- 1BSL-3 Laboratory (Guangdong), Guangdong Provincial Key Laboratory of Tropical Disease Research, Ministry of Education Key Laboratory of Infectious Diseases Research in South China, School of Public Health, Southern Medical University, Guangzhou, China
- 2School of Biomedical Engineering, Southern Medical University, Guangzhou, China
A Correction on
Identification, functional analysis, and clinical applications of defective viral genomes
by Yan, X., Pan, Y., Li, P., Zhu, L., Yu, J., Shen, C., Zhang, B., and Zhao, W. (2025). Front. Microbiol. 16:1642520. doi: 10.3389/fmicb.2025.1642520
In the published article, there was an error in positions of figures as published. Due to an oversight during the proofreading stage, Figure 2 and Figure 3 were swapped in the published version, resulting in a mismatch between the figure and their actual content. The corrected positions of figures and captions appear below.
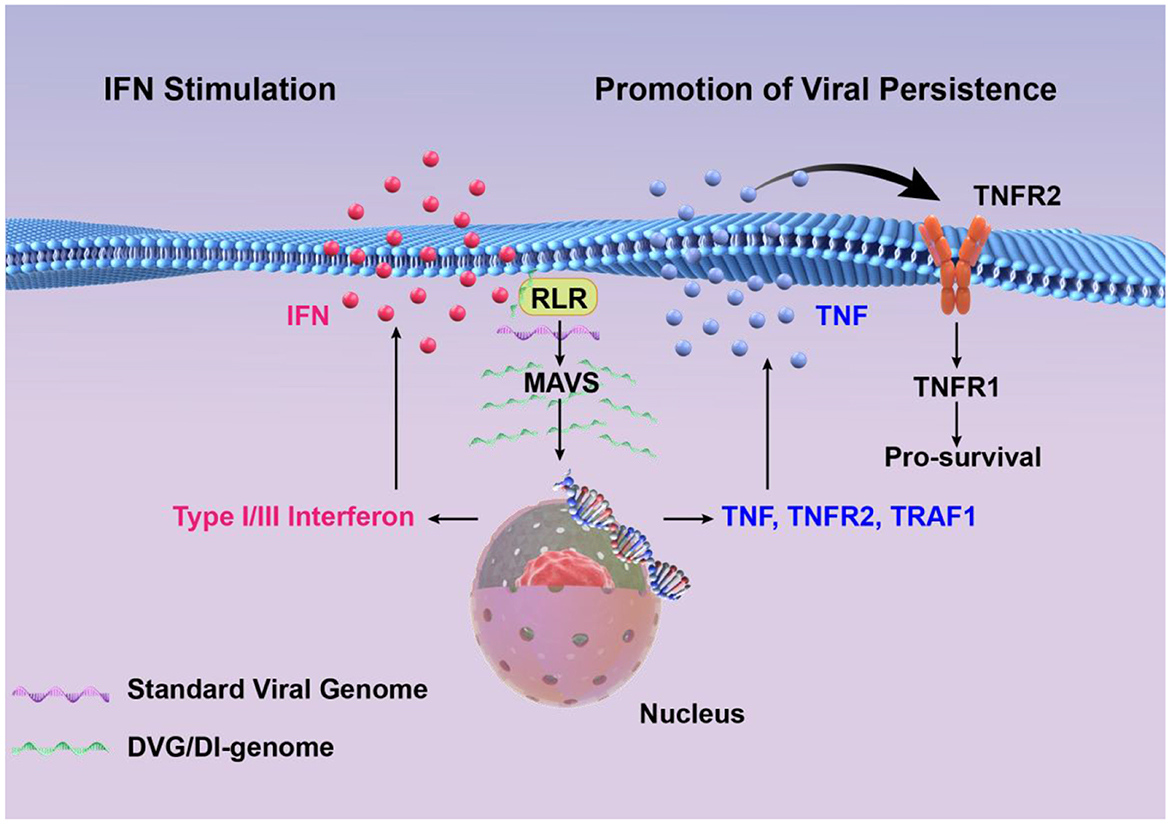
Figure 2. High concentrations of DVGs significantly enhance type I/III interferon signaling by detecting and activating RIG-I-like receptors and MAVS signaling pathways. DVGs-rich cells activate the cell survival pathway by up-regulating TNF/TNFR2/TRAF1, and combine with a small amount of standard viral genome to maintain a persistent infection state.
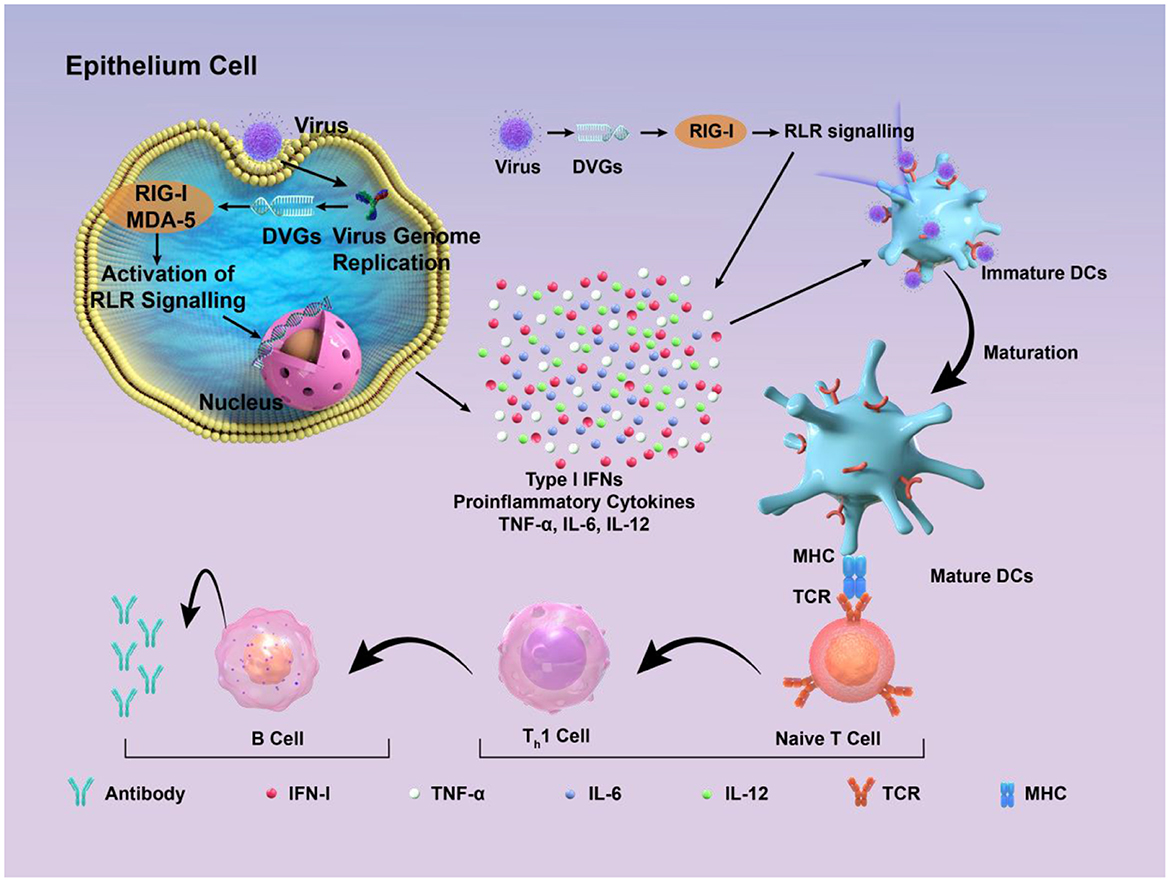
Figure 3. Schematic diagram of how DIPs activate host immune defense. DVGs are present in DIPs. Retinoic acid-inducible gene I (RIG-I) and melanoma differentiation-associated protein 5 (MDA-5) recognize copyback DVGs and then trigger the creation of interferons (IFNs) and other cytokines, which supports the activation of the innate immune response. These factors contribute to the maturation of dendritic cells, thus contributing to the adaptive immune response. Subsequently, DVGs can enhance the recognition between naive T cells and mature dendritic cells, promoting the cell-mediated immune process mediated by type I IFN signaling. This, in turn, stimulates B cells to produce corresponding antibodies, thereby exerting the function of humoral immunity.
The original article has been updated.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: interfering particle, defective viral genome, identification, function, application
Citation: Yan X, Pan Y, Li P, Zhu L, Yu J, Shen C, Zhang B and Zhao W (2025) Correction: Identification, functional analysis, and clinical applications of defective viral genomes. Front. Microbiol. 16:1670478. doi: 10.3389/fmicb.2025.1670478
Received: 21 July 2025; Accepted: 31 July 2025;
Published: 11 August 2025.
Approved by:
Frontiers Editorial Office, Frontiers Media SA, SwitzerlandCopyright © 2025 Yan, Pan, Li, Zhu, Yu, Shen, Zhang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhao, emhhb3dlaUBzbXUuZWR1LmNu; Bao Zhang, emhhbmdiQHNtdS5lZHUuY24=
†These authors have contributed equally to this work
 Xiaowei Yan1†
Xiaowei Yan1† Yitong Pan
Yitong Pan Li Zhu
Li Zhu Jianhai Yu
Jianhai Yu Chenguang Shen
Chenguang Shen Wei Zhao
Wei Zhao