- 1School of Life Science and Technology, Wuhan Polytechnic University, Wuhan, China
- 2Hubei Baiyunbian Liquor Industry Co., Ltd., Songzi, China
It is now understood that n-propanol, a higher alcohol constituent in Baijiu, significantly impacts its overall quality. Excessive levels can impart a spicy and bitter taste, necessitating strict control. Despite its importance, the precise mechanism underlying the production of high-content n-propanol in Baijiu brewing and the specific microorganisms responsible for this process remain unclear. This study isolated Lentilactobacillus diolivorans ZX6, known for its high n-propanol production, from Baijiu fermented grains using a modified MRS (SMRS) enrichment medium and gas chromatography detection. The pure strain ZX6 produced 4,399 mg/L of n-propanol in SMRS medium. Using the key enzyme gene sequence for 1,2-propanediol metabolism in L. diolivorans, we designed primers and established a quantitative PCR method to quantify L. diolivorans in first-round fermented grains from three different sauce-flavor Baijiu distilleries. The widespread presence of L. diolivorans in sauce-flavor Baijiu fermented grains indicates its potential role as one of the key microorganisms responsible for the high concentrations of n-propanol. L. diolivorans ZX6 directly utilized glucose fermentation to produce large amounts of n-propanol. High n-propanol synthesis from L. diolivorans ZX6 required high sugar concentrations, anaerobic conditions, and extended fermentation times. Transcriptome analysis revealed that pyruvate and lactic acid, likely major precursors, could enter the methylglyoxal pathway under the catalysis of lactaldehyde dehydrogenase, respectively. These compounds were subsequently metabolized to 1,2-propanediol and then to n-propanol. These findings provide insights into identifying n-propanol-producing microorganisms and establish a theoretical basis for elucidating the high-yield n-propanol mechanism in Baijiu brewing, along with strategies for regulating its content.
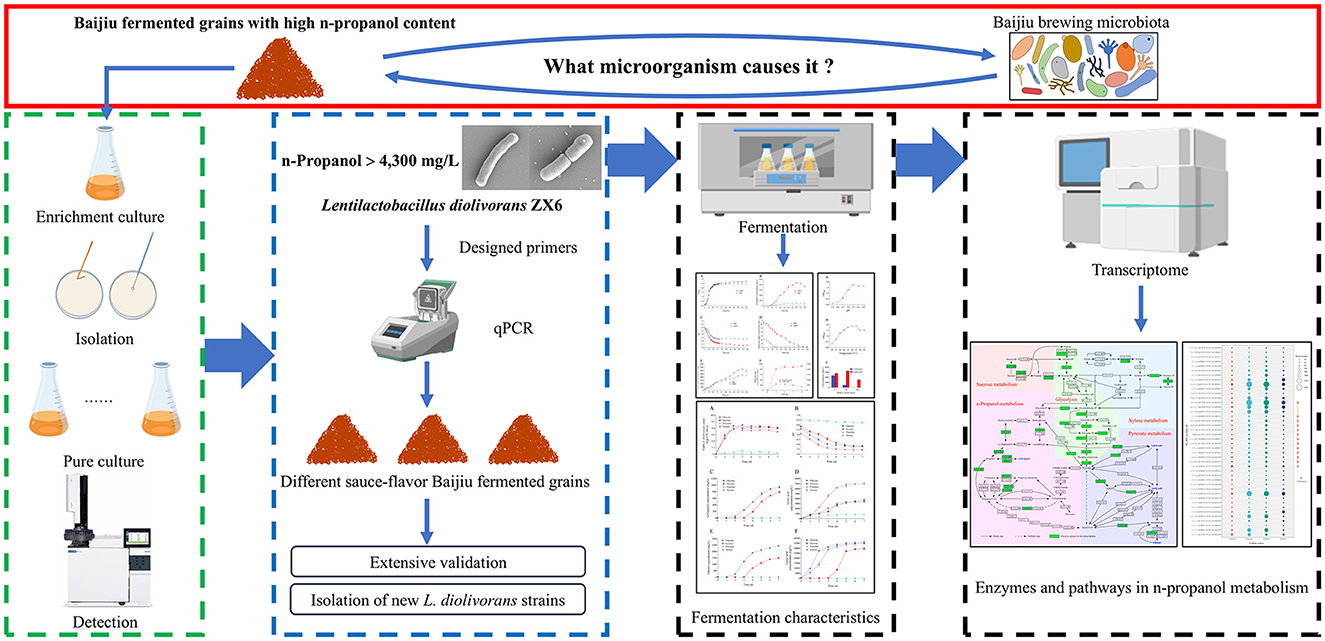
Graphical Abstract. Isolation, validation, and metabolic characterization of Lentilactobacillus diolivorans as a key producer of high n-propanol in Baijiu fermented grains.
1 Introduction
Baijiu (Chinese liquor), one of the world's six renowned distilled spirits, derives its quality from its flavor components (Jin et al., 2017). Higher alcohols, mainly including n-propanol, isobutanol, and isoamylol, are key flavor compounds in Baijiu (Gao et al., 2014; Tian et al., 2020). Their concentration significantly impacts Baijiu's flavor profile and drinking comfort. While moderate levels enhance flavor, excessive amounts can lead to off-flavors and discomfort (Mckarns et al., 1997; Strubelt et al., 1999). Therefore, higher alcohol content is strictly regulated in Baijiu. Among higher alcohols, n-propanol can impart a spicy and bitter taste at high concentrations (Dong et al., 2002; Luo et al., 2025). Various microorganisms can synthesize n-propanol, with yeast being the primary contributor to higher alcohols (Giudici et al., 1990; Wu et al., 2016; Hu et al., 2021; Wei et al., 2024). Bacteria and molds also possess limited n-propanol production capabilities (Janssen, 2004; Siebert and Wendisch, 2015; Matsubara et al., 2016; Walther and François, 2016). Traditional Chinese Baijiu's multi-batch solid-state fermentation and distillation process involves a diverse microbial community, including bacteria, yeast, and molds. This complexity has made the identification of the specific microorganisms responsible for target flavor substances challenging (Liu et al., 2017; Hao et al., 2021; Tu et al., 2022; Wang et al., 2025).
Sauce-flavor Baijiu (SFBJ), a representative Baijiu type, is characterized by its stacking fermentation and pit fermentation, which involve multiple fermentation rounds and nine rounds of steaming (Zhou et al., 2023; Chen et al., 2024b). A common issue in SFBJ production is the abnormally high n-propanol content in the first and second distillation raw liquors, significantly affecting Baijiu quality. Baiyunbian Liquor, a widely consumed sauce-strong-flavor Baijiu, employs the production techniques of SFBJ in the first seven rounds. Its n-propanol content in the first and second raw liquors reportedly reaches 25,690 mg/L and 18,011 mg/L, respectively, constituting 92% of total higher alcohols (Dong et al., 2002). Gas chromatography analysis of SFBJ in the Maotai town, Guizhou province, revealed that the n-propanol content in the third raw liquor was as high as 3,247 mg/L, accounting for 94.1% of total higher alcohols (n-propanol, isobutanol, isoamylol) (Guo et al., 2022). The production of such high n-propanol levels in Baijiu fermented grains cannot be explained solely by the n-propanol yields of existing pure or mixed strains of microorganisms. Studies indicate that pure yeast strains produce < 80 mg/L (kg) of n-propanol during liquid and solid-state fermentation, with n-propanol accounting for < 50% of total higher alcohols (Giudici et al., 1990; Qu et al., 2021). Our mixed fermentation experiments with three yeast species (Saccharomyces cerevisiae, Pichia kudriavzevii, Candida tropicalis) and three lactic acid bacteria (LAB) species (Lactiplantibacillus plantarum, Lentilactobacillus buchneri, Lactobacillus acetotolerans) isolated from SFBJ fermented grains did not reveal significant synergistic effects on n-propanol production. There was also a phenomenon of abnormally high levels of n-propanol during Xiaoqu Baijiu (XQBJ) fermentation (Qu et al., 2021). Therefore, the microorganisms or microbial communities capable of producing high levels of n-propanol in the Baijiu fermentation system remain to be elucidated.
Although the content 006Ff n-propanol in the first round of fermented grains of SFBJ was higher than that of XQBJ, the sampling time was strictly limited by the production cycle of SFBJ. To identify and isolate potential high n-propanol-producing microbial strains within the Baijiu brewing system, we initially collected high n-propanol-containing XQBJ fermented grains and inoculated them into a liquid medium for enrichment culture, which was then followed by dilution plate separation. After isolation and characterization of single colonies, their n-propanol fermentation yields were determined to screen for high-producing strains. Using the target high n-propanol-producing bacterium's genome information, we designed specific PCR primers to target n-propanol synthesis genes. We employed fluorescent quantitative PCR (qPCR) to quantify the target bacterium in the first round of fermented grains from representative SFBJ, and then isolated and identified new target bacterium strains. Furthermore, we investigated the effects of culture medium sugar concentration and fermentation conditions on n-propanol production by the target strain, aiming to elucidate the external factors contributing to high n-propanol formation during Baijiu brewing. Finally, transcriptomic analysis of the target strain grown on different carbon sources was performed, revealing its potential metabolic pathways for n-propanol biosynthesis.
2 Materials and methods
2.1 Sample collection and sources
Fermented grain samples from the first round of SFBJ pit fermentation (30 days) were collected from three distilleries in three regions: Songzi, Hubei (D1, D2); Shennongjia, Hubei (SNJ-1, SNJ-2); and Maotai town, Guizhou (GZ-1, GZ-2) (Supplementary Table S1). XQBJ fermented grains (sample X) were collected from Songzi, Hubei. For each fermentation pit, 2 kg of fermented grains were collected from the upper, middle, and lower layers and then thoroughly mixed to form the composite sample for analysis. Samples were stored at −20°C or 4°C for subsequent flavor and microbial analysis. DNA was extracted from fermented grains with high n-propanol content, and Lentilactobacillus diolivorans quantification was performed. Stacking-fermented (48 h) samples and Aged Zaopei (distilled fermented grains from XQBJ pit fermentation) were obtained from Hubei Baiyunbian Liquor Industry Co., Ltd. (Songzi, China).
2.2 Microorganisms and media
Limosilactobacillus panis strain CGMCC 1.3925 was obtained from China General Microbiological Culture Collection Center. The LAB strains L. diolivorans ZX6, ZD9, ZD92, and ZD93 were isolated and identified from Baijiu fermented grains in this study. S. cerevisiae SCY62 was isolated from SFBJ fermented grains and stored in our laboratory. Rhizopus Qu (Angel Rhizopus), used as a saccharifying agent in XQBJ fermentation, was purchased from Angel Yeast Co., Ltd. (Yichang, Hubei, China).
MRS medium (Hopebio Company, Qingdao, China), containing 10 g/L peptone, 8 g/L beef extract, 4 g/L yeast extract, 20 g/L glucose, 5 g/L sodium acetate, 1 g/L Tween-80, 2 g/L dipotassium phosphate, 2 g/L ammonium citrate, 0.2 g/L magnesium sulfate, 0.04 g/L manganese sulfate, and adjusted to pH 6.5 ± 0.2, was used for culturing and enumerating the total number of LAB. Yeast extract-peptone-dextrose (YPD) medium, consisting of 20 g/L glucose, 20 g/L peptone, and 10 g/L yeast extract, was used for yeast culturing. Rose Bengal Agar (Hopebio Company, Qingdao, China) was employed for culturing and counting yeast populations.
Sorghum extract medium (SS) was used for fermentation and enrichment culture preparation. The sorghum extract preparation method was previously described in the literature (Deng et al., 2020). The obtained sorghum extract was diluted with water to achieve a final sugar concentration of 12 °Bx and adjusted to a pH of 5.8 using a 1:1 mixture of lactic acid to acetic acid. Fermentation media were sterilized at 121°C for 20 min. The modified MRS (SMRS) enrichment medium, a 1:1 mixture of SS and MRS medium with a pH of 6.1 ± 0.1, was used for n-propanol-producing microorganism enrichment and isolation. Nystatin and ampicillin trihydrate (Macklin Biochemical Technology Co., Ltd., China) were added at concentrations of 100 mg/L and 80 mg/L, respectively. Xiaoqu Baijiu solid fermentation medium (XBSM) consisted of 40% (w/w) fresh steamed sorghum (sorghum cooked and cooled with pH 6.95) and 60% (w/w) sterilized aged Zaopei (with pH 3.75). This medium was used for solid-state mixed fermentation of yeast and LAB.
2.3 Laboratory fermentation
To evaluate n-propanol production by L. diolivorans in liquid co-culture systems, S. cerevisiae SCY62 and L. diolivorans ZX6 were first cultured separately under the following conditions: SCY62 in YPD medium at 28°C with 180 rpm shaking for 24 h, and ZX6 in MRS medium under static conditions at 30°C for 72 h. Mixed-culture fermentations were conducted in 250 mL triangular flasks containing 150 mL of SMRS medium. The inoculation ratios (cells/cells) of S. cerevisiae SCY62 to L. diolivorans ZX6 were 1:0, 1:1, 1:0.1, and 1:0.01, respectively. The initial cell concentration of S. cerevisiae SCY62 in fermentation medium was 1 × 106 cells/mL. Static cultures were maintained at 30°C for 6 days.
Five hundred milliliters of plastic bottles were used as containers for solid-state fermentation. XQBJ solid-state fermentation was conducted using an XBSM medium. Twelve XBSM medium samples were inoculated with 5% (w/w) of Rhizopus Qu, 5 × 105 CFU/g of S. cerevisiae SCY62, and varying amounts of L. diolivorans ZX6 (0, 104, 105, and 106 CFU/g). The fermentation material was initially fermented by stacking in a plastic box at 28°C for 24 h, then transferred to 500 mL plastic bottles for anaerobic fermentation at 28°C for 10–20 days.
To simulate the pit fermentation of SFBJ, fermented grains that had completed stacking fermentation were collected from the Baijiu distillery, individually packed into 500 mL plastic bottles, and incubated. Five fermented grain samples from the first round of SFBJ stacking fermentation at Baiyunbian Liquor Industry Co., Ltd. were randomly selected and separated. Five groups labeled Pit No. 1 through No. 5 were established, each containing 15 parallel plastic bottles. Fifteen kilograms of mixed samples were taken from each stacking fermentation mash and distributed equally among the 500 mL plastic bottles in the five groups. The bottles were sealed and incubated at 28°C for 28 days. Gas chromatography and qPCR analyses were conducted on samples from each fermentation pit at 0, 7, 14, 21, and 28 days. Three sample bottles were taken at each time point for analysis. Additionally, these samples were used for LAB isolation (refer to Section 2.6).
2.4 Enrichment and isolation of high n-propanol-producing bacteria
The sample X characterized by its high n-propanol content in XQBJ was selected and inoculated into 150 mL of MRS, SS, and SMRS liquid triangular flask media at a 5% (w/w) inoculation rate. Static cultures were maintained at 30°C for 5 days. The sample with the highest n-propanol content was serially diluted and plated onto SMRS agar plates containing 100 mg/L of nystatin. Plates were incubated at 30°C for 3 days, and single colony morphology was observed. Forty single bacterial colonies with distinct morphological characteristics were selected for 16S rRNA gene sequence analysis and strain identification. Twenty bacterial strains representing different species were selected and inoculated into 250 mL Erlenmeyer flasks containing 150 mL of SMRS medium. Static cultures were maintained at 30°C for 7 days. The strain exhibiting the highest n-propanol production ability was selected, and its morphological characteristics were captured using scanning electron microscopy.
2.5 Establishment of an absolute quantitative method for L. diolivorans
Total DNA was extracted from fermented grains following the method described in the literature (Liu et al., 2017). The genome sequence of L. diolivorans (GenBank GCA_007992335.1) was obtained from the National Center for Biotechnology Information (NCBI) database. Species-specific primers for L. diolivorans were designed based on the propanediol dehydratase large subunit pduC gene (GenBank Accession No. WP 004105132.1). We designed one primer to target a DNA sequence encoding a species-specific loop region absent in orthologous proteins, and positioned the second primer in a nearby low-similarity region to maintain the efficiency of the qPCR process. This dual strategy, exploiting both structural divergence and sequence dissimilarity, maximized specificity and amplification robustness. PCR was used to validate primer specificity using genomic DNA from the target organism and non-target organisms as templates (Du et al., 2020). A standard curve was generated as previously described in the literature (Du et al., 2020).
2.6 Isolation of LAB and rapid identification of L. diolivorans from SFBJ fermented grains
Two fermented grain samples exhibiting the highest n-propanol content after 7–14 days of bottle pit fermentation were selected from five stacked material samples for LAB separation. Following serial dilution of the fermented grains, LAB colonies were isolated by plating appropriate dilutions onto SMRS agar plates containing nystatin. Plates were incubated at 30°C for 3 days, and 120 colonies with distinct colony morphologies from different dilutions were selected and purified through repeated streaking. These purified colonies were numbered and used for rapid L. diolivorans identification and systematic bacterial classification.
For rapid L. diolivorans identification, the Ezup Column Bacteria Genomic DNA Purification Kit (Sangon Biotech, Shanghai, China) was used to extract DNA. Following PCR amplification with specific primers, detection was performed using 1% agarose gel electrophoresis and subsequent observation with an automated gel imaging analysis system.
2.7 Characterization of L. diolivorans ZX6
For seed cultures, L. diolivorans ZX6 was grown in MRS medium under static conditions at 30°C for 72 h. Then, different fermentation media were inoculated at 2% (v/v) for the study. The growth and metabolic curves of L. diolivorans ZX6 were measured in MRS and SMRS media under static culture conditions for 10 days. MRS media with varying pH values (3.5, 4.0, 4.5, 5.0, 5.5, 6.0, and 6.5) were adjusted using a mixed acid solution (lactic acid: acetic acid = 1:1) and sodium hydroxide. To investigate the effects of pH and temperature on L. diolivorans ZX6 growth, experiments were conducted in an MRS medium at different pH or temperatures (20, 25, 28, 30, 32, 34, 37, 40, and 42°C) under static culture conditions at 30°C for 24 h. The impact of shaker speed (0, 50, 180 rpm) on n-propanol production by L. diolivorans ZX6 was evaluated in SMRS medium at 30°C for 7 days.
MRS media containing varying glucose concentrations (2%, 5%, 9%, 12%, 15%, 20%, 30%, and 35% w/v) were used to assess the impact of glucose concentration on n-propanol production by L. diolivorans ZX6. Fermentation experimental media with different carbon sources were prepared by replacing glucose in MRS medium with an equivalent amount (5% w/v) of sucrose, fructose, and xylose, respectively. All fermentations were conducted under static culture conditions at 30°C for 7 days.
2.8 RNA extraction, sequencing, and analysis
L. diolivorans ZX6 was inoculated (2% v/v) into MRS medium containing individual sugars (5% w/v each of glucose, sucrose, fructose, and xylose) as the sole carbon sources. Fifty milliliters of each culture were collected during the n-propanol index production period (5 d) and centrifuged to obtain the sediment, frozen in liquid nitrogen for 30 min, and immediately transferred to −80°C for storage. Three independent repeat experiments were conducted for each treatment group. RNA extraction, quantification, and sequencing were performed by Shanghai Majorbio Bio-pharm Biotechnology Co., Ltd. (Shanghai, China).
The high-quality sequencing data were mapped to the L. diolivorans DSM 14421 genome (GenBank: GCA_001434255.1), and the original data were uploaded to NCBI (BioProject: PRJNA1225471). The expression level and quantify abundances of each gene were calculated based on previous literature (Chen et al., 2023). The pathways and enzymes of flavor metabolism related to n-propanol were obtained from the KEGG database and previous literature (Gao et al., 2024).
2.9 Physical and chemical analysis
To analyze higher alcohol, ethanol, and acetic acid content in fermented broth or fermented grains, 50 g or 50 mL samples were subjected to pretreatment. Distillation was employed as a pretreatment method to quantify these compounds (Cao et al., 2010). Higher alcohols, ethanol, and acetic acid were analyzed using an Agilent 7820 gas chromatograph equipped with flame ionization detection (Agilent, USA) (López-Vázquez et al., 2010). A J&W CP-Wax 57 CB Acidic column (50 m × 0.25 mm × 0.2 μm, Agilent, USA) was used.
To analyze propionic acid and lactic acid concentrations, fermentation broths were centrifuged at 12,000 rpm for 10 min to remove cells. The supernatant was filtered through a 0.22 μm syringe filter and mixed with methanol in a 1:2 ratio. The mixture was shaken and stored at −20°C overnight to precipitate proteins and polysaccharides. Following this incubation period, the solution was centrifuged at 12,000 rpm for 10 min, filtered through a 0.22 μm syringe filter, and transferred to a liquid phase bottle for detection. High-performance liquid chromatography analysis was performed using an Agilent 1260 Infinity II with an Agilent C18 column, following the method described in a previous study (Sugiura et al., 2023). The peak areas of standard samples were used to determine propionic acid and lactic acid concentrations in the fermentation samples.
Reducing sugars were assayed using the 3,5-dinitrosalicylic acid method (Solarbio, Beijing, China). pH was measured using a pH meter.
2.10 Statistical analysis
Data were reported as mean ± standard deviation (SD) based on three independent determinations of each sample. Statistical analyses were performed using Microsoft Excel for Windows. Statistical significance was considered at P < 0.05, and results were expressed as mean ± SD.
2.11 Data availability
The raw sequence data of the 16S rRNA amplicon sequencing have been deposited in NCBI with the accession numbers PQ136908, PQ136909, PQ136910, and PQ136911.
3 Results
3.1 Enrichment and isolation of high n-propanol-producing bacteria
Considering the convenience of sampling, we initially selected XQBJ fermented grains with high n-propanol content (sample X) as the material for enrichment culture. Sample X was added to liquid culture media, including MRS, SS, and SMRS, for enrichment cultivation. Subsequently, samples were analyzed for n-propanol content. As shown in Table 1, n-propanol content was relatively low when fermented with MRS and SS media. However, when fermented with SMRS mixed media, the n-propanol content increased significantly to 326.60 mg/L, which was 5.9 and 12.6 times higher than that of MRS and SS media, respectively. This suggests that SMRS medium could effectively enriches microorganisms with high n-propanol production capabilities. The n-propanol content of sample X-N4, supplemented with antifungal nystatin, reached 431.70 mg/L. In contrast, n-propanol was not detected in sample X-N5, supplemented with antibacterial ampicillin. These results indicated that the n-propanol-producing microorganisms in the SMRS fermentation broth were bacteria.
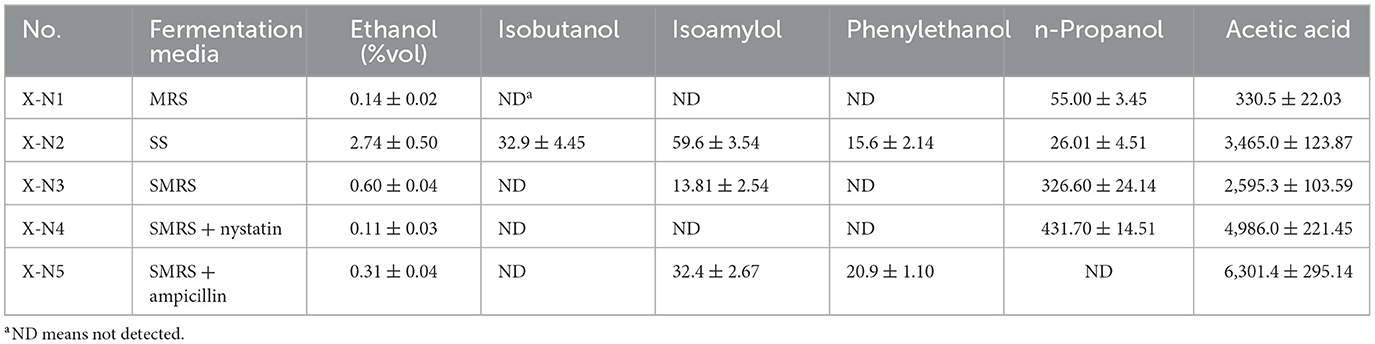
Table 1. Contents of main compounds after inoculation of fermented grain from XQBJ in different fermentation media (mg/L).
To isolate n-propanol-producing bacteria from sample X-N4, 40 bacterial colonies were selected based on morphological differences and purified through repeated streaking on SMRS plates. These strains, named ZX1 to ZX40, were classified into 9 species of LAB and one species of Acetobacter pasteurianus based on 16S rRNA gene sequence homology analyses. Twenty bacterial strains representing distinct species were inoculated into SMRS liquid medium to evaluate their n-propanol production capacity through fermentation experiments. Among these, strain ZX6 demonstrated the highest n-propanol yield, reaching 3,499.40 mg/L after a 7-day fermentation (Supplementary Table S2). No n-propanol was detected in the fermentation broth of the other 19 bacterial strains. Strain ZX6 was rod-shaped and Gram-positive. Its cell morphology was examined using scanning electron microscopy (Figure 1A). The 16S rRNA gene of strain ZX6 was amplified, resulting in a 1,450 bp single fragment that was subsequently sequenced (GenBank Accession No. PQ136908). BLAST analysis revealed a high similarity (>99.7%) between the ZX6 sequence and the 16S rRNA gene sequence of L. diolivorans LMG 19667T (AF264701) (Krooneman et al., 2002). A phylogenetic tree was constructed (Figure 1B). Based on colony morphology, morphological characteristics, and 16S rRNA gene sequence analysis, strain ZX6 was identified as L. diolivorans.
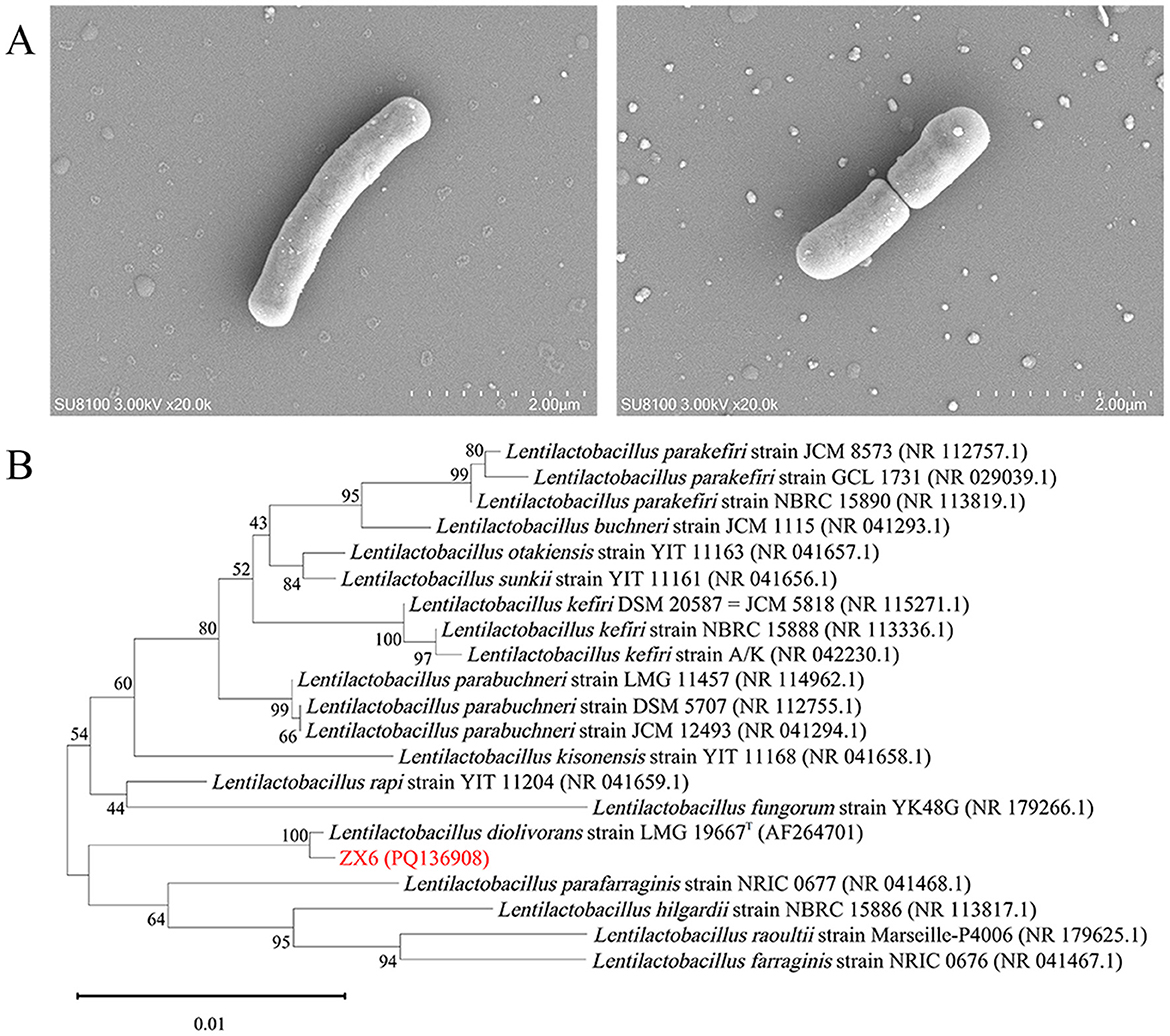
Figure 1. Identification of L. diolivorans ZX6. (A) The morphology of L. diolivorans ZX6 by SEM analysis, (B) phylogenetic tree of L. diolivorans ZX6 based on 16S rRNA gene sequences.
To evaluate the n-propanol production capability of L. diolivorans under brewing conditions, co-culture experiments with S. cerevisiae were conducted. Both liquid co-culture results (Supplementary Figure S1A) and solid-state co-culture results (Supplementary Figure S1B) of L. diolivorans ZX6 and S. cerevisiae SCY62 demonstrated a direct positive correlation between n-propanol content and the initial inoculation amount of L. diolivorans ZX6. When L. diolivorans ZX6 and S. cerevisiae SCY62 were mixed in a 1:1 (v/v) ratio, the n-propanol content in the SMRS fermentation broth was 246.01 mg/L. During solid-state fermentation, with an L. diolivorans ZX6 inoculation amount of 106 CFU/g, the n-propanol content in the fermented grain reached 1,762.05 mg/kg. Under solid-state fermentation conditions, high inoculation of L. diolivorans resulted in n-propanol content in laboratory-scale fermented grains equivalent to that in the first round of fermented grains from SFBJ distillery.
3.2 Absolute quantification of L. diolivorans in SFBJ fermented grains
Next, qPCR was conducted to quantify L. diolivorans in microbiome samples. Species-specific primers for L. diolivorans were designed: F: 5′-TTTCATTTGGTAAGAAGGATTCTTCTG-3′ and R: 5′-TAATATCACCATTTCCTGATGTGGAC-3′. These primers exhibited 100% exclusivity for the other 12 non-target microorganisms from Baijiu fermented grains (Supplementary Table S3), with no matches to other organisms in the NCBI database. We subsequently developed a qPCR method using these specific primers. The standard curve obtained was y = −3.4462x + 46.017 (R2 = 0.99), demonstrating a linear correlation (R2 = 0.99) over a range of 65.47 to 6.547 × 108 copies per reaction. This suggests that the qPCR method is suitable for quantifying L. diolivorans in the samples.
Significant differences were observed in the n-propanol content of the five fermented grains from the scaled-down pit fermentation experiment (Figure 2B). Sample No. 2, characterized by its high n-propanol content, was selected for the L. diolivorans bacterial count determination using qPCR. Initial L. diolivorans quantities were relatively low, at 5.51 × 105 copies/g, but rapidly increased to 2.50 × 107 copies/g by the 7th day of fermentation. Subsequently, the bacterial count decreased significantly, reaching 7.08 × 105 copies/g at the end of fermentation (Figure 2A). The quantities of L. diolivorans in sample No. 5 (which had an n-propanol content in the middle range) showed a similar trend, reaching a maximum of 4.98 × 104 copies/g on the 7th day and then gradually decreasing throughout the fermentation process. This decline might be attributed to the accumulation of acidity and alcohol content in the later stages of fermentation, which inhibits L. diolivorans growth. n-Propanol and ethanol concentrations in the fermented mash increased rapidly in the early stages of fermentation, reaching their peak by the 14th day and remaining relatively constant thereafter (Figures 2B, C). The pH of the fermented grains decreased significantly during the first 2 weeks of fermentation, from around pH 4.5 to pH 3.7, and then stabilized in the last 2 weeks (Figure 2D). Changes in the L. diolivorans bacterial count within the fermented grains generally aligned with the increasing trend of n-propanol content. However, the n-propanol content typically reached its peak around the 14th day of fermentation, exhibiting a slight lag compared to the peak L. diolivorans count.
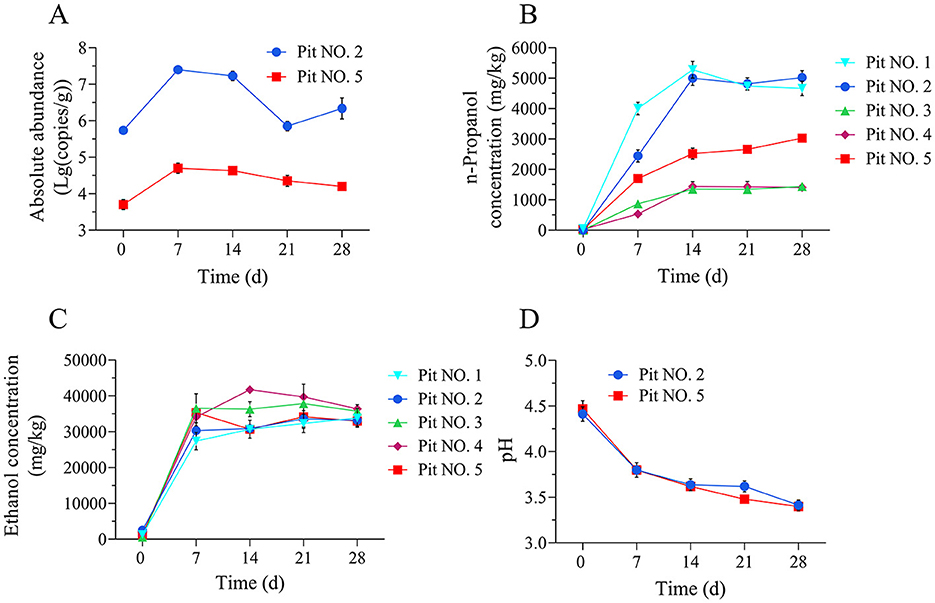
Figure 2. Changes in the quantities of L. diolivorans and the n-propanol content in different fermented grains during pit fermentation. (A) The absolute abundance of internal standard L. diolivorans in fermented grains of Pit No. 2 and Pit No. 5, (B) n-propanol content, (C) ethanol content, and (D) pH.
3.3 Rapid identification of L. diolivorans from SFBJ fermented grains
One hundred and seven LAB strains, numbered ZD1 to ZD107, were isolated and purified from SFBJ fermented grains with high n-propanol content. Three strains with positive gel electrophoresis bands were identified: ZD9 (GenBank Accession No. PQ136909), ZD92 (GenBank Accession No. PQ136910), and ZD93 (GenBank Accession No. PQ136911). BLAST analysis revealed that these three strains shared high similarity (>99.7%) with the 16S rRNA gene sequence of L. diolivorans LMG 19667T (AF264701), confirming their classification as L. diolivorans. This demonstrated the high reliability of the specific primer method for rapid L. diolivorans identification. 16S rRNA gene sequence analysis and homology alignment classified the 107 LAB strains into 7 genera and 12 species (Supplementary Figure S2A). The three most prevalent LAB species were L. buchneri, L. panis, and Lactobacillus pontis, accounting for 57.94%, 12.15%, and 10.28%, respectively. The other 9 species accounted for < 5%, and L. diolivorans accounted for only 2.80% (Supplementary Figure S2B). Fermentation tests of the three L. diolivorans strains from SFBJ fermented grain revealed n-propanol contents exceeding 2,400 mg/L, similar to L. diolivorans ZX6. However, no significant n-propanol production was detected in samples fermented by other LAB (Supplementary Table S4). This indicated that L. diolivorans was the primary microorganism isolated from SFBJ fermented grains with the capacity to synthesize high concentrations of n-propanol.
3.4 Characterization of L. diolivorans ZX6
3.4.1 Effects of different media and cultivation time on L. diolivorans ZX6
Two media, MRS and SMRS, were used to investigate the effects on L. diolivorans ZX6 growth and metabolism. The OD600 of L. diolivorans ZX6 in the SMRS medium was slightly higher than that in the MRS medium (Figure 3A). However, a significant difference was observed in n-propanol production during fermentation (Figure 3B). The n-propanol content in both media increased significantly after 48 h of fermentation. In SMRS, it reached a maximum of 4,399 mg/L at 192 h, while in MRS, it reached a maximum of 472 mg/L at 216 h. The decreasing trend of reducing sugars in the media aligned with the increasing n-propanol content (Figure 3D). The pH of both culture media rapidly decreased within the first 48 h of fermentation, from the initial pH of 6.0 to below pH 4.5. While the pH of MRS fermentation broth stabilized around pH 4.5, the pH of SMRS fermentation broth continued to decrease to around pH 3.5 (Figure 3C). After 216 h of fermentation, the acetic acid content peaked at 4,000 mg/L in MRS and 6,000 mg/L in SMRS (Figure 3E).
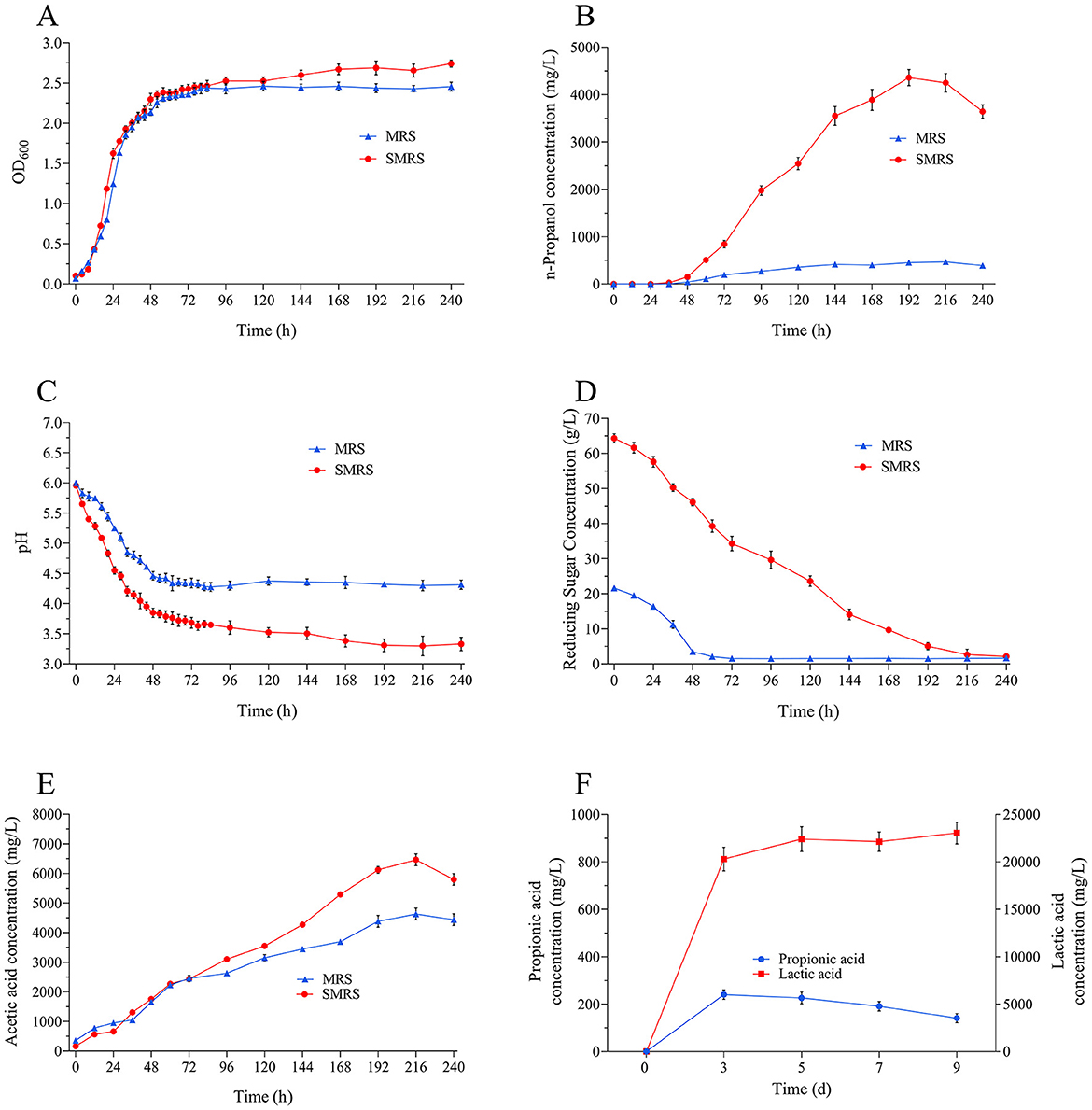
Figure 3. Growth curves and fermentation analysis of L. diolivorans ZX6 in different media. (A) OD600, (B) n-propanol, (C) pH, (D) reducing sugar, (E) acetic acid, (F) propionic acid and lactic acid in SMRS medium.
In the SMRS medium, lactic acid content increased rapidly during the early stages of fermentation. This growth pattern closely aligned with biomass accumulation, with lactic acid content reaching 23,053.5 mg/L after 9 days. Conversely, propionic acid content remained very low at 144.6 mg/L (Figure 3F).
3.4.2 Effects of fermentation conditions on L. diolivorans ZX6
The optimal growth pH of L. diolivorans ZX6 was 5.5, which aligned with the standard strain L. diolivorans LMG 19667T (Figure 4A). The optimal growth temperature of L. diolivorans ZX6 was 34°C (Figure 4B), which is similar to that of the LMG 19667T strain. However, L. diolivorans ZX6 strain exhibited greater heat resistance than the LMG 19667T strain. Fermentation experiments conducted at various shaking speeds revealed significantly higher n-propanol production by L. diolivorans ZX6 during static fermentation compared to shaking cultivation (Figure 4C). This suggests that L. diolivorans is more sensitive to oxygen during n-propanol synthesis.
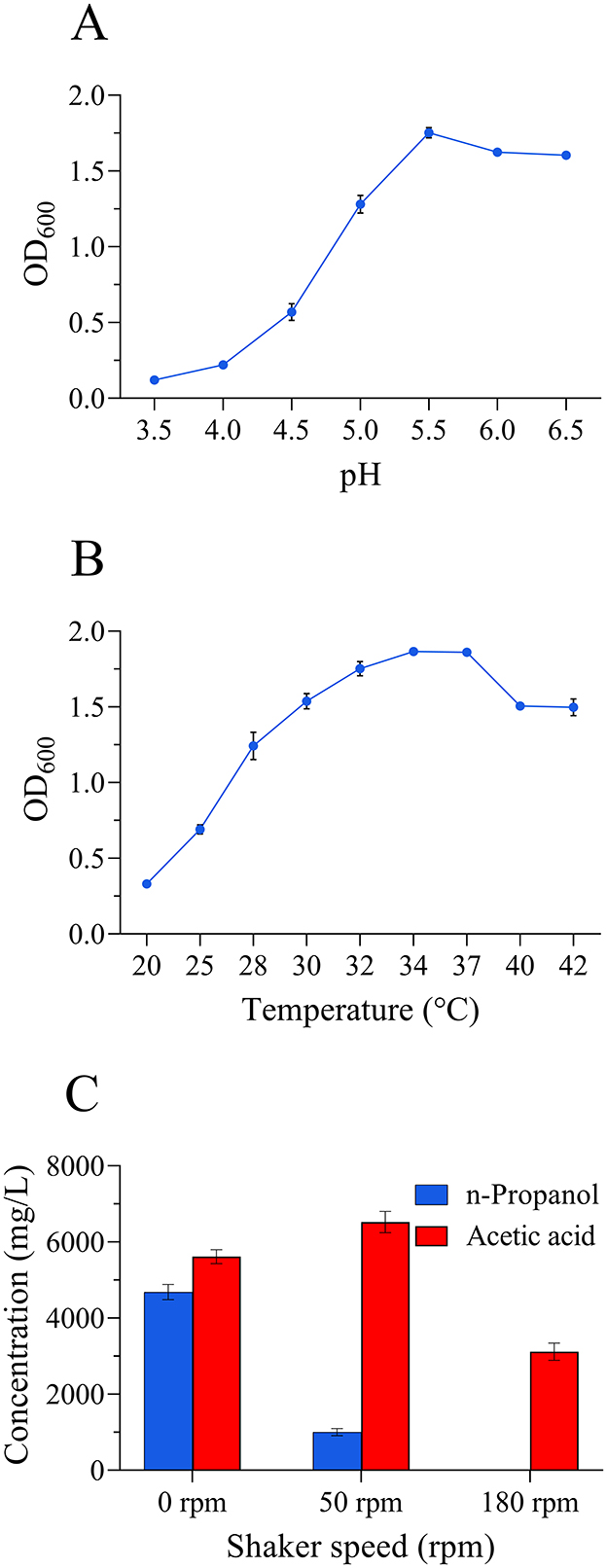
Figure 4. Effects of pH, temperature, and shaking speed on the growth of L. diolivorans ZX6. (A) pH, (B) temperature, and (C) shaker speed.
3.4.3 Effects of carbon source types and glucose concentration on L. diolivorans ZX6
The effect of glucose concentration on n-propanol content demonstrated that a 5% glucose concentration in MRS medium yielded the maximum n-propanol content of 1,555.9 mg/L. As the glucose concentration increased, n-propanol yield gradually decreased (Supplementary Figure S3).
L. diolivorans ZX6 demonstrated substantial biomass accumulation during cultivation in MRS medium containing glucose, xylose, or sucrose as the carbon sources. In contrast, biomass production decreased with fructose as the sole carbon source (Figure 5A). Fermentation with glucose, xylose, or sucrose as carbon sources led to progressive pH reduction, whereas fructose utilization resulted in minimal pH variation (Figure 5B). Cultures utilizing glucose or sucrose synthesized multiple metabolites, including n-propanol (Figure 5C), acetic acid (Figure 5D), ethanol (Figure 5E), and lactic acid (Figure 5F). Xylose-fed cultures, however, generated exclusively lactic acid and acetic acid.
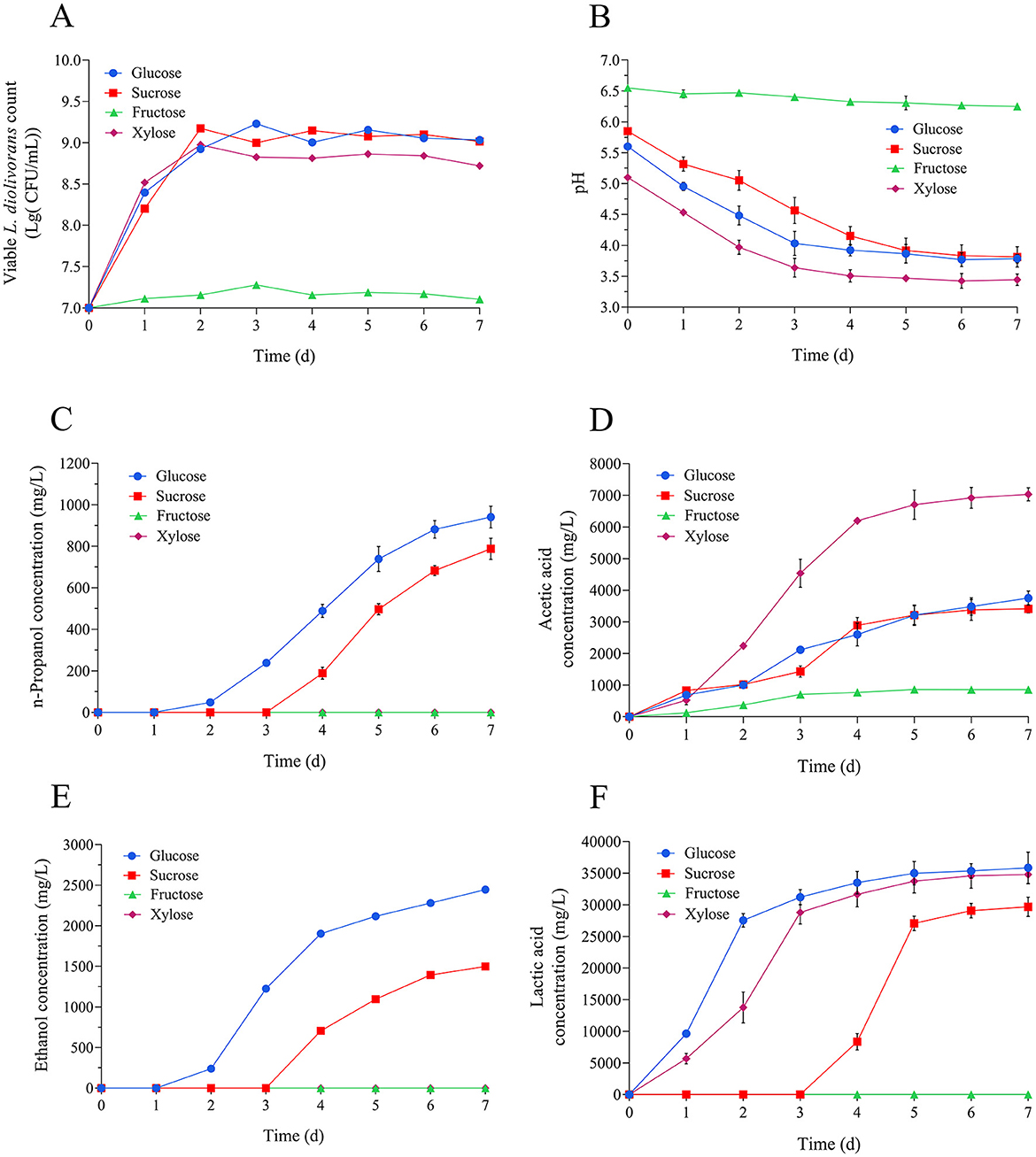
Figure 5. Effects of different carbon sources on the growth and metabolite production of L. diolivorans ZX6. (A) Viable cell counts, (B) pH, (C) n-propanol, (D) acetic acid, (E) ethanol, and (F) lactic acid.
3.4.4 Comparative analysis of transcriptome during fermentation of different carbon sources
When L. diolivorans ZX6 was cultured with four different carbon sources, the transcriptome analysis was performed on the 5th day of the rapid growth period of n-propanol. Based on the KEGG database and previous literature, we mapped the metabolic pathways and associated enzymes involved in the metabolism of n-propanol, lactic acid, ethanol, and acetic acid (Figure 6). A total of 94 enzymes were involved in the metabolic pathway, with 30 enzymes detected in the transcriptome. Among these, 24 enzymes exhibited significant differences in transcription levels, and 42 genes were associated with these enzymes (Supplementary Figure S4). Transcriptome data indicated that n-propanol is likely formed through the methylglyoxal pathway, in which methylglyoxal is converted to propanal via 1,2-propanediol and subsequently reduced to n-propanol. Three precursors entered the methylglyoxal pathway: glycerone-P, pyruvate, and lactic acid. Since methylglyoxal synthase (EC 4.2.3.3) was not present in the transcriptome and triosephosphate isomerase (EC 5.3.1.1) did not show significant differences in transcription levels across different carbon sources, glycerone-P was likely not the primary precursor for L. diolivorans ZX6 to enter the methylglyoxal pathway. Lactaldehyde dehydrogenase (EC 1.2.1.22) catalyzed the formation of lactaldehyde from lactic acid and the conversion of pyruvate to methylglyoxal. Additionally, the transcriptional level of its gene was significantly higher in cultures with glucose and sucrose than in those with fructose and xylose (P ≤ 0.001) (Figure 7 and Supplementary Figure S4, C1). Similar results were observed for the transcription levels of the three enzymes (EC 4.2.1.28, 1.1.1.102, and PduQ) involved in the conversion of 1,2-propanediol to n-propanol (Supplementary Figure S4 C5, C6, D1– D3). The transcriptional levels were consistent with the increase in n-propanol content: compared to cultures with fructose and xylose as carbon sources, n-propanol production significantly increased by day 5 when L. diolivorans ZX6 was cultured with glucose and sucrose as carbon sources (Figure 5C). Lactate dehydrogenase (EC 1.1.1.27) (Supplementary Figure S4, F1–F4) and pyruvate oxidase (EC 1.2.3.3) (Supplementary Figure S4, F5, F6, G1) were inferred as the key enzymes for L. diolivorans ZX6 to metabolize pyruvate into lactic acid and acetic acid, respectively.
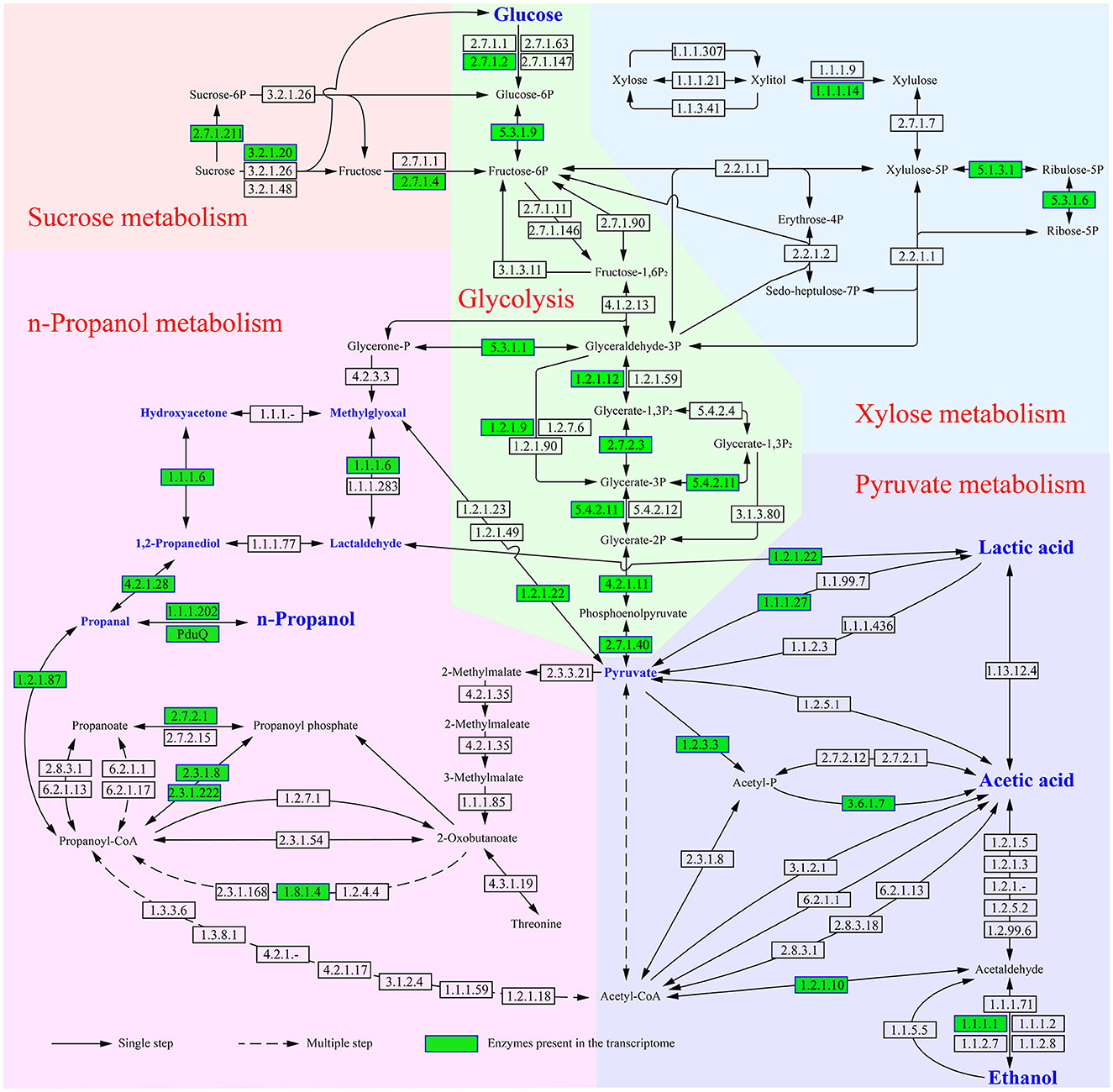
Figure 6. Metabolic pathways and enzymes involved in the conversion of four carbon sources into n-propanol, lactic acid, ethanol, and acetic acid.
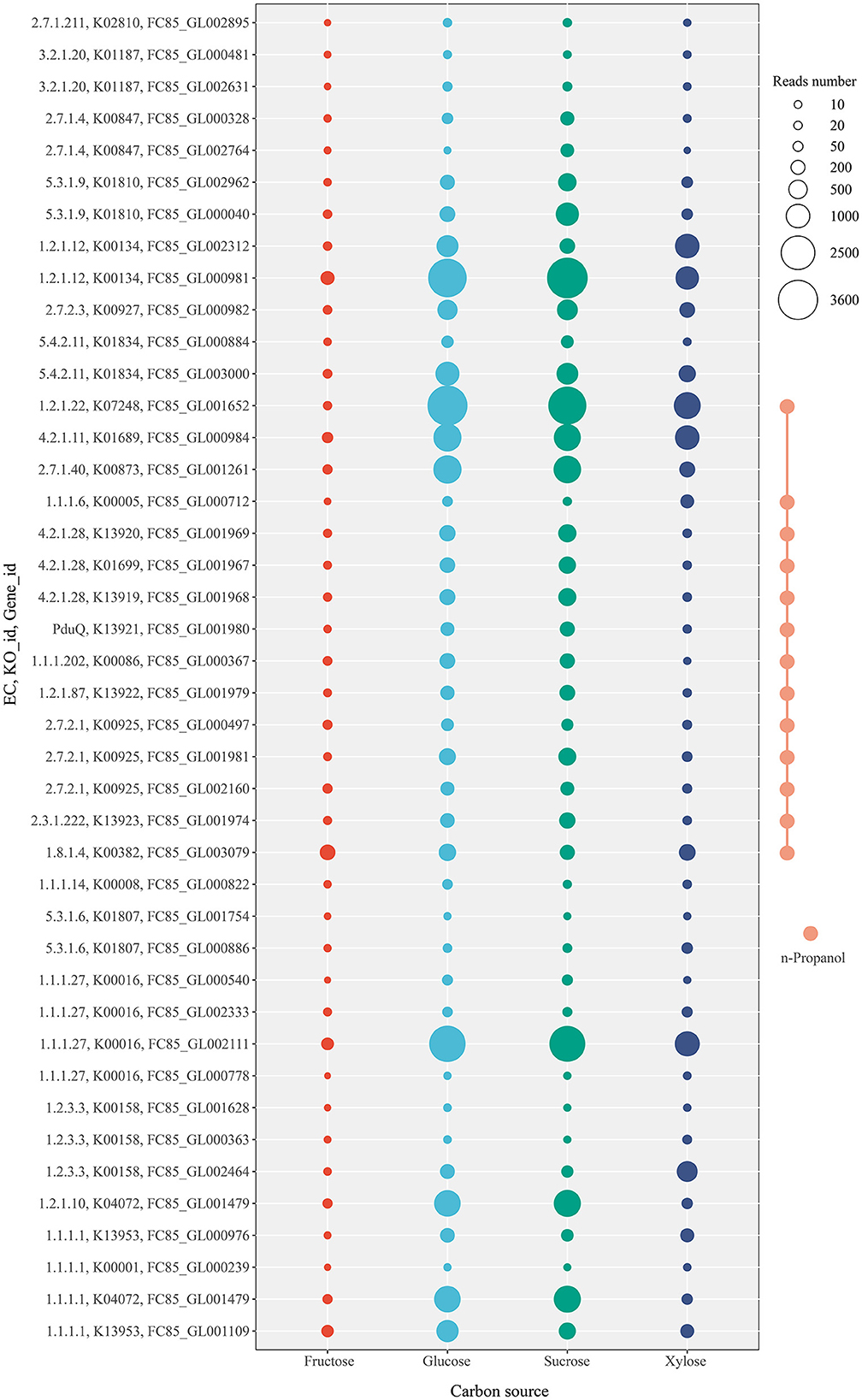
Figure 7. Transcriptional levels of genes in L. diolivorans ZX6 cultured with four different carbon sources.
4 Discussion
The mechanism underlying high n-propanol production in Baijiu brewing, particularly in SFBJ, has long been an industry enigma. In this study, a high n-propanol-producing strain of L. diolivorans was isolated from Baijiu fermented grains, which may be a key bacterium responsible for high n-propanol formation during Baijiu brewing. L. diolivorans has not been previously detected or reported in Baijiu fermentation systems, primarily due to its low abundance in Baijiu fermented grains. Even in SFBJ fermented grains with high n-propanol concentrations, L. diolivorans constitutes only about 2.8% of the total easily culturable LAB on aerobic plates (Supplementary Figure S2B), making it susceptible to dilution and potential exclusion from conventional culture methods. Moreover, the similar colony morphology of L. diolivorans to dominant LAB species like L. buchneri increases the likelihood of overlooking high n-propanol-producing strains during random colony selection. The complex microbial community in Baijiu fermentation, coupled with the diversity of species and n-propanol-producing microbial synthetic pathways (Pang et al., 2018; Du et al., 2019; Hao et al., 2021; Zhang et al., 2021), significantly complicates tracing the causal relationship between n-propanol and its producing microbial species (Liu and Miao, 2020; Xiao et al., 2021; Luo et al., 2023).
In addition, research on the microbial sources of high n-propanol production in SFBJ is constrained by the fermented grain materials. The fermentation process of SFBJ is not continuous; it involves a crucial preliminary solid-state stacking stage prior to pit fermentation (Chen et al., 2024a). Each round of fermented grains undergoes ~30 days of pit fermentation. Specifically, the fermentation and sampling period for the first round of fermented grains from the Baijiu distillery with the highest n-propanol content is limited to ~1 month annually. This limitation hinders the progression and deepening of research on the source of high concentrations of n-propanol in Baijiu. In contrast to SFBJ, the fermentation of XQBJ does not necessitate a high-temperature stacking process, and the fermentation period is relatively short (~10 days), making it convenient for factory sampling and small-scale fermentation experiments in the laboratory. More importantly, a significant amount of n-propanol is produced during the brewing process of XQBJ. Consequently, XQBJ fermented grains were selected as the material for enrichment culture and solid-state fermentation, thereby overcoming the limitations imposed by material availability and the challenges associated with conducting repeatable solid-state fermentation experiments of Baijiu at the laboratory scale. Furthermore, the L. diolivorans ZX6 strain, characterized by its high n-propanol production capacity, was successfully isolated from XQBJ fermented grains for the first time. Compared to MRS medium, the use of SMRS medium for the enrichment culture of XQBJ fermented grains and for fermentation research on L. diolivorans ZX6 can lead to a higher production of n-propanol. One contributing factor may be the increased glucose concentration. The specific reasons for this influence still require more detailed research in the future.
qPCR analysis revealed L. diolivorans counts ranging from 1.29 × 104 to 3.30 × 105 copies/g in fermented grains from three distilleries (Supplementary Table S1). This indicates L. diolivorans is widely distributed in SFBJ mash across different regional manufacturers. To our knowledge, this is the highest-yielding natural microorganism reported for fermentative n-propanol production directly from glucose. Furthermore, when L. diolivorans was co-cultured with Rhizopus and S. cerevisiae in solid-state fermentation, the n-propanol yield also reached the highest level reported to date. Therefore, its presence in fermented grains may be the main reason for the production of high n-propanol levels in Baijiu. Due to the limited number of samples collected, further in-depth research is required to investigate the origin of L. diolivorans, its population dynamics during Baijiu brewing, and the differences in n-propanol production capacity among strains from different regions.
Nutritional and environmental factors in Baijiu brewing process, such as the carbon-to-nitrogen ratio have important influence on n-propanol content (Jiang et al., 2019). Our research results show that high n-propanol synthesis from L. diolivorans ZX6 required high sugar concentrations (Supplementary Figure S3). Reducing sugar, acidity, alcohol, and temperature have been established as the primary driving forces in stacking and pit fermentation (Hao et al., 2021). In the present study, L. diolivorans abundance increased rapidly only in the early fermented grain, possibly due to its weak acid resistance. The optimal pH for L. diolivorans ZX6 was 5.5, and growth slowed below pH 4.0 (Figure 4A). This aligned with the detection of increasing acidity and decreasing n-propanol content in fermented grains with advancing fermentation rounds in SFBJ distilleries (Guo et al., 2022). As acidity increased, LAB with stronger acid resistance, such as L. acetotolerans and Acetilactobacillus jinshanensis, became dominant in the middle and later stages of fermented grains, replacing weaker acid-resistant LAB like L. diolivorans (Du et al., 2020; Liao et al., 2023).
L. diolivorans has been reported to coexist with various LAB in specific traditional fermented foods, including French sourdoughs (Lhomme et al., 2015), Mongolian fermented dairy products (Yu et al., 2011; Sánchez-Juanes et al., 2020), and other acidic fermented foods. This coexistence reflects the general adaptability of LAB to acidic growth environments. In silage feed and sourdough, L. diolivorans can utilize 1,2-propanediol, a metabolic product of L. buchneri, to synthesize propionic acid and n-propanol (Krooneman et al., 2002; Zhang et al., 2010). Our results demonstrated that L. diolivorans ZX6 possesses a complete metabolic pathway to metabolize glucose into n-propanol, lactic acid, acetic acid, and ethanol. Although transcriptome data suggest that n-propanol is likely formed through the methylglyoxal pathway, further validation through knockout mutant studies and overexpression studies remains to be explored. When fermenting with glucose as a substrate, L. diolivorans ZX6 produces substantial amounts of n-propanol. Is there a synergistic effect between the coexistence of L. diolivorans and L. buchneri in SFBJ fermented grains on the production of n-propanol? Follow-up research is needed. Additionally, the interactions of L. diolivorans with other dominant microbes, such as yeasts and LAB, within solid-state fermented grains, as well as their impact on n-propanol content and other flavor substances in Baijiu, are topics requiring in-depth exploration.
5 Conclusion
In conclusion, we isolated a rare L. diolivorans strain, ZX6, from Baijiu fermented grains. The pure strain exhibited n-propanol fermentation yields exceeding 4,300 mg/L in SMRS medium. Utilizing the pduC gene sequence of 1,2-propanediol dehydratase from L. diolivorans, we established a rapid PCR screening method for L. diolivorans and a qPCR method for its detection in fermented grains. L. diolivorans was detected in the first round of SFBJ fermented grains from three different regions, suggesting its potential role as one of the key microorganisms responsible for high n-propanol production. L. diolivorans ZX6 directly metabolized glucose to produce substantial amounts of n-propanol. High n-propanol synthesis occurred under high sugar concentrations, anaerobic environments, and relatively long fermentation times. Transcriptome analysis revealed that L. diolivorans ZX6 likely synthesized 1,2-propanediol via the methylglyoxal pathway, which was subsequently converted to n-propanol. L. diolivorans coexisted with the dominant L. buchneri in SFBJ fermented grains. Indeed, potential synergistic effects between these strains in n-propanol synthesis, as well as interactions between L. diolivorans and other dominant microorganisms, such as yeast and LAB, and their influence on n-propanol content, require further investigation. This study provides a theoretical foundation for developing processes to reduce n-propanol content in Baijiu brewing by artificially controlling the source and quantity of L. diolivorans.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1225471.
Author contributions
EZ: Writing – original draft, Methodology, Investigation. RG: Writing – review & editing, Investigation, Methodology, Writing – original draft, Validation. KZ: Writing – review & editing, Formal analysis, Investigation. PL: Methodology, Writing – review & editing. WL: Writing – review & editing, Methodology. TY: Resources, Writing – review & editing, Formal analysis. XL: Investigation, Data curation, Writing – review & editing. MY: Data curation, Investigation, Writing – review & editing. YM: Data curation, Writing – review & editing, Investigation. LM: Methodology, Project administration, Conceptualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Science and Technology Innovation Cooperation Project 2020 of Wuhan Polytechnic University and Hubei Baiyunbian Liquor Industry Co., Ltd. (whpu-2020-kj-009). The graphical abstract was created using BioGDP.com.
Conflict of interest
TY was employed by Hubei Baiyunbian Liquor Industry Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1624097/full#supplementary-material
References
Cao, Y. G., Hu, Y. G., Ma, Y. H., Du, X. W., Ma, E. B., Quan, Z. X., et al. (2010). Variation of aromatic components in solid phase fermented grains during fermentation of Fen liquor. Food Sci. 31, 367–371.
Chen, L. Q., Li, F. Z., Yang, F., Chen, B., Du, H., Wang, L., et al. (2024a). Microbiome dynamics and environment driving factors throughout pit fermentation of Jiang-flavor Baijiu: a multi-omics study. Food Biosci. 60:104363. doi: 10.1016/j.fbio.2024.104363
Chen, L. Q., Qin, X., Wang, G. Z., Teng, M. J., Zheng, Y. X., Yang, F., et al. (2024b). Oxygen influences spatial heterogeneity and microbial succession dynamics during Baijiu stacking process. Bioresour. Technol. 403:130854. doi: 10.1016/j.biortech.2024.130854
Chen, Y., Wan, Y., Cai, W., Che, X., Sun, X., Peng, H., et al. (2023). Transcriptomic and metabonomic to evaluate the effect mechanisms of the growth and aroma-producing of Pichia anomala under ethanol stress. Food Biosci. 56:103176. doi: 10.1016/j.fbio.2023.103176
Deng, N., Du, H., and Xu, Y. (2020). Cooperative response of Pichia kudriavzevii and Saccharomyces cerevisiae to lactic acid stress in Baijiu Fermentation. J. Agric. Food Chem. 68, 4903–4911. doi: 10.1021/acs.jafc.9b08052
Dong, Y. X., Guo, C. L., and Xiong, X. M. (2002). Investigation on the factors affecting the content of n-propanol in “Bai Yun bian” finishing liquor. Liquor Mak. 29, 30–31.
Du, H., Wang, X. S., Zhang, Y. H., and Xu, Y. (2019). Exploring the impacts of raw materials and environments on the microbiota in Chinese Daqu starter. Int. J. Food Microbiol. 297, 32–40. doi: 10.1016/j.ijfoodmicro.2019.02.020
Du, R. B., Wu, Q., and Xu, Y. (2020). Chinese liquor fermentation: Identification of key flavor-producing Lactobacillus spp. by quantitative profiling with indigenous internal standards. Appl. Environ. Microbiol. 86, e00456–e00420. doi: 10.1128/AEM.00456-20
Gao, R. J., Peng, P., Yu, L., Wan, B., Liang, X. T., Liu, P. L., et al. (2024). Metagenomic analysis reveals the correlations between microbial communities and flavor compounds during the brewing of traditional Fangxian huangjiu. Food Biosci. 58:103816. doi: 10.1016/j.fbio.2024.103816
Gao, W. J., Fan, W. L., and Xu, Y. (2014). Characterization of the key odorants in light aroma type Chinese liquor by gas chromatography-olfactometry, quantitative measurements, aroma recombination, and omission studies. J. Agric. Food Chem. 62, 5796–5804. doi: 10.1021/jf501214c
Giudici, P., Romano, P., and Zambonelli, C. (1990). A biometric study of higher alcohol production in Saccharomyces cerevisiae. Can. J. Microbiol. 36, 61–64. doi: 10.1139/m90-012
Guo, S. B., Xie, S. K., Zhang, J. J., Xue, X. X., Long, D. Z., Chen, Y. Q., et al. (2022). Analysis on the fermentation mechanism of sauce-flavor Dahui-jiu in Maotai region. China Brew. 41, 38–44. doi: 10.11882/j.issn.0254-5071.2022.07.008
Hao, F., Tan, Y. W., Lv, X. B., Chen, L. Q., Yang, F., Wang, H. Y., et al. (2021). Microbial community succession and its environment driving factors during initial fermentation of Maotai-flavor Baijiu. Front. Microbiol. 12:669201. doi: 10.3389/fmicb.2021.669201
Hu, Y. L., Yang, Q., Chen, D., Fu, B. A., Zhang, Y., Zhang, Y., et al. (2021). Study on microbial communities and higher alcohol formations in the fermentation of Chinese Xiaoqu Baijiu produced by traditional and new mechanical technologies. Food Res. Int. 140:109876. doi: 10.1016/j.foodres.2020.109876
Janssen, P. H. (2004). Propanol as an end product of threonine fermentation. Arch. Microbiol. 182, 482–486. doi: 10.1007/s00203-004-0732-y
Jiang, J., Liu, Y. C., Li, H. H., Yang, Q., Wu, Q., Chen, S. X., et al. (2019). Modeling and regulation of higher alcohol production through the combined effects of the C/N ratio and microbial interaction. J. Agric. Food Chem. 67, 10694–10701. doi: 10.1021/acs.jafc.9b04545
Jin, G. Y., Zhu, Y., and Xu, Y. (2017). Mystery behind Chinese liquor fermentation. Trends Food Sci. Technol. 63, 18–28. doi: 10.1016/j.tifs.2017.02.016
Krooneman, J., Faber, F., Alderkamp, A. C., Elferink, S., Driehuis, F., Cleenwerck, I., et al. (2002). Lactobacillus diolivorans sp. nov., a 1,2-propanediol-degrading bacterium isolated from aerobically stable maize silage. Int. J. Syst. Evol. Microbiol. 52, 639–646. doi: 10.1099/00207713-52-2-639
Lhomme, E., Lattanzi, A., Dousset, X., Minervini, F., De Angelis, M., Lacaze, G., et al. (2015). Lactic acid bacterium and yeast microbiotas of sixteen French traditional sourdoughs. Int. J. Food Microbiol. 215, 161–170. doi: 10.1016/j.ijfoodmicro.2015.09.015
Liao, W. F., Li, Y. P., Zhang, Y., Yang, Y. B., Yang, T. Y., and Miao, L. H. (2023). Comparative analysis of the transcriptional responses of Acetilactobacillus jinshanensis BJ01 to organic acids. Arch. Microbiol. 205:381. doi: 10.1007/s00203-023-03715-5
Liu, P. L., and Miao, L. H. (2020). Multiple batches of fermentation promote the formation of functional microbiota in Chinese miscellaneous-flavor Baijiu fermentation. Front. Microbiol. 11:75. doi: 10.3389/fmicb.2020.00075
Liu, P. L., Xiong, X. M., Wang, S., and Miao, L. H. (2017). Population dynamics and metabolite analysis of yeasts involved in a Chinese miscellaneous-flavor liquor fermentation. Ann. Microbiol. 67, 553–565. doi: 10.1007/s13213-017-1286-y
López-Vázquez, C., Bollaín, M. H., Berstsch, K., and Orriols, I. (2010). Fast determination of principal volatile compounds in distilled spirits. Food Control 21, 1436–1441. doi: 10.1016/j.foodcont.2010.03.008
Luo, L. J., Song, L., Han, Y., Zhen, P., Zhao, X., Zhou, X., et al. (2023). Microbial communities and their correlation with flavor compound formation during the mechanized production of light-flavor Baijiu. Food Res. Int. 172:113139. doi: 10.1016/j.foodres.2023.113139
Luo, X., Fan, W. L., Xu, Y., Cheng, P. Y., Sun, Y. L., Zhu, X. C., et al. (2025). Characterization of volatile bitter off-taste compounds in Maotai-flavor baijiu (Chinese liquor). LWT-Food Sci. Technol. 217:117363. doi: 10.1016/j.lwt.2025.117363
Matsubara, M., Urano, N., Yamada, S., Narutaki, A., Fujii, M., and Kataoka, M. (2016). Fermentative production of 1-propanol from D-glucose, L-rhamnose and glycerol using recombinant Escherichia coli. J. Biosci. Bioeng. 122, 421–426. doi: 10.1016/j.jbiosc.2016.03.011
Mckarns, S. C., Hansch, C., Caldwell, W. S., Morgan, W. T., Moore, S. K., and Doolittle, D. J. (1997). Correlation between hydrophobicity of short-chain aliphatic alcohols and their ability to alter plasma membrane integrity. Fundam. Appl. Toxicol. 36, 62–70. doi: 10.1006/faat.1996.2252
Pang, X. N., Han, B. Z., Huang, X. N., Zhang, X., Hou, L. F., Cao, M., et al. (2018). Effect of the environment microbiota on the flavour of light-flavour Baijiu during spontaneous fermentation. Sci. Rep. 8:3396. doi: 10.1038/s41598-018-21814-y
Qu, G. Y., Tang, J., Jiang, J., Yang, Q., Liu, Y. C., Wu, Q., et al. (2021). Metabolism characteristics of higher alcohols synthesized by microbiota in the fermentation process of light aroma type Baijiu started by Xiaoqu. Food Ferment. Ind. 47, 32–37.
Sánchez-Juanes, F., Teixeira-Martín, V., González-Buitrago, J. M., Velázquez, E., and Flores-Félix, J. D. (2020). Identification of species and subspecies of lactic acid bacteria present in Spanish cheeses type “Torta” by MALDI-TOF MS and pheS gene analyses. Microorg. 8:301. doi: 10.3390/microorganisms8020301
Siebert, D., and Wendisch, V. F. (2015). Metabolic pathway engineering for production of 1,2-propanediol and 1-propanol by Corynebacterium glutamicum. Biotechnol. Biofuels 8:91. doi: 10.1186/s13068-015-0269-0
Strubelt, O., Deters, M., Pentz, R., Siegers, C. P., and Younes, M. (1999). The toxic and metabolic effects of 23 aliphatic alcohols in the isolated perfused rat liver. Toxicol. Sci. 49, 133–142. doi: 10.1093/toxsci/49.1.133
Sugiura, J., Tsuchiyama, T., Taniguchi, M., Fukatsu, K., and Miyazaki, H. (2023). Novel SPE purification approach using the direct adsorption of vaporised propionic acid in food for rapid HPLC determination. Food Chem. 428:136799. doi: 10.1016/j.foodchem.2023.136799
Tian, Y., Kong, X. Y., and Fang, F. (2020). Microbial n-propanol synthesis during Luzhou-flavor liquor fermentation. Acta Microbiol. Sin. 60, 1421–1432. doi: 10.13343/j.cnki.wsxb.20190455
Tu, W. Y., Cao, X. N., Cheng, J., Li, L. J., Zhang, T., Wu, Q., et al. (2022). Chinese Baijiu: The perfect works of microorganisms. Front. Microbiol. 13:919044. doi: 10.3389/fmicb.2022.919044
Walther, T., and François, J. M. (2016). Microbial production of propanol. Biotechnol. Adv. 34, 984–996. doi: 10.1016/j.biotechadv.2016.05.011
Wang, C. L., Bin, Z. Q., Wang, L. Q., Zhu, G. J., Tang, S. P., Chen, Y. F., et al. (2025). Metagenomic and metabolomic profiling analyses to unravel the formation mechanism of n-propanol during the first and second round of Jiangxiangxing Baijiu fermentation. Food Res. Int. 200:115459. doi: 10.1016/j.foodres.2024.115459
Wei, J. L., Du, H., and Xu, Y. (2024). Revealing the key microorganisms producing higher alcohols and their assembly processes during Jiang-flavor Baijiu fermentation. Food Biosci. 61:104569. doi: 10.1016/j.fbio.2024.104569
Wu, Q., Kong, Y., and Xu, Y. (2016). Flavor profile of Chinese liquor is altered by interactions of intrinsic and extrinsic microbes. Appl. Environ. Microbiol. 82, 422–430. doi: 10.1128/AEM.02518-15
Xiao, C., Yang, Y., Lu, Z. M., Chai, L. J., Zhang, X. J., Wang, S. T., et al. (2021). Daqu microbiota exhibits species-specific and periodic succession features in Chinese baijiu fermentation process. Food Microbiol. 98:103766. doi: 10.1016/j.fm.2021.103766
Yu, J., Wang, W. H., Menghe, B. L. G., Jiri, M. T., Wang, H. M., Liu, W. J., et al. (2011). Diversity of lactic acid bacteria associated with traditional fermented dairy products in Mongolia. J. Dairy Sci. 94, 3229–3241. doi: 10.3168/jds.2010-3727
Zhang, C. G., Brandt, M. J., Schwab, C., and Gänzle, M. G. (2010). Propionic acid production by cofermentation of Lactobacillus buchneri and Lactobacillus diolivorans in sourdough. Food Microbiol. 27, 390–395. doi: 10.1016/j.fm.2009.11.019
Zhang, H. X., Wang, L., Tan, Y. W., Wang, H. Y., Yang, F., Chen, L. Q., et al. (2021). Effect of Pichia on shaping the fermentation microbial community of sauce-flavor Baijiu. Int. J. Food Microbiol. 336:108898. doi: 10.1016/j.ijfoodmicro.2020.108898
Zhou, Z. H., Liu, Z. H., Wen, S. Y., Ouyang, G. W., Shen, Y. X., Yang, Q., et al. (2023). Rare short- and medium-chain fatty acid-producing anaerobes from raw soil play vital roles in formation of diverse flavour compounds of Jiangxiangxing Baijiu. Food Microbiol. 112:104247. doi: 10.1016/j.fm.2023.104247
Keywords: Baijiu, n-propanol, isolation, Lentilactobacillus diolivorans, quantitative PCR, fermentation characteristics, transcriptome analysis
Citation: Zhang E, Gao R, Zhu K, Liu P, Liao W, Yang T, Liang X, Yang M, Ming Y and Miao L (2025) Isolation and characterization of Lentilactobacillus diolivorans: a high n-propanol-producing microorganism from Baijiu brewing. Front. Microbiol. 16:1624097. doi: 10.3389/fmicb.2025.1624097
Received: 07 May 2025; Accepted: 04 June 2025;
Published: 25 June 2025.
Edited by:
Yuanliang Hu, Hubei Normal University, ChinaReviewed by:
Wenchao Cai, Shihezi University, ChinaZhuang Guo, Hubei University of Arts and Science, China
Copyright © 2025 Zhang, Gao, Zhu, Liu, Liao, Yang, Liang, Yang, Ming and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lihong Miao, bWlhb3docHVAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Enhua Zhang1†
Enhua Zhang1† Ruijie Gao
Ruijie Gao Pulin Liu
Pulin Liu Weifang Liao
Weifang Liao Lihong Miao
Lihong Miao