- 1College of Chemistry and Life Sciences, Zhejiang Normal University, Jinhua, China
- 2Department of Science and Education, Jinhua Guangfu Oncology Hospital, Jinhua, China
Long non-coding RNAs (lncRNAs) are emerging as crucial regulators of gene expression and physiological processes. LncRNAs are a class of ncRNAs of 200 nucleotides in length. HOX transcript antisense RNA (HOTAIR), a trans-acting lncRNA with regulatory function on transcription, can repress gene expression by recruiting chromatin modifiers. HOTAIR is an oncogenic lncRNA, and numerous studies have determined that HOTAIR is highly upregulated in a wide variety of human cancers. In this review, we briefly summarize the impact of lncRNA HOTAIR expression and functions on different human solid tumors, and emphasize the potential of HOTAIR on tumor prognosis and therapy. Here, we review the recent studies that highlight the prognostic potential of HOTAIR in drug resistance and survival, and the progress of therapies developed to target HOTAIR to date. Furthermore, targeting HOTAIR results in the suppression of HOTAIR expression or function. Thus, HOTAIR knockdown exhibits great therapeutic potential in various cancers, indicating that targeting lncRNA HOTAIR may serve as a promising strategy for cancer therapy. We also propose that preclinical studies involving HOTAIR are required to provide a better understanding of the exact molecular mechanisms underlying the dysregulation of its expression and function in different human cancers and to explore effective methods of targeting HOTAIR and engineering efficient and targeted drug delivery methods in vivo.
Introduction
The ENCODE project revealed that the majority of the human genome is actively transcribed, but only a small minority of the genome encode proteins (1). Transcribed RNAs that do not encode proteins are known as non-coding RNAs (ncRNAs), which include a subgroup of ncRNAs classified as long ncRNAs (lncRNAs) based on their length of >200 nucleotides (2, 3). Many identified lncRNAs are transcribed by RNA polymerase II (RNA pol II) from different regions in the genome (4–6). Based on the genomic location, lncRNAs are mainly classified into four groups: intergenic lncRNAs, intronic lncRNAs, overlapping lncRNAs, and antisense lncRNAs. A comprehensive classification of lncRNAs can be obtained from the review by Jarroux et al. (7). Most lncRNAs can regulate gene expression regardless of the subtype of lncRNAs.
Accumulating evidence has shown that lncRNAs play a critical role both in physiological processes and in human disease development including cancer. LncRNAs are known as key epigenetic regulators for gene expression (8) and are involved in various processes of cellular homeostasis, including chromatin modification, chromatin silencing, transcriptional regulation for gene, and transcription/functional regulation of microRNAs (miRNAs). Several regulatory capacities of lncRNAs are achieved given the relatively complex structure of lncRNAs, which endow the lncRNAs with the ability to bind to DNA partners, protein, and RNA (9). Thereby, the aberrant expression of lncRNAs, especially lncRNA-mediated dysregulation of normal physiological process, may lead to human diseases including cancer (10). In reality, abnormal expression and function of lncRNAs in human cancers has been widely reported, highlighting their capacity to influence oncogenesis, metastatic progression, recurrence, prognosis, and therapeutic responses (11).
HOX transcript antisense RNA (HOTAIR), a trans-acting intergenic lncRNA, was first introduced by Rinn et al. as a polyadenylated and spliced RNA of 2,158 nucleotides in length (12, 13). In humans, HOTAIR is located on chromosome 12q13.13, between the HOXC11 and HOXC12 gene, and is transcribed in an antisense manner relative to the canonical HOXC genes, and partly overlapping with HOXC11 (Figure 1). Human HOTAIR is composed of seven exons, with the last two exons being nearly adjacent to each other; therefore, they are defined as two domains of exon 6 (14). HOTAIR can form a complex secondary structure, comprising several stem and loop structures (15). Evolutionarily, HOTAIR is highly conserved and has evolved faster than its neighboring HOXC gene (16).
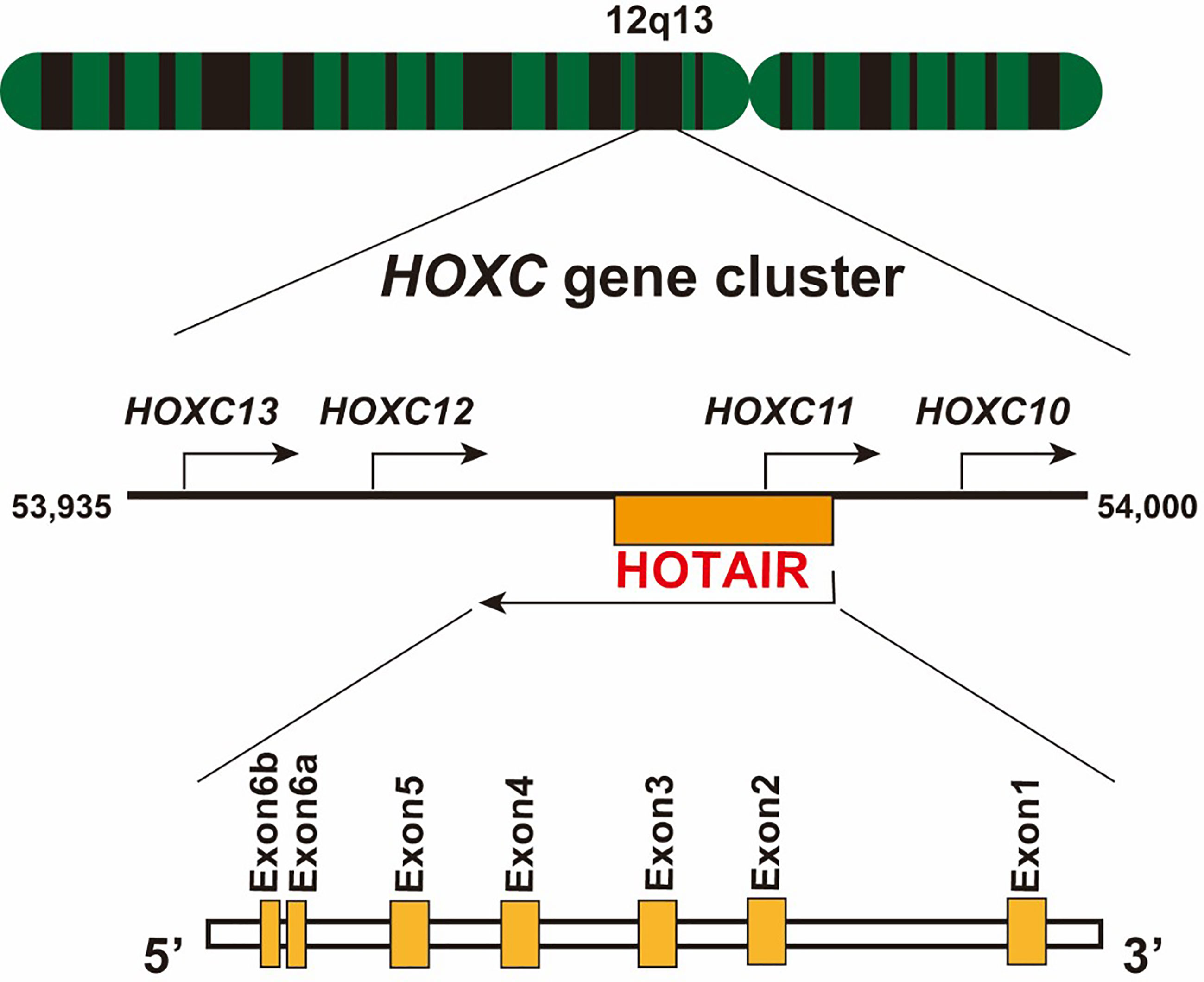
Figure 1 Schematic location of lncRNA HOTAIR. The HOTAIR gene is located in the center of the HOXC gene cluster on chromosome 12, specifically between HOXC11 and HOXC12, on the antisense strand. It consists of six exons, and exon 6 contains two domains.
HOTAIR is a crucial regulator of chromatin status and gene transcriptional silencing (17). Previous studies have largely determined that the mechanisms of HOTAIR, serving as an important epigenetic regulator, depends on interactions with protein or RNA partners. To date, four main molecules indispensable for HOTAIR’s function have been studied (18) (Figure 2). The most widely described partner is Polycomb repressive complex 2 (PRC2). PRC2 is a protein complex, which can mark a gene for transcriptional repression through tri-methylation of histone H3 Lys 27 (H3K27me3) (19, 20). The PRC2 complex contains four major subunits, namely, EZH2, EED, SUZ12, and RbAp46/48 (21). Although EZH2 is the critical subunit for the methyltransferase process, three other subunits are also essential for the EZH2 catalytic activity (22). Early studies have shown that HOTAIR is capable of binding to the PRC2 with an 89 bp fragment on the 5’ end (Figure 2) (12, 23, 24), and HOTAIR is necessary for PRC2 occupancy and H3K27me3 formation in different chromosomes (17). HOTAIR can bind a DNA polypurine motif to regulate gene transcription (25). Other researchers have proposed a different mechanism for the interaction between PRC2 and HOTAIR, in which the PRC2 complex interacts with HOTAIR through the short repeats of the consecutive guanines in the HOTAIR sequence, rather than with a specific structural domain (26–28). In addition to PRC2, the LSD1 complex is another vital partner of HOTAIR, in which lysine-specific demethylase 1(LSD1) is the key subunit (29, 30). The LSD1 complex consists of LSD1, CoREST, and REST, and it can lead to repression of gene expression by reducing the tri-methylation of histone H3 Lys 4(H3K4me3). H3K4me3 is a marker that can target a gene for transcriptional activation, so that H3K4 demethylation is associated with transcriptional inactivation. HOTAIR is capable of binding to the LSD1 complex through a 646 bp fragment in the last exon (Figure 2) (29). Intriguingly, HOTAIR binds to the PRC2 complex and the LSD1 complex through disparate domains: the 5′ end of HOTAIR (1–300 nt) binds to the RPC2 complex, and the 3′ end of HOTAIR (1,500–2,146 nt) binds to the LSD1 complex. In conclusion, HOTAIR provides a molecular scaffold for the assembly of a gene repressor complex consisting of PRC2 and LSD1, thereby silencing its target gene via H3K27 tri-methylation (PRC2 activity) and H3K4 demethylation (LSD1 activity) (17, 29, 31). Apart from functioning as a scaffold for chromatin modifications, HOTAIR also serves as a platform to control protein levels via the ubiquitin-proteasome pathway. Specifically, HOTAIR interacts with E3 ubiquitin ligases (Dzip3 and Mex3b) (Figure 2) and facilitates the ubiquitination of Ataxin-1 and Snurportin by Dzip3 and Mex3b, respectively, thereby contributing to their degradation (32, 33). Lastly, HOTAIR acts as a competitive endogenous RNA sponge for a wide variety of miRNAs (Figure 2) and thereby increases the expression of miRNA-targeted genes (34).
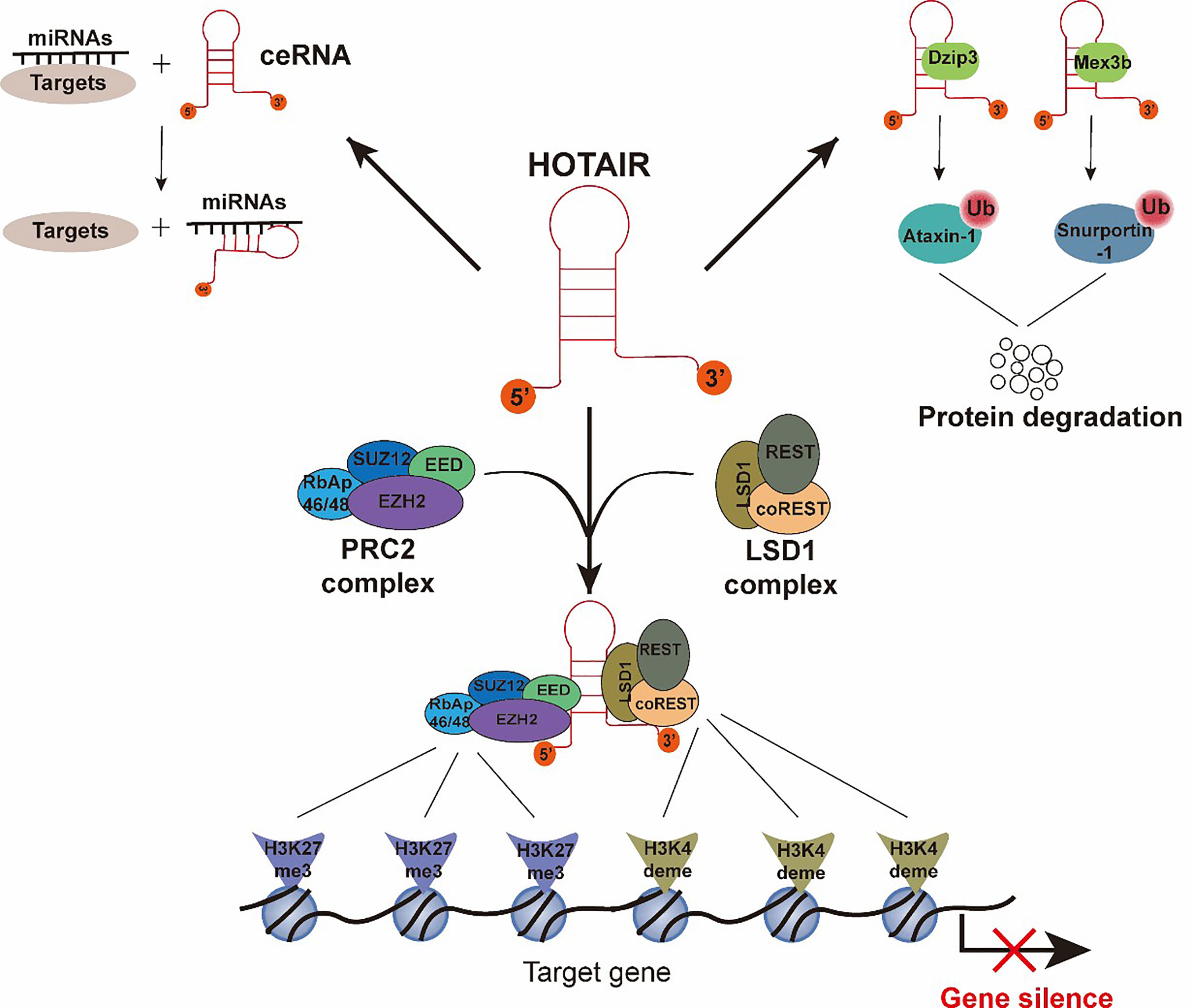
Figure 2 Functions and molecular mechanisms of HOTAIR. The interactions between HOTAIR and its several important partners are summarized. (1) The 5′-end of HOTAIR binds to the PRC2 complex. The 3′-end of HOTAIR binds to the LSD1 complex. H3K27 tri-methylation and H3K4me3 demethylation result from PRC2 complex and LSD1 complex activity, respectively, and cause gene silencing. (2) HOTAIR interacts with E3 ubiquitin ligases, Dzip3 and Mex3b, and facilitates the ubiquitination of Ataxin-1 and Snurportin-1, thereby contributing to their degradation. (3) HOTAIR interacts with miRNAs as a competitive endogenous RNA to promote the expression of miRNA-targeted genes.
HOTAIR Functions and Expression in Tumors
Considering HOTAIR can regulate gene expression and protein proteolysis, it has been reported that the lncRNA HOTAIR is dysregulated in the majority of human cancers (Figure 3). It has become increasingly obvious that HOTAIR dysregulation in several types of cancer is closely associated with the proliferation, metastasis, and invasion of tumor cells (35, 36). In this review, we describe 14 widely reported human solid tumors associated with HOTAIR dysregulation, and we briefly review the recent data focusing on breast, lung, liver, gastric, and pancreatic cancer and renal cell carcinoma.
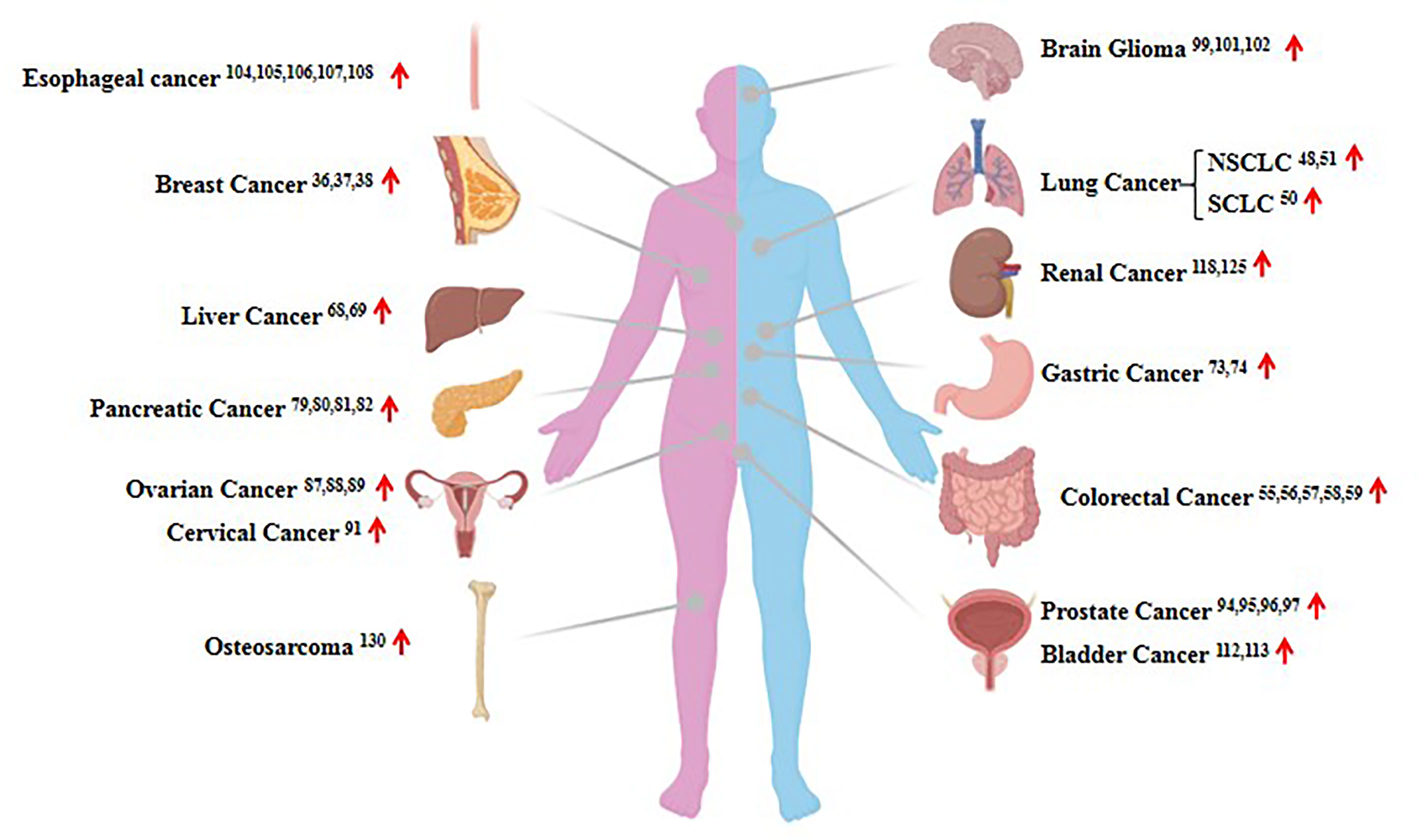
Figure 3 The expression of HOTAIR in human cancers. Shown are examples of the organ-specific expression of HOTAIR. The up red arrow indicates an increase in expression.
Breast Cancer
Breast cancer (BC) is the most prevalent cancer type in women (37, 38). There were more than two million new cases and over 626,000 deaths worldwide in 2018 (39, 40). HOTAIR expression is overexpressed in various types of breast cancer tissues and cells (41). Arshi (42) reported a significant increase in the level of HOTAIR in BC tissues compared to normal tissues using quantitative reverse transcription polymerase chain reaction (qRT-PCR). Xue et al. (43) reported that HOTAIR was elevated in tamoxifen-resistant BC tissues compared to control tissues. In addition, HOTAIR was also upregulated in serum exosomes of BC patients (44). A recent study revealed that higher HOTAIR was positively associated with BC malignancy but was negatively associated with the radiosensitivity of BC cells (45). These studies implicate the involvement of HOTAIR in the tumorigenesis and progression of BC.
Lung Cancer
Lung cancer (LC), a malignant tumor, is a serious public health concern worldwide, responsible for more than one million deaths every year (46). Non-small-cell LC (NSCLC) accounts for 80% of LC and is the main type of LC (47), while small-cell LC (SCLC) accounts for ~15% of the remaining cases. Accumulating evidence has suggested that HOTAIR plays a vital role in tumorigenesis, invasion, and metastasis of LC. Zheng et al. (48) reported that HOTAIR was markedly upregulated in NSCLC cells, and elevated HOTAIR promoted migration and invasion of NSCLC cells by increasing epithelial-mesenchymal transition (EMT). Similarly, Ono et al. (49) found that HOTAIR could mediate the invasive phenotype of SCLC cells by promoting EMT, through which HOTAIR could repress the expression of cell adhesion–related genes. These observations showed that higher HOTAIR expression could promote tumorigenesis, metastasis, and invasion of LCs by increasing EMT.
Liver Cancer
Hepatocellular carcinoma (HCC) is the fifth most prevalent human malignant cancer (50, 51), and the survival rate of HCC patients is still low, with only 30–40% achieving a 1-year survival after surgery (52). Gao et al. (53) reported that HOTAIR was upregulated in HCC tissues compared with adjacent non-cancerous tissues and that elevated HOTAIR contributed to poor tumor differentiation, metastasis, and early recurrence of HCC. Yang et al. (54) also found that the level of HOTAIR was significantly increased in HCC tissues and cell lines, including Hep3B, Huh7, HepG2, and MHCC97H, and elevated HOTAIR promoted migration and invasion of liver cancer cells by enhancing EMT. More importantly, Yang et al. (55) demonstrated that HOTAIR promoted exosome secretion from HCC cells, which subsequently influenced the microenvironment and resulted in tumor progression. Taken together, these observations suggested that HOTAIR could exert oncogenic activity to accelerate the progression of HCC through EMT and the microenvironment.
Gastric Cancer
Gastric cancer (GC), the second leading cause of cancer-related mortality, is one of the most prevalent cancers worldwide (56). Xiao et al. (57) showed that high levels of HOTAIR could promote proliferation and migration of GC cells through the miR-126/CXCR4 axis and downstream signaling pathways. Furthermore, high HOTAIR expression was closely associated with larger tumor size, extensive metastasis, and advanced pathological stages, and also correlated with shorter overall survival of GC patients. In addition, evidence showed that the suppression of HOTAIR decreased the invasion ability of GC cells by reversing EMT (58). Thus, apart from EMT, HOTAIR could also regulate the proliferation and migration of GC cells through miRNA-mediated signaling pathways.
Pancreatic Cancer
Pancreatic cancer (PCa) is one of the deadliest cancers. Given the concealed location, it is difficult to detect in its early stages, and the majority of PCa patients are diagnosed at advanced stages, which leads to poor prognosis and high mortality rates. HOTAIR was upregulated in PCa cells and promoted tumorigenesis through epigenetic regulation (29, 59, 60). HOTAIR is overexpressed in both PCa tissues and in cell lines (61). Kim et al. (62) found that HOTAIR was significantly increased in PCa tissues compared with non-tumor tissue, and higher levels of HOTAIR were associated with more aggressive tumors, supporting a pro-oncogenic function of HOTAIR in Pca. Further, elevated HOTAIR increased lactate production, glucose uptake, and ATP production and led to the promotion of PCa cell proliferation (63). Kim et al. (62) demonstrated that high HOTAIR expression could increase PCa cell invasion and proliferation. These observations provide us with new insight indicating HOTAIR increases glucose metabolism and promotes PCa cell proliferation.
Renal Cell Carcinoma
Renal cell carcinoma (RCC) is a common cancer type, which causes ~90,000 deaths worldwide annually (64). HOTAIR is significantly overexpressed in RCC cell lines and clinical tissues compared with normal cell lines and tissues, and HOTAIR has been associated with tumor progression and clinicopathological characteristics of patients (65). Higher HOTAIR expression promoted proliferation, migration, and invasion of RCC cell lines (66). Moreover, HOTAIR plays an essential role in RCC metastasis. Katayama et al. (67) found that HOTAIR could enhance RCC cell migration by regulating insulin growth factor-binding protein 2 (IGFBP2) expression, and HOTAIR was strongly associated with nuclear grade, lymph-node metastasis, and lung metastasis in RCC. Numerous studies have indicated that HOTAIR could promote RCC malignancy via different mechanisms (66, 68–70). More recently, a study (71) indicated that HOTAIR and androgen receptor synergistically promoted tumor angiogenesis and cancer stemness in RCC cells both in vitro and in vivo.
Other Solid Cancers
In addition to the six tumors described above, HOTAIR is consistently overexpressed in several other types of cancer, including colorectal cancer (72–75), ovarian cancer (76, 77), cervical cancer (78), prostate cancer (79, 80), brain glioma (81, 82), esophageal cancer (83–85), bladder cancer (86), and osteosarcoma (87–89). Elevated HOTAIR expression can be detected in both cancer cells and tumor tissues through qRT-PCR, in situ hybridization, and RNA-sequencing. For example, Tatangelo et al. (90) found that HOTAIR was upregulated in proximal colon cancers by in situ hybridization. Chang et al. (79) showed that HOTAIR was overexpressed in the neuroendocrine differentiation (NED) prostate cancer cells and in castration-resistant prostate cancer through RNA-sequencing. Moreover, numerous independent studies have unanimously reported HOTAIR was closely associated with tumorigenesis, tumor staging, metastasis, invasion, proliferation, and apoptosis in human solid cancers. In osteosarcoma, Wang et al. (89) found that HOTAIR was significantly associated with worse histological grade and advanced tumor stage. In the brain glioma, Yang et al. (81) verified that the knockdown of HOTAIR inhibited cell proliferation, promoted cell apoptosis, and suppressed cell invasion and migration during the development of brain glioma. Similarly, in bladder cancer, Yu et al. (86) found that the suppression of HOTAIR inhibited bladder cancer cell proliferation, invasion, and migration, but increased cell apoptosis. Mechanically, HOTAIR played an oncogene function to accelerate tumor progression mainly through epigenetic regulation, EMT, miRNA-mediated downstream signaling pathways, and regulation of other signaling pathways such as the Wnt/β-catenin signaling pathway (83). In addition, HOTAIR may be a reasonable biomarker for predicting tumor risk, diagnosis, and metastasis. Zhang et al. (80) clarified that the level of HOTAIR was significantly higher in bone metastasis tissues than that in the primary prostate cancer tissues, suggesting that HOTAIR could represent a reasonable biomarker for prostate cancer bone metastasis. Further, studies have identified tag single nucleotide polymorphisms (tagSNPs) in HOTAIR and demonstrated that the tagSNPs in HOTAIR were associated with bladder cancer risk in a Chinese population (91). In addition, HOTAIR could serve as a urinary liquid biopsy biomarkers to distinguish bladder cancer from chronic urocystitis (92).
HOTAIR Prognostic Potential
The correlation between HOTAIR expression and human cancers has been widely reported; however, there is lack of systematic reviews of the prognostic potential of HOTAIR in cancer. Thus, below we summarized recent data relative to the prognostic potential of HOTAIR in terms of drug resistance and survival.
Drug Resistance
Drug resistance is a major limiting factor in achieving a cure for cancer patients (93), and it also can serve as an indicator to evaluate prognosis. Drug resistance can be roughly divided into treatment drug resistance and chemoradiotherapy resistance. HOTAIR is closely related to the occurrence of drug resistance in several tumors. Fang et al. (94) reported that HOTAIR was overexpressed in SCLC multidrug resistance cells, and HOTAIR knockdown could increase cell sensitivity to anticancer drugs and inhibit tumor growth. HOTAIR was also involved in drug resistance of RCC. Li et al. (95) found that HOTAIR was highly upregulated in sunitinib-resistant cells compared with that in corresponding control cells, and HOTAIR could enhance sunitinib resistance in RCC cells by activating Beclin1-mediated cell autophagy. Moreover, the level of HOTAIR continually increased in prostate cancer cell lines upon treatment with enzalutamide (96), suggesting HOTAIR might serve as a biomarker indicating resistance against enzalutamide.
In addition, overexpressed HOTAIR reduced the sensitivity of tumor cells to chemoradiotherapy. HOTAIR was overexpressed in cisplatin (DDP)-resistant osteosarcoma cells and tissues and enhanced DDP resistance of osteosarcoma cells through the miR-106a-5p/STAT3 axis (97). HOTAIR was also upregulated in the serum exosomes of BC patients and was associated with poor prognosis and poor response to chemotherapy (44). Li et al. (98) demonstrated that high HOTAIR expression could contribute to fluorouracil (5-FU) resistance and was associated with poor response to 5FU treatment in colorectal cancer cells. Özeş et al. (99) revealed that high levels of HOTAIR induced platinum resistance in ovarian cancer. Moreover, the depletion of HOTAIR in HCC cell lines reduced cell susceptibility to TNF-α-induced apoptosis, while it increased the chemotherapeutic sensitivity of liver cancer cells to cisplatin and doxorubicin (100). Additionally, HOTAIR played a critical role in regulating the radiotherapy resistance of tumors. HOTAIR knockdown could increase radiosensitivity of PCa (101) and cervical cancer (102) by regulating autophagy and EMT. Similarly, Liu et al. (103) reported that HOTAIR knockdown potentiated radiosensitivity by regulating the miR-93/ATG12 axis in colorectal cancer.
Taken together, the level of HOTAIR is higher in drug-resistant cancer cells and tissues. Overexpressed HOTAIR could promote the occurrence of drug resistance in cancers and reduce the sensitivity of cancer cells to chemoradiotherapy, indicating HOTAIR may be a vital prognostic factor for predicting drug resistance of tumors.
Survival
Recently, the level of HOTAIR has been found to be associated with survival in several types of cancer. HOTAIR is closely associated with overall survival, disease-free survival, and survival rates of cancer patients. Martínez-Fernández et al. (104) reported that HOTAIR had prognostic value for bladder cancer progression, recurrence, and survival. Further, the aberrant expression of HOTAIR was associated with poor disease-free survival of bladder cancer (105). Lu et al. (106) measured circulating HOTAIR levels in the serum of 112 BC patients using RT-qRCR and found that BC patients with high circulating HOTAIR showed less clinical response and worse disease-free survival than those with low circulating HOTAIR. In addition, Kim et al. (107) first reported the association between HOTAIR gene polymorphisms and colorectal cancer mortality. They chose four HOTAIR polymorphisms (rs7958904G>C, rs920778T>C, rs4759314A>G, and rs1899663G>T), conducting genotype frequencies and Cox-regression analysis, and demonstrated that HOTAIR rs7958904G>C could be a potent prognostic biomarker for CRC, which was positively associated with CRC prevalence and mortality. These studies suggested overexpressed HOTAIR could decrease disease-free survival and survival rate in some tumors. However, HOTAIR might have the opposite effect. Wang et al. (108) found that HOTAIR was significantly downregulated in patients with primary and acquired resistance to EGFR-TKIs, and in clinical phenotype, they found that high HOTAIR expression was significantly associated with longer progression-free survival compared to low HOTAIR expression subgroup. Nevertheless, the opposite role whereby low HOTAIR expression is associated with unfavorable prognosis in EGFR-TKIs-resistance NSCLCs remains unclear and requires further study.
These studies have indicated that HOTAIR may be used to predict tumor survival, but HOTAIR has diverse effects in different cancers, which needs to be further clarified. The mechanisms underlying HOTAIR tumor survival still need to be further studied.
Therapeutic Potential of HOTAIR
Numerous studies have emphasized the impact of HOTAIR on tumorigenesis, progression, metastasis, and prognosis of various tumors. Therefore, many therapeutic strategies have been proposed for targeting HOTAIR including silencing HOTAIR expression or function. In terms of HOTAIR silencing, Kim et al. (109) showed that both antisense oligonucleotides (ASOs) and RNA interference (RNAi) could effectively suppress HOTAIR. Gupta et al. (59) used RNAi technology to target HOTAIR, which led to HOTAIR knockout. The loss of HOTAIR could inhibit cancer invasiveness. Besides, Bhan et al. (110) designed a synthetic oligonucleotide DNA as small interfering sense (aiSENSE) that is complementary to the HOTAIR transcript to reduce HOTAIR expression in breast cancer cells. Their results showed that siSENSE could knock down specifically and effectively HOTAIR transcript in breast cancer cells. Apart from directly targeting HOTAIR to reduce its expression, there are some inhibitors that inhibit HOTAIR function without changing HOTAIR level. For example, Li et al. (111) identified a small-molecule compound AC1Q3QWB (AQB) that could disrupt the interaction of HOTAIR-EZH2, and they verified that AQB could selectively and efficiently block PRC2 recruitment. Moreover, Jin et al. (112) studied a novel combination of AQB and CDK4/6 inhibitor palbociclib to evaluate its antitumor effects in glioblastoma. They found that the combination of AQB and Palbociclib had a stronger inhibitory effect on glioma cell growth and metastasis than that in the single drug. Similarly, peptide nucleic acid-PNA3 could disrupt the interaction between HOTAIR and EZH2. Wang et al. (113) disrupted HOTAIR-EZH2 with PNA3 in combination with DNMTi and found that the tumor initiation and stem cell frequency of ovarian cancer stem cells were inhibited, suggesting that dual inhibition of HOTAIR-EZH2 interaction and DNA methylation may be a potent strategy to eradicate ovarian cancer stem cells.
Taken together, the strategies for targeting HOTAIR are mainly through antisense oligonucleotides, RNAi, and small molecule inhibitor, in which antisense oligonucleotides and RNAi directly inhibit HOTAIR expression, while small-molecule inhibitor could block the HOTAIR function. Due to the important role of HOTAIR in tumors, several studies have reported that HOTAIR knockdown may be a potent approach for cancer treatment. HOTAIR knockdown could inhibit DDP resistance of GC cells through blocking the Wnt/β-catenin and PI3K/AKT signaling pathways by upregulating miR-34a (114). Guo et al. (102) reported that HOTAIR knockdown enhanced cervical cancer cell sensitivity to radiotherapy by autophagy reduction and reversal of EMT through inhibiting the Wnt signaling pathway. Similarly, Liu et al. (103) found that HOTAIR knockdown potentiated radiosensitivity of colorectal cancer through regulating the mi-93/ATG12 axis. Moreover, Jia et al. (115) revealed that the HOTAIR/miR-17-5p/PTEN axis might serve as the potential therapeutic strategy for GC. Thus, targeting lncRNA HOTAIR that lead to the suppression of HOTAIR expression or function could serve as a promising therapeutic strategy for several tumors.
Discussion and Future Perspectives
In recent years, there has been significant progress in clarifying the role of HOTAIR in various physiological and pathological processes. Increasing evidence has suggested that the HOTAIR is overexpressed in a variety of cancers and serves as a potent prognostic factor and therapeutic target in various cancers (Table 1). The level of HOTAIR is closely associated with tumor stage, proliferation, migration, and invasion in several human cancers. Moreover, HOTAIR has been demonstrated to affect the drug treatment response and correlates with drug resistance, including chemoradiotherapy resistance. Besides, overexpressed HOTAIR significantly decreased survival of patients in several tumors. These observations suggest that HOTAIR may serve as a potent prognostic factor to predict treatment response and survival rate.
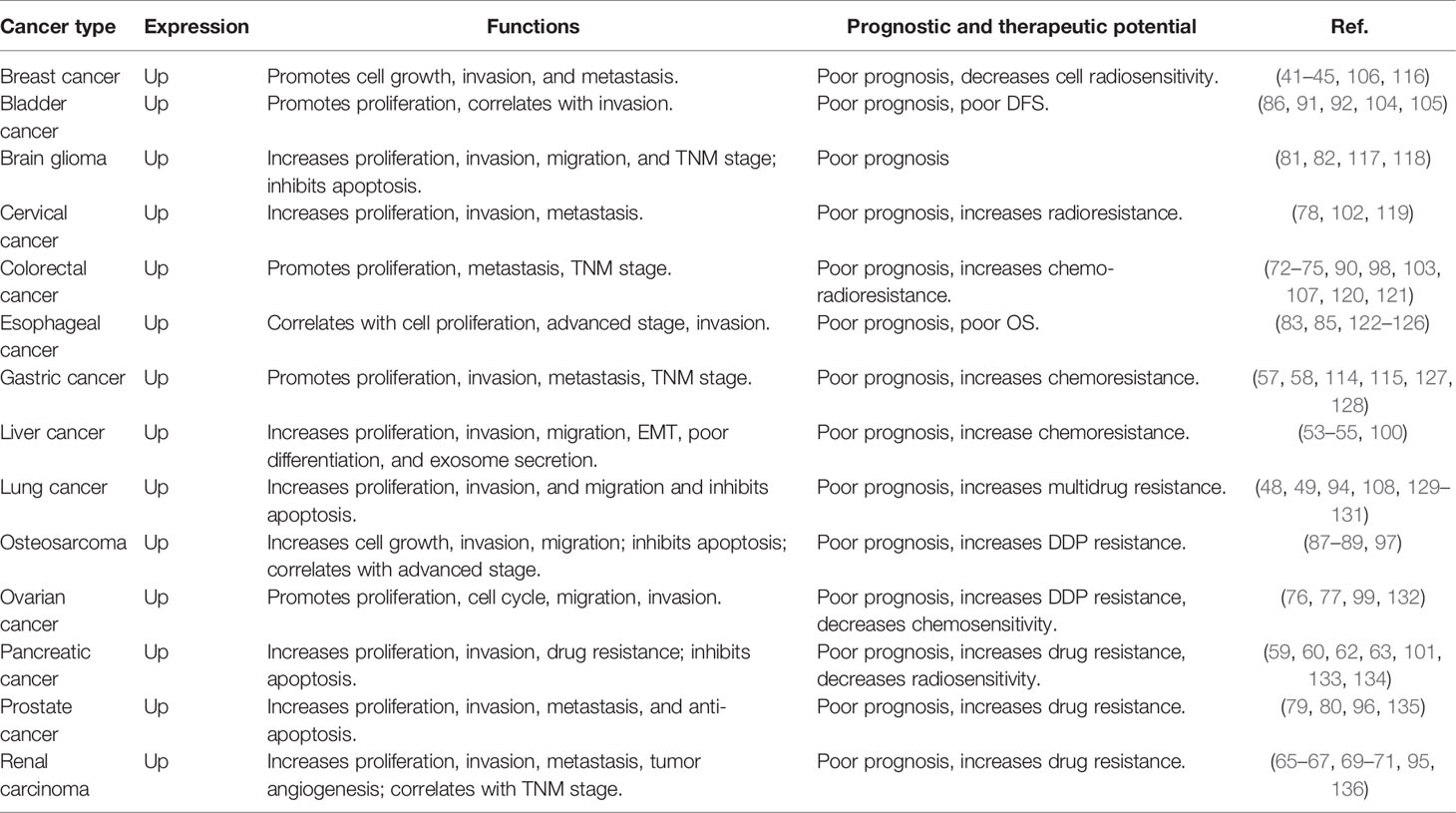
Table 1 HOTAIR expression, functions, and prognostic and therapeutic potential in different human cancers.
Based on the close association between HOTAIR and tumorigenesis, progression, and prognosis, targeting HOTAIR may serve as a novel strategy for cancer treatment. Until now, there have been three methods to target HOTAIR—synthetic antisense oligonucleotides, RNAi, and molecule inhibitors that block the interaction between HOTAIR and its partner. In particular, HOTAIR inhibitors mainly focus on blocking the interaction between HOTAIR-EZH2 at present. Thus, more HOTAIR inhibitors still need to be further studied. It has been reported that the HOTAIR knockdown can significantly slow the progression of several tumors and increase their sensitivity to drugs, while additional studies, especially preclinical studies, are needed to prove the therapeutic potential of HOTAIR.
In conclusion, HOTAIR plays a vital role in tumor crucial process such as occurrence, growth, invasion, metastasis, and drug resistance. For this reason, HOTAIR has been regarded as a potential new target for cancer prognosis and therapy. However, the understanding of HOTAIR’s clinical application still needs to be further evaluated to clarify the exact molecular mechanisms underlying dysregulation of its expression and function in different human cancers to provide novel molecules to repress HOTAIR activity in cancer cells. In addition, some studies have shown that HOTAIR can be used in conjunction with currently available drugs to sensitize tumors to the existing therapies, so finding an effective method to target HOTAIR and an efficient drug delivery method in vivo would be another critical point. Finally, with more and more studies emerging, lncRNAs such as HOTAIR will act as viable prognostic factors and therapeutic targets for treating human cancers shortly.
Author Contributions
All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Zhejiang Province [LQ21C060003], the Jin Hua Science and Technology Plan Project [2021-3-148] and the Doctoral Scientific Research Foundation of Zhejiang Normal University [YS304320122].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
LncRNA, Long non-coding RNA; HOTAIR, HOX transcript antisense RNA; RNA pol II, RNA polymerase II; miRNA, microRNA; PRC2, Polycomb repressive complex 2; LSD1, Lysine-specific demethylase 1; BC, Breast cancer; NSCLC, Non-small-cell lung cancer; SCLC, Small-cell lung cancer; EMT, Epithelial-mesenchymal transition; HCC, Hepatocellular carcinoma; GC, Gastric cancer; PCa, Pancreatic cancer; RCC, Renal cell carcinoma; IGFBP2, Insulin growth factor-binding protein 2; NED, Neuroendocrine differentiation; DDP, Cisplatin; 5-FU, Fluorouracil; ASOs, Antisense oligonucleotides; RNAi, RNA interference; DFS, Disease-free survival; OS, Overall survival.
References
1. Mudge JM, Frankish A, Harrow J. Functional Transcriptomics in the Post-ENCODE Era. Genome Res (2013) 23(12):1961–73. doi: 10.1101/gr.161315.113
2. Fatica A, Bozzoni I. Long Non-Coding RNAs: New Players in Cell Differentiation and Development. Nat Rev Genet (2013) 15(1):7–21. doi: 10.1038/nrg3606
3. Rinn JL, Chang HY. Genome Regulation by Long Noncoding RNAs. Annu Rev Biochem (2012) 81(1):145–66. doi: 10.1146/annurev-biochem-051410-092902
4. Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA Circles Function as Efficient microRNA Sponges. Nature (2013) 495(7441):384–8. doi: 10.1038/nature11993
5. Tam W. Identification and Characterization of Human BIC, a Gene on Chromosome 21 That Encodes a Noncoding RNA. Gene (2001) 274(1-2):157–67. doi: 10.1016/s0378-1119(01)00612-6
6. Consortium RGERGaGSGGNPCGatF. Antisense Transcription in the Mammalian Transcriptome. Science (2005) 309(5740):1564–6. doi: 10.1126/science.1112009
7. Jarroux J, Morillon A, Pinskaya M. History, Discovery, and Classification of lncRNAs. Adv Exp Med Biol (2017) 1008:1–46. doi: 10.1007/978-981-10-5203-3_1
8. Flynn Ryan A, Chang Howard Y. Long Noncoding RNAs in Cell-Fate Programming and Reprogramming. Cell Stem Cell (2014) 14(6):752–61. doi: 10.1016/j.stem.2014.05.014
9. Yang G, Lu X, Yuan L. LncRNA: A Link Between RNA and Cancer. Biochim Biophys Acta (2014) 1839(11):1097–109. doi: 10.1016/j.bbagrm.2014.08.012
10. Wapinski O, Chang HY. Long Noncoding RNAs and Human Disease. Trends Cell Biol (2011) 21(6):354–61. doi: 10.1016/j.tcb.2011.04.001
11. Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long Noncoding RNA in Prostate, Bladder, and Kidney Cancer. Eur Urol (2014) 65(6):1140–51. doi: 10.1016/j.eururo.2013.12.003
12. Zhang Y, Wei W, Cheng N, Wang K, Li B, Jiang X, et al. Hepatitis C Virus-Induced Up-Regulation of microRNA-155 Promotes Hepatocarcinogenesis by Activating Wnt Signaling. Hepatol (Baltimore Md) (2012) 56(5):1631–40. doi: 10.1002/hep.25849
13. Rinn JL. Functional Demarcation of Active and Silent Chromatin Domains in Human HOX Loci by Noncoding RNAs. NIH Public Access Author Manus (2007) 129(7):1311–23. doi: 10.1016/j.cell.2007.05.022
14. Cai B, Song XQ, Cai JP, Zhang S. HOTAIR: a Cancer-Related Long non-Coding RNA. Neoplasma (2014) 61(04):379–91. doi: 10.4149/neo_2014_075
15. Wang H, Zheng H, Wang C, Lu X, Zhao X, Li X. Insight Into HOTAIR Structural Features and Functions as Landing Pads for Transcription Regulation Proteins. Biochem Biophys Res Commun (2017) 485(3):679–85. doi: 10.1016/j.bbrc.2017.02.100
16. He S, Liu S, Zhu H. The Sequence, Structure and Evolutionary Features of HOTAIR in Mammals. BMC Evol Biol (2011) 11(1):102. doi: 10.1186/1471-2148-11-102
17. Bhan A, Mandal SS. LncRNA HOTAIR: A Master Regulator of Chromatin Dynamics and Cancer. Biochim Biophys Acta (BBA) - Rev Cancer (2015) 1856(1):151–64. doi: 10.1016/j.bbcan.2015.07.001
18. Qu X, Alsager S, Zhuo Y, Shan B. HOX Transcript Antisense RNA (HOTAIR) in Cancer. Cancer Lett (2019) 454:90–7. doi: 10.1016/j.canlet.2019.04.016
19. Onodera Y, Teramura T, Takehara T, Obora K, Mori T, Fukuda K. miR-155 Induces ROS Generation Through Downregulation of Antioxidation-Related Genes in Mesenchymal Stem Cells. Aging Cell (2017) 16(6):1369–80. doi: 10.1111/acel.12680
20. Al-Raawi D, Jones R, Wijesinghe S, Halsall J, Petric M, Roberts S, et al. A Novel Form of JARID2 Is Required for Differentiation in Lineage-Committed Cells. EMBO J (2018) 38(3):e98449. doi: 10.15252/embj.201798449
21. Margueron R, Reinberg D. The Polycomb Complex PRC2 and Its Mark in Life. Nature (2011) 469(7330):343–9. doi: 10.1038/nature09784
22. Rocha S, Wu H, Zeng H, Dong A, Li F, He H, et al. Structure of the Catalytic Domain of EZH2 Reveals Conformational Plasticity in Cofactor and Substrate Binding Sites and Explains Oncogenic Mutations. PloS One (2013) 8(12):e83737. doi: 10.1371/journal.pone.0083737
23. Wu L, Murat P, Matak-Vinkovic D, Murrell A, Balasubramanian S. Binding Interactions Between Long Noncoding RNA HOTAIR and PRC2 Proteins. Biochemistry (2013) 52(52):9519–27. doi: 10.1021/bi401085h
24. Rajagopal T, Talluri S, Akshaya RL, Dunna NR. HOTAIR LncRNA: A Novel Oncogenic Propellant in Human Cancer. Clin Chim Acta (2020) 503:1–18. doi: 10.1016/j.cca.2019.12.028
25. Chu C, Qu K, Zhong Franklin L, Artandi Steven E, Chang Howard Y. Genomic Maps of Long Noncoding RNA Occupancy Reveal Principles of RNA-Chromatin Interactions. Mol Cell (2011) 44(4):667–78. doi: 10.1016/j.molcel.2011.08.027
26. Wang X, Goodrich KJ, Gooding AR, Naeem H, Archer S, Paucek RD, et al. Targeting of Polycomb Repressive Complex 2 to RNA by Short Repeats of Consecutive Guanines. Mol Cell (2017) 65(6):1056–67.e5. doi: 10.1016/j.molcel.2017.02.003
27. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs With a Role in Cancer. Nat Rev Cancer (2006) 6(4):259–69. doi: 10.1038/nrc1840
28. Long Y. Conserved RNA-Binding Specificity of Polycomb Repressive Complex 2 Is Achieved by Dispersed Amino Acid Patches in EZH2. eLIFE (2017) 23:e31558. doi: 10.7554/eLife.31558.001
29. Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long Noncoding RNA as Modular Scaffold of Histone Modification Complexes. Science (2010) 329(5992):689–93. doi: 10.1126/science.1192002
30. Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone Demethylation Mediated by the Nuclear Amine Oxidase Homolog Lsd1. Cell (2004) 119(7):941–53. doi: 10.1016/j.cell.2004.12.012
31. Marchese FP, Huarte M. Long Non-Coding RNAs and Chromatin Modifiers. Epigenetics (2013) 9(1):21–6. doi: 10.4161/epi.27472
32. Yoon J-H, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, et al. Scaffold Function of Long Non-Coding RNA HOTAIR in Protein Ubiquitination. Nat Commun (2013) 4(1):2939. doi: 10.1038/ncomms3939
33. Hao Z. RNA Helicase DEAD Box Protein 5 Regulates Polycomb Repressive Complex 2_Hox Transcript Antisense Intergenic RNA Function in Hepatitis B Virus Infection and Hepatocarcinogenesis. Hepatology (2016) 16:1033–48. doi: 10.1002/hep.28698/suppinfo
34. Chen W, Han C, Zhang J, Song K, Wang Y, Wu T. Deletion of Mir155 Prevents Fas-Induced Liver Injury Through Up-Regulation of Mcl-1. Am J Pathol (2015) 185(4):1033–44. doi: 10.1016/j.ajpath.2014.12.020
35. Kogo R, Shimamura T, Mimori K, Kawahara K, Imoto S, Sudo T, et al. Long Noncoding RNA HOTAIR Regulates Polycomb-Dependent Chromatin Modification and Is Associated With Poor Prognosis in Colorectal Cancers. Cancer Res (2011) 71(20):6320–6. doi: 10.1158/0008-5472.can-11-1021
36. Li D, Feng J, Wu T, Wang Y, Sun Y, Ren J, et al. Long Intergenic Noncoding RNA HOTAIR Is Overexpressed and Regulates PTEN Methylation in Laryngeal Squamous Cell Carcinoma. Am J Pathol (2013) 182(1):64–70. doi: 10.1016/j.ajpath.2012.08.042
38. Semmler L, Reiter-Brennan C, Klein A. BRCA1 and Breast Cancer: A Review of the Underlying Mechanisms Resulting in the Tissue-Specific Tumorigenesis in Mutation Carriers. J Breast Cancer (2019) 22(1):1. doi: 10.4048/jbc.2019.22.e6
39. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer J Clin (2018) 68(6):394–424. doi: 10.3322/caac.21492
40. Benvenuto M, Focaccetti C, Izzi V, Masuelli L, Modesti A, Bei R. Tumor Antigens Heterogeneity and Immune Response-Targeting Neoantigens in Breast Cancer. Semin Cancer Biol (2021) 72:65–75. doi: 10.1016/j.semcancer.2019.10.023
41. Cantile M, Di Bonito M, Cerrone M, Collina F, De Laurentiis M, Botti G. Long Non-Coding RNA HOTAIR in Breast Cancer Therapy. Cancers (2020) 12(5):1197. doi: 10.3390/cancers12051197
42. Asghar Arshi MS. A Comparative Study of HOTAIR Expression in Breast Cancer Patient Tissues and Cell Lines. Orig Article (2019) 7. doi: 10.22074/cellj.2020.6543
43. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR Enhances ER Signaling and Confers Tamoxifen Resistance in Breast Cancer. Oncogene (2015) 35(21):2746–55. doi: 10.1038/onc.2015.340
44. Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D. Overexpression of Serum Exosomal HOTAIR Is Correlated With Poor Survival and Poor Response to Chemotherapy in Breast Cancer Patients. J Biosci (2019) 44(2):37. doi: 10.1007/s12038-019-9861-y
45. Zhang S, Wang B, Xiao H, Dong J, Li Y, Zhu C, et al. LncRNA HOTAIR Enhances Breast Cancer Radioresistance Through Facilitating HSPA1A Expression via Sequestering miR -449b-5p. Thorac Cancer (2020) 11(7):1801–16. doi: 10.1111/1759-7714.13450
46. Vachani A, Sequist LV, Spira A. AJRCCM: 100-YearAnniversary.The Shifting Landscape for Lung Cancer: Past, Present, and Future. Am J Respir Crit Care Med (2017) 195(9):1150–60. doi: 10.1164/rccm.201702-0433CI
47. Asano N, Matsuzaki J, Ichikawa M, Kawauchi J, Takizawa S, Aoki Y, et al. A Serum microRNA Classifier for the Diagnosis of Sarcomas of Various Histological Subtypes. Nat Commun (2019) 10(1):1299. doi: 10.1038/s41467-019-09143-8
48. Zheng F, Li J, Ma C, Tang X, Tang Q, Wu J, et al. Novel Regulation of miR-34a-5p and HOTAIR by the Combination of Berberine and Gefitinib Leading to Inhibition of EMT in Human Lung Cancer. J Cell Mol Med (2020) 24(10):5578–92. doi: 10.1111/jcmm.15214
49. Ono H, Motoi N, Nagano H, Miyauchi E, Ushijima M, Matsuura M, et al. Long Noncoding RNA HOTAIR Is Relevant to Cellular Proliferation, Invasiveness, and Clinical Relapse in Small-Cell Lung Cancer. Cancer Med (2014) 3(3):632–42. doi: 10.1002/cam4.220
50. Saran U, Humar B, Kolly P, Dufour J-F. Hepatocellular Carcinoma and Lifestyles. J Hepatol (2016) 64(1):203–14. doi: 10.1016/j.jhep.2015.08.028
51. Wallace MC, Preen D, Jeffrey GP, Adams LA. The Evolving Epidemiology of Hepatocellular Carcinoma: A Global Perspective. Expert Rev Gastroenterol Hepatol (2015) 9(6):765–79. doi: 10.1586/17474124.2015.1028363
52. Wang P, Ouyang L, Zheng L, Wang Z. Identifying Hepatocellular Carcinoma-Related Genes and Pathways by System Biology Analysis. Irish J Med Sci (1971 -) (2014) 184(2):357–64. doi: 10.1007/s11845-014-1119-y
53. Gao J-Z, Li JIA, Du J-L, Li X-L. Long Non-Coding RNA HOTAIR is a Marker for Hepatocellular Carcinoma Progression and Tumor Recurrence. Oncol Lett (2016) 11(3):1791–8. doi: 10.3892/ol.2016.4130
54. Yang T, He X, Chen A, Tan K, Du X. LncRNA HOTAIR Contributes to the Malignancy of Hepatocellular Carcinoma by Enhancing Epithelial-Mesenchymal Transition via Sponging miR-23b-3p From ZEB1. Gene (2018) 670:114–22. doi: 10.1016/j.gene.2018.05.061
55. Yang L, Peng X, Li Y, Zhang X, Ma Y, Wu C, et al. Long non-Coding RNA HOTAIR Promotes Exosome Secretion by Regulating RAB35 and SNAP23 in Hepatocellular Carcinoma. Mol Cancer (2019) 18(1):78. doi: 10.1186/s12943-019-0990-6
56. Karimi P, Islami F, Anandasabapathy S, Freedman ND, Kamangar F. Gastric Cancer: Descriptive Epidemiology, Risk Factors, Screening, and Prevention. Cancer Epidemiol Biomarkers Prev (2014) 23(5):700–13. doi: 10.1158/1055-9965.epi-13-1057
57. Xiao J, Lai H, Wei SH, Ye ZS, Gong FS, Chen LC. Lnc RNA HOTAIR Promotes Gastric Cancer Proliferation and Metastasis via Targeting miR-126 to Active CXCR 4 and RhoA Signaling Pathway. Cancer Med (2019) 8(15):6768–79. doi: 10.1002/cam4.1302
58. Xu Z-Y, Yu Q-M, Du Y-A, Yang L-T, Dong R-Z, Huang L, et al. Knockdown of Long Non-Coding RNA HOTAIR Suppresses Tumor Invasion and Reverses Epithelial-Mesenchymal Transition in Gastric Cancer. Int J Biol Sci (2013) 9(6):587–97. doi: 10.7150/ijbs.6339
59. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long Non-Coding RNA HOTAIR Reprograms Chromatin State to Promote Cancer Metastasis. Nature (2010) 464(7291):1071–6. doi: 10.1038/nature08975
60. Ishibashi M, Kogo R, Shibata K, Sawada G, Takahashi Y, Kurashige J, et al. Clinical Significance of the Expression of Long Non-Coding RNA HOTAIR in Primary Hepatocellular Carcinoma. Oncol Rep (2013) 29(3):946–50. doi: 10.3892/or.2012.2219
61. Rodriguez A, Vigorito E, Clare S, Warren MV, Couttet P, Soond DR, et al. Requirement of Bic/microRNA-155 for Normal Immune Function. Science (2007) 316(5824):608–11. doi: 10.1126/science.1139253
62. Kim K, Jutooru I, Chadalapaka G, Johnson G, Frank J, Burghardt R, et al. HOTAIR Is a Negative Prognostic Factor and Exhibits Pro-Oncogenic Activity in Pancreatic Cancer. Oncogene (2012) 32(13):1616–25. doi: 10.1038/onc.2012.193
63. Ma Y, Hu M, Zhou L, Ling S, Li Y, Kong B, et al. Long non−Coding RNA HOTAIR Promotes Cancer Cell Energy Metabolism in Pancreatic Adenocarcinoma by Upregulating Hexokinase−2. Oncol Lett (2019) 18(3):2212–9. doi: 10.3892/ol.2019.10551
64. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA: A Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
65. Dasgupta P, Kulkarni P, Majid S, Shahryari V, Hashimoto Y, Bhat NS, et al. MicroRNA-203 Inhibits Long Noncoding RNA HOTAIR and Regulates Tumorigenesis Through Epithelial-To-Mesenchymal Transition Pathway in Renal Cell Carcinoma. Mol Cancer Ther (2018) 17(5):1061–9. doi: 10.1158/1535-7163.mct-17-0925
66. Pan Y, Wu Y, Hu J, Shan Y, Ma J, Ma H, et al. Long Noncoding RNA HOTAIR Promotes Renal Cell Carcinoma Malignancy Through Alpha-2, 8-Sialyltransferase 4 by Sponging microRNA-124. Cell Prolif (2018) 51(6):e12507. doi: 10.1111/cpr.12507
67. Katayama H, Tamai K, Shibuya R, Nakamura M, Mochizuki M, Yamaguchi K, et al. Long non-Coding RNA HOTAIR Promotes Cell Migration by Upregulating Insulin Growth Factor–Binding Protein 2 in Renal Cell Carcinoma. Sci Rep (2017) 7(1):12016. doi: 10.1038/s41598-017-12191-z
69. Hong Q, Li O, Zheng W, Xiao W-z, Zhang L, Wu D, et al. LncRNA HOTAIR Regulates HIF-1α/AXL Signaling Through Inhibition of miR-217 in Renal Cell Carcinoma. Cell Death Dis (2017) 8(5):e2772–e. doi: 10.1038/cddis.2017.181
70. Ding J, Yeh C-R, Sun Y, Lin C, Chou J, Ou Z, et al. Estrogen Receptor β Promotes Renal Cell Carcinoma Progression via Regulating LncRNA HOTAIR-miR-138/200c/204/217 Associated CeRNA Network. Oncogene (2018) 37(37):5037–53. doi: 10.1038/s41388-018-0175-6
71. Bai J-Y, Jin B, Ma J-B, Liu T-J, Yang C, Chong Y, et al. HOTAIR and Androgen Receptor Synergistically Increase GLI2 Transcription to Promote Tumor Angiogenesis and Cancer Stemness in Renal Cell Carcinoma. Cancer Lett (2021) 498:70–9. doi: 10.1016/j.canlet.2020.10.031
72. Lin K, Jiang H, Zhang L-L, Jiang Y, Yang Y-X, Qiu G-D, et al. Down-Regulated LncRNA-HOTAIR Suppressed Colorectal Cancer Cell Proliferation, Invasion, and Migration by Mediating P21. Dig Dis Sci (2018) 63(9):2320–31. doi: 10.1007/s10620-018-5127-z
73. Pan S, Liu Y, Liu Q, Xiao Y, Liu B, Ren X, et al. HOTAIR/miR-326/FUT6 Axis Facilitates Colorectal Cancer Progression Through Regulating Fucosylation of CD44 via PI3K/AKT/mTOR Pathway. Biochim Biophys Acta (BBA) - Mol Cell Res (2019) 1866(5):750–60. doi: 10.1016/j.bbamcr.2019.02.004
74. Yang X-D, Xu H-T, Xu X-H, Ru GAN, Liu WEI, Zhu J-J, et al. Knockdown of Long Non-Coding RNA HOTAIR Inhibits Proliferation and Invasiveness and Improves Radiosensitivity in Colorectal Cancer. Oncol Rep (2016) 35(1):479–87. doi: 10.3892/or.2015.4397
75. Xiao Z, Qu Z, Chen Z, Fang Z, Zhou K, Huang Z, et al. LncRNA HOTAIR Is a Prognostic Biomarker for the Proliferation and Chemoresistance of Colorectal Cancer via MiR-203a-3p-Mediated Wnt/ß-Catenin Signaling Pathway. Cell Physiol Biochem (2018) 46(3):1275–85. doi: 10.1159/000489110
76. Zhang Z, Cheng J, Wu Y, Qiu J, Sun Y, Tong X. LncRNA HOTAIR Controls the Expression of Rab22a by Sponging miR-373 in Ovarian Cancer. Mol Med Rep (2016) 14(3):2465–72. doi: 10.3892/mmr.2016.5572
77. Chang L, Guo R, Yuan Z, Shi H, Zhang D. LncRNA HOTAIR Regulates CCND1 and CCND2 Expression by Sponging miR-206 in Ovarian Cancer. Cell Physiol Biochem (2018) 49(4):1289–303. doi: 10.1159/000493408
78. Kim HJ, Lee DW, Yim GW, Nam EJ, Kim S, Kim SW, et al. Long non-Coding RNA HOTAIR Is Associated With Human Cervical Cancer Progression. Int J Oncol (2015) 46(2):521–30. doi: 10.3892/ijo.2014.2758
79. Chang Y-T, Lin T-P, Tang J-T, Campbell M, Luo Y-L, Lu S-Y, et al. HOTAIR Is a REST-Regulated lncRNA That Promotes Neuroendocrine Differentiation in Castration Resistant Prostate Cancer. Cancer Lett (2018) 433:43–52. doi: 10.1016/j.canlet.2018.06.029
80. Zhang J-j, Zhou X-h, Zhou Y, Wang Y-g, Qian B-z, He A-n, et al. Bufalin Suppresses the Migration and Invasion of Prostate Cancer Cells Through HOTAIR, the Sponge of miR-520b. Acta Pharmacol Sin (2019) 40(9):1228–36. doi: 10.1038/s41401-019-0234-8
81. Yang B, Wei ZY, Wang BQ, Yang HC, Wang JY, Bu XY. Down-Regulation of the Long Noncoding RNA-HOX Transcript Antisense Intergenic RNA Inhibits the Occurrence and Progression of Glioma. J Cell Biochem (2017) 119(2):2278–87. doi: 10.1002/jcb.26390
82. Tan SK, Pastori C, Penas C, Komotar RJ, Ivan ME, Wahlestedt C, et al. Serum Long Noncoding RNA HOTAIR as a Novel Diagnostic and Prognostic Biomarker in Glioblastoma Multiforme. Mol Cancer (2018) 17(1):74. doi: 10.1186/s12943-018-0822-0
83. Ge X-S, Ma H-J, Zheng X-H, Ruan H-L, Liao X-Y, Xue W-Q, et al. HOTAIR, a Prognostic Factor in Esophageal Squamous Cell Carcinoma, Inhibits WIF-1 Expression and Activates Wnt Pathway. Cancer Sci (2013) 104(12):1675–82. doi: 10.1111/cas.12296
84. Gong GH, An FM, Wang Y, Bian M, Wang D, Wei CX. Comprehensive Circular RNA Profiling Reveals the Regulatory Role of the CircRNA-0067835/miR-155 Pathway in Temporal Lobe Epilepsy. Cell Physiol Biochem (2018) 51(3):1399–409. doi: 10.1159/000495589
85. Wang W, He X, Zheng Z, Ma X, Hu X, Wu D, et al. Serum HOTAIR as a Novel Diagnostic Biomarker for Esophageal Squamous Cell Carcinoma. Mol Cancer (2017) 16(1):75. doi: 10.1186/s12943-017-0643-6
86. Yu D, Zhang C, Gui J. RNA-Binding Protein HuR Promotes Bladder Cancer Progression by Competitively Binding to the Long Noncoding HOTAIR With miR-1. OncoTargets Ther (2017) 10:2609–19. doi: 10.2147/OTT.S132728
87. Wang B, Qu X-L, Liu J, Lu J, Zhou Z-Y. HOTAIR Promotes Osteosarcoma Development by Sponging miR-217 and Targeting ZEB1. J Cell Physiol (2019) 234(5):6173–81. doi: 10.1002/jcp.27394
88. Wang N, Meng X, Liu Y, Chen Y, Liang Q. LPS Promote Osteosarcoma Invasion and Migration Through TLR4/HOTAIR. Gene (2019) 680:1–8. doi: 10.1016/j.gene.2018.09.031
89. Wang B, Su Y, Yang Q, Lv D, Zhang W, Tang K, et al. Overexpression of Long Non-Coding RNA HOTAIR Promotes Tumor Growth and Metastasis in Human Osteosarcoma. Mol Cells (2015) 38(5):432–40. doi: 10.14348/molcells.2015.2327
90. Tatangelo F, Di Mauro A, Scognamiglio G, Aquino G, Lettiero A, Delrio P, et al. Posterior HOX Genes and HOTAIR Expression in the Proximal and Distal Colon Cancer Pathogenesis. J Trans Med (2018) 16(1):90. doi: 10.1186/s12967-018-1725-y
91. Wang X, Wang W, Zhang Q, Gu D, Zhang K, Ge Y, et al. Tagging SNPs in the HOTAIR Gene Are Associated With Bladder Cancer Risk in a Chinese Population. Gene (2018) 664:22–6. doi: 10.1016/j.gene.2018.04.039
92. Yu X, Wang R, Han C, Wang Z, Jin X. A Panel of Urinary Long Non-Coding RNAs Differentiate Bladder Cancer From Urocystitis. J Cancer (2020) 11(4):781–7. doi: 10.7150/jca.37006
93. Vasan N, Baselga J, Hyman DM. A View on Drug Resistance in Cancer. Nature (2019) 575(7782):299–309. doi: 10.1038/s41586-019-1730-1
94. Fang S, Gao H, Tong Y, Yang J, Tang R, Niu Y, et al. Long Noncoding RNA-HOTAIR Affects Chemoresistance by Regulating HOXA1 Methylation in Small Cell Lung Cancer Cells. Lab Invest (2015) 96(1):60–8. doi: 10.1038/labinvest.2015.123
95. Li D, Li C, Chen Y, Teng L, Cao Y, Wang W, et al. LncRNA HOTAIR Induces Sunitinib Resistance in Renal Cancer by Acting as a Competing Endogenous RNA to Regulate Autophagy of Renal Cells. Cancer Cell Int (2020) 20(1):338. doi: 10.1186/s12935-020-01419-0
96. Zhang A, Zhao Jonathan C, Kim J, Fong K-w, Yang Yeqing A, Chakravarti D, et al. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep (2015) 13(1):209–21. doi: 10.1016/j.celrep.2015.08.069
97. Guo J, Dou D, Zhang T, Wang B. HOTAIR Promotes Cisplatin Resistance of Osteosarcoma Cells by Regulating Cell Proliferation, Invasion, and Apoptosis via miR-106a-5p/STAT3 Axis. Cell Transplant (2020) 29:12. doi: 10.1177/0963689720948447
98. Li P, Zhang X, Wang L, Du L, Yang Y, Liu T, et al. lncRNA HOTAIR Contributes to 5FU Resistance Through Suppressing miR-218 and Activating NF-κb/TS Signaling in Colorectal Cancer. Mol Ther - Nucleic Acids (2017) 8:356–69. doi: 10.1016/j.omtn.2017.07.007
99. Özeş AR, Miller DF, Özeş ON, Fang F, Liu Y, Matei D, et al. NF-κb-HOTAIR Axis Links DNA Damage Response, Chemoresistance and Cellular Senescence in Ovarian Cancer. Oncogene (2016) 35(41):5350–61. doi: 10.1038/onc.2016.75
100. Yang Z, Zhou L, Wu L-M, Lai M-C, Xie H-Y, Zhang F, et al. Overexpression of Long Non-Coding RNA HOTAIR Predicts Tumor Recurrence in Hepatocellular Carcinoma Patients Following Liver Transplantation. Ann Surg Oncol (2011) 18(5):1243–50. doi: 10.1245/s10434-011-1581-y
101. Wu C, Yang L, Qi X, Wang T, Li M, Xu K. Inhibition of Long non-Coding RNA HOTAIR Enhances Radiosensitivity via Regulating Autophagy in Pancreatic Cancer. Cancer Manage Res (2018) 10:5261–71. doi: 10.2147/cmar.s174066
102. Guo X, Xiao H, Guo S, Li J, Wang Y, Chen J, et al. Long Noncoding RNA HOTAIR Knockdown Inhibits Autophagy and Epithelial-Mesenchymal Transition Through the Wnt Signaling Pathway in Radioresistant Human Cervical Cancer HeLa Cells. J Cell Physiol (2019) 234(4):3478–89. doi: 10.1002/jcp.26828
103. Liu Y, Chen X, Chen X, Liu J, Gu H, Fan R, et al. Long non-Coding RNA HOTAIR Knockdown Enhances Radiosensitivity Through Regulating microRNA-93/ATG12 Axis in Colorectal Cancer. Cell Death Dis (2020) 11(3):175. doi: 10.1038/s41419-020-2268-8
104. Martínez-Fernández M, Feber A, Dueñas M, Segovia C, Rubio C, Fernandez M, et al. Analysis of the Polycomb-Related lncRNAs HOTAIR and ANRIL in Bladder Cancer. Clin Epigenet (2015) 7(1):109. doi: 10.1186/s13148-015-0141-x
105. Quan J, Pan X, Zhao L, Li Z, Dai K, Yan F, et al. LncRNA as a Diagnostic and Prognostic Biomarker in Bladder Cancer: A Systematic Review and Meta-Analysis. OncoTargets Ther (2018) 11:6415–24. doi: 10.2147/ott.s167853
106. Lu R, Zhang J, Zhang W, Huang Y, Wang N, Zhang Q, et al. Circulating HOTAIR Expression Predicts the Clinical Response to Neoadjuvant Chemotherapy in Patients With Breast Cancer. Cancer Biomarkers (2018) 22(2):249–56. doi: 10.3233/cbm-170874
107. Kim JO, Jun HH, Kim EJ, Lee JY, Park HS, Ryu CS, et al. Genetic Variants of HOTAIR Associated With Colorectal Cancer Susceptibility and Mortality. Front Oncol (2020) 10:72. doi: 10.3389/fonc.2020.00072
108. Wang Q, Li X, Ren S, Su C, Li C, Li W, et al. HOTAIR Induces EGFR-TKIs Resistance in non-Small Cell Lung Cancer Through Epithelial-Mesenchymal Transition. Lung Cancer (2020) 147:99–105. doi: 10.1016/j.lungcan.2020.06.037
109. Lennox KA, Behlke MA. Cellular Localization of Long Non-Coding RNAs Affects Silencing by RNAi More Than by Antisense Oligonucleotides. Nucleic Acids Res (2016) 44(2):863–77. doi: 10.1093/nar/gkv1206
110. Bhan A, Hussain I, Ansari KI, Kasiri S, Bashyal A, Mandal SS. Antisense Transcript Long Noncoding RNA (lncRNA) HOTAIR is Transcriptionally Induced by Estradiol. J Mol Biol (2013) 425(19):3707–22. doi: 10.1016/j.jmb.2013.01.022
111. Li Y, Ren Y, Wang Y, Tan Y, Wang Q, Cai J, et al. A Compound AC1Q3QWB Selectively Disrupts HOTAIR-Mediated Recruitment of PRC2 and Enhances Cancer Therapy of DZNep. Theranostics (2019) 9(16):4608–23. doi: 10.7150/thno.35188
112. Shi J, Lv S, Wu M, Wang X, Deng Y, Li Y, et al. HOTAIR-EZH2 Inhibitor AC1Q3QWB Upregulates CWF19L1 and Enhances Cell Cycle Inhibition of CDK4/6 Inhibitor Palbociclib in Glioma. Clin Transl Med (2020) 10(1):182–98. doi: 10.1002/ctm2.21
113. Wang W, Fang F, Ozes A, Nephew KP. Targeting Ovarian Cancer Stem Cells by Dual Inhibition of HOTAIR and DNA Methylation. Mol Cancer Ther (2021) 20(6):1092–101. doi: 10.1158/1535-7163.MCT-20-0826
114. Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of Long Non-Coding RNA HOTAIR Inhibits Cisplatin Resistance of Gastric Cancer Cells Through Inhibiting the PI3K/Akt and Wnt/β-Catenin Signaling Pathways by Up-Regulating miR-34a. Int J Biol Macromol (2018) 107:2620–9. doi: 10.1016/j.ijbiomac.2017.10.154
115. Jia J, Zhan D, Li J, Li Z, Li H, Qian J. The Contrary Functions of lncRNA HOTAIR/miR-17-5p/PTEN Axis and Shenqifuzheng Injection on Chemosensitivity of Gastric Cancer Cells. J Cell Mol Med (2019) 23(1):656–69. doi: 10.1111/jcmm.13970
116. Zhang L, Song X, Wang X, Xie Y, Wang Z, Xu Y, et al. Circulating DNA of HOTAIR in Serum Is a Novel Biomarker for Breast Cancer. Breast Cancer Res Treat (2015) 152(1):199–208. doi: 10.1007/s10549-015-3431-2
117. Pastori C. The Bromodomain Protein BRD4 Controls HOTAIR, a Long Noncoding RNA Essential for Glioblastoma Proliferation. CrossMark (2015) 112(27):8326–31. doi: 10.1073/pnas.1424220112
118. Zhang J-X, Han L, Bao Z-S, Wang Y-Y, Chen L-Y, Yan W, et al. HOTAIR, a Cell Cycle–Associated Long Noncoding RNA and a Strong Predictor of Survival, Is Preferentially Expressed in Classical and Mesenchymal Glioma. Neuro-Oncology (2013) 15(12):1595–603. doi: 10.1093/neuonc/not131
119. Lee M. The Long Non-Coding RNA HOTAIR Increases Tumour Growth and Invasion in Cervical Cancer by Targeting the Notch Pathway. Oncotarget (2016) 7(28):44558–71. doi: 10.18632/oncotarget.10065
120. Liu B, Liu Q, Pan S, Huang Y, Qi Y, Li S, et al. The HOTAIR/miR-214/ST6GAL1 Crosstalk Modulates Colorectal Cancer Procession Through Mediating Sialylated C-Met via JAK2/STAT3 Cascade. J Exp Clin Cancer Res (2019) 38(1):455. doi: 10.1186/s13046-019-1468-5
121. Svoboda M, Slyskova J, Schneiderova M, Makovicky P, Bielik L, Levy M, et al. HOTAIR Long Non-Coding RNA Is a Negative Prognostic Factor Not Only in Primary Tumors, But Also in the Blood of Colorectal Cancer Patients. Carcinogenesis (2014) 35(7):1510–5. doi: 10.1093/carcin/bgu055
122. Chen F-J, Sun M, Li S-Q, Wu Q-Q, Ji L, Liu Z-L, et al. Upregulation of the Long non-Coding Rna Hotair Promotes Esophageal Squamous Cell Carcinoma Metastasis and Poor Prognosis. Mol Carcinog (2013) 52(11):908–15. doi: 10.1002/mc.21944
123. Lv X-B. Long Noncoding RNA HOTAIR Is a Prognostic Marker for Esophageal Squamous Cell Carcinoma Progression and Survival. PloS One (2013) 6:e63516. doi: 10.1371/journal.pone.0063516
124. Ren K, Li Y, Lu H, Li Z, Li Z, Wu K, et al. Long Noncoding RNA HOTAIR Controls Cell Cycle by Functioning as a Competing Endogenous RNA in Esophageal Squamous Cell Carcinoma. Trans Oncol (2016) 9(6):489–97. doi: 10.1016/j.tranon.2016.09.005
125. Zhang X, Zhou L, Fu G, Sun F, Shi J, Wei J, et al. The Identification of an ESCC Susceptibility SNP Rs920778 That Regulates the Expression of Lncrnahotairvia a Novel Intronic Enhancer. Carcinogenesis (2014) 35(9):2062–7. doi: 10.1093/carcin/bgu103
126. Wang W, Wu D, He X, Hu X, Hu C, Shen Z, et al. CCL18-Induced HOTAIR Upregulation Promotes Malignant Progression in Esophageal Squamous Cell Carcinoma Through the miR-130a-5p-ZEB1 Axis. Cancer Lett (2019) 460:18–28. doi: 10.1016/j.canlet.2019.06.009
127. Liu X-h, Sun M, Nie F-q, Ge Y-b, Zhang E-b, Yin D-d, et al. Lnc RNA HOTAIR Functions as a Competing Endogenous RNA to Regulate HER2 Expression by Sponging miR-331-3p in Gastric Cancer. Mol Cancer (2014) 13(1):92. doi: 10.1186/1476-4598-13-92
128. Feng X, Huang S. Effect and Mechanism of Long Noncoding RNAs HOTAIR on Occurrence and Development of Gastric Cancer. J Cell Biochem (2019) 120(5):6899–907. doi: 10.1002/jcb.26594
129. Liu X-h. The Long Non-Coding RNA HOTAIR Indicates a Poor Prognosis and Promotes Metastasis in Non-Small Cell Lung Cancer. BMC Cancer (2013) 13:464. doi: 10.1186/1471-2407-13-464
130. Guo F, Cao Z, Guo H, Li S. The Action Mechanism of lncRNA-HOTAIR on the Drug Resistance of Non-Small Cell Lung Cancer by Regulating Wnt Signaling Pathway. Exp Ther Med (2018) 15(6):4885–9. doi: 10.3892/etm.2018.6052
131. Ren MM, Xu S, Wei YB, Yang JJ, Yang YN, Sun SS, et al. Roles of HOTAIR in Lung Cancer Susceptibility and Prognosis. Mol Genet Genomic Med (2020) 8(7):e1299. doi: 10.1002/mgg3.1299
132. Liu S, Lei H, Luo F, Li Y, Xie L. The Effect of lncRNA HOTAIR on Chemoresistance of Ovarian Cancer Through Regulation of HOXA7. Biol Chem (2018) 399(5):485–97. doi: 10.1515/hsz-2017-0274
133. Cai H. LncRNA HOTAIR Acts as Competing Endogenous RNA to Control the Expression of Notch3 via Sponging miR-613 in Pancreatic Cancer. Oncotarget (2017) 8(20):32905–17. doi: 10.18632/oncotarget.16462
134. Yang S-z, Xu F, Zhou T, Zhao X, McDonald JM, Chen Y. The Long non-Coding RNA HOTAIR Enhances Pancreatic Cancer Resistance to TNF-Related Apoptosis-Inducing Ligand. J Biol Chem (2017) 292(25):10390–7. doi: 10.1074/jbc.M117.786830
135. Rishi A, Chiyomaru T, Yamamura S, Fukuhara S, Yoshino H, Kinoshita T, et al. Genistein Inhibits Prostate Cancer Cell Growth by Targeting miR-34a and Oncogenic HOTAIR. PloS One (2013) 8(8):e70372. doi: 10.1371/journal.pone.0070372
Keywords: HOTAIR, cancer, prognosis, therapy, potential, drug resistance, survival, knockdown
Citation: Xin X, Li Q, Fang J and Zhao T (2021) LncRNA HOTAIR: A Potential Prognostic Factor and Therapeutic Target in Human Cancers. Front. Oncol. 11:679244. doi: 10.3389/fonc.2021.679244
Received: 11 March 2021; Accepted: 08 July 2021;
Published: 22 July 2021.
Edited by:
Christina M. Annunziata, National Cancer Institute (NCI), United StatesReviewed by:
Chunyan Gu, Nanjing University of Chinese Medicine, ChinaMuhammad Umer, Griffith University, Australia
Copyright © 2021 Xin, Li, Fang and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tiejun Zhao, dGp6aGFvQHpqbnUuY24=
 Xiaoru Xin
Xiaoru Xin Qianan Li1
Qianan Li1 Jinyong Fang
Jinyong Fang Tiejun Zhao
Tiejun Zhao