- 1Department of Pathology, Microbiology and Forensic Medicine, School of Medicine, The University of Jordan, Amman, Jordan
- 2Department of Clinical Laboratories and Forensic Medicine, Jordan University Hospital, Amman, Jordan
1 Introduction
It is a well-established biological principle that exposure to antimicrobials imposes selective pressure on microbial populations (Charlebois, 2023; Hasan et al., 2021). This selective pressure favors the survival and propagation of resistant microbial sub-populations (Cantón and Morosini, 2011). The emergence of resistance under such conditions is both expected and extensively documented, supported by empirical evidence, mechanistic studies, and molecular analyses of genetic adaptation (Holmes et al., 2016; Muteeb et al., 2023). Accordingly, the phenomenon itself is not in question. Rather, it is the methodological framework through which resistance is defined and monitored that warrants critical examination, particularly in light of continued breakpoint changes.
A potentially consequential concern lies not in the biology of the organism itself, but in the interpretive frameworks through which laboratory data are classified. The widely accepted narrative of rising antimicrobial resistance (AMR) may reflect not only genuine microbial evolution, but also the cumulative impact of evolving interpretive standards—particularly revisions to minimum inhibitory concentration (MIC) breakpoints and zone diameter thresholds that determine categorical susceptibility (Hombach et al., 2012). These redefinitions, though grounded in scientific rationale, raise the possibility that some observed increases in AMR may result from shifting standards rather than true biologic changes of the tested infectious agents.
2 Breakpoint drift and its implications
In clinical microbiology, there is a foundational trust in the objectivity of diagnostic tools—the defined inhibition zone diameters, the broth microdilution assays that quantify minimum inhibitory concentrations (MICs), and the interpretive algorithms that generate categorical antimicrobial susceptibility testing (AST) results (Kowalska-Krochmal and Dudek-Wicher, 2021). Tools such as the Advanced Expert System (AES) further integrate these outputs into therapeutic recommendations (Winstanley and Courvalin, 2011). However, these tools produce raw data, not clinical meaning. Interpretation is contingent on breakpoints established by standards-setting organizations such as the Clinical and Laboratory Standards Institute (CLSI) and the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (Gaur et al., 2023). These interpretive thresholds evolve in response to emerging pharmacokinetic/pharmacodynamic (PK/PD), microbiologic, and clinical outcome data. Although such revisions are scientifically justified, they have significant implications for how AMR is defined, reported, and interpreted over time (Humphries et al., 2019).
The systematic analysis conducted by Hombach et al. (2012) provided compelling evidence of how shifts in interpretive breakpoints can significantly alter reported AMR rates—independent of any underlying biological change. The study demonstrated that applying updated CLSI and EUCAST breakpoints to a large collection of Gram-negative isolates led to substantial increases in reported resistance rates—solely due to changes in interpretive criteria (Hombach et al., 2012). Resistance classifications for several key pathogens and antimicrobial classes shifted significantly, despite no change in the underlying microbiology. These findings highlight how revised breakpoints alone can alter perceived resistance patterns, emphasizing the importance of accounting for such changes in longitudinal surveillance and trend analyses (Hombach et al., 2012).
Such changes in interpretive criteria constitute more than technical revisions—they represent fundamental shifts in the conceptual framework through which AMR is defined. For example, a bacterial isolate with an inhibition zone diameter previously categorized as susceptible under earlier guidelines may now be reported as resistant according to current standards—not because of recent adaptive change in the organism, but due to updated criteria that better reflect PK/PD evidence, patient factors, and contemporary clinical practice (Cardoso et al., 2025; Sader et al., 2023). Such reclassifications—illustrated by selected examples in Table 1—while improving the clinical accuracy of current susceptibility assessments, can nonetheless produce substantial shifts in reported AMR rates within surveillance datasets, cumulative antibiograms, and public health reports. Without explicit adjustment, these changes risk being misinterpreted as evidence of accelerated microbial evolution or dissemination, when in fact part of the observed change may arise from methodological redefinition rather than from changes in the underlying biology (GBD 2021 Antimicrobial Resistance Collaborators, 2024).
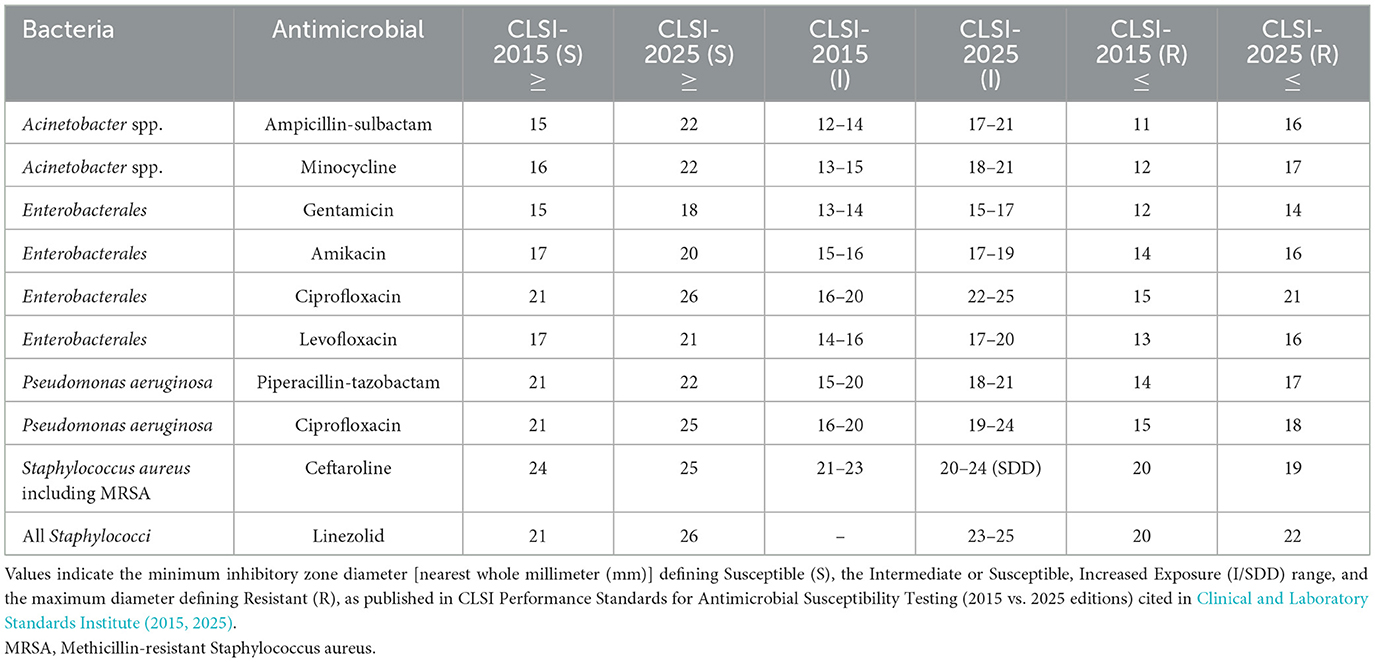
Table 1. Comparison of clinical and laboratory standards institute (CLSI) disk diffusion interpretive criteria (zone diameter, mm) for selected organism–antimicrobial combinations in 2015 and 2025.
3 Discussion, recommendations, and conclusion
Currently, there is no standardized method for harmonizing historical AST data with revised clinical breakpoints, nor is there a routine acknowledgment that increases in reported resistance may, at least in part, reflect methodological reclassification rather than true microbiological change. This creates an epistemological conflation: a biological phenomenon confounded by a shifting interpretive framework. The situation is akin to changing diagnostic thresholds in chronic disease classification—for example, redefining the blood pressure cutoff for hypertension inevitably increases disease prevalence, not because more individuals have developed pathology, but because the diagnostic criteria have shifted. Similarly, AMR may appear to rise, not necessarily due to increased pathogen resilience, but because the benchmarks used to define AMR have been made more stringent.
Failure to account for evolving interpretive criteria in the analysis of AMR trends carries direct implications for public health policy and clinical decision-making. Revisions to clinical breakpoints, even when grounded in robust PK/PD and clinical evidence, function as recalibrators of AMR rates; an effect acknowledged in some studies but not consistently adjusted for in surveillance analyses (GBD 2021 Antimicrobial Resistance Collaborators, 2024). The updated CLSI breakpoints for fluoroquinolones and cephalosporins in Enterobacterales, as well as EUCAST's redefinition of the “intermediate” category as “susceptible, increased exposure,” are emblematic of such changes (Nabal Díaz et al., 2022; Van et al., 2019). Such changes have substantial epidemiological consequences that are seldom integrated with appropriate granularity into AMR surveillance datasets.
Public discourse, meanwhile, has been saturated with urgent headlines—“Resistance is Rising,” “Superbugs on the March”—that often conflate microbial evolution with shifts in regulatory definitions (Arias and Murray, 2009; Capurro, 2020; Painuli et al., 2023). This is not a criticism of CLSI or EUCAST, whose breakpoint revisions are evidence-based and reflect scientific progress. However, for that progress to be meaningful, its evolution must be acknowledged. Unadjusted AMR data may lead to unnecessary broad-spectrum antibiotic use, misdirected investments, and flawed policy decisions. One contributing mechanism is that such data can make certain first-line agents appear less reliable for empirical therapy, prompting earlier escalation to agents such as carbapenems in place of β-lactam/β-lactamase inhibitor combinations (Lau et al., 2022). This perception, often shaped by institutional antibiograms and surveillance reports, while well-intentioned, can increase selection pressure on last-line drugs and accelerate the emergence of multidrug-resistant organisms (Zilberberg et al., 2017). Without accounting for shifting definitions, long-term AMR trends may capture changes in classification criteria rather than true microbial evolution, risking misinterpretation of resistance dynamics and potentially misleading clinical, epidemiological, and policy decisions.
What, then, are the necessary steps forward? First, AMR surveillance—whether global, national, or institutional—should routinely annotate their reports with breakpoint metadata. Just as genome browsers present annotation tracks alongside nucleotide sequences, and as widely circulated COVID-19 incidence and mortality plots annotated changes in diagnostic definitions or reporting criteria, so too should antibiograms and AMR trend analyses display the interpretive context used at the time of data acquisition, including the version and source of the breakpoints applied. Second, academic and regulatory bodies should establish a standardized nomenclature for breakpoint epochs. Resistance data labeled as “S[CLSI2012]” or “R[EUCAST2021],” for example, would clearly communicate the interpretive framework, facilitating both cross-sectional comparison and accurate longitudinal analysis. Third, when historical bacterial isolates are available, they should be retested using contemporary breakpoints to enable true retrospective trend evaluation. Although labor-intensive, this is feasible in reference laboratories and academic centers with bio-banked microbial strains. In cases where isolates are no longer available, modeling approaches should be employed to retrospectively adjust historical data based on known MIC distributions and classification changes. Fourth, modeling consortia—such as those behind the Global Burden of Disease (GBD) project—should incorporate breakpoint harmonization protocols into their methodologies. Adjustment factors or, at minimum, sensitivity analyses are essential to account for interpretive changes across time.
In the discourse surrounding AMR, a fundamental question must be asked: does our current narrative reflect true microbial evolution, or the cumulative effect of evolving diagnostic conventions? Increases in AMR prevalence may reflect not only bacterial adaptation but also redefinition through processes such as breakpoint drift; neglecting the latter risks incomplete or biased interpretation of surveillance data. The truth, as always, lies at the intersection of both. Microorganisms evolve, undoubtedly—but so too does the diagnostic lens through which we measure that evolution. Recognizing the profound implications of breakpoint recalibration and the confounding effects of breakpoint drift should not diminish the urgency of addressing AMR; rather, it should strengthen the foundation upon which our understanding rests. To address AMR effectively as a major medical challenge of the 21st century, we must ensure that our measurement tools keep pace with both microbial evolution and advances in diagnostic methodology, so that changes in reported resistance reflect true shifts in biology rather than artifacts of classification.
Author contributions
MS: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author declares that no financial support was received for the research and/or publication of this article.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author declares that Gen AI was used in the creation of this manuscript. Malik Sallam used OpenAI's ChatGPT-4o to assist in refining the language and improving the clarity of the manuscript. All scientific content, interpretations, and conclusions are the author's own.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arias, C. A., and Murray, B. E. (2009). Antibiotic-resistant bugs in the 21st century–a clinical super-challenge. N. Engl. J. Med. 360, 439–443. doi: 10.1056/NEJMp0804651
Cantón, R., and Morosini, M. I. (2011). Emergence and spread of antibiotic resistance following exposure to antibiotics. FEMS Microbiol. Rev. 35, 977–991. doi: 10.1111/j.1574-6976.2011.00295.x
Capurro, G. (2020). “Superbugs” in the risk society: assessing the reflexive function of North American newspaper coverage of antimicrobial resistance. SAGE Open 10:2158244020901800. doi: 10.1177/2158244020901800
Cardoso, A. M., Flores, V. R., do Rosario, G. G., Succar, J. B., Berbert, L. C., Oliveira, M. C. F., et al. (2025). Antimicrobial susceptibility of escherichia coli isolates causing community-acquired urinary tract infections: comparison of methods. Microorganisms 13:231. doi: 10.3390/microorganisms13020231
Charlebois, D. A. (2023). Quantitative systems-based prediction of antimicrobial resistance evolution. NPJ Syst. Biol. Appl. 9:40. doi: 10.1038/s41540-023-00304-6
Clinical and Laboratory Standards Institute (2015). M100-S25 Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI.
Clinical and Laboratory Standards Institute (2025). CLSI M100 Performance Standards for Antimicrobial Susceptibility Testing. Wayne, PA: CLSI.
Gaur, P., Hada, V., Rath, R. S., Mohanty, A., Singh, P., and Rukadikar, A. (2023). Interpretation of antimicrobial susceptibility testing using european committee on antimicrobial susceptibility testing (EUCAST) and clinical and laboratory standards institute (CLSI) breakpoints: analysis of agreement. Cureus 15:e36977. doi: 10.7759/cureus.36977
GBD 2021 Antimicrobial Resistance Collaborators (2024). Global burden of bacterial antimicrobial resistance 1990-2021: a systematic analysis with forecasts to 2050. Lancet 404, 1199–1226. doi: 10.1016/S0140-6736(24)01867-1
Hasan, C. M., Dutta, D., and Nguyen, A. N. T. (2021). Revisiting antibiotic resistance: mechanistic foundations to evolutionary outlook. Antibiotics 11:40. doi: 10.3390/antibiotics11010040
Holmes, A. H., Moore, L. S., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Hombach, M., Bloemberg, G. V., and Böttger, E. C. (2012). Effects of clinical breakpoint changes in CLSI guidelines 2010/2011 and EUCAST guidelines 2011 on antibiotic susceptibility test reporting of Gram-negative bacilli. J. Antimicrob. Chemother. 67, 622–632. doi: 10.1093/jac/dkr524
Humphries, R. M., Abbott, A. N., and Hindler, J. A. (2019). Understanding and addressing CLSI breakpoint revisions: a primer for clinical laboratories. J. Clin. Microbiol. 57:e00203–19. doi: 10.1128/JCM.00203-19
Kowalska-Krochmal, B., and Dudek-Wicher, R. (2021). The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens 10:165. doi: 10.3390/pathogens10020165
Lau, C. L., Periyasamy, P., Saud, M. N., Robert, S. A., Gan, L. Y., Chin, S. Y., et al. (2022). Plethora of antibiotics usage and evaluation of carbapenem prescribing pattern in intensive care units: a single-center experience of malaysian academic hospital. Antibiotics 11:1172. doi: 10.3390/antibiotics11091172
Muteeb, G., Rehman, M. T., Shahwan, M., and Aatif, M. (2023). Origin of antibiotics and antibiotic resistance, and their impacts on drug development: a narrative review. Pharmaceuticals 16:1615. doi: 10.3390/ph16111615
Nabal Díaz, S. G., Algara Robles, O., and García-Lechuz Moya, J. M. (2022). New definitions of susceptibility categories EUCAST 2019: clinic application. Rev. Esp. Quimioter. 35, 84–88. doi: 10.37201/req/s03.18.2022
Painuli, S., Semwal, P., Sharma, R., and Akash, S. (2023). Superbugs or multidrug resistant microbes: a new threat to the society. Health Sci. Rep. 6:e1480. doi: 10.1002/hsr2.1480
Sader, H. S., Mendes, R. E., Kimbrough, J. H., Kantro, V., and Castanheira, M. (2023). Impact of the recent clinical and laboratory standards institute breakpoint changes on the antimicrobial spectrum of aminoglycosides and the activity of plazomicin against multidrug-resistant and carbapenem-resistant enterobacterales from united states medical centers. Open Forum Infect. Dis. 10:ofad058. doi: 10.1093/ofid/ofad058
Van, T. T., Minejima, E., Chiu, C. A., and Butler-Wu, S. M. (2019). Don't get wound up: revised fluoroquinolone breakpoints for enterobacteriaceae and pseudomonas aeruginosa. J. Clin. Microbiol. 57:e02072–18. doi: 10.1128/JCM.02072-18
Winstanley, T., and Courvalin, P. (2011). Expert systems in clinical microbiology. Clin. Microbiol. Rev. 24, 515–556. doi: 10.1128/CMR.00061-10
Zilberberg, M. D., Nathanson, B. H., Sulham, K., Fan, W., and Shorr, A. F. (2017). Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect. Dis. 17:279. doi: 10.1186/s12879-017-2383-z
Keywords: guideline adherence, microbial sensitivity tests, drug resistance, AMR (antimicrobial resistance), AST (antibiotic susceptibility testing)
Citation: Sallam M (2025) Breakpoint drift: a hidden confounder in antimicrobial resistance surveillance? Front. Microbiol. 16:1674968. doi: 10.3389/fmicb.2025.1674968
Received: 28 July 2025; Accepted: 18 August 2025;
Published: 29 August 2025.
Edited by:
Taru Singh, Amity University, IndiaReviewed by:
Matthew J. Shepherd, The University of Manchester, United KingdomCopyright © 2025 Sallam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Malik Sallam, bWFsaWsuc2FsbGFtQGp1LmVkdS5qbw==
 Malik Sallam
Malik Sallam