- 1 Department of Neurology, Faculty of Medicine, Assiut University, Assiut, Egypt
- 2Institute of Global Health and Human Ecology, The American University in Cairo, Cairo, Egypt
- 3Department of Clinical Pharmacy, Faculty of Pharmacy, Assiut University, Assiut, Egypt
- 4Department of Pharmacology and Toxicology, Faculty of Pharmacy, The British University in Egypt, Cairo, Egypt
- 5Department of Neurology, Faculty of Medicine, Ain Shams University, Cairo, Egypt
- 6Department of Neurology, Faculty of Medicine, Sohag University, Sohag, Egypt
- 7Department of Neurology, Faculty of Medicine, Cairo University, Cairo, Egypt
- 8Department of Neuromuscular Diseases, UCL Queen Square Institute of Neurology, UCL, London, United Kingdom
- 9Department of Medical Physiology, Faculty of Medicine, Mansoura University, Dakahleya, Egypt
- 10Department of Internal Medicine, Rheumatology and Clinical Immunology Unit, Faculty of Medicine, Cairo University, Cairo, Egypt
- 11Faculty of Medicine, Newgiza University, Giza, Egypt
- 12Department of Neurology, Faculty of Medicine, Mansoura University, Dakahleya, Egypt
- 13Department of Neurology, Faculty of Medicine, Alexandria University, Alexandria, Egypt
- 14Department of Neurology, Mansoura International Hospital, Dakahleya, Egypt
- 15Forensic Medicine and Clinical Toxicology Department, Faculty of Medicine, Mansoura University, Dakahleya, Egypt
- 16Department of Neuropsychiatry, Faculty of Medicine, Kafr El Sheikh University, Egypt
Background: The apolipoprotein E (APOE) gene, encompassing three alleles (ε2, ε3, ε4), is a critical player in lipid metabolism and has been extensively studied for its role in neurodegenerative diseases. This study examines APOE genetic variability and its association with PD in an Egyptian cohort.
Methods: A total of 891 participants, including 422 PD patients and 469 healthy controls, were included in this study. APOE genotyping was performed using Kompetitive Allele Specific PCR (KASP) to detect the rs429358 and rs7412 SNPs, which define the APOE alleles. APOE alleles were categorized based on the genotypes into ε2, ε3, and ε4 groups. Clinical assessments of PD patients included age at onset, disease severity (MDS-UPDRS), and demographic factors. Statistical analyses compared APOE distributions between PD and control groups and examined associations with clinical variables.
Results: The ε3 allele was the most prevalent in the cohort (77.3%), aligning with global and African trends. The ε2 allele was observed in 11.4%, and the ε4 allele in 11.3%, with both frequencies being lower than reported African estimates. The ε3/ε3 genotype was predominant in both PD patients (72.51%) and controls (72.07%). The ε4/ε4 genotype was absent in PD cases and rare among controls (0.64%). No significant association was found between APOE genotypes and PD risk, age at onset, or disease severity.
Conclusion: Our findings do not support a significant role for APOE in PD susceptibility or severity in Egyptians.
Introduction
The apolipoprotein E (APOE) gene, located on chromosome 19q13.2, comprises three common polymorphic alleles (ε2, ε3, ε4), resulting in six possible genotypes (ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, ε4/ε4). APOE is crucial for lipid metabolism and is implicated in various neurodegenerative disorders, most notably Alzheimer’s disease (AD). The ε4 allele is recognized as a significant genetic risk factor for late-onset AD, associated with increased disease susceptibility and earlier onset, while the ε2 allele may provide some protective effects (Panza et al., 2012; Chartier-Hariln et al., 1994; Lambert et al., 2013). Such modulatory effects of APOE are strongly linked to its alleles differential modulation of amyloid-β metabolism, aggregation, and clearance, in addition to influencing tau pathology, neuroinflammation, and neuronal signaling (Kanekiyo et al., 2014; Baek et al., 2020).
Beyond AD, it has been suggested that intracellular α-synuclein contributes to Parkinson’s Disease (PD) pathology by partially activating extracellular signaling pathways involving APOE (Gallardo et al., 2008). The mechanisms underlying APOE’s potential involvement in PD, however, are less understood and likely distinct from those in AD, reflecting the different core pathologies. Several hypotheses attempt to bridge APOE function with PD pathophysiology. These include suggestions that APOE isoforms may differentially affect α-synuclein aggregation (Emamzadeh et al., 2016), propagation, or clearance (Kang et al., 2022; Liampas et al., 2024; Zhao et al., 2020), modulate microglial activation and neuroinflammatory responses in the context of synucleinopathy (Zhang et al., 2023), or influence lipid transport and cholesterol metabolism critical for dopaminergic neuron integrity and function (Fernández-Calle et al., 2022). Despite these plausible biological connections, establishing a definitive link between APOE variants and PD risk or progression has proven challenging.
Unlike in AD, the role of APOE in PD, especially the ε4 allele, remains less clearly defined and studies are yielding contradicting findings (Federoff et al., 2012; Li et al., 2018). Recent large genome-wide association studies (GWAS) on individuals of Northern European descent revealed no significant link between APOE genotype and PD status or age at onset. However, meta-analyses that included data from diverse ethnic groups—such as Europeans, Asians, and Latin Americans—demonstrate that the link between APOE genotype and PD risk may be ancestry dependent (Li et al., 2018; Sun et al., 2019).
Furthermore, the association between APOE and disease severity in PD is less straightforward. Although animal research suggests that the APOE genotype influences motor function in PD, with ε4 carriers showing worse motor performance and ε2 carriers experiencing less motor dysfunction (Davis et al., 2020), this association has not been established in clinical research (Kapan et al., 2023; Jo et al., 2021). This discrepancy highlights the need for further investigation into how APOE may differentially affect motor outcomes.
Despite considerable research on the APOE gene and its role in neurodegenerative diseases, studies focusing on PD in populations from the Middle East and Africa, including Egyptians, remain limited. This study aims to characterize the genetic variability of the APOE gene in Egyptians with and without PD. Additionally, it investigates APOE’s influence on genetic susceptibility to PD and explore how APOE variants may affect the age at onset and disease severity of PD within the study population.
Methods
The present study received ethical approval from the American University in Cairo review board (AUC-IRB) in Egypt, with the following reference numbers: Ethics Approval #2021–2022-058 and #2021–2022-203. Additionally, the ethical committee of University College London granted approval under REC #22/NE/0080. Written informed consent was obtained from all participants, in compliance with the Declaration of Helsinki and the Common Rule.
Participant recruitment and clinical assessments
A total of 891 Egyptians, comprising 422 PD patients and 469 unrelated healthy controls, were recruited through the Egyptian Network of Neurodegenerative Diseases (ENND) as a collaboration with International Parkinson’s Disease Genome Consortium- Africa (IPDGC-Africa). PD was diagnosed by neurologists using the UK Brain Bank Criteria (Hughes et al., 1992) and/or the criteria established by the Movement Disorder Society Task Force (Postuma et al., 2015). Basic demographic information, including age and gender, was collected from all subjects. Clinical information for PD cases included age at disease onset, age at diagnosis, family history, and history of consanguinity. Additionally, the Movement Disorder Society Unified Parkinson’s Disease Rating Scale (MDS-UPDRS) were assessed in all PD cases.
APOE genotyping
DNA was extracted from 10 mL samples of either venous whole blood or saliva using established protocols. Genotyping was performed using the Kompetitive Allele Specific Polymerase Chain Reaction (KASP) assay (LGC Genomics, Herts, United Kingdom), following the method outlined in Rizig et al. (2021). Two single-nucleotide polymorphisms (SNPs) from the APOE gene, specifically rs429358 and rs7412, were selected for genotyping. These SNPs are the primary genetic markers used to determine the APOE isoforms (ε2, ε3, ε4). Functionally, these SNPs influence the structure and function of the APOE protein, which plays a crucial role in neurodegeneration (Corbo and Scacchi, 1999). Both SNPs have validated KASP assays available, offering an efficient, cost-effective, and reliable method for genotyping our cohort.
Data analysis
APOE alleles were categorized based on the genotypes as follows: individuals with the ε2/ε2 genotype were assigned to the ε2 group; those with either ε2/ε3 or ε3/ε3 genotypes were classified into the ε3 group; and individuals with the ε2/ε4, ε3/ε4, or ε4/ε4 genotypes were grouped under the ε4 category.
Results
Cohort characteristics is summarized in Table 1.
APOE allelic and genotypic frequency proportions in Egyptians with PD and controls
In this study, the allele frequencies of APOE among all participants (n = 891) were as follows: ε3 (77.257%), ε4 (11.327%), and ε2 (11.416%). The genotypic frequencies were 72.28% for ε3/ε3, 0.34% for ε4/ε4, and (0.56%) for ε2/ε2 among the homozygotes, while the heterozygous frequencies were ε3/ε4 (12.91%), ε2/ε3 (12.79%) and ε2/ε4 (1.12%). The APOE allelic and genotypic distributions observed in this Egyptian cohort were generally consistent with reports from similar populations (Figure 1; Tables 2, 3).
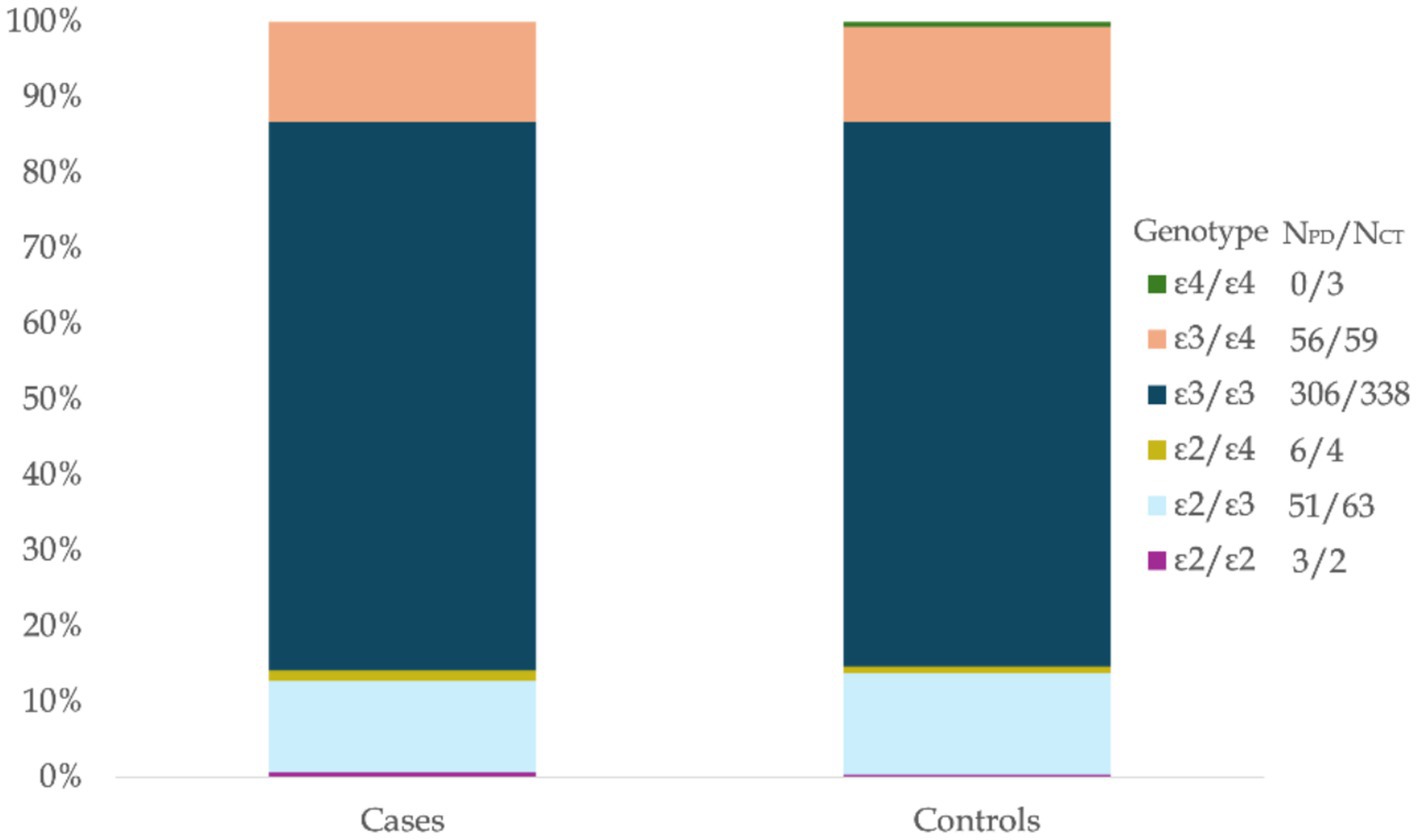
Figure 1. APOE genotype distribution in Egyptians with PD and controls. Percentage distribution of APOE genotypes in Egyptian Parkinson’s Disease (PD) patients (Cases, N = 422) and ethnically matched healthy controls (Controls, N = 469). The stacked bars illustrate the relative frequencies of the ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4 genotypes within each group. Absolute counts for each genotype are indicated in the legend for the cases and control groups.
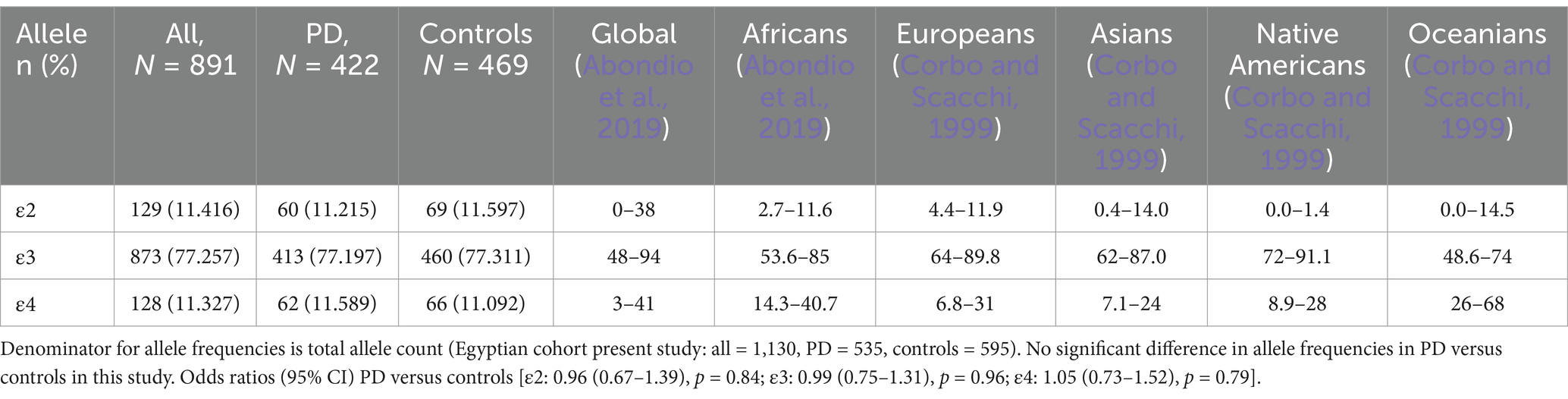
Table 2. APOE allele distribution in Egyptians with PD and controls in comparison to other normal global and ethnic populations.
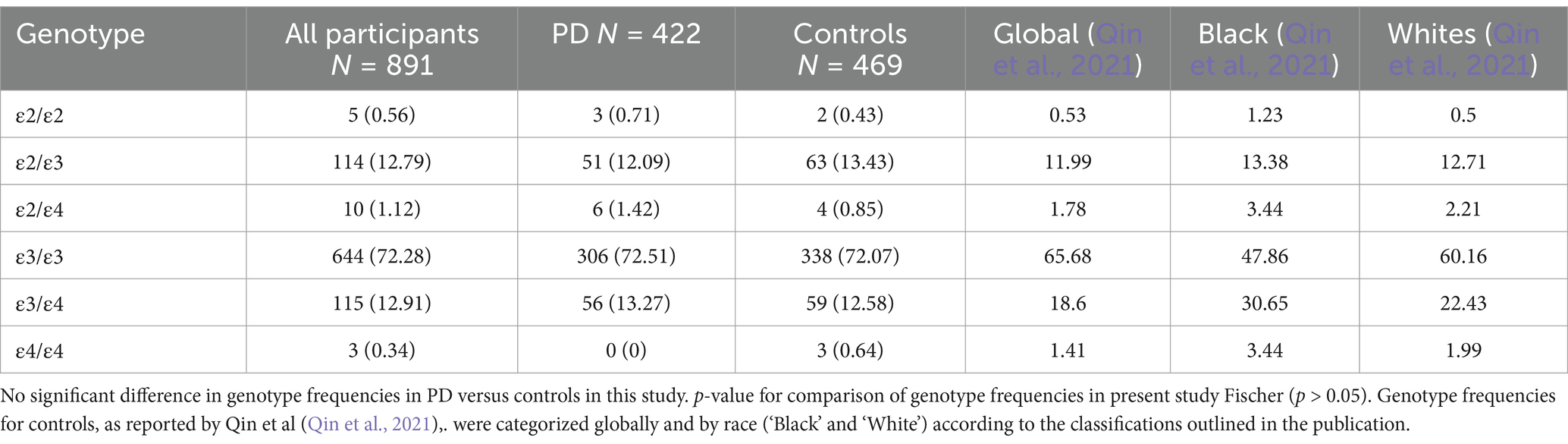
Table 3. APOE genotype distribution in Egyptians with PD and controls in comparison to other normal global and ethnic populations.
Association between APOE genotypes and PD age of onset, and disease severity
The link between APOE genotypes and Parkinson’s disease (PD) age of onset was assessed using linear regression. The analysis revealed that APOE genotypes explained only a small proportion of the variance in age of onset (R2 = 0.021), and this association was not statistically significant (F-statistic = 1.787, p = 0.114). Similarly, APOE genotypes were not significant predictors of disease severity, as measured by the MDS-UPDRS scores. In contrast, age, age of onset, and disease duration emerged as significant predictors of disease severity, underscoring their importance in the progression of PD.
Discussion
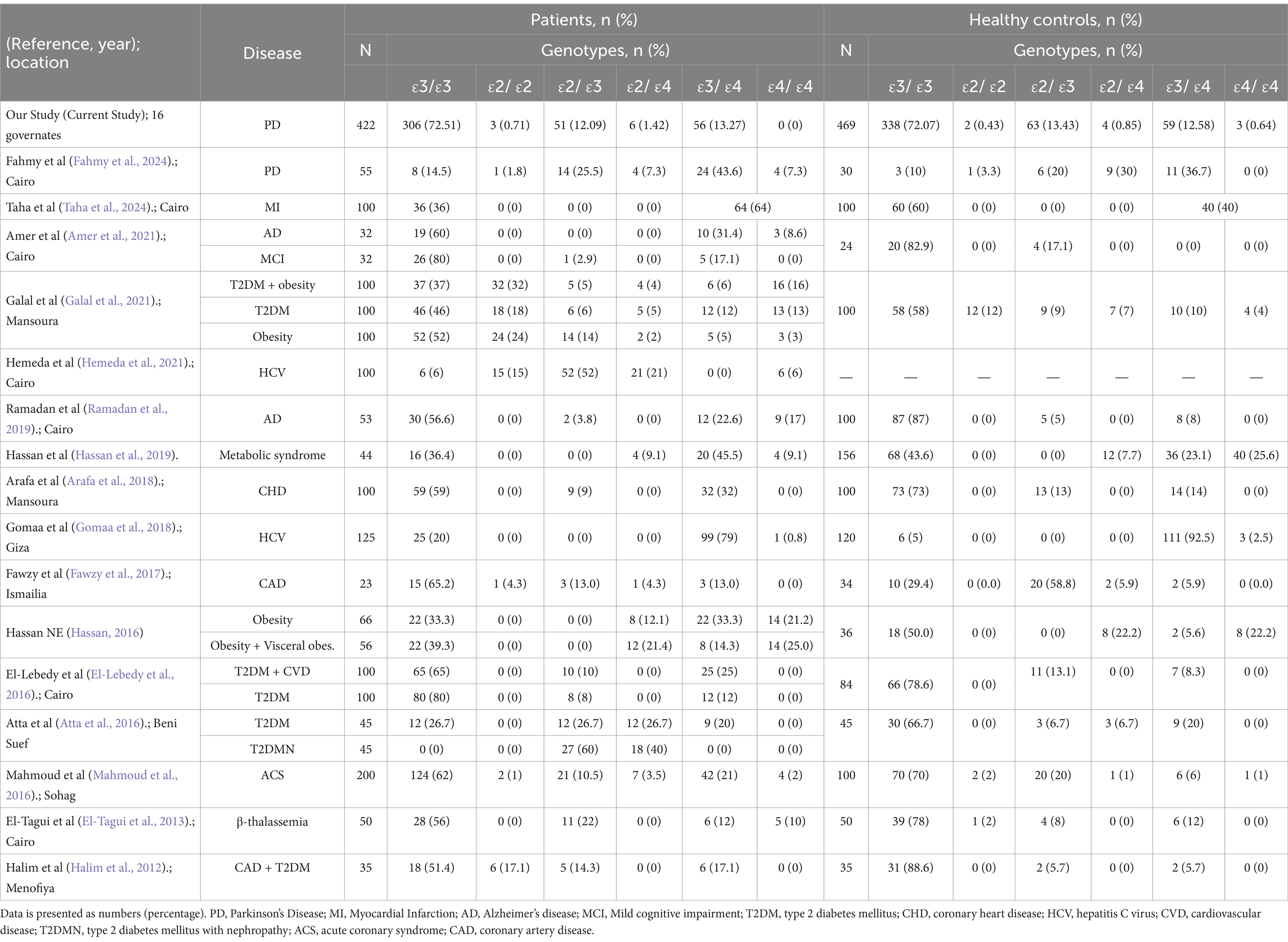
A detailed comparison of the herein communicated APOE genotype distribution findings with those from previous studies conducted in Egypt underscores the contributions of the present study (Table 4). Notably, our study represents the largest cohort to date examining APOE variants in Egyptian PD patients (N = 422) and controls (N = 469), significantly exceeding the sample size of the only other published study specifically focusing on PD in Egyptians (N = 55 patients, N = 30 controls) by Fahmy et al. Furthermore, our cohort encompasses a broader geographical representation, including participants from 16 different governorates, in contrast to previous studies which were often restricted to single locales such as Cairo, Mansoura, or Ismailia. This enhanced sample size and diversity strengthen the reliability and generalizability of our findings for the Egyptian population.
In terms of genotype distribution among PD patients, our results show a higher prevalence of the ε3/ε3 genotype (72.51%) compared to the Fahmy et al. study (14.5%). Conversely, the frequencies of the ε3/ε4 (13.27%) and ε4/ε4 (0%) genotypes in our PD cohort were lower than those previously reported (43.6 and 7.3%, respectively). Differences are also observed when comparing our control group’s genotype frequencies (e.g., ε3/ε3: 72.07%, ε3/ε4: 12.58%, ε4/ε4: 0.64%) to those in the aforementioned study’s controls (ε3/ε3: 10%, ε3/ε4: 36.7%, ε4/ε4: 0%) and other Egyptian studies focusing on different diseases such as AD, Myocardial Infarction (MI), or Type 2 Diabetes Mellitus (T2DM). These variations underscore the potential influence of cohort size, geographical location, and the specific disease context on APOE genotype distributions within the Egyptian population.
On the other hand, the APOE allelic and genotypic frequencies in our cohort align broadly with the distributions reported in populations of African ancestry, with some notable distinctions. Corroborating other studies, the ε3 allele was the most frequent, detected in ~ 77.3% of all participants, placing it within the mid-range of reported rates in African populations (53.6–85%) and consistent with other ethnic groups worldwide (48–94%; Abondio et al., 2019). The ε2 allele was present in 11.4% of the entire cohort, a prevalence that is consistent with previous reports from Africa, albeit at the higher end of the reported range (2.7–11.6%), and comparable to global frequencies (Abondio et al., 2019). In contrast, the frequency of the ε4 allele was lower than the African estimates (14.3–40.7; Abondio et al., 2019), with only 11.3% of our cohort carrying this allele. Within genotypic distributions, ε3/ε3 was the most prevalent in both the PD group (72.51%) and controls (72.07%), aligning with earlier studies (Galal et al., 2021; Ramadan et al., 2019) and mirroring global trends where the ε3 allele predominates (Qin et al., 2021). The ε2/ε2 genotype, observed in 0.71% of PD patients and 0.43% of controls, falls within global estimates, but lower than the frequencies typically observed in individuals of black ethnicity (Qin et al., 2021). The ε2/ε4 and ε3/ε4 combinations were relatively uncommon, with the ε3/ε4 genotype showing comparable frequencies between PD patients (13.27%) and controls (12.58%), which are lower than both global and regional estimates. Further, the ε4/ε4 genotype was not observed in PD cases and was found in only 0.64% of controls, a frequency much lower than the global estimates regardless of the ethnicity (Qin et al., 2021).
Significant gene flow from the Near East and Mediterranean/European populations, which generally have lower ε4 frequencies compared to ancestral Sub-Saharan African populations (Corbo and Scacchi, 1999), could have substantially shaped the Egyptian gene pool over millennia (Bassyouni et al., 2023). Additionally, while ε4 is the ancestral human allele and may have offered advantages in past environments (e.g., related to diet or pathogen resistance; Fujioka et al., 2013), the long history of agriculture in the Nile Valley might have favored the ε3 allele. Furthermore, the shift toward modern diets could potentially exert negative selective pressure on ε4 due to its association with increased risk for cardiovascular and metabolic diseases in contemporary settings (Huebbe and Rimbach, 2017). Therefore, the current ε4 frequency likely reflects a blend of ancient African history diluted by substantial admixture from regions with lower ε4 prevalence, and possibly finetuned by selective pressures related to historical changes in diet and lifestyle.
Regarding the secondary objectives of our study, we found no significant association between APOE genotypes and the risk of PD. This is consistent with prior research, which similarly reported no notable differences in APOE distribution, including ε4 carrier rates, between PD patients and healthy controls (Federoff et al., 2012; Okubadejo et al., 2022; Williams-Gray et al., 2009). Furthermore, we observed no significant relationship between APOE alleles or genotypes and the age of onset or disease severity of PD. The lack of association with disease severity, despite suggestive findings in some animal models (Davis et al., 2020; Fyfe, 2020), might stem from several factors in human studies: the complex interplay between genetic predisposition and diverse environmental influences throughout life, potential modification of APOE effects by other genetic loci not assessed here, or inherent differences between preclinical models and the multifaceted pathophysiology of human PD. Biologically, while APOE is crucial for lipid transport and neuronal maintenance (Yang et al., 2023; Husain et al., 2021), its specific role in the dopaminergic pathways primarily affected in PD may be less pronounced compared to its established impact in Alzheimer’s pathology, potentially explaining the divergence from findings in AD and the lack of strong association signals in many PD cohorts globally (Federoff et al., 2012; Okubadejo et al., 2022; Kurz et al., 2009; Wenjuan et al., 2016), including ours.
Notwithstanding the above, we acknowledge several limitations in our study. While 891 participants represent a relatively large dataset for a country-specific genetic study, this size may be insufficient to detect small effect sizes, or gene–gene/gene–environment interactions, particularly for rare alleles such as ε2 or ε4. This may partly explain the absence of significant associations between APOE and PD risk or severity in our study. Furthermore, our sample did not include any PD cases homozygous for the ε4 allele, which may have further limited our ability to assess its potential impact on disease risk or severity. For assessing PD severity, we relied on the total MDS-UPDRS score, as detailed sub-scores were unavailable. This limitation restricted our ability to explore more nuanced associations between APOE genotypes and specific symptom domains. Additionally, we were unable to examine the potential link between APOE and specific non-motor features like cognitive impairment, where APOE effects might be more relevant. Therefore, future studies with larger sample sizes and more detailed phenotypic data are needed to clarify APOE’s role in PD.
In addition, APOE is likely only one piece of a complex genetic landscape. Exploring alternative or complementary genetic pathways is crucial. Other PD-associated genes such as LRRK2, GBA, SNCA, and MAPT should be investigated in the Egyptian population to identify more relevant genetic risk factors. Furthermore, environmental contributors—including pesticide exposure, rural residency, dietary habits, and access to healthcare—may interact with genetic predispositions in shaping PD risk. Incorporating these variables into future studies may yield a more comprehensive understanding of PD etiology in this population.
Despite these limitations, our study provides valuable insights into the distribution of APOE genotypes in Egyptians with PD and contributes to the broader body of knowledge on genetic risk factors for PD in North African populations. Further research is needed to explore the role of APOE and other genetic and environmental factors in PD, with larger sample sizes and more comprehensive data collection.
Conclusion
This study provides the largest exploration to date of APOE genetic variability in an Egyptian cohort with PD, offering new insights into the distribution of APOE alleles and genotypes in populations of Middle Eastern and African descent, which are traditionally underrepresented in genetic studies. The findings suggest that APOE allelic and genotypic distributions in Egyptians broadly align with African and global trends, with some distinctions, particularly in the lower frequency of the ε4 allele and the strikingly low prevalence of the ε4/ε4 genotype. Importantly, our analysis found no significant association between APOE genotypes and the overall risk of PD, nor with the age of onset or severity of the disease within our cohort’s statistical power. While these results align with some large-scale studies in other populations suggesting APOE may not be a major driver of PD risk or progression, it is important to acknowledge that this does not preclude potential influences on other specific aspects of the disease not fully captured here, such as cognitive decline, or potential interactions with other genetic or environmental factors unique to this population. Therefore, future research should focus not only on larger sample sizes and diverse cohorts from the Middle East and Africa but also incorporate longitudinal study designs to track progression, detailed cognitive assessments stratified by genotype, and investigations into potential gene–environment interactions. Such focused approaches are warranted to fully elucidate the potentially subtle or context-dependent roles of APOE and to enhance our understanding of the complete genetic architecture contributing to PD in these populations.
Data availability statement
Anonymized genotyping and clinical data can be provided to bone fide researchers upon request from the corresponding author(s).
Ethics statement
The studies involving humans were approved by The Institutional Review Board of the American University in Cairo (AUC-IRB) and the ethics committee of University College London (UCL). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
EK: Investigation, Writing – review & editing. MW: Formal analysis, Investigation, Writing – original draft, Writing – review & editing. AE: Data curation, Formal analysis, Investigation, Writing – original draft. AS: Investigation, Writing – review & editing. GF: Investigation, Writing – review & editing. MY: Investigation, Writing – review & editing. SE-J: Investigation, Writing – review & editing. HL: Investigation, Writing – review & editing. AJ: Investigation, Writing – review & editing. MK: Investigation, Writing – review & editing. AH: Investigation, Writing – review & editing. YS: Investigation, Writing – review & editing. PG: Investigation, Writing – review & editing. NH: Investigation, Writing – review & editing. SN: Investigation, Writing – review & editing. AA: Investigation, Writing – review & editing. SaE: Investigation, Writing – review & editing. GR: Investigation, Writing – review & editing. MH: Investigation, Writing – review & editing. YE: Investigation, Writing – review & editing. AG: Investigation, Writing – review & editing. NS: Investigation, Writing – review & editing. LA: Investigation, Writing – review & editing. NA: Investigation, Writing – review & editing. TB: Investigation, Writing – review & editing. NE: Investigation, Writing – review & editing. ME-G: Investigation, Writing – review & editing. ShE: Investigation, Writing – review & editing. SR: Investigation, Writing – review & editing. JM: Investigation, Writing – review & editing. HH: Funding acquisition, Investigation, Writing – review & editing. MR: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing. MS: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. MR received funding from the University College London Grand Challenges Small Grants (Award ID: 177813), the MRC neurosciences and mental health research grant. April 2023—April 2026 and The Michael J Fox Foundation Genetic Diversity in Parkinson’s Disease 2019 (Grant ID: 17483). HH received funding from the Michael J Fox Foundation Genetic Diversity in Parkinson’s Disease (Grant ID: 17483). MS received funding from the Bartlett Fund for Critical Challenges-2021 (Agreement Number: 2—Cycle 3) and the American University in Cairo Faculty Support Grant 2021 awarded. The funders were not involved in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abondio, P., Sazzini, M., Garagnani, P., Boattini, A., Monti, D., Franceschi, C., et al. (2019). The genetic variability of APOE in different human populations and its implications for longevity. Genes (Basel) 10:222. doi: 10.3390/genes10030222
Amer, M. S., Hamza, S. A., Shaltoot, H. M., Zaki, O. K., Saad, W. E., Saab, A. A., et al. (2021). Interaction between apolipoprotein E genotyping, serum inflammatory biomarkers and cognitive functions in Egyptian elderly. Egypt J Immunol 28, 01–11. doi: 10.55133/eji.28.01.01
Arafa, S., Abdelsalam, S., El-Gilany, A. H., Mosaad, Y. M., and Abdel-Ghaffar, A. (2018). Endothelial nitric oxide synthase Glu 298 asp (G894T) and apolipoprotein E gene polymorphism as possible risk factors for coronary heart disease among Egyptians. Egyptian Heart J 70, 393–401. doi: 10.1016/j.ehj.2018.08.001
Atta, M. I., Abo Gabal, K., El-Hadidi, K., Swellam, M., Genina, A., and Zaher, N. F. (2016). Apolipoprotein E genotyping in Egyptian diabetic nephropathy patients. IUBMB Life 68, 58–64. doi: 10.1002/iub.1460
Baek, M. S., Cho, H., Lee, H. S., Lee, J. H., Ryu, Y. H., and Lyoo, C. H. (2020). Effect of APOE ε4 genotype on amyloid-β and tau accumulation in Alzheimer’s disease. Alzheimers Res Ther 12:140. doi: 10.1186/s13195-020-00710-6
Bassyouni, M., Mysara, M., Wohlers, I., Busch, H., Saber-Ayad, M., and El-Hadidi, M. (2023). A comprehensive analysis of genetic risk for metabolic syndrome in the Egyptian population via allele frequency investigation and Missense3D predictions. Sci Rep 13:20517. doi: 10.1038/s41598-023-46844-z
Chartier-Hariln, M. C., Parfitt, M., Legrain, S., Pérez-Tur, J., Brousseau, T., Evans, A., et al. (1994). Apolipoprotein E, ɛ4 allele as a major risk factor for sporadic early and late-onset forms of Alzheimer’s disease: analysis of the 19q13.2 chromosomal region. Hum Mol Genet 3, 569–574.
Corbo, R. M., and Scacchi, R. (1999). Apolipoprotein E (APOE) allele distribution in the world. Is APOE * 4 a ‘thrifty’ allele? Ann Hum Genet 63, 301–310.
Davis, A. A., Inman, C. E., Wargel, Z. M., Dube, U., Freeberg, B. M., Galluppi, A., et al. (2020). APOE genotype regulates pathology and disease progression in synucleinopathy. Sci Transl Med 12, 2–11. doi: 10.1126/scitranslmed.aay3069
El-Lebedy, D., Raslan, H. M., and Mohammed, A. M. (2016). Apolipoprotein E gene polymorphism and risk of type 2 diabetes and cardiovascular disease. Cardiovasc Diabetol 15:12. doi: 10.1186/s12933-016-0329-1
El-Tagui, M. H., Hamdy, M. M., Shaheen, I. A., Agha, H., and Abd-Elfatah, H. A. (2013). Apolipoprotein E gene polymorphism and the risk of left ventricular dysfunction among Egyptian β-thalassemia major. Gene 524, 292–295. doi: 10.1016/j.gene.2013.03.134
Emamzadeh, F. N., Aojula, H., McHugh, P. C., and Allsop, D. (2016). Effects of different isoforms of apoE on aggregation of the α-synuclein protein implicated in Parkinson’s disease. Neurosci Lett 618, 146–151. doi: 10.1016/j.neulet.2016.02.042
Fahmy, E. M., Rabah, A. M., Hashem, S. E., Rashed, L. A., Deraz, H. A., and Ismail, R. S. (2024). Serum Apo lipoprotein E, Apo lipoprotein E gene polymorphisms, and Parkinson’s disease. Neurol India 72, 319–325. doi: 10.4103/ni.ni_940_21
Fawzy, M. S., Toraih, E. A., Aly, N. M., Fakhr-Eldeen, A., Badran, D. I., and Hussein, M. H. (2017). Atherosclerotic and thrombotic genetic and environmental determinants in Egyptian coronary artery disease patients: a pilot study. BMC Cardiovasc Disord 17:26. doi: 10.1186/s12872-016-0456-3
Federoff, M., Jimenez-Rolando, B., Nalls, M. A., and Singleton, A. B. (2012). A large study reveals no association between APOE and Parkinson’s disease. Neurobiol Dis 46, 389–392. doi: 10.1016/j.nbd.2012.02.002
Fernández-Calle, R., Konings, S. C., Frontiñán-Rubio, J., García-Revilla, J., Camprubí-Ferrer, L., Svensson, M., et al. (2022). APOE in the Bullseye of neurodegenerative diseases: impact of the APOE genotype in Alzheimer’s disease pathology and brain diseases. Mol Neurodegener 17:62. doi: 10.1186/s13024-022-00566-4
Fujioka, H., Phelix, C. F., Friedland, R. P., Zhu, X., Perry, E. A., Castellani, R. J., et al. (2013). Apolipoprotein E4 prevents growth of malaria at the Intraerythrocyte stage: implications for diff erences in racial susceptibility to Alzheimer’s disease. J Health Care Poor Underserved 24, 70–78. doi: 10.1353/hpu.2014.0009
Fyfe, I. (2020). APOE*ε4 promotes synucleinopathy. Nat Rev Neurol 16:185. doi: 10.1038/s41582-020-0335-5
Galal, A. A., Abd Elmajeed, A. A., Elbaz, R. A., Wafa, A. M., and Elshazli, R. M. (2021). Association of Apolipoprotein E gene polymorphism with the risk of T2DM and obesity among Egyptian subjects. Gene 769:145223. doi: 10.1016/j.gene.2020.145223
Gallardo, G., Schlüter, O. M., and Südhof, T. C. (2008). A molecular pathway of neurodegeneration linking α-synuclein to ApoE and Aβ peptides. Nat Neurosci 11, 301–308. doi: 10.1038/nn2058
Gomaa, H. E., Mahmoud, M., Saad, N. E., Saad-Hussein, A., Ismail, S., Thabet, E. H., et al. (2018). Impact of Apo E gene polymorphism on HCV therapy related outcome in a cohort of HCV Egyptian patients. J Genetic Eng Biotechnol 16, 47–51. doi: 10.1016/j.jgeb.2017.10.008
Halim, E. F., Reda, A. A., Hendi, A. A. K., Zaki, S. A., Essa, E. S., and Khalifa, A. S. (2012). Apolipoprotein E gene variants as a risk factor for coronary artery disease in type 2 diabetic Egyptian patients. Egypt J Immunol 19, 1–10.
Hassan, N. E. (2016). Apo lipoprotein E polymorphism as risk factor for lipid profile disturbance among obese Egyptian females. Res J Pharm, Biol Chem Sci 7, 1826–1833.
Hassan, N. E., El Ashmawi, A. A., El-Masry, S. A., Zarouk, W. A., Mira, M. F., El-Saeed, G. S., et al. (2019). Metabolic syndrome in a sample of Egyptian adolescent girls and its association with apolipoprotein E. J Paediatr Child Health 55, 1344–1350. doi: 10.1111/jpc.14419
Hemeda, A. A., Ahmad Mohamed, A., Aziz, R. K., Abdel-Hakeem, M. S., and Ali-Tammam, M. (2021). Impact of IL10, MTP, SOD2, and APOE gene polymorphisms on the severity of liver fibrosis induced by HCV genotype 4. Viruses 13:714. doi: 10.3390/v13040714
Huebbe, P., and Rimbach, G. (2017). Evolution of human apolipoprotein E (APOE) isoforms: gene structure, protein function and interaction with dietary factors. Ageing Res Rev 37, 146–161. doi: 10.1016/j.arr.2017.06.002
Hughes, A. J., Daniel, S. E., Kilford, L., and Lees, A. J. (1992). Accuracy of clinical diagnosis of idiopathic Parkinson’s disease: a clinico-pathological study of 100 cases. J Neurol Neurosurg Psychiatry 55, 181–184.
Husain, M. A., Laurent, B., and Plourde, M. (2021). APOE and Alzheimer’s disease: from lipid transport to physiopathology and therapeutics. Front Neurosci 15:15. doi: 10.3389/fnins.2021.630502
Jo, S., Kim, S. O., Park, K. W., Lee, S. H., Hwang, Y. S., and Chung, S. J. (2021). The role of APOE in cognitive trajectories and motor decline in Parkinson’s disease. Sci Rep 11:7819. doi: 10.1038/s41598-021-86483-w
Kanekiyo, T., Xu, H., and Bu, G. (2014). ApoE and Aβ in Alzheimer’s disease: accidental encounters or partners? Neuron 81, 740–754. doi: 10.1016/j.neuron.2014.01.045
Kang, S. J., Kim, S. J., Noh, H. R., Kim, B. J., Kim, J. B., Jin, U., et al. (2022). Neuronal ApoE regulates the cell-to-cell transmission of α-Synuclein. Int J Mol Sci 23:8311. doi: 10.3390/ijms23158311
Kapan, A., Haider, S., Wakolbinger, M., and Spatt, J. (2023). Associations of apolipoprotein ε4 genotypes with motor and nonmotor symptoms in Parkinson’s disease: a cross-sectional study. Mov Disord Clin Pract 10, 1611–1619. doi: 10.1002/mdc3.13862
Kurz, M. W., Dekomien, G., Nilsen, O. B., Larsen, J. P., Aarsland, D., and Alves, G. (2009). APOE alleles in Parkinson disease and their relationship to cognitive decline: a population-based, longitudinal study. J Geriatr Psychiatry Neurol 22, 166–170. doi: 10.1177/0891988709332945
Lambert, J. C., Ibrahim-Verbaas, C. A., Harold, D., Naj, A. C., Sims, R., Bellenguez, C., et al. (2013). Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 45, 1452–1458. doi: 10.1038/ng.2802
Li, J., Luo, J., Liu, L., Fu, H., and Tang, L. (2018). The genetic association between apolipoprotein E gene polymorphism and Parkinson disease. Medicine 97:e12884. doi: 10.1097/MD.0000000000012884
Liampas, I., Kyriakoulopoulou, P., Siokas, V., Tsiamaki, E., Stamati, P., Kefalopoulou, Z., et al. (2024). Apolipoprotein E gene in α-Synucleinopathies: a narrative review. Int J Mol Sci 25:1795. doi: 10.3390/ijms25031795
Mahmoud, A. A., Yousef, L. M., and Zaki, N. A. E. (2016). Apolipoprotein E gene polymorphism in Egyptian acute coronary syndrome patients. Egyptian J Medical Human Genetics 17, 99–103. doi: 10.1016/j.ejmhg.2015.08.001
Okubadejo, N. U., Okunoye, O., Ojo, O. O., Arabambi, B., Akinyemi, R. O., Osaigbovo, G. O., et al. (2022). APOE E4 is associated with impaired self-declared cognition but not disease risk or age of onset in Nigerians with Parkinson’s disease. NPJ Parkinsons Dis 8:155. doi: 10.1038/s41531-022-00411-x
Panza, F., Frisardi, V., Seripa, D., D’Onofrio, G., Santamato, A., Masullo, C., et al. (2012). Apolipoprotein E genotypes and neuropsychiatric symptoms and syndromes in late-onset Alzheimer’s disease. Ageing Res Rev 11, 87–103. doi: 10.1016/j.arr.2011.06.005
Postuma, R. B., Berg, D., Stern, M., Poewe, W., Olanow, C. W., Oertel, W., et al. (2015). MDS clinical diagnostic criteria for Parkinson’s disease. Mov Disord 30, 1591–1601. doi: 10.1002/mds.26424
Qin, W., Li, W., Wang, Q., Gong, M., Li, T., Shi, Y., et al. (2021). Race-related association between APOE genotype and Alzheimer’s disease: a systematic review and Meta-analysis. J Alzheimers Dis 83, 897–906. doi: 10.3233/JAD-210549
Ramadan, A., Foda, B. M., Noha, A., Refaat, O., Saleh, A. A., and Fawzy, A. (2019). Association analysis of ApoE gene polymorphisms among Egyptian patients with Alzheimer’s disease. Gene Rep 14, 65–71. doi: 10.1016/j.genrep.2018.10.017
Rizig, M., Ojo, O. O., Athanasiou-Fragkouli, A., Agabi, O. P., Oshinaike, O. O., Houlden, H., et al. (2021). Negative screening for 12 rare LRRK2 pathogenic variants in a cohort of Nigerians with Parkinson’s disease. Neurobiol Aging 99, 101.e15–101.e19. doi: 10.1016/j.neurobiolaging.2020.09.024
Sun, R., Yang, S., Zheng, B., Liu, J., and Ma, X. (2019). Apolipoprotein E polymorphisms and Parkinson disease with or without dementia: a Meta-analysis including 6453 participants. J Geriatr Psychiatry Neurol 32, 3–15. doi: 10.1177/0891988718813675
Taha, M., Ibrahim, M. M. M., and Sedrak, H. (2024). Association of epistatic effects of MTHFR, ACE, APOB, and APOE gene polymorphisms with the risk of myocardial infarction and unstable angina in Egyptian patients. Gene 895:147976. doi: 10.1016/j.gene.2023.147976
Wenjuan, Y., Pingmin, W., Yang, X., Yan, G., Xiaoshan, L., Yue, S., et al. (2016). Relationship between apolipoprotein E gene polymorphism and Parkinson’s disease: a meta-analysis. nt J Clin Experimental Med 9, 5334–5346.
Williams-Gray, C. H., Goris, A., Saiki, M., Foltynie, T., Compston, D. A. S., Sawcer, S. J., et al. (2009). Apolipoprotein E genotype as a risk factor for susceptibility to and dementia in Parkinson’s disease. J Neurol 256, 493–498. doi: 10.1007/s00415-009-0119-8
Yang, L. G., March, Z. M., Stephenson, R. A., and Narayan, P. S. (2023). Apolipoprotein E in lipid metabolism and neurodegenerative disease. Trends Endocrinol Metab 34, 430–445. doi: 10.1016/j.tem.2023.05.002
Zhang, W., Xiao, D., Mao, Q., and Xia, H. (2023). Role of neuroinflammation in neurodegeneration development. Signal Transduct Target Ther 8:267. doi: 10.1038/s41392-023-01486-5
Keywords: Parkinson’s disease, genetics, APOE, KASP, Egyptian
Citation: Khedr EM, William MB, Elhosseiny AA, Shalash A, Fawi G, Yousef MH, El-Jaafary S, Lee H, Jama A, Koraym M, Helmy A, Salah Y, George P, Haridy NA, Nabhan S, Atputhavadivel A, Elfarrash S, Ragab G, Hegazy MT, Elsaid Y, Gabr AS, Shebl N, Aly L, Abdelwahhab N, Belal TM, Elsayed NAB, El-Gamal M, Elgamal S, Ragab S, Mekky J, Houlden H, Rizig M and Salama M (2025) APOE genetic variability in an Egyptian cohort of PD. Front. Neurosci. 19:1579968. doi: 10.3389/fnins.2025.1579968
Edited by:
Mounia Chami, Institut National de la Santé et de la Recherche Médicale (INSERM), FranceReviewed by:
Jerry Lorren Dominic, Jackson Memorial Hospital, United StatesAfef Achouri, Hôpital Charles-Nicolle, Tunisia
Copyright © 2025 Khedr, William, Elhosseiny, Shalash, Fawi, Yousef, El-Jaafary, Lee, Jama, Koraym, Helmy, Salah, George, Haridy, Nabhan, Atputhavadivel, Elfarrash, Ragab, Hegazy, Elsaid, Gabr, Shebl, Aly, Abdelwahhab, Belal, Elsayed, El-Gamal, Elgamal, Ragab, Mekky, Houlden, Rizig and Salama. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mie Rizig, bS5yaXppZ0B1Y2wuYWMudWs=; Mohamed Salama, bW9oYW1lZC1zYWxhbWFAYXVjZWd5cHQuZWR1
†These authors share senior authorship
‡These authors share last authorship
 Eman M. Khedr
Eman M. Khedr Martina B. William2,3
Martina B. William2,3 Aliaa A. Elhosseiny
Aliaa A. Elhosseiny Ali Shalash
Ali Shalash Mohamed H. Yousef
Mohamed H. Yousef Shaimaa El-Jaafary
Shaimaa El-Jaafary Sara Elfarrash
Sara Elfarrash Gaafar Ragab
Gaafar Ragab Mohamed Tharwat Hegazy
Mohamed Tharwat Hegazy Nourhan Shebl
Nourhan Shebl Mohamed El-Gamal
Mohamed El-Gamal Shimaa Elgamal
Shimaa Elgamal Jaidaa Mekky
Jaidaa Mekky Henry Houlden
Henry Houlden Mie Rizig
Mie Rizig Mohamed Salama
Mohamed Salama