- Department of Oncology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China
Brain metastasis, one of the common complications of lung cancer, is an important cause of death in patients with advanced cancer, despite progress in treatment strategies. Lung cancers with positive driver genes have higher incidence and risk of brain metastases, suggesting that driver events associated with these genes might be biomarkers to detect and prevent disease progression. Common lung cancer driver genes mainly encode receptor tyrosine kinases (RTKs), which are important internal signal molecules that interact with external signals. RTKs and their downstream signal pathways are crucial for tumor cell survival, invasion, and colonization in the brain. In addition, new tumor driver genes, which also encode important molecules closely related to the RTK signaling pathway, have been found to be closely related to the brain metastases of lung cancer. In this article, we reviewed the relationship between lung cancer driver genes and brain metastasis, and summarized the mechanism of driver gene-associated pathways in brain metastasis. By understanding the molecular characteristics during brain metastasis, we can better stratify lung cancer patients and alert those at high risk of brain metastasis, which helps to promote individual therapy for lung cancer.
Introduction
Lung cancer, accounting for 18.4% of the total cancer population, ranks first in cancer-associated mortality globally. Brain metastasis is one of the main causes of death in patients with lung cancer (1). With the advancement of cancer treatment strategies, cancer mortality has continued to decline since 1991. The decrease of mortality is particularly pronounced for lung cancer in recent years, which decreased by 5% per year (2013 to 2017) compared to 3% per year (2008 to 2013) in men, and in women by 4% per year compared to 2% per year (2). The prevalence of brain metastases in patients with advanced lung cancer is about 20~56%, accounting for 40~50% of all brain metastases (3–5). According to histopathology, lung cancer is classified into two types: non-small cell lung cancer (NSCLC) (85%) and small cell lung cancer (SCLC) (15%). SCLC has a single histological type and is highly aggressive. About 50% of patients presents with brain metastases at diagnosis and during treatment (6). At present, prophylactic cranial irradiation (PCI) can be used for SCLC, while for NSCLC, effective strategy to prevent brain metastases is still lacking (6). The RTOG 0214 trial of PCI in NSCLC showed that PCI reduces brain metastasis rates after 1 year (18 vs. 7.7%), but the overall survival is not improved (7). It revealed that identifying candidates who could benefit from PCI was difficult. Hence, for patients with NSCLC, it is important for clinician to detect the patient at high risk of brain metastasis.
Since Paget put forward the “seed-soil” hypothesis in 1889, theories such as cancer stem cells (CSCs), tumor microenvironment, and circulating tumor cells have been proposed successively, further supplementing the mechanism of tumor metastasis (8–12). In 2003, evolutionary geneticist Austin Burt proposed the concept of “gene drive,” thinking that cancer is a genetic disease, in which gene mutations eventually result in phenotype changes, leading to the occurrence of cancer (13). With the popularization of high-throughput sequencing technology, genetic cloning events have again caught people’s eye regarding tumorigenesis (14). Endogenous mutation process drives the occurrence of lung adenocarcinoma (15). Driver mutations can arise before and after subclonal diversification, and the subclonal mutations may be important for cancer progression (16, 17). By tracking the driving events of patients, researchers have found that genetic diversity is a determinant of patient outcome, and clonal evolution or chromosomal instability is suggested as a biomarker for detection and intervention of disease progression (18–21). Moreover, the Genotype-Tissue Expression project found that local genetic variation affects the expression level of most genes (22, 23). These findings suggested that driver genes could be used as biomarkers to predict tumor metastasis.
In lung cancer, patients with mutation of common driver genes (such as EGFR and ALK) can benefit from targeted therapy. Progress has also been achieved in research on rare mutations such as ERBB2, MET, RET, ROS1, and PIK3CA, and new inhibitors targeting the products of these genes are under development (24, 25). In recent years, several studies have found that driver genes have a predictive role in the occurrence of brain metastases (26–28). In this review, we summarized the current advances of lung cancer driver genes in the occurrence of brain metastasis and the related mechanisms, hoping to provide a general understanding for researchers and clinical doctors.
Lung Cancer Driver Genes and Brain Metastasis
In a study of 271 patients with lung adenocarcinoma, 85% of patients have driver gene mutations (29) (Table 1, Figure 1). A recent study reported that 74.4% of lung adenocarcinoma patients have at least one druggable mutation detected (30) (Table 1, Figure 1). In another study of 552 NSCLC patients, mutation of driver gene is present in 62% of patients, and those with driver gene mutations are more prone to brain metastases (26) (33 vs. 19%; Table 1, Figure 1). The three studies suggested that the incidence of driver gene mutations in NSCLC patient is high, and lung cancer driver genes, such as EGFR, ALK, and RET are risk factors for brain metastasis in advanced NSCLC patients. Tomasini et al. previously proved that EGFR and KRAS mutations have a predictive role on brain metastasis incidence, recurrence, and outcome in NSCLC patients (27). Patil confirmed that brain metastasis is common in patients with ROS1-positive advanced NSCLC (31). New brain metastasis driver genes, such as MYC, AXIN2, and NRG1, have also been discovered (32–35). At present, researchers have mainly studied the incidence of common driver genes and the rate of brain metastasis in NSCLC patients. New driver genes, due to their low incidence, have not been studied yet, although they may be involved in the occurrence and development of brain metastasis (Table 2).
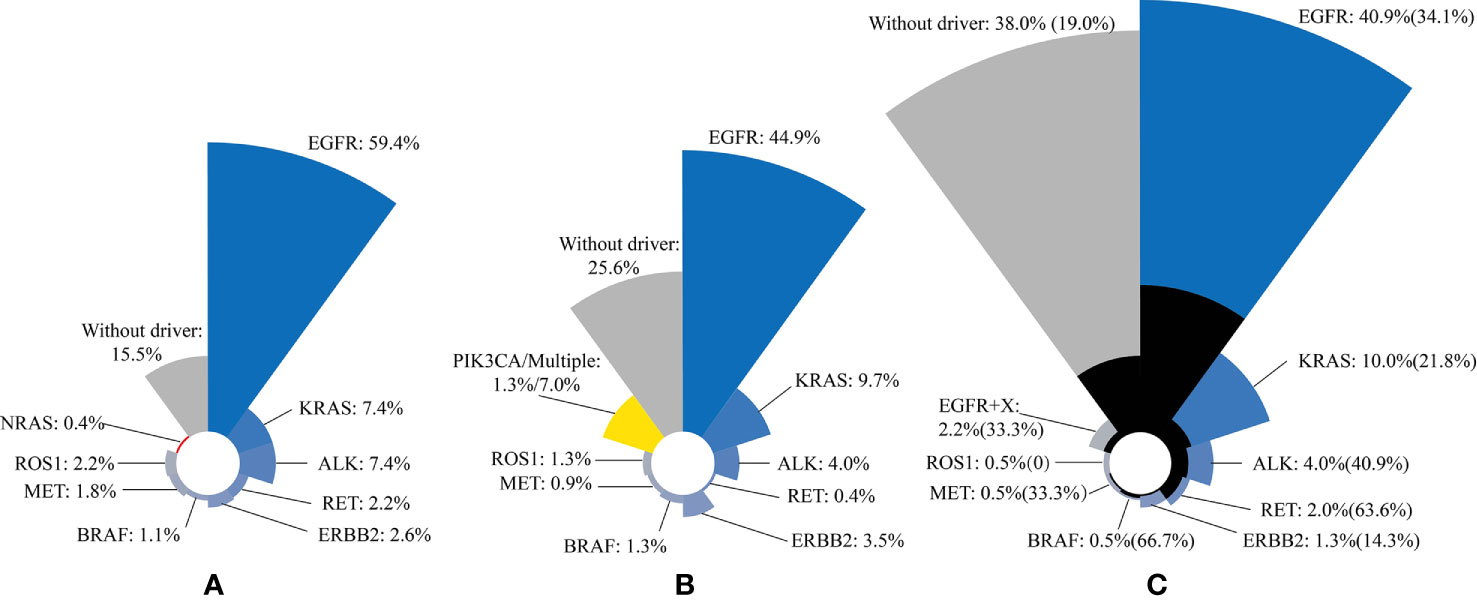
Figure 1 Three studies have shown that the most common type of lung cancer driver gene mutation is EGFR mutation, followed by KRAS mutation and ALK fusion mutation. The black area indicates the percentage of brain metastases in the total number of corresponding mutations. (A) n = 271; (B) n = 227; (C) n = 552. The incidence of brain metastases in patients with driver gene mutations is shown in brackets.
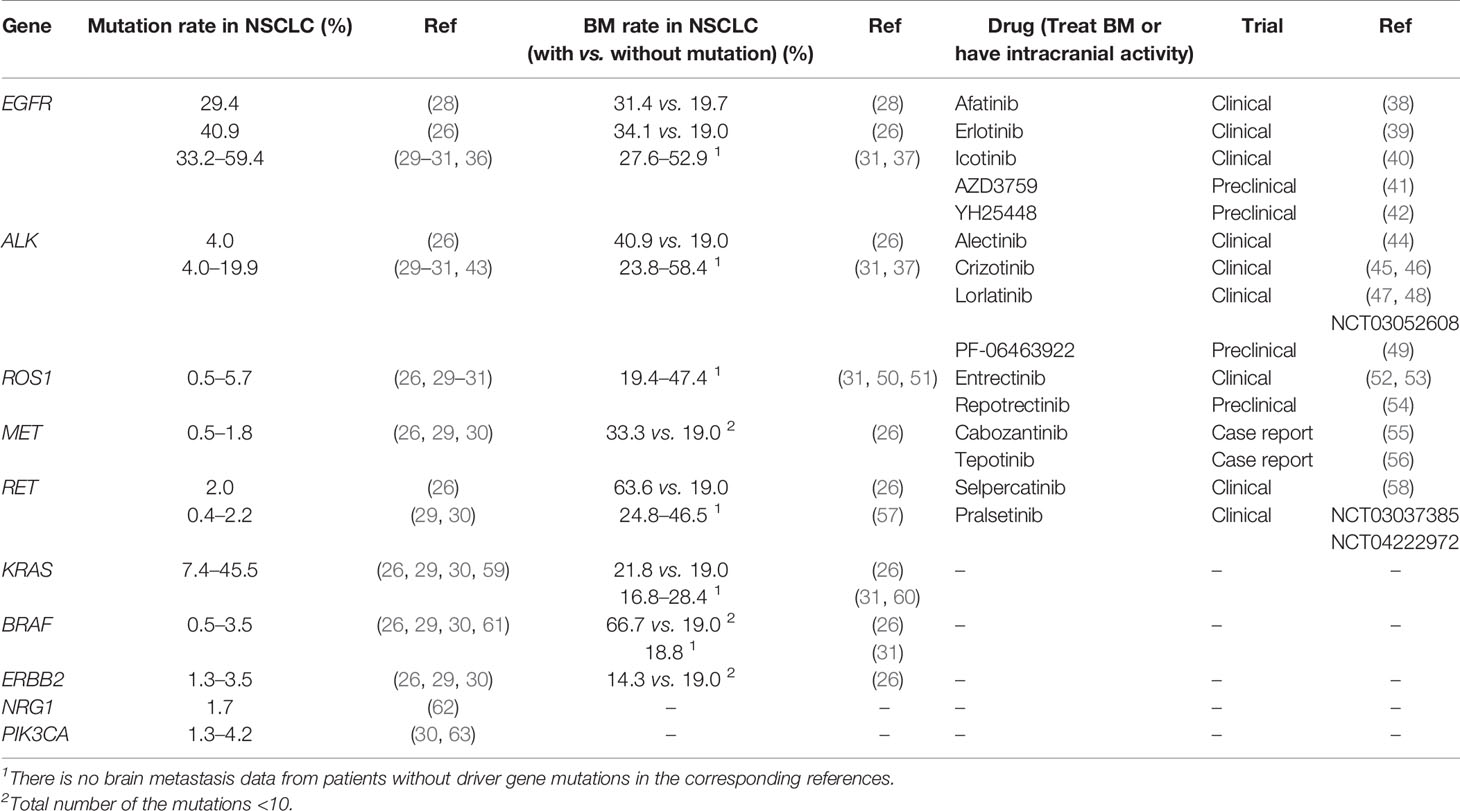
Table 2 Potential biomarkers in lung cancer driver genes and the targeted drugs in brain metastasis.
However, gene mutations in brain metastatic sites are inconsistent with the original lesions, and acquired mutations can occur during treatment, resulting in drug resistance and disease progression. For example, inconsistent mutation status of KRAS have been discovered between the primary and metastatic sites of lung adenocarcinoma, while the status of EGFR mutation is relatively consistent on the contrary (64). In Brastianos’s study, 86 matched brain metastases, primary tumors and normal tissues were sequenced with whole exome sequencing to check whether brain metastases had genetic changes different from the primary tumors. Changes were found only in brain metastases in about 53% of cases. Detected alteration was associated with the sensitivity to phosphatidylinositol 3 kinase (PI3K) pathway and epidermal growth factor receptor (EGFR) pathway inhibitors (65). Integrated genomic and transcriptomic analysis had identified crucial roles of EGFR signaling in brain metastasis (34). Furthermore, Paik and Wang H et al. also revealed a correlation of PI3K signaling with increased risks of brain metastasis in patients with NSCLC (32, 66). In Wang H’s study, mutations of EGFR, KRAS, and ALK are highly concordant between primary NSCLC and matched brain metastases, whereas discordance of PI3K signaling suggested the unique genomic evolution and oncogenic mechanisms of brain metastasis (32).
In addition, single nucleotide polymorphisms of driver genes are also closely related to the occurrence of lung cancer brain metastases. Li Q et al. proved for the first time that genetic variations in the transforming growth factor-β (TGF-β), PI3K/protein kinase B (AKT) pathways were associated with an increased risk of brain metastasis in NSCLC patients (67, 68). Recently, Xu Y et al. also proved that single nucleotide polymorphisms in the mammalian target of rapamycin complex 1 (mTORC1) signaling pathway are significantly associated with increased risk of brain metastasis (69). Activity of mTORC1/2 is higher in patients with lung cancer with brain metastases (70). Therefore, it is very necessary to clarify the status of lung cancer driver genes at the diagnosed with NSCLC, which can help predict the occurrence of brain metastasis. New driver genes may play a unique role in the mechanism of brain metastasis.
Signaling Pathway in Lung Cancer Driver Gene
Lung cancer driver genes mainly include EGFR, ERBB2, MET, RET, ALK, and ROS1, all encoding genes for receptor tyrosine kinases (RTKs). Abnormal RTKs activation in tumors mainly includes acquired mutations, genome amplification, chromosome rearrangement, and autocrine activation, which lead to the imbalance of RTK signals and promote cell proliferation, metabolism, cytoskeleton remodeling, cell migration, and anti-apoptosis effects (71). RTKs form dimers by binding to their corresponding ligands or closely combining with members of the same family to stabilize and enhance downstream signaling pathways. The downstream signaling pathways of RTKs mainly include RAS (a GTPase)/RAF (a kinase)/mitogen-activated protein kinase (MAPK) and PI3K/AKT/mTOR (72–74). Other genes encoding important molecules in these pathways are also lung cancer driver genes. Li D et al. reported that driver gene mutations can occur within tyrosine kinase domains and genetic alterations frequently occurs in genes of the MAPK signaling, WNT signaling and mTOR pathways in patients with lung adenocarcinoma (75).
Classical driver genes can also interact with other pathways, which have been shown to be activated in lung cancer. Proteomic data have uncovered an interdependence of PI3K and signal transducer and activator of transcription 3 (STAT3) (76). Tyrosine-759, located in Janus kinase (JAK), acts as a docking site for the adaptor molecule SHP2, which is crucial for the initiation of the PI3K and MAPK pathway (77). Govindan et al. proved that JAK/STAT pathway is significantly altered in patients with lung cancer (78). RAS can also cross-link with the WNT/β-catenin pathway to promote tumor invasion (79). In addition, the downstream pathways of RTKs can also act synergistically with TGF-β receptors. TGF-β receptors levels can differentially affect the activation of the MAPK pathway (80). In a transgenic mouse model of KRAS-induced lung cancer, invasive adenocarcinoma is modeled by the loss of the TGF-β receptors (81). Interfering with these pathways can suppress lung cancer with positive driver genes. For example, preclinical findings have identified that inhibition of the interleukin-6 (IL-6)/STAT3 pathway can also inhibit tumor growth with EGFR mutation in NSCLC and suppress KRAS-driven lung adenocarcinoma (82–85). Mohrherr et al. proved that JAK/STAT pathway inhibitors can attenuate the progression of lung cancer driven by KRAS in preclinical models (86). In addition, TGF-β receptor inhibitors may be an effective therapy in a subset of KRAS-mutant patients with NSCLC (87). Molecules in these pathways can be considered as potential biomarkers in preventing lung cancer driver gene-associated brain metastasis.
Mechanism of Lung Cancer Driver Genes in Brain Metastasis
Mutations introduced during primary tumor cell growth can result in clonal heterogeneity. Vogelstein put forward four types of genetic heterogeneity in tumors: intratumoral, intermetastatic, intrametastatic, and interpatient (88). Intratumoral heterogeneity provides the seeds for intermetastastic heterogeneity. Intratumoral heterogeneity mediated through chromosome instability is associated with an increased risk of recurrence or death in NSCLC and driven metastasis (19, 20, 89). Chromosome instability is increased in brain metastases and is a driver of metastasis (65, 90). Intermetastatic heterogeneity supports the idea that the genetic alterations required for metastasis are present before metastasis actually occurs (91). The founder clones are initiating events for lung cancer and other mutations are acquired later and perhaps are important for tumor progression (78). The outgrowth of distal colonizing cells necessitates further selection from subsequent genetic heterogeneity (92).
During the process of brain metastasis, abnormal genes drive tumor cells to escape normal regulatory mechanisms and change the microenvironment of lung cancer tumors through various signals, which continues to enhance tumor invasiveness, induce epithelial-mesenchymal transition (EMT) and accelerate vascular invasion (93). Metastatic tumor cells are arrest at vascular branch points, early extravasation, persistent close contacts to microvessels, and perivascular growth (94). Tumor cells cross the blood-brain barrier mediated by specific molecules and turn into dormancy/quiescence, laying the foundation for growth after several months or even longer (92, 95). Tissue remodeling creates a tumor microenvironment, affecting the homeostasis of the central nervous system and promoting tumor metastasis and growth (96). The following content mainly focuses on the mechanism of lung cancer driver genes and associated signaling pathway in the three main steps of brain metastasis (Figure 2).
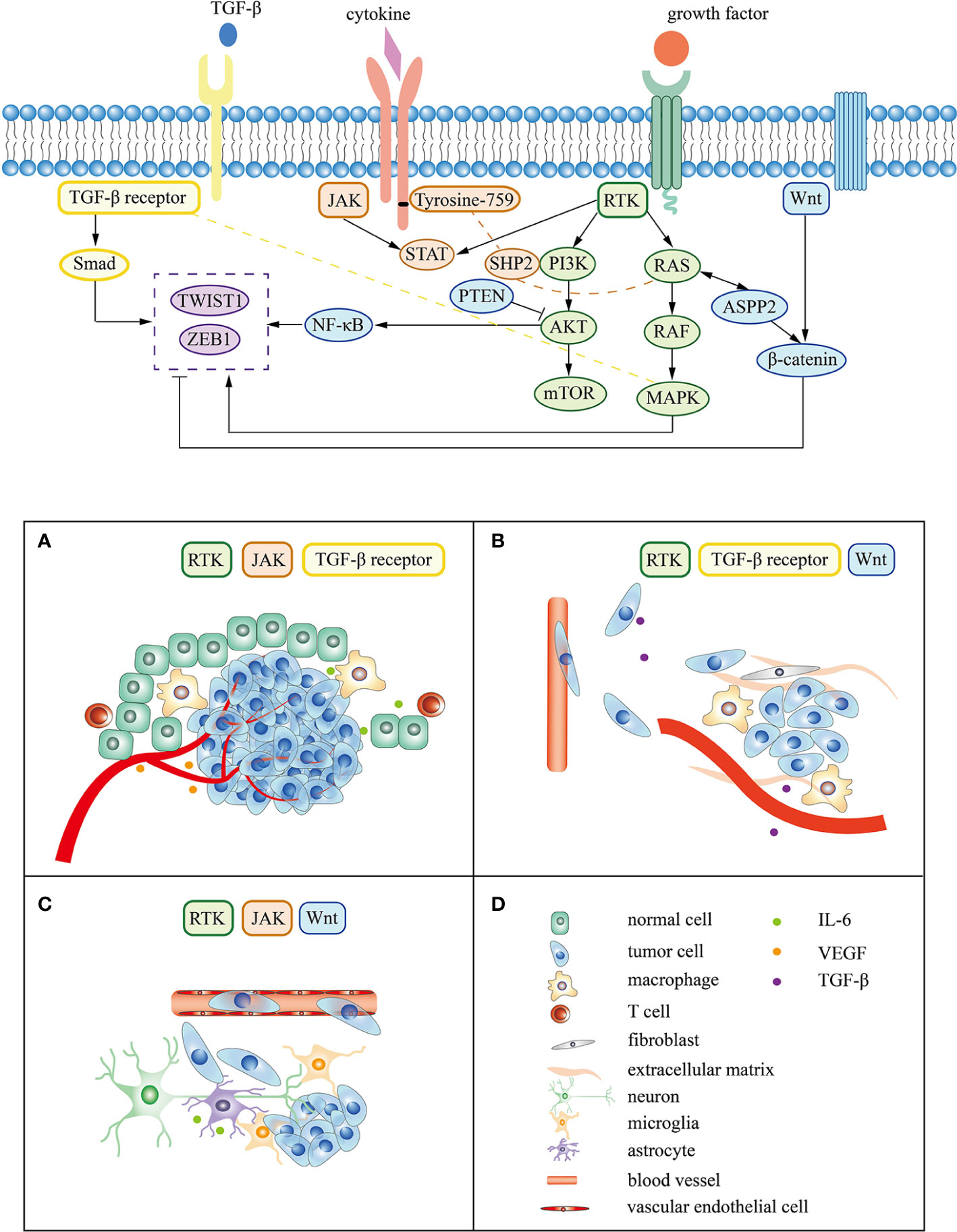
Figure 2 Signaling pathway and mechanism related to lung cancer driver gene in brain metastasis. (A–C) represent the tumor cell survival, EMT, and colonization respectively. (D) corresponds to the cell or molecule in (A–C).
Promote Tumor Cell Survival
Phenotypic and functional heterogeneity arise among cancer cells within the same tumor. Comparing with cancer cells without driver gene mutation, those with a mutation, such as EGFR, ERBB2, TGFbR2, MET, RAS, RAF, PIK3CA, and PTEN genes, can proliferate under limiting nutrient concentrations (88). Cells with driver gene mutations will have a selective growth advantage than others. For instance, mutations in KRAS or BRAF genes confer on cancer cells the ability to grow in lower glucose concentrations (97, 98). Some of driver genes encode RTKs to receive the growth factor signal, whereas others are signal transducers of RTK-related pathways (99). After ROS1 rearrangement, the extra-membrane part of the expressed protein is lost, leaving only the activated intra-membrane part, which fuses with other proteins and continuously transmits signals of growth and proliferation (100). Inhibition of these receptors or signals can interfere with cell growth and promote apoptosis. Inhibiting of EGFR activity increases apoptosis (101). Govindan et al. supposed that it is likely that driver gene mutations, such as EGFR and KRAS, are initiating events for lung cancer (78).
In cancer genome landscapes, driver genes regulate cell survival by the RAS/MAPK pathway, PI3K pathway, STAT pathway, and TGF-β pathway (88). Lysophosphatidylcholine acyltransferase 1 can up-regulate the PI3K/AKT pathway and promote EGFR mutation lung adenocarcinoma cell proliferation, invasion, and brain metastasis (102). Targeting lonidamine to mitochondria can inhibit AKT/mTOR signal, induce autophagic death of lung cancer cells with KRAS mutation and block tumorigenesis and brain metastasis (103). PTEN is an important gene that negatively regulates the AKT signaling pathway. Mutations in PTEN may have strong tumor-growth-promoting capability (75). In mouse models, tracheal epithelial cells lacking PTEN produce spontaneous tumors (104, 105). Abnormal activation of the above tumor driver genes leads to growth dominance and immortalization of lung cancer cells, opening the first step of tumor metastasis.
In the tumor microenvironment, driver gene-associated pathways are also involved in the formation of tumor immunosuppressive microenvironment, which is more conducive to the survival of tumor cells. In preclinical study, tumor cells use PI3K-hypoxia-inducible factor 1α axis to polarize macrophages into tumor-associated macrophages (TAMs), which produce IL-6 after engulfing particles released by tumor cells (106, 107). TAMs polarize towards M2 type through the IL-6/STAT3 signaling pathway to promote tumor metastasis and rejects immune cells from penetrating (108–110). Wu SY et al. proved that M2 macrophages are closely related to brain metastasis of lung cancer (111). On the other hand, by regulating mTOR, TAMs block normal glycolysis, induce excessive angiogenesis, and form abnormal blood vessels (112). These signals regulate the microenvironment of the primary tumor to escape from the immune system, and to create a microenvironment suitable for tumor growth and invasion. Moreover, activation of the EGFR pathway increases the production of tumor-derived vascular endothelial growth factor (VEGF), which acts on endothelial cells in a paracrine manner to promote angiogenesis (113). When driver gene is mutant or the coding molecule is activated, the above situation will be more likely to happen.
Promote Epithelial-Mesenchymal Transition
EMT is a temporary and reversible process characterized by epithelial cell dedifferentiation and migration to a distance site (114). Bakhoum et al. found that metastatic tumors contain a large number of differentially upregulated EMT- and inflammation-related genes (90). Markers of EMT, including E-cadherins and N-cadherin, can be used as biomarkers to predict brain metastasis (115, 116). Various internal signals (such as gene mutations) and external signals (such as growth factor signals) play an important role in this process (117). Yousefi et al. put forward CSCs may originate from somatic mutations of normal tissue stem cells or may dedifferentiate from cancer cells via EMT (118). In the absence of driver gene mutations, RTK, TGF-β, and WNT pathways play an important role in EMT via activating transcription factors, such as twist family bHLH transcription factor 1 (TWIST1) and zinc finger E-box binding homeobox 1 (ZEB1) protein (118, 119). These signal pathways interact with each other to promote EMT (120–124).
As crucial internal signals, driver gene mutations give tumor cells stronger capability of EMT. For instance, driver gene RAS is closely related to EMT, and TWIST promotes tumor initiation and progression in vivo only after interaction with activated RAS (125). Activation of RAS can stimulate apoptosis-stimulating protein of p53 2 (ASPP2) and β-catenin to translocate from the cell junction to the cytoplasm and nucleus, reducing the formation of ASPP2-β-catenin complex, leading to EMT of tumor cells (79). In addition, mutation of TGF-β receptor can lead to loss of cytostatic effects of TGF-β. Tumor cells in the absence of cytostatic response may undergo EMT in response to TGF-β, which helps to escape the immunosuppressive environment and induce angiogenesis as well as systemic spread in 3D Tissue Culture (126, 127).
Therefore, many molecules can affect the occurrence and development of tumors by promoting or interfering with EMT. TAM induces EMT through IL-6-mediated WNT pathway to promote the invasion of lung cancer cells (128). MicroRNA-330-3p and Insulin-like growth factor binding protein-3 affect EMT by regulating the TGF-β/Smad signaling pathway, thereby promoting brain metastasis in NSCLC (129, 130). Programmed death ligand-1 may induce EMT by activating the TGF-β/Smad signaling pathway, and this process contributes to the primary resistance of EGFR-mutant NSCLC cells to TKIs (131, 132). ASPP2 can stabilize the β-catenin–E-cadherin complex and prevent β-catenin from transactivating ZEB1 to limit the aggressiveness of RAS and inhibit tumor metastasis in vivo (133, 134). Another study suggested that apoptotic lung cancer cells can increase the level of phosphatase and tensin homolog (PTEN) in exosomes from TAMs, which results in reduction of ZEB1 and inhibition of EMT (135).
Affect the Colonization of Metastatic Tumor Cells Into the Brain
The PI3K/AKT, JAK/STST, and WNT signaling pathways are involved in vessel penetration and colonization of tumor cell to the brain (136). Attenuated WNT signaling is associated with the dormancy/quiescence of tumor cells in metastases (95). Ma SC et al. proved in vitro experiments that Claudin-5 regulates permeability of the blood-brain barrier by changing the proliferation, migration, and adhesion of brain microvascular endothelial cells, which resulted in decreased brain metastases from lung cancer (136). In preclinical study of breast cancer, heparin binding EGF, ligand of EGFR, can enhance the adhesion between tumor cells and brain endothelial cells, and help tumor cells penetrate the blood-brain barrier in breast cancer (92). Cathepsin S attenuates EGF-mediated EGFR degradation, which regulates EGFR signaling (137). Cathepsin S produced by tumor cells promotes tumor cell extravasation by accelerating the proteolysis of adhesion molecules between endothelial cells (138).
Tumor cells that enter the brain microenvironment interact with the original “residents” (mainly microglia and astrocytes), and grow autonomously in brain tissue through tumor-specific signaling pathways. Tumor cells interact with microglia and affect angiogenesis and survival through activation of STAT3 pathway in microglia (139). In multiple models of tumor metastasis, TAMs activate JAK/STAT signals to reverse EMT and promote metastatic colonization (140). Moreover, Chen Q et al. found that gap junctions between lung cancer cell and astrocyte triggers STAT1 survival signals in vivo and in vitro (141). Activated astrocytes produce IL-6, which in turn promotes lung cancer cell proliferation in Seike’s study (142). In brain metastases, astrocytes and tumor cells transduce bidirectional signals through endothelin and its receptors, as well as the AKT pathway, which produces chemotherapy protection (143). Unfortunately, this result is mainly verified in breast cancer cell lines.
However, despite the fact that driver genes are related to brain metastasis and driver gene-associated signaling pathways play a key role in colonization, there are still very few such preclinical studies on why lung cancer with driver gene mutation is more likely to develop brain metastasis than those without. At present, studies have focused on the relationship between patients with positive driver genes and the occurrence of brain metastasis, as well as the molecular mechanism of brain metastasis is not clear in patients with driver gene mutation. Preclinical models only center on the use of driver gene mutations in cell lines or animal models. For example, Nguyen demonstrated earlier that the WNT signaling pathway can enhance the ability of lung adenocarcinoma cells with KAS or EGFR mutation to colonize the brain (144). Adaptive loss of PTEN of breast cancer cells in brain metastasis, which was silenced by astrocyte-derived miRNAs, leads to increased secretion of chemokine (C-C motif) ligand 2, recruitment of myeloid cells, promotion of cell proliferation, and reduced apoptosis, which further enhances the growth of tumor cells in metastatic sites (104).
Why do tumor cells choose to “settled” in the brain? A reasonable explanation may be that the tumor cells that successfully grow, proliferate, and eventually form brain metastases have specific adaptations to the brain microenvironment. Transcriptome data of microarray hybridization showed that metastatic tumor cells are reprogrammed in the brain microenvironment to obtain neuronal cell characteristics (145). There is a similar situation in the lung cancer bone metastasis model. Tumor cells can acquire the characteristics of the metastatic microenvironment, which known as osteomimicry (146). Furthermore, residents in pre-metastasis microenvironment remodel the soil to promote seed growth in breast cancer lung metastasis model (147). In addition to the specific adaptations, tumor cells will also choose a more favorable microenvironment. Saunus’s research found that EGFR, ERBB2, and ERBB3 transcripts were abundantly expressed in lung cancer brain metastases, and ERBB3 transcript abundance correlated with its oncogenic partner ERBB2 (34). However, expression of neuregulin 1, which is the ligand for erb-b2 receptor tyrosine kinase 3, is very low in tumor cells and rich in brain microenvironment. This result suggests tumor cells are more likely colonized in more favorable microenvironment.
Therapy Prospects for Lung Cancer Driver Gene
The discovery of various driver gene mutations has greatly promoted targeted therapies for lung cancer. According to the National Comprehensive Cancer Network guidelines, most targeted therapies recommended for NSCLC are those targeting EGFR, ALK, ROS1, BRAF, RET, and MET (148). Genetic testing has become one of the routine diagnostic procedures for patients after confirmation of NSCLC diagnosis. A single biopsy can capture most functionally important mutations in metastatic tumors, thereby providing necessary information for treatment decisions (149). NSCLC patients with positive driver genes have good sensitivity to TKIs. Prolonged survival has been achieved with radiotherapy combined with TKIs-targeted therapy in patients with brain metastases (150). Although good clinical effects can be achieved with first- and second-generation of TKIs, recurrent metastasis can occur, with the brain being the most frequent metastatic site. This may be a result of the blood-brain barrier to make the brain a tumor “refuge” (151). At present, improving penetration into the blood-brain barrier and intracranial activity is one of the key points in developing the third-generation TKI and new drugs. The development of nano-targeted drug systems might also benefit patients with brain metastases (152).
With the in-depth study of the mechanism for driver genes in lung cancer with brain metastasis, driver gene-associated signaling pathways, such as RAS/RAF, PI3K/AKT/mTOR, WNT/β-catenin, and JAK/STAT, also provide new targets for the treatment of lung cancer with brain metastases. mTORC1/2 inhibitor, for example, have demonstrated inhibition effects on tumor growth, EMT, metastasis, and improvements in anti-tumor immunity in preclinical models of lung cancer (153). JAK1/2 inhibitors also have potential therapeutic effects in patients with KRAS mutations (86). The PI3K signaling pathway is also enriched in brain metastases, suggesting an association of this pathway with increased risk of brain metastasis, which is expected to become a new therapeutic target (66, 154). However, none of these inhibitors has been studied in brain metastasis models. Table 2 summarizes the potential biomarkers in lung cancer driver genes and lists the targeted drugs in brain metastasis. It is important to note that although targeted drugs of the rare driver gene associated with brain metastasis have not been studied in lung cancer, they have shown good results in other models of brain metastasis, such as breast cancer and melanoma (155, 156).
Moreover, the occurrence of secondary mutations in driver genes or other new mutations increases the complexity of the tumor genome, leading to drug resistance in targeted therapies and limiting patient’s survival (157). For NSCLC, more than 30% of patients with mutant EGFR undergo disease exacerbation due to brain metastasis during TKIs treatment (158, 159). On the other hand, the response to treatment is different between patients with or without driver gene mutation. For example, programmed death 1 (PD-1) inhibitor has played a role in treatment of NSCLC brain metastasis (160, 161). For patients without driver gene mutation, Ma ZY et al. found that the outcomes of patients with NSCLC presenting brain metastasis were comparable to patients without BMs when treated with nivolumab (PD-1 inhibitor) (162). This result suggests that driver genes are significant for the hierarchical management of patient treatment.
Outlook
This review summarizes for the first time the signaling pathways related to driver genes and the role of these signaling pathways in the mechanism of brain metastasis. Tumor driver gene-associated signaling pathways are important signals for lung cancer with brain metastasis, which promotes tumor cell survival, invasion, and colonization. Furthermore, various cytokines and chemokine signals can be released after interaction of tumor cells with original resident cells in the brain or lung. These signals promote metastasis by driving gene-related signaling pathways. However, it is worthy of attention to researchers that in recent studies, lung cancer driver genes are related to brain metastasis and can be used as biomarkers for predicting brain metastasis, but there is still a lack of molecular mechanisms of brain metastasis starting with driver genes.
With the popularization of genetic testing technology, when patients with non-small cell lung cancer are diagnosed, clarifying the driver mutation status or intratumoral heterogeneity of the primary lesion can not only guide medication, but also predict the subsequent development of the tumor, including brain metastasis. Currently, only clinicopathologic variables, such as patient age, disease stage, and tumor histology, are used to predict the risk of brain metastasis (163–165). In future studies, common and druggable lung cancer driver genes, which have been confirmed to predict brain metastasis, can be combined with high-risk clinical features through artificial intelligence algorithms to establish a brain metastasis prediction model. For patients at high risk of brain metastasis, more treatment strategies, including PCI, targeted therapy, and immunotherapy, can be chosen. And high-risk patients without neurological symptoms also need regular computed tomography or magnetic resonance imaging. New lung cancer driver gene, which involved in pathways associated with brain metastasis, can be studied as potential biomarkers. Last but not the least, understanding the molecular characteristics of primary tumor and brain metastases can provide more information about tumor driver genes in the clinic for precise treatment.
Author Contributions
YK performed a literature search, interpreted the data, and wrote the manuscript. YJ, QL, and XY supervised and contributed to the writing process. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 81773360).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin (2020) 70(1):7–30. doi: 10.3322/caac.21590
3. Tabouret E, Chinot O, Metellus P, Tallet A, Viens P, Goncalves A. Recent trends in epidemiology of brain metastases: an overview. Anticancer Res (2012) 32(11):4655–62.
4. Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, et al. Brain metastases. Nat Rev Dis Primers (2019) 5(1):5. doi: 10.1038/s41572-018-0055-y
5. Nayak L, Lee EQ, Wen PY. Epidemiology of brain metastases. Curr Oncol Rep (2012) 14(1):48–54. doi: 10.1007/s11912-011-0203-y
6. Arriagada R, Le Chevalier T, Riviere A, Chomy P, Monnet I, Bardet E, et al. Patterns of failure after prophylactic cranial irradiation in small-cell lung cancer: analysis of 505 randomized patients. Ann Oncol (2002) 13(5):748–54. doi: 10.1093/annonc/mdf123
7. Gore EM, Bae K, Wong SJ, Sun A, Bonner JA, Schild SE, et al. Phase III comparison of prophylactic cranial irradiation versus observation in patients with locally advanced non-small-cell lung cancer: primary analysis of radiation therapy oncology group study RTOG 0214. J Clin Oncol (2011) 29(3):272–8. doi: 10.1200/JCO.2010.29.1609
8. Coppes-Zantinga AR, Coppes MJ. Sir James Paget (1814-1889): a great academic Victorian. J Am Coll Surg (2000) 191(1):70–4. doi: 10.1016/s1072-7515(00)00250-7
9. Wicha MS, Liu S, Dontu G. Cancer stem cells: an old idea–a paradigm shift. Cancer Res (2006) 66(4):1883–1890; discussion 1895-1886. doi: 10.1158/0008-5472.CAN-05-3153
10. Budhu A, Wang XW. Transforming the microenvironment: a trick of the metastatic cancer cell. Cancer Cell (2012) 22(3):279–80. doi: 10.1016/j.ccr.2012.08.018
11. Bidard FC, Poupon MF. The metastatic process: history, models and recent advances. Med Sci (Paris) (2012) 28(1):89–95. doi: 10.1051/medsci/2012281022
12. Coghlin C, Murray GI. Current and emerging concepts in tumour metastasis. J Pathol (2010) 222(1):1–15. doi: 10.1002/path.2727
13. Leroi AM, Koufopanou V, Burt A. Cancer selection. Nat Rev Cancer (2003) 3(3):226–31. doi: 10.1038/nrc1016
14. Wu X, Northcott PA, Dubuc A, Dupuy AJ, Shih DJ, Witt H, et al. Clonal selection drives genetic divergence of metastatic medulloblastoma. Nature (2012) 482(7386):529–33. doi: 10.1038/nature10825
15. Lee JJ, Park S, Park H, Kim S, Lee J, Lee J, et al. Tracing Oncogene Rearrangements in the Mutational History of Lung Adenocarcinoma. Cell (2019) 177(7):1842–57.e1821. doi: 10.1016/j.cell.2019.05.013
16. de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science (2014) 346(6206):251–6. doi: 10.1126/science.1253462
17. Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science (2014) 346(6206):256–9. doi: 10.1126/science.1256930
18. Turajlic S, Xu H, Litchfield K, Rowan A, Horswell S, Chambers T, et al. Deterministic Evolutionary Trajectories Influence Primary Tumor Growth: TRACERx Renal. Cell (2018) 173(3):595–610.e511. doi: 10.1016/j.cell.2018.03.043
19. Jamal-Hanjani M, Wilson GA, McGranahan N, Birkbak NJ, Watkins TBK, Veeriah S, et al. Tracking the Evolution of Non-Small-Cell Lung Cancer. N Engl J Med (2017) 376(22):2109–21. doi: 10.1056/NEJMoa1616288
20. Biswas D, Birkbak NJ, Rosenthal R, Hiley CT, Lim EL, Papp K, et al. A clonal expression biomarker associates with lung cancer mortality. Nat Med (2019) 25(10):1540–8. doi: 10.1038/s41591-019-0595-z
21. Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell (2012) 150(6):1107–20. doi: 10.1016/j.cell.2012.08.029
22. Carithers LJ, Moore HM. The Genotype-Tissue Expression (GTEx) Project. Biopreserv Biobank (2015) 13(5):307–8. doi: 10.1089/bio.2015.29031.hmm
23. Consortium GT, Laboratory DA, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC, et al. Genetic effects on gene expression across human tissues. Nature (2017) 550(7675):204–13. doi: 10.1038/nature24277
24. Guo Y, Cao R, Zhang X, Huang L, Sun L, Zhao J, et al. Recent Progress in Rare Oncogenic Drivers and Targeted Therapy For Non-Small Cell Lung Cancer. Onco Targets Ther (2019) 12:10343–60. doi: 10.2147/OTT.S230309
25. Zhang Y, Xia M, Jin K, Wang S, Wei H, Fan C, et al. Function of the c-Met receptor tyrosine kinase in carcinogenesis and associated therapeutic opportunities. Mol Cancer (2018) 17(1):45. doi: 10.1186/s12943-018-0796-y
26. Wang H, Wang Z, Zhang G, Zhang M, Zhang X, Li H, et al. Driver genes as predictive indicators of brain metastasis in patients with advanced NSCLC: EGFR, ALK, and RET gene mutations. Cancer Med (2020) 9(2):487–95. doi: 10.1002/cam4.2706
27. Tomasini P, Serdjebi C, Khobta N, Metellus P, Ouafik L, Nanni I, et al. EGFR and KRAS Mutations Predict the Incidence and Outcome of Brain Metastases in Non-Small Cell Lung Cancer. Int J Mol Sci (2016) 17(12). doi: 10.3390/ijms17122132
28. Iuchi T, Shingyoji M, Itakura M, Yokoi S, Moriya Y, Tamura H, et al. Frequency of brain metastases in non-small-cell lung cancer, and their association with epidermal growth factor receptor mutations. Int J Clin Oncol (2015) 20(4):674–9. doi: 10.1007/s10147-014-0760-9
29. Li S, Choi YL, Gong Z, Liu X, Lira M, Kan Z, et al. Comprehensive Characterization of Oncogenic Drivers in Asian Lung Adenocarcinoma. J Thorac Oncol (2016) 11(12):2129–40. doi: 10.1016/j.jtho.2016.08.142
30. Zhang B, Zhang L, Yue D, Li C, Zhang H, Ye J, et al. Genomic characteristics in Chinese non-small cell lung cancer patients and its value in prediction of postoperative prognosis. Transl Lung Cancer Res (2020) 9(4):1187–201. doi: 10.21037/tlcr-19-664
31. Patil T, Smith DE, Bunn PA, Aisner DL, Le AT, Hancock M, et al. The Incidence of Brain Metastases in Stage IV ROS1-Rearranged Non-Small Cell Lung Cancer and Rate of Central Nervous System Progression on Crizotinib. J Thorac Oncol (2018) 13(11):1717–26. doi: 10.1016/j.jtho.2018.07.001
32. Wang H, Ou Q, Li D, Qin T, Bao H, Hou X, et al. Genes associated with increased brain metastasis risk in non-small cell lung cancer: Comprehensive genomic profiling of 61 resected brain metastases versus primary non-small cell lung cancer (Guangdong Association Study of Thoracic Oncology 1036). Cancer (2019) 125(20):3535–44. doi: 10.1002/cncr.32372
33. Shih DJH, Nayyar N, Bihun I, Dagogo-Jack I, Gill CM, Aquilanti E, et al. Genomic characterization of human brain metastases identifies drivers of metastatic lung adenocarcinoma. Nat Genet (2020) 52(4):371–7. doi: 10.1038/s41588-020-0592-7
34. Saunus JM, Quinn MC, Patch AM, Pearson JV, Bailey PJ, Nones K, et al. Integrated genomic and transcriptomic analysis of human brain metastases identifies alterations of potential clinical significance. J Pathol (2015) 237(3):363–78. doi: 10.1002/path.4583
35. Sun M, Behrens C, Feng L, Ozburn N, Tang X, Yin G, et al. HER family receptor abnormalities in lung cancer brain metastases and corresponding primary tumors. Clin Cancer Res (2009) 15(15):4829–37. doi: 10.1158/1078-0432.CCR-08-2921
36. Shin DY, Na II, Kim CH, Park S, Baek H, Yang SH. EGFR mutation and brain metastasis in pulmonary adenocarcinomas. J Thorac Oncol (2014) 9(2):195–9. doi: 10.1097/JTO.0000000000000069
37. Rangachari D, Yamaguchi N, VanderLaan PA, Folch E, Mahadevan A, Floyd SR, et al. Brain metastases in patients with EGFR-mutated or ALK-rearranged non-small-cell lung cancers. Lung Cancer (2015) 88(1):108–11. doi: 10.1016/j.lungcan.2015.01.020
38. Schuler M, Wu YL, Hirsh V, O’Byrne K, Yamamoto N, Mok T, et al. First-Line Afatinib versus Chemotherapy in Patients with Non-Small Cell Lung Cancer and Common Epidermal Growth Factor Receptor Gene Mutations and Brain Metastases. J Thorac Oncol (2016) 11(3):380–90. doi: 10.1016/j.jtho.2015.11.014
39. Liu J, Xing L, Meng X, Yue J, Meng X, Xie P, et al. Risk of brain metastasis reduced after erlotinib treatment in advanced pulmonary adenocarcinoma patients with sensitive EGFR mutation. Onco Targets Ther (2016) 9:671–9. doi: 10.2147/OTT.S100105
40. Zhao X, Zhu G, Chen H, Yang P, Li F, Du N. Efficacy of icotinib versus traditional chemotherapy as first-line treatment for preventing brain metastasis from advanced lung adenocarcinoma in patients with epidermal growth factor receptor-sensitive mutation. J Cancer Res Ther (2016) 12(3):1127–31. doi: 10.4103/0973-1482.194599
41. Yang Z, Guo Q, Wang Y, Chen K, Zhang L, Cheng Z, et al. AZD3759, a BBB-penetrating EGFR inhibitor for the treatment of EGFR mutant NSCLC with CNS metastases. Sci Transl Med (2016) 8(368):368ra172. doi: 10.1126/scitranslmed.aag0976
42. Yun J, Hong MH, Kim SY, Park CW, Kim S, Yun MR, et al. YH25448, an Irreversible EGFR-TKI with Potent Intracranial Activity in EGFR Mutant Non-Small Cell Lung Cancer. Clin Cancer Res (2019) 25(8):2575–87. doi: 10.1158/1078-0432.CCR-18-2906
43. Toyokawa G, Seto T, Takenoyama M, Ichinose Y. Insights into brain metastasis in patients with ALK+ lung cancer: is the brain truly a sanctuary? Cancer Metastasis Rev (2015) 34(4):797–805. doi: 10.1007/s10555-015-9592-y
44. Gadgeel S, Peters S, Mok T, Shaw AT, Kim DW, Ou SI, et al. Alectinib versus crizotinib in treatment-naive anaplastic lymphoma kinase-positive (ALK+) non-small-cell lung cancer: CNS efficacy results from the ALEX study. Ann Oncol (2018) 29(11):2214–22. doi: 10.1093/annonc/mdy405
45. Takeda M, Okamoto I, Nakagawa K. Clinical impact of continued crizotinib administration after isolated central nervous system progression in patients with lung cancer positive for ALK rearrangement. J Thorac Oncol (2013) 8(5):654–7. doi: 10.1097/JTO.0b013e31828c28e7
46. Costa DB, Shaw AT, Ou SH, Solomon BJ, Riely GJ, Ahn MJ, et al. Clinical Experience With Crizotinib in Patients With Advanced ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastases. J Clin Oncol (2015) 33(17):1881–8. doi: 10.1200/JCO.2014.59.0539
47. Akamine T, Toyokawa G, Tagawa T, Seto T. Spotlight on lorlatinib and its potential in the treatment of NSCLC: the evidence to date. Onco Targets Ther (2018) 11:5093–101. doi: 10.2147/OTT.S165511
48. Solomon BJ, Besse B, Bauer TM, Felip E, Soo RA, Camidge DR, et al. Lorlatinib in patients with ALK-positive non-small-cell lung cancer: results from a global phase 2 study. Lancet Oncol (2018) 19(12):1654–67. doi: 10.1016/s1470-2045(18)30649-1
49. Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, et al. PF-06463922, an ALK/ROS1 Inhibitor, Overcomes Resistance to First and Second Generation ALK Inhibitors in Preclinical Models. Cancer Cell (2015) 28(1):70–81. doi: 10.1016/j.ccell.2015.05.010
50. Gainor JF, Tseng D, Yoda S, Dagogo-Jack I, Friboulet L, Lin JJ, et al. Patterns of Metastatic Spread and Mechanisms of Resistance to Crizotinib in ROS1-Positive Non-Small-Cell Lung Cancer. JCO Precis Oncol (2017) 2017. doi: 10.1200/PO.17.00063
51. Park S, Ahn BC, Lim SW, Sun JM, Kim HR, Hong MH, et al. Characteristics and Outcome of ROS1-Positive Non-Small Cell Lung Cancer Patients in Routine Clinical Practice. J Thorac Oncol (2018) 13(9):1373–82. doi: 10.1016/j.jtho.2018.05.026
52. Sartore-Bianchi A, Pizzutilo EG, Marrapese G, Tosi F, Cerea G, Siena S. Entrectinib for the treatment of metastatic NSCLC: safety and efficacy. Expert Rev Anticancer Ther (2020) 20(5):333–41. doi: 10.1080/14737140.2020.1747439
53. Drilon A, Siena S, Dziadziuszko R, Barlesi F, Krebs MG, Shaw AT, et al. Entrectinib in ROS1 fusion-positive non-small-cell lung cancer: integrated analysis of three phase 1-2 trials. Lancet Oncol (2020) 21(2):261–70. doi: 10.1016/S1470-2045(19)30690-4
54. Yun MR, Kim DH, Kim SY, Joo HS, Lee YW, Choi HM, et al. Repotrectinib Exhibits Potent Antitumor Activity in Treatment-Naive and Solvent-Front-Mutant ROS1-Rearranged Non-Small Cell Lung Cancer. Clin Cancer Res (2020) 26(13):3287–95. doi: 10.1158/1078-0432.CCR-19-2777
55. Klempner SJ, Borghei A, Hakimian B, Ali SM, Ou SI. Intracranial Activity of Cabozantinib in MET Exon 14-Positive NSCLC with Brain Metastases. J Thorac Oncol (2017) 12(1):152–6. doi: 10.1016/j.jtho.2016.09.127
56. Blanc-Durand F, Alameddine R, Iafrate AJ, Tran-Thanh D, Lo YC, Blais N, et al. Tepotinib Efficacy in a Patient with Non-Small Cell Lung Cancer with Brain Metastasis Harboring an HLA-DRB1-MET Gene Fusion. Oncologist (2020) 5(11):916–20. doi: 10.1634/theoncologist.2020-0502
57. Drilon A, Lin JJ, Filleron T, Ni A, Milia J, Bergagnini I, et al. Frequency of Brain Metastases and Multikinase Inhibitor Outcomes in Patients With RET-Rearranged Lung Cancers. J Thorac Oncol (2018) 13(10):1595–601. doi: 10.1016/j.jtho.2018.07.004
58. Drilon A, Oxnard GR, Tan DSW, Loong HHF, Johnson M, Gainor J, et al. Efficacy of Selpercatinib in RET Fusion-Positive Non-Small-Cell Lung Cancer. N Engl J Med (2020) 383(9):813–24. doi: 10.1056/NEJMoa2005653
59. Gao W, Jin J, Yin J, Land S, Gaither-Davis A, Christie N, et al. KRAS and TP53 mutations in bronchoscopy samples from former lung cancer patients. Mol Carcinog (2017) 56(2):381–8. doi: 10.1002/mc.22501
60. Lohinai Z, Klikovits T, Moldvay J, Ostoros G, Raso E, Timar J, et al. KRAS-mutation incidence and prognostic value are metastatic site-specific in lung adenocarcinoma: poor prognosis in patients with KRAS mutation and bone metastasis. Sci Rep (2017) 7:39721. doi: 10.1038/srep39721
61. Marchetti A, Felicioni L, Malatesta S, Grazia Sciarrotta M, Guetti L, Chella A, et al. Clinical features and outcome of patients with non-small-cell lung cancer harboring BRAF mutations. J Clin Oncol (2011) 29(26):3574–9. doi: 10.1200/JCO.2011.35.9638
62. Fernandez-Cuesta L, Plenker D, Osada H, Sun R, Menon R, Leenders F, et al. CD74-NRG1 fusions in lung adenocarcinoma. Cancer Discovery (2014) 4(4):415–22. doi: 10.1158/2159-8290.CD-13-0633
63. Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, et al. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res (2012) 18(4):1167–76. doi: 10.1158/1078-0432.CCR-11-2109
64. Rau KM, Chen HK, Shiu LY, Chao TL, Lo YP, Wang CC, et al. Discordance of Mutation Statuses of Epidermal Growth Factor Receptor and K-ras between Primary Adenocarcinoma of Lung and Brain Metastasis. Int J Mol Sci (2016) 17(4):524. doi: 10.3390/ijms17040524
65. Brastianos PK, Carter SL, Santagata S, Cahill DP, Taylor-Weiner A, Jones RT, et al. Genomic Characterization of Brain Metastases Reveals Branched Evolution and Potential Therapeutic Targets. Cancer Discovery (2015) 5(11):1164–77. doi: 10.1158/2159-8290.CD-15-0369
66. Paik PK, Shen R, Won H, Rekhtman N, Wang L, Sima CS, et al. Next-Generation Sequencing of Stage IV Squamous Cell Lung Cancers Reveals an Association of PI3K Aberrations and Evidence of Clonal Heterogeneity in Patients with Brain Metastases. Cancer Discovery (2015) 5(6):610–21. doi: 10.1158/2159-8290.CD-14-1129
67. Li Q, Wu H, Chen B, Hu G, Huang L, Qin K, et al. SNPs in the TGF-beta signaling pathway are associated with increased risk of brain metastasis in patients with non-small-cell lung cancer. PloS One (2012) 7(12):e51713. doi: 10.1371/journal.pone.0051713
68. Li Q, Yang J, Yu Q, Wu H, Liu B, Xiong H, et al. Associations between single-nucleotide polymorphisms in the PI3K-PTEN-AKT-mTOR pathway and increased risk of brain metastasis in patients with non-small cell lung cancer. Clin Cancer Res (2013) 19(22):6252–60. doi: 10.1158/1078-0432.CCR-13-1093
69. Xu Y, Huang Y, Weng L, Zheng J, Huang Y, Lin Y, et al. Effects of single-nucleotide polymorphisms in the mTORC1 pathway on the risk of brain metastasis in patients with non-small cell lung cancer. J Cancer Res Clin Oncol (2020) 146(1):273–85. doi: 10.1007/s00432-019-03059-y
70. Krencz I, Sebestyen A, Fabian K, Mark A, Moldvay J, Khoor A, et al. Expression of mTORC1/2-related proteins in primary and brain metastatic lung adenocarcinoma. Hum Pathol (2017) 62:66–73. doi: 10.1016/j.humpath.2016.12.012
71. Du Z, Lovly CM. Mechanisms of receptor tyrosine kinase activation in cancer. Mol Cancer (2018) 17(1):58. doi: 10.1186/s12943-018-0782-4
72. Wee P, Wang Z. Epidermal Growth Factor Receptor Cell Proliferation Signaling Pathways. Cancers (Basel) (2017) 9(5). doi: 10.3390/cancers9050052
73. Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature (2007) 448(7153):561–6. doi: 10.1038/nature05945
74. Besset V, Scott RP, Ibanez CF. Signaling complexes and protein-protein interactions involved in the activation of the Ras and phosphatidylinositol 3-kinase pathways by the c-Ret receptor tyrosine kinase. J Biol Chem (2000) 275(50):39159–66. doi: 10.1074/jbc.M006908200
75. Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature (2008) 455(7216):1069–75. doi: 10.1038/nature07423
76. Vogt PK, Hart JR. PI3K and STAT3: a new alliance. Cancer Discovery (2011) 1(6):481–6. doi: 10.1158/2159-8290.CD-11-0218
77. Garbers C, Heink S, Korn T, Rose-John S. Interleukin-6: designing specific therapeutics for a complex cytokine. Nat Rev Drug Discovery (2018) 17(6):395–412. doi: 10.1038/nrd.2018.45
78. Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell (2012) 150(6):1121–34. doi: 10.1016/j.cell.2012.08.024
79. Wang Y, Godin-Heymann N, Dan Wang X, Bergamaschi D, Llanos S, Lu X. ASPP1 and ASPP2 bind active RAS, potentiate RAS signalling and enhance p53 activity in cancer cells. Cell Death Differ (2013) 20(4):525–34. doi: 10.1038/cdd.2013.3
80. Rojas A, Padidam M, Cress D, Grady WM. TGF-beta receptor levels regulate the specificity of signaling pathway activation and biological effects of TGF-beta. Biochim Biophys Acta (2009) 1793(7):1165–73. doi: 10.1016/j.bbamcr.2009.02.001
81. Borczuk AC, Sole M, Lu P, Chen J, Wilgus ML, Friedman RA, et al. Progression of human bronchioloalveolar carcinoma to invasive adenocarcinoma is modeled in a transgenic mouse model of K-ras-induced lung cancer by loss of the TGF-beta type II receptor. Cancer Res (2011) 71(21):6665–75. doi: 10.1158/0008-5472.CAN-11-1590
82. Cao W, Liu Y, Zhang R, Zhang B, Wang T, Zhu X, et al. Homoharringtonine induces apoptosis and inhibits STAT3 via IL-6/JAK1/STAT3 signal pathway in Gefitinib-resistant lung cancer cells. Sci Rep (2015) 5:8477. doi: 10.1038/srep08477
83. Miller A, McLeod L, Alhayyani S, Szczepny A, Watkins DN, Chen W, et al. Blockade of the IL-6 trans-signalling/STAT3 axis suppresses cachexia in Kras-induced lung adenocarcinoma. Oncogene (2017) 36(21):3059–66. doi: 10.1038/onc.2016.437
84. Brooks GD, McLeod L, Alhayyani S, Miller A, Russell PA, Ferlin W, et al. IL6 Trans-signaling Promotes KRAS-Driven Lung Carcinogenesis. Cancer Res (2016) 76(4):866–76. doi: 10.1158/0008-5472.CAN-15-2388
85. Caetano MS, Hassane M, Van HT, Bugarin E, Cumpian AM, McDowell CL, et al. Sex specific function of epithelial STAT3 signaling in pathogenesis of K-ras mutant lung cancer. Nat Commun (2018) 9(1):4589. doi: 10.1038/s41467-018-07042-y
86. Mohrherr J, Haber M, Breitenecker K, Aigner P, Moritsch S, Voronin V, et al. JAK-STAT inhibition impairs K-RAS-driven lung adenocarcinoma progression. Int J Cancer (2019) 145(12):3376–88. doi: 10.1002/ijc.32624
87. Sippel TR, Johnson AM, Li HY, Hanson D, Nguyen TT, Bullock BL, et al. Activation of PPARgamma in Myeloid Cells Promotes Progression of Epithelial Lung Tumors through TGFbeta1. Mol Cancer Res (2019) 17(8):1748–58. doi: 10.1158/1541-7786.MCR-19-0236
88. Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr., Kinzler KW. Cancer genome landscapes. Science (2013) 339(6127):1546–58. doi: 10.1126/science.1235122
89. Turajlic S, Swanton C. Metastasis as an evolutionary process. Science (2016) 352(6282):169–75. doi: 10.1126/science.aaf2784
90. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature (2018) 553(7689):467–72. doi: 10.1038/nature25432
91. Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet (2003) 33(1):49–54. doi: 10.1038/ng1060
92. Bos PD, Zhang XHF, Nadal C, Shu W, Gomis RR, Nguyen DX, et al. Genes that mediate breast cancer metastasis to the brain. Nature (2009) 459(7249):1005–9. doi: 10.1038/nature08021
93. Hanahan D, Weinberg RA. The hallmarks of cancer. Cell (2000) 100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9
94. Kienast Y, von Baumgarten L, Fuhrmann M, Klinkert WE, Goldbrunner R, Herms J, et al. Real-time imaging reveals the single steps of brain metastasis formation. Nat Med (2010) 16(1):116–22. doi: 10.1038/nm.2072
95. Malladi S, Macalinao DG, Jin X, He L, Basnet H, Zou Y, et al. Metastatic Latency and Immune Evasion through Autocrine Inhibition of WNT. Cell (2016) 165(1):45–60. doi: 10.1016/j.cell.2016.02.025
96. You H, Baluszek S, Kaminska B. Supportive roles of brain macrophages in CNS metastases and assessment of new approaches targeting their functions. Theranostics (2020) 10(7):2949–64. doi: 10.7150/thno.40783
97. Yun J, Rago C, Cheong I, Pagliarini R, Angenendt P, Rajagopalan H, et al. Glucose deprivation contributes to the development of KRAS pathway mutations in tumor cells. Science (2009) 325(5947):1555–9. doi: 10.1126/science.1174229
98. Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell (2012) 149(3):656–70. doi: 10.1016/j.cell.2012.01.058
99. Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer (2005) 5(5):341–54. doi: 10.1038/nrc1609
100. Davies KD, Doebele RC. Molecular pathways: ROS1 fusion proteins in cancer. Clin Cancer Res (2013) 19(15):4040–5. doi: 10.1158/1078-0432.CCR-12-2851
101. Yamaoka T, Arata S, Homma M, Homma T, Kusumoto S, Ando K, et al. Blockade of EGFR Activation Promotes TNF-Induced Lung Epithelial Cell Apoptosis and Pulmonary Injury. Int J Mol Sci (2019) 20(16). doi: 10.3390/ijms20164021
102. Wei C, Dong X, Lu H, Tong F, Chen L, Zhang R, et al. LPCAT1 promotes brain metastasis of lung adenocarcinoma by up-regulating PI3K/AKT/MYC pathway. J Exp Clin Cancer Res (2019) 38(1):95. doi: 10.1186/s13046-019-1092-4
103. Cheng G, Zhang Q, Pan J, Lee Y, Ouari O, Hardy M, et al. Targeting lonidamine to mitochondria mitigates lung tumorigenesis and brain metastasis. Nat Commun (2019) 10(1):2205. doi: 10.1038/s41467-019-10042-1
104. Zhang L, Zhang S, Yao J, Lowery FJ, Zhang Q, Huang WC, et al. Microenvironment-induced PTEN loss by exosomal microRNA primes brain metastasis outgrowth. Nature (2015) 527(7576):100–4. doi: 10.1038/nature15376
105. You R, DeMayo FJ, Liu J, Cho SN, Burt BM, Creighton CJ, et al. IL17A Regulates Tumor Latency and Metastasis in Lung Adeno and Squamous SQ.2b and AD.1 Cancer. Cancer Immunol Res (2018) 6(6):645–57. doi: 10.1158/2326-6066.CIR-17-0554
106. Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, et al. Cancer-Derived Succinate Promotes Macrophage Polarization and Cancer Metastasis via Succinate Receptor. Mol Cell (2020) 77(2):213–27.e215. doi: 10.1016/j.molcel.2019.10.023
107. Zhang H, Yu Y, Zhou L, Ma J, Tang K, Xu P, et al. Circulating Tumor Microparticles Promote Lung Metastasis by Reprogramming Inflammatory and Mechanical Niches via a Macrophage-Dependent Pathway. Cancer Immunol Res (2018) 6(9):1046–56. doi: 10.1158/2326-6066.CIR-17-0574
108. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol (2019) 19(6):369–82. doi: 10.1038/s41577-019-0127-6
109. Tan B, Shi X, Zhang J, Qin J, Zhang N, Ren H, et al. Inhibition of Rspo-Lgr4 Facilitates Checkpoint Blockade Therapy by Switching Macrophage Polarization. Cancer Res (2018) 78(17):4929–42. doi: 10.1158/0008-5472.CAN-18-0152
110. Jing B, Wang T, Sun B, Xu J, Xu D, Liao Y, et al. IL6/STAT3 Signaling Orchestrates Premetastatic Niche Formation and Immunosuppressive Traits in Lung. Cancer Res (2020) 80(4):784–97. doi: 10.1158/0008-5472.CAN-19-2013
111. Wu SY, Xing F, Sharma S, Wu K, Tyagi A, Liu Y, et al. Nicotine promotes brain metastasis by polarizing microglia and suppressing innate immune function. J Exp Med (2020) 217(8). doi: 10.1084/jem.20191131
112. Wenes M, Shang M, Di Matteo M, Goveia J, Martin-Perez R, Serneels J, et al. Macrophage Metabolism Controls Tumor Blood Vessel Morphogenesis and Metastasis. Cell Metab (2016) 24(5):701–15. doi: 10.1016/j.cmet.2016.09.008
113. Larsen AK, Ouaret D, El Ouadrani K, Petitprez A, Targeting EGFR. and VEGF(R) pathway cross-talk in tumor survival and angiogenesis. Pharmacol Ther (2011) 131(1):80–90. doi: 10.1016/j.pharmthera.2011.03.012
114. Brabletz T. EMT and MET in metastasis: where are the cancer stem cells? Cancer Cell (2012) 22(6):699–701. doi: 10.1016/j.ccr.2012.11.009
115. Yoo JY, Yang SH, Lee JE, Cho DG, Kim HK, Kim SH, et al. E-cadherin as a predictive marker of brain metastasis in non-small-cell lung cancer, and its regulation by pioglitazone in a preclinical model. J Neurooncol (2012) 109(2):219–27. doi: 10.1007/s11060-012-0890-8
116. Grinberg-Rashi H, Ofek E, Perelman M, Skarda J, Yaron P, Hajduch M, et al. The expression of three genes in primary non-small cell lung cancer is associated with metastatic spread to the brain. Clin Cancer Res (2009) 15(5):1755–61. doi: 10.1158/1078-0432.CCR-08-2124
117. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev (2009) 28(1-2):15–33. doi: 10.1007/s10555-008-9169-0
118. Yousefi M, Bahrami T, Salmaninejad A, Nosrati R, Ghaffari P, Ghaffari SH. Lung cancer-associated brain metastasis: Molecular mechanisms and therapeutic options. Cell Oncol (2017) 40(5):419–41. doi: 10.1007/s13402-017-0345-5
119. Xiao D, He J. Epithelial mesenchymal transition and lung cancer. J Thorac Dis (2010) 2(3):154–9. doi: 10.3978/j.issn.2072-1439.2010.02.03.7
120. Shin SY, Rath O, Zebisch A, Choo SM, Kolch W, Cho KH. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res (2010) 70(17):6715–24. doi: 10.1158/0008-5472.CAN-10-1377
121. Eger A, Stockinger A, Park J, Langkopf E, Mikula M, Gotzmann J, et al. beta-Catenin and TGFbeta signalling cooperate to maintain a mesenchymal phenotype after FosER-induced epithelial to mesenchymal transition. Oncogene (2004) 23(15):2672–80. doi: 10.1038/sj.onc.1207416
122. Su J, Morgani SM, David CJ, Wang Q, Er EE, Huang YH, et al. TGF-beta orchestrates fibrogenic and developmental EMTs via the RAS effector RREB1. Nature (2020) 577(7791):566–71. doi: 10.1038/s41586-019-1897-5
123. Janda E, Lehmann K, Killisch I, Jechlinger M, Herzig M, Downward J, et al. Ras and TGF[beta] cooperatively regulate epithelial cell plasticity and metastasis: dissection of Ras signaling pathways. J Cell Biol (2002) 156(2):299–313. doi: 10.1083/jcb.200109037
124. Bakin AV, Tomlinson AK, Bhowmick NA, Moses HL, Arteaga CL. Phosphatidylinositol 3-kinase function is required for transforming growth factor beta-mediated epithelial to mesenchymal transition and cell migration. J Biol Chem (2000) 275(47):36803–10. doi: 10.1074/jbc.M005912200
125. Morel AP, Hinkal GW, Thomas C, Fauvet F, Courtois-Cox S, Wierinckx A, et al. EMT inducers catalyze malignant transformation of mammary epithelial cells and drive tumorigenesis towards claudin-low tumors in transgenic mice. PloS Genet (2012) 8(5):e1002723. doi: 10.1371/journal.pgen.1002723
126. Seoane J, Gomis RR. TGF-beta Family Signaling in Tumor Suppression and Cancer Progression. Cold Spring Harb Perspect Biol (2017) 9(12). doi: 10.1101/cshperspect.a022277
127. Li R, Hebert JD, Lee TA, Xing H, Boussommier-Calleja A, Hynes RO, et al. Macrophage-Secreted TNFalpha and TGFbeta1 Influence Migration Speed and Persistence of Cancer Cells in 3D Tissue Culture via Independent Pathways. Cancer Res (2017) 77(2):279–90. doi: 10.1158/0008-5472.CAN-16-0442
128. Che D, Zhang S, Jing Z, Shang L, Jin S, Liu F, et al. Macrophages induce EMT to promote invasion of lung cancer cells through the IL-6-mediated COX-2/PGE2/beta-catenin signalling pathway. Mol Immunol (2017) 90:197–210. doi: 10.1016/j.molimm.2017.06.018
129. Wei C, Zhang R, Cai Q, Gao X, Tong F, Dong J, et al. MicroRNA-330-3p promotes brain metastasis and epithelial-mesenchymal transition via GRIA3 in non-small cell lung cancer. Aging (Albany NY) (2019) 11(17):6734–61. doi: 10.18632/aging.102201
130. Yang L, Li J, Fu S, Ren P, Tang J, Wang N, et al. Up-regulation of Insulin-like Growth Factor Binding Protein-3 Is Associated with Brain Metastasis in Lung Adenocarcinoma. Mol Cells (2019) 42(4):321–32. doi: 10.14348/molcells.2019.2441
131. Zhang Y, Zeng Y, Liu T, Du W, Zhu J, Liu Z, et al. The canonical TGF-beta/Smad signalling pathway is involved in PD-L1-induced primary resistance to EGFR-TKIs in EGFR-mutant non-small-cell lung cancer. Respir Res (2019) 20(1):164. doi: 10.1186/s12931-019-1137-4
132. Kurimoto R, Iwasawa S, Ebata T, Ishiwata T, Sekine I, Tada Y, et al. Drug resistance originating from a TGF-beta/FGF-2-driven epithelial-to-mesenchymal transition and its reversion in human lung adenocarcinoma cell lines harboring an EGFR mutation. Int J Oncol (2016) 48(5):1825–36. doi: 10.3892/ijo.2016.3419
133. Wang Z, Liu Y, Takahashi M, Van Hook K, Kampa-Schittenhelm KM, Sheppard BC, et al. N terminus of ASPP2 binds to Ras and enhances Ras/Raf/MEK/ERK activation to promote oncogene-induced senescence. Proc Natl Acad Sci USA (2013) 110(1):312–7. doi: 10.1073/pnas.1201514110
134. Wang Y, Bu F, Royer C, Serres S, Larkin JR, Soto MS, et al. ASPP2 controls epithelial plasticity and inhibits metastasis through beta-catenin-dependent regulation of ZEB1. Nat Cell Biol (2014) 16(11):1092–104. doi: 10.1038/ncb3050
135. Kim YB, Ahn YH, Jung JH, Lee YJ, Lee JH, Kang JL. Programming of macrophages by UV-irradiated apoptotic cancer cells inhibits cancer progression and lung metastasis. Cell Mol Immunol (2019) 16(11):851–67. doi: 10.1038/s41423-019-0209-1
136. Ma SC, Li Q, Peng JY, Zhouwen JL, Diao JF, Niu JX, et al. Claudin-5 regulates blood-brain barrier permeability by modifying brain microvascular endothelial cell proliferation, migration, and adhesion to prevent lung cancer metastasis. CNS Neurosci Ther (2017) 23(12):947–60. doi: 10.1111/cns.12764
137. Huang CC, Lee CC, Lin HH, Chang JY. Cathepsin S attenuates endosomal EGFR signalling: A mechanical rationale for the combination of cathepsin S and EGFR tyrosine kinase inhibitors. Sci Rep (2016) 6:29256. doi: 10.1038/srep29256
138. Sevenich L, Bowman RL, Mason SD, Quail DF, Rapaport F, Elie BT, et al. Analysis of tumour- and stroma-supplied proteolytic networks reveals a brain-metastasis-promoting role for cathepsin S. Nat Cell Biol (2014) 16(9):876–88. doi: 10.1038/ncb3011
139. Raza M, Prasad P, Gupta P, Kumar N, Sharma T, Rana M, et al. Perspectives on the role of brain cellular players in cancer-associated brain metastasis: translational approach to understand molecular mechanism of tumor progression. Cancer Metastasis Rev (2018) 37(4):791–804. doi: 10.1007/s10555-018-9766-5
140. Lee CC, Lin JC, Hwang WL, Kuo YJ, Chen HK, Tai SK, et al. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun (2018) 9(1):3763. doi: 10.1038/s41467-018-06268-0
141. Chen Q, Boire A, Jin X, Valiente M, Er EE, Lopez-Soto A, et al. Carcinoma-astrocyte gap junctions promote brain metastasis by cGAMP transfer. Nature (2016) 533(7604):493–8. doi: 10.1038/nature18268
142. Seike T, Fujita K, Yamakawa Y, Kido MA, Takiguchi S, Teramoto N, et al. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin Exp Metastasis (2011) 28(1):13–25. doi: 10.1007/s10585-010-9354-8
143. Kim SW, Choi HJ, Lee HJ, He J, Wu Q, Langley RR, et al. Role of the endothelin axis in astrocyte- and endothelial cell-mediated chemoprotection of cancer cells. Neuro Oncol (2014) 16(12):1585–98. doi: 10.1093/neuonc/nou128
144. Nguyen DX, Chiang AC, Zhang XH, Kim JY, Kris MG, Ladanyi M, et al. WNT/TCF signaling through LEF1 and HOXB9 mediates lung adenocarcinoma metastasis. Cell (2009) 138(1):51–62. doi: 10.1016/j.cell.2009.04.030
145. Park ES, Kim SJ, Kim SW, Yoon SL, Leem SH, Kim SB, et al. Cross-species hybridization of microarrays for studying tumor transcriptome of brain metastasis. Proc Natl Acad Sci USA (2011) 108(42):17456–61. doi: 10.1073/pnas.1114210108
146. Kan C, Vargas G, Pape FL, Clezardin P. Cancer Cell Colonisation in the Bone Microenvironment. Int J Mol Sci (2016) 17(10). doi: 10.3390/ijms17101674
147. Yan HH, Jiang J, Pang Y, Achyut BR, Lizardo M, Liang X, et al. CCL9 Induced by TGFbeta Signaling in Myeloid Cells Enhances Tumor Cell Survival in the Premetastatic Organ. Cancer Res (2015) 75(24):5283–98. doi: 10.1158/0008-5472.CAN-15-2282-T
148. Network NCC. NCCN Clinical Practice Guidelines in Oncology.Non-Small Cell Lung Cancer,Version 4.2020 (2020). Available at: https://www.nccn.org/professionals/physician_gls/f_guidelines.asp (Accessed May 15,2020).
149. Reiter JG, Makohon-Moore AP, Gerold JM, Heyde A, Attiyeh MA, Kohutek ZA, et al. Minimal functional driver gene heterogeneity among untreated metastases. Science (2018) 361(6406):1033–7. doi: 10.1126/science.aat7171
150. Johung KL, Yeh N, Desai NB, Williams TM, Lautenschlaeger T, Arvold ND, et al. Extended Survival and Prognostic Factors for Patients With ALK-Rearranged Non-Small-Cell Lung Cancer and Brain Metastasis. J Clin Oncol (2016) 34(2):123–9. doi: 10.1200/JCO.2015.62.0138
151. Kim ES. Olmutinib: First Global Approval. Drugs (2016) 76(11):1153–7. doi: 10.1007/s40265-016-0606-z
152. Yin W, Zhao Y, Kang X, Zhao P, Fu X, Mo X, et al. BBB-penetrating codelivery liposomes treat brain metastasis of non-small cell lung cancer with EGFR(T790M) mutation. Theranostics (2020) 10(14):6122–35. doi: 10.7150/thno.42234
153. Zhang Q, Zhang Y, Chen Y, Qian J, Zhang X, Yu K. A Novel mTORC1/2 Inhibitor (MTI-31) Inhibits Tumor Growth, Epithelial-Mesenchymal Transition, Metastases, and Improves Antitumor Immunity in Preclinical Models of Lung Cancer. Clin Cancer Res (2019) 25(12):3630–42. doi: 10.1158/1078-0432.CCR-18-2548
154. Wilson GD, Johnson MD, Ahmed S, Cardenas PY, Grills IS, Thibodeau BJ. Targeted DNA sequencing of non-small cell lung cancer identifies mutations associated with brain metastases. Oncotarget (2018) 9(40):25957–70. doi: 10.18632/oncotarget.25409
155. Ippen FM, Alvarez-Breckenridge CA, Kuter BM, Fink AL, Bihun IV, Lastrapes M, et al. The Dual PI3K/mTOR Pathway Inhibitor GDC-0084 Achieves Antitumor Activity in PIK3CA-Mutant Breast Cancer Brain Metastases. Clin Cancer Res (2019) 25(11):3374–83. doi: 10.1158/1078-0432.CCR-18-3049
156. Holbrook K, Lutzky J, Davies MA, Davis JM, Glitza IC, Amaria RN, et al. Intracranial antitumor activity with encorafenib plus binimetinib in patients with melanoma brain metastases: A case series. Cancer (2020) 126(3):523–30. doi: 10.1002/cncr.32547
157. Blakely CM, Watkins TBK, Wu W, Gini B, Chabon JJ, McCoach CE, et al. Evolution and clinical impact of co-occurring genetic alterations in advanced-stage EGFR-mutant lung cancers. Nat Genet (2017) 49(12):1693–704. doi: 10.1038/ng.3990
158. Heon S, Yeap BY, Britt GJ, Costa DB, Rabin MS, Jackman DM, et al. Development of central nervous system metastases in patients with advanced non-small cell lung cancer and somatic EGFR mutations treated with gefitinib or erlotinib. Clin Cancer Res (2010) 16(23):5873–82. doi: 10.1158/1078-0432.CCR-10-1588
159. Lee YJ, Choi HJ, Kim SK, Chang J, Moon JW, Park IK, et al. Frequent central nervous system failure after clinical benefit with epidermal growth factor receptor tyrosine kinase inhibitors in Korean patients with nonsmall-cell lung cancer. Cancer (2010) 116(5):1336–43. doi: 10.1002/cncr.24877
160. Goldberg SB, Gettinger SN, Mahajan A, Chiang AC, Herbst RS, Sznol M, et al. Pembrolizumab for patients with melanoma or non-small-cell lung cancer and untreated brain metastases: early analysis of a non-randomised, open-label, phase 2 trial. Lancet Oncol (2016) 17(7):976–83. doi: 10.1016/s1470-2045(16)30053-5
161. Goldberg SB, Schalper KA, Gettinger SN, Mahajan A, Herbst RS, Chiang AC, et al. Pembrolizumab for management of patients with NSCLC and brain metastases: long-term results and biomarker analysis from a non-randomised, open-label, phase 2 trial. Lancet Oncol (2020) 21(5):655–63. doi: 10.1016/s1470-2045(20)30111-x
162. Zhang G, Cheng R, Wang H, Zhang Y, Yan X, Li P, et al. Comparable outcomes of nivolumab in patients with advanced NSCLC presenting with or without brain metastases: a retrospective cohort study. Cancer Immunol Immunother (2020) 69(3):399–405. doi: 10.1007/s00262-019-02462-1
163. Ceresoli GL, Reni M, Chiesa G, Carretta A, Schipani S, Passoni P, et al. Brain metastases in locally advanced nonsmall cell lung carcinoma after multimodality treatment: risk factors analysis. Cancer (2002) 95(3):605–12. doi: 10.1002/cncr.10687
164. Ding X, Dai H, Hui Z, Ji W, Liang J, Lv J, et al. Risk factors of brain metastases in completely resected pathological stage IIIA-N2 non-small cell lung cancer. Radiat Oncol (2012) 7:119. doi: 10.1186/1748-717X-7-119
Keywords: brain metastasis, lung cancer, driver gene, receptor tyrosine kinase, epithelial-mesenchymal transition, colonization
Citation: Kang Y, Jin Y, Li Q and Yuan X (2021) Advances in Lung Cancer Driver Genes Associated With Brain Metastasis. Front. Oncol. 10:606300. doi: 10.3389/fonc.2020.606300
Received: 14 September 2020; Accepted: 01 December 2020;
Published: 18 January 2021.
Edited by:
Timothy F. Burns, University of Pittsburgh, United StatesReviewed by:
Anjali Rohatgi, University of Pittsburgh Medical Center, United StatesCarlos Pedraz, Germans Trias i Pujol Health Science Research Institute (IGTP), Spain
Copyright © 2021 Kang, Jin, Li and Yuan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xianglin Yuan, eXVhbnhpYW5nbGluQGh1c3QuZWR1LmNu
 Yalin Kang
Yalin Kang Yu Jin
Yu Jin Qianxia Li
Qianxia Li Xianglin Yuan
Xianglin Yuan