- 1Department of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Republic of Korea
- 2Division of Interdisciplinary Bioscience and Bioengineering, Pohang University of Science and Technology (POSTECH), Pohang, Republic of Korea
- 3Graduate School of Engineering Biology, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, Republic of Korea
Non-conventional yeasts exhibit exceptional genetic and functional diversity, serving as a largely untapped repertoire for biotechnological applications. Beyond the conventional yeast Saccharomyces cerevisiae, non-conventional yeasts are naturally more multifaceted, possessing the ability to utilize renewable and low-cost carbon sources while exhibiting robust physiology under challenging conditions. However, their vast potential remains largely unexplored, encompassing both challenges and opportunities for biotechnological advancements. Over the past decade, technological advancements in synthetic biology have unlocked new opportunities to harness their potential and overcome inherent limitations, enabling the full exploitation of their advantages across a broad spectrum of applications. In this review, we highlight recent advances in the synthetic biology of non-conventional yeasts, focusing on the development of new genetic building blocks (e.g., promoters and terminators), genome editing tools, and metabolic pathway engineering. Through these technologies, non-conventional yeasts are poised to emerge as pivotal next-generation workhorses tailored for specific applications in sustainable biomanufacturing, accelerating the transition to a bio-based economy.
1 Introduction
The convergence of environmental pollution, climate change, and resource scarcity is increasing intractable and compounding global challenges (Li W. et al., 2021; Karim et al., 2022; Liu et al., 2023). Microbial biotechnology provides a sustainable alternative (Raschmanova et al., 2018; Thorwall et al., 2020), enabling cost-effective and sustainable bioproduction across various medical, agricultural, food, and chemical industries. In particular, yeasts offer key advantages over other microbes in industrial biotechnology, including eukaryotic cellular machinery capable of post-translational modifications, the ability to utilize a wide range of inexpensive and renewable feedstocks, and robustness under harsh industrial conditions (Deparis et al., 2017; Nielsen, 2013; Qiu et al., 2019). Historically, yeasts have played crucial roles in the food industry and in the production of bulk and fine chemicals, as well as biofuels (Patra et al., 2021; Fabarius et al., 2021; Wang et al., 2025). Among them, Saccharomyces cerevisiae is the most extensively studied model yeast, known for its well-characterized genome and established molecular genetic engineering tools (Rainha et al., 2020; Moon et al., 2023). However, despite extensive research and engineering efforts, S. cerevisiae has inherent metabolic limitations, including low productivity, susceptibility to product toxicity, and an inability to convert alternative substrates into high-value products efficiently. These constraints restrict its commercial competitiveness and emphasize the need for alternative microbial platforms with superior metabolic capabilities (Park et al., 2022; Sibirny, 2023; Patra et al., 2021).
Potential solutions can be found in non-conventional yeasts, including Yarrowia lipolytica, Pichia pastoris, and Kluyveromyces marxianus. These yeasts exhibit superior metabolic flexibility, stress tolerance, and substrate utilization capabilities, making them highly attractive for industrial applications (Thorwall et al., 2020; Rebello et al., 2018; Wang et al., 2021; Nurcholis et al., 2020; Spohner et al., 2015; Yang and Zhang, 2018; Monteiro de Oliveira et al., 2021; Madhavan et al., 2017; Wagner and Alper, 2016). However, many non-conventional yeasts still suffer from limited genetic tractability, low transformation efficiency, and a lack of well-characterized regulatory parts, which constrain their broader application in biotechnology (Wagner and Alper, 2016; Lobs et al., 2017). In recent decades, the research on non-conventional yeasts has gained momentum, driven by rapid breakthroughs in synthetic biology. These advancements have significantly expanded their industrial utility by providing powerful tools for optimizing gene expression, metabolic pathways, and strain performance. This review covers a comprehensive overview of the attractive characteristics and role of non-conventional yeasts as versatile biotechnological workhorses, with a special emphasis on recent breakthroughs in synthetic biology that have enhanced their industrial applications. Key areas of focus include promoter and terminator engineering for precise gene regulation, CRISPR/Cas-based genome editing for efficient strain development, and pathway optimization strategies for improved biochemical production.
2 Non-conventional yeasts as biotechnological workhorses
Non-conventional yeasts have emerged as new potential workhorses for the overproduction of fuels, chemicals, and pharmaceuticals owing to their robust physiology, which includes high tolerances to bioprocess-induced stresses (e.g., low pH, high temperatures, and osmolarity), resistance to inhibitory toxic compounds, and ability to utilize non-conventional feedstocks and synthesize large amounts of metabolites and proteins (Rebello et al., 2018; Thorwall et al., 2020; Wang et al., 2021; Markham and Alper, 2018; Spohner et al., 2015; Yang and Zhang, 2018; Nurcholis et al., 2020). Notably, the ability of non-conventional yeasts to metabolize non-conventional substrates—such as lignocellulosic hydrolysates, waste oils, and methanol—offers substantial industrial benefits. These substrates are often derived from low-cost, renewable sources like agricultural residues and CO₂-based industrial waste streams, thereby supporting more sustainable and economically viable biomanufacturing process (Rerop et al., 2023; Cotton et al., 2020; Do et al., 2019). For instance, methanol, a key substrate for P. pastoris, can be industrially produced from synthesis gas derived from natural gas or biomass, enabling cost-effective and potentially carbon-neutral feedstock supply chains (Cai et al., 2022). Among non-conventioal yeasts, Y. lipolytica, P. pastoris (recently reclassified as Komagataella phaffii), and K. marxianus are particularly notable for their distinct and inherent advantages in lipid accumulation, heterologous protein production, and thermotolerance, respectively. These species have been relatively well-characterized and are frequently used as biotechnological workhorses (Figure 1).
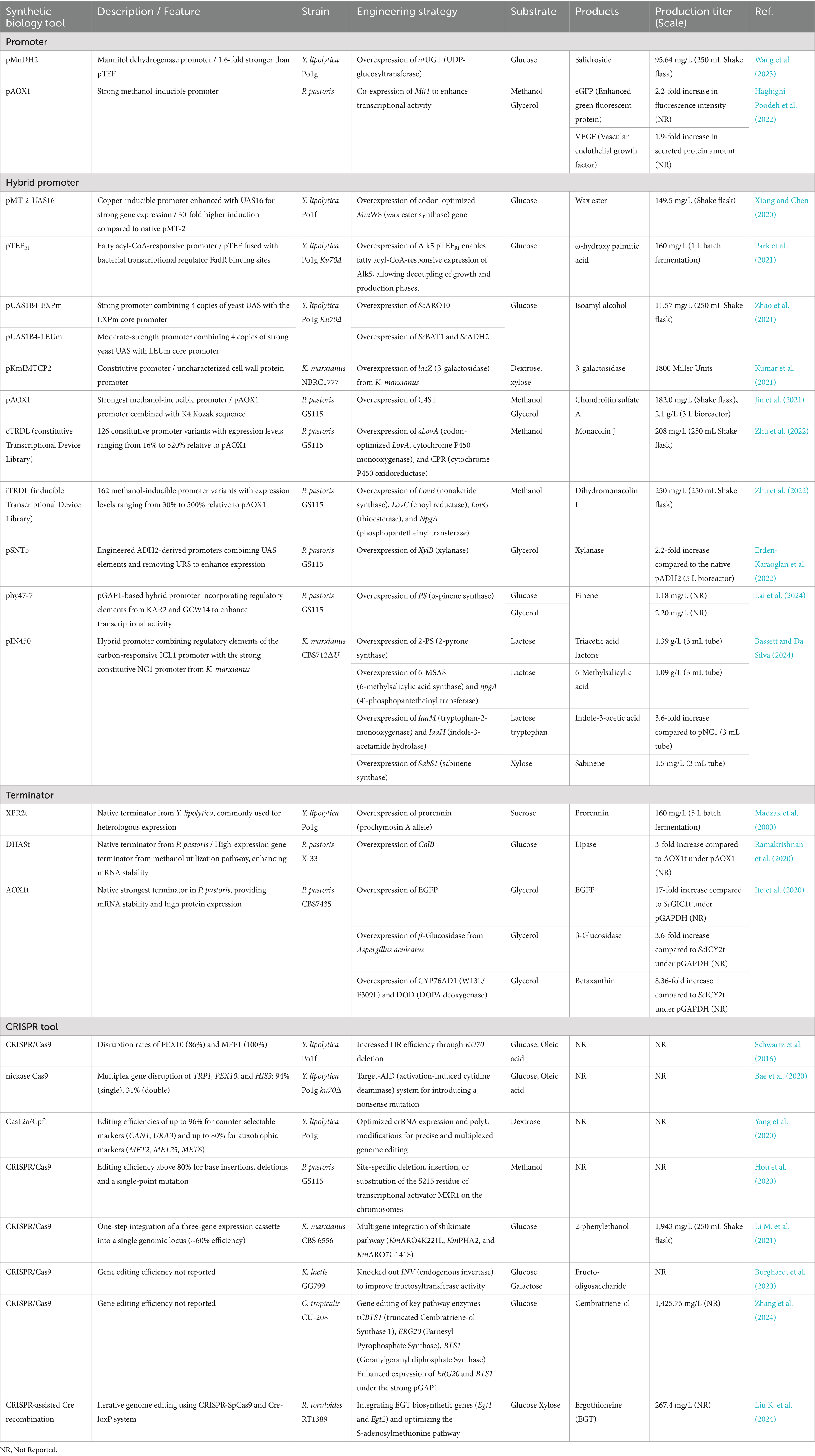
Figure 1. A key summary of synthetic biology tools and metabolic features of non-conventional yeasts for industrial biotechnology. This figure illustrates key synthetic biology strategies and the distinctive traits of non-conventional yeasts. Genetic components, including synthetic promoters, terminators, and transcription factors, enable fine-tuned control of gene expression. Genome editing tools such as homologous recombination via KU70 or KU80 deletion and CRISPR-associated systems (Cas9 and Cas12a) facilitate efficient and precise genome manipulation. Each non-conventional yeast exhibits unique metabolic traits suited for specific biotechnological uses. Y. lipolytica is characterized by high lipid accumulation and the ability to metabolize hydrophobic substrates, supporting the production of oleochemicals and carotenoids. P. pastoris exhibits strong protein expression and utilizes methanol and other cost-effective substrates, facilitating the industrial production of enzymes and pharmaceutical proteins. K. marxianus is known for thermotolerance, rapid growth, and broad substrate utilization, making it suitable for the biosynthesis of flavor compounds and industrial enzymes. S. stipitis efficiently ferments xylose derived from lignocellulosic biomass, contributing to bioethanol production. O. polymorpha metabolizes methanol to produce oleochemicals such as free fatty acids and fatty alcohols. K. lactis ferments lactose and is widely applied in dairy biotechnology. Collectively, these non-conventional yeasts offer complementary and adaptable platforms for sustainable and scalable bioproduction. UAS, upstream activating sequence; CDS, coding sequence; TF, transcription factor; HR, homologous recombination.
The oleaginous yeast Y. lipolytica is widely recognized for its exceptional lipid accumulation capacity, naturally reaching up to 30–50% of its cell dry weight, and its ability to metabolize diverse non-conventional substrates, including lignocellulosic hydrolysates, fatty acids, waste oils, crude glycerol, and acetate (Beopoulos et al., 2011; Madzak, 2021; Groenewald et al., 2014). Driven by efficient carbon flux through the acetyl-CoA and malonyl-CoA pathways, Y. lipolytica serves as a robust industrial chassis for commercial synthesis, supporting the production of lipids, advanced biofuels, and lipid-derived fine chemicals such as carotenoids and omega-3 fatty acids (Ma et al., 2020; Zhang et al., 2023; Xie et al., 2017; Shi and Zhao, 2017; Jiang et al., 2024; Ma et al., 2024; Liu Z. et al., 2024; Park et al., 2025; Sharpe et al., 2014). Additionally, its generally recognized as safe (GRAS) status makes Y. lipolytica a valuable platform for producing lipases widely used in the food, pharmaceutical, and environmental industries (Brígida et al., 2014; Hu et al., 2024).
Pichia pastoris is a methylotrophic yeast commonly used in the manufacture of industrial enzymes and pharmaceutical proteins owing to its various strengths. Notably, its strong protein expression and secretion capacity, along with its ability to perform post-translational modifications, make P. pastoris an ideal system for both cytosolic and secretory protein production (Gellissen et al., 2005; Karbalaei et al., 2020; Macauley-Patrick et al., 2005; Zha et al., 2023; Yang and Zhang, 2018). This feature enables the secretion of target proteins at high titers, simplifying downstream processing and facilitating the commercial production of pharmaceutical proteins, including human insulin and interferon, demonstrating the industrial relevance of this yeast (Patra et al., 2021; Nurdiani et al., 2024; Gao et al., 2021; Zha et al., 2023; Shrivastava et al., 2023). A further advantage of P. pastoris is its ability to utilize cost-effective substrates, tolerate high-stress conditions, and achieve higher cell densities than S. cerevisiae, making it a more suitable host for sustainable industrial processes. Owing to these attractive characteristics, P. pastoris has also been engineered to produce various value-added chemicals at low cultivation costs with high scalability (Lobs et al., 2017; Araya-Garay et al., 2012; Liu et al., 2015; Liu et al., 2018; Qian et al., 2022; Gao et al., 2023; Cai et al., 2022).
Kluyveromyces marxianus is also industrially relevant because of its beneficial traits, such as thermotolerance up to ~50°C, high growth rate, and broad substrate spectrum (e.g., hemi-cellulose hydrolysates, xylose, and dairy industry wastes) (Bilal et al., 2022; Lyu et al., 2021; Ha-Tran et al., 2020; Varela et al., 2017). Besides, K. marxianus serves as a natural producer of high-value ethyl acetate and other short-chain volatile esters, widely used as flavor and fragrance compounds (Lobs et al., 2018; Karim et al., 2020; Morrissey et al., 2015; Loser et al., 2013; Roy et al., 2023; Perpetuini et al., 2022). Its ability to grow at elevated temperatures facilitates simultaneous saccharification and fermentation of lignocellulosic and other polysaccharide-based feedstocks, reducing cooling costs, minimizing contamination risks, and improving bioprocess efficiency (Patra et al., 2021). Additionally, K. marxianus efficiently assimilates dairy byproducts such as lactose and cheese whey, taking a step toward more sustainable biomanufacturing (Bilal et al., 2022; Qiu et al., 2023).
Beyond these well-studied yeast species, several other non-conventional yeasts have recently attracted significant attention for their industrial potential. K. lactis, a GRAS yeast following S. cerevisiae, is a lactose-fermenting species frequently found in milk and dairy products. The β-galactosidase enzyme from K. lactis, which metabolizes milk constituents such as lactose, is widely used in the food industry to produce lactose-free dairy products (Becerra et al., 2001; Naumov et al., 2006). Furthermore, several metabolites are commercially produced in K. lactis, including lactate, D-gluconic acid derived from xylose, and D-arabitol produced from whey (Toivari et al., 2012; Toyoda and Ohtaguchi, 2011). While P. pastoris is primarily used for protein production, another methylotrophic yeast, Ogataea polymorpha, has attracted increasing attention as a promising chassis for producing various chemicals using methanol as the sole carbon source (Xie et al., 2024). Scheffersomyces stipitis, known for its superior xylose fermentation efficiency, holds promise for bioethanol production from lignocellulosic hydrolysates, as it converts xylose into ethanol with minimal or no xylitol byproduct formation, improving economic viability (Ruchala et al., 2020; Mastella et al., 2023; Kobayashi et al., 2022).
Still, beyond the natural advantages of these non-conventional yeasts, which make them more suited for tasks such as utilizing renewable and low-cost carbon sources or exhibiting high-stress tolerance, genetic engineering tailored to specific purposes can further enhance their functionality toward a robust bioeconomy in future biotechnological processes. To fully unlock the potential of these yeasts and facilitate their successful integration into biotechnological applications, genome engineering tools must be developed to pave the way for progress in metabolic engineering and synthetic biology to advance the application of new non-conventional yeasts rapidly. Below, we highlight recent synthetic biology tools and approaches that increase the industrial potential of non-conventional yeasts (Table 1). Key strategies include (1) promoter and terminator engineering for optimized gene expression and (2) CRISPR-based genome editing for efficient and multiplexed genetic modifications.
3 Synthetic biology tools and approaches to unlock the potential and function of non-conventional yeasts
3.1 Genetic building blocks for synthetic pathway engineering: promoters and terminators
The advancement and implementation of synthetic biology tools, combined with the expanding library of genetic building blocks, have significantly increased the utility of non-conventional yeasts as versatile systems and chassis cells in biotechnological applications. Recent studies have driven significant progress in their application, leveraging the availability of genetic elements such as promoters and terminators specifically tailored to non-conventional yeasts (Ji et al., 2024; Patra et al., 2021; Ma et al., 2020; Gao et al., 2021; Wang et al., 2023; Kumar et al., 2021; Teo and Chang, 2014; Qiu et al., 2023). These genetic elements play crucial roles in regulating transcription rates and mRNA stability, directly influencing protein expression levels and consequently enabling non-conventional yeasts to emerge as valuable platforms for synthetic biology and biomanufacturing (Patra et al., 2021; Ito et al., 2020; Wagner and Alper, 2016; Sun et al., 2022) (Figure 1).
3.1.1 Promoters
Promoters, in particular, are critical determinants of transcriptional regulation, as they govern the timing, strength, and spatial patterns of gene expression. This, in turn, profoundly shapes metabolic activities, enabling the precise modulation of cellular behaviors (Blazeck and Alper, 2013; Ji et al., 2024; Ma et al., 2020; Wang et al., 2023). Hence, discovering and selecting appropriate promoters, as well as engineering novel promoter elements, are fundamental steps in advancing metabolic engineering and synthetic biology for non-conventional yeasts (Madhavan et al., 2017; Rebello et al., 2018). Generally, a significant strategy for achieving high-expression of a given protein involved in synthetic pathways is using a strong and constitutive promoter. Strong promoters are typically derived from genes associated with the essential functions or unique metabolic traits of each yeast species. Representative examples include promoters driving translation (e.g., pTEF active across multiple yeast species), methanol utilization (e.g., pAOX1 in P. pastoris), or ethanol utilization (e.g., pADH2 in S. cerevisiae), taking advantage of the inherent metabolic capabilities and physiological traits of the respective species of yeast.
Among these promoters, the endogenous translation elongation factor-1α promoter (pTEF) is the most widely used because of its robust constitutive expression across diverse yeast species (Gu et al., 2023; Larroude et al., 2018; Ahn et al., 2007; Steiner and Philippsen, 1994; Kitamoto et al., 1998). In addition to the TEF1 promoter, to expand regulatory options in Y. lipolytica, a library of 81 endogenous promoters, primarily associated with carbon and nitrogen metabolism, has been systematically screened, offering expression strengths ranging from 0.0006- to 1.60-fold relative to pTEF. Notably, the MnDH2 promoter (encoding mannitol dehydrogenase) exhibited the highest strength, achieving an expression level 1.60-fold greater than pTEF. This promoter facilitated the production of the plant-derived aromatic compound salidroside in Y. lipolytica, reaching a titer of 95.64 mg/L, the highest reported to date (Wang et al., 2023).
Alternatively, artificial hybrid promoters, combining upstream activation sequences (UASs) with modified core promoter elements, have been developed to enhance gene expression control in Y. lipolytica, providing greater flexibility and dynamic regulation (Blazeck et al., 2011). A notable example is the development of fatty acid-sensitive hybrid promoters by combining pTEF with bacterial transcriptional regulator FadR binding sites. Fatty acyl-CoA binds to FadR, inducing a conformational change that inhibits FadR binding to its target sequences, thereby upregulating the expression of target genes. This mechanism allows FadR hybrid promoters to decouple the cell growth and production phases in response to intracellular fatty acyl-CoA concentrations, producing 160 mg/L of ω-hydroxy palmitic acid (Park et al., 2021).
Hybrid promoters incorporating UAS elements have also addressed the limited number of native Y. lipolytica promoters available, significantly expanding expression flexibility. For instance, a hybrid promoter incorporating sixteen copies of the UAS from the Y. lipolytica alkaline extracellular protease promoter into the copper-inducible MT-2 core promoter facilitated the efficient production of wax esters at a titer of 149.5 mg/L (Blazeck et al., 2011; Xiong and Chen, 2020). Similarly, another hybrid promoter, which incorporates four copies of the Y. lipolytica alkaline extracellular protease promoter UAS with the export protein EXP1 promoter, achieved an isoamyl alcohol titer of 11.57 mg/L (Zhao et al., 2021). These advancements highlight the versatility of hybrid promoters in Y. lipolytica, enabling the production of diverse valuable compounds and optimizing cell factory applications.
In the methylotrophic yeast P. pastoris, promoter engineering has advanced with the AOX1 promoter, a highly active methanol-inducible promoter widely used for recombinant protein production (Wu et al., 2023). To create pAOX1 variants with variable strengths, Zhu et al. (2022) fused bacterial DNA-binding proteins with yeast transactivation domains and linked bacterial binding sequences to the AOX1 core promoter. Consequently, 126 constitutive hybrid promoter libraries with expression strengths ranging from 16% to 520% and 162 methanol-inducible hybrid promoter libraries ranging from 30% to 500% were constructed relative to the native AOX1 promoter (Zhu et al., 2022).
In addition, in a recent study, various Kozak sequences were applied to the AOX1 promoter to enhance the intracellular expression of chondroitin-4-O-sulfotransferase (C4ST), a membrane-bound enzyme rarely expressed in microorganisms. Combined with chondroitin biosynthesis pathway genes, hybrid promoter-driven C4ST expression produced 182.0 mg/L of chondroitin sulfate A in P. pastoris (Jin et al., 2021). In another effort to improve the AOX1 promoter efficiency, the overexpression of methanol-induced transcription factor 1 (Mit1) strongly activated the AOX1 promoter and increased eGFP production by 2.2-fold. Doubling the methanol feed concentration further boosted the eGFP output by an additional 1.3-fold (Haghighi Poodeh et al., 2022). Similar to the activation of pAOX1 mediated by Mit1 overexpression, a separate study was dedicated to modifying the alcohol dehydrogenase 2 promoter (pADH2) based on its transcriptional regulatory mechanism. By replacing its repressor region with an activator region, pADH2 activity was enhanced by 2.2-fold compared with that of original pADH2 (Erden-Karaoglan et al., 2022).
Besides pAOX1, the glyceraldehyde-3-phosphate dehydrogenase promoter (pGAP) is commonly used as a constitutive promoter for protein expression in P. pastoris. Unlike pAOX1, pGAP does not require a toxic methanol inducer, making it suitable for continuous cultivation while maintaining stable cellular function (Wu et al., 2023; Vogl et al., 2016). Lai et al. (2024) developed a novel randomized hybrid promoter library derived from pGAP1 and demonstrated its potential by producing 1.18 mg/L of the biologically active natural monoterpene pinene, representing an 18% increase over the native GAP promoter.
Recent studies on K. marxianus have also been focused on identifying and optimizing native-derived promoters, from the weakest promoter REV1 (deoxycytidyl transferase) to the strongest promoter PDC1 (pyruvate decarboxylase), enabling 40-fold variation in gene expression (Qiu et al., 2023; Rajkumar et al., 2019). Among these efforts, novel expression toolkits were constructed by combining various promoters and terminators derived from K. marxianus. For example, pKmIMTCP2-KmIMTT1t, comprising an uncharacterized cell wall protein promoter (pIMTCP2) and a maltose transporter terminator (IMTT1t), demonstrated the highest activity in K. marxianus, producing approximately 1800 Miller units of β-galactosidase (Kumar et al., 2021).
Most recently, Bassett and Da Silva (2024) designed and built a novel carbon-responsive hybrid promoter, pIN450, by combining regulatory elements of the native K. marxianus carbon-responsive ICL1 promoter with the strong constitutive NC1 promoter from K. marxianus. The hybrid IN450 promoter exhibits carbon-responsive behavior in lactose and constitutive behavior in xylose, leading to over a 50% increase in the production of the high-value chemical triacetic acid lactone and a 6.6-fold increase in the production of the fungal polyketide 6-methylsalicylic acid compared to native pICL1 (Bassett and Da Silva, 2024).
3.1.2 Terminators
Terminators are also essential in transcriptional regulation, influencing mRNA stability, half-life, and abundance, directly affecting protein expression levels (Gu et al., 2023; Patra et al., 2021; Hu et al., 2024; Madzak, 2021). Despite their vital roles, only a few terminators in non-conventional yeasts have been systematically characterized. Meanwhile, several S. cerevisiae terminators have been successfully adapted to non-conventional yeasts, such as ScCYC1t in Y. lipolytica and P. pastoris, ScADH1t and ScPGK1t in K. marxianus, and ScADH1t in H. polymorpha (Madzak, 2021; Patra et al., 2021). Native and synthetic terminators from non-conventional yeasts have also demonstrated potential in modulating gene expression (Table 1). In Y. lipolytica, native terminators such as XPR2t (extracellular protease), LIP2t (extracellular lipase), and CyC1t (cytochrome C) have been identified and characterized (Madzak et al., 2000; Ma et al., 2020). In P. pastoris, DHASt (dihydroxyacetone synthase) enhanced the expression of Candida antarctica lipase B (CalB) by 3-fold compared with AOX1t under pAOX1 (Ramakrishnan et al., 2020). Additionally, a library of 72 terminators from S. cerevisiae, P. pastoris, and synthetic sources demonstrated a 17-fold tunable range of activity in P. pastoris (Ito et al., 2020). The IMTT1 (IMTCP1) and IMTT2 (IMTCP2) terminators from K. marxianus significantly increased β-galactosidase production (Kumar et al., 2021), highlighting the versatility of terminator engineering for optimizing gene expression (Ito et al., 2020).
3.2 Genome editing tool: CRISPR-based genome editing
Efficient genome editing tools for inserting, deleting, and altering target genes are critical for engineering non-conventional yeasts to reconstruct complex metabolism and thus enhance product synthesis for industrial applications. Homologous recombination (HR)-mediated tools are generally preferred in genetic engineering due to their ability to precisely control integration loci, minimizing the risk of disrupting essential genes (Flagfeldt et al., 2009; Cai et al., 2019; Donohoue et al., 2018; Xia et al., 2023). However, unlike S. cerevisiae, non-conventional yeasts face unique challenges because non-homologous end joining (NHEJ) dominates over HR, which often leads to imprecise integration of inserted DNA (Xia et al., 2023; Cai et al., 2019). Overcoming the natural dominance of NHEJ to increase HR efficiency in non-conventional yeasts remains a significant challenge. Nonetheless, modulation of the NHEJ or HR DNA repair pathways has shown promise in addressing this issue. For instance, deletion of native NHEJ-promoting genes such as Ku70 or Ku80 significantly increased HR efficiency in many non-conventional yeasts (Maassen et al., 2008; Kooistra et al., 2004; Verbeke et al., 2013; Naatsaari et al., 2012; Saraya et al., 2012; Choo et al., 2014). Furthermore, the overexpression of HR repair proteins such as RAD52, RAD59, MRE11, and SAE2 from S. cerevisiae has achieved multiplex gene integration efficiencies of 100%, ~98%, and ~81% at single, double, and triple loci, respectively, even with homology arms as short as 40 bp (Gao et al., 2022).
The advent of CRISPR/Cas9 technology has revolutionized genome editing in non-conventional yeasts, offering unparalleled precision, flexibility, multiplexing, and simplicity (Bai et al., 2023) (Figure 1, Table 1). The CRISPR/Cas9-mediated genome editing tool introduces targeted double-strand breaks (DSBs) at specific loci, enabling precise and programmable modifications guided by customized simple single-guide RNA (sgRNA), with the assistance of intracellular DNA repair pathways such as HR and NHEJ. CRISPR/Cas9 streamlines the editing workflow, improves accuracy, and accelerates strain engineering to achieve desired properties (Zha et al., 2023; Wu et al., 2023; Schwartz et al., 2016; Li M. et al., 2021; Burghardt et al., 2020; Zhang et al., 2024; Liu K. et al., 2024).
In Y. lipolytica, CRISPR/Cas9 was first adapted in 2016 using a codon-optimized Cas9 and sgRNA expression under a synthetic RNA polymerase III promoter to disrupt genes such as Ku70, as well as lipid oxidation-related Pex10 and Mfe1 (Schwartz et al., 2016). More recently, a base editor combining CRISPR/Cas9, cytidine deaminase, and uracil glycosylase inhibitor enabled targeted base modifications without introducing DSBs, further expanding the genome editing toolbox in Y. lipolytica. This system achieved editing efficiencies of 94% for single genes and 34% for dual genes, demonstrating its potential for precise genetic engineering (Bae et al., 2020). Additionally, the CRISPR-Cas12a/Cpf1 system has been implemented, allowing for the retention of PAM sites after NHEJ repair and enabling efficient multiplexed editing (Yang et al., 2020). Using this system, the single-gene disruption efficiencies reached 99%, while triplex edits achieved up to 30%, highlighting its utility for complex genetic modifications.
In P. pastoris, a highly efficient CRISPR/Cas9 system was developed through the systematic optimization of codon-optimized Cas9 DNA sequences, various sgRNA sequences, and promoters for the optimal expression of both Cas9 and sgRNA, achieving genome editing efficiencies approaching 100% (Weninger et al., 2016). This system enabled targeted editing of the methanol expression regulator MXR1 and facilitated base insertions and deletions at critical amino acid positions, allowing for studying this transcription factor and its targets (Hou et al., 2020). Additionally, the CRISPR/Cas12a system enabled the deletion of large DNA fragments (up to 20 kb) and one-step integration of multiplexed genes, exhibiting 99% efficiency for single-gene edits, 65–80% efficiency for duplex edits, and 30% efficiency for triplex integrations (Zhang et al., 2021).
CRISPR/Cas9 has also been applied in Kluyveromyces species, demonstrating its versatility for gene deletion and multiplexed gene integration. In K. marxianus, a CRISPR/Cas9-based multigene integration system was developed to engineer key genes in the shikimate pathway (KmARO4K221L, KmPHA2, and KmARO7G141S), resulting in a 2.8-fold increase in the production of the rose-scented flavor and fragrance compound 2-phenylethanol. Further optimization of the Ehrlich pathway through the overexpression of ARO10 and inactivation of EAT1 boosted 2-phenylethanol production to 1,943 ± 63 mg/L under fed-batch conditions (Li M. et al., 2021). In K. lactis, CRISPR/Cas9 was successfully applied to delete the endogenous invertase gene, resulting in a 66.9% increase in fructose transferase activity (Burghardt et al., 2020).
Similar advances have also been achieved in other non-conventional yeasts. In C. tropicalis, CRISPR/Cas9 enhanced the production of the plant-derived macrocyclic diterpene cembratriene-ol to 1,425.76 mg/L, a 1,602-fold increase, by integrating the codon-optimized cembratriene-ol synthase gene and optimizing metabolic flux (Zhang et al., 2024). Likewise, in Rhodotorula toruloides, the CRISPR-assisted Cre recombination system, which combines CRISPR/Cas9 with the site-specific recombinase Cre, enables iterative genome editing. This approach increased the production of ergothioneine (EGT), a high-value antioxidant and cytoprotectant, to 267.4 mg/L, a 1.5-fold improvement, by integrating EGT biosynthetic genes (Egt1 and Egt2) and optimizing the S-adenosylmethionine pathway (Liu K. et al., 2024).
4 Conclusions and future perspectives
Technological developments in synthetic biology, particularly in transcriptional regulation systems, CRISPR-based genome editing, and strain engineering, have greatly expanded the potential of non-conventional yeasts while overcoming their inherent limitations. Their applications are diverse, and they likely represent a crucial new means to address the looming challenges of biomanufacturing—for both narrow and broad product ranges—by enabling the production of a wide array of bio-based chemicals, fuels, and materials, thereby positioning them as valuable assets in the future of industrial biotechnology. In the near future, emerging technologies such as the design-build-test-learn cycle in synthetic biology will further accelerate this progress, driving the development of advanced and cost-effective methods for building, editing, and screening non-conventional yeasts with novel and optimized functions in a high-throughput manner, solidifying them as next-generation microbial workhorses for industrial biotechnology (Li X. et al., 2023; Whitford et al., 2021; Moon et al., 2024; Son et al., 2024).
The integration of automated genome synthesis, AI-assisted metabolic design, and omics-driven pathway optimization will further strengthen the potential of non-conventional yeasts, enabling precise metabolic fine-tuning for a wide range of applications (Patra et al., 2021; Darvishi et al., 2021; Li M. et al., 2023; Madhavan et al., 2017; Wang et al., 2025). Looking ahead, the convergence of synthetic biology, systems biology, and machine learning will be instrumental in streamlining strain engineering workflows, improving predictive modeling accuracy, and enhancing strain design efficiency. Additionally, expanding the molecular toolbox for non-conventional yeasts—including novel inducible promoters, tunable gene circuits, and genome-scale engineering strategies—will further enhance their versatility and adaptability. To fully realize the potential of non-conventional yeasts in biomanufacturing, future advances in synthetic biology should focus on addressing current limitations—such as the limited availability of species-specific regulatory elements and the narrow range of inducible promoters. Expanding modular, programmable, and scalable toolkits will enable more precise, flexible, and context-specific strain engineering, thereby accelerating the transition toward next-generation yeast-based production systems.
As synthetic biology continues to bridge the gap between a detailed understanding of non-conventional yeasts and their practical applications, these yeasts will undoubtedly play an expanding role in the global bioeconomy. Future research will likely be focused on harnessing automation, leveraging AI-driven metabolic design, and integrating multi-omics datasets to drive innovation in yeast engineering. Non-conventional yeasts will not only provide additional options alongside existing microbial platforms but also offer unique and tailored solutions for sustainable bioproduction, accelerating the transition toward a more sustainable bioeconomy.
Author contributions
SM: Writing – original draft, Writing – review & editing. N-YA: Investigation, Visualization, Writing – original draft. JL: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Bio & Medical Technology Development Program (RS-2024-00445145) and the Basic Science Research Program (RS-2025-00518119) through the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (MSIT), Republic of Korea.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahn, J., Hong, J., Lee, H., Park, M., Lee, E., Kim, C., et al. (2007). Translation elongation factor 1-alpha gene from Pichia pastoris: molecular cloning, sequence, and use of its promoter. Appl. Microbiol. Biotechnol. 74, 601–608. doi: 10.1007/s00253-006-0698-6
Araya-Garay, J. M., Ageitos, J. M., Vallejo, J. A., Veiga-Crespo, P., Sanchez-Perez, A., and Villa, T. G. (2012). Construction of a novel Pichia pastoris strain for production of xanthophylls. AMB Express 2:24. doi: 10.1186/2191-0855-2-24
Bae, S. J., Park, B. G., Kim, B. G., and Hahn, J. S. (2020). Multiplex gene disruption by targeted base editing of Yarrowia lipolytica genome using cytidine deaminase combined with the CRISPR/Cas9 system. Biotechnol. J. 15:e1900238. doi: 10.1002/biot.201900238
Bai, W., Huang, M., Li, C., and Li, J. (2023). The biological principles and advanced applications of DSB repair in CRISPR-mediated yeast genome editing. Synth. Syst. Biotechnol. 8, 584–596. doi: 10.1016/j.synbio.2023.08.007
Bassett, S., and Da Silva, N. A. (2024). Engineering a carbon source-responsive promoter for improved biosynthesis in the non-conventional yeast Kluyveromyces marxianus. Metab. Eng. Commun. 18:e00238. doi: 10.1016/j.mec.2024.e00238
Becerra, M., Prado, S. D., Siso, M. I., and Cerdan, M. E. (2001). New secretory strategies for Kluyveromyces lactis beta-galactosidase. Protein Eng. 14, 379–386. doi: 10.1093/protein/14.5.379
Beopoulos, A., Nicaud, J. M., and Gaillardin, C. (2011). An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl. Microbiol. Biotechnol. 90, 1193–1206. doi: 10.1007/s00253-011-3212-8
Bilal, M., Ji, L., Xu, Y., Xu, S., Lin, Y., Iqbal, H., et al. (2022). Bioprospecting Kluyveromyces marxianus as a robust host for industrial biotechnology. Front. Bioeng. Biotechnol. 10:851768. doi: 10.3389/fbioe.2022.851768
Blazeck, J., and Alper, H. S. (2013). Promoter engineering: recent advances in controlling transcription at the most fundamental level. Biotechnol. J. 8, 46–58. doi: 10.1002/biot.201200120
Blazeck, J., Liu, L., Redden, H., and Alper, H. (2011). Tuning gene expression in Yarrowia lipolytica by a hybrid promoter approach. Appl. Environ. Microbiol. 77, 7905–7914. doi: 10.1128/AEM.05763-11
Brígida, A. I., Amaral, P. F., Coelho, M. A., and Goncalves, L. R. (2014). Lipase from Yarrowia lipolytica: production, characterization and application as an industrial biocatalyst. J. Mol. Catal. B Enzym. 101, 148–158. doi: 10.1016/j.molcatb.2013.11.016
Burghardt, J. P., Fan, R., Baas, M., Eckhardt, D., Gerlach, D., and Czermak, P. (2020). Enhancing the heterologous Fructosyltransferase activity of Kluyveromyces lactis: developing a scaled-up process and abolishing invertase by CRISPR/Cas9 genome editing. Front. Bioeng. Biotechnol. 8:607507. doi: 10.3389/fbioe.2020.607507
Cai, P., Gao, J., and Zhou, Y. (2019). CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Factories 18:63. doi: 10.1186/s12934-019-1112-2
Cai, P., Wu, X., Deng, J., Gao, L., Shen, Y., Yao, L., et al. (2022). Methanol biotransformation toward high-level production of fatty acid derivatives by engineering the industrial yeast Pichia pastoris. Proc. Natl. Acad. Sci. USA 119:e2201711119. doi: 10.1073/pnas.2201711119
Choo, J. H., Han, C., Kim, J. Y., and Kang, H. A. (2014). Deletion of a Ku80 homolog enhances homologous recombination in the thermotolerant yeast Kluyveromyces marxianus. Biotechnol. Lett. 36, 2059–2067. doi: 10.1007/s10529-014-1576-4
Cotton, C. A., Claassens, N. J., Benito-Vaquerizo, S., and Bar-Even, A. (2020). Renewable methanol and formate as microbial feedstocks. Curr. Opin. Biotechnol. 62, 168–180. doi: 10.1016/j.copbio.2019.10.002
Darvishi, F., Blenner, M., and Ledesma-Amaro, R. (2021). Editorial: synthetic biology of yeasts for the production of non-native chemicals. Front. Bioeng. Biotechnol. 9:730047. doi: 10.3389/fbioe.2021.730047
Deparis, Q., Claes, A., Foulquie-Moreno, M. R., and Thevelein, J. M. (2017). Engineering tolerance to industrially relevant stress factors in yeast cell factories. FEMS Yeast Res. 17:fox036. doi: 10.1093/femsyr/fox036
Do, D. T. H., Theron, C. W., and Fickers, P. (2019). Organic wastes as feedstocks for non-conventional yeast-based bioprocesses. Microorganisms 7:229. doi: 10.3390/microorganisms7080229
Donohoue, P. D., Barrangou, R., and May, A. P. (2018). Advances in industrial biotechnology using CRISPR-Cas systems. Trends Biotechnol. 36, 134–146. doi: 10.1016/j.tibtech.2017.07.007
Erden-Karaoglan, F., Karaoglan, M., Yilmaz, G., Yilmaz, S., and Inan, M. (2022). Deletion analysis of Pichia pastoris alcohol dehydrogenase 2 (ADH2) promoter and development of synthetic promoters. Biotechnol. J. 17:e2100332. doi: 10.1002/biot.202100332
Fabarius, J. T., Wegat, V., Roth, A., and Sieber, V. (2021). Synthetic Methylotrophy in yeasts: towards a circular bioeconomy. Trends Biotechnol. 39, 348–358. doi: 10.1016/j.tibtech.2020.08.008
Flagfeldt, D. B., Siewers, V., Huang, L., and Nielsen, J. (2009). Characterization of chromosomal integration sites for heterologous gene expression in Saccharomyces cerevisiae. Yeast 26, 545–551. doi: 10.1002/yea.1705
Gao, J., Jiang, L., and Lian, J. (2021). Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products. Synth. Syst. Biotechnol. 6, 110–119. doi: 10.1016/j.synbio.2021.04.005
Gao, J., Ye, C., Cheng, J., Jiang, L., Yuan, X., and Lian, J. (2022). Enhancing homologous recombination efficiency in Pichia pastoris for multiplex genome integration using short homology arms. ACS Synth. Biol. 11, 547–553. doi: 10.1021/acssynbio.1c00366
Gao, J., Zuo, Y., Xiao, F., Wang, Y., Li, D., Xu, J., et al. (2023). Biosynthesis of catharanthine in engineered Pichia pastoris. Nat. Synth. 2, 231–242. doi: 10.1038/s44160-022-00205-2
Gellissen, G., Kunze, G., Gaillardin, C., Cregg, J. M., Berardi, E., Veenhuis, M., et al. (2005). New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica - a comparison. FEMS Yeast Res. 5, 1079–1096. doi: 10.1016/j.femsyr.2005.06.004
Groenewald, M., Boekhout, T., Neuveglise, C., Gaillardin, C., Van Dijck, P. W., and Wyss, M. (2014). Yarrowia lipolytica: safety assessment of an oleaginous yeast with a great industrial potential. Crit. Rev. Microbiol. 40, 187–206. doi: 10.3109/1040841X.2013.770386
Gu, Y., Lu, X., Liu, T., Song, Y., Sang, E., Ding, S., et al. (2023). Engineering the oleaginous yeast Yarrowia lipolytica to produce nutraceuticals: from metabolic design to industrial applications. Food Bioeng. 2, 187–199. doi: 10.1002/fbe2.12062
Haghighi Poodeh, S., Ranaei Siadat, S. O., Arjmand, S., and Khalifeh Soltani, M. (2022). Improving AOX1 promoter efficiency by overexpression of Mit1 transcription factor. Mol. Biol. Rep. 49, 9379–9386. doi: 10.1007/s11033-022-07790-7
Ha-Tran, D. M., Nguyen, T. T. M., and Huang, C.-C. (2020). Kluyveromyces marxianus: current state of omics studies, strain improvement strategy and potential industrial implementation. Fermentation 6:124. doi: 10.3390/fermentation6040124
Hou, C., Yang, Y., Xing, Y., Zhan, C., Liu, G., Liu, X., et al. (2020). Targeted editing of transcriptional activator MXR1 on the Pichia pastoris genome using CRISPR/Cas9 technology. Yeast 37, 305–312. doi: 10.1002/yea.3462
Hu, M., Ge, J., Jiang, Y., Sun, X., Guo, D., and Gu, Y. (2024). Advances and perspectives in genetic expression and operation for the oleaginous yeast Yarrowia lipolytica. Synth. Syst. Biotechnol. 9, 618–626. doi: 10.1016/j.synbio.2024.05.003
Ito, Y., Terai, G., Ishigami, M., Hashiba, N., Nakamura, Y., Bamba, T., et al. (2020). Exchange of endogenous and heterogeneous yeast terminators in Pichia pastoris to tune mRNA stability and gene expression. Nucleic Acids Res. 48, 13000–13012. doi: 10.1093/nar/gkaa1066
Ji, L., Xu, S., Zhang, Y., and Cheng, H. (2024). Screening of broad-host expression promoters for shuttle expression vectors in non-conventional yeasts and bacteria. Microb. Cell Factories 23:230. doi: 10.1186/s12934-024-02506-x
Jiang, D., Yang, M., Chen, K., Jiang, W., Zhang, L., Ji, X. J., et al. (2024). Exploiting synthetic biology platforms for enhanced biosynthesis of natural products in Yarrowia lipolytica. Bioresour. Technol. 399:130614. doi: 10.1016/j.biortech.2024.130614
Jin, X., Zhang, W., Wang, Y., Sheng, J., Xu, R., Li, J., et al. (2021). Biosynthesis of non-animal chondroitin sulfate from methanol using genetically engineered Pichia pastoris. Green Chem. 23, 4365–4374. doi: 10.1039/D1GC00260K
Karbalaei, M., Rezaee, S. A., and Farsiani, H. (2020). Pichia pastoris: a highly successful expression system for optimal synthesis of heterologous proteins. J. Cell. Physiol. 235, 5867–5881. doi: 10.1002/jcp.29583
Karim, A., Gerliani, N., and Aider, M. (2020). Kluyveromyces marxianus: an emerging yeast cell factory for applications in food and biotechnology. Int. J. Food Microbiol. 333:108818. doi: 10.1016/j.ijfoodmicro.2020.108818
Karim, N., Shishir, M. R. I., Gowd, V., and Chen, W. (2022). Hesperidin-an emerging bioactive compound against metabolic diseases and its potential biosynthesis pathway in microorganism. Food Rev. Intl. 38, 170–192. doi: 10.1080/87559129.2020.1858312
Kitamoto, N., Matsui, J., Kawai, Y., Kato, A., Yoshino, S., Ohmiya, K., et al. (1998). Utilization of the TEF1-alpha gene (TEF1) promoter for expression of polygalacturonase genes, pgaA and pgaB, in Aspergillus oryzae. Appl. Microbiol. Biotechnol. 50, 85–92. doi: 10.1007/s002530051260
Kobayashi, Y., Inokuma, K., Matsuda, M., Kondo, A., and Hasunuma, T. (2022). Resveratrol production of a recombinant Scheffersomyces stipitis strain from molasses. Biotechnol. Notes 3, 1–7. doi: 10.1016/j.biotno.2021.11.001
Kooistra, R., Hooykaas, P. J., and Steensma, H. Y. (2004). Efficient gene targeting in Kluyveromyces lactis. Yeast 21, 781–792. doi: 10.1002/yea.1131
Kumar, P., Sahoo, D. K., and Sharma, D. (2021). The identification of novel promoters and terminators for protein expression and metabolic engineering applications in Kluyveromyces marxianus. Metab. Eng. Commun. 12:e00160. doi: 10.1016/j.mec.2020.e00160
Lai, J., Song, L., Zhou, Y., Zong, H., Zhuge, B., and Lu, X. (2024). Fine-tuned gene expression elements from hybrid promoter libraries in Pichia pastoris. ACS Synth. Biol. 13, 310–318. doi: 10.1021/acssynbio.3c00534
Larroude, M., Rossignol, T., Nicaud, J. M., and Ledesma-Amaro, R. (2018). Synthetic biology tools for engineering Yarrowia lipolytica. Biotechnol. Adv. 36, 2150–2164. doi: 10.1016/j.biotechadv.2018.10.004
Li, M., Chu, Y., Dong, X., and Ji, H. (2023). General mechanisms of weak acid-tolerance and current strategies for the development of tolerant yeasts. World J. Microbiol. Biotechnol. 40:49. doi: 10.1007/s11274-023-03875-y
Li, X., Gadar-Lopez, A. E., Chen, L., Jayachandran, S., Cruz-Morales, P., and Keasling, J. D. (2023). Mining natural products for advanced biofuels and sustainable bioproducts. Curr. Opin. Biotechnol. 84:103003. doi: 10.1016/j.copbio.2023.103003
Li, M., Lang, X., Moran Cabrera, M., De Keyser, S., Sun, X., Da Silva, N., et al. (2021). CRISPR-mediated multigene integration enables shikimate pathway refactoring for enhanced 2-phenylethanol biosynthesis in Kluyveromyces marxianus. Biotechnol. Biofuels 14:3. doi: 10.1186/s13068-020-01852-3
Li, W., Shen, X., Wang, J., Sun, X., and Yuan, Q. (2021). Engineering microorganisms for the biosynthesis of dicarboxylic acids. Biotechnol. Adv. 48:107710. doi: 10.1016/j.biotechadv.2021.107710
Liu, Z., Huang, M., Chen, H., Lu, X., Tian, Y., Hu, P., et al. (2024). Metabolic engineering of Yarrowia lipolytica for high-level production of squalene. Bioresour. Technol. 394:130233. doi: 10.1016/j.biortech.2023.130233
Liu, H., Jin, Y., Zhang, R., Ning, Y., Yu, Y., Xu, P., et al. (2023). Recent advances and perspectives on production of value-added organic acids through metabolic engineering. Biotechnol. Adv. 62:108076. doi: 10.1016/j.biotechadv.2022.108076
Liu, X. B., Liu, M., Tao, X. Y., Zhang, Z. X., Wang, F. Q., and Wei, D. Z. (2015). Metabolic engineering of Pichia pastoris for the production of dammarenediol-II. J. Biotechnol. 216, 47–55. doi: 10.1016/j.jbiotec.2015.10.005
Liu, Y., Tu, X., Xu, Q., Bai, C., Kong, C., Liu, Q., et al. (2018). Engineered monoculture and co-culture of methylotrophic yeast for de novo production of monacolin J and lovastatin from methanol. Metab. Eng. 45, 189–199. doi: 10.1016/j.ymben.2017.12.009
Liu, K., Xiang, G., Li, L., Liu, T., Ke, J., Xiong, L., et al. (2024). Engineering non-conventional yeast Rhodotorula toruloides for ergothioneine production. Biotechnol. Biofuels Bioprod. 17:65. doi: 10.1186/s13068-024-02516-2
Lobs, A. K., Schwartz, C., Thorwall, S., and Wheeldon, I. (2018). Highly multiplexed CRISPRi repression of respiratory functions enhances mitochondrial localized ethyl acetate biosynthesis in Kluyveromyces marxianus. ACS Synth. Biol. 7, 2647–2655. doi: 10.1021/acssynbio.8b00331
Lobs, A. K., Schwartz, C., and Wheeldon, I. (2017). Genome and metabolic engineering in non-conventional yeasts: current advances and applications. Synth. Syst. Biotechnol. 2, 198–207. doi: 10.1016/j.synbio.2017.08.002
Loser, C., Urit, T., Stukert, A., and Bley, T. (2013). Formation of ethyl acetate from whey by Kluyveromyces marxianus on a pilot scale. J. Biotechnol. 163, 17–23. doi: 10.1016/j.jbiotec.2012.10.009
Lyu, Y., Wu, P., Zhou, J., Yu, Y., and Lu, H. (2021). Protoplast transformation of Kluyveromyces marxianus. Biotechnol. J. 16:e2100122. doi: 10.1002/biot.202100122
Ma, J., Gu, Y., Marsafari, M., and Xu, P. (2020). Synthetic biology, systems biology, and metabolic engineering of Yarrowia lipolytica toward a sustainable biorefinery platform. J. Ind. Microbiol. Biotechnol. 47, 845–862. doi: 10.1007/s10295-020-02290-8
Ma, Y., Shang, Y., and Stephanopoulos, G. (2024). Engineering peroxisomal biosynthetic pathways for maximization of triterpene production in Yarrowia lipolytica. Proc. Natl. Acad. Sci. USA 121:e2314798121. doi: 10.1073/pnas.2314798121
Maassen, N., Freese, S., Schruff, B., Passoth, V., and Klinner, U. (2008). Nonhomologous end joining and homologous recombination DNA repair pathways in integration mutagenesis in the xylose-fermenting yeast Pichia stipitis. FEMS Yeast Res. 8, 735–743. doi: 10.1111/j.1567-1364.2008.00383.x
Macauley-Patrick, S., Fazenda, M. L., Mcneil, B., and Harvey, L. M. (2005). Heterologous protein production using the Pichia pastoris expression system. Yeast 22, 249–270. doi: 10.1002/yea.1208
Madhavan, A., Jose, A. A., Binod, P., Sindhu, R., Sukumaran, R. K., Pandey, A., et al. (2017). Synthetic biology and metabolic engineering approaches and its impact on non-conventional yeast and biofuel production. Front. Energy Res. 5:8. doi: 10.3389/fenrg.2017.00008
Madzak, C. (2021). Yarrowia lipolytica strains and their biotechnological applications: how natural biodiversity and metabolic engineering could contribute to cell factories improvement. J. Fungi 7:548. doi: 10.3390/jof7070548
Madzak, C., Treton, B., and Blanchin-Roland, S. (2000). Strong hybrid promoters and integrative expression/secretion vectors for quasi-constitutive expression of heterologous proteins in the yeast Yarrowia lipolytica. J. Mol. Microbiol. Biotechnol. 2, 207–216.
Markham, K. A., and Alper, H. S. (2018). Synthetic biology expands the industrial potential of Yarrowia lipolytica. Trends Biotechnol. 36, 1085–1095. doi: 10.1016/j.tibtech.2018.05.004
Mastella, L., Senatore, V., Beltrani, T., and Branduardi, P. (2023). Scheffersomyces stipitis ability to valorize different residual biomasses for vitamin B(9) production. Microb. Biotechnol. 16, 392–403. doi: 10.1111/1751-7915.14177
Monteiro De Oliveira, P., Aborneva, D., Bonturi, N., and Lahtvee, P. J. (2021). Screening and growth characterization of non-conventional yeasts in a Hemicellulosic hydrolysate. Front. Bioeng. Biotechnol. 9:659472. doi: 10.3389/fbioe.2021.659472
Moon, S. Y., Son, S.-H., Baek, S.-H., and Lee, J. Y. (2024). Designing microbial cell factories for programmable control of cellular metabolism. Curr. Opin. Syst. Biol. 37:100493. doi: 10.1016/j.coisb.2023.100493
Moon, S. Y., Son, S.-H., Oh, S. S., and Lee, J. Y. (2023). Harnessing cellular organelles to bring new functionalities into yeast. Biotechnol. Bioprocess Eng. 28, 936–948. doi: 10.1007/s12257-022-0195-5
Morrissey, J. P., Etschmann, M. M., Schrader, J., and De Billerbeck, G. M. (2015). Cell factory applications of the yeast Kluyveromyces marxianus for the biotechnological production of natural flavour and fragrance molecules. Yeast 32, 3–16. doi: 10.1002/yea.3054
Naatsaari, L., Mistlberger, B., Ruth, C., Hajek, T., Hartner, F. S., and Glieder, A. (2012). Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One 7:e39720. doi: 10.1371/journal.pone.0039720
Naumov, G. I., Naumova, E. S., Barrio, E., and Querol, A. (2006). Genetic and molecular study of inability of the yeast Kluyveromyces lactis var drosophilarum to ferment lactose. Mikrobiologiia 75, 299–304. doi: 10.1134/S0026261706030027
Nielsen, J. (2013). Production of biopharmaceutical proteins by yeast: advances through metabolic engineering. Bioengineered 4, 207–211. doi: 10.4161/bioe.22856
Nurcholis, M., Lertwattanasakul, N., Rodrussamee, N., Kosaka, T., Murata, M., and Yamada, M. (2020). Integration of comprehensive data and biotechnological tools for industrial applications of Kluyveromyces marxianus. Appl. Microbiol. Biotechnol. 104, 475–488. doi: 10.1007/s00253-019-10224-3
Nurdiani, D., Putro, E. W., Septisetyani, E. P., Utami, N., Hariyatun, H., Prasetyaningrum, P. W., et al. (2024). Characterization of functional human insulin produced using the Pichia pastoris expression system. J. Appl. Pharm. Sci. 14, 216–226. doi: 10.7324/JAPS.2024.180473
Park, J., Kim, I. J., and Kim, S. R. (2022). Nonconventional yeasts engineered using the CRISPR-Cas system as emerging microbial cell factories. Fermentation 8:656. doi: 10.3390/fermentation8110656
Park, B. G., Kim, J., Kim, E. J., Kim, Y., Kim, J., Kim, J. Y., et al. (2021). Application of random mutagenesis and synthetic FadR promoter for de novo production of ω-Hydroxy fatty acid in Yarrowia lipolytica. Front. Bioeng. Biotechnol. 9:624838. doi: 10.3389/fbioe.2021.624838
Park, Y. K., Studena, L., Hapeta, P., Haddouche, R., Bell, D. J., Torres-Montero, P., et al. (2025). Efficient biosynthesis of beta-caryophyllene by engineered Yarrowia lipolytica. Microb. Cell Factories 24:38. doi: 10.1186/s12934-025-02660-w
Patra, P., Das, M., Kundu, P., and Ghosh, A. (2021). Recent advances in systems and synthetic biology approaches for developing novel cell-factories in non-conventional yeasts. Biotechnol. Adv. 47:107695. doi: 10.1016/j.biotechadv.2021.107695
Perpetuini, G., Tittarelli, F., Perla, C., and Tofalo, R. (2022). Influence of different aggregation states on volatile organic compounds released by dairy Kluyveromyces marxianus strains. Food Secur. 11:2910. doi: 10.3390/foods11182910
Qian, Z., Yu, J., Chen, X., Kang, Y., Ren, Y., Liu, Q., et al. (2022). De novo production of plant 4'-Deoxyflavones Baicalein and Oroxylin a from ethanol in Crabtree-negative yeast. ACS Synth. Biol. 11, 1600–1612. doi: 10.1021/acssynbio.2c00026
Qiu, Y., Lei, P., Wang, R., Sun, L., Luo, Z., Li, S., et al. (2023). Kluyveromyces as promising yeast cell factories for industrial bioproduction: from bio-functional design to applications. Biotechnol. Adv. 64:108125. doi: 10.1016/j.biotechadv.2023.108125
Qiu, X., Zhang, J., Zhou, J., Fang, Z., Zhu, Z., Li, J., et al. (2019). Stress tolerance phenotype of industrial yeast: industrial cases, cellular changes, and improvement strategies. Appl. Microbiol. Biotechnol. 103, 6449–6462. doi: 10.1007/s00253-019-09993-8
Rainha, J., Rodrigues, J. L., and Rodrigues, L. R. (2020). CRISPR-Cas9: a powerful tool to efficiently engineer Saccharomyces cerevisiae. Life (Basel) 11:13. doi: 10.3390/life11010013
Rajkumar, A. S., Varela, J. A., Juergens, H., Daran, J. G., and Morrissey, J. P. (2019). Biological parts for Kluyveromyces marxianus synthetic biology. Front. Bioeng. Biotechnol. 7:97. doi: 10.3389/fbioe.2019.00097
Ramakrishnan, K., Prattipati, M., Samuel, P., and Sankaranarayanan, M. (2020). Transcriptional control of gene expression in Pichia pastoris by manipulation of terminators. Appl. Microbiol. Biotechnol. 104, 7841–7851. doi: 10.1007/s00253-020-10785-8
Raschmanova, H., Weninger, A., Glieder, A., Kovar, K., and Vogl, T. (2018). Implementing CRISPR-Cas technologies in conventional and non-conventional yeasts: current state and future prospects. Biotechnol. Adv. 36, 641–665. doi: 10.1016/j.biotechadv.2018.01.006
Rebello, S., Abraham, A., Madhavan, A., Sindhu, R., Binod, P., Karthika Bahuleyan, A., et al. (2018). Non-conventional yeast cell factories for sustainable bioprocesses. FEMS Microbiol. Lett. 365. doi: 10.1093/femsle/fny222
Rerop, Z. S., Stellner, N. I., Graban, P., Haack, M., Mehlmer, N., Masri, M., et al. (2023). Bioconversion of a lignocellulosic hydrolysate to single cell oil for biofuel production in a cost-efficient fermentation process. Fermentation 9:189. doi: 10.3390/fermentation9020189
Roy, P., Gahlawat, V. K., Saravanan, C., and Singh, B. P. (2023). Enhancing bioflavor production by solid-state fermentation using Kluyveromyces marxianus and l-phenylalanine. J. Basic Microbiol. 63, 75–91. doi: 10.1002/jobm.202200503
Ruchala, J., Kurylenko, O. O., Dmytruk, K. V., and Sibirny, A. A. (2020). Construction of advanced producers of first- and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Ind. Microbiol. Biotechnol. 47, 109–132. doi: 10.1007/s10295-019-02242-x
Saraya, R., Krikken, A. M., Kiel, J. A., Baerends, R. J., Veenhuis, M., and Van Der Klei, I. J. (2012). Novel genetic tools for Hansenula polymorpha. FEMS Yeast Res. 12, 271–278. doi: 10.1111/j.1567-1364.2011.00772.x
Schwartz, C. M., Hussain, M. S., Blenner, M., and Wheeldon, I. (2016). Synthetic RNA polymerase iii promoters facilitate high-efficiency CRISPR-Cas9-mediated genome editing in Yarrowia lipolytica. ACS Synth. Biol. 5, 356–359. doi: 10.1021/acssynbio.5b00162
Sharpe, P. L., Rick, W. Y., and Zhu, Q. Q. (2014). Carotenoid production in a recombinant oleaginous yeast. United States: Google Patents.
Shi, S., and Zhao, H. (2017). Metabolic engineering of oleaginous yeasts for production of fuels and chemicals. Front. Microbiol. 8:2185. doi: 10.3389/fmicb.2017.02185
Shrivastava, A., Pal, M., and Sharma, R. K. (2023). Pichia as yeast cell factory for production of industrially important bio-products: current trends, challenges, and future prospects. J. Bioresour. Bioprod. 8, 108–124. doi: 10.1016/j.jobab.2023.01.007
Sibirny, A. A. (2023). Metabolic engineering of non-conventional yeasts for construction of the advanced producers of biofuels and high-value chemicals. BBA Adv. 3:100071. doi: 10.1016/j.bbadva.2022.100071
Son, S. H., Kang, J., Shin, Y., Lee, C., Sung, B. H., Lee, J. Y., et al. (2024). Sustainable production of natural products using synthetic biology: Ginsenosides. J. Ginseng Res. 48, 140–148. doi: 10.1016/j.jgr.2023.12.006
Spohner, S. C., Muller, H., Quitmann, H., and Czermak, P. (2015). Expression of enzymes for the usage in food and feed industry with Pichia pastoris. J. Biotechnol. 202, 118–134. doi: 10.1016/j.jbiotec.2015.01.027
Steiner, S., and Philippsen, P. (1994). Sequence and promoter analysis of the highly expressed TEF gene of the filamentous fungus Ashbya gossypii. Mol. Gen. Genet. 242, 263–271. doi: 10.1007/BF00280415
Sun, M. L., Shi, T. Q., Lin, L., Ledesma-Amaro, R., and Ji, X. J. (2022). Advancing Yarrowia lipolytica as a superior biomanufacturing platform by tuning gene expression using promoter engineering. Bioresour. Technol. 347:126717. doi: 10.1016/j.biortech.2022.126717
Teo, W. S., and Chang, M. W. (2014). Development and characterization of and-gate dynamic controllers with a modular synthetic Gal1 core promoter in Saccharomyces cerevisiae. Biotechnol. Bioeng. 111, 144–151. doi: 10.1002/bit.25001
Thorwall, S., Schwartz, C., Chartron, J. W., and Wheeldon, I. (2020). Stress-tolerant non-conventional microbes enable next-generation chemical biosynthesis. Nat. Chem. Biol. 16, 113–121. doi: 10.1038/s41589-019-0452-x
Toivari, M. H., Nygard, Y., Penttila, M., Ruohonen, L., and Wiebe, M. G. (2012). Microbial D-xylonate production. Appl. Microbiol. Biotechnol. 96, 1–8. doi: 10.1007/s00253-012-4288-5
Toyoda, T., and Ohtaguchi, K. (2011). D-arabitol production from lactose by Kluyveromyces lactis at different aerobic conditions. J. Chem. Technol. Biotechnol. 86, 217–222. doi: 10.1002/jctb.2497
Varela, J. A., Gethins, L., Stanton, C., Ross, P., and Morrissey, J. P. (2017). Applications of Kluyveromyces marxianus in biotechnology. Yeast Diver. Hum. Welf., 439–453. doi: 10.1007/978-981-10-2621-8_17
Verbeke, J., Beopoulos, A., and Nicaud, J. M. (2013). Efficient homologous recombination with short length flanking fragments in KU70 deficient Yarrowia lipolytica strains. Biotechnol. Lett. 35, 571–576. doi: 10.1007/s10529-012-1107-0
Vogl, T., Sturmberger, L., Kickenweiz, T., Wasmayer, R., Schmid, C., Hatzl, A. M., et al. (2016). A toolbox of diverse promoters related to methanol utilization: functionally verified parts for heterologous pathway expression in Pichia pastoris. ACS Synth. Biol. 5, 172–186. doi: 10.1021/acssynbio.5b00199
Wagner, J. M., and Alper, H. S. (2016). Synthetic biology and molecular genetics in non-conventional yeasts: current tools and future advances. Fungal Genet. Biol. 89, 126–136. doi: 10.1016/j.fgb.2015.12.001
Wang, C., Lin, M., Yang, Z., Lu, X., Liu, Y., Lu, H., et al. (2023). Characterization of the endogenous promoters in Yarrowia lipolytica for the biomanufacturing applications. Process Biochem. 124, 245–252. doi: 10.1016/j.procbio.2022.11.023
Wang, Y., Wang, Y., Cui, J., Wu, C., Yu, B., and Wang, L. (2025). Non-conventional yeasts: promising cell factories for organic acid bioproduction. Trends Biotechnol. doi: 10.1016/j.tibtech.2024.12.004
Wang, Z., Zhou, L., Lu, M., Zhang, Y., Perveen, S., Zhou, H., et al. (2021). Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl. Microbiol. Biotechnol. 105, 1745–1758. doi: 10.1007/s00253-021-11130-3
Weninger, A., Hatzl, A. M., Schmid, C., Vogl, T., and Glieder, A. (2016). Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 235, 139–149. doi: 10.1016/j.jbiotec.2016.03.027
Whitford, C. M., Cruz-Morales, P., Keasling, J. D., and Weber, T. (2021). The design-build-test-learn cycle for metabolic engineering of streptomycetes. Essays Biochem. 65, 261–275. doi: 10.1042/EBC20200132
Wu, X., Cai, P., Yao, L., and Zhou, Y. J. (2023). Genetic tools for metabolic engineering of Pichia pastoris. Eng. Microbiol. 3:100094. doi: 10.1016/j.engmic.2023.100094
Xia, Y., Li, Y., Shen, W., Yang, H., and Chen, X. (2023). CRISPR-Cas Technology for Bioengineering Conventional and non-Conventional Yeasts: Progress and new challenges. Int. J. Mol. Sci. 24:15310. doi: 10.3390/ijms242015310
Xie, D., Miller, E., Sharpe, P., Jackson, E., and Zhu, Q. (2017). Omega-3 production by fermentation of Yarrowia lipolytica: from fed-batch to continuous. Biotechnol. Bioeng. 114, 798–812. doi: 10.1002/bit.26216
Xie, L. F., Yu, W., Gao, J. Q., Wang, H. Y., and Zhou, Y. J. J. (2024). Ogataea polymorpha as a next-generation chassis for industrial biotechnology. Trends Biotechnol. 42, 1363–1378. doi: 10.1016/j.tibtech.2024.03.007
Xiong, X., and Chen, S. (2020). Expanding toolbox for genes expression of Yarrowia lipolytica to include novel inducible, repressible, and hybrid promoters. ACS Synth. Biol. 9, 2208–2213. doi: 10.1021/acssynbio.0c00243
Yang, Z., Edwards, H., and Xu, P. (2020). Crispr-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab. Eng. Commun. 10:e00112. doi: 10.1016/j.mec.2019.e00112
Yang, Z., and Zhang, Z. (2018). Engineering strategies for enhanced production of protein and bio-products in Pichia pastoris: a review. Biotechnol. Adv. 36, 182–195. doi: 10.1016/j.biotechadv.2017.11.002
Zha, J., Liu, D., Ren, J., Liu, Z., and Wu, X. (2023). Advances in metabolic engineering of Pichia pastoris strains as powerful cell factories. J. Fungi (Basel) 9:1027. doi: 10.3390/jof9101027
Zhang, L., Fan, C., Yang, H., Xia, Y., Shen, W., and Chen, X. (2024). Biosynthetic pathway redesign in non-conventional yeast for enhanced production of cembratriene-ol. Bioresour. Technol. 399:130596. doi: 10.1016/j.biortech.2024.130596
Zhang, X., Gu, S., Zheng, X., Peng, S., Li, Y., Lin, Y., et al. (2021). A novel and efficient genome editing tool assisted by CRISPR-Cas12a/Cpf1 for Pichia pastoris. ACS Synth. Biol. 10, 2927–2937. doi: 10.1021/acssynbio.1c00172
Zhang, T.-L., Yu, H.-W., and Ye, L.-D. (2023). Metabolic engineering of Yarrowia lipolytica for terpenoid production: tools and strategies. ACS Synth. Biol. 12, 639–656. doi: 10.1021/acssynbio.2c00569
Zhao, Y., Liu, S., Lu, Z., Zhao, B., Wang, S., Zhang, C., et al. (2021). Hybrid promoter engineering strategies in Yarrowia lipolytica: isoamyl alcohol production as a test study. Biotechnol. Biofuels 14:149. doi: 10.1186/s13068-021-02002-z
Keywords: non-conventional yeast, synthetic biology, gene editing tool, metabolic engineering, yeast biotechnology
Citation: Moon SY, An N-Y and Lee JY (2025) Transforming non-conventional yeasts into key players in biotechnology: advances in synthetic biology applications. Front. Microbiol. 16:1600187. doi: 10.3389/fmicb.2025.1600187
Edited by:
Dongsoo Yang, Korea University, Republic of KoreaReviewed by:
Nam Kyu Kang, Kyung Hee University, Republic of KoreaYong Hee Han, Chonnam National University, Republic of Korea
Copyright © 2025 Moon, An and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ju Young Lee, anV5b3VuZ2xlZUBrYWlzdC5hYy5rcg==
 Soo Young Moon
Soo Young Moon Nan-Yeong An
Nan-Yeong An Ju Young Lee
Ju Young Lee