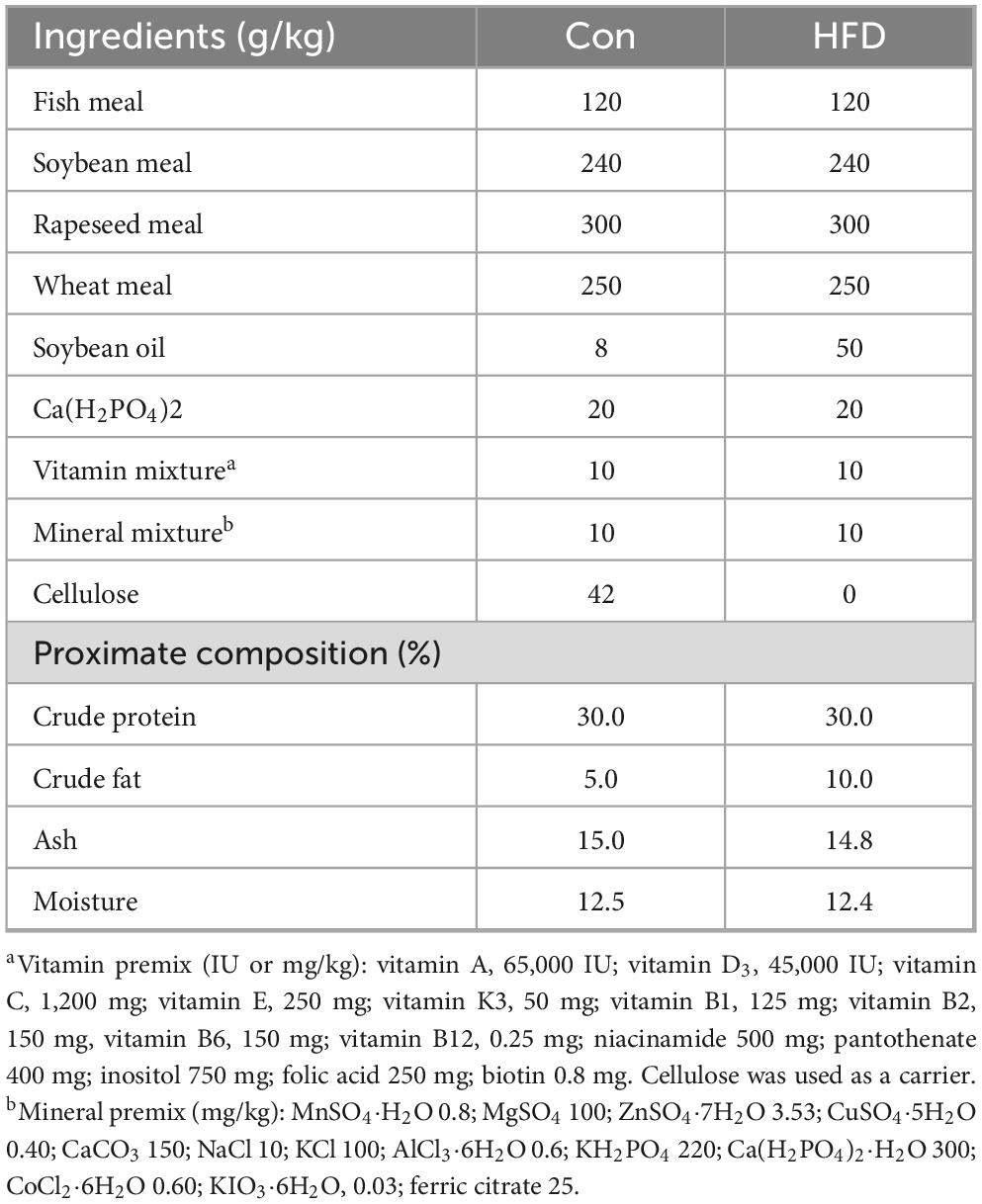
- 1National Engineering Research Center of Marine Facilities Aquaculture, Marine Science and Technology College, Zhejiang Ocean University, Zhoushan, China
- 2National Engineering Laboratory of Marine Germplasm Resources Exploration and Utilization, Marine Science and Technology College, Zhejiang Ocean University, Zhoushan, China
- 3Key Laboratory of Breeding Biotechnology and Sustainable Aquaculture (CAS), Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan, China
Fats have been widely applied in aquaculture to promote growth performance and substitute partial protein in fish feeds. However, excessive dietary fat levels induce metabolic disorders harming the health of cultured fish. Helminth infection in mammals was inversely correlated with metabolic syndrome, but its effect in aquatic animals is unknown yet. Here, we evaluated the impacts of Schyzocotyle acheilognathi infection on lipid metabolism of grass carp fed with high-fat diet (HFD). By comparison with the uninfected grass carp, helminth infection significantly increased the concentration of high-density lipoprotein (HDL) and condition factor (CF), and significantly decreased the concentration of low-density lipoprotein (LDL), the activity of AST, perimeter ratio (PR) and the thickness of muscularis mucosa (MM). Helminth infection also significantly lowered the lipid accumulation in liver, which may attribute to the significant up-regulated expression levels of apolipoprotein E (ApoE) and down-regulated expression of peroxisome proliferator-activated receptor-gamma (PPAR-γ) and lipoprotein lipase (LPL). Meanwhile in the grass carp infected by tapeworm, there was significant down-regulated expression of pro-inflammatory genes, interleukin-1beta (IL-1β) and tumor necrosis factor-alpha (TNF-α), and significant up-regulated expression of anti-inflammatory genes, transforming growth factor-beta 1 (TGF-β1) and interleukin-10 (IL-10). 16S rDNA sequencing results showed that helminth infection didn’t affect the α diversity of the intestinal microbiota, but increased the relative abundance of Cetobacterium, and significantly changed the structure of intestinal microbiota by PERMANOVA analysis. Correlation analysis showed the relative abundance of Cetobacterium was significant positively correlated with the helminth infection in grass carp fed HFD. PICRUST2 analysis indicated that several lipid metabolism-related pathways were significantly altered after helminth infection. Consequently, the above results indicated that tapeworm infection could ameliorate abnormal lipid metabolism through immune and gut microbiota regulation.
Introduction
Fat is one of the most important sources of nutrition for aquatic organisms, providing essential fatty acids, cholesterol, phospholipids and fat-soluble vitamins needed for normal growth, development and maintenance of the health of farmed fish (Malcolm, 2011). Previous studies indicated that increasing dietary fat content within proper range (5% for herbivorous fish, 8% for omnivorous fish and 10% for carnivorous fish) can boost growth performance, improve reproductive characteristics, exert a protein-sparing effect and decreased feed and production expenses (Boujard, 2004; Chou and Shiau, 1996; Li et al., 2012; Zhang et al., 2017). Thus, high fat diet (HFD) has been extensively utilized in intensive aquaculture. However, long-term excessively fat in the diet induced many adverse implications on farmed fish, increasing fat accumulation in liver, stimulation endoplasmic reticulum stress, impairment the intestinal mucosal barrier, triggering inflammatory responses, imbalance of microbiota and disturbance of metabolism (Cao et al., 2019; Jia et al., 2020a; Jia et al., 2020b; Jin et al., 2019; Tao et al., 2018; Yin et al., 2021; Yu et al., 2020). Therefore, addressing the metabolic imbalance and physiological disturbances induced by HFD will greatly advance the sustainable and healthy development of the aquaculture industry.
Parasitic helminths, mostly considered detrimental to host health, are common macrobiota in gastrointestinal (GI) tract of vertebrates (Peachey et al., 2017). However, several application researches of helminth in some chronic inflammation-related diseases demonstrated the positive effects of parasites on the health of host in recent years (Sobotkova et al., 2019). Infection with the nematode Heligmosomoides polygyrus has preventive and therapeutic roles on obesity caused by HFD in mice (Shimokawa et al., 2019). Transient infection with Nippostrongylus brasiliensis (nematode) in mice long-lasting improved insulin sensitivity and decreased adipose tissue mass in HFD obese mice (Wu et al., 2011; Yang et al., 2013). Chronic infection with the digenean Schistosoma mansoni or S. mansoni-soluble egg antigens (SEAs), a mixture of helminth-derived molecules, both improved insulin sensitivity and glucose homeostasis (Hussaarts et al., 2015). The above studies suggested that helminth infection or products derived from helminths promoted metabolic benefit for health of mammal hosts (Guigas and Molofsky, 2015). However, it remains unclear whether the protective effects of helminth against metabolic diseases also exists in aquatic animals.
Grass carp (Ctenopharyngodon idella) is one of the most important economic freshwater aquaculture species in China, and its production reached 5.9 million tons, accounting for 21.8% of the total annual production of freshwater farmed fish in 2023, according to the China Fishery Statistical Yearbook (Liu, 2023). Previous studies have suggested that a diet containing 4% lipids optimizes growth performance, feed efficiency, and the protein-sparing effect in juvenile grass carp (Du et al., 2005), while excess dietary fat level induced growth performance reduction, lipid deposition in liver, muscle and mesenteric tissue, damage of intestinal mucosal barrier and imbalance of intestinal microbiota (Du et al., 2006; Liu et al., 2022; Liu et al., 2023; Tang et al., 2019). Therefore, alleviating the detrimental impacts caused by HFD is crucial for the health of grass carp.
Schyzocotyle acheilognathi (syn. Bothriocephalus acheilognathi) is a common helminth species harboring in foregut of grass carp (Kuchta et al., 2018; Liao and Shi, 1956). In our previous study, S. acheilognathi infection altered the composition of gut microbiota (Fu et al., 2022). Studies in mammals have shown that helminth improved metabolic diseases through gut microbiota. The composition and diversity of the intestinal microbiota in vertebrates are always altered by helminth infection (Peachey et al., 2017). A limited number of consistent changes in the composition of the host’s gut microbiota have been repeatedly noted in animals infected with helminth (Peachey et al., 2017), called helminth-modified microbiota, which affects host immunity or metabolic capacity (Brosschot and Reynolds, 2018). The H. polygyrus infection protected against HFD-induced obesity by altering the composition of the gut microbiota, which led to an increase in norepinephrine (NE) concentration (Shimokawa et al., 2019), or elevated levels of short chain fatty acids (SCFAs) (Su et al., 2020). Thus, this study aims to elucidate the roles and underlying mechanisms of the protective effects of helminths against metabolic diseases in aquatic animals, using grass carp infected with the helminth S. acheilognathi, with the goal of offering novel strategies for the treatment of metabolic diseases in aquatic species.
Materials and methods
Experimental animals
The fry of grass carp (11 ± 1 cm, 11.4 ± 0.5 g) was purchased from an aquaculture pond in Jiangmen, Guangdong Province, where the prevalence and intensity of S. acheilognathi in grass carp were 40% and 3.2, respectively in previous survey. Grass carp were kept temporarily for 3 days prior to the formal experiment.
Feeding and samples collection
Grass carp were randomly divided into four buckets, fed with a normal diet (ND) with 5% fat content or a high-fat diet (HFD) with 10% fat content (Huaian Tongwei Feed Co., Ltd., China; Table 1) to apparent satiation twice daily (10: 00, 17: 00 oor ersity of se mationertebradomesticated in 100 L plastic buckets under a natural photoperiod (12 h light: 12 h dark), and kept at a water depth of 60 cm and a water temperature of 29°C. Following a 4-week feeding trial, the grass carp (n = 63) were anesthetized with eugenol (0.2 mL/L) before sampling.
The growth indices of the grass carp were measured, and blood samples were drawn from the caudal vein. Liver samples for gene expression analysis and oil red O staining were stored at −80°C and fixed in 4% paraformaldehyde (PFA) solution, respectively. The intestine was aseptically excised from visceral organs. The foregut and midgut were used for parasites checking under a stereomicroscope (Leica, Germany) to determine whether the host was infected with S. acheilognathi. Hindgut contents, intended for 16S rDNA high-throughput sequencing, were stored at −80°C for DNA extraction, and hindgut samples for hematoxylin and eosin (HE) staining were fixed in 4% PFA.
Based on the helminth infection status and the fat content in the feed, the samples were divided into four groups: ND (5% normal fat diet with no S. acheilognathi infection), ND + SA (5% normal fat diet with S. acheilognathi infection), HFD (10% high fat diet with no S. acheilognathi infection) and HFD + SA (10% high fat diet with S. acheilognathi infection). After dissection, it was recorded that the mean intensity of S. acheilognathi in grass carp was 3.9 (2–7).
Growth indices
The standard length (L) and body weight (W) of the grass carp were measured to calculate the condition factor (CF). The liver weight (Wl), viscera weight (Wv), and mesenteric fat weight (Wm) were measured individually to determine the hepatosomatic index (HSI), mesenteric fat index (MFI), and visceral index (VSI), respectively. The formulas for calculation of these indicators were shown as follows: CF = W/(L3) × 100%; HSI = Wl/W × 100%; VSI = Wv/W × 100%; MFI = Wm/W × 100%.
Serum biochemical index analysis
Blood samples were centrifuged at 3,000 rpm for 10 min at 4°C, after which the supernatant serum was carefully collected and stored at −80°C until used. The activity of alanine aminotransferase (ALT), aspartate aminotransferase (AST), as well as the concentration of high-density lipoprotein cholesterol (HDL), low-density lipoprotein cholesterol (LDL), total triglycerides (TG), and total cholesterol (TC) in serum were measured through a biochemical analyzer, using commercially available reagent kits (Seville Biotech Co., Ltd., Wuhan, China).
Histological analysis of intestine
Fresh hindgut tissues were immersed in 4% PFA solution for 48 h, and then processed routinely. These tissues were sectioned into 6 μm slices and stained with hematoxylin and eosin for examination under a light-microscopic (Zeiss, Germany). Photographs were analyzed using software ImageJ (National Institutes of Health, Bethesda, MD, United States). The thickness of muscularis mucosa (MM), the height of microvilli (MV) and perimeter ratio (PR) were measured, respectively. The PR was calculated using the following formulas: PR = the internal perimeter (IP) of the intestine lumen (villi and mucosal folding length)/the external perimeter (EP) of the intestine.
Oil red O staining
Liver samples from grass carp were excised and preserved in 4% PFA for 24 h. Subsequently, the tissues were trimmed to ensure smoothness and then dehydrated in 30% sucrose solution at 4°C for 24 h. Once the surface liquid of the liver tissue was evaporated, it was embedded in OCT embedding agent (SAKURA, United States). After freezing at −80°C, the tissue was then sectioned into 8 μm thick slices using a cryostat microtome (Leica, Germany) and stained with oil red O. Photomicrographs were captured using a light microscope (Zeiss, Germany). A total of six random fields of view were chosen from each sample, and the areas of lipid droplets were calculated using software ImageJ.
qPCR
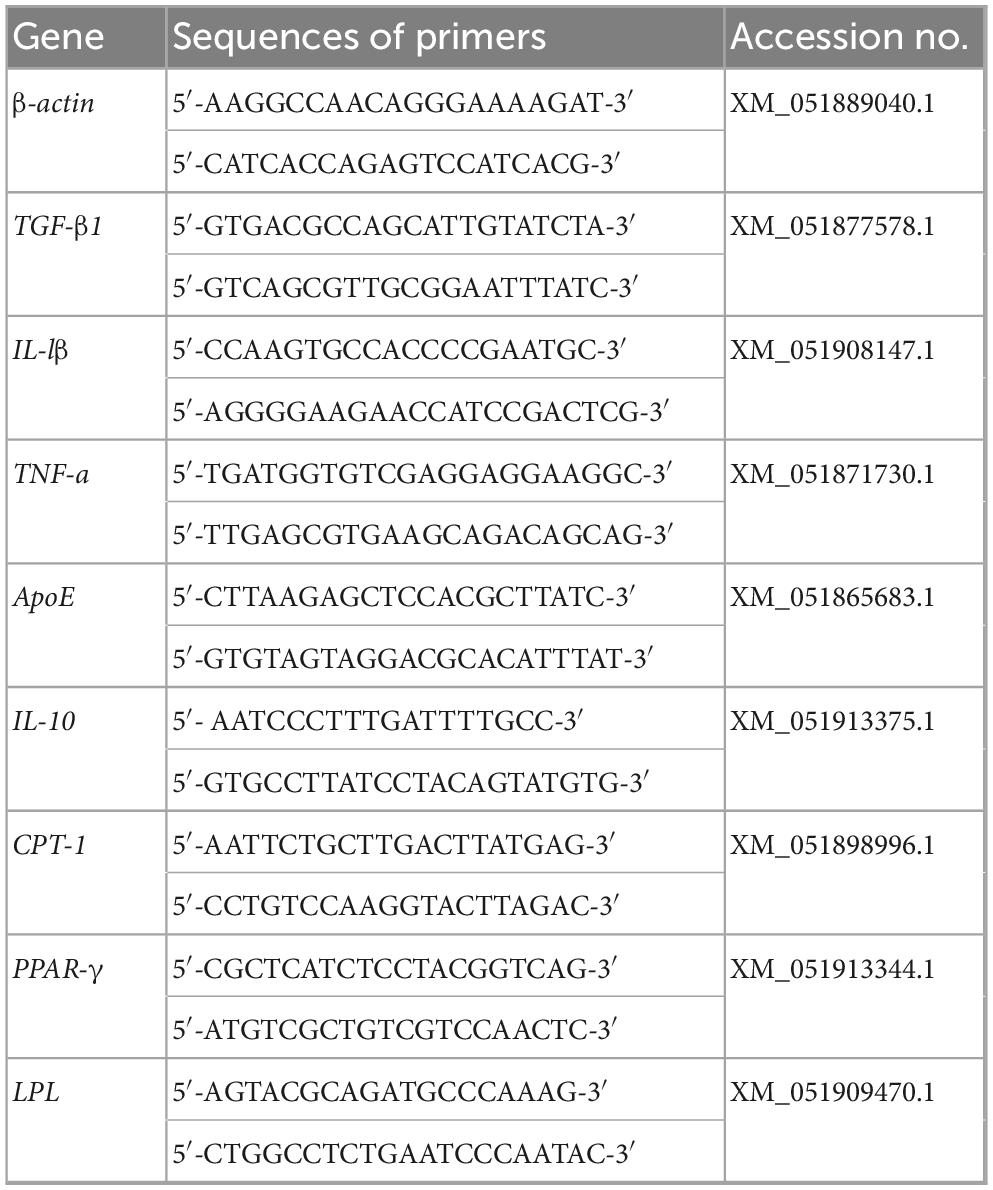
RNAiso Plus Reagent (Takara Bio Inc., Beijing, China) was used to extract total RNA. After being treated with RNase free DNase I (Promega, Wisconsin, United States), single-strand cDNA was synthesized using One−Step gDNA Remover kit (Servicebio, Wuhan, China). qPCR was carried out in a CFX96™ Real Time Detection System (BIO-RAD Bio Inc., Shanghai, China) using TB Green® Premix Ex Taq™ (Beijing, China). Gene-specific primers (Table 2) were used to amplify the target gene fragments. The β-actin gene of grass carp (Accession No. M25013.1) served as the reference gene. The qPCR cycle conditions were as follows: an initial denaturation at 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s, annealing at 60°C for 34 s, and a final Melt Curve analysis. The Ct method (2–ΔΔCT) (Livak and Schmittgen, 2001) was used to determine the relative expression levels of immune and lipid metabolic related genes in liver of grass carp.
DNA extraction, 16S rDNA amplification, and Illumina high throughput sequencing
The total bacterial DNA was extracted using the TGuide S96 magnetic bead method for soil/fecal genome DNA (Tiangen Biotech, Beijing, China), following the protocol provided by the manufacturer. The purity and concentration of genomic DNA were determined using the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer (Invitrogen, Thermo Fisher Scientific, Oregon, United States). The extracted DNA was preserved at −80°C. The V3-V4 hypervariable region of the bacterial 16S rDNA gene was amplified using the primers 338F (5′−ACT CCT ACG GGA GGC AGC A−3′) and 806R (5′−GGA CTA CHV GGG TWT CTA AT−3′) (Mori et al., 2013). The PCR amplification program was the same as previously reported (Fu et al., 2019). PCR products were purified with Agencourt AMPure XP Beads (Beckman Coulter, Indianapolis, IN) and quantified with the Qubit dsDNA HS Assay Kit and Qubit 4.0 Fluorometer. Following individual quantification, equimolar amounts of amplicons were combined into a single pool. Sequencing was conducted by Biomarker technologies (Qingdao, China) on the Illumina NovaSeq 6000 sequencing platform. The raw 16S rRNA sequence data can be obtained in the NCBI SRA database (Bioproject: PRJNA1045724).
Bioinformatics analysis of sequence data
The raw sequencing data were analyzed using the QIIME2 Pipeline, version 2021.4.1 The raw data were initially processed using Trimmomatic 0.35 (Bolger et al., 2014) for quality filtering, followed by the identification and removal of primer sequences with Cutadapt version 1.9.1 (Martin, 2011). DADA2 was subsequently employed to correct errors in the merged reads and to identify amplicon sequence variants (ASVs) (Callahan et al., 2016). The ASVs were then classified taxonomically using a native Bayes classifier (Wang et al., 2007), pre-trained on the SILVA 138 (Balvočiūtė and Huson, 2017).
Alpha diversity (ACE, Chao1, Simpson and Shannon index) of gut microbiota was calculated by QIIME2 and visualized in software R version 3.5. Beta diversity was used to evaluate the similarity between microbial communities across different samples in QIIME2. PERMANOVA analysis, non-metric multidimensional scaling (NMDS), Principal coordinate analysis (PCoA) and unweighted pair group mean algorithm (UPGMA) were used to visualize the beta diversity based on weighted Unifrac or Bray-curtis distance. The Pearson correlation coefficient was conducted using software PAST version 4.13 to examine the linear correlation between helminth infection status and the abundance of bacteria. Differences in taxa among groups were tested with Venn diagram and linear discriminant analysis coupled with effect size (Lefse). The metagenomic information of the samples was predicted from the 16S rDNA gene sequence data by employing PICRUST 2.0 in conjunction with the KEGG database (Langille et al., 2013). STAMP version 2.1.3 was used to perform all statistical analyses on the functional profiles (Parks et al., 2014).
Statistical analysis
Data analysis among the four groups utilized the One-Way ANOVA, supplemented by LSD post hoc testing. Students’ t-test was used to perform statistically analysis between two groups. All the statistical tests were performed in software SPSS 20 at the 0.05 significance threshold.
Results
Role of helminth infection in the effect of HFD on growth indices of grass carp
The growth indices of grass carp were presented in Figures 1A–D. Among the four groups, HFD + SA had the highest CF, MFI and VSI (Figures 1A,C,D). The CF of HFD + SA group was significantly higher than that of the HFD group (P = 0.015 < 0.05) and ND group (P = 0.004 < 0.05) (Figure 1A); the MFI of HFD group was merely higher than that of ND group (P = 0.039 < 0.05, Figure 1C); and the VSI was only showed significant difference between the HFD + SA group and the ND group (P = 0.025 < 0.05, Figure 1D). The HSI did not exhibit any significant differences across the four groups (P > 0.05, in all cases, Figure 1B).
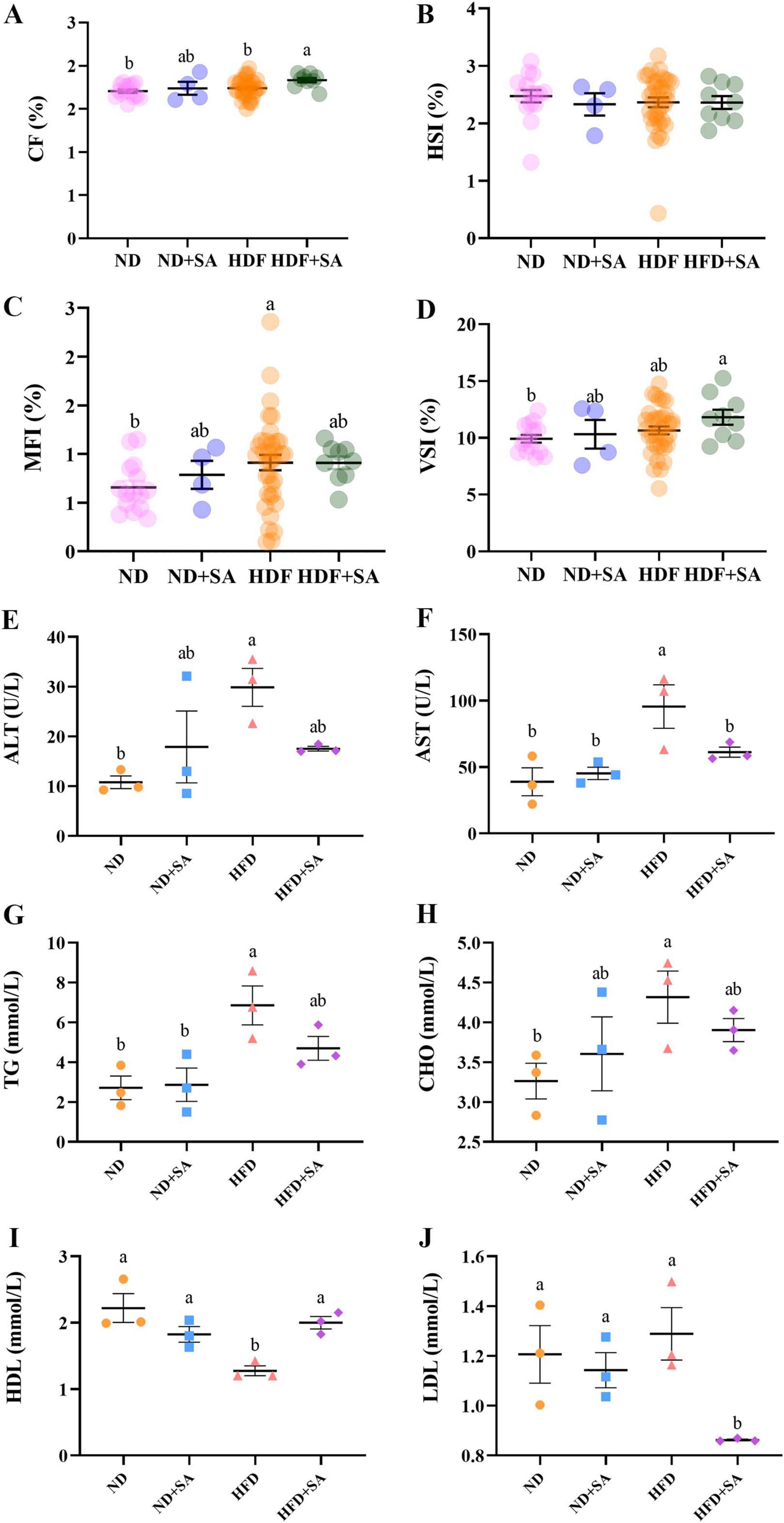
Figure 1. Effect of S. acheilognathi infection on growth performance and serum lipid parameters of C. idella fed on high-fat diet (HFD). (A) CF, condition factor; (B) HSI, hepatosomatic index; (C) VSI, visceral index; (D) MFI, mesenteric fat index; (E) ALT, alanine aminotransferase; (F) AST, aspartate aminotransferase; (G) TG, total triglycerides; (H) TC, total cholesterol; (I) HDL, high-density lipoprotein cholesterol (J) LDL, low-density lipoprotein cholesterol. (A–D) Values were presented as mean ± SEM (ND: n = 15; ND + SA: n = 4; HFD: n = 35; HFD + SA: n = 9); (E–J) values were presented as mean ± SEM (n = 3). a, bSignificant differences are indicated by different letters (P < 0.05). ND, 5% normal fat diet with no S. acheilognathi infection; ND + SA, 5% normal fat diet with S. acheilognathi infection; HFD, 10% high fat diet with no S. acheilognathi infection; HFD + SA, 10% high fat diet with S. acheilognathi infection.
Role of helminth infection in the effect of HFD on serum biochemical indices
The serum biochemical indices of grass carp were shown in Figures 1E–J. HFD significantly increased the activity of ALT (Figure 1E, P = 0.012) and AST (Figure 1F, P = 0.004), elevated serum levels of TG (Figure 1G, P = 0.005) and CHO (Figure 1H, P = 0.045), and reduced serum concentration of HDL (Figure 1I, P = 0.001) in comparison with normal diet. but the concentration of LDL between ND and HFD showed no significance (Figure 1J, P > 0.05). Comparing to HFD, helminth infection significantly reduced the activity of AST (Figure 1F, P = 0.044), decreased the concentration of LDL (Figure 1J, P = 0.008), and elevated the serum HDL content (Figure 1I, P = 0.006) in grass carp fed with HFD. The activity of ALT and the levels of TG and CHO in the serum showed a slight decrease in the HFD + SA group compared to the HFD group, but the differences were not significant (Figures 1E,G,H, P > 0.05 in all cases).
Role of helminth infection in the effect of HFD on intestinal structure and liver lipid content
The results of the hindgut sections stained with H&E were presented in Figures 2A–D. Helminth infection significantly decreased the MM (Figure 2F, P = 0.015) and PR (Figure 2G, P = 0.022) in grass carp fed on HFD, with no effect on MV (Figure 2E, P > 0.05). In the normal diet, helminth infection had no affection on the MM, MV, and PR (Figures 2E–G, P > 0.05 in all cases).
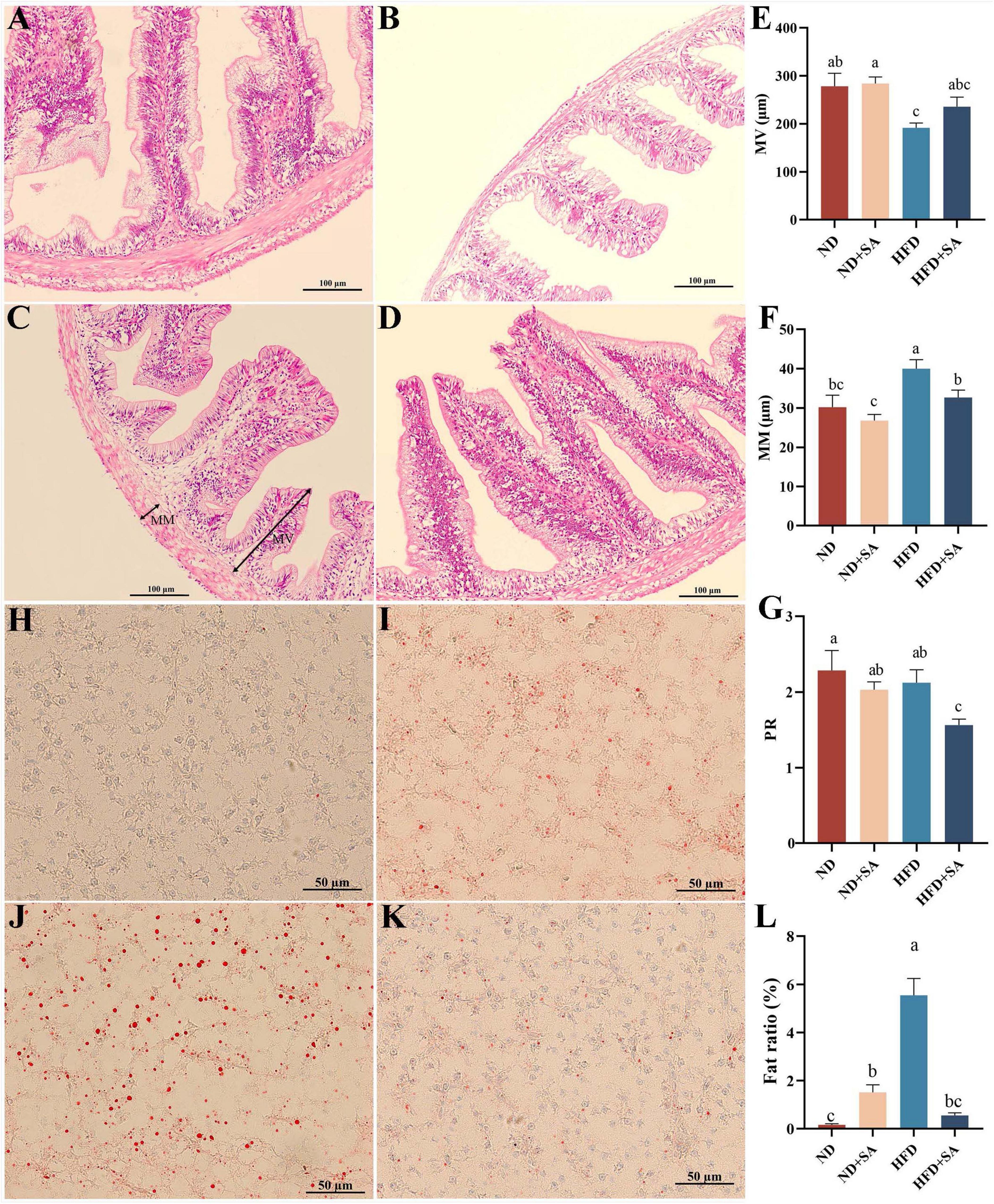
Figure 2. Effect of S. acheilognathi infection on intestinal morphology and liver lipid deposition of C. idella fed on HFD. (A–D) HE staining of intestine; (H–K) oil red O staining of liver. (A,H) ND; (B,I) ND + SA; (C,J) HFD; (D,K) HFD + SA; (E) MV, the height of microvilli; (F) MM, the thickness of muscularis mucosa; (G) PR, perimeter ratio; (L) Fat ratio. (E–G,L) Values were represented as mean ± SEM, (E) ND, n = 18; ND + SA, n = 23; HFD, n = 22; HFD + SA, n = 22; F: ND, n = 17; ND + SA, n = 24; HFD, n = 24; HFD + SA, n = 24; G: ND, n = 3; ND + SA, n = 4; HFD, n = 4; HFD + SA, n = 4; L: n = 5 for each group. a,b,cSignificant differences are indicated by different letters (P < 0.05). ND, 5% normal fat diet with no S. acheilognathi infection; ND + SA, 5% normal fat diet with S. acheilognathi infection; HFD, 10% high fat diet with no S. acheilognathi infection; HFD + SA, 10% high fat diet with S. acheilognathi infection.
The liver lipid content was determined using oil red O staining. The results indicated that the lipid droplets in the HFD group were larger and denser comparing with the other three groups (Figures 2H–K). Helminth infection significantly decreased the fat ratio value in liver of grass carp fed with HFD (P = 0.000, Figure 2L). However, helminth infection significantly increased the fat ratio value in grass carp fed normal diet (P = 0.025, Figure 2L).
Role of helminth infection in the effect of HFD on relative expression of immune and lipid metabolism related genes
The mRNA expression levels of immune and lipid metabolism related genes were showed in Figure 3. Helminth infection significantly promoted the expression of transforming growth factor-beta 1 (TGF-β1) (Figure 3C, P = 0.004), interleukin-10 (IL-10) (Figure 3D, P = 0.000) and apolipoprotein E (ApoE) (Figure 3G, P = 0.001), and significantly down-regulated the expression of interleukin-1beta (IL-1β) (Figure 3A, P = 0.009), tumor necrosis factor-alpha (TNF-α) (Figure 3B, P = 0.027), peroxisome proliferator-activated receptor-gamma (PPAR-γ) (Figure 3E, P = 0.003) and lipoprotein lipase (LPL) (Figure 3F, P = 0.001) in grass carp fed on HFD, but no affection on the expression of carnitine palmitoyltransferase 1(CPT1) (Figure 3H, P > 0.05). Meanwhile, helminth infection significantly elevated the relative expression levels of PPAR-γ (Figure 3E, P = 0.000), and significantly reduced the expression levels of ApoE (Figure 3G, P = 0.011) and CPT1 (Figure 3H, P = 0.007), but no impact on the expression levels of IL-1β, TNF-α, TGF-β1, IL-10, and LPL (Figures 3A–D,F; P > 0.05 in all cases) in grass carp fed with normal diet.
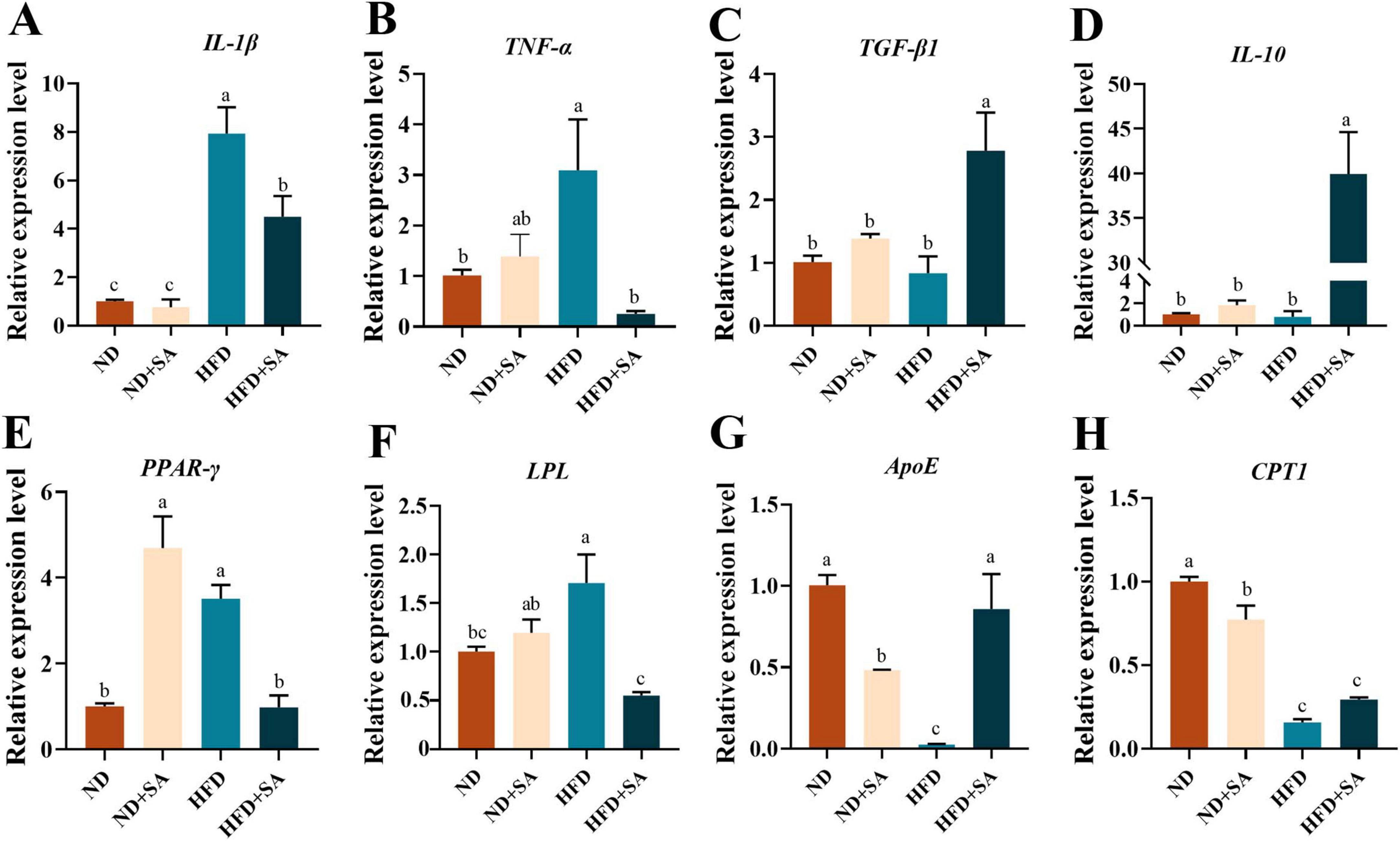
Figure 3. Effect of S. acheilognathi infection on the mRNA expression of immune and lipid metabolism related genes in C. idella fed on HFD. (A) IL-1β, interleukin-1beta; (B) TNF-α, tumor necrosis factor-alpha; (C) TGF-β1, transforming growth factor-beta 1; (D) IL-10, interleukin-10; (E) PPAR-γ, peroxisome proliferator-activated receptor-gamma; (F) LPL, lipoprotein lipase; (G) ApoE, apolipoprotein E; (H) CPT1, carnitine palmitoyltransferase 1; (A–H) Values were represented as mean ± SEM (n = 3); a,b,csignificant differences are indicated by different letters (P < 0.05). ND, 5% normal fat diet with no S. acheilognathi infection; ND + SA, 5% normal fat diet with S. acheilognathi infection; HFD, 10% high fat diet with no S. acheilognathi infection; HFD + SA, 10% high fat diet with S. acheilognathi infection.
Role of helminth infection in the effect of HFD on the intestinal microbiota
Composition of intestinal microbiota
The composition of intestinal microbiota was altered by helminth infection regardless of normal diet or high fat diet. At the phylum level (Figure 4A), the dominant taxa of the hindgut of grass carp were Fusobacteriota, Firmicutes, Proteobacteria and Bacteroidetes. Compared with the ND group, the relative abundance of Fusobacteria (72.59 ± 13.90% vs. 41.65 ± 29.51%) in the ND + SA was significantly increased (P = 0.019 < 0.05), and the relative abundance of Firmicutes (13.35 ± 6.73% vs. 25.75 ± 8.7%, P = 0.018 < 0.05), Proteobacteria (5.02 ± 2.61% vs. 11.84 ± 7.27%, P = 0.04 < 0.05) and Bacteroidetes (2.11 ± 1.06% vs. 10.82 ± 10.88%, P = 0.039 < 0.05) was significantly decreased. However, there was no significant difference in the relative abundance of dominant phyla of gut microbiota group between HFD and HFD + SA group in grass carp. Compared with HFD, the relative abundance of Fusobacteriota (91.70 ± 6.10% vs. 79.83 ± 9.72%) increased but not significant (P = 0.30 > 0.05) in HFD + SA group, and the relative abundance of Firmicutes (2.45 ± 1.40% vs. 9.24 ± 8.38%), Proteobacteria (4.27 ± 3.02% vs. 6.32 ± 3.06%), Bacteroidetes (1.12 ± 1.97% vs. 1.44 ± 0.78%), and Desulfobacterota (0.26 ± 0.26% vs. 2.62 ± 4.02%) in the HFD + SA group was also lower, but differences were not significant (P > 0.05 in all cases).
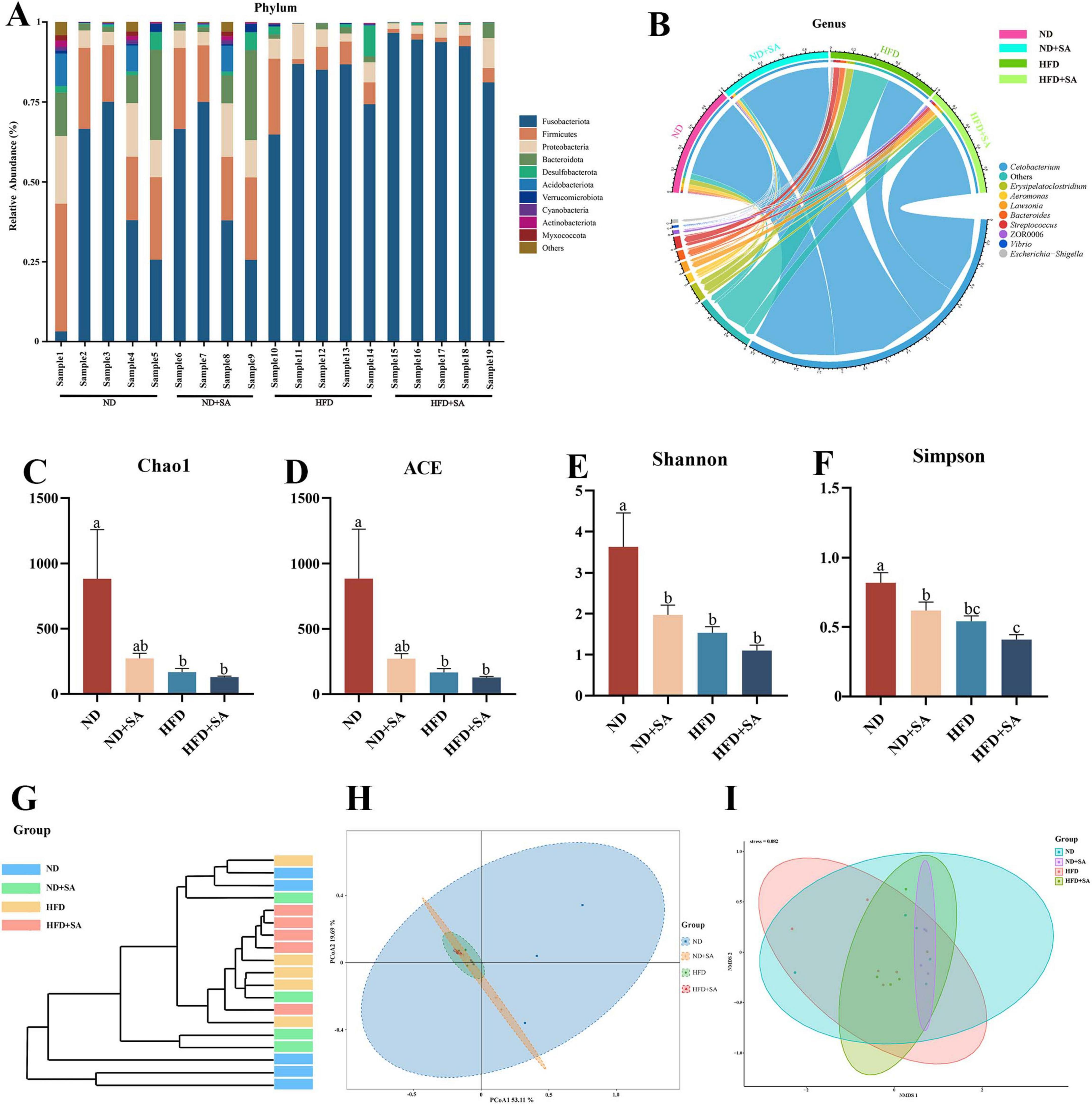
Figure 4. Effect of S. acheilognathi infection on the composition and diversity of intestinal microbiota in C. idella fed on HFD. (A,B) Microbiota composition; (C–F) α diversity indices; G-I: β diversity; (A) phylum level; (B) genus level; (G) cluster analysis; (H) PCoA, principal coordinates analysis; (I) NMDS, Non-metric multidimensional scaling; (A–I) ND, n = 5; ND + SA, n = 4; HFD, n = 5; HFD + SA, n = 5. a,b,cSignificant differences are indicated by different letters (P < 0.05). ND, 5% normal fat diet with no S. acheilognathi infection; ND + SA, 5% normal fat diet with S. acheilognathi infection; HFD, 10% high fat diet with no S. acheilognathi infection; HFD + SA, 10% high fat diet with S. acheilognathi infection.
At the genus level, Cetobacterium was the dominant taxa in the hindgut of grass carp (Figure 4B). Compared with ND, the relative abundance of Cetobacterium (72.58 ± 13.91% vs. 41.46 ± 29.48%, P = 0.018 < 0.05, Figure 4B) in ND + SA was significantly increased, the relative abundance of Aeromonas, Lawsonia, and ZOR0006 slightly increased (P > 0.05 in all cases, Figure 4B), and slightly decreased the relative abundance of Streptococcus, Bacteroides and Erysipelatoclostridium (P > 0.05 in all cases, Figure 4B). However, the relative abundance of Cetobacterium (91.58 ± 6.17% vs. 79.65 ± 9.86%, P = 0.30 > 0.05, Figure 4B) in HFD + SA group increased and the relative abundance of Lawsonia, Bacteroides, Aeromonas, and Erysipelatoclostridium all decreased comparing with HFD, but the differences were not significant (P > 0.05 in all cases, Figure 4B).
Diversity of gut microbiota
High fat diet significantly decreased the alpha diversity of hindgut microbiota in grass carp in comparison with normal diet (Figures 4C–F, P < 0.05 in all cases). However, helminth infection did not affect the alpha diversity of hindgut microbiota in grass carp fed on HFD (Figures 4C–F, P > 0.05 in all cases). Compared with HFD group, the HFD + SA group displayed a lower α diversity, but the differences were not significant (Figures 4C–F, P > 0.05 in all cases).
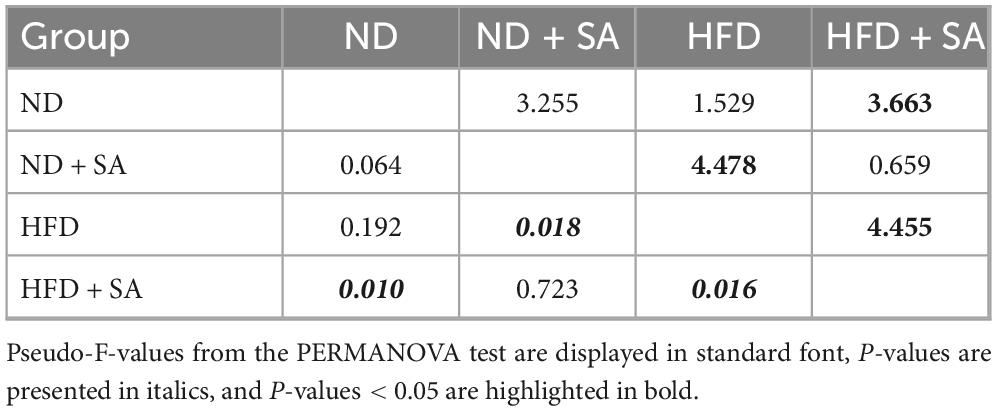
For the beta diversity, cluster analysis indicated that all samples were divided into two groups (Figure 4G), where almost all intestinal microbiota samples of grass carp fed on HFD clustered into one group, and samples that of feeding on ND clustered into a separate group. PERMANOVA (Table 3) analyses showed that the microbial communities of HFD + SA group was significantly different from HFD (P = 0.016). However, helminth infection had no affection on the microbial communities of grass carp fed normal diet (P = 0.064 > 0.05). The analyses of Principal coordinate analysis (PCoA) (Figure 4H) and non-metric multidimensional scaling (NMDS) (Figure 4I) showed that the microbial communities of the four groups could not be significantly distinguished from each other.
Differences in taxonomic abundance among groups
A total of 3,968 ASVs were obtained in this study. Venn plot showed that total ASVs of ND, ND + SA, HFD and HFD + SA groups were 3,352, 659, 474, and 299, respectively; unique ASVs in four groups were 2,855, 284, 166, and 94, respectively; and the four groups shared 96 ASVs (Figure 5A).
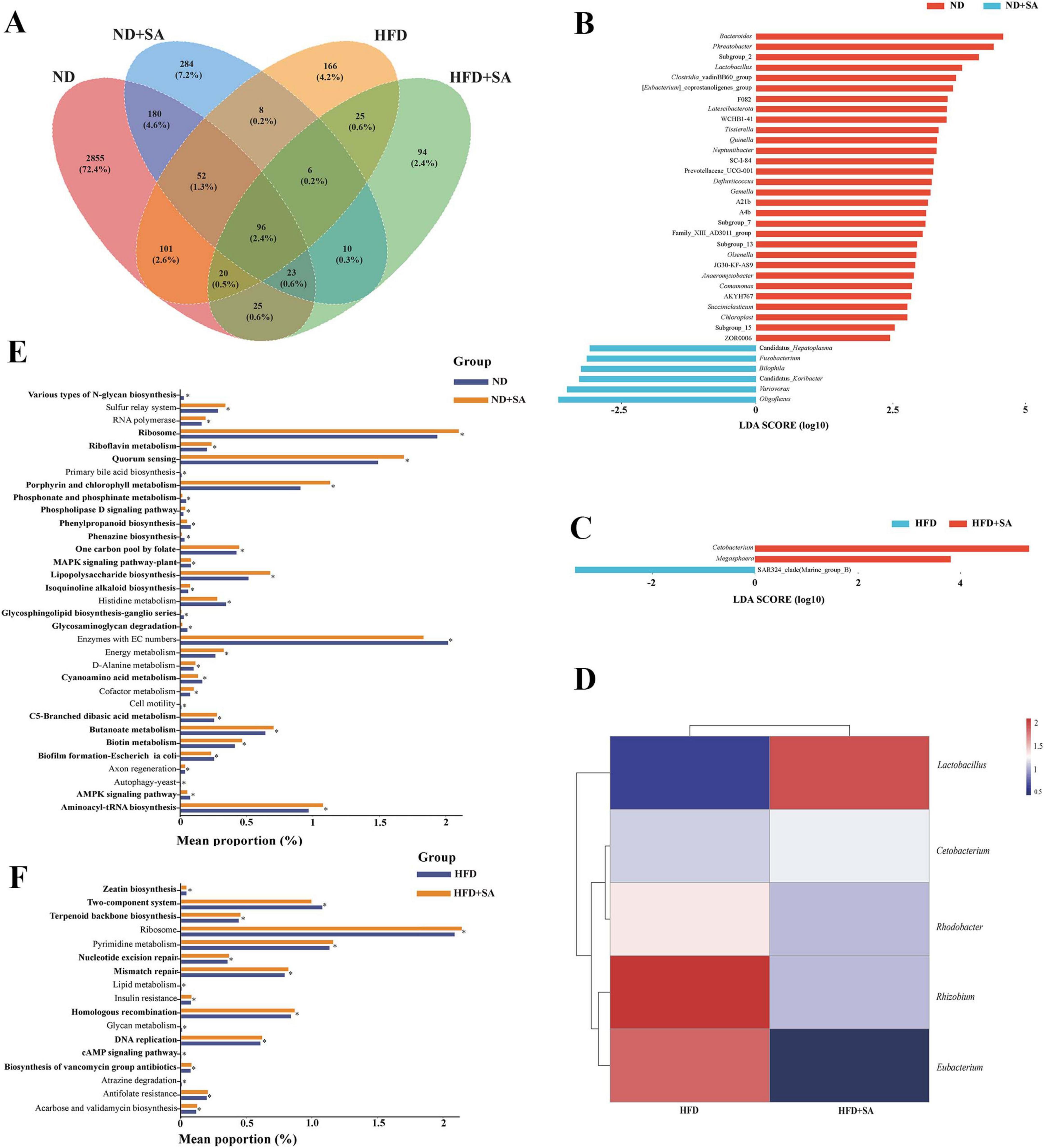
Figure 5. Differences and function of intestinal microbiota in C. idella fed on HFD. (A) Venn plot. (B) Lefse analysis between ND and ND + SA. (C) Lefse analysis between HFD and HFD + SA. (D) Heatmap of significant taxa in the intestine of HFD and HFD + SA. (E,F) Changes in the KEGG pathways predicted by PICRUST. (E) ND and ND + SA; (F) HFD and HFD + SA. (A–F) ND, n = 5; ND + SA, n = 4; HFD, n = 5; HFD + SA, n = 5. 0.01 < P < 0.05 values are marked with “*.” ND, 5% normal fat diet with no S. acheilognathi infection; ND + SA, 5% normal fat diet with S. acheilognathi infection; HFD, 10% high fat diet with no S. acheilognathi infection; HFD + SA, 10% high fat diet with S. acheilognathi infection.
Lefse analysis at the genus level indicated that there were thirty-six biomarkers between ND and ND + SA groups (Figure 5B), and only three biomarkers between HFD and HFD + SA groups (Figure 5C), including Cetobacterium, Megasphaera, and SAR324_clade.
Association between helminth infection and relative abundance microbiota in grass carp fed on HFD
Pearson correlation analysis indicated that helminth infection had a significant positive correlation with the relative abundance of Cetobacterium (P = 0.042, r = 0.65) and Lactobacillus (P = 0.001, r = 0.86), and three taxa, including Rhodobacter, Rhizobium, and Eubacterium, existed a significant negative correlation with the helminth infection (P < 0.05 in all cases; 0.63 ≤ |r| ≤ 0.71) (Figure 5D) in hindgut of grass carp fed on HFD (Table 4).
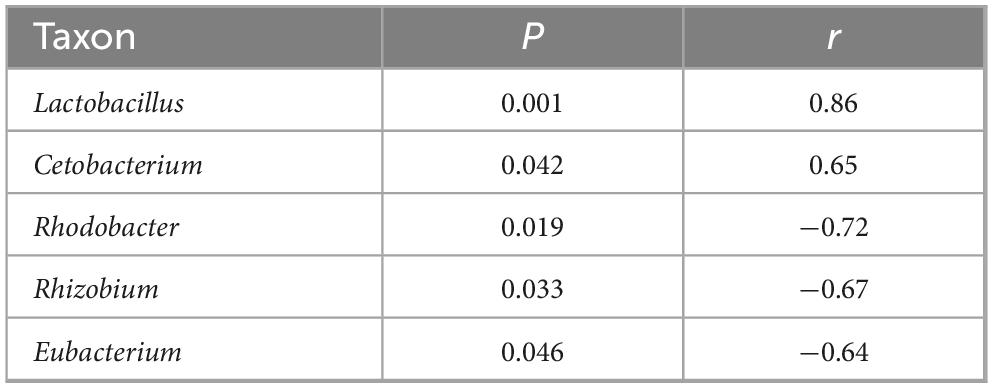
Table 4. Analysis of correlation coefficient between helminth infection and the taxonomic abundance of microbiota within the intestine of grass carp fed with HFD.
Functional alteration of gut microbiota
The PICRUST2 prediction identified thirty-three KEGG pathways at the L3 level with significant differences between the ND and ND + SA groups. Among these, twenty pathways pertained to metabolic processes, accounting for 60.6% (20/33), of which one pathway was associated with lipid metabolism, two with carbohydrate metabolism and one with energy metabolism (Figure 5E). In the comparison between HFD and HFD + SA, seventeen pathways exhibited notable differences. Of these, nine pathways were linked to metabolism functions (52.9%: 9/17), of which one KEGG pathway was related to lipid metabolism (Figure 5F).
Discussion
The protective effect of helminth on metabolic diseases had been reported in mammals including improved insulin resistance, enhancing glucose tolerance, reducing blood lipids, inhibiting proinflammatory response and M2 macrophage proliferation etc. (Kang et al., 2021; Obi et al., 2020; Rajamanickam et al., 2020; Shimokawa et al., 2019; Su et al., 2020; Su et al., 2018; Yang et al., 2013), but the existence of such a protective effect in aquatic fauna remains unknown. In this study, we attempted to explore the role of helminth on metabolic diseases in aquatic animals. From our experimental results, the role of helminth in metabolic diseases in aquatic animals was similar to that in mammals. Helminth infection effectively ameliorated the high-fat diet induced metabolic imbalance in grass carp.
The effect of S. acheilognathi infection on growth of grass carp fed on HFD
In growth performance, helminth infection with grass carp fed on HFD pointedly increased the CF, and slightly increased VSI and MFI. Numerous investigations on fish species, including Micropterus salmoides (Guo et al., 2019; Yin et al., 2021), Nibea japonica (Han et al., 2014), and Rachycentron canadum (Wang et al., 2005), have revealed a positive correlation between the VSI and MFI and the level of dietary lipids. The rise in CF observed in this study was associated with a concurrent upward trend in both VSI and MFI. The level of dietary fat usually causes no change of CF in fish (Guo et al., 2022; Han et al., 2014; Yin et al., 2021). Thus, the significant increasing of CF in HFD + SA group indicated that helminth infection boosted the growth of grass carp fed with HFD.
The effect of S. acheilognathi infection on serum biochemical indices of grass carp fed on HFD
Helminth infection with grass carp fed with HFD exhibited a notable rise in LDL level, while AST level and HDL concentration significantly declined. AST is primarily situated within hepatocytes, and an increase in its serum level is a response to hepatocellular damage and changes in plasma membrane permeability (Boone et al., 2005). The high-fat diet led to an elevation in serum AST level in grass carp, indicating potential damage to liver cells. However, helminth infection could possibly decrease the liver’s burden imposed by the high-fat diet and facilitate the recovery of liver function. HDL and LDL are typically used as indicators to reflect lipid metabolism in aquatic animals (Guo et al., 2019; Li et al., 2016; Zhao et al., 2016) and mammals (Lu et al., 2022; Teng et al., 2020). High serum LDL is always considered to be risk factors related to fatty livers in fish (Guo et al., 2019; Li et al., 2016; Zhao et al., 2016). HDL has been referred to as good cholesterol, inversely proportional to cardiovascular risk in many studies (Ertek, 2018). In present study, the increase in HDL and decrease in LDL indicated that helminth infection reversed the abnormal lipid metabolism in HFD of grass carp, which could improve health condition of cultured C. idella.
The effect of S. acheilognathi infection on lipid metabolism of grass carp fed on HFD
PPAR-γ, LPL, ApoE, and CPT1 are the key regulatory enzymes involved in lipid metabolism within the liver, playing roles in the uptake, transport and oxidation of fatty acids. PPAR-γ is a ligand-activated transcription factor that belongs to the nuclear hormone receptor superfamily and fosters lipogenesis by enhancing the expression of enzymes involved in lipid synthesis (Ahmadian et al., 2013). LPL catalyzes the hydrolysis of intravascular triglycerides contained within lipoproteins, such as chylomicrons and very low-density lipoprotein (VLDL), then the released fatty acids can be taken up by tissues for use in oxidation or for storage purposes (Wu et al., 2021). ApoE is a soluble apolipoprotein primarily synthesized in the liver and brain, and it has an important role in the metabolism of triglyceride rich lipoproteins (TRL) due to its high affinity binding to the LDL receptor (LDLR), the LDL receptor-related protein 1 (LRP1) and the VLDL receptor (VLDLR), which facilitates the hepatic clearance of remnant lipoprotein particles (Huang and Mahley, 2014). LPL-mediated triglyceride hydrolysis would reduce when ApoE concentration was high (Wu et al., 2021). CPT1 is regarded as a pivotal regulatory enzyme in mitochondrial fatty acid oxidation pathway. It catalyzes the conversion of fatty acyl-CoAs into fatty acyl-carnitine molecules, which are then transported into the mitochondrial matrix for further oxidation (Kerner and Hoppel, 2000). In the present study, helminth infection in grass carp fed with HFD resulted in a modest elevation of CPT1 expression, and caused a significantly reduction in the expression levels of PPAR-γ and LPL, while significantly increased the expression level of ApoE. These results indicated that helminth mainly regulate the expression of these lipid metabolism genes to inhibit the absorption of fatty acids, thereby reducing the deposition of lipid in the liver.
Additionally, the lipid content in the liver of the HFD + SA group was significantly lower than that of the HFD group, indicating that helminth infection mitigated the liver lipid deposition induced by the high-fat diet. The protective effect of helminth in grass carp was similar to that in mice (Su et al., 2018). Most of the digested lipids are absorbed by intestinal epithelial cells (Zhang et al., 2021). PR, one of the intestinal morphology parameters, was significantly decreased by helminth infection in this study, reducing the amount of lipid absorbed by intestine, thereby to decrease lipid deposition in the liver.
The effect of S. acheilognathi infection on immune response and gut microbiota of grass carp fed on HFD
Similarity to the protective effects of helminth in mammalian metabolic diseases, the primary regulatory pathways through which helminths exert positive effects on metabolic diseases in fish may include the following two aspects: (1) immune regulation, (2) alterations of gut microbiota. The type 2 immune response induced by helminth infection has been proved to ameliorate the side effects of HFD in mice (Su et al., 2020; Su et al., 2018). The intestinal nematode H. polygyrus could protect against obesity by triggering the production of Th2-mediated cytokines, such as IL-4, lL-10 and IL-13, and enhancing the anti-inflammatory M2 macrophages levels (Su et al., 2018). The increased M2 and IL-10 responses resulted in a reduction of the pro-inflammatory cytokine TNF-α expression, which has been shown to modulate lipid metabolism (Khovidhunkit et al., 2004). Similarly, in our study, helminth infection significantly elevated IL-10 levels while concurrently reducing decreased TNF-α expression, indicating that helminth infection protects against the HFD-induced abnormal lipid metabolism via activation of immune responses in teleost.
An increase of alpha diversity in intestinal microbiota is generally associated with a “healthy” gut homeostasis (Peachey et al., 2017). In this study, a high-fat diet significantly decreased the α diversity of intestinal microbiota in grass carp (Figures 4C–F), indicating that HFD disrupted gut microbial balance. Similar disruptions caused by HFD have also been reported in Monopterus albus (Peng et al., 2019). Additionally, there was no significant difference in the α diversity of gut microbiota between the HFD group and the HFD + SA group (Figures 4C–F), which may be attributed to the regulatory effect of helminth on intestine microbiota.
Microbiota and helminths occupy the same ecological niche within the host’s intestine, where they can interact with each other (Glendinning et al., 2014). Multiple investigations have been carried out to examine the effects of helminth infection on the gut microbiota in fish. Tapeworm and acanthocephalan infection caused alteration in the composition of fish gut microbiota (Fu et al., 2019; Fu et al., 2022; Jiang et al., 2019; Ling et al., 2020). Studies in mammals have shown that helminth infection protects against HFD-induced obesity through modifications to the gut microbiota (Shimokawa et al., 2019; Su et al., 2020).
helminth infection was found to enhance the relative abundance of Cetobacterium in hindgut of grass carp fed with HFD, and the proportion of Cetobacterium in the HFD + SA group was 91.6%. According to Spearman’s correlation analysis, Cetobacterium correlated positively with helminth infection. Cetobacterium, identified as an anaerobic, gram-negative bacterium (Finegold et al., 2003), constitutes a predominant member of the intestinal microbiota in freshwater fish (Di Maiuta et al., 2013; Fu et al., 2022; Larsen et al., 2014; van Kessel et al., 2011). The significant proliferation of Cetobacterium could perform fermentative metabolism of peptides and carbohydrates, resulting in the production of acetate, which in turn modifies glucose homeostasis (Wang et al., 2021). Additionally, this bacterium could produce vitamin B12 (Larsen et al., 2014), thereby enhancing the host couldsicrobioto pathogen infections (Qi et al., 2023). Incorporating Cetobacterium fermentation products into fish feed can effectively enhance intestinal and liver health and decrease liver lipid accumulation (Xie et al., 2022). In zebrafish, cypermethrin (CYP) exposure or CYP and microplastics (MPs) co-exposure increased the abundance of Cetobacterium, which was found to be positively associated with the majority of lipid metabolites (Xu et al., 2024). Consistently, helminth infection may ameliorate the lipid metabolism induced by HFD through increasing the high relative abundance of Cetobacterium to regulate lipid and maintain liver health of grass carp.
Lactobacillus, a group of gram-positive, facultative anaerobic bacteria widely used as probiotics, has been shown to enhance epithelial barrier function and modulate innate immune responses as well as cytokine profiles (Amdekar and Singh, 2016). Strongyloides venezuelensis infection in mice induced the increasing of Lactobacillus spp., which has a positive effect on the glucose metabolism of the host (Pace et al., 2018). In this study, helminth infection showed a significant positive correlation with Lactobacillus. This result indicated that helminth infection can improve host metabolic profile by regulating probiotics.
In study of Caenorhabditis elegans, reactive oxygen species produced by Rhizobium induce DNA damage, which caused abnormal intestinal nuclei divisions (Kniazeva and Ruvkun, 2019). Eubacterium, one of the main SCFAs-producing bacteria in humans (Parada Venegas et al., 2019). Rhodobacter is a gram-negative, purple non-sulfur photosynthetic bacteria. Orally given the Rhodobacter sphaeroides in mice increased the content of SCFAs (Yang et al., 2020). However, SCFA has a significant negative influence on established Oesophagostomum dentatum infection in pigs (Petkevicius et al., 2004). In this study, helminth infection showed significant positive correlation with Rhodobacter, Rhizobium, and Eubacterium, indicating that helminth may improve its survival in the intestine by altering the relative abundance of these three bacteria.
Conclusion
In conclusion, the findings of this study showed that helminth infection can ameliorate the abnormal lipid metabolism caused by a high-fat diet in grass carp. Helminth infection decreased the lipid deposition in liver, the concentration of LDL and activity of AST in serum, while increased the level of HDL. The synergistic effect of IL-10 produced by helminth mediated Th2 immune response and Cetobacterium alteration in intestine induced by helminth infection upregulated the expression level of ApoE and downregulated the expression level of PPAR-γ and LPL in liver of grass carp, thus improving lipid metabolism.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found at: https://www.ncbi.nlm.nih.gov, accession number PRJNA1045724.
Ethics statement
The animal study was approved by the Zhejiang Ocean University’s Animal Care and Use Ethics Committee in Zhoushan, China. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XY: Formal Analysis, Methodology, Software, Visualization, Writing – original draft. DZ: Investigation, Methodology, Writing – original draft. WL: Writing – original draft. PF: Conceptualization, Formal Analysis, Project administration, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the earmarked fund for CARS (CARS-45) and the General Research Project of Zhejiang Provincial Department of Education (Special Project for Reforming the Training Mode of Professional Degree Graduate Students) (Grant no. Y202352327).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Ahmadian, M., Suh, J. M., Hah, N., Liddle, C., Atkins, A. R., Downes, M., et al. (2013). PPARγ signaling and metabolism: The good, the bad and the future. Nat. Med. 19, 557–566. doi: 10.1038/nm.3159
Amdekar, S., and Singh, V. (2016). Studies on anti-inflammatory and analgesic properties of Lactobacillus rhamnosus in experimental animal models. J. Complement. Integr. Med. 13, 145–150. doi: 10.1515/jcim-2015-0087
Balvočiūtė, M., and Huson, D. H. (2017). SILVA, RDP, greengenes, NCBI and OTT - how do these taxonomies compare? BMC Genom. 18:114. doi: 10.1186/s12864-017-3501-4
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boone, L., Meyer, D., Cusick, P., Ennulat, D., Bolliger, A. P., Everds, N., et al. (2005). Selection and interpretation of clinical pathology indicators of hepatic injury in preclinical studies. Vet. Clin. Pathol. 34, 182–188. doi: 10.1111/j.1939-165x.2005.tb00041.x
Boujard, T. (2004). Regulation of feed intake, growth, nutrient and energy utilisation in European sea bass (Dicentrarchus labrax) fed high fat diets. Aquaculture 231, 529–545. doi: 10.1016/j.aquaculture.2003.11.010
Brosschot, T. P., and Reynolds, L. A. (2018). The impact of a helminth-modified microbiome on host immunity. Mucosal Immunol. 11, 1039–1046. doi: 10.1038/s41385-018-0008-5
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Cao, X. F., Dai, Y. J., Liu, M. Y., Yuan, X. Y., Wang, C. C., Huang, Y. Y., et al. (2019). High-fat diet induces aberrant hepatic lipid secretion in blunt snout bream by activating endoplasmic reticulum stress-associated IRE1/XBP1 pathway. BBA-Mol. Cell. Biol. Lipids 1864, 213–223. doi: 10.1016/j.bbalip.2018.12.005
Chou, B. S., and Shiau, S. Y. (1996). Optimal dietary lipid level for growth of juvenile hybrid tilapia, Oreochromis niloticus X Oreochromis aureus. Aquaculture 143, 185–195. doi: 10.1016/0044-8486(96)01266-5
Di Maiuta, N., Schwarzentruber, P., Schenker, M., and Schoelkopf, J. (2013). Microbial population dynamics in the faeces of wood-eating loricariid catfishes. Lett. Appl. Microbiol. 56, 401–407. doi: 10.1111/lam.12061
Du, Z. Y., Clouet, P., Zheng, W. H., Degrace, P., Tian, L. X., and Liu, Y. J. (2006). Biochemical hepatic alterations and body lipid composition in the herbivorous grass carp (Ctenopharyngodon idella) fed high-fat diets. Br. J. Nutr. 95, 905–915. doi: 10.1079/bjn20061733
Du, Z. Y., Liu, Y. J., Tian, L. X., Wang, J. T., Wang, Y., and Liang, G. Y. (2005). Effect of dietary lipid level on growth, feed utilization and body composition by juvenile grass carp (Ctenopharyngodon idella). Aquacult. Nutr. 11, 139–146. doi: 10.1111/j.1365-2095.2004.00333.x
Ertek, S. (2018). High-density lipoprotein (HDL) dysfunction and the future of HDL. Curr. Vasc. Pharmacol. 16, 490–498. doi: 10.2174/1570161115666171116164612
Finegold, S. M., Vaisanen, M. L., Molitoris, D. R., Tomzynski, T. J., Song, Y., Liu, C., et al. (2003). Cetobacterium somerae sp. nov. from human feces and emended description of the genus Cetobacterium. Syst. Appl. Microbiol. 26, 177–181. doi: 10.1078/072320203322346010
Fu, P. P., Xiong, F., Feng, W. W., Zou, H., Wu, S. G., Li, M., et al. (2019). Effect of intestinal tapeworms on the gut microbiota of the common carp. Cyprinus carpio. Parasit. Vect. 12:252. doi: 10.1186/s13071-019-3510-z
Fu, P. P., Xiong, F., Wu, S. G., Zou, H., Li, M., Wang, G. T., et al. (2022). Effects of Schyzocotyle acheilognathi (Yamaguti, 1934) infection on the intestinal microbiota, growth and immune reactions of grass carp (Ctenopharyngodon idella). PLoS One 17:e0266766. doi: 10.1371/journal.pone.0266766
Glendinning, L., Nausch, N., Free, A., Taylor, D. W., and Mutapi, F. (2014). The microbiota and helminths: Sharing the same niche in the human host. Parasitology 141, 1255–1271. doi: 10.1017/S0031182014000699
Guigas, B., and Molofsky, A. B. (2015). A worm of one’s own: How helminths modulate host adipose tissue function and metabolism. Trends Parasitol. 31, 435–441. doi: 10.1016/j.pt.2015.04.008
Guo, D., Xie, M., Xiao, H., Xu, L., Zhang, S., Chen, X., et al. (2022). Bacillus subtilis supplementation in a high-fat diet modulates the gut microbiota and ameliorates hepatic lipid accumulation in grass carp (Ctenopharyngodon idella). Fishes 7:94. doi: 10.3390/fishes7030094
Guo, J., Zhou, Y., Zhao, H., Chen, W., Chen, Y., and Lin, S. (2019). Effect of dietary lipid level on growth, lipid metabolism and oxidative status of largemouth bass. Micropterus salmoides. Aquaculture 506, 394–400. doi: 10.1016/j.aquaculture.2019.04.007
Han, T., Li, X., Wang, J., Hu, S., Jiang, Y., and Zhong, X. (2014). Effect of dietary lipid level on growth, feed utilization and body composition of juvenile giant croaker Nibea japonica. Aquaculture 434, 145–150. doi: 10.1016/j.aquaculture.2014.08.012
Huang, Y., and Mahley, R. W. (2014). Apolipoprotein E: Structure and function in lipid metabolism, neurobiology, and Alzheimer’s diseases. Neurobiol. Dis. 72(Pt A), 3–12. doi: 10.1016/j.nbd.2014.08.025
Hussaarts, L., García-Tardón, N., van Beek, L., Heemskerk, M. M., Haeberlein, S., van der Zon, G. C., et al. (2015). Chronic helminth infection and helminth-derived egg antigens promote adipose tissue M2 macrophages and improve insulin sensitivity in obese mice. FASEB J. 29, 3027–3039. doi: 10.1096/fj.14-266239
Jia, R., Cao, L. P., Du, J. L., He, Q., Gu, Z. Y., Jeney, G., et al. (2020a). Effects of high-fat diet on antioxidative status, apoptosis and inflammation in liver of tilapia (Oreochromis niloticus) via Nrf2, TLRs and JNK pathways. Fish Shellfish Immunol. 104, 391–401. doi: 10.1016/j.fsi.2020.06.025
Jia, R., Cao, L. P., Du, J. L., He, Q., Gu, Z. Y., Jeney, G., et al. (2020b). Effects of high-fat diet on steatosis, endoplasmic reticulum stress and autophagy in liver of tilapia (Oreochromis niloticus). Front. Mar. Sci. 7:363. doi: 10.3389/fmars.2020.00363
Jiang, M., Zhang, X., Yang, Y., Yin, D., Dai, P., Ying, C., et al. (2019). Effects of Acanthosentis cheni infection on microbiota composition and diversity in the intestine of Coilia nasus. J. Fish Sci. China 26, 577–585. doi: 10.3724/SP.J.1118.2019.18206
Jin, M., Pan, T., Tocher, D. R., Betancor, M. B., Monroig, O., Shen, Y., et al. (2019). Dietary choline supplementation attenuated high-fat diet-induced inflammation through regulation of lipid metabolism and suppression of NFκB activation in juvenile black seabream (Acanthopagrus schlegelii). J. Nutr. Sci. 8:e38. doi: 10.1017/jns.2019.34
Kang, S. A., Choi, J. H., Baek, K. W., Lee, D. I., Jeong, M. J., and Yu, H. S. (2021). Trichinella spiralis infection ameliorated diet-induced obesity model in mice. Int. J. Parasitol. 51, 63–71. doi: 10.1016/j.ijpara.2020.07.012
Kerner, J., and Hoppel, C. (2000). Fatty acid import into mitochondria. Biochim. Biophys. Acta 1486, 1–17. doi: 10.1016/s1388-1981(00)00044-5
Khovidhunkit, W., Kim, M. S., Memon, R. A., Shigenaga, J. K., Moser, A. H., Feingold, K. R., et al. (2004). Effects of infection and inflammation on lipid and lipoprotein metabolism: Mechanisms and consequences to the host. J. Lipid Res. 45, 1169–1196. doi: 10.1194/jlr.R300019-JLR200
Kniazeva, M., and Ruvkun, G. (2019). Rhizobium induces DNA damage in Caenorhabditis elegans intestinal cells. Proc. Natl. Acad. Sci. U. S. A. 116, 3784–3792. doi: 10.1073/pnas.1815656116
Kuchta, R., Choudhury, A., and Scholz, T. (2018). Asian fish tapeworm: The most successful invasive parasite in freshwaters. Trends Parasitol. 34, 511–523. doi: 10.1016/j.pt.2018.03.001
Langille, M. G. I., Zaneveld, J., Caporaso, J. G., McDonald, D., Knights, D., Reyes, J. A., et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 31, 814–821. doi: 10.1038/nbt.2676
Larsen, A. M., Mohammed, H. H., and Arias, C. R. (2014). Characterization of the gut microbiota of three commercially valuable warmwater fish species. J. Appl. Microbiol. 116, 1396–1404. doi: 10.1111/jam.12475
Li, A., Yuan, X., Liang, X.-F., Liu, L., Li, J., Li, B., et al. (2016). Adaptations of lipid metabolism and food intake in response to low and high fat diets in juvenile grass carp (Ctenopharyngodon idellus). Aquaculture 457, 43–49. doi: 10.1016/j.aquaculture.2016.01.014
Li, X., Jiang, Y., Liu, W., and Ge, X. (2012). Protein-sparing effect of dietary lipid in practical diets for blunt snout bream (Megalobrama amblycephala) fingerlings: Effects on digestive and metabolic responses. Fish Physiol. Biochem. 38, 529–541. doi: 10.1007/s10695-011-9533-9
Liao, X. H., and Shi, L. Z. (1956). Contribution to the biology and control of Bothriocephalus gowkongensis yeh, a tapeworm parasitic in the young grass carp (Ctenophapyngodon idellus C. and V.). Acta Hydrobiol. Sin. 2, 129–185.
Ling, F., Steinel, N., Weber, J., Ma, L., Smith, C., Correa, D., et al. (2020). The gut microbiota response to helminth infection depends on host sex and genotype. ISME J. 14, 1141–1153. doi: 10.1038/s41396-020-0589-3
Liu, S., Yu, H., Li, P., Wang, C., Liu, G., Zhang, X., et al. (2022). Dietary nano-selenium alleviated intestinal damage of juvenile grass carp (Ctenopharyngodon idella) induced by high-fat diet: Insight from intestinal morphology, tight junction, inflammation, anti-oxidization and intestinal microbiota. Anim. Nutr. 8, 235–248. doi: 10.1016/j.aninu.2021.07.001
Liu, S., Yu, H., Zhu, L., Zhang, X., Li, P., Wang, C., et al. (2023). Dietary nano-Se supplementation regulates lipid deposition, protein synthesis and muscle fibre formation in grass carp fed with high-fat diet. Br. J. Nutr. 130, 1678–1688. doi: 10.1017/s0007114523000892
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lu, S., Kuang, M., Yue, J., Hu, C., Sheng, G., and Zou, Y. (2022). Utility of traditional and non-traditional lipid indicators in the diagnosis of nonalcoholic fatty liver disease in a Japanese population. Lipids Health Dis. 21:95. doi: 10.1186/s12944-022-01712-z
Malcolm, J. (2011). National research council (Nrc): Nutrient requirements of fish and shrimp. Aquacult. Int. 20, 601–602. doi: 10.1007/s10499-011-9480-6
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17, 10–12. doi: 10.14806/EJ.17.1.200
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., Ohtsubo, Y., et al. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Obi, S., Shimokawa, C., Katsuura, M., Olia, A., Imai, T., Suzue, K., et al. (2020). IL-33 is essential to prevent high-fat diet-induced obesity in mice infected with an intestinal helminth. Parasite Immunol. 42:e12700. doi: 10.1111/pim.12700
Pace, F., Carvalho, B. M., Zanotto, T. M., Santos, A., Guadagnini, D., Silva, K. L. C., et al. (2018). Helminth infection in mice improves insulin sensitivity via modulation of gut microbiota and fatty acid metabolism. Pharmacol. Res. 132, 33–46. doi: 10.1016/j.phrs.2018.04.008
Parada Venegas, D., De la Fuente, M. K., Landskron, G., González, M. J., Quera, R., Dijkstra, G., et al. (2019). Short chain fatty acids (SCFAs)-mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front. Immunol. 10:277. doi: 10.3389/fimmu.2019.00277
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Peachey, L. E., Jenkins, T. P., and Cantacessi, C. (2017). This gut ain’t big enough for both of us. Or is it? Helminth-microbiota interactions in veterinary species. Trends Parasitol. 33, 619–632. doi: 10.1016/j.pt.2017.04.004
Peng, M., Xue, J., Hu, Y., Wen, C., Hu, B., Jian, S., et al. (2019). Disturbance in the homeostasis of intestinal microbiota by a high-fat diet in the rice field eel (Monopterus albus). Aquaculture 502, 347–355. doi: 10.1016/j.aquaculture.2018.12.062
Petkevicius, S., Murrell, K. D., Bach Knudsen, K. E., Jørgensen, H., Roepstorff, A., Laue, A., et al. (2004). Effects of short-chain fatty acids and lactic acids on survival of Oesophagostomum dentatum in pigs. Vet. Parasitol. 122, 293–301. doi: 10.1016/j.vetpar.2004.03.008
Qi, X., Zhang, Y., Zhang, Y., Luo, F., Song, K., Wang, G., et al. (2023). Vitamin B12 produced by Cetobacterium somerae improves host resistance against pathogen infection through strengthening the interactions within gut microbiota. Microbiome 11:135. doi: 10.1186/s40168-023-01574-2
Rajamanickam, A., Munisankar, S., Thiruvengadam, K., Menon, P. A., Dolla, C., Nutman, T. B., et al. (2020). Impact of helminth infection on metabolic and immune homeostasis in non-diabetic obesity. Front. Immunol. 11:2195. doi: 10.3389/fimmu.2020.02195
Shimokawa, C., Obi, S., Shibata, M., Olia, A., Imai, T., Suzue, K., et al. (2019). Suppression of obesity by an intestinal helminth through interactions with intestinal microbiota. Infect. Immun. 87:e00042-19. doi: 10.1128/iai.00042-19
Sobotkova, K., Parker, W., Leva, J., Ruzkova, J., Lukes, J., and Jirku Pomajbikova, K. (2019). Helminth therapy-from the parasite perspective. Trends Parasitol. 35, 501–515. doi: 10.1016/j.pt.2019.04.009
Su, C. W., Chen, C.-Y., Jiao, L., Long, S. R., Mao, T. Y., Ji, Q. R., et al. (2020). Helminth-induced and Th2-dependent alterations of the gut microbiota attenuate obesity caused by high-fat diet. Cell. Mol. Gastroenterol. Hepatol. 10, 763–778. doi: 10.1016/j.jcmgh.2020.06.010
Su, C. W., Chen, C. Y., Li, Y., Long, S. R., Massey, W., Kumar, D. V., et al. (2018). Helminth infection protects against high fat diet-induced obesity via induction of alternatively activated macrophages. Sci. Rep. 8:4607. doi: 10.1038/s41598-018-22920-7
Tang, T., Hu, Y., Peng, M., Chu, W., Hu, Y., and Zhong, L. (2019). Effects of high-fat diet on growth performance, lipid accumulation and lipid metabolism-related MicroRNA/gene expression in the liver of grass carp (Ctenopharyngodon idella). Comp. Biochem. Phys. B 234, 34–40. doi: 10.1016/j.cbpb.2019.04.006
Tao, Y. F., Qiang, J., Bao, J. W., Chen, D. J., Yin, G. J., Xu, P., et al. (2018). Changes in physiological parameters, lipid metabolism, and expression of micrornas in genetically improved farmed Tilapia (Oreochromis niloticus) with fatty liver induced by a high-fat diet. Front. Physiol. 9:1521. doi: 10.3389/fphys.2018.01521
Teng, B., Huang, C., Cheng, C. L., Udduttula, A., Yu, X. F., Liu, C., et al. (2020). Newly identified peptide hormone inhibits intestinal fat absorption and improves NAFLD through its receptor GPRC6A. J. Hepatol. 73, 383–393. doi: 10.1016/j.jhep.2020.02.026
van Kessel, M. A., Dutilh, B. E., Neveling, K., Kwint, M. P., Veltman, J. A., Flik, G., et al. (2011). Pyrosequencing of 16S rRNA gene amplicons to study the microbiota in the gastrointestinal tract of carp (Cyprinus carpio L.). AMB Exp. 1:41. doi: 10.1186/2191-0855-1-41
Wang, A. R., Zhang, Z., Ding, Q. W., Yang, Y., Bindelle, J., Ran, C., et al. (2021). Intestinal Cetobacterium and acetate modify glucose homeostasis via parasympathetic activation in zebrafish. Gut Microb. 13:e1900996. doi: 10.1080/19490976.2021.1900996
Wang, J. T., Liu, Y. J., Tian, L. X., Mai, K. S., Du, Z. Y., Wang, Y., et al. (2005). Effect of dietary lipid level on growth performance, lipid deposition, hepatic lipogenesis in juvenile cobia (Rachycentron canadum). Aquaculture 249, 439–447. doi: 10.1016/j.aquaculture.2005.04.038
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/aem.00062-07
Wu, D., Molofsky, A. B., Liang, H. E., Ricardo-Gonzalez, R. R., Jouihan, H. A., Bando, J. K., et al. (2011). Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 332, 243–247. doi: 10.1126/science.1201475
Wu, S. A., Kersten, S., and Qi, L. (2021). Lipoprotein lipase and its regulators: An unfolding story. Trends Endocrinol. Metab. 32, 48–61. doi: 10.1016/j.tem.2020.11.005
Xie, M., Hao, Q., Olsen, R. E., Ringo, E., Yang, Y., Zhang, Z., et al. (2022). Growth performance, hepatic enzymes, and gut health status of common carp (Cyprinus carpio) in response to dietary Cetobacterium somerae fermentation product. Aquacult. Rep. 23:101046. doi: 10.1016/j.aqrep.2022.101046
Xu, H., Wang, J., Wang, Q., Tu, W., and Jin, Y. (2024). Co-exposure to polystyrene microplastics and cypermethrin enhanced the effects on hepatic phospholipid metabolism and gut microbes in adult zebrafish. J. Hazard. Mater. 465:133051. doi: 10.1016/j.jhazmat.2023.133051
Yang, C., Luan, N., An, J., Zhang, M., Li, Z., Li, Q., et al. (2020). The effects of Rhodobacter sphaeroides on the composition of gut microbiota and short-chain fatty acids in mice. J. Food Nutr. Res. 8, 288–296. doi: 10.12691/jfnr-8-6-7
Yang, Z., Grinchuk, V., Smith, A., Qin, B., Bohl, J. A., Sun, R., et al. (2013). Parasitic nematode-induced modulation of body weight and associated metabolic dysfunction in mouse models of obesity. Infect. Immun. 81, 1905–1914. doi: 10.1128/iai.00053-13
Yin, P., Xie, S., Zhuang, Z., He, X., Tang, X., Tian, L., et al. (2021). Dietary supplementation of bile acid attenuate adverse effects of high-fat diet on growth performance, antioxidant ability, lipid accumulation and intestinal health in juvenile largemouth bass (Micropterus salmoides). Aquaculture 531:735864. doi: 10.1016/j.aquaculture.2020.735864
Yu, C., Zhang, J., Qin, Q., Liu, J., Xu, J., and Xu, W. (2020). Berberine improved intestinal barrier function by modulating the intestinal microbiota in blunt snout bream (Megalobrama amblycephala) under dietary high-fat and high-carbohydrate stress. Fish Shellfish Immunol. 102, 336–349. doi: 10.1016/j.fsi.2020.04.052
Zhang, Y., Zhang, T., Liang, Y., Jiang, L., and Sui, X. (2021). Dietary bioactive lipids: A review on absorption, metabolism, and health properties. J. Agric. Food Chem. 69, 8929–8943. doi: 10.1021/acs.jafc.1c01369
Zhang, Y. F., Sun, Z. Z., Wang, A. L., Ye, C. X., and Zhu, X. (2017). Effects of dietary protein and lipid levels on growth, body and plasma biochemical composition and selective gene expression in liver of hybrid snakehead (Channa maculata ♀× Channa argus ♂) fingerlings. Aquaculture 468, 1–9. doi: 10.1016/j.aquaculture.2016.09.052
Zhao, P. F., Li, F. J., Chen, X. R., Chen, Y. J., Lin, S. M., Zhang, L., et al. (2016). Dietary lipid concentrations influence growth, liver oxidative stress, and serum metabolites of juvenile hybrid snakehead (Channa argus × Channa maculata). Aquacult. Int. 24, 1353–1364. doi: 10.1007/s10499-016-9993-0
Keywords: Schyzocotyle acheilognathi, Ctenopharyngodon idella, high-fat diet, lipid metabolism, gut microbiota
Citation: Yang X, Zhu D, Li W and Fu P (2025) Intestinal helminth Schyzocotyle acheilognathi Yamaguti, 1934 infection ameliorate lipid metabolism of grass carp (Ctenopharyngodon idella) through immune and gut microbiota regulation. Front. Microbiol. 16:1538919. doi: 10.3389/fmicb.2025.1538919
Received: 03 December 2024; Accepted: 23 June 2025;
Published: 10 July 2025.
Edited by:
Hakdong Shin, Sejong University, Republic of KoreaReviewed by:
Shun Zhou, Chinese Academy of Fishery Sciences (CAFS), ChinaLingtong Ye, South China Sea Fisheries Research Institute (CAFS), China
Copyright © 2025 Yang, Zhu, Li and Fu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Peipei Fu, cGVpcGVpZnUwNzIwQDE2My5jb20=
 Xiaoao Yang
Xiaoao Yang Denghui Zhu
Denghui Zhu Wenxiang Li
Wenxiang Li Peipei Fu
Peipei Fu