- 1Key Lab of Biopesticide and Chemical Biology, Ministry of Education, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
- 2Department of Biological Sciences, University of Arkansas, Fayetteville, AR, United States
- 3Agricultural BioResources Institute, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 4Engineering Research Center of Edible and Medicinal Fungi, Ministry of Education, Jilin Agricultural University, Changchun, China
- 5College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, Fujian, China
Introduction: Species of Melanconiella include a diverse array of plant pathogens as well as endophytic fungi. Members of this genus have been frequently collected from the family Betulaceae (birches) in Europe and North America. Little, however, if known concerning the distribution of Melanconiella and/or their potential as pathogens of other plant hosts.
Methods: Fungi were noted and isolated from diseased leaves of Loropetalum chinense (Chinese fringe flower) and Camellia sinensis (tea) in Fujian Province, China. Genomic DNA was extracted from fungal isolates and the nucleotide sequences of four loci were determined and sued to construct phylogenetic trees. Morphological characteristics of fungal structures were determined via microscopic analyses.
Results: Four strains and two new species of Melanconiella were isolated from infected leaves of L. chinense and C. sinensis in Fujian Province, China. Based on morphology and a multi-gene phylogeny of the internal transcribed spacer regions with the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA (LSU), the second largest subunit of RNA polymerase II (RPB2), and the translation elongation factor 1-α gene (TEF1-α), Melanconiellaloropetali sp. nov. and Melanconiellacamelliae sp. nov. were identified and described herein. Detailed descriptions, illustrations, and a key to the known species of Melanconiella are provided.
Discussion: These data identify new species of Melanconiella, expanding the potential range and distribution of these dark septate fungi. The developed keys provide a reference source for further characterization of these fungi.
1. Introduction
Species of Melanconiella (Melanconiellaceae, Diaporthales) are important fungal wood pathogens and endophytes in Europe and North America (Du et al., 2017). Based on the dark-colored ascospore characteristics of the fungus, the genus Melanconiella was established and typified as M. spodiaea, with numerous revisions and adjustments occurring over the years (Saccardo, 1882; Voglmayr et al., 2012). These fungi belong within the broader category of dark septate fungi, that includes the dark septate root endophytes (DSE) which are conidial Ascomycetes (Jumpponen and Trappe, 1998; Ruotsalainen et al., 2022). DSE have been isolated from over 600 plant species, and have been distributed into >320 genera, occurring from the tropics to arctic and alpine habitats. These fungi comprise a heterogeneous group overlapping with soil, saprotrophic rhizoplane-inhabiting, mycorrhizal, and obligate/facultatively pathogenic fungi within ecosystems and in terms of their biology. Of note, many DSE have been examined in particular due to their effects on plant resistances to a wide range of biotic and abiotic stress (Santos et al., 2021). Within the past 30 years, 21 species in the genus Melanconiella have been recorded (Voglmayr et al., 2012). Wehmeyer (1926) indicated that most ascospores are arranged in a single row and only one type of conidia are needed to distinguish between Melanconiella and Melanconis. However, Wehmeyer (1941) considered Melanconiella to be a synonym of Melanconis, based on new classification standards. This viewpoint was supported by most scholars and led to confusion between Melanconiella and Melanconis (Voglmayr et al., 2012).
Since the development of modern genetic methods, molecular phylogenetic analysis has been applied to the taxonomy of Melanconiella. Based on the nucleotide sequence of the 28S large subunit of nuclear ribosomal RNA (LSU), the genus Melanconiella was separated from Melanconidaceae by Castlebury et al. (2002). Based on a multi-gene phylogeny (ITS, LSU, RPB2 and TEF1-α), Voglmayr et al. (2012) confirmed that Melanconiella was a special branch of Melanconis and suggested 13 accepted species. The genus Melanconiella was later accommodated into the Melanconiellaceae (Senanayake et al., 2017). A new species (M. syzygii) from diseased leaves of Syzygium sp. was added to Melanconiella by Crous et al. (2016). Du et al. (2017) reported a new species (M. cornuta) from Cornus controversa in Shaanxi Province. However, on the basis of phylogenetic analysis of combined ITS, LSU, CAL, RPB2 and TEF1-α sequence data, Fan et al. (2018) suggested that M.cornuta should be transferred from Melanconiella to Sheathospora. Meanwhile, two additional species (M. betulicola and M. corylina) were added to Melanconiella (Fan et al., 2018).
In the present study, four specimens of Melanconiella were collected from diseased leaves of Loropetalum chinense and Camellia sinensis in Fujian Province, China. Here, we sought to:
i. Determine/extend the host range and geographical distribution of Melanconiella;
ii. Report new species Melanconiella loropetali sp. nov. and Melanconiella camelliae sp. nov. with detailed descriptions and illustrations;
iii. Compare these new species with other species in the genus Melanconiella; and
iv. Provide a key to all known species of Melanconiella.
2. Materials and methods
2.1. Fungal isolates and morphology
Specimen samples were collected from the Wuyi Mountain National Nature Reserve, Fujian Province, China. Colonies of the two new species Melanconiella described herein were isolated from diseased leaves of Loropetalum chinense and Camellia sinensis using standard issue isolation methods (Senanayake et al., 2020; Jiang et al., 2022). Tissue fragments about 25 mm2 in total extent were taken from the margin of leaves with typical spot symptoms. These were sterilized by immersion in 75% ethanol solution for 60 s, placed in sterile deionized water for 45 s, transferred to 5% sodium hypochlorite solution for 30 s, and then rinsed three times in sterile deionized water for 60 s. The fragments were dried with sterilized filter paper and then transferred onto PDA plates (PDA medium: deionized water 1,000 mL, potato 200 g, agar 20 g, dextrose 20 g, pH ~7.0, available after sterilization) and incubated at 25°C for 5–7 days (Cai et al., 2009). Growing edges of fungal hyphae were removed to new PDA plates (at least two times) to obtain pure cultures. To promote sporulation and visualize the appearance of colonies, hyphae were inoculated onto the center of PDA prepared with pine needle and synthetic low nutrient agar SNA (SNA medium: deionized water 1,000 mL, KH2PO4 1 g, KNO3 1 g, MgSO4.7H2O 0.5 g, KCl 0.5 g, dextrose 0.2 g, sucrose 0.2 g, agar 12 g, available after sterilization) and incubated at 25°C under alternating conditions of 12 h near ultraviolet light and 12 h dark (Cai et al., 2009; Zhang et al., 2023).
Following 7–14 days of incubation, morphological characteristics of the (Melanconiella) isolates were recorded as per previous reports (Cai et al., 2009). Photographs of the colonies were taken at 7 days and 14 days after inoculation using a digital camera (Canon EOS 6D MarkII). Micromorphological characters of conidiomata were observed using a stereomicroscope (Nikon SMZ74), as well as by a compound microscope and by scanning electron microscopy (SEM, Nikon Ni-U; HITACHI SU3500). Measurements of micromorphological structures were determined using Digimizer software. All strains were stored in 10% sterilized glycerin and sterile water at 4°C for detailed studies in the future. The specimens were deposited in the Herbarium Mycologicum Academiae Sinicae, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS), accession numbers given in text. Living cultures were deposited in the China General Microbiological Culture Collection Center (CGMCC). Taxonomic information of the new taxa was submitted to MycoBank (http://www.mycobank.org; Crous et al., 2004).
2.2. DNA extraction and amplification
Genomic DNA was extracted from Melanconiella fungal mycelia grown on PDA for 14 days, using the Fungal DNA Mini Kit (OMEGA-D3390, Feiyang Biological Engineering Corporation, Guangzhou, China) according to the product manual. Nucleotide sequences were obtained from four gene loci including the internal transcribed spacer regions with the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA (LSU), the second largest subunit of RNA polymerase II (RPB2), and the translation elongation factor 1-α gene (TEF1-α). These were amplified by primer pairs and the polymerase chain reaction (PCR) programs as described (Table 1).
PCR was performed using a Bio-Rad Thermocycler (California, United States). Amplification reactions were performed in a 25 μL reaction volume which contained 12.5 μL of 2 × Rapid Taq Master Mix (Vazyme, Nanjing, China), 1 μL of each forward and reverse primer (10 μM) (Sangon, Shanghai, China), and 1 μL of template genomic DNA in the amplifier. These were adjusted with distilled deionized water to a total volume of 25 μL. PCR products were visualized on using 1% agarose gel electrophoresis. Bidirectional (both strand) sequencing was conducted by the Tsingke Company Limited (Fuzhou, China). Consensus sequences were assembled using MEGA 7.0 (Kumar et al., 2016).
2.3. Phylogenetic analysis
To construct the phylogenetic trees for Melanconiella, the sequences generated from the four strains considered in this study, and all available reference sequences were downloaded from GenBank. Multiple sequence alignments for ITS, LSU, RPB2 and TEF1-α were constructed and carried out using the MAFFT v.7.11 online program (http://mafft.cbrc.jp/alignment/server/; Katoh et al., 2019) and corrected manually using MEGA 7.0 (Kumar et al., 2016). Phylogenetic analyzes were based on maximum likelihood (ML) and Bayesian inference (BI) methods.
The ML was run on the CIPRES Science Gatewayportal by RaxML-HPC2 on XSEDE v. 8.2.12 (Miller et al., 2010; Stamatakis, 2014). Bayesian analysis was performed in MrBayes on XSEDE v. 3.2.7a (Huelsenbeck and Ronquist, 2001; Ronquist and Huelsenbeck, 2003; Ronquist et al., 2012) and the best evolutionary model for each partition was determined using MrModeltest v. 2.3 (Posada and Crandall, 1998; Nylander, 2004; Darriba et al., 2012) and included the analyzes (Dong et al., 2020). Four simultaneous Markov Chain Monte Carlo (MCMC) chains were run for 160,000 generations. In addition, a sampling frequency of 100 generations was performed in the test. The burn-in fraction was set to 0.25 and posterior probabilities (PP) were determined from the remaining trees (He et al., 2022). The consensus trees were plotted using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree; Rambaut, 2018) and edited using Adobe Illustrator CS 6.0 (Figure 1). New sequences generated in this study were deposited in GenBank (https://www.ncbi.nlm.nih.gov; Table 2).
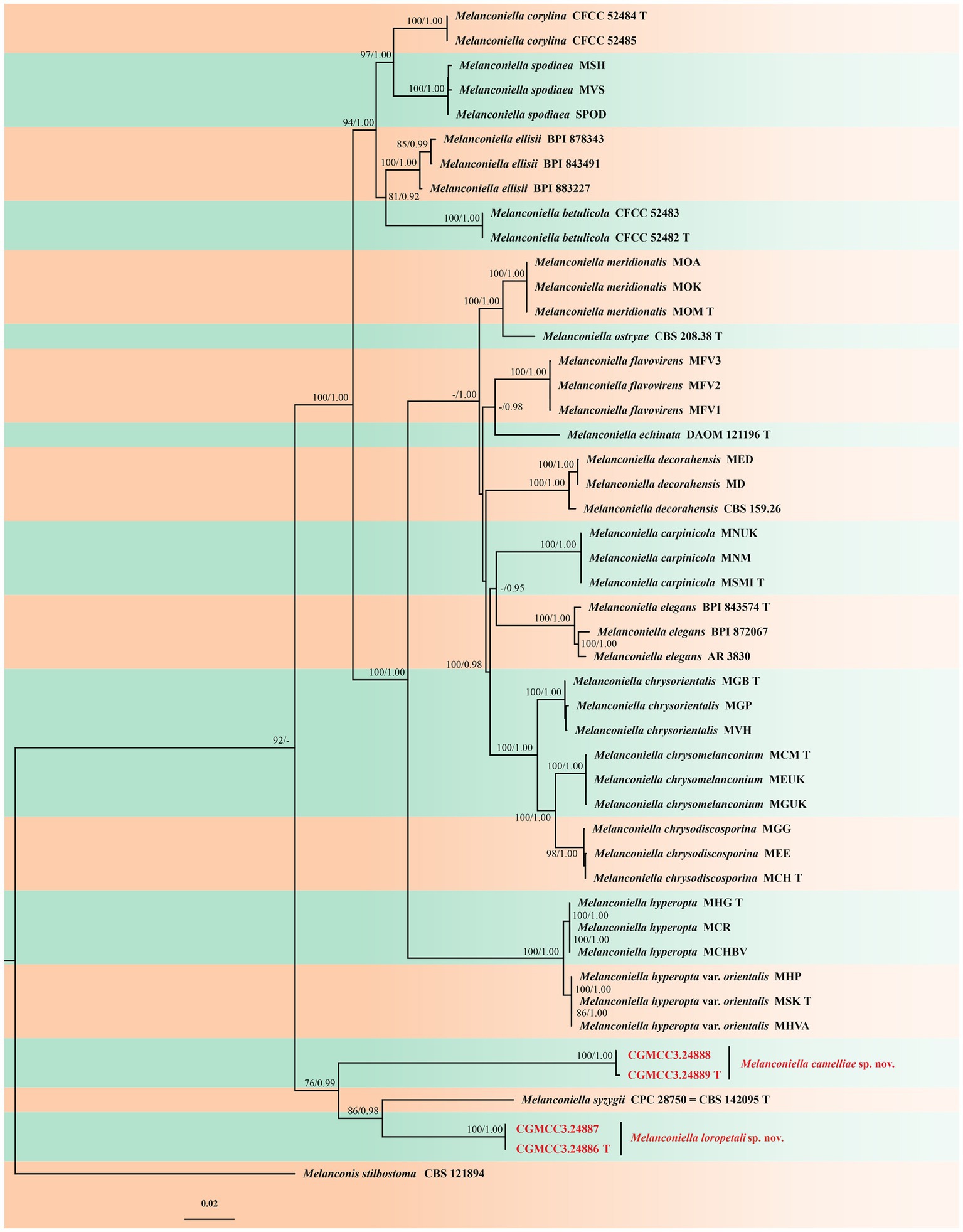
Figure 1. Phylogram of Melanconiella based on combined ITS, LSU, RPB2 and TEF1-α genes, with Melanconis stilbostoma (CBS 121894) as outgroup. The ML and BI bootstrap support values above 70% and 0.90 BYPP are shown at the first and second position, respectively. Strains marked with “T” are ex-type or ex-epitype. Strains from this study are shown in red. The scale bar at the left–bottom represents 0.02 substitutions per site.
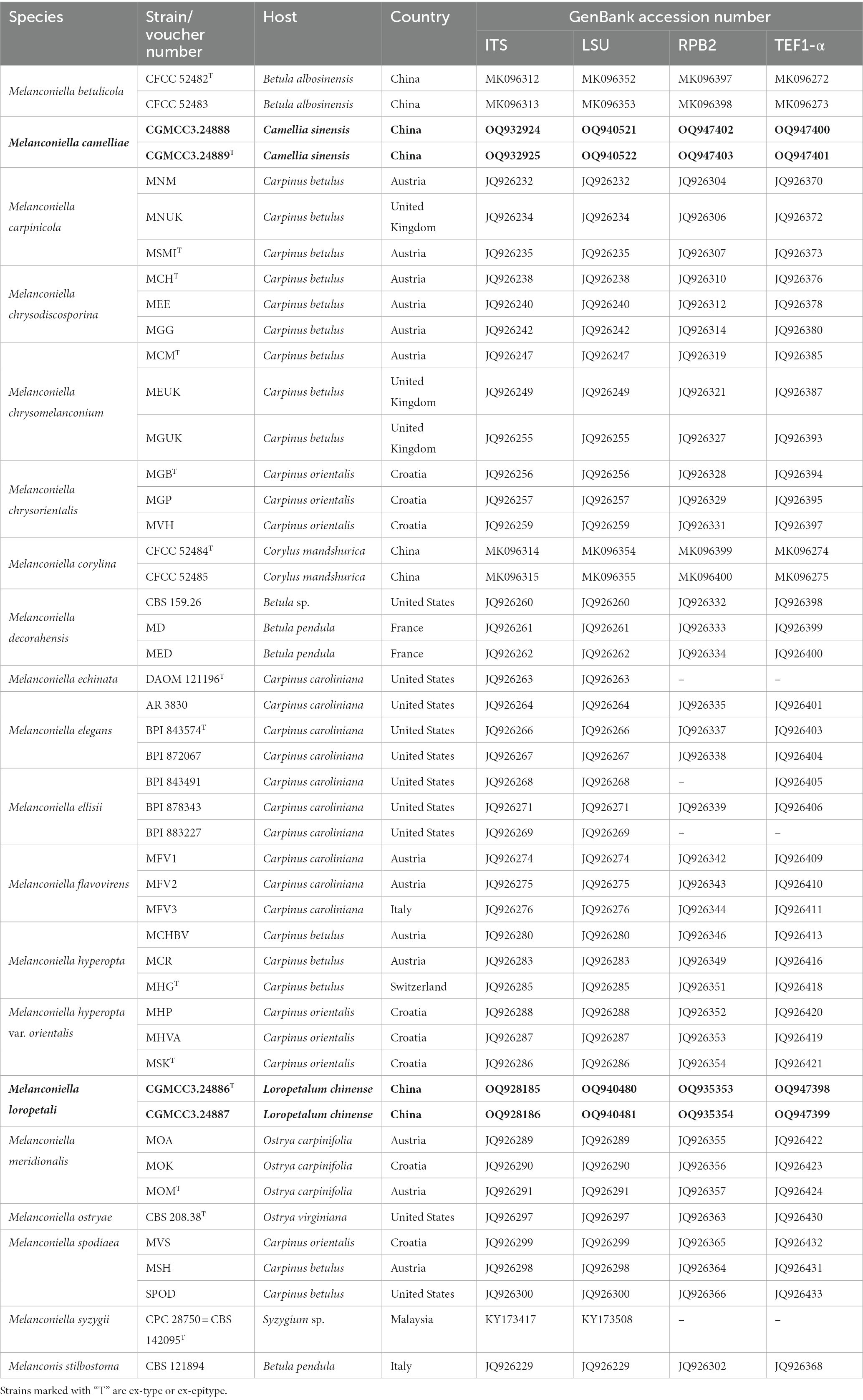
Table 2. Species and GenBank accession numbers of DNA sequences used in this study, with new sequences indicated in bold.
3. Results
3.1. Phylogeny
Four isolates of Melanconiella were identified as representing two new species based on an analysis of combined ITS, LSU, RPB2, and TEF1-α gene nucleotide sequences, and combined with 47 isolates of Melanconiella along with Melanconis stilbostoma (CBS 121894) as the outgroup taxon (Voglmayr et al., 2012). The dataset had an aligned length of 6,068 characters including gaps (i.e., ITS: 1–1723, LSU: 1724–3,416, RPB2: 3417–4,590, TEF1-α: 4591–6,068). Of these characters, 4,369 were constant, 357 were variable and parsimony-uninformative, and 1,342 were parsimony-informative.
The Bayesian analysis was performed for 160,000 generations, resulting in 3202 total trees, of which 2,402 trees were used to calculate the posterior probabilities. The BI posterior probabilities were plotted on the ML tree. For the ML and BI analyzes, GTR + I + G for ITS, RPB2, TEF1-α, and LSU [Lsetnst = 6, rates = invgamma; Prsetstatefreqpr = Dirichlet (1,1,1,1)], were selected and incorporated into the analysis. Bayesian analysis resulted in an average standard deviation of split frequencies = 0.009565. The topology of the ML tree was consistent with that of the Bayesian tree. Hence, the ML tree is presented (Figure 1).
ML bootstrap support values (≥70%) and Bayesian posterior probability (≥0.90) are shown as the first and second positions, respectively, above the nodes. The 48 strains were assigned to 20 species clades based on the four gene loci phylogeny (Figure 1). The four strains studied herein represented two novel species. The new species Melanconiella loropetali showed a close relationship to M. syzygii (CBS 142095), with good support (ML-BS:86% and BYPP:0.98). The new species M.camelliae (CGMCC3.24889) showed a close relationship to M. syzygii (CBS 142095) and M. loropetali (CGMCC3.24886) with good support (ML-BS:76% and BYPP:0.99).
3.2. Taxonomy
Melanconiella loropetali T.C. Mu and Jun Z. Qiu, sp. nov. (Figure 2).
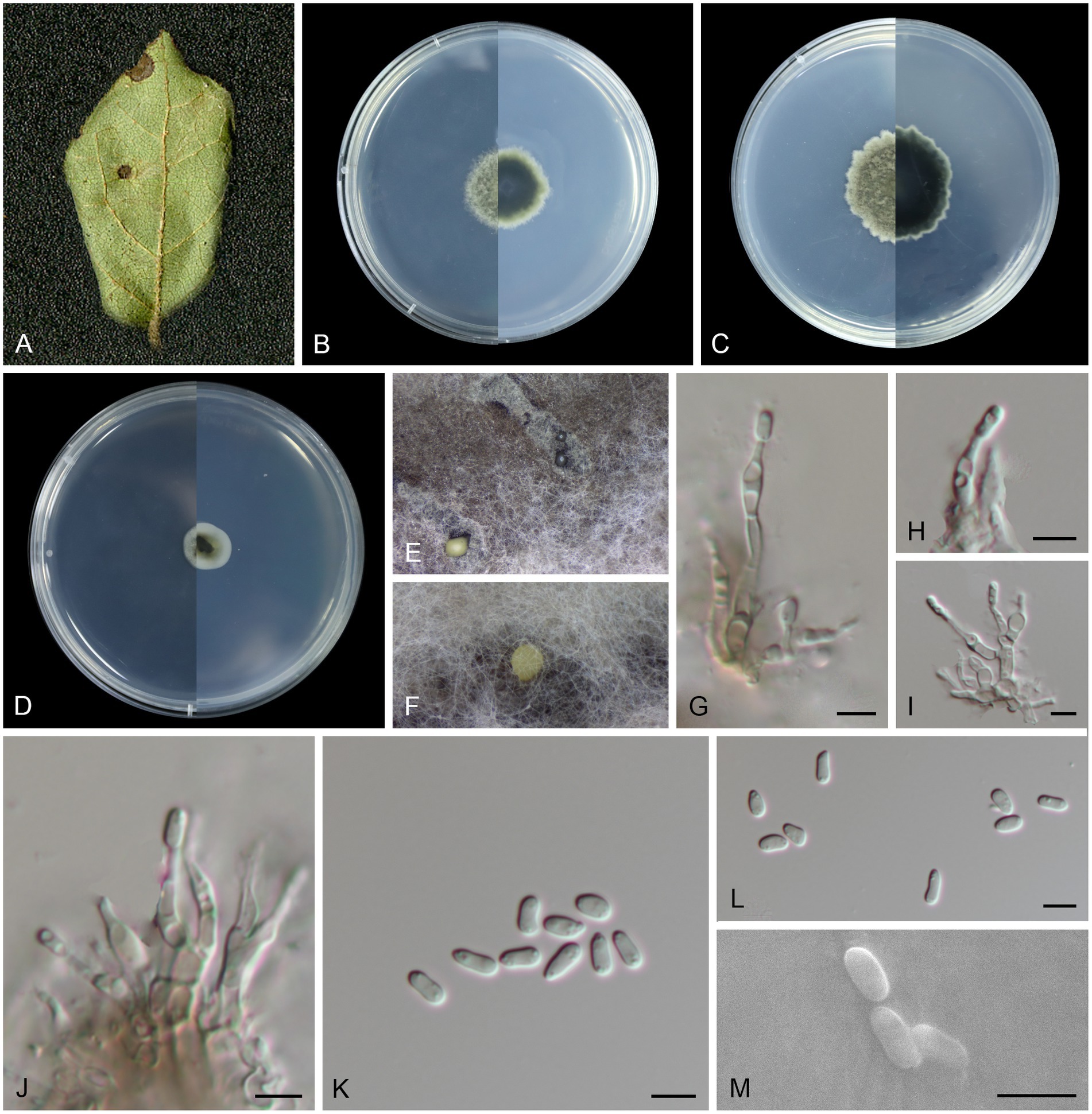
Figure 2. Morphological characteristics Melanconiella loropetali (holotype HMAS 257907) (A) Symptomatic leaves of Loropetalum chinense. (B) Surface and reverse sides of colony after incubation for 7 days on PDA (C) and 14 days. (D) Surface and reverse sides of colony after incubation for 7 days on SNA. (E,F) Conidiomata. (G-J) Conidiophores, conidiogenous cells and conidia. (K-M) Conidia. (G-M) Scale bars: 5 μm.
MycoBank number: MB848666.
Holotype: CHINA, Fujian Province, Fujian Wuyi Mountain National Nature Reserve, 117°41′19.82″E, 27°44′53.91″N, from diseased leaves of Loropetalum chinense, 7 September 2022, T.C. Mu (holotype HMAS 257907; ex-type living culture CGMCC3.24886).
Etymology: The epithet “loropetali” pertains to the generic name of the host plant Loropetalum chinense.
Description: Leaf spots circular, sunken in the middle, brown or tan, 4–10 mm diam. Conidiomata acervular to pycnidial, erumpent on agar, solitary, globose, black or creamy, 330–410 μm diam, black and cream conidial droplets exuding from the ostioles. Conidiophores hyaline to light brown, smooth, fusiform, subcylindrical, 1–5-septate, branched and septate at the base, 9.2–26.8 × 1.6–3.5 μm. Conidiogenous cells hyaline, smooth, phialidic, subglobular and globular, subcylindrical, 1.8–4.3 × 1.5–3.4 μm. Conidia unicellular, hyaline, smooth, narrowly ellipsoid to fusoid, subcylindrical, thin-walled, (3.3–6.6 × 1.6–3.0 μm, mean ± SD = 4.57 ± 0.75 × 2.17 ± 0.33 μm, L/W ratio = 2.2, n = 30). Sexual morph was not observed.
Culture characteristics: Colonies on PDA flat with feathery margin, aerial mycelium white or dark brick-red, floccose. On PDA surface pale mouse gray to mouse gray, reverse black. On SNA surface and reverse gray in the center and white margin. PDA attaining 18.5–22.8 mm in diameter after 7 days, at 25°C, with a calculated growth rate 2.6–3.2 mm/day. SNA attaining 9.8–13.6 mm in diameter after 7 days, at 25°C, slow growing, calculated growth rate 1.4–1.9 mm/day.
Other specimens examined: CHINA, Fujian Province, Fujian Wuyi Mountain National Nature Reserve, 117°41′19.82″E, 27°44′53.91″N, from diseased leaves of Loropetalum chinense, 7 September 2022, T.C. Mu (paratype HMAS 257908; ex-paratype living culture CGMCC3.24887).
Notes: In this study, a member of the genus Melanconiella was collected from the Hamamelidaceae. Two strains (CGMCC3.24886; CGMCC3.24887) were isolated from diseased leaves of Loropetalum chinense and identified as Melanconiella loropetali sp. nov. Phylogenetic analysis using four genes showed that M. loropetali formed an independent clade (Figure 1) and was phylogenetically distinct from M. syzygii (CBS 142095) with high statistical support (86% ML/0.98 PP, Figure 1). The nucleotide comparison of ITS sequences of M. syzygii revealed 58 bp (58/601 bp, 9.65%) nucleotide differences. The nucleotide comparison of LSU sequences of M. syzygii revealed 14 bp (14/817 bp, 1.71%) nucleotide differences. Morphologically, M. loropetali differs from M. syzygii in having smaller conidia and conidiophores (3.3–6.6 × 1.6–3.0 vs. 8.0–11.0 × 5.0–6.0 μm; 9.2–26.8 × 1.6–3.5 vs. 12.0–30.0 × 4.0–6.0 μm [Crous et al., 2016]). Therefore, we describe this fungus as a new species.
Melanconiella camelliae T.C. Mu and Jun Z. Qiu, sp. nov. (Figure 3).
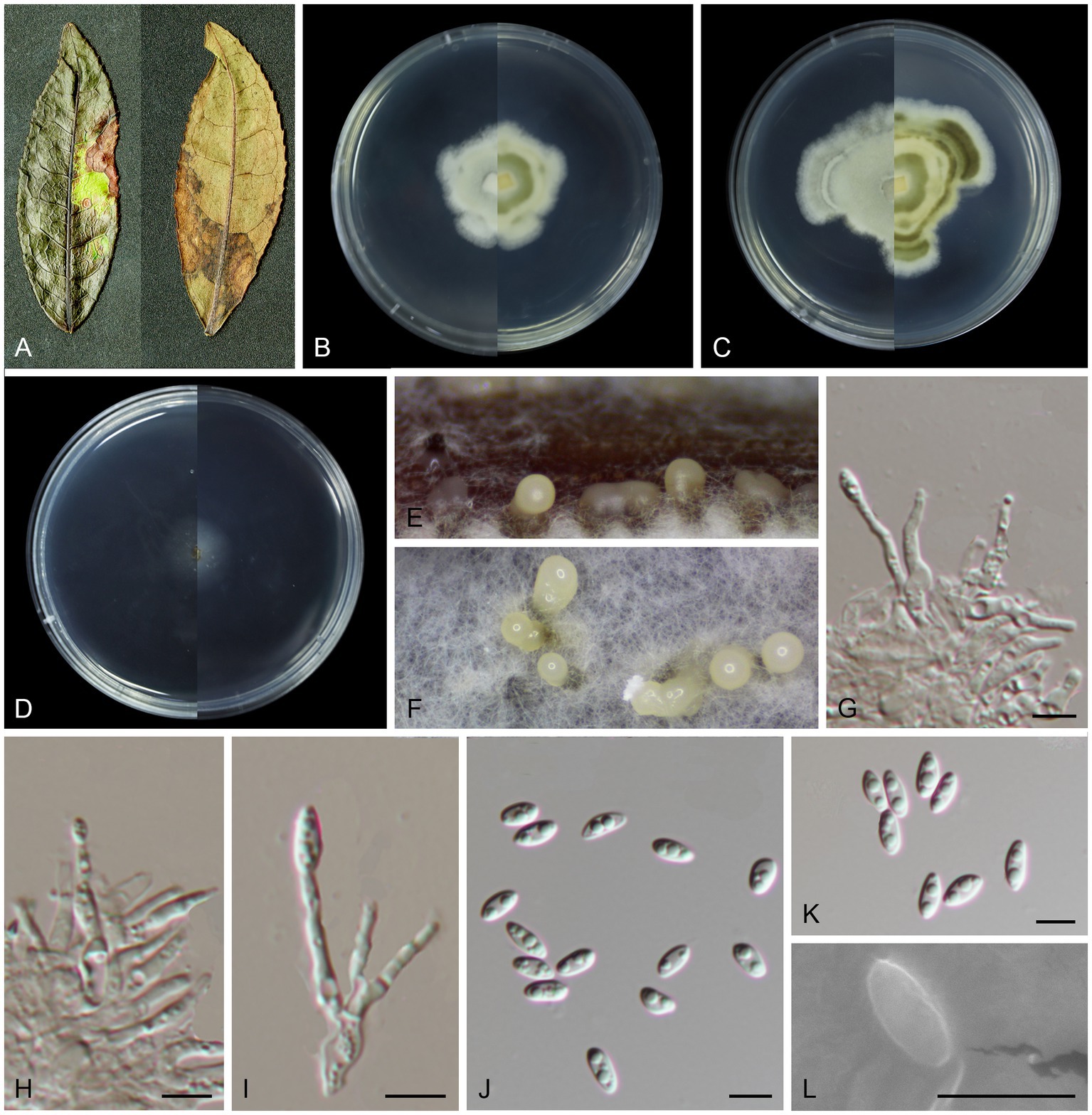
Figure 3. Morphological characteristics Melanconiella camelliae (holotype HMAS 257910) (A) Symptomatic leaves of Camellia sinensis. (B) Surface and reverse sides of colony after incubation for 7 days on PDA (C) and 14 days. (D) Surface and reverse sides of colony after incubation for 7 days on SNA. (E,F) Conidiomata. (G-I) Conidiophores, conidiogenous cells and conidia. (J-L) Conidia. (G-L) Scale bars: 5 μm.
MycoBank number: MB848667.
Holotype: CHINA, Fujian Province, Fujian Wuyi Mountain National Nature Reserve, 117°41′19.81″E, 27°44′53.92″N, from diseased leaves of Camellia sinensis, 7 September 2022, T.C. Mu (holotype HMAS 257910; ex-type living culture CGMCC3.24889).
Etymology: The epithet “camelliae” refers to the generic name of the host plant Camellia sinensis.
Description: Leaf spots irregular, brown or umber. Conidiomata acervular to pycnidial, solitary, 180–260 μm diam, with cream conidial droplets exuding from the ostioles. Conidiophores, hyaline, smooth, 1–3-septate, mostly straight, cylindrical-clavate, branched at the base, 10.4–18.6 × 1.4–2.1 μm. Conidiogenous cells hyaline, smooth, circular, elliptical, phialidic, 1.5–3.5 × 1.0–1.7 μm. Conidia hyaline, smooth, oblong elliptical and fusoid, multi-guttulate, (4.7–5.9 × 2.0–2.8 μm, mean ± SD = 5.36 ± 0.35 × 2.32 ± 0.23 μm, L/W ratio = 2.3, n = 30). Sexual morph was not observed.
Culture characteristics: Colonies on PDA flat with irregular stripes, aerial mycelium white, cottony. On PDA surface white, reverse yellowish and darker. Colonies on SNA sparse hyphae, slow growing. PDA attaining 26.3–31.9 mm in diameter after 7 days, at 25°C, with a calculated growth rate 3.7–4.5 mm/day. SNA attaining 13.8–18.8 mm in diameter after 7 days, at 25°C, slow growing, calculated growth rate 1.9–2.6 mm/day.
Other specimens examined: CHINA, Fujian Province, Fujian Wuyi Mountain National Nature Reserve, 117°41′19.81″E, 27°44′53.92″N, from diseased leaves of Camellia sinensis, 7 September 2022, T.C. Mu (paratype HMAS 257909; ex-paratype living culture CGMCC3.24888).
Notes: In this study a member of the Melanconiella was collected for the first time from the Theaceae. Two strains (CGMCC3.24888; CGMCC3.24889) were isolated from diseased leaves of Camellia sinensis and identified as Melanconiella camelliae sp. nov. Phylogenetic analysis of four genes showed that M. camelliae formed an independent clade (Figure 1) and was phylogenetically distinct from M. syzygii and M. loropetali with moderate statistical support (76% ML/0.99 PP, Figure 1). The nucleotide comparison of ITS sequences of M. syzygii revealed 116 bp (116/488 bp, 23.77%) nucleotide differences. The nucleotide comparison of LSU sequences of M. syzygii revealed 53 bp (53/821 bp, 6.46%) nucleotide differences. Morphologically, M. camelliae differs from M. syzygii in having smaller conidia and conidiophores (4.7–5.9 × 2.0–2.8 vs. 8.0–11.0 × 5.0–6.0 μm; 10.4–18.6 × 1.4–2.1 vs. 12.0–30.0 × 4.0–6.0 μm [Crous et al., 2016]). Melanconiella camelliae differs from M. loropetali in having smaller conidiophores (10.4–18.6 × 1.4–2.1 vs. 9.2–26.8 × 1.6–3.5 μm). Therefore, we describe this fungus as a new species.
3.3. Key to species of Melanconiella
1. Conidiophores septate……………2.
1. Conidiophores aseptate……………4.
2. On Camellia sinensis; conidiophores 1–3-septate; conidial size 4.7–5.9 × 2.0–2.8 μm, L/W ratio = 2.3……………M. camelliae sp. nov.
2. Conidiomata solitary, globose……………3.
3. On Syzygium; conidiophores ampulliform, unbranched; conidiogenous cells integrated, terminal; conidial size 8.0–11.0 × 5.0–6.0 μm……………M. syzygii.
3. On Loropetalum; conidiophores 1–5-septate; conidial size 3.3–6.6 × 1.6–3.0 μm……………M. loropetali sp.nov.
4. Ascospores dark brown; conidia dark brown……………5.
4. Ascospores hyaline; conidia hyaline or dark brown……………7.
5. Ascospores without appendages; conidia pip-shaped, multiguttulate when fresh, with a large guttule when dead; on Betula spp. in the north temperate zone……………M. decorahensis.
5. Conidia usually ellipsoid, guttulate……………6.
6. Perithecia 0.3–0.5 mm diam, up to 20 per stroma; conidia variable in shape, ovoid, obovoid, usually not distinctly pip-shaped; on Carpinus in Europe……………M. spodiaea.
6. Ectostromatic disc inconspicuous and cracked around the margin at maturity; conidia narrowly ellipsoid, 1–3 guttulate; on Corylus mandshurica in China……………M. corylina.
7. On Carpinus……………8.
7. On other hosts……………16.
8. Ascospores with obvious ascospore wall and with monomorphic, (sub) globose to bullet-shaped ascospore cells mostly larger than 7 μm in width; commonly with yellow ectostroma……………9.
8. Ascospores without obvious ascospore wall and with slightly to distinctly dimorphic cells mostly smaller than 7 μm in width; ectostroma gray, brownish, yellowish, pale brown, cream or yellow……………11.
9. Ascospores 16.0–27.5 × 7.5–15.5 μm; with obvious conidiomata containing dark brown conidia; on C. betulus……………M. chrysomelanconium.
9. Ascospores 15.5–22.0 × 6.0–15.0 μm; hyaline conidia……………10.
10. Entostroma crumbly, of subhyaline to yellowish hyphae; conidial size 12.5–19.0 × 4.5–6.0 μm; on C. betulus……………M. chrysodiscosporina.
10. Asci rarely biseriate ascospores; conidial size 11.5–15.5 × 5.5–7.5 μm; on C. orientalis……………M. chrysorientalis.
11.Ascospores were usually more than 5.5 μm in width and 18.5 μm in length……………12.
11. Ascospores were usually less than 5.5 μm in width and 18.5 μm in length……………14.
12. On Carpinus in Europe; ectostromatic discs concave, flat or slightly pustulate and little projecting above the perithecial level, paler to fawn; central column pale yellow to grayish brown……………13.
12. On C. caroliniana in North America; ectostromatic discs well-developed, distinctly projecting, often pulvinate, with circular, angular or ellipsoid outline, brownish, brown, cream or yellow; central column yellow when young, later green to brown; entostroma well-developed, of compacted hyphae, green or brown……………M. echinata.
13. Central column pale, yellowish, pale grayish brown or brownish; conidiogenous cells 10.0–30.0 × 1.5–3.0 μm; on C. betulus……………M. hyperopta.
13. Ascospores 2.5–3.6 × 1.8–2.3 μm; conidia often with one to three larger and few to numerous small guttules; On C. orientalis……………M. hyperopta var. orientalis.
14. Pseudostromata perithecia only rarely projecting, 0.8–1.7 mm diam; conidiogenous cells 10.0–18.0 × 1.5–2.5 μm; on C. betulus in Europe……………M. carpinicola.
14. On C. caroliniana or Betula……………15.
15. Ectostromatic discs distinctly projecting, mostly circular, angular or ellipsoid, often pulvinate, pale yellow to pale brown; or concealed by ostioles; ostioles evenly spaced in the disc; central column yellow, gray or brownish; conidiogenous cells 10.0–25.0 × 2.0–3.0 μm……………M. elegans.
15. Ectostromatic disc mostly circular; ostioles black……………16.
16. Ectostromatic disc mostly irregularly shaped, usually surrounded by irregular edges or flaps of bark, drab, light to dark gray or tanned; ostioles mostly marginal in the disc; central column gray or pale brown; conidial size 8.5–13.0 × 2.5–4.0 μm……………M. ellisii.
16. Ectostromatic disc usually buff to hazel; conidial size 9.5–15.0 × 2.0–5.5 μm; on Betula in China……………M. betulicola.
17. On Corylus; ascospores broadly fusoid; ascospore ends obviously subacute with persistent knob-like hyaline appendages; conidia often with one or two larger and numerous small guttules……………M. flavovirens.
17. On Ostrya; ascospores fusoid; ascospore ends narrowly rounded to subacute, without or with cap-like appendages……………18.
18. Ascospores 20.0–37.5 × 5.0–9.5 μm; conidia hyaline cylindrical to suballantoid; on Ostrya carpinifolia in Europe……………M. meridionalis.
18. Ascospores 14.5–23.0 × 4.0–8.5 μm; conidia dark brown with a light brown equatorial zone, without guttules or granules; on Ostrya virginiana in North America……………M. ostryae.
4. Discussion
In this study, fungal specimens were collected from diseased leaves of Loropetalum chinense and Camellia sinensis in China. Based on combined morphological and multi-gene phylogenetic analyzes, two new species, Melanconiella loropetali sp. nov. and Melanconiella camelliae sp. nov., were identified, with detailed descriptions and illustrations provided. To better clarify the relationship between species, as well as provide distinguishing characteristics useful for separating the isolates from similar species of Melanconiella, we provide a key to Melanconiella. These results extend the host range and geographical distribution of Melanconiella.
Melanconis spodiaea, the type species of the genus Melanconiella, was introduced by Saccardoin 1882. Currently, 40 names were recorded in the Index Fungorum and 43 species in MycoBank (http://www.indexfungorum.org/; https://www.mycobank.org/; accessed 22 April 2023).
In the late 19th and early 20th centuries, 21 species were classified within Melanconiella. Based on a multi-gene phylogeny (ITS, LSU, RPB2 and TEF1-α), Voglmayr et al. (2012) confirmed 13 species in the genus Melanconiella, including M. carpinicola (Fuckel) Voglmayr & Jaklitsch, M. chrysodiscosporina Voglmayr & Jaklitsch, M. chrysomelanconium Voglmayr & Jaklitsch, M. chrysorientalis Voglmayr & Jaklitsch, M. echinata Voglmayr & Jaklitsch, M. elegans Voglmayr & Jaklitsch, M. ellisii (Rehm ex Ellis & Everh.) Voglmayr & Jaklitsch, M. flavovirens (G.H. Otth) Voglmayr & Jaklitsch, M. hyperopta (Nitschke ex G.H. Otth) Voglmayr & Jaklitsch, M. hyperopta var. orientalis Voglmayr & Jaklitsch, M. meridionalis Voglmayr & Jaklitsch, M. ostryae (Dearn.) Voglmayr & Jaklitsch, and M. spodiaea (Tul. & C. Tul.) Sacc. (Voglmayr et al., 2012).
Voglmayr et al. (2012) considered species of Melanconiella to occur only in Europe and North America, and collected essentially from Betulaceae. Nevertheless, Crous et al. (2016) reported a new species (M. syzygii) from diseased leaves of Syzygium sp. in Malaysia, while M. betulicola and M. corylina were reported from symptomatic branches in China by Fan et al. (2018). In the present study, two new species are reported from diseased leaves of Loropetalum chinense and Camellia sinensis in China. It is likely that in Asia additional species of Melanconiella are present potentially from additional plant hosts. Both M. loropetali and M. camelliae were closely related to M. syzygii in their phylogenetic analyzes, with similar conidiomata characteristic. These reports, along with our own, may indicate a wider host specificity for Melanconiella than previously considered and may reflect local adaptations. However, Melanconiella may also exhibit opportunistic pathogenesis, with its main association with plants occurring as part of endophytic interactions (Du et al., 2017). Root inoculation of the cowpea plant (Vochysia divergens) with M. elegans (strain-21 W2) resulted in improved nutrition, growth, and photosynthesis under salt stress (Farias et al., 2020). However, research comparing endophytic vs. parasitic outcomes is lacking, and a more global analysis of the biology of Melanconiella is needed in order to better understand the ecology of these fungi.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TM and JQ designed the experimental plan. TM and JC analyzed the data with help from ZZ, XG, and WZ. TM, CY, and MZ collected the samples from the field. TM and HS wrote the manuscript. SS, PL, and JQ reviewed this manuscript. All authors contributed to the article and approved the submitted version.
Funding
The research was financed by the National Key R & D Program of China (nos. 2017YFE0122000 and 2022YFD1600300), the National Natural Science Foundation of China (nos. U1803232, 32270029, and 31670026), A Key Project from the Fujian Provincial Department of Science and Technology (no. 2020N5005), and Fujian Provincial Major Science and Technology Project (no. 2022NZ029017).
Acknowledgments
We would like to thank Prof. Nemat O. Keyhani at University of Florida, USA, for his carefully language editing. The authors would also like to thank China General Microbiological Culture Collection Center and Fungarium (HMAS), Institute of Microbiology, CAS for strain and sample storage help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1229705/full#supplementary-material
References
Cai, L., Hyde, K. D., Taylor, P. W. J., Weir, B. S., Waller, J. M., Abang, M. M., et al. (2009). A polyphasic approach for studying Colletotrichum. Fungal Divers. 26, 252–254. doi: 10.1016/j.riam.2009.11.001
Castlebury, L. A., Rossman, A. Y., Jaklitsch, W. J., and Vasilyeva, L. N. (2002). A preliminary overview of the Diaporthales based on large subunit nuclear ribosomal DNA sequences. Mycologia 94, 1017–1031. doi: 10.1080/15572536.2003.11833157
Chaverri, P., and Samuels, G. J. (2003). Hypocrea/Trichoderma (Ascomycota, Hypocreales, Hypocreaceae): species with green ascospores. Stud. Mycol. 48, 1–116. doi: 10.1023/B:MYCO.0000003579.48647.16
Crous, P. W., Gams, W., Stalpers, J. A., Robert, V., and Stegehuis, G. (2004). MycoBank: an online initiative to launch mycology into the 21stcentury. Stud. Mycol. 50, 19–22 doi: 10.1023/B:MYCO.0000012225.79969.29
Crous, P. W., Wingfield, M. J., Burgess, T. I., Hardy, G. E., Crane, C., Barrett, S., et al. (2016). Fungal planet description sheets: 469–557. Persoonia 37, 218–403. doi: 10.3767/003158516X694499
Darriba, D., Taboada, G. L., Doallo, R., and Posada, D. J. (2012). Model test 2: more models, new heuristics and parallel computing. Nat. Methods 9:772. doi: 10.1038/nmeth.2109
Dong, Z., Manawasinghe, I. S., Huang, Y., Shu, Y., Phillips, A. J. L., Dissanayake, A. J., et al. (2020). Endophytic Diaporthe associated with Citrus grandis cv. Tomentosa in China. Front. Microbiol. 11:609387. doi: 10.3389/fmicb.2020.609387
Du, Z., Fan, X. L., Yang, Q., and Tian, C. M. (2017). Host and geographic range extensions of Melanconiella, with a new species M. cornuta in China. Phytotaxa 327, 252–260. doi: 10.11646/phytotaxa.327.3.4
Fan, X., Du, Z., Bezerra, J. D. P., and Tian, C. (2018). Taxonomic circumscription of melanconis-like fungi causing canker disease in China. MycoKeys 42, 89–124. doi: 10.3897/mycokeys.42.29634
Farias, G. C., Nunes, K. G., Soares, M. A., De Siqueira, K. A., Lima, W. C., Neves, A. L. R., et al. (2020). Dark septate endophytic fungi mitigate the effects of salt stress on cowpea plants. Braz. J. Microbiol. 51, 243–253. doi: 10.1007/s42770-019-00173-4
He, J., Han, X., Luo, Z. L., Li, E. X., Tang, S. M., Luo, H. M., et al. (2022). Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microbiol. 13:1035434. doi: 10.3389/fmicb.2022.1035434
Huelsenbeck, J. P., and Ronquist, F. (2001). MRBAYES: Bayesian inference of phylogeny. Bioinformatics 17, 754–755. doi: 10.1093/bioinformatics/17.8.754
Jaklitsch, W. M., Komon, M., Kubicek, C. P., and Druzhinina, I. S. (2006). Hypocrea voglmayrii sp. nov. from the Austrian Alps represents a new phylogenetic clade in Hypocrea/Trichoderma. Mycologia 97, 1365–1378. doi: 10.1080/15572536.2006.11832743
Jiang, N., Voglmayr, H., Ma, C. Y., Xue, H., Piao, C. G., and Li, Y. (2022). A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys 92, 27–43. doi: 10.3897/mycokeys.92.86521
Jumpponen, A., and Trappe, J. M. (1998). Dark septate endophytes: a review of facultative biotrophic root-colonizing fungi. New Phytol. 140, 295–310. doi: 10.1046/j.1469-8137.1998.00265.x
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092
Miller, M. A., Pfeiffer, W., and Schwartz, T.. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE), New Orleans, LA, United States, pp. 1–8.
Nylander, J. A. A. (2004). MrModeltest 2.0; Program Distributed by the Author. Uppsala: Uppsala University.
Posada, D., and Crandall, K. A. (1998). Model test: testing the model of DN-A substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Rambaut, A. (2018). Molecular evolution, phylogenetics and epidemiology. FigTree ver.1.4.4 software (accessed October 2022).
Rehner, S. A., and Samuels, G. J. (1994). Taxonomy and phylogeny of Gliocladium analysed from nuclear large subunit ribosomal DNA sequences. Mycol. Res. 98, 625–634. doi: 10.1016/S0953-7562(09)80409-7
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Ruotsalainen, A. L., Kauppinen, M., Wali, P. R., Saikkonen, K., Helander, M., and Tuomi, J. (2022). Dark septate endophytes: mutualism from by-products? Trends Plant Sci. 27, 247–254. doi: 10.1016/j.tplants.2021.10.001
Santos, M., Cesanelli, I., Dianez, F., Sanchez-Montesinos, B., and Moreno-Gavira, A. (2021). Advances in the role of dark septate endophytes in the plant resistance to abiotic and biotic stresses. J. Fungi. 7:939. doi: 10.3390/jof7110939
Senanayake, I. C., Crous, P. W., Groenewald, J. Z., Maharachchikumbura, S. S. N., Jeewon, R., Phillips, A. J. L., et al. (2017). Families of Diaporthales based on morphological and phylogenetic evidence. Stud. Mycol. 86, 217–296. doi: 10.1016/j.simyco.2017.07.003
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Voglmayr, H., Rossman, A. Y., Castlebury, L. A., and Jaklitsch, W. M. (2012). Multigene phylogeny and taxonomy of the genus Melanconiella (Diaporthales). Fungal Divers. 57, 1–44. doi: 10.1007/s13225-012-0175-8
Wehmeyer, L. E. (1926). Cultural life histories of Melanconis and Pseudovalsa. II. Mycologia 18, 257–273. doi: 10.1080/00275514.1936.12017170
Wehmeyer, L. E. (1941). A revision of Melanconis, Pseudovalsa, Prosthecium and Titania. Univ. Michigan Stud., Sci. Ser. 14, 1–161.
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications. eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, NY: Academic), 315–322.
Keywords: leaf disease, Melanconiellaceae, multi-gene phylogeny, new species, taxonomy
Citation: Mu T, Chen J, Zhao Z, Zhang W, Stephenson SL, Yang C, Zhu M, Su H, Liu P, Guan X and Qiu J (2023) Morphological and phylogenetic analyzes reveal two new species of Melanconiella from Fujian Province, China. Front. Microbiol. 14:1229705. doi: 10.3389/fmicb.2023.1229705
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesReviewed by:
Rungtiwa Phookamsak, Shenzhen University, ChinaYusufjon Gafforov, Academy of Sciences Republic of Uzbekistan (UzAS), Uzbekistan
Hai-Sheng Yuan, Chinese Academy of Sciences (CAS), China
Copyright © 2023 Mu, Chen, Zhao, Zhang, Stephenson, Yang, Zhu, Su, Liu, Guan and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pu Liu, cHVsQGpsYXUuZWR1LmNu; Xiayu Guan, NDcxMjY5NDBAcXEuY29t; Junzhi Qiu, anVuemhpcWl1QDEyNi5jb20=
 Taichang Mu1
Taichang Mu1 Steven L. Stephenson
Steven L. Stephenson Junzhi Qiu
Junzhi Qiu