- 1Department of Obstetrics and Gynecology, Shenzhen Luohu Maternity and Child Health Hospital, Shenzhen, China
- 2Department of Paediatrics, Shenzhen University General Hospital, Shenzhen, China
- 3Department of Geriatrics, Chongqing General Hospital, Chongqing University, Chongqing, China
- 4Department of Endocrinology and Metabolism, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, China
Background: Hypertensive disorders of pregnancy (HDP) pose significant risks to both maternal and fetal health and have been associated with alterations in the maternal gut microbiota. However, the impact of HDP on neonatal microbiota remains poorly understood. This study aimed to characterize the gut microbiota of pregnant women with HDP and evaluate its potential influence on the meconium microbiota of their newborns.
Methods: A cohort of 67 pregnant women, including 36 diagnosed with HDP (HDP group) and 31 healthy, age-matched controls (HC group), along with their offspring, were recruited. Fecal samples collected during the third trimester and meconium samples from the newborns were subjected to microbial community profiling via 16S rRNA gene sequencing.
Results: Principal coordinate analysis (PCoA) based on Bray-Curtis distances revealed significant differences in microbial community composition between the HDP and HC groups in both maternal and neonatal samples. Subgroup analyses, stratified by HDP severity and medication use, further delineated distinct microbial profiles relative to controls. Notably, both maternal and neonatal microbiota in the HDP group exhibited increased abundances of Enterobacter, Klebsiella, and Sphingomonas, coupled with a reduction in Acidovorax, Azospirillum, Caulobacter, Flavobacterium, Magnetospirillum, and Rubrivivax compared to the HC group. Moreover, the P4-PWY pathway, which is involved in the biosynthesis of L-lysine, L-threonine, and L-methionine, was differentially represented in both maternal and neonatal microbiota in the HDP group. These parallel patterns suggest an intergenerational concordance associated with HDP.
Conclusion: This study demonstrates significant alterations in the microbial communities of both maternal fecal and neonatal meconium samples in the context of HDP. The findings highlight the importance of further research to elucidate the long-term health implications of HDP-associated microbiota shifts on offspring.
Introduction
Hypertensive disorder of pregnancy (HDP) represent a prevalent and serious medical complication that significantly contributes to maternal morbidity and mortality (Ramos Filho and Antunes, 2020). HDP encompasses a spectrum of conditions, including gestational hypertension, preeclampsia/eclampsia, chronic hypertension, and chronic hypertension complicated with preeclampsia/eclampsia (Brown et al., 2018; Dines and Kattah, 2020). The global incidence of HDP is rising, posing an increasing public health challenge (Garovic et al., 2020). In China, HDP affects approximately 7.3% of all pregnancies (Li et al., 2021). Beyond its immediate consequences, such as preterm delivery, low birth weight, and neonatal care unit admission (Abdelazim et al., 2020), HDP has enduring impacts on both maternal and offspring health. These long-term effects include an increased risk of high blood pressure, coronary heart disease, impaired neonatal brain development, and systemic vascular dysfunction (Garovic et al., 2020; Kanata et al., 2021). Elucidating the underlying mechanisms of HDP and its association with offspring health outcomes is essential for advancing clinical management and developing preventive strategies that benefit both mothers and their children.
Recent investigations have linked HDP with various maternal and placental factors, including inflammatory response, endothelial dysfunction, and alterations in the gut microbiota (Beckers and Sones, 2020; Yu et al., 2022). In particular, dysbiosis in the maternal gut microbiota has been reported in HDP cases, characterized by reduced microbial diversity, diminished short-chain fatty acid-producing bacteria, and an enrichment of potentially pathogenic genera, such as Enterobacter and Klebsiella (Beckers and Sones, 2020; Chang et al., 2020). This dysbiosis is hypothesized to contribute to the onset and progression of HDP by disrupting maternal physiological homeostasis (Zhang et al., 2015; Liu et al., 2017; Beckers and Sones, 2020).
The initial establishment of the infant gut microbiota, which plays a pivotal role in long-term health, is also influenced by maternal factors. Variables such as antibiotic exposure, delivery mode, and feeding practices significantly shape the neonatal microbial composition (Singh and Mittal, 2020). The maternal microbial reservoir, including the gut, oral, skin, and vaginal microbiota, critically contributes to the infant’s initial microbial colonization (Ferretti et al., 2018). For instance, the association between maternal conditions such as gestational diabetes and alterations in the meconium microbiota is well-documented (Gosalbes et al., 2013; Hu et al., 2013; Wang et al., 2018; Chen et al., 2021; Sililas et al., 2021). Neonates born to mothers with gestational diabetes exhibit reduced alpha diversity and notable shifts in the abundance of Firmicutes and Proteobacteria (Chen et al., 2021).
Despite these advances, the potential impact of HDP on the neonatal gut microbiota remains insufficiently explored. This gap in knowledge limits our understanding of the interplay between maternal HDP and the development of the offspring’s gut microbiota. Based on these considerations, we hypothesize that maternal HDP alters the neonatal meconium microbiota, contributing to microbial dysbiosis with potential long-term health implications for the offspring. Therefore, this study aims to comprehensively characterize the meconium microbiota of neonates born to mothers with HDP and elucidate the relationship between maternal HDP and alterations in the neonatal gut microbiota, thereby providing novel insights into the mechanisms by which HDP may affect offspring health.
Materials and methods
Study subjects
The study protocol was approved by the Ethical Committee of Shenzhen Luohu Maternity and Child Health Hospital. Written informed consent was obtained from all participants, and for neonates, consent was provided by their parents or legal guardians. Between April to November 2020, pregnant women in their third trimester who planned to deliver at our hospital and agreed to provide fecal samples were recruited. Exclusion criteria included pregnancy complications (e.g., gestational diabetes, chronic hypertension, intrahepatic cholestasis of pregnancy, premature delivery, or premature rupture of membrane), multiple gestations, antibiotic treatment within 1 month prior to sample collection, a history of smoking or alcohol consumption during pregnancy, or probiotic supplementation. Neonates with significant congenital anomalies, neurological dysfunction, chromosomal abnormalities, or metabolic diseases were also excluded. A total of 67 singleton pregnant women with full-term births were included in the final analysis. Of these, 36 women diagnosed with hypertensive disorders of pregnancy (HDP) were assigned to the HDP group, which comprised 23 with preeclampsia (PE subgroup) and 13 with gestational hypertension (GH subgroup). The remaining 31 normotensive women without complications served as the healthy control (HC) group. Diagnostic criteria for GH included a systolic blood pressure ≥ 140 mmHg and/or a diastolic blood pressure ≥ 90 mmHg, whereas PE was defined as a systolic blood pressure ≥ 140 mmHg and/or a diastolic blood pressure ≥ 90 mmHg in conjunction with proteinuria exceeding > 0.3 g/24 h after 20 weeks of gestation. Clinical information were recorded and verified by trained clinicians following standard procedures.
Sample collection
Fecal samples from pregnant women were collected during the third trimester, approximately 3 days before labor, using sterile swabs under the guidance of trained nursing staff. Meconium samples from the neonates were obtained within the first hours after birth using sterile swabs by experienced nurses in the labor ward. Approximately 200 mg of meconium was carefully collected directly from the interior of soiled diapers to ensure representative sampling and minimize potential contamination from external components (Chen et al., 2021). All samples were immediately transferred into sterile tubes, stored at −80°C to preserve their integrity, and subsequently processed for further analysis.
DNA extraction and 16S rRNA sequencing
Genomic DNA was extracted from both fecal and meconium samples using the QIAamp Fast DNA Stool Mini kit, following the manufacturer’s protocol. The V4 hypervariable region of the 16S rRNA gene was amplified by PCR using primers 515F and 806R, which included unique barcodes for sample identification (Kozich et al., 2013). PCR amplification was performed with the Phusion High-Fidelity PCR Master Mix, and the amplified products were pooled in equimolar concentrations and purified using the Qiagen Gel Extraction kit. Sequencing libraries were prepared with the TruSeq DNA PCR-Free Sample Preparation kit, and their quality was assessed using a Qubit@2.0 Fluorometer and Agilent Bioanalyzer 2,100. Libraries were sequenced on an Illumina HiSeq 2,500 platform (V3 chemistry) at Novogene Bioinformatics Technology Co., Ltd., generating 250-bp paired-end reads.
Bioinformatics and statistical analyses
Raw16S rRNA gene sequences were processed using QIIME2 (Bolyen et al., 2019). Paired-end reads were assembled and denoised with the DADA2 algorithm (Callahan et al., 2016). Taxonomic assignments were performed against the GreenGenes database using the Naïve Bayes classifier in QIIME2. Alpha and beta diversity metrics were calculated using QIIME2’s core diversity tools, and functional metabolic pathways were predicted using PICRUSt2.0 (Douglas et al., 2020).
Microbial community dissimilarities were evaluated through principal coordinate analysis (PCoA) based on Bray-Curtis distances, and statistical significance was assessed using permutational multivariate analysis of variance (PERMANOVA) implemented in the R package MicrobiotaProcess (Xu et al., 2023). Differentially abundant taxa were identified using LEfSe, with a linear discriminant analysis (LDA) score > 2.0 and p < 0.05 (Segata et al., 2011). Pathway differences between the HDP and HC groups were analyzed using STAMP with Bonferroni correction (Parks et al., 2014).
Clinical characteristics were summarized as means and standard deviations for continuous variables and proportions for categorical variables. Between-group differences were assessed using t-tests for continuous variables and chi-square tests for categorical variables. All statistical analyses were performed using R software (version 3.6.3), with significance set at p < 0.05.
Results
Characteristics of study participants
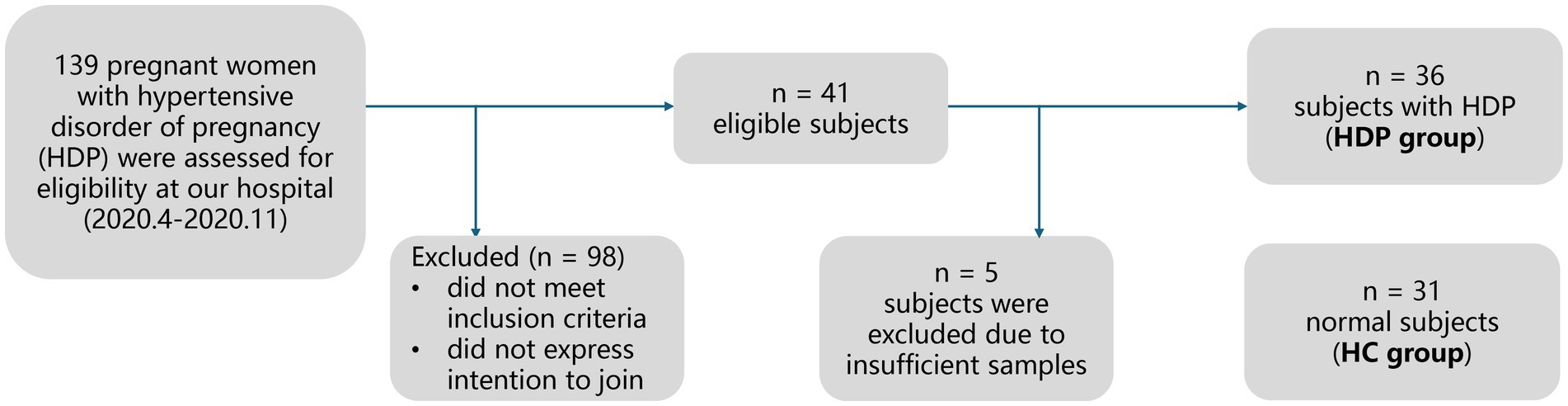
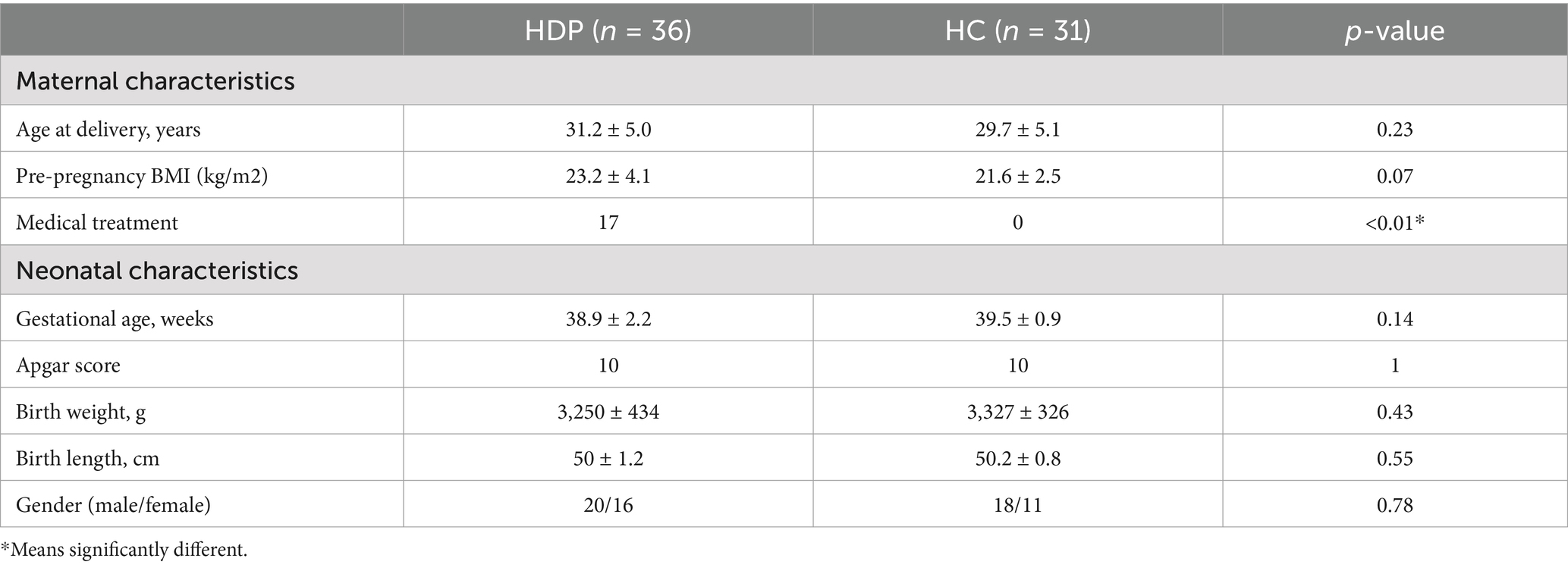
Figure 1 illustrates the recruitment and exclusion process. A total of 67 full-term neonate-mother pairs were included, comprising 36 pairs in the HDP group, and 31 pairs in the HC group. All participants were Han Chinese residing in Shenzhen, China. Table 1 summarizes the sociodemographic and clinical characteristics. There were no significant differences between the HDP and HC groups in terms of gestational age, birth weight, birth length, neonatal gender, maternal age, or maternal pre-pregnancy BMI. Among the mothers in the HDP group, 14 received labetalol, two were treated with nicardipine, and one received nifedipine during pregnancy.
Comparison of neonatal meconium microbiota between the HDP and HC groups
Sequencing of neonatal meconium samples yielded a total of 6,634,295 paired end reads, with an average of 96,128 per sample for the HDP group and 102,376 reads per sample for the HC group. And a total of 16,190 amplicon sequence variants were identified. Rarefaction analysis indicated that all samples reached an asymptote, suggesting that sufficient sequencing depth was achieved (data not shown).
Alpha diversity indices, including observed feature number, Pielou index, Chao1 index, Simpson index and Shannon index, were compared between the HDP and HC groups. While no significant differences were observed in the observed feature number, Chao1, Simpson and Shannon indices, the Pielou index was significantly lower in the HDP group compared to the HC group (p = 0.044; Figure 2a). Moreover, principal coordinate analysis based on Bray-Curtis distances revealed a distinct separation between the HDP and HC groups (R2 = 0.149, p = 0.0001; Figure 2b).
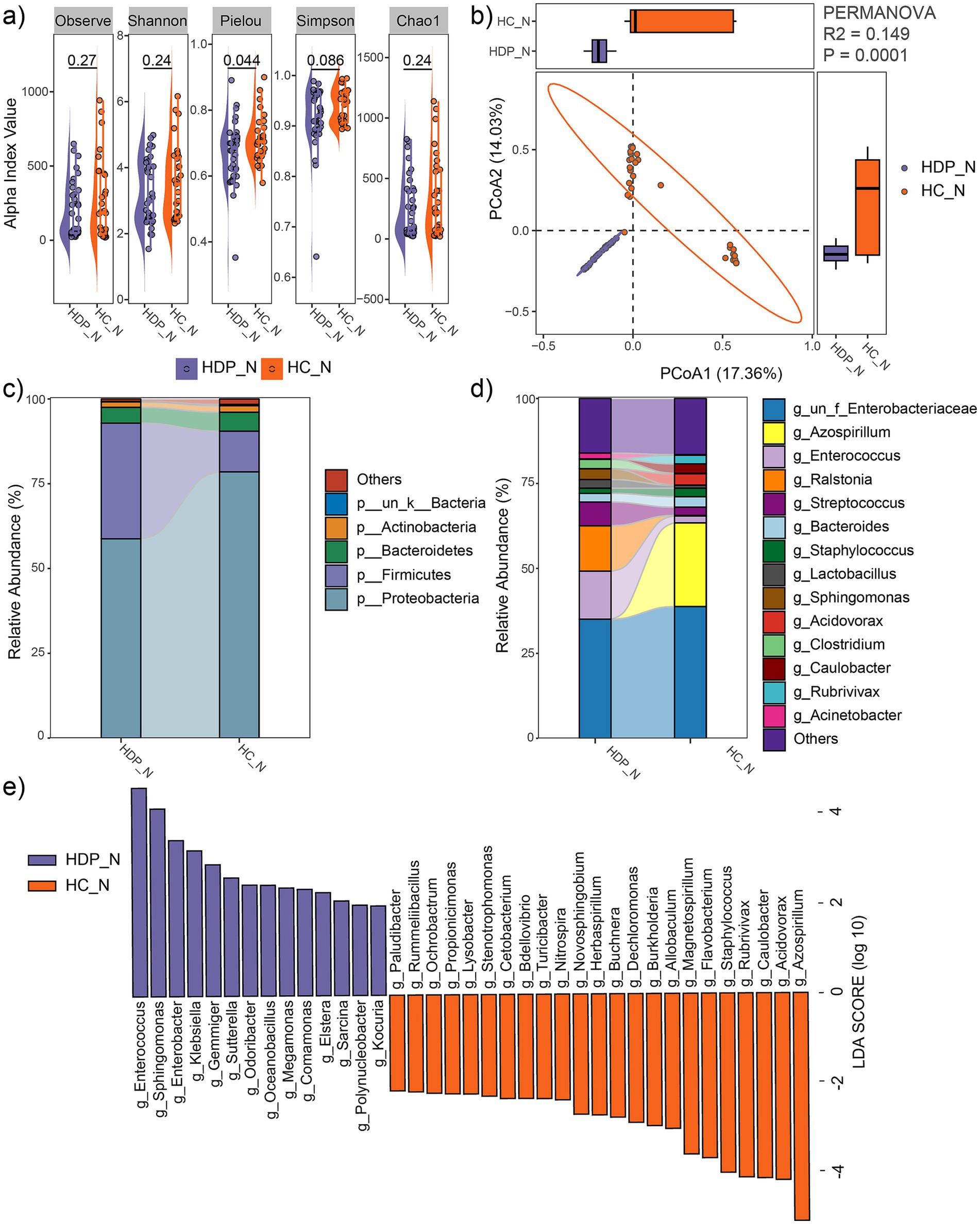
Figure 2. Comparison of neonatal meconium microbial communities between the HDP (HDP_N) and HC (HC_N) groups. (a) Comparisons of alpha diversity indices using two-tailed Student’s t-test. (b) PCoA plot illustrating differences in microbial community composition between groups, as assessed by PERMANOVA. (c) Relative abundances of the dominant phyla. (d) Relative abundances of the predominant genera. (e) Genera with significantly different relative abundances between the HDP_N and HC_N groups, identified using LEfSe software with an LDA score > 2.0 and p < 0.05.
At the phylum level, the predominant taxa in the neonatal meconium were Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria (Figure 2c), with Firmicutes being significantly more abundant in the HDP group (LDA = 4.96, p = 0.001). Notably, the combined relative abundances of Firmicutes and Proteobacteria exceeded 90% in both groups. At the genus level, the dominant taxa included Azospirillum, Enterococcus, Ralstonia, Streptococcus, Bacteroides, Staphylococcus, Lactobacillus, Sphingomonas, Acidovorax, Clostridium, Caulobacter, Rubrivivax, and Acinetobacter (Figure 2d). LEfSe analysis revealed significant differences in microbial composition between the groups. The HDP group showed higher relative abundances of Enterococcus, Sphingomonas, Enterobacter, Klebsiella, Gemmiger, Sutterella, Odoribacter, Megamonas, Comamonas, Elstera, Sarcina, Polynucleobacter, and Kocuria. In contrast, lower relative abundances in the HDP group were observed for Azospirillum, Acidovorax, Caulobacter, Rubrivivax, Staphylococcus, Flavobacterium, Magnetospirillum, Allobaculum, Burkholderia, Dechloromonas, Buchnera, Herbaspirillum, Novosphingobium, Nitrospira, Turicibacter, Bdellovibrio, Cetobacterium, Stenotrophomonas, Lysobacter, Propionicimonas, Ochrobactrum, Rummeliibacillus, and Paludibacter (Figure 2e).
Comparison of maternal fecal microbial community between the HDP and HC groups
Sequencing of maternal fecal samples generated a total of 6,135,597 paired end reads, with an average of 101,412 reads per sample in the HDP group and 80,153 reads per sample in the HC group. A total of 12,839 amplicon sequence variants were identified and rarefaction analysis confirmed that all samples reached an asymptote, indicating sufficient sequencing depth (data not shown).
No significant differences in alpha diversity metrics were observed between the HDP and HC groups (Figure 3a). However, PCoA based on Bray-Curtis distances demonstrated a significant separation between the two groups (R2 = 0.133, p = 0.0001; Figure 3b).
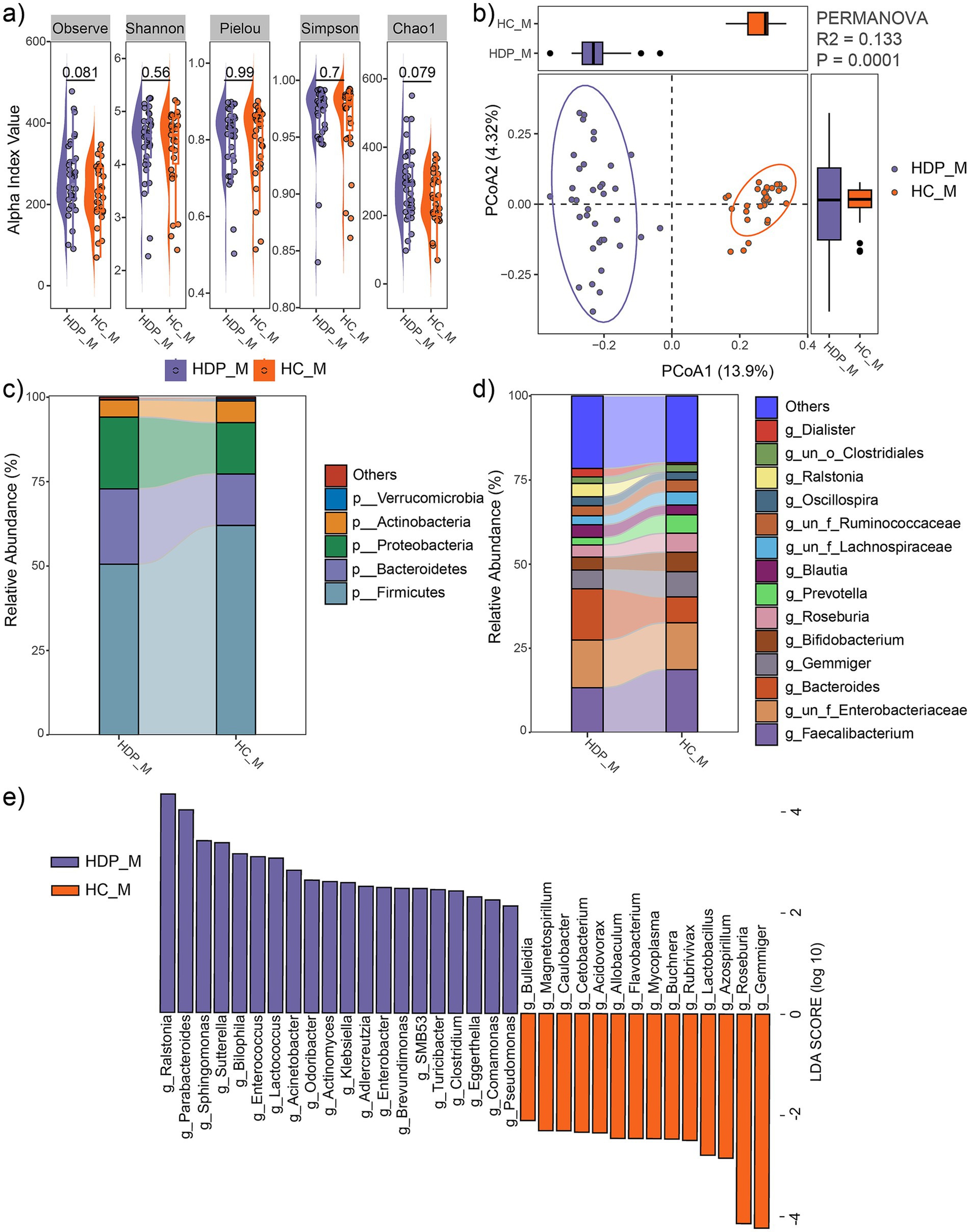
Figure 3. Comparison of maternal fecal microbial communities between the HDP (HDP_M) and HC (HDP_M) groups. (a) Comparisons of alpha diversity indices using t-tests. (b) PCoA plot illustrating the differences in microbial community composition between the two groups, as determined by PERMANOVA. (c) Relative abundances of the dominant phyla. (d) Relative abundances of the predominant genera. (e) Genera with significantly different relative abundances between the HDP_M and HC_M groups, as determined by LEfSe (LDA score > 2.0 and p < 0.05).
At the phylum level, the maternal fecal microbiota was predominantly composed of Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria (Figure 3c), with Proteobacteria being significantly more abundant in the HDP group (LDA = 4.65, p = 0.0064). At the genus level, dominant taxa included Faecalibacterium, Bacteroides, Gemmiger, Bifidobacterium, Roseburia, Prevotella, Blautia, Oscillospira, Ralstonia, and Dialister (Figure 3d). LEfSe analysis further revealed significant differences in microbial composition between the groups. In the HDP group, genera such as Ralstonia, Parabacteroides, Sphingomonas, Sutterella, Bilophila, Enterococcus, Lactococcus, Acinetobacter, Odoribacter, Actinomyces, Klebsiella, Adlercreutzia, Enterobacter, Brevundimonas, SMB53, Turicibacter, Clostridium, Eggerthella, Comamonas, and Pseudomonas were significantly enriched, whereas Bulleidia, Magnetospirillum, Caulobacter, Cetobacterium, Acidovorax, Allobaculum, Flavobacterium, Mycoplasma, Buchnera, Rubrivivax, Lactobacillus, Azospirillum, Roseburia, and Gemmiger were significantly depleted (Figure 3e).
Notably, an intergenerational concordance was evident, as genera such as Enterococcus, Sphingomonas, Enterobacter, Klebsiella, Sutterella, Odoribacter, and Comamonas were significantly increased in both the maternal fecal and neonatal meconium microbiota of the HDP group, while Azospirillum, Acidovorax, Caulobacter, Rubrivivax, Flavobacterium, Magnetospirillum, Allobaculum, Buchnera, Turicibacter, and Cetobacterium were significantly decreased in both compartments.
Comparison of functionally predicted metabolic pathways between the HDP and HC groups
Functional annotation was performed using PICRUSt2.0 against the MetaCyc database to assess the metabolic potential of the maternal fecal and neonatal meconium microbiota associated with HDP. In maternal fecal samples, 11 metabolic pathways exhibited significant differences between the HDP and HC groups (Figure 4a), while 22 metabolic pathways were significantly altered in the neonatal meconium microbiota (Figure 4b). Notably, the “superpathway of L-lysine, L-threonine and L-methionine biosynthesis I (P4-PWY)” and “Kdo transfer to lipid IVA III (Chlamydia; PWY-6467)” pathways were significantly altered in both maternal and neonatal samples, suggesting shared functional impairments linked to HDP.
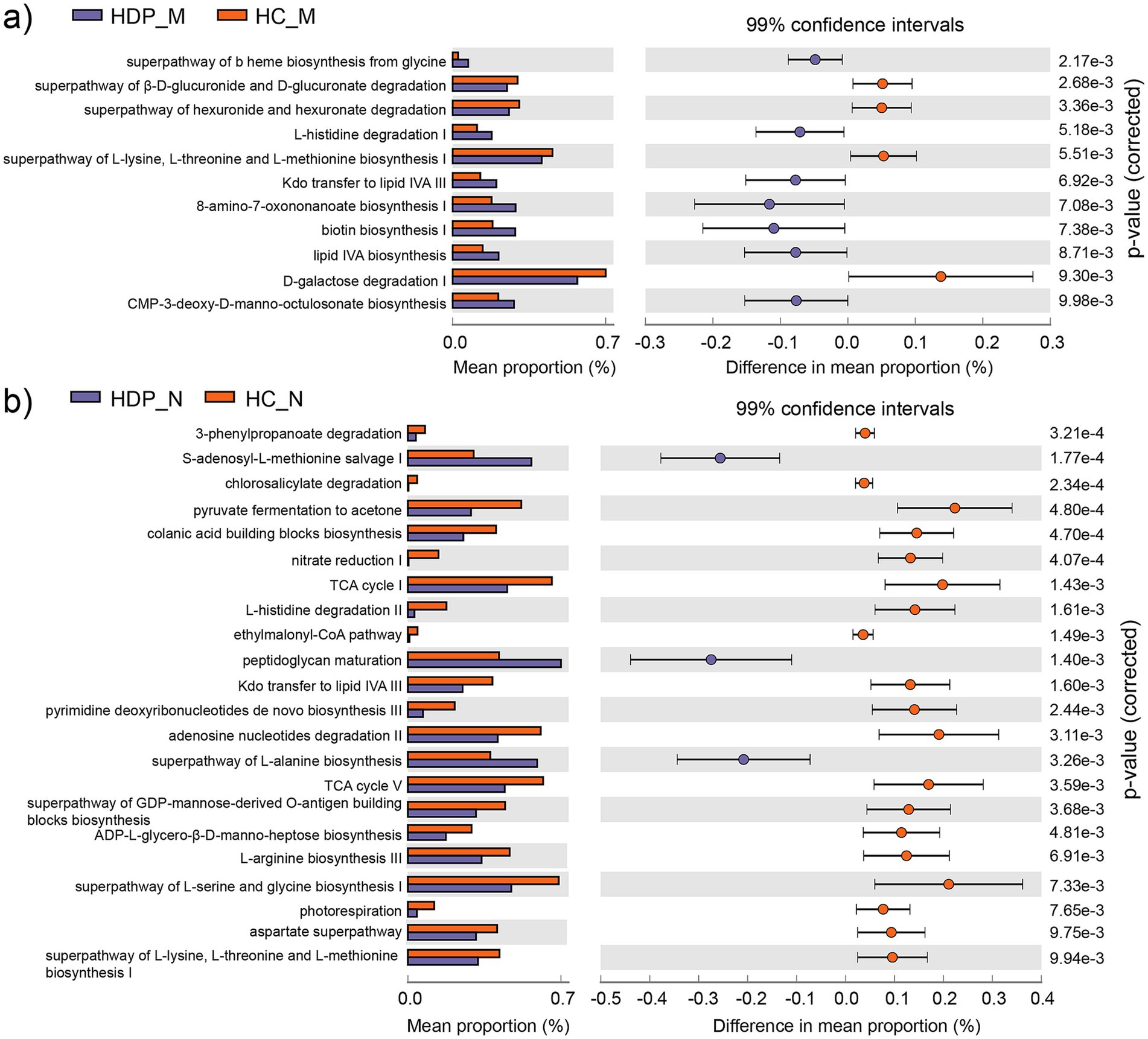
Figure 4. Differentially abundant metabolic pathways predicted by PICRUSt2 between the HDP and HC groups. (a) Differential metabolic pathways identified in the maternal gut microbiota between the HDP_M and HC_M groups with adjusted p value less than 0.01. (b) Differential metabolic pathways identified in the neonatal meconium microbiota between the HDP_N and HC_N groups with adjusted p value less than 0.01.
Comparison of microbial community between the sub-HDP groups and HC group
Based on clinical data, the HDP group was subdivided into the gestational hypertension (GH, n = 13) and preeclampsia (PE, n = 23) subgroups. PCoA with PERMANOVA revealed that the maternal fecal microbial community of the PE and GH subgroups were clustered closely (R2 = 0.03, p = 0.54), as did their neonatal meconium microbiota (R2 = 0.02, p = 0.85). However, when comparing the PE, GH, and HC groups, significant differences in microbial composition were observed. The maternal fecal microbial community showed marked separation among these groups (R2 = 0.147, p = 0.0001; Figure 5a), with similar findings in the neonatal meconium microbiota (R2 = 0.159, p = 0.0001; Figure 5b). Furthermore, subgroup analysis based on medication use confirmed significant variation between the HDP and HC groups for both maternal microbiota (R2 = 0.145, p = 0.0001; Figure 5c) and neonatal microbiota (R2 = 0.161, p = 0.0001; Figure 5d). Notably, within the HDP group, no significant differences were observed in microbial composition between mothers treated with antihypertensive drugs (labetalol, nicardipine, or nifedipine) and those who were untreated (R2 = 0.022, p = 0.646). These findings underscore the distinct microbial profiles associated with HDP compared to healthy controls.
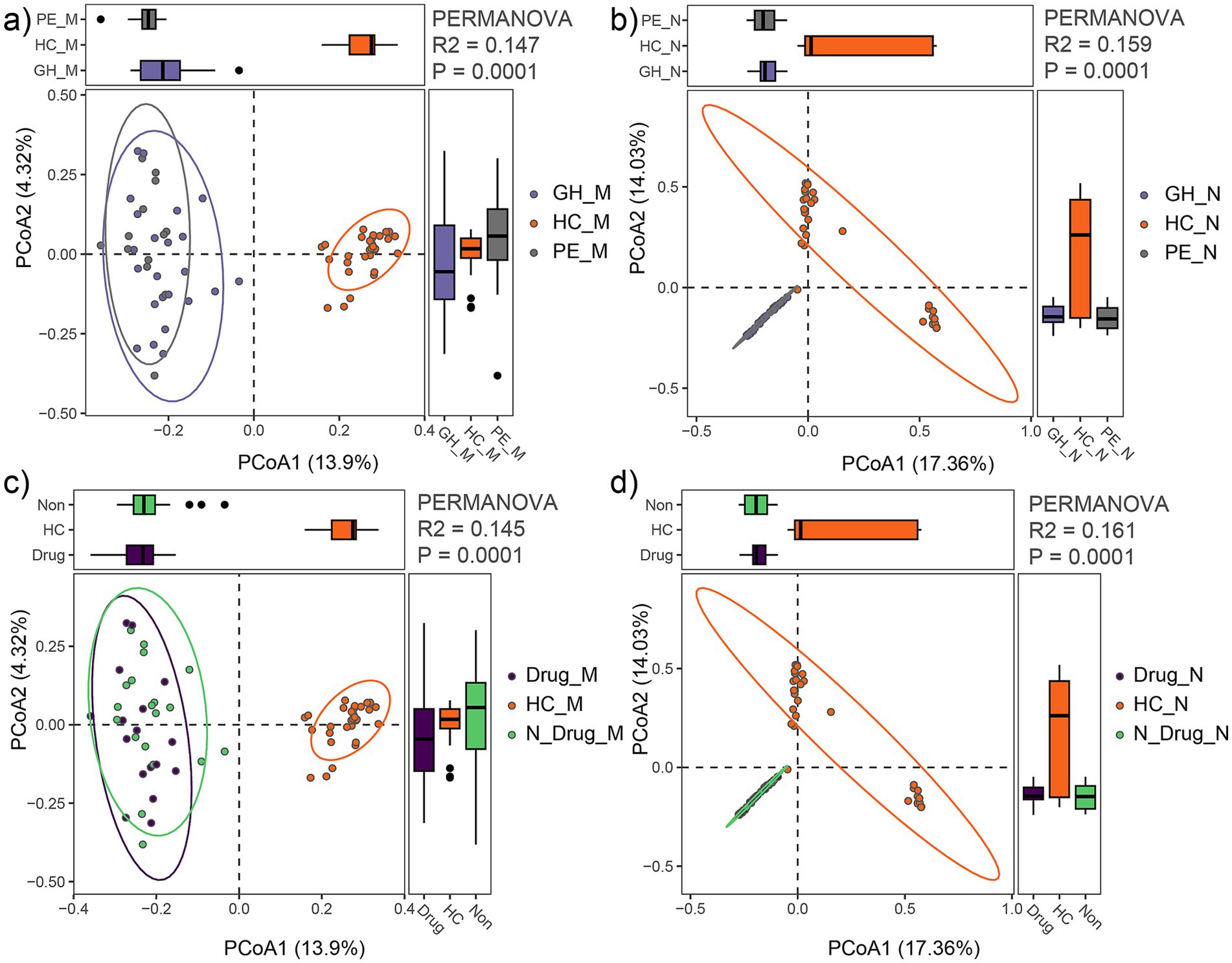
Figure 5. Comparison of gut microbial communities among HDP subgroups (GH, PE) and HC group. (a) PCoA plot showing differences in maternal fecal microbial communities among the GH_M, PE_M and HC_M groups. (b) PCoA plot showing differences in neonatal meconium microbial communities among the GH_N, PE_N and HC_N groups. (c) PCoA plot illustrating differences in maternal fecal microbial communities among mothers receiving drug treatment (Drug_M), those not receiving drug treatment (N_Drug_M), and HC (HC_M) groups. (d) PCoA plot illustrating differences in neonatal meconium microbial communities among neonates born to mothers receiving drug treatment (Drug_N), those not receiving drug treatment (N_Drug_N), and HC (HC_N) groups.
Discussion
In this study, we investigated the neonatal meconium microbiota in relation to the maternal fecal microbial community among mothers with HDP using high throughput 16S rRNA sequencing. Our analysis revealed that the meconium microbiota was predominantly composed of the phyla Proteobacteria and Firmicutes, consistent with previous reports (Bäckhed et al., 2015; Turunen et al., 2021). In contrast, the maternal fecal microbial community was mainly comprised of Firmicutes and Bacteroidetes, in agreement with earlier findings (Yu et al., 2022).
Principal coordinate analysis based on Bray-Curtis distances demonstrated a significant separation between the HDP and HC groups in the neonatal meconium microbiota. Moreover, we observed marked alterations in the maternal fecal microbial community of women with HDP, which corroborates previous studies (Gomez-Arango et al., 2016; Chang et al., 2020). These differences persisted across subgroups stratified by disease severity and medication use during pregnancy. Specifically, genera such as Klebsiella, Enterobacter, and Sphingomonas were significantly enriched in both the maternal and neonatal microbiota of the HDP group, whereas Acidovorax, Azospirillum, Caulobacter, Flavobacterium, Magnetospirillum, and Rubrivivax were more abundant in the HC group. Notably, previous research has associated certain species of Klebsiella (You et al., 2020; Mukherjee et al., 2023), Enterobacter (Ferry et al., 2020; Karambin and Zarkesh, 2011), and Sphingomonas (Mutlu et al., 2011; Bayram et al., 2013; Ranjan et al., 2016) with neonatal infection. Furthermore, a decrease in Azospirillum has been linked to helminth infections in preschool-aged children (Osakunor et al., 2020), while genera such as Acidovorax, Caulobacter, Flavobacterium, and Magnetospirillum have been detected in the gut microbiota (Simon et al., 2021; Dornelles et al., 2022; Xu et al., 2022). Increased abundance of Enterobacter and Klebsiella have also been reported in individuals with PE (Beckers and Sones, 2020; Chang et al., 2020). Both genera are known to secrete lipopolysaccharides, potent activators of the immune system that can elicit strong inflammatory responses (Holt et al., 2020). Such immune activation may contribute to an elevated risk of preterm labor and other complications. Although these microbial alterations may adversely affect neonatal health and development, the specific roles of these taxa in neonates born to mothers with HDP require further investigation.
Additionally, previous work by de Agüero MG et al. identified a high abundance of Lactobacillus spp. in the placental microbiota of healthy term deliveries (Gomez De Agüero et al., 2016), suggesting a beneficial role for Lactobacillus in pregnancy outcomes (Olaniyi et al., 2020). Consistent with these observations, our study found a significantly lower level of Lactobacillus in the maternal fecal microbial community of the HDP group compared to controls. Furthermore, Roseburia, an abundant genera associated with several diseases such as obesity, type 2 diabetes, nervous system conditions and allergies (Tamanai-Shacoori et al., 2017), was significantly reduced in the HDP group. Roseburia is a key producer of short-chain fatty acids, particularly butyrate, which is crucial for maintaining colonic motility, immunity function and anti-inflammatory responses (Tamanai-Shacoori et al., 2017; Yan et al., 2017; Calderón-Pérez et al., 2020). These findings suggest that reduced colonization by lactic acid and short-chain fatty acids producing bacteria may contribute to adverse pregnancy outcomes. However, larger prospective studies are needed to confirm this hypothesis.
Moreover, functional prediction analysis revealed that the abundance of the P4-PWY (superpathway of L-lysine, L-threonine and L-methionine biosynthesis I) was significantly lower in both the neonatal and maternal microbiota of the HDP group compared to the HC group. Considering the essential roles of L-lysine in protein synthesis and neonatal growth (van der Schoor et al., 2004), as well as the importance of maternal methionine supply for offspring development (Alharthi et al., 2018; Elolimy et al., 2019), these alterations in key metabolic pathways may have significant implications for neonatal health.
Despite these important insights, several limitations warrant consideration. The relatively small sample size and recruitment from a single geographic area may limit the generalizability of the findings. Additionally, the absence of long-term clinical follow-up data restricts our ability to assess the direct impact of gut microbial alterations on neonatal health outcomes. Future studies employing metagenomic sequencing, longitudinal follow-up, and animal models are necessary to elucidate the mechanistic links between HDP-related microbial dysbiosis and neonatal development. Such efforts will enhance the robustness and applicability of these findings and pave the way for more insightful research in this critical area.
In conclusion, our study demonstrates significant alterations in both the neonatal meconium microbiota and maternal fecal microbial community in the context of hypertensive disorders of pregnancy. These findings advance our understanding of the impact of HDP on neonatal gut microbiota. Further research is necessary to explore the potential implications of these microbial alterations on neonatal health outcomes and to develop strategies for modulating the gut microbiota in this vulnerable population.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://db.cngb.org/search/project/CNP0003728/, CNP0003728.
Ethics statement
The studies involving humans were approved by Ethical Committee of Shenzhen Luohu Maternity and Child Health Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin.
Author contributions
HY: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Resources, Writing – original draft, Writing – review & editing. ZH: Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing, Validation. JL: Methodology, Project administration, Writing – review & editing, Investigation. JY: Methodology, Project administration, Writing – review & editing, Investigation. QH: Methodology, Project administration, Writing – review & editing, Investigation. YC: Data curation, Formal analysis, Writing – review & editing, Funding acquisition. MT: Formal analysis, Methodology, Project administration, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. HH: Conceptualization, Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the Key Discipline of Luohu Obstetric Department (No. 2020005), Natural Science of Foundation of Chongqing (No. cstc2021jcyj-msxmX0517). The funder had no role in study design, data collection, analysis and interpretations, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank all the study subjects for their participation.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelazim, I. A., Bekmukhambetov, Y., Aringazina, R., Shikanova, S., Amer, O. O., Zhurabekova, G., et al. (2020). The outcome of hypertensive disorders with pregnancy. J. Family Med. Prim. Care 9, 1678–1683. doi: 10.4103/jfmpc.jfmpc_1054_19
Alharthi, A. S., Batistel, F., Abdelmegeid, M. K., Lascano, G., Parys, C., Helmbrecht, A., et al. (2018). Maternal supply of methionine during late-pregnancy enhances rate of Holstein calf development in utero and postnatal growth to a greater extent than colostrum source. J Anim Sci Biotechnol 9:83. doi: 10.1186/s40104-018-0298-1
Bäckhed, F., Roswall, J., Peng, Y., Feng, Q., Jia, H., Kovatcheva-Datchary, P., et al. (2015). Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703. doi: 10.1016/j.chom.2015.04.004
Bayram, N., Devrim, I., Apa, H., Gülfidan, G., Türkyılmaz, H. N., and Günay, I. (2013). Sphingomonas paucimobilis infections in children: 24 case reports. Mediterr J Hematol Infect Dis 5:e2013040. doi: 10.4084/MJHID.2013.040
Beckers, K. F., and Sones, J. L. (2020). Maternal microbiome and the hypertensive disorder of pregnancy, preeclampsia. Am. J. Phys. Heart Circ. Phys. 318, H1–H10. doi: 10.1152/ajpheart.00469.2019
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Brown, M. A., Magee, L. A., Kenny, L. C., Karumanchi, S. A., McCarthy, F. P., Saito, S., et al. (2018). Hypertensive disorders of pregnancy: ISSHP classification, diagnosis, and management recommendations for international practice. Hypertension 72, 24–43. doi: 10.1161/HYPERTENSIONAHA.117.10803
Calderón-Pérez, L., Gosalbes, M. J., Yuste, S., Valls, R. M., Pedret, A., Llauradó, E., et al. (2020). Gut metagenomic and short chain fatty acids signature in hypertension: a cross-sectional study. Sci. Rep. 10:6436. doi: 10.1038/s41598-020-63475-w
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chang, Y., Chen, Y., Zhou, Q., Wang, C., Chen, L., Di, W., et al. (2020). Short-chain fatty acids accompanying changes in the gut microbiome contribute to the development of hypertension in patients with preeclampsia. Clin. Sci. 134, 289–302. doi: 10.1042/CS20191253
Chen, T., Qin, Y., Chen, M., Zhang, Y., Wang, X., Dong, T., et al. (2021). Gestational diabetes mellitus is associated with the neonatal gut microbiota and metabolome. BMC Med. 19:120. doi: 10.1186/s12916-021-01991-w
Dines, V., and Kattah, A. (2020). Hypertensive disorders of pregnancy. Adv. Chronic Kidney Dis. 27, 531–539. doi: 10.1053/j.ackd.2020.05.006
Dornelles, L. V., Procianoy, R. S., Roesch, L. F. W., Corso, A. L., Dobbler, P. T., Mai, V., et al. (2022). Meconium microbiota predicts clinical early-onset neonatal sepsis in preterm neonates. J. Matern. Fetal Neonatal Med. 35, 1935–1943. doi: 10.1080/14767058.2020.1774870
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Elolimy, A., Alharthi, A., Zeineldin, M., Parys, C., Helmbrecht, A., and Loor, J. J. (2019). Supply of methionine during late-pregnancy alters fecal microbiota and metabolome in neonatal dairy calves without changes in daily feed intake. Front. Microbiol. 10:2159. doi: 10.3389/fmicb.2019.02159
Ferretti, P., Pasolli, E., Tett, A., Asnicar, F., Gorfer, V., Fedi, S., et al. (2018). Mother-to-infant microbial transmission from different body sites shapes the developing infant gut microbiome. Cell Host Microbe 24, 133–145.e5. doi: 10.1016/j.chom.2018.06.005
Ferry, A., Plaisant, F., Ginevra, C., Dumont, Y., Grando, J., Claris, O., et al. (2020). Enterobacter cloacae colonisation and infection in a neonatal intensive care unit: retrospective investigation of preventive measures implemented after a multiclonal outbreak. BMC Infect. Dis. 20:682. doi: 10.1186/s12879-020-05406-8
Garovic, V. D., White, W. M., Vaughan, L., Saiki, M., Parashuram, S., Garcia-Valencia, O., et al. (2020). Incidence and long-term outcomes of hypertensive disorders of pregnancy. J. Am. Coll. Cardiol. 75, 2323–2334. doi: 10.1016/j.jacc.2020.03.028
Gomez De Agüero, M., Ganal-Vonarburg, S. C., Fuhrer, T., Rupp, S., Uchimura, Y., Li, H., et al. (2016). The maternal microbiota drives early postnatal innate immune development. Science 351, 1296–1302. doi: 10.1126/science.aad2571
Gomez-Arango, L. F., Barrett, H. L., McIntyre, H. D., Callaway, L. K., Morrison, M., Nitert, M. D., et al. (2016). Increased systolic and diastolic blood pressure is associated with altered gut microbiota composition and butyrate production in early pregnancy. Hypertension 68, 974–981. Available at: https://pubmed.ncbi.nlm.nih.gov/27528065/
Gosalbes, M. J., Llop, S., Vallès, Y., Moya, A., Ballester, F., and Francino, M. P. (2013). Meconium microbiota types dominated by lactic acid or enteric bacteria are differentially associated with maternal eczema and respiratory problems in infants. Clin. Exp. Allergy 43, 198–211. doi: 10.1111/cea.12063
Holt, K. E., Lassalle, F., Wyres, K. L., Wick, R., and Mostowy, R. J. (2020). Diversity and evolution of surface polysaccharide synthesis loci in Enterobacteriales. ISME J. 14, 1713–1730. doi: 10.1038/s41396-020-0628-0
Hu, J., Nomura, Y., Bashir, A., Fernandez-Hernandez, H., Itzkowitz, S., Pei, Z., et al. (2013). Diversified microbiota of meconium is affected by maternal diabetes status. PLoS One 8:e78257. doi: 10.1371/journal.pone.0078257
Kanata, M., Liazou, E., Chainoglou, A., Kotsis, V., and Stabouli, S. (2021). Clinical outcomes of hypertensive disorders in pregnancy in the offspring during perinatal period, childhood, and adolescence. J. Hum. Hypertens. 35, 1063–1073. doi: 10.1038/s41371-021-00550-3
Karambin, M., and Zarkesh, M. (2011). Entrobacter, the most common pathogen of neonatal septicemia in Rasht, Iran. Iran. J. Pediatr. 21, 83–87. Available at: https://pubmed.ncbi.nlm.nih.gov/23056769/
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Li, F., Qin, J., Zhang, S., and Chen, L. (2021). Prevalence of hypertensive disorders in pregnancy in China: a systematic review and meta-analysis. Pregnancy Hypertension 24, 13–21. doi: 10.1016/j.preghy.2021.02.001
Liu, J., Yang, H., Yin, Z., Jiang, X., Zhong, H., Qiu, D., et al. (2017). Remodeling of the gut microbiota and structural shifts in preeclampsia patients in South China. Eur. J. Clin. Microbiol. Infect. Dis. 36, 713–719. doi: 10.1007/s10096-016-2853-z
Mukherjee, S., Bhadury, P., Mitra, S., Naha, S., Saha, B., Dutta, S., et al. (2023). Hypervirulent Klebsiella pneumoniae causing neonatal bloodstream infections: emergence of NDM-1-producing Hypervirulent ST11-K2 and ST15-K54 strains possessing pLVPK-associated markers. Microbiol Spectr 11:e0412122. doi: 10.1128/spectrum.04121-22
Mutlu, M., Bayramoglu, G., Yilmaz, G., Saygin, B., and Aslan, Y. (2011). Outbreak of Sphingomonas paucimobilis septicemia in a neonatal intensive care unit. Indian Pediatr. 48, 723–725. doi: 10.1007/s13312-011-0114-x
Olaniyi, K. S., Moodley, J., Mahabeer, Y., and Mackraj, I. (2020). Placental microbial colonization and its association with pre-eclampsia. Front. Cell. Infect. Microbiol. 10:413. doi: 10.3389/fcimb.2020.00413
Osakunor, D. N. M., Munk, P., Mduluza, T., Petersen, T. N., Brinch, C., Ivens, A., et al. (2020). The gut microbiome but not the resistome is associated with urogenital schistosomiasis in preschool-aged children. Commun Biol 3:155. doi: 10.1038/s42003-020-0859-7
Parks, D. H., Tyson, G. W., Hugenholtz, P., and Beiko, R. G. (2014). STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi: 10.1093/bioinformatics/btu494
Ramos Filho, F. L., and Antunes, C. M. D. F. (2020). Hypertensive disorders: prevalence, perinatal outcomes and cesarean section rates in pregnant women hospitalized for delivery. Rev. Bras. Ginecol. Obstet. 42, 690–696. doi: 10.1055/s-0040-1714134
Ranjan, R., Pandey, A., Kumar, R., and Chowdhary, P. (2016). Sphingomonas paucimobilis septicemia in a neonate: a rare case report. Indian J. Pathol. Microbiol. 59, 119–21. Available at: https://pubmed.ncbi.nlm.nih.gov/26960657/
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. Avaialble at: https://pubmed.ncbi.nlm.nih.gov/21702898/
Sililas, P., Huang, L., Thonusin, C., Luewan, S., Chattipakorn, N., Chattipakorn, S., et al. (2021). Association between gut microbiota and development of gestational diabetes mellitus. Microorganisms 9:1686. doi: 10.3390/microorganisms9081686
Simon, R. A., Ranasinghe, P. D., Barazanji, N., Jungeström, M. B., Xu, J., Bednarska, O., et al. (2021). Magnetotactic bacteria from the human gut microbiome associated with orientation and navigation regions of the brain. J. Oceanol. Limnol. 39, 2044–2052. doi: 10.1007/s00343-021-0495-1
Singh, A., and Mittal, M. (2020). Neonatal microbiome - a brief review. J. Matern. Fetal Neonatal Med. 33, 3841–3848. doi: 10.1080/14767058.2019.1583738
Tamanai-Shacoori, Z., Smida, I., Bousarghin, L., Loreal, O., Meuric, V., Fong, S. B., et al. (2017). Roseburia spp.: a marker of health? Future Microbiol. 12, 157–170. doi: 10.2217/fmb-2016-0130
Turunen, J., Tejesvi, M. V., Paalanne, N., Hekkala, J., Lindgren, O., Kaakinen, M., et al. (2021). Presence of distinctive microbiome in the first-pass meconium of newborn infants. Sci. Rep. 11:19449. doi: 10.1038/s41598-021-98951-4
Van Der Schoor, S. R. D., Reeds, P. J., Stellaard, F., Wattimena, J. D. L., Sauer, P. J. J., Büller, H. A., et al. (2004). Lysine kinetics in preterm infants: the importance of enteral feeding. Gut 53, 38–43. doi: 10.1136/gut.53.1.38
Wang, J., Zheng, J., Shi, W., Du, N., Xu, X., Zhang, Y., et al. (2018). Dysbiosis of maternal and neonatal microbiota associated with gestational diabetes mellitus. Gut 67, 1614–1625. doi: 10.1136/gutjnl-2018-315988
Xu, Y., Milburn, O., Beiersdorfer, T., Du, L., Akinbi, H., and Haslam, D. B. (2022). Antibiotic exposure prevents acquisition of beneficial metabolic functions in the preterm infant gut microbiome. Microbiome 10:103. doi: 10.1186/s40168-022-01300-4
Xu, S., Zhan, L., Tang, W., Wang, Q., Dai, Z., Zhou, L., et al. (2023). MicrobiotaProcess: a comprehensive R package for deep mining microbiome. Innovation 4:100388. doi: 10.1016/j.xinn.2023.100388
Yan, Q., Gu, Y., Li, X., Yang, W., Jia, L., Chen, C., et al. (2017). Alterations of the gut microbiome in hypertension. Front. Cell. Infect. Microbiol. 7:381. doi: 10.3389/fcimb.2017.00381
You, T., Zhang, H., Guo, L., Ling, K.-R., Hu, X.-Y., and Li, L.-Q. (2020). Differences in clinical characteristics of early-and late-onset neonatal sepsis caused by Klebsiella pneumoniae. Int. J. Immunopathol. Pharmacol. 34:205873842095058. doi: 10.1177/2058738420950586
Yu, J., Zhang, B., Miao, T., Hu, H., and Sun, Y. (2022). Dietary nutrition and gut microbiota composition in patients with hypertensive disorders of pregnancy. Front. Nutr. 9:862892. doi: 10.3389/fnut.2022.862892
Keywords: hypertensive disorders of pregnancy, pre-eclampsia, gestational hypertension, maternal fecal microbial community, neonatal meconium microbiota
Citation: Yang H, He Z, Lai J, Yang J, Huang Q, Chang Y, Tian M and Huang H (2025) Alterations of the paired maternal fecal microbiota and neonatal meconium microbiota in newborns from pregnant women with hypertensive disorders. Front. Microbiol. 16:1567721. doi: 10.3389/fmicb.2025.1567721
Edited by:
Lorena Coretti, University of Naples Federico II, ItalyReviewed by:
Muhammad Zohaib Aslam, Tuskegee University, United StatesYafei Duan, South China Sea Fisheries Research Institute, China
Erick Saul Sánchez Salguero, University of Oxford, United Kingdom
Copyright © 2025 Yang, He, Lai, Yang, Huang, Chang, Tian and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mingyuan Tian, bWluZ3l1YW50QGhvc3BpdGFsLmNxbXUuZWR1LmNu; Hongli Huang, OTMzMTk3NzlAcXEuY29t
†These authors have contributed equally to this work
 Heng Yang1†
Heng Yang1† Mingyuan Tian
Mingyuan Tian Hongli Huang
Hongli Huang