- Clinical Laboratory, Ganzhou Cancer Hospital, Ganzhou, Jiangxi, China
Background: Klebsiella pneumoniae (K. pneumoniae) is a major opportunistic pathogen associated with nosocomial infections, particularly in immunocompromised patients, including those with malignant cancer. The molecular epidemiology of K. pneumoniae isolated from distinct body sites from cancer patients remains poorly understood. This study aims to investigate the resistance genes, virulence factors, and sequence types of K. pneumoniae strains isolated from cancer patients to provide insights into their epidemiological and clinical characteristics.
Methods: A total of 105 K. pneumoniae isolates from malignant cancer patients were subjected to whole-genome sequencing. Resistance genes, virulence- associated genes, and multilocus sequence typing (MLST) were analyzed. The distribution of these genetic determinants was compared among isolates from distinct body sites from cancer patients. Whole-genome sequencing was performed on 105 K. pneumoniae isolates obtained from the blood and body fluids of patients with various cancers, including lung (n = 24), nasopharyngeal (n = 12), liver (n = 11), cervical (n = 8), and other cancer types (n = 50). The isolates were characterized in terms of antimicrobial resistance genes, virulence-associated genes, and MLST. Additionally, virulence was evaluated using a scoring system incorporating the virulence genes ybt, clb, and iuc.
Results: Among the isolates, 41 exhibited resistance to trimethoprim-sulfamethoxazole, predominantly harboring sul1, sul2, and dfrA genes. Resistance to tobramycin and levofloxacin was mainly associated with aadA16, aph(3″)-Ib, and AAC(6′)-Ib-cr genes. Eight isolates were identified as carbapenem-resistant K. pneumoniae (CRKP), carrying resistance genes bla_kpc-1, bla_ndm-5, and bla_oxa-10. Virulence genes included iron siderophores (entAB, ybt, iucABC), fimbriae (fimA, fimH), and OmpA (100%). Notably, isolates from cervical cancer patient samples had the lowest virulence scores, whereas those from lung and nasopharyngeal cancer patient samples demonstrated the highest virulence scores. MLST revealed 45 sequence types, with ST23 predominating in isolates from lung and nasopharyngeal cancer patient, while ST45 was the most frequent in cervical cancer patient isolates. Phylogenetic analysis demonstrated clustering of Klebsiella pneumoniae isolates from lung, nasopharyngeal, liver, and cervical cancer patient samples, with these isolates predominantly located within the third clade, accounting for 58.3, 66.7, 80, and 87.5%, respectively.
Conclusion: The K. pneumoniae isolates in this study demonstrate considerable diversity in their virulence genes, antimicrobial resistance genes, and sequence types. The findings highlight the importance of genomic surveillance to guide infection control and therapeutic strategies, particularly in high-risk oncology settings.
1 Introduction
Klebsiella pneumoniae is a significant pathogen responsible for severe infections in cancer patients, particularly those with compromised immune systems due to chemotherapy or immunosuppressive therapy (Chen et al., 2024; Rolston, 2017). Infections caused by K. pneumoniae in this vulnerable population often lead to increased morbidity and mortality, with higher risks of severe complications compared to non-cancer patients (Nanayakkara et al., 2021). Studies have shown that cancer patients are more susceptible to K. pneumoniae infections, including bloodstream infections, respiratory tract infections, and urinary tract infections, which are often difficult to treat due to the emergence of multidrug-resistant strains (Navon-Venezia et al., 2017). The widespread use of antibiotics in cancer patients for infection prevention and treatment has accelerated the development of antibiotic resistance in K. pneumoniae. Multidrug-resistant Klebsiella pneumoniae (MDR-KP) and CRKP have emerged as major global public health threats. MDR-KP employs a range of mechanisms to confer resistance to commonly used antibiotics, including β-lactams, aminoglycosides, fluoroquinolones, and even agents of last resort such as polymyxins and tigecycline (Li et al., 2023). The resistance mechanisms are complex and involve the production of inactivating enzymes (e.g., Extended-Spectrum Beta-Lactamases (ESBLs), AmpC, and carbapenemases), loss or mutation of outer membrane porins (e.g., OmpK35/36) that reduce antibiotic uptake, overexpression of efflux pumps (e.g., AcrAB-TolC, OqxAB), modifications of antibiotic target sites, and the formation of protective barriers like biofilms (Karampatakis et al., 2023). Carbapenem resistance is chiefly mediated by carbapenemases, including bla_kpc, bla_ndm, bla_vim, bla_imp, and bla_oxa-48 types, whose prevalence exhibits substantial geographic variation. For instance, in China, bla_kpc-2 is the dominant carbapenemase gene in CRKP and is significantly more prevalent than other carbapenemase genes, such as bla_ndm and bla_oxa-48 (Liao et al., 2022; Ge et al., 2024; Li et al., 2022). In contrast, bla_ndm-type metallo-β-lactamases are highly endemic in regions like South Asia, while bla_oxa-48 and its variants predominate in Europe and North Africa, often linked to high-risk clones including ST101 and ST405 (Budia-Silva et al., 2024). Infections caused by CRKP are associated with high mortality, increased treatment failure, and elevated healthcare costs. Effective therapeutic options are scarce, especially for critically ill or immunocompromised patients. Treatment often depends on novel β-lactam/β-lactamase inhibitor combinations (e.g., ceftazidime-avibactam) or polymyxin-based regimens. Nonetheless, the ongoing evolution of resistance poses a persistent challenge (Karampatakis et al., 2023). Beyond carbapenemase production, additional mechanisms—such as efflux pump overexpression, porin loss, and biofilm formation—collectively promote carbapenem resistance, further constraining clinical treatment options (Ma et al., 2023). The high prevalence and increasing multidrug resistance of K. pneumoniae in clinical oncology practice are of considerable concern (Nanayakkara et al., 2021). A study by Amanati et al. (2021) reported that K. pneumoniae accounted for 14.5% of Gram-negative bacteria isolated from bloodstream infections in cancer patients; among these isolates, 60.7% exhibited an ESBLs-positive phenotype and 13.2% were CRKP, underscoring a substantial antimicrobial resistance burden in this patient population. Infections caused by hypervirulent K. pneumoniae (hvKp) are also alarming. One documented case described a postoperative rectal cancer patient who succumbed to liver infarction and septic shock following hvKp infection, highlighting the potential for rare yet fatal outcomes in immunocompromised hosts (Li et al., 2024). Additionally, strains harboring the pks genomic island—which encodes the genotoxin colibactin—have been reported in cancer patients. According to Wang et al. (2025), pks-positive strains accounted for 12.54% of isolates from Chinese cancer patients, with many belonging to hypervirulent clones such as ST23 and K1, suggesting a possible role for genotoxicity in both infection progression and oncogenesis. Oncology and intensive care units are recognized high-risk environments for the transmission of K. pneumoniae. Notably, CRKP has been detected in up to 59% of pediatric cancer patients in some studies, with evidence of clonal dissemination of strains carrying carbapenemase genes such as bla_oxa-48 and bla_ndm-1 (Osama et al., 2021). Among patients with hematological malignancies, ESBLs-producing strains are predominantly of the bla_ctx-m genotype (91%) and frequently display a multidrug-resistant phenotype (Lubwama et al., 2024).
In addition to antibiotic resistance, the virulence of K. pneumoniae plays a crucial role in its pathogenicity in cancer patients. K. pneumoniae possesses various virulence factors, including fimbriae (e. g., fimA and fimH) that enhance adhesion to host cells, and siderophores (e. g., entA and entB) that facilitate iron acquisition under host immune pressure (Choby et al., 2020). These virulence factors allow K. pneumoniae to effectively colonize, invade, and survive in the host, leading to severe and persistent infections (Mendes et al., 2023). Cancer patients, due to their immunocompromised state, are particularly vulnerable to the effects of these virulence mechanisms (Mishra et al., 2024).
While there has been extensive research on K. pneumoniae in general populations, studies focusing on its genomic and molecular characteristics in cancer patients remain limited. Given the unique immune status and treatment regimens of cancer patients, K. pneumoniae strains isolated from this group might harbor specific strain characteristics worthy of investigation (Ntim et al., 2025; Touati et al., 2020). Understanding these features is essential for guiding targeted treatment strategies and developing effective infection control measures. This study aims to investigate the genomic characteristics of K. pneumoniae isolated from cancer patients, focusing on antibiotic resistance genes and virulence determinants. By utilizing whole-genome sequencing, we seek to identify the genetic basis underlying the resistance and virulence of K. pneumoniae in this population, providing insights into its molecular epidemiology. The findings will contribute to a better understanding of the pathogen’s behavior in immunocompromised hosts and support the development of precision medicine approaches for managing K. pneumoniae infections in cancer patients.
2 Methods and materials
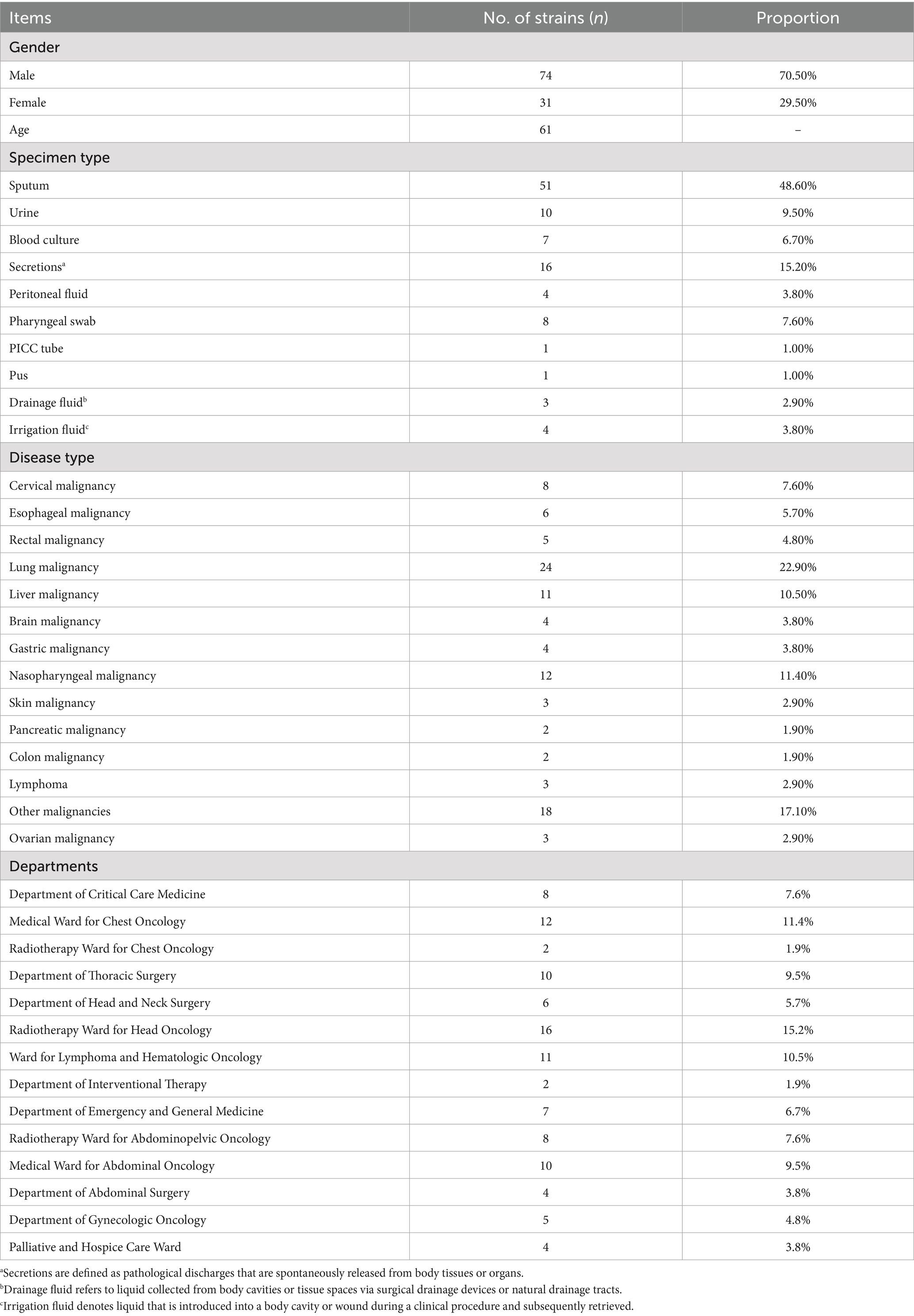
2.1 Bacterial isolation and identification
From October 2023 to September 2024, a total of 105 K. pneumoniae clinical isolates were obtained from the Department of Clinical Laboratory, Ganzhou Cancer Hospital. These isolates were derived from clinical specimens collected from 105 patients with various malignancies. The specimen types included sputum (48.6%), secretions (15.2%), urine (9.5%), blood cultures (6.7%), throat swabs (7.6%), and others (detailed in Supplementary material 1). The K. pneumoniae isolates incorporated in this study were selected based on the following criteria: (1) obtained from patients diagnosed with primary malignant tumors; (2) represented the first obtained isolate from each individual patient; (3) Serum inflammatory markers exceeding the following thresholds: C-reactive protein (CRP) > 10 μg/mL or procalcitonin (PCT) > 0.5 ng/mL. The isolates were identified as K. pneumoniae using matrix-assisted laser desorption ionization-time of flight mass spectrometry MALDI-TOF/MS (America, BEXS2600). Susceptibility testing was conducted using the AST-N334 card on the VITEK 2 Compact automated system (bioMérieux, France). This card tests the following antibiotics across the indicated concentration ranges: Amikacin (2–64 μg/mL), Amoxicillin/Clavulanic Acid (2/1–32/16 μg/mL), Cefepime (0.12–32 μg/mL), Cefoperazone/Sulbactam (8–64 μg/mL), Cefoxitin (4–64 μg/mL), Ceftazidime (0.12–64 μg/mL), Ceftriaxone (0.25–64 μg/mL), Cefuroxime (1–64 μg/mL), Ertapenem (0.12–8 μg/mL), Imipenem (0.25–16 μg/mL), Levofloxacin (0.12–8 μg/mL), Piperacillin/Tazobactam (4/4–128/4 μg/mL), Tigecycline (0.5–8 μg/mL), and Trimethoprim/Sulfamethoxazole (1/19–16/304 μg/mL). Furthermore, the Kirby-Bauer disk diffusion method was also performed with disks containing: Cefotaxime (30 μg), Ampicillin (10 μg), Ampicillin/Sulbactam (10/10 μg), Tobramycin (10 μg), Aztreonam (30 μg), and Cefazolin (30 μg). All isolates were cultured on blood agar plates, and antibiotic susceptibility tests were performed according to the Clinical and Laboratory Standards Institute 2024 guidelines. All procedures were carried out and results were interpreted based on the standards provided by Clinical and Laboratory Standards Institute (2024). Escherichia coli ATCC25922 was used as the quality control strain.
2.2 String test
A string test was performed to identify hypermucoviscous phenotypes. Bacterial colonies were grown overnight on blood agar plates at 37 °C. A loop was used to stretch a colony, and strains were considered positive for hypermucoviscosity if a viscous string >5 mm in length was observed (Kumabe and Kenzaka, 2014).
2.3 Whole-genome sequencing
The whole-genome sequencing of the K. pneumoniae isolates was performed to identify the distribution of antibiotic resistance genes and virulence factors. Colonies were inoculated into LB liquid medium (components: Tryptone (Oxoid, UK; manufactured in France), Yeast Extract (Oxoid, UK; manufactured in the United Kingdom), and Sodium Chloride [Macklin, China]). and incubated at 37 °C for 16 h, followed by centrifugation at 8,000 rpm for 10 min to collect bacterial pellets. Total DNA was extracted using a commercial kit (Beyotime, D0063) according to the manufacturer’s protocol. DNA purity was assessed with a NanoPhotometer® spectrophotometer (IMPLEN, USA), while concentration was quantified using a Qubit® 3.0 Fluorometer (Life Technologies, USA). DNA integrity and contamination were evaluated via 1% agarose gel electrophoresis (120 V, 45 min). Qualified samples (0.5 μg gDNA per reaction) underwent library preparation with the Annoroad® Universal DNA Fragmentase Kit v2.0 and Library Prep Kit v2.0 (AN200101-L) following standardized protocols. Cluster generation was performed on cBot using HiSeq PE Cluster Kit v4-cBot-HS (Illumina), and paired-end sequencing (150 bp) was conducted on the HiSeq platform with HiSeq X Ten Reagent Kit v2.5. Base calling was executed via WriteFQ software to generate raw sequencing reads (Sequenced Reads). Virulence factor genes were predicted by BLASTP analysis of the annotated protein sequences against the core dataset of the Virulence Factors Database (VFDB). Antimicrobial resistance genes were annotated using the Resistance Gene Identifier (RGI) tool against the Comprehensive Antibiotic Resistance Database (CARD), with a minimum identity cutoff of 80%. Sequence types were determined using the web-based PubMLST typing tool.1 Phylogenetic trees were constructed using the maximum likelihood method in MEGA11 software, based on multiple sequence alignments.
2.4 Virulence scoring
Virulence scoring was performed according to the system established by Lam et al. (2021), which assigns a quantitative virulence score based on the detection of three key virulence determinants: yersiniabactin (ybt), colibactin (clb), and aerobactin (iuc). The scoring system is designed to reflect increasing clinical risk associated with specific genetic profiles: a score of 0 indicates absence of all three genes; 1 indicates presence of ybt only (a siderophore linked to immune evasion but considered moderately virulent alone); 2 indicates presence of clb without iuc (irrespective of ybt, reflecting the genotoxic potential of clb); 3 indicates presence of iuc only (a hallmark of virulence plasmids associated with severe infections such as sepsis); 4 indicates coexistence of iuc and ybt in the absence of clb; and 5 indicates the presence of all three genes, representing the highest-risk profile. This hierarchical framework integrates established clinical and molecular insights into Klebsiella pneumoniae pathogenicity, translating complex genomic data into an interpretable and reproducible numerical score that facilitates consistent cross-study comparisons and enhances evaluative clarity.
2.5 Statistical analysis
Data visualization and statistical analyses were performed using GraphPad Prism 8.0. Quantitative data were expressed as mean ± standard deviation. Categorical data were presented as percentages (%), and comparisons were made using the Chi-square test or Fisher’s exact test. A p-value of <0.05 was considered statistically significant.
3 Results
3.1 Clinical characteristics of K. pneumoniae isolates
A total of 105 non-duplicate K. pneumoniae clinical isolates were obtained from patient specimens. Detailed clinical information is presented in Table 1. The majority of isolates were collected from patients with lung malignancies (22.9%, n = 24), followed by nasopharyngeal malignancies (11.4%, n = 12), liver malignancies (10.5%, n = 11), and cervical malignancies (7.6%, n = 8). The majority of isolates (85.7%, 90/105) were collected from specialized medical and surgical oncology wards, while a smaller proportion originated from the intensive care unit (7.6%, 8/105) and radiotherapy wards (9.5%, 10/105). The distribution of specimen types varied by malignancy type (Table 2). For lung malignancy patients, sputum was the predominant specimen type (83.3%). Similarly, sputum accounted for the majority of samples from nasopharyngeal malignancy patients (75.0%). In contrast, isolates from liver malignancy patients were primarily obtained from drainage fluid (27.3%), while cervical malignancy patients had a high proportion of isolates from secretions (62.5%).
3.2 Antibiotic susceptibility of K. pneumoniae isolates
The antibiotic susceptibility of 105 K. pneumoniae isolates to 20 antibiotics was determined using broth microdilution (Figure 1A). Resistance was observed for all tested antibiotics. The overall resistance rates for key antibiotics were as follows: ceftriaxone (39.0%, n = 41), ceftazidime (22.9% n = 23), trimethoprim-sulfamethoxazole (39.0%, n = 41), cefazolin (43.8%, n = 46), aztreonam (28.6%, n = 30), ampicillin (100%, n = 105), ampicillin-sulbactam (24.8%, n = 26), tobramycin (28.6%, n = 30), cefotaxime (41.0%, n = 43), cefepime (25.7%, n = 27), cefuroxime (38.1%, n = 40), levofloxacin (26.7%, n = 28), and piperacillin-tazobactam (20.0%, n = 21).
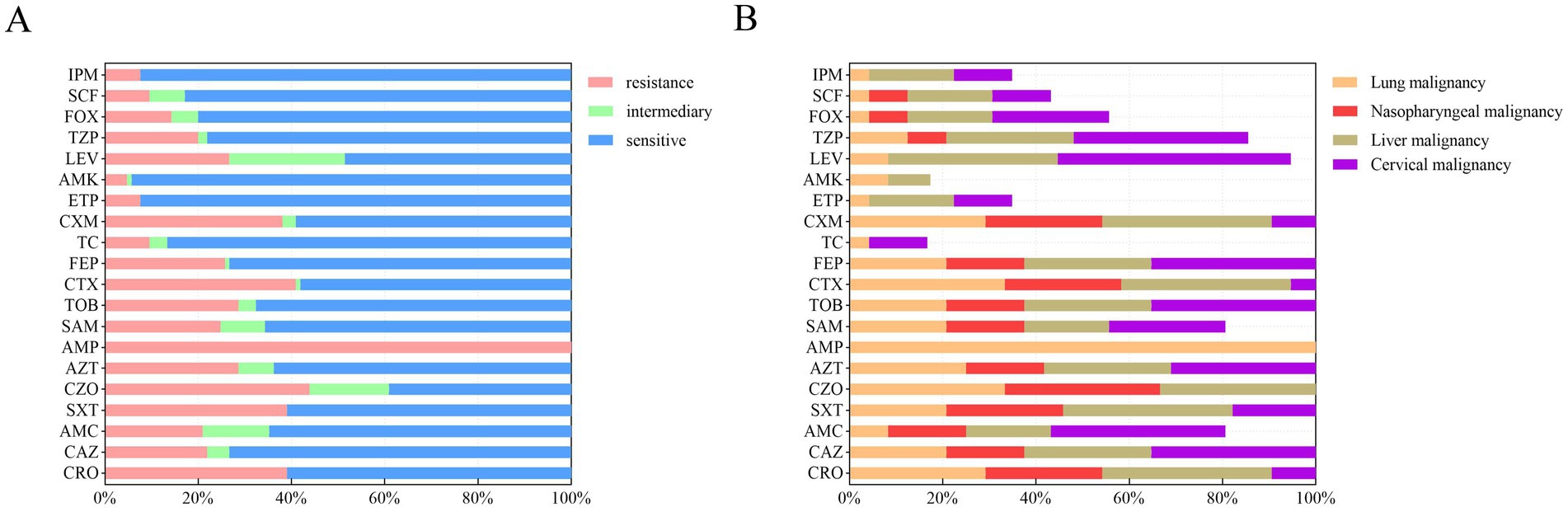
Figure 1. Antibiotic susceptibility of 105 K. pneumoniae isolates from patients with malignant tumors. (A) Overall resistance rates of all isolates to 20 antibiotics, presented as percentages. (B) Resistance rates of K. pneumoniae isolates stratified by four major cancer types: lung (N = 24), nasopharyngeal (N = 12), liver (N = 11), and cervical (N = 8) malignancies. Rates reflect the proportion of resistant isolates within each cancer group. IPM, imipenem; SCF, cefoperazone/sulbactam; FOX, cefoxitin; TZP, piperacillin/tazobactam; LEV, levofloxacin; AMK, amikacin; ETP, ertapenem; CXM, cefuroxime; TC, tigecycline; FEP, cefepime; CTX, cefotaxime; TOB, tobramycin; SAM, ampicillin/sulbactam; AMP, ampicillin; AZT, aztreonam; CZO, cefazolin; SXT, trimethoprim-sulfamethoxazole; AMC, amoxicillin/clavulanic acid; CAZ, ceftazidime; CRO, ceftriaxone.
Resistance patterns varied among isolates from patients with different cancer types (Figure 1B). Lung cancer patient samples: High resistance rates were observed for cefazolin (33%, n = 8), cefotaxime (33%, n = 8), and aztreonam (25%, n = 6). Nasopharyngeal cancer patient samples: Isolates exhibited notable resistance to cefazolin (33.3%, n = 4), trimethoprim- sulfamethoxazole (25%, n = 3), and aztreonam (16.7%, n = 2). Liver cancer patient samples: Resistance was highest for trimethoprim-sulfamethoxazole (36.4%, n = 4), cefotaxime (36.4%, n = 4), and levofloxacin (36.4%, n = 4). Cervical cancer patient samples: Isolates showed high resistance to cefotaxime (75%, n = 8), levofloxacin (50%, n = 4), and aztreonam (50%, n = 4). Across all malignancy types, resistance rates to cephalosporins were consistently >30%. Notably, isolates from liver and cervical malignancy patients exhibited ≥40% resistance to levofloxacin.
3.3 Whole-genome sequencing results
The quality of the raw sequencing data was assessed using FastQC. Adapters and low-quality bases were trimmed with Trimmomatic to ensure high-quality data for downstream analysis.
3.3.1 Antibiotic resistance genes
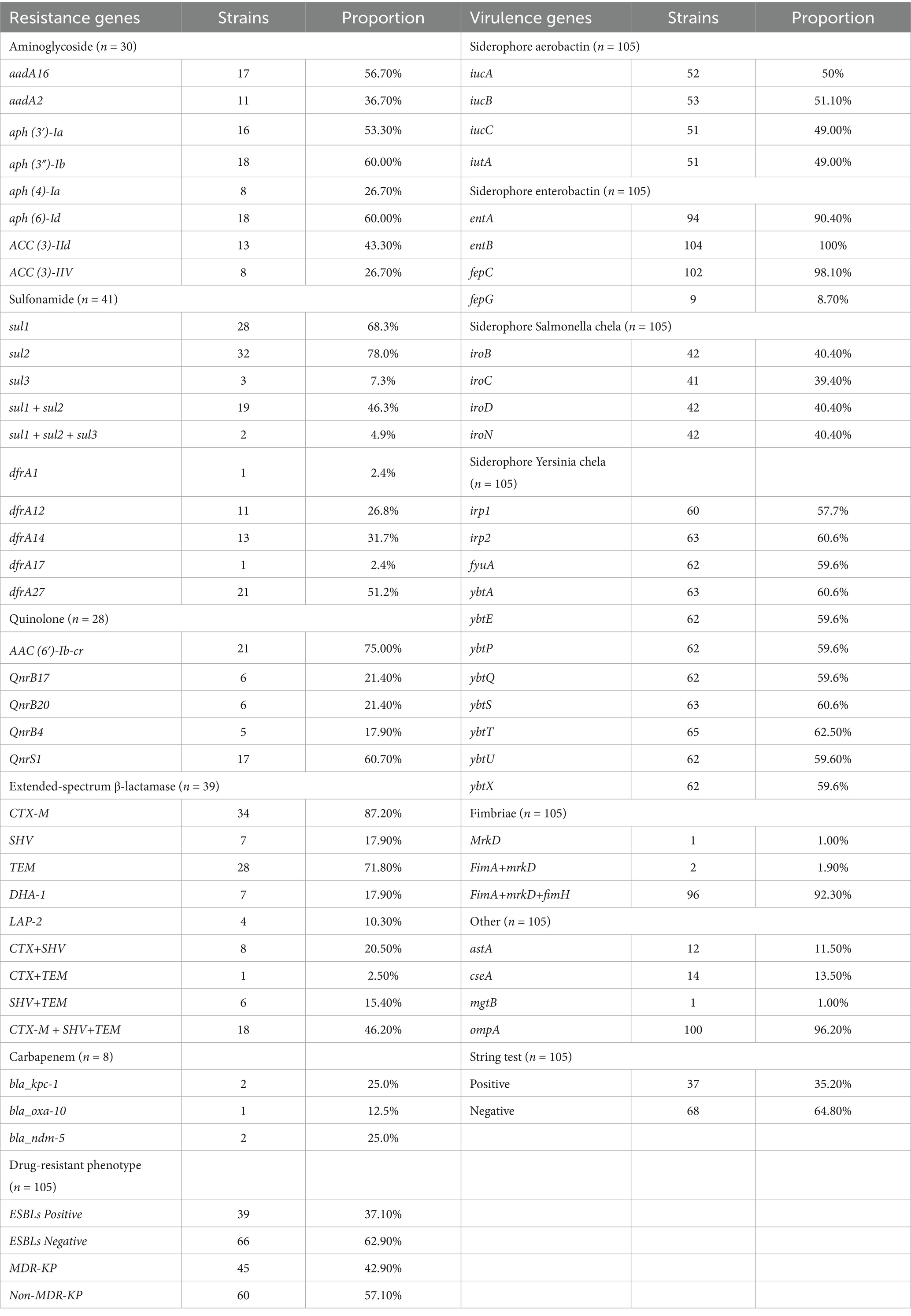
Whole-genome sequencing was performed on 105 K. pneumoniae isolates, and the distribution of antibiotic resistance genes is summarized in Table 3. Among the 41 trimethoprim-sulfamethoxazole-resistant isolates, the following resistance genes were identified: sul1 (70%), sul2 (80%), sul3 (7.5%), dfrA1 (2.5%), dfrA12 (27.5%), dfrA14 (32.5%), dfrA17 (2.5%), and dfrA27 (52.5%). Co-occurrence of resistance genes was noted in 19 isolates harboring sul1 + sul2 (48%) and 2 isolates carrying sul1 + sul2 + sul3 (5%).
Among the 30 isolates resistant to tobramycin, the predominant genes included aadA16 (56.7%), aadA2 (36.7%), aph(3′)-Ia (53.3%), aph(3″)-Ib (60%), aph(6)-Id (60%), aph(4)-Ia (26.7%), ACC(3)-IId (43.3%), and ACC(3)-IIV (26.7%). In the 28 isolates resistant to levofloxacin, the primary resistance genes identified were AAC(6′)-Ib-cr (75%), QnrS1 (60.7%), QnrB17 (21.4%), QnrB20 (21.4%), and QnrB4 (17.9%). CRKP was observed in 8 isolates, with harboring bla_kpc-1 (40%), bla_ndm-5 (40%), and bla_oxa-10 (20%). Among 39 extended-spectrum beta-lactamase (ESBLs)-producing isolates, the most common genes were CTX-M (87.2%), SHV (94.9%), TEM (71.8%), DHA-1 (17.9%), and LAP-2 (10.3%), with co-occurrence patterns including CTX-M + SHV (8 isolates), CTX-M + TEM (1 isolate), SHV + TEM (6 isolates), and CTX-M + SHV + TEM (18 isolates). Efflux pump genes were frequently detected, including acrB (100%), acrD (97.1%), acrA (100%), oqxB (97.1%), oqxA (99.0%), marA (100%), and ramA (100%). A detailed analysis showed that 40 isolates carried a single resistance gene, 25 carried four, and 23 carried five, with more than three resistance genes detected in 53.8% of isolates (Figure 2A).
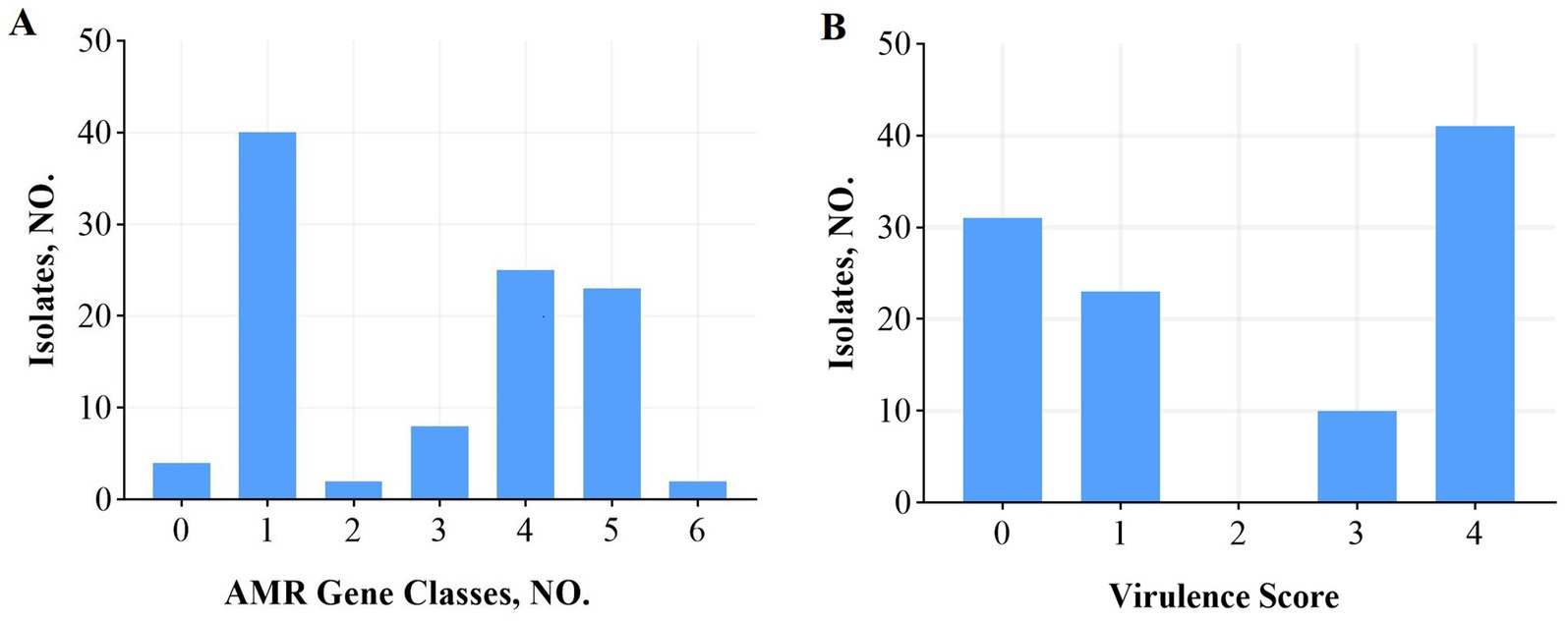
Figure 2. AMR gene categories and virulence scores among 105 K. pneumoniae isolates. (A) Number of antibiotic resistance gene categories per isolate. (B) Virulence scores (0–5) assigned using the Lam system based on the presence of yersiniabactin (ybt), colibactin (clb), and aerobactin (iuc) genes. A score ≥ 3 indicates high virulence potential. Nearly half of all isolates (48%) scored ≥3. The proportion of highly virulent isolates (score ≥ 3) varied across malignancy types. AMR, antimicrobial resistance.
The distribution of resistance genes varied across isolates from patients with different cancer types, as shown in Table 4. For quinolone resistance, isolates from lung, nasopharyngeal, liver, and cervical cancer patient samples predominantly harbored AAC(6′)-Ib-cr and QnrS1, with the highest prevalence of AAC(6′)-Ib-cr observed in cervical cancer patient samples. For aminoglycoside resistance, APH(3″)-Ib and APH(6)-Id were dominant, with cervical cancer patient samples isolates showing the highest frequency of aminoglycoside resistance genes (≥25%). For sulfonamide resistance, sul1 and sul2 were predominant, with nasopharyngeal cancer patient samples isolates most frequently carrying dfrA14 and sul2 (25%). For ESBLs genes, TEM-1 was most prevalent, with cervical cancer patient samples isolates showing the highest proportion of TEM-1 (62.5%).
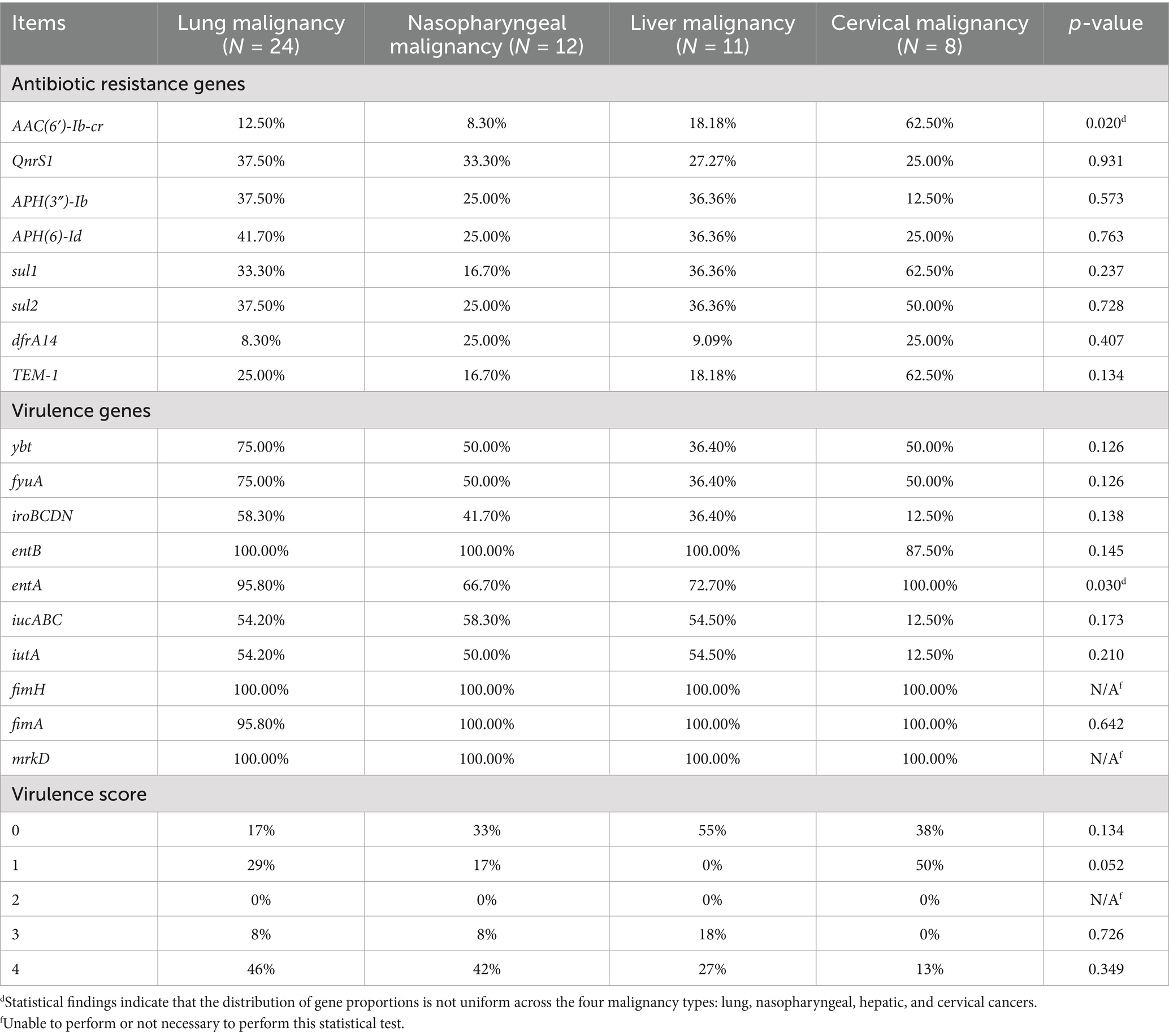
Table 4. Resistance genes, virulence genes and virulence scores of different K. pneumoniae isolates.
3.3.2 Virulence genes
Virulence gene profiles of the 105 K. pneumoniae isolates are summarized in Table 3, including siderophore and fimbrial genes such as fimA, fimH, and markD. The siderophore genes showed the following distribution: aerobactin (49%), yersiniabactin (32%), enterobactin (100%), and salmochelin (40%). A total of 96 isolates carried fimA + fimH + markD, 2 isolates carried fimA + markD, and 1 isolate carried only markD. The ompA gene was universally present (100%), while astA, cseA, and mgtB were less frequently detected.
Siderophore gene distribution varied among isolates from patients with different cancer types, as shown in Table 4. Isolates from nasopharyngeal cancer patient samples most frequently harbored the aerobactin-encoding iucABC genes, while lung cancer patient samples isolates showed the highest prevalence of yersiniabactin-related genes (ybt and fyuA). Conversely, cervical cancer patient samples isolates exhibited the lowest prevalence of iucABC and iroBCDN (salmochelin). Enterobactin genes (entAB) were present in ≥87.5% of isolates across all malignancies, with the highest prevalence of iroBCDN seen in lung cancer isolates.
Virulence scores, calculated using a system developed by Lam et al. that incorporates the number and type of virulence genes associated with clinical risk, ranged from 0 to 5. Scores ≥3, indicating high virulence potential, were found in 48% of isolates (Figure 2B). High scores (≥3) were more common in isolates from lung (50%), nasopharyngeal (50%), and liver cancer patients, whereas cervical cancer isolates showed the lowest prevalence of high virulence scores (13%).
3.3.3 Sequence types
Whole-genome sequencing identified 45 sequence types among the isolates. The most common types were ST23 (15.3%), followed by ST45 (6.7%), ST37 (4.8%), and ST25 (3.8%). Isolates from lung and nasopharyngeal cancer patient samples were predominantly ST23, whereas ST45 was most common among cervical cancer patient samples isolates, accounting for 37.5%.
3.3.4 Phylogenetic relationships and virulence gene analysis of K. pneumoniae isolates
Whole-genome sequencing was performed on all K. pneumoniae isolates, and a core single nucleotide polymorphism-based phylogenetic tree was constructed (Figure 3). Virulence gene profiles of each isolate were analyzed, including genes encoding enterobactin, adhesins, yersiniabactin, salmochelin, aerobactin, other virulence factors, fimbrial proteins, key components of the type III secretion system, lipopolysaccharide synthesis, and flagellar proteins. The phylogenetic analysis revealed three main branches. Strains in the second branch exhibited 84.6% positivity in the string test but had a low prevalence of ESBLs production (3.8%). These strains carried the most diverse siderophore virulence genes. In contrast, strains in the third branch demonstrated 18.9% positivity in the string test but a significantly higher ESBLs positivity rate (45.9%). The third branch predominantly included isolates from patients with lung (58.3%), nasopharyngeal (66.7%), liver (80%), and cervical (87.5%) malignant tumors. The most prevalent virulence genes were those encoding enterobactin (fepC: 101/105, entB: 100/105, entA: 91/105), adhesins (ykgK/ecpR: 103/105, yagZ/ecpA: 103/105, yagY/ecpB: 102/105, yagX/ecpC: 101/105, yagW/ecpD: 100/105, yagV/ecpE: 102/105), and fimbrial proteins (MrkD: 98/98, fimA: 96/98, fimH: 98/98). One MDR isolate, K-103, was identified from a patient with hypopharyngeal carcinoma. This ESBLs-producing strain was unique in carrying a complete set of key virulence genes, including those related to the type III secretion system, lipopolysaccharide synthesis, and flagellar proteins.
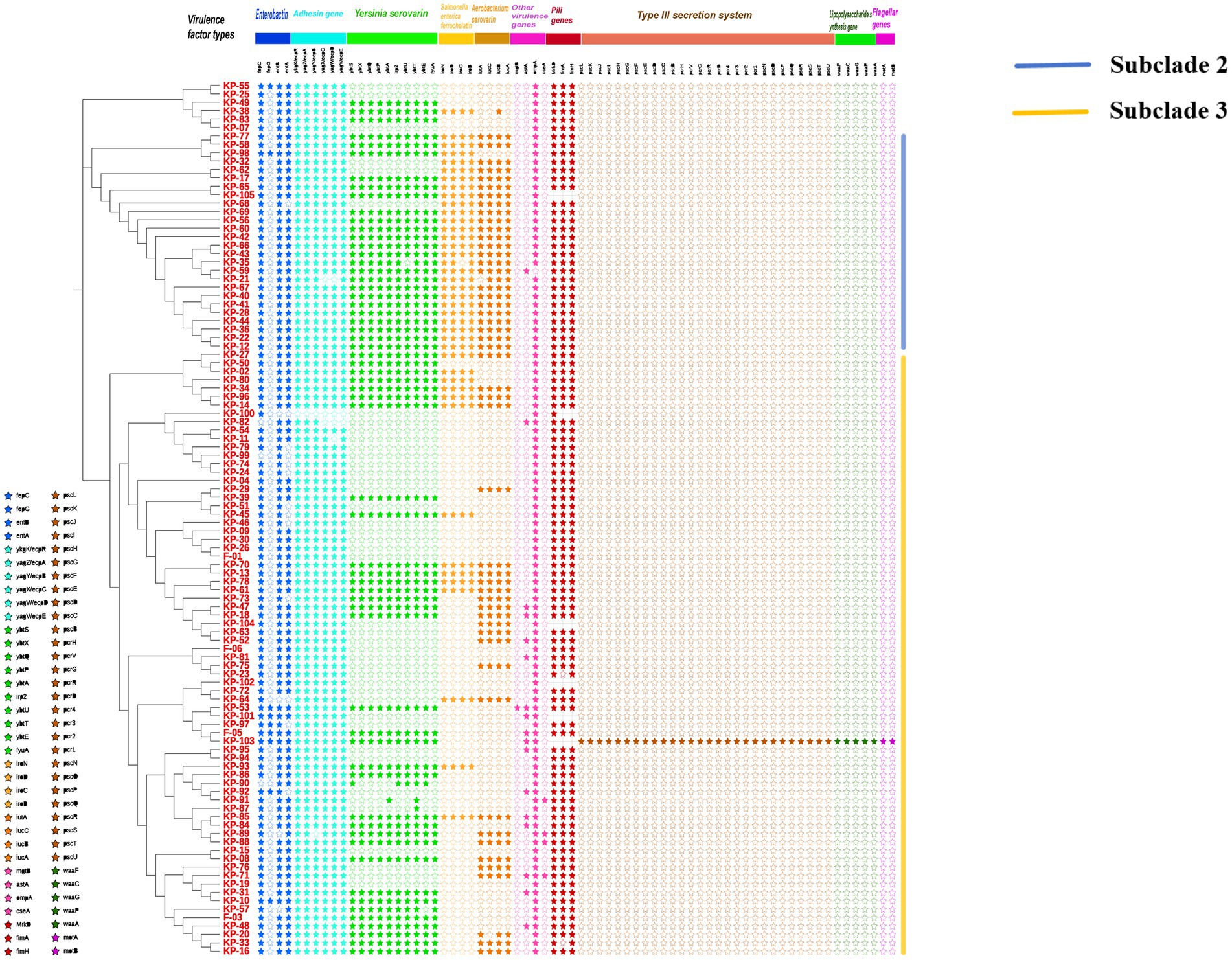
Figure 3. Phylogenetic relationships and virulence gene profiles of 105 K. pneumoniae isolates from patients with different malignant tumors. The maximum-likelihood phylogenetic tree (left) was constructed from whole-genome sequences. The accompanying heatmap (right) illustrates the distribution of key virulence genes across the strains. Genes are categorized by function. Symbols denote the presence (solid star, ★) or absence (hollow star, ☆) of each gene.
4 Discussion
Klebsiella pneumoniae is a significant pathogen capable of colonizing the respiratory, gastrointestinal, and urinary tracts and causing invasive infections, particularly in immunocompromised patients with malignant tumors (Lam et al., 2021). Neutropenia in these patients often necessitates prolonged antibiotic therapy, leading to the emergence of highly resistant strains. These circumstances complicate chemotherapy and treatment outcomes. This study characterized 105 K. pneumoniae isolates from malignant tumor patients, focusing on resistance profiles, virulence factors, and sequence typing. The specimens were primarily collected from sputum, secretions, and urine. In contrast, Victoria Ballén et al. (Holt et al., 2015) identified urine as the most common source, with 84.21% of urinary isolates carrying the uge virulence gene. Our study’s predominance of sputum specimens likely reflects the high prevalence of pulmonary and nasopharyngeal malignancies and the frequent use of ventilators in advanced-stage patients, which increases the risk of pulmonary infections.
Antibiotic susceptibility testing revealed resistance to all 20 tested antibiotics, with isolates from lung, nasopharyngeal, liver, and cervical cancer patient samples exhibiting particularly high resistance rates (≥25%) to β-lactams, sulfonamides, and quinolones. These results align with earlier findings, which report widespread MDR in K. pneumoniae, including resistance to β-lactams, quinolones, aminoglycosides, and sulfonamides (Ballén et al., 2021). Similarly, Anderson Lineu Siqueira dos Santos et al. (Abdelwahab et al., 2021) observed high resistance rates to ampicillin (95.3%), cefuroxime (67.2%), gentamicin (32.8%), and trimethoprim-sulfamethoxazole (12.5%) in oncology patients, consistent with our observations. These findings emphasize the growing challenge of MDR K. pneumoniae in malignant tumor settings.
Whole-genome sequencing revealed multiple resistance and virulence genes among the isolates. Common resistance genes included AAC(6′)-Ib-cr, QnrS1, APH(3″)-Ib, APH(6)-Id, sul1, and sul2, conferring resistance to quinolones, aminoglycosides, and sulfonamides. Notably, cervical cancer patient samples isolates exhibited the highest prevalence of AAC(6′)-Ib-cr and aminoglycoside resistance genes. Santos et al. (2020) and Enany et al. (2022) also identified similar resistance mechanisms in K. pneumoniae ST627 strains, corroborating our findings. The study also identified eight CRKP isolates, with bla_kpc-1 (40%), bla_ndm-5 (40%), and bla_oxa-10 (20%) being the predominant carbapenemase genes. These results differ from reports citing bla_vim-1, bla_ndm-1, bla_oxa-48, and bla_kpc-2 as common carbapenemase genes (Karampatakis et al., 2023), possibly due to the limited number of CRKP isolates. Additionally, 39 isolates were ESBLs-positive, with most carrying CTX-M (87.2%) and SHV (94.9%) genes, consistent with findings by Binzhi Dan et al. (Ma et al., 2023), who reported high co-occurrence of CTX-M, SHV, and TEM genes.
Efflux pump genes, which contribute to antibiotic resistance, were highly prevalent among the isolates. Key efflux pump genes detected included acrB (100%), acrD (97.1%), acrA (100%), oqxB (97.1%), oqxA (99.0%), marA (100%), and ramA (100%). These findings align with studies showing that efflux pumps are integral to the MDR phenotype of K. pneumoniae (Dan et al., 2023). Amir Mirzaie and Reza Ranjbar (Ren et al., 2024) reported a strong association between efflux pump gene expression and biofilm formation, a critical factor in recurrent infections. Our findings also highlight the significant presence of biofilm-associated genes, such as mrkA, further implicating their role in persistent infections in malignancy patients.
Virulence factors, particularly siderophores and fimbriae, were prevalent across the isolates. Iron acquisition, a critical factor for bacterial growth, is mediated by siderophores such as aerobactin, enterobactin, salmochelin, and yersiniabactin (Mirzaie and Ranjbar, 2021). Aerobactin, encoded by iucABCD, was detected in 49% of isolates, while enterobactin (entABCDE) was universally present, with individual gene carriage rates exceeding 90%. Salmochelin (iroBCDN) and yersiniabactin (ybt) were identified in 40 and 32% of isolates, respectively. Interestingly, cervical cancer patient samples isolates had the lowest prevalence of aerobactin (12.5%) and salmochelin (12.5%) genes. These siderophore gene carriage rates align with previous studies, underscoring their role as essential virulence factors for K. pneumoniae survival and pathogenesis (Mendes et al., 2023).
Virulence scoring, based on the system developed by Lam et al. (Kumabe and Kenzaka, 2014), showed that nearly 50% of isolates from malignant tumor patients scored ≥3, indicating high virulence potential. However, only 13% of cervical cancer patient samples isolates achieved this threshold, likely due to the smaller sample size. These virulence scores were higher than those reported by David J. Roach et al. (Jin et al., 2022), possibly reflecting the association between specific virulence factors and the malignancy-related infections studied here. The findings highlight the critical role of virulence determinants in the pathogenicity of K. pneumoniae in tumor patients.
Fimbriae play a crucial role in adhesion and biofilm formation, facilitating catheter-associated infections. The fimbrial genes fimA, fimH, and mrkD, encoding type 1 and type 3 fimbriae, were prevalent among the isolates. These fimbriae enhance K. pneumoniae’s ability to adhere to abiotic surfaces and form biofilms, critical for colonization and persistence in hospital settings (Roach et al., 2024; Arato et al., 2021). Wen-Ying Guo et al. (Schroll et al., 2010) reported that mrkABCDF and fimACDH operons, which encode these fimbriae, are often co-located with antibiotic resistance genes on plasmids, suggesting a linkage between virulence and resistance mechanisms. This co-occurrence further complicates treatment strategies for infections caused by these isolates.
Sequence typing involves analyzing the sequences of multiple housekeeping genes within bacterial genomes to determine genetic relationships between strains. In this study, whole-genome sequencing and database comparison identified 45 sequence types, with ST23 (15.3%) and ST45 (6.7%) being predominant. ST23, one of the major clones of hypervirulent Klebsiella pneumoniae (HV-KP) and a representative of the CC23 clonal lineage, is associated with high virulence and strong adaptability (Guo et al., 2022). Among ST23 isolates in this study, 87.5% were identified as HV-KP, consistent with previous reports. This suggests that most K. pneumoniae infections in cancer patients are caused by strains with high virulence and adaptability, contributing to recurrent, hard-to-cure infections and high mortality. Similarly, Siqin Zhang et al. (Lam et al., 2018) reported ST23 as the dominant type (38.7%) in K. pneumoniae strains causing pyogenic liver abscesses in southeastern China, aligning with our findings. In contrast, ST11 is commonly linked to hospital-acquired CRKP. Chen et al. (2024) reported ST11 as the predominant CRKP type (68.6%) in hematologic malignancy patients, while Yan Li et al. (Zhang et al., 2019) found ST11 in 66.7% of CRKP isolates from hospitalized patients across China, noting significant differences in antibiotic resistance between ST11 and non-ST11 strains. This study identified 8 CRKP isolates, with ST11 accounting for 25%, a lower proportion likely due to the limited sample size.
Phylogenetic analysis of 105 K. pneumoniae isolates revealed three distinct clades, with clustering patterns correlating to cancer types. K. pneumoniae strains isolated from lung, nasopharyngeal, liver, and cervical cancer patients were primarily grouped in the third clade, with liver and cervical cancer strains comprising 80 and 87.5%, respectively. This may reflect an association between infection dynamics and cancer type, as KP strains in respiratory tumors appear more transmissible. Previous studies have investigated KP strain evolution and dissemination. Liu et al. (2023); Li et al. (2024) analyzed the evolutionary relationships of 184 ST23-KP strains, identifying three clonal transmission events, mainly in ICUs, causing respiratory infections. These findings underscore the importance of preventing the hospital transmission of drug-resistant, hypervirulent KP strains. This study has several limitations. The relatively small sample size, particularly the limited number of cases for specific malignancy types, may constrain the generalizability of the findings. Furthermore, the absence of a non-cancer control group prevents definitive conclusions regarding whether the observed distribution of antimicrobial resistance genes and virulence factors is specific to cancer patients. Nevertheless, this work represents the first comprehensive genomic characterization of K. pneumoniae isolates from cancer patients in this region, providing valuable baseline data for future investigations. Subsequent studies should employ multi-center collaborations to expand the cohort and incorporate appropriately matched controls to verify and build upon these observations.
5 Conclusion
In conclusion, this study provides a comprehensive characterization of K. pneumoniae isolates from malignant tumor patients, revealing high levels of antibiotic resistance and virulence. The predominance of resistance genes, efflux pumps, siderophores, and fimbriae underscores the multifaceted mechanisms driving pathogenicity and persistence in these patients. The findings highlight the urgent need for targeted surveillance and effective infection control measures to manage K. pneumoniae infections in malignancy settings. Future studies should focus on larger sample sizes and explore the interplay between resistance and virulence mechanisms to develop novel therapeutic strategies.
Data availability statement
The raw sequencing data of 105 Klebsiella pneumoniae isolates generated in this study are publicly available in the NCBI BioProject database at: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1283480.
Ethics statement
The studies involving humans were approved by Medical Ethics Committee of Ganzhou Cancer Hospital. Affiliation: Ganzhou Cancer Hospital. The studies were conducted in accordance with the local legislation and institutional requirements. The human samples used in this study were acquired from a by-product of routine care or industry. Written informed consent for participation was not required from the participants or the participants’ legal guardians/next of kin in accordance with the national legislation and institutional requirements.
Author contributions
HL: Data curation, Writing – original draft, Visualization. WL: Conceptualization, Writing – original draft, Software, Formal analysis. XZ: Validation, Data curation, Supervision, Writing – review & editing. XW: Investigation, Methodology, Project administration, Writing – original draft. GH: Resources, Writing – review & editing, Funding acquisition, Methodology.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Grant support was provided by the Ganzhou Municipal Health Commission Science and Technology Project (GZWJW202402169), Guiding Science and Technology Plan Project of Ganzhou Science and Technology Bureau (20222ZDX8270).
Acknowledgments
We extend our sincere gratitude to the clinical departments and laboratory department of Ganzhou Cancer Hospital for their support with facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1676614/full#supplementary-material
Footnotes
References
Abdelwahab, R., Alhammadi, M. M., Hassan, E. A., Ahmed, E. H., Abu-Faddan, N. H., Daef, E. A., et al. (2021). Antimicrobial resistance and comparative genome analysis of Klebsiella pneumoniae strains isolated in Egypt. Microorganisms 9:1880. doi: 10.3390/microorganisms9091880
Amanati, A., Sajedianfard, S., Khajeh, S., Ghasempour, S., Mehrangiz, S., Nematolahi, S., et al. (2021). Bloodstream infections in adult patients with malignancy, epidemiology, microbiology, and risk factors associated with mortality and multi-drug resistance. BMC Infect. Dis. 21:636. doi: 10.1186/s12879-021-06243-z
Arato, V., Raso, M. M., Gasperini, G., Berlanda Scorza, F., and Micoli, F. (2021). Prophylaxis and treatment against Klebsiella pneumoniae: current insights on this emerging anti-microbial resistant global threat. Int. J. Mol. Sci. 22:4042. doi: 10.3390/ijms22084042
Ballén, V., Gabasa, Y., Ratia, C., Ortega, R., Tejero, M., and Soto, S. (2021). Antibiotic resistance and virulence profiles of Klebsiella pneumoniae strains isolated from different clinical sources. Front. Cell. Infect. Microbiol. 11:738223. doi: 10.3389/fcimb.2021.738223
Budia-Silva, M., Kostyanev, T., Ayala-Montaño, S., Bravo-Ferrer Acosta, J., Garcia-Castillo, M., Cantón, R., et al. (2024). International and regional spread of carbapenem-resistant Klebsiella pneumoniae in Europe. Nat. Commun. 15:5092. doi: 10.1038/s41467-024-49349-z
Chen, Y., Huang, J., Dong, L., Xu, B., Li, L., Zhao, Z., et al. (2024). Clinical and genomic characterization of carbapenem-resistant Enterobacterales bloodstream infections in patients with hematologic malignancies. Front. Cell. Infect. Microbiol. 14:1471477. doi: 10.3389/fcimb.2024.1471477
Choby, J. E., Howard-Anderson, J., and Weiss, D. S. (2020). Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J. Intern. Med. 287, 283–300. doi: 10.1111/joim.13007
Clinical and Laboratory Standards Institute (2024). Performance standards for antimicrobial susceptibility testing. 34th Edn. Wayne, PA: CLSI.
Dan, B., Dai, H., Zhou, D., Tong, H., and Zhu, M. (2023). Relationship between drug resistance characteristics and biofilm formation in Klebsiella Pneumoniae strains. Infect Drug Resist. 16, 985–998. doi: 10.2147/IDR.S396609
Enany, S., Zakeer, S., Diab, A. A., Bakry, U., and Sayed, A. A. (2022). Whole genome sequencing of Klebsiella pneumoniae clinical isolates sequence type 627 isolated from Egyptian patients. PLoS One 17:e0265884. doi: 10.1371/journal.pone.0265884
Ge, X., Zhou, Y., Jin, H., Liu, K., Zhu, K., Yu, Y., et al. (2024). Genomic insights and antimicrobial resistance profiles of CRKP and non-CRKP isolates in a Beijing geriatric medical center: emphasizing the blaKPC-2 carrying high-risk clones and their spread. Front. Microbiol. 15:1359340. doi: 10.3389/fmicb.2024.1359340
Guo, W. Y., Zhang, H., Cheng, M., Huang, M. R., Li, Q., Jiang, Y. W., et al. (2022). Molecular epidemiology of plasmid-mediated types 1 and 3 fimbriae associated with biofilm formation in multidrug resistant Escherichia coli from diseased food animals in Guangdong, China. Microbiol. Spectr. 10:e0250321. doi: 10.1128/spectrum.02503-21
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. USA 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Jin, Z., Wang, Z., Gong, L., Yi, L., Liu, N., Luo, L., et al. (2022). Molecular epidemiological characteristics of carbapenem-resistant Klebsiella pneumoniae among children in China. AMB Express 12:89. doi: 10.1186/s13568-022-01437-3
Karampatakis, T., Tsergouli, K., and Behzadi, P. (2023). Carbapenem-resistant Klebsiella pneumoniae: virulence factors, molecular epidemiology and latest updates in treatment options. Antibiotics (Basel) 12:234. doi: 10.3390/antibiotics12020234
Kumabe, A., and Kenzaka, T. (2014). String test of hypervirulent Klebsiella pneumonia. QJM 107:1053. doi: 10.1093/qjmed/hcu124
Lam, M. M. C., Wick, R. R., Watts, S. C., Cerdeira, L. T., Wyres, K. L., and Holt, K. E. (2021). A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat. Commun. 12:4188. doi: 10.1038/s41467-021-24448-3
Lam, M. M. C., Wyres, K. L., Duchêne, S., Wick, R. R., Judd, L. M., Gan, Y. H., et al. (2018). Population genomics of hypervirulent Klebsiella pneumoniae clonal-group 23 reveals early emergence and rapid global dissemination. Nat. Commun. 9:2703. doi: 10.1038/s41467-018-05114-7
Li, C., Jiang, X., Yang, T., Ju, Y., Yin, Z., Yue, L., et al. (2022). Genomic epidemiology of Carbapenemase-producing Klebsiella pneumoniae in China. Genomics Proteomics Bioinformatics 20, 1154–1167. doi: 10.1016/j.gpb.2022.02.005
Li, Y., Kumar, S., Zhang, L., Wu, H., and Wu, H. (2023). Characteristics of antibiotic resistance mechanisms and genes of Klebsiella pneumoniae. Open Med. (Wars) 18:20230707. doi: 10.1515/med-2023-0707
Li, Y., Xie, C., Zhang, Z., Liu, J., Chang, H., Liu, Y., et al. (2024). Molecular epidemiology and antimicrobial resistance profiles of Klebsiella pneumoniae isolates from hospitalized patients in different regions of China. Front. Cell. Infect. Microbiol. 14:1380678. doi: 10.3389/fcimb.2024.1380678
Li, Y., Yang, Y., Zheng, Y., Gao, Y., Shu, G., Gai, W., et al. (2024). Hypervirulent Klebsiella pneumoniae mediated hepatic infarction septic shock after rectal cancer surgery: a case report. Infect Drug Resist. 17, 1911–1918. doi: 10.2147/IDR.S452705
Liao, Q., Deng, J., Feng, Y., Zhang, W., Wu, S., Liu, Y., et al. (2022). Emergence of ceftazidime-avibactam resistance due to a novel blaKPC-2 mutation during treatment of carbapenem-resistant Klebsiella pneumoniae infections. J. Infect. Public Health 15, 545–549. doi: 10.1016/j.jiph.2022.04.002
Liu, Y., Jian, Z., Wang, Z., Yang, A., Liu, P., Tang, B., et al. (2023). Clinical characteristics and molecular epidemiology of ST23 Klebsiella pneumoniae in China. Infect Drug Resist. 16, 7597–7611. doi: 10.2147/IDR.S428067
Lubwama, M., Kateete, D. P., Katende, G., Kigozi, E., Orem, J., Phipps, W., et al. (2024). CTX-M, TEM, and SHV genes in Escherichia coli, Klebsiella pneumoniae, and Enterobacter spp isolated from hematologic cancer patients with bacteremia in Uganda. Infect Drug Resist. 17, 641–653. doi: 10.2147/IDR.S442646
Ma, J., Song, X., Li, M., Yu, Z., Cheng, W., Yu, Z., et al. (2023). Global spread of carbapenem-resistant Enterobacteriaceae: epidemiological features, resistance mechanisms, detection and therapy. Microbiol. Res. 266:127249. doi: 10.1016/j.micres.2022.127249
Mendes, G., Santos, M. L., Ramalho, J. F., Duarte, A., and Caneiras, C. (2023). Virulence factors in carbapenem-resistant hypervirulent Klebsiella pneumoniae. Front. Microbiol. 14:1325077. doi: 10.3389/fmicb.2023.1325077
Mirzaie, A., and Ranjbar, R. (2021). Antibiotic resistance, virulence-associated genes analysis and molecular typing of Klebsiella pneumoniae strains recovered from clinical samples. AMB Express 11:122. doi: 10.1186/s13568-021-01282-w
Mishra, R., Patel, A., Agarwal, A., Thaiyam, D., and Thaiyam, S. (2024). Infections and their outcomes in cancer patients with and without neutropenia: a single-center experience. Cureus 16:e51983. doi: 10.7759/cureus.51983
Nanayakkara, A. K., Boucher, H. W., Fowler, V. G. Jr., Jezek, A., Outterson, K., and Greenberg, D. E. (2021). Antibiotic resistance in the patient with cancer: escalating challenges and paths forward. CA Cancer J. Clin. 71, 488–504. doi: 10.3322/caac.21697
Navon-Venezia, S., Kondratyeva, K., and Carattoli, A. (2017). Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 41, 252–275. doi: 10.1093/femsre/fux013
Ntim, O. K., Awere-Duodu, A., Osman, A. H., and Donkor, E. S. (2025). Antimicrobial resistance of bacterial pathogens isolated from cancer patients: a systematic review and meta-analysis. BMC Infect. Dis. 25:296. doi: 10.1186/s12879-025-10481-w
Osama, D., El-Mahallawy, H., Mansour, M. T., Hashem, A., and Attia, A. S. (2021). Molecular characterization of Carbapenemase-producing Klebsiella pneumoniae isolated from Egyptian pediatric cancer patients including a strain with a rare gene-combination of β-lactamases. Infect Drug Resist. 14, 335–348. doi: 10.2147/IDR.S284455
Ren, J., Wang, M., Zhou, W., and Liu, Z. (2024). Efflux pumps as potential targets for biofilm inhibition. Front. Microbiol. 15:1315238. doi: 10.3389/fmicb.2024.1315238
Roach, D. J., Sridhar, S., Oliver, E., Rao, S. R., Slater, D. M., Hwang, W., et al. (2024). Clinical and genomic characterization of a cohort of patients with Klebsiella pneumoniae bloodstream infection. Clin. Infect. Dis. 78, 31–39. doi: 10.1093/cid/ciad507
Rolston, K. V. (2017). Infections in cancer patients with solid tumors: a review. Infect. Dis. Ther. 6, 69–83. doi: 10.1007/s40121-017-0146-1
Santos, A. L. S. D., Rodrigues, Y. C., Melo, M. V. H., Silva Dos Santos, P. A., Oliveira, T. N. D. C., Sardinha, D. M., et al. (2020). First insights into clinical and resistance features of infections by Klebsiella pneumoniae among oncological patients from a referral center in Amazon region, Brazil. Infect. Dis. Rep. 12, 110–120. doi: 10.3390/idr12030021
Schroll, C., Barken, K. B., Krogfelt, K. A., and Struve, C. (2010). Role of type 1 and type 3 fimbriae in Klebsiella pneumoniae biofilm formation. BMC Microbiol. 10:179. doi: 10.1186/1471-2180-10-179
Touati, A., Talbi, M., Mairi, A., Messis, A., Adjebli, A., Louardiane, M., et al. (2020). Fecal carriage of extended-spectrum β-lactamase and carbapenemase-producing Enterobacterales strains in patients with colorectal cancer in the oncology unit of Amizour hospital, Algeria: a prospective cohort study. Microb. Drug Resist. 26, 1383–1389. doi: 10.1089/mdr.2019.0350
Wang, Q., Wang, X., Xie, Z., Qu, J., Lin, Y., Hu, J., et al. (2025). Prevalence and molecular characteristics of Klebsiella pneumoniae harboring the Pks Island from cancer patients in China. Infect Drug Resist. 18, 3237–3246. doi: 10.2147/IDR.S522818
Zhang, S., Zhang, X., Wu, Q., Zheng, X., Dong, G., Fang, R., et al. (2019). Clinical, microbiological, and molecular epidemiological characteristics of Klebsiella pneumoniae-induced pyogenic liver abscess in southeastern China. Antimicrob. Resist. Infect. Control 8:166. doi: 10.1186/s13756-019-0615-2
Keywords: malignant tumor, Klebsiella pneumoniae, antibiotic resistance, virulence gene, whole genome sequencing
Citation: Liu H, Liu W, Zhou X, Wang X and Huang G (2025) Genomic characterization of Klebsiella pneumoniae clinical isolates from cancer patients: resistance profiles, virulence factors, and sequence typing. Front. Microbiol. 16:1676614. doi: 10.3389/fmicb.2025.1676614
Edited by:
Adriana Belas, Lusofona University, PortugalReviewed by:
Yasmine Hasanine Tartor, Zagazig University, EgyptMauricio Flores-Valdez, Secretaria de Salud, Mexico
Copyright © 2025 Liu, Liu, Zhou, Wang and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gang Huang, aHVhbmc1NjdnYW5nQDE2My5jb20=
†These authors have contributed equally to this work and share first authorship
 Hancong Liu
Hancong Liu Wan Liu†
Wan Liu†