- 1Department of Biochemistry, School of Basic and Applied Sciences, Central University of Punjab, Bathinda, India
- 2Department of Urology, Case Western Reserve University, Cleveland, OH, United States
- 3The Urology Institute, University Hospitals Cleveland Medical Center, Cleveland, OH, United States
- 4Department of Nutrition, Case Western Reserve University, Cleveland, OH, United States
- 5Divison of General Medical Sciences, Case Comprehensive Cancer Center, Cleveland, OH, United States
- 6Department of Urology, Louis Stokes Cleveland Veterans Affairs Medical Center, Cleveland, OH, United States
Tumor metastasis is a sequential event accounting for numerous cancer-related fatalities worldwide. The process of metastasis serially involves invasion, intravasation, extravasation, and tumor growth at the secondary site. Migration and invasion enhancer 1 (MIEN1) is a membrane associated protein overexpressed in various human cancers. Biological activity of MIEN1 is driven by geranylgeranyltransferase-I mediated prenylation at CAAX motif and methylation of the prenylated protein that anchors MIEN1 into the cellular membrane. Post-translationally modified MIEN1 interacts with Syk kinase and Annexin A2 protein; polymerizes G-actin and stabilizes F-actin filament; induces focal adhesion kinase phosphorylation and decrease cofilin phosphorylation implicated in both invasion and metastasis of different cancer types. In the present review, we discuss the structure, function, and involvement of MIEN1 in cancer progression. We also highlight the future prospects of MIEN1 as an emerging molecule and novel target in cancer cell invasion and metastasis.
Introduction
Metastasis is an intricate process of cell dissemination that primarily depends on the ability of tumor cells to detach from basement membrane through dynamic reorganization of the cytoskeleton proteins. MIEN1 has been identified as a primary regulator of cancer cell migration and invasion. The protein is overexpressed in human breast, prostate, colorectal, gastric, ovarian, squamous cell carcinoma and non-small cell lung cancer (NSCLC) (1–7). Emerging literature suggest MIEN1 as a new tumor-specific target protein as it facilitates cancer progression that plays key role in distinct processes of migration/invasion of cancer cells. MIEN1 protein has several aliases such as hepatitis B virus (HBV) X-Transactivated Gene 4 Protein, HBV XAg-Transactivated Protein 4, C17orf37 (Chromosome 17 Open Reading Frame 37), RDX12, XTP4, ORB3, and C35. In this review, we discuss the structure, function, expression and involvement of MIEN1 in cancer progression. We also highlight future prospects of MIEN1 as novel drug target against cancer cell invasion and metastasis.
Structure of MIEN1
MIEN1 is a membrane-anchored protein located in the 17q12 region of the human chromosome which is 505 nucleotide upstream to 3′ ERBB2 oncogene, inside the hot-spot locus of cancer (Figure 1a) (1, 8). MIEN1 encodes 115 amino acids and 12 kDa molecular weight protein. The spot contains multiple genes which are implicated in cancer initiation and development. Solution structure of MIEN1 reveals the presence of thioredoxin-like fold having a redox-active motif. Four stranded β-sheet and two α-helices forms a central α/β core domain of the protein (9). Two helices (α1 and α2) are comprised of Phe35-Gln49 and Glu86-Asn98 residues. Four β-strands (β1, β2, β3, and β4) ranges from Arg24 to Tyr29, Glu54 to Leu59, Phe65 to Glu68 and Val74 to Ser76 residues respectively (Figures 1b,c). MIEN1 hydrophobic core constitutes of Tyr39, Leu42, Val46, Tyr50, Phe65, Val74, Leu89, and Ile (53, 67, 69, 90, and 93 positions) residues. Approximately 15 residues at each N- (Met1-Glu16) and C- (Glu103-Leu115) terminal of the protein present an unordered structure. 1H-15N heteronuclear NOE experiments exhibited that MIEN1 backbone is flexible due to a lack of stabilizing long-range interactions with the rest of the protein (9). MIEN1 protein has reduced oxidative state, however, the oxidation state does not impede its protein activity. CXXC motif, present at C-terminus is directly involved in oxidation-reduction reaction. A cluster of some aromatic residues at MIEN1 active site might be responsible for overall protein structure stabilization and its binding interaction partners. The CXXC motif and aromatic amino acid cluster are located apart from each other (9). The backbone residues of MIEN1 in both oxidized and reduced state is observed to be within the same conformation, but differences in the active motif region were found due to amide resonance. MIEN1 possess a disulfide bond in its active sites. However, reduction of the active-site disulfide bond during reduction to oxidation state does not affect MIEN1 folding pattern (9). The MIEN1 oxidation-reduction potential plays critical role in AKT phosphorylation (9).
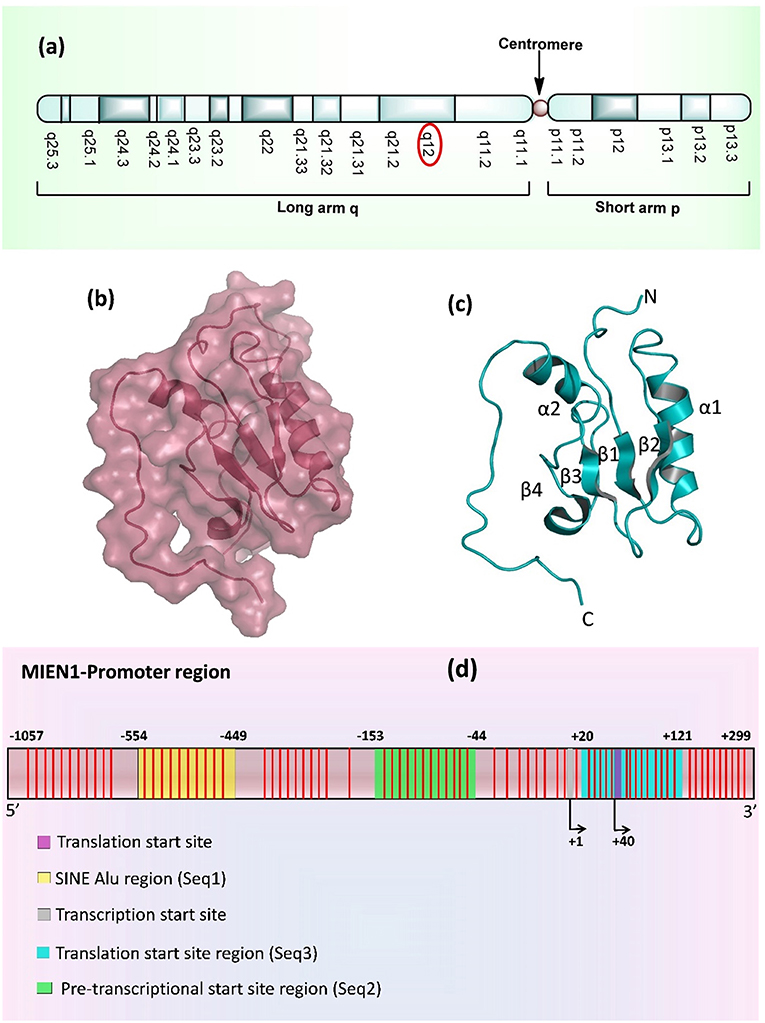
Figure 1. (a) Location of MIEN1 gene on chromosome 17 (q12 position) (b) Surface structure of the MIEN1 protein (PDB: 2LJK), (c) Ribbon representation of MIEN1 protein having α (α1 and α2) helices and β (β1, β2, β3, and β4) sheets; and N- and C-terminus (d) MIEN1 methylated promoter sequence. CpG islands are represented in red lines.
MIEN1 promoter region is comprised of several CpG islands, CpG dyads and a short interspersed nuclear element (SINE) Alu repeat. Alu are the short stretch of DNA, that form the Alu family of repetitive DNA elements. Alu elements of more than 300 base pairs are known as SINEs. Approximately, one-third of the total CpG sites (methylation sites) in the genome resides in Alu elements. Normally Alu elements are methylated and transcriptionally inactive (10, 11). Various sites in the Alu element of MIEN1 promoter region remain hypermethylated in the normal cells (12). The SINE Alu elements belong to a category of gene regulatory elements known as retrotransposons that have been shown to become less mobile upon genetic regulation (13–16). Genome wide study has established that SINEs are present in most human cancers near to transcriptional start sites (TSS), which are hypomethylated in cancer (15, 17). SINEs enriched regions present at about 550 bases from the TSS of MIEN1. The methylation landscape and promoter region of MIEN1 is shown in Figure 1d.
MIEN1 Expression
MIEN1 is predominantly present in the cytosol with little plasma membrane-associated expression. The biological function of MIEN is to increase cell migration by inducing filopodia formation at the leading edge of migrating cells (18). It also plays a key role in regulation of apoptosis, possibly through control of CASP3, and is involved in redox-related process. Microarray analysis demonstrates the expression of MIEN1 in various compartments in the blood, brain, liver, pancreas, lung, thyroid, and testis (Figure 2). Overexpression of MIEN1 was initially identified in breast and colon cancer. MIEN1 expression positively correlates with the grade and stage of breast cancer compared with minimal expression in normal tissues (1). Thus, the differerential expression of MIEN1 in normal vs. cancer cells proposes it to be a novel tumor biomarker. In patients with metastatic breast cancer, aberrant expression of MIEN1 has been noted in distant metastatic sites such as lungs and liver, suggesting a possible role of MIEN1 protein in metastatic dissemination of tumor cells (1, 7). In prostate cancer, MIEN1 is overexpressed in the higher grades of prostate adenocarcinoma compared with low expression in normal or benign prostatic tissue (2). Studies indicate that MIEN1 overexpression is linked to genomic amplification of the ERBB2 locus, since its sufficient expression is noted in the ERBB2 nonamplified breast and prostate tumors, suggesting that MIEN1 might have an independent functional promoter (19).
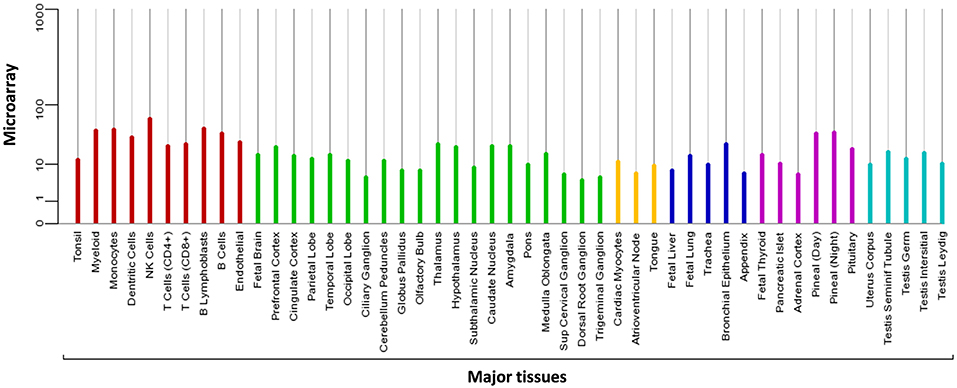
Figure 2. Microarray data representing the expression pattern of MIEN1 in various tissues. The measurements were obtained for 76 normal human tissues and compartments hybridized against HG-U133A. The Affymetrix MAS5 algorithm was used for array processing and probe sets were averaged per gene [https://www.genecards.org/cgi-bin/carddisp.pl?gene=MIEN1].
Posttranslational Modification of MIEN1
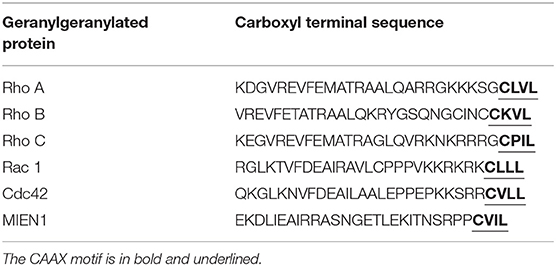
Modification of eukaryotic cellular protein with isoprenoid unit is known as prenylation. Protein prenylation involves transfer of either a farnesyl or a geranyl-geranyl moiety to C-terminal cysteine(s) of the target protein. This process mediates protein–protein interactions and protein–membrane interactions having decisive function in a biological system. Three different enzymes are known to be involved in protein prenylation, namely Ras converting CAAX endopeptidase 1 (RCE1), geranylgeranyltransferase (GGTase 1) and farnesyltransferase (FTase). The cysteine residue of CAAX motif is modified by either farnesylation (15 carbon chain addition by protein farnesyltransferase enzyme/FTase) or geranylgeranylation (addition of 20-carbon chain by GGTase-I) during prenylation (20, 21). Most of the prenylated proteins belong to the CAAX family, which are later post-translationally altered by the addition of isoprenyl groups. CAAX denotes a protein comprised of a CAAX motif at the C-terminal; where “C” is the cysteine amino acid that acts as the isoprenoid accessory site; “A” denotes one of the amino acid from Ile, Val, Leu, Ala, Met and Pro; and “X” signifies any of 22 amino acids (22). Farnesyltransferase (EC 2.5.1.58) is the prenyl transferase group enzyme, which adds fifteen-carbon isoprenoid (farnesyl group) to CAAX motif of the proteins. Geranylgeranyltransferase (GGTase 1) is another prenyl transferase group which adds twenty-carbon isoprenoid (geranylgeranyl group) to the CAAX motif of proteins (23–25). CAAX protein sequence, together with prenylated motif, determines the different localization of the protein, at plasma membrane/endomembrane or at a cytosolic location (26). Therefore, any modification at the C-terminus of these proteins might affect their stability, function, localization, and protein-protein interaction. The regulatory capacity of prenylated proteins plays confirmatory role in cellular signaling and pathophysiological conditions. The prenylation of several oncogenes including RAS GTPases, is known to be involved in cancer progression through migration, invasion, increased proliferation and filopodia formation (27).
MIEN1 has a functional isoprenylation “CAAX” motif at the C-terminal tail that is post-translationally modified by geranyl-geranyl transferase-I (GGTase-I), a cytosolic enzyme at Cys-112 (18). Geranylgeranylation of MIEN1 at the CAAX motif supports its anchorage to the cytosolic part of the plasma membrane. A study demonstrated that MIEN1 prenylation is accountable for the filopodia formation and potential directional migration (18). An interesting feature of membrane anchored MIEN1 is the existence of a consensus sequence for prenylation comprising of the last four amino acids, CVIL, at the C-terminal end (Table 1).
Addition of isoprenyl group in the MEIN1 protein leads to a two-step supplementary post-prenylation process including cleavage of the last three amino acid at C-terminus by an endoprotease enzyme, Rce1 (Ras-converting CAAX Endopeptidase 1) and prenylated-cysteine methylation executed by ICMT (isoprenylcysteine-O-carboxyl methyltransferase) enzyme (18) (Figure 3). Galectins are another family of proteins that bind to activated RAS proteins by interacting the farnesyl lipid and an adjoining RAS region thereby stabilizing their membrane anchorage and enhance signaling (28). Likewise the area is open to identification of protein(s) which may stabilize the membrane-MIEN1 association.
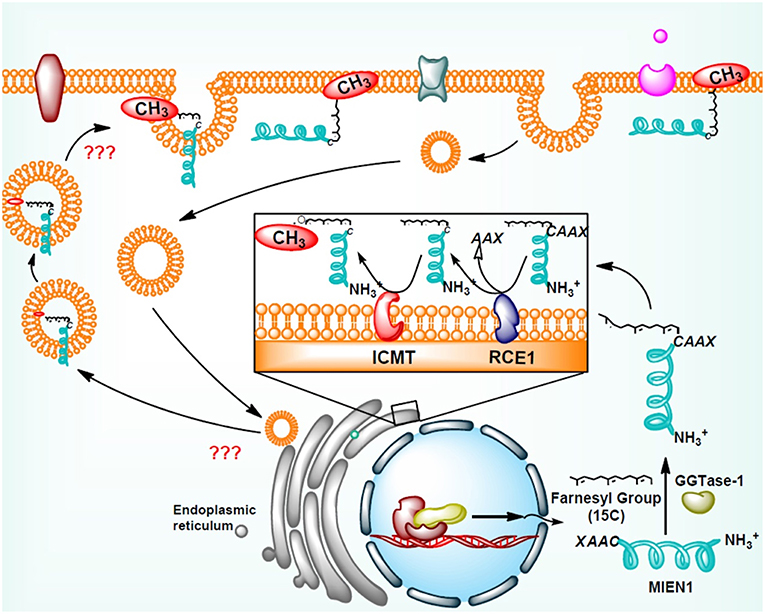
Figure 3. Post-translational modification of MIEN1 protein by protein geranylgeranyltransferase-I (GGTase-I).
Methylation and Demethylation of MIEN1
Dasgupta et al. demonstrate for the first time that MIEN1 protein undergoes prenylation during post translational modification (18). To-date, there is no information available regarding the methylation and demethylation of prenylated MIEN1 protein. It is known that MIEN1 protein, like RHO family and Cdc-42 proteins, have CAXX motif at the C-terminus. RCE1 enzyme removes AXX from the CAXX motif of these proteins, whereas the left C-terminus is methylated having a higher potential to attach to the cellular membrane. Reports suggest that MIEN1 is a membrane associated protein and its biological activity is dependent upon prenylation and subsequent methylation at the C-terminus of the CAXX motif (18). Post-translational modification of prenylated proteins having a CAAX motif changes the hydrophilic nature of the protein toward lipophilicity, which facilitates their attachment to plasma membrane (18). The study further reported that prenylation of MIEN1 protein is associated with filopodia formation and directional migration. Thus, the regulatory mechanism involved in the methylation/de-methylation of the protein might suggest it to be a potential target for different cancers.
MIEN1 and Metastasis
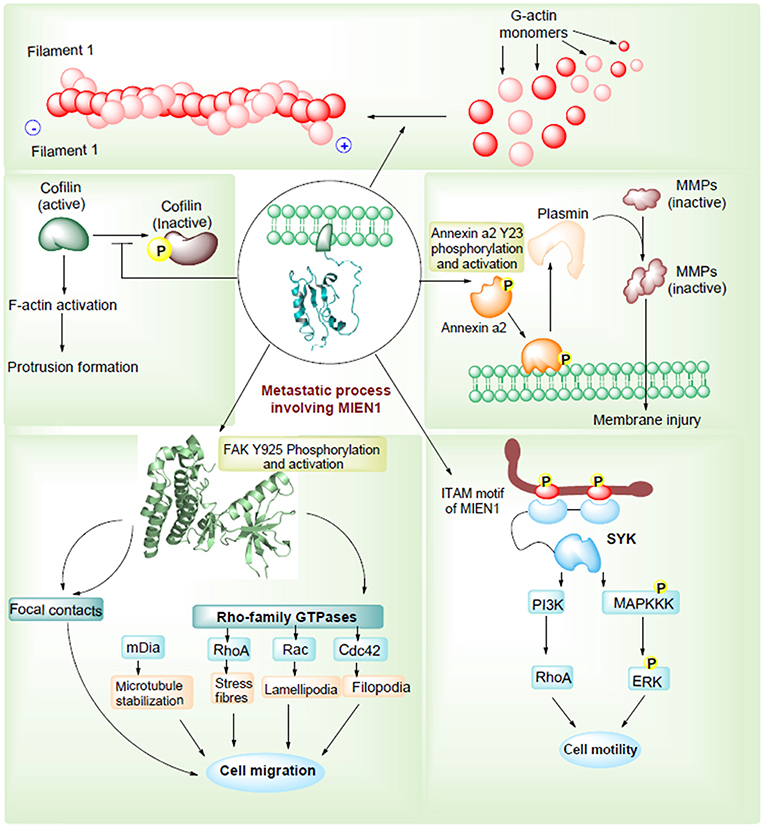
Epithelial-mesenchymal transition (EMT) permits tumor cells to dedifferentiate enhancing the invasive potential during the metastatic process. During EMT, expression of adhesion molecules, such as integrins and cadherins, and their receptors are involved in increased cellular motility in tumor cells (29). Besides, metalloproteases, cytokines and chemokines are important factors that regulate cell motility and metastasis. Annexin A2 (AnxA2) is a Ca2+ dependent phospholipid binding protein that acts as an extracellular proteolytic molecule regulating plasmin generation. Plasmin modifies the tumor microenvironment by enhancing the extracellular matrix protein degradation and release of growth factors, which are all associated with cancer progression (30). Higher levels of AnxA2, in association with STAT3, regulates proliferation, invasion, and migration of cancer cells. MIEN1 overexpression increases AnxA2 phosphorylation at Tyr23 residue, which stimulates cell surface translocation and catalyzes activation of proteolytic activity of AnxA2 (31). Thus, MIEN1-AnxA2 protein-protein interaction might be regarded as one of the mechanisms facilitating tumor metastasis (Figure 4). Katz et al. demonstrated that Syk kinase, a downstream mediator of MIEN1 signaling mediates breast epithelial cell transformation in 3D cultures (32). In the same study, high expression of MIEN protein was associated with various transformation events, such as colony formation, invasion and large acinar formation. Besides transformation, EMT changes were also observed including a decreased expression of E-cadherin and keratin-8 (33). Actin cytoskeleton has been extensively known for its importance in metastasis. Recently, MIEN1 protein was found to localize in a concerted manner beneath the action protrusion structures of migratory cancer cells (Figure 4). The study reported that MIEN1 levels regulate the actin-protrusive structures in migrating cells (34). MIEN1 is also involved in the F-actin polymerization (from G-actin monomers) and stabilization. MIEN1-mediated increase in focal adhesion kinase (FAK) and decrease in cofilin phosphorylation at Tyr-925 and Ser-3 are associated with actin dynamics and cellular adhesion in migratory cancer cells (34).
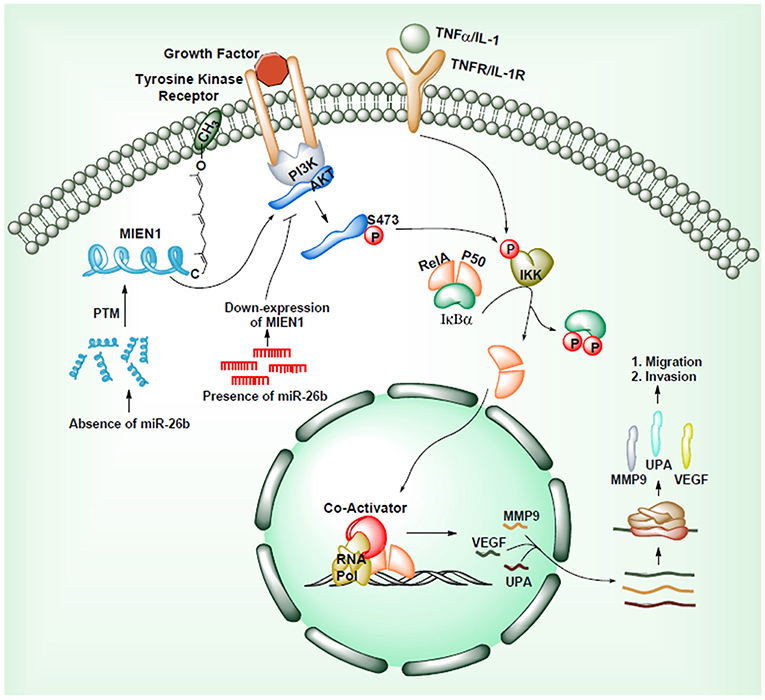
Another mechanism through which MIEN1 regulates invasion and metastasis is via the PI3K/Akt pathway (35). Activation of Akt can lead to NF-kB activation resulting in an increase in matrix metallopeptidase 9 (MMP-9), urokinase plasminogen activator (uPA) and vascular endothelial growth factor (VEGF). These are key proteases and angiogenic factors that are downstream of NF-kB pathway, facilitating migration and invasion of cancer cells. Studies have shown that the absence of miR-26b results to post translational modification of the MIEN1 to generate mature/functional protein. Overexpression of MIEN1 induces the PI3K/Akt pathway via phosphorylation of the S473 residue of Akt proteins. Ultimately phosphorylation of IκBα protein allows the release of RelA and P50 proteins, which can then translocate into the nucleus, interact with different coactivators and enhance the expression of invasion and migration gene. In the presence of miR-26b, decreased levels of MIEN1 functional protein results in reduced migration and invasion (Figure 5). Overall, in most studied cancers (1–7), MIEN1 plays a critical role in tumor dissemination and represents a potential target of metastasis in cancer cells.
Association of MIEN1 With Other Proteins
The STRING Database has been used to establish possible interactions between other proteins. In this analysis, 10 proteins were recognized by interaction analysis and a network mapping with MIEN1 (Figure 6). The possible interaction or association of MIEN1 is shown in Table 2. String database demonstrates direct interaction between MIEN1 and other proteins, such as TCAP, STARD3, PGAP3, GRB7, ERBB2, PRDX1, SEPW1, GPX1, VIMP, and SELT, respectively. It should be noted that these interactions are based on co-expression and text mining predictions (Figure 6). Further studies should be designed to evaluate the interactive response of MIEN1 with other proteins.
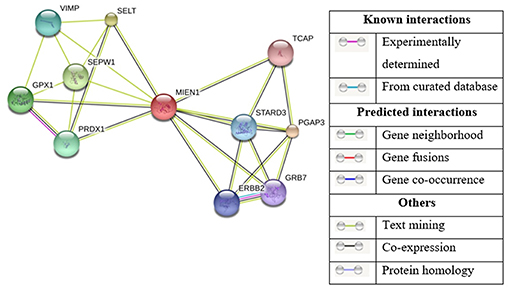
Figure 6. Based on the STRING database, the network of MIEN1 and other protein interactions. Ten differentially expressed proteins formed a network map with the MIEN1 protein.
MIEN1 and Drug Resistance
Little information is available regarding MIEN1 and drug resistance. Upregulated Dnp73, an isoform of p53 is associated with increased tumor potential. Leung et al. reported that MIEN1 in association with Dnp73 promotes tumor development and its involvement in cisplatin resistance in cancer cells (5).
MIEN1 and Cancer
MIEN1 is overexpressed in several human cancers including breast, prostate, colorectal, gastric, ovarian, squamous cell carcinoma and non-small cell lung cancer (NSCLC), therefore playing an important role in biological processes underlying tumor metastasis. Epigenetic and genetic studies performed on MIEN1 in various human cancers have been summarized in Table 3. Loss or gain of MIEN1 activity, and its relationship with metastasis and invasions in different cancer cells and tissue specimens is shown in Tables 4, 5, respectively. Below we summarize studies which shed light on the involvement of MIEN1 protein in various cancers.

Table 4. Effect of loss/gain of MIEN1 protein and its effects on metastasis and invasiveness in different cancer.
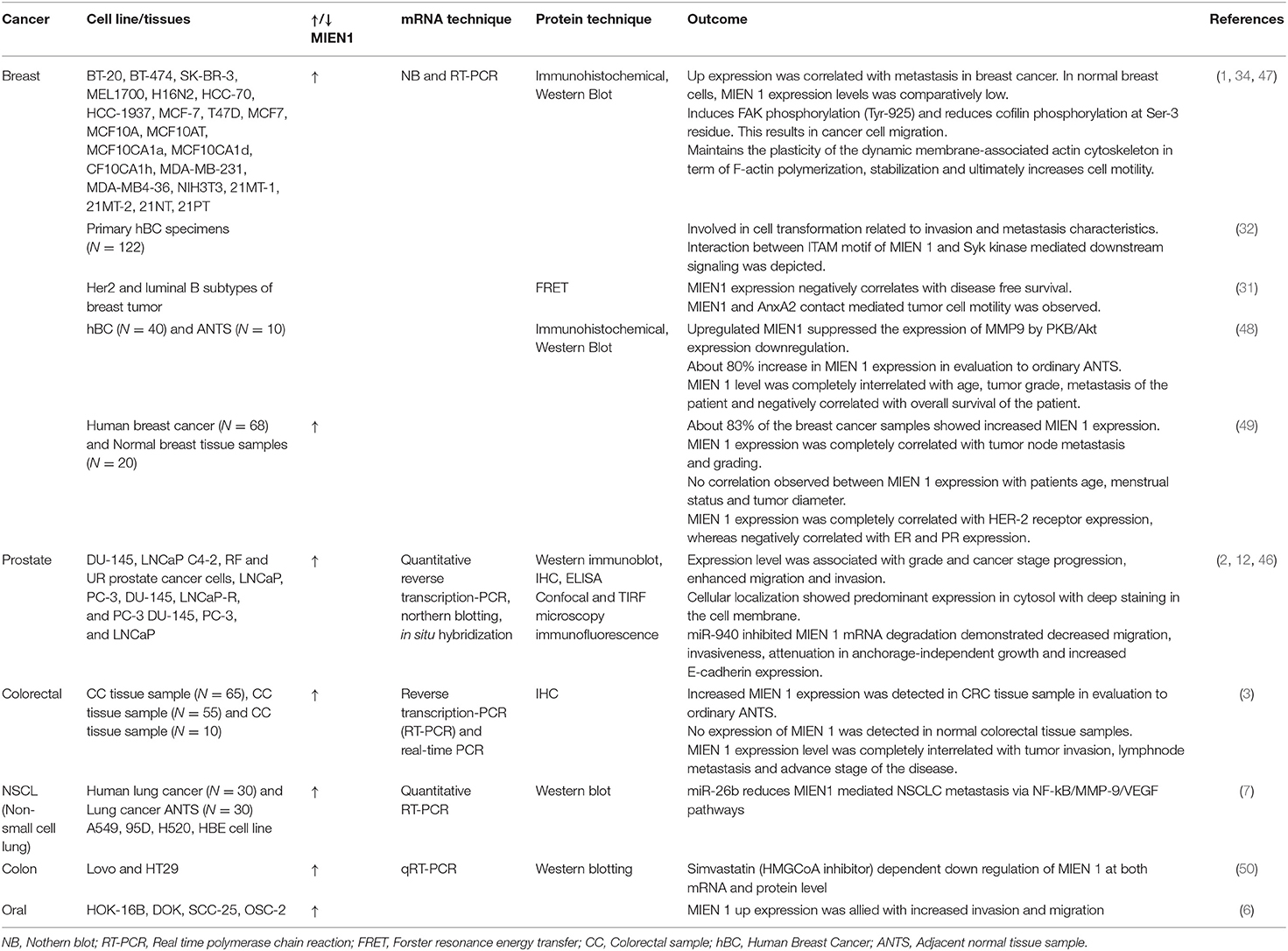
Table 5. MIEN 1 expression pattern in various cancer cell lines and tissue samples and their research outcomes.
Breast Cancer
Evans et al. used immunohistochemical staining technique to study C35 expression in human breast cancer and normal tissue specimens (1). The study revealed the excessive expression levels of the protein throughout, starting from tumor initiation indicated by DCIS (ductal carcinoma in situ) and LCIS (lobular carcinoma in situ) to late stage metastasis. Approximately 50% of the studied samples exhibited high MIEN1 levels especially in the distant metastasis specimens. These specimens also showed poor prognosis characterized by lymphocytic infiltration and ERBB2 positivity. The study indicates that MIEN1 might act as a potential early detection biomarker in breast cancer. In addition, MIEN1 levels were detected in distant tissues, mainly bone, brain, liver and lung which strengthen the role of MIEN1 during disease progression (1). Additional studies provide evidence for aberrant expression of MIEN1 in different stages of breast cancer with low and/or absence of expression in normal tissues, indicating its development as a potential biomarker. Previous studies describe the involvement of ITAM motif containing protein in mammary epithelial cell transformation and mammary carcinoma development (51–53). ITAM-transformed cells with the support of its downstream Syk signaling activation is closely linked to EMT progression in cancer cells (32). A recent report identified that MIEN1 protein contains ITAM motif and is involved in metastasis through interaction with Syk protein (32). Further screening of 122 invasive breast cancer specimens using microarray technique demonstrated high levels of MIEN1 protein in these tissues, having a positive correlation with HER2 expression (32). Other studies, such as association of MIEN1 with actin cytoskeletal dynamics, ITAM and AnxA2-mediated metastasis in breast cancer progression have been previously documented (31). Treuren et al. generated MIEN1 knockout (KO) breast cancer cell lines using CRISPR/Cas9 (Clustered Regularly Interspaced Short Palindromic Repeat–CRISPR associated protein 9) system. Authors reported that MIEN1 gene deletion has no effect on the proliferation and survival of breast cancer cells, necessitating further studies to understand the role of MIEN1 in breast cancer metastasis (54).
Prostate Cancer
Deregulation of MIEN1 protein level in prostate cancer was first reported in 2009 (2). The study demonstrated high expression of MIEN1 in prostate cancer cell lines and tumors in comparison to normal tissues (low expression) (2). The study established the role of MIEN1 overexpression with migration and invasion of prostate tumor cells mediated via Akt/NF-kB signaling and activation of downstream effector proteins. Akt phosphorylation at Ser473 subsequently phosphorylates IκB and IκBa in a sequential manner (Figure 5). Phosphorylation of IκBa leads to its degradation releasing NF-κB homo/heterodimers to translocate into the nucleus. MIEN1 overexpression enhances metastasis by augmenting NF-κB activation and DNA binding potential resulting in increased expression of its downstream target genes (2, 35).
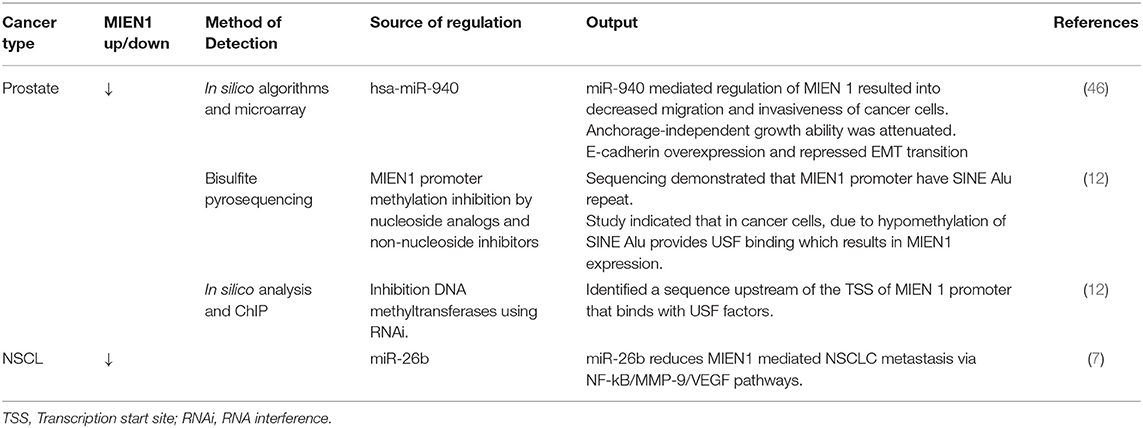
Non-coding RNA is a fully functional ribonucleic acid that does translate into functional protein. Non-coding RNAs are highly dysregulated in cancer and play an imperative role in disease progression (46). Rajendiran et al. demonstrated an association of miRNA and MIEN1 protein in prostate cancer (46). The study revealed that MIEN1 is an instantaneous target of miR-940 and its downstream effectors, resulting in an alteration in mesenchymal-to-epithelial transition. The study established mir-940 as a novel modulator of MIEN1 (46). In another study, Rajendiran et al. reported the DNA methylation is scattered in the SINE Alu region of the MIEN1 promoter (12). In normal cells, SINE Alu region is hypermethylated leading to subjugation of the MIEN1 gene. In cancer cells, tumor suppressor genes, including genes involved in apoptosis, cell-cycle regulation, and hormonal regulation are hypermethylated. Whereas, oncogenic genes are hypomethylated. The hypomethylation of a section of SINE Alu region opens the chromatin structure, facilitating the ligation of USF (upstream stimulatory factor) to MIEN1 promoter, thereby resulting in its expression. The role of MIEN1 expression affecting apoptosis in prostate cancer cells has not yet been determined.
Lung Cancer
Non-small cell lung cancer (NSCLC) is the major subgroup of lung cancer, which is one of the most common cancers among men and women. MIEN1 mRNA is shown to be overexpressed in NSCLC and correlates with poor prognosis in NSCLC patients, suggesting that MIEN1 expression could be developed as a prognostic marker (7). Li et al. reported that siRNA-mediated MIEN1 knockdown in lung cancer cells reduces the events of metastasis (7). Moreover it was established that miR-26b might be putative target of NF-κB/MMP-9/VEGF mediating metastasis in NSCLC cells (7).
Colorectal Cancer
Colorectal cancer (CRC) is a common cancer in which prognosis is associated with the degree of tumor invasion and metastasis. Dong et al. performed reverse qRT-PCR and immunohistochemistry for detection of MIEN1 mRNA and protein in colorectal cancer and normal/benign adjacent tissue (3). MIEN1 overexpression was associated with cancer aggressiveness and CRC metastasis more frequently in the lymph nodes (3). These results establish the role of MIEN1 expression in CRC, indicating that it and could be developed as a biomarker and/or therapeutic target.
Oral Cancer
Oral squamous cell carcinoma (OSCC) is a malignancy of epithelial cells. Higher MIEN1 expression was reported in OSCC which correlates with cell invasion, migration and filopodia formation (6). Rajendiran et al. demonstrated overexpression of MIEN1 in dysplastic oral keratinocyte cells which results in the up-regulation of Akt/NF-κB effectors viz.MMP-9, uPA and VEGF involved in metastasis and invasion (6). These results suggest an important role of MIEN1 overexpression in OSCC metastasis. Furthermore, differential immunohistochemical staining pattern of MIEN1 was observed in specimens derived from OSCC patient with intense staining in OSCC vs. normal tissue possessing minimal/weak staining. Findings from this study corroborated with TCGA dataset, revealing a positive association of MIEN1 expression with intermediate/high-grade cancer, low survival rate and smoking habit (6).
Gastric Cancer
Gastric cancer is the fifth most common disease worldwide and one of the leading causes of cancer-related deaths (4). Although incidence of gastric cancer has been on the decline over the past few years, resistance to chemotherapy and development of metastasis remains the major cause of patient mortality. Adenocarcinoma of the stomach and esophagus (upper gastrointestinal adenocarcinoma, UGC) is another important cause of cancer death. The 17q12 chromosomal region has been associated with various cancers including UGC. In a study, Maqani et al. noted amplified 17q12-q21 region in normal and UGC specimens where MIEN1 overexpression was frequently associated with invasive UGC tissue samples (55). Additional studies are required to substantiate these findings.
Ovarian Cancer
Leung et al. reported association of MIEN1 protein with ovarian cancer. High levels of MIEN1 are involved in ΔNp73-mediated cisplatin drug resistance in ovarian cancer (5). Reports on MIEN1 and ovarian cancer thus far are limited. There is a pressing need to design in-depth studies to determine the role of MIEN1 in pathophysiology of ovarian cancer in relation to metastasis.
MIEN1 and Other Diseases
Crohn's disease (CD) and ulcerative colitis (UC) are the two most important types of inflammatory bowel diseases (IBD). Out of total 163 loci genome wide known for IBD thus far; 110 loci are susceptibly shared by CD and UC. Studies prioritized the candidate risk gene occurring in these loci. Söderman et al. examined 13 genes within the genetic region of the shared CD and UC susceptibility locus rs2872507 and reported that reduced colonic expression of MIEN1 with other proteins such as ERBB2, GRB7, and PGAP3, resulted in inflammation (19).
Conclusion and Future Prospects
MIEN1 typically associates with the plasma membrane possessing ITAM motif that has the potential to strongly influence cell adhesion, migration, and invasion related to EMT. Limited information is available assessing the role of MIEN1 in various human malignancies, suggesting the demand for additional well-designed studies to evaluate its functional role in cancer. A possibility exists that MIEN1 proteins can directly or indirectly interact with soluble or cellular ligands. How closely MIEN1 function(s) is associates with pathophysiology of various human disease is unclear and remains a subject for further investigation. Higher MIEN1 expression mediates metastasis and invasion in malignant cells, indicating that the protein could be developed as a biomarker and/or therapeutic target. Although the solution structure of MIEN1 has been established, the discovery of MIEN1 inhibitors is not preferable. In silico drug discovery prefers crystal structure of proteins over solution structure as it provides the most stable protein conformation and high-resolution structure. This might be achieved through protein-protein interaction studies, in order to crystalize a particular protein (56). Lack of experimental evidence on MIEN1 protein-protein interaction (Figure 6) might be the reason behind lack of MIEN crystal structure to date.
Cancer drug resistance is a complex phenomenon that develops in response to chemotherapy. Studies on MIEN1 have shown its role in promoting drug resistance in some cancer types. Development of small molecule inhibitors specifically targeting MIEN1 in cancer has not yet been reported. However, as structural information becomes available, and as relevant molecular interactions are identified, this approach should become more feasible. We hereby recommend exploration and validation of natural and/or synthetic inhibitors of MIEN1 that might directly target this protein and/or its posttranslational modification. In this direction, the small size of MIEN1 protein might be one of the challenges associated with therapeutic drug targeting.
Use of antibodies or antibody-derived agents can be therapeutic target to cancer. In general, MIEN1 specific antibodies might have therapeutic potential in cancer treatment by suppressing important functions of MIEN1 or its associated proteins. It is still unknown whether MIEN1 has a role in normal cell development. Any association of MIEN1 with normal cell development will lead to challenges in acquiring tumor cell specificity. Another issue is the lack of information on cellular receptors for MIEN1 proteins. Consequently, downstream effects of MIEN1 knockdown, other than metastasis, requires further studies. As MIEN1 functional protein processing involves GGtase-1 enzyme, design of new novel inhibitor(s) of GGtase-1 may be another avenue to obstruct MIEN1 protein.
MIEN1 has been identified as a primary regulator of cancer cell migration and invasion. Emerging literature suggest that this protein is overexpressed in various human cancers including breast, prostate, colorectal, gastric, ovarian, squamous cell carcinoma and non-small cell lung cancer and in inflammatory bowel diseases. However, additional studies are needed to establish the involvement of MIEN1 in other types of cancers such as brain tumors, melanoma, hematological malignancies and other inflammatory disorders.
In conclusion, advances in the understanding of MIEN1 biochemistry and MIEN1 tumor biology over years, together with technological advancement in therapeutic targeting approaches, should lead to further in vivo validation of the anticancer benefits of targeting MIEN1 and associated proteins.
Author Contributions
SK designed and conceptualized the study. SK, SG, and PK have made substantial contributions to conception and writing of the manuscript. The figures and tables were developed by SK, SG, AS, and PK.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
PK and AS acknowledges financial support from University Grants Commission, India in the form of CSIR-UGC Senior Research fellowship and CSIR Junior Research Fellowship respectively. SK acknowledges Department of Science and Technology, India and University Grants Commission, India for providing financial support in the form of DST-SERB Grant [EEQ/2016/000350] and UGC-BSR Research Start-Up-Grant [No. F.30–372/2017 (BSR)] respectively.
References
1. Evans EE, Henn AD, Jonason A, Paris MJ, Schiffhauer LM, Borrello MA, et al. C35 (C17orf37) is a novel tumor biomarker abundantly expressed in breast cancer. Mol Cancer Ther. (2006) 5:2919–30. doi: 10.1158/1535-7163.MCT-06-0389
2. Dasgupta S, Wasson LM, Rauniyar N, Prokai L, Borejdo J, Vishwanatha JK. Novel gene C17orf37 in 17q12 amplicon promotes migration and invasion of prostate cancer cells. Oncogene. (2009) 28:2860–72. doi: 10.1038/onc.2009.145
3. Dong X, Huang Y, Kong L, Li J, Kou J, Yin L, et al. C35 is overexpressed in colorectal cancer and is associated tumor invasion and metastasis. Biosci Trends. (2015) 9:117–21. doi: 10.5582/bst.2015.01057
4. Fock KM. Review article: the epidemiology and prevention of gastric cancer aliment. Pharmacol Ther. (2014) 40:250–60. doi: 10.1111/apt.12814
5. Leung THY, Wong SCS, Chan KKL, Chan DW, Cheung ANY, Ngan HYS. The interaction between C35 and Δnp73 promotes chemo-resistance in ovarian cancer cells. Br J Cancer. (2013) 109:965–75. doi: 10.1038/bjc.2013.397
6. Rajendiran S, Kpetemey M, Maji S, Gibbs LD, Dasgupta S, Mantsch R, et al. MIEN1 promotes oral cancer progression and implicates poor overall survival. Cancer Biol Ther. (2015) 16:876–85. doi: 10.1080/15384047.2015.1040962
7. Li D, Wei Y, Wang D, Gao H, Liu K. MicroRNA-26b suppresses the metastasis of non-small cell lung cancer by targeting MIEN1 via NF- kB/MMP-9/VEGF pathways. Biochem Biophys Res Commun. (2016) 472:465–70. doi: 10.1016/j.bbrc.2016.01.163
8. Kauraniemi P, Kallioniemi A. Activation of multiple cancer-associated genes at the ERBB2 amplicon in breast cancer. Endocr Relat Cancer. (2006) 13:39–49. doi: 10.1677/erc.1.01147
9. Hsu CH, Shen TL, Chang CF, Chang YY, Huang LY. Solution structure of the oncogenic MIEN1 protein reveals a thioredoxin-like fold with a redox-active motif. PLoS ONE. (2012) 7:e52292. doi: 10.1371/journal.pone.0052292
10. Rubin CM, Vandevoort CA, Teplitz RL, Schmid CW. Alu repeated DNAs are differentially methylated in primate germ cells. Nucleic Acids Res. (1994) 22:5121–7. doi: 10.1093/nar/22.23.5121
11. Schmid CW. Human alu subfamilies and their methylation reveled by blot hybridzation. Nucleic Acids Res. (1991) 19:5613–7. doi: 10.1093/nar/19.20.5613
12. Rajendiran S, Gibbs LD, Van Treuren T, Klinkebiel DL, Vishwanatha JK. MIEN1 is tightly regulated by SINE Alu methylation in its promoter. Oncotarget. (2016) 7:65307–19. doi: 10.18632/oncotarget.11675
13. Häsler J, Strub K. Alu elements as regulators of gene expression. Nucleic Acids Res. (2006) 34:5491–7. doi: 10.1093/nar/gkl706
14. Tomilin NV. Control of genes by mammalian retroposons. Int Rev Cytol. (1999) 186:1–48. doi: 10.1016/S0074-7696(08)61050-5
15. Estécio MRH, Gallegos J, Vallot C, Castoro RJ, Chung W, Maegawa S, et al. Genome architecture marked by retrotransposons modulates predisposition to DNA methylation in cancer. Genome Res. (2010) 20:1369–82. doi: 10.1101/gr.107318.110
16. Zhang W, Edwards A, Fan W, Deininger P, Zhang K. Alu distribution and mutation types of cancer genes. BMC Genomics. (2011) 12:157. doi: 10.1186/1471-2164-12-157
17. Estécio MRH, Gallegos J, Dekmezian M, Lu Y, Liang S, Issa JPJ. SINE retrotransposons cause epigenetic reprogramming of adjacent gene promoters. Mol Cancer Res. (2013) 10:1332–42. doi: 10.1158/1541-7786.MCR-12-0351
18. Dasgupta S, Cushman I, Kpetemey M, Casey PJ, Vishwanatha JK. Prenylated C17orf37 induces filopodia formation to promote cell migration and metastasis. J Biol Chem. (2011) 286:25935–46. doi: 10.1074/jbc.M111.254599
19. Söderman J, Berglind L, Almer S. Gene expression-genotype analysis implicates GSDMA, GSDMB, and LRRC3C as contributors to inflammatory bowel disease susceptibility. BioMed Res Int. (2015) 2015:1–10. doi: 10.1155/2015/834805
20. Zhang FL, Casey PJ. Protein prenylation: molecular mechanisms and functional consequences. Annu Rev Biochem. (1996) 65:241–69. doi: 10.1146/annurev.bi.65.070196.001325
21. Casey PJ, Seabra MC. Protein Prenyltransferases. J Biol Chem. (1996) 271:5289–92. doi: 10.1074/jbc.271.10.5289
22. Wang M, Casey PJ. Protein prenylation: unique fats make their mark on biology. Nat Rev Mol Cell Biol. (2016) 17:110–22. doi: 10.1038/nrm.2015.11
23. Gelb MH, Brunsveld L, Hrycyna CA, Michaelis S, Van Voorhis WC, Waldmann H. Therapeutic intervention based on protein prenylation and associated modifications. Nat Chem Biol. (2006) 2:518–28. doi: 10.1038/nchembio818
24. Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. (2011) 11:775–91. doi: 10.1038/nrc3151
25. Taylor JS, Reid TS, Terry KL, Casey PJ, Beese LS. Structure of mammalian protein geranylgeranyltransferase type-I. EMBO J. (2003) 22:5963–74. doi: 10.1093/emboj/cdg571
26. Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, et al. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell. (2005) 16:1606–16. doi: 10.1091/mbc.e04-11-0960
27. Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. (2005) 5:405–12. doi: 10.1038/nrc1612
28. Arozarena I, Calvo F, Crespo P. Ras, an actor on many stages: posttranslational modifications, localization, and site-specified events. Genes Cancer. (2011) 2:182–94. doi: 10.1177/1947601911409213
29. Weber GF, Bjerke MA, DeSimone DW. Integrins and cadherins join forces to form adhesive networks. J Cell Sci. (2011) 124:1183–93. doi: 10.1242/jcs.064618
30. Deryugina EI, Quigley JP. Cell surface remodeling by plasmin: a new function for an old enzyme. J Biomed Biotechnol. (2012) 2012:1–21. doi: 10.1155/2012/564259
31. Kpetemey M, Dasgupta S, Rajendiran S, Das S, Gibbs LD, Shetty P, et al. MIEN1, a novel interactor of Annexin A2, promotes tumor cell migration by enhancing AnxA2 cell surface expression. Mol. Cancer. (2015) 14:1–13. doi: 10.1186/s12943-015-0428-8
32. Katz E, Sims AH, Faratian D, Li J, Smith ES, Quinn JA, et al. A gene on the HER2 amplicon, C35, is an oncogene in breast cancer whose actions are prevented by inhibition of Syk. Br J Cancer. (2010) 103:401–10. doi: 10.1038/sj.bjc.6605763
33. Polioudaki H, Agelaki S, Chiotaki R, Politaki E, Mavroudis D, Matikas A, et al. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC cancer. (2015) 15:399. doi: 10.1186/s12885-015-1386-7
34. Kpetemey M, Chaudhary P, Van Treuren T, Vishwanatha JK. MIEN1 drives breast tumor cell migration by regulating cytoskeletal-focal adhesion dynamics. Oncotarget. (2016) 7:54913–24. doi: 10.18632/oncotarget.10798
35. Bai D, Ueno L, Vogt P. Akt-mediated regulation of NFkB for the oncogenicity of PI3K and Akt. Int J Cancer. (2009) 125:2863–70. doi: 10.1002/ijc.24748
36. Fujita M, Kinoshita T. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. (2010) 584:1670–7. doi: 10.1016/j.febslet.2009.10.079
37. Boukhzar L, Hamieh A, Cartier D, Tanguy Y, Alsharif I, Castex M, et al. Selenoprotein T exerts an essential oxidoreductase activity that protects dopaminergic neurons in mouse models of parkinson's disease. Antioxid Redox Signal. (2016) 24:557–74. doi: 10.1089/ars.2015.6478
38. Dikiy A, Novoselov SV, Fomenko DE, Sengupta A, Carlson BA, Cerny RL, et al. SelT, SelW, SelH, and Rdx 12: genomics and molecular insights into the functions of selenoproteins of a novel thioredoxin-like family. Biochemistry. (2007) 46:6871–82. doi: 10.1021/bi602462q
39. Lubos E, Loscalzo J, Handy DE. Glutathione peroxidase-1 in health and disease : from molecular mechanisms to therapeutic opportunities. Antioxid Redox Signal. (2011) 15:1957–97. doi: 10.1089/ars.2010.3586
40. Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. (2004) 429:841–7. doi: 10.1038/nature02656
41. Jin D, Park S, Lee J, Kim K, Kim S, Kim H, et al. Copy number gains at 8q24 and 20q11-q13 in gastric cancer are more common in intestinal-type than diffuse-type. PLoS ONE. (2015) 10:e0137657. doi: 10.1371/journal.pone.0137657
42. Cuadros M, Cano C, Lopez FJ, Lopez-Castro R, Concha A. Expression profiling of breast tumors based on human epidermal growth factor receptor 2 status defines migration-related genes. Pathobiology. (2013) 80:32–40. doi: 10.1159/000339431
43. Grinchuk OV, Motakis E, Kuznetsov VA. Complex sense-antisense architecture of TNFAIP1/POLDIP2 on 17q11.2 represents a novel transcriptional structural-functional gene module involved in breast cancer progression. BMC Genomics. (2010) 11(Suppl. 1):S9. doi: 10.1186/1471-2164-11-S1-S9
44. Benusiglio PR, Pharoah PD, Smith PL, Lesueur F, Conroy D, Luben RN, et al. HapMap-based study of the 17q21 ERBB2 amplicon in susceptibility to breast cancer. Br J Cancer. (2006) 95:1689–95. doi: 10.1038/sj.bjc.6603473
45. Wieduwilt MJ, Moasser MM. The epidermal growth factor receptor family: biology driving targeted therapeutics. Cell Mol. Life Sci. (2008) 65:1566–84. doi: 10.1007/s00018-008-7440-8
46. Rajendiran S, Parwani AV, Hare RJ, Dasgupta S, Roby RK, Vishwanatha JK. MicroRNA-940 suppresses prostate cancer migration and invasion by regulating MIEN1. Mol Cancer. (2014) 13:1–15. doi: 10.1186/1476-4598-13-250
47. Liu QQ, Yin K, Zhu S, Zhang L, Wen P, Li C, et al. Inhibition of C35 gene expression by small interfering RNA induces apoptosis of breast cancer cells. Biosci. Trends. (2010) 4:254–9.
48. Zhao HB, Zhang XF, Wang HB, Zhang MZ. Migration and invasion enhancer 1 (MIEN1) is overexpressed in breast cancer and is a potential new therapeutic molecular target. Genet Mol Res. (2017) 16:1–9. doi: 10.4238/gmr16019380
49. Yin K, Ba Z, Li C, Xu C, Zhao G, Zhu S, et al. Overexpression of C35 in breast carcinomas is associated with tumor progression and lymphnode metastasis. Biosci Trends. (2015) 9:386–92. doi: 10.5582/bst.2015.01161
50. Li M, Huang Y, Dong X, Wei Q, Li J, Sun H, et al. Simvastatin downregulated C35 expression and inhibited the proliferation of colon cancer cells Lovo and HT29 in vitro. Biosci Trends. (2016) 10:227–30. doi: 10.5582/bst.2016.01025
51. Katz E, Lareef MH, Rassa JC, Grande SM, King LB, Russo J, et al. MMTV Env encodes an ITAM responsible for transformation of mammary epithelial cells in three-dimensional culture. J Exp Med. (2005) 201:431–9. doi: 10.1084/jem.20041471
52. Grande SM, Katz E, Crowley JE, Bernardini MS, Ross SR, Monroe JG. Cellular ITAM-containing proteins are oncoproteins in nonhematopoietic cells. Oncogene. (2006) 25:2748–57. doi: 10.1038/sj.onc.1209296
53. Ross SR, Schmidt JW, Katz E, Cappelli L, Hultine S, Gimotty P, et al. An immunoreceptor tyrosine activation motif in the mouse mammary tumor virus envelope protein plays a role in virus-induced mammary tumors. J Virol. (2006) 80:9000–8. doi: 10.1128/JVI.00788-06
54. Van Treuren T, Vishwanatha JK. CRISPR deletion of MIEN1 in breast cancer cells. PLoS ONE. (2018) 13:e0204976. doi: 10.1371/journal.pone.0204976
55. Maqani N, Belkhiri A, Moskaluk C, Knuutila S, Dar AA, El-Rifai W. Molecular dissection of 17q12 amplicon in upper gastrointestinal adenocarcinomas. Mol Cancer Res. (2006) 4:449–56. doi: 10.1158/1541-7786.MCR-06-0058
Keywords: MIEN1, cancer, metastasis, invasion, posttranslational modification
Citation: Kushwaha PP, Gupta S, Singh AK and Kumar S (2019) Emerging Role of Migration and Invasion Enhancer 1 (MIEN1) in Cancer Progression and Metastasis. Front. Oncol. 9:868. doi: 10.3389/fonc.2019.00868
Received: 11 March 2019; Accepted: 21 August 2019;
Published: 04 September 2019.
Edited by:
Francesco Fiorica, Azienda Ulss 9 Scaligera, ItalyReviewed by:
Pankaj Chaudhary, Queen's University Belfast, United KingdomSonia Missiroli, University of Ferrara, Italy
Copyright © 2019 Kushwaha, Gupta, Singh and Kumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shashank Kumar, c2hhc2hhbmtiaW9jaGVtYXVAZ21haWwuY29t; c2hhc2hhbmsua3VtYXJAY3VwYi5lZHUuaW4=
 Prem Prakash Kushwaha
Prem Prakash Kushwaha Sanjay Gupta
Sanjay Gupta Atul Kumar Singh
Atul Kumar Singh Shashank Kumar
Shashank Kumar