118
Moraru, C., Varsani, A., and Kropinski, A. M. (2020). VIRIDIC-A novel tool to calculate the intergenomic similarities of prokaryote-infecting viruses. Viruses 12:1268. doi: 10.3390/v12111268
Munro, J., Oakey, J., Bromage, E., and Owens, L. (2003). Experimental bacteriophage-mediated virulence in strains of Vibrio harveyi. Dis. Aquat. Org. 54, 187–194. doi: 10.3354/dao054187
Nachimuthu, R., Madurantakam Royam, M., Manohar, P., and Leptihn, S. (2021). Application of bacteriophages and endolysins in aquaculture as a biocontrol measure. Biol. Control 160:104678. doi: 10.1016/j.biocontrol.2021.104678
Nobrega, F. L., Costa, A. R., Kluskens, L. D., and Azeredo, J. (2015). Revisiting phage therapy: new applications for old resources. Trends Microbiol. 23, 185–191. doi: 10.1016/j.tim.2015.01.006
Phumkhachorn, P., and Rattanachaikunsopon, P. (2010). Isolation and partial characterization of a bacteriophage infecting the shrimp pathogen Vibrio harveyi. Afr. J. Microbiol. Res. 4, 1794–1800.
Pires, D. P., Melo, L., Vilas Boas, D., Sillankorva, S., and Azeredo, J. (2017). Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 39, 48–56. doi: 10.1016/j.mib.2017.09.004
Raghu Patil, J., Desai, S. N., Roy, P., Durgaiah, M., Saravanan, R. S., and Vipra, A. (2014). Simulated hatchery system to assess bacteriophage efficacy against Vibrio harveyi. Dis. Aquat. Org. 112, 113–119. doi: 10.3354/dao02806
Rombel, I. T., Sykes, K. F., Rayner, S., and Johnston, S. A. (2002). ORF-FINDER: a vector for high-throughput gene identification. Gene 282, 33–41. doi: 10.1016/S0378-1119(01)00819-8
Roux, S., Paez-Espino, D., Chen, I. A., Palaniappan, K., Ratner, A., Chu, K., et al. (2021). IMG/VR v3: an integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 49, D764–D775. doi: 10.1093/nar/gkaa946
Sano, T. (1998). Control of fish disease, and the use of drugs and vaccines in Japan. J. Appl. Ichthyol. 14, 131–137. doi: 10.1111/j.1439-0426.1998.tb00630.x
Sawabe, T., Kita-Tsukamoto, K., and Thompson, F. L. (2007). Inferring the evolutionary history of vibrios by means of multilocus sequence analysis. J. Bacteriol. 189, 7932–7936. doi: 10.1128/JB.00693-07
Shivu, M. M., Rajeeva, B. C., Girisha, S. K., Karunasagar, I., Krohne, G., and Karunasagar, I. (2007). Molecular characterization of Vibrio harveyi bacteriophages isolated from aquaculture environments along the coast of India. Environ. Microbiol. 9, 322–331. doi: 10.1111/j.1462-2920.2006.01140.x
Stalin, N., and Srinivasan, P. (2016). Molecular characterization of antibiotic resistant Vibrio harveyi isolated from shrimp aquaculture environment in the south east coast of India. Microb. Pathog. 97, 110–118. doi: 10.1016/j.micpath.2016.05.021
Stalin, N., and Srinivasan, P. (2017). Efficacy of potential phage cocktails against Vibrio harveyi and closely related Vibrio species isolated from shrimp aquaculture environment in the south east coast of India. Vet. Microbiol. 207, 83–96. doi: 10.1016/j.vetmic.2017.06.006
Surekhamol, I. S., Deepa, G. D., Somnath Pai, S., Sreelakshmi, B., Varghese, S., and Bright Singh, I. S. (2014). Isolation and characterization of broad spectrum bacteriophages lytic to Vibrio harveyi from shrimp farms of Kerala, India. Lett. Appl. Microbiol. 58, 197–204. doi: 10.1111/lam.12175
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thiyagarajan, S., Chrisolite, B., Alavandi, S. V., Poornima, M., Kalaimani, N., and Santiago, T. C. (2011). Characterization of four lytic transducing bacteriophages of luminescent Vibrio harveyi isolated from shrimp (Penaeus monodon) hatcheries. FEMS Microbiol. Lett. 325, 85–91. doi: 10.1111/j.1574-6968.2011.02415.x
Vinod, M. G., Shivu, M. M., Umesha, K. R., Rajeeva, B. C., Krohne, G., Karunasagar, I., et al. (2006). Isolation of Vibrio harveyi bacteriophage with a potential for biocontrol of luminous vibriosis in hatchery environments. Aquaculture 255, 117–124. doi: 10.1016/j.aquaculture.2005.12.003
Walchli, S., Espanel, X., Harrenga, A., Rossi, M., Cesareni, G., and Hooft van Huijsduijnen, R. (2004). Probing protein-tyrosine phosphatase substrate specificity using a phosphotyrosine-containing phage library. J. Biol. Chem. 279, 311–318. doi: 10.1074/jbc.M307617200
Wang, Y., Barton, M., Elliott, L., Li, X., Abraham, S., O'Dea, M., et al. (2017). Bacteriophage therapy for the control of Vibrio harveyi in greenlip abalone (Haliotis laevigata). Aquaculture 473, 251–258. doi: 10.1016/j.aquaculture.2017.01.003
Wu, L., Tian, Y., Pang, M., Yang, Z., Bao, H., Zhou, Y., et al. (2021). A novel vibriophage vB_VhaS_PcB-1G capable of inhibiting virulent Vibrio harveyi pathogen. Aquaculture 542:736854. doi: 10.1016/j.aquaculture.2021.736854
Yang, Y., Cai, L., Ma, R., Xu, Y., Tong, Y., Huang, Y., et al. (2017). A novel roseosiphophage isolated from the oligotrophic South China Sea. Viruses 9:109. doi: 10.3390/v9050109
Yen, M., Cairns, L. S., and Camilli, A. (2017). A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 8:14187. doi: 10.1038/ncomms14187
Keywords: Vibrio phage, phage therapy, genome, biological characteristics, broad host range
Citation: Cai L, Tian Y, Li Z, Yang Y, Ai C and Zhang R (2023) A broad-host-range lytic phage vB_VhaS-R18L as a candidate against vibriosis. Front. Microbiol. 14:1191157. doi: 10.3389/fmicb.2023.1191157
Edited by:
Jamshid Tanha, National Research Council Canada (NRC), CanadaReviewed by:
Prasanth Manohar, Texas A&M University, United StatesSwapnil Ganesh Sanmukh, University of Leicester, United Kingdom
Copyright © 2023 Cai, Tian, Li, Yang, Ai and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rui Zhang, cnVpemhhbmdAeG11LmVkdS5jbg==; Chunxiang Ai, Y2h1bnhhaUB4bXUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Lanlan Cai
Lanlan Cai Yuan Tian1
†
Yuan Tian1
† Yunlan Yang
Yunlan Yang Chunxiang Ai
Chunxiang Ai Rui Zhang
Rui Zhang