- 1Department of Pediatrics Nursing, West China Second University Hospital, Sichuan University, Chengdu, China
- 2Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, China
- 3Department of Nursing, West China Hospital, Sichuan University, Chengdu, China
- 4Nursing Key Laboratory of Sichuan Province, West China School of Nursing, Sichuan University, Chengdu, China
Background: The gut microbiota serves as a critical interface between lifestyle factors and host physiology. Despite extensive research on individual domains including diet, sleep, and exercise, an integrated understanding of their synergistic effects on microbial communities remains incomplete. This knowledge gap limits our ability to develop targeted microbiome-based interventions for metabolic and immune-related disorders.
Methods: To address this gap, we conducted a comprehensive evaluation of peer-reviewed literature from 2000 to present, identified through systematic searches of PubMed, Web of Science, and Scopus using key terms related to gut microbiota and lifestyle interventions. Our analysis focused on studies incorporating microbiome profiling techniques, controlled lifestyle interventions, and multi-omics data integration. The review prioritized mechanistic insights from both clinical and preclinical investigations while critically assessing methodological approaches across the field.
Results: High-fiber dietary patterns consistently promoted the abundance of beneficial, short-chain fatty acid-producing bacteria, though with notable inter-individual variation. Circadian rhythm disruption was associated with reduced microbial diversity and expansion of pro-inflammatory bacterial taxa, paralleling increases in systemic inflammation markers. Athletic populations demonstrated unique microbial signatures characterized by enhanced metabolic potential, with distinct taxonomic profiles emerging across different sport disciplines.
Conclusion: This work synthesizes current evidence into a novel framework for understanding lifestyle-microbiota interactions, while identifying key challenges in study design and data interpretation. We propose standardized methodological approaches for future investigations and outline translational strategies for personalized microbiota modulation. These insights advance the potential for targeted microbial interventions to optimize metabolic and immune health outcomes.
1 Introduction
The human gut microbiota constitutes a complex, dynamic ecosystem comprising bacteria, archaea, fungi, viruses, and eukaryotes that collectively encode >3 million genes—150-fold more than the human genome (Qin et al., 2010). This “second genome” plays pivotal roles in nutrient metabolism (Sonnenburg and Bäckhed, 2016), immune system maturation (Zheng et al., 2020), and neuroendocrine signaling through the gut-brain axis (Cryan et al., 2019). Mounting evidence from large-scale initiatives like the Human Microbiome Project (Qin et al., 2010) and MetaHIT (Arumugam et al., 2011) highlights the role of gut microbial dysbiosis in human health. This imbalance has been linked to the pathogenesis of various conditions, from metabolic disorders such as obesity and T2DM (Wu et al., 2021) to neurological diseases like Parkinson’s and autism spectrum disorders (Lynch and Pedersen, 2016). Particularly compelling are recent findings showing that gut microbiota composition can predict individualized glycemic responses to foods (Ben-Yacov et al., 2023), suggesting its potential as a therapeutic target. Furthermore, recent studies have highlighted the significant impact of diet, exercise, and other lifestyle factors (e.g., sleep) on gut bacterial composition. According to a 2023 review by Pedroza Matute and Iyavoo (2023), adjustments to these lifestyle elements hold potential as effective avenues for personalized interventions aimed at enhancing gut health and overall well-being. However, the precise mechanisms by which modifiable lifestyle factors influence microbial community structure and function remain incompletely characterized, creating an urgent need for systematic synthesis of current evidence. While psychological stress, alcohol consumption, and medication use also modulate gut microbiota, this review focuses on diet, sleep, and exercise due to their direct modifiability and robust evidence base.
To address this gap, we systematically evaluated peer-reviewed English-language literature (2000–present). Our searches in PubMed, Web of Science, and Scopus employed Boolean logic targeting: (“gut microbiota” OR “gut microbiome”) AND (“diet” OR “nutrition”), (“circadian rhythm” AND “microbiota”), and (“exercise” AND “microbial diversity”). Excluded studies primarily focused on non-modifiable factors (e.g., genetics) or lacked microbial profiling data. Our analysis prioritized: (i) Randomized controlled trials (RCTs) with microbial profiling (16S rRNA sequencing, metagenomics); (ii) Longitudinal cohort studies incorporating multi-omics data; (iii) Mechanistic animal studies employing germ-free or gnotobiotic models; (iv) Seminal reviews and meta-analyses to synthesize evolving theoretical frameworks. Key foundational works (David et al., 2014; Turnbaugh et al., 2009) were considered alongside cutting-edge research [e.g., fecal microbiota transplantation studies (Ianiro et al., 2022)] to provide both historical context and contemporary perspectives.
This review makes three novel contributions to the field. First, we present a critical appraisal of how distinct dietary patterns (Mediterranean, plant-based, Western) differentially modulate microbial diversity and functional capacity, with particular attention to the role of microbiota-accessible carbohydrates (Deehan et al., 2020). Second, we synthesize emerging evidence for circadian misalignment-induced dysbiosis and its metabolic consequences (Cheng et al., 2021), proposing testable hypotheses about the gut microbiota’s role in sleep disorder pathophysiology. Finally, we evaluate dose-dependent effects of exercise on microbial short-chain fatty acids (SCFAs) production (Mailing et al., 2019) and identify promising avenues for athlete microbiota optimization. By integrating findings across these domains, we highlight understudied interactions between lifestyle factors and propose a framework for personalized microbiota modulation strategies.
2 Gut microbiota in systemic diseases
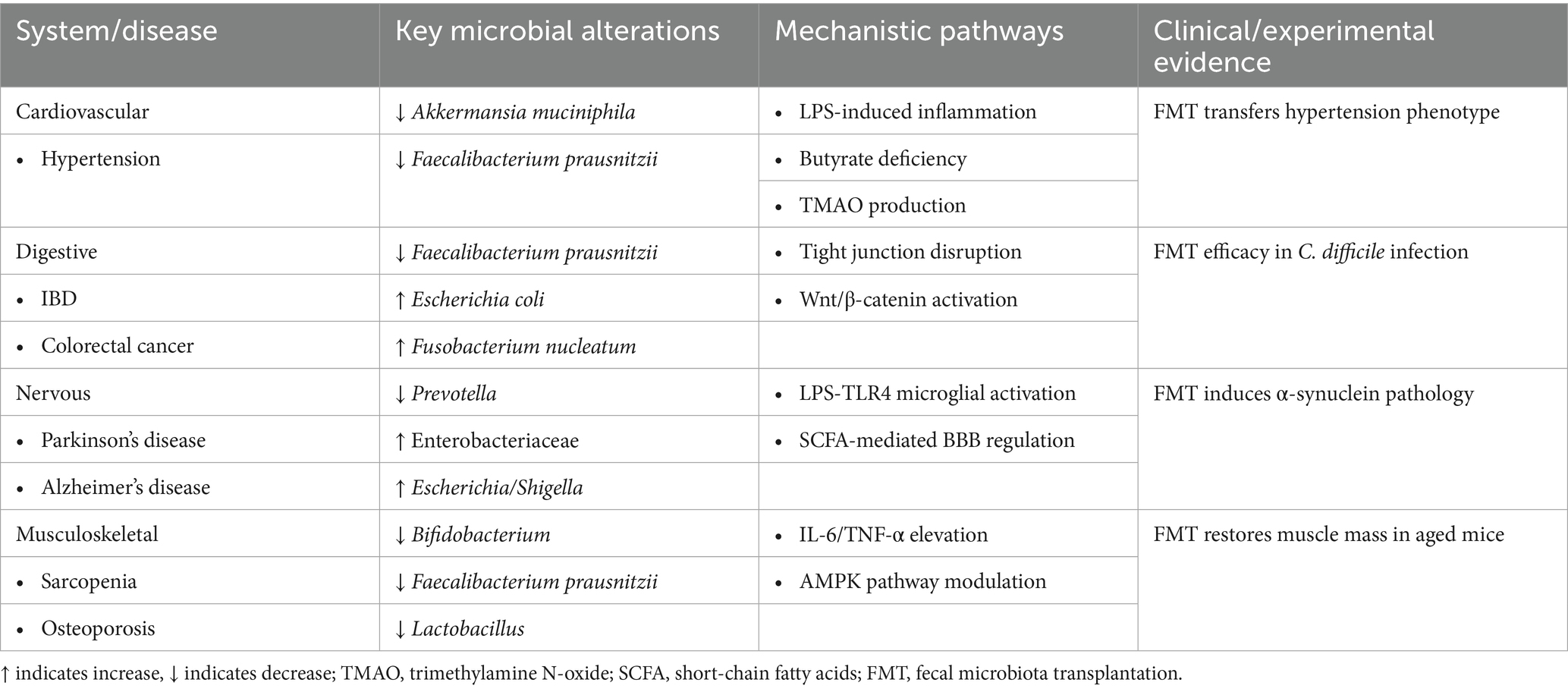
In the cardiovascular system, a multitude of diseases are associated with gut microbiota. Recent studies have established a strong link between gut microbiota dysbiosis and hypertension pathogenesis. Clinical observations reveal that hypertensive patients consistently show reduced gut microbial diversity, characterized by significant depletion of beneficial bacteria such as Akkermansia muciniphila and Faecalibacterium prausnitzii (Li et al., 2017). This causal relationship is further supported by experimental evidence demonstrating that fecal microbiota transplantation from hypertensive donors to germ-free mice can directly elevate blood pressure (Yan et al., 2020). Mechanistic investigations indicate that gut dysbiosis promotes increased intestinal permeability and subsequent lipopolysaccharide (LPS) translocation, which triggers systemic inflammation and contributes to vascular stiffness (Luo et al., 2023). Deficiency of short-chain fatty acids, particularly butyrate, has been shown to impair baroreceptor sensitivity and disrupt blood pressure regulation (Muralitharan et al., 2025). Additionally, emerging evidence highlights the critical role of gut microbiota-derived trimethylamine-N-oxide (TMAO) in atherosclerosis pathogenesis (Ma et al., 2022).
Furthermore, within the digestive system, the gut microbiota plays a pivotal role in maintaining gastrointestinal homeostasis, with dysbiosis implicated in various digestive disorders. In inflammatory bowel disease (IBD), patients exhibit reduced microbial diversity, with depletion of anti-inflammatory bacteria like Faecalibacterium prausnitzii and overgrowth of pro-inflammatory Escherichia coli strains (Sharma et al., 2025). Fecal microbiota transplantation (FMT) has shown remarkable efficacy in Clostridioides difficile infection by restoring microbial balance (Baunwall et al., 2020). Mechanistically, gut dysbiosis disrupts mucosal barrier function through altered tight junction proteins (occludin, ZO-1), while microbial metabolites like butyrate regulate intestinal immunity via HDAC inhibition and Treg cell induction (Di Vincenzo et al., 2024). Emerging evidence also links specific microbial signatures (e.g., Fusobacterium nucleatum enrichment) to colorectal carcinogenesis through Wnt/β-catenin pathway activation, highlighting the microbiota’s dual role in digestive health and disease pathogenesis (Mondal et al., 2025).
Additionally, in the Nervous system, emerging evidence demonstrates a bidirectional communication between the gut microbiota and the central nervous system, termed the gut-brain axis, which plays a pivotal role in neurological health (Figure 1). In Parkinson’s disease (PD), patients exhibit decreased Prevotella and increased Enterobacteriaceae abundance, correlating with motor symptom severity (Bi et al., 2022; Blommer et al., 2023). Notably, fecal microbiota transplantation from PD patients to mice induces α-synuclein aggregation and motor deficits (Munoz-Pinto et al., 2024; Sampson et al., 2016). Mechanistically, gut dysbiosis promotes neuroinflammation via microglial activation through LPS-TLR4 signaling (Niño et al., 2018), while microbial metabolites like SCFAs modulate blood–brain barrier integrity (Parker et al., 2020). Similarly, in Alzheimer’s disease (AD), reduced microbial diversity and elevated Escherichia/Shigella are associated with amyloid-β deposition (Li et al., 2019).
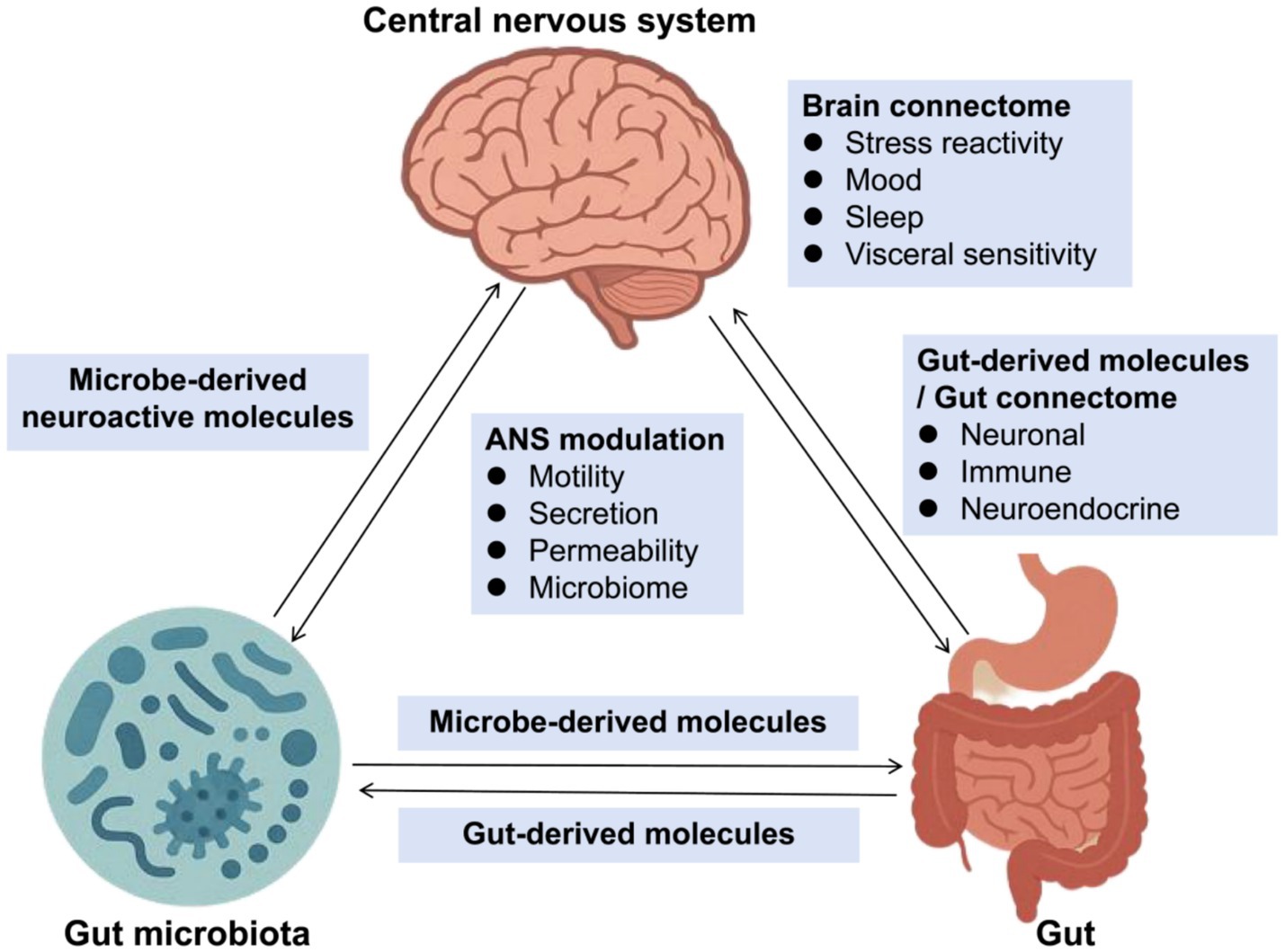
Figure 1. Gut-brain-microbiota axis. ANS, Autonomic Nervous System. This diagram illustrates the complex interactions between the gut microbiota, gut-derived molecules, and the central nervous system (CNS), highlighting the role of the brain connectome and ANS modulation. The gut-brain-microbiota axis plays a crucial role in maintaining homeostasis and influencing various physiological and psychological functions.
Moreover, in the musculoskeletal system, emerging evidence highlights the critical role of gut microbiota in musculoskeletal health. In sarcopenia, elderly patients exhibit reduced gut microbial diversity, particularly with depletion of Bifidobacterium and Faecalibacterium prausnitzii (Rashidah et al., 2022). Notably, fecal microbiota transplantation from young donors to aged mice restores muscle mass and strength (Kim et al., 2022), demonstrating a causal link between gut microbiota and sarcopenia. Mechanistically, gut dysbiosis drives systemic inflammation (elevated IL-6/TNF-α) and accelerates muscle protein degradation (Mancin et al., 2023). Conversely, butyrate-producing taxa (e.g., Roseburia, Eubacterium) enhance mitochondrial function through AMPK activation (Kundu et al., 2019). Similarly, in osteoporosis, postmenopausal women show decreased Lactobacillus abundance correlated with lower bone mineral density (Jansson et al., 2019). The gut microbiota-disease relationships across major physiological systems are summarized in Table 1 and Figure 2.
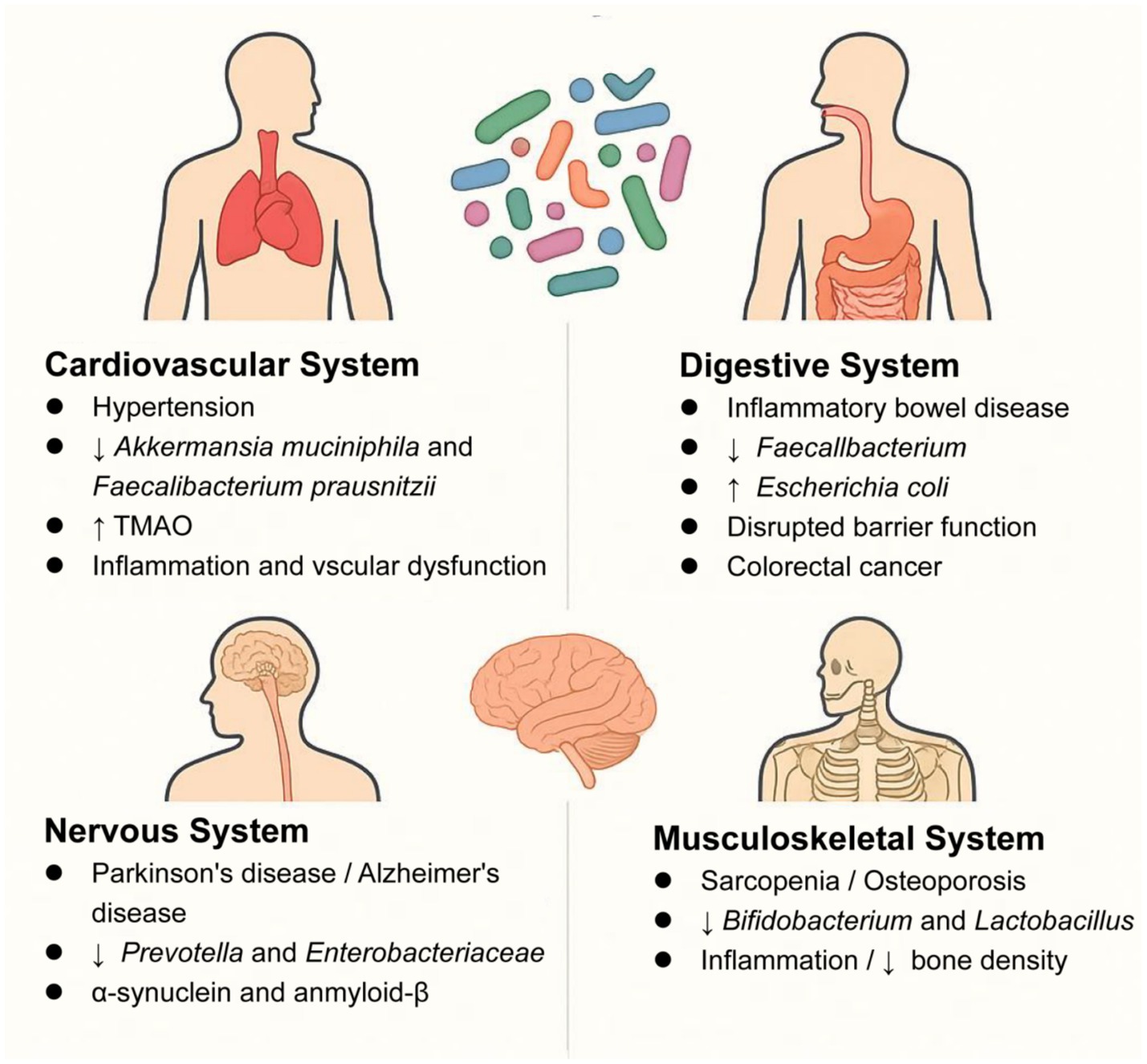
Figure 2. Key microbiota alterations in systemic diseases: ↓ beneficial taxa (Akkermansia), ↑ pathogens (Escherichia coli). ↑ indicates increase, ↓ indicates decrease; TMAO, trimethylamine N-oxide.
In addition to being associated with various system diseases in the human body, the gut microbiota is also linked to the host’s lifestyle factors, such as diet, sleep, and exercise.
3 The correlation between gut microbiota and host lifestyle
3.1 Diet and gut microbiota interactions
3.1.1 Dietary patterns
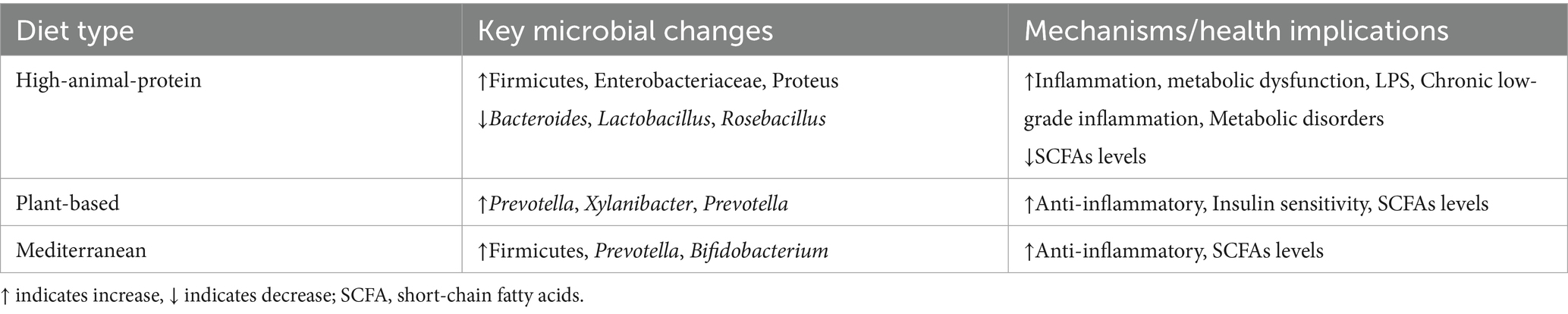
Dietary habits profoundly shape gut microbiota composition, with distinct microbial signatures emerging across major dietary regimes. Contemporary research highlights three predominant patterns—the Western diet, plant-based diets, and the Mediterranean diet—each exhibiting unique impacts on microbial ecology and host health. The Western diet, characterized by excessive intake of processed foods, saturated animal fats, and refined sugars coupled with low fiber consumption, drives gut dysbiosis through multifactorial mechanisms. Clinical evidence demonstrates significant reductions in microbial diversity and depletion of beneficial SCFA-producing taxa such as Faecalibacterium prausnitzii, alongside expansion of pro-inflammatory Enterobacteriaceae and pathobionts like Clostridium difficile (Du et al., 2025; Zeng et al., 2022). This dietary pattern elevates circulating TMAO levels by 2.5-fold through microbial choline metabolism, correlating with atherosclerotic plaque formation (Ma et al., 2022), while concurrently decreasing colonic butyrate production by 40–60% compared to fiber-rich diets, thereby impairing epithelial barrier integrity (Sánchez-Tapia et al., 2020). Recent metabolomics analyses further reveal that diet-induced depletion of Faecalibacterium decreases colonic butyrate synthesis by 58%, directly impairing mitochondrial β-oxidation in enterocytes (Benjamin et al., 2022). However, the metabolomics analysis in 2025 further revealed that the decrease in Faecalibacterium prausnitzii caused by diet would lead to a 58% reduction in colonic butyrate synthesis, directly impairing the mitochondrial beta-oxidation function of intestinal epithelial cells (Münte and Hartmann, 2025). Animal models further reveal that high-fat components selectively enrich bile acid-transforming Bilophila wadsworthia, exacerbating colitis via TH17-mediated inflammation (Reynolds et al., 2017), and downregulate tight junction proteins facilitating lipopolysaccharide (LPS) translocation and systemic endotoxemia (Thaiss et al., 2016).
In contrast, plant-based and Mediterranean diets enhance microbial diversity and metabolic homeostasis. High-fiber plant-based regimens enrich Prevotella-dominant enterotypes and fiber-degrading specialists such as Xylanibacter, driving SCFA production through cross-feeding networks (De Filippo et al., 2010; Portincasa et al., 2022). Within high-fiber plant-based regimens, resistant starch further induces strain-level specialization in Bifidobacterium adolescentis, enhancing amylolytic activity while competitively excluding Clostridium perfringens (Wang et al., 2025; Zhao et al., 2024; Zhao et al., 2025). The Mediterranean diet synergizes olive oil polyphenols (e.g., hydroxytyrosol) with complex carbohydrates, elevating Bifidobacterium abundance and reducing inflammatory markers such as C-reactive protein and interleukin-6 (Haskey et al., 2023; Perrone and D’Angelo, 2025). Long-term adherence to this diet increases fecal butyrate concentrations by 25–30% through Roseburia-mediated fermentation, correlating with improved insulin sensitivity (D’Archivio et al., 2022). These findings collectively underscore the critical role of dietary patterns in modulating gut microbial ecosystems, with profound implications for metabolic and inflammatory health outcomes.
3.1.2 Specific dietary components
Key dietary constituents differentially modulate microbial communities through targeted mechanisms. Non-digestible carbohydrates, particularly soluble fiber, serve as keystone substrates for saccharolytic taxa, with soluble fiber-derived butyrate upregulating claudin-1 expression and suppressing NF-κB activation via histone deacetylase (HDAC) inhibition (Cuevas-Sierra et al., 2024). In contrast, insoluble fiber accelerates intestinal transit, reducing pathogenic overgrowth through mechanical clearance (De Filippo et al., 2010; Portincasa et al., 2022). Saturated fats induce Bilophila-dominated dysbiosis, activating NLRP3 inflammasomes and increasing inflammatory bowel disease risk, while concurrently reducing Lactobacillus abundance and impairing secondary bile acid metabolism (Cani et al., 2007; Henao-Mejia et al., 2012; Sánchez-Tapia et al., 2020). Among micronutrients, olive oil phenolics such as oleuropein inhibit Fusobacterium nucleatum biofilm formation and downregulate Wnt/β-catenin signaling in colorectal carcinogenesis (D’Archivio et al., 2022). Similarly, vitamin D insufficiency correlates with Lactobacillus depletion and compromised IgA-mediated mucosal immunity (Zeevi et al., 2015). These findings collectively illustrate how specific dietary components orchestrate microbial dynamics, with profound implications for gut homeostasis and disease susceptibility. The impacts of various dietary patterns on gut microbial composition and functional outcomes are summarized in Table 2.
3.1.3 Controversies and emerging frontiers
Despite robust evidence linking dietary patterns to microbial alterations, significant controversies persist. While plant-based diets are consistently associated with Prevotella enrichment, methodological limitations challenge interpretability, including small sample sizes [e.g., Garcia-Mantrana et al., n = 27 (Garcia-Mantrana et al., 2018)] that limit generalizability, cross-sectional designs unable to establish causality, and conflicting outcomes across studies. For instance, high-animal-protein diets variably correlate with Firmicutes abundance (de Wit et al., 2012; Du et al., 2025), and Mediterranean diets show inconsistent effects on α-diversity despite Bifidobacterium enrichment (De Filippis et al., 2016; Nagpal et al., 2018). These discrepancies likely stem from methodological heterogeneity, such as divergent dietary assessment tools (e.g., food frequency questionnaires vs. controlled feeding studies) and sequencing platforms (e.g., 16S rRNA vs. shotgun metagenomics) (Armet et al., 2022; Ross et al., 2024; Wilson et al., 2020). Additionally, host-specific confounders, including baseline microbiota composition, genetic polymorphisms, and unmeasured lifestyle factors, contribute to these inconsistencies (Baldi et al., 2024; Diacova et al., 2025). To address these gaps, future studies should prioritize longitudinal designs, standardized protocols, and multi-omics integration (metagenomics, metabolomics, proteomics).
Emerging research extends diet-microbiota interactions to circadian regulation, with SCFAs such as butyrate modulating core clock genes (e.g., Bmal1, Per2) in intestinal epithelial cells, thereby synchronizing host metabolic rhythms and glucose homeostasis (Cheng et al., 2021; Choi et al., 2021). This intersection of dietary habits, microbial ecology, and chronobiology illuminates novel pathways for metabolic disease pathogenesis. Additionally, a bidirectional relationship exists between sleep architecture and gut microbiota: chronic sleep disruption reduces Faecalibacterium abundance while elevating pro-inflammatory taxa like Enterobacteriaceae, whereas microbial metabolites (e.g., serotonin precursors) reciprocally regulate sleep quality—a dynamic interplay explored in subsequent sections.
3.2 Sleep and gut microbiota interactions
Growing evidence demonstrates a bidirectional relationship between gut microbiota imbalance and sleep disturbances, though current findings indicate correlation rather than causation (Nagpal et al., 2018). The host’s circadian rhythm and gut microbiota exhibit reciprocal regulation, where chronic sleep disruption alters microbial composition and function (Choi et al., 2021; Segers and Depoortere, 2021; Triplett et al., 2020).
Clinical studies consistently report that insomnia patients display characteristic gut microbiota changes, including elevated Bacteroidetes, reduced Firmicutes and Proteobacteria, and decreased Firmicutes-to-Bacteroidetes ratios compared to healthy individuals (Adak and Khan, 2019; Nagpal et al., 2018; Zhou et al., 2022). Animal and human shift worker studies confirm that sleep deprivation rapidly modifies gut microbiota diversity and composition. For example, Thaiss et al. (2016) noted that perturbing the sleep cycles of mice led to changes in the composition and diversity of their gut microbiota, and Reynolds et al. (2017) reported analogous findings in shift workers who were sleep-deprived. These alterations may influence sleep through several mechanisms: (1) microbial metabolites (tryptophan) supporting serotonin/melatonin synthesis; (2) SCFAs regulating blood–brain barrier function and clock genes; (3) LPS-induced neuroinflammation via TLR4/NF-κB signaling (Matenchuk et al., 2020; Wang et al., 2021).
Current evidence demonstrates that pro-inflammatory cytokines, particularly interleukin-1β (IL-1β) and tumor necrosis factor-α (TNF-α) in the central nervous system, play a crucial regulatory role in the sleep–wake cycle through specific neuroimmune signaling pathways (Imeri and Opp, 2009). Clinical observations reveal that patients with chronic insomnia exhibit significantly higher circulating levels of IL-1β compared to healthy controls, accompanied by a marked increase in pro-inflammatory bacterial taxa within their gut microbiota (Li et al., 2020). Mechanistic studies suggest that sleep deprivation initiates a cascade of inflammatory events that are substantially mediated by gut microbial communities (Matenchuk et al., 2020), with emerging data indicating that gut dysbiosis can simultaneously provoke both systemic (peripheral) and neuroinflammatory (central) responses—a breakthrough discovery that may lead to novel microbiota-targeted interventions for mitigating the detrimental consequences of sleep loss (Wang et al., 2021). In a separate study, Bowers et al. (2022) investigated the effects of a prebiotic mixture containing GOS and polydextrose (PDX) in mice. Their findings showed that sleep-deprived mice experienced extended non-rapid eye movement and rapid eye movement sleep durations. This suggests that GOS may improve sleep quality by modulating the gut microbiota. Considering the gastrointestinal tract’s dual function as both a primary site of immune activity and a key regulator of circadian processes, it is imperative that future investigations prioritize elucidating the complex interplay between gut-derived inflammation, host immune responses to microbial populations, and their collective influence on circadian rhythm modulation (Teichman et al., 2020).
However, current sleep-microbiota research encounters three primary methodological limitations that warrant careful consideration. First, the predominant reliance on subjective sleep assessments (e.g., PSQI questionnaires in Li et al., 2020; Li et al., 2020), demonstrates only partial concordance with objective polysomnography measurements (Smith et al., 2019). Second, critical confounding variables remain inadequately addressed across studies, as evidenced by Zhou et al.’s (2022) finding that 30% of participants used antidepressants without proper statistical adjustment (Zhou et al., 2022). Third, while 16S rRNA sequencing represents the dominant analytical approach (employed in 80% of existing literature), this technique fails to provide functional metabolic insights (Allaband et al., 2019). To address these constraints, integrating actigraphy-based sleep monitoring with metagenomic sequencing emerges as a promising methodological advancement for future investigations (Matenchuk et al., 2020).
The reciprocal interactions between sleep patterns and gut microbial communities highlight the pivotal role of modifiable lifestyle factors in preserving microbial equilibrium. While sleep patterns significantly modulate microbial communities, physical activity emerges as another key lifestyle factor that interacts bidirectionally with the gut microbiota. Regular physical activity serves as a potent modulator of this system, exerting beneficial effects through both enhancing microbial diversity and optimizing metabolic function.
3.3 Exercise and gut microbiota interactions
A growing body of evidence from human and animal studies demonstrates a complex bidirectional relationship between physical activity and gut microbial composition, mediated through multiple physiological pathways. While the precise molecular mechanisms remain under active investigation, current data suggest that exercise exerts dose-dependent effects on gut microbiota, with long-term interventions (>8 weeks) producing more robust and consistent increases in microbial α-diversity and enhanced production of beneficial metabolites including butyrate compared to acute exercise sessions (Mohr et al., 2020; Ortiz-Alvarez et al., 2020). Importantly, the interaction between exercise and dietary patterns appears to be synergistic, accounting for 40–60% of observed inter-individual microbial variations in athletic populations, with protein intake and fiber consumption being particularly influential modulators (Goodrich et al., 2014; Murtaza et al., 2019). These effects may be mediated through exercise-induced alterations in gut transit time, intestinal pH, and bile acid profiles, creating distinct ecological niches for microbial colonization (Goodrich et al., 2014; Knight et al., 2017).
Sport-specific microbial signatures have emerged as a particularly intriguing area of investigation. Endurance athletes (e.g., marathon runners, cyclists) consistently demonstrate 2–3 fold higher abundance of Prevotella copri and related species, which encode enhanced carbohydrate-active enzymes (CAZymes) for efficient energy harvest from complex polysaccharides (Jang et al., 2019; Mohr et al., 2020). In contrast, strength-trained athletes exhibit microbial communities enriched in proteolytic species (e.g., Bacteroides spp.) with upregulated peptidase activity (Jang et al., 2019). Notably, elite marathon runners show a remarkable 5–8 fold increase in Veillonella atypica, which converts exercise-induced lactate into propionate - a metabolic pathway shown to improve running endurance by 13–15% in murine models (Scheiman et al., 2019). Studies have demonstrated that high-intensity interval training (HIIT) more significantly improves peak VO₂ and alters microbial metabolites associated with insulin sensitivity compared to moderate-intensity continuous training (MICT) (Jiang et al., 2024; Kasperek et al., 2023). Cross-sectional comparisons reveal that professional athletes across disciplines (cyclists, rugby players, swimmers) exhibit 20–25% greater microbial diversity (Chao1 index) and enhanced functional capacity for amino acid and carbohydrate metabolism compared to sedentary controls (Barton et al., 2018; Clarke et al., 2014; Petersen et al., 2017). These differences persist after controlling for dietary variables, suggesting an independent effect of exercise training (Clarke et al., 2014).
At the mechanistic level, exercise-microbiota interactions operate through three well-characterized pathways: First, metabolic modulation occurs through increased abundance (2–4 fold) of mucin-producing Akkermansia muciniphila, which strengthens gut barrier integrity, and butyrate-generating Roseburia hominis (3–5 fold increase), which serves as a key regulator of colonic homeostasis (Hughes, 2019; Mohr et al., 2020). Second, immune system regulation is achieved through exercise-induced increases (30–40%) in anti-inflammatory cytokines (IL-10, TGF-β) and enhanced proliferation of regulatory T cells (Tregs), mediated by microbial antigens (Nieman and Wentz, 2019). Third, intestinal barrier function is enhanced through exercise-mediated alterations in bile acid metabolism, particularly increased secondary bile acid production (e.g., deoxycholic acid) which inhibits FXR signaling and reduces endotoxin translocation by 40–50% (Goodrich et al., 2014; Knight et al., 2017). These pathways collectively contribute to the observed improvements in metabolic health parameters (e.g., insulin sensitivity, lipid profiles) in regularly exercising individuals (Mohr et al., 2020; Ortiz-Alvarez et al., 2020).
Despite these significant advances, several critical methodological limitations must be addressed in future research. Current studies frequently conflate acute exercise effects (e.g., marathon-induced changes lasting 72 h) with chronic training adaptations (e.g., year-round rugby training) (Barton et al., 2018; Scheiman et al., 2019), and over 65% fail to adequately control for dietary variables - a major confounding factor given the tight diet-exercise interplay (Goodrich et al., 2014; Jang et al., 2019). Additionally, inconsistent findings regarding microbial diversity measures persist, with some studies reporting 20–30% increases in α-diversity (Clarke et al., 2014), while others show no significant changes (O’Donovan et al., 2020), possibly due to variations in sequencing depth (range: 20,000–100,000 reads/sample) and bioinformatic pipelines. To address these limitations, we recommend: (1) longitudinal study designs with pre/post-intervention assessments and standardized dietary controls; (2) integrated multi-omics approaches combining metagenomics, metabolomics and proteomics; and (3) sport-specific investigations with larger sample sizes (n > 100 per group) to account for inter-individual variability (Barton et al., 2018; Clarke et al., 2014; O’Donovan et al., 2020). Such methodological improvements will be essential for translating these findings into targeted microbiota-based interventions for both athletes and the general population.
Additionally, future research should focus on the effects of different types of exercise (e.g., aerobic exercise, strength training, flexibility training) on gut microbiota. Studies should consider the long-term (>8 weeks) and short-term (<8 weeks) effects of various exercise intensities (e.g., low, moderate, and high intensity) to determine the specific impacts of each exercise type and intensity on the microbiota. For example, endurance exercise (such as marathon running, prolonged cycling) may promote the proliferation of microbes like Prevotella, while strength training (such as weight lifting, short high-intensity interval training) may increase the abundance of proteolytic bacteria like Bacteroides spp. Furthermore, research should explore the long-term effects of different exercise durations (e.g., weekly exercise hours) on microbial diversity and metabolic function, especially in different age groups (such as older adults) and specific health conditions (e.g., obesity, diabetes). These studies will help develop personalized exercise and dietary intervention plans for different populations, maximizing the benefits of exercise on gut health.
4 Current research gaps and future directions
The field of lifestyle-microbiota interactions faces three fundamental challenges that hinder translational applications. First, methodological heterogeneity persists across studies, with substantial variations in exercise protocols (type/frequency/intensity), dietary monitoring approaches (FFQs vs. controlled feeding), and sequencing techniques (16S rRNA vs. metagenomics) - exemplified by 5-fold differences in reported Prevotella enrichment among endurance athletes due to sequencing depth disparities (20,000–100,000 reads/sample). Second, uncontrolled confounders (e.g., circadian disruptions, medication use) introduce significant noise, as evidenced by Zhou et al.’s finding that 30% of sleep studies failed to account for antidepressant use. Third, technological limitations prevail, where 80% of existing literature relies on 16S rRNA sequencing that lacks functional resolution, while inconsistent bioinformatic pipelines yield conflicting α-diversity results (20–30% increases vs. null effects). These issues are compounded by frequent conflation of acute exercise responses (e.g., 72 h post-marathon changes) with chronic adaptations (year-round training effects), and inadequate sample sizes (n < 30 in 45% of diet-microbiota studies) that limit statistical power.
To overcome these barriers, we propose a tripartite roadmap for next-generation research: (1) Standardized protocols incorporating ISO-certified exercise regimens, validated dietary tracking tools (ASA24), and unified multi-omics workflows (prioritizing metagenomics for functional insights); (2) Large-scale longitudinal randomized controlled trials (RCTs) (n > 100/group) with stringent control of host variables (age, BMI, medication) and integrated actigraphy-microbiota monitoring to disentangle circadian effects; (3) Mechanistic synergy studies employing germ-free models and fecal microbiota transplantation to investigate how diet-exercise-sleep combinations (e.g., high-fiber diets + endurance training) modulate specific microbial functions (e.g., Roseburia-mediated butyrate production). Such approaches should be complemented by cross-validation of sequencing platforms (Illumina vs. Nanopore) and establishment of microbial “responder” thresholds (e.g., >10% Faecalibacterium increase) to enhance reproducibility.
Critical knowledge gaps demanding urgent attention include: (1) Exercise-type specificity- resolving how resistance training preferentially enriches proteolytic Bacteroides versus aerobic exercise-induced Prevotella through microbial lactate metabolism; (2) Population-tailored dynamics - determining optimal lifestyle prescriptions for elderly (probiotics + protein supplementation) versus metabolic syndrome patients (high-fiber diets + moderate exercise) to counteract age- or disease-related dysbiosis; (3) Circadian-microbiota crosstalk - elucidating how microbial metabolites (SCFAs, tryptophan) regulate clock genes in shift workers. As highlighted in Table 3, addressing these priorities through concerted multidisciplinary efforts will enable development of precision microbiota interventions targeting immune-metabolic disorders, bridging the current gap between mechanistic insights and clinical applications. Future studies should particularly focus on longitudinal monitoring of athlete cohorts and high-risk populations (T2DM, elderly) to establish causal timelines for microbial changes and their functional health impacts.
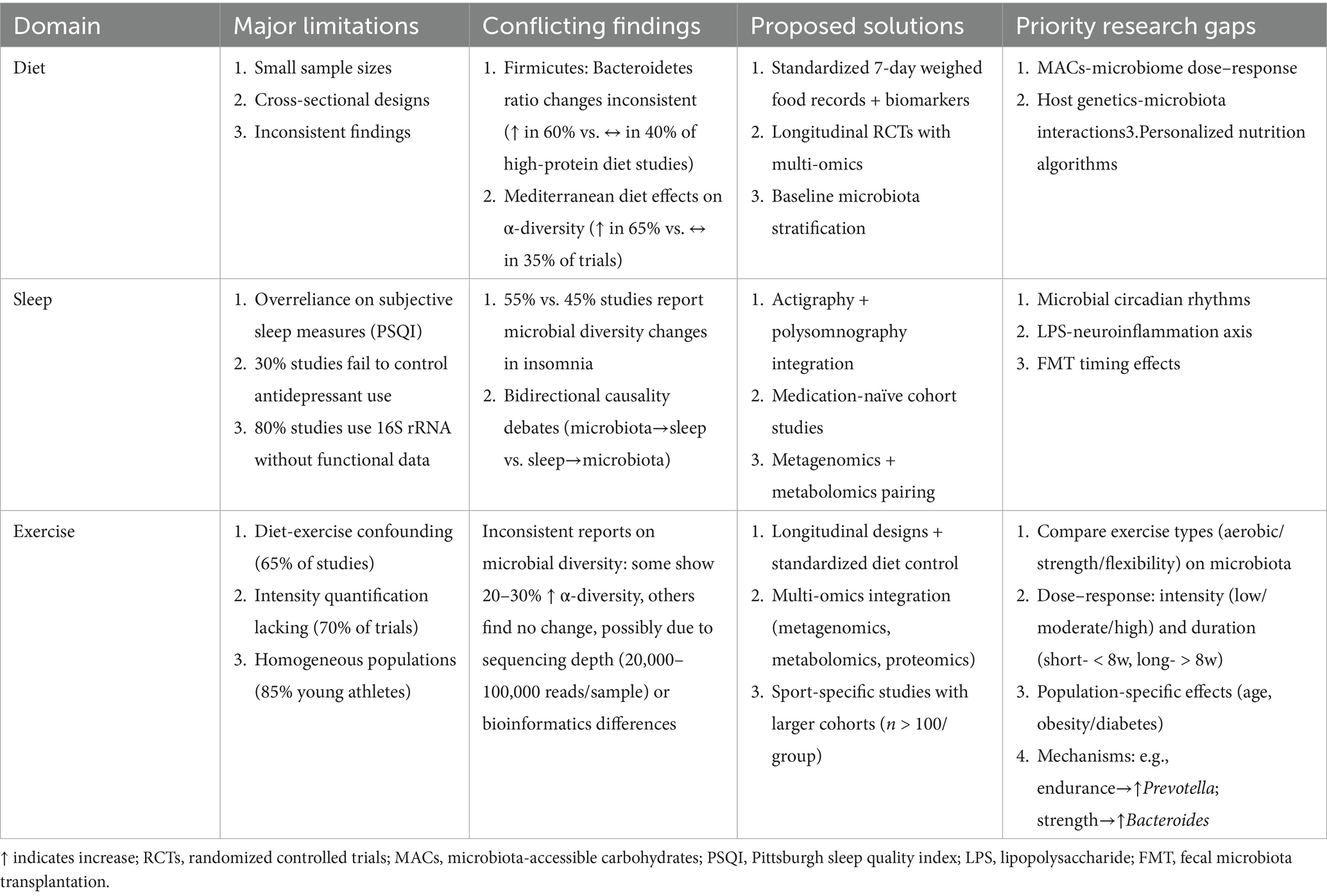
Table 3. Critical appraisal of microbiota research limitations and future directions across lifestyle domains.
5 Summary and translational perspectives
Collectively, the interplay of dietary habits, sleep architecture, and exercise regimens shapes gut microbial ecosystems in distinct yet interconnected ways, ultimately influencing host health outcomes. Having addressed the key research gaps and methodological challenges, this comprehensive synthesis establishes the gut microbiota as a pivotal mediator linking modifiable lifestyle factors (diet, sleep, and exercise) to host physiology and disease susceptibility. Our analysis demonstrates that targeted lifestyle interventions can reshape microbial communities to favor beneficial taxa (e.g., fiber-fermenting Roseburia and Faecalibacterium) while suppressing pro-inflammatory species, with measurable impacts on metabolic, immune, and neurological health outcomes. The accumulated evidence positions microbiota modulation as a promising strategy for chronic disease prevention and management, particularly for conditions like metabolic syndrome, IBD, and neurodegenerative disorders where dysbiosis plays an established pathogenic role.
Moving forward, the field must transition from observational correlations to mechanistic, causal understandings through: (1) standardized multi-omics protocols that resolve functional pathways beyond taxonomic profiling; (2) large-scale longitudinal interventions controlling for key confounders (genetics, medications, circadian rhythms); and (3) personalized approaches accounting for interindividual microbial variability. The integration of these strategies with emerging technologies - including AI-driven microbiota analysis and wearable monitoring devices - will accelerate the development of precision microbiota medicine. These advances promise to transform public health paradigms by enabling evidence-based, microbiota-conscious lifestyle recommendations tailored to individual risk profiles and health statuses.
Author contributions
QZ: Conceptualization, Methodology, Investigation, Writing – original draft, Writing – review & editing. XF: Supervision, Project administration, Writing – review & editing. YH: Data curation, Formal analysis, Visualization, Writing – review & editing. SS: Software, Validation, Resources, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors gratefully acknowledge all researchers whose work was cited in this review.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adak, A., and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Allaband, C., McDonald, D., Vázquez-Baeza, Y., Minich, J. J., Tripathi, A., Brenner, D. A., et al. (2019). Microbiome 101: studying, analyzing, and interpreting gut microbiome data for clinicians. Clin. Gastroenterol. Hepatol. 17, 218–230. doi: 10.1016/j.cgh.2018.09.017
Armet, A. M., Deehan, E. C., O’Sullivan, A. F., Mota, J. F., Field, C. J., Prado, C. M., et al. (2022). Rethinking healthy eating in light of the gut microbiome. Cell Host Microbe 30, 764–785. doi: 10.1016/j.chom.2022.04.016
Arumugam, M., Raes, J., Pelletier, E., Le Paslier, D., Yamada, T., Mende, D. R., et al. (2011). Enterotypes of the human gut microbiome. Nature 473, 174–180. doi: 10.1038/nature09944
Baldi, A., Braat, S., Hasan, M. I., Bennett, C., Barrios, M., Jones, N., et al. (2024). Effects of iron supplements and iron-containing micronutrient powders on the gut microbiome in Bangladeshi infants: a randomized controlled trial. Nat. Commun. 15:8640. doi: 10.1038/s41467-024-53013-x
Barton, W., Penney, N. C., Cronin, O., Garcia-Perez, I., Molloy, M. G., Holmes, E., et al. (2018). The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 67, 625–633. doi: 10.1136/gutjnl-2016-313627
Baunwall, S. M. D., Lee, M. M., Eriksen, M. K., Mullish, B. H., Marchesi, J. R., Dahlerup, J. F., et al. (2020). Faecal microbiota transplantation for recurrent Clostridioides difficile infection: an updated systematic review and meta-analysis. EClinicalMedicine 29-30:100642. doi: 10.1016/j.eclinm.2020.100642
Benjamin, D. I., Both, P., Benjamin, J. S., Nutter, C. W., Tan, J. H., Kang, J., et al. (2022). Fasting induces a highly resilient deep quiescent state in muscle stem cells via ketone body signaling. Cell Metab. 34, 902–918.e6. doi: 10.1016/j.cmet.2022.04.012
Ben-Yacov, O., Godneva, A., Rein, M., Shilo, S., Lotan-Pompan, M., Weinberger, A., et al. (2023). Gut microbiome modulates the effects of a personalised postprandial-targeting (PPT) diet on cardiometabolic markers: a diet intervention in pre-diabetes. Gut 72, 1486–1496. doi: 10.1136/gutjnl-2022-329201
Bi, M., Feng, L., He, J., Liu, C., Wang, Y., Jiang, H., et al. (2022). Emerging insights between gut microbiome dysbiosis and Parkinson’s disease: pathogenic and clinical relevance. Ageing Res. Rev. 82:101759. doi: 10.1016/j.arr.2022.101759
Blommer, J., Pitcher, T., Mustapic, M., Eren, E., Yao, P. J., Vreones, M. P., et al. (2023). Extracellular vesicle biomarkers for cognitive impairment in Parkinson’s disease. Brain 146, 195–208. doi: 10.1093/brain/awac258
Bowers, S. J., Summa, K. C., Thompson, R. S., González, A., Vargas, F., Olker, C., et al. (2022). A prebiotic diet alters the fecal microbiome and improves sleep in response to sleep disruption in rats. Front. Neurosci. 16:889211. doi: 10.3389/fnins.2022.889211
Cani, P. D., Amar, J., Iglesias, M. A., Poggi, M., Knauf, C., Bastelica, D., et al. (2007). Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772. doi: 10.2337/db06-1491
Cheng, W.-Y., Lam, K.-L., Li, X., Kong, A. P.-S., and Cheung, P. C.-K. (2021). Circadian disruption-induced metabolic syndrome in mice is ameliorated by oat β-glucan mediated by gut microbiota. Carbohydr. Polym. 267:118216. doi: 10.1016/j.carbpol.2021.118216
Choi, H., Rao, M. C., and Chang, E. B. (2021). Gut microbiota as a transducer of dietary cues to regulate host circadian rhythms and metabolism. Nat. Rev. Gastroenterol. Hepatol. 18, 679–689. doi: 10.1038/s41575-021-00452-2
Clarke, S. F., Murphy, E. F., O’Sullivan, O., Lucey, A. J., Humphreys, M., Hogan, A., et al. (2014). Exercise and associated dietary extremes impact on gut microbial diversity. Gut 63, 1913–1920. doi: 10.1136/gutjnl-2013-306541
Cryan, J. F., O’Riordan, K. J., Cowan, C. S. M., Sandhu, K. V., Bastiaanssen, T. F. S., Boehme, M., et al. (2019). The microbiota-gut-brain Axis. Physiol. Rev. 99, 1877–2013. doi: 10.1152/physrev.00018.2018
Cuevas-Sierra, A., Chero-Sandoval, L., Higuera-Gómez, A., Vargas, J. A., Martínez-Urbistondo, M., Castejón, R., et al. (2024). Modulatory role of Faecalibacterium on insulin resistance and coagulation in patients with post-viral long haulers depending on adiposity. IScience 27:110450. doi: 10.1016/j.isci.2024.110450
D’Archivio, M., Santangelo, C., Silenzi, A., Scazzocchio, B., Varì, R., and Masella, R. (2022). Dietary EVOO polyphenols and gut microbiota interaction: are there any sex/gender influences? Antioxidants 11:1744. doi: 10.3390/antiox11091744
David, L. A., Maurice, C. F., Carmody, R. N., Gootenberg, D. B., Button, J. E., Wolfe, B. E., et al. (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563. doi: 10.1038/nature12820
De Filippis, F., Pellegrini, N., Vannini, L., Jeffery, I. B., La Storia, A., Laghi, L., et al. (2016). High-level adherence to a Mediterranean diet beneficially impacts the gut microbiota and associated metabolome. Gut 65, 1812–1821. doi: 10.1136/gutjnl-2015-309957
De Filippo, C., Cavalieri, D., Di Paola, M., Ramazzotti, M., Poullet, J. B., Massart, S., et al. (2010). Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 107, 14691–14696. doi: 10.1073/pnas.1005963107
de Wit, N., Derrien, M., Bosch-Vermeulen, H., Oosterink, E., Keshtkar, S., Duval, C., et al. (2012). Saturated fat stimulates obesity and hepatic steatosis and affects gut microbiota composition by an enhanced overflow of dietary fat to the distal intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 303, G589–G599. doi: 10.1152/ajpgi.00488.2011
Deehan, E. C., Yang, C., Perez-Muñoz, M. E., Nguyen, N. K., Cheng, C. C., Triador, L., et al. (2020). Precision microbiome modulation with discrete dietary Fiber structures directs Short-chain fatty acid production. Cell Host Microbe 27, 389–404.e6. doi: 10.1016/j.chom.2020.01.006
Di Vincenzo, F., Del Gaudio, A., Petito, V., Lopetuso, L. R., and Scaldaferri, F. (2024). Gut microbiota, intestinal permeability, and systemic inflammation: a narrative review. Intern. Emerg. Med. 19, 275–293. doi: 10.1007/s11739-023-03374-w
Diacova, T., Cifelli, C. J., Davis, C. D., Holscher, H. D., Kable, M. E., Lampe, J. W., et al. (2025). Best practices and considerations for conducting research on diet-gut microbiome interactions and their impact on health in adult populations: an umbrella review. Adv. Nutr. (Bethesda, Md.) 16:100419. doi: 10.1016/j.advnut.2025.100419
Du, W., Zou, Z.-P., Ye, B.-C., and Zhou, Y. (2025). Gut microbiota and associated metabolites: key players in high-fat diet-induced chronic diseases. Gut Microbes 17:2494703. doi: 10.1080/19490976.2025.2494703
Garcia-Mantrana, I., Selma-Royo, M., Alcantara, C., and Collado, M. C. (2018). Shifts on gut microbiota associated to Mediterranean diet adherence and specific dietary intakes on general adult population. Front. Microbiol. 9:890. doi: 10.3389/fmicb.2018.00890
Goodrich, J. K., Di Rienzi, S. C., Poole, A. C., Koren, O., Walters, W. A., Caporaso, J. G., et al. (2014). Conducting a microbiome study. Cell 158, 250–262. doi: 10.1016/j.cell.2014.06.037
Haskey, N., Estaki, M., Ye, J., Shim, R. K., Singh, S., Dieleman, L. A., et al. (2023). A Mediterranean diet pattern improves intestinal inflammation concomitant with reshaping of the Bacteriome in ulcerative colitis: a randomised controlled trial. J. Crohns Colitis 17, 1569–1578. doi: 10.1093/ecco-jcc/jjad073
Henao-Mejia, J., Elinav, E., Jin, C., Hao, L., Mehal, W. Z., Strowig, T., et al. (2012). Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 482, 179–185. doi: 10.1038/nature10809
Hughes, R. L. (2019). A review of the role of the gut microbiome in personalized sports nutrition. Front. Nutr. 6:191. doi: 10.3389/fnut.2019.00191
Ianiro, G., Punčochář, M., Karcher, N., Porcari, S., Armanini, F., Asnicar, F., et al. (2022). Variability of strain engraftment and predictability of microbiome composition after fecal microbiota transplantation across different diseases. Nat. Med. 28, 1913–1923. doi: 10.1038/s41591-022-01964-3
Imeri, L., and Opp, M. R. (2009). How (and why) the immune system makes us sleep. Nat. Rev. Neurosci. 10, 199–210. doi: 10.1038/nrn2576
Jang, L.-G., Choi, G., Kim, S.-W., Kim, B.-Y., Lee, S., and Park, H. (2019). The combination of sport and sport-specific diet is associated with characteristics of gut microbiota: an observational study. J. Int. Soc. Sports Nutr. 16:21. doi: 10.1186/s12970-019-0290-y
Jansson, P.-A., Curiac, D., Lazou Ahrén, I., Hansson, F., Martinsson Niskanen, T., Sjögren, K., et al. (2019). Probiotic treatment using a mix of three Lactobacillus strains for lumbar spine bone loss in postmenopausal women: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet Rheumatol. 1, e154–e162. doi: 10.1016/S2665-9913(19)30068-2
Jiang, L., Liu, P., Wang, M., Deng, Q., Wang, J., Jiang, Y., et al. (2024). Effect of high-intensity intermittent rehabilitation training on physical function, gut microbiome and metabolite after percutaneous coronary intervention in patients with coronary heart disease. Front. Cardiovasc. Med. 11:1508456. doi: 10.3389/fcvm.2024.1508456
Kasperek, M. C., Mailing, L., Piccolo, B. D., Moody, B., Lan, R., Gao, X., et al. (2023). Exercise training modifies xenometabolites in gut and circulation of lean and obese adults. Physiol. Rep. 11:e15638. doi: 10.14814/phy2.15638
Kim, K. H., Chung, Y., Huh, J.-W., Park, D. J., Cho, Y., Oh, Y., et al. (2022). Gut microbiota of the young ameliorates physical fitness of the aged in mice. Microbiome 10:238. doi: 10.1186/s40168-022-01386-w
Knight, R., Callewaert, C., Marotz, C., Hyde, E. R., Debelius, J. W., McDonald, D., et al. (2017). The microbiome and human biology. Annu. Rev. Genomics Hum. Genet. 18, 65–86. doi: 10.1146/annurev-genom-083115-022438
Kundu, P., Lee, H. U., Garcia-Perez, I., Tay, E. X. Y., Kim, H., Faylon, L. E., et al. (2019). Neurogenesis and prolongevity signaling in young germ-free mice transplanted with the gut microbiota of old mice. Sci. Transl. Med. 11:eaau4760. doi: 10.1126/scitranslmed.aau4760
Li, B., He, Y., Ma, J., Huang, P., Du, J., Cao, L., et al. (2019). Mild cognitive impairment has similar alterations as Alzheimer’s disease in gut microbiota. Alzheimers Dement. 15, 1357–1366. doi: 10.1016/j.jalz.2019.07.002
Li, Y., Zhang, B., Zhou, Y., Wang, D., Liu, X., Li, L., et al. (2020). Gut microbiota changes and their relationship with inflammation in patients with acute and chronic insomnia. Nat. Sci. Sleep 12, 895–905. doi: 10.2147/NSS.S271927
Li, J., Zhao, F., Wang, Y., Chen, J., Tao, J., Tian, G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5:14. doi: 10.1186/s40168-016-0222-x
Luo, S., Zhao, Y., Zhu, S., Liu, L., Cheng, K., Ye, B., et al. (2023). Flavonifractor plautii protects against elevated arterial stiffness. Circ. Res. 132, 167–181. doi: 10.1161/CIRCRESAHA.122.321975
Lynch, S. V., and Pedersen, O. (2016). The human intestinal microbiome in health and disease. N. Engl. J. Med. 375, 2369–2379. doi: 10.1056/NEJMra1600266
Ma, S.-R., Tong, Q., Lin, Y., Pan, L.-B., Fu, J., Peng, R., et al. (2022). Berberine treats atherosclerosis via a vitamine-like effect down-regulating choline-TMA-TMAO production pathway in gut microbiota. Signal Transduct. Target. Ther. 7:207. doi: 10.1038/s41392-022-01027-6
Mailing, L. J., Allen, J. M., Buford, T. W., Fields, C. J., and Woods, J. A. (2019). Exercise and the gut microbiome: a review of the evidence, potential mechanisms, and implications for human health. Exerc. Sport Sci. Rev. 47, 75–85. doi: 10.1249/JES.0000000000000183
Mancin, L., Wu, G. D., and Paoli, A. (2023). Gut microbiota-bile acid-skeletal muscle axis. Trends Microbiol. 31, 254–269. doi: 10.1016/j.tim.2022.10.003
Matenchuk, B. A., Mandhane, P. J., and Kozyrskyj, A. L. (2020). Sleep, circadian rhythm, and gut microbiota. Sleep Med. Rev. 53:101340. doi: 10.1016/j.smrv.2020.101340
Mohr, A. E., Jäger, R., Carpenter, K. C., Kerksick, C. M., Purpura, M., Townsend, J. R., et al. (2020). The athletic gut microbiota. J. Int. Soc. Sports Nutr. 17:24. doi: 10.1186/s12970-020-00353-w
Mondal, T., Chattopadhyay, D., Saha Mondal, P., Das, S., Mondal, A., Das, A., et al. (2025). Fusobacterium nucleatum modulates the Wnt/β-catenin pathway in colorectal cancer development. Int. J. Biol. Macromol. 299:140196. doi: 10.1016/j.ijbiomac.2025.140196
Munoz-Pinto, M. F., Candeias, E., Melo-Marques, I., Esteves, A. R., Maranha, A., Magalhães, J. D., et al. (2024). Gut-first Parkinson’s disease is encoded by gut dysbiome. Mol. Neurodegener. 19:78. doi: 10.1186/s13024-024-00766-0
Münte, E., and Hartmann, P. (2025). The role of Short-chain fatty acids in metabolic dysfunction-associated Steatotic liver disease and other metabolic diseases. Biomol. Ther. 15:469. doi: 10.3390/biom15040469
Muralitharan, R., Zheng, T., Dinakis, E., Xie, L., Barbaro-Wahl, A., Jama, H. A., et al. (2025). Gut microbiota metabolites sensed by host GPR41/43 protect against hypertension. Circ. Res. 136, e20–e33. doi: 10.1161/CIRCRESAHA.124.325770
Murtaza, N., Burke, L. M., Vlahovich, N., Charlesson, B., O’Neill, H. M., Ross, M. L., et al. (2019). Analysis of the effects of dietary pattern on the oral microbiome of elite endurance athletes. Nutrients 11:614. doi: 10.3390/nu11030614
Nagpal, R., Shively, C. A., Appt, S. A., Register, T. C., Michalson, K. T., Vitolins, M. Z., et al. (2018). Gut microbiome composition in non-human primates consuming a western or Mediterranean diet. Front. Nutr. 5:28. doi: 10.3389/fnut.2018.00028
Nieman, D. C., and Wentz, L. M. (2019). The compelling link between physical activity and the body’s defense system. J. Sport Health Sci. 8, 201–217. doi: 10.1016/j.jshs.2018.09.009
Niño, D. F., Zhou, Q., Yamaguchi, Y., Martin, L. Y., Wang, S., Fulton, W. B., et al. (2018). Cognitive impairments induced by necrotizing enterocolitis can be prevented by inhibiting microglial activation in mouse brain. Sci. Transl. Med. 10:eaan0237. doi: 10.1126/scitranslmed.aan0237
O’Donovan, C. M., Madigan, S. M., Garcia-Perez, I., Rankin, A., O’ Sullivan, O., and Cotter, P. D. (2020). Distinct microbiome composition and metabolome exists across subgroups of elite Irish athletes. J. Sci. Med. Sport 23, 63–68. doi: 10.1016/j.jsams.2019.08.290
Ortiz-Alvarez, L., Xu, H., and Martinez-Tellez, B. (2020). Influence of exercise on the human gut microbiota of healthy adults: a systematic review. Clin. Transl. Gastroenterol. 11:e00126. doi: 10.14309/ctg.0000000000000126
Parker, A., Fonseca, S., and Carding, S. R. (2020). Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes 11, 135–157. doi: 10.1080/19490976.2019.1638722
Pedroza Matute, S., and Iyavoo, S. (2023). Exploring the gut microbiota: lifestyle choices, disease associations, and personal genomics. Front. Nutr. 10:1225120. doi: 10.3389/fnut.2023.1225120
Perrone, P., and D’Angelo, S. (2025). Gut microbiota modulation through Mediterranean diet foods: implications for human health. Nutrients 17:948. doi: 10.3390/nu17060948
Petersen, L. M., Bautista, E. J., Nguyen, H., Hanson, B. M., Chen, L., Lek, S. H., et al. (2017). Community characteristics of the gut microbiomes of competitive cyclists. Microbiome 5:98. doi: 10.1186/s40168-017-0320-4
Portincasa, P., Bonfrate, L., Vacca, M., De Angelis, M., Farella, I., Lanza, E., et al. (2022). Gut microbiota and Short chain fatty acids: implications in glucose homeostasis. Int. J. Mol. Sci. 23:1105. doi: 10.3390/ijms23031105
Qin, J., Li, R., Raes, J., Arumugam, M., Burgdorf, K. S., Manichanh, C., et al. (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464, 59–65. doi: 10.1038/nature08821
Rashidah, N. H., Lim, S. M., Neoh, C. F., Majeed, A. B. A., Tan, M. P., Khor, H. M., et al. (2022). Differential gut microbiota and intestinal permeability between frail and healthy older adults: a systematic review. Ageing Res. Rev. 82:101744. doi: 10.1016/j.arr.2022.101744
Reynolds, A. C., Paterson, J. L., Ferguson, S. A., Stanley, D., Wright, K. P., and Dawson, D. (2017). The shift work and health research agenda: considering changes in gut microbiota as a pathway linking shift work, sleep loss and circadian misalignment, and metabolic disease. Sleep Med. Rev. 34, 3–9. doi: 10.1016/j.smrv.2016.06.009
Ross, F. C., Patangia, D., Grimaud, G., Lavelle, A., Dempsey, E. M., Ross, R. P., et al. (2024). The interplay between diet and the gut microbiome: implications for health and disease. Nat. Rev. Microbiol. 22, 671–686. doi: 10.1038/s41579-024-01068-4
Sampson, T. R., Debelius, J. W., Thron, T., Janssen, S., Shastri, G. G., Ilhan, Z. E., et al. (2016). Gut microbiota regulate motor deficits and Neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12. doi: 10.1016/j.cell.2016.11.018
Sánchez-Tapia, M., Miller, A. W., Granados-Portillo, O., Tovar, A. R., and Torres, N. (2020). The development of metabolic endotoxemia is dependent on the type of sweetener and the presence of saturated fat in the diet. Gut Microbes 12:1801301. doi: 10.1080/19490976.2020.1801301
Scheiman, J., Luber, J. M., Chavkin, T. A., MacDonald, T., Tung, A., Pham, L.-D., et al. (2019). Meta-omics analysis of elite athletes identifies a performance-enhancing microbe that functions via lactate metabolism. Nat. Med. 25, 1104–1109. doi: 10.1038/s41591-019-0485-4
Segers, A., and Depoortere, I. (2021). Circadian clocks in the digestive system. Nat. Rev. Gastroenterol. Hepatol. 18, 239–251. doi: 10.1038/s41575-020-00401-5
Sharma, B., Agriantonis, G., Twelker, K., Ebelle, D., Kiernan, S., Siddiqui, M., et al. (2025). Gut microbiota serves as a crucial independent biomarker in inflammatory bowel disease (IBD). Int. J. Mol. Sci. 26:2503. doi: 10.3390/ijms26062503
Smith, R. P., Easson, C., Lyle, S. M., Kapoor, R., Donnelly, C. P., Davidson, E. J., et al. (2019). Gut microbiome diversity is associated with sleep physiology in humans. PLoS One 14:e0222394. doi: 10.1371/journal.pone.0222394
Sonnenburg, J. L., and Bäckhed, F. (2016). Diet-microbiota interactions as moderators of human metabolism. Nature 535, 56–64. doi: 10.1038/nature18846
Teichman, E. M., O’Riordan, K. J., Gahan, C. G. M., Dinan, T. G., and Cryan, J. F. (2020). When rhythms meet the blues: circadian interactions with the microbiota-gut-brain axis. Cell Metab. 31, 448–471. doi: 10.1016/j.cmet.2020.02.008
Thaiss, C. A., Levy, M., Korem, T., Dohnalová, L., Shapiro, H., Jaitin, D. A., et al. (2016). Microbiota diurnal rhythmicity programs host transcriptome oscillations. Cell 167, 1495–1510.e12. doi: 10.1016/j.cell.2016.11.003
Triplett, J., Ellis, D., Braddock, A., Roberts, E., Ingram, K., Perez, E., et al. (2020). Temporal and region-specific effects of sleep fragmentation on gut microbiota and intestinal morphology in Sprague Dawley rats. Gut Microbes 11, 706–720. doi: 10.1080/19490976.2019.1701352
Turnbaugh, P. J., Hamady, M., Yatsunenko, T., Cantarel, B. L., Duncan, A., Ley, R. E., et al. (2009). A core gut microbiome in obese and lean twins. Nature 457, 480–484. doi: 10.1038/nature07540
Wang, Z., Chen, W.-H., Li, S.-X., He, Z.-M., Zhu, W.-L., Ji, Y.-B., et al. (2021). Gut microbiota modulates the inflammatory response and cognitive impairment induced by sleep deprivation. Mol. Psychiatry 26, 6277–6292. doi: 10.1038/s41380-021-01113-1
Wang, X., Jia, R., Chen, W., Zheng, B., and Guo, Z. (2025). Structural characteristics of lotus seed starch-chlorogenic acid complexes during dynamic in vitro digestion and prebiotic activities. Food Chem. 467:142329. doi: 10.1016/j.foodchem.2024.142329
Wilson, A. S., Koller, K. R., Ramaboli, M. C., Nesengani, L. T., Ocvirk, S., Chen, C., et al. (2020). Diet and the human gut microbiome: an international review. Dig. Dis. Sci. 65, 723–740. doi: 10.1007/s10620-020-06112-w
Wu, J., Wang, K., Wang, X., Pang, Y., and Jiang, C. (2021). The role of the gut microbiome and its metabolites in metabolic diseases. Protein Cell 12, 360–373. doi: 10.1007/s13238-020-00814-7
Yan, X., Jin, J., Su, X., Yin, X., Gao, J., Wang, X., et al. (2020). Intestinal Flora modulates blood pressure by regulating the synthesis of intestinal-derived corticosterone in high salt-induced hypertension. Circ. Res. 126, 839–853. doi: 10.1161/CIRCRESAHA.119.316394
Zeevi, D., Korem, T., Zmora, N., Israeli, D., Rothschild, D., Weinberger, A., et al. (2015). Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094. doi: 10.1016/j.cell.2015.11.001
Zeng, X., Xing, X., Gupta, M., Keber, F. C., Lopez, J. G., Lee, Y.-C. J., et al. (2022). Gut bacterial nutrient preferences quantified in vivo. Cell 185, 3441–3456.e19. doi: 10.1016/j.cell.2022.07.020
Zhao, X., Jia, S., Zhao, H., Liu, P., Wu, Z., Tao, H., et al. (2025). The interaction between maize resistant starch III and Bifidobacterium adolescentis during in vitro fermentation. Food Chem. 463:140968. doi: 10.1016/j.foodchem.2024.140968
Zhao, M., Lu, C., Hu, X., and Ma, Z. (2024). Evolution of multi-scale structure and microbiota metabolism of lentil resistant starch during the dynamic fermentation in vitro. Food Chem. 461:140914. doi: 10.1016/j.foodchem.2024.140914
Zheng, D., Liwinski, T., and Elinav, E. (2020). Interaction between microbiota and immunity in health and disease. Cell Res. 30, 492–506. doi: 10.1038/s41422-020-0332-7
Keywords: gut microbiome, precision nutrition, circadian biology, exercise immunology, metabolic health
Citation: Zeng Q, Feng X, Hu Y and Su S (2025) The human gut microbiota is associated with host lifestyle: a comprehensive narrative review. Front. Microbiol. 16:1549160. doi: 10.3389/fmicb.2025.1549160
Edited by:
Guiguo Zhang, Shandong Agricultural University, ChinaReviewed by:
Catherine M. T. Sherwin, University of Western Australia, AustraliaHao Tian, Chengdu University of Traditional Chinese Medicine, China
Copyright © 2025 Zeng, Feng, Hu and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianqiong Feng, ZmVuZ3hpYW5xaW9uZzY2QDEyNi5jb20=
 Qin Zeng
Qin Zeng Xianqiong Feng
Xianqiong Feng Yanling Hu1,2
Yanling Hu1,2