- 1Department of Food Science, Faculty of Food Science and Technology, Universiti Putra Malaysia, Serdang, Malaysia
- 2Faculty of Agriculture, Forestry and Food Engineering, Yibin University, Yibin, China
- 3Solid-State Fermentation Resource Utilization Key Laboratory of Sichuan Province, Yibin University, Yibin, China
- 4College of Food Science, Sichuan Agricultural University, Ya’an, China
Introduction: The production of Nongxiangxing Baijiu (Chinese liquor) involves a complex interplay of microbial community metabolism and multi-microbial co-fermentation. The Nongxiangxing Baijiu pit mud is rich in anaerobic acid-producing microorganisms, and this study was designed to investigate the impact of multi-acid synergistic fermentation on feed quality.
Methods: Three Nongxiangxing Baijiu pit muds were subjected to selective serial passage (SSP) three times with four different media (GM, LM, GY, and LY). All samples fermented in GM exhibiting more microbial growth and higher total titratable acidity. Microbial composition analysis of these samples revealed the presence of three acid-producing microbiota (GMAS2, GMBS3, and GMCS3) which were then selected for bran fermentation with three times of SSP.
Results: The bran fermented with acid-producing microbiota was rich in Pediococcus and Lactobacillus and exhibited increased total titratable acidity and organic acid levels. Electronic nose and organic acid composition analysis revealed that GMAS2S3 (bran fermented with GMAS2 that underwent three times of SSP) had more pronounced flavor characteristics and a higher abundance of acids. Proximate and amino acid analyses confirmed that GMAS2S3 had a higher protein content (22.8%) than the conventional feed (22.8% vs. 16–18%) with abundant amino acids (229.41 mg/g). Palatability evaluation analysis revealed that GMAS2S3-supplemented groups initially showed significantly lower feed intake than the basal diet group, but exceeded basal diet intake during the later adaptation phase.
Discussion: In conclusion, multi-acid synergistic fermentation using anaerobic acid-producing microbiota from baijiu pit mud enhanced bran feed nutritional quality and organic acid content, while maintaining palatability, paving a way for a cost-effective alternative animal feed.
1 Introduction
Animal feed is crucial for animal-derived food production and serves as the foundation for the development of the breeding industry (Tolosa et al., 2021). However, with the continued rise in global feed prices, fermentation has been widely used to improve the nutritional value, absorption efficiency, as well as the digestibility of feed (Katu et al., 2025), enhancing nutrient utilization in animals and thus reducing the financial pressure (Dawood and Koshio, 2020). Fermented feed reduces antinutritional factors in feed and improves gut health, immune function, and overall animal growth performance (Sugiharto and Ranjitkar, 2019; Predescu et al., 2024).
In addition to nutritional components such as carbohydrates, proteins, and lipids, the acids in fermented feed also play important roles in animal growth and metabolism. They promote digestion and absorption, enhance nutrient utilization to improve weight gain and feed conversion rates, and ultimately, meat quality (Giorgino et al., 2023; Liu et al., 2023; Wang et al., 2023). Ng and Koh (2017) reported that the acidic compounds in the feed also inhibit the growth of harmful microorganisms, thereby improving the animals’ intestinal health. Furthermore, increasing the acidity in feed is an effective approach to replace antibiotics (Okoye et al., 2023).
The primary origin of acids in animal feed includes: (1) microbial fermentation (Chen et al., 2023; Hu et al., 2023); (2) the addition of acidifiers including inorganic acids such as phosphoric acid, hydrochloric acid, and sulfuric acid, as well as organic acids like formic acid, acetic acid, citric acid, and malic acid (Polycarpo et al., 2017); and (3) naturally occurring acidic substances such as distillers grains, whey, and fruit pomace (Roguski et al., 2023; Song et al., 2023). Fermented feed contains numerous organic acids depending on various factors such as the type and quantity of microorganisms involved, moisture, temperature, and fermentation duration (Cai et al., 2025; Polycarpo et al., 2017). For example, the use of Bifidobacterium for corn silage fermentation increases the acetic acid concentration (Huang et al., 2021), whereas Enterococcus increases the lactic acid content and decreases acetic acid and butyric acid levels (Marciňáková et al., 2008). Customized fermentation by adding selected microbes affects the composition of organic acids and improves the fermentation quality (Wang et al., 2016; Soundharrajan et al., 2017). Moreover, mixed-culture fermentation (Lu et al., 2020) provides numerous enzymes, potentially enhancing the yield and composition of organic acids (Xu et al., 2011). Therefore, selecting appropriate microbes is crucial for producing a high-quality fermented feed rich in organic acids.
Baijiu is a traditional Chinese distilled liquor, which is typically obtained from grains by solid-state fermentation using Daqu (an essential starter for Chinese baijiu, rich in microbial communities, functional enzyme systems, and flavor precursors), followed by distillation and aging (Ye et al., 2021). Nongxiangxing Baijiu production involves a complex interplay of microbial community metabolism and multi-microbial co-fermentation (Wang S. et al., 2024; Wang Y. et al., 2024; Zhang et al., 2024). The pit mud used in the Baijiu brewing process is a key source of microorganisms, primarily genera such as Lactobacillus, Pediococcus, Clostridium, Acetobacter, Sedimentibacter, and Methanobacterium (Ren et al., 2024), facilitating the production of a variety of short-chain fatty acids (such as acetic acid, propionic acid, lactic acid, butyric acid, and valeric acid), alcohols, aldehydes, esters, and other substances (Zhang Y. et al., 2021). However, these microbes can produce butyric acid, an organic acid that can negatively affect the feed palatability (Galfi and Bokori, 1990), so selecting the appropriate mix of microbes is essential for improving feed quality.
To address the growing need for sustainable and economical feed alternatives in livestock production, the current study was designed to investigate the microbial communities in Baijiu pit mud to establish an effective multi-acid synergistic fermentation system for optimizing organic acids in animal feed. A novel alternative feed was formulated by manipulating microbial community composition to produce beneficial organic acids during fermentation, transforming low-value wheat bran into a high-quality feed ingredient. This research focuses on duck applications due to their high feed conversion efficiency. Although this research primarily targets poultry applications, the fermentation approach has potential applications across various livestock species, such as swine and aquatic animals. The quality of fermented bran feed was comprehensively evaluated to verify its palatability and nutritional level, ensuring a safer and more sustainable feed alternative that promotes animal health while reducing the feeding costs. Future studies will examine meat quality effects and applications in swine and ruminants.
2 Materials and methods
2.1 Materials
Four different cultural media were used as follows: (1) glucose-based medium (GM) containing 20 g/L of glucose, 2.0 g/L of (NH₄)₂SO₄, 0.5 g/L K₂HPO₄, 10 g/L of peptone, 10 g/L of yeast extract powder, 0.1 g/L of MgSO₄·7H₂O, 0.015 g/L of FeSO₄·7H₂O, 0.01 g/L of CaCl₂, 0.01 g/L of MnSO4·H2O, 0.002 g/L of COCl₂, and 0.002 g/L of ZnSO4, pH 7.0; (2) lactic acid medium (LM) containing 10 g/L of lactic acid, 5 g/L of CH₃COONa, and all constituents as in GM, but without glucose, pH 7.0; (3) glucose + yellow water medium (GY) containing 1 g/L of yellow water and all constituents as in GM, pH 7.0; (4) lactic acid + yellow water medium (LY) containing 1 g/L of yellow water and all constituents as in LM, pH 7.0.
2.2 Microbial culture
Baijiu pit mud samples (100 g, labeled as A, B, and C, respectively) and yellow water, a nutrient-rich byproduct of the Baijiu fermentation process (Guo et al., 2024), were collected from Sichuan Yibin Gaozhou Liquor Co., Ltd. (Figure 1a). The basal diets consisted of commercially available standard feed purchased from Meishan Shuxia Feed Co., Ltd. (Meishan, China).
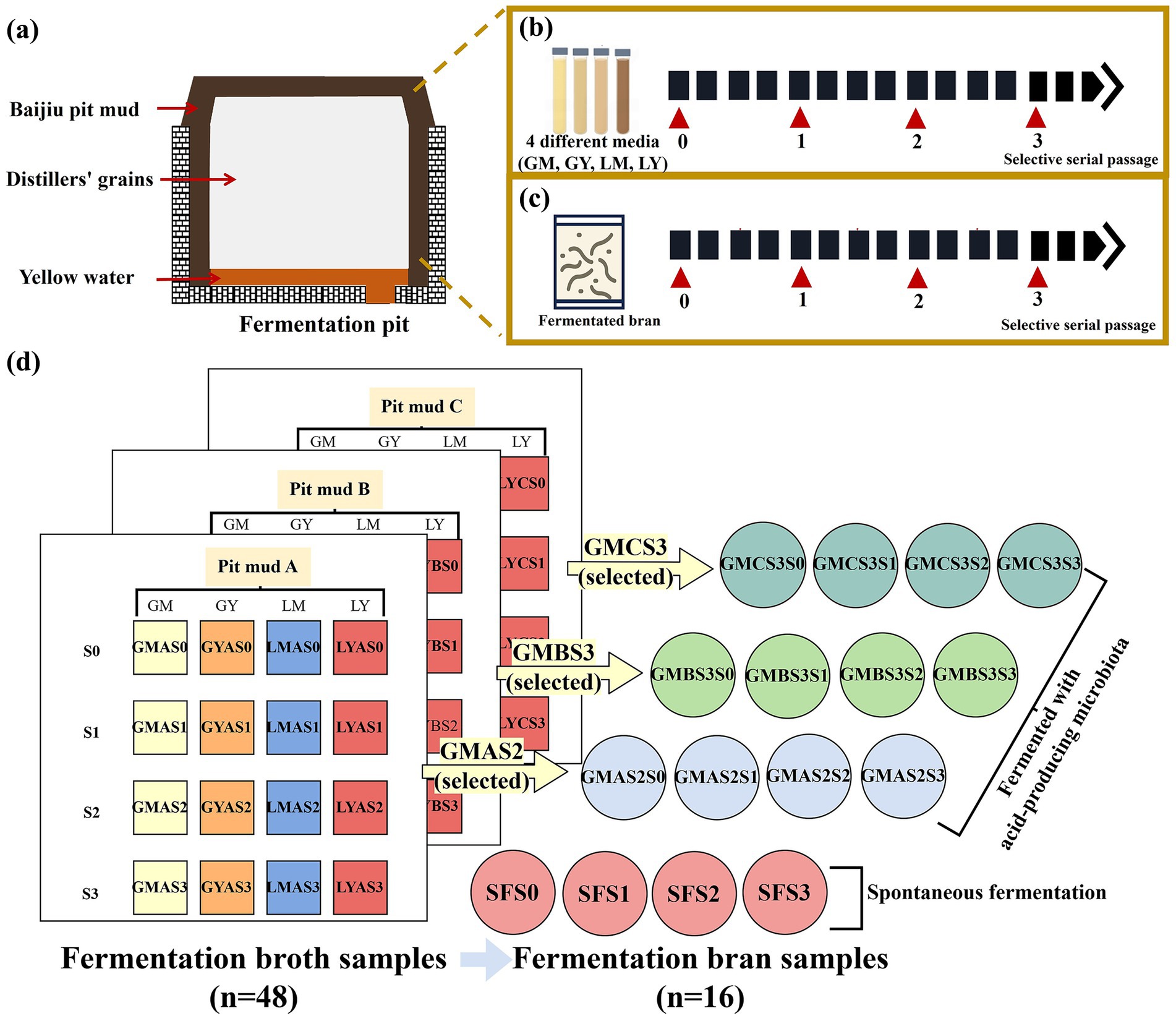
Figure 1. Overview of sample collection, microbial domestication, and samples: (a) diagram of the Nongxiangxing Baijiu fermentation pit; (b) fermentation broth (GM, GY, LM, and LY) preparation by pit mud microbiota with selective serial passage (SSP) (LM: Lactic acid medium; LY: Lactic acid + yellow water medium; GM: Glucose-based medium; GY: Glucose + yellow water medium); (c) bran fermentation by acid-producing microbiota with SSP; (d) summary of the fermentation broth samples and fermentation bran samples.
Fermentation broth samples (n = 48) were obtained through selective serial passage (SSP) of pit muds (A, B, and C) in the different culture media (GM, GY, LM, and LY) (Figure 1b), with acid-producing microbes used to ferment bran samples (n = 16) via three SSPs (Figures 1c,d).
Bajiu pit mud samples (30 g) were thoroughly mixed with 600 mL of culture media (referred to as S0, the sample before SSP) and anaerobically cultured at 25 °C for 48 h (referred to as S1, the first cycle of SSP). Fermentation was conducted in a cylindrical glass fermentation vessel (volume 660 mL; 30 cm height × 5.5 cm diameter; manufactured by Kunming Shanglin Plastic Packaging Co., Ltd., Kunming, China) The enriched cultures underwent two subsequent cycles of SSP, with the addition of 30 mL of culture medium in each cycle, designated as microbiota S2 and S3, respectively (Figure 1b). The microbial growth of fermentation broth from each SSP was quantified by measuring the OD600 using a UV–Vis spectrophotometer (UV-1500PC, Shanghai Macylab Instrument Co., LTD, Shanghai, China). The total titratable acidity (TTA) was quantified according to GB 12456–2021 (the State Standard of the People’s Republic of China) and determined by titration with 0.1 M NaOH and expressed as lactic acid equivalent. GMAS2 (GMA means the culture of Baijiu pit mud A are from glucose medium, S2 is after two times of selective serial passage.), GMBS3, and GMCS3 were selected for subsequent bran feed fermentation due to the higher TTA and desired bacterial composition.
The impact of three optimal acid-producing microbiotas on bran feed fermentation was evaluated by fermentation with SSP with spontaneous fermentation acting as the control group. Briefly, 324 g of bran was thoroughly mixed with 126 g of purified water and 50 mL of selected fermentation broth before the addition of 1% (w/w) sucrose and 0.5% (w/w) urea (S0). This formulation was based on pilot experiments and the selected fermentation broth was replaced with purified water in the control group. The mixtures were incubated at 25 °C for 72 h using a fermentation bag for the first cycle of SSP (S1), then two subsequent cycles of SSP (S2 and S3) were conducted using 5 g of fermented bran from the previous SSP, 135 g of purified water, 360 g of fresh bran, 1% (w/w) sucrose, and 0.5% (w/w) of urea anaerobically at 25 °C for 72 h for each cycle (Figure 1c).
The total plate counts (TPC) were determined according to GB/T 13093–2023 (National Standards of the People’s Republic of China for Determination of bacterial count in feeds). Briefly, 100 μL of serially diluted homogeneous samples (25 g of sample homogenized in 225 mL of sterile saline solution, followed by 10-fold serial dilutions up to 10−6) was plated on nutrient agar and incubated at 36 °C for 48 h. The TPC was reported as colony-forming units per g (CFU/g). The microbiota for bran feed fermentation with the highest TTA, TPC, and desired microbial composition were selected for subsequent analysis.
2.3 Microbial composition analysis
Microbial DNA was extracted from the fermentation broth and fermented bran samples using the cetyltrimethylammonium bromide (CTAB) method as described by Ren et al. (2024). The 16S rRNA genes of distinct regions (16S V3-V4) were amplified using specific primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACNNGGGTATCTAAT-3′). The PCR products were separated by 2% agarose gel electrophoresis and purified using the Universal DNA Kit (TianGen, China). Sequencing libraries were generated using NEB Next® Ultra DNA Library Prep Kit (Illumina, United States) and index codes were added according to the manufacturer’s instructions. Library quality was determined using the Agilent 5400 (Agilent Technologies Co. Ltd., United States). Finally, the library was sequenced using an Illumina NovaSeq platform generating 250 bp paired-end reads which were analyzed using Qiime2docs (Yang et al., 2022). The sequence data were analyzed by Wekemo Bioincloud (https://www.bioincloud.tech/) including linear discriminant analysis (LDA), effect size (LEfSe) and principal coordinates analysis (PCoA).
2.4 Composition analysis of fermented bran
2.4.1 Organic acid composition
The fermented bran samples (GMAS2S2: bran fermented with GMAS2 that underwent two times of SSP, GMAS2S3, GMBS3S2, GMBS3S3, GMCS3S2, GMCS3S3, and their corresponding spontaneous fermented bran) with the highest TTA and desired microbial composition were subjected to organic acid composition determination using low molecular weight organic acids (LMWOAs) following the method described by Hua et al. (2022). The samples were mixed with 500 μL of methanol/H2O/formic acid (30:70:0.1, v/v/v) with two steel balls, vortexed for 60 s and ground at 55 Hz for 1 min before centrifugation for 10 min (12,000 × g, 4 °C). The supernatant was filtered through a 0.22 μm membrane for LC–MS analysis on an ACQUITY Liquid chromatography (Waters, United States) and an AB5000 mass spectrometer (AB SCIEX, United States).
2.4.2 Flavor composition
An electronic nose was used to detect the flavor compounds in the fermented bran samples according to the method described by Lan et al. (2022) and Wang Y. et al. (2024). Briefly, 2 g of fermented bran was placed in a 20 mL headspace vial and the overall aroma was assessed using the PEN3 electronic nose (Beijing Ying Sheng Heng Tai Technology Co., Beijing, China) with a gas flow rate of 300 mL/min, a test time of 120 s, and a cleaning time of 100 s. The sensor information included W1C (aromatic and benzene components), W5S (nitrogen and oxygen compounds), W3C (aromatic and ammonia compounds), WGS (hydrogen selectivity), W5C (short-chain alkanes and aromatic components), W1S (methyl group), W1W (organic sulfide), W2S (alcohols, aldehydes, and ketones), W2W (aromatic and organic sulfides), and W3S (long-chain alkanes). The data were analyzed using WinMuster Software and Device Documentation Version 1.6.2.
2.5 Quality evaluation
The selected fermented bran (GMAS2S3) containing a high content of organic acids and a more favorable microbial community was subjected to quality evaluation.
2.5.1 Proximate analysis
The moisture content was determined using the direct drying method in Chinese National Standard GB/T 6435-2014. Crude protein content was analyzed based on the Kjeldahl nitrogen determination described in GB/T 6432-2018. Moreover, the crude fat content was quantified by the Soxhlet extraction method according to GB/T 6433-2006, while the crude fiber content was determined using the acid-alkaline digestion method in GB/T 6434-2006. The crude ash content was assessed based on the high-temperature burning method according to GB/T 6438-2007. The carbohydrate content was calculated based on the following formula: .
2.5.2 Amino acid quantification and data analysis
Free amino acids were analyzed by HPLC on a 1290 Infinity LC system (Agilent, California, United States) following the method described by Han et al. (2019). Briefly, 60 mg of GMAS2S3 was thoroughly mixed with 450 μL of cold methanol/ acetonitrile/H2O (4:4:1, v/v/v) and 16 isotope-labeled internal standards. The mixture was vortexed for 60 s and sonicated at a low temperature for 1 h. The proteins were precipitated at −20 °C for 2 h, collected by centrifugation for 20 min (14,000 × g, 4 °C) and dried in a vacuum centrifuge before subsequent analysis.
2.5.3 Palatability evaluation
The palatability of the fermented bran was evaluated using the method described by Pujaningsih and Mangisah (2020) using Sichuan Shelducks (Anas platyrhynchos domestica). A total of 96 ducks (not sexed), with the age (7-week-old), were reared on a farm at Yibin University, Yibin, China at an ambient temperature of 23 °C ± 3 °C and relative humidity of 50–60%. All animal procedures were approved by the Yibin University Animal Care and Use Committee (Approval No. 20231001001) and conducted following National Standard Guidelines for Laboratory Animal-Guideline for Ethical Review of Animal Welfare according to GB/T 35892-2008. Animals received a commercially available standard feed (Meishan Shuxia Feed Co., Ltd., Meishan, China) formulated primarily with soybean meal, corn, rapeseed meal, and wheat middlings. The diet contained crude protein 12.0–16.0%, crude fiber ≤ 10.0%, crude ash ≤ 20.0%, and moisture ≤ 14.0%. Animals had free access to feed and drinking water. The ducks were divided into four feeding groups: three experimental groups (T1: 20% of GMAS2S3 + 80% of basal diet, T2: 40% of GMAS2S3 + 60% of basal diet, T3: 60% of GMAS2S3 + 40% of basal diet) and a control group (T0: 100% of basal diet). Each group had three replicates, with eight ducks per replicate. The palatability test measured the feed intake during days 1–3 (initial response) and days 4–7 (adaptation phase).
2.6 Statistical analysis
The data were analyzed using one-way ANOVA in SPSS 23.0 (SPSS Inc., Chicago, IL, United States) with the post-hoc Duncan test. Correlations were determined by the R package and a p-value < 0.05 was considered significant.
3 Results and discussion
3.1 Microbial growth, composition, and acid production in different culture media
As shown in Figure 2a, the value of OD600 of pit mud microbes in GM at each SSP (S1: 2.66 ± 0.34, S2: 2.09 ± 0.12, S3: 2.25 ± 0.15), which has higher S2 and S3 values than GY (S1: 2.81 ± 0.16, S2: 1.74 ± 0.40, S3: 1.64 ± 0.41), LM (S1: 2.90 ± 0.18, S2: 0.35 ± 0.17, S3: 0.05 ± 0.01), and LY (S1: 2.17 ± 0.62, S2: 0.35 ± 0.17, S3: 0.032 ± 0.01) (Supplementary Table S1). This indicates that glucose media was a better culture medium than lactic acid, as glucose is the microbe’s preferred carbon source growth (Kiefer et al., 2002). Furthermore, GM supported greater microbial growth than GY for the three pit mud samples, indicating that yellow water does not effectively promote the growth of pit mud bacteria in selective culture media. This could be due to the presence of soluble starch, organic acids, and more alcohol that disrupt the original carbon-nitrogen balance in selective culture media, causing certain nutrients to be in excess or deficient, thereby inhibiting microbiota growth (Weng et al., 2022).
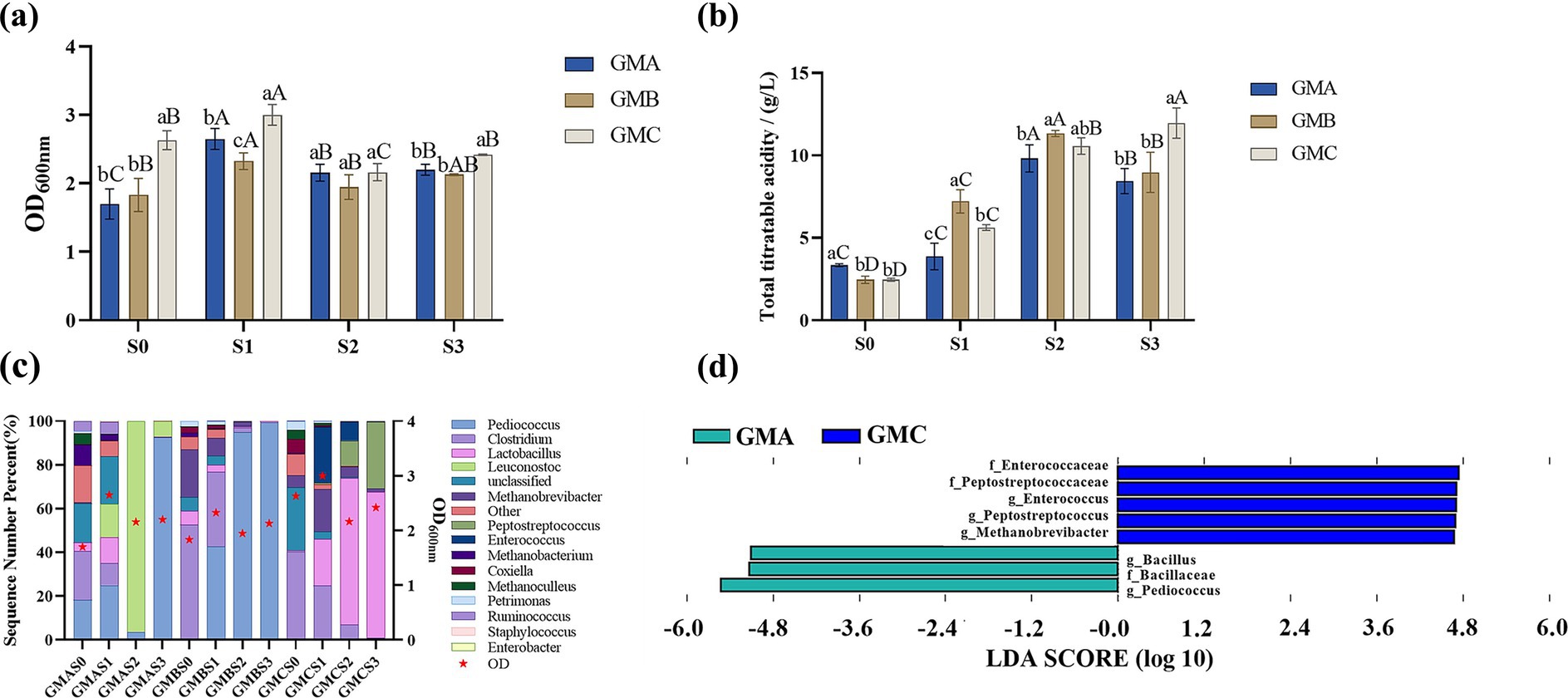
Figure 2. Growth of bacterial communities from Baijiu pit mud samples: A, B, and C cultured on GM medium (a). GM: Glucose-based medium. S0 represents the mixture of the initial Baijiu pit mud and culture media before fermentation. S1 represents the anaerobic fermentation products of S0 at 25 °C for 48 h; S2 and S3 were the selective serial passage (SSP) after the second and third cycles of culture, respectively. Acid production of Baijiu pit mud samples A, B and C in GM during SSP (b). Relative abundance of the bacterial community in the fermentation broth of each pit mud sample (c) and the characteristic taxa of GMA and GMC (d). Different letters indicate significant differences between groups (p < 0.05): lowercase letters (a–c) denote differences among different groups of samples within the same SSP; uppercase letters (A–D) denote differences among SSP within the same group of samples.
As shown in Figure 2b, the highest TTA levels were observed in GMAS2 (9.50 ± 0.82 g/L), GMBS2 (11.33 ± 0.19 g/L), and GMCS3 (11.61 ± 0.91 g/L). This elevated TTA with the increased selective serial passages may be due to the microbial growth. A moderate positive correlation was found between microbial biomass (OD value) and TTA (r = 0.49) in line with the study of Ke et al. (2022) which reported a correlation coefficient close to 1, suggesting that microbial growth promotes acid production. The lower TTA content in the first cycle might be due to a slower growth rate as the microbiota adapt to the new environment, with the microbes being more efficient at utilizing nutrients in the second and third cycles.
The bacterial community from Baijiu pit muds cultured in GM, with its higher microbial growth and superior acid-producing capabilities, was selected for microbial composition analysis identifying fifteen predominant genera (with relative abundance > 1%) (Figure 2c) including Pediococcus, Clostridium, Lactobacillus, Leuconostoc, Unclassified, Methanobrevibacter, Peptostreptococcus, Enterococcus, Methanobacterium, Coxiella, Methanoculleus, Petrimonas, Ruminococcus, Staphylococcus, and Enterobacter. LEfSe analysis revealed significant differences in abundance between the pit mud samples (Figure 2d), with the genera Enterococcus, Peptostreptococcus, and Methanobrevibacter being significantly enriched in GMC, while Bacillus and Pediococcus were significantly enriched in GMA. Enterococcus, a common pathogenic microorganism, also serves as a crucial reservoir for antibiotic resistance genes, potentially enabling horizontal gene transfer to other bacteria (Krawczyk et al., 2021). However, SSP in GM led to a significant reduction in the relative abundance of Enterococcus across all three pit mud samples, particularly in GMA and GMB, possibly due to the competitive inhibition of dominant microorganisms (Pediococcus and Leuconostoc). In addition, Peptostreptococcus has been linked to pro-inflammatory effects in mice and humans (Chen X. H. et al., 2019; Chen X. W. et al., 2019; Shen et al., 2024) and its relative abundance was reduced in the GMA and GMB pit mud samples after SSP culturing in GM medium. However, due to its high initial abundance in the microbial community of GMC, Peptostreptococcus in GMCS3 (30.419%) gradually became the dominant genus during SSP. Furthermore, the abundance of Staphylococcus (0.00–0.38%) and Coxiella (0.360–6.384%) reduced from S0 to S3 in all samples. This is desirable as Staphylococcus and Coxiella are detrimental to animal health and production efficiency, and thus negatively impact feed quality (Celina and Cerný, 2022; Ou et al., 2017).
Genera such as Clostridium and Ruminococcus belonging to the class Clostridia (Guo et al., 2020), Bacteroides and Petrimonas belonging to the class Bacteroidia (Ye et al., 2016), and Methanobrevibacter, a typical methanogen (Chaudhary et al., 2018), were identified in the pit mud samples. These microbes regulate the ecological balance of pit mud (Hu et al., 2016; Liu et al., 2017). After continuous passage, the different pit mud microbial communities became predominantly acid-producing such as Pediococcus in GMA and GMB, and Lactobacillus in GMC (Figure 2c).
Although GMBS2 showed superior acid-producing capability compared to GMBS3, it had higher proportions of Enterococcus and Coxiella known to be detrimental to the host and posing a risk to food safety (Krawczyk et al., 2021). Thus, GMBS3 was selected over GMBS2 for further analysis. Overall, based on the microbial growth, acid-production capability, and microbial community composition results, GMAS2, GMBS3, and GMCS3 were selected for subsequent bran fermentation.
3.2 Microbial growth, composition, and acid production capacity of the acid-producing microbes in bran
There was no significant difference in TPC among the GMAS2, GMBS3, and GMCS3 (Figure 3a) with the increasing to ideal levels (Yu et al., 2020) after three cycles of SSP (9.42 ± 0.61 log CFU/g, GMBS3S3: 9.47 ± 0.34 log CFU/g, and GMCS3S3: 9.43 ± 0.30 log CFU/g), indicating that the acid-producing microbes are capable of stable growth during bran fermentation.
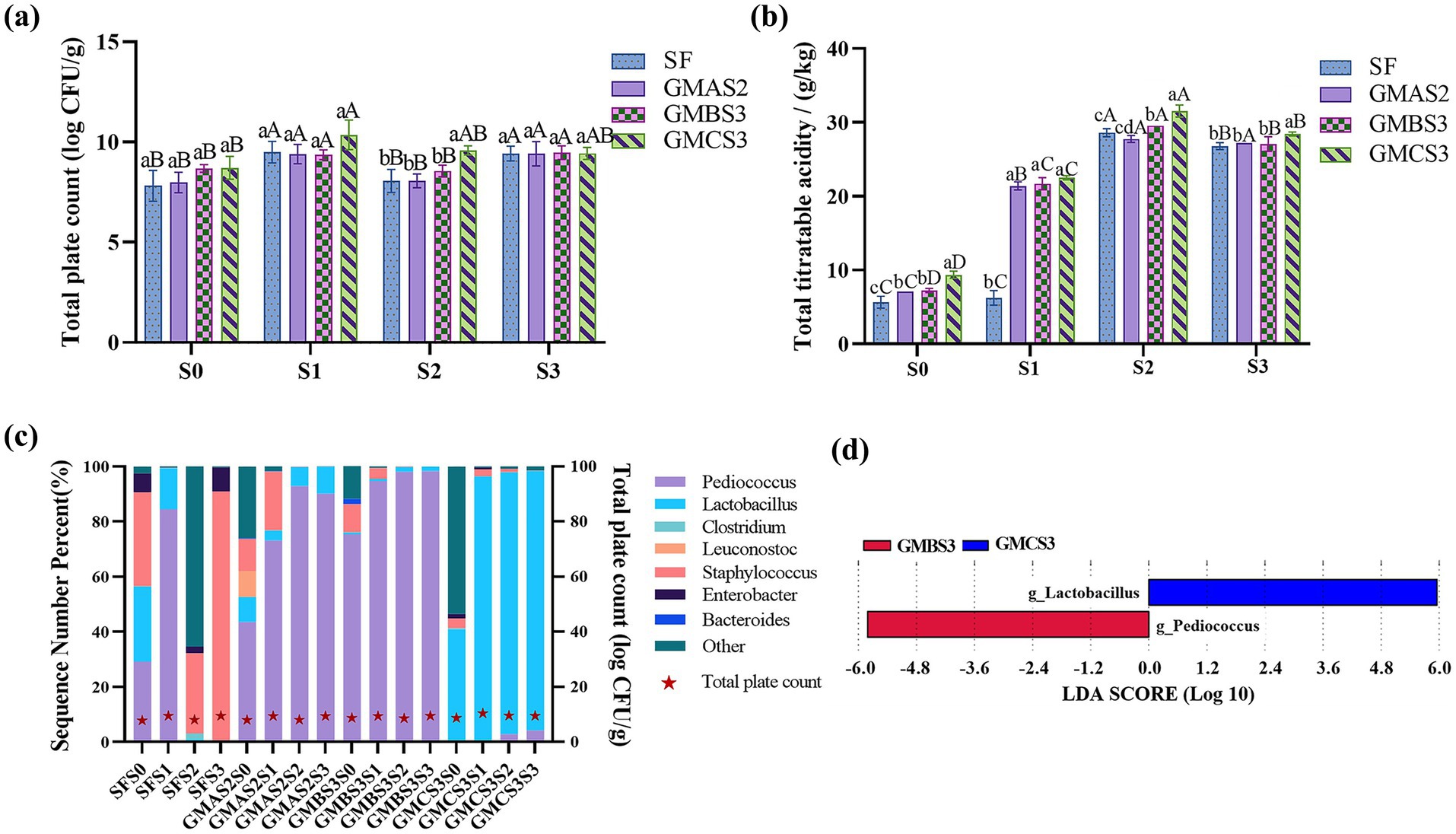
Figure 3. Total plate counts of GMAS2, GMBS3, and GMCS3 (a) in fermentation bran during selective serial passage (SSP). S0 is the initial bran sample after inoculation with the acid-producing microbes GMAS2, GMBS3, and GMCS3, respectively. S1, S2, and S3 are the fermentated bran samples after the first, second, and third SSP, respectively. SF is control group of spontaneous fermentation. (b) The total titratable acid of the bran samples fermented with GMAS2, GMBS3, and GMCS3 throughout SSP. (c) Relative abundance of the acid-producing microbes in the fermentation bran. (d) The characteristic taxa of GMBS3 and GMCS3 fermented bran (d). Different letters indicate significant differences between groups (p < 0.05): lowercase letters (a–d) denote differences among different groups of samples within the same SSP; uppercase letters (A–D) denote differences among SSP within the same group of samples.
The acid-producing microbiota fermented bran (APMFB) had a significantly higher TTA (GMAS2S1: 21.40 ± 0.55 g/kg, GMBS3S1: 21.69 ± 0.82 g/kg, and GMCS3S1: 22.50 ± 0.27 g/kg, respectively) compared to spontaneous fermented bran (SFB) (6.24 ± 0.97 g/kg) (Figure 3b), suggesting that the acid-producing microbes adapted more effectively to the bran fermentation environment, exhibiting stable growth and acid production compared to spontaneous fermentation. Acid production increased throughout SSP with a significantly higher TTA in S2 and S3 than in S1.
Seven predominant genera (with relative abundance >1%) were identified in the fermented bran including Pediococcus, Lactobacillus, Clostridium, Leuconostoc, Staphylococcus, Enterobacter, and Bacteroides (Figure 3c). The microbial composition of SFB shifted toward an undesirable microbial composition compared to APMFB, as evidenced by the abundance of Staphylococcus (90.596%) and Enterobacter (8.740%). Staphylococcus is a common pathogenic bacterium capable of causing food poisoning when consumed in animal-derived foods (Odetokun et al., 2023). Moreover, Enterobacter can adversely affect feed quality by competing to utilize fermentation substrates, increasing protein degradation, and affecting the feed utilization rate (Zhang Q. et al., 2021). Therefore, the presence of these genera in feed fermentation must be controlled to ensure the quality and safety of the final product.
Although the TTA of APMFB and SFB were not significantly different, the former had an increased abundance of acid-producing microbiota, such as Pediococcus and Lactobacillus (Balakrishnan and Agrawal, 2014). Furthermore, the acid-producing microbial community were stable after multiple passages. Specifically, GMBS3 (GMBS3S1, GMBS3S2, and GMBS3S3) was predominantly enriched with Pediococcus, whereas GMCS3 (GMCS3S1, GMCS3S2, and GMCS3S3) was dominated by Lactobacillus. This trend was consistent with the microbial composition observed in the corresponding culture media (GMBS3 and GMCS3, respectively). In contrast, GMAS2 fermented bran samples dominated by Pediococcus were inconsistent with the fermented broth GMAS2 dominated by Leuconostoc. This may be due to Leuconostoc not adapting well to the fermented bran environment, and thus was outgrown by Pediococcus.
Further LEfSe analysis revealed a significant difference between GMBS3 and GMCS3, as characterized by the dominance of Pediococcus and Lactobacillus (Figure 3d). Pediococcus can produce various organic acids and short-chain fatty acids during milk fermentation (Balakrishnan and Agrawal, 2014). Similarly, Lactobacillus can produce organic acids such as lactic acid and acetic acid as well as polypeptide enzymes which enhance the feed quality. These metabolites are environmental modulators that influence the colonization of fermented microbiota and promote the formation of unique microbiota (Ferguson et al., 2010). Thus, optimizing the microbial composition during fermentation to increase the abundance and microbial stability of Pediococcus and Lactobacillus in fermented bran results in a higher acid content.
3.3 Composition of the fermented bran products
3.3.1 Organic acids
Organic acids are often added to animal feed as acidifiers to improve gastrointestinal pH, regulate gut microbial composition, enhance nutrient absorption, and promote individual growth (Tugnoli et al., 2020). APMFB (GMAS2, GMBS3, and GMCS3) at S2 and S3 were selected for organic acid analysis due to the higher TTA and desired microbial composition. Compared to SFB, APMFB contained more lactic acid (5444.59–6244.11 μg/g), citric acid (0–1034.28 μg/g), pantothenic acid (864.99–1053.89 μg/g), pyroglutamic acid (60.45–112.39 μg/g), and phenyl lactic acid (51.52–102.28 μg/g) (Figure 4a) as expected due to the higher TTA. Most organic acids were present at concentrations exceeding 0.1%, a threshold known to influence fermentation (Asriqah et al., 2018; Haq et al., 2017). The citric acid content of bran fermented with GMAS2 (1029.64 ± 173.67 μg/g) and GMBS3 (1124.57 ± 127.68 μg/g) was 47 to 51 times higher than that of the spontaneously fermented group (21.94 ± 9.74 μg/g). Citric acid promotes nutrient absorption and improves poultry carcass quality, and the intestinal microecological environment (Khan et al., 2022). Furthermore, the lactic acid, pantothenic acid, and phenyl lactic acid contents in GMAS2S3 fermented bran were higher than the other samples. In addition, the total organic acid content in APMFB (8050.95 μg/g in GMAS2S2, 8902.82 μg/g in GMAS2S3, 8094.30 μg/g in GMBS3S2, and 8366.91 μg/g in GMBS3S3) was higher than in the SFB (7540.61 μg/g in SFS2 and 7598.11 μg/g in SFS3), indicating that the acid-producing microbiota enhanced the organic acid content of bran feed. GMAS2S3 fermented bran exhibited the highest total organic acid content with a greater acid composition than other groups, indicating its superior feeding potential.
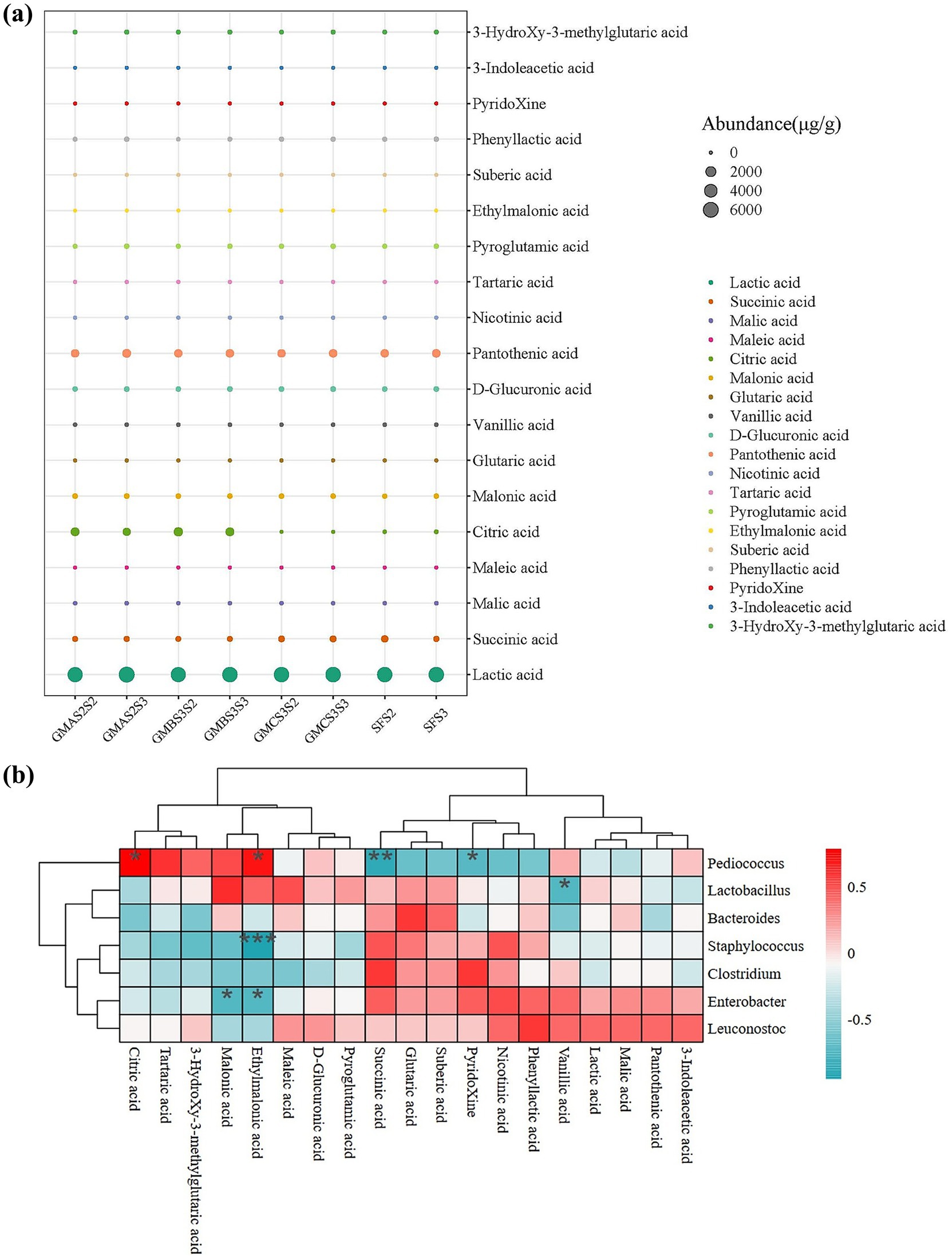
Figure 4. (a) Organic acid composition of bran samples fermented with acid-producing microbes at the second (S2) and third (S3) selective serial passage (SSP). (b) Correlation between organic acid composition and the microbial composition during bran fermentation. The correlation coefficient (r) is displayed in different colors. *p < 0.05, **p < 0.01, ***p < 0.001. SF: spontaneous fermented bran.
The correlation analysis between organic acid content and the bacterial community revealed that Pediococcus was significantly positively correlated with the production of ethylmalonic acid (r = 0.714) and citric acid (r = 0.786) and negatively correlated with the production of pyridoxine acid (r = −0.714) and succinic acid (r = −0.881) (Figure 4b). This is in agreement with the high citric acid production in Pediococcus-dominate fermented bran samples (GMBS3S2 and GMBS3S3), as shown in Figures 3c, 4a. In addition, there is a significant negative correlation (r = −0.738) between Lactobacillus and vanillic acid production, possibly due to the ability of Lactobacillus to convert vanillic acid into vanillin (Mostafa and Hashem, 2023), thereby reducing the vanillic acid content in fermented bran feed. Moreover, Staphylococcus (r = −0.952) and Enterobacter (r = −0.733) were negatively correlated with ethylmalonic acid production. Furthermore, there was a negative correlation (r = −0.733) between Enterobacter and malonic acid production which may be due to the antimicrobial effect of malonic acid against harmful microorganisms (Fei et al., 2023).
3.3.2 Flavor evaluation of fermented bran
The overall aroma of fermented bran was analyzed using an electronic nose (Figure 5), revealing the presence of non-polar organic compounds, aromatic organic compounds, ammonium, short-chain and long-chain alkanes (Kh et al., 2025). In contrast, Fomitopsis pinicola inoculation of wheat bran powder was reported to elicit low-level W2W and W2S responses (Tu et al., 2020) suggesting that acid-producing microbiota in this study might synthesize a wider range of substrates. PCA to determine the variations among the samples during the subculturing (Figures 5b–d) revealed that the samples fermented with GMAS2 were distinct from the other samples after three cycles of SSP. Furthermore, the radar plot (Figure 5e) revealed that the bran samples after three cycles of SSP exhibited relatively higher W1S and W1W responses, which correspond to methyl compounds and sulfides, respectively. Notably, GMAS2S3 exhibited higher acid production compared to other samples, possibly associated with its elevated W1W and W5S values. The degradation of proteins and ATP is related to the compounds detected by W1W and W5S, while acids are the byproduct of these processes (Li et al., 2024). The higher response of W1W and W5S in sample GMAS2S3 may have played a key role in driving the increased acid accumulation (Deng et al., 2023).
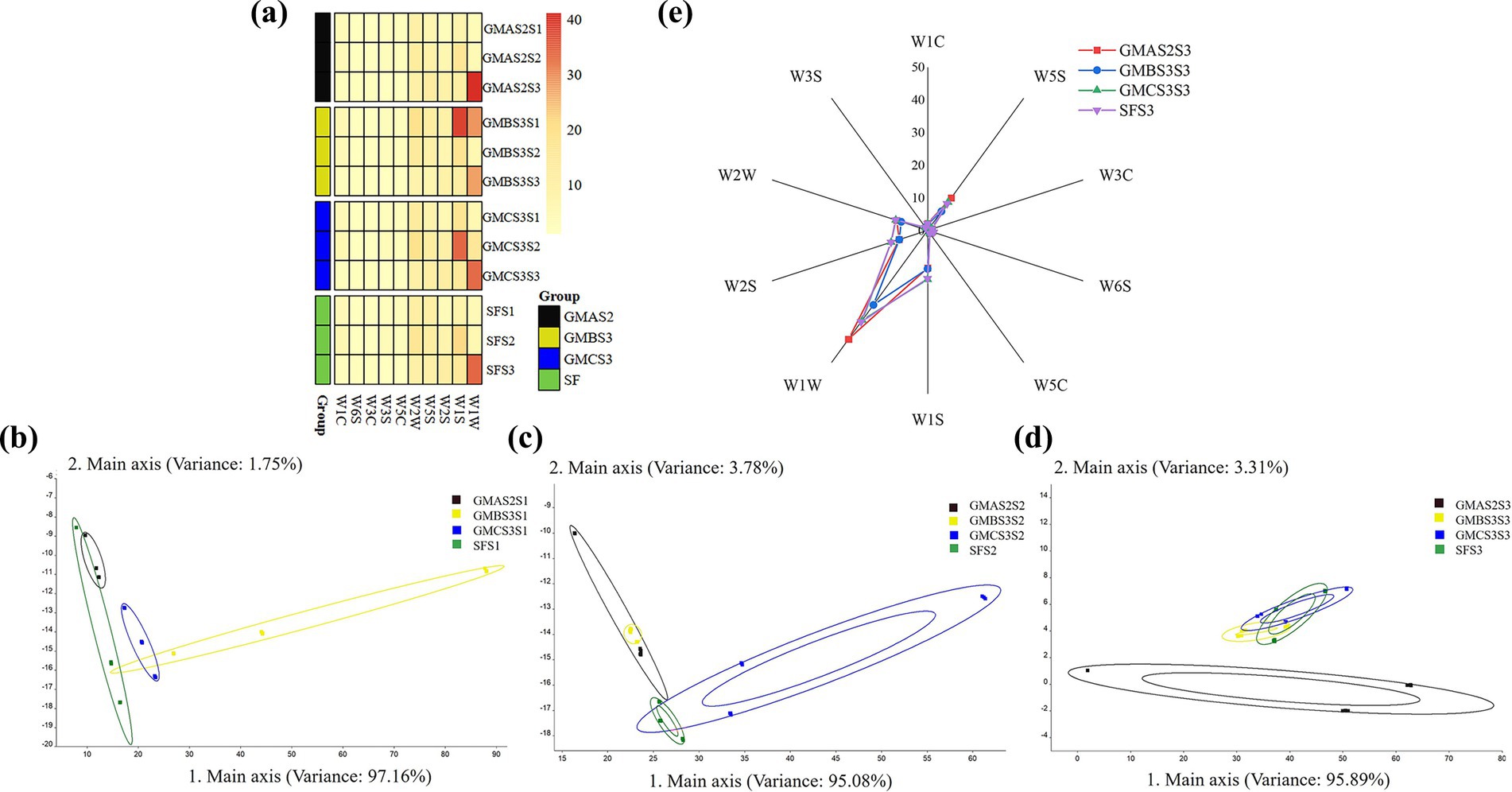
Figure 5. Characterization and analysis of the electronic nose responses: (a) sensor response heat map. Principal component analysis (PCA) of the fermented bran samples after the (b) first selective serial passage (SSP), (c) second SSP, and (d) third SSP. SF: spontaneous fermented bran. (e) The contour radar chart of the e-nose responses in S3.
3.4 Quality evaluation of GMAS2S3
3.4.1 Proximate and amino acid composition of GMAS2S3
GMAS2S3 was selected for further nutritional analysis due to its more pronounced flavor characteristics and high acid composition. As shown in Table 1a, GMAS2S3 exhibited a higher protein content (22.8%) and lower fat content (1.8%) than wheat bran (protein: 10.30% and fat: 3.97%) (Lin et al., 2019). The lower fat content in APMFB will not influence the duck’s body weight. This is supported by the fat content of the feed (0.9 to 7.83%) not affecting the body weight and body weight gain of the ducks (Bai et al., 2019). Furthermore, the lower fat content in the feed will prolong its shelf life as it will be less prone to oxidation. In addition, GMAS2S3 had a higher protein content (22.8% vs. 18.50%) compared to that reported by Zhang et al. (2022), possibly due to the enhanced nutrient composition (Márquez-Castillo and Vidal-Quintanar, 2011).
The composition and concentration of amino acids in the feed support the animal’s physiological activities and contribute to the feed quality (Mou et al., 2019). The total amino acid content in GMAS2S3 was 229.41 mg/g, including 50.21 mg/g of essential amino acids and 179.20 mg/g of non-essential amino acids (Table 1b). The free amino acid content in fermented bran was higher than in the fermented bean dregs and soybeans (Heng et al., 2022), suggesting that GMAS2S3 has a good feeding value.
3.4.2 Palatability evaluation
The palatability of the diet containing GMAS2S3 was evaluated based on the daily intake of Sichuan Shelducks (Anas platyrhunchos domestica). There was a pronounced decrease in daily intake as the proportion of APMFB in the diet increased (Table 2). During the initial response period (days 1–3), compared to the experimental groups, the control group (T0, 100% basal diet) exhibited the highest daily intake of 202.6 g/head/day, while the T3 group (40% basal diet + 60% GMAS2S3) had the lowest intake of 179.2 g/head/day, with the difference being statistically significant (p < 0.05). However, in the adaptation phase (days 4–7), the difference in the feed intake among groups were not statistically significant, and the intake levels in the experimental groups surpassed the control group. Although palatability is not the primary determinant of feed efficacy, maintaining acceptable palatability levels ensures that the feed does not negatively impact intake behavior. In this study, the feed provided enhanced nutrient composition without negatively affecting palatability, which not only supports its practical application but also highlights its potential as a more cost-effective option. While this evaluation is not entirely comprehensive, it provides an preliminary supporting evidence for the practicability of the feed.
4 Conclusion
Baijiu pit mud was utilized as a source of microbes to enhance the production and composition of acids in fermented bran. The GMAS2S3 bacterial community predominantly comprising Pediococcus and Lactobacillus after the third passage was selected as the optimal microbe to ferment bran, significantly increasing the total organic acid content with elevated concentrations of lactic acid, pantothenic acid, citric acid, and phenyl lactic acid. Moreover, the GMAS2S3 exhibited abundant nutritional content, with an amino acid profile that meets the physiological requirements of animals, relatively good palatability, and significant feeding value. Future research will focus on assessing the practical value of GMAS2S3 in conjunction with the growth performance of ducks to further explore its potential applications.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: NCBI, SAMN50675256-SAMN50675283.
Ethics statement
The animal study was approved by Yibin University Animal Care and Use Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
WP: Data curation, Writing – original draft. BL: Writing – original draft. KC: Writing – review & editing. SW: Writing – review & editing, Funding acquisition, Visualization, Data curation. LY: Data curation, Visualization, Writing – review & editing. XW: Writing – review & editing, Visualization, Data curation. MN-K: Writing – review & editing, Supervision. NM: Supervision, Writing – review & editing. NS: Writing – review & editing, Supervision. WW: Supervision, Writing – review & editing, Validation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Solid-state Fermentation Resource Utilization Key Laboratory of Sichuan Province (Grant No. 2022GTYY07), the Solid-state Fermentation Resource Utilization Key Laboratory of Sichuan Province (Grant. No. 2022GTYY01), and Scientific Research Project of Yibin University (Grant No. 2023QH21).
Acknowledgments
We greatly appreciate the reviewers whose comments and suggestions help improve and clarify this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1646911/full#supplementary-material
References
Asriqah, L., Nugroho, R. A., and Aryani, R. (2018). Effect of various organic acid supplementation diets on Clarias gariepinus BURCHELL, 1822: evaluation of growth, survival and feed utilization. F1000Res 7:1465. doi: 10.12688/f1000research.15954.1
Bai, W. Q., Zhang, K. Y., Ding, X. M., Bai, S. P., Wang, J. P., Peng, H. W., et al. (2019). High dietary energy content increases inflammatory markers after lipopolysaccharide challenge in meat ducks. Poult. Sci. 98, 164–171. doi: 10.3382/ps/pey380
Balakrishnan, G., and Agrawal, R. (2014). Antioxidant activity and fatty acid profile of fermented milk prepared by Pediococcus pentosaceus. J. Food Sci. Technol. 51, 4138–4142. doi: 10.1007/s13197-012-0891-9
Cai, F., Huang, M., Liu, W., Wan, X., Qiu, K., and Xu, X. (2025). Dietary addition of compound organic acids improves the growth performance, carcass trait, and body health of broilers. Front. Nutr. 12:1536606. doi: 10.3389/fnut.2025.1536606
Celina, S. S., and Cerný, J. (2022). Coxiella burnetii in ticks, livestock, pets and wildlife: a mini-review. Front. Vet. Sci. 9:1068129. doi: 10.3389/fvets.2022.1068129
Chaudhary, P. P., Conway, P. L., and Schlundt, J. (2018). Methanogens in humans: potentially beneficial or harmful for health. Appl. Microbiol. Biotechnol. 102, 3095–3104. doi: 10.1007/s00253-018-8871-2
Chen, X. H., Wang, A., Chu, A. N., Gong, Y. H., and Yuan, Y. (2019). Mucosa-associated microbiota in gastric cancer tissues compared with non-cancer tissues. Front. Microbiol. 10:1261. doi: 10.3389/fmicb.2019.01261
Chen, L., Wang, Y., Li, X., MacAdam, J. W., and Zhang, Y. (2023). Interaction between plants and epiphytic lactic acid bacteria that affect plant silage fermentation. Front. Microbiol. 14:1164904. doi: 10.3389/fmicb.2023.1164904
Chen, X. W., Wu, L., Luo, N., Mo, C. H., Wong, M. H., and Li, H. (2019). Arbuscular mycorrhizal fungi and the associated bacterial community influence the uptake of cadmium in rice. Geoderma 337, 749–757. doi: 10.1016/j.geoderma.2018.10.029
Dawood, M. A., and Koshio, S. (2020). Application of fermentation strategy in aquafeed for sustainable aquaculture. Rev. Aquac. 12, 987–1002. doi: 10.1111/raq.12368
Deng, Z., Ferreira, A. L. M., Spanjers, H., and van Lier, J. B. (2023). Anaerobic protein degradation: effects of protein structural complexity, protein concentrations, carbohydrates, and volatile fatty acids. Bioresour. Technol. Rep. 22:101501. doi: 10.1016/j.biteb.2023.101501
Fei, Y. C., Cheng, Q., Zhang, H., Han, C., Wang, X., Li, Y. F., et al. (2023). Maleic acid and malonic acid reduced the pathogenicity of Sclerotinia sclerotiorum by inhibiting mycelial growth, sclerotia formation and virulence factors. Stress Biol. 3, 1–12. doi: 10.1007/s44154-023-00122-0
Ferguson, R. M., Merrifield, D. L., Harper, G. M., Rawling, M. D., Mustafa, S., Picchietti, S., et al. (2010). The effect of Pediococcus acidilactici on the gut microbiota and immune status of on‐growing red tilapia (Oreochromis niloticus). J. Appl. Microbiol. 109, 851–862.
Galfi, P., and Bokori, J. (1990). Feeding trial in pigs with a diet containing sodium n-butyrate. Acta Vet. Hung. 38, 3–17.
Giorgino, A., Raspa, F., Valle, E., Bergero, D., Cavallini, D., Gariglio, M., et al. (2023). Effect of dietary organic acids and botanicals on metabolic status and milk parameters in mid–late lactating goats. Animals 13:797. doi: 10.3390/ani13050797
Guo, P., Zhang, K., Ma, X., and He, P. (2020). Clostridium species as probiotics: potentials and challenges. J. Anim. Sci. Biotechnol. 11, 1–10. doi: 10.1186/s40104-019-0402-1
Guo, Q., Zhao, J., Peng, J., Huang, Y., and Shao, B. (2024). Revealing the flavor compositions, microbial diversity, and biological functions of Huangshui from different production workshops. LWT 207:116683. doi: 10.1016/j.lwt.2024.116683
Han, X., Zhang, C., Wang, C., Huang, Y., and Liu, Z. (2019). Gadolinium inhibits cadmium transport by blocking non-selective cation channels in rice seedlings. Ecotoxicol. Environ. Saf. 179, 160–166. doi: 10.1016/j.ecoenv.2019.04.057
Haq, Z., Rastogi, A., Sharma, R., and Khan, N. (2017). Advances in role of organic acids in poultry nutrition: a review. J. Appl. Nat. Sci. 9, 2152–2157. doi: 10.31018/jans.v9i4.1502
Heng, X., Chen, H., Lu, C., Feng, T., Li, K., and Gao, E. (2022). Study on synergistic fermentation of bean dregs and soybean meal by multiple strains and proteases. LWT 154:112626. doi: 10.1016/j.lwt.2021.112626
Hu, X., Du, H., Ren, C., and Xu, Y. (2016). Illuminating anaerobic microbial community and cooccurrence patterns across a quality gradient in Chinese liquor fermentation pit muds. Appl. Environ. Microbiol. 82, 2506–2515. doi: 10.1128/AEM.03409-15
Hu, H., Wu, C., Ge, F., Ren, Y., Li, W., and Li, J. (2023). Poly-γ-glutamic acid-producing Bacillus velezensis fermentation can improve the feed properties of soybean meal. Food Biosci. 53:102503. doi: 10.1016/j.fbio.2023.102503
Hua, Y. J., Xie, F., Liu, X. Y., Liu, Y. K., Luo, Y. Y., and Ding, Y. J. (2022). Comprehensive metabolomics analysis of key taste components in different varieties of table grapes. J. Sep. Sci. 45, 3700–3713. doi: 10.1002/jssc.202200137
Huang, Z., Wang, M., Ke, W., and Guo, X. (2021). Screening of high 1, 2-propanediol production by Lactobacillus buchneri strains and their effects on fermentation characteristics and aerobic stability of whole-plant corn silage. Agriculture 11:590. doi: 10.3390/agriculture11070590
Katu, J. K., Tóth, T., and Varga, L. (2025). Enhancing the nutritional quality of low-grade poultry feed ingredients through fermentation: a review. Agriculture 15:476. doi: 10.3390/agriculture15050476
Ke, W., Zhang, H., Li, S., Xue, Y., Wang, Y., Dong, W., et al. (2022). Influence of condensed and hydrolysable tannins on the bacterial community, protein degradation, and fermentation quality of alfalfa silage. Animals 12:831. doi: 10.3390/ani12070831
Kh, A. A., Mi, S., Tian, H., Xu, X., Abdo, A. A. A., Aleryani, H., et al. (2025). Evaluation of flavor characteristics in Chinese wheat flour paste using electronic-nose, electronic-tongue, and headspace-gas chromatography-ion mobility spectrometry at different fermentation stages. J. Sci. Food Agric. 105, 2454–2465. doi: 10.1002/jsfa.14017
Khan, R. U., Naz, S., Raziq, F., Qudratullah, Q., Khan, N. A., Laudadio, V., et al. (2022). Prospects of organic acids as safe alternative to antibiotics in broiler chickens diet. Environ. Sci. Pollut. Res. 29, 32594–32604. doi: 10.1007/s11356-022-19241-8
Kiefer, P., Heinzle, E., and Wittmann, C. (2002). Influence of glucose, fructose and sucrose as carbon sources on kinetics and stoichiometry of lysine production by Corynebacterium glutamicum. J. Ind. Microbiol. Biotechnol. 28, 338–343. doi: 10.1038/sj.jim.7000252
Krawczyk, B., Wityk, P., Gałęcka, M., and Michalik, M. (2021). The many faces of Enterococcus spp.—commensal, probiotic and opportunistic pathogen. Microorganisms 9:1900. doi: 10.3390/microorganisms9091900
Lan, T., Wang, J., Yuan, Q., Lei, Y., Peng, W., Zhang, M., et al. (2022). Evaluation of the color and aroma characteristics of commercially available Chinese kiwi wines via intelligent sensory technologies and gas chromatography-mass spectrometry. Food Chem. X 15:100427. doi: 10.1016/j.fochx.2022.100427
Li, H., Deng, N., Cai, Y., Yang, J., Ouyang, F., Liu, M., et al. (2024). Dynamic changes in postmortem quality of grass carp (Ctenopharyngodon idella) muscle: from the perspectives of muscle degradation and flavor evolution. Food Chem. X 23:101751. doi: 10.1016/j.fochx.2024.101751
Lin, Y., Chen, K., Tu, D., Yu, X., Dai, Z., and Shen, Q. (2019). Characterization of dietary fiber from wheat bran (Triticum aestivum L.) and its effect on the digestion of surimi protein. LWT 102, 106–112. doi: 10.1016/j.lwt.2018.12.024
Liu, M., Tang, Y., Zhao, K., Gu, Y., Ren, D., Yao, W., et al. (2017). Recent advances in research on the community, isolation, and application of microbes in the pit mud used in manufacture of Chinese strong-flavor baijiu. Microbiol. China 44, 1222–1229. doi: 10.13344/j.microbiol.china.160559
Liu, Y., Zhang, Y., Bai, D., Li, Y., He, X., Ito, K., et al. (2023). Dietary supplementation with chlorogenic acid enhances antioxidant capacity, which promotes growth, jejunum barrier function, and cecum microbiota in broilers under high stocking density stress. Animals 13:303. doi: 10.3390/ani13020303
Lu, J., Lv, Y., Qian, X., Jiang, Y., Wu, M., Zhang, W., et al. (2020). Current advances in organic acid production from organic wastes by using microbial co-cultivation systems. Biofuels Bioprod. Biorefin. 14, 481–492. doi: 10.1002/bbb.2075
Marciňáková, M., Lauková, A., Simonová, M., Strompfová, V., Koréneková, B., and Naď, P. (2008). A new probiotic and bacteriocin-producing strain of Enterococcus faecium EF9296 and its use in grass ensiling. Czeh J. Anim. Sci. 53, 335–344. doi: 10.17221/348-CJAS
Márquez-Castillo, A., and Vidal-Quintanar, R. (2011). Improvements in the shelf life of commercial corn dry masa flour (CMF) by reducing lipid oxidation. J. Food Sci. 76, C236–C241. doi: 10.1111/j.1750-3841.2010.01983.x
Mostafa, H. S., and Hashem, M. M. (2023). Lactic acid bacteria as a tool for biovanillin production: a review. Biotechnol. Bioeng. 120, 903–916. doi: 10.1002/bit.28328
Mou, Q., Yang, H.-S., Yin, Y.-L., and Huang, P.-F. (2019). Amino acids influencing intestinal development and health of the piglets. Animals 9:302. doi: 10.3390/ani9060302
Ng, W. K., and Koh, C. B. (2017). The utilization and mode of action of organic acids in the feeds of cultured aquatic animals. Rev. Aquac. 9, 342–368. doi: 10.1111/raq.12141
Odetokun, I. A., Adetona, M. A., Ade-Yusuf, R. O., Adewoye, A. O., Ahmed, A. N., Ghali-Mohammed, I., et al. (2023). S taphylococcus aureus contamination of animal-derived foods in Nigeria: a systematic review, 2002—2022. Food Safety Risk 10:6. doi: 10.1186/s40550-023-00106-y
Okoye, C. O., Wang, Y., Gao, L., Wu, Y., Li, X., Sun, J., et al. (2023). The performance of lactic acid bacteria in silage production: a review of modern biotechnology for silage improvement. Microbiol. Res. 266:127212. doi: 10.1016/j.micres.2022.127212
Ou, Q., Peng, Y., Lin, D., Bai, C., Zhang, T., Lin, J., et al. (2017). A meta-analysis of the global prevalence rates of Staphylococcus aureus and methicillin-resistant S. aureus contamination of different raw meat products. J. Food Prot. 80, 763–774. doi: 10.4315/0362-028X.JFP-16-355
Polycarpo, G. V., Andretta, I., Kipper, M., Cruz-Polycarpo, V. C., Dadalt, J. C., Rodrigues, P. H. M., et al. (2017). Meta-analytic study of organic acids as an alternative performance-enhancing feed additive to antibiotics for broiler chickens. Poult. Sci. 96, 3645–3653. doi: 10.3382/ps/pex178
Predescu, N. C., Stefan, G., Rosu, M. P., and Papuc, C. (2024). Fermented feed in broiler diets reduces the antinutritional factors, improves productive performances and modulates gut microbiome—a review. Agriculture 14:1752. doi: 10.3390/agriculture14101752
Pujaningsih, R. I., and Mangisah, I. (2020). Physical characteristic management and palatability of processed mung bean sprout for duck. IOP Conference Series: Materials Science and Engineering (Vol. 845, No. 1, Article 012040). Bristol, UK: IOP Publishing Ltd.
Ren, H., Cai, Z., Du, C., Li, Z., Guo, X., Wang, Y., et al. (2024). Interrelated spatiotemporal variations between bacterial community and physicochemical factors in pit mud of Chinese strong-flavor baijiu. LWT 192:115630. doi: 10.1016/j.lwt.2023.115630
Roguski, M., Łozicki, A., Sońta, M., Bendowski, W., Niemiec, T., Zglińska, K., et al. (2023). Effect of using ensilaged corn wet distillers’ grains plus solubles (WDGS) as a partial replacement for concentrated feed for wet lot fed fatteners during fattening on growth performance, carcass characteristics and pork quality. Agriculture 13:2017. doi: 10.3390/agriculture13102017
Shen, X. H., Guan, J., Lu, D. P., Hong, S. C., Yu, L., and Chen, X. (2024). Peptostreptococcus Anaerobius enhances dextran sulfate sodium-induced colitis by promoting nf-κB-NLRP3-dependent macrophage pyroptosis. Virulence 15:2435391. doi: 10.1080/21505594.2024.2435391
Song, C., Zhang, T., Xu, D., Zhu, M., Mei, S., Zhou, B., et al. (2023). Impact of feeding dried distillers’ grains with solubles diet on microbiome and metabolome of ruminal and cecal contents in Guanling yellow cattle. Front. Microbiol. 14:1171563. doi: 10.3389/fmicb.2023.1171563
Soundharrajan, I., Kim, D. H., Srisesharam, S., Kuppusamy, P., Park, H. S., Yoon, Y. H., et al. (2017). Application of customised bacterial inoculants for grass haylage production and its effectiveness on nutrient composition and fermentation quality of haylage. 3 Biotech 7, 1–9. doi: 10.1007/s13205-017-0965-5
Sugiharto, S., and Ranjitkar, S. (2019). Recent advances in fermented feeds towards improved broiler chicken performance, gastrointestinal tract microecology and immune responses: a review. Anim. Nutr. 5, 1–10. doi: 10.1016/j.aninu.2018.11.001
Tolosa, J., Rodríguez-Carrasco, Y., Ruiz, M., and Vila-Donat, P. (2021). Multi-mycotoxin occurrence in feed, metabolism and carry-over to animal-derived food products: a review. Food Chem. Toxicol. 158:112661. doi: 10.1016/j.fct.2021.112661
Tu, J., Zhao, J., Liu, G., Tang, C., Han, Y., Cao, X., et al. (2020). Solid state fermentation by Fomitopsis pinicola improves physicochemical and functional properties of wheat bran and the bran-containing products. Food Chem. 328:127046. doi: 10.1016/j.foodchem.2020.127046
Tugnoli, B., Giovagnoni, G., Piva, A., and Grilli, E. (2020). From acidifiers to intestinal health enhancers: how organic acids can improve growth efficiency of pigs. Animals 10:134. doi: 10.3390/ani10010134
Wang, Z., He, Z., Beauchemin, K. A., Tang, S., Zhou, C., Han, X., et al. (2016). Evaluation of different yeast species for improving in vitro fermentation of cereal straws. Asian Australas. J. Anim. Sci. 29, 230–240. doi: 10.5713/ajas.15.0188
Wang, Y., Li, C., Ge, Q., Huo, X., Ma, T., Fang, Y., et al. (2024). Geographical characterization of wines from seven regions of China by chemical composition combined with chemometrics: quality characteristics of Chinese ‘Marselan’wines. Food Chem. X 23:101606. doi: 10.1016/j.fochx.2024.101606
Wang, S., Li, Z., Huang, D., and Luo, H. (2024). Contribution of microorganisms from pit mud to volatile flavor compound synthesis in fermented grains for nongxiangxing baijiu brewing. J. Sci. Food Agric. 104, 778–787. doi: 10.1002/jsfa.12968
Wang, L., Sagada, G., Wang, C., Liu, R., Li, Q., Zhang, C., et al. (2023). Exogenous bile acids regulate energy metabolism and improve the health condition of farmed fish. Aquaculture 562:738852. doi: 10.1016/j.aquaculture.2022.738852
Weng, X., Sui, X., Liu, Y., Yang, L., and Zhang, R. (2022). Effect of nitrogen addition on the carbon metabolism of soil microorganisms in a Calamagrostis angustifolia wetland of the Sanjiang plain, northeastern China. Ann. Microbiol. 72:18. doi: 10.1186/s13213-022-01674-8
Xu, W., Huang, Z., Zhang, X., Li, Q., Lu, Z., Shi, J., et al. (2011). Monitoring the microbial community during solid-state acetic acid fermentation of Zhenjiang aromatic vinegar. Food Microbiol. 28, 1175–1181. doi: 10.1016/j.fm.2011.03.011
Yang, X., Bao, L., Zhang, Y., Long, J., Li, Y., Wang, H., et al. (2022). Novel weight loss diet attenuates dietary-induced obesity in mice and might correlate with altered gut microbiota and metabolite profiles. Front. Nutr. 9:987955. doi: 10.3389/fnut.2022.987955
Ye, T., Cai, H., Liu, X., and Jiang, H.-L. (2016). Dominance of Oscillospira and Bacteroides in the bacterial community associated with the degradation of high-concentration dimethyl sulfide under iron-reducing condition. Ann. Microbiol. 66, 1199–1206. doi: 10.1007/s13213-016-1207-5
Ye, H., Wang, J., Shi, J., Du, J., Zhou, Y., Huang, M., et al. (2021). Automatic and intelligent technologies of solid-state fermentation process of baijiu production: applications, challenges, and prospects. Foods 10:680. doi: 10.3390/foods10030680
Yu, J., Hou, Q., Li, W., Huang, W., Mo, L., Yao, C., et al. (2020). Profiling of the viable bacterial and fungal microbiota in fermented feeds using single-molecule real-time sequencing. J. Anim. Sci. 98:skaa029. doi: 10.1093/jas/skaa029
Zhang, Q., Guo, X., Zheng, M., Chen, D., and Chen, X. (2021). Altering microbial communities: a possible way of lactic acid bacteria inoculants changing smell of silage. Anim. Feed Sci. Technol. 279:114998. doi: 10.1016/j.anifeedsci.2021.114998
Zhang, P., Liu, Y., Li, H., Hui, M., and Pan, C. (2024). Strategies and challenges of microbiota regulation in baijiu brewing. Foods 13:1954. doi: 10.3390/foods13121954
Zhang, A. R., Wei, M., Yan, L., Zhou, G. L., Li, Y., Wang, H. M., et al. (2022). Effects of feeding solid-state fermented wheat bran on growth performance and nutrient digestibility in broiler chickens. Poult. Sci. 101:101402. doi: 10.1016/j.psj.2021.101402
Keywords: Nongxiangxing Baijiu, acid-producing microbiota, multi-acid synergistic fermentation, fermented bran, feed
Citation: Pan W, Li B, Chong KH, Wang S, You L, Wang X, Nor-Khaizura MAR, Mustapha NA, Saari N and Wan Ibadullah WZ (2025) Multi-acid synergistic fermentation enhances the quality of bran feed. Front. Microbiol. 16:1646911. doi: 10.3389/fmicb.2025.1646911
Edited by:
E. Emma Tymczyszyn, National University of Quilmes, ArgentinaReviewed by:
Ayelen Amelia Hugo, CONICET, ArgentinaAna Florencia Moretti, National University of La Plata, Argentina
Copyright © 2025 Pan, Li, Chong, Wang, You, Wang, Nor-Khaizura, Mustapha, Saari and Wan Ibadullah. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wan Zunairah Wan Ibadullah, d2FuenVuYWlyYWhAdXBtLmVkdS5teQ==
 Wanshu Pan
Wanshu Pan Binbin Li4
Binbin Li4 Mahmud Ab Rashid Nor-Khaizura
Mahmud Ab Rashid Nor-Khaizura Nazamid Saari
Nazamid Saari

