- 1Department of Pediatrics, Shengjing Hospital of China Medical University, Shenyang, China
- 2School of Clinical Medicine, Qilu Medical University, Zibo, China
Pediatric asthma is a common chronic airway inflammatory disease that begins in childhood and its impact persists throughout all age stages of patients. With the continuous progress of detection technologies, numerous studies have firmly demonstrated that gut microbiota and respiratory microbiota are closely related to the occurrence and development of asthma, and related research is increasing day by day. This article elaborates in detail on the characteristics, composition of normal gut microbiota and lung microbiota at different ages and in different sites, as well as the connection of the gut—lung axis. Subsequently, it deeply analyzes various factors influencing microbiota colonization, including host factor, delivery mode, maternal dietary and infant feeding patterns, environmental microbial exposure and pollutants, and the use of antibiotics in early life. These factors are highly likely to play a crucial role in the onset process and disease progression of asthma. Research shows that obvious changes have occurred in the respiratory and gut microbiota of asthma patients, and these microbiomes exhibit different characteristics according to the phenotypes and endotypes of asthma. Finally, the article summarizes the microbiota—related treatment approaches for asthma carried out in recent years, including the application of probiotics, nutritional interventions, and fecal microbiota transplantation. These treatment modalities are expected to become new directions for future asthma treatment and bring new hope for solving the problem of childhood asthma.
1 Introduction
Bronchial asthma, a prevalent chronic inflammatory airway disorder in the pediatric population (Asher et al., 2021), emerges as a significant contributor to public health challenges, manifesting through elevated school absenteeism rates, increased emergency department utilization, and higher hospitalization frequencies (Naja et al., 2018).
The microbiota constitutes a complex and dynamic community of bacteria, viruses, and fungi, archaea, and other microorganisms, encompassing both commensal and pathogenic species, that colonize the surfaces or internal environments of hosts (humans, animals, plants) or external ecosystems. The microbiome primarily refers to the genomic repertoire of the microbiota, though these terms are frequently used interchangeably (Berg et al., 2020). Characterized by individual specificity, dynamic variability, and niche heterogeneity (Cho and Blaser, 2012), the human microbiota rapidly establishes colonization in the gastrointestinal tract, respiratory system, and other anatomical microenvironments after birth, with its composition continuously shaped by host factors and environmental exposures. A growing consensus emphasizes that these microbial communities and their metabolites participate in critical host biological processes, including maintaining immune homeostasis and modulating metabolic pathways, which promote health under homeostatic conditions while driving disease pathogenesis during dysbiosis. Notably, microbiota dysbiosis, defined as an imbalance between beneficial and pathogenic species or aberrant abundance of specific commensals, demonstrates significant associations with pulmonary disorders, including asthma (Barcik et al., 2020), bronchopulmonary dysplasia (Pammi et al., 2019), cystic fibrosis (Françoise and Héry-Arnaud, 2020) and COVID-19 (Ancona et al., 2023).
In recent times, with the remarkable advancements in microbiome technology, our comprehension of the relationship between the human microbiota and health—disease states has been continuously enhanced (Escobar-Zepeda et al., 2015; Davidson and Epperson, 2018). The gut microbiota and respiratory microbiota, being crucial constituents of the human microbiota (Clavijo-Salomon and Trinchieri, 2025), play a pivotal role in maintaining the body’s immune equilibrium and fending off pathogen incursions. A growing body of evidence suggests that the dysbiosis of gut microbiota and respiratory microbiota might be a key potential factor precipitating childhood asthma (Thorsen et al., 2023; Henrick et al., 2021). Through the gut—lung axis (Budden et al., 2017), the metabolites and immunomodulatory signals of gut microbes can influence the immune status and inflammatory responses within the respiratory tract. Nevertheless, our current knowledge regarding the characteristics of microbiota at diverse age phases and in different anatomical sites, along with their precise mechanisms of action in the initiation and progression of asthma, still requires substantial improvement. In—depth investigations into the associations among gut microbiota, respiratory microbiota, and childhood asthma not only contribute to unravelling the pathogenesis of asthma but also hold the promise of opening up novel avenues for the prevention and treatment of childhood asthma. Consequently, a systematic review of the research progress in this domain bears significant theoretical implications and clinical application value.
2 Healthy gut and respiratory microbiota
The establishment of the normal microbiota is intricately linked to the development of various systems in children and the onset and progression of diseases, particularly respiratory allergic disorders. The gut microbiota and respiratory microbiota vary significantly with age and across different distribution sites. The gut—lung axis likely exerts a subtle and far—reaching influence on the body through mechanisms such as metabolism and immune circulation. Moreover, diverse microbiota, including bacteria, fungi, and viruses, interact with one another to jointly maintain a healthy micro—ecological environment.
2.1 Healthy gut microbiota
The human gut inhabits ~1014 bacteria (Bäckhed et al., 2005), as the body’s most densely colonized site (Sender et al., 2016). The dynamic development of the gut microbiota constitutes a central biological determinant in early-life programming, with its interaction mechanisms through the gut-lung axis with distal organs (e.g., pulmonary system) emerging as a critical frontier in host immunometabolic regulation. Following birth, the intestinal mucosal surface undergoes rapid microbial colonization (Leiby et al., 2018), establishing itself as a primary site for microbiota development. Neonatal gut flora, dominated by Enterococcus, Escherichia, Streptococcus, and Rothia (Actinobacteria), reflects an aerobic environment. By 2–4 months, colonization shifts to Enterobacteriaceae, Bifidobacteriaceae, and Clostridiaceae, signaling oxygen reduction and lactic acid metabolism. These taxa gradually decline until 18 months under the “healthy microbiota maturation” framework (Ximenez and Torres, 2017), paralleling increased microbial diversity observed in neonatal fecal analyses (Casaburi et al., 2021). This dynamic developmental progression drives the establishment of unique gut microbial profiles during infancy. During this period, the gut microbiota exhibits lower microbial diversity, structural instability, and marked inter-individual variability. By approximately three years of age, this developmental trajectory transitions toward stabilization, converging with adult-like microbial composition while establishing a unique gut ecosystem characterized by host-specific features (Koenig et al., 2011; Solís et al., 2010). In humans, fecal microbiota maturation occurs predominantly within the first several years of life, marked by the dominance of Firmicutes and Bacteroidetes phyla typically observed by 3 years of age (Teo et al., 2018; Yatsunenko et al., 2012). Emerging evidence suggests early microbial interactions may begin prenatally: placental bacteria in some preterm infants share origins with maternal oral microbiota (Ye et al., 2020), implying fetal-microbe crosstalk during embryogenesis (Casaburi et al., 2021).
Although existing research has predominantly focused on bacterial components of the gut microbiome, it is crucial to recognize that the viral component also constitutes a significant member of this microbial community. During the initial months of life, bacteriophages, akin to bacteria, exhibit host-specific distribution within the gut and play a pivotal role in regulating bacterial growth through mechanisms such as lysis and lysogeny (Shah et al., 2023). In 2008, a groundbreaking study by Breitbart et al. (2008) on the infant gut virome first revealed that the intestinal viral community of neonates at one-week postpartum displays remarkably low biodiversity, with bacteriophages dominating the composition. This specific distribution pattern may regulate the population structure and quantitative dynamics of symbiotic microbiota through niche competition. Recent research, leveraging large-scale viral metagenomic analyses of fecal samples from 1-year-old infants, identified approximately 10,000 viral genomes spanning 248 viral family-level clades (VFCs). Notably, over half of these VFCs represent previously undocumented taxonomic units, predominantly classified under the Caudoviricetes class (Shah et al., 2023). Nevertheless, significant gaps persist in understanding the assembly mechanisms of viral communities during critical developmental stages in early life. Combined with the limited coverage of viral genome databases, these challenges substantially hinder efforts to decode the organizational principles of the human virome.
2.2 The gut-lung axis
The gut-lung axis refers to a bidirectional communication network between the gut and the lungs (Budden et al., 2017), mediated by microbiota, metabolites, immune signals, and neuroendocrine pathways. The gut and lung microbiota collectively influence respiratory health and diseases [such as asthma (Kim et al., 2021) and respiratory infections (Zhao et al., 2023) by regulating the host immune system, metabolic pathways, and inflammatory responses].
This intricate cross-talk is mechanistically underpinned by specific microbial-derived components and host signaling molecules that mediate bidirectional organ interactions. Imbalanced gut bacteria (e.g., low diversity or harmful overgrowth) disrupt lung immunity via blood or lymph, while lung microbes affect inflammation through immune cells like dendritic cells. Gut bacteria also boost regulatory T cells (Tregs) (Arpaia et al., 2013) and reduce harmful Th2/Th17 inflammation (Atarashi et al., 2015). Key metabolites (e.g., SCFAs) balance immune responses by targeting receptors or enzymes [e.g., HDAC (Wang N. et al., 2024)], linking gut health to lung diseases like asthma.
2.3 Healthy respiratory microbiota
Conventional microbiological dogma long maintained the sterility of pulmonary environments (lower respiratory tract), until paradigm-shifting molecular analyses circa 2010 (Hilty et al., 2010) fundamentally redefined our understanding of airway microbiota composition propelling respiratory microbiome research into scientific prominence. Building upon these revelations, this section critically examines the dynamic colonization mechanisms and signature microbial profiles within healthy respiratory ecosystems, including bacteria (bacteriome), virus (virome) and fungi (mycobiome).
2.3.1 Bacteria (bacteriome)
Respiratory bacterial colonization exhibits spatiotemporal heterogeneity across anatomical niches and age strata. Oropharyngeal specimens (via oral rinse sampling) demonstrate microbial predominance of Prevotella, Veillonella, and Streptococcus genera (Dickson et al., 2017). In contrast to the taxonomically rich ecosystems of gut and oropharyngeal microbiota, the lower respiratory tract (pulmonary microbiome) maintains reduced microbial biomass while exhibiting marked ecological heterogeneity. Developmental analyses reveal neonatal pulmonary colonization initiates with Staphylococcus or Corynebacterium predominance, undergoing successional displacement by Alloiococcus or Moraxella genera during microbiota maturation (Teo et al., 2015). The stabilized pulmonary microbiome in healthy individuals is characterized by tripartite dominance of the following bacterial phyla: Firmicutes (genus Streptococcus), Proteobacteria (genus Acinetobacter), and Actinobacteria (genus Corynebacterium) (Bassis et al., 2015; Segal et al., 2013; Venkataraman et al., 2015), with core genera (Streptococcus, Veillonella, Prevotella) maintaining trans-anatomical equilibrium (Dickson et al., 2017). Dolosigranulum pigrum was more abundant in younger individuals while remaining present across all age groups. In contrast, the nasopharyngeal microbiota of adolescents and adults (≥15 years) was characterized by a consortium of less common taxa, including Anaerococcus (octavius), Corynebacterium, Finegoldia magna, Lawsonella clevelandensis, and Peptoniphilus (Odendaal et al., 2024).
2.3.2 Virus (virome)
The groundbreaking advances in NGS have enabled scientists to systematically unravel the complex ecology of the human lung virome. Studies reveal distinct characteristics of respiratory viral composition in healthy populations: viral diversity is markedly reduced (Willner et al., 2009), particularly in pediatric cohorts where Anapoviridae dominates, with minimal presence of human herpesviruses. In healthy lung tissues, this viral family persists as the core eukaryotic virome, occasionally coexisting with herpesviruses, papillomaviruses, and retroviruses (Willner et al., 2009). Intriguingly, latent viral phases may confer protective benefits to hosts—by persistently stimulating interferon-gamma (IFN-γ) secretion and activating macrophages, thereby establishing a foundational immune defense barrier (MacDuff et al., 2015; Sun et al., 2015). This mechanism is experimentally validated: mice latently infected with murine herpesvirus exhibit enhanced resistance to Listeria monocytogenes infections (Barton et al., 2007). Notably, emerging evidence identifies a rich phage community within respiratory surfaces, hypothesized to comprise 19 core species (Willner et al., 2009; Lim et al., 2013). These phages orchestrate microbial equilibrium through dual strategies—precisely eliminating competitor bacterial strains via prophage release while dynamically regulating proliferation balance among niche-sharing bacteria, revealing intricate cross-kingdom interactions. Future investigations must elucidate the mechanisms underlying viral interactions with other pulmonary microbes, which promise to unlock novel therapeutic avenues for respiratory disease prevention and treatment.
2.3.3 Fungi (mycobiome)
Current mycological research primarily employs targeted sequencing approaches, including analysis of the internal transcribed spacer (ITS) region and 18S rRNA genes, complemented by shotgun metagenomic sequencing (Carney et al., 2020). Despite growing research interest in this field (Cui et al., 2013; Huffnagle and Noverr, 2013), pulmonary fungal studies face persistent technical challenges: extremely low biomass, limited taxonomic diversity, inefficient DNA extraction, 18S rRNA amplification bias, and inconsistent nomenclature standards, collectively hampering accurate fungal database annotation (Iliev et al., 2012; Marsland and Gollwitzer, 2014; Carmody et al., 2013). Compared to the well-characterized bacteriome, the functional significance of fungi in pulmonary ecosystems remains underexplored.
Emerging evidence reveals distinct diversity patterns in healthy pulmonary mycobiomes that significantly differ from pathological states (Fodor et al., 2012; Lim et al., 2014; Delhaes et al., 2012). Core fungal communities are dominated by Ascomycetes (phylum level) and Streptomyces (genus level), followed by Candida, Saccharomyces, Penicillium, Dictyostelium, and Fusarium (Huang et al., 2020; Sharma et al., 2019), with Candida demonstrating the highest relative abundance (Delhaes et al., 2012; Charlson et al., 2012; Ghannoum et al., 2010). Additional commensal species include Aspergillus, Davidiellaceae (family level), and Eurotium (Charlson et al., 2012). Notably, cross-kingdom interactions between fungi and bacteria exhibit regulatory mechanisms: Candida shows positive correlation with Lactobacillus but negative association with Helicobacter pylori. Experimental evidence confirms that Lactobacillus suppresses the epithelial adhesion capacity of both H. pylori and Candida albicans, thereby modulating their colonization dynamics (Dohlman et al., 2022). Previous studies have systematically delineated the colonization dynamics and characteristic microbiota composition of the gut and respiratory tract during early life, demonstrating the pivotal role of these symbiotic microorganisms in maintaining homeostasis for proper immune system development. Importantly, emerging evidence suggests that disruption of this delicate microbial equilibrium by various factors—including prenatal, perinatal, and postnatal influences—may compromise immune regulatory pathways, thereby substantially elevating the susceptibility to childhood asthma.
3 The factors that trigger changes in the microflora and trace asthma origin
The colonization process and change in species composition of respiratory and gut microbiome in early life are significantly associated with asthma susceptibility. This dynamic process is jointly regulated by the host’s age and multiple factors spanning prenatal to postnatal stages, including biological elements such as delivery mode, maternal dietary and infant feeding patterns; ecological components like environmental microbial exposure and pollutants; as well as critical regulatory factors including early life antibiotic use. These multifaceted influences collectively shape microbial colonization and immune development, ultimately affecting asthma risk.
3.1 Host
Recent studies have revealed significant variations in the composition of respiratory microbiota based on the host’s age (gestational age). Comparative studies have demonstrated gestational age-dependent variations in microbial community composition, revealing developmental-stage-specific differences (Bargheet et al., 2023). Extremely preterm infants exhibit delayed gut microbiota maturation with higher abundances of potentially pathogenic bacteria like Escherichia coli and Staphylococcus epidermidis compared to full-term infants, who are dominated by beneficial Bifidobacterium and Bacteroides species. Very preterm infants show intermediate microbiota profiles, characterized by reduced diversity and delayed colonization of symbiotic bacteria, influenced by gestational age. The nasopharynx of younger individuals is dominated by Dolosigranulum pigrum (D. pigrum), a species universally present across all age groups but exhibiting significantly higher abundance in pre-adolescent populations.
3.2 Delivery mode
The mode of delivery significantly influences the early colonization and developmental trajectory of both respiratory and gut microbiota in infants. Systematic reviews have demonstrated that vaginally delivered newborns exhibit significantly higher gut abundance of Actinobacteria, Bacteroides, and Bifidobacterium compared to cesarean-delivered counterparts (Rutayisire et al., 2016). Recent studies further reveal distinct gut microbial profiles at one week of age: cesarean-born infants predominantly harbor Citrobacter freundii, Clostridium spp., and Staphylococcus aureus, whereas vaginal delivery promotes preferential colonization by Escherichia coli (Stokholm et al., 2020).
The mode of delivery also influences the types of respiratory microbiota. In the respiratory tract, vaginally delivered neonates display early colonization with Corynebacterium and Dolosigranulum pigrum. The sustained presence of these commensal bacteria is associated with enhanced microbial maturity and a marked reduction in long-term asthma risk (Chen et al., 2023). Longitudinal studies further show that vaginally born infants maintain higher abundance of Bacteroides by one year of age, with its deficiency being strongly correlated with asthma susceptibility and delayed microbial development (Chen et al., 2023). Conversely, cesarean-delivered infants exhibit characteristic respiratory microbiota delays: early enrichment of Gemella and Streptococcus, followed by aberrant proliferation of oral bacterial genera such as Neisseria and Prevotella (Bosch et al., 2017). This dysbiotic profile, coupled with reduced colonization of health-associated commensals (e.g., Corynebacterium), may increase susceptibility to respiratory diseases through immune dysregulation (Bosch et al., 2016). Although the impact of delivery mode on respiratory microbiota is less pronounced than on gut microbiota (Chu et al., 2017; Biesbroek et al., 2014), their interplay remains clinically significant. For instance, early asymptomatic colonization of the respiratory tract with Streptococcus during infancy has been identified as a strong predictor of asthma development, likely mediated by disrupted immune education through host-microbial interactions in early life (Teo et al., 2015).
3.3 Maternal dietary and infant feeding patterns
Maternal dietary patterns during pregnancy, combined with postnatal feeding strategies, collectively shape offspring microbiota development trajectories, thereby influencing the risk of asthma and allergic diseases. A nested cross-sectional study (the MAMI cohort) demonstrated that maternal dietary patterns during pregnancy significantly influence neonatal gut microbiota development. Specifically, high intake of saturated fatty acids (SFAs) and monounsaturated fatty acids (MUFAs) leads to an abnormal enrichment of Firmicutes in the infant gut. This microbial dysbiosis is negatively associated with high consumption of fiber, proteins from vegetable sources, and vitamins during pregnancy (Selma-Royo et al., 2021). Arpaia et al. further support the protective role of dietary fiber, which modulates the Firmicutes: Bacteroidetes ratio to influences allergic airway disease (Arpaia et al., 2013). These findings underscore the importance of optimizing maternal fatty acid intake and increasing fiber-rich foods during pregnancy to mitigate asthma risk.
Infant feeding patterns (postnatal nutrition) also plays a critical role in shaping microbial trajectories that influence disease susceptibility. Breastfeeding promotes the early colonization of beneficial respiratory commensals (e.g., Corynebacterium and Dolosigranulum pigrum) and enhances gut Bifidobacterium abundance (Chen et al., 2023; Bosch et al., 2017). In contrast, formula feeding is associated with enrichment of Gemella, Streptococcus in the infant respiratory microbiota and oral-type anaerobic bacteria such as Prevotella and Neisseria species, disrupting microbial stability (Bosch et al., 2017). Moreover, breast milk-derived hereditary microbes may further protect against pediatric asthma by enhancing intestinal immune tolerance (Fang et al., 2024).
3.4 Environmental microbial exposure and pollutants
Epidemiological evidence demonstrates that residential microbial exposure critically modulates pediatric asthma risk, with cohort studies revealing an inverse correlation between indoor microbial diversity and asthma incidence across distinct living environments such as traditional farms versus urban communities (Sun et al., 2023; Ege et al., 2011). This protective effect may stem from dynamic microbial colonization during immune system maturation, where gut microbiota play a pivotal role in immune education (Abt et al., 2012). However, environmental pollutants can disrupt this balance. For instance, PM2.5 alters gut microbiota composition by increasing Bacteroidetes and decreasing Firmicutes, a shift mechanistically linked to asthma development (Zhao et al., 2023). Animal studies further demonstrate that chronic PM2.5 exposure induces persistent gut-lung microbiota dysbiosis, contributing to late-onset asthma progression (Zhao et al., 2023). Similarly, prenatal and postnatal tobacco smoke exposure elevates Enterobacteriaceae abundance, exacerbating respiratory symptom risks in infancy (Vardavas et al., 2016). Notably, urban microbial diversity may counteract allergic risks. High-allergen environments combined with indoor dust microbial richness—particularly the presence of protective taxa such as Prevotellaceae and Lachnospiraceae—attenuate wheezing, suggesting a potential buffering effect against asthma-related outcomes (Lynch et al., 2014). Conversely, crowding conditions, including the presence of young siblings or daycare attendance, reduce respiratory microbiota stability and promote Pasteurellaceae (e.g., Haemophilus) dominance (Bosch et al., 2017). These findings collectively highlight the delicate balance between protective microbial exposures and pollutant-driven dysbiosis in shaping asthma pathogenesis.
3.5 Early life antibiotic use
Recent studies have revealed that early-life antibiotic exposure profoundly influences asthma development by altering microbiome composition. The widespread use of antibiotics during pregnancy and infancy not only promotes the emergence of multidrug-resistant pathogens (Alfaqawi et al., 2021), complicating asthma treatment, but also directly disrupts the dynamic balance of respiratory and gut microbiota in affected individuals. Clinical evidence indicates that antibiotics significantly deplete key commensal bacteria in infant airways, such as Corynebacterium and Alloiococcus, leading to reduced microbial community stability (Bosch et al., 2017). This microbial dysbiosis extends to the gut, where antibiotic intervention promotes abnormal yeast proliferation, thereby exacerbating pulmonary allergic responses (Noverr et al., 2004). Animal studies provide direct evidence for the impact of early-life antibiotic exposure on asthma pathogenesis. Neonatal mice exposed to azithromycin or amoxicillin exhibited marked reductions in the diversity of core gut microbial taxa, including Lachnospiraceae and Muribaculaceae (Borbet et al., 2022). Such microbial disturbances have long-term consequences—when these mice were later exposed to house dust mite allergens, they displayed elevated IgE and IL-13 levels (hallmark biomarkers of allergic asthma), alongside hyperactivation of Th2/Th17 immune pathways and significantly enhanced airway hyperreactivity. Notably, microbiota transplantation experiments demonstrated that offspring of germ-free mice colonized with gut microbiota from antibiotic-exposed mice developed hyperactive immune responses and asthma-like symptoms despite no direct antibiotic exposure, suggesting that early-life microbial alterations can program immune development through transgenerational transmission. Animal studies have shown that antibiotic use not only causes intestinal bacterial dysbiosis increasing asthma risk, but also induces intestinal fungal dysbiosis. Kim et al. found that combined antibiotic use leads to overgrowth of the commensal fungus Candida in the gut. Candida promotes M2 macrophage polarization in the lungs by elevating plasma prostaglandin E2 (PGE2) levels, thereby increasing airway inflammatory cell infiltration and exacerbating tissue pathological changes (Kim et al., 2014).
4 Microbial alterations in pediatric asthma
Pediatric asthma demonstrates distinct gut-respiratory microbiota compositional differences compared to healthy controls. In this section, we review airway and gut microbial alterations in pediatric asthma, and microbial changes associated with asthma phenotypes and endotypes.
4.1 Intestinal microbita in pediatric asthma
The gut’s dysbiosis may disrupt immune homeostasis, and specific microbial shifts correlate with asthma development. Multicenter cohort studies revealed elevated Prevotella (P. bivia, P. disiens, P. oris) and Bacteroides fragilis colonization across oral-gut ecosystems, coupled with reduced Streptococcus thermophilus levels (Yan et al., 2024). The dynamic changes in gut microbiota during early life are closely associated with the progression of asthma (Rautava and Walker, 2009; Simonyte Sjödin et al., 2016). The Copenhagen Prospective Studies on Asthma in Childhood (COPSAC) (Stokholm et al., 2018) demonstrated that early-life gut microbial alterations in 1-year-old infants born to asthmatic mothers, characterized by dysregulated relative abundances of Veillonella, Lachnospiraceae, Bifidobacterium, and Ruminococcus, served as significant predictors of asthma development by age 5. The Canadian Healthy Infant Longitudinal Development (CHILD) Study (Arrieta et al., 2015) further identified the first 3 months of life as a critical window for gut microbiome-host interactions during which abnormal perturbations in gut microbiota structure exert profound long-term effects on airway health. Animal experiments using germ-free mice inoculated with stool from infants with atopic wheeze showed that supplementation with these depleted bacterial taxa reduced lung inflammation, elevated SCFA levels, and lowered proinflammatory cytokines in offspring (Cait et al., 2018; Trompette et al., 2014). SCFAs—produced through bacterial fermentation of dietary fiber —promote Treg differentiation (Roduit et al., 2019) via histone deacetylases (HDAC) inhibition, suppress M2 macrophage activation, and exert transgenerational protective effects (Calışkan et al., 2013; Wang et al., 2023) when supplemented during pregnancy (Arpaia et al., 2013; Trompette et al., 2014; Roduit et al., 2019; Calışkan et al., 2013; Huang et al., 2022). These findings collectively highlight the protective role of specific gut microbiota and their metabolites (e.g., SCFAs) in asthma pathogenesis.
Compared to the well-established gut microbiota-asthma research, studies on gut viruses and fungi are both fewer in number and more preliminary in nature. The virome comprises a small proportion of the overall gut microbiome. Recent study revealed associations between the infant gut virome composition and the risk of developing asthma (Leal Rodríguez et al., 2024). Bacteriophages, also known as phages, are viruses that infect and replicate within bacterial cells and are important determinants due to their ability to infect other bacteria, while they serve as mediators between pathogenic and nonpathogenic bacteria (Sweere et al., 2019). Specific temperate bacteriophage taxa, particularly 19 caudoviral families, were found to contribute to asthma risk. Children who later developed asthma exhibited lower relative abundances of these temperate phage families, which predominantly infect bacterial genera such as Faecalibacterium and Ruminococcus. Intriguingly, the virome-asthma association was independent of bacterial communities, with additive effects observed when combining virome and bacteriome signatures. The study also identified a potential interaction between the virome and the host immune system via the TLR9 rs187084 genetic variant, suggesting phage DNA may directly modulate immune responses. This study represents one of the few published investigations focusing on the intestinal virome in pediatric asthma pathogenesis. Although this is an observational study, its findings provide evidence for regulating gut phages through phage therapy or perinatal intervention and restoring the balanced temperate virome required for immune maturation as a novel preventive strategy for asthma. Future research should explore the diversity of gut viruses and fungi, their microbial interactions, and the underlying mechanisms in asthma development.
4.2 Respiratory microbiota in pediatric asthma
Significant differences exist in respiratory microbial composition between healthy children and asthma patients. In asthmatic children exhibit nasal microbiota enriched with Moraxella species [e.g., Moraxella catarrhalis (Raita et al., 2021)], whose relative abundance positively correlates with increased asthma exacerbation frequency (Zhou et al., 2019; McCauley et al., 2019; Durack et al., 2018). Conversely, enrichment of Corynebacterium and Dolosigranulum is associated with improved asthma control, reduced BAL eosinophil percentages, and decreased levels of pro-inflammatory factors such as IL-17 and IL-121 (McCauley et al., 2019; Durack et al., 2018). This microbial dysbiosis interacts closely with host immune responses: rhinovirus (RV) infection combined with colonization by M. catarrhalis or Streptococcus pneumoniae synergistically exacerbates asthma symptoms (Kloepfer et al., 2014), while early-life (1-month-old) upper respiratory colonization with S. pneumoniae, Haemophilus influenzae, or Moraxella significantly elevates asthma risk by age 5 (Thorburn et al., 2015). This effect exhibits atopy dependency—children with early atopic sensitization are more prone to developing “persistent wheeze” (Teo et al., 2018). Longitudinal studies demonstrate that seasonal fluctuations in nasal microbiota linked to virus-induced asthma exacerbations are further associated with the RV-C endotype, characterized by Moraxella-dominant communities, which significantly elevates recurrent wheeze risk (Raita et al., 2021; McCauley et al., 2022). In the lower respiratory tract, protective associations are observed in healthy children, where bronchial Actinomyces and nasal Corynebacterium negatively correlate with pro-inflammatory gene expression (Chun et al., 2020).
The respiratory virome also plays a critical role in asthma pathogenesis. Extensive research has demonstrated a strong association between pediatric asthma and respiratory viruses, with respiratory syncytial virus (RSV) and RV identified as key pathogenic drivers of asthma exacerbations (Korten et al., 2016; Sigurs et al., 2000; Rosas-Salazar et al., 2023). A prospective cohort study of 1,946 healthy term infants in the United States revealed that children uninfected with respiratory syncytial virus (RSV) during infancy had a lower incidence of asthma at age 5 compared to the infected group, and preventing RSV infection could reduce approximately 15% of pediatric asthma cases (Rosas-Salazar et al., 2023). RV is the most frequently detected pathogen in wheezing children over 1 year old, and its early-life infections (particularly repeated detection within the first 3 postnatal weeks) increase pre-2-year wheezing risk by 16% (Takashima et al., 2023). Lower respiratory RV infections within the first 3 years of life elevate asthma risk by 40-fold before age 6, far exceeding the effects of RSV infection (Jackson et al., 2008; Kusel et al., 2007; Psarras et al., 2006). Mechanistically, RV promotes airway remodeling by inducing growth factors (e.g., lumican, collagen I/V) and prolongs bronchial hyperreactivity via non-Th2-IFN pathways, with impaired Th1/IL-10 responses exacerbating this pathology in atopic individuals (Psarras et al., 2006; Skevaki et al., 2012; Spector et al., 2023; Xepapadaki et al., 2005; Megremis et al., 2018; Georgountzou et al., 2021). Additionally, asthmatic children display upper respiratory virome dysbiosis dominated by eukaryotic viruses and low bacteriophage abundance, while diminished antiviral cytokine responses correlate with high RV loads (Megremis et al., 2023; Rovira Rubió et al., 2023).
In fungal communities, the upper respiratory tract of asthmatic children is dominated by Malassezia globosa and Malassezia restricta. High baseline M. globosa abundance delays asthma control loss, whereas its reduced abundance during exacerbation correlates with heightened severe attack risk. Transition from well-controlled to uncontrolled asthma involves synchronized increases in fungal and bacterial diversity, suggesting fungal-bacterial interactions in disease progression (Yuan et al., 2023).
These findings highlight the multi-layered role of respiratory microbiota in asthma pathogenesis: bacterial dysbiosis modulates local immunity, viral colonization drives direct epithelial damage and immune remodeling, and fungal-bacterial crosstalk potentially shapes asthma phenotypes. These microbial signatures offer promising biomarkers for early prediction and targeted interventions in asthma management.
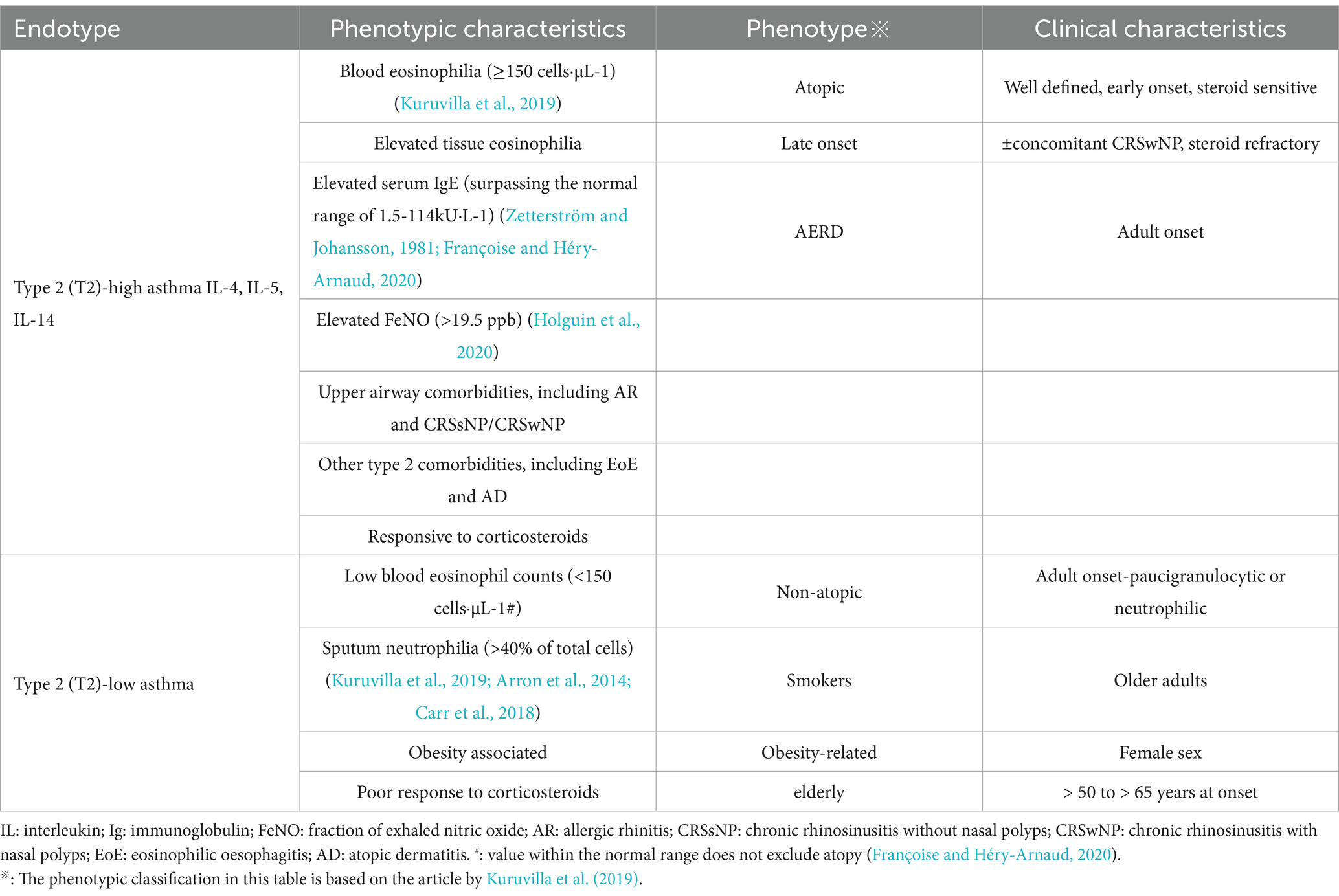
4.3 Gut and respiratory microbiota in pediatric asthma phenotypes and endotypes
Asthma, as a heterogeneous disease, encompasses different endotypic and phenotypic classifications (Akar-Ghibril et al., 2020), with corresponding variations in its microbial ecosystem dynamics. Table 1 presents an example of current asthma phenotypes as they relate to inflammatory endotypes (type 2-high or type 2-low) and phenotypic characteristics. Based on immune characteristics, it can be categorized into T-helper lymphocytes 2 high (Th2-high) and non-T-helper lymphocytes 2 high (non-Th2 endotypes) (Agache et al., 2021; Liang et al., 2022; Svenningsen and Nair, 2017). Based on inflammatory cell infiltration in induced sputum, asthma can be classified into four distinct inflammatory phenotypes: eosinophilic asthma, neutrophilic asthma, mixed granulocytic asthma, and paucigranulocytic asthma (Svenningsen and Nair, 2017). Th2-high endotype (often referred as eosinophilic asthma), driven by IL-4, IL-5, and IL-13-mediated Th2 polarization (Castan et al., 2020), exhibits selective enrichment of pathogenic genera within Firmicutes and Proteobacteria—including Haemophilus, Neisseria, Streptococcus, and Moraxella—alongside significant depletion of Lactobacillus (Durack et al., 2017). These microbial alterations correlate with elevated blood and sputum eosinophil levels (Zhang et al., 2016; Diver et al., 2022), with exacerbations marked by Haemophilus and Staphylococcus expansion paralleling Th2 cytokine surges, while stable phases favor anti-inflammatory commensals like Corynebacterium and Prevotella (Diver et al., 2022). Further investigations demonstrate seasonal variations in nasal microbiota linked to viral-induced asthma exacerbations, particularly highlighting that the RV-C endotype—characterized by Moraxella-dominated microbial composition, heightened Th2 cytokine levels, and modified lipid metabolites—significantly elevates the likelihood of recurrent wheezing episodes (Raita et al., 2021; McCauley et al., 2022). Experimental studies using Aspergillus fumigatus-induced allergic asthma models reveal that intestinal colonization by Candida albicans amplifies eosinophil and mast cell populations in bronchoalveolar lavage fluid (BALF), stimulates secretion of IL-5, IL-13, and interferon-gamma, and consequently intensifies pulmonary allergic immune reactions (Noverr et al., 2004).
Conversely, non-Th2 endotype, or non- eosinophilic asthma, encompassing neutrophilic asthma mediated through IL-6/IL-17-driven Th1/Th17 pathways and paucigranulocytic asthma in which neither eosinophils nor neutrophils are increased (Tliba and Panettieri, 2019; Hammad and Lambrecht, 2021). Neutrophilic asthma shows reduced microbial diversity with predominant Haemophilus and Moraxella enrichment in Proteobacteria, alongside dynamic Haemophilus-Staphylococcus fluctuations during exacerbations (Diver et al., 2022). A Proteobacteria-dominant microbial profile was associated with sputum neutrophilia, but also a longer duration of disease (Diver et al., 2022). The non-Th2 frequently displays corticosteroid resistance and severe clinical manifestations (Moore et al., 2014). Phenotypically, the enrichment of Haemophilus and Neisseria (phylum Proteobacteria) in the respiratory tract of children with asthma is associated with reduced post-bronchodilator FEV1/FVC ratio, mixed granulocytic asthma, and activation of the PD-L1/Th2 pathway (Kim et al., 2023). While nasal dominance of Moraxella and Alloiococcus coupled with bronchial Actinomyces-mediated anti-inflammatory regulation (Chun et al., 2020) further underscores microbial modulation of disease trajectories. Pediatric asthma highlights these interactions through heightened microbial-host crosstalk, where specific taxa influence inflammatory gene networks (Chun et al., 2020), collectively illustrating the respiratory microbiome’s multidimensional role in shaping asthma endotype-phenotype interrelationships.
Finally, we present Figure 1 to summarize key gut-respiratory microbiota dynamics in pediatric asthma. It delineates healthy colonization patterns, multifactorial influences (e.g., diet, antibiotics), and asthma-associated dysbiosis linked to phenotypes, emphasizing gut-lung crosstalk via immune-metabolic pathways.
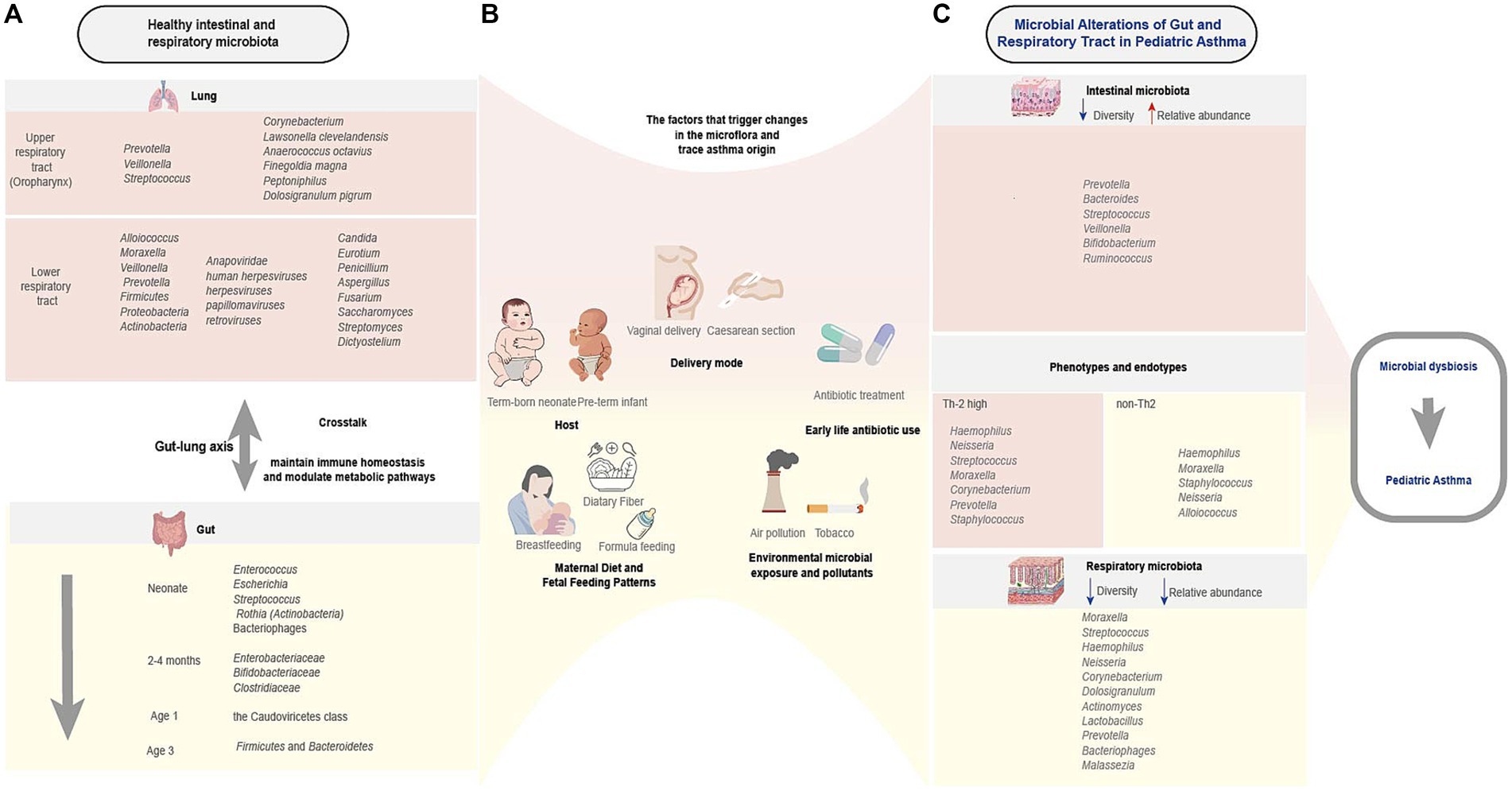
Figure 1. Characteristics of healthy and asthmatic gut-respiratory microbiota and associated influencing factors. This schematic illustrates the interplay between early-life microbial colonization, host-environment interactions, and asthma susceptibility. (A) Healthy gut microbiota (e.g., Firmicutes, Bacteroidetes) and respiratory microbiota (e.g., Streptococcus, Corynebacterium) undergo dynamic compositional shifts in early life, influencing immune development. (B) Multifactorial regulation spans prenatal to postnatal stages: biological factors (delivery mode, maternal diet, infant feeding), ecological exposures (microbial diversity, pollutants), and critical interventions (early antibiotic use), collectively shaping microbiota trajectories. (C) Microbial alterations in pediatric asthma and phenotypes and endotypes (Th2-high/non-Th2) correlate with microbial signatures and virome alterations. Bidirectional gut-lung crosstalk (arrows) via metabolic and immune pathways underlies asthma risk. This figure was created by the authors using Adobe illustrator.
5 Microbiota-modifying treatments of pediatric asthma
In recent years, groundbreaking advances in microbiome research have made the joint analysis of gut and respiratory microbiomes an important tool for identifying endotypes and patient subgroups in respiratory diseases, paving new avenues for personalized medicine. Based on the theory of the gut-lung axis, current microbiota intervention strategies focus on the regulation of gut microbiota, ranging from traditional probiotics and prebiotics to innovative approaches such as fecal microbiota transplantation (FMT), helminth immunoregulation, phage-targeted therapies, and CRISPR-Cas gene editing technology. These interventions remodel the balance of gut microbiota to regulate pulmonary immune responses, opening up a new path for precise treatment of respiratory diseases while laying a significant foundation for the future development of more targeted therapeutic options.
5.1 Probiotics
Probiotics, as foundational interventions, influence asthma progression by modulating gut microbiota balance through the bidirectional gut-lung axis. Representative probiotics such as Lactobacillus and Bifidobacterium exhibit dual efficacy in preclinical models. For instance, Lactobacillus species—particularly L. rhamnosus—prevent airway hyperreactivity by reducing eosinophil infiltration (a core pathological feature of asthma) while suppressing type 2 inflammation (Spacova et al., 2020). These effects align with the broader mechanisms by which probiotics (including Lactobacillus, Bifidobacterium, and Saccharomyces spp.) regulate gut-respiratory interactions (Ozdemir, 2010; Sanders et al., 2019). Bifidobacteria regulate early immune system development in infants by metabolizing human milk oligosaccharides (HMOs). Deficiency in HMO-metabolizing capacity within the gut microbiota correlates with heightened Th2- and Th17-driven inflammation (Henrick et al., 2021). Dairy products like yogurt, which enhance probiotic colonization in the gastrointestinal tract, serve as ideal delivery vehicles. Thus, it is recommended that expectant mothers increase their intake of fermented dairy products and prioritize breastfeeding to strengthen probiotic colonization and provide early protection for infant respiratory health.
5.2 Nutritional interventions
Nutritional interventions serve as a critical strategy for modulating the development and progression of asthma, encompassing two primary approaches: dietary fiber and nutritional supplements. Dietary fiber intake promotes the production of gut microbiota-derived metabolites, enhancing epithelial barrier function, driving regulatory T cell (Treg) differentiation, and suppressing Th2 polarization and mast cell hyperactivation, thereby synergistically alleviating asthma pathogenesis (Venter et al., 2022). Animal models further demonstrate that dietary fiber supplementation restructures the gut microbiota in asthmatic mice, accompanied by reduced eosinophilic inflammation, decreased Immunoglobulin E (IgE) levels, diminished Th2-associated mediators, and improved pulmonary function (Manni et al., 2021). Notably, maternal dietary fiber intake before and during pregnancy regulates offspring allergic sensitization and airway hyperreactivity through transgenerational microbial programming mechanisms (Thorburn et al., 2015). This vertical microbiota transmission effect provides innovative insights for early-life asthma prevention.
Another category of nutritional interventions—nutritional supplements (defined as bioactive components with health benefits beyond basic nutritional requirements)—exerts antioxidant and anti-inflammatory properties that are closely associated with pulmonary function maintenance (Tuna and Samur, 2025). For instance, prenatal supplementation with vitamin D or polyunsaturated fatty acids (PUFA) has been shown to reduce childhood croup incidence (Brustad et al., 2023), indicating their potential protective role against respiratory disorders. These interventions may indirectly influence asthma trajectories by modulating immune balance and inflammatory responses. Building on existing evidence, future research should further explore the application potential of targeted nutritional interventions, such as specific dietary fiber combinations or precision-based nutritional supplementation protocols, in primary prevention and clinical management of pediatric asthma. This approach aims to establish novel paradigms for asthma prevention and treatment grounded in nutritional regulation.
5.3 Fecal microbiota transplantation
In the realm of precision interventions, microbiota transplantation technologies show transformative potential. Fecal microbiota transplantation (FMT) enhances microbial diversity, reshapes gut microbiota composition by increasing Bacteroidetes (symbiotic bacteria) and reducing Proteobacteria (potential pathogens) (Seekatz et al., 2014; Zheng et al., 2025), and elevates SCFA levels, effectively alleviating airway inflammation in animal models (Lai et al., 2025). A novel whole-intestinal microbial intervention strategy combining healthy donor-derived intestinal fluid transplantation (HIFT) with FMT demonstrates translational potential for pediatric asthma, supported by clinical improvements observed in autism spectrum disorder (Ye et al., 2022). Additionally, emerging virome-targeted strategies reveal that specific temperate bacteriophage taxa in infant guts independently correlate with asthma risk via TLR9-mediated immune interactions (Leal Rodríguez et al., 2024), suggesting that phage therapy to restore temperate virome balance may become an innovative preventive approach.
5.4 Emerging microbiota-based therapies
Multiple lines of evidence suggest that helminth infections alter the composition of the intestinal microbiota (Rausch et al., 2018; Pillai et al., 2005). Studies have demonstrated that mice infected with Heligmosomoides polygyrus bakeri (a typical parasitic helminth) alter gut microbiota composition by increasing short-chain fatty acids (SCFAs), ultimately alleviating inflammatory responses in dust mite-induced asthma models (Zaiss et al., 2015). Studies have demonstrated that Heligmosomoides polygyrus bakeri infection alters gut microbiota composition by selectively promoting the proliferation of SCFA-producing Clostridiales while reducing the relative abundance of Bacteroidales and Lactobacillales, ultimately alleviating inflammatory responses in dust mite-induced asthma models (Zaiss et al., 2015). However, recent research highlights the direct immunomodulatory role of helminths rather than microbiota-dependent mechanisms. Specifically, helminths secrete specific proteins to establish an inhibitory microenvironment within the host, balancing the immune system to attenuate hypersensitivity to allergens (Sun et al., 2019). Therefore, helminth therapy not only provides novel insights into respiratory benefits via gut-lung axis microbiota modulation but also emerges as a promising avenue for asthma prevention and treatment.
As naturally occurring bacterial predators, bacteriophages and their lytic enzymes demonstrate targeted antimicrobial activity with minimal host toxicity, positioning them as viable alternatives against multidrug-resistant pathogens (Guo et al., 2023; MacNair et al., 2024; Wang H. et al., 2024). Research findings on the reduced bacteriophage abundance in asthma patients (Megremis et al., 2023; Choi et al., 2021) suggest that supplementation strategies hold promise for reestablishing microbiome homeostasis in affected individuals. The bacteriophage CRISPR-Cas system, as a defense mechanism against phages and other nucleic acids that invade bacteria and archaea (Dion et al., 2024), demonstrates unique potential through precise editing of bacterial or fungal genomes and targeted elimination of respiratory pathogens. Its delivery system has established a technical paradigm in gut microbiome research, exemplified by the bacteriophage-mediated CRISPR-Cas9 achieving in situ knockout of specific genes in Escherichia coli (Gencay et al., 2024). For chronic respiratory diseases such as asthma, this system could intervene in disease progression by modifying pathogen virulence genes or regulating host immune responses. Precision strategies combining bacteriophage-CRISPR technologies with patient-specific microbial profiles will advance personalized regulation of respiratory and gut microbiota, pioneering innovative pathways for asthma prevention and treatment.
These interventions collectively establish a microbiota-centric framework for asthma management, offering multi-target immunomodulation with minimal side effects. Future research should focus on optimizing probiotic formulations, advancing clinical translation of microbiota transplantation, elucidating virome regulatory mechanisms, and longitudinally tracking the impact of microbial interventions on disease progression, thereby strengthening the scientific foundation for personalized asthma therapies.
6 Conclusion
This study systematically elucidates the critical role of the gut-respiratory microbiome in the pathogenesis of pediatric asthma. It reveals the microbial colonization patterns in normal gut and respiratory tracts and their influencing factors (including host, delivery mode, feeding patterns, antibiotic use, and environmental exposures). The study further analyzes in depth the association mechanisms between specific gut and respiratory microbial dysbiosis and different asthma phenotypes/endotypes. Building on these findings, the study explores innovative microbiome-modulating therapeutic strategies, including probiotic interventions, dietary regulation, and microbiota transplantation. Additionally, it highlights the need for future research to focus on elucidating the precise molecular mechanisms of microbe-immune interactions, providing key insights for developing precision microbiome-based therapies for pediatric asthma.
Author contributions
LL: Writing – original draft, Writing – review & editing. WZ: Writing – original draft, Writing – review & editing. HZ: Supervision, Writing – review & editing. YS: Project administration, Writing – review & editing. WH: Supervision, Writing – review & editing. QC: Conceptualization, Data curation, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was funded by General Project in the Natural Science Foundation of China (No. 82470015), Liaoning Applied Basic Research Program (No. 2023JH2/101600022), and Liaoning Provincial Science and Technology Program Joint Program (No. 2024-MSLH-603).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abt, M. C., Osborne, L. C., Monticelli, L. A., Doering, T. A., Alenghat, T., Sonnenberg, G. F., et al. (2012). Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 37, 158–170. doi: 10.1016/j.immuni.2012.04.011
Agache, I., Eguiluz-Gracia, I., Cojanu, C., Laculiceanu, A., Del Giacco, S., Zemelka-Wiacek, M., et al. (2021). Advances and highlights in asthma in 2021. Allergy 76, 3390–3407. doi: 10.1111/all.15054
Akar-Ghibril, N., Casale, T., Custovic, A., and Phipatanakul, W. (2020). Allergic Endotypes and phenotypes of asthma. J Allergy Clin Immunol Pract 8, 429–440. doi: 10.1016/j.jaip.2019.11.008
Alfaqawi, M., Abuowda, Y., Böttcher, B., Alserr, K., and Elmassry, A. E. (2021). Antibiotic use in acute upper respiratory tract infections and uncomplicated lacerations in the Gaza strip: clinical audit and re-audit. Lancet (London, England) 398:S7. doi: 10.1016/S0140-6736(21)01493-8
Ancona, G., Alagna, L., Alteri, C., Palomba, E., Tonizzo, A., Pastena, A., et al. (2023). Gut and airway microbiota dysbiosis and their role in COVID-19 and long-COVID. Front. Immunol. 14:1080043. doi: 10.3389/fimmu.2023.1080043
Arpaia, N., Campbell, C., Fan, X., Dikiy, S., van der Veeken, J., deRoos, P., et al. (2013). Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504, 451–455. doi: 10.1038/nature12726
Arrieta, M. C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Arron, J. R., Choy, D. F., Laviolette, M., Kelsen, S. G., Hatab, A., Leigh, R., et al. (2014). Disconnect between sputum neutrophils and other measures of airway inflammation in asthma. Eur. Respir. J. 43, 627–629. doi: 10.1183/09031936.00117013
Asher, M. I., Rutter, C. E., Bissell, K., Chiang, C. Y., El Sony, A., Ellwood, E., et al. (2021). Worldwide trends in the burden of asthma symptoms in school-aged children: global asthma network phase I cross-sectional study. Lancet (London, England) 398, 1569–1580. doi: 10.1016/S0140-6736(21)01450-1
Atarashi, K., Tanoue, T., Ando, M., Kamada, N., Nagano, Y., Narushima, S., et al. (2015). Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell 163, 367–380. doi: 10.1016/j.cell.2015.08.058
Bäckhed, F., Ley, R. E., Sonnenburg, J. L., Peterson, D. A., and Gordon, J. I. (2005). Host-bacterial mutualism in the human intestine. Science (New York, N.Y.) 307, 1915–1920. doi: 10.1126/science.1104816
Barcik, W., Boutin, R. C. T., Sokolowska, M., and Finlay, B. B. (2020). The role of lung and gut microbiota in the pathology of asthma. Immunity 52, 241–255. doi: 10.1016/j.immuni.2020.01.007
Bargheet, A., Klingenberg, C., Esaiassen, E., Hjerde, E., Cavanagh, J. P., Bengtsson-Palme, J., et al. (2023). Development of early life gut resistome and mobilome across gestational ages and microbiota-modifying treatments. EBioMedicine 92:104613. doi: 10.1016/j.ebiom.2023.104613
Barton, E. S., White, D. W., Cathelyn, J. S., Brett-McClellan, K. A., Engle, M., Diamond, M. S., et al. (2007). Herpesvirus latency confers symbiotic protection from bacterial infection. Nature 447, 326–329. doi: 10.1038/nature05762
Bassis, C. M., Erb-Downward, J. R., Dickson, R. P., Freeman, C. M., Schmidt, T. M., Young, V. B., et al. (2015). Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio 6:e00037. doi: 10.1128/mBio.00037-15
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Biesbroek, G., Bosch, A. A., Wang, X., Keijser, B. J., Veenhoven, R. H., Sanders, E. A., et al. (2014). The impact of breastfeeding on nasopharyngeal microbial communities in infants. Am. J. Respir. Crit. Care Med. 190, 298–308. doi: 10.1164/rccm.201401-0073OC
Borbet, T. C., Pawline, M. B., Zhang, X., Wipperman, M. F., Reuter, S., Maher, T., et al. (2022). Influence of the early-life gut microbiota on the immune responses to an inhaled allergen. Mucosal Immunol. 15, 1000–1011. doi: 10.1038/s41385-022-00544-5
Bosch, A., de Steenhuijsen Piters, W. A. A., van Houten, M. A., Chu, M., Biesbroek, G., Kool, J., et al. (2017). Maturation of the infant respiratory microbiota, environmental drivers, and health consequences. A prospective cohort study. Am. J. Respir. Crit. Care Med. 196, 1582–1590. doi: 10.1164/rccm.201703-0554OC
Bosch, A., Levin, E., van Houten, M. A., Hasrat, R., Kalkman, G., Biesbroek, G., et al. (2016). Development of upper respiratory tract microbiota in infancy is affected by mode of delivery. EBioMedicine 9, 336–345. doi: 10.1016/j.ebiom.2016.05.031
Breitbart, M., Haynes, M., Kelley, S., Angly, F., Edwards, R. A., Felts, B., et al. (2008). Viral diversity and dynamics in an infant gut. Res. Microbiol. 159, 367–373. doi: 10.1016/j.resmic.2008.04.006
Brustad, N., Yang, L., Chawes, B. L., Stokholm, J., Gürdeniz, G., Bønnelykke, K., et al. (2023). Fish oil and vitamin D supplementations in pregnancy protect against childhood croup. J Allergy Clin Immunol Pract 11, 315–321. doi: 10.1016/j.jaip.2022.09.027
Budden, K. F., Gellatly, S. L., Wood, D. L., Cooper, M. A., Morrison, M., Hugenholtz, P., et al. (2017). Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 15, 55–63. doi: 10.1038/nrmicro.2016.142
Cait, A., Hughes, M. R., Antignano, F., Cait, J., Dimitriu, P. A., Maas, K. R., et al. (2018). Microbiome-driven allergic lung inflammation is ameliorated by short-chain fatty acids. Mucosal Immunol. 11, 785–795. doi: 10.1038/mi.2017.75
Calışkan, M., Bochkov, Y. A., Kreiner-Møller, E., Bønnelykke, K., Stein, M. M., Du, G., et al. (2013). Rhinovirus wheezing illness and genetic risk of childhood-onset asthma. N. Engl. J. Med. 368, 1398–1407. doi: 10.1056/NEJMoa1211592
Carmody, L. A., Zhao, J., Schloss, P. D., Petrosino, J. F., Murray, S., Young, V. B., et al. (2013). Changes in cystic fibrosis airway microbiota at pulmonary exacerbation. Ann. Am. Thorac. Soc. 10, 179–187. doi: 10.1513/AnnalsATS.201211-107OC
Carney, S. M., Clemente, J. C., Cox, M. J., Dickson, R. P., Huang, Y. J., Kitsios, G. D., et al. (2020). Methods in lung microbiome research. Am. J. Respir. Cell Mol. Biol. 62, 283–299. doi: 10.1165/rcmb.2019-0273TR
Carr, T. F., Zeki, A. A., and Kraft, M. (2018). Eosinophilic and noneosinophilic asthma. Am. J. Respir. Crit. Care Med. 197, 22–37. doi: 10.1164/rccm.201611-2232PP
Casaburi, G., Duar, R. M., Brown, H., Mitchell, R. D., Kazi, S., Chew, S., et al. (2021). Metagenomic insights of the infant microbiome community structure and function across multiple sites in the United States. Sci. Rep. 11:1472. doi: 10.1038/s41598-020-80583-9
Castan, L., Bøgh, K. L., Maryniak, N. Z., Epstein, M. M., Kazemi, S., O'Mahony, L., et al. (2020). Overview of in vivo and ex vivo endpoints in murine food allergy models: suitable for evaluation of the sensitizing capacity of novel proteins? Allergy 75, 289–301. doi: 10.1111/all.13943
Charlson, E. S., Bittinger, K., Chen, J., Diamond, J. M., Li, H., Collman, R. G., et al. (2012). Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One 7:e42786. doi: 10.1371/journal.pone.0042786
Chen, Y. C., Chen, Y., Lasky-Su, J., Kelly, R. S., Stokholm, J., Bisgaard, H., et al. (2023). Environmental and genetic associations with aberrant early-life gut microbial maturation in childhood asthma. J. Allergy Clin. Immunol. 151, 1494–502.e14. doi: 10.1016/j.jaci.2023.01.006
Cho, I., and Blaser, M. J. (2012). The human microbiome: at the interface of health and disease. Nat. Rev. Genet. 13, 260–270. doi: 10.1038/nrg3182
Choi, S., Sohn, K. H., Jung, J. W., Kang, M. G., Yang, M. S., Kim, S., et al. (2021). Lung virome: new potential biomarkers for asthma severity and exacerbation. J. Allergy Clin. Immunol. 148, 1007–15.e9. doi: 10.1016/j.jaci.2021.03.017
Chu, D. M., Ma, J., Prince, A. L., Antony, K. M., Seferovic, M. D., and Aagaard, K. M. (2017). Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat. Med. 23, 314–326. doi: 10.1038/nm.4272
Chun, Y., Do, A., Grishina, G., Grishin, A., Fang, G., Rose, S., et al. (2020). Integrative study of the upper and lower airway microbiome and transcriptome in asthma. JCI Insight 5:e133707. doi: 10.1172/jci.insight.133707
Clavijo-Salomon, M. A., and Trinchieri, G. (2025). Unlocking the power of the microbiome for successful cancer immunotherapy. J. Immunother. Cancer 13:e011281. doi: 10.1136/jitc-2024-011281
Cui, L., Morris, A., and Ghedin, E. (2013). The human mycobiome in health and disease. Genome Med. 5:63. doi: 10.1186/gm467
Davidson, R. M., and Epperson, L. E. (2018). Microbiome sequencing methods for studying human diseases. Methods Molec Biol. 1706, 77–90. doi: 10.1007/978-1-4939-7471-9_5
Delhaes, L., Monchy, S., Fréalle, E., Hubans, C., Salleron, J., Leroy, S., et al. (2012). The airway microbiota in cystic fibrosis: a complex fungal and bacterial community—implications for therapeutic management. PLoS One 7:e36313. doi: 10.1371/journal.pone.0036313
Dickson, R. P., Erb-Downward, J. R., Freeman, C. M., McCloskey, L., Falkowski, N. R., Huffnagle, G. B., et al. (2017). Bacterial topography of the healthy human lower respiratory tract. MBio 8:e02287-16. doi: 10.1128/mBio.02287-16
Dion, M. B., Shah, S. A., Deng, L., Thorsen, J., Stokholm, J., Krogfelt, K. A., et al. (2024). Escherichia coli CRISPR arrays from early life fecal samples preferentially target prophages. ISME J. 18:wrae005. doi: 10.1093/ismejo/wrae005
Diver, S., Haldar, K., McDowell, P. J., Busby, J., Mistry, V., Micieli, C., et al. (2022). Relationship between inflammatory status and microbial composition in severe asthma and during exacerbation. Allergy 77, 3362–3376. doi: 10.1111/all.15425
Dohlman, A. B., Klug, J., Mesko, M., Gao, I. H., Lipkin, S. M., Shen, X., et al. (2022). A pan-cancer mycobiome analysis reveals fungal involvement in gastrointestinal and lung tumors. Cell 185, 3807–22.e12. doi: 10.1016/j.jaci.2014.02.030
Durack, J., Huang, Y. J., Nariya, S., Christian, L. S., Ansel, K. M., Beigelman, A., et al. (2018). Bacterial biogeography of adult airways in atopic asthma. Microbiome 6:104. doi: 10.1073/pnas.1000081107
Durack, J., Lynch, S. V., Nariya, S., Bhakta, N. R., Beigelman, A., Castro, M., et al. (2017). Features of the bronchial bacterial microbiome associated with atopy, asthma, and responsiveness to inhaled corticosteroid treatment. J. Allergy Clin. Immunol. 140, 63–75. doi: 10.1016/j.jaci.2016.08.055
Ege, M. J., Mayer, M., Normand, A. C., Genuneit, J., Cookson, W. O., Braun-Fahrländer, C., et al. (2011). Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 364, 701–709. doi: 10.1056/NEJMoa1007302
Escobar-Zepeda, A., Vera-Ponce de León, A., and Sanchez-Flores, A. (2015). The road to metagenomics: from microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 6:348. doi: 10.3389/fgene.2015.00348
Fang, Z. Y., Stickley, S. A., Ambalavanan, A., Zhang, Y., Zacharias, A. M., Fehr, K., et al. (2024). Networks of human milk microbiota are associated with host genomics, childhood asthma, and allergic sensitization. Cell Host Microbe 32, 1838–52.e5. doi: 10.1016/j.chom.2024.08.014
Fodor, A. A., Klem, E. R., Gilpin, D. F., Elborn, J. S., Boucher, R. C., Tunney, M. M., et al. (2012). The adult cystic fibrosis airway microbiota is stable over time and infection type, and highly resilient to antibiotic treatment of exacerbations. PLoS One 7:e45001. doi: 10.1371/journal.pone.0045001
Françoise, A., and Héry-Arnaud, G. (2020). The microbiome in cystic fibrosis pulmonary disease. Genes 11. doi: 10.3390/genes11050536
Gencay, Y. E., Jasinskytė, D., Robert, C., Semsey, S., Martínez, V., Petersen, A., et al. (2024). Engineered phage with antibacterial CRISPR-Cas selectively reduce E. coli burden in mice. Nat. Biotechnol. 42, 265–274. doi: 10.1038/s41587-023-01759-y
Georgountzou, A., Kokkinou, D., Taka, S., Maggina, P., Lakoumentas, J., Papaevangelou, V., et al. (2021). Differential maturation trajectories of innate antiviral immunity in health and atopy. Pediatric Aller Immunol 32, 1843–1856. doi: 10.1111/pai.13601
Ghannoum, M. A., Jurevic, R. J., Mukherjee, P. K., Cui, F., Sikaroodi, M., Naqvi, A., et al. (2010). Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog. 6:e1000713. doi: 10.1371/journal.ppat.1000713
Guo, Z., Liu, M., and Zhang, D. (2023). Potential of phage depolymerase for the treatment of bacterial biofilms. Virulence 14:2273567. doi: 10.1080/21505594.2023.2273567
Hammad, H., and Lambrecht, B. N. (2021). The basic immunology of asthma. Cell 184, 1469–1485. doi: 10.1016/j.cell.2021.02.016
Henrick, B. M., Rodriguez, L., Lakshmikanth, T., Pou, C., Henckel, E., Arzoomand, A., et al. (2021). Bifidobacteria-mediated immune system imprinting early in life. Cell 184, 3884–98.e11. doi: 10.1016/j.cell.2021.05.030
Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS One 5:e8578. doi: 10.1371/journal.pone.0008578
Holguin, F., Cardet, J. C., Chung, K. F., Diver, S., Ferreira, D. S., Fitzpatrick, A., et al. (2020). Management of severe asthma: a European Respiratory Society/American Thoracic Society guideline. Eur. Respir. J. 55:1900588. doi: 10.1183/13993003.00588-2019
Huang, C., Du, W., Ni, Y., Lan, G., and Shi, G. (2022). The effect of short-chain fatty acids on M2 macrophages polarization in vitro and in vivo. Clin. Exp. Immunol. 207, 53–64. doi: 10.1093/cei/uxab028
Huang, C., Yu, Y., Du, W., Liu, Y., Dai, R., Tang, W., et al. (2020). Fungal and bacterial microbiome dysbiosis and imbalance of trans-kingdom network in asthma. Clin. Transl. Allergy 10:42. doi: 10.1186/s13601-020-00345-8
Huffnagle, G. B., and Noverr, M. C. (2013). The emerging world of the fungal microbiome. Trends Microbiol. 21, 334–341. doi: 10.1016/j.tim.2013.04.002
Iliev, I. D., Funari, V. A., Taylor, K. D., Nguyen, Q., Reyes, C. N., Strom, S. P., et al. (2012). Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science (New York, N.Y.) 336, 1314–1317. doi: 10.1126/science.1221789
Jackson, D. J., Gangnon, R. E., Evans, M. D., Roberg, K. A., Anderson, E. L., Pappas, T. E., et al. (2008). Wheezing rhinovirus illnesses in early life predict asthma development in high-risk children. Am. J. Respir. Crit. Care Med. 178, 667–672. doi: 10.1164/rccm.200802-309OC
Kim, Y. H., Park, M. R., Kim, S. Y., Kim, M. Y., Kim, K. W., and Sohn, M. H. (2023). Respiratory microbiome profiles are associated with distinct inflammatory phenotype and lung function in children with asthma. J Investig Allergol Clin Immunol 34, 246–256. doi: 10.18176/jiaci.0918
Kim, Y. G., Udayanga, K. G., Totsuka, N., Weinberg, J. B., Núñez, G., and Shibuya, A. (2014). Gut dysbiosis promotes M2 macrophage polarization and allergic airway inflammation via fungi-induced PGE₂. Cell Host Microbe 15, 95–102. doi: 10.1016/j.chom.2013.12.010
Kim, Y. J., Womble, J. T., Gunsch, C. K., and Ingram, J. L. (2021). The gut/lung microbiome Axis in obesity, asthma, and bariatric surgery: a literature review. Obesity (Silver Spring, Md) 29, 636–644. doi: 10.1002/oby.23107
Kloepfer, K. M., Lee, W. M., Pappas, T. E., Kang, T. J., Vrtis, R. F., Evans, M. D., et al. (2014). Detection of pathogenic bacteria during rhinovirus infection is associated with increased respiratory symptoms and asthma exacerbations. J. Allergy Clin. Immunol. 133, 1301–7, 7.e1-3.
Koenig, J. E., Spor, A., Scalfone, N., Fricker, A. D., Stombaugh, J., Knight, R., et al. (2011). Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 108, 4578–4585.
Korten, I., Mika, M., Klenja, S., Kieninger, E., Mack, I., Barbani, M. T., et al. (2016). Interactions of respiratory viruses and the nasal microbiota during the first year of life in healthy infants. mSphere 1:e00312-16. doi: 10.1128/mSphere.00312-16
Kuruvilla, M. E., Lee, F. E., and Lee, G. B. (2019). Understanding asthma phenotypes, Endotypes, and mechanisms of disease. Clin. Rev. Allergy Immunol. 56, 219–233. doi: 10.1007/s12016-018-8712-1
Kusel, M. M., de Klerk, N. H., Kebadze, T., Vohma, V., Holt, P. G., Johnston, S. L., et al. (2007). Early-life respiratory viral infections, atopic sensitization, and risk of subsequent development of persistent asthma. J. Allergy Clin. Immunol. 119, 1105–1110. doi: 10.1016/j.jaci.2006.12.669
Lai, Y., Qiu, R., Zhou, J., Ren, L., Qu, Y., and Zhang, G. (2025). Fecal microbiota transplantation alleviates airway inflammation in asthmatic rats by increasing the level of short-chain fatty acids in the intestine. Inflammation. doi: 10.1007/s10753-024-02233-w
Leal Rodríguez, C., Shah, S. A., Rasmussen, M. A., Thorsen, J., Boulund, U., Pedersen, C. T., et al. (2024). The infant gut virome is associated with preschool asthma risk independently of bacteria. Nat. Med. 30, 138–148. doi: 10.1038/s41591-023-02685-x
Leiby, J. S., McCormick, K., Sherrill-Mix, S., Clarke, E. L., Kessler, L. R., Taylor, L. J., et al. (2018). Lack of detection of a human placenta microbiome in samples from preterm and term deliveries. Microbiome 6:196. doi: 10.1186/s40168-018-0575-4
Liang, J., Liu, X. H., Chen, X. M., Song, X. L., Li, W., and Huang, Y. (2022). Emerging roles of non-coding RNAs in childhood asthma. Front. Pharmacol. 13:856104. doi: 10.3389/fphar.2022.856104
Lim, Y. W., Evangelista, J. S. 3rd, Schmieder, R., Bailey, B., Haynes, M., Furlan, M., et al. (2014). Clinical insights from metagenomic analysis of sputum samples from patients with cystic fibrosis. J. Clin. Microbiol. 52, 425–437. doi: 10.1128/JCM.02204-13
Lim, Y. W., Schmieder, R., Haynes, M., Willner, D., Furlan, M., Youle, M., et al. (2013). Metagenomics and metatranscriptomics: windows on CF-associated viral and microbial communities. J. Cyst. Fibr. 12, 154–164. doi: 10.1016/j.jcf.2012.07.009
Lynch, S. V., Wood, R. A., Boushey, H., Bacharier, L. B., Bloomberg, G. R., Kattan, M., et al. (2014). Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J. Allergy Clin. Immunol. 134, 593–601.e12. doi: 10.1016/j.jaci.2014.04.018
MacDuff, D. A., Reese, T. A., Kimmey, J. M., Weiss, L. A., Song, C., Zhang, X., et al. (2015). Phenotypic complementation of genetic immunodeficiency by chronic herpesvirus infection. eLife 4:4. doi: 10.7554/eLife.04494
MacNair, C. R., Rutherford, S. T., and Tan, M. W. (2024). Alternative therapeutic strategies to treat antibiotic-resistant pathogens. Nat. Rev. Microbiol. 22, 262–275. doi: 10.1038/s41579-023-00993-0
Manni, M. L., Heinrich, V. A., Buchan, G. J., O'Brien, J. P., Uvalle, C., Cechova, V., et al. (2021). Nitroalkene fatty acids modulate bile acid metabolism and lung function in obese asthma. Sci. Rep. 11:17788. doi: 10.1038/s41598-021-96471-9
Marsland, B. J., and Gollwitzer, E. S. (2014). Host-microorganism interactions in lung diseases. Nat. Rev. Immunol. 14, 827–835. doi: 10.1038/nri3769
McCauley, K., Durack, J., Valladares, R., Fadrosh, D. W., Lin, D. L., Calatroni, A., et al. (2019). Distinct nasal airway bacterial microbiotas differentially relate to exacerbation in pediatric patients with asthma. J. Allergy Clin. Immunol. 144, 1187–1197. doi: 10.1016/j.jaci.2019.05.035
McCauley, K. E., Flynn, K., Calatroni, A., DiMassa, V., LaMere, B., Fadrosh, D. W., et al. (2022). Seasonal airway microbiome and transcriptome interactions promote childhood asthma exacerbations. J. Allergy Clin. Immunol. 150, 204–213. doi: 10.1016/j.jaci.2022.01.020
Megremis, S., Constantinides, B., Xepapadaki, P., Yap, C. F., Sotiropoulos, A. G., Bachert, C., et al. (2023). Respiratory eukaryotic virome expansion and bacteriophage deficiency characterize childhood asthma. Sci. Rep. 13:8319. doi: 10.1038/s41598-023-34730-7
Megremis, S., Niespodziana, K., Cabauatan, C., Xepapadaki, P., Kowalski, M. L., Jartti, T., et al. (2018). Rhinovirus species-specific antibodies differentially reflect clinical outcomes in health and asthma. Am. J. Respir. Crit. Care Med. 198, 1490–1499. doi: 10.1164/rccm.201803-0575OC
Moore, W. C., Hastie, A. T., Li, X., Li, H., Busse, W. W., Jarjour, N. N., et al. (2014). Sputum neutrophil counts are associated with more severe asthma phenotypes using cluster analysis. J. Allergy Clin. Immunol. 133, 1557–63.e5. doi: 10.1016/j.jaci.2013.10.011
Naja, A. S., Permaul, P., and Phipatanakul, W. (2018). Taming asthma in school-aged children: a comprehensive review. J Allergy Clin Immunol Pract 6, 726–735. doi: 10.1016/j.jaip.2018.01.023
Noverr, M. C., Noggle, R. M., Toews, G. B., and Huffnagle, G. B. (2004). Role of antibiotics and fungal microbiota in driving pulmonary allergic responses. Infect. Immun. 72, 4996–5003. doi: 10.1128/IAI.72.9.4996-5003.2004
Odendaal, M. L., de Steenhuijsen Piters, W. A. A., Franz, E., Chu, M., Groot, J. A., van Logchem, E. M., et al. (2024). Host and environmental factors shape upper airway microbiota and respiratory health across the human lifespan. Cell 187, 4571–85.e15. doi: 10.1016/j.cell.2024.07.008
Ozdemir, O. (2010). Various effects of different probiotic strains in allergic disorders: an update from laboratory and clinical data. Clin. Exp. Immunol. 160, 295–304. doi: 10.1111/j.1365-2249.2010.04109.x
Pammi, M., Lal, C. V., Wagner, B. D., Mourani, P. M., Lohmann, P., Luna, R. A., et al. (2019). Airway microbiome and development of bronchopulmonary dysplasia in preterm infants: a systematic review. J. Pediatr. 204, 126–33.e2. doi: 10.1016/j.jpeds.2018.08.042
Pillai, A., Ueno, S., Zhang, H., Lee, J. M., and Kato, Y. (2005). Cecropin P1 and novel nematode cecropins: a bacteria-inducible antimicrobial peptide family in the nematode Ascaris suum. Biochem. J. 390, 207–214. doi: 10.1042/BJ20050218
Psarras, S., Volonaki, E., Skevaki, C. L., Xatzipsalti, M., Bossios, A., Pratsinis, H., et al. (2006). Vascular endothelial growth factor-mediated induction of angiogenesis by human rhinoviruses. J. Allergy Clin. Immunol. 117, 291–297. doi: 10.1016/j.jaci.2005.11.005
Raita, Y., Camargo, C. A. Jr., Bochkov, Y. A., Celedón, J. C., Gern, J. E., Mansbach, J. M., et al. (2021). Integrated-omics endotyping of infants with rhinovirus bronchiolitis and risk of childhood asthma. J. Allergy Clin. Immunol. 147, 2108–2117. doi: 10.1016/j.jaci.2020.11.002
Rausch, S., Midha, A., Kuhring, M., Affinass, N., Radonic, A., Kühl, A. A., et al. (2018). Parasitic nematodes exert antimicrobial activity and benefit from microbiota-driven support for host immune regulation. Front. Immunol. 9:2282. doi: 10.3389/fimmu.2018.02282
Rautava, S., and Walker, W. A. (2009). Academy of breastfeeding medicine Founder's lecture 2008: breastfeeding—an Extrauterine link between mother and Child. Breastfeed. Med. 4, 3–10. doi: 10.1089/bfm.2009.0004
Roduit, C., Frei, R., Ferstl, R., Loeliger, S., Westermann, P., Rhyner, C., et al. (2019). High levels of butyrate and propionate in early life are associated with protection against atopy. Allergy 74, 799–809. doi: 10.1111/all.13660
Rosas-Salazar, C., Chirkova, T., Gebretsadik, T., Chappell, J. D., Peebles, R. S. Jr., Dupont, W. D., et al. (2023). Respiratory syncytial virus infection during infancy and asthma during childhood in the USA (INSPIRE): a population-based, prospective birth cohort study. Lancet (London, England) 401, 1669–1680. doi: 10.1016/S0140-6736(23)00811-5
Rovira Rubió, J., Megremis, S., Pasioti, M., Lakoumentas, J., Constantinides, B., Xepapadaki, P., et al. (2023). Respiratory virome profiles reflect antiviral immune responses. Allergy 78, 1258–1268. doi: 10.1111/all.15634
Rutayisire, E., Huang, K., Liu, Y., and Tao, F. (2016). The mode of delivery affects the diversity and colonization pattern of the gut microbiota during the first year of infants' life: a systematic review. BMC Gastroenterol. 16:86. doi: 10.1186/s12876-016-0498-0
Sanders, M. E., Merenstein, D. J., Reid, G., Gibson, G. R., and Rastall, R. A. (2019). Probiotics and prebiotics in intestinal health and disease: from biology to the clinic. Nat. Rev. Gastroenterol. Hepatol. 16, 605–616. doi: 10.1038/s41575-019-0173-3
Seekatz, A. M., Aas, J., Gessert, C. E., Rubin, T. A., Saman, D. M., Bakken, J. S., et al. (2014). Recovery of the gut microbiome following fecal microbiota transplantation. MBio 5, e00893–e00814. doi: 10.1128/mBio.00893-14
Segal, L. N., Alekseyenko, A. V., Clemente, J. C., Kulkarni, R., Wu, B., Gao, Z., et al. (2013). Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 1:19. doi: 10.1186/2049-2618-1-19
Selma-Royo, M., García-Mantrana, I., Calatayud, M., Parra-Llorca, A., Martínez-Costa, C., and Collado, M. C. (2021). Maternal diet during pregnancy and intestinal markers are associated with early gut microbiota. Eur. J. Nutr. 60, 1429–1442. doi: 10.1007/s00394-020-02337-7
Sender, R., Fuchs, S., and Milo, R. (2016). Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 164, 337–340. doi: 10.1016/j.cell.2016.01.013
Shah, S. A., Deng, L., Thorsen, J., Pedersen, A. G., Dion, M. B., Castro-Mejía, J. L., et al. (2023). Expanding known viral diversity in the healthy infant gut. Nat. Microbiol. 8, 986–998. doi: 10.1038/s41564-023-01345-7
Sharma, A., Laxman, B., Naureckas, E. T., Hogarth, D. K., Sperling, A. I., Solway, J., et al. (2019). Associations between fungal and bacterial microbiota of airways and asthma endotypes. J. Allergy Clin. Immunol. 144, 1214–27.e7. doi: 10.1016/j.jaci.2019.06.025
Sigurs, N., Bjarnason, R., Sigurbergsson, F., and Kjellman, B. (2000). Respiratory syncytial virus bronchiolitis in infancy is an important risk factor for asthma and allergy at age 7. Am. J. Respir. Crit. Care Med. 161, 1501–1507. doi: 10.1164/ajrccm.161.5.9906076
Simonyte Sjödin, K., Vidman, L., Rydén, P., and West, C. E. (2016). Emerging evidence of the role of gut microbiota in the development of allergic diseases. Curr. Opin. Allergy Clin. Immunol. 16, 390–395. doi: 10.1097/ACI.0000000000000277
Skevaki, C. L., Psarras, S., Volonaki, E., Pratsinis, H., Spyridaki, I. S., Gaga, M., et al. (2012). Rhinovirus-induced basic fibroblast growth factor release mediates airway remodeling features. Clin. Transl. Allergy 2:14. doi: 10.1186/2045-7022-2-14
Solís, G., de Los Reyes-Gavilan, C. G., Fernández, N., Margolles, A., and Gueimonde, M. (2010). Establishment and development of lactic acid bacteria and bifidobacteria microbiota in breast-milk and the infant gut. Anaerobe 16, 307–310. doi: 10.1016/j.anaerobe.2010.02.004
Spacova, I., Van Beeck, W., Seys, S., Devos, F., Vanoirbeek, J., Vanderleyden, J., et al. (2020). Lactobacillus rhamnosus probiotic prevents airway function deterioration and promotes gut microbiome resilience in a murine asthma model. Gut Microbes 11, 1729–1744. doi: 10.1080/19490976.2020.1766345
Spector, C., De Sanctis, C. M., Panettieri, R. A. Jr., and Koziol-White, C. J. (2023). Rhinovirus induces airway remodeling: what are the physiological consequences? Respir. Res. 24:238. doi: 10.1186/s12931-023-02529-9
Stokholm, J., Blaser, M. J., Thorsen, J., Rasmussen, M. A., Waage, J., Vinding, R. K., et al. (2018). Maturation of the gut microbiome and risk of asthma in childhood. Nat. Commun. 9:141. doi: 10.1038/s41467-017-02573-2
Stokholm, J., Thorsen, J., Blaser, M. J., Rasmussen, M. A., Hjelmsø, M., Shah, S., et al. (2020). Delivery mode and gut microbial changes correlate with an increased risk of childhood asthma. Sci. Transl. Med. 12:eaax9929. doi: 10.1126/scitranslmed.aax9929
Sun, S., Li, H., Yuan, Y., Wang, L., He, W., Xie, H., et al. (2019). Preventive and therapeutic effects of Trichinella spiralis adult extracts on allergic inflammation in an experimental asthma mouse model. Parasit. Vectors 12:326. doi: 10.1186/s13071-019-3561-1
Sun, L., Miyoshi, H., Origanti, S., Nice, T. J., Barger, A. C., Manieri, N. A., et al. (2015). Type I interferons link viral infection to enhanced epithelial turnover and repair. Cell Host Microbe 17, 85–97. doi: 10.1016/j.chom.2014.11.004
Sun, Y., Tang, H., Du, S., Chen, Y., Ou, Z., Zhang, M., et al. (2023). Indoor metabolites and chemicals outperform microbiome in classifying childhood asthma and allergic rhinitis. Eco-Environ. Health 2, 208–218. doi: 10.1016/j.eehl.2023.08.001
Svenningsen, S., and Nair, P. (2017). Asthma Endotypes and an overview of targeted therapy for asthma. Front. Med. 4:158. doi: 10.3389/fmed.2017.00158
Sweere, J. M., Van Belleghem, J. D., Ishak, H., Bach, M. S., Popescu, M., Sunkari, V., et al. (2019). Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science (New York, N.Y.) 363:eaat9691. doi: 10.1126/science.aat9691
Takashima, M. D., Grimwood, K., Sly, P. D., Lambert, S. B., and Ware, R. S. (2023). Association of rhinovirus and potentially pathogenic bacterial detections in the first 3 months of life with subsequent wheezing in childhood. Pediatr. Pulmonol. 58, 3428–3436. doi: 10.1002/ppul.26667
Teo, S. M., Mok, D., Pham, K., Kusel, M., Serralha, M., Troy, N., et al. (2015). The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe 17, 704–715. doi: 10.1016/j.chom.2015.03.008
Teo, S. M., Tang, H. H. F., Mok, D., Judd, L. M., Watts, S. C., Pham, K., et al. (2018). Airway microbiota dynamics uncover a critical window for interplay of pathogenic Bacteria and allergy in childhood respiratory disease. Cell Host Microbe 24, 341–52.e5. doi: 10.1016/j.chom.2018.08.005
Thorburn, A. N., McKenzie, C. I., Shen, S., Stanley, D., Macia, L., Mason, L. J., et al. (2015). Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat. Commun. 6:7320. doi: 10.1038/ncomms8320
Thorsen, J., Li, X. J., Peng, S., Sunde, R. B., Shah, S. A., Bhattacharyya, M., et al. (2023). The airway microbiota of neonates colonized with asthma-associated pathogenic bacteria. Nat. Commun. 14:6668. doi: 10.1038/s41467-023-42309-z
Tliba, O., and Panettieri, R. A. Jr. (2019). Paucigranulocytic asthma: uncoupling of airway obstruction from inflammation. J. Allergy Clin. Immunol. 143, 1287–1294. doi: 10.1016/j.jaci.2018.06.008
Trompette, A., Gollwitzer, E. S., Yadava, K., Sichelstiel, A. K., Sprenger, N., Ngom-Bru, C., et al. (2014). Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166. doi: 10.1038/nm.3444
Tuna, T., and Samur, G. (2025). The role of nutrition and nutritional supplements in the prevention and treatment of malnutrition in chronic obstructive pulmonary disease: current approaches in nutrition therapy. Curr. Nutr. Rep. 14:21. doi: 10.1007/s13668-025-00613-8
Vardavas, C. I., Hohmann, C., Patelarou, E., Martinez, D., Henderson, A. J., Granell, R., et al. (2016). The independent role of prenatal and postnatal exposure to active and passive smoking on the development of early wheeze in children. Eur. Respir. J. 48, 115–124. doi: 10.1183/13993003.01016-2015
Venkataraman, A., Bassis, C. M., Beck, J. M., Young, V. B., Curtis, J. L., Huffnagle, G. B., et al. (2015). Application of a neutral community model to assess structuring of the human lung microbiome. MBio 6:e02284-14. doi: 10.1128/mBio.02284-14
Venter, C., Meyer, R. W., Greenhawt, M., Pali-Schöll, I., Nwaru, B., Roduit, C., et al. (2022). Role of dietary fiber in promoting immune health-an EAACI position paper. Allergy 77, 3185–3198. doi: 10.1111/all.15430
Wang, L., Cai, Y., Garssen, J., Henricks, P. A. J., Folkerts, G., and Braber, S. (2023). The bidirectional gut-lung Axis in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 207, 1145–1160. doi: 10.1164/rccm.202206-1066TR
Wang, N., Li, C., and Zhang, Z. (2024). Arctigenin ameliorates high-fat diet-induced metabolic disorders by reshaping gut microbiota and modulating GPR/HDAC3 and TLR4/NF-κB pathways. Phytomedicine 135:156123. doi: 10.1016/j.phymed.2024.156123
Wang, H., Liu, Y., Bai, C., and Leung, S. S. Y. (2024). Translating bacteriophage-derived depolymerases into antibacterial therapeutics: challenges and prospects. Acta Pharm. Sin. B 14, 155–169. doi: 10.1016/j.apsb.2023.08.017
Willner, D., Furlan, M., Haynes, M., Schmieder, R., Angly, F. E., Silva, J., et al. (2009). Metagenomic analysis of respiratory tract DNA viral communities in cystic fibrosis and non-cystic fibrosis individuals. PLoS One 4:e7370. doi: 10.1371/journal.pone.0007370
Xepapadaki, P., Papadopoulos, N. G., Bossios, A., Manoussakis, E., Manousakas, T., and Saxoni-Papageorgiou, P. (2005). Duration of postviral airway hyperresponsiveness in children with asthma: effect of atopy. J. Allergy Clin. Immunol. 116, 299–304. doi: 10.1016/j.jaci.2005.04.007
Ximenez, C., and Torres, J. (2017). Development of microbiota in infants and its role in maturation of gut mucosa and immune system. Arch. Med. Res. 48, 666–680. doi: 10.1016/j.arcmed.2017.11.007
Yan, T., Bao, Y., Cao, S., Jiang, P., Zhang, Z., Li, L., et al. (2024). The investigation of the role of oral-originated Prevotella-induced inflammation in childhood asthma. Front. Microbiol. 15:1400079. doi: 10.3389/fmicb.2024.1400079
Yatsunenko, T., Rey, F. E., Manary, M. J., Trehan, I., Dominguez-Bello, M. G., Contreras, M., et al. (2012). Human gut microbiome viewed across age and geography. Nature 486, 222–227. doi: 10.1038/nature11053
Ye, C., Chen, Q. Y., Yan, Y. M., Lv, X. Q., Ma, C. L., Li, N., et al. (2022). Establishment and preliminary clinical application of human intestinal fluid transplantation. Zhonghua wei chang wai ke za zhi = Chin. J. Gastr. Surg. 25, 819–825. doi: 10.3760/cma.j.cn441530-20220601-00239
Ye, C., Katagiri, S., Miyasaka, N., Kobayashi, H., Khemwong, T., Nagasawa, T., et al. (2020). The periodontopathic bacteria in placenta, saliva and subgingival plaque of threatened preterm labor and preterm low birth weight cases: a longitudinal study in Japanese pregnant women. Clin. Oral Investig. 24, 4261–4270. doi: 10.1007/s00784-020-03287-4
Yuan, H., Liu, Z., Dong, J., Bacharier, L. B., Jackson, D., Mauger, D., et al. (2023). The fungal microbiome of the upper airway is associated with future loss of asthma control and exacerbation among children with asthma. Chest 164, 302–313. doi: 10.1016/j.chest.2023.03.034
Zaiss, M. M., Rapin, A., Lebon, L., Dubey, L. K., Mosconi, I., Sarter, K., et al. (2015). The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43, 998–1010. doi: 10.1016/j.immuni.2015.09.012
Zetterström, O., and Johansson, S. G. (1981). IgE concentrations measured by PRIST in serum of healthy adults and in patients with respiratory allergy. A diagnostic approach. Allergy 36, 537–547. doi: 10.1111/j.1398-9995.1981.tb01871.x
Zhang, Q., Cox, M., Liang, Z., Brinkmann, F., Cardenas, P. A., Duff, R., et al. (2016). Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS One 11:e0152724. doi: 10.1371/journal.pone.0152724
Zhao, X., Hu, M., Zhou, H., Yang, Y., Shen, S., You, Y., et al. (2023). The role of gut microbiome in the complex relationship between respiratory tract infection and asthma. Front. Microbiol. 14:1219942. doi: 10.3389/fmicb.2023.1219942
Zhao, L., Li, B., Zhou, L., Song, C., Kang, T., Xu, Y., et al. (2023). PM(2.5) exposure promotes asthma in aged Brown-Norway rats: implication of multiomics analysis. Ecotoxicol. Environ. Saf. 263:115393. doi: 10.1016/j.ecoenv.2023.115393
Zheng, J., Huang, Y., Zhang, L., Liu, T., Zou, Y., He, L., et al. (2025). Role of the gut-lung microbiome Axis in airway inflammation in OVA-challenged mice and the effect of azithromycin. J. Inflamm. Res. 18, 2661–2676. doi: 10.2147/JIR.S506688
Keywords: respiratory microbiota, intestinal microbiota, microbiome, pediatric asthma, gut-lung axis
Citation: Liu L, Zhao W, Zhang H, Shang Y, Huang W and Cheng Q (2025) Relationship between pediatric asthma and respiratory microbiota, intestinal microbiota: a narrative review. Front. Microbiol. 16:1550783. doi: 10.3389/fmicb.2025.1550783
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Ignacio Ramos-Tapia, Universidad del Desarrollo, ChileYen-Chu Huang, Taichung Veterans General Hospital, Taiwan
Vinícius de Molla, Federal University of São Paulo, Brazil
Copyright © 2025 Liu, Zhao, Zhang, Shang, Huang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qi Cheng, cWNoZW5nQGNtdS5lZHUuY24=; Wanjie Huang, d2podWFuZ0BjbXUuZWR1LmNu
†These authors have contributed equally to this work
 Lian Liu
Lian Liu Wenqi Zhao
Wenqi Zhao Han Zhang1
Han Zhang1 Yunxiao Shang
Yunxiao Shang